Abstract
Background:
Spectral library searching is currently the most common approach for compound annotation in untargeted metabolomics. Spectral libraries applicable to liquid chromatography mass spectrometry have grown in size over the past decade to include hundreds of thousands to millions of mass spectra and tens of thousands of compounds, forming an essential knowledge base for the interpretation of metabolomics experiments.
Aim of Review:
We describe existing spectral library resources, highlight different strategies for compiling spectral libraries, and discuss quality considerations that should be taken into account when interpreting spectral library searching results. Finally, we describe how spectral libraries are empowering the next generation of machine learning tools in computational metabolomics, and discuss several opportunities for using increasingly accessible large spectral libraries.
Key Scientific Concepts of Review:
This review focuses on the current state of spectral libraries for untargeted LC-MS/MS based metabolomics. We show how the number of entries in publicly accessible spectral libraries has increased more than 60-fold in the past eight years to aid molecular interpretation and we discuss how the role of spectral libraries in untargeted metabolomics will evolve in the near future.
Keywords: untargeted metabolomics, mass spectrometry, spectral library, compound identification
Introduction
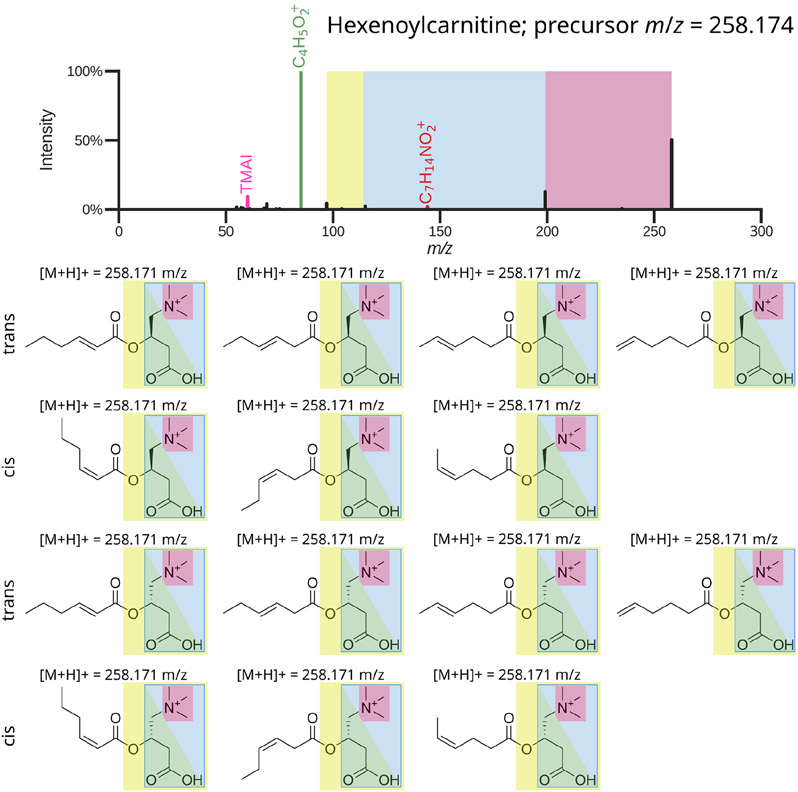
Spectral library searching is currently the most common approach for compound identification in untargeted metabolomics, with the earliest historical spectral libraries that can be traced back to the 1950s (Zemany, 1950). Metabolite annotation using spectral library searching is based on the concept that molecules undergo fragmentation that creates a reproducible “fingerprint.” Matching against a spectral library of ground truth MS/MS spectra collected with chemical standards of known molecules can then be used to narrow down structural hypotheses. During library searching, experimental MS/MS spectra are annotated by matching against the library MS/MS spectra and transferring compound labels from library to experimental spectra when a high-scoring match is achieved. This is the gold standard for metabolite annotation from MS/MS data only, and it forms a level 2 or level 3 annotation based on the guidelines of the Metabolomics Standards Initiative (Sumner et al., 2007). A level 2 annotation corresponds to library searching resulting in a structural hypothesis for a specific molecule, while a level 3 annotation is a match hypothesis to a molecular family. Especially isomeric compounds with identical precursor mass may result in more than one structural match. For example, it is impossible to distinguish between various stereoisomers of hexenoylcarnitine by MS/MS matching only (Figure 1). To promote such a level 3 match to a level 1 identification, complementary analytical approaches, such as nuclear magnetic resonance (NMR), are needed, or all possible isomers in the molecular family have to be tested under the same mass spectrometry conditions to best determine MS/MS spectrum similarity, in addition to liquid chromatography (LC) co-migration of the compound of interest with the chemical standards to validate whether it elutes with the same peak shape and retention time.
Figure 1:
Representative example of a molecular family level annotation from spectral library searching that matches to hexenoylcarnitine. The MS/MS spectrum contains several diagnostic fragments and neutral losses that make it possible to assign it to the acylcarnitines molecular family, as indicated on the molecular structures (Yan et al., 2020). However, routine spectral library matching cannot distinguish between the 14 potential stereo- and regioisomers, resulting in a level 3 annotation. This highlights the need for new strategies to communicate the results from spectral library searching, as narrowing down to the molecular family, even when the exact molecular identity is unknown, can often already be valuable for biological interpretation. Top is the experimental observed MS/MS spectrum, with a precursor m/z deviation of 11.6 ppm compared to the calculated m/z of the protonated ions.
Although in proteomics, sequence database searching is the dominant strategy to annotate MS/MS spectra (Eng et al., 2011), the usage of spectral libraries has become increasingly popular for the analysis of peptide MS/MS data as well in recent years (Griss, 2016; Shao & Lam, 2017; Deutsch et al., 2018). Spectral library searching is more sensitive than sequence database searching, achieving a higher rate of spectrum identifications (Zhang et al., 2011), and results from spectral library searching and sequence database searching can be combined to maximize the number of identified MS/MS spectra (Shteynberg et al., 2013). This increased sensitivity is especially relevant for the analysis of data-independent acquisition (DIA) experiments, where mixtures of analytes within large, pre-specified mass ranges are measured, in contrast to data-dependent acquisition (DDA), which attempts to isolate and measure individual analytes (Hu et al., 2016). The resulting complex DIA spectra contain signals from multiple peptides, and most DIA analysis tools require detailed MS/MS fragmentation patterns from reference spectral libraries to annotate peptides.
As the authors of this perspective believe that open and transparent science has strong cascading benefits for the larger scientific community (Wilson et al., 2021) and are most familiar with the GNPS/MassIVE platform (M. Wang et al., 2016), most of the following discussion is contextualized in reference to this resource for untargeted metabolomics analysis. In this context, we discuss the state of spectral libraries for untargeted metabolomics in 2022, describe the essential role of spectral libraries in the development of computational tools, and highlight some open challenges and opportunities for the metabolomics community to address in the coming years.
Impact of growing and freely accessible spectral libraries
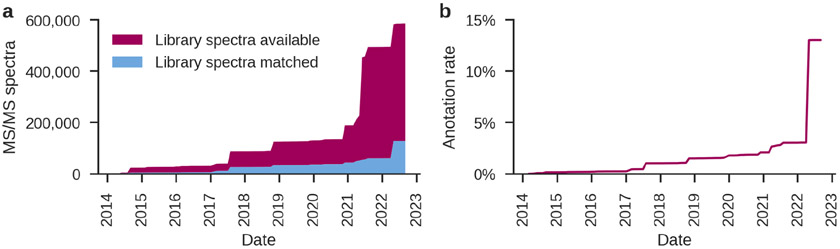
Over the past decade, MS/MS small molecule spectral libraries have steadily increased in size to include hundreds of thousands to millions of MS/MS spectra and hundreds of thousands of compounds (Figure 1a). Some of the largest experimental small molecule spectral libraries that are currently available include both commercial libraries, such as the National Institute of Standards and Technology (NIST) tandem mass spectral library (https://chemdata.nist.gov/) and the METLIN Gen2 spectral library (Xue et al., 2020), and open spectral libraries, which also serve as aggregation sites for third-party community spectral libraries, such as the Global Natural Products Social Molecular Networking (GNPS) community spectral libraries (M. Wang et al., 2016) and Massbank of North America (MoNA; https://mona.fiehnlab.ucdavis.edu/). Additionally, mass spectrometry instrument vendors also provide commercial spectral libraries, such as mzCloud (https://www.mzcloud.org/). Excitingly, publicly and freely accessible MS/MS spectral libraries recently saw explosive growth (Figure 2a).
Figure 2:
Advances in spectral libraries for LC-MS/MS based untargeted metabolomics. (a) The GNPS community spectral libraries (non-commercial only) have grown from 23,790 MS/MS spectra in 2014 to 586,647 MS/MS spectra in 2022 (September 2022). Concurrently, the number of library spectra that matched to public data has grown from 4,727 MS/MS spectra in 2014 to 127,405 MS/MS spectra in 2022 (22% of the publicly available library spectra have matches to experimental MS/MS spectra in public data). (b) Fueled by growing spectral libraries, the MS/MS spectrum annotation rate for the GNPS continuous identification mode as part of living data (M. Wang et al., 2016), which periodically reanalyses all public datasets on GNPS/MassIVE with the latest spectral libraries, has increased from 2% of MS/MS spectra on average in 2014 to 13% in 2022.
There also exist many other, often subject-specific spectral libraries, including Massbank (Horai et al., 2010) and Massbank EU (https://massbank.eu/MassBank/), the Human Metabolome Database (HMDB) (Wishart et al., 2021), the RIKEN tandem mass spectral database (ReSpect) (Sawada et al., 2012), the monoterpene indole alkaloid database (MIADB) (Fox Ramos et al., 2019), the Critical Assessment of Small Molecule Identification (CASMI) contest libraries (Schymanski & Neumann, 2013), European Molecular Biology Laboratory–Metabolomics Core Facility (EMBL-MCF) (Phapale et al., 2021), the Pacific Northwest National Lab lipids library (Kyle et al., 2017), the National Institutes of Health natural products library (Huang et al., 2019), the Lichen Database (LDB) (Olivier-Jimenez et al., 2019), fungal dereplication (El-Elimat et al., 2013), Chemicalsoft (Dresen et al., 2009), WEIZMASS (Shahaf et al., 2016), MSforID (Oberacher et al., 2011), the reverse metabolomics libraries (Gentry et al., 2021), and many others. Barring access restrictions, these spectral libraries are also often integrated into the previous spectral library aggregation resources, such as GNPS and MoNA. In this case, the SPLASH (SPectraL hASH) mechanism, which assigns unambiguous, database-independent hashed identifiers to MS/MS spectra, can be a useful tool for provenance of spectral data and detection of duplicate spectra that are shared across multiple data resources (Wohlgemuth et al., 2016), similar to how InChIKeys are used as chemical identifiers.
Several large proteomics spectral libraries exist as well. These include peptide MS/MS spectral libraries for multiple organisms (human, mouse, rat, yeast, etc.) from NIST (https://chemdata.nist.gov/dokuwiki/doku.php?id=peptidew:start), the ProteomeTools project of synthetic human peptide MS/MS spectra (Zolg et al., 2017), and the MassIVE Knowledge Base (MassIVE-KB) of the human proteome (M. Wang et al., 2018). Different strategies for compiling spectral libraries are exemplified by the ProteomeTools (Zolg et al., 2017) and MassIVE-KB peptide spectral libraries (M. Wang et al., 2018). On the one hand, ProteomeTools followed the traditional approach to generate a spectral library by synthesizing unique tryptic peptides from the human proteome and acquiring MS/MS data on multiple instrument platforms (Zolg et al., 2017). This was subsequently expanded to include additional tryptic peptides and modified peptides (Zolg et al., 2018), non-tryptic peptides (Wilhelm et al., 2021), and isobarically labeled peptides (Gabriel et al., 2022) to currently consist of more than one million unique synthetic peptides and over 14 million MS/MS spectra. In contrast, MassIVE-KB employed a data-driven approach towards spectral library creation by re-analyzing hundreds of millions to billions of public MS/MS spectra on the MassIVE data repository using sequence database searching (M. Wang et al., 2018). The most confidently identified MS/MS spectra and their peptide labels were then extracted to create the MassIVE-KB human peptide spectral library, which currently contains 2.5 million unique peptides and 6 million MS/MS spectra (version 2.0.15). Although an equivalent strategy to sequence database searching in proteomics currently does not exist for metabolomics, approaches employed by ProteomeTools and MassIVE-KB demonstrate how alternative strategies can be used to create valuable collections of reference MS/MS spectra. Furthermore, as it is not uncommon to observe peptides in metabolomics data, it is conceivable that proteomics libraries can be repurposed to also inform a subset of metabolomics data through creative use of algorithms that find analogs of peptides or peptidic molecules.
Similarly, in untargeted metabolomics, each of the libraries provides complementary MS/MS data and pieces of information. For example, the commercial NIST small molecule spectral library predominantly contains human and plant metabolites, ReSpect contains plant metabolites, and the commercial METLIN library historically contained a significant proportion of lipids and dipeptides (full details on the current composition after its explosive growth (Xue et al., 2020) are unknown as the library and information on the molecules that are part of the library have not been released publicly). The GNPS libraries historically focused on natural products, but they have since grown to include many major publicly available reference libraries, including lipids, drugs, pesticides, primary metabolites, food derived metabolites, common contaminants, and microbial metabolites. Furthermore, these libraries are exchanged with MoNA, MassBank EU, and other resources, such that they are not only leveraged in the GNPS analysis ecosystem but also by other analysis systems such as MZmine (Pluskal et al., 2010), MS-DIAL (Tsugawa et al., 2020), and others. This broad sharing of spectral libraries ensures that untargeted metabolomics analyses can be performed against the largest possible spectral libraries, irrespective of the analysis platform. It should be noted that some spectral libraries, such as NIST and METLIN, are exclusively obtained in a single lab under more consistent experimental conditions, whereas other spectral libraries, such as MoNA and GNPS, are aggregated from community contributions and contain data that has been acquired in multiple labs, using different instruments, instrument platforms, and experimental protocols, and thus are more heterogeneous.
Some metabolomics spectral library resources do not only include direct experimental MS/MS data from pure reference compounds, but also MS/MS spectra that were obtained using computational tools. For example, the MoNA and HMDB spectral libraries are augmented with in silico MS/MS spectra that were simulated using e.g. LipidBlast (MoNA) (Kind et al., 2013) and CFM-ID (HMDB) (F. Wang et al., 2021). Additionally, NIST provides smaller spectral libraries focused on specific types of molecules, such as oligosaccharides (Remoroza et al., 2018, 2020) and acylcarnitines (Yan et al., 2020), that were annotated using analog searching (Burke et al., 2017)—a strategy to identify structurally related molecules that differ by a modification by using a very wide precursor mass window (on the order of 100s Da)—rather than by measuring pure reference standards. GNPS contains secondary reference MS/MS spectra that have been annotated by high-quality matching against the NIST spectral library and “nearest neighbor suspect” MS/MS spectra (Bittremieux, Avalon, et al., 2022) that were obtained by propagating annotations using molecular networking (Aron et al., 2020) across all public untargeted metabolomics data in the GNPS/MassIVE repository. By propagating annotations from existing spectral libraries to related MS/MS spectra it becomes possible to provide annotations that would otherwise not be accessible to the community. Therefore, these strategies expand the set of putative annotations that can be obtained in untargeted metabolomics experiments. This is especially relevant for molecules for which pure standards are not available, because their structures have never been synthesized or isolated from biological material, or because they cannot be described as a structure (e.g. sodium formate clusters or a specific modification of unknown regio- or stereochemistry). However, because the MS/MS spectra are not directly measured from pure reference material, additional care should be taken when interpreting annotations that match such library spectra. In other words, the user has to verify whether the annotations match the data and whether they make sense in the context of the experiment before investing precious time and resources to perform additional validation experiments.
Besides these traditional spectral libraries for untargeted metabolomics that focus on fragmentation data, other libraries that include complementary information or for different data acquisition methods are starting to become available. For example, some spectral libraries contain LC retention time information as well (Stanstrup et al., 2015; Tada et al., 2019), such as the METLIN small molecule retention time dataset (Domingo-Almenara et al., 2019). Additionally, with the increasing integration of ion mobility functionality in modern mass spectrometry instruments, ion mobility libraries that contain reference collision cross section (CCS) measurements are emerging (Zheng et al., 2017; Hernández-Mesa et al., 2018; Righetti et al., 2018; Picache et al., 2018; Schroeder et al., 2019; Z. Zhou et al., 2020). This availability of retention time and CCS reference measurements provides orthogonal information for metabolite annotation from untargeted MS/MS data. Additionally, spectral libraries for alternative data acquisition methods exist. For example, mzCloud organizes MSn spectra into “fragmentation trees,” and the METLIN-MRM spectral library is a multiple-reaction monitoring (MRM) transition repository for small-molecule quantitative mass spectrometry that contains MRM transitions for more than 15,500 unique molecules (Domingo-Almenara et al., 2018).
With the growing commodification of advanced instrumentation capabilities, there is a need for further expansion of alternative spectral libraries. Whereas most LC-MS/MS spectral libraries use collision-induced dissociation (CID) or higher-energy C-trap dissociation (HCD), various other fragmentation techniques, such as electron-induced dissociation (X. Chen et al., 2018), ultraviolet photodissociation (Bowers et al., 1984), charge transfer dissociation (W. D. Hoffmann & Jackson, 2014), and others (Heiles, 2021), can now be used as well. Because different fragmentation techniques can result in dramatically different MS/MS fragmentation patterns, traditional spectral libraries might not be suitable for MS/MS spectral matching of such data and custom libraries will be needed. Even when using CID/HCD fragmentation, different instrument platforms or employing different collision energies can produce MS/MS data that exhibit dissimilar fragmentation behavior. Consequently, it is not always possible to get a spectral match when data is collected differently. Nevertheless, we recommend searching experimental MS/MS data against the broadest possible relevant spectral libraries, irrespective of instrument platform details. Even if the MS/MS spectra differ to some extent, it can still be possible to obtain relevant matches, especially with modern algorithmic techniques that preprocess spectra to try to minimize the effects of experimental variability. Furthermore, some advanced MS/MS fragmentation strategies might enable synergies between previously disparate library generation efforts. For example, CID spectra can contain a non-negligible number of radical fragment ions (K. Chen et al., 2008; Xing & Huan, 2022), and fragmentation mechanisms from electron-induced dissociation techniques show significant similarity to fragmentation events under electron ionization, which is commonly used in gas chromatography mass spectrometry (GC-MS) (Ducati et al., 2021). This suggests that it could be possible to repurpose the information content from large amounts of historical spectral libraries that have been generated for GC-MS.
The increasing availability of large-scale and open spectral libraries is driving their growing role in computational mass spectrometry (Stein, 2012; Vinaixa et al., 2016; Aksenov et al., 2017; Tsugawa, 2018). Whereas in untargeted metabolomics experiments, using all commercial and openly available spectral libraries, only 2% of MS/MS spectra could be successfully annotated by spectral library searching less than a decade ago (M. Wang et al., 2016), in 2022 the spectrum annotation rate for untargeted metabolomics on the GNPS platform has increased to 13% (Figure 2b). This increase by up to an order of magnitude in the number of unique MS/MS spectrum annotations that can be obtained is essential in advancing the amount of biological knowledge that can be achieved using untargeted metabolomics, and has only been possible by tremendous and continued efforts of various stakeholders—both academic and industry—and the metabolomics community at large.
Interpreting spectral library searching results
When interpreting spectrum annotations from spectral library searching, it is essential to have a clear understanding of the information that mass spectrometry can and cannot provide (Stein, 2012). For example, mass spectrometry may not always distinguish between isomeric molecules. Although the Metabolomics Standards Initiative provides guidelines to denote the level of identification rigor for reported metabolite identifications (Sumner et al., 2007), these do not fully capture the ambiguity related to isomers (e.g. using an ontology) and do not provide a system to build provenance into the confidence of spectrum annotations. Additionally, MS/MS spectra might not contain sufficiently discriminative information to annotate specific molecules if there are too few fragment ions or no unique fragment ions. Analyzing non-discriminative MS/MS spectra is equivalent to searching a genetic sequence database with a two-mer oligonucleotide, which would result in an excessive number of non-specific matches. Instead, when few ions are available or the sample contains multiple isomers, spectral library annotations might only go up to the molecular family if fragment ions correspond to similar (sub)structures that are shared by related molecules. Therefore, it is recommended that users do not restrict themselves to only the top MS/MS match obtained using spectral library searching, but carefully consider lower ranked MS/MS matches that fall within the user defined inclusion criteria of acceptable errors of MS and MS/MS ions, and minimum number of matching fragment ions. If there are multiple annotations with similar MS/MS match scores that correspond to isomeric molecules or belong to the same molecular family—which usually consists of isomeric structures—additional information is needed to further refine the most likely candidate structures. At present, verifying such ambiguity often still involves careful manual investigation by expert users, isolation and NMR confirmation, or purchase or synthesis of all possible structures to validate the assignments. In the future, we anticipate that a new generation of computational mass spectrometry tools that can directly communicate this information to the user will be developed, for example by rolling up spectrum annotations to the family level or indicating spectral evidence of the (sub)structures that can be unambiguously explained. The goal of these tools should be to clearly communicate the maximum amount of knowledge that can be derived from the mass spectral data and then follow up with additional experiments to differentiate among all possible annotations.
In the best case, a library MS/MS spectrum should be measured from only a single, pure reference compound. In practice, during large-scale spectral library generation efforts multiple reference compounds are measured simultaneously to minimize the data acquisition time that is needed. Although it is typically ensured that no near-isobaric compounds are simultaneously measured during such multiplexing of reference compounds to avoid potential confusion when annotating the library spectra, interference during MS data acquisition might still occur. Additionally, other typical quality considerations for mass spectrometry experiments (Bittremieux, Tabb, et al., 2018), such as the presence of contaminants, carry-over, and other factors that can influence the data can impact spectral library generation.
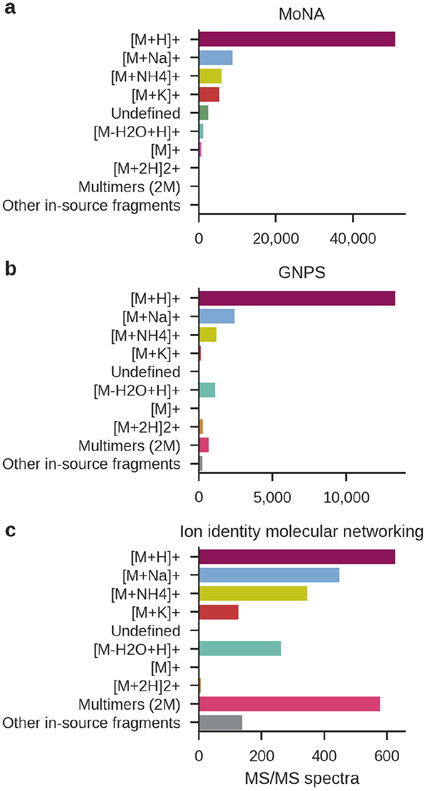
However, it is also important to be mindful of the biases associated with using pure reference compounds to generate spectral libraries. First, this requires a physical specimen of the pure compound, obtained from commercial sources or through laborious purification of biological samples. Unfortunately the majority of biological molecules whose structures have been elucidated are not readily available for purchase. An example of this bias is the disproportionately large number of unique matches to medicines and drugs when analyzing human fecal samples, while there are much fewer matches to microbial metabolites, which are not well represented in reference spectral libraries. A second type of bias is via the adduct that is chosen for fragmentation. For example, protonated and sodiated adducts are most frequently considered, with two thirds of positively charged MS/MS spectra in the MoNA and GNPS spectral libraries corresponding to protonated adducts (Figure 3a-b). However, many other adducts can be formed as well, especially during analysis of heterogeneous biological samples. Therefore, unless a complex background matrix is added to the pure standards, it is likely that an adduct that is observed in an experiment may not have been measured while generating library spectra from a reference compound. This is illustrated by the “ion identity molecular networking” approach, which was recently used to create a propagated spectral library that exhibits a broader coverage of different adducts, multimers, and in-source fragments (Figure 3c) (Schmid et al., 2021). Nevertheless, because ion identity molecular networking can only find predefined ion forms, and we generally do not know the distribution and diversity of all ion forms that exist yet, several unanswered questions remain. For example, how many ions are protonated, sodiated, or acetonitrile-ammonia ion forms? How many ions are magnesium adducts, heterodimers, or other ion forms that are currently not considered? To alleviate these biases, although this is typically not performed, library spectra could be acquired by running pure reference compounds with a more representative background or in a biological matrix and unbiased searches need to be performed to find all ion forms of the standards. Alternatively, as is possible on the GNPS ecosystem, researchers that are experts in the biological systems under investigation can annotate experimental MS/MS spectra directly and add them to the reference libraries.
Figure 3.
Distribution of ion adducts in public spectral libraries. The majority of positive ion mode MS/MS spectra in MoNA (a) and GNPS (b) are protonated, while other adducts, in-source fragments, multiply charged species, and multimers are minimally represented. (c) Ion identity molecular networking was used to extract novel reference MS/MS spectra that exhibit overall broader coverage of different adducts, multimers, and in-source fragments (Schmid et al., 2021). Note that these ion forms are found with a predefined inclusion list, rather than a comprehensive search for all ion forms that might be present in untargeted metabolomics data of a biological sample.
Another important, yet often overlooked aspect when evaluating spectral library searching results is the confidence that is ascribed to the original library annotation. If an original library spectrum is incorrectly annotated, this error will propagate through all future studies that find a match to this library spectrum. Consequently, even when a match is obtained, the researcher should still make sure that this makes sense in the context of their experiment. Therefore, it is paramount that library spectra are of the highest possible quality and that their provenance is tracked, so that the end user can understand the origin of their spectral library annotations. Having a clear understanding of the provenance of reference MS/MS spectra is especially relevant when spectral libraries are crowd-sourced, with spectral data coming from heterogeneous sources with potentially differing quality levels, although mistakes have also been found in commercial spectral libraries.
To assign quality levels to MS/MS library spectra, several community resources, including GNPS and MoNA, use a rating system. For example, on GNPS, library spectra are categorized based on the source of the MS/MS spectra. “Gold” spectra are derived from synthetic samples that have been characterized using mass spectrometry and an orthogonal analytical method, such as NMR or crystallography, and can only be contributed by privileged users; “silver” spectra are obtained from an isolated or lysate/crude sample with a scientific publication confirming the presence of the molecule in the sample; and “bronze” spectra are other experimental MS/MS spectra that provide evidence for putative or partial annotations. Finally, there are “in silico” spectra that have been produced using computational approaches. The latter are not selected by default when performing spectral library searching using GNPS, however, as we believe that such spectra should be used with extreme caution and generally only give insights into molecular families rather than specific identities. GNPS also allows users to update spectral library annotations if the original submission contained limited details (e.g. someone may have denoted the spectrum as a saccharide but further insights revealed that the specific molecule is azithromycin, or the original submission did not include the molecular structure which was subsequently added) or to correct previously misassigned library spectra. In these cases, the GNPS system always retains a complete record of the full annotation history. Additionally, GNPS allows users to rate the quality of MS/MS matches from spectral library searching using four star (correct), three star (likely correct, e.g. could also be isomers with similar fragmentation patterns), two star (unable to confirm the annotation due to limited information), and one star (incorrect) ratings. MoNA assigns a five-star quality rating to all spectra based on the amount of metadata that was provided (ionization mode, instrument model, collision energy, liquid chromatography details, etc.), and top rated MS/MS spectra and a leaderboard of their submitting users are advertised on the MoNA homepage. Additionally, users can rate spectra as being either “clean” or “noisy.” These rating approaches allow users to manage expectations based on the evaluation of the library spectra so that they can make informed decisions based on the veracity of an MS/MS spectrum match, as well as provide feedback to help improve library annotations.
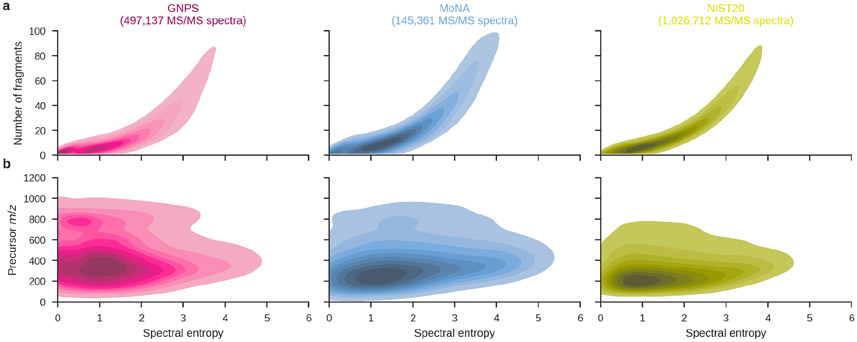
As an example of spectral library curation, a strategy for inter-library comparison was described to detect mis-annotated outliers by visual inspection based on an extensive checklist of potential issues (Wallace et al., 2017). Manual quality annotations have limited scalability, however, as they depend on scarce expert user knowledge and require a significant time investment. Because such domain experts often produce very trustworthy manual spectrum annotations and their expertise is not (yet) translated into community knowledge, this represents a unique opportunity to further improve the quality of spectral libraries. Alternatively, some computational approaches for MS/MS spectral library assessment have been proposed, including spectral entropy. Entropy is often likened to the disorder of a system. For example, there are more disorderly states in which a deck of cards can occur in random order (high entropy) than those in which the deck occurs in sorted order (low entropy). Spectral entropy (Li et al., 2021) was recently proposed as a measure to assess the quality of MS/MS spectra, with lower-quality spectra receiving higher spectral entropies. For example, there are small differences in the spectral entropy distributions of the highly curated NIST spectral library and more heterogeneous spectral libraries from MoNA and GNPS (Figure 4). Nevertheless, we would argue against a simple maximum spectral entropy cut-off to determine whether MS/MS spectra are of sufficient quality. There is a strong (nonlinear) relationship between spectral entropy and the number of fragment ions, with MS/MS spectra that contain only a few fragment ions getting low spectral entropy scores (Li et al., 2021). Although such spectra might be arguably of higher quality and more “clean,” this could also indicate that some of the spectra with low spectral entropy contain insufficiently discriminative fragmentation information to achieve sensitive MS/MS annotation. Nevertheless, spectral entropy is an interesting criterion to support the automated quality assessment of MS/MS spectra, which forms an open challenge that warrants additional research.
Figure 4:
Spectral entropy distributions for the GNPS, MoNA, and NIST20 spectral libraries. GNPS consists of 497,137 MS/MS spectra from the “ALL_GNPS_NO_PROPOGATED” library (downloaded on 2022-09-08), MoNA contains 145,361 MS/MS spectra from the “LC-MS/MS Spectra” collection (downloaded on 2022-09-08), and NIST20 consists of 1,026,712 MS/MS spectra (high-resolution MS/MS collection). Spectra were processed by removing noise peaks below 1% of the base peak intensity and normalizing fragment intensities to sum to one. (a) There is a strong relationship between spectral entropy and the number of fragment ions (Spearman correlation 0.963). (b) Although the NIST20 library contains smaller molecules than GNPS and MoNA, the difference in entropy distributions cannot be directly explained by the weight of the molecules (Spearman correlation 0.095).
Besides the quality of the library spectra, the veracity of the matches between library spectra and experimental spectra is also essential in determining whether to accept spectrum annotations. Typically, valid spectrum annotations are accepted based on common heuristics, such as a minimum cosine similarity threshold of 0.7 and minimum 6 matching peaks (M. Wang et al., 2016; Scheubert et al., 2017). However, such heuristics do not provide a statistical confidence estimate of the spectrum annotations, and as such, the number of false positives (i.e. incorrectly accepted high-scoring annotations) and false negatives (i.e. missed low-scoring annotations) are unknown. Although not widely used yet at this time, there are emerging strategies for estimating the false discovery rate of MS/MS spectrum annotations. For example, the Passatuto approach constructs a decoy library by modifying MS/MS library spectra based on re-rooted fragmentation trees to enable estimating false discovery rates using a target–decoy strategy (Scheubert et al., 2017). This allows the researcher to accept spectrum annotations with a controlled false discovery rate such that they can decide how many incorrect matches they are willing to include in their results. Although a few other methods to control false discovery rates in metabolomics have been introduced (Palmer et al., 2016; X. Wang et al., 2018; Alka et al., 2022), none are currently routinely used. Statistical control of MS/MS spectrum annotations is an important area of research to explore further and advance untargeted metabolomics into a highly scalable quantitative technique, and we anticipate that such tools will become routinely accessible in emerging MS/MS-based spectrum annotation software.
Spectral libraries as a source of machine learning training data
Besides their primary function for spectrum annotation, spectral libraries are also an extremely valuable resource to develop machine learning approaches for the analysis of mass spectrometry data (Kelchtermans et al., 2014). In proteomics, the availability of high-quality spectral libraries that can be used as large-scale training data has spurred the development of several innovative deep learning tools. For example, Prosit is a deep neural network that was trained on the ProteomeTools library to learn peptide fragmentation patterns and predict MS/MS fragment intensities with high fidelity (Gessulat et al., 2019). MS/MS spectra predicted by Prosit, as well as related tools that were developed in a similar fashion (Tiwary et al., 2019; Xu et al., 2020; X.-X. Zhou et al., 2017, p. 201), are now regularly used in lieu of experimental spectral libraries, for example, for the analysis of DIA data without the need to acquire a custom spectral library in advance. This illustrates how the important effort of synthesizing and measuring peptide standards provides continuing benefits outside of the original study by enabling the development of deep learning methods that can be used to simulate highly accurate MS/MS spectra for novel peptides to complement experimental spectral libraries. Similarly, MassIVE-KB was recently used to develop the GLEAMS neural network that can efficiently process hundreds of millions of MS/MS spectra at the repository scale to explore the dark proteome (Bittremieux, May, et al., 2022).
Small molecule spectral libraries are also used as the basis of computational tool and resource development in metabolomics (Krettler & Thallinger, 2021). For example, fragmentation patterns of acylcarnitines were derived from the NIST spectral library using the hybrid search strategy, which could then be used to extract and validate additional related acylcarnitine MS/MS spectra (Yan et al., 2020). The GNPS nearest neighbor suspect spectral library was created in a data-driven fashion by molecular networking of hundreds of millions of public MS/MS spectra on the GNPS repository in combination with reference MS/MS spectra in the GNPS community spectral libraries, and is a unique resource that provides insights into common modifications that molecules can undergo (Bittremieux, Avalon, et al., 2022). Additionally, high-quality annotated MS/MS spectra in open spectral libraries are increasingly being used to train and validate machine learning methods in metabolomics (Krettler & Thallinger, 2021; Liu et al., 2021). For example, they can be used to learn relationships between MS/MS patterns and molecular (sub)structures (e.g. MS2LDA (van der Hooft et al., 2016), MESSAR (Liu et al., 2020)), develop machine learning-inspired spectrum similarity scores (e.g. Spec2Vec (Huber, Ridder, et al., 2021), MS2DeepScore (Huber, van der Burg, et al., 2021), SIMILE (Treen et al., 2022)), simulate MS/MS spectra (e.g. CFM-ID (F. Wang et al., 2021)), and predict spectrum annotations (e.g. CSI:FingerID (Dührkop et al., 2015), COSMIC (M. A. Hoffmann et al., 2021), MassGenie (Shrivastava et al., 2021)).
These are inspiring examples of computational advances that are beginning to define the next generation of metabolomics analysis capabilities, which could not have been developed without the availability of comprehensive and high-quality open spectral libraries. Although these are already exciting advances in their own right, we believe that this is only the beginning of a more data-driven approach to computational metabolomics. Especially with the emergence and commodification of deep learning approaches, the availability of large training data is paramount to achieve optimal performance. Deep learning is an extremely powerful class of machine learning models that especially excels in deriving complex patterns from massive amounts of data and discovering otherwise hidden data structures (LeCun et al., 2015). However, care must be taken—as with any learning approach—that the analyses are reproducible and findings are carefully validated using follow-up studies (Gibney, 2022). In other words, computational tools, including those based on statistics or machine learning, can help investigators formulate hypotheses, but it is critical that any discoveries made with such tools are confirmed using follow-up experiments designed to refute the hypotheses. However, as public spectral libraries continue to grow, we excitedly anticipate that this will further power the development of creative machine learning and other computational solutions to provide further tools in the researcher’s arsenal to understand the rich data content that untargeted metabolomics provides.
Discussion
Spectral libraries are essential knowledge bases that form a bridge between the past and future of metabolomics: they capture the historical achievements of the metabolomics community in structure elucidation to empower the next generation of biological insights. Currently there are two prevalent strategies towards spectral library dissemination: as a commercial product or freely available for public use. Although commercializing spectral libraries can be appealing to offset the significant costs associated with generating them, open spectral libraries that can freely be used and reused provide a larger community benefit to advance science, by enabling biological discoveries and supporting the development of the next generation of computational and machine learning tools. We anticipate that with the ongoing shift towards open science and data FAIRness (Findable, Accessible, Interoperable, Reusable), open spectral libraries will keep growing in the near future to form increasingly comprehensive resources for the metabolomics community.
There are still some challenges associated with generating and using spectral libraries in metabolomics, however. Many spectral libraries have a considerable amount of missing information. When compiling crowd-sourced spectral libraries, there is a trade-off between requiring that all metadata has been unambiguously specified, which entails an additional time commitment and complexity for users submitting their data, and freely accepting contributions. The former results in a higher barrier towards contributing data to community spectral libraries, leading to smaller spectral libraries, while the latter results in less defined spectral libraries. As it is very challenging to completely eliminate all mistakes from spectral libraries, it is of the utmost importance to understand the provenance of spectral library matches. This allows the end user to make informed judgment calls to decide whether the matches should be followed up in subsequent experiments. Popular community spectral libraries, such as GNPS and MoNA, address this dichotomy by using a multi-faceted ranking system to rate individual MS/MS spectra, contributing users, and MS/MS assignments. Furthermore, a critical evaluation of any results by the user, irrespective of the spectral library source, is essential.
Despite their impressive growth in the past few years (Stein, 2012; Vinaixa et al., 2016; Kind et al., 2018; Peisl et al., 2018; Xue et al., 2020), spectral libraries still only cover a minor part of the known chemical space. For example, PubChem (Kim et al., 2021) currently contains information for 112 million unique compounds (September 2022), whereas all metabolomics spectral libraries combined account for less than 1% of those molecules. As spectral library searching can only annotate known molecules with reference MS/MS spectra or related molecules using analog searching, “unknown unknowns,” where experimental MS/MS spectra did not match any of the reference spectra included in the spectral library, cannot be identified (Stein, 2012). Some spectral library providers have started to integrate in silico MS/MS spectra alongside experimental MS/MS spectra to partially address this issue. Especially as spectrum prediction tools are getting increasingly better, this could be a viable strategy to expand the coverage of spectral libraries. At present, however, we strongly urge caution when accepting annotations based on simulated spectra only. It is still easiest to assess whether an MS/MS spectrum match is acceptable based on the user’s search criteria through manual inspection of experimental MS/MS data. Rather than being able to simulate MS/MS spectra for all 112 million compounds in PubChem, we anticipate that in silico spectra could be a valuable addition for a subset of specific molecular families for which the performance and quality of spectrum prediction tools is well understood and has been carefully validated. For example, high-fidelity peptide mass spectra simulated by deep learning-powered spectrum prediction tools are being increasingly incorporated into various proteomics bioinformatics workflows (Gessulat et al., 2019), and the LipidBlast library, which consists of approximately half a million simulated MS/MS spectra, is available through MoNA to annotate lipids (Kind et al., 2013).
Furthermore, there is a mismatch between the compounds included in spectral libraries and the MS/MS spectra observed in experimental data. For example, out of 586,647 MS/MS spectra present in the GNPS community spectral libraries, 22% have been found in experimental datasets deposited to GNPS (Figure 2a). This indicates that many of the compounds represented in reference libraries are not observed in metabolomics experiments, or that the library MS/MS spectra were created in a different fashion than for experimental data, such as when the preferred ion form is not included (Figure 3). Notably, even as the public libraries have grown spectacularly over the previous decade, the rate of matched library spectra has remained relatively consistent. This illustrates the previously described bias in the commercial availability of pure reference compounds that are typically used for spectral library creation efforts. It also indicates that many relevant biological compounds are currently still missing from available spectral libraries, and that careful prioritization of the reference compounds to include is an important aspect of generating spectral libraries that provide maximum benefit.
An emerging approach towards creating comprehensive metabolomics knowledge bases is to expand upon traditional spectral libraries by integrating controlled and structured metadata information alongside the mass spectral data. “Reference data-driven metabolomics” uses not only annotated MS/MS spectra, but also all unannotated spectra in combination with metadata-annotated source data (e.g. were the samples derived from foods, personal care products, medications, etc.) as a pseudo spectral library (Gauglitz et al., 2022). This strategy was exemplified by linking approximately 100,000 MS/MS spectra to 3,600 foods. A key aspect of this approach is that the foods are organized in a hierarchical ontology to enable granular downstream analyses of the food origins. For instance, an example path in the food hierarchy consists of “fruit → citrus → lemon → pink lemon.” This enables performing analyses akin to microbiome science, in which the data may be interpreted at the class, genus, species, or even strain level depending on the research question at hand. Although this approach does not produce exact molecular identities, it provides essential insights into the origin of the data by matching against the reference source data, such as food. Reference data-driven metabolomics using the GNPS platform can increase the number of interpreted MS/MS spectra by up to an order of magnitude, and it has been used to obtain empirical assessments of dietary patterns from untargeted metabolomics data (Gauglitz et al., 2022). Metadata-driven analyses can be broadly applied beyond diet readouts to also investigate other exposures (e.g. medications, personal care products, agrichemicals), disease phenotypes, organ system distributions, taxonomic matching, and many other uses. The key aspect that empowers reference data-driven metabolomics is that the spectral data are linked to controlled and curated metadata to be used as a pseudo-reference library. A less flexible but related metadata system is available for GC-MS data using BinBase, which covers a limited set of metadata (Lai et al., 2017).
Although applied to proteomics, a related approach consists of “spectral archives,” which include MS/MS spectra that have been repeatedly observed, irrespective of whether they could be annotated (Frank et al., 2011). Spectral archives can be built by large-scale clustering of MS/MS spectra across multiple datasets or in an entire repository (Frank et al., 2008; Griss et al., 2013, 2016; Bittremieux, May, et al., 2022). In this fashion, commonly observed spectra can be grouped and unannotated spectra can be linked across multiple experiments to find correlations with identified compounds (Stein, 2012).
There are also challenges associated with the ever-increasing size of spectral libraries. First, this makes it more difficult to process the data, and better compute infrastructure and optimized algorithms are necessary to process large spectral libraries (Bittremieux et al., 2019; Bittremieux, Meysman, et al., 2018). Cloud-based solutions, such as the GNPS analysis platform, have the potential to be extremely scalable while hiding this complexity from the user. For example, GNPS allows users to query their data against 1.2 billion open MS/MS spectra using the Mass Spectrometry Search Tool (MASST) to discover public datasets that contain similar MS/MS spectra (M. Wang et al., 2020; West et al., 2022). Developing and maintaining such platforms requires suitable, continued investments and a team willing to maintain the resources for the benefit of the community. The same is also true for MetaboLights (Haug et al., 2013), Metabolomics Workbench (Sud et al., 2015), HMDB (Wishart et al., 2021), MetaboAnalyst (Pang et al., 2022), MZmine (Pluskal et al., 2010), MS-DIAL (Tsugawa et al., 2020, p. 4), and other popular untargeted metabolomics resources. In response, to overcome some of these challenges, subscription models or commercial libraries such as NIST, mzCloud, or METLIN Gen2 continue to be needed. Second, interoperability of various tools and resources is important. There currently does not exist an official data standard for spectral libraries yet. Frequently used spectral library file formats include the mzML (Martens et al., 2011), mzXML (Pedrioli et al., 2004), Mascot Generic Format (MGF), and the NIST MSP formats. Unfortunately, some of these formats are only loosely defined, change over time, often without explicit versioning, and spectrum metadata can be encoded in various non-standardized ways, limiting the usability and portability of such spectral library files. The Proteomics Standards Initiative of the Human Proteome Organization (HUPO-PSI) (Deutsch et al., 2017), which has previously developed fundamental mass spectrometry data standards such as the mzML peak file format (Martens et al., 2011), is currently working on a specification for spectral libraries (https://github.com/HUPO-PSI/mzSpecLib/). Although the HUPO-PSI primarily develops data standards for proteomics, many of their efforts are relevant for any application of biological mass spectrometry. The HUPO-PSI working groups are open to any community contributions, and interested parties are encouraged to engage in the development of this nascent spectral library format to ensure its full compatibility with applications in metabolomics.
In conclusion, we want to re-emphasize the exciting times ahead for spectral libraries in metabolomics. The community has become increasingly aware that capturing metabolomics knowledge in the form of reference MS/MS spectra accelerates discoveries. Existing spectral libraries have grown tremendously in the past few years, and we expect this growth to continue. Bigger libraries, especially those that are freely available for community use, will enable researchers to get more and better annotations from their data and achieve important biological insights. Additionally, it will be possible to develop increasingly powerful machine learning algorithms by training them on large spectral libraries. As some of these machine learning tools will improve the annotation rate in metabolomics and derive more value from existing and new data, this will make it possible to annotate new high-quality MS/MS spectra for inclusion in the next iteration of spectral libraries. As such, the growth of open spectral libraries and development of machine learning tools will proceed in lockstep to power a virtuous cycle and advance metabolomics in the upcoming years.
Acknowledgments
This research was supported by BBSRC-NSF award 2152526 and National Institutes of Health award U19 AG063744.
Footnotes
Conflict of interests statement
PCD is an advisor to Cybele and co-founder and scientific advisor to Ometa and Enveda, with prior approval by UC San Diego. MW is a co-founder of Ometa Labs LLC.
Compliance with ethical standards
This article does not contain any studies with human and/or animal participants performed by any of the authors.
References
- Aksenov AA, da Silva R, Knight R, Lopes NP, & Dorrestein PC (2017). Global chemical analysis of biology by mass spectrometry. Nature Reviews Chemistry, 1(7), 0054. 10.1038/s41570-017-0054 [DOI] [Google Scholar]
- Alka O, Shanthamoorthy P, Witting M, Kleigrewe K, Kohlbacher O, & Röst HL (2022). DIAMetAlyzer allows automated false-discovery rate-controlled analysis for data-independent acquisition in metabolomics. Nature Communications, 13(1), 1347. 10.1038/s41467-022-29006-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron AT, Gentry EC, McPhail KL, Nothias L-F, Nothias-Esposito M, Bouslimani A, Petras D, Gauglitz JM, Sikora N, Vargas F, van der Hooft JJJ, Ernst M, Kang KB, Aceves CM, Caraballo-Rodríguez AM, Koester I, Weldon KC, Bertrand S, Roullier C, … Dorrestein PC (2020). Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nature Protocols, 15(6), 1954–1991. 10.1038/S41596-020-0317-5 [DOI] [PubMed] [Google Scholar]
- Bittremieux W, Avalon NE, Thomas SP, Kakhkhorov SA, Aksenov AA, Gomes PWP, Aceves CM, Caraballo Rodriguez AM, Gauglitz JM, Gerwick WH, Jarmusch AK, Kaddurah-Daouk RF, Kang KB, Kim HW, Kondic T, Mannochio-Russo H, Meehan MJ, Melnik A, Nothias L-F, … Dorrestein PC, (2022). Open access repository-scale propagated nearest neighbor suspect spectral library for untargeted metabolomics. BioRxiv. 10.1101/2022.05.15.490691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittremieux W, Laukens K, & Noble WS (2019). Extremely fast and accurate open modification spectral library searching of high-resolution mass spectra using feature hashing and graphics processing units. Journal of Proteome Research, 18(10), 3792–3799. 10.1021/acs.jproteome.9b00291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittremieux W, May DH, Bilmes J, & Noble WS (2022). A learned embedding for efficient joint analysis of millions of mass spectra. Nature Methods, 19, 675–678. 10.1038/S41592-022-01496-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittremieux W, Meysman P, Noble WS, & Laukens K (2018). Fast open modification spectral library searching through approximate nearest neighbor indexing. Journal of Proteome Research, 17(10), 3463–3474. 10.1021/acs.jproteome.8b00359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittremieux W, Tabb DL, Impens F, Staes A, Timmerman E, Martens L, & Laukens K (2018). Quality control in mass spectrometry-based proteomics. Mass Spectrometry Reviews, 37(5), 697–711. 10.1002/mas.21544 [DOI] [PubMed] [Google Scholar]
- Bowers WD, Delbert SS, Hunter RL, & McIver RT (1984). Fragmentation of oligopeptide ions using ultraviolet laser radiation and Fourier transform mass spectrometry. Journal of the American Chemical Society, 106(23), 7288–7289. 10.1021/ja00335a094 [DOI] [Google Scholar]
- Burke MC, Mirokhin YA, Tchekhovskoi DV, Markey SP, Heidbrink Thompson J, Larkin C, & Stein SE (2017). The hybrid search: A mass spectral library search method for discovery of modifications in proteomics. Journal of Proteome Research. 10.1021/acs.jproteome.6b00988 [DOI] [PubMed] [Google Scholar]
- Chen K, Rannulu NS, Cai Y, Lane P, Liebl AL, Rees BB, Corre C, Challis GL, & Cole RB (2008). Unusual odd-electron fragments from even-electron protonated prodiginine precursors using positive-ion electrospray tandem mass spectrometry. Journal of the American Society for Mass Spectrometry, 19(12), 1856–1866. 10.1016/j.jasms.2008.08.002 [DOI] [PubMed] [Google Scholar]
- Chen X, Wang Z, Wong Y-LE, Wu R, Zhang F, & Chan T-WD (2018). Electron-ion reaction-based dissociation: A powerful ion activation method for the elucidation of natural product structures. Mass Spectrometry Reviews, 37(6), 793–810. 10.1002/mas.21563 [DOI] [PubMed] [Google Scholar]
- Deutsch EW, Orchard S, Binz P-A, Bittremieux W, Eisenacher M, Hermjakob H, Kawano S, Lam H, Mayer G, Menschaert G, Perez-Riverol Y, Salek RM, Tabb DL, Tenzer S, Vizcaíno JA, Walzer M, & Jones AR (2017). Proteomics Standards Initiative: Fifteen years of progress and future work. Journal of Proteome Research, 16(12), 4288–4298. 10.1021/acs.jproteome.7b00370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch EW, Perez-Riverol Y, Chalkley RJ, Wilhelm M, Tate S, Sachsenberg T, Walzer M, Käll L, Delanghe B, Böcker S, Schymanski EL, Wilmes P, Dorfer V, Kuster B, Volders P-J, Jehmlich N, Vissers JPC, Wolan DW, Wang AY, … Röst H (2018). Expanding the use of spectral libraries in proteomics. Journal of Proteome Research, 17(12), 4051–4060. 10.1021/acs.jproteome.8b00485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo-Almenara X, Guijas C, Billings E, Montenegro-Burke JR, Uritboonthai W, Aisporna AE, Chen E, Benton HP, & Siuzdak G (2019). The METLIN small molecule dataset for machine learning-based retention time prediction. Nature Communications, 10(1), 5811. 10.1038/s41467-019-13680-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo-Almenara X, Montenegro-Burke JR, Ivanisevic J, Thomas A, Sidibé J, Teav T, Guijas C, Aisporna AE, Rinehart D, Hoang L, Nordström A, Gómez-Romero M, Whiley L, Lewis MR, Nicholson JK, Benton HP, & Siuzdak G (2018). XCMS-MRM and METLIN-MRM: A cloud library and public resource for targeted analysis of small molecules. Nature Methods, 15(9), 681–684. 10.1038/S41592-018-0110-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresen S, Gergov M, Politi L, Halter C, & Weinmann W (2009). ESI-MS/MS library of 1,253 compounds for application in forensic and clinical toxicology. Analytical and Bioanalytical Chemistry, 395(8), 2521–2526. 10.1007/s00216-009-3084-2 [DOI] [PubMed] [Google Scholar]
- Ducati AO, Ruskic D, Sosnowski P, Baba T, Bonner R, & Hopfgartner G (2021). Improved metabolite characterization by liquid chromatography – Tandem mass spectrometry through electron impact type fragments from adduct ions. Analytica Chimica Acta, 1150, 338207. 10.1016/j.aca.2021.338207 [DOI] [PubMed] [Google Scholar]
- Dührkop K, Shen H, Meusel M, Rousu J, & Böoker S (2015). Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proceedings of the National Academy of Sciences, 112(41), 12580–12585. 10.1073/pnas.1509788112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Elimat T, Figueroa M, Ehrmann BM, Cech NB, Pearce CJ, & Oberlies NH (2013). High-resolution MS, MS/MS, and UV database of fungal secondary metabolites as a dereplication protocol for bioactive natural products. Journal of Natural Products, 76(9), 1709–1716. 10.1021/np4004307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, Searle BC, Clauser KR, & Tabb DL (2011). A face in the crowd: Recognizing peptides through database search. Molecular & Cellular Proteomics, 10(11), R111.009522. 10.1074/mcp.R111.009522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox Ramos AE, Le Pogam P, Fox Alcover C, Otogo N’Nang E, Cauchie G, Hazni H, Awang K, Bréard D, Echavarren AM, Frédérich M, Gaslonde T, Girardot M, Grougnet R, Kirillova MS, Kritsanida M, Lémus C, Le Ray A-M, Lewin G, Litaudon M, … Beniddir MA (2019). Collected mass spectrometry data on monoterpene indole alkaloids from natural product chemistry research. Scientific Data, 6(1), 15. 10.1038/s41597-019-0028-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank AM, Bandeira N, Shen Z, Tanner S, Briggs SP, Smith RD, & Pevzner PA (2008). Clustering millions of tandem mass spectra. Journal of Proteome Research, 7(1), 113–122. 10.1021/pr070361e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank AM, Monroe ME, Shah AR, Carver JJ, Bandeira N, Moore RJ, Anderson GA, Smith RD, & Pevzner PA (2011). Spectral archives: Extending spectral libraries to analyze both identified and unidentified spectra. Nature Methods, 8(7), 587–591. 10.1038/nmeth.1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel W, The M, Zolg DP, Bayer FP, Shouman O, Lautenbacher L, Schnatbaum K, Zerweck J, Knaute T, Delanghe B, Huhmer A, Wenschuh H, Reimer U, Médard G, Kuster B, & Wilhelm M (2022). Prosit-TMT: Deep learning boosts identification of TMT-labeled peptides. Analytical Chemistry, 94(20), 7181–7190. 10.1021/acs.analchem.1C05435 [DOI] [PubMed] [Google Scholar]
- Gauglitz JM, West KA, Bittremieux W, Williams CL, Weldon KC, Panitchpakdi M, Di Ottavio F, Aceves CM, Brown E, Sikora NC, Jarmusch AK, Martino C, Tripathi A, Meehan MJ, Dorrestein K, Shaffer JP, Coras R, Vargas F, Goldasich LD, … Dorrestein PC (2022). Enhancing untargeted metabolomics using metadata-based source annotation. Nature Biotechnology. 10.1038/S41587-022-01368-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry E, Collins S, Panitchpakdi M, Belda-Ferre P, Stewart A, Wang M, Jarmusch A, Avila-Pacheco J, Plichta D, Aron A, Vlamakis H, Ananthakrishnan A, Clish C, Xavier R, Baker E, Patterson A, Knight R, Siegel D, & Dorrestein PC (2021). A synthesis-based reverse metabolomics approach for the discovery of chemical structures from humans and animals. Research Square. 10.21203/rs.3.rs-820302/v1 [DOI] [Google Scholar]
- Gessulat S, Schmidt T, Zolg DP, Samaras P, Schnatbaum K, Zerweck J, Knaute T, Rechenberger J, Delanghe B, Huhmer A, Reimer U, Ehrlich H-C, Aiche S, Kuster B, & Wilhelm M (2019). Prosit: Proteome-wide prediction of peptide tandem mass spectra by deep learning. Nature Methods, 16(6), 509–518. 10.1038/S41592-019-0426-7 [DOI] [PubMed] [Google Scholar]
- Gibney E. (2022). Could machine learning fuel a reproducibility crisis in science? Nature, d41586-022-02035-w. 10.1038/d41586-022-02035-w [DOI] [PubMed] [Google Scholar]
- Griss J. (2016). Spectral library searching in proteomics. PROTEOMICS, 16(5), 729–740. 10.1002/pmic.201500296 [DOI] [PubMed] [Google Scholar]
- Griss J, Foster JM, Hermjakob H, & Vizcaíno JA (2013). PRIDE Cluster: Building a consensus of proteomics data. Nature Methods, 10(2), 95–96. 10.1038/nmeth.2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griss J, Perez-Riverol Y, Lewis S, Tabb DL, Dianes JA, del-Toro N, Rurik M, Walzer M, Kohlbacher O, Hermjakob H, Wang R, & Vizcaíno JA (2016). Recognizing millions of consistently unidentified spectra across hundreds of shotgun proteomics datasets. Nature Methods, 13(8), 651–656. 10.1038/nmeth.3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug K, Salek RM, Conesa P, Hastings J, de Matos P, Rijnbeek M, Mahendraker T, Williams M, Neumann S, Rocca-Serra P, Maguire E, González-Beltrán A, Sansone S-A, Griffin JL, & Steinbeck C (2013). MetaboLights—An open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Research, 41(D1), D781–D786. 10.1093/nar/gks1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiles S. (2021). Advanced tandem mass spectrometry in metabolomics and lipidomics—Methods and applications. Analytical and Bioanalytical Chemistry, 413(24), 5927–5948. 10.1007/s00216-021-03425-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Mesa M, Le Bizec B, Monteau F, García-Campaña AM, & Dervilly-Pinel G (2018). Collision cross section (CCS) database: An additional measure to characterize steroids. Analytical Chemistry, 90(7), 4616–4625. 10.1021/acs.analchem.7b05117 [DOI] [PubMed] [Google Scholar]
- Hoffmann MA, Nothias L-F, Ludwig M, Fleischauer M, Gentry EC, Witting M, Dorrestein PC, Dührkop K, Böcker S (2021). High-confidence structural annotation of metabolites absent from spectral libraries. Nature Biotechnology, 40(3), 411–421. 10.1038/S41587-021-01045-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann WD, & Jackson GP(2014). Charge transfer dissociation (CTD) mass spectrometry of peptide cations using kiloelectronvolt helium cations. Journal of the American Society for Mass Spectrometry, 25(11), 1939–1943. 10.1007/s13361-014-0989-6 [DOI] [PubMed] [Google Scholar]
- Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, Oda Y, Kakazu Y, Kusano M, Tohge T, Matsuda F, Sawada Y, Hirai MY, Nakanishi H, Ikeda K, … Nishioka T (2010). MassBank: A public repository for sharing mass spectral data for life sciences. Journal of Mass Spectrometry, 45(7), 703–714. 10.1002/jms.1777 [DOI] [PubMed] [Google Scholar]
- Hu A, Noble WS, & Wolf-Yadlin A (2016). Technical advances in proteomics: New developments in data-independent acquisition. F1000Research, 5(F1000 Faculty Rev), 419. 10.12688/f1000research.7042.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Zhu H, Shinn P, Ngan D, Ye L, Thakur A, Grewal G, Zhao T, Southall N, Hall MD, Simeonov A, & Austin CP (2019). The NCATS Pharmaceutical Collection A 10-year update. Drug Discovery Today, 24(12), 2341–2349. 10.1016/j.drudis.2019.09.019 [DOI] [PubMed] [Google Scholar]
- Huber F, Ridder L, Verhoeven S, Spaaks JH, Diblen F, Rogers S, & van der Hooft JJJ (2021). Spec2Vec: Improved mass spectral similarity scoring through learning of structural relationships. PLOS Computational Biology, 17(2), e1008724. 10.1371/journal.pcbi.1008724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber F, van der Burg S, van der Hooft JJJ, & Ridder L (2021). MS2DeepScore: A novel deep learning similarity measure to compare tandem mass spectra. Journal of Cheminformatics, 13(1), 84. 10.1186/s13321-021-00558-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelchtermans P, Bittremieux W, De Grave K, Degroeve S, Ramon J, Laukens K, Valkenborg D, Barsnes H, & Martens L (2014). Machine learning applications in proteomics research: How the past can boost the future. PROTEOMICS, 14(4–5), 353–366. 10.1002/pmic.201300289 [DOI] [PubMed] [Google Scholar]
- Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, Zaslavsky L, Zhang J, & Bolton EE (2021). PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Research, 49(D1), D1388–D1395. 10.1093/nar/gkaa971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind T, Liu K-H, Lee DY, DeFelice B, Meissen JK, & Fiehn O (2013). LipidBlast in silico tandem mass spectrometry database for lipid identification. Nature Methods, 10(8), 755–758. 10.1038/nmeth.2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind T, Tsugawa H, Cajka T, Ma Y, Lai Z, Mehta SS, Wohlgemuth G, Barupal DK, Showalter MR, Arita M, & Fiehn O (2018). Identification of small molecules using accurate mass MS/MS search. Mass Spectrometry Reviews, 37(4), 513–532. 10.1002/mas.21535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krettler CA, & Thallinger GG (2021). A map of mass spectrometry-based in silico fragmentation prediction and compound identification in metabolomics. Briefings in Bioinformatics, 22(6), bbab073. 10.1093/bib/bbab073 [DOI] [PubMed] [Google Scholar]
- Kyle JE, Crowell KL, Casey CP, Fujimoto GM, Kim S, Dautel SE, Smith RD, Payne SH, & Metz TO (2017). LIQUID: an-open source software for identifying lipids in LC-MS/MS-based lipidomics data. Bioinformatics, 33(11), 1744–1746. 10.1093/bioinformatics/btx046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Tsugawa H, Wohlgemuth G, Mehta S, Mueller M, Zheng Y, Ogiwara A, Meissen J, Showalter M, Takeuchi K, Kind T, Beal P, Arita M, & Fiehn O (2017). Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nature Methods, 15(1), 53–56. 10.1038/nmeth.4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCun Y, Bengio Y, & Hinton G (2015). Deep learning. Nature, 521(7553), 436–444. 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- Li Y, Kind T, Folz J, Vaniya A, Mehta SS, & Fiehn O (2021). Spectral entropy outperforms MS/MS dot product similarity for small-molecule compound identification. Nature Methods, 18(12), 1524–1531. 10.1038/s41592-021-01331-z [DOI] [PubMed] [Google Scholar]
- Liu Y, De Vijlder T, Bittremieux W, Laukens K, & Heyndrickx W (2021). Current and future deep learning algorithms for MS/MS-based small molecule structure elucidation. Rapid Communications in Mass Spectrometry, e9120. 10.1002/rcm.9120 [DOI] [PubMed] [Google Scholar]
- Liu Y, Mrzic A, Meysman P, De Vijlder T, Romijn EP, Valkenborg D, Bittremieux W, & Laukens K (2020). MESSAR: Automated recommendation of metabolite substructures from tandem mass spectra. PLOS ONE, 15(1), e0226770. 10.1371/journal.pone.0226770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens L, Chambers M, Sturm M, Kessner D, Levander F, Shofstahl J, Tang WH, Römpp A, Neumann S, Pizarro AD, Montecchi-Palazzi L, Tasman N, Coleman M, Reisinger F, Souda P, Hermjakob H, Binz P-A, & Deutsch EW (2011).MzML—a community standard for mass spectrometry data. Molecular & Cellular Proteomics, 10(1), R110.000133–R110.000133. 10.1074/mcp.R110.000133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberacher H, Weinmann W, & Dresen S (2011). Quality evaluation of tandem mass spectral libraries. Analytical and Bioanalytical Chemistry, 400(8), 2641–2648. 10.1007/S00216-010-4598-3 [DOI] [PubMed] [Google Scholar]
- Olivier-Jimenez D, Chollet-Krugler M, Rondeau D, Beniddir MA, Ferron S, Delhaye T, Allard P-M, Wolfender J-L, Sipman HJM, Lücking R, Boustie J, & Le Pogam P (2019). A database of high-resolution MS/MS spectra for lichen metabolites. Scientific Data, 6(1), 294. 10.1038/s41597-019-0305-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer A, Phapale P, Chernyavsky I, Lavigne R, Fay D, Tarasov A, Kovalev V, Fuchser J, Nikolenko S, Pineau C, Becker M, & Alexandrov T (2016). FDR-controlled metabolite annotation for high-resolution imaging mass spectrometry. Nature Methods, 14(1), 57–60. 10.1038/nmeth.4072 [DOI] [PubMed] [Google Scholar]
- Pang Z, Zhou G, Ewald J, Chang L, Hacariz O, Basu N, & Xia J, (2022). Using MetaboAnalyst 5.0 for LC–HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nature Protocols. 10.1038/S41596-022-00710-w [DOI] [PubMed] [Google Scholar]
- Pedrioli PGA, Eng JK, Hubley R, Vogelzang M, Deutsch EW, Raught B, Pratt B, Nilsson E, Angeletti RH, Apweiler R, Cheung K, Costello CE, Hermjakob H, Huang S, Julian RK, Kapp E, McComb ME, Oliver SG, Omenn G, … Aebersold R (2004). A common open representation of mass spectrometry data and its application to proteomics research. Nature Biotechnology, 22(11), 1459–1466. 10.1038/nbt1031 [DOI] [PubMed] [Google Scholar]
- Peisl BYL, Schymanski EL, & Wilmes P (2018). Dark matter in host-microbiome metabolomics: Tackling the unknowns–A review. Analytica Chimica Acta, 1037, 13–27. 10.1016/j.aca.2017.12.034 [DOI] [PubMed] [Google Scholar]
- Phapale P, Palmer A, Gathungu RM, Kale D, Brügger B, & Alexandrov T (2021). Public LC-Orbitrap Tandem Mass Spectral Library for Metabolite Identification. Journal of Proteome Research, 20(4), 2089–2097. 10.1021/acs.jproteome.0c00930 [DOI] [PubMed] [Google Scholar]
- Picache JA, Rose BS, Balinski A, Leaptrot KL, Sherrod SD, May JC, & McLean JA (2018). Collision cross section compendium to annotate and predict multi-omic compound identities. Chemical Science, 10(4), 983–993. 10.1039/C8SC04396E [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluskal T, Castillo S, Villar-Briones A, & Orešič M (2010). MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics, 11(1), 395. 10.1186/1471-2105-11-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remoroza CA, Liang Y, Mak TD, Mirokhin Y, Sheetlin SL, Yang X, San Andres JV, Power ML, & Stein SE (2020). Increasing the coverage of a mass spectral library of milk oligosaccharides using a hybrid-search-based bootstrapping method and milks from a wide variety of mammals. Analytical Chemistry, 92(15), 10316–10326. 10.1021/acs.analchem.0c00342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remoroza CA, Mak TD, De Leoz MLA, Mirokhin YA, & Stein SE (2018). Creating a mass spectral reference library for oligosaccharides in human milk. Analytical Chemistry, 90(15), 8977–8988. 10.1021/acs.analchem.8b01176 [DOI] [PubMed] [Google Scholar]
- Righetti L, Bergmann A, Galaverna G, Rolfsson O, Paglia G, & Dall’Asta C (2018). Ion mobility-derived collision cross section database: Application to mycotoxin analysis. Analytica Chimica Acta, 1014, 50–57. 10.1016/j.aca.2018.01.047 [DOI] [PubMed] [Google Scholar]
- Sawada Y, Nakabayashi R, Yamada Y, Suzuki M, Sato M, Sakata A, Akiyama K, Sakurai T, Matsuda F, Aoki T, Hirai MY, & Saito K (2012). RIKEN tandem mass spectral database (ReSpect) for phytochemicals: A plant-specific MS/MS-based data resource and database. Phytochemistry, 82, 38–45. 10.1016/j.phytochem.2012.07.007 [DOI] [PubMed] [Google Scholar]
- Scheubert K, Hufsky F, Petras D, Wang M, Nothias L-F, Dührkop K, Bandeira N, Dorrestein PC, & Böcker S (2017). Significance estimation for large scale metabolomics annotations by spectral matching. Nature Communications, 8(1), 1494. 10.1038/s41467-017-01318-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid R, Petras D, Nothias L-F, Wang M, Aron AT, Jagels A, Tsugawa H, Rainer J, Garcia-Aloy M, Dührkop K, Korf A, Pluskal T, Kameník Z, Jarmusch AK, Caraballo-Rodríguez AM, Weldon KC, Nothias-Esposito M, Aksenov AA, Bauermeister A, … Dorrestein PC (2021). Ion identity molecular networking for mass spectrometry-based metabolomics in the GNPS environment. Nature Communications, 12(1), 3832. 10.1038/s41467-021-23953-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder M, Meyer SW, Heyman HM, Barsch A, & Sumner LW (2019). Generation of a collision cross section library for multi-dimensional plant metabolomics using UHPLC-trapped ion mobility-MS/MS. Metabolites, 10(1), 13. 10.3390/metabo10010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymanski E, & Neumann S (2013). The Critical Assessment of Small Molecule Identification (CASMI): Challenges and solutions. Metabolites, 3(3), 517–538. 10.3390/metabo3030517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahaf N, Rogachev I, Heinig U, Meir S, Malitsky S, Battat M, Wyner H, Zheng S, Wehrens R, & Aharoni A (2016). The WEIZMASS spectral library for high-confidence metabolite identification. Nature Communications, 7(1), 12423. 10.1038/ncomms12423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W, & Lam H (2017). Tandem mass spectral libraries of peptides and their roles in proteomics research. Mass Spectrometry Reviews, 36(5), 634–648. 10.1002/mas.21512 [DOI] [PubMed] [Google Scholar]
- Shrivastava AD, Swainston N, Samanta S, Roberts I, Wright Muelas M, & Kell DB (2021). MassGenie: A transformer-based deep learning method for identifying small molecules from their mass spectra. Biomolecules, 11(12), 1793. 10.3390/biom11121793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shteynberg D, Nesvizhskii AI, Moritz RL, & Deutsch EW (2013). Combining results of multiple search engines in proteomics. Molecular & Cellular Proteomics, 12(9), 2383–2393. 10.1074/mcp.R113.027797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanstrup J, Neumann S, & Vrhovšek U (2015). PredRet: Prediction of retention time by direct mapping between multiple chromatographic systems. Analytical Chemistry, 87(18), 9421–9428. 10.1021/acs.analchem.5b02287 [DOI] [PubMed] [Google Scholar]
- Stein S. (2012). Mass spectral reference libraries: An ever-expanding resource for chemical identification. Analytical Chemistry, 84(17), 7274–7282. 10.1021/ac301205z [DOI] [PubMed] [Google Scholar]
- Sud M, Fahy E, Cotter D, Azam K, Vadivelu I, Burant C, Edison A, Fiehn O, Higashi R, Nair KS, Sumner S, & Subramaniam S (2015). Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Research, 44(D1), D463–D470. 10.1093/nar/gkv1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW-M, Fiehn O, Goodacre R, Griffin JL, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane AN, Lindon JC, Marriott P, Nicholls AW, … Viant MR (2007). Proposed minimum reporting standards for chemical analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics, 3(3), 211–221. 10.1007/s11306-007-0082-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada I, Tsugawa H, Meister I, Zhang P, Shsu R, Katsumi R, Wheselock CE, Arita M, Chaleckis R (2019). Creating a reliable mass spectral-retention time library for all ion fragmentation-based metabolomics. Metabolites, 9(11), 251. 10.3390/metabo9110251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwary S, Levy R, Gutenbrunner P, Salinas Soto F, Palaniappan KK, Deming L, Berndl M, Brant A, Cimermancic P, & Cox J (2019). High-quality MS/MS spectrum prediction for data-dependent and data-independent acquisition data analysis. Nature Methods, 16(6), 519–525. 10.1038/s41592-019-0427-6 [DOI] [PubMed] [Google Scholar]
- Treen DGC, Wang M, Xing S, Louie KB, Huan T, Dorrestein PC, Northen TR, & Bowen BP (2022). SIMILE enables alignment of tandem mass spectra with statistical significance. Nature Communications, 13(1), 2510. 10.1038/s41467-022-30118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugawa H. (2018). Advances in computational metabolomics and databases deepen the understanding of metabolisms. Current Opinion in Biotechnology, 54, 10–17. 10.1016/j.copbio.2018.01.008 [DOI] [PubMed] [Google Scholar]
- Tsugawa H, Ikeda K, Takahashi M, Satoh A, Mori Y, Uchino H, Okahashi N, Yamada Y, Tada I, Bonini P, Higashi Y, Okazaki Y, Zhou Z, Zhu Z-J, Koelmel J, Cajka T, Fiehn O, Saito K, Arita M, & Arita M (2020). A lipidome atlas in MS-DIAL 4. Nature Biotechnology, 38(10), 1159–1163. 10.1038/s41587-020-0531-2 [DOI] [PubMed] [Google Scholar]
- van der Hooft JJJ, Wandy J, Barrett MP, Burgess KEV, & Rogers S (2016). Topic modeling for untargeted substructure exploration in metabolomics. Proceedings of the National Academy of Sciences, 113(48), 13738–13743. 10.1073/pnas.1608041113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinaixa M, Schymanski EL, Neumann S, Navarro M, Salek RM, & Yanes O (2016). Mass spectral databases for LC/MS- and GC/MS-based metabolomics: State of the field and future prospects. TrAC Trends in Analytical Chemistry, 78, 23–35. 10.1016/j.trac.2015.09.005 [DOI] [Google Scholar]
- Wallace WE, Ji W, Tchekhovskoi DV, Phinney KW, & Stein SE (2017). Mass spectral library quality assurance by inter-library comparison. Journal of the American Society for Mass Spectrometry, 28(4), 733–738. 10.1007/s13361-016-1589-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Liigand J, Tian S, Arndt D, Greiner R, & Wishart DS (2021). CFM-ID 4.0: More accurate ESI-MS/MS spectral prediction and compound identification. Analytical Chemistry, 93(34), 11692–11700. 10.1021/acs.analchem.1c01465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T, Porto C, Bouslimani A, Melnik AV, Meehan MJ, Liu W-T, Crüsemann M, Boudreau PD, Esquenazi E, Sandoval-Calderón M, … Bandeira N (2016). Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nature Biotechnology, 34(8), 828–837. 10.1038/nbt.3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Jarmusch AK, Vargas F, Aksenov AA, Gauglitz JM, Weldon K, Petras D, da Silva R, Quinn R, Melnik AV, van der Hooft JJJ, Caraballo-Rodríguez AM, Nothias LF, Aceves CM, Panitchpakdi M, Brown E, Di Ottavio F, Sikora N, Elijah EO, … Dorrestein PC (2020). Mass spectrometry searches using MASST. Nature Biotechnology. 10.1038/s41587-019-0375-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wang J, Carver J, Pullman BS, Cha SW, & Bandeira N (2018). Assembling the community-scale discoverable human proteome. Cell Systems, 7(4), 412–421.e5. 10.1016/j.cels.2018.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Jones DR, Shaw TI, Cho J-H, Wang Y, Tan H, Xie B, Zhou S, Li Y, & Peng J (2018). Target-decoy-based false discovery rate estimation for large-scale metabolite identification. Journal of Proteome Research, 17(7), 2328–2334. 10.1021/acs.jproteome.8b00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West KA, Schmid R, Gauglitz JM, Wang M, & Dorrestein PC (2022). FoodMASST a mass spectrometry search tool for foods and beverages. Npj Science of Food, 6(1), 22. 10.1038/S41538-022-00137-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Zolg DP, Graber M, Gessulat S, Schmidt T, Schnatbaum K, Schwencke-Westphal C, Seifert P, de Andrade Krätzig N, Zerweck J, Knaute T, Bräunlein E, Samaras P, Lautenbacher L, Klaeger S, Wenschuh H, Rad R, Delanghe B, Huhmer A, … Kuster B (2021). Deep learning boosts sensitivity of mass spectrometry-based immunopeptidomics. Nature Communications, 12(1), 3346. 10.1038/s41467-021-23713-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SL, Way GP, Bittremieux W, Armache J-P, Haendel MA, & Hoffman MM (2021). Sharing biological data: Why, when, and how. FEBS Letters, 595(7), 847–863. 10.1002/1873-3468.14067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Guo A, Oler E, Wang F, Anjum A, Peters H, Dizon R, Sayeeda Z, Tian S, Lee BL, Berjanskii M, Mah R, Yamamoto M, Jovel J, Torres-Calzada C, Hiebert-Giesbrecht M, Lui VW, Varshavi D, Varshavi D, … Gautam V (2021). HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Research, 50(D1), D622–D631. 10.1093/nar/gkab1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth G, Mehta SS, Mejia RF, Neumann S, Pedrosa D, Pluskal T, Schymanski EL, Willighagen EL, Wilson M, Wishart DS, Arita M, Dorrestein PC, Bandeira N, Wang M, Schulze T, Salek RM, Steinbeck C, Nainala VC, Mistrik R, … Fiehn O (2016). SPLASH, a hashed identifier for mass spectra. Nature Biotechnology, 34(11), 1099–1101. 10.1038/nbt.3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, & Huan T (2022). Radical fragment ions in collision-induced dissociation-based tandem mass spectrometry. Analytica Chimica Acta, 1200, 339613. 10.1016/j.aca.2022.339613 [DOI] [PubMed] [Google Scholar]
- Xu R, Sheng J, Bai M, Shu K, Zhu Y, & Chang C (2020). A comprehensive evaluation of MS/MS spectrum prediction tools for shotgun proteomics. PROTEOMICS, 1900345. 10.1002/pmic.201900345 [DOI] [PubMed] [Google Scholar]
- Xue J, Guijas C, Benton HP, Warth B, & Siuzdak G (2020). METLIN MS2 molecular standards database: A broad chemical and biological resource. Nature Methods, 17(10), 953–954. 10.1038/s41592-020-0942-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Markey SP, Marupaka R, Dong Q, Cooper BT, Mirokhin YA, Wallace WE, & Stein SE (2020). Mass spectral library of acylcarnitines derived from human urine. Analytical Chemistry, 92(9), 6521–6528. 10.1021/acs.analchem.0c00129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemany PD (1950). Punched card catalog of mass spectra useful in qualitative analysis. Analytical Chemistry, 22(7), 920–922. 10.1021/ac60043a021 [DOI] [Google Scholar]
- Zhang X, Li Y, Shao W, & Lam H (2011). Understanding the improved sensitivity of spectral library searching over sequence database searching in proteomics data analysis. PROTEOMICS, 11(6), 1075–1085. 10.1002/pmic.201000492 [DOI] [PubMed] [Google Scholar]
- Zheng X, Aly NA, Zhou Y, Dupuis KT, Bilbao A, Paurus VL, Orton DJ, Wilson R, Payne SH, Smith RD, & Baker ES (2017). A structural examination and collision cross section database for over 500 metabolites and xenobiotics using drift tube ion mobility spectrometry. Chemical Science, 8(11), 7724–7736. 10.1039/C7SC03464D [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X-X, Zeng W-F, Chi H, Luo C, Liu C, Zhan J, He S-M, & Zhang Z (2017). pDeep: Predicting MS/MS spectra of peptides with deep learning. Analytical Chemistry, 89(23), 12690–12697. 10.1021/acs.analchem.7b02566 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Luo M, Chen X, Yin Y, Xiong X, Wang R, & Zhu Z-J (2020). Ion mobility collision cross-section atlas for known and unknown metabolite annotation in untargeted metabolomics. Nature Communications, 11(1), 4334. 10.1038/S41467-020-18171-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolg DP, Wilhelm M, Schmidt T, Médard G, Zerweck J, Knaute T, Wenschuh H, Reimer U, Schnatbaum K, & Kuster B (2018). ProteomeTools: Systematic characterization of 21 post-translational protein modifications by liquid chromatography tandem mass spectrometry (LC-MS/MS) using synthetic peptides. Molecular & Cellular Proteomics, 17(9), 1850–1863. 10.1074/mcp.TIR118.000783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolg DP, Wilhelm M, Schnatbaum K, Zerweck J, Knaute T, Delanghe B, Bailey DJ, Gessulat S, Ehrlich H-C, Weininger M, Yu P, Schlegl J, Kramer K, Schmidt T, Kusebauch U, Deutsch EW, Aebersold R, Moritz RL, Wenschuh H, … Kuster B (2017). Building ProteomeTools based on a complete synthetic human proteome. Nature Methods. 10.1038/nmeth.4153 [DOI] [PMC free article] [PubMed] [Google Scholar]