Abstract
Analysis of circulating tumor cells (CTCs) as a tool for monitoring metastatic cancers, early diagnosis, and evaluation of disease prognosis paves the way toward personalized cancer treatment. Developing an effective, feasible, and low-cost method to facilitate CTC isolation is, therefore, vital. In the present study, we integrated magnetic nanoparticles (MNPs) with microfluidics and used them for the isolation of HER2-positive breast cancer cells. Iron oxide MNPs were synthesized and functionalized with the anti-HER2 antibody. The chemical conjugation was verified by Fourier transform infrared spectroscopy, energy-dispersive X-ray spectroscopy, and dynamic light scattering/zeta potential analysis. The specificity of the functionalized NPs for the separation of HER2-positive from HER2-negative cells was demonstrated in an off-chip test setting. The off-chip isolation efficiency was 59.38%. The efficiency of SK-BR-3 cell isolation using a microfluidic chip with a S-shaped microchannel was considerably enhanced to 96% (a flow rate of 0.5 mL/h) without chip clogging. Besides, the analysis time for the on-chip cell separation was 50% faster. The clear advantages of the present microfluidic system offer a competitive solution in clinical applications.
1. Introduction
Cancer is one of the leading causes of death worldwide.1,2 Tumor metastasis is the main cause of high cancer mortality,3 and preventing that by early diagnosis can reduce the death rate. Currently, cancer diagnosis mostly relies on tissue biopsy and imaging techniques.4 The former is invasive, and the latter is rather expensive. Thus, more versatile tools for the early detection of cancer, particularly in poor developing countries, are required. In this respect, liquid biopsy as a non-invasive method can potentially facilitate the identification and isolation of cancer-related biomarkers, such as circulating tumor cells (CTCs). Liquid biopsy provides in-depth information about cancer, although it has not been translated into the clinic. Therefore, technologies that can capture CTCs from the bloodstream are highly desired for early cancer detection.5−7 Some cancer cell isolation techniques include cell density-based isolation,8,9 size-based filtration isolation,10 and immunomagnetic isolation.11 These methods have the limitations of low yield and purity, the requirement of expensive instruments, and a large sample size. Also, they have a long processing time that lowers the chance of capturing viable CTCs.
Microfluidic techniques have shown great prospects for solving bio-related problems due to their automation, high-throughput nature, high sensitivity, low processing time, and small sample volume. One of the microfluidic-based techniques for CTC isolation is micro-filtration, which is a size-based method.12−15 It provides rapid filtration, and it is label-free. Nevertheless, difficulty in the separation of similar-sized cells, a large sample volume, and chip clogging are the limitations associated with micro-filtration. Contrarily, dielectrophoresis, which relies on dielectric properties of cells, is clog-free and capable of label-free isolation of CTCs.16 However, complicated device fabrication and the use of high electric fields are the challenges that limit the use of this method. Another microfluidic technique is the inertial microfluidic approach that separates CTCs based on their specific size and deformability. The stiffness of CTCs ranges from 200 to 2000 Pa, whereas the stiffness of neutrophils ranges from 69 to 243 Pa.17,18 Previous works based on this method19−22 reported the successful label-free isolation of CTCs without the need for an external electric field. However, they cannot separate CTCs from other cells of the same size.
Unlike the above-mentioned microfluidic techniques, immunoaffinity offers higher selectivity as it involves the use of antibodies coated on the interior side of microchannels. The capture efficiency, however, is low because the laminar flow of the cells decreases the cell–antibody interaction. The efficiency has increased with the employment of pillar-based microchannel shapes. However, the complex 3D structure of microposts makes subsequent analysis of captured cells, like the evaluation of their gene expression profiles by imaging assays like FISH, challenging. Alternatively, immunomagnetic separation inside a microfluidic device using antibody-modified magnetic nanoparticles (MNPs) is an appropriate choice of CTC isolation.23−25 To this end, magnetic microbeads, or MNPs, are chemically bound with antibodies. Unlike microbeads, NPs provide high surface-to-volume ratios and establish stronger interaction with cells during the incubation time; thus, they are preferred over microbeads.
Anti-EpCAM and anti-HER2 are the two common antibodies employed in the existing immunomagnetic microfluidic systems. Cancer cells that overexpress EpCAM, however, undergo epithelial to mesenchymal transition (EMT), leading to the downregulation of several epithelial markers.26,27 As a result, EpCAM protein levels on the surface of cancer cells reduce, leading to the loss of a portion of CTCs’ population due to the disappearance of epithelial markers. HER2 expression, on the other hand, is not affected by the EMT process.28 Meanwhile, HER2 is overexpressed in around 20–30% of breast cancer tumors and is also expressed in bladder cancer, gastric cancer, pancreatic cancer, and ovarian cancer,29 all of which make it a better cell membrane target than EpCAM for CTC isolation. In the present study, we isolated HER2-positive breast cancer cells with high efficiency by combining the high specificity of HER2 antibodies on the surface of MNPs, the superparamagnetic property of MNPs, and the miniaturized nature of microfluidic systems. We synthesized MNPs by the coprecipitation method and functionalized their surface by HER2 through covalent amide linkages. Co-injection of HER2-MNPs and cells into a microfluidic system enabled us to study the recovery of HER2-positive breast cancer cells. The nanosize of MNPs and the repetitive S-shaped pattern of the mixing region allowed for the high interaction of cancer cells’ surface proteins and HER2-MNPs. The proposed on-chip immunomagnetic separation outperformed the conventional magnetic isolation of cancer cells with respect to separation time, sample volume, and isolation efficiency.
2. Materials and Instrumentation
The X-ray diffraction (XRD) pattern of MNPs was recorded by a STOE diffractometer using Cu Kα radiation (λ = 0.15406). Energy-dispersive X-ray spectroscopy (EDS) analysis was conducted for the identification of the elemental composition of MNPs and HER2-MNPs. The Fourier transform infrared (FTIR) spectra were measured with a PerkinElmer (Spectrum 65, United States) spectrometer. A Horiba (SZ-100, Japan) Zetasizer calculated the zeta potential value and hydrodynamic size of MNPs, CEPA-MNPs, and HER2-MNPs. Transmission electron microscopy (TEM) images were achieved by a 100 KV Philips (CM300, United States) electron microscope. Attune NxT flow cytometry and conventional FlowJo 10.8 software were used for cell counting.
Iron(II) chloride tetrahydrate (FeCl2), iron(III) chloride hexahydrate (FeCl3), 3-phosphopropionic acid (CEPA), N-(3-dimethylaminpropyl)-N′-etylcarbodiimide hydrochloride (EDC), N-hydroxysulfosuccinimide sodium salt (Sulfo-NHS), 2-(N-morpholino) ethanesulfonic acid (MES), ammonia solution, hydrochloric acid (HCl), sodium hydroxide (NaOH), phosphate-buffered saline (PBS), and polydimethylsiloxane (PDMS) were purchased from Sigma-Aldrich. Lyophilized herceptin was obtained from the Aryogen company. Fetal bovine serum (FBS), penicillin/streptomycin, trypsin/EDTA, and Dulbecco’s modified Eagle’s medium (DMEM) were obtained from Gibco (Grand Island, NY, USA). All reagents were used without further purification.
2.1. Synthesis of MNPs
MNPs were synthesized by the co-precipitation method.30 Briefly, 2.0 M FeCl2 in 2 M HCl (39.67 g of FeCl2·4H2O in 100 mL of 2 M HCl), 1.0 M FeCl3 in 2 M HCl (67.58 g of FeCl3·6H2O in 250 mL of 2 M HCl), and 50 mL of 0.7 M NH3 were prepared. 1.0 mL of FeCl2 and 4.0 mL of FeCl3 were mixed, followed by the drop-wise addition of 0.7 M NH3. During the addition of a base, the mixture was stirred vigorously. MNPs in the form of a black precipitate are formed. MNPs were subjected to centrifugal washing at 1000 rpm several times and dried in an oven for 24 h. They were characterized by XRD, TEM, vibrating sample magnetometry (VSM), dynamic light scattering (DLS), EDS, and FTIR.
2.1.1. Bio Conjugation of MNPs with Anti-HER2
A two-step procedure was used to conjugate HER2 to the surface of MNPs.31 First, MNPs were coated with 3-phosphopropionic acid (CEPA). 50 mL of 0.1 M NaOH was mixed with 50 mL of 25 mg/mL CEPA, followed by the drop-wise addition of 10 mL of 2.5 mg/mL MNPs during ultrasonication. The mixture was sonicated for 2 h to achieve a fully dispersed suspension. Unreacted CEPA was separated by successive centrifugal washing with deionized water. In the second step, conjugation of anti-HER2 to the surface of CEPA-coated MNPs was conducted through the formation of covalent amide bonds between the carboxyl functional group of CEPA and the amino functional group of the antibody. To this end, 5 mL of 0.5 M MES buffer at pH 6.0 was mixed with 121 mg of sulfo-NHS and 40 mg of EDC. The mixture was added to 15 mL of CEPA-coated MNPs and stirred for 45 min. The colloidal suspension was centrifuged at 4000 rpm to remove the unreacted sulfo-NHS and EDC molecules in the supernatant. Activated MNPs were suspended in 10 mL of PBS (1 mM, pH 8.0) by ultrasonication. Then, a solution of anti-HER2 (17 nmol in 10 mL PBS) was added slowly to the activated MNPs and kept at 4 °C overnight. Unreacted anti-HER2 molecules were removed after the final conjugation step. The HER2-MNPs were redispersed in PBS for further use.
2.2. Design and Fabrication of the Microfluidic Chip
Figure S1 online shows the microfluidic chip comprising a micromixer and resembling a repetitive s-shaped pattern. Magnetophoresis accounts for particles’ isolation in the chip. The specific pattern of the micromixer maximized the interaction between MNPs and cells. The chip included two inlets, one outlet, a mixing region, and a trap region (Figure S1 online). The magnetic field gradient at the trap region of the chip was generated by two neodymium magnets (42MGOe). Figure S1 online shows two magnets located at the trapping region to collect cells which were bound with MNPs.
Soft lithography was used for the fabrication of microfluidic chips (Figure S2 online). Briefly, PDMS was cast by a previously made mold. For creating the mold, a silicon wafer was washed with acetone and isopropyl alcohol three times. Next, 2 g of the SU-8 photoresist was spin-coated at 3000 rpm on the silicon wafer to achieve a thickness of around 30 μm. The wafer was then soft-baked at 95 °C for 10 min and exposed to a laser (Model micro PG101, Heidelberg, Germany) using 68% of 8 mW power. The wafer was post-baked with the same protocol as the soft-baked one. Then, it was dipped into the developer, followed by washing with isopropyl alcohol several times. The treated wafer was hard-baked at 150 °C for 20 min as the final step of mold preparation. In the next step, PDMS was mixed with the curing agent at an 8:1 ratio and transferred to a vacuum chamber for the elimination of bubbles. The mixture was poured on the mold and baked for 1 h at 60 °C. After the mold removal, PDMS was manually cut to the pre-determined size and bonded on a 150 μm-thick glass substrate by oxygen plasma. Prior to the bonding process, the inlets and outlets were punched with an inner diameter of 1.5 mm.
2.3. Cell Culture
SK-BR-3 and MDA-MB-231 cells as HER2-positive and triple-negative cell lines were used, respectively. Cells were cultured in Dulbecco’s modified Eagle’s minimal essential medium-high glucose (DMEM; Life Technologies, Inc., Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) and incubated at 37 °C under a 5% CO2 atmosphere. When 80% confluency was observed, the cells were trypsinized and re-suspended in PBS (Sigma-USA). Before each experiment, the cells were stained using a solution of 4% trypan blue (Sigma-Germany), and their viability was examined by a hemocytometer.
2.4. Cancer Cell Isolation
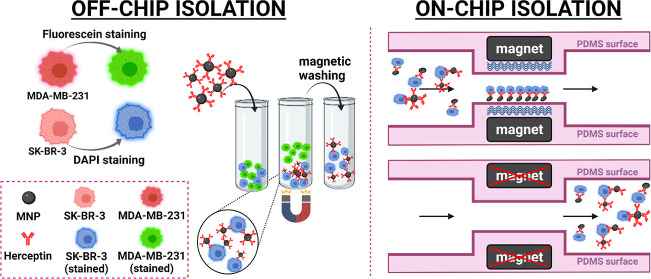
2.4.1. Off-Chip Cell Isolation
The off-chip experiment was conducted on individual cells and cell cocktail (SK-BR-3 + MDA-MB-231). To this end, SK-BR-3 cells (1 × 105) were stained by DAPI and incubated with HER2-MNPs (250 μg/mL). After 1 h, magnetic separation was conducted to collect cells that were attached to the surface of HER2-MNPs. Unattached cells were removed by three-time PBS washing. The captured cells were observed using a fluorescence microscope. In another experiment, cell cocktails comprising SK-BR-3 (HER2-positive) and MDA-MB-231 (HER2-negative) cells of different ratios (1:1, 1:3, and 1:5) were incubated with HER2-MNPs. SK-BR-3 and MDA-MB-231 cells were stained with DAPI and fluorescein dyes, respectively. Magnetic separation was accomplished according to the same protocol used for SK-BR-3 cells. Incubation of bare MNPs with SK-BR-3 cells and HER2-MNPs with MDA-MB-231 cells were the negative control experiments. The total cell number, incubation time, and washing steps were conducted as described for SK-BR-3 cells. Flow cytometry (Attune NxT by applying FlowJo 10.8 software) was adapted for the quantitation of off-chip isolation efficiency. Fluorescein-colored SK-BR-3 cells were mixed by uncolored MDA-MB-231 in different ratios (1:0, 1:2, 1:4, 1:9, 1:19, and 1:99). The total cell number was constant (5 × 105). The fluorescence signal associated with fluorescein inside the SK-BR-3 cells was recorded for each mixture, and a calibration curve was achieved (Figure S4 online). The signal correlated linearly to the number of SK-BR-3 cells over the range of 5 × 103 to 5 × 105 (y = 0.1927x + 2.092, R2 = 0.9926). The off-chip capture efficiency was calculated using the linear equation, in which cell number was the independent variable and fluorescein percentage was the dependent variable. To this end, three identical cell mixtures containing fluorescein-colored SK-BR-3 cells and uncolored MDA-MB-231 in a 1:1 ratio (5 × 105 cells in total) were prepared. The cells were incubated with MNPs-HER2, MNPs, and HER2 for 1 h at room temperature. In each case, after magnetic separation, the supernatant was removed, and the captured cells were washed three times with PBS to ensure the elution of physically attached cells. The separated cells were redispersed in PBS, and their fluorescence signal associated with internalized fluorescein was analyzed by flow cytometry. The number of isolated cells was quantified using the linear correlation between the fluorescein percentage and cell number. Off-chip isolation efficiency was calculated according to (1).
| 1 |
2.4.2. On-Chip Cell Isolation
The microfluidic device was autoclaved before use. Prior to each test, microfluidic channels were filled with BSA 0.5% w/v to decrease nonspecific cell binding. HER2-MNPs were dispersed in 1 mL PBS with the final concentration of 83 μg/mL and ultrasonicated before the test. SK-BR-3 cells (1 × 104) were stained by fluorescein and suspended in 1 mL of PBS. Stained cells and MNPs-HER2 were introduced to the device from inlets 1 and 2, respectively, by the flow rates of 2, 1, 0.75, 0.5, and 0.25 mL/h. Before each run, the device was placed on the neodymium magnets. At the end of the on-chip experiment, captured cells were released by turning off the magnetic field and rinsing the channels with PBS (Figure 1). The collected mixture was centrifuged at 8000 rpm for 5 min, and the supernatant was slowly pipetted out and discarded. The pellet was re-dispersed in 50 μL of PBS and counted using a hemocytometer. Equation 2 was used to determine the capture efficiency in the off-chip test setting.
| 2 |
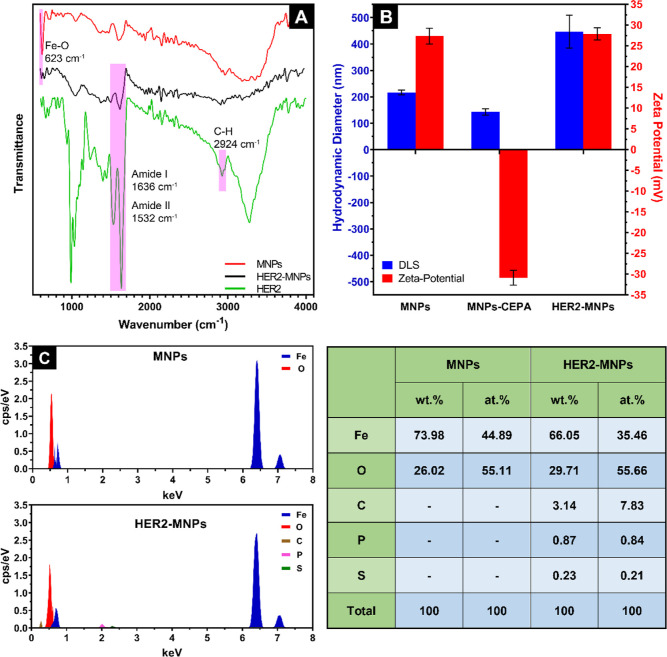
Figure 1.
MNPs’ surface modification by HER2 investigated through (A) FTIR, (B) DLS/zeta potential analysis, and (C) EDS analysis.
3. Results and Discussion
In the present study, HER2-functionalized MNPs combined with a microfluidic device were employed for the capture and isolation of HER2-positive breast cancer cells. MNPs are among the most commonly used nanomaterials in a host of research endeavors, including biology. The synthesis process of MNPs is straightforward. MNPs in the present study were prepared by co-precipitation of iron(II) and iron(III) salts. Modification of HER2 on the NPs’ surface was accomplished using a linker named CEPA. The modified NPs were characterized by TEM, FTIR, DLS, zeta potential, and EDS analyses. The isolation of HER2-positive cancer cells was conducted through off- and on-chip approaches, and the results were compared. In the former, HER2-MNPs and cells were incubated in a microtube for an hour, followed by magnetic separation and washing. In the latter approach, mixing of cells and NPs proceeded in the microfluidic channel. The attached cells to HER2-MNPs were retained at the trap region of the chip using two magnets on both sides of the microfluidic channel. Compared with the conventional off-chip method, the capture efficiency of microfluidic-based separation was considerably higher. In the optimized experimental conditions, the capture efficiency of the on-chip method reached 96.5%.
3.1. Characterization of MNPs
MNPs were synthesized via a co-precipitation method.30 The result of TEM analysis in Figure S3 online showed that MNPs were almost spherical, with an average size of around 20 nm. XRD measurement was used to look into the crystalline structure of the MNPs (Figure S3 online). The peaks coincided with the standard data of magnetite (JCPDS 19-0629), proving that the particles were Fe3O4. Debye–Scherrer equation (D = kλ/β cos φ) determined the crystallites’ size range of 58–97 nm based on the X-ray line broadening. Figure S3 online shows the field-dependent magnetization of bare MNPs studied by a VSM. The measurement was performed with the maximum applied field of 14 kOe in both directions. The absence of hysteresis, i.e., remanence and coercivity close to zero, verifies the superparamagnetic nature and single domain of MNPs. The saturation magnetization (Ms), remanence (Mr), and coercivity (Hc) are 18.633 emu/g, 4.147 emu/g, and 120 Oe, respectively. The ratio of remanence to saturation magnetization (Mr/Ms) below 25% indicates superparamagnetism.32 The Mr/Ms ratio in the present study is 0.22, proving the superparamagnetic property of the as-synthesized MNPs.
3.2. Conjugation of Herceptin to MNPs
Functionalization of MNPs with HER2 was conducted in two steps. The first was to functionalize the surface of MNPs with CEPA as the linker, and the second was the conjugation of HER2 to the linker. FTIR, EDS, and DLS/zeta potential analyses were conducted to verify the covalent bonding of HER2 to MNPs. The FTIR spectrum of MNPs was recorded before (red line) and after (black line) HER2 conjugation (Figure 1A). Bare MNPs displayed a sharp peak at 623 cm–1 associated with Fe–O bonds. The peak was weakened, however, after HER2 covalent attachment. Two sharp peaks at 1636 and 1532 cm–1 in the FTIR spectrum of free HER2 ascribe to the amide I (C=O stretching) and amide II (N–H bending) bands of the protein, respectively. The FTIR spectrum of HER2-MNPs shows the same amide bands at the same locations. Altogether, it is deduced from the FTIR data that HER2 was bonded to the surface of MNPs.
At each step of MNPs’ functionalization, the hydrodynamic diameter and zeta potential of the particles were measured (Figure 1B). The size of bare MNPs determined by DLS (∼200 nm) was larger than that of TEM (∼20 nm). The discrepancy is due to the different measurement modalities of DLS and TEM. DLS measures the hydrodynamic diameter, which includes the particle and the hydrated layer around it. TEM analysis, however, is accompanied by the dehydration of the NPs’ surface, leading to a smaller size than the one recorded by DLS. In the first step of modification, a slight size reduction was observed after conjugation of CEPA to MNPs. Moreover, the charge of bare MNPs flipped from a positive value to a negative one. It is inspected to see whether the carboxyl functional groups of CEPA are hydrolyzed. The resulting negative charges on the particles’ surface induce electrostatic repulsion, leading to less aggregation. The addition of the protein layer on the surface of MNPs-CEPA increased the hydrodynamic size of the particles. Exposure of the protein’s amine group on the particles’ surface resulted in a positive surface charge. Elemental analysis by EDS further revealed the chemical composition of MNPs and HER2-MNPs. Iron and oxygen were observed in both samples. According to the results shown in Figure 1C, the atomic ratio of Fe/O in bare MNPs was 3/3.68, close to the ratio in magnetite (3/4). The EDS data of HER2-MNPs showed additional signals of carbon, phosphor, and sulfur, proving the chemical conjugation of HER2 on the NPs’ surface.
3.3. Off-Chip Cell Isolation
In the present study, we chose SKBR3 cancer cells as they are more likely to represent HER2-positive CTCs. The most important similarity between HER2-positive CTCs and SKBR3 cancer cells is the overexpression of HER2 proteins on their surface, which is why different studies aiming to determine the HER2 status of CTCs use SKBR3 for device calibration.33,34 There might be differences in genetic content, other surface markers, size, deformability, etc., between HER2-positive CTCs and SKBR3 cells. However, our choice of SKBR3 as model cells is aligned with the goal of our study, which is the microfluidic-assisted isolation of HER2-positive cells using the antiHER2 antibody. The functionality of the HER2-MNPs was approved by the off-chip capture of SK-BR-3 cells. To this end, cells were stained with DAPI (Figure 2A) and incubated with HER2-MNPs for an hour. As shown in Figure 2B, aggregates of the NPs are attached to the cells. After 1 h of incubation with HER2-MNPs, followed by magnetic separation and washing with PBS, the pellet was redispersed in PBS and observed using a fluorescence microscope. The number of DAPI-stained SK-BR-3 cells after magnetic separation (Figure 2C) decreased due to the low capture efficiency of the off-chip method. It is worth mentioning that we maintained consistent sample preparation before and after the washing and redispersion steps, including identical sample volumes and the amount loaded onto the microscopy slide. Our procedures were conducted with high repeatability, ensuring a reliable comparison of the samples before and after the cell isolation experiment. While we acknowledge that the off-chip method may inherently lose some aggregates during the washing steps, we have accounted for this by keeping experimental conditions consistent across all samples. Therefore, the observed differences in aggregate concentrations between Figure 2B,C primarily reflect the off-chip method’s lower capture efficiency than variations in experimental conditions.
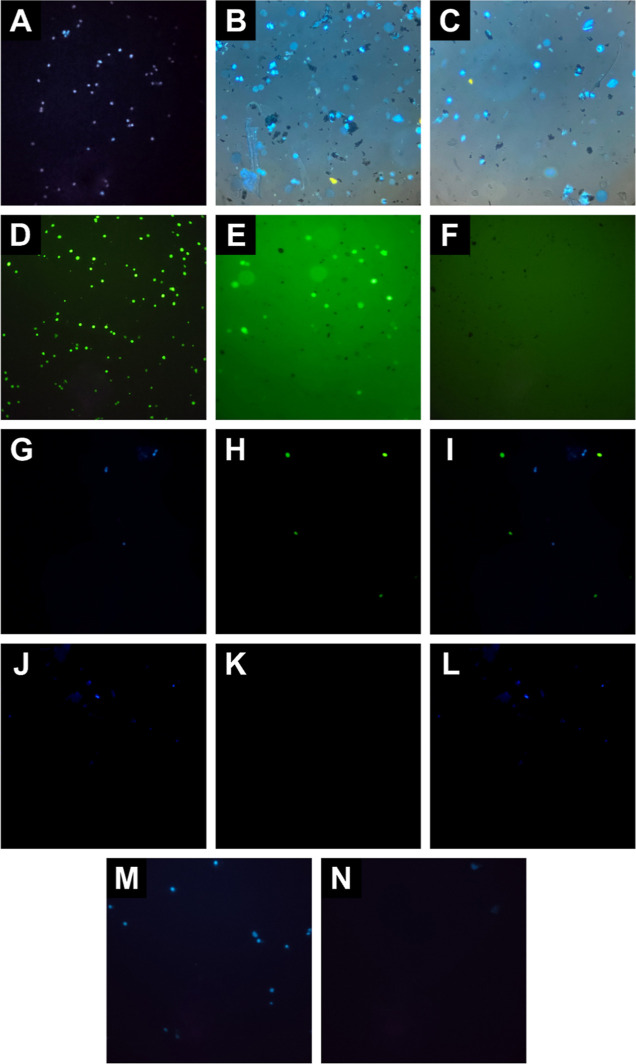
Figure 2.
Representative fluorescent microscopy images of off-chip cell isolation, (A) SKBR3 cells stained with DAPI, (B,C) stained SKBR3 cells incubated with HER2-MNPs before and after magnetic washing, (D) MDA-MB-231 cells stained with fluorescein dye, (E,F) stained MDA-MB-231 cells incubated with HER2-MNPs before and after magnetic washing, (G–I) blue, green, and merged fluorescent images of stained SKBR3 and MDA-MB-231 cell cocktail incubated with HER2-MNPs before magnetic washing, (J–L) blue, green, and merged fluorescent images of stained SKBR3 and MDA-MB-231 cell cocktail incubated with HER2-MNPs after magnetic washing, aand (M,N) stained SKBR3 cells incubated with bare MNPs before and after magnetic washing.
As a negative control experiment, HER2-negative MDA-MB-231 breast cancer cells were colored with fluorescein (Figure 2D) and incubated with HER2-MNPs. The off-chip isolation method was proceeded for the experiment. Figure 2E, which is taken before magnetic separation and washing, shows that the NPs and cells are dispersed separately in the medium. No NP aggregate is observed around the green-colored MDA-MB-231 cells. After magnetic separation and washing with PBS, the pellet only contains MNPs, shown as black spots (Figure 2F). The absence of MDA-MB-231 in panel F of Figure 2 is due to the absence of affinity binding between HER2-MNPs and HER2-negative cells. As a result, MDA-MB-231 cells did not attach to HER2-MNPs and were removed upon magnetic washing. The efficiency of HER2-MNPs in cell separation was reliably demonstrated by the selective separation of HER2-positive cells in a mixture of HER2-positive and HER2-negative cells.
In addition, the ability of functionalized NPs to capture and separate HER2-positive cells from a mixture of HER2-positive and HER2-negative cells was evaluated. A mixture of DAPI-stained SK-BR-3 and fluorescein-stained MDA-MB-231 cells was incubated with HER2-MNPs. The fluorescence signals of DAPI and fluorescein located inside the SK-BR-3 and MDA-MB-231 cell lines are observed as blue and green, respectively (Figure 2G–I). After the washing step, the cell population attached to HER2-MNPs was free from HER2-negative cells, indicating the ability of HER2-MNPs for selective off-chip isolation of HER2-positive cells from cell cocktail (Figure 2J–L). As a control experiment, bare MNPs were incubated with HER2-positive cells (Figure 2M). Microscopic images of the redispersed pellet did not show SK-BR-3 cells demonstrating low nonspecific adsorption of cells on the surface of bare MNPs (Figure 2N). More accurate and reliable analysis of cell adsorption was conducted using flow cytometry, as described in the following paragraph.
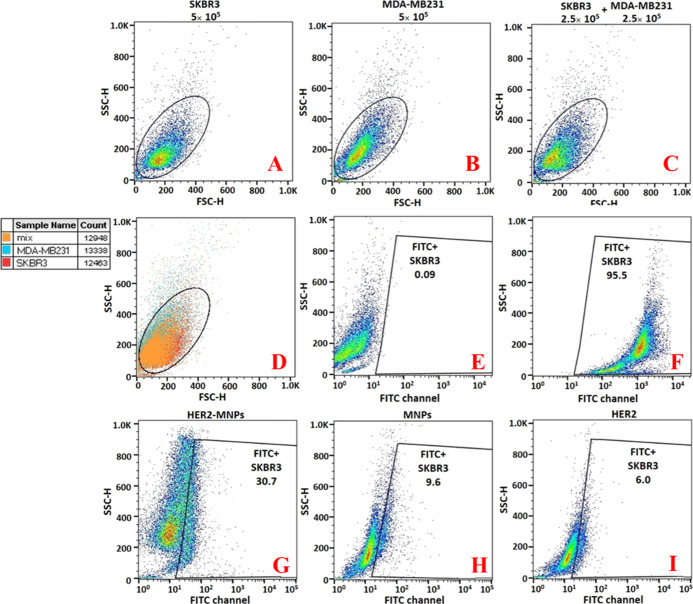
The off-chip capture efficiency was calculated by flow cytometry. To this end, a standard curve for the enumeration of HER2-positive cells was achieved (Figure S4 online). Different ratios of fluorescein-stained SK-BR-3 and unstained MDA-MB-231 (1:0, 1:2, 1:4, 1:9, 1:19, and 1:99) were prepared. The green fluorescence intensity of each sample was analyzed by flow cytometry in the FITC channel. Mixing HER2-positive and HER2-negative cells allowed for the compensation of the effect of HER2-negative cells on the target cells. The standard curve showed a linear correlation between the fluorescein percentage and the SK-BR-3 cell number (Figure S4 online). The capture efficiency was measured using the standard curve. 2.5 × 105 fluorescein-stained SK-BR-3 cells were mixed with 2.5 × 105 unstained MDA-MB-231, followed by HER2-MNP incubation. The magnetic separation was performed, and the pellet was analyzed by flow cytometry. The result revealed that 30.7% of the separated cells by HER2-MNPs were HER2-positive, which is equivalent to 148,460 cells (Figure 3G). The capture efficiency was calculated as 59.38%. In addition, two control experiments were conducted to ensure the specificity of HER2-MNPs for the off-chip cell isolation (Figure 3H,I). In the first, bare MNPs were incubated with 2.5 × 105-stained SK-BR-3 and 2 × 105-unstained MDA-MB-231. Using the standard curve, it was revealed that bare MNPs separated 38,900 out of 2.5 × 105 SK-BR-3 cells through nonspecific adsorption, which is equivalent to the capture efficiency of 15.5% (Figure 3H). In the second experiment, free antibody was mixed with SK-BR-3 + MDA-MB-231 cells for 1 h. The whole suspension was poured out of the microtube. The empty microtube was washed with PBS. In the final step, the microtube was filled with fresh PBS, and the solution was analyzed by flow cytometry. The fluorescence signal associated with 20,500 out of 2.5 × 105 SK-BR-3 cells was detected, corresponding to the capture value of 8.2% (Figure 3I). The second control experiment shows that part of the capture efficiency in the first control experiment is associated with the adsorption of the cell-antibody complex to the walls of microtubes.
Figure 3.
Capture efficiency of the off-chip isolation by flow cytometry. Forward and side scatter density plots of (A) 5 × 105 unstained SK-BR-3, (B) 5 × 105 unstained MDA-MB-231, (C) 2.5 × 105 unstained SK-BR-3 mixed with 2.5 × 105 unstained MDA-MB-231, (D) the overlay of (A–C), the fluorescein signal of (E) 5 × 105 unstained SK-BR-3, (F) 5 × 105 stained SK-BR-3, 2.5 × 105 stained SK-BR-3 + 2.5 × 105 unstained MDA-MB-231 incubated with (G) HER2-MNPs, (H) bare MNPs, and (I) HER2.
3.4. On-Chip Cell Isolation
The on-chip cell isolation process is shown in Figure 1. The slow movement of cells and HER2-MNPs inside the chip was tracked using a fluorescence microscope (Figure 4). 1 mL of stained SK-BR-3 cells (1 × 104) were injected into the chip from inlet 1, and 1 mL of HER2-MNPs (83 μg/mL) were entered from inlet 2 using a syringe pump with a flow rate of 0.5 mL/h. The whole experiment took 2 h. Figure 4A,B shows that HER2-MNPs and SK-BR-3 cells are attached and move together inside the mixing zone. The mixing region was long enough to allow binding between HER2-MNPs and cells. Upon reaching the trap region, an external magnetic field was applied, under which HER2-MNPs accompanied by the attached cells stopped and accumulated close to the channel walls. Figure 4C shows the respected region. Due to the high accumulation of MNPs, cells were barely visible. Under higher magnification and in the green channel of the fluorescence microscope, however, some cells trapped in the NP aggregates were observed (Figure 4D). The magnetic field was removed, and captured cells came out of the outlet. The colloidal suspension was centrifuged at 1500 rpm for 5 min, and the supernatant was removed. The precipitate was redispersed in PBS and dropped on a glass slide. As shown in Figure 4E, more stained cells are observed. A hemocytometer was used to count the captured cells. It is noteworthy that the analysis of a small sample volume is a general characteristic of microfluidic approaches. However, to be applicable in CTC isolation, large amounts of blood samples need to be analyzed. To solve the challenge, some studies have enlarged the size of the chip35 or used multiple chips.36 As another strategy, Fischer employed leukapheresis to preconcentrate CTCs in patients’ blood.37 Similarly, Ficoll–Hypaque was used to collect CTCs from an anticoagulated blood sample in a tiny fraction (buffy coat) and separate them from plasma and RBCs. The on-chip analysis of CTCs in buffy coats was quicker than processing the entire blood sample.38
Figure 4.
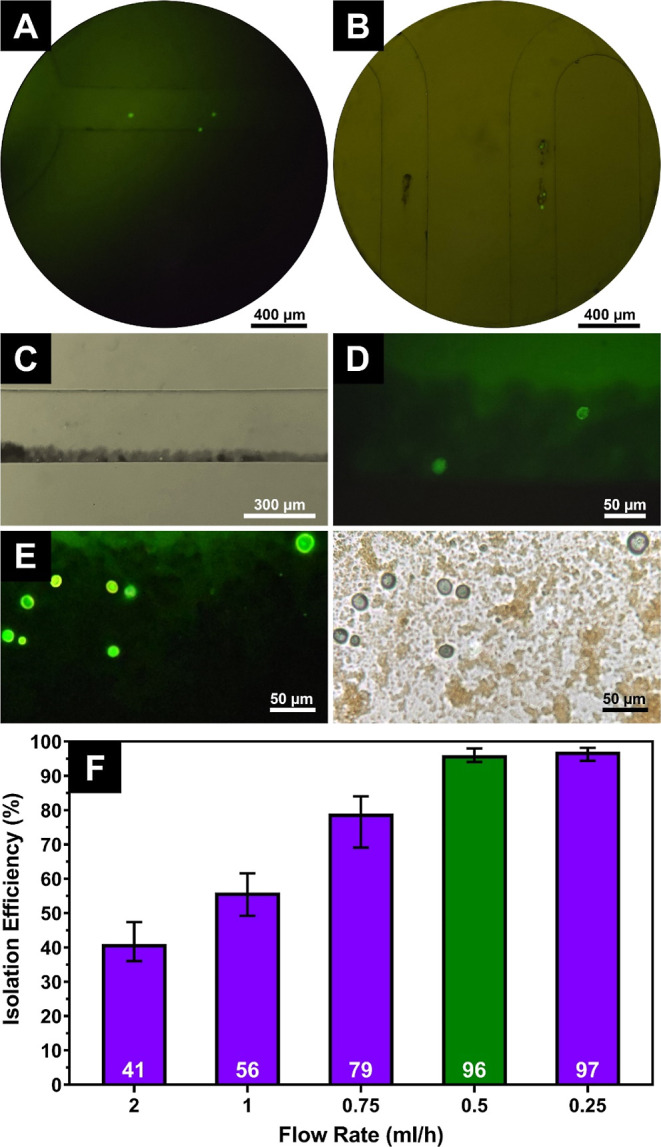
Fluorescence microscopy images of on-chip cell isolation, (A,B) movement of stained SK-BR-3 cells and HER2-MNPs inside the chip, (C) magnetic enrichment of the cells attached to the HER2-MNPs in the trap region, (D) high magnification of fluorescent SK-BR-3 cells in the trap region, (E) fluorescence and bright-field images of SK-BR-3 cells separated after magnetic washing, and (F) SK-BR-3 cell isolation efficiency versus fluid’s flow rate inside the chip (the green bar shows the optimum flow rate value).
3.5. Effect of Flow Rate on the Capture Efficiency
The capture efficiency was highly affected by the fluids’ flow rate inside the chip. Several flow rates were tested. The number of cells was fixed at 10,000. By decreasing the flow rate from 2 to 0.25 mL/h, the capture efficiency increased substantially from 41 to 97% at the flow rate of 0.5 mL/h and then reached a plateau (Figure 4F). It is proposed that as the incubation time of HER2-MNPs and cells in the mixing region increases, the attachment of cells to antibodies on the MNPs enhances due to a higher probability of their collision, as demonstrated in previous studies.39 Microfluidic devices’ capture efficiency has been found to be affected by the fluid flow rate.40 When flow rates are low, particles and cells in the mixing region incubate longer, which increases their chances of capturing. This is mainly due to the decrease in the diffuse interface width as the Reynolds number increases at higher flow rates.41 Also, it is widely reported that at high flow rates, the shear stress on the particles and cells increases, which hinders the capture process.42 This is because high shear stress can cause the detachment of the particles and cells in the microchannel. Therefore, we believe that the increase in capture efficiency at lower flow rates observed in our study is a combined effect of incubation time and shear rate. The flow rate of 0.5 mL/h was selected as the optimum value, as it provides a high capture rate for SK-BR-3 cells. Further decreasing the flow rate did not change the capture efficiency. Meanwhile, the operation time was doubled.
4. Conclusions
In the present study, a microfluidic chip and HER2-MNPs were employed to selectively isolate HER2-positive SK-BR-3 cells from a mixture of HER2-positive and HER2-negative cells. The isolation experiment was conducted off- and on-chip, and the results were compared. The off-chip method had limitations, including a large sample volume, a high process time, and low efficiency. The capacity of our microfluidic chip from inlets to the outlet was 0.9488 μL, whereas in the off-chip isolation method, at least 150 μL of the sample was required to run each experiment. The off-chip capture efficiency was calculated as 58%. Contrarily, the cell isolation in the microfluidic chip allowed a more efficient antibody–cell interaction, leading to an increase of capture efficiency to 96% at 0.5 mL/h. The analysis time by our microfluidic chip (2 h) was half of that required for the off-chip analysis (around 4 h). No channel clogging by HER2-MNPs was observed, as the zeta potential value of HER2-MNPs was negative enough to induce electrostatic repulsion and NPs’ colloidal stability. Altogether, our results demonstrated that the proposed setup may work in clinical settings. However, subtle changes in the experimental conditions may be required due to the inherent complexity of biological fluids.
Acknowledgments
This work was supported by the Motamed Cancer Institute and the University of Tehran.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c01287.
Schematic representation of the chip configuration and the fabrication process; characterization of MNPs; and standard curve of the fluorescein signal versus SKBR3 cell number (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Ruddon R. W.Characteristics of Human Cancer. Cancer Biology; Oxford University Press, 2007; pp 3–15. [Google Scholar]
- Jemal A.; Siegel R.; Xu J.; Ward E. Cancer Statistics, 2010. Ca-Cancer J. Clin. 2010, 60, 277–300. 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Miller K. D.; Nogueira L.; Mariotto A. B.; Rowland J. H.; Yabroff K. R.; Alfano C. M.; Jemal A.; Kramer J. L.; Siegel R. L. Cancer Treatment and Survivorship Statistics, 2019. Ca-Cancer J. Clin. 2019, 69, 363–385. 10.3322/caac.21565. [DOI] [PubMed] [Google Scholar]
- Ferlay J.; Soerjomataram I.; Dikshit R.; Eser S.; Mathers C.; Rebelo M.; Parkin D. M.; Forman D.; Bray F. Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- Alix-Panabières C.; Pantel K. Circulating Tumor Cells: Liquid Biopsy of Cancer. Clin. Chem. 2013, 59, 110–118. 10.1373/clinchem.2012.194258. [DOI] [PubMed] [Google Scholar]
- Rolfo C.; Russo A. Liquid Biopsy for Early Stage Lung Cancer Moves Ever Closer. Nat. Rev. Clin. Oncol. 2020, 17, 523–524. 10.1038/s41571-020-0393-z. [DOI] [PubMed] [Google Scholar]
- Freitas C.; Sousa C.; Machado F.; Serino M.; Santos V.; Cruz-Martins N.; Teixeira A.; Cunha A.; Pereira T.; Oliveira H. P.; Costa J. L.; Hespanhol V. The Role of Liquid Biopsy in Early Diagnosis of Lung Cancer. Front. Oncol. 2021, 11, 634316. 10.3389/fonc.2021.634316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg R.; Gertler R.; Friederichs J.; Fuehrer K.; Dahm M.; Phelps R.; Thorban S.; Nekarda H.; Siewert J. R. Comparison of Two Density Gradient Centrifugation Systems for the Enrichment of Disseminated Tumor Cells in Blood. Cytometry 2002, 49, 150–158. 10.1002/cyto.10161. [DOI] [PubMed] [Google Scholar]
- Gertler R.; Rosenberg R.; Fuehrer K.; Dahm M.; Nekarda H.; Siewert J. R.. Detection of Circulating Tumor Cells in Blood Using an Optimized Density Gradient Centrifugation. Molecular Staging of Cancer; Springer Berlin Heidelberg: Berlin, Heidelberg, 2003; pp 149–155. [DOI] [PubMed] [Google Scholar]
- Xu T.; Lu B.; Tai Y.-C.; Goldkorn A. A Cancer Detection Platform Which Measures Telomerase Activity from Live Circulating Tumor Cells Captured on a Microfilter. Cancer Res. 2010, 70, 6420–6426. 10.1158/0008-5472.CAN-10-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talasaz A. H.; Powell A. A.; Huber D. E.; Berbee J. G.; Roh K.-H.; Yu W.; Xiao W.; Davis M. M.; Pease R. F.; Mindrinos M. N.; Jeffrey S. S.; Davis R. W. Isolating Highly Enriched Populations of Circulating Epithelial Cells and Other Rare Cells from Blood Using a Magnetic Sweeper Device. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 3970–3975. 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.; Lin H. K.; Lu B.; Williams A.; Datar R.; Cote R. J.; Tai Y.-C. 3D Microfilter Device for Viable Circulating Tumor Cell (CTC) Enrichment from Blood. Biomed. Microdevices 2011, 13, 203–213. 10.1007/s10544-010-9485-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. J.; Yobas L.; Lee G. Y. H.; Ong C. N.; Lim C. T. Microdevice for the Isolation and Enumeration of Cancer Cells from Blood. Biomed. Microdevices 2009, 11, 883–892. 10.1007/s10544-009-9305-9. [DOI] [PubMed] [Google Scholar]
- Kim T.-H.; Lim M.; Park J.; Oh J. M.; Kim H.; Jeong H.; Lee S. J.; Park H. C.; Jung S.; Kim B. C.; Lee K.; Kim M.-H.; Park D. Y.; Kim G. H.; Cho Y.-K. FAST: Size-Selective, Clog-Free Isolation of Rare Cancer Cells from Whole Blood at a Liquid–Liquid Interface. Anal. Chem. 2017, 89, 1155–1162. 10.1021/acs.analchem.6b03534. [DOI] [PubMed] [Google Scholar]
- Qin X.; Park S.; Duffy S. P.; Matthews K.; Ang R. R.; Todenhöfer T.; Abdi H.; Azad A.; Bazov J.; Chi K. N.; Black P. C.; Ma H. Size and Deformability Based Separation of Circulating Tumor Cells from Castrate Resistant Prostate Cancer Patients Using Resettable Cell Traps. Lab Chip 2015, 15, 2278–2286. 10.1039/C5LC00226E. [DOI] [PubMed] [Google Scholar]
- Chou W.-P.; Wang H.-M.; Chang J.-H.; Chiu T.-K.; Hsieh C.-H.; Liao C.-J.; Wu M.-H. The Utilization of Optically-Induced-Dielectrophoresis (ODEP)-Based Virtual Cell Filters in a Microfluidic System for Continuous Isolation and Purification of Circulating Tumour Cells (CTCs) Based on Their Size Characteristics. Sens. Actuators, B 2017, 241, 245–254. 10.1016/j.snb.2016.10.075. [DOI] [Google Scholar]
- Vona G.; Sabile A.; Louha M.; Sitruk V.; Romana S.; Schütze K.; Capron F.; Franco D.; Pazzagli M.; Vekemans M.; Lacour B.; Bréchot C.; Paterlini-Bréchot P. Isolation by Size of Epithelial Tumor Cells. Am. J. Pathol. 2000, 156, 57–63. 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W.; Mezencev R.; Kim B.; Wang L.; McDonald J.; Sulchek T. Cell Stiffness Is a Biomarker of the Metastatic Potential of Ovarian Cancer Cells. PLoS One 2012, 7, e46609 10.1371/journal.pone.0046609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivedita N.; Garg N.; Lee A. P.; Papautsky I. A High Throughput Microfluidic Platform for Size-Selective Enrichment of Cell Populations in Tissue and Blood Samples. Analyst 2017, 142, 2558–2569. 10.1039/C7AN00290D. [DOI] [PubMed] [Google Scholar]
- Sollier E.; Go D. E.; Che J.; Gossett D. R.; O’Byrne S.; Weaver W. M.; Kummer N.; Rettig M.; Goldman J.; Nickols N.; McCloskey S.; Kulkarni R. P.; Di Carlo D. Size-Selective Collection of Circulating Tumor Cells Using Vortex Technology. Lab Chip 2014, 14, 63–77. 10.1039/C3LC50689D. [DOI] [PubMed] [Google Scholar]
- Warkiani M. E.; Guan G.; Luan K. B.; Lee W. C.; Bhagat A. A. S.; Kant Chaudhuri P.; Tan D. S.-W.; Lim W. T.; Lee S. C.; Chen P. C. Y.; Lim C. T.; Han J. Slanted Spiral Microfluidics for the Ultra-Fast, Label-Free Isolation of Circulating Tumor Cells. Lab Chip 2014, 14, 128–137. 10.1039/C3LC50617G. [DOI] [PubMed] [Google Scholar]
- Park E. S.; Jin C.; Guo Q.; Ang R. R.; Duffy S. P.; Matthews K.; Azad A.; Abdi H.; Todenhöfer T.; Bazov J.; Chi K. N.; Black P. C.; Ma H. Continuous Flow Deformability-Based Separation of Circulating Tumor Cells Using Microfluidic Ratchets. Small 2016, 12, 1909–1919. 10.1002/smll.201503639. [DOI] [PubMed] [Google Scholar]
- Pankhurst Q. A.; Connolly J.; Jones S. K.; Dobson J. Applications of Magnetic Nanoparticles in Biomedicine. J. Phys. D: Appl. Phys. 2003, 36, R167–R181. 10.1088/0022-3727/36/13/201. [DOI] [Google Scholar]
- Hoshino K.; Huang Y.-Y.; Lane N.; Huebschman M.; Uhr J. W.; Frenkel E. P.; Zhang X. Microchip-Based Immunomagnetic Detection of Circulating Tumor Cells. Lab Chip 2011, 11, 3449. 10.1039/c1lc20270g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-H.; Huang Y.-Y.; Chen P.; Hoshino K.; Liu H.; Frenkel E. P.; Zhang J. X. J.; Sokolov K. V. Versatile Immunomagnetic Nanocarrier Platform for Capturing Cancer Cells. ACS Nano 2013, 7, 8816–8823. 10.1021/nn403281e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieuwerts A. M.; Kraan J.; Bolt J.; van der Spoel P.; Elstrodt F.; Schutte M.; Martens J. W. M.; Gratama J.-W.; Sleijfer S.; Foekens J. A. Anti-Epithelial Cell Adhesion Molecule Antibodies and the Detection of Circulating Normal-Like Breast Tumor Cells. JNCI, J. Natl. Cancer Inst. 2009, 101, 61–66. 10.1093/jnci/djn419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld J.-W.; Meder S.; Wohlberg M.; Friedrich R. E.; Rau T.; Riethdorf L.; Löning T.; Pantel K.; Riethdorf S. Overexpression of TACE and TIMP3 MRNA in Head and Neck Cancer: Association with Tumour Development and Progression. Br. J. Cancer 2011, 104, 138–145. 10.1038/sj.bjc.6606017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai M.; Liang K.; Lu Y.; Qiu S.; Fan Z. Brk/PTK6 Cooperates with HER2 and Src in Regulating Breast Cancer Cell Survival and Epithelial-to-Mesenchymal Transition. Cancer Biol. Ther. 2013, 14, 237–245. 10.4161/cbt.23295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard S.; Pupa S. M.; Campiglio M.; Tagliabue E. Biologic and Therapeutic Role of HER2 in Cancer. Oncogene 2003, 22, 6570–6578. 10.1038/sj.onc.1206779. [DOI] [PubMed] [Google Scholar]
- Berger P.; Adelman N. B.; Beckman K. J.; Campbell D. J.; Ellis A. B.; Lisensky G. C. Preparation and Properties of an Aqueous Ferrofluid. J. Chem. Educ. 1999, 76, 943–948. 10.1021/ed076p943. [DOI] [Google Scholar]
- Cędrowska E.; Pruszyński M.; Gawęda W.; Zuk M.; Krysiński P.; Bruchertseifer F.; Morgenstern A.; Karageorgou M. A.; Bouziotis P.; Bilewicz A. Trastuzumab Conjugated Superparamagnetic Iron Oxide Nanoparticles Labeled with 225Ac as a Perspective Tool for Combined α-Radioimmunotherapy and Magnetic Hyperthermia of HER2-Positive Breast Cancer. Molecules 2020, 25, 1025. 10.3390/molecules25051025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjithkumar V.; Sangeetha S.; Vairam S. Synthesis of magnetic activated carbon/α-Fe2O3 nanocomposite and its application in the removal of acid yellow 17 dye from water. J. Hazard. Mater. 2014, 273, 127–135. 10.1016/j.jhazmat.2014.03.034. [DOI] [PubMed] [Google Scholar]
- Riethdorf S.; Müller V.; Zhang L.; Rau T.; Loibl S.; Komor M.; Roller M.; Huober J.; Fehm T.; Schrader I.; Hilfrich J.; Holms F.; Tesch H.; Eidtmann H.; Untch M.; Von Minckwitz G.; Pantel K. Detection and HER2 Expression of Circulating Tumor Cells: Prospective Monitoring in Breast Cancer Patients Treated in the Neoadjuvant GeparQuattro Trial. Clin. Cancer Res. 2010, 16, 2634–2645. 10.1158/1078-0432.CCR-09-2042. [DOI] [PubMed] [Google Scholar]
- Jaeger B. A. S.; Neugebauer J.; Andergassen U.; Melcher C.; Schochter F.; Mouarrawy D.; Ziemendorff G.; Clemens M.; Abel E. V.; Heinrich G.; Schueller K.; Schneeweiss A.; Fasching P.; Beckmann M. W.; Scholz C. H.; Friedl T. W. P.; Friese K.; Pantel K.; Fehm T.; Janni W.; Rack B. The HER2 Phenotype of Circulating Tumor Cells in HER2-Positive Early Breast Cancer: A Translational Research Project of a Prospective Randomized Phase III Trial. PLoS One 2017, 12, e0173593 10.1371/journal.pone.0173593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edd J. F.; Mishra A.; Dubash T. D.; Herrera S.; Mohammad R.; Williams E. K.; Hong X.; Mutlu B. R.; Walsh J. R.; Machado De Carvalho F.; Aldikacti B.; Nieman L. T.; Stott S. L.; Kapur R.; Maheswaran S.; Haber D. A.; Toner M. Microfluidic Concentration and Separation of Circulating Tumor Cell Clusters from Large Blood Volumes. Lab Chip 2020, 20, 558–567. 10.1039/c9lc01122f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro-Samy S.; Oliveira M. I.; Pereira-Veiga T.; Muinelo-Romay L.; Carvalho S.; Gaspar J.; Freitas P. P.; López-López R.; Costa C.; Diéguez L. Fast and Efficient Microfluidic Cell Filter for Isolation of Circulating Tumor Cells from Unprocessed Whole Blood of Colorectal Cancer Patients. Sci. Rep. 2019, 9, 8032. 10.1038/s41598-019-44401-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J. C.; Niederacher D.; Topp S. A.; Honisch E.; Schumacher S.; Schmitz N.; Zacarias Föhrding L.; Vay C.; Hoffmann I.; Kasprowicz N. S.; Hepp P. G.; Mohrmann S.; Nitz U.; Stresemann A.; Krahn T.; Henze T.; Griebsch E.; Raba K.; Rox J. M.; Wenzel F.; Sproll C.; Janni W.; Fehm T.; Klein C. A.; Knoefel W. T.; Stoecklein N. H. Diagnostic Leukapheresis Enables Reliable Detection of Circulating Tumor Cells of Nonmetastatic Cancer Patients. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 16580–16585. 10.1073/pnas.1313594110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmanian M.; Sartipzadeh Hematabad O.; Askari E.; Shokati F.; Bakhshi A.; Moghadam S.; Olfatbakhsh A.; Al Sadat Hashemi E.; Khorsand Ahmadi M.; Morteza Naghib S.; Sinha N.; Tel J.; Eslami Amirabadi H.; den Toonder J. M. J.; Majidzadeh-A K. A Micropillar Array-Based Microfluidic Chip for Label-Free Separation of Circulating Tumor Cells: The Best Micropillar Geometry?. J. Adv. Res. 2023, 47, 105–121. 10.1016/j.jare.2022.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian R.; Li X.; Zhang H.; Ma L.; Zhang H.; Wang Z. Ulex Europaeus Agglutinin-I-Based Magnetic Isolation for the Efficient and Specific Capture of SW480 Circulating Colorectal Tumor Cells. ACS Omega 2022, 7, 30405–30411. 10.1021/acsomega.2c03702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.; Georgiev T. Z.; Sheng W.; Zheng X.; Varillas J. I.; Zhang J.; Hugh Fan Z. Tumor Cell Capture Patterns around Aptamer-Immobilized Microposts in Microfluidic Devices. Biomicrofluidics 2017, 11, 054110. 10.1063/1.5000707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamholz A. E.; Weigl B. H.; Finlayson B. A.; Yager P. Quantitative Analysis of Molecular Interaction in a Microfluidic Channel: The T-Sensor. Anal. Chem. 1999, 71, 5340–5347. 10.1021/ac990504j. [DOI] [PubMed] [Google Scholar]
- Sarioglu A. F.; Aceto N.; Kojic N.; Donaldson M. C.; Zeinali M.; Hamza B.; Engstrom A.; Zhu H.; Sundaresan T. K.; Miyamoto D. T.; Luo X.; Bardia A.; Wittner B. S.; Ramaswamy S.; Shioda T.; Ting D. T.; Stott S. L.; Kapur R.; Maheswaran S.; Haber D. A.; Toner M. A Microfluidic Device for Label-Free, Physical Capture of Circulating Tumor Cell Clusters. Nat. Methods 2015, 12, 685–691. 10.1038/nmeth.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.