1. Introduction
Interactions between gut microbiota and the host play an important role in central nervous system function and behavior, primarily mediated through immune and neuroendocrine pathways (i.e., the gut-brain axis) (Cryan and Dinan, 2012; Liang et al., 2018). Over the past decade, clinical studies and animal models have suggested that chronic distress-related conditions, such as depression and anxiety disorders, are associated with altered gut microbiome composition. For instance, fecal samples from individuals with depression exhibit altered species diversity and taxonomy (Cheung et al., 2019; Horne and Foster, 2018). Various neuropsychiatric conditions are also characterized by under-representation of bacteria that produce anti-inflammatory metabolites (reviewed in Dalile et al., 2019), one potential mechanism by which gut-mediated immune alterations affect behavior. Furthermore, interventions that influence gut-brain interactions may improve depression and anxiety-related symptoms (Bruce-Keller et al., 2018) by altering immune and neuroendocrine pathways, and microbial profiles have been shown to predict treatment responses in mental illness (Mantere et al., 2017).
Our current understanding of these relationships is primarily informed by stool-derived specimens, which, though useful, can be limited by practical sampling issues in certain contexts, such as one or fewer specimens per day based on intestinal motility. By comparison, the oral microbiome (e.g., salivary, supragingival) is relatively straightforward to collect multiple times per day and can be sampled ‘on-demand,’ enabling its investigation in acute laboratory manipulations such as stress reactivity. Although tissue or body-site specific microbiome differences are recognized and such data remain limited, initial reports show modest correlations between oral and gut microbiome composition (Ding and Schloss, 2014; Huttenhower et al., 2012). Oral microbiomes have also been reported to differ in patients with gastrointestinal diseases, such as inflammatory bowel (Said et al., 2014) and celiac disease (Francavilla et al., 2014), as well as inflammatory disorders such as rheumatoid arthritis (Du Teil Espina et al., 2019), and neuropsychiatrie disorders, including schizophrenia (Castro-Nallar et al., 2015; Yolken et al., 2015) and Parkinson’s disease (Pereira et al., 2017), as well as obesity (Tam et al., 2018), indicating that diverse pathologies may be reflected in the oral microbiome. What remains unresolved, however, is whether the oral microbiome also serves as a biomarker of subclinical psychological distress or host inflammation, which would provide proof-of-concept for experimental, or observational, investigations of host-microbiome interactions not currently feasible using gut-derived samples.
Studies of the oral microbiome, particularly of saliva, suggest that considerable diurnal variability exists both within and between individuals (Takayasu et al., 2017), similar to the gut (Flores et al., 2014). Part of this variation may be due to host behaviors, such as feeding (Collado et al., 2018) or tooth brushing (Morton et al., 2019). However, emerging evidence suggests that microbiome compositional oscillations may be entrained by and dependent upon host circadian biology (reviewed in Nobs et al., 2019), the disruption of which can affect microbial proliferation and survival. While the mechanisms of circadian cross-talk across physiological systems are poorly understood in the gut or mouth, neuroendocrine and immunological factors impacted by psychological distress may play a role within the oral microenvironment, which could in turn impact oral and/or systemic health. For example, chronic psychological distress, including depressed mood and anxiety, blunts diurnal patterns of glucocorticoid and catecholamine secretion in saliva (e.g., cortisol (Miller et al., 2007) and alpha-amylase (Nater et al., 2007), respectively), both of which are known to modulate gut microbes (Huang et al., 2015; Lyte et al., 2011) and thus, may also blunt diurnal rhythms in relative abundance and/or functional pathways of microbial features within the mouth (Poole et al., 2019).
Chronic distress is also associated with immunosuppression, which can manifest in decreased salivary antibody production (e.g., IgA) (Engeland et al., 2016), an important host factor in oral microbiome homeostasis (reviewed in Bosch et al., 2002). Elevation of proinflammatory cytokines in peripheral blood, such as tumor necrosis factor-alpha (TNF-α), interleukin-6 (IL-6), and interleukin 1-beta (IL-1β), is also observed in chronically distressed individuals (Slavich and Irwin, 2014), and are correlated with salivary cytokine levels during acute psychological stress (La Fratta et al., 2018). Salivary cytokines have been associated with salivary microbial composition in the context of periodontal disease (Lundmark et al., 2019), but relationships with peripheral cytokines in non-clinical samples remain unexplored. However, inter-individual variation in stimulated host cytokine responses is partially explained by gut microbiome composition and may be mediated by immunomodulatory microbial metabolites (Schirmer et al., 2016), which also highlights the need for parallel salivary microbiome data in understanding complex host-microbiome interactions. Little is known about the relationships between the host’s psychological or inflammatory state, salivary microbiome composition, and its diurnal changes.
The present study sought to examine whether salivary microbiome composition was associated with or predictive of distress- and inflammation-derived host profiles in a non-clinical adult population. Furthermore, we hypothesized that individuals with higher distress or inflammation would exhibit altered temporal variability in microbial community structure (e.g., alpha and beta diversity) and less variation in microbial abundance over a single day.
2. Materials and methods
2.1. Participants
All participants gave informed consent to the protocol, approved by the University of California, San Diego Institutional Review Board. Data from 68 healthy, non-smoking adults between 20–65 years who were recruited from the local community for a larger study of the role of obesity on vascular inflammation and immune cell activation in normotension vs. stage 1 hypertension (SBP: 130–140 mmHg; DBP: 80–90 mmHg), were included in this investigation. Initial screening via telephone interviews, followed by face-to-face confirmation of health history, established the absence of the following exclusion criteria: diabetes, current or recent history (<6 months) of tobacco smoking or substance abuse, history of cardiovascular disease (e.g., symptomatic coronary or cerebrovascular disease, arrhythmia, myocardial infarction, cardiomyopathy, heart failure), history of bronchospastic pulmonary disease, inflammatory disorders or health-related factors affecting immune function (e.g., vaccinations within 10 days of study visit, active infections/illness, immunomodulatory medication such as steroids or antibiotics, uncontrolled thyroid disease), psychosis, major depressive disorder, and stage 2 clinical hypertension indicated by use of anti-hypertensive medication or laboratory-assessed BP ≥145/90 mmHg. Sociodemographic characteristics (i.e., age, sex, and race) were determined through participant interview. Average resting systolic blood pressure was calculated from two sets of three consecutive measurements at 5-min intervals on two separate days, using a Dinamap Compact BP monitor (Critikon, Tampa, FL). BMI was based on height and weight assessment (kg/m2). All procedures were approved by the UCSD Human Research Protections Program, and all participants provided written informed consent.
2.2. Psychological distress assessment
Given prior evidence from humans and animal models supporting a link between gut microbiome composition and host anxiety, depression, and stress, we generated a psychological distress profile for each participant based on six valid and widely-studied self-report assessments to capture anxiety, depressive, and stress-related symptoms. Each participant’s measurements were then, converted to z-scores and averaged to create a unitary distress score, from which participants were categorized into ‘high’ or ‘low’ distress groups (see section 2.5.1). Anxiety measures were derived from the state (STAI-S) and trait (STAI-T) subscales of the 40-item State-Trait Anxiety Inventory (STAI) (Spielberger, 1983), as well as the 9-item tension-anxiety POMS subscale (PoMs-T). Depressive symptomatology was assessed using the 21-item self-report Beck Depression Inventory (BDI-Ia) (Beck and Steer, 1993) and the 15-item depression subscale (POMS-D) of the Profile of Mood States (POMS) (McNair et al., 1971). General psychological distress was appraised using the 10-item Perceived Stress Scale (PSS) (Cohen et al., 1983).
2.3. Plasma cytokines and in vitro response to LPS
Blood for plasma interleukin-6 (IL-6), interleukin-1β (IL-1β), and tumor necrosis factor-alpha (TNF-α) was drawn into EDTA-treated vacutainers and immediately placed on ice until centrifugation. These three cytokines have been shown to be associated with psychological states such as stress and depression (Rohleder, 2014) as well as psychological manipulations such as acute laboratory stress tests (Marsland et al., 2017). Plasma was stored at −80C until assays were performed. Plasma cytokines were assessed using commercially available multiplex immunoassay kits (MesoScale Discovery, Gaithersburg, MD). The intra- and inter-assay coefficients of variation were 8.5% and 7.8% for IL-6, 8.4% and 6.0% for IL-1β, and 6.8% and 6.4% for TNF-α, respectively. For IL-1β, 15 of 66 sample values fell below the manufacturer-supplied lowest calibration standard (0.1 pg/mL) and thus imputed to a value of 0.0 pg/mL for downstream analyses.
In addition to basal plasma cytokine levels, the in vitro inflammatory response to endotoxin was assessed in peripheral blood monocytes to achieve comprehensive profiling of host’s inflammatory system. Briefly, heparinized blood (300uL) was combined with lipopolysaccharide (LPS; 200 pg/mL) (E.coli 0111:B4, #L4391, Sigma-Aldrich, St. Louis, MO) and incubated with 10ug/mL Brefeldin A (#B6751, Sigma-Aldrich) for 3.5 hrs at 37C with 5% CO2 in 96-well plates, along with a non-LPS-treated sample. Cells were surface stained using anti-CD14/APC (#301808, BioLegend, San Diego, CA) and anti-HLA-DR/PE (#307605, BioLegend), and intracellularly stained post-permeabilization and fixation using anti-TNFa/FITC (#502906, BioLegend). The proportion of CD14+/dimHLA-DR+ cells that were TNF-α+ was determined using FlowJo software (v10, TreeStar, Ashland, OR), and used for analysis (Dimitrov et al., 2017; Kohn et al., 2019).
2.4. Saliva collection and sequencing
Participants were provided with saliva collection kits with written instructions (“Salivettes”, Sarstedt, #51.1534, Nümbrecht, Germany), and were instructed to saturate the provided cotton roll with saliva by rolling it inside the mouth before placing the roll back into the tube. Participants were cautioned not to handle the cotton roll with hands but to place the roll in the mouth directly from and back into the salivette tube. The procedure was repeated at five time points across a single day: waking, mid-morning (10:00 hrs), midday (12:00 hrs), afternoon (14:00 hrs), and evening (17:00 hr). Notably, all waking samples were collected prior to ingestion of food or drink, and prior to tooth brushing or other oral hygiene behaviors. In addition, participants were instructed to not consume food or drinks other than plain water for at least 30 min and to rinse their mouth with water prior to collection at all other time points. Saliva samples were refrigerated until participants returned samples in-person at their next laboratory visit generally in 3 – 7 days. Returned salivette tubes were centrifuged at 1,000 x g for 2 minutes for saliva collection, and recovered saliva was frozen at −80°C until DNA extraction.
DNA extraction, amplification, and sequencing were performed as follows. Frozen samples were thawed and transferred into 96-well plates containing garnet beads and extracted using the Qiagen PowerSoil DNA kit adapted for magnetic bead purification as previously described (Marotz et al., 2017). DNA was eluted in 100 μL Qiagen elution buffer. 16S rRNA gene amplification was performed according to the Earth Microbiome Project protocol (Thompson et al., 2017). Briefly, Illumina primers with unique reverse primer barcodes (Caporaso et al., 2012) were used to target the V4 region of the 16S rRNA gene (Walters et al., 2016). Amplicons were quantified by PicoGreen™ and 240 ng of each sample was pooled for sequencing. The pooled library was sequenced on the Illumina MiSeq sequencing platform with a MiSeq Reagent Kit v2 and paired-end 150 bp cycles. Raw data and associated feature tables are publicly available in Qiita (qiita.ucsd.edu) (Gonzalez et al., 2018) as study ID 11259.
2.5. Bioinformatics and statistical analyses
Sequence data were processed using QIIME2 (Rideout et al., 2018) (v.2018.4). Raw sequencing results were demultiplexed and microbial ASVs (Amplicon Sequence Variants) were identified using the Deblur algorithm (Amir et al., 2017). Taxonomic annotation was performed using QIIME2 feature-classifier plugin using a naïve-Bayes approach with a pre-fit classifier (Bokulich et al., 2018). Mitochondrial and chloroplast-derived sequences were filtered out based on taxonomic annotation. The output feature table contained an average of 19,911 ± 9,687 16S rRNA sequences per sample and 2,231 unique features in total. Sequences were queried against the Expanded Human Oral Microbiome Database (eHOMD; version 15.1), which identified 485 unique taxa in the present dataset with >99% identity to the reference database. For downstream analyses, samples from participants without psychological and inflammation data were omitted (N=14 samples dropped), and low abundance features with fewer than 10 reads across the entire experiment (N=89 features) were filtered out. All samples were rarefied to a depth of 3,200 reads based on visual inspection of alpha rarefaction curves (N=22 samples dropped due to insufficient reads) for analysis of community structure, yielding a final dataset containing 284 samples (87.9% of all samples) and 2,007 unique features (90.0% of original feature set).
Microbial community structure was characterized using measures of alpha- and beta-diversity. Alpha-diversity represents the number and distribution of taxa within each sample, and included the number of observed features (i.e. species richness), Shannon diversity index (i.e., species evenness), and Faith’s Phylogenetic Diversity, which measures the total length of branches in a reference phylogenetic tree for all species in a given sample. Beta-diversity, which reflects differences in taxonomic composition based upon either presence-absence or quantitative abundance data, was calculated using Bray-Curtis dissimilarity and weighted UniFrac, respectively. Output matrices were ordinated using principal coordinates analysis (PCoA) and visualized in Emperor (Vázquez-Baeza et al., 2013).
2.5.1. Data analysis
Statistical analyses were conducted using R (v.3.5.1). Descriptive data are presented as mean±SD. Missing psychological (<1.0% missing) and immune data (4.3% missing) were imputed using an iterative algorithm based on minimizing mean squared error (Josse and Husson, 2016). In order to generate equivalently-sized, heterogeneous groups based on psychological and immune characteristics, all six psychological measures and all four immune measures were converted to z-scores, as aforementioned and separately averaged for each participant. Thus, each participant had a single composite z-score for psychological measures, whereby higher values indicate greater distress, and a single composite z-score for immune measures, whereby higher values indicate greater inflammation. Participants were assigned as either high or low distress (34 subjects per group), and either high or low inflammation (33 subjects per group), based on a median split of study-wide composite z-scores. Assignments were used as predictors of microbial composition.
To assess the temporal dynamics of alpha diversity as a function of host distress and inflammatory status, each alpha diversity metric was regressed on time of day and its interaction with distress and inflammation category using a single linear mixed-effects model (LMM) fit using restricted maximum likelihood (REML) with a random-intercept by participant to account for repeated measurement. Time of day was included as a categorical, rather than continuous factor, to model temporal trends non-linearly. Age, sex, BMI, and race were included as covariates. Statistical significance of predictors of alpha-diversity was determined by type-II analysis of variance (ANOVA) Wald F-tests using the car package (Fox and Weisberg, 2011), which applies Kenward-Roger denominator degrees of freedom approximation and controls Type I error (Luke, 2017). Model residuals were visually inspected for normality using quantile-quantile plots, and influence statistics (e.g., df-beta, Cook’s distance) were computed to identify observations with exceedingly high leverage. Time-by-distress and time-by-inflammation group interaction contrasts were assessed for significance by Tukey-adjusted t-ratios of estimated marginal means using Ismeans (Lenth, 2016). Statistical significance of temporal dynamics of beta-diversity distances between groups was assessed using PERMANOVA with 999 permutations constrained by participant, and between-group differences in within-group dispersion were assessed using PERMDISP2 with the same constraints, implemented in vegan (Oksanen, 2017). Pairwise posthoc tests were conducted to test for time-dependent differences in beta-diversity relative to waking samples using pairwiseAdonis.
In order to enhance the reliability of the relative abundance findings, the Songbird method (Morton et al., 2019) was used to calculate the differential ranking (DR) of taxa and pathways (see below) that were associated with distress and inflammation, as well as those that were time-varying relative to the data from the waking time point. Songbird computes relative differentials using count-based multinomial regression. Relative differential ranks were computed for each taxa for time of day, psychological distress, and inflammation. Age, sex, and BMI were entered as covariates. Log-ratios for time-varying effects were established by selecting as numerator the top 20-ranked taxa for each time point relative to waking (i.e., strongest positive association with time), and selecting as the reference frame (denominator), the bottom-20 ranked taxa (i.e., strongest negative association with time). For distress and inflammation effects, log-ratios were computed by selecting as numerator the 40 top-ranked taxa, and as denominator the 40 bottom-ranked taxa. Statistical tests were performed by regressing the computed log-ratios (3 separate LMMs) on time of day, distress, and inflammation z-scores, adjusted for age, sex, BMI, and race, with subject as random intercept.
Machine learning preparation and analyses were conducted using calour (Xu et al., 2019). Two separate Random Forest classification models based on microbiome data and demographic information were constructed to predict high- versus low- distress and inflammation profiles. In all cases, age, sex, BMI, and race were one-hot encoded as features. Normalized microbiome counts were used as remaining features. In each case, a minimum of 1,000 reads per sample was used as a cut-off and per-sample data was normalized to 100,000 counts. Then, features were filtered by abundance to include features with at least 1,000 counts across all samples (at least 1% prevalence). For both models, all available time points were used, but time point information was one hot encoded as an additional feature. All data were CLR transformed prior to training. Grid search sampling was employed for each model to find optimal model parameters. Due to a limited number of samples, 5-fold stratified cross-validation was performed, and a separate test set was not constructed.
Metagenomic pathways were predicted from 16S rRNA sequence data rarefied to 3,200 reads using the PICRUSt2 (Langille et al., 2013) package in QIIME2. Relative abundance of pathway features was used in LMMs for alpha diversity analysis (e.g., richness and Shannon index), and PERMANOVA for beta diversity analysis (e.g., Bray-Curtis), as described above in feature-level analyses. Differential relative pathway abundances were also computed using Songbird for the 384 unique pathways that passed prevalence filtration. The top- and bottom-15 differentially ranked pathways for associations with distress and inflammation scores were used in downstream analyses, and a similar approach as performed above for taxa was applied for time-varying pathways. Pathway parent class ontologies were derived from the MetaCyc database (version 23.1) (Caspi et al., 2016) and data visualization was performed in R.
Jupyter notebooks reproducing the analyses are deposited on GitHub (https://github.com/knightlab-analyses/oral-microbiome-hong), and differentials tables from the Songbird analyses are provided as Supplemental Tables.
3. Results
3.1. Psychological and inflammatory characteristics of participants
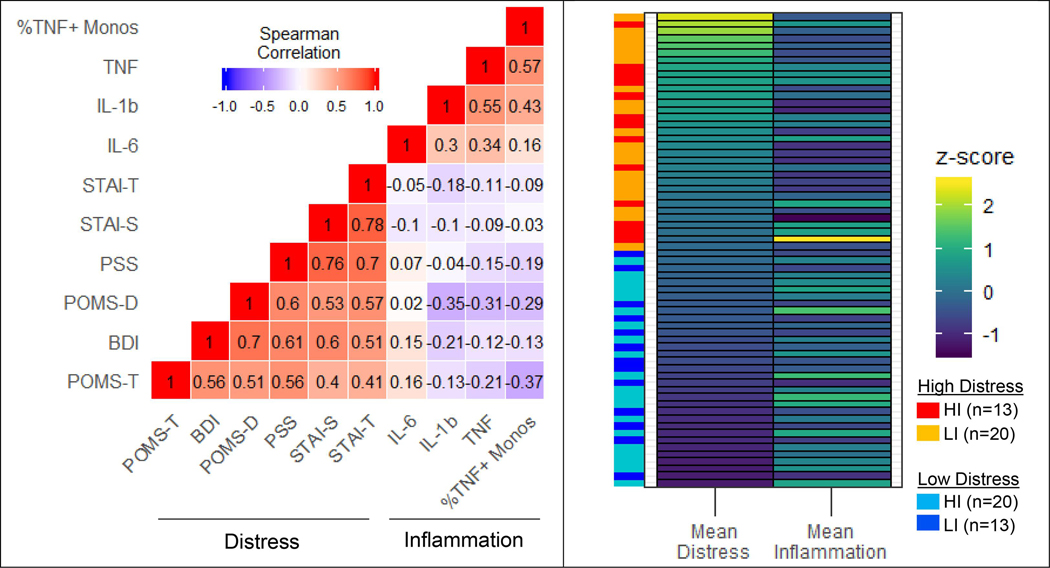
Psychological characteristics and immune biomarkers for groups are presented in Table 1. As expected, symptoms related to depressed mood, stress, and anxiety were moderately-to-strongly correlated between each other across all subjects (Spearman’s ρ: range=0.40–0.78; Fig. 1). Similarly, inflammation measures were positively correlated between each other, though somewhat less strongly (ρ: range=0.16–0.57). Univariate correlations between psychological and inflammatory measures were weak (ρ: range=−0.37–0.16) and somewhat unexpectedly, tended to be negatively correlated in our sample (mean ρ=−0.12±0.03). Therefore, of the 33 participants with the highest inflammatory measurements based on a median split of averaged z-scores (i.e., HI: high inflammation), 20 (61%) were among the bottom half of psychological distress scores based on median split (i.e., LD: low distress) (Fig. 1). The HI group had larger BMI than the low inflammation (LI) group (X2=4.56, p=0.03), as expected, but fewer depressive symptoms (POMS-D: X2=8.84, p=0.003). The high distress (HD) group was over-represented by women (70%, X2=10.2, p=0.001). No other group differences were observed according to age, sex, BMI, or race (Table 1).
Table 1.
Demographic, psychological, and inflammatory characteristics of study participants.
| Variable | Psychological Distress | Inflammation | ||
|---|---|---|---|---|
|
|
||||
| Low (N = 34) | High (N = 34) | Low (N = 33) | High (N = 33) | |
|
| ||||
| Age (yrs) | 41.3 ± 12 | 40.1 ± 12 | 41.3 ± 12 | 40.3 ± 12 |
| Sex (%female) | 30 | 70 | 61 | 39 |
| Race (%C/AA/Asn) | 64/21/15 | 52/27/21 | 48/27/24 | 67/21/12 |
| BM1 (kg/m2) | 28.6 ± 6.0 | 29.9 ± 6.0 | 27.9 ± 6.2 | 30.7 ± 5.6 |
| SBP (mmHg) | 121 ± 13.3 | 121 ± 14.4 | 118 = 13.2 | 123 ± 14.4 |
| BDI | 1.2 ± 1.7 | 7.1 ± 5.8 | 5.4 ± 6.1 | 3.1 ± 3.8 |
| STAI-S | 23.2 ± 4.1 | 35.0 ± 9.6 | 30.0 ± 10.0 | 28.3 ± 9.2 |
| STAI-T | 25.9 ± 6.2 | 37.9 ± 8.9 | 32.3 ± 9.8 | 30.9 ± 9.9 |
| POMS-D | 0.76 ± 1.5 | 6.1 ± 4.1 | 5.2 ± 7.8 | 1.7 ± 4.0 |
| POMS-T | 4.0 ± 2.2 | 8.6 ± 4.8 | 7.2 ± 4.9 | 5.5 ± 3.7 |
| PSS | 7.0 ± 3.8 | 17.1 ± 5.7 | 13.2 ± 6.7 | 11.2 ± 7.3 |
| Plasma TNF | 6.7 ± 4.3 | 5.4 ± 4.4 | 3.2 ± 1.9 | 8.8 ± 4.3 |
| Plasma IL-1β+ | 0.55 ± 0.5 | 0.49 ± 0.7 | 0.17 ± 0.2 | 0.86 ± 0.6 |
| Plasma IL-6 | 1.2 ± 1.6 | 1.6 ± 2.6 | 0.64 ± 0.5 | 2.1 ± 2.8 |
| TNF+ monocytes (%) | 50.9 ± 16.3 | 58.7 ± 15.8 | 44.3 ± 9.7 | 64.9 ± 15.5 |
Values presented as mean ± SD. Bold indicates p < 0.05, uncorrected. P-values derived from Mann-Whitney or chi-squared tests on raw values for between-group differences (i.e., low versus high distress, low versus high inflammation). Abbreviations: C = Caucasian: AA = African-American: Asn = Asian-American; BMI = body mass index; BDI = Beck Depression inventory-II total score; STAI-S = State anxiety inventory total score; STAI-T = Trait anxiety inventory total score; POMS-D = Profile of Mood States depressive subscale; POMS-T = Profile of Mood States tension subscale; PSS = Perceived Distress Scale total score; TNF = tumor necrosis factor alpha; IL-1β = Interleukin-1β: IL-6 = Interleukin-6; TNF+ monocytes = %TNF+ monocytes 3.5hr post-lipopolysaccharide stimulation. +IL-1β levels (15 of 66 total samples) below the lowest manufacturer standard (0.1 pg/mL) were imputed as 0.0 pg/mL.
Fig. 1.
(left) Spearman correlation matrix. Values reflect Spearman’s rank correlation coefficients between standardized (z-score) scores on psychological distress measures and inflammatory biomarkers across all study participants. (right) Heat map illustrating subject-wise mean distress and inflammation z-scores. Each horizontal bar represents an individual subject. The vertical bars on the left indicates z-score-based groupings. Abbreviations: BDI = Beck Depression Inventory-II total score; STAI-S = State anxiety inventory total score; STAI-T = Trait anxiety inventory total score; POMS-D = Profile of Mood States depressive subscale; POMS-T = Profile of Mood States tension subscale; PSS = Perceived Distress Scale total score; TNF = plasma tumor necrosis factor alpha; IL-1b = plasma interleukin-1b; IL-6 = plasma interleukin-6; %TNF + Monos = %TNF+ monocytes 3.5hr post-lipopolysaccharide stimulation; HI = high inflammation; LI = low inflammation, based on median split of sample-wide inflammation z-scores.
3.2. Microbial community structure
Alpha-diversity analysis of multinomial models adjusted for age, sex, BMI and race indicated that across the sampling day, from waking to evening (19:00 hr), evenness and phylogenetic diversity were generally unchanged (Table S1), though community richness tended to decrease (F=1.98, p=0.098).
Comparing high- and low-distress groups, HD individuals had greater alpha diversity, as measured by richness (t=2.79, p=0.007), evenness (t=2.16, p=0.034), and phylogenetic diversity (t=2.29, p=0.025), compared to LD individuals (Fig. S1). Despite increased overall diversity in the HD group, their time-specific community structure tended to vary less throughout the day (Faith’s: Ftime*distress=2.31, p=0.058). More specifically, phylogenetic diversity in HD individuals was essentially unchanged between waking and evening samples (Faith’s: 13.3±0.6 to 13.6±0.6; t=0.52, p=0.99), but was significantly reduced from waking to evening in LD individuals (13.1 ±0.6 to 11.0±0.6; t=−3.38, p=0.008).
Comparing high- and low-inflammation groups, alpha diversity did not differ by inflammation status (all p>0.05). Comparing other sociodemographic variables between groups, larger BMI was associated with greater phylogenetic diversity (F=4.60, p=0.036) and somewhat greater richness (F=3.71, p=0.059; Table S1).
Beta-diversity analysis indicated significant community-level separation as a function of time of day according to weighted UniFrac (wUF: pseudo-F=2.36, p=0.001) and Bray-Curtis (BC) dissimilarity (pseudo-F=1.35, p=0.013) metrics (Fig. S2). Pairwise contrasts revealed that waking samples were most distinct from other timepoints, and in fact, waking samples significantly differed from all other time points according to wUF, while no other contrasts were significantly significant (all FDR-corrected p<0.05; Table S4). While there were no significant time-by-distress or time-by-inflammation interactions, suggesting that temporal community dynamics did not differ by host distress or inflammation status, HD and LD groups differed significantly by wUF (pseudo-F=3.55, p=0.008) and BC (pseudo-F=3.34, p=0.001). HD individuals also clustered more tightly than LD individuals, who were dispersed more widely across the PCoA space (within-group distance comparison; wUF: p=0.007; BC: p=0.003), indicating greater homogeneity in microbial community structure among HD individuals. Inflammation groups also exhibited community-level separation according to the BC metric (pseudo-F=2.27, p=0.003), but not wUF (pseudo-F=1.55, p=0.11), and were not differentially dispersed (within-group distance comparison; BC: p=0.05; wUF: p=0.24). Across groups, significant beta-diversity differences were also observed by age and sex, but not BMI or race, according to wUF (Table S2).
3.3. Differentially ranked taxonomic features
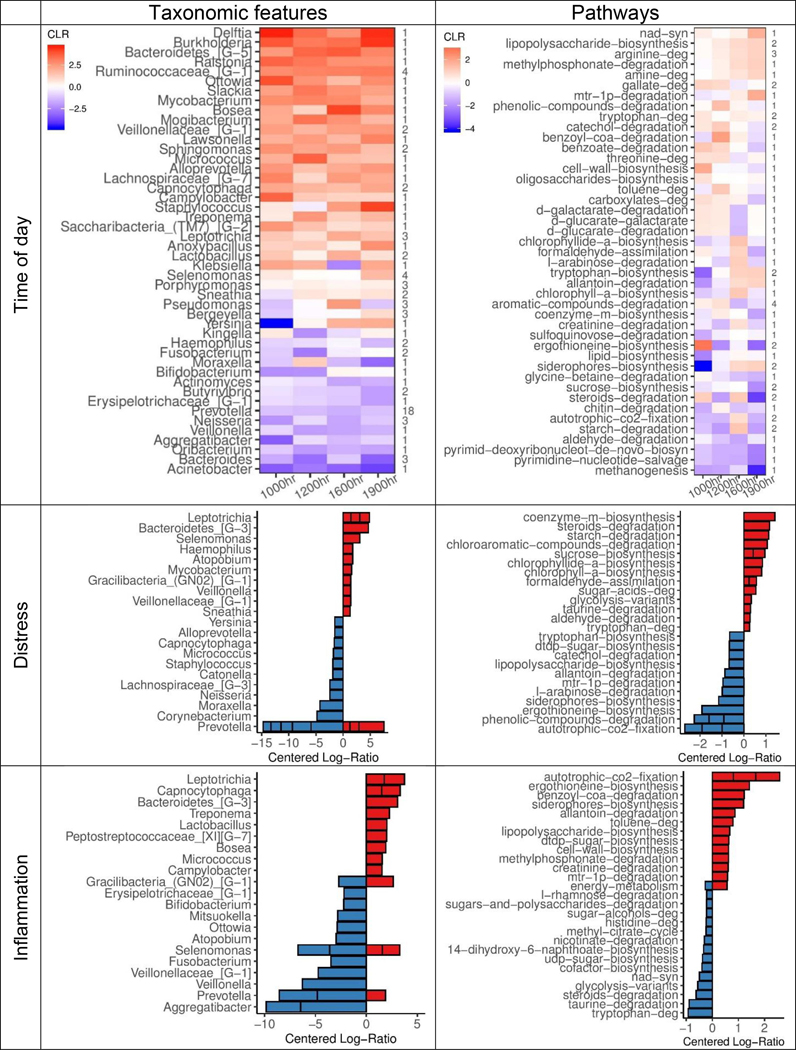
A differential ranking (DR) procedure was used to identify taxa that were most strongly associated with psychological distress and host inflammation (Morton et al., 2019), and ratio-based analyses were performed on the top- and bottom-40 ranked taxa for each host factor. LMMs indicated significant relative differences between top and bottom-ranked taxa associated with host distress (βstd=0.58, 95% CI: [0.42, 0.75]), and host inflammation (βstd=0.67, 95% CI: [0.50, 0.84]). Top-ranked taxa that were positively associated with host distress included Leptotrichia, Bacteroidetes, Selenomonas, and Haemophilus, while bottom-ranked, negatively associated taxa were predominately of the Prevotella genus. Taxa that were positively associated with host inflammation included Leptotrichia, Capnocytophaga, Treponema, and Bacteroidetes, whereas negatively associated taxa included Aggregatibacter, Bifidobacterium, Prevotella, and Veillonella. Of these 131 unique taxa, 29 (22%) were top- or bottom-40 ranked by both distress and inflammation, and included features from Leptotrichia and Bacteroidetes genera (Table S5), and 16 taxa shared the same directionality of association.
Surprisingly, log-ratios of the 90 unique time-varying taxa were also associated with host inflammatory status (βstd=0.18, 95% CI: [0.00, 0.35]), but not psychological distress (βstd=−0.01, 95% CI: [−0.15, 0.18]), suggesting that taxa with increased relative abundance throughout the day relative to waking were associated with greater host inflammation, but not with distress. Neither age nor BMI were associated with DR taxa, based on the magnitude of their differential rankings relative to time point, distress, or inflammation; though, there was evidence of DR taxa by host sex (Table S5).
Ranked differentials of taxa were largely directionally consistent across time (Fig. 2). For instance, of the top- and bottom-20 ranked taxa at each time point relative to waking (160 total; 90 unique taxa), 44 taxa were highly ranked at 2 or more time points in the same direction, whereas none were highly ranked in the opposite direction (Table S5). Ratio-based analysis revealed that these 90 unique taxa (37 positively associated, 53 negatively associated) were significantly different from waking samples across time, relative to each other (10:00 hr: βstd=0.27, 95% CI: [0.13, 0.40]; 12:00 hr: βstd=0.23 [0.10, 0.37]; 16:00 hr: βstd=0.25 [0.11, 0.38]; 19:00 hr: βstd=0.30 [0.16, 0.43]). These results corroborate evidence for a diurnal pattern in salivary microbial composition across time. As a whole, Prevotella, Bacertoides, and Neisseria were most strongly associated with waking samples, whereas Ruminococcaceae, Veillonellaceae, and Sphingomonas were associated with later timepoints (Fig. 2).
Fig. 2.
Centered log-ratios (CLR) of top-ranked taxonomic features (left) and metabolic pathways (right) derived from two multinomial regression models (e.g., one taxonomic feature-based model, one pathway-based model) of differentially ranked (DR) relative abundances. (Top row) Mean CLRs for all time-associated features (90 unique of 485 in feature set) and top- and bottom-ranked pathways (59 unique of 384 in feature set), averaged and grouped by genus or parent ontology, at each sampling time point relative to waking. Number of unique features per genus or pathway shown on right side of plot. (Middle and bottom rows) Mean CLRs for 15 top- and bottom-DR taxa and pathways by distress (middle) and inflammation (bottom) scores, grouped by genus or ontology. Genera or pathways containing more than 1 unique feature are shown as stacked bars. Age, sex, BMI, and race were included as covariates in all models. Parent pathway ontologies were derived from MetaCyc v23.1.
3.4. Predictive group classification
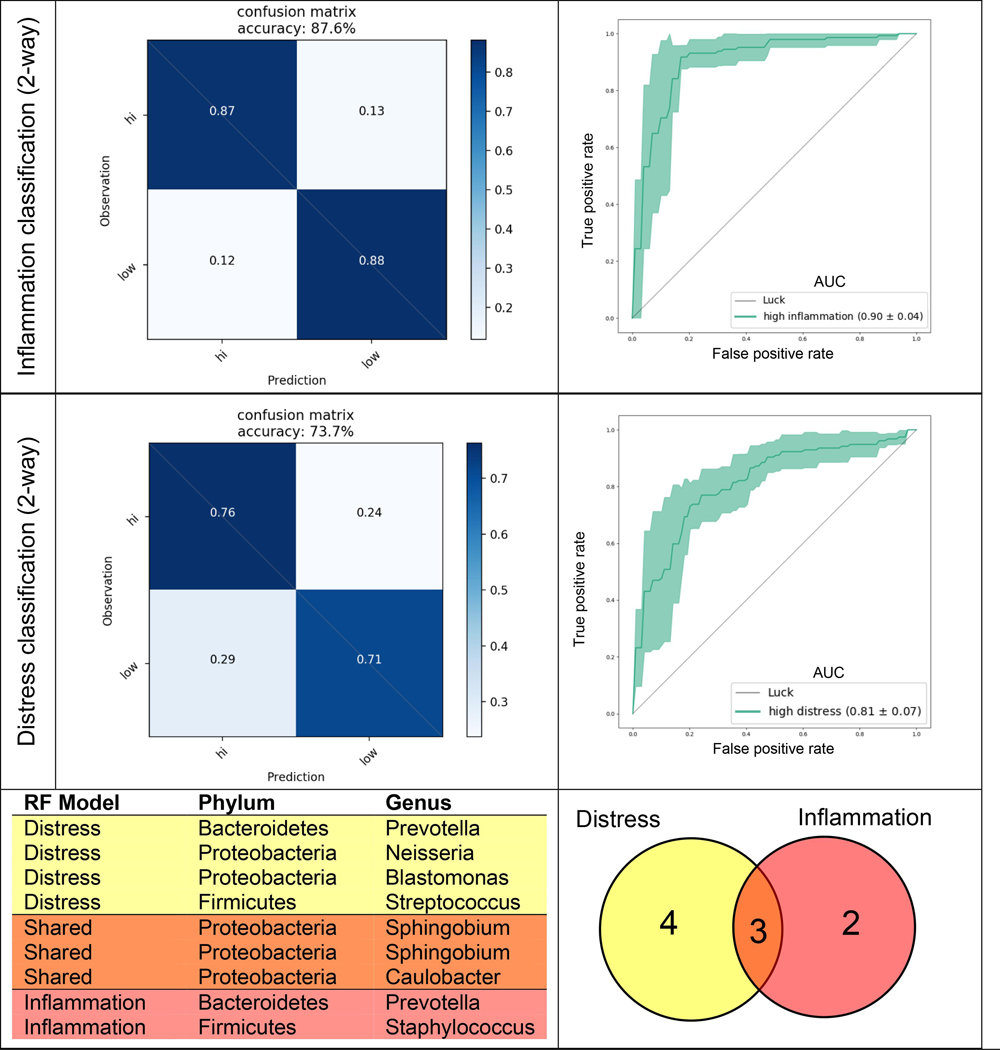
Two separate random forest models were implemented to determine whether samples could be accurately predicted as originating from participants with either high or low distress or inflammation based on relative microbial abundances alone. The models were able to classify individuals into distress and inflammation groups with 73.7% (AUC: 0.81) and 87.6% (AUC: 0.90) accuracy, respectively (Fig. 3). Examination of the features that were most informative for each prediction model (feature importance > 0.01) revealed seven ASVs from the distress model and five ASVs from the inflammation model. Within the distress classifier, three features were from the Sphingomonadaceae family, in addition to taxa from Prevotella, Neisseria, Streptococcus, and Caulobacter genera. The inflammation classifier contained two features of the Sphingomonadaceae family, as well as Prevotella, Caulobacter, and Staphylococcus. Of these key features, 3 were shared between the two classifiers: 2 from the Sphingobium genus and one from Caulobacter (Fig. 3). However, these features were not enriched within taxa identified as highly-ranked by DR analysis (Section 3.3) (only 1 of 7 in distress, and 1 of 5 in inflammation classifier).
Fig. 3.
Confusion matrices (left) and receiver operating characteristic (ROC) curves (right) derived from 2 random forest (RF) classification models: (top) 2-way inflammation classification (high versus low), (middle) 2-way distress classification (high versus low). Age, sex, BMI, and race were one hot encoded as features in all models. Filtered (> 1% prevalence), normalized (100,000 counts) microbiome counts sampled at all five time points were used, with time point information encoded as an additional feature. All data were CLR transformed prior to training, and grid search sampling employed for each model to optimize parameters. Five-fold stratified cross-validation was performed without test set construction. (Bottom) Table and venn diagram of amplicon sequence variants (ASVs) with feature importance > 0.01 from both RF classifiers. Area under the curve (AUC) values shown in ROC curve legend.
3.5. Predictive metagenomic profile diversity
In order to determine whether functional pathways detectable in the salivary microbiome differed as a function of time-of-day or host profiles, we used PICRUSt2 (Langille et al., 2013) to extract abundance data of 384 unique pathways from our 16S rRNA data. LMMs of alpha diversity metrics indicated that functional pathway diversity did not fluctuate across the day (all p>0.05). However, pathways themselves shifted and were somewhat dynamic throughout the day (Fig. 2), with increasingly ranked LPS biosynthesis and decreased ranking of methanogenesis. Individuals in the HD group had greater pathway evenness across the day (Shannon=5.35±0.01; F=4.67, p=0.035) compared to the LD group (Shannon=5.31 ±0.01), consistent with the community structure results (Section 3.2) indicating higher alpha diversity in HD versus LD individuals (Fig. S3). Notably, distress scores were associated with more highly-ranked steroid and tryptophan degradation, and bottom-ranked tryptophan biosynthesis (Table S6).
Time-dependent changes in alpha diversity also differed as a function of inflammation profile (Ftime*infiammation=2.97, p=0.02), whereby richness had declined in the high inflammation group at 16:00 relative to other time points (Observed: 16:00 hr=308±4.6 versus 10:00 hr=330±4.5, t=3.58, p=0.004; 12:00 hr=327±4.6, t=3.07, p=0.02; 19:00 hr=325±4.5, t=2.90, p=0.03), but this mid-day difference was not observed in low-inflammation individuals (p>0.05, all contrasts). Host inflammation was also associated with bottom-ranked steroid and tryptophan degradation pathways, and highly-ranked LPS biosynthesis (Table S6). Notably, of the 12 top-ranked pathways shared between distress and inflammation sets, all had opposing relationships with each host factor (e.g., positive association with inflammation, negative with distress).
Larger BMI was associated with lower pathway richness (F=5.37, p=0.024) and evenness (F=5.79, p=0.02), but no effects were observed for age, sex, or race. Beta diversity analysis indicated pathway-level separation as a function of distress group (BC: F=3.23, p=0.008), but not inflammation group (F=1.74, p=0.12). Time-by-distress or time-by-inflammation interactions were not significant, though across groups, beta diversity differences were observed by age, sex, and BMI (Table S3). Larger BMI was associated with lower pathway richness (F=5.37, p=0.024) and evenness (F=5.79, p=0.02), but no effects were observed for age, sex, or race.
4. Discussion
In this study, we identified differences in overall composition and diurnal patterns of relative microbial abundance and functional pathways in the salivary microbiome as a function of psychological distress and inflammatory profiles in healthy adults. To our knowledge, this study provides the first evidence that psychological distress and affect in a non-clinical population is associated with greater microbial diversity in saliva, and with specific microbial taxa and functional pathways, and that proinflammatory cytokine levels in the periphery are associated with salivary microbial composition. Importantly, these differences were only slightly attenuated when adjusted for key demographic factors, specifically age, sex, race, and BMI. Although measures of inflammation and distress were largely unrelated in our sample of healthy adults, our classification was able to differentiate individuals of (i) high-inflammation from low-inflammation, and (ii) high-distress from low-distress by their oral microbial community with high predictability such that at the ASV level, individuals were classified as either high or low inflammation, or either high or low distress with 88% and 74% accuracy, respectively. The most important features were derived from the Sphingomonadaceae family, but also included taxa such as Streptococcus and Staphylococcus, which may have pathogenic potential, but these features were not specifically enriched for within highly ranked taxa for host distress or inflammation according to DR analysis. This analysis also identified taxa that were positively and negatively associated with host distress and inflammation, and those that were associated with both factors, which included Leptotrichia and Bacteroidetes.
Our findings of higher alpha diversity among individuals with greater distress are unexpected based on the existing gut microbiome diversity findings. Meanwhile, unlike the gut microbiome findings, where greater alpha diversity tends to be associated with better psychological and somatic health, lower alpha diversity in the salivary microbiome may in fact be associated with better oral health (Abusleme et al., 2013; Chen et al., 2018; Takeshita et al., 2016). Our findings offer additional insight into potentially divergent organ-specific microbiome diversity in relation to health, and further investigation is warranted. Our findings are in line with the previous studies of the associations between oral microbiome diversity and neuropsychiatric conditions, specifically Parkinson’s Disease (Pereira et al., 2017), schizophrenia (Castro-Nallar et al., 2015; Yolken et al., 2015), and autism spectrum disorders (Hicks et al., 2018), reporting significant microbial community differences (i.e., beta diversity) between clinical and non-clinical subjects. In contrast, these reports did not find significant differences in alpha diversity. Furthermore, the circadian variation in salivary microbiome in relation to psychiatric conditions remained unknown based on previous studies.
Notably, our analyses included multiple saliva samples per individual across a single day, which provide further evidence for diurnal changes in the salivary microbiome and their relationship to the host. Our analyses revealed that phylogenetic diversity decreased from morning to evening in low-distress individuals, but not in the high-distress group, which to our knowledge is the first report that diurnal features of the oral microbiome differ by host psychological status. Chronic psychological distress is associated with other circadian markers such as blunted diurnal cortisol (Adam et al., 2017) and proinflammatory cytokine rhythms (Reinhardt et al., 2019) in saliva, and represent possible mechanisms by which psychological factors affect the oral microbiome. In support of this hypothesis, brief in vitro treatment of dental plaques with exogenous cortisol has been shown to acutely shift their transcriptome toward increased Leptotrichia abundance, virulence factor production, and pathways associated with periodontal disease (Duran-Pinedo et al., 2018). We found that, collectively, the Leptotrichia taxa present in our sample were generally higher-ranked in association with distress (21 of 31 unique Leptotrichia) and represented 5 of the top-20 ranked taxa, but whether elevated endogenous glucocorticoids upregulate microbes or their metabolic function within the oral environment in vivo is unknown. In vivo studies also indicate that chronic, systemic glucocorticoid infusion, which mimics HPA axis dysregulation, profoundly shifts commensal gut bacteria, particularly toward increased Actinobacteria abundance (Huang et al., 2015), and that oral prebiotics can attenuate the salivary cortisol awakening response (Schmidt et al., 2015) by affecting gut microbial proliferation. Interestingly, steroid degradation was one of the top-ranked distress-associated pathways in our analysis. Others have described the role of gut microbes in endogenous corticosteroid metabolism and subsequent effects on host physiology, such as blood pressure (Morris and Brem, 2019). Lastly, salivary flow rate is also associated with chronic distress (Bulthuis et al., 2018) and may impact the oral microbiome (Proctor et al., 2018).
Importantly, waking samples alone were insufficient to identify host factor-associated differences in taxa DR or microbial diversity (visually represented in Fig. S1, wherein group differences only emerge across the sampling day), highlighting the benefits of repeated measurement of the oral microbiome as a biomarker of host psychological distress or peripheral inflammatory status.
Although we found blunted microbial oscillations in high-distress individuals, diurnal variation in functional pathway diversity was in fact present in the high-inflammation group, but not low-inflammation group (Morris and Brem, 2019). This result is unlikely to have resulted from overlapping group membership between low-distress and high-inflammation individuals, and distress group membership was included as a predictor in all models. One possibility is that higher peripheral levels of proinflammatory cytokines are reflected in larger diurnal cytokine variation, which has been reported for plasma IL-6 in arthritis patients (Cutolo et al., 2008), but not in post-traumatic stress (Agorastos et al., 2019). Whether peripheral and salivary cytokine levels are well-correlated also remains unclear. However, gut microbial composition has been linked to ex vivo pathogen-stimulated inflammatory cytokine production (Schirmer et al., 2016) via immunomodulatory bacterial metabolites, such as tryptophan. The reverse mechanism is also likely: that host cytokines modulate bacterial virulence (Mahdavi et al., 2013) and proliferation (Lee et al., 2003), though these host-microbe interactions are unexplored in the context of commensal and pathogenic bacteria of the oral microbiome.
In addition to host-microbiome interactions occurring within the mouth, the oral cavity may serve as an important bacterial source for the gut-brain axis. The oral environment may shed 3–4 × 108 bacterial cells per minute to the stomach (lorgulescu, 2009; Sender et al., 2016), and although fecal and oral microbial compositions are dramatically distinct (Caporaso et al., 2011) oral bacteria were found to be enriched in the feces of patients with various diseases, suggesting that the oral microbiome serves as a reservoir of opportunistic colonizers should the gut microbiome become dysbiotic (Lira-Junior and Bostrom, 2018). Recent work by Atarashi et al. demonstrated that strains of Klebsiella from the salivary microbiome ectopically colonize the gut and induce chronic intestinal TH1 cell-mediated inflammation (Atarashi et al., 2017). The gut microbiome has also been implicated in periodontal diseases (Lourengo et al., 2018), which tend to be more prevalent in depression and anxiety disorders (Kisely et al., 2016), and in neurodegenerative diseases, such as Alzheimer’s dementia (Ad) (Kamer et al., 2015), suggesting that oral microbes may affect the gut-brain axis by “seeding” the gut microbiome. An alternative mechanism by which the oral microbiome can affect host immunity was recently demonstrated through transmigration of pathogenic oral microbes into the bloodstream and brain, thereby potentiating neuroinflammation and AD-associated neuropathology (Dominy et al., 2019) in older adults.
4.1. Limitations and Strengths
We note following limitations in the current study and future considerations. First, meal timing and composition, which are known to impact salivary microbial rhythmicity (Collado et al., 2018), were not recorded for this study. It is therefore possible that unknown differences in meal timing or feeding behaviors between groups mediated some of the observed effects, or conversely obscured true effects of host profiles. Consistent with prior findings of increased relative abundance of Actinomyces relative to Haemophilus taxa (Morton et al., 2019), we observed similar changes at 10:00 hr compared to waking in our samples (βstd=0.11, 95% CI: [0.01, 0.22]), suggesting that time-dependent changes our sample were related in part to oral hygiene behaviors. Future replications should capture this information to allow for these factors to be included in modeling the observed changes. Second, while chronic distress and depressed mood can be associated with poorer oral hygiene and are risk factors for periodontal disease (Benatti et al., 2007), the specific oral and periodontal health history of our participants was not known. Thus, distress-related differences we identified in diversity, taxa, and pathways may reflect underlying periodontal disease, rather than other sequelae of distress-related physiology. Future investigations of oral microbiome should aim to characterize the relationship between psychological distress as well as periodontal health, salivary neuroendocrine factors, metabolites, and flow rate to the oral microbiome. Third, while our saliva sampling method (using a cotton roll) performs similarly to another widely used sampling method (i.e., passive drool) for host DNA collection (Koni et al., 2011), its performance for microbial DNA collection has not been formally assessed. However, others have found that a 1 min saline “oral rinse” procedure for collection yielded greater microbial alpha-diversity than drool or spit methods (Lim et al., 2017), which reflects the complexity of methodological impacts and differing microbial community dependent upon sample types. Though, the degree to which the microbial ‘snapshot’ provided with our Salivette method is similar to or different from other sampling procedures is unknown, and its exploration is warranted. Fourth, weak associations between psychological distress and inflammation scores within our sample were somewhat unexpected, but may reflect the sub-syndromal status of the population studied. Lastly, a consideration should be given to the physiological implications of the link between a systemic inflammatory profile and oral microbiome for which current knowledge is limited. Some salivary inflammatory biomarker levels, such as IL-6, undergo a diurnal rhythm in healthy adults (Izawa et al., 2013) but exhibit a blunted pattern in night shift workers (Reinhardt et al., 2019), a chronic psychosocial stressor that disrupts circadian biology. However, whether salivary cytokines accurately reflect peripheral (i.e., plasma, serum) levels is unclear. Future investigations should test for simultaneous associations between diurnal patterns of salivary and systemic markers of inflammation and their relationship to the oral microbiome.
Despite these limitations, the present study possesses several strengths. Firstly, our findings add novel knowledge of salivary microbial community, diversity, and diurnal variations in association with psychological and inflammatory profile to the current paucity of literature. Second, no current cigarette smokers or tobacco users were included in the analysis. Smoking is a significant driver of oral microbiome composition (Belstrøm et al., 2014), is more prevalent in psychologically distressed individuals (Cook et al., 2014), and is associated with higher proinflammatory biomarker levels in peripheral blood (Levitzky et al., 2008). Thus, host-related microbiome compositional differences in our study are not attributable to smoking behavior, nor can they be quantified relative to the size of the effect of smoking. Lastly, by testing proinflammatory cytokine levels in the periphery, we were able to evaluate whether the salivary microbiome, an easily accessible, on-demand biological substrate, is associated with a valid biomarker of systemic immune status, which is known to influence the brain and behavior. These findings lay the groundwork for future investigations in further elucidating the role of host psychological status and circadian neuroendocrinology and immunology in oral microbiome composition and vice versa. The oral microbiome holds promise as a biomarker of high accessibility and utility for host health and disease and as a possible therapeutic target to mitigate the morbidity of psychiatric and immunological conditions.
Supplementary Material
Acknowledgements
We would like to thank Gregory Humphrey, Lindsay DeRight-Goldasich, Carolina Carpenter, and Tara Schwartz for sample processing, Gail Ackermann for assistance with metadata curation and data handling, and Daniel Freed for his contributions to the project. The authors would like to express their gratitude to all individuals who participated in this study.
Funding Sources
This work was supported by NIH grants R01 HL090975 and HL090975S1 (SH), 1TL1TR001443 (JNK), and a Seed Grant from the UC San Diego Center for Microbiome Innovation (SH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, Gamonal J, Diaz PI, 2013. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 7, 1016–1025. 10.1038/ismej.2012.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam EK, Quinn ME, Tavernier R, McQuillan MT, Dahlke KA, Gilbert KE, 2017. Diurnal cortisol slopes and mental and physical health outcomes: A systematic review and meta-analysis. Psychoneuroendocrinology 83, 25–41. 10.1016/j.psyneuen.2017.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agorastos A, Hauger RL, Barkauskas DA, Lerman IR, Moeller-Bertram T, Snijders C, Haji U, Patel PM, Geracioti TD, Chrousos GP, Baker DG, 2019. Relations of combat stress and posttraumatic stress disorder to 24-h plasma and cerebrospinal fluid interleukin-6 levels and circadian rhythmicity. Psychoneuroendocrinology 100, 237–245. 10.1016/j.psyneuen.2018.09.009 [DOI] [PubMed] [Google Scholar]
- Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Zech Xu Z, Kightley EP, Thompson LR, Hyde ER, Gonzalez A, Knight R, 2017. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems 2, 1–7. 10.1128/mSystems.00191-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, Kiguchi Y, Yasuma K, Watanabe E, Tanoue T, Thaiss CA, Sato M, Toyooka K, Said HS, Yamagami H, Rice SA, Gevers D, Johnson RC, Segre JA, Chen K, Kolls JK, Elinav E, Morita H, Xavier RJ, Hattori M, Honda K, 2017. Ectopic colonization of oral bacteria in the intestine drives T H 1 cell induction and inflammation. Science (80-. ). 358, 359–365. 10.1126/science.aan4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck AT, Steer RA, 1993. Manual for the Beck Depression Inventory. [Google Scholar]
- Belstrøm D, Holmstrup P, Nielsen CH, Kirkby N, Twetman S, Heitmann BL, Klepac-Ceraj V, Paster BJ, Fiehn NE, 2014. Bacterial profiles of saliva in relation to diet, lifestyle factors, and socioeconomic status. J. Oral Microbiol 6, 1–9. 10.3402/jom.v6.23609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benatti BB, Sallum EA, Peruzzo DC, Casati MZ, Nociti FH, Ambrosano GMB, Nogueira-Filho GR, 2007. A Systematic Review of Stress and Psychological Factors as Possible Risk Factors for Periodontal Disease. J. Periodontol 78, 1491–1504. 10.1902/jop.2007.060371 [DOI] [PubMed] [Google Scholar]
- Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Gregory Caporaso J, 2018. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 1–17. 10.1186/s40168-018-0470-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch JA, Ring C, de Geus EJC, Veerman ECI, Nieuw Amerongen AV, 2002. Stress and secretory immunity, in: Encyclopedia of Stress. pp. 213–253. 10.1016/S0074-7742(02)52011-0 [DOI] [PubMed] [Google Scholar]
- Bruce-Keller AJ, Salbaum JM, Berthoud HR, 2018. Harnessing Gut Microbes for Mental Health: Getting From Here to There. Biol. Psychiatry 83, 214–223. 10.1016/j.biopsych.2017.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulthuis MS, Jan Jager DH, Brand HS, 2018. Relationship among perceived stress, xerostomia, and salivary flow rate in patients visiting a saliva clinic. Clin. Oral Investig 22, 3121–3127. 10.1007/s00784-018-2393-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, Knights D, Gajer P, Ravel J, Fierer N, Gordon JI, Knight R, 2011. Moving pictures of the human microbiome. Genome Biol. 12, R50. 10.1186/gb-2011-12-5-r50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R, 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Billington R, Ferrer L, Foerster H, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Subhraveti P, Weaver DS, Karp PD, 2016. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 44, D471–D480. 10.1093/nar/gkv1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Nallar E, Perez-Losada M, Dickerson FB, Sabuncyan S, Schroeder JR, Severance EG, Yolken RH, Crandall KA, Bendall ML, 2015. Composition, taxonomy and functional diversity of the oropharynx microbiome in individuals with schizophrenia and controls. PeerJ 3, e1140. 10.7717/peerj.1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Hemme C, Beleno J, Shi ZJ, Ning D, Qin Y, Tu Q, Jorgensen M, He Z, Wu L, Zhou J, 2018. Oral microbiota of periodontal health and disease and their changes after nonsurgical periodontal therapy. ISME J. 12, 1210–1224. 10.1038/s41396-017-0037-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung SG, Goldenthal AR, Uhlemann AC, Mann JJ, Miller JM, Sublette ME, 2019. Systematic review of gut microbiota and major depression. Front. Psychiatry 10. 10.3389/fpsyt.2019.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Kamarck T, Mermeistein R, 1983. Perceived Stress Scale. J. Health Soc. Behav 24, 386–396. [PubMed] [Google Scholar]
- Collado MC, Engen PA, Bandín C, Cabrera-Rubio R, Voigt RM, Green SJ, Naqib A, Keshavarzian A, Scheer FAJL, Garaulet M, 2018. Timing of food intake impacts daily rhythms of human salivary microbiota: A randomized, crossover study. FASEB J. 32, 2060–2072. 10.1096/fj.201700697RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook BL, Wayne GF, Kafali EN, Liu Z, Shu C, Flores M, 2014. Trends in Smoking Among Adults With Mental Illness and Association Between Mental Health Treatment and Smoking Cessation. JAMA 311, 172. 10.1001/jama.2013.284985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan JF, Dinan TG, 2012. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci 13, 701–712. 10.1038/nrn3346 [DOI] [PubMed] [Google Scholar]
- Cutolo M, Straub RH, Buttgereit F, 2008. Circadian rhythms of nocturnal hormones in rheumatoid arthritis: translation from bench to bedside -- Cutolo et al. 67 (7): 905 -- Annals of the Rheumatic Diseases. Ann. Rheum. Dis 67, 905–8. 10.1136/ard.2008.088955 [DOI] [PubMed] [Google Scholar]
- Dalile B, Van Oudenhove L, Vervliet B, Verbeke K, 2019. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat. Rev. Gastroenterol. Hepatol 10.1038/s41575-019-0157-3 [DOI] [PubMed] [Google Scholar]
- Dimitrov S, Hulteng E, Hong S, 2017. Inflammation and exercise: Inhibition of monocytic intracellular TNF production by acute exercise via β2-adrenergic activation. Brain. Behav. Immun 61, 60–68. 10.1016/j.bbi.2016.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding T, Schloss PD, 2014. Dynamics and associations of microbial community types across the human body. Nature 509, 357–360. 10.1038/nature13178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominy SS, Lynch C, Ermini F, Benedyk M, Marczyk A, Konradi A, Nguyen M, Haditsch U, Raha D, Griffin C, Holsinger LJ, Arastu-Kapur S, Kaba S, Lee A, Ryder MI, Potempa B, Mydel P, Hellvard A, Adamowicz K, Hasturk H, Walker GD, Reynolds EC, Faull RLM, Curtis MA, Dragunow M, Potempa J, 2019. Porphyromonas gingivalis in Alzheimer’s disease brains: Evidence for disease causation and treatment with small-molecule inhibitors. Sci. Adv 5, eaau3333. 10.1126/sciadv.aau3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Teil Espina M, Gabarrini G, Harmsen HJM, Westra J, Van Winkelhoff AJ, Van Dijl JM, 2019. Talk to your gut: The oral-gut microbiome axis and its immunomodulatory role in the etiology of rheumatoid arthritis. FEMS Microbiol. Rev 43, 1–18. 10.1093/femsre/fuy035 [DOI] [PubMed] [Google Scholar]
- Duran-Pinedo AE, Solbiati J, Frias-Lopez J, 2018. The effect of the stress hormone cortisol on the metatranscriptome of the oral microbiome. npj Biofilms Microbiomes 4, 1–4. 10.1038/s41522-018-0068-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engeland CG, Hugo FN, Hilgert JB, Nascimento GG, Junges R, Lim H-J, Marucha PT, Bosch JA, 2016. Psychological distress and salivary secretory immunity. Brain. Behav. Immun 52, 11–17. 10.1016/j.bbi.2015.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores GE, Caporaso JG, Henley JB, Rideout JR, Domogala D, Chase J, Leff JW, Vazquez-Baeza Y, Gonzalez A, Knight R, Dunn RR, Fierer N, 2014. Temporal variability is a personalized feature of the human microbiome. Genome Biol. 15, 531. 10.1186/s13059-014-0531-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J, Weisberg S, 2011. An R Companion to Applied Regression, Second Edition [WWW Document]. Sage, Thousand Oaks CA. URL http://socserv.socsci.mcmaster.ca/jfox/ Books/Companion. [Google Scholar]
- Francavilla R, Ercolini D, Piccolo M, Vannini L, Siragusa S, De Filippis F, De Pasquale I, Di Cagno R, Di Toma M, Gozzi G, Serrazanetti DI, De Angelis M, Gobbetti M, 2014. Salivary microbiota and metabolome associated with celiac disease. Appl. Environ. Microbiol 80, 3416–3425. 10.1128/AEM.00362-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Navas-molina JA, Kosciolek T, Mcdonald D, Vazquez-baeza Y, Ackermann G, Dereus J, Janssen S, Swafford AD, Orchanian SB, Sanders JG, Shorenstein J, Holste H, Petrus S, Robbins-pianka A, Brislawn CJ, Wang M, Rideout JR, Bolyen E, Dillon M, Caporaso JG, Dorrestein PC, Knight R, 2018. Qiita: rapid, web-enabled microbiome meta-analysis. Nat. Methods 15. 10.1038/s41592-018-0141-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks SD, Uhlig R, Afshari P, Williams J, Chroneos M, Tierney-Aves C, Wagner K, Middleton FA, 2018. Oral microbiome activity in children with autism spectrum disorder. Autism Res. 11, 1286–1299. 10.1002/aur.1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne R, Foster JA, 2018. Metabolic and Microbiota Measures as Peripheral Biomarkers in Major Depressive Disorder. Front. Psychiatry 9, 1–8. 10.3389/fpsyt.2018.00513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EY, Inoue T, Leone VA, Dalal S, Touw K, Wang Y, Musch MW, Theriault B, Higuchi K, Donovan S, Gilbert J, Chang EB, 2015. Using corticosteroids to reshape the gut microbiome: Implications for inflammatory bowel diseases. Inflamm. Bowel Dis 21, 963–972. 10.1097/MIB.0000000000000332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, Fitzgerald MG, Fulton RS, Giglio MG, Hallsworth-Pepin K, Lobos EA, Madupu R, Magrini V, Martin JC, Mitreva M, Muzny DM, Sodergren EJ, Versalovic J, Wollam AM, Worley KC, Wortman JR, Young SK, Zeng Q, Aagaard KM, Abolude OO, Allen-Vercoe E, Alm EJ, Alvarado L, Andersen GL, Anderson S, Appelbaum E, Arachchi HM, Armitage G, Arze CA, Ayvaz T, Baker CC, Begg L, Belachew T, Bhonagiri V, Bihan M, Blaser MJ, Bloom T, Bonazzi V, Paul Brooks J, Buck GA, Buhay CJ, Busam DA, Campbell JL, Canon SR, Cantarel BL, Chain PSG, Chen IMA, Chen L, Chhibba S, Chu K, Ciulla DM, Clemente JC, Clifton SW, Conlan S, Crabtree J, Cutting MA, Davidovics NJ, Davis CC, Desantis TZ, Deal C, Delehaunty KD, Dewhirst FE, Deych E, Ding Y, Dooling DJ, Dugan SP, Michael Dunne W, Scott Durkin A, Edgar RC, Erlich RL, Farmer CN, Farrell RM, Faust K, Feldgarden M, Felix VM, Fisher S, Fodor AA, Forney LJ, Foster L, Di Francesco V, Friedman J, Friedrich DC, Fronick CC, Fulton LL, Gao H, Garcia N, Giannoukos G, Giblin C, Giovanni MY, Goldberg JM, Goll J, Gonzalez A, Griggs A, Gujja S, Kinder Haake S, Haas BJ, Hamilton HA, Harris EL, Hepburn TA, Herter B, Hoffmann DE, Holder ME, Howarth C, Huang KH, Huse SM, Izard J, Jansson JK, Jiang H, Jordan C, Joshi V, Katancik JA, Keitel WA, Kelley ST, Kells C, King NB, Knights D, Kong HH, Koren O, Koren S, Kota KC, Kovar CL, Kyrpides NC, La Rosa PS, Lee SL, Lemon KP, Lennon N, Lewis CM, Lewis L, Ley RE, Li K, Liolios K, Liu B, Liu Y, Lo CC, Lozupone CA, Dwayne Lunsford R, Madden T, Mahurkar AA, Mannon PJ, Mardis ER, Markowitz VM, Mavromatis K, McCorrison JM, McDonald D, McEwen J, McGuire AL, McInnes P, Mehta T, Mihindukulasuriya KA, Miller JR, Minx PJ, Newsham I, Nusbaum C, Oglaughlin M, Orvis J, Pagani I, Palaniappan K, Patel SM, Pearson M, Peterson J, Podar M, Pohl C, Pollard KS, Pop M, Priest ME, Proctor LM, Qin X, Raes J, Ravel J, Reid JG, Rho M, Rhodes R, Riehle KP, Rivera MC, Rodriguez-Mueller B, Rogers YH, Ross MC, Russ C, Sanka RK, Sankar P, Fah Sathirapongsasuti J, Schloss JA, Schloss PD, Schmidt TM, Scholz M, Schriml L, Schubert AM, Segata N, Segre JA, Shannon WD, Sharp RR, Sharpton TJ, Shenoy N, Sheth NU, Simone GA, Singh I, Smillie CS, Sobel JD, Sommer DD, Spicer P, Sutton GG, Sykes SM, Tabbaa DG, Thiagarajan M, Tomlinson CM, Torralba M, Treangen TJ, Truty RM, Vishnivetskaya TA, Walker J, Wang L, Wang Z, Ward DV, Warren W, Watson MA, Wellington C, Wetterstrand KA, White JR, Wilczek-Boney K, Wu Y, Wylie KM, Wylie T, Yandava C, Ye L, Ye Y, Yooseph S, Youmans BP, Zhang L, Zhou Y, Zhu Y, Zoloth L, Zucker JD, Birren BW, Gibbs RA, Highlander SK, Methe BA, Nelson KE, Petrosino JF, Weinstock GM, Wilson RK, White O, 2012. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorgulescu G, 2009. Saliva between normal and pathological. Important factors in determining systemic and oral health. J. Med. Life 2, 303–7. [PMC free article] [PubMed] [Google Scholar]
- Izawa S, Miki K, Liu X, Ogawa N, 2013. The diurnal patterns of salivary interleukin-6 and C-reactive protein in healthy young adults. Brain. Behav. Immun 27, 38–41. 10.1016/j.bbi.2012.07.001 [DOI] [PubMed] [Google Scholar]
- Josse J, Husson F, 2016. missMDA: A Package for Handling Missing Values in Multivariate Data Analysis. J. Stat. Softw 70, 1–31. 10.18637/jss.v070.i01 [DOI] [Google Scholar]
- Kamer AR, Pirraglia E, Tsui W, Rusinek H, Vallabhajosula S, Mosconi L, Yi L, McHugh P, Craig RG, Svetcov S, Linker R, Shi C, Glodzik L, Williams S, Corby P, Saxena D, de Leon MJ, 2015. Periodontal disease associates with higher brain amyloid load in normal elderly. Neurobiol. Aging 36, 627–633. 10.1016/j.neurobiolaging.2014.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisely S, Sawyer E, Siskind D, Lalloo R, 2016. The oral health of people with anxiety and depressive disorders - A systematic review and meta-analysis. J. Affect. Disord 200, 119–132. 10.1016/jjad.2016.04.040 [DOI] [PubMed] [Google Scholar]
- Kohn JN, Cabrera Y, Dimitrov S, Guay-Ross N, Pruitt C, Shaikh FD, Hong S, 2019. Sex-specific roles of cellular inflammation and cardiometabolism in obesity-associated depressive symptomatology. Int. J. Obes 10.1038/s41366-019-0375-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koni AC, Scott RA, Wang G, Bailey MES, Peplies J, Bammann K, Pitsiladis YP, 2011. DNA yield and quality of saliva samples and suitability for large-scale epidemiological studies in children. Int. J. Obes 35, S113–S118. 10.1038/ijo.2011.43 [DOI] [PubMed] [Google Scholar]
- La Fratta I, Tatangelo R, Campagna G, Rizzuto A, Franceschelli S, Ferrone A, Patruno A, Speranza L, De Lutiis MA, Felaco M, Grilli A, Pesce M, 2018. The plasmatic and salivary levels of IL-1β, IL-18 and IL-6 are associated to emotional difference during stress in young male. Sci. Rep 8, 1–10. 10.1038/s41598-018-21474-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C, 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol 31, 814–821. 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Del Sorbo L, Khine AA, De Azavedo J, Low DE, Bell D, Uhlig S, Slutsky AS, Zhang H, 2003. Modulation of Bacterial Growth by Tumor Necrosis Factor-α In Vitro and In Vivo. Am. J. Respir. Crit. Care Med 168, 1462–1470. 10.1164/rccm.200302-303OC [DOI] [PubMed] [Google Scholar]
- Lenth RV, 2016. Least-Squares Means: The R Package lsmeans. J. Stat. Softw. 69 10.18637/jss.v069.i01 [DOI] [Google Scholar]
- Levitzky YS, Guo C-Y, Rong J, Larson MG, Walter RE, Keaney JF, Sutherland PA, Vasan A, Lipinska I, Evans JC, Benjamin EJ, 2008. Relation of smoking status to a panel of inflammatory markers: The Framingham offspring. Atherosclerosis 201, 217–224. 10.1016/j.atherosclerosis.2007.12.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Wu X, Jin F, 2018. Gut-Brain Psychology: Rethinking Psychology From the Microbiota-Gut-Brain Axis. Front. Integr. Neurosci 12, 1–24. 10.3389/fnint.2018.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y, Totsika M, Morrison M, Punyadeera C, 2017. The saliva microbiome profiles are minimally affected by collection method or DNA extraction protocols. Sci. Rep 7, 1–10. 10.1038/s41598-017-07885-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira-Junior R, Boström EA, 2018. Oral-gut connection: One step closer to an integrated view of the gastrointestinal tract? Mucosal Immunol. 11, 316–318. 10.1038/mi.2017.116 [DOI] [PubMed] [Google Scholar]
- Lourenςo TGB, Spencer SJ, Alm EJ, Colombo APV, 2018. Defining the gut microbiota in individuals with periodontal diseases: an exploratory study. J. Oral Microbiol 10. 10.1080/20002297.2018.1487741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke SG, 2017. Evaluating significance in linear mixed-effects models in R. Behav. Res. Methods 49, 1494–1502. 10.3758/s13428-016-0809-y [DOI] [PubMed] [Google Scholar]
- Lundmark A, Hu YOO, Huss M, Johannsen G, Andersson AF, Yucel-Lindberg T, 2019. Identification of salivary microbiota and its association with host inflammatory mediators in periodontitis. Front. Cell. Infect. Microbiol 9, 1–13. 10.3389/fcimb.2019.00216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyte M, Vulchanova L, Brown DR, 2011. Stress at the intestinal surface: Catecholamines and mucosa-bacteria interactions. Cell Tissue Res. 343, 23–32. 10.1007/s00441-010-1050-0 [DOI] [PubMed] [Google Scholar]
- Mahdavi J, Royer P-J, Sjolinder HS, Azimi S, Self T, Stoof J, Wheldon LM, Brannstrom K, Wilson R, Moreton J, Moir JWB, Sihlbom C, Boren T, Jonsson A-B, Soultanas P, Ala’Aldeen DAA, 2013. Pro-inflammatory cytokines can act as intracellular modulators of commensal bacterial virulence. Open Biol. 3, 130048–130048. 10.1098/rsob.130048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantere O, Orešič M, Saarela M, Suvisaari J, Maukonen J, Hyytiäinen T, Yolken R, Sabunciyan S, Kieseppä T, Schwarz E, 2017. Analysis of microbiota in first episode psychosis identifies preliminary associations with symptom severity and treatment response. Schizophr. Res 192, 398–403. 10.1016/j.schres.2017.04.017 [DOI] [PubMed] [Google Scholar]
- Marotz C, Amir A, Humphrey G, Gaffney J, Gogul G, Knight R, 2017. DNA extraction for streamlined metagenomics of diverse environmental samples. Biotechniques 62, 290–293. 10.2144/000114559 [DOI] [PubMed] [Google Scholar]
- Marsland AL, Walsh C, Lockwood K, John-Henderson NA, 2017. The effects of acute psychological stress on circulating and stimulated inflammatory markers: A systematic review and meta-analysis. Brain. Behav. Immun 64, 208–219. 10.1016/j.bbi.2017.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNair DM, Lorr M, Droppleman LF, 1971. Manual for the POMS. Educ. Ind. Test. Serv [Google Scholar]
- Miller GE, Chen E, Zhou ES, 2007. If it goes up, must it come down? Chronic stress and the hypothalamic-pituitary-Adrenocortical axis in humans. Psychol. Bull 133, 25–45. 10.1037/0033-2909.133.1.25 [DOI] [PubMed] [Google Scholar]
- Morris DJ, Brem AS, 2019. Role of gut metabolism of adrenal corticosteroids and hypertension: Clues gut-cleansing antibiotics give us. Physiol. Genomics 51, 83–89. 10.1152/physiolgenomics.00115.2018 [DOI] [PubMed] [Google Scholar]
- Morton JT, Marotz C, Washburne A, Silverman J, Zaramela LS, Edlund A, Zengler K, Knight R, 2019. Establishing microbial composition measurement standards with reference frames. Nat. Commun 10. 10.1038/s41467-019-10656-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nater UM, Rohleder N, Schlotz W, Ehlert U, Kirschbaum C, 2007. Determinants of the diurnal course of salivary alpha-amylase. Psychoneuroendocrinology 32, 392–401. 10.1016/j.psyneuen.2007.02.007 [DOI] [PubMed] [Google Scholar]
- Nobs SP, Tuganbaev T, Elinav E, 2019. Microbiome diurnal rhythmicity and its impact on host physiology and disease risk. EMBO Rep. e47129, e47129. 10.15252/embr.201847129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, 2017. Vegan: ecological diversity. R Packag. Version 2.5–4. 10.1029/2006JF000545 [DOI] [Google Scholar]
- Pereira PAB, Aho VTE, Paulin L, Pekkonen E, Auvinen P, Scheperjans F, 2017. Oral and nasal microbiota in Parkinson’s disease. Park. Relat. Disord 38, 61–67. 10.1016/j.parkreldis.2017.02.026 [DOI] [PubMed] [Google Scholar]
- Poole AC, Goodrich JK, Youngblut ND, Luque GG, Ruaud A, Sutter JL, Waters JL, Shi Q, El-Hadidi M, Johnson LM, Bar HY, Huson DH, Booth JG, Ley RE, 2019. Human Salivary Amylase Gene Copy Number Impacts Oral and Gut Microbiomes. Cell Host Microbe 25, 553–564.e7. 10.1016/j.chom.2019.03.001 [DOI] [PubMed] [Google Scholar]
- Proctor Di.M., Fukuyama JA, Loomer PM, Armitage GC, Lee SA, Davis NM, Ryder MI, Holmes SP, Relman DA, 2018. A spatial gradient of bacterial diversity in the human oral cavity shaped by salivary flow. Nat. Commun 9. 10.1038/s41467-018-02900-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt ÉL, Fernandes PACM, Markus RP, Fischer FM, 2019. Night work effects on salivary cytokines TNF, IL-1β and IL-6. Chronobiol. Int 36, 11–26. 10.1080/07420528.2018.1515771 [DOI] [PubMed] [Google Scholar]
- Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Gabriel A, Ghalith A, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Benjamin J, Mauricio A, Rodríguez C, Chase J, Cope EK, Da Silva R, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibson DL, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Benjamin D, Kang K.Bin Keefe CR, Keim P, Kelley ST, Knights D, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, Metcalf JL, Morgan SC, Jamie T, Nothias LF, Orchanian SB, Segata N, Willis AD, 2018. QIIME 2 : Reproducible, interactive, scalable, and extensible microbiome data science. PeerJ. 10.7287/peerj.preprints.27295v2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohleder N, 2014. Stimulation of systemic low-grade inflammation by psychosocial stress. Psychosom. Med 76, 181–189. 10.1097/PSY.0000000000000049 [DOI] [PubMed] [Google Scholar]
- Said HS, Suda W, Nakagome S, Chinen H, Oshima K, Kim S, Kimura R, Iraha A, Ishida H, Fujita J, Mano S, Morita H, Dohi T, Oota H, Hattori M, 2014. Dysbiosis of Salivary Microbiota in Inflammatory Bowel Disease and Its Association With Oral Immunological Biomarkers. DNA Res. 21, 15–25. 10.1093/dnares/dst037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, Jansen T, Jacobs L, Bonder MJ, Kurilshikov A, Fu J, Joosten LAB, Zhernakova A, Huttenhower C, Wijmenga C, Netea MG, Xavier RJ, 2016. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell 167, 1125–1136.e8. 10.1016/j.cell.2016.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K, Cowen PJ, Harmer CJ, Tzortzis G, Errington S, Burnet PWJ, 2015. Prebiotic intake reduces the waking cortisol response and alters emotional bias in healthy volunteers. Psychopharmacology (Berl). 232, 1793–1801. 10.1007/s00213-014-3810-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R, Fuchs S, Milo R, 2016. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLOS Biol. 14, e1002533. 10.1371/journal.pbio.1002533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavich GM, Irwin MR, 2014. From stress to inflammation and major depressive disorder: a social signal transduction theory of depression. Psychol. Bull 140, 774–815. 10.1037/a0035302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielberger CD, 1983. Manual for State-Trait Anxiety Inventory. Palo Alto, CA Consult. Psychol. Press. 10.1016/jjbi.2014.03.016 [DOI] [Google Scholar]
- Takayasu L, Suda W, Takanashi K, Iioka E, Kurokawa R, Shindo C, Hattori Y, Yamashita N, Nishijima S, Oshima K, Hattori M, 2017. Circadian oscillations of microbial and functional composition in the human salivary microbiome. DNA Res. 24, 261–270. 10.1093/dnares/dsx001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita T, Kageyama S, Furuta M, Tsuboi H, Takeuchi K, Shibata Y, Shimazaki Y, Akifusa S, Ninomiya T, Kiyohara Y, Yamashita Y, 2016. Bacterial diversity in saliva and oral health-related conditions: The Hisayama Study. Sci. Rep 6, 1–11. 10.1038/srep22164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam J, Hoffmann T, Fischer S, Bornstein S, Gräßler J, Noack B, 2018. Obesity alters composition and diversity of the oral microbiota in patients with type 2 diabetes mellitus independently of glycemic control. PLoS One 13, e0204724. 10.1371/journal.pone.0204724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, Prill RJ, Tripathi A, Gibbons SM, Ackermann G, Navas-Molina JA, Janssen S, Kopylova E, Vázquez-Baeza Y, González A, Morton JT, Mirarab S, Zech Xu Z, Jiang L, Haroon MF, Kanbar J, Zhu Q, Jin Song S, Kosciolek T, Bokulich NA, Lefler J, Brislawn CJ, Humphrey G, Owens SM, Hampton-Marcell J, Berg-Lyons D, McKenzie V, Fierer N, Fuhrman JA, Clauset A, Stevens RL, Shade A, Pollard KS, Goodwin KD, Jansson JK, Gilbert JA, Knight R, 2017. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463. 10.1038/nature24621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R, 2013. EMPeror: A tool for visualizing high-throughput microbial community data. Gigascience 2, 2–5. 10.1186/2047-217X-2-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, Gilbert JA, Jansson JK, Caporaso JG, Fuhrman JA, Apprill A, Knight R, Improved Bacterial 16S rRNA Gene (V4 and V4–5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems 1, 1–10. 10.1128/mSystems.00009-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZZ, Amir A, Sanders J, Zhu Q, Morton JT, Bletz MC, Tripathi A, Huang S, McDonald D, Jiang L, Knight R, 2019. Calour: an Interactive, Microbe-Centric Analysis Tool. mSystems 4, 1–12. 10.1128/mSystems.00269-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yolken RH, Severance EG, Sabunciyan S, Gressitt KL, Chen O, Stallings C, Origoni A, Katsafanas E, Schweinfurth LAB, Savage CLG, Banis M, Khushalani S, Dickerson FB, 2015. Metagenomic Sequencing Indicates That the Oropharyngeal Phageome of Individuals With Schizophrenia Differs From That of Controls. Schizophr. Bull 41, 1153–1161. 10.1093/schbul/sbu197 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.