Abstract
Personalized medicine tailors therapies, disease prevention, and health maintenance to the individual, with pharmacogenomics serving as a key tool to improve outcomes and prevent adverse effects. Advances in genomics have transformed pharmacogenetics, traditionally focused on single gene-drug pairs, into pharmacogenomics, encompassing all “-omics” fields (e.g., proteomics, transcriptomics, metabolomics, and metagenomics). This review summarizes basic genomics principles relevant to translation into therapies, assessing pharmacogenomics’ central role in converging diverse elements of personalized medicine. We discuss genetic variations in pharmacogenes (drug-metabolizing enzymes, drug transporters, and receptors), their clinical relevance as biomarkers, and the legacy of decades of research in pharmacogenetics. All types of therapies, including proteins, nucleic acids, viruses, cells, genes, and irradiation, can benefit from genomics, expanding the role of pharmacogenomics across medicine. Food and Drug Administration approvals of personalized therapeutics involving biomarkers increase rapidly, demonstrating the growing impact of pharmacogenomics. A beacon for all therapeutic approaches, molecularly targeted cancer therapies highlight trends in drug discovery and clinical applications. To account for human complexity, multicomponent biomarker panels encompassing genetic, personal, and environmental factors can guide diagnosis and therapies, increasingly involving artificial intelligence to cope with extreme data complexities. However, clinical application encounters substantial hurdles, such as unknown validity across ethnic groups, underlying bias in health care, and real-world validation. This review address the underlying science and technologies germane to pharmacogenomics and personalized medicine, integrated with economic, ethical, and regulatory issues, providing insights into the current status and future direction of health care.
Significance Statement
Personalized medicine aims to optimize health care for the individual patients with use of predictive biomarkers to improve outcomes and prevent adverse effects. Pharmacogenomics drives biomarker discovery and guides the development of targeted therapeutics. This review addresses basic principles and current trends in pharmacogenomics, with large-scale data repositories accelerating medical advances. The impact of pharmacogenomics is discussed, along with hurdles impeding broad clinical implementation, in the context of clinical care, ethics, economics, and regulatory affairs.
I. Introduction
A. Personalized Medicine
Successful treatment and prevention of disease is guided by personal characteristics, including age, sex, disease status, body weight, socioeconomic status, and common diagnostic parameters such as cholesterol and glucose levels. Recent scientific and technological advances, including genomics, have drastically enhanced our ability to optimize therapies tailored to well-defined patient subgroups or even uniquely to individual patients, resulting in a new understanding of personalized medicine. Addressing confusion about the definition of personalized medicine, Simmons et al. (2012) state on p.85: “personalized healthcare is an approach to care that utilizes personalized medicine tools to deliver patient-centered, predictive care within the context of coordinated service delivery.” The terms “personalized medicine” and “personalized health care” are often used interchangeably; however, “health care” applies more broadly to include maintenance of optimal health, focusing on resilience rather than disease risk factors, requiring conceptually distinct approaches.
Genomic medicine has become an important aspect of personalized medicine, with some promoting the term “precision medicine” targeting uniquely an individual patient. Yet translating genomics into clinical practice has met substantial hurdles, due to complex relationships between genetic factors and phenotypes and obstacles to practical implementation. Today, genomics encompasses many -omics areas including proteomics, metabolomics, and more, each yielding biomarkers useful as therapeutic guides. Applied to drug therapies, pharmacogenomics has emerged as a driver of personalized medicine with numerous personalized therapies moving into clinical use; yet similar hurdles need to be overcome. This review broadly addresses the promise and limitations of pharmacogenomics.
B. Brief History of Pharmacogenetics
In 1909, Archibald Garrod proposed that inborn errors in metabolism provide a genetic basis of some diseases and predicted that “every active drug is a poison, when taken in large enough doses, and in some subjects, a dose which is innocuous to the majority of people has toxic effects, whereas others show exceptional tolerance to the drug.” Demonstration of genetic factors in response to xenobiotics came from the observation by Arthur L. Fox in 1932 that some individuals are unable to sense the bitter taste of phenylthiocarbamide, present in broccoli, and identified it as an inherited trait, subsequently associated with polymorphisms in the bitter-taste receptor TAS2R38, implicated in food acceptance (Keller and Adise, 2016). Friedrich Vogel coined the term “pharmacogenetics” in 1959, after discoveries of genetic defects in N-acetyl transferase (which metabolizes niacin), glucose-6-phosphate dehydrogenase deficiency (primaquine hemolysis), and plasma cholinesterase deficiency (prolonged apnea caused by succinylcholine), followed by CYP2D6 deficiency causing severe adverse effects to debrisoquin and sparteine (reviewed in Ma and Lu, 2011; Mueller et al., 2019). This series of discoveries triggered an intensive search for genetic factors in drug response, cloning the responsible genes, and identifying the causative genetic variants. Subsequent studies discovered numerous drug metabolizing enzymes and drug transporters, followed by molecular cloning of the genes and uncovering the mutations affecting the encoded protein’s functions, with potential clinical relevance. For example, a CYP2D6 mutation was found to abrogate debrisoquine metabolism, leading to severe adverse effects (Skoda et al., 1988). Expanding knowledge of underlying genetic causes led to genetic bio-markers predicting drug exposure and response, the goal of pharmacogenetics. However, as we discuss, numerous factors determine drug exposure and response; therefore, any such biomarker needs to be embedded into a broad decision framework to optimize personal therapeutics.
Drug levels in the blood can serve as an intermediate phenotype that integrates genetic factors with all other factors, including enzyme induction and inhibition, renal functions, and more. Already in use for decades, therapeutic drug monitoring as a measure of drug exposure requires pharmacokinetic modeling often with sparse temporal data (Derendorf et al., 2000). Novel drug-level assays and data analytics render drug-level monitoring still relevant today, as valuable biomarkers in several areas including cancer (Knezevic and Clarke, 2020), inflammatory diseases (Shmais et al., 2021), and depression (Papamichael and Cheifetz, 2019).
A similar historical path led to the discovery of receptors mediating drug response. Among these, a large group of genes encoding G protein-coupled receptors, remains the dominant target of currently used drugs (Hauser et al., 2017). Drug effects are assessed with pharmacodynamics, designed to optimize drug response across diverse groups of patients (Derendorf et al., 2000). However, the expectation that genetic variants in drug-receptor genes would be equally applicable to optimizing therapy is confounded by the multifactorial nature of complex biologic response pathways. Complexity is enhanced by receptor proteins aggregating into large complexes with multiple functions, for example, heterocomplex formation between dopamine and N-methyl-D-aspartate receptors (Tevzadze et al., 2022). Nevertheless, clinically useful biomarkers have been developed, for example genetic variants in β1-adrenergic receptors in therapy of heart failure (Thomas and Johnson, 2020). The genetics of G protein-coupled receptors remains a promising area of research and clinical utility. Pharmacogenomics of cancer has evolved rapidly, with both germline mutations and tumor-driving somatic mutations guiding targeted therapies, a signature topic discussed separately further later.
Pharmacogenomic biomarkers applied to improve therapeutic efficacy and avoid adverse drug reactions (ADRs) are listed in Table 1. While drug efficacy is often polygenic, serious ADRs tend to be less frequent and more likely to arise from specific genetic factors. ADRs are considered a major cause of hospitalizations and death, defined by the World Health Organization to arise from standard accepted therapy, excluding overdoses and errors. To reduce the incidence of ADRs, countless studies continue in search of causative genetic variants that affect susceptibility to ADRs (Phillips et al., 2001; Cacabelos et al., 2019), defined as type A reactions (for example to warfarin and thiopurines) and type B reactions, the latter involving immune reactions of skin, liver, and heart (idiosyncratic drug reactions) (Osanlou et al., 2018). Overlaying the literature of ADRs with known pharmacogenetic variants provided early evidence that genetics play a significant role in ADRs, and their prospective use could help avoid ADRs (Phillips et al., 2001). This approach revealed variants in genes encoding drug-metabolizing enzymes displaying significant associations with ADRs of type A, their level of activity being directly related to drug levels in the body (Table 1).
TABLE 1.
Pharmacogenetic biomarkers with guidelines for clinical use from at least one of the pharmacogenomics societies in different countries (CPIC, DPWG, RNPGx, CPNDS, SEFF/SEOM, AusNZ)
Data are from Pharmacogenomics Knowledge Base (https://www.pharmgkb.org/guidelineAnnotations). Only common variants are listed.
| Gene | Alleles tested | Molecular Mechanisms | Related Medications |
|---|---|---|---|
| ABCG2 | rs2231142 (c.421 G>T) | Missense | Allopurinol, rosuvastatin |
| CACNA1S | rs772226819 (C520C>T), rs1800559 (c.3257G>A) | Missense | Enflurane, desflurane, isoflurane, methoxyflurane, sevoflurane, succinylcholine, halothane |
| CFTR | 38 rare variants | Missense | Ivacaftor |
| CYP2B6 | rs3745274 and rs2279343 containing alleles, rare variants | Disrupt intronic splicing site (rs2279343), missense (rs3745274) | Efavirenz |
| CYP2C19 | rs12769205(*2), rs4986893(*3), rs28399504 (*4), rs12248560(*17), rare variants | Splicing defect (*2), create stop codon (*3), delete start codon (*4), promoter transcription (*17) | Amitriptyline, citalopram, clomipramine, clopidogrel, dexlansoprazole, doxepin, escitalopram, imipramine, lansoprazole, omeprazole, pantoprazole, sertraline, trimipramine, voriconazole |
| CYP2C9 | rs1799853 (*2), rs1057910 (*3), rs28371686 (*5), rs7900194 (*8), rs28371685 (*11), rare variants | Missense | Acenocoumarol, celecoxib, fluindione, flurbiprofen, Fluvastatin, fosphenytoin, ibuprofen, lornoxicam, meloxicam, phenytoin, piroxicam, siponimod, tenoxicam, warfarin |
| CYP2D6 | rs35742686 (*3), rs3892097 (*4), rs5030655 (*6), rs5030656 (*9), rs1065852 (*10), rs28371706 (*17), rs59421388 (*29), rs28371725 (*41), structure and copy number variants | Indels (*3, *6), splicing defect (*4), missense (*9, *10, *17, *29), gene expression (*41) | Amitriptyline, aripiprazole, atomoxetine, brexpiprazole, clomipramine, codeine, desipramine, doxepin, eliglustat, flecainide, fluvoxamine, haloperidol, hydrocodone, imipramine, metopolol, nortriptyline, ondansetron, paroxetine, pimozide, propafenone, risperidone, tamoxifen, tramadol, trimipramine, venlafaxine, zuclopenthixol |
| CYP3A4 | rs35599367 (*22) | Aberrant splicing (*22) | Atorvastatin, quetiapine, many drugs |
| CYP3A5 | rs776746 (*3), rs10264272 (*6), rs41303343 (*7) | Aberrant splicing (*3), splicing defect (6), indel (*7) | Tacrolimus |
| CYP4F2 | rs2108622 | Missense | Warfarin |
| DPYD | 90 rare variants | Splicing defect (*2A), indels (*3, *7), missense | Capecitabine, flucytosine, fluorouracil, tegafur |
| F5 | rs6025 | Missense | Hormonal contraceptives for systemic use |
| G6PD | >150 rare variants | Missense, indels | Rasburicase |
| HLA-A | HLA-A*31.01 | Missense | Carbamazepine |
| HLA-B | HLA-B*15.01 | Missense | Abacavir, flu-+cloxacillin |
| HLA-B | HLA-B*15.02 | Missense | Carbamazepine, fosphenytoin, lamotrigine, oxcarbazepine, phenytoin |
| HLA-B | HLAB*57-01 | Missense | Abacavir, flucloxacillin |
| HLA-B | HLAB*58-01 | Missense | Allopurinol |
| IFNL3 | rs12979860, rs8099917 | Noncoding, regulatory | Peginterferon alfa-2a, peginterferon alfa-2b, ribavirin |
| MT-RNR1 | m.1095T>C, m.1494C>T, m.155A>G | Affecting mitochondrial 12S rRNA function | Amikacin, gentamicin, kanamycin, paromomycin, plazomicin, streptomycin, tobramycin |
| NUDT15 | rs746071566(*2, *6), rs116855232(*3) rs186364861(*5) | Missense (*3, *5), indels (*2, *6) | Azathioprine, mercaptopurine, thioguanine |
| RARG | rs2229774 | Missense | Daunorubicin, doxorubicin |
| RYR1 | 50 rare variants | Missense | Desflurane, enflurane, halothane, isoflurane, methoxyflurane |
| SLC28A3 | rs7853758 | Missense | Daunorubicin, doxorubicin |
| SLCO1B1 | rs4149056 and rs59502379 containing alleles | Missense | Atorvastatin, fluvastatin, lovastatin, pitavastatin, pravastatin, rosuvastatin, simvastatin |
| TPMT | rs1800462(*2), rs1142345(*3), rs1800584(*4), rs75543815(*6), rs72552736(*7), rs56161402(*8) | Missense (most), splicing defect (*4) | Azathioprine, cisplatin, mercaptopurine, thioguanine |
| UGT1A1 | rs3064744 (*28, *36, *37), rs4148323(*6), rs887829(*80) | Promoter variants, gene expression (*28, *36, *37, *80), missense (*6) | Atazanavir, itinotecan |
| UGT1A6 | rs17863783 (*4) | Missense | Daunorubicin, doxorubicin |
| VKORC1 | rs9923231 | Promoter variant, gene expression | Acenocoumarol, fluindione, warfarin |
DPWG, Royal Dutch Association for the Advancement of Pharmacy-Pharmacogenetics working Group; CPNDS, Canadian Pharmacogenomics Network for Drug Safety; indel, insertions/deletions; RNPGx, French National Network of Pharmacogenetics; SEFF/SEOM, Spanish Pharmacogenetics and Pharmacogenomics Society/Spanish Society of Medical Oncology; AusNZ, Australian and New Zealand consensus guideline.
Over time, numerous highly penetrant pathogenic variants have been discovered in different pharmacogene classes. Drug-induced QT prolongation increases the risk of torsade de pointes, with potentially lethal ventricular arrhythmias (Roden, 2016), with many drug classes involved, including antibiotics, cardiovascular drugs, antipsychotics, and anticancer drugs. All new drugs are tested for QT prolongation before Food and Drug Administration (FDA) approval is attained. However, these ADRs can be rare when associated with rare mutations in cardiac ion channels, a main cause of removal from clinical use after FDA approval. Several genetic variants have been identified to enhance the risk of drug-induced long QT, including KCNE1-D85N and KCNE2-T8A (Lopez-Medina et al., 2022).
Among the type B ADRs, human leukocyte antigens (HLAs) have shown highly significant associations with severe ADRs (see Table 1 for examples), such as drug-induced skin injury and drug-induced liver injury (Yip et al., 2015). For example, carriers of the HLA-B*15:02 are highly susceptible to carbamazepine hypersensitivity reactions leading to Stevens-Johnson syndrome. As such ADRs are relatively rare, depending on ethnicity, prospective genotyping is not universally performed or only in select ethnic groups with increased allele frequency (HLA-B*15:02 in Asians), even though clinical outcomes can be severe.
Pharmacogenetics historically focuses on single drug–single gene interactions that can affect therapeutic decisions, with few examples of two or more gene interactions (Dalle Fratte et al., 2023) rapidly increasing complexity of biomarker assays. Clearly, human physiology and response to therapies involves countless factors beyond genetics. Integration of these elements, each of high complexity in itself, has emerged as a compelling trend, in part driven by advances in genomics, reflecting the entire genome rather than individual genes.
C. Survey of Drug-Metabolizing Enzymes and Transporters
Pharmacogenetics has traditionally focused on genes encoding proteins directly interacting with small molecular weight drugs, specifically metabolizing enzymes, transporters, and receptors, already extensively reviewed (Weinshilboum and Wang, 2017; Roden et al., 2019; Cecchin and Stocco, 2020). Therefore, this review includes only a few concepts relevant to understanding the potential impact of genetic variants on drug absorption, distribution, metabolism, and excretion. Physicochemical properties such as polarity and molecular weight determine whether a drug can readily penetrate cell membranes composed of lipid bilayers that represent robust barriers to polar compound such as amino acids, nucleosides, sugars, and quaternary ammonium ions. To bring essential nutrients into the cell, several hundred genes encode transmembrane transporters that recognize nutrients and ions, and molecules including drugs with similar structure, by facilitating diffusion across the membrane or by an active mechanism via cotransport of ions such as Na+ (Jetter and Kullak-Ublick, 2020). On the other hand, primary active transporters driven by ATP typically act as extrusion pumps, for example ABCB1, also known as multidrug resistance protein 1 (MDR1). Absent any substantive interaction with transporters, polar drugs do not distribute extensively into tissues and are largely excreted into the urine, as shown for digoxin, which is slowly metabolized and relies mostly on renal function for elimination from the body. As such, renal function can guide drug dosing. On the other hand, lipophilic drugs distribute extensively into tissues, are only minimally excreted into the urine unchanged, and depend on metabolism for functional elimination.
Numerous enzymes contribute to the metabolism of xenobiotics by oxidation, reduction, hydrolysis, and more. In this “phase I metabolism” of lipophilic drugs, functional groups such as -OH are introduced that enable conjugation with polar substituents such as glucuronide, sulfate, phosphate, and amino acids. This “phase II metabolism” enables excretion into the urine or feces via biliary processing. Most prominent among phase I enzymes, cytochrome P450 oxygenases (CYPs) are encoded by the large CYP family of genes and metabolize a majority of clinically used drugs, with the following CYPs listed in order of the number of clinically used drug substrates: 3A4(5) > 2D6 > 2C9 > 1A2 > 2C19 = 2B6 > 2C8 > 2A6 = 2E1 = 2J2 (Zhao et al., 2021). Many drugs are inactivated by metabolism, but both phase I and II metabolism can also lead to active metabolites, activate an administered inactive drug (e.g., codeine to morphine), or lead to toxic metabolites or intermediates, such as aryl epoxides. To assess the potential impact of genetic variants in enhancing or reducing enzyme activity, one needs to consider whether the metabolic steps are drug activating or inactivating. Therefore, a detailed assessment of the activity of the parent drug and all its metabolites is essential. For example, the anticancer drug irinotecan, inactive per se, is activated by an esterase to the active SN38, which subsequently is inactivated by the glucuronidation enzyme UGT1A1, encoded by a gene carrying a frequent promoter repeat that reduces activity and can lead to unwanted drug toxicity (alleles UGT1A1*6 and *28) (Campbell et al., 2017). Multiple factors determine whether UGT1A1 genetic biomarkers can improve treatment outcomes with irinotecan, including frequency and penetrance of the variants, additional metabolic pathways that can bypass UGT1A1 dependency, therapeutic means to reduce the severity of ADRs, and issues involving access to testing and cost (Campbell et al., 2017). These factors must be addressed with each pharmacogenetic biomarker test.
Multiple elements, in addition to genetic variants, can contribute to variable drug metabolizing activity. Diverse substances and conditions can cause CYP450 induction or inhibition, including exposure to cigarette smoke, pathophysiological conditions, age, and sex. Drug-induced enzyme inhibition leads to ‘phenoconversion,” resulting in a mismatch between genetically predicted and observed enzyme activity and drug-drug-gene-gene interactions with potentially severe clinical impact (reviewed by Klomp et al., 2020). For each enzyme, the relative impact of genetic and other factors needs to be assessed in interpreting biomarker results.
D. Transition to Pharmacogenomics
Sequencing the human genome in 2001 has led to rapid advances in genomic sciences, focusing our attention not just on single genes but on the ensemble of all genes and genomic functions. This broader view has led to the term “pharmacogenomics,” defined by the National Human Genome Research Institute as “Pharmacogenomics is a branch of pharmacology concerned with using DNA and amino acid sequence data to inform drug development and testing.” Multiple reviews cover pharmacogenomics but often still embrace mainly pharmacogenetics concepts, reflecting the influence of one or only a few genes on drug response (Weinshilboum and Wang, 2017; Roden et al., 2019; Patrinos and Shuldiner, 2022). Reviews also have addressed the pharmacogenomics of select complex diseases, including cardiovascular disorders (Duarte and Cavallari, 2021), diabetes (Venkatachalapathy et al., 2021), psychiatric diseases (Hoehe and Morris-Rosendahl, 2018), and cancer (Morganti et al., 2019). Targeted cancer therapies mainly rely on somatic mutations in oncogenes, but when the therapeutic window is narrow, germline mutations in drug-metabolizing enzymes and transporters also play a key role in drug response (Miteva-Marcheva et al., 2020). Next-generation sequencing (NGS) and other massively parallel methods have opened the door to interrogate the entire genome, protein coding exome, transcriptome [RNA sequencing (seq), single-cell RNA-seq], methylome, and epigenome (ChIP-seq, ATAC-seq) to predict disease risk and therapy response (Rabbani et al., 2016). Yet such large-scale data generate a complex landscape that poses hurdles in defining robust genome-phenome relationships and obstacles to clinical implementation (Ji et al., 2018). The potential of genomics in clinical applications is often highlighted, but few examples go beyond the influence of one or only a few genes on treatments. In this review, we analyze challenges and advances that can overcome hurdles in clinical implementation.
New treatment modalities beyond traditional drugs further expand the field of pharmacogenomics (Brown and Wobst, 2021). Novel therapies include RNA and DNA molecules, gene therapy, gene editing, proteins, live cells, tissues, bacteria, and viruses, supplemented with radiation therapy, diet, and exercise. Treatment strategies then are adjusted to personal characteristics (e.g., age, sex, ethnicity, health status, socioeconomic status, culture, and more), illustrating the highly complex therapeutic landscape. RNA pharmaceuticals (messenger RNAs, antisense RNAs, RNAs addressing splicing defects, microRNAs) promise effective means for vaccination and therapies for infections, inherited disorders, cancer, and cardiovascular disease, made feasible by genomics insights with biomarkers (Servick, 2020). In addition, the National Institutes of Health (NIH) promotes “precision nutrition”—including the microbiome as a target—as an alternative or complement to medical therapies (Kaiser, 2021). The role of microbiota in cancer and immune response has become evident (Mager et al., 2020; Woelk and Snyder, 2021). Combined, these features coalesce in the field of personalized medicine, requiring the integration of diverse datasets to arrive at optimal personalized therapies and prevention strategies.
II. Principles of Genetics and Genomics
A. Complexity of Genes and the Genome—Integration of Diverse Large Datasets
Pharmacogenetics first focused on genetic variants that change the protein sequence, for example, nonsynonymous single nucleotide polymorphism (nsSNPs), generating predictive biomarker panels of drug response. nsSNPs are changes in the DNA coding sequence readily detectable by whole exome sequencing (Silgado-Guzman et al., 2022). However, genome-wide association studies (GWAS) have revealed that most variants associated with clinical phenotypes reside in untranscribed domains, in noncoding RNAs and noncoding regions of protein genes, or are synonymous single nucleotide polymorphism (SNPs) in coding regions that do not alter the protein sequence (e.g., rs3435 in ABCB1, encoding MDR1, destabilizing the RNA) (Wang et al., 2005). Any variant in a transcribed region can affect RNA functions, such as processing, splicing, stability, intracellular sequestration, and translation (Sadee et al., 2011; Mascarenhas et al., 2015). These variants are presumed to have regulatory functions affecting the expression of the encoded proteins or alter the folding and function of noncoding RNA transcripts (Collins et al., 2022). Tens of thousands of long noncoding RNAs (lncRNAs) have been identified [e.g., lncRNA networks affecting CYP3A expression (Huang et al., 2022)], but their functions remain mostly unknown. RNA processing further gives rise to short RNAs, for example, the conversion of long microRNA precursors to short microRNAs that regulate the expression of numerous pharmacogenes, including CYP enzymes (Manikandan and Nagini, 2018). Some microRNA precursors can have functions per se (Morelli et al., 2023), illustrating functional complexity. Also, several thousand open reading frames previously thought to be noncoding are now suspected of encoding short proteins mostly with as yet poorly defined functions (Schlesinger and Elsasser, 2022), implying we are still far from a comprehensive understanding of the human genome. Issues with GWAS data arising with the analysis of highly polygenic traits, such as body weight, across ethnic populations include varying allele frequencies and linkage disequilibrium (LD), discussed by (Hui et al., 2022). GWAS combined with massively parallel reporter assays has served to identify countless noncoding regulatory variants and derived gene networks, with the potential to account for Alzheimer’s disease as an example (Cooper et al., 2022). Generating functional networks strengthens associations of multiple individual regulatory variants, revealing pathways underlying pathophysiology.
The genomic architecture of single genes also displays high complexity, a steep hurdle for use of clinical biomarkers. First, enhancer and suppressor elements are distributed over large genomic regions—sometimes 0.1 to >1 million base pairs distant from the transcription initiation site, interacting with the proximate promoter via DNA looping [e.g., CYP2D6 (Wang et al., 2014; Ray et al., 2019)]. Second, structurally homologous genes generated by gene duplication often reside in contiguous multigene clusters (e.g., CHNR5, CHNR3, CHNB2) (Lee et al., 2018). The CYP3A multigene locus similarly contains interacting regulatory elements that harbor genetic variants (Collins and Wang, 2020; Collins et al., 2022). Genetic factors are thought to account for a substantial portion of intersubject variability in functional expression, for example, approximately 60% estimated for CYP3A4; however, known variants still account for only a fraction of overall heritability (reviewed by Zhai et al., 2022). Multigene clusters also express noncoding and antisense RNAs as potential regulatory factors (Hartmann et al., 2022). Genetic variation includes not only SNPs and structural variants (large and small deletions and duplications) but also transversions from adjacent pseudogenes (Gallou et al., 2001), leading to complex genome-phenotype relationships.
Genetic variants of the germline reside in and are inherited as genomic regions that can be disrupted by generational crossing over. Any variants in such regions are in some degree of LD; that is, they tend to reside on the same chromosome. As a result, multiple GWAS SNPs are often significantly associated with a given trait, and the degree of significance correlates with the degree of LD with the causative variant (Hartmann et al., 2022). If two null variants (abrogating function of the encoded protein) reside in a given gene locus, it becomes imperative to understand the LD between them and determine the phasing—whether both null variants detected in an individual are on the same or opposite strands, forming functionally distinct haplotypes and diplotypes (combination of two haplotypes in autosomal genes and in females with two X chromosomes). In the former case, one allele can be fully functional, whereas in the latter both alleles are nonfunctional. In the case of CYP2D6 (discussed in more detail later), haplotype structures between variants of different causalities can be deduced statistically from large-scale population sequence data, reflecting distinct LD patterns between ethnic groups, or by long-range single-molecular sequencing (Hernandez et al., 2014; Budd et al., 2016).
To translate genetic variants into valid biomarkers for drug response, one must further consider the complexity of pharmacokinetic and pharmacodynamic networks, involving multiple genes. The pharmacokinetic-pharmacodynamic network of thiopurines, comprising anticancer and immunosuppressant drugs, illustrates the multifactorial nature of drug response (Fig. 1). Each key gene must be studied for relevant functional variants, in particular for the “very important pharmacogene” TPMT, and the subsequently added NUDT15 (Moriyama et al., 2016), each with complex genetics. Any errors in assigning correct LD structures across the entire gene locus reduce clinical biomarker utility and even can cause harm.
Fig. 1.
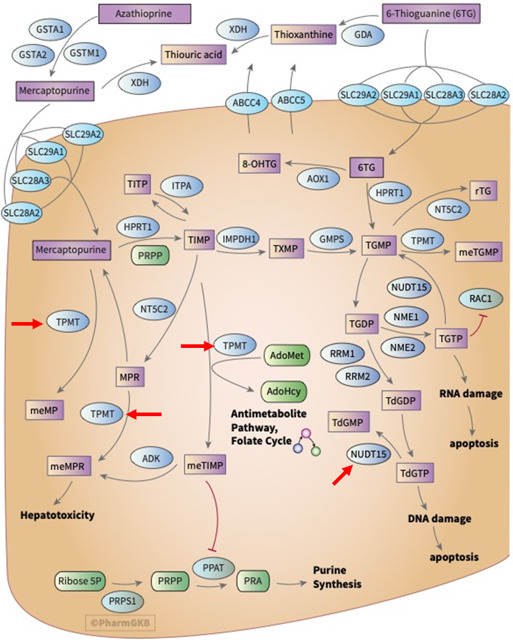
Combined pharmacokinetic-pharmacodynamic network of thiopurines (thioguanine, mercaptopurine, azathioprine) displayed in the PharmGKB (https://www.pharmgkb.org/pathway/PA2040). Marked with red arrows are two key genes labeled as “very important pharmacogene” are TPMT (thiopurine methyl transferase), inactivating thiopurines, and NUDT15 (nucleoside diphosphate linked moiety X-type motif 15), mediating dephosphorylation of thioguanine phosphates. When deficient, both genes can cause severe toxicity. Clinical biomarker testing is recommended but not yet broadly implemented. PharmGKB grants use of its data and contents under the Creative Commons Attribution-ShareAlike 4.0 International License. PharmGKB, Pharmacogenomics Knowledge Base.
Genetic variants and mutations in the X-chromosome result in pronounced differences between males and females. Whereas either X chromosome is epigenetically and randomly inactivated in females, chromatin states can vary between females (Allis and Muir, 2011), up to the point that only one of the two X-chromosomes is largely active throughout all cells (approaching presence of a single X-chromosome in males without a Y chromosome)—a point typically neglected as to clinical relevance in X-linked traits. In addition, some X regions escape silencing in females, for example, the MAOA region with clinically relevant regulatory variants (Pinsonneault et al., 2006), a confounding factor in the interpretation of biomarkers. In general, sex differences arise from multiple factors and need to be included in preclinical and clinical research (Becker et al., 2005).
Variants in the maternally inherited mitochondrial genome can also affect disease risk and response to drugs. For example, gentamicin ototoxicity can occur at normal therapeutic blood levels in carriers of a mutation in the mitochondrial gene MT-RTNR1, leading to clinical guidelines for the use of aminoglycosides (McDermott et al., 2022). An overview of the role of mitochondrial DNA in drug response includes antiviral drugs, anticancer agents, and antimicrobials but cautions that the results thus far are sporadic and more research is required (Jones et al., 2021).
B. Role of Evolution in Shaping Genomic Architecture
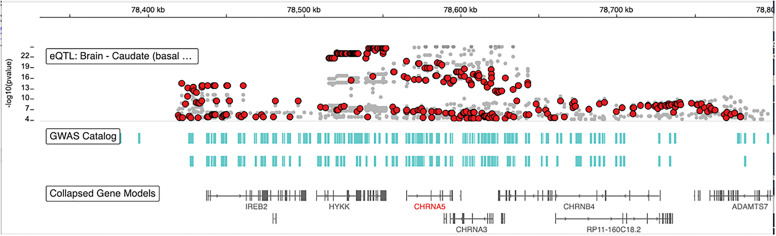
Medical genetics has traditionally emphasized deleterious variants identified as disease risk factors, often with low allele frequency if under negative purifying selection, including mutations implicated in approximately 7,000 monogenic disorders. On the other hand, variants under positive selection can accumulate rapidly and reside in long unbroken LD blocks—a feature of positive selection. Drug target genes are likely candidates for hub genes in networks critical to cell physiology and thus also likely to be under evolutionary selection pressure. In addition, genes encoding drug metabolism enzymes rapidly evolve to protect against external toxins, resulting in varying drug responses across ethnogeographic populations (Ramamoorthy et al., 2022). Studying the evolutionary landscape of CYPP450 enzymes has revealed extensive selection pressures that differ between ethnogeographic groups, often affecting regulatory variants of gene expression (A. Richard-St-Hilaire et al., 2023, preprint, DOI: 10.1101/2023.02.23.529697). To detect regulatory variants under selection pressure, we developed precise assays to measure allelic expression RNA imbalance (AEI, the RNA allele ratio deviates from the genomic DNA ratio), including genome-wide analysis (Smith et al., 2013), a powerful method detecting cis-acting regulatory variants even over long distances in heterozygous carriers. This approach revealed frequent variants regulating transcription, RNA processing and function, and translation in a majority of genes analyzed, including drug-metabolizing enzymes (e.g., CYP3A4, CYP2D6, NAT1), transporters (ABCB1, DAT, SERT) and receptors (e.g., DRD2, 5HT2A, VKORC1) (reviewed in Sadee et al., 2014). Variants associated with RNA transcript and protein abundance are termed expression quantitative trait loci and protein quantitative trait loci, respectively. Many of these regulatory variants reside in long LD blocks suggestive of positive evolutionary selection. Long overlapping LD blocks with hundreds of SNPs reveal a genomic domain regulating the CHRNA5/CHNA3/CHNB4 multigene locus (Fig. 2) (Barrie et al., 2017), including nonlinear epistatic interactions (Lee et al., 2018). SNPs representing each LD block reveal significant associations with smoking behavior; yet these associations alone are insufficient as clinical biomarkers because of the complexity of this polygenic trait. Similar findings apply to other substance use disorders (Peng et al., 2021), presenting barriers to guiding personalized therapies.
Fig. 2.
CHRNA5, CHNA3, CHNB4 gene cluster. This screenshot of the GTEx IGV Browser displays the alignment of the three nicotinic receptors, with GWAS hits (green bars) and RNA eQTLs (red dots: CHRNA5, gray dots: mostly CHRNB4, some for the noncoding RP11-160C18.2). Top CHRNA5 eQTLS are highly significant (p<e-22), and together with GWAS hits line up over 400,000 bps in several very long overlapping LD blocks. The LD blocks each carry one or more functional variants, including the functionally relevant nsSNP rs16969968. The data used for the analyses described in this manuscript were obtained from the GTEx Portal on January 20, 2023 (https://www.gtexportal.org/home/browseEqtls?location=CHRNA5) (GTEx Consortium, 2020). eQTLs, expression quantitative expression loci; GTEx, Genotype-Tissue Expression.
Genomic regions under shared regulatory control—characterized by extensive DNA looping and promoter/enhancer/suppressor interactions—are termed topologically associated domains (Gong et al., 2022), characterized by epigenetic markers with regulatory variants that alter epigenetic marks and transcription of one or more target genes. In pharmacogenomics, topologically associated domains have been assessed for influence on the response to valproic acid therapy in brain injury (Higgins et al., 2017), but clinically valid biomarkers have yet to be developed. Coevolution and coregulation of the gene clusters CYP3A and CYP4F exemplify the concept that regulatory variants exert effect over large genomic regions with several genes and can be revealed with use of evolutionary methods (A. Richard-St-Hilaire et al., 2023, preprint, DOI: 10.1101/2023.02.23.529697).
Positive evolutionary selection often arises initially from reproductive advantages occurring in heterozygous carriers, then continuing to a complete sweep of the variant. If, however, homozygosity is deleterious, such variants will rise to intermediate stable frequency. This process is referred to as balancing selection, documented for the CYP4F gene cluster (A. Richard-St-Hilaire et al., 2023, preprint, DOI: 10.1101/2023.02.23.529697). In addition, new variants counteracting deleterious effects can emerge, for example in CYP2D6 discussed further later. As another example, the hepatic enzyme CYP7A1 catalyzes a main pathway for removal of cholesterol (protect against cardiovascular diseases) to generate deoxycholic acid (risk of diabetes) (Zaborska et al., 2018). A frequent variant in the proximate promoter rs3808607 robustly reduces CYP7A1 transcription but failed to reveal significant association with coronary artery disease or diabetes. Scanning the gene locus revealed a second frequent variant, rs9297794 approximately 10kb downstream of CYP7A1, in an enhancer domain interacting with the promoter via long-range DNA looping (Wang et al., 2018). This variant robustly reduces CYP7A1 transcription but resides mostly in a long LD block in strong LD with the promoter SNP, masking effects on clinical phenotypes. Testing their combined effect in a two-SNP model in electronic clinical databases revealed significant associations with risk of coronary artery disease and diabetes, in opposite directions as expected (Wang et al., 2018). This example demonstrates how GWAS analyses can be buttressed by functional studies to reveal hidden causative relationships that can vary between different ethnic groups (Patel et al., 2022).
C. Molecular Genetics/Genomics of Pharmacogenes
Frequent variants in at least 26 pharmacogenes serve as biomarkers for therapy with more than 90 medications (Table 1). Knowledge of genetic mechanisms, interactions between more than one variant per gene locus, and ethnogeographic differences in frequencies support clinical utility. Numerous additional variants can contribute to the overall genetic effect and are assessed with polygenic SNP panels, mostly using surrogate markers for unknown causative variants (Zhou and Lauschke, 2022). Most current clinically applicable variants are located in transcribed regions (exons and exon-intron boundary), including SNPs, small insertions/deletions, gene copy number variation, and structure variations, affecting the function of encoded proteins through changing the amino acid sequence (e.g., nsSNPs), disrupting splicing, shifting open reading frames, and changing the translation start or stop codons. However, GWAS reveal that nearly 90% of trait- or disease-associated SNPs are noncoding variants (Hindorff et al., 2009), with regulation of gene expression a main mechanism.
Genetic variants in CYP genes can alter metabolism of >60% of clinically used drugs (Ingelman-Sundberg, 2022), cataloged in PharmVar (https://www.pharmvar.org/). Exon sequencing and GWAS have led to discovery of additional rare coding variants (Amin et al., 2012; Holzinger et al., 2012; Krumsiek et al., 2012; Perera et al., 2013; Loukola et al., 2015). In addition, whole genome sequencing documents numerous variants outside transcribed regions associated with CYP activity (Zhou and Lauschke, 2022). Many of the GWAS implicated variants are still under study and could account in part for the “missing heritability” (Johnson et al., 2008; Sadee et al., 2011). Arguably the most important drug metabolizing enzyme, CYP3A4 displays large interindividual differences in expression, with substantial hereditary influence, but the known CYP3A4 variants account for only a small portion of variance across a population. Nevertheless, new drugs are often designed to be metabolized by CYP3A4.
Regulatory variants can affect transcription or RNA processing (Sadee et al., 2011), such as splicing, RNA protein interactions, and translation as reported for the μ opioid receptor (Mascarenhas et al., 2015). Genetic variants can also act indirectly via lncRNAs, for example regulating CYP3A4 activity (Collins and Wang, 2022). In CYP3A4, an intronic SNP (CYP3A4*22, rs35599367, MAF 2%–6%) affects alternative splicing, reducing CYP3A4 hepatic expression and statin dose requirements (Wang et al., 2011). CYP3A4*22 has emerged as a clinically relevant variant in CYP3A4 (Elens et al., 2013; Zanger and Schwab, 2013; Werk and Cascorbi, 2014). However, CYP3A4*22 accounts for only portion of CYP3A4 variance across populations, due to its relatively low allele frequency.
SNPs regulating transcription, a mechanism for nearly 50% of GWAS hits (GTEx Consortium, 2020), can serve as biomarkers, for example promoter SNPs in VCORK1 (Wang et al., 2008) and UGT1A1 (Campbell et al., 2017). However, transcriptional regulation remains only partially characterized for most pharmacogenes including CYPs. Gene transcription is controlled by a core promoter and proximal or distal enhancers and suppressors. The core promoter and proximal enhancer (<4kb from transcription start sites) are extensively studied for most CYP genes (Kuehl et al., 2001; Lin et al., 2002; Wang et al., 2011), but distal regulatory elements and their interactions are less well explored. Reporter gene assays have identified several putative distal enhancers (5–10kb) upstream of CYP3A4 (Goodwin et al., 1999; Matsumura et al., 2004; Martínez-Jiménez et al., 2007; Tegude et al., 2007). However, subsequent studies failed to detect any in vivo effect (Wang et al., 2011), indicating reporter gene assays alone cannot reliably identify regulatory regions, lacking interacting regulatory elements (Bu and Gelman, 2007; Kato et al., 2007; Wang et al., 2008). A substantial portion of transcriptional genetics remains uncertain.
Emerging functional genomics technologies facilitate discovery of distal regulatory elements. Publicly accessible genomic databases, including the Encyclopedia of DNA Elements, reveal that proximity is not the rule for enhancer-promoter communication. Many distal enhancers interact over 100kb to 1Mb (Maas and Fallon, 2005; Sanyal et al., 2012) targeting gene promoters via long-range DNA looping (Sanyal et al., 2012; Zhang et al., 2013; Heidari et al., 2014). Chromatin conformation capture technologies allow rapid identification of interacting chromatin regions (Dekker et al., 2002; Tolhuis et al., 2002), revealing complex connectivity maps of the human genome, with numerous interactions between promoters and distal enhancers (Sanyal et al., 2012; Zhang et al., 2013; Heidari et al., 2014). Moreover, clustered regularly interspaced short palindromic repeat technologies (Hsu et al., 2014) allow alteration of the genome in live cells, assessing enhancer function and interactions in native chromatin context.
Applying these genomic technologies to pharmacogenes, we identified a distant enhancer region, located 115kb downstream of the CYP2D6 promoter, critical for CYP2D6 transcription. This region carries a frequent SNP (rs5758550) that further increases CYP2D6 transcription at least twofold (Wang et al., 2014, 2015). rs5758550 is in high LD over a span of >100 kb with the even more frequent rs16947 (core SNP designating CYP2D6*2 allele), shown to reduce CYP2D6 expression (Wang et al., 2014). Such a long-distance LD block suggests positive evolutionary selection pressure. Together, rs16947 and rs5758550 form haplotypes with varying frequencies in different populations, accounting for variable CYP2D6 expression levels predicted by CYP2D6*2 containing haplotypes (Ray et al., 2019). However, additional unrecognized regulatory variants confound this interpretation (unpublished observation).
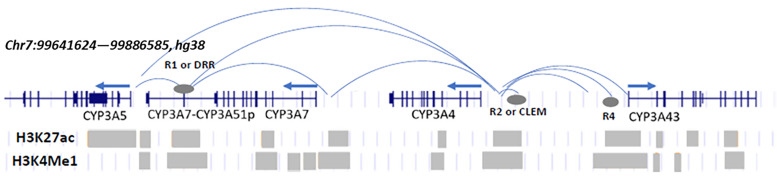
Transcriptional regulation of CYP3A4 involves a multigene CYP3A cluster (CYP3A4, CYP3A5, CYP3A7, and CYP3A43) of complex genomic architecture with shared regulatory domains, interdependent domain-domain interactions, and regulatory variants (Fig. 3) (Collins and Wang, 2020). Several SNPs in distal enhancers regulate CYP3A expression and interact with each other: SNP rs62471956 upstream of CYP3A4 exhibits opposite effects on CYP3A43 and CYP3A4 expression, possibly via competitive interactions (Collins and Wang, 2020). rs62471956 is in complete LD with CYP3A4*22 (Wang et al., 2011) and is associated with ticagrelor plasma levels in a GWAS (Varenhorst et al., 2015), potentially contributing to reduced expression of CYP3A4*22. Two SNPs (rs115025140 and rs776744) in a shared enhancer region approximately 90kb downstream of CYP3A4 associate with increased expression of CYP3A4 and CYP3A5, respectively (Collins et al., 2022). rs115025140 is unique to African populations with an allele frequency of approximately 8%, the most frequent functional CYP3A4 variant compared with low frequency of CYP3A4*22 (<1%). Both CYP2D6 and CYP3A4 highlight genomic complexity under evolutionary selection pressures, likely caused by environmental exposure.
Fig. 3.
Chromatin interactions and histone marks within the CYP3A locus in hepatic tissue or cells. Arrows indicate the orientation of each gene. Blue arced lines represent contact interactions and gray ovals regulatory regions identified in Collins and Wang (2020) and Collins et al. (2022). Gray bars represent histone mark intervals from public databases.
In addition to CYP enzymes and nicotinic receptors (Figs. 2 and 3), countless genes reside in clusters with homologous genes, pseudogenes, and various noncoding genes, regulated by enhancer networks and epistasis conferring functional robustness (Lin et al., 2022). Dissecting the regulatory elements and genetic variants, epistasis, and epigenetic chromatin modifications is challenging but can reveal the composite genetic and epigenetic influences. Accounting for such complexity could fill in the missing heritability encountered with polygenic risk scores and improve clinical utility of pharmacogenetic biomarkers.
III. Integration of Diverse Large Datasets, Large-Scale Data Analytics
Large-scale repositories of genomic and health care data have evolved into essential research tools readily accessible to all without the need for deep informatics expertise. Such datasets of large cohorts with genome-wide sequencing and deep phenotyping serve to address questions about specific genes or gene-drug interactions. Large cohorts are needed to detect functional variants affecting common polygenic disorders when individual variants have only modest effects and to integrate multiple factors with diverse phenotypes. Biobanks such as the UK Biobank (https://www.ukbiobank.ac.uk/) redefined “large-scale” with nearly half a million participants, all with genomic sequencing and deep phenotyping (Sudlow et al., 2015; Backman et al., 2021). Intimate connections between biobanks and electronic health records, as in All of Us (https://allofus.nih.gov/), blur the lines between data gathered for clinical care versus research, with an infrastructure that facilitates integration of clinical practice with research (Ramirez et al., 2022). These large-scale cohorts are just beginning to be explored within pharmacogenomics, with initial studies detecting known and new associations between 200 drugs and 9 genes while quantifying the prevalence of allele and phenotype frequencies at a population level (McInnes and Altman, 2021). Cross-ancestry GWAS studies can reveal genetic associations more accurately than studies with limited ethnic diversity, for example validation of the CYP2D6 locus as principal genetic determinant of endoxifen, the main active metabolite of tamoxifen (Khor et al., 2023). Yet genetics accounts for only a portion of interperson variability, which can be addressed with drug-level monitoring (Buijs et al., 2023).
Large cohorts have also emerged by harmonizing data from smaller studies, a paradigm that has become possible with ongoing cultural shifts in data sharing. The NIH, other funding agencies, and journals have long promoted data sharing, with more recent NIH policies requiring investigators to develop plans for data sharing. With such mandates, the NIH has developed substantial infrastructure to support these initiatives, such as the Database of Genotypes and Phenotypes (dbGaP) (https://www.ncbi.nlm.nih.gov/gap/). dbGaP provides a mechanism by which studies can be uploaded, stored, and downloaded with additional protections when controlled access is required (i.e., data requiring institutional review board approval or additional protections). dbGaP also provides a standardized approach to facilitating data interoperability and exchange. The platform now hosts more than 2,000 studies with close to 1 billion individual-level values with phenotype variables to molecular data, medical imaging, research protocols, and questionnaires (Sayers et al., 2023). These initiatives have rendered massive data analytics accessible in the public domain.
Technological revolutions in sequencing and data storage have evolved in tandem. Sequencing cost has dropped significantly, falling from $300 million for sequencing of the first human genome in 2003 to approximately $100 per genome (Pennisi, 2022). As whole-genome sequencing can be completed within 8 hours, inclusion into clinical workflow has become feasible (Gorzynski et al., 2022). Embedding cloud-based computational tools into large datasets such as All of Us has enabled access to vast computing resources (Alvarez et al., 2021), increasingly accessible to all researchers. As a result, large-scale data analytics, including machine learning techniques, are beginning to reveal novel relationships (Mooney, 2015).
Data sharing enables meta-analyses using individual-level data and testing questions not accessible to the primary investigators. Data sharing can be viewed as an ethical imperative as participants have taken on risks by participating in research (Bauchner et al., 2016), expecting optimal use to benefit health care broadly. Yet a number of challenges remain: 1) Protecting patient privacy remains a priority and becomes more challenging with increased large-scale data sharing. Protecting patient data and privacy is imperative (Jagadeesh et al., 2017). 2) Previous studies had targeted predominately European populations, limiting interpretation of findings for non-European ancestry individuals and assessment of traits with low allele frequencies in European populations. Current initiatives such as All of Us make a concerted effort to recruit patients from diverse backgrounds to address this gap. 3) Reproducibility remains an important question, requiring robust methods to harmonize data across studies, representing real-world data acquired through electronic health records, and replicate findings by sharing original datasets. To realize the promise of large-scale data mining in personalized medicine, international legal reforms will be needed to overcome a patchwork of copyright laws and restrictions across jurisdiction limits (Fiil-Flynn et al., 2022).
In addition to the wealth of patient data in the public domain, molecular data from tissue samples, cell lines, and animal models has increased exponentially. For example, the Encyclopedia of DNA Elements (https://www.encodeproject.org/) functionally characterizes the genome using information about transcription factor binding, chromatin structure, and histone markers (ENCODE Project Consortium, 2012). Both raw and summary-level data from genome-wide experiments in a variety of cell lines are available in the public domain and easily viewed using the UCSC Genome Browser (https://genome.ucsc.edu/). The Genotype and Tissue Expression Project (https://gtexportal.org/home/) (GTEx Consortium, 2020) captures tissue-specific differences in RNA expression paired with DNA sequencing to generate tissue-specific expression quantitative trait loci (Battle et al., 2017). The most recent version of the Genotype and Tissue Expression Project includes >17,000 RNA-seq experiments from approximately 1,000 individuals and additional analysis such as allele-specific RNA expression, protein quantification, and single-cell sequencing.
These resources allow researchers ready access to diverse data that had previously required deep technical expertise, predicting functional variants, transcription factor binding, and more (Jayaram et al., 2016; Yazar and Özbek, 2021), much of which is available in user-friendly browsers. Availability of these resources in the public domain has profoundly changed how researchers pursue scientific questions, generating preliminary data and initial hypotheses with little or no cost to address previously unimaginable questions—highly relevant to pharmacogenomics but still underused.
IV. Clinical Translation
A. Clinical Utility of Pharmacogenomic Biomarkers
Numerous reviews address clinical relevance of genetic biomarkers in drug therapy (Relling and Evans, 2015; Garcia-Gonzalez et al., 2016), replacing in part therapeutic drug-level monitoring (Salman and Al-Khabori, 2021; Haidar et al., 2022), while combining genetic biomarkers and drug levels could further guide optimal dosing, for example, for warfarin. A causative relationship between drug response and variants in genes encoding drug-metabolizing enzymes, membrane transporters, and receptors is intuitively assumed. For drug receptors, signaling pathways involve multiple components, each with the potential to introduce variance in drug response, diluting the effect of drug receptor variants alone. On the other hand, if a drug is eliminated from the body primarily by a single drug-metabolizing enzyme such as CYP2D6 and an individual carries two null alleles (no enzymatic function), the exposure to a common drug dosage, for example doxepin, can be multiplied (measured by the area under the blood level time course) (Kirchheiner et al., 2005). Drastically increased exposure increases risk of adverse reactions. Poor CYP2D6 metabolizer status is prevalent in Caucasian populations (approximately 7%) and has been linked to substantially increased hospitalization costs and treatment outcomes across the board, supporting a pervasive role of CYP2D6 in outcomes (Phillips et al., 2001). However, each gene carries multiple functional variants that affect the encoded protein’s function, and not all variants have been characterized, or the LD between null mutations could be uncertain in compound heterozygotes, introducing error in genetically predicted outcomes. As a result, drug blood levels predicted by genetic biomarkers can vary over a broad range (e.g., doxepin) (Kirchheiner et al., 2005), reducing clinical utility of genetic biomarkers. In addition, enzyme induction and drug-drug interactions (DDI) introduce additional variables. Whereas enzyme induction is thought to play a minor role in determining CYP2D6 activity, it serves as a major confounding factor for CYP3A4. Because only a few relatively infrequent nsSNPs (<2% allele frequency) had been identified for CYP3A4, new drug discovery tends to favor drugs metabolized by CYP3A4, even though intersubject variability is large—in part defeating the purpose of this drug design paradigm.
Biomarker panels of CYP2D6 are widely used. As codeine needs to be converted by CYP2D6 to morphine to be active, codeine has little or no analgesic effect in CYP2D6 poor metabolizers (Carranza-Leon et al., 2021). Similarly, the platelet inhibitor clopidogrel is activated by CYP2C19, with poor metabolizer status most prevalent in Asian populations (Sun et al., 2020). Proactive CYP2C19 genotyping to avoid treating coronary stent patients having CYP2C19 poor metabolizer status with clopidogrel significantly reduces the incidence of restenosis and is mandatory in some countries. On the other hand, prasugrel is not metabolized by CYP2C19 and can be substituted for clopidogrel, as well as other platelet inhibitors with different mechanisms of action (Norgard and Abu-Fadel, 2009). Therapeutic decisions must judiciously balance efficacy with and without genetic testing, efficacy for the intended clinical use, cost, and feasibility of genetic testing, a task often left to therapy committees in each hospital setting, with variable outcomes.
Transporters can deliver the drug or prevent access to the site of action. SCLO1B1 delivers certain statins (e.g., simvastatin) into hepatocytes, inhibiting HMG-CoA reductase to lower cholesterol biosynthesis. Genetic variants of SLCO1B1 conveying reduced transport activity require higher simvastatin doses to reach the cholesterol-lowering goal, leading to increased risk of myelotoxicity of simvastatin at higher doses because elevated drug levels in the circulation are needed to reach cholesterol reduction targets (Turongkaravee et al., 2021). The transporter MDR1 (encoded by ABCB1) recognizes a large number of drugs (Huang et al., 2004) reducing access to intracellular drug targets, one cause of drug resistance in cancer chemotherapy, while also excluding drug access through the blood-brain barrier to the central nervous system. As the opioid agonist loperamide is a strong substrate of MDR1, its action is limited peripherally as an antidiarrhea medication. While functional genetic variants of ABCB1 have only limited effects, upregulation of MDR1 in cancer tissues can thwart chemotherapy (Huang et al., 2004).
Discussed earlier, occurrence of ADRs has been associated with polymorphic drug-metabolizing enzymes (Phillips et al., 2001). Numerous companies developing pharmacogenetic biomarkers offer biomarker panels affecting drug metabolism. However, many drugs are metabolized by more than one enzyme, leading to the development multigene biomarker panels. While successful in some applications, for example in improving outcomes with antidepressants (Corponi et al., 2019; Forester et al., 2021), integrating the effect of multiple genes does not necessarily improve personalized therapies, due to the complexity of each gene locus. Nevertheless, attempts have been made to assess combined effects of drug metabolizing enzymes and transporters, for example to guide diabetes therapy with nateglinide (Naushad et al., 2022), and predict pharmacokinetics of rosuvastatin with three transporter genes (Lehtisalo et al., 2023). Without full knowledge of the genetic variation of each gene across ethnic groups, errors can be compounded, leading to incorrect assessment of the combined genetic influence in any given patient.
The primary goals of pharmacogenomic biomarkers are selection of the suitable treatment modality or optimal drug and drug-dosage regimen. Drugs that are mainly metabolized by a single enzyme should be avoided in poor metabolizers (null alleles), while partially reduced or enhanced activity can be used to adjust dosages. However, biomarker predictions of graded enzyme activity often display large variance even when reflecting the latest genomics insights, reducing clinical utility—for example CYP2D6 variants. Again, treatment decisions must include multiple personal factors, including issues related to compliance, which can be a significant factor, for example with antipsychotics.
One can assume that inclusion of pharmacogenomic biomarkers has the potential to result in fewer ADRs and improve outcomes. Yet variants in transporters and receptors, as well as gene networks affecting disease status, introduce confounding factors that are difficult to resolve, limiting clinical utility to specific conditions. Whether the benefit of introducing a biomarker outweighs the additional effort and cost of using a biomarker is the critical question to be addressed. Inclusion of pharmacogenomics data in electronic health records coupled with clinical decision support will be essential to expand personalized therapy (Hicks et al., 2016). Access to genetic information is most effective if available when prescribing the drug; therefore, prospective genotyping of most pharmacogenetic variants, or eventually general whole-genome sequencing, is being implemented but meets logistical concerns, such as reimbursement, reporting preemptive results over an individual's lifetime, data portability, and privacy (Haidar et al., 2022). Clinically actionable genotypes for at least one “pharmacogene” are estimated to be present in 90% to 95% of individuals, while implementation remains relatively low (Haidar et al., 2022). A recent survey in Florida reports that 27% of responding physicians apply pharmacogenomics information; absence of guidelines or protocols is cited as a main barrier to broad implementation (Ho et al., 2022), even when national and international consortia guidelines for implementation of pharmacogenomics are widely accessible (Nicholson et al., 2021).
Preemptive pharmacogenomic testing for in-patient care with point of care decision support is still largely unavailable. An ongoing study using a genotyping panel for all “actionable” pharmacogenes, “Implementation of Point-of-Care Pharmacogenomic Decision Support Accounting for Minority Disparities” (Chen et al., 2023), provides insights for implementation in general medicine, in particular for African American populations, and guidelines for workflow in a hospital setting.
B. Multicomponent Biomarkers in Clinical Medicine
Diverse genetic and nongenetic factors influence maintenance of health, disease risk and progression, and therapy response. While genetic variation represents the fundamental basis of pharmacogenomics, other -omics disciplines are also relevant: transcriptomics, proteomics, metabolomics, methylomics, epigenomics, phenomics, and metagenomics, each providing intermediate phenotypes more proximal to therapeutic outcomes. In turn, RNAs, proteins, and metabolites are established bio-markers per se. For example, blood cholesterol levels serve to titrate statin therapy, as an indicator of drug response and risk of cardiovascular disease.
Each genetic variant must be assessed in the context of the genomic background of an individual that can mask or enhance the variant’s effect (epistasis) (Bakerlee et al., 2022). Medical genetics mostly orients itself to risk factors of disease or ADRs. However, genetic variants conveying resilience likely differ from those carrying risk, typically with opposing evolutionary positive or purifying selection. In contrast to Mendelian monogenic disorders, common complex diseases including cardiovascular, metabolic, neurologic, and psychiatric, disorders and cancer, are associated with multiple genes, mostly involving noncoding regulatory variants (Sadee et al., 2014) that interact with each other and environmental factors. GWAS results suggest the involvement of countless genes in schizophrenia, diabetes, autoimmune disorders, and more. Targeted NGS can detect variants in select genes involved in pathogenesis and drug resistance (e.g., cisplatin) (Hattinger et al., 2022), but combining variants associated with phenotypes into a clinical diagnostic panel remains a formidable hurdle.
To detect associations between any given variant in GWAS, very large patient-control cohorts are needed to reach genome-wide significance in multigenic traits, since single SNPs gleaned from GWAS account for only a small portion of the expected heritability (e.g., determined from twin studies). Searching for epistatic interactions between two variants or factors (gene-gene and gene-environment interactions) in GWAS imposes formidable additional statistical obstacles. Wang et al. (2021) have identified gene pairs displaying epistasis, proposing that an epistasis risk score can identify individuals at risk of early-onset Alzheimer’s disease. Multifactorial risk estimates have been generated to capture the sum of small contributions, for example the use of RNA profiles in MammaPrint and Oncotype Dx for predicting outcome and aiding adjunct therapy decision-making in patients with ER-positive, HER2-negative breast cancers (Nicolini et al., 2018). This approach has been successful in accounting for a portion of outcome results but suffers from several confounding factors, with RNA panels representing complex expression networks with uncertain causal links to outcomes. Most polygenic GWAS scores represent SNPs in noncoding regions that are not the causative variants but rather in LD with them. Applying multigenic biomarker panels to distinct ethnic populations is confounded by distinct LD patterns between the GWAS hit and causative variant. Also, the contributions of rare versus common functional variants remains to be resolved, a question that can be addressed only with use of very large subject cohorts. For CYP genes, rare alleles appear to contribute 1.5% to 15% to the overall genetically encoded functional variability (Zhou and Lauschke, 2022), but clinical utility remains to be determined. On the other hand, rare variants may contribute substantially to narrow sense heritability, for example of smoking behaviors (Jang et al., 2022). Further confounding factors include epistatic interactions, for example in the CHRNA5/A3/B4 cluster (Lee et al., 2018). Difficulties arise from exponential growth not only of the signal but also the noise in overall association estimates with the addition of each new factor, confounded by limited understanding of the causative factors.
A majority of patients receive multiple drugs; therefore, we must consider DDIs as another critical factor in drug response. DDIs can occur through inhibition of drug-metabolizing enzymes or transporters, or enzyme induction, as detailed for drugs approved by the FDA in 2020 (Yu et al., 2022), most frequently involving interactions with CYP3A enzymes—a preferred drug target. Interactions between a genetic deficiency and drug-induced inhibition of two CYPs—such as CYP2D6 and CYP2C19—can cause profound response changes in drugs that are eliminated by these two enzymes, a result of drug-drug-gene interactions (Bahar et al., 2017; Malki and Pearson, 2020). A survey of drug-drug, drug-gene, and drug-gene-gene interactions finds up to 30% of cases with clinically relevant outcomes (Ashcraft et al., 2022). Therefore, inclusion of DDIs in predictive therapeutic decision tools is essential. On the other hand, a directed DDI can serve to enhance drug response, for example the inclusion of ritonavir (an inhibitor of several CYP enzymes, including CYP3A4) in anti-HIV and anti-SARS-CoV-2 medications. For the latter, Plaxlovid (nirmatrelvir + ritonavir) has proven effective in reducing severe COVID symptoms (Najjar-Debbiny et al., 2023). However, inclusion of ritonavir affects the metabolism of numerous other drugs, such as atorvastatin, which is metabolized by CYP3A4 and requires therapeutic adjustments.
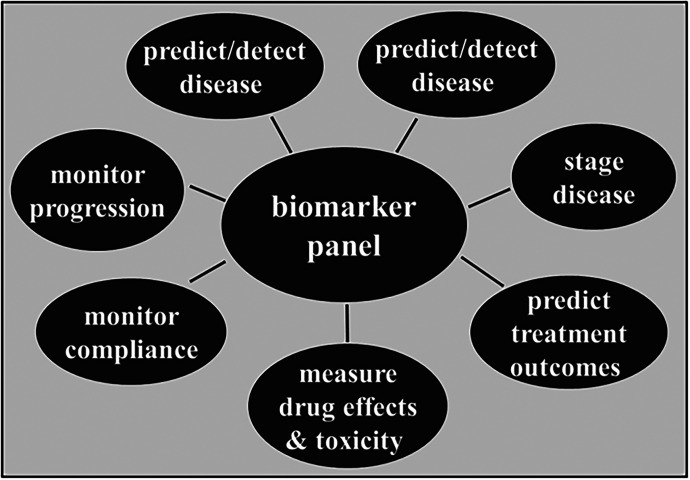
The rich landscape of genetic, clinical, and environmental markers and phenotypes, including personal variables (sex, age, ancestry, socioeconomic status, sociodeterminants of health, weight, nutritional status) will require large-scale integration with use of artificial intelligence (AI) to generate clinically valid overall guidance of personalized medicine. Figure 4 illustrates diverse applications of biomarkers in clinical medicine. While single biomarkers such as genetic variants in drug-metabolizing enzymes can assist in optimizing therapies, only a portion of response variance can be covered. Broadening the use of diverse markers suffers from uncertain interactions, leading to black-box therapeutic recommendations, lacking complete understanding of the decision process. This challenge presents a hurdle for clinical implementation and must be overcome in the future of personalized medicine.
Fig. 4.
Use of biomarkers in personalized medicine. Panels include phenotypic and genomic markers. Note that germline mutations can be detected at any time, whereas other markers, including somatic mutations, vary with disease state, age, drug treatment, and other factors.
C. Clinical Guidelines
Translation of pharmacogenenetic/genomic biomarkers into clinical practice depends on multiple factors and encounters robust hurdles. Above all, any biomarker predicts only a portion of disease risk or treatment outcome, with substantial variability between patients. Thus, overall impact such as genetic penetrance and intersubject variance, together with cost, are the main criteria that define cost/benefit ratios and determine clinical feasibility. The FDA table of Pharmacogenomic Biomarkers in Drug Labels (https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling) lists drugs that are associated with genetic biomarkers, with links to complete public drug label information, including type of genetic variant and allele frequencies between ethnic groups (reviewed in Mehta et al., 2020). The number of drugs with FDA-declared bio-markers has grown dramatically over the past 20 years; examples are shown in Table 2. As a function of a biomarker’s clinical impact, drug-biomarker pairs are either provided on advice only, for consideration by therapists, or further highlighted with a boxed warning when ADRs can be severe. For example, tamoxifen for treating breast cancer largely depends on CYP2D6 for activation to the main active metabolite endoxifen; therefore, CYP2D6 poor metabolizers are less likely to respond and may need other therapies (Chan et al., 2020). However, variability in response derives from involvement of several metabolizing enzymes, dietary factors, and patients' compliance. Further research is needed to optimize clinical utility for CYP2D6-guided therapy (Chan et al., 2020).
TABLE 2.
FDA CDER: Abbreviated Table of Pharmacogenomic Biomarkers in Drug Labeling
For each of the 517 gene-drug pairs in the FDA (2022) table, the drug labels include clinically actionable pharmacogenomics information. Some of these combinations are mandatory: the test must be taken before the drug can be dispensed (or reimbursed by the insurer); however, the FDA table does not specify how the biomarkers are to be applied. Drugs are marked with stars to indicate how stringent test requirements are; absence of a * marker indicates that testing may be considered on case-by-case basis. (https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling).
| Drug Link in Drugs@FDA |
Therapeutic Area | Biomarker | Label Sections with Pharmacogenomic Information |
|---|---|---|---|
| Abacavir** | Antivirals | HLA-B*5701 | Boxed Warning, Contraindications Warnings and Precautions Patient Counseling |
| Atezoliozumab* | Oncology | PD-L1, gene signature, GFR, ALK, BRAF | Indications and Usage, Dosage and Administration, Adverse Reactions |
| Carbamazepine** | Neurology | HLA-B*1502 | Boxed Warning, Warnings, Precautions |
| Cetuximab* | Oncology | EGFR KRAS |
Indications and Usage Warnings and Precautions Indications and Usage |
| Clopidogrel | Cardiology | CYP2C19 | Boxed Warning, Warnings and Precautions |
| Clozapine | Psychiatry | CYP2D6 | Dosage and Administration, Use in Specific Populations, Clinical Pharmacology |
| Codeine | Anesthesiology | CYP2D6 | Boxed Warning, Warnings and Precautions, Use in Specific Populations, Patient Counseling Information |
| Dasatinib* | Oncology | Ph Chromosome | Indications and Usage |
| Doxepin | Psychiatry | CYP2D6 | Precautions |
| Elosulfase* | Inborn Errors of Metabolism | GALNS | Indications and Usage, Warnings and Precautions, Use in Specific Populations |
| Imatinib* | Oncology | C-KIT, PH Chrom., PDGFR, FIP1L1-PDGFRa | Indications and Usage, Adverse Reactions, Use in Specific Populations |
| Ivacaftor* | Pulmonary | CFTR | Indications and Usage, Adverse Reactions, Use in Specific Populations |
| Maraviroc* | Antivirals | CCR5 | Warnings and Precautions |
| Nitrofurantoin | Infectious Diseases | G6PD | Warnings, Adverse Reactions |
| Tamoxifen | Oncology | ESR, F5, F2, CYP2D6 | Indications and Usage |
| Thioguanine | Oncology | TPMT, NUDT15 | Dosage and Administration, Warnings, Precautions, Clinical Pharmacology |
| Trastuzumab**,** | Oncology | ERB2 (HER2) | Indications and Usage, Dosage and Administration |
CDER, Center for Drug Evaluation and Research.
*biomarker testing required; ** biomarker testing strongly advised; ***Herceptin.
Lastly, the biomarker test can be classified as mandatory without which the drug cannot be prescribed (examples in Table 2). Mandatory testing is indicated when the drug is efficacious in severe or life-threatening disease when a positive response depends on a genetic variant [e.g., trastuzumab (Herceptin)] or severe toxicity can occur (Jacobs et al., 2022). The FDA table does not specify whether and how a biomarker should be applied, even when clinical evidence supports the notion that biomarker use can be beneficial with an acceptable cost-benefit ratio. When clear evidence links a genetic variant to a potentially severe ADR, the FDA drug label includes a “boxed warning,” for example for Abacavir (Fig. 5). This type of idiosyncratic ADR has been linked to HLA variants, involved in immune response multiple dugs and disease risk (Jaruthamsophon et al., 2022; Moyer and Gandhi, 2022). Between 2000 and 2020, pharmacogenetic information in drug labels has increased from 10% to 28% for newly approved drugs (Kim et al., 2021). The Personalized Medicine Coalition has termed such therapies defined by a biomarker as “personalized medicine” benefitting select patients (as may be the case in rare genetic disorders) or well-defined subgroups of patients with common disorders.
Fig. 5.
Boxed warning in the FDA drug label for Abacavir. HLA-B*-5701carriers are at high risk of fatal hypersensitivity reactions. Drug label boxed warning established in 1998, text box copied from the 11.24.2020 label. (https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm?event=overview.process&varApplNo=020977).
To provide guidance on pharmacogenetic biomarker use, several organizations have developed databases, criteria for assessing clinical utility, and directives for drug selection and dosing strategies. The Pharmacogenomics Knowledge Base is a comprehensive database providing information for numerous drugs, developed out of the Pharmacogenomics Research Network (Whirl-Carrillo et al., 2012). Building on the Pharmacogenomics Knowledge Base database, members of the Clinical Pharmacogenomics Implementation Consortium (CPIC; https://cpicpgx.org/) have developed standardized methods and terms to write reports for those drug treatments that can benefit from biomarker guidance (Caudle et al., 2017). These recommendations are supported by exhaustive literature reviews of clinical trials, metanalyses, underlying mechanism of action, and more. Published reports highlight the complexity of certain genes such as CYP2D6, addressing mostly single gene-drug interactions. Complexity of the algorithms leading to recommended dosing regimens is greatly increased if two genes are contributing substantially to outcomes, for example dosing tables for the anticoagulant warfarin as a function of genetic variants in both CYP2C9 and VKORC1; however, the algorithms have been optimized predominantly for Caucasian and Asian populations (Asiimwe et al., 2021).
Published CPIC reports serve as implementation tools for biotech companies and clinical laboratories to translate the biomarker test results into therapeutic recommendations. CPIC members have developed a Pharmacogenomics Clinical Annotation Tool, providing automatic reports of patient genetic data, extracted from recommendations by CPIC and Dutch Pharmacogenetics Working Group (Li et al., 2022). In a growing number of cases, multigene pharmacogenomics panels are performed, providing a wealth of information for immediate use and for future therapies, but also require careful interpretation to avoid errors (Ho et al., 2022).
If an acceptable cost benefit has been demonstrated in clinical trials, the test can be accepted by insurance carriers, Medicare, and others for reimbursement. However, prospective polygene biomarker panels—even if cost-effective over time—are mostly not or only partially covered by insurance carriers unless the combined panel assessment is proven superior in a specific application, for example treatment of major depression (Forester et al., 2021).
V. Applications in Cancer Therapy—A Beacon for Common Complex Disorders
A. Early Molecular Markers
Traditional cancer treatments including surgery and aggressive cytotoxic chemotherapies are encumbered with prominent adverse effects. The hunt for molecularly targeted therapies has come into focus since the 1980s, defined as treatments guided by molecular genetic markers and biologic consequences such as pathway activation or protein overexpression. Tamoxifen is considered the first targeted cancer therapy, adopted in the 1990s as an adjuvant therapy in women with ER-positive breast cancer (Jordan, 2021), decades after its efficacy against ER-positive breast cancer was proposed in 1971 (Cole et al., 1971; Jordan, 2021). The discovery rate of drug-biomarker pairs has increased exponentially since then, with hundreds of targeted therapies approved for nearly every cancer type (Aleksakhina and Imyanitov, 2021; Jacobs et al., 2022).
Among the first molecular cancer biomarkers, proteins or genes showing amplification, translocation, or overexpression were assayed in tumors using immunohistochemistry, fluorescent in situ hybridization, or karyotyping. These genes and proteins mostly promote tumor growth or aggressive behavior through aberrant protein activation or increased activity. A classic example from the late 1980s is the finding in a subset of breast tumors that amplification of the HER2 gene resulting in increased protein expression correlates with worse outcomes (Slamon et al., 1987; Hudziak et al., 1989). In 1998, the antibody against the HER2/neu protein trastuzumab (Herceptin) was approved by the FDA for treating specifically HER2/neu-positive breast tumors because of the enhanced efficacy when linked to the biomarker test (Jacobs et al., 2022).
B. Next-Generation Sequencing and Proteomics
Widespread adoption of NGS has led to standard-of-care targeted sequencing panels, detecting overexpressed genes, germline and somatic variants, or response markers such as tumor mutational burden (TMB) and microsatellite instability (MSI) (Freedman et al., 2018; Luchini et al., 2019; Mosele et al., 2020). Genetic/genomic biomarkers include gene amplification, translocation, activating mutations in oncogenes, mutational signatures, and TMB. With decreasing NGS costs, more patients receive tumor testing with tissue-agnostic cancer gene panels (Aleksakhina and Imyanitov, 2021). While many tumors carrying a somatic mutation lack treatment options, targeted therapies have shown clinical efficacy in both solid and hematologic cancers. Targets include the oncogenes EGFR, ALK, BRAF, MET, KIT, lacking traditional drug binding pockets and most recently KRAS (p.Gly12Cys) (Lee and Pant, 2021; Ji et al., 2022; Turpin et al., 2022), encoding a protein previously deemed not amenable to drug inhibition. Inactivating germline mutations in tumor-suppressor genes are found mostly in heterozygous carriers and promote cancer upon somatic loss of the wild-type allele (Aaltonen et al., 1993; Gilson et al., 2021).
Large-scale protein sequencing has emerged to integrate genomic sequence data with mass spectrometry-based proteomics (Subbannayya et al., 2021; Mani et al., 2022). Proteogenomics detects protein fusions, novel splice forms, and neo-antigens, promising biomarkers as molecular targets or of response to therapy (Fu et al., 2017; Subbannayya et al., 2021).
C. Molecular Signatures and Pathways
Genetic and genomic variation can result in molecular signatures of perturbed cancer pathways as potential biomarkers (Gilson et al., 2021). One such biomarker is MSI. Tumors with deficient mismatch repair have random insertions or deletions at repetitive regions of DNA, such as homopolymers or microsatellite repeats, leading to somatic mutations of key cancer-related genes and a genomic MSI signature. MSI was first recognized in endometrial and colorectal tumors arising in individuals with Lynch syndrome, a hereditary cancer syndrome caused by germline pathogenic variants in mismatch repair genes (Aaltonen et al., 1993). Recent studies suggest that MSI is a promising biomarker for response to immune-based checkpoint inhibitors (Le et al., 2015; Ludford et al., 2021; Cercek et al., 2022).
Tumors arising in individuals with germline BRCA1 or BRCA2 pathogenic alleles typically show loss of homology-directed repair (HDR) proficiency, important in double-strand break repair, upon somatic loss of the wild-type allele. Poly-adenosine diphosphate-ribose polymerase (PARP) inhibitors (PARPi) were developed as a synthetic lethality approach to exploit reliance of BRCA-deficient tumors on base excision and single-strand break repair, which is mediated through PARPs. Failure to repair single-strand breaks in tumors treated with PARPi leads to irreparable double-strand breaks in BRCA-deficient tumors and cell death (Chen, 2011). The first clinical trials for PARPi were for advanced BRCA-deficient ovarian cancers with germline or somatic mutations in BRCA1 or BRCA2 leading to FDA approval in 2014 (Ledermann et al., 2012), followed by BRCA-deficient metastatic breast (2018), pancreatic (2019), and prostate (2020) tumors (Sigorski et al., 2020). Current research seeks to expand the PARPi approach to additional HDR genes, such as RAD51C/D, CHEK1, ATR, and PALB2(Stover et al., 2020). To broaden use of PARP-inhibitor therapies, assessing “BRCAness” phenotypes and HDR genomic scoring methods have been developed, including differential gene expression profiling, DNA copy-number alterations, chromosomal rearrangements, genomic instability scores, and mutational signatures (Watkins et al., 2014; Stover et al., 2020; Beinse et al., 2022). Studies are underway to test PARPi in tumors with somatic mutations in any DNA damage repair gene (Zhang et al., 2022).
Mutational signatures also serve as biomarkers of resistance to therapies. A common mutational signature is caused by dysregulated activity of apolipoprotein b mRNA-editing enzyme, catalytic polypeptide-like (APOBEC) (Mertz et al., 2022). Causing deamination of cytidines at specific sequence motifs, APOBEC dysregulated tumors display a characteristic mutational pattern associated with therapeutic resistance (Venkatesan et al., 2018). For example, APOBEC mutational signatures occur frequently in receptor-tyrosine kinase driven lung cancer and in tumors treated with osimertinib, which targets EGFR-mutant lung cancers (Selenica et al., 2022).
D. Emerging Role of “Basket Trials”
Molecularly targeted therapies were originally tested against tissue-specific cancers, but insufficient patient numbers carrying a target mutation impeded design of clinical trials. As molecular targets can occur in multiple tumor types, “basket trials” have been implemented for tissue-agnostic therapy of tumors that share the molecular biomarker (FDA 2022: https://www.fda.gov/media/120721/download), assuming that drug efficacy depends primarily on the tumor driver mutation. One of the first basket trials evaluated imatinib to treat tumors with high expression or activating mutations of receptor tyrosine kinases such as KIT, PDGFRA, or PDGFRB (Heinrich et al., 2008). Of over 40 tumor types tested, 6 malignancies showed evidence of clinical responses. Subsequent studies revealed that some therapies (such as PARPi) are effective against a broad range of tumor types (Domchek et al., 2020), whereas others (BRAF inhibitors) work better for melanoma and non–small cell lung cancer than for colorectal cancer, because of pathway-escape mechanisms (Planchard et al., 2016; Corcoran et al., 2018; Robert et al., 2019).
E. Molecular Biomarkers for Response to Onco-Immune Therapy
The immune system plays a key role in keeping cancers in check (Mellman et al., 2011) by recognizing foreign tumor-specific antigens and proteins not normally made by adult cells. This immunosurveillance can lead to elimination of tumor cells by cytotoxic immune cells (Dunn et al., 2004). In turn, cancer cells can escape death by upregulating immune checkpoint proteins such as PD-1 and PD-L1 or CTLA-4, which competes for binding of T-cells to B7 molecules. Antibodies against these immune checkpoint proteins (singly or in combination) have shown promise for treating melanoma and non–small cell lung cancer by blocking inhibitory tumor tissue interaction with T-cells and enabling recognition and elimination of tumors cells (Overman et al., 2018; Qin et al., 2019). Immune checkpoint-based antibody therapies work only in a fraction of patients that overexpress these proteins (Lantuejoul et al., 2020). The hunt is on for molecular biomarkers that predict response and effective combination therapies. Tumors with a high mutational burden (TMB) tend to respond best to checkpoint inhibitors, likely because more neoantigens are created. However, TMB has shown less than robust predictive value (Sholl et al., 2020; Marcus et al., 2021; Mino-Kenudson et al., 2022). Ongoing studies to improve biomarkers for immune checkpoint inhibitor response are assessing immune gene signatures, selective polyadenylation signals, somatic copy number alterations in CD274 (the gene encoding PD-L1), and polymorphisms in CD274 and PDCD1 (the gene encoding PD-1) (Yoshida et al., 2021; Chen et al., 2022; Murugesan et al., 2022; Wu et al., 2022), promising enhanced prediction of responses to onco-immune therapies.
F. Acquired Resistance to Targeted Therapy and Counterstrategies
1. Somatic Mutations in Parallel Pathway Genes
Targeted cancer therapies can result in initial dramatic responses, but tumors tend to develop resistance. For example, vismodegib inhibits the Hedgehog pathway and was originally approved for individuals with metastatic or locally advanced basal cell carcinoma with PTCH1 mutations. Early trial results showed dramatic elimination of both metastatic lesions and primary basal cell carcinomas (Von Hoff et al., 2009) over 6 to 12 months of treatments (Dessinioti et al., 2019). Yet tumors in about 20% of patients came back at about a year’s time (Chang and Oro, 2012). In addition, a large subset of patients suffers from considerable side effects (Leavitt et al., 2019). New mutations in other parts of the Hedgehog pathway enable tumors to escape inhibition, such as activating mutations in SMO or amplification of GLI genes (Yauch et al., 2009; Buonamici et al., 2010; Chang and Oro, 2012). Secondary mutations in other pathway genes are common mechanisms of resistance to targeted therapies. Multiple other examples of secondary mutations leading to therapeutic resistance include activating mutations in ESR1 in estrogen receptor positive breast tumors treated with aromatase inhibitors or estrogen receptor ligands such as tamoxifen (Brett et al., 2021). ESR1 mutations are rare in treatment-naïve tumors suggesting that their emergence is primarily due to selective pressure from the therapy. Tumor resistance to targeted therapies has led to a call for clinical trials evaluating intermittent dosing or combination therapies that may reduce side effects as well as decrease resistance (Leavitt et al., 2019).
2. Reversion Mutations
Resistance to therapies can be due to reversion mutations that correct the initial defect as observed for breast tumors in individuals with germline pathogenic variants in BRCA1 or BRCA2 treated with PARPi (Pettitt et al., 2020; Zong et al., 2022). Although rare, tumors have been identified with secondary mutations that “correct” the original germline variant by, for example, restoring the reading frame of the gene. Reversion mutations in BRCA1 and BRCA2 are more common in platinum-resistant or PARPi refractory tumors by repairing the drug-induced DNA damage (Weigelt et al., 2017; Pettitt et al., 2020; Vidula et al., 2020).
3. Tumor Heterogeneity
Tumor heterogeneity arises from both genetic (e.g., mutations) and nongenetic factors (epigenetic, immune, and stromal cell type diversity) and is associated with worse overall outcomes and higher frequency of treatment resistance (Biswas and De, 2021). NGS technology applied to intratumoral heterogeneity has revealed subclones in untreated tumors that contain secondary mutations fostering resistance (Shah et al., 2009; Gerlinger et al., 2012). Single-cell NGS has revealed rapid tumor evolution in response to therapy, in part due to tumor heterogeneity (Kuipers et al., 2017; Maynard et al., 2020). Tumors with high heterogeneity and high neoantigen diversity may also be less responsive to immune checkpoint therapies (Craig et al., 2022).
Cancer treatment strategies are moving from reliance on primary molecular targets to combination therapies to account for predicted routes of tumor resistance. Combination therapies target both the main driver event and driver genes in subclones to increase the likelihood of durable response (Plana et al., 2022). This strategy with molecularly targeted and immune-checkpoint therapies has been successfully applied to multiple cancers (Golay and Andrea, 2020; Varayathu et al., 2021).
Real-time evaluation of tumor changes can be assessed through liquid biopsies—a noninvasive means of monitoring tumors (Nikanjam et al., 2022)—to identify resistance before clinical presentation. Most commonly, blood is monitored for circulating tumor cells, cell-free DNA, or circulating tumor DNA (ctDNA), while urine, ascites fluid, breast milk, and pleural fluid are also clinically used. Tumor DNA is released into the circulation from tumor cells undergoing apoptosis. Circulating DNA (cell-free DNA) can be interrogated using NGS to detect clonal tumor diversity, targetable mutations, response to therapies, minimal residual disease after surgery, and new mutations that may confer treatment resistance (Siravegna et al., 2019). These technologies could replace tumor-imaging technologies especially for tumors with high ctDNA levels. ctDNA monitoring has been FDA approved, showing efficacy in guiding clinical trials for lung and colorectal cancers (Chabon et al., 2016; Malla et al., 2022) and to identify reversion mutations in tumors being treated with PARPi (Goodall et al., 2017; Kondrashova et al., 2017; Quigley et al., 2017). Detecting how tumors are evading therapies in near real time can provide critical information on guiding therapeutic strategies.
G. The Future of Molecular Targets to Guide Therapy for Cancer and Other Diseases
Cancer therapy guided by molecular biomarkers has served as a paradigm of personalized therapeutics. Combinatorial strategies are emerging that target a main molecular driver and additional molecular events causing treatment failure. Multiomics approaches combining proteomics, genomics, methylomics, metabolomics, and metatranscriptomics need to be merged with individual characteristics (age, sex, ancestry, immune status, sociodeterminants of health) to refine personalized cancer treatments (Ye et al., 2021; Mani et al., 2022). Surveillance of treatment response and early tumor detection using liquid biopsies may improve therapy outcomes. As molecular detection of cancer cells and risk factors improve, therapy will focus on targeting preclinical tumors.
While somatic tumor mutations have served as molecular targets for more than 30 years, expansion of somatic mutations in noncancerous tissues are now recognized risk factors for numerous other diseases, often associated with aging, for example in Alzheimer’s disease (Downey et al., 2022). Clonal expansion of noncancerous blood cells driven by genetic variants in DNMT3A, TET2, JAK2, and ASXL1 have been linked to age-related cardiovascular risk and inflammation and may also lead to molecularly guided treatments (Haring et al., 2022).
VI. Pharmacogenomics in Drug Discovery and Development
Genomics has become an integral part of drug discovery and development, shepherding new drugs into clinical use. Detection of valid drug targets benefits from integration of genomic sequence, transcriptomes in affected tissues, and the proteome and metabolome (Plenge, 2019). For example, genetics had revealed PCSK9 deficiency as protective of cardiovascular diseases by reducing cholesterol load, leading to effective therapies using antibodies to or RNA blockers of PCSK9 (White, 2018). As discussed, targeted anticancer drug discovery focuses on oncogenic mutations. On the other hand, most common disorders, such as diabetes and cardiovascular and psychiatric diseases, have a polygenic origin. In principle, these common disorders each could be classified into subtypes on the basis of distinct pathophysiology and genetics, but progress has been slow because of gene network complexity. Nevertheless, biomarkers can serve to identify patient subgroups benefitting most from a given treatment or being at increased risk of ADRs.
Defining subgroups of patients affected with a common disorder who are more likely to respond has proven effective in curtailing cost of bringing a drug to clinical use—currently estimated to reach into the billion-dollar range. While an attractive approach to personalized medicine, the additional burden of developing the biomarker test and validating its clinical utility had previously impeded successful implementation. However, advances in genomics, clinical sciences, and regulatory measures have led to a surge of such “theranostics.”
An increasing number of drugs are now in clinical use that can be prescribed only in conjunction with an obligatory biomarker, required to assure sufficient efficacy or prevent serious harm. Without use of a biomarker, Herceptin would not have been approved (Jacobs et al., 2022). Theranostics have proliferated in cancer therapy, accounting for 75% of all biomarker drug pairs with required pharmacogenomic testing (Kim et al., 2021). Use of radiolabeled pharmaceuticals that bind to a specific molecular target enables imaging of the distribution of disease with one type of radiopharmaceutical and treatment with another, higher energy molecule. For example, prostate specific membrane antigen can be imaged with gallium-68–labeled prostate-specific membrane antigen positron emission tomography scans and treated with Lutetium-177 (177Lu)-PSMA-617 (Sartor et al., 2021). If indeed required by regulatory agencies (e.g., the FDA), the biomarker assumes the same value as the drug itself. Table 3 lists examples of recent personalized medications. For yearly overviews of “personalized therapeutics” approved by the FDA, the Personalized Medicine Coalition publishes comprehensive reports that mirror the broad scope of such medicines, accounting for approximately 30% of all new drug approvals (Personalized Medicine at the FDA, Scope and Significance of Progress in 2021; www.PersonalizedMedicine.org).
TABLE 3.
U.S. FDA Novel Orphan Drug Approvals in 2020–2021
Only those drugs considered first-in-class are shown: 26 of 50 (52%). Some 21 of those drugs (81%) reflected first-in-class approvals, an indication of the strong innovation. 2020: CDER approved 53 novel drugs, 31 orphan drugs (58%) (Source: FDA; taken from https://globalgenes.org/raredaily/a-group-of-innovative-orphan-drugs-win-fda-approval-in-2021/?gclid=CjwKCAiAyfybBhBKEiwAgtB7fmCXF8VAW0aHnwGHogVoFWwiRqkm GXkSPD-dZLyIaB6QXdFwwrw-FBoCMVcQAvD_BwE).
| Proprietary Name | Active Ingredient | Indication | |
|---|---|---|---|
| Besremi | Ropeginterferon alfa-2b-njft | To treat polycythemia vera | |
| Bylvay | Odevixibat | To treat pruritus in progressive familial intrahepatic cholestasis |
|
| Cytalux | Pafolacianine | To use for ovarian cancer imaging | |
| Empaveli | Pegcetacoplan | To treat paroxysmal nocturnal hemoglobinuria | |
| Evkeeza | Evinacumab-dgnb | To treat homozygous familial hypercholesterolemia | |
| Livtencity | Maribavir | To treat cytomegalovirus infection | |
| Lumakras | Sotorasib | To treat types of non–small cell lung cancer | |
| Nulibry | Fosdenopterin | To reduce the risk of mortality in molybdenum cofactor deficiency type A | |
| Rezurock | Belumosudil | To treat chronic graft-versus-host disease after failure of at least two prior lines of systemic therapy | |
| Tavneos | Avacopan | To treat severe active antineutrophil cytoplasmic autoantibody-associated vasculitis in combination with standard therapy, including glucocorticoids | |
| Voxzogo | Vosoritide | To improve growth in children 5 years of age and older with achondroplasia and open epiphyses |
|
| Vyvgart | Efgartigimod alfa-fcab | To treat generalized myasthenia gravis | |
| Welireg | Belzutifan | To treat von Hippel-Lindau disease | |
| Zynlonta | Loncastuximab tesirine-lpyl | To treat types of relapsed or refractory large B-cell lymphoma |
Drug discovery in the pharmaceutical industry traditionally aimed at developing therapies for common disorders, with large economic return. However, successfully bringing such drugs into the clinic under the motto “one drug fits all” has become increasingly difficult, requiring huge investments. To incentivize the development of therapies against less common diseases, the U.S. Congress passed the Orphan Drug Act in 1983. Orphan drug regulations followed in Japan (1993), Australia (1998), the European Union, and many more countries (Haffner, 2016). The act provides substantial benefits to industry, such as tax credits, fee waivers, and 7 years market exclusivity, to encourage targeting of diseases affecting a patient population limited to <200,000 in the United States. After a slow start, driven largely by genomics and novel drug discovery tools and supporting novel treatment modalities (including gene therapy), the act has led to a spectacular rise in approved orphan drugs (Mueller et al., 2019; Brown and Wobst, 2021). Many of these drugs require a biomarker test to select eligible patients (so-called precision drugs). Until 2019, 44% of orphan drugs targeted cancer, accounting for 88% of all precision drugs (Mueller et al., 2019). While cancer is a common disease, biomarkers can identify distinct subgroups of cancer patients with shared mutations, recognizing that cancer is uniquely individual and requires personalized therapies, thereby qualifying for orphan drug status. Orphan drug status, combined with required pharmacogenetic testing, is also advancing in other diseases, mostly relatively rare monogenic disorders. For example, cystic fibrosis is caused by multiple mutations in encoding cystic fibrosis transmembrane conductance regulator (CFTR), affecting proper protein folding, channel function, and transport to the plasma membrane. As each CFTR mutation has varying effects on CFTR function, different drugs may optimally overcome the defect. A three-drug combination of tezacaftor (folding corrector), ivacaftor (channel potentiator), and elaxacaftor (dual function modulator) synergistically and fully rescues Δ508 CFTR function (Fiedorczuk and Chen, 2022) and is potentially effective for other CFTR mutations as well.
One might predict that for common polygenic diseases other than cancer, defining smaller subpopulations with defined genotypes might also open opportunities for novel orphan drugs, but this goal is difficult to achieve because of their polygenic nature. To assure return on investment with small patient populations, orphan drugs tend to be costly, adding a substantial burden to the health care system. Given strident advances in developing drug-biomarker combinations, with potentially substantial cost reduction feasible, the Orphan Drug Act provisions may need revision in the era of personalized medicine.
VII. Convergence of Technological and Scientific Advances
A. Drivers of Fundamental Changes in Health Sciences, Medicine, and Health Care
NGS ranks among the top disruptive technologies changing our understanding of biology, moving from analysis of single genes to assessment of the functions embedded in the entire genome—encompassing genomics and transcriptomics. Parallel developments have enabled complete analysis of the proteome, metabolome, and metagenome (including intestinal bacteria and viruses). A compelling extension of this trend lies in the use of all clinical and pharmacological phenotypes—referred to as the phenome—taking advantage of large-scale clinical electronic databases (Liu and Crawford, 2022). These clinical datasets can be compared between diseases to reveal overlapping causality. In combination with GWAS data, each clinical trait can be linked to variants in one or more genes or gene networks as a guide for therapies. Drug treatment outcomes and adverse effects can be classified by symptoms as biomarkers, while genetic variants can be tested for impact on phenotypic biomarkers such as metabolites. Adding more complexity, environmental factors impinge on biologic systems and modify genetic influence. Discerning patterns across large-scale diverse interrelated datasets requires increasing computational power, combined with AI and related technologies such as machine learning (Abdelhalim et al., 2022). Applied to complex diseases such as psychiatric disorders (Hoehe and Morris-Rosendahl, 2018), genomic and medical data can be merged to guide therapy, but the AI approach still encounters multiple hurdles as outcome predictions lack a cohesive causative foundation and cannot be readily validated in the clinic.
Further technology advances such as nanomaterials, organoid cultures, and gene editing are expanding emerging novel therapies. Taken together, convergence of technologies and integration of large-scale comprehensive datasets lead to new concepts in biomedical research and drug discovery, exemplified in the fields of systems biology and systems pharmacology (Plenge, 2019). In this context, pharmacogenomics serves as an integrative science for the purpose of translation into clinical use, encompassing a broad spectrum of disciplines. Those engaged in pharmacogenomics must also recognize the relevance of economic, ethical, and legal issues—critical elements for successful translation in health care.
B. Implementation of Pharmacogenomics: Regulatory Hurdles, Economics, and Ethics
Countless pharmacogenomics biomarker tests continue to emerge, but approval by regulatory agencies and acceptance by health insurance companies and institutions require valid evidence of clinical utility. The FDA has developed guidelines that must be met for approval, often relying on clinical trials to test efficacy in the targeted population compared with an equivalent biomarker-negative control. Considering the cost of such trials, potentially matching that of traditional drug trials, cost-benefit analyses are essential, showing that the biomarker-drug combination is superior, or the drug per se would not meet criteria for FDA approval in the general patient population, as demonstrated with trastuzumab in the treatment of breast cancer (Jacobs et al., 2022).
Pharmacoeconomics assesses the cost-benefit ratios for therapeutic inventions, including application of pharmacogenomics compared with health care as usual. The application of NGS to guide therapeutic decisions raises additional issues such as incidental findings of pathogenic variants (Deverka et al., 2020). Ultimately, it is important to consider alternative strategies in reaching a clinical decision that might be more cost-effective than pharmacogenomics. For example, in the case of simvastatin and genetic variants of SLCO1B1 when high-dose simvastatin is required to lower cholesterol levels, causing potential toxicity, prescribing alternative statins obviates the need for SLCO1B1 genetic data. Real-life data are still sparse and will require standardized real-world evidence design. Thus far, few studies address evidence review standards, payer engagement in real-world evidence study design, and education of payers and providers concerning the use of NGS (Deverka et al., 2020).
Use of genetic data invokes sensitive issues regarding confidentiality, misuse by third parties, and questions with relevance to the patient’s family if deleterious mutations are discovered. Concerns about misuse of genetic information and discrimination had been allayed in the United States with passage in 2008 of the Genetic Information Nondiscrimination Act, ensuring that genetic information not be used in health insurance and employment decisions. This critical civil right bill has paved the way for genetic information to gain traction in health care. For patients, the right to know or to be shielded from findings of deleterious variants must be clarified by signed informed consent in clinical trials.
The main ethical principle in medicine is to do no harm and to adhere to the main guiding principles: beneficence, justice, equality. Substantial discrepancies in health care access exist between various populations in the United States. Despite scientific advances in the medical sciences, life expectancy has been declining for a number of years, only in part accounted for by the COVID pandemic, with Black and U.S. indigenous populations disproportionally affected. Social exclusion of minority populations in health care remains a pernicious problem. Bracic et al. (2022) argue that attempts to overcome such disparities in the context of dominant-group and minoritized-group social behaviors tend to reinforce “exclusion cycles.” Expanding use of big data and AI-based systems in medicine to solve these issues carry a danger of reinforcing such cycles when founded on biased information (Bracic et al., 2022). Nevertheless, personalized medicine inevitably progresses toward convergence and integration of all medical, genomics, personal, cultural, and socioeconomic factors, using widely accessible personal electronic health care records (Hicks et al., 2016). To what extent this approach can supplant the intuitive judgments of experienced health care providers remains an open question, but observing beneficence, justice, and equality is imperative.
Despite medical advances, death rates for the two leading causes, heart disease and cancer, have remained at a high rate (approximately 600,000 per year), and life expectancy has declined in the United States over the past several years. Genomics medicine promises improved outcomes but still has to be judiciously integrated into common clinical practice to become fully effective.
To summarize, progress in personalized health care requires convergence of multiple disciplines and technologies to reveal connections between diverse elements that are entangled and influence each other. Understanding such complexities may emerge from artificial intelligence and machine learning in the future. As pharmacogenomics serves as an important link, basic genomics research must strive to integrate key aspects of personalized medicine to enhance clinical translation. Yet, as stated by impressionist painter Vincent van Gough: “We are far from the time when we can understand the peculiar relationships between one or the other fragment of nature which however complement and reinforce each other.”
Abbreviations
- ADR
adverse drug reaction
- AEI
allelic expression imbalance
- AI
artificial intelligence
- APOBEC
apolipoprotein b mRNA-editing enzyme, catalytic polypeptide-like
- CFTR
cystic fibrosis transmembrane conductance regulator
- CPIC
Clinical Pharmacogenomics Implementation Consortium
- CYP
CYP450 cytochrome P450 monooxygenase
- ctDNA
circulating tumor DNA
- dbGaP
Database of Genotypes and Phenotypes
- DDI
drug-drug interactions
- ER
estrogen receptor
- FDA
Food and Drug Administration
- GWAS
genome-wide association studies
- HDR
homology-directed repair
- HLA
human leukocyte antigen
- LD
linkage disequilibrium
- lncRNA
long noncoding RNA
- MDR1
multidrug resistance protein 1
- MSI
microsatellite instability
- NGS
next-generation sequencing
- NIH
National Institutes of Health
- nsSNPS
nonsynonymous single nucleotide polymorphisms
- OMIM
Online Mendelian Inheritance in Man
- PARPi
poly-adenosine diphosphate-ribose polymerase (PARP) inhibitor
- seq
sequencing
- SNP
single nucleotide polymorphism
- TMB
tumor mutational burden
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Sadee, Wang, Hartmann, Toland.
Footnotes
This work was supported by the National Institutes of Health, National Institute of General Medical Sciences [R35-GM140845] (D.W.), and National Cancer Institute [R01-CA215151-01A1] (A.E.T.). The Genotype-Tissue Expression Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health and by National Cancer Institute, National Human Genome Research Institute, National Heart, Lung, and Blood Institute, National Institute on Drug Abuse, National Institute of Mental Health, and National Institute of Neurological Disorders and Stroke.
No author has an actual or perceived conflict of interest with the contents of this article.
References
- Aaltonen LAPeltomäki PLeach FSSistonen PPylkkänen LMecklin JPJärvinen HPowell SMJen JHamilton SR, et al. (1993) Clues to the pathogenesis of familial colorectal cancer. Science 260:812–816. [DOI] [PubMed] [Google Scholar]
- Abdelhalim HBerber ALodi MJain RNair APappu APatel KVenkat VVenkatesan CWable R, et al. (2022) Artificial intelligence, healthcare, clinical genomics, and pharmacogenomics approaches in precision medicine. Front Genet 13:929736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleksakhina SN, Imyanitov EN (2021) Cancer therapy guided by mutation tests: current status and perspectives. Int J Mol Sci 22:10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, Muir TW (2011) Spreading chromatin into chemical biology. ChemBioChem 12:264–279. [DOI] [PubMed] [Google Scholar]
- Alvarez RV, Mariño-Ramírez L, Landsman D (2021) Transcriptome annotation in the cloud: complexity, best practices, and cost. Gigascience 10:giaa163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin NByrne EJohnson JChenevix-Trench GWalter SNolte IMVink JMRawal RMangino MTeumer A, et al. ; kConFab Investigators (2012) Genome-wide association analysis of coffee drinking suggests association with CYP1A1/CYP1A2 and NRCAM. Mol Psychiatry 17:1116–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashcraft K, Grande K, Bristow SL, Moyer N, Schmidlen T, Moretz C, Wick JA, Blaxall BC (2022) Validation of pharmacogenomic interaction probability (PIP) scores in predicting drug–gene, drug–drug–gene, and drug–gene–gene interaction risks in a large patient population. J Pers Med 12:1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asiimwe IG, Zhang EJ, Osanlou R, Jorgensen AL, Pirmohamed M (2021) Warfarin dosing algorithms: a systematic review. Br J Clin Pharmacol 87:1717–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman JDLi AHMarcketta ASun DMbatchou JKessler MDBenner CLiu DLocke AEBalasubramanian S, et al. ; Regeneron Genetics Center; DiscovEHR (2021) Exome sequencing and analysis of 454,787 UK Biobank participants. Nature 599:628–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar MA, Setiawan D, Hak E, Wilffert B (2017) Pharmacogenetics of drug-drug interaction and drug-drug-gene interaction: a systematic review on CYP2C9, CYP2C19 and CYP2D6. Pharmacogenomics 18:701–739. [DOI] [PubMed] [Google Scholar]
- Bakerlee CW, Nguyen Ba AN, Shulgina Y, Rojas Echenique JI, Desai MM (2022) Idiosyncratic epistasis leads to global fitness-correlated trends. Science 376:630–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrie ES, Hartmann K, Lee SH, Frater JT, Seweryn M, Wang D, Sadee W (2017) The CHRNA5/CHRNA3/CHRNB4 nicotinic receptor regulome: genomic architecture, regulatory variants, and clinical associations. Hum Mutat 38:112–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle A, Brown CD, Engelhardt BE, Montgomery SB; GTEx Consortium (2017) Genetic effects on gene expression across human tissues. Nature 550:204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauchner H, Golub RM, Fontanarosa PB (2016) Data sharing: an ethical and scientific imperative. JAMA 315:1237–1239. [DOI] [PubMed] [Google Scholar]
- Becker JBArnold APBerkley KJBlaustein JDEckel LAHampson EHerman JPMarts SSadee WSteiner M, et al. (2005) Strategies and methods for research on sex differences in brain and behavior. Endocrinology 146:1650–1673. [DOI] [PubMed] [Google Scholar]
- Beinse GJust PALe Frere Belda MALaurent-Puig PJacques SKoual MGarinet SLeroy KDelanoy NBlons H, et al. (2022) Discovery and validation of a transcriptional signature identifying homologous recombination-deficient breast, endometrial and ovarian cancers. Br J Cancer 127:1123–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas A, De S (2021) Drivers of dynamic intratumor heterogeneity and phenotypic plasticity. Am J Physiol Cell Physiol 320:C750–C760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracic A, Callier SL, Price WN 2nd (2022) Exclusion cycles: reinforcing disparities in medicine. Science 377:1158–1160. [DOI] [PubMed] [Google Scholar]
- Brett JO, Spring LM, Bardia A, Wander SA (2021) ESR1 mutation as an emerging clinical biomarker in metastatic hormone receptor-positive breast cancer. Breast Cancer Res 23:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DG, Wobst HJ (2021) A decade of FDA-approved drugs (2010-2019): trends and future directions. J Med Chem 64:2312–2338. [DOI] [PubMed] [Google Scholar]
- Bu Y, Gelman IH (2007) v-Src-mediated down-regulation of SSeCKS metastasis suppressor gene promoter by the recruitment of HDAC1 into a USF1-Sp1-Sp3 complex. J Biol Chem 282:26725–26739. [DOI] [PubMed] [Google Scholar]
- Budd WTMJR, Dilts JR, O’Hanlon K, Woody JR, Bostwick DG, Drury JR, Reynolds T (2016) Next generation sequencing reveals disparate population frequencies among cytochrome P450 genes: clinical pharmacogenmoics of the CYP2 family. Int J Comput Biol Drug Des 9:34. [Google Scholar]
- Buijs SM, Hoop EO, Braal CL, van Rosmalen MM, Drooger JC, van Rossum-Schornagel QC, Vastbinder MB, Koolen SLW, Jager A, Mathijssen RHJ (2023) The impact of endoxifen-guided tamoxifen dose reductions on endocrine side-effects in patients with primary breast cancer. ESMO Open 8:100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonamici SWilliams JMorrissey MWang AGuo RVattay AHsiao KYuan JGreen JOspina B, et al. (2010) Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Sci Transl Med 2:51ra70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacabelos R, Cacabelos N, Carril JC (2019) The role of pharmacogenomics in adverse drug reactions. Expert Rev Clin Pharmacol 12:407–442. [DOI] [PubMed] [Google Scholar]
- Campbell JM, Stephenson MD, Bateman E, Peters MD, Keefe DM, Bowen JM (2017) Irinotecan-induced toxicity pharmacogenetics: an umbrella review of systematic reviews and meta-analyses. Pharmacogenomics J 17:21–28. [DOI] [PubMed] [Google Scholar]
- Carranza-Leon D, Dickson AL, Gaedigk A, Stein CM, Chung CP (2021) CYP2D6 genotype and reduced codeine analgesic effect in real-world clinical practice. Pharmacogenomics J 21:484–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudle KE, Dunnenberger HM, Freimuth RR, Peterson JF, Burlison JD, Whirl-Carrillo M, Scott SA, Rehm HL, Williams MS, Klein TE,, et al. (2017) Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet Med 19:215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchin E, Stocco G (2020) Pharmacogenomics and personalized medicine. Genes (Basel) 11:679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cercek ALumish MSinopoli JWeiss JShia JLamendola-Essel MEl Dika IHSegal NShcherba MSugarman R, et al. (2022) PD-1 blockade in mismatch repair-deficient, locally advanced rectal cancer. N Engl J Med 386:2363–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabon JJSimmons ADLovejoy AFEsfahani MSNewman AMHaringsma HJKurtz DMStehr HScherer FKarlovich CA, et al. (2016) Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 7:11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CWH, Law BMH, So WKW, Chow KM, Waye MMY (2020) Pharmacogenomics of breast cancer: highlighting CYP2D6 and tamoxifen. J Cancer Res Clin Oncol 146:1395–1404. [DOI] [PubMed] [Google Scholar]
- Chang AL, Oro AE (2012) Initial assessment of tumor regrowth after vismodegib in advanced basal cell carcinoma. Arch Dermatol 148:1324–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A (2011) PARP inhibitors: its role in treatment of cancer. Chin J Cancer 30:463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lin R, Lin W, Chen Q, Ye D, Li J, Feng J, Cheng W, Zhang M, Qi Y (2022) An immune gene signature to predict prognosis and immunotherapeutic response in lung adenocarcinoma. Sci Rep 12:8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TO’Donnell PHMiddlestadt MRuhnke GWDanahey Kvan Wijk XMRChoksi AKnoebel RHartman SYeo KJ, et al. (2023) Implementation of pharmacogenomics into inpatient general medicine. Pharmacogenet Genomics 33:19–23. [DOI] [PubMed] [Google Scholar]
- Cole MP, Jones CT, Todd ID (1971) A new anti-oestrogenic agent in late breast cancer. An early clinical appraisal of ICI46474. Br J Cancer 25:270–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JMNworu ACMohammad SJLi LLi CLi CSchwendeman ECefalu MAbdel-Rasoul MSun JW, et al. (2022) Regulatory variants in a novel distal enhancer regulate the expression of CYP3A4 and CYP3A5. Clin Transl Sci 15:2720–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JM, Wang D (2020) Cis-acting regulatory elements regulating CYP3A4 transcription in human liver. Pharmacogenet Genomics 30:107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JM, Wang D (2022) Regulation of CYP3A4 and CYP3A5 by a lncRNA: a potential underlying mechanism explaining the association between CYP3A4*1G and CYP3A metabolism. Pharmacogenet Genomics 32:16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper YATeyssier NDräger NMGuo QDavis JESattler SMYang ZPatel AWu SKosuri S, et al. (2022) Functional regulatory variants implicate distinct transcriptional networks in dementia. Science 377:eabi8654. [DOI] [PubMed] [Google Scholar]
- Corcoran RBAndré TAtreya CESchellens JHMYoshino TBendell JCHollebecque AMcRee AJSiena SMiddleton G, et al. (2018) Combined BRAF, EGFR, and MEK inhibition in patients with BRAFV600E-mutant colorectal cancer. Cancer Discov 8:428–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corponi F, Fabbri C, Serretti A (2019) Pharmacogenetics and depression: a critical perspective. Psychiatry Investig 16:645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig DJ, Bailey MM, Noe OB, Williams KK, Stanbery L, Hamouda DM, Nemunaitis JJ (2022) Subclonal landscape of cancer drives resistance to immune therapy. Cancer Treat Res Commun 30:100507. [DOI] [PubMed] [Google Scholar]
- Dalle Fratte CGagno SRoncato RPolesel JZanchetta MBuzzo MPosocco BDe Mattia EBorsatti RPuglisi F, et al. (2023) CYP2D6 and CYP2C8 pharmacogenetics and pharmacological interactions to predict imatinib plasmatic exposure in GIST patients. Br J Clin Pharmacol 89:1089–1098. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N (2002) Capturing chromosome conformation. Science 295:1306–1311. [DOI] [PubMed] [Google Scholar]
- Derendorf H, Lesko LJ, Chaikin P, Colburn WA, Lee P, Miller R, Powell R, Rhodes G, Stanski D, Venitz J (2000) Pharmacokinetic/pharmacodynamic modeling in drug research and development. J Clin Pharmacol 40:1399–1418. [PubMed] [Google Scholar]
- Dessinioti C, Plaka M, Soura E, Mortaki D, Papaxoinis G, Gogas H, Stratigos AJ (2019) A practical guide for the follow-up of patients with advanced basal cell carcinoma during treatment with Hedgehog pathway inhibitors. Oncologist 24:e755–e764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deverka PA, Douglas MP, Phillips KA (2020) Use of real-world evidence in US payer coverage decision-making for next-generation sequencing-based tests: challenges, opportunities, and potential solutions. Value Health 23:540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domchek SMPostel-Vinay SIm SAPark YHDelord JPItaliano AAlexandre JYou BBastian SKrebs MG, et al. (2020) Olaparib and durvalumab in patients with germline BRCA-mutated metastatic breast cancer (MEDIOLA): an open-label, multicentre, phase 1/2, basket study. Lancet Oncol 21:1155–1164. [DOI] [PubMed] [Google Scholar]
- Downey J, Lam JCK, Li VOK, Gozes I (2022) Somatic mutations and Alzheimer’s disease. J Alzheimers Dis 90:475–493. [DOI] [PubMed] [Google Scholar]
- Duarte JD, Cavallari LH (2021) Pharmacogenetics to guide cardiovascular drug therapy. Nat Rev Cardiol 18:649–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn GP, Old LJ, Schreiber RD (2004) The three Es of cancer immunoediting. Annu Rev Immunol 22:329–360. [DOI] [PubMed] [Google Scholar]
- Elens L, van Gelder T, Hesselink DA, Haufroid V, van Schaik RH (2013) CYP3A4*22: promising newly identified CYP3A4 variant allele for personalizing pharmacotherapy. Pharmacogenomics 14:47–62. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedorczuk K, Chen J (2022) Molecular structures reveal synergistic rescue of Δ508 CFTR by Trikafta modulators. Science 378:284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiil-Flynn SMButler BCarroll MCohen-Sasson OCraig CGuibault LJaszi PJütte BJKatz AQuintais JP, et al. (2022) Legal reform to enhance global text and data mining research. Science 378:951–953. [DOI] [PubMed] [Google Scholar]
- Forester BPParikh SVWeisenbach SAjilore OVahia IRothschild AJThase MEDunlop BWDeBattista CConway CR, et al. (2021) Combinatorial pharmacogenomic testing improves outcomes for older adults with depression. Focus Am Psychiatr Publ 19:76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman ANKlabunde CNWiant KEnewold LGray SWFilipski KKKeating NLLeonard DGBLively TMcNeel TS, et al. (2018) Use of next-generation sequencing tests to guide cancer treatment: results from a nationally representative survey of oncologists in the United States. JCO Precis Oncol 2:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S, Liu X, Luo M, Xie K, Nice EC, Zhang H, Huang C (2017) Proteogenomic studies on cancer drug resistance: towards biomarker discovery and target identification. Expert Rev Proteomics 14:351–362. [DOI] [PubMed] [Google Scholar]
- Gallou CLonguemaux SDeloménie CMéjean AMartin NMartinet SPalais GBouvier RDroz DKrishnamoorthy R, et al. (2001) Association of GSTT1 non-null and NAT1 slow/rapid genotypes with von Hippel-Lindau tumour suppressor gene transversions in sporadic renal cell carcinoma. Pharmacogenetics 11:521–535. [DOI] [PubMed] [Google Scholar]
- García-González X, Cabaleiro T, Herrero MJ, McLeod H, López-Fernández LA (2016) Clinical implementation of pharmacogenetics. Drug Metab Pers Ther 31:9–16. [DOI] [PubMed] [Google Scholar]
- Gerlinger MRowan AJHorswell SMath MLarkin JEndesfelder DGronroos EMartinez PMatthews NStewart A, et al. (2012) Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366:883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson P, Merlin JL, Harlé A (2021) Detection of microsatellite instability: state of the art and future applications in circulating tumour DNA (ctDNA). Cancers (Basel) 13:1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golay J, Andrea AE (2020) Combined anti-cancer strategies based on anti-checkpoint inhibitor antibodies. Antibodies (Basel) 9:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H, Yang Y, Zhang X, Li M, Zhang S, Chen Y (2022) CASPIAN: a method to identify chromatin topological associated domains based on spatial density cluster. Comput Struct Biotechnol J 20:4816–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall JMateo JYuan WMossop HPorta NMiranda SPerez-Lopez RDolling DRobinson DRSandhu S, et al. ; TOPARP-A Investigators (2017) Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov 7:1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin B, Hodgson E, Liddle C (1999) The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol Pharmacol 56:1329–1339. [DOI] [PubMed] [Google Scholar]
- Gorzynski JEGoenka SDShafin KJensen TDFisk DGGrove MESpiteri EPesout TMonlong JBaid G, et al. (2022) Ultrarapid nanopore genome sequencing in a critical care setting. N Engl J Med 386:700–702. [DOI] [PubMed] [Google Scholar]
- GTEx Consortium (2020) The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369:1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner ME (2016) History of orphan drug regulation—United States and beyond. Clin Pharmacol Ther 100:342–343. [DOI] [PubMed] [Google Scholar]
- Haidar CE, Crews KR, Hoffman JM, Relling MV, Caudle KE (2022) Advancing pharmacogenomics from single-gene to preemptive testing. Annu Rev Genomics Hum Genet 23:449–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring B, Wissel S, Manson JE (2022) Somatic mutations and clonal hematopoiesis as drivers of age-related cardiovascular risk. Curr Cardiol Rep 24:1049–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann K, Seweryn M, Sadee W (2022) Interpreting coronary artery disease GWAS results: a functional genomics approach assessing biological significance. PLoS One 17:e0244904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattinger CM, Casotti C, Patrizio MP, Luppi S, Fantoni L, Scotlandi K, Ibrahim T, Serra M (2022) Pharmacogenomic profiling of cisplatin-resistant and -sensitive human osteosarcoma cell lines by multimodal targeted next generation sequencing. Int J Mol Sci 23:4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser AS, Attwood MM, Rask-Andersen M, Schiöth HB, Gloriam DE (2017) Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov 16:829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidari N, Phanstiel DH, He C, Grubert F, Jahanbani F, Kasowski M, Zhang MQ, Snyder MP (2014) Genome-wide map of regulatory interactions in the human genome. Genome Res 24:1905–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich MCJoensuu HDemetri GDCorless CLApperley JFletcher JASoulieres DDirnhofer SHarlow ATown A, et al. ; Imatinib Target Exploration Consortium Study B2225 (2008) Phase II, open-label study evaluating the activity of imatinib in treating life-threatening malignancies known to be associated with imatinib-sensitive tyrosine kinases. Clin Cancer Res 14:2717–2725. [DOI] [PubMed] [Google Scholar]
- Hernandez WGamazon ERAquino-Michaels KPatel SO’Brien TJHarralson AFKittles RABarbour ATuck MMcIntosh SD, et al. (2014) Ethnicity-specific pharmacogenetics: the case of warfarin in African Americans. Pharmacogenomics J 14:223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks JK, Dunnenberger HM, Gumpper KF, Haidar CE, Hoffman JM (2016) Integrating pharmacogenomics into electronic health records with clinical decision support. Am J Health Syst Pharm 73:1967–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins GA, Georgoff P, Nikolian V, Allyn-Feuer A, Pauls B, Higgins R, Athey BD, Alam HE (2017) Network reconstruction reveals that valproic acid activates neurogenic transcriptional programs in adult brain following traumatic injury. Pharm Res 34:1658–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA 106:9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TT, Gift M, Alexander E (2022) Prioritizing pharmacogenomics implementation initiates: a survey of healthcare professionals. Per Med 19:15–23. [DOI] [PubMed] [Google Scholar]
- Hoehe MR, Morris-Rosendahl DJ (2018) The role of genetics and genomics in clinical psychiatry. Dialogues Clin Neurosci 20:169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzinger ERGrady BRitchie MDRibaudo HJAcosta EPMorse GDGulick RMRobbins GKClifford DBDaar ES, et al. (2012) Genome-wide association study of plasma efavirenz pharmacokinetics in AIDS Clinical Trials Group protocols implicates several CYP2B6 variants. Pharmacogenet Genomics 22:858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Zhang S, Wen X, Sadee W, Wang D, Yang S, Li L (2022) Transcription factors and ncRNAs associated with CYP3A expression in human liver and small intestine assessed with weighted gene co-expression network analysis. Biomedicines 10:3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Anderle P, Bussey KJ, Barbacioru C, Shankavaram U, Dai Z, Reinhold WC, Papp A, Weinstein JN, Sadée W (2004) Membrane transporters and channels: role of the transportome in cancer chemosensitivity and chemoresistance. Cancer Res 64:4294–4301. [DOI] [PubMed] [Google Scholar]
- Hudziak RM, Lewis GD, Winget M, Fendly BM, Shepard HM, Ullrich A (1989) p185HER2 monoclonal antibody has antiproliferative effects in vitro and sensitizes human breast tumor cells to tumor necrosis factor. Mol Cell Biol 9:1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui DXiao BDikilitas OFreimuth RRIrvin MRJarvik GPKottyan LKullo ILimdi NALiu C, et al. (2022) Quantifying factors that affect polygenic risk score performance across diverse ancestries and age groups for body mass index, in Biocomputing 2023 (Altman RB, Hunter L, Ritchie MD, Murray T, Klein TE eds) pp 437–448, World Scientific, Hackensack, NJ. [PMC free article] [PubMed] [Google Scholar]
- Ingelman-Sundberg M (2022) Cytochrome P450 polymorphism: from evolution to clinical use. Adv Pharmacol 95:393–416. [DOI] [PubMed] [Google Scholar]
- Jacobs AT, Martinez Castaneda-Cruz D, Rose MM, Connelly L (2022) Targeted therapy for breast cancer: an overview of drug classes and outcomes. Biochem Pharmacol 204:115209. [DOI] [PubMed] [Google Scholar]
- Jagadeesh KA, Wu DJ, Birgmeier JA, Boneh D, Bejerano G (2017) Deriving genomic diagnoses without revealing patient genomes. Science 357:692–695. [DOI] [PubMed] [Google Scholar]
- Jang SKEvans LFialkowski AArnett DKAshley-Koch AEBarnes KCBecker DMBis JCBlangero JBleecker ER, et al. (2022) Rare genetic variants explain missing heritability in smoking. Nat Hum Behav 6:1577–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaruthamsophon K, Thomson PJ, Sukasem C, Naisbitt DJ, Pirmohamed M (2022) HLA allele-restricted immune-mediated adverse drug reactions: framework for genetic prediction. Annu Rev Pharmacol Toxicol 62:509–529. [DOI] [PubMed] [Google Scholar]
- Jayaram N, Usvyat D, R Martin AC (2016) Evaluating tools for transcription factor binding site prediction. BMC Bioinformatics 17:547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetter A, Kullak-Ublick GA (2020) Drugs and hepatic transporters: a review. Pharmacol Res 154:104234. [DOI] [PubMed] [Google Scholar]
- Ji J, Wang C, Fakih M (2022) Targeting KRAS G12C-mutated advanced colorectal cancer: research and clinical developments. OncoTargets Ther 15:747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Si Y, McMillin GA, Lyon E (2018) Clinical pharmacogenomics testing in the era of next generation sequencing: challenges and opportunities for precision medicine. Expert Rev Mol Diagn 18:411–421. [DOI] [PubMed] [Google Scholar]
- Johnson AD, Zhang Y, Papp AC, Pinsonneault JK, Lim JE, Saffen D, Dai Z, Wang D, Sadée W (2008) Polymorphisms affecting gene transcription and mRNA processing in pharmacogenetic candidate genes: detection through allelic expression imbalance in human target tissues. Pharmacogenet Genomics 18:781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SW, Ball AL, Chadwick AE, Alfirevic A (2021) The role of mitochondrial DNA variation in drug response: a systematic review. Front Genet 12:698825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan VC (2021) 50th anniversary of the first clinical trial with ICI 46,474 (tamoxifen): then what happened? Endocr Relat Cancer 28:R11–R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser J (2021) NIH’s “precision nutrition” bet aims for individualized diets. Science 371:552. [DOI] [PubMed] [Google Scholar]
- Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R (2007) MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci USA 104:3432–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller KL, Adise S (2016) Variation in the ability to taste bitter thiourea compounds: implications for food acceptance, dietary intake, and obesity risk in children. Annu Rev Nutr 36:157–182. [DOI] [PubMed] [Google Scholar]
- Khor CCWinter SSutiman NMürdter TEChen SLim JSLLi ZLi JSim KSGanchev B, et al. (2023) Cross-ancestry genome-wide association study defines the extended CYP2D6 locus as the principal genetic determinant of endoxifen plasma concentrations. Clin Pharmacol Ther 113:712–723. [DOI] [PubMed] [Google Scholar]
- Kim JA, Ceccarelli R, Lu CY (2021) Pharmacogenomic biomarkers in US FDA-approved drug labels (2000-2020). J Pers Med 11:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchheiner J, Henckel HB, Franke L, Meineke I, Tzvetkov M, Uebelhack R, Roots I, Brockmöller J (2005) Impact of the CYP2D6 ultra-rapid metabolizer genotype on doxepin pharmacokinetics and serotonin in platelets. Pharmacogenet Genomics 15:579–587. [DOI] [PubMed] [Google Scholar]
- Klomp SD, Manson ML, Guchelaar HJ, Swen JJ (2020) Phenoconversion of cytochrome P450 metabolism: a systematic review. J Clin Med 9:2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezevic CE, Clarke W (2020) Cancer chemotherapy: the case for therapeutic drug monitoring. Ther Drug Monit 42:6–19. [DOI] [PubMed] [Google Scholar]
- Kondrashova ONguyen MShield-Artin KTinker AVTeng NNHHarrell MIKuiper MJHo GYBarker HJasin M, et al. ; AOCS Study Group (2017) Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov 7:984–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumsiek JSuhre KEvans AMMitchell MWMohney RPMilburn MVWägele BRömisch-Margl WIllig TAdamski J, et al. (2012) Mining the unknown: a systems approach to metabolite identification combining genetic and metabolic information. PLoS Genet 8:e1003005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehl PZhang JLin YLamba JAssem MSchuetz JWatkins PBDaly AWrighton SAHall SD, et al. (2001) Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet 27:383–391. [DOI] [PubMed] [Google Scholar]
- Kuipers J, Jahn K, Beerenwinkel N (2017) Advances in understanding tumour evolution through single-cell sequencing. Biochim Biophys Acta Rev Cancer 1867:127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantuejoul SSound-Tsao MCooper WAGirard NHirsch FRRoden ACLopez-Rios FJain DChou TYMotoi N, et al. (2020) PD-L1 testing for lung cancer in 2019: perspective from the IASLC Pathology Committee. J Thorac Oncol 15:499–519. [DOI] [PubMed] [Google Scholar]
- Le DTUram JNWang HBartlett BRKemberling HEyring ADSkora ADLuber BSAzad NSLaheru D, et al. (2015) PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 372:2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt E, Lask G, Martin S (2019) Sonic Hedgehog pathway inhibition in the treatment of advanced basal cell carcinoma. Curr Treat Options Oncol 20:84. [DOI] [PubMed] [Google Scholar]
- Ledermann JHarter PGourley CFriedlander MVergote IRustin GScott CMeier WShapira-Frommer RSafra T, et al. (2012) Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med 366:1382–1392. [DOI] [PubMed] [Google Scholar]
- Lee MS, Pant S (2021) Personalizing medicine with germline and somatic sequencing in advanced pancreatic cancer: current treatments and novel opportunities. Am Soc Clin Oncol Educ Book 41:1–13. [DOI] [PubMed] [Google Scholar]
- Lee SH, Ahn WY, Seweryn M, Sadee W (2018) Combined genetic influence of the nicotinic receptor gene cluster CHRNA5/A3/B4 on nicotine dependence. BMC Genomics 19:826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtisalo MTaskinen STarkiainen EKNeuvonen MViinamaki JPaile-Hyvarinen MLilius TOTapaninen TBackman JTTornio A, et al. (2023) A comprehensive pharmacogenomic study indicates roles for SLCO1B1, ABCG2 and SLCO2B1 in rosuvastatin pharmacokinetics. Br J Clin Pharmacol 89:242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BSangkuhl KKeat KWhaley RMWoon MVerma SDudek STuteja SVerma AWhirl-Carrillo M, et al. (2022) How to run the Pharmacogenomics Clinical Annotation Tool (PharmCAT). Clin Pharmacol Ther DOI: 10.1002/cpt.2790 [published ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Liu Y, Liu S, Zhu X, Wu L, Zhu Y, Zhao D, Xu X, Chemparathy A, Wang H, et al. (2022) Nested epistasis enhancer networks for robust genome regulation. Science 377:1077–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YS, Dowling AL, Quigley SD, Farin FM, Zhang J, Lamba J, Schuetz EG, Thummel KE (2002) Co-regulation of CYP3A4 and CYP3A5 and contribution to hepatic and intestinal midazolam metabolism. Mol Pharmacol 62:162–172. [DOI] [PubMed] [Google Scholar]
- Liu S, Crawford DC (2022) Maturation and application of phenome-wide association studies. Trends Genet 38:353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Medina AI, Chahal CAA, Luzum JA (2022) The genetics of drug-induced QT prolongation: evaluating the evidence for pharmacodynamic variants. Pharmacogenomics 23:543–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukola ABuchwald JGupta RPalviainen THällfors JTikkanen EKorhonen TOllikainen MSarin APRipatti S, et al. (2015) A genome-wide association study of a biomarker of nicotine metabolism. PLoS Genet 11:e1005498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchini CBibeau FLigtenberg MJLSingh NNottegar ABosse TMiller RRiaz NDouillard JYAndre F, et al. (2019) ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: a systematic review-based approach. Ann Oncol 30:1232–1243. [DOI] [PubMed] [Google Scholar]
- Ludford KCohen RSvrcek MFoo WCColle RParc YThomas JVMorris VKKopetz SChang GJ, et al. (2021) Pathological tumor response following immune checkpoint blockade for deficient mismatch repair advanced colorectal cancer. J Natl Cancer Inst 113:208–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Lu AY (2011) Pharmacogenetics, pharmacogenomics, and individualized medicine. Pharmacol Rev 63:437–459. [DOI] [PubMed] [Google Scholar]
- Maas SA, Fallon JF (2005) Single base pair change in the long-range Sonic hedgehog limb-specific enhancer is a genetic basis for preaxial polydactyly. Dev Dyn 232:345–348. [DOI] [PubMed] [Google Scholar]
- Mager LFBurkhard RPett NCooke NCABrown KRamay HPaik SStagg JGroves RAGallo M, et al. (2020) Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 369:1481–1489. [DOI] [PubMed] [Google Scholar]
- Malki MA, Pearson ER (2020) Drug-drug-gene interactions and adverse drug reactions. Pharmacogenomics J 20:355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malla M, Loree JM, Kasi PM, Parikh AR (2022) Using circulating tumor DNA in colorectal cancer: current and evolving practices. J Clin Oncol 40:2846–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani DR, Krug K, Zhang B, Satpathy S, Clauser KR, Ding L, Ellis M, Gillette MA, Carr SA (2022) Cancer proteogenomics: current impact and future prospects. Nat Rev Cancer 22:298–313. [DOI] [PubMed] [Google Scholar]
- Manikandan P, Nagini S (2018) Cytochrome P450 structure, function and clinical significance: a review. Curr Drug Targets 19:38–54. [DOI] [PubMed] [Google Scholar]
- Marcus LFashoyin-Aje LADonoghue MYuan MRodriguez LGallagher PSPhilip RGhosh STheoret MRBeaver JA, et al. (2021) FDA approval summary: pembrolizumab for the treatment of tumor mutational burden-high solid tumors. Clin Cancer Res 27:4685–4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Jiménez CP, Jover R, Donato MT, Castell JV, Gómez-Lechón MJ (2007) Transcriptional regulation and expression of CYP3A4 in hepatocytes. Curr Drug Metab 8:185–194. [DOI] [PubMed] [Google Scholar]
- Mascarenhas R, Pietrzak M, Smith RM, Webb A, Wang D, Papp AC, Pinsonneault JK, Seweryn M, Rempala G, Sadee W (2015) Allele-selective transcriptome recruitment to polysomes primed for translation: protein-coding and noncoding RNAs, and RNA isoforms. PLoS One 10:e0136798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura K, Saito T, Takahashi Y, Ozeki T, Kiyotani K, Fujieda M, Yamazaki H, Kunitoh H, Kamataki T (2004) Identification of a novel polymorphic enhancer of the human CYP3A4 gene. Mol Pharmacol 65:326–334. [DOI] [PubMed] [Google Scholar]
- Maynard AMcCoach CERotow JKHarris LHaderk FKerr DLYu EASchenk ELTan WZee A, et al. (2020) Therapy-induced evolution of human lung cancer revealed by single-cell RNA sequencing. Cell 182:1232–1251.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott JHWolf JHoshitsuki KHuddart RCaudle KEWhirl-Carrillo MSteyger PSSmith RJHCody NRodriguez-Antona C, et al. (2022) Clinical pharmacogenetics implementation consortium guideline for the use of aminoglycosides based on MT-RNR1 genotype. Clin Pharmacol Ther 111:366–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes G, Altman RB (2021) Drug response pharmacogenetics for 200,000 UK Biobank participants. Pac Symp Biocomput 26:184–195. [PMC free article] [PubMed] [Google Scholar]
- Mehta DUber RIngle TLi CLiu ZThakkar SNing BWu LYang JHarris S, et al. (2020) Study of pharmacogenomic information in FDA-approved drug labeling to facilitate application of precision medicine. Drug Discov Today 25:813–820. [DOI] [PubMed] [Google Scholar]
- Mellman I, Coukos G, Dranoff G (2011) Cancer immunotherapy comes of age. Nature 480:480–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz TM, Collins CD, Dennis M, Coxon M, Roberts SA (2022) APOBEC-induced mutagenesis in cancer. Annu Rev Genet 56:229–252. [DOI] [PubMed] [Google Scholar]
- Mino-Kenudson MSchalper KCooper WDacic SHirsch FRJain DLopez-Rios FTsao MSYatabe YBeasley MB, et al. ; IASLC Pathology Committee (2022) Predictive biomarkers for immunotherapy in lung cancer: perspective from the International Association for the Study of Lung Cancer Pathology Committee. J Thorac Oncol 17:1335–1354. [DOI] [PubMed] [Google Scholar]
- Miteva-Marcheva NN, Ivanov HY, Dimitrov DK, Stoyanova VK (2020) Application of pharmacogenetics in oncology. Biomark Res 8:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney SD (2015) Progress towards the integration of pharmacogenomics in practice. Hum Genet 134:459–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli EFulciniti MSamur MKRibeiro CWert-Lamas LHenninger JEGulla AAktas Samur ATodoerti KTalluri S, et al. (2023) A MIR17HG-derived long noncoding RNA provides an essential chromatin scaffold for protein interaction and myeloma growth. Blood 141:391–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morganti S, Tarantino P, Ferraro E, D’Amico P, Duso BA, Curigliano G (2019) Next generation sequencing (NGS): a revolutionary technology in pharmacogenomics and personalized medicine in cancer. Adv Exp Med Biol 1168:9–30. [DOI] [PubMed] [Google Scholar]
- Moriyama TNishii RPerez-Andreu VYang WKlussmann FAZhao XLin TNHoshitsuki KNersting JKihira K, et al. (2016) NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat Genet 48:367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosele FRemon JMateo JWestphalen CBBarlesi FLolkema MPNormanno NScarpa ARobson MMeric-Bernstam F, et al. (2020) Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol 31:1491–1505. [DOI] [PubMed] [Google Scholar]
- Moyer AM, Gandhi MJ (2022) Human leukocyte antigen (HLA) testing in pharmacogenomics, in Pharmacogenomics in Drug Discovery and Development (Yan Q ed) pp 21–45, Springer US, New York. [DOI] [PubMed] [Google Scholar]
- Mueller CM, Rao GR, Miller Needleman KI (2019) Precision medicines’ impact on orphan drug designation. Clin Transl Sci 12:633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugesan KJin DXComment LAFabrizio DHegde PSElvin JAAlexander BLevy MAFrampton GMMontesion M, et al. (2022) Association of CD274 (PD-L1) copy number changes with immune checkpoint inhibitor clinical benefit in non-squamous non-small cell lung cancer. Oncologist 27:732–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najjar-Debbiny R, Gronich N, Weber G, Khoury J, Amar M, Stein N, Goldstein LH, Saliba W (2023) Effectiveness of paxlovid in reducing severe COVID-19 and mortality in high risk patients. Clin Infect Dis 76:e342–e349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naushad SM, Hussain T, Alrokayan SA, Kutala VK (2022) Development of pharmacogenomic algorithm to optimize nateglinide dose for the treatment of type 2 diabetes mellitus. Pharmacol Rep 74:1083–1091. [DOI] [PubMed] [Google Scholar]
- Nicholson WT, Formea CM, Matey ET, Wright JA, Giri J, Moyer AM (2021) Considerations when applying pharmacogenomics to your practice. Mayo Clin Proc 96:218–230. [DOI] [PubMed] [Google Scholar]
- Nicolini A, Ferrari P, Duffy MJ (2018) Prognostic and predictive biomarkers in breast cancer: Past, present and future. Semin Cancer Biol 52:56–73. [DOI] [PubMed] [Google Scholar]
- Nikanjam M, Kato S, Kurzrock R (2022) Liquid biopsy: current technology and clinical applications. J Hematol Oncol 15:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgard NB, Abu-Fadel M (2009) Comparison of prasugrel and clopidogrel in patients with acute coronary syndrome undergoing percutaneous coronary intervention. Vasc Health Risk Manag 5:873–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osanlou O, Pirmohamed M, Daly AK (2018) Pharmacogenetics of adverse drug reactions. Adv Pharmacol 83:155–190. [DOI] [PubMed] [Google Scholar]
- Overman MJLonardi SWong KYMLenz HJGelsomino FAglietta MMorse MAVan Cutsem EMcDermott RHill A, et al. (2018) Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol 36:773–779. [DOI] [PubMed] [Google Scholar]
- Papamichael K, Cheifetz AS (2019) Therapeutic drug monitoring in inflammatory bowel disease: for every patient and every drug? Curr Opin Gastroenterol 35:302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RAMusharoff SASpence JPPimentel HTcheandjieu CMostafavi HSinnott-Armstrong NClarke SLSmith CJDurda PP, et al. ; V.A. Million Veteran Program (2022) Genetic interactions drive heterogeneity in causal variant effect sizes for gene expression and complex traits. Am J Hum Genet 109:1286–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrinos GP, Shuldiner AR (2022) Pharmacogenomics: the low-hanging fruit in the personalized medicine tree. Hum Genet 141:1109–1111. [DOI] [PubMed] [Google Scholar]
- Peng Q, Wilhelmsen KC, Ehlers CL (2021) Common genetic substrates of alcohol and substance use disorder severity revealed by pleiotropy detection against GWAS catalog in two populations. Addict Biol 26:e12877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E (2022) Upstart DNA sequencers could be a “game changer.” Science 376:1257–1258. [DOI] [PubMed] [Google Scholar]
- Perera MACavallari LHLimdi NAGamazon ERKonkashbaev ADaneshjou RPluzhnikov ACrawford DCWang JLiu N, et al. (2013) Genetic variants associated with warfarin dose in African-American individuals: a genome-wide association study. Lancet 382:790–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt SJ, Frankum JR, Punta M, Lise S, Alexander J, Chen Y, Yap TA, Haider S, Tutt ANJ, Lord CJ (2020) Clinical BRCA1/2 reversion analysis identifies hotspot mutations and predicted neoantigens associated with therapy resistance. Cancer Discov 10:1475–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips KA, Veenstra DL, Oren E, Lee JK, Sadee W (2001) Potential role of pharmacogenomics in reducing adverse drug reactions: a systematic review. JAMA 286:2270–2279. [DOI] [PubMed] [Google Scholar]
- Pinsonneault JK, Papp AC, Sadée W (2006) Allelic mRNA expression of X-linked monoamine oxidase a (MAOA) in human brain: dissection of epigenetic and genetic factors. Hum Mol Genet 15:2636–2649. [DOI] [PubMed] [Google Scholar]
- Plana D, Palmer AC, Sorger PK (2022) Independent drug action in combination therapy: implications for precision oncology. Cancer Discov 12:606–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchard DBesse BGroen HJMSouquet PJQuoix EBaik CSBarlesi FKim TMMazieres JNovello S, et al. (2016) Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. Lancet Oncol 17:984–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plenge RM (2019) Priority index for human genetics and drug discovery. Nat Genet 51:1073–1075. [DOI] [PubMed] [Google Scholar]
- Qin W, Hu L, Zhang X, Jiang S, Li J, Zhang Z, Wang X (2019) The diverse function of PD-1/PD-L pathway beyond cancer. Front Immunol 10:2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley DAlumkal JJWyatt AWKothari VFoye ALloyd PAggarwal RKim WLu ESchwartzman J, et al. (2017) Analysis of circulating cell-free DNA identifies multiclonal heterogeneity of BRCA2 reversion mutations associated with resistance to PARP inhibitors. Cancer Discov 7:999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani B, Nakaoka H, Akhondzadeh S, Tekin M, Mahdieh N (2016) Next generation sequencing: implications in personalized medicine and pharmacogenomics. Mol Biosyst 12:1818–1830. [DOI] [PubMed] [Google Scholar]
- Ramamoorthy A, Kim HH, Shah-Williams E, Zhang L (2022) Racial and ethnic differences in drug disposition and response: review of new molecular entities approved between 2014 and 2019. J Clin Pharmacol 62:486–493. [DOI] [PubMed] [Google Scholar]
- Ramirez AHSulieman LSchlueter DJHalvorson AQian JRatsimbazafy FLoperena RMayo KBasford MDeflaux N, et al. ; All of Us Research Program (2022) The All of Us Research Program: Data quality, utility, and diversity. Patterns (N Y) 3:100570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray B, Ozcagli E, Sadee W, Wang D (2019) CYP2D6 haplotypes with enhancer single-nucleotide polymorphism rs5758550 and rs16947 (*2 allele): implications for CYP2D6 genotyping panels. Pharmacogenet Genomics 29:39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relling MV, Evans WE (2015) Pharmacogenomics in the clinic. Nature 526:343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert CGrob JJStroyakovskiy DKaraszewska BHauschild ALevchenko EChiarion Sileni VSchachter JGarbe CBondarenko I, et al. (2019) Five-year outcomes with dabrafenib plus trametinib in metastatic melanoma. N Engl J Med 381:626–636. [DOI] [PubMed] [Google Scholar]
- Roden DM (2016) Predicting drug-induced QT prolongation and torsades de pointes. J Physiol 594:2459–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden DM, McLeod HL, Relling MV, Williams MS, Mensah GA, Peterson JF, Van Driest SL (2019) Pharmacogenomics. Lancet 394:521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadee W, Hartmann K, Seweryn M, Pietrzak M, Handelman SK, Rempala GA (2014) Missing heritability of common diseases and treatments outside the protein-coding exome. Hum Genet 133:1199–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadee W, Wang D, Papp AC, Pinsonneault JK, Smith RM, Moyer RA, Johnson AD (2011) Pharmacogenomics of the RNA world: structural RNA polymorphisms in drug therapy. Clin Pharmacol Ther 89:355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salman B, Al-Khabori M (2021) Applications and challenges in therapeutic drug monitoring of cancer treatment: a review. J Oncol Pharm Pract 27:693–701. [DOI] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J (2012) The long-range interaction landscape of gene promoters. Nature 489:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor Ode Bono JChi KNFizazi KHerrmann KRahbar KTagawa STNordquist LTVaishampayan NEl-Haddad G, et al. ; VISION Investigators (2021) Lutetium-177-PSMA-617 for metastatic castration-resistant prostate cancer. N Engl J Med 385:1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EWBolton EEBrister JRCanese KChan JComeau DCFarrell CMFeldgarden MFine AMFunk K, et al. (2023) Database resources of the National Center for Biotechnology Information in 2023. Nucleic Acids Res 51:D29–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger D, Elsässer SJ (2022) Revisiting sORFs: overcoming challenges to identify and characterize functional microproteins. FEBS J 289:53–74. [DOI] [PubMed] [Google Scholar]
- Selenica PMarra AChoudhury NJGazzo AFalcon CJPatel JPei XZhu YNg CKYCurry M, et al. (2022) APOBEC mutagenesis, kataegis, chromothripsis in EGFR-mutant osimertinib-resistant lung adenocarcinomas. Ann Oncol 33:1284–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servick K (2020) mRNA’s next challenge: will it work as a drug? Science 370:1388–1389. [DOI] [PubMed] [Google Scholar]
- Shah SPMorin RDKhattra JPrentice LPugh TBurleigh ADelaney AGelmon KGuliany RSenz J, et al. (2009) Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461:809–813. [DOI] [PubMed] [Google Scholar]
- Shmais M, Regueiro M, Hashash JG (2021) Proactive versus reactive therapeutic drug monitoring: why, when, and how? Inflamm Intest Dis 7:50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl LMHirsch FRHwang DBotling JLopez-Rios FBubendorf LMino-Kenudson MRoden ACBeasley MBBorczuk A, et al. (2020) The promises and challenges of tumor mutation burden as an immunotherapy biomarker: a perspective from the International Association for the Study of Lung Cancer Pathology Committee. J Thorac Oncol 15:1409–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigorski D, Iżycka-Świeszewska E, Bodnar L (2020) Poly(ADP-ribose) polymerase inhibitors in prostate cancer: molecular mechanisms, and preclinical and clinical data. Target Oncol 15:709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silgado-Guzmán DF, Angulo-Aguado M, Morel A, Niño-Orrego MJ, Ruiz-Torres DA, Contreras Bravo NC, Restrepo CM, Ortega-Recalde O, Fonseca-Mendoza DJ (2022) Characterization of ADME gene variation in Colombian population by exome sequencing. Front Pharmacol 13:931531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LA, Dinan MA, Robinson TJ, Snyderman R (2012) Personalized medicine is more than genomic medicine: confusion over terminology impedes progress towards personalized healthcare. Per Med 9:85–91. [DOI] [PubMed] [Google Scholar]
- Siravegna GMussolin BVenesio TMarsoni SSeoane JDive CPapadopoulos NKopetz SCorcoran RBSiu LL, et al. (2019) How liquid biopsies can change clinical practice in oncology. Ann Oncol 30:1580–1590. [DOI] [PubMed] [Google Scholar]
- Skoda RC, Gonzalez FJ, Demierre A, Meyer UA (1988) Two mutant alleles of the human cytochrome P-450db1 gene (P450C2D1) associated with genetically deficient metabolism of debrisoquine and other drugs. Proc Natl Acad Sci USA 85:5240–5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL (1987) Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235:177–182. [DOI] [PubMed] [Google Scholar]
- Smith RM, Webb A, Papp AC, Newman LC, Handelman SK, Suhy A, Mascarenhas R, Oberdick J, Sadee W (2013) Whole transcriptome RNA-Seq allelic expression in human brain. BMC Genomics 14:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover EH, Fuh K, Konstantinopoulos PA, Matulonis UA, Liu JF (2020) Clinical assays for assessment of homologous recombination DNA repair deficiency. Gynecol Oncol 159:887–898. [DOI] [PubMed] [Google Scholar]
- Subbannayya Y, Di Fiore R, Urru SAM, Calleja-Agius J (2021) The role of omics approaches to characterize molecular mechanisms of rare ovarian cancers: recent advances and future perspectives. Biomedicines 9:1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudlow CGallacher JAllen NBeral VBurton PDanesh JDowney PElliott PGreen JLandray M, et al. (2015) UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Lu Q, Tao X, Cheng B, Yang G (2020) Cyp2C19*2 polymorphism related to clopidogrel resistance in patients with coronary heart disease, especially in the Asian population: a systematic review and meta-analysis. Front Genet 11:576046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegude H, Schnabel A, Zanger UM, Klein K, Eichelbaum M, Burk O (2007) Molecular mechanism of basal CYP3A4 regulation by hepatocyte nuclear factor 4alpha: evidence for direct regulation in the intestine. Drug Metab Dispos 35:946–954. [DOI] [PubMed] [Google Scholar]
- Tevzadze G, Zhuravliova E, Okriashvili N, Narmania N, Barbakadze T, Mikeladze D (2022) Different arrangement of dopamine receptors/NMDA receptors heterocomplexes in the brain regions of a healthy male, female and audiogenic seizure-prone male rats. Am J Biochem Biotechnol 18:195–204. [Google Scholar]
- Thomas CD, Johnson JA (2020) Pharmacogenetic factors affecting β-blocker metabolism and response. Expert Opin Drug Metab Toxicol 16:953–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W (2002) Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell 10:1453–1465. [DOI] [PubMed] [Google Scholar]
- Turongkaravee S, Jittikoon J, Lukkunaprasit T, Sangroongruangsri S, Chaikledkaew U, Thakkinstian A (2021) A systematic review and meta-analysis of genotype-based and individualized data analysis of SLCO1B1 gene and statin-induced myopathy. Pharmacogenomics J 21:296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpin A, Neuzillet C, Colle E, Dusetti N, Nicolle R, Cros J, de Mestier L, Bachet JB, Hammel P (2022) Therapeutic advances in metastatic pancreatic cancer: a focus on targeted therapies. Ther Adv Med Oncol 14:17588359221118019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varayathu H, Sarathy V, Thomas BE, Mufti SS, Naik R (2021) Combination strategies to augment immune check point inhibitors efficacy—implications for translational research. Front Oncol 11:559161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varenhorst CEriksson NJohansson ÅBarratt BJHagström EÅkerblom ASyvänen ACBecker RCJames SKKatus HA, et al. ; PLATO Investigators (2015) Effect of genetic variations on ticagrelor plasma levels and clinical outcomes. Eur Heart J 36:1901–1912. [DOI] [PubMed] [Google Scholar]
- Venkatachalapathy PPadhilahouse SSellappan MSubramanian TKurian SJMiraj SSRao MRaut AAKanwar RKSingh J, et al. (2021) Pharmacogenomics and personalized medicine in type 2 diabetes mellitus: potential implications for clinical practice. Pharm Genomics Pers Med 14:1441–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan S, Rosenthal R, Kanu N, McGranahan N, Bartek J, Quezada SA, Hare J, Harris RS, Swanton C (2018) Perspective: APOBEC mutagenesis in drug resistance and immune escape in HIV and cancer evolution. Ann Oncol 29:563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidula NRich TASartor OYen JHardin ANance TLilly MBNezami MAPatel SPCarneiro BA, et al. (2020) Routine plasma-based genotyping to comprehensively detect germline, somatic, and reversion BRCA mutations among patients with advanced solid tumors. Clin Cancer Res 26:2546–2555. [DOI] [PubMed] [Google Scholar]
- Von Hoff DDLoRusso PMRudin CMReddy JCYauch RLTibes RWeiss GJBorad MJHann CLBrahmer JR, et al. (2009) Inhibition of the Hedgehog pathway in advanced basal-cell carcinoma. N Engl J Med 361:1164–1172. [DOI] [PubMed] [Google Scholar]
- Wang D, Chen H, Momary KM, Cavallari LH, Johnson JA, Sadée W (2008) Regulatory polymorphism in vitamin K epoxide reductase complex subunit 1 (VKORC1) affects gene expression and warfarin dose requirement. Blood 112:1013–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Guo Y, Wrighton SA, Cooke GE, Sadee W (2011) Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics 11:J 11:274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Hartmann K, Seweryn M, Sadee W (2018) Interactions between regulatory variants in CYP7A1 (cholesterol 7α-hydroxylase) promoter and enhancer regions regulate CYP7A1 expression. Circ Genom Precis Med 11:e002082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Johnson AD, Papp AC, Kroetz DL, Sadée W (2005) Multidrug resistance polypeptide 1 (MDR1, ABCB1) variant 3435C>T affects mRNA stability. Pharmacogenet Genomics 15:693–704. [PubMed] [Google Scholar]
- Wang D, Papp AC, Sun X (2015) Functional characterization of CYP2D6 enhancer polymorphisms. Hum Mol Genet 24:1556–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Poi MJ, Sun X, Gaedigk A, Leeder JS, Sadee W (2014) Common CYP2D6 polymorphisms affecting alternative splicing and transcription: long-range haplotypes with two regulatory variants modulate CYP2D6 activity. Hum Mol Genet 23:268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Bennett DA, De Jager PL, Zhang QY, Zhang HY (2021) Genome-wide epistasis analysis for Alzheimer’s disease and implications for genetic risk prediction. Alzheimers Res Ther 13:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins JA, Irshad S, Grigoriadis A, Tutt AN (2014) Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res 16:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt BComino-Méndez Ide Bruijn ITian LMeisel JLGarcía-Murillas IFribbens CCutts RMartelotto LGNg CKY, et al. (2017) Diverse BRCA1 and BRCA2 reversion mutations in circulating cell-free dna of therapy-resistant breast or ovarian cancer. Clin Cancer Res 23:6708–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinshilboum RM, Wang L (2017) Pharmacogenomics: precision medicine and drug response. Mayo Clin Proc 92:1711–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werk AN, Cascorbi I (2014) Functional gene variants of CYP3A4. Clin Pharmacol Ther 96:340–348. [DOI] [PubMed] [Google Scholar]
- Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, Altman RB, Klein TE (2012) Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther 92:414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CM (2018) The pharmacologic role and clinical utility of PCSK9 inhibitors for the treatment of hypercholesterolemia. J Cardiovasc Pharmacol Ther 23:301–308. [DOI] [PubMed] [Google Scholar]
- Woelk CH, Snyder A (2021) Modulating gut microbiota to treat cancer. Science 371:573–574. [DOI] [PubMed] [Google Scholar]
- Wu LZhong YYu XWu DXu PLv LRuan XLiu QFeng YLiu J, et al. (2022) Selective poly adenylation predicts the efficacy of immunotherapy in patients with lung adenocarcinoma by multiple omics research. Anticancer Drugs 33:943–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch RLDijkgraaf GJAlicke BJanuario TAhn CPHolcomb TPujara KStinson JCallahan CATang T, et al. (2009) Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science 326:572–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazar M, Özbek P (2021) In silico tools and approaches for the prediction of functional and structural effects of single-nucleotide polymorphisms on proteins: an expert review. OMICS 25:23–37. [DOI] [PubMed] [Google Scholar]
- Ye M, Lin Y, Pan S, Wang ZW, Zhu X (2021) Applications of multi-omics approaches for exploring the molecular mechanism of ovarian carcinogenesis. Front Oncol 11:745808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip VL, Alfirevic A, Pirmohamed M (2015) Genetics of immune-mediated adverse drug reactions: a comprehensive and clinical review. Clin Rev Allergy Immunol 48:165–175. [DOI] [PubMed] [Google Scholar]
- Yoshida HNomizo TOzasa HTsuji TFunazo TYasuda YAjimizu HYamazoe MKuninaga KOgimoto T, et al. (2021) PD-L1 polymorphisms predict survival outcomes in advanced non-small-cell lung cancer patients treated with PD-1 blockade. Eur J Cancer 144:317–325. [DOI] [PubMed] [Google Scholar]
- Yu J, Wang Y, Ragueneau-Majlessi I (2022) Pharmacokinetic drug-drug interactions with drugs approved by the US Food and Drug Administration in 2020: mechanistic understanding and clinical recommendations. Drug Metab Dispos 50:1–7. [DOI] [PubMed] [Google Scholar]
- Zaborska KE, Lee SA, Garribay D, Cha E, Cummings BP (2018) Deoxycholic acid supplementation impairs glucose homeostasis in mice. PLoS One 13:e0200908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanger UM, Schwab M (2013) Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 138:103–141. [DOI] [PubMed] [Google Scholar]
- Zhai Q, van der Lee M, van Gelder T, Swen JJ (2022) Why we need to take a closer look at genetic contributions to CYP3A activity. Front Pharmacol 13:912618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Xu X, Wei Y, Chen X, Li G, Lu Z, Zhang X, Ren X, Wang S, Qin C (2022) Prognostic role of DNA damage response genes mutations and their association with the sensitivity of olaparib in prostate cancer patients. Cancer Contr 29:10732748221129451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YWong CHBirnbaum RYLi GFavaro RNgan CYLim JTai EPoh HMWong E, et al. (2013) Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature 504:306–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MMa JLi MZhang YJiang BZhao XHuai CShen LZhang NHe L, et al. (2021) Cytochrome P450 enzymes and drug metabolism in humans. Int J Mol Sci 22:12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Lauschke VM (2022) The genetic landscape of major drug metabolizing cytochrome P450 genes-an updated analysis of population-scale sequencing data. Pharmacogenomics J 22:284–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong HZhang JXu ZPan JNWang RHan JJiang MRen RZang LWang H, et al. (2022) Comprehensive analysis of somatic reversion mutations in homologous recombination repair (HRR) genes in a large cohort of Chinese pan-cancer patients. J Cancer 13:1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]