Abstract
Background
The COVID-19 pandemic exerted significant impacts on public health and global economy. Research efforts to develop vaccines at warp speed against SARS-CoV-2 led to novel mRNA, viral vectored, and inactivated vaccines being administered. The current COVID-19 vaccines incorporate the full S protein of the SARS-CoV-2 Wuhan strain but rapidly emerging variants of concern (VOCs) have led to significant reductions in protective efficacies. There is an urgent need to develop next-generation vaccines which could effectively prevent COVID-19.
Methods
PubMed and Google Scholar were systematically reviewed for peer-reviewed papers up to January 2023.
Results
A promising solution to the problem of emerging variants is a DNA vaccine platform since it can be easily modified. Besides expressing whole protein antigens, DNA vaccines can also be constructed to include specific nucleotide genes encoding highly conserved and immunogenic epitopes from the S protein as well as from other structural/non-structural proteins to develop effective vaccines against VOCs. DNA vaccines are associated with low transfection efficiencies which could be enhanced by chemical, genetic, and molecular adjuvants as well as delivery systems.
Conclusions
The DNA vaccine platform offers a promising solution to the design of effective vaccines. The challenge of limited immunogenicity in humans might be solved through the use of genetic modifications such as the addition of nuclear localization signal (NLS) peptide gene, strong promoters, MARs, introns, TLR agonists, CD40L, and the development of appropriate delivery systems utilizing nanoparticles to increase uptake by APCs in enhancing the induction of potent immune responses.
Keywords: DNA vaccine, SARS-CoV-2, Variants, Vaccine efficacy, Plasmid design
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the etiological agent of coronavirus disease 2019 (COVID-19). It has caused an alarming pandemic associated with 766,895,075 infections and 6,935,889 deaths as of 15 May 2023 [1]. The pandemic has also resulted in severe economic losses with the imposition of lockdowns, causing a debilitating burden on healthcare systems. Novel vaccines such as those based on the mRNA, viral-vectored platforms, and traditional inactivated vaccines were given emergency approval for global vaccinations. Each of the vaccine platforms has its own strengths and weaknesses. However, the emergence of SARS-CoV-2 variants of concern (VOCs) which caused continuing waves of infections and reduced protective efficacies of current vaccines against the B.1.1.7 (Alpha), B.1.351 (Beta), B.1.617.2 (Delta), and (B.1.1.529) (Omicron) variants were reported [2]. The prospect of using DNA vaccines to immunize against SARS-CoV-2 is promising because of the ease of production and reduced cost over other vaccine platforms. They can easily be modified to incorporate different antigenic targets simply by changing the gene sequences of the DNA plasmid, making the DNA vaccine platform ideal for rapidly developing vaccines against emerging SARS-CoV-2 VOCs. Therefore, this review discusses the advantages and limitations of DNA vaccines and summarizes current DNA vaccine development against SARS-CoV-2. Furthermore, an insight into the design of DNA vaccines and the strategies to increase the immunological potency of plasmid DNA vaccines in terms of adjuvants, delivery systems, and administration routes are thoroughly reviewed.
2. Review
2.1. Mechanism of DNA vaccines
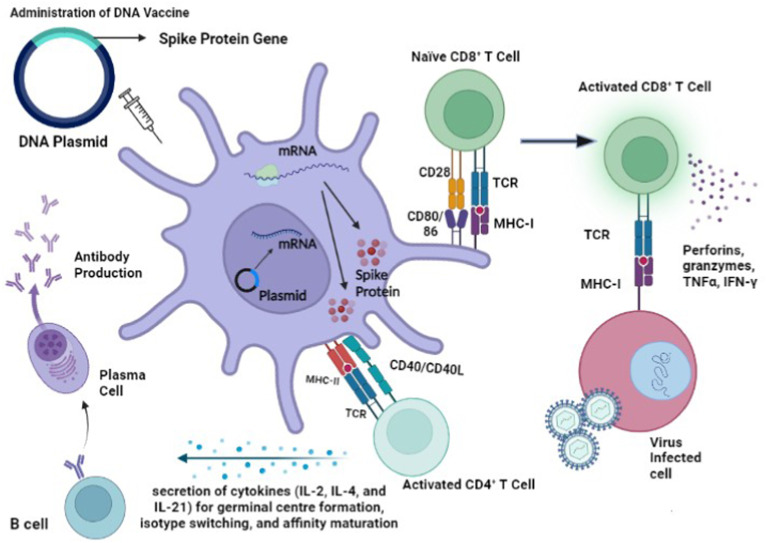
Current DNA vaccines against SARS-CoV-2 are able to elicit both humoral and cellular immune responses against the main antigen, the Spike (S) glycoprotein of the SARS-CoV-2. Eiz-Vesper et al. [3] explained that DNA vaccines which target and transfect muscle cells would likely result in weak antigen presentation due to a lack of co-stimulatory molecules since antigen presenting cells (APCs) allow antigen presentation to major histocompatibility complex (MHC) molecules. The transfection of the recombinant DNA plasmid into professional APCs such as dendritic cells (DCs) would result in a much more potent immune response. Inside the APC, the recombinant DNA plasmid enters the nucleus so that it is transcribed to mRNA. The single stranded RNA would then exit the nucleus to undergo translation to produce the S protein in the cytoplasm which is processed inside the cell to smaller peptides to be presented to naïve B and T cells [4].
DNA vaccines can elicit cellular immune responses by binding to naïve CD4+ T cells to activate them. Degradation of the S viral protein would result in the production of peptide fragments which could be presented to MHC class II molecules to form the MHC class II-peptide complex. T cell receptors (TCRs) of specific CD4+ T cells are able to recognize and bind to these MHC class II-peptide complexes. This resulted in the activation of DCs through the interaction of CD40 with CD40 ligand (CD40L) on the DC. The activation of DCs led to the upregulation of CD80/CD86 which interacted with CD28 to activate naïve CD8+ T cells [5]. Together with the help of CD4+ T cells, activation of CD8+ T cells is induced. Viral peptides are also taken up by APCs that were presented on MHC class I molecules which were recognized and bind to TCRs of CD8+ T cells. The activated CD8+ effector T cells then released cytotoxic granules containing perforins and granzymes and produced cytokines such as tumor necrosis factor α (TNF-α) and interferon-gamma (IFN-γ) [6].
A specialized subset of CD4+ T cells known as T follicular helper (Tfh) cells aid in B cell antibody production through the formation of germinal centres which are specialized microstructures involved in the production of long-lived antibody secreting plasma cells and memory B cells resulting from the expression of signaling lymphocyte activation molecules (SLAM)-associated protein (SAP). Cytokine-polarized CD4+ memory T cell subsets such as Th1, Th2 and Th17 cells can each develop into Tfh to influence the B cell response. Tfh also promotes B cell activation through cell–cell communication such as CD40L (expressed on activated CD4+ T cells) and CD40 (expressed by B cell) interactions, leading to the release of cytokines [7,8] (Fig. 1 ).
Fig. 1.
The elicitation of humoral and cellular responses to DNA vaccines. CD4+ T cells, such as T follicular helper (Tfh) and Foxp3+ T follicular regulatory (Tfr) cells, provide specialized assistance and serve as important mediators towards the formation of germinal centre B cells through T and B cell interactions. Tfh cells also provide help to B cells through CD40L– CD40 interactions, and lead to the release of cytokines (IL-2, IL-4, and IL-21, IFN-γ). Stimulation by cytokines further promotes germination centre formation, and maturation into plasma cells which produce memory B cells and long-lived antibody secreting plasma cells. CD8+ T cells directly target and kill infected cells through the production of perforin and granyzymes, limiting the spread of the pathogen within the body [7,8]. Fig. 1 was produced using the graphical software Biorender (https://www.biorender.com/).
Tfh cells are involved in giving specialized assistance to germinal centre B cells through B and T cell interactions. Another set of cells known as Foxp3+ T follicular regulatory (Tfr) cells also serve as important mediators of germinal centre regulation. Furthermore, memory CD4+ T cells were also implicated in providing help to B cells in promoting earlier B cell proliferation, higher antibody levels and earlier class-switching responses when compared to naïve CD4+ T cells. Memory CD4+ T cells produce higher amounts and a more polarised profile of cytokines, which is believed to encourage more robust B cell antibody response and determine the antibody isotype. Current research suggests that Tfh cells produce interleukin (IL)-4 and IFN which play a key role in regulating B cell affinity development as well as immunoglobulin class switching in the germinal centre [7,8] (Fig. 1).
2.2. Advantages of DNA vaccines
The prospect of designing and constructing DNA vaccines targeting SARS-CoV-2 is attractive for a number of reasons. Firstly, the DNA vaccine platform shows great potential in eliciting both humoral and cellular immune responses [9]. Antibodies elicited by the DNA vaccine were shown to neutralize the virus before it could gain entry into the host cell [10]. Current DNA vaccines targeting the S protein of the SARS-CoV-2 have shown effective humoral immune responses by eliciting neutralizing antibodies and cellular immune responses in terms of production of IFN-γ and IL-2 [[10], [11], [12], [13]].
DNA vaccines are easier to produce in large quantities compared to the complexities of producing whole inactivated virus and mRNA vaccines [14]. Smith et al. [10] reported that DNA-based vaccines could be developed quickly since a variety of vaccine candidates could be prepared and tested using high-throughput approaches. DNA vaccines offered convenience in vaccine development. For example, a single DNA plasmid vector could be used to construct recombinant DNA plasmids containing genes encoding the whole antigen or immunogenic peptides. It is also feasible to construct a DNA plasmid expressing highly conserved peptides specifying multi-valent epitopes against the SARS-CoV-2 prototype (Wuhan strain) and VOCs.
It is relatively easy to produce the DNA vaccine in large quantities for distribution and large-scale immunizations [15]. The recombinant plasmid DNA could be conveniently produced in large quantities in bacteria such as E. coli and expressed in eukaryotic cells such as HEK-293 T cells [15].
The use of DNA vaccines is associated with concerns that the administered plasmid DNA might lead to possible insertional mutagenesis. However, no insertional mutagenesis activity through integration into the host genome has been reported in the development of numerous DNA vaccines against SARS-CoV-2 [16]. Insertion sequence (IS) elements which are characterized as small DNA segments encoding for proteins necessary for the mobility of the IS element are ubiquitously found in the form of bacterial mobile genetic elements and are capable of causing deleterious, neutral, or beneficial mutations [17]. Indeed, de Visser et al. [18] reported a total of 9 IS-mediated mutations occurring in the form of insertions and genetic recombination deletions to conditions of growth and starvation in Lactococcus lactis. Nevertheless, there have been no reports of the existence of IS elements in HEK-293 T cells which could potentially induce mutations in DNA plasmids after transfection in eukaryotic cells.
Efficient and cost-effective production of DNA vaccines for large-scale immunizations offers an attractive practical advantage. Nevertheless, Wang et al. [19] indicated that glycosylation, the binding site and adjacent amino acids could impact the binding of the S protein and the angiotensin-converting enzyme 2 (ACE2) host cell receptor. Therefore, the selection of expression systems must be carefully evaluated for the effective development of DNA vaccines against SARS-CoV-2. In a study to evaluate N-linked and O-linked glycosylations of the S protein of SARS-CoV-2 expressed in HEK-293 T cells, the expressed SARS-CoV-2 S protein showed extensive N-linked and O-linked glycosylations with the formation of N-glycans and sialylated O-glycans. Mass spectrometry using matrix-assisted laser desorption-ionization (MALDI-MS) analysis revealed that expression in HEK-293 T cells also produced α2,3- and α2,6-linked sialic acids. Various groups have successfully expressed recombinant DNA plasmids encoding the S protein in HEK-293 T cells for in vitro characterization of the expressed recombinant protein without the need for modifications to the recombinant DNA plasmid [[10], [11], [12],20]. This suggested that HEK-293 T cells served as an excellent host for the expression of the SARS-CoV-2 S protein.
Vaccine storage is an important issue since it is directly associated with vaccine stability and the maintenance of its quality. Therefore, in order to ensure that the efficacy of mRNA vaccines is not negatively affected, cold storage at −70 °C is necessary for the preservation of the BNT162b2 mRNA-based vaccine. DNA vaccines are highly stable, less prone to degradation unlike mRNA vaccines, and do not require an extremely low temperature for storage. DNA vaccines could be stored at room temperature. Since they do not require specific cold chain storage conditions, DNA vaccines would be ideal for immunizing populations in third world countries which lack the financial resources and infrastructures to maintain cold chain distribution and storage conditions [21,22].
DNA vaccines could be stored at room temperature for as long as 6 months [23]. This presented DNA vaccines with an advantage over other vaccine platforms such as mRNA and viral-vectored vaccines since the high stability at room temperature meant that cold chain conditions were not required for transport and storage [24].
2.3. Current DNA vaccine candidates
Inovio Pharmaceuticals developed the vaccine candidate, INO-4800, a DNA based vaccine against SARS-CoV-2 which incorporated the full-length S gene in the pGX0001 vector [10]. Protein expression was confirmed by Western blot and immunofluorescence studies in the transfected HEK-293T cells. Sera from immunized mice and guinea pigs showed IgGs specific for the SARS-CoV-2 S protein. The IgG binding end-point titers showed that the SARS-CoV-2 S region (S1 + S2) as well as the receptor-binding domain (RBD) region were capable of eliciting higher levels of IgG than the IgG elicited by the S1 region alone. The antibodies in the sera derived from immunized animals could prevent the binding of the S protein to the ACE2 receptor by pseudotyped virus and confirmed the effectiveness of the neutralizing antibodies in preventing an infection. An evaluation of live virus neutralization activities conducted in C57BL/6 mice showed that the immunization with INO-4800 was able to elicit antibodies that neutralized wildtype SARS-CoV-2 virus with an average ND50 titer of 340. Bronchoalveolar lavage (BAL) fluid from immunized mice also showed a significant increase in IgG antibodies against the S protein from sera of immunized mice, demonstrating that such antibodies in the lungs could protect the host from lower respiratory disease. Cellular immune responses were associated with IFN-γ production and an increase in CD4+ and CD8+ T cells. Flow cytometric analysis of splenocytes harvested from BALB/c mice immunized with a single dose of INO-4800 showed 0.04% CD4+ and 0.32% CD8+ T cells producing IFN-γ after stimulation with the SARS-CoV-2 S protein antigen. Epitope mapping revealed that strong CD4+ and CD8+ T cell responses were associated with several epitopes from the RBD and the S2 domain [10]. INO-4800 has shown good safety and tolerability in phase 1 clinical trial. Both humoral and cellular immune responses were elicited in the 401 vaccinated human participants who received the vaccine. Recorded adverse effects were mainly grade 1 and 2 and they were not exacerbated by the higher dosage of 2.0 mg of the DNA vaccine. Levels of binding and neutralizing antibodies as well as ELISpot analysis of T cell immune responses were higher when the vaccine dose was increased from 1.0 mg to 2.0 mg [25].
Yu et al. [13] reported the development of multiple DNA vaccine candidates encoding different regions of the S protein and evaluation of immunogenicity in rhesus macaque models by Barouch's group at Harvard University. Immunizations of macaques resulted in S-specific binding antibodies and neutralizing antibodies. The levels of neutralizing antibodies against the S protein were similar to those from convalescent humans. S and RBD-specific antibodies with effector functions such as antibody-dependent neutrophil phagocytosis (ADNP), antibody-dependent complement deposition (ADCD), antibody-dependent monocyte cellular phagocytosis (ADCP), and antibody-dependent NK cell activation (IFN-γ production, CD107a degranulation, and MIP-1β expression) were observed. High levels of IFN-γ+, CD4+, and CD8+ T cell immune responses against the whole S protein were recorded. Moreover, cellular responses of lower magnitudes were observed in macaques immunized with the S1 and RBD vaccine candidates. In challenge studies, macaques vaccinated with the S protein showed minor symptoms as well non-existing and lower SARS-CoV-2 RNA levels in the plasma, BAL, and nasopharyngeal swabs (NS), respectively when compared to controls which did not receive the vaccine. The deletion of the transmembrane domain and cytoplasmic region of the S protein elicited no protection in challenge models, demonstrating that the prefusion ectodomain stabilization was significant in eliciting immune response against SARS-CoV-2 [13].
Other DNA vaccine candidates against SARS-CoV-2 have also focused on the S protein. Chai et al. [11] reported the immunogenicity of different DNA vaccine candidates based on the S protein. The S genes and its derivatives were cloned into the DNA plasmid vector, pVAX1, and following confirmation of expression in HEK-293T cells, the recombinant plasmid DNA was administered via electroporation in mice and Syrian hamsters. Sera from the SARS-CoV-2 S protein immunized mice demonstrated antibody titers against the RBD that was able to neutralize infections caused by SARS-CoV-2. The binding of ACE2 with the RBD was observed to be blocked by the immunized mice sera. IgG and neutralizing antibody titers following SARS-CoV-2 S DNA immunization were associated with protection at 20 weeks. Moreover, the SARS-CoV-2 S recombinant DNA vaccine elicited neutralizing antibody titers against the S1 protein carrying the D614G mutation. Upon stimulation with SARS-CoV-2 S protein, high levels of Th1 type cytokines like IFN-γ and IL-2 were observed with significantly lower levels of the Th2 type cytokines such as IL-5 and IL-13 in both BALB/c and C57BL/6 mice, demonstrating that CD4+ T cell responses were Th1 polarized. Challenge studies demonstrated the superiority of the DNA vaccine incorporating the full-length S protein over the RBD in terms of the significantly higher levels of anti-Spike and neutralizing antibodies. Immunizations with the recombinant S DNA vaccine resulted in 2.29 log10 reductions of the infectious virus titer and 1.37 log10 reductions in the number of viral RNA copies when compared to the vector control group [11].
Similarly, Prompetchara et al. [12] constructed DNA vaccine candidates carrying the full-length S, S1(pCMVkan-S1) or S2 (pCMVkan-S2). All vaccine candidates elicited high levels of S-specific binding IgGs that exhibited a balance of IgG1and IgG2a. Sera from mice immunized with S and S1 vaccine candidates were able to effectively neutralize RBD-ACE2 binding. In particular, the sera from S immunized mice demonstrated significant inhibition of RBD-ACE2 binding when compared to sera from the S1 immunizations. The levels of neutralization antibodies were the highest in the sera of S immunized mice (GMT:2551) when compared to S1 (GMT:1005) and S2 (GMT:291) immunizations. S2 immunizations were associated with non-detectable levels of inhibition of RBD-ACE2 binding as well as the lowest levels of neutralizing antibodies when compared to S1 and S immunizations. Splenocytes from mice immunized with the S DNA vaccine produced IFN-γ in response to peptide pools from the S1 or S2 regions while splenocytes from mice immunized with the S1 or S2 vaccines elicited IFN-γ in response to peptide pools from their respective regions. The highest levels of IFN-γ induced was from the S immunized mice (2991 SCF/106 splenocytes), followed by the S2 immunized mice (1885 SCF/106 splenocytes), and lastly by the S1 immunized mice (1376 SCF/106 splenocytes). Moreover, peptide pools associated with potent IFN-γ based on ELISpot assays were from the RBD region and the heptad repeat (HR) 1 region from the S1 and S2 regions [12].
Researchers from Osaka University presented a preclinical study to develop a recombinant DNA vaccine for SARS-CoV-2, with the S protein as the main antigen and it was designated as pVAX-1- SAR-CoV-2 S [20]. The DNA vaccine was administered to rats and humoral responses were assessed in the presence and absence of the alum adjuvant. The alum adjuvant formulated with the DNA vaccine could provide potent humoral immune response, with 666.6 μg of recombinant plasmid DNA carrying the SARS-CoV-2 S gene formulated with 66.7 μl of alum adjuvant. It was able to induce high levels of antibodies against the S and RBD proteins at week 4 through to week 16. These antibodies were able to recognize the S1 subunit carrying the D614G mutation of SARS-CoV-2. IgG2a and IgG2b were the main subclasses of produced IgG, indicating Th1 polarization. Intramuscular administration of the DNA vaccine in rats showed that IFN-γ production was significantly increased while IL4 was only slightly increased in response to immunizations with the DNA vaccines carrying the S and RBD. It was observed that DNA immunizations in rats produced sera which could decrease binding of ACE2 with the S1+S2 protein. Immunized sera when diluted 5-fold could inhibit 50% of the binding of ACE2 with the RBD protein. Sera dilutions from immunized rats demonstrated neutralizing activities when tested with the pseudo-typed vesicular stomatitis virus (VSV) carrying the luciferase gene in Vero E6 cells. Neutralizing titers remained at an average of 98.4 at 8 weeks after immunization with the first dose of the DNA vaccine. Furthermore, body tissues and serum biochemical parameters showed that there were no toxic effects of the DNA vaccine [20].
Many other research groups are currently developing similar DNA vaccines. For example, researchers from the King Abdulaziz University developed a DNA vaccine consisting of a codon optimized S protein sequence [26]. Intramuscular immunization of BALB/c and C57BL/6J mice with 100 μg of the DNA vaccine elicited S-specific IgG antibodies and neutralizing antibodies. Moreover, the use of needle-free intradermal administration of the vaccine induced a potent Th1-biased humoral response including binding IgG antibodies, and neutralizing antibodies as well as IFN-γ, TNF-α and IL-2 cytokine production from memory CD4+ and CD8+ T cells when administered to BALB/c mice [26]. A DNA vaccine consisting of the S1 subunit was also able to elicit a Th1 polarized response specifically associated with IgG and neutralizing antibodies along with potent CD4+ and CD8+ T cell responses [27]. DNA vaccine development is also associated with different routes of administrations other than the intramuscular route. For example, the use of the Pyro-drive Jet Injector for intradermal administration of a DNA vaccine expressing the S protein resulted in potent humoral responses consisting of neutralizing antibodies without any safety concerns [28].
An evaluation of the literature on preclinical and clinical development of DNA vaccines showed that most of these vaccines were focusing on the S protein or the RBD of the S protein as the main antigenic target. The S protein of SARS-CoV-2 plays a significant role in the attachment of the virus to the ACE2 cell receptor as well as viral and host cell membrane fusion to promote the entry of the virus into the cells. Furthermore, the S protein was shown to be a prime target for eliciting neutralizing antibodies [29]. Vaccinations against SARS-CoV-2 using the full-length S protein also produced potent CD4+ and CD8+ T cell responses [30]. Table 1 shows the preclinical development of DNA vaccines against SARS-CoV-2 while Table 2 shows an overview of the clinical development of DNA vaccines against SARS-CoV-2.
Table 1.
Preclinical development of DNA vaccines against SARS-CoV-2.
| Name of Vaccine | Developer | Plasmid vector | Main antigenic region | Adjuvant | Mode of Administration | Reference |
|---|---|---|---|---|---|---|
| INO-4800 | Inovio Pharmaceuticals | pGX0001 | full length S gene | – | Electroporation using CELLECTRA® delivery device | [10] |
| SARS-CoV-2 DNA vaccine tested in Syrian hamsters | Taiwan Group | pVAX1 | spike genes of SARS-CoV and SARS-CoV-2 | – | Intramuscular electroporation with a BTX electroporator (ECM830) | [11] |
| DNA Vaccine candidates developed by Chulalongkorn University | Thailand Group | pCMVkan | full length S, S1, and S2 | – | Intramuscular electroporation using TriGrid delivery system | [12] |
| SARS-CoV-2 Spike glycoprotein DNA plasmid vaccine | Osaka University (Japan group) | pVAX1 | full length S gene | alum | Intramuscular injection | [20] |
| IgE-spike-S1/S2-D614G-6P-foldon | Southern University of Science and Technology, China | PCDNA3.1 | S-protein S1+S2 | – | Intramuscular injection | [31] |
| pSARS2-S | National Institute of Infectious Diseases and Vaccinology Taiwan | pVAX1 | full length S gene | alum | Electroacupuncture | [32] |
| VIU-1005 | King Abdulaziz University | pVAX1 | full length S gene | – | intramuscular needle injection | [26] |
| pVAX-S1 | King Abdulaziz University | pVAX1 | S1 subunit | – | intramuscular administration using customized needle-free Tropis system | [27] |
| pVAX1-SARS-CoV2-co | Osaka University | pVAX1 | S protein | – | Intradermal using a pyro-drive jet injector | [28] |
Table 2.
An Overview of the clinical development of DNA vaccines against SARS-CoV-2.
| # | Type of Vaccine Candidate | Number of Doses | Route of administration | Developers | Clinical Trial Phase |
|---|---|---|---|---|---|
| 1. | nCov vaccine | 3 | ID | Zydus Cadila | Phase 4 |
| 2. | INO-4800+electroporation | 2 | ID | Inovio Pharmaceuticals + International Vaccine Institute + Advaccine (Suzhou) Biopharmaceutical Co., Ltd | Phase 3 |
| 3. | AG0301-COVID19 | 2 | IM | AnGes + Takara Bio + Osaka University | Phase 2/3 |
| 4. | GX-19 N | 2 | IM | Genexine Consortium | Phase 1/2 |
| 5. | GLS-5310 | 2 | ID | GeneOne Life Science, Inc. | Phase 1/2 |
| 6. | COVID-eVax | 2 | IM | Takis + Rottapharm Biotech | Phase 1/2 |
| 7. | AG0302-COVID19 | 2–3 | IM | AnGes Inc. | Phase 1/2 |
| 8. | VB10.2129 | 1–2 | IM | Vaccibody AS | Phase 1/2 |
| 9. | VB10.2210 | 2 | IM | Vaccibody AS | Phase 1/2 |
| 10. | Covigenix VAX-001 | 2 | IM | Entos Pharmaceuticals Inc. | Phase 1 |
| 11. | CORVax 12 | 2 | ID | Providence Health & Services | Phase 1 |
| 12. | bacTRL-Spike | 1 | Oral | Symvivo Corporation | Phase 1 |
| 13. | COVIGEN | 2 | ID or IM | University of Sydney, Bionet Co., Ltd Technovalia" | Phase 1 |
| 14. | COVIDITY | 2 | ID | Scancell Ltd | Phase 1 |
| 15. | SARS-CoV-2 DNA vaccine | 2 | IM | The University of Hong Kong; Immuno Cure 3 Limited | Phase 1 |
| 16. | Prophylactic pDNA Vaccine | 3 | IM | Imam Abdulrahman Bin Faisal University | Phase 1 |
Abbreviations: ID - Intradermal; IM - Intramuscular.
∖Adapted from COVID-19 vaccine tracker and landscape as of September 9, 2022.https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines
2.4. Reduced vaccine efficacies against SARS-CoV-2 VOCs
While the vaccine candidates under emergency approval offer protection from the original SARS-CoV-2 Wuhan strain, they have been shown to exhibit reduced protective efficacies against VOCs. Indeed, current viral vectored and mRNA vaccines such as ChAdOx1 nCoV-19, BNT162b2, and mRNA-1273, which utilize the full length S protein as the main antigenic target, have been reported to demonstrate reduced protective efficacies against SARS-CoV-2 VOCs. For example, when the sera from individuals immunized with the Pfizer BNT162b2 vaccine were tested for neutralizing ability upon reinfection with SARS-CoV-2, it was observed that the neutralizing activity was reduced by 1- to 3-folds against a pseudovirus containing mutations such as E484K, N501Y, and a combination of K417 N, E484K, and N501Y mutations. In contrast, there was no reduction in neutralizing activity in response to the wild type strain and the variant with the K417 N mutation [33]. Furthermore, humoral response elicited by the mRNA vaccines was significantly reduced in response to the B.1.351 VOC. This was seen by a 9.4 fold and 10.3–12.4-fold reductions in neutralizing activities in the sera of convalescent patients and those who had received the mRNA vaccines [34]. This demonstrated that SARS-CoV-2 VOCs with mutations in the RBD were not completely neutralized by neutralizing antibodies elicited by the mRNA vaccines approved under emergency use against SARS-CoV-2.
Harvey et al. [35] also reported that since the S protein gene from the Wuhan strain was present in all of the licensed vaccines under emergency use, any mutation in the S gene of the VOC would potentially impact its interaction with neutralizing antibodies elicited by the Wuhan strain. Pseudoviruses carrying the mutations associated with the B.1.1.7 variant along with the E484K mutation showed a 6.7-fold reduction in neutralization activity when sera from individuals immunized with the Pfizer BNT162b2 vaccine were tested [36]. Live virus neutralizations also showed that neutralizing titers in the sera of those immunized with the AstraZeneca ChAdOx1 nCoV-19 vaccine were 9 fold lower against the B.1.1.7 SARS-CoV-2 variant when compared to sera from vacinees immunized with the original Wuhan strain [37]. Similarly, a 6.4-fold reduction in neutralization response to the B.1.351 variant was observed in the sera of individuals vaccinated with two doses of Moderna mRNA-1273 [38].
The efficacies of the current SARS-CoV-2 vaccines were particularly challenged by mutations in the RBD which were present in variants such as B.1. 351 (Beta) and P.1 (Gamma) that could escape neutralizations. Besides immune escape mediated by B-cell epitopes, SARS-CoV-2 variants might also evade cytotoxic T lymphocyte (CTL) immunity through T cell mutations in viral epitopes which could lead to reduced CTL responses. It was reported that SARS-CoV-2 variants had a less than marginal effect on the elicitation of CD4+ and CD8+ T cell responses in convalescent patients and vacinees receiving the mRNA vaccines [39]. Moreover, T cell responses were found to be similar to those elicited from the original SARS-CoV-2 Wuhan strain [40]. Noh et al. [40] concluded that CD4+ and CD8+ T cell epitopes were conserved. However, through mutations in the MHC-I restricted viral epitope genes, there is likelihood for SARS-CoV-2 variants to escape CD8+ T cell surveillance. Agerer et al. [41] reported that some mutations in SARS-CoV-2 reduced the binding of peptides to major histocompatibility complex Class I which ultimately resulted in reduced productions of IFN-γ as well as lower cytotoxic activity of CD8+ T cells. In conclusion, humoral and cellular responses elicited by the mRNA vaccine against the SARS-CoV-2 Wuhan strain offered protective immunity but VOCs were reported to reduce the neutralizing activities of the humoral responses elicited by the mRNA vaccines. Nevertheless, some studies have reported a marginal effect of VOCs on cellular responses as mutations in the MHC-I epitope genes might lower the potency of T cell responses [42].
The most recent VOC is the Omicron variant (B.1.1.529) which has emerged in multiple countries. A recent study conducted with 19 individuals in South Africa demonstrated that the protective efficacy of the Pfizer-BioNTech vaccine was significantly reduced against the omicron variant. In individuals immunized with the BNT162b2 vaccine, a 22-fold reduction in the levels of neutralizing antibodies in response to the Omicron variant (Pango lineage B.1.1.529) was observed when compared to the prototype Wuhan strain [43].
An evaluation of neutralizing antibody titers against the SARS-CoV-2 Wuhan strain and the Omicron BA.1, BA.2, BA.2.12.1, and BA.4 or BA.5 subvariants showed that when compared to the Wuhan strain, the neutralizing antibody titers were reduced by a factor of 6.4 against BA.1, by a factor of 7.0 against BA.2, by a factor of 14.1 against BA.2.12.1, and by a factor of 21.0 against BA.4 or BA.5. Omicron subvariants BA.2.12.1, BA.4 and BA.5 were shown to be more likely to escape neutralizations than the BA.1 subvariant. The median neutralizing antibody titer was lowered by a factor of 2.2 against the BA.2.12.1 subvariant and by a factor of 3.3 against the BA.4 or BA.5 subvariant when compared to the BA.1 subvariant [44]. It was also asserted that the emergence of Omicron was met with concerns regarding reduced vaccine-induced neutralizing activities [45]. In comparison with the Delta variant, the Omicron variant significantly reduced the protective neutralizing activities of sera derived from those receiving immunizations with the current approved vaccines.
2.4.1. DNA vaccine-induced humoral and cellular immune responses against VOCs
Andrade et al. [46] evaluated the humoral and cellular responses against SARS-CoV-2 variants such as B.1.1.7 (Alpha), B.1.351 (Beta), and P.1 (Gamma) VOCs in vacinees who were immunized with the INO-4800 DNA vaccine. The evaluation of humoral responses in terms of the levels of serum IgG titers to the SARS-CoV-2 S protein from the B.1.1.7, B.1.351, and P.1 variants showed that serum IgG titers were comparable for the Wuhan, and the B.1.1.7 and B.1.351 variants. However, a 1.9-fold reduction was reported against the P.1 variant at week 8 in vacinees who received two doses of INO-4800 DNA vaccine. Pseudovirus neutralization assays showed 2.1 and 6.9-fold reductions against the B.1.1.7 and B.1.351 variants, respectively. There was no reduction in neutralizing antibodies for the P.1 variant when compared with the levels of neutralizing antibodies elicited against the Wuhan strain. However, T cell responses in terms of IFNγ and production of cytokines by CD8+ cytotoxic T cells showed similar responses between the Wuhan strain and the B.1.1.7, B.1.351, and P.1 variants.
Azevedo et al. [47] investigated the efficacy of pCTV-WS, a Spike-based DNA vaccine against the Wuhan strain and the Gamma, Delta, and Omicron VOCs in transgenic (K18-hACE2) mice and hamsters. The levels of neutralizing antibodies were similar against the Wuhan and the Delta VOC. However, a 10-fold reduction in the levels of neutralizing antibodies was observed when immunized mice were challenged with the Gamma and Omicron VOCs. The T cell response was generally conserved across the different strains and played a prominent role in the reduction of viral loads and disease severity in immunized mice challenged with the Gamma or Omicron VOCs.
There are also reports showing incorporation of conserved regions from the SARS-CoV-2 into the design of the DNA vaccine that could lead to more universal and sustained immune responses in the face of emerging variants. For example, a universal SARS-CoV-2 DNA vaccine comprising antigens from immunogenic epitopes from the RBD, membrane, and nucleoprotein (NP) from the Wuhan strain, the Alpha, and the Beta variants was able to elicit cross-reactive neutralizing antibodies that successfully neutralized the Wuhan strain, Beta, Delta, and Omicron viruses in vitro [48]. Challenge studies showed that mice were protected from lethal infection with the SARS-CoV-2 Beta variant and NP-specific T cells alone were responsible for 60% of the conferred protection [48].
Furthermore, the administration of a pan-Spike vaccine known as INO-4802 in animal models showed the elicitation of neutralizing antibodies and T cell responses against the Wuhan strain as well as B.1.1.7, P.1, and B.1.351 VOCs with potent humoral responses induced against VOCs in priming with a heterologous wild type vaccine, and followed by INO-4802 boost administration [49].
In view of the emergence of SARS-CoV-2 variants, there is a need to incorporate highly conserved epitopes to confer broad protection against variants. DNA vaccines could offer a promising approach to develop effective vaccines, particularly against the rapidly mutating SARS-CoV-2 VOCs. DNA vaccines can easily be modified to incorporate different genes simply by changing the gene sequences of the recombinant DNA plasmid. Multiple cloning sites incorporated into the recombinant plasmid DNA can easily be used to incorporate different nucleotides encoding peptides specifying conserved immunogenic epitopes as these can be predicted from bioinformatics or through an analysis of binding sites of monoclonal antibodies found in convalescent sera [[50], [51], [52]]. Therefore, the DNA vaccine platform is considered an attractive and promising approach to immunize against SARS-CoV-2 due to the advantages that it offers over other vaccine platforms [10].
2.5. Limitations of DNA vaccines
DNA vaccines against infections caused by the West Nile Virus in horses and canine melanoma have been approved by the Food and Drug Administration (FDA) and the United States Department of Agriculture (USDA) for veterinary immunizations. With the exception of ZyCoV-D, no DNA vaccines have been authorized for human use. This suggests that there are limitations which have to be overcome before DNA vaccine candidates could progress to the clinic. It has been reported that preclinical studies demonstrated the ability of DNA vaccine candidates to elicit potent humoral and cellular responses in small animals such as mice, rats, and guinea pigs and even in non-human primates [[10], [11], [12], [13]] but there are challenges in eliciting the same potency of immune responses in larger animals and humans. It was reasoned that low immunogenicity is due to the non-feasibility of upscaling plasmid DNA amounts administered in small animal models to humans [53]. Indeed, as much as 5–25 mg plasmid DNA would need to be administered to achieve the same effect [54].
Another challenge is the route of administration of DNA vaccines. All current DNA vaccines in clinical development against SARS-CoV-2 used either electroporation, intradermal, oral, or intramuscular routes to deliver the naked plasmid DNA. Another reason for low immunogenicity is the low transfection efficiency of naked plasmid DNA. The administration of naked plasmid DNA was associated with low transfection efficiency due to degradation. Indeed, the in vivo half-life of naked plasmid DNA was shown to be reduced in a few minutes following administration since it was vulnerable to degradation and removal by the reticular endothelial system (RES) [55]. DNA vaccines harbouring the DNA plasmid would need to target APCs to ensure the elicitation of potent cellular immunity [56]. The administration of naked DNA did not lead to uptake by DCs. Unsuccessful APC targeting resulted in low transfection efficiency and low immunogenicity. Moreover, several barriers have to be overcome such as penetration through the negatively charged phospholipid membrane, escape from the endosome, and entry into the nucleus.
2.6. Design of DNA vaccine and formulation improvements
Considering the potential of the SARS-CoV-2 prototype and VOCs to cause debilitating consequences to public health and wellbeing, it is essential to develop strategies to overcome the limitations of DNA vaccines so that the advantages of the platform can be used to develop effective vaccines against rapidly emerging variants. These include optimizations that might be made to the plasmid with the use of molecular or chemical adjuvants as well as exploring alternative delivery systems and routes.
2.6.1. Plasmid optimization for gene expressions
Effective construction of a recombinant DNA plasmid needs to incorporate certain genetic elements to ensure adequate protein expressions. These could include specific nucleic acid sequences that would upregulate transcriptions. An evaluation of these genetic elements is crucial to establish optimum recombinant plasmid design.
2.6.1.1. Strong promoters
It is imperative to select a promoter for efficient plasmid construction so that the transfection of the recombinant DNA plasmid into a mammalian cell line would result in both high-level and long-term transgene expressions. A promoter is defined as a specific set of nucleotides upstream of the gene of interest where RNA polymerase binds to initiate transcription. Several plasmid vectors such as pcDNA™3.1(+) and pVAX1™ were designed to contain the cytomegalovirus (CMV) promoter and enhancer for high level expression of the protein of interest when transfected into mammalian cells. However, it was observed that transcriptional silencing resulted in a reduction in the expression of recombinant gene of interest controlled by the CMV promoter following transfection as evidenced by the reduction in expression of exogenous genes driven by the CMV promoter [57]. A solution to this issue is to utilize the human elongation factor-1 alpha (hEF1α) promoter to initiate and regulate gene expression. Kim et al. [58] demonstrated high levels of gene expression in a variety of cell types using the pEF-CAT plasmid whereby the bacterial CAT gene of interest was ligated to specific sites within the hEF1α gene such as at the end of the TATA box, exon 1, and exon 2. The hEF1α promoter was also effective in driving gene expression when viral promoters did not drive the expression of downstream genes. Indeed, the hEF1α promoter drove the stable and high level expression of the bacterial neo gene more efficiently than the viral promoter of the simian virus 40 (SV40) early gene. Moreover, the CHO-derived elongation factor-1 promoter (CHEF-1) was reported to serve as a promising element to regulate high-level expression of recombinant DNA plasmids in mammalian cell lines. Expression vectors containing 5′ and 3′ flanking sequences from the CHEF1 gene had greater than 10-fold expression levels of the chemokine receptor CCR4 when compared to the CMV promoter [59]. Even though the use of such flanking sequences could enhance expression of the gene of interest, their inclusions into the vector increased the vector size which might not be ideal for transfections. Furthermore, the SV40 promoter was described to be a fairly strong promoter for the expression of therapeutic proteins in mammalian cell lines. Although it might result in comparatively lower expression, it had greater stability than the hEF1α and CMV promoters [60].
The CMV promoter was shown to be primarily responsible for the transient transcriptional expression of recombinant genes in the human embryonic kidney 293 (HEK293) cells [61]. Johari et al. [62] reported that the CMV promoter functioned by directing transcriptional expression which depended on the different components of the promoter such as the transcription factor regulatory elements (TFREs) which included AhR:ARNT, CREB, E4F, Sp1, ZBED1, JunB, c-Rel, and NF-κB. The CMV promoter could be modified or engineered to incorporate optimized binding sites. The TRFEs might also be used for the construction of synthetic promoters. The researchers found multiple suboptimal TF binding sequences including MYBL1, Oct, and E2F which might be used to further improve the transcriptional activity of the CMV promoter. The use of high-throughput parallel screening methods could be utilized to screen several hundred TFREs and evaluate their binding affinities [62].
2.6.1.2. Kozak sequence
The Kozak consensus sequence is characterized as a specific set of nucleotides that serves as the initiation site where protein translation begins in eukaryotic mRNA produced from transcription. The sequence is important for the initiation of translation and for the regulation of protein production [63]. The Kozak sequence ensures accurate translation of the protein in terms of ribosome assembly and translation initiation, considering that an incorrect initiation site might lead to the expression of non-functional proteins [64]. Plasmid vectors such as pcDNA™3.1(+) and pVAX1 are designed to require the insert to contain a Kozak consensus sequence (−6 GCCA/GCCAUGG +4) located around the initiation codon to ensure the accurate and specific initiation of translation.
An example of a Kozak sequence found in a recombinant DNA plasmid constructed using the plasmid vector pcDNA™3.1(+) is GCCACC with the ATG initiation codon located downstream of the 6-nucleotide sequence. More specifically, for a strong consensus sequence that could strongly express the recombinant protein, the nucleotides at the +4 and −3 position (relative to the +1 assigned to the A of the initiation codon) would need to match the consensus. For effective translation, the −3 position should contain a purine base. In the absence of a purine base, a guanine should be present at +4 [65]. Recognition of AUG and alternative initiator codons is augmented by a G at position +4 but is not generally affected by the nucleotides at position +4 and + 6 [66].
2.6.1.3. Matrix attachment regions (MARs)
A series of scientific discoveries have led to the incorporation of the matrix attachment regions (MARs) into mammalian expression vectors. It was observed that chromatin could be divided into topologically constrained domains separated by elements such as scaffolds of MARs [67]. Moreover, MARs were reported to be able to serve as insulator elements which prevented the spread of heterochromatin as well as gene silencing [68]. Therefore, MARs were associated with the production of an anti-silencing effect by shielding the gene of interest from the suppressive impacts of heterochromatin [69]. Mammalian expression vectors might be optimized by incorporating a variety of genetic elements to produce the right combination for optimum gene expression. MARs might be used in conjunction with strong promoters for enhancing gene expressions. For example, the incorporation of the CMV promoter to drive the expression of green fluorescent protein flanked by two different MARs or the use of SV40 promoter with two β-globin MARs substantially increased expression and stability of the transgene [70].
2.6.1.4. Introns
Intron-mediated enhancement refers to the higher level of expression of a DNA construct containing a specific intron as compared to the expression of the construct when the intron was not included [71]. The enhancement of gene expression as a result of the inclusion of certain introns has been known to occur in eukaryotes such as mammals, plants, yeast, and insects. It was reported that introns could enhance transcript levels by affecting the rate of transcription, nuclear export, transcript stability, and mRNA translation [72].
There is considerable evidence for intron mediated enhancement of gene expressions. The rate of transcription in transgenic mice was increased by 10- to 100-fold when compared to the same genes not containing introns [73]. Moreover, in eukaryotic organisms such as humans and Saccharomyces cerevisiae, genes that contained introns produced significantly more copies of RNA than genes lacking introns [74]. Genes that were prominently expressed were shown to contain a larger intron density in terms of the number of introns per kilobase of coding sequence when compared to genes that were weakly expressed [74].
There are also other ways in which introns could enhance gene expression. Spliceosomal introns increased gene expressions at numerous stages from transcription to translation [75]. Splicing was considered to be the crucial factor leading to the enhancement of expression of the leader intron of the Arabidopsis AtMHX gene [76]. The intron sequence showed only weak enhancement without splicing. Moreover, enhancement of gene expression was found to be strongly dependent upon intron splicing based on the analysis of mutated genes contained in rice mutants [77]. Introns are involved in enhancing gene expression and increasing gene products during multiple processes such as splicing, transcription, polyadenylation, mRNA export, and translation. Introns could contain enhancer elements to increase expression as well as sequences to enhance translation of mRNA into proteins [78].
Some genes are fully dependent on introns for their expressions and they could remain undetectable when the gene was expressed without the intron [79]. In fact, the impact of intron sequences on the expression of plasmids could be quite significant; exceeding 10-fold in some cases and depending on the right combination of factors such as the type of gene and intron in the recombinant plasmid [80]. Several studies have reported enhanced gene expression upon the inclusion of introns. A motif derived from an intron was able to substantially enhance gene expression even when it was placed significantly downstream of the transcriptional start site [71]. Baier et al. [81] selected a group of 33 native and 13 non-native introns from highly expressed genes of Chlamydomonas reinhardtii with the potential to significantly increase abundance of transcripts as an efficient solution to the low transcript levels of nuclear transgenes in C. reinhardtii. An SV40 intron was identified as a strong intron element that effectively enhanced the expression levels of erythropoietin protein in Chinese hamster ovary (CHO) cells [82].
While it is true that the introduction of introns was reported to have a positive impact on plasmid optimization in terms of increased gene expression, there is also evidence that a larger size of the insert in the DNA plasmid vector might have a negative impact on the levels of gene expression. In an experiment where two vectors of differing plasmid lengths containing a firefly luciferase encoding gene insert were compared for levels of gene expression, it was observed that 2-fold decrease in the size of the plasmid vector backbone could increase gene expression levels by more than 10-fold in rat tenocytes in vitro, and rat myocardium in vivo [83]. Moreover, the administration of varying sizes of plasmid DNA into Bacillus subtilis ISW1214 showed that transformation efficiency was reduced with increasing size of the plasmid DNA [84]. Gene transfection efficiency was reported to be highest for the DNA plasmid shortest in size [85].
2.6.1.5. Nuclear localization signal peptides
Nuclear localization signal (NLS) peptides are characterized as short signaling molecules that facilitate the transport of substances such as proteins from the cytoplasm to the nucleus [86]. Data from several experimental investigations have shown that the addition of nuclear localization signal peptides might aid in gene delivery by enhancing the translocation of DNA to the nucleus through the nuclear membrane. The development of a capped 3.3kbp CMV-Luciferase-NLS gene incorporating a single nuclear localization signal peptide led to a remarkable enhancement of transfection efficiency [87]. This 10 to 10,000-fold increase in transfection efficiency was attributable to the NLS peptide since the introduction of a substitution mutation in the third amino acid in the NLS peptide was able to reduce the transfection efficiency to lower levels. Zanta et al. [87] reasoned that the DNA in the cytoplasm was translocated to the nucleus first by docking and then translocating through a pore in the nuclear envelope. The exogenous DNA would be present in the form of a chromatin-shaped structure inside the nucleus. The incorporation of NLS peptides into DNA vaccine has also been documented and was shown to demonstrate increased transfection efficiency of DNA in in vitro testing. For example, the addition of four NLS peptides, namely SV40 large T-antigen derived NLS, nucleoplasmin targeting signal, M9 sequence, and the reverse SV40 derived NLS was shown to enhance the transfection efficiency of the DNA delivery vector, LAH4, after transfection into slow-dividing epithelial cancer cells (Calu-3), macrophages (RAW264.7), DCs (JAWSII), and thymidine-induced growth-arrested cells [88]. Therefore, the use of NLS peptides in DNA vaccines against SARS-CoV-2 is a feasible option which might enhance transfection efficiency during clinical trials to elicit stronger immune responses.
2.6.1.6. Linear minimalistic (MIDGE) vectors
Current plasmid DNA vectors consist of antibiotic resistance genes that act as selection markers and allow the detection of successful transformation of plasmids for bacterial expression. However, this approach is likely to be associated with the spread of antibiotic resistance genes as the recombinant plasmid DNAs are administered to animals or humans. A suitable alternative is the use of minimalistic, immunologically-defined gene expression (MIDGE) vectors which were described as linear vectors that only consist of the sequence associated with the direct expression of the antigen which can be chemically altered to enhance the resulting immune response [89]. MIDGE vectors expressing the Leishmania homologue of receptors for activated C-kinase (LACK) antigen were shown to confer a high degree of protection against Leishmania infection in BALB/c mice [89]. It was also observed that high levels of protection could be achieved through the use of much lower doses than what is required for traditional recombinant plasmid DNAs. Protective efficacy may be further improved through the addition of an NLS peptide to the MIDGE vector. Furthermore, MIDGE vectors could be used to elicit high levels of antigen expression both in cell cultures as well as in animal models. In particular, the development of MIDGE and MIDGE-NLS vectors encoding for the hepatitis B surface antigen (HBsAg) and subsequent immunogenicity testing showed strong humoral and cellular immune responses [90].
2.6.2. Chemical adjuvants
To date, most of the experimental preclinical DNA vaccines developed against SARS-CoV-2 were administered on their own as naked plasmid DNAs. However, it has been reported that the administration of DNA vaccines against SARS-CoV-2 together with chemical adjuvants such as Montanide, Alum, Vaxfectin might improve immunogenicity. Indeed, there is significant evidence to suggest this since the use of such chemical adjuvants showed a positive impact on immunogenicity of vaccines in the past [91,92].
2.6.2.1. Montanide
Montanide as a chemical adjuvant was shown to enhance both the humoral and the cellular immune response in murine and bovine models [93]. Moreover, DNA vaccines incorporating Montanide have been successfully commercialized for protection against the foot-and-mouth disease virus (FMDV). Furthermore, a DNA vaccine consisting of the glycoprotein D chemically adjuvanted with Montanide 903110 against the Bovine herpesvirus-1 (BoHV-1) was shown to protect against bovine infectious rhinotracheitis [92]. Cows immunized with the vaccine formulation containing Montanide effectively enhanced both the humoral and cellular response, ameliorated clinical symptoms, and significantly reduced viral excretions [92].
2.6.2.2. Alum
Considering the development of DNA vaccines against SARS-CoV-2, the only example of the use of alum administered along with recombinant SARS-CoV-2 S DNA vaccine is by researchers from Osaka University [20]. The use of aluminum salts (alum) in optimizing the humoral response in licensed DNA vaccines is well established. When a DNA vaccine encoding HBsAg was formulated with aluminum phosphate, the antibody titers were increased by 10–100 fold. The adjuvant aluminum phosphate was able to decrease the antigen dosage required by 10-fold. Boosting the adjuvanted DNA vaccine with a HBs protein was found to elicit HBs-specific IgG2a which reflected a Th1 response. The adjuvanting effect was suggested to be due to an increase in the number of T cells secreting HBs peptide antigen-specific IFN-γ and IL-2 [94].
Furthermore, a DNA vaccine against rabies adjuvanted with alum was demonstrated to provide 80% protective immunity of BALB/c mice challenged with a rabies virus strain. The immune response was observed to be Th2 polarized with increased IgG antibody titers [95]. Adjuvanting the DNA vaccine with alum was shown to elicit more potent immune responses demonstrated by an increase in the IgG antibody titer and neutralizing antibodies against rabies virus.
However, there are instances where the administration of alum with DNA vaccines failed to provide protective immunity. This was the case with the alum adjuvanted DNA vaccine against the human immunodeficiency virus (HIV) type 1 DNA vaccine. Considering that alum was present in adequately detectable levels at the injection depot, the failure to elicit sufficient levels of immunogenic humoral and cellular responses was attributed to a lack of effective interaction between the DNA vaccine and alum adjuvant [96].
2.6.2.3. Vaxfectin
Another well-known chemical adjuvant is the cationic lipid formulation, Vaxfectin, in enhancing the immunogenicity of DNA vaccines by improving delivery efficiency. This increased immunogenicity might be attributable to Vaxfectin being directly involved in regulating immune response pathways. Several studies have also documented Vaxfectin to successfully optimize the immune response in several animal models. For example, the intradermal or intramuscular administrations of a DNA vaccine encoding the hemagglutinin (H) and fusion (F) proteins of measles virus successfully conferred protection in rhesus macaques [97]. This was demonstrated mainly by the elicitation of higher levels of neutralizing antibodies upon the administration of the DNA vaccine adjuvanted with Vaxfectin when compared to the unadjuvanted DNA vaccine. The immune response was also characterized by the rapid induction of T cells but production of IFN-γ did not increase upon treatment with Vaxfectin. Challenge studies protected the macaques from developing rash or viremia but did not confer protection against infection [97]. Nevertheless, efforts to develop a more balanced immune response consisting of the elicitation of both humoral and cellular immune response were carried out by Pan et al. [98]. In the murine model, both humoral and cellular immune responses were demonstrated by the administration of a Vaxfectin adjuvanted DNA vaccine against measles virus encoding the H and F proteins. The DNA vaccine also conferred protection against intratracheal infection and prevented the development of viremia and rashes [98].
Vaxfectin has also been incorporated as a chemical adjuvant in the development of DNA vaccines against diseases such as dengue and influenza. For example, the administration of a tetravalent dengue DNA vaccine administered with a cationic lipid-based adjuvant in a phase 1 clinical trial resulted in good safety and immunogenicity [99]. It was observed that the group of mice which were immunized three times with the high dose of the tetravalent dengue DNA vaccine adjuvanted with Vaxfectin showed the most potent IFN-γ T cell responses when compared to mice which were immunized with the DNA vaccine on its own. Moreover, a 60 μg dose of Vaxfectin adjuvanted recombinant DNA plasmid encoding highly conserved antigens namely, the NP and ion channel protein (M2) from influenza virus conferred 100% protection in mice against influenza. An evaluation of the efficacy of Vaxfectin as a chemical adjuvant suggested that on its own, Vaxfectin might be effective to optimize immunogenicity in only smaller animal models such as mice but not in larger non-human primates (NHPs) [99].
2.6.2.4. AS03
The use of emulsion based adjuvants could be administered in conjunction with DNA vaccines to enhance humoral and cellular immune responses. An investigation into the impact of the use of emulsion-based and α-tocopherol containing adjuvant Diluvac Forte® on the immunogenicity of a recombinant DNA plasmid, which encoded hemagglutinin and a non-glycosylated NP, against influenza in the murine model showed that when the naked recombinant plasmid was administered with Diluvac Forte® or an emulsion containing α-tocopherol, the humoral response was enhanced in terms of significantly elevated levels of immunoglobulin G (IgG)1 and IgG2c [100]. While AS03 has not been used for adjuvanting the current DNA vaccines against SARS-CoV-2, several recombinant protein vaccines against SARS-CoV-2 employed the use of AS03 as a vaccine adjuvant [101]. Future DNA vaccine development against SARS-CoV-2 should consider the possibility of administering DNA vaccines in a 1:1 emulsion together with AS03 which might serve as an effective adjuvant to augment the immune response elicited.
In conclusion, a number of chemical adjuvants have been incorporated in DNA vaccines and the resulted increase in immunogenicity was demonstrated in animal models. However, a number of observations were elucidated. Firstly, the immune response elicited by the administration of chemically adjuvanted DNA vaccines resulted in strong humoral responses. Secondly, the immunogenic potential of chemical adjuvants in optimizing the immune response was higher in smaller animals which brought into question their applications in DNA vaccine development and immunogenicity testing in humans. Therefore, future efforts need to focus on incorporating novel adjuvants into DNA vaccine formulations to elicit more balanced humoral and cellular immune responses, whereby its efficacy could be evaluated in larger animal models.
Implications for the development of vaccines against the SARS-CoV-2 involve evaluating and exploring strategies that could elicit both strong antibodies and potent cellular immune responses. In light of the recent COVID-19 pandemic and the need for effective vaccines that could offer sufficient protection against the disease, the immune response must involve both the humoral and cellular immune response. It was reported that humoral response against SARS-CoV-2 in terms of IgG, total and neutralizing antibodies declined after 6 months [102]. Full resolution of SARS-CoV-2 required a Th1 polarized cellular immune response as well as the elicitation of memory B cells. An evaluation of the use of chemical adjuvants in DNA vaccines in the past showed that immune responses were mainly Th2 biased. Therefore, additional incorporation of novel adjuvants which have similar effects as the monophosphoryl lipid A (MPLA) adjuvant by shifting the immune response towards a more Th1 biased response must be explored for the future development of chemically adjuvanted DNA vaccines against SARS-CoV-2.
2.6.3. Molecular adjuvants
An understanding of the use of chemical adjuvants in formulations with DNA vaccines is needed to explain the failure to elicit optimum immunogenicity. It must be noted that chemical adjuvants are simply mixed with the DNA vaccine which might have limited interactions with the adjuvant. The use of molecular adjuvants fused to the sequence of the target gene coding for the antigen in the recombinant plasmid might overcome this problem by ensuring that the adjuvant and antigen are both expressed simultaneously, resulting in an increased interaction. Alternatively, the molecular adjuvant could be encoded by a separate plasmid which might encode Toll-like receptor agonists, cytokines, and chemokines.
CD40 is a costimulatory molecule that plays an important role in the effective functioning of the immune system. The receptor is expressed by cells of the immune system such as B cells, APCs, and macrophages [103]. It can also be expressed by other non-immune cells such as endothelial cells, smooth muscle cells, fibroblasts and epithelial cells [104]. CD4F cells could activate DCs by the interaction of its CD40 with the CD40L on the DC. Activation of DCs would result in upregulation of CD80/CD86 which interacts with CD28 on naïve CD8+ T cells. The CD40L is reported to serve as an effective adjuvant capable of enhancing the immune response elicited when it is expressed as part of the DNA vaccine. For example, Tamming et al. [105] reported the protective efficacy of a DNA vaccine encoding the SARS-CoV-2 Spike glycoprotein expressed in conjunction with CD40L. It was observed that CD40L served not only as the target ligand for its costimulatory molecule but also as a molecular adjuvant. The administration of the vaccine in Syrian hamsters was associated with potent humoral responses in terms of the production of neutralizing antibodies. In the group immunized with the Spike-CD40L DNA vaccine, lung pathology was more successfully ameliorated when compared to the group of mice immunized with the DNA vaccine not adjuvanted with CD40L. Moreover, clinical trials to test the immunogenicity of GX-19, a recombinant DNA plasmid expressing the S protein together with CD40L, demonstrated the elicitation of broad binding antibody responses [106]. Moreover, when the recombinant protein SARS-CoV-2 RBD vaccine was adjuvanted with a TLR7/8 agonist formulation, alum-3M − 052, it was able to elicit a neutralizing antibody response 100 fold higher than that elicited upon the administration of the vaccine with alum alone [107].
Moreover, other sequences that might serve as molecular adjuvants to enhance the immunogenicity of DNA vaccines have been reported. For example, a novel RBD based DNA vaccine against SARS-CoV-2 elicited far more potent immune responses in terms of the production of inflammatory cytokines IL-6 and TNF-α when linked to the N-terminal inclusion of a 33-bp (11 aa) preS1 sequence of the HBV W4P variant (N-terminal HBV preS1) comparing to the weaker immune response elicited in response to the RBD-based vaccine alone. This demonstrated that the N-terminal HBV preS1 acted as an adjuvant which enhanced immunogenicity [108].
2.6.4. Delivery systems
Apart from the type of antigen and the incorporation of an adjuvant in the vaccine design and development, the protective efficacy of the vaccine and the potency of the immune responses it could elicit depends on the type of vaccine delivery platform [109]. A promising delivery method could involve using the intranasal route to administer DNA vaccines. Currently, none of the 16 DNA vaccine candidates in clinical development have utilized the intranasal route to deliver the DNA vaccine. Nevertheless, nasal delivery could overcome problems of dependency on needles, needle-stick injuries, disposal, inconvenience, and cost. Since most SARS-CoV-2 infections originate at mucosal surfaces, the intranasal route is a promising approach to induce immune responses since it could provide a convenient and accessible route to the mucosal immune system. Mucosal surfaces are strongly associated with immune responses of the lymphoid tissues. The intranasal route of vaccination involves the mucosa-associated lymphoid tissue (MALT), known as the nasopharynx-associated lymphoid tissue (NALT). NALT is characterized as the immune system present in the nasal mucosa involving lymphoid tissue, B cells, T cells, APCs, and an epithelial layer of memory cells which are responsible to carry the antigen across the epithelium. Depending on the type of antigen, it is transported across the epithelium to interact with macrophages and DCs. The antigen is taken up by APCs and transported to the lymph nodes for presentation to T cells. Soluble antigens are endocytosed by APCs directly [110].
2.6.5. Encapsulation of DNA plasmid in poly (lactic-co-glycolic acid) PLGA
Although the DNA vaccine platform has been extensively studied for the last three decades, its clinical application is impeded by the main challenge of the vaccine antigen in not being able to reach target APCs. Different methods have been used to deliver naked DNA vaccines. Electroporation was used by Inovio pharmaceuticals to deliver the DNA vaccine INO-4800 using the CELLECTRA® delivery device [10]. This created an electrical impulse using an electroporation device to stimulate permeability of cell membranes for enhanced uptake of the DNA vaccine [111]. Although transfection efficiency is high, and the method is quick and easy, disadvantages include cell death at the site of administration and pain to the patient when the DNA vaccine is administered. Moreover, certain considerations such as the electrode shape, size, and the formulation of the DNA vaccine would need to be optimized to enhance immunogenicity [112]. Oral delivery is an interesting mode of delivery adopted by only one vaccine developer, Symvivo Corporation (Australia), to manufacture the bacTRL-Spike oral DNA vaccine. In order to be successfully delivered orally, DNA vaccines would need to be specially formulated to survive against the gastric conditions of the digestive system [113]. The Needle-Free Injection System (NFIS) was also used by Zydus Cadila Healthcare (India) to administer the ZyCoV-D DNA vaccine. Specifically, the Pharmajet Tropis® device was used to deliver the DNA vaccine intradermally through a narrow and precise fluid stream. The ZyCoV-D vaccine is the only DNA vaccine to progress to phase 4 clinical trial under Emergency Use Authorization (EUA) in India as it had previously demonstrated humoral neutralizing antibody responses and Th1 polarized cellular responses characterized by IFN-γ production in animal models [114]. Furthermore, adequate humoral and cellular immune responses were observed at day 70 after the third dose in phase 1 clinical trial. Although needle-free intradermal administration could result in efficient and painless administrations, it required expensive and sophisticated devices which might not be available in resource poor countries [115]. Mechanical delivery for plasmid DNA could also be mediated by a gene gun. Heavy metallic particles were coated with plasmid DNA and significantly reduced DNA doses which could be directly injected into the cytosol of target cells. However, the cost of the gene gun system and gold particles is high [116]. Viral vectors could be used for delivery of DNA vaccines but they were associated with safety issues such as the elicitation of unwanted immune response against the viral vector, potential reversion to virulence, and potential insertional mutagenesis. Therefore, non-viral vectors are increasingly being recognized as potential carriers for DNA vaccine delivery.
The prospect of developing nanomaterials as carriers of DNA vaccines offers several advantages over traditional naked DNA administration in terms of delivery to the target cells in the lymphoid tissues, high transfection efficiency, induction of DC maturation and antigen presentation [116,117].
Nanoparticles as carriers can prevent the DNA plasmid from potential degradation during delivery to target APCs [118]. Nanoparticle carriers which encapsulate the DNA vaccine were reported to have a size range of 10–500 nm and they were small enough to be effectively edocytosed [119]. Nanoparticles with a size less than 100 nm were observed to have increased lymphatic uptake and improved transfections of the APCs in the lymph nodes [120]. Thus, encapsulation of DNA vaccines in nanoparticles could lead to increased immunogenicity, reduced toxicity and reactogenicity, effective presentations to APCs, improved endocytosis of DNA and transport to the nucleus.
PLGA was reported to break down into lactic acid and glycolic acid. Lactic acid was reported to be further metabolized into carbon dioxide and water which were excreted from the body [121]. This feature is an added advantage in terms of safety when compared to viral vectors [116].
The synthesis of plasmid DNA is convenient and inexpensive. However, due to reports of low transfection efficiency, it is important to improve the immunogenicity of DNA vaccines through the use of different delivery systems and adjuvants. Although sophisticated equipment has been available that used different routes of administration, such as the Pharmajet Tropis® device, which used NFIS for intradermal delivery of the ZyCoV-D DNA vaccine, and the CELLECTRA® delivery device, which used electroporation to deliver the INO-4800 DNA vaccine, these were more expensive options and may not be feasible for mass vaccination [116]. In contrast, the use of PLGA NPs to encapsulate DNA could serve as a relatively inexpensive method to increase immunogenicity of administered plasmid DNA. PLGA encapsulated plasmid DNA could be conveniently produced in the laboratory using the simple manual double emulsion method or mass produced using a microfluidics system [122,123].
There are various materials that can be used as nanocarriers to encapsulate DNA vaccines. For example, plasmid DNAs were encapsulated by either proteolipid nanoparticles formulated with neutral lipid and fusion associated trans-membrane. Another prominent and safe approach involves the use of polymers such as PLGA and chitosan. Choosing the appropriate antigen to be expressed and the material for nanoparticle design, the nanoparticle-based vaccine could result in a timely release of the DNA payload [116].
The use of PLGA as a nanodelivery carrier is increasingly being employed to facilitate the delivery of DNA-based vaccines. PLGA is an FDA-approved biopolymer, well-known for its biodegradability, biocompatibility, and minimal toxicity which made it ideal for use as delivery vehicle. Encapsulation by PLGA also protected the DNA vaccine antigen from degradation by DNases of the host. It could also promote the long-lasting release of the DNA vaccine. This occurred through a sophisticated mechanism of hydrolysis in which the payload was released slowly [124]. This meant that special modifications are required to ensure sustained release of the payload [116].
Extensive work has been conducted involving the use of PLGA nanoparticles to encapsulate DNA plasmids. For example, Zhao et al. [125] reported the encapsulation of a recombinant DNA plasmid expressing the F gene of the Newcastle Disease Virus (pFNDV) DNA Vaccine in PLGA nanoparticles. The constructed pFNDV-PLGA nanoparticle-based DNA vaccine had a diameter of 433.5 ± 7.5 nm and a Zeta potential of +2.7 mV. The recombinant plasmid DNA demonstrated high sustainable release (93.14% of the total amount) from the pFNDV-PLGA nanoparticles. Superior and more potent immune responses were elicited from immunization of chickens with pFNDV-PLGA nanoparticles as compared to nanoparticles containing pFNDV alone. Therefore, pFNDV-PLGA nanoparticles were associated with the elicitation of more potent cellular, humoral, and mucosal immune responses with a sustained release of the DNA payload [125].
PLGA could serve as an effective nanomaterial to encapsulate DNA vaccines which were that have been administered intranasally to elicit potent protective immune responses. Wang et al. [126] utilized chitosan-coated PLGA to encapsulate recombinant DNA plasmid encoding the FMDV capsid protein as the antigenic target and the bovine IL-6 gene to enhance the mucosal immune response. Animal models such as rats and guinea pigs were intranasally immunized with the DNA vaccine. Out of the three different expression vectors used, the recombinant plasmid constructed using the recombinant pc-P12AIL3C plasmid which contained IL-6 located between the P12A and 3C genes elicited the most potent antigen-specific serum IgG and IgA responses and the strongest titers of secretory IgA in mucosal tissues. The recombinant plasmid, pc-P12AIL3C, was able to elicit the highest levels of neutralizing antibodies and also produced the strongest cellular immune responses. Cellular immune responses were associated with T cell proliferations in response to target antigens and high levels of IFN-γ produced by CD4+ and CD8+ splenic T cells. Challenge studies in animal models showed that 3/5 mice were protected against FMDV infection after immunization with the pc-IL2AP12A3C DNA vaccine. This approach is particularly attractive since it points towards the increased efficacy of DNA vaccines especially when encapsulated in chitosan-coated PLGA nanoparticles, with the use of IL-6 as a molecular adjuvant to optimize the immune response. The importance of incorporating DNA vaccines into chitosan-coated PLGA nanoparticles was further demonstrated by the intranasal administration of a naked DNA vaccine against pseudorabies virus, which elicited local and systemic immune responses. A combination of the DNA vaccine with PLGA-polyethylenimine (PEI) nanoparticles further increased the time during which mucosal IgA was detectable in pigs [127].
PLGA nanoparticles could be formulated with the cationic polymer, PEI which is a synthetic polymer regarded as an efficient nanodelivery carrier due to its ability to complex with DNA and deliver the plasmid DNA into the nucleus [128]. Properties that make PEI an ideal biomaterial to be formulated in complex with DNA include its high molecular weight and branched structure. The larger molecular weight resulted in densely populated amine groups which in turn increased the cationic characteristics. This allowed the PEI complex to achieve a higher rate of transfection through the proton sponge effect associated with endosomal escape. The use of PEIs with higher molecular weight was found to induce greater toxicity, and those with lower molecular weights induced lower toxicity but with reduced transfection efficiency. Therefore, certain modifications such as increasing branching and molecular weights could be made to PEIs to incorporate polysaccharides, polymers, and disulphide bridges. Previous studies to develop DNA vaccines utilizing the PLGA-PEI complex as the nanodelivery carrier reported the induction of humoral immunity utilizing the pseudorabies glycoprotein B as the target antigen, and a H1N1 DNA vaccine was delivered through intranasal delivery [127,129].
The use of PLGA-PEI nanoparticles was reported to effectively induce DC maturation and the production of cytokines such as IL-2 and TNF-α. Approaches such as a prime boost with recombinant protein vaccines showed that the DNA vaccine was able to elicit cellular immune responses in the form of T cell mediated immunity through the production of IFN-γ [130].
3. Conclusions
The rapid emergence of SARS-CoV-2 VOCs such as the highly infectious Omicron variant has contributed to new waves of SARS-CoV-2 infections globally since late 2021. There is an urgent need to develop strategies to curb the further spread of COVID-19 as well as to curb the threat of variants that might arise in the future. The development of next generation vaccine platforms such as DNA vaccines appears to be a promising solution since DNA vaccines can be quickly developed to incorporate antigenic regions other than the full length S protein which might become ineffective in eliciting neutralizing antibodies against new VOCs. In particular, the DNA vaccine platform is associated with attributes such as its ability to incorporate different genes representing highly conserved B and T cell epitopes, high stability at room temperature, and accelerated developmental timelines to speed up their progression to clinical trials. Genetic modifications of a plasmid vector such as the insertion of the NLS peptide gene could enhance the transfection and translocation of recombinant DNA. Additional genetic modifications to increase expressions of the recombinant plasmid include the use of strong promoters, insertion of MARs, and introns. Vectors could be modified to carry molecular adjuvants such as TLR agonists and CD40L to increase immunogenicity of the DNA vaccine. With the exception of ZyCoV-D vaccine in India, no DNA vaccines have been given emergency approval under phase 4 clinical development due to limited immunogenicity in humans. This challenge might be solved through the use of chemical/molecular adjuvants and the development of appropriate delivery systems utilizing nanoparticles to increase uptake by APCs in enhancing the induction of potent immune responses.
Financial disclosure
The authors have no funding to disclose.
The author contribution
Study Design: Chit Laa Poh.
Data Collection: Kanwal Khalid.
Statistical Analysis: n/a.
Data Interpretation: Kanwal Khalid, Chit Laa Poh.
Manuscript Preparation: Kanwal Khalid.
Literature Search: Kanwal Khalid.
Funds Collection: n/a.
Declaration of competing interest
The authors declare no conflict of interests.
References
- 1.WHO WHO coronavirus (COVID-19) https://covid19.who.int/ Dashboard 2022 [Available from:
- 2.Bian L., Gao F., Zhang J., He Q., Mao Q., Xu M., et al. Effects of SARS-CoV-2 variants on vaccine efficacy and response strategies. Expert Rev Vaccines. 2021;20(4):365–373. doi: 10.1080/14760584.2021.1903879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eiz-Vesper B., Schmetzer H.M. Antigen-presenting cells: potential of proven und new players in immune therapies. Transfus Med Hemotherapy. 2020;47(6):429–431. doi: 10.1159/000512729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shafaati M., Saidijam M., Soleimani M., Hazrati F., Mirzaei R., Amirheidari B., et al. A brief review on DNA vaccines in the era of COVID-19. Future Virol. 2022;17(1):49–66. doi: 10.2217/fvl-2021-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim T.S., Goh J.K., Mortellaro A., Lim C.T., Hämmerling G.J., Ricciardi-Castagnoli P. CD80 and CD86 differentially regulate mechanical interactions of T-cells with antigen-presenting dendritic cells and B-cells. PLoS One. 2012;7(9) doi: 10.1371/journal.pone.0045185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosendahl Huber S., van Beek J., de Jonge J., Luytjes W., van Baarle D. T cell responses to viral infections - opportunities for Peptide vaccination. Front Immunol. 2014;5:171. doi: 10.3389/fimmu.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stebegg M., Kumar S.D., Silva-Cayetano A., Fonseca V.R., Linterman M.A., Graca L. Regulation of the germinal center response. Front Immunol. 2018;9 doi: 10.3389/fimmu.2018.02469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swain S.L., McKinstry K.K., Strutt T.M. Expanding roles for CD4+ T cells in immunity to viruses. Nat Rev Immunol. 2012;12(2):136–148. doi: 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee L.Y.Y., Izzard L., Hurt A.C. A review of DNA vaccines against influenza. Front Immunol. 2018;9:1568. doi: 10.3389/fimmu.2018.01568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith T.R.F., Patel A., Ramos S., Elwood D., Zhu X., Yan J., et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat Commun. 2020;11(1):2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chai K.M., Tzeng T.-T., Shen K.-Y., Liao H.-C., Lin J.-J., Chen M.-Y., et al. DNA vaccination induced protective immunity against SARS CoV-2 infection in hamsterss. PLoS Neglected Trop Dis. 2021;15(5) doi: 10.1371/journal.pntd.0009374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prompetchara E., Ketloy C., Tharakhet K., Kaewpang P., Buranapraditkun S., Techawiwattanaboon T., et al. DNA vaccine candidate encoding SARS-CoV-2 spike proteins elicited potent humoral and Th1 cell-mediated immune responses in mice. PLoS One. 2021;16(3) doi: 10.1371/journal.pone.0248007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369(6505):806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poland G.A., Ovsyannikova I.G., Kennedy R.B. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet. 2020;396(10262):1595–1606. doi: 10.1016/S0140-6736(20)32137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586(7830):516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 16.Liu M.A. A comparison of plasmid DNA and mRNA as vaccine technologies. Vaccines (Basel) 2019;7(2) doi: 10.3390/vaccines7020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Consuegra J., Gaffé J., Lenski R.E., Hindré T., Barrick J.E., Tenaillon O., et al. Insertion-sequence-mediated mutations both promote and constrain evolvability during a long-term experiment with bacteria. Nat Commun. 2021;12(1):980. doi: 10.1038/s41467-021-21210-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Visser J.A.G.M., Akkermans A.D.L., Hoekstra R.F., de Vos W.M. Insertion-sequence-mediated mutations isolated during adaptation to growth and starvation in Lactococcus lactis. Genetics. 2004;168(3):1145–1157. doi: 10.1534/genetics.104.032136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y., Wu Z., Hu W., Hao P., Yang S. Impact of expressing cells on glycosylation and glycan of the SARS-CoV-2 spike glycoprotein. ACS Omega. 2021;6(24):15988–15999. doi: 10.1021/acsomega.1c01785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi H., Sun J., Yanagida Y., Otera T., Kubota-Koketsu R., Shioda T., et al. Preclinical study of a DNA vaccine targeting SARS-CoV-2. Curr Res Transl Med. 2022;70(4) doi: 10.1016/j.retram.2022.103348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silveira M.M., Moreira G., Mendonça M. DNA vaccines against COVID-19: perspectives and challenges. Life Sci. 2021;267 doi: 10.1016/j.lfs.2020.118919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tebas P., Kraynyak K.A., Patel A., Maslow J.N., Morrow M.P., Sylvester A.J., et al. Intradermal SynCon® ebola GP DNA vaccine is temperature stable and safely demonstrates cellular and humoral immunogenicity advantages in healthy volunteers. J Infect Dis. 2019;220(3):400–410. doi: 10.1093/infdis/jiz132. [DOI] [PubMed] [Google Scholar]
- 23.Song X., Xu L., Yan R., Huang X., Shah M.A.A., Li X. The optimal immunization procedure of DNA vaccine pcDNA–TA4–IL-2 of Eimeria tenella and its cross-immunity to Eimeria necatrix and Eimeria acervulina. Vet Parasitol. 2009;159(1):30–36. doi: 10.1016/j.vetpar.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 24.Conforti A., Marra E., Palombo F., Roscilli G., Ravà M., Fumagalli V., et al. COVID-eVax, an electroporated DNA vaccine candidate encoding the SARS-CoV-2 RBD, elicits protective responses in animal models. Mol Ther. 2022;30(1):311–326. doi: 10.1016/j.ymthe.2021.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mammen M.P., Tebas P., Agnes J., Giffear M., Kraynyak K.A., Blackwood E., et al. Safety and immunogenicity of INO-4800 DNA vaccine against SARS-CoV-2: a preliminary report of a randomized, blinded, placebo-controlled, Phase 2 clinical trial in adults at high risk of viral exposure. medRxiv. 2021:2021. 05.07.21256652. [Google Scholar]
- 26.Alamri S.S., Alluhaybi K.A., Alhabbab R.Y., Basabrain M., Algaissi A., Almahboub S., et al. Synthetic SARS-CoV-2 spike-based DNA vaccine elicits robust and long-lasting th1 humoral and cellular immunity in mice. Front Microbiol. 2021:12. doi: 10.3389/fmicb.2021.727455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alluhaybi K.A., Alharbi R.H., Alhabbab R.Y., Aljehani N.D., Alamri S.S., Basabrain M., et al. Cellular and humoral immunogenicity of a candidate DNA vaccine expressing SARS-CoV-2 spike subunit 1. Vaccines (Basel) 2021;9(8) doi: 10.3390/vaccines9080852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishikawa T., Chang C.Y., Tai J.A., Hayashi H., Sun J., Torii S., et al. Anti-CoVid19 plasmid DNA vaccine induces a potent immune response in rodents by Pyro-drive Jet injector intradermal inoculation. bioRxiv. 2021;13 doi: 10.1080/25785826.2022.2111905. 2021.01. [DOI] [PubMed] [Google Scholar]
- 29.Yang Y., Du L. SARS-CoV-2 spike protein: a key target for eliciting persistent neutralizing antibodies. Curr Signal Transduct Ther. 2021;6(1):95. doi: 10.1038/s41392-021-00523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisher E., Padula L., Podack K., O'Neill K., Seavey M.M., Jayaraman P., et al. Induction of SARS-CoV-2 protein S-specific CD8+ T cells in the lungs of gp96-Ig-S vaccinated mice. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.602254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X., Rcheulishvili N., Cai J., Liu C., Xie F., Hu X., et al. Development of DNA vaccine candidate against SARS-CoV-2. Viruses. 2022;14:1049. doi: 10.3390/v14051049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzeng T.-T., Chai K.M., Shen K.-Y., Yu C.-Y., Yang S.-J., Huang W.-C., et al. A DNA vaccine candidate delivered by an electroacupuncture machine provides protective immunity against SARS-CoV-2 infection. NPJ Vaccines. 2022;7(1):60. doi: 10.1038/s41541-022-00482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592(7855):616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 35.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collier D.A., De Marco A., Ferreira I.A.T.M., Meng B., Datir R.P., Walls A.C., et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593(7857):136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 variant. N Engl J Med. 2021;384(20):1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu K., Werner A.P., Moliva J.I., Koch M., Choi A., Stewart-Jones G.B.E., et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv. 2021;25 2021.01. [Google Scholar]
- 39.Tarke A., Sidney J., Methot N., Yu E.D., Zhang Y., Dan J.M., et al. Impact of SARS-CoV-2 variants on the total CD4(+) and CD8(+) T cell reactivity in infected or vaccinated individuals. Cell Rep Med. 2021;2(7) doi: 10.1016/j.xcrm.2021.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noh J.Y., Jeong H.W., Shin E.-C. SARS-CoV-2 mutations, vaccines, and immunity: implication of variants of concern. Curr Signal Transduct Ther. 2021;6(1):203. doi: 10.1038/s41392-021-00623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agerer B., Koblischke M., Gudipati V., Montaño-Gutierrez L.F., Smyth M., Popa A., et al. SARS-CoV-2 mutations in MHC-I-restricted epitopes evade CD8(+) T cell responses. Sci Immunol. 2021;6(57) doi: 10.1126/sciimmunol.abg6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moss P. The T cell immune response against SARS-CoV-2. Nat Immunol. 2022;23(2):186–193. doi: 10.1038/s41590-021-01122-w. [DOI] [PubMed] [Google Scholar]
- 43.Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602(7898):654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hachmann N.P., Miller J., Collier A.Y., Ventura J.D., Yu J., Rowe M., et al. Neutralization escape by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4, and BA.5. NEJM. 2022;387(1):86–88. doi: 10.1056/NEJMc2206576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilhelm A., Widera M., Grikscheit K., Toptan T., Schenk B., Pallas C., et al. Limited neutralisation of the SARS-CoV-2 Omicron subvariants BA.1 and BA.2 by convalescent and vaccine serum and monoclonal antibodies. EBioMedicine. 2022:82. doi: 10.1016/j.ebiom.2022.104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andrade V.M., Christensen-Quick A., Agnes J., Tur J., Reed C., Kalia R., et al. INO-4800 DNA vaccine induces neutralizing antibodies and T cell activity against global SARS-CoV-2 variants. NPJ Vaccines. 2021;6(1):121. doi: 10.1038/s41541-021-00384-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Azevedo P.O., Hojo-Souza N.S., Faustino L.P., Fumagalli M.J., Hirako I.C., Oliveira E.R., et al. Differential requirement of neutralizing antibodies and T cells on protective immunity to SARS-CoV-2 variants of concern. NPJ Vaccines. 2023;8(1):15. doi: 10.1038/s41541-023-00616-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Appelberg S., Ahlén G., Yan J., Nikouyan N., Weber S., Larsson O., et al. A universal SARS-CoV DNA vaccine inducing highly cross-reactive neutralizing antibodies and T cells. EMBO Mol Med. 2022;14(10) doi: 10.15252/emmm.202215821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reed C.C., Schultheis K., Andrade V.M., Kalia R., Tur J., Schouest B., et al. Design, immunogenicity and efficacy of a Pan-SARS-CoV-2 synthetic DNA vaccine. bioRxiv. 2021;5(11):443592. [Google Scholar]
- 50.Lim H.X., Masomian M., Khalid K., Kumar A.U., MacAry P.A., Poh C.L. Identification of B-cell epitopes for eliciting neutralizing antibodies against the SARS-CoV-2 spike protein through bioinformatics and monoclonal antibody targeting. Int J Mol Sci. 2022;23(8) doi: 10.3390/ijms23084341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Montes-Grajales D., Olivero-Verbel J. Bioinformatics Prediction of SARS-CoV-2 epitopes as vaccine candidates for the Colombian population. Vaccines (Basel) 2021;9(7) doi: 10.3390/vaccines9070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Polyiam K., Phoolcharoen W., Butkhot N., Srisaowakarn C., Thitithanyanont A., Auewarakul P., et al. Immunodominant linear B cell epitopes in the spike and membrane proteins of SARS-CoV-2 identified by immunoinformatics prediction and immunoassay. Sci Rep. 2021;11(1) doi: 10.1038/s41598-021-99642-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suschak J.J., Williams J.A., Schmaljohn C.S. Advancements in DNA vaccine vectors, non-mechanical delivery methods, and molecular adjuvants to increase immunogenicity. Hum Vaccines Immunother. 2017;13(12):2837–2848. doi: 10.1080/21645515.2017.1330236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dupuis M., Denis-Mize K., Woo C., Goldbeck C., Selby M.J., Chen M., et al. Distribution of DNA vaccines determines their immunogenicity after intramuscular injection in mice. J Immunol J. 2000;165(5):2850–2858. doi: 10.4049/jimmunol.165.5.2850. [DOI] [PubMed] [Google Scholar]
- 55.Hobernik D., Bros M. DNA vaccines-how far from clinical use? Int J Mol Sci. 2018;19(11) doi: 10.3390/ijms19113605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Braathen R., Spång H.C.L., Hinke D.M., Blazevski J., Bobic S., Fossum E., et al. A DNA vaccine that encodes an antigen-presenting cell-specific heterodimeric protein protects against cancer and influenza. Mol Ther Methods Clin Dev. 2020;17:378–392. doi: 10.1016/j.omtm.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grozdanov P.N., MacDonald C.C. Generation of plasmid vectors expressing FLAG-tagged proteins under the regulation of human elongation factor-1α promoter using Gibson assembly. J Vis Exp. 2015;96 doi: 10.3791/52235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim D.W., Uetsuki T., Kaziro Y., Yamaguchi N., Sugano S. Use of the human elongation factor 1α promoter as a versatile and efficient expression system. Gene. 1990;91(2):217–223. doi: 10.1016/0378-1119(90)90091-5. [DOI] [PubMed] [Google Scholar]
- 59.Deer J.R., Allison D.S. High-level expression of proteins in mammalian cells using transcription regulatory sequences from the Chinese hamster ef-1α gene. Biotechnol Prog. 2004;20(3):880–889. doi: 10.1021/bp034383r. [DOI] [PubMed] [Google Scholar]
- 60.Qin J.Y., Zhang L., Clift K.L., Hulur I., Xiang A.P., Ren B.Z., et al. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS One. 2010;5(5) doi: 10.1371/journal.pone.0010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Backliwal G., Hildinger M., Chenuet S., Wulhfard S., De Jesus M., Wurm F.M. Rational vector design and multi-pathway modulation of HEK 293E cells yield recombinant antibody titers exceeding 1 g/l by transient transfection under serum-free conditions. Nucleic Acids Res. 2008;36(15):e96. doi: 10.1093/nar/gkn423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johari Y.B., Scarrott J.M., Pohle T.H., Liu P., Mayer A., Brown A.J., et al. Engineering of the CMV promoter for controlled expression of recombinant genes in HEK293 cells. Nat Biotechnol. 2022;17(8) doi: 10.1002/biot.202200062. [DOI] [PubMed] [Google Scholar]
- 63.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299(1–2):1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234(2):187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 65.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 66.Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997;16(9):2482–2492. doi: 10.1093/emboj/16.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Girod P.-A., Mermod N. Use of scaffold/matrix-attachment regions for protein production. N Compr Biochem. 2003;38:359–379. Elsevier. [Google Scholar]
- 69.Kim J.D., Yoon Y., Hwang H.-Y., Park J.S., Yu S., Lee J., et al. Efficient selection of stable Chinese hamster ovary (CHO) cell lines for expression of recombinant proteins by using human interferon β SAR element. Biotechnol Prog. 2005;21(3):933–937. doi: 10.1021/bp049598v. [DOI] [PubMed] [Google Scholar]
- 70.Zhao C.-P., Guo X., Chen S.-J., Li C.-Z., Yang Y., Zhang J.-H., et al. Matrix attachment region combinations increase transgene expression in transfected Chinese hamster ovary cells. Sci Rep. 2017;7(1) doi: 10.1038/srep42805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gallegos J.E., Rose A.B. An intron-derived motif strongly increases gene expression from transcribed sequences through a splicing independent mechanism in Arabidopsis thaliana. Sci Rep. 2019;9(1) doi: 10.1038/s41598-019-50389-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shaul O. How introns enhance gene expression. Int J Biochem Cell Biol. 2017;91(Pt B):145–155. doi: 10.1016/j.biocel.2017.06.016. [DOI] [PubMed] [Google Scholar]
- 73.Brinster R.L., Allen J.M., Behringer R.R., Gelinas R.E., Palmiter R.D. Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci U S A. 1988;85(3):836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Juneau K., Miranda M., Hillenmeyer M.E., Nislow C., Davis R.W. Introns regulate RNA and protein abundance in yeast. Genetics. 2006;174(1):511–518. doi: 10.1534/genetics.106.058560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Niu D.-K., Yang Y.-F. Why eukaryotic cells use introns to enhance gene expression: splicing reduces transcription-associated mutagenesis by inhibiting topoisomerase I cutting activity. Biol Direct. 2011;6(1):24. doi: 10.1186/1745-6150-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Akua T., Shaul O. The Arabidopsis thaliana MHX gene includes an intronic element that boosts translation when localized in a 5' UTR intron. J Exp Bot. 2013;64(14):4255–4270. doi: 10.1093/jxb/ert235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morello L., Gianì S., Troina F., Breviario D. Testing the IMEter on rice introns and other aspects of intron-mediated enhancement of gene expression. J Exp Bot. 2011;62(2):533–544. doi: 10.1093/jxb/erq273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rose A.B. Introns as gene regulators: a brick on the accelerator. Front Genet. 2019;9 doi: 10.3389/fgene.2018.00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buchman A.R., Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol. 1988;8(10):4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gallegos J.E., Rose A.B. The enduring mystery of intron-mediated enhancement. Plant Sci. 2015;237:8–15. doi: 10.1016/j.plantsci.2015.04.017. [DOI] [PubMed] [Google Scholar]
- 81.Baier T., Jacobebbinghaus N., Einhaus A., Lauersen K.J., Kruse O. Introns mediate post-transcriptional enhancement of nuclear gene expression in the green microalga Chlamydomonas reinhardtii. PLoS Genet. 2020;16(7) doi: 10.1371/journal.pgen.1008944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu D-h, Wang X-y, Jia Y-l, Wang T-y, Tian Z-w, Feng X., et al. SV40 intron, a potent strong intron element that effectively increases transgene expression in transfected Chinese hamster ovary cells. J Cell Mol Med. 2018;22(4):2231–2239. doi: 10.1111/jcmm.13504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boye C., Arpag S., Francis M., DeClemente S., West A., Heller R., et al. Reduction of plasmid vector backbone length enhances reporter gene expression. Bioelectrochemistry. 2022;144 doi: 10.1016/j.bioelechem.2021.107981. [DOI] [PubMed] [Google Scholar]
- 84.Ohse M., Takahashi K., Kadowaki Y., Kusaoke H. Effects of plasmid DNA sizes and several other factors on transformation of Bacillus subtilis ISW1214 with plasmid DNA by electroporation. Biosci Biotechnol Biochem. 1995;59(8):1433–1437. doi: 10.1271/bbb.59.1433. [DOI] [PubMed] [Google Scholar]
- 85.Kreiss P., Mailhe P., Scherman D., Pitard B., Cameron B., Rangara R., et al. Plasmid DNA size does not affect the physicochemical properties of lipoplexes but modulates gene transfer efficiency. Nucleic Acids Res Spec Publ. 1999;27(19):3792–3798. doi: 10.1093/nar/27.19.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lu J., Wu T., Zhang B., Liu S., Song W., Qiao J., et al. Types of nuclear localization signals and mechanisms of protein import into the nucleus. Cell Commun Signal. 2021;19(1):60. doi: 10.1186/s12964-021-00741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zanta M.A., Belguise-Valladier P., Behr J.P. Gene delivery: a single nuclear localization signal peptide is sufficient to carry DNA to the cell nucleus. Proc Natl Acad Sci U S A. 1999;96(1):91–96. doi: 10.1073/pnas.96.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu Y., Liang W., Qiu Y., Cespi M., Palmieri G.F., Mason A.J., et al. Incorporation of a nuclear localization signal in pH responsive LAH4-L1 peptide enhances transfection and nuclear uptake of plasmid DNA. Mol Pharm. 2016;13(9):3141–3152. doi: 10.1021/acs.molpharmaceut.6b00338. [DOI] [PubMed] [Google Scholar]
- 89.López-Fuertes L., Pérez-Jiménez E., Vila-Coro A.J., Sack F., Moreno S., Konig S.A., et al. DNA vaccination with linear minimalistic (MIDGE) vectors confers protection against Leishmania major infection in mice. Vaccine. 2002;21(3–4):247–257. doi: 10.1016/s0264-410x(02)00450-4. [DOI] [PubMed] [Google Scholar]
- 90.Moreno S., López-Fuertes L., Vila-Coro A.J., Sack F., Smith C.A., Konig S.A., et al. DNA immunisation with minimalistic expression constructs. Vaccine. 2004;22(13–14):1709–1716. doi: 10.1016/j.vaccine.2003.09.051. [DOI] [PubMed] [Google Scholar]
- 91.Jin H., Li Y., Ma Z., Zhang F., Xie Q., Gu D., et al. Effect of chemical adjuvants on DNA vaccination. Vaccine. 2004;22(21):2925–2935. doi: 10.1016/j.vaccine.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 92.Quattrocchi V., Soria I., Langellotti C.A., Gnazzo V., Gammella M., Moore D.P., et al. A DNA vaccine formulated with chemical adjuvant provides partial protection against bovine Herpes virus infection in cattle. Front Immunol. 2017;8 doi: 10.3389/fimmu.2017.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Di Giacomo S., Quattrocchi V., Zamorano P. Use of adjuvants to enhance the immune response induced by a DNA vaccine against bovine Herpesvirus-1. Viral Immunol. 2015;28(6):343–346. doi: 10.1089/vim.2014.0113. [DOI] [PubMed] [Google Scholar]
- 94.Wang S., Liu X., Fisher K., Smith J.G., Chen F., Tobery T.W., et al. Enhanced type I immune response to a hepatitis B DNA vaccine by formulation with calcium- or aluminum phosphate. Vaccine. 2000;18(13):1227–1235. doi: 10.1016/s0264-410x(99)00391-6. [DOI] [PubMed] [Google Scholar]
- 95.Garg R., Kaur M., Saxena A., Prasad R., Bhatnagar R. Alum adjuvanted rabies DNA vaccine confers 80% protection against lethal 50 LD(50) rabies challenge virus standard strain. Mol Immunol. 2017;85:166–173. doi: 10.1016/j.molimm.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 96.Grunwald T., Ulbert S. Improvement of DNA vaccination by adjuvants and sophisticated delivery devices: vaccine-platforms for the battle against infectious diseases. Clin Exp Vaccine Res. 2015;4(1):1–10. doi: 10.7774/cevr.2015.4.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lin W.H., Vilalta A., Adams R.J., Rolland A., Sullivan S.M., Griffin D.E. Vaxfectin adjuvant improves antibody responses of juvenile rhesus macaques to a DNA vaccine encoding the measles virus hemagglutinin and fusion proteins. J Virol. 2013;87(12):6560–6568. doi: 10.1128/JVI.00635-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pan C.-H., Jimenez Gretchen S., Nair N., Wei Q., Adams Robert J., Polack Fernando P., et al. Use of Vaxfectin adjuvant with DNA vaccine encoding the measles virus hemagglutinin and fusion proteins protects juvenile and infant rhesus macaques against measles virus. Clin Vaccine Immunol. 2008;15(8):1214–1221. doi: 10.1128/CVI.00120-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Danko J.R., Kochel T., Teneza-Mora N., Luke T.C., Raviprakash K., Sun P., et al. Safety and immunogenicity of a tetravalent Dengue DNA vaccine administered with a cationic lipid-based adjuvant in a phase 1 clinical trial. Am J Trop Med Hyg. 2018;98(3):849–856. doi: 10.4269/ajtmh.17-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Karlsson I., Borggren M., Nielsen J., Christensen D., Williams J., Fomsgaard A. Increased humoral immunity by DNA vaccination using an α-tocopherol-based adjuvant. Hum Vaccines Immunother. 2017;13(8):1823–1830. doi: 10.1080/21645515.2017.1321183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Grigoryan L., Lee A., Walls A.C., Lai L., Franco B., Arunachalam P.S., et al. Adjuvanting a subunit SARS-CoV-2 vaccine with clinically relevant adjuvants induces durable protection in mice. NPJ Vaccines. 2022;7(1):55. doi: 10.1038/s41541-022-00472-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bayart J.L., Douxfils J., Gillot C., David C., Mullier F., Elsen M., et al. Waning of IgG, total and neutralizing antibodies 6 months post-vaccination with BNT162b2 in healthcare workers. Vaccines (Basel) 2021;9(10) doi: 10.3390/vaccines9101092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Elgueta R., Benson M.J., de Vries V.C., Wasiuk A., Guo Y., Noelle R.J. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev. 2009;229(1):152–172. doi: 10.1111/j.1600-065X.2009.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chatzigeorgiou A., Lyberi M., Chatzilymperis G., Nezos A., Kamper E. CD40/CD40L signaling and its implication in health and disease. Biofactors. 2009;35(6):474–483. doi: 10.1002/biof.62. [DOI] [PubMed] [Google Scholar]
- 105.Tamming L.A., Duque D., Tran A., Zhang W., Pfeifle A., Laryea E., et al. DNA Based Vaccine Expressing SARS-CoV-2 Spike-CD40L Fusion protein confers protection against challenge in a Syrian hamster model. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.785349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ahn J.Y., Lee J., Suh Y.S., Song Y.G., Choi Y.-J., Lee K.H., et al. Safety and immunogenicity of two recombinant DNA COVID-19 vaccines containing the coding regions of the spike or spike and nucleocapsid proteins: an interim analysis of two open-label, non-randomised, phase 1 trials in healthy adults. Lancet Microbe. 2022;3(3):e173–e183. doi: 10.1016/S2666-5247(21)00358-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Routhu N.K., Cheedarla N., Bollimpelli V.S., Gangadhara S., Edara V.V., Lai L., et al. SARS-CoV-2 RBD trimer protein adjuvanted with Alum-3M-052 protects from SARS-CoV-2 infection and immune pathology in the lung. Nat Commun. 2021;12(1):3587. doi: 10.1038/s41467-021-23942-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jeong H., Choi Y.-M., Seo H., Kim B.-J. A Novel DNA Vaccine against SARS-CoV-2 encoding a chimeric protein of its receptor-binding domain (RBD) fused to the amino-terminal region of Hepatitis B virus preS1 with a W4P mutation. Front Immunol. 2021:12. doi: 10.3389/fimmu.2021.637654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Park K.S., Sun X., Aikins M.E., Moon J.J. Non-viral COVID-19 vaccine delivery systems. Adv Drug Deliv Rev. 2021;169:137–151. doi: 10.1016/j.addr.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu H.Y., Nguyen H.H., Russell M.W. Nasal lymphoid tissue (NALT) as a mucosal immune inductive site. Scand J Immunol. 1997;46(5):506–513. doi: 10.1046/j.1365-3083.1997.d01-159.x. [DOI] [PubMed] [Google Scholar]
- 111.Lambricht L., Lopes A., Kos S., Sersa G., Préat V., Vandermeulen G. Clinical potential of electroporation for gene therapy and DNA vaccine delivery. Expet Opin Drug Deliv. 2016;13(2):295–310. doi: 10.1517/17425247.2016.1121990. [DOI] [PubMed] [Google Scholar]
- 112.Sardesai N.Y., Weiner D.B. Electroporation delivery of DNA vaccines: prospects for success. Curr Opin Immunol. 2011;23(3):421–429. doi: 10.1016/j.coi.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vela Ramirez J.E., Sharpe L.A., Peppas N.A. Current state and challenges in developing oral vaccines. Adv Drug Deliv Rev. 2017;114:116–131. doi: 10.1016/j.addr.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dey A., Chozhavel Rajanathan T.M., Chandra H., Pericherla H.P.R., Kumar S., Choonia H.S., et al. Immunogenic potential of DNA vaccine candidate, ZyCoV-D against SARS-CoV-2 in animal models. Vaccine. 2021;39(30):4108–4116. doi: 10.1016/j.vaccine.2021.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ravi A.D., Sadhna D., Nagpaal D., Chawla L. Needle free injection technology: a complete insight. Int J Pharm Investig. 2015;5(4):192–199. doi: 10.4103/2230-973X.167662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lim M., Badruddoza A.Z.M., Firdous J., Azad M., Mannan A., Al-Hilal T.A., et al. Engineered nanodelivery systems to improve DNA vaccine technologies. Pharmaceutics. 2020;12(1):30. doi: 10.3390/pharmaceutics12010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Farris E., Brown D.M., Ramer-Tait A.E., Pannier A.K. Micro- and nanoparticulates for DNA vaccine delivery. Exp Biol Med. 2016;241(9):919–929. doi: 10.1177/1535370216643771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pati R., Shevtsov M., Sonawane A. Nanoparticle vaccines against infectious diseases. Front Immunol. 2018;9:2224. doi: 10.3389/fimmu.2018.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sousa de Almeida M., Susnik E., Drasler B., Taladriz-Blanco P., Petri-Fink A., Rothen-Rutishauser B. Understanding nanoparticle endocytosis to improve targeting strategies in nanomedicine. Chem Soc Rev. 2021;50(9):5397–5434. doi: 10.1039/d0cs01127d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Barua S., Mitragotri S. Challenges associated with penetration of nanoparticles across cell and tissue barriers: a review of current status and future prospects. Nano Today. 2014;9(2):223–243. doi: 10.1016/j.nantod.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Crotts G., Park T.G. Protein delivery from poly(lactic-co-glycolic acid) biodegradable microspheres: release kinetics and stability issues. J Microencapsul. 1998;15(6):699–713. doi: 10.3109/02652049809008253. [DOI] [PubMed] [Google Scholar]
- 122.Santhanes D., Wilkins A., Zhang H., John Aitken R., Liang M. Microfluidic formulation of lipid/polymer hybrid nanoparticles for plasmid DNA (pDNA) delivery. Int J Pharm. 2022;627 doi: 10.1016/j.ijpharm.2022.122223. [DOI] [PubMed] [Google Scholar]
- 123.Zhao K., Li W., Huang T., Luo X., Chen G., Zhang Y., et al. Preparation and efficacy of Newcastle disease virus DNA vaccine encapsulated in PLGA nanoparticles. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0082648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Makadia H.K., Siegel S.J. Poly Lactic-co-Glycolic Acid (PLGA) as biodegradable controlled drug delivery carrier. Polymers. 2011;3(3):1377–1397. doi: 10.3390/polym3031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhao K., Zhang Y., Zhang X., Li W., Shi C., Guo C., et al. Preparation and efficacy of Newcastle disease virus DNA vaccine encapsulated in chitosan nanoparticles. Int J Nanomed. 2014;9:389–402. doi: 10.2147/IJN.S54226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang G., Pan L., Zhang Y., Wang Y., Zhang Z., Lü J., et al. Intranasal delivery of cationic PLGA nano/microparticles-loaded FMDV DNA vaccine encoding IL-6 elicited protective immunity against FMDV challenge. PLoS One. 2011;6(11) doi: 10.1371/journal.pone.0027605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Souci L., Jaunet H., Le Diguerher G., Guionnet J.-M., Béven V., Paboeuf F., et al. Intranasal inoculations of naked or PLGA-PEI nanovectored DNA vaccine induce systemic and mucosal antibodies in pigs: a feasibility study. Res Vet Sci. 2020;132:194–201. doi: 10.1016/j.rvsc.2020.06.018. [DOI] [PubMed] [Google Scholar]
- 128.Bivas-Benita M., Romeijn S., Junginger H.E., Borchard G. PLGA-PEI nanoparticles for gene delivery to pulmonary epithelium. Eur J Pharm Biopharm. 2004;58(1):1–6. doi: 10.1016/j.ejpb.2004.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhou Y., Fang F., Chen J., Wang H., Chang H., Yang Z., et al. Electroporation at low voltages enables DNA vaccine to provide protection against a lethal H5N1 avian influenza virus challenge in mice. Intervirology. 2008;51(4):241–246. doi: 10.1159/000156483. [DOI] [PubMed] [Google Scholar]
- 130.Shen C., Li J., Zhang Y., Li Y., Shen G., Zhu J., et al. Polyethylenimine-based micro/nanoparticles as vaccine adjuvants. Int J Nanomed. 2017;12:5443–5460. doi: 10.2147/IJN.S137980. [DOI] [PMC free article] [PubMed] [Google Scholar]