IMPORTANCE:
CD4+ T cells contribute to lung inflammation in acute respiratory distress syndrome. The CD4+ T-cell response in pediatric acute respiratory distress syndrome (PARDS) is unknown.
OBJECTIVES:
To identify differentially expressed genes and networks using a novel transcriptomic reporter assay with donor CD4+ T cells exposed to the airway fluid of intubated children with mild versus severe PARDS.
DESIGN:
In vitro pilot study.
SETTING:
Laboratory-based study using human airway fluid samples admitted to a 36-bed university-affiliated pediatric intensive care unit.
PATIENTS/SUBJECTS:
Seven children with severe PARDS, nine children with mild PARDS, and four intubated children without lung injury as controls.
INTERVENTIONS:
None.
MEASUREMENTS AND MAIN RESULTS:
We performed bulk RNA sequencing using a transcriptomic reporter assay of CD4+ T cells exposed to airway fluid from intubated children to discover gene networks differentiating severe from mild PARDS. We found that innate immunity pathways, type I (α and β), and type II (γ) interferon response and cytokine/chemokine signaling are downregulated in CD4+ T cells exposed to airway fluid from intubated children with severe PARDS compared with those with mild PARDS.
CONCLUSIONS:
We identified gene networks important to the PARDS airway immune response using bulk RNA sequencing from a novel CD4+ T-cell reporter assay that exposed CD4+ T cells to airway fluid from intubated children with severe and mild PARDS. These pathways will help drive mechanistic investigations into PARDS. Validation of our findings using this transcriptomic reporter assay strategy is needed.
Keywords: pediatric acute respiratory distress syndrome, pediatric intensive care, ribonucleic acid sequencing, T cells, tracheal aspirate, transcriptomics
KEY POINTS
Question: What is the CD4+ helper T-cell response to the tracheal aspirate fluid from intubated children with severe, mild, and no pediatric acute respiratory distress syndrome (PARDS)?
Findings: Innate immunity pathways, type I (α and β), and type II (γ) interferon response and cytokine/chemokine signaling pathways are downregulated in CD4+ helper T cells exposed to the tracheal aspirate fluid from children with severe PARDS compared with those with mild or no PARDS.
Meaning: Use of a transcriptomic reporter assay with RNA sequencing may provide a clinically useful strategy to understand the complex heterogeneity of the airway immune response of PARDS.
Pediatric acute respiratory distress syndrome (PARDS) is a heterogenous disorder (1) affecting approximately 6% of all mechanically ventilated children (2). One in three children with severe PARDS die (2). Despite the clinical impact, we know little about the underlying biological pathways associated with PARDS severity. While we know that innate immune cells contribute to the pathobiology of acute respiratory distress syndrome (ARDS), we now recognize that helper T-cell subsets, including regulatory T cells, play a crucial role in regulating lung inflammation in ARDS (3–6) and mouse models of acute lung injury (7–11). To date, helper T-cell responses have not been examined in children with PARDS.
Primary T cells are limited in airway fluid samples from intubated children, necessitating the use of a transcriptomic reporter assay to elucidate the T-cell transcriptome in PARDS. Transcriptomic reporter assays are based on the premise that biological fluids, such as airway fluid or plasma, contain immunologic mediators that can stimulate responses of a human donor immune cell population ex vivo (12–16). Herein, we use donor CD4+ T cells exposed to tracheal aspirate fluid from intubated children to compare the differential transcriptomic responses from intubated children with mild, severe, and no PARDS. We hypothesized that donor CD4+ T cells exposed to tracheal aspirate fluid from intubated children with severe versus mild or no PARDS would have differentially expressed genes that would enable the discovery of novel immune pathways important to PARDS severity.
METHODS
Patient Cohort and Ethics Statement
The airway fluid samples from children enrolled in this study are part of an ongoing, prospective observational cohort study conducted in the Children’s Healthcare of Atlanta PICU which is a 36-bed unit affiliated with the Emory University School of Medicine. The Emory University School of Medicine Institutional Review Board (IRB00034236: Prevalence of Oxidative Stress in Critically Ill Children and Its Relationship to Immune Function: A Pilot Study. Approved: February 14, 2018 and IRB00113035: Airway Immune Response in Critically Ill Children: Precision Medicine in Children at Risk for Acute Respiratory Distress Syndrome. Approved: July 29, 2019) approved the study. Informed consent from a parent or legal guardian was performed by a trained study coordinator prior to enrollment. All study procedures were performed according to the relevant guidelines and regulations in the Declaration of Helsinki. Participants were enrolled between September 2018 and March 2020. Children were eligible for enrollment if they were admitted to the PICU, greater than 2 days old and with a corrected gestational age of 40 weeks, were less than 18 years old, and were within 72 hours of endotracheal intubation. Children were excluded if they had perinatal-related lung disease, chronic respiratory failure requiring mechanical ventilation via a tracheostomy or RAM cannula, immunodeficiency, were receiving immunosuppression from chemotherapy for an oncologic disease, were chronically immunosuppressed as a hematologic or solid-organ transplant recipient, there was no parent or legal guardian to provide in-person written informed consent, or the attending physician did not wish the patient to participant in the study. Severity of hypoxia was classified according to the 2015 Pediatric Acute Lung Injury Consensus Conference-1 definitions for having PARDS (17). PARDS severity was calculated at the time of sample collection. Severity of illness scores were determined using the Pediatric Risk of Mortality (PRISM)-III, calculated at the time of PICU admission, and Pediatric Logistic Organ Dysfunction (PELOD-2) score calculated within 24 hours of study enrollment (18, 19).
Tracheal Aspirate Collection and Sample Processing
Tracheal aspirate samples were collected within 72 hours of endotracheal intubation at the time of study enrollment with an inline Ballard suction catheter connected to a sterile Lukens trap using up to 5 mL of sterile saline and processed according to published protocols (20–22). The Ballard inline suction catheter was passed once to obtain a sample. If 50% of the instilled volume was not returned, then a repeat passage of the Ballard was performed. The Ballard suction catheter could be flushed with additional saline to move the aspirate into the Lukens trap. The airway fluid was separated from the cells by centrifugation at 800 × g for 15 minutes at 4°C. Airway fluid supernatant was collected into a fresh tube, centrifuged at 3,000 × g for 15 minutes at 4°C, and the resulting cell-free supernatant was stored at –80°C until use.
Cell Culture
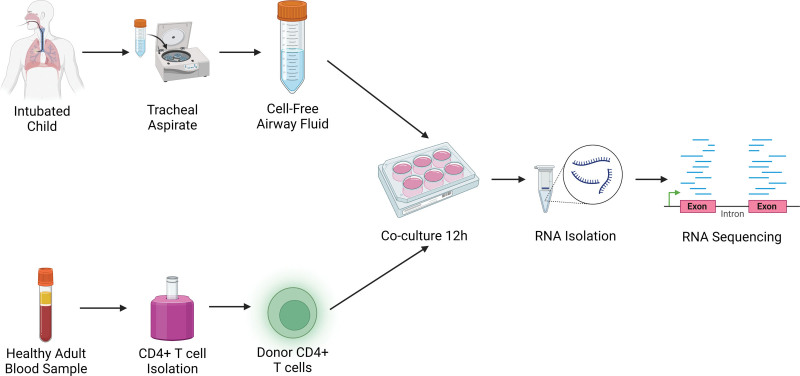
Commercially available single donor CD4+ T cells were purchased from Lonza (Greenwood, SC). CD4+ T cells were resuspended in a 1:1 vol:vol mixture of airway supernatant from individual patients in Roswell Park Memorial Institute 1640 medium with L-glutamine (Corning, Corning, NY), 10% fetal calf serum supplemented with penicillin, streptomycin, and gentamicin and cultured for 12 hours in a humidified 37°C, 5% Co2 incubator. Following overnight culture, CD4+ T cells were pelleted by centrifugation at 400 × g at 4°C for 15 minutes, resuspended in 500 μL of RNAlater, and stored at –80°C until RNA was isolated. A schematic of the tracheal aspirate sample processing and CD4+ T cells transcriptomic reporter assay using airway fluid is shown in Figure 1.
Figure 1.
CD4+ T-cell transcriptomic reporter assay with patient airway fluid from children with no, mild, and severe pediatric acute respiratory distress syndrome. Cell-free airway fluid from tracheal aspirate samples was obtained within 72 hr of intubation. CD4 T cells were isolated from the blood of healthy adults using negative magnetic bead selection. Airway fluid was used to stimulate healthy donor CD4+ T cells in media (50% volume/volume) for 12 hr. Following in vitro stimulation, RNA was isolated from CD4+ T cells and RNA sequencing was performed.
RNA Preparation
RNA was isolated from CD4+ T cells using the Nucleospin RNA II kit with on-column genomic DNA digestion according to the manufacturer’s protocol (Takara, Mountain View, CA). RNA sizing quantification and quality control was performed in the Emory Integrated Genomics Core (Atlanta, GA) on an Agilent (Santa Clara, CA) 2100 bioanalyzer using Pico and Nano Agilent kits and a Tecan (Morrisville, NC) optical density plate reader to measure the concentration of the RNA (21).
RNA Sequencing
RNA was quality assessed using the 2100 Bioanalyzer (Agilent). Complementary DNA was generated using the SMART-Seq v4 Ultra Low Input RNA Kit Takara Bio (San Jose, CA), and the final RNA sequencing library was generated using the Nextera XT kit Illumina (San Diego, CA). RNA sequencing libraries were sequenced on the Illumina NovaSeq 6000 with a sequencing depth of 30M PE100 reads per sample.
Data Preprocessing
Fastp was used to perform quality control and adapter trimming and the results were filtered by quality. HISAT2 (Version: 2.2.1) (23) was used to map RNA sequencing reads prior to building the index using the human reference genome Genome Reference Consortium Human Build 38. We then used indices to map our sequential reads to the human genome. We applied featureCounts (Version: 2.0.2) (24) to generate a matrix of row counts of each of the genes in all the samples. Genes were kept in the analysis that had at least 10 reads in at least half of the samples.
Differential Gene Expression Analysis
The R package Deseq2 (Version: 1.38.3) (25) was used to determine differential gene expression. We used variance stabilizing transformations for raw data transformation. We next transformed data on the log2 scale and normalized to library size. We used the Anan empirical Bayes shrinkage methods to detect and correct for dispersion and calculate log2[fold change] for the genes. Differentially expressed genes were defined as having a |log2[fold change]| of greater than 1. The p values were corrected for multiple testing using the Benjamini-Hochberg procedure. A false discovery rate (FDR) less than 0.05 was considered significant.
Pathway Analysis
Reactome (https://reactome.org) was used to obtain the relevant biological pathways compared with different groups derived from the differentially expressed genes (26–31). The report generated is an overrepresentation analysis based on a hypergeometric distribution test that determines whether certain Reactome pathways are enriched in the submitted differentially gene expression data compared with that by chance alone. The statistical test produces a probability score corrected for an FDR using the Benjamini-Hochberg method. Only pathways with an FDR less than 0.05 were used for the analysis.
RESULTS
Characteristics of the Study Participants
The demographics and clinical characteristics of the 20 children (four critically ill without PARDS, nine mild PARDS, and seven severe PARDS) are summarized in Table 1. All but three tracheal aspirate samples were collected within 48 hours of intubation. Four of the seven children with severe PARDS received life-support with extracorporeal membrane oxygenation (ECMO) during their hospital course, and none were on ECMO at the time of sample collection. No participants died. A flow diagram of the participant samples used in this study is shown in Supplemental Figure 1 (http://links.lww.com/CCX/B212).
TABLE 1.
Demographic and Clinical Features of the Participants by Pediatric Acute Respiratory Distress Syndrome Status and Pediatric Acute Respiratory Distress Syndrome Severity
| Feature | Overall (n = 20) | Control (n = 4) | Mild PARDS (n = 9) | Severe PARDS (n = 7) |
|---|---|---|---|---|
| Age, mo | 24 (18–36) | 35 (24–49) | 21 (7–24) | 24 (22–33) |
| Male | 12 (60) | 3 (75) | 5 (55.6) | 4 (57.1) |
| Ethnicity | ||||
| Hispanic or Latino | 1 (5) | 0 (0) | 0 (0) | 1 (14.2) |
| Not Hispanic or Latino | 19 (95) | 4 (100) | 9 (100) | 6 (85.7) |
| Race | ||||
| White | 8 (40) | 3 (75) | 3 (33.3) | 2 (28.6) |
| Black | 10 (50) | 1 (25) | 5 (55.6) | 4 (57.1) |
| More than one race | 2 (10) | 0 | 1 (11.1) | 1 (14.3) |
| Bacterial respiratory culture | ||||
| Positive | 12 (60) | 0 (0) | 8 (88.9) | 4 (57.1) |
| Negative | 3 (15) | 0 (0) | 0 (0) | 3 (42.6) |
| Not performed | 5 (25) | 4 (100) | 1 (11.1) | 0 (0) |
| Respiratory viral panel | ||||
| Positive | 10 (50) | 0 | 5 (55.6) | 5 (71.4) |
| Negative | 3 (15) | 0 | 1 (11.1) | 2 (28.6) |
| Not performed | 7 (35) | 4 (100) | 3 (33.3) | 0 |
| Pediatric Risk of Mortality-III | 13 (11–17) | 9 (8–11) | 13 (12–18) | 14 (12–18) |
| Pediatric Logistic Organ Dysfunction-2 | 7 (5–8) | 8 (6–9) | 6 (5–8) | 7 (5–11) |
| Hours to sample collection | 25 (2–57) | 12 (10–14) | 26 (9–51) | 30 (2–57) |
| ICU days | 7 (6–12) | 4 (2–6) | 7 (6–12) | 15 (9–29)a |
| Ventilator daysc | 6 (4–9) | 2 (2–3) | 5 (4–8) | 7 (7–23)a |
| Extracorporeal membrane oxygenation | 4 (20) | 0 (0) | 0 (0) | 4 (57.1)b |
PARDS = pediatric acute respiratory distress syndrome.
p < 0.01 compared with mild and control by Kruskal-Wallis test.
p < 0.05 compared with mild by Fisher exact test.
No deaths occurred in any of the groups.
Transcriptional Profiles of Mild and Severe PARDS
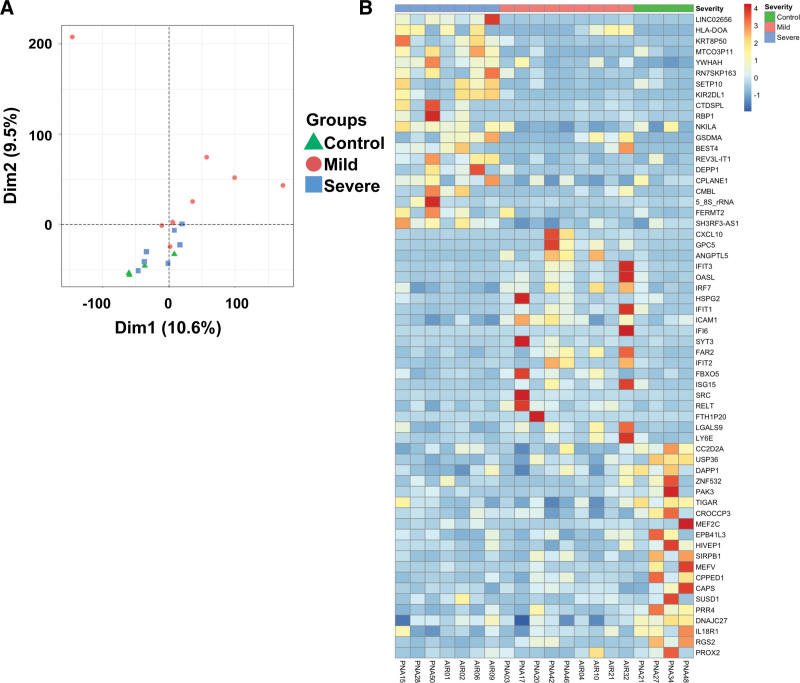
We performed a principal component analysis (PCA) (Fig. 2A) and generated a heatmap (Fig. 2B) of the differentially expressed genes of T cells exposed to airway fluid from children without PARDS and with mild and severe PARDS. The first- and second-dimensions accounted for 10.6% and 9.5% of the variance in the differentially expressed genes among the control, mild and severe PARDS groups. There was no change in variance when clinical features such as age at admission, severity of PARDS or control status, PRISM-III score, PELOD-2 score, viral lower respiratory tract infection, and positive respiratory culture were included (PCA-1: 10.6% and PCA-2: 9.5%). A heatmap of the top 20 up-regulated and down-regulated differentially expressed genes for among the three groups is shown in Figure 2B.
Figure 2.
Cluster and heat map of differentially expressed genes. A, Principal component analysis of differentially expressed genes in donor CD4+ T cells exposed to airway fluid from control children (no pediatric acute respiratory distress syndrome [PARDS] in green) and those with mild PARDS (red) and severe PARDS (blue). B, Heat maps of the top 60 differentially expressed genes in naive donor CD4+ T cells exposed to airway fluid from children with no PARDS (control) (green), mild PARDS (red), and severe PARDS (blue). Dim = dimension.
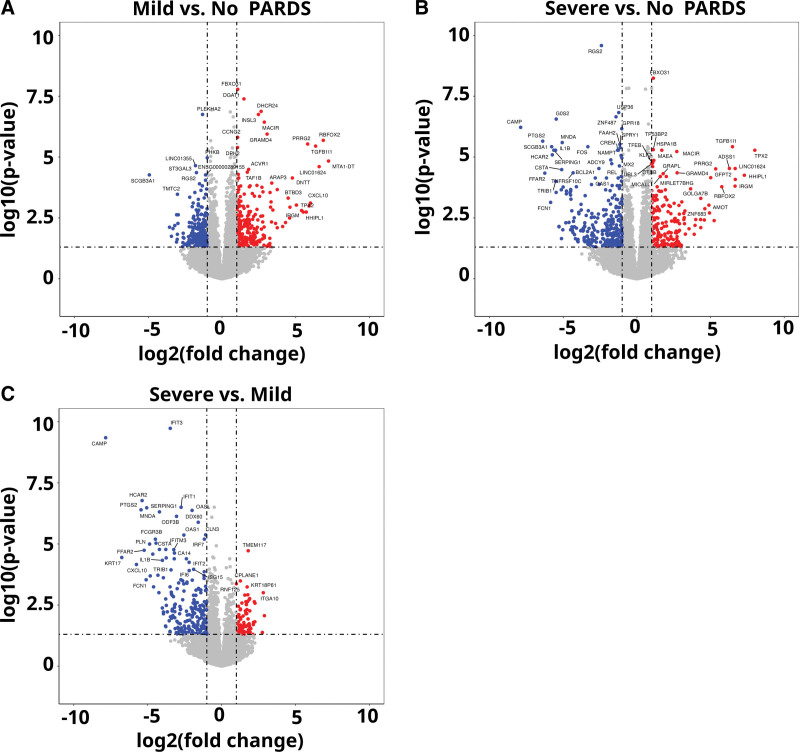
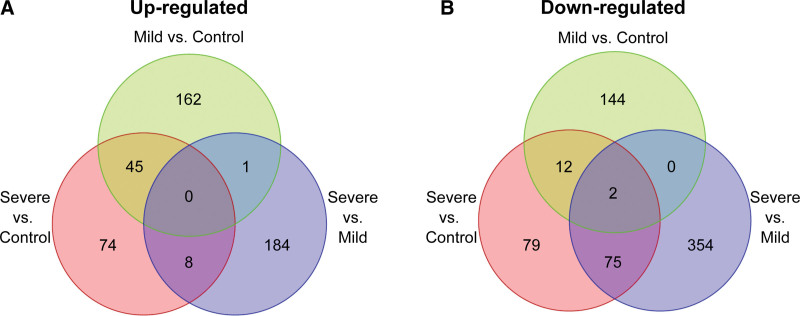
Volcano plots of the differentially expressed genes for cells in the mild versus no PARDS groups and the severe versus mild and no PARDS groups are shown in Figure 3A–C. Lists of differentially expressed genes for each comparison are publicly available (https://github.com/mjripple/Tcell-RNA-seq.git) and in the Supplemental Digital Content E1–E6 (http://links.lww.com/CCX/B211). Transcripts were selected if there was at least a two-fold difference in response to airway fluid stimulation compared with the respective mild PARDS or control comparison group. Venn diagrams of the up-regulated (Fig. 4A; and Supplemental Digital Content E2, E4, and E6, http://links.lww.com/CCX/B211) and down-regulated (Fig. 4B; and Supplemental Digital Content E1, E3, and E5, http://links.lww.com/CCX/B211) among the three groups are shown.
Figure 3.
Volcano plots. Volcano plots of differentially expressed genes in donor CD4+ T cells exposed to airway fluid from children with mild versus no pediatric acute respiratory distress syndrome (PARDS) (A), severe versus no PARDS (B), and severe verses mild PARDS (C). Each circle is a gene. Blue circles represent downregulated genes that have at least a two-fold change with a false discovery rate of less than 0.05. Red circles represent up-regulated genes that have at least a two-fold change with a false discovery rate of less than 0.05. Dashed lines indicate the x-axis threshold of |log2[fold change]| > 1 and the y-axis threshold of log10[p] < 0.05.
Figure 4.
Venn diagrams. Venn diagrams indicate the number of up-regulated (A) and down-regulated (B) differentially expressed genes in naive donor CD4+ T cells exposed to airway fluid from children with no pediatric acute respiratory distress syndrome (PARDS) (control) (green), mild PARDS (red), and severe PARDS (blue).
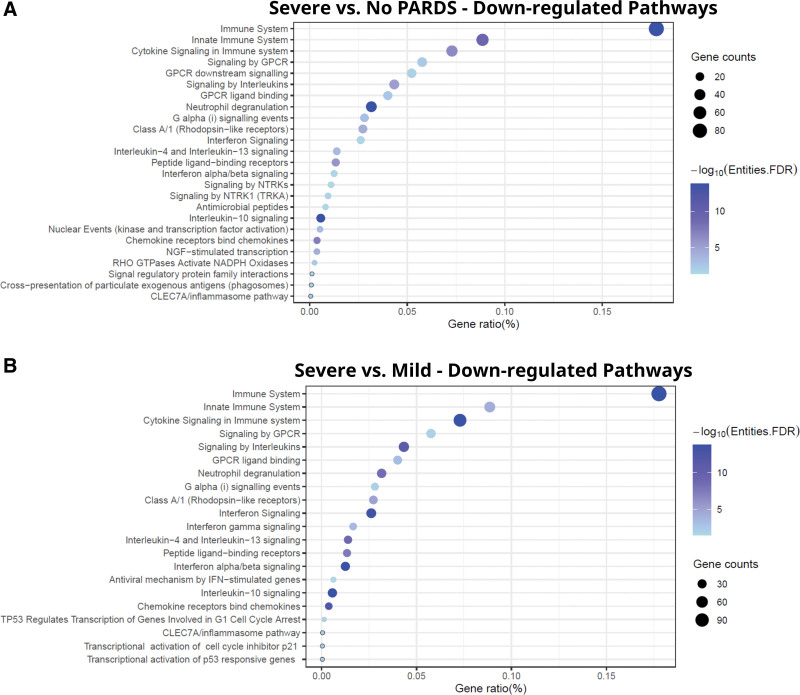
Pathway Analysis of Severe PARDS
Most of the significantly enriched functional networks were relevant to immune processes and were down-regulated in samples exposed to airway fluid from patients with severe PARDS compared with those with mild PARDS (Fig. 5A) or controls (Fig. 5B) including the innate immune system, cytokine/interleukin signaling, G protein-coupled receptors signaling, and neutrophil degranulation. There were 25 significant Reactome pathways found using the down-regulated genes when comparing severe PARDS versus control groups (Fig. 5A). There were no significant pathways that differed in the up-regulated genes in the severe versus no PARDS groups. There were 21 significant Reactome pathways found using the down-regulated genes when comparing the severe versus mild PARDS groups (Fig. 5B). A full list of enriched down-regulated pathways in Reactome for severe versus mild PARDS groups (Supplemental Digital Content E7, http://links.lww.com/CCX/B211) and severe PARDS versus control groups (Supplemental Digital Content E8, http://links.lww.com/CCX/B211) is publicly available (https://github.com/mjripple/Tcell-RNA-seq.git). The only significant pathways that differed in the up-regulated genes in the severe versus no PARDS groups were the synthesis of phosphatidylinositol. There were no significant pathways that differed when comparing the differentially expressed genes in the mild versus no PARDS groups.
Figure 5.
Gene pathway analysis. Reactome pathways listed by gene ratio (%) for the significantly downregulated genes in donor CD4+ T cells exposed to airway fluid from children with severe versus no pediatric acute respiratory distress syndrome (PARDS) (A), and severe versus mild PARDS (B). Size of the circle indicated gene counts in the pathway. Darker blue indicates a lower adjusted p value. A false discovery rate (FDR) of less than 0.05 was considered significant. CLEC7A = C-type lectin domain containing 7A, GPCR = G-protein coupled receptor, GTPases = guanosine triphosphatases, IFN = interferon, NGF = nerve growth factor, NADPH = nicotinamide adenine dinucleotide phosphate, NTRK = neurotrophic tyrosine receptor kinase gene fusion, RHO = rhodopsin, TRKA = tropomyosin receptor kinase A, TP53 = tumor protein p53.
Transcriptional Profiles and Pathway Analysis of Viral Versus Nonviral Triggered PARDS
Volcano plots of the differentially expressed genes for cells in the viral versus nonviral triggered PARDS are shown in Supplemental Figure 2A (http://links.lww.com/CCX/B212). The viral-triggered PARDS group had significantly up-regulated pathways related to interferon (IFN) signaling and anti-viral responses compared with the nonviral triggered PARDS group (Supplemental Fig. 2B, http://links.lww.com/CCX/B212).
DISCUSSION
In this study, we used a transcriptomic reporter assay with bulk RNA sequencing to analyze the CD4+ T-cell gene signature response to airway fluid from patients with PARDS. We found that activation of the antiviral, type I (α and β), and type II (γ) IFN response and cytokine/chemokine signaling are downregulated in donor CD4+ T cells exposed to tracheal aspirate fluid from intubated children with severe PARDS compared with those with mild PARDS or those with acute respiratory failure without PARDS.
Dysregulated host responses, including impaired type I IFN activity in viral lower respiratory tract infections, have been shown to contribute to disease severity (32, 33). Specifically, having no IFN-β and low IFN-α production and activity was associated with persistent viremia and high levels of tumor necrosis factor-α and interleukin-6 signaling in severe COVID-19 infections (33). In a secondary analysis of adults with COVID-19 ARDS (n = 8) compared with those with non-COVID-19 viral lower respiratory tract infection ARDS, Sarma et al (34) found that IFN-related gene expression was lower in the COVID-19 versus viral (not COVID-19) induced ARDS. In a mouse model of cytokine release syndrome-associated extrapulmonary acute lung injury, neutralization or genetic knockout of IFN-γ resulted in worse lung injury (35). Gene expression analysis of nasopharyngeal aspiration samples from infants intubated with severe respiratory syncytial virus (RSV) bronchiolitis had lower type I IFN levels compared with moderately ill nonintubated hospitalized children with RSV bronchiolitis (36). Children hospitalized with severe RSV expressed higher levels of neutrophil and inflammatory genes and lower type I and II IFN genes compared with nonhospitalized children with mild RSV (32). A low type I and III IFN response with high systemic proinflammatory cytokine and chemokine levels is a hallmark of severe COVID-19 (37, 38).
As shown by the heatmaps, there is a wide variation in gene expression for individual patients within a PARDS severity classification. One explanation for this is the inherent heterogeneity in the underlying immune response in children with PARDS regardless of severity. There is poor separation on the PCA plot of the controls and severe PARDS group that is likely due to the small sample size and an outlier in the mild PARDS group that further highlights the heterogeneity within PARDS severity groups. Other explanations for the heterogeneity among samples with the same PARDS severity include differences in the timing of sample collection from intubation, the initial trigger inciting PARDS, variation in sample collection and dilution, and having other organ dysfunctions besides lung failure. Demographic and clinical variables considered in the PCA did not explain the observed heterogeneity suggesting that the differentially expressed genes determined the T-cell response to tracheal aspirate fluid among the control, mild and severe PARDS groups.
This study is limited to a small sample of patients at a single children’s hospital, which may limit generalizability. Because the study relied on healthy donor T cells, some of the findings may be attributable to genetic or epigenetic influences of the donor cells. Variability was limited in this context as all experiments were performed with the same aliquot of donor cells. We grouped samples by PARDS severity as calculated on the day of sample collection and therefore cannot correlate clinical trajectory or subsequent changes in severity with transcriptome responses. Given the complexities of performing bronchoalveolar lavage in children with PARDS, our study relied on the use of tracheal aspirates. Samples were collected by endotracheal tube suctioning for convenience and safety and may not recapitulate the immune system mediators present at the level of the alveolus. While we attempted to be consistent in the lavage procedure, we acknowledge that there are no good alternatives to controlling the volume instilled by age and the variability in the amounts of fluid returned with the tracheal aspirate sampling method. Furthermore, we do not know the impact that sample collection outside of a 24-hour window following intubation has on the results. Without more data that pairs clinical trajectory with biological sampling, we can only speculate that children with sustained severe PARDS may have a different donor CD4+ T transcriptional profile after exposure to tracheal aspirate samples compared with those who initially have severe PARDS, but rapidly recover. The trajectory of both the airway and systemic immune responses of children with PARDS—to answer why some children are resilient and recover, while others do not recover—should be a focus of future studies.
In conclusion, we report the use of a CD4+ T-cell transcriptomic reporter assay using airway fluid from children with severe, mild, and no PARDS. We found a dampened type I IFN response from children with severe compared with mild or no PARDS. Use of a transcriptomic reporter assay may provide a clinically useful strategy to understand the complex heterogeneity of the airway immune response of PARDS. Further testing of this strategy in a larger group of children to confirm our findings and determine the utility of a transcriptomic reporter assay for predictive and prognostic enrichment into clinical trials for PARDS.
Supplementary Material
Footnotes
This work was performed at Emory University School of Medicine, Children’s Healthcare of Atlanta, and Georgia Institute of Technology.
Funding was provided by National Institutes of Health (NIH) grants K12HD072245 (Atlanta Pediatric Scholars Program), K23 HL151897-01, and an Emory University Pediatrics Research Alliance Junior Faculty Focused Pilot award to Dr. Grunwell. Funding was provided by the NIH grant K24 NR018866 to Dr. Fitzpatrick. This study was supported, in part, by the Emory Integrated Genomics Core, which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. Additional support was provided by the Georgia Clinical & Translational Science Alliance of NIH under Award Number UL1TR002378.
The authors have disclosed that they do not have any potential conflicts of interest.
Drs. Fitzpatrick, Kamaleswaran, and Grunwell conceived and developed the study, supervised the acquisition of the biological data, and analyzed and interpreted the data. Dr. Grunwell drafted and edited the article. Drs. Ripple, Fitzpatrick, and Kamaleswaran assisted with drafting and editing the article. Dr. Stephenson and Mr. Mohammad helped with patient sample processing, performed experiments, and edited the article. Ms. Tidwell assisted in identifying, consenting, acquiring patient samples, and assisted in collecting clinical information about the patients. All authors edited and approved the final version of this article.
The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/ccejournal).
REFERENCES
- 1.Ware LB, Matthay MA: The acute respiratory distress syndrome. N Engl J Med 2000; 342:1334–1349 [DOI] [PubMed] [Google Scholar]
- 2.Khemani RG, Smith L, Lopez-Fernandez YM, et al. ; Pediatric Acute Respiratory Distress syndrome Incidence and Epidemiology (PARDIE) Investigators: Paediatric acute respiratory distress syndrome incidence and epidemiology (PARDIE): An international, observational study. Lancet Respir Med 2019; 7:115–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nie L, Wu W, Lu Z, et al. : CXCR3 may help regulate the inflammatory response in acute lung injury via a pathway modulated by IL-10 secreted by CD8 + CD122+ regulatory T cells. Inflammation 2016; 39:526–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connors TJ, Ravindranath TM, Bickham KL, et al. : Airway CD8(+) T cells are associated with lung injury during infant viral respiratory tract infection. Am J Respir Cell Mol Biol 2016; 54:822–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakaguchi R, Chikuma S, Shichita T, et al. : Innate-like function of memory Th17 cells for enhancing endotoxin-induced acute lung inflammation through IL-22. Int Immunol 2016; 28:233–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mikacenic C, Hansen EE, Radella F, et al. : Interleukin-17A is associated with alveolar inflammation and poor outcomes in acute respiratory distress syndrome. Crit Care Med 2016; 44:496–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li JT, Melton AC, Su G, et al. : Unexpected role for adaptive alphabetaTh17 cells in acute respiratory distress syndrome. J Immunol 2015; 195:87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan W, Zhang C, Liu J, et al. : Regulatory T-cells promote pulmonary repair by modulating T helper cell immune responses in lipopolysaccharide-induced acute respiratory distress syndrome. Immunology 2019; 157:151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garibaldi BT, D’Alessio FR, Mock JR, et al. : Regulatory T cells reduce acute lung injury fibroproliferation by decreasing fibrocyte recruitment. Am J Respir Cell Mol Biol 2013; 48:35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mock JR, Dial CF, Tune MK, et al. : Impact of regulatory T cells on type 2 alveolar epithelial cell transcriptomes during resolution of acute lung injury and contributions of IFN-gamma. Am J Respir Cell Mol Biol 2020; 63:464–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mock JR, Tune MK, Dial CF, et al. : Effects of IFN-gamma on immune cell kinetics during the resolution of acute lung injury. Physiol Rep 2020; 8:e14368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grunwell JR, Rad MG, Stephenson ST, et al. : Machine learning-based discovery of a gene expression signature in pediatric acute respiratory distress syndrome. Crit Care Explor 2021; 3:e0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khaenam P, Rinchai D, Altman MC, et al. : A transcriptomic reporter assay employing neutrophils to measure immunogenic activity of septic patients’ plasma. J Transl Med 2014; 12:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cabrera SM, Chen YG, Hagopian WA, et al. : Blood-based signatures in type 1 diabetes. Diabetologia 2016; 59:414–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YG, Cabrera SM, Jia S, et al. : Molecular signatures differentiate immune states in type 1 diabetic families. Diabetes 2014; 63:3960–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurram B, Salzman NH, Kaldunski ML, et al. : Plasma-induced signatures reveal an extracellular milieu possessing an immunoregulatory bias in treatment-naive paediatric inflammatory bowel disease. Clin Exp Immunol 2016; 184:36–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khemani RG, Smith LS, Zimmerman JJ, et al. ; Pediatric Acute Lung Injury Consensus Conference Group: Pediatric acute respiratory distress syndrome: Definition, incidence, and epidemiology: Proceedings from the Pediatric Acute Lung Injury Consensus Conference. Pediatr Crit Care Med 2015; 16(5 Suppl 1):S23–S40 [DOI] [PubMed] [Google Scholar]
- 18.Leteurtre S, Duhamel A, Salleron J, et al. ; Groupe Francophone de Réanimation et d’Urgences Pédiatriques (GFRUP): PELOD-2: An update of the Pediatric Logistic Organ Dysfunction score. Crit Care Med 2013; 41:1761–1773 [DOI] [PubMed] [Google Scholar]
- 19.Pollack MM, Patel KM, Ruttimann UE: PRISM III: An updated Pediatric Risk of Mortality score. Crit Care Med 1996; 24:743–752 [DOI] [PubMed] [Google Scholar]
- 20.Grunwell JR, Giacalone VD, Stephenson S, et al. : Neutrophil dysfunction in the airways of children with acute respiratory failure due to lower respiratory tract viral and bacterial coinfections. Sci Rep 2019; 9:2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grunwell JR, Stephenson ST, Mohammad AF, et al. : Differential type I interferon response and primary airway neutrophil extracellular trap release in children with acute respiratory distress syndrome. Sci Rep 2020; 10:19049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ripple MJ, Mohammad AF, Stephenson ST, et al. : Expression patterns of airway fluid cytokines from intubated children with pediatric acute respiratory distress syndrome. Crit Care Explor 2022; 4:e0819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim D, Paggi JM, Park C, et al. : Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 2019; 37:907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao Y, Smyth GK, Shi W: featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014; 30:923–930 [DOI] [PubMed] [Google Scholar]
- 25.Love MI, Huber W, Anders S: Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fabregat A, Jupe S, Matthews L, et al. : The reactome pathway knowledgebase. Nucleic Acids Res 2018; 46:D649–D655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fabregat A, Sidiropoulos K, Garapati P, et al. : The reactome pathway knowledgebase. Nucleic Acids Res 2016; 44:D481–D487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fabregat A, Sidiropoulos K, Viteri G, et al. : Reactome pathway analysis: A high-performance in-memory approach. BMC Bioinf 2017; 18:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fabregat A, Sidiropoulos K, Viteri G, et al. : Reactome diagram viewer: Data structures and strategies to boost performance. Bioinformatics 2018; 34:1208–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jassal B, Matthews L, Viteri G, et al. : The reactome pathway knowledgebase. Nucleic Acids Res 2020; 48:D498–D503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sidiropoulos K, Viteri G, Sevilla C, et al. : Reactome enhanced pathway visualization. Bioinformatics 2017; 33:3461–3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinonen S, Velazquez VM, Ye F, et al. : Immune profiles provide insights into respiratory syncytial virus disease severity in young children. Sci Transl Med 2020; 12:eaaw0268. [DOI] [PubMed] [Google Scholar]
- 33.Hadjadj J, Yatim N, Barnabei L, et al. : Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020; 369:718–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarma A, Christenson SA, Byrne A, et al. ; COMET Consortium: Tracheal aspirate RNA sequencing identifies distinct immunological features of COVID-19 ARDS. Nat Commun 2021; 12:5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Y, Hu B, Stanley G, et al. : IFN-gamma is protective in cytokine release syndrome-associated extrapulmonary acute lung injury. Am J Respir Cell Mol Biol 2023; 68:75–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thwaites RS, Coates M, Ito K, et al. : Reduced nasal viral load and IFN responses in infants with respiratory syncytial virus bronchiolitis and respiratory failure. Am J Respir Crit Care Med 2018; 198:1074–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Z, John Wherry E: T cell responses in patients with COVID-19. Nat Rev Immunol 2020; 20:529–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blanco-Melo D, Nilsson-Payant BE, Liu WC, et al. : Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 2020; 181:1036–1045.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.