Abstract
There is evidence that RNA editing is related to plant cellular stress as well as electron transport organelles, such as mitochondria. The mitochondrial atp1 gene encodes the alpha-subunit of Atp synthase. Control as well as two periods of drought stress treatments were analyzed in the cDNAs generated from the mitochondrial atp1 gene of two cultivars of Triticum aestivum [Giza 168 (G168) and Gemmiza 10 (GM10)]. Following RNA-seq data assembly, atp1 cDNAs from the control (acc. no. OQ129415), 2-hour (acc. no. OQ129416), and 12-hour (acc. no. OQ129417) time points of the T. aestivum cultivar G168 were obtained. Control (acc. no. OQ129419), 2-hour (acc. no. OQ129420), and 12-hour (acc. no. OQ129421) samples all included reconstructed atp1 transcripts from Gemmiza 10. Atp1 transcripts were assembled using the wheat atp1 gene (acc. no. NC_036024). RNA-seq raw data was utilized to identify 11 RNA editing sites in atp1 in the tolerant cultivar Giza168 and 6 in the sensitive cultivar Gemmiza10. The significant difference in RNA editing observed between control and drought stress conditions in sites led to synonymous amino acids. This led to no change in tertiary structure between tolerant and sensitive cultivars. But the change was focused between produced protein and its correspondence sequence on DNA.
Keywords: RNA editing, Mitochondrial atp1 gene, Tertiary structure, Triticum aestivum
1. Introduction
RNA editing is a process that modifies the sequence of RNA molecules in either a coding or noncoding way. A post-transcriptional modification can occur after transcription by removing, substituting, or altering nucleotides (Ramadan et al., 2021). RNA-editing processes in mitochondrial trypanosomes were first discovered in 1986 (Benne et al., 1986), who found that the frame-shifted coxII gene was processed in an unnatural manner. Humans, viruses, fungi, animals, and plants have all been found to have different types of RNA editing (Yan et al., 2018, Gott and Emeson, 2000, Popitsch et al., 2020). Particular nucleotides in plant organelles are converted from cytidine to uridine via RNA editing, while in animal cells they are converted from adenosine to inosine (Terajima et al., 2017). Coding regions of mRNAs are more likely to be edited than introns and structural RNAs (Zheng et al., 2016). Most flowering plants modify their RNA in both their plastids and their mitochondria (Small et al., 2020, Alhamdan et al., 2020), changing 30–40 Cs in plastid mRNA and more than 400 Cs in mitochondria. According to studies (Ruwe et al., 2013, Bentolila et al., 2013), the plastid genome of Arabidopsis contains 43 editing sites, while the mitochondrial genome includes 619 editing sites.
A membrane-bound protein complex called ATP synthase produces ATP, the primary energy currency of the cell. There are four different types of ATPases that play critical roles in cellular physiology, ranging from ion homeostasis and acidification of intracellular compartments to ATP synthesis, drug resistance, and bacterial pathogenesis. ATPase types include F-ATPases, V-ATPases, A-ATPases, and P-ATPases. F-type ATPases can be located in chloroplasts, mitochondria, and bacteria. They function as both ATP synthases and proton-transporting ATPases, playing key roles in the production of ATP. Because of the rotational mechanism they employ to generate ATP from the transmembrane proton gradient, F-type ATPases are also known as rotary motors. The F1 sector, which is in charge of ATP production, and the Fo sector, which is in charge of forming the proton channel across the membrane, are the two primary parts. There have been studies on this topic (Sobti et al., 2021, Vinothkumar et al., 2016).
ATP1 is an integral membrane protein subunit that forms a critical part of the mitochondrial ATP synthase complex. This complex consists of two main sectors, the F1 and Fo sectors, with the F1 sector located in the mitochondrial matrix and containing various subunits, including ATP1. The ATP1 subunit acts as a proton channel in the F1 sector, facilitating protons passage through the mitochondrial inner membrane, which is essential for ATP synthesis (Zancani et al., 2020). The energy derived from this proton translocation drives ATP synthesis in the mitochondrial matrix. The ATP1 subunit is vital for the proper functioning of the ATP synthase complex and for cellular energy metabolism.
The ATP1 subunit in wheat is essential for the survival and growth of the plant, notably in harsh environments where water is scarce, salt levels are high and temperatures are high. Under such conditions, the ATP1 subunit plays a vital role in ensuring the reliability of the energy supply to the plant cells, which is necessary for their survival. The ATP1 subunit plays an important part in several facets of plant growth, including germination, root expansion, and flowering. Studies have shown that alteration or alterations in the ATP1 subunit gene in wheat can affect the energy metabolism of the plant cells, leading to hampered plant development and growth by decreasing ATP generation. In addition, changes in the expression levels of the ATP1 subunit gene in wheat have been observed under various environmental stresses, implicating this subunit in plant response regulation to stress (Li et al., 2021). Further research on the role of the ATP1 subunit in wheat and other plant species can provide valuable insights into the plant's energy metabolism and its response to environmental stresses.
The alpha subunit in its mature form comprises three main domains: a beta-barrel at the N-terminus, a central domain for binding nucleotides, and a helix bundle located at the C-terminal end. Wheat's alpha subunit of mitochondrial ATP synthase is an intricate protein with a crucial function in the production of ATP. The alpha subunit in wheat, like that in other species, is divided into an N-terminal domain and a C-terminal domain, each of which includes its own unique collection of essential amino acids. The location of these vital amino acids in the alpha subunit is important for its overall function in ATP synthesis. The alpha subunit interacts with other subunits of the ATP synthase complex to form a proton channel, which drives ATP synthesis by producing a proton gradient across the inner mitochondrial membrane. As a result of numerous environmental challenges, including cold and heat stress (Nakajima and Mulligan, 2001, Kurihara-Yonemoto and Kubo, 2010), RNA editing in chloroplast transcripts has also been shown to occur in mitochondrial genes. It is crucial to understand how plants respond to environmental stressors since it is believed that RNA editing aids in plant defence in response to some environmental conditions.
2. Material and methods
2.1. Data from RNA-sequencing experiments
NCBI's SRA databank of T. aestivum for drought treatment with 12% PEG 6000 was retrieved and downloaded as follows: genotype Giza168; SRR3098724, SRR3098725, SRR3089142 and SRR3089143 (control), SRR3098728, SRR3098729, SRR3089146 and SRR3089147 (2 h, post-drought treatment), SRR3098730, SRR3098731, SRR3089148 and SRR3089149 (12 h, post-drought treatment). For genotype Gemmiza10; SRR3098732, SRR3098733, SRR3089150 and SRR3089151 (control), SRR3098734, SRR3098735, SRR3089152 and SRR3089153 (2 h, post- drought treatment), SRR3098726, SRR3098727, SRR3089144 and SRR3089145 (12 h, post-drought treatment).
2.2. Bioinformatics analysis of RNA editing
Modified CLC Genomic Workbench 3.6.5 (Ishii et al., 2013) was used to identify RNA editing sites. The length percentage and similarity of the mapping were both set to 0.98 in order to eliminate false positives. According to Hisano et al., (Hisano et al., 2016), the readings were mapped to the NC_036024 accession number for the T. aestivum mitochondrial atp1 gene. While 50% was the bar for minimum reading length, 80% was the limit for minimum similarity. The nucleotide editing settings are 4% minimum count, 5% minimum frequency, 20% minimum coverage, and 5% low-frequency variant. Then, the overall reading counts, coverage depth counts, and RNA editing sites were determined. Nucleotide conversion frequency was calculated for each site using the number of reads divided by the total number of reads for all drought exposure periods and the control (Wang et al., 2015).
2.3. Amino acid analysis of the atp1 gene
cDNAs and correspondence atp1 gene from the genome were analyzed with CLC Genomic workbench 3.5.6 for evidence of RNA editing sites and amino acid substitutions. Alterations to proteins' secondary structures were also predicted with this tool.
2.4. QRT-PCR for the confirmation of RNA editing sites
In this investigation, two drought-tolerant and -sensitive Triticum aestivum cultivars (G168 and GM10) were employed. After two weeks of growth in a greenhouse with 14 h of light per day, 80% humidity, and a temperature of 22° C, seeds were germinated in trays of vermiculite: perlite (1:1) potting mix and subjected to drought stress using Hoagland solution containing polyethylene glycol (PEG-6000 12% w/v) at varying times (2 and 12 h). Wheat leaves were collected from individual plants at each time point in three independent experiments. After being quickly frozen in liquid nitrogen, all of the tissues were kept at −80° C. RNA-seq data were used to predict where editing would happen, and biological triplicates of leaves from each dose were used to check if this was true. All of the samples' total RNA was taken out with Qiazol (Qiagen, Cat No. 79306). Real-time polymerase chain reaction (PCR) was used to verify the presence of RNA editing sites using Stratagene's Mx3005P qPCR equipment. For first-strand cDNA synthesis, M−MuLV reverse transcriptase (MIR BIOTECH cat. no. chb20, 004), 1 µg of total RNA, and 0.05 µg of reverse primers for each gene were used (Slugina et al., 2019). All that went into the reaction (24 ul) was RT2 SYBR® Green qPCR Mastermix (12.5 ul), 0.2 mM of forward and reverse primers for each gene (Table S1). To complete the reaction, 1 ul of diluted cDNA template was added. Protocol for PCR was 40 cycles, each of which lasted 15 s at 94 °C, 30 s at 55 °C, and 45 s at 72 °C. Amplified plots of Rn vs cycle number were used for data analysis. Rodrigues et al. formula (Rodrigues et al., 2017) was used as a reference for the RNA editing percentage calculations.
2.5. Data analysis
Statistical analysis was performed using analysis of variance (ANOVA) using the SPSS software, and Tukey's HSD was used for multiple comparisons (Tukey, 1949).
2.6. 3D protein analysis
The modeling of all protein sequences were performed through, AlphaFold using default parameters (Jumper et al., 2021). The RMSD between the two models was calculated using TM-align (Zhang & Skolnick, 2005). To analyze RNA editing events potential affinity in the ATP1 alpha subunit, we used MutPred2 (Pejaver et al., 2020). The MutPred2 scores were interpreted with caution, considering other available evidence. TM-align algorithm using dynamic programming iterations to perform optimized residue to residue alignment based on structural similarity. Protein stability was measured using DynaMut2 (Rodrigues et al., 2021) and I-Mutant+ (Capriotti and Fariselli, 2017, Capriotti et al., 2005) servers. To obtain conservation scores of ATP1 amino acid residues, we used the CONSURF tool (Armon et al., 2001, Ashkenazy et al., 2016) webserver. It indicates the distribution of structural and functional residues over the structure. The ATP1 protein sequence was submitted with the PDB file and ClustalW alignment method. The resulting scores were displayed with a color-coded scheme. Conservation scores were interpreted based on the level of conservation across species.
3. Results
3.1. Detection of wheat atp1 transcripts
T. aestivum cultivar G168 atp1 cDNAs were reprocessed from the control (acc. no. OQ129415), after 2 h (acc. no. OQ129416), and after 12 h (acc. no. OQ129417). A total of 170,200,000 pair-end RNA sequencing reads were used to obtain atp1 transcripts. Additionally, 172,700,000 for 12 h and 170,450,000 for 2 h of recovery were made. After 2 h (acc. no. OQ129420), after 12 h (acc. no. OQ129421), and while GM10 was reprocessed from the control (acc. no. OQ129419). In order to produce atp1 transcripts, 172,880,000 pair-end long RNA sequence reads were retrieved. Also retrieved were 173,800,000 for 2 h and 172,900,000 for 12 h. The wheat atp1 gene (acc. no. NC_036024) was used to assemble atp1 transcripts.
3.2. RNA editing and amino acid modifications
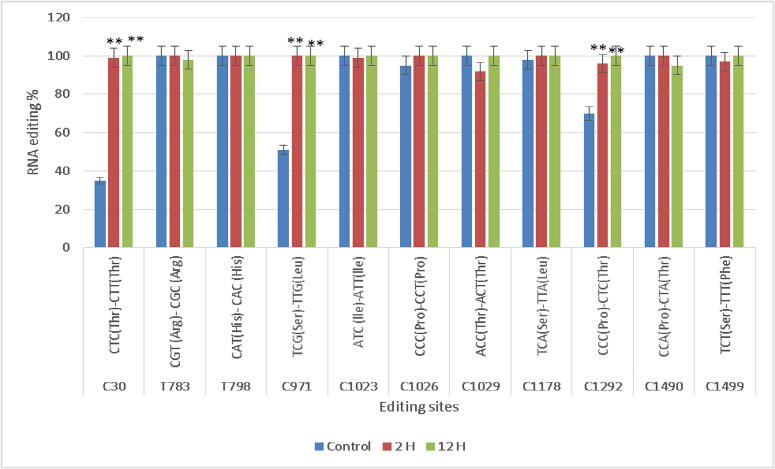
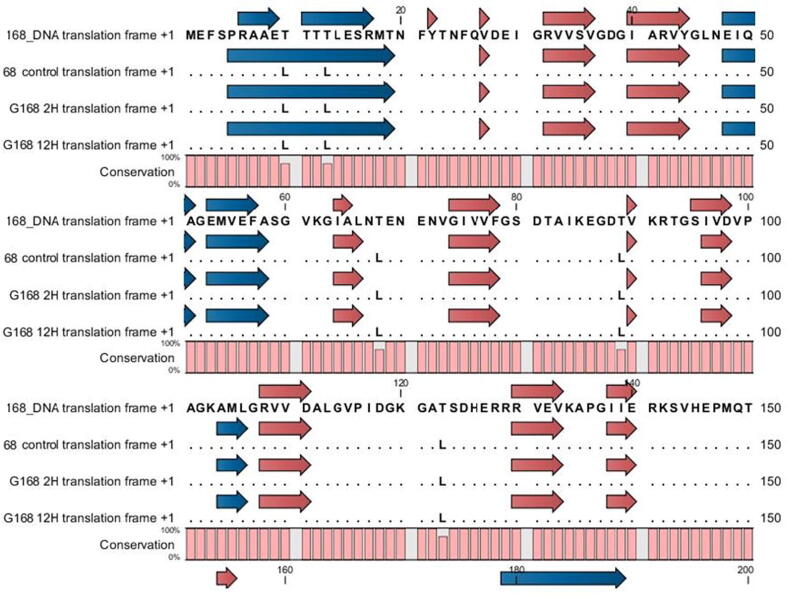
G168 mitochondrial DNA atp1 sequence was compared to cDNA sequences for the three treatments and evidence of preferential editing of C to T was found in 9 RNA editing sites (C30, C971, C1023, C1026, C1029, C1178, C1292, C1490, and C1499) and two T to C sites (T783 and T798) (Fig. 1, Fig. S1 and Table S2). In the 2 and 12 h treatments, each of the eleven sites was conserved. Moreover, eight of them were conserved at all times (T783, T798, C1023, C1026, C1029, C1178, C1490, C1499). However, significant RNA editing was only observed at three locations in 2 and 12 h treatments. It was noted from the obtained results that, amino acids are largely unaffected by the majority of RNA editing sites (C30, T783, T798, C1023, C1026, and C1029). Two RNA modifications are responsible for the transition from proline to threonine. (C1292, C1490) and serine to leucine.
Fig. 1.
Efficiency and distinction using data from total RNA-seq, G168 atp1 RNA editing was compared to the control. (C or T) nucleotide position, (Phe) phenylalanine, (Pro) proline, (Thr) threonine, (Ser) serine, and (Leu) leucine. Three biological replicates' means and standard deviations (black bars) are used to express the data. ** denotes a significant distinction between the treatments. P < 0.01.
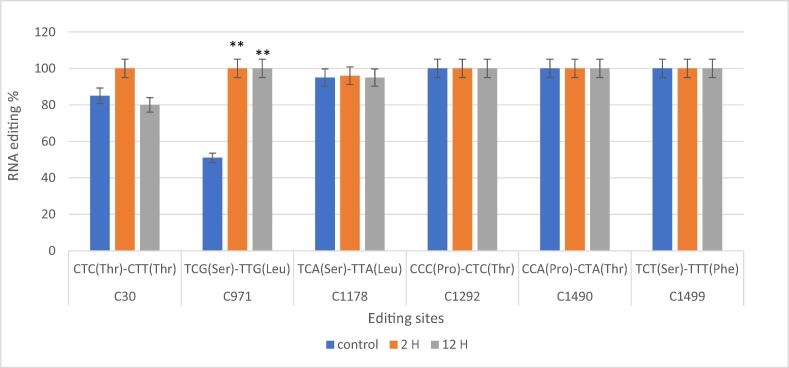
On the other hand, by comparing atp1 sequences of the cultivar Gemmiza 10 mitochondrial DNA and the three treatments cDNA sequence (Fig. 2), six RNA editing sites showed deferential editing of C to T (C30, C971, C1178, C1292, C1490, C1499) and the majority of them are stable after being treated for 2 or 12 h.C1178, C1292, C1490, and C1499 were conserved at all times. The RNA edits for C30 did not show any conserved ratio between treatments. Moreover, the position, C971 showed significant RNA editing in 2 and 12 h treatments due to drought stress. Proline was changed to threonine by two RNA editing (C1292, C1490), whereas serine was changed to lucine by C971 and C1178 editing sites. However, in one instance, RNA editing has no effect on amino acids (C30). Some sites' editing ratios clearly vary with drought and exposure time (Fig. 2, Fig. S2, TableS2).
Fig. 2.
Efficiency and distinction using data from total RNA-seq, GM10 atp1 RNA editing was compared to the control. (C or T) nucleotide position, (Phe) phenylalanine, (Pro) proline, (Thr) threonine, (Ser) serine, and (Leu) leucine. Three biological replicates' means and standard deviations (black bars) are used to express the data. ** denotes a significant distinction between the treatments. P < 0.01.
3.3. Atp1 gene modification validation
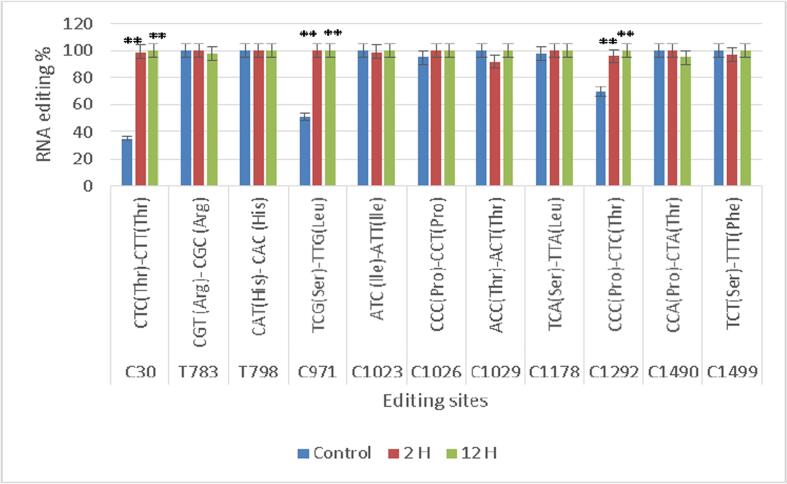
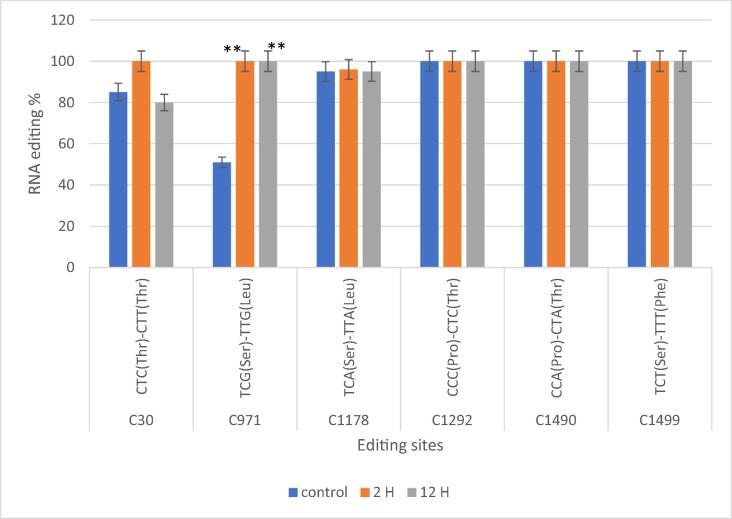
All editing sites of nucleotide alterations was selected in Giza 168 and Gemmiza 10 and evaluated them using RT-qPCR to guarantee the Validation and precision of the RNA-seq method for characterising editing sites. Quantification and measurement of atp1editing locations at three different exposure times were performed for both G1 (C30, T783, T798, C1023, C1026, 1029, C1178, C1292, C1490, C1499) and G2 (C30, C971, C1178, C1292, C1490, C1499) (Fig. 3 and Figs. 4).
Fig. 3.
Triticum aestivum cultivar G168 editing sites were confirmed by qRT-PCR in three different drought conditions (control, two hours after exposure to drought, and twelve hours after exposure to drought). Three biological replicates' means and standard deviations (black bars) are used to express the data. ** denotes a significant distinction between the treatments. P < 0.01.
Fig. 4.
Triticum aestivum cultivar GM10 editing sites were confirmed by qRT-PCR in three different drought conditions (control, two hours after exposure to drought, and twelve hours after exposure to drought). Three biological replicates' means and standard deviations (black bars) are used to express the data. ** denotes a significant distinction between the treatments. P < 0.01.
3.4. Secondary structure of atp1 protein
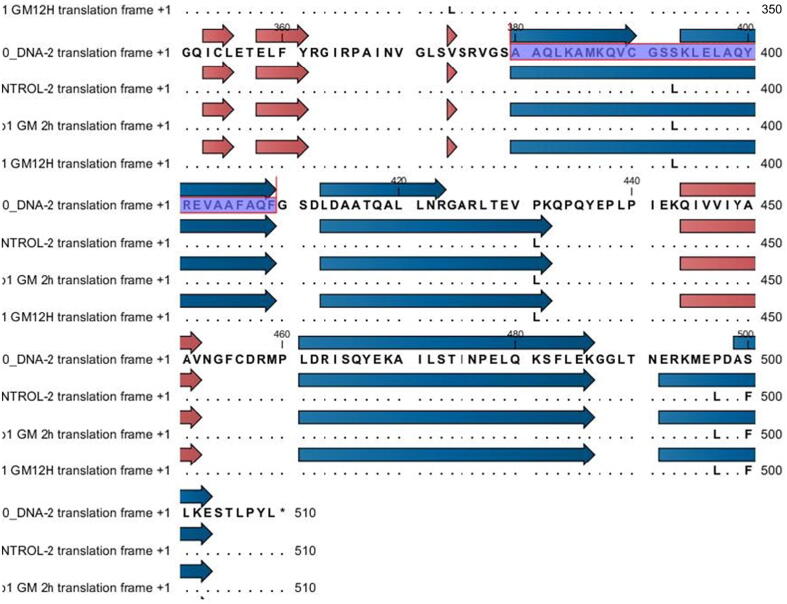
Using CLC Genomic work bench version 3.6.5, changes in the length and absence of the alpha helix and beta sheets were detected in the atp1 protein at control as well as drought stress due to RNA editing but no change in quantity (13 alpha helices and 24 beta sheets for each before and after editing). For instance in cultivar Giza168, the two alpha helices (6…9; 11…17) in original sequence (at DNA correspondence) changed to one long alpha helices after editing (5…19) however, beta sheet (position 22) was absent after editing. Alpha helices (53…57) is increased in size (53…58) after editing. Alpha helices (104…106) is created after editing, however, not found before. More changed is clarified in Fig. 5, Fig. S3. In addition to change in length in cultivar Gemmiza 10, there are difference in quantity due to merge two alpha sheet (380…390; 394…409) before editing to become one region after editing (380…409) (Fig. 6, Fig. S4). These changes in secondary structure may be indicative of changes in the protein's stability and function.
Fig. 5.
Change secondary structure regions in cultivar G168 atpase subunit 1 before and after RNA editing due to drought treatment as well as correspondence sequence at DNA. Alpha helix (blue) and beta sheet (brown).
Fig. 6.
Change secondary structure regions in cultivar Gemmiza 10 (GM 10) atpase subunit 1 before and after RNA editing e to drought treatment as well as correspondence sequence at DNA. Alpha helix (blue) and beta sheet (brown).
3.5. 3D structure of ATP1 protein
3.5.1. Structural insights in alpha-subunit
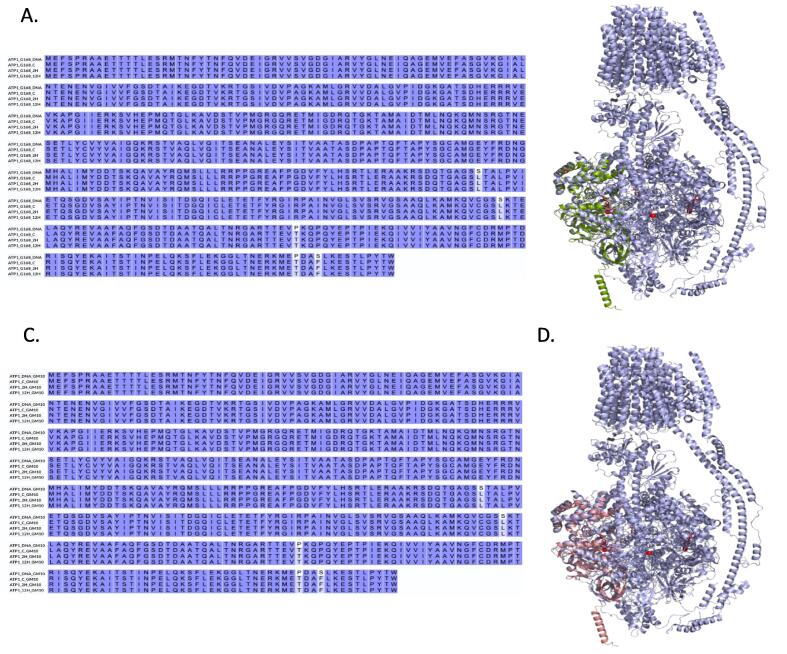
The protein sequence alignment was done for both GM10 and G186 cultivars. Five amino acids substitutions were reported at positions S324L, S393L, P431T, P497T and S500F (Fig. 7). The ATP1 protein models were built for both wheat cultivars at different treatments; Control (C), 2 h (2H) and 12 h (12H) (Fig. 7).
Fig. 7.
RNA editing sites of ATP1. A. The protein sequence alignment between all treatments and the predicted ATP1 wild type (ATP1_WT) in G168. A total five edits were introduced during the stress. B. the ATP1_complex in light purple while the ATP1 subunit in olive green. C. RNA editing sites of ATP1. C. The protein sequence alignment between all treatments and the predicted ATP1 wild type (ATP1_WT) in GM10. D. The ATP1_complex in light purple while the ATP1 subunit in salmon.
3.5.2. Stability and structural effect of RNA edits
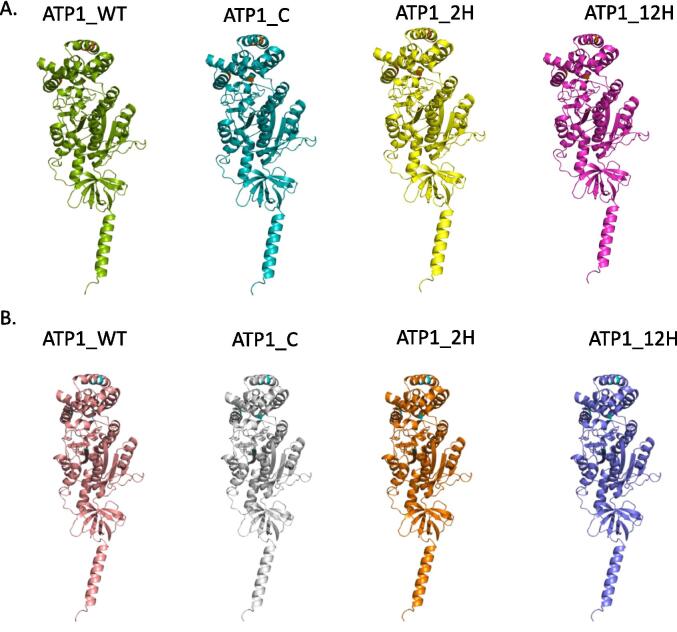
The five edits were introduced in near places of the binding sites. The overall structure of alpha subunit of ATP1 did not show a critical change in the protein folding. The best protein models from AlphFold were visualized using Pymol (Fig. 8). In the context of the ATP1 alpha subunit gene and drought stress, the alterations S324L, S393L, P431T, P497T, and S500F were analyzed using MutPred2, resulting in the scores of 0.084, 0.157, 0.187, 0.087, and 0.041, respectively. These scores suggest varying levels of predicted model for each alteration. S324L and S500F alteration have the lowest scores, indicating a relatively low predicted affinity. However, this does not necessarily mean that they are benign alterations or that they have no effect on the protein function. The S393L alteration has a slightly higher score, indicating a higher predicted affinity than S324L and S500F. P431T and P497T alterations have even higher scores, suggesting a greater likelihood of being deleterious. These MutPred2 scores suggest that the alteration in ATP1 alpha subunit under drought stress could potentially have varying effects on the protein function and affinity. However, further experimental validation is necessary to confirm these predictions and to fully understand the functional impact of the alterations under drought stress.
Fig. 8.
RNA editing sites of ATP1 at different drought treatment. A. Represents all the PDB models of ATP1 wild type (ATP1_WT), ATP1 control (ATP1_C), ATP1 2 h (ATP1_2H) and ATP1 12 h (ATP1_12H) in G168. B. Represents all PDB models of ATP1 wild type (ATP1_WT), ATP1 control (ATP1_C), ATP1 2 h (ATP1_2H) and ATP1 12 h (ATP1_12H) in GM10.
The TM-align results were similar in both G168 and GM10 with RMSD of 0.00 that indicates that the root-mean-square deviation between the two protein structures is minimal, suggesting that the overall protein fold is very similar. Additionally, the Seq_ID score of 1.000 indicates that all residues in the aligned regions are identical between the two protein structures. This suggests that the two protein structures are not only structurally similar, but also share a high degree of sequence identity.
DynaMut2 was used to predict the effects of RNA editing events in the ATP1 alpha subunit gene, resulting in the prediction of ΔΔG stability scores of −2.89 for alteration S324L, S393L, P431T, P497T, and S500F.
3.5.3. Conservation of RNA edits and their effect on the structure
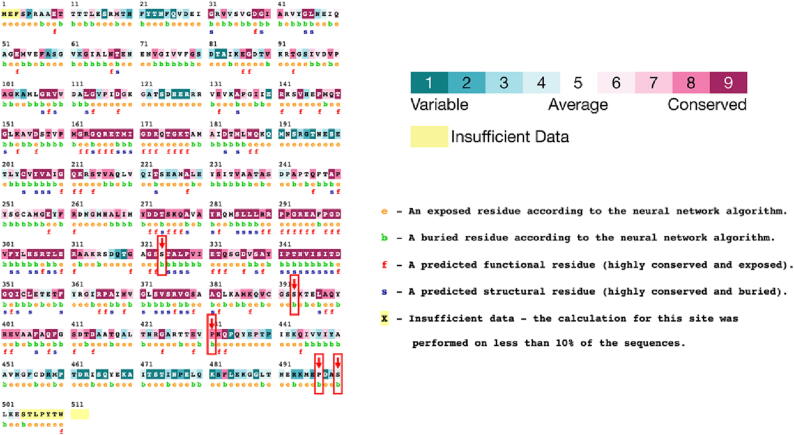
The atp1 gene encodes an alpha subunit of a plasma membrane ATPase in plants, which is involved in the transport of ions across the membrane and is important for maintaining cellular homeostasis (Morth et al., 2011). Under drought stress, plants undergo various molecular adaptations, including changes in gene expression and RNA editing (Mahmood et al., 2019). The conservation scores of specific amino acid residues in the atp1 gene were analyzed using the CONSURF tool, with lower scores indicating greater variability and higher scores indicating greater conservation (Fig. 9). Residues at positions 324, 393, and 431 had relatively low conservation scores, suggesting they may be more susceptible to adaptation or functional changes under drought stress. In contrast, residues at positions 497 and 500 had high conservation scores, suggesting they may be functionally important and less susceptible to changes under drought stress.
Fig. 9.
Conservation of RNA editing sites of ATP1 in both G168 and GM10. Residues with high conservation scores are colored purple, while residues with low conservation scores are colored green. Amino acid residues 324, 393, and 431 have relatively low conservation scores, while residues 497 and 500 have high conservation scores.
4. Discussion
The SRA data provide a valuable resource for further studies on the molecular mechanisms of drought stress tolerance in Triticum aestivum and can also contribute to the understanding of mitochondrial functions and evolution in plants. The identification of these RNA editing sites provides new insights into the regulation of the atp1 gene in Triticum aestivum and may have implications for understanding how this plant responds to stress conditions. Functional protein expression relies heavily on RNA editing, and this is notably true in mitochondria (Castandet and Araya, 2011, Bentolila et al., 2013) and chloroplasts (Takenaka et al., 2013, Ichinose and Sugita, 2017). Multiple studies have demonstrated a correlation between abiotic stress and RNA editing, showing the significance of this mechanism in preserving cellular homeostasis (Yuan and Liu, 2012, Bentolila et al., 2013). The ATP1 subunit, a component of complex V (atp synthase) subunits, participates in the electron transport chain. Free oxygen (ROS) production is increased in Arabidopsis when RNA editing is disrupted, increasing drought sensitivity (Yuan & Liu 2012). These reports highlight the critical role of RNA editing in plant biology and its potential impact on plant responses to environmental stress.
Different editing sites have been reported in different plants for the atp1 gene, depending on the species. Arabidopsis had six editing sites, onion had thirteen, cucumber had five, wild carrot had five, soybean had two, and sunflower had six (Edera et al., 2018).
In this study, we identified 11 RNA editing sites (nucleotide nos. C30, T783, T798, C971, C1023, C1026, C1029, C1178, C1292, C1490 and C1499) in tolerant cultivar Giza168 and 6 in the sensitive tolerant Gemmiza 10 (C30, C971, C1178, C1292, C1490 and C1499). Most editing sites were fully edited in both control and drought stress conditions except positions C30, C971 and C1292, as confirmed by real-time PCR validation (Fig. 3 and Table S2). These findings contribute to a better understanding of the RNA editing mechanisms and their potential role in plant responses to abiotic stress, in addition to repair alteration to restore conserved proteins (Edera et al., 2018, Ramadan et al., 2023).
In the current study, several observations were made regarding RNA editing in Triticum aestivum. Firstly, it was found that not all editing sites were affected by drought stress—just three sites in tolerant cultivar Giza 168 (C30, C971 and C1292) and two in sensitive cultivar Gemmiza 10 (C30, C971). This suggests that RNA editing at most sites in the atp1 gene may be aimed at restoring the conserved protein, indicating that it may have evolved as a means to correct past alteration and emphasizing the importance of this editing process. Furthermore, it was observed that synonymous amino acids were present in two cultivars due to RNA editing, like threonine in C30, which is in contrast to previous reports investigating RNA editing in other species (Hajrah et al., 2017, Edera et al., 2018).
An important observation is that the synonymous edited in tolerant cultivar Giza 168 in positions T783, T798, C971, C1023, C1026 and C1029 are not found in sensitive cultivar Gemmiza 10. As a result of drought stress, synonymous amino acids were significantly altered. In these results, some codons are unable to produce the same amino acid under drought stress, indicating that something changes in tRNA under stress that causes a change to another tRNA for different codons to produce the same amino acid (Ramadan et al., 2021).
The analysis of RNA editing events using MutPred2 provides insights into the functional impact of RNA editing events in the ATP1 alpha subunit gene. These MutPred2 scores suggest that the alteration in the ATP1 alpha subunit under drought stress could potentially have varying effects on protein function and affinity. However, further experimental validation is necessary to confirm these predictions and fully understand the functional impact of the alteration under drought stress. TM-align scores suggest that the two cultivars protein structures are highly similar in both structure and sequence.
The DynaMut2 negative ΔΔG stability score of −2.89 for the RNA edits S324L, S393L, P431T, P497T, and S500F suggests that these alterations are predicted to stabilize the protein structure of the ATP1 alpha subunit gene. This suggests that these RNA edits may not be deleterious and may even be beneficial to the protein's stability.
Structurally, the alpha subunits are composed of several domains, including the N-terminal domain, the central domain, and the C-terminal domain (Vonck et al., 2009). The N-terminal domain contains a helix bundle that interacts with the other subunits of ATP synthase, while the central domain contains a coiled-coil structure that forms the central stalk (Moore et al., 2008). The C-terminal domain contains the catalytic site for ATP synthesis and binds to the beta subunits.
The crystal structure of the alpha subunits reveals several important insights into their function. For example, the central stalk structure is formed by the coiled-coil arrangement of the alpha subunits, with each alpha subunit forming a dimer that interacts with the neighboring subunits (Lee et al., 2015). This arrangement allows for the efficient transfer of energy from the proton gradient to the mechanical rotation necessary for ATP synthesis (Ruhle & Leister 2015).
Additionally, the catalytic site in the C-terminal domain of the alpha subunits contains several key residues that are critical for ATP synthesis (Cingollani & Duncan 2011). These residues, including lysine and arginine, are involved in the binding and transfer of phosphate groups, which are necessary for the synthesis of ATP (Mnatsakanyan et al., 2019).
The N-terminal domain of the alpha subunit in wheat contains several critical amino acids that are important for the interaction of the subunit with other subunits in the ATP synthase complex. A conserved position in the atp1 gene refers to a position in the nucleotide sequence that is highly conserved across different species, suggesting that it is functionally important. If an RNA edit occurs in a conserved position of the atp1 gene under drought stress, it may be an adaptive response that alters the function of the protein in a way that is beneficial for the plant under these conditions.
An RNA edit in a conserved position P431T of the atp1 gene might lead to a change in the amino acid sequence of the protein that alters its ion transport activity or its sensitivity to regulatory signals such as phosphorylation. Alternatively, the RNA edit might affect the stability or localization of the protein, leading to changes in its overall abundance or activity.
Overall, the occurrence of RNA editing in a conserved position of the atp1 gene under drought stress could represent an important molecular adaptation that helps plants survive in challenging environments. However, further research is needed to fully understand the functional significance of such edits and their impact on plant physiology and fitness. These results suggest that the latter residues are functionally important and less susceptible to changes under drought stress, while the former residues may be more variable and prone to adaptation.
CRediT authorship contribution statement
Mona I M Ibrahim: . Ahmed M Ramadan: Writing – review & editing. Marwa Amer: Writing – review & editing. Thana K Khan: . Nermin G. Mohamed: . Osama A.M. Said: .
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This research work was funded by Institutional Fund Projects under grant no. (IFPIP: 273-130-1443). Therefore, the authors gratefully acknowledge technical and financial support from the Ministry of Education and King Abdulaziz University, Jeddah, Saudi Arabia.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sjbs.2023.103703.
Contributor Information
Mona I.M. Ibrahim, Email: mona.ibrahim@must.edu.eg.
Ahmed M. Ramadan, Email: aamara@kau.edu.sa.
Marwa Amer, Email: Marwa.amer@must.edu.eg.
Thana K. Khan, Email: tkhan@kau.edu.sa.
Nermin G. Mohamed, Email: nermine.imran@must.edu.eg.
Osama A. Said, Email: osama.said@must.edu.eg.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Alhamdan H., Alshammari W.B., Khan T., Edris S., Hassanein S.E., Ramadan A.M. Differential RNA editing of mitochondrial atp1 gene in Catharanthus roseus tissues. Adv. Environ. Biol. 2020;14(3) [Google Scholar]
- Armon A., Graur D., Bental N. Consurf: an algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J. Mol. Biol. 2001;307:447–463. doi: 10.1006/jmbi.2000.4474. [DOI] [PubMed] [Google Scholar]
- Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., Ben-tal N. Consurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:w344–w350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne R., Van den Burg J., Brakenhoff J.P., Sloof P., Van Boom J.H., Tromp M.C. Major transcript of the frame shifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46(6):819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- Bentolila S., Oh J., Hanson M.R., Bukowski R. Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E., Fariselli P. Phd-snpg: a webserver and lightweight tool for scoring single nucleotide variants. Nucleic Acids Res. 2017;45:w247–w252. doi: 10.1093/nar/gkx369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E., Fariselli P., Casadio R. I-mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33:w306–w310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castandet B., Araya A. RNA editing in plant organelles. Why make it easy? Biochemistry (Mosc) 2011;76(8):924–931. doi: 10.1134/S0006297911080086. [DOI] [PubMed] [Google Scholar]
- Cingollani G., Duncan T.M. Structure of the ATP synthase catalytic complex (F(1)) from Escherichia coli in an autoinhibited conformation. Nat. Struct. Mol. Biol. 2011;18:701–707. doi: 10.1038/nsmb.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edera A.A., Gandini C.L., Sanchez-Puerta M.V. Towards a comprehensive picture of C-to-U RNA editing sites in angiosperm mitochondria. Plant Mol. Biol. 2018;97(3):215–231. doi: 10.1007/s11103-018-0734-9. [DOI] [PubMed] [Google Scholar]
- Gott J.M., Emeson R.B. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- Hajrah N.H., Obaid A.Y., Atef A., Ramadan A.M., Arasappan D., Nelson C.A., Edris S., Mutwakil M.Z., Alhebshi A., Gadalla N.O., et al. Transcriptomic analysis of salt stress responsive genes in Rhazya stricta. PLoS One. 2017;12(5) doi: 10.1371/journal.pone.0177589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisano H., Tsujimura M., Yoshida H., Terachi T., Sato K. Mitochondrial genome sequences from wild and cultivated barley (Hordeum vulgare) BMC Genomics. 2016;17(1) doi: 10.1186/s12864-016-3159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii S., Suzuki S., Norden-Krichmar T.M., Tenney A., Chain P.S., Scholz M.B., Nealson K.H., Bretschger O. A novel metatranscriptomic approach to identify gene expression dynamics during extracellular electron transfer. Nat. Commun. 2013;4 doi: 10.1038/ncomms2615. [DOI] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A., Bridgland A., Meyer C., Kohl S.A.A., Ballard A.J., Cowie A., Romera-paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A.W., Kavukcuoglu K., Kohli P., Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara-Yonemoto S., Kubo T. Increased accumulation of intron-containing transcripts in rice mitochondria caused by low temperature: is cold-sensitive RNA editing implicated? Curr. Genet. 2010;56(6):529–541. doi: 10.1007/s00294-010-0320-4. [DOI] [PubMed] [Google Scholar]
- Lee J., Ding S., Walpole T.B., Holding A.N., Montgomery M.G., Fearnley I.M., Walker J.E. Organization of subunits in the membrane domain of the bovine F-ATPase revealed by covalent cross-linking. J. Biol. Chem. 2015;290:13308–13320. doi: 10.1074/jbc.M115.645283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Song J., Zhu G., Hou Z., Wang L., Wu X., Fang Z., Liu Y., Gao C. Genome-wide identification and expression analysis of ADP-ribosylation factors associated with biotic and abiotic stress in wheat (Triticum aestivum L.) PeerJ. 2021;9 doi: 10.7717/peerj.10963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood T., Khalid S., Abdullah M., Ahmed Z., Shah M.K.N., Ghafoor A., Du X. Insights into drought stress signaling in plants and the molecular genetic basis of cotton drought tolerance. Cells. 2019;9 doi: 10.3390/cells9010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnatsakanyan N., Li Y., Weber J. Identification of two segments of the gamma subunit of ATP synthase responsible for the different affinities of the catalytic nucleotide-binding sites. J. Biol. Chem. 2019;294:1152–1160. doi: 10.1074/jbc.RA118.002504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K.J., Angevine C.M., Vincent O.D., Schwem B.E., Fillingame R.H. The cytoplasmic loops of subunit a of Escherichia coli ATP synthase may participate in the proton translocating mechanism. J. Biol. Chem. 2008;283:13044–13052. doi: 10.1074/jbc.M800900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morth J.P., Pedersen B.P., Buch-pedersen M.J., Andersen J.P., Vilsen B., Palmgren M.G., Nissen P. A structural overview of the plasma membrane Na+, K+-ATPase and H+-ATPase ion pumps. Nat. Rev. Mol. Cell Biol. 2011;12:60–70. doi: 10.1038/nrm3031. [DOI] [PubMed] [Google Scholar]
- Nakajima Y., Mulligan R.M. Heat stress results in incomplete C-to-U editing of maize chloroplast mRNAs and correlates with changes in chloroplast transcription rate. Curr. Genet. 2001;40(3):209–213. doi: 10.1007/s002940100249. [DOI] [PubMed] [Google Scholar]
- Pejaver V., Urresti J., Lugo-martinez J., Pagel K.A., Lin G.N., Nam H.J., Mort M., Cooper D.N., Sebat J., Iakoucheva L.M., Mooney S.D., Radivojac P. Inferring the molecular and phenotypic impact of amino acid variants with MutPred2. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-19669-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popitsch N., Huber C.D., Buchumenski I., Eisenberg E., Jantsch M., von Haeseler A., et al. A-to-I RNA editing uncovers hidden signals of adaptive genome evolution in animals. Genome Biol. Evol. 2020;12:345–357. doi: 10.1093/gbe/evaa046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan A.M., Alnufaei A.A., Khan T.K., et al. The first report of RNA U to C or G editing in the mitochondrial NADH dehydrogenase subunit 5 (Nad5) transcript of wild barley. Mol. Biol. Rep. 2021 doi: 10.1007/s11033-021-06609-1. [DOI] [PubMed] [Google Scholar]
- Ramadan A., Alnufaei A.A., Fiaz S., Khan T.K., Hassan S.M. Effect of salinity on ccmfn gene RNA editing of mitochondria in wild barley and uncommon types of RNA editing. Funct. Integr. Genomics. 2023;23(1) doi: 10.1007/s10142-023-00978-5. PMID: 36707470. [DOI] [PubMed] [Google Scholar]
- Rodrigues N.F., Christoff A.P., Da Fonseca G.C., Kulcheski F.R., Margis R. Unveiling chloroplast RNA editing events using next generation small RNA sequencing data. Front. Plant Sci. 2017;8 doi: 10.3389/fpls.2017.01686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues C.H.M., Pires D.E.V., Ascher D.B. Dynamut2: assessing changes in stability and flexibility upon single and multiple point missense mutations. Proteinsci. 2021;30:60–69. doi: 10.1002/pro.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhle T., Leister D. Assembly of F1F0-ATP synthases. Biochim. Biophys. Acta. 2015;1847(849–60) doi: 10.1016/j.bbabio.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Ruwe H., Castandet B., Schmitz-Linneweber C., Stern D.B. Arabidopsis chloroplast quantitative editotype. FEBS Lett. 2013;587:1429–1433. doi: 10.1016/j.febslet.2013.03.022. [DOI] [PubMed] [Google Scholar]
- Slugina M.A., Shchennikova A.V., Kochieva E.Z. The expression pattern of the Pho1a genes encoding plastidic starch phosphorylase correlates with the degradation of starch during fruit ripening in green-fruited and red-fruited tomato species. Funct. Plant Biol. 2019;46(12):1146–1157. doi: 10.1071/FP18317. [DOI] [PubMed] [Google Scholar]
- Ichinose, M, Sugita, M., 2017. RNA editing and its molecular mechanism in plant organelles. Genes 2016, 8, 1. doi.org/10.3390/genes8010005. [DOI] [PMC free article] [PubMed]
- Small, I., Schallenberg‐Rüdinger, M., Takenaka, M., Mireau, H., Ostersetzer‐Biran, O., 2020 Plant organellar RNA editing: What 30 years of research has revealed. The Plant Journal 101(5) 1040-1056. doi.org/10.1111/tpj.14578. [DOI] [PubMed]
- Sobti M., Ueno H., Noji H., Stewart A.G. The six steps of the complete f (1)-atpase rotary catalytic cycle. Nat. Commun. 2021;12 doi: 10.1038/s41467-021-25029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M., Zehrmann A., Verbitskiy D., Härtel B., Brennicke A. RNA editing in plants and its evolution. Annu. Rev. Genet. 2013;47:335–352. doi: 10.1146/annurev-genet-111212-133519. [DOI] [PubMed] [Google Scholar]
- Terajima H., Yoshitane H., Ozaki H., Suzuki Y., Shimba S., Kuroda S.I., wasaki W., Fukada Y. ADARB1 catalyzes circadian A-to-I editing and regulates RNA rhythm. Nat. Genet. 2017;49:146–151. doi: 10.1038/ng.3731. [DOI] [PubMed] [Google Scholar]
- Tukey J. Comparing individual means in the analysis of variance. Biometrics. 1949;5(2):99–114. [PubMed] [Google Scholar]
- Vinothkumar K.R., Montgomery M.G., Liu S., Walker J.E. Structure of the mitochondrial atp synthase from pichia angusta determined by electron cryo-microscopy. Proc. Natl. Acad. Sci. USA. 2016;113:12709–12714. doi: 10.1073/pnas.1615902113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonck J., Pisa K.Y., Morgner N., Brutschy B., Muller V. Three-dimensional structure of A1A0 ATP synthase from the hyperthermophilic archaeon Pyrococcus furiosus by electron microscopy. J. Biol. Chem. 2009;284:10110–10119. doi: 10.1074/jbc.M808498200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cui L., Feng K., Deng P., Du X., Wan F., Weining S., Nie X. Comparative analysis of Asteraceae chloroplast genomes: structural organization, RNA editing and evolution. Plant Mol. Biol. Rep. 2015;33(5):1526–1538. [Google Scholar]
- Yan J., Zhang Q., Yin P. RNA editing machinery in plant organelles. Sci. China Life Sci. 2018;61:162–169. doi: 10.1007/s11427-017-9170-3. [DOI] [PubMed] [Google Scholar]
- Yuan H., Liu D. Functional disruption of the pentatricopeptide protein SLG1affects mitochondrial RNA editing, plant development, and responses to abiotic stresses in Arabidopsis. Plant J. 2012;70:432–444. doi: 10.1111/j.1365-313X.2011.04883.x. [DOI] [PubMed] [Google Scholar]
- Zancani M., Braidot E., Filippi A., Lippe G. Structural and functional properties of plant mitochondrial F-ATP synthase. Mitochondrion. 2020;53:178–193. doi: 10.1016/j.mito.2020.06.001. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Skolnick, J., 2005. Tm-align: a protein structure alignment algorithm based on the tm-score. Nucleic acids res, 33, 2302-9. [DOI] [PMC free article] [PubMed]
- Zheng Y., Ji B., Song R., Wang S., Li T., Zhang X., Chen K., Li J. Accurate detection for a wide range of mutation and editing sites of microRNAs from small RNA high-throughput sequencing profiles. Nucleic Acids Res. 2016;44 doi: 10.1093/nar/gkw471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.