Summary
Background
Early detection of cancer aims to reduce cancer deaths. Unfortunately, many established cancer screening technologies are not suitable for use in low- and middle-income countries (LMICs) due to cost, complexity, and dependency on extensive medical infrastructure. We aimed to assess the performance and robustness of a protein assay (OncoSeek) for multi-cancer early detection (MCED) that is likely to be more practical in LMICs.
Methods
This observational study comprises a retrospective analysis on the data generated from the routine clinical testings at SeekIn and Sun Yat-sen Memorial Hospital. 7565 participants (954 with cancer and 6611 without) from the two sites were divided into training and independent validation cohort. The second validation cohort (1005 with cancer and 812 without) was from Johns Hopkins University School of Medicine. Patients with cancer prior to therapy were eligible for inclusion in the study. Individuals with no history of cancer were enrolled from the participating sites as the non-cancer group. One tube of peripheral blood was collected from each participant and quantified a panel of seven selected protein tumour markers (PTMs) by a common clinical electrochemiluminescence immunoassay analyser. An algorithm named OncoSeek was established using artificial intelligence (AI) to distinguish patients with cancer from those without cancer by calculating the probability of cancer (POC) index based on the quantification results of the seven PTMs and clinical information including sex and age of the individuals and to predict the possible affected tissue of origin (TOO) for those who have been detected with cancer signals in blood.
Findings
Between November 2012 and May 2022, 7565 participants were enrolled at SeekIn and Sun Yat-sen Memorial Hospital. The conventional clinical method, which relies only on a single threshold for each PTM, would suffer from a high false positive rate that accumulates as the number of markers increased. OncoSeek was empowered by AI technology to significantly reduce the false positive rate, increasing the specificity from 56.9% (95% confidence interval [CI]: 55.8–58.0) to 92.9% (92.3–93.5). In all cancer types, the overall sensitivity of OncoSeek was 51.7% (49.4–53.9), resulting in 84.3% (83.5–85.0) accuracy. The performance was generally consistent in the training and the two validation cohorts. The sensitivities ranged from 37.1% to 77.6% for the detection of the nine common cancer types (breast, colorectum, liver, lung, lymphoma, oesophagus, ovary, pancreas, and stomach), which account for ∼59.2% of global cancer deaths annually. Furthermore, it has shown excellent sensitivity in several high-mortality cancer types for which routine screening tests are lacking in the clinic, such as the sensitivity of pancreatic cancer which was 77.6% (69.3–84.6). The overall accuracy of TOO prediction in the true positives was 66.8%, which could assist the clinical diagnostic workup.
Interpretation
OncoSeek significantly outperforms the conventional clinical method, representing a novel blood-based test for MCED which is non-invasive, easy, efficient, and robust. Moreover, the accuracy of TOO facilitates the follow-up diagnostic workup.
Funding
The National Key Research and Development Programme of China.
Keywords: Protein tumour markers (PTMs), Artificial intelligence (AI), Multi-cancer early detection (MCED), Liquid biopsy, Low- and middle-income countries (LMICs)
Research in context.
Evidence before this study
Many patients with cancer can be cured if diagnosed early and treated effectively. Yet cancer remains an important public health issue worldwide, especially in low- and middle-income countries (LMICs) due to lack of resources and limited healthcare infrastructure. At present the majority of high-mortality cancers do not have standard-of-care screening methods available. Therefore, a non-invasive and efficient multi-cancer early detection (MCED) test is a highly unmet need. Importantly, the test should be simple and affordable, which is also suitable for LMICs.
Added value of this study
Here, we developed and validated a blood-based MCED test named OncoSeek. This assay integrated the measurement of a panel of seven selected protein tumour markers (PTMs) and clinical information of the individual for MCED and predicting affected tissue of origin (TOO), empowered by artificial intelligence (AI) technology. This large study (n = 9382) containing more than nine common cancer types and dominated by early-stage patients (63.2% stage I and II) showed that the performance of OncoSeek was significantly superior to the conventional clinical method. The test achieved a sensitivity of 51.7% with a specificity of 92.9%, resulting in 84.3% accuracy, with 49.5% sensitivity in stage I and II patients. In addition, the overall accuracy of TOO prediction in the true positives was 66.8%.
Implications of all the available evidence
If used alongside the existing screening approaches, OncoSeek could offer the potential to find more types of cancer at earlier stages using one tube of blood, to improve patient outcomes by treating the disease when it is typically most responsive to therapy, and ultimately to have a notable impact on public health. Further large-scale prospective studies are required to provide more evidence to support the application of OncoSeek in population-scale cancer screening.
Introduction
Cancer is an important public health issue worldwide. The global cancer burden is increasing rapidly, and nearly 19.3 million new cases and 10.0 million cancer deaths were estimated in 2020. It is estimated that more than two-thirds of annual cancer deaths in the world occur in LMICs. The global cancer burden is expected to be 28.4 million cases in 2040, a 47% rise from 2020, with a larger increase in transitioning (64%–95%) versus transitioned (32%–56%) countries due to demographic changes, although this may be further exacerbated by increasing risk factors associated with limited medical infrastructure in LMICs.1 It is well acknowledged that cancer early detection offers a higher cure rate and 5-year survival rate as well as a reduction in treatment cost and loss of economic productivity.2 Various cancer screening techniques are currently available in clinical practice. Examples include low-dose computed tomography (LDCT) for lung cancer screening,3 mammogram used to detect breast cancer,4 HPV test or cytology combined with colposcopy for early detection of cervical cancer,5 faecal occult blood test (FOBT) combined with colonoscopy for colorectal cancer screening,6 and prostate-specific antigen (PSA) for prostate cancer.7 However, the high cost of these screening methods and their need for specialised infrastructure and skilled technicists limit their application, and this is why there are alternatives of screening tests in LMICs: visual inspection with acetic acid (VIA) for cervical cancer and clinical breast examination (CBE) for breast cancer screening. Moreover, these methods are individually designed for screening for specific cancer types, hindering their widespread use as screening tools. In addition, liquid biopsy methods, which detect blood-based analytes such as cancer-derived DNA, are now being adopted in human medicine to simultaneously screen for multiple types of cancer; these MCED tests, such as Galleri,8 CancerSEEK,9,10 and SeekInCare,11 represent a paradigm shift in cancer screening and promise to significantly increase the number of patients with cancer that are detected at earlier stages. However, these tests are not suitable for using in LMICs due to cost, complexity, and dependency on high-end infrastructure and a rigorous laboratory. Taken together, these factors contribute to the fact that cancer is often diagnosed at a later stage and to inequalities in health care. In order to perform large-scale cancer screening among apparently healthy individuals in the future, especially among the population in LMICs, the development and validation of a more general, robust, and affordable MCED test are essential.
Immunological measurement of blood-based PTMs has been performed over decades in clinical for cancer screening of apparently healthy individuals on large-scale; for example, alpha-fetoprotein (AFP) for hepatocellular carcinoma,12 CA125 for ovarian cancer,13, 14, 15 CA15-3 for breast cancer,16,17 CA19-9 for pancreatic cancer,18 CA72-4 for ovarian cancer,19,20 carcinoembryonic antigen (CEA) for cancers in digestive tract,21 and CYFRA 21-1 for breast carcinoma.22 Such methods have significant advantages including their non-invasive nature, automation, and relatively low cost compared with many other clinical detection methods (endoscopy, imaging, etc.).23 However, the low sensitivity of these methods for early cancer detection limits their widespread use for screening purposes in a general population setting.
Previous studies have shown that PTM panels are diagnostically superior to single marker for the early detection of colorectal cancer,24,25 lung cancer,26 breast cancer,27 liver cancer,28 gastric cancer,29,30 pancreatic cancer,31 ovarian cancer,32 and oesophagus cancer.33 Several reports have also demonstrated that a combined PTM panel could be used for detecting several cancer types at the same time.34,35 However, different cancer types normally show different serological characteristics. Test results can also become more complex as the sample size increases, and traditional statistical methods may not be able to handle such big data. In addition, conventional clinical methods detect multiple PTMs at the same time and use a single threshold to evaluate the results, which may cause the accumulation of false-positive rates and lead to unnecessary clinical diagnostic workups. Hence, they were not suitable for asymptomatic large-scale population screening. AI is a good analytical method for solving classification challenges by identifying implicit patterns from complex data. Over the last decade, the significant contribution of AI techniques to this advanced technology has played a critical role in medicine and healthcare research. AI is considered a valuable tool in transforming the future of healthcare and precision oncology. Several novel algorithms have shown promising results for the accurate detection and characterisation of suspected lesions.36, 37, 38
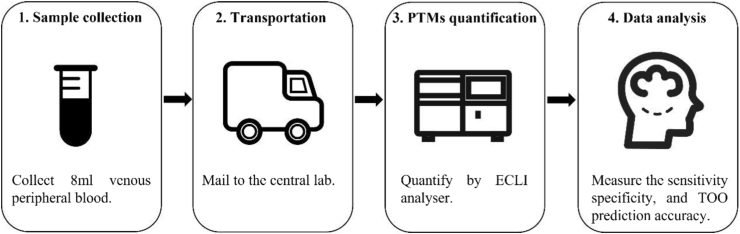
In this study, we assessed the performance and robustness of a protein assay named OncoSeek for MCED by integrating the measurement of a panel of seven selected PTMs and clinical information of the individuals, dramatically empowered by AI technology, which is more practical in LMICs (Fig. 1).
Fig. 1.
Schematic representation of clinical implementation workflow of OncoSeek test. 8 ml peripheral blood sample was collected from the individual in a cell-free DNA blood collection tube and mailed to the central lab. This is a special tube that proteins are stabilised stored at room temperature for seven days which makes it remote accessible as long as there's a local nurse who can draw blood. After plasma separation by centrifugation in the lab, PTM levels were measured by an electrochemiluminescence immunoassay analyser. OncoSeek was established using AI to distinguish cancer from non-cancer individuals by calculating the probability of cancer (POC) index based on the plasma levels of seven PTMs and clinical information including sex and age of the individuals. Then using another model to predict the possible affected TOO who has been detected with a cancer signal. PTMs, protein tumour markers. ECLI, electrochemiluminescence immunoassay. TOO, tissue of origin.
Methods
Participants
591 patients with cancer and 1055 non-cancer individuals were recruited as SeekIn laboratory-developed test (training cohort). 363 patients with cancer and 5556 non-cancer individuals were enrolled in Sun Yat-sen Memorial Hospital, Sun Yat-sen University as independent validation cohort 1. This observational study was a retrospective analysis on the data generated from the routine clinical testings at SeekIn and Sun Yat-sen Memorial Hospital from November 2012 to May 2022. The data were anonymised and all participants provided written informed consent upon enrolment. It was approved by the ethics committee of Sun Yat-sen Memorial Hospital (SYSKY-2023-435-01). Eligible patients with cancer were diagnosed with pathological confirmation and treatment-naïve prior to blood draw. Cancer stage was assigned by the attending physicians according to the American Joint Committee on Cancer (AJCC) Staging Manual (8th edition).39 Non-cancer individuals had no history of cancer. Cancer participants were also excluded for a prior diagnosis of cancer. Clinical data and PTMs quantification data of 1005 patients with cancer and 812 non-cancer individuals previously published by Cohen et al. were included as independent validation cohort 2 for analysis in this study.9
Quantification of PTMs
Peripheral blood from Sun Yat-sen Memorial Hospital was collected using a serum collection tube (BD Biosciences, San Jose, USA). Serum was separated within 4∼6 h using a centrifuge at 1300×g for 10 min at 4 °C. Samples from SeekIn were collected using a Cell-Free DNA BCT (Streck, La Vista, USA). Plasma samples were separated by centrifugation at 1600×g for 10 min at 4 °C within 3∼5 days. A total of 500 μL of plasma or serum from each blood sample was used to measure the levels of seven designated PTMs, including AFP, CA125, CA15-3, CA19-9, CA72-4, CEA, and CYFRA 21-1, using Roche cobas e411/e601 analyser (Roche Diagnostics GmbH, Mannheim, Germany) and commercially available reagent kits following manufacturer's instructions. The selection of these seven protein markers was based on our previous publication.40 The cut-off values for each biomarker were as follows: 5.8 IU/ml for AFP, 35.0 U/ml for CA125, 26.4 U/ml for CA15-3, 27.0 U/ml for CA19-9, 6.9 U/ml for CA72-4, 4.7 ng/ml for CEA, and 3.3 ng/ml for CYFRA21-1, as recommended by the manufacturer, which is set in advance based on a large-scale normal population. This test was calibrated as per the manufacturer's instructions using a two-point calibration, and quality control was performed. Plasma samples of independent validation cohort 2 from Johns Hopkins University School of Medicine used the Bioplex 200 platform (Bio-Rad, Hercules CA) for quantification of only six PTMs without CA72-4.9
Construction of the models of cancer detection and TOO prediction
The study utilised seven protein markers and two clinical characteristics (age and sex) as input features for an AI algorithm. Supplementary Figure S1 showed the detailed modelling process. The whole method includes two parts: one is a cancer detection model to determine whether the individual has cancer risk; another one is a TOO model for cancer location for tested positive groups. The first part is the development and validation of the cancer detection model. Different AI methods, including Gradient Boosting Machine (GBM), Generalised Linear Model (GLM), Random Forest (RF), and Support Vector Machine (SVM), were employed to create the model that could differentiate between cancer and non-cancer individuals. For the assessments, we compared the area under the curve (AUC) values and sensitivity/specificity of different models by the R package “pROC (1.18.0)”. Based on the performance of the model on the training set and two independent validation sets and the complexity of the model, as well as the positive correlation between the risk of cancer and protein expression, the GLM algorithm was finally chosen to establish the model to distinguish cancer from non-cancer individuals. The final model was built using GLM and 10-fold cross-validation was repeated 30 times. The average prediction value from these GLM models was defined as the probability of cancer (POC). POC value at 90.0% specificity was selected as cut-off value. When the test result was greater than cut-off, it indicated that cancer signals were detected. Otherwise, no cancer signal was detected.
For the prediction of TOO, the true positive patients of three cohorts were used to develop the model through the RF and GBM methods according to the publication.9,41 Due to imbalanced sample size for each cancer type, the downsampling method was employed to balance sample size of each cancer type. The top two organs with the highest prediction probability were considered as the potential TOO.
Statistical analysis
The Buderer's method42 was employed to estimate the sample size for our study. The objective was to achieve a sensitivity of 50% at a specificity of 90%. The confidence level was set at 95%, and the ratio of cases to controls was 1:2. Based on the calculations, n1 and n2 were determined to be 1165 and 207, and the larger value was chosen as the number of samples. Accounting for a 5% dropout rate, the final sample size was determined to be 1227. As we were able to collect a total of 1646 samples in the training cohort, that exceeded the required sample size. This larger sample size should provide sufficient statistical power for the study and enhance the reliability of the results.
We used the most common approach known as One-at-a-time (OAT) to perform the sensitivity analysis. We systematically excluded one PTM marker at a time while keeping the other PTM markers unchanged. By employing the same method (i.e., GLM), we calculated the sensitivity achieved when a particular PTM marker was excluded. This process was repeated for each of the remaining PTM markers in a similar fashion.
We conducted post-hoc subgroup analysis43 to compare the POC values between patients with cancer and individuals without cancer, considering factors such as gender and age. Additionally, we compared the POC values among individuals without cancer within three different source cohorts.
All statistical analyses were performed using R statistical software (https://www.r-project.org, version 4.1.2). pROC package version 1.18.0 was used to estimate the performance of the receiver operating characteristic in classifying cancer versus healthy individuals. We conducted Shapiro–Wilk tests on each protein biomarker of the healthy individuals in the training cohort and all P-values were less than 0.0001, indicating that the data is not a Gaussian distribution. This can also be observed through QQ plots, such as the example in Supplementary Figure S2 depicting the QQ plot of AFP expression from the healthy individuals. So, the Wilcoxon rank-sum test was used to compare the expression of each protein marker between the patients with cancer and the healthy individuals. Sensitivity, specificity, and predictive values were calculated by epiR software package version 2.0.40 (https://cran.r-project.org/web/packages/epiR/index.html). We also applied Delong test for statistical comparison of the AUCs.
Role of the funding source
The funder had no role in study design, data collection, data analyses, interpretation, or writing of the report.
Results
Study design and participants’ demographic characteristics
Between November 2012 and May 2022, 7565 participants were enrolled at SeekIn and Sun Yat-sen Memorial Hospital. The demographics and clinical characteristics of all participants are summarised in Table 1. Samples were divided into training (n = 1646, the samples from SeekIn) and independent validation cohort 1 (n = 5919, the samples from Sun Yat-sen Memorial Hospital). Independent validation cohort 2 (n = 1817) was from Johns Hopkins University School of Medicine. The cancer group included 496 cases of colorectal cancer, 300 cases of lung cancer, 291 cases of breast cancer, 244 cases of liver cancer, 159 cases of lymphoma, 149 cases of stomach cancer, 125 cases of pancreatic cancer, 82 cases of ovarian cancer, 66 cases of oesophageal cancer and 47 cases of cancers of the other origins. It should be noted that the selection of these cancer patients was not random, and there might be potential bias across different cancer types. As a case–control study to evaluate an MCED test, we intentionally selected the cancer types which were prevalent globally.1
Table 1.
Clinical and demographic characteristics of participants.
| Training cohort (SeekIn) |
Independent validation cohort 1 (SYSMH) |
Independent validation cohort 2 (JHUSM) |
||||
|---|---|---|---|---|---|---|
| Cancer |
Non-cancer |
Cancer |
Non-cancer |
Cancer |
Non-cancer |
|
| (n = 591) | (n = 1055) | (n = 363) | (n = 5556) | (n = 1005) | (n = 812) | |
| Platform | Roche cobas e411 | Roche cobas e601 | Bio-Rad Bioplex 200 platform | |||
| Sample type | Plasma | Serum | Plasma | |||
| Age | ||||||
| Mean(SD) | 55.3 (13.6) | 47.8 (12.4) | 58.5 (16.2) | 53.8 (17.4) | 62.9 (12.4) | 49.3 (19.5) |
| ≤55 years, n (%) | 289 (48.9%) | 788 (74.7%) | 143 (39.4%) | 2840 (51.1%) | 279 (22.8%) | 408 (50.2%) |
| >55 years, n (%) | 302 (51.1%) | 267 (25.3%) | 220 (60.6%) | 2716 (48.9%) | 726 (72.2%) | 404 (49.8%) |
| Sex | ||||||
| Female, n (%) | 256 (43.3%) | 543 (51.5%) | 140 (38.6%) | 2495 (44.9%) | 543 (54.0%) | 378 (46.6%) |
| Male, n (%) | 335 (56.7%) | 512 (48.5%) | 223 (61.4%) | 3061 (55.1%) | 462 (46.0%) | 434 (53.4%) |
| Cancer type | ||||||
| Lymphoma, n (%) | 159 (26.9%) | 0 (0.0%) | 0 (0.0%) | |||
| Liver, n (%) | 128 (21.7%) | 72 (19.8%) | 44 (4.4%) | |||
| Breast, n (%) | 66 (11.2%) | 16 (4.4%) | 209 (20.8%) | |||
| Colorectum, n (%) | 62 (10.5%) | 46 (12.7%) | 388 (38.6%) | |||
| Stomach, n (%) | 56 (9.5%) | 25 (6.9%) | 68 (6.8%) | |||
| Lung, n (%) | 41 (6.9%) | 155 (42.7%) | 104 (10.3%) | |||
| Pancreas, n (%) | 17 (2.9%) | 15 (4.1%) | 93 (9.3%) | |||
| Oesophagus, n (%) | 10 (1.7%) | 11 (3.0%) | 45 (4.5%) | |||
| Ovary, n (%) | 5 (0.8%) | 23 (6.3%) | 54 (5.4%) | |||
| Others, n (%) | 47 (8.0%) | 0 (0.0%) | 0 (0.0%) | |||
SYSMH, Sun Yat-sen Memorial Hospital, Sun Yat-sen University. JHUSM, Johns Hopkins University School of Medicine.
The performance of the conventional clinical method for cancer detection based on the PTMs
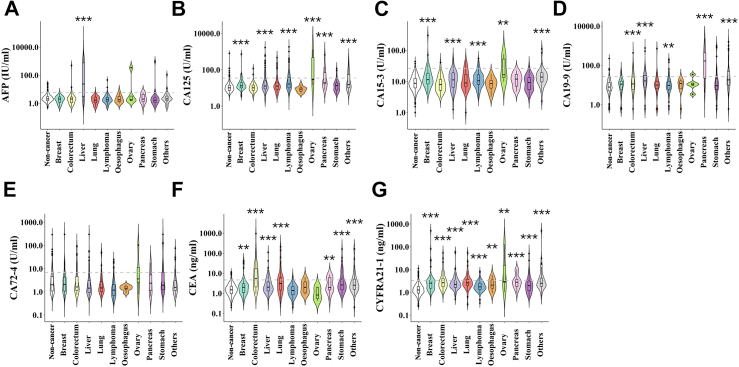
Except for the samples from Sun Yat-sen Memorial Hospital, which were serum, the other two cohorts had plasma samples. The SeekIn set and Sun Yat-sen Memorial Hospital set are using Roche cobas analysers, while the third cohort from Johns Hopkins University School of Medicine published data used the Bioplex 200 platform. Considering that the data from Johns Hopkins University only contained six tumour markers and lacked the value of CA72-4, we used the mean value of CA72-4 from the non-cancer sample group in the training cohort to replace the missing value. The values of these seven PTMs for the training cohort are shown in Fig. 2. The performance of these seven PTMs in individual tumour types in the training cohort is depicted in Supplementary Table S1. Almost all PTMs had a specificity of more than 95.0%, except CA72-4, which was 85.7%, whereas the sensitivity of an isolated PTM for the detection of individual malignancies was very low (ranging from 0 to 70.6%, the median value was 16.1%). In addition, when a single tumour marker was analysed, the specificity was high, while multiple tumour markers were analysed according to the conventional clinical method, and the false-positive rate was cumulative. The conventional clinical method mentioned here is a method based on the quantification of PTMs and assesses the results merely by a single threshold, which is based on pre-determined reference ranges for each PTM recommended by the manufacturer [i.e., the manufacturer-suggested cut-off value (MSCV)]. The sensitivity elevated when the number of PTMs increased in the conventional clinical method but, as a trade-off, the specificity decreased at the same time (Supplementary Figure S3). A total of 705 cases had at least one positive biomarker. The data in Supplementary Table S2 showed that simultaneous detection of these seven PTMs in all samples, the specificity was only 69.4% (95% CI: 66.5%–72.2%). Taken together, these results indicated that the conventional clinical method using multiple tumour marker panels has a very high false-positive rate.
Fig. 2.
Quantification of PTMs in different cancer types. Quantification value of each PTM (y-axis) based on healthy or individual cancer types (x-axis). The black horizontal lines are cut-off values that are recommended by the manufacturer. ∗∗ indicates P-value <0.01, ∗∗∗ indicates P-value <0.001.
The performance characteristics comparison between OncoSeek and the conventional clinical method
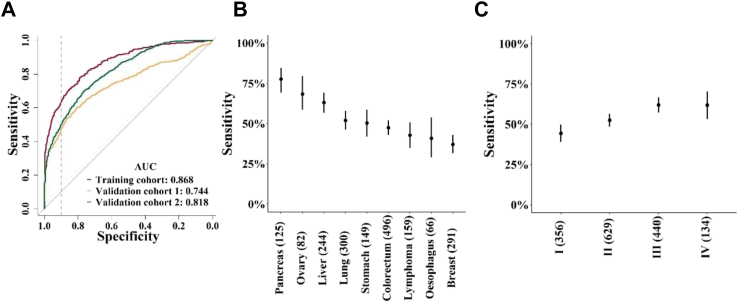
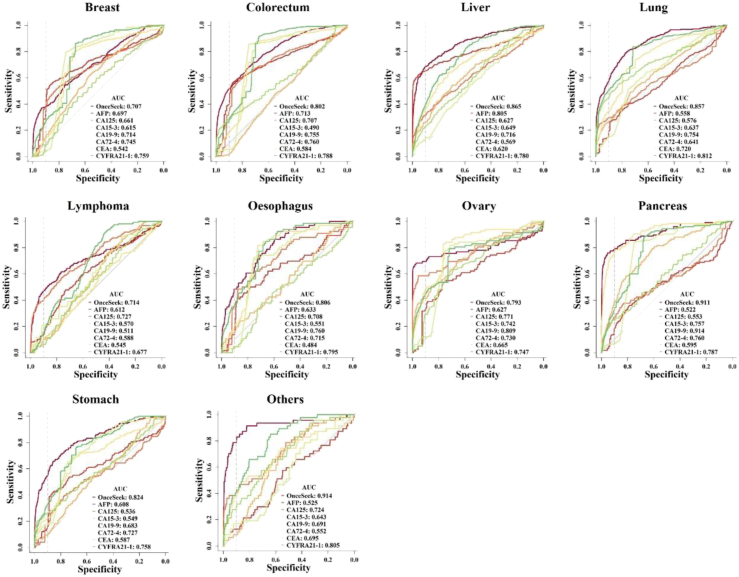
Considering the high false-positive rate of the conventional clinical method, a highly specific and robust MCED method is very essential in clinic. An algorithm named OncoSeek was established using AI to distinguish cancer from non-cancer individuals by calculating the probability of cancer (POC) index based on the expression of the seven PTMs and clinical basic information including sex and age of the individuals and to predict the possible affected TOO for those with a high-risk cancer signal. Four different machine learning algorithms (GBM, GLM, RF, and SVM) are adopted, no matter whether GBM (AUC = 0.869), GLM (AUC = 0.868), or RF (AUC = 0.869) algorithm was used, the model has similar performance, but all of them significantly outperformed the SVM (AUC = 0.817) algorithm across these three cohorts (Supplementary Figure S4A and B). However, GBM and RF are more complex than GLM models because they are composed of multiple decision trees and they take into consideration the correlations between features and their interactions. As a result, these algorithms are often able to capture more complex relationships and carry a greater risk of overfitting. Finally, the GLM algorithm was chosen to establish the OncoSeek model as a single locked algorithm to apply across the three cohorts, which could distinguish cancer from non-cancer controls in the training (AUC = 0.868) and the two independent validation cohorts (AUC = 0.744 and 0.818, respectively; P < 0.001 by Delong test; Fig. 3A). With the specificity at ∼90.0%, the sensitivity across these three cohorts was 58.2% (training cohort, 95% CI: 54.1%–62.2%), 47.4% (independent validation cohort 1, 95% CI: 42.1%–52.7%), and 49.3% (independent validation cohort 2, 95% CI: 46.1%–52.4%), respectively (Supplementary Table S3). Based on the subgroup analysis, we observed no significant difference in the POC values among individuals without cancer within any two out of the three cohorts (Wilcoxon rank-sum test, P > 0.05). Meanwhile, we found a significant increase in the POC values of patients with cancer compared to individuals without cancer across different gender and age groups (Wilcoxon rank-sum test, P < 0.001). Among these tumour types, pancreas had the highest sensitivity of 77.6% (95% CI: 69.3%–84.6%), followed by ovary [68.3% (95% CI: 58.5%–79.5%)], liver [63.1% (95% CI: 56.7%–69.2%)], lung [52.0% (95% CI: 46.2%–57.8%)], stomach [50.3% (95% CI: 42.0% to 58.6%)], colorectum [47.4% (95% CI: 42.9%–51.9%)], lymphoma [42.8% (95% CI: 35.0%–50.8%)], oesophagus [40.9% (95% CI: 29.0%–53.7%)], and breast [37.1% (95% CI: 31.5%–42.9%)]. The sensitivity basically increased with increasing clinical stage [stage I (n = 356), 44.4% (95% CI: 39.1%–49.7%); stage II (n = 629), 52.5% (95% CI: 48.5%–56.4%); stage III (n = 440), 62.0% (95% CI: 57.3%–66.6%); stage IV (n = 134), 61.9% (95% CI: 53.2%–70.2%)]. The sensitivities of OncoSeek in individual tumour types and each cancer stage are depicted in Fig. 3B and C. The probabilistic nature of the approach used to determine a positive sample was evident from Supplementary Figure S5, where each panel represented the sensitivity of OncoSeek when a specific protein marker was excluded from the analysis. The difference in sensitivity, compared to that achieved by OncoSeek, reflected the relative contribution of each PTM marker to the performance of OncoSeek test. To compare the performance of the conventional clinical method and OncoSeek, we evaluated the sensitivity and specificity of both methods in all samples. The sensitivity of the two methods was 63.8% (conventional clinical method, 95% CI: 61.6%–65.9%) and 51.7% (OncoSeek, 95% CI: 49.4%–53.9%) respectively, but the specificity was significantly different, 56.9% (conventional clinical method, 95% CI: 55.8%–58.0%) and 92.9% (OncoSeek, 95% CI: 92.3%–93.5%), respectively (Table 2). This reflected that OncoSeek had a relatively high specificity level, which was very important for a population screening test and indicated that it could avoid burdening the population with false-positive results. Meanwhile, OncoSeek achieved 84.3% (95% CI: 83.5%–85.0%) accuracy. We then evaluated the performance of OncoSeek and each PTM in individual tumour types and the data is reported in Fig. 4, which indicated that tumour markers had TOO information. Furthermore, OncoSeek has shown excellent performance in several high-mortality cancer types for which routine screening tests are lacking in the clinic, such as the AUC value of pancreatic cancer which was 0.911.
Fig. 3.
The performance of OncoSeek assay. (A) The receiver operating characteristic (ROC) curve evaluated the performance of OncoSeek in the training and independent validation cohorts. The area under the curve (AUC) of the three cohorts was depicted in the figure. The dotted vertical line in the ROC figures represents a 90.0% specificity. (B) The sensitivity of OncoSeek in individual tumour types. Sensitivity (y-axis) by cancer class based on individual cancer classes (x-axis), including multiple cancer types. Cancer classes are ordered based on sensitivity reducing, bars indicate 95% CI. The numbers in parentheses indicate the samples for each cancer class. (C) The sensitivity of OncoSeek in each clinical stage. Sensitivity (y-axis) based on individual cancer stage (x-axis), bars indicate 95% CI. The numbers in parentheses indicate the samples for each clinical stage.
Table 2.
The comparison of performance between the conventional clinical method and OncoSeek.
| Conventional clinical method |
OncoSeek |
|||
|---|---|---|---|---|
| Cancer | Non-cancer | Cancer | Non-cancer | |
| Predict cancer | 1249a | 3200a | 1012 | 527 |
| Predict non-cancer | 710 | 4223 | 947 | 6896 |
| Sensitivity (95% CI) | 63.8% (61.6%, 65.9%) | 51.7% (49.4%, 53.9%) | ||
| Specificity (95% CI) | 56.9% (55.8%, 58.0%) | 92.9% (92.3%, 93.5%) | ||
| PPV (95% CI) | 28.1% (26.8%, 29.4%) | 65.8% (63.3%, 68.1%) | ||
| NPV (95% CI) | 85.6% (84.6%, 86.6%) | 87.9% (87.2%, 88.6%) | ||
PPV, Positive predictive value. NPV, Negative predictive value.
Individuals with at least one of the markers included in the panel showing values above the cut-off point were considered as being positive.
Fig. 4.
Receiver operating characteristic (ROC) curves of these seven PTMs and OncoSeek in individual cancer types. The area under the curve (AUC) of these seven PTMs and OncoSeek were depicted in the figure. The dotted vertical line in the ROC figures represents a 90.0% specificity.
Predicting affected TOO
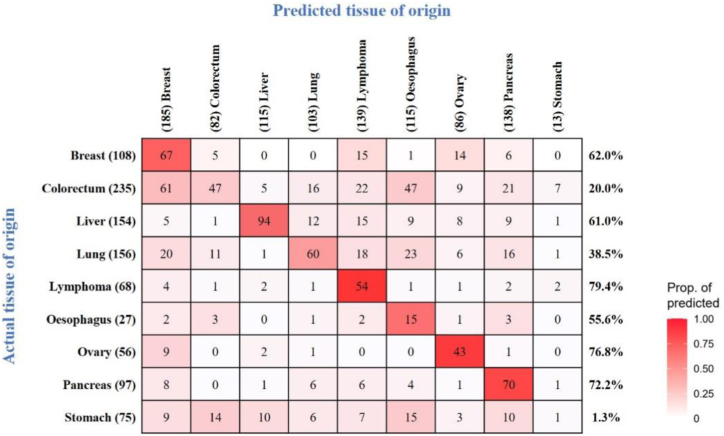
Most liquid biopsies are unable to identify affected TOO in patients who test positive, especially those based on genetic mutations, because the same gene mutations drive multiple tumour types. However, a critical attribute of a blood-based multi-cancer detection test is the ability to localise the TOO to direct the diagnostic workup. Based on the tissue-specific characteristics of PTMs, we used supervised AI algorithm to predict the underlying cancer type in patients with true positive tests. We then used this algorithm to study 976 patients with cancer scoring true positives from these three cohorts in the OncoSeek test to predict the possible affected organ system. The overall accuracy of the top two most possible organ systems was 66.8% (Fig. 5).
Fig. 5.
TOO accuracy by individual cancer type. Confusion matrices representing the accuracy of TOO localisation. Agreement between the actual (x-axis) and predicted (y-axis) TOO per sample using the OncoSeek model was depicted. Colour corresponds to the proportion of predicted TOO calls. Included 976 participants were those with cancer predicted as having cancer at 92.9% specificity.
Discussion
Herein, we report an efficient, easier, and affordable MCED approach, named as OncoSeek, which is based on seven PTMs and clinical information of the individual and empowered by AI. In this large multicentre study (n = 9382) containing more than nine cancer types and dominated by early-stage patients (63.2% stage I and II), 51.7% sensitivity was achieved at 92.9% specificity, resulting in 84.3% accuracy. The accuracy of the top two most possible organ systems was 66.8%. The validation cohort 1 was using serum. The validation cohort 2 was a non-Chinese cohort (a mix of Caucasians, Asians, blacks and Latinos), only had six PTMs and was on Bioplex 200. All these variables (sample type, platform, number of PTMs and race) demonstrated the robustness of OncoSeek and would also make our findings generalisable.
A wealth of literature has evaluated the utility of PTMs in cancer detection and monitoring.12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22 These tests are now common practice in clinics. Our results proved that individual PTMs could contribute to cancer detection. For example, AFP, as a specific marker for liver cancer,12 resulting a sensitivity of 62.5% at a specificity of 95.4% in the training cohort. At the same time, CEA as a multi-cancer PTM,44 and its quantitative level in multiple tumour types is significantly higher than that in the non-cancer group. Almost all PTMs had high specificity, whereas the sensitivity of an isolated PTM for the detection of individual malignancies was very low. Overall, the performance of each biomarker included in our study, when analysed individually, was similar to what has previously been reported in the literatures.12,15,17, 18, 19,22
However, tumour heterogeneity is a well-known concept in tumour biology, which is also an important reason for the difficulty of tumour diagnosis and treatment.45 Therefore, multiple cancer detection cannot be based on a single tumour marker. More and more literature has confirmed that the performance of PTM panels is better than that of single tumour marker.24, 25, 26, 27, 28, 29, 30, 31, 32, 33 However, the conventional clinical method usually uses a single threshold (i.e., MSCV), which is set in advance based on a large-scale normal population. In a selected PTM panel, when any one of the tumour markers exceeds the cut-off value, the individual is considered to be positive. Our results, as well as previous literature,24 suggest that false-positive rates accumulate as the number of tumour markers increases. In all samples of these three cohorts, the sensitivity of the conventional clinical method based on MSCV was 63.8%, but the specificity was only 56.9%. The conventional clinical method, which simply combines the results of different tumour markers tests, would generally have a cumulative false-positive rate higher than the individual tests, which led to unnecessary imaging, invasive procedures, or other diagnostic workups and potentially increasing unnecessary medical costs and anxiety. By contrast, OncoSeek used AI to learn and identify specific patterns of tumour markers and clinical factors and their interdependencies to distinguish cancer from non-cancer individuals, thus greatly increasing its specificity, and achieving a specificity of 92.9%. A sufficiently high specificity to ensure a low rate of false positives is a fundamental characteristic of asymptomatic population screening. AI algorithms play a key role in our approach, which makes it possible to do multiple cancer screening based on PTMs. Recent reports have demonstrated that AI has already been leveraged to improve the performance of different liquid biopsy assays and might facilitate their future integration into the clinical workflow,41,46 which is based on its excellent ability to handle complex interactions between large numbers of data and information. We have tried several AI algorithms including GBM, GLM, RF, and SVM. There was no difference in the performance of the first three algorithms, but the performance of SVM was significantly worse than others. The SVM algorithm was therefore excluded while the remaining three algorithms all have different advantages, disadvantages, and characteristics. GBM and RF are two machine learning algorithms based on decision trees, while a model based on a normal distribution is typically referred to as a GLM. GBM and RF are more complex than GLM models because they are composed of multiple decision trees and they take into consideration the correlations between features and their interactions. As a result, GBM and RF algorithms are often able to capture more complex relationships in the data and perform better in prediction compared to GLM models. Moreover, GBM and RF algorithms also carry a greater risk of overfitting, due to their complexity causing them to better adapt to the training data, leading to a decline in performance on test data. Therefore, we finally used the GLM algorithm for modelling.
Our study included two independent validation cohorts, which may lower the possibility of overestimation of the performance. With the specificity at ∼90.0%, the sensitivity of OncoSeek across the three cohorts was 58.2%, 47.4%, and 49.3%, respectively, which was similar, although the sample types and the platforms of these three cohorts were different (see Table 1), even in independent validation cohort 2, only containing six tumour markers thereinto. In addition, the sample size of the whole study reached nearly 10,000 cases, which is a very large data set. These data strongly proved that our method was robust. In addition, though the overall sensitivity of the OncoSeek test was only 51.7%, less than that of previously reported single-cancer detection tests,47,48 as an MCED test, the overall incidence of cancer was higher than that of any single cancer. The nine cancer types studied here account for ∼59.2% of global cancer deaths annually.1 This means that multiple cancer tests with moderate sensitivity may yield higher cancer detection rates than single cancer tests with very high sensitivity. Furthermore, the OncoSeek assay showed a relatively high sensitivity for several cancers for which there is no clinical screening method. For example, the OncoSeek assay showed a sensitivity of 77.6% for pancreatic cancer, which has a high mortality. The application of this assay may lead to earlier detection of pancreatic cancer and improve the therapeutic effect, thus potentially reducing mortality.
Another part of the data in this study is the accuracy of predicting affected organ systems. Accurate TOO localisation is critical to direct the diagnostic workup; in its absence, patients with a positive test may be subjected to unnecessary diagnostic tests. TOO is a multiple classification and non-linear relationship between the independent (cancer types) and the dependent (PTMs expression) variables. However, SVM and GLM are linear classifiers and often used for binary classification, so only GBM and RF were used to predict TOO in our study. Meanwhile, the publications of CancerSEEK and DELFI assays also used RF and GBM algorithms for the prediction of the cancer type, respectively.9,46 We found that the overall accuracy of top-two most possible organ systems from GBM was 60.9%, lower than RF (66.8%). Taken together, we finally chose RF for TOO analysis. The function of TOO may also be helpful for some patients with unknown primary lesions.
In addition, it should be emphasised that the OncoSeek method also has the following advantages. This is an MCED method that could detect multiple cancer types at once. PTMs are quantified by an automated electrochemiluminescence immunoassay analyser that is easy to perform and does not require sophisticated training and expertise to be deployed. The turnaround time of this test is very fast. It takes less than 10 min to quantify these seven tumour markers, making it possible to report on the same day samples are received. The newly developed MCED assays, such as Galleri,8 can achieve similar sensitivity (51.5%) and higher specificity (99.5%). However, they require high-end and complex infrastructure like next-generation sequencing instruments, and are very expensive (∼$1000). Based on these factors, OncoSeek assay is easier to implement in large-scale populations, even in LMICs.
We would like to acknowledge some limitations in this study. First, a prospective study shall provide more compelling evidence to support the application of OncoSeek in clinic. Second, the accuracy of TOO needs to be further improved to limit the scope, cost, and complexity of clinical evaluation of asymptomatic patients. Third, the selection of these patients was not random in this study, and the percentage of cancers was different from the real world incidence of cancers. This study was a case–control study, and as such, was not reflective of performance in real world. Finally, despite the broad range of cancer types captured in this study, for some cancer types the sample size was small, precluding a full representation of heterogeneity within some cancer types.
Taken together, this retrospective study demonstrates that OncoSeek, a blood-based MCED test, is significantly superior to the conventional clinical method based on MSCV in the detection of multiple cancer types, with a high specificity and provides accurate TOO prediction that may inform patients’ clinical management. The nine cancer types studied here account for ∼59.2% of global cancer deaths in 2020,1 and the majority of cancer types currently lack clinical screening tests, so their earlier detection could conceivably reduce deaths from these cancers. The results also support that OncoSeek, as an MCED assay, is also affordable (less than $25) and requires nothing more than a blood draw, which makes it more practical in LMICs.
Contributors
M.M. and C.D. designed the study; Y.L. and X.L. collected participant samples and clinical information; G.Z., D.Z., Y.F., and Y.Z. performed experiments; S.L. and W.W. designed bioinformatics pipelines and analysed results with the help of M.M.; G.Z. wrote the manuscript, with critical review from M.M. Y.L. and S.L. accessed and verified the underlying data.
Data sharing statement
We uploaded the protein quantification data with age and sex information into the National GeneBank Data Center in China (https://ngdc.cncb.ac.cn) with accession number PRJCA017145. Code for analyses is available at https://github.com/seekincancer/OncoSeek.
Declaration of interests
G.Z. is a full-time employee of and holds stock options in SeekIn Inc. S.L. is a full-time employee of and holds stock options in SeekIn Inc. W.W. is a full-time employee of and holds stock options in SeekIn Inc. Y.F. is a full-time employee of and holds stock options in SeekIn Inc. Y.Z. is a full-time employee of and holds stock options in SeekIn Inc. M.M. is a full-time employee of and holds stock options in SeekIn Inc. D.Z. is a full-time employee of Shenyou Bio, a wholly-owned subsidiary of SeekIn Inc, and holds stock options in SeekIn Inc. All other authors declare no competing interest.
Acknowledgements
This study was supported by the National Key Research and Development Programme of China (approval number: 2020YFC2004505). The authors would like to thank all individuals who participated in this study. The authors also thank the Johns Hopkins University School of Medicine for the clinical data and PTMs quantification data of independent validation cohort 2 in this study. The following Inspire2Live colleagues are acknowledged for helping to explore the use of OncoSeek in Africa: Peter Kapitein, Eltjo Heddema, Khama Rogo, Arthur Ajwang, and Franklin Mtei. We thank Drs. Catherine Sauvaget and Jun Zhou for helpful comments on the manuscript. We would like to acknowledge the English editing provided by Anthea Bull.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.eclinm.2023.102041.
Contributor Information
Chaohui Duan, Email: duanchh@mail.sysu.edu.cn.
Mao Mao, Email: maomao@yuhs.ac.
Appendix A. Supplementary data
Figure S1: The process of model development and validation. GBM, Gradient Boosting Machine. GLM, Generalised Linear Model. RF, Random Forest. SVM, Support Vector Machine. TOO, tissue of origin. +, means prediction positive case.
Figure S2: QQ plot of AFP expression fromindividuals without cancer.
Figure S3: A big problem with the conventional clinical method. The performance of a different number of PTMs was evaluated. When only one PTM is tested, the sensitivity is low, but the specificity is high. In contrast, the sensitivity increases, and specificity decreases when the number of PTMs is increased. PTMs, protein tumour markers.
Figure S4: Performance comparison of different algorithms used in the model. (A) The receiver operating characteristic (ROC) curve evaluates the performance of different algorithms in the training cohort. The area under the curve (AUC) was depicted in the figure. The dotted vertical line in the ROC figures represents a 90.0% specificity. (B) The sensitivity (orange dots) and specificity (blue dots) of these four algorithms in the two independent validation cohorts. The dotted vertical line in the figures was used to divide the data of two cohorts, bars indicate 95% CI. GBM, Gradient Boosting Machine. GLM, Generalised Linear Model. RF, Random Forest. SVM, Support Vector Machine.
Figure S5: Effect of each protein tumor marker on sensitivity. (A) Sensitivity of OncoSeek for each tumor type was shown, same as Fig. 3B. (B–H) Each panel showed the sensitivity achieved when a particular PTM marker was excluded through the same method (GLM).
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Hung M.C., Liu M.T., Cheng Y.M., Wang J.D. Estimation of savings of life-years and cost from early detection of cervical cancer: a follow-up study using nationwide databases for the period 2002–2009. BMC Cancer. 2014;14:505. doi: 10.1186/1471-2407-14-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitchell E.P. Screening for lung cancer. J Natl Med Assoc. 2021;113:239–240. doi: 10.1016/j.jnma.2021.05.012. [DOI] [PubMed] [Google Scholar]
- 4.Siu A.L. On behalf of the U.S. Preventive services task force. Screening for breast cancer: U.S. Preventive services task force recommendation statement. Ann Intern Med. 2016;164:279. doi: 10.7326/M15-2886. [DOI] [PubMed] [Google Scholar]
- 5.US Preventive Services Task Force. Curry S.J., Krist A.H., et al. Screening for cervical cancer: US preventive services task force recommendation statement. JAMA. 2018;320:674. doi: 10.1001/jama.2018.10897. [DOI] [PubMed] [Google Scholar]
- 6.US Preventive Services Task Force. Davidson K.W., Barry M.J., et al. Screening for colorectal cancer: US preventive services task force recommendation statement. JAMA. 2021;325:1965. doi: 10.1001/jama.2021.6238. [DOI] [PubMed] [Google Scholar]
- 7.US Preventive Services Task Force. Grossman D.C., Curry S.J., et al. Screening for prostate cancer: US preventive services task force recommendation statement. JAMA. 2018;319:1901. doi: 10.1001/jama.2018.3710. [DOI] [PubMed] [Google Scholar]
- 8.Klein E.A., Richards D., Cohn A., et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann Oncol. 2021;32:1167–1177. doi: 10.1016/j.annonc.2021.05.806. [DOI] [PubMed] [Google Scholar]
- 9.Cohen J.D., Li L., Wang Y., et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lennon A.M., Buchanan A.H., Kinde I., et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science. 2020;369 doi: 10.1126/science.abb9601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao M., Li S., Ren Q., et al. Poster at the early detection of cancer conference. 2022. Integrating multi-omics features for blood-based pan-cancer early detection. [Google Scholar]
- 12.Gervain J. Symptoms of hepatocellular carcinoma. Laboratory tests used for its diagnosis and screening. Orv Hetil. 2010;151:1415–1417. doi: 10.1556/OH.2010.28945. [DOI] [PubMed] [Google Scholar]
- 13.Zhang B., Cai F.F., Zhong X.Y. An overview of biomarkers for the ovarian cancer diagnosis. Eur J Obstet Gynecol Reprod Biol. 2011;158:119–123. doi: 10.1016/j.ejogrb.2011.04.023. [DOI] [PubMed] [Google Scholar]
- 14.Scholler N., Urban N. CA125 in ovarian cancer. Biomark Med. 2007;1:513–523. doi: 10.2217/17520363.1.4.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Calster B., Timmerman D., Bourne T., et al. Discrimination between benign and malignant adnexal masses by specialist ultrasound examination versus serum CA-125. J Natl Cancer Inst. 2007;99:1706–1714. doi: 10.1093/jnci/djm199. [DOI] [PubMed] [Google Scholar]
- 16.Safi F., Kohler I., Beger H.G., Röttinger E. The value of the tumor marker CA 15-3 in diagnosing and monitoring breast cancer. A comparative study with carcinoembryonic antigen. Cancer. 1991;68:574–582. doi: 10.1002/1097-0142(19910801)68:3<574::aid-cncr2820680322>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 17.Dnistrian A.M., Schwartz M.K., Greenberg E.J., Smith C.A., Schwartz D.C. Evaluation of CA M26, CA M29, CA 15-3 and CEA as circulating tumor markers in breast cancer patients. Tumor Biol. 1991;12:82–90. doi: 10.1159/000217692. [DOI] [PubMed] [Google Scholar]
- 18.Gu Y.L., Lan C., Pei H., Yang S.N., Liu Y.F., Xiao L.L. Applicative value of serum CA19-9, CEA, CA125 and CA242 in diagnosis and prognosis for patients with pancreatic cancer treated by concurrent chemoradiotherapy. Asian Pac J Cancer Prev. 2015;16:6569–6573. doi: 10.7314/apjcp.2015.16.15.6569. [DOI] [PubMed] [Google Scholar]
- 19.Xu Y., Zhang P., Zhang K., Huang C. The application of CA72-4 in the diagnosis, prognosis, and treatment of gastric cancer. Biochim Biophys Acta Rev Cancer. 2021;1876 doi: 10.1016/j.bbcan.2021.188634. [DOI] [PubMed] [Google Scholar]
- 20.Yanqing H., Cheng D., Ling X. Serum CA72-4 as a biomarker in the diagnosis of colorectal cancer: a meta-analysis. Open Med. 2018;13:164–171. doi: 10.1515/med-2018-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang S., Zhou F., Sun Y., et al. CEA in breast ductal secretions as a promising biomarker for the diagnosis of breast cancer: a systematic review and meta-analysis. Breast Cancer. 2016;23:813–819. doi: 10.1007/s12282-016-0680-9. [DOI] [PubMed] [Google Scholar]
- 22.Yuan J., Sun Y., Wang K., et al. Development and validation of reassigned CEA, CYFRA21-1 and NSE-based models for lung cancer diagnosis and prognosis prediction. BMC Cancer. 2022;22:686. doi: 10.1186/s12885-022-09728-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duffy M.J. Tumor markers in clinical practice: a review focusing on common solid cancers. Med Princ Pract. 2013;22:4–11. doi: 10.1159/000338393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Y., Wang J., Zhou Y., Sheng S., Qian S.Y., Huo X. Evaluation of serum CEA, CA19-9, CA72-4, CA125 and ferritin as diagnostic markers and factors of clinical parameters for colorectal cancer. Sci Rep. 2018;8:2732. doi: 10.1038/s41598-018-21048-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo H., Shen K., Li B., Li R., Wang Z., Xie Z. Clinical significance and diagnostic value of serum NSE, CEA, CA19-9, CA125 and CA242 levels in colorectal cancer. Oncol Lett. 2020;20:742–750. doi: 10.3892/ol.2020.11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulpa J., Wójcik E., Reinfuss M., Kołodziejski L. Carcinoembryonic antigen, squamous cell carcinoma antigen, CYFRA 21-1, and neuron-specific enolase in squamous cell lung cancer patients. Clin Chem. 2002;48:1931–1937. [PubMed] [Google Scholar]
- 27.Wang W., Xu X., Tian B., et al. The diagnostic value of serum tumor markers CEA, CA19-9, CA125, CA15-3, and TPS in metastatic breast cancer. Clin Chim Acta. 2017;470:51–55. doi: 10.1016/j.cca.2017.04.023. [DOI] [PubMed] [Google Scholar]
- 28.Si Y.Q., Wang X.Q., Fan G., et al. Value of AFP and PIVKA-II in diagnosis of HBV-related hepatocellular carcinoma and prediction of vascular invasion and tumor differentiation. Infect Agents Cancer. 2020;15:70. doi: 10.1186/s13027-020-00337-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Z., Zhang N. Clinical evaluation of CEA, CA19-9, CA72-4 and CA125 in gastric cancer patients with neoadjuvant chemotherapy. World J Surg Oncol. 2014;12:397. doi: 10.1186/1477-7819-12-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He C.Z., Zhang K.H., Li Q., Liu X.H., Hong Y., Lv N.H. Combined use of AFP, CEA, CA125 and CAl9-9 improves the sensitivity for the diagnosis of gastric cancer. BMC Gastroenterol. 2013;13:87. doi: 10.1186/1471-230X-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang P., Zou M., Wen X., et al. Development of serum parameters panels for the early detection of pancreatic cancer: zhang et al. Int J Cancer. 2014;134:2646–2655. doi: 10.1002/ijc.28584. [DOI] [PubMed] [Google Scholar]
- 32.Yurkovetsky Z.R., Linkov F.Y., E Malehorn D., Lokshin A.E. Multiple biomarker panels for early detection of ovarian cancer. Future Oncol. 2006;2:733–741. doi: 10.2217/14796694.2.6.733. [DOI] [PubMed] [Google Scholar]
- 33.Zheng Q., Zhang L., Tu M., et al. Development of a panel of autoantibody against NSG1 with CEA, CYFRA21-1, and SCC-Ag for the diagnosis of esophageal squamous cell carcinoma. Clin Chim Acta. 2021;520:126–132. doi: 10.1016/j.cca.2021.06.013. [DOI] [PubMed] [Google Scholar]
- 34.Tsao K.C., Wu T.L., Chang P.Y., Hong J.H., Wu J.T. Detection of carcinomas in an asymptomatic Chinese population: advantage of screening with multiple tumor markers. J Clin Lab Anal. 2006;20:42–46. doi: 10.1002/jcla.20102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wen Y.H., Chang P.Y., Hsu C.M., Wang H.Y., Chiu C.T., Lu J.J. Cancer screening through a multi-analyte serum biomarker panel during health check-up examinations: results from a 12-year experience. Clin Chim Acta. 2015;450:273–276. doi: 10.1016/j.cca.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 36.Sechopoulos I., Teuwen J., Mann R. Artificial intelligence for breast cancer detection in mammography and digital breast tomosynthesis: state of the art. Semin Cancer Biol. 2021;72:214–225. doi: 10.1016/j.semcancer.2020.06.002. [DOI] [PubMed] [Google Scholar]
- 37.Mitsala A., Tsalikidis C., Pitiakoudis M., Simopoulos C., Tsaroucha A.K. Artificial intelligence in colorectal cancer screening, diagnosis and treatment. A new era. Curr Oncol. 2021;28:1581–1607. doi: 10.3390/curroncol28030149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pantanowitz L., Quiroga-Garza G.M., Bien L., et al. An artificial intelligence algorithm for prostate cancer diagnosis in whole slide images of core needle biopsies: a blinded clinical validation and deployment study. Lancet Digit Health. 2020;2:e407–e416. doi: 10.1016/S2589-7500(20)30159-X. [DOI] [PubMed] [Google Scholar]
- 39.Amin M.B., Greene F.L., Edge S.B., et al. The Eighth Edition AJCC Cancer Staging Manual: continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA Cancer J Clin. 2017;67:93–99. doi: 10.3322/caac.21388. [DOI] [PubMed] [Google Scholar]
- 40.Ji X., Li J., Huang Y., et al. Identifying occult maternal malignancies from 1.93 million pregnant women undergoing noninvasive prenatal screening tests. Genet Med. 2019;21:2293–2302. doi: 10.1038/s41436-019-0510-5. [DOI] [PubMed] [Google Scholar]
- 41.Cristiano S., Leal A., Phallen J., et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. 2019;570:385–389. doi: 10.1038/s41586-019-1272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buderer N.M. Statistical methodology: I. Incorporating the prevalence of disease into the sample size calculation for sensitivity and specificity. Acad Emerg Med. 1996;9:895–900. doi: 10.1111/j.1553-2712.1996.tb03538.x. [DOI] [PubMed] [Google Scholar]
- 43.Adams K.F., Jr. Post hoc subgroup analysis and the truth of a clinical trial. Am Heart J. 1998;5:753–758. doi: 10.1016/s0002-8703(98)70116-4. [DOI] [PubMed] [Google Scholar]
- 44.Hao C., Zhang G., Zhang L. Progress in molecular biology and translational science. Elsevier; 2019. Serum CEA levels in 49 different types of cancer and noncancer diseases; pp. 213–227. [DOI] [PubMed] [Google Scholar]
- 45.Seoane J., De Mattos-Arruda L. The challenge of intratumour heterogeneity in precision medicine. J Intern Med. 2014;276:41–51. doi: 10.1111/joim.12240. [DOI] [PubMed] [Google Scholar]
- 46.Wan N., Weinberg D., Liu T.Y., et al. Machine learning enables detection of early-stage colorectal cancer by whole-genome sequencing of plasma cell-free DNA. BMC Cancer. 2019;19:832. doi: 10.1186/s12885-019-6003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imperiale T.F., Ransohoff D.F., Itzkowitz S.H., et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 48.The PreCar Team. Chen L., Abou-Alfa G.K., et al. Genome-scale profiling of circulating cell-free DNA signatures for early detection of hepatocellular carcinoma in cirrhotic patients. Cell Res. 2021;31:589–592. doi: 10.1038/s41422-020-00457-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: The process of model development and validation. GBM, Gradient Boosting Machine. GLM, Generalised Linear Model. RF, Random Forest. SVM, Support Vector Machine. TOO, tissue of origin. +, means prediction positive case.
Figure S2: QQ plot of AFP expression fromindividuals without cancer.
Figure S3: A big problem with the conventional clinical method. The performance of a different number of PTMs was evaluated. When only one PTM is tested, the sensitivity is low, but the specificity is high. In contrast, the sensitivity increases, and specificity decreases when the number of PTMs is increased. PTMs, protein tumour markers.
Figure S4: Performance comparison of different algorithms used in the model. (A) The receiver operating characteristic (ROC) curve evaluates the performance of different algorithms in the training cohort. The area under the curve (AUC) was depicted in the figure. The dotted vertical line in the ROC figures represents a 90.0% specificity. (B) The sensitivity (orange dots) and specificity (blue dots) of these four algorithms in the two independent validation cohorts. The dotted vertical line in the figures was used to divide the data of two cohorts, bars indicate 95% CI. GBM, Gradient Boosting Machine. GLM, Generalised Linear Model. RF, Random Forest. SVM, Support Vector Machine.
Figure S5: Effect of each protein tumor marker on sensitivity. (A) Sensitivity of OncoSeek for each tumor type was shown, same as Fig. 3B. (B–H) Each panel showed the sensitivity achieved when a particular PTM marker was excluded through the same method (GLM).