Abstract
Nonalcoholic fatty liver disease (NAFLD) is characterised by hepatic steatosis, inflammation, and insulin resistance. The role of long noncoding RNA (lncRNA)-regulated pyroptosis in NAFLD development remains largely unknown. This study aimed to investigate whether NAFLD development is controlled by lncRNA growth-arrest specific transcript 5 (GAS5)/miR-28a-5p/membrane associated ring-CH-type finger 7 (MARCH7)-mediated pyroptosis using in vivo and in vitro models. First, GAS5 expression was decreased but miR-28a-5p expression was increased in the livers of NAFLD patients, high-fat diet (HFD)-fed mice and leptin-deficient obese (Ob/Ob) mice. Furthermore, GAS5 suppressed while miR-28a-5p promoted NAFLD development, and overexpression of miR-28a-5p reversed the GAS5 overexpression-induced attenuation of NAFLD. Mechanistically, GAS5 served as a sponge of miR-28a-5p, and miR-28a-5p enhanced pyroptosis by targeting the 3′ untranslated region (UTR) of the E3 ligase MARCH7 during NAFLD development. MARCH7 interacted with the NOD-like receptor protein 3 (NLRP3) protein, resulting in proteasomal degradation of NLRP3 to inhibit pyroptosis. As expected, MARCH7 knockdown abolished the miR-28a-5p knockdown-induced inhibition of NAFLD development, and the ubiquitin E3 ligase-inactive mutant (W589A/I556A) of MARCH7 failed to inhibit NAFLD development. In conclusion, GAS5 protected against NAFLD development by binding to miR-28a-5p, miR-28a-5p promoted NAFLD development by targeting MARCH7, and MARCH7 ameliorated NAFLD by suppressing NLRP3-mediated pyroptosis. The GAS5/miR-28a-5p/MARCH7/NLRP3 axis plays an important role in NAFLD progression, and it might be a biomarker for NAFLD.
Subject terms: Endocrine system and metabolic diseases, Epigenetics
Introduction
Nonalcoholic fatty liver disease (NAFLD) frequently leads to chronic liver disease, and effective therapies remain lacking [1, 2]. NAFLD pathogenesis is characterised by excessive lipid deposition in hepatocytes, which causes the dysfunction of lipid metabolism (hepatic steatosis), failure to properly use insulin (insulin resistance), and release of inflammatory factors (inflammation) [1, 2]. NAFLD often results in liver fibrosis, cirrhosis and hepatocellular carcinoma [2]. Based on recent epidemiological studies, NAFLD is the second most common liver disease in China, and that its incidence is up to 40% in Europe, North America, and Australia [3], indicating that NAFLD is a serious threat to human health. However, the mechanisms of NAFLD pathogenesis have not been thoroughly explored.
Increasing evidence suggests that long noncoding RNAs (lncRNAs), a class of RNAs longer than 200 nt, play a vital role in NAFLD development [4, 5]. Growth-arrest specific transcript 5 (GAS5) has multiple functions, such as participating in cellular growth arrest and apoptosis. It has also been identified as a potential tumour suppressor, with its downregulation associated with cancer in multiple different tissues [6]. Recently, GAS5 was reported to promote NAFLD via the miR-29a-3p/NOTCH2 axis [7]. Shizan Xu et al. also proved that GAS5 is activated in in vitro and in vivo fatty liver models and that knockdown of GAS5 abrogates hepatic lipid accumulation [8]. However, some evidence has demonstrated that GAS5 ameliorates liver fibrosis by serving as a sponge of miRNAs: GAS5 inhibits liver fibrogenesis by acting as a competing endogenous RNA of miR-222 [9], and GAS5 suppresses CCL4-induced hepatic fibrosis by sponging miR-23a [10]. MiR-28a-5p was proven to inhibit malignant tumour progression [11]. Interestingly, downregulation of miR-28a-5p was found to attenuate liver fibrosis in mice [12] These findings suggest that the lncRNA GAS5 and miR-28a-5p play critical roles in liver disorders.
The membrane-associated RING-CH (MARCH) family includes MARCH1 to MARCH11, and the members of the MARCH family are diverse, participating in multiple physiological functions, such as sperm formation, membrane transfer, and lipid synthesis [13]. MARCH proteins belong to the subfamily of RING-type E3 ubiquitin ligases and regulate posttranslational modifications, which play a key role in regulating the stability, localisation, protein‒protein interactions, and other properties of their substrates [14]. As a noncanonical MARCH family member, MARCH7 plays critical roles in immune tolerance by polyubiquitinating NLRP3 to target it for degradation [14, 15]. Although the NLRP3 inflammasome has been proven to mediate nonischaemic dilated cardiomyopathy [16], Parkinson’s disease [17] and NAFLD [18], the detailed regulation of NLRP3-dependent pyroptosis still needs further study.
Usually, NLRP3 inflammasome activation triggers pyroptosis by increasing caspase-1 activity [19], and then activated caspase-1 cleaves its downstream effector GSDMD into GSDMD-N, an executor of pyroptosis [20]. Pyroptosis may be activated by lipid toxicity, inflammation, and oxidative stress during NAFLD development [20, 21]. In addition, pyroptosis may increase lipogenesis in hepatocytes to promote NAFLD development by upregulating the lipogenic gene srebp1c and downregulating the expression of lipolytic genes, including pparα, Aco, lcad, cyp4a10 and cypa14 [20, 22]. Pyroptosis can also induce inflammation in hepatocytes to enhance NAFLD progression by elevating the expression of proinflammatory cytokines [20, 22]. Collectively, these observations indicate that abnormal activation of pyroptosis could facilitate lipid deposition and inflammation, resulting in NAFLD development. Consistent with these observations, knockout of NLRP3 or GSDMD-N could constitute a beneficial strategy to inhibit the development of steatohepatitis [19, 20, 23]. In this study, we evaluated whether GAS5/miR-28a-5p/MARCH7-mediated NLRP3-dependent pyroptosis is involved in the development of NAFLD using both in vivo and in vitro models. HFD-fed mice and leptin-deficient obese (Ob/Ob) mice were used to simulate NAFLD development for in vivo experiments, and AML12 normal mouse hepatocytes and human hepatoblastoma HepG2 cells were selected for in vitro experiments. The results of this study will facilitate the development of novel strategies for the clinical treatment of NAFLD.
Results
GAS5 interacts with miR-28a-5p in the livers of mice
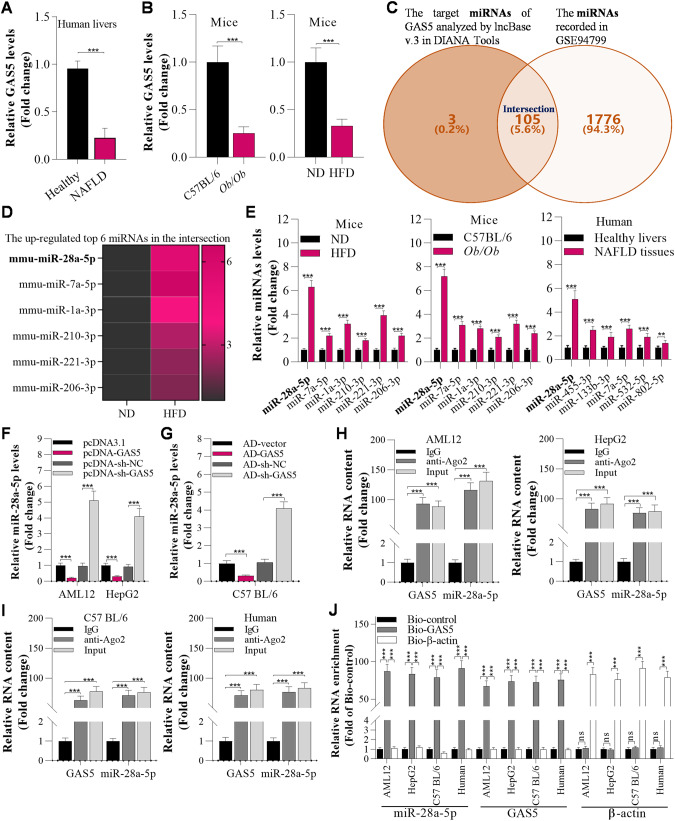
As lncRNAs have crucial roles in NAFLD development, we first downloaded 2 human NAFLD-related microarray datasets, GSE48452 (containing 14 healthy control and 32 NAFLD tissue samples) and GSE107231 (containing 5 normal liver and 5 NAFLD biopsy tissue samples), from Gene Expression Omnibus (GEO) and analysed the downregulated lncRNAs in both datasets. Here, we reconfirmed the top 10 downregulated lncRNAs by RT‒qPCR in our collected clinical specimens (Fig. S1A). GAS5 was significantly downregulated in human NAFLD specimens (Fig. 1A). Furthermore, GAS5 expression was decreased in the livers of NAFLD mice (Fig. S1B). Then, we observed that GAS5 expression was markedly decreased in the livers of HFD and Ob/Ob mice, as evidenced by RT‒qPCR analysis (Fig. 1B). In addition, a Venn diagram was constructed to determine the intersection between the potential target miRNAs of GAS5 (catalogued in lncBase v.3 in DIANA Tools) and the miRNAs in GSE94799. This Venn diagram showed that 105 GAS5-binding miRNAs were upregulated in the livers of HFD mice (Fig. 1C). Among them, miR-28a-5p was the most significantly upregulated miRNA in the livers of HFD mice, Ob/Ob mice and NAFLD patients (Fig. 1D, E). Therefore, we assumed that miR-28a-5p was the target of GAS5. Furthermore, GAS5 suppressed miR-28a-5p expression in AML12 and HepG2 cells and in the livers of C57BL/6 mice (Fig. 1F, G; Fig. S2A–H). Moreover, we observed that GAS5 interacted with miR-28a-5p, as evidenced by the RIP assay and biotinylated GAS5 pull-down assay, in AML12 and HepG2 cells and in the liver tissue of C57BL/6 mice and humans (Fig. 1H, I, J).
Fig. 1. GAS5 interacts with miR-28a-5p in hepatic cell lines and the livers of mice.
A RT‒qPCR analysis of lncRNA GAS5 expression in the liver in healthy humans and humans with NAFLD (n = 16/group). B RT‒qPCR analysis of GAS5 expression in the livers of mice fed a ND or HFD at the12th week (n = 8/group). C The Venn diagram was generated by taking the intersection of the target miRNAs of GAS5 (catalogued in lncBase v.3 in DIANA Tools) and the miRNAs in GSE94799. D Heatmap showing the top 6 upregulated overlapping miRNAs in the livers of HFD-fed mice. E RT‒qPCR analysis of the top 6 miRNAs in the livers of healthy humans and humans with NAFLD (n = 16/group) and in the livers of ND-fed and HFD-fed mice at the 12th week (n = 8/group); #p < 0.05 versus healthy livers or ND-fed mice. F RT‒qPCR analysis of miR-28a-5p levels in AML12 and HepG2 cells 12 h after transfection with pcDNA3.1, pcDNA-GAS5, pcDNA-sh-NC, and pcDNA-sh-GAS5. G RT‒qPCR analysis of miR-28a-5p levels in the livers of mice 7 days after transfection with AD-vector, AD-GAS5 AD-sh-NC, or AD-sh-GAS5. H RT‒qPCR analysis of GAS5 and miR-28a-5p levels in AML12 and HepG2 cell lysates incubated with an anti-Ago2 antibody for RIP. I RT‒qPCR analysis of GAS5 and miR-28a-5p levels in C57BL/6 mouse or human tissue lysates incubated with an anti-Ago2 antibody for RIP assay. J RT‒qPCR analysis of GAS5 and miR-28a-5p enrichment using a biotinylated GAS5 pull-down assay in AML12 cells, HepG2 cells, mouse livers and human livers. The data are presented as the mean ± S.D. of three independent experiments. Two-tailed Student’s t test was used to determine the significance of differences between two groups (A, B, E, F, G), and ANOVA with the Bonferroni post hoc test was conducted for comparisons among more than two groups (H, I, J). A p value of <0.05 was considered statistically significant; and *p < 0.05, **p < 0.01, ***p < 0.001; nsp > 0.05, ns not significant.
Knockdown of miR-28a-5p ameliorates HFD-induced hepatic steatosis, inflammation, and insulin resistance
To examine the role of miR-28a-5p in NAFLD development, we knocked down miR-28a-5p in in vitro models (AML12 and HepG2) (Fig. S2A). Because miR-28a-5p expression was increased in the livers of NAFLD mice as well as in FA-treated hepatocytes, we knocked down miR-28a-5p to highlight the more pronounced changes in our experiments. We observed that interfering with miR-28a-5p expression significantly decreased lipid deposition in FA-treated AML12 and HepG2 cells (Fig. S3A, B). In addition, knockdown of miR-28a-5p inhibited FA-induced inflammation by downregulating proinflammatory factors and upregulating the anti-inflammatory factor IL-10 (Fig. S3C).
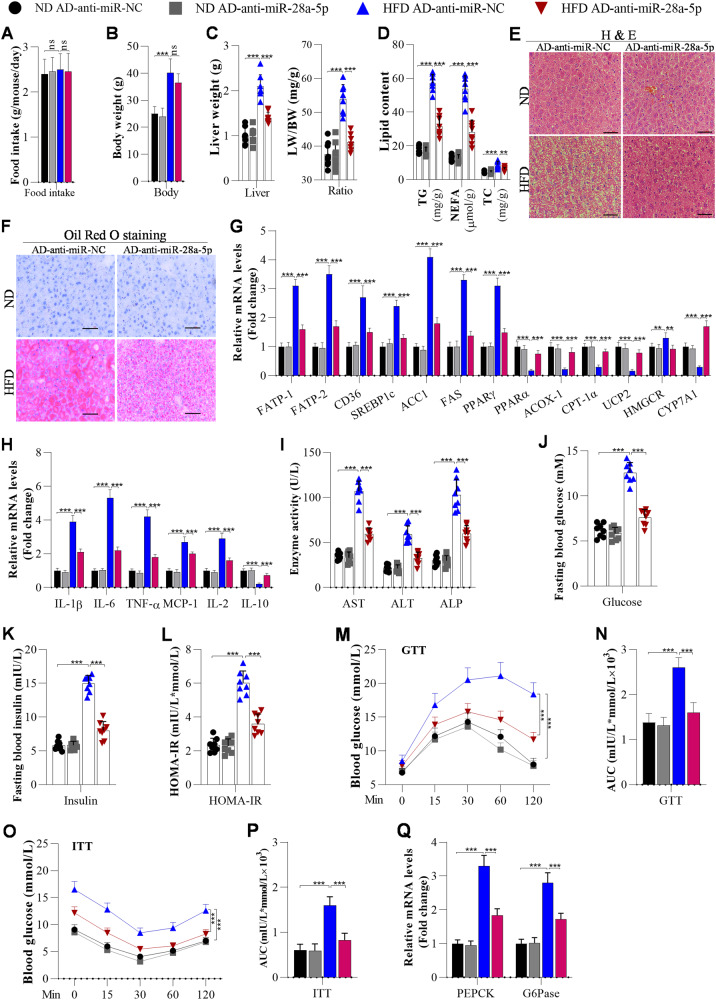
The role of miR-28a-5p in NAFLD development was investigated in mice. We observed no significant differences in food intake and body weight between AD-anti-miR-28a-5p-injected HFD-fed mice and control HFD-fed mice (Fig. 2A, B). However, we observed lower liver weights and liver weight-to-body weight (LW/BW) ratios in miR-28a-5p-knockdown HFD-fed mice than in anti-miR-NC-injected HFD-fed mice (Fig. 2C). The hepatic TG, NEFA, and TC contents were decreased in the livers of miR-28a-5p-knockdown mice compared to AD-anti-miR-NC mice post HFD feeding (Fig. 2D). Accordingly, H&E and Oil Red O staining demonstrated that HFD feeding resulted in less lipid deposition in the livers of miR-28a-5p-knockdown mice than in the livers of the control mice (Fig. 2E, F). Furthermore, we measured the mRNA expression of lipid metabolism genes. The data showed that the expression levels of genes related to fatty acid uptake (FATP1, FABP-1 and CD36) and genes related to fatty acid synthesis (SREBF1, FAS, ACCα and PPAR-γ) were markedly decreased by miR-28a-5p knockdown, whereas the mRNA levels of a cholesterol efflux-related gene (CYP7A1) and fatty acid β-oxidation-related genes (PPARα, ACOX-1, CPT-1α, UCP2) were increased in the miR-28a-5p-knockdown mice compared with the control mice post HFD feeding (Fig. 2G).
Fig. 2. Knockdown of miR-28a-5p attenuates HFD-induced hepatic steatosis, inflammation, and insulin resistance in HFD-fed mice.
Mice were fed a ND or HFD for 12 weeks. Then, they were injected with adenoviruses every 10 days, and these mice were continuously fed a ND or HFD. After the first injection for 3 weeks, the mice were used for further analysis. A Food intake was measured every day during ND or HFD feeding (n = 8/group). B The body weight of mice measured after 3 weeks of the first injections (n = 8/group). C Liver weights and LW/BW ratios of mice were measured at the end of ND or HFD feeding (n = 8/group). D Changes in TG, NEFA, and TC levels were determined by ELISA in the livers of mice after injection with AD-anti-miR-NC or AD-anti-miR-28a-5p for 3 weeks (n = 8/group). E, F Histological images of liver sections from mice injected with AD-anti-miR-NC or AD-anti-miR-28a-5p for 3 weeks (n = 8/group) stained by H&E and with Oil Red O to visualise lipid droplets; Scale bar, 50 µM. G The mRNA levels of lipid metabolism-associated genes were measured by RT‒qPCR analysis (n = 8/group). H The mRNA levels of inflammation-related genes analysed by RT‒qPCR (n = 8/group). I Liver function, manifested by serum AST, ALT, and ALP levels, was evaluated using ELISA in adenovirus-injected mice (n = 8/group). J Fasting blood glucose levels of mice (n = 8/group). K, L Fasting insulin levels and HOMA-IR scores of adenovirus-injected mice (n = 8/group). M–P GTT and ITT of adenovirus-injected mice post ND or HFD feeding, and the areas under the curve (AUCs) for the GTT and ITT (N = 8/group). Q RT‒qPCR analysis of PEPCK and G6Pase expression in the livers of mice (N = 8/group). The data are presented as the mean ± S.D. of three independent experiments. A p value of <0.05 was considered statistically significant; and *p < 0.05, **p < 0.01, ***p < 0.001 assessed via a ANOVA with the Bonferroni post hoc test for comparisons among more than two groups (A–Q); ns not significant.
Previous studies have suggested that excessive lipid accumulation always significantly induces inflammatory responses during NAFLD development. We examined the impacts of miR-28a-5p on HFD-triggered inflammation. After HFD feeding, the hepatic levels of proinflammatory cytokines (such as IL-1β, IL-6, TNF-α, MCP-1, and IL-2) were obviously lower in the miR-28a-5p-knockdown mice than in the control mice, but that of the anti-inflammatory cytokine IL-10 was much higher in the miR-28a-5p-knockdown mice (Fig. 2H). Similarly, the serum levels of liver function indicators (AST, ALT, ALP) were decreased in the miR-28a-5p-knockdown mice compared with the control mice after HFD feeding (Fig. 2I).
In examining the role of miR-28a-5p in hepatic insulin resistance, we observed that miR-28a-5p-knockdown HFD-fed mice exhibited lower fasting blood glucose levels, insulin resistance, and HOMA-IR scores than the AD-anti-miR-NC-injected HFD-fed mice (Fig. 2J–L). Further study showed that miR-28a-5p-knockdown HFD-fed mice exhibited decreased glucose tolerance and insulin sensitivity compared with the AD-anti-miR-NC-injected HFD-fed mice (Fig. 2M–P). In addition, the miR-28a-5p-knockdown HFD-fed mice exhibited lower levels of gluconeogenesis-associated genes (PEPCK and G6Pase) than the control HFD-fed mice (Fig. 2Q).
Overexpression of miR-28a-5p partially abolishes the effects of GAS5 on hepatic steatosis, inflammation, and insulin resistance
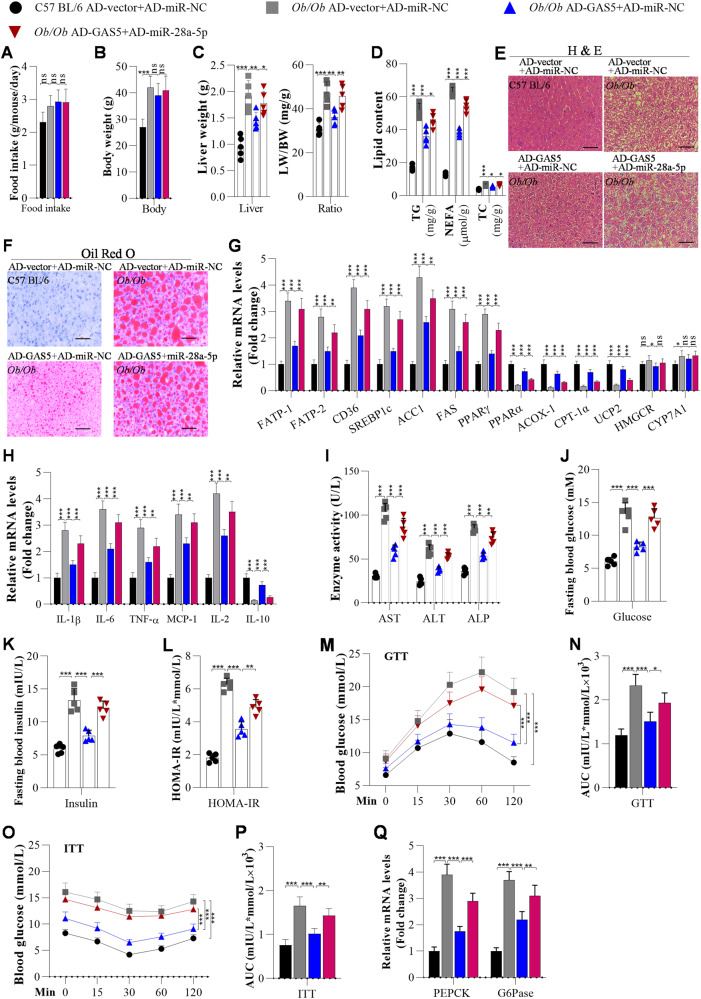
To examine the role of the GAS5/miR-28a-5p interaction in NAFLD pathogenesis, we overexpressed GAS5 and miR-28a-5p in hepatocytes (Fig. S2). The data showed that overexpression of miR-28a-5p obviously reversed the GAS5 overexpression-induced suppression of lipid accumulation and inflammation in AML12 and HepG2 cells (Fig. S3D–F). Moreover, we overexpressed GAS5 and miR-28a-5p in the livers of mice to investigate whether miR-28a-5p can counter the function of GAS5 in NAFLD development manifested by hepatic steatosis, inflammation, and insulin resistance in Ob/Ob mice and HFD-fed mice. We found that overexpression of GAS5 did not significantly affect the food intake or body weight of Ob/Ob mice (Fig. 3A, B). Overexpression of GAS5 obviously decreased the liver weight and LW/BW ratio of the Ob/Ob mice (Fig. 3C). The hepatic TG, NEFA, and TC contents were reduced in GAS5-overexpressing Ob/Ob mice compared with control Ob/Ob mice (Fig. 3D). Furthermore, H&E and Oil Red O staining showed that GAS5 overexpression significantly inhibited lipid deposition in the livers of Ob/Ob mice (Fig. 3E, F), and RT‒qPCR analysis proved that GAS5 overexpression markedly downregulated the mRNA expression of FATP1, FABP-1, CD36, SREBF1, FAS, ACC1 and PPAR-γ but upregulated the mRNA expression of PPARα, ACOX-1, CPT-1α, UCP2, and CYP7A1 in the livers of Ob/Ob mice (Fig. 3G). However, overexpression of miR-28a-5p partially abolished the GAS5-controlled suppression of hepatic steatosis in Ob/Ob mice (Fig. 3A–G).
Fig. 3. Overexpression of miR-28a-5p significantly abolishes GAS5 overexpression-mediated suppression of hepatic steatosis, inflammation, and insulin resistance in Ob/Ob mice.
C57BL/6 and Ob/Ob mice were fed a chow diet. When they were 10 weeks old, they were injected with adenoviruses every 10 days. After 3 weeks of the first injection, the mice were used for further analysis. A Food intake was measured every day during ND or HFD feeding (n = 5/group). B The body weight of mice was measured after 3 weeks of the first injection of adenoviruses (n = 5/group). C Liver weights and LW/BW ratios of C57BL/6 or Ob/Ob mice after 3 weeks of the first injection (n = 5/group). D TG, NEFA, and TC levels were measured using ELISA in the livers of adenovirus-injected C57BL/6 and Ob/Ob mice (n = 5/group). E, F H&E and Oil Red O staining of liver sections from adenovirus-injected C57BL/6 and Ob/Ob mice (n = 5/group); Scale bar, 50 µM. G The mRNA expression of lipid metabolism-related genes in the livers of C57BL/6 or Ob/Ob mice was measured using RT‒qPCR analysis (n = 5/group). H The mRNA levels of inflammation-related genes were analysed by RT‒qPCR (n = 5/group). I Serum AST, ALT, and ALP contents were analysed using ELISA in mice (n = 5/group). J Fasting blood glucose levels of C57BL/6 and Ob/Ob mice (n = 5/group). K, L Fasting insulin levels and HOMA-IR scores of adenovirus-injected C57BL/6 and Ob/Ob mice (n = 5/group). M–P GTT and ITT of C57BL/6 and Ob/Ob mice after 3 weeks of adenovirus injection and the areas under the curve (AUCs) for the GTT and ITT (N = 5/group). Q RT‒qPCR analysis of PEPCK and G6Pase expression in the livers of mice (N = 5/group). The data are presented as the mean ± S.D. of three independent experiments. A p value of <0.05 was considered statistically significant; and *p < 0.05, **p < 0.01, ***p < 0.001 assessed via a ANOVA with the Bonferroni post hoc test for comparisons among more than two groups (A–Q); ns not significant.
The effects of the interaction between GAS5 and miR-28a-5p on hepatic inflammation were determined. Overexpression of GAS5 decreased the mRNA levels of IL-1β, IL-6, TNF-α, MCP-1, and IL-2 and increased that of IL-10 in the livers of Ob/Ob mice, as evidenced by RT‒qPCR (Fig. 3H). However, overexpression of miR-28a-5p increased the levels of IL-1β, IL-6, TNF-α, MCP-1, and IL-2 and reduced the level of IL-10 (Fig. 3H). In addition, overexpression of miR-28a-5p significantly increased the levels of AST, ALT, and ALP in the Ob/Ob mice (Fig. 3I). These results suggested that overexpression of miR-28a-5p abolished the effect of GAS5 on ameliorating inflammation and improving liver function in Ob/Ob mice.
The influence of the binding between GAS5 and miR-28a-5p on insulin resistance was examined. Overexpression of GAS5 reduced the fasting glucose level, fasting insulin level, and HOMA-IR score in Ob/Ob mice (Fig. 3J–L). Overexpression of GAS5 improved glucose tolerance and insulin resistance in Ob/Ob mice (Fig. 3M–P). Then, overexpression of GAS5 was found to significantly reduce the mRNA expression of PEPCK and G6Pase expression in the livers of Ob/Ob mice (Fig. 3Q). Interestingly, the effect of GAS5 on insulin resistance was reversed by overexpression of miR-28a-5p (Fig. 3J–Q), suggesting that overexpression of miR-28a-5p can block the effect of GAS5 on insulin resistance.
We also investigated the role of the GAS5/miR-28a-5p interaction in NAFLD development in HFD-fed mice. We observed similar results: overexpression of miR-28a-5p significantly reversed the GAS5-induced decreases in hepatic lipid deposition (Fig. S4A–D), inflammation (Fig. S4E), fasting blood glucose, and fasting blood insulin (Fig. S4F–H) and improvements in glucose tolerance and insulin resistance in HFD-fed mice (Fig. S4I–L).
In addition, there are two conserved binding sites between GAS5 and miR-28a-5p in humans and mice (Fig. S2I). We found that overexpression of human GAS5 in AML12 cells and mouse livers significantly downregulated miR-28a-5p (Fig. S5A). Then, a biotinylated GAS5 pull-down assay showed that human GAS5 can bind to miR-28a-5p in mouse livers and AML12 cells (Fig. S5B). Functionally, overexpression of human GAS5 in AML12 cells and mouse livers markedly inhibited hepatic lipid deposition and inflammation (Fig. S5C–E). Similarly, overexpression of mouse GAS5 in HepG2 cells also decreased miR-28a-5p levels and reduced hepatic lipid accumulation and inflammation (Fig. S5F–J).
miR-28a-5p promotes NLRP3-associated hepatocyte pyroptosis by directly targeting the MARCH7 3’UTR
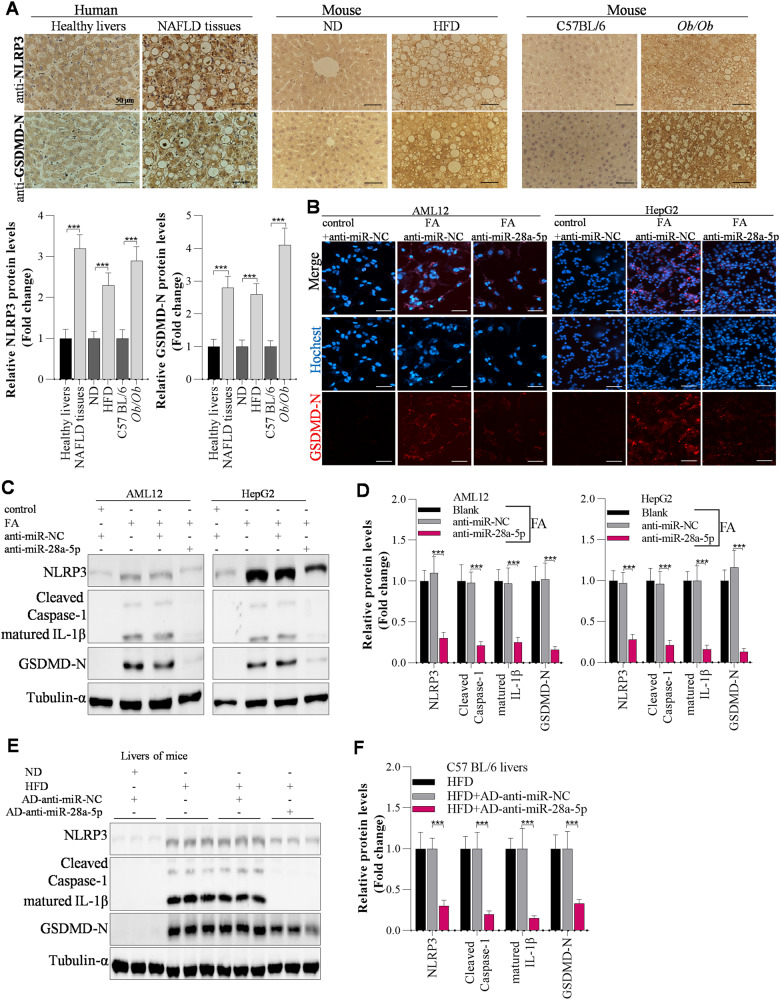
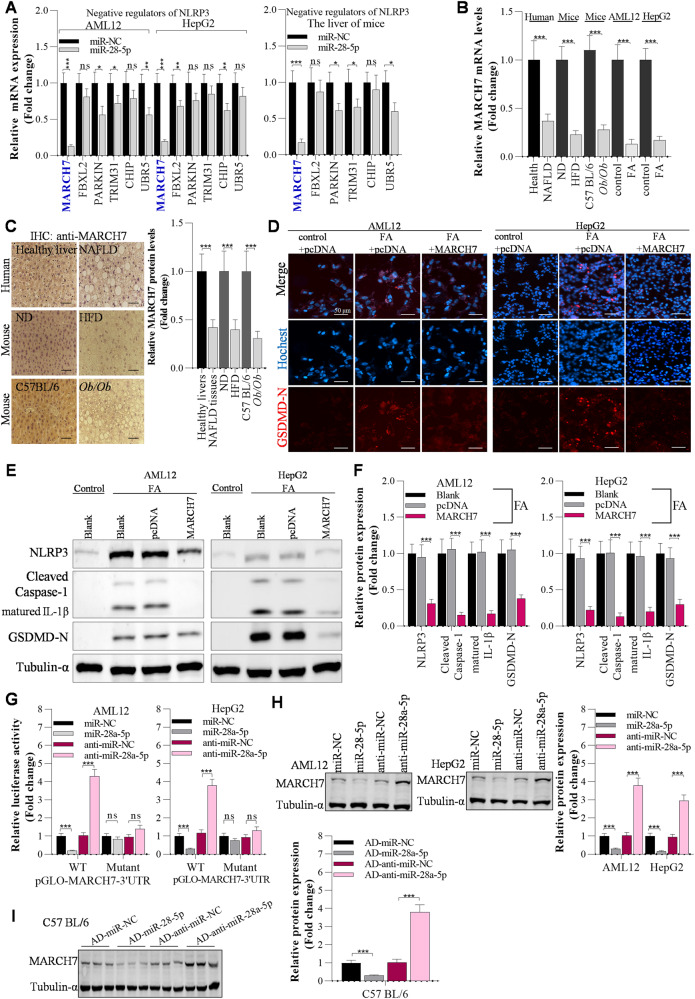
It has been reported that activation of pyroptosis promotes NAFLD development, whereas inhibition of pyroptosis can alleviate NAFLD [24, 25]. To further explore the potential mechanisms of GAS5 and miR-28a-5p in NAFLD pathogenesis, we focused on the targets of miR-28a-5p in hepatocyte pyroptosis. First, we observed that pyroptosis was activated in the livers of NAFLD patients, HFD-fed mice and Ob/Ob mice, as evidenced by IHC analysis of the pyroptosis markers NLRP3 and GSDMD-N (Fig. 4A). Then, we explored the potential mechanisms of GAS5/miR-28a-5p in NAFLD development by investigating the effects of miR-28a-5p on hepatocyte pyroptosis. By immunofluorescence staining, we observed that knockdown of miR-28a-5p markedly inhibited FA-induced upregulation of GSDMD-N protein expression in AML12 and HepG2 cells (Fig. 4B). Using Western blot analysis, we found that FA significantly activated pyroptosis by upregulating the expression of proteins involved in NLRP3/caspase-1/GSDMD signalling in FA-treated AML12 and HepG2 cells; however, knockdown of miR-28a-5p obviously inhibited NLRP3-mediated pyroptosis by downregulating NLRP3 expression (Fig. 4C, D). Accordingly, HFD feeding markedly elevated hepatic pyroptosis in mice, and interfering with miR-28a-5p expression attenuated hepatic pyroptosis in HFD-fed mice by decreasing NLRP3 levels (Fig. 4E, F). Most interestingly, interfering with NLRP3 expression significantly abolished the miR-28a-5p overexpression-induced activation of pyroptosis in AML12 and HepG2 cells (Fig. S6A).
Fig. 4. miR-28a-5p modulates NLRP3 expression to affect pyroptosis during hepatic steatosis.
A IHC analysis of NLRP3 and GSDMD-N protein levels in the livers of humans or mice; Scale bar, 50 µM. B Immunofluorescence staining of the GSDMD-N protein in anti-miR-NC- or anti-miR-28a-5p-transfected AML12 and HepG2 cells after 24 h of FA treatment; Scale bar, 50 µM. C, D Western blot showing pyroptosis-related gene expression in anti-miR-NC- or anti-miR-28a-5p-transfected AML12 and HepG2 cells after 24 h of FA treatment. E, F Western blot showing pyroptosis-related gene expression in the livers of adenovirus-injected mice after ND or HFD feeding. The data are presented as the mean ± S.D. of three independent experiments. A p value of <0.05 was considered statistically significant; and *p < 0.05, **p < 0.01, ***p < 0.001 assessed via a two-tailed t test for examining the significance of differences between two groups (A) or ANOVA with the Bonferroni post hoc test for comparisons among more than two groups (D, F); ns not significant.
Because the NLRP3 protein is the key initiator of pyroptosis, we investigated how miR-28a-5p upregulates the NLRP3 protein to further deeply expose the role of miR-28a-5p in pyroptosis. We found that miR-28a-5p did not markedly affect NLRP3 mRNA levels and it also did not reduce luciferase activity of NLRP3 3′UTR (Fig. S6B, C). However, miR-28a-5p significantly supressed Cycloheximide (CHX)-induced downregulation of NLRP3 protein, suggesting that miR-28a-5p modulates NLRP3 expression mainly at the protein levels (Fig. S6D). Because miRNAs always exert their functions by targeting the 3′UTRs of downstream genes to reduce the expression of these target genes, it is reasonable to propose the hypothesis that miR-28a-5p increases NLRP3 protein levels by targeting the 3′UTRs of its negative regulators, including MARCH7, FBXL2, PARKIN, TRIM31, CHIP, and UBR5, which are reported to be E3 ligases involved in proteasome- or lysosome-mediated degradation of the NLRP3 protein [26]. First, we determined the potential targets of miR-28a-5p among these negative regulators of NLRP3 protein expression. Among the negative regulators, MARCH7 had the most significantly changed expression after overexpression of miR-28a-5p, as evidenced by RT‒qPCR, in AML12 and HepG2 cells as well as in the livers of mice (Fig. 5A), suggesting that MARCH7 is the target of miR-28a-5p. Accordingly, MARCH7 mRNA expression was decreased in FA-treated AML12 and HepG2 cells compared with the control cells, and MARCH7 mRNA expression was reduced in the livers of NAFLD patients, HFD-fed mice and Ob/Ob mice compared with the healthy controls (Fig. 5B). The MARCH7 protein was downregulated in the livers of NAFLD patients, as evidenced by IHC analysis (Fig. 5C). Using immunofluorescence staining, we found that overexpression of MARCH7 significantly suppressed the FA-induced increase in GSDMD-N protein levels in AML12 and HepG2 cells (Fig. 5D). In line with our expectations, overexpression of MARCH7 decreased NLRP3 expression, leading to repression of hepatic pyroptosis (Fig. 5E, F). Furthermore, the luciferase reporter assay showed that overexpression of miR-28a-5p significantly suppressed the luciferase activity of pGLO-MARCH7-3′UTR, while knockdown of miR-28a-5p increased the luciferase activity of pGLO-MARCH7-3′UTR in AML12 and HepG2 cells (Fig. 5G, Fig. S6E, Fig. S2). Moreover, Western blotting showed that overexpression of miR-28a-5p obviously downregulated MARCH7 expression and that interfering with miR-28a-5p expression upregulated MARCH7 expression in both the in vitro and in vivo models (Fig. 5H, I).
Fig. 5. MARCH7 is the direct target of miR-28a-5p that regulates NLRP3 protein expression.
A RT‒qPCR analysis of negative regulators of NLRP3, including MARCH7, FBXL2, PARKIN, TRIM31, CHIP, and UBR5, in AML12 cells, HepG2 cells, and mouse livers. B RT‒qPCR analysis of MARCH7 mRNA expression in the in vivo and in vitro models in the livers of humans (N = 16/group) and mice (N = 8/group). C IHC analysis of MARCH7 protein levels in the livers of humans and mice. Then, AML12 and HepG2 cells were transfected with the overexpression plasmids for 12 h and incubated with FAs for another 12 h; Scale bar, 50 µM. D Immunofluorescence staining of the GSDMD-N protein in pcDNA- or MARCH7-transfected AML12 and HepG2 cells after 24 h of FA treatment; Scale bar, 50 µM. E, F Western blot analysis of pyroptosis-related gene expression. G The effects of miR-28a-5p on the luciferase activity of pGLO-MARCH7-3’UTR were determined by a luciferase reporter assay. H Western blot analysis of MARCH7 expression in AML12 and HepG2 cells after 24 h of transfection. I Western blot analysis of MARCH7 expression in the livers of C57BL/6 mice after 7 days of injection with adenoviruses. The data are presented as the mean ± S.D. of three independent experiments. A p value of <0.05 was considered statistically significant; and *p < 0.05, **p < 0.01, ***p < 0.001 assessed via a two-tailed t-test for examining the significance of differences between two groups (A–C, G–I) or ANOVA with the Bonferroni post hoc test for comparisons among more than two groups (F); ns not significant.
In addition, overexpression of MARCH7 reversed the effects of miR-28a-5p overexpression-activated pyroptosis in AML12 and HepG2 cells (Fig. S7A, B) and in the livers of mice (Fig. S7C, D). As expected, overexpression of miR-28a-5p significantly reversed the effects of GAS5 overexpression on MARCH7-controlled pyroptosis in AML12 and HepG2 cells and the livers of HFD-fed mice (Fig. S7E–H). Thus, we can conclude that miR-28a-5p promotes NLRP3-regulated hepatic pyroptosis by targeting the MARCH7 3’UTR.
Interfering with MARCH7 blocks anti-miR-28a-5p-mediated suppression of hepatic steatosis, inflammation, and insulin resistance
Here, we explored the function of the miR-28a-5p/MARCH7 axis in NAFLD development using in vitro and in vivo models. We observed that overexpression of MARCH7 significantly inhibited lipid accumulation in AML12 and HepG2 cells (Fig. S8A–C, Fig. S2). Consistent with this finding, interfering with MARCH7 expression partially reversed the miR-28a-5p knockdown-induced suppression of lipid accumulation and inflammation in AML12 and HepG2 cells (Fig. S8D–F). Then, Ob/Ob mice and HFD-fed mice were used as the in vivo model.
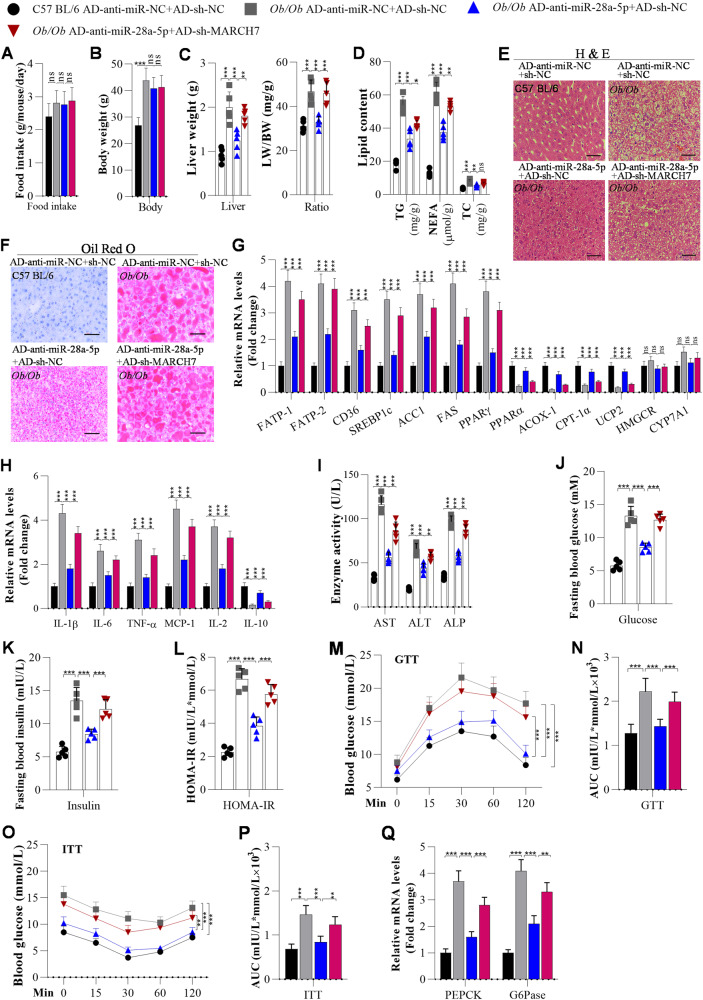
First, we also examined the role of the miR-28a-5p/MARCH7 axis in NAFLD development in Ob/Ob mice. Knockdown of miR-28a-5p did not significantly affect food intake or body weight in Ob/Ob mice (Fig. 6A, B). However, knockdown of miR-28a-5p obviously attenuated hepatic steatosis, inflammation, and insulin resistance in Ob/Ob mice (Fig. 6) as well as in HFD-fed mice (Fig. 2). However, interfering with MARCH7 expression partially abolished the miR-28a-5p knockdown-induced decreases in liver weight and LW/BW ratios in Ob/Ob mice (Fig. 6C). Interfering with MARCH7 expression obviously reversed the miR-28a-5p knockdown-induced inhibition of lipid accumulation, as determined by ELISA, H&E staining, and Oil Red O staining, in the livers of Ob/Ob mice (Fig. 6D–F). It was also found that interfering with MARCH7 expression markedly blocked the regulatory effect of miR-28a-5p knockdown on the expression of lipid metabolism-associated genes, including FATP1, FABP-1, CD36, SREBF1c, ACC1, FAS, PPAR-γ, PPARα, ACOX-1, CPT-1α, UCP2, HMGCR, and CYP7A1, in the livers of Ob/Ob mice, as evidenced by RT‒qPCR analysis (Fig. 6G).
Fig. 6. Interference with MARCH7 expression blocks anti-miR-28a-5p-mediated suppression of hepatic steatosis, inflammation, and insulin resistance in Ob/Ob mice.
Mice were fed a chow diet, and when they were 10 weeks old, they were injected with adenoviruses every 10 days. After the first injection for 3 weeks, the mice were used for further analysis. A Food intake was measured every day during ND or HFD feeding (n = 5/group). B The body weight of mice after 3 weeks of the first injection (n = 5/group). C Liver weights and LW/BW ratios of C57BL/6 or Ob/Ob mice after 3 weeks of the first injection of adenovirus (n = 5/group). D TG, NEFA, and TC levels were measured by ELISA in the livers of adenovirus-injected C57BL/6 and Ob/Ob mice (n = 5/group). E, F H&E and Oil Red O staining of liver sections from adenovirus-injected C57BL/6 and Ob/Ob mice (n = 5/group); Scale bar, 50 µM. G The mRNA levels of lipid metabolism-associated genes in the livers of C57BL/6 and Ob/Ob mice were analysed by RT‒qPCR (n = 5/group). H The mRNA levels of inflammation-related genes were measured by RT‒qPCR (n = 5/group). I Liver function, as indicated by serum AST, ALT, and ALP, was analysed by ELISA in adenovirus-injected C57BL/6 and Ob/Ob mice (n = 5/group). J Fasting blood glucose levels of C57BL/6 and Ob/Ob mice (n = 5/group). K, L Fasting insulin levels and HOMA-IR scores of adenovirus-injected C57BL/6 and Ob/Ob mice (n = 5/group). M–P GTT and ITT of C57BL/6 and Ob/Ob mice after 3 weeks of adenovirus injection and the areas under the curve (AUCs) for the GTT and ITT (N = 5/group). Q RT‒qPCR analysis of PEPCK and G6Pase expression in the livers of mice (N = 5/group). The data are presented as the mean ± S.D. of three independent experiments. A p value of < 0.05 was considered statistically significant; and *p < 0.05, **p < 0.01, ***p < 0.001 assessed via a ANOVA with the Bonferroni post hoc test for comparisons among more than two groups (A–D, G–Q); ns not significant.
Second, we investigated the role of the miR-28a-5p/MARCH7 axis in hepatic inflammation in Ob/Ob mice. RT‒qPCR analysis showed that interfering with MARCH7 expression significantly blocked the miR-28a-5p-knockdown-mediated upregulation of IL-10 and downregulation of IL-1β, IL-6, TNF-α, MCP-1, and IL-2 in the livers of Ob/Ob mice (Fig. 6H). ELISA showed that interfering with MARCH7 expression reversed the miR-28a-5p knockdown-induced decrease in ALT, AST, and ALP in the serum of Ob/Ob mice (Fig. 6I).
Third, the role of the miR-28a-5p/MARCH7 axis in insulin resistance in the livers of Ob/Ob mice was examined. We observed that interfering with MARCH7 expression markedly abolished the miR-28a-5p-knockdown-induced improvements in fasting glucose, fasting insulin, and HOMA-IR scores in Ob/Ob mice (Fig. 6J–L). Interestingly, interfering with MARCH7 expression significantly reversed the miR-28a-5p knockdown-induced improvements in glucose tolerance and insulin resistance in Ob/Ob mice (Fig. 6M–P). In addition, interfering with MARCH7 expression abolished the effects of miR-28a-5p on glucose metabolism by modulating the expression of PECK and G6Pase in the livers of Ob/Ob mice (Fig. 6Q).
As expected, we also observed similar effects in HFD-fed mice: knockdown of MARCH7 significantly attenuated the anti-miR-28a-5p-mediated improvements in hepatic lipid deposition (Fig. S9A–D), inflammation (Fig. S9E), fasting blood glucose, fasting blood insulin (Fig. S9F–H), glucose tolerance and insulin resistance in HFD-fed mice (Fig. S9I–L).
The E3 ligase MARCH7 serves as a negative regulator of NLRP3 protein expression
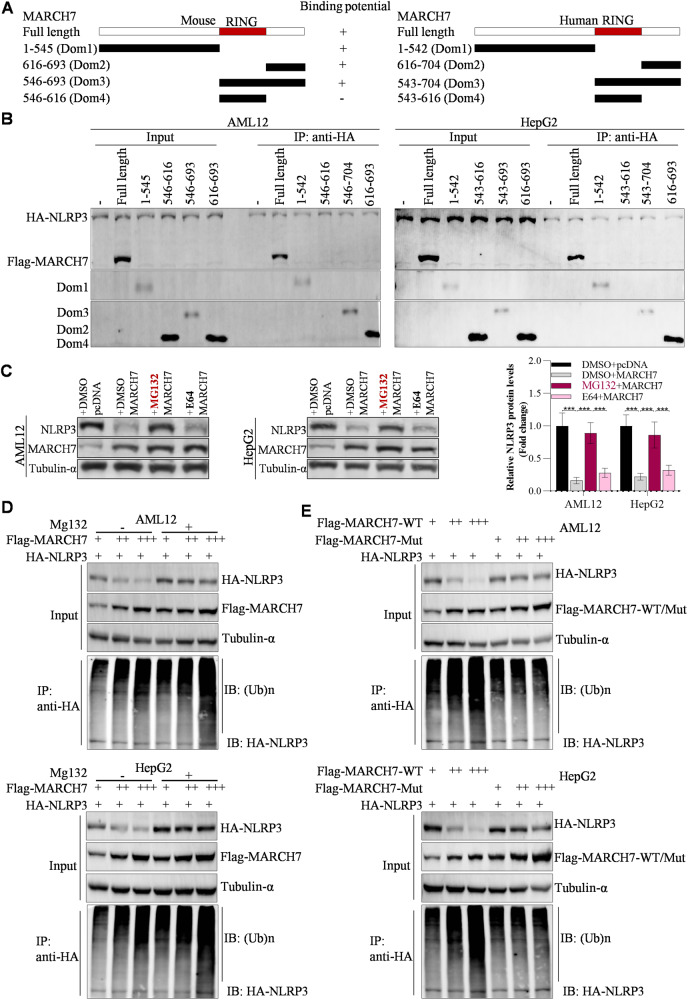
From the above findings in this study, we concluded that miR-28a-5p increased NLRP3 expression by targeting the MARCH7 3′UTR. We also found that miR-28a-5p could not significantly increase NLRP3 protein levels in MARCH7-knockdown hepatocytes compared with normal hepatocytes (Fig. S10A) and that miR-28a-5p could not obviously increase hepatic lipid deposition and inflammation in MARCH7-knockdown hepatocytes compared to normal hepatocytes (Fig. S10B, C). Overexpression of MARCH7 obviously blocked the effects of miR-28-5p on both the NLRP3 protein level and NLRP3 ubiquitination (Fig. S10D). Accordingly, these findings indicate that MARCH7 is required for miR-28a-5p functions in the liver. By conducting a GST pull-down assay, we observed that the purified GST-MARCH7 protein interacted not only with HA-NLRP3 but also with Flag-GSDMD-N and Myc-caspase-1 (Fig. S10E). In addition, GST-MARCH7 decreased HA-NLRP3 levels by inducing ubiquitination-mediated degradation of HA-NLRP3, whereas GSR-MARCH7 did not decrease the Flag-GSDMD-N and Myc-caspase-1 levels (Fig. S10F, G), suggesting that MARCH7 could serve as the E3 ligase for NLRP3 in the liver. Although the E3 ligase MARCH7 interacts with NLRP3 to catalyse NLRP3 ubiquitination in neuroinflammation [26], the details of this interaction remain unclear in liver diseases. To identify the detailed regions of MARCH7 that are responsible for its interaction with NLRP3, we constructed a panel of MARCH7 deletion mutants (Fig. 7A). By conducting an IP assay, we observed that full-length MARCH7, Dom1, Dom2, and Dom3 bound strongly to NLRP3 in AML12 and HepG2 cells, while the RING domain (Dom4) did not bind to NLRP3 (Fig. 7B), suggesting that both the N- and C- regions (Dom1 and Dom2) of MARCH7 are required for its interaction with NLRP3. Then, we found that the proteasome inhibitor MG132 but not the lysosome inhibitor E64 significantly suppressed the MARCH7 overexpression-induced downregulation of the NLRP3 protein in AML12 and HepG2 cells (Fig. 7C), indicating that MARCH7 decreases NLRP3 expression through the proteasome pathway. Furthermore, when MARCH7 was coexpressed with NLRP3 in AML12 and HepG2 cells, the levels of NLRP3 were decreased by MARCH7 in a dose-dependent manner, while MARCH7 did not reduce NLRP3 levels in the presence of the proteasome pathway inhibitor MG132 (Fig. 7D). Unlike wild-type MARCH7, the ubiquitin E3 ligase-inactivating mutation (W589A/I556A) in the RING domain of MARCH7 failed to result in downregulation of NLRP3 expression (Fig. 7E), indicating that the E3 ligase activity of MARCH7 is the essential component that regulates NLRP3 expression.
Fig. 7. MARCH7 serves as an E3 ligase of NLRP3.
A Mapping of interaction regions between MARCH7 and NLRP3. B IP analysis of interaction regions between MARCH7 and NLRP3. C Western blot analysis of NLRP3 expression after overexpression of MARCH7 and another 6 h of incubation with MG132 and E64. D Western blot analysis of NLRP3 expression and detection of NLRP3 ubiquitination by IP. AML12 and HepG2 cell lines were transfected with HA-NLRP3 alone or together with increasing amounts of Flag-MARCH7. Twenty-four hours later, these cells were treated with or without 20 μM MG132 for another 12 h. Then, the cells were collected for IP with an anti-HA antibody to detect NLRP3 ubiquitination. E The cell lines were transfected with HA-NLRP3 alone or together with the indicated Flag-MARCH7 constructs; WT wild type, mut mutant (W589A/I556A). Thirty-six hours later, these cells were prepared for detection of NLRP3 ubiquitination using an IP assay. The data are presented as the mean ± S.D. of three independent experiments. A p value of < 0.05 was considered statistically significant; and *p < 0.05, **p < 0.01, ***p < 0.001 assessed via a ANOVA with the Bonferroni post hoc test for comparisons among more than two groups (C); ns not significant.
The ligase-inactive mutant of MARCH7 fails to inhibit hepatic steatosis, inflammation, and insulin resistance
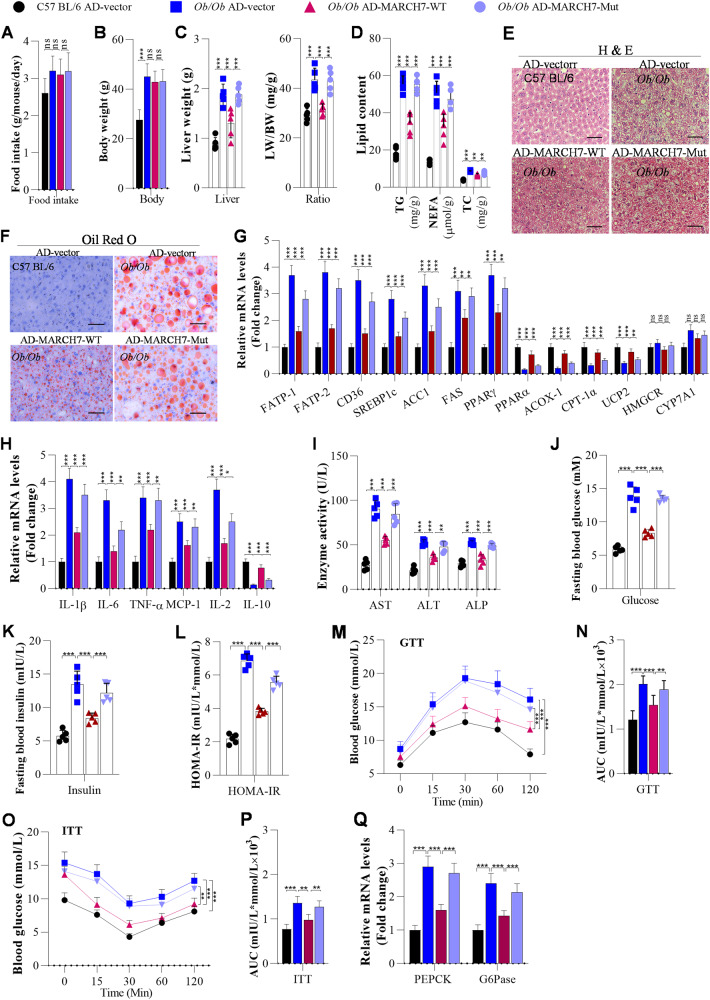
Although the ubiquitin E3 ligase-inactive mutant (W589A/I556A) of MARCH7 retained NLRP3-binding ability, the mutant failed to inhibit NLRP3 expression. To verify the function of the E3 ligase-inactive mutant of MARCH7 in NAFLD development, we overexpressed wild-type or mutant MARCH7 in the livers of Ob/Ob mice. We observed that overexpression of MARCH7 did not markedly affect the food intake or body weight of Ob/Ob mice (Fig. 8A, B). However, overexpression of MARCH7 significantly reduced the liver weight and LW/BW ratio in Ob/Ob mice (Fig. 8C). Overexpression of MARCH7 obviously suppressed lipid accumulation in the livers of Ob/Ob mice by decreasing the expression of lipid metabolism-associated genes, including FATP1, FABP-1, CD36, SREBF1c, ACC1, FAS, and PPAR-γ, in the livers of Ob/Ob mice, and increasing the expression of PPARα, ACOX-1, CPT-1α, UCP2, HMGCR, and CYP7A1 (Fig. 8D–G). We also observed that overexpression of MARCH7 significantly attenuated hepatic inflammation by downregulating the expression of IL-1β, IL-6, TNF-α, MCP-1, and IL-2 and upregulating IL-10 expression in the livers of Ob/Ob mice (Fig. 8H). Similarly, overexpression of MARCH7 markedly reduced the serum levels of ALT, AST, and ALP in Ob/Ob mice (Fig. 8I). Furthermore, MARCH7 overexpression obviously decreased fasting glucose, fasting insulin, and HOMA-IR scores in Ob/Ob mice (Fig. 8J–L), and MARCH7 overexpression significantly improved glucose tolerance and insulin resistance in the Ob/Ob mice (Fig. 8M–P). In addition, MARCH7 affected glucose metabolism by inhibiting the expression of PECK and G6Pase in the livers of Ob/Ob mice (Fig. 8Q). Similarly, the results in HFD-fed mice demonstrated that the MARCH7 mutant obviously failed to improve hepatic lipid deposition (Fig. S11A–D), inflammation (Fig. S11E), fasting blood glucose, fasting blood insulin (Fig. S11F–H), glucose tolerance and insulin resistance (Fig. S11I–L).
Fig. 8. The ubiquitin E3 ligase-inactive mutant of MARCH7 fails to suppress hepatic steatosis, inflammation, and insulin resistance in Ob/Ob mice.
Ten-week-old C57BL/6 and Ob/Ob mice were injected with adenoviruses once a week. After the first injection for 3 weeks, these mice were used for further experiments. A The body weight of mice measured after 3 weeks of injection of adenoviruses (n = 5/group). B Food intake was measured every day in C57BL/6 or Ob/Ob mice (n = 5/group). C Liver weights and LW/BW ratios of C57BL/6 and Ob/Ob mice after 3 weeks of adenovirus injection (n = 5/group). D The contents of TG, NEFA, and TC were measured using ELISA in the livers of adenovirus-injected C57BL/6 and Ob/Ob mice (n = 5/group). E, F H&E and Oil Red O staining of liver sections from adenovirus-injected C57BL/6 and Ob/Ob mice (n = 5/group); Scale bar, 50 µM. G The mRNA expression of lipid metabolism-related genes in the livers of C57BL/6 and Ob/Ob mice was measured using RT‒qPCR analysis (n = 5/group). H The mRNA levels of inflammation-related genes were analysed by RT‒qPCR (n = 5/group). I Serum AST, ALT, and ALP contents were measured using ELISA in mice (n = 5/group). J Fasting blood glucose levels of C57BL/6 and Ob/Ob mice (n = 5/group). K, L Fasting insulin levels and HOMA-IR scores of adenovirus-injected C57BL/6 and Ob/Ob mice (n = 5/group). M–P GTT and ITT of C57BL/6 and Ob/Ob mice after 3 weeks of adenovirus injection and the areas under the curve (AUCs) for the GTT and ITT (N = 5/group). Q RT‒qPCR analysis of PEPCK and G6Pase expression in the livers of mice (N = 5/group). The data are presented as the mean ± S.D. of three independent experiments. One-way analysis of variance (ANOVA) with the Bonferroni post hoc test was conducted for comparisons among more than two groups (A–D, G–Q). A p value of < 0.05 was considered statistically significant; and *p < 0.05, **p < 0.01, ***p < 0.001; nsp > 0.05, ns not significant.
Interestingly, to investigate the potential clinical significance of the GAS5/miR-28a-5p/MARCH7 signalling pathway in the clinical treatment of NAFLD, we treated C57BL/6 mice and hepatocytes (AML12 and HepG2) with metformin because metformin has been considered a potential drug for NAFLD treatment [27–29], and it was listed in our NAFLD drug screening database (data not shown). We observed that metformin significantly decreased lipid accumulation and inflammation in the livers of HFD-fed mice as well as in FA-treated hepatocytes (Fig. S12A–C, Fig. S12D–F). Furthermore, metformin increased the levels of GAS5 and MARCH7 but decreased the levels of miR-28a-5p and NLRP3 in the livers of HFD-fed mice and in FA-treated hepatocytes (Fig. S12G, H). Moreover, metformin obviously restored the expression of the MARCH7 protein to attenuate NLRP3-mediated pyroptosis in the livers of HFD-fed mice and in FA-treated hepatocytes (Fig. S12I, J). In addition, knockdown of GAS5 significantly inhibited the effects of metformin on miR-28a-5p expression and pyroptosis (Fig. S13A–D). Subsequently, knockdown of GAS5 significantly blocked the inhibitory effect of metformin on hepatic lipid deposition (Fig. S13E, F). These findings implied that GAS5, miR-28a-5p, MARCH7, and NLRP3 might be potential targets for developing drugs to ameliorate clinical NAFLD.
Discussion
NAFLD has become a serious threat to human health worldwide [3]. No effective drugs have been found for NAFLD treatment, whereas a daily diet can be recommended to protect against NAFLD [27]. NAFLD pathogenesis involves a complex interplay of multiple processes, including hepatic steatosis, inflammatory responses, and insulin resistance [1, 2]. Thus, it is an urgent need to expose the molecular mechanisms of NAFLD pathogenesis and to identify effective drugs for NAFLD treatment. Although pyroptosis has been considered a promoting factor of NAFLD development [24, 25], the regulatory mechanisms of pyroptosis in NAFLD development remain unclear. In this study, we identified the role of the GAS5/miR-28a-5p/MARCH7/NLRP3 axis in pyroptosis and facilitated the understanding of the mechanisms underlying NAFLD development and strategies to develop novel clinical therapies for NAFLD. By using in vivo and in vitro models, we found that GAS5 significantly suppressed NAFLD development by binding to miR-28a-5p, that miR-28a-5p inhibited NAFLD development by targeting MARCH7, and that MARCH7 inhibited NAFLD development by inducing proteasomal degradation of NLRP3, resulting in suppression of hepatic steatosis, whereas the ubiquitin E3 ligase-inactive mutant of MARCH7 failed to inhibit NAFLD development due to its failure to induce the ubiquitin‒proteasome pathway-mediated degradation of the NLRP3 protein (the key initiator of pyroptosis). Overexpression of miR-28a-5p partially blocked GAS5 overexpression-induced inhibition of NAFLD development, and MARCH7 knockdown significantly abolished the miR-28a-5p knockdown-mediated effects on NAFLD development. Moreover, we observed that metformin obviously upregulated the expression of GAS5 and MARCH7 but downregulated the expression of miR-28a-5p and suppressed pyroptosis in NAFLD development. Consequently, GAS5/miR-28a-5p/MARCH7/NLRP3 signalling-mediated pyroptosis might be the underlying mechanism of NAFLD pathogenesis. GAS5, miR-28a-5p, MARCH7, NLRP3, and pyroptosis might be potential biomarkers for NAFLD diagnosis and treatment.
LncRNAs play a vital role in NAFLD development [30, 31]. Although GAS5 was reported to be upregulated in NAFLD and inhibit NAFLD progression [7, 8], GAS5 plays complex roles in NAFLD, as determined by analysis of online microarray datasets (GSE48452 and GSE107231) and our own microarray data of hepatic lncRNAs. GAS5 also attenuates liver fibrogenesis [9, 10, 32]. As expected, we found that overexpression of GAS5 obviously suppressed HFD-induced hepatic steatosis, insulin resistance and inflammation, indicating that GAS5 can serve as an anti-NAFLD regulator.
Here, a Venn diagram was generated to screen for the potential miRNAs binding to GAS5 to determine the molecular mechanisms of GAS5 in NAFLD development. Among these overlapping miRNAs identified by the Venn diagram, miR-28a-5p had the most significantly increased expression in the livers of HFD-fed mice, Ob/Ob mice and NAFLD patients, suggesting that GAS5 may inhibit NAFLD development by sponging miR-28a-5p. miR-28a-5p may enhance liver fibrosis in mice [12], implying that miR-28a-5p may act as a pro-NAFLD regulator. Interestingly, we found that knockdown of miR-28a-5p significantly alleviated hepatic steatosis and improved insulin resistance in HFD-fed mice. Thus, we identified miR-28a-5p as the target miRNA of GAS5, although our preliminary experiments showed that GAS5 may inhibit lipid deposition by targeting miR-21/PTEN (data not shown). Accordingly, GAS5 may interact with miR-28a-5p, and overexpression of miR-28a-5p may obviously abolish the GAS5 overexpression-induced amelioration of NAFLD. More importantly, due to the conserved binding sites between GAS5 and miR-28a-5p between mice and humans, the biological functions of GAS5/miR-28a-5p in hepatic lipid deposition and inflammation are also conserved between mice and humans. Subsequently, our findings proved that GAS5 exerted its anti-NAFLD functions by sponging miR-28a-5p. Considering these results collectively, it is reasonable to conclude that increasing GAS5 expression and decreasing miR-28a-5p expression could constitute novel treatment strategies for alleviating clinical NAFLD.
To reveal the potential mechanisms of GAS5 and miR-28a-5p in NAFLD pathogenesis, we focused our attention on NLRP3-mediated hepatic pyroptosis, which is activated in NAFLD [24, 25]. The NLRP3 inflammasome can trigger pyroptosis by activating caspase-1, which cleaves GSDMD into GSDMD-N (an executor of pyroptosis), resulting in pyroptosis [19, 20, 24, 25]. Abnormal pyroptosis may facilitate lipogenesis of hepatocytes and inflammation to promote NAFLD development by affecting the levels of lipogenesis-related genes and inflammation factors [20, 22]. Thus, knockout of NLRP3 or GSDMD-N could counter steatohepatitis [19, 20, 23]. Our data showed that miR-28a-5p markedly elevated pyroptosis by upregulating NLRP3 protein expression but NLRP3 mRNA levels, implying that miR-28a-5p modulates NLRP3 expression at the protein level but not at the mRNA level. Because the protein level is often modulated via negative regulators, including the ubiquitin‒proteasome pathway or lysosomal pathway [26], and miRNAs always exert their functions by targeting the 3′UTRs of downstream genes to reduce target gene expression, we assumed that miR-28a-5p elevates NLRP3 protein expression by binding to the 3′UTRs of the negative regulators of NLRP3 protein expression, including the E3 ligases MARCH7, FBXL2, PARKIN, TRIM31, CHIP, and UBR5. Among these proteins, MARCH7 had the most significantly change in expression after miR-28a-5p overexpression in AML12 cells, HepG2 cells, and the livers of mice; overexpression of MARCH7 significantly abolished the effects of miR-28a-5p on both the protein levels and ubiquitination of NLRP3; and knockdown of MARCH7 obviously reversed the miR-28a-5p knockdown-induced attenuation of HFD-induced NAFLD. These findings suggest that MARCH7 is required for the functions of miR-28a-5p in NAFLD development. Thus, we concluded that miR-28a-5p promotes NAFLD development by targeting the MARCH7 3’UTR to suppress degradation of the NLRP3 protein, leading to enhancement of NLRP3-mediated pyroptosis. In addition, miR-28a-5p and GAS5 may reciprocally inhibit each other’s effects on MARCH7 expression and MARCH7-controlled hepatic pyroptosis. These findings explicitly exhibit the potential clinical significance of overexpression of MARCH7 in the development of novel therapeutic strategies for improving clinical NAFLD by reversing the abnormal hepatic lipid deposition, insulin resistance, and inflammation induced by upregulation of miR-28a-5p or downregulation of GAS5.
In this study, MARCH7 inhibited pyroptosis by interacting with NLRP3 but not GSDMD-N and cleaved caspase-1, resulting in NAFLD inhibition. MARCH7 induces ubiquitination of NLRP3 for degradation, leading to suppression of pyroptosis in neuroinflammation [15, 26]. However, the details of the interaction between MARCH7 and NLRP3 in NAFLD remain unclear, and the function of MARCH7 in NAFLD remains completely unknown. In our study, MG132 obviously inhibited MARCH7-mediated degradation of the NLRP3 protein, suggesting that MARCH7 mediates NLRP3 protein degradation via the proteasome pathway in NAFLD development. In contrast, a previous study showed that MARCH7 promotes NLRP3 degradation via the lysosome pathway in neuroinflammation [18, 33], indicating that the mechanisms of MARCH7 vary in different diseases. Then, the N- and C- regions (Dom1 and Dom2) but not the RING domain (Dom4) of MARCH7 were found to be required for its interaction with NLRP3, whereas the E3 ligase-inactive mutant of MARCH7 (harbouring mutations in the RING domain) could not induce the degradation of NLRP3, implying that the E3 ligase activity of MARCH7 is necessary for its ability to degrade the NLRP3 protein, although this mutant retained its NLRP3-binding ability. Accordingly, overexpression of MARCH7 significantly inhibited NAFLD development, whereas the E3 ligase-inactive mutant of MARCH7 failed to inhibit NAFLD development due to its failure to mark the NLRP3 protein for degradation, suggesting that the E3 ligase activity of MARCH7 is required for its function as an anti-NAFLD molecule. Therefore, GAS5 overexpression eventually suppresses NAFLD development by sponging miR-28a-5p to downregulate MARCH7 expression, resulting in the suppression of NLRP3-mediated hepatic pyroptosis. In summary, improving the abnormal expression of MARCH7, miR-28a-5p, and GAS5 could constitute novel strategies for the treatment of NAFLD development by inhibiting pyroptosis.
Our data demonstrate that GAS5 and its downstream molecules play critical roles in NAFLD, and we screened a chemical library to identify agonists or inducers of GAS5 (data not shown here). In FA-treated hepatocytes, metformin was found to significantly inhibit hepatic pyroptosis. In addition, it increased the levels of GAS5 and MARCH7 but decreased the levels of miR-28a-5p, NLRP3, GSDMD-N and cleaved caspase-1 in the livers of NAFLD mice. Consistent with our findings, metformin was found to induce GAS5 expression in breast cancer cells [34]. Dagan Zhang, Xia He et al. reported that metformin can induce hepatocarcinoma cell apoptosis and pyroptosis [35]. Combining previous reports and our findings, we suggest that metformin is a promising drug for NAFLD via regulation of GAS5-mediated cell death. In conclusion, we identified GAS5 as a suppressor of NAFLD. This lncRNA can interact with miR-28a-5p, which is a pro-NAFLD miRNA. Moreover, miR-28a-5p targets the MARCH7 3’UTR, and MARCH7 exerts its anti-NAFLD activity by inducing proteasomal degradation of NLRP3 to inhibit hepatic pyroptosis. In addition, metformin affects the expression of GAS5, miR-28a-5p, MARCH7, NLRP3, and pyroptosis-related factors. We can conclude that GAS5 functions in NAFLD development by modulating miR-28a-5p/MARCH7/NLRP3-associated hepatic pyroptosis. These findings suggest that GAS5, miR-28a-5p, MARCH7, and NLRP3 might be biomarkers for NAFLD intervention.
Limitations of this study
Notably, several of our findings need to be discussed. First, although the results of our cell experiments basically confirmed the results of the animal studies, it is more appropriate to prepare organ-on-chip (OOC), precision-cut liver slice (PCLS), and organoid models as in vitro models, because both AML12 and HepG2 cells are different from the fully differentiated hepatocytes in in vivo models. Pyroptosis signalling in this study was active in the livers of both humans and mice, even though the sequence homology of GAS5 between humans and mice is low, whereas miR-28a-5p, MARCH7 and NLRP3 are highly conserved. Third, because only several negative regulators of the NLRP3 protein have been reported, it appears to be a limitation to regard MARCH7 as the only target of miR-28a-5p that promotes hepatic pyroptosis. Fourth, although 12 weeks of HFD feeding has been used for NAFLD investigation in mice previously [36–38], we consider that our findings seem to be more appropriate for interpreting the early phases of NAFLD development. Fifth, although we evaluated the nonalcoholic fatty liver disease activity scores in Table 1, we did not clarify the role of GAS5/miR-28a-5p/MARCH7/NLRP3 signalling in liver fibrosis. Finally, further assays are needed to elucidate the potential application of GAS5/miR-28a-5p/MARCH7/NLRP3 signalling for the diagnosis and treatment of clinical patients with NAFLD.
Table 1.
Nonalcoholic fatty liver disease activity scores.
| Liver samples | Histologic feature | Fibrosis grade | |
|---|---|---|---|
| Human | Healthy liver | 0 | 0 |
| NAFLD liver | 6 ± 1 | 3 ± 1 | |
| Mouse | ND AD-anti-miR-NC | 0 | 0 |
| ND AD-anti-miR-28a-5p | 0 | 0 | |
| HFD AD-anti-miR-NC | 5 ± 1 | 2 ± 1 | |
| HFD AD-anti-miR-28a-5p | 2 ± 1 | 1 ± 1 | |
| C57 BL/6 AD-vector + AD-miR-NC | 0 | 0 | |
| Ob/Ob AD-vector + AD-miR-NC | 6 ± 1 | 2 ± 1 | |
| Ob/Ob AD-GAS5 + AD-miR-NC | 2 ± 1 | 1 ± 1 | |
| Ob/Ob AD-GAS5 + AD-miR-28a-5p | 5 ± 1 | 2 ± 1 | |
| C57 BL/6 AD-anti-miR-NC + AD-sh-NC | 0 | 0 | |
| Ob/Ob AD-anti-miR-NC + AD-sh-NC | 6 ± 1 | 2 ± 1 | |
| Ob/Ob AD-sh-miR-28a-5p + AD-miR-NC | 2 ± 1 | 1 ± 1 | |
| Ob/Ob AD-sh-miR-28a-5p + AD-sh-MARCH7 | 4 ± 1 | 2 ± 1 | |
| C57 BL/6 AD-vector | 0 | 0 | |
| Ob/Ob AD-vector | 6 ± 1 | 2 ± 1 | |
| Ob/Ob AD-MARCH7-WT | 2 ± 1 | 1 ± 1 | |
| Ob/Ob AD-MARCH7-Mut | 5 ± 1 | 2 ± 1 |
Materials and methods
Cells and clinical specimens
HepG2 and AML12 cells were purchased from the Cell Bank of Type Culture Collection, Chinese Academy of Sciences with STR profiling report (Shanghai, China). HepG2 cells were maintained in DMEM (Thermo Fisher Scientific, Massachusetts, USA) containing 10% FBS (#SH30084.03, HyClone, Logan, Utah, USA), 100 IU/mL penicillin, and 100 μg/mL streptomycin (#P1400, Solarbio, Beijing, China) in a 37 °C incubator with 5% CO2. AML12 cells were maintained in DMEM/F12 (#11320033, Thermo Fisher Scientific) containing FBS, penicillin, and streptomycin as described above, and the medium was also supplemented with ITS (#ITSS-10201-10, Cyagen, Guangzhou, Guangdong, China). Then, the cells were treated with a mixture of oleic acid (OA) and palmitic acid (PA) (Sigma‒Aldrich, St. Louis, MO, USA) for 24 h to induce in vitro lipogenesis to establish the in vitro model of NAFLD. The mixture was prepared with OA and PA at an equal molar ratio as described previously [33]. In addition, to determine the effects of metformin on NAFLD development in the in vitro models, the cell lines were incubated with FAs for 6 h, and they were then incubated with both 10 nM metformin and FA for another 6 h. All the cells are checked for mycoplasma contamination by polymerase chain reaction (PCR) reaction every 3 months.
Obese adults who underwent bariatric surgery in the Department of Gastroenterology, Second Affiliated Hospital of Chongqing Medical University between February 2020 and October 2020 were enrolled. Clinical specimen collection was approved by the Medical Ethics Committee of the Second Affiliated Hospital of Chongqing Medical University (Chongqing, China), and all participants provided written informed consent. Standard-of-care wedge liver biopsies were performed intraoperatively, and these biopsies were evaluated in a blinded manner by three independent pathologists (Dr Zhihang Zhou, Dr Lingfeng Wan and Ms. Xueru Song) for the presence of NAFLD. In total, 16 normal liver specimens were defined as those with <5% of hepatocytes exhibiting macrovesicular steatosis and 16 NAFLD specimens as those with >5% of hepatocytes exhibiting macrovesicular steatosis, as previously reported, and all the nonalcoholic fatty liver disease activity scores are shown in Table 1 according to previous review [39].
Plasmids and adenoviruses
First, the following sequences were constructed: miR-NC, 5′-UUC UCC GAA CGU GUC ACG UUU-3′; miR-28a-5p mimic, 5′-AAG GAG CUCA CAG UCU AUU GAG-3′; anti-miR-NC, 5′-AAG UGG CUA CGU CUG AGC UUU-3′; and anti-miR-28a-5p, 5′-GAG UUA UCU GAC ACU CGA GGA AGA GUU AUC UGA CAC UCG AGG AAG AGU UAU CUG ACA CUC GAG GAA-3′. The sequences of human GAS5 (ENST00000651080.1) and mouse GAS5 (ENSMUST00000161005.7) were obtained from the UCSC database (http://genome.ucsc.edu/index.html). The sequences of human MARCH7 (NM_001282805.2) and mouse MARCH7 (NM_020575.2) were obtained from the NCBI Reference Sequence database (https://www.ncbi.nlm.nih.gov/), and the sequences used for knockdown of MARCH7 were 5′-GGC UCA AGG UGG AAG AAA U-3′ and 5′-GCG UCA UCA UCU AAU UUA U-3′. Second, plasmids for overexpressing or knocking down GAS5 and MARCH7 were constructed in pcDNA3.1 (+) in accordance with the above sequences by Sangon (Shanghai, China). The mouse and human ubiquitin E3 ligase-inactive mutant (W589A/I556A or W587A/I554A) of MARCH7 are denoted MARCH7-Mut according to a previous study [40]. The adenoviruses for overexpressing or knocking down GAS5, miR-28a-5p, and MARCH7 were generated according to the manufacturer’s instructions in the pAd-Easy system (#240009, Invitrogen, California, USA), and they were purified by Sangon. These viruses were diluted in PBS and injected into the mice through the tail vein at a dose of 3*1010 plaque-forming units (PFU) per mouse. Third, as shown in Supplementary Fig. 2 and Fig. S6, the wild-type and mutant MARCH7 3′UTRs were cloned into the pmir-GLO control luciferase reporter vector (#E1300, Promega, Wisconsin, USA) by Sangon. Finally, the plasmids or RNA interference fragments were transfected into cells by using Transfectamine™ 5000 following the manufacturer’s instructions (#60021, AATbio, California, USA).
Bioinformatic analysis
First, we downloaded 2 human NAFLD-related microarray datasets, GSE48452 (containing 14 healthy control and 32 NAFLD tissue samples) and GSE107231 (containing 5 normal liver and 5 NAFLD biopsy tissue samples), from Gene Expression Omnibus (GEO) and analysed the downregulated lncRNAs in both datasets. Then, we reconfirmed the top 10 downregulated lncRNAs by RT‒qPCR in our collected clinical specimens, and afterwards, we confirmed the expression of the selected lncRNA in the livers of mice. Second, to determine the target miRNA of GAS5, we generated a Venn diagram by using the online tool “VEENY 2.1” (https://bioinfogp.cnb.csic.es/tools/venny/). The Venn diagram shows the intersection between the target miRNAs of GAS5 (catalogued in lncBase v.3 in DIANA Tools) and the miRNAs in GSE94799. The overlapping miRNAs were the potential binding miRNAs of GAS5, and the expression of these miRNAs was increased in livers with NAFLD. After extracting total RNA from the liver tissues from mice and humans, we measured the expression of the top 6 upregulated miRNAs by RT‒qPCR analysis. The miRNA with the most significant change in expression was considered the potential binding miRNA of GAS5. In addition, we performed sequence alignment analysis to determine the binding sites between GAS5 and miR-28a-5p and between miR-28a-5p and the MARCH7 3′UTR.
Experimental animals
Eight-week-old male C57BL/6 mice and 10-week-old male leptin-deficient obese (Ob/Ob) male mice were purchased from the Model Animal Research Center of Nanjing University (Nanjing, China). All animal protocols were approved by the Experimental Animal Ethics Committee of Nantong University. Mice were housed in a 23 ± 2 °C environment with a 12 h light/dark cycle. They had free access to food and water. All animals were cared for and sacrificed in accordance with ethical guidelines. C57BL/6 and Ob/Ob mice were fed a chow diet (10.2% fat, 18.3% protein, and 71.3% carbohydrate). HFD-fed mice were C57BL/6 mice fed a high-fat diet (61.6% fat, 18.1% protein, and 20.3% carbohydrate; FBSH, Shanghai, China) for 12 weeks. Then, these mice were injected every 10 days with the above adenoviruses via the tail vein at a dose of 3*1010 plaque-forming units (PFU) in 150 μL PBS per mouse, and these mice were continuously fed a ND or HFD. After 3 weeks of the first injection, the mice and their tissues were used for further analysis. In addition, metformin (#625-24-9, MedChemExpress, New Jersey, USA) or placebo was administered to HFD-fed mice intragastrically at 200 mg/kg/day [29] for 1 month, accompanied by 12 weeks of ND or HFD feeding. Metformin was dissolved in sodium carboxymethylcellulose (#A501427, Sangon). The NFD, HFD, or Ob/Ob mice were randomly assigned into different experiment groups. Sample sizes of all experiments were predetermined according to our experience and published literatures.
Mouse metabolic assays
The mice were fasted for 6 h for assessment of metabolic status. Fasting blood glucose and insulin were measured using a glucometer. The glucose tolerance test (GTT) and insulin tolerance test (ITT) were performed using a glucometer (#582, Yuwell, Danyang, Jiangsu, China) 30 min after the mice were administered 1 g/kg glucose (#A600219, Sangon) and 0.75 U/kg insulin (#11061-68-0, Sigma) via intraperitoneal injection. All experiments were performed independently in triplicate at least.
RNA isolation and real-time PCR
Total RNA from clinical specimens, mouse tissues and cells was extracted using TRIzol reagent (#15596026, Thermo Fisher Scientific). The obtained RNA was reverse transcribed into cDNA using the PrimeScriptTM RT Reagent Kit with gDNA Eraser (#RR047A, Takara, Dalian, Liaoning, China). For miRNA determination, total RNA was reverse transcribed using the miScript II RT kit (#243584, QIAGEN, Shanghai, China). Next, real-time PCR was conducted by applying SYBR Green Premix Ex Taq (#RR390L, Takara). To normalise gene expression, β-actin was used as an internal control for mRNAs and lncRNAs, and U6 was used as an internal control for miRNAs. Finally, analysis of the real-time PCR data was performed by the 2-∆∆CT method. The primers used herein are listed in Table 2.
Table 2.
The used primers in this study.
| Gene name | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| m-GAS5 | TCTCACAGCCAGTTCTGTGG | CTTGTCACAGGAGCCCTTTC |
| m-FATP-1 | TGCACAGCAGGTACTACCGCAT | TGCGCAGTACCACCGTCAAC |
| m-FATP-2 | GTGCAACACACCGCAGAAAC | TGCCTTCAGTGGATGCGTAG |
| m-CD36 | TGGCCAAGCTATTGCGACAT | AGGCATTGGCTGGAAGAACA |
| m-SREBP1c | GGAGCCATGGATTGCACATT | GCTTCCAGAGAGGAGGCCAG |
| m-ACC1 | AGCTGATCCTGCGAACCT | GCCAAGCGGATGTAAACT |
| m-FAS | ATCCTGGAACGAGAACACGATCT | AGAGACGTGTCACTCCTGGACTT |
| m-PPARγ | TGAACGTGAAGCCCATCGAG | CTTGGCGAACAGCTGAGAGG |
| m-PPARα | ACGGCAATGGCTTTATCA | CGCTGCGTCGGACTCGGT |
| m-ACOX-1 | GTCTCCGTCATGAATCCCGA | TGCGATGCCAAATTCCCTCA |
| m-CPT-1α | AGGACCCTGAGGCATCTATT | ATGACCTCCTGGCATTCTCC |
| m-UCP2 | GCTGGTGGTGGTCGGAGATA | ACTGGCCCAAGGCAGAGTT |
| m-HMGCR | ATCATGTGCTGCTTCGGCTGCAT | AAATTGGACGACCCTCACGGCT |
| m-CYP7A1 | TCAAAGAGCGCTGTCTGGGTCA | TTTCCCGGGCTTTATGTGCGGT |
| m-IL-1β | CCGTGGACCTTCCAGGATGA | GGGAACGTCACACACCAGCA |
| m-IL-6 | AGTTGCCTTCTTGGGACTGA | TCCACGATTTCCCAGAGAAC |
| m-TNF-α | CATCTTCTCAAAATTCGAGTGACAA | TGGGAGTAGACAAGGTACAACCC |
| m-MCP-1 | TAAAAACCTGGATCGGAACCAAA | GCATTAGCTTCAGATTTACGGGT |
| m-IL-2 | TGCGGCATGTTCTGGATTTG | CAAATGTGTTGTCAGAGCCCTTT |
| m-IL-10 | CCAAGCCTTATCGGAAATGA | TTTTCACAGGGGAGAAATCG |
| m-PEPCK | TGCCCCAGGCAGTGAGGAAGTT | GTCAGTGAGAGCCAGCCAACAGT |
| m-G6Pase | TCTGTCCCGGATCTACCTTG | GCTGGCAAAGGGTGTAGTGT |
| m-MARCH7 | GCTCATGCAAATGAACAAGCTG | AATCGGACACGAGTGCTTGG |
| m-actin-β | CGCAGCCACTGTCGAGTC | TATCGTCATCCATGGCGAACTGG |
| m-FBXL2 | AGTCGGGGACTCCTCTTTGA | TAAGGCTGTAACACGTGCTGT |
| m-PARKIN | AGCCAGAGGTCCAGTTAAACC | CACCACTCATCCGGTTTGGA |
| m-TRIM31 | GACGCCTTGAGCAGGGGTAA | ATGATGGACTCATGCCTTGCT |
| m-CHIP | CCTGATAAGAGCCCGAGTGC | AAGTGGGTTCCGAGTGATGG |
| m-UBR5 | TGCAGAGCCTGGATCGATTT | ATCGCGATCGACCTCTAGTGA |
| m-U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
| h-GAS5 | CAGTGTGGCTCTGGATAGCA | TTAAGCTGGTCCAGGCAAGT |
| h-IL-1β | AGCCATGGCAGAAGTACCTG | CCTGGAAGGAGCACTTCATCT |
| h-TNF-α | TAGCCCATGTTGTAGCAAACC | ATGAGGTACAGGCCCTCTGAT |
| h-IL-10 | GGCACCCAGTCTGAGAACAG | GGCAACCCAGGTAACCCTTA |
| h-MARCH7 | GAGAAAGGCGGCGGAATTCAT | AGGAAAAACCAGCTCTGACCA |
| h-actin-β | ACCCGCCGCCAGCTC | CATCACGCCCTGGTGCC |
| h-FBXL2 | GCTCTTCGGCCATGGTTTTC | GTGCACATCGGCACAAAGTT |
| h-PARKIN | GGAAGGACTCACCACCAGC | CAGCAAGATGGACCCTGG |
| h-TRIM31 | GTCGTCTTGTGCAGAAGTGAAG | AGGCTCCCTGTGATGGAGT |
| h-CHIP | AGGGCAATCGTCTGTTCGTG | CGGGTTCCGGGTGATCG |
| h-UBR5 | CCTCAGAAAAGGTACAGCAGGA | CTTGCCAGGCCTTGGAGTAT |
| h-U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
| miR-28a-5p | AAGGAGCTCACAGTCT | GTGCAGGGTCCGAGGT |
| miR-455-3p | GCAGTCCATGGGCAT | GTGCAGGGTCCGAGGT |
| miR-133b-3p | TTTGGTCCCCTTCAAC | GTGCAGGGTCCGAGGT |
| miR-7a-5p | TGGAAGACTAGTGATTT | GTGCAGGGTCCGAGGT |
| miR-532-5p | CAUGCCTTGAGTGTAG | GTGCAGGGTCCGAGGT |
| miR-802-5p | TCAGTAACAAAGATTC | GTGCAGGGTCCGAGGT |
m mouse, h human.
Western blot
The liver tissues and cell lines were lysed using radioimmunoprecipitation assay (RIPA) buffer (#P1003B, Beyotime). Approximately 30 μg of total protein was added to 8% SDS‒PAGE gels using Bio-Rad equipment (Shanghai, China). After separation, the proteins were transferred to PVDF membranes (#ISEQ00010, Millipore, Massachusetts, USA). The protein-loaded PVDF membranes were blocked with 5% skim milk (#A600669, Sangon), immersed in solutions of the primary antibodies at 4 °C overnight, and then incubated with Alexa Fluor 680-conjugated secondary antibodies (Thermo Fisher Scientific). Tubulin-α or GAPDH was used as an internal control. The used primary antibodies were listed (anti-NLRP3, #IMG-6668A, Novus Biologicals, Colorado, USA; anti-MARCH7, #PA5-54572, Sigma‒Aldrich; anti-Cleaved caspase-1, #PA5-99390, Thermo Fisher Scientific; anti-Matured IL-1β, #AF401, R&D Systems, City of Emeryville, USA; anti-GSDMD-N, #GSDMD antibody Abcam EPR 19828, Abcam, Cambridge, UK; anti-Tubulin-α, #NB100-690, Novus Biologicals; anti-GAPDH, #MA1-16757, Thermo Fisher Scientific; anti-actin-β, #NB600-501, Novus Biologicals; anti-HA, #NB600-363, Novus Biologicals; anti-Flag, #MA1-91878, Thermo Fisher Scientific; anti-Ubiquitin, #NB300-130, Novus Biologicals; anti-GST, #13-6700, Thermo Fisher Scientific; anti-His, #NBP2-61482, Novus Biologicals; anti-Myc, #2276, Cell Signaling Technology, Boston, USA).
Immunoprecipitation assay
To detect the interaction between MARCH7 and NLRP3, we conducted an immunoprecipitation assay in AML12 and HepG2 cells. First, the pRK5-HA/Flag vector was used to construct plasmids for expressing NLRP3 or the deletion mutants of MARCH7. These plasmids were transfected into AML12 and HepG2 cells using Tranfectamine 5000 reagent (#60021, AATbio). The cells were lysed with ice-cold lysis buffer (#P0013, Beyotime, Shanghai). The lysates were centrifuged, and the supernatants were immunoprecipitated with anti-HA antibodies at 4 °C overnight. Then, these samples were incubated with Protein G-agarose beads (#P2108, Beyotime) at 4 °C for 2 h. Afterwards, the beads were washed with the above lysis buffer three times. After adding 4× loading buffer, the final samples were heated at 98 °C for 10 min. Finally, the beads were pelleted by centrifugation and removed, and the remaining immunoprecipitated samples were collected and subjected to immunoblotting (Western blot) as described above. Similarly, purified proteins were prepared by Sangon, and the interactions between MARCH7 and NLRP3 and between GSDMD-N, and cleaved caspase-1 were detected by immunoprecipitation assays. The used primary antibodies here were showed above.
We also used an immunoprecipitation assay to detect the ubiquitination of NLRP3 according to a previous study [41]. In brief, HA-NLRP3 and Flag-MARCH7 were co-transfected into AML12 and HepG2 cells. After 36 h of transfection, 20 μM MG132 (HY13259, MedChemExpress) was added for another 12 h of incubation. The cells were collected and boiled for 10 min, and the cells were lysed with a sonication device. Then, the supernatant was incubated with anti-HA antibody for 12 h with rotation and with protein A- or G-agarose beads for another 2 h. Next, the bead-containing samples were washed with a buffer containing 10 mM Tris-HCl, pH 8.0, 1% NP-40, 1 mM EDTA, and 1 M NaCl three times. The beads were centrifuged at 20,000 × g for 3 min, the residual supernatant was aspirated, and the resin was boiled with 4× SDS loading buffer. Finally, the prepared samples were loaded onto SDS‒PAGE gels for immunoblot analysis with the respective antibodies to detect ubiquitin and other proteins. In addition, in vitro ubiquitination assays were conducted to determine the effects of MARCH7 on the ubiquitination of NLRP3. The other molecular biology grade agents were from Sangon. All experiments were performed independently in triplicate at least.
Immunofluorescence staining
Here, we conducted immunofluorescence staining to determine GSDMD-N protein levels in AML12 and HepG2 cells. First, hepatocytes were plated in a 12-well plate, and we transfected the hepatocytes with anti-miR-NC, anti-miR-28a-5p, pcDNA, or MARCH7 for 24 h. Subsequently, the hepatocytes were incubated with FFA for 24 h. Afterwards, the hepatocytes were washed with 4 °C cold PBS three times. The cells were incubated with an anti-GSDMD-N antibody (#GSDMD antibody Abcam EPR 19828, Abcam) overnight in a 4 °C refrigerator. After washing with PBS, they were incubated with DyLight633 (#35563, Thermo Fisher Scientific) for 1 h. Then, the AML12 and HepG2 cells were stained with Hoechst (#C1011, Beyotime) for 10 min to visualise the nuclei. Finally, they were washed with PBS and observed under a Zeiss fluorescence microscope (Carl Zeiss AG, Oberkochen, Germany). In addition, we bought Cy5-labled miR-28a-5p probe and FITC-labled GAS5 probe (GenePharm, Shanghai, China) to determine the transduction efficiency of adenoviruses in liver of mice by using RNA fluorescent in situ hybridisation (FISH) kit (# C10910, RiboBio, GuangZhou, Guangdong, China). Briefly, the frozen liver sections were washed with PBS and treated with 0.5% Trition X-100 for 10 min at 4 °C. Next, they were hybridised with Cy5-labled miR-28a-5p probe and FITC-labled GAS5 probe for 30 min at 37 °C, and washed with hybridisation buffer at 42 °C in dark room. Afterwards, they incubated with DAPI for 5 min. Finnaly, they were washed with PBS and observed under a Zeiss fluorescence microscope.
Luciferase reporter assay
AML12 and HepG2 cells were transfected with miR-NC, miR-28a-5p, anti-miR-NC, or anti-miR-28a-5p. They were co-transfected with the empty pmir-GLO vector (#E1300, Promega, Wisconsin, USA) and pmir-GLO containing the wild-type or mutant MARCH7 3′ UTR using Transfectamine™ 5000 for 36 h. In addition, they were transfected with miR-NC, miR-28a-5p, and co-transfected with the empty pmir-GLO vector and pmir-GLO containing the wild-type NLRP3 3′ UTR. Finally, luciferase activity was measured using the Dual-Glo luciferase assay system (Promega). All experiments were performed independently in triplicate at least.
RNA immunoprecipitation (RIP)
We performed RIP assays to examine the potential interaction between GAS5 and miR-28a-5p using a Magna RIPTM RNA‐Binding Protein Immunoprecipitation Kit (#17-700, Millipore). The protein solution was prepared by lysing liver tissues and cell lines, and then an anti-Ago2 antibody (NBP2-59159, Novus) or IgG was added to the protein solution for the RIP assay. Then, we applied TRIzol reagent to purify RNA from the immunoprecipitated samples. Finally, GAS5 and miR-28a-5p expression levels were measured by RT‒qPCR.
Biotinylated GAS5 pull-down assay
Biotin-labelled human and mouse GAS5 sequences and the corresponding negative control sequences were prepared by GenePharma (Shanghai, China). The sequences are listed below: human GAS5, 5′-Bio-AGG TAT GGT GCT GGG TGC AGA TGC AGT GTG GCT CTG GAT AGC ACC TTA TGG ACA GTT GTG TCC CCA AGG AAG GAT GAG-3′; mouse GAS5, 5′-Bio-GAC AGC CCG GAG CCA AAG GGG CGG AAG GTC GCC GAG TGC TGG GAG GGG AGG TGG GGC AGC GAC TGA GCG CCG CAG GAG-3′; a negative control sequence for human GAS5, 5′-Bio-AAA ATA CAA TTG CCG ACA TCG CGA GAT GTG GCT CTG GAT AGC ACC TAC CCC TCT AAA CTC AGC GTA CTT TCG TGT CTT-3′; negative control sequence for mouse GAS5, 5′-Bio-TCA TCA AGA CGA TTC GGC AAC CAG TTG TCC TTC GCA CGG CTC GGG CAA CCA GTT GTC TGG GTT CCA TGC TAG CTC AGG-3′. The human and mouse actin-β were used also as a negative control, human actin-β, 5′-Bio-ATG GAT GAT GAT ATC GCC GCG CTC GTC GTC GAC AAC GGC TCC GGC ATG TGC AAG GCC GGC TTC GCG GGC GAC GAT GC; mouse actin-β, 5′-Bio-ATG GAT GAC GAT ATC GCT GCG CTG GTC GTC GAC AAC GGC TCC GGC ATG TGC AAA GCC GGC TTC GCG GGC GAC GAT GC. The biotin-labelled sequences were mixed gently with magnetic beads (#11206D, Invitrogen) for 3 h following the manufacturer’s instructions. Then, the beads were washed with buffer and incubated with cell lysates or liver tissue lysates. After incubation overnight at 4 °C, the beads were collected and washed with buffer, and RNA was extracted for measurement of GAS5 and miR-28a-5p expression by RT‒qPCR.
Histological analysis, immunohistochemistry, and Oil Red O staining
We applied haematoxylin and eosin (H & E) staining kit (#C0105M, Beyotime) to determine the histological changes in livers, and we used immunohistochemistry (IHC) to measure the indicated protein expression levels in the liver as described previously [42], and the used primary antibodies were showed above. Briefly, the liver tissues were fixed in 4% paraformaldehyde overnight and embedded in paraffin. The liver sections were stained with haematoxylin and eosin and dehydrated in graded alcohol solutions. For Oil Red O staining, liver cryosections or cells were stained with Oil Red O solution (#O1391, Sigma) for 30 min, and then, the samples were washed with phosphate buffered saline (PBS) three times. Micrographs were acquired under an Olympus microscope (Tokyo, Japan). All other molecular biology grade agents were bought from Sangon.
ELISAs for hepatic lipid analyses and liver function assays
The contents of hepatic triglyceride (TG), total cholesterol (TC) and nonesterified fatty acid (NEFA) were determined by using commercial ELISA kits (#E-BC-K261-M, #E-BC-K109-M, #E-BC-K013-M, Elabscience, Wuhan, China). Liver function in the mice was evaluated by measuring the concentrations of aspartate aminotransferase (AST), alanine aminotransferase (ALT), and alkaline phosphatase (ALP) in the serum of mice using ELISA kits (#ab263882, #ab282882, #ab267583, Abcam). All experiments were performed independently in triplicate at least. All the other molecular biology grade agents were from Sangon.
Statistics
All statistical analyses were conducted using GraphPad Prism 8.0 (GraphPad Software, CA, USA). All results are expressed as the mean ± standard deviation (S.D.) values from at least three independent biological experiments. Two-tailed Student’s t test was used to determine the significance of differences between two groups, and one-way analysis of variance (ANOVA) with the Bonferroni post hoc test was conducted for comparisons among more than two groups. In the respective figure legends statistical tests are declared, and a p value of < 0.05 was considered statistically significant, and *p < 0.05, **p < 0.01, ***p < 0.001, nsp > 0.05. Biological replicates gave comparable results, and no technical or biological replicates were excluded, and the sample sizes of all experiments were predetermined following our experience and published literatures. Investigators were blinded to the group allocation during the experiment and outcome assessment.
Supplementary information
Acknowledgements
We thank Prof. Donghua Yang for constructive discussions.
Author contributions
WZ and TC conceptualised and supervised the overall project. TC, ZZ, HL, LW, AK, WG, KR, and XS performed experiments. ZZ, HL, LW, AK and YC performed validation studies. YC, WG, KR and XS performed formal data analysis. WZ, TC and ZZ drafted the paper, and all authors contributed to the methodology and revisions.
Funding
This study was supported by the National Natural Science Foundation of China (81602636, 31600269 and 82174292), the Sichuan Science and Technology Program (2019YJ0369 and 2020YJ0401), the China Postdoctoral Science Foundation (2021M693953), the Scientific Research Project of Sichuan Medical Association (S18018), the Postdoctoral Foundation of Jiangsu (2021K385C), Disciplinary Construction Innovation Team Foundation of Chengdu Medical College (No. CMC-XK-2103), Natural Science Foundation of Sichuan Province (2022NSFSC0725) and the Collaborative Innovation Center of Sichuan for Elderly Care and Health (19Z02).
Data availability
The original data presented in this study are included in the paper/Supplementary Material, and further inquiries can be directed to the corresponding authors.
Competing interests
The authors declare no competing interests.
Ethics approval
In this study, the clinical specimen collection was approved by the Medical Ethics Committee of the Second Affiliated Hospital of Chongqing Medical University, and all participants provided written informed consent. All animal protocols were approved by the Experimental Animal Ethics Committee of Nantong University.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-023-01183-4.
References
- 1.Arshad T, Paik JM, Biswas R, Alqahtani S, Henry L, Younossi ZM. Trends in the Prevalence of Nonalcoholic Fatty Liver Disease (Nafld) among Adolescents and Young Adults in the United States, 2007-2016. Hepatology. 2020;72:886a–7a. doi: 10.1002/hep4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wruck W, Graffmann N, Kawala MA, Adjaye J. Concise Review: Current Status and Future Directions on Research Related to Nonalcoholic Fatty Liver Disease. Stem Cells. 2017;35:89–96. doi: 10.1002/stem.2454. [DOI] [PubMed] [Google Scholar]
- 3.Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 4.Zhao XY, Xiong XL, Liu TY, Mi L, Peng XL, Rui C, et al. Long noncoding RNA licensing of obesity-linked hepatic lipogenesis and NAFLD pathogenesis. Nat Commun. 2018;9:2986. [DOI] [PMC free article] [PubMed]
- 5.Ilieva M, Dao J, Miller HE, Madsen JH, Bishop AJR, Kauppinen S, et al. Systematic Analysis of Long Non-Coding RNA Genes in Nonalcoholic Fatty Liver Disease. Noncoding RNA. 2022;8:56. [DOI] [PMC free article] [PubMed]
- 6.Lin G, Wu T, Gao X, He Z, Nong W. Research Progress of Long Non-Coding RNA GAS5 in Malignant Tumors. Front Oncol. 2022;12:846497. doi: 10.3389/fonc.2022.846497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui J, Wang Y, Xue H. Long non-coding RNA GAS5 contributes to the progression of nonalcoholic fatty liver disease by targeting the microRNA-29a-3p/NOTCH2 axis. Bioengineered. 2022;13:8370–81. doi: 10.1080/21655979.2022.2026858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu S, Wang Y, Li Z, Hua Q, Jiang M, Fan X. LncRNA GAS5 Knockdown Mitigates Hepatic Lipid Accumulation via Regulating MiR-26a-5p/PDE4B to Activate cAMP/CREB Pathway. Front Endocrinol. 2022;13:889858. [DOI] [PMC free article] [PubMed]
- 9.Yu F, Zheng J, Mao Y, Dong P, Lu Z, Li G, et al. Long Non-coding RNA Growth Arrest-specific Transcript 5 (GAS5) Inhibits Liver Fibrogenesis through a Mechanism of Competing Endogenous RNA. J Biol Chem. 2015;290:28286–98. doi: 10.1074/jbc.M115.683813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong Z, Li S, Wang X, Si L, Ma R, Bao L, et al. lncRNA GAS5 restrains CCl4-induced hepatic fibrosis by targeting miR-23a through the PTEN/PI3K/Akt signaling pathway. Am J Physiol Gastrointest Liver Physiol. 2019;316:G539–G550. doi: 10.1152/ajpgi.00249.2018. [DOI] [PubMed] [Google Scholar]
- 11.Rizzo M, Berti G, Russo F, Evangelista M, Pellegrini M, Rainaldi G. The miRNA Pull Out Assay as a Method to Validate the miR-28-5p Targets Identified in Other Tumor Contexts in Prostate Cancer. Int J Genom. 2017;2017:5214806. doi: 10.1155/2017/5214806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brea R, Motino O, Frances D, Garcia-Monzon C, Vargas J, Fernandez-Velasco M, et al. PGE(2) induces apoptosis of hepatic stellate cells and attenuates liver fibrosis in mice by downregulating miR-23a-5p and miR-28a-5p. Biochim et Biophys Acta Mol Basis Dis. 2018;1864:325–37. doi: 10.1016/j.bbadis.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Jahnke M, Trowsdale J, Kelly AP. Ubiquitination of HLA-DM by different MARCH family E3 ligases targets different endocytic pathways. J Biol Chem. 2012;287:7256–64. [DOI] [PMC free article] [PubMed]
- 14.Lin H, Li S, Shu HB. The Membrane-Associated MARCH E3 Ligase Family: Emerging Roles in Immune Regulation. Front Immunol. 2019;10:1751. doi: 10.3389/fimmu.2019.01751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan Y, Jiang W, Liu L, Wang X, Ding C, Tian Z, et al. Dopamine controls systemic inflammation through inhibition of NLRP3 inflammasome. Cell. 2015;160:62–73. doi: 10.1016/j.cell.2014.11.047. [DOI] [PubMed] [Google Scholar]
- 16.Zeng C, Duan F, Hu J, Luo B, Huang B, Lou X, et al. NLRP3 inflammasome-mediated pyroptosis contributes to the pathogenesis of non-ischemic dilated cardiomyopathy. Redox Biol. 2020;34:101523. doi: 10.1016/j.redox.2020.101523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Araujo FM, Cuenca-Bermejo L, Fernandez-Villalba E, Costa SL, Silva VDA, Herrero MT. Role of Microgliosis and NLRP3 Inflammasome in Parkinson’s Disease Pathogenesis and Therapy. Cell Mol Neurobiol. 2022;42:1283–300. doi: 10.1007/s10571-020-01027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu L, Hong W, Lu S, Li Y, Guan Y, Weng X, et al. The NLRP3 Inflammasome in Non-Alcoholic Fatty Liver Disease and Steatohepatitis: Therapeutic Targets and Treatment. Front Pharmacol. 2022;13:780496. doi: 10.3389/fphar.2022.780496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wree A, Eguchi A, McGeough MD, Pena CA, Johnson CD, Canbay A, et al. NLRP3 inflammasome activation results in hepatocyte pyroptosis, liver inflammation, and fibrosis in mice. Hepatology. 2014;59:898–910. doi: 10.1002/hep.26592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beier JI, Banales JM. Pyroptosis: An inflammatory link between NAFLD and NASH with potential therapeutic implications. J Hepatol. 2018;68:643–5. doi: 10.1016/j.jhep.2018.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gautheron J, Gores GJ, Rodrigues CMP. Lytic cell death in metabolic liver disease. J Hepatol. 2020;73:394–408. doi: 10.1016/j.jhep.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Carvalho Ribeiro M, Szabo G. Role of the Inflammasome in Liver Disease. Annu Rev Pathol. 2022;17:345–65. doi: 10.1146/annurev-pathmechdis-032521-102529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koh EH, Yoon JE, Ko MS, Leem J, Yun JY, Hong CH, et al. Sphingomyelin synthase 1 mediates hepatocyte pyroptosis to trigger non-alcoholic steatohepatitis. Gut. 2021;70:1954–64. [DOI] [PMC free article] [PubMed]
- 24.Xu B, Jiang M, Chu Y, Wang W, Chen D, Li X, et al. Gasdermin D plays a key role as a pyroptosis executor of non-alcoholic steatohepatitis in humans and mice. J Hepatol. 2018;68:773–82. doi: 10.1016/j.jhep.2017.11.040. [DOI] [PubMed] [Google Scholar]
- 25.Wree A, McGeough MD, Pena CA, Schlattjan M, Li H, Inzaugarat ME, et al. NLRP3 inflammasome activation is required for fibrosis development in NAFLD. J Mol Med (Berl) 2014;92:1069–82. doi: 10.1007/s00109-014-1170-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez-Castejon G. Control of the inflammasome by the ubiquitin system. FEBS J. 2020;287:11–26. doi: 10.1111/febs.15118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sumida Y, Yoneda M. Current and future pharmacological therapies for NAFLD/NASH. J Gastroenterol. 2018;53:362–76. doi: 10.1007/s00535-017-1415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wabitsch S, McCallen JD, Kamenyeva O, Ruf B, McVey JC, Kabat J, et al. Metformin treatment rescues CD8(+) T cell response to immune checkpoint inhibitor therapy in mice with NAFLD. J Hepatol. 2022;77:748–60. [DOI] [PMC free article] [PubMed]
- 29.Liu ZQ, Song XM, Chen QT, Liu T, Teng JT, Zhou K, et al. Effect of metformin on global gene expression in liver of KKAy mice. Pharmacol Rep. 2016;68:1332–8. doi: 10.1016/j.pharep.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 30.Liu CN, Yang ZH, Wu JG, Zhang L, Lee SM, Shin DJ, et al. Long noncoding RNA H19 interacts with polypyrimidine tract-binding protein 1 to reprogram hepatic lipid homeostasis. Hepatology. 2018;67:1768–83. doi: 10.1002/hep.29654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atanasovska B, Rensen SS, van der Sijde MR, Marsman G, Kumar V, Jonkers I, et al. A liver-specific long noncoding RNA with a role in cell viability is elevated in human nonalcoholic steatohepatitis. Hepatology. 2017;66:794–808. doi: 10.1002/hep.29034. [DOI] [PubMed] [Google Scholar]
- 32.Lu X, Jiang M, Tian J, Liu W, Wu F, Yu L, et al. Growth Arrest-Specific Transcript 5 (GAS5) Exerts Important Roles on the Treatment of BM45 Cells of Liver Cirrhosis. Mol Ther Nucleic Acids. 2020;22:1154–63. doi: 10.1016/j.omtn.2020.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Zhou LK, Xu L, Ye J, Li D, Wang WS, Li XH, et al. Cidea promotes hepatic steatosis by sensing dietary fatty acids. Hepatology. 2012;56:95–107. doi: 10.1002/hep.25611. [DOI] [PubMed] [Google Scholar]
- 34.Jiang Y, Qian T, Li S, Xie Y, Tao M. Metformin reverses tamoxifen resistance through the lncRNA GAS5-medicated mTOR pathway in breast cancer. Ann Transl Med. 2022;10:366. doi: 10.21037/atm-22-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen Z, Zhou H, Li A, Wu T, Ji X, Guo L, et al. Metformin inhibits hepatocellular carcinoma development by inducing apoptosis and pyroptosis through regulating FOXO3. Aging. 2021;13:22120–33. doi: 10.18632/aging.203464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guzman S, Dragan M, Kwon H, de Oliveira V, Rao S, Bhatt V, et al. Targeting hepatic kisspeptin receptor ameliorates non-alcoholic fatty liver disease in a mouse model. J Clin Investig. 2022;132:e145889. [DOI] [PMC free article] [PubMed]
- 37.Luo P, Qin C, Zhu L, Fang C, Zhang Y, Zhang H, et al. Ubiquitin-specific Peptidase 10 (USP10) Inhibits Hepatic Steatosis, Insulin Resistance and Inflammation via Sirt6. Hepatology. 2018;68:1786–803. [DOI] [PubMed]
- 38.Personnaz J, Piccolo E, Dortignac A, Iacovoni JS, Mariette J, Rocher V, et al. Nuclear HMGB1 protects from nonalcoholic fatty liver disease through negative regulation of liver X receptor. Sci Adv. 2022;8:eabg9055. doi: 10.1126/sciadv.abg9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheka AC, Adeyi O, Thompson J, Hameed B, Crawford PA, Ikramuddin S. Nonalcoholic Steatohepatitis: A Review. Jama. 2020;323:1175–83. doi: 10.1001/jama.2020.2298. [DOI] [PubMed] [Google Scholar]
- 40.Zhao K, Yang Y, Zhang G, Wang C, Wang D, Wu M, et al. Regulation of the Mdm2-p53 pathway by the ubiquitin E3 ligase MARCH7. EMBO Rep. 2018;19:305–19. doi: 10.15252/embr.201744465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choo YS, Zhang Z. Detection of protein ubiquitination. J Vis Exp. 2009;1293. [DOI] [PMC free article] [PubMed]
- 42.Chen T, Yan D, Cheng X, Ji X, Bian J, Yin W. miR-1224-5p Enhances Hepatic Lipogenesis by Targeting Adenosine Monophosphate-Activated Protein Kinase alpha1 in Male Mice. Endocrinology. 2018;159:2008–21. doi: 10.1210/en.2017-03231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original data presented in this study are included in the paper/Supplementary Material, and further inquiries can be directed to the corresponding authors.