Abstract
Circular RNAs (circRNAs) play an important regulatory role in the pathogenesis and progression of nasopharyngeal carcinoma (NPC), which have not been thoroughly elucidated. In this study, we revealed for the first time that circRILPL1 was upregulated in NPC, weakened adhesion and decreased stiffness of NPC cells, and promoted NPC proliferation and metastasis in vitro and in vivo. Mechanistically, circRILPL1 inhibited the LATS1-YAP kinase cascade by binding to and activating ROCK1, resulting in decrease of YAP phosphorylation. Binding and cooperating with transport receptor IPO7, circRILPL1 promoted the translocation of YAP from the cytoplasm to the nucleus, where YAP enhanced the transcription of cytoskeleton remodeling genes CAPN2 and PXN. By which, circRILPL1 contributed to the pathogenesis of NPC. Our results demonstrated that circRILPL1 promoted the proliferation and metastasis of NPC through activating the Hippo-YAP signaling pathway by binding to both ROCK1 and IPO7. Highly expressed circRILPL1 in NPC may serve as an important biomarker for tumor diagnosis and may also be a potential therapeutic target.
Subject terms: Metastasis, Tumour biomarkers, Cancer genomics, Protein-protein interaction networks, Tight junctions
Introduction
Nasopharyngeal carcinoma (NPC) is a polygenic hereditary malignancy originating from nasopharyngeal epithelial cells. NPC is especially common in East and Southeast Asia with obvious regional clustering and ethnic predilection [1, 2]. Genetic components and environmental factors such as Epstein-Barr virus (EBV) infection are considered to be the synergic pathogenic contributors to NPC, which exhibits strong tendency of local invasion and lymph node metastasis [3, 4]. However, during the pathogenesis of NPC, the oncogenic genomic changes and the relevant signaling processes remain far from clear [5–7].
Circular RNAs (circRNAs) are covalently closed non-coding RNAs formed by back-splicing at the 3′ and 5’ ends with lengths of hundreds or even thousands of bases. They have tissue-specific expression patterns, conserved and stable structures [8–11]. Studies have shown that circRNAs play important regulatory roles during tumorigenesis by acting as miRNA sponges, binding to proteins, or encoding small peptides [12–14]. For instance, circSETD3 competitively adsorbs miR-615-5p and miR-1538, thereby upregulating the expression of MAPRE1 [15]. CircARHGAP12 directly binds to the 3′ UTR of EZR mRNA, promoting its stability and thus enhancing the invasiveness of NPC cells [16]. In recent years, several unique circRNAs have been identified in NPC cells using high-throughput sequencing technology and bioinformatics techniques. They are significantly differentially expressed between NPC cells and normal cells, closely correlated with NPC proliferation, differentiation levels, lymph node and distant metastases, TNM stages, and overall survival rates [17, 18], suggesting that circRNAs can be used as potential clinical diagnostic and prognostic indicators. However, there are still many circRNAs whose specific roles and pathological mechanisms in the development of NPC remain largely unknown.
In this study, we discovered that a circRNA, circRILPL1, is highly expressed in NPC which promotes NPC progression through activating the Hippo signaling pathway. The Hippo pathway is an evolutionarily conserved signaling pathway that plays a biological role through its core transcriptional co-activators YAP/TAZ to initiate downstream gene expression. YAP/TAZ are mainly regulated by LATS1/2, which directly phosphorylate YAP/TAZ, leading to its destabilization and restricting its entry into the nucleus [19]. Many upstream signals affect the activation of the Hippo pathway, such as the extracellular matrix, cell polarity, mechanical forces, and cell adhesion [20]. The Rho/ROCK pathway is involved in cancer metastasis through regulating cellular actin dynamics [21]. Actin polymerization and microtubule assembly also play an important role in regulating the Hippo pathway [22, 23]. We found that in this study circRILPL1 bound to and activated ROCK1 to inhibit the LATS1 kinase, thereby inhibited the phosphorylation of YAP at Ser127 and Ser397 by LATS1, and enhanced the YAP activity. Therefore, circRILPL1 promoted YAP translocation into the nucleus by enhancing the interaction between YAP and IPO7, a nuclear transport receptor. In the nucleus, activated YAP promoted the transcription of CAPN2 and PXN. Through which, circRILPL1 was shown to promote the proliferation and metastasis of NPC in vitro and in vivo.
Results
CircRILPL1 is highly expressed in NPC and associated with poor prognosis of NPC patients
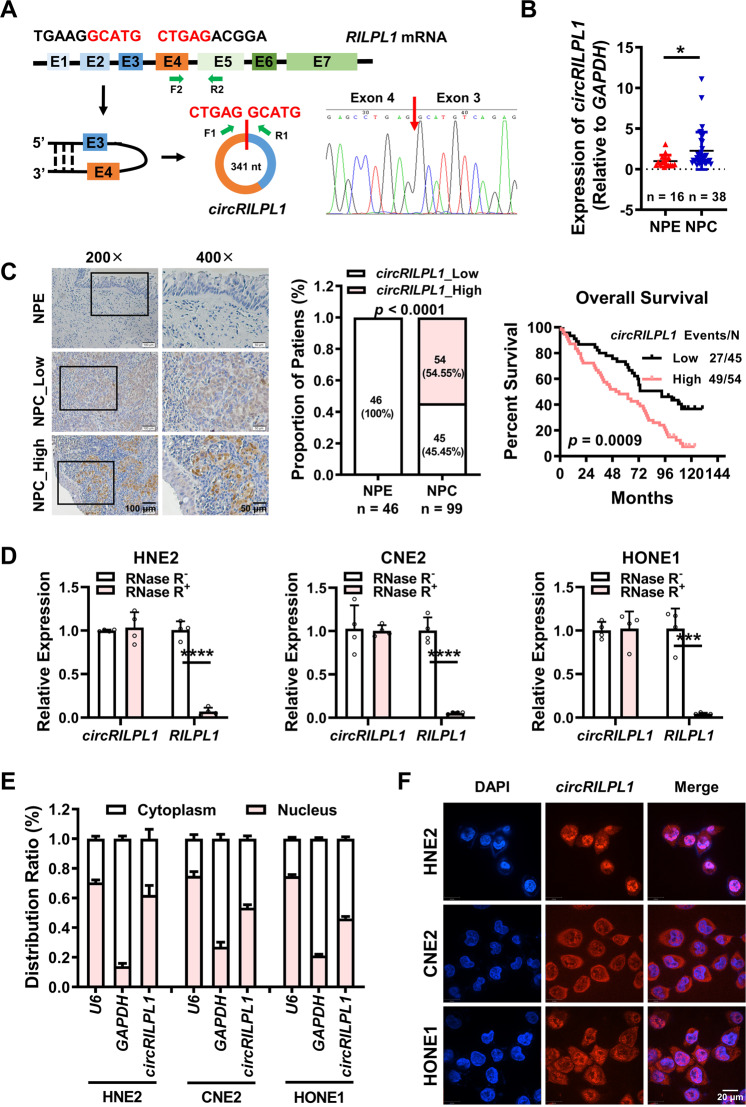
To evaluate the role of circRNAs in the pathogenesis of NPC, we searched the RNA-seq dataset for NPC (GSE68799) in the GEO database, which includes data from 4 chronically inflamed nasopharyngeal epithelial tissues and 41 NPC tissues. We analyzed the expression levels of circRNAs in these RNA-Seq data and identified 30 circRNAs with high expression in NPC tissues by extensive analysis (Table S1), among which circRILPL1 (hsa_circ_0007552) that located on chromosome 12 (chr12:123983090-123984082) was not reported in cancer before (Fig. S1A). Results of qRT-PCR and Sanger sequencing confirmed that it was formed by circular splicing of the exons 3 and 4 of the Rab interacting lysosomal protein like 1 (RILPL1) gene (NM_178314), with a total length of 341 nt (Fig. 1A). To examine the expression level of circRILPL1 in NPC, 38 NPC tissues and 16 non-cancerous tissues (chronic rhinitis epithelial tissues, as controls) were collected for qRT-PCR detection, and the results showed that the expression of circRILPL1 was significantly elevated in NPC tissues than that in the controls (Fig. 1B, Fig. S1B). The results of ISH on paraffin sections of 99 NPCs and 46 non-cancer nasopharyngeal epithelial (NPE) tissues also confirmed that the expression of circRILPL1 in NPCs was significantly higher than that in the NPE samples. Importantly, Kaplan-Meier analysis revealed that the overall survival rate of patients with high circRILPL1 expression was lower than that of patients with low circRILPL1 expression (Fig. 1C). RNase R digestion assays showed that circRILPL1 was more resistant to RNase R digestion than RILPL1 mRNA (Fig. 1D, Fig. S1C). Actinomycin D treatment further showed that the stability of circRILPL1 was much higher than that of RILPL1 linear mRNA (Fig. S1D), which is consistent with the stable circular RNA structure of circRILPL1. Results of RNA cytosolic/nuclear fraction assay and FISH showed that circRILPL1 was distributed both in the cytoplasm and the nucleus (Fig. 1E, F). All these data demonstrated that circRILPL1, a new circRNA that is highly expressed in NPC, may play a role in NPC progression.
Fig. 1. CircRILPL1 is highly expressed in NPC and associated with poor prognosis of NPC patients.
A Schematic diagram of the structure of circRILPL1. Sanger sequencing verified that circRILPL1 was formed by reverse splicing of exons 3 and 4 of RILPL1 mRNA. F1, R1, F2, and R2: primer sites for detecting circRILPL1 and RILPL1 mRNA are indicated. B The differential expression of circRILPL1 in 16 chronic rhinitis epithelial tissues and 38 NPC tissues was detected by qRT-PCR. C ISH assays indicated that circRILPL1 was highly expressed and correlated with the prognosis of NPC patients in 99 NPC paraffin tissue sections, as compared with 46 adjacent non-cancerous NPEs. Left, representative images of ISH. Magnification: 200×, Scale bar = 100 μm; Magnification: 400×, Scale bar = 50 μm. Middle, statistical analysis of ISH scores. Right, Kaplan-Meier survival analysis showed that the overall survival rate of NPC patients with high circRILPL1 expression was significantly lower than that of NPC patients with low circRILPL1 expression, p = 0.0009. D The relative expression levels of circRILPL1 and RILPL1 mRNA in NPC cells were detected after treating with RNase R for 30 min. E The intracellular localization of circRILPL1 was detected by RNA cytoplasm/nucleus fractionation assay in NPC cells. U6 was used as the nuclear internal reference, and GAPDH was used as the cytoplasmic internal control. F The intracellular localization of circRILPL1 was detected by FISH in NPC cells. Nuclei were stained with DAPI (blue). Scale bar = 20 μm. Data were presented as the means ± SD. *p < 0.05, ***p < 0.001, ****p < 0.0001.
CircRILPL1 promotes the proliferation and metastasis of NPC cells, and alters the mechanical properties of NPC cells in vitro
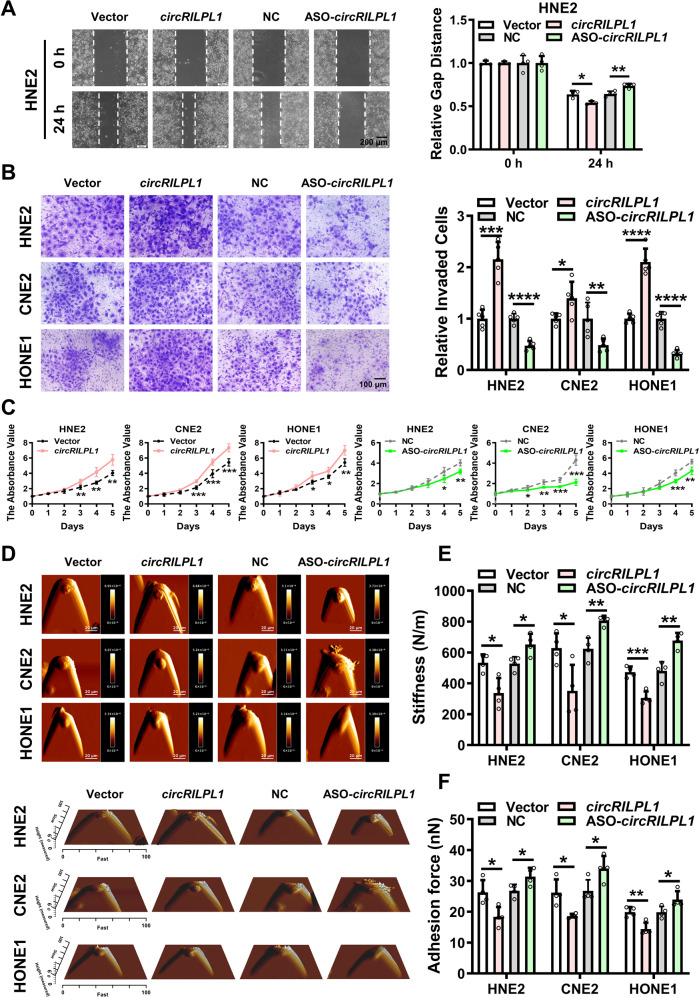
To investigate the biological functions of circRILPL1, circRILPL1 was overexpressed in NPC cell lines HNE2, CNE2, and HONE1 (Fig. S2A, B), or its expression was knocked down by using the specific ASO targeting to circRILPL1 splice site without affecting RILPL1 linear mRNA (Fig. S2C). Conversely, knockdown of RILPL1 linear mRNA had no effect on the expression of circRILPL1 (Fig. S2D). Wound healing and transwell assays showed that circRILPL1 could significantly enhance the migration and invasion of NPC cells (Fig. 2A, B and Fig. S3A). MTT and colony formation assays showed that overexpression of circRILPL1 promoted NPC cells proliferation, whereas knockdown of circRILPL1 exerted opposite effects (Fig. 2C, Fig. S3B). The process of carcinogenesis is also reflected by changes in the biophysical properties of cells [24]. Atomic force microscopy (AFM) assays showed that overexpression of circRILPL1 resulted in weakened adhesion and decreased stiffness of NPC cells, suggesting that cells were easier to detach from the surrounding tissues and their deformability was enhanced, which is consistent with the elevated capabilities of invasion and metastasis in NPC cells with high expression of circRILPL1. And opposite results were obtained after knockdown of circRILPL1 (Fig. 2D–F). Cytoskeleton remodeling and pseudopod formation are closely associated with cancer invasion and metastasis. Immunofluorescence with phalloidin-labeled F-actin showed that overexpression of circRILPL1 promoted the polymerization of actin filaments, whereas knockdown of circRILPL1 resulted in reduced microfilaments and loss of cytoskeleton integrity in NPC cells (Fig. S3C). The above results indicated that circRILPL1 regulated the biophysical properties of NPC cells and promoted their proliferation, invasion, and migration in vitro.
Fig. 2. CircRILPL1 promotes NPC cells migration, invasion, proliferation and alters the mechanical properties of NPC cells in vitro.
A The migration ability of circRILPL1 was examined in HNE2 cells after overexpression or knockdown of circRILPL1 by wound healing assay. Images were acquired at 0 and 24 h. Scale bars = 200 μm. B The invasion ability of circRILPL1 was assessed in NPC cells after overexpression or knockdown of circRILPL1 by transwell assay. Scale bars =100 μm. C The effect of circRILPL1 on proliferation was evaluated in NPC cells after overexpression or knockdown of circRILPL1 by MTT assay. D Representative AFM deflection images (top) and representative three-dimensional height distribution images (bottom) of NPC cells. E, F The stiffness (N/m) and adhesion force (nN) of NPC cells were measured and analyzed by using JPK image processing software. All experiments were performed in at least triplicate samples. Data were presented as the means ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
CircRILPL1 promotes the proliferation and metastasis of NPC cells in vivo
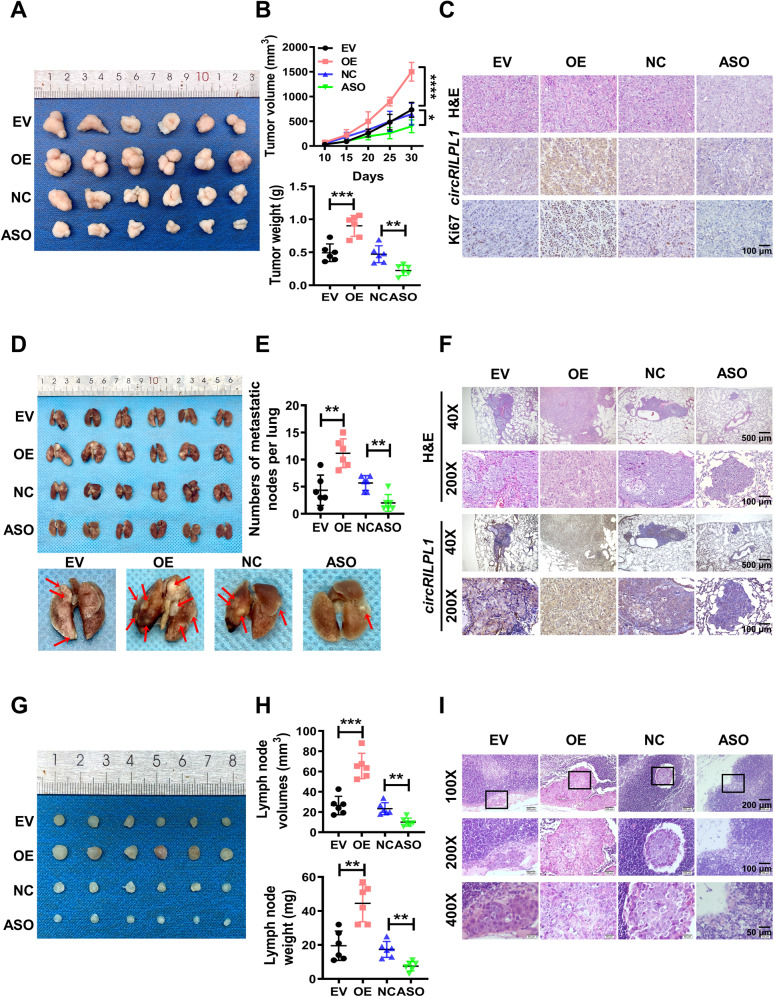
To explore the tumor-promoting capability of circRILPL1 in vivo, xenograft tumor models were established by injection of CNE2 cells transfected with circRILPL1 overexpression plasmid or ASO-circRILPL1 into 4-week-old nude mice. In the subcutaneous tumor model, tumor volume and tumor weight in the circRILPL1 overexpression group increased significantly compared to that of the control group. In contrast, knockdown of circRILPL1 resulted in significantly lower tumor volume and weight (Fig. 3A, B). IHC data revealed that, when compared with the control group, the expression of Ki67 was higher in the circRILPL1 overexpression group but lower in the circRILPL1 knockdown group (Fig. 3C, Fig. S4A), indicating that circRILPL1 promoted NPC cells proliferation in nude mice. In the tail vein-lung metastasis model, the number and area of lung metastatic nodules were significantly increased in the circRILPL1 overexpression group, while there were fewer and smaller lung metastatic nodules in the circRILPL1 knockdown group when compared with the control group (Fig. 3D–F), indicating that circRILPL1 significantly promoted lung metastasis of NPC cells in mice. NPC cells commonly spread through lymphoid vessels [25, 26]. In the footpad-lymph node metastasis model, the inguinal lymph nodes were dissected and analyzed after 28 days as the tumor volume in situ gradually increased (Fig. S4B, C). The data showed the weight and volume of metastatic inguinal lymph nodes in the circRILPL1 overexpression group were significantly higher than that in control group, whereas circRILPL1 knockdown produced the opposite effect (Fig. 3G, H). H&E staining confirmed that the area and number of metastatic tumor cells in lymph nodes were significantly increased after overexpression of circRILPL1, while there were fewer metastatic tumor cells in lymph nodes in the circRILPL1 knockdown group compared to the control group (Fig. 3I). All of these data demonstrated that circRILPL1 promoted NPC proliferation and metastasis in vivo.
Fig. 3. CircRILPL1 promotes the proliferation and metastasis of NPC cells in vivo.
CNE2 cells (2 × 106) transfected with the empty vector, the circRILPL1 overexpression plasmid, antisense oligonucleotides against circRILPL1 (ASO-circRILPL1), or the scramble negative control were injected subcutaneously, through the tail vein, or footpad of nude mice (n = 6 per group). A Representative images of subcutaneous tumor tissues (n = 6 per group). B The tumor volume growth curves (top) and the tumor weights (bottom) of subcutaneous tumors in nude mice. C Representative images of H&E staining, circRILPL1 staining, and Ki67 staining of subcutaneous tumor sections. Magnification: 200×, scale bar = 100 μm. D Representative images of lung tissues dissected from nude mice (top, n = 6 per group). Representative images of metastatic nodules on the lung surface as indicated by arrows (bottom). E Quantification of the number of metastatic nodes in the lungs. F Representative images of H&E staining of metastatic lung lesions and the expression of circRILPL1 by ISH assay. Magnification: 40×, scale bar = 500 μm; Magnification: 200×, scale bar = 100 μm. G Images of inguinal lymph nodes dissected from nude mice 28 days after injection via footpad (n = 6 per group). H The volume (top) and weight (bottom) of lymph nodes were measured for each group. I Representative images of H&E staining of inguinal lymph nodes showing metastatic tumor cells. Magnification: 100×, scale bar = 200 μm; Magnification: 200×, scale bar = 100 μm; Magnification: 400×, scale bar = 50 μm. All experiments were technically repeated six times. Data were presented as the means ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
CircRILPL1 activates the Hippo-YAP signaling pathway
An interesting phenomenon is the recent discovery that circRNAs can regulate biological processes and influence tumorigenesis by encoding small peptides [12]. There is a potential open reading frame (ORF) on circRILPL1 by prediction using circRNADb (http://reprod.njmu.edu.cn/circrnadb). However, when this potential ORF was tagged with flag and transfected into HNE2 cells, we failed to detect any flag tagged protein by western blot, thus excluding the possibility that circRILPL1 is a peptide-encoding circRNA (Fig. S5).
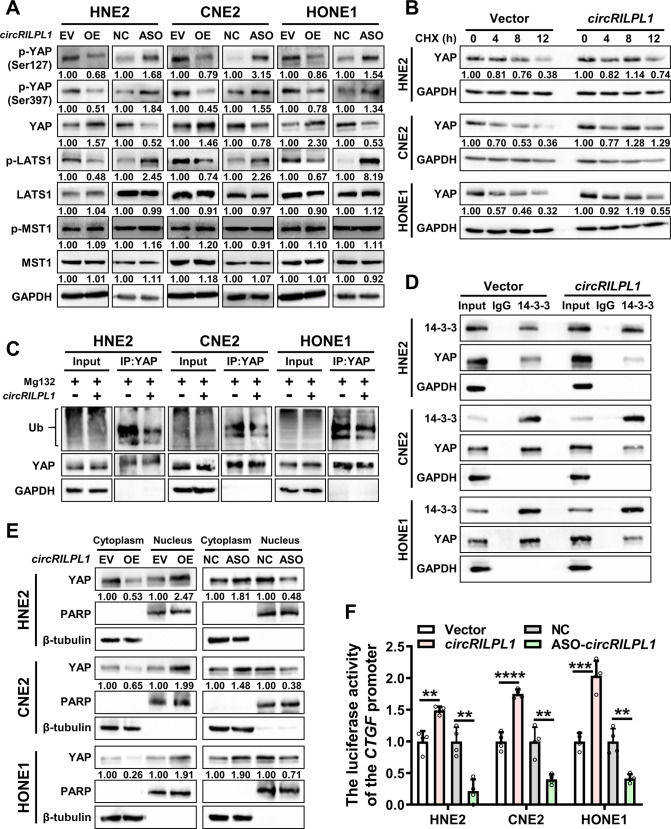
To investigate the molecular mechanism underlying circRILPL1-mediated NPC proliferation and metastasis, the liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) was performed to identify differentially expressed proteins upon overexpression of circRILPL1 in HNE2 cells (Table S2). DAVID bioinformatics site (https://david.ncifcrf.gov/) was used to classify the function categories of differentially expressed proteins and the data showed that 11 proteins were enriched in the Hippo signaling pathway (Table S3). Dysregulation of the Hippo pathway leads to cancer development. The core of the Hippo pathway consists of a kinase cascade, kinases MST1/2 and LATS1/2, as well as their downstream effectors such as transcriptional coactivators YAP and TAZ [27, 28]. Phosphorylation of Ser127 and Ser397 inactivates YAP as Ser397 phosphorylation triggering its ubiquitination and proteasomal degradation, whereas Ser127 phosphorylation mediating the interaction with 14-3-3 proteins and cytoplasmic retention [29]. Western blotting confirmed that the phosphorylation levels of YAP (Ser127 and Ser397), and phosphorylation level of LATS1 in NPC cells were decreased by overexpression of circRILPL1, concomitant with the increase of total YAP protein level. In contrast, knockdown of circRILPL1 in NPC cells exerted opposite effects on phosphorylated YAP, phosphorylated LATS1 and total YAP. CircRILPL1 had no significant effect on MST1 (Fig. 4A). While the mRNA expression levels of them were not significantly altered (Fig. S6A). Cycloheximide (CHX) treatment and ubiquitination assays confirmed that circRILPL1 inhibited the ubiquitin-mediated degradation of YAP and increased its protein stability, leading to the upregulation of YAP protein level (Fig. 4B, C and Fig. S6B). Moreover, immunoprecipitation assay showed that circRILPL1 inhibited the binding of YAP to 14-3-3 (Fig. 4D). Cytosolic/nuclear protein fractionation assay and immunofluorescence staining further showed that circRILPL1 enhanced the abundance of YAP in nucleus while reduced the amount of YAP in cytoplasm, consistent with the reduction of binding of YAP to 14-3-3 by circRILPL1 and also suggesting that circRILPL1 may function as a regulator for YAP-mediated transcription (Fig. 4E, Fig. S6C). The CTGF luciferase reporter assay revealed that overexpression of circRILPL1 significantly enhanced the transcriptional activity of YAP, while knockdown of circRILPL1 had the opposite effects (Fig. 4F). Furthermore, qRT-PCR assays indicated that circRILPL1 upregulated the mRNA levels of YAP target genes, including CTGF, cysteine-rich angiogenic inducer 61 (CYR61), and ankyrin repeat domain 1 (ANKRD1) (Fig. S6D). IHC data also confirmed that the expression of YAP was higher in the circRILPL1 overexpression group but lower in the circRILPL1 knockdown group in subcutaneous xenograft tumors and lung metastases (Fig. S6E).
Fig. 4. CircRILPL1 activates the Hippo-YAP signaling pathway.
A The effect of circRILPL1 on the expression level of key proteins of the Hippo signaling pathway was examined by western blotting in NPC cells. B The stability of YAP protein was measured in NPC cells after overexpression of circRILPL1. Cells were treated with cycloheximide (CHX, 50 μg/mL) for 0, 4, 8, 12 h and western blotting was used to measure the expression of YAP protein. The experiments were repeated three times. C The effect of circRILPL1 on the ubiquitination level of YAP protein was detected in NPC cells treated with MG132 for 12 h after overexpression of circRILPL1 plasmid for 48 h. Cell lysates were subjected to immunoprecipitation using anti-YAP antibody followed by western blotting using anti-ubiquitin antibody. D The interaction between YAP and 14-3-3 proteins in NPC cells after overexpression of circRILPL1 was detected by immunoprecipitation using anti-14-3-3 antibody, followed by western blotting using YAP antibody. GAPDH was used as a negative control. E The abundance of YAP protein in nucleus and cytoplasm was examined in NPC cells after overexpression or knockdown of circRILPL1. PARP was used as a nuclear marker and β-tubulin as a cytoplasmic marker. F The result of CTGF luciferase reporter assay showed that circRILPL1 increased the transcriptional activity of YAP in NPC cells. Data were presented as the means ± SD. *p < 0.05, **p < 0.01, ****p < 0.0001.
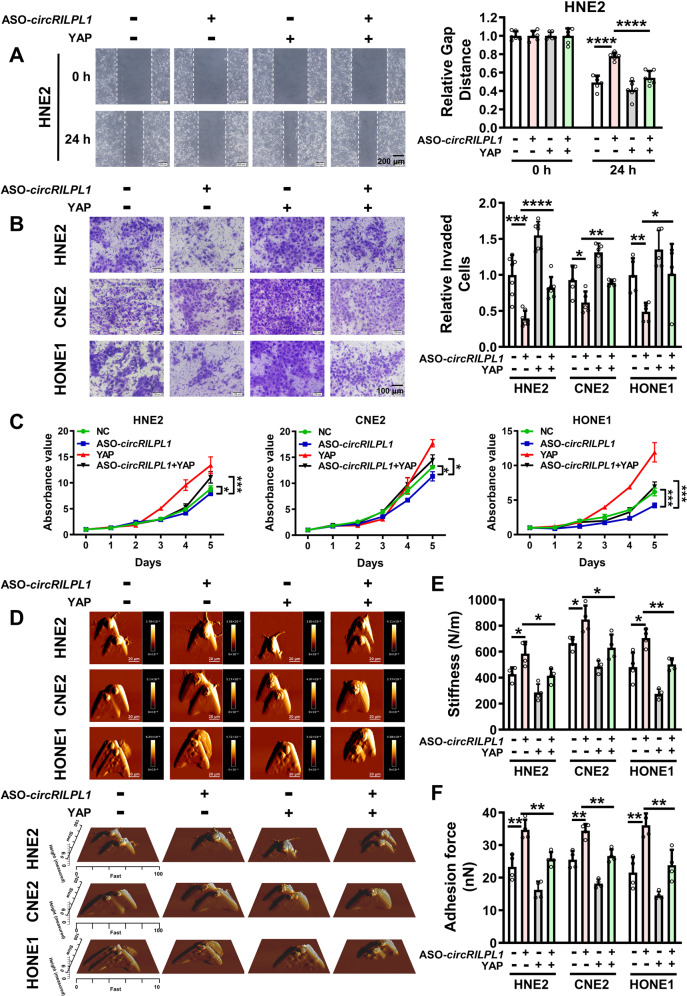
We further verified whether YAP participated in circRILPL1-mediated proliferation and migration of NPC cells. Overexpression of YAP partially restored the inhibitory effect of circRILPL1 knockdown on migration (Fig. 5A and Fig. S7A, B), invasion (Fig. 5B), and proliferation (Fig. 5C, Fig. S7C) of NPC cells. AFM assays revealed that overexpression of YAP partially reversed the circRILPL1 knockdown-induced changes of cell biophysical properties (Fig. 5D–F). Additionally, knockdown of YAP significantly impaired the promotive effects of circRILPL1 overexpression on migration (Fig. S7D), invasion (Fig. S7E), and proliferation (Fig. S7F, G) in NPC cells. Alterations of mechanical properties induced by circRILPL1 overexpression in NPC cells were also reversed by knockdown of YAP (Fig. S7H–J). These data suggested that the function of circRILPL1 in NPC cells was dependent on activation of the YAP signaling pathway.
Fig. 5. CircRILPL1 regulates NPC cells migration, invasion, proliferation and mechanical properties through YAP signaling.
Wound healing assay (A), transwell assay (B), MTT assay (C), and AFM assay (D–F) were performed to assess the effects of YAP on NPC cells migration, invasion, proliferation and biophysical properties that regulated by circRILPL1. All experiments were performed in at least triplicate samples. Data were presented as the means ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
CircRILPL1 relieves LATS1-mediated YAP inhibition by binding to ROCK1
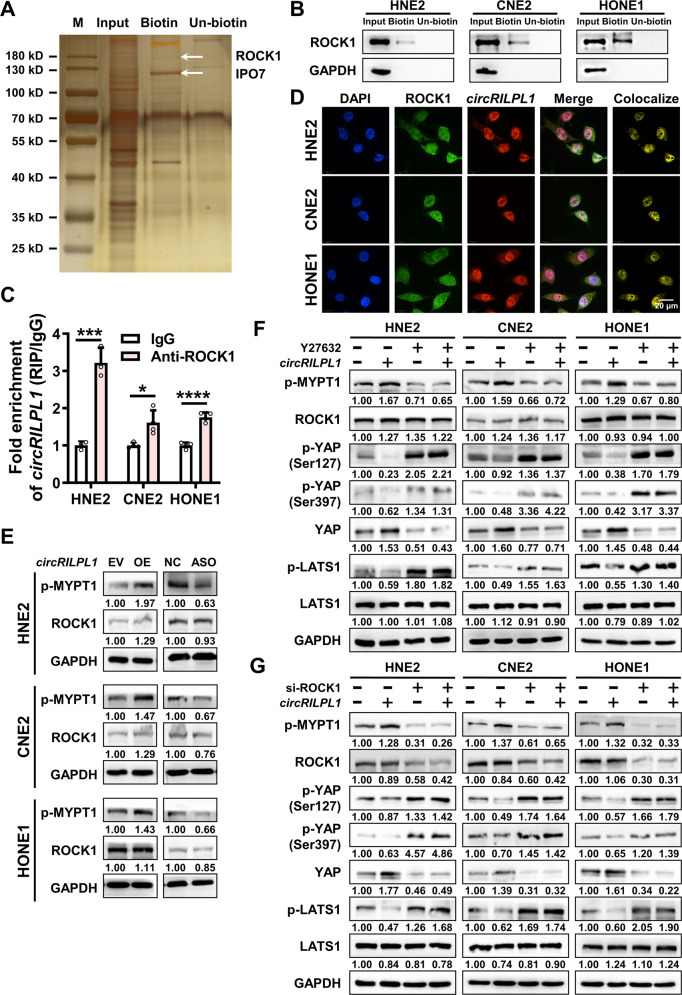
To explore the mechanism by which circRILPL1 regulates the Hippo-YAP signaling pathway, RNA pull-down assay was performed and the result showed that circRILPL1 did not directly bind to LATS1 or YAP (Fig. S8A, B). Then, the RNA pull-down product was further analyzed by mass spectrometry and a total of 69 peptides were identified (Fig. 6A, Table S4). Among these interacting candidates, Rho associated coiled-coil containing protein kinase 1 (ROCK1) was selected for further analysis as it plays essential roles in cytoskeleton remodeling and cell polarity regulation, contributing to cancer cells proliferation and metastasis, while actin and cytoskeleton regulate YAP activity by stimulating of LATS1/2 kinase [30, 31]. RNA pull-down assay confirmed the binding of circRILPL1 to ROCK1 (Fig. 6B). On the other hand, RIP assay showed that circRILPL1 was significantly enriched in the precipitated complex using anti-ROCK1 antibody compared to IgG control (Fig. 6C). The co-localization of circRILPL1 and ROCK1 was demonstrated by IF-FISH (Fig. 6D). The above data indicated that there was an interaction between circRILPL1 and ROCK1.
Fig. 6. CircRILPL1 inhibits the LATS1-YAP kinase cascade by binding to ROCK1.
A The proteins pulled-down by circRILPL1 were separated by the SDS-PAGE, followed by silver staining and identification by LC-MS/MS. Biotin: biotinylated probe targeting the circRILPL1 back-spliced site; Un-biotin: unbiotinylated probe, as control. Bands corresponding to ROCK1 (158 kDa) and IPO7 (119 kDa) were marked. B The interaction between circRILPL1 and ROCK1 protein was detected in NPC cells using RNA pull-down assay with a biotin-labeled circRILPL1 probe. GAPDH was used as a negative control. C The interaction between circRILPL1 and ROCK1 protein was detected in NPC cells by RIP assay using anti-ROCK1 antibody. The fold enrichment of circRILPL1 was relative to that of the control IgG. Data were presented as the means ± SD. *p < 0.05, ***p < 0.001, ****p < 0.0001. D The co-localization between circRILPL1 and ROCK1 protein was detected by IF-FISH. DAPI: blue; ROCK1: green; CircRILPL1: red; Scale bar = 20 μm. E The effects of circRILPL1 on MYPT1 (Thr696) phosphorylation and ROCK1 protein were measured by western blotting in NPC cells after overexpression or knockdown of circRILPL1. F. The expressions of p-MYPT1 (Thr696), ROCK1, p-YAP (Ser127 and Ser397), YAP, p-LATS1, and LATS1 were measured in the circRILPL1 overexpressed NPC cells treated with the ROCK1 inhibitor Y27632 (10 uM) for 2 h by western blotting. G The expressions of p-MYPT1 (Thr696), ROCK1, p-YAP (Ser127 and Ser397), YAP, p-LATS1, and LATS1 were measured in NPC cells after co-transfection of the circRILPL1 overexpression plasmid and ROCK1 siRNA by western blotting.
To investigate the role of ROCK1 in circRILPL1 regulated pathway, phosphorylation of Thr 696 on MYPT1, a well-known substrate of ROCK1 [32], was examined in NPC cells using western blotting after overexpression or knockdown of circRILPL1. The data showed that circRILPL1 promoted the phosphorylation of MYPT1 (Fig. 6E), but had no effect on the expression of ROCK1 at the protein and mRNA levels (Fig. 6E, Fig. S8C). Treatment with ROCK1 inhibitor Y27632 or siRNA targeting ROCK1 in NPC cells significantly inhibited dephosphorylation of LATS1, which in turn inhibited dephosphorylation of YAP and resulted in decrease of YAP protein level (Fig. S8D, E). When Y27632 or ROCK1 siRNA were used in circRILPL1-overexpressing NPC cells, the dephosphorylation of LATS1 and YAP, and the stability of YAP induced by circRILPL1 were significantly reduced (Fig. 6F, G). These results suggested that circRILPL1-mediated ROCK1 activation was critical for the regulation of LATS1-YAP kinase cascade. Transwell and MTT assays demonstrated that knockdown of ROCK1 partially reversed the promoting effects of circRILPL1 overexpression on the invasion and proliferation capabilities of NPC cells (Fig. S8F, G). Together, these data demonstrated that ROCK1, as an important downstream effector of circRILPL1, is essential for the activation of YAP and promoting the proliferation and migration of NPC cells.
CircRILPL1 promotes YAP protein nuclear translocation by binding to IPO7
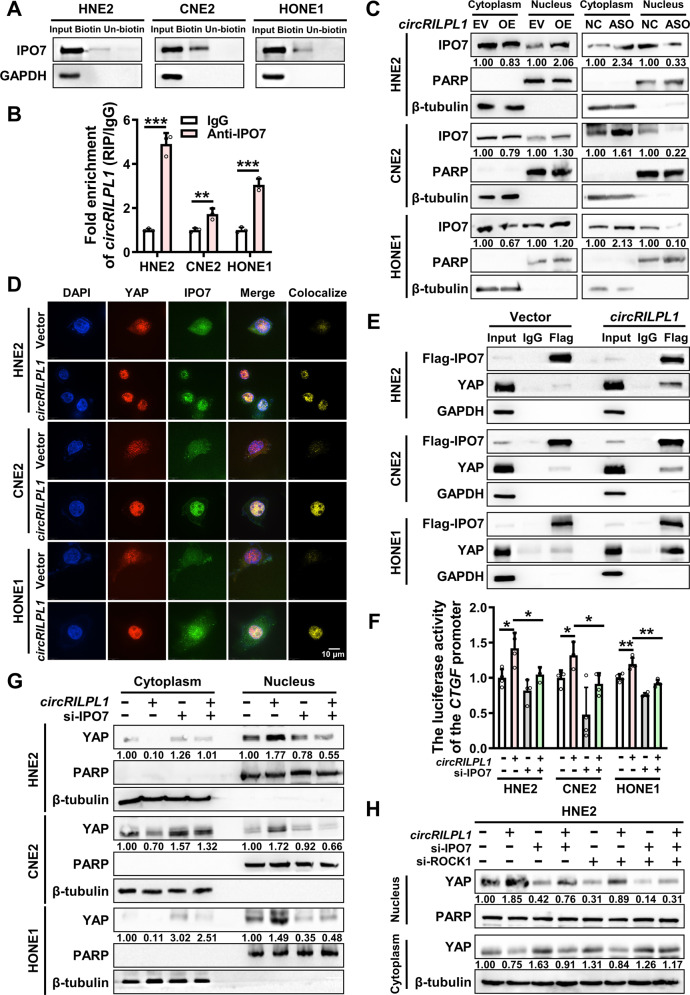
According to mass spectrometry analysis, we noticed that importin 7 (IPO7), a nuclear transport receptor [33, 34], was pulled down by circRILPL1. The interaction between circRILPL1 and IPO7 was further confirmed by RNA pull-down, RIP assay, and IF-FISH assays (Fig. 7A, B and Fig. S9A). Immunoprecipitation assays showed that ROCK1 did not bind to IPO7 (Fig. S9B). To test whether circRILPL1 regulated the nuclear import of IPO7, cytosolic/nuclear protein fractionation and immunofluorescence were performed and the results showed that overexpression of circRILPL1 facilitated the entry of IPO7 into nucleus and knockdown of circRILPL1 reduce its entry (Fig. 7C, Fig. S9C). Results of qRT-PCR and western blotting showed that circRILPL1 had no effect on the protein and mRNA expression of IPO7 (Fig. S9D, E). In addition, a weak interaction existed between endogenous IPO7 and YAP. But IPO7 did not regulate the expression and phosphorylation of YAP (Fig. S9F, G). Immunoprecipitation and immunofluorescence assay showed that the binding between IPO7 and YAP was significantly enhanced following overexpression of circRILPL1 (Fig. 7D, E). These results suggested that circRILPL1 promoted the binding between IPO7 and YAP proteins. Notably, knockdown of IPO7 hindered circRILPL1-induced translocation of YAP from the cytoplasm to the nucleus and inhibited YAP transcriptional activity (Fig. 7F, G and Fig. S9H), indicating that circRILPL1 increasing the YAP nuclear import was partially IPO7-dependent. And this inhibition was more pronounced after simultaneous knockdown of IPO7 and ROCK1 (Fig. 7H).
Fig. 7. CircRILPL1 promotes YAP nuclear translocation through mediating its binding to IPO7.
A The binding between circRILPL1 and IPO7 protein was detected in NPC cells by RNA pull-down with biotin-labeled circRILPL1 probe. GAPDH was used as a negative control. B The binding between circRILPL1 and IPO7 protein was examined in NPC cells using anti-IPO7 antibody by RIP assay. The fold enrichment of circRILPL1 is relative to that of the control IgG. C CircRILPL1 promotes the nuclear translocation of IPO7 in NPC cells as examined by cytosolic/nuclear protein fractionation. PARP was used as a nuclear marker and β-tubulin as a cytoplasmic marker. D. Co-localization of YAP and IPO7 in NPC cells after overexpression of circRILPL1. DAPI: blue; YAP: red; IPO7: green; Scale bar = 10 μm. E The effect of circRILPL1 on the interaction between YAP and IPO7 was detected by immunoprecipitation using anti-Flag (IPO7) antibody, followed by western blotting using YAP antibody in NPC cells transfected with Flag-IPO7 plasmid, co-transfected with circRILPL1 overexpression plasmid or empty vector. GAPDH was used as a negative control. F The transcriptional activity of YAP was measured by luciferase reporter assay after overexpression or knockdown of circRILPL1 or IPO7 in NPC cells. G Knockdown of IPO7 blocked the nuclear import of YAP induced by circRILPL1 in NPC cells. PARP was used as a nuclear marker and β-tubulin as a cytoplasmic marker. H The effects of knockdown of IPO7 or ROCK1 on circRILPL1-induced YAP nuclear translocation in HNE2 cells. Data were presented as the means ± SD. *p < 0.05, **p < 0.01, ***p < 0.001.
CircRILPL1 interacts with ROCK1 and IPO7 through nucleotides 136-189
Using catRAPID (http://s.tartaglialab.com/page/catrapid_group) and RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) bioinformatics tools, two well-characterized stem loop structures (136–189 nt and 251–291 nt) on circRILPL1 were predicted to bind with ROCK1 and IPO7 proteins (Fig. S10A, B). RNA pull-down and RIP assays showed that the deletion mutant DEL1 lack of 136–189 nt failed to bind with ROCK1 and IPO7 proteins, whereas deletion mutant DEL2 lack of 251–291 nt still possessed the binding ability, suggesting that the 136–189 nt region was essential for circRILPL1 binding with ROCK1 and IPO7 proteins (Fig. S10C, D). Functionally, the deletion mutant circRILPL1-DEL1 had no effect on the migration and proliferation of HNE2 cells (Fig. S10E, F).
To further illustrate whether nucleotides 136–189 is critical for regulating YAP activation by circRILPL1, we overexpressed wild-type circRILPL1 or the deletion mutant circRILPL1-DEL1 in HNE2 cells. The data showed circRILPL1-DEL1 failed to activate the Hippo-YAP signaling pathway (Fig. S11A) and could not stabilize and promote nuclear translocation of YAP protein (Fig. S11B, C), when compared with wild-type circRILPL1. Also, circRILPL1-DEL1 failed to promote the activation of ROCK1 (Fig. S11D) and lost the potential to facilitate the binding between YAP and IPO7 (Fig. S11E). Wound healing and MTT assays showed that wild-type circRILPL1, rather than the mutant circRILPL1-DEL1, promoted the migration and proliferation of HNE2 cells (Fig. S11F, G). Collectively, these data further confirmed that binding with ROCK1 and IPO7 through nucleotides 136–189 is crucial for circRILPL1 to promote tumor progression in NPC cells.
The expression levels of YAP, ROCK1 and IPO7 in the NPC clinical samples were detected by IHC assays, and the results showed that YAP and ROCK1 protein levels were higher in NPC tissues when compared to that in NPE tissues, while levels of IPO7 protein in NPC and NPE tissues had no difference (Fig. S12A, B).
CircRILPL1-YAP signaling promotes the transcription of CAPN2 and PXN
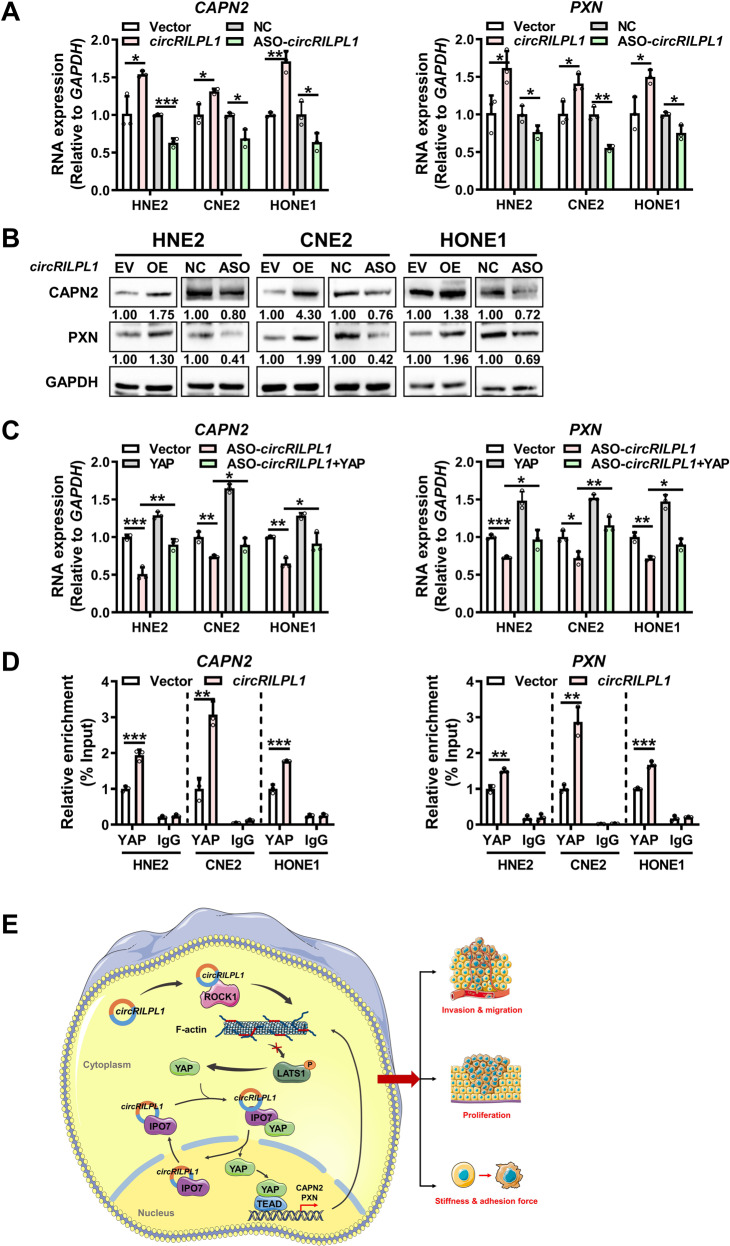
To search for the key downstream effectors activated by the circRILPL1-YAP axis in NPC cells, those upregulated proteins identified by mass spectrometry analysis following overexpression of circRILPL1 and associated with cytoskeleton remodeling and malignant behaviors of cancers were further analyzed (Table S5). Results of qRT-PCR and western blotting confirmed that circRILPL1 up-regulated the mRNA and protein expression of CAPN2 and PXN in NPC cells (Fig. 8A, B). Overexpression of YAP promoted the transcription of CAPN2 and PXN, while knockdown of YAP inhibited their transcription (Fig. S13A, B). Overexpression of YAP partially reversed the decrease of CAPN2 and PXN mRNA induced by circRILPL1 knockdown (Fig. 8C). ChIP experiment showed that circRILPL1 prompted the enrichment of YAP on the CAPN2 and PXN promoters in NPC cells (Fig. 8D). These results suggested that circRILPL1 promoted the transcription of CAPN2 and PXN through activation of YAP.
Fig. 8. CircRILPL1-YAP signaling promotes the transcription of CAPN2 and PXN.
A The expression levels of CAPN2 and PXN mRNA were examined by qRT-PCR in NPC cells after overexpression or knockdown of circRILPL1. B The expressions of CAPN2 and PXN protein were examined by western blotting in NPC cells after overexpression or knockdown of circRILPL1. C The effects of circRILPL1 and YAP on the expressions of CAPN2 and PXN mRNA in NPC cells were detected by qRT-PCR. D The effect of circRILPL1 on the enrichment of YAP on CAPN2 and PXN promoters was examined by ChIP assay. E Schematic diagram of the signaling pathways regulated by circRILPL1 in NPC cells. CircRILPL1 inhibits the LATS1 kinase through binding to and activating ROCK1, thereby suppressing the phosphorylation of YAP. In cooperation with the nuclear transport receptor IPO7, circRILPL1 prompts loading of YAP onto IPO7 and thus enhances YAP translocation into nucleus, where YAP activates the transcription of cytoskeleton remodeling genes CAPN2 and PXN. CircRILPL1 activates the Hippo-YAP signaling pathway, ultimately resulting in biomechanical differences, proliferation, and metastasis of NPC cells. Data were presented as the means ± SD. *p < 0.05, **p < 0.01, ***p < 0.001.
In summary, circRILPL1, as a tumor activator, bound to and activated ROCK1 to inhibit the LATS1 kinase, which led to reduction of YAP phosphorylation at Ser127 and Ser397 sites. Binding and cooperating with nuclear transport receptor IPO7, circRILPL1 promoted YAP nuclear translocation by enhancing its interaction with IPO7. CircRILPL1 activated the Hippo-YAP signaling pathway to promote the transcription of CAPN2 and PXN and NPC progression (Fig. 8E).
Discussion
CircRNAs are a new class of non-coding RNAs with covalently closed structure that have attracted much attention recently. CircRNAs regulate the occurrence and progression of a large variety of cancers [35]. For example, circRNF13 directly binds to and stabilizes SUMO2 mRNA and promotes GLUT1 degradation through SUMOylation and ubiquitination, thereby inhibiting the glycolysis [36]. EBV-encoded CircBART2.2 promotes the expression of PD-L1 in NPC cells by binding with RIG-I and activating the transcription factors IRF3 and NF-κB, leading to tumor immune escape [37]. In this study, circRILPL1, which is derived from reverse splicing of exons 3 and 4 of RILPL1 gene, was identified to be elevated in NPC tissues. RILPL1 regulates cell morphology and polarity, and studies have shown that RILPL1 plays an important role in LRRK2-mediated pathogenic interference with centrosomal cohesion and ciliogenesis [38, 39]. Previous studies showed that circRILPL1 acts as a sponge for miR-145 to activate IGF1R/PI3K/AKT signaling, thus promoting bovine muscle proliferation [40]. However, the role of either RILPL1 or circRILPL1 in tumors has not been reported. Our study confirmed for the first time that circRILPL1 promotes the proliferation and metastasis of NPC.
By using proteomic approaches, we found proteins involved in Hippo signaling pathway were significantly perturbed by overexpression of circRILPL1 in NPC cells. The Hippo pathway is an evolutionarily conserved signaling pathway that regulates cell morphology, proliferation, and migration in response to extracellular stimuli such as mechanical sensation or cell density. The activity of YAP, a core transcription factor of the Hippo pathway, is directly regulated by its upstream kinase LATS1/2 [41]. Phosphorylation of Ser397 (annotated as Ser381 by Zhao et al. [42]) by LATS1/2 triggers β-TrCP-mediated YAP ubiquitination and proteasomal degradation, while phosphorylation of YAP at Ser127 is essential for binding of 14-3-3 and cytoplasmic retention [43]. The roles of Hippo signaling pathway in NPC development are largely unknown. Our data showed that circRILPL1 activated the Hippo-YAP signaling pathway through stabilizing YAP and facilitating its nuclear translocation, thereby transcriptionally activating CAPN2 and PXN, which are functional genes involved in cytoskeleton-remodeling and cancer metastasis. The mechanical properties of cancer cells are influenced by the dynamic assembly of the cytoskeleton, which reflects their potential of malignant transformation, proliferation, and migration. Cells with different degrees of malignancy have different biophysical properties such as stiffness and adhesion tendency [44, 45]. By employing AFM, we accurately measured biophysical features of cancer cells, including adhesion force and stiffness. Our work demonstrated that circRILPL1 reduced the adhesion and stiffness of NPC cells, making them prone to invasion and migration, ultimately promoting the malignant progression of NPC.
In addition, we found that circRILPL1 regulates the Hippo-YAP signaling pathway by binding with ROCK1 and IPO7 proteins. As a downstream effector of Rho, ROCK1 phosphorylates and regulates downstream protein kinases including MYPT, ERM, and LIMK to modulate actin polymerization. Actin and cytoskeleton remodeling stimulates LATS1/2 kinase activity, leading to YAP phosphorylation [22, 46]. Xin Zhou et al. demonstrated that GPER stimulation activated YAP and TAZ via the Gαq-11, PLCβ/PKC, and Rho/ROCK signaling pathways, leading to actin reorganization in breast cancer [47]. Yang XM et al. found that RACGAP1 inhibited Hippo signaling and activated YAP in HCC cells by increasing the activity of RhoA, leading to polymerization of filamentous actin [48]. SPON2 regulates the activities of RhoA and Rac1 through unique integrin signaling, affecting the F-actin reorganization, LATS1-Hippo pathway, and liver cancer cell migration [49]. Here we discovered that circRILPL1 modulated the LATS1-YAP kinase cascade through binding to and activating ROCK1, resulting in decreased YAP phosphorylation. Of course, the exact mechanism by which ROCK1 inhibits the kinase activity of LATS1 still needs to be further investigated.
IPO7 functions in protein nuclear import through acting as a nuclear transport receptor or as an adapter-like protein in association with the importin-beta subunit KPNB1. FOXM1 induced nuclear import and transcription of GLI1 by promoting the expression of IPO7, which contributes to cells proliferation and invasion in glioma [50]. KRT19 promotes the nuclear localization of Egr1 by mediating the binding between Egr1 and IPO7, thereby negatively regulating Akt signaling pathway in breast cancer [34]. Many proteins were discovered to shuttle between the nucleus and the cytoplasm in response to cellular stress signals [51, 52]. María García-García et al. newly found that IPO7 drives nuclear import of YAP in response to mechanical signals, whereas YAP governs the mechanoresponse of Imp7 by forming a YAP/Imp7 complex in response to mechanical cues via the kinases MST1/2 [53]. IPO7 usually binds to protein substrates containing nuclear localization signal (NLS) [54, 55]. A conserved bipartite NLS was identified in the N-terminal region of NUAK1, which is responsible for its nuclear import [56]. However, many proteins lacking the typical NLS have also been reported to undergo nuclear translocation. For instance, the sequence (Ser-Pro-Ser, SPS) in ERKs is phosphorylated upon stimulation. The phosphorylated ERK then binds to IPO7 and leads to its nuclear translocation [57]. In addition, IPO7 has been reported to be responsible for nuclear import of plasmid DNA and human mitochondrial DNA, suggesting that IPO7 may represent a key molecule for nuclear import of DNA [58–60]. In this study, circRILPL1 was demonstrated to bind with IPO7 and facilitate the interaction between YAP and IPO7, thereby promoting the nuclear translocation of YAP. Our study extended the understanding of non-coding RNA-mediated YAP activation during cancer progression. It would be of great interest to unveil the exact mechanism by which circRILPL1 facilitates IPO7 nuclear translocation.
Collectively, we characterized a circRNA circRILPL1 playing a pivotal oncogenic role in NPC progression through synergically regulating the activation of YAP and its nuclear translocation by binding to both ROCK1 and IPO7. This finding highlights the diagnostic and therapeutic potential of targeting circRILPL1 in NPC, uncovers a direct interaction between a circRNA and signaling proteins, and emphasizes the significance of circRNA functions in NPC proliferation and metastasis.
Materials and methods
Patient samples
Two cohorts of NPC tissue samples were collected at Hunan Cancer Hospital, Changsha. The first cohort included 38 NPC tissues and 16 chronic nasopharyngeal inflammation tissues (Table S6) which were used for quantitative real-time polymerase chain reaction (qRT-PCR). The second cohort included 99 NPC tissues and 46 adjacent non-NPC tissues (Table S7) for in situ hybridization (ISH). This study was approved by the Joint Ethics Committee of Central South University, and informed consents were obtained from all participants.
Cell culture and cell transfection
Cell lines (HNE2, CNE2, and HONE1) were obtained from the Cell Center of Central South University, and cultured in RPMI-1640 (Gibco, USA) containing 10% fetal bovine serum (Gibco, USA) at 37 °C and 5% CO2.
The full-length circRILPL1 (hsa_circ_0007552) was amplified by PCR and then inserted into pcDNA3.1(+) circRNA Mini Vector, which was a gift from Professor Li Yong at Baylor College of Medicine. The YAP cDNA was cloned into pcDNA3.1(+). The construct for overexpression of IPO7 (IPO7 pcDNA3.1-3xFlag-C) was purchased from YouBio Company (Changsha, China). All constructs were confirmed by sequencing. Antisense oligonucleotides (ASO) specifically targeting circRILPL1, siRNAs targeting RILPL1, YAP, IPO7, and ROCK1, and the controls were purchased from RiboBio Co., Ltd. (Guangzhou, China). The sequences for siRNA were listed in Table S8. Neofect (Neofect biotech Co., Ltd. China) was used for plasmid transfection. Hiperfect (Qiagen, Hilden, Germany) was used to transfect ASOs and siRNAs. The ROCK1 inhibitor Y-27632 (Selleck, Shanghai, China) was used in cells with a concentration of 10 uM.
RNA extraction and qRT-PCR
Total RNA was extracted from cells or tissues using TRIzol (Life, USA). Reverse transcription was performed using the HiScript cDNA Synthesis kit (Vazyme, Nanjing, China). Then, qRT-PCR was performed on CFX96™ Real-Time PCR Detection System using 2×SYBR Green qPCR Master Mix (Bimake, USA). All primers used were listed in Table S8.
Northern blot
10–20 μg of RNA was electrophoresed in a 1.5% formaldehyde-agarose gel in MOPS buffer. RNA was transferred to Hybond-N + membranes (Amersham, UK) with 10 x SSC buffer by capillary transfer. After UV cross-linking and fixation, the membranes were hybridized with digoxigenin-labelled circRILPL1 or RILPL1 mRNA probes (Table S8) at 65 °C for 16–18 h and detected by Anti-Digoxigenin-AP (Roche). Then the signal was developed with the chemiluminescent substrate NBT/BCIP.
Western blotting
RIPA buffer (Beyotime Biotechnology, Shanghai, China) and Protease Inhibitor cocktail (Roche Applied Sciences, Mannheim, Germany) were used for cell lysis and protein extraction. Protein samples (30–50 μg) were separated via 10–12% SDS-PAGE and transferred onto 0.2 μm PVDF membrane (Millipore, Billerica, MA, USA). After blocking with 5% nonfat milk for 1 h, the membrane was incubated with primary and secondary antibodies sequentially following the manufacturer’s instruction. After washing with tris-buffered saline (TBS) or phosphate-buffered saline (PBS) supplemented with 0.1% Tween 20 for 3 times, the target protein bands were detected using the ECL detection system (Millipore, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. All antibodies used are listed in Table S9.
RNase R and actinomycin D treatment
Actinomycin D (Sigma, USA) was added into NPC cells at a final concentration of 2 μg/ml, and RNA was collected at 0 h, 8 h, 16 h, and 24 h for qRT-PCR. For RNase R (RNR07250, Epicentre, USA) digestion, 20 U/μl RNase R was incubated with RNA extracted from NPC cells at 37 °C for 30 min, and then the enzyme was inactivated by heating at 70 °C for 10 min.
Cytosolic/nuclear fraction assay
Cytosolic and nuclear RNAs were isolated using the PARIS™ Protein and RNA Isolation System (Invitrogen, USA) following to the manufacturer’s instructions. Cytosolic and nuclear proteins were separated using the NE-PER Nuclear and Cytoplasmic Extraction Reagent (Thermo Scientific, USA).
Immunohistochemistry (IHC) and in situ hybridization (ISH)
IHC was done using an immunohistochemical kit (KIT-9720, MXB Biotechnologies, Fuzhou, China). For ISH, digoxigenin-labeled circRILPL1-specific probes were synthesized by Sangon Biotech (Shanghai, China) and the expression of circRILPL1 in NPC tissue was detected specimens using the Enhanced Sensitive ISH Detection kit I (POD) (MK1030, BOSTER, China) following the manufacturer’s instruction. The probes used were listed in Table S8. All sections were independently scored by two pathologists who were blind to the clinicopathological features of the samples (Table S7). A semi-quantitative scoring criterion was used based on the staining intensity and the proportion of positive cells. When the tissue was not stained, scoring 0. When the tissue was pale yellow, scoring 1; light brown, scoring 2; dark brown, scoring 3. On the other hand, when less than 25% of cells were positive, scoring 0; 25% - 50% positive, scoring 1; 50%–75% positive, scoring 2; and more than 75% positive, scoring 3. Finally, a comprehensive score was calculated as the product of the staining intensity score and the positive ratio score. Scores greater than 5 were determined as high expression; others were considered as low expression.
RNA fluorescence in situ hybridization (FISH)
Cells were fixed with 4% paraformaldehyde and penetrated with 0.1% Triton-100. After incubation with pre-hybridization solution for 2–4 h at 37 °C, cells were incubated with digoxigenin-labeled circRILPL1-specific probes (Sangon Biotech, China) at 37 °C overnight. After washing with SSC buffer, cells were incubated with biotinylated mouse anti-digoxigenin antibody for 1 h. Red fluorescence-labeled mouse secondary antibody (LIFE, USA) was added at a dilution ratio of 1:200, and further incubated in the dark at 37 °C for 1 h. DAPI (Invitrogen, USA) was added to counterstain the nuclei. Cells were imaged and analyzed using confocal microscope Ultra-View Vox (Perkin-Elmer, USA).
Wound healing and transwell assays
For wound healing assay, transfected cells were plated in 6-well plateand scratched evenly using sterilized mini tips. Then cells in each well were photographed and counted at 0 h and 24 h. For transwell assay, the diluted Matrigel (BD, Shanghai, China) was added to the upper chamber of Transwell chamber (Millipore, USA). Then transfected cells were added to the upper chamber and RPMI-1640 medium containing 20% FBS was added to the bottom chamber. Two days later, cells in the bottom chamber were photographed and counted using an inverted phase-contrast microscope after fixation and staining with 0.1% crystal violet.
MTT and colony formation assays
For MTT assay, 800 transfected cells were seeded in 96-well plates with 5 replicates in each group. After cell adhesion, 20 ul MTT (Beyotime, China) was added and incubated in the dark for 4 h at 37 °C. Then, the plate was scanned at 490 nm using a microplate reader from day 0 to day 6. For colony formation assay, 2,000 transfected cells were seeded in 12-well plate and cultured for a week. The colonies in each well were photographed and counted after washing with PBS, fixation with 4% paraformaldehyde, and staining with 0.1% crystal violet.
Atomic Force Microscopy (AFM)
AFM (JPK NanoWizard 4 BioScience, JPK Instruments, Germany) was used to measure the biophysical properties of NPC cells. The cells were fixed in 2% glutaraldehyde for 45 s, 4% paraformaldehyde for 20 min, and then washed and maintained in appropriate amount of PBS for AFM scanning. The probe HYDRA6V-100NG (AppNano, CA, USA) with a spring constant of 0.292 N/m was used in the experiments. The indentation process was performed at a loading and retraction rate of approximately 2.5 μm/s with an indentation depth of at least 1 mm. Images were captured under QI mode and analyzed with JPK software to obtain data on cell adhesion, stiffness, and Rq.
Immunofluorescence (IF)
Cells were fixed with pre-warmed 4% paraformaldehyde, penetrated with 0.1% Triton-100, and blocked with 5% calf serum albumin for 30 min at room temperature. Then cells were incubated with primary antibodies at 4 °C overnight. After washing with PBS, they were incubated with fluorescently labeled secondary antibody (LIFE, USA) at 37 °C for 1 h. Images were captured with laser confocal microscope Ultra-View Vox (Perkin-Elmer, USA) after counterstaining the nuclei with DAPI.
Luciferase reporter assay
Cells were co-transfected with the YAP luciferase reporter plasmid and the pRL-TK plasmid (which expresses Renilla luciferase as an internal control) after overexpression or knockdown of circRILPL1. The luciferase activity was measured using the Dual-Luciferase® Reporter Assay System (E1910, Promega, USA) 48 hours later. Relative luciferase activity was obtained by normalizing with the Renilla luciferase activity.
RNA immunoprecipitation (RIP) and chromatin immunoprecipitation (ChIP)
RIP was performed to analyze the interaction between circRILPL1 and ROCK1 or IPO7 using the Magna RIPTM Kit (17-701, Millipore, USA) according to the manufacturer’s instruction. ChIP was performed using the ChIP Assay Kit (P2078, Beyotime, China) following the manufacturer’s instruction.
RNA pull-down
After transfection of biotin-labeled circRILPL1 probes for 24 hours, cells were lysed with the RIP buffer (150 mM KCl, 25 mM Tris-HCl, 0.5 mM DTT, 0.5% NP40) and incubated with 50 ul of Streptavidin Dynabeads (M-280, Invitrogen, USA) overnight at 4 °C with rotation. Then the RNA-protein complexes were examined by western blotting.
Immunoprecipitation
Cell lysates were incubated with antibodies (or the control IgG) and 50 ul of protein A/G magnetic beads (Bimake, Houston, Texas, USA) overnight at 4 °C with rotation. After washing with GLB+ buffer (10 mM NaCl, 10 mM Tris-HCl, 10 mM EDTA, 0.5% Triton-100) for 3 times, the precipitates were analyzed by western blotting.
Liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS)
To identify circRILPL1 interacting proteins, cell lysates were incubated with biotin-labeled probes and biotin-affinity magnetic beads overnight at 4 °C to pull-down circRILPL1-associated proteins. The purified proteins were separated by SDS-PAGE gels and followed by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) using Ultimate 3000 RSLC Nano System (Dionex, CA, USA) coupled with LTQ Orbitrap Velos Pro mass spectrometer (Thermo Scientific). Whole proteomic analysis was performed by searching the UniProt KB/Swiss-Prot database using the Proteome Discoverer 1.4 software. A fold change of ≥1.68 or ≤0.62 was used to define differentially expressed proteins. DAVID bioinformatics site (https://david.ncifcrf.gov/) was used to classify the function categories of differentially expressed proteins.
Animal experiments
Female BALB/C nude mice were randomly divided into four groups. CNE2 cells (2 × 106) transfected with the vector, the circRILPL1 overexpression plasmid, antisense oligonucleotides against circRILPL1 (ASO-circRILPL1) or the scramble negative control were injected subcutaneously, via tail vein, or footpad, respectively. For the subcutaneous tumor model, mice (6 per group) were sacrificed 30 days after inoculation. The size of the subcutaneous tumor nodule was measured every 5 days and histological examinations were done after 30 days of inoculation. For the tail vein-lung metastasis model, mice (6 per group) were sacrificed 8 weeks after inoculation. The number and area of lung surface metastatic nodules in each mouse were recorded. The lungs were removed, imaged, and embedded in paraffin. Then, the tissues were sectioned for hematoxylin-eosin (H&E) staining, ISH experiments, and metastatic evaluation. For footpad-lymph node metastasis, mice (6 per group) were sacrificed 28 days after inoculation and the ipsilateral inguinal lymph nodes were excised for analysis. All experimental protocols involving animals were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee of Central South University (Changsha, China).
Statistical analysis
Statistical analysis was performed using the GraphPad Prism 8.0. Student’s t-test (two-tailed) to compare the difference between two groups of data. All data were represented as mean ± standard deviation (SD). P value < 0.05 was considered statistically significant.
Supplementary information
Acknowledgements
We thank Prof. Yong Li for providing pcDNA3.1(+) CircRNA Mini Vectors.
Author contributions
BX conceived and designed the project. PW completed the majority of experiments and wrote the manuscript. XH, MP, XD, QY, CF, YM, YW, ZL performed some of the experiments. FW, CG, MZ, ZZ and GL revised the manuscript. QL, HW and WJ collected tissue samples. WX and BX is responsible for research supervision and funding acquisition. All authors read and approved the final manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (U21A20382, 82072374, 82002239, and 82272631), the Natural Science Foundation of Hunan Province (2023JJ30732 and 2021JJ41043).
Data availability
All data that support the findings of this study are available from the corresponding authors upon reasonable request.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Approval for the use of clinical samples and information was obtained from each patient and the Research Ethics Committee of Central South University. Written informed consent was received prior to patient participation. The animal experiments were conducted according to the protocol approved by the Animal Welfare Committee of Central South University.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-023-01171-8.
References
- 1.Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet. 2019;394:64–80. doi: 10.1016/S0140-6736(19)30956-0. [DOI] [PubMed] [Google Scholar]
- 2.Wong KCW, Hui EP, Lo KW, Lam WKJ, Johnson D, Li L, et al. Nasopharyngeal carcinoma: an evolving paradigm. Nat Rev Clin Oncol. 2021;18:679–95. doi: 10.1038/s41571-021-00524-x. [DOI] [PubMed] [Google Scholar]
- 3.Chua MLK, Wee JTS, Hui EP, Chan ATC. Nasopharyngeal carcinoma. Lancet. 2016;387:1012–24. doi: 10.1016/S0140-6736(15)00055-0. [DOI] [PubMed] [Google Scholar]
- 4.Xiong W, Zeng ZY, Xia JH, Xia K, Shen SR, Li XL, et al. A susceptibility locus at chromosome 3p21 linked to familial nasopharyngeal carcinoma. Cancer Res. 2004;64:1972–4. doi: 10.1158/0008-5472.CAN-03-3253. [DOI] [PubMed] [Google Scholar]
- 5.Kang Y, He W, Ren C, Qiao J, Guo Q, Hu J, et al. Advances in targeted therapy mainly based on signal pathways for nasopharyngeal carcinoma. Signal Transduct Target Ther. 2020;5:245. doi: 10.1038/s41392-020-00340-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin DC, Meng X, Hazawa M, Nagata Y, Varela AM, Xu L, et al. The genomic landscape of nasopharyngeal carcinoma. Nat Genet. 2014;46:866–71. doi: 10.1038/ng.3006. [DOI] [PubMed] [Google Scholar]
- 7.Zeng Z, Huang H, Zhang W, Xiang B, Zhou M, Zhou Y, et al. Nasopharyngeal carcinoma: advances in genomics and molecular genetics. Sci China Life Sci. 2011;54:966–75. doi: 10.1007/s11427-011-4223-5. [DOI] [PubMed] [Google Scholar]
- 8.Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA. 2015;6:563–79. doi: 10.1002/wrna.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He AT, Liu J, Li F, Yang BB. Targeting circular RNAs as a therapeutic approach: current strategies and challenges. Signal Transduct Target Ther. 2021;6:185. doi: 10.1038/s41392-021-00569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–8. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng X, et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol Cancer. 2020;19:22. doi: 10.1186/s12943-020-1147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19:188–206. doi: 10.1038/s41571-021-00585-y. [DOI] [PubMed] [Google Scholar]
- 14.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–91. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 15.Tang L, Xiong W, Zhang L, Wang D, Wang Y, Wu Y, et al. circSETD3 regulates MAPRE1 through miR-615-5p and miR-1538 sponges to promote migration and invasion in nasopharyngeal carcinoma. Oncogene. 2021;40:307–21. doi: 10.1038/s41388-020-01531-5. [DOI] [PubMed] [Google Scholar]
- 16.Fan C, Qu H, Xiong F, Tang Y, Tang T, Zhang L, et al. CircARHGAP12 promotes nasopharyngeal carcinoma migration and invasion via ezrin-mediated cytoskeletal remodeling. Cancer Lett. 2021;496:41–56. doi: 10.1016/j.canlet.2020.09.006. [DOI] [PubMed] [Google Scholar]
- 17.Yang M, Huang W. Circular RNAs in nasopharyngeal carcinoma. Clin Chim Acta. 2020;508:240–8. doi: 10.1016/j.cca.2020.05.029. [DOI] [PubMed] [Google Scholar]
- 18.Zhou DN, Ye CS, Yang QQ, Deng YF. Integrated analysis of transcriptome profiling predicts potential lncRNA and circRNA targets in human nasopharyngeal carcinoma. Oncol Lett. 2020;19:3123–36. doi: 10.3892/ol.2020.11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–57. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 20.Dey A, Varelas X, Guan KL. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat Rev Drug Discov. 2020;19:480–94. doi: 10.1038/s41573-020-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narumiya S, Tanji M, Ishizaki T. Rho signaling, ROCK and mDia1, in transformation, metastasis and invasion. Cancer Metastasis Rev. 2009;28:65–76. doi: 10.1007/s10555-008-9170-7. [DOI] [PubMed] [Google Scholar]
- 22.Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–83. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 24.Deng X, Xiong F, Li X, Xiang B, Li Z, Wu X, et al. Application of atomic force microscopy in cancer research. J Nanobiotechnology. 2018;16:102. doi: 10.1186/s12951-018-0428-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho FC, Tham IW, Earnest A, Lee KM, Lu JJ. Patterns of regional lymph node metastasis of nasopharyngeal carcinoma: a meta-analysis of clinical evidence. BMC Cancer. 2012;12:98. doi: 10.1186/1471-2407-12-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong X, Liu N, Liang Y, He Q, Yang X, Lei Y, et al. Circular RNA CRIM1 functions as a ceRNA to promote nasopharyngeal carcinoma metastasis and docetaxel chemoresistance through upregulating FOXQ1. Mol Cancer. 2020;19:33. doi: 10.1186/s12943-020-01149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13:877–83. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the Roots of Cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Q, Wang M, Hu Y, Zhao E, Li J, Ren L, et al. MYBL2 disrupts the Hippo-YAP pathway and confers castration resistance and metastatic potential in prostate cancer. Theranostics. 2021;11:5794–812. doi: 10.7150/thno.56604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pocaterra A, Santinon G, Romani P, Brian I, Dimitracopoulos A, Ghisleni A, et al. F-actin dynamics regulates mammalian organ growth and cell fate maintenance. J Hepatol. 2019;71:130–42. doi: 10.1016/j.jhep.2019.02.022. [DOI] [PubMed] [Google Scholar]
- 32.Boyle ST, Poltavets V, Kular J, Pyne NT, Sandow JJ, Lewis AC, et al. ROCK-mediated selective activation of PERK signalling causes fibroblast reprogramming and tumour progression through a CRELD2-dependent mechanism. Nat Cell Biol. 2020;22:882–95. doi: 10.1038/s41556-020-0523-y. [DOI] [PubMed] [Google Scholar]
- 33.Yao X, Chen X, Cottonham C, Xu L. Preferential utilization of Imp7/8 in nuclear import of Smads. J Biol Chem. 2008;283:22867–74. doi: 10.1074/jbc.M801320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ju JH, Yang W, Lee KM, Oh S, Nam K, Shim S, et al. Regulation of cell proliferation and migration by keratin19-induced nuclear import of early growth response-1 in breast cancer cells. Clin Cancer Res. 2013;19:4335–46. doi: 10.1158/1078-0432.CCR-12-3295. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Mo Y, Gong Z, Yang X, Yang M, Zhang S, et al. Circular RNAs in human cancer. Mol Cancer. 2017;16:25. doi: 10.1186/s12943-017-0598-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mo Y, Wang Y, Zhang S, Xiong F, Yan Q, Jiang X, et al. Circular RNA circRNF13 inhibits proliferation and metastasis of nasopharyngeal carcinoma via SUMO2. Mol Cancer. 2021;20:112. doi: 10.1186/s12943-021-01409-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ge J, Wang J, Xiong F, Jiang X, Zhu K, Wang Y, et al. Epstein-Barr virus-encoded circular RNA circBART2.2 promotes immune escape of nasopharyngeal carcinoma by regulating PD-L1. Cancer Res. 2021;81:5074–88. doi: 10.1158/0008-5472.CAN-20-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobu Y, Wawro PS, Dhekne HS, Yeshaw WM, Pfeffer SR. Pathogenic LRRK2 regulates ciliation probability upstream of tau tubulin kinase 2 via Rab10 and RILPL1 proteins. Proc Natl Acad Sci USA. 2021;118:e2005894118. doi: 10.1073/pnas.2005894118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lara Ordonez AJ, Fernandez B, Fdez E, Romo-Lozano M, Madero-Perez J, Lobbestael E, et al. RAB8, RAB10 and RILPL1 contribute to both LRRK2 kinase-mediated centrosomal cohesion and ciliogenesis deficits. Hum Mol Genet. 2019;28:3552–68. doi: 10.1093/hmg/ddz201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen X, Tang J, Jiang R, Wang X, Yang Z, Huang Y, et al. CircRILPL1 promotes muscle proliferation and differentiation via binding miR-145 to activate IGF1R/PI3K/AKT pathway. Cell Death Dis. 2021;12:142. doi: 10.1038/s41419-021-03419-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma S, Meng Z, Chen R, Guan KL. The Hippo pathway: biology and pathophysiology. Annu Rev Biochem. 2019;88:577–604. doi: 10.1146/annurev-biochem-013118-111829. [DOI] [PubMed] [Google Scholar]
- 42.Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao T, Wang Z, Fang J, Cheng W, Zhang Y, Huang J, et al. HTLV-1 activates YAP via NF-kappaB/p65 to promote oncogenesis. Proc Natl Acad Sci USA. 2022;119:e2115316119. doi: 10.1073/pnas.2115316119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwon T, Gunasekaran S, Eom K. Atomic force microscopy-based cancer diagnosis by detecting cancer-specific biomolecules and cells. Biochim Biophys Acta Rev Cancer. 2019;1871:367–78. doi: 10.1016/j.bbcan.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 45.Deng X, Xiong W, Jiang X, Zhang S, Li Z, Zhou Y, et al. LncRNA LINC00472 regulates cell stiffness and inhibits the migration and invasion of lung adenocarcinoma by binding to YBX1. Cell Death Dis. 2020;11:945. doi: 10.1038/s41419-020-03147-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang YC, Wu JW, Wang CW, Jang AC. Hippo signaling-mediated mechanotransduction in cell movement and cancer metastasis. Front Mol Biosci. 2019;6:157. doi: 10.3389/fmolb.2019.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou X, Wang S, Wang Z, Feng X, Liu P, Lv XB, et al. Estrogen regulates Hippo signaling via GPER in breast cancer. J Clin Invest. 2015;125:2123–35. doi: 10.1172/JCI79573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang XM, Cao XY, He P, Li J, Feng MX, Zhang YL, et al. Overexpression of Rac GTPase activating protein 1 contributes to proliferation of cancer cells by reducing hippo signaling to promote cytokinesis. Gastroenterology. 2018;155:1233–49.e22. doi: 10.1053/j.gastro.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 49.Zhang YL, Li Q, Yang XM, Fang F, Li J, Wang YH, et al. SPON2 promotes M1-like macrophage recruitment and inhibits hepatocellular carcinoma metastasis by distinct Integrin-Rho GTPase-Hippo pathways. Cancer Res. 2018;78:2305–17. doi: 10.1158/0008-5472.CAN-17-2867. [DOI] [PubMed] [Google Scholar]
- 50.Xue J, Zhou A, Tan C, Wu Y, Lee HT, Li W, et al. Forkhead Box M1 is essential for nuclear localization of glioma-associated oncogene Homolog 1 in Glioblastoma Multiforme cells by promoting Importin-7 expression. J Biol Chem. 2015;290:18662–70. doi: 10.1074/jbc.M115.662882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kodiha M, Chu A, Matusiewicz N, Stochaj U. Multiple mechanisms promote the inhibition of classical nuclear import upon exposure to severe oxidative stress. Cell Death Differ. 2004;11:862–74. doi: 10.1038/sj.cdd.4401432. [DOI] [PubMed] [Google Scholar]
- 52.Stochaj U, Rassadi R, Chiu J. Stress-mediated inhibition of the classical nuclear protein import pathway and nuclear accumulation of the small GTPase Gsp1p. FASEB J. 2000;14:2130–2. doi: 10.1096/fj.99-0751fje. [DOI] [PubMed] [Google Scholar]
- 53.Garcia-Garcia M, Sanchez-Perales S, Jarabo P, Calvo E, Huyton T, Fu L, et al. Mechanical control of nuclear import by Importin-7 is regulated by its dominant cargo YAP. Nat Commun. 2022;13:1174. doi: 10.1038/s41467-022-28693-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen J, Liu MY, Parish CR, Chong BH, Khachigian L. Nuclear import of early growth response-1 involves importin-7 and the novel nuclear localization signal serine-proline-serine. Int J Biochem Cell Biol. 2011;43:905–12. doi: 10.1016/j.biocel.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 55.Panagiotopoulos AA, Polioudaki C, Ntallis SG, Dellis D, Notas G, Panagiotidis CA, et al. The sequence [EKRKI(E/R)(K/L/R/S/T)] is a nuclear localization signal for importin 7 binding (NLS7) Biochim Biophys Acta Gen Subj. 2021;1865:129851. doi: 10.1016/j.bbagen.2021.129851. [DOI] [PubMed] [Google Scholar]
- 56.Palma M, Riffo EN, Suganuma T, Washburn MP, Workman JL, Pincheira R, et al. Identification of a nuclear localization signal and importin beta members mediating NUAK1 nuclear import inhibited by oxidative stress. J Cell Biochem. 2019;120:16088–107. doi: 10.1002/jcb.28890. [DOI] [PubMed] [Google Scholar]
- 57.Chuderland D, Konson A, Seger R. Identification and characterization of a general nuclear translocation signal in signaling proteins. Mol Cell. 2008;31:850–61. doi: 10.1016/j.molcel.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 58.Dhanoya A, Wang T, Keshavarz-Moore E, Fassati A, Chain BM. Importin-7 mediates nuclear trafficking of DNA in mammalian cells. Traffic. 2013;14:165–75. doi: 10.1111/tra.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller AM, Munkonge FM, Alton EW, Dean DA. Identification of protein cofactors necessary for sequence-specific plasmid DNA nuclear import. Mol Ther. 2009;17:1897–903. doi: 10.1038/mt.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zaitseva L, Cherepanov P, Leyens L, Wilson SJ, Rasaiyaah J, Fassati A. HIV-1 exploits importin 7 to maximize nuclear import of its DNA genome. Retrovirology. 2009;6:11. doi: 10.1186/1742-4690-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support the findings of this study are available from the corresponding authors upon reasonable request.