ABSTRACT
Human health/socioeconomic development is closely correlated to environmental pollution, highlighting the need to monitor contaminants in the real environment with reliable devices such as biosensors. Recently, variety of biosensors gained high attention and employed as in-situ application, in real-time, and cost-effective analytical tools for healthy environment. For continuous environmental monitoring, it is necessary for portable, cost-effective, quick, and flexible biosensing devices. These benefits of the biosensor strategy are related to the Sustainable Development Goals (SDGs) established by the United Nations (UN), especially with reference to clean water and sources of energy. However, the relationship between SDGs and biosensor application for environmental monitoring is not well understood. In addition, some limitations and challenges might hinder the biosensor application on environmental monitoring. Herein, we reviewed the different types of biosensors, principle and applications, and their correlation with SDG 6, 12, 13, 14, and 15 as a reference for related authorities and administrators to consider. In this review, biosensors for different pollutants such as heavy metals and organics were documented. The present study highlights the application of biosensor for achieving SDGs. Current advantages and future research aspects are summarized in this paper.
Abbreviations: ATP: Adenosine triphosphate; BOD: Biological oxygen demand; COD: Chemical oxygen demand; Cu-TCPP: Cu-porphyrin; DNA: Deoxyribonucleic acid; EDCs: Endocrine disrupting chemicals; EPA: U.S. Environmental Protection Agency; Fc-HPNs: Ferrocene (Fc)-based hollow polymeric nanospheres; Fe3O4@3D-GO: Fe3O4@three-dimensional graphene oxide; GC: Gas chromatography; GCE: Glassy carbon electrode; GFP: Green fluorescent protein; GHGs: Greenhouse gases; HPLC: High performance liquid chromatography; ICP-MS: Inductively coupled plasma mass spectrometry; ITO: Indium tin oxide; LAS: Linear alkylbenzene sulfonate; LIG: Laser-induced graphene; LOD: Limit of detection; ME: Magnetoelastic; MFC: Microbial fuel cell; MIP: Molecular imprinting polymers; MWCNT: Multi-walled carbon nanotube; MXC: Microbial electrochemical cell-based; NA: Nucleic acid; OBP: Odorant binding protein; OPs: Organophosphorus; PAHs: Polycyclic aromatic hydrocarbons; PBBs: Polybrominated biphenyls; PBDEs: Polybrominated diphenyl ethers; PCBs: Polychlorinated biphenyls; PGE: Polycrystalline gold electrode; photoMFC: photosynthetic MFC; POPs: Persistent organic pollutants; rGO: Reduced graphene oxide; RNA: Ribonucleic acid; SDGs: Sustainable Development Goals; SERS: Surface enhancement Raman spectrum; SPGE: Screen-printed gold electrode; SPR: Surface plasmon resonance; SWCNTs: single-walled carbon nanotubes; TCPP: Tetrakis (4-carboxyphenyl) porphyrin; TIRF: Total internal reflection fluorescence; TIRF: Total internal reflection fluorescence; TOL: Toluene-catabolic; TPHs: Total petroleum hydrocarbons; UN: United Nations; VOCs: Volatile organic compounds
KEYWORDS: Biosensor, bioavailability, sustainable development goals (SDGs), heavy metals, organic pollutants
Graphical abstract
Highlights
Biosensors are robust in both specific and total responses to environmental pollutants
Promising prospects for achieving on-site monitoring and real-time environmental data
Biosensors illustrate a reliable, simple, effective, and fast method for monitoring pollution
Biosensors consume less energy and leave a smaller carbon footprint
High performing biosensors make a solid contribution to SDGs
1. Introduction
Detection and monitoring of pollutants in the environment is crucial for assessing the harmful effects of potential toxicants to people, flora, and fauna [1–3]. Chemical analysis using instruments such as high performance liquid chromatography (HPLC), gas chromatography (GC), and inductively coupled plasma-mass spectrometry (ICP-MS) has been traditionally used for monitoring environmental water or soil samples for their sensitivity and accuracy [4]. To further understand the bioavailability of environmental pollutants, biosensors using biological sensor to detect such pollutants have been invented, and they have features of low-cost, energy-saving, and feasibility for real-time in situ monitoring [4,5]. Different types of biosensors were successfully applied for specific or nonspecific detection of heavy metals [6,7] and organic pollutants [1,8]. In groundwater and river water samples, the bioavailable and toxic fractions of metals and organic pollutants such as Cd and toluene could be successfully evaluated using biosensors [9,10]. Moreover, bioavailability of the pollutants such as Hg and phenanthrene was revealed by the use of biosensors [11,12], suggesting the useful application of biosensors on bioavailability assessment of pollutants in the environmental samples.
Biosensor is an analytical strategy for pollutants based on biotechnology, and consists of elements for signal transducer producing detectable or quantifiable signals when sensing pollutants [6,13]. Types of biosensor include cell-free-based and whole-cell-based, nonspecific and specific ones according to the biological elements and sensing ability [14]. The bioavailability and toxic effects can be established by whole-cell-based biosensor compared with a cell-free one [14,15]. For example, microorganisms possess various responsive mechanisms to cope with environmental stress, organic pollutants, and heavy metals, which are associated with different regulatory genes and proteins. The Cd detecting protein cadC regulator could be used in biosensor for Cd detection [16]. In addition, the benzene metabolizing regulatory protein would be useful in recognizing benzene-like organic pollutants [17]. Those regulatory systems interacting with environmental pollutants are the key factors for distinguishing specific pollutants from others [16,17]. However, pollutants with similar structure might hamper the selectivity of biosensors, such as the benzene regulatory protein targeting toluene, ethylbenzene, toluene, and xylene [17]. Therefore, the choice of regulatory systems or recognizing elements would greatly affect the selectivity of specific biosensors. Currently, biosensors targeting heavy metals, pesticides, or pharmaceuticals show the advantages of biosensor such as portability, ease of use, and saving time [18,19]. Recent studies have highlighted the advances made in sensitivity, stability, selectivity, and their contribution to environmental monitoring [1,8].
It has been shown that in clinical laboratories, the consumption of electricity, water, and gas as well as the waste produced are the key factors generating large carbon footprints [20]. Energy use was also the major contributor to greenhouse gases (GHGs) emissions [21,22]. It is suggested that potential carbon reduction could be achieved by partially using biosensors in pollutants analysis due to their low energy requirements. One review has suggested that the contribution of using biosensor for human health on SDGs [23]. However, discussions about the application of biosensor on environmental monitoring and its relationship between sustainable development goals (SDGs) are scarce. Biosensors technology is anticipated to be powerful tool for monitoring the environmental pollutants for its advantages on sustainable development. In this article, different types of biosensors, principles of biosensor construction as well as the application and performance are reviewed. The relationship between biosensors and SDGs is also noted. This review will also help to achieve the SDGs since biosensors play a significant role to achieve the SDG 6, 12, 13, 14, and 15.
2. Types of biosensors
2.1. Cell-free biosensors
The general mechanism of biosensors for environmental monitoring is displayed in Figure 1. The different types of biosensors and the application on SDGs achievement were shown in Figure 2. The sensing elements of cell-free biosensors include deoxyribonucleic acid (DNA), protein, and aptamer. DNA-based biosensors could be used for monitoring heavy metals such as arsenic and lead made possible by changes in the specific DNA structure and oxidative damage [14,24]. Sulfur on the protein structure reacts easily with toxicants like arsenic and the resulting inhibition of enzyme activities can help detect toxic metals like arsenic [25,26]. Other enzyme such as acetylcholinesterase could also be used in inhibition-based biosensor, which is contributed from the binding of organic P pesticides with acetylcholinesterase inhibiting the further signal producing from the reaction of acetylcholinesterase [27]. However, the response of DNA and protein would involve other toxic substances and might lack selectivity to specific pollutants. To enhance the selectivity of target chemicals, aptamer, an artificial single-strand DNA or ribonucleic acid (RNA), was developed for specific pollutants detection based on selective bonding [28]. Currently, aptamer-based biosensors coupled with nanomaterials have been employed in several studies for detecting heavy metals and organic pollutants such as pesticides [29].
Figure 1.
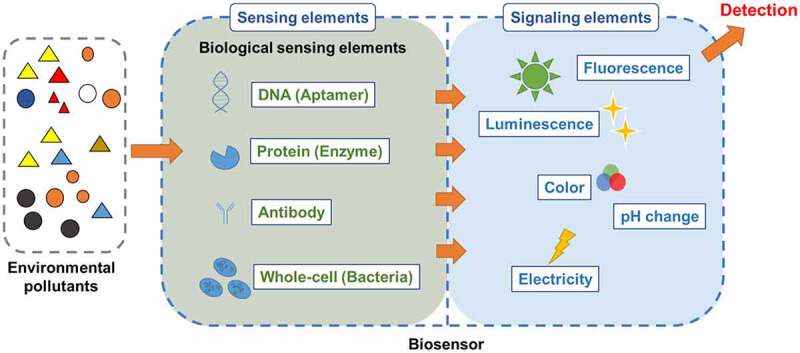
General mechanisms of biosensor for pollutants detection. The basic construction of biosensor contains sensing elements and signaling elements. The biological-sensing elements including DNA (aptamer), protein (enzyme), antibody, and whole-cell (bacteria) are able to recognize the environmental pollutants such as heavy metals and organic pollutants. The signaling elements would be triggered by sensing elements and produce the different signals such as fluorescence, luminescence, color, pH change, or electricity that could be measured or detected by the operators.
Figure 2.
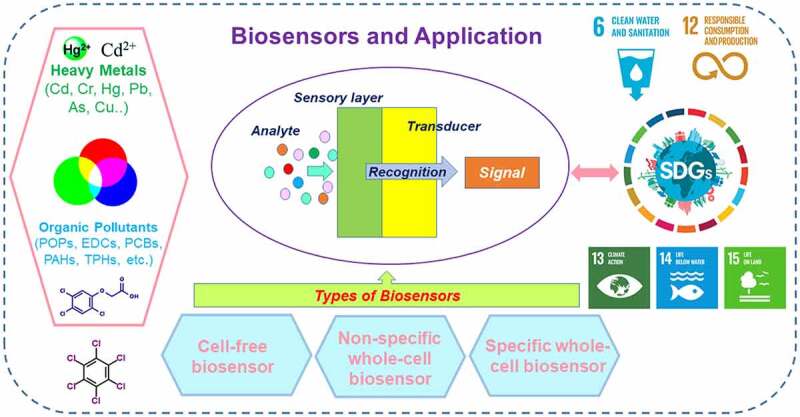
The types of biosensors and the applications on SDGs achievements. The different environmental pollutants including heavy metals and organic pollutants could trigger the sensing, transducing, and signaling of biosensors. The different types of biosensors include cell-free biosensor, nonspecific whole-cell biosensor, and specific whole-cell biosensor, which is categorized mainly by their sensing elements and selectivity. Those biosensors detecting the environmental pollutants could help achieving SDG 6, 12, 13, 14, 15.
2.2. Nonspecific whole-cell biosensors
Nonspecific whole-cell biosensors are constructed mainly by stress responsive genetic regulation, such as heat shock and DNA damage responses [6]. Information about bioavailability and toxicity can be revealed by nonspecific cell-based biosensors. The damage done to protein will trigger heat shock stress-related genes, which could be used as a sensing element of nonspecific cell-based biosensors for pollutants that damage protein [30–32]. The expression of SOS response genes enhanced by DNA damage could be incorporated in nonspecific sensing elements [33–36]. Indigenous bacteria and their activity as far as environmental stress is concerned could also serve as a nonspecific whole-cell biosensor. For example, sulfur oxidizing and iron oxidizing abilities in certain types of bacteria would be inhibited by toxic pollutants and could be used for acid samples [37–39]. As well, indigenous bacterium isolated from soil environments was successfully engineered with green fluorescent protein (GFP) signaling gene, which could help assess bioavailable heavy metals in the soils by the GFP signals [40]. The nonspecific biosensor based on the stress response provides an early warning of hazard presence in samples that could be harmful for microorganisms despite the lack of selectivity.
2.3. Specific whole-cell biosensors
Specific whole-cell biosensors are mostly based on the metabolism or detoxification genes in microorganisms [8,41]. For organic pollutants, toluene and xylene can be metabolized by toluene-catabolic (TOL) plasmid in bacteria harboring those regulatory genes [42]. The xylR and xylS genes on the TOL plasmid can serve in biosensors specifically designed for benzene-related compounds [43–45]. Other operons such as nah, alkBAC, and DntR were applied in a specific cell-based biosensor for naphthalene, linear alkanes, and pesticides [46–49]. For heavy metals, the sensing elements of specific cell-based biosensor belong to resistance genes instead of metabolized genes, which are located in the plasmid in microorganisms that could survive in toxic metal-rich environments [50]. Expression of the resistance genes including redox transformation, active transport, and intracellular/extracellular precipitation is regulated by intracellular metal level [51]. Both toxic metals and excessive essential metals could trigger the response of resistance or detoxification genes [52,53]. The arsR or arsD in ars operon for arsenic [14], the cadC/A in cad operon for cadmium [54], the merR in mer operon for mercury [6], and the chrB in chr operon for chromium were used in specific cell-based biosensors [55]. Cell-based biosensors for cadmium and mercury were able to detect other metal ions, such as copper, zinc, or lead, which reduces their selectivity [6,54]. zntA and copA promoter/operator evaluated the bioavailability of Zn and Cu [56]. zntA accounts for the removal of intracellular Zn and responds to Cd and Pb [57,58]. copA is responsible for the concentration of Cu and Ag in cells [59]. Regulatory proteins of zntA and copA belong to the MerR homologue, thereby making them responsible for Hg [60,61]. The cnrYXH gene was used to detect the bioavailability of Co and Ni in soil [62]. In addition, the Pb(II)-binding regulator PbrR in MerR family has the ability to recognize lead after being engineered in whole-cell biosensor [63]. The characteristics of biosensors of different types are summarized in Table 1. In addition, the advantages of biosensors on SDGs achievement are displayed in Figure 3.
Table 1.
Characteristics of various type biosensors.
| Biosensors | Selectivity | Response time | Sensitivity | Bioavailability |
|---|---|---|---|---|
| Cell-free biosensor | High | Fast | High | No |
| Nonspecific whole-cell biosensor | Low | Medium | Medium | Yes |
| Specific whole-cell biosensor | High | Medium | Medium | Yes |
Figure 3.
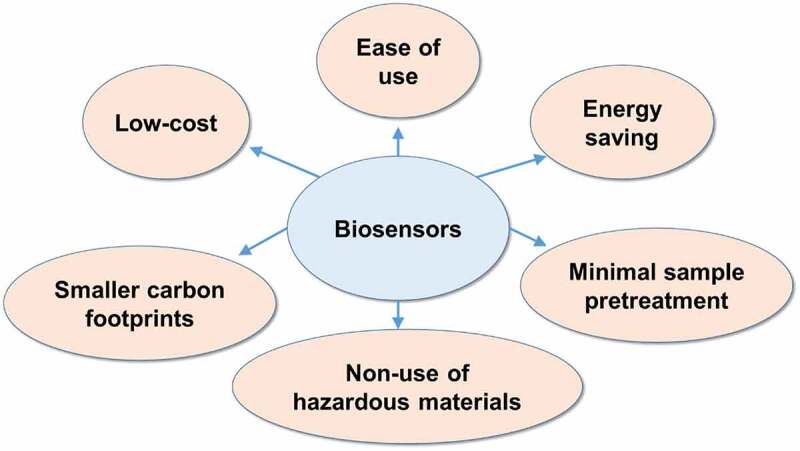
The advantages of biosensors on SDGs achievements. Several advantages of biosensors include low-cost, ease of use, saving energy, smaller carbon footprints, nonuse of hazardous materials, and minimal sample pretreatment compared with traditional physicochemical methods. These advantages are helpful to achieving SDGs such as SDG 6, 12, 13, 14, and 15, which relate to clean water, responsible consumption and production, climate action, and marine and terrestrial life.
3. Principle of whole-cell biosensor construction
3.1. Toxicity-based nonspecific biosensor
The mechanisms of nonspecific biosensors based on toxicity include Microtox and adenosine triphosphate (ATP)-bioluminescence system, bacterial functions such as iron and sulfur oxidizing, and DNA damage or heat shock protein-related genes. The Microtox test has been devised using the bacterium Vibrio fischeri whose bioluminescence would diminish due to the toxicity and disrupted metabolism [64,65]. The mechanisms of bioluminescence-based inhibitory biosensors might be associated with the ATP reduction resulted from stress responsive mechanisms or cell death resulted from excessive environmental stress, which further decreased the bioluminescence by lower ATP and cell viability [17,66]. It is worth noting that the genes of bioluminescence can be genetically engineered in other lab-cultivable bacteria such as E. coli. Similarly, the constitutively expressed bioluminescence of these toxicity-based nonspecific biosensors would diminish upon the exposure of toxicants, thereby making them capable of quantifying the general toxicity equivalent of multiple pollutants [66]. As well, the ATP level reflected by bioluminescence could serve as an indicator of toxicity and metabolic disruption.
The modified Saccharomyces cerevisiae with substrate luciferin could serve as a reporter system based on ATP-bioluminescence for detecting nonspecific toxicity [67]. The metabolic cost might increase for stress response when the bacterial functions are inhibited by environmental pollutants, thereby reducing the amount of products of iron and sulfur oxidizing [37–39]. For example, the pH decrease and conductivity increase resulted from sulfur oxidation was inhibited by the toxicants [37], which might be due to the bacterial death or disrupted microbial functions. Based on the inhibition level, toxicity could be quantified by using iron and sulfur oxidizing bacteria [37–39]. The recA and its promoter responsible for DNA repair and maintenance were widely used in DNA damage-sensing biosensors for their response to genotoxic compounds and pharmaceuticals [34,68,69]. Recent studies have applied both recA-based and metabolic inhibition-based bioluminescence bacterial biosensor for pollutants such as toxic metals in urban rivers and seawater [70,71]. Similarly, the stress-inducible heat shock genes like hsp70 and its promoter were used in biosensors for detecting nonspecific toxicity since the heat shock protein systems will be triggered not only by heat shock but also other environmental stressors, and then induce the downstream fused signals [72,73].
3.2. Metabolic inhibition-based bioluminescence bacterial biosensors
The metabolic inhibition-based bioluminescence bacterial biosensors have been developed and utilized in several pollutants detection [74]. Different signaling elements producing bioluminescence such as luciferase were used in the construction of bacterial biosensor, or naturally presented in bacteria that could be directly used in sensing [74]. The luciferase genes from bacteria and firefly have been suggested to be highly sensitive and broadly used as a reporter gene. The luxCDABE from bacterial operon is conservative in many bacteria species [75]. The luxAB is enough for producing the luminescence, while using the whole luxCDABE operon could reduce the amount of additional substrates that is needed for luminescence reaction [15]. The protein from luxAB is responsible for catalyzing long-chain aldehydes shown in the reaction below:
FMNH2 + R – CHO + O2 → FMN + H2O + RCOOH + hν (490 nm)
The lucFF gene from the firefly requires luciferin, O2, and ATP for bioluminescence [76]:
Firefly luciferin + O2 + ATP → oxyluciferin + AMP + Pi + hν (550–575 nm)
The bioluminescence would diminish in the presence of environmental pollutants due to the inhibited bioluminescence reaction. The ATP-dependent bioluminescence could be used in developing the metabolic inhibition-based biosensors for it can reflect the ATP decrease caused by environmental toxicants [67,74]. On the other hand, the energy-consuming lux operon reaction in Vibro spp. could also reflect the metabolic state of the microorganisms [66]. Therefore, the bioluminescence biosensors with lux operon such as Microtox (Vibrio fischeri) were applied in assessing the toxicity of environmental pollutants in river and sediment samples [71,77,78]. In addition to Vibro spp., the recombinant Escherichia coli harboring the lux operon could successfully detect the environmental pollutants, suggesting the feasibility of lux operon engineered in other bacterial species that could be easily cultured [8,79].
3.3. Microbial fuel cell-based biosensors
Microbial fuel cell (MFC)-based biosensors with single- or dual-chamber design were developed based on the oxidation of organic compounds and the corresponding electrical currents [80,81]. The dual chambers of MFC-based biosensors consist of an anaerobic anodic chamber where the oxidation happens and aerobic cathodic chamber, transferring electrons through circuits and protons through the ion-exchange membrane from anode to cathode [82]. MFC-based biosensors can detect the water quality and wastewater parameters such as biological oxygen demand (BOD) and toxins [81]. Compared with nonspecific whole-cell biosensors, MFC-based biosensors save more energy and are portable; they produce the electrical current and do not need signal transducers [82]. It has been reported that heavy metals such as copper and arsenic in wastewater were detected by MFC-based biosensor based on the toxicity associated inhibition [83]. The bacteria strains for organics degradation could be mixed culture from activated sludge, and the toxicants in wastewater could be sensed by the inhibited degradation [82]. In addition to pollutant-degrading bacteria, the electroactive bacteria such as Geobacter and Shewanella were shown to facilitate the current production in MFC-based biosensor [84]. Other bacterial strains in MFC-based biosensors for sensing specific pollutants were also reported in recent studies. For example, the linear alkylbenzene sulfonate could be simultaneously quantified and degraded dominantly by Pseudomonas species, generating electrical power using a MFC-based biosensor [85]. To increase the selectivity for specific detection, Cr(VI)-reducing bacterium Exiguobacterium aestuarii YC211 was cultured in a MFC-based biosensor system [86]. It can detect in situ Cr(VI) in industrial wastewater via the electrical currents generated from the reduction of Cr(VI) to Cr(III) [86], which suggests the possibility of specific detection and indigenous bacterium application in MFC-based biosensors. In addition, the genetically engineered E. coli was developed in MFC-based biosensors for detecting Zn(II) and Cu(II), which is accomplished by the recombining metal-sensing promoter, and the synthesis genes of riboflavin and porin [87,88]. The riboflavin and porin are responsible for facilitating electron transfer and improving cell membrane permeability, respectively [87,88].
3.4. Genetic modification using detoxification and metabolism genes
To detect specific pollutants, genetic methods using detoxification and metabolism genes to differentiate environmental pollutants have been developed. For organic compounds, the metabolic and utilization mechanisms and their regulatory genes were mainly used [8]. Recent research has combined genes and hosts from different bacterial species and used AtzR regulatory protein to detect cyanuric acid [89]. Biodegradable organic pollutants in the whole-cell biosensors trigger metabolic mechanisms and the downstream utilizing proteins or signals such as bioluminescence [8,90]. Subsequently, it is difficult to detect biodegradable organic pollutants since the metabolism mechanisms are unclear in the microorganisms. However, recent research has developed a whole-cell biosensor to detect persistent polybrominated diphenyl ethers (PBDEs) using genome-wide screening. It can find regulatory networks in the microorganisms [91], shedding light on the application of biosensors on organic pollutants that can only be degraded with difficulty.
For heavy metals, the resistance and detoxification mechanisms were applied in the sensing elements of specific whole-cell biosensors [92]. The regulatory systems consisted of regulatory protein that could bind metals and promoter regions that trigger downstream detoxification or signaling responses [92,93]. For example, arsR and the promoter region from Geobacter sulfurreducens could be genetically combined with a fluorescence reporter, usable for arsenic screening with the detection limit down to the ppb level [41]. Basically, the detoxification and metabolism operons were activated when the toxicants permeated in the cells, but it has been suggested that the two-component regulatory systems could be incorporated in bacterial biosensors for detecting extracellular pollutants [94]. The regulatory membrane protein has the ability to bind pollutants and then trigger downstream phosphorylation of the other regulator proteins that activate the signal transduction [94].
4. Application of biosensor to environmental monitoring
4.1. Heavy metals monitoring
Heavy metals pollution is usually discharged as a result of anthropogenic and industrial activities (e.g., refineries, metal works, mining, cement factories, smelting plants, etc.), and their pollution endangers human health and the environment [95–97]. Elements such as Fe, Cu, Mn, and Zn, then highly toxic Pb, Cr, Cd, As, Hg, etc, are very resistant to biodegradation. Heavy metals are transported into the environment, especially water sources and easily absorbed by living organisms. Due to their pronounced toxicity, these heavy metals are important for monitoring the environment (water, wastewater, air, solid waste, and organisms) [95,97,98]. Monitoring water contamination is essential for environmental conservation and the prevention of illnesses. Due to the accumulation in the environment (e.g., water, soil, sediment) and wildlife such as plants, animals over a long period of time, heavy metals pose a serious danger. Several methods have been devised to detect their concentrations and presence in the environmental matrix. Especially, biosensors can detect the levels of heavy metals and determine how much pollution they cause. Biosensors can easily detect the presence of heavy metals in order to regulate and manage water safety and quality.
The DNA probe is used as the element recognition to detect heavy metal elements/ions and is based on the principles as follows. The first is the formation of a stable DNA-duplex due to the selective binding of specific DNA bases with heavy metal ions; for the second, heavy metal ions aid in the breaking DNAzymes (deoxyribozymes), and finally, the guanine-rich probe switches to a stable G-quadruplex structure [99,100]. Similar to the design for heavy metals detection in environmental monitoring, microbial whole-cell biosensors detect concentrations based on the genetic elements that respond to chemical species. Their effectiveness is determined by the regulatory protein types connected with these promoters, as well as the reporter genes for transcriptional pollutants control [101,102]. A reporter gene in living cells (i.e., microorganisms) is employed as a sensor to convert its biological response into detectable physicochemical signals.
The whole-cell biosensors illustrate a prospective technique for the detection/monitoring of heavy metals and are critical to selectivity and sensitivity [70,100,103]. Using biosensors for continuous and/or individual detection and measurements normally depends on the biologically active element types. Also, biosensors (i.e., organisms) can be integrated into pollutants exposure and used as a method for predicting chemical pollution in the environment. The literature review demonstrated that Shen et al. [104] designed a toxicity MFC system for detecting Cu concentrations of 5–7 mg L–1 in a quick speedy response, and Wu et al. [105] also found at a level of 12 mg L–1 in their study. Recently, by using the decrease in the cell voltage, the MFC biosensor could monitor arsenic (0.5–5.0 mg L–1) and copper (1.0–10 mg L–1) with added Cu/As to the anolyte solution [83]. Wu et al. [86] inoculated a facultative anaerobe bacterium into an MFC to investigate an in-situ Cr6+ detection sensor. In their research, Wu et al. [105] developed a new sediment MFC-based toxic sensor for online and in-situ Cu2+ monitoring by using bacteria species such as Clostridium and Geobacter. A similar study was conducted using Pseudomonas and Geobacter for in-situ Cr6+ detection in real-time from industrial wastewater [106]. Researchers have recently become interested in electro-active biofilms because of their potential applications in environmental monitoring as amperometric biosensors, i.e., in-situ toxicant detection. For the assessment of heavy metal contamination in tap water, an O2 reducing microbial cathode-based MFC biosensor was designed, with detection limits in the 1.0 to 10 mg L–1 range for ions, including Cr6+, Hg2+ and Pb2+ [107]. As a result, biosensors can accurately reflect the harmful effects of numerous contaminants in the environment. This understanding provides strong technical solutions for online, direct pollutant detection (i.e., heavy metals) and the establishment of an early warning sensor system. Table 2 summarizes the materials and performance of biosensors for heavy metals monitoring.
Table 2.
Biosensors for heavy metal monitoring with detection limits and response time.
| Heavy metals | Configuration of biosensors | Electrodes/Materials |
Environmentalmatrix | Detection limits | Response times | Remarks | Refs. | |
|---|---|---|---|---|---|---|---|---|
| Anode | Cathode | |||||||
| Cu, As, Cr, Hg | Double chamber MFC | Carbon fiber | Carbon fiber | Municipal wastewater | 0.5–10 mg L–1 | 5 min | MFC as a heavy metal biosensor | [83] |
| Pb, etc. | A dual-chamber self-powered MFC | Graphite felts | Graphite felts | Water containing heavy metal ions | 2 mg L–1 | N/A | A simple, cheap and sensitive | [101] |
| Cr | Sediment MFC | Carbon cloth | Graphite felt | Industrial wastewater | 0.2–0.7 mg L–1 | 18.3 min | Real-time in-situ detection | [106] |
| Hg | Doublechamber MFC | Graphitefelt | Graphitepaper | Artificial water | 200 mg L–1 | N/A | Environmentally friendly method | [108] |
| Pb, Cd, Cu | Magnetoelastic (ME) biosensor | ME ribbon | Heavy metal ions solutions | 0.23–0.33 µM | 5 min | High-sensitive | [109] | |
| Pb | Micromechanicalbiosensor | A silicon membrane | Lead ions solutions | 10 µM | N/A | Based on 8–17 DNAzyme | [130] | |
| Hg, Ag, Cu, Pb | DNA-based electrochemical biosensors | Gold, SPGE, PGE, ITO, magnetic beads, etc. | Environmental matrices | 0.0001–300 nM | N/A | Providing a platform for heavy metal detection | [99] | |
| Cu, Cr, Zn, Ni | Reusable single-chambered cylindrical MFC | Graphite sheet | Graphite sheet | Industrial wastewater | 5–20 mg L–1 | N/A | Cost-effective | [100] |
| Cd, Cu,Zn | Whole-cell bacterial biosensor | Sol-gel matrices (a thin film of sol-gel) | Soilenvironment | 1.42 × 10−4–3.16 × 10−4 mg L–1 | 4–7 h | A green fluorescent protein | [40] | |
| Hg | DNA-based electrodes | Gold nanoparticles, MWCNT | Water samples | 1 × 10−6 nM | N/A | A time and cost-effective | [127] | |
| Se, Sn, Cr | Bacterial molecular biosensors | Acinetobacter baylyi ADP1 Tox2 | River water | Low | 30 min | Respond at low levels | [71] | |
| Hg, Zn, Cu, Cd | A bacterial whole-cell biosensor | A. baylyi Tox2biosensor | Seawater | 0.02–1.0 mg L–1 | A quick | Rapid detection, sensitive biosensor | [70] | |
| Cu, Hg, Zn, Fe, Cr, Cd | MFC-based biosensors | Graphite, carbon cloth, stainless steel | Graphite, carbon cloth, platinum | Water, wastewater, sewage sludge | 0.0125–200 mg L–1 | Short time | Cost-effective | [110] |
| Cu | A single-chamber MFC | Carbon brush | Carbon cloth | Artificial solution | 0.5–12.5 mg L–1 | N/A | A cost-effective | [105] |
| Pb | Light-up biosensor | Micro-spin column (Bio-Rad) | Water, serum samples | 60.7 nM | N/A | A simple, facile, and cost-effective | [111] | |
| Hg | SERS-based biosensor | Magnetic nanoparticles, SWCNT substrate and CoFe3O4@Ag | Aqueous solution | 8.4 × 10−4 nM | N/A | Good sensitivity, selectivity, simplicity, and rapidity | [112] | |
| Pb, Ni, Cd, etc. | Biosensor cells | A novel marine luminescentbacterium Vibrio sp. MM1 | Aquatic environments | 0.97–14.54 mg L–1 | N/A | High sensitivity | [78] | |
| Hg | DNAzyme-based biosensors | Nanomaterial-assisted DNAzymes | Tap, river, lake, wastewater, etc. | 5 × 10−5–100 nM | N/A | Highly selective and sensitive recognition | [103] | |
| Pb | A self-cleaning electrochemical biosensor | Cu-TCPP nanofilm and DNAzymes | Artificial solution | 1.7 nM | N/A | High sensitive and selective | [123] | |
| Hg, Ag | Novel conductometric biosensor | Three-enzyme system (invertase, mutarotase, glucose oxidase) | Artificial solution | 1.25 mM | 10–20 min | High sensitivity | [113] | |
| Cd | A novel signal amplification biosensor | Agarose gel electrophoresis, oligonucleotides DNA | Artificial solution | 19.93 nM | N/A | A satisfactory recovery rate, high sensitive | [138] | |
| Hg | Whole-cell biosensors | A gas reporting biosensor system | Artificially contaminated soil | 5 μM | 45 min | On-site detection, saving time and costs | [102] | |
Remarks: MFC: Microbial fuel cell, PGE: Polycrystalline gold electrode, SPGE: Screen-printed gold electrode, ITO: Indium tin oxide, ME: Magnetoelastic, DNA: Deoxyribonucleic acid, MWCNT: Multi-walled carbon nanotube, SERS: Surface enhancement Raman spectrum, SWCNTs: single-walled carbon nanotubes, TCPP: Tetrakis (4-carboxyphenyl) porphyrin.
Several biosensors based on electrochemical, colorimetric, and fluorescence measurements were successfully devised for heavy metal detection [114–116]. It has been suggested that the nanomaterials such as metal oxide and nanostructured carbon could be effectively used in biosensors development [5,117–119]. In addition, several nanomaterials with heterostructure such as α-Fe2O3–g–C3N4, V2O5 g–C3N4, and CuS–WuO3 were developed, indicating the potential use in electrochemical biosensors [120–122]. For electrochemical sensors, nano-scale structures are attractive materials [123], and their huge specific surface can enhance enzyme, signaling molecule, and catalyst immobilization. Applying electrochemical DNA biosensors for heavy metals detection includes implementing core-shell nanoparticles, nicking enzyme–assisted amplification, and nanocomposites modification [124,125]. Apart from these, applications of nanomaterials based on electrochemical nucleic acid (NA) biosensors for quantitative and qualitative analysis/monitoring of environmental pollutants, e.g., heavy metals have been discussed [126,127]. Table 2 demonstrates how biosensors were classified depending on the recognition component that was used for detecting heavy metals. For example, the bioavailable copper ions in synthetic samples were monitored by biosensor used as an optical transducer and immobilized engineered bacterium Alcaligenes eutrophus (AE1239), with the limit of detection (LOD) being 1 μM [128]. DNA-based electrodes biosensor is an excellent candidate for Hg detection, with a LOD of 3 fM [127]. Another DNA biosensor using electrode modification with Fe3O4@3D-GO reached an excellent LOD for Ag+ ion detection, which was equal to 2.0 pM and in the wide range of 0.01–100 nM [129]. A self-cleaning electrochemical biosensor based on DNA nanomotors, novel super G-quadruplex (G4), and 2D Cu-porphyrin (Cu-TCPP) metal-organic nanofilms was designed for the cyclic detection of Pb2+ ions, which the LOD was equal to 1.7 nM, and the linear range of 5 nM–5 μM [123]. The synergistic effects of G4-hemin DNAzymes and Cu-TCPP, which has efficient and catalytic H2O2 reduction, resulting in better performance of electrochemical biosensors [123].
DNA biosensors focusing on the recent design of selective and sensitive detection of heavy metal elements/ions by electrochemical transduction are depicted in Table 2. DNA biosensors used electrochemical transduction to provide a suitable platform for early monitoring/detection of heavy metal ions. Many effective DNA biosensors based on various electrochemical redox indicators have been applied for Ag+, Hg2+, Pb2+ ions detection [99]. A bacterial whole-cell biosensor (Acinetobacter baylyi Tox2) was designed to detect heavy metal cytotoxicity in the polluted seawater sources [70]. Consequently, A. baylyi Tox2 exhibits excellent application as a sensitive and quick biosensor for investigating cytotoxicity in the marine environment. Many detection methods have been assessed using the 8–17 DNAzyme specific for lead ions [115,130,131]. Shen et al. [132] used DNA-Au bio-barcode based on a signal amplification assay for Pb2+ ion detection. Thus, biosensors have attracted much interest for biomonitoring purposes, especially of heavy metal ions.
Results found that Hg2+ concentrations analyzed by biosensor could be confirmed by standard methods (e.g., Inductively coupled plasma mass spectrometry, ICP-MS), showing that the biosensor was reliable and relevant for Hg2+ ion detection in real samples [133]. For instance, autocatalytic DNA circuit based on exonuclease III and G-quadruplex DNAzyme for mercury ions detection achieved high selectivity and sensitivity. For an LOD, it was 10 fM and the linear range was from 10 fM to 100 nM [134]. Shi et al. [133] used a biosensing system for detecting Hg2+ in tap and lake water samples with a 3D graphene/gold electrode and a reporter probe attached to Au nanoparticles. An excellent widely linear range from 0.1 fM to 0.1 µM was reached. In contrast, Hg2+, Pb2+ and Cd2+ levels are strictly monitored by the European Union [103] due to these substances’ highly toxic, bioaccumulative properties, and their impact on human health and the environment [133]. According to the U.S. Environmental Protection Agency (EPA) regulations, the maximum levels of heavy metals such as Hg2+, Pb2+, and Cd2+ in drinking water are 10 nM, 72 nM and 45 nM, respectively, so these described biosensors could effectively monitor their concentrations [103,116]. The LOD of these heavy metals are detected by biosensors lower than the toxicity safety levelfor example, in drinking water to monitor the quality. Shown here is the critical role of advanced biosensors for heavy metals detection/monitoring to meet requirements, such as monitoring Hg2+ concentration in drinking water (i.e., level of 10 nM), following the U.S. EPA standard [134].
Several studies have used eukaryotic microorganisms to investigate whole-cell biosensors for environmental pollution monitoring of heavy metals in aquatic habitats or soils [135]. Yeasts, ciliated protozoa, and microalgae are generally selected as the three main eukaryotic taxonomic groups. For example, a new conductometric biosensor based on alkaline phosphatase activity was developed utilizing immobilized whole-cell microalgae to detect cadmium ions in aquatic environments and habitats [136]. A novel whole-cell microbial biosensor was invented for rapid on-site detection related to Hg contaminated soil with gas signals [102]. This technique could detect bioavailable mercury levels within 45 min and a range from 5 to 500 μM, effectively showing how much pollution there was in the soil. Guo et al. [137] illustrated that a fluorescent whole-cell biosensor could detect mercury (Hg2+) contamination in cosmetics with the detection range from 50 nM to 10 μM with incubating time for two hours. It means that biosensors can be used for many purposes and in many industries.
Biosensors could detect and provide information quickly about the toxic pollutants and contamination zone, which is necessary for good environmental management and monitoring. Another advantage of biosensors over traditional analytic techniques is related to their mobility, making possible the measurement of in-situ pollutant levels without added chemical agents and sample preparation. Biosensors can detect and conduct single measurements or continuous real-time monitoring during analysis processes. DNA biosensors based on electrochemical transduction, constitute an sensitive technique and affordable method for detection and monitoring of heavy metal elements [99,138]. Biological sensors are promising in heavy metal ions detection, especially DNA. DNA and DNAzymes are biodegradable compositions, exhibit high selectivity, the advantage of a portable analytical device and in-vitro technique, as well. Based on the highly selectivity of DNAzymes (as a biological recognition element), electrochemical biosensors can determine heavy metal compounds. DNA is easy to synthesize, low-cost, stable at room or higher temperatures, and is an ideal material for biosensors [134]. It shows novel advantages with excellent potential in toxicity determination, i.e., sensitive, cost-effective devices, rapid response to the toxin and short life cycle [71,139,140].
The excellent sensitivity of DNA-based or DNAzyme-based biosensors on heavy metals was indicated by its low LOD (below nM), especially those integrating nanomaterials (Table 2). However, the cell-free biosensor could not provide the information of bioavailable metals in the environment. In addition, the high cost and requirements of advanced technology of nanomaterials might impede the production and application of these cell-free biosensors in developing countries. On the other hand, although the whole-cell or MFC-based biosensors could reveal the bioavailable heavy metals in the samples with lower cost, the sensitivity of them are mostly lower than DNA-based biosensors, limiting their commercial feasibility.
4.2. Organic pollutants monitoring
Anthropogenic activities ensure that the natural environment is contaminated by organic pollutants. A wide range of contaminants originates from different industrial, household and agricultural activities. Agricultural waste organic herbicides and pesticides contain toxic compounds mostly found in wastewater, and the compounds are widely used to remove weeds, pests and unwanted vegetation. Industrial wastewater contains organic matter carrying various hydrocarbons, chlorine compounds, aromatic substances and surfactants. Persistent organic pollutants (POPs) including polychlorinated biphenyls (PCBs), polybrominated diphenyl ethers (PBDEs), polybrominated biphenyls (PBBs), phthalate esters (PAEs), and other hazardous pollutants exist in industrial wastewater [141–143]. Household wastes also contribute to organic pollutants in wastewater such as phenyl ether which is found in everyday household products such as soaps, deodorants, plastics and cosmetics [144]. Monitoring the particular organic matter in wastewater is an important aspect of human health and the environment, particularly wastewater treatment and water reclamation processes. The application of biosensor for detecting organic contaminants in wastewater achieves the fastest and most accurate results compared to other traditional methods [145]. Biosensor application can be used for environmental organic pollutants including endocrine disrupting chemicals (EDCs), PCBs, POPs, total petroleum hydrocarbons (TPHs), polycyclic aromatic hydrocarbons (PAHs) and volatile organic compounds (VOCs) (see Table 3).
Table 3.
Summary of organic pollutant biosensors with detection limits.
| Pollutants | Classification | Configuration of biosensors |
Detection limit | Remarks | Refs. | |
|---|---|---|---|---|---|---|
| Sensing element | Signaling element | |||||
| Bisphenol A | EDC | Enzyme (tyrosinase) | Amperometry/MWCNT modified | 0.01 nM | Promising reliable | [146] |
| Bisphenol A | EDC | Enzyme biosensor | Tyr – AuNPs/Graphite oxide | 1 nM | Excellent performanceLong-term stabilityPromising on-site analysis | [147] |
| Bisphenol A | EDC | Labeled antibody | Fluorimetry | 0.014 μg L−1 | Satisfactory repeatabilityGood agreement | [148] |
| Bisphenol A | EDC | Sol–gel films | Tyr/TiO2-MWCNTs-PDDA- Nafion/graphite | 66 nM | High sensitivity Good precision | [149] |
| Estrogen Estradiol Phenol Nonylphenol Bisphenol A |
EDCs | Enzyme (tyrosinase) | Amperometry | 4.4 mg L−14.06 mg L−10.94 mg L−12.2 mg L−15.2 mg L−1 | FastGood sensitivity | [150] |
| Estrone | EDC | Labeled antibody | Fluorimetry | 0.2 ng L−1 | Fast, sensitive, and cost-effective detectionA powerful tool in aquatic analytics | [151] |
| Phenol | EDC | Whole-cell (Acinetobacter sp. DF4) | Light emission/bioluminescence | 2.5 mg L−1 | Response to phenol-contaminated soils | [152] |
| Triclosan | EDC | Molecular imprinting polymers (MIP) | Surface plasmon resonance (SPR) | 0.017 μg L−1 | Excellent performanceFast, sensitive and simple nanosensor | [144] |
| AtrazineBisphenol AEstroneIsoproturon | HerbicideEDCEDCHerbicide | Labeled antibodies | Total internal reflection fluorescence (TIRF) | 0.002 μg L−10.005 μg L−10.019 μg L−10.016 μg L−1 | Low-costEarly warning application | [153] |
| Propanil | Herbicide | Polyclonal IgG antibody | Fluorimetry | 0.6 ng L−1 | Fast, sensitive, and cost-effective detection | [154] |
| Acetamiprid | Pesticide | Electrochemical DNA Aptasensor | Ferrocene (Fc)- based hollow polymeric nanospheres (Fc-HPNs) | 3.3 × 10−7 nM | Excellent sensitivity | [155] |
| Carbaryl | Pesticide | Monoclonal antibody | Surface plasmon resonance (SPR) | 1.38 μg L−1 | Useful and portable for on-line monitoring | [156] |
| Organophosphorus (OPs) | Pesticide | A portable LIG-based electrochemical biosensor | MnO2-bridged enzyme-aided signal amplification | 1.2 μg L−1 | Good performance for pesticideApplications in the environment and food safety fields | [157] |
| Pyrethroid | Pesticide | Whole-cell and antibody | Calorimetry | 3 μg L−1 | Simple and portable for on-site colorimetric applicationFunctional at least 90 days after lyophilization | [169] |
| NaphthalenePhenanthrene | PAHs | DNA/Sol–gel array | Fluorimetry | 0–10 mg L−1 | Effective PAHs detection in water, and biological samples | [158] |
| Polybrominated diphenyl ethers (PBDEs) | POP | Whole-cell (Sphingobium xenophagum) | Bioluminescence | 10 nM | Novel PBDE-specific sensing element | [91] |
| PCB-28PCB-101 | PCB | Enzyme (peroxidase)/immobilized polyaniline modified Pt electrode | Amperometry | 0.016 μg L−10.019 μg L−1 | Useful in POPs detection in landfill leachates | [159] |
| TPH | TPH | Antibody and enzyme | Immunoassay | 25 mg kg−1 | Quick on-site useDetect TPH in soil and water | [160] |
Remarks: TIRF: Total internal reflection fluorescence, SPR: Surface plasmon resonance, OPs: Organophosphorus, LIG: Laser-induced graphene, TIRF: Total internal reflection fluorescence, MIP: Molecular imprinting polymers, MWCNT: Multi-walled carbon nanotube, EDC: Endocrine disrupting chemical, PCBs: Polychlorinated biphenyls, POPs: Persistent organic pollutants, PAHs: Polycyclic aromatic hydrocarbons, TPH: Total petroleum hydrocarbons.
Advances in the field of biotechnology have enabled biological materials to function as receptors, so these bioreceptors can analyze VOCs with high selectivity and sensitivity [161]. Some applications can detect VOCs based on protein and peptides biosensors, such as sensitive olfactory biosensors having a remarkable performance for high selectivity and sensitivity at very low concentrations by using odorant-binding protein (OBP) operated as a sensing component [162]. In addition, the nonspecific biosensor based on microbial electrochemical cell-based (MXC) structure can be successfully used to monitor PCBs and PAHs such as toluene [163]. For specific targeting of benzene, phenol, toluene, and other related pollutants, engineered whole-cell biosensors based on the metabolic genes as sensing elements were developed [164,165]. Other substances including pesticides and EDCs could be detected in river water and wastewater by biosensors especially nanomaterials-based ones, suggesting their feasibility in real environmental samples [166].
Currently, the successful application on VOCs and PAHs such as alkanes, toluene, naphthalene in wastewater, seawater, and soil using whole-cell luminescence biosensor has been reported [8]. However, limited information about the key sensing element based on the metabolic mechanisms of microorganisms constrain the development of biosensors for specific organic pollutants such as emerging contaminants [91]. Recent research has highlighted the importance of high-throughput sequencing and synthetic biology tools on biosensors for organic pollutants since these techniques provide more genetic information about sensing elements [91,167]. Additionally, the usage of two different fluorescence as signaling elements enables the biosensor to detect different types of hydrocarbon simultaneously [165]. For simple and rapid on-site use, the colorimetric biosensors for pyrethroid insecticide and halogenated hydrocarbons performed well in testing samples [168,169]. So it can be states that a biosensor for detecting organic pollutants in the environment is now more efficient and feasible in rapid on-site screening. It complements traditional chemical analysis for large-scale monitoring.
Nowadays widespread industrial spillage is the major problem causing environmental degradation or destruction. For unique applications, biosensors can be personalized such as TPHs screening kit which is a favored tool for monitoring analytes on site in different media (soil, water and vapor) [170]. The immunoassay-based TPHs kit founded on antigen-antibody and enzyme is a biosensor due to its biological sensing elements. It has been suggested that TPHs immunoassay is one of the most widely used methods to quickly detect petroleum compounds especially in soil samples [171,172]. The low-cost and quick on-site utility are the main strengths of using the TPHs immunoassay kit [172]. However, the drawbacks of interference of media and nonspecific target were reported in TPHs assay kit, which is similar to other biosensors [171,172]. For this reason, the TPHs immunoassay could be used as large-scale screening method prior to precise quantification of TPHs using instruments such as GC. It could reduce the amount of energy consumed and carbon footprint as required by the SDGs.
An overview of the current achievements of biosensors and those devised to monitor organic pollutants was done in this section. Monitoring of organic contaminants plays an important role in the protection of human health and the ecosystem. By applying a biosensor, the rapid detection and screening of organic contaminants can be achieved. These biosensors are quite inexpensive, portable, available for on-site use, and no waste is generated during analysis. However, there are some limitations of these biosensors such as sensitivity and selectivity of the target pollutants, limiting applicability. Some challenges need to be overcome for biosensors including interference of humidity, effect of pH on sensitivity and selectivity, enhancing the sustainability of sensing components of biosensors, and ensuring the adaptation of nanomaterials, which can improve the performance of biosensors. More knowledge of metabolic genes for POPs is needed for further improvement of sensing elements. Thus, current biosensors technology could be utilized in monitoring EDCs, PCBs, POPs, TPHs, PAHs, and VOCs, while those difficult-to-degrade organic pollutants might not be efficiently monitored.
Similar to the biosensor detecting heavy metals, the cell-free biosensors using enzyme, DNA, and aptamer showed high sensitivity to organic pollutants including EDCs, pesticides, and PCBs (Table 3). However, the lack of bioavailability data and higher requirements of nanomaterials are also the limitations of cell-free biosensors. The whole-cell biosensors detecting organic pollutants are relatively few, which is due to the low biodegradability of the organic pollutants that hinders their development. In addition, most of the whole-cell biosensors have higher LOD than cell-free biosensors, showing the lower sensitivity (Table 3). Currently, the commercial whole-cell biosensors are mainly nonspecific and toxicity-based with the bioluminescence signals, whereas the biosensors specific to heavy metals and organic pollutants (TPHs) are all cell-free biosensors with DNAzyme, enzyme, and antibody (Table 4).
Table 4.
Commercial biosensors and the product details.
| Product name | Analyte | Sensing element | Signaling element | Manufacturer (country) |
|---|---|---|---|---|
| Microtox | Toxicity | Whole-cell | Bioluminescence | Azur Environmental (US) |
| LUMIStox | Toxicity | Whole-cell | Bioluminescence | Hach (US) |
| ToxAlert | Toxicity | Whole-cell | Bioluminescence | Merck (Germany) |
| Catalytic DNA Sensors | Metals | DNAzyme | Fluorescence | Quasar Instruments (US) |
| PETRO RISc Soil Test | TPHs | Enzyme and antibody | Color | Ensys Energy Systems (US) |
| EnvironGard Petroleum Fuels in Soil | TPHs | Enzyme and antibody | Color | Merck Millipore (Germany) |
In short, establishing monitoring activities based on biosensors can be applied for the significant recognition/detection of heavy metals, organic chemicals, microorganisms, etc. Biosensors are robust environmental monitoring solutions with important novel characteristics and advantages [173]. Biosensors are early detection techniques with much promise in achieving on-site monitoring and continuous real-time environmental data. Wastewater and water monitoring biosensors are an innovative approach for the ultimate goal of on-site monitoring within the framework of the UN’s Sustainable Development Goals (SDGs).
4.3. Wastewater quality monitoring and environmental viability
Wastewater discharge is the one of the source of environmental pollution, and the wastewater quality monitoring could be also achieved by the biosensors [145]. The biosensors especially MFC-based were applied in wastewater monitoring globally. In Asia Pacific countries, it has been reported that Au3+ ions in wastewater from Taiwan could be detected by engineered bacteria [174]. In addition, The MFC-based BOD biosensor was used in monitoring swine wastewater in Japan, which minimizes the energy cost for aeration [175]. The multiple heavy metals and phenols in real wastewater samples from China were able to be detected by MFC-based biosensors [176]. The immobilized engineered bacteria were used in screening PAHs in industrial wastewater in India [177]. The toxic metals (Cr6+ and Fe3+), NO3−, and sodium acetate in the wastewater from Connecticut (USA) could be assessed by the batch mode MFC-based biosensors [178]. Similarly, the brewery wastewater obtained from Canada containing COD and others such as NH4+ was shown to successfully monitored by MFC-based biosensors [179]. The MFC-based biosensors were also used in measuring the BOD in rice washed wastewater in Ecuador [180]. In South Africa, it has been shown that the influents and effluents from wastewater treatment plants were assessed by nonspecific whole-cell bioluminescence biosensors [181]. In Europe countries, the MFC-based biosensors were applied in detecting the BOD in domestic and brewery wastewaters in Hungary [182]. The COD in domestic wastewater from Spain discharged into constructed wetlands could be monitored by MFC-based biosensors [183]. Taken together, the MFC-based biosensors are broadly applied in wastewater quality monitoring due to its strengths of saving energy and continuous monitoring, which could correlated to the SDGs 6 (clean water and sanitation). However, further studies on detecting specific pollutants such as toxic metals in industrial wastewater are required for better wastewater quality. The performance and application of biosensors in real environmental samples such as river, wastewater, and soil samples were suggested in recent studies (Tables 2 and 3). For better commercial and environmental feasibility, more research about the real environmental sample tests is needed.
5. Biosensor for Sustainable Development Goals (SDGs)
Biosensors can detect impurities in the air, soil or water and in this way lead to finding the main sources of pollutants by implementing activities that mitigate impurities and help realize the SDGs, especially SDG 6 (clean water and sanitation), 12 (responsible consumption and production), 13 (climate action), 14 (life below water), and 15 (life on land). In developing countries, the environmental pollution might raise the health concern such as As contamination in groundwater, drinking water, and irrigation water, highlighting the needs for screening and monitoring the contaminants [184]. Compared with traditional environmental analysis methods, biosensors for environmental monitoring possess characteristics such as low-cost, minimal technical expertise and sample pretreatment, and feasibility for on-site use, saving energy and nonuse of hazardous materials [167]. For instance, the color-based arsenic biosensor on paper strip can be easily used in drinking water by naked eyes without complicate instruments [14]. Therefore, applying the biosensors with such advantages on environmental monitoring could help achieving SDG 6 (clean water and sanitation) by accomplishing large screening of pollutants for clean water and improving the access to affordable drinking water especially in developing countries (Table 5). Furthermore, the SDG 6 (clean water and sanitation) and its target 6.3 (water quality and wastewater) could be achieved by using biosensors in simultaneous monitoring of multiple toxic metals in polluted rivers before human health is affected [71]. Water quality monitoring such as BOD, COD, and toxicants using the MFC-based biosensors and the toxicity-based nonspecific biosensors reflects not only the toxicity in drinking water but also the extent of eutrophication [23,81]. Targeted here are both SDGs 6 and 14 (life below water) which refers to detecting the potential pollution in freshwater and marine ecosystems as well as drinking water. For terrestrial ecosystems this refers to SDG 15 (life on land), and the soil contaminants-detecting biosensor could be applied to screen potential soil pollution, which can guide what happens in terrestrial ecosystems [185,186].
Table 5.
The relationship of SDGs and biosensor application.
| SDGs | Contributions of biosensor application |
|---|---|
| SDG 6: clean water and sanitation |
|
| SDG 12: responsible consumption and production |
|
| SDG 13: climate action |
|
| SDG 14: life below water |
|
| SDG 15: life on land |
|
Furthermore, the results of biosensors provide more information about bioavailability and toxicity, which can complement traditional chemical analysis methods. From the sustainability point of view, biosensors’ deployment reduces the amount of hazardous chemicals that are used in operational and sample pretreatment processes, which is also related to SDG targets 6.3 and 12.4 (reducing the hazardous waste produced) [167]. Portability and on-site monitoring feasibility using the biosensors reduce the carbon footprint which is a consequence of transportation. In addition, the large-scale screening of biosensor environmental analysis helps with precise measurements and saves electricity and energy [167]. It has been reported that chemicals and energy used in laboratories did make a substantial contribution to the carbon footprint and green GHGs [20–22]. A summary of the relationship between SDGs and biosensor application is shown in Table 5. The complementary use of biosensors and traditional chemical analysis methods could realize SDG 13 (climate action) by reducing energy consumption and the carbon footprint.
6. Challenges of biosensor application on SDGs achievement
To provide reliable environmental monitoring results with low carbon footprint and less hazardous waste for achieving SDGs, biosensors need to be more feasible for quick-screening and on-site usage. The progress of biosensors which involves better selectivity, limits of detection and sensitivity is improving in the way that contaminants in the environments are detected, promoting a clean and green environment, while the challenges of sustainability, portability, and reusability still remain. Researchers want to resolve these challenges to enhance the biosensor application at advanced levels [187]. Currently, several advanced nanomaterials have been applied in the electrodes of cell-free and whole-cell biosensor to improve pollutants detection [188]. For cell-free biosensor, aptamer coupled with nanomaterials biosensors were commonly used for their good selectivity and reliability [29]. However, the cost for nanomaterials and aptamer-based biosensor production means they are still not commercially feasible, limiting their environmental application. The energy and carbon footprint from manufacturing these biosensors with nanomaterials and high performance might compromise their contribution to the SDGs. The possible risk of environmental nanomaterial use is still unclear so further information about the safety of nanomaterial-based biosensor and lowering the costs and energy consumption is needed. Recent studies mostly focused on sensing and signaling elements optimization, whereas the practicality tests using real wastewater samples are still rarely undertaken. More studies using biosensors in real environmental samples to assess the influence of other interfering substances such as organic matter chelation are warranted.
7. Conclusions and future prospects
Recognizing the growing call for more environmentally, economically, and socially responsible societies, emerging remediation technologies and governing strategies are being developed in alignment to the sustainable future. Biosensors using biological sensing elements coupled with signaling elements can be used to detect impurities in environmental samples so that the responsible sources of pollutants are known. By implementing remedial activities to remove harmful impurities from the sources, the UNSDG goals can be achieved. Despite the potential contribution to SDG achievements, few research reported the importance of biosensor application on SDGs, and the correlation of environmental monitoring using biosensors and SDGs. Herein, this article reviewed the current progress being made in biosensors for the purposes of environmental monitoring and their contribution to achieving the UNSDGs, including clean water and sanitation (SDG 6), responsible consumption and production (SDG 12), climate action (SDG 13), saving life below water (SDG 14), and saving life on land (SDG 15). While bio-sensing technologies have advanced significantly in the past decades, further research on how to improve the sensitivity, stability, safety, and portability is warranted for environmental monitoring biosensors and their future applications.
Cell-free biosensors are powerful in terms of their quick and specific response to environmental pollutants, whereas a whole-cell biosensor can provide additional information about the bioavailability and toxicity that cannot be analyzed by cell-free biosensors. The results from nonspecific whole-cell biosensors directly reflect the total impact of pollutants on the test microorganisms, which is mostly based on the inhibition of microbial functions. However, nonspecificity is the main weakness when designing a specific targeting policy. On the other hand, the specific whole-cell biosensor can provide good information about the target pollutants, yet the sensitivity and stability still need to be improved for feasible commercial application. Recently, genetic methods have been developed in whole-cell biosensor to improve its sensing and signaling elements for better sensitivity. However, research into environmental sample tests, storage time such as shelf life, and safety of genetically modified biosensors is needed.
Acknowledgements
The authors would like to thank the Office of Marine Science and Technology (OMST), National Kaohsiung University of Science and Technology (NKUST) for the financial support of the project #OMST-111E15. The authors sincerely thank the editor and four anonymous reviewers for their constructive suggestion and contribution to enhance this manuscript.
Funding Statement
This work was supported by the National Kaohsiung University of Science and Technology (NKUST) under Grant #OMST-111E15.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Ethical statement
No ethics approval was required for this study as it involved no human participants or animals.
References
- [1].Justino CIL, Duarte AC, Rocha-Santos TAP.. Recent progress in biosensors for environmental monitoring: a review. Sensors (Basel). 2017;17(12):2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Theerthagiri J, Lee SJ, Karuppasamy K, et al. Application of advanced materials in sonophotocatalytic processes for the remediation of environmental pollutants. J Hazard Mater. 2021;412:125245. [DOI] [PubMed] [Google Scholar]
- [3].Tran H-T, Dang B-T, Thuy LTT, et al. Advanced treatment technologies for the removal of organic chemical sunscreens from wastewater: a review. Curr Poll Rep. 2022. DOI: 10.1007/s40726-022-00221-y [DOI] [Google Scholar]
- [4].Zhang Y, Zhu Y, Zeng Z, et al. Sensors for the environmental pollutant detection: are we already there? Coord. Chem Rev. 2021;431:213681. [Google Scholar]
- [5].Theerthagiri J, Salla S, Senthil RA, et al. A review on ZnO nanostructured materials: energy, environmental and biological applications. Nanotechnology. 2019;30(39):392001. [DOI] [PubMed] [Google Scholar]
- [6].Daunert S, Barrett G, Feliciano JS, et al. Genetically engineered whole-cell sensing systems: coupling biological recognition with reporter genes. Chem Rev. 2000;100(7):2705–2738. [DOI] [PubMed] [Google Scholar]
- [7].Zhan S, Wu Y, Wang L, et al. A mini-review on functional nucleic acids-based heavy metal ion detection. Biosens Bioelectron. 2016;86:353–368. [DOI] [PubMed] [Google Scholar]
- [8].Voon CH, Yusop NM, Khor SM. The state-of-the-art in bioluminescent whole-cell biosensor technology for detecting various organic compounds in oil and grease content in wastewater: from the lab to the field. Talanta. 2022;241:123271. [DOI] [PubMed] [Google Scholar]
- [9].Wang G-H, Tsai T-H, Kui -C-C, et al. Analysis of bioavailable toluene by using recombinant luminescent bacterial biosensors with different promoters. J Biol Eng. 2021;15(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hui C-Y, Guo Y, Gao C-X, et al. A tailored indigoidine-based whole-cell biosensor for detecting toxic cadmium in environmental water samples. Environ Technol Inno. 2022;27:102511. [Google Scholar]
- [11].He W, Hu ZH, Yuan S, et al. Bacterial bioreporter‐based mercury and phenanthrene assessment in Yangtze river delta soils of China. J Environ Qual. 2018;47(3):562–570. [DOI] [PubMed] [Google Scholar]
- [12].Brányiková I, Lucáková S, Kuncová G, et al. Estimation of Hg(II) in soil samples by bioluminescent bacterial bioreporter E. coli ARL1, and the effect of humic acids and metal ions on the biosensor performance. Sensors. 2020;20(11):3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rogers KR. Recent advances in biosensor techniques for environmental monitoring. Anal Chim Acta. 2006;568(1–2):222–231. [DOI] [PubMed] [Google Scholar]
- [14].Kaur H, Kumar R, Babu JN, et al. Advances in arsenic biosensor development–a comprehensive review. Biosens Bioelectron. 2015;63:533–545. [DOI] [PubMed] [Google Scholar]
- [15].Gu MB, Mitchell RJ, Kim BC. Whole-cell-based biosensors for environmental biomonitoring and application. Adv Biochem Eng Biotechnol. 2004;87:269–305. [DOI] [PubMed] [Google Scholar]
- [16].Ali SA, Mittal D, Kaur G. In-situ monitoring of xenobiotics using genetically engineered whole-cell-based microbial biosensors: recent advances and outlook. World J Microbiol Biotechnol. 2021;37(5):81. [DOI] [PubMed] [Google Scholar]
- [17].Bilal M, Iqbal HMN. Microbial-derived biosensors for monitoring environmental contaminants: recent advances and future outlook. Process Saf Environ Prot. 2019;124:8–17. [Google Scholar]
- [18].Chadha U, Bhardwaj P, Agarwal R, et al. Recent progress and growth in biosensors technology: a critical review. J Ind Eng Chem. 2022;109:21–51. [Google Scholar]
- [19].Sohrabi H, Hemmati A, Majidi MR, et al. Recent advances on portable sensing and biosensing assays applied for detection of main chemical and biological pollutant agents in water samples: a critical review. TrAC, Trends Anal Chem. 2021;143:116344. [Google Scholar]
- [20].Ni K, Hu Y, Ye X, et al. Carbon footprint modeling of a clinical lab. Energies. 2018;11(11):3105. [Google Scholar]
- [21].Sangwan KS, Bhakar V, Arora V, et al. Measuring carbon footprint of an Indian university using life cycle assessment. Procedia CIRP. 2018;69:475–480. [Google Scholar]
- [22].Larsen HN, Pettersen J, Solli C, et al. Investigating the varbon footprint of a university - the case of NTNU. J Clean Prod. 2013;48:39–47. [Google Scholar]
- [23].Nakamura H. Biosensors: monitoring human health for the SDGs sustainable development goals. Biomed J Sci Tech Res. 2018;9:6953–6956. [Google Scholar]
- [24].Yang Y, Li W, Liu J. Review of recent progress on DNA-based biosensors for Pb2+ detection. Anal Chim Acta. 2021;1147:124–143. [DOI] [PubMed] [Google Scholar]
- [25].Sarkar P, Banerjee S, Bhattacharyay D, et al. Electrochemical sensing systems for arsenate estimation by oxidation of L-cysteine. Ecotoxicol Environ Saf. 2010;73(6):1495–1501. [DOI] [PubMed] [Google Scholar]
- [26].Stoytcheva M, Sharkova V, Panayotova M. Electrochemical approach in studying the inhibition of acetylcholinesterase by arsenate(III): analytical characterisation and application for arsenic determination. Anal Chim Acta. 1998;364(1–3):195–201. [Google Scholar]
- [27].Vinotha Alex A, Mukherjee A. Review of recent developments (2018–2020) on acetylcholinesterase inhibition based biosensors for organophosphorus pesticides detection. Microchem J. 2021;161:105779. [Google Scholar]
- [28].Wu Y, Zhan S, Wang F, et al. Cationic polymers and aptamers mediated aggregation of gold nanoparticles for the colorimetric detection of arsenic(III) in aqueous solution. Chem Commun (Camb). 2012;48(37):4459–4461. [DOI] [PubMed] [Google Scholar]
- [29].McConnell EM, Nguyen J, Li Y. Aptamer-based biosensors for environmental monitoring. Front Chem. 2020;8:434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Van Dyk TK, Majarian WR, Konstantinov KB, et al. Rapid and sensitive pollutant detection by induction of heat shock gene-bioluminescence gene fusions. Appl Environ Microbiol. 1994;60(5):1414–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cha HJ, Srivastava R, Vakharia VN, et al. Green fluorescent protein as a noninvasive stress probe in resting Escherichia coli cells. Appl Environ Microbiol. 1999;65(2):409–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Eltzov E, Marks RS, Voost S, et al. Flow-through real time bacterial biosensor for toxic compounds in water. Sens Actuators B: Chem. 2009;142(1):11–18. [Google Scholar]
- [33].Belkin S, Smulski DR, Vollmer AC, et al. Oxidative stress detection with Escherichia coli harboring a katG’::lux fusion. Appl Environ Microbiol. 1996;62:2252–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kostrzynska M, Leung KT, Lee H, et al. Green fluorescent protein-based biosensor for detecting SOS-inducing activity of genotoxic compounds. J Microbiol Methods. 2002;48(1):43–51. [DOI] [PubMed] [Google Scholar]
- [35].Mitchell RJ, Gu MB. An Escherichia coli biosensor capable of detecting both genotoxic and oxidative damage. Appl Microbiol Biotechnol. 2004;64(1):46–52. [DOI] [PubMed] [Google Scholar]
- [36].Chen Z, Lu M, Zou D, et al. An E. coli SOS-EGFP biosensor for fast and sensitive detection of DNA damaging agents. J Environ Sci (China). 2012;24(3):541–549. [DOI] [PubMed] [Google Scholar]
- [37].Van Ginkel SW, Hassan SHA, Ok YS, et al. Detecting oxidized contaminants in water using sulfur-oxidizing bacteria. Environ Sci Technol. 2011;45(8):3739–3745. [DOI] [PubMed] [Google Scholar]
- [38].Hassan SH, Van Ginkel SW, Oh SE. Effect of organics and alkalinity on the sulfur oxidizing bacteria (SOB) biosensor. Chemosphere. 2013;90(3):965–970. [DOI] [PubMed] [Google Scholar]
- [39].Yang S-H, Cheng K-C, Liao VH-C. A novel approach for rapidly and cost-effectively assessing toxicity of toxic metals in acidic water using an acidophilic iron-oxidizing biosensor. Chemosphere. 2017;186:446–452. [DOI] [PubMed] [Google Scholar]
- [40].Rathnayake IVN, Megharaj M, and Naidu R. Green fluorescent protein based whole cell bacterial biosensor for the detection of bioavailable heavy metals in soil environment. Environ Technol Inno. 2021;23:101785. [Google Scholar]
- [41].Li P, Wang Y, Yuan X, et al. Development of a whole-cell biosensor based on an ArsR-Pars regulatory circuit from Geobacter sulfurreducens. Environ Sci Ecotech. 2021;6:100092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Assinder SJ, Williams PA. The TOL plasmids: determinants of the catabolism of toluene and the xylenes. Adv Microb Physiol. 1990;31:1–69. [DOI] [PubMed] [Google Scholar]
- [43].Ikariyama Y, Nishiguchi S, Koyama T, et al. Fiber-optic-based biomonitoring of benzene derivatives by recombinant E . coli bearing luciferase gene-fused TOL-plasmid immobilized on the fiber-optic end. Anal Chem. 1997;69(13):2600–2605. [DOI] [PubMed] [Google Scholar]
- [44].Willardson BM, Wilkins JF, Rand TA, et al. Development and testing of a bacterial biosensor for toluene-based environmental contaminants. Appl Environ Microbiol. 1998;64(3):1006–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Li YF, Li FY, Ho CL, et al. Construction and comparison of fluorescence and bioluminescence bacterial biosensors for the detection of bioavailable toluene and related compounds. Environ Pollut. 2008;152(1):123–129. [DOI] [PubMed] [Google Scholar]
- [46].Smirnova IA, Dian C, Leonard GA, et al. Development of a bacterial biosensor for nitrotoluenes: the crystal structure of the transcriptional regulator DntR. J Mol Biol. 2004;340(3):405–418. [DOI] [PubMed] [Google Scholar]
- [47].Hay AG, Rice JF, Applegate BM, et al. A bioluminescent whole-cell reporter for detection of 2, 4-dichlorophenoxyacetic acid and 2,4-dichlorophenol in soil. Appl Environ Microbiol. 2000;66(10):4589–4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Heitzer A, Malachowsky K, Thonnard JE, et al. Optical biosensor for environmental on-line monitoring of naphthalene and salicylate bioavailability with an immobilized bioluminescent catabolic reporter bacterium. Appl Environ Microbiol. 1994;60(5):1487–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sticher P, Jaspers MC, Stemmler K, et al. Development and characterization of a whole-cell bioluminescent sensor for bioavailable middle-chain alkanes in contaminated groundwater samples. Appl Environ Microbiol. 1997;63(10):4053–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Silver S. Bacterial resistances to toxic metal ions–a review. Gene. 1996;179(1):9–19. [DOI] [PubMed] [Google Scholar]
- [51].Bruins MR, Kapil S, Oehme FW. Microbial resistance to metals in the environment. Ecotoxicol Environ Saf. 2000;45(3):198–207. [DOI] [PubMed] [Google Scholar]
- [52].Bontidean I, Lloyd JR, Hobman JL, et al. Bacterial metal-resistance proteins and their use in biosensors for the detection of bioavailable heavy metals. J Inorg Biochem. 2000;79(1–4):225–229. [DOI] [PubMed] [Google Scholar]
- [53].Bontidean I, Mortari A, Leth S, et al. Biosensors for detection of mercury in contaminated soils. Environ Pollut. 2004;131(2):255–262. [DOI] [PubMed] [Google Scholar]
- [54].Nucifora G, Chu L, and Misra TK, et al. Cadmium resistance from Staphylococcus aureus plasmid pI258 cadA gene results from a cadmium-efflux ATPase. Proc Natl Acad Sci USA. 1989;86(10):3544–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Branco R, Cristovao A, Morais PV. Highly sensitive, highly specific whole-cell bioreporters for the detection of chromate in environmental samples. PloS One. 2013;8(1):e54005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Riether KB, Dollard MA, Billard P. Assessment of heavy metal bioavailability using Escherichia coli zntAp::lux and copAp::lux-based biosensors. Appl Microbiol Biotechnol. 2001;57(5–6):712–726. [DOI] [PubMed] [Google Scholar]
- [57].Beard SJ, Hashim R, Membrillo-Hernández J, et al. Zinc(II) tolerance in Escherichia coli K-12: evidence that the zntA gene (o732) encodes a cation transport ATPase. Mol Microbiol. 1997;25(5):883–891. [DOI] [PubMed] [Google Scholar]
- [58].Rensing C, Mitra B, and Rosen BP. The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc Natl Acad Sci USA. 1997;94(26):14326–14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Rensing C, Fan B, Sharma R, et al. CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc Natl Acad Sci USA. 2000;97(2):652–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Outten CE, Outten FW, O’Halloran TV. DNA distortion mechanism for transcriptional activation by ZntR, a Zn(II)-responsive MerR homologue in Escherichia coli. J Biol Chem. 1999;274(53):37517–37524. [DOI] [PubMed] [Google Scholar]
- [61].Outten FW, Outten CE, Hale J, et al. Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue. cueR J Biol Chem. 2000;275(40):31024–31029. [DOI] [PubMed] [Google Scholar]
- [62].Tibazarwa C, Corbisier P, Mench M, et al. A microbial biosensor to predict bioavailable nickel in soil and its transfer to plants. Environ Pollut. 2001;113(1):19–26. [DOI] [PubMed] [Google Scholar]
- [63].Hui CY, Guo Y, Liu LS, et al. Genetic control of violacein biosynthesis to enable a pigment-based whole-cell lead biosensor. RSC Adv. 2020;10(47):28106–28113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Hao OJ, Shin C-J, Lin C-F, et al. Use of microtox tests for screening industrial wastewater toxicity. Water Sci Technol. 1996;34(10):43–50. [Google Scholar]
- [65].LeBlond JB, Duffy LK. Toxicity assessment of total dissolved solids in effluent of Alaskan mines using 22-h chronic microtox® and selenastrum capricornatum assays. Sci Total Environ. 2001;271(1–3):49–59. [DOI] [PubMed] [Google Scholar]
- [66].Kurvet I, Ivask A, Bondarenko O, et al. LuxCDABE—Transformed constitutively bioluminescent Escherichia coli for toxicity screening: comparison with naturally luminous vibrio fischeri. Sensors. 2011;11(8):7865–7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Välimaa A-L, Kivistö A, Virta M, et al. Real-time monitoring of non-specific toxicity using a Saccharomyces cerevisiae reporter system. Sensors. 2008;8(10):6433–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Matejczyk M, Rosochacki SJ. Potential applications of sos-gfp biosensor to in vitro rapid screening of cytotoxic and genotoxic effect of anticancer and antidiabetic pharmacist residues in surface water. J of Ecol Eng. 2015;16:116–121. [Google Scholar]
- [69].Alhadrami HA, Paton GI. The potential applications of SOS- lux biosensors for rapid screening of mutagenic chemicals. FEMS Microbiol Lett. 2013;344(1):69–76. [DOI] [PubMed] [Google Scholar]
- [70].Cui Z, Luan X, Jiang H, et al. Application of a bacterial whole cell biosensor for the rapid detection of cytotoxicity in heavy metal contaminated seawater. Chemosphere. 2018;200:322–329. [DOI] [PubMed] [Google Scholar]
- [71].Rampley CPN, Whitehead PG, Softley L, et al. River toxicity assessment using molecular biosensors: heavy metal contamination in the Turag-Balu-buriganga river systems, Dhaka, Bangladesh. Sci Total Environ. 2020;703:134760. [DOI] [PubMed] [Google Scholar]
- [72].Yagi K. Applications of whole-cell bacterial sensors in biotechnology and environmental science. Appl Microbiol Biotechnol. 2007;73(6):1251–1258. [DOI] [PubMed] [Google Scholar]
- [73].Akboga D, Saltepe B, Bozkurt EU, et al. A recombinase-based genetic circuit for heavy metal monitoring. Biosensors (Basel). 2022;12(2):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Hurtado-Gallego J, Pulido-Reyes G, and González-Pleiter M, et al. Luminescent microbial bioassays and microalgal biosensors as tools for environmental toxicity evaluation. Handb of Cell Biosens. 2022;1:767–824. [Google Scholar]
- [75].Billard P, DuBow MS. Bioluminescence-based assays for detection and characterization of bacteria and chemicals in clinical laboratories. Clin Biochem. 1998;31(1):1–14. [DOI] [PubMed] [Google Scholar]
- [76].Lewis JC, Feltus A, Ensor CM, et al. Applications of reporter genes. Anal Chem. 1998;70(17):579A–585A. [DOI] [PubMed] [Google Scholar]
- [77].Sahli L, Belhiouani H, Burga Pérez KF, et al. Assessment of freshwater sediment quality: potential ecological risk and ecotoxicity tests as complementary approaches. Chem Ecol. 2021;37(3):219–233. [Google Scholar]
- [78].Mohseni M, Abbaszadeh J, Maghool -S-S, et al. Heavy metals detection using biosensor cells of a novel marine luminescent bacterium Vibrio sp. MM1 isolated from the Caspian Sea. Ecotoxicol Environ Saf. 2018;148:555–560. [DOI] [PubMed] [Google Scholar]
- [79].Li Y, He X, Zhu W, et al. Bacterial bioluminescence assay for bioanalysis and bioimaging. Anal Bioanal Chem. 2022;414(1):75–83. [DOI] [PubMed] [Google Scholar]
- [80].Cui Y, Lai B, and Tang X. Microbial fuel cell-based biosensors. Biosensors (Basel). 2019;9:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Do MH, Ngo HH, Guo W, et al. Microbial fuel cell-based biosensor for online monitoring wastewater quality: a critical review. Sci Total Environ. 2020;712:135612. [DOI] [PubMed] [Google Scholar]
- [82].Sun JZ, Peter Kingori G, Si RW, et al. Microbial fuel cell-based biosensors for environmental monitoring: a review. Water Sci Technol. 2015;71(6):801–809. [DOI] [PubMed] [Google Scholar]
- [83].Do MH, Ngo HH, Guo W, et al. A dual chamber microbial fuel cell based biosensor for monitoring copper and arsenic in municipal wastewater. Sci Total Environ. 2022;811:152261. [DOI] [PubMed] [Google Scholar]
- [84].Xiao N, Selvaganapathy PR, Wu R, et al. Influence of wastewater microbial community on the performance of miniaturized microbial fuel cell biosensor. Bioresour Technol. 2020;302:122777. [DOI] [PubMed] [Google Scholar]
- [85].Askari A, Vahabzadeh F, Mardanpour MM. Quantitative determination of linear alkylbenzene sulfonate (LAS) concentration and simultaneous power generation in a microbial fuel cell-based biosensor. J Clean Prod. 2021;294:126349. [Google Scholar]
- [86].Wu LC, Tsai TH, and Liu MH, et al. A green microbial fuel cell-based biosensor for in situ chromium (VI) measurement in electroplating wastewater. Sensors (Basel). 2017;17:2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Khan A, Salama E-S, Chen Z, et al. A novel biosensor for zinc detection based on microbial fuel cell system. Biosens Bioelectron. 2020;147:111763. [DOI] [PubMed] [Google Scholar]
- [88].Zhou T, Li R, Zhang S, et al. A copper-specific microbial fuel cell biosensor based on riboflavin biosynthesis of engineered Escherichia coli. Biotechnol Bioeng. 2021;118(1):210–222. [DOI] [PubMed] [Google Scholar]
- [89].Liu X, Silverman AD, Alam KK, et al. Design of a transcriptional biosensor for the portable, on-demand detection of cyanuric acid. ACS Synth Biol. 2020;9(1):84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Zeng N, Wu Y, Chen W, et al. Whole-cell microbial bioreporter for soil contaminants detection. Front Bioeng Biotechnol. 2021;9:622994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Chen X, Yao H, Song D, et al. Extracellular chemoreceptor of deca-brominated diphenyl ether and its engineering in the hydrophobic chassis cell for organics biosensing. Chem Eng J. 2022;433:133266. [Google Scholar]
- [92].Kannappan S, Ramisetty BCM. Engineered whole-cell-based biosensors: sensing environmental heavy metal pollutants in water—a review. Appl Biochem Biotechnol. 2021;194(4):1814–1840. [DOI] [PubMed] [Google Scholar]
- [93].Singh A, Kumar V. Recent advances in synthetic biology–enabled and natural whole-cell optical biosensing of heavy metals. Anal Bioanal Chem. 2021;413(1):73–82. [DOI] [PubMed] [Google Scholar]
- [94].Ravikumar S, Baylon MG, Park SJ, et al. Engineered microbial biosensors based on bacterial two-component systems as synthetic biotechnology platforms in bioremediation and biorefinery. Microb Cell Fact. 2017;16(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Vareda JP, Valente AJM, Durães L. Assessment of heavy metal pollution from anthropogenic activities and remediation strategies: a review. J Environ Manage. 2019;246:101–118. [DOI] [PubMed] [Google Scholar]
- [96].Sanità Di Toppi L, Gabbrielli R. Response to cadmium in higher plants. Environ Exp Bot. 1999;41(2):105–130. [Google Scholar]
- [97].Phan CC, Nguyen TQH, Nguyen MK, et al. Aerosol mass and major composition characterization of ambient air in Ho Chi Minh City. Vietnam Int J Environ Sci Technol. 2020;17(6):3189–3198. [Google Scholar]
- [98].Karthik V, Karuna B, and Kumar PS, et al. Development of lab-on-chip biosensor for the detection of toxic heavy metals: a review. Chemosphere. 2022;299:134427. [DOI] [PubMed] [Google Scholar]
- [99].Saidur MR, Aziz ARA, Basirun WJ. Recent advances in DNA-based electrochemical biosensors for heavy metal ion detection: a review. Biosens Bioelectron. 2017;90:125–139. [DOI] [PubMed] [Google Scholar]
- [100].Naik S, Jujjavarapu SE. Self-powered and reusable microbial fuel cell biosensor for toxicity detection in heavy metal polluted water. J Environ Chem Eng. 2021;9(4):105318. [Google Scholar]
- [101].Yu D, Bai L, Zhai J, et al. Toxicity detection in water containing heavy metal ions with a self-powered microbial fuel cell-based biosensor. Talanta. 2017;168:210–216. [DOI] [PubMed] [Google Scholar]
- [102].Liu Y, Guo M, Du R, et al. A gas reporting whole-cell microbial biosensor system for rapid on-site detection of mercury contamination in soils. Biosens Bioelectron. 2020;170:112660. [DOI] [PubMed] [Google Scholar]
- [103].Cheng Z, Wei J, and Gu L, et al. DNAzyme-based biosensors for mercury (II) detection: rational construction, advances and perspectives. J Hazard Mater. 2022;431:128606. [DOI] [PubMed] [Google Scholar]
- [104].Shen Y, Wang M, Chang IS, et al. Effect of shear rate on the response of microbial fuel cell toxicity sensor to Cu(II). Bioresour Technol. 2013;136:707–710. [DOI] [PubMed] [Google Scholar]
- [105].Wu Y, Zhao X, Jin M, et al. Copper removal and microbial community analysis in single-chamber microbial fuel cell. Bioresour Technol. 2018;253:372–377. [DOI] [PubMed] [Google Scholar]
- [106].Zhao S, Liu P, and Niu Y, et al. A novel early warning system based on a sediment microbial fuel cell for in situ and real time hexavalent chromium detection in industrial wastewater. Sensors. 2018;18(2):642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Prévoteau A, Rabaey K. Electroactive biofilms for sensing: reflections and perspectives. ACS Sens. 2017;2(8):1072–1085. [DOI] [PubMed] [Google Scholar]
- [108].Wang Z, Lim B, and Choi C. Removal of Hg2+ as an electron acceptor coupled with power generation using a microbial fuel cell. Bioresour Technol. 2011;102(10):6304–6307. [DOI] [PubMed] [Google Scholar]
- [109].Guo X, Sang S, Jian A, et al. A bovine serum albumin-coated magnetoelastic biosensor for the wireless detection of heavy metal ions. Sens Actuators B: Chem. 2018;256:318–324. [Google Scholar]
- [110].Kumar T, Naik S, and Jujjavarappu SE. A critical review on early-warning electrochemical system on microbial fuel cell-based biosensor for on-site water quality monitoring. Chemosphere. 2021;291:133098. [DOI] [PubMed] [Google Scholar]
- [111].Chen Y, Li H, Gao T, et al. Selection of DNA aptamers for the development of light-up biosensor to detect Pb(II). Sens Actuators B: Chem. 2018;254:214–221. [Google Scholar]
- [112].Yang X, He Y, and Wang X, et al. A SERS biosensor with magnetic substrate CoFe2O4@Ag for sensitive detection of Hg2+. Appl Surf Sci. 2017;416:581–586. [Google Scholar]
- [113].Soldatkin OO, Kucherenko IS, Pyeshkova VM, et al. Novel conductometric biosensor based on three-enzyme system for selective determination of heavy metal ions. Bioelectrochemistry. 2012;83:25–30. [DOI] [PubMed] [Google Scholar]
- [114].Liu J, Cao Z, and Lu Y, et al. Functional Nucleic Acid Sensors. Chem Rev. 2009;109:1948–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Xiao Y, Rowe AA, Plaxco KW. Electrochemical detection of parts-per-billion lead via an electrode-bound DNAzyme assembly. J Am Chem Soc. 2007;129(2):262–263. [DOI] [PubMed] [Google Scholar]
- [116].Chen S-Y, Li Z, Li K, et al. Small molecular fluorescent probes for the detection of lead, cadmium and mercury ions. Coord Chem Rev. 2021;429:213691. [Google Scholar]
- [117].Khalid K, Tan X, Mohd Zaid HF, et al. Advanced in developmental organic and inorganic nanomaterial: a review. Bioengineered. 2020;11(1):328–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Jayaraman T, Murthy AP, Elakkiya V, et al. Recent development on carbon based heterostructures for their applications in energy and environment: a review. J Ind Eng Chem. 2018;64:16–59. [Google Scholar]
- [119].Theerthagiri J, Chandrasekaran S, Salla S, et al. Recent developments of metal oxide based heterostructures for photocatalytic applications towards environmental remediation. J Solid State Chem. 2018;267:35–52. [Google Scholar]
- [120].Theerthagiri J, Senthil RA, Malathi A, et al. Synthesis and characterization of a CuS–WO3 composite photocatalyst for enhanced visible light photocatalytic activity. RSC Adv. 2015;5:52718–52725. [Google Scholar]
- [121].Theerthagiri J, Senthil RA, and Priya A, et al. Photocatalytic and photoelectrochemical studies of visible-light active α-Fe2O3-g-C3N4 nanocomposites. RSC Adv. 2014;4(72):38222–38229. [Google Scholar]
- [122].Jayaraman T, Raja SA, and Priya A, et al. Synthesis of a visible-light active V2O5-g-V3N4 heterojunction as an efficient photocatalytic and photoelectrochemical material. New J Chem. 2015;39(2):1367–1374. [Google Scholar]
- [123].Ma J, Bai W, Zheng J. A novel self-cleaning electrochemical biosensor integrating copper porphyrin-derived metal-organic framework nanofilms, G-quadruplex, and DNA nanomotors for achieving cyclic detection of lead ions. Biosens Bioelectron. 2022;197:113801. [DOI] [PubMed] [Google Scholar]
- [124].Sun J, Gan Y, Liang T, et al. Signal enhancement of electrochemical DNA biosensors for the detection of trace heavy metals. Curr Opin Electrochem. 2019;17:23–29. [Google Scholar]
- [125].Lee SJ, Lee H, Begildayeva T, et al. Nanogap-tailored Au nanoparticles fabricated by pulsed laser ablation for surface-enhanced Raman scattering. Biosens Bioelectron. 2022;197:113766. [DOI] [PubMed] [Google Scholar]
- [126].Hashem A, Hossain MAM, Al Mamun M, et al. Nanomaterials based electrochemical nucleic acid biosensors for environmental monitoring: a review. Appl Surf Sci Adv. 2021;4:100064. [Google Scholar]
- [127].Martín-Yerga D, Costa-García A. Recent advances in the electrochemical detection of mercury. Curr Opin Electrochem. 2017;3(1):91–96. [Google Scholar]
- [128].Leth S, Maltoni S, Simkus R, et al. Engineered bacteria based biosensors for monitoring bioavailable heavy metals. Electroanalysis: An Int J Devoted to Fundam and Pract Aspects of Electroanalysis. 2002;14(1):35–42. [Google Scholar]
- [129].Yang Y, Kang M, and Fang S, et al. A feasible C-rich DNA electrochemical biosensor based on Fe3O4@3D-GO for sensitive and selective detection of Ag+. J Alloys Compd. 2015;652:225–233. [Google Scholar]
- [130].Tsekenis G, Filippidou MK, Chatzipetrou M, et al. Heavy metal ion detection using a capacitive micromechanical biosensor array for environmental monitoring. Sens Actuators B: Chem. 2015;208:628–635. [Google Scholar]
- [131].Breaker RR, Joyce GF. A DNA enzyme that cleaves RNA. Chem Biol. 1994;1(4):223–229. [DOI] [PubMed] [Google Scholar]
- [132].Shen L, Chen Z, Li Y, et al. Electrochemical DNAzyme sensor for lead based on amplification of DNA−Au bio-bar codes. Anal Chem. 2008;80(16):6323–6328. [DOI] [PubMed] [Google Scholar]
- [133].Shi L, Wang Y, Ding S, et al. A facile and green strategy for preparing newly-designed 3D graphene/gold film and its application in highly efficient electrochemical mercury assay. Biosens Bioelectron. 2017;89:871–879. [DOI] [PubMed] [Google Scholar]
- [134].Zhou D, Zeng L, Pan J, et al. Autocatalytic DNA circuit for Hg2+ detection with high sensitivity and selectivity based on exonuclease III and G-quadruplex DNAzyme. Talanta. 2020;207:120258. [DOI] [PubMed] [Google Scholar]
- [135].Gutiérrez JC, Amaro F, Martín-González A. Heavy metal whole-cell biosensors using eukaryotic microorganisms: an updated critical review. Front Microbiol. 2015;6:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Chouteau C, Dzyadevych S, Chovelon J-M, et al. Development of novel conductometric biosensors based on immobilised whole cell Chlorella vulgaris microalgae. Biosens Bioelectron. 2004;19(9):1089–1096. [DOI] [PubMed] [Google Scholar]
- [137].Guo M, Wang J, Du R, et al. A test strip platform based on a whole-cell microbial biosensor for simultaneous on-site detection of total inorganic mercury pollutants in cosmetics without the need for predigestion. Biosens Bioelectron. 2020;150:111899. [DOI] [PubMed] [Google Scholar]
- [138].Peng Y, Xu M, and Guo Y, et al. A novel signal amplification biosensor for detection of Cd2+ based on asymmetric PCR. Spectrochim Acta A Mol Biomol Spectrosc. 2022;271:120885. [DOI] [PubMed] [Google Scholar]
- [139].Li J, Yu Y, Qian J, et al. A novel integrated biosensor based on co-immobilizing the mediator and microorganism for water biotoxicity assay. Analyst. 2014;139(11):2806–2812. [DOI] [PubMed] [Google Scholar]
- [140].Yu D, Zhai J, Yong D, et al. A rapid and sensitive p-benzoquinone-mediated bioassay for determination of heavy metal toxicity in water. Analyst. 2013;138(11):3297–3302. [DOI] [PubMed] [Google Scholar]
- [141].Weber R, Watson A, Forter M, et al. Persistent organic pollutants and landfills-a review of past experiences and future challenges. Waste Manage Res. 2011;29(1):107–121. [DOI] [PubMed] [Google Scholar]
- [142].Tuan Tran H, Lin C, Bui X-T, et al. Phthalates in the environment: characteristics, fate and transport, and advanced wastewater treatment technologies. Bioresour Technol. 2022;344:126249. [DOI] [PubMed] [Google Scholar]
- [143].Begildayeva T, Lee SJ, Yu Y, et al. Production of copper nanoparticles exhibiting various morphologies via pulsed laser ablation in different solvents and their catalytic activity for reduction of toxic nitroaromatic compounds. J Hazard Mater. 2021;409:124412. [DOI] [PubMed] [Google Scholar]
- [144].Atar N, Eren T, Yola ML, et al. A sensitive molecular imprinted surface plasmon resonance nanosensor for selective determination of trace triclosan in wastewater. Sens Actuators B: Chem. 2015;216:638–644. [Google Scholar]
- [145].Ejeian F, Etedali P, Mansouri-Tehrani H-A, et al. Biosensors for wastewater monitoring: a review. Biosens Bioelectron. 2018;118:66–79. [DOI] [PubMed] [Google Scholar]
- [146].Zehani N, Fortgang P, Lachgar MS, et al. Highly sensitive electrochemical biosensor for bisphenol A detection based on a diazonium-functionalized boron-doped diamond electrode modified with a multi-walled carbon nanotube-tyrosinase hybrid film. Biosens Bioelectron. 2015;74:830–835. [DOI] [PubMed] [Google Scholar]
- [147].Pan D, Gu Y, Lan H, et al. Functional graphene-gold nano-composite fabricated electrochemical biosensor for direct and rapid detection of bisphenol A. Anal Chim Acta. 2015;853:297–302. [DOI] [PubMed] [Google Scholar]
- [148].Rodriguez-Mozaz S, De Alda ML, Barceló D. Analysis of bisphenol A in natural waters by means of an optical immunosensor. Water Res. 2005;39(20):5071–5079. [DOI] [PubMed] [Google Scholar]
- [149].Kochana J, Wapiennik K, Kozak J, et al. Tyrosinase-based biosensor for determination of bisphenol A in a flow-batch system. Talanta. 2015;144:163–170. [DOI] [PubMed] [Google Scholar]
- [150].Dempsey E, Diamond D, Collier A. Development of a biosensor for endocrine disrupting compounds based on tyrosinase entrapped within a poly (thionine) film. Biosens Bioelectron. 2004;20(2):367–377. [DOI] [PubMed] [Google Scholar]
- [151].Tschmelak J, Proll G, Gauglitz G. Optical biosensor for pharmaceuticals, antibiotics, hormones, endocrine disrupting chemicals and pesticides in water: assay optimization process for estrone as example. Talanta. 2005;65(2):313–323. [DOI] [PubMed] [Google Scholar]
- [152].Abd-El-Haleem D, Ripp S, and Scott C, et al. A luxCDABE-based bioluminescent bioreporter for the detection of phenol. J Ind Microbiol Biotechnol. 2002;29(5):233–237. [DOI] [PubMed] [Google Scholar]
- [153].Tschmelak J, Proll G, Gauglitz G. Verification of performance with the automated direct optical TIRF immunosensor (River analyser) in single and multi-analyte assays with real water samples. Biosens Bioelectron. 2004;20(4):743–752. [DOI] [PubMed] [Google Scholar]
- [154].Tschmelak J, Proll G, Gauglitz G. Ultra-sensitive fully automated immunoassay for detection of propanil in aqueous samples: steps of progress toward sub-nanogram per liter detection. Anal Bioanal Chem. 2004;379(7–8):1004–1012. [DOI] [PubMed] [Google Scholar]
- [155].Chen P, Qiao X, Liu J, et al. Dual-signaling amplification electrochemical aptasensor based on hollow polymeric nanospheres for acetamiprid detection. ACS Appl Mater Interfaces. 2019;11(16):14560–14566. [DOI] [PubMed] [Google Scholar]
- [156].Mauriz E, Calle A, Abad A, et al. Determination of carbaryl in natural water samples by a surface plasmon resonance flow-through immunosensor. Biosens Bioelectron. 2006;21(11):2129–2136. [DOI] [PubMed] [Google Scholar]
- [157].Liu X, Cheng H, Zhao Y, et al. Portable electrochemical biosensor based on laser-induced graphene and MnO2 switch-bridged DNA signal amplification for sensitive detection of pesticide. Biosens Bioelectron. 2022;199:113906. [DOI] [PubMed] [Google Scholar]
- [158].Doong R-A, Shih H-M, Lee S-H. Sol–gel-derived array DNA biosensor for the detection of polycyclic aromatic hydrocarbons in water and biological samples. Sens Actuators B: Chem. 2005;111:323–330. [Google Scholar]
- [159].Nomngongo PN, Ngila JC, Msagati TA, et al. Determination of selected persistent organic pollutants in wastewater from landfill leachates, using an amperometric biosensor. Phys Chem Earth Parts A/B/C. 2012;50:252–261. [Google Scholar]
- [160].Lambert P, Fingas M, Goldthorp M. An evaluation of field total petroleum hydrocarbon (TPH) systems. J Hazard Mater. 2001;83(1–2):65–81. [DOI] [PubMed] [Google Scholar]
- [161].Wasilewski T, Gębicki J, Kamysz W. Advances in olfaction-inspired biomaterials applied to bioelectronic noses. Sens Actuators B: Chem. 2018;257:511–537. [Google Scholar]
- [162].Brito NF, Oliveira DS, Santos TC, et al. Current and potential biotechnological applications of odorant-binding proteins. Appl Microbiol Biotechnol. 2020;104(20):8631–8648. [DOI] [PubMed] [Google Scholar]
- [163].Chung HT, Meshref MNA, Ranjan Dhar B. A review and roadmap for developing microbial electrochemical cell-based biosensors for recalcitrant environmental contaminants, emphasis on aromatic compounds. Chem Eng J. 2021;424:130245. [Google Scholar]
- [164].Roy R, Ray S, Chowdhury A, et al. Tunable multiplexed whole-cell biosensors as environmental diagnostics for ppb-level detection of aromatic pollutants. ACS Sens. 2021;6(5):1933–1939. [DOI] [PubMed] [Google Scholar]
- [165].Patel R, Zaveri P, Mukherjee A, et al. Development of fluorescent protein-based biosensing strains: a new tool for the detection of aromatic hydrocarbon pollutants in the environment. Ecotoxicol Environ Saf. 2019;182:109450. [DOI] [PubMed] [Google Scholar]
- [166].Verma ML, Rani V. Biosensors for toxic metals, polychlorinated biphenyls, biological oxygen demand, endocrine disruptors, hormones, dioxin, phenolic and organophosphorus compounds: a review. Environ Chem Lett. 2021;19(2):1657–1666. [Google Scholar]
- [167].Thavarajah W, Verosloff MS, Jung JK, et al. A primer on emerging field-deployable synthetic biology tools for global water quality monitoring. NPJ Clean Water. 2020;3(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Shahar H, Tan LL, Ta GC, et al. Detection of halogenated hydrocarbon pollutants using enzymatic reflectance biosensor. Sens Actuators B: Chem. 2019;281:80–89. [Google Scholar]
- [169].Riangrungroj P, Bever CS, Hammock BD, et al. A label-free optical whole-cell Escherichia coli biosensor for the detection of pyrethroid insecticide exposure. Sci Rep. 2019;9(1):12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Brewer R, Nagashima J, Kelley M, et al. Risk-based evaluation of total petroleum hydrocarbons in vapor intrusion studies. Int J Env Res Public Health. 2013;10:2441–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Kuppusamy S, Maddela NR, Megharaj M, et al. Methodologies for analysis and identification of total petroleum hydrocarbons. In: Kuppusamy S, Maddela NR, Megharaj M, et al, editors. Total petroleum hydrocarbons: environmental fate, toxicity, and remediation. Cham: Springer International Publishing; 2020. p. 29–55. [Google Scholar]
- [172].Okparanma RN, Mouazen AM. Determination of total petroleum hydrocarbon (TPH) and polycyclic aromatic hydrocarbon (PAH) in soils: a review of spectroscopic and nonspectroscopic techniques. Appl Spectrosc Rev. 2013;48(6):458–486. [Google Scholar]
- [173].Mao K, Zhang H, Wang Z, et al. Nanomaterial-based aptamer sensors for arsenic detection. Biosens Bioelectron. 2020;148:111785. [DOI] [PubMed] [Google Scholar]
- [174].Tsai S-T, Cheng W-J, and Zhang Q-X, et al. Gold-specific biosensor for monitoring wastewater using genetically engineered Cupriavidus metallidurans CH34. ACS Synth Biol. 2021;10(12):3576–3582. [DOI] [PubMed] [Google Scholar]
- [175].Yamashita T, Hasegawa T, Hayashida Y, et al. Energy savings with a biochemical oxygen demand (BOD)- and pH-based intermittent aeration control system using a BOD biosensor for swine wastewater treatment. Biochem Eng J. 2022;177:108266. [Google Scholar]
- [176].Gao G, Fang D, Yu Y, et al. A double-mediator based whole cell electrochemical biosensor for acute biotoxicity assessment of wastewater. Talanta. 2017;167:208–216. [DOI] [PubMed] [Google Scholar]
- [177].Patel R, Mukherjee A, Zaveri P, et al. Optimization of immobilization process and survival study of microbial sensing strains used for aromatic hydrocarbon detection in industrial wastewater. Water Environ J. 2020;34(S1):937–948. [Google Scholar]
- [178].Liu B, Lei Y, Li B. A batch-mode cube microbial fuel cell based “shock” biosensor for wastewater quality monitoring. Biosens Bioelectron. 2014;62:308–314. [DOI] [PubMed] [Google Scholar]
- [179].Adekunle A, Raghavan V, Tartakovsky B. A comparison of microbial fuel cell and microbial electrolysis cell biosensors for real-time environmental monitoring. Bioelectrochemistry. 2019;126:105–112. [DOI] [PubMed] [Google Scholar]
- [180].Logroño W, Guambo A, Pérez M, et al. A terrestrial single chamber microbial fuel cell-based niosensor for biochemical oxygen demand of synthetic rice washed wastewater. Sensors. 2016;16(1):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Olaniran AO, Hiralal L, Pillay B. Whole-cell bacterial biosensors for rapid and effective monitoring of heavy metals and inorganic pollutants in wastewater. J Environ Monit. 2011;13(10):2914–2920. [DOI] [PubMed] [Google Scholar]
- [182].Tardy GM, Lóránt B, Gyalai-Korpos M, et al. Microbial fuel cell biosensor for the determination of biochemical oxygen demand of wastewater samples containing readily and slowly biodegradable organics. Biotechnol Lett. 2021;43(2):445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Corbella C, Hartl M, Fernandez-gatell M, et al. MFC-based biosensor for domestic wastewater COD assessment in constructed wetlands. Sci Total Environ. 2019;660:218–226. [DOI] [PubMed] [Google Scholar]
- [184].Berberich J, Li T, and Sahle-Demessie E. Chapter 11 - Biosensors for monitoring water pollutants: a case study with arsenic in groundwater. In: Ahuja S, editor. Sep. Sci. Technol. Vol. 11. Massachusetts, United States: Academic Press; 2019. p. 285–328. [Google Scholar]
- [185].Rajkumar P, Ramprasath T, and Selvam GS. 12 - A simple whole cell microbial biosensors to monitor soil pollution. In: New pesticides and soil sensors, A.M. Grumezescu. (editor) Massachusetts, United States: Academic Press;2017. 437–481. [Google Scholar]
- [186].Babapoor A, Hajimohammadi R, Jokar SM, et al. Biosensor design for detection of mercury in contaminated soil using rhamnolipid biosurfactant and luminescent bacteria. J Chem. 2020;2020:9120959. [Google Scholar]
- [187].Hashwan SSB, Khir MHBM, Al-Douri Y, et al. Recent progress in the development of biosensors for chemicals and pesticides detection. IEEE Access. 2020;8:82514–82527. [Google Scholar]
- [188].Naresh V, Lee N. A review on biosensors and recent development of nanostructured materials-enabled biosensors. Sensors (Basel). 2021;21(4):1109. [DOI] [PMC free article] [PubMed] [Google Scholar]


