Abstract
DNA is highly vulnerable to spontaneous and environmental timely damage in living cells. DNA damage may cause genetic instability and increase cancer risk if the damages are not repaired timely and efficiently. Human cells possess several DNA damage response (DDR) mechanisms to protect the integrity of their genome. Clarification of the mechanisms underlying the DNA damage response following lethal damage will facilitate the identification of therapeutic targets for cancers. Histone post-translational modifications (PTMs) have been indicated to play different roles in the repair of DNA damage. In this context, histone PTMs regulate recruitment of downstream effectors, and facilitate appropriate repair response. This review outlines the current understanding of different histone PTMs in response to DNA damage repair, besides, enumerates the role of new type PTMs such as histone succinylation and crotonylation in regulating DNA damage repair processes.
Keywords: DNA damage, DNA damage response, Histone, Post-translational modifications (PTMs)
Introduction
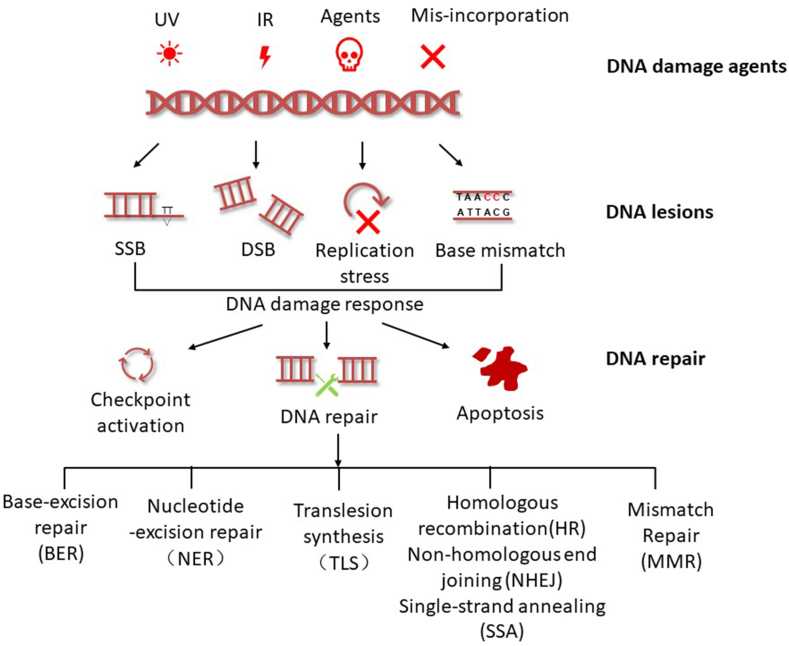
DNA reserves crucial genetic information and regulates essential pathways including accurate DNA replication, transcription, and translation. However, genomic DNA in living organisms is continuously threatened by DNA damage agents such as ultraviolet (UV) light, ionizing radiation (IR), alkylating agents or crosslink agents.1 Those damages interfere with DNA transcription and replication, resulting in senescence, genomic instability, mutations, malignant transformation and even cell death ultimately. To sustain regular cellular events, multiple DNA damage response (DDR) pathways have been involved to maintain the integrity of genomic DNA, including cell cycle checkpoint activation, cellular apoptosis pathway and precise cellular damage repair mechanisms. Different types of DNA damages require specific DNA repair mechanisms to fix the damage sites for conducting routine genetic information transmitting (Fig. 1). For example, UV-induced DNA lesions or other bulky lesions are repaired by nucleotide excision repair (NER). In contrast, base-excision repair (BER) and DNA mismatch repair (MMR) are commonly proceeded to fix the basic site and the oxidized site, or correct insertion loops, respectively.2, 3, 4 Double stand breaks (DSBs) are the most severe and lethal damages which lead to genome instability due to the highly cytotoxicity. There are two major pathways to repair DSBs in mammalian cells: homologous recombination (HR) and non-homologous end-joining (NHEJ). In addition, single-strand annealing (SSA) is reported to be involved in DSBs repair.5,6 The invalid DNA repair mechanisms might induce disorders of cellular environmental homeostasis and cause severe diseases such as ataxia telangiectasia syndrome (A-T) induced by ATM gene mutatione,7 hereditary disease xeroderma pigmentosum variant (XP-V)8 and Fanconi anemia (FA).9
Figure 1.
DNA damage response framework.
In eukaryotes, chromatin is composed of DNA and protein complexes called nucleosomes. Each two of four core histones, H2A, H2B, H3, and H4, are assembled into an octamer to form the core particle of a nucleosome with 147 base pairs of DNA wrapped around. The linker histone, known as H1, serves as linker to stabilize nucleosomes.10 Histones play a critical role in sustaining the structure of DNA, maintaining the stability of genetic information, and regulating gene expression. Post-translational modifications (PTMs) of histone are involved in many biology processes and closely related to gene transcription, DNA replication, chromatin condensation and DNA damage repair.11, 12, 13 Histone modifications include methylation, acetylation, phosphorylation, ubiquitination and SUMOylation, as well as novel PTMs crotonylation, succinylation and so on. PTMs of histone are triggered at different DNA damage sites induced by endogenous or exogenous stimulus, through which the DNA damage repair factors are recruited to complete DNA damage repair14 as well as chromatin remodeling (Fig. 2).
Figure 2.
Distributions of histones post-translational modifications (PTMs). The mainly identified PTMs, including lysine acetylation, ubiquitination, methylation, succinylation and crotonylation sites on histones are listed here.
In recent years, multifarious histone modification sites and advanced modification types are investigated (Table S1), which provide us a novel insight on how histone PTMs mediated gene transcription, chromatin remodeling and DNA damage repair.
Histone ubiquitination
Protein ubiquitination is an enzymatic process by adding ubiquitin onto a lysine amino acid in the substrate protein, which requires sequential actions of three enzymes, ubiquitin activating enzyme (E1), ubiquitin conjugating enzyme (E2), and ubiquitin ligase (E3). To reverse the ubiquitination reaction, the deubiquitinating enzymes (DUBs) remove ubiquitin molecules or polyubiquitin chains from substrates. Protein ubiquitination is strongly demonstrated to contribute in multiple cellular process. K48-linked polyubiquitin chain is one of the most common types, which is associated with proteasomal degradation of the target protein, whereas K63-linked polyubiquitin chains can regulate DNA damage response but not ubiquitin-dependent degradation. Moreover, other types of chains assembled through K6, K11, K27, K29 and K33 residues capture equal attention in the latest studies.15, 16, 17, 18 Pioneering studies reported that K6-linked ubiquitination chains formulated in replication stress and DSBs.
H2A/B ubiquitination
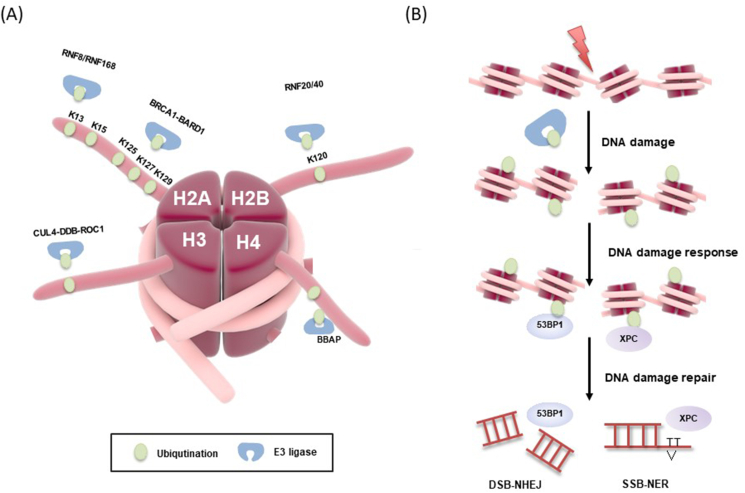
RNF8 and RNF168 are two crucial E3 ligases that promote H2AK15ub for 53BP1 recruitment during DNA damage repair.19 KMT5A is identified as the substrate of RNF8 in vitro and in vivo. RNF8-induced KMT5A ubiquitination increases the binding capacity of KMT5A to RNF168, which further promotes the activity of RNF168 in catalyzing H2A ubiquitination. Simultaneously, KMT5A could also increase H4K20 mono-methylation at DSBs sites, which highlights a new insight into the modification crosstalk during DNA damage repair.20,21
Histone ubiquitinated by RNF168 at DSBs site recruits BRCA1 and 53BP1, which are key factors of the HR and NHEJ repair pathways, respectively. RNF168 catalyzes H2A ubiquitination at K13/15, forming a binding module for 53BP1. BRCC36 and POH1, two DUBs, display strong deubiquitination activity for RNF8/RNF168-mediated K63 ubiquitination at DSB sites.19 Other models have been proposed that BRCA1 partners with BARD1 and mobilizes 53BP1 through its E3 ligase activity. BRCA1-BARD1 performs H2A K125/127/129 ubiquitination to enhance the recruitment of chromatin remodeler SMARCAD1, which recruits 53BP1 allowing the completion of DNA damage repair. Ultimately, these events promote end resection and successful completion of NHEJ-mediated repair.22 Literally, RNF8/RNF168 was shown to repair DSBs through HR and NHEJ. However, USP48 deubiquitinates H2AK125/127/129 and inhibits the function of the BRCA1 E3 ligase. Loss of USP48 and 53BP1 increases retention at the break site and DNA resection lengths are extended.
The mono-ubiquitinated residue was first mapped to K119 (H2AK119ub1) and was initially speculated to be involved in transcriptional regulation. RNF2/BMI1 was specifically shown to ubiquitinate H2AX at K119/120 and the expression of this mutant increases sensitivity to IR, which further identified that loss of RNF2/BMI1-mediated H2AXK120ub may impair γH2AX and phosphorylated ATM foci, as well as the accumulation of DSBs mediators including MDC1, BRCA1, and 53BP1.23
Beyond that, a non-canonical role of H2B ubiquitination in DNA damage repair has also been investigated. Bre1 is the homolog of RNF20/RNF40 dimer in yeast. Upon the induction of DSBs, Bre1 is recruited to DSB ends to stimulate H2Bub which allows efficient Rad51 loading and performs HR repair. Bre1-mediated H2Bub is regulated by specific interaction with RPA, suggesting that the distribution of Bre1-H2Bub on chromatin is spatiotemporally regulated.24
H2B ubiquitination has been demonstrated to be essential for maintaining the functionality of p53 tumor suppressor protein.25 RNF20/RNF40 dimer catalyzes the H2B mono-ubiquitination at K120 (H2BK120ub). WAC, one functional partner of RNF20/RNF40 dimer, further mediates the interaction of RNF20/RNF40 dimer to recruit the complex for H2Bub to target the p53 gene loci.26,27
Not like the consensus pathway, histone ubiquitination is recruited by γH2AX when suffering the DNA damage. IR-induced H2B ubiquitination is not required for conventional γH2AX as antecedent. This notion may declare that RNF20/RNF40-mediated H2B ubiquitination in response to DSBs probably represent an independent yet uncharacterized pathway for DSBs signaling.28 Several studies have attempted to illuminate the added functional consequence of H2B ubiquitination after DNA damage. One potential function could be to recruit a specific subset of DSB repair factors to the lesion as aforementioned, whereas another rational speculation for H2Bub in DSBs repair is to promote or restrain other histone PTMs to exert their effects in DSBs repair.29 Several studies have proposed that RNF20/RNF40-mediated H2Bub is a prerequisite for H3K4/K79 methylation in order to promote transcription.30
H3, H4 ubiquitination
Histone ubiquitination tends to occur on H2A and H2B, whereas ubiquitination of H3 and H4 play a significant role in regulating chromatin function as well as DNA damage response despite the levels of H3/H4 ubiquitination are limited (0.05%–0.3%).31
H3 and H4 were ubiquitinated by CUL4-DDB-ROC1, which subsequently interacted with CUL4 to effectively repair UV-induced thymine dimers via recruiting downstream repair factors XPC to the damage sites for NER. In addition, ubiquitinated H3/H4 also changed nucleosome stability to release histones, allowing the naked DNA to tightly bind to XPC, thus further enhancing repair ability.32 More convincing results regarding the role of H4 ubiquitination acting on DDR have been investigated. The E3 ligase BBAP directly interacted and mono-ubiquitinated H4 in vivo, specifically at H4K91. Additionally, an H4K91 mutant yeast strain had increased sensitivity to hydroxyurea and doxorubicin, pointing to a role for H4K91ub in regulating the response to DSB-inducing agents.33,34
H1 ubiquitination
The most essential ubiquitination event is the E3 ligase RNF168-RNF8-mediated histone ubiquitination. Exogenous damage agent UV and IR could result in DSBs, while H2AX instantly is phosphorylated to generated γH2AX as the DSBs marker. γH2AX recruits the mediator protein MDC1 to the chromatin, subsequently, ATM phosphorylates MDC1 to recruit the E2-E3 complex RNF8-UBC13.35 H2A/H2AX is ubiquitinated by the RNF8-UBC13 complex, which is prerequisite to accumulate downstream DNA damage repair factors to the damage sites, such as 53BP1 and BRCA1.36
RAD6A and RAD6B, two novel partners of RNF168, were shown involved in IR-induced DNA damage repair.37,38 In RAD6-deficient cells, the key repair factors 53BP1 and BRCA1 are impaired, in line with the report from Liu et al which indicated RNF168-RAD6 complex can ubiquitinate H3 and H4 in vitro as well as H1.2ub in response to the IR-induced DNA damage.39
Future researches are needed to establish a clear correlation of multiple histone ubiquitination in response to DNA damage repair. Since H3 and H4 ubiquitination involves in DNA repair, it is vital to elaborate on the mechanistic details to clarify the role of H3/H4 ubiquitination in the DDR (Fig. 3).
Figure 3.
Model of site-specific ubiquitination in the DNA damage response (DDR). (A) Different E3 ligases play a role in DNA damage response, each modifying a specific site on the histone. (B) DNA damage-induced histone ubiquitination triggered the activation of NHEJ (53BP1) and NER (XPC) pathway.
Histone methylation
Compared with phosphorylation and acetylation, methylation modifications are diverse and more complex due to their diversiform modification types. Histone lysine methylation can undergo three types of modifications containing me1 (mono-methyl), me2 (di-methyl) and me3 (tri-methyl), while arginine can just undergo both mono- and di-methyl modifications. Distinct methylation varieties harbor multifarious functions, containing activation/inhibition of gene expression together with chromatin properties regulation.40
H3K4me2/3
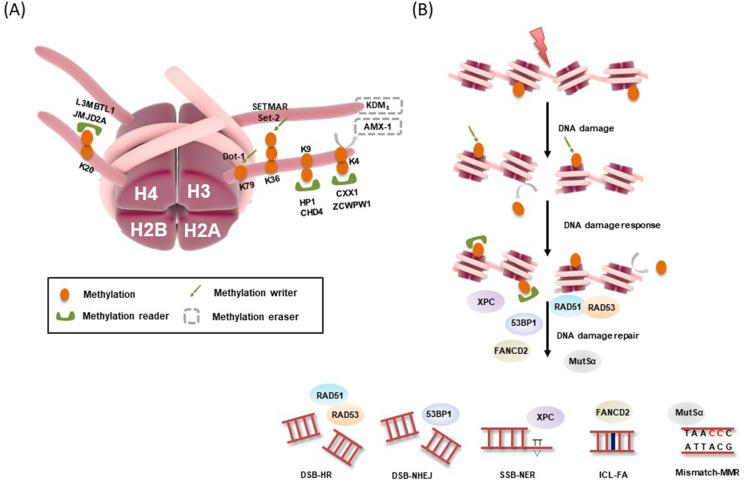
DSBs evoked a series of reactions in downstream, triggering the poor DSB predictor H3K4me3 at the earliest stages of damage repair process, despite H3K4me3 constitutively higher close to DSB sites.41 In yeast and mammals, H3K4me3 marks preferred sites, called hotspots, of DSB formation. DSBs initiate recombination within hotspots where PRDM9 binds, whereafter H3K4me3 and H3K36me3 are deposited.42 The histone methylation reader ZCWPW1 is recruited by PRDM9 to its binding sites and catalyzes dual H3K4me3-H3K36me3 mark for DSBs positioning, further enabling homologous chromosome pairing.43,44 Although it has been suggested that ZCWPW1 might recruit the DSBs machinery to PRDM9 binding sites, but antipodal notion suggests that DSBs positioning is completely unaffected by loss of ZCWPW1. Spp1, a PHD finger containing methyltransferase in yeast, recognized H3K4me3 to initiate meiotic recombination. During meiosis, Spp1 binds to H3K4me3 and Mer2 to promote DSBs formation closing to gene promoters.45 Opposite perspective has also been investigated in mammalians. H3K4me3 is deposited by a family of histone methyltransferases, known as SET1.46
CXXC1, one DNA-binding component of the SETD1 methyltransferase complex, is essential in regulating H3K4me3 for proper DSBs repair and meiotic cell cycle progression during spermatogenesis. Lack of CXXC1 might interrupt the recruitment of H3K4me3 to chromosome axis to form DSBs, especially in hotspots. Thus, other chromatin loops without H3K4me3 modification were randomly tethered to form DSBs resulting in disorder of the canonical hotspots.47
Close liaison with DSBs, evidence suggests that H3K4 methylation memory, which is normally associated with increased transcriptional activity, interferes with cell reprogramming, and the development of somatic cell nuclear transfer embryos.48 Therefore, the functional significance of H3K4me in response to DNA damage warrants further investigation.
De-H3K4/9me
Heterochromatin protein 1 (HP1) is a conserved factor critical for heterochromatin organization and gene silencing. It is recruited to chromatin by its direct interaction with H3K9me, an epigenetic mark for silenced chromatin.49 In direct vicinity of DSB sites, BRCA1 is recruited to DSB sites through BARD1 interaction with HP1 associated with H3K9me2.
Demethylation of H3K4/9 identically helps orchestrate events such as transcription, DNA damage repair, and meiotic crossover recombination.50 Histone lysine demethylases KDM1A, KDM5A, KDM5B, as well as KDM4B and KDM4D, are suggested to be involved in the DNA damage response process.51 H3K4me2 levels are important for recovery from DNA damage repair due to global gene expression changes. Meanwhile, H3K4me2 is modulated by the FA repair pathway, which serves as a connection between global methylation and DSBs repair.52 Recent study by Wang et al53 indicated that mutant of AMX-1, one of the C. elegans histone demethylase, increased the resistance of C. elegans to UV irradiation while UV-induced H3K4me2 was irrespective of NER. Otherwise, AMX-1 mutants activate the ATR/CHK-1 pathway and p53/CEP-1-mediated apoptosis, which supports the idea that AMX-1 is engaged in inter-strand crosslink damage except for UVC.54 Additionally, KDM5B-mediated demethylation of H3K4 facilitates the access of DNA repair enzymes, and increases radio-resistance in cancer cells via its attenuation or pharmacological inhibition.55
KDM7C can demethylate both H3K9me1/2 and H4K20me3, and promote the expression of these genes likely via H3K9 demethylation on the respective gene promoters. Accordingly, knockdown of KDM7C induced R-loop accumulation which led to DSB damage. Thus, KDM7C could prevent DSB damage in neural progenitor cells by regulating gene expression through its H3K9 demethylase activity.56
H3K36me1/2/3
Methyltransferase Set2-mediated methylation of H3K36 in yeast, which involves H3K36me1/2/3, has been demonstrated to intervene in numerous chromatin-coupled events including but not limited to transcription, mRNA splicing and DNA repair.57 SETMAR-mediated H3K36me2 but not H3K36me3 promotes NHEJ in human cells.58 H3K36me2 near DSBs can be enhanced through local generation of fumarate and be counteracted by expression of the KDM2 histone demethylases. Once accumulated, H3K36me2 improves the association of early DNA repair components, including Ku70, with DSBs. In yeast, Set2-mediated H3K36me3 is enriched in the DSB sites, subsequently promoting NHEJ repair. Lacking of H3K36me exhibits highly sensitivity to DNA damage agents, thereby inhibiting H2AX phosphorylation and DNA damage checkpoint activation, ultimately resulting in cell growth inhibition and even cell death.59 In contrast to yeast, H3K36me3 promotes MMR by recruiting the mismatch recognition protein MutSalpha through its PWWP domain in mammalian.60 Genome-wide mutational analyses have shown that the presence of H3K36me3 reduces local mutation rate, operating more efficiently MMR in H3K36me3-enriched exons.61 Upon DNA damage, multifunctional chromatin protein PSIP1 is recruited by H3K36me3, subsequently recruiting the important repair factors CtIP and RAD51 to promote HR repair via efficient resection, which protects these vulnerable regions of the genome from DNA damage.62 Recent reports by Sundarraj et al63 demonstrated two isoforms of PSIP1:p52/p75, who interact with H3K36me3 to take effect further. Interestingly, there is a specific association of a DNA damage marker γH2AX with PSIP/p75, but not the p52 isoform. These results confirm that both isoforms of PSIP1 have different protein partners and cellular functions, also suggesting a wider role of H3K36me3/PSIP1 axis in DNA damage.
H3K79me
In yeast, 90% of H3K79 undergoes methylation modifications, which is deemed to activate transcription and chromatin rearrangement.64 Recent genome-wide mapping of H3K79 methylation revealed that promoter regions of the yeast genome, where most of the meiotic DSBs occur, tend to exhibit reduced levels of H3K9me.65 Consistent reports by Usui et al66 demonstrated that meiotic cells rely more on Dot1-dependent H3K79me to activate Rad53 in response to exogenous DSBs, while reduction of H3K79 methylation around DSB sites might partially explain the weak binding of Rad9 to meiotic DSBs. Ultimately, DOT1L-mediates H3K79me2 further recruit 53BP1 to the DSBs sites for the NEHJ repair process.
Chromatin structure has a profound influence on the repair of UV lesions by restricting access of the NER machinery to sites of DNA damage. The genome of quiescent cells is present in condensed, and deacetylated chromatin with H3K36me or H3K79me3 marked mutation site of exons.67 However, these modifications may generally alter chromatin structure to allow access to NER repair factors since the basilic role of H3K79me in emblemizing during NER in quiescent cells.64 In proliferating cells, Dot1-mediated H3K79me has been reported to play a key role in promoting NER by the GGR-NER pathway, which appears to be a significant NER pathway in quiescent cells.68 Conversely, Zhang et al69 found that polycyclic aromatic hydrocarbons (PAHs) exposure destroys intracellular homeostasis, symbolized by decreased level of H3K79me2. Their results have revealed that aberrant H3K79me2 may contribute to genomic instability and DNA mutation accumulation following long-term exposure to PAHs, which may initiate and promote carcinogenesis. Hitherto, H3K79me has been confirmed to be involved in various types of cellular process, which may indicate its pivot role. How the H3K79me facilitating chromatin rearrangement and DDR is an attractive question to investigate in future.
H4me
H4K20 methylation oscillates during the cell cycle with crucial implications for chromosome replication, condensation, and stability.70 It has been reported that the H4K20me1/2 remains intact after IR exposure, whereas H4K20me3 was significantly pronounced, which may imply the potential effects of H4K20 methylation in regulating DNA damage.71 Recently, it has been shown that H4K20me2 is most abundant during G1 phase, but decrease 2-fold during S phase, and unmodified histones are deposited onto nascent DNA. H4K20me2 levels are subsequently restored during G2 phase upon expression of lysine methyltransferase 5A (KMT5A). Decreased levels of H4K20me2 during S phase are highly likely to impact DNA repair pathway choice.72 Consistently, restoration of H4K20me2 levels during G2 phase coincides with 53BP1 nuclear foci formation.73
H4K20me2 is bound by TUDOR domain-containing proteins including JMJD2A and L3MBTL1. L3MBTL1 is sent for proteasomal degradation following its RNF8-mediated K48-linked polyubiquitination in DNA damage sites. The removal of L3MBTL1 from chromatin exposes H4K20me2 and enables 53BP1 binding. Likewise, deletion of JMJD2A triggered recruitment of 53BP1 to the damage sites. These findings give us respectable insight that DNA damage not only leads to de novo H4K20me2 synthesis but the unmasking of this mark as well.74 The current model suggests that the combination of poly-ubiquitinated L3MBTL1 and p97 leads to the extraction of L3MBTL1 from chromatin to allow for proper 53BP1 assembly at H4K20me2 marks. Paquin et al75 performed that H4K20me2 could interact with FANCD2, sequentially involved in FA repair pathway. Disruption of the methyl-binding domain resulted in the loss of FANCD2 foci formation but increasing the interaction between 53BP1 and H4K20me2, which suggested that the chromosomal aberrations were associated with loss of HR and repaired via NHEJ. Disparate perspectives support that MMSET-mediated H4K20me2 is also involved in NER.76
However, the mechanistic details for how specific site histone methylation triggering downstream repair factors is unknown. Understanding the inter-relationship between methylation and demethylation would provide a clear clue to precisely regulate DDR (Fig. 4).
Figure 4.
Schematic of a site-specific methylation in the DNA damage response (DDR). (A) Histone methylation sites involved in the DDR are shown. Histone methylation writer catalyzed the addition of methyl groups to histone lysine and eraser reversed this reaction. (B) DNA damage-induced histone ubiquitination triggered the activation of NHEJ (53BP1), HR (RAD51/53), NER (XPC), FA repair (FANCD2) and MMR (MutSα) pathways, respectively.
Histone acetylation
Histone acetylation (Kac) was firstly verified to be associated with gene transcription regulation. Increasing levels of histone acetylation promotes gene transcription, whereas insufficient levels of acetylation causing gene silencing.77 Histone acetylase (HAT) and histone deacetylase (HDAC) are key components to regulate histone acetylation levels, whose alteration in their expression may cause abnormal gene expression, furthermore leading to disease production.78
H1ac
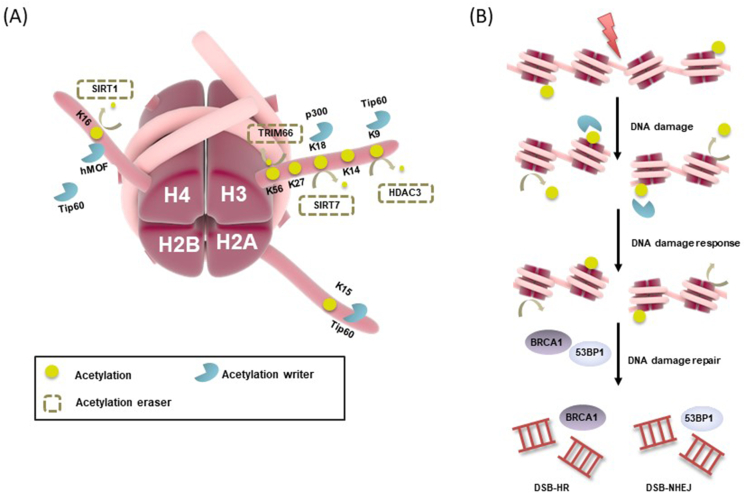
Linker histone H1 is essential for maintaining chromatin and dynamically acetylated by the acetyltransferase PCAF in response to DNA damage.79 H1K85 is one of the PTM-hotpots of the globular domain, which can be modified by acetylation, crotonylation and 2-hydroxyisobutyrylation.80 Intriguingly, DNA damage-induced dynamic changes of H1K85ac permit chromatin relaxation and restoration to ensure genome stability. H1K85ac is deemed to increase H1 binding affinity to result in chromatin condensation, promoting the recruitment of HP1, thus leading to chromatin compaction. Nevertheless, H1 binding affinity and recruitment of HP1 are rapidly reduced due to decreased level of H1K85ac, thus resulting in chromatin decondensation. PCAF is gradually recruited to chromatin in response to DNA damage, restoring H1K85ac levels and chromatin structure after DNA repair.79
H3K9ac
In DSBs site, CK2 phosphorylation releases HP1β from chromatin to expose H3K9me3 and triggers the interaction with TIP60, a HAT that acts as a core factor for the assembly of the repair complex.81 Transforming from H3K9ac to H3K9me3 is one of the key histone modifications that contributes to the DDR and acts as the primary step in DSBs repair either in HR or NHEJ.82 The defect in the H3K9ac/H3K9me3 transition following HDAC3 inactivation impairs TIP60 binding to DSBs foci, further disturbs the assembly of the DDR complex upon DNA damage, thereby results in DNA damage accumulation. Ji et al83 demonstrated that HDAC3 was essential for DNA damage repair, in which HDACs were knocked-out, further identifying H3K9 was exclusively targeted by HDAC3. HDAC1 and HDAC2 have also been reported to play critical roles in DNA damage repair,84 but blockage of their function did not impair DNA damage repair in the mouse livers. In addition, elevated HDAC1 and HDAC2 levels in the liver did not facilitate DNA repair in response to iron exposure, most likely because they do not target H3K9 in the liver.85 In summary, the HDAC3-mediacted H3K9ac/H3K9me3 transition serves as a critical intersection that controls both DNA damage repair and the transcription of many tumor-related genes.
The previous study86 indicated that Rtt109 could acetylate the residues H3K9, H3K27, and H3K56 in yeast. Researches87 found that Rtt109 was a bona fide HAT for acetylation of both H3K9 and H3K56. Sun et al88 indicated that Rtt109 could activate the DNA damage repair function of Aspergillus flavus upon methyl methanesulfonate (MMS)-induced DNA damage, whereas deletion of Rtt109 may vitiate DNA damage repair efficiency.
Recent biochemical and structural studies have suggested that the site-specific efficiency of Rtt109 activity is increased by binding to histone chaperones, Vps75 and Asf1.89 Rtt109-Vps75 specifically acetylates H3K9 and H3K23, whereas Rtt109-Asf1 is tested to favor the acetylation of H3K56 in vitro. Additionally, Rtt109-Asf1 inhibits acetylation at other sites on H3 or H3/H4 compared to Rtt109-Vps75, thereby altering the selectivity.90 Contrary opinion proposed that deletion of Rtt109 or Asf1 in vivo resulted in the same reduction of H3K9ac, which suggested that Asf1was required for efficient H3K9ac.91 Taken together, Vps75 and Asf1 enhance Rtt109 acetylation on H3/H4, although via different mechanisms, but have little impact on the residue selectivity. Importantly, these results provide evidences that histone chaperones can work together via interactions with either the enzyme or the substrate to acetylate histones more efficiently.
H3K56ac and De-H3K56ac
H3K56ac is a core domain acetylation mark that is actively involved in transcription, DNA replication and DDR.92 Current studies report that the dynamics of H3K56ac upon DNA damage depend on the initial acetylation status of the cell, which is also dependent on the level of metabolites in the extracellular environment.93 The low initial acetylation status increases H3K56ac levels in low density cells but decreases H3K56ac content in high cell density cells. These results highlight that H3K56ac levels are dependent on the metabolites in the extracellular milieu which impact chromatin structure via regulating chromatin-modifying enzymes.94 As H3K56ac increasing in tumors, lactic acid and low pH might alter H3K56ac status in tumors, leading to deregulated gene expression, and contributing to tumor progression.94
However, previous studies indicated that H3K56ac is deacetylated at the damage sites, which is a prerequisite for the subsequent DDR.95 TRIM66, as one reader of H3K56ac, negatively correlated with the expression of H3K56ac in embryonic stem cells (ESCs). Wild type TRIM66, rather than TRIM66-depleted mutants, rescued the up-regulated levels of H3K56ac,96 which relieved blockage of the recruitment of downstream DNA repair factors at the damage sites. Further studies showed that TRIM66 might associate with SIRT6, which was responsible for deacetylating H3K56ac in mammalians in response to DNA damage.95,96
H3K56ac deacetylation is down-regulated by HDAC1 and HDAC2 at DSB sites. Cells become more sensitive to the DNA damage agent when H3K56ac is impaired, eventually resulting in chromatin dissociation.97 Simultaneously, another study demonstrated that cells performed recombination-related DNA damage repair typically in H3K56ac impaired cells. Thus, H3K56 acetylation is strictly selective for the type of DNA damage, exercising different damage response capabilities.98
H4ac
In humans, MOF (hMOF), a member of the MYST family of HATs, acetylates histone H4 at lysine 16 (H4K16ac).99 Depleting hMOF renders global reduction of H4K16ac and DNA repair defects in yeast and mammalian cells.100 Another HAT TIP60 acetylates two different histones at H4K16 and H2AK15 after DNA damage. The acetylation of H4K16 provides a steric obstruction to 53BP1 for its binding to the adjacent methylation of H4K20. H2AK15ac directly blocks ubiquitination by modifying the same residue. Horikoshi et al101 showed that higher HR repair efficiency in gene-rich chromatin regions was associated with higher H4K16ac levels as compared with the different gene-poor chromosomal regions. The genomic regions with elevated pre-existing H4K16ac levels are associated with preferential recruitment of HR-related DSB repair proteins and an increased frequency of DSB repair by HR. Then again, one major histone deacetylase SIRT1, which can deacetylate H4K16ac as well as H3K9/14ac, is required for DNA damage repair and maintaining genomic stability in either yeast or mammal.102 Zhong et al103 proved following exposure to H2O2, H4K16ac is significantly decreased to the acute oxidative stress, which indicates that H4K16ac plays a key effect in regulating gene expression in response to DNA damage. Simultaneously, the decreased H4K16ac trendy is closely regulated by SIRT1, which may support the hypothesis that SIRT1 may modulate H4K16ac via two mechanisms. First, SIRT1 may directly deacetylate H4K16ac in response to oxidative stress. Second, SIRT1 may regulate chromatin affinity and the activity of hMOF to promote hypo-acetylation of H4K16 indirectly.
Acetylome analyses revealed a large number of acetylated proteins, including H4K16ac, which are involved in plentiful metabolic processes in the filamentous fungi B.cinerea. Latest study revealed104 that gene BcSAS2, an ortholog of S. cerevisiae Sas2, might be one of the acetyltransferases of B.cinerea. BcSAS2 significantly affected the level of H4K16ac. Chromatin immunoprecipitation (ChIP) and quantitative real-time PCR (qPCR) assay showed that BcSAS2 and H4K16ac could be enriched in the promoter regions of oxidative stress–related genes, which indicating that BcSAS2 and H4K16ac were involved in gene transcriptional regulation in B.cinerea.
HAT, not only the acetylase
Moreover, several HATs also play crucial roles in repairing of DSBs. Specificity protein 1 (Sp1) is ubiquitously known as a transcription factor that regulating genes, taking effect in cell proliferation, apoptosis, and DNA repair.105 Swift et al106 testified that Sp1 acts a pivotal role in modulating p300-mediated chromatin marks involved in NHEJ, but only in G1 phase. Subsequently, p300 performs in DSBs repair by transcriptionally activating BRCA1 to facilitate HR.107 Sp1 is necessary for p300-mediated H3K18ac, which is a prerequisite for SWI/SNF and Ku70 recruitment to DSBs sites for NHEJ.106 Previous studies108 demonstrated that Sp1 was necessary to recruit p300 and SWI/SNF complex to relax the chromatin surrounding the break sites, potentially then interacted with SIRT7 to deacetylate H3K18. Thus, SIRT7-mediated de-H3K18ac was required for 53BP1 recruitment (Fig. 5).
Figure 5.
Schematic of a site-specific acetylation in the DNA damage response (DDR). (A) Histone acetylation sites involved in the DDR are shown. (B) DNA damage-induced histone ubiquitination triggered the activation of NHEJ (53BP1) and HR (BRCA1) pathway.
In brief, histone acetylation is a vital and extensive modification to disassemble chromatin structure, promoting DNA repair machinery accessibly. However, further studies are needed to clarify the detailed mechanism of histone acetylation in DDR, which is essential to insight into selective approaches to target translational application.
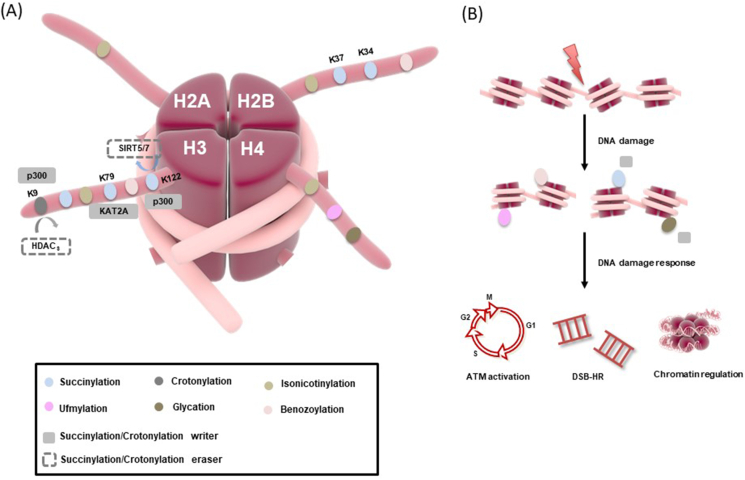
Histone succinylation
Lysine succinylation (Ksucc) was first described by Zhang et al109 Subsequently, the succinylation of H3K122 (H3K122succ) was identified through mass spectrometry immediately.110 In contrast to the majority of other acylation, Ksuc converts the positive charge of the lysine residue to a negative charge and increases the steric hindrance due to its larger volume accordingly affecting interactions into the lysine side chain, and the subsequent chromatin-based processes.111 Jing et al112 preliminary synthesized site-specific succinylation H4 at K77 at a stoichiometric level, which destabilizes the nucleosome and significantly accelerates unwrapping rate of the nucleosome under physiologically relevant condition.
Typical histone succinylation in DDR
Several studies suggested that specific histone acetyltransferase could also catalyze histone succinylation, which might imply the similar effect of Ksucc with Kac. Previously, the site-directed succinylation of H2BK34 in vitro showed that Ksucc destabilized the nucleosome by affecting the interaction of DNA and histone,113 which was consistent with the structural defects of global chromatin in H2BK37E mutant cells. In the report from Zorro and coworkers, succinyl-lysine modification sites in chromatin were enriched at the gene promoter region detected by ChIP-seq, suggesting the role of Ksucc in transcriptional regulation. Recently, it was also found that H3K122succ was enriched in transcription start sites acting as the mark of active genes, stimulating transcription in vitro, and changing the structure of the stable nucleosome.114 Cavalier et al115 demonstrated that p300 could conduct as a lysine succinyl transferase in vitro, which was also confirmed by Zorro et al, as well as ensuring the co-localization between H3K122succ and p300 enrichment at transcription start sites.
Histone desuccinylation and DDR
As described earlier, SIRT7 is one histone deacetylase, more recently, Li et al116 found that SIRT7 can regulate Ksucc in a catalytic activity-dependent manner, supporting a notion that SIRT7 functions as a potential histone desuccinylase. Remarkably, camptothecin (CPT)-induced H3K122succ decline was functionally linked with SIRT7. However, SIRT7-mediated H3K122 desuccinylation after micro-irradiation is PARP1-dependent and specific to DSB sites, which is an essential factor in the priming stage of DNA damage. Besides, H3K122desucc can enhance the binding of H3 with DNA, thereby strengthening chromatin condensation and DNA damage repair.114 Moreover, Li et al revealed that SIRT7 significantly compromised cell survival in response to IR treatment, further catalyzing H3K122desucc to regulate DNA damage repair and cell survival. The results provided novel evidences that histone desuccinylation is important in modulating gene expression.116 SIRT5, another histone deacetylase, has also been reported to desuccinylate H3K122. SIRT5-mediated H3K122desucc is crucial in regulating fatty acid metabolism in multiple tissues.117
Intriguingly, chromatin succinylation represents a novel information-carrying mark of gene activation. ChIP-seq firstly characterizes for epigenome-wide distribution of succinyl-lysine marks in chromatin, revealing a potential role of chromatin succinylation in modulating gene expression.118 Several reports support the notion that chromatin hyper-succinylation may impede DNA repair activities, which is essential for repairing genotoxic DNA damage (Fig. 6).119
Figure 6.
Schematic of a site-specific novel modification in the DNA damage response (DDR). (A) Histone novel modification sites involved in the DDR are shown. (B) Upon DNA damage, histone PTMs trigger ATM activation or homologous recombination (HR) pathway, initiating DNA damage response. In addition, some histone modification could perform chromatin regulation.
Histone crotonylation
As one novel histone PTM, histone crotonylation (Kcr) was first identified in 2011, which is highly evolutionary conserved and completely different type of histone modification from acetylation.120 Previous reports initially identified that histone Kcr enriched in the promoter and enhancer regions in both human somatic and male germinal cells, indicating Kcr is an indicator of gene expression.121,122 Similar to acetylation, Kcr is a reversible modification that is catalyzed by the activity of p300 “writer” protein and removed predominantly by HDACs “eraser” proteins.123 Furthermore, it has been also reported that SIRT1-3 exhibit modest decrotonylase activity.124
Since gene expression is switched off at DNA damage sites and given the emerging role of Kcr in regulating gene expression, histone Kcr in DNA damage repair also exists. Abu-Zhayia et al125 showed a transient local reduction in pan Kcr at laser-micro-irradiated sites marked by γH2AX. The expression level of H3K9cr showed rapid reduction upon IR, UV or damaging agent VP16, which indicated the recovery of H3K9cr after DNA damage is influenced by the type of genotoxic agents.
SIRT-specific inhibitor nicotinamide (NAM), results in a mild increase in Kcr, while HDACs inhibition using a specific inhibitor trichostatin A (TSA) leads to a severely increase in the levels of H3K9cr, suggesting that HDACs are the major lysine decrotonylases.126 The damage-induced reduction in Kcr was dependent on the decrotonylase activity of HDACs, which are known to accumulate at DNA damage sites. Interestingly, DNA damage induction is accompanied by dynamic changes, consisting of increase and decrease in the levels of H3K9me3. The reduction of Kcr might be prerequisite for the alteration in H3K9me3 levels during DNA damage. Due to the dual enzymatic activity of HDACs in regulating both Kac and Kcr, it is unfeasible to decipher the alleged crosstalk between Kcr and Kac, thus the development of a strategy to selectively target either activity is required to elucidate the contribution of each modification to DDR.127,128
Histone novel modification and DDR
In the last decade, multiple novel types of histone modifications and new candidates of PTMs have been reported, including ufmylation, benozoylation, isonicotinylation and glycation.
Ufmylation, a novel identified ubiquitin-like modification, was firstly recognized by Komatsu and Tatsumi et al.129,130 However, there is limited understanding and investigations on function and mechanisms of ufmylation. UFL1, a ubiquitin-fold modifier (UFM1) E3 ligase, is ubiquitously expressed in metazoans. In humans, UFL1-deficient cells are linked to diverse pathologies, including inflammatory, autophagy and ER stress-induced apoptosis, which indicates that ufmylation is essential to regulate multiple cellular processes.131, 132, 133
Recently, Qin et al134 performed that UFL1 was recruited to IR-induced damage sites via the MRE11-RAD50-NBS1 (MRN) complex. Using in vitro system, histone H2A, H2B and H3 could not be modified by UFL1, except for H4. Further experiments confirmed that UFL1 preferentially ufmylated H4 at K31 following DNA damage. This modification enhanced the H3K9me3 expression, which is the prerequisite for subsequent TIP60 recruitment, finally triggering ATM activation. Inhibition of ATM also impaired the recruitment of UFL1 to DSB sites, these results strongly suggested that UFL1 and ATM is closely related to each other and ufmylation of histone is important for regulating the DDR.135
Using Liquid chromatograph Mass spectrometer (LC-MS) and PTMap analysis, Huang et al first identified histone benzoylation (Kbz).136 Benzoylation is one novel and unique histone modification compared with other lysine modifications due to its larger molecular volume and stronger hydrophobicity. In vitro studies demonstrated that SIRT2 as the Kbz eraser, not long afterward, Ren et al137 detected downstream readers of histone Kbz, further confirmed the potential cellular function of histone Kbz. DPF and YEATS, two prominent acylation reader families, were identified as Kbz reader. However, DPF displayed a clear preference for Kcr than Kbz, whereas YEATS2 show totally contrary function. As aforementioned, H3K9cr is one crucial site to respond to DNA damage, H3K9bz is also recognized by AF9YEATS, which may be one potential mechanism to regulate the further DDR process.
Histone PTMs regulate the nucleosome assembly, thus regulating gene expression levels and downstream processes.138 However, histone Kbz is expected to have a more pronounced regulatory effect on genes with more significant structural impact, thus we speculate that specifical histone Kbz may influence gene expression and chromatin conformation.
In the meantime, Jiang et al139 reported one new type histone PTM named lysine isonicotinylation (Kinic). Isoniazid (INH)-induced histone Kinic is dynamically regulated by acetyltransferases CBP/p300 and deacetylase HDAC3. INH-induced Kinic leads to higher sensitivity of nucleosomal DNA, further trigger higher chromatin accessibility. Sat-2 is a marker of altered heterochromatin structure, and the transcription of Sat-2 gene was increased when the mitotic region was relaxed. Similar with another acylation, INH-induced histone Kinic caused heterochromatin relaxation and promoted gene transcription by unfasten the chromatin structure in the genome.
Histone methylglyoxal (MGO) glycation is firstly found by Zheng et al.140 MGO is one vital glycolysis by-product and one most reactive reducing sugar.141 H3 and H4 are prime targets for glycation in vitro recombinant histone system since H3 and H4 glycation quickly rearrange to covalent cross-linking of histone-DNA. These modifications disrupt nucleosome assembly and stability as well as disrupting both local and global chromatin architecture. At high concentrations of MGO or long-term reaction after prolonged exposure, MGO is capable of causing histone-DNA cross-coupling, which can block gene transcription and cause apoptosis. While it is yet to be determined whether histone MGO glycation participates in further DDR process, histone glycation could offer insight into these changes in cell fate through epigenetic regulation (Fig. 6).
Conclusion and future perspectives
Although individual histone PTMs have been reported to involve in DNA damage repair. Defacto, relevant histone PTMs usually undergo intricate crosstalk to modulate multiple cellular processes. In DNA damage sites, H2BK120 undergoes a switch from ubiquitination to acetylation, which is required for H3K79 methylation. Finally H2B ubiquitination stimulates H3K4me3.142 DSBs-mediated TIP60 activation requires the interaction between TIP60 and H3K9me3, thus transition from H3K9ac to H3K9me3 is the key histone modifications in DSB repair either in HR or NHEJ.82 More than that, the expression level of H3K9cr show rapid reduction upon DNA damage agents, thus there must be spatial-temporal rule in regulating DNA damage repair. Upon DSB induction, H4K16ac elicits an open chromatin environment that is permissive for DNA DSB repair. In addition, elevated acetylation of H4K16 diminishes the binding of 53BP1 to H4K20me2-decorated chromatin, which enhances the binding of BRCA1 to chromatin and HR repair. Besides, the discovery that H3K36me3 and H4K16ac are mechanistically coupled in HR-mediated repair of DNA DSBs provides important new knowledge about the modulation of DNA repair by histone epigenetic marks.143 53BP1 as a bivalent histone modification reader that recognizes nucleosomes modified with H4K20me2 and the DNA damage-inducible H2AK15ub mark. As aforementioned, H4K16ac provides a steric obstruction to 53BP1 for its binding to the adjacent methylation of H4K20. H2AK15ac directly blocks ubiquitylation by modifying the same residue. H2AK15 as one hotpot site, the transformation from acetylation to ubiquitination is crucial for routine DNA damage repair performing. With the rapid evolution of novel types of histone PTMs, how to regulate the interactions among revised modifications requires further investigation. Understanding the interrelationship among multiple histone PTMs would provide pivotal insights into how to regulate the DDR.
Not just histone modifications participate in regulating the DDR, moreover, several histone modification enzymes also play crucial roles in repairing of DSBs. In histone acetylation parts, we discussed Sp1 interacts with p300 to modulate the DSBs repair. In addition, DUBs are also involved in DDR. USP13 is an important DUB which is phosphorylated by ATM to facilitate USP13 recruitment to DSBs. In turn, USP13 deubiquitinates RAP80 and promotes RAP80 recruitment to DSBs following DNA damage. Taken together, USP13 regulates RAP80-BRCA1 foci formation by a phosphorylation–deubiquitination axis.144 USP48 deubiquitinates H2AK125/127/129ub and inhibits the function of the BRCA1 E3 ligase. Loss of USP48 and 53BP1 increases retention at the break site and DNA resection lengths are extended.145 In addition, following DNA damage, USP38 interacts with and deubiquitinates HDAC1 directly, which in turn increases the deacetylase activity of HDAC1 and promotes NHEJ.146
With continuing development of technology, increasing new kinds of detecting approaches and PTMs database come into being, providing a basis for sorting and exploiting multiple novel PTMs type from a higher level. Nowadays, computational approaches, particularly artificial intelligence (AI), machine learning (ML) and protein-protein docking, have been applied successfully to resolve bulky PTMs proteomic data analysis. These advancements in AI, ML and protein-protein docking may lead to a completely different future of histone PTMs. Computational techniques have overtaken experimental methods in popularity but need much refinement for the former to replace the later. These methods analyze the physicochemical and geometric properties of proteins to predict probable near-native structures. Precise predicate and data analysis could be performed by these computational approaches to increase realizability from fundamental researcher to clinical transformation.
Recently, numerous studies have shown that PTMs crosstalk can regulate the resistance to radiotherapy of cancer diseases. Though interfering with cellular DNA damage response, HDAC inhibitor, one important epigenetic modulation drug, inhibits DNA damage repair ability and enhances the level of apoptosis, which increases cellular radio-sensitivity.147 Histone PTMs have multiple types to regulate DDR, however, the detailed reports of explicit function in radiosensitivity is still scarce. Therefore, it is crucial to identify and screen out the crosstalk between other PTMs and radiosensitivity, which might be a potential method to assist tumor radiotherapy.
In this review, we discuss the typical or novel histone PTMs in regulating in DDR. However, more investigations on histone modification are still needed, in addition, more novel modifications have yet to be characterized. Insight into these novel modifications will further expand our knowledge of histone PTMs in DNA damage repair and other cellular processes. With progressive method and sophisticated database, it will be much more vital to look at the precise molecular architecture of single DSB site in the future. Solving these problems will be imperative in understanding the detailed molecular mechanism of DDR.
Author contributions
Conception and design of study: Degui Wang, Haoyun Song, Rong Shen, Xiangwen Liu, Xuguang Yang, Kun Xie, Zhao Guo
Drafting the manuscript: Haoyun Song
Revising the manuscript critically for important intellectual content: Rong Shen, Degui Wang, Zhao Guo.
Approval of the version of the manuscript to be published: Degui Wang, Haoyun Song, Rong Shen, Xiangwen Liu, Xuguang Yang, Kun Xie, Zhao Guo
Conflict of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding
This work was financially supported by National Natural Science Foundation of China (No. 82071695 and 82060535), Natural Science Foundation of Gansu Province, China (No. 21JR7RA450).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.04.002.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Chatterjee N., Walker G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ Mol Mutagen. 2017;58(5):235–263. doi: 10.1002/em.22087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullenders L.H.F. Solar UV damage to cellular DNA: from mechanisms to biological effects. Photochem Photobiol Sci. 2018;17(12):1842–1852. doi: 10.1039/c8pp00182k. [DOI] [PubMed] [Google Scholar]
- 3.Kumar N., Raja S., Van Houten B. The involvement of nucleotide excision repair proteins in the removal of oxidative DNA damage. Nucleic Acids Res. 2020;48(20):11227–11243. doi: 10.1093/nar/gkaa777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ijsselsteijn R., Jansen J.G., de Wind N. DNA mismatch repair-dependent DNA damage responses and cancer. DNA Repair. 2020;93:102923. doi: 10.1016/j.dnarep.2020.102923. [DOI] [PubMed] [Google Scholar]
- 5.Ui A., Chiba N., Yasui A. Relationship among DNA double-strand break (DSB), DSB repair, and transcription prevents genome instability and cancer. Cancer Sci. 2020;111(5):1443–1451. doi: 10.1111/cas.14404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ceccaldi R., Rondinelli B., D'Andrea A.D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26(1):52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amirifar P., Ranjouri M.R., Yazdani R., Abolhassani H., Aghamohammadi A. Ataxia-telangiectasia: a review of clinical features and molecular pathology. Pediatr Allergy Immunol. 2019;30(3):277–288. doi: 10.1111/pai.13020. [DOI] [PubMed] [Google Scholar]
- 8.Lambert W.C., Lambert M.W. Development of effective skin cancer treatment and prevention in xeroderma pigmentosum. Photochem Photobiol. 2015;91(2):475–483. doi: 10.1111/php.12385. [DOI] [PubMed] [Google Scholar]
- 9.Nepal M., Che R., Zhang J., Ma C., Fei P. Fanconi Anemia signaling and cancer. Trends Cancer. 2017;3(12):840–856. doi: 10.1016/j.trecan.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammond C.M., Strømme C.B., Huang H., Patel D.J., Groth A. Histone chaperone networks shaping chromatin function. Nat Rev Mol Cell Biol. 2017;18(3):141–158. doi: 10.1038/nrm.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., Sun Z., Jia J., et al. Overview of histone modification. Adv Exp Med Biol. 2021;1283:1–16. doi: 10.1007/978-981-15-8104-5_1. [DOI] [PubMed] [Google Scholar]
- 12.Audia J.E., Campbell R.M. Histone modifications and cancer. Cold Spring Harbor Perspect Biol. 2016;8(4) doi: 10.1101/cshperspect.a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrence M., Daujat S., Schneider R. Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 2016;32(1):42–56. doi: 10.1016/j.tig.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Van H.T., Santos M.A. Histone modifications and the DNA double-strand break response. Cell Cycle. 2018;17(21–22):2399–2410. doi: 10.1080/15384101.2018.1542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faktor J., Pjechová M., Hernychová L., Vojtěšek B. Protein ubiquitination research in Oncology. Klin Onkol. 2019;32(Supplementum 3):56–64. doi: 10.14735/amko20193S. [DOI] [PubMed] [Google Scholar]
- 16.van Wijk S.J., Fulda S., Dikic I., Heilemann M. Visualizing ubiquitination in mammalian cells. EMBO Rep. 2019;20(2) doi: 10.15252/embr.201846520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mansour M.A. Ubiquitination: friend and foe in cancer. Int J Biochem Cell Biol. 2018;101:80–93. doi: 10.1016/j.biocel.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Tracz M., Bialek W. Beyond K48 and K63:non-canonical protein ubiquitination. Cell Mol Biol Lett. 2021;26(1):1. doi: 10.1186/s11658-020-00245-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horn V., Uckelmann M., Zhang H., et al. Structural basis of specific H2A K13/K15 ubiquitination by RNF168. Nat Commun. 2019;10(1):1751. doi: 10.1038/s41467-019-09756-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu X., Xu M., Zhu Q., et al. RNF8-ubiquitinated KMT5A is required for RNF168-induced H2A ubiquitination in response to DNA damage. Faseb J. 2021;35(4) doi: 10.1096/fj.202002234R. [DOI] [PubMed] [Google Scholar]
- 21.Kelliher J.L., West K.L., Gong Q., Leung J.W.C. Histone H2A variants alpha1-extension helix directs RNF168-mediated ubiquitination. Nat Commun. 2020;11(1):2462. doi: 10.1038/s41467-020-16307-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Densham R.M., Garvin A.J., Stone H.R., et al. Human BRCA1-BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat Struct Mol Biol. 2016;23(7):647–655. doi: 10.1038/nsmb.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ismail I.H., Andrin C., McDonald D., Hendzel M.J. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J Cell Biol. 2010;191(1):45–60. doi: 10.1083/jcb.201003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng Z.H., Ai H.S., Lu C.P., Li J.B. The Bre1/Rad6 machinery: writing the central histone ubiquitin mark on H2B and beyond. Chromosome Res. 2020;28(3–4):247–258. doi: 10.1007/s10577-020-09640-3. [DOI] [PubMed] [Google Scholar]
- 25.Liu J., Li X., Zhou G., et al. Silica nanoparticles induce spermatogenesis disorders via L3MBTL2-DNA damage-p53 apoptosis and RNF8-ubH2A/ubH2B pathway in mice. Environ Pollut. 2020;265(Pt A):114974. doi: 10.1016/j.envpol.2020.114974. [DOI] [PubMed] [Google Scholar]
- 26.So C.C., Ramachandran S., Martin A. E3 ubiquitin ligases RNF20 and RNF40 are required for double-stranded break (DSB) repair: evidence for monoubiquitination of histone H2B lysine 120 as a novel axis of DSB signaling and repair. Mol Cell Biol. 2019;39(8) doi: 10.1128/MCB.00488-18. e00488-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang F., Yu X. WAC, a functional partner of RNF20/40, regulates histone H2B ubiquitination and gene transcription. Mol Cell. 2011;41(4):384–397. doi: 10.1016/j.molcel.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hung S.H., Wong R.P., Ulrich H.D., Kao C.F. Monoubiquitylation of histone H2B contributes to the bypass of DNA damage during and after DNA replication. Proc Natl Acad Sci U S A. 2017;114(11):E2205–E2214. doi: 10.1073/pnas.1612633114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu G., Yan J., Wang X., et al. RPA-mediated recruitment of Bre1 couples histone H2B ubiquitination to DNA replication and repair. Proc Natl Acad Sci U S A. 2021;118(8) doi: 10.1073/pnas.2017497118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valencia-Sánchez M.I., De Ioannes P., Wang M., et al. Regulation of the Dot1 histone H3K79 methyltransferase by histone H4K16 acetylation. Science. 2021;371(6527) doi: 10.1126/science.abc6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi Y.K., He Q.Q., Ai H.S., Guo J., Li J.B. The convergent chemical synthesis of histone H3 protein for site-specific acetylation at Lys56 and ubiquitination at Lys122. Chem Commun. 2017;53(29):4148–4151. doi: 10.1039/c7cc01721a. [DOI] [PubMed] [Google Scholar]
- 32.Wang H., Zhai L., Xu J., et al. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell. 2006;22(3):383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 33.Johnson D.P., Spitz G.S., Tharkar S., et al. HDAC1, 2 inhibition impairs EZH2- and BBAP-mediated DNA repair to overcome chemoresistance in EZH2 gain-of-function mutant diffuse large B-cell lymphoma. Oncotarget. 2015;6(7):4863–4887. doi: 10.18632/oncotarget.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tessadori F., Giltay J.C., Hurst J.A., et al. Germline mutations affecting the histone H4 core cause a developmental syndrome by altering DNA damage response and cell cycle control. Nat Genet. 2017;49(11):1642–1646. doi: 10.1038/ng.3956. [DOI] [PubMed] [Google Scholar]
- 35.Smeenk G., Mailand N. Writers, readers, and erasers of histone ubiquitylation in DNA double-strand break repair. Front Genet. 2016;7:122. doi: 10.3389/fgene.2016.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aleksandrov R., Hristova R., Stoynov S., Gospodinov A. The chromatin response to double-strand DNA breaks and their repair. Cells. 2020;9(8):1853. doi: 10.3390/cells9081853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Z., Tian Y., Guo Y., et al. RAD6B plays a critical role in neuronal DNA damage response to resist neurodegeneration. Front Cell Neurosci. 2019;13:392. doi: 10.3389/fncel.2019.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang D., Tian Y., Wei D., et al. DNA damage-induced foci of E2 ubiquitin-conjugating enzyme are detectable upon co-transfection with an interacting E3 ubiquitin ligase. Biochem Genet. 2016;54(2):147–157. doi: 10.1007/s10528-015-9707-8. [DOI] [PubMed] [Google Scholar]
- 39.Liu C., Wang D., Wu J., Keller J., Ma T., Yu X. RNF168 forms a functional complex with RAD6 during the DNA damage response. J Cell Sci. 2013;126(Pt 9):2042–2051. doi: 10.1242/jcs.122945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gong F., Miller K.M. Histone methylation and the DNA damage response. Mutat Res Rev Mutat Res. 2019;780:37–47. doi: 10.1016/j.mrrev.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faucher D., Wellinger R.J. Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet. 2010;6(8) doi: 10.1371/journal.pgen.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diagouraga B., Clément J.A.J., Duret L., Kadlec J., de Massy B., Baudat F. PRDM9 methyltransferase activity is essential for meiotic DNA double-strand break formation at its binding sites. Mol Cell. 2018;69(5):853–865. doi: 10.1016/j.molcel.2018.01.033. e6. [DOI] [PubMed] [Google Scholar]
- 43.Huang T., Yuan S., Gao L., et al. The histone modification reader ZCWPW1 links histone methylation to PRDM9-induced double-strand break repair. Elife. 2020;9 doi: 10.7554/eLife.53459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wells D., Bitoun E., Moralli D., et al. ZCWPW1 is recruited to recombination hotspots by PRDM9 and is essential for meiotic double strand break repair. Elife. 2020;9 doi: 10.7554/eLife.53392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adam C., Guérois R., Citarella A., et al. The PHD finger protein Spp1 has distinct functions in the Set1 and the meiotic DSB formation complexes. PLoS Genet. 2018;14(2) doi: 10.1371/journal.pgen.1007223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sze C.C., Ozark P.A., Cao K., et al. Coordinated regulation of cellular identity-associated H3K4me3 breadth by the COMPASS family. Sci Adv. 2020;6(26):eaaz4764. doi: 10.1126/sciadv.aaz4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang Y., Zhang H.Y., Lin Z., et al. CXXC finger protein 1-mediated histone H3 lysine-4 trimethylation is essential for proper meiotic crossover formation in mice. Development. 2020;147(6):dev183764. doi: 10.1242/dev.183764. [DOI] [PubMed] [Google Scholar]
- 48.Hörmanseder E., Simeone A., Allen G.E., et al. H3K4 methylation-dependent memory of somatic cell identity inhibits reprogramming and development of nuclear transfer embryos. Cell Stem Cell. 2017;21(1):135–143. doi: 10.1016/j.stem.2017.03.003. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ball A.R., Jr., Yokomori K. Revisiting the role of heterochromatin protein 1 in DNA repair. J Cell Biol. 2009;185(4):573–575. doi: 10.1083/jcb.200904033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li J., Ma L., Wang X., et al. Modifications of H3K9me2, H3K36me3 and H4K20me2 may be involved in arsenic-induced genetic damage. Toxicol Res (Camb) 2016;5(5):1380–1387. doi: 10.1039/c6tx00117c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glanzner W.G., Gutierrez K., Rissi V.B., et al. Histone lysine demethylases KDM5B and KDM5C modulate genome activation and stability in porcine embryos. Front Cell Dev Biol. 2020;8:151. doi: 10.3389/fcell.2020.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim H.M., Beese-Sims S.E., Colaiácovo M.P. Fanconi Anemia FANCM/FNCM-1 and FANCD2/FCD-2 are required for maintaining histone methylation levels and interact with the histone demethylase LSD1/SPR-5 in Caenorhabditis elegans. Genetics. 2018;209(2):409–423. doi: 10.1534/genetics.118.300823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang S., Meyer D.H., Schumacher B. H3K4me2 regulates the recovery of protein biosynthesis and homeostasis following DNA damage. Nat Struct Mol Biol. 2020;27(12):1165–1177. doi: 10.1038/s41594-020-00513-1. [DOI] [PubMed] [Google Scholar]
- 54.Zhang X., Tian S., Beese-Sims S.E., et al. Histone demethylase AMX-1 is necessary for proper sensitivity to interstrand crosslink DNA damage. PLoS Genet. 2021;17(7) doi: 10.1371/journal.pgen.1009715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu Y.D., Huang M.J., Guo J.W., et al. Targeting histone demethylase KDM5B for cancer treatment. Eur J Med Chem. 2020;208:112760. doi: 10.1016/j.ejmech.2020.112760. [DOI] [PubMed] [Google Scholar]
- 56.Jeon H.Y., Hussain A., Qi J. Role of H3K9 demethylases in DNA double-strand break repair. J Cancer Biol. 2020;1(1):10–15. doi: 10.46439/cancerbiology.1.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mei Y.C., Feng J., He F., et al. Set2-mediated H3K36 methylation states redundantly repress the production of antisense transcripts: role in transcription regulation. FEBS Open Bio. 2021;11(8):2225–2235. doi: 10.1002/2211-5463.13226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaur E., Nair J., Ghorai A., et al. Inhibition of SETMAR-H3K36me2-NHEJ repair axis in residual disease cells prevents glioblastoma recurrence. Neuro Oncol. 2020;22(12):1785–1796. doi: 10.1093/neuonc/noaa128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pai C.C., Deegan R.S., Subramanian L., et al. A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nat Commun. 2014;5:4091. doi: 10.1038/ncomms5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang Y., Gu L., Li G.M. H3K36me3-mediated mismatch repair preferentially protects actively transcribed genes from mutation. J Biol Chem. 2018;293(20):7811–7823. doi: 10.1074/jbc.RA118.002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aska E.M., Dermadi D., Kauppi L. Single-cell sequencing of mouse thymocytes reveals mutational landscape shaped by replication errors, mismatch repair, and H3K36me3. iScience. 2020;23(9):101452. doi: 10.1016/j.isci.2020.101452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niu N., Lu P., Yang Y., et al. Loss of Setd2 promotes Kras-induced acinar-to-ductal Metaplasia and epithelia-mesenchymal transition during pancreatic carcinogenesis. Gut. 2020;69(4):715–726. doi: 10.1136/gutjnl-2019-318362. [DOI] [PubMed] [Google Scholar]
- 63.Sundarraj J., Taylor G.C.A., von Kriegsheim A., Pradeepa M.M. H3K36me3 and PSIP1/LEDGF associate with several DNA repair proteins, suggesting their role in efficient DNA repair at actively transcribing loci. Wellcome Open Res. 2021;2:83. doi: 10.12688/wellcomeopenres.11589.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wood K., Tellier M., Murphy S. DOT1L and H3K79 methylation in transcription and genomic stability. Biomolecules. 2018;8(1):11. doi: 10.3390/biom8010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fu H., Maunakea A.K., Martin M.M., et al. Methylation of histone H3 on lysine 79 associates with a group of replication origins and helps limit DNA replication once per cell cycle. PLoS Genet. 2013;9(6) doi: 10.1371/journal.pgen.1003542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Usui T., Shinohara A. Rad9, a 53BP1 ortholog of budding yeast, is insensitive to Spo11-induced double-strand breaks during meiosis. Front Cell Dev Biol. 2021;9:635383. doi: 10.3389/fcell.2021.635383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Long L.J., Lee P.H., Small E.M., Hillyer C., Guo Y., Osley M.A. Regulation of UV damage repair in quiescent yeast cells. DNA Repair. 2020;90:102861. doi: 10.1016/j.dnarep.2020.102861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu B., Chen S., Wang H., et al. The protective role of DOT1L in UV-induced melanomagenesis. Nat Commun. 2018;9(1):259. doi: 10.1038/s41467-017-02687-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Z., Xing X., Jiang S., et al. Global H3K79 di-methylation mediates DNA damage response to PAH exposure in Chinese coke oven workers. Environ Pollut. 2021;268(Pt B):115956. doi: 10.1016/j.envpol.2020.115956. [DOI] [PubMed] [Google Scholar]
- 70.Evertts A.G., Manning A.L., Wang X., Dyson N.J., Garcia B.A., Coller H.A. H4K20 methylation regulates quiescence and chromatin compaction. Mol Biol Cell. 2013;24(19):3025–3037. doi: 10.1091/mbc.E12-07-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Svobodová Kovaříková A., Legartová S., Krejčí J., Bártová E. H3K9me3 and H4K20me3 represent the epigenetic landscape for 53BP1 binding to DNA lesions. Aging. 2018;10(10):2585–2605. doi: 10.18632/aging.101572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Audry J., Wang J., Eisenstatt J.R., Berkner K.L., Runge K.W. The inhibition of checkpoint activation by telomeres does not involve exclusion of dimethylation of histone H4 lysine 20 (H4K20me2) F1000Res. 2018;7:1027. doi: 10.12688/f1000research.15166.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Simonetta M., de Krijger I., Serrat J., et al. H4K20me2 distinguishes pre-replicative from post-replicative chromatin to appropriately direct DNA repair pathway choice by 53BP1-RIF1-MAD2L2. Cell Cycle. 2018;17(1):124–136. doi: 10.1080/15384101.2017.1404210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mallette F.A., Mattiroli F., Cui G., et al. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 2012;31(8):1865–1878. doi: 10.1038/emboj.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Paquin K.L., Howlett N.G. Understanding the histone DNA repair code: H4K20me2 makes its mark. Mol Cancer Res. 2018;16(9):1335–1345. doi: 10.1158/1541-7786.MCR-17-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chitale S., Richly H. H4K20me2:Orchestrating the recruitment of DNA repair factors in nucleotide excision repair. Nucleus. 2018;9(1):212–215. doi: 10.1080/19491034.2018.1444327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Koprinarova M., Schnekenburger M., Diederich M. Role of histone acetylation in cell cycle regulation. Curr Top Med Chem. 2016;16(7):732–744. doi: 10.2174/1568026615666150825140822. [DOI] [PubMed] [Google Scholar]
- 78.Tang J., Zhuang S. Histone acetylation and DNA methylation in ischemia/reperfusion injury. Clin Sci (Lond) 2019;133(4):597–609. doi: 10.1042/CS20180465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Y., Li Z., Dong L., et al. Histone H1 acetylation at lysine 85 regulates chromatin condensation and genome stability upon DNA damage. Nucleic Acids Res. 2018;46(15):7716–7730. doi: 10.1093/nar/gky568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Andrés M., García-Gomis D., Ponte I., Suau P., Roque A. Histone H1 post-translational modifications: update and future perspectives. Int J Mol Sci. 2020;21(16):5941. doi: 10.3390/ijms21165941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ayoub N., Jeyasekharan A.D., Bernal J.A., Venkitaraman A.R. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453(7195):682–686. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 82.Ayrapetov M.K., Gursoy-Yuzugullu O., Xu C., Xu Y., Price B.D. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc Natl Acad Sci U S A. 2014;111(25):9169–9174. doi: 10.1073/pnas.1403565111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ji H., Zhou Y., Zhuang X., et al. HDAC3 deficiency promotes liver cancer through a defect in H3K9ac/H3K9me3 transition. Cancer Res. 2019;79(14):3676–3688. doi: 10.1158/0008-5472.CAN-18-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu M., Tu H.Q., Chang Y., et al. USP19 deubiquitinates HDAC1/2 to regulate DNA damage repair and control chromosomal stability. Oncotarget. 2017;8(2):2197–2208. doi: 10.18632/oncotarget.11116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ji H., Zhou Y., Zhuang X., et al. Correction: HDAC3 deficiency promotes liver cancer through a defect in H3K9ac/H3K9me3 transition. Cancer Res. 2020;80(4):923. doi: 10.1158/0008-5472.CAN-19-3887. [DOI] [PubMed] [Google Scholar]
- 86.Albaugh B.N., Arnold K.M., Lee S., Denu J.M. Autoacetylation of the histone acetyltransferase Rtt109. J Biol Chem. 2011;286(28):24694–24701. doi: 10.1074/jbc.M111.251579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pointer B.R., Schmidt M. Boric acid-dependent decrease in regulatory histone H3 acetylation is not mutagenic in yeast. FEMS Microbiol Lett. 2016;363(13):fnw124. doi: 10.1093/femsle/fnw124. [DOI] [PubMed] [Google Scholar]
- 88.Sun R., Wen M., Wu L., Lan H., Yuan J., Wang S. The fungi-specific histone acetyltransferase Rtt109 mediates morphogenesis, aflatoxin synthesis and pathogenicity in Aspergillus flavus by acetylating H3K9. IMA Fungus. 2021;12(1):9. doi: 10.1186/s43008-021-00060-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.D'Arcy S., Luger K. Understanding histone acetyltransferase Rtt109 structure and function: how many chaperones does it take? Curr Opin Struct Biol. 2011;21(6):728–734. doi: 10.1016/j.sbi.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Radovani E., Cadorin M., Shams T., et al. The carboxyl Terminus of Rtt109 functions in chaperone control of histone acetylation. Eukaryot Cell. 2013;12(5):654–664. doi: 10.1128/EC.00291-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kuo Y.M., Henry R.A., Huang L., Chen X., Stargell L.A., Andrews A.J. Utilizing targeted mass spectrometry to demonstrate Asf1-dependent increases in residue specificity for Rtt109-Vps75 mediated histone acetylation. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0118516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vempati R.K., Haldar D. DNA damage in the presence of chemical genotoxic agents induce acetylation of H3K56 and H4K16 but not H3K9 in mammalian cells. Mol Biol Rep. 2012;39(1):303–308. doi: 10.1007/s11033-011-0739-9. [DOI] [PubMed] [Google Scholar]
- 93.Rajagopalan M., Balasubramanian S., Ioshikhes I., Ramaswamy A. Structural dynamics of nucleosome mediated by acetylations at H3K56 and H3K115, 122. Eur Biophys J. 2017;46(5):471–484. doi: 10.1007/s00249-016-1191-5. [DOI] [PubMed] [Google Scholar]
- 94.Vadla R., Chatterjee N., Haldar D. Cellular environment controls the dynamics of histone H3 lysine 56 acetylation in response to DNA damage in mammalian cells. J Biosci. 2020;45:19. [PubMed] [Google Scholar]
- 95.Costelloe T., Lowndes N.F. Chromatin assembly and signalling the end of DNA repair requires acetylation of histone H3 on lysine 56. Subcell Biochem. 2010;50:43–54. doi: 10.1007/978-90-481-3471-7_3. [DOI] [PubMed] [Google Scholar]
- 96.Chen J., Wang Z., Guo X., et al. TRIM66 reads unmodified H3R2K4 and H3K56ac to respond to DNA damage in embryonic stem cells. Nat Commun. 2019;10(1):4273. doi: 10.1038/s41467-019-12126-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhu Q., Battu A., Ray A., et al. Damaged DNA-binding protein down-regulates epigenetic mark H3K56Ac through histone deacetylase 1 and 2. Mutat Res. 2015;776:16–23. doi: 10.1016/j.mrfmmm.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fu I., Geacintov N.E., Broyde S. Molecular dynamics simulations reveal how H3K56 acetylation impacts nucleosome structure to promote DNA exposure for lesion sensing. DNA Repair. 2021;107:103201. doi: 10.1016/j.dnarep.2021.103201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sheikh B.N., Bechtel-Walz W., Lucci J., et al. MOF maintains transcriptional programs regulating cellular stress response. Oncogene. 2016;35(21):2698–2710. doi: 10.1038/onc.2015.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sharma G.G., So S., Gupta A., et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol. 2010;30(14):3582–3595. doi: 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Horikoshi N., Sharma D., Leonard F., et al. Pre-existing H4K16ac levels in euchromatin drive DNA repair by homologous recombination in S-phase. Commun Biol. 2019;2:253. doi: 10.1038/s42003-019-0498-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hajji N., Wallenborg K., Vlachos P., Füllgrabe J., Hermanson O., Joseph B. Opposing effects of hMOF and SIRT1 on H4K16 acetylation and the sensitivity to the topoisomerase II inhibitor etoposide. Oncogene. 2010;29(15):2192–2204. doi: 10.1038/onc.2009.505. [DOI] [PubMed] [Google Scholar]
- 103.Zhong J., Ji L., Chen H., et al. Acetylation of hMOF modulates H4K16ac to regulate DNA repair genes in response to oxidative stress. Int J Biol Sci. 2017;13(7):923–934. doi: 10.7150/ijbs.17260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang G., Song L., Bai T., Liang W. BcSas2-mediated histone H4K16 acetylation is critical for virulence and oxidative stress response of Botrytis cinerea. Mol Plant Microbe Interact. 2020;33(10):1242–1251. doi: 10.1094/MPMI-06-20-0149-R. [DOI] [PubMed] [Google Scholar]
- 105.Swift M.L., Beishline K., Flashner S., Azizkhan-Clifford J. DSB repair pathway choice is regulated by recruitment of 53BP1 through cell cycle-dependent regulation of Sp1. Cell Rep. 2021;34(11):108840. doi: 10.1016/j.celrep.2021.108840. [DOI] [PubMed] [Google Scholar]
- 106.Swift M.L., Beishline K., Azizkhan-Clifford J. Sp1-dependent recruitment of the histone acetylase p300 to DSBs facilitates chromatin remodeling and recruitment of the NHEJ repair factor Ku70. DNA Repair. 2021;105:103171. doi: 10.1016/j.dnarep.2021.103171. [DOI] [PubMed] [Google Scholar]
- 107.Pao G.M., Janknecht R., Ruffner H., Hunter T., Verma I.M. CBP/p300 interact with and function as transcriptional coactivators of BRCA1. Proc Natl Acad Sci U S A. 2000;97(3):1020–1025. doi: 10.1073/pnas.97.3.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang P.Y., Li G., Deng Z.J., et al. Dicer interacts with SIRT7 and regulates H3K18 deacetylation in response to DNA damaging agents. Nucleic Acids Res. 2016;44(8):3629–3642. doi: 10.1093/nar/gkv1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang Z., Tan M., Xie Z., Dai L., Chen Y., Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol. 2011;7(1):58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xu Y.M., Du J.Y., Lau A.Y. Posttranslational modifications of human histone H3:an update. Proteomics. 2014;14(17–18):2047–2060. doi: 10.1002/pmic.201300435. [DOI] [PubMed] [Google Scholar]
- 111.Liu J., Shangguan Y., Tang D., Dai Y. Histone succinylation and its function on the nucleosome. J Cell Mol Med. 2021;25(15):7101–7109. doi: 10.1111/jcmm.16676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jing Y., Ding D., Tian G., et al. Semisynthesis of site-specifically succinylated histone reveals that succinylation regulates nucleosome unwrapping rate and DNA accessibility. Nucleic Acids Res. 2020;48(17):9538–9549. doi: 10.1093/nar/gkaa663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jing Y., Liu Z., Tian G., Bao X., Ishibashi T., Li X.D. Site-specific installation of succinyl lysine analog into histones reveals the effect of H2BK34 succinylation on nucleosome dynamics. Cell Chem Biol. 2018;25(2):166–174. doi: 10.1016/j.chembiol.2017.11.005. e7. [DOI] [PubMed] [Google Scholar]
- 114.Zorro Shahidian L., Haas M., le Gras S., et al. Succinylation of H3K122 destabilizes nucleosomes and enhances transcription. EMBO Rep. 2021;22(3) doi: 10.15252/embr.202051009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cavalieri V. The expanding constellation of histone post-translational modifications in the epigenetic landscape. Genes. 2021;12(10):1596. doi: 10.3390/genes12101596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li L., Shi L., Yang S., et al. SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat Commun. 2016;7:12235. doi: 10.1038/ncomms12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sadhukhan S., Liu X., Ryu D., et al. Metabolomics-assisted proteomics identifies succinylation and SIRT5 as important regulators of cardiac function. Proc Natl Acad Sci U S A. 2016;113(16):4320–4325. doi: 10.1073/pnas.1519858113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Alleyn M., Breitzig M., Lockey R., Kolliputi N. The dawn of succinylation: a posttranslational modification. Am J Physiol Cell Physiol. 2018;314(2):C228–C232. doi: 10.1152/ajpcell.00148.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hang T., Chen W., Wu M., et al. Structural insights into the molecular mechanism underlying Sirt5-catalyzed desuccinylation of histone peptides. Biochem J. 2019;476(2):211–223. doi: 10.1042/BCJ20180745. [DOI] [PubMed] [Google Scholar]
- 120.Tan M., Luo H., Lee S., et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146(6):1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wan J., Liu H., Chu J., Zhang H. Functions and mechanisms of lysine crotonylation. J Cell Mol Med. 2019;23(11):7163–7169. doi: 10.1111/jcmm.14650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ntorla A., Burgoyne J.R. The regulation and function of histone crotonylation. Front Cell Dev Biol. 2021;9:624914. doi: 10.3389/fcell.2021.624914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Huang H., Wang D.L., Zhao Y. Quantitative crotonylome analysis expands the roles of p300 in the regulation of lysine crotonylation pathway. Proteomics. 2018;18(15) doi: 10.1002/pmic.201700230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kelly R.D.W., Chandru A., Watson P.J., et al. Histone deacetylase (HDAC) 1 and 2 complexes regulate both histone acetylation and crotonylation in vivo. Sci Rep. 2018;8(1):14690. doi: 10.1038/s41598-018-32927-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Abu-Zhayia E.R., Machour F.E., Ayoub N. HDAC-dependent decrease in histone crotonylation during DNA damage. J Mol Cell Biol. 2019;11(9):804–806. doi: 10.1093/jmcb/mjz019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wei W., Liu X., Chen J., et al. Class I histone deacetylases are major histone decrotonylases: evidence for critical and broad function of histone crotonylation in transcription. Cell Res. 2017;27(7):898–915. doi: 10.1038/cr.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Machour F.E., Ayoub N. Transcriptional regulation at DSBs: mechanisms and consequences. Trends Genet. 2020;36(12):981–997. doi: 10.1016/j.tig.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 128.Li K., Wang Z. Histone crotonylation-centric gene regulation. Epigenet Chromatin. 2021;14(1):10. doi: 10.1186/s13072-021-00385-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Komatsu M., Chiba T., Tatsumi K., et al. A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J. 2004;23(9):1977–1986. doi: 10.1038/sj.emboj.7600205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tatsumi K., Sou Y.S., Tada N., et al. A novel type of E3 ligase for the Ufm1 conjugation system. J Biol Chem. 2010;285(8):5417–5427. doi: 10.1074/jbc.M109.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xiao J., Liu S., Yu T., et al. UFMylation is associated with LPS-induced inflammatory response in goat endometrial epithelial cells. Reprod Domest Anim. 2020;55(12):1725–1734. doi: 10.1111/rda.13832. [DOI] [PubMed] [Google Scholar]
- 132.Stephani M., Picchianti L., Gajic A., et al. A cross-kingdom conserved ER-phagy receptor maintains endoplasmic Reticulum homeostasis during stress. Elife. 2020;9 doi: 10.7554/eLife.58396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang X., Yu T., Guo X., et al. Ufmylation regulates granulosa cell apoptosis via ER stress but not oxidative stress during goat follicular atresia. Theriogenology. 2021;169:47–55. doi: 10.1016/j.theriogenology.2021.04.009. [DOI] [PubMed] [Google Scholar]
- 134.Qin B., Yu J., Nowsheen S., et al. UFL1 promotes histone H4 ufmylation and ATM activation. Nat Commun. 2019;10(1):1242. doi: 10.1038/s41467-019-09175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fang Z., Pan Z. Essential role of ubiquitin-fold modifier 1 conjugation in DNA damage response. DNA Cell Biol. 2019;38(10):1030–1039. doi: 10.1089/dna.2019.4861. [DOI] [PubMed] [Google Scholar]
- 136.Huang H., Zhang D., Wang Y., et al. Lysine benzoylation is a histone mark regulated by SIRT2. Nat Commun. 2018;9(1):3374. doi: 10.1038/s41467-018-05567-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ren X., Zhou Y., Xue Z., et al. Histone benzoylation serves as an epigenetic mark for DPF and YEATS family proteins. Nucleic Acids Res. 2021;49(1):114–126. doi: 10.1093/nar/gkaa1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Daskalaki M.G., Tsatsanis C., Kampranis S.C. Histone methylation and acetylation in macrophages as a mechanism for regulation of inflammatory responses. J Cell Physiol. 2018;233(9):6495–6507. doi: 10.1002/jcp.26497. [DOI] [PubMed] [Google Scholar]
- 139.Jiang Y., Li Y., Liu C., et al. Isonicotinylation is a histone mark induced by the anti-tuberculosis first-line drug isoniazid. Nat Commun. 2021;12(1):5548. doi: 10.1038/s41467-021-25867-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zheng Q., Omans N.D., Leicher R., et al. Reversible histone glycation is associated with disease-related changes in chromatin architecture. Nat Commun. 2019;10(1):1289. doi: 10.1038/s41467-019-09192-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bellier J., Nokin M.J., Lardé E., et al. Methylglyoxal, a potent inducer of AGEs, connects between diabetes and cancer. Diabetes Res Clin Pract. 2019;148:200–211. doi: 10.1016/j.diabres.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 142.Worden E.J., Hoffmann N.A., Hicks C.W., Wolberger C. Mechanism of cross-talk between H2B ubiquitination and H3 methylation by Dot1L. Cell. 2019;176(6):1490–1501. doi: 10.1016/j.cell.2019.02.002. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li L., Wang Y. Cross-talk between the H3K36me3 and H4K16ac histone epigenetic marks in DNA double-strand break repair. J Biol Chem. 2017;292(28):11951–11959. doi: 10.1074/jbc.M117.788224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li Y., Luo K., Yin Y., et al. USP13 regulates the RAP80-BRCA1 complex dependent DNA damage response. Nat Commun. 2017;8:15752. doi: 10.1038/ncomms15752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Uckelmann M., Densham R.M., Baas R., et al. USP48 restrains resection by site-specific cleavage of the BRCA1 ubiquitin mark from H2A. Nat Commun. 2018;9(1):229. doi: 10.1038/s41467-017-02653-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yang Y., Yang C., Li T., et al. The deubiquitinase USP38 promotes NHEJ repair through regulation of HDAC1 activity and regulates cancer cell response to genotoxic insults. Cancer Res. 2020;80(4):719–731. doi: 10.1158/0008-5472.CAN-19-2149. [DOI] [PubMed] [Google Scholar]
- 147.Rivera S., Leteur C., Mégnin F., et al. Time dependent modulation of tumor radiosensitivity by a pan HDAC inhibitor: abexinostat. Oncotarget. 2017;8(34):56210–56227. doi: 10.18632/oncotarget.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.