Abstract
Solid-like protein deposits found in aged and diseased human brains have revealed a relationship between insoluble protein accumulations and the resulting deficits in neurologic function. Clinically diverse neurodegenerative diseases, including Alzheimer’s disease, Parkinson’s disease, frontotemporal lobar degeneration, and amyotrophic lateral sclerosis, exhibit unique and disease-specific biochemical protein signatures and abnormal protein depositions that often correlate with disease pathogenesis. Recent evidence indicates that many pathologic proteins assemble into liquid-like protein phases through the highly coordinated process of liquid-liquid phase separation. Over the last decade, biomolecular phase transitions have emerged as a fundamental mechanism of cellular organization. Liquid-like condensates organize functionally related biomolecules within the cell, and many neuropathology-associated proteins reside within these dynamic structures. Thus, examining biomolecular phase transitions enhances our understanding of the molecular mechanisms mediating toxicity across diverse neurodegenerative diseases. This Review explores the known mechanisms contributing to aberrant protein phase transitions in neurodegenerative diseases, focusing on tau and TDP-43 proteinopathies and outlining potential therapeutic strategies to regulate these pathologic events.
Introduction
Neurodegenerative diseases (NDDs) are a heterogeneous class of incurable and debilitating disorders characterized by the progressive degeneration of vulnerable cell populations in the central nervous system (CNS). Decades of research investigating the most common NDDs, such as Alzheimer’s disease (AD), Parkinson’s disease (PD), frontotemporal lobar degeneration (FTLD), and amyotrophic lateral sclerosis (ALS), revealed clinical and neuropathologic heterogeneity between, and within, these diseases (1–3). However, NDDs display a fundamental commonality — proteins soluble under physiologic conditions accumulate into solid-like pathologic protein inclusions, and this is associated with clinical progression (4, 5). Furthermore, disease-causing mutations in genes that encode proteins that pathologically accumulate, such as amyloid-β (APP) in AD, tau (MAPT) in FTLD-tau, TDP-43 (TARDBP) in ALS/FTLD–TDP-43, and α-synuclein (SNCA) in PD, cause familial forms of each disease (6–8). Sporadic-NDD patients with unclear familial inheritance and no genetic mutation in the genes that encode these proteins similarly present with neuropathologic deposits of the wild-type protein in the CNS. Furthermore, these sporadic NDDs often display remarkably similar clinical syndromes when compared with the familial form of the disease (2, 9).
Multidisciplinary efforts have gone into understanding mechanisms through which tau and TDP-43 proteins regulate neuronal homeostasis and contribute to NDDs (10–13). These efforts revealed considerable clinical overlap between tau and TDP-43 proteinopathies (14–16). In addition to AD, the most common NDD, solid self-assemblies of tau are found in related dementias termed “tauopathies,” including FTLD-tau, corticobasal degeneration (CBD), Pick’s disease, progressive supranuclear palsy (PSP), and chronic traumatic encephalopathy (CTE) (17). While several mutations in the TARDBP gene contribute to a small percentage of ALS and FTLD–TDP-43 cases, mislocalized and insoluble TDP-43 self-assemblies are found in up to about 97% of individuals with sporadic ALS, up to about 85% with CTE, about 45% with FTLD, and about 40%–60% with AD (18, 19). Several recent studies characterized the ability of these proteins to undergo liquid-like phase separation under physiologic conditions, often into membraneless organelles (13, 20–24). Accordingly, the incidence of tau and TDP-43 pathology across genetic and sporadic NDDs likely highlights a convergence of several upstream mechanisms driving aberrant protein phase transitions and disease progression.
In the following sections, we will explore the relationship between protein structure, biological phase transitions, protein self-assembly, and the organization of multicomponent condensates using tau and TDP-43 as representative proteins. Later sections will survey diverse targeting strategies proposed for tau and TDP-43 proteinopathies, focusing on how protein phase transitions and condensate assembly mechanisms can be leveraged as potential therapeutic avenues of intervention.
Protein self-assembly through homotypic phase transitions
Intracellular NDD-associated proteins self-assemble into diverse polymeric structures and liquid-like protein phases capable of organizing functionally related proteins, nucleic acids, and various biomolecules (13, 22, 25–27). Physiologically, most proteins are soluble and exist in a liquid-like state. In a simple system, e.g., a purified protein in solution, a protein will be soluble when the attractive interactions between different molecules are low enough to maintain a well-mixed state (26, 28, 29). Raising the protein concentration or modifying the balance of attractive and repulsive forces may exceed a protein’s saturation concentration (Csat) and precipitate a new and denser phase. In this context, where the protein is lacking complex biomolecular interactions, homotypic interactions regulate a segregative protein phase transition, which results in a new, denser phase (Cdense) coexisting within the dilute phase (28, 30, 31). Strong driving forces for a given phase transition are generated by lowering of the Csat, a context-dependent property affected by factors intrinsic to a protein’s sequence, localized concentration, cell size changes, temperature, pH, and ionic environments (28, 32–35).
Phase transitions that give rise to two coexisting phases can be liquid-like or display solid-like properties. The appropriate prefix (liquid-like, solid-like) depends on the material properties of the emerging phase. Defining characteristics that distinguish liquid-like protein phases include rapid reversibility, interior molecular diffusion, and the ability to exchange molecules with the surrounding phase (i.e., cytosol) (26, 36). Notably, liquids may transition into solid-like states that emerge through several processes, including gelation, requiring networks of interactions (gel-like), or age-dependent increases in viscosity (glass-like). Solids can also emerge from liquid-like phases by forming fibrils or crystal-like aggregates such as amyloids.
The human proteome is a continuum of protein structures ranging from intrinsically folded proteins to intrinsically disordered proteins (IDPs), with most containing both ordered domains and intrinsically disordered regions (IDRs) (28, 37–39). IDRs are regions of a protein sequence that lack well-defined secondary and tertiary structures. Phase separation and condensate-promoting features include modular interaction domains and stretches of low-complexity sequences found within IDRs. Tau and TDP-43 are modular, multivalent proteins with IDRs that enable and regulate homotypic and heterotypic interactions to generate complex and context-dependent molecular interactions (10, 23, 40–42). These protein architectures tune the concentration required for phase separation and dictate the resultant assembly and material states. Importantly, proteins with IDRs exist in a dynamic equilibrium of conformationally distinct states, and the structural properties of IDRs can quickly adjust owing to changes in solution conditions, posttranslational modifications (PTMs), or interactions with other molecules (26, 34, 36, 43–45).
The generation of neurotoxic self-assemblies represents a fundamental transformation during the pathogenesis of NDDs (4, 5, 17, 46–48). In vitro experiments using purified proteins, work in transgenic animal disease models, and studies with postmortem human brain tissue show that both tau and TDP-43 self-assemble into polymeric states with varying structures and material properties (17, 49–59). Self-polymerization can occur under dilute conditions or within liquid-like droplets. However, pathogenic tau and TDP-43 self-assemblies rely on the exposure of small aggregation-prone regions, including steric zippers or low-complexity, aromatic-rich, kinked segments (LARKS) (60, 61). Disruption in protein conformation due to intrinsic factors (NDD-causing missense mutations, PTMs) or extrinsic factors (biomolecular interactions, cellular environment) may expose these buried short, aggregation-prone sequences in proteins (24, 62). Importantly, specific conformational transformations of protein monomers that are capable of nucleating stable self-interactions with other monomers are required (48, 63). These structural conformations drive unique assembly pathways specific to that protein, which ultimately translates to distinct pathologies observed in NDDs (64). Recent groundbreaking cryo–electron microscopy studies found that specific conformations underlie clinical subtypes of tau and TDP-43 pathologies. Remarkably, an increasing number of clinical presentations and neuropathologic findings correlate with structurally specific fibrils of varying biophysical properties and cellular effects (51–53, 56–58).
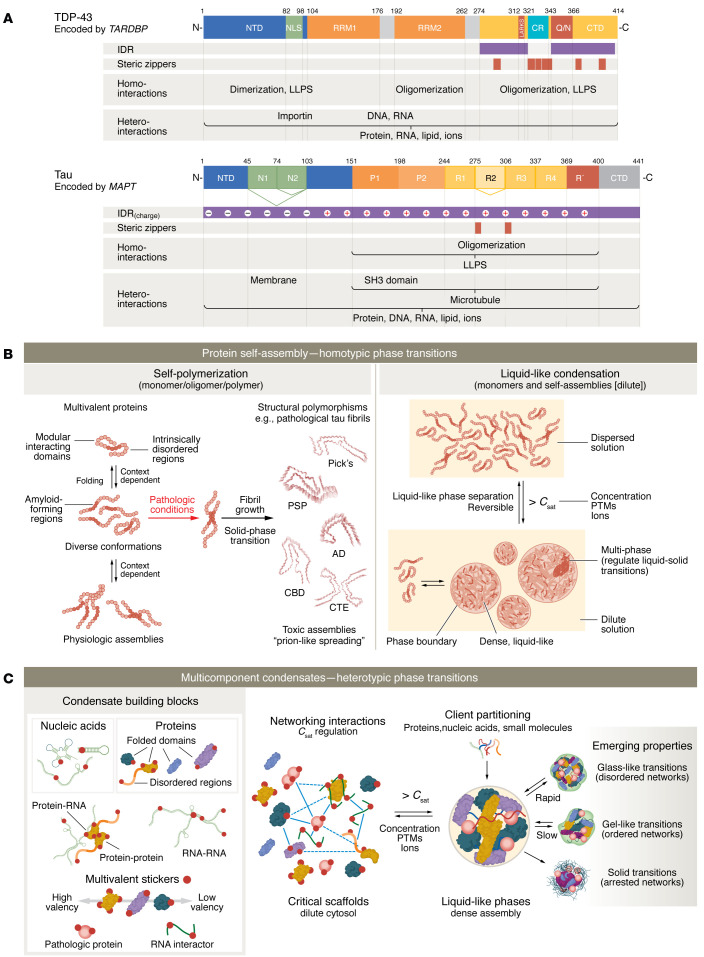
While short aggregation-prone sequences necessary for pathologic self-assemblies have been discovered in tau and TDP-43 (steric zippers and LARKS), several other regions within these two proteins can regulate phase transition behavior. TDP-43 contains a C-terminal domain (CTD) that comprises two disordered regions (IDR1, IDR2) and a short α-helical fold (CR helix) that is stabilized by adjacent homomeric contacts between other TDP-43 CR helices (Figure 1A) (23, 65). This CTD, also defined as a low-complexity domain (LCD), is sufficient for liquid-like phase separation and likely important for physiologic functions, including RNA splicing (24, 66, 67). However, current experimental evidence supports a TDP-43 oligomerization model where physically distant regions regulate the ability to nucleate self-assembly. While the N-terminal domain (NTD) drives the physiologic self-assembly necessary for RNA binding through two canonical RNA recognition motifs, the C-terminal LCD, a region where most ALS/FTLD-associated mutations are found, mediates dysfunctional assemblies (24, 68–71). Segments of the TDP-43 LCD can form both steric zippers and LARKS. Interestingly, the NTD appears to resist self-assemblies regulated by pathologic (LCD) self-interactions (72). Similarly, tau protein can be divided into distinct motifs: the negatively charged NTD and CTD and the positively charged proline-rich domain (PRD) and microtubule-binding domain (MTBD) (Figure 1B). Six different tau isoforms are generated in the human brain by alternative splicing containing varying NTD inserts (zero, one, or two) and three or four MTBD repeats. Importantly, tau contains two aggregation-prone steric zipper motifs within the MTBD (10), and while both the PRD and MTBD domains are capable of phase separation, the PRD has a prominent role in regulating tau liquid-liquid phase separation in cells (42).
Figure 1. Phase transitions of NDD-related proteins.
(A) Domain structure and interaction features of TDP-43 and tau. TDP-43 contains three domains: an N-terminal domain (NTD) including a nuclear localization sequence (NLS); two RNA recognition motifs (RRM1 and RRM2); and a C-terminal domain (CTD) with a short α-helical fold (CR helix) and a glutamine/arginine-rich region (Q/N). Tau contains four domains: the negatively charged NTD and CTD, and the positively charged proline-rich domain (P1–P2) and microtubule-binding domain (MTBD; R1–R4). Six different tau isoforms are generated by alternative splicing containing zero, one, or two NTD inserts and three or four MTBD repeats. The intrinsically disordered regions (IDRs), aggregation-prone steric zippers, and domain-dependent homo/heterotypic biomolecular interactions of TDP-43 and tau are shown accordingly. LLPS, liquid-liquid phase separation. (B) Aberrant protein conformations, toxic polymeric self-assemblies, and solid accumulations of proteins are found across the most common NDDs. TDP-43 and tau are modular, multivalent proteins exhibiting conformational flexibility, allowing diverse monomeric conformations, polymeric assemblies, and liquid-like phase behaviors in normal physiology and pathology. Sequence-specific properties found within distinct protein domains (modular interaction domains, intrinsically disordered regions, and amyloid-forming regions) are influenced by intrinsic (isoforms, mutations, PTMs) and extrinsic factors (molecular interactions, environmental conditions), ultimately regulating phase behavior and unique polymerization pathways. While increased homotypic interactions drive protein self-polymerization and the phase separation of proteins into liquid-like droplets, they are independent processes regulated by overlapping conditions. (C) TDP-43 and tau reside within multicomponent biomolecular condensates and thus are subjected to diverse homo/heterotypic biomolecular interactions, ultimately regulating physiologic and pathologic phase transitions. Biomolecules necessary for condensate assembly (scaffolds) spatially organize and concentrate functionally related biomolecules (clients) through liquid-like phase transitions. A sticker and spacer model has been proposed in which sticker sequences regulate multivalent networking interactions and spacer sequences regulate the solubilities of individual biomolecules and emerging networks.
Condensate assembly through heterotypic phase transitions
Membrane-bound structures were historically considered the established systems of intracellular organization. However, emerging research has since highlighted the role of dynamic biomolecular condensates, commonly referred to as membraneless organelles, as another process underlying cellular compartmentalization. These are dynamic assemblies formed through phase transitions consisting of homotypic/heterotypic interactions between proteins, nucleic acids, and cofactors (26, 28, 36). Hundreds or thousands of intracellular biomolecular interactions (“heterotypic buffering effect”) that occur under physiologic conditions prevent deleterious homotypic protein interactions observed in NDDs (22, 28). Thus, a better understanding of liquid-like phases and their liquid-to-solid transitions is important for understanding NDD pathogenesis.
A leading hypothesis for condensate assembly defines the condensate components as either scaffolds or clients. Scaffolds ultimately regulate the incorporation of various client biomolecules, which are not necessary for condensate assembly but essential for condensate dynamics and function. Subsequently, necessary scaffold-client interactions drive the assembly and tune the dynamic compositions of biomolecular condensates (33, 43, 73, 74). RNA species are integral components of many described condensates and, like proteins, are capable of scaffolding condensates through multivalent interactions (24, 75–83). Furthermore, RNA can promote or dissolve condensates scaffolded by RNA-binding proteins (RBPs), likely depending on their sequence, structure, and valence (23, 75, 83, 84).
Scaffolding molecules encode structural elements that drive and regulate phase transitions, including the generation of pre-assemblies such as small clusters and/or liquid-like phases. Multivalence is a common feature of scaffolding molecules and acts as a critical regulator of heterotypic phase transitions (35, 39, 73, 74, 79). Multivalence can be achieved in several ways, though it generally involves weak, transient contacts through modular interaction domains. Scaffolding protein motifs participating in specific interactions can occur on folded domains, low-complexity motifs, and sometimes even single residues. These interaction motifs form reversible cross-links through various chemical interactions, referred to as stickers (73, 74, 85). Sticker motifs are the same cross-links that drive protein folding, fold-specific recognition motifs, and many “classic” molecular assemblies known in biology (74). Additionally, while stickers engage in physical cross-links, various spacer sequences within the protein impact its overall solubility.
Biomolecular condensation can be coupled to both segregative phase separation (density transitions) and percolation, an associative phase transition (or networking transitions), as well as cooperative density-driven network transitions (phase separation coupled to percolation) (28). In a percolated network, physically cross-linked networks form via liquid-to-gel transitions, leading to network-spanning structures (28). The valence of stickers and their interaction strengths define the intrinsic concentration, or “percolation threshold” (Cperc), necessary for networking phase transitions (28, 74). Client-scaffold binding significantly alters saturation concentrations required for assembly and dissolution, providing switch-like, rapid behavior. Therefore, a networking transition is enabled by specific interactions between biomolecules (scaffolds) with a multivalence of interaction motifs (stickers). Regardless of the mechanisms driving their assembly, the ability to locally concentrate specific biomolecules is a classic description of all discovered biomolecular condensate structures (Figure 1C).
Cellular functions of biomolecular condensates
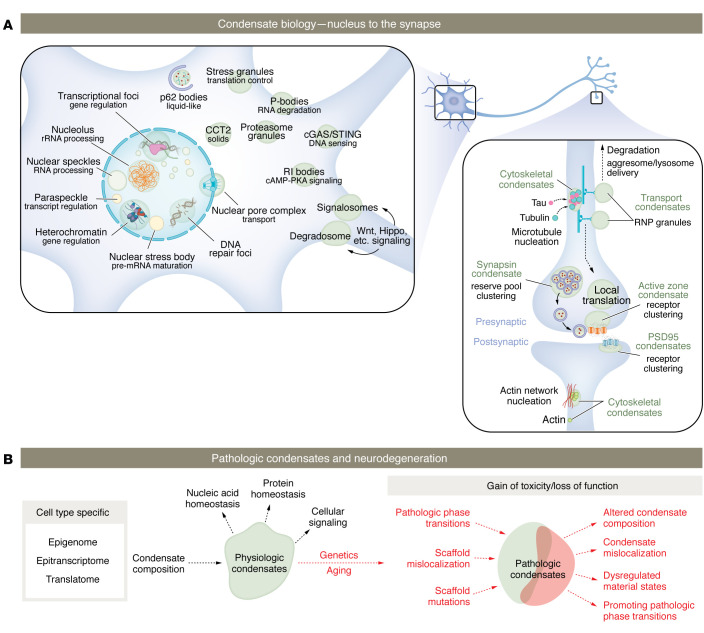
Today, biomolecular condensates are thought to spatially organize related processes in compartments ranging from the nucleus to the end of neuronal synapses (Figure 2A) (86–88). Primarily, condensates act as organization hubs, allowing spatiotemporal control of a variety of localized functions. They can also act as reaction crucibles, where the concentration of molecules in a condensed state promotes dynamic exchanges of products/reactants and sequesters biomolecules for storage or degradation. By spatiotemporally organizing biomolecules, unique biomolecular condensates dictate the biosynthesis, transport, regulation, and function of the basic building blocks necessary for cellular homeostasis (28, 30, 32, 36, 89, 90).
Figure 2. Hallmarks of neurodegeneration involve functions related to diverse biomolecular condensates.
(A) Schematic diagram showing the localization of various biomolecular condensates in a neuronal cell. Various biomolecular condensates are associated with many cellular processes that influence the homeostasis of nucleic acids and proteins from the nucleus to the end of synapses. (B) Neurodegeneration is accompanied by genetic, transcriptomic, and translational disruptions within vulnerable, cell type–specific neuronal populations. Imbalances in nucleic acid and protein homeostasis will directly affect the compositions, localization, and function of condensates (loss of function), additionally leading to aberrant phase transitions occurring within pathologic condensates (gain of function). Additionally, pathologic protein assemblies lead to downstream disruptions of physiologic condensates. RI, type I regulatory subunity of cAMP-dependent protein kinase (PKA).
In the absence of membrane-bound elements, distinct condensates can regulate nuclear functions, including chromatin compaction, DNA repair, RNA transcription, processing, transport, and decay (29, 89, 91–95). The most widely known subnuclear biomolecular condensate is the nucleolus. Nucleoli are multiphase condensates present within all eukaryotic organisms and are known as the site of rRNA transcription and ribosome assembly. The liquid-like state of nucleoli allows for a rapid exchange of newly transcribed/processed rRNA and ribosomal subunits between subcompartments of the nucleolus, permitting proper assembly and export of ribosomes from the nucleus (89, 94, 96–98). Phase-separated condensates are also implicated in driving gene activation through transcriptional condensates assembled at enhancer-rich gene clusters (95, 99–101). Properties inherent to chromatin, including the spacing of nucleosomes, allow it to phase-separate within the nucleoplasm, thus enabling the establishment and maintenance of distinct chromatin subcompartments (91, 102). Other well-studied nuclear condensates worth mentioning include Cajal bodies (associated with maturation of spliceosomal RNA and small nuclear ribonucleoprotein complexes), paraspeckles (involved in RNA editing and a protein buffering reservoir), nuclear speckles (“assembly line” involved in transcription-splicing mRNA export), and promyelocytic leukemia (PML) bodies (implicated in DNA damage and telomere maintenance) (89, 92).
Another well-known biomolecular condensate is cytoplasmic stress granules (SGs). SGs are a considerable focus in the field of neurodegeneration following the discovery that several disease-linked RBPs, including TDP-43 and tau, can localize to and modify SG assembly and dynamics (103). This micrometer-sized condensate assembles RNA and RBPs under various cellular stressors, and these structures regulate RNA stability and triage non-essential protein translation until the stress is removed (103–109). Recent studies show that the initial pre-assembly of G3BP1/2 dimers (which promote liquid-liquid phase separation) and the newly released mRNAs from polysomes during translational inhibition provide a physical platform for SG assembly. This initial assembly process is then followed by the subsequent recruitment of client molecules required for SG condensation and function necessary during cellular stress (79, 109).
During the last several years, studies demonstrate the importance of liquid-like condensation with regard to the spatiotemporal organization of neurons (86–88, 110). This growing group of structures includes synaptic active zones, synaptic vesicles, and excitatory/inhibitory pre- and postsynaptic densities (87, 111–114). Further, to maintain active signaling complexes necessary for electrical signaling homeostasis and physiologic function, neurons rely on localized protein translation in axons/dendrites/synapses, which can be up to 1 meter in distance from the cell body (28, 115). Notably, a fraction of the intracellular RNA is associated with RBPs in condensates termed ribonucleoprotein (RNP) granules. Once these silenced RNA granules arrive at axons/dendrites/synapses, signaling-dependent PTMs regulate condensate properties, resulting in the release of RNA for either degradation or translation; this is particularly important for maintaining dendritic plasticity and regulating axon growth, regeneration, and maintenance (45, 115–117).
Tau and TDP-43 regulate biological processes within liquid-like condensates in various cellular compartments, from the nucleoplasm to the synapse (12, 76, 118–121). Interestingly, tau and TDP-43 share many functions, as revealed by an extensively similar interactome embodied by RNP complexes, RNA/protein metabolism, molecular transport, and the neuronal stress response (122–124). Tau is usually a cytosolic axonal protein, and under disease conditions, tau accumulates in postsynaptic compartments, presynaptic terminals, and the nucleus (10, 125–127). Physiologically, tau can undergo phase separation to enhance the polymerization of microtubules by condensing tubulin dimers (118, 120). This drives microtubule polymerization, after which tau dissipates onto the microtubule surface. TDP-43 exerts multiple functions, including the regulation of splicing, trafficking, and stabilization of RNA (40, 123, 128, 129). While TDP-43 typically resides in the nucleus, it also shuttles from the nucleus to the cytoplasm and is found mislocalized to the cytoplasm of diseased neurons (40, 128). Notably, TDP-43 is a component of several RNP granules, including paraspeckles, nuclear stress bodies, and RNA transport granules in neurons (12, 69, 130).
Biomolecular condensate dysfunction
Given the essential roles that biomolecular condensates have in regulating cellular processes, one expects that many condensates are dysregulated in related diseases. Current evidence suggests that condensate dysregulation is a prevalent pathogenic mechanism underlying a broad spectrum of human diseases best described across NDDs and cancer (26, 27, 131). NDD phenotypes resulting from aging/disease-related insults include genomic DNA damage, defects in nucleocytoplasmic transport, and altered protein and RNA homeostasis (3, 16, 132–135). Under such conditions, dysregulated gene expression, alternative splicing events, disrupted RNA/protein transport, abnormal RNA/protein PTMs, and a loss in RNA/protein quality control have been observed. Many of these changes will directly impact threshold concentrations for phase separation, resulting in aberrant compositions and potential loss- and gain-of-function toxicity mechanisms. Consistent with this, pathogenic mutations across NDDs and cancer are increasingly associated with condensate dysregulation (25, 27, 136–139).
The relationship between pathogenic mutations and dysregulated condensates may be best understood by studying RBPs. Many RBPs, including TDP-43 and non-canonical RBPs like tau, are genetically linked to NDDs (22, 25, 83, 107, 140–143). Such mechanisms include enhanced driving forces for liquid-liquid phase separation and liquid-to-solid transitions, as well as altered material properties and localization of the condensates they reside within. Consistent with this notion, prolonged residency time within dense liquid-like phases was shown to increase the likelihood of liquid-to-solid phase transitions for tau, TDP-43, and other NDD-related proteins using in vitro model systems (20, 22, 23, 27, 68, 71, 108, 137, 139). However, disease-causing mutations in RBPs shift the balance of interactions between RNP assemblies, which, regardless of the mutation’s impact on liquid-to-solid phase transitions, ultimately alters condensate composition, material properties, and function (22, 23, 27, 29, 107, 144). This discovery has uncovered potentially novel mechanisms of toxicity and prompted a reexamination of loss- and gain-of-function mutations in solid-phase transitions (22, 27, 29, 38, 137, 145). Notably, the altered subcellular localization of critical condensate scaffolds can change the behavior of the scaffold and condensate components, leading to dysfunctional condensate assembly and toxicity. Additionally, disease-causing mutations may perturb the selective partitioning/exclusion of critical clients necessary for condensate assembly, localization, material properties, and subsequent function (Figure 2B).
Collectively, these discoveries have led to an exciting new framework for understanding the cellular biology underlying, and the potential molecular mechanisms driving, NDDs. Future research into dysregulated soluble protein phases containing tau/TDP-43 and other pathologic proteins will likely reveal additional links between aberrant condensates and neurotoxic mechanisms.
Targeting aberrant phase transitions
Extensive knowledge regarding alterations to the localization and biophysical properties of tau and TDP-43 in disease has provided fundamental examples linking aberrant phase transition behaviors with toxicity and potential targets for therapeutic intervention. Review of current and potential therapeutic targeting strategies directed at tau and TDP-43 proteinopathies highlights three potential therapeutic avenues that utilize the phase transition– and condensate-based hypotheses of NDDs (Figure 3). We will first examine strategies that directly target tau and TDP-43 (Table 1). Based on the residency of these pathologic proteins within various biomolecular condensates or “pathologic condensates,” we will discuss unique strategies that leverage properties of condensate biology and the critical cellular pathways regulating biomolecular condensates (Table 2).
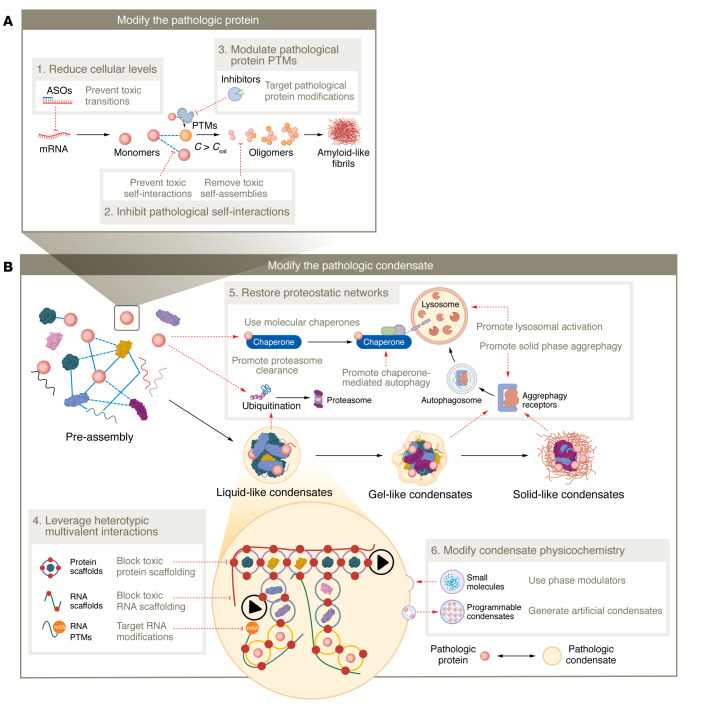
Figure 3. Drug discovery avenues for targeting aberrant phase transitions associated with neurodegeneration.
Three major avenues for targeting pathologic protein phase transitions in NDDs are proposed. (A) Modify the pathologic protein. The phase behavior of a pathologic protein may be directly modified by modulation of pathologic protein levels and PTMs, and by direct targeting of toxic homotypic interactions. (B) Modify the pathologic condensate. With the inherent limitations of direct targeting of a single protein, modifying pathologic condensates vastly extends the pool of drug targets. The aberrant condensate features may be altered by leveraging of heterotypic multivalent interactions and physicochemical properties, and by restoration of cellular proteostatic networks.
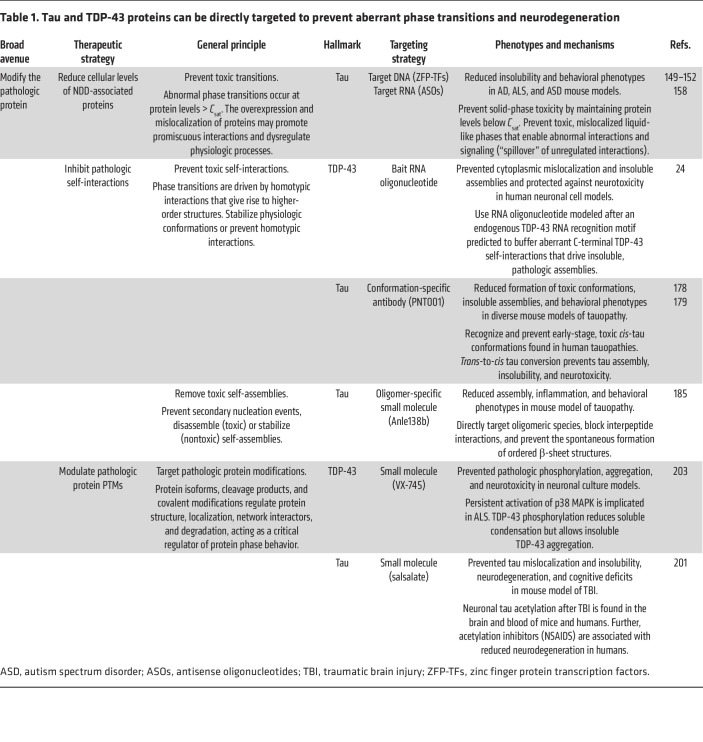
Table 1. Tau and TDP-43 proteins can be directly targeted to prevent aberrant phase transitions and neurodegeneration.
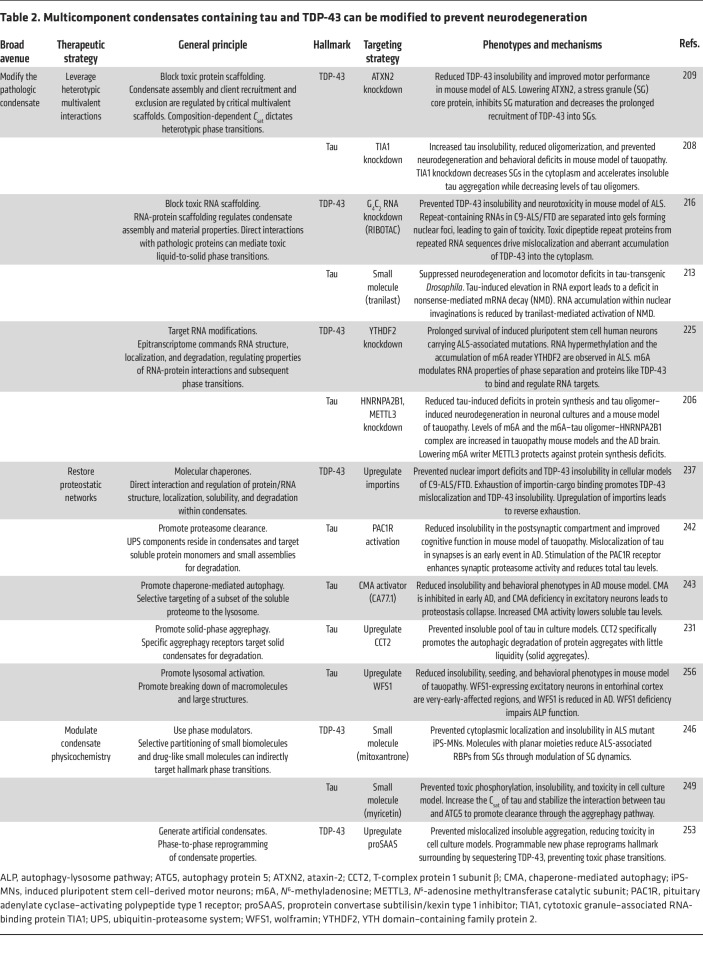
Table 2. Multicomponent condensates containing tau and TDP-43 can be modified to prevent neurodegeneration.
Modify the pathologic protein
Reduce cellular accumulation of NDD-associated proteins.
As previously mentioned, protein phase transitions can be described by local saturation concentrations (Csat) and strongly influenced by protein concentration (28, 33). Therefore, it is unsurprising that both tau and TDP-43 overexpression in cellular and animal models results in neurodegeneration and is further exacerbated by disease-causing mutations (10, 146, 147). Thus, targeting RNA to reduce the cellular accumulation of NDD proteins, such as through GAPmer antisense oligonucleotides (ASOs), bypasses the many unresolved questions regarding the toxicity of specific protein conformations, modifications, and polymeric assemblies and effectively prevents downstream toxicity. For TDP-43, both motor deficits and embryonic lethality have been described after partial and complete knockdown in animal models, respectively (147, 148). Therefore, reduction of wild-type TDP-43 levels does not appear viable for clinical translation. Tau knockdown, however, has proven tolerable in many experimental models and repeatedly demonstrated cognitive protection in AD and FTD-taumut animal models (149–152). More recent work extends this protection to neuronal cultures treated with ALS synaptoneurosomes (153). Tau-lowering strategies include tau-targeting immunotherapies and RNA-targeting MAPT ASOs, which are currently in clinical trials for tauopathies (154). Additionally, MAPT isoform–specific ASOs and small molecules targeting MAPT RNA splicing regulatory elements have demonstrated therapeutic potential by targeting overabundant tau isoforms in rodent models of genetic forms of frontotemporal dementia (FTD) (155–157). DNA-targeting zinc finger protein transcription factors (ZFP-TFs) capable of directly targeting and lowering specific protein-coding sequences provide long-lasting reductions in tau expression following a viral-mediated introduction in disease models of tauopathy (151, 158). Embedded within most genes encoding IDPs, an endogenous mechanism exists that controls translation through the expression of natural antisense transcripts (NATs) that contain mammalian-wide interspersed repeats (MIRs) (159). These MIR-NAT sequences compete for rRNA pairing and transcript translation and may act as a potential avenue for therapeutic intervention. For example, silencing of the MIR-NAT MAPT-AS1 led to increased tau levels, and its expression correlated with aggregated tau in the human brain (159).
Inhibit pathologic self-interactions.
As discussed in previous sections, intrinsic and extrinsic factors govern the energy state of intramolecular interactions, orming physiologic protein conformations and preventing aggregation-prone conformations (5, 53, 62). Therefore, designing small molecules that stabilize physiologic protein conformations and prevent pathologic conformations, self-assembly, and subsequent deleterious phase transitions is a viable therapeutic strategy. Attempts to design IDR small-molecule modulators have proven difficult, and no clinically approved small-molecule therapeutics targeting disease-related IDPs/IDRs currently exist (160). However, studies did identify small molecules that recognize monomeric tau and TDP-43, thus supporting the possibility of this approach for future investigation (161–164). Tau monomers may occupy distinct conformational ensembles, where some conformations are relatively inert, while others have the intrinsic ability to self-assemble and are seed-competent (63, 165). The initiation of tau self-assembly likely begins with a stable transition of tau monomer from an inert to a seed-competent monomeric form. One of the most well-studied tau-interacting ligands, the small molecule methylene blue (MB) and its derivative TRx0237, has been through several phase III clinical trials (163, 166). MB and its derivatives directly interact with tau monomer, thus blocking tau-tau interactions to prevent and reverse tau aggregation in vitro (167, 168). Investigation of TRx0237 and other tau-binding small molecules highlights the potential of binding and sequestering IDPs in monomeric, soluble states. Similarly, a small molecule, nTRD22, targeting the N-terminal domain of TDP-43 was recently shown to be an allosteric modulator of TDP-43–RNA binding and conferred protection against motor deficits in an ALS-Drosophila model that overexpresses TDP-43 (169). These highlighted examples suggest that further research targeting monomeric forms of pathologic proteins with small molecules is a viable and promising approach to prevent and/or reverse aberrant phase transitions.
Recent work by us and others also highlights the ability of specific RNAs to regulate protein phase transitions through specific RNA-protein interactions. In the case of TDP-43, homotypic low-complexity domain (LCD) interactions initiate its pathologic aggregation through aberrant liquid-liquid phase separation, and this homotypic interaction is antagonized by RNA binding (23, 24, 76). An RNA-dependent mechanism of pathologic interaction was also shown for other NDD-associated RBPs, including FUS and tau (120, 142, 170, 171). This mechanism highlights an intriguing RNA-based targeting strategy in which an RNA aptamer or “bait oligonucleotide” might be able to engage RNA-deficient TDP-43 in the cytoplasm and prevent or reverse pathologic phase transitions. In the case of TDP-43, a bait oligonucleotide (Clip_34) comprising the TARDBP mRNA 3′-UTR autoregulatory domain engages the TDP-43 RNA recognition motifs and prevents neurotoxic TDP-43 self-interactions, phase transitions, and associated in vitro neurotoxicity (24, 172).
Immunotherapies to disrupt existing pathologic homotypic assemblies also showed promise for both tau and TDP-43 (161, 173–175). Tau-based immunotherapies have gone from proof-of-concept studies to clinical trials for AD and other tauopathies (154). Several notable disease-conformation-specific tau antibodies have since been developed, presenting promising results for reducing tau aggregation in preclinical models of tauopathy (176, 177). For example, the PNT001 antibody is capable of recognizing a toxic, trans-to-cis conformational change occurring early in tauopathies (178–180). PNT1001 prevents tau aggregation, neuropathology, and cognitive impairment in several preclinical tauopathy models, including models of CTE. PNT1001 is currently entering clinical trials in patients with various tauopathies, including traumatic brain injury (TBI). Similarly, a rationally developed antibody targeting an RNA recognition domain of TDP-43 was shown to successfully reduce insoluble TDP-43 inclusions, inflammation, and cognitive impairment in a transgenic ALS mouse model expressing the familial ALS TDP-43G348C protein (173).
Soluble oligomeric protein assemblies of tau and TDP-43 are synaptotoxic and, in the case of tau, capable of propagating self-assembly through connected neural networks (46, 50, 54, 181–183). Recently, many rational designs leveraging stable structures mediated by LCDs/IDRs through aberrant phase transitions and the accumulation of homotypic self-assemblies have brought exciting opportunities for structure-specific targeting. Targeting the neurotoxic and misfolded protein structure and not the protein monomers should limit interference with the physiologic function of the protein when in its proper conformation. This is notable since the physiologic phase separation of IDR-containing proteins into biomolecular condensates is critical for various cellular processes. For example, physiologic phase transitions of TDP-43 into reversible biomolecular condensates is hypothesized to be essential for the binding of specific RNA sequences (23, 24, 76, 184). Thus, the development of strategies that target pathologic but not physiologic phase-separated assemblies is a powerful approach.
Regarding tau, multiple in vitro studies identified small molecules that inhibit tau assembly with various mechanisms of action. These include molecules that block inducer-specific fibril growth, preventing fibril growth by initializing nontoxic, off-pathway assemblies, and those capable of disassembling preformed fibrils (185–188). Use of cryo–electron microscopic structures of human AD tau filaments bound to small molecules has allowed the identification of novel, drug-like molecules capable of disaggregating brain-derived tau fibrils in vitro (189). One example is the small molecule Anle138b, which is currently in clinical trials for Parkinson’s disease and multiple-system atrophy and has previously been shown to reduce tau aggregation and behavioral deficits in numerous cellular and animal models of tauopathy (188). Experimental evidence demonstrated that Anle138 avoids tau monomer binding and selectively binds oligomeric tau assemblies, preventing the formation of amyloidogenic fibrils (188). Furthermore, crystal structures of tau steric zippers led to the rational design of small steric zipper–binding peptides, referred to as “fibril capping” peptides (190).
Modulate pathologic protein PTMs.
PTMs, including covalent modifications and cleavage events, offer a fine-tuned response to diverse extracellular stimuli and intracellular signaling pathways (45, 191–193). PTMs substantially alter the intrinsic properties of a sequence and thus regulate intra- and intermolecular interactions (62). Therefore, covalent modifications can act as potent regulators of protein/RNA conformations and, consequently, the properties of biomolecular condensates. In disease, tau and TDP-43 are often found heavily modified by PTMs (phosphorylation, acetylation, ubiquitination, etc.) and cleaved into fragments (10, 53, 193–197). While the effect of PTMs on biomolecular phase behavior is only beginning to be understood, PTMs may directly regulate phase behavior by altering either intra- or intermolecular interactions, leading to an altered Csat. Lysine-modifying acetylation in tau and TDP-43 significantly reduces critical lysine-RNA interactions, resulting in altered phase behaviors (198–200). Targeting of tau acetylation after TBI using acetylation-inhibiting drugs (salsalate) is associated with reduced neurodegeneration in humans and prevents tau mislocalization, insolubility, and cognitive deficits in preclinical models (201). TDP-43 acetylation, which mitigates RNA binding, enhances its phase separation into complex nuclear droplets called anisomes that colocalize with HSP70 and can promote aberrant phase transitions when localized to the cytoplasm. This results in gel-like and insoluble assemblies and highlights the role of RNA binding as a modulator of TDP-43 liquid-liquid phase separation (24, 200, 202). Importantly, pairings of PTMs may have distinct effects on downstream modifications, either stimulating or inhibiting hallmark phase transitions (53, 197, 203, 204). PTM-modifying therapies will require extensive study with regard to the complex interplay between single and combinatorial PTMs and how PTMs alter biomolecular interacting partners, resulting phase behaviors, and subsequent neurotoxicity.
Modify pathologic condensates
Leverage heterotypic multivalent interactions.
The growing knowledge regarding condensate assembly and regulation opens avenues for interfering with pathologic phase transitions. With the inherent limitations of direct targeting of pathogenic phase transitions of a single protein (tau or TDP-43), targeting biomolecular condensates that might drive aberrant phase transitions vastly extends the pool of drug targets. The residency and scaffolding potential of pathologic proteins in critical cellular condensates are intriguing. Modifying condensate scaffolds would significantly affect condensates’ stability, including assembly, dissolution, material properties, and composition of scaffold/ligands/etc. (28, 43, 73, 79, 205). Thus, disrupting specific components and regulatory pathways of biomolecular condensates to indirectly modify abnormal hallmark phase transitions and cellular toxicity may be a therapeutic approach. The ultimate goal of this is to shift tau or TDP-43 Csat and phase transition behaviors.
Scaffold modulation can be achieved in several ways. Approaches may include preventing or stabilizing protein-protein, protein-RNA, and RNA-RNA interactions that contribute to condensate scaffolding. Intriguingly, the genetic manipulation of RBPs often alters the rate of tau and TDP-43 aggregation in several model systems (206–209). TIA1 is an RBP and a major component of stress granules (SGs). Previous studies indicate that TIA1 interacts with tau, and this interaction modulates tau aggregation and toxicity (208, 210, 211). TIA1 knockdown prevents tau-mediated toxicity, reduces toxic soluble tau oligomers, and increases insoluble tau fibrils (208). Importantly, tau fibrils isolated from the diseased brain contain numerous RNA species. Recent research has demonstrated tau-mediated disruptions in RNA metabolism, leading to tau-RNA accumulations building on the nuclear envelope (212, 213). Remarkably, promoting nonsense-mediated mRNA decay with a small molecule, tranilast, disrupts these tau-RNA accumulations, suppressing neurodegeneration and locomotor deficits in a tau-transgenic Drosophila model.
Ataxin-2 (ATXN2) is an RBP found in mature SGs, and intermediate CAG expansions within the ATXN2 gene are found in subsets of ALS cases (214). Recent work found that ATXN2 knockdown reduces abnormal SG formation and is neuroprotective in both in vitro and in vivo rodent models with elevated levels of TDP-43 (209). Additionally, ATXN2 reduction significantly reduces TDP-43 pathology. Further, ALS’s most common genetic cause (expansions of C9orf72) leads to the overexpression of expanded GC RNA repeats, leading to TDP-43 mislocalization, assembly, and toxicity (215). Recently, a small-molecule-guided ribonuclease-targeting chimera (RIBOTAC) method capable of directly targeting the removal of G4C2 duplications prevented TDP-43 insolubility and neurotoxicity in animal models (216). Additionally, recent work has demonstrated that upregulating an endogenous TDP-43–interacting noncoding RNA, NEAT1_1, lowered TDP-43 insolubility and toxicity in Drosophila and yeast models of TDP-43 proteinopathy (217). Together, this suggests that pathogenic interactions within biomolecular condensates may promote aberrant TDP-43 and tau phase transitions and that modulating these interactions might confer neuroprotection and be a potential therapeutic approach.
While considerable attention has been focused on protein modifications, recent work highlights a long list of covalent nucleic acid modifications that may alter TDP-43 and tau phase transitions (78, 218–222). DNA and RNA methylation are potent regulators of nucleic acid phase separation and affect the condensation properties of specific protein–nucleic acid complexes (221, 223, 224). Interestingly, the knockdown of the canonical RNA N6-methyladenosine (m6A) reader YTHDF2 was recently shown to prolong the survival of induced pluripotent stem cell human neurons carrying ALS-associated mutations (225). Consistent with this, knockdown of the canonical RNA m6A reader HNRNPA2B1 and the m6A writer METTL3 rescued tau-oligomer-induced neurodegeneration in models of tauopathy (206). Thus, the targeting of these RNA modifications is slowly being revealed as a novel approach capable of regulating pathologic protein phase transitions.
Restore proteostatic networks.
The proteostasis network, a protein quality control (PQC) system, regulates and balances protein synthesis, folding, transport, and degradation (226–228). Impairment of one or several PQC mechanisms can result in aberrant phase transitions and the accumulation of protein aggregates inside neurons. The PQC system is an integrated network of molecular chaperones, co-chaperones, and two degradative systems, the ubiquitin-proteasome system (UPS) and autophagy, a lysosome-mediated bulk degradation pathway (226, 229). Traditionally, autophagy was believed to preferentially clear protein aggregates with a certain amount of “liquidity” in a process referred to as aggrephagy (230–232). An arm of aggrephagy was recently discovered and selectively targets protein aggregates with little liquidity (solids) for lysosomal degradation, thus highlighting critical cellular mechanisms that interact with biomolecular condensates with specific intrinsic material properties (231). While aggrephagy was thought to process condensates with some liquidity, recent work demonstrated that the CCT2 autophagy receptor allows for the selective targeting of solid condensates. Notably, the upregulation of CCT2 cleared several solid protein aggregates from cells, including mutant tau protein (231).
Several pharmacologic agents that modulate the ATPase activity of HSP70, a core chaperone, have been designed and tested in NDD models (233, 234). Interestingly, reduction of HSP70 ATPase activity transforms TDP-43 liquid phases into gel-like structures, leading to insoluble TDP-43 assemblies and increased toxicity (200). Substantial efforts found that non-core chaperones, including a specific class, the peptidyl-prolyl cis-trans isomerases (PPIases), protected against aberrant tau phase transitions (235). Specifically, Pin1 catalyzes proline cis-to-trans isomerization, a conformational change that protects against the stabilization of toxic conformations that lead to pathologic tau fibrils (176, 177, 179). Increasing evidence shows that nuclear-import receptors chaperone and disaggregate RBPs, including TDP-43 (40, 236, 237). Not only do nuclear localization sequences (NLSs) mediate the nuclear import of NLS-containing proteins, but they also inhibit deleterious phase transitions and promote the disaggregation of solid assemblies. In the cytoplasm, specific nuclear-import receptors that engage the TDP-43 NLS, importin-α and -β, prevent and reverse TDP-43 aggregation n models of C9orf72 ALS/FTLD (236).
The UPS predominantly regulates soluble tau and TDP-43, and the accumulation of these species can lead to protein nucleation (238–242). While macroautophagy pathways can directly sequester and degrade larger condensates, soluble protein monomers can be degraded by chaperone-mediated autophagy (CMA). The inability to remove accumulating soluble proteins eventually promotes the aggregation of the CMA-regulated proteome. Consistent with this, CMA deficiency in the aging brain is an aggravating factor in the onset of NDD (243). Activation of CMA with small molecules has proven neuroprotective in animal models of tauopathy (243, 244).
Modulate condensate physicochemistry.
While the direct engagement of condensate components may allow a prospective drug to occupy a condensate, a drug may also concentrate within a condensate due to a network of transient contacts without high affinity toward a specific target (101, 245–249). Therefore, a small molecule, through interactions with the chemical environment of the condensate, may strongly influence condensate properties regulating the formation or dissolution of condensates. Therefore, using small-molecule ligands to target condensates may be a promising therapeutic strategy. This premise is clearly illustrated by cellular metabolites like ATP, cAMP, glucose, and many others, which were previously demonstrated to modulate condensate properties (34, 43, 73, 250–252). Several known small molecules can alter the phase behaviors of tau and TDP-43 proteins by either directly interfering with the ability of the pathologic protein to self-condense into liquid-like phase, or interfering with their recruitment to biomolecular condensates (i.e., SGs) (245–248). Specifically, molecules with planar moieties, such as mitoxantrone, were shown to prevent TDP-43 cytoplasmic localization and prolonged residency in SGs (246). Further, the compound myricetin can slow the liquid-like phase separation of tau, shifting its phase boundary while stabilizing the interaction of tau protein within the aggrephagy clearance pathway (249). Besides regulating the properties of existing hallmark condensates with small molecules, interest in generating artificial condensate systems to engage with endogenous condensates is growing. Interestingly, the cytoplasmic expression of the neuronal chaperone proSAAS created micron-scale membraneless spheres with condensate features that selectively encapsulated and sequestrated TDP-43 aggregates and reduced their toxicity in cell culture models (253). Further work is under way designing programmable condensates capable of sequestering pathologic aggregates, stabilizing the pathologic proteins’ normal physiology, and facilitating drug delivery and enrichment toward specific condensates.
Conclusion
It is believed that the biochemical changes responsible for initiating NDDs begin decades before the clinical presentation (2, 9, 26, 226, 228, 254). Furthermore, there are fundamental challenges to differentiating “normal” age-related events from pathologic biochemical processes that drive the earliest stages of neurodegeneration or distinguishing primary causes from a cascade of secondary insults. Aberrant protein conformations, oligomers, and fibrils composed of neuropathologic protein depositions may symbolize both a symptom and a cause of the underlying disease. As new discoveries emerge describing structure-specific protein assemblies in NDD subtypes, a thorough understanding of the cellular conditions driving these unique self-assemblies will prove important to develop disease-modifying therapies (56, 57, 64, 255). Condensate biology is fundamental to numerous cellular processes, and a growing understanding of these mechanisms is already transforming our understanding of how cells spatiotemporally organize biomolecules to regulate critical cell functions. As the formation of biomolecular condensates involves and influences all levels of macromolecular organization, condensate biology can profoundly expand our understanding of the pathologic conditions that lead to toxic protein assemblies, resulting downstream cellular dysfunction, and subsequent neurodegeneration. Targeting of aberrant phase transitions as a therapeutic intervention for neurodegenerative disorders will require substantial work to better characterize the diverse condensate subtypes and their components, physicochemical properties, assembly mechanisms, and physiologic function.
Author contributions
BTH and LX conceptualized and outlined the contents of this review, figures, and tables. BTH wrote the initial full draft with input from LX. BTH and LX both addressed reviewers’ comments.
Acknowledgments
LX is a Tsinghua MD, PhD Scholar working in the CJD laboratory and supported by a partnership between the University of Pittsburgh School of Medicine and the Tsinghua University School of Medicine. The CJD laboratory is supported by grants from the NIH (R01NS127187, L30AG048607, R01NS105756), the Target ALS Foundation, the LiveLikeLou Center for ALS Research at the University of Pittsburgh Brain Institute, and LiveLikeLou at the Pittsburgh Foundation.
Version 1. 07/03/2023
Electronic publication
Footnotes
Conflict of interest: CJD is cofounder of Confluence Therapeutics Inc.
Copyright: © 2023, Hurtle et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: J Clin Invest. 2023;133(13):e168549. https://doi.org/10.1172/JCI168549.
Contributor Information
Bryan T. Hurtle, Email: Hurtle.Bryan@medstudent.pitt.edu.
Longxin Xie, Email: XIEL@pitt.edu.
Christopher J. Donnelly, Email: chrisdonnelly@pitt.edu.
References
- 1.Fu H, et al. Selective vulnerability in neurodegenerative diseases. Nat Neurosci. 2018;21(10):1350–1358. doi: 10.1038/s41593-018-0221-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elahi FM, Miller BL. A clinicopathological approach to the diagnosis of dementia. Nat Rev Neurol. 2017;13(8):457–476. doi: 10.1038/nrneurol.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson DM, et al. Hallmarks of neurodegenerative diseases. Cell. 2023;186(4):693–714. doi: 10.1016/j.cell.2022.12.032. [DOI] [PubMed] [Google Scholar]
- 4.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148(6):1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soto C, Pritzkow S. Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat Neurosci. 2018;21(10):1332–1340. doi: 10.1038/s41593-018-0235-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellenguez C, et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat Genet. 2022;54(4):412–436. doi: 10.1038/s41588-022-01024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gan L, et al. Converging pathways in neurodegeneration, from genetics to mechanisms. Nat Neurosci. 2018;21(10):1300–1309. doi: 10.1038/s41593-018-0237-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karch CM, et al. Selective genetic overlap between amyotrophic lateral sclerosis and diseases of the frontotemporal dementia spectrum. JAMA Neurol. 2018;75(7):860–875. doi: 10.1001/jamaneurol.2018.0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arvanitakis Z, et al. Diagnosis and management of dementia: review. JAMA. 2019;322(16):1589–1599. doi: 10.1001/jama.2019.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Mandelkow E. Tau in physiology and pathology. Nat Rev Neurosci. 2016;17(1):5–21. doi: 10.1038/nrn.2015.1. [DOI] [PubMed] [Google Scholar]
- 11.Chang CW, et al. Tau: enabler of diverse brain disorders and target of rapidly evolving therapeutic strategies. Science. 2021;371(6532):eabb8255. doi: 10.1126/science.abb8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Portz B, et al. FUS and TDP-43 phases in health and disease. Trends Biochem Sci. 2021;46(7):550–563. doi: 10.1016/j.tibs.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zbinden A, et al. Phase separation and neurodegenerative diseases: a disturbance in the force. Dev Cell. 2020;55(1):45–68. doi: 10.1016/j.devcel.2020.09.014. [DOI] [PubMed] [Google Scholar]
- 14.Nelson PT, et al. Limbic-predominant age-related TDP-43 encephalopathy (LATE): consensus working group report. Brain. 2019;142(6):1503–1527. doi: 10.1093/brain/awz099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chornenkyy Y, et al. Tau and TDP-43 proteinopathies: kindred pathologic cascades and genetic pleiotropy. Lab Invest. 2019;99(7):993–1007. doi: 10.1038/s41374-019-0196-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ling SC, et al. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79(3):416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung DEC, et al. Cellular and pathological heterogeneity of primary tauopathies. Mol Neurodegener. 2021;16(1):57. doi: 10.1186/s13024-021-00476-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKee AC, et al. TDP-43 proteinopathy and motor neuron disease in chronic traumatic encephalopathy. J Neuropathol Exp Neurol. 2010;69(9):918–929. doi: 10.1097/NEN.0b013e3181ee7d85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tremblay C, et al. Accumulation of transactive response DNA binding protein 43 in mild cognitive impairment and Alzheimer disease. J Neuropathol Exp Neurol. 2011;70(9):788–798. doi: 10.1097/NEN.0b013e31822c62cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wegmann S, et al. Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J. 2018;37(7):e98049. doi: 10.15252/embj.201798049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ambadipudi S, et al. Liquid-liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat Commun. 2017;8(1):275. doi: 10.1038/s41467-017-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathieu C, et al. Beyond aggregation: pathological phase transitions in neurodegenerative disease. Science. 2020;370(6512):56–60. doi: 10.1126/science.abb8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hallegger M, et al. TDP-43 condensation properties specify its RNA-binding and regulatory repertoire. Cell. 2021;184(18):4680–4696. doi: 10.1016/j.cell.2021.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mann JR, et al. RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron. 2019;102(2):321–338. doi: 10.1016/j.neuron.2019.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nedelsky NB, Taylor JP. Bridging biophysics and neurology: aberrant phase transitions in neurodegenerative disease. Nat Rev Neurol. 2019;15(5):272–286. doi: 10.1038/s41582-019-0157-5. [DOI] [PubMed] [Google Scholar]
- 26.Alberti S, Hyman AA. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat Rev Mol Cell Biol. 2021;22(3):196–213. doi: 10.1038/s41580-020-00326-6. [DOI] [PubMed] [Google Scholar]
- 27.Alberti S, Dormann D. Liquid-liquid phase separation in disease. Annu Rev Genet. 2019;53:171–194. doi: 10.1146/annurev-genet-112618-043527. [DOI] [PubMed] [Google Scholar]
- 28.Mittag T, Pappu RV. A conceptual framework for understanding phase separation and addressing open questions and challenges. Mol Cell. 2022;82(12):2201–2214. doi: 10.1016/j.molcel.2022.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017;357(6357):eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 30.Li P, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483(7389):336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brangwynne CP, et al. Polymer physics of intracellular phase transitions. Nat Phys. 2015;11(11):899–904. doi: 10.1038/nphys3532. [DOI] [Google Scholar]
- 32.Lyon AS, et al. A framework for understanding the functions of biomolecular condensates across scales. Nat Rev Mol Cell Biol. 2021;22(3):215–235. doi: 10.1038/s41580-020-00303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimobayashi SF, et al. Nucleation landscape of biomolecular condensates. Nature. 2021;599(7885):503–506. doi: 10.1038/s41586-021-03905-5. [DOI] [PubMed] [Google Scholar]
- 34.Snead WT, Gladfelter AS. The control centers of biomolecular phase separation: how membrane surfaces, PTMs, and active processes regulate condensation. Mol Cell. 2019;76(2):295–305. doi: 10.1016/j.molcel.2019.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banani SF, et al. Compositional control of phase-separated cellular bodies. Cell. 2016;166(3):651–663. doi: 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Banani SF, et al. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18(5):285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uversky VN, et al. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 38.Darling AL, et al. Intrinsic disorder-based emergence in cellular biology: physiological and pathological liquid-liquid phase transitions in cells. Polymers (Basel) 2019;11(6):990. doi: 10.3390/polym11060990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peran I, Mittag T. Molecular structure in biomolecular condensates. Curr Opin Struct Biol. 2020;60:17–26. doi: 10.1016/j.sbi.2019.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tziortzouda P, et al. Triad of TDP43 control in neurodegeneration: autoregulation, localization and aggregation. Nat Rev Neurosci. 2021;22(4):197–208. doi: 10.1038/s41583-021-00431-1. [DOI] [PubMed] [Google Scholar]
- 41.Wang X, et al. The proline-rich domain and the microtubule binding domain of protein tau acting as RNA binding domains. Protein Pept Lett. 2006;13(7):679–685. doi: 10.2174/092986606777790566. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X, et al. The proline-rich domain promotes Tau liquid-liquid phase separation in cells. J Cell Biol. 2020;219(11):e202006054. doi: 10.1083/jcb.202006054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruff KM, et al. Polyphasic linkage and the impact of ligand binding on the regulation of biomolecular condensates. Biophys Rev (Melville) 2021;2(2):021302. doi: 10.1063/5.0050059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boija A, et al. Biomolecular condensates and cancer. Cancer Cell. 2021;39(2):174–192. doi: 10.1016/j.ccell.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hofweber M, Dormann D. Friend or foe—post-translational modifications as regulators of phase separation and RNP granule dynamics. J Biol Chem. 2019;294(18):7137–7150. doi: 10.1074/jbc.TM118.001189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng C, et al. Protein transmission in neurodegenerative disease. Nat Rev Neurol. 2020;16(4):199–212. doi: 10.1038/s41582-020-0333-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiti F, Dobson CM. Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu Rev Biochem. 2017;86:27–68. doi: 10.1146/annurev-biochem-061516-045115. [DOI] [PubMed] [Google Scholar]
- 48.Knowles TPJ, et al. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15(6):384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 49.Dujardin S, et al. Tau molecular diversity contributes to clinical heterogeneity in Alzheimer’s disease. Nat Med. 2020;26(8):1256–1263. doi: 10.1038/s41591-020-0938-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gibbons GS, et al. Mechanisms of cell-to-cell transmission of pathological tau: a review. JAMA Neurol. 2019;76(1):101–108. doi: 10.1001/jamaneurol.2018.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fitzpatrick AWP, et al. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature. 2017;547(7662):185–190. doi: 10.1038/nature23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao Q, et al. Cryo-EM structures of four polymorphic TDP-43 amyloid cores. Nat Struct Mol Biol. 2019;26(7):619–627. doi: 10.1038/s41594-019-0248-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arakhamia T, et al. Posttranslational modifications mediate the structural diversity of tauopathy strains. Cell. 2020;180(4):633–644. doi: 10.1016/j.cell.2020.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fang YS, et al. Full-length TDP-43 forms toxic amyloid oligomers that are present in frontotemporal lobar dementia-TDP patients. Nat Commun. 2014;5:4824. doi: 10.1038/ncomms5824. [DOI] [PubMed] [Google Scholar]
- 55.Lye YS, Chen Y-R. TAR DNA-binding protein 43 oligomers in physiology and pathology. IUBMB Life. 2022;74(8):794–811. doi: 10.1002/iub.2603. [DOI] [PubMed] [Google Scholar]
- 56.Li D, Liu C. Hierarchical chemical determination of amyloid polymorphs in neurodegenerative disease. Nat Chem Biol. 2021;17(3):237–245. doi: 10.1038/s41589-020-00708-z. [DOI] [PubMed] [Google Scholar]
- 57.Shi Y, et al. Structure-based classification of tauopathies. Nature. 2021;598(7880):359–363. doi: 10.1038/s41586-021-03911-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arseni D, et al. Structure of pathological TDP-43 filaments from ALS with FTLD. Nature. 2022;601(7891):139–143. doi: 10.1038/s41586-021-04199-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laferrière F, et al. TDP-43 extracted from frontotemporal lobar degeneration subject brains displays distinct aggregate assemblies and neurotoxic effects reflecting disease progression rates. Nat Neurosci. 2019;22(1):65–77. doi: 10.1038/s41593-018-0294-y. [DOI] [PubMed] [Google Scholar]
- 60.Hughes MP, et al. Prevalence and species distribution of the low-complexity, amyloid-like, reversible, kinked segment structural motif in amyloid-like fibrils. J Biol Chem. 2021;297(4):101194. doi: 10.1016/j.jbc.2021.101194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guenther EL, et al. Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation. Nat Struct Mol Biol. 2018;25(6):463–471. doi: 10.1038/s41594-018-0064-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Houben B, et al. Protein structure and aggregation: a marriage of necessity ruled by aggregation gatekeepers. Trends Biochem Sci. 2022;47(3):194–205. doi: 10.1016/j.tibs.2021.08.010. [DOI] [PubMed] [Google Scholar]
- 63.Eschmann NA, et al. Signature of an aggregation-prone conformation of tau. Sci Rep. 2017;7:44739. doi: 10.1038/srep44739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fitzpatrick AW, Saibil HR. Cryo-EM of amyloid fibrils and cellular aggregates. Curr Opin Struct Biol. 2019;58:34–42. doi: 10.1016/j.sbi.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Staderini T, et al. Biophysical characterization of full-length TAR DNA-binding protein (TDP-43) phase separation. Protein Sci. 2022;e4509(12):e4509. doi: 10.1002/pro.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li HR, et al. The physical forces mediating self-association and phase-separation in the C-terminal domain of TDP-43. Biochim Biophys Acta Proteins Proteom. 2018;1866(2):214–223. doi: 10.1016/j.bbapap.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 67.Babinchak WM, et al. The role of liquid-liquid phase separation in aggregation of the TDP-43 low-complexity domain. J Biol Chem. 2019;294(16):6306–6317. doi: 10.1074/jbc.RA118.007222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Conicella AE, et al. ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure. 2016;24(9):1537–1549. doi: 10.1016/j.str.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alami NH, et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014;81(3):536–543. doi: 10.1016/j.neuron.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prasad A, et al. Molecular mechanisms of TDP-43 misfolding and pathology in amyotrophic lateral sclerosis. Front Mol Neurosci. 2019;12:25. doi: 10.3389/fnmol.2019.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gopal PP, et al. Amyotrophic lateral sclerosis-linked mutations increase the viscosity of liquid-like TDP-43 RNP granules in neurons. Proc Natl Acad Sci U S A. 2017;114(12):E2466–E2475. doi: 10.1073/pnas.1614462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zeng YT, et al. Different intermolecular interactions drive nonpathogenic liquid-liquid phase separation and potentially pathogenic fibril formation by TDP-43. Int J Mol Sci. 2022;23(23):15227. doi: 10.3390/ijms232315227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruff KM, et al. Ligand effects on phase separation of multivalent macromolecules. Proc Natl Acad Sci U S A. 2021;118(10):e2017184118. doi: 10.1073/pnas.2017184118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choi J-M, et al. Physical principles underlying the complex biology of intracellular phase transitions. Annu Rev Biophys. 2020;49:107–133. doi: 10.1146/annurev-biophys-121219-081629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Van Treeck B, Parker R. Emerging roles for intermolecular RNA-RNA interactions in RNP assemblies. Cell. 2018;174(4):791–802. doi: 10.1016/j.cell.2018.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grese ZR, et al. Specific RNA interactions promote TDP-43 multivalent phase separation and maintain liquid properties. EMBO Rep. 2021;22(12):e53632. doi: 10.15252/embr.202153632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garcia-Jove Navarro M, et al. RNA is a critical element for the sizing and the composition of phase-separated RNA-protein condensates. Nat Commun. 2019;10(1):3230. doi: 10.1038/s41467-019-11241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lewis CJT, et al. RNA modifications and structures cooperate to guide RNA-protein interactions. Nat Rev Mol Cell Biol. 2017;18(3):202–210. doi: 10.1038/nrm.2016.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sanders DW, et al. Competing protein-RNA interaction networks control multiphase intracellular organization. Cell. 2020;181(2):306–324. doi: 10.1016/j.cell.2020.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van Treeck B, et al. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc Natl Acad Sci U S A. 2018;115(11):2734–2739. doi: 10.1073/pnas.1800038115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roden C, Gladfelter AS. RNA contributions to the form and function of biomolecular condensates. Nat Rev Mol Cell Biol. 2021;22(3):183–195. doi: 10.1038/s41580-020-0264-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aarum J, et al. Enzymatic degradation of RNA causes widespread protein aggregation in cell and tissue lysates. EMBO Rep. 2020;21(10):e49585. doi: 10.15252/embr.201949585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mann JR, Donnelly CJ. RNA modulates physiological and neuropathological protein phase transitions. Neuron. 2021;109(17):2663–2681. doi: 10.1016/j.neuron.2021.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang X, et al. RNA stores tau reversibly in complex coacervates. PLoS Biol. 2017;15(7):e2002183. doi: 10.1371/journal.pbio.2002183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martin EW, et al. Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science. 2020;367(6478):694–699. doi: 10.1126/science.aaw8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu X, et al. Liquid-liquid phase separation in neuronal development and synaptic signaling. Dev Cell. 2020;55(1):18–29. doi: 10.1016/j.devcel.2020.06.012. [DOI] [PubMed] [Google Scholar]
- 87.McDonald NA, et al. Assembly of synaptic active zones requires phase separation of scaffold molecules. Nature. 2020;588(7838):454–458. doi: 10.1038/s41586-020-2942-0. [DOI] [PubMed] [Google Scholar]
- 88.Ryan VH, Fawzi NL. Physiological, pathological, and targetable membraneless organelles in neurons. Trends Neurosci. 2019;42(10):693–708. doi: 10.1016/j.tins.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lacroix E, Audas TE. Keeping up with the condensates: the retention, gain, and loss of nuclear membrane-less organelles. Front Mol Biosci. 2022;9:998363. doi: 10.3389/fmolb.2022.998363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Su Q, et al. Liquid-liquid phase separation: orchestrating cell signaling through time and space. Mol Cell. 2021;81(20):4137–4146. doi: 10.1016/j.molcel.2021.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shin Y, et al. Liquid nuclear condensates mechanically sense and restructure the genome. Cell. 2018;175(6):1481–1491. doi: 10.1016/j.cell.2018.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sabari BR, et al. Biomolecular condensates in the nucleus. Trends Biochem Sci. 2020;45(11):961–977. doi: 10.1016/j.tibs.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ishov AM, et al. Coordination of transcription, processing, and export of highly expressed RNAs by distinct biomolecular condensates. Emerg Top Life Sci. 2020;4(3):281–291. doi: 10.1042/ETLS20190160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Frottin F, et al. The nucleolus functions as a phase-separated protein quality control compartment. Science. 2019;365(6451):342–347. doi: 10.1126/science.aaw9157. [DOI] [PubMed] [Google Scholar]
- 95.Henninger JE, et al. RNA-mediated feedback control of transcriptional condensates. Cell. 2021;184(1):207–225. doi: 10.1016/j.cell.2020.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lafontaine DLJ, et al. The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol. 2021;22(3):165–182. doi: 10.1038/s41580-020-0272-6. [DOI] [PubMed] [Google Scholar]
- 97.Audas TE, et al. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell. 2012;45(2):147–157. doi: 10.1016/j.molcel.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 98.Feric M, et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165(7):1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sharp PA, et al. RNA in formation and regulation of transcriptional condensates. RNA. 2022;28(1):52–57. doi: 10.1261/rna.078997.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Guo YE, et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature. 2019;572(7770):543–548. doi: 10.1038/s41586-019-1464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Klein IA, et al. Partitioning of cancer therapeutics in nuclear condensates. Science. 2020;368(6497):1386–1392. doi: 10.1126/science.aaz4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gibson BA, et al. Organization of chromatin by intrinsic and regulated phase separation. Cell. 2019;179(2):470–484. doi: 10.1016/j.cell.2019.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wolozin B, Ivanov P. Stress granules and neurodegeneration. Nat Rev Neurosci. 2019;20(11):649–666. doi: 10.1038/s41583-019-0222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Molliex A, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163(1):123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Glauninger H, et al. Stressful steps: progress and challenges in understanding stress-induced mRNA condensation and accumulation in stress granules. Mol Cell. 2022;82(14):2544–2556. doi: 10.1016/j.molcel.2022.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gasset-Rosa F, et al. Cytoplasmic TDP-43 de-mixing independent of stress granules drives inhibition of nuclear import, loss of nuclear TDP-43, and cell death. Neuron. 2019;102(2):339–357. doi: 10.1016/j.neuron.2019.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mackenzie IR, et al. TIA1 mutations in amyotrophic lateral sclerosis and frontotemporal dementia promote phase separation and alter stress granule dynamics. Neuron. 2017;95(4):808–816. doi: 10.1016/j.neuron.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Markmiller S, et al. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell. 2018;172(3):590–604. doi: 10.1016/j.cell.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang P, et al. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell. 2020;181(2):325–345. doi: 10.1016/j.cell.2020.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hayashi Y, et al. Liquid-liquid phase separation in physiology and pathophysiology of the nervous system. J Neurosci. 2021;41(5):834–844. doi: 10.1523/JNEUROSCI.1656-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Milovanovic D, De Camilli P. Synaptic vesicle clusters at synapses: a distinct liquid phase? Neuron. 2017;93(5):995–1002. doi: 10.1016/j.neuron.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wu X, et al. Interactions between membraneless condensates and membranous organelles at the presynapse: a phase separation view of synaptic vesicle cycle. J Mol Biol. 2022;167629(1):167629. doi: 10.1016/j.jmb.2022.167629. [DOI] [PubMed] [Google Scholar]
- 113.Milovanovic D, et al. A liquid phase of synapsin and lipid vesicles. Science. 2018;361(6402):604–607. doi: 10.1126/science.aat5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zeng M, et al. Phase transition in postsynaptic densities underlies formation of synaptic complexes and synaptic plasticity. Cell. 2016;166(5):1163–1175. doi: 10.1016/j.cell.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bourke AM, et al. De-centralizing the central dogma: mRNA translation in space and time. Mol Cell. 2023;83(3):452–468. doi: 10.1016/j.molcel.2022.12.030. [DOI] [PubMed] [Google Scholar]
- 116.Parker DM, et al. It’s just a phase: exploring the relationship between mRNA, biomolecular condensates, and translational control. Front Genet. 2022;13:931220. doi: 10.3389/fgene.2022.931220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kim TH, et al. Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science. 2019;365(6455):825–829. doi: 10.1126/science.aax4240. [DOI] [PubMed] [Google Scholar]
- 118.Tan R, et al. Microtubules gate tau condensation to spatially regulate microtubule functions. Nat Cell Biol. 2019;21(9):1078–1085. doi: 10.1038/s41556-019-0375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hernández-Vega A, et al. Local nucleation of microtubule bundles through tubulin concentration into a condensed tau phase. Cell Rep. 2017;20(10):2304–2312. doi: 10.1016/j.celrep.2017.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hochmair J, et al. Molecular crowding and RNA synergize to promote phase separation, microtubule interaction, and seeding of tau condensates. EMBO J. 2022;41(11):e108882. doi: 10.15252/embj.2021108882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Maina MB, et al. The involvement of tau in nucleolar transcription and the stress response. Acta Neuropathol Commun. 2018;6(1):70. doi: 10.1186/s40478-018-0565-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tracy TE, et al. Tau interactome maps synaptic and mitochondrial processes associated with neurodegeneration. Cell. 2022;185(4):712–728. doi: 10.1016/j.cell.2021.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Freibaum BD, et al. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J Proteome Res. 2010;9(2):1104–1120. doi: 10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kavanagh T, et al. Tau interactome and RNA binding proteins in neurodegenerative diseases. Mol Neurodegener. 2022;17(1):66. doi: 10.1186/s13024-022-00572-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ittner A, Ittner LM. Dendritic tau in Alzheimer’s disease. Neuron. 2018;99(1):13–27. doi: 10.1016/j.neuron.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 126.DeVos SL, et al. Synaptic tau seeding precedes tau pathology in human Alzheimer’s disease brain. Front Neurosci. 2018;12:267. doi: 10.3389/fnins.2018.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McInnes J, et al. Synaptogyrin-3 mediates presynaptic dysfunction induced by tau. Neuron. 2018;97(4):823–835. doi: 10.1016/j.neuron.2018.01.022. [DOI] [PubMed] [Google Scholar]
- 128.Lee EB, et al. Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nat Rev Neurosci. 2012;13(1):38–50. doi: 10.1038/nrn3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Afroz T, et al. Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation. Nat Commun. 2017;8(1):45. doi: 10.1038/s41467-017-00062-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sekar D, et al. TDP-43 and NEAT long non-coding RNA: roles in neurodegenerative disease. Front Cell Neurosci. 2022;16:954912. doi: 10.3389/fncel.2022.954912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Banani SF, et al. Genetic variation associated with condensate dysregulation in disease. Dev Cell. 2022;57(14):1776–1788. doi: 10.1016/j.devcel.2022.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hou Y, et al. Ageing as a risk factor for neurodegenerative disease. Nat Rev Neurol. 2019;15(10):565–581. doi: 10.1038/s41582-019-0244-7. [DOI] [PubMed] [Google Scholar]
- 133.Soto-Palma C, et al. Epigenetics, DNA damage, and aging. J Clin Invest. 2022;132(16):158446. doi: 10.1172/JCI158446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Donnelly CJ, et al. Aberrant RNA homeostasis in amyotrophic lateral sclerosis: potential for new therapeutic targets? Neurodegener Dis Manag. 2014;4(6):417–437. doi: 10.2217/nmt.14.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Spead O, et al. Nuclear pore dysfunction in neurodegeneration. Neurotherapeutics. 2022;19(4):1050–1060. doi: 10.1007/s13311-022-01293-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mehta S, Zhang J. Liquid-liquid phase separation drives cellular function and dysfunction in cancer. Nat Rev Cancer. 2022;22(4):239–252. doi: 10.1038/s41568-022-00444-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tsang B, et al. Phase separation as a missing mechanism for interpretation of disease mutations. Cell. 2020;183(7):1742–1756. doi: 10.1016/j.cell.2020.11.050. [DOI] [PubMed] [Google Scholar]
- 138.Zhang H, et al. Liquid-liquid phase separation in biology: mechanisms, physiological functions and human diseases. Sci China Life Sci. 2020;63(7):953–985. doi: 10.1007/s11427-020-1702-x. [DOI] [PubMed] [Google Scholar]
- 139.Zhou X, et al. Mutations linked to neurological disease enhance self-association of low-complexity protein sequences. Science. 2022;377(6601):eabn5582. doi: 10.1126/science.abn5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nedelsky NB, Taylor JP. Pathological phase transitions in ALS-FTD impair dynamic RNA-protein granules. RNA. 2022;28(1):97–113. doi: 10.1261/rna.079001.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kim HJ, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495(7442):467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Maharana S, et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 2018;360(6391):918–921. doi: 10.1126/science.aar7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Maziuk B, et al. Dysregulation of RNA binding protein aggregation in neurodegenerative disorders. Front Mol Neurosci. 2017;10:89. doi: 10.3389/fnmol.2017.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Niaki AG, et al. Loss of dynamic RNA interaction and aberrant phase separation induced by two distinct types of ALS/FTD-linked FUS mutations. Mol Cell. 2020;77(1):82–94. doi: 10.1016/j.molcel.2019.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Aguzzi A, Altmeyer M. Phase separation: linking cellular compartmentalization to disease. Trends Cell Biol. 2016;26(7):547–558. doi: 10.1016/j.tcb.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 146.Joel Z, et al. Improving mouse models for dementia. Are all the effects in tau mouse models due to overexpression? Cold Spring Harb Symp Quant Biol. 2018;83:151–161. doi: 10.1101/sqb.2018.83.037531. [DOI] [PubMed] [Google Scholar]
- 147.Wood A, et al. Molecular mechanisms underlying TDP-43 pathology in cellular and animal models of ALS and FTLD. Int J Mol Sci. 2021;22(9):4705. doi: 10.3390/ijms22094705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Brown AL, et al. TDP-43 loss and ALS-risk SNPs drive mis-splicing and depletion of UNC13A. Nature. 2022;603(7899):131–137. doi: 10.1038/s41586-022-04436-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.DeVos SL, et al. Tau reduction in the presence of amyloid-β prevents tau pathology and neuronal death in vivo. Brain. 2018;141(7):2194–2212. doi: 10.1093/brain/awy117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tai C, et al. Tau reduction prevents key features of autism in mouse models. Neuron. 2020;106(3):421–437. doi: 10.1016/j.neuron.2020.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Roberson ED, et al. Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer’s disease mouse model. Science. 2007;316(5825):750–754. doi: 10.1126/science.1141736. [DOI] [PubMed] [Google Scholar]
- 152.DeVos SL, et al. Tau reduction prevents neuronal loss and reverses pathological tau deposition and seeding in mice with tauopathy. Sci Transl Med. 2017;9(374):eaag0481. doi: 10.1126/scitranslmed.aag0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Petrozziello T, et al. Targeting tau mitigates mitochondrial fragmentation and oxidative stress in amyotrophic lateral sclerosis. Mol Neurobiol. 2022;59(1):683–702. doi: 10.1007/s12035-021-02557-w. [DOI] [PubMed] [Google Scholar]
- 154.Jabbari E, Duff KE. Tau-targeting antibody therapies: too late, wrong epitope or wrong target? Nat Med. 2021;27(8):1341–1342. doi: 10.1038/s41591-021-01465-9. [DOI] [PubMed] [Google Scholar]
- 155.Chen JL, et al. Design, optimization, and study of small molecules that target tau pre-mRNA and affect splicing. J Am Chem Soc. 2020;142(19):8706–8727. doi: 10.1021/jacs.0c00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schoch KM, et al. Increased 4R-tau induces pathological changes in a human-tau mouse model. Neuron. 2016;90(5):941–947. doi: 10.1016/j.neuron.2016.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Espíndola SL, et al. Modulation of tau isoforms imbalance precludes tau pathology and cognitive decline in a mouse model of tauopathy. Cell Rep. 2018;23(3):709–715. doi: 10.1016/j.celrep.2018.03.079. [DOI] [PubMed] [Google Scholar]
- 158.Wegmann S, et al. Persistent repression of tau in the brain using engineered zinc finger protein transcription factors. Sci Adv. 2021;7(12):eabe1611. doi: 10.1126/sciadv.abe1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Simone R, et al. MIR-NATs repress MAPT translation and aid proteostasis in neurodegeneration. Nature. 2021;594(7861):117–123. doi: 10.1038/s41586-021-03556-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Heller GT, et al. Targeting disordered proteins with small molecules using entropy. Trends Biochem Sci. 2015;40(9):491–496. doi: 10.1016/j.tibs.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 161.Francois-Moutal L, et al. Direct targeting of TDP-43, from small molecules to biologics: the therapeutic landscape. RSC Chem Biol. 2021;2(4):1158–1166. doi: 10.1039/D1CB00110H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pickhardt M, et al. Identification of small molecule inhibitors of tau aggregation by targeting monomeric tau as a potential therapeutic approach for tauopathies. Curr Alzheimer Res. 2015;12(9):814–828. doi: 10.2174/156720501209151019104951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang L, et al. Small molecule therapeutics for tauopathy in Alzheimer’s disease: walking on the path of most resistance. Eur J Med Chem. 2021;209:112915. doi: 10.1016/j.ejmech.2020.112915. [DOI] [PubMed] [Google Scholar]
- 164.Baggett DW, Nath A. The rational discovery of a tau aggregation inhibitor. Biochemistry. 2018;57(42):6099–6107. doi: 10.1021/acs.biochem.8b00581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mirbaha H, et al. Inert and seed-competent tau monomers suggest structural origins of aggregation. Elife. 2018;7:e36584. doi: 10.7554/eLife.36584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Taniguchi S, et al. Inhibition of heparin-induced tau filament formation by phenothiazines, polyphenols, and porphyrins. J Biol Chem. 2005;280(9):7614–7623. doi: 10.1074/jbc.M408714200. [DOI] [PubMed] [Google Scholar]
- 167.Akoury E, et al. Mechanistic basis of phenothiazine-driven inhibition of tau aggregation. Angew Chem Int Ed Engl. 2013;52(12):3511–3515. doi: 10.1002/anie.201208290. [DOI] [PubMed] [Google Scholar]
- 168.Schafer KN, et al. Structural determinants of tau aggregation inhibitor potency. J Biol Chem. 2013;288(45):32599–32611. doi: 10.1074/jbc.M113.503474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mollasalehi N, et al. An allosteric modulator of RNA binding targeting the N-terminal domain of TDP-43 yields neuroprotective properties. ACS Chem Biol. 2020;15(11):2854–2859. doi: 10.1021/acschembio.0c00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Abskharon R, et al. Cryo-EM structure of RNA-induced tau fibrils reveals a small C-terminal core that may nucleate fibril formation. Proc Natl Acad Sci U S A. 2022;119(15):e2119952119. doi: 10.1073/pnas.2119952119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhang H, et al. RNA controls PolyQ protein phase transitions. Mol Cell. 2015;60(2):220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bhardwaj A, et al. Characterizing TDP-43 interaction with its RNA targets. Nucleic Acids Res. 2013;41(9):5062–5074. doi: 10.1093/nar/gkt189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Pozzi S, et al. Virus-mediated delivery of antibody targeting TAR DNA-binding protein-43 mitigates associated neuropathology. J Clin Invest. 2019;129(4):1581–1595. doi: 10.1172/JCI123931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Abskharon R, et al. Crystal structure of a conformational antibody that binds tau oligomers and inhibits pathological seeding by extracts from donors with Alzheimer’s disease. J Biol Chem. 2020;295(31):10662–10676. doi: 10.1074/jbc.RA120.013638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Jicha GA, et al. Alz-50 and MC-1, a new monoclonal antibody raised to paired helical filaments, recognize conformational epitopes on recombinant tau. J Neurosci Res. 1997;48(2):128–132. doi: 10.1002/(SICI)1097-4547(19970415)48:2<128::AID-JNR5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 176.Nakamura K, et al. Proline isomer-specific antibodies reveal the early pathogenic tau conformation in Alzheimer’s disease. Cell. 2012;149(1):232–244. doi: 10.1016/j.cell.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lu KP et al. Potential of the antibody against cis-phosphorylated tau in the early diagnosis, treatment, and prevention of Alzheimer disease and brain injury. JAMA Neurol. 2016;73(11):1356–1362. doi: 10.1001/jamaneurol.2016.2027. [DOI] [PubMed] [Google Scholar]
- 178.Qiu C, et al. Cis P-tau underlies vascular contribution to cognitive impairment and dementia and can be effectively targeted by immunotherapy in mice. Sci Transl Med. 2021;13(596):eaaz7615. doi: 10.1126/scitranslmed.aaz7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kondo A, et al. Antibody against early driver of neurodegeneration cis P-tau blocks brain injury and tauopathy. Nature. 2015;523(7561):431–436. doi: 10.1038/nature14658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Albayram O, et al. Cis P-tau is induced in clinical and preclinical brain injury and contributes to post-injury sequelae. Nat Commun. 2017;8(1):1000. doi: 10.1038/s41467-017-01068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Porta S, et al. Patient-derived frontotemporal lobar degeneration brain extracts induce formation and spreading of TDP-43 pathology in vivo. Nat Commun. 2018;9(1):4220. doi: 10.1038/s41467-018-06548-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Jucker M, Walker LC. Propagation and spread of pathogenic protein assemblies in neurodegenerative diseases. Nat Neurosci. 2018;21(10):1341–1349. doi: 10.1038/s41593-018-0238-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Vaquer-Alicea J, et al. Tau strains shape disease. Acta Neuropathol. 2021;142(1):57–71. doi: 10.1007/s00401-021-02301-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Koehler LC, et al. TDP-43 oligomerization and phase separation properties are necessary for autoregulation. Front Neurosci. 2022;16:818655. doi: 10.3389/fnins.2022.818655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wagner J, et al. Anle138b: a novel oligomer modulator for disease-modifying therapy of neurodegenerative diseases such as prion and Parkinson’s disease. Acta Neuropathol. 2013;125(6):795–813. doi: 10.1007/s00401-013-1114-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ingham DJ, et al. Fungally derived isoquinoline demonstrates inducer-specific tau aggregation inhibition. Biochemistry. 2021;60(21):1658–1669. doi: 10.1021/acs.biochem.1c00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Baggett DW, Nath A. Structure-activity relationships of novel tau ligands: passive fibril binders and active aggregation inhibitors. ACS Chem Biol. 2022;17(3):701–708. doi: 10.1021/acschembio.2c00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wagner J, et al. Reducing tau aggregates with anle138b delays disease progression in a mouse model of tauopathies. Acta Neuropathol. 2015;130(5):619–631. doi: 10.1007/s00401-015-1483-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Seidler PM, et al. Structure-based discovery of small molecules that disaggregate Alzheimer’s disease tissue derived tau fibrils in vitro. Nat Commun. 2022;13(1):5451. doi: 10.1038/s41467-022-32951-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Seidler PM, et al. Structure-based inhibitors of tau aggregation. Nat Chem. 2018;10(2):170–176. doi: 10.1038/nchem.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Owen I, Shewmaker F. The role of post-translational modifications in the phase transitions of intrinsically disordered proteins. Int J Mol Sci. 2019;20(21):5501. doi: 10.3390/ijms20215501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Jeon P, et al. Regulation of cellular ribonucleoprotein granules: from assembly to degradation via post-translational modification. Cells. 2022;11(13):2063. doi: 10.3390/cells11132063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sternburg EL, et al. Post-translational modifications on RNA-binding proteins: accelerators, brakes, or passengers in neurodegeneration? Trends Biochem Sci. 2022;47(1):6–22. doi: 10.1016/j.tibs.2021.07.004. [DOI] [PubMed] [Google Scholar]
- 194.Wesseling H, et al. Tau PTM profiles identify patient heterogeneity and stages of Alzheimer’s disease. Cell. 2020;183(6):1699–1713. doi: 10.1016/j.cell.2020.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Farina S, et al. Post-translational modifications modulate proteinopathies of TDP-43, FUS and hnRNP-A/B in amyotrophic lateral sclerosis. Front Mol Biosci. 2021;8:693325. doi: 10.3389/fmolb.2021.693325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Rahman S. Posttranslational modifications of TDP-43. In: Kumar V, Jaiswal MK, eds. TDP-43 and Neurodegeneration. Elsevier; 2022:45–79. [Google Scholar]
- 197.Gruijs da Silva LA, et al. Disease-linked TDP-43 hyperphosphorylation suppresses TDP-43 condensation and aggregation. EMBO J. 2022;41(8):e108443. doi: 10.15252/embj.2021108443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ukmar-Godec T, et al. Lysine/RNA-interactions drive and regulate biomolecular condensation. Nat Commun. 2019;10(1):2909. doi: 10.1038/s41467-019-10792-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Cohen TJ, et al. The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nat Commun. 2011;2:252. doi: 10.1038/ncomms1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Yu H, et al. HSP70 chaperones RNA-free TDP-43 into anisotropic intranuclear liquid spherical shells. Science. 2021;371(6529):eabb4309. doi: 10.1126/science.abb4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Shin MK, et al. Reducing acetylated tau is neuroprotective in brain injury. Cell. 2021;184(10):2715–2732. doi: 10.1016/j.cell.2021.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cohen TJ, et al. An acetylation switch controls TDP-43 function and aggregation propensity. Nat Commun. 2015;6:5845. doi: 10.1038/ncomms6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Aikio M, et al. Opposing roles of p38α-mediated phosphorylation and arginine methylation in driving TDP-43 proteinopathy [preprint]. Posted on bioRxiv August 4, 2021. [DOI]
- 204.Stefanoska K, et al. Alzheimer’s disease: ablating single master site abolishes tau hyperphosphorylation. Sci Adv. 2022;8(27):eabl8809. doi: 10.1126/sciadv.abl8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Riback JA, et al. Composition-dependent thermodynamics of intracellular phase separation. Nature. 2020;581(7807):209–214. doi: 10.1038/s41586-020-2256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Jiang L, et al. Interaction of tau with HNRNPA2B1 and N6-methyladenosine RNA mediates the progression of tauopathy. Mol Cell. 2021;81(20):4209–4227. doi: 10.1016/j.molcel.2021.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wheeler JM, et al. Activity of the poly(A) binding protein MSUT2 determines susceptibility to pathological tau in the mammalian brain. Sci Transl Med. 2019;11(523):eaao6545. doi: 10.1126/scitranslmed.aao6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Apicco DJ, et al. Reducing the RNA binding protein TIA1 protects against tau-mediated neurodegeneration in vivo. Nat Neurosci. 2018;21(1):72–80. doi: 10.1038/s41593-017-0022-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Becker LA, et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature. 2017;544(7650):367–371. doi: 10.1038/nature22038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Vanderweyde T, et al. Interaction of tau with the RNA-binding protein TIA1 regulates tau pathophysiology and toxicity. Cell Rep. 2016;15(7):1455–1466. doi: 10.1016/j.celrep.2016.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ash PEA, et al. TIA1 potentiates tau phase separation and promotes generation of toxic oligomeric tau. Proc Natl Acad Sci U S A. 2021;118(9):e2014188118. doi: 10.1073/pnas.2014188118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lester E, et al. Tau aggregates are RNA-protein assemblies that mislocalize multiple nuclear speckle components. Neuron. 2021;109(10):1675–1691. doi: 10.1016/j.neuron.2021.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Zuniga G, et al. Tau-induced deficits in nonsense-mediated mRNA decay contribute to neurodegeneration. Alzheimers Dement. 2023;19(2):405–420. doi: 10.1002/alz.12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Elden AC, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466(7310):1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Zhang K, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525(7567):56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bush JA, et al. Ribonuclease recruitment using a small molecule reduced c9ALS/FTD r(G4C2) repeat expansion in vitro and in vivo ALS models. Sci Transl Med. 2021;13(617):eabd5991. doi: 10.1126/scitranslmed.abd5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Matsukawa K, et al. Long non-coding RNA NEAT1_1 ameliorates TDP-43 toxicity in in vivo models of TDP-43 proteinopathy. RNA Biol. 2021;18(11):1546–1554. doi: 10.1080/15476286.2020.1860580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Chatterjee B, et al. RNA modifications and RNA metabolism in neurological disease pathogenesis. Int J Mol Sci. 2021;22(21):10. doi: 10.3390/ijms222111870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Jiapaer Z, et al. Regulation and roles of RNA modifications in aging-related diseases. Aging Cell. 2022;21(7):e13657. doi: 10.1111/acel.13657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Roundtree IA, et al. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169(7):1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Drino A, Schaefer MR. RNAs, phase separation, and membrane-less organelles: are post-transcriptional modifications modulating organelle dynamics? Bioessays. 2018;40(12):e1800085. doi: 10.1002/bies.201800085. [DOI] [PubMed] [Google Scholar]
- 222.Edupuganti RR, et al. N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol. 2017;24(10):870–878. doi: 10.1038/nsmb.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Lee J-H, et al. Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol Cell. 2021;81(16):3368–3385. doi: 10.1016/j.molcel.2021.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zaccara S, et al. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20(10):608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 225.McMillan M, et al. RNA methylation influences TDP43 binding and disease pathogenesis in models of amyotrophic lateral sclerosis and frontotemporal dementia. Mol Cell. 2023;83(2):219–236. doi: 10.1016/j.molcel.2022.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Kaushik S, Cuervo AM. Proteostasis and aging. Nat Med. 2015;21(12):1406–1415. doi: 10.1038/nm.4001. [DOI] [PubMed] [Google Scholar]
- 227.Alberti S, Carra S. Quality control of membraneless organelles. J Mol Biol. 2018;430(23):4711–4729. doi: 10.1016/j.jmb.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 228.Labbadia J, Morimoto RI. The biology of proteostasis in aging and disease. Annu Rev Biochem. 2015;84:435–464. doi: 10.1146/annurev-biochem-060614-033955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19(8):983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 230.Vargas JNS, et al. The mechanisms and roles of selective autophagy in mammals. Nat Rev Mol Cell Biol. 2023;24(3):167–185. doi: 10.1038/s41580-022-00542-2. [DOI] [PubMed] [Google Scholar]
- 231.McMillan M, et al. RNA methylation influences TDP43 binding and disease pathogenesis in models of amyotrophic lateral sclerosis and frontotemporal dementia. Mol Cell. 2023;83(2):219–236. doi: 10.1016/j.molcel.2022.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Yamasaki A, et al. Liquidity is a critical determinant for selective autophagy of protein condensates. Mol Cell. 2020;77(6):1163–1175. doi: 10.1016/j.molcel.2019.12.026. [DOI] [PubMed] [Google Scholar]
- 233.Jinwal UK, et al. Hsp70 ATPase modulators as therapeutics for Alzheimer’s and other neurodegenerative diseases. Mol Cell Pharmacol. 2010;2(2):43–46. [PMC free article] [PubMed] [Google Scholar]
- 234.Chiang AN, et al. Synthesis and evaluation of esterified Hsp70 agonists in cellular models of protein aggregation and folding. Bioorg Med Chem. 2019;27(1):79–91. doi: 10.1016/j.bmc.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Babu M, et al. Peptidyl prolyl isomerase a modulates the liquid-liquid phase separation of proline-rich IDPs. J Am Chem Soc. 2022;144(35):16157–16163. doi: 10.1021/jacs.2c07149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Hutten S, et al. Nuclear import receptors directly bind to arginine-rich dipeptide repeat proteins and suppress their pathological interactions. Cell Rep. 2020;33(12):108538. doi: 10.1016/j.celrep.2020.108538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Guo L, et al. Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell. 2018;173(3):677–692. doi: 10.1016/j.cell.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Caballero B, et al. Interplay of pathogenic forms of human tau with different autophagic pathways. Aging Cell. 2018;17(1):e12692. doi: 10.1111/acel.12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Carrettiero DC, et al. Stress routes clients to the proteasome via a BAG2 ubiquitin-independent degradation condensate. Nat Commun. 2022;13(1):3074. doi: 10.1038/s41467-022-30751-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Scotter EL, et al. Differential roles of the ubiquitin proteasome system and autophagy in the clearance of soluble and aggregated TDP-43 species. J Cell Sci. 2014;127(pt 6):1263–1278. doi: 10.1242/jcs.140087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Yan Y, et al. X-linked ubiquitin-specific peptidase 11 increases tauopathy vulnerability in women. Cell. 2022;185(21):3913–3930. doi: 10.1016/j.cell.2022.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Schaler AW, et al. PAC1 receptor-mediated clearance of tau in postsynaptic compartments attenuates tau pathology in mouse brain. Sci Transl Med. 2021;13(595):eaba7394. doi: 10.1126/scitranslmed.aba7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Bourdenx M, et al. Chaperone-mediated autophagy prevents collapse of the neuronal metastable proteome. Cell. 2021;184(10):2696–2714. doi: 10.1016/j.cell.2021.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Caballero B, et al. Acetylated tau inhibits chaperone-mediated autophagy and promotes tau pathology propagation in mice. Nat Commun. 2021;12(1):2238. doi: 10.1038/s41467-021-22501-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Babinchak WM, et al. Small molecules as potent biphasic modulators of protein liquid-liquid phase separation. Nat Commun. 2020;11(1):5574. doi: 10.1038/s41467-020-19211-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Fang MY, et al. Small-molecule modulation of TDP-43 recruitment to stress granules prevents persistent TDP-43 accumulation in ALS/FTD. Neuron. 2019;103(5):802–819. doi: 10.1016/j.neuron.2019.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Kilgore HR, Young RA. Learning the chemical grammar of biomolecular condensates. Nat Chem Biol. 2022;18(12):1298–1306. doi: 10.1038/s41589-022-01046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Jonchhe S, et al. Small molecules modulate liquid-to-solid transitions in phase-separated tau condensates. Angew Chem Int Ed. 2022;61(23):e202113156. doi: 10.1002/anie.202113156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Dai B, et al. Myricetin slows liquid-liquid phase separation of tau and activates ATG5-dependent autophagy to suppress tau toxicity. J Biol Chem. 2021;297(4):101222. doi: 10.1016/j.jbc.2021.101222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Gray MJ, et al. Polyphosphate is a primordial chaperone. Mol Cell. 2014;53(5):689–699. doi: 10.1016/j.molcel.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Patel A, et al. ATP as a biological hydrotrope. Science. 2017;356(6339):753–756. doi: 10.1126/science.aaf6846. [DOI] [PubMed] [Google Scholar]
- 252.Zhang JZ, et al. Phase separation of a PKA regulatory subunit controls cAMP compartmentation and oncogenic signaling. Cell. 2020;182(6):1531–1544. doi: 10.1016/j.cell.2020.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Peinado JR, et al. Sequestration of TDP-43216-414 aggregates by cytoplasmic expression of the proSAAS chaperone. ACS Chem Neurosci. 2022;13(11):1651–1665. doi: 10.1021/acschemneuro.2c00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Alberti S, Hyman AA. Are aberrant phase transitions a driver of cellular aging? Bioessays. 2016;38(10):959–968. doi: 10.1002/bies.201600042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Lashuel HA. Rethinking protein aggregation and drug discovery in neurodegenerative diseases: why we need to embrace complexity? Curr Opin Chem Biol. 2021;64:67–75. doi: 10.1016/j.cbpa.2021.05.006. [DOI] [PubMed] [Google Scholar]
- 256.Chen S, et al. Wolframin is a novel regulator of tau pathology and neurodegeneration. Acta Neuropathol. 2022;143(5):547–569. doi: 10.1007/s00401-022-02417-4. [DOI] [PubMed] [Google Scholar]