Abstract
Congenital heart diseases often involve maldevelopment of the evolutionarily recent right heart chamber. To gain insight into right heart structure and function, we fine-tuned deep learning models to recognize the right atrium, right ventricle, and pulmonary artery, measuring right heart structures in 40,000 individuals from the UK Biobank with magnetic resonance imaging. Genome-wide association studies identified 130 distinct loci associated with at least one right heart measurement, of which 72 were not associated with left heart structures. Loci were found near genes previously linked with congenital heart disease, including NKX2–5, TBX5/TBX3, WNT9B, and GATA4. A genome-wide polygenic predictor of right ventricular ejection fraction was associated with incident dilated cardiomyopathy (HR 1.33 per standard deviation; P = 7.1 × 10−13) and remained significant after accounting for a left ventricular polygenic score. Harnessing deep learning to perform large-scale cardiac phenotyping, our results yield insights into the genetic determinants of right heart structure and function.
The heart evolved hundreds of millions of years ago as a tubular organ1. Septation of the main pumping chamber of the heart into distinct left and right ventricles evolved later in birds, mammals, and some reptiles, and is under the control of conserved transcription factors such as TBX52. Enhanced delivery of oxygen to the systemic circulation—and to the heart itself—is the putative advantage of this separation of the circulatory system into a left heart-driven systemic circuit and a right heart-driven pulmonary circuit3.
Left and right heart structures are derived from different progenitor cell populations and operate under different pressure regimes: the left heart faces high afterload, while the right heart generally faces relatively low afterload. During embryogenesis, the left ventricle (LV) forms from the first heart field, while the right ventricle (RV), the outflow tract, and portions of the atria form from the second heart field4–7. Septation of the bilateral ventricular outflow tracts and the truncus arteriosus into the aorta and pulmonary artery (PA) also requires neuroectodermal neural crest cells8–10.
The distinct embryological origins of the right and left ventricles likely contribute to the occurrence of right heart-predominant pathologies. These include various types of arrhythmogenic right ventricular cardiomyopathy (ARVC)11–16, Brugada syndrome17, and pulmonary hypertension. In addition, right ventricular dysfunction can be an important determinant of outcomes for individuals with heart failure syndromes18–20.
To date, a large-scale epidemiological analysis of right ventricular structure and function has been conducted using deep learning-derived cardiac measurements21,22. The distinct pathologies, embryology, and physiology of the right heart motivated our efforts to quantify right heart structure and function, and to probe the common genetic basis for their variation.
In this work, we developed deep learning models to determine the dimensions and function of the right atrium (RA), the RV, and the PA in up to 45,000 UK Biobank participants. We then evaluated the epidemiologic associations, pathologic outcomes, and common genetic basis for variation in these right heart structures.
Results
Deep learning with cardiovascular magnetic resonance images.
We derived right heart measurements in the UK Biobank imaging substudy of over 45,000 people23–25 using deep learning models26,27 trained on magnetic resonance images that were manually annotated by cardiologists (Fig. 1)24. We randomly selected 714 short axis images (out of over 24 million) and 445 four-chamber long axis images (out of over 2.2 million) for annotation. U-Net derived deep learning models were then trained from these data26,28–30. The deep learning models were then used to produce pixel labels for the remaining images. Model construction, training, and quality assessment are detailed in the Online Methods and Supplementary Note31,32.
Figure 1 |. Right heart measurement with deep learning.
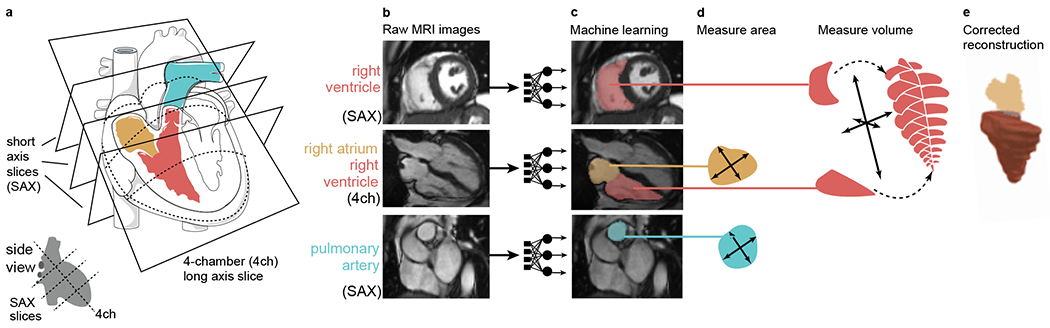
Measurement of right heart structures from cardiovascular magnetic resonance imaging using deep learning. In all panels, the PA is colored turquoise, the RV is colored red, and the RA is colored yellow. The magnetic resonance images in this figure are reproduced by kind permission of UK Biobank ©. a, Graphical depictions of the right heart structures in a cutaway view of the heart. The art in this panel is derived from Servier Medical Art (licensed under creative commons by attribution, CC-BY-3.0). b, Cardiovascular magnetic resonance imaging. SAX, short axis view; 4ch, four-chamber long axis view. The RV is visible in the SAX and 4ch views, the RA in the 4ch view, and the PA in the basal SAX view. c, The raw images are fed into the trained deep learning model, producing pixel-by-pixel output (here, colorized and laid on top of the raw images). d, The deep learning models are applied to all images, allowing measurement of the right heart structures. e, The right ventricular surface is reconstructed by combining data from SAX and 4ch images (see Online Methods), allowing a volumetric measurement.
Reconstruction of right heart structures from deep learning.
The deep learning model output was then post-processed to extract measurements of the RA, the ventricles, and the PA (detailed in Supplementary Note). In total, we were able to measure at least one cardiac structure in 45,504 individuals, of whom 41,135 contributed to at least one genome-wide association study after genotyping quality control and exclusion for prevalent disease (Table 1 and Supplementary Fig. 1). The mean and standard deviation (s.d.) of the right atrial area measurements, right ventricular volumes, and PA diameters are visualized in Supplementary Figure 2. Standard values aggregated by sex for each of the phenotypes are reported in Supplementary Table 1, and by age bands and sex in Supplementary Table 2. Cross-correlation between left- and right-heart structures is represented in Supplementary Figure 3 and described in the Supplementary Note.
Table 1 |.
Participant characteristics
| Women | Men | All | |
|---|---|---|---|
| n | 21,946 | 19,189 | 41,135 |
| Age at time of MRI | 63.9 (7.6) | 65.0 (7.8) | 64.4 (7.7) |
| BMI (kg/m2) | 26.0 (4.7) | 26.9 (3.9) | 26.4 (4.4) |
| Height (cm) | 163 (6) | 176 (7) | 169 (9) |
| Weight (kg) | 68.9 (13.1) | 83.5 (13.3) | 75.7 (15.0) |
| Systolic blood pressure (mmHg) | 136 (19) | 142 (17) | 139 (19) |
| Diastolic blood pressure (mmHg) | 77.1 (10.0) | 80.9 (9.8) | 78.9 (10.1) |
| Drinking status | |||
| Current | 20,134 (92 %) | 18,020 (94 %) | 38,154 (93 %) |
| Never | 901 (4 %) | 439 (2 %) | 1,340 (3 %) |
| Prefer not to answer | 7 (0 %) | 12 (0 %) | 19 (0 %) |
| Previous | 747 (3 %) | 604 (3 %) | 1,351 (3 %) |
| Standard drinks/week | 4.72 (5.32) | 5.63 (6.80) | 5.15 (6.08) |
| Smoking status | |||
| Current | 606 (3 %) | 797 (4 %) | 1,403 (3 %) |
| Never | 14,343 (65 %) | 11,296 (59 %) | 25,639 (62 %) |
| Prefer not to answer | 85 (0 %) | 49 (0 %) | 134 (0 %) |
| Previous | 6,755 (31 %) | 6,933 (36 %) | 13,688 (33 %) |
| Smoking quantity (pack years) | 3.40 (9.11) | 5.43 (12.59) | 4.35 (10.92) |
Clinical characteristics of the 41,135 participants whose data contributed to at least one GWAS. For quantitative phenotypes, values shown represent mean (s.d.). For count data, values shown represent count (%). Cardiovascular phenotypes are detailed by age and sex in Supplementary Table 1.
To provide a direct comparison to left heart structures within the same sample, we also measured the left ventricle from short axis images. Left ventricular measurements included end diastolic volume (LVEDV), end systolic volume (LVESV), stroke volume (LVSV), and ejection fraction (LVEF). We compared PA measurements with previously reported aortic diameter measurements (Supplementary Note)33.
Prevalent cardiovascular diseases linked to the right heart.
We tested for correlations between the right heart phenotypes and diseases. These included analyses of hundreds of PheCode-based diseases prevalent at the time of imaging (Fig. 2 and Supplementary Table 3) and an analysis of three curated diseases with established chamber-specific links to the right heart diagnosed after imaging (atrial fibrillation, congestive heart failure, and pulmonary hypertension, defined in Supplementary Table 4)20,34,35. We also probed the properties of right ventricular volume throughout the cardiac cycle in the presence of congestive heart failure, pulmonary hypertension, or non-cardiac disease (Fig. 3 and Supplementary Fig. 4). Pre-existing pulmonary hypertension was associated with elevated right ventricular volumes even after accounting for the corresponding left ventricular volume, yielding a reduced right ventricular ejection fraction (RVEF; two-tailed P = 3.9 × 10−4 against the null hypothesis of no effect). Each of these findings is detailed in the Supplementary Note.
Figure 2 |. Right heart structures are associated with PheCode-based disease definitions.
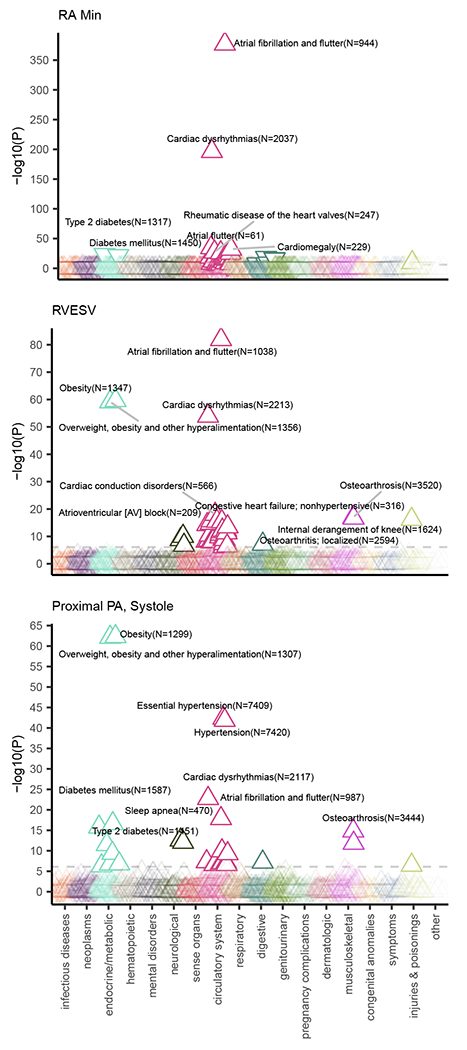
PheCode-based disease labels (x-axis) are plotted against a transformation of their association P-value (y-axis) with three right heart phenotypes: minimum right atrial area, RVESV, and proximal PA diameter. The values are derived from a linear model that associates the presence or absence of a PheCode-based disease with the right heart measurement, after adjustment for anthropometric covariates and genetic principal components. The direction of the arrow indicates whether the presence of the disease is associated with an increase (upward arrow) or a decrease (downward arrow) of the right heart measurement. The color indicates the disease grouping (as labeled on the x-axis). All values are available in Supplementary Table 3.
Figure 3 |. Alterations in right ventricular volume with prevalent disease.
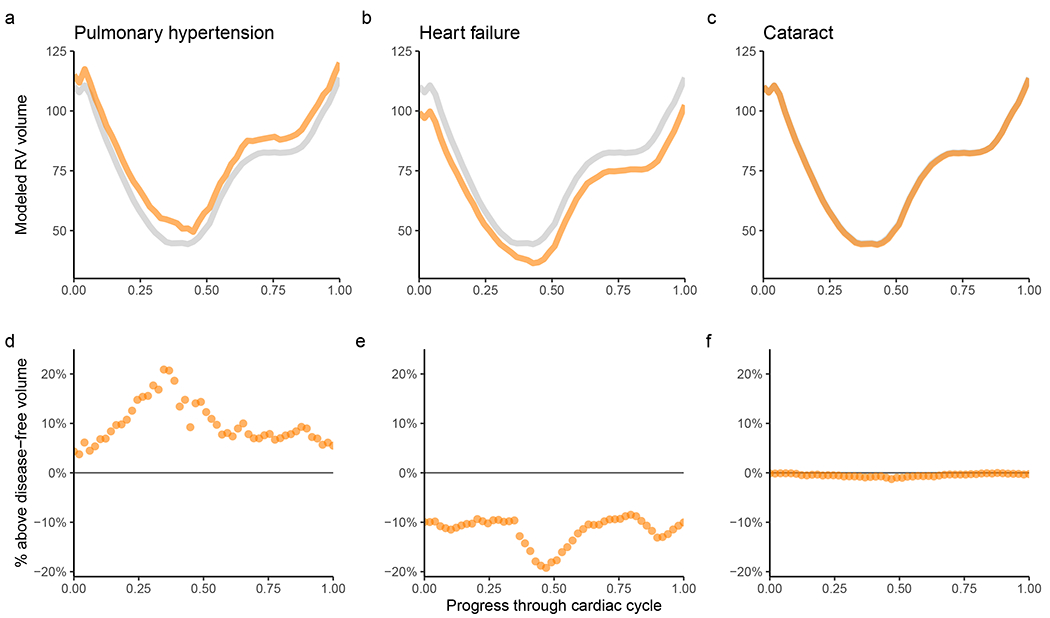
Disease diagnoses that occur prior to the date of MRI are linked with distinct changes in the volume of the RV throughout the cardiac cycle. For all panels, the x-axis represents fractions of a cardiac cycle (divided evenly into 50 components, starting at end-diastole). a-c, The y-axis represents volume in mL. Values are generated with a linear model for each time point accounting for the left ventricular volume at that time point, as well as clinical covariates; the gray line represents the population without disease, while the orange line represents the population with disease. In the UK Biobank, participants with pulmonary hypertension (a) have elevated right ventricular volume throughout the cardiac cycle, even after accounting for left ventricular volume. Those with heart failure (b) predominantly have elevated left ventricular volume, with relative sparing of their right ventricular volume (see Supplementary Fig. 4 for right ventricular volume without adjustment for left ventricular volume). Cataract (c) is used as a control to demonstrate little association between a non-cardiovascular disease and the volume of the RV. d-f, For pulmonary hypertension (d), heart failure (e), and cataract (f), at each time point the right ventricular volume of individuals with disease is subtracted from the volume without disease and divided by the volume without disease. This represents the percentage above or below the disease-free right ventricular volume for those with disease.
Heritability and genetic correlation of the right heart.
The size-related phenotypes showed significant heritability using BOLT-REML (as high as 0.36 for the maximum right atrial area, 0.41 for RV end diastolic volume (RVEDV), and 0.44 for the pulmonary root diameter)36,37. Heritabilities were lower for measurements of right heart function, such as RVEF, which had a heritability of 0.24 (Supplementary Table 5).
We found strong genetic correlation between the right- and left-ventricular measurements (rg = 0.90 between RVEDV and LVEDV; rg = 0.76 between right ventricular end systolic volume (RVESV) and LVESV; and rg = 0.55 between RVEF and LVEF)37. The proximal PA diameter had a genetic correlation of 0.63 with the ascending aortic diameter (Supplementary Table 6 and Supplementary Fig. 5).
Common variant genetic analysis of the right heart.
To conduct genome-wide association studies (GWAS) of each trait, we excluded participants with diagnoses of heart failure, atrial fibrillation, or myocardial infarction prior to their magnetic resonance imaging study (participant characteristics in Table 1; sample exclusion flowchart in Supplementary Fig. 1). We conducted ten primary right heart GWAS: maximum and minimum right atrial area; RA fractional area change (FAC); RVESV, RVEDV, RVSV, and RVEF; pulmonary root diameter; and proximal PA diameter in systole and diastole (Manhattan plots in Fig. 4; quantile-quantile plots in Supplementary Fig. 6). We also evaluated PA strain, and the body surface area (BSA)-indexed versions of all traits except for those which are dimensionless. Where paired left heart traits were available (such as LVEDV and RVEDV), we conducted within-sample GWAS of the left heart traits, and GWAS of right heart traits divided by their left heart counterparts. In total, we conducted five GWAS of right atrial phenotypes (Supplementary Fig. 7), 11 GWAS of right ventricular phenotypes (Supplementary Fig. 8), and nine GWAS of pulmonary trunk phenotypes (Supplementary Fig. 9).
Figure 4 |. Manhattan plots of right heart traits.
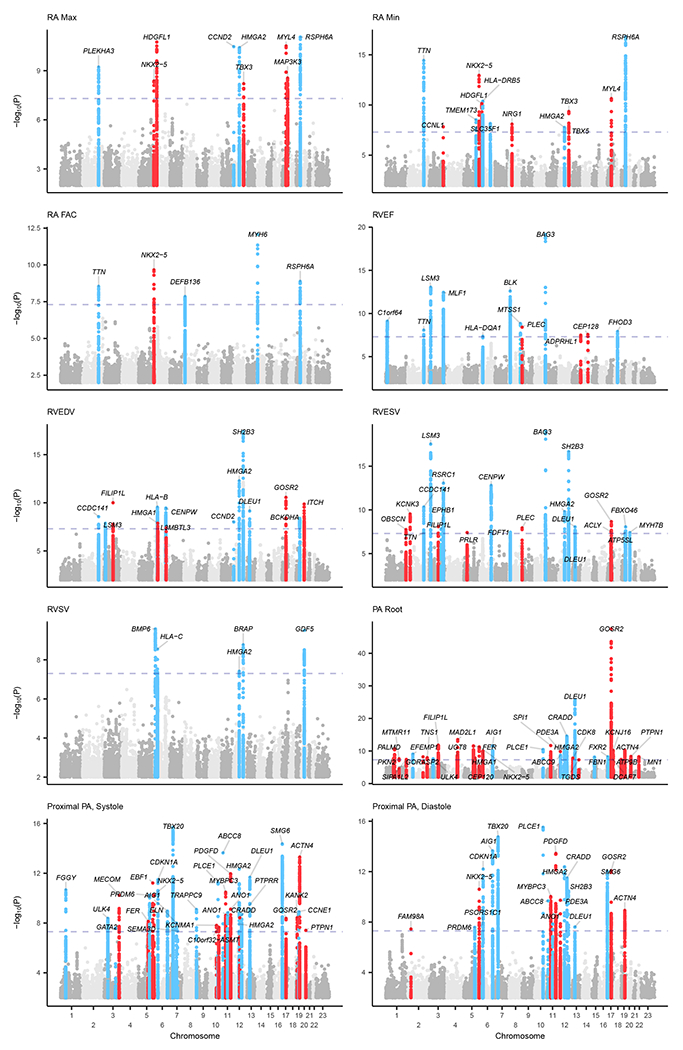
Manhattan plots show the chromosomal position (x-axis) and the strength of association (−log10 of the P-value, y-axis) for all non-BSA-indexed phenotypes. The X-chromosome is represented as “Chromosome 23.” Loci that contain SNPs with P < 5 × 10−8 were labeled with the name of the nearest gene; genes may appear multiple times for the same trait when multiple variants at the same locus are in linkage equilibrium with one another (r2 < 0.001). Loci were colored blue if they were also associated with left heart phenotypes with P < 5 × 10−8, and red otherwise. SNPs with P > 0.01 are not plotted. Manhattan plots by compartment are available in Supplementary Figure 7 (RA), Supplementary Figure 8 (RV), and Supplementary Figure 9 (pulmonary root and PA).
Up to 39,766 participants were included in the right heart GWAS (Supplementary Fig. 1), and we tested 11.6 million imputed SNPs with minor allele frequency (MAF) > 0.005. Additional GWAS quality control results are detailed in the Supplementary Note. Several loci were shared by multiple traits; counting each locus only once, we identified 130 independent loci associated with one or more right heart traits at a commonly used significance threshold of P < 5 × 10−8 (Supplementary Table 7). 71 loci were associated with at least two right heart traits, and one locus (near WNT9B/GOSR2/MYL4) was associated with 14 right heart phenotypes.
We conducted within-sample GWAS analyses of left ventricular and aortic traits, allowing us to identify that 58 of the 130 right heart loci were also associated at P < 5 × 10−8 with left heart phenotypes, while 72 were right heart-specific (Table 2). Of the 72 right heart-specific loci, 12 came to significance only after adjusting the right heart traits for their left heart counterparts (Supplementary Fig. 10). 48 of the 72 loci were associated with dimensionless right heart phenotypes (e.g., RVEF and the RVEDV/LVEDV ratio) or right heart phenotypes that accounted for BSA (Supplementary Table 8), while 24 loci were significant only prior to accounting for body size, no longer remaining significant after BSA-indexing (Supplementary Table 9). In gene set enrichment analyses38,39, the 48 loci that remained significant after accounting for body size were enriched for genes involved in cardiac proliferation, chamber development, and septum morphogenesis (Supplementary Table 10).
Table 2 |.
Loci specific to the right atrium and right ventricle
| Trait | CHR | BP | SNP | Effect allele | Other allele | EAF | BETA | SE | P-value | Nearest gene |
|---|---|---|---|---|---|---|---|---|---|---|
| RVEDV | 3 | 99779984 | rs57848867 | A | T | 0.526 | −0.034 | 0.0053 | 9.90E-11 | FILIP1L |
| RVEDV | 6 | 34205465 | rs202228093 | G | GGAGCCC | 0.106 | 0.05 | 0.0087 | 1.30E-08 | HMGA1 |
| RVEDV | 6 | 130349119 | rs6569648 | C | T | 0.238 | 0.034 | 0.0062 | 3.80E-08 | L3MBTL3 |
| RVEDV | 17 | 45128762 | rs1056064 | T | C | 0.833 | −0.047 | 0.0071 | 2.60E-11 | GOSR2 |
| RVEDV | 20 | 32987687 | rs62212171 | T | C | 0.859 | 0.05 | 0.0076 | 1.30E-10 | ITCH |
| RVESV | 1 | 228556788 | rs3738685 | C | T | 0.626 | −0.031 | 0.0056 | 1.80E-08 | OBSCN |
| RVESV | 2 | 26922062 | rs1314982 | G | A | 0.261 | 0.039 | 0.0062 | 2.80E-10 | KCNK3 |
| RVESV | 3 | 99779984 | rs57848867 | A | T | 0.526 | −0.031 | 0.0055 | 7.70E-09 | FILIP1L |
| RVESV | 5 | 35191701 | rs67209755 | T | C | 0.813 | 0.038 | 0.007 | 3.80E-08 | PRLR |
| RVESV | 8 | 145018354 | rs11786896 | C | T | 0.951 | 0.071 | 0.0126 | 1.10E-08 | PLEC |
| RVESV | 17 | 40023617 | rs781797066 | T | TA | 0.826 | −0.041 | 0.0073 | 3.10E-08 | ACLY |
| RVESV | 17 | 45013271 | rs17608766 | T | C | 0.858 | −0.046 | 0.0077 | 2.20E-09 | GOSR2 |
| RVEF | 8 | 145018354 | rs11786896 | C | T | 0.951 | −0.095 | 0.0159 | 3.60E-09 | PLEC |
| RVEF | 13 | 114075109 | rs76382172 | G | C | 0.964 | −0.101 | 0.0185 | 3.10E-08 | ADPRHL1 |
| RVEF | 14 | 81171138 | rs34540535 | T | C | 0.958 | 0.098 | 0.0175 | 2.60E-08 | CEP128 |
| RA Max | 5 | 172664163 | rs6882776 | G | A | 0.712 | −0.041 | 0.0071 | 4.20E-09 | NKX2–5 |
| RA Max | 6 | 22613847 | rs7757005 | G | A | 0.642 | −0.046 | 0.0067 | 1.70E-11 | HDGFL1 |
| RA Max | 12 | 115162091 | GTGTGCCCC | G | 0.623 | 0.04 | 0.0067 | 6.10E-09 | TBX3 | |
| RA Max | 17 | 45280802 | rs117154502 | T | G | 0.94 | −0.089 | 0.0134 | 3.10E-11 | MYL4 |
| RA Max | 17 | 61772449 | GA | G | 0.636 | −0.041 | 0.007 | 2.80E-09 | MAP3K3 | |
| RA Min | 3 | 156827227 | rs11928162 | C | T | 0.53 | −0.037 | 0.0065 | 1.40E-08 | CCNL1 |
| RA Min | 5 | 172662024 | rs2277923 | T | C | 0.703 | −0.053 | 0.0071 | 1.10E-13 | NKX2–5 |
| RA Min | 6 | 22613847 | rs7757005 | G | A | 0.642 | −0.045 | 0.0068 | 7.00E-11 | HDGFL1 |
| RA Min | 8 | 32413240 | rs112852637 | T | C | 0.529 | −0.038 | 0.0065 | 7.60E-09 | NRG1 |
| RA Min | 12 | 114835428 | rs1895602 | G | T | 0.545 | −0.037 | 0.0067 | 4.90E-08 | TBX5 |
| RA Min | 12 | 115162091 | GTGTGCCCC | G | 0.623 | 0.043 | 0.0068 | 4.30E-10 | TBX3 | |
| RA Min | 17 | 45280802 | rs117154502 | T | G | 0.94 | −0.091 | 0.0136 | 2.30E-11 | MYL4 |
| RA FAC | 5 | 172644017 | rs12652726 | C | T | 0.856 | 0.066 | 0.0105 | 2.10E-10 | NKX2–5 |
| RVEDV Indexed | 3 | 99779984 | rs57848867 | A | T | 0.525 | −0.046 | 0.0063 | 3.20E-13 | FILIP1L |
| RVEDV Indexed | 10 | 30332445 | rs4749523 | A | G | 0.634 | 0.037 | 0.0066 | 3.60E-08 | KIAA1462 |
| RVEDV Indexed | 11 | 57771538 | rs10526240 | T | A | 0.704 | 0.044 | 0.007 | 2.50E-10 | OR9Q1 |
| RVEDV Indexed | 17 | 45013271 | rs17608766 | T | C | 0.858 | −0.064 | 0.0089 | 6.30E-13 | GOSR2 |
| RVESV Indexed | 1 | 228556788 | rs3738685 | C | T | 0.626 | −0.038 | 0.0064 | 7.50E-10 | OBSCN |
| RVESV Indexed | 2 | 26922062 | rs1314982 | G | A | 0.26 | 0.046 | 0.0071 | 2.60E-11 | KCNK3 |
| RVESV Indexed | 3 | 99779984 | rs57848867 | A | T | 0.525 | −0.038 | 0.0062 | 1.50E-09 | FILIP1L |
| RVESV Indexed | 4 | 169847115 | TA | T | 0.2 | 0.044 | 0.0079 | 1.70E-08 | PALLD | |
| RVESV Indexed | 8 | 9287587 | rs28549922 | G | A | 0.861 | −0.05 | 0.0089 | 4.20E-09 | TNKS |
| RVESV Indexed | 8 | 145018354 | rs11786896 | C | T | 0.951 | 0.083 | 0.0144 | 7.00E-09 | PLEC |
| RVESV Indexed | 14 | 81171138 | rs34540535 | T | C | 0.958 | −0.095 | 0.0158 | 8.40E-10 | CEP128 |
| RVESV Indexed | 17 | 45013271 | rs17608766 | T | C | 0.858 | −0.056 | 0.0088 | 7.40E-11 | GOSR2 |
| RVSV Indexed | 3 | 99779984 | rs57848867 | A | T | 0.525 | −0.039 | 0.007 | 2.40E-08 | FILIP1L |
| RVSV Indexed | 11 | 57771538 | rs10526240 | T | A | 0.704 | 0.045 | 0.0077 | 8.10E-09 | OR9Q1 |
| RA Max Indexed | 3 | 71599571 | rs7640614 | C | G | 0.606 | −0.044 | 0.0075 | 7.40E-09 | FOXP1 |
| RA Max Indexed | 8 | 32413240 | rs112852637 | T | C | 0.529 | −0.042 | 0.0074 | 1.10E-08 | NRG1 |
| RA Max Indexed | 8 | 106379363 | rs201748964 | T | G | 0.691 | −0.043 | 0.0079 | 4.20E-08 | ZFPM2 |
| RA Max Indexed | 12 | 115162091 | GTGTGCCCC | G | 0.623 | 0.045 | 0.0077 | 5.20E-09 | TBX3 | |
| RA Max Indexed | 17 | 45280802 | rs117154502 | T | G | 0.94 | −0.092 | 0.0154 | 3.10E-09 | MYL4 |
| RA Min Indexed | 5 | 172664163 | rs6882776 | G | A | 0.712 | −0.052 | 0.0081 | 1.50E-10 | NKX2–5 |
| RA Min Indexed | 8 | 32413240 | rs112852637 | T | C | 0.529 | −0.045 | 0.0074 | 6.80E-10 | NRG1 |
| RA Min Indexed | 12 | 115164024 | rs11067264 | G | A | 0.623 | 0.05 | 0.0076 | 6.60E-11 | TBX3 |
| RA Min Indexed | 17 | 45280802 | rs117154502 | T | G | 0.94 | −0.094 | 0.0153 | 8.60E-10 | MYL4 |
| RVEDV/LVEDV Ratio | 2 | 42145432 | rs2374381 | T | C | 0.7 | 0.044 | 0.0077 | 6.20E-09 | C2orf91 |
| RVEDV/LVEDV Ratio | 6 | 126068914 | rs1935983 | C | T | 0.391 | −0.048 | 0.0072 | 3.80E-11 | HEY2 |
| RVEDV/LVEDV Ratio | 7 | 136636260 | rs112206296 | A | C | 0.985 | 0.178 | 0.0299 | 4.30E-09 | CHRM2 |
| RVEDV/LVEDV Ratio | 9 | 73049120 | rs61634638 | G | GT | 0.41 | −0.041 | 0.0072 | 1.30E-08 | KLF9 |
| RVEDV/LVEDV Ratio | 12 | 123639539 | rs67657805 | T | TA | 0.238 | −0.049 | 0.0087 | 1.50E-08 | MPHOSPH9 |
| RVESV/LVESV Ratio | 3 | 123105119 | rs62262391 | C | T | 0.777 | −0.053 | 0.0086 | 7.60E-10 | ADCY5 |
| RVESV/LVESV Ratio | 6 | 73906746 | rs10943078 | A | T | 0.757 | −0.047 | 0.0083 | 1.50E-08 | KCNQ5 |
| RVESV/LVESV Ratio | 6 | 126090377 | rs9388451 | T | C | 0.486 | −0.04 | 0.0072 | 4.10E-08 | HEY2 |
| RVESV/LVESV Ratio | 10 | 76089763 | TA | T | 0.263 | −0.046 | 0.0083 | 2.60E-08 | ADK | |
| RVESV/LVESV Ratio | 12 | 123493123 | rs12820906 | A | G | 0.755 | 0.05 | 0.0083 | 1.80E-09 | PITPNM2 |
| RVEF/LVEF Ratio | 3 | 123110581 | rs55968914 | C | G | 0.777 | 0.057 | 0.0088 | 1.40E-10 | ADCY5 |
Shown are clumped SNPs with BOLT-LMM P < 5 × 10−8 from the right atrial and right ventricular GWAS, excluding those that were also found in left ventricular or aortic GWAS within the same participants. PA and pulmonary root loci are too numerous to represent here; all right and left-heart loci with P < 5 × 10−8 can be found in Supplementary Table 7. For ratio phenotypes (e.g., “RVEDV/LVEDV,” which represents the RVEDV-to-LVEDV ratio), the SNPs listed here must additionally not be found within 500 kb of SNPs from a non-ratio phenotype. When multiple SNPs with P < 5 × 10−8 are found within 500 kb of one another and are in linkage equilibrium (r2 < 0.001), each independent SNP is displayed; an example is at the TBX5/TBX3 locus for the ‘RA Min’ phenotype. In the Trait column, suffix -S represents “systole,” -D represents “diastole,” and Idx represents “indexed to body surface area”. CHR, chromosome; BP, GRCh37 position; SNP, single nucleotide polymorphism (where a dbSNP ID was not available, this field was left empty because the SNP is uniquely identified by its position and alleles); EAF, effect allele frequency; BETA, effect size; SE, standard error of effect size; P-value, BOLT-LMM P value.
All lead SNPs associated at P < 5 × 10−8 with any left or right heart phenotype in this analysis, after clumping within each GWAS to remove SNPs in linkage disequilibrium (r2 > 0.001), are reported in Supplementary Table 7, where they are assigned a study-wide locus identifier to facilitate comparison between phenotypes. Those SNPs that are within 500 kb of one another are considered to be at the same locus and assigned the same study-wide identifier, and the strongest associated SNP at that locus is termed the lead SNP.
Gene-based analyses, including a transcriptome-wide association study (TWAS), exome sequencing-based rare variant analysis, and OpenTargets gene set enrichment analyses, are detailed in the Supplementary Note.
Right ventricular loci.
Among the right ventricular phenotypes, RVESV was linked with the greatest number of loci (20). Of these, seven loci were also associated with RVESV’s left heart counterpart (LVESV) at genome-wide significance. The effects of each SNP on right and left ventricular phenotypes are depicted in Figure 5.
Figure 5 |. Right heart loci.
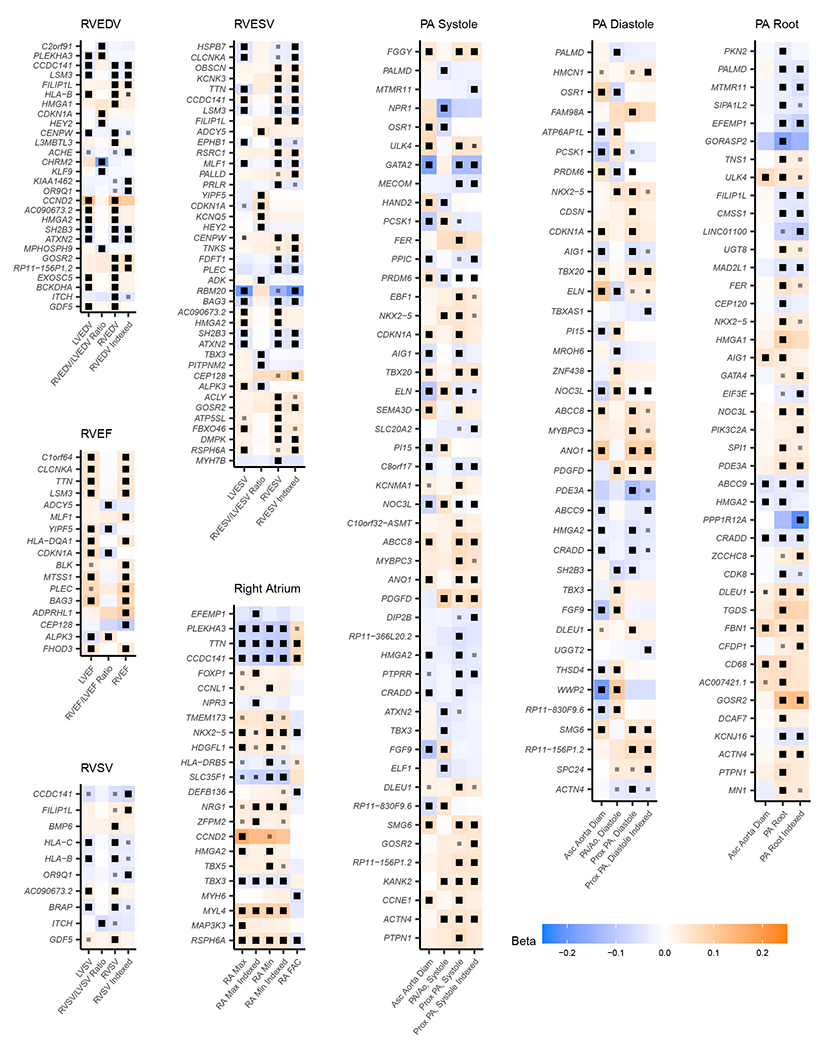
Right heart loci are shown grouped by trait. Where a paired left-heart phenotype is available, the effect size and P-value for the same SNP are shown next to its corresponding right heart trait. Each grid region represents the lead SNP (sorted in chromosomal order and tagged by its nearest gene, labeled on the y-axis) for each trait (x-axis). The effect magnitude (Beta) is represented with shades of orange (increase) and blue (decrease), and the effect direction is oriented with respect to the minor allele within the study population. Black boxes within a grid region indicate that the association between the SNP and the trait has BOLT-LMM P < 5 × 10−8; those with a smaller gray box indicate BOLT-LMM P < 5 × 10−6. Exact effect sizes and P values are provided in Supplementary Table 7 for traits with BOLT-LMM P < 5 × 10−8, and in the publicly available summary statistics where BOLT-LMM P ≥ 5 × 10−8. “PA/Ao” is the ratio of the PA diameter to the ascending aortic diameter. “Indexed” traits have been divided by body surface area. Genes may appear multiple times for the same trait when multiple variants at the same locus are in linkage equilibrium with one another (r2 < 0.001).
The strongest common variant association with RVESV was from a variant near BAG3; this same variant (rs72840788) was also the SNP with the strongest association with LVESV, with concordant effects. The rs72840788 variant is in near perfect linkage disequilibrium with rs2234962, which leads to the missense change p.Cys151Arg in the BAG3 protein (Supplementary Fig. 11).
Two SNPs at the TTN locus had association P < 5 × 10−8 with RVESV and were in linkage equilibrium (r2 = 0.001) with one another: rs955738 (P = 4.4 × 10−11) and rs2562845 (P = 4.2 × 10−8). Interestingly, while both SNPs were also associated with LVESV, the pattern of association strength was reversed when compared with the RVESV (rs2562845 was more strongly associated with LVESV than rs955738; Supplementary Fig. 12). It is possible that this distinction between primary association signals in the two ventricles is associated with differences in the regulation of TTN between the first (LV) and second (RV) heart fields, but establishing this will require additional investigation.
Among loci that were significantly associated with RVESV but not LVESV, some, like the GATA4/CTSB/FDFT1 locus on chromosome 8, had a cluster of sub-threshold SNPs for LVESV. At this locus, the RVESV lead SNP (rs34015932, P = 3.4 × 10−8) was only weakly correlated (r2 = 0.16) with the strongest LVESV-associated SNP near the locus (rs750190198, P = 1.1 × 10−6), also suggesting allelic heterogeneity (Supplementary Fig. 13). Other loci, such as that of OBSCN—encoding obscurin, a giant sarcomeric protein in the same family as titin—appeared to be right-ventricle specific, showing very little evidence of association with left ventricular phenotypes (Supplementary Fig. 14).
Finally, some loci achieved P < 5 × 10−8 only after adjustment of the right ventricular phenotype for its left ventricular counterpart (Supplementary Fig. 15). These include variants near ADCY5, which encodes the major isoform of adenylyl cyclase in the heart; pathogenic variation in this gene has previously been associated with heart failure40.
Pulmonary artery and pulmonary root loci.
Counting SNPs once for each associated trait, there were 172 trait-locus pairs associated with proximal PA diameter or pulmonary root diameter. 82 distinct genomic loci were associated with at least one PA or pulmonary root phenotype. Of these, 40 were exclusive to these tissues and were not associated with phenotypes from the other right or left heart compartments. Seven loci were shared by both the PA and pulmonary root, 16 were exclusive to PA diameter measurements, and 17 were exclusive to pulmonary root measurements. No loci were significantly associated with PA strain. Of 28 lead SNPs for PA diameter that were identified in the Framingham Heart Study (FHS), 25 had concordant effect direction (binomial test two-tailed P = 2.7 × 10−5; Supplementary Table 11 and Supplementary Fig. 16). This external replication is detailed in the Supplementary Note.
Several loci had putative connections to vascular tone. A locus associated with both pulmonary root and PA diameter, but via distinct SNPs (r2 between artery-associated rs79013608 and root-associated rs10770612 = 0.006) was found in an intergenic region whose nearest protein-coding gene is PDE3A. The protein product of this gene is inhibited by milrinone and cilostazol, which are in clinical use and have been shown in humans to reduce PA pressure41,42. A locus associated exclusively with PA diameter was tagged by a lead SNP intronic to KCNMA1, which encodes the channel-forming alpha subunit of the BKCa or the large conductance calcium- and voltage-activated potassium channels43. In a rat model, activation of endothelial BKCa channels in pulmonary endothelial cells was previously reported to cause pulmonary vasodilation44.
Some of the loci that were predominantly associated with the pulmonary root rather than the PA had previously been associated with aortic root or aortic valve phenotypes. For example, a pulmonary root-specific locus near CFDP1 from our study has previously been linked to aortic valve stenosis45. Another locus near GOSR2 has an association with the pulmonary root that is 35 orders of magnitude stronger than its association with the PA; it has previously been linked to aortic valve area46. The PALMD locus has been previously associated with the diameter of the aortic root and with aortic valve stenosis in humans47,48. The SNPs at the PALMD locus identified in our analysis were in tight linkage disequilibrium with those from Wild et al. (r2 = 0.96–1.0)49. In fact, of the 12 aortic root-associated loci in Wild et al. achieving P < 5 × 10−8 in their discovery analysis, seven were significantly associated with the pulmonary root in our present analysis, including loci near CDFP1, CEP120 (previously CCDC100), GOSR2, PALMD, HMGA2, PDE3A, and the KCNRG/DLEU1 locus.
Right atrial loci.
There were 42 trait-locus pairs associated with right atrial size and function. Accounting for multiple phenotypes having an association at the same locus, 20 genomic loci were associated with at least one right atrial phenotype. Of these, five were only identified in association with right atrial phenotypes and not other compartments: the lead SNPs at these loci were nearest to HDGLF1, CCNL1, NRG1, FOXP1, and ZFPM2. The latter three are notable for their established roles in cardiovascular development and disease.
The protein product of ZFPM2 interacts with GATA transcription factors, particularly GATA4, and plays a role in cardiac development50,51. Variants in ZFPM2 have previously been linked to congenital heart defects52–54. In mice, Foxp1 has been shown to play a role in cardiac morphogenesis, and in humans FOXP1 variants have been linked to congenital heart defects55–57. Finally, NRG1 encodes neuregulin-1, which participates in signaling through receptor tyrosine kinases and ErbB signaling in particular58–60. Clinical trials are ongoing to assess the effects of recombinant neuregulin on heart failure61. The rs112852637-T allele is associated with reduced right atrial area during atrial systole; reduced atrial area is inversely associated with arrhythmias and heart failure (Fig. 2 and Supplementary Table 3). Interestingly, this same allele is directionally associated with increased NRG1 expression in the right atrial appendage, although this expression signal is not statistically significant in GTEx v862.
As a sensitivity analysis, we also assessed right atrial volumes. The GWAS results from those analyses are reported in Supplementary Table 12.
Right heart polygenic score analyses and external validation.
We used PRScs-auto to compute ~1.1-million SNP polygenic scores for RVEDV, RVESV, and RVEF63, finding the RVEF score to be most strongly correlated with dilated cardiomyopathy (DCM; Supplementary Table 13). There were 603 DCM events and 359,296 non-events among UK Biobank participants unrelated to the MRI cohort; hazard ratio (HR) 1.33 per s.d. decrease; P = 7.1 × 10−13. Even after adjustment for a 1.1-million SNP polygenic score derived from the previously reported BSA-indexed left ventricular end systolic volume (LVESVi) GWAS, the RVEF score remained significantly associated with DCM (HR 1.21 per s.d. decrease; P = 1.2 × 10−5; Fig. 6). These findings were replicated, with attenuation, in the Mass General Brigham (MGB) Biobank and BioBank Japan (BBJ; Supplementary Note)64–66.
Figure 6 |. Cumulative incidence of dilated cardiomyopathy stratified by genetic prediction of RVEF.
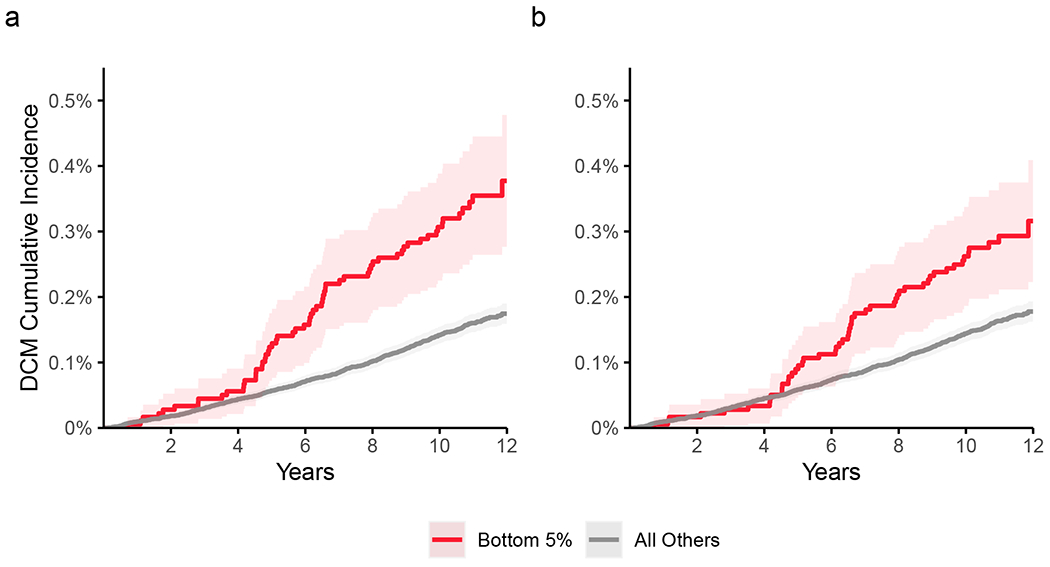
359,899 UK Biobank participants were unrelated within 3 degrees of the participants who underwent MRI. 603 participants were diagnosed with dilated cardiomyopathy (DCM) after enrollment. Those in the bottom 5% of genetically predicted RVEF are depicted in red, and the remaining 95% are depicted in gray. The darker shades of red and gray represent the central estimate of the cumulative incidence (defined as 1 minus the Kaplan-Meier survival estimate). The lighter shades of red and gray represent the respective 95% confidence intervals (based on the standard error). The x-axis depicts years since enrollment in the UK Biobank; the y-axis depicts cumulative incidence of dilated cardiomyopathy. a, Strata based on genetic prediction of RVEF. Those in the bottom 5% had an elevated risk of DCM (62 incident cases; Cox HR 2.2; P = 7.6 × 10−9). b, Strata based on genetic prediction of RVEF after residualization for genetic prediction of LVESVi. Those in the bottom 5% had an elevated risk of DCM (52 incident cases; Cox HR 1.8; P = 5.4 × 10−5). The Schoenfeld global P value for both models was 0.26, indicating no violation of the proportional hazards assumption.
We performed the same PRScs-auto procedure to generate 1.1-million SNP scores for the PA phenotypes. Of these, only the score for the proximal PA diameter in systole was associated with pulmonary hypertension in the UK Biobank (1,405 cases and 371,985 controls; HR 1.09 per s.d.; P = 2.2 × 10−3; Supplementary Table 13). This association remained significant in MGB, but not in BBJ (Supplementary Note). This polygenic score explained approximately 6.5% of the heritability of PA diameter based on an external analysis in FHS (Supplementary Note).
A PRScs-auto polygenic score for RA FAC was weakly inversely associated with the risk of atrial fibrillation or flutter (for 13,928 events and 353,311 non-events; HR 0.98 per s.d.; P = 1.9 × 10−2). The evidence was slightly stronger when considering only atrial flutter as the outcome of interest (841 atrial flutter events and 372,565 non-events; HR 0.91 per s.d.; P = 4.9 × 10−3).
Limitations.
This study is subject to limitations. The study population is largely of European ancestries, similar to the remainder of UK Biobank, limiting generalizability of the findings to other populations. In future work, genetic analyses of these phenotypes in people of globally diverse ancestries will be important. Future work will be required to understand whether polygenic scores are associated with disease progression. The transcriptional data used in the TWAS came from left-sided structures (aortic gene expression for the PA and left ventricular gene expression for the RV), which may not capture right-sided expression patterns. The disease gene enrichment analyses account for local genomic context and gene density, but not for other features such as chromatin interactions. We focused on the genes nearest to the strongest association signals; future work will be required to determine the causal factors driving each association. OpenTargets scores are algorithmically determined and can change between versions. Because we have used hospital-based International Classification of Diseases, Tenth Revision (ICD-10) and procedural codes to identify individuals with disease, our study lacks an ARVC-specific analysis (which does not have a unique ICD-10 code), and our disease definitions are susceptible to misclassification. We describe technical limitations related to MRI acquisition and deep learning in the Supplementary Note.
Discussion
We produced measurements of the right heart, including the RA, RV, and PA, analyzed their relationships with their left heart counterparts and with cardiovascular diseases, and identified 130 distinct genetic loci that were associated with these right heart measurements. We drew several conclusions from these findings.
First, right heart phenotypes, including structural and functional measurements of the RA, RV, and PA, are heritable. While they share strong epidemiological and genetic correlation with the corresponding left heart structures, our findings of partial genetic correlation and distinct genome-wide significant loci also imply distinct drivers of variation between right and left heart structures. 48 of the 72 right heart-specific loci remained significant after accounting for the corresponding left heart structure or overall body size via BSA-indexing (Supplementary Table 8) and were associated with dozens of gene sets involved in cardiac morphogenesis and cardiomyocyte proliferation (Supplementary Table 10). Twelve loci achieved significance in neither left nor right heart GWAS in isolation, but instead only after indexing the right heart phenotype for its left heart counterpart (Supplementary Fig. 10). Developing a better understanding of these distinct drivers of right and left heart structure may ultimately permit more targeted therapies for RV-predominant heart failure syndromes and primary cardiomyopathies such as ARVC.
Second, we found that the GWAS loci were enriched for genes associated with developmental diseases. In addition to the GATA4, ZFPM2, FOXP1, and NRG1 loci addressed above, several others were notable for connections to cardiovascular development. Right heart structures were associated with SNPs near NKX2–5, which plays a key role in maintaining the progenitor pool of cells of the secondary heart field resulting in outflow tract defects in people with NKX2–5 variants5,67; MYL4, which encodes atrial light chain 1, missense variants in which have been linked to familial atrial fibrillation68; and TBX3, which controls the formation of the sinus node and loss of which leads to outflow tract malformations and septal defects69–71. The TBX5/TBX3 locus also stands out because of the diversity of signals revealed in the data, with links to variation in the RA, the RV, and the PA (Supplementary Fig. 17). Two distinct signals drive the observed associations with right atrial size (rs1895602 near TBX5, and rs71447956/rs11067264 near TBX3). A third set of SNPs (rs4767282/rs10850409/rs35514224) is associated with the right-vs-left proportions of the ventricles and outflow tract. For example, at rs4767282, the C allele is associated with a slightly smaller RVESV and a slightly larger LVESV, achieving P < 5 × 10−8 only for the ratio of RVESV/LVESV. Given the proximity of these SNPs to TBX5, which plays a key role in atrial and ventricular septal placement2,72,73, and TBX3, which is required for outflow tract development and variants in which cause conotruncal defects74,75, it is tempting to speculate that these signals may influence the left-right localization of the site of septation during development. Indeed, the right heart-specific loci are enriched for genes that play roles in cardiac septum development (near NKX2–5, TBX3, TBX5, HEY2, and ZFPM2; Supplementary Table 10). We hypothesize that chamber-specific associations may be attributable to the different embryological origins of the right and left ventricles and their respective proximal conduction systems76, the distinct afterload regimes they face, or differences in physiological inputs during adult life. Future studies across the human lifespan will be helpful to answer this question.
Third, we observed links between cardiovascular disease and right ventricular measurements—as well as polygenic predictions of these measurements. Individuals with pre-existing diagnoses of pulmonary hypertension had enlarged right ventricular volumes throughout the cardiac cycle even after accounting for left ventricular volumes (Fig. 3). In UK Biobank participants, a polygenic predictor of RVEF was associated with incident dilated cardiomyopathy (Fig. 6). Notably, the RVEF polygenic score remained significantly associated with incident dilated cardiomyopathy even after accounting for a left ventricular polygenic score—implying a shared genetic basis for right ventricular dysfunction and dilated cardiomyopathy. These results were validated in external biobanks including MGB and BBJ, an external biobank of Japanese-ancestry participants. The role of right ventricular size and function as prognostic markers in individuals with dilated cardiomyopathy is well established77. Consistent with emerging clinical evidence, right ventricular structure and function are not merely of anthropometric interest, but instead represent endophenotypes for cardiomyopathy. Our findings suggest that earlier consideration of abnormalities of right ventricular function may afford the opportunity for earlier diagnosis of ensuing left ventricular dysfunction.
Fourth, we found epidemiological and genetic associations between proximal PA diameter and pulmonary hypertension. We produced a genome-wide polygenic prediction of PA diameter that was modestly associated with incident pulmonary hypertension in the UK Biobank; this finding replicated externally in MGB. The genetic prediction of PA diameter accounted for approximately 6.5% of the heritability of PA diameter in an external cohort (FHS). A previous version of this score produced from only clumped, genome-wide significant SNPs did not find a significant association with pulmonary hypertension; we suspect that this discrepancy may be because variation in PA diameter is most strongly driven by genetic variants affecting size during development, and more weakly by the pressure of the pulmonary circuit—whose contributions may be mostly sub-genome-wide significant at the current sample size. In future work, distinguishing the anatomical and developmental drivers of variation in cardiovascular structures from pathophysiological drivers may assist in the development of more clinically relevant polygenic scores78. As demonstrated by the lack of replication of the association between the PA score and pulmonary hypertension in BBJ, future ancestrally diverse discovery efforts will also be critical.
Finally, machine learning enables the derivation of complex traits in a manner that is scalable. This permits biobank-wide investigation of previously understudied human phenotypes, and promises to accelerate our understanding of cardiovascular disease.
Methods
Study design.
Except where otherwise stated, all analyses were conducted in the UK Biobank, which is a richly phenotyped, prospective, population-based cohort that recruited 500,000 individuals aged 40–69 in the UK via mailer from 2006–201025. We analyzed 487,283 participants with genetic data who had not withdrawn consent as of February 2020. Informed consent was obtained from all participants. Access to UK Biobank was provided under application #7089 and approved by the Mass General Brigham institutional review board (IRB; protocol 2019P003144). Mass General Brigham Biobank analyses were approved by the MGB IRB. Framingham Heart Study participants were ascertained and enrolled with written informed consent as described previously and approved by the institutional review boards of the Boston University Medical Center and the Massachusetts General Hospital79. BioBank Japan analyses were approved by the Institute of Medical Science, the University of Tokyo, as well as the cooperating hospitals65. Here we provide an overview of the methods used in this manuscript that are explained in more detail below.
We manually annotated pixels from magnetic resonance images from the UK Biobank: the pulmonary artery and the left and right ventricles were annotated in the short axis view, and the RA and RV were annotated in the four-chamber long axis view. We then trained two deep learning models (one for each of the views) with our manual annotations, and applied this model to the remaining images in the UK Biobank. For the RV, we integrated the data from the four chamber view and the short axis view to generate a surface mesh and derived the ventricular volumes from this mesh. We analyzed the relationships between each of these derived quantitative measurements of the right heart. We also analyzed their relationships with diseases and other phenotypes in the UK Biobank.
Then, we excluded people with prevalent heart failure, pulmonary hypertension, atrial fibrillation or coronary artery disease at time of enrollment and conducted genome-wide association studies of the right heart phenotypes. We performed transcriptome-wide association studies (TWAS) that incorporated publicly available gene expression data with our GWAS results to prioritize genes at most genomic loci. We analyzed the GWAS results in light of the four-chamber single nucleus sequencing data that is publicly available. We also performed a rare-variant association test in UK Biobank participants with both imaging and exome sequencing data. Polygenic scores produced from SNPs associated with right heart phenotypes in the UK Biobank GWAS were used to predict incident atrial fibrillation or flutter, dilated cardiomyopathy, and pulmonary hypertension in the UK Biobank participants whose data did not contribute to the GWAS. Replication of the polygenic analysis was pursued in external biobanks.
Statistical analyses were conducted with R version 3.6 (R Foundation for Statistical Computing, Vienna, Austria).
Semantic segmentation and deep learning model training.
Semantic segmentation is the process of assigning labels to pixels of an image. Here, we labeled pixels within specific anatomical structures (the right atrial blood pool, the right ventricular blood pool, and the PA blood pool), using a process similar to that described in our prior work evaluating the thoracic aorta33. Segmentation of cardiovascular structures was manually annotated in four-chamber and short axis images from the UK Biobank by a cardiologist (J.P.P.). To produce the model used in this manuscript, 714 short axis images were chosen, manually segmented, and used to train a deep learning model with PyTorch v1.6 and fastai v1.0.6126,27. The same was done separately with 445 four-chamber images.
An earlier developmental model was produced from 250 training samples for the short axis images, and the errors produced by that model informed the structures that we segmented in the 714 short axis training examples; see the Supplementary Note for additional detail. For both the short axis and the four-chamber long axis views, the models were based on the U-Net-derived architecture from fastai v1.0.61 constructed with a ResNet34 encoder, which was pre-trained on ImageNet28,30,80,81. The U-Net design incorporates skip connections between downsampling and upsampling layers, allowing more precise pixel labeling. During training, random perturbations of the input images, known as augmentations, were applied; these included affine rotation, zooming, and modification of the brightness and contrast. The Adam optimizer was used82. The models were trained with a cyclic learning rate training policy83. 80% of the samples were used to train the model, and 20% were used for validation.
For the short axis images, all images were resized initially to 104×104 pixels during the first half of training, and then to 224×224 pixels during the second half of training. The model was trained with a mini-batch size of 16 (with small images) or 8 (with large images). Maximum weight decay was 1 × 10−3. The maximum learning rate was 1 × 10−3, chosen based on the learning rate finder26,84. Because the RV and PA blood pools occupied very little of the overall short axis image area, a focal loss function was used (with alpha 0.7 and gamma 0.7), which can improve performance in the case of imbalanced labels85. When training with small images, 60% of iterations were permitted to have an increasing learning rate during each epoch, and training was performed over 30 epochs while keeping the weights for all but the final layer frozen. Then, all layers were unfrozen, the learning rate was decreased to 1 × 10−7, and the model was trained for an additional 10 epochs. When training with large images, 30% of iterations were permitted to have an increasing learning rate, and training was done for 30 epochs while keeping all but the final layer frozen. Finally, all layers were unfrozen, the learning rate was decreased to 1 × 10−7, and the model was trained for an additional 10 epochs.
For the four-chamber long axis images, all images were resized initially to 76×104 pixels during the first half of training, and then to 150×208 pixels during the second half of training. The model was trained with a mini-batch size of 4 (with small images) or 2 (with large images). Maximum weight decay was 1 × 10−2. Cross entropy loss was used86. 30% of iterations were permitted to have an increasing learning rate during each epoch. When training with small images, the maximum learning rate was initially 1 × 10−3, and training was performed over 50 epochs while keeping all weights frozen except for the final layer. Then, all layers were unfrozen, the learning rate was decreased to 3 × 10−5, and the model was trained for an additional 15 epochs. When training with large images, the maximum learning rate was set to 3 × 10−4, and the model was trained for 50 epochs while keeping all but the final layer frozen. Finally, all layers were unfrozen, the learning rate was decreased to 1 × 10−7, and the model was retrained for an additional 15 epochs.
Held-out test sets that were not used for training or validation were used to assess the final quality of both models (detailed in the Supplementary Note). The final short axis and four-chamber long axis models were then applied, respectively, to all available short axis images and four-chamber long axis images available in the UK Biobank as of November 2020. The techniques used to post-process the deep learning output to measure right atrial area and PA diameter, and to perform Poisson surface reconstruction to compute right ventricular volume, are detailed in the Supplementary Note.
Genotyping, imputation, and genetic quality control.
UK Biobank samples were genotyped on either the UK BiLEVE or UK Biobank Axiom arrays and imputed into the Haplotype Reference Consortium panel and the UK10K+1000 Genomes panel87. Variant positions were keyed to the GRCh37 human genome reference. Genotyped variants with genotyping call rate < 0.95 and imputed variants with INFO score < 0.3 or minor allele frequency ≤ 0.005 in the analyzed samples were excluded. After variant-level quality control, 11,631,796 imputed variants remained for analysis.
Participants without imputed genetic data, or with a genotyping call rate < 0.98, mismatch between self-reported sex and sex chromosome count, sex chromosome aneuploidy, excessive third-degree relatives, or outliers for heterozygosity were excluded from genetic analysis87. Participants were also excluded from genetic analysis if they had a history of pulmonary hypertension, atrial fibrillation, heart failure, or coronary artery disease documented by ICD code or procedural code from the inpatient setting prior to the time they underwent cardiovascular magnetic resonance imaging at a UK Biobank assessment center. Our definitions of these diseases in the UK Biobank are provided in Supplementary Table 4.
Heritability and genome-wide association analyses.
For the RA, we assessed maximum area, minimum area, and fractional area change. For the RV, we assessed end diastolic volume, end systolic volume, stroke volume, and ejection fraction. For the pulmonary system, we assessed the diameter of the proximal PA diameter in systole and in diastole, strain, and the pulmonary root diameter. In addition, we analyzed body surface area-indexed values for all areas and volumes (i.e., excluding strain, RA FAC, and RVEF, which are dimensionless). Where paired left heart phenotypes were available, we also analyzed those left heart phenotypes alone, as well as the ratio of the right heart phenotype to its corresponding left heart phenotype. The ascending aortic diameter was paired with the PA diameter; the left ventricular end diastolic volume, end systolic volume, stroke volume, and ejection fraction were paired with their corresponding right ventricular counterparts.
BOLT-REML v2.3.4 was used to assess the SNP-heritability of the phenotypes, as well as their genetic correlation with one another using the directly genotyped variants in the UK Biobank36.
Before conducting genome-wide association studies, a rank-based inverse normal transformation was applied to the quantitative right heart traits88. Therefore, effect estimates are reported in dimensionless units that represent approximately 1 standard deviation of the underlying trait. All traits were adjusted for age at enrollment, age and age2 at the time of MRI, the first 10 principal components of ancestry, sex, the genotyping array, and the MRI scanner’s unique identifier.
Genome-wide association studies for each phenotype were conducted using BOLT-LMM version 2.3.4 to account for cryptic population structure and sample relatedness36,37. We used the full autosomal panel of 714,558 directly genotyped SNPs that passed quality control to construct the genetic relationship matrix (GRM), with covariate adjustment as noted above. Associations on the X chromosome were also analyzed, using all autosomal SNPs and X chromosomal SNPs to construct the GRM (n = 732,193 SNPs), with the same covariate adjustments and significance threshold as in the autosomal analysis. In this analysis mode, BOLT treats individuals with one X chromosome as having an allelic dosage of 0/2 and those with two X chromosomes as having an allelic dosage of 0/1/2. Variants with association P < 5 × 10−8, a commonly used threshold, were considered to be genome-wide significant.
We used the following procedure to identify distinct GWAS loci and lead SNPs for each trait. Linkage disequilibrium (LD) clumping was performed with PLINK-1.989 using the same participants used for the GWAS, rather than a generic reference panel. We outlined a 5-Mb window (--clump-kb 5000) and used a stringent LD threshold (--r2 0.001) in order to account for long LD blocks such as those near the Williams-Beuren locus on chromosome 7 and the Noonan syndrome locus on chromosome 1290–92. With the independently significant clumped SNPs, distinct genomic loci were then defined by starting with the SNP with the strongest P value, excluding other SNPs within 500 kb, and iterating until no SNPs remained. The independently significant SNP with the strongest association P value at each genomic locus are termed lead SNPs.
Lead SNPs were tested for deviation from Hardy-Weinberg equilibrium (HWE) at a threshold of P < 1 × 10−6 using the exact test89,93. To assess whether the HWE violations affected the association signals, SNPs with HWE P < 1 × 10−6 were re-analyzed with R’s glm after excluding samples that were not within the UK Biobank’s centrally-adjudicated subset of individuals who self-reported British ancestry and were found to be genetic inliers for the European-ancestry cluster, using the same covariates as were used in the BOLT-LMM model.
Linkage disequilibrium (LD) score regression analysis was performed using ldsc version 1.0.094. With ldsc, the genomic control factor (lambda GC) was partitioned into components reflecting polygenicity and inflation, using the software’s defaults.
Locus plots were produced with LocusZoom95.
Gene set enrichment analysis.
Gene set enrichment analysis was conducted with the online GSEA platform39 including all nine major Molecular Signatures Database (MSigDB) collections38. The nearest gene to each locus from Supplementary Table 8 was input into the online platform at https://www.gsea-msigdb.org/gsea/msigdb/annotate.jsp and the top 100 results were returned. The same procedure was repeated for the nearest genes from Supplementary Table 9.
Stratified LD score regression.
To identify putative cell types most relevant for each GWAS trait, we performed stratified linkage disequilibrium (LD) score regression analysis using single nucleus RNA-sequencing data from Tucker et al.94,96,97. Cell type specific markers within the RA and RV were calculated separately for the nine main cell types using a limma-voom differential expression model on aggregated counts per individual98. Only individuals with greater than 25 nuclei of a given cell type were considered. Genes were sorted by t statistic per cell type and the top 90% of genes were used to generate LD Score Regression annotations96. SNPs within 100 kb of any gene from a specific cell type were annotated for the respective cell type using 1000 Genomes European individuals99. We then performed stratified LD score regression with these annotations in combination with the baseline model described in Finucane et al.100, only including high quality HapMap3 SNPs. We used the RA cell type specific annotations and RV cell type specific annotations for the RA and RV specific GWAS traits, respectively.
Replication of PA diameter GWAS results in the Framingham Heart Study (FHS).
For external replication of the UK Biobank GWAS results, we analyzed SNP associations with PA diameter in FHS, measured on computed tomography (CT) images. The genetic profiles of FHS participants were measured by the Affymetrix GeneChip 500k Array Set & 50K Human Gene Focused Panel, and genotyping was called using BRLMM as previously described101,102. Variants with call rate < 0.97, HWE P < 10−6, n > 100 Mendelian errors, or MAF < 0.01 were excluded. The remaining variants were then imputed to the TOPMed imputation panel using Michigan Imputation Server (https://imputationserver.sph.umich.edu/index.html)103.
A multi-detector computed tomography (CT) scanner (General Electric Lightspeed + 8 detector scanner) was used to assess the PA in FHS participants79,104. The PA measurements and genotyping data were available from dbGaP (#phs000007.v32.p13 and #phs000342.v20.p13, respectively). The association between each genetic variant and CT traits was tested with linear mixed effects models using the kinship package in R, and adjusted for sex, age, age2, cohort (original cohort, offspring cohort, or third generation cohort), and first five principal components of ancestry.
SNPs from the UK Biobank PA diameter in systole GWAS were clumped based on in-sample LD within the UK Biobank using a linkage disequilibrium r2 cutoff of 0.001 to identify independent signals in PLINK-1.989. A lookup of SNP effect sizes in FHS was then conducted. We applied a variety of UK Biobank GWAS P-value cutoffs (from 5 × 10−3 to 5 × 10−10) and then used SNPs below those cutoffs in linear models, recording the correlation between the FHS and UK Biobank SNP effect sizes and plotting the results.
For the SNPs associated in UK Biobank at P < 5 × 10−8, a two-tailed binomial test was performed to compare the number of directionally concordant SNP effects with that expected by chance (an expectation of 0.5 probability of concordance at each SNP).
Polygenic score analysis.
For RVEDV, RVESV, and RVEF, we computed polygenic scores using the software program PRScs (version sha1@43128be) with a UK Biobank European ancestry linkage disequilibrium panel made publicly available by the software authors63. The PRScs method applies a continuous shrinkage prior to the SNP weights. PRScs was run in ‘auto’ mode on a per-chromosome basis. This mode places a standard half-Cauchy prior on the global shrinkage parameter and learns the global scaling parameter from the data; as a consequence, PRScs-auto does not require a validation data set for tuning. Based on the software default settings, only the 1,117,425 SNPs found at HapMap3 sites that were also present in the UK Biobank were permitted to contribute to the score. These scores were applied to the entire UK Biobank.
The three RV scores were tested for association with dilated cardiomyopathy using Cox proportional hazards models as implemented by the R survival package105. Participants related within 3 degrees of kinship to those who had undergone MRI, based on the precomputed relatedness matrix from the UK Biobank, were excluded from analysis87. We conducted these analyses in individuals who were “genetic inliers” for European ancestry based on the first three pairs of genetic principal components (PC1&2, PC3&4, PC5&6) by using the aberrant package as described previously87,106. We also excluded individuals with disease that was diagnosed prior to enrollment in the UK Biobank. We counted survival as the number of years between enrollment and disease diagnosis (for those who developed disease) or death, loss to follow-up, or end of follow-up time (for those who did not develop disease). We adjusted for covariates including sex, the cubic basis spline of age at enrollment, the interaction between the cubic basis spline of age at enrollment and sex, the genotyping array, the first five principal components of ancestry, and the cubic basis splines of height (cm), weight (kg), BMI (kg/m2), diastolic blood pressure, and systolic blood pressure.
The single strongest right ventricular score was also analyzed jointly in a model that additionally accounted for a polygenic score produced using the same PRScs-auto method for the left ventricular end systolic volume indexed to body surface area (LVESVi), chosen because this phenotype was the trait that produced the strongest left ventricular polygenic score for predicting dilated cardiomyopathy107.
The same procedure, including the application of PRScs-auto, was repeated to produce polygenic scores for the PA phenotypes, which were tested for association with pulmonary hypertension. It was also repeated to produce polygenic scores for right atrial phenotypes, which were tested for association with atrial fibrillation and flutter.
Cumulative incidence curves were plotted in order to demonstrate the relationship between the RVEF polygenic score and dilated cardiomyopathy, using the survminer 3.1-8 package. The population was split into the top 5% of the score and the remaining 95%. Another plot was produced after residualizing the RVEF polygenic score for the LVESVi polygenic score and then splitting into the top 5% and the remaining 95%. To identify violations of the proportional hazards assumptions, Schoenfeld residuals were computed for both of these models.
Supplementary Material
Acknowledgements
We thank all participants of UK Biobank, MGB, BBJ, and FHS. We acknowledge the staff of BBJ for their assistance. Cardiac magnetic resonance images in Figure 1 are reproduced by kind permission of UK Biobank ©. We acknowledge Servier Medical Art (smart.servier.com) for the right heart illustration in Figure 1, which is licensed under a Creative Commons Attribution 3.0 Unported License (CC-BY-3.0). We also acknowledge Mary O’Reilly, from Pattern at the Broad Institute, for modifying the right heart illustration and for creating the remaining graphical illustrations in Figure 1.
This work was supported by grants from the National Institutes of Health K08HL159346 (J.P.P.), R01HL092577 (P.T.E.), K24HL105780 (P.T.E.), R01HL134893 (J.E.H.), R01HL140224 (J.E.H.), K24HL153669 (J.E.H.), 5T32HL007604-35 (V.N.), T32HL007208 (S. Khurshid), R01HL128914 (E.J.B.), R01HL092577 (E.J.B.), R01HL141434 (E.J.B.), U54HL120163 (E.J.B.), and R01HL139731 (S.A.L.). This work was supported by the Fondation Leducq 14CVD01 (P.T.E.). This work was supported by a John S LaDue Memorial Fellowship (J.P.P.) and the Sarnoff Cardiovascular Research Foundation Scholar Award (J.P.P.). This work was supported by the Tailor-Made Medical Treatment Program of the Ministry of Education, Culture, Sports, Science, and Technology (BBJ). This work was supported by the Japan Agency for Medical Research via JP17km0305002 (BBJ), JP17km0305001 (BBJ), JP20km0405209 (BBJ, S. Koyama, K.I., I.K.) and JP20ek0109487 (BBJ, S. Koyama, K.I., I.K.). This work was supported by student scholarships from the Dutch Heart Foundation (S.J.) and the Amsterdams Universiteitsfonds (S.J.). This work was supported by American Heart Association grants 18SFRN34110082 (E.J.B.), 18SFRN34250007 (S.A.L.), and a Strategically Focused Research Networks grant (P.T.E.). This work was supported by the Fredman Fellowship for Aortic Disease (M.E.L.) and the Toomey Fund for Aortic Dissection Research (M.E.L.). This work was funded by a collaboration between the Broad Institute and IBM Research.
Competing Interests
J.P.P. has served as a consultant for Maze Therapeutics. P.B. is supported by grants from Bayer AG and IBM applying machine learning in cardiovascular disease. S.A.L. receives sponsored research support from Bristol Myers Squibb / Pfizer, Bayer AG, Boehringer Ingelheim, and Fitbit, and has consulted for Bristol Myers Squibb / Pfizer and Bayer AG, and participates in a research collaboration with IBM. K.N. is employed by IBM Research. J.E.H. is supported by a grant from Bayer AG focused on machine-learning and cardiovascular disease and a research grant from Gilead Sciences. J.E.H. has received research supplies from EcoNugenics. A.A.P. is employed as a Venture Partner at GV; he is also supported by a grant from Bayer AG to the Broad Institute focused on machine learning for clinical trial design. P.T.E. received sponsored research support from Bayer AG and IBM Research. P.T.E. has also served on advisory boards or consulted for Bayer AG, Quest Diagnostics, MyoKardia and Novartis. The Broad Institute has filed for a patent on an invention from P.T.E., M.E.L., and J.P.P. related to a genetic risk predictor for aortic disease. All remaining authors report no competing interests.
BioBank Japan Consortium
Koichi Matsuda23,24, Yuji Yamanashi25, Yoichi Furukawa26, Takayuki Morisaki27, Yoshinori Murakami28, Yoichiro Kamatani24,29, Kaori Mutu30, Akiko Nagai30, Wataru Obara31, Ken Yamaji32, Kazuhisa Takahashi33, Satoshi Asai34,35, Yasuo Takahashi36, Takao Suzuki37, Nobuaki Sinozaki37, Hiroki Yamaguchi38, Shiro Minami39, Shigeo Murayama40, Kozo Yoshimori41,42, Satoshi Nagayama43, Daisuke Obata44, Masahiko Higashiyama45, Akihide Masumoto46, Yukihiro Koretsune47
23 Laboratory of Genome Technology, Human Genome Center, Institute of Medical Science, The University of Tokyo, Tokyo, Japan
24 Laboratory of Clinical Genome Sequencing, Graduate School of Frontier Sciences, The University of Tokyo, Tokyo, Japan
25 Division of Genetics, The Institute of Medical Science, The University of Tokyo, Tokyo, Japan
26 Division of Clinical Genome Research, Institute of Medical Science, The University of Tokyo, Tokyo, Japan
27 Division of Molecular Pathology IMSUT Hospital, Department of Internal Medicine Project Division of Genomic Medicine and Disease Prevention The Institute of Medical Science, The University of Tokyo, Tokyo, Japan
28 Department of Cancer Biology, Institute of Medical Science, The University of Tokyo, Tokyo, Japan
29 Laboratory of Complex Trait Genomics, Graduate School of Frontier Sciences, The University of Tokyo, Tokyo, Japan
30 Department of Public Policy, Institute of Medical Science, The University of Tokyo, Tokyo, Japan
31 Department of Urology, Iwate Medical University, Iwate, Japan
32 Department of Internal Medicine and Rheumatology, Juntendo University Graduate School of Medicine, Tokyo, Japan
33 Department of Respiratory Medicine, Juntendo University Graduate School of Medicine, Tokyo, Japan
34 Division of Pharmacology, Department of Biomedical Science, Nihon University School of Medicine, Tokyo, Japan
35 Division of Genomic Epidemiology and Clinical Trials, Clinical Trials Research Center, Nihon University School of Medicine, Tokyo, Japan
36 Division of Genomic Epidemiology and Clinical Trials, Clinical Trials Research Center, Nihon University School of Medicine, Tokyo, Japan
37 Tokushukai Group, Tokyo, Japan
38 Department of Hematology, Nippon Medical School, Tokyo, Japan
39 Department of Bioregulation, Nippon Medical School, Kawasaki, Japan
40 Tokyo Metropolitan Geriatric Hospital and Institute of Gerontology, Tokyo, Japan
41 Fukujuji Hospital, Tokyo, Japan
42 Japan Anti-Tuberculosis Association, Tokyo, Japan
43 The Cancer Institute Hospital of the Japanese Foundation for Cancer Research, Tokyo, Japan
44 Center for Clinical Research and Advanced Medicine, Shiga University of Medical Science, Shiga, Japan
45 Department of General Thoracic Surgery, Osaka International Cancer Institute, Osaka, Japan
46 Iizuka Hospital, Fukuoka, Japan
47 National Hospital Organization Osaka National Hospital, Osaka, Japan
Footnotes
Code availability
The code used to perform Poisson surface reconstruction from segmentation output is located at https://github.com/broadinstitute/ml4h and is available under an open-source BSD license. The code used to perform permutation testing to assess enrichment of disease-related genes near GWAS loci is located at https://github.com/carbocation/genomisc and is available under an open-source BSD license. The code used to annotate magnetic resonance images is located at https://github.com/carbocation/traceoverlay and is available under an open-source BSD license.
Data availability
UK Biobank data are made available to researchers from research institutions with genuine research inquiries, following IRB and UK Biobank approval. GWAS summary statistics are available at the Broad Institute Cardiovascular Disease Knowledge Portal (http://www.broadcvdi.org). Single nucleus RNA sequencing data are publicly available at the Single Cell Portal (https://singlecell.broadinstitute.org/single_cell accession #SCP498). The dbGAP study accession numbers used for FHS replication were #phs000007.v32.p13 for PA diameter measurement and #phs000342.v20.p13 for genotyping. BioBank Japan data are available to bona fide researchers for approved research by application to the Japanese Genotype-phenotype Archive. Mass General Brigham data are available to Mass General Brigham investigators. All other data are contained within the article and its supplementary information, or are available upon reasonable request to the corresponding author.
References
- 1.Olson EN Gene regulatory networks in the evolution and development of the heart. Science 313, 1922–1927 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koshiba-Takeuchi K et al. Reptilian heart development and the molecular basis of cardiac chamber evolution. Nature 461, 95–98 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farmer CG Evolution of the vertebrate cardio-pulmonary system. Annual Review of Physiology 61, 573–592 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Galli D et al. Atrial myocardium derives from the posterior region of the second heart field, which acquires left-right identity as Pitx2c is expressed. Development 135, 1157–1167 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Meilhac SM & Buckingham ME The deployment of cell lineages that form the mammalian heart. Nature Reviews Cardiology 15, 705–724 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Verzi MP, McCulley DJ, De Val S, Dodou E & Black BL The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev Biol 287, 134–145 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Zaffran S, Kelly RG, Meilhac SM, Buckingham ME & Brown NA Right ventricular myocardium derives from the anterior heart field. Circ Res 95, 261–268 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Jiang X, Rowitch DH, Soriano P, McMahon AP & Sucov HM Fate of the mammalian cardiac neural crest. Development 127, 1607–1616 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Li J, Chen F & Epstein JA Neural crest expression of Cre recombinase directed by the proximal Pax3 promoter in transgenic mice. genesis 26, 162–164 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Lin C-J, Lin C-Y, Chen C-H, Zhou B & Chang C-P Partitioning the heart: mechanisms of cardiac septation and valve development. Development 139, 3277–3299 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gotschy A et al. Right ventricular outflow tract dimensions in arrhythmogenic right ventricular cardiomyopathy/dysplasia-a multicentre study comparing echocardiography and cardiovascular magnetic resonance. Eur Heart J Cardiovasc Imaging 19, 516–523 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Marcus FI et al. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia. Circulation 121, 1533–1541 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKoy G et al. Identification of a deletion in plakoglobin in arrhythmogenic right ventricular cardiomyopathy with palmoplantar keratoderma and woolly hair (Naxos disease). The Lancet 355, 2119–2124 (2000). [DOI] [PubMed] [Google Scholar]
- 14.McNally E, MacLeod H & Dellefave-Castillo L Arrhythmogenic Right Ventricular Cardiomyopathy. in GeneReviews® (eds. Adam MP et al.) (University of Washington, Seattle, 1993). [Google Scholar]
- 15.Protonotarios N & Tsatsopoulou A Naxos disease: cardiocutaneous syndrome due to cell adhesion defect. Orphanet Journal of Rare Diseases 1, 4 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romero J, Mejia-Lopez E, Manrique C & Lucariello R Arrhythmogenic right ventricular cardiomyopathy (ARVC/D): a systematic literature review. Clin Med Insights Cardiol 7, CMC.S10940 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behr ER, Ben-Haim Y, Ackerman MJ, Krahn AD & Wilde AAM Brugada syndrome and reduced right ventricular outflow tract conduction reserve: a final common pathway? Eur Heart J 42, 1073–1081 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Ghio S et al. Independent and additive prognostic value of right ventricular systolic function and pulmonary artery pressure in patients with chronic heart failure. J Am Coll Cardiol 37, 183–188 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Kjaergaard J et al. Right ventricular dysfunction as an independent predictor of short- and long-term mortality in patients with heart failure. Eur J Heart Fail 9, 610–616 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Melenovsky V, Hwang S-J, Lin G, Redfield MM & Borlaug BA Right heart dysfunction in heart failure with preserved ejection fraction. Eur Heart J 35, 3452–3462 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai W et al. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. Journal of cardiovascular magnetic resonance : official journal of the Society for Cardiovascular Magnetic Resonance 20, 65 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bai W et al. A population-based phenome-wide association study of cardiac and aortic structure and function. Nature Medicine 26, 1654–1662 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petersen SE et al. Imaging in population science: cardiovascular magnetic resonance in 100,000 participants of UK Biobank - rationale, challenges and approaches. Journal of Cardiovascular Magnetic Resonance 15, 46 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petersen SE et al. UK Biobank’s cardiovascular magnetic resonance protocol. J Cardiovasc Magn Reson 18, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sudlow C et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Medicine 12, e1001779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howard J & Gugger S Fastai: a Layered API for deep learning. Information 11, 108 (2020). [Google Scholar]
- 27.Paszke A et al. PyTorch: an imperative style, high-performance deep learning library. arXiv:1912.01703 [cs, stat] (2019). [Google Scholar]
- 28.Deng J et al. ImageNet: a large-scale hierarchical image database. in 2009 IEEE Conference on Computer Vision and Pattern Recognition 248–255 (2009). doi: 10.1109/CVPR.2009.5206848. [DOI] [Google Scholar]
- 29.Krizhevsky A, Sutskever I & Hinton GE ImageNet classification with deep convolutional neural networks. Commun. ACM 60, 84–90 (2017). [Google Scholar]
- 30.Ronneberger O, Fischer P & Brox T U-Net: convolutional networks for biomedical image segmentation. arXiv:1505.04597 [cs] (2015). [Google Scholar]
- 31.Dice LR Measures of the amount of ecologic association between species. Ecology 26, 297–302 (1945). [Google Scholar]
- 32.Sørensen TJ A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commons. (I kommission hos E. Munksgaard, 1948). [Google Scholar]
- 33.Pirruccello JP et al. Deep learning enables genetic analysis of the human thoracic aorta. Nat Genet 54, 40–51 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edwards PD, Bull RK & Coulden R CT measurement of main pulmonary artery diameter. BJR 71, 1018–1020 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Sanfilippo AJ et al. Atrial enlargement as a consequence of atrial fibrillation. A prospective echocardiographic study. Circulation 82, 792–797 (1990). [DOI] [PubMed] [Google Scholar]
- 36.Loh P-R et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nature Genetics 47, 284–290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loh P-R, Kichaev G, Gazal S, Schoech AP & Price AL Mixed-model association for biobank-scale datasets. Nature Genetics 50, 906–908 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liberzon A et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y-Z Autosomal dominant familial dyskinesia and facial myokymia: single exome sequencing identifies a mutation in adenylyl cyclase 5. Arch Neurol 69, 630 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Givertz MM, Hare JM, Loh E, Gauthier DF & Colucci WS Effect of bolus milrinone on hemodynamic variables and pulmonary vascular resistance in patients with severe left ventricular dysfunction: a rapid test for reversibility of pulmonary hypertension. J Am Coll Cardiol 28, 1775–1780 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Sahin M et al. The effect of cilostazol on right heart function and pulmonary pressure. Cardiovasc Ther 31, e88–93 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Singh H et al. mitoBKCa is encoded by the Kcnma1 gene, and a splicing sequence defines its mitochondrial location. Proc Natl Acad Sci U S A 110, 10836–10841 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vang A, Mazer J, Casserly B & Choudhary G Activation of endothelial BKCa channels causes pulmonary vasodilation. Vascular Pharmacology 53, 122–129 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Helgadottir A et al. Genome-wide analysis yields new loci associating with aortic valve stenosis. Nat Commun 9, 987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Córdova-Palomera A et al. Cardiac imaging of aortic valve area From 34 287 UK Biobank participants reveals novel genetic associations and shared genetic comorbidity with multiple disease phenotypes. Circulation: Genomic and Precision Medicine 13, e003014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thériault S et al. A transcriptome-wide association study identifies PALMD as a susceptibility gene for calcific aortic valve stenosis. Nat Commun 9, 988 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wild PS et al. Large-scale genome-wide analysis identifies genetic variants associated with cardiac structure and function. J. Clin. Invest 127, 1798–1812 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Machiela MJ & Chanock SJ LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–3557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu J et al. FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Molecular and Cellular Biology 19, 4495–4502 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Svensson EC, Tufts RL, Polk CE & Leiden JM Molecular cloning of FOG-2: A modulator of transcription factor GATA-4 in cardiomyocytes. Proc Natl Acad Sci U S A 96, 956–961 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.D’Alessandro LCA et al. Exome sequencing identifies rare variants in multiple genes in atrioventricular septal defect. Genet Med 18, 189–198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pu T et al. Identification of ZFPM2 mutations in sporadic conotruncal heart defect patients. Mol Genet Genomics 293, 217–223 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Qian Y et al. Multiple gene variations contributed to congenital heart disease via GATA family transcriptional regulation. J Transl Med 15, 69 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang S-W et al. Genetic abnormalities in FOXP1 are associated with congenital heart defects. Hum Mutat 34, 1226–1230 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lozano R et al. FOXP1 syndrome: a review of the literature and practice parameters for medical assessment and monitoring. Journal of Neurodevelopmental Disorders 13, 18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang B et al. Foxp1 regulates cardiac outflow tract, endocardial cushion morphogenesis and myocyte proliferation and maturation. Development 131, 4477–4487 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Meyer D & Birchmeier C Multiple essential functions of neuregulin in development. Nature 378, 386–390 (1995). [DOI] [PubMed] [Google Scholar]
- 59.Rentschler S et al. Neuregulin-1 promotes formation of the murine cardiac conduction system. Proc Natl Acad Sci U S A 99, 10464–10469 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rupert CE & Coulombe KL The roles of Neuregulin-1 in cardiac development, homeostasis, and disease. Biomark Insights 10, 1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zensun Sci. & Tech. Co., Ltd. A Multi-center, Randomized, Double-blind, Placebo-controlled Phase III Clinical Trial to Evaluate the Effect of Injectable Neucardin on the Cardiac Function of Subjects With Chronic Systolic Heart Failure on Standard Heart Failure Therapy. https://clinicaltrials.gov/ct2/show/NCT04468529 (2021).
- 62.Lonsdale J et al. The Genotype-Tissue Expression (GTEx) project. Nature Genetics 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ge T, Chen C-Y, Ni Y, Feng Y-CA & Smoller JW Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nature Communications 10, 1776 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karlson EW, Boutin NT, Hoffnagle AG & Allen NL Building the Partners HealthCare Biobank at Partners Personalized Medicine: informed consent, return of research results, recruitment lessons and operational considerations. J Pers Med 6, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nagai A et al. Overview of the BioBank Japan Project: study design and profile. J Epidemiol 27, S2–S8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sakaue S et al. Dimensionality reduction reveals fine-scale structure in the Japanese population with consequences for polygenic risk prediction. Nat Commun 11, 1569 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McElhinney DB, Geiger E, Blinder J, Benson DW & Goldmuntz E NKX2.5 mutations in patients with congenital heart disease. J Am Coll Cardiol 42, 1650–1655 (2003). [DOI] [PubMed] [Google Scholar]
- 68.Orr N et al. A mutation in the atrial-specific myosin light chain gene (MYL4) causes familial atrial fibrillation. Nat Commun 7, 11303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bakker Martijn L et al. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circulation Research 102, 1340–1349 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Bruneau BG Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harbor Perspectives in Biology 5, a008292 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hoogaars WMH et al. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev 21, 1098–1112 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boogerd CJ & Evans SM TBX5 and NuRD divide the heart. Dev Cell 36, 242–244 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mori AD & Bruneau BG TBX5 mutations and congenital heart disease: Holt-Oram syndrome revealed. Curr Opin Cardiol 19, 211–215 (2004). [DOI] [PubMed] [Google Scholar]
- 74.Mesbah K, Harrelson Z, Théveniau-Ruissy M, Papaioannou VE & Kelly RG Tbx3 is required for outflow tract development. Circulation Research 103, 743–750 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xie H et al. Identification of TBX2 and TBX3 variants in patients with conotruncal heart defects by target sequencing. Human Genomics 12, 44 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Eif VWW, Devalla HD, Boink GJJ & Christoffels VM Transcriptional regulation of the cardiac conduction system. Nat Rev Cardiol 15, 617–630 (2018). [DOI] [PubMed] [Google Scholar]
- 77.Juillière Y et al. Additional predictive value of both left and right ventricular ejection fractions on long-term survival in idiopathic dilated cardiomyopathy. Eur Heart J 18, 276–280 (1997). [DOI] [PubMed] [Google Scholar]
- 78.Udler MS et al. Type 2 diabetes genetic loci informed by multi-trait associations point to disease mechanisms and subtypes: a soft clustering analysis. PLoS Medicine 15, e1002654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 79.Is R et al. Distribution, determinants, and normal reference values of thoracic and abdominal aortic diameters by computed tomography (from the Framingham Heart Study). Am J Cardiol 111, 1510–1516 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.He K, Zhang X, Ren S & Sun J Deep residual learning for image recognition. arXiv:1512.03385 [cs] (2015). [Google Scholar]
- 81.Krizhevsky A, Sutskever I & Hinton GE ImageNet classification with deep convolutional neural networks. Commun. ACM 60, 84–90 (2017). [Google Scholar]
- 82.Kingma DP & Ba J Adam: a method for stochastic optimization. arXiv:1412.6980 [cs] (2017). [Google Scholar]
- 83.Smith LN Cyclical learning rates for training neural networks. arXiv:1506.01186 [cs] (2015). [Google Scholar]
- 84.Smith LN A disciplined approach to neural network hyper-parameters: Part 1 -- learning rate, batch size, momentum, and weight decay. arXiv:1803.09820 [cs, stat] (2018). [Google Scholar]
- 85.Lin T-Y, Goyal P, Girshick R, He K & Dollár P Focal loss for dense object detection. arXiv:1708.02002 [cs] (2018). [DOI] [PubMed] [Google Scholar]
- 86.Cox DR The regression analysis of binary sequences. Journal of the Royal Statistical Society: Series B (Methodological) 20, 215–232 (1958). [Google Scholar]
- 87.Bycroft C et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang J et al. FTO genotype is associated with phenotypic variability of body mass index. Nature 490, 267–272 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Osborne LR & Mervis CB Rearrangements of the Williams–Beuren syndrome locus: molecular basis and implications for speech and language development. Expert Rev Mol Med 9, 1–16 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pober BR Williams-Beuren syndrome. N. Engl. J. Med 362, 239–252 (2010). [DOI] [PubMed] [Google Scholar]
- 92.Tartaglia M et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet 29, 465–468 (2001). [DOI] [PubMed] [Google Scholar]
- 93.Wigginton JE, Cutler DJ & Abecasis GR A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet 76, 887–893 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nature Genetics 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Boughton AP et al. LocusZoom.js: interactive and embeddable visualization of genetic association study results. Bioinformatics 37, 3017–3018 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Finucane HK et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet 50, 621–629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tucker NR et al. Transcriptional and cellular diversity of the human heart. Circulation 142, 466–482 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Law CW, Chen Y, Shi W & Smyth GK voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biology 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Auton A et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Finucane HK et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet 47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Benjamin EJ et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet 41, 879–881 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hong H et al. Assessing batch effects of genotype calling algorithm BRLMM for the Affymetrix GeneChip Human Mapping 500 K array set using 270 HapMap samples. BMC Bioinformatics 9, S17 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Das S et al. Next-generation genotype imputation service and methods. Nature Genetics 48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Qazi S et al. Increased aortic diameters on multidetector computed tomographic scan are independent predictors of incident adverse cardiovascular events: the Framingham Heart Study. Circ Cardiovasc Imaging 10, e006776 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Therneau TM & Grambsch PM Modeling Survival Data: Extending the Cox Model. (Springer-Verlag, 2000). doi: 10.1007/978-1-4757-3294-8. [DOI] [Google Scholar]
- 106.Bellenguez C et al. A robust clustering algorithm for identifying problematic samples in genome-wide association studies. Bioinformatics 28, 134–135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pirruccello JP et al. Analysis of cardiac magnetic resonance imaging in 36,000 individuals yields genetic insights into dilated cardiomyopathy. Nature Communications 11, 2254 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
UK Biobank data are made available to researchers from research institutions with genuine research inquiries, following IRB and UK Biobank approval. GWAS summary statistics are available at the Broad Institute Cardiovascular Disease Knowledge Portal (http://www.broadcvdi.org). Single nucleus RNA sequencing data are publicly available at the Single Cell Portal (https://singlecell.broadinstitute.org/single_cell accession #SCP498). The dbGAP study accession numbers used for FHS replication were #phs000007.v32.p13 for PA diameter measurement and #phs000342.v20.p13 for genotyping. BioBank Japan data are available to bona fide researchers for approved research by application to the Japanese Genotype-phenotype Archive. Mass General Brigham data are available to Mass General Brigham investigators. All other data are contained within the article and its supplementary information, or are available upon reasonable request to the corresponding author.


