Abstract
KRAS is the most commonly mutated RAS family gene and is a primary cause of the occurrence of several types of cancer. However, KRAS mutations have several unique and diverse molecular identities, making it difficult to find specific treatments. Here, we developed universal pegRNAs which can correct all types of G12 and G13 oncogenic KRAS mutations with CRISPR-mediated prime editors (PEs). The universal pegRNA successfully corrected 12 types of KRAS mutations, accounting for 94% of all known KRAS mutations, by up to 54.8% correction frequency in HEK293T/17 cells. We also applied the universal pegRNA to correct endogenous KRAS mutations in human cancer cells and found that G13D KRAS mutation was successfully corrected to wild-type KRAS sequences with up to 40.6% correction frequency without indel mutations. We propose prime editing with the universal pegRNA as a ‘one–to–many’ potential therapeutic strategy for KRAS oncogene variants.
Subject terms: Targeted gene repair, CRISPR-Cas9 genome editing
A CRISPR prime editing approach corrects all types of G12 and G13 oncogenic KRAS mutations using a universal pegRNA, demonstrating the potential of applying this system to KRAS gene therapy through a oneto-many approach.
Introduction
RAS family genes are involved in tumor formation in many human cancers, and there are three RAS family genes: HRAS, NRAS, and KRAS1,2. Ras proteins are monomeric GTPases and regulate cell differentiation, proliferation, and survival, and can deliver cell growth signals by regulating either guanosine diphosphate (GDP) inactivation or guanosine triphosphate (GTP) activation3. When a genetic mutation occurs in RAS, GTP remains active and cell growth signals are sent into the nucleus to induce tumorigenesis. KRAS mutations account for most of the RAS family gene mutations4, and intensive efforts have been made over the past 40 years to inhibit the KRAS mutants. Although direct inhibition of KRAS mutants is a desirable approach for treating KRAS-associated tumors, it is still challenging to target mutant KRAS proteins. The structure of KRAS has proven difficult to target due to its smooth surface that hinders the binding by small molecules5, and a picomolar affinity for GTP, which prevents the development of effective inhibitors6. To date, there are two types of inhibitors that have been developed for KRAS in a mutant-specific manner: covalent inhibitors targeting the mutated cysteine residue in KRAS G12C7–10 and a selective non-covalent inhibitor of KRAS G12D11,12. However, direct inhibitors of other KRAS mutations, present in over 50% of KRAS-associated tumors, have not been developed. Therefore, an alternative approach is needed to directly inhibit KRAS mutants.
CRISPR-Cas9 mediated genome editing tools are widely used in a variety of biomedical applications including KRAS gene therapy13–16. In this system, Cas9 and gRNA complexes of the CRISPR-Cas9 system generate double-strand breaks (DSBs) at target sequences of KRAS mutants, and the disrupted KRAS mutants inhibit tumor growth14,15. However, Cas9 proteins and gRNAs should be carefully adopted for each KRAS mutant to reduce unwanted gene disruption in wild-type KRAS, and specific gRNAs targeting each KRAS mutant should be used for each target mutation (i.e., G12V targeting gRNAs could be used for disrupting only G12V mutation). It has been shown that prolonged Cas9 expression induces other oncogenic KRAS variants that are resistant to Cas9 cleavage16. CRISPR-Cas9-based base editors (BEs) have been developed to induce base conversions without generating DSBs and have recently been applied to correct KRAS G12S, G12D, and G13D mutations16. However, each gRNA for base editing should be designed to correct each target KRAS mutation and BEs can correct transition mutations such as C:G to T:A and A:T to G:C as well as transversion mutations such as C:G to A:T or G:C and A:T to C:G or T:A, however, BEs can only correct cytosine or adenine within the base editing window, and often induce undesired bystander substitutions within the base editing window17–20.
Recently, a revolutionary genome-editing technique, called a prime editor (PE), was developed to induce accurate point mutations in the genome without requiring DSBs or donor DNA templates21. PE2 is a fusion protein composed of an engineered reverse transcriptase (RT) and a catalytically inactivated SpCas9 nickase (SpCas9-H840A). PEs are directed to target sites by a prime editing guide RNA (pegRNA) containing primer binding sites (PBS) and a reverse transcriptase template (RTT), which allows the RT to transcribe the additional sequences, which is then introduced at the target sites. The PE3 and PE3b systems increase prime editing efficiency by introducing additional gRNA to nick the non-edited DNA strand, facilitating the edited strand to be used as a repair template.
In this study, we applied the PE system to correct G12 and G13 KRAS mutations, which account for most KRAS mutations. We designed universal pegRNAs which can correct all types of G12 and G13 KRAS mutations and confirmed that the universal pegRNA can correct endogenous KRAS mutations in HEK293T/17 cells and three human cancer cell lines. Taken together, these results show the potential of applying the PE system to KRAS gene therapy.
Results
Design of universal pegRNAs for KRAS gene correction
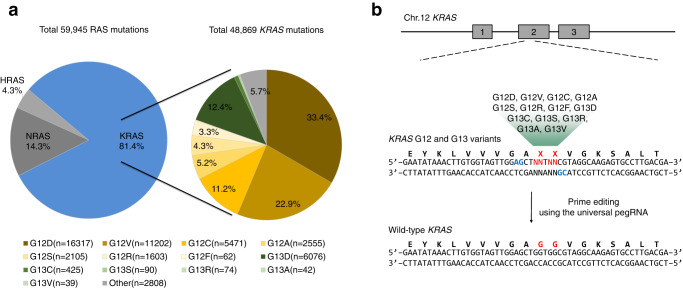
We first investigated the mutation frequency of three RAS family genes, HRAS, NRAS, and KRAS, which are involved in tumorigenesis. According to the COSMIC (Catalog Of Somatic Mutations In Cancer) database (https://cancer.sanger.ac.uk/)22, KRAS mutations account for the majority (81.4%) of RAS mutations, followed by NRAS mutations and HRAS mutations, 14.3% and 4.3%, respectively (Fig. 1a). The majority of KRAS mutations consist of missense mutations in the G12 and G13 amino acid residues, 80.4% and 13.8%, respectively (Fig. 1a and Table 1).
Fig. 1. Analysis of Ras mutations and design of universal pegRNAs to correct the KRAS mutations.
a Classification of all RAS gene family mutations, the left side, and subdivision of KRAS mutations, the right side. b Schematic overviews of KRAS corrections by universal pegRNAs. The G12 and G13 positions of KRAS and the PAM sequences of two universal pegRNAs are highlighted in red and blue, respectively.
Table 1.
Targetability of 12 KRAS mutations by ABE and PE systems.
| KRAS mutations | Counts | Ratio (%) | Targetability | ||
|---|---|---|---|---|---|
| Amino acids | Nucleotides | ABE | PE | ||
| p.G12D | c.35 G > A | 16317 | 35.4 | ✓ | ✓ |
| p.G12V | c.35 G > T | 11202 | 24.3 | ✓ | |
| p.G13D | c.38 G > A | 6076 | 13.2 | ✓ | ✓ |
| p.G12C | c.34 G > T | 5471 | 11.9 | ✓ | |
| p.G12A | c.35 G > C | 2555 | 5.5 | ✓ | |
| p.G12S | c.34 G > A | 2105 | 4.6 | ✓ | ✓ |
| p.G12R | c.34 G > C | 1603 | 3.5 | ✓ | |
| p.G13C | c.37 G > T | 425 | 0.9 | ✓ | |
| p.G13S | c.37 G > A | 90 | 0.2 | ✓ | ✓ |
| p.G13R | c.37 G > C | 74 | 0.2 | ✓ | |
| p.G12F | c.34_35delinsTT | 62 | 0.1 | ✓ | |
| p.G13A | c.38 G > C | 42 | 0.1 | ✓ | |
| p.G13V | c.38 G > T | 39 | 0.1 | ✓ | |
| Total counts | 46,061 | 24,588 | 46,061 | ||
Because the PE system can introduce various point mutations at target sites using the RTT sequences of pegRNAs, we designed universal pegRNAs to manipulate the majority of KRAS missense mutations. Previous studies show that prime editing frequencies are dependent on the length between the Cas9-mediated nicking site and the altering site of desired mutation21,23. Therefore, we selected two adjacent protospacer sequences with altered NG PAM sequences, KRAS-#1 and KRAS-#2, to edit G12 and G13 missense mutations and used PAM flexible PE variant, PE2-SpG24 (Fig. 1b). The length of PBS and RTT in pegRNAs was changed from 9 to 13 bp and 10 to 16 bp, respectively, to generate a 3′ flap of different lengths. In total, we designed nine universal pegRNAs containing RTT sequences to introduce wild-type KRAS sequences to the G12 and G13 amino acid residue positions.
Optimization of universal pegRNAs for KRAS gene correction in pooled library
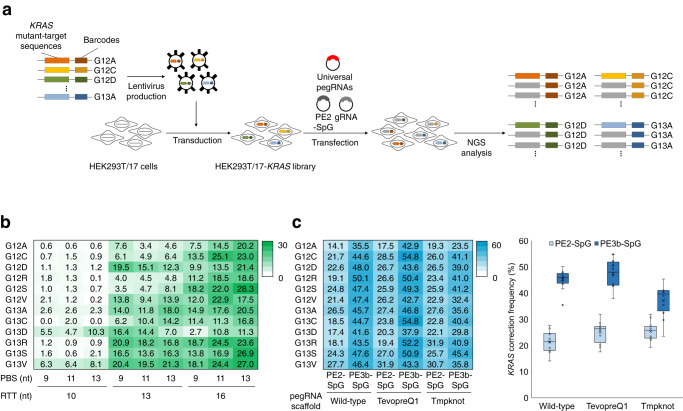
To evaluate the KRAS correction activity of universal pegRNAs, we established HEK293T/17 library cell lines containing 12 selected KRAS mutations (HEK293T/17-KRAS library). We cloned each of the 12 KRAS mutant sequences and barcode sequences into lentiviral plasmids. Lentivirus particles were produced and infected into HEK293T/17 cells with a low multiplicity of infection to introduce only one type of KRAS mutation per cell (Fig. 2a). To measure KRAS correction activity for each mutation, PCR amplicons were obtained from genomic DNA and subjected to targeted deep sequencing analysis. Using the barcode sequence of the amplicon, sequencing results were classified as the original KRAS mutations and the KRAS correction activity of each mutation was calculated. Using the HEK293T/17-KRAS library cell lines, we examined KRAS correction activities of universal pegRNAs using PE2-SpG and revealed that universal pegRNAs including KRAS-#2 protospacer sequences could correct all 12 types of KRAS mutations with higher activity compared to universal pegRNAs including KRAS-#1 protospacer sequences (Fig. 2b and Supplementary Fig. 1). The pegRNAs with longer RTTs tended to have higher KRAS correction activities and a pegRNA including 13-nt PBS and 16-nt RTT had 28.3% KRAS G12S correction activity. We also found that none of the pegRNAs with PE2-SpG induced any substitutions or indels at endogenous KRAS sites (Supplementary Table 1).
Fig. 2. Construction of HEK293T/17-KRAS library cells and correction of KRAS mutations using universal pegRNAs.
a Schematic overview for correcting KRAS mutations in HEK293T/17-KRAS library cells. Lentivirus particles containing 12 types of KRAS mutant-target sequences and barcode sequences were delivered in HEK293T/17 cells to generate HEK293T/17-KRAS library cells. After editing KRAS mutations using universal pegRNAs, the KRAS-corrected cells were collected and subjected to NGS analysis to measure the KRAS correction frequencies of each KRAS mutation. b Nine universal pegRNAs containing various lengths of PBS and RTT and PE2-SpG were delivered in HEK293T/17-KRAS library cells, and the KRAS correction frequency of each KRAS mutation is shown in the heatmap. c Universal pegRNAs containing 13-nt PBS and 16-nt RTT were engineered to have two types of RNA motif, tevopreQ1 and tmpknot, and subjected to KRAS correction in PE2-SpG or PE3b-SpG systems. The KRAS correction frequencies are shown in the heatmap and box-whisker plot, respectively. The experiments were conducted with biological triplicate (n = 3).
To further improve the KRAS correction efficiency, the universal pegRNA containing 13-nt PBS and 16-nt RTT was subjected to the PE3b system (hereafter named, PE3b-SpG) which uses additional gRNAs to nick the opposite strand of the edited strand to increase the editing frequency21. As shown in Fig. 2c, the KRAS correction activity was improved up to 2.6-fold in the G12R mutation by the PE3b-SpG, and in the G12R KRAS mutation, the correction efficiency was improved to 50.1% without any mutations at endogenous KRAS sites. In addition, we further constructed engineered pegRNAs (epegRNAs) containing either tevopreQ1 or tmpknot RNA motif at the 3′ end of pegRNAs, which is known to enhance prime editing activity25. Compared with the original universal pegRNA, universal epegRNAs had improved KRAS correction activities, which were further enhanced by PE3b-SpG. We found that the universal epegRNA containing tevopreQ1 RNA motif could edit KRAS G12C and G13C mutation by 54.8% (Fig. 2c). We also confirmed that there were no substitutions or indel mutations at endogenous KRAS sites (Supplementary Table 1). Overall, these results indicated that the universal pegRNA could correct the majority of KRAS mutations, and epegRNA and PE3b-SpG could further improve the KRAS correction efficiency without altering the wild-type KRAS sequence.
Correction of endogenous KRAS mutations using universal pegRNAs
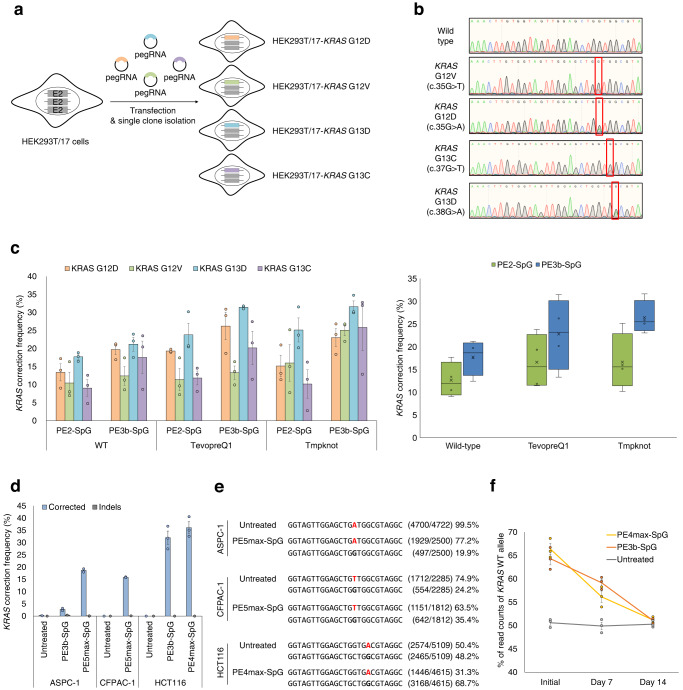
We then decided to correct endogenous KRAS mutations using the universal epegRNA. To generate cell lines containing endogenous KRAS mutations, we designed several pegRNAs to introduce KRAS G12D, G12V, G13C, or G13D mutations at the wild-type endogenous KRAS sequence (Fig. 3a). We delivered plasmids encoding the pegRNAs and PE2 into HEK293T/17 cells and prime editing frequencies were measured by targeted-deep sequencing. As shown in Supplementary Fig. 2, prime editing frequencies varied from 0.3% to 36.0% and we selected pegRNAs with the highest prime editing frequency per target KRAS mutation. We then transfected the plasmid encoding each pegRNA and PE2 into HEK293T/17 cells and isolated single clones containing KRAS G12D, G12V, G13C, or G13D mutations at the endogenous KRAS gene. The Sanger sequencing results showed that each clone had heterozygous KRAS mutations (Fig. 3b). As the KRAS gene is located on chromosome 12, which exists as a triploid in HEK293T/17 cells26, each clone had heterogenous KRAS sequences, with two copies of the wild-type KRAS sequence and one copy of the mutated KRAS sequence.
Fig. 3. Correction of endogenous KRAS mutations in HEK293T/17 cells and three human cancer cell lines.
a Schematic for the construction of HEK293T/17 cell lines containing four types of KRAS mutations at endogenous KRAS sites. The pegRNAs and PE2 were delivered into HEK293T/17 cells and single clones were analyzed to obtain HEK293T/17 cells with endogenous KRAS mutations. b Sanger sequencing results for each HEK293T/17 cell line containing endogenous KRAS mutations. c Endogenous KRAS correction frequency in KRAS mutated-HEK293T/17 cells. The universal pegRNAs and epegRNAs were delivered into each KRAS mutated-HEK293T/17 cell by PE2-SpG or PE3b-SpG systems. The KRAS correction frequency was calculated from read counts of NGS sequencing results described in Table S2. Error bars mean s.e.m. of biological triplicate samples (n = 3). d Endogenous KRAS correction in three human cancer cell lines, ASPC-1, CFPAC-1 and HCT116 cells. The KRAS correction frequencies of KRAS heterogenous cell lines, CFPAC-1 and HCT116, were calculated from read counts of NGS sequencing results. Error bars mean s.e.m. of biological duplicate or triplicate samples. e The representative NGS sequencing results of KRAS correction in three human cancer cell lines. KRAS mutations in each cell line (G12D, 12 V, and G13D) were highlighted in red. f The KRAS correction frequency of HCT116 was measured over time. Error bars mean s.e.m. of biological triplicate samples (n = 3).
To measure the endogenous KRAS correction frequency, we delivered the universal pegRNA containing 13-nt PBS and 16-nt RTT and PE2-SpG into each HEK293T/17-KRAS G12D, G12V, G13C, and G13D cell line. We also examined epegRNAs and PE3b-SpG to determine whether endogenous KRAS correction frequency could be enhanced. The KRAS correction frequency of KRAS heterogenous cells was calculated using the reads counts from targeted-deep sequencing as described in the methods section. As in HEK293T/17-KRAS library cell lines in Fig. 2b, c, we identified that the PE3b-SpG showed higher KRAS correction activity than the PE2-SpG in four endogenous KRAS mutated HEK293T/17 cells (Fig. 3c and Supplementary Fig. 3). The tevopreQ1 and tmpknot RNA motifs could improve KRAS correction activity, and the epegRNA containing tmpknot RNA motif could correct KRAS G13D mutation by 31.6% in PE3b-SpG. We found that there were no detectable indel mutations in the heterogenous cell lines (Supplementary Table 2). We also constructed a universal pegRNA capable of inducing a silent mutation to confirm that the KRAS corrections were indeed induced by prime editing and confirmed that the KRAS corrected alleles also contained the silent mutation (Supplementary Fig. 4).
Furthermore, we examined whether the universal pegRNA could correct the endogenous KRAS mutations in three types of human cancer cells: two pancreatic cancer cell lines, CFPAC-1 and ASPC-1, and one colon cancer cell line, HCT11627,28. We confirmed that CFPAC-1 cells have three KRAS G12V alleles and one KRAS wild-type allele, ASPC-1 cells have two KRAS G12D mutation-bearing alleles, and HCT116 cells have one KRAS G13D allele and one KRAS wild-type allele by targeted-deep sequencing. We first examined the KRAS correction activity of the universal epegRNA with tevopreQ1 motif in PE3b-SpG, but the KRAS correction activity was only 2.7% in ASPC-1 cells, much lower than that of the HEK293T/17 cells (Fig. 3d). To further improve the prime editing frequency, we applied PE5max system29, which utilized PEmax-SpG-P2A-hmLH1dn (hereafter named, PE5max-SpG) instead of PE2-SpG, and found that the KRAS mutations in 13.2% of CFPAC-1 cells and 18.7% of ASPC-1 cells were corrected by the universal epegRNAs (Fig. 3d, e). In the case of HCT116, PE3b-SpG showed 32.0% KRAS correction frequency and PEmax-SpG (hereafter named, PE4max-SpG) showed 36.1% KRAS correction frequency (Fig. 3d, e). We measured the KRAS correction frequency for two weeks to evaluate the functional effect of KRAS gene correction and found that the KRAS correction frequency decreased over time, reflecting that the KRAS corrected HCT116 cells have a growth disadvantage (Fig. 3f). Taken together, we demonstrated that endogenous KRAS mutations could be corrected by the universal pegRNA in HEK293T/17 cells, pancreatic cancer cell lines, and colon cancer cell lines.
Discussion
Although KRAS mutations are the most frequent oncogenic alterations, these mutations have remained an intractable therapeutic target. In this study, we developed the universal pegRNA to correct most types of KRAS mutations and corrected these KRAS mutations without unwanted mutations using the PE system in the library cells. Furthermore, we successfully corrected endogenous KRAS mutations in human cells.
Previously, the Cas9 nuclease and base editors were successfully applied to target KRAS14,15. Cas9 nuclease with mutant–specific gRNAs enable KRAS mutant-specific disruption. Although Cas9 nuclease disrupts KRAS mutations with high efficiency, this method requires careful selection of mutant–specific gRNAs and Cas9 protein at each KRAS variant16. The adenine base editors which introduce A:T to G:C conversion in the target locus could also correct several types of KRAS mutations including G12S, G12D, and G13D, however, gRNAs for each mutation should be designed individually for the BE-meditated correction of KRAS mutations. Furthermore, adenine base editors can target only 4 types of KRAS mutations in KRAS G12 and G13 mutations (Table 1). As unwanted bystander editing often occurs in BE system due to the base editing active window30, it is necessary to confirm the performance of each gRNA, which is laborious.
The PE system has advantages over the Cas9 nuclease and base editors in editing KRAS mutations. Theoretically, the PE system can manipulate all types of KRAS G12 and G13 mutations using only one type of pegRNA, making it simple to use in biomedical applications. Although the PE system has advantages in correcting KRAS mutations, the correction efficiencies of two pancreatic cancer cell lines were much lower than those of HEK293T/17 cell lines and HCT116 colon cancer cells as previously described29. As low editing frequencies made it difficult to detect the effects of KRAS gene correction, such as aberrant growth rate and functional analysis of KRAS downstream pathways, It is necessary to improve the prime editing activity in vitro and in vivo for the PE system to be used in therapeutic approaches. Previously, it has shown that the PE system does not induce appreciable off-target mutations, however, off-target effects should be considered to be used in gene therapy strategy31–34. We selected five potential off-target sites based on the number of mismatches with the universal epegRNA and analyzed whether mutation was induced at those five sites (Supplementary Table 3). We confirmed that there was no remarkable mutation at potential off-target sites by targeted-deep sequencing, and high-throughput analysis, such as transcriptome analysis, might be necessary to elucidate the potential off-target effects (Supplementary Fig. 5). Our finding showed that the PE system could introduce and correct the KRAS mutations, thereby demonstrating the potential of the system for developing KRAS-specific targeted therapies.
Methods
Cell culture and cell line generation
CFPAC-1 cells and ASPC-1 cells were gifted by Dr. Eunsung Jun (Asan Medical Center) and maintained in Roswell Park Memorial Institute 1640 Medium and HEK293T/17 cells (ATCC CRL-11268) and HCT116 cells (ATCC CCL-247) were maintained in Dulbecco’s modified Eagle’s medium, supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin.
To generate the HEK293T-KRAS library stable cell line, lentiviral particles were generated and transduced into HEK293T/17 cells. Briefly, 1 μg of plasmids (500 ng of KRAS mutant sequence containing vectors, 300 ng of psPAX2, and 200 ng of pMD2.G) were transfected into 2 × 105 HEK293T/17 cells using Lipofectamine 2000 (Thermo Fisher Scientific) by the manufacturer’s instructions, and the medium was changed 24 h after transfection23. The supernatants containing lentiviral particles were harvested and filtered with 0.45 μm filter 48 h after transfection and stored at –70 °C until use. The lentivirus particles were then transduced into HEK293T/17 cells with a low multiplicity of infection and infected cells were selected for 3 days with 1 μg/ml of puromycin. The KRAS mutant sequences were listed in Supplementary Table 4.
Plasmid construction
The pRG2 plasmid (Addgene plasmids #104174) was used for gRNA cloning and pU6-pegRNAGG-acceptor (Addgene plasmids #132777), pU6-tevopreq1-GG-acceptor (Addgene plasmids #174038), and pU6-tmpknot-GG-acceptor (Addgene plasmids #174039) plasmids were used for pegRNA or epegRNA cloning. The PCR amplicon of epegRNAs and PE3b gRNAs were cloned into the lentiGuide-puro plasmid (Addgene plasmids #52963) to construct an all-in-one vector for use in human cancer cells. To construct a lentiviral vector containing KRAS mutant target sequences to generate HEK293T/17-KRAS library stable cell lines, KRAS mutant target sequences were cloned into pCW-Cas9 plasmids (Addgene # 50661). The sequences of target sequences, PBS, and RTT are listed in Supplementary Table 5. The pCMV-PE2-SpG-V3, which was constructed by fusing two nuclear localization signals to both of C- and N-terminal of pCMV-PE2-SpG (Addgene plasmid #159978), and pCMV-PEmax-SpG-P2A-hMLH1dn, which was constructed by site-directed mutagenesis from pCMV-PEmax-P2A-hMLH1dn (Addgene plasmid #174828), were used for prime editing experiments.
Transfection
For the plasmid transfections, HEK293T/17 cells (0.6 × 105), HEK293T/17-KRAS library cells (0.6 × 105), CFPAC-1 cells (3 × 104), ASPC-1 cells (3 × 104), or HCT116 cells (2 × 104) were seeded onto a TC-treated 48-well plate and transfection was performed the next day when cell confluency reached 50–60%. A total of 500 ng of plasmids (250 ng of PEs with 250 ng of pegRNAs or epegRNAs) were delivered using 1.5 μL of Lipofectamine 2000 (Thermo Fisher Scientific) or 1.5 μL Lipofectamine 3000 (Thermo Fisher Scientific) with 1 μL of P3000 reagent according to the manufacturer’s protocol. For the PE3b experiments in HEK293T/17 cells, 83 ng of plasmids encoding gRNAs were additionally delivered to the cells. For the PE3b experiments in CFPAC-1, ASPC-1, or HCT116 cells, 250 ng of all-in-one vector was used for the plasmid transfection, and transfected cells were selected one day after transfection with 1 μg/mL of puromycin.
Targeted-deep sequencing
Genomic DNA was extracted 72 h after transfection using cell lysis buffer (0.05% SDS in pH 7.5 of 100 nM Tris-HCl and 100 μg/ml proteinase K) and subjected to PCR amplification. The target sites were amplified using Phusion High-Fidelity DNA Polymerase (New England Biolabs) with target-specific primer pairs and the Illumina TruSeq HT dual index adaptor primer (Supplementary Table 6)35. The libraries were sequenced at 150-bp paired-end using the Illumina MiniSeq or iSeq 100 sequencing equipment to analyze the mutation frequencies and Cas-Analyzer (http://www.rgenome.net/cas-analyzer)36 was used to analyze the sequencing data. KRAS correction frequencies in homozygous KRAS mutant cells including HEK293T/17-KRAS library cell lines and ASPC-1 cells were directly measured as prime editing frequencies, and those in heterozygous KRAS mutant cells including HEK293T/17 mutant clones, CFPAC-1 and HCT116 cells, were calculated as follows:
| 1 |
Sanger sequencing
Genomic DNA was isolated using the DNA Blood & Tissue kit (Qiagen) in accordance with the manufacturer’s instructions and target sites were amplified using phusion high-fidelity DNA polymerase (New England Biolabs) to verify the endogenous KRAS sequences of single clones. The PCR products were purified with the QIAquick PCR Purification Kit (Qiagen) and analyzed by Sanger sequencing.
Statistics and reproducibility
All statistical analyzes were performed independently on two or three biological experiments. Error bars represent standard error of the mean.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank Dr. Eunsung Jun (Asan Medical Center) for providing CFPAC-1 cells and ASPC-1 cells. Y.K. supervised the research. G.J. and J.K. performed the experiments and analyzed the data. G.J., J.K. and Y.K. wrote the manuscript. All authors approved the manuscript. This work was supported by the National Research Foundation of Korea [2021R1C1C100716212, 2018R1A5A2020732 to Y.K.].
Author contributions
G.J. and Y.K. conceived and designed this study. G.J. and J.K. carried out the experiments and analyzed the data. G.J. and Y.K. wrote the manuscript. Y.K. supervised the research.
Peer review
Peer review information
Communications Biology thanks Frank Buchholz and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: [Anam Akhtar and George Inglis]. A peer review file is available.
Data availability
DNA sequencing data have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database with BioProject accession code PRJNA972834. Source data for the plots and graphs in the figures is available as Supplementary Data 1 and any remaining information can be obtained from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-023-05052-1.
References
- 1.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 2.Cox AD, Der CJ. Ras history: the saga continues. Small GTPases. 2010;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: post-translational modification of RAS. Nat. Rev. Mol. Cell Biol. 2011;13:39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prior IA, Hood FE, Hartley JL. The frequency of Ras mutations in cancer. Cancer Res. 2020;80:2969–2974. doi: 10.1158/0008-5472.CAN-19-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwan AK, Piazza GA, Keeton AB, Leite CA. The path to the clinic: a comprehensive review on direct KRAS(G12C) inhibitors. J. Exp. Clin. Cancer Res. 2022;41:27. doi: 10.1186/s13046-021-02225-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: mission possible? Nat. Rev. Drug Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canon J, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217–223. doi: 10.1038/s41586-019-1694-1. [DOI] [PubMed] [Google Scholar]
- 9.Hallin J, et al. The KRAS(G12C) inhibitor MRTX849 Provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov. 2020;10:54–71. doi: 10.1158/2159-8290.CD-19-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lito P, Solomon M, Li LS, Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science. 2016;351:604–608. doi: 10.1126/science.aad6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, et al. Identification of MRTX1133, a noncovalent, potent, and selective KRAS(G12D) inhibitor. J. Med. Chem. 2022;65:3123–3133. doi: 10.1021/acs.jmedchem.1c01688. [DOI] [PubMed] [Google Scholar]
- 12.Hallin J, et al. Anti-tumor efficacy of a potent and selective non-covalent KRASG12D inhibitor. Nat. Med. 2022;28:2171–2182. doi: 10.1038/s41591-022-02007-7. [DOI] [PubMed] [Google Scholar]
- 13.Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020;38:824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- 14.Kim W. et al. Targeting mutant KRAS with CRISPR-Cas9 controls tumor growth. Genome Res. 28, 374–382 (2018). [DOI] [PMC free article] [PubMed]
- 15.Lee W, Lee JH, Jun S, Lee JH, Bang D. Selective targeting of KRAS oncogenic alleles by CRISPR/Cas9 inhibits proliferation of cancer cells. Sci. Rep. 2018;8:11879. doi: 10.1038/s41598-018-30205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sayed S, et al. Efficient correction of oncogenic KRAS and TP53 mutations through CRISPR base editing. Cancer Res. 2022;82:3002–3015. doi: 10.1158/0008-5472.CAN-21-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tong H. et al. Programmable A-to-Y base editing by fusing an adenine base editor with an N-methylpurine DNA glycosylase. Nat. Biotechnol. 10.1038/s41587-022-01595-6 (2023). [DOI] [PubMed]
- 18.Zhao D, et al. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol. 2021;39:35–40. doi: 10.1038/s41587-020-0592-2. [DOI] [PubMed] [Google Scholar]
- 19.Kurt IC, et al. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol. 2021;39:41–46. doi: 10.1038/s41587-020-0609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaudelli NM, et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anzalone AV, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tate JG, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim HK, et al. Predicting the efficiency of prime editing guide RNAs in human cells. Nat. Biotechnol. 2021;39:198–206. doi: 10.1038/s41587-020-0677-y. [DOI] [PubMed] [Google Scholar]
- 24.Kweon J, et al. Engineered prime editors with PAM flexibility. Mol. Ther. 2021;29:2001–2007. doi: 10.1016/j.ymthe.2021.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson JW, et al. Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. 2022;40:402–410. doi: 10.1038/s41587-021-01039-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Binz RL, et al. Identification of novel breakpoints for locus- and region-specific translocations in 293 cells by molecular cytogenetics before and after irradiation. Sci. Rep. 2019;9:10554. doi: 10.1038/s41598-019-47002-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sirivatanauksorn V, et al. Non-random chromosomal rearrangements in pancreatic cancer cell lines identified by spectral karyotyping. Int. J. Cancer. 2001;91:350–358. doi: 10.1002/1097-0215(200002)9999:9999<::AID-IJC1049>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 28.Burgess MR, et al. KRAS allelic imbalance enhances fitness and modulates MAP kinase dependence in cancer. Cell. 2017;168:817–829.e815. doi: 10.1016/j.cell.2017.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen PJ, et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell. 2021;184:5635–5652.e5629. doi: 10.1016/j.cell.2021.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HS, Jeong YK, Hur JK, Kim JS, Bae S. Adenine base editors catalyze cytosine conversions in human cells. Nat. Biotechnol. 2019;37:1145–1148. doi: 10.1038/s41587-019-0254-4. [DOI] [PubMed] [Google Scholar]
- 31.Kim DY, Moon SB, Ko JH, Kim YS, Kim D. Unbiased investigation of specificities of prime editing systems in human cells. Nucleic Acids Res. 2020;48:10576–10589. doi: 10.1093/nar/gkaa764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwon J, et al. TAPE-seq is a cell-based method for predicting genome-wide off-target effects of prime editor. Nat. Commun. 2022;13:7975. doi: 10.1038/s41467-022-35743-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang, S. Q. et al. Genome-wide profiling of prime editor off-target sites in vitro and in vivo using PE-tag. Nat. Methods20, 898–907(2023). [DOI] [PubMed]
- 34.Schene IF, et al. Prime editing for functional repair in patient-derived disease models. Nat. Commun. 2020;11:5352. doi: 10.1038/s41467-020-19136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin HR, et al. Small-molecule inhibitors of histone deacetylase improve CRISPR-based adenine base editing. Nucleic Acids Res. 2021;49:2390–2399. doi: 10.1093/nar/gkab052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park J, Lim K, Kim JS, Bae S. Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics. 2017;33:286–288. doi: 10.1093/bioinformatics/btw561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
DNA sequencing data have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database with BioProject accession code PRJNA972834. Source data for the plots and graphs in the figures is available as Supplementary Data 1 and any remaining information can be obtained from the corresponding author upon reasonable request.