Abstract
Purpose and scope
The aim of this position statement is to provide recommendations for clinicians regarding the use of genetic and metabolic investigations for patients with neurodevelopmental disorders (NDDs), specifically, patients with global developmental delay (GDD), intellectual disability (ID) and/or autism spectrum disorder (ASD). This document also provides guidance for primary care and non-genetics specialists caring for these patients while awaiting consultation with a clinical geneticist or metabolic specialist.
Methods of statement development
A multidisciplinary group reviewed existing literature and guidelines on the use of genetic and metabolic investigations for the diagnosis of NDDs and synthesised the evidence to make recommendations relevant to the Canadian context. The statement was circulated for comment to the Canadian College of Medical Geneticists (CCMG) membership-at-large and to the Canadian Pediatric Society (Mental Health and Developmental Disabilities Committee); following incorporation of feedback, it was approved by the CCMG Board of Directors on 1 September 2022.
Results and conclusions
Chromosomal microarray is recommended as a first-tier test for patients with GDD, ID or ASD. Fragile X testing should also be done as a first-tier test when there are suggestive clinical features or family history. Metabolic investigations should be done if there are clinical features suggestive of an inherited metabolic disease, while the patient awaits consultation with a metabolic physician. Exome sequencing or a comprehensive gene panel is recommended as a second-tier test for patients with GDD or ID. Genetic testing is not recommended for patients with NDDs in the absence of GDD, ID or ASD, unless accompanied by clinical features suggestive of a syndromic aetiology or inherited metabolic disease.
Keywords: genetics, medical; clinical decision-making; genetics; genetic testing
Introduction
Neurodevelopmental disorders (NDDs) are a group of conditions first manifesting early in childhood, characterised by impairments of personal, social, academic or occupational functioning.1 Examples of NDDs include autism spectrum disorder (ASD), global developmental delay (GDD), intellectual disability (ID), learning disability (LD) and attention-deficit hyperactivity disorder (ADHD). NDDs have a combined prevalence of ~17% in children aged 3–17 years, making them the most common chronic medical conditions encountered in paediatric primary care.2 Given their clinical and aetiological heterogeneity, the cause of NDDs can be challenging to diagnose. Comprehensive clinical care includes investigations aimed at elucidating the underlying cause, such as brain imaging and laboratory tests, including genetic testing.
Identifying the genetic aetiology of an NDD is of clinical and personal utility.3 Increasingly, accurate genetic diagnosis can lead to tailored therapy4–8 (and may provide information about prognosis that in turn can guide medical care and therapy.8 Definitive diagnosis may avoid unnecessary testing or treatments,5 9 10 enable surveillance for known comorbidities5 9–16 and help families access additional disease-specific support.9 17–19 Accurate diagnosis of the cause of NDDs facilitates genetic counselling and informs recurrence risks5 9 10 12 13 16 20 that guides reproductive decision making and enables prenatal diagnosis.6 When an inherited aetiology is identified, this can provide diagnoses to other family members with NDD.21 Finally, by ending the diagnostic odyssey for families, identifying an aetiology can also provide psychological benefits (eg, alleviation of guilt, acceptance, closure, empowerment)5 13 17 18 22 23 and improve parental quality of life.24
The diagnostic process begins with a thorough clinical assessment of the patient, which includes a medical, developmental and family history with physical examination. The presence or absence of certain clinical features may provide clues as to the underlying cause and therefore direct the appropriate investigations. If a known genetic condition is recognised, targeted testing should be done first. For example, if a 2-year-old girl presents with classical features of Rett syndrome, it is prudent to do MECP2 sequencing prior to other investigations. However, it is more often the case that the clinical presentation is non-specific, and thus there are a large number of potential genetic aetiologies. It is therefore important to broaden the testing approach, which could result in earlier diagnosis.
When a known syndrome is not readily apparent, there are a variety of testing options, including chromosomal microarray (CMA), fragile X syndrome (FXS) testing, testing for a range of inherited metabolic diseases (‘metabolic testing’) and tests that use next-generation sequencing such as multigene panels and exome sequencing (ES). Professional organisations from a range of disciplines recommend CMA as a first-tier test for children with GDD, ID and ASD.25–30 While fragile X testing is generally recommended as a first-tier test for these NDDs, criteria for testing are inconsistent.25 27 28 31–33 Recommendations vary considerably for metabolic testing,25–28 32–35 while the use of gene panels and ES is relatively new and typically not included in recommendations aimed at non-geneticists. Not surprisingly, a recent survey of Canadian genetic and metabolic physicians found that CMA and fragile X testing are the most often used tests for patients with NDDs, while the use of metabolic tests, gene panels and ES was highly variable.36 Therefore, Canadian recommendations for genetic investigation of NDDs are urgently needed.
Definitions
Clinical definitions
ADHD: a persistent pattern of inattention and/or hyperactivity impulsivity that interferes with functioning or development.
ASD: an NDD characterised by persistent deficits in social communication and social interaction and the presence of restricted, repetitive patterns of behaviour, interests or activities. May be diagnosed with or without accompanying intellectual or language impairment.
Dysmorphic features: visible morphological findings that differ from those commonly seen in the general population or same genetic ancestry.
GDD: an NDD characterised by significant delay (at least two SD below the mean using standardised testing) in achieving at least two developmental domains, including gross or fine motor, speech and language, cognition, social and personal functioning, and activities of daily living. A diagnosis of GDD applies to individuals under the age of 5 years who are too young to participate in standardised testing to assess intellectual functioning.
IMDs: genetic disorders that result in metabolic defects due to deficiency of enzymes, membrane transporters or other functional proteins.
ID: an NDD characterised by deficits in intellectual and adaptive functioning. Intellectual functioning includes reasoning, problem solving, planning, abstract thinking, judgement, academic learning and experiential learning. Adaptive functioning are the skills needed to live independently and responsibly and include communication, social skills, personal independence at home or in the community and school or work functioning.
LD: also known as specific learning disorder; an NDD characterised by deficits in the ability to learn or use foundational academic skills (reading, writing or arithmetic), which is not better accounted for by ID.
NDD: a group of conditions manifesting early in development, characterised by impairments of personal, social, academic or occupational functioning.
Syndromic: for an individual with an NDD, refers to the presence of additional clinical features such as dysmorphisms, congenital malformations, medical comorbidities etc, which may suggest an underlying genetic cause (refer to table 1).
Table 1.
Clinical features that may be suggestive of a syndromic aetiology for patients with neurodevelopmental disorders
| Clinical feature | Definitions and/or examples |
| Dysmorphic features | Visible morphological findings that differ from those commonly seen in the general population or same genetic ancestry. For example, hypertelorism and syndactyly. |
| Congenital malformations | A non-progressive morphological anomaly of a single organ or body part that is present at birth. For example, cleft palate, tetralogy of Fallot and polydactyly. |
| Abnormal head size | Occipitofrontal circumference less than or greater than 2 SD from the mean for age, sex and ethnicity. For example, microcephaly and macrocephaly. |
| Unexplained growth abnormalities | Growth parameters greater than or less than 2 SD from the mean for age, sex and ethnicity, particularly if parental heights are average. For example, prenatal growth restriction, postnatal failure to thrive, short stature and overgrowth. |
| Family history consistent with Mendelian inheritance | There are similarly affected individuals related to the proband in a pattern suggestive of an autosomal recessive, autosomal dominant or X-linked condition For example, siblings with similar phenotype and consanguineous parents (may suggest autosomal recessive condition); affected males related through maternal line with similar phenotype (may suggest X-linked inheritance). |
| Additional medical comorbidities | Medical conditions, particularly when multiple or uncommon, that are not expected to be present secondary to the neurodevelopmental disorder itself. For example, sensorineural hearing loss, vision impairment, renal disease, epilepsy, ataxia and neuromotor deficits. |
Technical definitions
Next-generation sequencing (NGS): high-throughput sequencing technology used to determine nucleotide sequences and genome dosage at numerous loci simultaneously.
-
Genome-wide sequencing (GWS): sequence analysis of all or a large part of the genome performed by NGS; includes exome and ES.
ES: an NGS approach that determines the DNA sequence of most of the protein-coding exons found in the genome of an individual. As of 2022, this test is available in most provinces for patients meeting specific clinical criteria.
Genome sequencing (GS): an NGS approach that determines the sequence of most of the DNA content encompassing the entire genome of an individual. As of 2022, this test is only available on a research basis or through clinical pilots within Canada.
Multigene panel: simultaneous sequencing of multiple genes associated with a specific clinical presentation.
First-tier test: a laboratory test or other diagnostic investigation that should be pursued prior to all others.
Second-tier test: a laboratory test or other diagnostic investigation that should be pursued if first-tier tests are negative or inconclusive.
Metabolic testing: blood and/or urine tests that measure analytes (such as amino and organic acids) in order to identify patterns indicating an inherited metabolic disease.
Methods
To develop recommendations, the Canadian College of Medical Geneticists (CCMG) formed an NDDs working group of physicians and clinical scientists from medical genetics and genomics, developmental paediatrics, paediatric neurology, biochemical genetics, cytogenetics and molecular genetics. The CCMG is a Canadian organisation responsible for certifying medical geneticists and clinical laboratory geneticists and for establishing professional and ethical standards for clinical genetics services in Canada.
The working group considered laboratory tests currently clinically available in Canada to investigate the aetiology of NDDs, organised into three subgroups: (1) CMA and FXS testing, (2) metabolic testing and (3) NGS-based testing including ES and multigene panels. Each subgroup, in consultation with the broader working group, identified key questions, conducted a literature search, reviewed and discussed the evidence and drafted recommendations based on quality and level of evidence. The working group reviewed all draft recommendations and reached consensus.
The draft manuscript was reviewed by subcommittees of the CCMG (laboratory practice, clinical practice and metabolics committees). It was then circulated for comment to the CCMG membership-at-large and, following incorporation of feedback, approved by the CCMG Board of Directors. This position statement was also circulated for comment to the Mental Health and Developmental Disabilities committee of the Canadian Pediatric Society.
Considerations
Target audience and scope
These recommendations apply to patients with NDDs for whom no specific aetiological diagnosis is suspected after a thorough history and physical examination. If a specific genetic disorder is suspected with a high degree of clinical certainty, targeted testing should be done first, and if negative, then one should proceed with the testing recommended in this position statement. If no diagnosis is reached after the recommended investigations, periodic re-evaluation of the patient by a geneticist is recommended, as testing technology and genomic knowledge is rapidly evolving. The time interval for re-evaluation is specific to the patient’s unique situation (eg, age and testing done) and should be determined by the geneticist.
We acknowledge that the demand for genetic consultation is high, and waitlists are often long for non-urgent referrals. Thus, we provide recommendations for first-tier testing that primary care and/or non-genetic specialist clinicians can order while the patient awaits a clinical genetics or metabolic assessment to ensure faster access to genetic testing. The non-geneticist should have a good understanding of the benefits and limitations of first-tier tests and should obtain informed consent for testing from the patient or their guardian(s). Should first-tier testing be abnormal, a referral to the appropriate specialist is indicated, and many genetic/metabolic clinics will expedite consultation for patients referred with an abnormal result. We also provide guidance as to which patients should be referred to genetics if first-tier testing is not diagnostic. The role of the geneticist is to determine whether second-tier testing is indicated to ensure that the patient and family have an opportunity to discuss the test results and to make recommendations for follow-up care if a diagnosis is made.
The evidence reviewed for the development of this position statement applies specifically to patients with GDD, ID and/or ASD. The existing evidence was highly heterogeneous with respect to the clinical phenotypes of the patients studied, so it was not possible to make separate recommendations for patients with and without syndromic features, for patients with different severities of impairment or for patients in different age groups (paediatric or adult). Evidence was also reviewed for patients with other NDDs (eg, ADHD and LD), but there are no high-quality studies that enable distinction between patients with and without comorbid GDD or ID. Therefore, until such evidence emerges, we do not recommend genetic investigations for patients without GDD, ID or ASD unless there are other clinical features suggestive of a genetic (‘syndromic’; see table 1) or metabolic aetiology.
The recommendations herein apply to probands of all ages. The urgency of testing is typically higher for younger children, particularly if the parents are planning a pregnancy. This does not preclude testing and/or genetic/metabolic specialist referral for adolescents and adults, particularly if the patient has not been tested or evaluated in 5 years or more.
The recommendations herein also apply to probands with ID regardless of severity, as there was insufficient evidence to exclude probands from such testing based on level of disability. Further studies addressing this are required.
There may be specific clinical scenarios that demand a different approach to that suggested herein, so clinicians should always use their judgement as to whether the recommended investigations are appropriate. Patient and family preferences should always be considered. This position statement is not intended to be a comprehensive guide to the medical investigation of individuals with NDDs, and as such, we do not make recommendations regarding neuroimaging or health surveillance (eg, thyroid screening, vision assessment, etc).
The Canadian context
Canada’s population of nearly 39 million is concentrated in the southern parts of Ontario, Quebec and Alberta, with relatively sparse population density in the rest of the country. Twenty-one per cent of the population lives in rural or remote areas with limited access to specialist medical care (Canadian Medical Association Policy Statement, 2014). The Canada Health Act stipulates that all Canadians should have universal insurance coverage for medically necessary healthcare service. Healthcare is a provincial and territorial responsibility, and nationwide there is variability regarding the types of genetic and metabolic tests that are publicly funded. There is also some variability in the conditions screened for by newborn screening. The most expensive and resource-intensive genetic tests, such as ES, are not always accessible due to provincial healthcare insurance exclusions or limited access to specialists qualified to order and interpret this testing. The working group has taken these inequities into consideration, and where possible, we have offered alternatives, such as recommending multigene panel testing when ES is not available.
Limitations
The working group formulated the recommendations using the best available evidence at the time of writing (2022). The quality of the evidence, however, generally falls at a level II–IV (moderate) based on the GRADE system,37 so these recommendations should be considered conditional on completion of higher quality studies. Expert opinion was used to fill in the gaps where evidence was lacking.
It was beyond the scope of the working group to conduct an independent cost-effectiveness analysis as part of the development of this position statement. Recognising that resources are finite, these recommendations attempt to strike a balance between avoiding over-testing and the risk of missing a diagnosis that may have treatment or recurrence risk implications for the patient and their family.
Finally, input from patients and their families was not solicited in the development of these recommendations, but for future iterations of this statement, this will be an important addition.
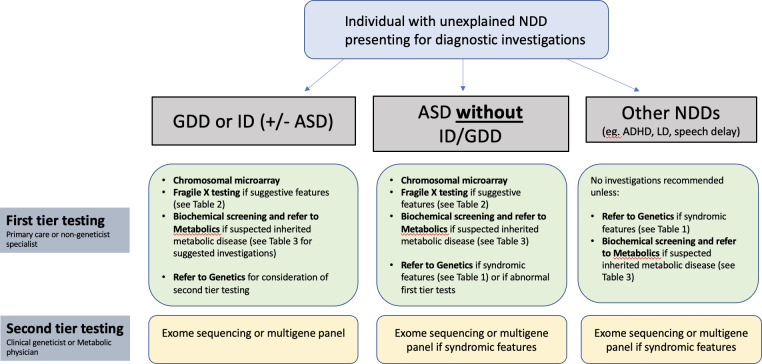
The sections further will briefly summarise the evidence for each test modality and list recommendations for use of the test as first or second tier for patients with NDDs. It should be understood that first tier tests are, for the most part, able to be ordered by any qualified clinician and therefore should generally be done prior to (or at the same time as) referral for genetics consultation. Figure 1 summarises the recommendations and provides guidance as to when a patient should be referred to genetics or metabolics for further evaluation.
Figure 1.
Summary of first and second tier testing recommendations for individuals with neurodevelopmental disorders presenting for diagnostic investigations. ADHD, attention-deficit hyperactivity disorder; ASD, autism spectrum disorder; GDD, global developmental delay; ID, intellectual disability; LD, learning disability.
Fragile X testing (FMR1 CGG repeat analysis)
FXS) is an X-linked condition caused by the unstable expansion of a CGG repeat in the 5′UTR of the FMR1 gene and subsequent hypermethylation, preventing gene expression. It is a common monogenic cause of NDDs, with a prevalence of 1.4 per 10 000 males and 0.9 per 10 000 females.38 FXS testing is widely available and can be ordered by a variety of clinicians. FXS cannot, at this time, be diagnosed by CMA or ES.
The ‘classic’ FXS phenotype in males includes moderate-to-severe ID, macro-orchidism and distinctive facial features (long face, large ears and prominent jaw).39 Common comorbid conditions include ASD, anxiety and hyperactivity.40 Deviation from the classical phenotype is influenced by age and sex (with females and prepubescent children manifesting fewer physical characteristics) and the presence of DNA methylation or repeat size mosaicism.41 The phenotype in females is highly variable, with IQ ranging from normal to moderate ID, likely due to the effects of random X-inactivation.42 FXS has a high recurrence risk in families, as children with a full mutation have mothers with a premutation that confers a risk for expansion in the oocyte,43 or with a full mutation. If the diagnosis of FXS is delayed, parents may not have the opportunity to have prenatal diagnosis in subsequent pregnancies, which may result in multiple affected children.44 For these reasons, a variety of professional organisations recommend FXS testing as a first-line diagnostic test (concurrent with CMA) for GDD, ID and ASD.27 45 46 These recommendations were largely based on expert opinion and/or published prior to publication of larger studies examining the diagnostic yield of NGS-based testing for NDDs.
The diagnostic yield of fragile X testing varies between studies, based on the study design and characteristics of the study population. Taken together, the sex-specific prevalence of FXS in individuals with NDD is 2.2%–2.5% for males and 1.3%–1.6% in females.38 47–53 Retrospective reviews of laboratory databases demonstrate a diagnostic yield of 0%–2.5% for FXS testing.54–57 The diagnostic yield of FXS testing increases (9.5%–17%) when inclusion criteria are restricted to males with NDD and characteristic physical and behavioural features or family history suggestive of FXS.58–60 Use of a diagnostic ‘checklist’ to determine likelihood of FXS in males generally has high sensitivity, but only if a low threshold is used.58 60–64 There is insufficient data to determine whether this approach yields a higher diagnostic rate in females with NDDs.60–62 No studies have evaluated the cost-effectiveness of selective FXS testing based on clinical criteria.
Recommendations for fragile X testing
-
FXS testing is recommended as a first-tier diagnostic test for individuals presenting with:
GDD, ID or ASD and a clinical presentation or family history suggestive of FXS (see table 2).
Any NDD and a family history of FXS or other FMR1-related disorder.
FXS testing is not recommended for individuals with GDD or ID who do not meet the above criteria and have a complex clinical presentation that is not consistent with FXS (eg, multiple congenital anomalies, profound neurological impairment).
Table 2.
Clinical features that may be suggestive of fragile X syndrome
| Proband history and physical exam | Family history |
| Macro-orchidism (may not be present until after puberty). Relative or mild (+2–3 SD) macrocephaly. Facial features: large or prominent ears, long or narrow face, tall forehead, high-arched palate and prominent jaw. Connective tissue findings: soft or velvety skin, redundant skin on dorsum of hands, hyperextensible joints, pes planus and mitral valve prolapse. Behaviour: ASD or autistic features, hyperactivity, shyness, gaze avoidance, hand biting, tactile defensiveness and anxiety. |
In relatives on the maternal side:
|
ASD, autism spectrum disorder; GDD, global developmental delay; ID, intellectual disability.
Chromosomal microarray
CMA uses comparative genomic hybridisation or SNP array to detect gains and losses of chromosomal material, known as CNVs. SNP-based arrays are also able to detect areas of long contiguous stretches of homozygosity, which may suggest parental consanguinity or uniparental disomy. While CMA can detect CNVs as small as approximately 20–50 kb,29 resolution varies depending on the technology used and density of DNA probes. CMA is widely available and can be ordered by a variety of clinicians.
CNVs can be classified as pathogenic, likely pathogenic, uncertain significance, likely benign or benign.65 CNVs of uncertain significance require interpretation from a clinical geneticist and may warrant additional investigations. CMA may detect ‘susceptibility CNVs’: recurrent CNVs with variable expressivity and/or incomplete penetrance.66–68 The phenotype resulting from these CNVs is variable and likely influenced by other genetic and environmental factors. Detection of these variants in an individual with a neurodevelopmental disorder may not entirely explain the observed phenotypes, so additional investigations may be warranted; however, this is not well studied.
A variety of professional organisations recommend CMA as a first-tier test for individuals with GDD, ID or ASD,27 28 32 33 69 based on its high diagnostic yield compared with standard karyotype.29 70 In general, the diagnostic yield of CMA is lower in studies in which the probands have mild ID (12%–19% vs 20%–30% for moderate to severe ID71–73) or ASD without syndromic features (4%–5% for non-syndromic ASD vs up to 25% for those with syndromic features74 75).
There are no studies that investigate the diagnostic yield of CMA for isolated speech or motor delay or LD. Although three studies report a detection rate of 8%–9% for rare CNVs in individuals with ADHD,76–78 they lack sufficient clinical information about the probands to determine whether ADHD was isolated or associated with GDD/ID or syndromic features. Furthermore, in these studies, many of the detected rare CNVs were inherited and the pathogenicity and clinical impact of these CNVs were unclear. Another study found that children with isolated ADHD had similar detection of rare CNVs compared with controls.79
There is strong evidence for the utility of CMA as a first-tier test for individuals with GDD, ID or ASD. There is insufficient evidence for the utility of CMA as a first-tier test for other NDDs such as ADHD without GDD, ID or ASD unless there are other clinical features suggestive of a syndromic aetiology (see table 1).
Recommendations for CMA testing
-
CMA is recommended as a first-tier diagnostic test for individuals presenting with:
GDD, ID or ASD.
Other NDDs when syndromic features are present (see table 1).
Metabolic testing
Inherited metabolic disorders (IMDs) comprise a group of genetic conditions that can present with NDDs as part of the clinical spectrum. Metabolic testing can detect the presence or absence of biochemical markers that lead to a diagnosis of IMD. While individually rare, there can be clinical value in diagnosing IMDs: for some disorders, early identification and treatment can improve outcomes, and most of the conditions are autosomal recessive and therefore have a high recurrence risk. According to a systematic literature review, over 100 IMDs with ID as a major feature have potential treatments.80 81 Examples of IMDs causing NDD that have disease-modifying treatments are urea cycle disorders,82 83 maple syrup urine disease,82 homocystinurias, including cobalamin-related conditions,84–86 and some creatine disorders including guanidinoacetate methyltransferase deficiency.82 84 86 87
Some authors suggest that all individuals presenting with GDD/ID/ASD should be screened for IMDs,27 28 32–35 88 while others recommend screening only for individuals with additional clinical features suggestive of an IMD.25 26 84 There is also a lack of consensus among experts as to which metabolic investigations comprise a comprehensive screen for IMDs. Indeed, there is wide variability in Canadian clinical practice.36
Both the Canadian Pediatric Society27 and the American Academy of Pediatrics28 reference a systematic literature review80 as the rationale to screen for treatable IMDs in all children presenting with GDD/ID; however, the diagnostic yield of this approach was unknown at the time. The protocol suggested by the Treatable Intellectual Disability Endeavor (TIDE) (www.tidebc.org) is extensive; yet, not all the tests in this protocol are widely available, and clinical utility and cost-effectiveness have not been evaluated.
The reported diagnostic yield of metabolic testing for patients with NDDs can vary depending on the population studied. However, using the best evidence available (more recent publications in populations with expanded newborn screening, larger case series with clear case definitions), the overall diagnostic yield of metabolic testing for patients with NDDs is 0.25%–0.42% for GDD/ID in non-consanguineous populations.84 87 Retrospective reviews of the medical records of patients with diagnosed IMDs show that most of these patients have clinical features suggestive of an IMD in addition to GDD/ID.84 87 89 90 In a prospective study by Campistol et al 83 in 2016, an IMD diagnosis was rarely identified in children with non-syndromic ASD. A review of the pilot implementation of the TIDE protocol for patients with GDD/ID, with and without suggestive neurological features,34 demonstrated no significant increase in diagnoses of IMD during time periods before and after implementation despite a greater than fourfold increase in test volumes.84 Metabolic testing early in the diagnostic process should theoretically improve ‘time to diagnosis’ and clinical outcomes for IMDs not detected by newborn screening; however, this has not been rigorously studied.
Reviews, commentaries and guidelines that quote diagnostic yields of metabolic investigations for NDDs as high as 5% should be interpreted with caution. Literature examining the diagnostic yield of metabolic testing for patients with NDDs is generally of low quality due to lack of peer review, small sample sizes, variability in tests performed and lack of robust case definitions. Furthermore, some publications pre-date the expansion of newborn screening to detect more treatable IMDs.87 Although most children with treatable IMDs are now diagnosed by routine provincial newborn screening programmes in Canada, newborn screening cannot detect all treatable IMDs, neonatal screening programmes vary among provinces/territories and false-negative screens are possible.27 84
For children with NDDs, most IMD diagnoses are made in those with clinical features such as regression or plateauing of development, acute encephalopathy or altered level of consciousness, movement disorders, intractable seizures, multisystemic involvement, specific ophthalmological findings, organomegaly and/or features suggestive of a storage disease. Patients with such ‘red flags’ should be referred for specialist consultation without delay. Table 3 provides guidance on additional investigations that should be ordered, where possible, for patients with these features while they await consultation.
Table 3.
Clinical features suggestive of inherited metabolic disorders (IMDs) and suggested metabolic testing that could be done while awaiting metabolics (or other specialist) consultation
| Clinical feature | Major groups of IMDs to consider | Targeted metabolic testing If URGENT, clearly indicate on the requisitions when ordering tests |
| Developmental plateau or regression in the context of an abnormal neurological exam | LSD, peroxisomal disorders (X-ALD), UCD, HCYS | Ammonia, blood gas, lactate, PAA, UOA, TPH, urine MPS, VLCFA† |
| Altered level of consciousness, especially if episodic; stroke-like episodes | UCD, MSUD, HCYS, organic acidurias, mitochondrial disorders | Ammonia, blood gas, glucose, lactate, electrolytes, anion gap, PAA, TPH, ACP, UOA |
| Movement disorder (ataxia, dystonia, choreoathetosis, myoclonus, tremor) | Organic acidurias, HCYS, creatine disorders, LSD, Wilson disease | Blood gas, lactate, glucose, ACP, PAA, UOA, TPH, urine MPS, creatine panel†, copper, ceruloplasmin |
| MRI/MRS brain abnormality (eg, white matter change, abnormal cerebellum / basal ganglia) | Peroxisomal disorders, organic acidurias, LSD | Metabolic testing tailored to MRI findings by metabolic/genetic specialist |
| Hepatomegaly, splenomegaly* | LSD, peroxisomal disorders | Urine MPS (if other systemic abnormalities noted*), VLCFA† |
| Specific food aversions: avoiding high protein foods | UCD, MSUD and organic acidurias | PAA, ammonia, UOA, ACP |
| Ophthalmological findings (most common: cataracts, dislocated lens, corneal depositions, retinopathy, cherry red spot) | Galactosaemia, Lowe disease, sulfite oxidase deficiency, HCYS, LSD, peroxisomal disorders | Metabolic testing tailored to ophthalmological findings by metabolic/genetic specialist |
| Seizures: drug resistant, myoclonic or neonatal | UCD, organic acidurias, HCYS, creatine disorders, vitamin-dependent epilepsies, peroxisomal disorders, Menkes disorder | Ammonia, blood gas, glucose, lactate, electrolytes, anion gap, PAA, UOA, TPH, creatine panel†, VLCFA† |
| Abnormal tone (hypotonia or spasticity) | Organic acidurias, HCYS, creatine disorders, UCD (eg, arginase deficiency and HHH), biotinidase deficiency, peroxisomal disorders, LSD | Ammonia, blood gas, glucose, lactate, electrolytes, anion gap, PAA, UOA, TPH, VLCFA†, creatine panel† |
| Coarse facial features and/or skeletal abnormalities on X-ray* | MPS and other storage disorders | Urine MPS |
| Multisystemic involvement | LSD, mitochondrial disorders, peroxisomal disorders, CDG, SLOS | Metabolic testing tailored to specific findings by metabolic/genetic specialist |
X-linked ALD (adrenoleukodystrophy).135–137
Highly specialised tests that are not recommended for initial workup are not included.
*For mucopolysaccharidoses (MPS), look for noisy breathing, short stature, macrocephaly and sensorineural hearing impairment. For MPS III (Sanfilippo syndrome), aggressive behaviour, sleep disturbance and hirsutism may be early features.
†Test ordering may be restricted to certain specialists.
ACP, acylcarnitine profile; CDG, congenital disorders of glycosylation; HCYS, homocystinuria and remethylation/cobalamin disorders; HHH, hyperornithinaemia hyperammonaemia homocitrullinuria syndrome; LSD, lysosomal storage disorder; MRS, Magnetic resonance spectroscopy; MSUD, maple syrup urine disease; PAA, plasma amino acids; SLOS, Smith-Lemli-Opitz syndrome; TPH, total plasma homocysteine; UCD, urea cycle disorder; UOA, urine organic acids; VLCFA, very long chain fatty acids.
Given its very low yield, routine metabolic testing is not recommended for patients with GDD/ID/ASD without suggestive clinical features listed in table 3. Rarely, a treatable IMD may be missed by not performing metabolic testing for such patients; for example, patients with homocystinurias, including cobalamin-related conditions, can occasionally present with isolated NDDs.85 However, these conditions are very rare, and those with time-sensitive treatments are detected by newborn screening. Creatine transporter disorder, which is X-linked, may rarely present with isolated NDD.84 90 91 Treatment does not alter clinical outcomes for this condition; however, diagnosis might be helpful for future family planning.
Clinicians should be aware of the conditions tested for via newborn screening in their jurisdiction. Children who have not had newborn screening may be at increased risk for having an undiagnosed IMD, so clinicians should determine what newborn screening was done and, if limited, consider referring to a metabolic specialist.
Recommendations for metabolic testing
-
Metabolic testing is recommended as a first-tier diagnostic test for individuals presenting with:
GDD, ID, or ASD and clinical features suggestive of an IMD. Metabolic testing should be tailored to the presentation (see table 3) and a referral made to a metabolic specialist promptly.
Metabolic testing is not recommended for individuals presenting with ASD/GDD/ID without suggestive features unless newborn screening was not performed or the test panel was not comparable with standard provincial programmes (eg, newcomers to Canada).
ES and multigene panels
Massively parallel ‘next-generation’ sequencing technology has allowed for the simultaneous analysis of hundreds to thousands of genes from a single DNA sample. Currently, ES is widely available as a clinical diagnostic test. An alternative to ES is the use of comprehensive multigene panels, which are targeted to a specific phenotype. For NDDs, commercially available panels vary widely in the number of genes analysed; the largest ones include over 2500 genes.92 In future, GS is poised to replace both ES and CMA, but as it is not yet publicly funded in Canada, these recommendations consider the use of panels and ES only.
The main advantage of ES and large panels is the ability to simultaneously interrogate large numbers of genes, which means that the clinician need not have a particular genetic condition in mind to direct the diagnosis. However, they have important limitations that means that a negative test does not rule out a genetic cause. Depending on the methodology used, all coding exons may not be completely sequenced and, thus, some variants may be missed.93 ES is less able to detect mosaicism or exon-level deletions compared with GS or panels that include deletion/duplication analysis.94 ES and panels do not reliably assess repetitive DNA sequences (such as trinucleotide repeats), intronic or non-coding variants, methylation, epigenetic or mitochondrial DNA variants or balanced chromosomal rearrangements.94
These tests can identify multiple rare variants in a patient’s sample, the clinical relevance of which can be challenging to decipher. They can identify misattributed parentage (when done as a ‘trio’ with both declared parents),95 96 as well as incidental and secondary findings (pathogenic variants in medically actionable genes unrelated to the indication for testing).97 Furthermore, they can identify misattributed parentage (when done, as they often are, as a ‘trio’ with both declared parents) and secondary findings (pathogenic variants in medically actionable genes unrelated to the indication for testing).95 96 Testing of the proband only (vs proband-parent trio) results in a lower diagnostic yield98 99 and more labour-intensive downstream analysis and/or additional testing to resolve variants, so a ‘trio’ approach is the most useful, time-efficient and informative approach.98 100 Given these considerations, these tests require robust pretest and post-test genetic counselling and input from a clinical geneticist.97 101
ES and comprehensive multigene panels were not widely available as routine clinical tests when the practice guidelines for genetic testing for patients with NDDs were published by the American Association of Neurology in 201126 and the American Association of Pediatrics in 2014.28 However, a 2015 CCMG position statement recommended clinical GWS (either exome or genome sequencing) for patients with moderate-to-severe or syndromic ID, when appropriate first-tier genetic testing (such as CMA) is non-diagnostic.102 The 2021 ACMG practice guidelines103 strongly recommend that GWS be considered as a first-tier or second-tier test for paediatric patients with developmental delays and/or ID.
Diagnostic yield of ES for patients with NDDs
The reported diagnostic yield of ES for patients with GDD/ID demonstrates wide variation (8%–68%),9 16 74 98 103–118 likely owing to differences in cohort size and composition, whether ES was performed clinically or under a research protocol and whether a proband-parent trio or proband-only approach was used. When considering the largest studies from clinical laboratories,105 106 the diagnostic yield of ES is 26%–31%, consistent with results from a meta-analysis that showed pooled estimates of 29%.104 It is difficult to tease out the diagnostic yield in specific patient subgroups (such as ASD without GDD/ID, or GDD/ID without syndromic features). A few studies dedicated to patients with ASD, most of whom had ID (with or without other features), reported that ES had a diagnostic yield of 8%–25.8%.74 106 109–111 119 The few studies that have compared the diagnostic yield of ES for GDD/ID patients with and without syndromic features have not found statistically significant differences.107 120 121
Diagnostic yield of NGS panels for patients with NDDs
A few studies have examined comprehensive panels for patients with NDDs, reporting a diagnostic yield of 11%–39%.118 122–124 Two studies directly compared a targeted NDD gene panel to ES, and both demonstrated slightly lower diagnostic yields for the panel117 120
Clinical utility of ES for individuals with NDDs
The clinical utility of ES for the diagnosis of patients with suspected monogenic disorders has been well established. A 2020 Ontario Health Technology Assessment examined the clinical utility of ES in unexplained developmental disabilities and multiple congenital anomalies, concluding that ES allows for changes in clinical management, provides insight into the natural history of the patient’s condition, informs reproductive planning and optimises the patient’s ability to access disease-specific supports.99 Similar findings were reported in a 2020 ACMG systematic evidence review that examined the health, clinical, reproductive and psychosocial outcomes resulting from ES for the investigation of congenital anomalies, developmental delay or ID.125 It also considered possible negative impacts such as insurance discrimination, financial burden and psychological impact on patients and found that return of ES test results incur no clinically significant psychological harms with low levels of test-related distress and positive psychological effects. Genetic counselling and an appropriate consenting process can help mitigate any associated risks.103
Cost-effectiveness of ES
The significant cost of ES has historically been a barrier to its implementation within the Canadian healthcare system as standard-of-care testing for NDDs. Recent evidence shows that performing ES early in the diagnostic testing pathway is cost-effective,99 113 126–131 although other studies have been inconclusive.132–134 In Ontario, a 2020 cost analysis of ES testing before or concurrently with CMA for patients with developmental disabilities supported ES as a second-tier test (when CMA is non-diagnostic) as the most cost-effective approach99; this study is based on ES analysis that does not include duplication/deletion analysis.
In summary, the evidence strongly supports using ES as a diagnostic test for patients with ID/GDD due to its high diagnostic yield, cost-effectiveness and demonstrable clinical utility. The diagnostic yield for patients with GDD/ID is significant even in the absence of syndromic features. For patients with ASD without GDD/ID, there is insufficient data to make evidence-based recommendations, although evidence suggests a higher diagnostic yield for patients with syndromic features (table 1). A trio-based testing approach is recommended. While ES provides a higher diagnostic yield, a comprehensive multigene panel for NDDs is a suitable substitute when ES is not available, especially when done as a trio-based test. When GS becomes a widely available clinical test, it is anticipated to replace fragile X, CMA and ES as a single test of choice for NDDs.
In most cases ES is recommended as a second-tier test after CMA, as the latter has a high diagnostic yield, is more widely available, less costly and can be ordered by non-geneticists. However, it may be appropriate to do GWS before or concurrently with CMA; for example, when the proband is highly likely to have an autosomal recessive condition (based on parental consanguinity and/or affected siblings), or when a diagnosis must be rapidly obtained, such as to inform the management of an ongoing pregnancy (for a sibling of the proband), or to guide medical care in a patient who is acutely ill or whose clinical status is deteriorating (eg, neurological regression).
Recommendations for ES or multigene panels
-
Recommended as a second-tier diagnostic test for individuals presenting with:
GDD or ID (with or without ASD).
ASD and/or other NDD and clinical features suggestive of a syndrome (see table 1).
Conclusions
Patients with NDDs in Canada require a thorough evaluation for the underlying cause of their condition. The CCMG Neurodevelopmental Disorders working group recommends an approach to the diagnostic evaluation that is tailored to the patient and based on current evidence. These recommendations are based on best available evidence as of 2022 and may evolve as new evidence emerges. Therefore, they should be reviewed every 2–3 years.
An overview of these recommendations are found in figure 1. We recommend a tiered testing strategy, such that first-tier testing is ordered by non-geneticists (family doctors, paediatricians, neurologists, etc), to allow patients faster access to testing and potentially a rapid diagnosis. The provision of genetic testing by non-specialists should be supported with enhanced genetics education at all levels of medical training. We provide guidelines for when a patient should be referred to a specialist in genetics or metabolics: specifically, anyone with GDD/ID (regardless of other features), as well as patients with ASD or other NDDs if they have syndromic features (table 1) or features suggestive of an inherited metabolic disease (table 3).
Footnotes
Twitter: @hvallance
Correction notice: This article has been corrected since it was published online first. The placement of tables 2 and 3 has been corrected within the article.
Contributors: MTC, MS and HV were responsible for identifying the literature, summarising subgroup meetings and drafting their test-specific sections. MTC, HV and HH were responsible for drafting the manuscript. All authors contributed to literature review, the conception and design of the position statement, interpretation of data, writing and critical revisions contributing to the intellectual content and approval of the final version of the manuscript.
Funding: HH was supported with funding from the Canadian College of Medical Geneticists. SMEL is supported by an Investigator Grant Award through BC Children’s Hospital Research Institute.
Competing interests: TNN: spouse employed by Illumina.
Provenance and peer review: Not commissioned; externally peer reviewed.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
Not applicable.
References
- 1. American Psychiatric Association . Diagnostic and statistical manual of mental disorders. 5th edn. American Psychiatric Association, 2013. 10.1176/appi.books.9780890425596 [DOI] [Google Scholar]
- 2. Zablotsky B, Black LI, Maenner MJ, et al. Prevalence and trends of developmental disabilities among children in the United States: 2009-2017. Pediatrics 2019;144:e20190811. 10.1542/peds.2019-0811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Savatt JM, Myers SM. Genetic testing in neurodevelopmental disorders. Front Pediatr 2021;9:526779. 10.3389/fped.2021.526779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dixon-Salazar TJ, Silhavy JL, Udpa N, et al. Exome sequencing can improve diagnosis and alter patient management. Sci Transl Med 2012;4:138ra78. 10.1126/scitranslmed.3003544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. ACMG Board of Directors . Clinical utility of genetic and genomic services: a position statement of the American College of medical genetics and genomics. Genet Med 2015;17:505–7. 10.1038/gim.2015.41 [DOI] [PubMed] [Google Scholar]
- 6. Fung JLF, Yu MHC, Huang S, et al. A three-year follow-up study evaluating clinical utility of exome sequencing and diagnostic potential of reanalysis. NPJ Genom Med 2020;5:37. 10.1038/s41525-020-00144-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tărlungeanu DC, Novarino G. Genomics in neurodevelopmental disorders: an avenue to personalized medicine. Exp Mol Med 2018;50:100. 10.1038/s12276-018-0129-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Narcisa V, Discenza M, Vaccari E, et al. Parental interest in a genetic risk assessment test for autism spectrum disorders. Clin Pediatr (Phila) 2013;52:139–46. 10.1177/0009922812466583 [DOI] [PubMed] [Google Scholar]
- 9. Iglesias A, Anyane-Yeboa K, Wynn J, et al. The usefulness of whole-exome sequencing in routine clinical practice. Genet Med 2014;16:922–31. 10.1038/gim.2014.58 [DOI] [PubMed] [Google Scholar]
- 10. Niguidula N, Alamillo C, Shahmirzadi Mowlavi L, et al. Clinical whole-exome sequencing results impact medical management. Mol Genet Genomic Med 2018;6:1068–78. 10.1002/mgg3.484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coulter ME, Miller DT, Harris DJ, et al. Chromosomal microarray testing influences medical management. Genet Med 2011;13:770–6. 10.1097/GIM.0b013e31821dd54a [DOI] [PubMed] [Google Scholar]
- 12. Henderson LB, Applegate CD, Wohler E, et al. The impact of chromosomal microarray on clinical management: a retrospective analysis. Genet Med 2014;16:657–64. 10.1038/gim.2014.18 [DOI] [PubMed] [Google Scholar]
- 13. Hens K, Peeters H, Dierickx K. Genetic testing and counseling in the case of an autism diagnosis: a caregivers perspective. Eur J Med Genet 2016;59:452–8. 10.1016/j.ejmg.2016.08.007 [DOI] [PubMed] [Google Scholar]
- 14. Saam J, Gudgeon J, Aston E, et al. How physicians use array comparative genomic hybridization results to guide patient management in children with developmental delay. Genet Med 2008;10:181–6. 10.1097/GIM.0b013e3181634eca [DOI] [PubMed] [Google Scholar]
- 15. Riggs ER, Wain KE, Riethmaier D, et al. Chromosomal microarray impacts clinical management. Clin Genet 2014;85:147–53. 10.1111/cge.12107 [DOI] [PubMed] [Google Scholar]
- 16. Srivastava S, Love-Nichols JA, Dies KA, et al. Meta-Analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet Med 2019;21:2413–21. 10.1038/s41436-019-0554-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hayeems RZ, Babul-Hirji R, Hoang N, et al. Parents’ experience with pediatric microarray: transferrable lessons in the era of genomic counseling. J Genet Couns 2016;25:298–304. 10.1007/s10897-015-9871-3 [DOI] [PubMed] [Google Scholar]
- 18. Reiff M, Giarelli E, Bernhardt BA, et al. Parents’ perceptions of the usefulness of chromosomal microarray analysis for children with autism spectrum disorders. J Autism Dev Disord 2015;45:3262–75. 10.1007/s10803-015-2489-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stivers T, Timmermans S. The actionability of exome sequencing testing results. Sociol Health Illn 2017;39:1542–56. 10.1111/1467-9566.12614 [DOI] [PubMed] [Google Scholar]
- 20. Hayeems RZ, Hoang N, Chenier S, et al. Capturing the clinical utility of genomic testing: medical recommendations following pediatric microarray. Eur J Hum Genet 2015;23:1135–41. 10.1038/ejhg.2014.260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Trosman JR, Weldon CB, Slavotinek A, et al. Perspectives of US private payers on insurance coverage for pediatric and prenatal exome sequencing: results of a study from the program in prenatal and pediatric genomic sequencing (P3EGS). Genet Med 2020;22:283–91. 10.1038/s41436-019-0650-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mollison L, O’Daniel JM, Henderson GE, et al. Parents’ perceptions of personal utility of exome sequencing results. Genet Med 2020;22:752–7. 10.1038/s41436-019-0730-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krabbenborg L, Vissers LELM, Schieving J, et al. Understanding the psychosocial effects of WES test results on parents of children with rare diseases. J Genet Couns 2016;25:1207–14. 10.1007/s10897-016-9958-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lingen M, Albers L, Borchers M, et al. Obtaining a genetic diagnosis in a child with disability: impact on parental quality of life. Clin Genet 2016;89:258–66. 10.1111/cge.12629 [DOI] [PubMed] [Google Scholar]
- 25. Schaefer GB, Mendelsohn NJ, Professional Practice and Guidelines Committee . Clinical genetics evaluation in identifying the etiology of autism spectrum disorders: 2013 guideline revisions. Genet Med 2013;15:399–407. 10.1038/gim.2013.32 [DOI] [PubMed] [Google Scholar]
- 26. Michelson DJ, Shevell MI, Sherr EH, et al. Evidence report: genetic and metabolic testing on children with global developmental delay: report of the Quality Standards Subcommittee of the American Academy of Neurology and the practice Committee of the child Neurology Society. Neurology 2011;77:1629–35. 10.1212/WNL.0b013e3182345896 [DOI] [PubMed] [Google Scholar]
- 27. Bélanger SA, Caron J. Evaluation of the child with global developmental delay and intellectual disability. Paediatr Child Health 2018;23:403–19. 10.1093/pch/pxy093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moeschler JB, Shevell M, Committee on Genetics . Comprehensive evaluation of the child with intellectual disability or global developmental delays. Pediatrics 2014;134:e903–18. 10.1542/peds.2014-1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 2010;86:749–64. 10.1016/j.ajhg.2010.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Waggoner D, Wain KE, Dubuc AM, et al. Yield of additional genetic testing after chromosomal microarray for diagnosis of neurodevelopmental disability and congenital anomalies: a clinical practice resource of the american college of medical genetics and genomics (ACMG). Genet Med 2018;20:1105–13. 10.1038/s41436-018-0040-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sherman S, Pletcher BA, Driscoll DA. Fragile X syndrome: diagnostic and carrier testing. Genet Med 2005;7:584–7. 10.1097/01.gim.0000182468.22666.dd [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mithyantha R, Kneen R, McCann E, et al. Current evidence-based recommendations on investigating children with global developmental delay. Arch Dis Child 2017;102:1071–6. 10.1136/archdischild-2016-311271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. O’Byrne JJ, Lynch SA, Treacy EP, et al. Unexplained developmental delay/learning disability: guidelines for best practice protocol for first line assessment and genetic/metabolic/radiological investigations. Ir J Med Sci 2016;185:241–8. 10.1007/s11845-015-1284-7 [DOI] [PubMed] [Google Scholar]
- 34. van Karnebeek CD, Stockler-Ipsiroglu S. Early identification of treatable inborn errors of metabolism in children with intellectual disability: the treatable intellectual disability endeavor protocol in British Columbia. Paediatr Child Health 2014;19:469–71. 10.1093/pch/19.9.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Silove N, Collins F, Ellaway C. Update on the investigation of children with delayed development. J Paediatr Child Health 2013;49:519–25. 10.1111/jpc.12176 [DOI] [PubMed] [Google Scholar]
- 36. Carter MT, Cloutier M, Tsampalieros A, et al. Genetic and metabolic investigations for individuals with neurodevelopmental disorders: a survey of Canadian geneticists’ practices. Am J Med Genet A 2021;185:1757–66. 10.1002/ajmg.a.62167 [DOI] [PubMed] [Google Scholar]
- 37. Guyatt GH, Oxman AD, Vist GE, et al. Grade: an emerging consensus on rating quality of evidence and strength of recommendations. BMJ 2008;336:924–6. 10.1136/bmj.39489.470347.AD [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hunter J, Rivero-Arias O, Angelov A, et al. Epidemiology of fragile X syndrome: a systematic review and meta-analysis. Am J Med Genet A 2014;164A:1648–58. 10.1002/ajmg.a.36511 [DOI] [PubMed] [Google Scholar]
- 39. Kidd SA, Lachiewicz A, Barbouth D, et al. Fragile X syndrome: a review of associated medical problems. Pediatrics 2014;134:995–1005. 10.1542/peds.2013-4301 [DOI] [PubMed] [Google Scholar]
- 40. Abbeduto L, McDuffie A, Thurman AJ. The fragile X syndrome-autism comorbidity: what do we really know? Front Genet 2014;5:355. 10.3389/fgene.2014.00355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cohen IL, Nolin SL, Sudhalter V, et al. Mosaicism for the FMR1 gene influences adaptive skills development in fragile X-affected males. Am J Med Genet 1996;64:365–9. [DOI] [PubMed] [Google Scholar]
- 42. Bartholomay KL, Lee CH, Bruno JL, et al. Closing the gender gap in fragile X syndrome: review on females with FXS and preliminary research findings. Brain Sci 2019;9:11. 10.3390/brainsci9010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nolin SL, Glicksman A, Ding X, et al. Fragile X analysis of 1112 prenatal samples from 1991 to 2010. Prenat Diagn 2011;31:925–31. 10.1002/pd.2815 [DOI] [PubMed] [Google Scholar]
- 44. Gabis LV, Hochberg O, Leon Attia O, et al. Prolonged time lag to final diagnosis of fragile X syndrome. J Pediatr 2018;193:217–21. 10.1016/j.jpeds.2017.10.008 [DOI] [PubMed] [Google Scholar]
- 45. Hyman SL, Levy SE, Myers SM, et al. Identification, evaluation, and management of children with autism spectrum disorder. Pediatrics 2020;145:e20193447. 10.1542/peds.2019-3447 [DOI] [PubMed] [Google Scholar]
- 46. Siegel M, McGuire K, Veenstra-VanderWeele J, et al. Practice parameter for the assessment and treatment of psychiatric disorders in children and adolescents with intellectual disability (intellectual developmental disorder). J Am Acad Child Adolesc Psychiatry 2020;59:468–96. 10.1016/j.jaac.2019.11.018 [DOI] [PubMed] [Google Scholar]
- 47. Kanwal M, Alyas S, Afzal M, et al. Molecular diagnosis of fragile X syndrome in subjects with intellectual disability of unknown origin: implications of its prevalence in regional Pakistan. PLoS One 2015;10:e0122213. 10.1371/journal.pone.0122213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sofocleous C, Kitsiou S, Fryssira H, et al. 10 years’ experience in fragile X testing among mentally retarded individuals in greece: a molecular and epidemiological approach. In Vivo 2008;22:451–5. [PubMed] [Google Scholar]
- 49. Rauch A, Hoyer J, Guth S, et al. Diagnostic yield of various genetic approaches in patients with unexplained developmental delay or mental retardation. Am J Med Genet A 2006;140:2063–74. 10.1002/ajmg.a.31416 [DOI] [PubMed] [Google Scholar]
- 50. Chen X, Wang J, Xie H, et al. Fragile X syndrome screening in chinese children with unknown intellectual developmental disorder. BMC Pediatr 2015;15:77. 10.1186/s12887-015-0394-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shen Y, Dies KA, Holm IA, et al. Clinical genetic testing for patients with autism spectrum disorders. Pediatrics 2010;125:e727–35. 10.1542/peds.2009-1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mordaunt D, Gabbett M, Waugh M, et al. Uptake and diagnostic yield of chromosomal microarray in an Australian child development clinic. Children (Basel) 2014;1:21–30. 10.3390/children1010021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Roesser J. Diagnostic yield of genetic testing in children diagnosed with autism spectrum disorders at a regional referral center. Clin Pediatr (Phila) 2011;50:834–43. 10.1177/0009922811406261 [DOI] [PubMed] [Google Scholar]
- 54. Borch LA, Parboosingh J, Thomas MA, et al. Re-evaluating the first-tier status of fragile X testing in neurodevelopmental disorders. Genet Med 2020;22:1036–9. 10.1038/s41436-020-0773-x [DOI] [PubMed] [Google Scholar]
- 55. Hartley T, Potter R, Badalato L, et al. Fragile X testing as a second-tier test. Genet Med 2017;19:gim.2017.147. 10.1038/gim.2017.147 [DOI] [PubMed] [Google Scholar]
- 56. Weinstein V, Tanpaiboon P, Chapman KA, et al. Do the data really support ordering fragile X testing as a first-tier test without clinical features? Genet Med 2017;19:1317–22. 10.1038/gim.2017.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mullegama SV, Klein SD, Nguyen DC, et al. Is it time to retire fragile X testing as a first-tier test for developmental delay, intellectual disability, and autism spectrum disorder? Genet Med 2017;19:gim.2017.146. 10.1038/gim.2017.146 [DOI] [PubMed] [Google Scholar]
- 58. Christofolini DM, Abbud EM, Lipay MVN, et al. Evaluation of clinical checklists for fragile X syndrome screening in Brazilian intellectually disabled males: proposal for a new screening tool. J Intellect Disabil 2009;13:239–48. 10.1177/1744629509348429 [DOI] [PubMed] [Google Scholar]
- 59. Giangreco CA, Steele MW, Aston CE, et al. A simplified six-item checklist for screening for fragile X syndrome in the pediatric population. J Pediatr 1996;129:611–4. 10.1016/s0022-3476(96)70130-0 [DOI] [PubMed] [Google Scholar]
- 60. Lubala TK, Lumaka A, Kanteng G, et al. Fragile X checklists: a meta-analysis and development of a simplified universal clinical checklist. Mol Genet Genomic Med 2018;6:526–32. 10.1002/mgg3.398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Guruju MR, Lavanya K, Thelma BK, et al. Assessment of a clinical checklist in the diagnosis of fragile X syndrome in India. J Clin Neurosci 2009;16:1305–10. 10.1016/j.jocn.2008.12.018 [DOI] [PubMed] [Google Scholar]
- 62. de Vries BB, van den Ouweland AM, Mohkamsing S, et al. Screening and diagnosis for the fragile X syndrome among the mentally retarded: an epidemiological and psychological survey. Collaborative fragile X Study Group. Am J Hum Genet 1997;61:660–7. 10.1086/515496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Arvio M, Peippo M, Simola KO. Applicability of a checklist for clinical screening of the fragile X syndrome. Clin Genet 1997;52:211–5. 10.1111/j.1399-0004.1997.tb02549.x [DOI] [PubMed] [Google Scholar]
- 64. Nolin SL, Snider DA, Jenkins EC, et al. Fragile X screening program in New York state. Am J Med Genet 1991;38:251–5. 10.1002/ajmg.1320380218 [DOI] [PubMed] [Google Scholar]
- 65. Riggs ER, Andersen EF, Cherry AM, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of medical genetics and genomics (ACMG) and the clinical genome resource (clingen). Genet Med 2020;22:245–57. 10.1038/s41436-019-0686-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gillentine MA, Lupo PJ, Stankiewicz P, et al. An estimation of the prevalence of genomic disorders using chromosomal microarray data. J Hum Genet 2018;63:795–801. 10.1038/s10038-018-0451-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rosenfeld JA, Coe BP, Eichler EE, et al. Estimates of penetrance for recurrent pathogenic copy-number variations. Genet Med 2013;15:478–81. 10.1038/gim.2012.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Smajlagić D, Lavrichenko K, Berland S, et al. Population prevalence and inheritance pattern of recurrent CNVs associated with neurodevelopmental disorders in 12,252 newborns and their parents. Eur J Hum Genet 2021;29:205–15. 10.1038/s41431-020-00707-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Manning M, Hudgins L, Professional Practice and Guidelines Committee . Array-Based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet Med 2010;12:742–5. 10.1097/GIM.0b013e3181f8baad [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. McGrew SG, Peters BR, Crittendon JA, et al. Diagnostic yield of chromosomal microarray analysis in an autism primary care practice: which guidelines to implement? J Autism Dev Disord 2012;42:1582–91. 10.1007/s10803-011-1398-3 [DOI] [PubMed] [Google Scholar]
- 71. Bartnik M, Wiśniowiecka-Kowalnik B, Nowakowska B, et al. The usefulness of array comparative genomic hybridization in clinical diagnostics of intellectual disability in children. Dev Period Med 2014;18:307–17. [PubMed] [Google Scholar]
- 72. Fan Y, Wu Y, Wang L, et al. Chromosomal microarray analysis in developmental delay and intellectual disability with comorbid conditions. BMC Med Genomics 2018;11:49. 10.1186/s12920-018-0368-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. D’Arrigo S, Gavazzi F, Alfei E, et al. The diagnostic yield of array comparative genomic hybridization is high regardless of severity of intellectual disability/developmental delay in children. J Child Neurol 2016;31:691–9. 10.1177/0883073815613562 [DOI] [PubMed] [Google Scholar]
- 74. Tammimies K, Marshall CR, Walker S, et al. Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. JAMA 2015;314:895–903. 10.1001/jama.2015.10078 [DOI] [PubMed] [Google Scholar]
- 75. Ho KS, Wassman ER, Baxter AL, et al. Chromosomal microarray analysis of consecutive individuals with autism spectrum disorders using an ultra-high resolution chromosomal microarray optimized for neurodevelopmental disorders. Int J Mol Sci 2016;17:2070. 10.3390/ijms17122070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zarrei M, Burton CL, Engchuan W, et al. A large data resource of genomic copy number variation across neurodevelopmental disorders. NPJ Genom Med 2019;4:26. 10.1038/s41525-019-0098-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Baccarin M, Picinelli C, Tomaiuolo P, et al. Appropriateness of array-CGH in the ADHD clinics: a comparative study. Genes Brain Behav 2020;19:e12651. 10.1111/gbb.12651 [DOI] [PubMed] [Google Scholar]
- 78. Lionel AC, Crosbie J, Barbosa N, et al. Rare copy number variation discovery and cross-disorder comparisons identify risk genes for ADHD. Sci Transl Med 2011;3:95ra75. 10.1126/scitranslmed.3002464 [DOI] [PubMed] [Google Scholar]
- 79. Elia J, Gai X, Xie HM, et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry 2010;15:637–46. 10.1038/mp.2009.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. van Karnebeek CDM, Stockler S. Treatable inborn errors of metabolism causing intellectual disability: a systematic literature review. Mol Genet Metab 2012;105:368–81. 10.1016/j.ymgme.2011.11.191 [DOI] [PubMed] [Google Scholar]
- 81. Hoytema van Konijnenburg EMM, Wortmann SB, Koelewijn MJ, et al. Treatable inherited metabolic disorders causing intellectual disability: 2021 review and digital APP. Orphanet J Rare Dis 2021;16:170. 10.1186/s13023-021-01727-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sayson B, Popurs MAM, Lafek M, et al. Retrospective analysis supports algorithm as efficient diagnostic approach to treatable intellectual developmental disabilities. Mol Genet Metab 2015;115:1–9. 10.1016/j.ymgme.2015.03.001 [DOI] [PubMed] [Google Scholar]
- 83. Campistol J, Díez-Juan M, Callejón L, et al. Inborn error metabolic screening in individuals with nonsyndromic autism spectrum disorders. Dev Med Child Neurol 2016;58:842–7. 10.1111/dmcn.13114 [DOI] [PubMed] [Google Scholar]
- 84. Vallance H, Sinclair G, Rakic B, et al. Diagnostic yield from routine metabolic screening tests in evaluation of global developmental delay and intellectual disability. Paediatr Child Health 2021;26:344–8. 10.1093/pch/pxaa112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Çakar NE, Yilmazbaş P. Cases of inborn errors of metabolism diagnosed in children with autism. Ideggyogy Sz 2021;74:67–72. 10.18071/isz.74.0067 [DOI] [PubMed] [Google Scholar]
- 86. Wasim M, Khan HN, Ayesha H, et al. Biochemical screening of intellectually disabled patients: a stepping stone to initiate a newborn screening program in pakistan. Front Neurol 2019;10:762. 10.3389/fneur.2019.00762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sempere A, Arias A, Farré G, et al. Study of inborn errors of metabolism in urine from patients with unexplained mental retardation. J Inherit Metab Dis 2010;33:1–7. 10.1007/s10545-009-9004-y [DOI] [PubMed] [Google Scholar]
- 88. Eun S-H, Hahn SH. Metabolic evaluation of children with global developmental delay. Korean J Pediatr 2015;58:117–22. 10.3345/kjp.2015.58.4.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Djordjevic D, Tsuchiya E, Fitzpatrick M, et al. Utility of metabolic screening in neurological presentations of infancy. Ann Clin Transl Neurol 2020;7:1132–40. 10.1002/acn3.51076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Bahl S, Cordeiro D, MacNeil L, et al. Urine creatine metabolite panel as a screening test in neurodevelopmental disorders. Orphanet J Rare Dis 2020;15:339. 10.1186/s13023-020-01617-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Thurm A, Himelstein D, DʼSouza P, et al. Creatine transporter deficiency: screening of males with neurodevelopmental disorders and neurocognitive characterization of a case. J Dev Behav Pediatr 2016;37:322–6. 10.1097/DBP.0000000000000299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hoang N, Buchanan JA, Scherer SW. Heterogeneity in clinical sequencing tests marketed for autism spectrum disorders. NPJ Genom Med 2018;3:27. 10.1038/s41525-018-0066-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kong SW, Lee I-H, Liu X, et al. Measuring coverage and accuracy of whole-exome sequencing in clinical context. Genet Med 2018;20:1617–26. 10.1038/gim.2018.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Corominas J, Smeekens SP, Nelen MR, et al. Clinical exome sequencing-mistakes and caveats. Hum Mutat 2022;43:1041–55. 10.1002/humu.24360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Eno C, Bayrak-Toydemir P, Bean L, et al. Misattributed parentage as an unanticipated finding during exome/genome sequencing: current clinical laboratory practices and an opportunity for standardization. Genet Med 2019;21:861–6. 10.1038/s41436-018-0265-4 [DOI] [PubMed] [Google Scholar]
- 96. Hercher L, Jamal L. An old problem in a new age: revisiting the clinical dilemma of misattributed paternity. Appl Transl Genom 2016;8:36–9. 10.1016/j.atg.2016.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013;15:565–74. 10.1038/gim.2013.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Bowling KM, Thompson ML, Amaral MD, et al. Genomic diagnosis for children with intellectual disability and/or developmental delay. Genome Med 2017;9:43. 10.1186/s13073-017-0433-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ontario Health (Quality) . Genome-wide sequencing for unexplained developmental disabilities or multiple congenital anomalies: a health technology assessment. Ont Health Technol Assess Ser 2020;20:1–178. [PMC free article] [PubMed] [Google Scholar]
- 100. Ewans LJ, Schofield D, Shrestha R, et al. Whole-exome sequencing reanalysis at 12 months boosts diagnosis and is cost-effective when applied early in mendelian disorders. Genet Med 2018;20:1564–74. 10.1038/gim.2018.39 [DOI] [PubMed] [Google Scholar]
- 101. American College of Medical Genetics and Genomics . Incidental findings in clinical genomics: a clarification. Genet Med 2013;15:664–6. 10.1038/gim.2013.82 [DOI] [PubMed] [Google Scholar]
- 102. Boycott K, Hartley T, Adam S, et al. The clinical application of genome-wide sequencing for monogenic diseases in Canada: position statement of the Canadian College of medical geneticists. J Med Genet 2015;52:431–7. 10.1136/jmedgenet-2015-103144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Manickam K, McClain MR, Demmer LA, et al. Exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability: an evidence-based clinical guideline of the american college of medical genetics and genomics (ACMG). Genet Med 2021;23:2029–37. 10.1038/s41436-021-01242-6 [DOI] [PubMed] [Google Scholar]
- 104. Washington State Health Care Authority . Whole exome sequencing: final evidence report. Olympia, WA, 2018. [Google Scholar]
- 105. Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 2014;312:1870–9. 10.1001/jama.2014.14601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Retterer K, Juusola J, Cho MT, et al. Clinical application of whole-exome sequencing across clinical indications. Genet Med 2016;18:696–704. 10.1038/gim.2015.148 [DOI] [PubMed] [Google Scholar]
- 107. Baldridge D, Heeley J, Vineyard M, et al. The exome clinic and the role of medical genetics expertise in the interpretation of exome sequencing results. Genet Med 2017;19:1040–8. 10.1038/gim.2016.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Evers C, Staufner C, Granzow M, et al. Impact of clinical exomes in neurodevelopmental and neurometabolic disorders. Mol Genet Metab 2017;121:297–307. 10.1016/j.ymgme.2017.06.014 [DOI] [PubMed] [Google Scholar]
- 109. Lee H, Deignan JL, Dorrani N, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA 2014;312:1880–7. 10.1001/jama.2014.14604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Codina-Solà M, Rodríguez-Santiago B, Homs A, et al. Integrated analysis of whole-exome sequencing and transcriptome profiling in males with autism spectrum disorders. Mol Autism 2015;6:21. 10.1186/s13229-015-0017-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Rossi M, El-Khechen D, Black MH, et al. Outcomes of diagnostic exome sequencing in patients with diagnosed or suspected autism spectrum disorders. Pediatr Neurol 2017;70:34–43. 10.1016/j.pediatrneurol.2017.01.033 [DOI] [PubMed] [Google Scholar]
- 112. Srivastava S, Cohen JS, Vernon H, et al. Clinical whole exome sequencing in child neurology practice. Ann Neurol 2014;76:473–83. 10.1002/ana.24251 [DOI] [PubMed] [Google Scholar]
- 113. Vissers LELM, van Nimwegen KJM, Schieving JH, et al. A clinical utility study of exome sequencing versus conventional genetic testing in pediatric neurology. Genet Med 2017;19:1055–63. 10.1038/gim.2017.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Palmer EE, Schofield D, Shrestha R, et al. Integrating exome sequencing into a diagnostic pathway for epileptic encephalopathy: evidence of clinical utility and cost effectiveness. Mol Genet Genomic Med 2018;6:186–99. 10.1002/mgg3.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Sawyer SL, Hartley T, Dyment DA, et al. Utility of whole-exome sequencing for those near the end of the diagnostic odyssey: time to address gaps in care. Clin Genet 2016;89:275–84. 10.1111/cge.12654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Soden SE, Saunders CJ, Willig LK, et al. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci Transl Med 2014;6:265ra168. 10.1126/scitranslmed.3010076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Snoeijen-Schouwenaars FM, van Ool JS, Verhoeven JS, et al. Diagnostic exome sequencing in 100 consecutive patients with both epilepsy and intellectual disability. Epilepsia 2019;60:155–64. 10.1111/epi.14618 [DOI] [PubMed] [Google Scholar]
- 118. Martínez F, Caro-Llopis A, Roselló M, et al. High diagnostic yield of syndromic intellectual disability by targeted next-generation sequencing. J Med Genet 2017;54:87–92. 10.1136/jmedgenet-2016-103964 [DOI] [PubMed] [Google Scholar]
- 119. Arteche-López A, Gómez Rodríguez MJ, Sánchez Calvin MT, et al. Towards a change in the diagnostic algorithm of autism spectrum disorders: evidence supporting whole exome sequencing as a first-tier test. Genes (Basel) 2021;12:560. 10.3390/genes12040560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Dillon OJ, Lunke S, Stark Z, et al. Exome sequencing has higher diagnostic yield compared to simulated disease-specific panels in children with suspected monogenic disorders. Eur J Hum Genet 2018;26:644–51. 10.1038/s41431-018-0099-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Gieldon L, Mackenroth L, Kahlert A-K, et al. Diagnostic value of partial exome sequencing in developmental disorders. PLoS One 2018;13:e0201041. 10.1371/journal.pone.0201041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Grozeva D, Carss K, Spasic-Boskovic O, et al. Targeted next-generation sequencing analysis of 1,000 individuals with intellectual disability. Hum Mutat 2015;36:1197–204. 10.1002/humu.22901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Pekeles H, Accogli A, Boudrahem-Addour N, et al. Diagnostic yield of intellectual disability gene panels. Pediatr Neurol 2019;92:32–6. 10.1016/j.pediatrneurol.2018.11.005 [DOI] [PubMed] [Google Scholar]
- 124. Chérot E, Keren B, Dubourg C, et al. Using medical exome sequencing to identify the causes of neurodevelopmental disorders: experience of 2 clinical units and 216 patients. Clin Genet 2018;93:567–76. 10.1111/cge.13102 [DOI] [PubMed] [Google Scholar]
- 125. Malinowski J, Miller DT, Demmer L, et al. Systematic evidence-based review: outcomes from exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability. Genet Med 2020;22:986–1004. 10.1038/s41436-020-0771-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Tan TY, Dillon OJ, Stark Z, et al. Diagnostic impact and cost-effectiveness of whole-exome sequencing for ambulant children with suspected monogenic conditions. JAMA Pediatr 2017;171:855–62. 10.1001/jamapediatrics.2017.1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Monroe GR, Frederix GW, Savelberg SMC, et al. Effectiveness of whole-exome sequencing and costs of the traditional diagnostic trajectory in children with intellectual disability. Genet Med 2016;18:949–56. 10.1038/gim.2015.200 [DOI] [PubMed] [Google Scholar]
- 128. Stavropoulos DJ, Merico D, Jobling R, et al. Whole genome sequencing expands diagnostic utility and improves clinical management in pediatric medicine. NPJ Genom Med 2016;1:15012. 10.1038/npjgenmed.2015.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Córdoba M, Rodriguez-Quiroga SA, Vega PA, et al. Whole exome sequencing in neurogenetic odysseys: an effective, cost- and time-saving diagnostic approach. PLoS One 2018;13:e0191228. 10.1371/journal.pone.0191228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Li C, Vandersluis S, Holubowich C, et al. Cost-Effectiveness of genome-wide sequencing for unexplained developmental disabilities and multiple congenital anomalies. Genet Med 2021;23:451–60. 10.1038/s41436-020-01012-w [DOI] [PubMed] [Google Scholar]
- 131. Dong X, Liu B, Yang L, et al. Clinical exome sequencing as the first-tier test for diagnosing developmental disorders covering both CNV and SNV: a Chinese cohort. J Med Genet 2020;57:558–66. 10.1136/jmedgenet-2019-106377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Schwarze K, Buchanan J, Taylor JC, et al. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet Med 2018;20:1122–30. 10.1038/gim.2017.247 [DOI] [PubMed] [Google Scholar]
- 133. Dragojlovic N, Elliott AM, Adam S, et al. The cost and diagnostic yield of exome sequencing for children with suspected genetic disorders: a benchmarking study. Genet Med 2018;20:1013–21. 10.1038/gim.2017.226 [DOI] [PubMed] [Google Scholar]
- 134. Yuen T, Carter MT, Szatmari P, et al. Cost-Effectiveness of genome and exome sequencing in children diagnosed with autism spectrum disorder. Appl Health Econ Health Policy 2018;16:481–93. 10.1007/s40258-018-0390-x [DOI] [PubMed] [Google Scholar]
- 135. Scriver CR, Beaudet AL, Sly WS, et al. The metabolic and molecular basis of inherited disease. McGraw-Hill, 2001. [Google Scholar]
- 136. Saudubray J-M. Clinical approach to inborn errors of metabolism in paediatrics. In: Inborn Metabolic Diseases Diagnosis and Treatment. Berlin Heidelberg: Springer-Verlag, 2012. [Google Scholar]
- 137. Hoffmann GF, Zschocke J, Nyhan WL. Inherited metabolic diseases. Berlin, Heidelberg, 2017. 10.1007/978-3-662-49410-3 [DOI] [Google Scholar]