Abstract
Introduction
Posttraumatic stress disorder (PTSD) is a complex multifactorial disorder influenced by the interaction of genetic and environmental factors. Analyses of epigenomic and transcriptomic modifications may help to dissect the biological factors underlying the gene-environment interplay in PTSD. To date, most human PTSD epigenetics studies have used peripheral tissue, and these findings have complex and poorly understood relationships to brain alterations. Studies examining brain tissue may help characterize the brain-specific transcriptomic and epigenomic profiles of PTSD. In this review, we compiled and integrated brain-specific molecular findings of PTSD from humans and animals.
Methods
A systematic literature search according to the PRISMA criteria was performed to identify transcriptomic and epigenomic studies of PTSD, focusing on brain tissue from human postmortem samples or animal-stress paradigms.
Results
Gene- and pathway-level convergence analyses revealed PTSD-dysregulated genes and biological pathways across brain regions and species. A total of 243 genes converged across species, with 17 of them significantly enriched for PTSD. Chemical synaptic transmission and signaling by G-protein-coupled receptors were consistently enriched across omics and species.
Discussion
Our findings point out dysregulated genes highly replicated across PTSD studies in humans and animal models and suggest a potential role for the corticotropin-releasing hormone/orexin pathway in PTSD’s pathophysiology. Further, we highlight current knowledge gaps and limitations and recommend future directions to address them.
Keywords: Posttraumatic stress disorder, DNA methylation, Gene expression, Brain, Central nervous system
Introduction
Posttraumatic stress disorder (PTSD) is a mental illness that develops after exposure to an extremely stressful and often life-threatening event. According to the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-5) definition, PTSD is characterized by heterogeneous symptom clusters such as re-experiencing, avoidance, numbing, and hyperarousal [1]. Predominant PTSD typologies include anxious re-experiencing, dysphoria, and high symptoms [2, 3]. Among trauma-exposed individuals, only a small percentage develop PTSD [4–8], with a reported lifetime prevalence of 6–7% [9–13]. This suggests the presence of biological factors underlying individual differences in PTSD risk [14–18].
Genetic factors may confer susceptibility to PTSD. Twin studies have reported a heritability rate of approximately 49% [19]. Further, genome-wide association studies (GWAS) estimated a 6–20% variance explained by common genetic variations (h2SNP 0.06–0.2) [19–23]. Though recent large-scale GWAS have successfully identified genetic risk variants associated with PTSD [21, 23], the function of most of these remains unknown, making their biological interpretation difficult. Transcriptomic profiling can shed light on the potential function of these risk variants and may identify risk biomarkers. For example, PTSD risk variants identified in GWAS are mapped to genes that themselves can influence gene regulation, such as METTL15, which is involved in RNA methylation, and AUTS2 or ZNF813, which regulate transcriptional processes [21, 23]. By integrating GWAS and transcriptomic data in transcriptome-wide association studies (TWAS), there are now reports of genotype-dependent effects of PTSD risk variants on differential gene expression. For example, a previous TWAS comparing resilient versus chronic symptom trajectories in blood samples showed an association of GRIN3B with PTSD, showing lower mRNA expression levels. These effects were dependent on genotype variation at rs10401454 and were associated with differential expression levels of GRIN3B in brain tissue [24].
Epigenetic modifications also participate in the interplay of genetics and environmental factors in PTSD, influencing gene regulation without altering the DNA sequence. DNA methylation (DNAm) is an epigenetic modification that results from the addition of a methyl group in the 5’ position of a cytosine ring (5-methylcytosine [5mC]). It occurs mainly in CpG islands and is commonly associated with transcriptional repression at the promoter region. Whole-genome DNAm studies, also called epigenome-wide association studies (EWAS), performed on human peripheral samples have identified genes likely involved in PTSD, such as AKT, ANK3, BDNF, CNR1, COMT, CREB, DRD2, DMRTA2, DOCK2, EFS, ELK1, ETS-2, and GATA3[25–31]. Studies utilizing genomic, epigenomic, and transcriptomic analyses in peripheral samples of PTSD cases and trauma-exposed controls can help map gene-regulatory processes involved in disease risk and reveal putative molecular phenotypes associated with PTSD.
The degree to which specific epigenomic and transcriptomic changes in peripheral tissues correspond to the diverse profiles of changes in specific cell types in the brain is uncertain [29, 30, 32]. Thus, examining peripheral tissue may be of limited applicability as a strategy for understanding the brain [31, 33]. Considering the importance of assessing human brain samples in PTSD, the US Department of Veterans Affairs has diligently worked to create and curate the National PTSD Brain Bank (NPBB), a biorepository of human postmortem brains from individuals with a PTSD diagnosis and healthy controls. However, the scarcity of curated brain banks housing samples from PTSD individuals is still evident. To investigate brain-specific molecular changes and further characterize the mechanisms involved in PTSD, the evaluation of brain tissue from animal models represents a complementary strategy to human studies. Several animal-stress paradigms have been used for the study of PTSD, including immobilization, electric shock, predator stress, single prolonged stress, and stress-enhanced fear learning (SEFL) [34–37]. These models are used as physical stressors to develop conditioned fear memory, a central mechanism in PTSD [34–37].
Studying the molecular convergence in brain tissue across humans and animal models may help to replicate and prioritize molecular findings in PTSD. Since molecular epigenomic and transcriptomic signatures can be tissue or cell-type specific [38, 39], more recent work in both humans and animal models has focused on examining brain tissue. In this review, we summarize brain-specific epigenomics and transcriptomics findings in PTSD across species and report convergent genes and biological pathways likely involved in the pathophysiology of PTSD.
Methods
Search Strategy and Study Selection
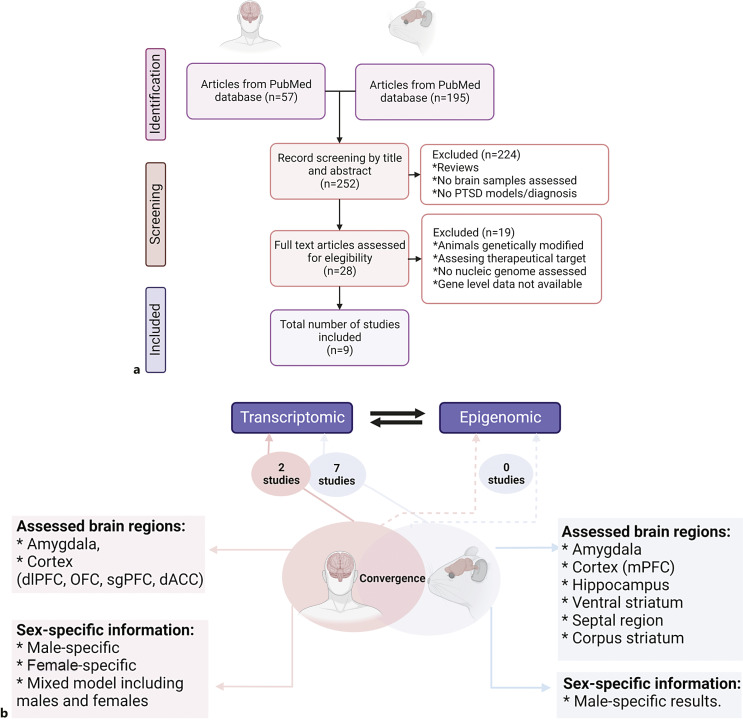
A systematic literature search was conducted using the PubMed database between September 2021 and September 2022. We focused on genome-wide studies that investigated the epigenomic or transcriptomic profiles associated with PTSD by examining brain tissue from human postmortem samples or animal models. To reduce bias in the selection of studies, we followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) criteria (Fig. 1a; online suppl. Table 1; for all online suppl. material, see www.karger.com/doi/10.1159/000529536). Our inclusion criteria were as follows: (1) full-text original research articles written in English; (2) studies evaluating tissue from at least one brain region; (3) studies using genome-wide approaches; and (4) studies using animal-stress paradigms including exposure to electric shock, predator stress, single prolonged stress, and SEFL, all of which have been suggested to model PTSD behaviors.
Fig. 1.
Study overview. a Flow diagram of the PRISMA study selection workflow in our systematic literature review. The identification and screening process of selected reports, focused on brain-specific transcriptomic and epigenomic studies of PTSD in humans and animal models, is shown. b Cross-species convergence in PTSD. The number of identified transcriptomic and epigenomic studies in animal models and humans is depicted. A high variability is observed in the brain regions examined across species. Subgenual prefrontal cortex (sgPFC), orbitofrontal cortex (OFC), dorsolateral prefrontal cortex (dlPFC), dorsolateral anterior cingulate cortex (dACC), ventromedial prefrontal cortex (vmPFC), amygdala (AMY), hippocampus (Hippo), hemibrain (Hemibr), medial prefrontal cortex (mPFC), ventral striatum (VS), septal region (SE), corpus striatum (ST).
Exclusion criteria included (1) studies examining genetically modified (knock-down or -out) animal models, (2) studies assessing changes related to therapeutic intervention, (3) studies evaluating a trait other than PTSD (in humans), (4) studies with gene-level data not available, and (5) review articles. Exclusion criteria 1 and 2 were utilized to reduce methodological variability across species.
For human studies, we used (“traumatic” OR “PTSD” OR “post-traumatic stress disorder” OR “posttraumatic stress disorder”) AND (“epigenetics” OR “methylation” OR “histone” OR “gene expression” OR “transcriptomic” OR “RNAseq”) AND (“postmortem brain” OR “post-mortem brain” OR “postmortem” OR “post mortem”) as keywords. Among 57 records identified, 50 were excluded after screening by title or abstract. A full-text review was performed on seven records to confirm that they met inclusion or exclusion criteria. Five records were excluded because they corresponded to studies assessing mitochondrial genome, studies with gene-level data not available, or those assessing features related to PTSD such as body mass index (BMI) or epigenetic age. Only two records were left after applying inclusion and exclusion criteria.
For animal models (“PTSD” OR “PTSD model” OR “PTSD-like”) AND (“epigenetics” OR “methylation” OR “histone” OR “gene expression” OR “transcriptomic” OR “RNAseq”) AND (“animal model” OR “mouse” OR “rat” OR “mice”) AND (“brain” OR “cortex” OR “amygdala” OR “hippocampus”) were used as keywords. Among the 195 records identified, after screening by title and abstract, 174 were excluded because they were associated with review articles, targeted gene studies, studies assessing a therapeutic target, studies examining genetically modified animals, and studies assessing traits other than PTSD. A full-text review was performed on 21 records. Fourteen were excluded because they were assessing the mitochondrial genome, using targeted approaches, or examining therapeutic targets. Seven records were left after applying the inclusion and exclusion criteria.
Data Extraction and Convergence Analysis
After an exhaustive full-text review, gene-level data (including fold change and significance value) publicly available in the main text or supplementary information were collected from each of the nine studies identified. Collected data were organized according to brain region, sex, and species and later used as input in convergence analyses. Metascape, a comprehensive web-based tool (https://metascape.org/gp/index.html#/main/step1) useful for analyzing and integrating multiple and orthogonal gene lists [40], was utilized to assess convergence across species. Metascape automatically recognizes different gene identifier types (such as Entrez ID or gene symbol) from various organisms’ datasets (including H. sapiens, M. musculus, and R. norvegicus), which are then mapped to a unique Entrez ID for further annotation analyses [40]. Gene names directly collected from each paper (“as-is”) were used as input in Metascape, with the “any species” option selected for the gene-identifier-conversion step [40]. Comparative functional enrichment analyses were carried out across the independent gene lists by species, sex, and brain region [40]. Annotated terms from ontology sources such as GO and KEGG were hierarchically clustered, thereby avoiding enrichment-term redundancy and eliminating confounding data interpretation [40].
To evaluate how each study affects convergent findings and to confirm that the robustness of our convergence findings across species was not driven by one single study, we performed a leave-one-out sensitivity analysis [41]. Genes and biological pathways that remained convergent after leaving one out were defined as “convergent.” A protein-protein interaction (PPI) enrichment analysis was also performed using the Molecular Complex Detection (MCODE) algorithm implemented in Metascape [40] to identify the networks associated with the identified convergent genes. Figures and statistical analyses were carried out using the R packages ggpubr [42] and ggplot2 [43] (R version 4.1.0) [44]. A Venn diagram web-based tool (http://bioinformatics.psb.ugent.be/webtools/Venn/) was used to assess gene-level convergence after leave-one-out sensitivity analysis. An ideogram was built using the Phenogram [45] web-based tool. BioRender was used to design the proposed model of convergent genes in PTSD [46].
Results
For transcriptomics, we identified nine studies, all examining bulk brain tissue without considering potential cell-type specificity (Fig. 1b; online suppl. Table 2). In terms of species and sex, the seven studies conducted on animal models were performed on rodents, five using only males [47–51] and two including both males and females [52, 53]. The two transcriptomic studies identified in humans performed both combined and sex-specific analyses. Among the brain regions analyzed, the amygdala was the most commonly assessed in animal models [48–50, 52, 53] and was also evaluated in one of the two identified human studies [54]. In humans, the cortex was the most commonly assessed brain region, but only one such study was identified in animals [48]. No studies examining epigenetics of PTSD at the genome-wide scale in the brain were identified in our review (Fig. 1b; online suppl. Table 2). Below, we provide a brief description of each identified study, followed by a cross-species convergence analysis to identify common genes and biological pathways in PTSD.
Human Studies
We identified three genome-wide studies assessing human postmortem brain samples: two transcriptomic studies using RNA sequencing (RNAseq) [54–56] and one epigenomic study using the Illumina Infinium Methylation EPIC array. Both transcriptomic studies examined differential expression using combined and sex-specific models (Fig. 1b; online suppl. Table 2). The epigenomic study was limited to the evaluation of epigenetic aging.
Transcriptomic Studies
The first transcriptomic study [55] of PTSD examined human postmortem brain samples from the NPBB, including 52 PTSD cases, 46 healthy controls, and 45 individuals with major depressive disorder. This study evaluated four different brain regions: the subgenual prefrontal cortex (sgPFC), the orbitofrontal cortex (OFC), the dorsolateral prefrontal cortex (dlPFC), and the dorsolateral anterior cingulate cortex (dACC) [55]. In the combined-sex model, a total of 635 differentially expressed genes (DEGs) were identified across four brain regions: the sgPFC (n = 1), OFC (n = 170), dlPFC (n = 390), and dACC (n = 74). Specifically, 234 genes were downregulated and mainly involved in gliogenesis; in contrast, 401 were upregulated and mainly implicated in synapse and neuronal development (online suppl. Table 3). Among all the identified DEGs across the four brain regions, ELK1 and ADAMTS2 were found in the dlPFC, OFC, and dACC, and 43 genes were found in at least two of the four evaluated regions (online suppl. Table 3).
A follow-up sex-specific analysis comparing PTSD cases with controls found that the number of DEGs was significantly higher in women than in men in all four brain regions, particularly the OFC and the sgPFC (online suppl. Table 3). Enrichment analysis of DEGs showed decreased expression of GABA-related genes, although this low expression is observed predominantly in women, suggesting sex-specific effects in gene expression. In a TWAS, ELFN1, GJC1, LRRC37A, MAPT, MST1R, RBM6, and ZHX3 were the seven cortical hits associated with PTSD, of which ELFN1 is a GABA-related gene. Of note, ELFN1 encodes a synaptic adhesion protein required for the recruitment of presynaptic GRM7 (metabotropic glutamate receptor 7) and GRIK2 (glutamate receptor kainate 2) [55]. The significant genes identified in the combined and sex-specific models were collected for convergent analyses (online suppl. Table 3).
A more recent transcriptomic study evaluated the basolateral amygdala (BLA), medial amygdala, dlPFC, and dACC of human postmortem brain samples from 107 PTSD cases and 109 controls collected from several US medical examiners’ offices in Maryland, the District of Columbia and Virginia [54]. In this study, types of trauma included combat-related, childhood maltreatment, and any assaultive violence. With a false discovery rate (FDR) <0.05 and a combined-sex model, 41 DEGs were identified in the cortex, including CORT, HDAC4, and SPRED1. CRHBP was the only DEG identified in the amygdala. In the sex-specific analysis, authors report potential sex-specific effects, but gene-level statistical data were not provided. A comparison between combat-exposed individuals with and without PTSD showed CHI3L2, GALNT15, and TJP2 to be significant DEGs in the amygdala. No significant genes among combat-exposed individuals were observed in the cortex [54]. For our convergent analyses, we used the significant genes identified in the combined-sex model (online suppl. Table 3).
Epigenomic Studies
The only epigenetic study of PTSD in the human brain is an epigenetic analysis of the effect of the rs9315202 SNP on the KL gene. This study was also conducted using samples from the NPBB. The rs9315202 SNP interacts with PTSD to predict decreased expression of KL and advanced epigenetic age in the motor cortex of individuals older than 45 years [56]. This is consistent with results obtained in blood samples from PTSD patients, where the minor allele A of this SNP was associated with PTSD symptom severity and advanced epigenetic age [57]. A related study from the same group, not identified in our literature search, evaluated the relationship of advanced epigenetic age with PTSD, alcohol use disorder, and gene expression in the dlPFC, ventromedial prefrontal cortex, and motor cortex [58]. It identified 11 genes with differential expression related to epigenetic age residuals in both disorders, including SNORA73B, COL6A3, ADGRG6, IL1B, NUTM2A-AS1, GCNT1, GPRIN3, CES3, ADAMTS18, LINC00643, and RCOR2. They highlighted IL1B, RCOR2, and GCNT1, which participate in the inflammatory response, and suggested that inflammatory processes may accelerate cellular aging and participate in the development of PTSD and alcohol use disorder [58]. Because differential methylation at the genome-wide scale was not assessed, these studies were not included in our convergent analyses. In summary, we identified two human transcriptomic studies of PTSD examining brain tissue, which were later included in our convergence analyses (online suppl. Table 3).
Animal Models
Seven full-text transcriptomic studies of animal-stress paradigms of PTSD were identified (Fig. 1b; online suppl. Table 2), five using RNA sequencing [49–53] and two using expression arrays [47, 48]. Four studies evaluated acute and chronic consequences of trauma in animal-stress paradigms of PTSD [47, 48, 50, 51]. Brain-specific epigenomic changes were also evaluated, but gene-level data on significantly differentially methylated genes were not provided [47]. The animal models of PTSD-related stress paradigms utilized in these studies include fear conditioning, inescapable electric foot shocks, SEFL, aggressor exposure, and immobilization stress [34–37] in mice or rats.
Transcriptomic Studies
Electric foot shock is a physical stressor useful for evaluating learning, memory, and traumatic fear in animals and is often implemented in fear-conditioning paradigms. In this model, re-exposure to shock context or conditioned stimuli may reproduce PTSD symptoms [34–37]. A recent sex-specific transcriptomic study using fear conditioning as a stress paradigm and RNA sequencing examined the central and BLA in mice [53]. A total of 190 upregulated and 401 downregulated genes were reported as significant DEGs in fear-conditioned male mice. Nervous system development, axonogenesis, and neurotransmitter regulation were among the most significant pathways enriched [53]. These 591 significant genes were used for the convergence analyses (online suppl. Table 3).
Long intergenic noncoding RNA (lincRNA) profiles and mRNA differential expression were assessed in the left dorsal hippocampus of male rats exposed to fear conditioning using electric foot shocks (a series of ten single shocks followed by re-exposure to the chamber without shock) [51]. Differential expression was assessed in rodents that received D-cycloserine, a partial N-methyl-D-aspartate receptor agonist that facilitates extinction learning [51]. Based on our inclusion criteria, we focused on DEGs identified among drug-free animals. A total of 392 genes showed differential expression between non-trauma-exposed and trauma-exposed rats, of which 352 were downregulated and 40 upregulated. These DEGs were enriched for several pathways, including central nervous system, inflammation, neurodegeneration, and abnormal blood-brain barrier function [51]. They were used in our convergence analyses (online suppl. Table 3).
Another RNA-sequencing study evaluated gene-expression profiles in the amygdala and blood samples of male mice exposed to fear conditioning (electric foot shock) and fear conditioning combined with previous immobilization stress, as well as control mice. A comparison between the control group and mice exposed to fear conditioning showed 607 DEGs in the amygdala, mainly involved in memory formation and consolidation (online suppl. Table 3), while the 352 DEGs found in the blood were related to immune-system processes and homophilic cell adhesion of plasma membranes of adjacent cells [24]. A comparison between mice exposed to fear conditioning versus mice exposed to fear conditioning with previous immobilization stress identified 516 DEGs in the amygdala, which were involved mainly in cell proliferation and cellular response to drugs, and 468 in the blood, which were enriched in the immune response [49]. Some of the DEGs identified in the amygdala had been previously associated with PTSD, such as DRD2 and HTR2A. The lists of 607 and 516 significant genes identified in these records were collected for our convergence analyses (online suppl. Table 3).
SEFL is another stress-paradigm model that recapitulates core PTSD symptoms such as hypervigilance, insomnia, and impaired attention and consists of exposure to unpredictable foot shocks followed by exposure to fear-conditioning context [34–37]. Sillivan et al., 2017 [52] aimed to elucidate the transcriptomic differences in the resilient and susceptible behavioral phenotypes of PTSD in the BLA of mice exposed to SEFL using RNA sequencing. Sixty-one DEGs were identified (52 downregulated and 9 upregulated). Genes with the greatest fold change were involved in cognition, memory, and learning [52]. The list of significant genes identified in this study (19 DEGs) was collected for our convergence analyses (online suppl. Table 3).
A transcriptomic study of fear conditioning with electric foot shock in mice focused on identifying brain region- and time-dependent patterns of gene expression in the amygdala and anterior cingulate cortex (ACC). At two and 5 weeks post-stress, the number of DEGs in the ACC was 3,233 and 43, respectively. In the amygdala, the number of DEGs at two and 5 weeks was comparable (1,022 and 1,063, respectively) [50]. The list of significant genes available in this record (352 DEGs in the amygdala) was collected for our convergence analyses (online suppl. Table 3). Gene ontology (GO) analysis in the ACC showed that genes deregulated at 2 weeks participate in hormone responses (including oxytocin, thyroid, gonadotropin, and estrogen signaling) and synapse signaling (particularly cholinergic, dopaminergic, and glutamatergic synapses). For the amygdala, negative regulation of transcription was observed at 2 and 5 weeks; in contrast, positive regulation was observed only at 5 weeks. GO analysis revealed that genes participating in synapse formation and activity were enriched at 2 and 5 weeks, while genes participating in synapse-mediated signaling were only enriched at 5 weeks. Further, excitatory and inhibitory neurotransmission deregulation under stressful conditions was observed in the amygdala at 5 weeks, with a high enrichment of GABAergic- and glutamatergic-related genes [50].
The impaired gene expression in the amygdala reported by Tanaka et al. [50] is consistent with other reports suggesting an increased glutamate release (probably mediated by glucocorticoids) and an extensive neuronal proliferation in the same region, which is involved in fear memory [59, 60]. Further, this supports that an excitatory-inhibitory imbalance in the amygdala after trauma exposure may be implicated in the pathophysiology of PTSD [61, 62].
Social-defeat stress induced by a series of unprotected and protected exposures to an aggressor animal is a model used to evaluate resilience and susceptibility to PTSD [34–37]. Early and long-term responses to social-defeat stress were evaluated in a study that conducted a microarray expression analysis on mice exposed to a resident aggressor versus control (never exposed to aggressor) [47, 48]. This group performed their analysis in two phases. The first evaluated the hippocampus, amygdala, and medial prefrontal cortex (mPFC), ventral striatum (VS), septal region (SE), and corpus striatum (ST) [48]. The second phase evaluated the hemibrain, peripheral blood, and spleen [47]. Comparing peripheral and central tissues, 3,631 DEGs (1,874 upregulated and 1,757 downregulated) overlapped among all evaluated tissues and were mainly associated with the immune response. The list of nominally significant genes (p < 0.05) reported per brain region was collected for our convergence analyses (online suppl. Table 3). Brain region-specific analysis revealed that inflammatory responses, neurogenesis, and synaptic plasticity were activated during the early response to trauma but inhibited during the long-term response in the amygdala. In the hippocampus and mPFC, inflammatory responses were activated and remained constant in early and long-term responses, while neurogenesis and synaptic plasticity were reduced in the long-term response. The authors concluded that neuroinflammation may have an inhibitory impact on neurogenesis and synaptic plasticity and suggested that blood samples could be a useful source to evaluate the trauma response [47, 48], particularly when assessing early and long-term effects.
While this animal-model study suggests activation of the brain inflammatory system in the early and long-term responses to trauma, a recent study in humans combining neuroimaging and gene expression with postmortem brain data found an opposite effect. By using 11C-PBR28 positron emission tomography brain imaging, low prefrontal-limbic availability of the microglial marker TSPO was observed in PTSD participants [63]. A lower expression of TSPO and microglia-associated genes TNFRSF14 and TSPOAP1 was also found in the female PTSD group. In the same study, authors also found that neuroimmune suppression was accompanied by an immune activation in the peripheral system, as indicated by higher C-reactive protein levels. These inconsistent findings across humans and animal models may be due to differences in type, intensity, and duration between the trauma exposure in humans and the stress-paradigm approaches used in animal models. More research is needed to confirm the association between peripheral immune activation and neuroimmune suppression in PTSD.
Epigenomic Studies
The study by Muhie et al. [47] evaluating transcriptomic changes in the hemibrain in an animal model of PTSD also investigated methylomic changes followed by an integrative multi-omic analysis, but gene-level data on significant results were not provided. This study reported increased promoter-associated DNAm and decreased gene expression in genes involved in learning and memory processes and synaptic plasticity. They also identified genes involved in the inflammatory response that showed increased gene expression and DNAm in the promoter region [47]. In summary, seven transcriptomic studies with available information in rodents were used in our convergence analyses, including findings from the amygdala, cortex, hippocampus, and other brain regions, including hemibrain, VS, SE, and ST (online suppl. Table 3).
Cross-Species Brain-Specific Convergence in PTSD
While there has been evident progress in recent years in studying transcriptomic and epigenomic profiles at the genome-wide scale, there are still many challenges and limitations in terms of sample size, study design (e.g., sex-specific vs. combined-sex), brain tissues examined, and heterogeneity in methodological approaches (e.g., different platforms used) (Fig. 1b; online suppl. Table 2). In terms of brain regions analyzed, the amygdala was the main region assessed in rodents, with hundreds of significant DEGs [27, 48–50, 52, 53], while the human amygdala was assessed in only one study, with only one significant DEG [54]. The cortex was the main brain region evaluated in human postmortem brain studies, with hundreds of DEGs reported [54, 55], while in rodents, only one study assessed the cortex, reporting only nominal findings [48]. In terms of sex, five of the seven identified animal studies evaluated only males [47–51]. The two studies evaluating both sexes in animal models only reported male-specific DEGs [52, 53]. Two studies using human postmortem brain samples carried out both sex-specific and combined-sex analyses, but the list of significant DEGs was available in only one of these [55]. In terms of study design, four animal-model studies examined fear response at different time points [47–50], while PTSD studies in humans are postmortem [54, 55], meaning that the tissue is collected at the time of death and data on the time of trauma exposure are not available. Cross-species convergence analysis was performed by combining all datasets, five from animal models (amygdala, cortex, hippocampus, hemibrain, and other regions) and three from humans (female and male specific, as well as combined-sex analyses in the cortex). We additionally explored cortex-specific and male-specific convergence.
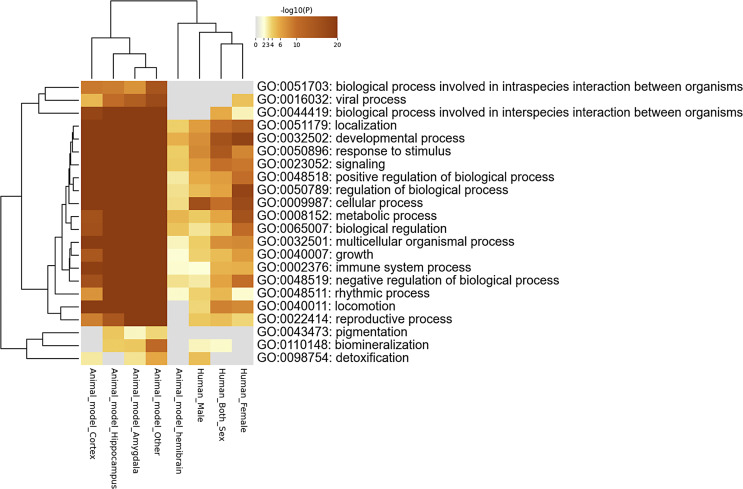
At the pathway level, cross-species convergence analysis identified enrichment for parental pathways (FDR <0.05) such as developmental process (GO:0032502), including cell morphogenesis; signaling (GO:0023052), including chemical synaptic transmission; and multicellular organismal process (GO:0032501), including behavior (Fig. 2; online suppl. Table 4). To demonstrate the robustness of our convergence findings, we conducted a leave-one-out analysis by performing four rounds of convergence analysis with each round missing one study in the following order: (1) [47, 48] (online suppl. Table 5), (2) [49] (online suppl. Table 6), (3) [50] (online suppl. Table 7), and (4) [53] (online suppl. Table 8). After the leave-one-out analysis, the top enriched biological processes convergent across all studies remained significant (Table 1), including developmental process (GO:0032502), multicellular organismal process (GO:0032501), and regulation of biological process (GO:0050789).
Fig. 2.
Pathway-level convergence. Heatmap showing biological pathways in which human and rodent datasets converge.
Table 1.
Top 10 convergent pathways dysregulated in animal-stress paradigms and humans with PTSD after publication bias analyses
| GO number | Parent_Go | Description | Across all studies Log(q value) | Excluding Muhie study Log(q value) | Excluding Lori study Log(q value) | Excluding Tanaka study Log(q value) | Excluding Reis study Log(q value) |
|---|---|---|---|---|---|---|---|
| GO:0000902 | 19_GO:0032502 developmental process | Cell morphogenesis | −96.56 | −82.58 | −96.97 | −96.97 | −96.50 |
| GO:0000904 | 19_GO:0032502 developmental process | Cell morphogenesis involved in differentiation | −96.99 | −68.96 | −96.97 | −96.97 | −96.97 |
| GO:0007268 | 19_GO:0023052 signaling | Chemical synaptic transmission | −96.99 | −40.80 | −96.97 | −96.97 | −96.97 |
| GO:0007409 | 19_GO:0032502 developmental process | Axonogenesis | −96.99 | −55.78 | −96.97 | −96.97 | −96.97 |
| GO:0007610 | 19_GO:0032501 multicellular organismal process | Behavior | −96.99 | −46.50 | −96.97 | −96.97 | −96.97 |
| GO:0030155 | 19_GO:0050789 regulation of biological process | Regulation of cell adhesion | −96.56 | −43.12 | −96.97 | −96.56 | −96.97 |
| GO:0031175 | 19_GO:0032502 developmental process | Neuron projection development | −96.56 | −76.86 | −96.97 | −96.56 | −96.50 |
| GO:0032989 | 19_GO:0032502 developmental process | Cellular component morphogenesis | −96.99 | −68.96 | −96.97 | −96.97 | −96.97 |
| GO:0032990 | 19_GO:0032502 developmental process | Cell part morphogenesis | −96.99 | −63.66 | −96.97 | −96.97 | −96.97 |
| GO:0048667 | 19_GO:0032502 developmental process | Cell morphogenesis involved in neuron differentiation | −96.99 | −59.76 | −96.97 | −96.97 | −96.97 |
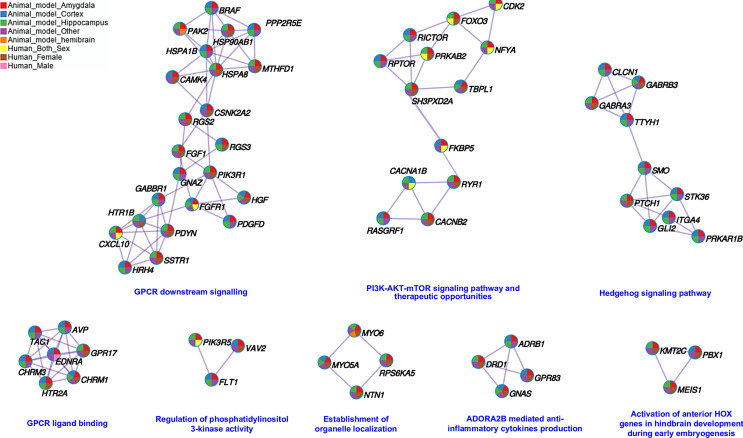
At the gene level, a total of 656 DEGs converged across species. Figure 3 depicts the PPI network and shows the subset of proteins with physical interactions from the convergent genes identified across all studies. After the leave-one-out analysis, 243 genes remained convergent across species (online suppl. Fig. 1; Table 9). Proteoglycans in cancer (hsa05205), calcium signaling pathway (hsa04020), signaling by G-protein-coupled receptor (GPCR) (hsa372790), and oxytocin-signaling pathway (hsa04921) were among the top pathways enriched by these 243 convergent genes (Table 2; online suppl. Table 10).
Fig. 3.
Protein-protein interaction (PPI) enrichment analysis. PPI analysis was performed using the Molecular Complex Detection (MCODE) algorithm implemented in Metascape. This PPI contains the subset of proteins that interact in common pathways colored by counts identified in each study.
Table 2.
Top 10 enriched pathways by the 243 convergent genes across species in animal-stress paradigms and humans with PTSD
| Biological_process | Counts | Log(q value) |
|---|---|---|
| Proteoglycans in cancer | 20 | −11.10 |
| Calcium signaling pathway | 21 | −11.10 |
| Signaling by GPCR | 31 | −9.79 |
| GPCR downstream signaling | 29 | −9.54 |
| head development | 32 | −9.54 |
| brain development | 31 | −9.52 |
| Intracellular signaling by second messengers | 21 | −9.47 |
| Signaling by receptor tyrosine kinases | 26 | −9.35 |
| Focal adhesion: PI3K-Akt-mTOR-signaling pathway | 20 | −8.59 |
| PI3K-Akt signaling pathway | 21 | −8.48 |
Counts correspond to the number of convergent genes enriched in that pathway.
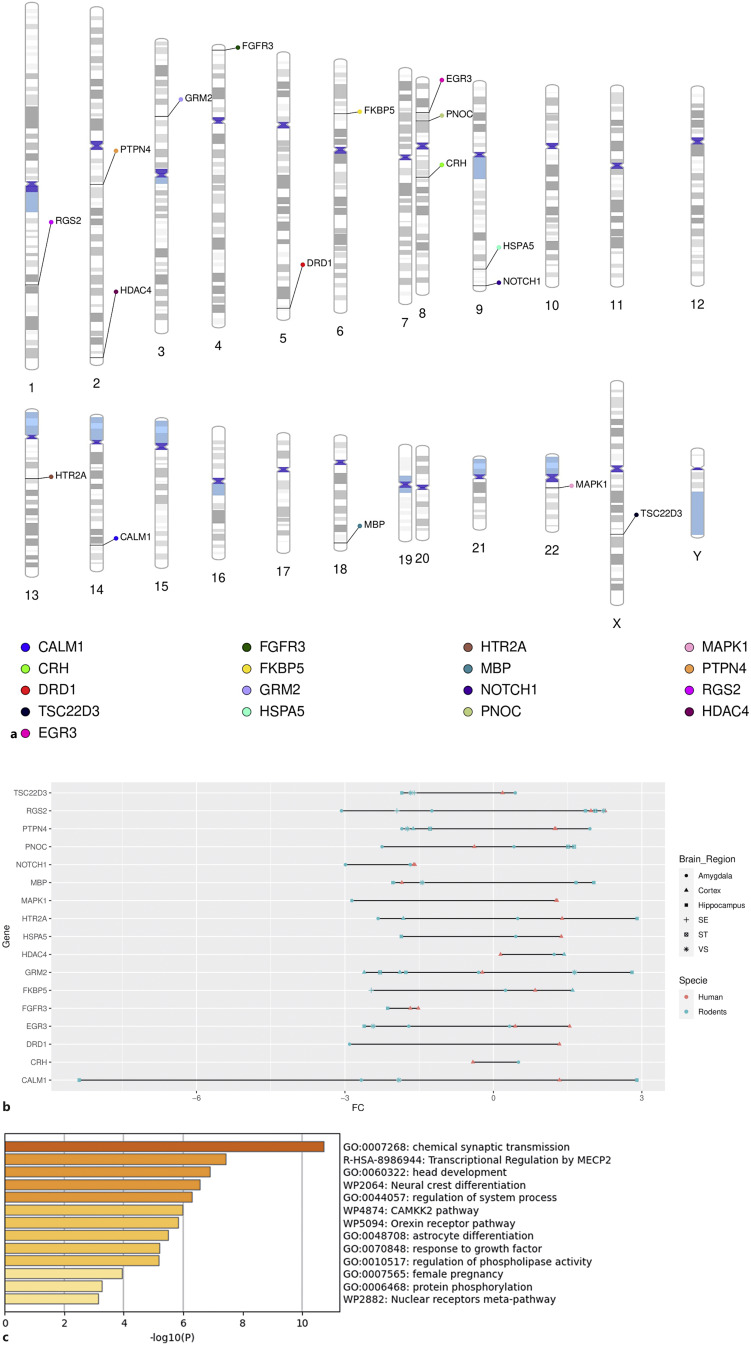
Among the 243 convergent genes identified, using the DisGeNET database in Metascape, CALM1, CRH, DRD1, TSC22D3, EGR3, FGFR3, FKBP5, GRM2, HSPA5, HTR2A, MBP, NOTCH1, PNOC, MAPK1, PTPN4, RGS2, and HDAC4 were found to be significantly enriched in PTSD (FDR <0.05) (Fig. 4a, b; online suppl. Table 11). GO analyses of these 17 convergent genes enriched for PTSD showed enrichment for synaptic signaling, GPCR signaling, brain development, and the orexin-receptor pathway (Fig. 4c; online suppl. Table 12).
Fig. 4.
Convergent PTSD genes across species. a This ideogram illustrates the 17 convergent genes identified in our cross-species analysis that were significantly enriched for PTSD. b Cleveland plot showing fold chance (FC) direction of the 17 convergent PTSD genes across species. c GO enrichment analysis for the 17 convergent PTSD genes across species.
We explored male-specific convergence by evaluating the DEGs reported in the seven identified animal-model studies [47–53] as well as in the male-specific findings reported in the human study [55]. Regulation of biological process (GO:0050789) and response to stimulus (GO:0050896) were the top parental convergent pathways across species (online suppl. Table 13). At the gene level, we identified 26 male-specific convergent genes across species, such as CFH, ADAMTS2, PER2, EDNRA, IGF2, RND2, CDKN1A, TGM2, ADAMTS1, THBD, PENK, FOXC2, NFIL3, CCL5, SYNDIG1L, ARNTL, DSP, ELK1, FRZB, HEYL, FMOD, COL1A2, KDR, DNMT1, SLC13A4, and COL1A1.
Cross-species convergence was also examined for cortex-specific transcriptomic dysregulation. At the gene level, we found that 15, 50, and three DEGs reported in the rodent mPFC were also identified in at least one of the four human cortex subregions (dlPFC, OFC, dACC, and sgPFC) of the combined-sex, female-specific, and male-specific analyses, respectively (online suppl. Table 14; Fig. 2–4). Correlation of convergent genes in the cortex was explored across cortex subregions and sexes. A significant negative correlation was observed between the mPFC of male rodents and the female human OFC (R = −0.41, p = 0.0014) (online suppl. Fig. 5). No additional significant correlation was observed across the datasets. In terms of direction effect, we observed the same expression pattern in several genes enriched for PTSD identified in the combined-sex analysis in humans (online suppl. Fig. 6). HDAC4 was upregulated in human dACC and rodent mPFC (online suppl. Fig. 6). FKBP5 was upregulated in rodent mPFC and human dlPFC (online suppl. Fig. 6). GRM2 was downregulated in rodent mPFC and human dlPFC (online suppl. Fig. 6). Comparing the rodent mPFC with the human-female-specific datasets, downregulation of FOXO4 and upregulation of CHRNA7 were observed in the rodent mPFC and female OFC (online suppl. Fig. 7). Lastly, upregulation of EDNRA and CFH is reported in the mPFC of rodents and dlPFC of human males (online suppl. Fig. 8). In summary, the cortex-specific analyses showed convergence at the gene level, as well as a same-direction effect of some convergent genes across different cortex subregions.
Discussion
Most of the growing research studying the transcriptomics and epigenomics of PTSD in human samples has focused on peripheral tissues such as blood and saliva. In contrast, studies examining brain tissues have mainly been conducted in rodents by using animal-stress paradigms to replicate PTSD-like behaviors. Previous work has discussed the ability of animal models to mimic PTSD features in certain animal-stress paradigms [34–37]. A recent review reported gene-level transcriptomic convergence across studies assessing different stress paradigms in mice [64]. Here, we conducted a systematic review on brain-specific transcriptomic and epigenomic studies of PTSD and assessed gene- and pathway-level convergence across species. A total of 243 DEGs identified in a human cohort were also found in animal models mimicking PTSD behaviors. Fear conditioning, either using electric foot shock or aggressor exposure, was the main stress paradigm implemented across the identified studies in this systematic review. Among these 243 genes, TSC22D3, FKBP5, and DRD1 were also enriched in PTSD. Our results showing cross-species convergence help to prioritize genes or pathways involved in PTSD but also provide evidence on how animal models can inform human work (and vice versa) in the context of PTSD.
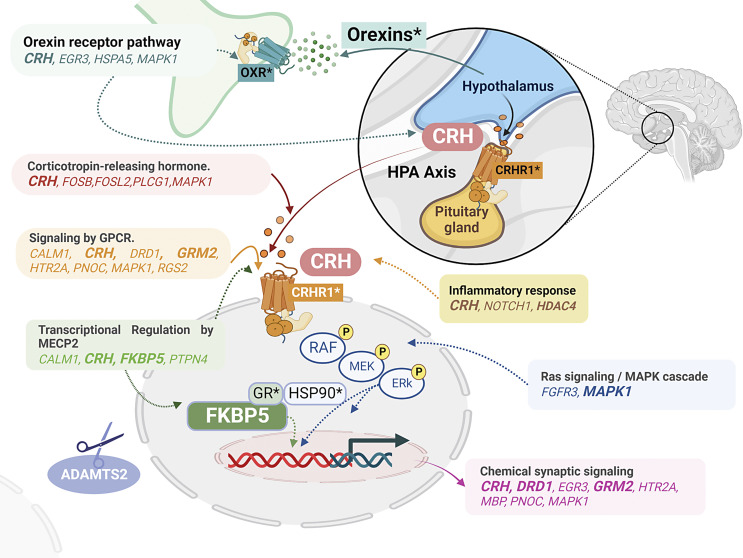
Dysregulation of the hypothalamic-pituitary-adrenal (HPA) axis has been closely implicated in the development and maintenance of PTSD. Briefly, during stressful events, the hypothalamic paraventricular nucleus (PVN) secretes corticotropin-releasing hormone (CRH), which then activates the anterior pituitary to release adrenocorticotrophic hormone (ACTH). ACTH induces cortisol release (corticosterone in rodents) in the adrenal cortex [65]. Cortisol binds to the glucocorticoid receptor (GR, encoded by NR3C1) and other co-chaperones (such as FKBP5 and HSP90) to mediate the nuclear translocation of GR. Within the nucleus, GR binds to glucocorticoid-response elements in the promoter region of several genes and regulates their expression [12, 66, 67]. Epigenetic studies have highlighted the role of HPA-axis-related genes NR3C1 and FKBP5 in PTSD [68–72]. Here we found that FKBP5 was upregulated in the human dlPFC (in the combined-sex analysis) as well as in the mPFC, amygdala, and SE of animal models. CRH was downregulated in the human dlPFC in the combined-sex analysis and upregulated in the rodent amygdala. This shows CRH and FKBP5 to be convergent genes across species, reinforcing the important role of the HPA axis in the pathophysiology of PTSD.
CRH and FKBP5, key regulators of the HPA axis, were also enriched in the orexin-receptor pathway (WP5094) [73]. Recently, the orexin-signaling pathway has also been involved in the pathophysiology and clinical features of PTSD [74–79]. Orexin signaling was recently implicated in contextual generalization of social-stress learning in the BLA [79] and in conditioned fear [80] and emotional behaviors [81] in the central amygdala. The orexin A and B neuropeptides and the G-protein-coupled orexin receptors type 1 (OX1R) and type 2 (OX2R) are members of the orexinergic circuit. Orexin A and B are produced by neurons located in the hypothalamus that project widely to multiple brain regions including the cortex, thalamus, brainstem, and spinal cord [82–84]. Orexin A equally binds to OX1R and OX2R, while orexin B preferentially binds to OX2R [85–87]. Orexin receptors exhibit distinctly different distribution patterns in the brain. OX1R has been described in the bed nucleus of the stria terminalis, amygdala, locus coeruleus, laterodorsal tegmental nucleus, and pedunculopontine tegmental nucleus, whereas OX2R is predominantly expressed in the tuberomammillary nucleus and nucleus accumbens. Both OX1R and OX2R are found in the mPFC, hippocampus, hypothalamic PVN, PVN of the thalamus, dorsal raphe, and ventral tegmental area.
An interaction between OX1R/OXR2 and the HPA axis has been suggested based on findings from animal models. In mice, exogenous administration of orexins was related to HPA-axis activation, increasing ACTH and corticosterone release [88]. Further, inhibiting OX2R reduces stress-related ACTH release [89]. Using mouse pituitary tumor cell line AtT20, treatment with orexin A can activate the CRH receptor type 1 (CRHR1)-signaling pathway [90]. Reciprocally, CRH can stimulate orexin release [91]. Increased orexin mRNA levels and activation of the orexin system have been associated with acute and chronic stress [85]. Additional reports have also supported the role of orexin in PTSD, particularly in mediating memory and appetite dysfunction [75] and promoting resilience in the predator-scent stress model [74]. Inhibition of the orexin receptor (using suvorexant as an orexin antagonist) can attenuate PTSD-like symptoms in the stress/re-stress model [76]. Levels of orexin A are reduced in the plasma and cerebrospinal fluid of combat-exposed veterans with a history of PTSD [77]. In human postmortem brains, a study reported a dysregulation of orexigenic neuropeptides in PTSD and suggested that changes in gene expression may be impacted by BMI [78]. Our cross-species convergence analyses further support the role of the orexinergic system in the pathophysiology of PTSD and the interrelation between orexin signaling and the HPA axis in the brain, but further analysis is needed to confirm and disentangle this relationship.
Additional molecular mechanisms pointed out in this review that may play a key role in PTSD are chemical synaptic transmission (GO:0007268) and signaling by GPCR (R-HSA-372790), which are enriched by 243 convergent genes, 17 of which show disease enrichment for PTSD. Changes in synaptic plasticity have been previously described in the BLA [92], PFC, and hippocampus [60] of PTSD patients. A recent in vivo study scanning the PFC with magnetic resonance spectroscopy reported decreased glutamatergic synaptic strength in PTSD [93]. Neurotransmitter and neurohormone receptors coupled to G proteins may modulate several cellular processes in response to stress [94, 95]. Some of the most well-known GPCRs implicated in the stress response and PTSD are the corticotropin-releasing factor (CRF) receptors, which are closely related to behavioral, neuroendocrine, and immune responses to stress [94, 95]. The CRH-signaling pathway (WP2355), including CRH, FOSB, FOSL2, PLCG1, and MAPK1, was significantly enriched by the 243 convergent genes identified in this systematic review. Moreover, CRH, DRD1, GRM2, HTR2A, PNOC, and MAPK1 were among the 17 convergent genes enriched in chemical synaptic transmission (GO:0007268) and signaling by GPCR pathways. These findings suggest that GPCR signaling, via CRHR1, OX1R, or OX2R, may regulate molecular mechanisms in response to stress, including synaptic processes.
Histone deacetylase 4 (HDAC4) and group II metabotropic glutamate receptor (GRM2) were among the 17 identified convergent genes enriched for PTSD. HDAC4 encodes for histone deacetylase 4 enzyme, which is involved in epigenetic gene-regulatory processes by removing acetyl groups in histone proteins [96, 97]. A previous study assessing blood samples of women diagnosed with PTSD found that differential DNAm of HDAC4 was associated with estradiol levels. The same study confirmed that differential expression of HDAC4 is observed in the amygdala of female rodents exposed to fear conditioning, as well as those with low estrogen levels [96]. In our systematic review, HDAC4 did not show significant gene-expression differences in the human-female-specific analysis, but this gene was found to be upregulated in the male rodent mPFC and the human dACC in the combined-sex analysis. GRM2 was downregulated in the rodent mPFC and human dlPFC (combined-sex analysis). A study using both knockout Grm2−/− mice or pharmacological antagonists for this receptor found that decreased GRM2 activity may regulate stress-induced behaviors and promote resilience [98].
ELK1 and ADAMTS2 were the only two genes in the human combined-sex analysis differentially expressed in three different human brain regions: dlPFC, OFC, and dACC [55]. ADAMTS2 was upregulated in the combined-sex, male-specific, and female-specific analyses [55]. Upregulation of ADAMTS2 was also observed in a recent human transcriptomic study in males and females [54]. In animal models, ADAMTS2 was upregulated in the amygdala, ST, and VS but downregulated in the hippocampus. In the cortex, ADAMTS2 was upregulated in the early response to trauma and downregulated in the long-term trauma response [47–53]. ADAMTS2, a disintegrin and metalloproteinase with thrombospondin motifs 2, encodes for a protein that cleaves Reelin, a glycoprotein involved in synaptic plasticity [99, 100]. A previous study reported an upregulation of ADAMTS2 in peripheral blood mononuclear cells of patients with schizophrenia, as well as downregulation in clinical responders to antipsychotic treatments [99]. The same study showed that ADAMTS2 expression was modulated by dopaminergic signaling, including DRD1 gene, another identified convergent gene enriched in PTSD, cAMP/CREB, and MAP/ERK signaling [99]. The identified convergence of ADAMTS2 across brain regions, sex, and species, as well as its functional role in synaptic plasticity, strongly supports a critical role in the development of PTSD and represents a potential candidate for future treatment strategies.
The sex-specific transcriptional landscape, mainly in the cortex, has been recently reviewed in the context of PTSD [101]. The methodological heterogeneity across the studies selected in this review limited our ability to conduct well-powered region-specific and sex-specific meta-analyses in PTSD across species. Exploratory cortex-specific, male-specific convergence analyses of PTSD across species identified EDNRA and CFH as upregulated genes in the cortex of males, but this should be confirmed in additional studies.
We show that identified convergent genes across brain regions and species exhibit shared functional processes including synaptic plasticity, response to stimulus, and behavior. At the gene level, we also show that genes reported as dysregulated in the human cortex have also been reported, in animal models, in the mPFC, amygdala, and hippocampus. Considering the well-established interconnection at the circuit level among the cerebral cortex, amygdala, and hippocampus in PTSD and thread learning [9], the convergence observed in this review [9] suggests that gene-regulatory mechanisms in response to trauma may impact key biological processes in the amygdala-hippocampus-medial prefrontal circuit that are involved in the pathophysiology of PTSD.
Figure 5 shows a proposed model of the gene and pathway cross-species convergence in PTSD identified in this systematic review. Briefly, the orexin system may interact with the HPA axis by regulating the release of CRH in the brain. CRH binding to GPCRs such as CRHR1 can orchestrate downstream regulatory mechanisms including regulation of gene expression mediated by pathways such as Ras/MAP kinases. FKBP5, a co-chaperone that modulates GR activity in the stress response, may stimulate gene transcription through nuclear GR translocation. Inflammatory response and synaptic plasticity, both closely implicated in PTSD, were among the CRH-related pathways that converged across species. Lastly, ADAMTS2, a metalloproteinase, was consistently convergent across brain regions and species and may act as a key regulator of synaptic plasticity in response to trauma. Future studies are warranted to confirm the proposed model and disentangle the relationship between these identified pathways in the pathophysiology of PTSD.
Fig. 5.
A schematic diagram of the proposed model of convergent genes in PTSD. In this diagram, we show a proposed model of how identified convergent genes may interact in the context of PTSD. The orexin pathway may mediate CRH release in the hypothalamus, activating the CRH pathway. CRH binding to GPCRs may modulate downstream regulatory mechanisms including gene expression and chemical synaptic signaling.
Conclusion
Here we conducted a systematic review of the current evidence for brain-specific transcriptomics and epigenomics of PTSD in both humans and animal models. Our convergence analysis identified 243 dysregulated genes across species. They included genes well-known in the PTSD literature such as CRH, FKBP5, HDAC4, TSC22D3, EGR3, GRM2, MBP, and PNOC. ADAMTS2 was consistently reported across brain regions, sex, and species. Enrichment analysis revealed the orexin-receptor system and the CRH pathway, as well as GPCR signaling and synaptic signaling, to be the top convergent pathways. Based on the convergent findings reported here and the current literature, we propose that the interaction between the orexin system and the CRH pathway may play a key role in the pathophysiology of PTSD. Additional studies are needed, however, to support and disentangle the potential bidirectional orexin-CRH relationship in the pathophysiology of PTSD.
Gaps, Limitations, and Future Directions
Our review unmasks some important knowledge gaps and limitations when evaluating brain-specific convergence of transcriptomic and epigenomic studies across species in PTSD. These include a) the limited number of PTSD studies examining brain tissue, particularly in humans, which is mainly driven by the scarcity of curated brain banks housing samples from individuals with PTSD, with NPBB as the only biorepository focused on this disorder; b) the lack of epigenomic studies of PTSD in brain samples; c) the insufficient number of well-powered studies, which impedes the evaluation of sex- and population-specific differences in PTSD; d) the lack of information about stress effects at different PTSD phases in terms of type and severity; e) the limited overlap in the brain regions examined; f) the inability to conduct brain-specific meta-analyses because of limited summary- and gene-level data availability; and g) the high variability in physical stressors used to model PTSD behaviors in animal-stress paradigms, as well as the limited capacity of these models to fully recapitulate PTSD symptoms (including comorbidities), which can make potential translation of findings challenging. Moreover, as epigenomic and transcriptomic markers are cell-type specific [38, 39], this review also revealed the lack of studies assessing cell-type specificity in PTSD.
Future work should consider the following recommendations to help move the field forward. In humans, research should focus on conducting transcriptomic and epigenomic studies in well-powered samples, considering both sex- and population-specific effects. Transcriptomic and epigenomic modifications are not only tissue specific but cell-type specific. Because the brain is a complex interconnected circuit that is highly heterogeneous in terms of cell types [102, 103], studies applying single-cell approaches are warranted to unravel cell-type-specific disease markers that may be undetectable in bulk-tissue analyses. Considering the inherent limitations of animal models and human studies, research utilizing organoids or induced pluripotent stem cells is also needed. In recent years, these approaches have been successfully applied to model various neuropsychiatric disorders [104–108]. As the number of PTSD studies evaluating different omics increases, integrative multi-omics analyses can help confirm and prioritize genes, gene networks, and molecular pathways. Further, exploration of additional gene-regulatory mechanisms such as histone modifications, microRNAs, and DNA hydroxymethylation may provide a better understanding of the epigenomic landscape of PTSD [109–111]. Taken together, brain- and cell-type-specific multi-omic investigation may help uncover the biological mechanisms and molecular phenotypes underlying the etiology of PTSD and identify novel treatments. Finally, detailed phenotypic information about individuals with PTSD and behavioral studies on animal-stress paradigms may help to confirm the accuracy of animal models to evaluate PTSD pathophysiology.
Acknowledgments
The authors would like to thank Sarah Beck, Research Associate 2 at Yale University – MEDPSY Psych Divisions-CNRU., for English editing of the manuscript.
Statement of Ethics
An ethics statement is not applicable because this study is based exclusively on published literature.
Conflict of Interest Statement
John H. Krystal, MD participated as a consultant in Aptinyx, Inc., Biogen, Idec, MA, Bionomics, Limited (Australia), Boehringer Ingelheim International, Epiodyne, Inc. , EpiVario, Inc., Janssen Research & Development, Jazz Pharmaceuticals, Inc., Otsuka America Pharmaceutical, Inc.,Spring Care, Inc., Sunovion Pharmaceuticals, Inc., Freedom Biosciences, Inc., Biohaven Pharmaceuticals, BioXcel Therapeutics, Inc. (Clinical Advisory Board), Cerevel Therapeutics, LLC, Delix Therapeutics, Inc., Eisai, Inc., EpiVario, Inc., Jazz Pharmaceuticals, Inc., Neumora Therapeutics, Inc., Neurocrine Biosciences, Inc., Novartis Pharmaceuticals Corporation, PsychoGenics, Inc., Takeda Pharmaceuticals, Tempero Bio, Inc., Terran Biosciences, Inc., Biohaven Pharmaceuticals, Freedom Biosciences, Spring Health, Inc., Biohaven Pharmaceuticals Medical Sciences, Cartego Therapeutics, Damona Pharmaceuticals, Delix Therapeutics, EpiVario, Inc., Neumora Therapeutics, Inc., Rest Therapeutics, Tempero Bio, Inc., Terran Biosciences, Inc., Tetricus, Inc. Other authors have no conflicts of interest to declare.
Funding Sources
This work is supported by the Department of Veterans Affairs via 1IK2CX002095-01A1 (JLMO), NIDA R21DA050160 (JLMO), and the US Department of Veterans Affairs National Center for Posttraumatic Stress Disorder.
Author Contributions
Diana L Núñez-Rios: conceptualization, investigation, methodology, and writing (original draft preparation). José Jaime Martinez-Magaña, Sheila T. Nagamatsu, John H. Krystal, Karen G. Martínez-González, and Paola Giusti-Rodríguez: review and editing. Janitza L. Montalvo-Ortiz: conceptualization, writing, review, and editing.
Funding Statement
This work is supported by the Department of Veterans Affairs via 1IK2CX002095-01A1 (JLMO), NIDA R21DA050160 (JLMO), and the US Department of Veterans Affairs National Center for Posttraumatic Stress Disorder.
Data Availability Statement
All data generated or analyzed during this study are included in this article and its online supplementary materials. Further inquiries may be directed to the corresponding author.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
References
- 1. APA. American Psychiatric Association, 2013 . Diagnostic and statistical manual of mental disorders. 2013. [Google Scholar]
- 2. Pietrzak RH, Tsai J, Harpaz-Rotem I, Whealin JM, Southwick SM. Support for a novel five-factor model of posttraumatic stress symptoms in three independent samples of Iraq/Afghanistan veterans: a confirmatory factor analytic study. J Psychiatr Res. 2012;46(3):317–22. 10.1016/j.jpsychires.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 3. Pietrzak RH, Tsai J, Armour C, Mota N, Harpaz-Rotem I, Southwick SM. Functional significance of a novel 7-factor model of DSM-5 PTSD symptoms: results from the national health and resilience in veterans study. J Affect Disord. 2015;174:522–6. 10.1016/j.jad.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 4. Ogińska-Bulik N, Kobylarczyk M. Association between resiliency and posttraumatic growth in firefighters: the role of stress appraisal. Int J Occup Saf Ergon. 2016;22(1). 40–8. 10.1080/10803548.2015.1109372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Elam T, Taku K. Differences between posttraumatic growth and resiliency: their distinctive relationships with empathy and emotion recognition ability. Front Psychol. 2022;13:825161. 10.3389/fpsyg.2022.825161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ryan M, Ryznar R. The molecular basis of resilience: a narrative review. Front Psychiatry. 2022;13:856998. 10.3389/fpsyt.2022.856998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wrenn GL, Wingo AP, Moore R, Pelletier T, Gutman AR, Bradley B, et al. The effect of resilience on posttraumatic stress disorder in trauma-exposed inner-city primary care patients. J Natl Med Assoc. 2011;103(7):560–6. 10.1016/s0027-9684(15)30381-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Habib A, Stevelink SAM, Greenberg N, Williamson V. Post-traumatic growth in (ex-) military personnel: review and qualitative synthesis. Occup Med. 2018;68(9):617–25. 10.1093/OCCMED/KQY140. [DOI] [PubMed] [Google Scholar]
- 9. Alexandra Kredlow M, Fenster RJ, Laurent ES, Ressler KJ, Phelps EA. Prefrontal cortex, amygdala, and threat processing: implications for PTSD. Neuropsychopharmacology. 2022;47(1):247–59. 10.1038/s41386-021-01155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Flores Á, Saravia R, Maldonado R, Berrendero F. Orexins and fear: implications for the treatment of anxiety disorders. Trends Neurosci. 2015;38(9):550–9. [DOI] [PubMed] [Google Scholar]
- 11. Joseph NM, Benedick A, Flanagan CD, Breslin MA, Simpson M, Ragone C, et al. Prevalence of posttraumatic stress disorder in acute trauma patients. OTA Int. 2020;3(1):e056. 10.1097/oi9.0000000000000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Girgenti MJ, Duman RS. Transcriptome alterations in posttraumatic stress disorder. Biol Psychiatry. 2018;83(10):840–8. 10.1016/J.BIOPSYCH.2017.09.023. [DOI] [PubMed] [Google Scholar]
- 13. Smith SM, Goldstein RB, Grant BF. The association between post-traumatic stress disorder and lifetime DSM-5 psychiatric disorders among veterans: data from the National Epidemiologic Survey on Alcohol and Related Conditions-III (NESARC-III). J Psychiatr Res. 2016;82:16–22. 10.1016/j.jpsychires.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zannas AS, Provençal N, Binder EB. Epigenetics of posttraumatic stress disorder: current evidence, challenges, and future directions. Biol Psychiatry. 2015;78(5):327–35. 10.1016/J.BIOPSYCH.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 15. Brady KT, Back SE. Childhood trauma, posttraumatic stress disorder, and alcohol dependence. Alcohol Res. 2012;34(4):408–13. [PMC free article] [PubMed] [Google Scholar]
- 16. Thakur GS, Daigle BJ Jr, Dean KR, Zhang Y, Rodriguez-Fernandez M, Hammamieh R, et al. Systems biology approach to understanding post-traumatic stress disorder. Mol Biosyst. 2015;11(4):980–93. 10.1039/C4MB00404C. [DOI] [PubMed] [Google Scholar]
- 17. Gillespie CF, Bradley B, Mercer K, Smith AK, Conneely K, Gapen M, et al. Trauma exposure and stress-related disorders in inner city primary care patients. Gen Hosp Psychiatry. 2009;31(6):505–14. 10.1016/j.genhosppsych.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee J-Y, Kim S-W, Kim J-M. The impact of community disaster trauma: a focus on emerging research of PTSD and other mental health outcomes. Chonnam Med J. 2020;56(2):99–107. 10.4068/cmj.2020.56.2.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wolf EJ, Miller MW, Sullivan DR, Amstadter AB, Mitchell KS, Goldberg J, et al. A classical twin study of PTSD symptoms and resilience: evidence for a single spectrum of vulnerability to traumatic stress. Depress Anxiety. 2018;35(2):132–9. 10.1002/DA.22712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mundy J, Hübel C, Gelernter J, Levey D, Murray RM, Skelton M, et al. Psychological trauma and the genetic overlap between posttraumatic stress disorder and major depressive disorder. Psychol Med. 2021;52(16):1–10. 10.1017/S0033291721000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nievergelt CM, Maihofer AX, Klengel T, Atkinson EG, Chen C-Y, Choi KW, et al. International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat Commun. 2019;10(1):4558. 10.1038/s41467-019-12576-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mundy SS, Foss SLW, Poulsen S, Hjorthøj C, Carlsson J. Sex differences in trauma exposure and symptomatology in trauma-affected refugees. Psychiatry Res. 2020;293:113445. 10.1016/J.PSYCHRES.2020.113445. [DOI] [PubMed] [Google Scholar]
- 23. Stein MB, Levey DF, Cheng Z, Wendt FR, Harrington K, Pathak GA, et al. Genome-wide association analyses of post-traumatic stress disorder and its symptom subdomains in the Million Veteran Program. Nat Genet. 2021;53(2):174–84. 10.1038/s41588-020-00767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lori A, Schultebraucks K, Galatzer-Levy I, Daskalakis NP, Katrinli S, Smith AK, et al. Transcriptome-wide association study of post-trauma symptom trajectories identified GRIN3B as a potential biomarker for PTSD development. Neuropsychopharmacology. 2021;46(10):1811–20. 10.1038/s41386-021-01073-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mehta D, Bruenig D, Carrillo-Roa T, Lawford B, Harvey W, Morris CP, et al. Genomewide DNA methylation analysis in combat veterans reveals a novel locus for PTSD. Acta Psychiatr Scand. 2017;136(5):493–505. 10.1111/acps.12778. [DOI] [PubMed] [Google Scholar]
- 26. Rutten BPF, Vermetten E, Vinkers CH, Ursini G, Daskalakis NP, Pishva E, et al. Longitudinal analyses of the DNA methylome in deployed military servicemen identify susceptibility loci for post-traumatic stress disorder. Mol Psychiatry. 2018;23(5):1145–56. 10.1038/mp.2017.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hammamieh R, Chakraborty N, Gautam A, Muhie S, Yang R, Donohue D, et al. Whole-genome DNA methylation status associated with clinical PTSD measures of OIF/OEF veterans. Transl Psychiatry. 2017;7:e1169. 10.1038/tp.2017.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morrison FG, Miller MW, Logue MW, Assef M, Wolf EJ. DNA methylation correlates of PTSD: recent findings and technical challenges. Prog Neuropsychopharmacol Biol Psychiatry. 2019;90:223–34. 10.1016/J.PNPBP.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Montalvo-Ortiz JL, Gelernter J, Cheng Z, Girgenti MJ, Xu K, Zhang X, et al. Epigenome-wide association study of posttraumatic stress disorder identifies novel loci in U.S. military veterans. Transl Psychiatry. 2022;12(1):65. 10.1038/S41398-022-01822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smith AK, Ratanatharathorn A, Maihofer AX, Naviaux RK, Aiello AE, Amstadter AB, et al. Epigenome-wide meta-analysis of PTSD across 10 military and civilian cohorts identifies novel methylation changes in AHRR. Nat Commun. 2019;11(1):5965. 10.1101/585109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Duman RS, Girgenti MJ. Molecular and cellular studies of PTSD: postmortem transcriptome analysis and novel therapeutic targets. J Neurosci Res. 2019;97(3):292–9. 10.1002/JNR.24306. [DOI] [PubMed] [Google Scholar]
- 32. Logue MW, Miller MW, Wolf EJ, Huber BR, Morrison FG, Zhou Z, et al. An epigenome-wide association study of posttraumatic stress disorder in US veterans implicates several new DNA methylation loci. Clin Epigenetics. 2020;12(1):46. 10.1186/s13148-020-0820-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang J, Kaye AP, Wang J, Girgenti MJ. Transcriptomics of the depressed and PTSD brain. Neurobiol Stress. 2021;15:100408. 10.1016/J.YNSTR.2021.100408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zoladz PR, Diamond DM. Predator-based psychosocial stress animal model of PTSD: preclinical assessment of traumatic stress at cognitive, hormonal, pharmacological, cardiovascular and epigenetic levels of analysis. Exp Neurol. 2016;284(Pt B):211–9. 10.1016/J.EXPNEUROL.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 35. Richter-Levin G, Stork O, Schmidt MV. Animal models of PTSD: a challenge to be met. Mol Psychiatry. 2018;24(8):1135–56. 10.1038/s41380-018-0272-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Verbitsky A, Dopfel D, Zhang N. Rodent models of post-traumatic stress disorder: behavioral assessment. Transl Psychiatry. 2020;10(1):132. 10.1038/s41398-020-0806-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Borghans B, Homberg JR. Animal models for posttraumatic stress disorder: an overview of what is used in research. World J Psychiatry. 2015;5(4):387–96. 10.5498/wjp.v5.i4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bissiere S, Gasnier M, Alvarez YD, Plachta N. Cell fate decisions during preimplantation mammalian development. Curr Top Dev Biol. 2018;128:37–58. 10.1016/bs.ctdb.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 39. Burton A, Torres-Padilla ME. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat Rev Mol Cell Biol. 2014;15(11):723–34. 10.1038/nrm3885. [DOI] [PubMed] [Google Scholar]
- 40. Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lin L, Chu H, Hodges JS. Alternative measures of between-study heterogeneity in meta-analysis: reducing the impact of outlying studies. Biometrics. 2017;73(1):156–66. 10.1111/biom.12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kassambara A. ggpubr: “ggplot2” based publication ready plots. R package version 0.2. 2020. https://CRANR-project.org/package=ggpubr.
- 43. Wickham H. ggplot2. Wiley Interdiscip Rev Comput Stat. 2011;3(2):180–5. 10.1002/wics.147. [DOI] [Google Scholar]
- 44. Team RCR. A language and environment for statistical computing. R Found Stat Comput. 2021. [Google Scholar]
- 45. Wolfe D, Dudek S, Ritchie MD, Pendergrass SA. Visualizing genomic information across chromosomes with PhenoGram. BioData Min. 2013;6(1):18. 10.1186/1756-0381-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aoki S. Biorender. 2017. [Google Scholar]
- 47. Muhie S, Gautam A, Chakraborty N, Hoke A, Meyerhoff J, Hammamieh R, et al. Molecular indicators of stress-induced neuroinflammation in a mouse model simulating features of post-traumatic stress disorder. Transl Psychiatry. 2017;7(5):e1135. 10.1038/TP.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Muhie S, Gautam A, Meyerhoff J, Chakraborty N, Hammamieh R, Jett M. Brain transcriptome profiles in mouse model simulating features of post-traumatic stress disorder. Mol Brain. 2015;8:14. 10.1186/S13041-015-0104-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lori A, Maddox SA, Sharma S, Andero R, Ressler KJ, Smith AK. Dynamic patterns of threat-associated gene expression in the amygdala and blood. Front Psychiatry. 2018;9:778. 10.3389/fpsyt.2018.00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tanaka M, Li H, Zhang X, Singh J, Dalgard CL, Wilkerson M, et al. Region- and time-dependent gene regulation in the amygdala and anterior cingulate cortex of a PTSD-like mouse model. Mol Brain. 2019;12(1):25. 10.1186/S13041-019-0449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Malan-Müller S, De Souza VBC, Daniels WMU, Seedat S, Robinson MD, Hemmings SMJ. Shedding light on the transcriptomic dark matter in biological psychiatry: role of long noncoding RNAs in D-cycloserine-induced fear extinction in posttraumatic stress disorder. Disorder. 2020;24(6):352–69. 10.1089/OMI.2020.0031. [DOI] [PubMed] [Google Scholar]
- 52. Sillivan SE, Joseph NF, Jamieson S, King ML, Chévere-Torres I, Fuentes I, et al. Susceptibility and resilience to posttraumatic stress disorder-like behaviors in inbred mice. Biol Psychiatry. 2017;82(12):924–33. 10.1016/J.BIOPSYCH.2017.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Reis ALM, Hammond JM, Stevanovski I, Arnold JC, McGregor IS, Deveson IW, et al. Sex-specific transcriptomic and epitranscriptomic signatures of PTSD-like fear acquisition. iScience. 2022;25(9):104861. 10.1016/j.isci.2022.104861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jaffe AE, Tao R, Page SC, Maynard KR, Pattie EA, Nguyen CV, et al. Decoding shared versus divergent transcriptomic signatures across cortico-amygdala circuitry in PTSD and depressive disorders. Am J Psychiatry. 2022;179(9):673–86. 10.1176/appi.ajp.21020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Girgenti MJ, Wang J, Ji D, Cruz DA; Traumatic Stress Brain Research Group, Stein MB, et al. Transcriptomic organization of the human brain in post-traumatic stress disorder. Nat Neurosci. 2021;24(1):24–33. 10.1038/s41593-020-00748-7. [DOI] [PubMed] [Google Scholar]
- 56. Wolf EJ, Chen CD, Zhao X, Zhou Z, Morrison FG, Daskalakis NP, et al. Klotho, PTSD, and advanced epigenetic age in cortical tissue. Neuropsychopharmacology. 2021;46(4):721–30. 10.1038/S41386-020-00884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wolf EJ, Morrison FG, Sullivan DR, Logue MW, Guetta RE, Stone A, et al. The goddess who spins the thread of life: klotho, psychiatric stress, and accelerated aging. Brain Behav Immun. 2019;80:193–203. 10.1016/j.bbi.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wolf EJ, Zhao X, Hawn SE, Morrison FG, Zhou Z, Fein-Schaffer D, et al. Gene expression correlates of advanced epigenetic age and psychopathology in postmortem cortical tissue. Neurobiol Stress. 2021;15:100371. 10.1016/j.ynstr.2021.100371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cohen H, Kozlovsky N, Matar MA, Zohar J, Kaplan Z. Distinctive hippocampal and amygdalar cytoarchitectural changes underlie specific patterns of behavioral disruption following stress exposure in an animal model of PTSD. Eur Neuropsychopharmacol. 2014;24(12):1925–44. 10.1016/j.euroneuro.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 60. Popoli M, Yan Z, McEwen BS, Sanacora G. The stressed synapse: the impact of stress and glucocorticoids on glutamate transmission. Nat Rev Neurosci. 2011;13(1):22–37. 10.1038/nrn3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Han K, Lee M, Lim HK, Jang MW, Kwon J, Lee CJ, et al. Excitation-inhibition imbalance leads to alteration of neuronal coherence and neurovascular coupling under acute stress. J Neurosci. 2020;40(47):9148–62. 10.1523/JNEUROSCI.1553-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fang Q, Li Z, Huang G, Zhang HH, Chen YY, Zhang LB, et al. Traumatic stress produces distinct activations of GABAergic and glutamatergic neurons in amygdala. Front Neurosci. 2018;12:387. 10.3389/fnins.2018.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bhatt S, Hillmer AT, Girgenti MJ, Rusowicz A, Kapinos M, Nabulsi N, et al. PTSD is associated with neuroimmune suppression: evidence from PET imaging and postmortem transcriptomic studies. Nat Commun. 2020;11(1):2360. 10.1038/S41467-020-15930-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Flati T, Gioiosa S, Chillemi G, Mele A, Oliverio A, Mannironi C, et al. A gene expression atlas for different kinds of stress in the mouse brain. Sci Data. 2020;7(1):437. 10.1038/s41597-020-00772-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chen Y, Andres AL, Frotscher M, Baram TZ. Tuning synaptic transmission in the hippocampus by stress: the CRH system. Front Cell Neurosci. 2012;6:13. 10.3389/fncel.2012.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dunlop BW, Wong A. The hypothalamic-pituitary-adrenal axis in PTSD: pathophysiology and treatment interventions. Prog Neuropsychopharmacol Biol Psychiatry. 2019;89:361–79. 10.1016/J.PNPBP.2018.10.010. [DOI] [PubMed] [Google Scholar]
- 67. Nishi M. Effects of early-life stress on the brain and behaviors: implications of early maternal separation in rodents. Int J Mol Sci. 2020;21(19):7212. 10.3390/IJMS21197212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM, et al. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat Neurosci. 2013;16(1):33–41. 10.1038/NN.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wilker S, Pfeiffer A, Kolassa S, Elbert T, Lingenfelder B, Ovuga E, et al. The role of FKBP5 genotype in moderating long-term effectiveness of exposure-based psychotherapy for posttraumatic stress disorder. Transl Psychiatry. 2014;4(6):e403. 10.1038/TP.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bustamante AC, Aiello AE, Galea S, Ratanatharathorn A, Noronha C, Wildman DE, et al. Glucocorticoid receptor DNA methylation, childhood maltreatment and major depression. J Affect Disord. 2016;206:181–8. 10.1016/j.jad.2016.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Alexander N, Kirschbaum C, Wankerl M, Stauch BJ, Stalder T, Steudte-Schmiedgen S, et al. Glucocorticoid receptor gene methylation moderates the association of childhood trauma and cortisol stress reactivity. Psychoneuroendocrinology. 2018;90:68–75. 10.1016/j.psyneuen.2018.01.020. [DOI] [PubMed] [Google Scholar]
- 72. Palma-Gudiel H, Córdova-Palomera A, Leza JC, Fañanás L. Glucocorticoid receptor gene (NR3C1) methylation processes as mediators of early adversity in stress-related disorders causality: a critical review. Neurosci Biobehav Rev. 2015;55:520–35. 10.1016/J.NEUBIOREV.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 73. Martens M, Ammar A, Riutta A, Waagmeester A, Slenter DN, Hanspers K, et al. WikiPathways: connecting communities. Nucleic Acids Res. 2021;49(D1):D613–21. 10.1093/nar/gkaa1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cohen S, Matar MA, Vainer E, Zohar J, Kaplan Z, Cohen H. Significance of the orexinergic system in modulating stress-related responses in an animal model of post-traumatic stress disorder. Transl Psychiatry. 2020;10(1):10. 10.1038/s41398-020-0698-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Han D, Han F, Shi Y, Zheng S, Wen L. Mechanisms of memory impairment induced by orexin-A via orexin 1 and orexin 2 receptors in post-traumatic stress disorder rats. Neuroscience. 2020;432:126–36. 10.1016/J.NEUROSCIENCE.2020.02.026. [DOI] [PubMed] [Google Scholar]
- 76. Prajapati SK, Krishnamurthy S. Non-selective orexin-receptor antagonist attenuates stress-re-stress-induced core PTSD-like symptoms in rats: behavioural and neurochemical analyses. Behav Brain Res. 2021;399:113015. 10.1016/J.BBR.2020.113015. [DOI] [PubMed] [Google Scholar]
- 77. Strawn JR, Pyne-Geithman GJ, Ekhator NN, Horn PS, Uhde TW, Shutter LA, et al. Low cerebrospinal fluid and plasma orexin-A (hypocretin-1) concentrations in combat-related posttraumatic stress disorder. Psychoneuroendocrinology. 2010;35(7):1001–7. 10.1016/j.psyneuen.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 78. Stone LA, Girgenti MJ, Wang J, Ji D, Zhao H, Krystal JH, et al. Cortical transcriptomic alterations in association with appetitive neuropeptides and body mass index in posttraumatic stress disorder. Int J Neuropsychopharmacol. 2021;24(2):118–29. 10.1093/ijnp/pyaa072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yaeger JDW, Krupp KT, Summers TR, Summers CH. Contextual generalization of social stress learning is modulated by orexin receptors in basolateral amygdala. Neuropharmacology. 2022;215:109168. 10.1016/j.neuropharm.2022.109168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dustrude ET, Caliman IF, Bernabe CS, Fitz SD, Grafe LA, Bhatnagar S, et al. Orexin depolarizes central amygdala neurons via orexin receptor 1, phospholipase C and sodium-calcium exchanger and modulates conditioned fear. Front Neurosci. 2018;12:934. 10.3389/fnins.2018.00934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Pan YP, Liu C, Liu MF, Wang Y, Bian K, Xue Y, et al. Involvement of orexin-A in the regulation of neuronal activity and emotional behaviors in central amygdala in rats. Neuropeptides. 2020;80:102019. 10.1016/j.npep.2020.102019. [DOI] [PubMed] [Google Scholar]
- 82. Broberger C, De Lecea L, Sutcliffe JG, H-kfelt T. Hypocretin/orexin- and melanin-concentrating hormone-expressing cells form distinct populations in the rodent lateral hypothalamus: relationship to the neuropeptide y and agouti gene-related protein systems. J Comp Neurol. 1998;402(4):460–74. [PubMed] [Google Scholar]
- 83. Van Den Pol AN, PolVan Den AN. Hypothalamic hypocretin (orexin): robust innervation of the spinal cord. J Neurosci. 1999;19(8):3171–82. 10.1523/jneurosci.19-08-03171.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Peyron C, Tighe DK, Van Den Pol AN, De Lecea L, Heller HC, Sutcliffe JG, et al. Neurons containing hypocretin (orexin) project to multiple neuronal systems. J Neurosci. 1998;18(23):9996–10015. 10.1523/jneurosci.18-23-09996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sargin D. The role of the orexin system in stress response. Neuropharmacology. 2019;154:68–78. 10.1016/J.NEUROPHARM.2018.09.034. [DOI] [PubMed] [Google Scholar]
- 86. Flores Á, Maldonado R, Berrendero F. Cannabinoid-hypocretin cross-talk in the central nervous system: what we know so far. Front Neurosci. 2013;7:256. 10.3389/fnins.2013.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bahammam AS, Neubauer DN, Pandi-Perumal SR, Bahammam AS, Neubauer DN, Pandi-Perumal SR. Sodium oxybate (Xyrem<Superscript>®</Superscript>): a new and effective treatment for narcolepsy with cataplexy. Milestones Drug Ther. 2015;49:231–48. 10.1007/978-3-319-11514-6_11. [DOI] [Google Scholar]
- 88. Berridge CW, España RA, Vittoz NM. Hypocretin/orexin in arousal and stress. Brain Res. 2010;1314:91–102. 10.1016/j.brainres.2009.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Yun S, Wennerholm M, Shelton JE, Bonaventure P, Letavic MA, Shireman BT, et al. Selective inhibition of orexin-2 receptors prevents stress-induced ACTH release in mice. Front Behav Neurosci. 2017;11:83. 10.3389/fnbeh.2017.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Fujisawa S, Komatsubara M, Tsukamoto-Yamauchi N, Iwata N, Nada T, Wada J, et al. Orexin a enhances pro-opiomelanocortin transcription regulated by bmp-4 in mouse corticotrope att20 cells. Int J Mol Sci. 2021;22(9):4553. 10.3390/ijms22094553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Winsky-Sommerer R, Yamanaka A, Diano S, Borok E, Roberts AJ, Sakurai T. Interaction between the corticotropin-releasing factor system and hypocretins (orexins): a novel circuit mediating stress response. J Neurosci. 2004;24(50):11439. 10.1523/JNEUROSCI.3459-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhang HH, Meng SQ, Guo XY, Zhang JL, Zhang W, Chen YY, et al. Traumatic stress produces delayed alterations of synaptic plasticity in basolateral amygdala. Front Psychol. 2019;10:2394. 10.3389/fpsyg.2019.02394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Averill LA, Jiang L, Purohit P, Coppoli A, Averill CL, Roscoe J, et al. Prefrontal glutamate neurotransmission in PTSD: a novel approach to estimate synaptic strength in vivo in humans. Chronic Stress. 2022;6:24705470221092734. 10.1177/24705470221092734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tiwari P, Fanibunda SE, Kapri D, Vasaya S, Pati S, Vaidya VA. GPCR signaling: role in mediating the effects of early adversity in psychiatric disorders. FEBS J. 2021;288(8):2602–21. 10.1111/febs.15738. [DOI] [PubMed] [Google Scholar]
- 95. Hauger RL, Olivares-Reyes JA, Dautzenberg FM, Lohr JB, Braun S, Oakley RH. Molecular and cell signaling targets for PTSD pathophysiology and pharmacotherapy. Neuropharmacology. 2012;62(2):705–14. 10.1016/j.neuropharm.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Maddox SA, Kilaru V, Shin J, Jovanovic T, Almli LM, Dias BG, et al. Estrogen-dependent association of HDAC4 with fear in female mice and women with PTSD. Mol Psychiatry. 2018;23(3):658–65. 10.1038/mp.2016.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zhang L, Chen C, Qi J. Activation of HDAC4 and GR signaling contributes to stress-induced hyperalgesia in the medial prefrontal cortex of rats. Brain Res. 2020;1747:147051. 10.1016/j.brainres.2020.147051. [DOI] [PubMed] [Google Scholar]
- 98. Highland JN, Zanos P, Georgiou P, Gould TD. Group II metabotropic glutamate receptor blockade promotes stress resilience in mice. Neuropsychopharmacology. 2019;44(10):1788–96. 10.1038/s41386-019-0380-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ruso-Julve F, Pombero A, Pilar-Cuéllar F, García-Díaz N, Garcia-Lopez R, Juncal-Ruiz M, et al. Dopaminergic control of ADAMTS2 expression through cAMP/CREB and ERK: molecular effects of antipsychotics. Transl Psychiatry. 2019;9(1):306. 10.1038/s41398-019-0647-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Yamakage Y, Kato M, Hongo A, Ogino H, Ishii K, Ishizuka T, et al. A disintegrin and metalloproteinase with thrombospondin motifs 2 cleaves and inactivates Reelin in the postnatal cerebral cortex and hippocampus, but not in the cerebellum. Mol Cell Neurosci. 2019;100:103401. 10.1016/j.mcn.2019.103401. [DOI] [PubMed] [Google Scholar]
- 101. Wang J, Zhao H, Girgenti MJ. Posttraumatic stress disorder brain transcriptomics: convergent genomic signatures across biological sex. Biol Psychiatry. 2022;91(1):6–13. 10.1016/j.biopsych.2021.02.012. [DOI] [PubMed] [Google Scholar]
- 102. Chen Y, Wang Y, Ertürk A, Kallop D, Jiang Z, Weimer RM, et al. Activity-induced Nr4a1 regulates spine density and distribution pattern of excitatory synapses in pyramidal neurons. Neuron. 2014;83(2):431–43. 10.1016/J.NEURON.2014.05.027. [DOI] [PubMed] [Google Scholar]
- 103. Hashem S, Nisar S, Bhat AA, Yadav SK, Azeem MW, Bagga P, et al. Genetics of structural and functional brain changes in autism spectrum disorder. Transl Psychiatry. 2020;10(1):229. 10.1038/s41398-020-00921-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. McNeill RV, Ziegler GC, Radtke F, Nieberler M, Lesch KP, Kittel-Schneider S. Mental health dished up-the use of iPSC models in neuropsychiatric research. J Neural Transm. 2020;127(11):1547–68. 10.1007/S00702-020-02197-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Di Lullo E, Kriegstein AR, Di Lullo E, Kriegstein AR. The use of brain organoids to investigate neural development and disease. Nat Rev Neurosci. 2017;18(10):573–84. 10.1038/nrn.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Vadodaria KC, Amatya DN, Marchetto MC, Gage FH. Modeling psychiatric disorders using patient stem cell-derived neurons: a way forward. Genome Med. 2018;10:1–3. 10.1186/s13073-017-0512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Sawada T, Chater TE, Sasagawa Y, Yoshimura M, Fujimori-Tonou N, Tanaka K, et al. Developmental excitation-inhibition imbalance underlying psychoses revealed by single-cell analyses of discordant twins-derived cerebral organoids. Mol Psychiatry. 2020;25(11):2695–711. 10.1038/s41380-020-0844-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Quadrato G, Brown J, Arlotta P. The promises and challenges of human brain organoids as models of neuropsychiatric disease. Nat Med. 2016;22(11):1220–8. 10.1038/nm.4214. [DOI] [PubMed] [Google Scholar]
- 109. Mellén M, Ayata P, Heintz N. 5-Hydroxymethylcytosine accumulation in postmitotic neurons results in functional demethylation of expressed genes. Proc Natl Acad Sci U S A. 2017;114(37):E7812–21. 10.1073/pnas.1708044114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Irwin RE, Thakur A, O’ Neill KM, Walsh CP. 5-Hydroxymethylation marks a class of neuronal gene regulated by intragenic methylcytosine levels. Genomics. 2014;104(5):383–92. 10.1016/j.ygeno.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 111. Engel M, Eggert C, Kaplick PM, Eder M, Röh S, Tietze L, et al. The role of m6A/m-RNA methylation in stress response regulation. Neuron. 2018;99(2):389–403.e9. 10.1016/J.NEURON.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article and its online supplementary materials. Further inquiries may be directed to the corresponding author.