Abstract
Background
Fusobacterium nucleatum (F. nucleatum) is a vital pro-oncogenic bacterium. Our previous study revealed that a high abundance of F. nucleatum in head and neck squamous cell carcinoma (HNSCC) is correlated with poor patient prognosis. However, the impact of F. nucleatum on metabolic reprogramming and tumor progression in HNSCC awaits more exploration.
Methods
Liquid chromatography‒mass spectrometry (LC‒MS) was applied to analyze the altered metabolites in a head and neck carcinoma cell line (AMC-HN-8) after coculture with F. nucleatum for 24 hrs and 48 hrs. Both univariate and multivariate analyses were used to screen for differential metabolites. Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathway enrichment analysis was further used to explore the metabolic changes.
Results
We observed a significantly altered metabolic profile in AMC-HN-8 cells over time after coculture with F. nucleatum. Among the several enriched pathways, the purine metabolic pathway was the most significantly enriched (P = 0.0005), with downregulation of purine degradation. Furthermore, uric acid, the end product of purine metabolism, significantly reversed F. nucleatum-triggered tumor progression and altered the intracellular reactive oxygen species (ROS) level. Moreover, the negative correlation between the serum uric acid level and the abundance of F. nucleatum was verified in 113 HNSCC patients (P = 0.0412, R = − 0.1924).
Conclusions
Our study revealed obviously aberrant purine metabolism driven by F. nucleatum in HNSCC, which was closely related to tumor progression and patient prognosis. These findings indicate the possibility of targeting F. nucleatum-induced purine metabolism reprogramming in the future treatment of HNSCC.
Keywords: Head and neck carcinoma, Fusobacterium nucleatum, Metabolic reprogramming, Purine metabolic pathway, Prognosis
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the seventh most common cancer worldwide, with more than 500,000 newly diagnosed cases and 400,000 related deaths annually [1]. Moreover, its incidence shows an increasing trend and is estimated to increase 30% by 2030 [2]. In the past several years, advances have been made in the therapeutic approaches for HNSCC, including surgery, radiotherapy and systemic therapy [3]. However, these promising treatments, either alone or in combination, have not significantly improved patient prognosis, and the five-year survival rate of HNSCC has remained approximately 50% [4]. Hence, it is still vital to reveal the molecular mechanisms involved in disease progression and to provide more evidence for identifying new therapeutic targets.
In recent years, the involvement of the microbiota in cancer progression has gained increasing attention. The head and neck is a part of the upper respiratory airway, which is a natural shelter for various microbial floras [5]. Among these microbes, Fusobacterium have a higher abundance in HNSCC tissues than in noncancerous tissues, as verified in our previous studies [6–9]. Fusobacterium nucleatum (F. nucleatum), an important species of Fusobacterium, is considered a pro-oncogenic bacterium that mediates the initiation, progression and chemoresistance of several alimentary and respiratory cancers [10, 11]. However, the mechanism underlying these findings in HNSCC awaits more exploration.
We observed altered ethanol metabolism in HNSCC induced by F. nucleatum accumulation and validated the negative effect of this feed-forward loop on patient survival [12]. As F. nucleatum-induced metabolic reprogramming plays an important role in tumor progression, we hypothesized that F. nucleatum may alter metabolic profiles in addition to ethanol metabolism in HNSCC. Thus, in this study, we further elucidate F. nucleatum-induced metabolic reprogramming of HNSCC and explore the potential impact of this metabolic crosstalk on disease development and patient prognosis.
Method
Tissue sample collection and F. nucleatum detection
The study was approved by the ethics committee of the Eye & ENT Hospital, Fudan University. A total of 113 tissue samples were obtained from HNSCC patients undergoing surgery at the Eye & ENT Hospital, Fudan University. Patients were pathologically diagnosed with HNSCC, the classification was confirmed according to the American Joint Committee on Cancer (eighth edition) cancer staging manual. The exclusion criteria were as follows: (i) distant metastasis, (ii) a concurrent second primary tumor, (iii) active infection, (iv) antibiotic use within the three months before surgery, and (v) preoperative treatment, including chemotherapy or radiotherapy. The patients’ clinical data are shown in Table 1.
Table 1.
Patient clinical characteristics
| Number (%) | |
|---|---|
| Total | 113 (100) |
| Age | |
| > 65 years | 44 (38.9) |
| ≤ 65 years | 69 (61.1) |
| Sex | |
| Male | 111 (98.2) |
| Female | 2 (1.8) |
| Smoking | |
| Yes | 102 (90.3) |
| No | 11 (9.7) |
| Drinking | |
| Yes | 67 (59.3) |
| No | 46 (40.7) |
| Subregion | |
| Supraglottis | 44 (38.9) |
| Glottis | 65 (57.5) |
| Subglottis | 4 (3.5) |
| T classification | |
| T1/2 | 44 (38.9) |
| T3/4 | 69 (61.1) |
| N classification | |
| N0 | 75 (66.4) |
| N1 | 11 (9.7) |
| N2/3 | 27 (23.9) |
| C stage | |
| C1/2 | 34 (30.1) |
| C3/4 | 79 (69.9) |
By using the primer set for the gene prostaglandin transporter (PGT) extracted from fresh tissues and paraffin embedded tissues as the reference, the cycle threshold (Ct) values of F. nucleatum DNA were normalized to the amount of human genomic DNA (gDNA) as described previously (Castellarin et al. [13]). F. nucleatum DNA was amplified and detected in a 96-well optical PCR plate with an ABI 7500 Real-Time PCR System (Thermo Fisher, Massachusetts, USA) as previously described (Hsueh et al. [9]). Then, the -Δ Ct method was applied for relative quantification.
Cell and bacterial culture
Head and neck carcinoma cell lines, including AMC-HN-8 and FaDu, were generously given by Stem Cell Bank, Chinese Academy of Sciences and were cultured in RPMI-1640 medium (HyClone, Utah, USA) supplemented with 1% (v/v) penicillin‒streptomycin and 10% fetal bovine serum (FBS) under standard cell culture conditions (37 °C, 5% CO2). The F. nucleatum strain was purchased from the American Type Culture Collection (ATCC 25586, Virginia, USA) and was maintained overnight at 37 °C under anaerobic conditions on Columbia blood agar supplemented with five mg/mL hemin, 5% defibrinated sheep blood, and one mg/mL vitamin K1. AMC-HN-8 cells and F. nucleatum were then cocultured for 24 hs (Fn_24hr group) and 48 hs (Fn_48hr group) at a cell: bacteria ratio of 1:500 with six biological duplicates.
Cell collection and extraction
Cells were washed with ice-cold PBS. After trypsin digestion and centrifugation, detached cells were collected and then immediately stored at − 80 °C until analysis. Stored samples were thawed slowly on ice. To extract intracellular metabolites, 500 µL of pure methanol (Merck, Germany) was added to the sample. The mixture was vortexed for 5 min (Jingxin MIX-200, Shanghai, China), and subjected to three freeze‒thaw cycles. After centrifugation at 4 °C for 10 min (Eppendorf 5427R, Germany), 300 µL of methanol containing the internal standard was added to the sample. The mixture was then vortexed for one min and placed in a − 20 °C freezer for one hr. Then, the sample was centrifuged at 12,000 rpm at 4 °C for 10 min. 200 µL of the supernatant was retained and allowed to stand for 30 min. Then, centrifugation was repeated under the same conditions. The collected supernatant was then stored in a sample injection bottle for further analysis.
Liquid chromatography‒mass spectrometry (LC‒MS) analysis
Chromatographic analysis was performed using an ultra-high-performance liquid chromatography (UHPLC) system (Agilent 1290 INFINITY, USA). The mobile phases consisted of water with 0.04% acetic acid (solution A) (Thermo Fisher, USA) and acetonitrile (Merck, Germany) with 0.04% acetic acid (solution B). The column temperature was maintained at 40 °C. The flow rate was 0.35 mL/min, and the sample injection volume was 2 µL. Analysis was performed in both negative and positive ion modes with a Q-TOF mass spectrometer (Agilent 6545, USA). The voltages were 250 and 1500 in ESI − and ESI + modes, respectively. The other parameters were the same in bodes modes, and the settings were as follows: gas flow rate, 8; fragmentor voltage, 135; gas temperature, 325; sheath temperature, 325; sheath flow rate, 11; nebulizer gas pressure, 40. To examine the repeatability of the samples with the same treatment, we prepared samples for quality control by mixing equal amounts of extracts from the experimental samples. To further monitor the analysis’ stability, A quality control sample was inserted into every ten samples throughout the whole analysis.
Data extraction and compound identification
The original mass spectrometry data were converted into mzML format with ProteoWizard. The XCMS program was used for peak identification, peak alignment and retention time correction. The peak area was corrected by the SVR method. For every sample group, the peaks were filtered based on a deletion rate of greater than 50%. Then, metabolic information was identified by searching the MetWare database (MetWare Biotechnology, Wuhan, China) and a public metabolite information database combined with the MetDNA algorithm.
Cell proliferation, invasion, migration and intracellular ROS assays
The malignance of cancer cells is commonly evaluated by assessing their proliferation, migration and invasion. HNSCC cells were harvested at logarithmic growth phase and seeded into a 96-well plate at 3000 cells per well. Then, 10 μL Cell Counting Kit-8 (CCK8, Dojindo, Kumamoto, Japan) reagent was added to the culture medium and incubated with the sample for two hs. The optical density value at 450 nm were measured using a microplate reader (BioTek Instruments Inc., Vermont, USA). For the migration assay, 1 × 106 HSCC cells were first cultured with or without F. nucleatum or 0.7 g/L uric acid for 72 hrs. Then, they were resuspended in serum-free medium and seeded into the upper chamber of Transwell inserts (Corning, New York, USA) containing a membrane with an eight µm pore size. The bottom chamber contained RPMI-1640 medium or BEGM supplemented with 10% FBS. After an additional 24 hrs of incubation, the inserts were removed. The cells were washed with PBS, fixed with 4% methanol, and stained using crystal violet. Three fields/insert were randomly selected under a light microscope to count the stained cells. The cell invasion assay was performed using Transwell chambers containing Matrigel-coated membranes (BD Biosciences). Intracellular ROS levels were measured using a fluorometric intracellular ROS assay kit (Sigma). One microliter of ROS detection reagent stock solution was added to the cell medium per mL of cells, and the cells were incubated in a 5% CO2 incubator at 37 °C for 30 min. Then, images were acquired using fluorescence microscopy, and fluorescence intensity values were determined using ImageJ software (National Institutes of Health, USA).
Statistical analysis
To identify differential metabolites between the groups, both univariate and multivariate analyses were performed. The univariate analyses included Student’s t test and fold change analysis, and the multivariate analyses included principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA). PCA is an unsupervised multidimensional statistical analysis that was applied to visualize the overall metabolic differences and the variability within the groups. The quality control samples are labeled as “mix” in the PCA plot. OPLS-DA is a supervised multivariate analysis that maximizes the variance between groups. The variable importance in projection (VIP) value was calculated to summarize the relative importance of each X variable to the association between the X and Y variables. A VIP value of ≥ 1 indicated that the metabolite had a statistically significant contribution to the model. For correlation analysis, Pearson or Spearman correlation analysis was applied depending on the normality of the data. P < 0.05 was considered statistically significant.
Results
F. nucleatum significantly alters purine metabolism in AMC-HN-8 cells
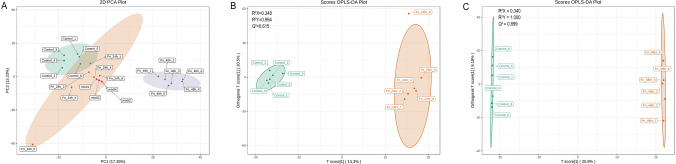
We explored changes in the overall metabolic profile of AMC-HN-8 cells after coculture with F. nucleatum. The PCA results showed evident separation among the three groups without outliers detected in the overview (Fig. 1A). Additionally, there were no obvious differences among the quality control samples, indicating that the system was stable for analysis with good repeatability. OPLS-DA was then performed to further track the metabolic changes in AMC-HN-8 cells after coculture with F. nucleatum. The OPLS-DA model achieved a good overall model fit (R2Y = 0.994 and 1.000 for the Fn_24hr group and Fn_48hr group, respectively) and prediction ability (Q2 = 0.615 and 0.899 for the Fn_24hr group and Fn_48hr group, respectively) (Fig. 1B, C). Therefore, in the initial assessment, we observed obviously different metabolic profiles in the Fn_48hr group, Fn_24hr group and control group, indicating that F. nucleatum can interfere with metabolism in AMC-HN-8 cells.
Fig. 1.
Overall metabolic profiles of control, Fn_24hr group and Fn_48hr groups. A principal component analysis (PCA) plot for three groups was performed to visualize the overall metabolic differences and the variability within the groups. B orthogonal partial least squares discriminant analysis (OPLS-DA) plot of Fn_24hr group and control group, and C OPLS-DA of Fn_48hr group and control group were performed to further track the metabolic changes, which maximizes the variance between groups. Fn_24hr group: AMC-HN-8 cells and F. nucleatum were cocultured for 24hs. Fn_48hr group: AMC-HN-8 cells and F. nucleatum were cocultured for 48hs
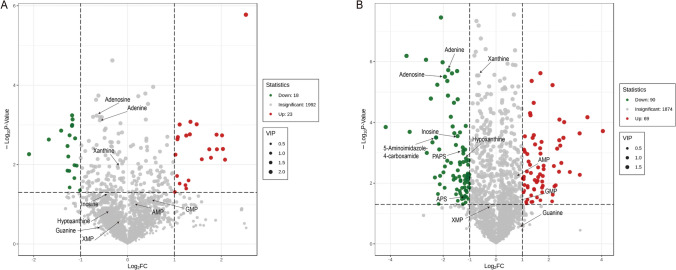
After data extraction and processing, we detected and identified a total of 2033 metabolites in all samples. Significant metabolites were confirmed by combining the results of both univariate and multivariate analyses. Only metabolites with P < 0.05 on the t test, a fold change (FC) of ≥ 2 or ≤ 0.5 and a VIP of ≥ 1 were considered significantly altered. According to these criteria, 41 metabolites in the Fn_24hr group, namely, 23 upregulated and 18 downregulated metabolites, were identified in the comparison with the control group (Fig. 2A). More significantly altered metabolites, including 69 upregulated and 90 downregulated metabolites, were observed in the comparison of the Fn_48hr and control groups (Fig. 2B). Thus, we verified the significantly altered metabolites in AMC-HN-8 cells after coculture with F. nucleatum, as directly shown in the volcano plots.
Fig. 2.
Volcano plot showing metabolic alterations of (A) Fn_24hr group vs. control group and (B) Fn_48hr group vs. control group. Arrows point out critical metabolites related to the purine metabolic pathway. Fn_24hr group: AMC-HN-8 cells and F. nucleatum were cocultured for 24 hs. Fn_48hr group: AMC-HN-8 cells and F. nucleatum were cocultured for 48 hs
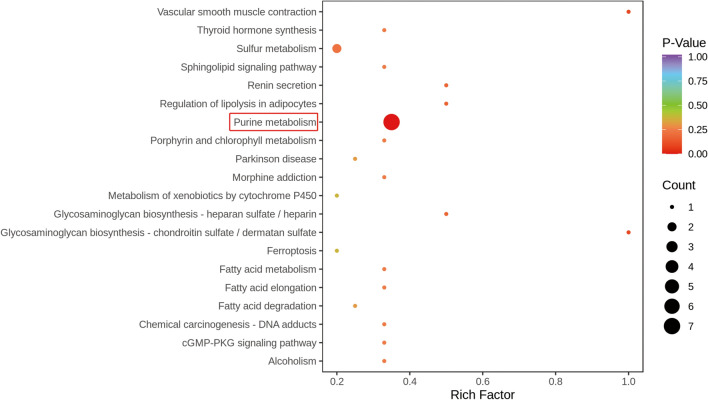
Candidate metabolites were then imported into the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis tool for metabolic pathway enrichment analysis. A total of 33 metabolic pathways were affected in AMC-HN-8 cells by F. nucleatum after 48 hs of coculture compared with control cells (Fig. 3). The top five metabolic pathways were purine metabolism, glycosaminoglycan biosynthesis–chondroitin sulfate/dermatan sulfate, vascular smooth muscle contraction, renin secretion and regulation of lipolysis in adipocytes (Table 2). The most significant metabolic pathway (P = 0.0005), purine metabolism, was enriched with 20 identifiable metabolites and seven significantly altered metabolites, namely, adenine, adenosine, hypoxanthine, inosine, phosphoadenosine phosphosulfate (PAPS), adenosine-5’-phosphosulfate (APS) and 5-aminoimidazole-4-carboxamide. These results indicate that purine metabolic reprogramming plays a dominant role in the metabolic crosstalk of F. nucleatum with AMC-HN-8 cells.
Fig. 3.
Metabolic pathway enrichment analysis for significantly altered metabolites for Fn_48hr group and control group. Purine metabolism was the most significant metabolic pathway, which enriched seven significantly altered metabolites
Table 2.
Metabolic pathway enrichment distinguished by Fn_48hr and control
| KEGG pathway | Cluster frequency | P value | |
|---|---|---|---|
| Total | Significant metabolites | All metabolites | |
| 19 (100.00%) | 217 (100.00%) | ||
| Purine metabolism | 7 (36.84%) | 20 (9.22%) | 0.0005 |
| Glycosaminoglycan biosynthesis—chondroitin sulfate/dermatan sulfate | 1 (5.26%) | 1 (0.46%) | 0.0876 |
| Vascular smooth muscle contraction | 1 (5.26%) | 1 (0.46%) | 0.0876 |
| Renin secretion | 1 (5.26%) | 2 (0.92%) | 0.1678 |
| Regulation of lipolysis in adipocytes | 1 (5.26%) | 2 (0.92%) | 0.1678 |
*: The table showed the top five metabolic pathways
F. nucleatum causes downregulation of purine degradation in AMC-HN-8 cells
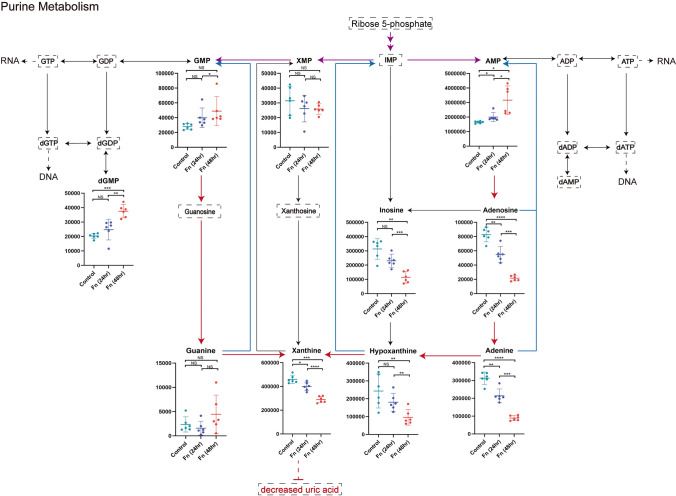
To further explore the altered purine metabolic pathways, we summarized all detected purine-related metabolites in Table 3 and showed the relationships among critical metabolites in Fig. 4. We found that compared to those in the control group, the levels of adenosine 5’-monophosphate (AMP) and guanosine 5’-phosphate (GMP), which are not only two key end-products of purine synthesis but also substrates for further synthesis of DNA and RNA, were increased over time (P = 0.0338 and 0.0101 for AMP, P = 0.1010 and 0.0274 for GMP, respectively). In addition to upregulation of purine synthesis, the downregulation of purine degradation is also observed in our study. Although uric acid could not be detected, the levels of its upstream metabolites involved in purine degradation, including xanthine (P = 0.0181 and 0.0002), hypoxanthine (P = 0.1597 and 0.0089) and adenine (P = 0.0067 and < 0.0001), were significantly decreased in a time-dependent manner compared to those in the control group. This result reveals that F. nucleatum can cause significant decreases in the levels of metabolites in the purine degradation pathway, which may provide essential purine nucleotides to support cancer cell proliferation.
Table 3.
Difference analysis of metabolites related to purine metabolism pathway
| Metabolites | Fn_24hr vs. control | Fn_48hr vs. control | Fn_48hr vs. Fn_24hr | |||
|---|---|---|---|---|---|---|
| P value* | FC | P value | FC | P value | FC | |
| Adenine | 0.0067 | 0.68 | < 0.0001 | 0.28 | 0.0001 | 0.42 |
| Adenosine | 0.0095 | 0.66 | < 0.0001 | 0.26 | 0.0001 | 0.39 |
| Hypoxanthine | 0.1597 | 0.74 | 0.0089 | 0.39 | 0.0022 | 0.53 |
| Inosine | 0.0627 | 0.74 | 0.0028 | 0.36 | 0.0002 | 0.50 |
| PAPS | 0.0485 | 0.65 | 0.0040 | 0.44 | 0.0626 | 0.68 |
| APS | 0.6943 | 0.92 | 0.0312 | 0.41 | 0.0092 | 0.44 |
| AMP | 0.0338 | 1.22 | 0.0101 | 1.92 | 0.0278 | 1.58 |
| AppppA | 0.0413 | 1.50 | 0.0091 | 1.95 | 0.2587 | 1.30 |
| Sulfate | 0.1383 | 1.21 | 0.9233 | 1.02 | 0.0542 | 0.84 |
| Xanthine | 0.0181 | 0.87 | 0.0002 | 0.63 | < 0.0001 | 0.72 |
| XMP | 0.4004 | 0.83 | 0.2067 | 0.82 | 0.9208 | 0.99 |
| Guanine | 0.3620 | 0.67 | 0.0929 | 1.89 | 0.1056 | 2.82 |
| GMP | 0.1010 | 1.43 | 0.0274 | 1.75 | 0.4194 | 1.23 |
| dGMP | 0.2015 | 1.22 | 0.0002 | 1.85 | 0.0017 | 1.52 |
| Allantoic acid | 0.0633 | 1.07 | 0.5780 | 0.98 | 0.0112 | 0.91 |
| 5-Aminoimidazole-4-carboxamide | 0.1549 | 0.70 | 0.0004 | 0.21 | 0.0039 | 0.30 |
| ppGpp | 0.2117 | 0.71 | 0.3996 | 1.14 | 0.0605 | 1.60 |
| ADP-ribose | 0.1783 | 0.90 | 0.0686 | 1.29 | 0.0149 | 1.43 |
| 5-Amino-4-imidazole carboxylate | 0.1812 | 0.87 | < 0.0001 | 0.57 | 0.0116 | 0.65 |
| IDP | 0.4088 | 1.31 | 0.2108 | 1.34 | 0.8791 | 1.03 |
*: P value was obtained from t-test
Fig. 4.
Overview of changed metabolites in purine metabolic pathway. Purple arrow: de novo biosynthetic pathway. Blue arrow: complementary salvage pathway. Red arrow: purine degradation pathway. Metabolites surrounded by dashed line were undetectable ones. *: P < 0.05. **: P < 0.01. ***: P < 0.001. ****: P < 0.0001
F. nucleatum-triggered tumor progression and ROS accumulation can be reversed by uric acid
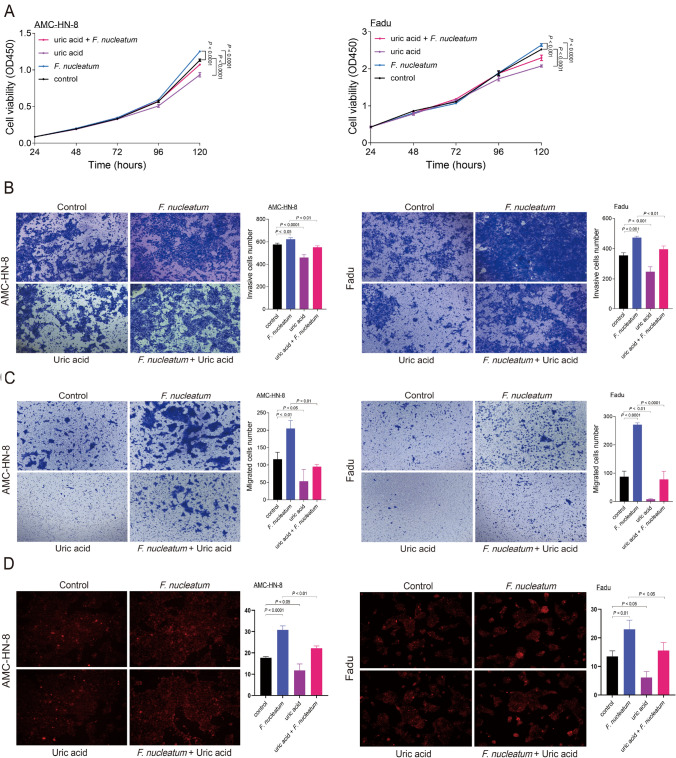
We then explored the roles of F. nucleatum and uric acid in HNSCC progression. Our results showed that F. nucleatum significantly promoted the proliferation (P < 0.0001 and P < 0.001 in AMC-HN-8 and FaDu cells, respectively) and facilitated the invasion (P = 0.0262, P = 0.0005) and migration (P = 0.0043, P < 0.0001) of both AMC-HN-8 and FaDu cells. In contrast, uric acid significantly inhibited tumor cell proliferation (P < 0.0001 and P < 0.0001 in AMC-HN-8 and FaDu cells, respectively), invasion (P < 0.0001, P = 0.0009) and migration (P = 0.0274, P = 0.0017). F. nucleatum-triggered tumor progression was reversed by uric acid, as indicated by the decreases in proliferation (P < 0.0001 and P < 0.0001 in AMC-HN-8 and FaDu cells, respectively), invasion (P = 0.0024, P = 0.0076) and migration (P = 0.0011, P < 0.0001) (Fig. 5A, B, C). Furthermore, F. nucleatum led to ROS accumulation (P < 0.0001 and P = 0.0056 in AMC-HN-8 and FaDu cells, respectively), whereas uric acid decreased intracellular ROS levels (P = 0.0161, P = 0.0217). F. nucleatum-triggered ROS accumulation was reversed by uric acid (P = 0.0016, P = 0.0212) (Fig. 5D). Thus, uric acid may reverse F. nucleatum-triggered tumor progression by decreasing intracellular ROS levels.
Fig. 5.
F. nucleatum–triggered tumor progression and ROS accumulation could be reversed by uric acid: A cell proliferation measured using Cell Counting Kit-8 assay (CCK8), B invasion assay performed using transwell chambers, C migration assay performed using transwell chambers and D Intracellular ROS detection using fluorometric intracellular ROS assay kit. Left: AMC-HN-8 cells. Right: Fadu cells
F. nucleatum is negatively correlated with the serum uric acid level and leads to poor prognosis in patients
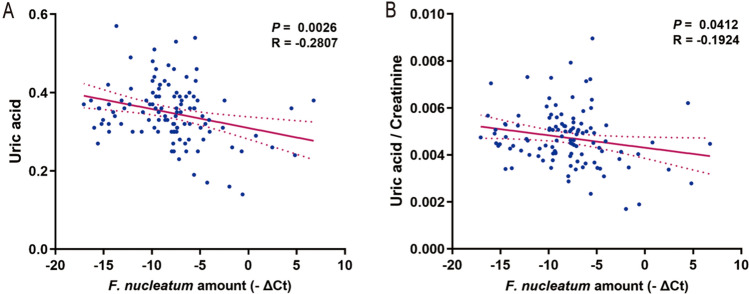
We then explored the correlation between the serum uric acid level and F. nucleatum abundance in 113 patients diagnosed with HNSCC. Patients’ serum was collected within one month before surgery. The results showed that the uric acid level in serum was significantly negatively correlated with the abundance of F. nucleatum (P = 0.0026, R = -0.2807), as shown in Fig. 6A. Furthermore, to exclude the possible impact of patients’ renal function, we studied the relationship between the uric acid-to-creatinine ratio and the abundance of F. nucleatum. Similarly, a significant negative correlation was observed between the uric acid-to-creatinine ratio and the F. nucleatum abundance (P = 0.0412, R = − 0.1924), as shown in Fig. 6B. These results support the hypothesis that F. nucleatum can lead to a decreased level of uric acid in serum, which is strongly associated with unfavorable survival outcomes in patients.
Fig. 6.
Correlation analysis of F. nucleatum abundance and uric acid level in head and neck squamous cell carcinoma (HNSCC) patients. A F. nucleatum and uric acid correlation analysis, B F. nucleatum and uric acid/creatinine correlation analysis
Discussion
Metabolic reprogramming is vital for cancer cells with rapid proliferation to meet the energy demand for survival and to adapt to the surrounding microenvironment, which is a hallmark of cancer [14]. Emerging evidence has revealed that F. nucleatum can induce metabolic shifts in cancer cells, such as increases in glucose metabolism and glutamine metabolism, which are closely related to tumor occurrence and development [15, 16]. In this study, we showed an altered metabolic profile in HNSCC cells induced by F. nucleatum using LS-MS. This alteration was enhanced over time after coculture. The results of KEGG pathway enrichment analysis showed that these significantly changed metabolites were related to several critical metabolic pathways, with purine metabolism as the most significantly enriched pathway. These results supported our previous assumption that the involvement of F. nucleatum is vital to the mechanism underlying metabolic reprogramming and poor clinical outcomes in patients with HNSCC.
Uric acid is the end product of purine degradation and is a well-known potent antioxidant in bodily fluids [17]. Although there is still controversy regarding the antitumor effect of uric acid in different types of cancer, our previous study in 814 HNSCC patients revealed that uric acid could serve as an independent prognostic factor for HNSCC [18]. Higher uric acid levels in serum before surgery were significantly correlated with favorable survival outcomes in patients [18]. In line with our previous findings, large prospective population-based studies have shown that a high uric acid level is inversely associated with the risk of cancer and, furthermore, that a high uric acid level can predict better prognosis in patients with cancer [19, 20]. The present study revealed one possible explanation for this phenomenon. F. nucleatum could lead to a decreasing trend in purine degradation and thus a decreased uric acid level in HNSCC, which then facilitates tumor progression and results in poor patient outcomes.
Downregulated purine degradation means that more purine nucleotides are available to support cell proliferation. Purine metabolism plays a paramount role in providing substrates for DNA and RNA biosynthesis. Additionally, it generates essential energy materials and cofactors to support cell survival and proliferation [21]. Previous studies have noted aberrant purine metabolism in tumor cells, and antitumor drugs targeting purine metabolism were among the first drugs to be widely used in clinical practice [22, 23]. One feature of this aberrant metabolism is enhanced purine biosynthesis to provide fundamental purine nucleotides to support the uncontrolled proliferation characterizing tumor cells [24]. In our study, F. nucleatum induced increases in the AMP and GMP levels in AMC-HN-8 cells, indicating that F. nucleatum increases purine supplementation to support cancer proliferation.
Downregulation of the purine degradation pathway is closely related to the altered level of reactive oxygen species (ROS) in cells, which may disrupt the balance between the amounts of ROS generated and scavenged. ROS play a dual role as both tumor suppressors and tumor promoters [25]. From one aspect, ROS can exert their tumor-suppressive effects and serve as a general antitumor mechanism in all treatment modalities except surgery [25, 26]. During purine degradation, hypoxanthine is first oxidized into xanthine, which is the main substrate that is further oxidized into uric acid. It has been verified by in vitro studies that ROS generated via this xanthine oxidase system can exert inhibitory effects on proliferation [27]. From another aspect, as ROS may lead to DNA, RNA, protein and lipid damage, they promote tumor generation and progression [28]. Uric acid is a well-known potent antioxidant that can react with ROS and prevent this damage. Thus, a decreased uric acid level indicates an increased chance for cancer cells to be exposed to ROS, which confers tumor growth advantages. According to the results of our study, the significant increase in the intracellular ROS level triggered by F. nucleatum was reversed by uric acid, which may indicate the mechanism underlying the protective role of uric acid in HNSCC.
Our study also indicated that F. nucleatum upregulated the de novo biosynthetic pathway in HNSCC. There are two main sources in purine metabolism from which mammalian cells derive purine nucleotides, namely, the complementary salvage pathway and the de novo biosynthetic pathway, and a balance is maintained between these pathways in cells [29]. As described in previous studies, when there is an emerging requirement for purine nucleotides, as occurs in tumor cells, the de novo biosynthetic pathway is the basis for replenishing the purine pool [30]. Elevated levels of enzymes involved in the de novo biosynthetic pathway, such as PPAT and PAICS, have been observed in lung cancer and may serve as potential prognostic markers [31]. Although metabolites related to the de novo biosynthetic pathway were not detected in our study, we discovered that the levels of three critical metabolites in the complementary salvage pathway—hypoxanthine, adenosine and adenine—were significantly decreased, indicating that increased AMP synthesis may originate via the de novo biosynthetic pathway.
Conclusion
Overall, our study has significant implications for understanding the mechanism by which F. nucleatum alters the metabolic profile in HNSCC. Most importantly, F. nucleatum triggers obviously aberrant purine metabolism and an imbalance in intracellular ROS, leading to tumor progression and poor prognosis in patients, which can be reversed by uric acid. These findings indicate the possibility of targeting F. nucleatum-induced purine metabolism reprogramming in the future treatment of HNSCC.
Acknowledgements
The authors would like to thank all participating investigators.
Author contributions
Conceptualization and data curation: MZ and HG formal analysis: HH and JX funding acquisition: MZ, HG and CH investigation and methodology: FL and CH project administration: MZ, HG and CH resources, software and supervision: LT and LZ validation: FL, HH and JX visualization: HG and CH writing—original draft: FL, HH and JX writing—review and editing: MZ, HG, CH, LT and LZ.
Funding
This study was supported by the Natural Science Foundation of Shanghai (No. 21ZR1411900), the National Natural Science Foundation of China (82273097 and 81972529), Shanghai Municipal Key Clinical Specialty (shslczdzk00801), the Science and Technology Commission of Shanghai Municipality (21Y11900100 and 19411961300), Shanghai Sailing Program (22YF1405700).
Data availability
Data availability on request from the author
Declarations
Ethics approval
This study was approved by the ethical committee of Eye & ENT Hospital, Fudan University (ethical committees or internal review boards: the ethical committee of Eye & ENT Hospital, Fudan University; guidelines: Declaration of Helsinki and relevant regulation of the National Health Commission and National Medical Products Administration).
Consent to participate and publication
All authors have approved the submission and publishment of this manuscript.
Competing interests
The authors have no conflicts of interest to disclose.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Feiran Li, Huiying Huang and Jing Xu are first author.
Contributor Information
Chiyao Hsueh, Email: hsuehchiyao@gmail.com.
Hongli Gong, Email: gonghlent@126.com.
Ming Zhang, Email: ent_zhm@126.com.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries [J] CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Johnson DE, Burtness B, Leemans CR, et al. Head and neck squamous cell carcinoma [J] Nat Rev Dis Primers. 2020;6(1):92. doi: 10.1038/s41572-020-00224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mody MD, Rocco JW, Yom SS, et al. Head and neck cancer [J] Lancet. 2021;398(10318):2289–2299. doi: 10.1016/S0140-6736(21)01550-6. [DOI] [PubMed] [Google Scholar]
- 4.Pignon J-P, le Maître A, Maillard E, Bourhis J, et al. Meta-analysis of chemotherapy in head and neck cancer (MACH-NC): an update on 93 randomised trials and 17,346 patients [J] Radiother Oncol. 2009;92(1):4–14. doi: 10.1016/j.radonc.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 5.Li K, Bihan M, Yooseph S, et al. Analyses of the microbial diversity across the human microbiome [J] PLoS ONE. 2012;7(6):e32118. doi: 10.1371/journal.pone.0032118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong HL, Shi Y, Zhou L, et al. The composition of microbiome in larynx and the throat biodiversity between laryngeal squamous cell carcinoma patients and control population [J] PLoS ONE. 2013;8(6):e66476. doi: 10.1371/journal.pone.0066476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gong H, Shi Y, Zhou X, et al. Microbiota in the throat and risk factors for laryngeal carcinoma [J] Appl Environ Microbiol. 2014;80(23):7356–7363. doi: 10.1128/AEM.02329-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong H, Shi Y, Xiao X, et al. Alterations of microbiota structure in the larynx relevant to laryngeal carcinoma [J] Sci Rep. 2017;7(1):5507. doi: 10.1038/s41598-017-05576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsueh CY, Gong H, Cong N, et al. Throat microbial community structure and functional changes in postsurgery laryngeal carcinoma patients [J] Appl Environ Microbiol. 2020 doi: 10.1128/AEM.01849-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium [J] Nat Rev Microbiol. 2019;17(3):156–166. doi: 10.1038/s41579-018-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han YW. Fusobacterium nucleatum: a commensal-turned pathogen [J] Curr Opin Microbiol. 2015;23:141–147. doi: 10.1016/j.mib.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsueh CY, Huang Q, Gong H, et al. A positive feed-forward loop between Fusobacterium nucleatum and ethanol metabolism reprogramming drives laryngeal cancer progression and metastasis [J] iScience. 2022;25(2):103829. doi: 10.1016/j.isci.2022.103829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma [J] Genome Res. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism [J] Cell Metab. 2016;23(1):27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong J, Guo F, Lu S-Y, et al. F. nucleatum targets lncRNA ENO1-IT1 to promote glycolysis and oncogenesis in colorectal cancer [J] Gut. 2021;70(11):2123–2137. doi: 10.1136/gutjnl-2020-322780. [DOI] [PubMed] [Google Scholar]
- 16.Ternes D, Tsenkova M, Pozdeev VI, et al. The gut microbial metabolite formate exacerbates colorectal cancer progression [J] Nat Metab. 2022;4(4):458–475. doi: 10.1038/s42255-022-00558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ames BN, Cathcart R, Schwiers E, et al. Uric acid provides an antioxidant defense in humans against oxidant- and radical-caused aging and cancer: a hypothesis [J] Proc Natl Acad Sci U S A. 1981;78(11):6858–6862. doi: 10.1073/pnas.78.11.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsueh CY, Shao M, Cao W, et al. Pretreatment serum uric acid as an efficient predictor of prognosis in men with laryngeal squamous cell cancer: a retrospective cohort study [J] Oxid Med Cell Longev. 2019 doi: 10.1155/2019/1821969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taghizadeh N, Vonk JM, Boezen HM. Serum uric acid levels and cancer mortality risk among males in a large general population-based cohort study [J] Cancer Causes Control. 2014;25(8):1075–1080. doi: 10.1007/s10552-014-0408-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kühn T, Sookthai D, Graf ME, et al. Albumin, bilirubin, uric acid and cancer risk: results from a prospective population-based study [J] Br J Cancer. 2017;117(10):1572–1579. doi: 10.1038/bjc.2017.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin J, Ren W, Huang X, et al. Potential mechanisms connecting purine metabolism and cancer therapy [J] Front Immunol. 2018 doi: 10.3389/fimmu.2018.01697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fridley BL, Batzler A, Li L, et al. Gene set analysis of purine and pyrimidine antimetabolites cancer therapies [J] Pharmacogenet Genomics. 2011;21(11):701–712. doi: 10.1097/FPC.0b013e32834a48a9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parker WB. Enzymology of purine and pyrimidine antimetabolites used in the treatment of cancer [J] Chem Rev. 2009;109(7):2880–2893. doi: 10.1021/cr900028p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu L, Aa J, Xu J, et al. Metabolomic phenotype of gastric cancer and precancerous stages based on gas chromatography time-of-flight mass spectrometry [J] J Gastroenterol Hepatol. 2011;26(8):1290–1297. doi: 10.1111/j.1440-1746.2011.06724.x. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Yi J. Cancer cell killing via ROS: to increase or decrease, that is the question [J] Cancer Biol Ther. 2008;7(12):1875–1884. doi: 10.4161/cbt.7.12.7067. [DOI] [PubMed] [Google Scholar]
- 26.Renschler MF. The emerging role of reactive oxygen species in cancer therapy [J] Eur J Cancer. 2004;40(13):1934–1940. doi: 10.1016/j.ejca.2004.02.031. [DOI] [PubMed] [Google Scholar]
- 27.Sakuma S, Abe M, Kohda T, et al. Hydrogen peroxide generated by xanthine/xanthine oxidase system represses the proliferation of colorectal cancer cell line Caco-2 [J] J Clin Biochem Nutr. 2015;56(1):15–19. doi: 10.3164/jcbn.14-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farhadi A, Fields J, Banan A, et al. Reactive oxygen species: are they involved in the pathogenesis of GERD, Barrett's esophagus, and the latter’s progression toward esophageal cancer? [J] Am J Gastroenterol. 2002;97(1):22–26. doi: 10.1111/j.1572-0241.2002.05444.x. [DOI] [PubMed] [Google Scholar]
- 29.Yamada S, Sato A, Sakakibara SI. Nwd1 Regulates Neuronal Differentiation and Migration through Purinosome Formation in the Developing Cerebral Cortex [J] iScience. 2020;23(5):101058. doi: 10.1016/j.isci.2020.101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamaoka T, Kondo M, Honda S, et al. Amidophosphoribosyltransferase limits the rate of cell growth-linked de novo purine biosynthesis in the presence of constant capacity of salvage purine biosynthesis [J] J Biol Chem. 1997;272(28):17719–17725. doi: 10.1074/jbc.272.28.17719. [DOI] [PubMed] [Google Scholar]
- 31.Goswami MT, Chen G, Chakravarthi BV, et al. Role and regulation of coordinately expressed de novo purine biosynthetic enzymes PPAT and PAICS in lung cancer [J] Oncotarget. 2015;6(27):23445–23461. doi: 10.18632/oncotarget.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data availability on request from the author