Abstract
Protein quality control pathways have evolved to ensure the fidelity of protein synthesis and efficiently clear potentially toxic protein species. Defects in ribosome-associated quality control and its associated factors have been implicated in the accumulation of aberrant proteins and neurodegeneration. C9orf72 repeat-associated non-AUG translation has been suggested to involve inefficient translation elongation, lead to ribosomal pausing and activation of ribosome-associated quality control pathways. However, the role of the ribosome-associated quality control complex in the processing of proteins generated through this non-canonical translation is not well understood. Here we use reporter constructs containing the C9orf72-associated hexanucleotide repeat, ribosome-associated quality control complex deficient cell models and stain for ribosome-associated quality control markers in C9orf72-expansion carrier human tissue to understand its role in dipeptide-repeat protein pathology.
Our studies show that canonical ribosome-associated quality control substrates products are efficiently cleared by the ribosome-associated quality control complex in mammalian cells. Furthermore, using stalling reporter constructs, we show that repeats associated with the C9orf72-expansion induce ribosomal stalling when arginine (R)-rich dipeptide-repeat proteins are synthesized in a length-dependent manner. However, despite triggering this pathway, these arginine-rich dipeptide-repeat proteins are not efficiently processed by the core components of the ribosome-associated quality control complex (listerin, nuclear-export mediator factor and valosin containing protein) partly due to lack of lysine residues, which precludes ubiquitination. Deficient processing by this complex may be implicated in C9orf72-expansion associated disease as dipeptide-repeat protein inclusions were observed to be predominantly devoid of ubiquitin and co-localize with nuclear-export mediator factor in mutation carriers’ frontal cortex and cerebellum tissue. These findings suggest that impaired processing of these arginine-rich dipeptide-repeat proteins derived from repeat-associated non-AUG translation by the ribosome-associated quality control complex may contribute to protein homeostasis dysregulation observed in C9orf72-expansion amyotrophic lateral sclerosis and frontotemporal degeneration neuropathogenesis.
Keywords: ribosome-associated quality control, C9orf72, ALS/FTD, RAN translation, NEMF
Viera Ortiz et al. show that the ribosome-associated quality control (RQC) complex cannot degrade C9orf72 dipeptide-repeat proteins (DPR), despite arginine-dependent ribosome stalling. Co-accumulation of the RQC factor NEMF with DPR aggregates in cells and human brain suggests RQC dysfunction may contribute to C9orf72-associated pathogenesis.
Introduction
Protein homeostasis dysregulation and protein aggregation are hallmarks of neurodegenerative diseases. Many protein quality control pathways have evolved to ensure the fidelity of protein synthesis from genetic material. Faulty mRNAs are continuously degraded by tightly monitored mRNA decay pathways while their defective and potentially harmful protein products are eliminated by ribosome-associated quality control (RQC) pathways.1–4 Repeat-associated non-AUG (RAN) translation is a non-canonical translation process associated with repeat expansions.5–7 These repeat expansions are involved in a variety of neurogenetic disorders like fragile X-associated tremor/ataxia syndrome, Huntington's disease, amyotrophic lateral sclerosis (ALS) and frontotemporal degeneration (FTD).7–10 RAN translation generates proteins that accumulate in patient brains and may contribute to toxicity.5–8,11–13 How these aberrant protein species may be cleared or escape cellular quality control pathways is not well understood and may play an important role in the pathobiology of these diseases. This study focused on the role of the RQC complex in the clearance of proteins that arise from RAN translation of the C9orf72 (Chromosome 9 open reading frame 72) repeat expansion associated with ALS and FTD.
The G4C2 repeat expansion in the first intron of the C9orf72 gene is the most prevalent pathogenetic variant of both ALS and FTD.14,15 Potential mechanisms linked to this mutation include C9orf72 haploinsufficiency through reduced mRNA and protein levels, RNA toxicity mediated by sequestering of RNA-binding proteins and dipeptide-repeat (DPR) proteins that are produced through RAN translation.6,10,13,16–19 These DPR proteins are generated by RAN translation of the sense and antisense strand of the repeat-containing RNA at different reading frames. All five different DPR products generated from the C9orf72 repeat accumulate in patient brains and have been suggested to disrupt multiple cellular processes.6,10,20–22 The G4C2 RNA has been shown to be able to form stable RNA secondary structures in vitro and has been associated with impaired translation elongation.23–28 Currently, the regulation of C9orf72 RAN translation and its DPR protein products remains poorly understood.
Ribosome stalling during elongation results in the activation of mRNA decay and RQC pathways. The RQC complex recycles stalled ribosomal subunits by facilitating efficient extraction and degradation of the nascent polypeptide.2,29–35 Binding of the RQC complex component nuclear-export mediator factor (NEMF) to the ribosomal subunit/nascent polypeptide recruits the E3 ubiquitin ligase, listerin (LTN1) and can also mediate the addition of residues to the C terminus of the nascent polypeptide (named C-terminal extensions, CTE) thought to promote availability of lysine residues for ubiquitination.29–31,33–37 Following tRNA release, the nascent polypeptide is extracted by valosin containing protein (VCP) for downstream proteasomal degradation.30,38,39 Lack of efficient ubiquitination and degradation of the nascent polypeptide has been shown to lead to CTE-mediated aggregation in yeast.30,34,36,40 Furthermore, mutations in core RQC complex components, such as listerin and NEMF, and RQC dysfunction have been associated with neurodegeneration and ageing.41–46 Therefore, impaired RQC, RQC-mediated CTEs and sequestration of RQC-associated factors may contribute to the pathobiology of proteins derived from the stalling prone translation of repetitive elements characteristic of RAN translation.
Recent studies have shown that translation of R-rich (arginine-rich) DPR proteins results in ribosome stalling and can lead to dysregulation of RQC-associated factors.28,47–49 Furthermore, some studies have implicated that C-terminally extended poly-GR (glycine-arginine) species may contribute to mitochondrial stress.48 To further expand our understanding of the mechanism by which R-rich DPRs induce ribosome stalling, we used stalling reporters containing the C9orf72-associated repeats and DPR protein encoding sequences to assess elongation dynamics in cell and in vitro neuronal models. We evaluated the functional role of the core RQC complex in the expression of DPR proteins in mammalian cells. Moreover, we characterized the distribution of core RQC complex components in frontal cortex and cerebellum from C9orf72-expansion carriers and assessed the interaction of the RQC complex and DPR inclusions. Here, we provide further insights into the mechanisms by which R-rich RAN translation products trigger RQC pathways and how impaired processing of these proteins by the RQC may contribute to dysfunction of protein quality control in disease.
Materials and methods
Cloning and plasmid constructs
Stalling reporter plasmid with (AAA)20 sequence (poly-A) was a gift from Ramanujan Hegde (#105688, Addgene) and was previously described by Juszkiewicz et al.50 Control stalling reporter without an insert was generated by digestion of poly-A plasmid with SalI and KpnI, and subsequent ligation with annealed oligos (Genosys Oligo, Sigma-Aldrich) to circularize vector and remove the insert. Stalling reporters with C9orf72 repeat sequences were generated by modification of the multiple cloning site of vector using annealed oligos to generate each reading frame after repeat insertion. Repeat inserts were generated by sequential digestion with BssHII and ApaI of plasmids containing two or 28 repeats (previously described in Cali et al.51) or 66 repeats (kindly gifted by Dr Aaron Gitler, Stanford University) and ligated into modified vector. Due to repeat instability during cloning of the sense orientation repeat at a length of 66, the origin of replication sequence orientation was inverted by digestion of this sequence with BsaI and PciI, followed by blunting using DNA Polymerase I, Large (Klenow) Fragment (New England BioLabs) and subsequent ligation. Note that plasmids with 66 repeats contain a substitution (G→A) at repeat no. 15 and (C→T) at repeat 27 (sense orientation). Tri-nucleotide repeat inserts (CTG100, CAG40 and CAG70) and codon-optimized DPR sequences for GA (glycine-alanine)66, GA101, PR (proline-arginine)66, PR101, GP (glycine-proline)66, GP101, GR66 and GR101 were obtained from plasmids synthesized by GenScript by digestion with SalI and KpnI and ligated into vector.47 pEGFP-C1 plasmid was used to generate enhanced green fluorescent protein (EGFP) reporter constructs. pEGFP-NS was generated by HincII blunt digestion and ligation to remove vector stop codons, followed by digestion with SacII and blunting using DNA Polymerase I, Large (Klenow) Fragment (New England BioLabs) for subsequent ligation to remove downstream in frame stop codon within poly-adenylation signal. pEGFP-3tag plasmid was generated by EcoRI digestion of pEGFP-C1 plasmid and insertion of 3-tag sequence from annealed oligos with the corresponding overhangs. EGFP reporters with codon-optimized DPR sequences were generated by Eco53KI digestion of pEGFP-3tag vector and inserts were obtained by SmaI digestion of plasmids containing DPR101 codon-optimized sequences synthesized by GenScript.47 EGFP reporter with GR101 codon-optimized sequence followed by 165 nt of C9orf72 intron 1 sequence downstream of hexanucleotide repeat in GR reading frame were generated by digestion of pEGFP-GR101-3tag reporter with HindIII and SalI, and subsequent ligation of annealed oligos containing the intron 1 sequence with the corresponding overhangs. Lysine interrupted poly-GR reporter [EGFP-(G/K-R)100] was synthesized and cloned into a pcDNA3.1 + N-eGFP vector by GenScript. Insert sequences for all repeat and codon-optimized reporters are listed in Supplementary Table 1. For listerin rescue experiments, the LTN1_pCSdest plasmid used was a gift from Roger Reeves (#53855, Addgene).52 Plasmids expressing VCP wild-type (WT) or dominant-negative mutant (D2) were previously described.53 For NEMF antibody validation experiments, a plasmid expressing C-terminally HA tagged NEMF synthesized by GenScript was used. pDsRed-monomer-N1 plasmid was used as a transfection control.
Cell culture, differentiation, drug treatment and transfection
HEK 293T cell culture maintenance and siRNA/DNA plasmid transfection
Human embryonic kidney (HEK) 293T (ATCC CRL-11268) and HEK 293T LTN1 knock-out (KO) cell lines were maintained in Dulbecco’s modified Eagle media containing 10% foetal bovine serum (FBS). Transfection of DNA plasmids (CRISPR–Cas9 plasmid, EGFP and stalling reporters) was done using Fugene 6 transfection reagent (E2691, Promega) in a 1:3 ratio following the manufacturer’s recommendations. Cell lines were regularly tested for mycoplasma using a PCR-based detection assay. For siRNA-mediated knock-down (KD), HEK 293T cells were seeded 24 h before transfection and Dhamacon ON-TARGETplus SMARTpool Human NEMF siRNA (9147) or ON-TARGET plus non-targeting pool (D-001810-10) were transfected using Lipofectamine RNAiMAX (13778100, Thermo Fisher) following the manufacturer’s recommended protocol. Transfected cells were pooled and seeded for plasmid reporter transfection 24 h post-siRNA transfection and transfected with DNA plasmids as described before. For listerin rescue or VCP overexpression experiments, listerin or VCP-mutant plasmids were cotransfected with EGFP reporter plasmids in a 1:3 ratio using Fugene 6 transfection reagent as described before. Cells were analysed 24 h (stalling reporters) or 48 h (EGFP) after transfection unless otherwise specified.
Epoxomicin treatment
Proteasome inhibition was done by treating HEK 293T cells with 2 µM epoxomicin (E3652, Sigma-Aldrich) or media with DMSO (vehicle) for 6 h before analysis.
Generation of LTN1 and NEMF knock-out cell lines
Single guide RNAs for targeting LTN1 gene were designed using the Broad Institute GPP sgRNA design tools (LTN1 sgRNA: AAGAACAAGCAGCGAACTAA). sgRNAs were cloned into pSpCas9(BB)-2A-Puro (PX459) V2.0 vector (a gift from Feng Zhang; #62988, Addgene).54 HEK 293T cells were transfected with these plasmids and selected with puromycin (631305, Takara Bio) for 48 h. Monoclonal cell lines were isolated by using limiting dilution methods and subsequently validated by PCR Sanger sequencing (DNA), quantification of LTN1 mRNA levels by quantitative PCR (qPCR) and immunoblotting against listerin. Plasmid with an sgRNA targeting the NEMF gene (NEMF sgRNA: CTAGACGGCATCATATTCTT) in a pSpCas9(BB)-2A-GFP (PX458) vector was a gift from Thomas Tuschl (#127126, Addgene). Validation of NEMF depletion was done by either immunoblotting or immunofluorescence using two antibodies against NEMF protein.
Human induced pluripotent stem cell culture maintenance
CRISPRi-i3N iPS cell line described previously (male WTC11 background, kindly gifted by Dr Michael Ward, NIH) was maintained in mTesR1 medium (85850, STEMCELL Technologies) on plates coated with hESC-Qualified, LDEV-Free, Matrigel Matrix (354277, Corning).55–57 mTesR1 medium was replaced every day and cells were passaged using Versene (15040066, Gibco) when 80–90% confluent. Briefly, cells were washed with Dulbecco’s phosphate buffered saline (DPBS) and incubated with Versene at 37°C for 7 min. Cells were lifted by washing with mTesR1 medium supplemented with 10 nM Y-27632 dihydrochloride ROCK inhibitor (125410, Tocris) and gently scraping. Colonies were broken up by gently triturating the cell mixture and transferring to a Matrigel-coated plate.
Differentiation of human i3N-induced pluripotent stem cells into neurons and DNA transfection of i3Ns
CRISPRi-i3N induced pluripotent stem (iPS) cells were differentiated using doxycycline-induced expression of NGN2 based on methods previously described.55,57 Briefly, iPS cells were collected using Accutase (A1110501, Gibco) and washed with DPBS. Suspension was diluted with mTeSR1 medium supplemented with 10 nM ROCK inhibitor and pelleted. Cells were resuspended in Neuronal Medium [Neurobasal Plus Medium (A3582901, Gibco), N2 supplement (A1370701, Gibco), B27 Plus supplement (A3582801, Gibco), MEM Non-Essential Amino Acids (11140-050, Gibco) and GlutaMAX supplement (35050-061, Gibco)] supplemented with 10 nM ROCK inhibitor and 2 mg/ml doxycycline hydrochloride (D3072, Sigma-Aldrich) to induce expression of NGN2 and plated in Matrigel-coated plates. After 3 days, predifferentiated cells were collected as detailed before and replated in base Neuronal Medium supplemented with 10 nM ROCK inhibitor, 2 mg/ml doxycycline hydrochloride, CultureONE supplement (A3320201, Gibco), 10 ng/ml NT-3 (450-03, PeproTech), 10 ng/ml BDNF (450-02, PeproTech), 10 ng/ml GDNF (450-10, PeproTech) and 200 ng/ml L-ascorbic acid (A8960, Sigma-Aldrich) on plates coated with 100 µg/ml poly-L-ornithine (P3655, Sigma-Aldrich). After cells attached, additional media volume was added without ROCK inhibitor and supplemented with 1 µg/ml laminin (23017-015, Gibco). Thereafter, half media changes were performed once or twice per week with base Neuronal Medium supplemented as detailed before for predifferentiated cells without doxycycline. Transfection of DNA plasmids (EGFP and stalling reporters) was done on Day 8 of differentiation using NeuroMag transfection reagent (NM50200, OZ Biosciences) in a 1:3 ratio following the manufacturer’s recommendations. Cells were collected 72 h post transfection for analysis by flow cytometry or processed for immunofluorescence staining.
Immunofluorescence and human tissue staining
Immunofluorescence staining of HEK 293T cells and i3Ns
HEK 293T cells were seeded onto 12 mm coverslips coated with 0.1 mg/ml poly-D-lysine (P6407, Sigma-Aldrich). I3Ns were seeded onto 12 mm coverslips coated with 100 µg/ml poly-L-ornithine (P3655, Sigma-Aldrich). Cells were transfected with the corresponding reporters as detailed before. Coverslips were washed with DPBS and cells were fixed in 4% paraformaldehyde for 15 min at room temperature. Cells were permeabilized with 0.1% Triton X-100 in PBS, blocked with a 5% FBS solution and incubated with primary antibody overnight at 4°C. Primary antibodies are listed in Supplementary Table 3. Coverslips were washed and incubated for 1 h at room temperature with the corresponding secondary antibody (Supplementary Table 3). Nuclei were stained using DAPI (D9564, 300 nM, Thermo Fisher) for 5 min at room temperature. Coverslips were mounted with ProLong Glass Antifade Mountant (P36980, Thermo Fisher).
Immunofluorescence staining of paraffin-embedded tissue
Autopsy tissues were obtained from the Center for Neurodegenerative Disease Research Brain Bank at the University of Pennsylvania that are maintained as previously described.58 Local rules and regulations indicate that brain autopsy studies are not considered human subject research. However, legal consent for research autopsies were obtained from next of kin at time of death in all instances. Dual immunofluorescence was performed on 6 µm formalin-fixed paraffin-embedded cerebellum tissue sections from cases with the C9orf72-expansion mutation. Tissue sections were deparaffinized and rehydrated followed by antigen retrieval by microwaving in a citrate-based antigen unmasking solution (H-3300, Vector Laboratories). Sections were blocked in 2% FBS in 0.1 M Tris buffer and incubated with primary antibodies overnight at 4°C in a humidified chamber (Supplementary Table 3). Sections were washed in 0.1 M Tris buffer, blocked in 2% FBS in 0.1 M Tris buffer and incubated for 2 h at room temperature with corresponding secondary antibodies (Supplementary Table 3). Sections were washed in 0.1 M Tris buffer, stained using DAPI to visualize nuclei for 10 min at room temperature and then coverslipped with ProLong Glass Antifade Mountant.
Confocal microscopy
Confocal images were obtained using a Leica TCS SPE laser scanning confocal microscope with a ×40 objective (numerical aperture 1.15) or ×63 objective (numerical aperture 1.30) using the 405, 488, 561 and 635 nm lasers to visualize DAPI, Alexa Fluor 488, autofluorescence and Alexa Fluor 647, respectively. Images were processed using the Leica AF software (Leica Biosystems). Images were black-balanced and brightness and contrast was adjusted for the entire image using Adobe Photoshop software.
Sequential protein extraction and immunoblotting
For analysis of protein from cells expressing EGFP reporters, transfected cells were washed with DPBS and lysed in ice-cold 1% Triton X-100 cell lysis buffer (50 mM Tris, 150 mM NaCl, 5 mM EDTA) supplemented with protease inhibitor (PI) cocktail (Leupeptin, 11017128001, Roche; Pepstatin, P4265; TPCX, T4376; Trypsin inhibitor, T9003; Sigma-Aldrich and EDTA, 15575020, Invitrogen) for 15 min on ice. Lysates were sonicated three times for 1 s at 40% amplitude (630-0423, 2 mm microtip; VCX130, Sonics Vibra-cell) and centrifuged at 55 000 rpm on a Beckman TLA 55 rotor for 30 min at 4°C. Pellets were then resuspended in 2% SDS cell lysis buffer supplemented with PI for 5 min at room temperature, sonicated and centrifuged at 55 000 rpm on a Beckman TLA 55 rotor for 30 min at 22°C. Concentration of protein lysates was determined from the pre-cleared triton soluble fraction using a BCA assay (Pierce BCA Protein Assay Kit, 23225, Thermo Fisher). For LTN1 KO validation, triton soluble fractions from control or LTN1 KO cells were analysed by immunoblotting. For NEMF KD validation, cells were lysed on RIPA buffer supplemented with PI for 15 min on ice and subsequently sonicated as described before. Lysates were centrifuged at 20 000g for 30 min at 4°C. Supernatant was collected as the RIPA-soluble fraction and analysed. Samples were prepared using 4× Lammeli sample buffer with dithiothreitol (1610747, BioRad) and boiled at 99°C for 5 min. Samples were run on either 8 or 12.5% polyacrylamide gels or pre-cast MiniProtean TGX 4–20% or 4–15% gels (4561094 or 4561086, BioRad). Protein was transferred to a 0.2 µm nitrocellulose membrane using the BioRad Turbo Transfer apparatus. Alternatively, SDS-insoluble fractions were blotted on a 0.2 µm nitrocellulose membrane using a BioRad Bio-Dot 96-well apparatus (1706545, BioRad). Membranes were washed and blocked in 5% non-fat milk in tris-buffered solution. Blots were incubated with primary antibodies (Supplementary Table 3) at the corresponding dilution in tween-20 containing tris-buffered solution overnight at 4°C. Membranes were washed at least three times in tween-20 containing tris-buffered solution and incubated with secondary antibodies for 1 h (Supplementary Table 3). Blots were developed with Pierce ECL 2 Western Blotting Substrate (80196, Thermo Fisher) and exposed to film or scanned on a Typhoon FLA 7000 (GE Healthcare Bio-Sciences AB). Alternatively, blots were imaged on a Licor Odyssey (LI-COR Biosciences).
RNA isolation and qPCR
Total RNA was isolated from cells using Qiagen RNeasy Mini kit (74104, Qiagen) following manufacturer’s recommendations including DNAse I digestion. Quantification of isolated RNA was done using a Qubit 3.0 fluorometer (Q33216, Invitrogen) and quality was assessed by NanoDrop absorbance measurements. cDNA was prepared using the High Capacity RNA-to-cDNA Kit (4387406, Applied Biosystems). qPCR analysis was done using Power Up SYBR Green master mix (A25742, Applied Biosystems) on a StepOne Plus Real-Time PCR Machine (Applied Biosystems). RNA levels for the gene of interest were normalized to the geometric mean of ACTB and GPS1 using the ΔΔCt method. Primers used for qPCR are listed in Supplementary Table 2.
Flow cytometry and data analysis
HEK 293T cells were collected using trypsin and spun down at 300g for 3 min. i3N neurons were collected using a papain dissociation solution containing: papain (LK003176, Worthington), DNAse I (LS00633, Worthington) and magnesium chloride (BP214-500, Fisher). Dissociation solution was neutralized in Dulbecco’s modified Eagle media containing 10% FBS and neural sheet was triturated 10 times and spun down at 200g for 10 min. Cell pellets were resuspended in DPBS, 2% FBS, 0.5 mM EDTA and filtered through a BD tube with cell strainer (352235, BD Biosciences) for flow cytometry analysis on a BD LSR II instrument (BD Biosciences). Data analysis was done using FlowJo v.10 software. For experiments with EGFP reporters, gating for green fluorescent protein (GFP)-positive population was done based on a mock transfected control. For stalling reporter experiments, gating of the GFP-positive population was done based on a mock transfected control and calculations of the ratio of the median fluorescence intensity of red fluorescent protein (RFP) and GFP were done for the transfected population (GFP-positive). Similarly, for DsRed and EGFP co-transfection experiments, gating for DsRed-positive and GFP-positive population was done based on a mock transfected control. Graphs were generated using GraphPad Prism 7.
Statistical analysis
Student’s t-test with Welch’s correction, one- or two-way ANOVA statistical analyses, Dunnett’s multiple comparison and Fisher’s exact tests were done using GraphPad Prism 7. Linear mixed effects regression models were done using the nlme package in R. A P-value <0.05 was considered significant: P values were * ≤ 0.05, ** ≤ 0.01, *** ≤ 0.001, **** ≤ 0.0001 and ns, not significant.
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
Results
Non-stop mRNA protein products are degraded in a listerin-dependent manner in mammalian cells
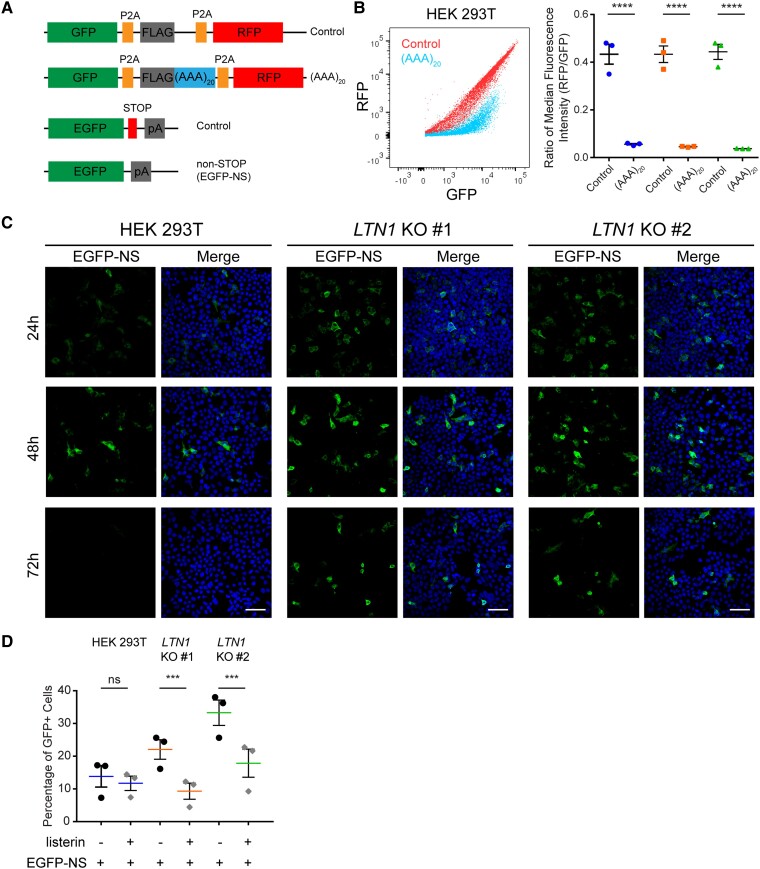
As there are relatively few studies on the RQC pathway in mammalian cells, we sought to demonstrate that core RQC complex components were involved in the clearance of a canonical RQC substrate. To study the role of listerin, E3 ubiquitin ligase, monoclonal LTN1 KO HEK 293T cell lines were generated using CRISPR–Cas9. Lowered levels of LTN1 mRNA and protein were observed in LTN1 KO lines by qPCR and immunoblotting (Supplementary Fig. 1A and B). We also used a dual-fluorescent protein stalling reporter described previously by Juszkiewicz.50 This reporter expresses poly-lysine encoded by an (AAA)20 sequence that mimics the poly-A tail of non-stop mRNA, a canonical RQC substrate that is known to cause ribosome stalling during elongation. The poly-lysine sequence is placed between two fluorescent proteins reporters (an upstream GFP and a downstream RFP) with each coding sequence separated by P2A ribosomal skipping sequences, resulting in three independent proteins during translation of the reporter RNA. Monitoring the expression of the downstream RFP compared to GFP by flow cytometry provides a quantitative measurement of ribosome stalling during elongation through the internal poly-A sequence. When the (AAA)20 reporter construct was expressed in parental HEK 293T or LTN1 KO cells, flow cytometry dot plots of GFP and RFP fluorescence showed a decrease in RFP expression compared to a control reporter without an insert (Fig. 1B, left). Comparison of the ratio of median fluorescence intensity of RFP and GFP showed a significant decrease in RFP fluorescence in all cell lines with no effect of LTN1 KO (Fig. 1B, right). This demonstrated that poly-A sequences trigger ribosomal stalling independent of LTN1 as expected, since listerin acts downstream of ribosome stalling.
Figure 1.
Non-stop mRNA protein products are degraded by the RQC in a Listerin-dependent manner in mammalian cells. (A) Schematic of stalling reporters and EGFP constructs. (B) GFP versus RFP expression profile of stalling reporter with poly-A sequences compared to a no insert control (left). Ratio of median fluorescence intensity of RFP to GFP shows that poly-A sequences induce stalling independent of listerin. Data are shown as mean ± standard error of the mean (SEM) with all data points (two-way ANOVA, n = 3, reporter P < 0.0001, LTN1 KO P = 0.9771, interaction P = 0.8548) (C) Confocal images of HEK 293T and LTN1 KO cells expressing EGFP-NS reporter show accumulation and reduced clearance of NS protein in LTN1 KO cells over time (scale bar = 50 µm). (D) Listerin overexpression in LTN1 KO cells significantly reduces levels of NS reporter protein in cells quantified by flow cytometry. Percentage is shown as mean ± SEM (mixed effects model, n = 3, Ltn1:HEK 293T P = 0.1246, Ltn1:LTN1 KO #1 P = 0.0001, Ltn1:LTN1 KO #2 P < 0.0001).
While the dual-fluorescence reporter is useful for demonstrating ribosome stalling, the fate of the protein product translated from the internal stalling sequence is not directly assessed. Therefore, an EGFP DNA plasmid reporter lacking a stop codon was generated as a reporter for the translated protein product expressed from non-stop mRNA (referred to as ‘EGFP-NS’) (Fig. 1A). This EGFP-NS reporter was transfected and expressed in parental HEK 293T and LTN1 KO cells, and GFP fluorescence was used to assess protein expression levels. Confocal imaging of GFP fluorescence from cells expressing EGFP-NS reporter at 24, 48 and 72 h after transfection showed that GFP expression is diminished by 72 h in parental cells (Fig. 1C, left). In contrast, a higher number of GFP-positive cells were observed at 48 and 72 h in LTN1 KO cells (Fig. 1C, middle and right). Analysis of an extended GFP fluorescence time-course shows LTN1 KO monoclonal cell lines retained higher levels of GFP-positive cells when expressing EGFP-NS reporter at later time points (Supplementary Fig. 1C, left). This higher percentage of GFP-positive cells was observed in both LTN1 KO monoclonal lines despite the lower transfection efficiency seen for LTN1 KO #1 (Supplementary Fig. 1C, right). To control for differences in transfection efficiency, EGFP reporters were cotransfected with a DsRed expressing plasmid as an internal transfection control in parental and LTN1 KO cells. LTN1 KO resulted in a significantly higher percentage of GFP-positive cells in the transfected DsRed-positive population for EGFP-NS reporter (Supplementary Fig. 1D). Similarly, accumulation of EGFP-NS protein was observed in LTN1 KO cells by immunoblotting against GFP (Supplementary Fig. 1E).
As additional evidence that listerin promotes turnover of EGFP-NS protein, overexpression of listerin in LTN1 KO cells significantly rescued the LTN1 KO phenotype as evidenced by a reduction in the percentage of GFP-positive cells (Fig. 1D). In contrast, no significant change in percentage of GFP-positive cells was observed in the control line on listerin expression (Fig. 1D). These results indicate that listerin assists in the degradation of aberrant translational protein products.
RQC complex degrades non-stop mRNA protein in mammalian cells
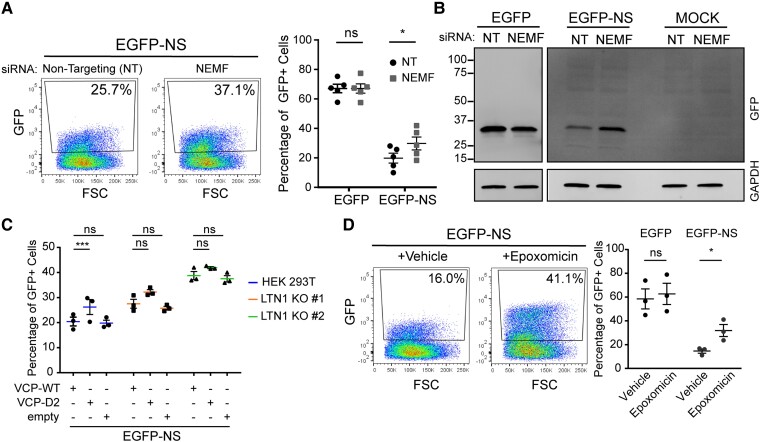
To assess the role of NEMF, another core RQC complex factor, HEK 293T cells were transfected with siRNAs that target NEMF versus non-targeting siRNA controls and subsequently transfected with the EGFP-NS plasmid reporter. Levels of this protein that is subject to listerin-mediated degradation that were quantified by flow cytometry and compared to a control EGFP reporter an intact 3′ stop codon. KD efficiency was assessed by quantifying NEMF mRNA levels by qPCR and immunoblotting for NEMF (Supplementary Fig. 2A). Representative dot plots of EGFP-NS fluorescence from NEMF versus non-targeting siRNA transfected cells showed an increase in the percentage of GFP-positive cells upon NEMF KD (Fig. 2A, left). Quantification of EGFP expression revealed a significant increase in the percentage of GFP-positive population of cells expressing EGFP-NS on NEMF depletion but not when expressing the control EGFP reporter (Fig. 2A, right). To verify flow cytometry results, protein was extracted from NEMF or non-targeting KD cells expressing EGFP-NS reporter and immunoblotted using antibodies that recognize GFP. Higher levels of GFP protein were detected for NEMF KD cells compared to control non-targeting siRNAs (Fig. 2B).
Figure 2.
RQC complex degrades non-stop mRNA protein in mammalian cells. (A) KD of NEMF in HEK 293T was done using siRNAs and EGFP-NS reporter expression was analysed using flow cytometry. Representative dot plot of GFP fluorescence in NEMF and control siRNAs cells is shown (left). Percentage of GFP-positive cells expressing GFP control or EGFP-NS reporter from five biological replicates (right). Percentage is shown as mean ± SEM with all data points (mixed effects model, n = 5, EGFP:NEMF KD P = 0.9941, NS:NEMF KD P = 0.0213). (B) Triton soluble protein fraction from cells transfected with NEMF and non-targeting siRNAs expressing either EGFP control, EGFP-NS reporter or Mock control was analysed by immunoblotting against GFP. (C) Effect of co-expression of VCP-mutant proteins in EGFP-NS reporter in parental HEK 293T and LTN1 KO cells was assessed by flow cytometry. Graph shows the percentage of GFP-positive cells as mean ± SEM with all data points (mixed effects model, n = 3, parental:D2 P = 0.0039, parental:empty P = 0.7173, LTN1 KO #1:D2 P = 0.6569, LTN1 KO #1:empty P = 0.6473, LTN1 KO #2:D2 P = 0.2946, LTN1 KO #1:empty P = 0.7977). (D) Proteasome activity was inhibited by treating the cells with 2 µM epoxomicin or vehicle and percentage of GFP-positive cells was quantified by flow cytometry 6 h after treatment. Data are shown as mean ± SEM with all data points and expanded in Supplementary Fig. 2B (mixed effects model, n = 3, EGFP:Epo P = 0.2468, NS:Epo P = 0.0176).
To demonstrate the role of VCP in EGFP-NS protein clearance, either wild-type VCP (VCP-WT), a dominant-negative VCP mutant (VCP-D2) or an empty vector was co-expressed with the EGFP-NS reporter in parental HEK 293T and LTN1 KO cells. Overexpression was used because prolonged inhibition of VCP resulted in significant cytotoxicity. Dominant-negative mutant VCP-D2 expression in parental HEK 293T cells resulted in a significant increase in the percentage of GFP-positive cells compared to VCP-WT (Fig. 2C). In contrast, expression of the dominant-negative VCP-D2 construct in LTN1 KO cells had no significant effect on the percentage of GFP-positive population (Fig. 2C). Therefore, VCP appears to facilitate degradation of EGFP-NS protein in an LTN1-dependent manner, consistent with the fact that listerin is a ubiquitin ligase and that VCP requires poly-ubiquitination of substrates for target recognition.
To determine whether this canonical RQC substrate is ultimately degraded by the proteasome in mammalian cells, proteasomal activity was inhibited by treatment with 2 µM of epoxomicin for 6 h before analysis. Proteasome inhibition resulted in accumulation of GFP-positive cells as seen on representative dot plots (Fig. 2D, left). Quantification of GFP-positive population of cells expressing EGFP-NS reporter shows a significant increase on epoxomicin treatment compared to vehicle (Fig. 2D, right).
Translation of R-rich dipeptide-repeat proteins induces ribosomal stalling in mammalian cells
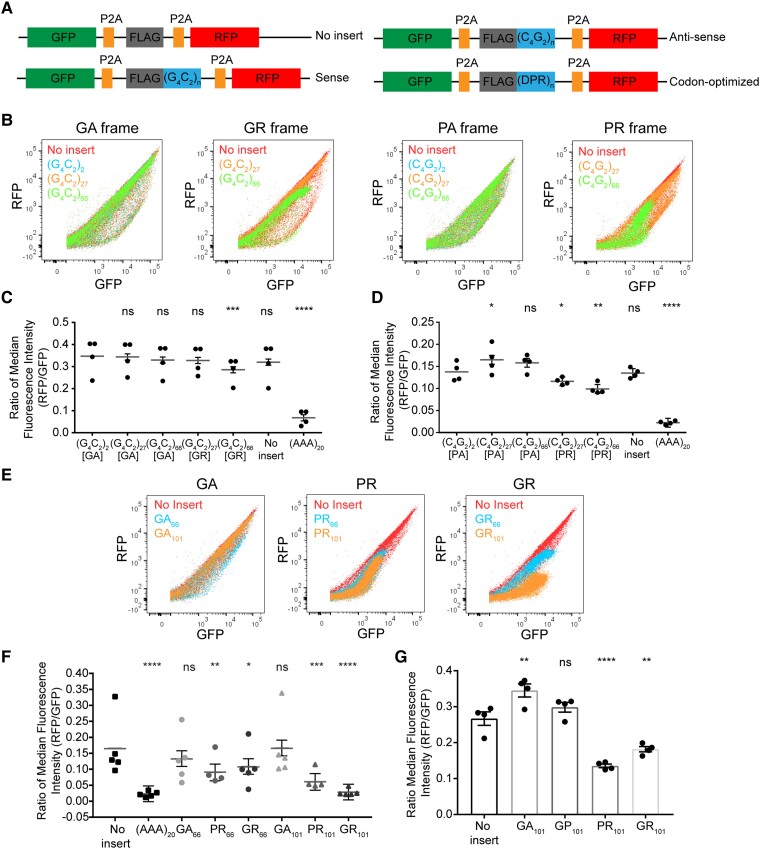
Having validated that the previous fluorescence reporter constructs are sensitive to perturbations of the RQC pathway, we sought to determine whether ribosome stalling occurs during elongation through the C9orf72-expansion repeat. Dual-fluorescent protein stalling reporters with the hexanucleotide repeat sequence in both sense and antisense orientations were generated and analysed as described previously for poly-A sequences (Fig. 3A). These reporters were expressed in HEK 293T cells and expression of RFP and GFP was monitored by flow cytometry. Dot plots of GFP and RFP fluorescence signal from reporters with G4C2 repeats (sense orientation) at short lengths of two or 27 were similar to cells expressing control constructs without an insert for either GA or GR-encoding reading frames (Fig. 3B, left). A reduction of RFP signal was observed for reporters with repeats at intermediate lengths (66) in the GR reading frame but not for GA (Fig. 3B, left). Representative dot plots of cells expressing reporters with C9orf72 repeats in the antisense orientation showed that translation in the PA frame does not reduce RFP expression at short (two, 27) or intermediate (66) lengths (Fig. 3B, right). In contrast, elongation in the PR frame resulted in a mild decrease in RFP levels at short repeat lengths (27) and loss of RFP expression at intermediate lengths of 66 units (Fig. 3B, right). These findings were quantified by the ratio of the median fluorescence intensity of RFP compared to GFP as a measure of ribosome stalling during elongation through C9orf72-associated repeats. Reporters with repeats in the sense orientation showed no significant difference when comparing two and 27 G4C2 repeats irrespective of reading frame and a significant decrease at intermediate lengths in the GR frame (Fig. 3C). Similarly, reporters with repeats in the antisense orientation showed that elongation in the PR frame at 27 and 66 units results in a significant decrease in the RFP to GFP ratio (Fig. 3D). In contrast, an increase was observed for the reporter with 27 antisense repeats in the PA reading frame (Fig. 3D).
Figure 3.
Arginine-rich DPR proteins induce ribosomal stalling in cells. (A) Schematic of stalling reporters with C9orf72-associated sequences [(G4C2) n = 2, 27 or 66, (C4G2) n = 2, 27 or 66 and codon-optimized n = 66 or 101]. (B) Dot plot of GFP and RFP expression profile for stalling reporters with C9orf72 repeats in different reading frames. (C) Ratio of median fluorescence intensity of RFP to GFP of C9orf72-associated repeats in the sense orientation for four biological replicates shows stalling in GR reading frame at 66 units. Ratio is shown as β (grey line) with all data points and error bars represent standard error [mixed effects model, n = 4, (G4C2)27-GA P = 0.8254, (G4C2)66-GA P = 0.2318, (G4C2)27-GR P = 0.1890, (G4C2)66-GR P = 0.0004, no insert P = 0.0705, (AAA)20P < 0.0001]. (D) Ratio of median fluorescence intensity of RFP to GFP of C9orf72-associated repeats in the antisense orientation for four biological replicates shows stalling in PR reading frame at 66 units. Ratio is shown as β (grey line) with all data points and error bars represent standard error [mixed effects model, n = 4, (C4G2)27-PA P = 0.0135, (C4G2)66-PA P = 0.0553, (C4G2)27-PR P = 0.0466, (C4G2)66-PR P = 0.0011, no insert P = 0.7911, (AAA)20 P < 0.0001]. (E) Expression profile of GFP and RFP for stalling reporters with codon-optimized DPR protein sequences expressed in HEK 293T cells. (F) Ratio of RFP to GFP median fluorescence intensity of five biological replicates of codon-optimized stalling reporters expressed in HEK 293T cells shows stalling in PR and GR-encoding sequences. Ratio is shown as β (grey line) with all data points and error bars represent standard error [mixed effects model, n = 5, GA66P = 0.1972, PR66P = 0.0076, GR66P = 0.0271, GA101P = 0.9730, PR101P = 0.0004, GR101P < 0.0001, (AAA)20P < 0.0001]. (G) Ratio of RFP to GFP median fluorescence intensity of four biological replicates of codon-optimized stalling reporters expressed in induced neuronal cells. Ratio is shown as mean ± SEM with all data points (Dunnett’s multiple comparisons test, n = 4, GA101P = 0.0039, GP101P = 0.3361, PR101P = 0.0001, GR101P = 0.0019).
To determine whether ribosomal stalling occurs in tri-nucleotide repeats associated to other repeat-expansion mutations, either CAG or CTG repeats at disease-relevant lengths were cloned into the stalling reporters (Supplementary Fig. 3A). These were expressed in cells and no significant effect in the RFP to GFP expression ratio was observed (Supplementary Fig. 3B).
Previous studies have suggested that ribosome stalling within lysine-encoding poly-A sequences is primarily driven by its mRNA sequence in mammalian cells. 50,59–61 In contrast, our data indicate that translational stalling of C9orf72 repeats is dependent on the G4C2 reading frame and not the RNA sequence per se. Thus, we generated stalling reporters with codon-optimized DPR encoding sequences. To assess whether R-rich DPR proteins induce stalling during elongation, reporters with codon-optimized DPR encoding sequences at intermediate (66) or long (101) lengths were expressed in HEK 293T cells (Fig. 3A). Representative flow cytometry dot plots from stalling reporters with GA encoding sequences showed no loss of RFP expression at any length tested (Fig. 3E, left). In contrast, elongation through sequences that generate PR led to a reduction in RFP protein starting at intermediate lengths of 66 and 101 (Fig. 3E, middle). GR-encoding sequences moderately reduced RFP expression at 66 units and dramatically reduced RFP expression at 101 (Fig. 3E, right). Quantification of the ratio of median fluorescence intensity of RFP to GFP of these reporters showed a significant reduction in RFP expression for PR66, PR101, GR66 and GR101, but no significant effect for GA at any length (Fig. 3F). In addition, no stalling was observed in reporters encoding for GP at any length tested in HEK 293T cells (Supplementary Fig. 3C). Similarly, a significant decrease in the RFP to GFP ratio was observed in stalling reporters with PR and GR-encoding sequences expressed in i3N neurons compared to a control without an insert (Fig. 3G). In contrast, an increase in RFP levels was observed for GA encoding sequences while no effect was seen for the GP reporter. The reason for the observed increase in RFP:GFP ratio for GA codon-optimized sequences and antisense repeats in the PA frame is unknown but may indicate that these sequences contain a cryptic internal ribosome entry site that drives RFP expression independent of GFP expression. Regardless, these results indicate that R-rich DPR protein products trigger ribosome stalling during elongation in cell models and neurons.
While the loss of RFP expression appears likely to be due to ribosome stalling, it remained possible that reduced RFP expression was due to frameshifting during elongation. To assess whether there was evidence of frameshifting, these codon-optimized DPR sequences were cloned into EGFP reporters with C-terminal protein tags (FLAG [+1], myc [0] or HA [−1]) in all three reading frames (Supplementary Fig. 3D). Protein was extracted from HEK 293T cells expressing these EGFP-DPR101-3tag reporters or a mock transfected control and immunoblotted with antibodies that recognize GFP and each C-terminal protein tag. Blot of the detergent insoluble fraction showed that the EGFP-GA101 protein was myc positive when compared to mock control and negative for the other C-terminal tags suggesting that minimal to no frameshifting was occurring for this sequence. In contrast, EGFP-PR101 and EGFP-GR101 proteins showed low myc positive signal and appeared negative for other C-terminal protein tags when compared to the mock control (Supplementary Fig. 3E). Therefore, complete readthrough of PR- and GR-encoding sequences was relatively low, and we could not find definite evidence of frameshifting although low levels cannot be ruled out. These results are consistent with R-rich DPR proteins causing ribosome stalling and indicates that loss of RFP expression in stalling reporters is likely not due to frameshifting within the PR- and GR-encoding sequences.
C9orf72-associated DPR proteins are not degraded by the RQC complex
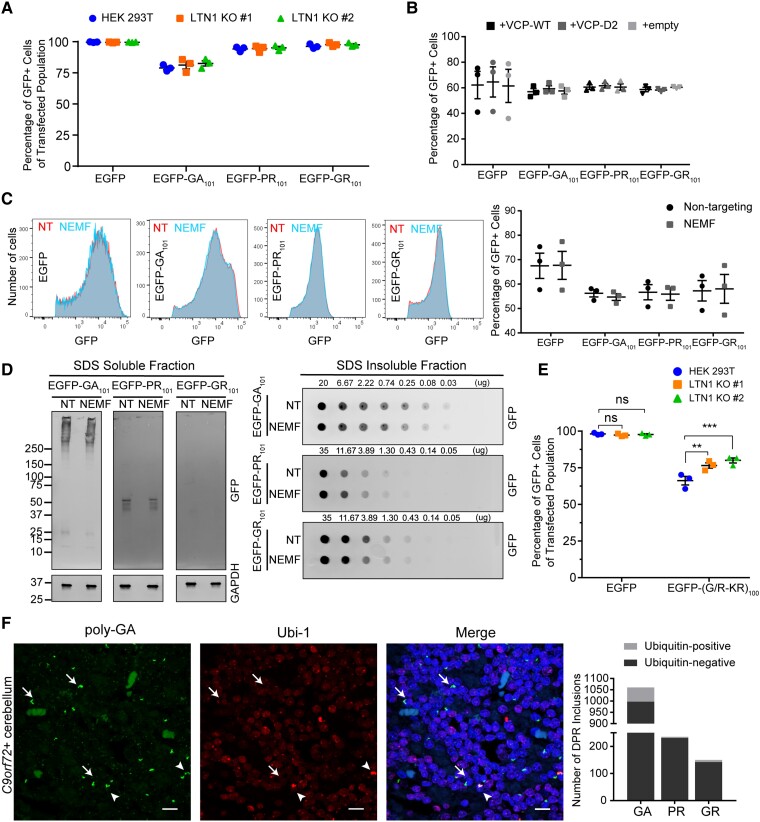
As R-rich DPR proteins appeared to induce ribosome stalling, we assessed whether listerin plays a role in clearing these proteins. EGFP-DPR101 reporters (Supplementary Fig. 3D) were cotransfected with a DsRed plasmid (transfection internal control) into parental HEK 293T and LTN1 KO cells. The expression level of EGFP reporter proteins within the transfected population (DsRed-positive) was quantified by flow cytometry. LTN1 KO did not have a significant effect on the percentage of GFP-positive cells for any EGFP-DPR101 construct or for an EGFP control (Fig. 4A). To study the role of the RQC factor VCP in DPR protein levels, a VCP dominant-negative mutant was co-expressed with the EGFP reporters as described before. Co-expression of VCP-D2 mutant had no significant effect on the percentage GFP-positive cells expressing GA-, PR- or GR-encoding reporters or an EGFP control (Fig. 4B). To determine whether these DPR proteins may be cleared by the proteasome, the GFP-positive population of cells expressing EGFP-DPR constructs was monitored after treatment with epoxomicin as described before. No significant increase in the percentage of GFP-positive cells was detected for any of the EGFP-DPR constructs (Supplementary Fig. 2B).
Figure 4.
C9orf72-associated DPR proteins are not degraded by the RQC complex. (A) Levels of expression of EGFP-DPR101 reporter proteins was assessed in HEK 293T and LTN1 KO by flow cytometry using an internal transfection control (DsRed). Percentages are shown as mean ± SEM with all data points (two-way ANOVA, n = 3, reporter P < 0.0001, LTN1 KO P = 0.3476, interaction P = 0.8988). (B) Effect of VCP-mutant overexpression in the levels of EGFP-DPR101 reporter proteins in HEK 293T assessed by flow cytometry. Percentage is shown as mean ± SEM with all data points (mixed effects model, n = 3, EGFP:D2 P = 0.7516, EGFP:empty P = 0.9264, GA:D2 P = 0.9901, GA:empty P = 0.9083, PR:D2 P = 0.9009, PR:empty P = 0.9479, GR:D2 P = 0.7891 GR:empty P = 0.9479). (C) Effect of KD of NEMF by siRNAs in the expression of EGFP-DPR101 was analysed by flow cytometry. GFP fluorescence histogram of GFP-positive population (left) and quantification of percentage GFP-positive population (right). Data are shown as mean ± SEM with all data points (two-way ANOVA, n = 3, reporter P = 0.0305, NEMF KD P = 0.9114, interaction P = 0.9919). (D) Depletion of NEMF in HEK 293T cells effect on EGFP-DPR101 protein solubility was tested by sequential protein extraction and immunoblotting of detergent soluble and insoluble fractions of reporter protein. Soluble fraction was blotted for GAPDH as a loading control. (E) Effect of LTN1 KO in the expression of interrupted EGFP-GR reporter with lysine residues every 20 GR units was analysed by flow cytometry. Percentage of GFP-positive population of transfected cells (DsRed-positive, transfection internal control) is shown as mean ± SEM with all data points [Dunnett’s multiple comparisons, n = 3, EGFP: HEK 293T-LTN1 KO #1 P = 0.9064, HEK 293T-LTN1 KO #2 P = 0.9773, EGFP-(G/K-R)100: HEK 293T-LTN1 KO #1 P = 0.0012, HEK 293T-LTN1 KO #2 P = 0.0001]. (F) Representative dual immunofluorescence of poly-GA inclusions and ubiquitin (Ubi-1) of cerebellum sections from C9orf72-expansion carrier (left). Arrows denote poly-GA inclusions and arrowheads Ubi-1 + poly-GA inclusions. Quantification of ubiquitin-positive and negative DPR inclusions (right; scale bar = 10 µm).
To evaluate whether NEMF plays a role in the overall expression of DPR proteins in mammalian cells, NEMF was depleted in cells using siRNAs targeting this factor and subsequently transfected EGFP reporters with DPR encoding sequences. Representative histograms of GFP fluorescence showed no effect of NEMF depletion in the number of GFP-positive cells or fluorescence intensity (Fig. 4C, left). Furthermore, NEMF KD did not significantly affect the percentage of GFP-positive cells for any of the EGFP reporters (EGFP control, GA101, PR101 or GR101; Fig. 4C, right). Previous studies have suggested that NEMF-mediated CTEs result in decreased solubility of RQC substrates, and that poly-GR proteins can be C-terminally extended with highly hydrophobic residues by the RQC complex in cells.30,42,48,62 To assess whether NEMF alters DPR solubility, immunoblotting of sequentially extracted proteins was performed from cells expressing EGFP-DPR proteins but depleted of NEMF by siRNAs. When blotting for GFP, most EGFP-DPR101 proteins were detected in the detergent insoluble fraction (Fig. 4D). No major effect of NEMF depletion in the solubility profiles of EGFP-GA101, EGFP-PR101 or EGFP-GR101 protein could be observed (Fig. 4D). Triton soluble fractions were blotted for GAPDH as a loading control. This suggested that NEMF deficiency and limiting CTEs does not significantly alter the solubility of DPR proteins in our model.
These results indicate that DPR proteins are not efficiently processed by the RQC complex despite the strong induction of ribosome stalling by R-rich proteins. To assess whether this could be due to lack of lysine residues within R-rich DPR encoding sequences, we generated an interrupted poly-GR reporter where several glycine residues were substituted with lysine-encoding codons while total length was maintained at 101 dipeptide units. Expression of this interrupted reporter in parental HEK 293T and LTN1 KO cells was monitored by quantification of the GFP-positive population. Listerin-deficiency resulted in a significant increase in the percentage of GFP-positive cells when expressing the interrupted EGFP-GR reporter but not an EGFP control (Fig. 4E). Previous studies have reported that DPR inclusions in patient tissue may contain the endogenous C-terminus sequence that results from translation past the repeat sequence.17,63 Of all DPR C termini, the endogenous C terminus for poly-GR reading frame in the sense strand encodes for a C-terminal lysine. To determine whether the poly-GR C terminus would affect the susceptibility of this protein to RQC-mediated degradation in the rare event of complete readthrough, an EGFP reporter with codon-optimized GR101 sequence with the endogenous C terminus was cloned and expressed in parental HEK 293T and LTN1 KO cells. Protein levels of EGFP reporter for poly-GR with endogenous C terminus was monitored by flow cytometry. No significant increase in the percentage of GFP-positive cells of this reporter relative to EGFP control were observed in LTN1 KO compared to parental cells (Supplementary Fig. 3F).
To determine whether DPR proteins in C9orf72-mutation carriers’ tissue are marked by ubiquitin for degradation, dual-immunofluorescence staining for DPR inclusions (poly-GA, poly-PR and poly-GR) and ubiquitin (using the Ubi-1 antibody, which preferentially stains poly-ubiquitin chains) of cerebellar tissue sections was performed. Representative confocal images of stained sections collected within the cerebellar cell granule layer (where DPR aggregates are known to accumulate) showed that there are few ubiquitin-positive poly-GA inclusions (arrowheads) and numerous ubiquitin-negative poly-GA inclusions (arrows) (Fig. 4F, left). Quantification of ubiquitin-positive and -negative DPR inclusions (poly-GA, poly-GR and poly-PR) from six C9orf72-expanded ALS/FTD cases was done by manual counting of ×40 microscopic fields (Fig. 4F, right). For poly-GA and poly-GR, 5.9 and 5.3% of DPR inclusions were observed to be ubiquitin positive by confocal imaging, respectively. For the antisense product poly-PR, only 1.7% of inclusions were observed to be ubiquitin positive in expansion carriers. In summary, DPR inclusions are rarely ubiquitinated in C9orf72-mutation carriers’ cerebellum and do not appear to be efficiently degraded by the canonical RQC complex in cell models.
NEMF is recruited to DPR inclusions in C9orf72-expansion post-mortem tissue and neurons
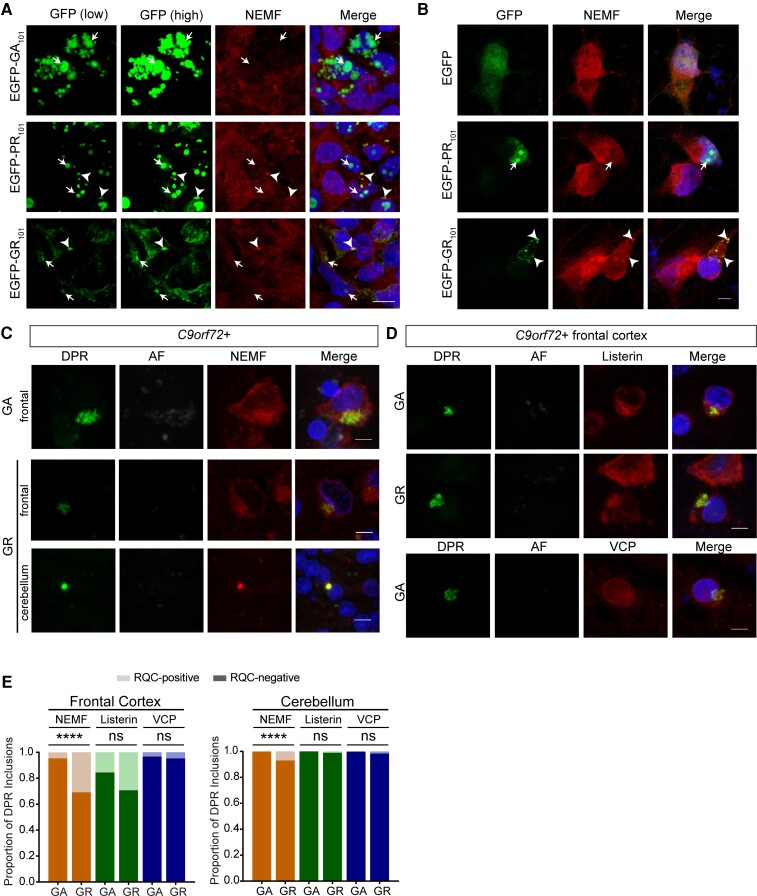
Previous studies have suggested that DPR proteins can be C-terminally extended and interact with some RQC factors.28,47,48,64,65 Thus, the interaction of the core RQC factor NEMF and DPR proteins was assessed in cell and neuronal models, and post-mortem brain tissue by immunofluorescence staining. The NEMF antibodies used for this study were validated for immunofluorescence in cells depleted of NEMF using CRISPR–Cas9 or overexpressing C-terminally HA tagged NEMF protein (Supplementary Fig. 4A). Dual-immunofluorescence staining of HEK 293T cells expressing EGFP-DPR101 reporters and NEMF shows its colocalization with a subset of GR and PR aggregates but not GA (Fig. 5A and Supplementary Fig. 4B). Notably, we observed that NEMF only colocalized to cytoplasmic but not nuclear R-rich DPR aggregates in HEK 293T cells. Similarly, we observed colocalization of cytoplasmic poly-GR aggregates and NEMF in a neuronal model (Fig. 5B and Supplementary Fig. 4C, top). We note that expression of the EGFP-PR101 reporter in I3N neurons mainly resulted in nuclear aggregates so we did not observe colocalization between PR and NEMF in this model.
Figure 5.
NEMF is recruited to DPR inclusions. (A) Representative confocal images of colocalization of NEMF and R-rich DPR proteins (arrowheads) expressed in HEK 293T cells (scale bar = 10 µm). (B) Representative confocal images of colocalization of NEMF and R-rich EGFP-DPR reporters (arrowheads) in I3N neurons (scale bar = 5 µm). (C) Confocal images show colocalization of DPR inclusions (poly-GA and poly-GR) and NEMF in frontal cortex and cerebellar granular layer of C9orf72 human tissue (scale bar, 5 µm). (D) Representative double immunofluorescence confocal images of DPR inclusions (poly-GA and poly-GR) and RQC factors (listerin and VCP) in frontal cortex of C9orf72-mutation carriers (scale bar = 5 µm). (E) Quantification of proportion of DPR inclusions (poly-GA and poly-GR) that co-localize with RQC complex factors NEMF, listerin and VCP in frontal cortex and cerebellum from five C9orf72-expansion cases (total number of inclusions counted for poly-GA n = 90–130 or n = 1049–1201 and for poly-GR n = 41–55 or n = 62–114 in frontal cortex and cerebellum, respectively; frontal cortex: Fisher’s exact test, NEMF P < 0.0001, listerin P = 0.0668 and VCP P = 0.6507; cerebellum: Fisher’s exact test, NEMF P < 0.0001, listerin P = 0.0767 and VCP P = 0.1403).
Next, to assess the potential interaction of this core RQC complex-associated factor and DPR inclusions in human brain tissue, dual-immunofluorescence staining for NEMF and DPR proteins (poly-GA and poly-GR) was done on frontal cortex and cerebellar tissue sections from five C9orf72-expansion cases. NEMF was observed as granular cytoplasmic staining in cortical neurons and numerous inclusion-like structures within the cerebellar granule cell layer (Fig. 5C and Supplementary Fig. 4C, bottom). Representative confocal double immunofluorescence images showed that rare poly-GA and poly-GR inclusions appeared to co-localize with NEMF in frontal cortex and cerebellum of C9orf72-expansion cases (Fig. 5C and Supplementary Fig. 4C, bottom). Given that we did not observe NEMF accumulation in GA aggregates in cultured cells, which contrasts with what we observed in human brain tissues, we assessed whether there was any evidence of chimeric DPR species by immunostaining cerebellum from C9orf72 expansion cases for poly-GA and poly-GP or poly-GR proteins. We observed that rare poly-GA inclusions are GR-positive (4.40%) or GP-positive (6.59%) suggestive of the presence of chimeric species in C9orf72 inclusions as reported previously (Supplementary Fig. 4D).66,67
To assess whether other core RQC complex-associated factors listerin and VCP co-localize with DPR proteins, dual-immunofluorescence staining of these factors and DPR proteins (poly-GA and poly-GR) was done in C9orf72 ALS/FTD tissues. For listerin, co-accumulation with cytoplasmic poly-GA and poly-GR inclusions was observed in frontal cortex of repeat-expansion carriers (Fig. 5D, top). In contrast, VCP staining appeared to show diffuse nuclear and cytoplasmic immunoreactivity, which overlapped with poly-GA inclusion staining in frontal cortex, although VCP did not appear to noticeably accumulate within poly-GA aggregates (Fig. 5D, bottom). Quantification of the number of DPR inclusions (poly-GA and poly-GR) that colocalized with core RQC complex factors (NEMF, listerin and VCP) was done for frontal cortex and cerebellum of five C9orf72 expansion cases by manual counting of ×63 microscopic fields (Fig. 5E). NEMF seemed to preferentially co-localize and accumulate within poly-GR inclusions in frontal cortex and cerebellum compared to poly-GA (Fig. 5E; NEMF-positive percentage in frontal cortex: GA 4.62%, GR 30.91% and in cerebellum: GA 0.10%, GR 7.02%). On the other hand, listerin seemed to co-localize and accumulate within both poly-GA and poly-GR inclusions in frontal cortex but not cerebellum of mutation carriers (Fig. 5E; listerin-positive percentage in frontal cortex: GA 15.53%, GR 29.27% and in cerebellum: GA 0.00%, GR: 1.04%). VCP immunoreactivity rarely overlapped with DPR inclusions in either region assessed (Fig. 5E; VCP-positive percentage in frontal cortex: GA 3.3%, GR 4.8% and in cerebellum: GA 0.002%, GR 0.016%). These observations suggest that there may be an association between DPR proteinopathy and NEMF dysfunction in the setting of the C9orf72 repeat-expansion mutation.
Discussion
Dysregulation of protein homeostasis and aggregation is known to be a hallmark of neurodegenerative diseases. Protein quality control pathways ensure the fidelity of protein synthesis and clearance of defective potentially toxic products. How the aberrant protein species generated from RAN translation in repeat-expansion diseases disrupt and bypass these quality control pathways is not well characterized. Dysfunctional RQC pathways have been associated with neurodegeneration and ageing.41–46,68,69 Furthermore, it has been shown to contribute to the hallmark protein aggregation in Huntington’s, Parkinson’s and Alzheimer’s disease.42,46,70,71 This study focused on the role of the RQC complex in the regulation of DPR protein aggregates associated to the C9orf72 repeat expansion in ALS and FTD.
RQC pathways have been shown to be conserved from bacteria to higher eukaryotes.30–34,46,72 Although RQC pathways have been extensively studied in yeast and many RQC components have been shown to be conserved in eukaryotes, the degradation of substrates by the RQC complex in mammalian cells is not well characterized. Recent studies using reporter constructs have further characterized the mammalian RQC showing that CTEs may be more complex than in yeast, may involve redundant mechanisms for degradation of nascent polypeptides and its function is crucial for neuronal survival.32,46,62,73,74 In this study, using an EGFP reporter lacking a STOP codon, we demonstrate that the aberrant non-STOP protein product is efficiently degraded by the canonical RQC complex composed of listerin, NEMF and VCP. Our data support previous findings that deficiency of these factors does not affect the trigger event for RQC complex recruitment of ribosome stalling within a poly-A sequence. Our studies cannot discard the contribution of alternate RQC factors that may add redundancy in mammalian cells as previously described.32,75 More work is needed to identify endogenous mammalian RQC substrates as most studies use overexpression reporters. A study identified mitochondrial outer membrane-associated complex-I 30 kD subunit as an endogenous RQC substrate on mitochondrial damage but additional endogenous substrates have not been described in mammalian cells.46 However, our data showed that clearance of the known canonical RQC substrate, non-STOP mRNA, seems primarily dependent on the main factors identified as the core RQC complex.
C9orf72 RAN translation and its derived DPR proteins have been shown to affect translation dynamics, elicit cellular stress responses and lead to dysregulation of multiple cellular protein quality control processes.11,20,22,76–79 Recent work has suggested that translation of the repeat in the poly-GR-encoding reading frame results in ribosome stalling in cells.28,47,48 RQC pathways are essential in the sensing and clearance of ribosome stalled complexes that form from the translation of defective RNAs.1–3 Our work further expands the understanding of the mechanism by which repetitive sequences associated with RAN translation may induce ribosome stalling in cells. We showed that translation of R-rich DPR proteins induces ribosomal stalling in a length-dependent manner. Use of codon-optimized sequences compared to G4C2/C4G2 sequences suggests that stalling is primarily dependent on the composition of the protein being synthesized rather than the RNA sequence. Of note, we observed that the threshold for inducing ribosome stalling differs for each R-rich DPR protein. Poly-PR seemed to mildly reduce RFP expression of stalling reporters at PR27, strongly inhibited RFP expression at 66 repeats, and showed no further loss of RFP at longer lengths. In contrast, poly-GR seems to require at least 66 poly-GR units to induce stalling but showed even stronger stalling at longer lengths, as previously reported.48 We did not observe induction of ribosome stalling in tri-nucleotide repeats associated with other repeat-expansion diseases suggesting that RQC may not be activated. However, our studies cannot discard the contribution of endogenous sequences flanking these other repeat expansions in elongation dynamics in RAN translation. Notably, as the reporters used here rely on the presence of a canonical AUG start codon for expression of fluorescent proteins, these studies cannot determine the effect of non-AUG-mediated initiation, if any, on the downstream elongation and stalling of G4C2 RNA. In general, C9orf72 RAN translation has been shown to affect translation dynamics and our data showed that R-rich DPR proteins induce ribosome stalling during elongation, which appears to be at least in part due to the co-translational engagement of arginine residues within the ribosome tunnel. This is consistent with a growing literature showing that R-rich DPR proteins interact and sequester ribosomes, which may contribute to global translational inhibition.47,49,77,80–82 However, the poly-GR and poly-PR length thresholds for stalling are longer than the length of the ribosomal exit tunnel suggesting that electrostatic interactions by arginine residues within the ribosome tunnel alone may not account for the mechanism of stalling in cells as previously suggested for poly-GR.48,49,83 Indeed, the relationship between translational initiation, translational elongation rate and ribosome stalling is likely to be complex.
Dysfunction in the RQC pathway leads to neurodegeneration and the accumulation of aberrant protein species through the C-terminal extension of these nascent proteins.41–44,70,71 However, the role of the RQC complex and CTE in proteins generated from RAN translation observed in repeat-expansion diseases remains poorly characterized. Previous studies have implicated a role for CTE in the distribution of poly-Q aggregates in yeast and suggested that CTE of poly-GR proteins may enhance its aggregation.48,70,71 In this study, we focused on understanding if dysfunctional processing of DPR proteins by the RQC complex function plays a role in C9orf72 DPR protein biology. We showed that despite the strong ribosome stalling induced by R-rich DPR sequences, these proteins are not efficiently processed by the canonical RQC complex. The functional deficiency of listerin, NEMF or VCP did not affect the expression of DPR proteins as quantified by the fluorescent signal from reporters. DPR proteins are known to be aggregation prone and highly insoluble, quantification of fluorescent signal provides a way to monitor the overall protein levels regardless of aggregation state. However, NEMF KD did not alter solubility of DPR proteins, which suggests either that the inherently high insolubility of these proteins masks the effects of CTEs on biochemical solubility in our models, or perhaps that NEMF is unable to add CTEs to these proteins due to the inability to dislodge R-rich DPR proteins trapped within the translating ribosome. We hypothesize that the lack of lysine residues within the DPR sequence precludes efficient ubiquitination by listerin and subsequent recruitment of VCP. This is supported by the observation that poly-GR reporters become more amenable to listerin-dependent clearance when poly-GR units are interrupted with lysine residues. Although we have observed that poly-GR units strongly induce ribosome stalling in a length-dependent manner, we assessed whether the endogenous C terminus of the GR frame from repeats in the sense strand would make poly-GR reporters amenable to clearance by the canonical RQC complex. The functional deficiency of listerin did not affect the levels of reporters with codon-optimized GR-encoding sequences with the C terminus probably due to lack of complete ribosome readthrough. We also note that poly-GR reporters depicted in Fig. 4 contain lysine residues on the N-terminal EGFP tag and the C-terminal myc tag but are not susceptible to the canonical RQC complex, probably due to stalling within the GR sequence. Therefore, our data support previous studies that propose listerin-mediated ubiquitination efficiency may be influenced by the positioning of available lysine residues within the substrate.34,84 Moreover, the paucity of ubiquitination of DPR inclusions in human brain tissues with the C9orf72 expansion suggests that DPR inclusions are resistant to proteasomal degradation. Previous studies have reported association of the RQC factor VCP with DPR aggregates.64,65 It is well established that VCP primarily recognizes its substrates through poly-ubiquitin modifications, and we observed that DPR inclusions appear to be primarily devoid of poly-ubiquitination in C9orf72 post-mortem tissue. This is consistent with our results showing a lack of VCP co-accumulation within DPR inclusions from post-mortem brain tissue, similar to previous findings in a neuronal model indicating that VCP is excluded from GA aggregates.85 Taken together, these findings suggest that the lack of internal lysine residues within the R-rich DPR encoding sequences that cause ribosome stalling may preclude their ubiquitination and clearance by the RQC.
Our work highlights the importance of protein quality control and how aberrant protein species generated through RAN translation may lead to their dysregulation. Interestingly, we observed that R-rich DPR proteins co-localize with NEMF in cells but not with poly-GA which does not induce stalling in cells. This observation is congruent with canonical RQC complex recruitment to nascent proteins as a consequence of ribosome stalling. We also observed that NEMF appears to co-localize with DPR inclusions in frontal cortex and cerebellum tissue of C9orf72-expanded cases. While expression patterns of R-rich DPR (PR and GR) in cellular models have been shown to primarily form nuclear aggregates; NEMF seemed to co-accumulate with cytoplasmic R-rich DPR aggregates in HEK 293T cells and human neurons. This distinct interaction may reflect what is observed in the endogenous repeat expansion as intranuclear DPR inclusions are extremely rare and are mainly cytoplasmic in C9orf72 pathology. We hypothesize that NEMF recruitment to DPR inclusions and their impaired processing may perhaps be leading to sequestration of this core RQC factor. More work is needed to identify endogenous RQC substrates to further determine whether the C9orf72 repeat expansion is associated with RQC dysfunction including the accumulation of endogenous RQC substrates. NEMF also has other less characterized molecular functions outside of the RQC complex including the nuclear export of proteins that could be affected.86 Finally, further work is needed to continue to characterize the mammalian RQC and identify if alternate or redundant factors may participate in clearance of aberrant translation products including DPR proteins. Overall, these findings suggest that impaired processing by the canonical RQC complex of these R-rich DPR proteins may contribute to protein homeostasis dysregulation observed in C9orf72-expansion ALS and FTD neuropathogenesis.
Supplementary Material
Acknowledgements
We would like to thank the Translational Neuropathology Research Laboratory for their support and technical assistance. Special thanks to Dennis Zhang and Elena Isasi Theus for contributing to the cloning of plasmids generated for this study. We thank Dr Aaron Gitler (Stanford University) for kindly providing the original plasmid containing the (G4C2)66 repeats. We would like to thank Dr Michael Ward (NIH) for kindly gifting the CRISPRi-i3N iPS cell line used in this study. We thank Dr Corey McMillan and Dr Barbara Spencer (University of Pennsylvania) for their helpful input and discussions.
Contributor Information
Ashley P Viera Ortiz, Biochemistry and Molecular Biophysics Graduate Group, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA 19104, USA; Translational Neuropathology Research Laboratory, Department of Pathology and Laboratory Medicine, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA 19104, USA.
Gregory Cajka, Center for Cellular and Molecular Therapeutics, Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA; Department of Genetics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA 19104, USA.
Olamide A Olatunji, Translational Neuropathology Research Laboratory, Department of Pathology and Laboratory Medicine, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA 19104, USA.
Bailey Mikytuck, Translational Neuropathology Research Laboratory, Department of Pathology and Laboratory Medicine, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA 19104, USA.
Ophir Shalem, Center for Cellular and Molecular Therapeutics, Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA; Department of Genetics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA 19104, USA.
Edward B Lee, Translational Neuropathology Research Laboratory, Department of Pathology and Laboratory Medicine, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA 19104, USA.
Funding
This work was supported by grants from the NIH including P30AG072979 (E.B.L.), P01AG066597 (E.B.L.), R01NS095793 (E.B.L.), RF1AG065341 (E.B.L.) and DP2GM137416 (O.S.), in addition to the CURE grant (O.S.) from the Pennsylvania Department of Health.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Joazeiro CAP. Ribosomal stalling during translation: Providing substrates for ribosome-associated protein quality control. Annu Rev Cell Dev Biol. 2017;33:343–368. [DOI] [PubMed] [Google Scholar]
- 2. Joazeiro CAP. Mechanisms and functions of ribosome-associated protein quality control. Nat Rev Mol Cell Biol. 2019;20:368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sitron CS, Brandman O. Detection and degradation of stalled nascent chains via ribosome-associated quality control. Annu Rev Biochem. 2020;89:417–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Filbeck S, Cerullo F, Pfeffer S, Joazeiro CAP. Ribosome-associated quality-control mechanisms from bacteria to humans. Mol Cell. 2022;82:1451–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zu T, Gibbens B, Doty NS, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011;108:260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Malik I, Kelley CP, Wang ET, Todd PK. Molecular mechanisms underlying nucleotide repeat expansion disorders. Nat Rev Mol Cell Biol. 2021;22:589–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Banez-Coronel M, Ranum LPW. Repeat-associated non-AUG (RAN) translation: Insights from pathology. Lab Investig. 2019;99:929–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Todd PK, Oh SY, Krans A, et al. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron. 2013;78:440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bañez-Coronel M, Ayhan F, Tarabochia AD, et al. RAN translation in Huntington disease. Neuron. 2015;88:667–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Balendra R, Isaacs AM. C9orf72-mediated ALS and FTD: Multiple pathways to disease. Nat Rev Neurol. 2018;14:544–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang YJ, Jansen-West K, Xu YF, et al. Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol. 2014;128:505–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gendron TF, Bieniek KF, Zhang YJ, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126:829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. MacKenzie IR, Arzberger T, Kremmer E, et al. Dipeptide repeat protein pathology in C9ORF72 mutation cases: Clinico-pathological correlations. Acta Neuropathol. 2013;126:859–879. [DOI] [PubMed] [Google Scholar]
- 14. DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haeusler AR, Donnelly CJ, Rothstein JD. The expanding biology of the C9orf72 nucleotide repeat expansion in neurodegenerative disease. Nat Rev Neurosci. 2016;17:383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zu T, Liu Y, Banez-Coronel M, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A. 2013;110:E4968–E4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shi Y, Lin S, Staats KA, et al. Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat Publ Gr. 2018;24:313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sareen D, O’Rourke JG, Meera P, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med. 2013;5:208ra149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Freibaum BD, Lu Y, Lopez-Gonzalez R, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525:129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mackenzie IRA, Frick P, Grässer FA, et al. Quantitative analysis and clinico-pathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers. Acta Neuropathol. 2015;130:845–861. [DOI] [PubMed] [Google Scholar]
- 22. Sun Y, Eshov A, Zhou J, Isiktas AU, Guo JU. C9orf72 arginine-rich dipeptide repeats inhibit UPF1-mediated RNA decay via translational repression. Nat Commun. 2020;11:3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reddy K, Zamiri B, Stanley SYR, Macgregor RB, Pearson CE. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J Biol Chem. 2013;288:9860–9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dodd DW, Tomchick DR, Corey DR, Gagnon KT. Pathogenic C9ORF72 antisense repeat RNA forms a double helix with tandem C:C mismatches. Biochemistry. 2016;55:1283–1286. [DOI] [PubMed] [Google Scholar]
- 25. Endoh T, Sugimoto N. Mechanical insights into ribosomal progression overcoming RNA G-quadruplex from periodical translation suppression in cells. Sci Rep. 2016;6(June 2015):6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang X, Goodrich KJ, Conlon EG, et al. C9orf72 and triplet repeat disorder RNAs: G-quadruplex formation, binding to PRC2 and implications for disease mechanisms. Rna. 2019;25:935–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mori K, Gotoh S, Yamashita T, et al. The porphyrin TMPyP4 inhibits elongation during the noncanonical translation of the FTLD/ALS-associated GGGGCC repeat in the C9orf72 gene. J Biol Chem. 2021;297:101120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Park J, Lee J, Kim J, Lee J, Park H, Lim C. ZNF598 co-translationally titrates poly(GR) protein implicated in the pathogenesis of C9ORF72-associated ALS/FTD. Nucleic Acids Res. 2021;49:11294–11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bengtson MH, Joazeiro CAP. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467:470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Defenouillère Q, Zhang E, Namane A, Mouaikel J, Jacquier A, Fromont-Racine M. Rqc1 and Ltn1 prevent C-terminal alanine-threonine tail (CAT-tail)-induced protein aggregation by efficient recruitment of Cdc48 on stalled 60 s subunits. J Biol Chem. 2016;291:12245–12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Doamekpor SK, Lee JW, Hepowit NL, et al. Structure and function of the yeast listerin (Ltn1) conserved N-terminal domain in binding to stalled 60S ribosomal subunits. Proc Natl Acad Sci U S A. 2016;113:E4151–E4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thrun A, Garzia A, Kigoshi-Tansho Y, et al. Convergence of mammalian RQC and C-end rule proteolytic pathways via alanine tailing. Mol Cell. 2021;81:2112–2122.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shen PS, Park J, Qin Y, et al. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science. 2015;347:75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kostova KK, Hickey KL, Osuna BA, et al. CAT-tailing as a fail-safe mechanism for efficient degradation of stalled nascent polypeptides. Science. 2017;357:414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sitron CS, Brandman O. CAT tails drive degradation of stalled polypeptides on and off the ribosome. Nat Struct Mol Biol. 2019;26:450–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yonashiro R, Tahara EB, Bengtson MH, et al. The Rqc2/Tae2 subunit of the ribosome-associated quality control (RQC) complex marks ribosome-stalled nascent polypeptide chains for aggregation. eLife. 2016;5(e11794):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shao S, Brown A, Santhanam B, Hegde RS. Structure and assembly pathway of the ribosome quality control complex. Mol Cell. 2015;57:433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Defenouillere Q, Yao Y, Mouaikel J, et al. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc Natl Acad Sci U S A. 2013;110:5046–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Verma R, Reichermeier KM, Burroughs AM, et al. Vms1 and ANKZF1 peptidyl-tRNA hydrolases release nascent chains from stalled ribosomes. Nature. 2018;557:446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sitron CS, Park JH, Giafaglione JM BO. Aggregation of CAT tails blocks their degradation and causes proteotoxicity in S. cerevisiae. PLoS ONE. 2019;15:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martin PB, Kigoshi-Tansho Y, Sher RB, et al. NEMF mutations that impair ribosome-associated quality control are associated with neuromuscular disease. Nat Commun. 2020;11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rimal S, Li Y, Vartak R, et al. Inefficient quality control of ribosome stalling during APP synthesis generates CAT-tailed species that precipitate hallmarks of Alzheimer’s disease. Acta Neuropathol Commun. 2021;9:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chu J, Hong NA, Masuda CA, et al. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc Natl Acad Sci U S A. 2009;106:2097–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ahmed A, Wang M, Bergant G, et al. Biallelic loss-of-function variants in NEMF cause central nervous system impairment and axonal polyneuropathy. Hum Genet. 2021;140:579–592. [DOI] [PubMed] [Google Scholar]
- 45. Stein KC, Morales-polanco F, Van Der Lienden J, Rainbolt TK. Ageing exacerbates ribosome pausing to disrupt cotranslational proteostasis. Nature. 2022;601:637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wu Z, Tantray I, Lim J, et al. MISTERMINATE Mechanistically links mitochondrial dysfunction with proteostasis failure. Mol Cell. 2019;75:835–848.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Radwan M, Ang CS, Ormsby AR, et al. Arginine in C9ORF72 dipolypeptides mediates promiscuous proteome binding and multiple modes of toxicity. Mol Cell Proteomics. 2020;19:640–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li S, Wu Z, Tantray I, et al. Quality-control mechanisms targeting translationally stalled and C-terminally extended poly(GR) associated with ALS/FTD. Proc Natl Acad Sci U S A. 2020;117:25104–25115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Loveland AB, Svidritskiy E, Susorov D, et al. Ribosome inhibition by C9ORF72-ALS/FTD-associated poly-PR and poly-GR proteins revealed by cryo-EM. Nat Commun. 2022;13:2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Juszkiewicz S, Hegde RS. Initiation of quality control during poly(A) translation requires site-specific ribosome ubiquitination. Mol Cell. 2017;65:743–750.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cali CP, Park DS, Lee EB. Targeted DNA methylation of neurodegenerative disease genes via homology directed repair. Nucleic Acids Res. 2019;47:11609–11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Edie S, Zaghloul NA, Leitch CC, et al. Survey of human chromosome 21 gene expression effects on early development in Danio rerio. G3 Genes|Genomes|Genetics. 2018;8:2215–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Darwich NF, Phan JM, Kim B, et al. Autosomal dominant VCP hypomorph mutation impairs disaggregation of PHF-tau. Science. 2020;370:eaay8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang C, Ward ME, Chen R, et al. Scalable production of iPSC-derived human neurons to identify tau-lowering compounds by high-content screening. Stem Cell Rep. 2017;9:1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tian R, Gachechiladze MA, Ludwig CH, et al. CRISPR interference-based platform for multimodal genetic screens in human iPSC-derived neurons. Neuron. 2019;104:239–255.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fernandopulle MS, Prestil R, Grunseich C, Wang C, Gan L, Ward ME. Transcription factor–mediated differentiation of human iPSCs into neurons. Curr Protoc Cell Biol. 2018;79:e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Toledo JB, Van Deerlin VM, Lee EB, et al. A platform for discovery: The University of Pennsylvania Integrated Neurodegenerative Disease Biobank. Alzheimers Dement. 2014;10:477–484.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Arthur L, Pavlovic-Djuranovic S, Smith-Koutmou K, Green R, Szczesny P DS. Translational control by lysine-encoding A-rich sequences. Sci Adv. 2015;1:e1500154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chandrasekaran V, Juszkiewicz S, Choi J, et al. Mechanism of ribosome stalling during translation of a poly(A) tail. Nat Struct Mol Biol. 2019;26:1132–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sundaramoorthy E, Leonard M, Mak R, Liao J, Fulzele A, Bennett EJ. ZNF598 and RACK1 regulate mammalian ribosome-associated quality control function by mediating regulatory 40S ribosomal ubiquitylation. Mol Cell. 2017;65:751–760.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Udagawa T, Seki M, Okuyama T, et al. Failure to degrade CAT-tailed proteins disrupts neuronal morphogenesis and cell survival. Cell Rep. 2021;34:108599. [DOI] [PubMed] [Google Scholar]
- 63. Mori K, Arzberger T, Grässer FA, et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 2013;126:881–893. [DOI] [PubMed] [Google Scholar]
- 64. Božič J, Motaln H, Janež AP, et al. Interactome screening of C9orf72 dipeptide repeats reveals VCP sequestration and functional impairment by polyGA. Brain. 2022;145:684–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liu F, Morderer D, Wren MC, et al. Proximity proteomics of C9orf72 dipeptide repeat proteins identifies molecular chaperones as modifiers of poly-GA aggregation. Acta Neuropathol Commun. 2022;10:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tabet R, Schaeffer L, Freyermuth F, et al. CUG initiation and frameshifting enable production of dipeptide repeat proteins from ALS/FTD C9ORF72 transcripts. Nat Commun. 2018;9:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. McEachin ZT, Gendron TF, Raj N, et al. Chimeric peptide species contribute to divergent dipeptide repeat pathology in c9ALS/FTD and SCA36. Neuron. 2020;107:292–305.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Terrey M, Adamson SI, Gibson AL, et al. GTPBP1 resolves paused ribosomes to maintain neuronal homeostasis. eLife. 2020;9:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bertoli-Avella AM, Garcia-Aznar JM, Brandau O, et al. Biallelic inactivating variants in the GTPBP2 gene cause a neurodevelopmental disorder with severe intellectual disability. Eur J Hum Genet. 2018;26:592–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yang J, Hao X, Cao X, Liu B, Nyström T. Spatial sequestration and detoxification of huntingtin by the ribosome quality control complex. eLife. 2016;5(e11792):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zheng J, Yang J, Choe YJ, et al. Role of the ribosomal quality control machinery in nucleocytoplasmic translocation of polyQ-expanded huntingtin exon-1. Biochem Biophys Res Commun. 2017;493:708–717. [DOI] [PubMed] [Google Scholar]
- 72. Lytvynenko I, Paternoga H, Thrun A, et al. Alanine tails signal proteolysis in bacterial ribosome-associated quality control. Cell. 2019;178:76–90.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Saito S, Hosoda N, Hoshino SI. The Hbs1-Dom34 protein complex functions in non-stop mRNA decay in mammalian cells. J Biol Chem. 2013;288:17832–17843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gogakos T, Morozov P, Garzia A, et al. The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. 2017; (May). doi: 10.1038/ncomms16056. [DOI] [PMC free article] [PubMed]
- 75. Kuroha K, Zinoviev A, Hellen CUT, Pestova T V. Release of ubiquitinated and non-ubiquitinated nascent chains from stalled mammalian ribosomal complexes by ANKZF1 and Ptrh1. Mol Cell. 2018;72:286–302.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lee KH, Zhang P, Kim HJ, et al. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell. 2016;167:774–788.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kanekura K, Yagi T, Cammack AJ, et al. Poly-dipeptides encoded by the C9ORF72 repeats block global protein translation. Hum Mol Genet. 2016;25:1803–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sonobe Y, Aburas J, Krishnan G, et al. A C. elegans model of C9orf72-associated ALS/FTD uncovers a conserved role for eIF2D in RAN translation. Nat Commun. 2021;12:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Green KM, Glineburg MR, Kearse MG, et al. RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nat Commun. 2017;8:2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hartmann H, Hornburg D, Czuppa M, et al. Proteomics and C9orf72 neuropathology identify ribosomes as poly-GR/PR interactors driving toxicity. Life Sci Alliance. 2018;1:e201800070-–13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Moens TG, Niccoli T, Wilson KM, et al. C9orf72 arginine-rich dipeptide proteins interact with ribosomal proteins in vivo to induce a toxic translational arrest that is rescued by eIF1A. Acta Neuropathol. 2019;137:487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhang YJ, Gendron TF, Ebbert MTW, et al. Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat Med. 2018;24:1136–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol. 2009;16:589–597. [DOI] [PubMed] [Google Scholar]
- 84. Osuna BA, Howard CJ KCS, Frost A, Weinberg DE. In vitro analysis of RQC activities provides insights into the mechanism and function of CAT tailing. Hinnebusch AG, ed. eLife. 2017;6:e27949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Guo Q, Lehmer C, Martínez-Sánchez A, et al. In situ structure of neuronal C9orf72 poly-GA aggregates reveals proteasome recruitment. Cell. 2018;172:696–705.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bi X, Jones T, Abbasi F, et al. Drosophila caliban, a nuclear export mediator, can function as a tumor suppressor in human lung cancer cells. Oncogene. 2005;24:8229–8239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.