Abstract
The WAVE regulatory complex (WRC), composed of five components—Cyfip1/Sra1, WAVE/Scar, Abi, Nap1/Nckap1, and Brk1/HSPC300—is essential for proper actin cytoskeletal dynamics and remodeling in eukaryotic cells, likely by matching various patterned signals to Arp2/3-mediated actin nucleation. Accumulating evidence from recent studies has revealed diverse functions of the WRC in neurons, demonstrating its crucial role in dictating the assembly of molecular complexes for the patterning of various trans-synaptic signals. In this review, we discuss recent exciting findings on the physiological role of the WRC in regulating synaptic properties and highlight the involvement of WRC dysfunction in various brain disorders.
Subject terms: Neuroscience, Synaptic transmission
Brain disorders: Multiple roles of protein complex need further exploration
Extensive research is needed to understand how the components of a critical protein complex at synapses influence the properties of neural circuits. Jaewon Ko and Kyung Ah Han, Daegu Gyeongbuk Institute of Science and Technology, South Korea, reviewed the roles of the five protein components in the WAVE Regulatory Complex (WRC), and how they organize synaptic properties and neural circuits. The WRC plays key roles in the dynamics and remodeling of actin, a major cytoskeletal protein critical for synapse formation and maintenance. The WRC also activates other protein complexes significantly involved in controlling actin activity. Each WRC component has distinct functions at synapses, such as regulating the growth of dendritic spines (protrusions on dendrites, the extensions of neurons that receive stimuli) and presynaptic assembly. Undoubtedly, dysfunction of the WRC components are associated with various brain disorders.
Introduction
Actin, a major cytoskeletal element, is essential for a range of fundamental cellular processes in eukaryotes. Actin plays a key role in neuronal morphogenesis and synapse formation during nervous system development1. In mature neurons, actin is the most prominent cytoskeletal component in both presynaptic and postsynaptic compartments, including dendritic spines, which mediate most excitatory synaptic transmission in the brain2–4. In addition, actin cytoskeletal remodeling has been implicated in structural synaptic plasticity and is intimately linked to the proper operation of molecular machineries critical for synaptic functions involving a cohort of various actin-binding proteins that regulate the assembly and disassembly of actin filaments5–7. Notably, it has been established that the Arp2/3 (actin-related proteins-2 and -3) complex, an evolutionarily conserved actin nucleating hub, acts in conjunction with dozens of other nucleation-promoting factors to drive polymerization, organization, and recycling of the actin filament network8.
Among nucleation-promoting factors, members of the WASP (Wiskott-Aldrich syndrome protein), neuronal WASP, and WAVE (WASP family verprolin homologous protein; also known as SCAR [suppressor of cyclic AMP [cAMP] receptor] family have been highlighted as ubiquitous regulators of actin cytoskeletal remodeling9–11. These proteins exist in a heteropentameric macromolecular complex (~400 kDa) known as the WRC (WAVE regulatory complex)11,12. The WRC is assembled from five different proteins—CYFIP (cytoplasmic FMR1-interacting protein; also known as Sra1), NAP (NCK-associated protein; also known as Nckap1), ABI (Abelson-interacting protein), HSPC300 (hematopoietic stem progenitor cell 300; also known as Brk1), and WAVE—that are all essential for WRC functions11 (Fig. 1a). Remarkably, for each component protein, there are homologous proteins that likely exhibit tissue-specific and/or cell-type-specific expression, employ distinct regulatory mechanism(s) and/or differentially activate Arp2/3-mediated actin polymerization, as elaborated below. The same is true for orthologous subunits in different organisms. The WRC is intrinsically inactive but is activated upon interaction with numerous cytosolic proteins, small GTPases, and transmembrane receptors, causing its translocation to the plasma membrane, where it activates the Arp2/3 complex11. Notable regulators include the Rho-family GTPase Rac1 and Arf GTPase Arf1, which allosterically relieve autoinhibition of the WRC by releasing the WCA (WASP homology 2-central-acidic) domain13,14 (Fig. 1b). Moreover, PIP3 [phosphatidylinositol-(3,4,5)-triphosphate] further enhances Rac1-mediated WRC activation15,16. Although our understanding is still incomplete, extensive research on the basic biology and regulatory mechanisms of the WRC has significantly contributed to our view of how actin networks are organized in eukaryotic cells.
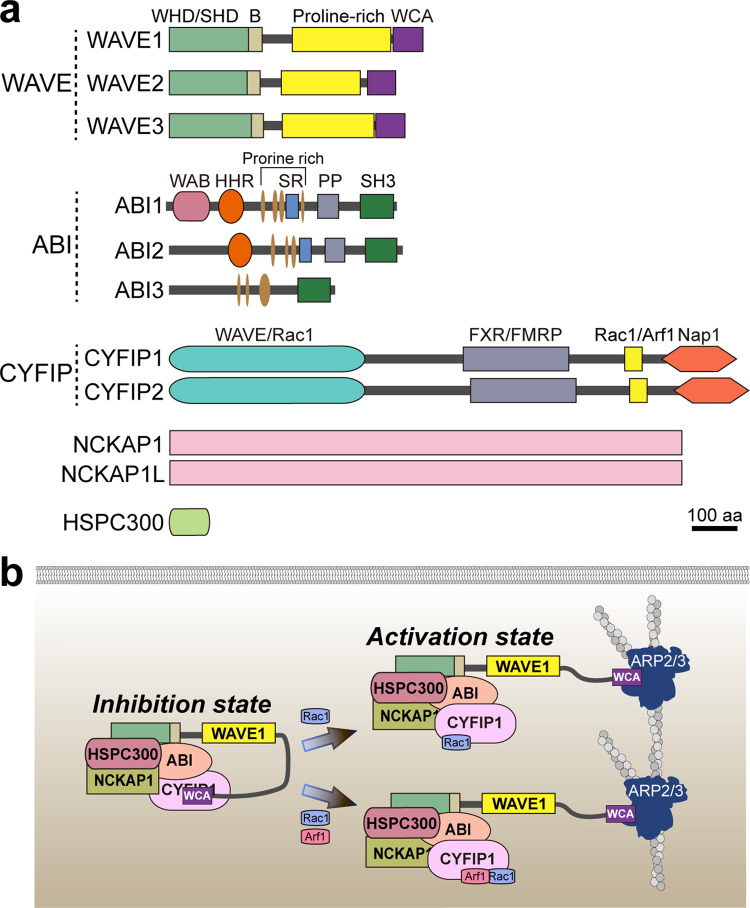
Fig. 1. WRC subunit composition and assembly mechanism.
a Schematic depicting WRC subunits and their homologs. The WRC is a five-subunit complex comprising the following protein families: ABI (ABI1, ABI2 or ABI3), WAVE (WAVE1, WAVE2, or WAVE3), Nap1 (Nckap1 or Nckap1L), CYFIP (CYFIP1 or CYFIP2), and HSCP300. aa amino acid, B basic domain, HHR Hox homology region, PP polyproline structure, SH3 Src homology 3 domain, SHD SCAR homology domain, SR serine/threonine-rich region, WAB WAVE-binding domain, WCA WASP homology 2-central-acidic, WHD WAVE homology domain. b Schematic illustration of two modes of WRC activation in actin polymerization. In the absence of Rac1 binding, WRCs exist in an autoinhibited state. Rac1 binding to the A site located at the N-terminus of CYFIP1 induces WRC activation. This destabilizes the meander sequence of WAVE1, which is critical for autoinhibition, inducing a conformal change that triggers the release of the WCA sequence, making it accessible to the ARP2/3 complex. In contrast, Rac1 binding to the D site, located in the C-terminal region of CYFIP1, does not directly activate WRC but does increase the affinity for ARF1 binding between the D site of CYFIP1 and the W helix of the WCA domain of WAVE, allowing the WCA region of WAVE1 to activate ARP2/3.
In the current review, we describe the known functions of WRC components at neuronal synapses. We then discuss the role of the WRC in orchestrating key aspects of synapse development. Finally, we highlight the association of WRC dysfunctions with certain brain disorders and consider their ramifications. Owing to space constraints, we limit our presentation of background information on the WRC to neuroscience-related topics; recent review articles are available for those interested in greater detail11,17.
The role of WRC components during nervous system development
Although the significance of the WRC in regulating actin dynamics and remodeling has gradually come to be appreciated, the roles of individual WRC components in various processes during nervous system development have not yet been convincingly defined. Because each of the five components—Cyfip1/Sra1, WAVE/Scar, Abi, Nap1/Nckap1, and HSPC300/Brk1—also mediates complex formation with other distinct proteins, it is possible to speculate that distinct functions of each WRC component shape concerted Arp2/3-mediated F-actin polymerization. In this section, we describe key observations involving each WRC component and their relation to actin remodeling processes in various contexts. We also highlight a subset of issues that require further clarification.
Cyfip
The two evolutionarily conserved proteins, Cyfip1/Sra1 and Cyfip2/Pir121, that constitute the Cyfip protein family were initially identified as interacting partners of FMRP (fragile X mental retardation protein), an RNA-binding protein involved in the GTP-dependent translational control of synaptic proteins18. It was subsequently shown that Cyfip1 and Cyfip2, as Rac1 effectors, constitute the WRC19–21 and form a complex with eIF4E (eukaryotic translation initiation Factor 4E) modulated by BC1 (brain cytoplasmic RNA 1)22, suggesting that Cyfip might provide a bridge between actin remodeling and translation. Moreover, BDNF (brain-derived neurotrophic factor) activates Rac1, causing Cyfip1 to dissociate from the FMRP-eIF4E complex (thereby antagonizing FMR1 [fragile X messenger ribonucleoprotein 1] functions) and associate with Rac1-WRC, leading to inhibition of translational repression22.
Both Cyfip1 and Cyfip2 are enriched at synapses of excitatory postsynaptic spines, where they regulate F-actin dynamics and dendritic spine development23–25. Overexpression of Cyfip1 or Cyfip2 enhances dendritic complexity and outgrowth, whereas haploinsufficiency of either result in abnormal dendritic spines26,27. Another study showed that both Cyfip1 and Cyfip2 are also localized at GABAergic synapses28. Strikingly, overexpression of Cyfip1 or Cyfip2 decreases inhibitory synapse structure and transmission, disrupting the excitatory-to-inhibitory (E/I) synaptic balance28. Similarly, postsynaptic loss of Cyfip1 increases inhibitory synapse size and strength, concomitant with the upregulation of GABAA receptor β2/3 subunits and the synaptic adhesion protein Nlgn3 (neuroligin−3)28. Studies on Cyfip1 heterozygous (Cyfip1+/-) mice illustrate the postsynaptic role of Cyfip1, demonstrating altered synapse composition in the hippocampus of these mice, with decreased levels of SynGAP1 (a synaptic Ras GTPase activating protein 1) and GluA1 (an AMPAR [α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor] subunit) and increased levels of mGluR1/5 [metabotropic glutamate receptor 1 and 5], GluN2B (an NMDAR [N-methyl-D-aspartate receptor] subunit), and F-actin29,30. Cyfip1 also participates in the regulation of presynaptic nerve terminal size and vesicle release probability through its actions on the WRC downstream of Rac131, whereas Cyfip2 regulates presynaptic short-term plasticity through its actions on presynaptic mitochondria in medial prefrontal cortical neurons32. However, it remains unclear how Cyfip proteins, which are present at both excitatory and inhibitory synapses, form complexes with Rac1, WAVE1, FMRP, and other proteins to bidirectionally regulate the synaptic E/I ratio. In addition, it is unknown whether the identified presynaptic roles of Cyfip proteins are universal across diverse brain regions.
Each Cyfip paralog also performs distinct functions. Analyses of Cyfip1/2-haploinsufficient mice showed that Cyfip2+/−, but not Cyfip1+/−, mice exhibit cell-type–specific defects in spine morphogenesis27. In line with this observation, Cyfip1 and Cyfip2 distinctively regulate retinal ganglion cell axon growth and guidance in zebrafish33. In particular, upon axon-axon contact, Cyfip2 is cotransported with RNPs (ribonucleoprotein particles) to the growth cone periphery, where it switches its association, dissociating from RNPs and associating with the WRC to regulate actin polymerization and filopodial dynamics33. Expression of Cyfip1 fails to rescue the axon-sorting defect phenotype associated with Cyfip2 deletion33, indicating the nonredundant function of Cyfip paralogs during retinal development. This axon-segregating function of Cyfip2 likely relies on its binding to a subset of transmembrane receptors involved in processes that govern proper axon sorting34. It is also possible that Cyfip2 functions are distinct across diverse animal species (e.g., see ref. 35). Intriguingly, single-cell RNA sequencing analyses revealed that Cyfip1 is expressed in both neurons and astrocytes, whereas Cyfip2 is predominantly expressed in neurons36, a finding that warrants future studies using conditional KO (knockout) lines to systematically analyze the effects of neuron-specific or astrocyte-specific deletion. It is also important to consider differences in the post-transcriptional regulatory activities of Cyfip paralogs in analyses of phenotypes arising from cell-type-specific deletion37.
Abi
Members of the Abi protein family, which includes Abi1, Abi2, and Abi3/NESH, were initially identified as substrates of c-Abl tyrosine kinase (with the exception of Abi3)38–40. Abi proteins form complexes with various proteins (e.g., Eps8, Sos1, or WAVE2) linked to the Rac1-orchestrated actin remodeling pathway that leads to enhanced Arp2/3-mediated actin nucleation40–42. Abi1 promotes tyrosine phosphorylation of several proteins, including Mena (mammalian enabled), BCAP (B-cell adaptor for PI3-kinase), Cdc3 and WAVE2, promoting localization of these proteins to specific subcellular compartments43–46. Intriguingly, Abi1 itself is phosphorylated at serine 88 by CaMKIIα (Ca2+/calmodulin-dependent kinase IIα), which is essential for Abi1-dependent modulation of spine morphogenesis47. Abi1, initially enriched in growth cones, is relocated to filopodia and dendritic spines and becomes restricted to the postsynaptic density matrix through interactions with ProSAP2/Shank3 (SH3 and multiple ankyrin repeat domains 3). Upon synaptic activation, it translocates to the nucleus through interactions with heterogeneous nuclear ribonucleoprotein K or is retransported to excitatory synaptic sites through interaction with the motor protein Kif26B48–50. In the nucleus, Abi1 forms a trimeric complex with Myc and Max transcription factors that facilitates the transcription of E-box-regulated genes, including epidermal growth factor receptors48. All three Abi proteins are characterized by their involvement in dynamic actin cytoskeleton remodeling, as demonstrated in various model organisms42,48,51–56, but whether and how each Abi paralog exerts distinct actions in the regulation of actin dynamics at synaptic sites remain unexplored.
WAVE/SCAR
Three members of the WAVE protein family (WAVE1–3), also known as the SCAR family, possess modular domains that interact with the small GTPases Cdc42 or Rac1 or the adapter protein IRSp53, leading to activation of the Arp2/3 complex and subsequent actin remodeling and branching at the leading edges of cells57. WAVE1 is highly expressed in the brain, and its deletion abnormally alters the size of both presynaptic terminals and postsynaptic spines58. Moreover, Wave1-KO mice exhibit deficits in sensorimotor function, including impaired motor coordination and balance, reduced anxiety levels, and deficits in hippocampus-dependent learning and memory59. Because removal of WAVE1 impedes assembly of the WRC, it is likely that WAVE1 and its binding proteins, such as WRP (WAVE1-associated RacGAP protein) and profilin, form a localized hub that serves to properly regulate Rac signaling. Notably, as a kinase-anchoring protein, WAVE1 is linked to various protein kinases, including cAMP-dependent PKA (protein kinase A) and Cdk5 (cyclin-dependent kinase 5)60,61. Cdk5 phosphorylates three residues in WAVE1, an action that is reversed by stimulation of D1-type dopamine receptors and NMDARs62,63. An acute challenge of mice with cocaine following a 2-week course of cocaine administration and subsequent 2-week withdrawal period similarly dephosphorylates residues phosphorylated by Cdk560. Moreover, WAVE1 is expressed in MSNs (medium spiny projection neurons) in the striatum; further analyses using D1-MSN-expressing, neuron-specific Wave1-KO mice showed that WAVE1 is essential for activity-dependent regulation of dendritic spine density and excitatory synaptic transmission selectively in D1-MSN neurons60.
Similar to WAVE1, WAVE2 is involved in maintaining dendritic spine density and size through a mechanism that involves an IRSp53-dependent pathway64; it also controls dysbindin-1-mediated dendritic morphogenesis65. Moreover, WAVE2 interacts with c-Abl tyrosine kinase, which is modulated by Abi166. Notably, c-Abl-mediated tyrosine phosphorylation of WAVE2 is required for the actin remodeling activity of WAVE266. WAVE3 is also phosphorylated by c-Abl, and this phosphorylated form of WAVE3 regulates lamellipodia formation and cell migration67. Puzzlingly, Abi1 is crucial for c-Abl-mediated tyrosine phosphorylation of WAVE2 but not WAVE366,67, implying the presence of additional intermediate(s) that could be uniquely associated with WAVE3-containing WRCs. Because each WAVE paralog exhibits differential localization in growth cones68, it is plausible that complexes containing distinct WAVE orthologs have different roles during synapse development. Future analyses of the interactomes of each WAVE paralog in vivo will be required to better understand the conserved and divergent functions of the WRC.
Nap1/NCKAP1
Nap1, also termed NCKAP1 (Nck-associated protein 1), was demonstrated to interact directly with Rac1 and the other WRC components, Cyfip1 and Abi1, to regulate WAVE1 activity, which is required for Arp2/3-mediated actin polymerization and branching at protrusive membrane edges and subsequent lamellipodial extension54,69,70. Nap1 is localized along lamellipodia and mediates cell migration and laminar-specific neuronal differentiation in the developing neocortex55,71–73. It is also responsible for remodeling the motility and adhesion machinery by forming complexes with OL-protocadherin72. Moreover, Nap1 expression in cortical neurons is upregulated by BDNF73, and its stability is modulated by interaction with HSP90 (heat shock protein 90)74. These studies clearly establish a role for Nap1 in developing neurons, but the function of Nap1 at mature stages of synapse development and details of the underlying mechanism remains unclear.
BRK1/HSPC300
Brk1 (BRICK1)/HSPC300, the smallest component (~8 kDa) of the WRC, has been most extensively studied in the context of cytoskeletal remodeling in Arabidopsis75,76, with these studies suggesting a crucial role of HSPC300 in promoting Arp2/3 activity77. However, a study employing RNAi (RNA interference) showed that the effects of HSPC300 knockdown in cultured Drosophila S2 cells are modest relative to those observed with RNAi-mediated ablation of other WRC components54. Moreover, other studies have shown that HSPC300 is not required for the assembly of the WAVE complex in vitro or Arp2/3-mediated actin polymerization42,78. In the only currently available study, Drosophila HSPC300 was shown to regulate synaptic morphology at NMJs (neuromuscular junctions) by forming a complex with Rac1-WAVE proteins79. However, the lack of studies targeting vertebrate HSPC300 orthologs has hindered the establishment of the critical roles of these proteins in the WRC.
Synaptic functions of the WRC
Postsynaptic spine morphogenesis
Dendritic spines are morphologically diverse, protrusive structures studding from dendritic shafts that receive the most excitatory synaptic inputs2,80,81. They are almost exclusively enriched with F-actin, a polymerized form of actin filaments; notably, remodeling of F-actin governs excitatory synapse physiology. Numerous actin regulators that mediate the tight control of assembly and signaling in dendritic spines have been identified and shown to collectively orchestrate actin remodeling dynamics in conjunction with a host of spine-enriched activators and/or inhibitors that tune the activity of various small GTPases82–84. Arp2/3 is the most notable central regulator, and together with other F-actin regulators, such as formins and profilins, it drives actin filament assembly, dendritic spine maturation, and dendrite branching81,85,86. A subset of protein kinases provides phosphorylation-based regulation of the WRC87, altering its conformation in a manner that facilitates interactions with Arp2/3. The WRC is activated and relocated to membranes by the concerted actions of upstream factors, including Rac1 and other proteins, as well as by PIP311,88. Rac1-mediated promotion of spine growth involves NMDA receptor activation and direct interaction with the WRC component Cyfip125. In addition, PIP3 binds to the WRC component WAVE2 to regulate the formation of filopodia-like protrusion structures that project from spines during structural plasticity in hippocampal CA1 pyramidal neurons89,90. Furthermore, the WRC physically interacts with diverse synaptic receptors that likely transduce patterned extracellular signals into the intracellular compartment34 (Table 1; see also below for details). Whether the WRC-Arp2/3 axis operates similarly across diverse cell types and brain regions needs to be systematically investigated in future studies.
Table 1.
WIRS-containing transmembrane proteins.
| Gene name | Protein name | Sequences and positions of WIRS |
|---|---|---|
| PTPRS | Receptor-type tyrosine-protein phosphatase S |
LATFCV (aa 1526–1531) LGSFDH (aa 1940–1945) |
| PTPRD | Receptor-type tyrosine-protein phosphatase delta | LGSFDH (aa 1904–1909) |
| PTPRF | Receptor-type tyrosine-protein phosphatase F | LGSFDH (aa 1899–1904) |
| EPHA3 | Ephrin type-A receptor 3 |
LDSFLR (aa 707–712) ITTFRT (aa 910–915) |
| EPHA5 | Ephrin type-A receptor 5 | LDTFLK (aa 761–766) |
| EPHA6 | Ephrin type-A receptor 6 |
LDSFLR (aa 759–764) IVSFLD (aa 926–931) FTTFDL (aa 987–992) |
| EPHA7 | Ephrin type-A receptor 7 | FTTFCS (aa 922–927) |
| EPHA8 | Ephrin type-A receptor 8 |
LDTFLR (aa 721–726) FRTFSS (aa 812–817) |
| EPHA10 | Ephrin type-A receptor 10 |
FSTFPS (aa 929–934) FPSFGS (aa 932–937) |
| EPHB1 | Ephrin type-B receptor 1 | LDSFLR (aa 705–710) |
| EPHB2 | Ephrin type-B receptor 2 |
LDSFLR (aa 707–712) YTSFNT (aa 912–917) FTSFDV (aa 939–944) |
| EPHB3 | Ephrin type-B receptor 3 |
LDSFLR (aa 719–724) YTTFTT (aa 924–929) FASFDL (aa 951–956) |
| EPHB4 | Ephrin type-B receptor 4 |
LDSFLR (aa 701–706) FGSFEL (aa 933–938) |
| EPHB6 | Ephrin type-B receptor 6 |
LDSFLR (aa 756–761) LSSFAF (aa 786–791) LCTFSD (aa 974–979) |
| BAI2 | Adhesion G protein-coupled receptor B2 |
YPSFLS (aa 1395–1400) FHTFDR (aa 1492–1497) WSTFKS (aa 1544–1549) |
| BAI3 | Adhesion G protein-coupled receptor B3 | WDTFKN (aa 1470–1475) |
| NLGN1 | Neuroligin-1 |
LHTFNT (aa 834–839) FNTFTG (aa 837–842) |
| NLGN3 | Neuroligin-3 | YNTFAA (aa 827–832) |
| NLGN4X | Neuroligin-4 |
LHTFNT (aa 792–797) FNTFSG (aa 795–800) |
| LPHN1 | Latrophilin-1 | ISTFCF (aa 879–884) |
| UNC5A | Netrin receptor UNC5A | YGTFNF (aa 444–449) |
| UNC5C | Netrin receptor UNC5C | FGSFNS (aa 533–538) |
| UNC5D | Netrin receptor UNC5D |
FQTFNF (aa 424–429) LDSFGT (aa 670–675) |
| SLITRK6 | SLIT and NTRK-like protein 6 | FLSFQD (aa 746–751) |
| DSC2 | Desmocollin-2 | WHSFTQ(aa 826–831) |
| DSC3 | Desmocollin-3 | WHSFTQ(aa 821–826) |
| PCDHA1 | Protocadherin alpha-1 | FITFGK (aa 913–918) |
| PCDHA2 | Protocadherin alpha-2 | FITFGK (aa 911–916) |
| PCDHA5 | Protocadherin alpha-5 | FITFGK (aa 899–904) |
| PCDHA6 | Protocadherin alpha-6 | FITFGK (aa 913–918) |
| PCDHA7 | Protocadherin alpha-7 | FITFGK (aa 900–905) |
| PCDHA8 | Protocadherin alpha-8 | FITFGK (aa 913–918) |
| PCDHA9 | Protocadherin alpha-9 | FITFGK (aa 913–918) |
| PCDHA10 | Protocadherin alpha-10 | FITFGK (aa 911–916) |
| PCDHA11 | Protocadherin alpha-11 | FITFGK (aa 912–917) |
| PCDHA12 | Protocadherin alpha-12 | FITFGK (aa 904–909) |
| PCDHA13 | Protocadherin alpha-13 | FITFGK (aa 913–918) |
| PCDHAC1 | Protocadherin alpha-C1 | FITFGK (aa 926–931) |
| PCDHAC2 | Protocadherin alpha-C2 | FITFGK (aa 970–975) |
| PCDH9 | Protocadherin-9 | LSTFAP (aa 1158–1163) |
| PCDH11X | Protocadherin-11X | LTTFTP (aa 1323–1328) |
| PCDH11Y | Protocadherin-11Y | LTTFAP (aa 1316–1321) |
| PCDH8 | Protocadherin-8 | MSTFCK (aa 984–989) |
| PCDH10 | Protocadherin-10 |
MPSFVP (aa 955–960) FSTFGK (aa 1000–1005) |
| PCDH17 | Protocadherin-17 | FCTFGK (aa 998–1003) |
| PCDH18 | Protocadherin-18 | FSTFGK (aa 983–988) |
| PCDH19 | Protocadherin-19 | FATFGK (aa 1015–1020) |
| PCDH12 | Protocadherin-12 | FQTFGK (aa 1087–1092) |
| FAT2 | Protocadherin Fat 2 | LVTFGP (aa 4128–4133) |
| FAT3 | Protocadherin Fat 3 |
MTTFHP (aa 4263–4268) LSSFQS (aa 4339–4344) FSTFAV (aa 4482–4487) |
| CELSR3 | Flamingo homolog 1 | LASFNS (aa 3250–3255) |
| TMEM132A | Transmembrane protein 132A | FVTFAP (aa 965–970) |
| TMEM132C | Transmembrane protein 132C | FTTFTT (aa 1056–1061) |
| TMEM132D | Transmembrane protein 132D | FTTFTA (aa 1047–1052) |
| TMEM132E | Transmembrane protein 132E | FTTFTT (aa 1015–1020) |
| ROBO1 | Roundabout homolog 1 | MKTFNS (aa 1050–1055) |
| ROBO3 | Roundabout homolog 3 | LQTFHG (aa 1029–1034) |
| KIRREL1 | Kin of IRRE-like protein 1 | YSSFKD (aa 572–577) |
| NEO1 | Neogenin | LKSFAV (aa 1359–1364) |
aa amino acid.
The indicated residue numbers are based on human protein sequences.
Axon guidance
Accurate formation of synaptic connections during nervous system development requires that axonal growth cones detect a vast array of guidance cues that direct them to their appropriate synaptic targets. These extracellular cues lead to coordinated regulation of actin and microtubule networks. The Arp2/3 complex negatively regulates growth cone translocation and pathfinding but not growth cone morphology91,92. In addition, the Arp2/3 complex is required for axon guidance and initiation of growth cone filopodia in Caenorhabditis elegans (C. elegans) through multiple actin modulatory pathways, including the WRC93–98. A number of transmembrane molecules are also linked to the Arp2/3-mediated polymerization of branched actin needed for proper axon guidance in Drosophila embryos. Robo1 (roundabout guidance receptor 1) directly interacts with the WRC, which is further modulated by the presence of Slit (a repulsive ligand for the Robo family of receptors in Drosophila) secreted from midline glia99. The Robo-WRC interaction is essential for Robo1-mediated growth cone repulsion at the midline99. In addition, neogenin, a well-known axon guidance regulator, recruits the WRC-Arp2/3 complex to promote actin nucleation and thereby maintain adherens junction dynamics and tension100 (Fig. 2). Overall, these studies suggest that by regulating actin cytoskeletal dynamics, the WRC-Arp2/3 axis could act as a central hub for axon guidance. However, whether other axon guidance regulators are also coupled to the WRC-Arp2/3 complex and how they functionally interact in a combinatorial manner in vertebrate neurons remains to be determined.
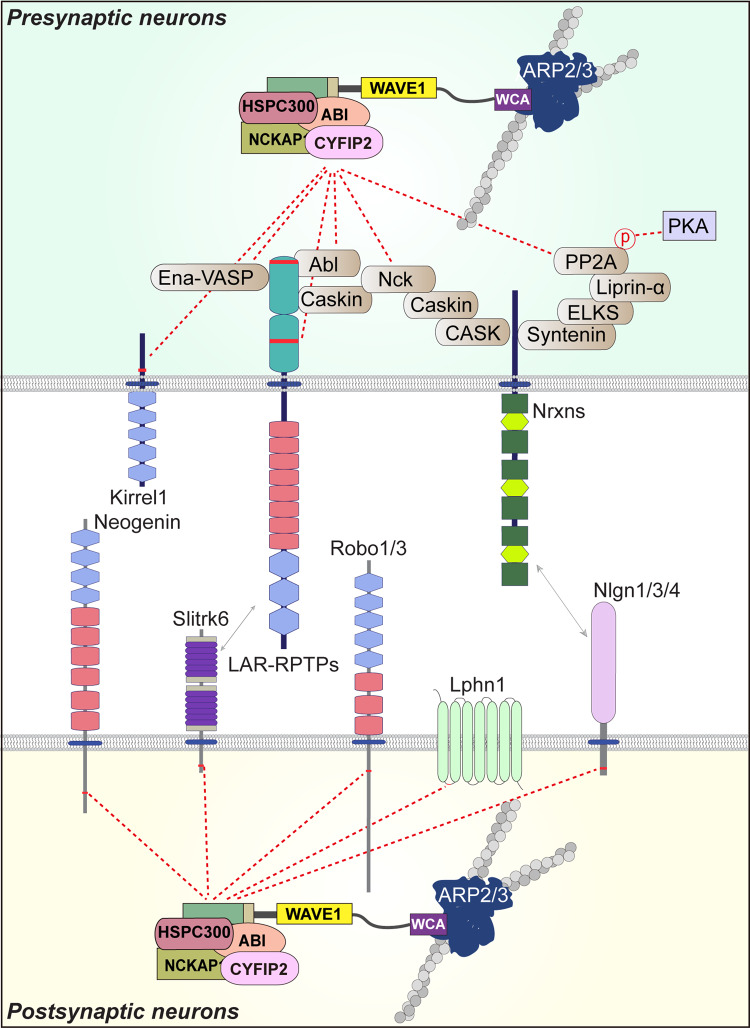
Fig. 2. Molecular model of WRC recruitment at neuronal synapses.
WRC recruitment is regulated by cell-adhesion proteins and their interacting scaffold proteins at synapses. Various scaffold proteins present in the presynaptic active zone play important roles in WRC recruitment and function. Prominent among them are LAR-RPTPs and Kirrel1. Neurexins (Nrxns) can also bind to the WRC but do not do so directly; instead, they bind indirectly through presynaptic scaffolding proteins, including syntenin, ELKS, liprin-α, PP2A, CASK, Caskins and Nck. In addition to interacting directly with the WRC through their WIRS motif, LAR-RPTPs can also indirectly bind the WRC through interactions with Ena, Abl, and Caskins. Robo receptors, Nlgns, neogenin, Slitrks, and latrophilins are also located at the postsynapse and might recruit the WRC through their WIRS motif.
Synaptic vesicle clustering and presynaptic assembly
F-actin is highly and preferentially localized around presynaptic vesicle clusters and enriched at the presynaptic active zone, providing a structural substrate for synaptic vesicle clustering, nerve terminal arborization, and axon guidance101–103. Intriguingly, synaptic activity promotes presynaptic F-actin assembly, which is essential for proper development of the presynaptic active zone and synaptic vesicle pools103. Arp2/3-dependent F-actin networks coordinate the concomitant activation of diverse pathways involving synaptic vesicle proteins, active zone proteins, and/or adapters that dictate synaptic vesicle localization and clustering102 (Fig. 2). This WRC component also appears to be involved in synaptic vesicle clustering at nascent synapses through netrin- and Rac1-dependent pathways51,104 and synaptic cell-adhesion signaling pathways105 in C. elegans. Moreover, orthologs of WRC components strongly accumulate in axons, and functional studies using fly lines harboring a null mutation of each component have delineated common abnormalities in axonal and synaptic morphology—specifically, reduced synaptic terminal length at the NMJ—in Drosophila20,70,106–108. These studies reinforce the idea that the cooperative operation of WRC components in recruiting functional presynaptic machinery involving Arp2/3-mediated actin remodeling is evolutionarily conserved. However, understanding whether and how the WRC contributes to presynaptic assembly, particularly in vertebrate presynaptic neurons, downstream of presynaptic F-actin organization, will require further investigation.
Transduction of extracellular synaptic adhesion signals to intracellular machinery
Actin dynamics are intimately involved in the regulation of cell surface expression and trafficking of synaptic receptors109. The WRC interacts with a host of transmembrane receptors that likely orchestrate trans-synaptic adhesion pathways11,34. Intriguingly, a highly conserved consensus peptide motif composed of six residues (Φ-x-T/S-F-X-X; Φ = hydrophobic amino acid and X = any amino acid), termed the WIRS (WRC interacting receptor sequence), is present in the cytoplasmic regions of ~115 transmembrane proteins34. The WIRS binding surface is occupied by both the Cyfip and Abi subunits of the WRC and is present only in the fully assembled complex34. Further analyses have also established that WIRS motifs are prevalent in other potential candidate WRC ligands (Table 1). Because many WIRS-containing proteins are also considered synaptic cell-adhesion molecules (CAMs), these studies suggest the tantalizing concept that various synaptic adhesion pathways, in collaboration with other types of WRC ligands, are involved in the recruitment of the WRC to cellular membranes. Indeed, the WRC forms complexes with the cell-adhesion protein SYG-1 at presynaptic sites and regulates both synaptogenesis and axonal branching in egg-laying motor neurons of C. elegans105.
Another study reported that Drosophila Nlgn1 directly interacts with the WRC through its WIRS motif and regulates NMJ growth and synaptic transmission through its WRC-binding activity110. Because vertebrate Nlgns also possess the WIRS motif in their cytoplasmic regions110 (Table 1), the regulation of F-actin assembly through the Nlgn-mediated synaptic adhesion pathway is likely an evolutionarily conserved mechanism. Intriguingly, the WRC regulatory pathway appears to integrate cues from other signaling pathways encompassing PKA62,63,111. Moreover, a subset of protein kinase pathways, including PKA and postsynaptic cAMP signaling, is essential for compartmentalized signaling at excitatory synapses112–114, suggesting the fascinating hypothesis that differentially activated WRC pathways determine the strength of intracellular signals in postsynaptic neurons (Fig. 2). This hypothesis is particularly compelling because synaptic CAMs are thought to mediate the specificity of neural circuit architecture113,115,116. Another avenue for future research would be to investigate whether specific paralogs of WRC components align with specific synaptic adhesion pathways. To this end, it would be invaluable to determine the expression patterns of each WRC component in distinct cell types, ideally at single-cell resolution.
The association of WRC dysfunction with brain disorders
Considering the broad ramifications of the WRC and its interacting networks in governing actin-regulatory machinery in cells, it should come as no surprise that dysfunction in the WRC is associated with a variety of genetic disorders. Because of the universal significance of actin dynamics and remodeling and the fact that mutations affecting the expression and/or other biochemical properties of a single WRC component also influence those of other components, dysregulation of WRC components could manifest as neurological disorders, immune deficiencies, or cancer11,117. Here, in keeping with the focus of this review, we primarily discuss links between the dysfunction of WRC components and brain disorders (Fig. 3).
Fig. 3. Implication of WRC components in various neurological disorders.
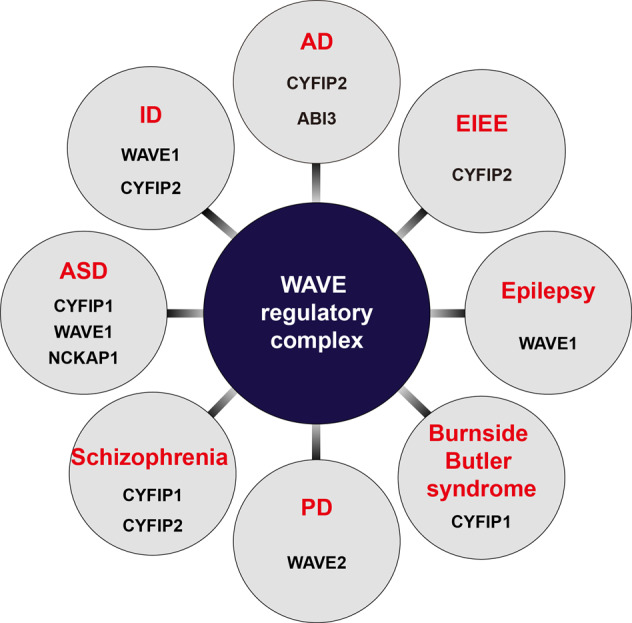
Schematic of the neurological disorders discussed in the current paper that are related to dysfunctions of each WRC component. Dysfunction of WAVE1/2, NAP1, ABI3 and CYFIP1/2 might cause neurodevelopmental, neuropsychiatric, and neurodegenerative disorders. ASD autism spectrum disorder, AD Alzheimer’s disease, EIEE early infantile epileptic encephalopathy, ID intellectual disability, PD Parkinson’s disease.
Disturbance of Cyfip1 functions linked to neurodevelopmental and neuropsychiatric diseases
Deletions in the 15q11.2 region of the human genome (15q11.2 microdeletion), also called Burnside Butler syndrome, are a rare chromosomal anomaly clinically associated with developmental delay, mental retardation, epilepsy, autism spectrum disorder (ASD), schizophrenia, and congenital heart defects118. One of the genes within this chromosomal locus is CYFIP1119,120. Because homozygous deletion of Cyfip1 in mice is embryonic lethal, most animal studies on this syndrome have primarily been performed using Cyfip1+/− mice. These mice exhibit impairments in motor learning and coordination, altered sensory motor gating, aberrant sensory perception/novelty seeking, and enhanced extinction of inhibitory avoidance—phenotypes that are frequently found in individuals with ASD and schizophrenia26,121–123. Conversely, transgenic mice overexpressing Cyfip1 exhibit a series of distinct behavioral phenotypes, including increased fear with mild learning and memory deficits but no ASD-like behavioral abnormalities124. These studies suggest that altered CYFIP1 dosage contributes to divergent endophenotypes associated with diverse neurodevelopmental and neuropsychiatric disorders. Although incompletely understood, abnormalities in NMDAR or mTOR (mammalian target of rapamycin) signaling and altered white matter changes (e.g., thinning of myelin sheath in the corpus callosum) have been proposed as possible pathophysiological mechanisms underlying Cyfip1 dysfunction-associated brain diseases26,30,122,125. However, it remains to be determined how Cyfip1 dysfunction-related mechanisms cause alterations in specific neural circuits responsible for specific behavioral domains associated with the indicated neurological disorders.
Cyfip2 mutations in early infantile epileptic encephalopathy
As described in the previous section (Role of WRC components during nervous system development), Cyfip1 and Cyfip2 perform different neuronal functions, as supported by distinct interactome profiles and spatiotemporal expression patterns in the brain36,126. Consistent with this, deletions in the chromosomal region (5q33.3–5q35.1) harboring CYFIP2 have been observed in patients with symptoms distinct from those with CYFIP1 mutations117. In addition, reduced CYFIP2 levels were reported in patients with schizophrenia and AD (Alzheimer’s disease)127,128. Intriguingly, de novo Arg87-residue CYFIP2 variants are associated with various facets of neurodevelopmental disorders, including EIEE (early infantile epileptic encephalopathy)129–131. Structural studies have demonstrated that Arg87 CYFIP2 variants likely disrupt hydrogen bonding between CYFIP2 and WAVE1 or Nap1, leading to structural instability of the WRC and dysregulation of Rac1-mediated WRC activity132. Intriguingly, Cyfip2+/R87C knock-in mice recapitulate a variety of neurological phenotypes that resemble symptoms of patients with West syndrome133. Because West syndrome and Ohtahara syndrome are categorized as subtypes of EIEE134—both of which are known to involve presynaptic defects135—it will be interesting to examine how the reported functions of CYFIP2 are linked to their pathogenesis mechanisms. Other CYFIP2 missense variants, notably including presumably pathogenic variants at the Asp724 residue, exhibit variable clinical phenotypes130. Moreover, mice with a Cyfip2 deficiency display visual impairments that are frequently observed in individuals with intellectual disability136. Determining how these phenotypes are caused by Cyfip2 dysfunction will require future studies that integrate the various cellular and clinical phenotypes observed in Cyfip2 KO and/or missense variants.
Dysfunctions of WAVE are linked to neurodevelopmental and neurodegenerative disorders
Exome sequencing and whole-genome sequencing have also identified a number of de novo truncating or missense variants of WAVE1 in patients with various neurodevelopmental symptoms137–140. Certain WAVE1 variants were predicted to disrupt the WCA domain11. However, systematic analyses to determine whether the reported WAVE1 variants are pathogenic in the regulation of actin polymerization processes in the nervous system are currently lacking. WAVE2 is also suggested to be involved in PD (Parkinson’s disease)141. WAVE2 interacts with LRRK2 (leucine-rich repeat kinase 2), a key culprit involved in the pathogenesis of PD141. LRRK2 phosphorylates WAVE2, stabilizes its levels and modulates the dynamics of WAVE2-mediated phagocytic activity of macrophage cells141. Several subsequent reports appear to support the association of WAVE2 with PD142,143, although the mechanistic basis of WAVE2 action in the pathogenesis of PD is still unclear.
Association of Abi3 with Alzheimer’s disease
Rare coding variants of ABI3 were initially shown to be linked to AD144, a relationship that appears to find consistent support in follow-up studies145,146. Moreover, deletion of Abi3 exacerbates various pathophysiological features in a mouse model of AD147. ABI3 was also recently proposed as an early biomarker for AD148. Overall, a series of these studies have strongly implicated ABI3 in AD pathogenesis, likely through its involvement in microglial motility and/or phagocytosis, in relation to microglial migration into amyloid plaques149,150. Whether ABI3 is also involved in other neurodegenerative disorders remains to be determined.
Implication of NAP1/NCKAP1 in neurodevelopmental disorders
Several heterozygous de novo and ultrarare deleterious variants of NAP1 have been reported in individuals with various symptoms found in neurodevelopmental disorders151. In support of this observation, NAP1 loss of function induces defective neuronal differentiation and abnormal neuronal migration in mice73,151. Although these observations provide compelling evidence that Nap1 could be a contributing factor to neurodevelopmental disorders with an ASD core, it is unclear whether NAP1 variants indeed lead to dysregulation of the WRC. Intriguingly, NAP1 appears to exhibit cell-type–specific expression in the developing human brain151. Given the low correlation between mRNA and protein levels in the brain, future work is warranted to determine whether cell-type–specific expression patterns of Nap1 mRNA are recapitulated at the protein level.
Outlook and future avenues
The significance of the WRC in regulating various structural and functional aspects of nervous system development and synapse formation has been clearly recognized. However, to fully understand WRC mechanisms, researchers will need to address a number of questions. For example, how is the WRC activated and/or clustered in response to different patterns of extracellular signals in neurons? In particular, do interactions of the WRC with various synaptic adhesion proteins produce distinct trans-synaptic signals that converge on a downstream pathway? Does the WRC act similarly in both pre- and postsynaptic neurons? What determines the composition of the WRC in different types of neurons? Does the distinct composition of the presynaptic WRC relate to distinct modes of synaptic transmission by influencing release probability? Do other actin-regulatory pathways (e.g., formins) universally crosstalk with the WRC-mediated Arp2/3 pathway across diverse cell types in various brain areas? Answering these questions will enable researchers to develop various tools for controlling the localization and/or strength of WRC signaling activities, providing an unprecedented opportunity to elucidate the role of the WRC in mediating the specificity and diversity of neural circuit architectures. Given that the Arp2/3 complex is also expressed in nonneuronal cells152, it will be illuminating to investigate the nonneuronal roles of each WRC component, ideally using conditional KO mouse lines.
Acknowledgements
This study was supported by the National Creative Research Initiative Program (NRF-2022R1A3B1077206 to J.K.) and the National Research Foundation of Korea (NRF) grant funded by the Korea Government (MSIT) (2021R1C1C2010767 to K.A.H.).
Author contributions
J.K. conceptualized the review; K.A.H. created the figures, and J.K. edited the figures; J.K. wrote an initial version of the manuscript; and K.A.H. edited, revised, and reviewed the manuscript. Both authors have read and agreed to the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cingolani LA, Goda Y. Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nat. Rev. Neurosci. 2008;9:344–356. doi: 10.1038/nrn2373. [DOI] [PubMed] [Google Scholar]
- 2.Matus A. Actin-based plasticity in dendritic spines. Science. 2000;290:754–758. doi: 10.1126/science.290.5492.754. [DOI] [PubMed] [Google Scholar]
- 3.Matus A, Brinkhaus H, Wagner U. Actin dynamics in dendritic spines: a form of regulated plasticity at excitatory synapses. Hippocampus. 2000;10:555–560. doi: 10.1002/1098-1063(2000)10:5<555::AID-HIPO5>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 4.Yuste R, Bonhoeffer T. Genesis of dendritic spines: insights from ultrastructural and imaging studies. Nat. Rev. Neurosci. 2004;5:24–34. doi: 10.1038/nrn1300. [DOI] [PubMed] [Google Scholar]
- 5.Dillon C, Goda Y. The actin cytoskeleton: integrating form and function at the synapse. Annu. Rev. Neurosci. 2005;28:25–55. doi: 10.1146/annurev.neuro.28.061604.135757. [DOI] [PubMed] [Google Scholar]
- 6.Kennedy MB, Beale HC, Carlisle HJ, Washburn LR. Integration of biochemical signalling in spines. Nat. Rev. Neurosci. 2005;6:423–434. doi: 10.1038/nrn1685. [DOI] [PubMed] [Google Scholar]
- 7.Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev. Biophys. Biomol. Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 8.Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 9.Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- 10.Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rottner K, Stradal TEB, Chen B. WAVE regulatory complex. Curr. Biol. 2021;31:R512–R517. doi: 10.1016/j.cub.2021.01.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z, et al. Structure and control of the actin regulatory WAVE complex. Nature. 2010;468:533–538. doi: 10.1038/nature09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang S, et al. Arf GTPase activates the WAVE regulatory complex through a distinct binding site. Sci. Adv. 2022;8:eadd1412. doi: 10.1126/sciadv.add1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding B, et al. Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase. Nat. Commun. 2022;13:5444. doi: 10.1038/s41467-022-33174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oikawa T, et al. PtdIns(3,4,5)P3 binding is necessary for WAVE2-induced formation of lamellipodia. Nat. Cell Biol. 2004;6:420–426. doi: 10.1038/ncb1125. [DOI] [PubMed] [Google Scholar]
- 16.Yang Y, et al. Leep1 interacts with PIP3 and the Scar/WAVE complex to regulate cell migration and macropinocytosis. J. Cell Biol. 2021;220:e202010096. doi: 10.1083/jcb.202010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaks M, Giannone G, Rottner K. Actin dynamics in cell migration. Essays Biochem. 2019;63:483–495. doi: 10.1042/EBC20190015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc. Natl Acad. Sci. USA. 2001;98:8844–8849. doi: 10.1073/pnas.151231598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cory GO, Ridley AJ. Cell motility: braking WAVEs. Nature. 2002;418:732–733. doi: 10.1038/418732a. [DOI] [PubMed] [Google Scholar]
- 20.Schenck A, et al. CYFIP/Sra-1 controls neuronal connectivity in Drosophila and links the Rac1 GTPase pathway to the fragile X protein. Neuron. 2003;38:887–898. doi: 10.1016/S0896-6273(03)00354-4. [DOI] [PubMed] [Google Scholar]
- 21.Derivery E, et al. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev. Cell. 2009;17:712–723. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 22.Napoli I, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 23.Pathania M, et al. The autism and schizophrenia associated gene CYFIP1 is critical for the maintenance of dendritic complexity and the stabilization of mature spines. Transl. Psychiatry. 2014;4:e374. doi: 10.1038/tp.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abekhoukh S, et al. New insights into the regulatory function of CYFIP1 in the context of WAVE- and FMRP-containing complexes. Dis. Model Mech. 2017;10:463–474. doi: 10.1242/dmm.025809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Rubeis S, et al. CYFIP1 coordinates mRNA translation and cytoskeleton remodeling to ensure proper dendritic spine formation. Neuron. 2013;79:1169–1182. doi: 10.1016/j.neuron.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bozdagi O, et al. Haploinsufficiency of Cyfip1 produces fragile X-like phenotypes in mice. PLoS ONE. 2012;7:e42422. doi: 10.1371/journal.pone.0042422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han K, et al. Fragile X-like behaviors and abnormal cortical dendritic spines in cytoplasmic FMR1-interacting protein 2-mutant mice. Hum. Mol. Genet. 2015;24:1813–1823. doi: 10.1093/hmg/ddu595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davenport EC, et al. Autism and schizophrenia-associated CYFIP1 regulates the balance of synaptic excitation and inhibition. Cell Rep. 2019;26:2037–2051. doi: 10.1016/j.celrep.2019.01.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sahasrabudhe A, et al. Cyfip1 regulates SynGAP1 at hippocampal synapses. Front. Synaptic Neurosci. 2020;12:581714. doi: 10.3389/fnsyn.2020.581714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim NS, et al. CYFIP1 dosages exhibit divergent behavioral impact via diametric regulation of NMDA receptor complex translation in mouse models of psychiatric disorders. Biol. Psychiatry. 2022;92:815–826. doi: 10.1016/j.biopsych.2021.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsiao K, Harony-Nicolas H, Buxbaum JD, Bozdagi-Gunal O, Benson DL. Cyfip1 regulates presynaptic activity during development. J. Neurosci. 2016;36:1564–1576. doi: 10.1523/JNEUROSCI.0511-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim GH, et al. Altered presynaptic function and number of mitochondria in the medial prefrontal cortex of adult Cyfip2 heterozygous mice. Mol. Brain. 2020;13:123. doi: 10.1186/s13041-020-00668-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cioni JM, et al. Axon-axon interactions regulate topographic optic tract sorting via CYFIP2-dependent WAVE complex function. Neuron. 2018;97:1078–1093. doi: 10.1016/j.neuron.2018.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen B, et al. The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell. 2014;156:195–207. doi: 10.1016/j.cell.2013.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marsden KC, et al. A Cyfip2-dependent excitatory interneuron pathway establishes the innate startle threshold. Cell Rep. 2018;23:878–887. doi: 10.1016/j.celrep.2018.03.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Kang HR, Han K. Differential cell-type-expression of CYFIP1 and CYFIP2 in the adult mouse hippocampus. Anim. Cells Syst. 2019;23:380–383. doi: 10.1080/19768354.2019.1696406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biembengut IV, Silva ILZ, Souza T, Shigunov P. Cytoplasmic FMR1 interacting protein (CYFIP) family members and their function in neural development and disorders. Mol. Biol. Rep. 2021;48:6131–6143. doi: 10.1007/s11033-021-06585-6. [DOI] [PubMed] [Google Scholar]
- 38.Dai Z, Pendergast AM. Abi-2, a novel SH3-containing protein interacts with the c-Abl tyrosine kinase and modulates c-Abl transforming activity. Genes Dev. 1995;9:2569–2582. doi: 10.1101/gad.9.21.2569. [DOI] [PubMed] [Google Scholar]
- 39.Shi Y, Alin K, Goff SP. Abl-interactor-1, a novel SH3 protein binding to the carboxy-terminal portion of the Abl protein, suppresses v-abl transforming activity. Genes Dev. 1995;9:2583–2597. doi: 10.1101/gad.9.21.2583. [DOI] [PubMed] [Google Scholar]
- 40.Ichigotani Y, Fujii K, Hamaguchi M, Matsuda S. In search of a function for the E3B1/Abi2/Argbp1/NESH family (Review) Int J. Mol. Med. 2002;9:591–595. [PubMed] [Google Scholar]
- 41.Innocenti M, et al. Phosphoinositide 3-kinase activates Rac by entering in a complex with Eps8, Abi1, and Sos-1. J. Cell Biol. 2003;160:17–23. doi: 10.1083/jcb.200206079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Innocenti M, et al. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat. Cell Biol. 2004;6:319–327. doi: 10.1038/ncb1105. [DOI] [PubMed] [Google Scholar]
- 43.Tani K, et al. Abl interactor 1 promotes tyrosine 296 phosphorylation of mammalian enabled (Mena) by c-Abl kinase. J. Biol. Chem. 2003;278:21685–21692. doi: 10.1074/jbc.M301447200. [DOI] [PubMed] [Google Scholar]
- 44.Leng Y, et al. Abelson-interactor-1 promotes WAVE2 membrane translocation and Abelson-mediated tyrosine phosphorylation required for WAVE2 activation. Proc. Natl Acad. Sci. USA. 2005;102:1098–1103. doi: 10.1073/pnas.0409120102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maruoka M, et al. Identification of B cell adaptor for PI3-kinase (BCAP) as an Abl interactor 1-regulated substrate of Abl kinases. FEBS Lett. 2005;579:2986–2990. doi: 10.1016/j.febslet.2005.04.052. [DOI] [PubMed] [Google Scholar]
- 46.Lin TY, Huang CH, Chou WG, Juang JL. Abi enhances Abl-mediated CDC2 phosphorylation and inactivation. J. Biomed. Sci. 2004;11:902–910. doi: 10.1007/BF02254375. [DOI] [PubMed] [Google Scholar]
- 47.Park E, Chi S, Park D. Activity-dependent modulation of the interaction between CaMKIIalpha and Abi1 and its involvement in spine maturation. J. Neurosci. 2012;32:13177–13188. doi: 10.1523/JNEUROSCI.2257-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Proepper C, et al. Abelson interacting protein 1 (Abi-1) is essential for dendrite morphogenesis and synapse formation. EMBO J. 2007;26:1397–1409. doi: 10.1038/sj.emboj.7601569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Proepper C, et al. Heterogeneous nuclear ribonucleoprotein k interacts with Abi-1 at postsynaptic sites and modulates dendritic spine morphology. PLoS ONE. 2011;6:e27045. doi: 10.1371/journal.pone.0027045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heinrich J, et al. The postsynaptic density protein Abelson interactor protein 1 interacts with the motor protein Kinesin family member 26B in hippocampal neurons. Neuroscience. 2012;221:86–95. doi: 10.1016/j.neuroscience.2012.06.055. [DOI] [PubMed] [Google Scholar]
- 51.Stavoe AK, et al. Synaptic vesicle clustering requires a distinct MIG-10/Lamellipodin isoform and ABI-1 downstream from Netrin. Genes Dev. 2012;26:2206–2221. doi: 10.1101/gad.193409.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rogers SL, Wiedemann U, Stuurman N, Vale RD. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 2003;162:1079–1088. doi: 10.1083/jcb.200303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McShea MA, et al. Abelson interactor-1 (ABI-1) interacts with MRL adaptor protein MIG-10 and is required in guided cell migrations and process outgrowth in C. elegans. Dev. Biol. 2013;373:1–13. doi: 10.1016/j.ydbio.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kunda P, Craig G, Dominguez V, Baum B. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr. Biol. 2003;13:1867–1875. doi: 10.1016/j.cub.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 55.Steffen A, et al. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 2004;23:749–759. doi: 10.1038/sj.emboj.7600084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Innocenti M, et al. Abi1 regulates the activity of N-WASP and WAVE in distinct actin-based processes. Nat. Cell Biol. 2005;7:969–976. doi: 10.1038/ncb1304. [DOI] [PubMed] [Google Scholar]
- 57.Pollitt AY, Insall RH. WASP and SCAR/WAVE proteins: the drivers of actin assembly. J. Cell Sci. 2009;122:2575–2578. doi: 10.1242/jcs.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hazai D, et al. Ultrastructural abnormalities in CA1 hippocampus caused by deletion of the actin regulator WAVE-1. PLoS ONE. 2013;8:e75248. doi: 10.1371/journal.pone.0075248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soderling SH, et al. Loss of WAVE-1 causes sensorimotor retardation and reduced learning and memory in mice. Proc. Natl Acad. Sci. USA. 2003;100:1723–1728. doi: 10.1073/pnas.0438033100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ceglia I, et al. WAVE1 in neurons expressing the D1 dopamine receptor regulates cellular and behavioral actions of cocaine. Proc. Natl Acad. Sci. USA. 2017;114:1395–1400. doi: 10.1073/pnas.1621185114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Westphal RS, Soderling SH, Alto NM, Langeberg LK, Scott JD. Scar/WAVE-1, a Wiskott-Aldrich syndrome protein, assembles an actin-associated multi-kinase scaffold. EMBO J. 2000;19:4589–4600. doi: 10.1093/emboj/19.17.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim Y, et al. Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature. 2006;442:814–817. doi: 10.1038/nature04976. [DOI] [PubMed] [Google Scholar]
- 63.Ceglia I, Kim Y, Nairn AC, Greengard P. Signaling pathways controlling the phosphorylation state of WAVE1, a regulator of actin polymerization. J. Neurochem. 2010;114:182–190. doi: 10.1111/j.1471-4159.2010.06743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Choi J, et al. Regulation of dendritic spine morphogenesis by insulin receptor substrate 53, a downstream effector of Rac1 and Cdc42 small GTPases. J. Neurosci. 2005;25:869–879. doi: 10.1523/JNEUROSCI.3212-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ito H, et al. Dysbindin-1, WAVE2 and Abi-1 form a complex that regulates dendritic spine formation. Mol. Psychiatry. 2010;15:976–986. doi: 10.1038/mp.2010.69. [DOI] [PubMed] [Google Scholar]
- 66.Stuart JR, Gonzalez FH, Kawai H, Yuan ZM. c-Abl interacts with the WAVE2 signaling complex to induce membrane ruffling and cell spreading. J. Biol. Chem. 2006;281:31290–31297. doi: 10.1016/S0021-9258(19)84041-3. [DOI] [PubMed] [Google Scholar]
- 67.Sossey-Alaoui K, Li X, Cowell JK. c-Abl-mediated phosphorylation of WAVE3 is required for lamellipodia formation and cell migration. J. Biol. Chem. 2007;282:26257–26265. doi: 10.1074/jbc.M701484200. [DOI] [PubMed] [Google Scholar]
- 68.Nozumi M, Nakagawa H, Miki H, Takenawa T, Miyamoto S. Differential localization of WAVE isoforms in filopodia and lamellipodia of the neuronal growth cone. J. Cell Sci. 2003;116:239–246. doi: 10.1242/jcs.00233. [DOI] [PubMed] [Google Scholar]
- 69.Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- 70.Bogdan S, Klambt C. Kette regulates actin dynamics and genetically interacts with Wave and Wasp. Development. 2003;130:4427–4437. doi: 10.1242/dev.00663. [DOI] [PubMed] [Google Scholar]
- 71.Ibarra N, Pollitt A, Insall RH. Regulation of actin assembly by SCAR/WAVE proteins. Biochem Soc. Trans. 2005;33:1243–1246. doi: 10.1042/BST0331243. [DOI] [PubMed] [Google Scholar]
- 72.Nakao S, Platek A, Hirano S, Takeichi M. Contact-dependent promotion of cell migration by the OL-protocadherin-Nap1 interaction. J. Cell Biol. 2008;182:395–410. doi: 10.1083/jcb.200802069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yokota Y, Ring C, Cheung R, Pevny L, Anton ES. Nap1-regulated neuronal cytoskeletal dynamics is essential for the final differentiation of neurons in cerebral cortex. Neuron. 2007;54:429–445. doi: 10.1016/j.neuron.2007.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiong Y, et al. Nck-associated protein 1 associates with HSP90 to drive metastasis in human non-small-cell lung cancer. J. Exp. Clin. Cancer Res. 2019;38:122. doi: 10.1186/s13046-019-1124-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vadde BVL. Tip growth: SPIRRIG and BRICK1 regulate root hair development by modulating the spatiotemporal dynamics of actin. Plant Cell. 2021;33:2106–2107. doi: 10.1093/plcell/koab129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Le J, Mallery EL, Zhang C, Brankle S, Szymanski DB. Arabidopsis BRICK1/HSPC300 is an essential WAVE-complex subunit that selectively stabilizes the Arp2/3 activator SCAR2. Curr. Biol. 2006;16:895–901. doi: 10.1016/j.cub.2006.03.061. [DOI] [PubMed] [Google Scholar]
- 77.Djakovic S, Dyachok J, Burke M, Frank MJ, Smith LG. BRICK1/HSPC300 functions with SCAR and the ARP2/3 complex to regulate epidermal cell shape in Arabidopsis. Development. 2006;133:1091–1100. doi: 10.1242/dev.02280. [DOI] [PubMed] [Google Scholar]
- 78.Gautreau A, et al. Purification and architecture of the ubiquitous Wave complex. Proc. Natl Acad. Sci. USA. 2004;101:4379–4383. doi: 10.1073/pnas.0400628101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qurashi A, et al. HSPC300 and its role in neuronal connectivity. Neural Dev. 2007;2:18. doi: 10.1186/1749-8104-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yuste R. Dendritic spines and distributed circuits. Neuron. 2011;71:772–781. doi: 10.1016/j.neuron.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Spence EF, Soderling SH. Actin out: regulation of the synaptic cytoskeleton. J. Biol. Chem. 2015;290:28613–28622. doi: 10.1074/jbc.R115.655118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tolias KF, Duman JG, Um K. Control of synapse development and plasticity by Rho GTPase regulatory proteins. Prog. Neurobiol. 2011;94:133–148. doi: 10.1016/j.pneurobio.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hedrick NG, Yasuda R. Regulation of Rho GTPase proteins during spine structural plasticity for the control of local dendritic plasticity. Curr. Opin. Neurobiol. 2017;45:193–201. doi: 10.1016/j.conb.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 84.Penzes P, Rafalovich I. Regulation of the actin cytoskeleton in dendritic spines. Adv. Exp. Med. Biol. 2012;970:81–95. doi: 10.1007/978-3-7091-0932-8_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Spence EF, Kanak DJ, Carlson BR, Soderling SH. The Arp2/3 complex is essential for distinct stages of spine synapse maturation, including synapse unsilencing. J. Neurosci. 2016;36:9696–9709. doi: 10.1523/JNEUROSCI.0876-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sturner T, et al. Transient localization of the Arp2/3 complex initiates neuronal dendrite branching in vivo. Development. 2019;146:dev113397. doi: 10.1242/dev.171397. [DOI] [PubMed] [Google Scholar]
- 87.Mendoza MC. Phosphoregulation of the WAVE regulatory complex and signal integration. Semin Cell Dev. Biol. 2013;24:272–279. doi: 10.1016/j.semcdb.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hume PJ, Humphreys D, Koronakis V. WAVE regulatory complex activation. Methods Enzymol. 2014;540:363–379. doi: 10.1016/B978-0-12-397924-7.00020-0. [DOI] [PubMed] [Google Scholar]
- 89.Ueda Y, Hayashi Y. PIP(3) regulates spinule formation in dendritic spines during structural long-term potentiation. J. Neurosci. 2013;33:11040–11047. doi: 10.1523/JNEUROSCI.3122-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Basu S, Lamprecht R. The role of actin cytoskeleton in dendritic spines in the maintenance of long-term memory. Front. Mol. Neurosci. 2018;11:143. doi: 10.3389/fnmol.2018.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Strasser GA, Rahim NA, VanderWaal KE, Gertler FB, Lanier LM. Arp2/3 is a negative regulator of growth cone translocation. Neuron. 2004;43:81–94. doi: 10.1016/j.neuron.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 92.San Miguel-Ruiz JE, Letourneau PC. The role of Arp2/3 in growth cone actin dynamics and guidance is substrate dependent. J. Neurosci. 2014;34:5895–5908. doi: 10.1523/JNEUROSCI.0672-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Norris AD, Dyer JO, Lundquist EA. The Arp2/3 complex, UNC-115/abLIM, and UNC-34/Enabled regulate axon guidance and growth cone filopodia formation in Caenorhabditis elegans. Neural Dev. 2009;4:38. doi: 10.1186/1749-8104-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shakir MA, et al. The Arp2/3 activators WAVE and WASP have distinct genetic interactions with Rac GTPases in Caenorhabditis elegans axon guidance. Genetics. 2008;179:1957–1971. doi: 10.1534/genetics.108.088963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Demarco RS, Lundquist EA. RACK-1 acts with Rac GTPase signaling and UNC-115/abLIM in Caenorhabditis elegans axon pathfinding and cell migration. PLoS Genet. 2010;6:e1001215. doi: 10.1371/journal.pgen.1001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alan JK, Struckhoff EC, Lundquist EA. Multiple cytoskeletal pathways and PI3K signaling mediate CDC-42-induced neuronal protrusion in C. elegans. Small GTPases. 2013;4:208–220. doi: 10.4161/sgtp.26602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wallace AG, Raduwan H, Carlet J, Soto MC. The RhoGAP HUM-7/Myo9 integrates signals to modulate RHO-1/RhoA during embryonic morphogenesis in Caenorhabditis elegans. Development. 2018;145:dev168724. doi: 10.1242/dev.168724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mohamed AM, Boudreau JR, Yu FP, Liu J, Chin-Sang ID. The Caenorhabditis elegans Eph receptor activates NCK and N-WASP, and inhibits Ena/VASP to regulate growth cone dynamics during axon guidance. PLoS Genet. 2012;8:e1002513. doi: 10.1371/journal.pgen.1002513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chaudhari K, Gorla M, Chang C, Kania A, Bashaw GJ. Robo recruitment of the Wave regulatory complex plays an essential and conserved role in midline repulsion. eLife. 2021;10:e64474. doi: 10.7554/eLife.64474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee NK, et al. Neogenin recruitment of the WAVE regulatory complex maintains adherens junction stability and tension. Nat. Commun. 2016;7:11082. doi: 10.1038/ncomms11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Murthy VN, De Camilli P. Cell biology of the presynaptic terminal. Annu. Rev. Neurosci. 2003;26:701–728. doi: 10.1146/annurev.neuro.26.041002.131445. [DOI] [PubMed] [Google Scholar]
- 102.Nelson JC, Stavoe AK, Colon-Ramos DA. The actin cytoskeleton in presynaptic assembly. Cell Adhes. Migr. 2013;7:379–387. doi: 10.4161/cam.24803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rust MB, Maritzen T. Relevance of presynaptic actin dynamics for synapse function and mouse behavior. Exp. Cell Res. 2015;335:165–171. doi: 10.1016/j.yexcr.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 104.Stavoe AK, Colon-Ramos DA. Netrin instructs synaptic vesicle clustering through Rac GTPase, MIG-10, and the actin cytoskeleton. J. Cell Biol. 2012;197:75–88. doi: 10.1083/jcb.201110127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chia PH, Chen B, Li P, Rosen MK, Shen K. Local F-actin network links synapse formation and axon branching. Cell. 2014;156:208–220. doi: 10.1016/j.cell.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schenck A, et al. WAVE/SCAR, a multifunctional complex coordinating different aspects of neuronal connectivity. Dev. Biol. 2004;274:260–270. doi: 10.1016/j.ydbio.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 107.Hummel T, Leifker K, Klambt C. The Drosophila HEM-2/NAP1 homolog KETTE controls axonal pathfinding and cytoskeletal organization. Genes Dev. 2000;14:863–873. doi: 10.1101/gad.14.7.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zallen JA, et al. SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. J. Cell Biol. 2002;156:689–701. doi: 10.1083/jcb.200109057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hanley JG. Actin-dependent mechanisms in AMPA receptor trafficking. Front. Cell Neurosci. 2014;8:381. doi: 10.3389/fncel.2014.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xing G, et al. Neurexin-Neuroligin 1 regulates synaptic morphology and functions via the WAVE regulatory complex in Drosophila neuromuscular junction. eLife. 2018;7:e30457. doi: 10.7554/eLife.30457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yamashita H, Ueda K, Kioka N. WAVE2 forms a complex with PKA and is involved in PKA enhancement of membrane protrusions. J. Biol. Chem. 2011;286:3907–3914. doi: 10.1074/jbc.M110.145409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liang M, et al. Oligomerized liprin-alpha promotes phase separation of ELKS for compartmentalization of presynaptic active zone proteins. Cell Rep. 2021;34:108901. doi: 10.1016/j.celrep.2021.108901. [DOI] [PubMed] [Google Scholar]
- 113.Lim D, Kim D, Um JW, Ko J. Reassessing synaptic adhesion pathways. Trends Neurosci. 2022;45:517–528. doi: 10.1016/j.tins.2022.04.004. [DOI] [PubMed] [Google Scholar]
- 114.Sando R, Ho ML, Liu X, Südhof TC. Engineered synaptic tools reveal localized cAMP signaling in synapse assembly. J. Cell Biol. 2022;221:e202109111. doi: 10.1083/jcb.202109111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim HY, Um JW, Ko J. Proper synaptic adhesion signaling in the control of neural circuit architecture and brain function. Prog. Neurobiol. 2021;200:101983. doi: 10.1016/j.pneurobio.2020.101983. [DOI] [PubMed] [Google Scholar]
- 116.Südhof TC. The cell biology of synapse formation. J. Cell Biol. 2021;220:e202103052. doi: 10.1083/jcb.202103052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang Y, Lee Y, Han K. Neuronal function and dysfunction of CYFIP2: from actin dynamics to early infantile epileptic encephalopathy. BMB Rep. 2019;52:304–311. doi: 10.5483/BMBRep.2019.52.5.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cox DM, Butler MG. Distal partial trisomy 15q26 and partial monosomy 16p13.3 in a 36-year-old male with clinical features of both chromosomal abnormalities. Cytogenet Genome Res. 2015;145:29–34. doi: 10.1159/000381293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Clifton NE, Thomas KL, Wilkinson LS, Hall J, Trent S. FMRP and CYFIP1 at the synapse and their role in psychiatric vulnerability. Complex Psychiatry. 2020;6:5–19. doi: 10.1159/000506858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hosak L. New findings in the genetics of schizophrenia. World J. Psychiatry. 2013;3:57–61. doi: 10.5498/wjp.v3.i3.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bachmann SO, et al. Behavioral training rescues motor deficits in Cyfip1 haploinsufficiency mouse model of autism spectrum disorders. Transl. Psychiatry. 2019;9:29. doi: 10.1038/s41398-018-0338-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Oguro-Ando A, et al. Increased CYFIP1 dosage alters cellular and dendritic morphology and dysregulates mTOR. Mol. Psychiatry. 2015;20:1069–1078. doi: 10.1038/mp.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dominguez-Iturza N, et al. The autism- and schizophrenia-associated protein CYFIP1 regulates bilateral brain connectivity and behaviour. Nat. Commun. 2019;10:3454. doi: 10.1038/s41467-019-11203-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fricano-Kugler C, et al. CYFIP1 overexpression increases fear response in mice but does not affect social or repetitive behavioral phenotypes. Mol. Autism. 2019;10:25. doi: 10.1186/s13229-019-0278-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Silva AI, et al. Cyfip1 haploinsufficient rats show white matter changes, myelin thinning, abnormal oligodendrocytes and behavioural inflexibility. Nat. Commun. 2019;10:3455. doi: 10.1038/s41467-019-11119-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bonaccorso CM, et al. Fragile X mental retardation protein (FMRP) interacting proteins exhibit different expression patterns during development. Int. J. Dev. Neurosci. 2015;42:15–23. doi: 10.1016/j.ijdevneu.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 127.Focking M, et al. Proteomic and genomic evidence implicates the postsynaptic density in schizophrenia. Mol. Psychiatry. 2015;20:424–432. doi: 10.1038/mp.2014.63. [DOI] [PubMed] [Google Scholar]
- 128.Tiwari SS, et al. Alzheimer-related decrease in CYFIP2 links amyloid production to tau hyperphosphorylation and memory loss. Brain. 2016;139:2751–2765. doi: 10.1093/brain/aww205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zweier M, et al. Spatially clustering de novo variants in CYFIP2, encoding the cytoplasmic FMRP interacting protein 2, cause intellectual disability and seizures. Eur. J. Hum. Genet. 2019;27:747–759. doi: 10.1038/s41431-018-0331-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Begemann A, et al. New insights into the clinical and molecular spectrum of the novel CYFIP2-related neurodevelopmental disorder and impairment of the WRC-mediated actin dynamics. Genet. Med. 2021;23:543–554. doi: 10.1038/s41436-020-01011-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nakashima M, et al. De novo hotspot variants in CYFIP2 cause early-onset epileptic encephalopathy. Ann. Neurol. 2018;83:794–806. doi: 10.1002/ana.25208. [DOI] [PubMed] [Google Scholar]
- 132.Schaks, M., Reinke, M., Witke, W. & Rottner, K. Molecular dissection of neurodevelopmental disorder-causing mutations in CYFIP2. Cells9, 1355 (2020). [DOI] [PMC free article] [PubMed]
- 133.Kang M, et al. CYFIP2 p.Arg87Cys causes neurological defects and degradation of CYFIP2. Ann. Neurol. 2023;93:155–163. doi: 10.1002/ana.26535. [DOI] [PubMed] [Google Scholar]
- 134.Pavone P, et al. West syndrome: a comprehensive review. Neurol. Sci. 2020;41:3547–3562. doi: 10.1007/s10072-020-04600-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Patzke C, et al. Analysis of conditional heterozygous STXBP1 mutations in human neurons. J. Clin. Invest. 2015;125:3560–3571. doi: 10.1172/JCI78612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chaya T, et al. Deficiency of the neurodevelopmental disorder-associated gene Cyfip2 alters the retinal ganglion cell properties and visual acuity. Hum. Mol. Genet. 2022;31:535–547. doi: 10.1093/hmg/ddab268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Srivastava S, et al. Expansion of the genotypic and phenotypic spectrum of WASF1-related neurodevelopmental disorder. Brain Sci. 2021;11:931. doi: 10.3390/brainsci11070931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhao A, et al. Trio exome sequencing identified a novel de novo WASF1 missense variant leading to recurrent site substitution in a Chinese patient with developmental delay, microcephaly, and early-onset seizures: a mutational hotspot p.Trp161 and literature review. Clin. Chim. Acta. 2021;523:10–18. doi: 10.1016/j.cca.2021.08.030. [DOI] [PubMed] [Google Scholar]
- 139.Ito Y, et al. De novo truncating mutations in WASF1 cause intellectual disability with seizures. Am. J. Hum. Genet. 2018;103:144–153. doi: 10.1016/j.ajhg.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shimojima Yamamoto K, et al. Recurrent de novo pathogenic variant of WASF1 in a Japanese patient with neurodevelopmental disorder with absent language and variable seizures. Hum. Genome Var. 2021;8:43. doi: 10.1038/s41439-021-00176-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kim KS, et al. Regulation of myeloid cell phagocytosis by LRRK2 via WAVE2 complex stabilization is altered in Parkinson’s disease. Proc. Natl Acad. Sci. USA. 2018;115:E5164–E5173. doi: 10.1073/pnas.1718946115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dwyer Z, et al. Leucine-rich repeat kinase-2 (LRRK2) modulates microglial phenotype and dopaminergic neurodegeneration. Neurobiol. Aging. 2020;91:45–55. doi: 10.1016/j.neurobiolaging.2020.02.017. [DOI] [PubMed] [Google Scholar]
- 143.Fenner BM, Fenner ME, Prowse N, Hayley SP. LRRK2 and WAVE2 regulate microglial-transition through distinct morphological phenotypes to induce neurotoxicity in a novel two-hit in vitro model of neurodegeneration. J. Cell Physiol. 2022;237:1013–1032. doi: 10.1002/jcp.30588. [DOI] [PubMed] [Google Scholar]
- 144.Sims R, et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat. Genet. 2017;49:1373–1384. doi: 10.1038/ng.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Conway OJ, et al. ABI3 and PLCG2 missense variants as risk factors for neurodegenerative diseases in Caucasians and African Americans. Mol. Neurodegener. 2018;13:53. doi: 10.1186/s13024-018-0289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Castillo E, et al. Comparative profiling of cortical gene expression in Alzheimer’s disease patients and mouse models demonstrates a link between amyloidosis and neuroinflammation. Sci. Rep. 2017;7:17762. doi: 10.1038/s41598-017-17999-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Karahan H, et al. Deletion of Abi3 gene locus exacerbates neuropathological features of Alzheimer’s disease in a mouse model of Abeta amyloidosis. Sci. Adv. 2021;7:eabe3954. doi: 10.1126/sciadv.abe3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cao M, et al. ABI3 is a novel early biomarker of Alzheimer’s disease. J. Alzheimers Dis. 2022;87:335–344. doi: 10.3233/JAD-215635. [DOI] [PubMed] [Google Scholar]
- 149.Turner AK, Shaw BC, Simpson JF, Estus S. Identification and quantitation of novel ABI3 isoforms relative to Alzheimer’s disease genetics and neuropathology. Genes. 2022;13:1607. doi: 10.3390/genes13091607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Satoh JI, et al. Microglia express ABI3 in the brains of Alzheimer’s disease and Nasu-Hakola disease. Intractable Rare Dis. Res. 2017;6:262–268. doi: 10.5582/irdr.2017.01073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Guo H, et al. NCKAP1 disruptive variants lead to a neurodevelopmental disorder with core features of autism. Am. J. Hum. Genet. 2020;107:963–976. doi: 10.1016/j.ajhg.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Murk K, Blanco Suarez EM, Cockbill LM, Banks P, Hanley JG. The antagonistic modulation of Arp2/3 activity by N-WASP, WAVE2 and PICK1 defines dynamic changes in astrocyte morphology. J. Cell Sci. 2013;126:3873–3883. doi: 10.1242/jcs.125146. [DOI] [PMC free article] [PubMed] [Google Scholar]