Abstract
There is a growing interest in 3D printing to fabricate culture substrates; however, the surface properties of the scaffold remain pertinent to elicit targeted and expected cell responses. Traditional 2D polystyrene (PS) culture systems typically require surface functionalization (oxidation) to facilitate and encourage cell adhesion. Determining the surface properties which enhance protein adhesion from media and cellular extracellular matrix (ECM) production remains the first step to translating 2D PS systems to a 3D culture surface. Here we show that the presence of carbonyl groups to PS surfaces correlated well with successful adhesion of ECM proteins and sustaining ECM production of deposited human mesenchymal stem cells, if the surface has a water contact angle between 50° and 55°. Translation of these findings to custom-fabricated 3D PS scaffolds reveals carbonyl groups continued to enhance spreading and growth in 3D culture. Cumulatively, these data present a method for 3D printing PS and the design considerations required for understanding cell-material interactions
Keywords: polystyrene, surface chemical modification, plasma treatment, protein adhesion, 3D printing
INTRODUCTION
Polystyrene (PS) has served as the fundamental substrate for adherent cell culture for more than 50 years.1 However, the lack of surface functionality which cells recognize and spread on has dictated the need to modify PS surfaces to facilitate anchorage and develop 3D models to more accurately mimic real biological systems.2 Typical tissue culture polystyrene (TCPS) exhibits high surface oxidation, providing a functionalized surface for cells to adhere and proliferate.3 A number of plasma chemistries have been used to functionalize PS surfaces, including air,4 ammonia,5 oxygen,6 and multiple organic compounds.7 Surface activation with plasma has been leveraged to incorporate biofunctional groups and increase surface wettability, a mechanism thought to facilitate cell and protein adhesion.8 Linking together surface chemistry, material properties, and biological response provides a viable path to translating 2D in vitro culture surfaces to 3D scaffolds. The fundamental basis of these protein–surface interactions lies in engineering the surface chemistry to direct protein adsorption.9 Building in vitro surfaces to mimic tissue niches through protein adhesion, cell patterning, and guiding differentiation provides a unique opportunity to sequester cells and generate functional tissues.10,11
The widespread use of PS as a 2D culture substrate, and demonstrated improvements 3D culture provides, suggests transitioning PS to a 3D culture platform. However, the applications of 3D-printed PS as a cell-contacting growth surface have been limited,12 likely due to the difficulty in liquefying PS without thermally degrading the polymer.13 Previous approaches tend to rely on fabricating large objects by sintering smaller objects (e.g., a packed-bead bioreactor)14 or microfluidic approaches.15-17 Transforming the standard PS culture surface to a complex bioreactor scaffold would aid in expanding culture populations for clinical and research applications.18 However, transitioning PS from 2D to 3D cell culture requires surface modification to facilitate protein and cell adhesion, where understanding surface chemical properties and cell response could lead to both universal and targeted culture constructs.
The combination of 3D printing and surface chemical modification provides the potential for a highly tunable process to select culture environments for select cell types. Directed surface modification has been shown to influence human mesenchymal stem cell (hMSC) differentiation, but further exploration would fully develop the link between surface chemical cues and cell phenotype.19 Variable phenotype expression of hMSCs has been established through surface modification, where carbonyl (carboxyl) containing surfaces showed evidence for preference to the chondrogenic phenotype and high ECM protein association with the surfaces (particularly fibronectin).20 In addition, fibronectin and integrin have been seen to associate preferentially with carbonyl and amine containing surfaces.21 Surface wettability influences the adhesive force between proteins and the substrate, resulting in conformational changes at the interface22 which can be sensed by adhered cells and influences spreading and proliferation.23 Building surface chemistry and properties to selectively elicit and pattern protein adhesion provides a viable path to engineering cell–material interactions.24-26 Amine groups have been used to direct hMSC osteogenic differentiation, attributed to differential activation of focal adhesion kinases by preferentially associating ECM proteins.27
Significant progress in developing 2D surfaces targeting growth and differentiation has been achieved, but the biomimicry of 3D scaffolds and ability to control the physiological environment through fluid dynamics,28 mechanical stresses,29 oxygen content,30 cellular distribution,31 and overall geometry32 warrant a transition to 3D culture. Transforming PS from standard flat cell culture dishes to a 3D scaffold could provide the means to better replicate the in vivo environment by improving cell–cell interactions and enabling higher culture densities.33 3D printing provides a unique avenue to rapidly manufacture and test models with biologically relevant structures; a platform that could establish clinical niches in vitro for highly replicative screening of novel pharmaceuticals, cancer therapies, and disease models.34-38 The main objective presented in this work explores engineering surface chemistry for 3D-printed PS scaffolds to target hMSC expansion. First, we sought to establish the surface characteristics generated by our plasma system. Next, we determined the link between surface properties and protein adhesion. Then, we investigated hMSC spreading, growth, and ECM expression on the surfaces and ultimately show the concept translates to a 3D model. We demonstrate that carbonyls on moderately hydrophilic surfaces help to promote hMSC proliferation and phenotype maintenance, likely through associated surface ECM proteins expressed by the cells and deposited from media. These strategies present a viable 3D PS scaffold capable of supporting hMSC expansion with wide applicability.
MATERIALS AND METHODS
2D substrate preparation
Silicon wafers of 15 mm2 were wiped clean with ethanol (EtOH), sonicated for 5 min in acetone, then methanol, and dried with nitrogen. Fifty microliters of 10% (w/w) PS (>50,000 molecular weight atactic flakes, Polysciences, Inc., Warrington, PA)/toluene was spun cast at 314.2 rad/s (3000 RPM) for 40 s with a spin coater. These samples were used for surface characterization. For cell and protein analyses, cover glass slides were wiped clean with EtOH and coated 10% PS solution following the same spin casting method: 22 mm × 40 mm and 25 × 40 mm used 150 μL of solution and 24 mm × 50 mm used 250 μL of solution. Protein analyses used only the smallest glass slides.
Surface activation
A low-temperature plasma activated samples by creating a dielectric barrier discharge (DBD) in a 4 mm ID glass capillary tube attached to a 20 kHz AC high-voltage source [Figure 1(A)]. Ultrapure helium was used as a discharge gas with acetonitrile vapor or oxygen added at a concentration of 2.5% (v/v). Ignition frequency was visually tuned with the lowest power selected achieving uniform and consistent plasma ignition (0.14–0.34 kV). Samples were translated under the plasma jet with servo motors. For 3D-printed objects, each side was exposed to plasma treatment.
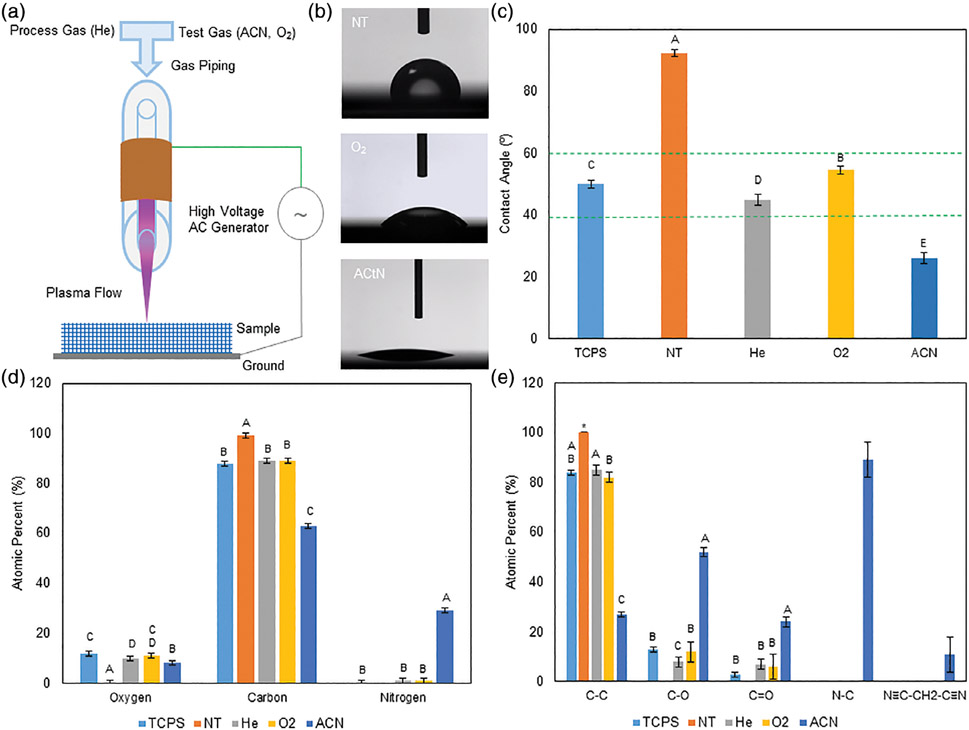
FIGURE 1.
A: Diagram of the DBD system with plasma flowing over the sample. B: Example contact angle images of the NT-, O2-, and ACN-treated surfaces. C: Quantified contact angles. Activating the surfaces with He, O2, and ACN reduced contact angles. Groups with different letters indicate statistical difference (p < 0.05), data are shown as mean ± standard deviation, n = 5. D: Survey XPS data shows surface oxygen and nitrogen presence on the DBD-treated surfaces and TCPS and nitrogen on the DBD-treated surfaces. Nitrogen presence in the DBD surfaces is likely a by-product of plasma treatment in ambient atmosphere. The greatest nitrogen presence was seen on the ACN-treated surfaces. Oxygen and nitrogen presence was accompanied by a reduction in carbon. E: High-resolution DBD-treated surfaces showed single bonded oxygen and carbonyl groups. TCPS showed both single bonded oxygen and carbonyl groups. NT surfaces remained purely carbon. Nitrogen presence on the ACN-treated surfaces were primarily bound to carbon as imine groups, with 13% of nitrogen on the surface was present as malononitrile. XPS data are shown as mean ± standard deviation; six scans per sample; single sample for NT, ACN, and TCPS; two samples for He and O2. Groups with different letters indicate statistical difference; asterisk indicates identical values (Tukey multiple comparison, p < 0.05).
Goniometer wettability measurements (water contact angle)
Two microliters of deionized water was dispensed onto leveled samples with an inverted syringe pump. A video camera recorded the pendant droplet touching and stabilizing on the surface. The baseline water–surface interface was determined and Young-Laplace (Sessile) contact angle measured on at least three drops.
X-ray photoelectron spectroscopy (XPS)
Quantitative XPS measurements determined elemental compositions with a survey scan of 0–1100 eV, a pass energy of 160 eV, and a resolution of 1.0 eV. The binding stoichiometry was determined from high-resolution scans with a pass energy of 40 eV and a resolution of 0.1 eV. A Gaussian–Lorentzian distribution was used for peak fitting, with a full width at half maximum constraint of 1.7 eV on six spectra per sample.
Total protein determination
Samples were covered in fetal bovine serum (FBS) for 1 h at 37°C, then rinsed with phosphate-buffered saline (PBS), placed on ice, and covered with cold radioimmunoprecipiration assay (RIPA) buffer for 5 min. Isolated protein was centrifuged at 14,000g for 15 min at 4°C. The supernatant was tested on three samples in technical triplicate with a Pierce BCA Protein Assay following standard microplate procedures.
Polyacrylamide gel electrophoresis (PAGE) protein separation and Coomassie staining
Protein was isolated following the methods explained above with Halt Protease Inhibitor (ThermoFisher, Waltham, MA) added to RIPA, including a 1:100 FBS:RIPA control. Mini-Protean TGX (Bio-Rad, Hercules, CA) gels were loaded with Laemmli buffer and samples, as described.39 Gels were run for 45 min at 120 V in 1× Tris-Glycine buffer and fixed in 5% acetic acid, 45% deionized distilled water (ddH2O, >18.2 MΩ resistivity), and 50% methanol for 30 min. Gels were washed three times for 5 min in ddH2O with protein bands visualized and imaged with Coomassie blue.
Proteomics through mass spectrometry (MS)
Protein was isolated through electrophoresis, as described. Gel lanes were separated, cut into 10 equal pieces, digested with sequencing grade trypsin, and peptides extracted with acetonitrile-formic acid buffer.40 Liquid-chromatography (LC)-MS/MS was performed using a nano-LC system (Easy nLC1000) connected to Q Exactive mass spectrometer (ThermoFisher). Peptides were eluted at 300 nL/min using a series of linear gradients of acetonitrile in 0.1% formic acid. Data sets were searched against UniProt human database using MaxQuant software (version 1.5.5.1).41 Groups with peptide counts less than the total number of test groups (6), nonspecies, or obvious contaminants were eliminated.
Cell culture practices
hMSCs (RoosterBio, Frederick, MD) were thawed and expanded in Dulbecco’s Modified Eagle Medium with 10% FBS, MEM non-essential amino acids 0.1 mmol/L, and penicillin (100 U/mL)–streptomycin (100 μg/mL) (growth media). 90% confluent hMSCs were washed with pH 7.4 PBS and lifted with a 5 min incubation with 0.25% trypsin–EDTA and neutralized with growth media. Passage 2–4 hMSCs were used during experiments, with cells treated as passage 1 upon receipt. FIfty thousand hMSCs per 30 minute UV sterilized sample were used unless noted. Cell counting was accomplished using the Trypan Blue exclusion method with a hemocytometer.
Confocal imaging and image processing
Cells were lifted as described and resuspended in the media of interest (growth media) or RoosterBio High-Performance Basal Media (no serum media). hMSCs were seeded onto surfaces and grown overnight. Cytochalasin-D-treated hMSCs had their media replaced with growth media containing 10 μmol/L cytochalasin-D for 1 h prior to fixing.42-45 Samples were chemically fixed using 4% formaldehyde and 1% sucrose solution and permeabilized with a 300 mmol/L sucrose, 50 mmol/L sodium chloride, 6 mmol/L magnesium chloride, 20 mmol/L HEPES, and 0.5% Triton-X-100 solution. Actin was stained with 2.5% AlexaFluor 594 Phalloidin in PBS, counterstained diamidino-2-phenylindole (DAPI) or VectaSheild (Vector Labs, Burlingame, CA). A confocal microscope (SP5 X, Leica Microsystems, Buffalo Grove, IL) captured Z-stack images, which were processed with a MATLAB (MathWorks, Natick, MA) program. The program read in image files from the specified directory, and separated the image stacks into individual image locations (i.e., groups of individual z-stacks). Small debris and back-ground fluorescence were eliminated with size and intensity exclusion thresholds. For each image stack, the average spread area was calculated by dividing the total area covered by phalloidin (red channel) by the number of detected nuclei (blue channel). Running this process for each stack was used to generate statistics (Supporting Information, Appendix 1).
Flow cytometry
hMSCs were seeded as described and grown for 6 days, with media replaced on the third day. Cells were lifted with StemPro Accutase (ThermoFisher), diluted with an equal volume of growth media, and resuspended in 1% FBS in PBS (flow buffer). To stain, a PE-Negative Cocktail (CD45, CD34, CD11b, CD19, and HLA-DR) and -Positive Cocktail (FITC CD90, PerCPCy 5.5 CD105, and APC CD73) from a Human MSC Analysis kit (BD Biosciences, Franklin Lakes, NJ) was used, following manufacturer’s recommendations. hMSC Isotype controls were run and UltraComp eBeads (ThermoFisher) single color controls were used for compensation. Data collection was performed on an FACS Canto II (BD Biosciences) and analysis was performed with Cytobank Community software (Cytobank Inc. Santa Clara, CA).
DNA quantification
Cells were dissociated from three samples with Trypsin and 0.25% EDTA and resuspended in PBS. DNA was isolated following the procedures for DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) and quantified in technical triplicate using a Quant-iT PicoGreen (ThermoFisher) kit, following the manufacture’s protocol. Fluorescence measurements were read on an M5 SpectraMax plate reader (Molecular Devices, San Jose, CA) with an excitation wavelength of 490 nm and emission read at 538 nm.
RT-PCR
RNA was isolated with a Trizol phase separation technique. Samples were covered with Trizol, gently mixed, and transferred to a microcentrifuge tube. Chloroform was added and vigorously mixed. Phase layers were separated by centrifugation at 12,000g for 15 min at 4°C, with the aqueous phase transferred. 70% EtOH was added to 35% of the final volume. Samples were transferred to a RNeasy mini column (Qiagen). Complete RNA isolation followed kit instructions. cDNA conversion was completed using a High Capacity cDNA Archive Kit (ThermoFisher) following manufacturers procedures. A ΔΔCT gene expression assay was completed, comparing genes of interest to GAPDH (endogenous control). A 7900HT RT-PCR system (Applied Biosystems, Foster City, CA) cycled samples: 2 min at 50°C, 10 min at 95°C, 40 cycles of 15 s at 95°C, and 1 min at 60°C.
3D scaffold fabrication
A 3D Bioplotter (EnvisionTEC GmbH, Gladbeck, Germany) fabricated scaffolds. A sugar–glass support material (EnvisionTEC GmbH) was heated to 150°C and extruded through a 0.3 mm ID needle tip at 70–80 kPa at 10–30 mm/s. The PS was melted at 155°C and extruded through a 0.4 mm ID needle tip at 900 kPa at 3–5 mm/s. The build surface was maintained at 60°C. The extrusion temperature of the PS is below the thermal decomposition temperature of the material (~163°C13) to avoid degradation of the material. At the completion of prints, the support was removed with ddH2O, scaffolds washed overnight in ddH2O, cleaned in 100% EtOH for 30 min, and air dried.
Scanning electron microscopy (SEM)
A FEI Quanta 200 FEG-ESEM (Hillsboro, OR) was used to image PS scaffolds under high vacuum at 5.0 keV. Samples were coated with Au–Pd prior to imaging to reduce charging artifacts.
Gel permeation chromatography (GPC)
The molecular weight of printed and stock PS was determined by GPC (Waters Alliance Separations Module e2695, Waters 2414 Refractive Index Detector, and Waters HSPgel columns in series (HR MB-L and HR 3.0 columns, 6.0 mm ID × 15 cm), Waters, Milford, MA). Three samples of each were dissolved in tetrahydrofuran with molecular weight determined against PS standards in at least technical duplicate.
Differential scanning calorimetry (DSC)
The glass transition temperature was determined by measuring the thermal energy change as a function of temperature change with a TA DSC Q100 (TA Instruments, New Castle, DE). Three ~10 mg samples of printed and stock PS were sealed in hermetic aluminum pans. Samples were rapidly equilibrated at 160°C and kept isothermal for 5 min. The temperature was ramped at 10°C per min to −20°C and back to 160°C and heat flow recorded.
Statistical analysis
Statistical significance was determined through a one-way two-sided ANOVA at the 95% confidence interval using the Games-Howell comparison method in Minitab 18 (Minitab, Inc. State College, PA) unless noted.
RESULTS
Surface characterization of 2D treated surfaces
In this work, we sought to develop a surface modification method which facilitates hMSC adhesion and growth on custom PS scaffolds. This work was performed on 2D surfaces to validate our method. Wettability analysis [Figure 1(B,C)] shows that treated surfaces experience a significant increase in wettability as compared to the no treatment (NT) control (92.3°). 2.5% oxygen (O2) and 2.5% acetonitrile (ACN) plasmas reduced contact angles to 54.6° and 26.4°, respectively, while TCPS surfaces maintained a native contact angle of 50.0°. Further surface characterization, accomplished through XPS, confirmed the NT surface as primarily carbon containing, with moderate oxygen and little nitrogen content observed in TCPS [Figure 1(D)]. Nitrogen incorporation in O2 and He surfaces were similar. Total oxygen incorporation was similar in TCPS-, 100% helium plasma (He)-, and O2 plasma-treated samples, followed by ACN. ACN-treated samples had much greater nitrogen incorporation. Specific bonding stoichiometry indicate that oxygen present in He- and O2-treated surfaces contained both single-bonded oxygen and carbonyl groups [Figure 1(E) and Supporting Information Table SI]. The carbonyl content of He- and O2-treated surfaces exceeded, though not statistically different, the quantities present in TCPS. ACN-treated surfaces contained the greatest quantity of single- and double-bonded oxygen, as well as potential surface amines groups and malononitrile.
Characterization of adhered surface protein
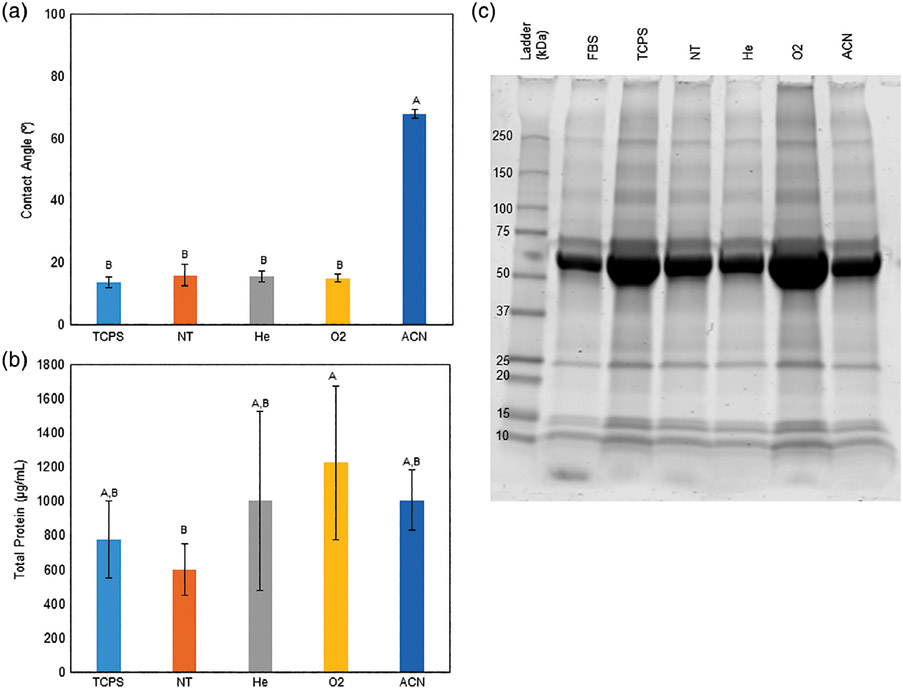
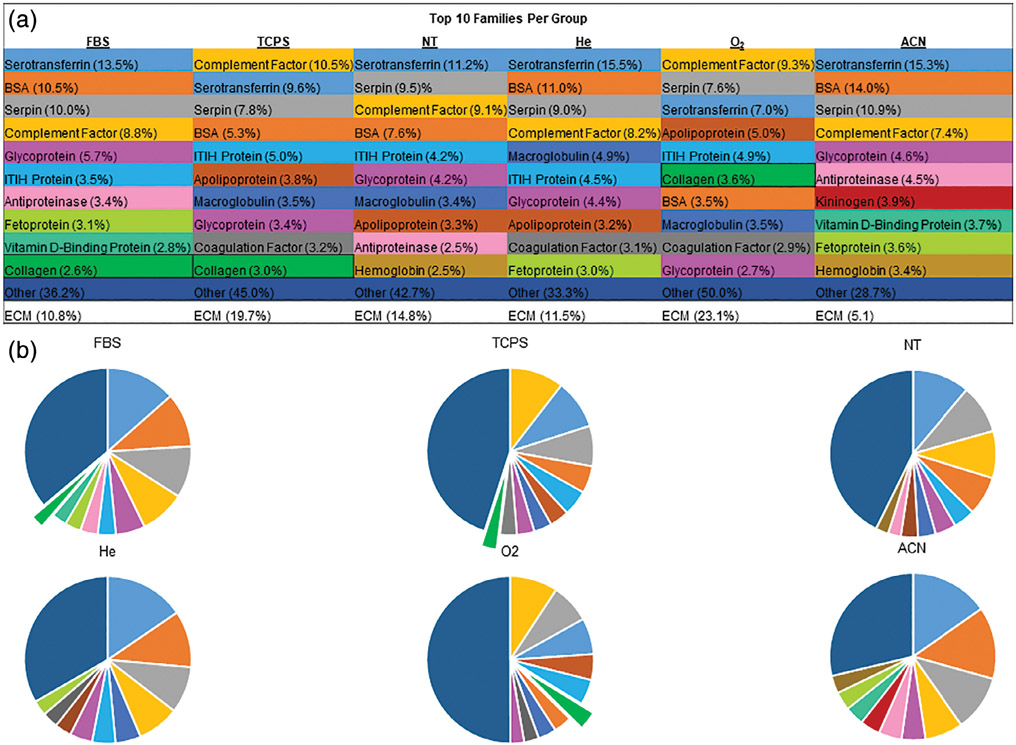
The contact angle decreased relative to initial values following serum exposure, and was statistically similar across all surfaces, except for ACN-treated surfaces [Figure 2(A)]. ACN-treated surfaces were observed to have an increase in contact angle. All contact angle changes were statistically different from initial values (two-sample t test, p < 0.001 for all test surfaces at the 95% confidence interval). A Pierce BCA analysis revealed O2-treated samples were the only group to contain a statistically greater quantity of adhered protein relative to the NT control, but no different from the other surfaces [Figure 2(B)]. Coomassie staining confirmed the adhered protein quantities similarities, where the darker bands of the O2- and TCPS-treated samples indicate greater protein presence [Figure 2(C)]. Additionally, darker high-molecular-weight bands appear present in these samples. To better quantify protein differences, mass spectrometry was performed on the PAGE gel separated proteins. The proteins have been grouped by family type and the top 10 protein families present ranked [Figure 3(A,B)]. Based on peptide counts (e.g., instances of matching protein fragments), O2-treated surfaces tend to associate better with (1) proteins generally, attracting more and a wider variety and (2) greater relative amounts of ECM proteins (23.1% of O2, 19.7% of TCPS, 14.8% of NT, 11.5% of He, 10.8% of FBS, and 5.1% of ACN). Specifically, collagen family proteins were only found to be a top 10 present protein by peptide count in the O2 (#6, 3.6%) surfaces and TCPS surfaces (#10, 3.0%). These both represent a greater fraction than in the diluted FBS control (#10, 2.6%).
FIGURE 2.
A: Quantified contact angles across the surfaces following serum exposure. Contact angles were reduced for all surfaces except on ACN-treated surfaces. This indicates increased surface wettability and surface energy following exposure to serum. Groups with different letters indicate statistical difference (p < 0.05), data are shown as mean ± standard deviation, n = 5 except TCPS n = 4, ACN n = 3. B: Total protein content following coating. O2 surfaces showed statistically greater protein adhesion than the NT surface, but not different from any other surface. Data are shown as mean ± standard deviation, n = 3. C: Coomassie stained gel. Visually, the O2 surface band appears darker than the NT surface across the whole lane. Similar bands are observed across all treatment samples and the source serum.
FIGURE 3.
A: Table showing the top 10 proteins present by peptide count, grouped by identified protein family. ECM proteins are called out separately and represent percent of total peptides counted. Collagen was only present in the top 10 for FBS, TCPS, and O2, indicating a possible correlation between carbonyls on surfaces and ECM protein association. Conversely, ACN surfaces had the lowest percentage of ECM proteins present. B: Graphical representation of the percentages of counted peptides. Colors in figures match those in the table for each group.
Cellular interaction with protein-coated surfaces
Surfaces containing the highest quantities of carbonyl groups tended to facilitate the greatest spreading (Figure 4). Protein containing media and coating the surfaces with media increased cell spreading, while the use of a serum-free media and an actin inhibitor (cytochalasin-D) reduced cell spreading. NT surfaces tended to prevent cellular spreading, where cells remained rounded. Quantified hMSC phenotype (Table I) shows that the cells generally expressed (>95%) of the positive markers and lacked expression of negative markers (83%–90%). O2-treated samples exhibited the highest amount of phenotypically expected hMSCs. To contrast this, He-treated samples, with lower wettability and similar oxygen content, exhibited higher ratios of hMSCs expressing the differentiation markers in the Negative Cocktail Mix. The ACN-treated surfaces showed decreases in the positive and negative markers.
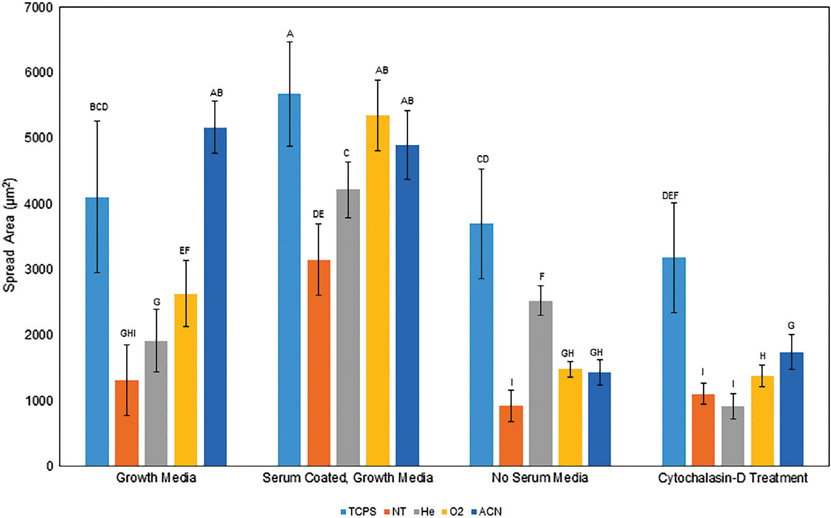
FIGURE 4.
A: Average spread area of hMSCs on treated surfaces in various media. The data indicates that proteins tend to associate with the carbonyl containing surfaces, facilitating cell spreading through an actin-related mechanism. Surfaces precoated with serum generally had greater spread areas, where media lacking protein showed less spread cells. The addition of cytochalasin-D, an actin inhibitor, to the media prior to imaging generally reduced spread area of the hMSCs. Note: ANOVA was performed over the whole dataset, groups with different letters indicate statistical difference (p < 0.05), and data are shown as mean ± standard deviation, n = 16 except NT cytochalasin-D treatment, n = 15.
TABLE I.
hMSC Phenotype Assessment Through Flow Cytometry
| Condition | % CD90 (+) | % CD73 (+) | % CD105 (+) | % Negative (−) |
|---|---|---|---|---|
| TCPS | 94.8 | 95.5 | 96.1 | 83.3 |
| NT | 97.9 | 97.3 | 95.3 | 84.0 |
| He | 99.4 | 98.4 | 98.3 | 74.3 |
| O2 | 94.1 | 97.2 | 95.6 | 89.9 |
| ACN | 74.4 | 73.2 | 80.5 | 58.5 |
Carbonyl containing surfaces tended to have greater percentages of hMSCs displaying expected surface markers, where ACN-treated surfaces tended to have reduce the percentage of expected phenotype hMSCs.
Cellular response to 2D surfaces
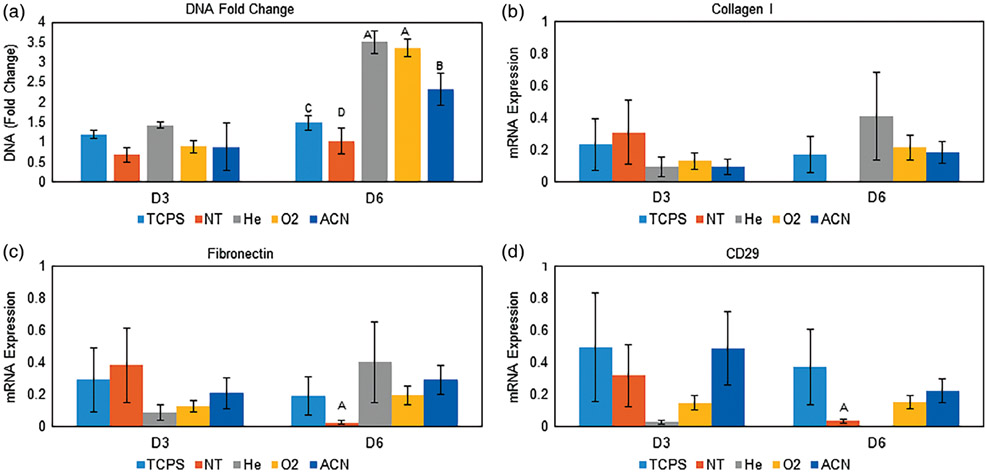
DNA quantification showed significantly higher growth (DNA fold change) after 6 days [Figure 5(A)]. The He- and O2-treated surfaces showed the greatest increase in DNA, followed by treatment with ACN. PCR showed similar collagen I [Figure 5(B)] and fibronectin [Figure 5(C)] mRNA expression of hMSCs attached to the plasma-treated samples and TCPS over the course of 6 days. Expression levels were significantly greater than on the NT surface (collagen I, expression on the NT surface was inconclusive). As well, CD29 [Figure 5(D)] expression was significantly greater from hMSCs on the treated surfaces relative to the NT surfaces (except for He, where expression was inconclusive).
FIGURE 5.
A: Normalized DNA content to each D0 value. The He and O2 surfaces showed the greatest DNA content fold change, followed by ACN, TCPS, and NT surfaces. This indicates surface activation successfully facilitated hMSC proliferation. Data are shown as mean ± standard deviation, n = 3. B: mRNA expression of collagen I. All surfaces showed detectable expression except for the NT surface. C: mRNA expression of fibronectin. All surfaces showed greater expression than the NT surface on day 6. D: mRNA expression of CD29. All surfaces showed detectable expression except for the He surface. Statistically greater expression was seen on TCPS, O2, and ACN surfaces compared to the NT surface at day 6. For ΔΔCT mRNA expression, depressed expression on the NT surface at day 6 may indicate reduced ability to modify the surrounding environment and express ECM proteins, likely related to the underlying protein content directed by the surface chemistry. Note: groups with different letters indicate statistical difference (p < 0.05) for ANOVA was performed on each time point. For B-D, data are in technical triplicate and displayed as mean ± standard deviation.
Translation of results to 3D model
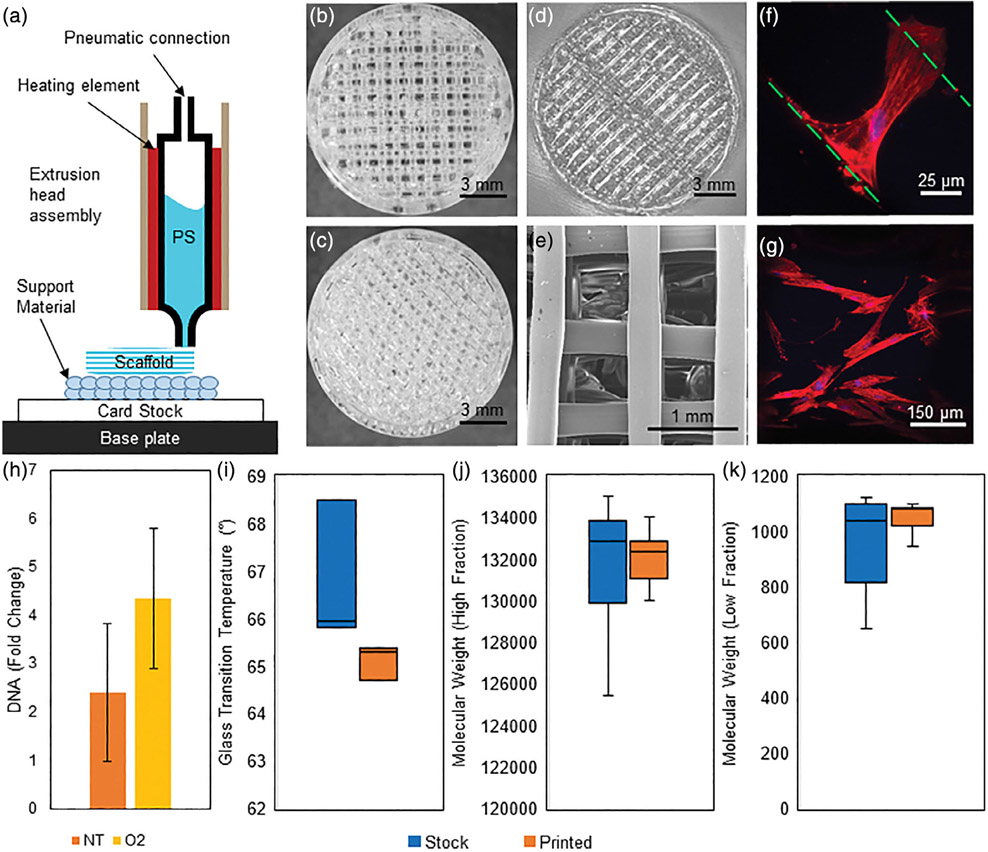
To explore the applicability of this method on 3D-printed PS, we developed a method for extrusion deposition modeling of PS without thermally degrading the polymer [Figure 6(A)]. Scaffolds with 90° [Figure 6(B)], 60° [Figure 6(C)], and 85.5° [Figure 6(D)] rotations between individual layers were printed. SEM imaging of the 90° matching demonstrates open internal pore structure [Figure 6(E)]. When the 85.5° surface was treated with the O2 plasma, hMSCs adhered and spread within the scaffolds. At 3 days, hMSCs were observed to bridge fibers [Figure 6(F)] and remained spread on fibers with expected morphology after 6 days [Figure 6(G)]. hMSC seeded at 5000 cells/cm2 continued to expand in static culture up to 7 days [two-sample t test p value between NT and O2 scaffolds = 0.011, Figure 6(H)]. No differences in glass transition temperature [two-sample t test, p = 0.212, Figure 6(I)], high or low fraction molecular weights [two-sample t test, p = 0.825, 0.210, Figure 6(J,K)], or molecular weight area fractions (two-sample t test, p = 0.905; Supporting Information Table SII) were observed between the printed and stock material, indicating our method did not thermally degrade the material during extrusion.
FIGURE 6.
A: A schematic of the 3D printing method. Card stock is placed onto the temperature controlled base plate with the support material printed on top of this. The extrusion head is then replaced automatically and PS is extruded onto the support material. 3D-printed PS scaffolds with layer offset angles of (B) 90°, (C) 60°, and (D) 85.5°. E: An SEM image of the 90° inner geometry of the PS scaffold. F: A single hMSC is seen bridging two PS fibers in a 85.5° scaffold at 3 days. The green lines indicate the edges of the PS fibers, DNA is stained blue with DAPI, and F-actin is stained red with phalloidin. G: hMSCs on a 85.5° scaffold display expected morphology on a O2-treated scaffold at 6 days. The image is a projected z-stack over the curved surface of a single PS fiber. DNA is stained blue with DAPI and F-actin is stained red with phalloidin. H: DNA fold change from day 0 after 7 days of growth. hMSCs were initially seeded at 5,000 cells/cm2. Trends indicate that the 85.5° scaffold O2 scaffold supports hMSC growth throughout the scaffold and warrants further development (p = 0.011, data shown as mean ± standard deviation n = 3). To characterize the PS before and after printing, matching batches of source (stock) and extruded (printed) material were evaluated. I: Differential scanning calorimetry was used to determine the glass transition temperature of the material. No difference (p = 0.212) was observed between samples (n = 3). (J) High molecular weight fraction and (K) low molecular weight fraction of stock and printed polystyrene as determined by gel permeation chromatography (n = 3). Samples were run against a standard curve to determine molecular weight. No difference was found between high (p = 0.825) and low (p = 0.210) fractions of the stock or printed material.
DISCUSSION
Our first goal was to decouple the 3D geometry and surface chemistry by understanding the wetting and chemical properties generated by the DBD plasma treatment. The He test group was included as a process control with O2 and ACN surfaces intended to increase wettability and incorporate carbonyl and amine groups, respectively. NT samples set a baseline for the DBD groups and TCPS provided a comparison to standard culture surfaces. Targeting contact angles within the 40°−60° range have been shown to best promote cell adhesion [Figure 1 (B,C)], independent of surface chemistry.8 TCPS, He, and O2 surfaces achieved this target range and ACN and NT surfaces were statistically outside this range. As well, the differences in contact angle between the He- and O2-treated surfaces may be attributable different, subtle (10s of nm) structures on each23,46 or differences in surface charge.47,48
Previous work suggests that carbonyl incorporation on surfaces tends to correlate positively with cell growth7 and surface wettability influences protein conformation.49 The moderate oxygen and low nitrogen content in TCPS is consistent with the manufacturing process: a corona discharge plasma in an oxygen rich environment.50 O2 and He chemical similarity [Figure 1(D,E)] likely due to the experimental DBD system depositing reactive species from the ambient atmosphere. However, significant malononitrile deposition was observed on ACN substrates, likely a by-product of plasma polymerization of the monomer as well as degradation of the vaporized acetonitrile in the energetic plasma discharge. Malononitrile is thought to inhibit metabolic rate of mammalian cells and form thiocyanate in tissues,51 causing detrimental effects on seeded cells. The developed custom DBD system promotes surface changes, controlled by changing the input gasses, and effectively modifies the PS targets with reactive ionized species.
Having established surface chemical differences between the groups, we next investigated how these surface properties impacted protein adhesion. It is well established that proteins will tend to adhere on surfaces, regardless of underlying composition through nonspecific adsorption.52 However, protein conformational differences, types of bound proteins, and relative concentrations may differ due to surface chemistry.53 The interaction between the surface chemistry, wettability, and proteins likely dictates preferences toward individual protein adhesion types and conformation.54 By engineering surfaces to attract or repel proteins of interest, targeted cell expansion and niche generation may be possible. The first step to realizing differences in our treated surfaces included repeating wettability measurements on serum-coated surfaces [Figure 2(A)]. We found the surface wettability significantly increased on all surfaces, except the ACN surfaces. These data indicate that the adhered proteins may undergo a conformational change depending on the surface chemistry, as seen previously.22,53 Total protein quantification [Figure 2(B)], PAGE gel separation [Figure 2(C)], and MS analysis (Figure 3) support differences in total protein content and composition. As with all protein interacting surfaces, the Vroman Effect is certainly in play.55 We sought to investigate how hMSCs will begin to interact with the surface. It has been previously observed that at an hour, ~10% of seeded cells will begin to attach to TCPS,56 beginning to spread on the surfaces.57 As well, this time is near the peak (areal mass) of attached protein, forming a uniform layer, and cells may sense this attached protein as a single uniform structure.58 The protein layer is fully formed, though it may change over time, with higher molecular weight material generally replacing lower weight material.59 Particular proteins of interest contain the peptide sequence arginine–glycine–aspartic acid (RGD), help to promote cell adhesion through integrin proteins.60 ECM proteins, such as collagen, contain the RGD sequence and have been shown to influence proliferation and osteoblastic differentiation of hMSCs through cellular pathway activation and mechanical substrate deformation.61-64 Determining the surface proteins informs ultimate hMSC response. Other than collagen, the remaining proteins mentioned in the table are not known to have definitively beneficial cell adhesion affects. Kininogen and apolipoprotein have been observed to inhibit cell adhesion processes.65,66 ITIH proteins work to stabilize hyaluronic acid.67 Glycoproteins play a role in cell-cell interactions, but not ECM interactions directly.68,69 Serum albumin works to inhibit endothelial apoptosis.70 The serpin superfamily inhibit enzymes, such as those in the coagulation cascade.71 Macroglobulin and antiproteinases act in the foreign body response.72,73 Serotransferrin, vitamin D-binding protein, hemoglobin, and fetoprotein are all linked to carrying metabolites.74-77 Collagen stands apart from this group, as the only ECM component protein. Improved protein adhesion on the carbonyl containing surfaces could be due to their electronegativity relative to single-bonded oxygen, encouraging hydrogen bond formation associating with proteins.78 Conversely, ACN-treated surfaces appear to resist protein adhesion, specifically ECM proteins, which may be linked to the extremely high surface wettability79 or surface chemistry and likely acidic surface environment.80
Protein adhesion differences were compared against hMSC spread area, a measure of surface attractiveness, binding, and phenotype. Quantified cell spreading (Figure 4) suggests the carbonyl groups on surfaces with contact angles between 50° and 55° act with proteins either in the media or during a coating to facilitate attachment, potentially through an amenable conformational preference or increased presence of ECM proteins.49 Surfaces lacking these traits may lack the native biological residues necessary for optimal protein adhesion and cell anchorage, attachment, and spreading. All the treated surfaces should allow hMSCs to initially attach and grow, and that the carbonyls on surfaces are likely coordinating with proteins to encourage the cell attachment and spreading. For example, more ECM proteins were observed on O2 surfaces and these had higher spreading as compared to He and NT surfaces. Both the use of a serum free media and treating the cells with an actin inhibitor (cytochalasin-D) reduced cell spread area as compared to both growth media conditions. This correlation indicates cell spreading is influenced by an actin-mediated mechanism with the underlying surface proteins, guided by the underlying surface properties. Though the cells likely do not sense the chemistry directly, the coordination of surface chemistry and protein adhesion certainly facilitates cell adhesion, patterning, and growth.81,82 The underlying surface properties can be used to influence hMSC adhesion characteristics by modulating underlying adhered protein.
To ensure the hMSCs maintained their stemness, flow cytometry was performed with a set of standard positive and negative multipotency markers (Table I). Moderately hydrophilic surfaces (TCPS, O2) supported a high fraction of phenotypically expected cells, suggesting some interaction between the underlying surface chemistry, adhered protein, and differentiation potential. The adhered hMSCs on these surfaces were found to align with previously set guidelines of surface marker expression.83 In contrast, the ACN surface showed decreased positive and increased lacking of negative markers, indicating a reduction in multipotency.
The next objective was to evaluate hMSC growth and ECM protein expression differences of hMSCs grown on the test surfaces. Cumulative results suggest that adding oxygen to the plasma tends to improve hMSC–surface interactions, where treatment with ACN tends to diminish these (Figure 5). The hMSCs expanded significantly more on the treated surfaces versus the untreated [Figure 5(A)] and continued to express collagen I, fibronectin, and CD29 [Figure 5(B-D)]. Collagen I and fibronectin are two representative ECM matrix proteins.84 CD29 (Integrin β1), in part, helps to regulate hMSC ECM adhesion, migration, and focal adhesion kinase pathway activation.85,86 The diminishing presence of ECM production suggests the cells may lack the ability to modify the surrounding environment on the NT surface.87 This may be caused by the underlying protein layer directed by the treated PS surface properties. The carbonyl containing surfaces tended to allow continued matrix production over the time studied.
The final goal was to evaluate the translatability of our 2D findings to a 3D model, working toward bringing PS from a 2D surface to 3D scaffold with directed functionalization. The 2D work details the importance of the O2 treatment and was tested in 3D as a proof-of-concept The method [Figure 6 (A)] is capable of producing a wide range of geometries and heights [Figure 6(B-D)] with an open internal architecture [Figure 6(E)] without thermally degrading the polymer [Figure 6(I-K) and Supporting Information Table SII]. hMSCs were observed to spread and interact with the scaffold [Figure 6(F,G)], utilizing both the polymer filaments and filling in the void spaces between the fibers. The trends indicate that the plasma treatment improved growth [Figure 6(H)]. O2-treated scaffolds demonstrate a method to transform PS to a 3D culture surface which hMSCs attach and grow with future work seeking to investigate this further. Extrusion 3D-printed PS scaffolds have previously isolated and expanded lymphoma cancer cells, with the scaffold isolating the cells of interest36 and PS punched scaffold have expanded hMSCs in a perfusion bioreactor.12 Continued customization of PS scaffolds would define surface chemistry, geometry, and flow properties for individual cell populations. Further work seeks to understand how hMSCs sense these 3D culture platform, and the influences on directed differentiation, leading to highly specialized PS bioreactors.
CONCLUSION
Here we propose a set of criteria to transform PS from a 2D to 3D culture surface. Incorporating carbonyls on surfaces with water contact angles between 50° and 55° tended to facilitate ECM protein and hMSC adhesion, produce greater numbers of cells, and an increased proportion of phenotypically expected cells. The data suggest that protein–cell coordination at the surface was influenced by the underlying surface properties, where the high electronegativity of the carbonyl bonds and moderate wettability likely improves hMSC response to adhered proteins. Extremely wetting surface detracted from these findings. Transitioning from 2D to 3D, namely a surface containing ~6% carbonyl groups on an O2-treated PS base with water contact angle between 50° and 55°, would serve well as a growth substrate in 3D. Initial demonstration shown here presents a viable path for 3D PS bioreactors. Transitioning PS from a 2D to 3D culture surface would allow for culture systems utilizing flow, complex geometry, and densified culture, better replicating the in vivo environment and potential to generate clinically relevant population sizes for transplantation and research. Particularly, understanding how surface chemistry influences protein and cell adhesion could be used for building complex engineered environments to elicit targeted responses from cells.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Maryland Stem Cell Research Fund (MSCRF) under Award #2015-MSCRFI1717 (JPF), Maryland Industrial Partnerships grant (JPF), MSCRF/TEDCO Postdoctoral Fellowship under Award #2017-MSCRF-9320 (JL), NIST fellowship program 2014-NIST-MSE-01 (MJL), and by and the National Institute of Biomedical Imaging and Bioengineering/National Institutes of Health (NIBIB/NIH) Center for Engineering Complex Tissues P41 EB023833 (JPF). Proteomics analysis was supported by Award Number 1U54HD090257 from NIH, District of Columbia Intellectual and Developmental Disabilities Research Center Award Program. Certain commercial equipment, instruments, or materials are identified to adequately specify the experimental procedure. Such identification does not imply recommendation or endorsement by NIST. Any opinions, findings, conclusions, or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of NIST. The authors thank MTECH ASPIRE program and Madelyn Golding, Dr. Jesse Placone, Dr. Marco Santoro, Dr. Hannah Baker, Dr. Aswini Panigrahi, and John Daristotle for their assistance. The authors thank Amy Beaven, the UMD Imaging Core, the Functional Macromolecular Laboratory, and the UMD NanoCenter for the use of their equipment.
Contract grant sponsor: Maryland Stem Cell Research Fund; contract grant number: 2045-MSCRFI1717
Contract grant sponsor: Material Measurement Laboratory; contract grant number: 2014-NIST-MSE-01
Contract grant sponsor: National Institutes of Health; contract grant number: 1U54HD090257
Contract grant sponsor: National Institute of Biomedical Imaging and Bioengineering; contract grant number: P41 EB023833
Contract grant sponsor: NIST fellowship; contract grant number: 2014-NIST-MSE-01
Contract grant sponsor: MSCRF/TEDCO Postdoctoral Fellowship; contract grant number: #2017-MSCRF-9320
Footnotes
CONFLICT OF INTEREST
JPF is a founding member of, and has a financial interest in, 3DBioWorks, which focuses on developing bioreactor technology.
REFERENCES
- 1.Rubin H. Altering bacteriological plastic petri dishes for tissue culture use. Public Health Rep 1966;81:843–844. [PMC free article] [PubMed] [Google Scholar]
- 2.Lerman MJ, Lembong J, Muramoto S, Gillen G, Fisher JP. The evolution of polystyrene as a cell culture material. Tissue Eng Part B Rev 2018;24:359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Kooten TG, Spijker HT, Busscher HJ. Plasma-treated polystyrene surfaces: Model surfaces for studying cell-biomaterial interactions. Biomaterials 2004;25:1735–1747. [DOI] [PubMed] [Google Scholar]
- 4.Lee J-H, Kwon J-S, Kim Y-H, Choi E-H, Kim K-M, Kim K-N. The effects of enhancing the surface energy of a polystyrene plate by air atmospheric pressure plasma jet on early attachment of fibroblast under moving incubation. Thin Solid Films 2013;547:99–105. [Google Scholar]
- 5.Kleinhans C, Barz J, Wurster S, Willig M, Oehr C, Müller M, Walles H, Hirth T, Kluger PJ. Ammonia plasma treatment of polystyrene surfaces enhances proliferation of primary human mesenchymal stem cells and human endothelial cells. Biotechnol J 2013;8:327–337. [DOI] [PubMed] [Google Scholar]
- 6.Beaulieu I, Geissler M, Mauzeroll J. Oxygen plasma treatment of polystyrene and zeonor: Substrates for adhesion of patterned cells. Langmuir 2009;25:7169–7176. [DOI] [PubMed] [Google Scholar]
- 7.Ertel SI, Chilkoti A, Horbetti TA, Ratner BD. Endothelial cell growth on oxygen-containing films deposited by radio-frequency plasmas: The role of surface carbonyl groups. J Biomater Sci Polym Ed 1992;3:163–183. [DOI] [PubMed] [Google Scholar]
- 8.Arima Y, Iwata H. Effect of wettability and surface functional groups on protein adsorption and cell adhesion using well-defined mixed self-assembled monolayers. Biomaterials 2007;28:3074–3082. [DOI] [PubMed] [Google Scholar]
- 9.Horbett TA. The role of adsorbed proteins in animal cell adhesion. Colloids Surf B Biointerfaces 1994;2:225–240. [Google Scholar]
- 10.Soares da Costa D, Márquez-Posadas M d C, Araujo AR, Yang Y, Merino S, Groth T, Reis RL, Pashkuleva I. Adhesion of adipose-derived mesenchymal stem cells to glycosaminoglycan surfaces with different protein patterns. ACS Appl Mater Interfaces 2015;7:10034–10043. [DOI] [PubMed] [Google Scholar]
- 11.Shukla A, Slater JH, Culver JC, Dickinson ME, West JL. Biomimetic surface patterning promotes mesenchymal stem cell differentiation. ACS Appl Mater Interfaces 2016;8:21883–21892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar A, Lau W, Starly B. Human mesenchymal stem cells expansion on three-dimensional (3D) printed poly-styrene (PS) scaffolds in a perfusion bioreactor. Proc CIRP 2017;65:115–120. [Google Scholar]
- 13.Beyler CL, Hirschler MM. Thermal Decomposition of Polymers. SFPE Handbook of Fire Protection Engineering, 2nd ed. Quincy, MA: National Fire Projection Association; 1995. pp. 110–131. [Google Scholar]
- 14.Osiecki MJ, Michl TD, Kul Babur B, Kabiri M, Atkinson K, Lott WB, Griesser HJ, Doran MR. Packed bed bioreactor for the isolation and expansion of placental-derived mesenchymal stromal cells. PLoS One 2015;10:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young EWK, Berthier E, Guckenberger DJ, Sackmann E, Lamers C, Meyvantsson I, Huttenlocher A, Beebe DJ. Rapid prototyping of arrayed microfluidic systems in polystyrene for cell-based assays. Anal Chem 2011;83:1408–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Fan Y, Kodzius R, Foulds IG. Fabrication of polystyrene microfluidic devices using a pulsed CO2 laser system. Microsyst Technol 2012;18:373–379. [Google Scholar]
- 17.Nargang TM, Brockmann L, Nikolov PM, Schild D, Helmer D, Keller N, Sachsenheimer K, Wilhelm E, Pires L, Dirschka M, Kolew A, Schneider M, Worgull M, Giselbrecht S, Neumann C, Rapp BE. Liquid polystyrene: A room-temperature photocurable soft lithography compatible pour-and-cure-type polystyrene. Lab Chip 2014;14:2698–2708. [DOI] [PubMed] [Google Scholar]
- 18.dos Santos FF, Andrade PZ, da Silva CL, Cabral JMS. Bioreactor design for clinical-grade expansion of stem cells. Biotechnol J 2013;8:644–654. [DOI] [PubMed] [Google Scholar]
- 19.Curran JM, Chen R, Hunt JA. The guidance of human mesenchymal stem cell differentiation in vitro by controlled modifications to the cell substrate. Biomaterials 2006;27:4783–4793. [DOI] [PubMed] [Google Scholar]
- 20.Curran JM, Chen R, Hunt JA. Controlling the phenotype and function of mesenchymal stem cells in vitro by adhesion to silane-modified clean glass surfaces. Biomaterials 2005;26:7057–7067. [DOI] [PubMed] [Google Scholar]
- 21.Keselowsky BG, Collard DM, García AJ. Surface chemistry modulates fibronectin conformation and directs integrin binding and specificity to control cell adhesion. J Biomed Mater Res Part A 2003;66A: 247–259. [DOI] [PubMed] [Google Scholar]
- 22.Xu LC, Siedlecki CA. Effects of surface wettability and contact time on protein adhesion to biomaterial surfaces. Biomaterials 2007;28: 3273–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dowling DP, Miller IS, Ardhaoui M, Gallagher WM. Effect of surface wettability and topography on the adhesion of osteosarcoma cells on plasma-modified polystyrene. J Biomater Appl 2011;26:327–347. [DOI] [PubMed] [Google Scholar]
- 24.McFarland CD, Thomas CH, DeFilippis C, Steele JG, Healy KE. Protein adsorption and cell attachment to patterned surfaces. J Biomed Mater Res 2000;49:200–210. [DOI] [PubMed] [Google Scholar]
- 25.Ostuni E, Chen CS, Ingber DE, Whitesides GM. Selective deposition of proteins and cells in arrays of microwells. Langmuir 2001;17:2828–2834. [Google Scholar]
- 26.Richter B, Hahn V, Bertels S, Claus TK, Wegener M, Delaittre G, Barner-Kowollik C, Bastmeyer M. Guiding cell attachment in 3D microscaffolds selectively functionalized with two distinct adhesion proteins. Adv Mater 2017;29:1–6. [DOI] [PubMed] [Google Scholar]
- 27.Griffin MF, Ibrahim A, Seifalian AM, Butler PEM, Kalaskar DM, Ferretti P. Chemical group-dependent plasma polymerisation preferentially directs adipose stem cell differentiation towards osteogenic or chondrogenic lineages. Acta Biomater 2017;50:450–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartnikowski M, Klein TJ, Melchels FPW, Woodruff MA. Effects of scaffold architecture on mechanical characteristics and osteoblast response to static and perfusion bioreactor cultures. Biotechnol Bioeng 2014;111:1440–1451. [DOI] [PubMed] [Google Scholar]
- 29.Guo T, Yu L, Lim CG, Goodley AS, Xiao X, Placone JK, Ferlin KM, Nguyen B-NB, Hsieh AH, Fisher JP. Effect of dynamic culture and periodic compression on human mesenchymal stem cell proliferation and Chondrogenesis. Ann Biomed Eng 2016;44:2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bauwens C, Yin T, Dang S, Peerani R, Zandstra PW. Development of a perfusion fed bioreactor for embryonic stem cell-derived cardiomyocyte generation: Oxygen-mediated enhancement of cardiomyocyte output. Biotechnol Bioeng 2005;90:452–461. [DOI] [PubMed] [Google Scholar]
- 31.Yeatts AB, Geibel EM, Fears FF, Fisher JP. Human mesenchymal stem cell position within scaffolds influences cell fate during dynamic culture. Biotechnol Bioeng 2012;109:2381–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen B-NB, Ko H, Moriarty RA, Etheridge JM, Fisher JP. Dynamic bioreactor culture of high volume engineered bone tissue. Tissue Eng Part A 2016;22:263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rumpler M, Woesz A, Dunlop JWC, van Dongen JT, Fratzl P. The effect of geometry on three-dimensional tissue growth. J R Soc Interface 2008;5:1173–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferlin KM, Prendergast ME, Miller ML, Nguyen B-NB, Kaplan DS, Fisher JP. Development of a dynamic stem cell culture platform for mesenchymal stem cell adhesion and evaluation. Mol Pharm 2014;11:2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gelinsky M, Bernhardt A, Milan F. Bioreactors in tissue engineering: Advances in stem cell culture and three-dimensional tissue constructs. Eng Life Sci 2015;15:670–677. [Google Scholar]
- 36.Caicedo-Carvajal CE, Liu Q, Remache Y, Goy A, Suh KS. Cancer tissue engineering: A novel 3D polystyrene scaffold for in vitro isolation and amplification of lymphoma cancer cells from heterogeneous cell mixtures. J Tissue Eng 2011;2011:362326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fong ELS, Lamhamedi-Cherradi S-E, Burdett E, Ramamoorthy V, Lazar AJ, Kasper FK, Farach-Carson MC, Vishwamitra D, Demicco EG, Menegaz BA, Amin HM, Mikos AG, Ludwig JA. Modeling Ewing sarcoma tumors in vitro with 3D scaffolds. Proc Natl Acad Sci 2013;110:6500–6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D’Avanzo C, Chen H, Hooli B, Asselin C, Muffat J, Klee JB, Zhang C, Wainger BJ, Peitz M, Kovacs DM, Woolf CJ, Wagner SL, Tanzi RE, Kim DY. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature 2014;515:274–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bracaglia LG, Messina M, Winston S, Kuo C-Y, Lerman M, Fisher JPJP. 3D printed pericardium hydrogels to promote wound healing in vascular applications. Biomacromolecules 2017;18:3802–3811. [DOI] [PubMed] [Google Scholar]
- 40.Kuo C-Y, Guo T, Cabrera-Luque J, Arumugasaamy N, Bracaglia L, Garcia-Vivas A, Santoro M, Baker H, Fisher J, Kim P. Placental basement membrane proteins are required for effective cytotrophoblast invasion in a three-dimensional bioprinted placenta model. J Biomed Mater Res Part A 2018;106:1476–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008;26:1367–1372. [DOI] [PubMed] [Google Scholar]
- 42.Resutek L, Hsieh AH. The vacuolated morphology of chordoma cells is dependent on cytokeratin intermediate filaments. J Cell Physiol 2019;234:3458–3468. [DOI] [PubMed] [Google Scholar]
- 43.Lembong J, Lerman MJ, Kingsbury TJ, Civin CI, Fisher JP. A fluidic culture platform for spatially patterned cell growth, differentiation, and cocultures. Tissue Eng Part A 2018;24:1715–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goddette DW, Frieden C. Actin polymerization. The mechanism of action of cytochalasin D. J Biol Chem 1986;261:15974–15980. [PubMed] [Google Scholar]
- 45.Casella JF, Flanagan MD, Lin S. Cytochalasin D inhibits actin polymerization and induces depolymerization of actin filaments formed during platelet shape change. Nature 1981;293:302–305. [DOI] [PubMed] [Google Scholar]
- 46.Biazar E, Heidari M, Asefnejad A, Asefnezhad A, Montazeri N. The relationship between cellular adhesion and surface roughness in polystyrene modified by microwave plasma radiation. Int J Nanomedicine 2011;6:631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drygiannakis AI, Papathanasiou AG, Boudouvis AG. On the connection between dielectric breakdown strength, trapping of charge, and contact angle saturation in electrowetting. Langmuir 2009;25:147–152. [DOI] [PubMed] [Google Scholar]
- 48.Digilov R. Charge-induced modification of contact angle: The secondary electrocapillary effect. Langmuir 2000;16:6719–6723. [Google Scholar]
- 49.Baujard-Lamotte L, Noinville S, Goubard F, Marque P, Pauthe E. Kinetics of conformational changes of fibronectin adsorbed onto model surfaces. Colloids Surf B Biointerfaces 2008;63:129–137. [DOI] [PubMed] [Google Scholar]
- 50.Barker SL, LaRocca PJ. Method of production and control of a commercial tissue culture surface. J Tissue Cult Methods 1994;16:151–153. [Google Scholar]
- 51.Stern J, Weil-Malherbe H, Green RH. The effects and the fate of malononitrile and related compounds in animals tissues. Biochem J 1952;52:114–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brash JL, Horbett TA. Proteins at interfaces. J Dispers Sci Technol 1995;18:1–23. [Google Scholar]
- 53.Tidwell CD, Castner DG, Golledge SL, Ratner BD, Meyer K, Hagenhoff B, Benninghoven A. Static time-of-flight secondary ion mass spectrometry and x-ray photoelectron spectroscopy characterization of adsorbed albumin and fibronectin films. Surf Interface Anal 2001;31:724–733. [Google Scholar]
- 54.Ho Y-C, Huang F-M, Chang Y-C. Cytotoxicity of formaldehyde on human osteoblastic cells is related to intracellular glutathione levels. J Biomed Mater Res Part B Appl Biomater 2007;83B:340–344. [DOI] [PubMed] [Google Scholar]
- 55.Vroman L, Adams AL, Fischer GC, Munoz PC. Interaction of high molecular weight kininogen, factor XII, and fibrinogen in plasma at interfaces. Blood 1980;55:156–159. [PubMed] [Google Scholar]
- 56.Liu W, Zhan J, Su Y, Wu T, Wu C, Ramakrishna S, Mo X, Al-Deyab SS, El-Newehy M. Effects of plasma treatment to nanofibers on initial cell adhesion and cell morphology. Colloids Surf B Biointerfaces 2014;113:101–106. [DOI] [PubMed] [Google Scholar]
- 57.Athanassiou G, Deligianni D. Adhesion strength of individual human bone marrow cells to fibronectin. Integrin beta1-mediated adhesion. J Mater Sci Mater Med 2001;12:965–970. [DOI] [PubMed] [Google Scholar]
- 58.Hirsh SL, McKenzie DR, Nosworthy NJ, Denman JA, Sezerman OU, Bilek MMM. The Vroman effect: Competitive protein exchange with dynamic multilayer protein aggregates. Colloids Surf B Biointerfaces 2013;103:395–404. [DOI] [PubMed] [Google Scholar]
- 59.Latour R. Biomaterials: Protein-surface interactions. Encyclopedia of Biomaterials and Biomedical Engineering, Boca Raton, FL, Vol. 1 Editors: Wnek Gary E., Bowlin Gary L., 2nd ed. CRC Press; 2008. pp. 270–284. [Google Scholar]
- 60.Ruoslahti E, Pierschbacher MD. New perspectives in cell adhesion: RGD and integrins. Science 1987;238:491–497. [DOI] [PubMed] [Google Scholar]
- 61.Salasznyk RM, Williams WA, Boskey A, Batorsky A, Plopper GE. Adhesion to vitronectin and collagen I promotes osteogenic differentiation of human mesenchymal stem cells. J Biomed Biotechnol 2004;1:24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Linsley C, Wu B, Tawil B. The effect of fibrinogen, collagen type I, and fibronectin on mesenchymal stem cell growth and differentiation into osteoblasts. Tissue Eng Part A 2013;19:1416–1423. [DOI] [PubMed] [Google Scholar]
- 63.Li B, Moshfegh C, Lin Z, Albuschies J, Vogel V. Mesenchymal stem cells exploit extracellular matrix as mechanotransducer. Sci Rep 2013;3:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsai K-S, Kao S-Y, Wang C-Y, Wang Y-J, Wang J-P, Hung S-C. Type I collagen promotes proliferation and osteogenesis of human mesenchymal stem cells via activation of ERK and Akt pathways. J Biomed Mater Res Part A 2010;94A:673–682. [DOI] [PubMed] [Google Scholar]
- 65.Asakura S, Hurley RW, Skorstengaard K, Ohkubo I, Mosher DF. Inhibition of cell adhesion by high molecular weight kininogen. J Cell Biol 1992;116:465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stannard AK, Riddell DR, Sacre SM, Tagalakis AD, Langer C, Von Eckardstein A, Cullen P, Athanasopoulos T, Dickson G, Owen JS. Cell-derived apolipoprotein E (ApoE) particles inhibit vascular cell adhesion molecule-1 (VCAM-1) expression in human endothelial cells. J Biol Chem 2001;276:46011–46016. [DOI] [PubMed] [Google Scholar]
- 67.Bost F, Diarra-Mehrpour M, Martin JP. Inter-alpha-trypsin inhibitor proteoglycan family—A group of proteins binding and stabilizing the extracellular matrix. Eur J Biochem 1998;252:339–346. [DOI] [PubMed] [Google Scholar]
- 68.Ocklind C, Forsum U, Obrink B. Cell surface localization and tissue distribution of a hepatocyte cell-cell adhesion glycoprotein (cell-CAM 105). J Cell Biol 1983;96:1168–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eidelman S, Damsky CH, Wheelock MJ, Damjanov I. Expression of the cell-cell adhesion glycoprotein cell-CAM 120/80 in normal human tissues and tumors. Am J Pathol 1989;135:101–110. [PMC free article] [PubMed] [Google Scholar]
- 70.Zoellner H, Höfler M, Beckmann R, Hufnagl P, Vanyek E, Bielek E, Wojta J, Fabry A, Lockie S, Binder BR. Serum albumin is a specific inhibitor of apoptosis in human endothelial cells. J Cell Sci 1996;109:2571–2580. [DOI] [PubMed] [Google Scholar]
- 71.Law RHP, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W, Rosado CJ, Langendorf CG, Pike RN, Bird PI, Whisstock JC. An overview of the serpin superfamily. Genome Biol 2006;7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Borth W. Alpha 2-macroglobulin, a multifunctional binding protein with targeting characteristics. FASEB J 1992;6:3345–3353. [DOI] [PubMed] [Google Scholar]
- 73.Sallenave JMJ-M. Antimicrobial activity of antiproteinases. Biochem Soc Trans 2002;30:111–115. [DOI] [PubMed] [Google Scholar]
- 74.Castellano AC, Barteri M, Castagnola M, Bianconi A, Borghi E, Dellalonga S. Structure-function relationship in the serotransferrin: The role of the pH on the conformational change and the metal ions release. Biochem Biophys Res Commun 1994;198:646–652. [DOI] [PubMed] [Google Scholar]
- 75.White P, Cooke N. The multifunctional properties and characteristics of vitamin D-binding protein. Trends Endocrinol Metab 2000;11:320–327. [DOI] [PubMed] [Google Scholar]
- 76.Pappano AJ, Gil WW. Overview of the circulation and blood. Cardiovascular Physiology, 10th ed. Philadelphia, PA: Elisevier, Inc.; 2013. pp. 1–9. [Google Scholar]
- 77.Mizejewski GJ. Alpha-fetoprotein structure and function: Relevance to isoforms, epitopes, and conformational variants. Exp Biol Med 2001;226:377–408. [DOI] [PubMed] [Google Scholar]
- 78.Lee MH, Ducheyne P, Lynch L, Boettiger D, Composto RJ. Effect of biomaterial surface properties on fibronectin-α5β1integrin interaction and cellular attachment. Biomaterials 2006;27:1907–1916. [DOI] [PubMed] [Google Scholar]
- 79.Lestelius M, Liedberg B, Tengvall P. In vitro plasma protein adsorption on ω-functionalized alkanethiolate self-assembled monolayers. Langmuir 1997;13:5900–5908. [Google Scholar]
- 80.Xu Z, Grassian VH. Bovine serum albumin adsorption on TiO2 nanoparticle surfaces: Effects of pH and coadsorption of phosphate on protein-surface interactions and protein structure. J Phys Chem C 2017;121:21763–21771. [Google Scholar]
- 81.Mitchell SA, Emmison N, Shard AG. Spatial control of cell attachment using plasma micropatterned polymers. Surf Interface Anal 2002;33:742–747. [Google Scholar]
- 82.Anselme K, Ploux L, Ponche A. Cell/material interfaces: Influence of surface chemistry and surface topography on cell adhesion. J Adhes Sci Technol 2010;24:831–852. [Google Scholar]
- 83.Krampera M, Galipeau J, Shi Y, Tarte K, Sensebe L. Immunological characterization of multipotent mesenchymal stromal cells—The International Society for Cellular Therapy (ISCT) working proposal. Cytotherapy 2013;15:1054–1061. [DOI] [PubMed] [Google Scholar]
- 84.Silva WA, Covas DT, Panepucci RA, Proto-Siqueira R, Siufi JLC, Zanette DL, Santos ARD, Zago MA. The profile of gene expression of human marrow mesenchymal stem cells. Stem Cells 2003;21:661–669. [DOI] [PubMed] [Google Scholar]
- 85.Veevers-Lowe J, Ball SG, Shuttleworth A, Kielty CM. Mesenchymal stem cell migration is regulated by fibronectin through a5 b1-integrin-mediated activation of PDGFR- and potentiation of growth factor signals. J Cell Sci 2011;124:1288–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Olivares-Navarrete R, Rodil SE, Hyzy SL, Dunn GR, Almaguer-Flores A, Schwartz Z, Boyan BD. Role of integrin subunits in mesenchymal stem cell differentiation and osteoblast maturation on graphitic carbon-coated microstructured surfaces. Biomaterials 2015;51:69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mann BK, Tsai AT, Scott-Burden T, West JL. Modification of surfaces with cell adhesion peptides alters extracellular matrix deposition. Biomaterials 1999;20:2281–2286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.