Summary
The Forkhead Box P3 (FOXP3) protein is an essential transcription factor for the development and function of regulatory T cells (Tregs), involved in the maintenance of immunological tolerance. Although extensive research over the last decade has investigated the critical role of FOXP3+ cells in preserving immune homeostasis, our understanding of their specific functions remains limited. Therefore, unveiling the molecular mechanisms underpinning the up- and downstream transcriptional regulation of and by FOXP3 is crucial for developing Treg-targeted therapeutics. Dysfunctions in FOXP3+ Tregs have also been found to be inherent drivers of autoimmune disorders and have been shown to exhibit multifaceted functions in the context of cancer. Recent research suggests that these cells may also be involved in tissue-specific repair and regeneration. Herein, we summarize current understanding of the thymic-transcriptional regulatory landscape of FOXP3+ Tregs, their epigenetic modulators, and associated signaling pathways. Finally, we highlight the contributions of FOXP3 on the functional development of Tregs and reflect on the clinical implications in the context of pathological and physiological immune responses.
Keywords: FOXP3, regulatory T cells, thymus, autoimmunity, therapeutics
A subset of CD4+ T cells, recognized as FOXP3+ regulatory T (Treg) cells, are involved in preserving immune tolerance. Defects in these cells drive the pathogenesis of autoimmune diseases. In this review, we discuss the functions and dysfunctions in the developmental pathways of FOXP3+ Tregs, their emerging roles, and explore potential therapeutic targets.
Graphical Abstract
Graphical Abstract.
Introduction
In the context of peripheral self-tolerance maintenance, regulatory T cells (Tregs) were originally defined as a subset of T cells expressing CD4 and CD25. However, it was later discovered that Tregs specifically expressed the Forkhead Box P3 (FOXP3) transcription factor, a master regulator of Treg differentiation and function, which is crucial for the maintenance of immune tolerance. Abnormalities in FOXP3 were found to cause a wide range of immunopathological diseases. Deficiency in the Foxp3 gene can lead to the development of fatal lymphoproliferative autoimmune disease, likely due to the absence in FOXP3+ Treg-mediated immune-suppressive mechanisms [1]. Similarly, mutations, in human FOXP3, have been shown to cause immune dysregulation, polyendocrinopathy enteropathy, and X-linked syndrome (IPEX) [2]. Using a scurfy mouse model deficient in CD4+ FOXP3+ Tregs, an adoptive transfer of CD4+ T cells demonstrated that these cells were critical mediators of disease development [3].
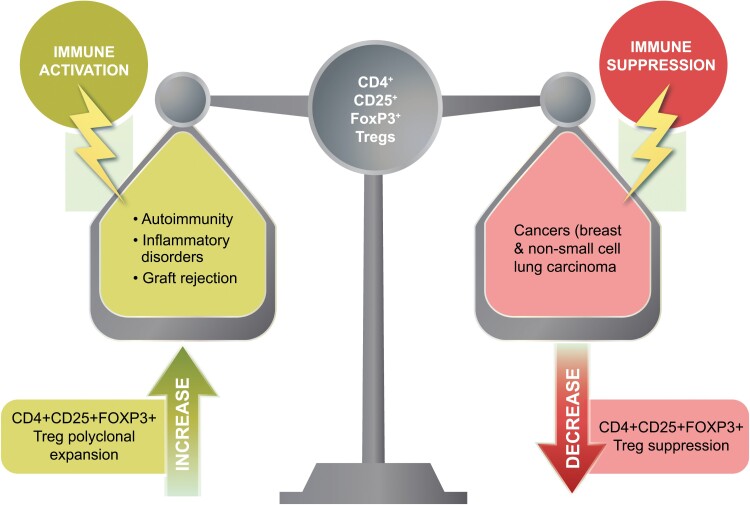
After years of research establishing FOXP3+ Tregs as distinct regulators of CD4+ T cells, recent evidence suggests that they play a multifaceted role beyond immunosuppression. As the onset of FOXP3 expression was used as a standard marker of Tregs, it became clear that these cells have diverse functions in regulating immune responses. These recent advances highlight a promising avenue for potential therapeutics using biologicals targeting CD4+FOXP3+ Tregs. In certain pathologies, such as autoimmune diseases, inflammatory disorders, and cases of graft rejection, the polyclonal expansion of FOXP3+ Tregs can enhance their immunomodulatory capacity. However, in other cases, such as certain types of cancers where FOXP3+ Tregs accumulate and are associated with poor prognosis [4], a potential antitumor immune response may involve suppressing Treg function (Fig. 1). Here, we present current literature on the development and function of FOXP3+ Tregs, including their emerging implications in tissue regeneration, and discuss prospects for harnessing these cells in clinical settings.
Figure 1.
Therapeutic targeting of Tregs. The targeting of Tregs for therapeutic purposes has been proposed for various pathologies. Experimental models have provided evidence showing that reducing Tregs can result in a decrease in tumor growth and enhanced efficacy of tumor immunotherapy. In such cases, suppressing the CD4+CD25+FOXP3+ Treg population could prove beneficial (right side of the scale). However, in the context of autoimmune disease, inflammatory disorders, and graft rejection, polyclonal expansion and proliferation of CD4+CD25+FOXP3+ Tregs may have potential utility by mitigating autoimmune attack and augmenting Treg-mediated immunosuppression (left side of the scale).
Development and differentiation of Tregs
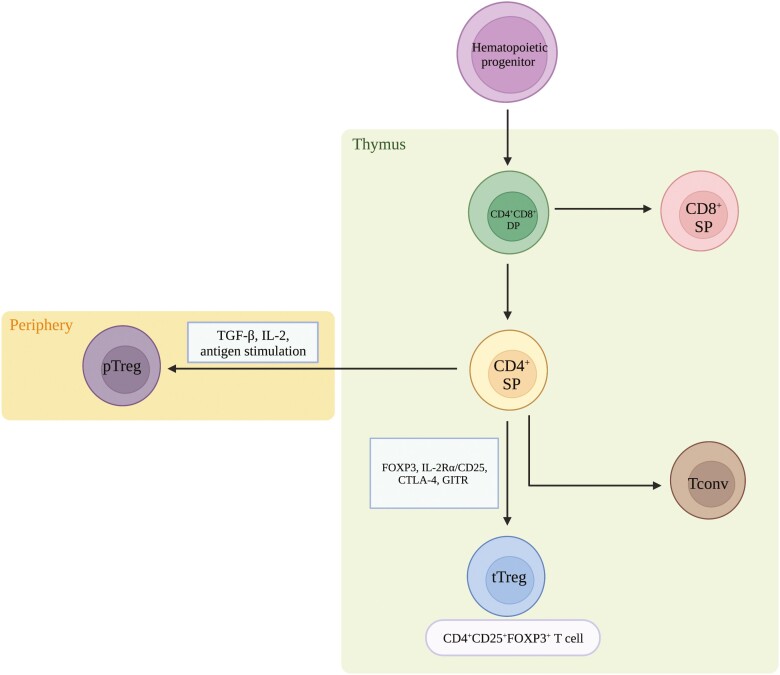
The thymus is an indispensable organ for the development and maturation of Tregs. During fetal thymic development (Fig. 2), T-cell receptor (TCR+) CD4+ CD8+ double positive thymocytes undergo negative selection, when they are presented with high-affinity or high-avidity self-peptides via major histocompatibility complex class II (MHC II) by thymic epithelial cells or dendritic cells (DCs). A small subset of these thymocytes develop into thymic Tregs (tTreg), which can detect self-antigens via the expression of a biased self-reactive TCR repertoire that results in the induction of FOXP3 expression [5]. In addition to the signaling mediated by the TCR, CD28 is also essential in the development of tTregs. This is supported by evidence from mouse models deficient in CD28-CD80/86, which exhibit reduced frequency of Tregs [6]. Although engagement of TCR-CD28 leads to the activation of the phosphatidylinositol-3-kinase (PI3K) signaling pathway, the differentiation of FOXP3+ Tregs appears to require a limited duration of this signaling cascade [7]. In contrast, extended activation of the PI3K pathway in conventional T (Tconv) cells appears to counter the induction of FOXP3 expression.
Figure 2.
Schematic of Treg development. Derived from hematopoietic progenitor cells, TCR+CD4+CD8+ double-positive (DP) thymocytes differentiate to become either CD8+ single-positive (SP) or CD4+ SP thymocytes. Once in the periphery, upon antigen stimulation, with the expression of TGF-β and IL-2, CD4+ T cells differentiate into pTregs. In the thymus, however, there is upregulation of IL-2Rα/CD25, FOXP3, CTLA-4, and GITR, all of which denote the differentiation into tTregs. In the absence of AIRE by mTECs, developing T cells are not negatively selected and can escape into the periphery as potentially pathogenic T cells.
As Treg development progresses, there is an upregulation of the interleukin (IL)-2 receptor alpha chain, (IL-2Rα/CD25), which is a marker of tTregs that have undergone TCR signaling [8, 9]. The expression of CD25 is concurrent with the expression of FOXP3 and is also associated with the expression of additional Treg markers, such as cytotoxic T-lymphocyte–associated antigen 4 (CTLA-4) and glucocorticoid-induced TNFR-related protein (GITR) [8, 9]. Moving forward in the developmental process, medullary thymic epithelial cells (mTECs) that express the transcription factor autoimmune regulator (AIRE) contribute to negative selection and the prevention of autoimmunity by presenting tissue-restricted self-antigens to developing T cells [10]. In the absence of AIRE, potentially self-reactive CD4+ T cells are then able to exit to the periphery and become pathogenic, leading to peripheral autoimmunity. Thus, AIRE is critical in maintaining self-tolerance to a vast array of autoantigens and ensuring proper development of FOXP3+ Tregs.
In the presence of IL-2 and transforming growth factor β (TGF-β), antigenic stimulation can convert naïve CD4+ Tconv cells into peripheral Tregs (pTregs) cells. Although pTregs make up a small fraction of the overall Treg population in the blood, they have a tendency to accumulate in specific organs, such as the gut, and are crucial for preserving maternal–fetal tolerance [11]. Within the small intestine, pTregs recognize foreign antigens such as those found in food and commensal bacteria. In addition to maintaining tolerance to commensal bacteria, pTregs can also play an essential role in preserving the expression of FOXP3 via the production of TGF-β [12, 13]. Furthermore, short-chain fatty acids, including butyrate and propionate, generated by commensal microorganisms, have been shown to induce FOXP3 expression in Tregs and expand their population in the colon [14].
During the course of Treg development, signature markers, including CTLA4 and CD25, have recently been found to be influenced by the insulin receptor substrate 1 (IRS1) signaling pathway. In vitro studies by Lee et al. identified that over-expression of IRS1 in Tregs led to a reduced expression of FOXP3 and Treg phenotype markers. Moreover, in an adoptive cell transfer model of colitis, IRS1-overexpressing Tregs demonstrated reduced immunosuppressive capacity and were unable to inhibit the pathogenic effects of cotransferred Tconv cells. Further investigation revealed that IRS1 led to Treg instability through activation of the mTORC1 pathway and upregulation of IFN-γ and glucose transporter 1 (Glut1) [15].
To investigate the heterogeneity and functional diversity of Tregs, Zemmour et al. employed a multimodal approach consisting of single-cell RNA sequencing (scRNA-seq), activation reporter, and TCR-seq analysis. They obtained gene expression profiles of distinct Treg subsets in mice and humans and identified that resting Tregs had similarities to Tconv cells, with similar gene and protein expression profiles. Moreover, all Tregs were found to express a fundamental set of FOXP3-dependent transcripts, with additional gene expressions or pathways activated nonuniformly across the different subtypes. Genes required for Treg function, including Il2ra and Ctla4, were revealed to be consistently expressed across the different Tregs subsets, while inhibitory cytokines were more variably expressed. Finally, it was also revealed that TCR signaling strength appeared to influence the gene expression patterns of activated Tregs [16].
Functional Tregs can, however, be distinguished based on their levels of FOXP3 expression, with continuous and elevated FOXP3 transcription levels leading to their maturation [17]. Nonetheless, stable expression of FOXP3 is essential for the development and function of Tregs and is partly dependent on the epigenetic modifications of the Treg-specific demethylated region (TSDR) [18, 19]. This non-coding region, located in the first intron of the FOXP3 gene locus, has become a defining marker of true tTregs, distinguishing them from activated CD4+CD25+FOXP3+ Tconv cells, which lack suppressive function and only transiently express FOXP3. Therefore, the presence of DNA hypomethylation at the TSDR is critical for lineage stability, ensuring stable FOXP3 expression and long-term Treg function. However, in the event of FOXP3 expression loss, two phenotypic outcomes are possible: the induction of either a memory Treg cell or an inflammatory effector T cell [20, 21].
Tregs beyond immunosuppression
As part of the regulatory pathway, certain Tregs (tTregs or pTregs) express the CC chemokine receptor 4 and respond to its ligand, CC chemokine ligand 22 (CCL22). However, in some pathologies, including cancer, this mechanism allows Tregs to migrate toward CCL22-expressing tumor-associated macrophages and cancer cells. Within the tumor stroma, Tregs are induced to proliferate and expand by IL-2 and TCR stimulation, as they recognize tumor antigens presented by DCs [22]. In addition, TGF-β from the tumor microenvironment can induce differentiation of Tconv cells into Tregs [23]. In certain cancers, such as non-small-cell lung carcinoma and breast cancer, inhibiting the expansion and proliferation of Tregs may enhance therapeutic outcomes [22, 23].
Interestingly, evidence also points towards a potentially immunosupportive role of Tregs. In the context of immunological memory of antiviral responses, Tregs facilitate rapid movement of effector cells to the site of infection. In addition, they regulate the differentiation of memory CD8+ T cells via the production of IL-10, TGF-β, and downregulation of IL-2. Emerging data have shown that specific Treg-derived cytokines are crucial for the diversification of memory T cell populations. For instance, the proliferation and optimal maintenance of memory CD8+ T cells is dependent on Treg-derived IL-15, while KLRG1-IL-7Rα+ memory CD8+ T cells rely on Treg-derived IL-10 [24]. By highlighting the diverse functions of Tregs beyond immunosuppression and revealing their underlying mechanisms in different pathological states, it may allow them to be used for applications beyond their immunoregulatory function.
Transcriptional regulation of Treg function by FOXP3
The FOXP3 gene consists of 12 exons that code for a 431 amino acid FOXP3 protein. The protein includes a C2H2 zinc finger (Cys2-His2), a central leucine zipper domain, and a C-terminal forkhead (FKH) domain [25]. The FHK domain is essential for DNA binding, nuclear localization, and interaction with other transcription factors, such as nuclear factor of activated T cells (NFAT). The central leucine zipper domain and zinc finger domains create a structural scaffold, which plays a pivotal role in facilitating protein–protein interactions and the formation of FOXP3 complexes required for regulatory activity [25]. A recent study on the dimerization states of FOXP3 identified that it can fold into two distinct structures, with the functional one being a head-to-head dimer, which uses the RUNX1-binding region as a link for the forkhead domains [26]. This dimerization state allows for FOXP3 monomers to interact with one another and bind DNA to facilitate transcriptional regulation of gene expression.
To further facilitate Treg development, FOXP3 can bind to distinct genomic sites and interact with various binding partners depending on the extracellular environmental cues. Moreover, driving FOXP3 programming of Tregs are numerous domains of complexes that act as either repressors or activators [27, 28]. An alanine-scanning mutagenesis analysis identified that FOXP3 was able to interact with certain binding partners to form an “operative” complex, active in both target gene transcriptional activation and repression. FOXP3 was also able to form a “non-operative” complex with reduced activation and repressive capacity [28]. Other factors affecting FOXP3 binding interactions include mutations in the N-terminus, leading to inefficient Treg-mediated suppression of T helper 1 (Th1) responses, but increased suppression of Th2 and Th17 [29]. Other binding partners of FOXP3, including GATA3 and enhancer of zeste homolog 2 (EZH2), were found to interact following TCR activation [27]. These findings emphasize the requirement for FOXP3 interactions in the regulation of Treg responses to external stimuli and ultimately demonstrate functional adaptability.
Emerging evidence has revealed that the loss of FOXP3 expression by Tregs is regulated through various transcriptional, epigenetic, and post-translational regulatory mechanisms. Transcription factors including signal transducer and activator of transcription 5 (STAT5), NFAT, and forkhead box protein O1 (FOXO1), directly bind to the FOXP3 promoter region to regulate its transcriptional activation. In addition, conserved non-coding sequence (CNS) elements upstream and within the FOXP3 gene locus interact with multiple transcription factors to regulate gene expression. For instance, CNS1 binds to SMAD3, which is activated downstream of TGF-β signaling [30]; CNS2 interacts with STAT5, NFAT, RUNX1, and CREB [31, 32]; and, CNS3 binds to the NF-κB signaling molecule c-Rel [33]. CNS2 also contains the Treg-specific TSDR, which is demethylated in tTregs and partially methylated in pTregs [34]. The demethylated TSDR stabilizes FOXP3 expression through the recruitment of various transcription factors, including FOXP3 itself, CREB, and Ets1 [35, 36]. Moreover, FOXP3 stability is also impacted by protein acetylation, phosphorylation, and ubiquitination [37].
In addition, metabolism has been reported to play a crucial role in the regulation of Treg stability through mechanisms that intersect with transcriptional, epigenetic, and post-translational modifications of FOXP3 expression. For instance, hypoxia-inducible factor 1α (HIF-1α) has been implicated in Treg differentiation, wherein HIF-1α can interact with FOXP3, leading to its degradation in hypoxic conditions. In models of HIF-1α deficiency, Tregs exhibited efficient suppressive function and reduced levels of IFNγ. In contrast, continuous T cell expression of HIF-1α led to an upregulation of IFNγ+ effector T cells as compared to Tregs. Treg stability was also dependent on the expression of the HIF-1α repressor, and E3 ligase, Deltex1 [38, 39]. However, further research is necessary to fully elucidate the various mechanisms by which metabolism and hypoxic factors contribute to the transcriptional regulation of Tregs.
Treg stability and adaptability
Treg stability is defined as the ability to maintain FOXP3 expression and suppress the pro-inflammatory effector functions during immunogenic challenges. However, recent studies have shown that some FOXP3+ Tregs can lose their expression of FOXP3 and acquire effector-like functions, indicative of lineage instability. Despite this, Tregs exhibit functional adaptability and increase expression of transcription factors and chemokine receptors associated with Th1 [40], Th17 [41], and T follicular helper (Tfh) cells [42]. Perhaps, this may be a mechanism that allows for greater homing and suppression of T-effector cells at sites of inflammation
Epigenetic regulation of the FOXP3 locus is important for the stability of Treg function by diminishing their conversion into “pathogenic” cells. However, moderate exposure to inflammatory cytokines can destabilize Tregs. The presence of repressive histone markers at the promoter of IL-12Rβ2 can impair the responsiveness of Tregs to IL-12, which prevents activation of the Th1 pathway and subsequent upregulation of T-bet (T-box expressed in T cells), encoded by Tbx21, CXC chemokine receptor 3 (CXCR3), and IFNγ [43]. Tregs maintain FOXP3 expression but acquire various effector T cell functions, which has been referred to as Treg plasticity. Given this potential plasticity, it is critical for Tregs to fine-tune their response to inflammatory and environmental challenges. For instance, in Th1-driven responses, Tregs accumulate at the site of inflammation and are induced to express T-bet, which helps to enhance the immunoregulatory function of Tregs in this context. Consistent with this notion, depletion of T-bet-expressing Tregs was shown to result in Th1 autoimmunity [40]. However, for Th2-dependent responses, Tregs acquire expression of interferon regulatory factor 4, and in Th17, signal transducer and activator of transcription 3 (STAT3) [40, 41, 43, 44]. In addition, suppressor of cytokine signaling 1 protects Tregs from responding to excessive pro-inflammatory cytokines by the inhibition of STAT1/3 signaling in response to IFN-γ and IL-17 [45].
In vitro work has demonstrated that Tregs can lose stability upon stimulation with pro-inflammatory cytokines, including IL-6 and IL-4 [46], to become FOXP3- “exTregs.” In adoptive transfer experiments using purified FOXP3+ Tregs transplanted into recipients lacking T cells, there was a loss of FOXP3 expression [46, 47], which led to the expression of inflammatory cytokines [48]. As the majority of unstable Treg subsets have been found to be CD25loFOXP3+, this may explain their inherent underlying instability in vivo. In addition, fate-mapping in experimental mouse models have shown that these exTregs derive from activated T cells that failed to undergo Treg differentiation and only transiently expressed FOXP3 [20]. Perhaps, Treg stability is dependent on the host context to modulate inflammation and induce peripheral tolerance.
FOXP3 dysfunction and autoimmunity
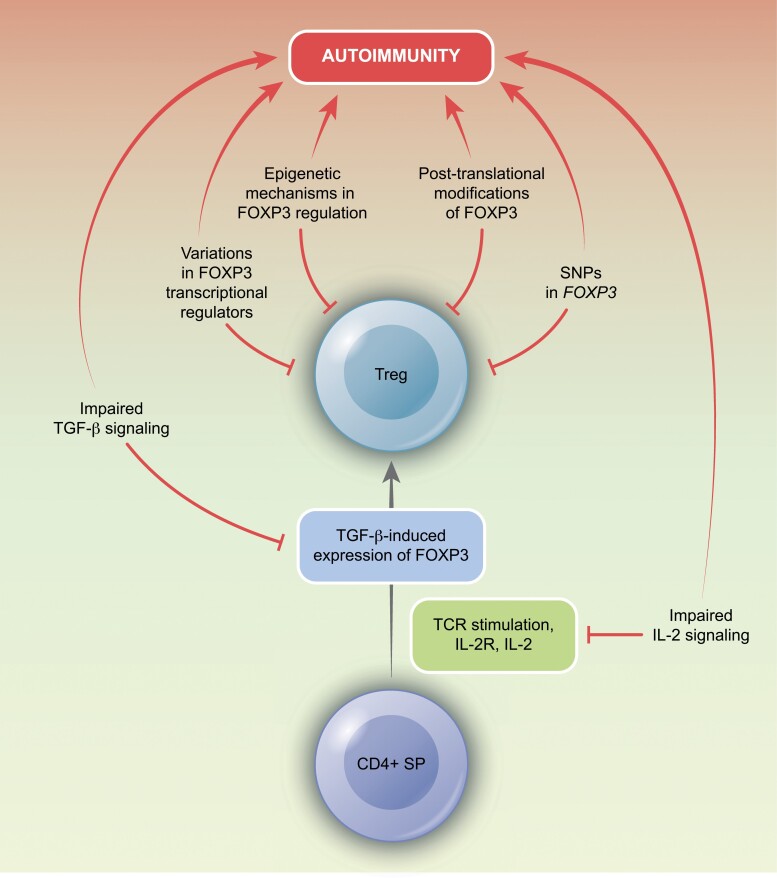
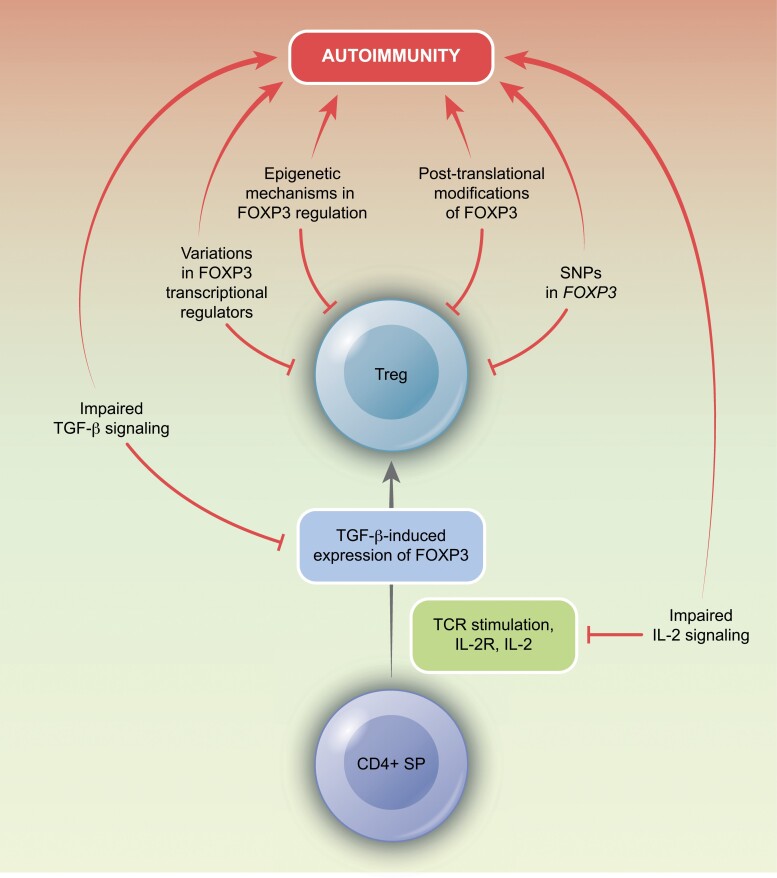
Although the etiology of autoimmunity is multifactorial, changes in FOXP3 expression and function are thought to underlie mechanisms relating to its development [28]. Dysregulated FOXP3 expression in autoimmune diseases may be attributed to variations in genomic regulation, transcriptional regulators, and post-translational modifications. In the thymus, potentially self-reactive Treg precursors, following TCR stimulation, express the CD25 to induce activation of STAT5 [49]. This, together with TGF-β signaling, results in the expression of FOXP3 [50]. The signal received from IL-2 is, therefore, a critical requirement for their differentiation [49, 50], and dysfunctions in IL-2 and TGF-β signaling have been implicated in the development of autoimmunity. Additionally, several dysfunctional immune pathways can contribute to the pathogenesis of self-reactive disorders (Fig. 3).
Figure 3.
Disruptions in Treg development can contribute to autoimmune pathologies. This can occur through a number of mechanisms, including the loss of FOXP3 transcriptional regulation, altered epigenetic mechanisms, post-translational modifications, single-nucleotide polymorphisms, and dysfunctions in upstream signaling pathways of IL-2 or TGF-β.
Molecular mechanisms of FOXP3 regulation in autoimmunity
Single-nucleotide polymorphisms (SNPs) in coding and non-coding gene regions of the FOXP3 gene have been linked to autoimmune diseases and allergies [51] and been found to influence mRNA stability in affected children [51]. Additionally, SNPs in genes regulating IL-2 responses, including IL2RA (CD25), PTPN2, and PTPN22, have also been associated with autoimmunity [52–54]. These genes are thought to mediate their function through activation of a STAT5 feedback loop that positively reinforces FOXP3 expression [52]. However, SNPs can interfere with this loop to affect FOXP3 expression. For example, in Type 1 Diabetes (T1D), a SNP in PTPN2 has been shown to reduce levels of FOXP3 and inhibit Treg activity through the downregulation of IL-2-mediated STAT5 activation [55]. Similarly, in Primary Sclerosing Cholangitis and Multiple Sclerosis, reduced FOXP3 and Treg suppression are observed, due to downregulated activation of STAT5 [53, 54]. While polymorphisms in PTPN2 are implicated in various signaling mechanisms, their regulation of FOXP3 may be mediated by an increase in IL-2 sensitivity following TCR activation [56]. For instance, decreased expression of PTPN2 reduces the differentiation of Tregs in conditions of strong TCR signaling but leads to an increase in FOXP3 expression in weak TCR activity [57].
Micro RNAs (miRNAs) also play a key role in regulating FOXP3 mRNA expression by binding to stress granules. The absence of miRNAs, including miR-155, can result in failure of Treg function [58]. Specifically, miR-155 has been shown to positively regulate Treg differentiation and development [59], and is involved in mediating pregnancy tolerance [60]. In addition, miR-146a has been identified as a key regulatory of Treg suppressor function, and a deficiency in this miRNA can result in a loss of immune tolerance mechanisms [61]. Notably, activated but naïve Tregs from at-risk T1D patients have been found to have elevated levels of miR-26a, a miRNA which disrupts the transcriptional repressive function of FOXP3 by reducing levels of the histone methyltransferase, EZH2 [62]. In the presence of miR-26a, a reduction in EZH2 expression impairs FOXP3 function [63].
In autoimmune diseases such as Juvenile Idiopathic Arthritis (JIA) and psoriasis, Treg dysfunction has also been identified as a contributing factor. In JIA, CD25+ FOXP3lo Tregs were present in afflicted joints, which surprisingly showed hypomethylation of their TSDRs [64]. These FOXP3lo Tregs also showed impaired IL-2R signaling [63] and given the central role of IL-2 signaling in core pathological mechanisms, improving sensitivity to IL-2 may be a promising avenue for therapeutic intervention in autoimmunity. In psoriasis, Tregs appear to have impaired suppressive capacity, perhaps due to elevated levels of phospho-STAT3 (pSTAT3) resulting from increased IL-6 and IL-21 [65]. However, in vitro studies have shown that a reduction in Treg FOXP3 levels from JIA synovial fluid can be corrected through IL-6R-mediated activation of STAT3 [64]. These findings highlight the complex downstream effects of FOXP3 dysfunction, and the involvement of related pathways.
Other molecules have also been implicated in regulation of FOXP3 stability and Treg activity. For instance, cyclin-dependent kinase 2 (CDK2) has been identified as a negative regulator of FOXP3+ Treg function and stability [66]. Mutations of serine/threonine to alanine residues at every CDK motif resulted in increased stability of FOXP3 protein in CD4+ T cells and enhanced ability of T cells to inhibit Tconv proliferation in vitro. Furthermore, this mutation was effective at mitigating colitis in a mouse model of inflammatory bowel disease [66]. CDK2, along with lymphocyte-specific protein tyrosine kinase (LCK) [67] and proto-oncogene serine/threonine-protein kinase (PIM)-1 [68], were all able to phosphorylate FOXP3 on Tyr, and Ser and Thr residues, respectively, leading to impaired function [69] and disabled Treg suppression [66, 68, 69]. Therefore, manipulation of kinases through the use of targeted inhibitors may provide potential therapeutic approaches in countering the loss of Treg function [69]. Identification of the various inter-connected pathways and molecular targets in the functional regulation of Tregs demonstrates the complex and dynamic nature of immune regulation.
Emerging roles of Tregs in wound healing and regeneration
Growing evidence suggests that in addition to their role in immune suppression and tolerance, Tregs may also be involved in tissue regeneration and wound repair processes [70]. For example, in skin wound healing, the activation of epidermal and dermal regeneration is mediated by epidermal growth factor receptor (EGFR) signaling. Tregs express the EGFR ligand and are involved in tissue repair and maintenance [71]. Studies by Nosbaum et al. have shown that following skin injury, activated Tregs in the skin reduce levels of IFNγ and pro-inflammatory macrophages, while a deficiency in Tregs impedes wound repair, as evidenced by a delay in wound re-epithelialization and closure [70].
Tregs have also been demonstrated to contribute to tissue repair in lung injury. In an experimental model, FOXP3+ Tregs were shown to promote increased proliferation of alveolar epithelial cells [72]. These effects were also observed in acute lipo-polysaccharide (LPS)-induced models of lung inflammation, wherein Tregs helped to resolve inflammation by suppressing the activity of alveolar macrophages and neutrophils [73].
Tregs have also been proposed to play a role in neural repair. In a mouse model of ischemic stroke, Ito et al. observed an accumulation of Tregs in the brain. In addition, it was found that these Tregs can inhibit astrogliosis by producing amphiregulin [74]. The recent discoveries regarding the immune tolerogenic-independent functions of Tregs in various tissues suggest their potential use as broad therapeutic agents. However, additional research is required to identify the tissue-specific mechanisms underlying these effects for precise therapeutic targeting.
Conclusion and future directions
In the years since the discovery of Tregs, research has progressed to recognize them as a vital component of immune homeostasis, particularly in relation to self-tolerance, anti-tumor responses, autoimmunity, and more recently, wound healing and regeneration. However, targeting Tregs presents a significant challenge as a precise understanding of the tissue-specific environmental cues will be needed for effective strategies. In the context of autoimmunity, alterations in various levels of FOXP3 activity can result in a loss of effective Treg function. Fluctuations in IL-2 signaling or modifications of FOXP3 at the post-translational or transcriptional levels can all contribute to a breakdown in self-tolerance. As a result, it is critical to identify and evaluate the interconnected immune pathways of FOXP3+ Tregs for immunological and pharmacological manipulation.
There remains a need for the development of new methods to selectively target Tregs with minimal adverse effects. This challenge can potentially be overcome through the use of cytokine partial agonists, which function at a lower capacity than full agonists. In a recent study by Glassman et al., they took advantage of the underlying differences in cytokine sensitivities to generate an IL-2 partial agonist that exclusively retained its ability to activate Tregs [75]. Future clinical translation of these approaches will pave the path toward developing cell-specific immunotherapies.
Representing a potential strategy in the treatment of autoimmune diseases and transplantation may also involve the use of a combination therapy consisting of costimulation blockade and IL-2. Although CD28 costimulation blockade leads to a loss of Tregs, administration with IL-2 in humanized mice was shown to counteract this effect by selectively disabling T cell effector responses while preserving Tregs [76]. These findings provide promising guidance for current clinical implementation; potentially introducing the use of a supplementary immunotherapy to counteract the limitations of the primary treatment.
Further elaboration on methods for expanding FOXP3+ Tregs in vivo are clearly needed. However, due to constraints in identifying FOXP3 expression levels in humans, it would be difficult to assess Treg manipulation success rates. Thus, methods to measure infused or in vivo administered Tregs are required to better understand the half-life and points of accumulation to track any phenotypic alterations [77]. Recent efforts in the clinical setting include attempts to expand the population of Tregs in vivo by adoptively transferring ex vivo-expanded Tregs, rather than relying solely on the expansion of endogenous Tregs. Low doses of IL-2 have been used to expand Tregs in a way that prevents natural killer and effector T cell activation and expansion [77]. However, this therapy depends on the assumption that polyclonal Tregs maintain their ability to recruit to sites of inflammation and remain antigen-specific at a population level. Another approach has been the development of chimeric antigen receptors, which use CAR Tregs with an antibody Fab specific for a self-antigen to prevent the autoimmune activation of T cells [78].
Increasing efforts for Treg TCR sequencing will be monumental in expanding the understanding of Treg heterogeneity and functionality. Through increased sequencing, clinicians can better comprehend inter-individual immune diversity and identify effective Treg clones for potential immunological manipulation. In addition, identifying the differential protein expression patterns of different Treg populations can be used to better understand their functions and associated features. This has recently been executed through a proteomic analysis wherein iTregs were found to share similar expression of signaling and metabolic-related proteins with Tconv cells but were distinct from tTregs [79]. Further research is needed to investigate the functional differences among iTregs, tTregs, and Tconv cells.
Continued exploration and discoveries relating to FOXP3+ Tregs, their immunosuppressive role, and emerging functions, represent a promising avenue for clinical advancements. Undoubtedly, focused research on the transcriptional and molecular dynamics underpinning FOXP3+ Tregs will allow for a greater understanding of the regulation of T cell-mediated immune responses.
Acknowledgments
This paper is part of the 'Unconventional T Cells in Health and Disease' series. Figures were made using BioRender.
Glossary
Abbreviations
- AIRE
autoimmune regulator
- C
cysteine
- CAR
chimeric antigen receptor
- CCR4
C-C chemokine receptor 4
- CCL22
C-C chemokine ligand 22
- CD
cluster of differentiation
- CDK2
cyclin-dependent kinase 2
- CNS
conserved non-coding sequence
- Cys2-His2
C2H2 zinc finger
- CTLA4
cytotoxic T-lymphocyte-associated antigen 4
- CXCR3
C-X-C chemokine receptor 3
- DC
dendritic cells
- DP
double positive
- EGFR
epidermal growth factor receptor
- EZH2
enhancer of zeste homolog 2
- FKH
forkhead domain
- FOXO1
forkhead box protein 01
- FOXP3
forkhead Box P3
- GITR
glucocorticoid-induced TNFR-related protein
- Glut1
glucose transporter 1
- HIF-1α
hypoxia-inducible factor 1α
- IL-2
interleukin 2
- IL-2R
IL-2 receptor
- IFNγ
interferon γ
- iTreg
induced Tregs
- IPEX
immune dysregulation, polyendocrinopathy enteropathy and X-linked syndrome
- IRF4
interferon regulatory factor 4
- IRS1
insulin receptor substrate 1
- JIA
juvenile idiopathic arthritis
- LCK
lymphocyte-specific protein tyrosine kinase
- LPS
lipopolysaccharide
- MHC
major histocompatibility complex
- miRNA
micro RNA
- mTEC
medullary TEC
- NFAT
nuclear factor of activated T cells
- NSCLC
non-small-cell lung carcinoma
- pSTAT
phospho-STAT
- PI3K
phosphatidylinositol 3-kinase
- PIM
proto-oncogene serine/threonine-protein kinase
- pTreg
peripheral Treg
- RBR
Runx1-binding region
- RNA
ribonucleic acid
- SCFA
short-chain fatty acid
- scRNAseq
single-cell RNA sequencing
- SNP
single-nucleotide polymorphism
- SP
single positive
- STAT
signal transducer and activator transcription
- SOCS1
suppressor of cytokine signaling 1
- T1D
type 1 diabetes
- T-bet
T-box expressed in T cells
- Tconv
conventional T cells
- TCR
T cell receptor
- TEC
thymic epithelial cell
- Tfh
T follicular helper
- TGFβ
transforming growth factor β
- Th
T helper
- TNFR
tumor necrosis factor receptor
- Treg
regulatory T cells
- tTreg
thymic Tregs
- TSDR
Treg-specific demethylated region.
Contributor Information
Mahdieh Golzari-Sorkheh, Department of Immunology, University of Toronto, Toronto, ON, Canada.
Juan Carlos Zúñiga-Pflücker, Department of Immunology, University of Toronto, Toronto, ON, Canada; Biological Sciences, Sunnybrook Research Institute, Toronto, ON, Canada.
Ethical approval
Not applicable.
Conflict of interests
None declared.
Funding
This research was funded by the Canadian Institutes of Health Research (CIHR; FND-154332). J.C.Z.P. was supported by a Canada Research Chair in Developmental Immunology.
Author contributions
Writing—original draft preparation: M.G.S. Writing—review and editing: M.G.S. and J.C.Z.P. Supervision: J.C.Z.P. All authors contributed to the article and approved the submitted version.
Data availability
Not applicable.
References
- 1. Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet 2001, 27, 68–73. doi: 10.1038/83784 [DOI] [PubMed] [Google Scholar]
- 2. Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet 2001, 27, 20–1. doi: 10.1038/83713 [DOI] [PubMed] [Google Scholar]
- 3. Blair PJ, Bultman SJ, Haas JC, Rouse BT, Wilkinson JE, Godfrey VL.. CD4+CD8- T cells are the effector cells in disease pathogenesis in the scurfy (sf) mouse. J Immunol 1994, 153, 3764–74. [PubMed] [Google Scholar]
- 4. Hermans C, Anz D, Engel J, Kirchner T, Endres S, Mayr D.. Analysis of FoxP3+ T-regulatory cells and CD8+ T-cells in ovarian carcinoma: location and tumor infiltration patterns are key prognostic markers. PLoS One 2014, 9, e111757. doi: 10.1371/journal.pone.0111757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jordan MS, Boesteanu A, Reed AJ, Petrone AL, Holenbeck AE, Lerman MA, et al. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat Immunol 2001, 2, 301–6. doi: 10.1038/86302 [DOI] [PubMed] [Google Scholar]
- 6. Tai X, Cowan M, Feigenbaum L, Singer A.. CD28 costimulation of developing thymocytes induces Foxp3 expression and regulatory T cell differentiation independently of interleukin 2. Nat Immunol 2005, 6, 152–62. doi: 10.1038/ni1160 [DOI] [PubMed] [Google Scholar]
- 7. Sauer S, Bruno L, Hertweck A, Finlay D, Leleu M, Spivakov M, et al. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc Natl Acad Sci USA 2008, 3, 7797–802. doi: 10.1073/pnas.0800928105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Darrasse-Jèze G, Marodon G, Salomon BL, Catala M, Klatzmann D.. Ontogeny of CD4+CD25+ regulatory/suppressor T cells in human fetuses. Blood 2005, 105, 4715–21. doi: 10.1182/blood-2004-10-4051 [DOI] [PubMed] [Google Scholar]
- 9. Michaëlsson J, Mold JE, McCune JM, Nixon DF.. Regulation of T cell responses in the developing human fetus. J Immunol 2006, 176, 5741–8. doi: 10.4049/jimmunol.176.10.5741 [DOI] [PubMed] [Google Scholar]
- 10. Malchow S, Leventhal DS, Lee V, Nishi S, Socci ND, Savage PA.. Aire enforces immune tolerance by directing autoreactive T cells into the regulatory T cell lineage. Immunity 2016, 44, 1102–13. doi: 10.1016/j.immuni.2016.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Samstein RM, Josefowicz SZ, Arvey A, Treuting PM, Rudensky AY.. Extrathymic generation of regulatory T cells in placental mammals mitigates maternal-fetal conflict. Cell 2012, 150, 29–38. doi: 10.1016/j.cell.2012.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pandiyan P, Bhaskaran N, Zou M, Schneider E, Jayaraman S, Huehn J.. Microbiome dependent regulation of Tregs and Th17 cells in mucosa. Front Immunol 2019, 10, 426. doi: 10.3389/fimmu.2019.00426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim KS, Hong SW, Han D, Yi J, Jung J, Yang BG, et al. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science 2016, 351, 858–63. doi: 10.1126/science.aac5560 [DOI] [PubMed] [Google Scholar]
- 14. Park J, Kim M, Kang SG, Jannasch AH, Cooper B, Patterson J, et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol 2015, 8, 80–93. doi: 10.1038/mi.2014.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee WH, Kim GE, Hong KJ, Kim HS, Lee GR.. Insulin receptor substrate 1 signaling inhibits Foxp3 expression and suppressive functions in Treg Cells through the mTORC1 pathway. Int J Mol Sci 2023, 24, 2551. doi: 10.3390/ijms24032551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zemmour D, Zilionis R, Kiner E, Klein AM, Mathis D, Benoist C.. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat Immunol 2018, 19, 291–301. doi: 10.1038/s41590-018-0051-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bending D, Paduraru A, Ducker CB, Prieto Martín P, Crompton T, Ono M.. A temporally dynamic Foxp3autoregulatory transcriptional circuit controls the effector Treg programme. EMBO J 2018, 37, e99013. doi: 10.15252/embj.201899013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol 2007, 5, e38. doi: 10.1371/journal.pbio.0050038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Polansky JK, Kretschmer K, Freyer J, Floess S, Garbe A, Baron U, et al. DNA methylation controls Foxp3 gene expression. Eur J Immunol 2008, 38, 1654–63. doi: 10.1002/eji.200838105 [DOI] [PubMed] [Google Scholar]
- 20. Miyao T, Floess S, Setoguchi R, Luche H, Fehling HJ, Waldmann H, et al. Plasticity of Foxp3(+) T cells reflects promiscuous Foxp3 expression in conventional T cells but not reprogramming of regulatory T cells. Immunity 2012, 36, 262–75. doi: 10.1016/j.immuni.2011.12.012 [DOI] [PubMed] [Google Scholar]
- 21. Bailey-Bucktrout SL, Martinez-Llordella M, Zhou X, Anthony B, Rosenthal W, Luche H, et al. Self-antigen-driven activation induces instability of regulatory T cells during an inflammatory autoimmune response. Immunity 2013, 39, 949–62. doi: 10.1016/j.immuni.2013.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gobert M, Treilleux I, Bendriss-Vermare N, Bachelot T, Goddard-Leon S, Arfi V, et al. Regulatory T cells recruited through CCL22/CCR4 are selectively activated in lymphoid infiltrates surrounding primary breast tumors and lead to an adverse clinical outcome. Cancer Res 2009, 69, 2000–9. doi: 10.1158/0008-5472.CAN-08-2360 [DOI] [PubMed] [Google Scholar]
- 23. Devi-Marulkar P, Fastenackels S, Karapentiantz P, Goc J, Germain C, Kaplon H, et al. Regulatory T cells infiltrate the tumor-induced tertiary lymphoïd structures and are associated with poor clinical outcome in NSCLC. Commun Biol 2022, 5, 1416. doi: 10.1038/s42003-022-04356-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Madi A, Wu J, Ma S, Weisshaar N, Mieg A, Hering M, et al. Regulatory T cell-derived interleukin-15 promotes the diversity of immunological memory. Eur J Immunol 2023, 53, e2149400. doi: 10.1002/eji.202149400 [DOI] [PubMed] [Google Scholar]
- 25. Georgiev P, Charbonnier LM, Chatila TA.. Regulatory T cells: the many faces of Foxp3. J Clin Immunol 2019, 39, 623–40. doi: 10.1007/s10875-019-00684-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leng F, Zhang W, Ramirez RN, Leon J, Zhong Y, Hou L, et al. The transcription factor FoxP3 can fold into two dimerization states with divergent implications for regulatory T cell function and immune homeostasis. Immunity 2022, 55, 1354–1369.e8. doi: 10.1016/j.immuni.2022.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rudra D, deRoos P, Chaudhry A, Niec RE, Arvey A, Samstein RM, et al. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat Immunol 2012, 13, 1010–9. doi: 10.1038/ni.2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kwon HK, Chen HM, Mathis D, Benoist C.. Different molecular complexes that mediate transcriptional induction and repression by FoxP3. Nat Immunol 2017, 18, 1238–48. doi: 10.1038/ni.3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Van Gool F, Nguyen MLT, Mumbach MR, Satpathy AT, Rosenthal WL, Giacometti S, et al. A mutation in the transcription factor Foxp3 drives T helper 2 effector function in regulatory T cells. Immunity 2019, 50, 362–377.e6. doi: 10.1016/j.immuni.2018.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu L, Kitani A, Stuelten C, McGrady G, Fuss I, Strober W.. Positive and negative transcriptional regulation of the Foxp3 gene is mediated by access and binding of the Smad3 protein to enhancer I. Immunity 2010, 33, 313–25. doi: 10.1016/j.immuni.2010.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li X, Liang Y, LeBlanc M, Benner C, Zheng Y.. Function of a Foxp3 cis-element in protecting regulatory T cell identity. Cell 2014, 158, 734–48. doi: 10.1016/j.cell.2014.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rudra D, Egawa T, Chong MM, Treuting P, Littman DR, Rudensky AY.. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat Immunol 2009, 10, 1170–7. doi: 10.1038/ni.1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Visekruna A, Volkov A, Steinhoff U.. A key role for NF-κB transcription factor c-Rel in T-lymphocyte-differentiation and effector functions. Clin Dev Immunol 2012, 2012, 239368. doi: 10.1155/2012/239368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kressler C, Gasparoni G, Nordström K, Hamo D, Salhab A, Dimitropoulos C, et al. Targeted de-methylation of the FOXP3-TSDR is sufficient to induce physiological foxp3 expression but not a functional Treg phenotype. Front Immunol 2021, 11, 609891. doi: 10.3389/fimmu.2020.609891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim HP, Leonard WJ.. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med 2007, 204, 1543–51. doi: 10.1084/jem.20070109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Polansky JK, Schreiber L, Thelemann C, Ludwig L, Krüger M, Baumgrass R, et al. Methylation matters: binding of Ets-1 to the demethylated Foxp3 gene contributes to the stabilization of Foxp3 expression in regulatory T cells. J Mol Med (Berl) 2010, 88, 1029–40. doi: 10.1007/s00109-010-0642-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Deng G, Song X, Fujimoto S, Piccirillo CA, Nagai Y, Greene MI.. Foxp3 post-translational modifications and Treg suppressive activity. Front Immunol 2019, 10, 2486. doi: 10.3389/fimmu.2019.02486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hsu TS, Lin YL, Wang YA, Mo ST, Chi PY, Lai AC, et al. HIF-2α is indispensable for regulatory T cell function. Nat Commun 2020, 11, 5005. doi: 10.1038/s41467-020-18731-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hsiao HW, Hsu TS, Liu WH, Hsieh WC, Chou TF, Wu Y-J, et al. Deltex1 antagonizes HIF-1α and sustains the stability of regulatory T cells in vivo. Nat Commun 2015, 6, 6353. doi: 10.1038/ncomms7353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Levine AG, Mendoza A, Hemmers S, Moltedo B, Niec RE, Schizas M, et al. Stability and function of regulatory T cells expressing the transcription factor T-bet. Nature 2017, 546, 421–5. doi: 10.1038/nature22360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science 2009, 326, 986–91. doi: 10.1126/science.1172702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med 2011, 17, 983–8. doi: 10.1038/nm.2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Koch MA, Thomas KR, Perdue NR, Smigiel KS, Srivastava S, Campbell DJ.. T-bet(+) Treg cells undergo abortive Th1 cell differentiation due to impaired expression of IL-12 receptor β2. Immunity 2012, 37, 501–10. doi: 10.1016/j.immuni.2012.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature 2009, 458, 351–6. doi: 10.1038/nature07674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Takahashi R, Nishimoto S, Muto G, Sekiya T, Tamiya T, Kimura A, et al. SOCS1 is essential for regulatory T cell functions by preventing loss of Foxp3 expression as well as IFN-{gamma} and IL-17A production. J Exp Med 2011, 208, 2055–67. doi: 10.1084/jem.20110428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kastner L, Dwyer D, Qin FX.. Synergistic effect of IL-6 and IL-4 in driving fate revision of natural Foxp3+ regulatory T cells. J Immunol 2010, 185, 5778–86. doi: 10.4049/jimmunol.0901948 [DOI] [PubMed] [Google Scholar]
- 47. Duarte JH, Zelenay S, Bergman ML, Martins AC, Demengeot J.. Natural Treg cells spontaneously differentiate into pathogenic helper cells in lymphopenic conditions. Eur J Immunol 2009, 39, 948–55. doi: 10.1002/eji.200839196 [DOI] [PubMed] [Google Scholar]
- 48. Attias M, Al-Aubodah T, Piccirillo CA.. Mechanisms of human FoxP3+ Treg cell development and function in health and disease. Clin Exp Immunol 2019, 197, 36–51. doi: 10.1111/cei.13290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hemmers S, Schizas M, Azizi E, Dikiy S, Zhong Y, Feng Y, et al. IL-2 production by self-reactive CD4 thymocytes scales regulatory T cell generation in the thymus. J Exp Med 2019, 216, 2466–78. doi: 10.1084/jem.20190993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zheng SG, Wang J, Wang P, Gray JD, Horwitz DA.. IL-2 is essential for TGF-beta to convert naive CD4+CD25- cells to CD25+Foxp3+ regulatory T cells and for expansion of these cells. J Immunol 2007, 178, 2018–27. doi: 10.4049/jimmunol.178.4.2018 [DOI] [PubMed] [Google Scholar]
- 51. Pacheco-Gonzalez RM, Avila C, Dávila I, García-Sánchez A, Hernández-Hernández L, Benito-Pescador D, et al. Analysis of FOXP3 gene in children with allergy and autoimmune diseases. Allergol Immunopathol (Madr) 2016, 44, 32–40. doi: 10.1016/j.aller.2015.01.012 [DOI] [PubMed] [Google Scholar]
- 52. Yang JH, Cutler AJ, Ferreira RC, Reading JL, Cooper NJ, Wallace C, et al. Natural variation in interleukin-2 sensitivity influences regulatory T-cell frequency and function in individuals with long-standing type 1 diabetes. Diabetes 2015, 64, 3891–902. doi: 10.2337/db15-0516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sebode M, Peiseler M, Franke B, Schwinge D, Schoknecht T, Wortmann F, et al. Reduced FOXP3(+) regulatory T cells in patients with primary sclerosing cholangitis are associated with IL2RA gene polymorphisms. J Hepatol 2014, 60, 1010–6. doi: 10.1016/j.jhep.2013.12.027 [DOI] [PubMed] [Google Scholar]
- 54. Cerosaletti K, Schneider A, Schwedhelm K, Frank I, Tatum M, Wei S, et al. Multiple autoimmune-associated variants confer decreased IL-2R signaling in CD4+ CD25(hi) T cells of type 1 diabetic and multiple sclerosis patients. PLoS One 2013, 8, e83811. doi: 10.1371/journal.pone.0083811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li CW, Concepcion E, Tomer Y.. Dissecting the role of the FOXP3 gene in the joint genetic susceptibility to autoimmune thyroiditis and diabetes: a genetic and functional analysis. Gene 2015, 556, 142–8. doi: 10.1016/j.gene.2014.11.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Flosbach M, Oberle SG, Scherer S, Zecha J, von Hoesslin M, Wiede F, et al. PTPN2 deficiency enhances programmed T cell expansion and survival capacity of activated T cells. Cell Rep 2020, 32, 107957. doi: 10.1016/j.celrep.2020.107957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fousteri G, Jofra T, Debernardis I, Stanford SM, Laurenzi A, Bottini N, et al. The protein tyrosine phosphatase PTPN22 controls forkhead box protein 3 T regulatory cell induction but is dispensable for T helper type 1 cell polarization. Clin Exp Immunol 2014, 178, 178–89. doi: 10.1111/cei.12393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E.. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol 2009, 182, 2578–82. doi: 10.4049/jimmunol.0803162 [DOI] [PubMed] [Google Scholar]
- 59. Yao R, Ma YL, Liang W, Li HH, Ma ZJ, Yu X, et al. MicroRNA-155 modulates Treg and Th17 cells differentiation and Th17 cell function by targeting SOCS1. PLoS One 2012, 7, e46082. doi: 10.1371/journal.pone.0046082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schjenken JE, Moldenhauer LM, Zhang B, Care AS, Groome HM, Chan HY, et al. MicroRNA miR-155 is required for expansion of regulatory T cells to mediate robust pregnancy tolerance in mice. Mucosal Immunol 2020, 13, 609–25. doi: 10.1038/s41385-020-0255-0 [DOI] [PubMed] [Google Scholar]
- 61. Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell 2010, 142, 914–29. doi: 10.1016/j.cell.2010.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang Y, Feng ZP, Naselli G, Bell F, Wettenhall J, Auyeung P, et al. MicroRNAs in CD4(+) T cell subsets are markers of disease risk and T cell dysfunction in individuals at risk for type 1 diabetes. J Autoimmun 2016, 68, 52–61. doi: 10.1016/j.jaut.2015.12.006 [DOI] [PubMed] [Google Scholar]
- 63. DuPage M, Chopra G, Quiros J, Rosenthal WL, Morar MM, Holohan D, et al. The chromatin-modifying enzyme Ezh2 is critical for the maintenance of regulatory T cell identity after activation. Immunity 2015, 42, 227–38. doi: 10.1016/j.immuni.2015.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bending D, Pesenacker AM, Ursu S, Wu Q, Lom H, Thirugnanabalan B, et al. Hypomethylation at the regulatory T cell-specific demethylated region in CD25hi T cells is decoupled from FOXP3 expression at the inflamed site in childhood arthritis. J Immunol 2014, 193, 2699–708. doi: 10.4049/jimmunol.1400599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yang L, Li B, Dang E, Jin L, Fan X, Wang G.. Impaired function of regulatory T cells in patients with psoriasis is mediated by phosphorylation of STAT3. J Dermatol Sci 2016, 81, 85–92. doi: 10.1016/j.jdermsci.2015.11.007 [DOI] [PubMed] [Google Scholar]
- 66. Morawski PA, Mehra P, Chen C, Bhatti T, Wells AD.. Foxp3 protein stability is regulated by cyclin-dependent kinase 2. J Biol Chem 2013, 288, 24494–502. doi: 10.1074/jbc.M113.467704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nakahira K, Morita A, Kim NS, Yanagihara I.. Phosphorylation of FOXP3 by LCK downregulates MMP9 expression and represses cell invasion. PLoS One 2013, 8, e77099. doi: 10.1371/journal.pone.0077099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li Z, Lin F, Zhuo C, Deng G, Chen Z, Yin S, et al. PIM1 kinase phosphorylates the human transcription factor FOXP3 at serine 422 to negatively regulate its activity under inflammation. J Biol Chem 2014, 289, 26872–81. doi: 10.1074/jbc.M114.586651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Deng G, Nagai Y, Xiao Y, Li Z, Dai S, Ohtani T, et al. Pim-2 kinase influences regulatory T cell function and stability by mediating Foxp3 protein N-terminal phosphorylation. J Biol Chem 2015, 290, 20211–20. doi: 10.1074/jbc.M115.638221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nosbaum A, Prevel N, Truong HA, Mehta P, Ettinger M, Scharschmidt TC, et al. Cutting edge: regulatory T cells facilitate cutaneous wound healing. J Immunol 2016, 196, 2010–4. doi: 10.4049/jimmunol.1502139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Arpaia N, Green JA, Moltedo B, Arvey A, Hemmers S, Yuan S, et al. A distinct function of regulatory T cells in tissue protection. Cell 2015, 162, 1078–89. doi: 10.1016/j.cell.2015.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mock JR, Garibaldi BT, Aggarwal NR, Jenkins J, Limjunyawong N, Singer BD, et al. Foxp3+ regulatory T cells promote lung epithelial proliferation. Mucosal Immunol 2014, 7, 1440–51. doi: 10.1038/mi.2014.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang L, Zhao L, Lv J, Yin Q, Liang X, Chu Y, et al. BLT1-dependent alveolar recruitment of CD4(+)CD25(+) Foxp3(+) regulatory T cells is important for resolution of acute lung injury. Am J Respir Crit Care Med 2012, 186, 989–98. doi: 10.1164/rccm.201202-0261OC [DOI] [PubMed] [Google Scholar]
- 74. Ito M, Komai K, Mise-Omata S, Iizuka-Koga M, Noguchi Y, Kondo T, et al. Brain regulatory T cells suppress astrogliosis and potentiate neurological recovery. Nature 2019, 565, 246–50. doi: 10.1038/s41586-018-0824-5 [DOI] [PubMed] [Google Scholar]
- 75. Glassman CR, Su L, Majri-Morrison SS, Winkelmann H, Mo F, Li P, et al. Calibration of cell-intrinsic interleukin-2 response thresholds guides design of a regulatory T cell biased agonist. Elife 2021, 10, e65777. doi: 10.7554/eLife.65777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wang CJ, Petersone L, Edner NM, Heuts F, Ovcinnikovs V, Ntavli E, et al. Costimulation blockade in combination with IL-2 permits regulatory T cell sparing immunomodulation that inhibits autoimmunity. Nat Commun 2022, 13, 6757. doi: 10.1038/s41467-022-34477-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pesenacker AM, Cook L, Levings MK.. The role of FOXP3 in autoimmunity. Curr Opin Immunol 2016, 43, 16–23. doi: 10.1016/j.coi.2016.07.004 [DOI] [PubMed] [Google Scholar]
- 78. Ferreira LMR, Muller YD, Bluestone JA, Tang Q.. Next-generation regulatory T cell therapy. Nat Rev Drug Discov 2019, 18, 749–69. doi: 10.1038/s41573-019-0041-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mensink M, Schrama E, Cuadrado E, Amsen D, de Kivit S, Borst J.. Proteomics reveals unique identities of human TGF-β-induced and thymus-derived CD4+ regulatory T cells. Sci Rep 2022, 12, 20268. doi: 10.1038/s41598-022-23515-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.