Abstract
New drug discovery is under growing pressure to satisfy the demand from a wide range of domains, especially from the pharmaceutical industry and healthcare services. Assessment of drug efficacy and safety prior to human clinical trials is a crucial part of drug development, which deserves greater emphasis to reduce the cost and time in drug discovery. Recent advances in microfabrication and tissue engineering have given rise to organ-on-a-chip, an in vitro model capable of recapitulating human organ functions in vivo and providing insight into disease pathophysiology, which offers a potential alternative to animal models for more efficient pre-clinical screening of drug candidates. In this review, we first give a snapshot of general considerations for organ-on-a-chip device design. Then, we comprehensively review the recent advances in organ-on-a-chip for drug screening. Finally, we summarize some key challenges of the progress in this field and discuss future prospects of organ-on-a-chip development. Overall, this review highlights the new avenue that organ-on-a-chip opens for drug development, therapeutic innovation, and precision medicine.
Key words: Microfluidics, Drug discovery, In vitro models, Microphysiological systems, Toxicity assessment, Bioprinting, Drug safety, Human-on-a-chip
Graphical abstract
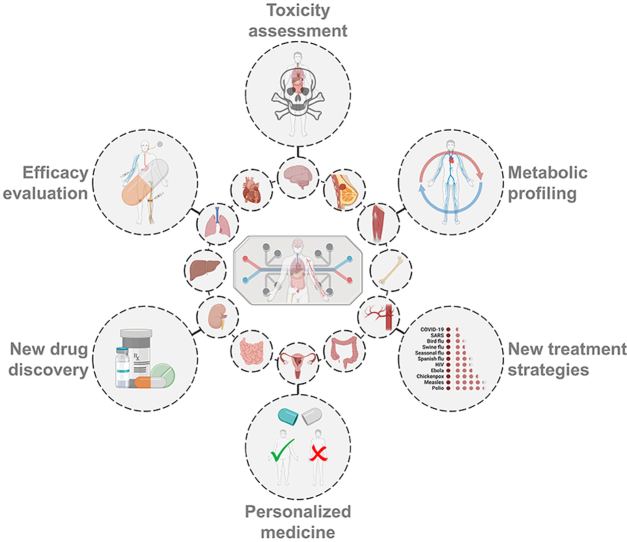
This review summarizes the recent research works on various organ-on-a-chip systems for drug screening mainly based on drug-induced toxicity, therapeutic efficacy and drug metabolism.
1. Introduction
New drugs are continually being developed in the pharmacological industry to meet urgent unmet needs in different therapeutic areas. However, launching a new drug to market is a lengthy and expensive process1: it takes 13–15 years and up to $2.8 billion for a new drug to be approved and commercialized, with 90% of drug candidates failing during clinical trials2, which is the most concerning problem of the modern pharmaceutical industry3. The new drug development procedure typically includes three major stages: early-stage drug discovery, preclinical drug development, and clinical trials. One of the key issues in the drug development procedure is drug screening4, which aims to select the appropriate drug candidates from a large pool of available lead compounds based on their toxicity, safety, efficacy, clearance, etc. With efficient drug screening prior to expensive human clinical trials, it is possible to dramatically reduce the cost and time of new drug development.
Many approaches have been developed to achieve efficient drug screening, among which animal-based in vivo models and cell-based in vitro models5,6 are the most commonly applied methods. Animal testing allows for systemic in vivo drug responses through drug metabolism and pharmacokinetic studies7,8. However, it does not allow comprehensive prediction of drug-induced toxicity, potential side effects, and treatment efficiency on the human body due to interspecies differences in physiology and metabolism. Furthermore, animal studies are always time-consuming, labor-intensive, expensive, and limited by ethical controversies. Using human cells could, to a great extent, avoid cross-species differences9. Most in vitro models, which rely on 2D cell monolayers or 3D cell cultures, are a cost-effective and simplified method for drug screening. Nevertheless, they are incapable of recapitulating the complex structure of native tissue and systemic physiological processes, leading to poor predictive ability.
With the development of microfabrication technology, microfluidics has experienced explosive advances and extensive applications in a wide range of biological domains, such as the isolation of extracellular vesicles10, biomarker quantification11,12, cancer diagnosis13, and multiplex assays11,14. The combination of microfluidics, tissue engineering, and cell biology has contributed to the birth of a microengineered organ-on-a-chip (OOAC) platform15, a novel in vitro bionic on-chip system that mimics the in vivo structures and primary functions of human organs. In addition, multiple organs and tissues could be connected to create multi-organ-on-a-chip (multi-OOAC) or even human-on-a-chip, which is capable of emulating complex organ–organ interactions, thus permitting a more systematic study of drug metabolism and pharmacokinetics16,17. Fabricated by using human cells and allowing for organ-level or systemic physiological simulations, OOACs exhibit significant advantages over traditional animal-based models and cell-based models, in terms of prediction capability, fabrication cost, operation complexity, test duration, ethical issue, etc. OOAC technique is considered a next-generation in vitro model, demonstrating tremendous potential in disease research and drug screening as an alternative or supplement to animal models or even as part of clinical trials.
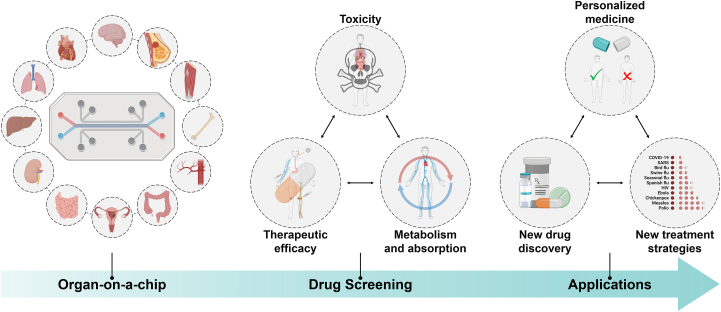
In this review, we summarize recent innovations and advanced research in organ-on-a-chip devices for drug screening (Fig. 1). First, we provide a snapshot of organ-on-a-chip engineering, focusing on its definition and design considerations, including cell sources, materials and fabrication techniques, stimulations, and sensing techniques. Then, we provide an overview of a variety of organ-on-a-chip categories, mainly including the heart-, liver-, kidney-, brain-, other single-organ-, multi-organ-, and tumor-on-a-chip. The main characteristics of each organ-on-a-chip system are introduced, and the emphasis is placed on their contributions to drug-induced toxicity, drug therapeutic efficiency, and drug metabolism toward drug screening. Finally, we discuss the challenges to be addressed in further studies and formulate a view on the prospects of this field.
Figure 1.
Scheme of organ-on-a-chip for drug screening.
2. Organ-on-a-chip engineering
OOAC, first proposed in 201018, is an artificial physiological system created on a tiny chip through tissue engineering and microfluidic technologies to mimic the essential functions of human organs. According to their modality and functionality, OOACs can be divided into four major types: i) single-organ-on-a-chip emulating key functions of unique tissue or organ, such as heart19, liver20, kidney21, lung22, and gut-on-a-chip23; ii) multi-organ-on-a-chip combining multiple tissues or organs to reproduce the systemic interactions occurring in vivo, e.g., liver-heart24, liver-heart-lung25, heart-vessels26, intestine-liver-cancer-on-a-chip27; iii) tumor-on-a-chip mimicking the structure and microenvironment of tumor tissues, e.g., breast-cancer28, colorectal-tumor29, glioblastoma30, and metastasis-on-a-chip31; and iv) body-on-chip or human-on-a-chip aiming at deciphering the human body system32.
Owning advantages of traditional microfluidics, such as ultimate miniaturization, high integration, and low cost, OOACs also enable precise control of cellular and tissue architecture, co-culture of various cells, in vivo-like characterization avoiding animal models, incorporation of microsensors for real-time monitoring of tissue functions, personalized estimation of physiological responses to drugs, etc33,34. The past decade has witnessed the rapid development of OOACs and their expanded employment in disease modeling, disease progression study, drug discovery, risk assessment, metastasis investigation, etc35, 36, 37. Herein, we focus on the drug screening applications of OOACs. Other excellent papers are available for readers interested in topics beyond the scope of this review38, 39, 40, 41.
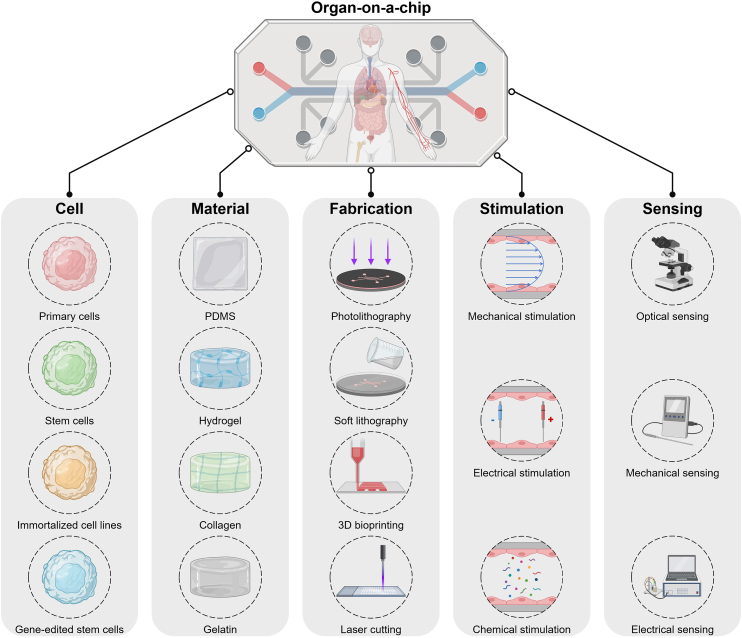
Briefly, a typical OOAC for drug screening consists of four main parts: 3D cell tissues, microfluidic systems, stimuli, and sensing components42 (Fig. 2). Numerous aspects should be taken into consideration when developing an application-specific OOAC43. First, cell sources are the first issue to address in the concept design of OOAC, which ensures the reconstruction of 3D organ-level tissue structures with functionality satisfying the desired context of use44. Then, microfluidic systems providing support for cell culture, tissue construction, and microenvironment recapitulation should be precisely designed to accurately manipulate dynamic physiological parameters, including flow rate, shear stress, substance delivery, and chemical concentration gradients45. Next, diverse stimuli, such as electrical, physical, or chemical signals and media flow, are applied to physiological microsystems to maintain the in vivo biological environment or to promote microtissue maturation and function. Finally, the integration of output components, such as microsensors, is essential to detect organ-related metabolites, assess cellular responses to drugs, and monitor real-time organ functionality. Apart from these major design considerations, context of use, linkage of multiple components, flow control, dimension and shape, universal culture medium, etc., should also be carefully considered in order to implement a validated OOAC device.
Figure 2.
Schematic representation of key components for organ-on-a-chip engineering.
2.1. Cell sources
The human cell types used in OOACs are determined by a series of factors, including cell availability, viability, culture difficulty, and the ability to form functional tissues. The most commonly used cell sources include human primary cells, immortalized cells, and stem cells46,47. In addition, along with advancements in genetic engineering technologies, gene-edited stem cells have emerged as an innovative cell source for OOACs, paving the way for disease-targeted personalized and precision medicine. Each cell type has its own advantages and limitations (Table 1).
Table 1.
Cell sources for OACC engineering.
| Cell type | Advantage | Limitation |
|---|---|---|
| Primary cells | Derived from living organ | Less available |
| Patient-specific | Long pre-culture period | |
| Mature phenotype | Finite lifespan | |
| Difficult for long-term culture | ||
| Immortalized cell lines | Derived from adult | Different sources for each tissue type |
| Mature phenotype | Nonrepresentative of in vivo physiology | |
| Noninvasive | Not patient-specific | |
| Established culture protocols | Time-dependent genotypic and phenotypic change | |
| Readily available | Homogeneous population | |
| ESC | Long-term culture | Derived from embryos |
| Pluripotent differentiation ability | Ethically regulated | |
| Complex differentiation protocols | ||
| Immature phenotypes | ||
| Limited quantity | ||
| ASC | Derived from adult | Complex differentiation protocols |
| Patient-specific | Immature phenotypes | |
| Less-invasive | Difficult for long-term culture | |
| Multipotent differentiation ability | ||
| iPSC | Derived from adult | Complex differentiation protocols |
| Patient-specific | Immature phenotypes | |
| Pluripotent differentiation ability | Viral reprogramming | |
| Noninvasive | ||
| Gene-edited iPSC | Derived from adult | Complex differentiation protocols |
| Patient-specific | Immature phenotypes | |
| Disease-specific | Time-consuming and expensive | |
| Noninvasive |
2.1.1. Human primary cells
Primary cells are directly extracted from living organs or tissues (e.g., biopsy material or solid tumor tissue) without any modification. These cells are phenotypically mature and fully functional, with high bioactivity and heterogeneity, and carry genetic information of the donors. Compared with other cell types, primary cells are most representative of the in vivo state of the functional organ and promise a more reliable and personalized drug screening result. However, primary cells are difficult to obtain, have limited quantity, display declined functionality with time, and require a long pre-culture period and specialized culture media to retain their phenotypes, which prevents their extensive employment in OOACs. Recently, a few accessible primary human cells, such as lung cells48, intestinal cells49, and hepatic cells50, have been used in OOAC fabrications.
2.1.2. Immortalized cell lines
Immortalized cell lines generally refer to standardized cell lines that can proliferate indefinitely over generations. These cells are readily available, genetically identical, and easy to culture, enabling massive assays and ensuring reproducible results. However, immortalized cells show several fatal disadvantages that limit their use in OOACs. First, they are just approximations of the in vivo primary cells and incapable of accurately recapitulating the functional characteristics of the organ that they intend to represent. Second, they experience genotypic and phenotypic changes during passages, leading to inconsistent drug screening results with clinical data51. Finally, they are unable to represent heterogeneous cellular responses in vivo due to their homogeneity in genetics, epigenetics, and phenotypes. These cells are usually used in the design and optimization stages of OOAC development, in the initial screening steps, or when better options such as primary cells or stem cells are not available.
2.1.3. Stem cells
Stem cells are self-renewing cells possessing the potential to differentiate into specialized cell types of a tissue or organ in vivo52. Based on their differentiation potential, they can be divided into three major types: totipotent cells that can produce an entire organism; pluripotent cells that can give rise to all cell types found in the organism; and multipotent cells that are capable of developing into limited cell types53. Stem cells can also be classified according to their sources and mainly include embryonic stem cells (ESCs) from embryos of 3–5 days old, adult stem cells (ASCs) found in adult tissues such as bone marrow or fat, and induced pluripotent stem cells (iPSCs) reprogrammed in the laboratory54.
2.1.3.1. Embryonic stem cells
Embryonic stem cells (ESCs) are cells derived from preimplantation-stage embryos55. Human ESCs can be either totipotent or pluripotent, depending on their “age” after fertilization. Cells during the first few divisions are capable of generating a viable embryo along with its extraembryonic tissues, such as the placenta56, being totipotent. After a few days (usually 3–5 days for humans), the embryo becomes a blastocyst, a hollow sphere containing an inner cell mass (ICM) with approximately 150 cells, from which human ESCs are primarily obtained57,58. These cells can differentiate into any type of human cells but not extraembryonic tissues and are pluripotent.
ESCs have unlimited differentiation potential and allow long-term culture and proliferation, making them excellent cell sources for in vitro studies of early embryonic development, cell-based drug testing, regenerative therapy of damaged cells and tissues, etc59. However, as derived from human embryos, the use of ESCs has long been an ethically controversial issue and is therefore strongly regulated. In addition, the generation of large amounts of diverse cell lines and the control of ESC differentiation to obtain desired cell types are still technically challenging. The resulting cells often have barely functional and immature structural phenotypes46. It is still difficult to apply ESCs in precision medicine for disease modeling and therapeutic drug evaluations47.
2.1.3.2. Adult stem cells
Adult stem cells (ASCs) are found in many adult tissues, such as bone marrow, blood, brain, heart, lung, skin, fat, muscle, and intestine. They are multipotent stem cells and can differentiate into a limited number of mature cell types within their organ of origin. Their capability to regenerate the tissue's structures and functions in the case of injury ensures the maintenance of tissue homeostasis and makes ASCs particularly useful in therapeutic applications60. One of the most famous ASCs is hematopoietic stem cells, which have been used in the therapy for chronic myeloid leukemia owing to their capacity to generate entire hematopoietic lineages.
The main advantage of ACSs is that they can be directly extracted from human tissues with little harm to the donor and allow for autologous transplantation to avoid the risk of rejection. In addition, it is possible to transform ASCs into pluripotent stem cells as an alternative to human embryos. Although ASCs show great promise in OOAC development, challenges remain in the obtention of sufficient quantities due to their rarity in tissues and difficulty in long-term culture in vitro.
2.1.3.3. Induced pluripotent stem cells
Induced pluripotent stem cells (iPSCs) are somatic cells endowed with pluripotency through reprogramming61, 62, 63, processing the ability to give rise to all cell types in the body. The generation of patient-specific iPSCs provides a competent approach to fabricate personalized OOACs for precision medicine (including tissue regeneration and cell transplantation), which offers tremendous opportunities to shape the future of healthcare64,65.
iPSCs that are also pluripotent show several advantages over ESCs. For example, abundant somatic cells are available from volunteer donors, which ensures an unlimited quantity of iPSCs and avoids ethical implications, as the harvesting procedure is usually harmless to human individuals. A variety of iPSC-derived cells, such as brain microvascular endothelial cells66 and cardiomyocytes67,68, have shown remarkable applications in OOAC construction. Nevertheless, the widespread use of iPSCs is hindered by obstacles, including the lack of technology to differentiate immature iPSCs into any mature somatic cells and the use of retroviruses associated with cancer for iPSC generation.
2.1.4. Gene-edited stem cells
Gene-edited stem cells have come to the fore along with the advances of novel gene-editing technologies, notably the CRISPR-Cas system69, which allowed removing, inserting, or modifying genetic information in a DNA sequence. This innovative and attractive strategy has revolutionized both biological and pharmaceutical research, and offered novel opportunities for the tracking of gene-associated diseases and cancers, from diverse aspects, including drug development, drug screening, gene therapy, therapeutic improvement, and immune response strategy70,71.
The combination of genome editing and iPSCs72 has given rise to a variety of patient-derived genome-corrected cells targeted for specific diseases, such as β-thalassemia, Alzheimer's disease, HIV infection, and Duchenne muscular dystrophy73. The employment of gene-edited iPSCs in OOACs, taking advantage of the outstanding merits of gene editing, iPSCs, and OOACs, opens a new route to implement personalized in vitro human tissue models instead of animal models and to promote precision medicine74, 75, 76. Nevertheless, these cells may not fully recapitulate all features of mature cells in humans77, and the selection of correlated clones is often labor-intensive and time-consuming.
2.2. Materials and fabrications
The microfluidic chip in an OOAC, providing support for cell culture and tissue manufacturing, should be fabricated with materials that are nontoxic to cells, gas-permeable for cellular respiration, and optically transparent for observation. Besides, other criteria, such as material cost, fabrication complexity, and chemical and physical stability, should also be taken into consideration. A great variety of materials have been employed in chip fabrication, including inorganic materials such as silicon and glass, elastomers such as polydimethylsiloxane (PDMS), polyurethane methacrylate (PUMA) and thermoset polyester (TPE), and plastics such as polycarbonate (PC), poly (methyl methacrylate) (PMMA), and polystyrene (PS)78. Among these materials, PDMS has been the most widely used, benefiting from its biocompatibility, oxygen permeability, transparency, flexibility, low cost, and simplicity of processing. Nevertheless, PDMS can absorb small hydrophobic molecules79 and sometimes drug molecules, which may disrupt the drug screening results80. Recent studies and future trends are focused on surface modification methods, complementary materials, or substitutions of PDMS81 to overcome this issue.
Biomaterials play an important role in tissue engineering to construct the 3D scaffold architecture and artificial extracellular matrix (ECM), mimicking the native ECM to ensure the cellular assemblage and formation of functional tissue structure82,83. The ideal biomaterials must be biocompatible, noncytotoxic to cells, adequately porous, permeable, biodegradable, mechanically strengthened to support cells, etc. A great variety of biomaterials, including protein-based materials such as collagen, gelatin, fibrin, and hyaluronic acid, polysaccharide-based materials such as chitosan and alginate, and synthetic materials such as hydrogels, have been reported to be widely used in OOACs, each having its own benefits and limitations46 (Table 2). Recently, researchers have also made efforts to develop scaffold-free OOACs, maintaining the main organ functions84, which significantly simplifies the fabrication procedure.
Table 2.
Common materials for OOAC fabrication.
| Biomaterial | Advantage | Disadvantage |
|---|---|---|
| Collagen | Natural origin | Sensitive degradability to enzyme attack |
| High tensile strength | Cross-linking to enhance stability | |
| High flexibility | ||
| Gelatin | Natural origin | Weak thermostability |
| Cost-effective | Poor mechanical stability | |
| Water absorbent | Lack of immune responses | |
| Increased cell adhesion | ||
| Fibrin | Natural origin | Tunable structure and function |
| Rapid biodegradability | ||
| Easy fabrication | ||
| Hyaluronic acid | Natural origin | Weak cell adhesion |
| High hydrophilicity | Weak mechanical properties | |
| High viscosity | Limited immunogenicity | |
| High porosity | ||
| Easy to form hydrogel | ||
| Chitosan | Natural origin | Weak mechanical strength |
| Abundant quantity on earth | ||
| Inherent antimicrobial properties | ||
| Alginate | Natural origin | Lack of biologic recognition sites |
| Gentle gelling property | Uncontrollable mechanic properties | |
| Synthetic biomaterial | Tunable chemical, physical, and mechanical properties | Lack of cell adhesion ligands |
| Tunable degradation rates | Poor hydrophilicity | |
| Adjustable crosslinking level | Undesirable degradation products | |
| Reproducibility | ||
| Controllable fabrication process |
There are two main strategies to fabricate an OOAC. The first strategy is a multiple-step procedure, mainly including the manufacturing of microfluidic chips by hot embossing, injection molding, laser-cutting, photolithography, lithography, etc., followed by on-chip cell culture and tissue reconstruction. The second strategy is a one-step technique to continuously fabricate an OOAC as an entirety, realized by 3D bioprinting85. Bioprinting allows layer-by-layer deposition of cells, biomaterials, biomolecules, etc., to generate a complex OOAC according to computer design with great accuracy86. It is among the most advanced technologies for producing biomimetic cellular constructions and the most promising candidates for OOAC fabrication. However, cellular physiological performance may be affected during bioprinting due to exposure to mechanical or thermal stresses, which is the most important challenge for bioprinting87.
2.3. Stimulations
In vivo, cells and tissues are subjected to a combination of various mechanical, electrical, and chemical stimuli88. Applying appropriate stimulations is essential to induce the development of the cell phenotype, mimic the appropriate organ functions, and guarantee the correct response to a drug candidate in OOAC.
All cells and tissues in the body experience varied mechanical forces, such as fluid shear force, tensile stretch, compression, hydrostatic pressure, interstitial fluid flow, and contraction89. Mechanical stimulation is well established to determine tissue function in many major biological systems, such as the cardiovascular system, respiratory system, and digestive system90. The duration, frequency, and amplitude of mechanical forces applied to an OOAC device through external pumping, integrated pumping, gravity flow, etc., are important to mimic in vivo biomechanical cues and regulate cellular behaviors and pharmaceutical responses.
Electrical stimulation plays an essential role in electroactive tissues, such as neuronal, muscle, and cardiac tissues. One typical example of applying electrical stimulation in the development of OOACs concerns the heart. Electrical conduction in the heart ensures stable ventricular myocyte contractibility, and on-chip electrical stimulation is necessary to facilitate cardiomyocytes (CM) maturation and to develop their conductive and contractile properties91 by precisely regulating the time, amplitude, and frequency of the stimulation.
In vivo, cells that are in close contact with each other are surrounded by complex ECM and follow chemical stimulation, such as continual oxygen, ions, and nutrient supplies from blood flow, to maintain cellular homeostasis and tissue growth. The most familiar biochemical stimulation is growth factor (GF)92, which plays a significant role in skeletal muscle tissue engineering by influencing cell migration, proliferation, differentiation, and apoptosis. Chemical stimulation is usually delivered to an OOAC through fluid flows93,94.
2.4. Sensing techniques
The characterization of drug-induced responses of cells and organs is the endpoint of drug screening and plays a critical role in an OOAC platform. Traditional off-chip analytical techniques, such as polymerase chain reaction (PCR), enzyme-linked immunosorbent assay (ELISA), and mass spectrometry (MS)95, are not capable of continuously evaluating the physiological and metabolic behaviors of microtissues, which may change dynamically under the effect of administered drugs. Recently, the integration of microsensors into an OOAC system has gained enormous attention, since it allows for not only sensitive detection but also real-time monitoring of the performances of cells, microtissues, and ECM in a minimally invasive way96,97. A great variety of integrated sensors have been developed for the measurement of physical parameters such as heart-beating intervals, for the assessment of cellular characteristics such as viability and morphology, and for the quantification of organ-related biomarkers such as cytokine, cholesterol, and microRNA, to name a few98. Biosensors compatible with OOACs can be mainly classified into optical, mechanical, and electrical sensors, according to their sensing principle99.
Optical biosensors, based on the measurement of optical signals (such as absorption, reflection, infrared, fluorescence, and chemiluminescence) produced during biological reactions, are the most commonly used sensing technique in OOAC systems. They allow for both real-time culture monitoring and endpoint detection of numerous metabolic states, such as pH, level of glucose or lactate, and dissolved gases. The optical sensing method to estimate the contractility of a heart-on-a-chip system is also one of the most widespread and basic techniques for recording the contraction of beating cells100.
Mechanical biosensors characterize forces, displacements, mass changes, and cellular mechanical properties such as stress and strain101. These sensors are mainly used in OOACs for the analysis of membranes and the assessment of drug-induced responses of certain cell types, e.g., cardiomyocytes and hepatocytes102.
Electrical sensors refer to a group of sensing devices that transduce biological events to electrical signals and are more suitable for recording and processing signals than optical sensors since no light source or optical detector is needed. Numerous electrical sensors have been developed, including transepithelial electrical resistance (TEER) impedance-based biosensors, photoelectric pH sensors, and electrochemical affinity-based biosensors97, to name a few, and are widely used in the monitoring of metabolites, cell viability, etc.
Additionally, many other sensors have been shown to be successfully integrated into OOACs for more efficient drug screening. Readers interested in this topic could refer to other excellent reviews for more details103,104.
3. Drug screening based on organ-on-a-chip platforms
This section is dedicated to providing an overview of the recent advances in OOACs for drug screening, mainly focusing on the assessment of therapeutic effect, toxicity (including direct toxicity caused by the drug and indirect toxicity related to its metabolites), and metabolism. Although a considerable variety of OOACs have been developed, the heart-, liver-, kidney-, and brain-on-a-chip are the most commonly investigated organs in the domain of drug screening, as they are the four major target organs of drug toxicity. Therefore, herein, a strong emphasis is placed on these four OOAC systems. Moreover, several representative studies on other single-organ-on-a-chip systems, such as lung-, intestine-, skin- and blood-vessel-on-a-chip systems, as well as multi-organ-on-a-chip systems, are briefly discussed. Finally, recent works on tumor-on-a-chip for anticancer drug screening are also highlighted.
3.1. Heart-on-a-chip
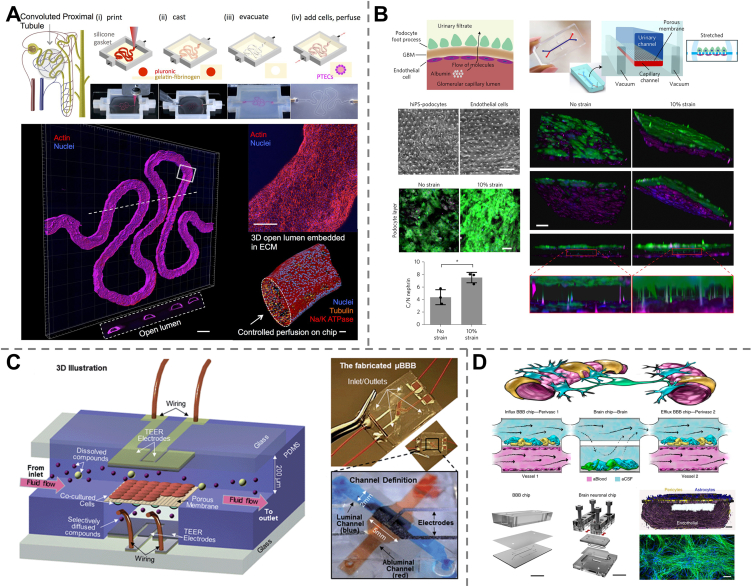
Cardiovascular diseases (heart diseases) are reported to be the leading cause of mortality in the world, leading to approximately 17.9 million deaths annually and representing 32% of all global deaths105. Besides, drug-induced cardiotoxicity is one of the most common causes of drug failure and drug attrition, suggesting the limitations of current drug-evaluation approaches in predicting cardiac effects106. Therefore, there is an urgent need to discover better drugs for heart diseases and to test the cardiac safety of other drugs before clinical testing, which requires an efficient and accurate tool for drug-induced cardiotoxicity assessment. Heart-on-a-chip (heart-OAC), an in vitro device recapitulating cardiac tissue-level physiology and functionality, is emerging for this purpose.
The heart is the central organ of the human circulatory system, pumping blood to the whole body as it beats. The main cell types of the heart and cardiovascular system include CMs, fibroblasts (FBs), endothelial cells (ECs), pericytes (PCs), and smooth muscle cells (SMCs), whose morphology and physiology may change with a variety of stimuli, including mechanical, biological, chemical, and electrical stimulations107. Among these cells, CMs, which are responsible for a heart pumping, are the most popular cardiac cells used in OOACs since the beating of CMs can directly reflect drug effects on heart contractions. Cardiomyoblasts (the precursors of CMs), capable of generating transverse striated heart muscle cells108, are also often utilized in OOACs. However, it is difficult to obtain a sufficient quantity of primary CMs from human hearts and to expand them in vitro, which is the major challenge for developing heart-OACs and could be addressed by applying human iPSC-derived CMs (hiPSC-CMs)87. In addition, electrodes are usually employed in contact with the cells to ensure electrical simulations, since electrical signaling is significantly important in tissue construction, maintenance, and regeneration. A variety of characterization techniques, such as electrochemical and optical sensors, are employed to monitor the functionality of heart-OACs based on cell viability, cell morphology, cell proliferation, contraction and beating frequency, level of calcium ions, expression of cardiac markers, etc100.
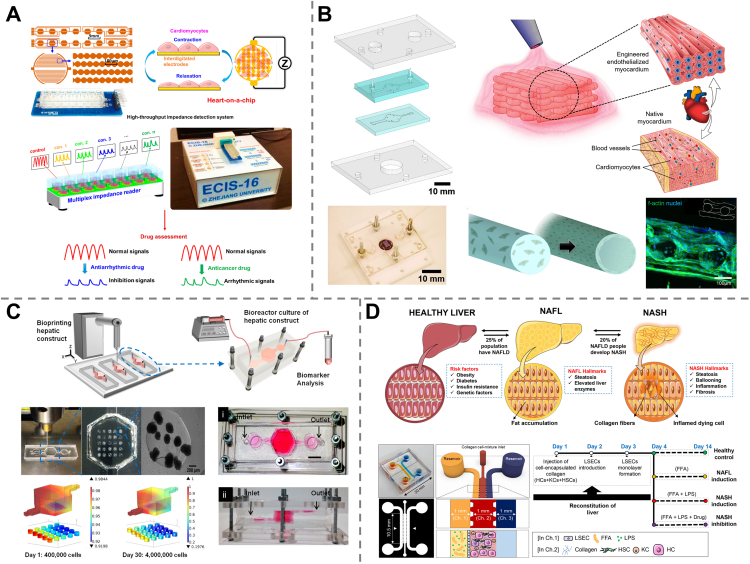
Screening of cardiovascular agents based on their therapeutic efficacy could be performed on heart-OACs. For example, a heart-OAC device was fabricated using primary neonatal rat cardiomyocytes along with a high-speed impedance detection component to evaluate the responses of CMs to drugs. After treatment with verapamil (an antiarrhythmic drug), both the contractility and beating rate of CMs were observed to be decreased, which matched well with the approved effect of verapamil, proving the function of the drug efficacy test of the heart-OAC109 (Fig. 3A). In contrast to verapamil, isoproterenol has been well characterized for the treatment of bradycardia, and its positive inotropic effect on cardiac contractility has also been demonstrated using a heart-OAC platform110. Besides, many non-cardiovascular drugs (e.g., antipsychotics, analgesic drugs, and anticancer drugs) exhibit high cardiotoxicity or can cause severe cardiac side effects, such as cardiac inflammation, delay of cellular depolarization, and tachycardia. The development of the heart-OAC shows great promise in screening such drugs according to their cardiotoxicity. A heart-OAC created with human iPSC-derived CMs and endothelial cells through a 3D bioprinting technique was reported to engineer endothelialized myocardial tissues and enabled the cardiotoxicity assessment of a well-known chemotherapeutic drug, doxorubicin, based on the measurement of the CM beating rate via optical microscopy111 (Fig. 3B). Another Heart-OAC was developed by culturing human iPSC-derived CMs on micromolded gelatin to form laminar cardiac tissues and was applied to study tissue-level electrophysiological responses based on cardiac field potentials. The drug responses of the cardiotoxic prodrug terfenadine along with its metabolite fexofenadine (non-cardiotoxic) were tested in this device, which showed correlated results with clinical data, demonstrating the capability of heart-OACs for drug cardiotoxicity screening112.
Figure 3.
Heart- and liver-on-a-chip device for drug screening. (A) A heart-OAC measuring the responses of CMs to drugs and proving the antiarrhythmic efficacy of verapamil. Reprinted with the permission from Ref. 109. Copyright © 2016 MDPI (Basel, Switzerland). (B) A 3D bioprinted heart-OAC to assess doxorubicin-induced cardiotoxicity based on CM beating rate. Reprinted with the permission from Ref. 111. Copyright © 2016 Elsevier. (C) A liver-OAC demonstrating the hepatoxicity of APFP. Reprinted with the permission from Ref. 113. Copyright © 2016 IOP Publishing Ltd. (D) A liver-OAC recapitulating nonalcoholic steatohepatitis for testing the therapeutic effect of elafibranor. Reprinted with the permission from Ref. 114. Copyright © 2021 Wiley Periodicals LLC.
3.2. Liver-on-a-chip
The liver is the central organ of drug metabolism and plays a crucial role in detoxification, thereby serving as the primary target of drug toxicity. Drug-induced liver injury may account for 50% of all acute liver failures115 and 10% of deaths and has been one of the most frequent causes of drug failure and withdrawal. In vivo hepatotoxicity accounted for 90% of the drug failures after phase I clinical trials and accounted for 32% of all cases of post-approval drug withdrawals between 1975 and 2007116. Moreover, the liver may suffer severe injury due to chronic diseases and viral infections. Liver-on-a-chip (liver-OAC) is an advanced in vitro model that provides better a prediction of hepatoxicity than traditional animal models, and the extended use of liver-OACs in the pharmaceutical industry offers opportunities to increase the success rate for new drug discovery.
The liver is composed of parenchymal and nonparenchymal cells. Parenchymal cells, referring to hepatocytes, are the main constituent cells of the liver and maintain most liver functions. Nonparenchymal cells, such as hepatic stellate cells, sinusoidal endothelial cells, and Kupffer cells, communicate closely with hepatocytes to form extracellular matrix proteins, produce liver growth mediators, dominate liver regeneration, etc. In terms of cell sources, most current liver-OACs use primary hepatocytes for drug screening and toxicological testing117, and hepatic stellate cells, as a neural companion of hepatocytes118, are also often co-cultured. Hepatic microenvironment mimicking, such as perfusion of fresh physiological flow media, is also a crucial parameter for liver-OACs, as adequate blood flow and oxygen tension play important roles in maintaining liver functionality119. Besides, a variety of advanced technologies, such as spheroidal culture and 3D bioprinting, have been applied in liver-OAC engineering to make it more realistic120,121. A range of cytotoxicity biomarkers, such as ATP, albumin, miR-122, and α-GST, were further analyzed to evaluate the functions of liver-OACs.
Multiple liver-OAC devices have been developed and used to study hepatotoxicity. Acetaminophen (APFP) is one of the most frequently prescribed analgesic and antipyretic drugs worldwide. However, it can potentially cause fetal liver damage and is therefore often chosen as a model drug122 for toxicity screening in liver-OAC devices. A microfluidic biochip lined with HepG2/C3A cells was proposed to investigate the APFP injury pathway. Calcium homeostasis perturbation, lipid peroxidation, and cell death in the presence of APFP were observed, which was the first example of liver-OAC for toxicity assessment123,124. More recently, by using a bioprinting technique, another HepG2/C3A cell-based liver-OAC was developed and utilized to demonstrate the hepatoxicity of APFP leading to a reduction in cell density, metabolic activity, and biomarker production113 (Fig. 3C). Research works making use of other drug models, such as chlorpromazine (an antipsychotic drug) and tacrine (a medicine for Alzheimer's disease), to demonstrate the capability of liver-OAC devices in drug-induced hepatotoxicity screening have also been reported117.
Other works have been reported to perform drug screening based on therapeutic efficacy against liver diseases, such as nonalcoholic fatty liver disease (NAFLD). A liver-OAC comprising hepatocytes and white adipocytes was developed to emulate the NFLAD in different human metabolic states, including healthy, diabetic, obese, and proinflammatory states. This platform was used to evaluate the preclinical efficacy of metformin and the revealed inhibition of hepatic steatosis by metformin at increased concentrations125. Another liver-OAC was implemented by co-culturing four human primary liver cells (hepatocytes, Kupffer cells, liver sinusoidal endothelial cells, and hepatic stellate cells) and exposing them to a lipotoxic environment to recapitulate nonalcoholic steatohepatitis (NASH) to further investigate the therapeutic effect of Elafibranor, a new anti-NASH drug still under development. Inhibition of disease progression and fibrosis was observed, which is consistent with clinical trials, suggesting the potential of liver OACs for understanding disease pathogenesis and developing therapies114 (Fig. 3D).
The liver plays a crucial role in drug metabolism, and liver-OACs that mimic the main functions of the liver are expected to be capable of predicting not only drug hepatoxicity and efficacy but also drug metabolism and clearance126. A liver-OAC co-culturing cryopreserved human primary hepatocytes and inflamed human Kupffer cells was developed to investigate the anti-inflammatory effect of glucocorticoids by stimulating the metabolism of hydrocortisone. Both phase I and phase II metabolites, as well as pharmacokinetic parameters such as half-life, elimination rate, and clearance, were evaluated and found to be correlated with clinical data127. Another interesting work integrating a cancer-on-a-chip module with a liver-OAC to investigate the metabolism of simvastatin by the liver. Cancer cell viability was obviously reduced when simvastatin was perfused into the cancer-on-a-chip after flowing through the artificial liver, demonstrating the hepatic metabolism effect of the liver-OAC to convert prodrug simvastatin into the active drug atorvastatin128.
3.3. Kidney-on-a-chip
The kidney regulates blood filtration and urine production to remove certain metabolites, wastes, and toxins from the blood. It is also an important organ for metabolic activities and drug clearance, thus is also a major target of drug toxicity. Drug-induced nephrotoxicity (DIN) is the cause of approximately 20% of acute renal failure cases. DIN also accounts for approximately 25% of the reported severe adverse side effects and approximately 19% of total drug attrition during the phase III trial129. The identification of nephrotoxic therapeutic agents in vitro would allow the minimization of severe kidney injury in the clinical stage. To more efficiently predict nephrotoxicity in the preclinical stage, kidney-on-a-chip (kidney-OAC) has been developed as a novel in vitro model and has shown great potential in drug-induced nephrotoxicity assessment for drug screening130.
The kidney is one of the most sophisticated organs and is composed of more than ten cell types (glomerular cells, proximal tubule cells, renal endothelial cells, etc.). iPSCs131 have been demonstrated to be successfully employed for kidney-OAC construction. Co-culture of various cell types, which enables the investigation of intercellular interactions, specific signaling pathways, and immune cell recruitment, is required to recapitulate renal function and physiology. In addition to the cell type, liquid composition, substance delivery method, fluid dynamics and fluid shear stress132 are also important parameters for a kidney-OAC. Apart from cell death, nephrotoxicity could also be evaluated based on the different statuses of cell polarity, membrane integrity, and mitochondrial function133.
The clinical use of various drugs, especially antimicrobials, could cause kidney damage. The proximal tubule is the major target of many nephrotoxicants, and numerous kidney-OAC platforms have been developed as tubule-on-a-chip for the assessment of drug-induced nephrotoxicity. A 3D bioprinted kidney proximal-tubule-chip was developed with long-term perfusion of cell media, recapitulating the in vivo phenotype and function of proximal tubules, for more accurate nephrotoxicity prediction. The introduction of cyclosporine A, a common drug against transplant rejection, into the chip led to the disruption of epithelial barrier function, suggesting the promising capability of the device in drug screening based on proximal tubule-related toxicity134 (Fig. 4A). Primary kidney proximal tubule cells were seeded in a Nortis device to mimic the kidney tubule microphysiological system for toxicity evaluation of polymyxin B and its two structural analogs that are still in clinical development, showing great promise of kidney-OAC platforms for safety testing of new chemical entities135. Other drug models, such as gentamicin, cisplatin, tenofovir, tobramycin, and cyclosporin A, were also used to demonstrate drug screening by tubule-on-a-chip136, 137, 138.
Figure 4.
Kidney- and brain-on-a-chip for drug screening. (A) A 3D bioprinted kidney proximal-tubule-chip for nephrotoxicity prediction caused by cyclosporine A. Reprinted with the permission from Ref. 134. Copyright © 2016 Springer Nature. (B) A glomerular-on-a-chip mimicking the podocyte injury caused by the anticancer drug adriamycin. Reprinted with the permission from Ref. 140. Copyright © 2017 Springer Nature. (C) A μBBB model-on-chip used to predict the delivery rate of dextrans and propidium iodide. Reprinted with the permission from Ref. 142. Copyright © 2012 The Royal Society of Chemistry. (D) An NVU-on-a-chip to investigate the metabolic role of brain vasculature by intravascular administration of methamphetamine. Reprinted with the permission from Ref. 143. Copyright © 2018 Springer Nature.
The glomerulus is one of the most essential functional units of the kidney that serves to filter circulating blood, and glomerulus-on-a-chip were also expected to be applicable in assessing drug-induced nephrotoxicity139. However, their development was limited by the lack of functional podocytes, the main components of glomerulus capillaries that regulate permselectivity. The first glomerulus-on-a-chip was implemented by Musah et al.131 when they succeeded in the controlled differentiation of human iPSCs into mature podocytes. These podocytes were further co-cultured with human glomerular endothelial cells to produce a glomerular-on-a-chip that recapitulated the in vivo glomerular function of blood filtration and urinary clearance, and mimicked the podocyte injury caused by the anticancer drug adriamycin140 (Fig. 4B). More recently, a glomerular microfluidic chip was modeled by seeding human podocytes and glomerular endothelial cells into Organoplates™ (MIMETAS), and podocyte injury induced by a nephrotoxic agent, puromycin aminonucleoside, was observed141. These advanced works highlighted the potential of glomerular-on-a-chip in nephrotoxicity studies toward the screening of new drug compounds and kidney disease treatment.
3.4. Brain-on-a-chip
According to the World Health Organization (WHO), over one billion people suffer from neurological disorders, and this number is rising with the increasing age of the population144. However, the average success rate of developing new drugs to treat central nervous system (CNS) disorders is only approximately 8%, which is lower than the 11% average rate for all therapeutic areas145. This is mainly because of the insufficient predictive capabilities of current animal-based toxicity testing methods. In fact, neurodegenerative diseases, such as Alzheimer's disease (AD), rarely exist in other animal species146. Besides, neurotoxicity caused by drugs such as antibiotics147, anticancer drugs148,149, and anesthetics is widely recognized and has become one of the leading causes of toxicity-related clinical trial failures150 and pharmaceutical product withdrawals from the market145. As a result, the development of adequate in vitro models, such as 3D tissue engineering151 and especially brain-on-a-chip (brain-OAC) mimicking human brain functions, are urgently needed for both the discovery of new CNS drugs and the neurotoxicity assessment of other drugs.
The brain is the most complicated organ in the human body, comprising a number of topologically organized regions that exhibit specific behaviors and interplay with each other to ensure the proper functioning of the brain. Neurons and glial cells (such as astroglia, oligoglia, and microglia) are the two major cell types in the human brain that maintain brain function, with electrical and chemical signals passing between neurons. At present, the development of brain-OACs is still in its infancy, and it is technically challenging to recapitulate the complete structure and function of the brain. Therefore, the design of a brain-OAC device is generally based on the use of iPSCs152 and mainly focuses on the recapitulation of certain specific tissue components of the brain, such as the blood–brain barrier (BBB) and neurovascular unit (NVU), based on the integration of neuronal cells, nonneuronal cells, fluid flows, stimulations, etc.
The BBB is a highly selective semipermeable border of endothelial cells that protects the central nervous system against toxins or pathogens from circulating systems153. Its function to hinder the passage of most compounds to the CNS complicates drug development. In vitro BBB models are particularly useful for studying drug delivery and assessment. Booth et al.142 developed a μBBB model-on-chip by bonding PDMS layers, electrode layers, and polycarbonate membranes together, with endothelial and astrocytic cells seeded on both sides of the membrane. The system was used to predict the delivery rates of dextrans 4 k, 20 k, 70 k, and propidium iodide and showed potential in preclinical drug testing (Fig. 4C).
The neurovascular unit (NVU) is a multicellular component in the brain that serves to transfer nutrients, metabolites, and drugs between the systemic circulation and brain parenchyma, thus regulating metabolic homeostasis as well as the drug PK-PD in the central nervous system. In vitro NVU models could help investigate the interactions between multiple cellular populations and their role in NVU functions. An innovative NVU-on-a-chip was reported and fabricated by connecting two BBB chips on each side of a brain parenchymal compartment to emulate an NVU. The coupled system was used to mimic the metabolic role of brain vasculature by intravascular administration of the psychoactive drug methamphetamine, providing an in vitro way to probe drug transport, efficacy, toxicity, and action mechanism143 (Fig. 4D).
3.5. Other single-organ-on-a-chip
Although the heart, liver, kidneys, and brain are the major targets of drug-induced toxicity, other organs, such as the lungs, intestine, skin, and vessels, may also suffer from adverse drug side effects. Meanwhile, the development of new drugs for treating specific diseases in these organs is also of great importance for human healthcare. Both rely on in vivo models that mimic the functions of organs of interest. Herein, we provide a snapshot of several other single-organ-on-a-chip (single-OOAC) developments for drug screening.
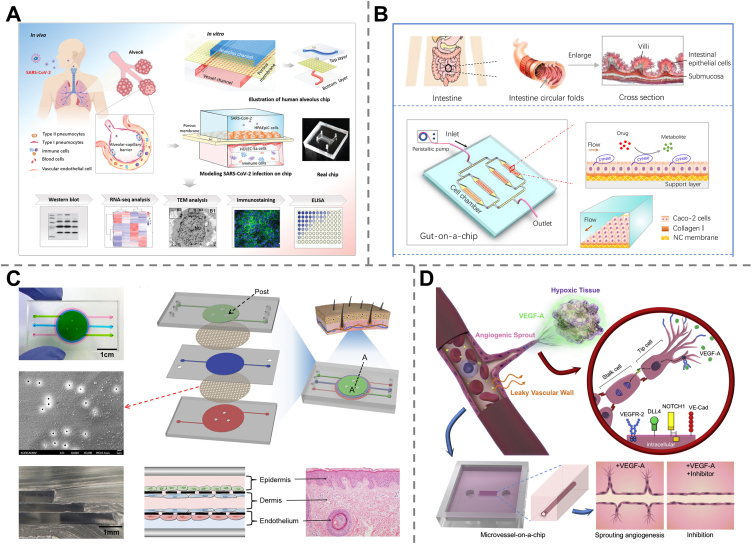
The lungs serve the vital purpose of gas exchange in the respiratory system. Severe acute or chronic respiratory diseases are among the leading causes of death worldwide. Lung-on-a-chip (lung-OAC) was the first reported concept of OOAC154 and has rapidly advanced physiological and pathophysiological studies in lungs, disease models, and drug screening. An innovative work dedicated to constructing a human lung small airway-on-a-chip was carried out by taking advantage of living, fully differentiated, pseudostratified, mucocutaneous human bronchiolar epithelium along with an underlying microvascular endothelium. Furthermore, asthma and human pulmonary inflammation were modeled on a chip to assess the therapeutic responses to interleukin-13. The device offered great prospects in human pathophysiology studies and preclinical drug evaluation155. More recently, Zhang et al.156 fabricated a human alveolus chip based on the co-culture of the human alveolar epithelium, microvascular endothelium, and circulating immune cells to reproduce the main functions of the alveolar-capillary barrier. Then, lung injury caused by SARS-CoV-2 infection was mimicked on a chip to explore the immune response and antiviral responses of the cells. The results demonstrated that viral replication was inhibited and barrier disruption was alleviated under remdesivir, suggesting that the device could be a promising platform for research on drug candidates against COVID-19 (Fig. 5A).
Figure 5.
Lung-, intestine-, skin- and blood-vessel-on-a-chip for drug screening. (A) A human alveolus chip reproducing the lung injury caused by SARS-CoV-2 infection to explore the treatment efficacy of remdesivir. Reprinted with the permission from Ref. 156. Copyright © 2021 The Authors. Advanced Science published by Wiley-VCH GmbH. (B) A gut-on-a-chip mimicking the intestinal microenvironment to assess the metabolism of ifosfamide and verapamil. Reprinted with the permission from Ref. 158. Copyright © 2018 John Wiley and Sons. (C) A skin-on-a-chip model to evaluate the anti-inflammatory effect of dexamethasone. Reprinted with the permission from Ref. 162. Copyright © 2016 Springer Nature. (D) A blood-vessel-on-a-chip to study the anti-angiogenic effects of sorafenib and sunitinib. Reprinted with the permission from Ref. 166. Copyright © 2018 Elsevier.
The intestines are responsible for digestion and are the main absorption sites of orally administered drugs157. Biomimetic human intestine-on-a-chip is highly desirable for in vitro modeling of drug metabolism and intestinal absorption. A gut-on-a-chip was developed to mimic the intestinal microenvironment by adopting Caco-2 cells, porous nitrocellulose membranes, and collagen I under constant fluid flow. Two drug models, ifosfamide and verapamil, were used to assess the metabolic activity of the biomimetic intestine, which offers a simple and robust platform for intestinal metabolism studies and preclinical drug evaluation158 (Fig. 5B). Another microfluidic human organ chip was lined with human intestinal epithelial cells and microvascular endothelial cells, which was then exposed to γ-radiation to mimic radiation injury. The model demonstrated the capability of assessing the radiation-protecting effects of dimethyloxalylglycine (DMOS), a potential radiation countermeasure drug159. More recent work was carried out in the context of the COVID-19 pandemic by seeding human primary intestinal epithelium in emulate organ chips followed by NL63 coronavirus infection to investigate coronavirus-related intestinal pathology. This platform was used to test potential antiviral drugs and showed that the approved protease inhibitor drug nafamostat exhibited an efficient antiviral effect by inhibiting viral entry, while remdesivir was found to be toxic to the intestinal endothelium, although it has been newly approved for SARS-CoV-2 virus infection160.
The human skin is the largest organ and the first physiological barrier that protects other organs and tissues from harmful environmental conditions, such as bacteria, viruses, inflammation, chemical toxicants, and ultraviolet radiation. Skin-on-a-chip is highly desirable as a novel drug testing approach to investigate therapies for skin diseases161. An inflammatory skin-on-a-chip model was developed by co-culturing human keratinocytes (HaCaTs), fibroblasts (HS27) and human umbilical vein endothelial cells (HUVECs) to form the separate layers (epidermal, dermal, and vascular layers), which was followed by the perfusion of TNF-α to induce inflammation. The model was used to evaluate the anti-inflammatory effects of dexamethasone based on the evaluation of proinflammatory cytokines (IL-1β, IL-6) and chemokine (IL-8) levels162 (Fig. 5C).
Vascular networks play a vital role in maintaining the life and function of all organs by transporting blood throughout the body. Besides, it has been found that cancer metastasis is strongly related to blood vessel angiogenesis. The blood-vessel-on-a-chip enabled mimicking main functions of blood vessels, such as permeability, has been widely employed in vascular disease modeling163, drug diffusion164, and drug screening165. An initial human blood vessel was created within collagen gel in a PDMS chip by using primary HUVECs, to mimic the vascular endothelial growth factor (VEGF)-dependent angiogenesis. The vessel-on-a-chip model has been applied to study the anti-angiogenic effects of sorafenib and sunitinib, as well as the endothelial barrier function after treatment with the two angiogenic inhibitors166 (Fig. 5D).
In addition, human tissues such as nerve167,168, bone marrow169,170, and fat171 were also modeled on chips for the assessment of drug toxicity and efficacy, as well as for the discovery of new drugs.
3.6. Multi-organ-on-a-chip and human-on-a-chip
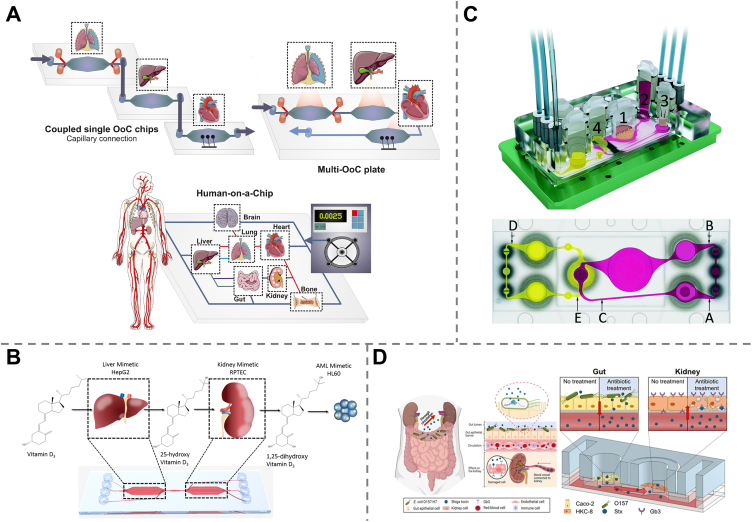
Multi-organ-on-a-chip (Multi-OOAC) refers to the biometric microphysiological system containing several different organoids or tissues in a single microfluidic device, which can mimic not only the functions of isolated single organs but also inter-organ communications. Multi-OOACs are mainly realized through two distinct engineering approaches: the connection of single-OOACs via capillary tubes or microfluidic motherboards, and the integration of spatially separated multiple organ models on a single plate with microfluidic channels serving as a “vascular” system172 (Fig. 6A). As all in vivo organs are communicated through blood and lymphatic circulation, and in most cases, the metabolic activities of one organ may induce effects in other organs, multi-OOACs are capable of providing systemic insight into the therapeutic efficiency of drugs as well as the drug-induced side effects are more advantageous than single-OOAC for the retrieval and optimization of drug candidates.
Figure 6.
Multi-organ-on-a-chip for drug screening. (A) Schematic representation of the two engineering approaches of multi-OOAC. Reprinted with the permission from Ref. 172. Copyright © 2021 Elsevier. (B) A liver-kidney-on-a-chip to investigate the hepatic metabolism and renal activation of vitamin D3. Reprinted with the permission from Ref. 177. Copyright © 2019 Springer Nature. (C) A skin-liver-kidney-intestine-on-a-chip for drug absorption, distribution, metabolism, and excretion (ADME) model. Reprinted with the permission from Ref. 182. Copyright © 2015 The Royal Society of Chemistry. (D) A kidney-gut-on-a-chip for nephrotoxicity assessment related to ciprofloxacin and gentamicin treatment. Reprinted with the permission from Ref. 184. Copyright © 2021 MDPI (Basel, Switzerland).
To date, various multi-OOACs with specific organ combinations have been developed as models in different biomedical applications, for example, tumor–vasculature combinations for metastasis studies, tumor–liver combinations as PK-PD models173, and many other combinations for the assessment of drug safety and efficacy172.
Specifically, the liver is often integrated into a multi-organ-on-a-chip device targeting systemic drug effect assessment, as it is the central organ for in vivo drug metabolism174,175. A functional liver-heart-on-a-chip system lined with iPSC-CMs and primary hepatocytes has been proposed and used for pharmacokinetic studies of two drugs, cyclophosphamide and terfenadine. It was demonstrated that the presence of the liver model induced or reduced the cardiotoxicity from cyclophosphamide or terfenadine, respectively, in accordance with both drugs pharmacology176. A liver-kidney-on-a-chip was fabricated by cultivating HepG2 and RPTEC cells in interconnected chambers to investigate the hepatic metabolism of vitamin D3 as well as its bio-activation by the kidneys177 (Fig. 6B). A four-organ system composed of heart, muscle, neuron, and liver modules in a continuous recirculation system was developed to investigate inter-organ communication and systemic drug-induced toxicity screening. Doxorubicin, atorvastatin, valproic acid, and APAP were selected as model drugs, and their reported effects, such as doxorubicin-induced cardiotoxicity and hepatoxicity, atorvastatin-related myotoxicity, protective effects on neurons of valproic acid and liver failure caused by APFP, were demonstrated178. Another microfluidic chip consisting of three organoids (liver, heart, and lung) was fabricated, enabling both the recapitulation of capecitabine metabolism in the liver and the assessment of cytotoxicity induced by its metabolite 5-fluorouracil (5-FU) in the lung and heart. The device was then expanded to integrate the endothelium, brain, and testis on the same platform, with the six organoids positioned in the order following in vivo blood flow direction. Similarly, hepatic metabolism of the prodrug ifosfamide and the release of the neurotoxin chloracetaldehyde were demonstrated179. Other liver-modeled multi-organ chips, such as liver-kidney- and liver-lung-on-a-chip, have also been proposed for testing different drugs, such as aflatoxin B1 and benzaldehyde (BαP)180,181.
Moreover, the absorption of oral drugs in the intestine is also related to cytotoxicity, as reported in clinical practice. For example, a four-organ-chip with skin-liver-kidney-intestine combination was reported for the drug absorption, distribution, metabolism, and excretion (ADME) model182 (Fig. 6C). An intestine-kidney-on-a-chip was developed, which co-cultured intestinal and glomerular endothelial cells in compartmentalized microchambers to explore drug absorption and subsequent nephrotoxicity by adopting digoxin combined with cholestyramine or verapamil as a drug model183. More recently, a kidney-gut chip was proposed to study nephrotoxicity related to antibiotic treatment by ciprofloxacin and gentamicin184 (Fig. 6D).
3.7. Tumor-on-a-chip
Cancer has become one of the most significant global healthcare problems and one of the leading causes of human mortality, taking more than 10 million lives worldwide in 2020 alone185. It was reported that tumor growth, invasion, and metastasis are closely related to the tumor microenvironment (TME), which is characterized by dense ECM, irregular vessels, limited blood supply, hypoxia, acidic pH, etc. The main objective of oncology research is to understand the tumor biology and metastasis mechanisms related to the TME, thereby providing faithful guidance toward efficient cancer therapy and anticancer drug discovery. To this end, tumor-on-a-chip (tumor-OAC) has been proposed as an in vitro TME model, reproducing the key features of the in vivo TME (such as complex 3D tissue structure, biochemical gradients, and dynamic cell–cell interactions) and has shown great promise as a novel technology for studying both cancer biology and therapeutic strategies.
Similar to OOAC, tumor-OAC also consisted of four key elements, i.e., microtissue, microfluidic system, stimulation, and sensor. Meanwhile, tumors are complex systems with various functional components and factors, including different cell types (tumor cells, cancer-associated fibroblasts, and endothelial cells) along with their interactions, shear stress induced by dynamic flow, and chemical factors such as chemotaxis, oxygen tension, and hypoxia gradient. Which should also be taken into consideration in the design of tumor-OACs186. A variety of reviews have been published focusing on tumor-OACs from different perspectives187, 188, 189, 190, 191. Here, we specifically emphasize their applications in screening anticancer drug candidates.
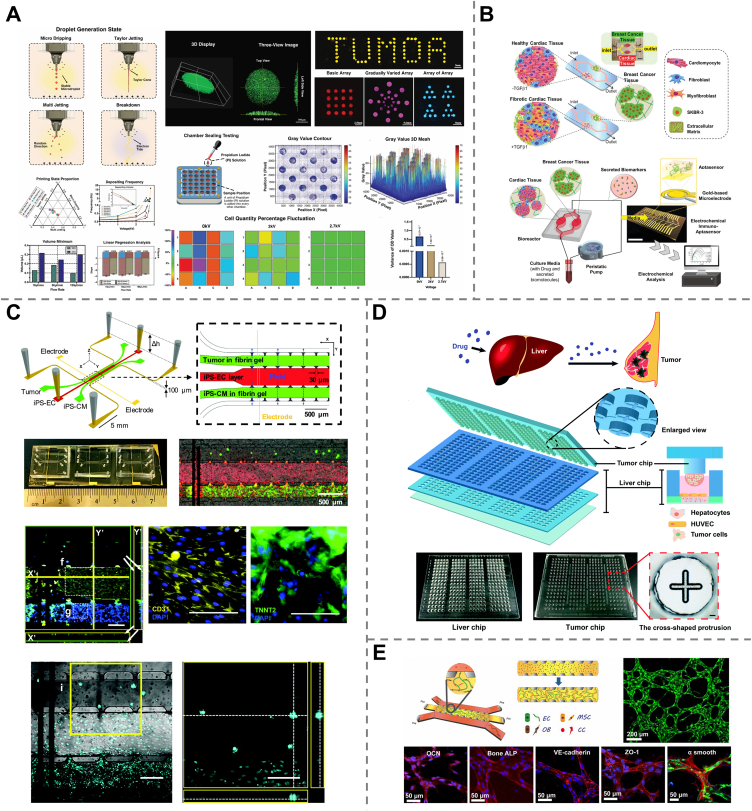
A lung cancer-on-a-chip equipped with a pH sensor, a TEER impedance sensor and a fluorescence microscope was developed for real-time monitoring of cellular responses to different concentrations of the anticancer drugs, doxorubicin, and docetaxel. The results revealed that higher drug concentrations led to increased cell death, and doxorubicin exhibited stronger toxicity than docetaxel, suggesting that the system allowed initial cytotoxicity evaluation for drug screening192. Similarly, another tumor-on-a-chip fabricated by electrohydrodynamic 3D bioprinting with Gelatin methacryloyl (GelMA) droplets containing breast tumor cells was reported and applied for screening epirubicin and paclitaxel at different concentrations193 (Fig. 7A).
Figure 7.
Tumor-on-a-chip devices for drug screening. (A) A breast-tumor-on-a-chip for screening the concentration-dependent toxicity of epirubicin and paclitaxel. Reprinted with the permission from Ref. 193. Copyright © 2020 Springer Nature. (B) A heart-breast-cancer-on-a-chip to investigate chemotherapy-induced cardiotoxicity (CIC) after treatment with doxorubicin. Reprinted with the permission from Ref. 197. Copyright © 2021 John Wiley and Sons. (C) A heart-colon-tumor chip for simultaneous evaluation of the anticancer effect and dose-dependent cardiotoxicity of doxorubicin and oxaliplatin. Reprinted with the permission from Ref. 198. Copyright © 2020 Mary Ann Liebert, Inc. (D) A biomimetic array chip combining liver and tumor microtissues to evaluate the hepatic metabolism-related anticancer efficacy of the prodrugs capecitabine and irinotecan, as well as their hepatoxicity. Reprinted with the permission from Ref. 200. Copyright © 2020 The Royal Society of Chemistry. (E) A 3D vascularized organotypic chip to investigate the anti-metastatic role of adenosine in breast cancer cell extravasation. Reprinted with the permission from Ref. 203. Copyright © 2000 National Academy of Science.
Chemotherapy is one of the most common anticancer treatments194,195, which makes use of certain drugs traveling throughout the whole body to kill cancer cells or to prevent their growth and metastasis. However, its extensive application may be limited by the fact that healthy cells could also be damaged, leading to chemotherapy toxicity196. Connecting tumor tissues and healthy organs onto a chip allows mimicking organ responses to anticancer drugs. For example, a microfluidic chip was realized by the interconnection of human iPSC-derived cardiac spheroids and SK-BR-3 cell-generated breast cancer spheroids to investigate chemotherapy-induced cardiotoxicity (CIC). Based on the level of cell-secreted biomarkers (such as troponin T, CK-MB, and HER-2) monitored with integrated electrochemical immuno-aptasensors and beating frequency, notable cardiac toxicity was observed in both healthy and fibrotic cardiac tissues after doxorubicin treatment197 (Fig. 7B). Another chip integrating iPSC-derived heart tissues and colon tumors was fabricated to simultaneously evaluate the anticancer effect of doxorubicin and oxaliplatin, as well as their dose-dependent cardiotoxicity. The results revealed that cardiotoxicity of doxorubicin occurred around the half maximal inhibitory concentration (IC50) determined with the colon tumor on-a-chip, while oxaliplatin-induced cardiotoxicity was observed at concentrations dramatically higher than its IC50, which is consistent with the in vivo reports198 (Fig. 7C).
Sometimes toxicity or anticancer bioactivity arises from the metabolized form of the drug rather than the drug itself. Integration of liver cells on a chip makes it possible to evaluate organ toxicity induced by the hepatic metabolites of many anticancer drugs. An integrated heart-liver cancer-on-a-chip was proposed based on human healthy heart cells and liver cancer cells (HepG2) to recapitulate the metabolism of doxorubicin, a common chemotherapy drug, by liver cancer and its side effects on the heart. The metabolite of doxorubicin, doxorubicin, was shown to be released through hepatic metabolism from HepG2 cells and caused heart damage199. An integrated biomimetic array chip was designed to construct 3D liver and tumor microtissues, allowing for not only evaluation of the anticancer bioactivity of hepatic metabolites of several prodrugs but also their hepatoxicity. This platform offers an opportunity for anticancer drug screening based on both efficacy and safety200 (Fig. 7D). Another work combining primary human liver microtissues and tumor microtissues on a single chip was reported to evaluate the metabolic competence of the liver to convert the anticancer prodrugs cyclophosphamide (CP) and ifosfamide (IFF) and their drug–drug interactions with the antiretroviral drug ritonavir201.
Moreover, various tumor-on-a-chip have been reported as promising platforms for screening anticancer drug efficacy. Multidrug-resistant (MDR) cancer refers to cancer that develops resistance to a wide variety of chemotherapy drugs, which is a major factor in chemotherapy failure and accounts for over 90% of deaths in patients receiving traditional chemotherapeutics. Tumor-on-a-chip can be used as a novel approach to evaluate the potential efficacy of new drugs or therapeutic strategies for MRC treatment. A liver-bone marrow-uterine cancer chip was fabricated by culturing HepG2/C3A, MEG-01, MES-SA and MES-SA/DX-5 cell lines in four individual but interconnected organ compartments, representing liver, bone marrow, uterine cancer, and multidrug-resistant uterine cancer. The device demonstrated enhanced efficacy of doxorubicin with cyclosporine and nicardipine for the treatment of MDR cancers202. Another human 3D microfluidic in vitro model was developed using primary human bone marrow-derived mesenchymal stem cells (hBM-MSCs), osteo-differentiated primary hBM-MSCs, and primary GFP-human umbilical vein endothelial cells to construct a microvascular network enclosed in a bone-mimicking microenvironment to investigate the extravasation of breast cancer cells. This device was used as a drug screening platform to study the role of adenosine in cancer cell extravasation and demonstrated its promising anti-metastatic effect203 (Fig. 7E).
4. Current challenges and future perspectives
Since the first proposal of the OOAC concept in 2010, the past decade has witnessed a rapid breakthrough in this field. This technology was on the list of the “top ten emerging technologies” in 2016 by the World Economic Forum (WEF). A large number of OOAC devices based on various single organs (heart, liver, kidney, brain, gut, lung, skin, vessel, bone, etc.), multi-organs (heart-liver, and kidney-liver, to name a few), and tumor tissues have been developed to emulate the unique physiology and main functions of organs/tissues with accurate spatiotemporal regulation. Allowing for cellular behaviors in response to exogenous substances, OOACs have emerged as the most attractive in vitro drug assessment approach and are particularly advantageous in terms of cost, inter-species differences, and ethical issues when compared to conventional animal tests. To date, OOACs have demonstrated successful applications not only in drug screening (Table 3) as summarized in this review, but also in disease modeling and cancer metastasis studies. However, this technology is still in its infancy of development. As summarized in the previous sections, the capability of OOACs to evaluate therapeutic efficiency, side effects and cytotoxicity has been verified mainly through clinically approved drugs, such as APFP, verapamil, doxorubicin, and gentamicin. There is still a long way to go and a plethora of obstacles to overcome before OOAC devices can be devoted to faithfully screening potential drug candidates in drug discovery. Herein, we would like to discuss the challenges and our points of view on the future development of OOACs toward drug screening.
Table 3.
Various organ-on-a-chip systems for drug screening.
| Organ/tissue type | Drug model | Cell source | Chip material | ECM material | Fabrication technique | Readout signal | Application | Ref. |
|---|---|---|---|---|---|---|---|---|
| Heart | Verapamil | Primary neonatal CM | Quartz | Gelatin | Electron beam lithography | CM contraction status | Therapeutic efficacy evaluation | 109 |
| Gold | ||||||||
| Isoproterenol | Rat CM | PIPAAm | N/A | Laser engraver | Contractility | Therapeutic efficacy evaluation | 110 | |
| PDMS | ||||||||
| Doxorubicin | Human iPSC-CM | PDMS | Hydrogel | 3D bioprinting | Beating rate | Toxicity assessment | 111 | |
| PMMA | ||||||||
| Isoproterenol | Human iPSC-CM | PDMS | Gelatin | Soft lithography | Cardiac field potentials | Toxicity assessment | 112 | |
| Terfenadine | ||||||||
| Fexofenadine | ||||||||
| Liver | Acetaminophen | HepG2/C3A | PDMS | Fibronectin | N/A | Transcriptomics, proteomics and metabolomic profiles | Toxicity assessment | 123 |
| Acetaminophen | HepG2/C3A | PDMS | GelMA hydrogel | 3D bioprinting | Secretion rates of hepatocyte markers | Toxicity assessment | 113 | |
| PMMA | ||||||||
| Acetaminophen | Primary mouse hepatocytes | Silicon | N/A | Photolithography | Liver-related biomarkers | Toxicity assessment | 117 | |
| Chlorpromazine | PDMS | |||||||
| Tacrine | ||||||||
| Metformin | Primary human hepatocytes | PMMA | N/A | Laser cut | Cell viability | Therapeutic efficacy evaluation | 125 | |
| White adipocytes | PDMS | Insulin resistant biomarkers | ||||||
| Elafibranor | Human primary hepatocyte | Collagen | Hydrogel | Standard photolithography | Cell morphology | Therapeutic efficacy evaluation | 114 | |
| Kupffer cells | Hepatic biomarker level | |||||||
| Liver sinusoidal endothelial cells | ||||||||
| Hepatic stellate cells | ||||||||
| Hydrocortisone | Human primary hepatocytes | Commercialized LiverChip (CNBio Innovations) | N/A | N/A | Pharmacokinetic parameters | Metabolism study | 127 | |
| Kupffer cells | ||||||||
| Simvastatin | Primary rat hepatocytes | PDMS | Collagen | Photolithography | Cell viability | Metabolism study | 128 | |
| Atorvastatin | Human prostatic cancer cells | Glass | Soft lithography | Transcriptomics | ||||
| Metabolomics | ||||||||
| Kidney | Cyclosporine A | Proximal tubule epithelial cells | Silicon | Hydrogel | 3D bioprinting | Cell morphology | Toxicity assessment | 134 |
| Hydrogel | Cytoskeleton organization | |||||||
| Polymyxin B | Human primary proximal tubule epithelial cells | Nortis device | Collagen | N/A | Gene expression | Toxicity assessment | 135 | |
| Injury biomarkers | ||||||||
| Cholesterol concentrations | ||||||||
| Gentamicin | Madin–Darby canine kidney cells | PDMS | Fibronectin | UV polymerization | Cell viability | Toxicity assessment | 136 | |
| Kidney injury marker | ||||||||
| Cisplatin | Proximal tubule epithelial cells lines (ciPTEC-OAT1 and RPTEC) | OrganoPlate® | Collagen | N/A | Cell viability | Toxicity assessment | 137 | |
| Tenofovir | Biomarker release | |||||||
| Tobramycin | Barrier integrity | |||||||
| Cyclosporin A | Gene expression | |||||||
| Cisplatin | Renal proximal tubular epithelial cells | PDMS | Collagen | Laser-cutting | Cell viability | Toxicity assessment | 138 | |
| Gentamicin | Peritubular capillary endothelial cells | |||||||
| Cyclosporine A | ||||||||
| Adriamycin | Human iPSC-derived podocytes | PDMS | N/A | Stereolithography | Podocyte layer integrity | Toxicity assessment | 140 | |
| Human glomerular endothelial cells | Cell viability | |||||||
| Puromycin | Human podocytes | OrganoPlate™ | N/A | N/A | Podocyte morphology | Therapeutic efficacy evaluation | 141 | |
| Aminonucleoside | Glomerular endothelial cells | |||||||
| Brain | Dextrans 4 k, 20 k, 70 k | Endothelial cell line (b. End3) | PDMS | Fibronectin | Piranha etch | Cell viability | Drug delivery study | 142 |
| Propidium iodide | Astrocyte cell line (C8D1A) | Glass | Laser patterning | Astrocytic morphology | ||||
| Polycarbonate | TEER levels | |||||||
| Permeability | ||||||||
| Methamphetamine | Primary human brain microvascular endothelial cells | PDMS | Mixture of fibronectin and collagen | Soft lithograph | BBB permeability | Metabolism study | 143 | |
| Primary brain microvascular pericytes | PTE | Metabolites expression | ||||||
| Astrocytes | ||||||||
| Primary human neural cells | ||||||||
| Lung | Interleukin-13 | Primary human airway epithelial cells | PDMS | Collagen | Stereolithography | Gene expression | Therapeutic efficacy evaluation | 155 |
| Chemokines and cytokines concentrations | ||||||||
| Neutrophil adhesion | ||||||||
| Remdesivir | Human alveolar epithelial type II cell line | PDMS | Collagen | Conventional soft lithography | Alveolar-capillary barrier permeability | Therapeutic efficacy evaluation | 156 | |
| Lung microvasculature cell line | Inflammatory cytokines concentration | |||||||
| Gene expression | ||||||||
| Intestine | Ifosfamide | Human intestinal epithelial cells (Caco-2) | PDMS | Nitrocellulose | Soft photolithography | Drug metabolite level | Metabolism study | 158 |
| Verapamil | Collagen | |||||||
| Dimethyloxaloylglycine | Human intestinal epithelial cells (Caco-2) | PDMS | Collagen | Soft lithography | Intestinal permeability | Therapeutic efficacy evaluation | 159 | |
| Matrigel | ROS generation and lipid peroxidation | |||||||
| Injury-biomarker expression | ||||||||
| Nafamostat | Intestinal epithelium | Emulate™ | N/A | N/A | Viral load | Therapeutic efficacy evaluation | 160 | |
| Remdesivir | Cytokine secretion | |||||||
| Skin | Dexamethasone | Human keratinocyte cell line (HaCaTs) | PDMS | N/A | Soft lithography | Proinflammatory cytokine (IL-1β, IL-6) and chemokine (IL-8) levels | Therapeutic efficacy evaluation | 162 |
| HS27 Fibroblasts | ||||||||
| HUVEC | ||||||||
| Blood vessel | Sorafenib and sunitinib | HUVEC | PDMS | Collagen | 3D printing | Morphology of the angiogenic sprouts | Therapeutic efficacy evaluation | 166 |
| Liver/heart | Cyclophosphamide | Human iPSC-CM | PDMS | Collagen | Standard photolithography | Beat frequency | Metabolism-associated toxicity assessment | 176 |
| Terfenadine | Human primary hepatocytes | PMMA | Deep reactive ion etching | Conduction velocity | ||||
| HepG2/C3A | QT-interval | |||||||
| Contractile force | ||||||||
| Liver/kidney | Vitamin D3 | HepG2 | Device from ChipShop | N/A | N/A | Level of mRNA expression | Metabolism study | 177 |
| RPTEC | ||||||||
| HL60 cells | ||||||||
| Liver/muscle/neuron/heart | Doxorubicin | HepG2/C3A | Silicon on insulator (SOI) wafer | N/A | Photolithography | Cell viability | Toxicity assessment | 178 |
| Atorvastatin | iPSC-CM | Cell functionality | ||||||
| Valproic acid | Skeletal myofiber | |||||||
| Acetaminophen | Motoneurons | |||||||
| N-Acetyl-m-aminophenol | iPSC-neurons | |||||||
| Liver/heart/lung | Capecitabine | Primary human hepatocytes | Adhesive film | Hydrogel | Rapid-prototyping of patterned adhesive films | Cell viability | Metabolism-associated toxicity assessment | 179 |
| Ifosfamide | Hepatic stellate cells | Glass | ||||||
| Kupffer cells | PMMA | |||||||
| Liver-derived endothelial cells | ||||||||
| iPSC-CM | ||||||||
| Human cardiac fibroblast | ||||||||
| Cardiac endothelium cells | ||||||||
| A549 | ||||||||
| CC-2512 | ||||||||
| CC-2540 | ||||||||
| Lung/liver | Aflatoxin B1 | Human bronchial epithelial (NHBE) cells | Polyetheretherketone (PEEK) | Collagen | N/A | Cell viability | Metabolism-associated toxicity assessment | 180 |
| HepaRG™ cells | Transepithelial electrical resistance | |||||||
| Gene expression | ||||||||
| Liver/kidney | Aflatoxin B1 | HepG2 | Device from ChipShop | Collagen | N/A | Cell viability | Metabolism-associated toxicity assessment | 181 |
| Benzoalphapyrene | Hek293 | Cell survival curve | ||||||
| Intestines/kidney | Digoxin combined with colestyramine or verapamil | Caco-2 cells | PDMS | Collagen | Soft lithography | Cell apoptosis | Absorption-associated toxicity assessment | 183 |
| Primary rat glomerular microtissues | Micromolding | Cell viability | ||||||
| Lactate dehydrogenase leakage | ||||||||
| Gut/kidney | Ciprofloxacin | Caco-2 | Polycarbonate | N/A | Computer numerical control machining | Cell viability | Therapeutic efficacy | 184 |
| Gentamicin | HKC-8 | PDMS | Soft lithography | Transepithelial electrical resistance | Toxicity assessment | |||
| Glass | ||||||||
| Lung cancer | Doxorubicin | Lung cancer NCI–H1437 cells | Nusil medical grade silicone | Collagen | 3D printing | Impedance | Toxicity assessment | 192 |
| Docetaxel | Elastomer | pH | ||||||
| Glass | Cell viability | |||||||
| Breast cancer | Epirubicin | MDA-MB-231 | GelMA microdroplets | GelMA hydrogel | Electrohydrodynamic 3D printing | Cell viability | Toxicity assessment | 193 |
| Paclitaxel | Cell morphology | |||||||
| Heart/breast cancer | Doxorubicin | Human iPSC-CM | PDMS | Gelatin | Conventional photolithography | Cardiac biomarkers (Troponin T and CK-MB) | Chemotherapy-induced toxicity assessment | 197 |
| Fibroblasts | GelMA hydrogel | Breast cancer biomarker (HER-2) | ||||||
| Myofibroblasts | ||||||||
| SK-BR-3 | ||||||||
| Heart/colon cancer | Doxorubicin | iPSC-CM | PDMS | Fibrin gel | Soft lithography | Heart beating | Chemotherapy-induced toxicity assessment | 198 |
| Oxaliplatin | iPSC-EC | Replica molding | ||||||
| Colon adenocarcinoma cell line (SW620) | ||||||||
| Heart/liver cancer | Doxorubicin | Primary human CM | PDMS | Fibronectin | Multilayer soft lithography replica molding | Cell viability | Metabolism-associated toxicity assessment | 199 |
| HepG2 | Bovine gelatin | Release of lactate dehydrogenase (LDH) | ||||||
| Matrigel | ||||||||
| Liver/breast tumor | Capecitabine | HepG2 | PDMS | Alginate | Soft lithography | Cell viability | Metabolism-associated therapeutic efficacy evaluation | 200 |
| Irinotecan | HUVEC | PMMA | Agarose | |||||
| Adriamycin | HCT116 | Collagen | ||||||
| Epirubicin | MCF7 | Matrigel | ||||||
| Plumbagin | ||||||||
| Liver/colorectal cancer | Cyclophosphamide | Primary human hepatocytes | Polystyrene | N/A | Injection molding | Tumor microtissue diameters | Metabolism study | 201 |
| Ifosfamide | HCT116 | Toxicity assessment | ||||||
| Ritonavir | ||||||||
| Liver/bone marrow/uterine cancer | Mixture of doxorubicin, cyclosporine, and nicardipine | HepG2/C3A | Silicon, Plexiglass | Human blood plasma fibronectin | Photolithography | Cell viability | Therapeutic efficacy evaluation | 202 |
| MEG-01 | Deep reactive ion etching | |||||||
| MES-SA | ||||||||
| MES-SA/DX-5 | ||||||||
| Breast cancer/vascularized microenvironments | Adenosine | hBM-MSC | PDMS | Fibrin gel | Soft lithography | Cell viability | Therapeutic efficacy evaluation | 203 |
| OD hBM-MSC | Vessel permeability | |||||||
| GFP-HUVEC | Expression of antimetastatic marker A3AR | |||||||
| Cancer cell migration distance |
N/A, not applicable; CM, cardiomyocytes; ECM, extracellular matrix; BBB, blood–brain barrier; GelMA, gelatin methacryloyl; HUVEC, human umbilical vein endothelial cells; iPSC, induced pluripotent stem cells; PDMS, polydimethylsiloxane; PIPAAm, poly(N-isopropylacrylamide); PMMA, poly(methyl methacrylate); ROS, reactive oxygen species; TEER, transepithelial electrical resistance.
4.1. Challenges
Since their birth in 2007, OOACs have attracted widespread attention from scientists, have rapidly progressed during the past decade, have achieved huge success in drug discovery, and have demonstrated great promise for future personalized medicine. However, the development of functional and efficient OOACs is a complicated task involving extensive exchanges between biologists and engineers, since living organisms do not perform in a programmed or controlled way. OOACs are still in their infancy period, and their future development meets several key challenges in terms of cell source, cell culture medium, chip design and materials, and implementation of human-on-a-chip.
Unlimited and renewable cell sources are one of the universal issues to be addressed in OOAC development. As already described, immortalized cells, primary cells, and stem cells are commonly used cell types in OOACs, each having proper advantages and limitations. Immortalized cells are commercially available, convenient to use, and cost-effective, but usually lack in vivo phenotype and functionality; therefore, they are generally used in the design and optimization stages of OOAC development and initial screening steps. Primary cells are theoretically capable of recapitulating all functions in vivo, but their time-dependent phenotypic modification, difficulty of in vitro proliferation, and limited quantity hinder their widespread use in OOACs. Embryo stem cells are pluripotent and capable of differentiating into any type of cell to reconstruct the desired organ or tissue but suffer from ethical conflicts that strongly restrict their use. Adult stem cells extracted from the targeted patient may contribute to promoting precision/personal medicine; however, hurdles lie in isolation and long-term in vitro culture. iPSCs, not only holding the potential to generate all cell types but also avoiding ethical issues and limited quantity, are now the most promising cell source for predictive drug screening, with an increasing number of successful employments in OOACs. Nevertheless, the commonly used reprogramming methods are based on virus genes, and the phenotype of many iPSC-derived differentiated cells is immature. Non-viral gene reprogramming and standardized differentiation protocols are highly desirable.
In addition to addressing cell sourcing challenges, cell culture medium is also an essential concern that should be taken into consideration, especially when designing multi-OOACs. A universal blood substitute media that can support all cell types in the multi-OOAC system is needed. Actually, the culture medium varies from one cell type to another, as specific growth factors are required for different cells to maintain viability and phenotype. In a multi-OOAC, different organ compartments are interconnected in a single circulatory system. The development of a universal medium with all well-defined factors, including chemical composition, oxygen tension, and nutrient delivery, to ensure the functionality of all tissues and organs37 is an urgent need but also challenging.
Another critical issue is chip design and materials. On the one hand, the manufacturing cost of traditional lithography and soft lithography is relatively high, which limits their widespread applications, especially commercialization. It will be preferable to consider the utilization of novel techniques, materials, and integrated components of lower cost to reduce the fabrication cost. For example, 3D bioprinting has been used by many researchers, which could lead to the more cost-efficient fabrication of OOACs. Incorporation of diverse modules for pumping, media perfusion, stimulation, bioreaction, sample collection, biomarker analysis, etc., in a compact system could significantly reduce the reagent consumption and manipulation complexity, thus reducing the operation cost of OOACs. Such a high level of integration requires more appropriate designs and more advanced manufacturing technologies, being one of the challenges that will be met by future OOAC development. On the other hand, according to our survey, PDMS is currently the predominant material for chip fabrication (see Table 2), as it exhibits a variety of advantages, including biocompatibility, permeability, flexibility, cost-effectiveness, transparency, ease of processing, to name a few. However, PDMS tends to absorb small hydrophobic molecules, which may cause disordered on-chip pharmacological activities of drugs204. Optimizations should be carried out to mitigate this drawback of PDMS, for example, by surface modification205 or by adopting alternative suitable materials206.
Although OOACs are capable of emulating organ functions and could be a promising alternative to animal models in preclinical tests, human-on-a-chip will be widely required to systemically study pharmacokinetics and pharmacodynamics for more reliable drug candidate screening. Apart from the aforementioned challenges encountered by single- and multiple-OOAC development, a variety of specific difficulties should be overcome to realize reliable, reproducible, and standardized human-on-a-chip. Biological scaling is one of the most important aspects to be taken into consideration. In most recently reported multi-OOACs, although organ functions and organ–organ interactions have been modeled, the relative size of organ components has not been considered, which may undermine drug effects. To create a more physiologically relevant system, the bioscaling issue must be addressed based on various factors, such as cell number, cell surface area, metabolic rate, and blood residence time of each organ. Besides, sterility, media perfusion, metabolic waste removal, simulation of missing organs, etc., also remain ongoing challenges. In short, human-on-a-chip is a promising concept but involves a variety of sophisticated biological and engineering problems to be solved before its widespread application.
4.2. Perspectives
Currently, the majority of reported studies on OOACs for drug screening utilized commercialized medicines such as verapamil, APFP, capecitabine, and doxorubicin to demonstrate the capability and potential of OOACs for drug-induced toxicity assessment, side-effect evaluation, efficacy testing, etc. Future studies will focus on the discovery of new therapeutic effects of existing approved drugs and the development of new drugs. For example, in the face of the ongoing global pandemic COVID-19, which has caused over 612 million infections and 6.5 million mortalities as of September 2022 (the numbers are still increasing), there is an urgent need for efficient drugs and reliable therapies. However, it is impossible to find novel drugs in such a short period, especially when so little is known about this new SARS-CoV-2 virus. OOACs have attracted the attention of many scientists who have made efforts to test the efficacy of existing drugs against SARS-CoV-2207. In 2020, Emulate signed a collaboration agreement with the US Food and Drug Administration (FDA) to apply lung-OACs to evaluate the safety and protective immunity of COVID-19 vaccines. The Wyss Institute fabricated a bronchial airway-on-a-chip by taking advantage of highly differentiated human bronchial airway epithelium and pulmonary endothelium to model viral infection and identified that the clinically relevant doses of amodiaquine could inhibit infection and may be repurposed for COVID-19 treatment208.
When you get ill and receive a treatment, you are truly unsure whether the therapy is effective, ineffective, or even harmful in your particular case, since the therapeutic efficiency and side effects of traditional general medicine may vary from patient to patient. The concept of precision medicine, whereby each patient will receive better-tailored treatments based on their unique genetic characteristics, is becoming increasingly important today for the improvement of therapeutic efficacy and reduction of potential healthcare costs. With the advancement of iPSC technologies, personalized OOACs can be manufactured209 based on primary cells or stem cells from the person who receives the treatment, offering new opportunities to realize precision medicine in terms of new drug discovery, safety evaluation, and efficacy assessment. To this end, the acquisition of individual tissue samples, the obtention of corresponding health data, and the cooperation of biologists, clinicians, and patients are required to implement the transition from personalized OOAC to precision medicine.
The majority of the current OOACs are developed to emulate some specific tissue components or certain functions of in vivo organs, such as renal proximal tubules, kidney glomeruli, small lung airways, lung alveoli, BBB, and NVU. Reconstruction of whole organs with intact structure and functions in vitro is still impossible due to limited technologies. The recapitulation of the entire organ, the linkage of multiple organ models, and the construction of a miniaturized human body on a single chip are needed to study the organ–organ interactions, ADME profiling, pharmacokinetics and pharmacodynamics of drugs, and the body's systemic responses to drugs, especially for the study of prodrugs that are metabolized in vivo to release active compounds. Many efforts have been undertaken to meet these aspirations; for example, funding from the US Defense Advanced Research Project Agency (DARPA), the US National Institutes of Health (NIH), and the US FDA was allocated to the development of body-on-a-chip. We believe that the concept of body-on-a-chip holds fascinating prospects for wider applications of OOACs in the future.
The lack of human physiologically relevant in vitro models for efficient drug testing is a major reason for the high cost and long duration of new drug discovery, plaguing pharmaceutical companies around the world. The invention of OOACs made them shine, and in 2011, the president of the United States announced the establishment of the “Microphysiological System” research project. To date, a large number of companies, including Emulate Inc. in the United States, TissUse GmbH in Germany, MIMETAS in the Netherlands, and CN Bio in the United Kingdom, have been launched and are committed to the development and commercialization of OOACs. The global organ-on-a-chip market is estimated at $54.6 million in 2021 and is expected to reach $697.7 million by 2028, growing at a CAGR of 37.6%, according to the latest report from Research Dive. Commercialization is likely to occur in a stepwise way and finally integrated into the early stages of the drug development pipeline for lead compound validation and optimization.
The transition from the laboratory to the market requires low-cost and large-scale manufacturing of OOACs in a repeatable and standardized manner. Specifically, the standardization of technology in multiple aspects is becoming critically important210. Several general standards are already available or used as guidelines for establishing new standards in the OOAC fields, for example, the ISO 100991–2009 on micro-process engineering vocabulary for OOAC definitions, CEN/ISO on medical devices for OOAC sterilization and packaging, ASTM or VDI standards for medical grade plastic materials for biocompatible material control, ISO standards for Tissue Engineered Medical Products and CEN/ISO standards for in vitro diagnostics. Standardization plays a significant role in promoting the future advancement of this field.
Acknowledgments
We acknowledge the financial support from the National Key R&D Program of China (2019YFA0709200), the National Natural Science Foundation of China (21874066, and 61804076), the Key Research and Development Program of Jiangsu Province (BE2021373, China), the Natural Science Foundation of Jiangsu Province (BK20180700, and BK20200336, China), the Fundamental Research Funds for Central Universities (China), and the Program for Innovative Talents and Entrepreneur in Jiangsu (China).
Author contributions
Yanping Wang and Yanfeng Gao wrote the draft manuscript with feedback from other authors. Yongchun Pan, Dongtao Zhou, Yuta Liu, Yi Yin, Jingjing Yang, Yuzhen Wang and Yujun Song contributed to the discussion and revision. All of the authors have read and approved the final manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Contributor Information
Yanfeng Gao, Email: gaoyanfeng@nju.edu.cn.
Yuzhen Wang, Email: iamyzwang@njtech.edu.cn.
Yujun Song, Email: ysong@nju.edu.cn.
References
- 1.Hughes J., Rees S., Kalindjian S., Philpott K. Principles of early drug discovery. Br J Pharmacol. 2011;162:1239–1249. doi: 10.1111/j.1476-5381.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun D.X., Gao W., Hu H.X., Zhou S. Why 90% of clinical drug development fails and how to improve it?. Acta Pharm Sin B. 2022;12:3049–3062. doi: 10.1016/j.apsb.2022.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wouters O.J., McKee M., Luyten J. Estimated research and development investment needed to bring a new medicine to market, 2009‒2018. JAMA. 2020;323:844–853. doi: 10.1001/jama.2020.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith A. Screening for drug discovery: the leading question. Nature. 2002;418:453–455. doi: 10.1038/418453a. [DOI] [PubMed] [Google Scholar]
- 5.Zhai J., Yi S.H., Jia Y.W., Mak P.-I., Martins R.P. Cell-based drug screening on microfluidics. Trends Analyt Chem. 2019;117:231–241. [Google Scholar]
- 6.Liu X.Y., Zheng W.F., Jiang X.Y. Cell-based assays on microfluidics for drug screening. ACS Sens. 2019;4:1465–1475. doi: 10.1021/acssensors.9b00479. [DOI] [PubMed] [Google Scholar]
- 7.Li Y.H., Meng Q., Yang M.B., Liu D.Y., Hou X.Y., Tang L., et al. Current trends in drug metabolism and pharmacokinetics. Acta Pharm Sin B. 2019;9:1113–1144. doi: 10.1016/j.apsb.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mak K.-K., Epemolu O., Pichika M.R. The role of DMPK science in improving pharmaceutical research and development efficiency. Drug Discov Today. 2022;27:705–729. doi: 10.1016/j.drudis.2021.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Freires I.A., Sardi J.D.C.O., De Castro R.D., Rosalen P.L. Alternative animal and non-animal models for drug discovery and development: bonus or burden? Pharm Res (N Y) 2017;34:681–686. doi: 10.1007/s11095-016-2069-z. [DOI] [PubMed] [Google Scholar]
- 10.Yang J.J., Pan B., Zeng F., He B.S., Gao Y.F., Liu X.L., et al. Magnetic colloid antibodies accelerate small extracellular vesicles isolation for point-of-care diagnostics. Nano Lett. 2021;21:2001–2009. doi: 10.1021/acs.nanolett.0c04476. [DOI] [PubMed] [Google Scholar]
- 11.Sun L., Zhao Q., Liu X.L., Pan Y.C., Gao Y.F., Yang J.J., et al. Enzyme-mimicking accelerated signal enhancement for visually multiplexed quantitation of telomerase activity. Chem Commun. 2020;56:6969–6972. doi: 10.1039/d0cc01951h. [DOI] [PubMed] [Google Scholar]
- 12.Liu X.L., Wang Y.P., Gao Y.F., Song Y.J. Gas-propelled biosensors for quantitative analysis. Analyst. 2021;146:1115–1126. doi: 10.1039/d0an02154g. [DOI] [PubMed] [Google Scholar]
- 13.Xu Q., Pan Y.C., Liu X.L., Gao Y.F., Luan X.W., Zeng F., et al. Hypoxia-responsive platinum supernanoparticles for urinary microfluidic monitoring of tumors. Angew Chem Int Ed Engl. 2022;61 doi: 10.1002/anie.202114239. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y.P., Gao Y.F., Yin Y., Pan Y.C., Wang Y.Z., Song Y.J. Nanomaterial-assisted microfluidics for multiplex assays. Mikrochim Acta. 2022;189:139. doi: 10.1007/s00604-022-05226-4. [DOI] [PubMed] [Google Scholar]
- 15.Aziz A., Geng C., Fu M., Yu X., Qin K., Liu B. The role of microfluidics for organ on chip simulations. Bioengineering. 2017;4:39. doi: 10.3390/bioengineering4020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huh D., Kim H.J., Fraser J.P., Shea D.E., Khan M., Bahinski A., et al. Microfabrication of human organs-on-chips. Nat Protoc. 2013;8:2135–2157. doi: 10.1038/nprot.2013.137. [DOI] [PubMed] [Google Scholar]
- 17.Jalili-Firoozinezhad S., Miranda C.C., Cabral J.M.S. Modeling the human body on microfluidic chips. Trends Biotechnol. 2021;39:838–852. doi: 10.1016/j.tibtech.2021.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Huh D., Matthews Benjamin D., Mammoto A., Montoya-Zavala M., Hsin Hong Y., Ingber Donald E. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsano A., Conficconi C., Lemme M., Occhetta P., Gaudiello E., Votta E., et al. Beating heart on a chip: a novel microfluidic platform to generate functional 3D cardiac microtissues. Lab Chip. 2016;16:599–610. doi: 10.1039/c5lc01356a. [DOI] [PubMed] [Google Scholar]
- 20.Yoon No D., Lee K.H., Lee J., Lee S.-H. 3D liver models on a microplatform: well-defined culture, engineering of liver tissue and liver-on-a-chip. Lab Chip. 2015;15:3822–3837. doi: 10.1039/c5lc00611b. [DOI] [PubMed] [Google Scholar]
- 21.Nieskens T.T.G., Wilmer M.J. Kidney-on-a-chip technology for renal proximal tubule tissue reconstruction. Eur J Pharmacol. 2016;790:46–56. doi: 10.1016/j.ejphar.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 22.Huang D., Liu T., Liao J., Maharjan S., Xie X., Pérez M., et al. Reversed-engineered human alveolar lung-on-a-chip model. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2016146118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashammakhi N., Nasiri R., Barros N.R.D., Tebon P., Thakor J., Goudie M., et al. Gut-on-a-chip: current progress and future opportunities. Biomaterials. 2020;255 doi: 10.1016/j.biomaterials.2020.120196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pires de Mello C.P., Carmona-Moran C., McAleer C.W., Perez J., Coln E.A., Long C.J., et al. Microphysiological heart–liver body-on-a-chip system with a skin mimic for evaluating topical drug delivery. Lab Chip. 2020;20:749–759. doi: 10.1039/c9lc00861f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skardal A., Murphy S.V., Devarasetty M., Mead I., Kang H.W., Seol Y.J., et al. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci Rep. 2017;7:8837. doi: 10.1038/s41598-017-08879-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schimek K., Busek M., Brincker S., Groth B., Hoffmann S., Lauster R., et al. Integrating biological vasculature into a multi-organ-chip microsystem. Lab Chip. 2013;13:3588. doi: 10.1039/c3lc50217a. [DOI] [PubMed] [Google Scholar]
- 27.Satoh T., Sugiura S., Shin K., Onuki-Nagasaki R., Ishida S., Kikuchi K., et al. A multi-throughput multi-organ-on-a-chip system on a plate formatted pneumatic pressure-driven medium circulation platform. Lab Chip. 2018;18:115–125. doi: 10.1039/c7lc00952f. [DOI] [PubMed] [Google Scholar]
- 28.Aung A., Kumar V., Theprungsirikul J., Davey S.K., Varghese S. An engineered tumor-on-a-chip device with breast cancer–immune cell interactions for assessing T-cell recruitment. Cancer Res. 2020;80:263–275. doi: 10.1158/0008-5472.CAN-19-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carvalho M.R., Barata D., Teixeira L.M., Giselbrecht S., Reis R.L., Oliveira J.M., et al. Colorectal tumor-on-a-chip system: a 3D tool for precision onco-nanomedicine. Sci Adv. 2019;5:eaaw1317. doi: 10.1126/sciadv.aaw1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yi H.G., Jeong Y.H., Kim Y., Choi Y.-J., Moon H.E., Park S.H., et al. A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat Biomed Eng. 2019;3:509–519. doi: 10.1038/s41551-019-0363-x. [DOI] [PubMed] [Google Scholar]
- 31.Seo J., Kim K.S., Park J.W., Cho J.Y., Chang H., Fukuda J., et al. Metastasis-on-a-chip reveals adipocyte-derived lipids trigger cancer cell migration via HIF-1α activation in cancer cells. Biomaterials. 2021;269 doi: 10.1016/j.biomaterials.2020.120622. [DOI] [PubMed] [Google Scholar]
- 32.Luni C., Serena E., Elvassore N. Human-on-chip for therapy development and fundamental science. Curr Opin Biotechnol. 2014;25:45–50. doi: 10.1016/j.copbio.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y.S., Aleman J., Shin S.R., Kilic T., Kim D., Shaegh S.A.M., et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc Natl Acad Sci U S A. 2017;114:E2293–E2302. doi: 10.1073/pnas.1612906114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Low L.A., Mummery C., Berridge B.R., Austin C.P., Tagle D.A. Organs-on-chips: into the next decade. Nat Rev Drug Discov. 2021;20:345–361. doi: 10.1038/s41573-020-0079-3. [DOI] [PubMed] [Google Scholar]
- 35.Sosa-Hernández J.E., Villalba-Rodríguez A.M., Romero-Castillo K.D., Aguilar-Aguila-Isaías M.A., García-Reyes I.E., Hernández-Antonio A., et al. Organs-on-a-chip module: a review from the development and applications perspective. Micromachines. 2018;9:536. doi: 10.3390/mi9100536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rothbauer M., Rosser J.M., Zirath H., Ertl P. Tomorrow today: organ-on-a-chip advances towards clinically relevant pharmaceutical and medical in vitro models. Curr Opin Biotechnol. 2019;55:81–86. doi: 10.1016/j.copbio.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 37.Polini A., Prodanov L., Bhise N.S., Manoharan V., Dokmeci M.R., Khademhosseini A. Organs-on-a-chip: a new tool for drug discovery. Expet Opin Drug Discov. 2014;9:335–352. doi: 10.1517/17460441.2014.886562. [DOI] [PubMed] [Google Scholar]
- 38.Dhiman N., Kingshott P., Sumer H., Sharma C.S., Rath S.N. On-chip anticancer drug screening – recent progress in microfluidic platforms to address challenges in chemotherapy. Biosens Bioelectron. 2019;137:236–254. doi: 10.1016/j.bios.2019.02.070. [DOI] [PubMed] [Google Scholar]
- 39.Sun W.J., Luo Z.M., Lee J.M., Kim H.J., Lee K.J., Tebon P., et al. Organ-on-a-chip for cancer and immune organs modeling. Adv Healthc Mater. 2019;8 doi: 10.1002/adhm.201801363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quan Y., Sun M., Tan Z.Y., Eijkel J.C.T., van den Berg A., van der Meer A., et al. Organ-on-a-chip: the next generation platform for risk assessment of radiobiology. RSC Adv. 2020;10:39521–39530. doi: 10.1039/d0ra05173j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vulto P., Joore J. Adoption of organ-on-chip platforms by the pharmaceutical industry. Nat Rev Drug Discov. 2021;20:961–962. doi: 10.1038/s41573-021-00323-0. [DOI] [PubMed] [Google Scholar]
- 42.Wu Q.R., Liu J.F., Wang X.H., Feng L.Y., Wu J.B., Zhu X.L., et al. Organ-on-a-chip: recent breakthroughs and future prospects. Biomed Eng Online. 2020;19:9. doi: 10.1186/s12938-020-0752-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leung C.M., De Haan P., Ronaldson-Bouchard K., Kim G.-A., Ko J., Rho H.S., et al. A guide to the organ-on-a-chip. Nat Rev Methods Primers. 2022;2:33. [Google Scholar]
- 44.Ronaldson-Bouchard K., Vunjak-Novakovic G. Organs-on-a-chip: a fast track for engineered human tissues in drug development. Cell Stem Cell. 2018;22:310–324. doi: 10.1016/j.stem.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian C., Tu Q., Liu W., Wang J. Recent advances in microfluidic technologies for organ-on-a-chip. Trends Analyt Chem. 2019;117:146–156. [Google Scholar]
- 46.Ahadian S., Civitarese R., Bannerman D., Mohammadi M.H., Lu R., Wang E., et al. Organ-on-a-chip platforms: a convergence of advanced materials, cells, and microscale technologies. Adv Healthc Mater. 2018;7 doi: 10.1002/adhm.201800734. [DOI] [PubMed] [Google Scholar]
- 47.Wnorowski A., Yang H., Wu J.C. Progress, obstacles, and limitations in the use of stem cells in organ-on-a-chip models. Adv Drug Deliv Rev. 2019;140:3–11. doi: 10.1016/j.addr.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stucki J.D., Hobi N., Galimov A., Stucki A.O., Schneider-Daum N., Lehr C.M., et al. Medium throughput breathing human primary cell alveolus-on-chip model. Sci Rep. 2018;8 doi: 10.1038/s41598-018-32523-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dutton J.S., Hinman S.S., Kim R., Wang Y., Allbritton N.L. Primary cell-derived intestinal models: recapitulating physiology. Trends Biotechnol. 2019;37:744–760. doi: 10.1016/j.tibtech.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner I., Materne E.-M., Brincker S., Süßbier U., Frädrich C., Busek M., et al. A dynamic multi-organ-chip for long-term cultivation and substance testing proven by 3D human liver and skin tissue co-culture. Lab Chip. 2013;13:3538. doi: 10.1039/c3lc50234a. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y., Yang Q.Z., Zhang H., Han S., Liu N., Ren H., et al. Construction of cancer-on-a-chip for drug screening. Drug Discov Today. 2021;26:1875–1890. doi: 10.1016/j.drudis.2021.03.006. [DOI] [PubMed] [Google Scholar]
- 52.van der Kooy D., Weiss S. Why stem cells? Science. 2000;287:1439–1441. doi: 10.1126/science.287.5457.1439. [DOI] [PubMed] [Google Scholar]
- 53.Solter D. From teratocarcinomas to embryonic stem cells and beyond: a history of embryonic stem cell research. Nat Rev Genet. 2006;7:319–327. doi: 10.1038/nrg1827. [DOI] [PubMed] [Google Scholar]
- 54.Yu J., Vodyanik Maxim A., Smuga-Otto K., Antosiewicz-Bourget J., Frane Jennifer L., Tian S., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 55.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 56.Baker C.L., Pera M.F. Capturing totipotent stem cells. Cell Stem Cell. 2018;22:25–34. doi: 10.1016/j.stem.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 57.Hoffman L.M., Carpenter M.K. Characterization and culture of human embryonic stem cells. Nat Biotechnol. 2005;23:699–708. doi: 10.1038/nbt1102. [DOI] [PubMed] [Google Scholar]
- 58.Rippon H.J., Bishop A.E. Embryonic stem cells. Cell Prolif. 2004;37:23–34. doi: 10.1111/j.1365-2184.2004.00298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wobus A.M. Potential of embryonic stem cells. Mol Aspect Med. 2001;22:149–164. doi: 10.1016/s0098-2997(01)00006-1. [DOI] [PubMed] [Google Scholar]
- 60.Perin E.C., Geng Y.J., Willerson J.T. Adult stem cell therapy in perspective. Circulation. 2003;107:935–938. doi: 10.1161/01.cir.0000057526.10455.bd. [DOI] [PubMed] [Google Scholar]
- 61.Takahashi K., Okita K., Nakagawa M., Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc. 2007;2:3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- 62.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 63.Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 64.Robinton D.A., Daley G.Q. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park I.-H., Arora N., Huo H.G., Maherali N., Ahfeldt T., Shimamura A., et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Workman M.J., Svendsen C.N. Recent advances in human iPSC-derived models of the blood–brain barrier. Fluids Barriers CNS. 2020;17:30. doi: 10.1186/s12987-020-00191-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ellis B.W., Acun A., Can U.I., Zorlutuna P. Human iPSC-derived myocardium-on-chip with capillary-like flow for personalized medicine. Biomicrofluidics. 2017;11 doi: 10.1063/1.4978468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vatine G.D., Barrile R., Workman M.J., Sances S., Barriga B.K., Rahnama M., et al. Human iPSC-derived blood-brain barrier chips enable disease modeling and personalized medicine applications. Cell Stem Cell. 2019;24:995–1005. doi: 10.1016/j.stem.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 69.Jackson Simon A., McKenzie Rebecca E., Fagerlund Robert D., Kieper Sebastian N., Fineran Peter C., Brouns Stan J.J. CRISPR-Cas: adapting to change. Science. 2017;356 doi: 10.1126/science.aal5056. [DOI] [PubMed] [Google Scholar]
- 70.De Masi C., Spitalieri P., Murdocca M., Novelli G., Sangiuolo F. Application of CRISPR/Cas9 to human-induced pluripotent stem cells: from gene editing to drug discovery. Forensic Genom. 2020;14:25. doi: 10.1186/s40246-020-00276-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Valverde M.G., Faria J., Sendino Garví E., Janssen M.J., Masereeuw R., Mihăilă S.M. Organs-on-chip technology: a tool to tackle genetic kidney diseases. Pediatr Nephrol. 2022;37:2985–2996. doi: 10.1007/s00467-022-05508-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bassett A.R. Editing the genome of hiPSC with CRISPR/Cas9: disease models. Mamm Genome. 2017;28:348–364. doi: 10.1007/s00335-017-9684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hendriks D., Clevers H., Artegiani B. CRISPR-Cas tools and their application in genetic engineering of human stem cells and organoids. Cell Stem Cell. 2020;27:705–731. doi: 10.1016/j.stem.2020.10.014. [DOI] [PubMed] [Google Scholar]
- 74.Artegiani B., Hendriks D., Beumer J., Kok R., Zheng X., Joore I., et al. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR–Cas9 precision genome editing. Nat Cell Biol. 2020;22:321–331. doi: 10.1038/s41556-020-0472-5. [DOI] [PubMed] [Google Scholar]
- 75.Schwank G., Koo B.K., Sasselli V., Johanna Heo I., Demircan T., et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 76.Kim J., Koo B.K., Yoon K.J. Modeling host-virus interactions in viral infectious diseases using stem-cell-derived systems and CRISPR/Cas9 technology. Viruses. 2019;11:124. doi: 10.3390/v11020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tian R., Gachechiladze M.A., Ludwig C.H., Laurie M.T., Hong J.Y., Nathaniel D., et al. CRISPR interference-based platform for multimodal genetic screens in human iPSC-derived neurons. Neuron. 2019;104:239–255. doi: 10.1016/j.neuron.2019.07.014. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ding C.Z., Chen X., Kang Q.S., Yan X.H. Biomedical application of functional materials in organ-on-a-chip. Front Bioeng Biotechnol. 2020;8:823. doi: 10.3389/fbioe.2020.00823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Toepke M.W., Beebe D.J. PDMS absorption of small molecules and consequences in microfluidic applications. Lab Chip. 2006;6:1484–1486. doi: 10.1039/b612140c. [DOI] [PubMed] [Google Scholar]
- 80.van Meer B.J., de Vries H., Firth K.S.A., van Weerd J., Tertoolen L.G.J., Karperien H.B.J., et al. Small molecule absorption by PDMS in the context of drug response bioassays. Biochem Biophys Res Commun. 2017;482:323–328. doi: 10.1016/j.bbrc.2016.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Radisic M., Loskill P. Beyond PDMS and membranes: new materials for organ-on-a-chip devices. ACS Biomater Sci Eng. 2021;7:2861–2863. doi: 10.1021/acsbiomaterials.1c00831. [DOI] [PubMed] [Google Scholar]
- 82.Eisenbarth E. Biomaterials for tissue engineering. Adv Eng Mater. 2007;9:1051–1060. [Google Scholar]
- 83.Keane T.J., Badylak S.F. Biomaterials for tissue engineering applications. Semin Pediatr Surg. 2014;23:112–118. doi: 10.1053/j.sempedsurg.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 84.Weng Y.S., Chang S.F., Shih M.C., Tseng S.H., Lai C.H. Scaffold-free liver-on-a-chip with multiscale organotypic cultures. Adv Mater. 2017;29 doi: 10.1002/adma.201701545. [DOI] [PubMed] [Google Scholar]
- 85.Lee H., Cho D.W. One-step fabrication of an organ-on-a-chip with spatial heterogeneity using a 3D bioprinting technology. Lab Chip. 2016;16:2618–2625. doi: 10.1039/c6lc00450d. [DOI] [PubMed] [Google Scholar]
- 86.Yang Q.Z., Lian Q., Xu F. Perspective: fabrication of integrated organ-on-a-chip via bioprinting. Biomicrofluidics. 2017;11 doi: 10.1063/1.4982945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Faulkner-Jones A., Zamora V., Hortigon-Vinagre M.P., Wang W., Ardron M., Smith G.L., et al. A bioprinted heart-on-a-chip with human pluripotent stem cell-derived cardiomyocytes for drug evaluation. Bioengineering. 2022;9:32. doi: 10.3390/bioengineering9010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang B., Korolj A., Lai B.F.L., Radisic M. Advances in organ-on-a-chip engineering. Nat Rev Mater. 2018;3:257–278. [Google Scholar]
- 89.Kaarj K., Yoon J.Y. Methods of delivering mechanical stimuli to organ-on-a-chip. Micromachines. 2019;10:700. doi: 10.3390/mi10100700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thompson C.L., Fu S., Heywood H.K., Knight M.M., Thorpe S.D. Mechanical stimulation: a crucial element of organ-on-chip models. Front Bioeng Biotechnol. 2020;8 doi: 10.3389/fbioe.2020.602646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nunes S.S., Miklas J.W., Liu J., Aschar-Sobbi R., Xiao Y., Zhang B., et al. Biowire: a platform for maturation of human pluripotent stem cell–derived cardiomyocytes. Nat Methods. 2013;10:781–787. doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen F.M., Zhang M., Wu Z.F. Toward delivery of multiple growth factors in tissue engineering. Biomaterials. 2010;31:6279–6308. doi: 10.1016/j.biomaterials.2010.04.053. [DOI] [PubMed] [Google Scholar]
- 93.Phan D.T.T., Wang X.L., Craver B.M., Sobrino A., Zhao D., Chen J.C., et al. A vascularized and perfused organ-on-a-chip platform for large-scale drug screening applications. Lab Chip. 2017;17:511–520. doi: 10.1039/c6lc01422d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang Y.Q., Wang L., Guo Y.Q., Zhu Y.J., Qin J.H. Engineering stem cell-derived 3D brain organoids in a perfusable organ-on-a-chip system. RSC Adv. 2018;8:1677–1685. doi: 10.1039/c7ra11714k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li X., Tian T. Recent advances in an organ-on-a-chip: biomarker analysis and applications. Anal Methods. 2018;10:3122–3130. [Google Scholar]
- 96.Zhu Y., Mandal K., Hernandez A.L., Kawakita S., Huang W., Bandaru P., et al. State of the art in integrated biosensors for organ-on-a-chip applications. Curr Opin Biomed Eng. 2021;19 doi: 10.1016/j.cobme.2021.100309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Aleman J., Kilic T., Mille L.S., Shin S.R., Zhang Y.S. Microfluidic integration of regeneratable electrochemical affinity-based biosensors for continual monitoring of organ-on-a-chip devices. Nat Protoc. 2021;16:2564–2593. doi: 10.1038/s41596-021-00511-7. [DOI] [PubMed] [Google Scholar]
- 98.Ferrari E., Palma C., Vesentini S., Occhetta P., Rasponi M. Integrating biosensors in organs-on-chip devices: a perspective on current strategies to monitor microphysiological systems. Biosensors. 2020;10:110. doi: 10.3390/bios10090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fuchs S., Johansson S., Tjell A.Ø., Werr G., Mayr T., Tenje M. In-line analysis of organ-on-chip systems with sensors: integration, fabrication, challenges, and potential. ACS Biomater Sci Eng. 2021;7:2926–2948. doi: 10.1021/acsbiomaterials.0c01110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cho K.W., Lee W.H., Kim B.S., Kim D.H. Sensors in heart-on-a-chip: a review on recent progress. Talanta. 2020;219 doi: 10.1016/j.talanta.2020.121269. [DOI] [PubMed] [Google Scholar]
- 101.Arlett J.L., Myers E.B., Roukes M.L. Comparative advantages of mechanical biosensors. Nat Nanotechnol. 2011;6:203–215. doi: 10.1038/nnano.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ju Lind, Busbee T.A., Valentine A.D., Pasqualini F.S., Yuan H.Y., Yadid M., et al. Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nat Mater. 2017;16:303–308. doi: 10.1038/nmat4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Clarke G.A., Hartse B.X., Niaraki Asli A.E., Taghavimehr M., Hashemi N., Abbasi Shirsavar M., et al. Advancement of sensor integrated organ-on-chip devices. Sensors. 2021;21:1367. doi: 10.3390/s21041367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kilic T., Navaee F., Stradolini F., Renaud P., Carrara S. Organs-on-chip monitoring: sensors and other strategies. Microphysiol Syst. 2018;2:1–32. [Google Scholar]
- 105.Nawaz M.S., Shoaib B., Ashraf M.A. Intelligent cardiovascular disease prediction empowered with gradient descent optimization. Heliyon. 2021;7 doi: 10.1016/j.heliyon.2021.e06948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ferri N., Siegl P., Corsini A., Herrmann J., Lerman A., Benghozi R. Drug attrition during pre-clinical and clinical development: understanding and managing drug-induced cardiotoxicity. Pharmacol Ther. 2013;138:470–484. doi: 10.1016/j.pharmthera.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 107.Simmons C.S., Petzold B.C., Pruitt B.L. Microsystems for biomimetic stimulation of cardiac cells. Lab Chip. 2012;12:3235–3248. doi: 10.1039/c2lc40308k. [DOI] [PubMed] [Google Scholar]
- 108.Chen J.F., Mandel E.M., Thomson J.M., Wu Q., Callis T.E., Hammond S.M., et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang X., Wang T.X., Wang P., Hu N. High-throughput assessment of drug cardiac safety using a high-speed impedance detection technology-based heart-on-a-chip. Micromachines. 2016;7:122. doi: 10.3390/mi7070122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Agarwal A., Goss J.A., Cho A., McCain M.L., Parker K.K. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab Chip. 2013;13:3599. doi: 10.1039/c3lc50350j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang Y.S., Arneri A., Bersini S., Shin S.R., Zhu K., Goli-Malekabadi Z., et al. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials. 2016;110:45–59. doi: 10.1016/j.biomaterials.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kujala V.J., Pasqualini F.S., Goss J.A., Nawroth J.C., Parker K.K. Laminar ventricular myocardium on a microelectrode array-based chip. J Mater Chem B. 2016;4:3534–3543. doi: 10.1039/c6tb00324a. [DOI] [PubMed] [Google Scholar]
- 113.Bhise N.S., Manoharan V., Massa S., Tamayol A., Ghaderi M., Miscuglio M., et al. A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication. 2016;8 doi: 10.1088/1758-5090/8/1/014101. [DOI] [PubMed] [Google Scholar]
- 114.Freag M.S., Namgung B., Reyna Fernandez M.E., Gherardi E., Sengupta S., Jang H.L. Human nonalcoholic steatohepatitis on a chip. Hepatol Commun. 2021;5:217–233. doi: 10.1002/hep4.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee W.M. Acetaminophen (APAP) hepatotoxicity—isn’t it time for APAP to go away? J Hepatol. 2017;67:1324–1331. doi: 10.1016/j.jhep.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stevens J.L., Baker T.K. The future of drug safety testing: expanding the view and narrowing the focus. Drug Discov Today. 2009;14:162‒7. doi: 10.1016/j.drudis.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 117.Delalat B., Cozzi C., Rasi Ghaemi S., Polito G., Kriel F.H., Michl T.D., et al. Microengineered bioartificial liver chip for drug toxicity screening. Adv Funct Mater. 2018;28 [Google Scholar]
- 118.Du Y., Li N., Yang H., Luo C.H., Gong Y.X., Tong C.F., et al. Mimicking liver sinusoidal structures and functions using a 3D-configured microfluidic chip. Lab Chip. 2017;17:782–794. doi: 10.1039/c6lc01374k. [DOI] [PubMed] [Google Scholar]
- 119.Beckwitt C.H., Clark A.M., Wheeler S., Taylor D.L., Stolz D.B., Griffith L., et al. Liver ‘organ on a chip. Exp Cell Res. 2018;363:15–25. doi: 10.1016/j.yexcr.2017.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee S.-A., No D.Y., Kang E., Ju J., Kim D.S., Lee S.H. Spheroid-based three-dimensional liver-on-a-chip to investigate hepatocyte–hepatic stellate cell interactions and flow effects. Lab Chip. 2013;13:3529. doi: 10.1039/c3lc50197c. [DOI] [PubMed] [Google Scholar]
- 121.Ma L., Wu Y.T., Li Y.T., Aazmi A., Zhou H.Z., Zhang B., et al. Current advances on 3D-bioprinted liver tissue models. Adv Healthc Mater. 2020;9 doi: 10.1002/adhm.202001517. [DOI] [PubMed] [Google Scholar]
- 122.Jaeschke H., Adelusi O.B., Akakpo J.Y., Nguyen N.T., Sanchez-Guerrero G., Umbaugh D.S., et al. Recommendations for the use of the acetaminophen hepatotoxicity model for mechanistic studies and how to avoid common pitfalls. Acta Pharm Sin B. 2021;11:3740–3755. doi: 10.1016/j.apsb.2021.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Prot J.M., Bunescu A., Elena-Herrmann B., Aninat C., Snouber L.C., Griscom L., et al. Predictive toxicology using systemic biology and liver microfluidic “on chip” approaches: application to acetaminophen injury. Toxicol Appl Pharmacol. 2012;259:270–280. doi: 10.1016/j.taap.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 124.Knowlton S., Tasoglu S. A bioprinted liver-on-a-chip for drug screening applications. Trends Biotechnol. 2016;34:681–682. doi: 10.1016/j.tibtech.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 125.Slaughter V.L., Rumsey J.W., Boone R., Malik D., Cai Y., Sriram N.N., et al. Validation of an adipose-liver human-on-a-chip model of NAFLD for preclinical therapeutic efficacy evaluation. Sci Rep. 2021;11 doi: 10.1038/s41598-021-92264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Starokozhko V., Groothuis G.M.M. Judging the value of ‘liver-on-a-chip’ devices for prediction of toxicity. Expet Opin Drug Metabol Toxicol. 2017;13:125–128. doi: 10.1080/17425255.2017.1246537. [DOI] [PubMed] [Google Scholar]
- 127.Sarkar U., Rivera-Burgos D., Large E.M., Hughes D.J., Ravindra K.C., Dyer R.L., et al. Metabolite profiling and pharmacokinetic evaluation of hydrocortisone in a perfused three-dimensional human liver bioreactor. Drug Metab Dispos. 2015;43:1091. doi: 10.1124/dmd.115.063495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen P.Y., Hsieh M.J., Liao Y.H., Lin Y.C., Hou Y.T. Liver-on-a-chip platform to study anticancer effect of statin and its metabolites. Biochem Eng J. 2021;165 [Google Scholar]
- 129.Paoli R., Samitier J. Mimicking the kidney: a key role in organ-on-chip development. Micromachines. 2016;7:126. doi: 10.3390/mi7070126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wilmer M.J., Ng C.P., Lanz H.L., Vulto P., Suter-Dick L., Masereeuw R. Kidney-on-a-chip technology for drug-induced nephrotoxicity screening. Trends Biotechnol. 2016;34:156–170. doi: 10.1016/j.tibtech.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 131.Musah S., Dimitrakakis N., Camacho D.M., Church G.M., Ingber D.E. Directed differentiation of human induced pluripotent stem cells into mature kidney podocytes and establishment of a glomerulus chip. Nat Protoc. 2018;13:1662. doi: 10.1038/s41596-018-0007-8. 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xu Y.Y., Qin S., Niu Y.N., Gong T., Zhang Z.R., Fu Y. Effect of fluid shear stress on the internalization of kidney-targeted delivery systems in renal tubular epithelial cells. Acta Pharm Sin B. 2020;10:680–692. doi: 10.1016/j.apsb.2019.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Faria J., Ahmed S., Gerritsen K.G.F., Mihaila S.M., Masereeuw R. Kidney-based in vitro models for drug-induced toxicity testing. Arch Toxicol. 2019;93:3397–3418. doi: 10.1007/s00204-019-02598-0. [DOI] [PubMed] [Google Scholar]
- 134.Homan K.A., Kolesky D.B., Skylar-Scott M.A., Herrmann J., Obuobi H., Moisan A., et al. Bioprinting of 3d convoluted renal proximal tubules on perfusable chips. Sci Rep. 2016;6 doi: 10.1038/srep34845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Weber E.J., Lidberg K.A., Wang L., Bammler T.K., Macdonald J.W., Li M.J., et al. Human kidney on a chip assessment of polymyxin antibiotic nephrotoxicity. JCI Insight. 2018;3 doi: 10.1172/jci.insight.123673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kim S., LesherPerez S.C., Yamanishi C., Labuz J.M., Leung B., Takayama S. Pharmacokinetic profile that reduces nephrotoxicity of gentamicin in a perfused kidney-on-a-chip. Biofabrication. 2016;8 doi: 10.1088/1758-5090/8/1/015021. [DOI] [PubMed] [Google Scholar]
- 137.Vormann M.K., Vriend J., Lanz H.L., Gijzen L., van den Heuvel A., Hutter S., et al. Implementation of a human renal proximal tubule on a chip for nephrotoxicity and drug interaction studies. J Pharm Sci. 2021;110:1601–1614. doi: 10.1016/j.xphs.2021.01.028. [DOI] [PubMed] [Google Scholar]
- 138.Yin L., Du G.R., Zhang B., Zhang H.B., Yin R.X., Zhang W.J., et al. Efficient drug screening and nephrotoxicity assessment on co-culture microfluidic kidney chip. Sci Rep. 2020;10:6568. doi: 10.1038/s41598-020-63096-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Allison S.J. Kidney glomerulus-on-a-chip. Nat Rev Nephrol. 2017;13:382. doi: 10.1038/nrneph.2017.79. [DOI] [PubMed] [Google Scholar]
- 140.Musah S., Mammoto A., Ferrante T.C., Jeanty S.S.F., Hirano-Kobayashi M., Mammoto T., et al. Mature induced-pluripotent-stem-cell-derived human podocytes reconstitute kidney glomerular-capillary-wall function on a chip. Nat Biomed Eng. 2017;1 doi: 10.1038/s41551-017-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Petrosyan A., Cravedi P., Villani V., Angeletti A., Manrique J., Renieri A., et al. A glomerulus-on-a-chip to recapitulate the human glomerular filtration barrier. Nat Commun. 2019;10:3653. doi: 10.1038/s41467-019-11577-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Booth R., Kim H. Characterization of a microfluidic in vitro model of the blood-brain barrier (μBBB) Lab Chip. 2012;12:1784. doi: 10.1039/c2lc40094d. [DOI] [PubMed] [Google Scholar]
- 143.Maoz B.M., Herland A., FitzGerald E.A., Grevesse T., Vidoudez C., Pacheco A.R., et al. A linked organ-on-chip model of the human neurovascular unit reveals the metabolic coupling of endothelial and neuronal cells. Nat Biotechnol. 2018;36:865–874. doi: 10.1038/nbt.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Feigin Vlnichols Ealam Tbannick Msbeghi Eblake N., et al. Global, regional, and national burden of neurological disorders, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019;18:459–480. doi: 10.1016/S1474-4422(18)30499-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kola I., Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3:711–716. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 146.Amirifar L., Shamloo A., Nasiri R., de Barros N.R., Wang Z.Z., Unluturk B.D., et al. Brain-on-a-chip: recent advances in design and techniques for microfluidic models of the brain in health and disease. Biomaterials. 2022 doi: 10.1016/j.biomaterials.2022.121531. [DOI] [PubMed] [Google Scholar]
- 147.Chatellier D., Jourdain M., Mangalaboyi J., Ader F., Chopin C., Derambure P., et al. Cefepime-induced neurotoxicity: an underestimated complication of antibiotherapy in patients with acute renal failure. Crit Care Explor. 2002;28:214. doi: 10.1007/s00134-001-1170-9. 7. [DOI] [PubMed] [Google Scholar]
- 148.James S.E., Burden H., Burgess R., Xie Y.M., Yang T., Massa S.M., et al. Anti-cancer drug induced neurotoxicity and identification of Rho pathway signaling modulators as potential neuroprotectants. Neurotoxicology. 2008;29:605–612. doi: 10.1016/j.neuro.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Stone J.B., DeAngelis L.M. Cancer-treatment-induced neurotoxicity—focus on newer treatments. Nat Rev Clin Oncol. 2016;13:92–105. doi: 10.1038/nrclinonc.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Walker A.L., Imam S.Z., Roberts R.A. Drug discovery and development: biomarkers of neurotoxicity and neurodegeneration. Exp Biol Med. 2018;243:1037–1045. doi: 10.1177/1535370218801309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jensen G., Morrill C., Huang Y. 3D tissue engineering, an emerging technique for pharmaceutical research. Acta Pharm Sin B. 2018;8:756–766. doi: 10.1016/j.apsb.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Harberts J., Fendler C., Teuber J., Siegmund M., Silva A., Rieck N., et al. Toward brain-on-a-chip: human induced pluripotent stem cell-derived guided neuronal networks in tailor-made 3d nanoprinted microscaffolds. ACS Nano. 2020;14:13091–13102. doi: 10.1021/acsnano.0c04640. [DOI] [PubMed] [Google Scholar]
- 153.Bang S., Jeong S., Choi N., Kim H.N. Brain-on-a-chip: a history of development and future perspective. Biomicrofluidics. 2019;13 doi: 10.1063/1.5120555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Baker M. A living system on a chip. Nature. 2011;471:661–665. doi: 10.1038/471661a. [DOI] [PubMed] [Google Scholar]
- 155.Benam K.H., Villenave R., Lucchesi C., Varone A., Hubeau C., Lee H.H., et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses. in vitro. Nat Methods. 2016;13:151–157. doi: 10.1038/nmeth.3697. [DOI] [PubMed] [Google Scholar]
- 156.Zhang M., Wang P., Luo R.H., Wang Y.Q., Li Z.Y., Guo Y.Q., et al. Biomimetic human disease model of SARS-CoV-2-induced lung injury and immune responses on organ chip system. Adv Sci. 2021;8 doi: 10.1002/advs.202002928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhang X.T., Han Y., Huang W., Jin M., Gao Z. The influence of the gut microbiota on the bioavailability of oral drugs. Acta Pharm Sin B. 2021;11:1789–1812. doi: 10.1016/j.apsb.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Guo Y.Q., Li Z.Y., Su W.T., Wang L., Zhu Y.J., Qin J.H. A biomimetic human gut-on-a-chip for modeling drug metabolism in intestine. Artif Organs. 2018;42:1196–1205. doi: 10.1111/aor.13163. [DOI] [PubMed] [Google Scholar]
- 159.Jalili-Firoozinezhad S., Prantil-Baun R., Jiang A., Potla R., Mammoto T., Weaver J.C., et al. Modeling radiation injury-induced cell death and countermeasure drug responses in a human Gut-on-a-Chip. Cell Death Dis. 2018;9:233. doi: 10.1038/s41419-018-0304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bein A., Kim S., Goyal G., Cao W.J., Fadel C., Naziripour A., et al. Enteric coronavirus infection and treatment modeled with an immunocompetent human intestine-on-a-chip. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.718484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang Q., Sito L., Mao M., He J.K., Zhang Y.S., Zhao X. Current advances in skin-on-a-chip models for drug testing. Microphysiol Syst. 2018;1:1. doi: 10.21037/mps.2018.08.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wufuer M., Lee G.H., Hur W., Jeon B.J., Kim B.J., Choi T.H., et al. Skin-on-a-chip model simulating inflammation, edema and drug-based treatment. Sci Rep. 2016;6 doi: 10.1038/srep37471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kim S., Kim W., Lim S., Jeon J. Vasculature-on-a-chip for in vitro disease models. Bioengineering. 2017;4:8. doi: 10.3390/bioengineering4010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cao X., Ashfaq R., Cheng F., Maharjan S., Li J., Ying G.L., et al. A tumor-on-a-chip system with bioprinted blood and lymphatic vessel pair. Adv Funct Mater. 2019;29 doi: 10.1002/adfm.201807173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Osaki T., Sivathanu V., Kamm R.D. Vascularized microfluidic organ-chips for drug screening, disease models and tissue engineering. Curr Opin Biotechnol. 2018;52:116–123. doi: 10.1016/j.copbio.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pauty J., Usuba R., Cheng I.G., Hespel L., Takahashi H., Kato K., et al. A vascular endothelial growth factor-dependent sprouting angiogenesis assay based on an in vitro human blood vessel model for the study of anti-angiogenic drugs. EBioMedicine. 2018;27:225–236. doi: 10.1016/j.ebiom.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Spijkers X.M., Pasteuning-Vuhman S., Dorleijn J.C., Vulto P., Wevers N.R., Pasterkamp R.J. A directional 3D neurite outgrowth model for studying motor axon biology and disease. Sci Rep. 2021;11:2080. doi: 10.1038/s41598-021-81335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ruiz A., Joshi P., Mastrangelo R., Francolini M., Verderio C., Matteoli M. Testing Aβ toxicity on primary CNS cultures using drug-screening microfluidic chips. Lab Chip. 2014;14:2860–2866. doi: 10.1039/c4lc00174e. [DOI] [PubMed] [Google Scholar]
- 169.Dhami S.P.S., Kappala S.S., Thompson A., Szegezdi E. Three-dimensional ex vivo co-culture models of the leukaemic bone marrow niche for functional drug testing. Drug Discov Today. 2016;21:1464–1471. doi: 10.1016/j.drudis.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 170.Houshmand M., Soleimani M., Atashi A., Saglio G., Abdollahi M., Nikougoftar Zarif M. Mimicking the acute myeloid leukemia niche for molecular study and drug screening. Tissue Eng C Methods. 2016;23:72–85. doi: 10.1089/ten.TEC.2016.0404. [DOI] [PubMed] [Google Scholar]
- 171.McCarthy M., Brown T., Alarcon A., Williams C., Wu X., Abbott R.D., et al. Fat-on-a-chip models for research and discovery in obesity and its metabolic comorbidities. Tissue Eng B Rev. 2020;26:586–595. doi: 10.1089/ten.teb.2019.0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Picollet-D’hahan N., Zuchowska A., Lemeunier I., Le Gac S. Multiorgan-on-a-chip: a systemic approach to model and decipher inter-organ communication. Trends Biotechnol. 2021;39:788–810. doi: 10.1016/j.tibtech.2020.11.014. [DOI] [PubMed] [Google Scholar]
- 173.Shinha K., Nihei W., Ono T., Nakazato R., Kimura H. A pharmacokinetic–pharmacodynamic model based on multi-organ-on-a-chip for drug–drug interaction studies. Biomicrofluidics. 2020;14 doi: 10.1063/5.0011545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ferrari E., Rasponi M. Liver–Heart on chip models for drug safety. APL Bioeng. 2021;5 doi: 10.1063/5.0048986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Boeri L., Izzo L., Sardelli L., Tunesi M., Albani D., Giordano C. Advanced organ-on-a-chip devices to investigate liver multi-organ communication: focus on gut, microbiota and brain. Bioengineering. 2019;6:91. doi: 10.3390/bioengineering6040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Oleaga C., Riu A., Rothemund S., Lavado A., McAleer C.W., Long C.J., et al. Investigation of the effect of hepatic metabolism on off-target cardiotoxicity in a multi-organ human-on-a-chip system. Biomaterials. 2018;182:176–190. doi: 10.1016/j.biomaterials.2018.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Theobald J., Abu El Maaty M.A., Kusterer N., Wetterauer B., Wink M., Cheng X., et al. In vitro metabolic activation of vitamin D3 by using a multi-compartment microfluidic liver-kidney organ on chip platform. Sci Rep. 2019;9:4616. doi: 10.1038/s41598-019-40851-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Oleaga C., Bernabini C., Smith A.S.T., Srinivasan B., Jackson M., McLamb W., et al. Multi-Organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci Rep. 2016;6 doi: 10.1038/srep20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Rajan S.A.P., Aleman J., Wan M., Pourhabibi Zarandi N., Nzou G., Murphy S., et al. Probing prodrug metabolism and reciprocal toxicity with an integrated and humanized multi-tissue organ-on-a-chip platform. Acta Biomater. 2020;106:124–135. doi: 10.1016/j.actbio.2020.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bovard D., Sandoz A., Luettich K., Frentzel S., Iskandar A., Marescotti D., et al. A lung/liver-on-a-chip platform for acute and chronic toxicity studies. Lab Chip. 2018;18:3814–3829. doi: 10.1039/c8lc01029c. [DOI] [PubMed] [Google Scholar]
- 181.Theobald J., Ghanem A., Wallisch P., Banaeiyan A.A., Andrade-Navarro M.A., Taškova K., et al. Liver-kidney-on-chip to study toxicity of drug metabolites. ACS Biomater Sci Eng. 2018;4:78–89. doi: 10.1021/acsbiomaterials.7b00417. [DOI] [PubMed] [Google Scholar]
- 182.Maschmeyer I., Lorenz A.K., Schimek K., Hasenberg T., Ramme A.P., Hübner J., et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip. 2015;15:2688–2699. doi: 10.1039/c5lc00392j. [DOI] [PubMed] [Google Scholar]
- 183.Li Z.Y., Su W.T., Zhu Y.J., Tao T.T., Li D., Peng X.J., et al. Drug absorption related nephrotoxicity assessment on an intestine-kidney chip. Biomicrofluidics. 2017;11 doi: 10.1063/1.4984768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lee Y., Kim M.-H., Alves D.R., Kim S., Lee L.P., Sung J.H., et al. Gut–kidney axis on chip for studying effects of antibiotics on risk of hemolytic uremic syndrome by shiga toxin-producing. Escherichia Coli. Toxins. 2021;13:775. doi: 10.3390/toxins13110775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 186.Wan L., Neumann C.A., Leduc P.R. Tumor-on-a-chip for integrating a 3D tumor microenvironment: chemical and mechanical factors. Lab Chip. 2020;20:873–888. doi: 10.1039/c9lc00550a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Subia B., Dahiya U.R., Mishra S., Ayache J., Casquillas G.V., Caballero D., et al. Breast tumor-on-chip models: from disease modeling to personalized drug screening. J Control Release. 2021;331:103–120. doi: 10.1016/j.jconrel.2020.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhang Y.S., Zhang Y.N., Zhang W. Cancer-on-a-chip systems at the frontier of nanomedicine. Drug Discov Today. 2017;22:1392–1399. doi: 10.1016/j.drudis.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Portillo-Lara R., Annabi N. Microengineered cancer-on-a-chip platforms to study the metastatic microenvironment. Lab Chip. 2016;16:4063–4081. doi: 10.1039/c6lc00718j. [DOI] [PubMed] [Google Scholar]
- 190.Sleeboom J.J.F., Eslami Amirabadi H., Nair P., Sahlgren C.M., den Toonder J.M.J. Metastasis in context: modeling the tumor microenvironment with cancer-on-a-chip approaches. Dis Model Mech. 2018;11 doi: 10.1242/dmm.033100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kashaninejad N., Nikmaneshi M., Moghadas H., Kiyoumarsi Oskouei A., Rismanian M., Barisam M., et al. Organ-tumor-on-a-chip for chemosensitivity assay: a critical review. Micromachines. 2016;7:130. doi: 10.3390/mi7080130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Khalid M.A.U., Kim Y.S., Ali M., Lee B.G., Cho Y.J., Choi K.H. A lung cancer-on-chip platform with integrated biosensors for physiological monitoring and toxicity assessment. Biochem Eng J. 2020;155 [Google Scholar]
- 193.Xie M.J., Gao Q., Fu J.Z., Chen Z., He Y. Bioprinting of novel 3D tumor array chip for drug screening. Biodes Manuf. 2020;3:175–188. [Google Scholar]
- 194.Hevener K., Verstak T.A., Lutat K.E., Riggsbee D.L., Mooney J.W. Recent developments in topoisomerase-targeted cancer chemotherapy. Acta Pharm Sin B. 2018;8:844–861. doi: 10.1016/j.apsb.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Jiang W., Cai G., Hu P., Wang Y. Personalized medicine of non-gene-specific chemotherapies for non-small cell lung cancer. Acta Pharm Sin B. 2021;11:3406–3416. doi: 10.1016/j.apsb.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hoekman K., van der Vijgh W.J.F., Vermorken J.B. Clinical and preclinical modulation of chemotherapy-induced toxicity in patients with cancer. Drugs. 1999;57:133–155. doi: 10.2165/00003495-199957020-00002. [DOI] [PubMed] [Google Scholar]
- 197.Lee J., Mehrotra S., Zare-Eelanjegh E., Rodrigues R.O., Akbarinejad A., Ge D., et al. A heart-breast cancer-on-a-chip platform for disease modeling and monitoring of cardiotoxicity induced by cancer chemotherapy. Small. 2021;17 doi: 10.1002/smll.202004258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Weng K.C., Kurokawa Y.K., Hajek B.S., Paladin J.A., Shirure V.S., George S.C. Human induced pluripotent stem-cardiac-endothelial-tumor-on-a-chip to assess anticancer efficacy and cardiotoxicity. Tissue Eng C Methods. 2020;26:44–55. doi: 10.1089/ten.tec.2019.0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kamei K.I., Kato Y., Hirai Y., Ito S., Satoh J., Oka A., et al. Integrated heart/cancer on a chip to reproduce the side effects of anti-cancer drugs. in vitro. RSC Adv. 2017;7:36777–36786. [Google Scholar]
- 200.Hou Y., Ai X.N., Zhao L., Gao Z., Wang Y.J., Lu Y.Y., et al. An integrated biomimetic array chip for high-throughput co-culture of liver and tumor microtissues for advanced anticancer bioactivity screening. Lab Chip. 2020;20:2482–2494. doi: 10.1039/d0lc00288g. [DOI] [PubMed] [Google Scholar]
- 201.Lohasz C., Bonanini F., Hoelting L., Renggli K., Frey O., Hierlemann A. Predicting metabolism-related drug–drug interactions using a microphysiological multitissue system. Adv Biosyst. 2020;4 doi: 10.1002/adbi.202000079. [DOI] [PubMed] [Google Scholar]
- 202.Tatosian D.A., Shuler M.L. A novel system for evaluation of drug mixtures for potential efficacy in treating multidrug resistant cancers. Biotechnol Bioeng. 2009;103:187–198. doi: 10.1002/bit.22219. [DOI] [PubMed] [Google Scholar]
- 203.Jeon Jessie S., Bersini S., Gilardi M., Dubini G., Charest Joseph L., Moretti M., et al. Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proc Natl Acad Sci U S A. 2015;112:214–219. doi: 10.1073/pnas.1417115112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Berthier E., Young E.W.K., Beebe D. Engineers are from PDMS-land, biologists are from polystyrenia. Lab Chip. 2012;12:1224. doi: 10.1039/c2lc20982a. [DOI] [PubMed] [Google Scholar]
- 205.Kuddannaya S., Chuah Y.J., Lee M.H.A., Menon N.V., Kang Y., Zhang Y. Surface chemical modification of poly(dimethylsiloxane) for the enhanced adhesion and proliferation of mesenchymal stem cells. ACS Appl Mater Interfaces. 2013;5:9777–9784. doi: 10.1021/am402903e. [DOI] [PubMed] [Google Scholar]
- 206.Campbell S.B., Wu Q., Yazbeck J., Liu C., Okhovatian S., Radisic M. Beyond polydimethylsiloxane: alternative materials for fabrication of organ-on-a-chip devices and microphysiological systems. ACS Biomater Sci Eng. 2021;7:2880–2899. doi: 10.1021/acsbiomaterials.0c00640. [DOI] [PubMed] [Google Scholar]
- 207.Adhikary P.P., Ul Ain Q., Hocke A.C., Hedtrich S. COVID-19 highlights the model dilemma in biomedical research. Nat Rev Mater. 2021;6:374–376. doi: 10.1038/s41578-021-00305-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Si L.L., Bai H.Q., Rodas M., Cao W.J., Oh C.Y., Jiang A., et al. A human-airway-on-a-chip for the rapid identification of candidate antiviral therapeutics and prophylactics. Nat Biomed Eng. 2021;5:815–829. doi: 10.1038/s41551-021-00718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.van den Berg A., Mummery C.L., Passier R., van der Meer A.D. Personalised organs-on-chips: functional testing for precision medicine. Lab Chip. 2019;19:198–205. doi: 10.1039/c8lc00827b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Piergiovanni M., Leite S.B., Corvi R., Whelan M. Standardisation needs for organ on chip devices. Lab Chip. 2021;21:2857–2868. doi: 10.1039/d1lc00241d. [DOI] [PubMed] [Google Scholar]