Abstract
Combined multi-omics analysis of proteomics, polar metabolomics, and lipidomics requires separate liquid chromatography–mass spectrometry (LC–MS) platforms for each omics layer. This requirement for different platforms limits throughput and increases costs, preventing the application of mass spectrometry-based multi-omics to large scale drug discovery or clinical cohorts. Here, we present an innovative strategy for simultaneous multi-omics analysis by direct infusion (SMAD) using one single injection without liquid chromatography. SMAD allows quantification of over 9,000 metabolite m/z features and over 1,300 proteins from the same sample in less than five minutes. We validated the efficiency and reliability of this method and then present two practical applications: mouse macrophage M1/M2 polarization and high throughput drug screening in human 293T cells. Finally, we demonstrate relationships between proteomic and metabolomic data are discovered by machine learning.
Introduction
Multi-omics analysis and integration, which involves the simultaneous analysis of at least a pair of genomic, epigenomic, transcriptomic, proteomic, lipidomic, and metabolomic data, has become increasingly essential for gaining a comprehensive understanding of biological processes and disease progression.1, 2 Currently, the combined use of LC and MS is the prevailing technology for proteome, metabolome and lipidome analysis. Separation of analytes with LC before MS is important to increase sensitivity and coverage, which enables detection of over 10,000 protein groups or thousands of metabolites from separate injection.3–6 However, despite advances in LC to enable shorter gradients, LC ultimately limits throughput of MS-based omics because of separation time followed by time for column washing and equilibration, and also due to requirements for different LC configurations for each omics layer.
The logical extreme of shorter LC is to remove LC completely and analyze molecules directly by direct infusion, which has already been demonstrated for both proteome and metabolome analysis.7–9 However, there exist two key challenges that restrict the coverage and depth of direct infusion mass spectrometry (DI-MS) methods: (1) ion suppression at the ion source caused by variations in ionization efficiency or abundance of ionizable analytes, and (2) ion competition effect in the mass analyzer, where high abundant ions conceal lower abundance ones.
Recent improvements in MS instrumentation (acquisition speed, mass resolution and sensitivity),10–12 advancements in mass spectrum interpretation software,13–17 and the integration of MS and ion mobility techniques (FAIMS, TIMS)18–21 have encouraged us to revisit the potential of DI-MS. For example, a spectral-stitching DI-MS method, which measures data as a series of mass-to-charge (m/z) intervals that are subsequently ‘stitched’ together to create a full mass spectrum, realized a total of ~9,000 lipidome and metabolome m/z features in ~5 min.22 An additional innovative technique for direct infusion metabolomic analysis involves using computational analysis to determine the optimal scan ranges that would yield the greatest number of m/z features.23 For direct infusion proteomic analysis, we originally described a rapid quantitative proteome analysis method by using two gas-phase separations by ion mobility and quadrupole selection, which identified over 500 proteins and quantified over 300 proteins in up to 3 min of acquisition time per sample.24 In a series of subsequent works, combined with our newly developed software CsoDIAq,25 we further improved the performance of this technology to more than 2,000 protein identifications, of which 1100 were quantified.26, 27 There are also several previous instances of proteome analysis using direct infusion.28–30 However, the methods described above all focus on a single omics layer and are insufficient to satisfy current demands for multi-omics analysis, namely, rapid analysis of multiple omics components originating from the same sample simultaneously.31–33
Because proteomics, polar metabolomics, and lipidomics each require different LC configurations, the removal of LC presents an opportunity for combined analysis of all three omics layers. In this article, we describe a simultaneous multi-omics analysis by direct infusion mass spectrometry (SMAD-MS) that integrates polar metabolome, lipidome, and proteome analysis in a single shot from the same sample. We replace LC with an extra gas-phase separation by high field asymmetric waveform ion mobility spectrometry (FAIMS). We applied data-independent acquisition mass spectrometry (DIA-MS) for proteome analysis and spectral-stitching quadrupole slices with MS1 measurement for metabolome analysis, respectively. As we demonstrated previously for proteomics, we found that quadrupole slices plus FAIMS separation enabled detection of the most unique metabolite features. With this protocol, we realized more than 1,300 protein identifications and detected over 9,000 metabolite m/z features from the same sample in only 5 min of total data acquisition per sample. In two proof-of-principle studies, we first demonstrate the profiling of multi-omics variation of macrophages after different polarization, revealing significant multi-omics dysregulation and interaction. Secondly, we perform high throughput screening of human cellular multi-omics responses to different drug treatments with 96-well plates, followed by further machine learning integration of metabolome and proteome changes. Our approach offers an option for simple, high-throughput and cost-effective multi-omics analysis.
Results
Overview of Simultaneous Multi-Omics approach by Direct Infusion Mass Spectrometry (SMAD-MS)
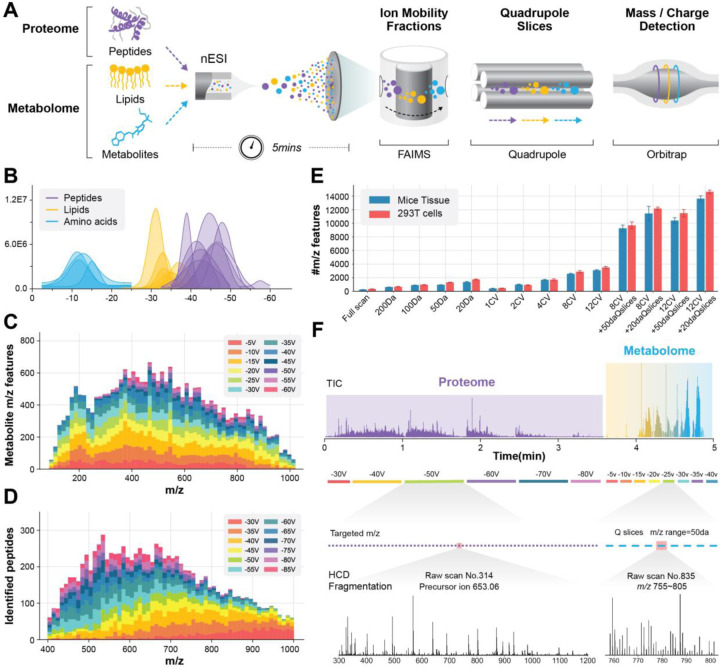
The principled schematic and workflow of the SMAD method are shown in Fig. 1a. In brief, different omics isolated from the same sample were mixed and then directly infused to the ion source without any separation in liquid chromatography. Following the ionization, two gas-phase separation techniques were utilized with the aim to reduce the complexity of gas-phase ions prior to their entry into the mass analyzer, namely, (1) FAIMS separates ions depending on the compensation voltage and (2) the quadrupole separates ions according to their mass-to-charge ratio (m/z). Ultimately, all gas-phase ions are sequentially detected by the orbitrap.
Fig. 1|. Overview of SMAD-MS workflow.
A, Scheme of our high-throughput approach for simultaneous analysis of peptides and metabolites using SMAD-MS. B, Typical compensation voltage (CV) intervals of peptide, amino acid, and lipid standards in FAIMS CV space. The raw data traces over CV space were smoothed to approximate gaussian distributions. C, d, Number of detected features in different m/z intervals and FAIMS compensation voltages (CV) from a real metabolome (c) and proteome (d) sample derived from 293T cells. e, Number of detected metabolite features under various compensation voltages and quadrupole slices of samples extracted from mouse tissue and 293T cells. f, Schematic diagram of typical experimental settings for SMAD, including a typical TIC diagram, CV settings, targeted m/z, Q slices, and typical mass spectrum of proteome and metabolome, respectively. The entire data collection requires an acquisition time of approximately 5 minutes per sample.
We first tested the compensation voltage (CV) range of different standard molecules and results demonstrated that lipids, amino acids, and peptides occupy different optimum CV intervals (Fig. 1b), for example, amino acids transmit through FAIMS in the range of −5V to −20V, lipids transmit in −20V to −35V, and peptides transmit from −30V to −70V. A more complexed real metabolome and proteome sample derived from 293T cells further proved that the optimum CV interval for metabolome (metabolites, lipids) and proteome (peptides) are 10–35V, and 40–60V, respectively (Fig. 1c, d). This result enables the fractionation of molecules originating from distinct omic sources, thereby minimizing interference between them.
Next we assessed the impact of gas phase separation using FAIMS compensation voltages and/or quadrupole slices on the number of detected metabolite m/z features from samples extracted from mouse tissue and 293T cells as we did previously for peptides from the proteome.24 The number of m/z features detected increased with the number of FAIMS CVs or with decreasing quadrupole isolation window width (Fig 1E). Surprisingly, the use of both FAIMS and quadrupole slices further increases the number of unique detectable metabolite m/z features, which provided confirmation of significant ion competition in the orbitrap and highlighted the necessity of multiple gas-phase separations before mass analysis.
Based on these findings we established the final experimental settings of SMAD as depicted in Fig. 1f. We applied data-independent acquisition mass spectrometry (DIA-MS) for proteome analysis and spectral-stitching quadrupole slices with MS1 measurement for metabolome analysis. We utilized six FAIMS CVs ranging from −30V to −80V in a step of 10V for proteome acquisition, and −5V to −40V with a 5V step for metabolome analysis. It should be noted that the instrument parameters, such as mass resolution, compensation voltages, and the number of target peptides, are adjustable and can be modified to match the specific requirements of the experiment and sample type. In general, the total acquisition time does not surpass five minutes excluding the 0.5 min sample loading time.
Data processing, performance optimization and quantitative evaluation of SMAD
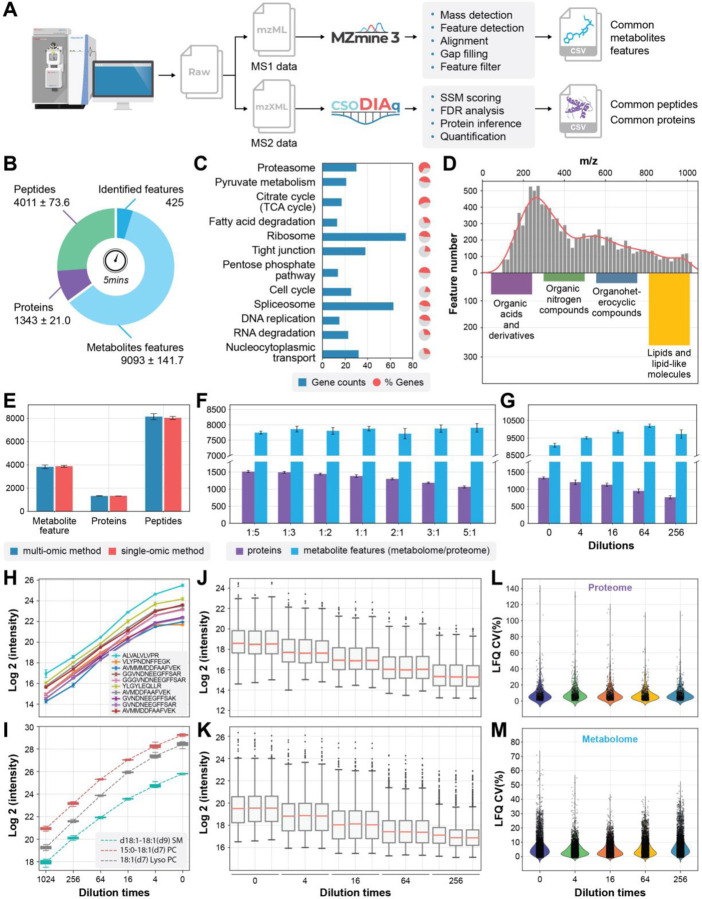
Fig. 2a summarizes our data processing workflow for raw files produced by SMAD. MS1 data from the metabolome part are extracted from the RAW file during transformation to mzML.34 The direct infusion data is formatted such that retention time corresponds to decreasing FAIMS compensation voltage, allowing extraction of extracted ion mobiligrams (XIMs) for quantification using MZmine3 (Fig. S1).14 MS/MS data from the proteome part is converted to mzXML for analysis using csoDIAq software 25, 26 to identify and quantify peptides and proteins. This data analysis workflow was used to assess the performance of SMAD.
Fig. 2|. Data processing, performance optimization and quantitative evaluation of SMAD.
a, Scheme showing data processing flow started from a typical raw file produced by SMAD. b, Number of detected metabolite features, peptides, and proteins in 5 minutes acquisition time by SMAD of 293T cells. c, KEGG pathway enrichment analysis of the proteins identified by SMAD of 293T cells. The bars indicate how many proteins in the pathway were identified, and the colored proportion of the circle reflects the coverage of the proteins in each pathway. d, m/z distribution of detected metabolite features and classes of identified metabolite features by SMAD of 293T cells. e, The comparison of detected molecule features (metabolites, peptides, proteins) between simultaneously multi-omics acquisition method and separately single-omic acquisition method. f, The effect of different mixing proportions of peptides and metabolites on detected molecule features by SMAD. (metabolome/proteome) g, Performance of SMAD in detecting of different concentrations. Original concentration is proteome samples dissolved in 40ul metabolome extraction (produced by adding 500ul solvent to 2million cells) and maintain the final proteome concentration to 4ug/Ul. h,i, label free quantification curve of standard peptides from MS-QCAL protein (h) and standard lipids from Avanta (i) at different concentrations. j,k, Untargeted quantification of detected proteins and metabolite features was performed by SMAD (each ratio was measured in triplicate). l,m, coefficient of variation of all quantified proteins and metabolite features.
SMAD collected in only five minutes per 293T sample produced an average of 4011 peptides, 1343 protein groups, and 9093 metabolite m/z features (out of which 425 were identified) (Fig. 2b). For the identified protein groups, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis revealed numerous important cellular pathways, including central carbon metabolism (i.e., tricarboxylic acid cycle, pyruvate metabolism, pentose phosphate pathway), protein synthesis and degradation (i.e., ribosome, spliceosome and proteasome), and nucleic acid replication and transport (DNA replication, RNA degradation and nucleocytoplasmic transport). For TCA cycle and proteasome pathways, the identified proteins accounted for more than 50% of all related genes in those specific pathways (Fig. 2c and Fig S2). The metabolite feature m/z distribution shows that most signals are within the interval from 200 to 400 m/z (Fig. 2d). We separately performed MS/MS of same sample attempting to identify some metabolite features quantified by SMAD, and results demonstrated many lipids, but also organic acids and organic nitrogen compounds were found (Fig. 2d). Similarity to data from 293T cells, data from macrophages revealed same level of protein identifications (more than 1300) and metabolite m/z features (~9000 metabolite m/z features), as well as abundant cellular pathways (Fig. S3).
We wondered whether SMAD would result in a reduction in the number of detected molecule features compared to separately analyzing the metabolome and proteome fractions. The number of detected peptides, proteins, and metabolite features is essentially the same between two different methods (Fig. 2e), as well as protein species and m/z distribution of metabolite features (Fig. S4a-c). In addition, we investigated whether variations in the mixing ratio of two different omics samples could impact the performance of SMAD. Particularly, we examined seven samples with different mixing ratios across 25-fold (metabolome/proteome volume ratio) and observed that, as the proportion of metabolome increased, the number of identified proteins decreased by up to 30%. However, the quantity of detected metabolite features remained relatively constant (Fig. 2f). Furthermore, we also tested the influence of the total concentration of the sample. The results show that as the concentration of the sample decreases, the number of detected proteins also decreases, but the detected metabolites show a trend of increasing first and then decreasing (Fig. 2g). These results indicate that protein identifications, namely, the proteome part, is more sensitive to sample mixing ratios and concentrations. The potential reason is that the signal intensity of tandem mass spectrometry in proteomic analysis is much lower than that of the precursor signal of metabolites, resulting in a more sensitive phenomenon to factors such as concentration. The influences of these factors for protein groups, protein functions and metabolite m/z distribution were illustrated in Fig. S4 d-i.
The direct infusion strategy completely omitted liquid chromatography, leading to the absence of retention time or elution profiles that could be utilized for peak integration. Here, we implemented a label free quantitative strategy for both metabolome and proteome results based on XIM area and peptide fragments, respectively (Fig. S1). Starting with a mixture of 3 lipids and 10 QCAL peptide standards, the results showed an excellent linearity across a broad range of concentrations (Fig 2h, i). We further spiked these mixture standards into real multi-omics sample derived from 293T cells and surprisingly, the standards remain good linearity across different concentrations (Fig. S5a-d). Furthermore, we applied the same strategy for the whole proteome and metabolome, quantification results exhibit excellent repeatability (Fig. S5e, f) and linearity across all 437 proteins and 3970 metabolites m/z features (Fig. 2j, k) and typical proteins and metabolites were also shown in Fig. S5g, h. The coefficient of variance analysis at different concentrations demonstrated that 90.6% proteins and 93.4% metabolite m/z features had CV less than 0.2, which further proved the reliability and robustness of our LFQ strategy of SMAD (Fig 2l, m).
SMAD reveals multi-omics responses of macrophages after M1/M2 polarization
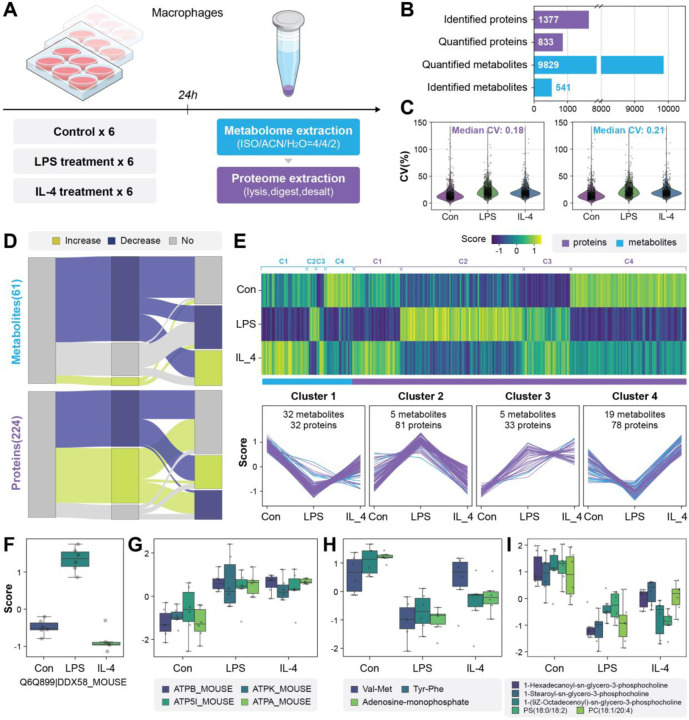
To demonstrate the potential of SMAD for real biological samples, here we conducted the first case study focused on the multi-omics responses of macrophages following polarization. Macrophages are well known to differentiate into the pro-inflammatory M1 or anti-inflammatory M2 polarization states after LPS and IL-4 treatment, respectively.35. 36 To assess multi-omic differences in M1 and M2 macrophage subsets using SMAD, we treated mouse bone-marrow derived macrophages (BMDMs) with immune stimulators LPS and IL-4 in 10 cm dishes for 24 hours. After washing with PBS, lipids and metabolites were extracted by a mixed solvent (ISO/ACN/H2O, 4:4:2, volume ratio), then the precipitated protein pellet from the same sample was further processed (lysis, digest and desalt) for proteomics into peptides (Fig. 3a). We measured the multi-omics by SMAD and an average of 1,377 protein groups were identified, 833 of which were quantified in all 18 samples (only protein groups found in all replicates and treatments were quantified to exclude missing values, Fig. 3b and Fig. S6a). We also detected 9,829 metabolite m/z features and 541 of them were identified using GNPS15 (Fig. S6b-f, Supplementary Table 1). The coefficient of variance analysis of quantified proteins (median coefficient of variation: 0.18) and metabolite m/z features (median coefficient of variation: 0.21) confirms the stability and robustness of SMAD method (Fig. 3b).
Fig. 3|. Application of SMAD for rapid multi-omics screening of macrophage polarization.
a, Schematic diagram of experimental design. (LPS: lipopolysaccharide, IL-4: Interleukin 4). b, Overview of molecules monitored by SMAD. c, Changes of significant and identified molecules (metabolites and proteins) between two treatments and control. (two-sided Wilcoxon rank test). d, The Heatmap and clustering of dysregulated molecules following immune activation. (K-means clustering). e,f Typical dysregulated proteins of macrophages after immune stimulator treatment. g,h Typical dysregulated metabolites and lipids after LPS treatment.
Both proteins and metabolites were subjected to ANOVA analysis to identify molecules that were significantly altered with either treatment. Two hundred and twenty-four proteins and 3,595 metabolite m/z features (61 were identified) exhibited significant alterations (Benjamini–Hochberg (BH)-adjusted P values <0.05) (Fig. S7a, Supplementary Table 1). KEGG pathway enrichment demonstrated that the significant changed proteins are mainly related to pathways including glycolysis, TCA cycle, oxidative phosphorylation, spliceosome, lysosome and protein processing in endoplasmic reticulum (Fig. S7b). The Wilcoxon rank-sum test was further applied to determine which of these molecules were different between each treatment and control. Interestingly, the Sankey plot shows that the multi-omics response of macrophages varies greatly for LPS and IL-4 treatments (Fig. 3c). Further PCA analysis of proteome, metabolome and multi-omics data also demonstrated that different treatments were separated with each other significantly (Fig. S8), highlighting the difference for different polarization states, which is consistent with current knowledge.37
To better integrate and interpret these variations of metabolites and proteins, we utilized K-means clustering to analyze the identified significant molecule features from ANOVA and gain insight into the changes associated with immune activation. The significantly dysregulated molecules were categorized into four primary clusters (Fig. 3d). Specifically, cluster 2 primarily consisted of molecules (81 proteins and 5 metabolites) that were upregulated in response to LPS. These molecules were associated with significant pathways, including lysosome, ribosome, energy metabolism, immune response receptors, and fatty acid metabolism (Fig. 3e, Fig. S9), which is consistent with previous studies regarding the changes in macrophages under M1 polarization.38 In addition, in cluster 3, which included 5 metabolites and 33 proteins that exhibited an increase in both LPS and IL-4 treatment. The identified proteins are mainly related to pathways such as amino acid metabolism, TCA cycle, oxidative phosphorylation (ATPK, ATPB, ATPA), ribosome and lysosome (Fig. 3f). This reflects an increase in energy expenditure of macrophages following polarization. Noteworthy, further metabolome results revealed that metabolites related to these pathways such as TCA cycle (AMP) decreased in both two different polarizations, which reflect an enhanced ATP consumption (Fig. 3g). Moreover, changes in other metabolites, such as the significant decline of dipeptides in LPS treatment, and lipids showing a significant decrease in both treatments, could be attributed to the increased energy consumption following macrophage polarization, resulting in more catabolism of phospholipids and peptides for providing energy (Fig. 3g, h). We also noticed that LPS treatment was associated with decreases in spermidine and glutathione (Fig. S10). Clustering of all dysregulated proteins with unfiltered metabolite m/z features (including unidentified metabolites) is shown in Fig. S11, as well as histograms of metabolite feature m/z distributions in each cluster.
SMAD enables high throughput screening of human cellular multi-omics responses to drugs
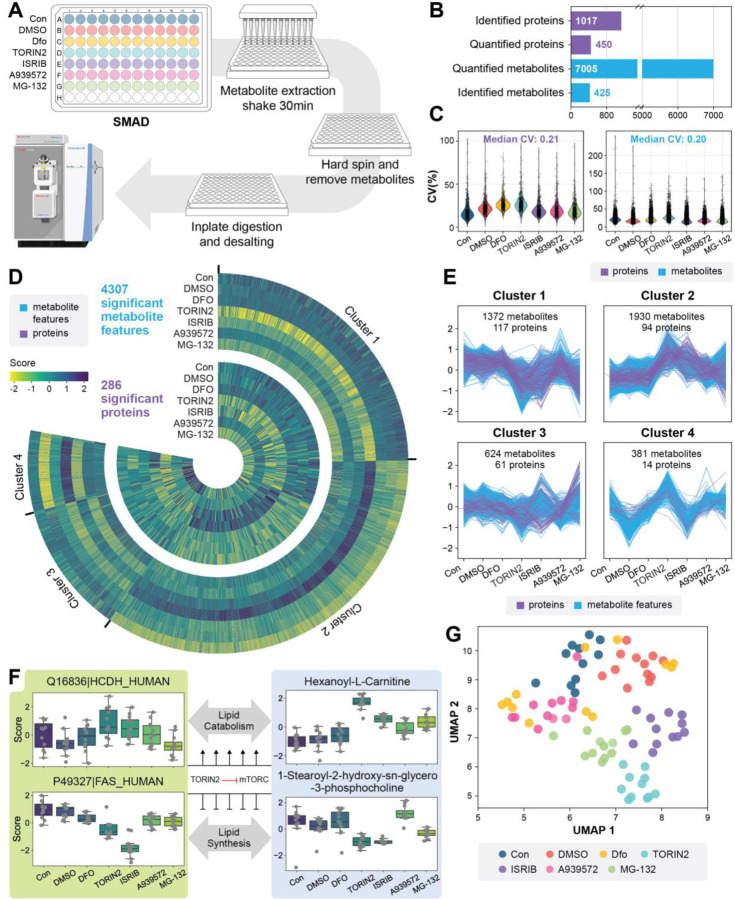
Rapid and high throughput profiling of multi-omics responses of cells to drugs holds immense significance in drug discovery and chemical biology. Here also we applied SMAD for high throughput monitoring of human cellular multi-omics responses to drugs. We cultured 293T cells in a 96-well plate and treated them with five different drugs including deferoxamine (chelator), Torin2 (mTOR inhibitor), ISRIB (integrated stress response inhibitor), MG132 (proteasome inhibitor) and A939572 (SCD1 inhibitor). Samples were processed for SMAD using a protocol adapted for 96 well plates (Fig. 4a). The MS acquisition time of a whole 96 well plate was around 7 hours. After data acquisition by SMAD, we identified an average of 1,017 proteins, and 450 proteins were quantified (only protein groups exist in all replicates and treatments were quantified to exclude missing values) for all replicates (Fig. S12a). A total of 7,005 metabolite features were quantified and 425 of them were identified (Fig. 4b, Fig. S12b-f, Supplementary Table 2). Over 80% of quantified features has a coefficient of variance lower than 0.5 and the median coefficient of variance for proteomics and metabolomics are 0.21 and 0.20, respectively (Fig 4c).
Fig. 4|. A high-throughput application of SMAD for rapid screening of human cellular multi-omics responses to drugs.
a, Overview of experimental design and sample processing flow in 96-well plates. b, Molecular information and data quality of multi-omics dataset acquired by SMAD. including identified, quantified feature, coefficient of Variances. c, The Heatmap showing significant dysregulated molecules among drug treatments. d, Four clusters of significant metabolites and proteins (K-means clustering). e, UMAP analysis of multi-omics responses among all drugs and controls. (All metabolite features and proteins included). f, Examples of proteins and metabolites for each cluster. g, pathway network for xxx.
Similarity, ANOVA analysis was first applied to determine which proteins and metabolites were altered by the drug treatments. As shown in Fig. 4c, a total of 4,307 metabolite features and 286 proteins were significantly dysregulated (Supplementary Table 2). Further analysis with t-tests comparing each protein to the control revealed TORIN2, ISRIB and MG-132 induced the greatest changes to both proteins and metabolites (Fig. S13), while very limited variations were observed between the control, DMSO, and DFO treatment. We previously established that the minimum concentration for DFO to cause an effect in cells is 100 μM, and since we used 10 μM here these results makes sense.26
To better illustrate the variations of proteins and metabolites across various treatments, we consolidated all significantly dysregulated molecules and categorized them into four clusters utilizing K-means clustering (Fig. 4d). The distinct clusters reflect differences in the multi-omics responses of 293T cells to diverse drug treatments and many interesting changes were revealed. For example, cluster 2, which represents the profiles of 94 proteins and 1,930 metabolite m/z features, generally peaked at TORIN2 or ISRIB treatment. Further in-depth analysis revealed that the disrupted proteins in this cluster are mainly related to pentose phosphate pathway, TCA cycle, cell cycle, spliceosome, fatty acid degradation and protein processing in endoplasmic reticulum (Fig. S14a, b). This may be due to the repair of some protein synthesis processes and the increase of cellular energy consumption after ISRIB treatment. On the other hand, cluster 1, enriched by 117 proteins and 1,372 metabolite m/z features, demonstrated a opposite trend that downregulated at either of these two treatments. Proteomic results revealed that the downregulated proteins are mainly related to pathways like glycolysis, HIF-1 signaling pathway, amino acid metabolism and ribosome (Fig. S14c, d), which can be attributed to the inhibition of cell growth by TORIN2. Cluster 3, which represents the profiles of 61 proteins and 624 metabolite m/z features, generally peaked at MG-132 treatment. MG132 is a proteasome inhibitor that blocks the activity of the proteasome. Here we demonstrated that proteins associated with cell cycle, ribosome, spliceosome, and DNA replication were significantly upregulated in MG-132 (Fig. S14e, f). What’s more interesting is that a multi-omics response related to fatty acid and lipid metabolism in the TORIN2 treatment group was discovered by SMAD. We observed a significant upregulation of mitochondrial fatty acid beta-oxidation enzyme (Q16836, HADH) and correspondingly increased carnitine, acetyl-carnitine, and hexanoyl-carnitine the enhanced lipid catabolism (Fig. 4e, S15). We also observed a notably decrease of fatty acid synthetase (P49327, FAS) in TORIN2 treated cells and typically lipids were accordingly downregulated in this treatment, which represents the inhibited lipid synthesis process (Fig. 4e). Notably, ISRIB treatment was associated with molecular changes suggestive of decreased lipid synthesis but not decreased lipid catabolism.
UMAP dimension reduction of the multi-omics data from each replicate demonstrates that different groups can be finely separated between each other, which demonstrated the distinct multi-omics profile among various treatments and controls (Fig. 4f). To further explore the relationships on cellular multi-omics response after different drug treatment, we calculated the correlations by using the differences in multi-omics data between each treatment group and the control group. The results showed that TORIN2 and ISRIB had the strongest correlation for proteome variations (Pearson Corr = 0.701). TORIN2 and MG-132 had the strongest correlation for metabolome changes (Pearson Corr = 0.729) (Fig. S16).
Machine learning of multi-omics interaction
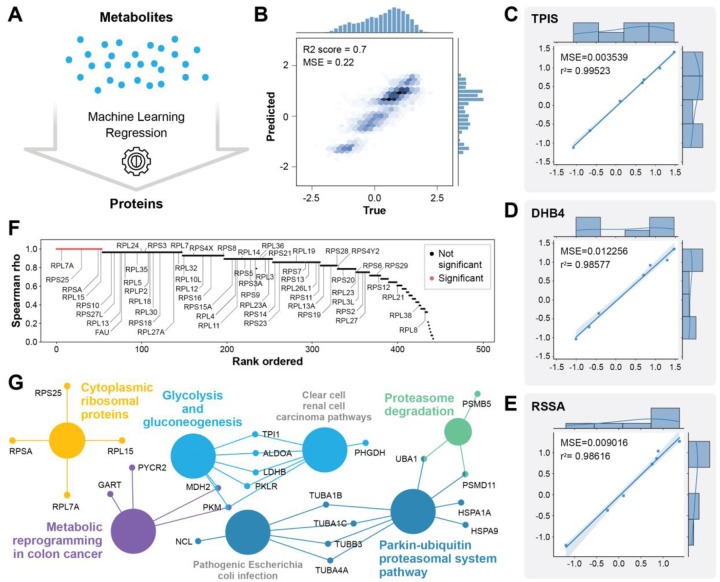
To understand relationships between the metabolomic and proteomic data we used an unbiased machine learning approach similar to our previous work.39 Data from the drug screening experiment was used to predict changes in proteins from changes in metabolites using an optimized extratrees model 40 (Fig. 5a). Overall, the prediction of most proteins was very effective with an R2 score of 0.7 between true and predicted values (Fig. 5b). Several proteins from diverse pathways were predicted with remarkable accuracy with R2 scores over 0.98 (Fig. 5c, d, e). Given the task of predicting proteins from metabolites we were not surprised to see metabolic proteins predicted well, but we were surprised to see a ribosomal protein among the top well predicted proteins (Fig. 5e). In fact, the distribution of all proteins predicted from metabolites indicates that most proteins were predicted with rho over 0.8, and most ribosome proteins in our dataset were well predicted (Fig. 5f). To determine which proteins were statistically significantly well predicted by machine learning from metabolites measured from the same sample by SMAD, we used a strict Bonferroni correction of the p-values from Spearman correlation between the true and predicted protein values. This left us with 54 statistically well predicted proteins. Term enrichment analysis of the most well predicted proteins indicated that diverse pathways beyond metabolic proteins are accurately predicted from the metabolomic data, which indicates that there are strong relationships between the metabolome and multiple cellular pathways including protein synthesis and degradation. This makes sense because most cellular energy is used for protein regulation.41
Fig. 5|. Machine learning predicts proteomic changes from metabolomic changes.
a, Scheme showing the ML task of predicting proteins from metabolomic data measured in parallel by SMAD. b, Overall performance of all proteins true versus ML predicted values. c,d,e, Examples of three of the most well predicted proteins from different pathways: TPIS from glycolysis, DHB4 from fatty acid oxidation, and RSSA from the ribosome. f, plot of the Spearman rho between true versus predicted protein quantities for all 450 proteins. Red indicates statistically significantly predicted proteins according to a Bonferroni corrected p-value from Spearman correlation analysis. g, WikiPathways term enrichment analysis of the 54 significant proteins from (f).
Discussion
Multi-omics integration and analysis is a technology frontier whose application could accelerate biomedical research and clinical diagnosis. However, we still need more time efficient methods to realize the full potential. In this study, we demonstrate a high throughput direct infusion mass spectrometry strategy that enables simultaneous analysis of peptides, polar metabolites, and lipids all together by leveraging ion mobility. We demonstrated that we could identify more than 1,300 proteins and detect over 9,000 metabolite m/z features in only 5 minutes of total data collection time, which is equivalent to identifying an average of 4.3 proteins and detecting 30 metabolic m/z features per second. The successful implementation of this method stems from the following three major factors: 1) multiple gas phase separation by ion mobility and quadrupole, which reduced the complexity of ions in mass analyzer and improved the coverage, 2) high sensitivity ion source and mass spectrometry instruments, and 3) computational strategies capable of analyzing this complicated data.
We demonstrated that even in highly complex ionization systems resulting from the removal of liquid chromatography, simultaneous multi-omics analysis can still be achieved by adding an additional dimension of gas-phase separation. This approach avoids the time-consuming and economic cost associated with current LC-based methodologies, which require long separation time and frequent changing of mobile phases and chromatographic columns, as well as extensive washing, cleaning, and other maintenance when analyzing different omics samples. We optimized the method and validated its robustness by testing different ratios of mixed omics samples, combinations of compensation voltages, and quadrupole window intervals. Additionally, we further developed protein and metabolite quantification methods based on peptide fragment intensities and ion mobility peak areas, respectively. Further, machine learning of the data from SMAD shows that the metabolome is connected to most if not all proteome pathways.
It is undeniable that the current SAMD method still lags behind traditional liquid chromatography in terms of identification depth and coverage, and there are still shortcomings in the identification of m/z features in metabolome. However, this issue can be overcome in the future by developing new software that incorporates the additional gas-phase dimension and establishing mass spectrum libraries from metabolite standards. It is worth noting that the quantitative multi-omics data produced by SMAD now is sufficient depth and quality to monitor cellular phenotypes and cellular responses to drugs, which provides a window into important pathways such as amino acid metabolism, glycolysis, fatty acids and lipids synthesis, and protein synthesis. This approach can therefore lead to a wealth of testable hypotheses for validation.
Currently, for mass spectrometry-based multi-omics analysis, our focus is primarily on establishing stable, high-throughput multi-omics methods with the aim of its truly widespread application in biomedical research and clinical diagnostics. However, when promoting this technology comprehensively, have we fallen into a certain stereotype or is it possible to have other perspectives? (1) Do we really need to detect such breadth and depth of metabolites and proteins every time, especially for diseases that are already well-understood and have established biomarkers? Combined with specific sample preparation methods, would a simple, robust, flexible, fast, and low-cost approach be more suitable for such applications? (2) Against the background of rapid iterations in mass spectrometry instrumental and data analysis software, are we underestimating the analytical capabilities of mass spectrometry without chromatographic separation, particularly when combined with other techniques such as ion mobility? In our previous direct infusion methodological studies in proteomics, we have improved protein identifications from approximately 300 to over 2,000 proteins. We expect further improvements in the performance of this technology in metabolomics analysis or its application on other instruments.
In summary, here we demonstrate for the first time the ability to simultaneously analyze complex multi-omics mixtures without LC. Together with a series of data analysis and integration strategies, we have explored the potential of the SMAD method in cellular phenotyping and drug screening through two practical applications. We anticipate advances in all aspects, including sample preparation, mass spectrometry instruments, and data analysis software, to collectively advance the practical application of this method.
Methods
Materials.
Angiotensin I (Sigma, A9650–1MG), QCAL Peptide Mix (Sigma, MSQC2) and Hela digest standard (Thermo Fisher Scientific, Catalog number: 88328) were dissolved into different concentrations with 50% acetonitrile (ACN) in 0.2% formic acid (FA). Lipid standards (product No. 330707) was purchased form Avanti. Drugs including deferoxamine mesylate salt (Product No. D9533), mTOR inhibitor torin2 (Product No. SML1224), integrated stress response inhibitor ISRIB (Product No. SML0843), proteasome inhibitor MG-132 (Product No. 474790) and SCD1 inhibitor A939572 (Product No. SML2356) were purchased from Sigma-Aldrich.
Mass spectrometry and data acquisition
SMAD analysis was performed on an Orbitrap Lumos (Thermo Fisher Scientific) mass spectrometer coupled with the FAIMS Pro Interface. Different compensation voltages were applied for metabolome (−5V to −40V in steps of 5V) and proteome (−30V to −80V in steps of 10V) analysis. A nano-ESI source (“Nanospray Flex”) and LOTUS nESI emitters from Fossiliontech were used for ionization. The ultimate 3000 HPLC system (Thermo Fisher Scientific) was used to control automated sample loading, flow rate, and mobile phase composition. Flow rate was maintained at 1.4ul/min at the first 0.5 min for transferring samples to nano emitter and then maintained a 0.3ul/min flowrate to the end of the acquisition. Mobile phase composition is ACN/H2O (70:30) in 0.1% formic acid (FA) for the whole acquisition process. Data acquisition was conducted at positive mode with 2200V. AGC was set at 100% and ion injection time was set at auto. For proteome acquisition, targeted MS2 mode was used for each compensation voltage from −30 V to −80 V in a step of 10V. For metabolome acquisition, tSIM mode with a quadrupole window of 50da was used to scan full m/z range from 100 to 1100 for each compensation voltage from −5V to −40 V in a step of 5V.
Quantitative Evaluation of SMAD
Three lipid standards (d18:1–18:1(d9) SM, 29.6 μg/ml; 15:0–18:1(d7) PC, 150.6 μg/ml; 18:1(d7) Lyso PC, 23.8 μg/ml) and QCAL proteins (0.25 μg/μl) was mixed and diluted every four times. The mixed standard sample was directly analyzed by targeting their accurate m/z with SMAD. Lipid standards and MS-QCAL peptides were quantified with python by manually extracting MS1 intensity and representative y-ion fragments intensity, respectively. For real samples, original metabolome samples were produced by adding 500ul metabolite extraction solvent (ISO/ACN/H2O, 4:4:2) to 2 million 293T cells and then the supernatant was collected. Proteome samples was derived from 293T cells with same proteome preparation protocol used for two case studies. Then the multi-omics sample was produced by mixture of metabolome and proteome samples at 1:1 volume ratio. The sample was diluted every four times to produce concentration gradients and analyzed by SMAD.
Cell culture and sample preparation for macrophages
BMDMs were derived from bone marrow extracted from the femurs of euthanized mice (11-week-old male C57BL/6J) and plated at 3 × 106 cells per 10-cm dish in 10 ml of macrophage growth medium (complete RPMI containing 25% M-CSF containing L929-conditioned medium). Cells were cultured for 7 days to differentiate and were supplemented with 5 ml of macrophage growth medium on day 5. On day 7, 7×106 BMDMs were counted and replated on 10 cm dishes in macrophage growth medium overnight prior to experiments. On the day of experiments, BMDMs were treated with 100ng/ml LPS (Invivogen, LPS-EK Ultrapure) and 10ng/ml recombinant murine IL-4 (Peprotech) for 24 hours. Then the cells were washed twice with cold PBS and harvested from the plate by scraping. The cells were pelleted into 1.5 ml centrifuge tubes and snap frozen. Then the metabolites and lipids were extracted from samples with 500μl isopropanol/acetonitrile/water 4:4:2 for 20 min, following by a hard spin of 10 minutes in 12000 rpm and all metabolome supernatant was removed to new centrifuge tubes and stored in −80℃. The precipitated proteome pellet was dry out and then lysed by addition of 8 M urea with 50 mM TEAB buffer at pH 8.5. The tubes were vortexed and sonicated until homogenous with lysis buffer. Then TCEP and chloroacetamide were each added to 10 mM final concentration to reduce protein disulfide bonds and alkylate the free cysteines in the dark for 30min. Then lysis buffer was diluted to 2 M urea using 50 mM TEAB, and catalytic hydrolysis of proteins was initiated by trypsin (Promega) at a weight ratio of 1:50 protease:substrate. Proteome proteolysis was incubated in a 37℃ incubator for six hours. Peptides were desalted using Strata reversed-phase cartridges from Phenomonex, and then dried completely in Speed-Vac. Peptides were resuspended with ACN/Water/FA (50%/49.9%/0.1%, volume ratio) and mixed with metabolome samples for SMAD analysis.
293T Cell culture and sample preparation for drug screening
T293 cells were cultured in a 96-well plate to 70% confluency and then treated with different drugs. The treated concentrations of drugs were deferoxamine (10μM), Torin2 (1μM), ISRIB (1μM), MG132 (1μM) and A939572 (1μM). After 24 hours incubation, cell culture media was removed and washed with PBS for twice. Then 100ul metabolite extraction solvent (IPA/ACN/H2O, 4:4:2) was added to each well of the plate and vortexed for ten minutes. After vertexing, the plate was centrifuged in 4,000 rpm and all metabolome supernatant was removed to a new 96-well plate and stored in −80℃. The precipitated proteome pellet in original 96-well plate was dry out first and then lysed by addition of 8 M urea with 50 mM TEAB buffer at pH 8.5. The plate was vortexed at 900 rpm around 5 minutes until homogenous with lysis buffer and sonicated 5 min in a Covaris water bath maintained at 4°C. After sonication, TCEP and chloroacetamide were added to a 10 mM final concentration to reduce protein disulfide bonds and alkylate the free cysteines in the dark for 30 min. Then, lysis buffer was diluted to 2 M urea using 50 mM TEAB, and catalytic hydrolysis of proteins was initiated by trypsin (Promega) at a weight ratio of 1:50 protease:substrate. Then Proteome proteolysis was incubated in 37℃ incubator for six hours. Peptides were desalted using 96-well µElution Plate from Waters (Oasis HLB 96-well µElution Plate, 2 mg Sorbent per Well, 30 µm), and then dried completely in Speed-Vac. Peptides were resuspended in 10 ul ACN/Water/FA (50%/49.9%/0.1%, volume ratio) and mixed with 10ul metabolome samples for SMAD analysis. To prepare the 293T proteome for experimental parameter analysis, we followed a consistent protocol for cell lysis and digestion except that cells were cultured in a 12cm plate and subsequently desalted using Strata reversed-phase cartridges from Phenomonex.
Proteome library generation
The building of proteome library used for CsoDIAq generally including three steps: 1) performing LC-MS/MS analysis of Macrophage and 293T samples with DDA for eleven compensation voltages from −30V to −80V in a step of 5V. 2) Using Fragpipe to produce pepxml files of peptides and proteins with appropriate fasta database and add decoys (50%). 3) Building library for CsoDIAq with SpectraST.
Identification and quantification of proteome
Peptides and proteins were identified with CsoDIAq. CsoDIAq is a python software package designed to enhance usability and sensitivity of the projected spectrum−spectrum match scoring concept. The process was described as the following steps: Firstly, the original Thermo .RAW files were converted to mzXML files using msconvert with default settings. Then the output mzXML files were input to CsoDIAq GUI and the appropriate spectral library for that sample was selected with the default settings keeping the fragment mass tolerance at 20ppm and the “Label free quantification” should be selected. CsoDIAq produces three output files for each input mzXML file that report spectra, peptides, and proteins filtered to <1% FDR. In each case, CsoDIAq sorts peptide identifications by match count and cosine (MaCC) score, calculates the FDR for each identification using a modification of the target-decoy approach where FDR at score S = number of decoys/number of targets, and removes SSMs below a 0.01 FDR threshold. The peptide FDR calculations only use the highest-scoring instance among all SSMs for each peptide. CsoDIAq uses the IDPicker algorithm to identify protein groups from the list of discovered peptides and adds them as an additional column in the output. A detailed description about data processing, FDR calculation and protein inference was listed in our previous paper26. Peptides and proteins were quantified using CsoDIAq to extract the sum of all detected fragment ion intensities of common peptides in all input files.
Identification and quantification of metabolome
Thermo .RAW files were converted to mzML files using msconvert. All peaks are picked according to the sequence of Q slices. The produced mzML files was analyzed with MZmine3 software for mass detection, feature detection, alignment, gap filling and feature filter. The output files contain m/z and quantification by FAIMS peak area. For metabolite annotation, we applied a direct infusion based DDA tandem mass spectrometry analysis with the same sample and same compensation voltages applied in SMAD analysis. Then the MS2 spectrum of metabolites was compared with library and analyzed through GNPS website.15
Consensus clustering and dimensionality reduction
In two applications of macrophage polarization and drug screening, the unsupervised k-means consensus clustering of all treatments was performed with the python packages “sklearn”. The significantly dysregulated molecules that were discovered among different treatments were used for clustering. The number of groups for clustering was determined by “Elbow Method”. PCA and UMAP analysis was performed with the python packages “sklearn” and “UMAP”, respectively.
Pathway enrichment analysis
The UniProt IDs from CsoDIAq outputs were converted to gene IDs. The KEGG_2022_Human gene set library was applied and pathway enrichment analysis was done in Cytoscape 42 with the plugin clueGO.43 GO Term/Pathway network connectivity (Kappa score) was set at 0.5. Additional settings included GO Term grouping and two-sided hypergeometric tests, and leading group term ranking based on highest significance.
Statistical Analysis
Data normalization, analysis, and visualization were performed in Python version 3.9.7. One-way ANOVA was applied for group analysis and select the significant dysregulated molecules (Benjamini–Hochberg (BH)-adjusted P values <0.05) among control and two treatments. Wilcoxon rank-sum test was used to compare each treatment with control and significant features were defined as a p-value less than 0.05 after Benjamini–Hochberg (BH)-adjustment.
Machine learning
Machine learning was performed using scikit-learn in python. Data was split into training and test sets where one sample from each of the 7 conditions was stratified into the test set. An extratrees model was optimized on the training data using 5-fold cross validation. A single model to predict each protein was trained using the best parameters from 5-fold cross validation and then the model performance was evaluated by predicting the quantity of that proteins in the test set by computing the mean squared error and Spearman’s rank correlation.
Data Availability
All raw mass spectrometry data is available from ftp://MSV000092273@massive.ucsd.edu with password “smaddata”. All code for data analysis and data visualization are provided open source via xomicsdatascience github.
Supplementary Material
Acknowledgements
We thank Dasom Hwang for help with graphic design. We thank the NIH for funding (R35GM142502 and R21AG074234).
Reference
- 1.Hasin Y., Seldin M. & Lusis A. Multi-omics approaches to disease. Genome Biol 18, 83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Subramanian I., Verma S., Kumar S., Jere A. & Anamika K. Multi-omics Data Integration, Interpretation, and Its Application. Bioinform Biol Insights 14, 1177932219899051 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawashima Y. et al. Single-Shot 10K Proteome Approach: Over 10,000 Protein Identifications by Data-Independent Acquisition-Based Single-Shot Proteomics with Ion Mobility Spectrometry. J Proteome Res 21, 1418–1427 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breitkopf S.B. et al. A relative quantitative positive/negative ion switching method for untargeted lipidomics via high resolution LC-MS/MS from any biological source. Metabolomics 13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao Q. et al. High-throughput proteomics and AI for cancer biomarker discovery. Adv Drug Deliv Rev 176, 113844 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Fuhrer T. & Zamboni N. High-throughput discovery metabolomics. Curr Opin Biotechnol 31, 73–78 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Sidoli S. et al. One minute analysis of 200 histone posttranslational modifications by direct injection mass spectrometry. Genome Res 29, 978–987 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kretschy D. et al. High-throughput flow injection analysis of labeled peptides in cellular samples - -ICP-MS analysis versus fluorescence based detection. Int J Mass Spectrom 307, 105–111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chekmeneva E. et al. Optimization and Application of Direct Infusion Nanoelectrospray HRMS Method for Large-Scale Urinary Metabolic Phenotyping in Molecular Epidemiology. J Proteome Res 16, 1646–1658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Messner C.B. et al. Ultra-fast proteomics with Scanning SWATH. Nat Biotechnol 39, 846–854 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meier F. et al. diaPASEF: parallel accumulation-serial fragmentation combined with data-independent acquisition. Nat Methods 17, 1229–1236 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Scigelova M. & Makarov A. Orbitrap mass analyzer--overview and applications in proteomics. Proteomics 6 Suppl 2, 16–21 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Wang J. et al. MSPLIT-DIA: sensitive peptide identification for data-independent acquisition. Nat Methods 12, 1106–1108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmid R. et al. Integrative analysis of multimodal mass spectrometry data in MZmine 3. Nat Biotechnol 41, 447–449 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M. et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol 34, 828–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Washburn M.P., Wolters D. & Yates J.R. 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol 19, 242–247 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Stancliffe E., Schwaiger-Haber M., Sindelar M. & Patti G.J. DecoID improves identification rates in metabolomics through database-assisted MS/MS deconvolution. Nat Methods 18, 779–787 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hebert A.S. et al. Comprehensive Single-Shot Proteomics with FAIMS on a Hybrid Orbitrap Mass Spectrometer. Anal Chem 90, 9529–9537 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ridgeway M.E., Lubeck M., Jordens J., Mann M. & Park M.A. Trapped ion mobility spectrometry: A short review. International Journal of Mass Spectrometry 425, 22–35 (2018). [Google Scholar]
- 20.Winter D.L., Wilkins M.R. & Donald W.A. Differential Ion Mobility-Mass Spectrometry for Detailed Analysis of the Proteome. Trends Biotechnol 37, 198–213 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Chen X. et al. Trapped ion mobility spectrometry-mass spectrometry improves the coverage and accuracy of four-dimensional untargeted lipidomics. Anal Chim Acta 1210, 339886 (2022). [DOI] [PubMed] [Google Scholar]
- 22.Southam A.D., Weber R.J., Engel J., Jones M.R. & Viant M.R. A complete workflow for high-resolution spectral-stitching nanoelectrospray direct-infusion mass-spectrometry-based metabolomics and lipidomics. Nat Protoc 12, 310–328 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Sarvin B. et al. Fast and sensitive flow-injection mass spectrometry metabolomics by analyzing sample-specific ion distributions. Nat Commun 11, 3186 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer J.G., Niemi N.M., Pagliarini D.J. & Coon J.J. Quantitative shotgun proteome analysis by direct infusion. Nat Methods 17, 1222–1228 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cranney C.W. & Meyer J.G. CsoDIAq Software for Direct Infusion Shotgun Proteome Analysis. Anal Chem 93, 12312–12319 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Jiang Y., Hutton A., Cranney C.W. & Meyer J.G. Label-Free Quantification from Direct Infusion Shotgun Proteome Analysis (DISPA-LFQ) with CsoDIAq Software. Anal Chem 95, 677–685 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trujillo E.A. et al. Rapid Targeted Quantitation of Protein Overexpression with Direct Infusion Shotgun Proteome Analysis (DISPA-PRM). Anal Chem (2022). [DOI] [PMC free article] [PubMed]
- 28.Valentine S.J. et al. Developing IMS–IMS–MS for rapid characterization of abundant proteins in human plasma. International Journal of Mass Spectrometry 283, 149–160 (2009). [Google Scholar]
- 29.Tang R. et al. Repeat-Enhancing Featured Ion-Guided Stoichiometry for Identification and Quantification of Direct Infusion Proteome. J Proteome Res 22, 1947–1958 (2023). [DOI] [PubMed] [Google Scholar]
- 30.Chen S. Rapid protein identification using direct infusion nanoelectrospray ionization mass spectrometry. Proteomics 6, 16–25 (2006). [DOI] [PubMed] [Google Scholar]
- 31.He Y. et al. Multi-Omic Single-Shot Technology for Integrated Proteome and Lipidome Analysis. Anal Chem 93, 4217–4222 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen X. et al. Multi-omics microsampling for the profiling of lifestyle-associated changes in health. Nat Biomed Eng (2023). [DOI] [PMC free article] [PubMed]
- 33.Shen B. et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell (2020). [DOI] [PMC free article] [PubMed]
- 34.Chambers M.C. et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol 30, 918–920 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray P.J. Macrophage Polarization. Annu Rev Physiol 79, 541–566 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Yunna C., Mengru H., Lei W. & Weidong C. Macrophage M1/M2 polarization. Eur J Pharmacol 877, 173090 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Zhu L., Zhao Q., Yang T., Ding W. & Zhao Y. Cellular metabolism and macrophage functional polarization. Int Rev Immunol 34, 82–100 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Li P. et al. Comparative Proteomic Analysis of Polarized Human THP-1 and Mouse RAW264.7 Macrophages. Front Immunol 12, 700009 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dickinson Q., Aufschnaiter A., Ott M. & Meyer J.G. Multi-omic integration by machine learning (MIMaL). Bioinformatics 38, 4908–4918 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geurts P., Ernst D. & Wehenkel L. Extremely randomized trees. Machine Learning 63, 3–42 (2006). [Google Scholar]
- 41.Hardie D.G. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol 8, 774–785 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Shannon P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bindea G. et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw mass spectrometry data is available from ftp://MSV000092273@massive.ucsd.edu with password “smaddata”. All code for data analysis and data visualization are provided open source via xomicsdatascience github.