SUMMARY:
Activity-dependent expression of immediate early genes (IEG) is critical for long-term synaptic remodeling and memory. It remains unknown how IEGs are maintained for memory despite rapid transcript and protein turnover. To address this conundrum, we monitored Arc, an IEG essential for memory consolidation. Using a knock-in mouse where endogenous Arc alleles were fluorescently tagged, we imaged Arc mRNA dynamics in individual neurons in real time, in cultures and brain tissue. Unexpectedly, a single burst stimulation was sufficient to induce cycles of transcriptional reactivation in the same neuron. Subsequent transcription cycles required translation; whereby new Arc proteins engaged in autoregulatory positive feedback to reinduce transcription. The ensuing Arc mRNAs preferentially localized at sites marked by previous Arc protein, assembling a “hotspot” of translation, and consolidating “hubs” of dendritic Arc. These cycles of transcription-translation coupling sustain protein expression and provide a mechanism by which a short-lived event may support long-term memory.
Keywords: Immediate early gene, Arc, transcription, local translation, protein hubs, translation hotspots, long term memory, memory consolidation, single mRNA imaging, cyclical gene expression
eTOC
How are short-lived mRNAs and proteins maintained over time to impact long-term memory? Das et al. report that repetitive cycles of transcription and local translation of the immediate early gene Arc amplifies a single burst stimulation over time, to assemble Arc protein hubs, revealing a molecular mechanism supporting memory consolidation.
Graphical Abstract
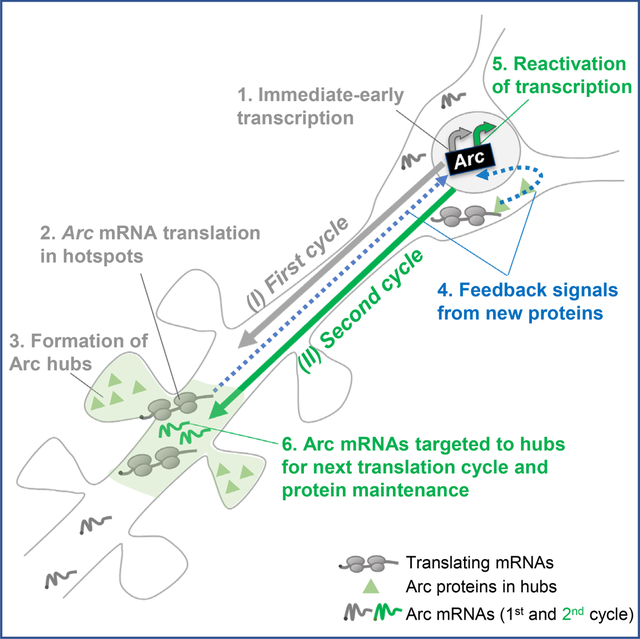
INTRODUCTION
The ability to learn new information and store it for long periods is one of the most remarkable features of the brain. Memory consolidation requires modifications of synaptic connectivity and stabilization of these changes 1,2. Both transcription and translation are required for long term changes at the synapses 3,4, with growing evidence indicating the role of local translation 5–7. However, the molecular mechanisms underlying transcription and translation regulation to drive persistent synaptic remodeling for memory consolidation remains unknown. To meet the demands of sustained local protein synthesis for long term memory 8,9, the dendritic levels of mRNAs encoding plasticity proteins must be maintained over time. A precise control of local RNA concentrations in the dendrites ensures that the right amount of proteins are synthesized for proper synapse remodeling 8. For example, constitutive, long-lived β-actin mRNAs can persist in the dendrites for several hours and undergo multiple rounds of translation for structural changes at spines 10,11. However, this model would not apply for inducible transcripts with short half-lives, raising the conundrum of how these mRNAs, often encoding short-lived proteins, could facilitate long-term physiological effects. Immediate early genes (IEGs) have established roles in transducing experiences (activity) into long lasting molecular changes for memory 12–14. Despite the importance of IEGs in memory storage, these mRNAs and proteins have short half-lives between 30–60 min 15,16. One such IEG is the Activity Regulated Cytoskeletal Associated (Arc) gene, which is critical for consolidating long-term memory 13,17,18 and implicated in several neurodegenerative and cognitive disorders 19–21. Arc protein is multifunctional and regulates different forms of synaptic plasticity: long term potentiation (LTP), long term depression (LTD), and homeostatic plasticity 13,22–25. A precise temporal regulation of Arc expression is critical for normal cognitive functions including memory consolidation and recall 24,26–28. Both Arc mRNA and proteins have rapid turnover (half-lives approx. 60 mins) because of non-sense mediated mRNA decay 29, and rapid ubiquitination of the protein 30. Therefore, the transient nature of Arc expression is inconsistent with the requirement of Arc protein for long term plasticity maintenance and memory consolidation occurring over several hours to days 13,17,24,28. We addressed this temporal paradox by utilizing genetic approaches to tag endogenous Arc mRNAs and proteins combined with high resolution imaging in cultures and in tissue. Recently, we generated a knock-in mouse (Arc-PBS), where the endogenous Arc gene was tagged with stem loops and detected by a fluorescent binding protein 31,32, allowing us to follow single Arc mRNAs from synthesis to transport and decay in living neurons.
In this study, we identified an unprecedented reactivation of the Arc gene at single neuron resolution in situ that provides a mechanistic explanation of how an IEG can drive long-term changes in neurons and synapses. The Arc gene underwent cycles of transcription and translation in the same neuron in response to an initial stimulus via an autoregulatory feedback loop wherein the Arc protein reinitiates its own transcription. Reactivating the gene furnished a steady supply of mRNAs to distal dendrites necessary to replenish unstable Arc transcripts and proteins and maintain the dendritic protein levels in restricted “hubs”. The coupling of transcription and local translation with similar periodicity ensures that unstable mRNAs can produce the required amount of proteins at synapses at the right time. This could be a general mechanism by which short-lived mRNAs and proteins support persistent synaptic remodeling necessary for memory consolidation.
RESULTS
Reactivation of Arc but not actin drives transcriptional cycling after a single stimulation
Arc mRNA levels are tightly regulated by transcription 18,33. To investigate the long-term transcriptional behavior of the Arc gene and compare it to the constitutive β-actin gene in response to a brief burst of neuronal activity, we used tetrodotoxin withdrawal/washout, TTX-w 31,33, and monitored transcription for four hours in individual hippocampal neurons in culture. Surprisingly, immediate early (IE) activation of Arc and its subsequent decay was followed by a second transcriptionally active (“ON”) state without further stimulation. This reactivation of transcription was re-initiated in the same neuron after a prolonged OFF-period (Figure 1A, B; Video S1, S2). The average intensity trace of transcribing alleles from multiple neurons revealed a coordinated cycling of Arc transcription with an OFF period of 72 ± 9.3 min between cycles (Figure 1C). β-actin transcription did not exhibit such distinct cycling with reactivation (Figure 1D).
Figure 1: Reactivation of Arc transcription drives subsequent transcription cycle.
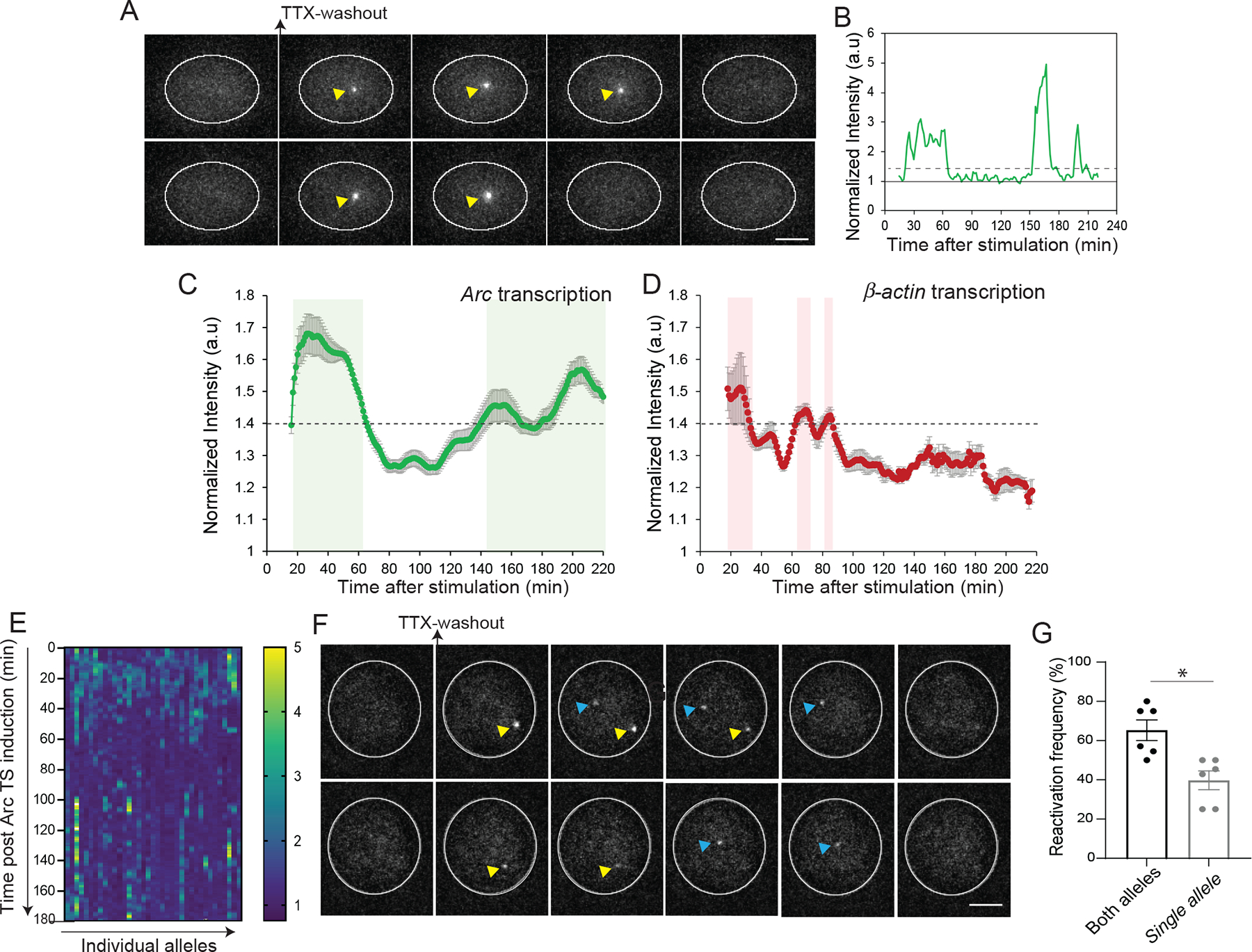
(A) Representative images showing the PCP-GFP labeled nucleus of a single neuron. Arc transcribing alleles (yellow arrows) after evoked activity upon TTX-washout. (B) Intensity trace of the transcribing Arc allele in A. Solid line shows normalization to nuclear background. Dashed line indicates threshold for inclusion criteria as active transcription. (C) Average intensity trace of Arc transcription from multiple neurons. Shaded areas indicate active transcription (n= 58 neurons from 6 independent experiments). (D) Average intensity trace of β-actin transcription from MBS-tagged β-actin allele after TTX-w. Dashed line indicates threshold for inclusion criteria as active transcription. n = 22 cells from 3 independent experiments. (E) Heat map of transcription from individual Arc alleles after induction. Each column shows a single allele, and the rows represent time. Warmer colors indicate higher transcription amplitude. IE-activation is followed by shutdown (darker colors) and then reactivation (warmer colors). n = 37 alleles, 5 independent experiments. (F) Representative images showing IE- and reactivation of both Arc alleles albeit with different onset times. Arrows indicate transcribing alleles. (G) Frequency of reactivation from both alleles or from a single Arc allele (n = 45 neurons, 6 independent experiments, each circle represents one experiment). Scale bar is 5 μm. Error bars indicate SEM.
Arc transcriptional events beyond the IE-phase (100 min post stimulation) were further categorized as either sustained, reactivated, or simply delayed. “Sustained” transcription occurred when an allele was constitutively active, “reactivated” when re-initiation of transcription occurred after a shutdown of ≥30 min post IE-event, and “delayed” when transcription initiation occurred after a latency period (> 90 min) post stimulation. A detailed examination of individual neurons over time demonstrated that the reactivation of the gene (59.5 ± 3.1 %) was the predominant feature driving the second Arc cycle (Figure S1A). This feature was conserved in alternate stimulation condition (chemical long-term potentiation, cLTP) with comparable reactivation onset times (154.3 ± 4.2 min for TTX-w; 141 ± 6.0 min for cLTP) (Figure S1B). Of note, ~60% of the transcriptionally active neurons from the IE-phase underwent reactivation in either stimulation condition (Figure S1C). Remarkably, transcription reactivation events were observed for 6 hours post stimulation, indicating multiple cycles of Arc mRNA synthesis after a single stimulation (Figure S1D).
A heat map of intensities from individual alleles in the neurons undergoing reactivation revealed that reactivation occurred often at the same allele in the population (Figure 1E). Furthermore, if two alleles were induced in IE stage, there was a significant chance that both would be reactivated (62% for both vs 38% for single, p < 0.05, paired t-test) (Figures 1F–G, Video S2). Thus, the long-term transcriptional signature revealed a novel cycling of the inducible Arc gene, which was distinguishable from constitutive β-actin mRNA transcription.
Two cycles of Arc transcription in the hippocampus after a single burst of stimulation
Our understanding of how neuronal activity regulates the expression of Arc in tissue derives from studies that mostly focused on the immediate early stages (within two hours after activity) 14,25,34. We investigated the long-term transcriptional behavior of the Arc gene in situ in response to a brief burst of neuronal activity. An optogenetic stimulation approach was developed to deliver a specific stimulus strength, along with real-time monitoring of the transcriptional response from individual dentate granule cells (GCs) in acute hippocampal slices. The Arc-PBS (ArcP/P) mouse was crossed with the PCP-GFP transgenic mouse for cell specific detection of Arc transcription in brain tissue32. A cocktail of two viruses, AAVDJ-FLEX-ChIEF-tdTomato and AAV5-mCherry-Cre, was injected into the dentate gyrus to achieve Cre-specific expression of fast channel rhodopsin ChIEF and PCP-GFP in GCs (Figure 2A, B). To activate GCs, a 25 Hz optical stimulation was delivered to the GC layer of dentate gyrus (Figure 2C), and transcription from both alleles in individual neurons was imaged for over four hours using two-photon microscopy (Figure 2D). Following an initial stimulation, immediate early Arc transcription peaked at 30 min (14.6 ± 2.2 % at 30 min vs 5.7 ± 1.1% at basal) and declined to basal levels at 100 min (Figure 2E). Similar to the observations in cultured neurons, a second peak of transcribing GCs occurred at 150 min without additional stimulation and was maintained till 180 min (9.7 ± 0.8 % at 150 min; 9.1 ± 0.9 % at 180 min). The second transcriptional cycle primarily consisted of GCs undergoing reactivation and sustained transcription (Figure 2F). Importantly, the reactivation onset times (120–180 min) were similar to that observed in cultures, suggesting that the temporal regulation of Arc gene transcription was recapitulated in brain tissue.
Figure 2: Cycles of Arc transcription in the dentate gyrus.
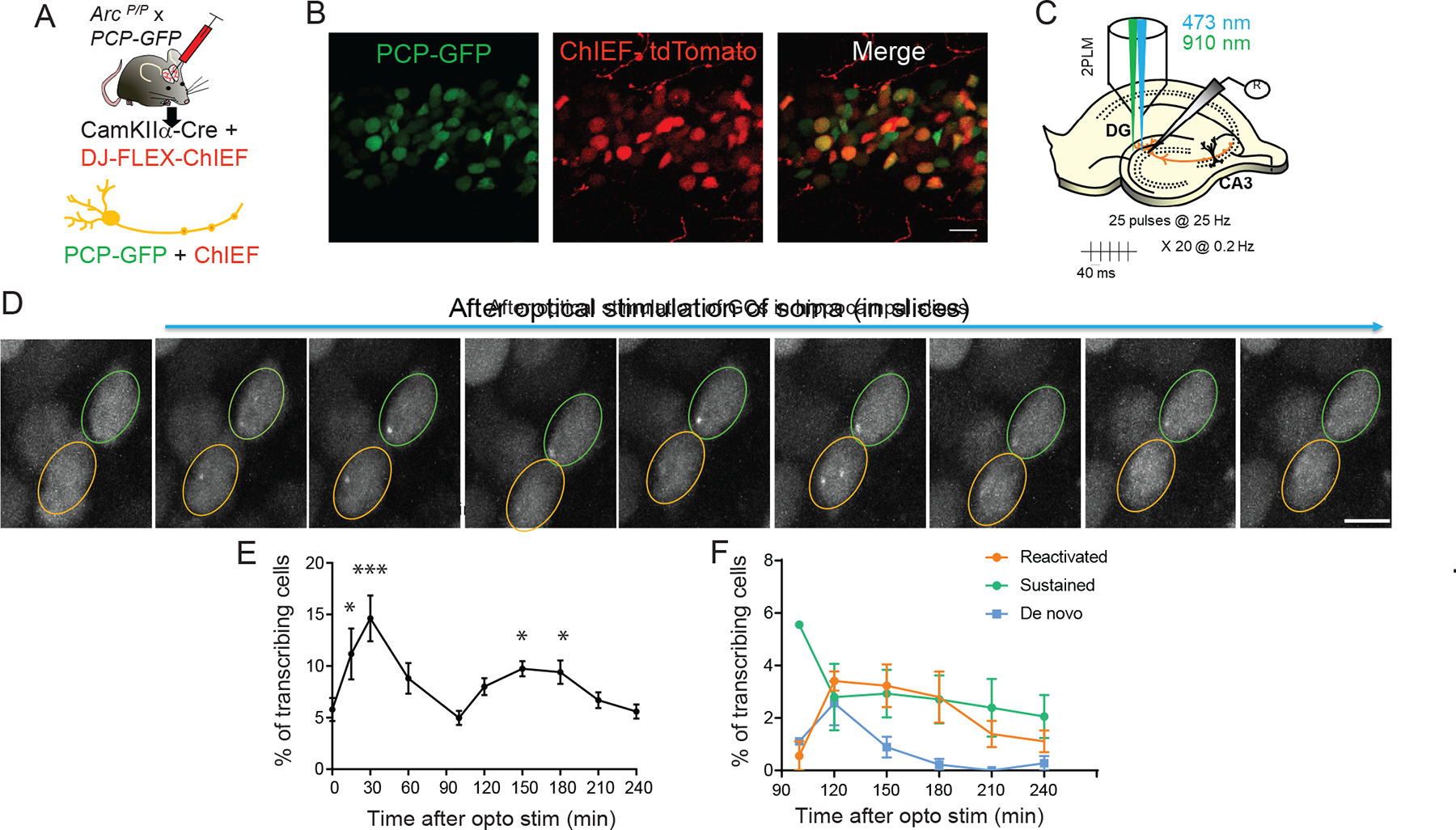
(A) Approach for Cre-dependent expression of PCP-GFP and ChIEF in the same neuron by injecting a cocktail of two viruses-CaMKIIα-Cre and DJ-FLEX-ChIEF-mCherry to the dentate gyrus of Arc-PBS x PCP-GFP mice. (B) Image showing co-expression of PCP-GFP and ChIEF-mCherry in the same granule cells of DG. (C) Stimulation paradigm for ChIEF (473nm) and two-photon imaging of GCs (910 nm illumination). (D) Representative images of two GC nuclei displaying transcription after optical stimulation. Orange outline shows neuron with transcriptional reactivation, green outline displays sustained activation. Scale bar 10 μm. (E) Total percentage of transcribing GC neurons after optical stimulation revealed two cycles (14.6 ± 2.2 % at 30 min, 9.7 ± 0.74 % at 150 min, 9.1 ± 0.9 % at 180 min, *** p = 0.003 at 30 min, *p = 0.01 at 150 min, *p= 0.04 at 180 min, compared to baseline 0 min, one-way ANOVA). (F) Distribution of the different transcriptional states during the second cycle (100 min post stimulation). n = 5 slices, 5 animals.
The transcriptional response using an optogenetic stimulation was further assessed in cultured hippocampal neurons from the Arc-PBS mouse (Figure S2). Two trains of 20 Hz pulses (25 pulses/train) separated by a two-minute interval were used to selectively stimulate the soma only once, and this resulted in robust increase of Ca2+ transients (CaTs). (Figure S2A–C). Cyclical transcription was observed in individual neurons with reactivation probability 54.2 ± 4.2 % and average onset time of 146.4 ± 7.1 min (Figure S2 D–F). During reactivation, the transcriptional output was reduced in the amount of nascent mRNA compared to that during initial activation. This occurred after global and single neuron optogenetic stimulation (Figure S2 G–I). Likewise, the duration of the reactivation was equivalently reduced (Figure S2 J–K), suggesting a possible dampening of transcription over time.
Reactivation was independent from the increase in nuclear calcium
Elevated levels of somatic and nuclear calcium have been implicated for induction of IEG transcription14. We therefore tested whether transcription reactivation could depend on Ca2+ activity. To this end, we measured nuclear Ca2+ levels using the indicator NLS-tagged jRGECO1a at different time points after stimulation (Figure S3A). An immediate increase in the frequency of Ca2+ transients (CaTs) at 10 min post TTX-w was observed, which peaked at 30 min and remained until 60 min (Figure S3B, C). These initial CaTs were followed by a significant reduction at 120 min, and negligible activity was observed at 180 min. The amplitude of CaTs showed a similar profile, that is, a rapid increase followed by a decrease at 120 min (Figure S3D). These findings showed a severe dampening of nuclear CaTs at the two-hour time point when reactivation was induced.
To confirm that reactivation occurred independently of Ca2+ rise, neuronal activity was silenced by reapplying TTX to the imaging media at 90 min (Figure 3A). All IE-events typically occur within 90 min post stimulation, and therefore we chose this time point to modulate subsequent cycles. Co-imaging of nuclear Ca2+ and Arc transcription was performed by expressing NLS-jRGECO1a and PCP-GFP in the same neuron (Figure 3B, C). Increase in nuclear Ca2+ levels and concomitant Arc transcription were observed in the same neuron in the IE-phase. After reapplication of TTX (Figure 3B), CaTs were abolished. However, reactivation of Arc transcription occurred in those same neurons even though nuclear Ca2+ levels were undetectable (Figure 3C, Video S3). Despite being lower than TTX-w, a notable 45 ± 2.9 % of neurons displayed reactivation after TTX reapplication (Figure 3D), with no significant difference in transcription onset times (154.3 ± 4.2 for TTX-w versus 145.3 ± 6.4 for TTX-w + TTX, Figure 3E). Therefore, while the IE-phase involved a synchronous Ca2+ elevation, the reactivation onset appeared uncorrelated with activity, suggesting that the later transcriptional cycle was governed by a mechanism distinct from the conventional excitation-transcription coupling involving Ca2+ rise in neurons.
Figure 3: Reactivation of Arc transcription is independent of Ca2+ rise.
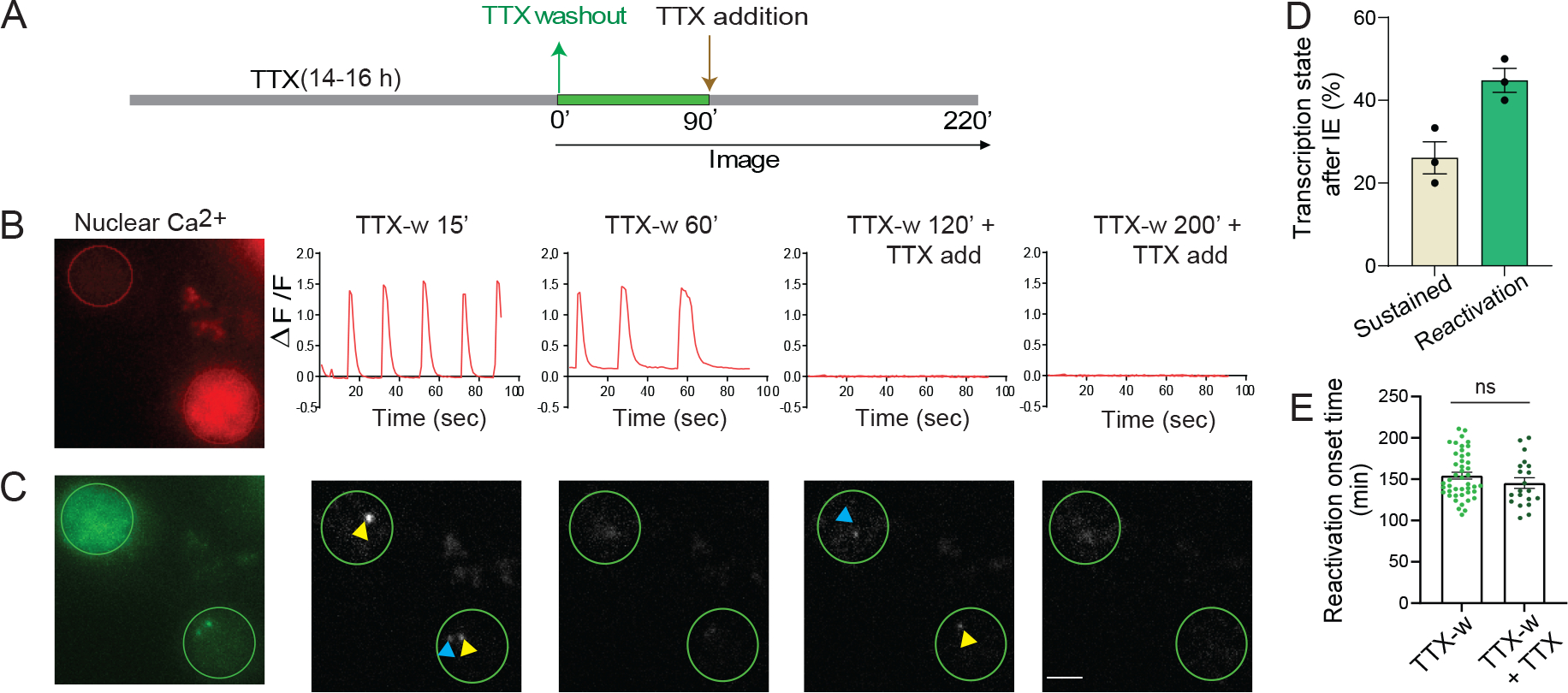
(A) Schematic of stimulation paradigm. (B-C) Neurons co-expressing red-shifted NLS-jRGECO1a and PCP-GFP were imaged for nuclear Ca2+ levels and Arc transcription. Nuclear CaTs triggered by TTX-w ceased after reapplying TTX (B). Imaging TS in the same neuron showed reactivation even after TTX addition (C). (D) Different transcriptional states after TTX reapplication in neurons activated in IE-phase (n = 43 neurons from 3 independent experiments). Delayed transcription induction was not observed. (E) Comparison of reactivation onset times (n = 20 neurons for TTX-w + TTX; 45 neurons for TTX-w, p = 0.24, unpaired t-test). Error bars indicate SEM. p > 0.05 non-significant. Scale bar 5μm.
Transcriptional reactivation required protein synthesis including Arc
Classically, IEGs including Arc transcribe rapidly in response to stimulation, without the requirement for new protein synthesis 15,35. We therefore assessed the role of protein synthesis for the second transcription cycle. Adding the elongation inhibitor cycloheximide (CHX) 90 min after stimulation (Figure 4A) reduced the reactivation efficiency (7.8 ± 2.1% in CHX vs 59.5 ± 3.1 % in TTX-w) (Figures 4B–D). Importantly, CHX removal resulted in robust induction of transcription (Figures 4B–D) with synchronous initiation times (median = 20 min after washout) (Figure 4E, Video S4). By inhibiting elongation, CHX blocked new protein synthesis but retained mRNAs with stalled ribosomes that were readily translated upon CHX washout. Many of these new proteins fed back into the nucleus to reactivate transcription. Notably, the impact of protein synthesis on transcriptional reactivation was recapitulated with another inhibitor puromycin (7.8 ± 2.1 % in CHX vs 20.5 ± 2.7 % in Puro; 78 ± 4.8 % in CHX-w vs 67.5 ± 2.3 % in Puro-w; Figure 4D, E). The dependence of the second transcription cycle on protein synthesis was evaluated in tissue as well. Acute hippocampal slices were briefly depolarized with KCl and maintained with or without CHX, followed by CHX-washout in one set (Figure S4A). The transcribing GC fraction at 120 min post depolarization was significantly reduced with CHX application compared to no CHX but were rescued upon washout (Figures S4B and S4C). Moreover, transcription from both alleles was also restored (Figure S4D). Taken together, these findings indicate that later Arc transcriptional cycles required protein synthesis.
Figure 4: Autoregulatory feedback by new protein synthesis reactivates Arc transcription.
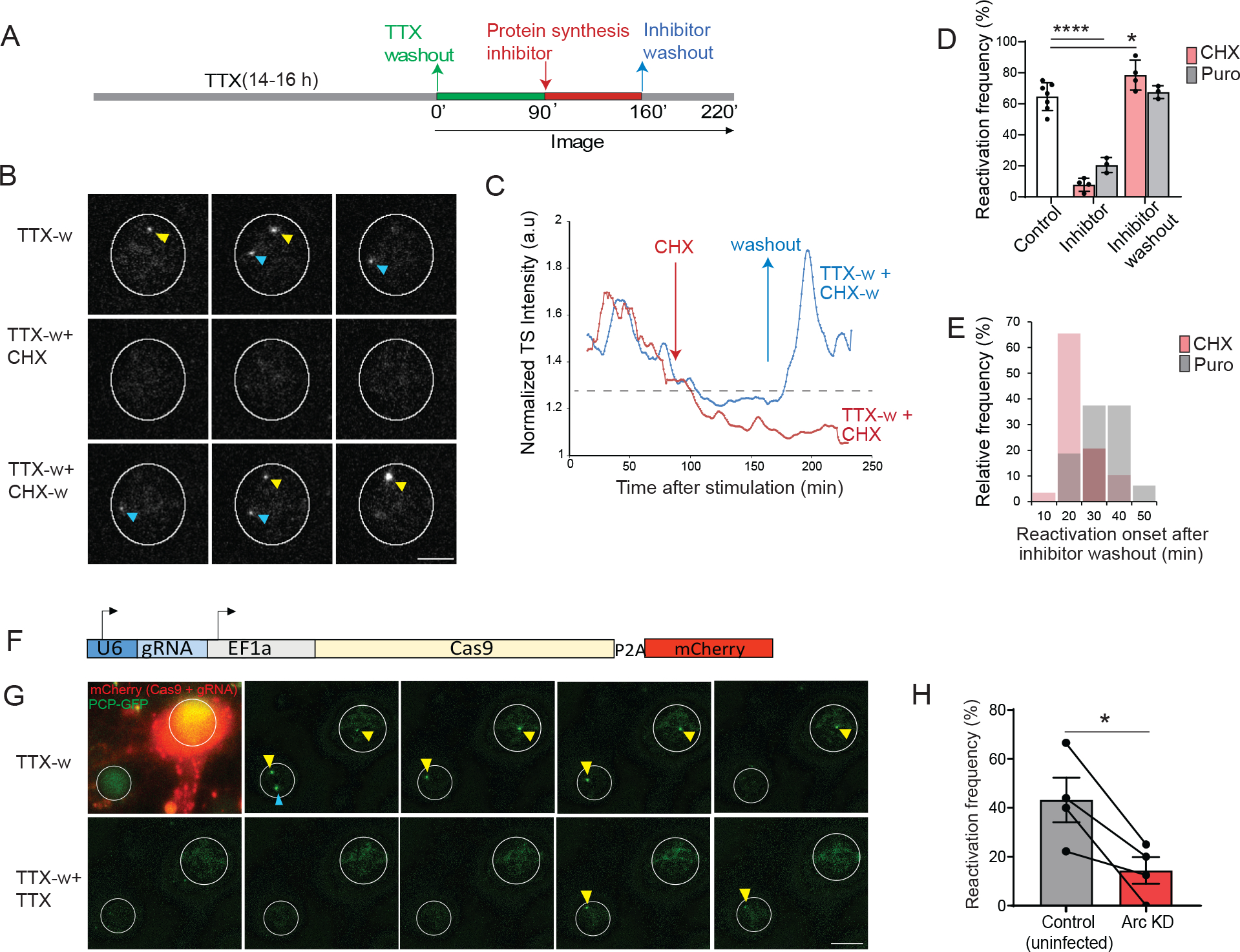
(A) Schematic of stimulation paradigm to monitor the effect of protein synthesis. Inhibitor of protein synthesis (CHX, 50 μg/ml or Puromycin 50 μg/ml) was added at 90 min (TTX-w + CHX or TTX-w + Puro). In another set of experiments, inhibitor was incubated for 70 min and then washed out (TTX-w + CHX-w; TTX-w + Puro-w). (B) Representative images showing IE-transcription from both alleles, followed by shutdown maintained with CHX addition. Washout of CHX restored transcription. (C) Intensity trace of transcribing alleles from two conditions-CHX addition (TTX-w + CHX), and washout (TTX-w + CHX-w). (D) Percentage of reactivation across conditions, each circle represents one experiment (TTX-w vs TTX-w + CHX, TTX-w vs TTX-w + Puro **** p < 0.001; TTX-w vs TTX-w + CHX-w, * p = 0.02; TTX-w vs TTX-w + Puro-w, p = 0.98, two-way ANOVA; n = 21 neurons for TTX-w + CHX, n = 47 neurons for TTX-w + CHX-w, n = 26 neurons for TTX-w + Puro and TTX-w + Puro-w from 3 independent experiments, TTX-w from Figure 1E). (E) Frequency distribution of reactivation onset times after translation inhibitor washout (Median = 20 min for CHX-w, 31.5 min for Puro-w). (F) All-in one lentiviral construct used for Arc KD. (G) Representative images showing neurons expressing PCP-GFP with or without Cas9 + gRNA (mCherry). Note that the IE-transcription occurred in both neurons (arrows indicate TS). Neuron with Arc KD do not exhibit reactivation (after TTX reapplication at 90 min). (H) Quantification of transcriptional reactivation frequency (n = 26 neurons for control, n= 24 neurons for Arc KD from 4 independent experiments. * p = 0.03, paired t-test). Error bars represent SEM. **** denotes p < 0.001, * denotes p < 0.05. Scale bar 10μm.
Several proteins synthesized in the IE-phase are putative transcription regulators, and we sought to examine whether Arc protein itself could participate in controlling subsequent transcription. To test this possibility, knockdown (KD) of Arc protein was performed by CRISPR-Cas9 using guide RNAs (gRNAs) targeting the coding region of the gene, and a specific gRNA (gRNA-2) was chosen based on the KD efficiency (Figure S5A–B). A lentiviral construct expressing gRNA and Cas9 with mCherry (Figure 4F) was used alongside PCP-GFP to co-infect neurons from Arc-PBS. Expression of mCherry identified the neurons with Arc KD (Figure 4G). Comparative analysis of PCP-GFP neurons with and without mCherry revealed that the reactivation efficiency was significantly decreased upon Arc KD (43.2 ± 9.1 % in control vs 14.4 ± 5.4 % in Arc KD, Figure 4H). These results were validated using a shRNA mediated KD strategy, which showed a similar decrease in reactivation frequency in Arc KD cells compared to the WT uninfected neurons or scrambled shRNAs (69.4 ± 10.01 % in control vs 14.1 ± 8.7 % in Arc shRNA; Figure S5 C–D). In summary, the findings indicated that Arc protein levels were a critical determinant of subsequent cycling of the gene, highlighting an autoregulatory positive feedback mechanism.
Arc mRNAs and proteins were maintained in dendrites long after an initial stimulation
We assessed whether cycles of transcription were reflected in Arc mRNA levels in dendrites. Time-lapse imaging of single mRNAs showed (Figure S6A, B) that the RNA number in dendrites was not constant but displayed fluctuations over time (Figure S6C). The RNA density plot in dendrites across multiple neurons displayed an initial rise at 90 min followed by a plateau, and a second peak at 210 min (Figure S6D). Since the residence time of the Arc mRNAs in the dendrites was short (average 7.6 ± 1.2 min) (Figure S6E), we propose the increase in RNA density during the second phase was not due to the long-term persistence of mRNAs but resulted from new Arc mRNAs populating the dendrites.
Additional mRNAs transported and localized in the dendrites would result in local protein synthesis. To examine whether Arc proteins in the dendrites were replenished over time, a reporter was designed to identify the Arc proteins from different cycles. An activity-regulated SARE promoter 36 drove the expression of a HaloTag fused to the Arc coding sequence (CDS) followed by the 3’UTR comprising the cis-acting regulatory elements (Figure 5A). Cell permeable Halo-ligand conjugated to JF dyes 37 were used to label the Halo-tagged Arc proteins. Specifically, pulse-chase imaging with spectrally distinct JF dyes- JF646 and JF549 dyes detected Arc proteins synthesized from the first (IE) and the second cycle respectively (Figure S6F). Since inducible Arc translation peaks at two hours 25, labeling with JF646 was performed 150 min post stimulation to detect all Arc proteins from the IE-phase. Notably, the labeled Arc protein was not evenly distributed, but displayed discrete puncta along the dendrites (Figure S6G). A subsequent chase with JF549 for 60 min not only revealed a second cycle of Arc protein synthesis but surprisingly, the new proteins were in close proximity or overlapped with the previous 646 signals. The distances between the brightest JF646 puncta to the nearest JF549 signal were measured and the majority of these puncta (75 %) were within 3.4 μm, indicating the enrichment of Arc proteins from the first and second cycle in discrete dendritic domains termed “hubs” (Figure S6H). These hubs potentially represent sites where dendritic Arc proteins accumulate and are maintained over time to influence long-term spine remodeling.
Figure 5: Detection of Arc protein hubs in dendrites and accumulation of mRNAs in the hubs.
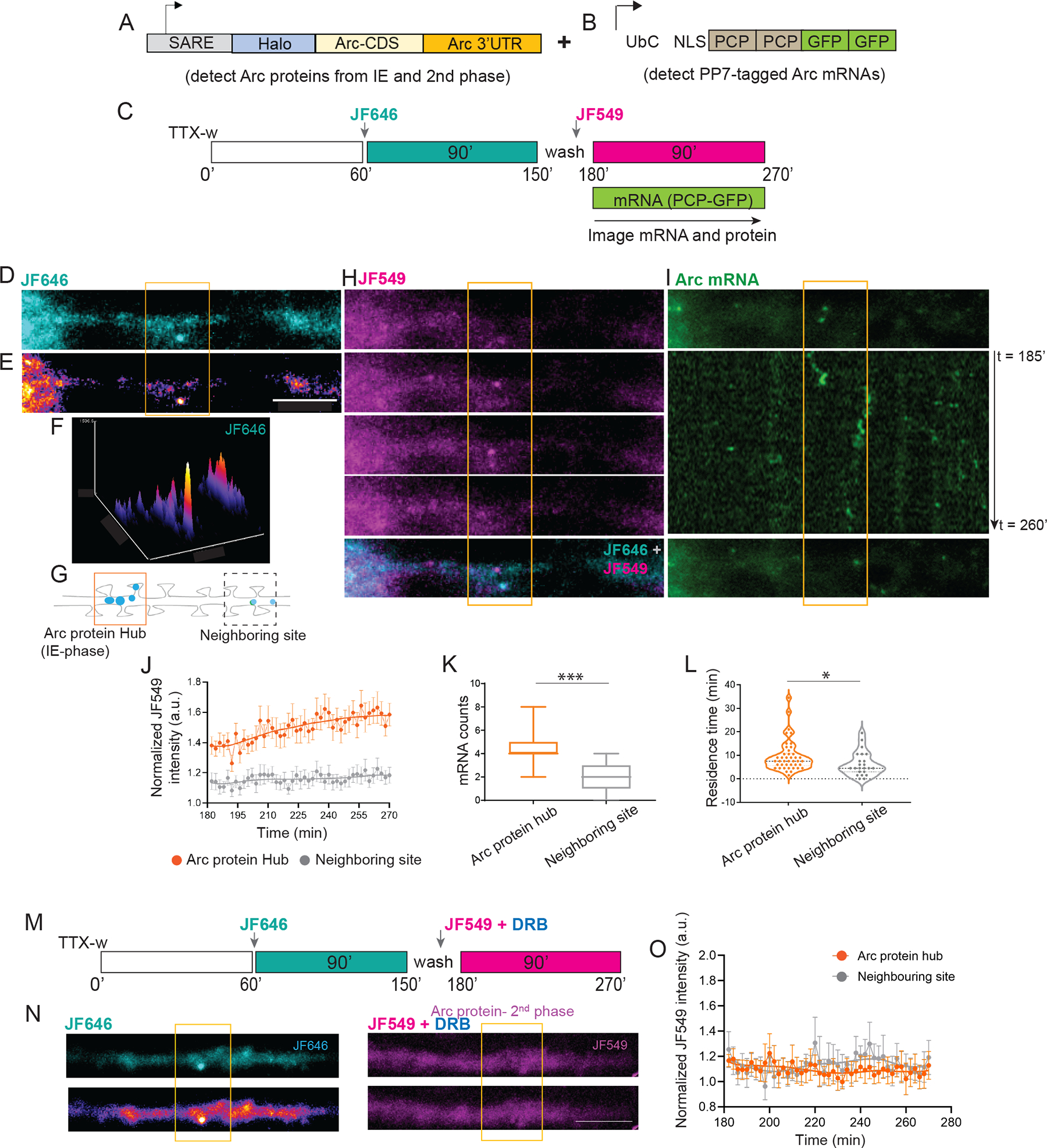
(A) Schematic of the Halo-Arc reporter to detect proteins from the two cycles. (B) PCP-GFP construct to visualize endogenous Arc mRNAs. A cocktail of lentiviruses expressing A and B was used to image Arc proteins and mRNAs in the same neuron. (C) Schematic of JF646/JF549 labeling timeline after stimulation. Arc mRNAs and proteins from the second cycle were imaged 3 hr onwards. (D-E) Representative image indicating dendritic Arc protein from first cycle (JF646). Differential intensity of JF646 signal shown in E. (F) Intensity profile of JF646 intensity showed distinct peaks along the dendrite. (G) A 6 μm segment around the local maxima in F was used to designate Arc protein hub from IE-stage (orange outline). A ROI of same dimension was used for a neighboring site (dashed outline). (H) Time lapse imaging of Arc proteins synthesized in the second phase (JF549). Bottom panel shows a merged image of JF549 with JF646, indicating close proximity of both puncta within the hub. (I) Single molecule imaging of Arc mRNAs in the same dendrite shows localization in the hub. Middle panel shows a kymograph, number of localized mRNAs indicated. (J) Normalized intensity trace of new Arc protein (JF549 signal) over time in hub versus neighboring site as defined in G. (K) Comparison of Arc mRNA counts populating the hubs vs neighboring sites (*** p = 0.002, Wilcoxon signed rank test). (L) Comparison of the residence times of Arc mRNAs in the hubs vs neighboring sites (* p = 0.029, unpaired t-test). n =12 neurons from 3 independent experiments (J, K). n = 43 mRNAs for hub versus n = 23 mRNAs for neighboring site (L). (M) Labeling scheme of Arc proteins after blocking the second transcription cycle with DRB. (N) Representative images show JF646 and JF549 label in the same dendrite. Lack of distinct JF549 puncta was observed with DRB treatment. (O) Normalized intensity trace of new Arc protein (JF549 signal) in the Arc hub versus in neighboring site. n =10 dendrites from 2 independent experiments. Scale bar is 6 microns. *** denotes p < 0.005, * denotes p < 0.05. Error bars represent SEM. Scale bar is 6 μm.
Arc protein hubs were consolidated over time and accumulated Arc mRNAs from the second transcription cycle
If the Arc protein hubs were selectively consolidated compared to other dendritic regions, then repeated enrichment of Arc mRNAs and proteins should occur in these hubs. To test this possibility, the localization kinetics of mRNAs and proteins from the second cycle were assessed relative to the hubs with high temporal resolution. A three-color real-time imaging approach was developed, where neurons from the Arc-PBS mouse were infected with two lentiviruses: one expressing PCP-GFP and the other expressing the Halo-Arc protein reporter (Figures 5A and 5B). The timeline of labeling and imaging has been depicted in the scheme (Figure 5C). Arc protein from the first cycle (JF646) was spatially concentrated along the dendrite as shown by the local maxima of JF646 puncta intensities (Figures 5D and 5E). A segment around the centroid of the peak (3 μm on either side, based on Figure 5H) was used to designate the Arc hub (Figures 5F and 5G). Time lapse imaging with JF549 showed that Arc proteins translated in the second cycle congregated in this same region of the existing hub (Figure 5H). However, these JF549 puncta were not long-lasting, suggesting possible degradation of the Halo-Arc protein. Importantly, the endogenous Arc mRNAs in the same dendrite preferentially localized at or in the vicinity of the existing hubs, as shown by the kymographs (Figure 5I).
Further analysis showed that the levels of Arc protein from the second cycle (JF549) specifically increased in the hub over time compared to a neighboring dendritic segment (Figure 5J). The peak of new Arc enrichment in the hub occurred at 252 mins after a latency period, supporting the idea that these hubs were reinforced by cycles of new protein synthesis. Notably, the localization of the Arc mRNAs from the second cycle spatially correlated with the protein hubs from IE; exhibiting a two-fold higher mRNA enrichment in the initial hub versus the neighboring regions (Figure 5K). Once localized, the mRNAs persisted at these hubs as calculated from their residence times (11.9 ± 1.9 min in hub versus 6.3 ± 1.1 min in neighboring site) (Figure 5L). To establish that consolidation of the hub was driven by newly transcribed Arc mRNAs from the second cycle, transcription was blocked using 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB) after the IE-phase. We found that inhibition of the second transcriptional cycle prevented the Arc protein enrichment in the hub over time (Figure 5M–O). These results revealed distinct spatial and temporal character of dendritic Arc mRNAs and its cognate proteins: i) a biphasic regulation of Arc protein synthesis, and ii) the emergence of local hubs of Arc proteins, which are reinforced over time by repetitive localization of Arc mRNA from later transcription cycles.
Cycles of local Arc translation in dendritic hotspots supports the maintenance of Arc protein hubs
Since the Arc protein hubs were tightly controlled in space and in time, we hypothesized that local translation is the underlying mechanism permitting nascent protein accumulation in select dendritic domains to form the hubs. To test this hypothesis, translation was imaged in real time with multimerized epitopes (“Suntag”) on the Arc protein and detected with a genetically encoded single chain antibody (scFV) fused to a fluorescent protein 38–40. The Suntag-Arc reporter contained 24 repeats of the epitope in the N-terminus of Arc CDS with Arc mRNA 5′ and 3′UTRs for translation regulation (Figure 6A). The ability of the Suntag-Arc construct to report translating Arc mRNAs was tested by performing single molecule FISH (smFISH) for the mRNA and immunofluorescence (IF) against the Suntag protein in fixed neurons (Figure 6B, left panel). Co-localization of smFISH spots with bright Suntag IF-signal showed mRNAs undergoing translation at two hours post stimulation. This was significantly reduced upon addition of the translation initiation inhibitor Harringtonine (37.2 ± 2.4 % TTX-w 2h vs 16 ± 2.3 % Harringtonine, Figures 6C and 6D). Of note, the translating Arc mRNAs were maintained at 4 hours post stimulation, indicating that the mRNAs from early and late cycles were translationally competent.
Figure 6: Long-term imaging of Arc translation reveals hotspots and biphasic dynamics.
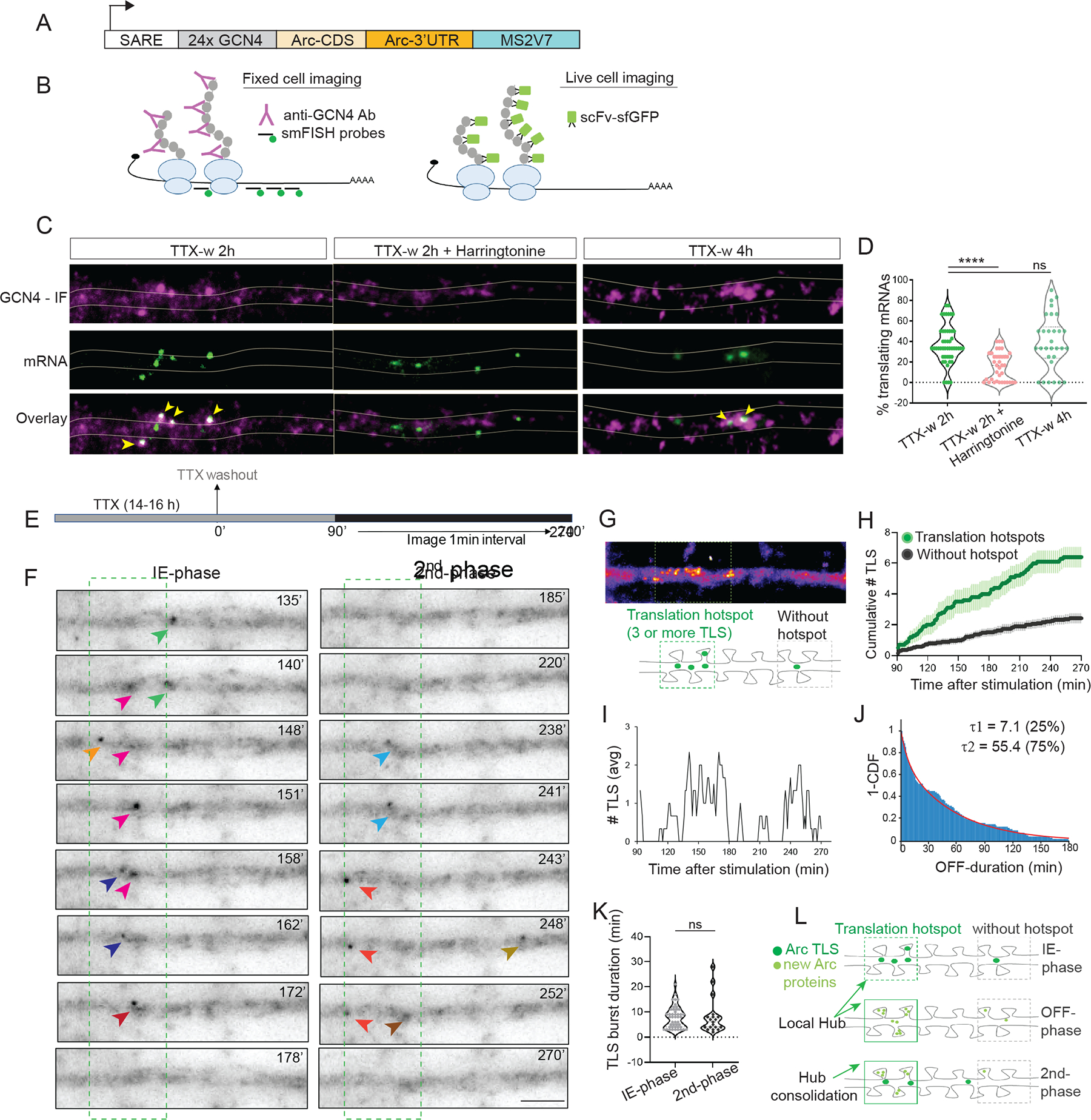
(A) Schematic of the Suntag-Arc translation reporter. The reporter is driven by SARE (activity-regulated promoter) and contains the 24X GCN4 epitopes (Suntag) upstream of the Arc CDS and followed by the 3′ UTR and stem loops MS2V7. (B) Translating mRNAs are detected by fixed-cell imaging with antibodies against GCN4 and by smFISH with probes against GCN4 and stem loop sequence. In live cells, translation sites (TLS) are detected by binding of the single chain antibody against GCN4 (scFV) fused to superfolder GFP (sfGFP). (C) Images of dendrites showing both nascent peptides and mRNAs in stimulated and after inhibition with Harringtonine. Co-localization of IF-smFISH spots indicate translating mRNAs (yellow arrows). (D) Comparison of translating mRNAs after stimulation and translation inhibition (TTX-w 2h vs TTX-w 2h + Harringtonine, **** p < 0.001, TTXw 2h vs TTXw 4h, p = 0.23, one-way ANOVA; n = 49 dendrites for TTX-w 2h, n = 34 dendrites for TTX-w 4h, n = 37 dendrites for TTX-w + Harringtonine from 2 independent experiments). (E) Stimulation and imaging paradigm to capture long-term dynamics of Arc TLS. (F) Time-lapse images from a dendrite show de novo Arc translation (arrows). Different colors represent different translation sites. Time was binned into two 90 min segments to represent IE (90–180 min) and second (181–270 min) phase. (G) A time-projected image of (F) shows spatial clustering of TLS to form a translation hotspot. Schematic in lower panel shows inclusion criterion for a hotspot. (H) Cumulative TLS count in hotspots versus regions without hotspots (n = 20 dendrites, 3 independent experiments). (I) Average TLS counts show biphasic dynamics. (J) Inverse cumulative distribution of time duration without de novo translation (OFF-period) fitted to a 2-component exponential. Relative percentage of events in τ1, τ2 indicated (n = 90 events). (K) Duration of each translation event during the IE and second phase (p = 0.5, Kolmogorov-Smirnov test, n= 46 events in IE, n=20 events in second phase). (L) Proposed model of Arc hub formation and maintenance by local translation in the hotspots. Arc translation in hotspots (IE-phase) increases local protein density to form the “hubs”. During the OFF-phase, translation is low due to limited mRNA availability. Transcriptional reactivation supplies new mRNAs, which visit the hubs resulting in a second translation phase, thereby consolidating the hub. Error bars indicate SEM. **** denotes p < 0.001, * denotes p < 0.05, ns for p > 0.05.
To investigate the spatio-temporal dynamics of local translation over time in living neurons, Suntag-Arc reporter and scFV-GFP (Figure 6B, right panel) were co-expressed and imaged for several hours after stimulation (Figure 6E). Discrete foci, much brighter than the faster diffusing proteins were detected in the dendrites (Figure 6F, Video S5), indicative of translation sites (TLS). Spatial analysis of TLS distribution showed that they were not homogeneous but clustered along the dendrites (Figure 6G), forming hotspots that could represent sites with increased translation efficiency 41. Accordingly, the appearance of multiple TLS (3 or more) in an 8 μm dendritic segment within two-hour post stimulation was used as a criterion to define a translation hotspot. A cumulative increase of Arc TLS continued in these hotspots after IE-phase at a considerably higher rate than in a region without a hotspot (Figure 6H), resembling the patterns of spatially selective Arc protein enrichment in Figure 5. Therefore, the congregation of translating mRNAs creates an abundance of Arc proteins within the hotspots and could facilitate the formation of local hubs.
Tracking these TLS revealed that their numbers were not constant over time but displayed cycles of increase and decay (Figure 6I). Distribution of the OFF periods where no TLS are detected revealed a long t2 component of 55.4 ± 8.3 min (Figure 6J). These measurements closely resembled the OFF periods of transcription (~1hr), suggesting a temporal coordination of transcription and translation cycles.
Further analysis of TLS kinetics in these hotspots was performed by binning the graph in Figure 6H into two 90-minute bins, congruent with the two phases of Arc protein detection in Figure 5. Higher TLS counts indicative of more translation events during the IE phase were observed compared to the second phase (Figures S7A and S7B), mimicking the mRNA output measurements during transcriptional cycles (Figure S2G–K). Furthermore, tracking the position and the integrated intensity of individual TLS from the two phases demonstrated that translation occurred in bursts lasting an average of ~8 min/burst, irrespective of the phase (Figure 6K). Hence, intermittent bursts of local protein synthesis at translation hotspots could be a mechanism by which Arc proteins are organized in the hubs and maintained over time (Figure 6L).
Arc translation hotspots were associated with clustering of initiation factors and high spine density
Like many mammalian mRNAs, Arc undergoes cap-dependent translation and the RNA binding protein eIF4E is a critical rate-limiting step determining translation initiation 42,43 and exhibits localization to the dendrites 44. Therefore, we hypothesized that the initiation factors may define the translation hotspot regions. To visualize this process in real time, we co-expressed Halo-eIF4E (labeled with JF646) along with Suntag-Arc reporter and scFV-sfGFP in the same neuron. Following stimulation, dendritic segments with at least one TLS were selected and co-imaged with JF646 labeled Halo-eIF4E. Slow diffusion of eIF4E along with mRNAs likely represent translation initiation 43, hence, we imaged every minute starting at 90 min post stimulation to determine the spatial distributions of Halo-eIF4E in relation to the translation hotspots. Clusters of eIF4E were observed in the hotspots, including co-localization of Arc TLS with the Halo-eIF4E (Figure 7A). Quantification of eIF4E puncta revealed an enrichment of eIF4E and formation of larger clusters in the hotspots compared to the neighboring dendritic regions (Figure 7B, C). Notably, the clustering in the hotspots were stable over time (18.4 ± 1.9 min), often outlasting the Arc translation event (Figure 7D, E).
Figure 7: Arc translation hotspots display clustering of eIF4E and situated in dendritic regions with high spine density.
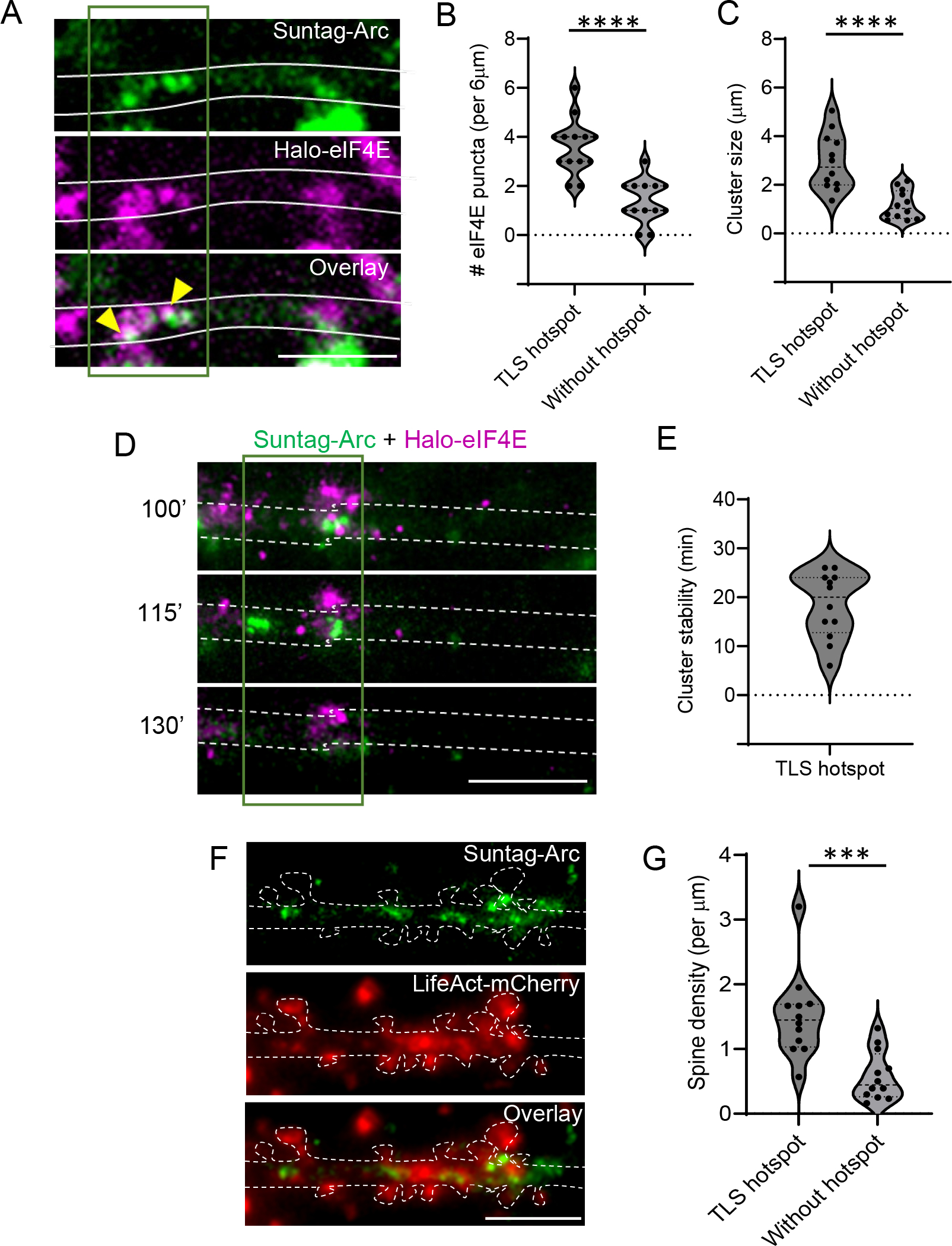
(A) Representative image showing de novo Arc translation (green) and JF646-labeled Halo-eIF4E (magenta). Arrows indicate co-localization. Dark green outline indicates the TLS hotspot, with evident clustering of Halo-eIF4E. (B - C) Comparison of eIF4E puncta number (B) and the total size of eIF4E clusters (C) in the TLS hotspot versus a neighboring non-hotspot dendritic region (p < 0.001, paired t-test, n= 12 dendrites from 4 experiments). (D) Time-lapse images from a dendrite with overlay of Arc TLS (green) and JF646-labeled Halo-eIF4E (magenta) post stimulation (min). Images time-averaged for 5 min. (E) Stability of Halo-eIF4E cluster in the hotspot measured as persistence time (min). (F) Image of a dendritic segment and associated spines (red, LifeAct - mCherry) showing Arc TLS clustering (green puncta) in regions with high spine density. (G) Quantification of spine density in TLS hotspot versus neighboring region without hotspot (p = 0.001, paired t-test, n =12 dendrites from 3 experiments). Scale bar is 6 microns. ***p < 0.005, **** denotes p < 0.001.
Next, we assessed the spatial correlation between the hotspots and the spine density and observed that the hotspots displayed significantly higher density of spines compared to a region lacking without a hotspot in individual dendritic segments (Figure 7 F, G). To summarize, local Arc protein synthesis occurs in hotspots, which exhibit distinct clustering of initiation factors and are typically formed around dendritic regions with high spine density.
DISCUSSION
In this study, we identified a unique cyclical regulation of Arc, an IEG with critical roles in long term memory. High-resolution imaging of the Arc gene from transcription to translation in live neurons demonstrated that temporally coordinated phases of mRNA and protein synthesis were critical for the maintenance of dendritic Arc protein. The initial protein buildup occurred in selective hubs by local translation in dendritic hotspots and remarkably, mRNAs from the later cycles find and populate these hotspots and undergo translation to maintain protein hubs over time. Our findings may resolve the conundrum of how short-lived mRNAs maintain local protein levels and provide a mechanism by which transient changes from a single stimulation can persist, consistent with the molecular events during memory consolidation.
One of the cellular mechanisms underlying memory formation, is the stabilization of synaptic contacts, and compelling evidence indicates that mRNA localization to activated spines leads to localized translation of proteins necessary for the structural and functional integrity of synaptic structures 4,6,10,42,45–47. To support memory consolidation, the activity-driven changes at synapses need to persist, by maintaining the local pool of plasticity proteins 48,49. For a structural protein such as β-actin, its mRNA is constitutively expressed, abundant, long-lived and can persist at, or revisit sites of activity to translate multiple rounds and promote stability of the cytoskeletal architecture within the postsynaptic region 10,11. The plasticity protein, Arc, is synthesized from mRNAs transcribed upon activity 15,25, after they traffic to sites of activity to be translated locally 34,50,51. However, in contrast to actin, both Arc mRNAs and its proteins are transient and degrade within a couple of hours 16,29,30,52. For synaptic structures and physiology to be maintained, Arc must be concentrated over time scales of consolidation 24,28, and this likely occurs via an alternate mechanism: cycles of transcription and translation in response to an initial stimulation. Given the importance of Arc for long term memory 13,17, this cyclical regulation begets additional rounds of transcription and protein synthesis, hence creating a repetitive feedback loop that may enable memory consolidation.
Sustained levels of Arc mRNAs and proteins have been observed in cultured neurons 36, and in the hippocampus after spatial learning 28,53. However, it has been challenging to distinguish whether different cell populations expressed Arc at different time points. Using the Arc-PBS mouse and the new Arc translation reporters, we characterized the different phases of gene expression with unprecedented temporal resolution. Reactivation of transcription in the same neuron predominantly accounted for the transcriptional events beyond the IE phase in both cultures and in tissue (Figures 1 and 2) with different forms of neuronal activity, suggesting a conserved intrinsic feature of the Arc gene. Since additional depolarization is not a requirement (Figure 3), transcriptional reactivation may occur by i) signaling cascades activated during the IE-phase that can sustain over long periods, or, ii) factors synthesized in the IE phase that do not require additional Ca2+-mediated activation. Signaling pathways such as extracellular signal-regulated kinases (ERK1/2 or MAPK) and neurotrophic factors such as BDNF, TGF-β have been implicated in LTP and memory consolidation 54–56. These synapse-to-nuclear signaling controls persistent activation of transcription factors and activators and may generate a permissive state for sustained transcriptional activation of plasticity genes 57,58. Our findings on the dependency of Arc reactivation on protein synthesis (Figure 4) including Arc protein itself favors the existence of an autoregulatory feedback loop. Since Arc is both a synaptic and nuclear protein, it will be important to delineate how each of these fractions contribute to transcription, and the mechanism governing autoregulation. Moreover, several canonical transcription factors capable of regulating Arc expression, such as zif268 59,60, Npas4 61 are rapidly translated IEGs that may have synergistic roles in reactivation. Thus, the molecular regulation of the two transcriptional cycles is distinct owing to their differential requirement of Ca2+ and de novo protein synthesis.
The phenomenon of reactivation is not just restricted to transcription; local translation of Arc mRNAs in the dendrites also occurs with a periodicity of ~2 hours. Reactivation could have several benefits. First, it allows production of short bursts of mRNAs and proteins without saturating the system. This is important for Arc, the levels of which need to be strictly regulated for cognitive flexibility 26. Second, reactivation maintains the RNA levels and efficiently replenishes the Arc hubs over long term after an initial stimulation. Finally, reactivation of Arc expression in a subset of neurons could favor their recruitment to a neuronal ensemble supporting the memory trace 1. Therefore, reactivation could potentially identify neurons involved in circuit strengthening during memory consolidation.
The formation and maintenance of the local Arc hubs by periodic translation in hotspots highlights that the maintenance of Arc protein along the dendrites occurs in selective dendritic regions. These hotspots may either indicate regions of elevated synaptic activity where spine remodeling is persistent, or regions of increased ribosome density 62 corresponding to high rates of translation 41,63; reviewed in 46,64. A distinct clustering of translation initiation factor, eIF4E was observed with the Arc translation hotspots (Figure 7). This finding is in agreement with a recent study measuring the binding dynamics of endogenous eIF4E to mRNAs and uncovering restricted diffusion and clustering in neurons after stimulation 43. The spatial distribution of the translation toolkit in response to synaptic activity may define the hotspot boundaries, promoting nascent protein synthesis in a restricted dendritic space to form the hubs. At the cellular level, Arc translation hotspots displayed a tight relationship with increased spine density. Such anatomical and functional grouping of synapses has been reported to be critical for plasticity 65,66 and our findings on translation hotspots potentially support this model.
In the last decades, studies have independently focused on the contribution of transcription and local translation to long-term memory 1,3,42,45,67. Little is known about how these two molecular events converge to regulate memory consolidation, with recent studies indicating long lasting transcriptional and translational programs for memory persistence 49,67. By providing a high-resolution spatio-temporal insight into activity-dependent gene expression, we show that coupling between transcription and translation is maintained for subsequent cycles several hours after stimulation to stabilize activity-driven changes. The long-term dynamics of Arc gene expression may serve as a template for studying other IEGs involved in memory and determining the ubiquitous or specific nature of these transcription-translation cycles. Future studies characterizing the persistence of Arc cycles not only during memory consolidation but also during memory storage are needed to delineate the dependence of memory events on gene expression. Improvements in technologies to image mRNAs and proteins in vivo and single cell approaches to manipulate gene expression will pave the way towards understanding transcription-translation coupling after a learning task, and whether perturbations of these cycles affect memory consolidation and storage.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact:
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sulagna Das, email: sulagna.das@einsteinmed.edu
Materials Availability:
All unique reagents and materials used in this study are available from Lead Contact, but we may require a completed Materials Transfer Agreement. Plasmids generated in this study will be deposited to Addgene in the future.
Data and Code Availability:
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Mice were group housed in a standard 12 hr light/12 hr dark cycle. Experimental procedures adhered to NIH and Albert Einstein College of Medicine Institutional Animal Care and Use Committee guidelines. Three mouse strains were used- Arc-PBS KI (ArcP/P) 31, ArcP/P × PCP-GFP 32, and Actin-MBS 68. Acute hippocampal slices were prepared from male and female mice (ArcP/P × PCP-GFP), but no significant differences were observed, so data from both sexes were combined.. The PCP-GFP mice were generated by introducing a neocassette containing CAG-stop/flox-NLS-PCP-GFP in the ROSA 26 locus, where PCP-GFP expression is Cre-inducible. The Arc-PBS mouse (Arc P/P) were crossed with the PCP-GFP animals to generate the ArcP/P × PCP-GFP line. All animals were maintained at homozygosity with routine genotyping at Transnetyx. Genotyping primers and the conditions have been described in earlier studies 31,32,68.
Primary Mouse Hippocampal Neurons
Mouse hippocampi were isolated from ArcP/P or ArcP/P × PCP-GFP animals at post-natal day 0 or 1 (P0/P1), both sexes were used and combined in the cultures. The tissue was digested in 0.25% tryspin for 15 min at 37 °C, triturated and plated onto Poly-D-lysine (Sigma) coated glass bottom Mattek dishes at a density of 75,000 cells/dish for live imaging and 60,000 cells/dish for fixed cell imaging. Primary neurons were cultured in Neurobasal A media supplemented with B-27, GlutaMax and primocin (InvivoGen). Viral transduction was usually done at or after DIV 7, and neurons were imaged between DIV 16–19.
METHOD DETAILS
Plasmids and Viruses
All lentiviral constructs were cloned into the phage-ubc-RIG lentiviral vector. For Arc-translation construct, 24X Suntag or GCN4 repeats were PCR-amplified from the SINAPs construct 38. The promoter was the minimal ESARE promoter previously described 36 and synthesized as a gene block along with the 5’UTR sequences. The Arc coding sequence was synthesized based on the sequence (NM_001276684.1), and the 3’UTR sequence was a kind gift from Xiaowei Zhuang 39. The 3’UTR sequence was inserted before the woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) in the lentiviral backbone. Lentivirus expressing Halo-eIF4E 43 as previously characterized was a kind gift from Valentina Gandin, Janelia Research Campus, VA. The Halo-Arc protein reporter was designed based off the Halo-Actin-reporter 10, where the β-actin coding sequence and β-actin 3′UTR were replaced by the Arc CDS and Arc 3′UTR respectively. For PBS coat protein, PCP, we used the synonymously transformed version, stdPCP-stdGFP 31.
The construct for red-shifted nuclear calcium indicators, NLS-jRGECO1a were cloned into the p323 backbone. The sequence for jRGECO1a was obtained from Addgene #61563, and PCR amplified with primers to add NLS to the N-terminus. ChR2-mCherry construct was obtained from Addgene (#20938) and subcloned into p323 backbone. For Arc KD, all-in-one construct expressing sgRNAs and Cas9-P2A-mCherry was obtained from Addgene (#99154) and specific single guide RNAs were cloned to generate pLenti-Arc sgRNA-CRISPRv2-mCherry. The following sgRNA sequences were used: sgRNA-1 (5’- ATGGGCGGCAAATACCCAGT-3’), sgRNA-2 (5’- GTTGACCGAAGTGTCCAAGC −3’). Lentiviral particles were produced by transfecting the expression vector with accessory plasmids, ENV, REV, VSVG and GAG in HEK 293T cells. Collected lentiviral particles were purified with lenti-X concentrator (Clontech, Mountain View, CA). Arc shRNAs (5’-TAACGGTATAGTCATAGCC - 3’) and non-targeting controls (scrambled; Clone ID VSM11618) with Turbo RFP reporter were designed with Horizon Discovery. High-titer lentiviral particles in SMARTvector format were obtained from Horizon Discovery.
Stereotaxic Surgery and Hippocampal slice preparation
ArcP/P × PCP-GFP mice at postnatal day 27 (P27) were anesthetized with oxygen-isoflurane flowing at 1.5 ml/min and positioned into a Kopf stereotaxic instrument. A beveled Hamilton syringe injected 1–1.5 μl of 1:2 mix of AAV5-CaMKII-mcherry-Cre/AAV-DJ-FLEX-ChIEF-tdTomato virus at coordinates targeting the dentate gyrus (−2.1 mm A/P, 1.7 mm M/L, 2.5 mm D/V). Animals were sacrificed 5-weeks post-surgery using anesthesia (4% Isoflurane) followed by decapitation, and slices were prepared for two-photon microscopy. No differentiation of sex was done. Briefly, the animals were perfused with 20 mL of cold N-Methyl-D-glucamine (NMDG) solution containing in (mM): 93 NMDG, 2.5 KCl, 1.25 NaH2PO4, 30 NaHCO3, 20 HEPES, 25 glucose, 5 sodium ascorbate, 2 Thiourea, 3 sodium pyruvate, 10 MgCl2, 0.5 CaCl2, maintained at pH 7.35. The hippocampi were isolated and cut (300 μm thick) using a VT1200s microslicer in cold NMDG solution. Acute hippocampal slices were placed in a chamber with artificial cerebral spinal fluid solution (ACSF) solution composed of 124 NaCl, 2.5 KCl, 26 NaHCO3, 1 NaH2PO4, 2.5 CaCl2, 1.3 MgSO4 and 10 glucose (in mM), and incubated in a 33–34°C water bath. All solutions were equilibrated with 95% O2 and 5% CO2 (pH 7.4). Post-sectioning, acute slices were allowed to recover at room temperature for at least 45 min prior to experiments.
2-photon imaging in slices and optical stimulation
An Ultima 2P laser scanning microscope (Bruker Corp.) equipped with an Insight Deep See laser (Spectra-Physics) tuned to 910–930 nm was used to image Arc transcription in DG with 512 × 512 pixel resolution using 4 mW laser power measured at the 60X objective (Nikon, 1.0 NA). GCs expressing the PCP-GFP coat protein were imaged at 1X magnification to detect at least one transcribing neuron, which was then chosen as the region of interest (ROI) for 2X magnification. A Z-stack of 25 μm thickness with 0.5 μm steps was taken to assess baseline Arc transcription signals before optical stimulation.
Acute slices showing optimal ChIEF-tdTomato reporter expression (at least 75% of DG was fluorescent) were selected for optical stimulation and imaging. The Invitro Ultima 2P microscope (Bruker Corp.) contains a Coherent 473 nm laser path that delivered optical stimulation of 25 pulses at 25 Hz repeated 20 times every 5 s (8 mW, 2–4 ms pulse duration)32. The stimulation area was specifically defined using customized Mark Point software (Bruker Corp.) and was empirically determined based on at least one transcribing neuron in the field of view. After stimulation, Z-stack images of 25 μm thickness with 0.5 μm steps were acquired every 15 min for 4–5 hour.
Depolarization of hippocampal slices and imaging of fixed slices
ArcP/P × PCP-GFP animals injected with AAV5-CaMKII-mcherry, were sacrificed 3-weeks post injection and acute hippocampal slices were prepared. The slices were briefly depolarized with 90 mM KCl for 3 mins and returned to ACSF at room temperature (RT). After 90 mins, slices were grouped into three treatment conditions: first group in ACSF, second group was incubated with CHX (100 mg/ml) for 1 hour, and in the third group, CHX was added for 1 hour followed by washout. The first two groups were fixed at 2.5 hours, and the group with CHX-washout was fixed 45 min later to allow complete washout. Fixation was done with 4% PFA in PBS overnight, and washed thrice with PBS, and then mounted onto slides with Prolong Diamond (Invitrogen). Imaging was performed on a wide-field fluorescence microscope built around an IX-81 stand (Olympus) and illumination was with 488nm laser, captured on EMCCD camera (Andor, iXon3 DU-897E-CS0-#BV). 300nm z-stacks were acquired, and max-projected to obtain the images of GCs.
Stimulation paradigms for primary cultured neurons
The paradigm for TTX-washout was used as described before 31,33. Neurons were treated overnight (14–16 hours) with TTX (2 μM), followed by washes, and fresh Hibernate A media (Brainbits) was added. In another set of experiments, a chemical LTP paradigm was used 69. Briefly, neuronal cultures were incubated with 50 μM APV (Tocris) for 12 hours, followed by induction with 200 μM glycine (Sigma) and 100 μM picrotoxin (Tocris) in Mg2+-free Hibernate A media for 5 min. Cells were then washed twice and returned to Hibernate A media with calcium and magnesium for imaging. Live neuron imaging was performed for six hours for optimum neuronal health and to avoid any phototoxic damage.
Pulse-chase labeling with JF646 and JF549
Primary hippocampal neurons from ArcP/P pups were infected with a cocktail of lentiviruses expressing SARE-HaloTag-ArcCDS-Arc3′UTR reporter and stdPCP-stdGFP at DIV10. After 60 min of stimulation (TTX-w), neurons were labeled with JF646, added at a final concentration of 20nM for 1.5 hr in Hibernate A media. Mattek dishes were transferred to the microscope (descriptions below) and Z-stacks of dendrites were acquired at 150 min post TTX-w. The cells were then washed 3X, and kept in Hibernate A for 20 min, followed by addition of chase dye JF549 dye (1nM). After 10 min, images of the same dendrite were acquired with JF549 in the Hibernate A imaging media. Two-color imaging was performed every 2 min for reporter protein (JF549) and Arc mRNA (PCP-GFP) till 270 min post TTX-w. Images of dendrites were max-projected and straightened for puncta analysis.
Imaging in cultured hippocampal neurons
Hippocampal neurons were imaged in Hibernate A media (Brainbits) at 35°C in a humidified chamber. Time-lapse imaging of transcription and translation was performed on a fluorescence microscope built around an IX-81 stand (Olympus) as described before 31,38. The following lasers were used for illumination: 491 nm laser (Calypso-25, Cobolt, San Jose, CA), 561 nm line (LASOS-561-50, Lasertechnik GmbH, Germany) and a 640 nm line (CUBE 640-40C, Coherent Inc, Santa Clara, CA) were combined, expanded and delivered through the back port. The power for all lasers were controlled by an acousto-optic tunable filter (AOTF) (AOTFnC-400.650-TN, AA Opto-electronic).
The lasers were reflected by a four-band excitation dichroic mirror (Di01-R405/488/561/635, Semrock) to a 150× 1.45 N.A. oil immersion objective (Olympus). For transcription imaging, a 60× 1.40 NA oil immersion objective was used. The fluorescence collected by the same objective, were recorded on an EMCCD camera (Andor, iXon3 DU-897E-CS0-#BV). The emission filters (FF01-525/50 for green and FF01-605/64 (Semrock) for red respectively) were mounted on a motorized filter wheel (FW-1000, Applied Scientific Instrumentation) for fast switching between wavelengths. For stimulation of single ChR2-expressing neurons, a size-adjustable pinhole was used in the excitation light path to restrict the illumination to an area of approximately 10 μm in diameter and prevent cross-activation of neighboring cells. The microscope is equipped with an automated XY-stage (ASI, MS2000-XY) and a piezo-Z stage (ASI) for fast z-stack acquisition. The microscope was controlled and image acquisition was done on the Metamorph platform. The ChR2 stimulation paradigm was automated with custom journals in Metamorph. Briefly, the 10 μm illumination spot was recorded and the ChR2 expressing neuron was moved to that area, and 5 images were taken. Next, 2 trains of stimulation were delivered using 491 nm laser at 20 Hz for 20 times at power density (7 mW/mm2) and switched back to wide-field illumination without the pinhole in the light path. For most experiments, a total of 11 z-stacks with 400 nm distance between stacks were acquired. Arc mRNAs and translation sites were imaged as z-stacks with 300 nm step size. The stacks were z-projected and used for analysis. For nuclear calcium, imaging was performed at single z-plane with 50ms exposure times at 1Hz acquisition rate.
Single molecule fluorescence in situ hybridization (smFISH) and immunofluorescence (IF)
Hippocampal neurons plated at 60,000 cells/Mattek dish were transduced with Suntag-Arc translation reporter for 10 days, then stimulated with TTX-w paradigm at DIV 19 and fixed. The neurons were then permeabilized and smFISH-IF was performed according to the protocol described in 70. Briefly, 100nM probes against 24X Suntag sequence and primary antibodies against GCN4 epitopes (Clone C11L34, Ab00436-1.4, Absolute antibody, Wilton, UK) was used in hybridization buffer for 3 hours at 37°C. After washes with 10% Formamide in 2x SSC buffer, cells were incubated with Alexa Fluor 647 secondary antibody (Invitrogen) and mounted using ProLong diamond antifade reagent with DAPI (Invitrogen). Images were taken in a custom up-right widefield Olympus BX-63 microscope equipped with a Lumencor SOLA-Light engine, ORCA-R2 Digital CCD camera (Hamamatsu), SuperApochromatic 60x/1.35 NA Olympus objective (UPLSAPO60XO) and zero pixel shift filter sets: DAPI-5060C-Zero, Cy3-4040C-Zero and Cy5-4040C-Zero (Semrock). Image pixel size: XY, 107.5 nm; Z-steps, 200 nm. The sequences for FISH probes have been described in 38.
Immunocytochemistry
Hippocampal neurons plated at 60,000 cells/Mattek dish were used as controls or infected with lentiviruses encoding pLenti-Arc sgRNA-CRISPRv2-mCherry and shRNAs against Arc. Neurons were stimulated using TTX-w and then fixed with 4%PFA in PBS along with MgCl2 and CaCl2. Following a permeabilization step with 0.2% Triton-X for 10 min on ice, cells were incubated in blocking buffer (5% goat serum in 1XPBS + 2% BSA + 0.1% Tx-100) for 1 hr at RT. Primary antibodies against Arc (Synaptic Systems, 1:700) was diluted directly in blocking buffer and incubated overnight at 4°C. After 3 washes with 1XPBS, cells were incubated in secondary antibodies (Alexa Fluor 647 or Alexa Fluor 488, Invitrogen) diluted 1:1500 in blocking buffer for 45 min at RT. After 4X washes with 1XPBS, neurons were mounted using ProLong diamond antifade reagent with DAPI (Invitrogen).
Fixed cell Image analysis:
smFISH - IF images were analyzed using FISH Quant 71. Briefly, the FISH spots in the dendrites were filtered and fit to a 3D Gaussian to determine the coordinates of the mRNAs in Cy3 channel. The intensity and the width of the 3D Gaussian were thresholded to exclude non-specific and autofluorescent particles. Similarly, independent analysis of Suntag spots were performed using FISH-Quant. Co-localization analysis was done using a criterion of 300nm distance between mRNA and Suntag signal.
Live-cell imaging data analysis:
Transcription site analysis:
Time-lapse images of transcription were obtained after maximum intensity projection of the z-series. The transcription sites (TS) in time lapse images were tracked and their fluorescence intensities were quantified with custom programs written in Matlab (Mathworks). The intensity of TS was normalized to the diffusive PCP-GFP signal in the nucleus. Intensity traces were subjected to a rolling average of 3 frames to remove fast fluctuations in fluorescence signal. A value ≥ 1.4 was considered ON-state of the gene 31. Values below 1.4 were considered OFF-state. An OFF-period of at least 30 mins between two transcriptional bursts, when the second burst was after 90 min post stimulation was used as criterion to designate reactivation. If any of the alleles was transcriptionally active beyond the IE phase and continued till 100 min post stimulation, then the transcription was considered sustained. Induction of de novo transcription after 100 min of stimulation was marked as a delayed (de novo) event. To determine the transcriptional bursting parameters in each cycle, the images were binned into two time segments: IE (15–75 min) and reactivation phase (105–200 min). Each phase can be composed of multiple transcriptional bursts 72. The duration of the ON-state, and the area under each burst from the different cycles were quantified.
Translation site analysis:
To track translation sites (TLS) in neurons, dendritic segments >30 μm from soma were chosen. The presence of at least one TLS at the start of imaging was used as a criterion for choosing a particular dendrite. Semi-automated tracking of TLS was performed using Trackmate and custom-built program on MATLAB, with a gap of 2 frames allowed to be treated as the same site. TLS which lasted for at least 3 frames were used for analysis. The intensity of TLS was normalized to the diffusive scFv-sfGFP signal in the dendrites, and a threshold of 1.5 was used as a cutoff to be considered a TLS. Gradual increase in TLS intensity indicates more nascent peptide synthesis and therefore deemed to be translating mRNA. The OFF-periods between active translation bursts across multiple TLS were plotted as an inverse cumulative distribution function (1-CDF) and fitted to a 2-component exponential fit. The parameters were determined by a maximal likelihood analysis (goodness of fit test performed in MATLAB).
Single mRNA analysis:
Single particle tracking of mRNAs was performed using Trackmate plugin on Fiji. The dendrites were straightened before analysis. Tracks shorter than 3 frames were not considered. mRNA counts were normalized to the length of the dendrites to determine RNA density. The track lengths of both stationary and moving mRNAs were used to calculate the residence times of mRNAs in dendrites. For Figure 5, kymographs were plotted for straightened dendrites. The number of mRNAs which last for ≥2 frames (3 min) were counted for analysis in Figure 5 K,L.
Pulse-chase assay:
The puncta from JF646 and JF549 channels were localized with Analyze particles plugin on Image J. The puncta were diffraction limited spots, which could be fitted to a 2D-Gaussian. JF646 puncta whose brightness were above 20% above background fluorescence (diffusive signal in dendrites) was used for analysis. The distances to JF549 signal were calculated from the brightest JF646 puncta using the nearest neighbor analysis and plotted as a frequency distribution. 75 percentile was used as the cutoff to designate protein enrichment in space. Accordingly, we used a segment of 6 μm length (3 μm on either side of the centroid of the brightest 646 puncta) to define the Arc protein hub from IE-phase.
Puncta and cluster analysis:
Images depicting Halo-eIF4E puncta were set for intensity threshold and determined using Analyze particles plugin on Fiji. In the same dendrite, a 6 μm segment corresponding to translation hotspot (3 or more TLS) and a neighboring non-hotspot region were designated and particle tracking were performed using the same parameters for both the ROIs. The total area of the Halo-eIF4E puncta was used as the cluster size.
Spine density analysis was performed using the max-projected images and using the Plugin Dendritic spine counter on Fiji.
Nuclear Ca2+ imaging analysis:
Time series images were acquired at 1Hz frequency on single focal planes. The ROIs for the nuclei were detected in a semi-automated manner in Fiji. The same ROIs with required correction for x-y drift were used to quantify fluorescence at different time points. The average value from the first 3 frames was treated as baseline fluorescence (F), and the following change in fluorescence (ΔF) was measured. The values in the traces are represented as ΔF/F. Traces were fitted to a peak-fitting algorithm and the maximum likelihood analysis was performed for peak assignment. The frequency and amplitude of the peaks were calculated.
Analysis of transcription imaging in slices
Images of GC nuclei were filtered, and then semi-automated detection of transcription sites was performed using ROIs of 28–32 pixels for each transcribing allele. The intensity of each transcription site was normalized to the background signal of the nucleus with an ROI of same dimension. The normalized intensity value of 1 represents that no transcription sites are detected. An intensity threshold of 10% change (i.e. values 1.1 or higher) was used to designate a transcription site. The same transcription site was followed over time to measure the change in normalized intensity values, and below 1.1 was considered as transcriptional shutdown. Quantification of total transcribing cells were performed as follows:
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are presented as mean ± s.e.m. Normality test for all conditions was performed using the Shapiro-Wilk test. Wilcoxon Signed Ranks test was used for not normally distributed data. One-way ANOVA (Dunett’s multiple comparison) was used to determine statistical significance for more comparison between more than two groups. Student’s t-test determined statistical significance for all other experimental conditions. Paired t-test was performed when comparing the same neuron or the same allele from the IE and reactivation phases. Statistical parameters reported in figure legends, p < 0.05 was considered significant. All statistical tests were performed on Graph Pad Prism.
Supplementary Material
Video S1: Long term imaging of Arc transcription shows transcriptional reactivation after the immediate early phase, Related to Figure 1. Time lapse images acquired after a brief stimulation (TTX-w) of neurons from ArcP/P animals expressing PCP-GFP. Time in min.
Video S2: Cycling of transcription at both Arc alleles, Related to Figure 1. Time lapse imaging shows transcriptional induction and reactivation from both Arc alleles (yellow and cyan arrows) after a brief stimulation (TTX-w) of neurons from ArcP/P animals. Time in min.
Video S3: Effect of TTX on Arc transcriptional reactivation, Related to Figure 3. Hippocampal neurons from ArcP/P coexpressing PCP-GFP and NLS-jRGECO1a show reactivation of Arc transcription even though Ca2+ transients were negligible (in Figure 3B). Time in min.
Video S4: Effect of translation inhibition on Arc transcriptional reactivation, Related to Figure 4. Hippocampal neurons from ArcP/P imaged over time after TTX-w, followed by CHX addition at 90 min, and then washout of CHX at 160min. Time in min.
Video S5: Arc translation imaging in dendrites shows hotspots and biphasic dynamics, Related to Figure 6. Hippocampal neurons infected with Arc-Suntag reporter and scFV-sfGFP were imaged 90 min post TTX-w. Arc TLS (green circles) shows TLS clustering forming a hotspot (dark green box, represented in Figure 6G). Time in min. Scale bar 5μm.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Primary Antibody anti-GCN4 | Absolute Antibody | Cat# Ab00436–1.4; Clone ID C11L34 |
| Primary Antibody anti-Arc | Synaptic Systems | Cat# 156111; RRID:AB_2631221 |
| Secondary Antibody Goat Anti-Rabbit Alexa FluorTM 647 | Invitrogen | Cat#A21244; RRID: AB_2535812 |
| Secondary Antibody Goat Anti-Mouse Alexa FluorTM 647 | Invitrogen | Cat#A32728; RRID: AB_2633277 |
| Chemicals, peptides, and recombinant proteins | ||
| D-APV | Tocris Bioscience | Cat# 0106/1 |
| TTX | Tocris Bioscience | Cat# 1069/1 |
| Picrotoxin | Tocris Bioscience | Cat# 1128 |
| Anisomycin | Tocris Bioscience | Cat# 1290 |
| Cycloheximide (CHX) | Tocris Bioscience | Cat# 0970 |
| Glycine hydrochloride | Sigma | G2879 |
| Janelia Flour 549-Halotag ligand | Janelia Research Campus, Ashburn, VA, USA | N/A |
| Janelia Flour 646-Halotag ligand | Janelia Research Campus, Ashburn, VA, USA | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: ArcP/P | Dr. Robert H. Singer, Albert Einstein College of Medicine | N/A |
| Mouse: ArcP/P × PCP-GFP | Dr. Robert H. Singer, Albert Einstein College of Medicine | N/A |
| Mouse: ActinM/M | Dr. Robert H. Singer, Albert Einstein College of Medicine | N/A |
| Oligonucleotides | ||
| Arc sgRNA1: ATGGGCGGCAAATACCCAGT | This paper | N/A |
| Arc sgRNA2: GTTGACCGAAGTGTCCAAGC | This paper | N/A |
| Arc shRNA 1: TAACGGTATAGTCATAGCC | This paper, custom made from Horizon Discovery | N/A |
| Scrambled shRNA | Horizon Discovery | Clone ID VSM11618 |
| Recombinant DNA | ||
| pUbc-stdPCP-stdGFP | Singer lab | N/A |
| AAV1/9-hSyn-Cre | Janelia Research Campus, Ashburn, VA, USA | N/A |
| phage-SARE-HaloTag-ArcCDS-Arc3′UTR | This paper | N/A |
| phage-SARE-24xGNC4-ArcCDS-Arc3′UTR-MS2V7 | This paper | N/A |
| pHR-scFv-GCN4-sfGFP-GB1-dWPRE | Dr. Ron Vale, University of California San Francisco | Addgene Plasmid #60907 |
| pLV-Halotag-eIF4E | Dr. Valentina Gandin, Janelia Research Campus, VA, previously in 43. | N/A |
| p323-Ubc-NLS-jRGECO1a | This paper | N/A |
| pLenti-Arc sgRNA-CRISPRv2-mCherry | This paper | N/A |
| ChR2-mCherry | Karl Deisseroth, Stanford University | Addgene Plasmid #61563 |
| AAV5-CamKII-mCherry-Cre | UNC Vector Core | N/A |
| AAV-DJ-FLEX-ChIEF-tdTomato | Dr. Pascal Kaeser, Harvard University, custom order UNC Vector core | N/A |
| Software and algorithms | ||
| ImageJ FIJI | ImageJ | http://imagej.net/Welcome |
| Matlab | Mathworks | https://www.mathworks.com/products/matlab.html |
| FISH-QUANT | Github, published in 71 | https://fish-quant.github.io/ |
| TrackMate | Image J | https://imagej.net/plugins/trackmate/ |
| Graphpad Prism 8 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
Highlights:
Reactivation of transcription drives cycling of the Arc gene in individual neurons
Feedback from new proteins reinduce Arc transcription in the next cycle
Arc mRNAs from later cycles localize to sites marked with previous Arc protein
Repetitive translation in hotspots consolidates dendritic Arc in selective hubs
Acknowledgments:
We thank Chiso Nwokafor for animal maintenance, Xiuhua Meng for help with cloning, and Melissa Lopez-Jones for technical assistance. We thank all members of Singer and Castillo laboratories for helpful discussions, and Dong-woo Hwang and Weihan Li for critical reading of the manuscript. We are grateful to Valentina Gandin from Janelia Research Campus (JRC) for gifting the Halo-eIF4E lentivirus, and to Luke Lavis from JRC for providing the JF dyes. We thank the members of the Program in RNA biology, particularly U. Thomas Meier, and Robert Coleman for thoughtful suggestions on the project.
The work is supported by National Institutes of Health grant R21 MH122961 and a pilot grant from Rose F. Kennedy IDDRC Center to S.D.; R01 NS083085 to R.H.S. and S.D.; R01 MH125772, MH116673, NS115543, NS113600 (PEC); and Ruth L. Kirschstein NRSA Fellowship MH109267 to P.J.L.
Footnotes
Declaration of interests: The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Asok A, Leroy F, Rayman JB, and Kandel ER (2019). Molecular Mechanisms of the Memory Trace. Trends in neurosciences 42, 14–22. 10.1016/j.tins.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey CH, Kandel ER, and Harris KM (2015). Structural Components of Synaptic Plasticity and Memory Consolidation. Cold Spring Harbor perspectives in biology 7, a021758. 10.1101/cshperspect.a021758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alberini CM, and Kandel ER (2014). The regulation of transcription in memory consolidation. Cold Spring Harbor perspectives in biology 7, a021741. 10.1101/cshperspect.a021741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang DO, Kim SM, Zhao Y, Hwang H, Miura SK, Sossin WS, and Martin KC (2009). Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science 324, 1536–1540. 10.1126/science.1173205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richter JD, and Klann E (2009). Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes Dev 23, 1–11. 10.1101/gad.1735809. [DOI] [PubMed] [Google Scholar]
- 6.Holt CE, Martin KC, and Schuman EM (2019). Local translation in neurons: visualization and function. Nat Struct Mol Biol 26, 557–566. 10.1038/s41594-019-0263-5. [DOI] [PubMed] [Google Scholar]
- 7.Holt CE, and Schuman EM (2013). The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron 80, 648–657. 10.1016/j.neuron.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelleher RJ 3rd, Govindarajan A, and Tonegawa S (2004). Translational regulatory mechanisms in persistent forms of synaptic plasticity. Neuron 44, 59–73. 10.1016/j.neuron.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 9.Shrestha P, and Klann E (2022). Spatiotemporally resolved protein synthesis as a molecular framework for memory consolidation. Trends in neurosciences 45, 297–311. 10.1016/j.tins.2022.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoon YJ, Wu B, Buxbaum AR, Das S, Tsai A, English BP, Grimm JB, Lavis LD, and Singer RH (2016). Glutamate-induced RNA localization and translation in neurons. Proceedings of the National Academy of Sciences of the United States of America 113, E6877–E6886. 10.1073/pnas.1614267113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buxbaum AR, Wu B, and Singer RH (2014). Single beta-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science 343, 419–422. 10.1126/science.1242939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tischmeyer W, and Grimm R (1999). Activation of immediate early genes and memory formation. Cellular and molecular life sciences : CMLS 55, 564–574. 10.1007/s000180050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, Mao X, Engelsberg A, Mahlke C, Welzl H, et al. (2006). Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron 52, 437–444. 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 14.Yap EL, and Greenberg ME (2018). Activity-Regulated Transcription: Bridging the Gap between Neural Activity and Behavior. Neuron 100, 330–348. 10.1016/j.neuron.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tyssowski KM, DeStefino NR, Cho JH, Dunn CJ, Poston RG, Carty CE, Jones RD, Chang SM, Romeo P, Wurzelmann MK, et al. (2018). Different Neuronal Activity Patterns Induce Different Gene Expression Programs. Neuron 98, 530–546 e511. 10.1016/j.neuron.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao VR, Pintchovski SA, Chin J, Peebles CL, Mitra S, and Finkbeiner S (2006). AMPA receptors regulate transcription of the plasticity-related immediate-early gene Arc. Nature neuroscience 9, 887–895. 10.1038/nn1708. [DOI] [PubMed] [Google Scholar]
- 17.Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, and Barnes CA (2000). Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. The Journal of neuroscience : the official journal of the Society for Neuroscience 20, 3993–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, and Worley PF (1995). Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron 14, 433–445. 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- 19.Wu J, Petralia RS, Kurushima H, Patel H, Jung MY, Volk L, Chowdhury S, Shepherd JD, Dehoff M, Li Y, et al. (2011). Arc/Arg3.1 regulates an endosomal pathway essential for activity-dependent beta-amyloid generation. Cell 147, 615–628. 10.1016/j.cell.2011.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, Flavell SW, Kim TK, Griffith EC, Waldon Z, Maehr R, et al. (2010). The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell 140, 704–716. 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park S, Park JM, Kim S, Kim JA, Shepherd JD, Smith-Hicks CL, Chowdhury S, Kaufmann W, Kuhl D, Ryazanov AG, et al. (2008). Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron 59, 70–83. 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, Huganir RL, and Worley PF (2006). Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron 52, 475–484. 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, and Worley PF (2006). Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron 52, 445–459. 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Messaoudi E, Kanhema T, Soule J, Tiron A, Dagyte G, da Silva B, and Bramham CR (2007). Sustained Arc/Arg3.1 synthesis controls long-term potentiation consolidation through regulation of local actin polymerization in the dentate gyrus in vivo. The Journal of neuroscience : the official journal of the Society for Neuroscience 27, 10445–10455. 10.1523/JNEUROSCI.2883-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shepherd JD, and Bear MF (2011). New views of Arc, a master regulator of synaptic plasticity. Nature neuroscience 14, 279–284. 10.1038/nn.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wall MJ, Collins DR, Chery SL, Allen ZD, Pastuzyn ED, George AJ, Nikolova VD, Moy SS, Philpot BD, Shepherd JD, et al. (2018). The Temporal Dynamics of Arc Expression Regulate Cognitive Flexibility. Neuron 98, 1124–1132 e1127. 10.1016/j.neuron.2018.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramirez-Amaya V, Vazdarjanova A, Mikhael D, Rosi S, Worley PF, and Barnes CA (2005). Spatial exploration-induced Arc mRNA and protein expression: evidence for selective, network-specific reactivation. The Journal of neuroscience : the official journal of the Society for Neuroscience 25, 1761–1768. 10.1523/JNEUROSCI.4342-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama D, Iwata H, Teshirogi C, Ikegaya Y, Matsuki N, and Nomura H (2015). Long-delayed expression of the immediate early gene Arc/Arg3.1 refines neuronal circuits to perpetuate fear memory. The Journal of neuroscience : the official journal of the Society for Neuroscience 35, 819–830. 10.1523/JNEUROSCI.2525-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, and Moore MJ (2007). The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell 130, 179–191. 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 30.Soule J, Alme M, Myrum C, Schubert M, Kanhema T, and Bramham CR (2012). Balancing Arc synthesis, mRNA decay, and proteasomal degradation: maximal protein expression triggered by rapid eye movement sleep-like bursts of muscarinic cholinergic receptor stimulation. The Journal of biological chemistry 287, 22354–22366. 10.1074/jbc.M112.376491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das S, Moon HC, Singer RH, and Park HY (2018). A transgenic mouse for imaging activity-dependent dynamics of endogenous Arc mRNA in live neurons. Science advances 4, eaar3448. 10.1126/sciadv.aar3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lituma PJ, Singer RH, Das S, and Castillo PE (2022). Real-time imaging of Arc/Arg3.1 transcription ex vivo reveals input-specific immediate early gene dynamics. Proceedings of the National Academy of Sciences of the United States of America 119, e2123373119. 10.1073/pnas.2123373119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saha RN, Wissink EM, Bailey ER, Zhao M, Fargo DC, Hwang JY, Daigle KR, Fenn JD, Adelman K, and Dudek SM (2011). Rapid activity-induced transcription of Arc and other IEGs relies on poised RNA polymerase II. Nature neuroscience 14, 848–856. 10.1038/nn.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steward O, Wallace CS, Lyford GL, and Worley PF (1998). Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron 21, 741–751. [DOI] [PubMed] [Google Scholar]
- 35.Greenberg ME, and Ziff EB (1984). Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature 311, 433–438. 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 36.Kawashima T, Okuno H, Nonaka M, Adachi-Morishima A, Kyo N, Okamura M, Takemoto-Kimura S, Worley PF, and Bito H (2009). Synaptic activity-responsive element in the Arc/Arg3.1 promoter essential for synapse-to-nucleus signaling in activated neurons. Proceedings of the National Academy of Sciences of the United States of America 106, 316–321. 10.1073/pnas.0806518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grimm JB, Muthusamy AK, Liang Y, Brown TA, Lemon WC, Patel R, Lu R, Macklin JJ, Keller PJ, Ji N, and Lavis LD (2017). A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nature methods 14, 987–994. 10.1038/nmeth.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu B, Eliscovich C, Yoon YJ, and Singer RH (2016). Translation dynamics of single mRNAs in live cells and neurons. Science 352, 1430–1435. 10.1126/science.aaf1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C, Han B, Zhou R, and Zhuang X (2016). Real-Time Imaging of Translation on Single mRNA Transcripts in Live Cells. Cell 165, 990–1001. 10.1016/j.cell.2016.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, and Vale RD (2014). A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646. 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Job C, and Eberwine J (2001). Identification of sites for exponential translation in living dendrites. Proceedings of the National Academy of Sciences of the United States of America 98, 13037–13042. 10.1073/pnas.231485698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Costa-Mattioli M, Sossin WS, Klann E, and Sonenberg N (2009). Translational control of long-lasting synaptic plasticity and memory. Neuron 61, 10–26. 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gandin V, English BP, Freeman M, Leroux LP, Preibisch S, Walpita D, Jaramillo M, and Singer RH (2022). Cap-dependent translation initiation monitored in living cells. Nature communications 13, 6558. 10.1038/s41467-022-34052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gindina S, Botsford B, Cowansage K, LeDoux J, Klann E, Hoeffer C, and Ostroff L (2021). Upregulation of eIF4E, but not other translation initiation factors, in dendritic spines during memory formation. J Comp Neurol 529, 3112–3126. 10.1002/cne.25158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sutton MA, and Schuman EM (2006). Dendritic protein synthesis, synaptic plasticity, and memory. Cell 127, 49–58. 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 46.Das S, Vera M, Gandin V, Singer RH, and Tutucci E (2021). Intracellular mRNA transport and localized translation. Nat Rev Mol Cell Biol 22, 483–504. 10.1038/s41580-021-00356-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swanger SA, and Bassell GJ (2011). Making and breaking synapses through local mRNA regulation. Curr Opin Genet Dev 21, 414–421. 10.1016/j.gde.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kandel ER, Dudai Y, and Mayford MR (2014). The molecular and systems biology of memory. Cell 157, 163–186. 10.1016/j.cell.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 49.Bekinschtein P, Cammarota M, Igaz LM, Bevilaqua LR, Izquierdo I, and Medina JH (2007). Persistence of long-term memory storage requires a late protein synthesis- and BDNF- dependent phase in the hippocampus. Neuron 53, 261–277. 10.1016/j.neuron.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 50.Steward O, Farris S, Pirbhoy PS, Darnell J, and Driesche SJ (2014). Localization and local translation of Arc/Arg3.1 mRNA at synapses: some observations and paradoxes. Front Mol Neurosci 7, 101. 10.3389/fnmol.2014.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Na Y, Park S, Lee C, Kim DK, Park JM, Sockanathan S, Huganir RL, and Worley PF (2016). Real-Time Imaging Reveals Properties of Glutamate-Induced Arc/Arg 3.1 Translation in Neuronal Dendrites. Neuron 91, 561–573. 10.1016/j.neuron.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Farris S, Lewandowski G, Cox CD, and Steward O (2014). Selective localization of arc mRNA in dendrites involves activity- and translation-dependent mRNA degradation. The Journal of neuroscience : the official journal of the Society for Neuroscience 34, 4481–4493. 10.1523/JNEUROSCI.4944-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramirez-Amaya V, Angulo-Perkins A, Chawla MK, Barnes CA, and Rosi S (2013). Sustained transcription of the immediate early gene Arc in the dentate gyrus after spatial exploration. The Journal of neuroscience : the official journal of the Society for Neuroscience 33, 1631–1639. 10.1523/JNEUROSCI.2916-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Panja D, Kenney JW, D’Andrea L, Zalfa F, Vedeler A, Wibrand K, Fukunaga R, Bagni C, Proud CG, and Bramham CR (2014). Two-stage translational control of dentate gyrus LTP consolidation is mediated by sustained BDNF-TrkB signaling to MNK. Cell reports 9, 1430–1445. 10.1016/j.celrep.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 55.Rosenblum K, Futter M, Voss K, Erent M, Skehel PA, French P, Obosi L, Jones MW, and Bliss TV (2002). The role of extracellular regulated kinases I/II in late-phase long-term potentiation. The Journal of neuroscience : the official journal of the Society for Neuroscience 22, 5432–5441. 10.1523/JNEUROSCI.22-13-05432.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shobe J, Philips GT, and Carew TJ (2016). Transforming growth factor beta recruits persistent MAPK signaling to regulate long-term memory consolidation in Aplysia californica. Learning & memory 23, 182–188. 10.1101/lm.040915.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bambah-Mukku D, Travaglia A, Chen DY, Pollonini G, and Alberini CM (2014). A positive autoregulatory BDNF feedback loop via C/EBPbeta mediates hippocampal memory consolidation. The Journal of neuroscience : the official journal of the Society for Neuroscience 34, 12547–12559. 10.1523/JNEUROSCI.0324-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Campbell RR, and Wood MA (2019). How the epigenome integrates information and reshapes the synapse. Nature reviews. Neuroscience 20, 133–147. 10.1038/s41583-019-0121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li L, Carter J, Gao X, Whitehead J, and Tourtellotte WG (2005). The neuroplasticity-associated arc gene is a direct transcriptional target of early growth response (Egr) transcription factors. Molecular and cellular biology 25, 10286–10300. 10.1128/MCB.25.23.10286-10300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Penke Z, Chagneau C, and Laroche S (2011). Contribution of Egr1/zif268 to Activity-Dependent Arc/Arg3.1 Transcription in the Dentate Gyrus and Area CA1 of the Hippocampus. Frontiers in behavioral neuroscience 5, 48. 10.3389/fnbeh.2011.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun X, and Lin Y (2016). Npas4: Linking Neuronal Activity to Memory. Trends in neurosciences 39, 264–275. 10.1016/j.tins.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun C, Nold A, Fusco CM, Rangaraju V, Tchumatchenko T, Heilemann M, and Schuman EM (2021). The prevalence and specificity of local protein synthesis during neuronal synaptic plasticity. Science advances 7, eabj0790. 10.1126/sciadv.abj0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chouaib R, Safieddine A, Pichon X, Imbert A, Kwon OS, Samacoits A, Traboulsi AM, Robert MC, Tsanov N, Coleno E, et al. (2020). A Dual Protein-mRNA Localization Screen Reveals Compartmentalized Translation and Widespread Co-translational RNA Targeting. Dev Cell 54, 773–791 e775. 10.1016/j.devcel.2020.07.010. [DOI] [PubMed] [Google Scholar]
- 64.Job C, and Eberwine J (2001). Localization and translation of mRNA in dendrites and axons. Nature reviews. Neuroscience 2, 889–898. 10.1038/35104069. [DOI] [PubMed] [Google Scholar]
- 65.Govindarajan A, Kelleher RJ, and Tonegawa S (2006). A clustered plasticity model of long-term memory engrams. Nature reviews. Neuroscience 7, 575–583. 10.1038/nrn1937. [DOI] [PubMed] [Google Scholar]
- 66.Kastellakis G, Cai DJ, Mednick SC, Silva AJ, and Poirazi P (2015). Synaptic clustering within dendrites: an emerging theory of memory formation. Prog Neurobiol 126, 19–35. 10.1016/j.pneurobio.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen MB, Jiang X, Quake SR, and Sudhof TC (2020). Persistent transcriptional programmes are associated with remote memory. Nature 587, 437–442. 10.1038/s41586-020-2905-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, and Singer RH (2011). A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nature methods 8, 165–170. 10.1038/nmeth.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Donlin-Asp PG, Polisseni C, Klimek R, Heckel A, and Schuman EM (2021). Differential regulation of local mRNA dynamics and translation following long-term potentiation and depression. Proceedings of the National Academy of Sciences of the United States of America 118. 10.1073/pnas.2017578118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eliscovich C, Shenoy SM, and Singer RH (2017). Imaging mRNA and protein interactions within neurons. Proceedings of the National Academy of Sciences of the United States of America 114, E1875–E1884. 10.1073/pnas.1621440114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mueller F, Senecal A, Tantale K, Marie-Nelly H, Ly N, Collin O, Basyuk E, Bertrand E, Darzacq X, and Zimmer C (2013). FISH-quant: automatic counting of transcripts in 3D FISH images. Nature methods 10, 277–278. 10.1038/nmeth.2406. [DOI] [PubMed] [Google Scholar]
- 72.Larsson AJM, Johnsson P, Hagemann-Jensen M, Hartmanis L, Faridani OR, Reinius B, Segerstolpe Å, Rivera CM, Ren B, and Sandberg R (2019). Genomic encoding of transcriptional burst kinetics. Nature. 10.1038/s41586-018-0836-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1: Long term imaging of Arc transcription shows transcriptional reactivation after the immediate early phase, Related to Figure 1. Time lapse images acquired after a brief stimulation (TTX-w) of neurons from ArcP/P animals expressing PCP-GFP. Time in min.
Video S2: Cycling of transcription at both Arc alleles, Related to Figure 1. Time lapse imaging shows transcriptional induction and reactivation from both Arc alleles (yellow and cyan arrows) after a brief stimulation (TTX-w) of neurons from ArcP/P animals. Time in min.
Video S3: Effect of TTX on Arc transcriptional reactivation, Related to Figure 3. Hippocampal neurons from ArcP/P coexpressing PCP-GFP and NLS-jRGECO1a show reactivation of Arc transcription even though Ca2+ transients were negligible (in Figure 3B). Time in min.
Video S4: Effect of translation inhibition on Arc transcriptional reactivation, Related to Figure 4. Hippocampal neurons from ArcP/P imaged over time after TTX-w, followed by CHX addition at 90 min, and then washout of CHX at 160min. Time in min.
Video S5: Arc translation imaging in dendrites shows hotspots and biphasic dynamics, Related to Figure 6. Hippocampal neurons infected with Arc-Suntag reporter and scFV-sfGFP were imaged 90 min post TTX-w. Arc TLS (green circles) shows TLS clustering forming a hotspot (dark green box, represented in Figure 6G). Time in min. Scale bar 5μm.
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.


