Abstract
The actin cytoskeleton impacts practically every function of a eukaryotic cell. Historically, the best-characterized cytoskeletal activities are in cell morphogenesis, motility, and division. The structural and dynamic properties of the actin cytoskeleton are also crucial for establishing, maintaining, and changing the organization of membrane-bound organelles and other intracellular structures. Such activities are important in nearly all animal cells and tissues, although distinct anatomical regions and physiological systems rely on different regulatory factors. Recent work indicates that the Arp2/3 complex, a broadly expressed actin nucleator, drives actin assembly during several intracellular stress response pathways. These newly described Arp2/3-mediated cytoskeletal rearrangements are coordinated by members of the Wiskott-Aldrich Syndrome Protein (WASP) family of actin nucleation-promoting factors. Thus, the Arp2/3 complex and WASP-family proteins are emerging as crucial players in cytoplasmic and nuclear activities including autophagy, apoptosis, chromatin dynamics, and DNA repair. Characterizations of the functions of the actin assembly machinery in such stress response mechanisms are advancing our understanding of both normal and pathogenic processes, and hold great promise for providing insights into organismal development and interventions for disease.
Keywords: Actin, Arp2/3 complex, Apoptosome, Apoptosis, Autophagy, Caspase, Cytoskeleton, DNA repair, JMY, Macropinocytosis, Mitochondria, N-WASP, Proteostasis, Transcription, WASH complex, WASP, WAVE complex, WHAMM, WHIMP, Wiskott-Aldrich Syndrome
1. Introduction
The basic mechanics underlying actin-associated cellular behaviors are simple: globular (G-) actin monomers assemble into filamentous (F-) actin polymers that can disassemble back into monomers. Actin polymerization generates protrusive forces, while the filaments themselves provide physical structure, serve as scaffolds, and act as tracks for motor proteins. To ensure that actin assembles when and where it is needed, sophisticated regulatory mechanisms control the formation of dimeric, trimeric, or tetrameric actin ‘nuclei’ that elongate into filaments (Pollard, 2016). Efficient initiation of polymer assembly de novo occurs through the actions of proteins called nucleators. Mammals encode 3 main classes of actin nucleators. One class consists of ≥ 8 monomer-aligning proteins that instigate linear actin assembly (Dominguez, 2016; Shekhar et al., 2016). The second is composed of ~15 Formin-family proteins that stabilize actin dimers and elongate linear filaments (Breitsprecher and Goode, 2013; Courtemanche, 2018). The third comprises a 7-subunit macromolecular nucleator called the Arp2/3 complex and its ~12 activators, termed the nucleation-promoting factors (Alekhina et al., 2017; Campellone and Welch, 2010). The Arp2/3 complex cooperates with these factors at the sides of existing actin filaments to nucleate new branched networks that direct diverse cellular activities.
Most mammalian Arp2/3 activators are members of the Wiskott-Aldrich Syndrome Protein (WASP) family: WASP, N-WASP, WAVE1, WAVE2, WAVE3, WASH, WHAMM, JMY, and WHIMP (Campellone et al., 2008b; Derry et al., 1994; Kabrawala et al., 2020; Linardopoulou et al., 2007; Machesky and Insall, 1998; Miki et al., 1998b, 1996; Shikama et al., 1999; Suetsugu et al., 1999; Symons et al., 1996; Zuchero et al., 2009). Other Arp2/3-binding factors include Cortactin and HS1, which stabilize F-actin branchpoints (Helgeson and Nolen, 2013; Schnoor et al., 2018), and WISH/DIP1/SPIN90, which cooperates with Arp2/3 to assemble linear instead of branched filaments (Cao et al., 2020; Wagner et al., 2013). The structural and biochemical mechanisms of Arp2/3 activation by nucleation-promoting factors have been reviewed elsewhere (Gautreau et al., 2022; Kadzik et al., 2020; Lappalainen et al., 2022) and recently described at higher resolution (Shaaban et al., 2020; Zimmet et al., 2020; et al., 2022; Ding et al., 2022). The current review provides a more cell biological perspective on the functions of the Arp2/3 complex and its WASP-family activators. We first summarize the plasma membrane remodeling and endocytic trafficking activities in which the Arp2/3 complex and WASP-family members are well characterized before focusing on newly emerging functions for these nucleation factors in homeostatic and stress response pathways including autophagy, apoptosis, chromatin dynamics, and DNA repair.
2. Cell morphogenesis and movement
Cell motility and directional migration are integral to organismal development, responses to extracellular stimuli, and execution of immune system functions (Pollard, 2018; SenGupta et al., 2021; Swaney and Li, 2016). Morphological changes that allow for the cell to effectively control its movement rely on the coordinated assembly and disassembly of actin filaments beneath the plasma membrane at sites of protrusion and adhesion (Schaks et al., 2019; Svitkina, 2018). This polymerization is dictated by the Arp2/3 complex, originally purified from Acanthamoeba castellanii and shown to localize to the cell cortex (Machesky et al., 1994). The complex is composed of 7 subunits: Arp2, Arp3, ArpC1, ArpC2, ArpC3, ArpC4, and ArpC5, and is highly conserved across eukaryotes (Machesky et al., 1997; Welch et al., 1997). The ability of the Arp2/3 complex to nucleate actin was realized upon the discovery of proteins that activate the complex – the nucleation-promoting factors. Notably, the first Arp2/3 activator to be identified was not of human origin but was bacterial. ActA, uncovered due to its requirement in actin ‘comet tail’ assembly by pathogenic Listeria monocytogenes (Kocks et al., 1992), was shown to promote Arp2/3 complex-mediated actin nucleation in vitro (Welch et al., 1998).
The number of known Arp2/3-activating factors, both bacterial and endogenous mammalian, has since greatly increased (Campellone and Welch, 2010; Rotty et al., 2013). The founding member of the human nucleation-promoting factor family, WASP, was originally identified as the protein product of the WAS gene, which is mutated in patients with the immune disorder Wiskott-Aldrich Syndrome (Derry et al., 1994). Soon thereafter, WASP was observed to regulate the actin cytoskeleton (Symons et al., 1996). The crucial link to Arp2/3 was then made when WASP and a homologous protein, WAVE1 (related to the Dictyostelium protein SCAR (Bear et al., 1998)), were shown to interact with the Arp2/3 complex (Machesky and Insall, 1998). The key shared feature of WASP, WAVE1, and all subsequently-identified mammalian WASP-family members is their possession of a conserved C-terminal ‘WCA’ domain, consisting of one or more WASP-homology 2 (WH2) motifs that bind actin monomers, and connector (C) and acidic (A) sequences responsible for engaging the Arp2/3 complex during branched actin nucleation (Machesky and Insall, 1998; Mullins et al., 1998).
The next major advances in our understanding of the Arp2/3 complex and its WASP-family activators came when investigators began to explore the functions of these actin assembly factors at the plasma membrane. The greatest depth of work characterizing the cellular functions of the Arp2/3 complex and WASP-family proteins has been related to their roles in protrusive structures (Fig. 1A). Similar to how the polymerization of actin filaments into comet tails can physically propel bacterial pathogens through the cytosol, the force of actin assembly also drives the plasma membrane forward during protrusion and cell motility.
Fig. 1.
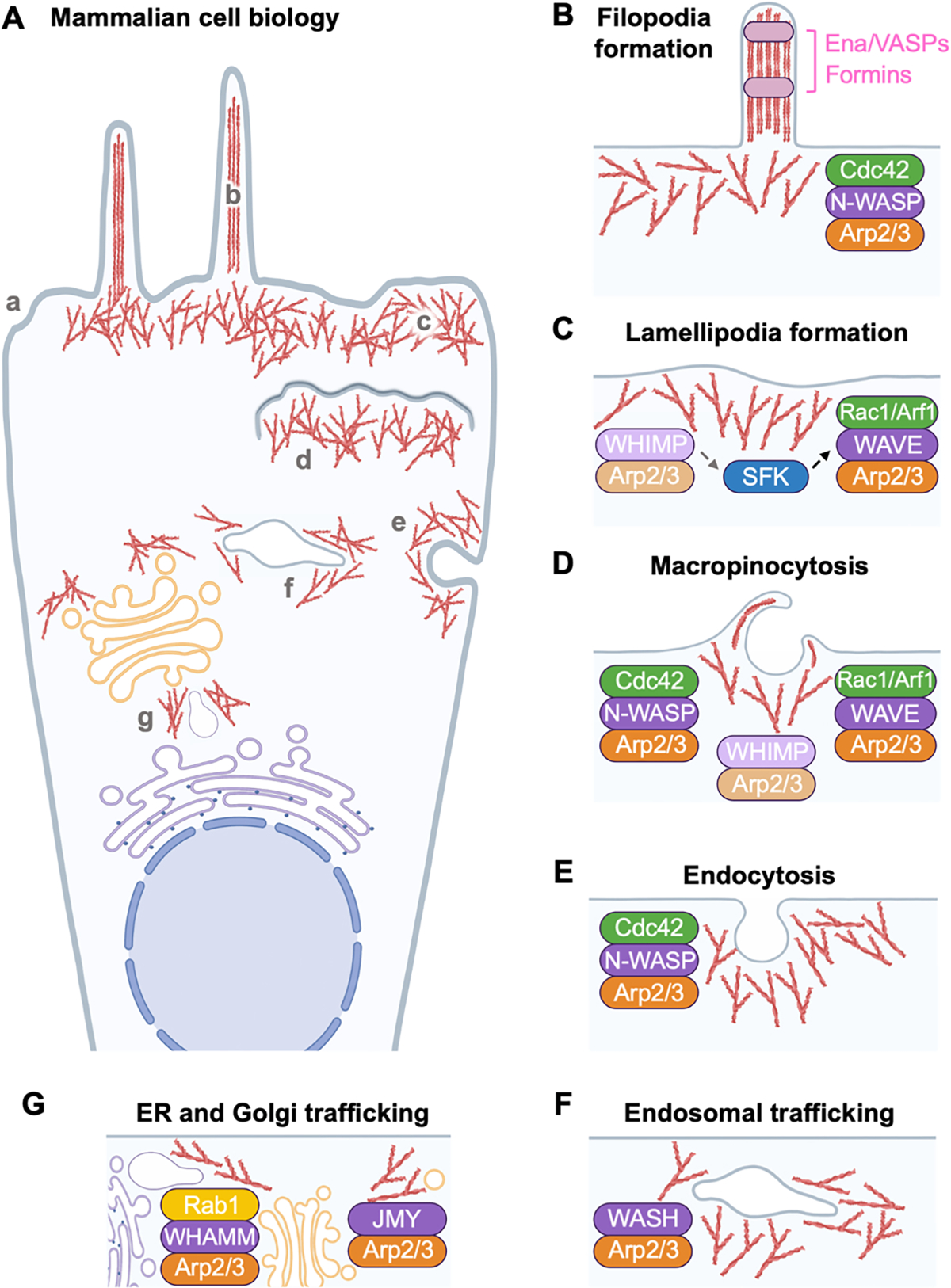
Cellular functions for the Arp2/3 complex and WASP-family actin nucleation factors. (A) Many aspects of mammalian cell biology are impacted by the actin cytoskeleton. F-actin structures are assembled in (a) a generic mammalian cell at (b) filopodia, (c) lamellipodia, (d) sites of membrane ruffling or macropinocytosis, (e) sites of endocytosis, (f) tubular endosomes, and (g) the ER-Golgi intermediate compartment (ERGIC) and Golgi, among other locations. (B) Filopodia may form following activation of a Cdc42-N-WASP-Arp2/3 pathway in some experimental systems, but filopodial biogenesis generally involves multiple nucleation and elongation factors from the Formin and Ena/VASP protein families. (C) Lamellipodia protrude via a highly conserved Rac1/Arf1-WAVE-Arp2/3 signaling mechanism to allow for cell migration. Protrusions in mice may involve WHIMP-Arp2/3 interactions and Src-family kinase (SFK) activation (dashed arrows). The muted color of WHIMP indicates its presence in only a subset of animals. (D) Macropinocytosis at locations of dorsal membrane ruffling can be stimulated by several WASP-family members and small G-proteins. (E) Endocytosis involves N-WASP-Arp2/3-mediated actin assembly and membrane constriction. (F) Endosomal tubule scission and cargo sorting is controlled by WASH-Arp2/3-assembled actin filaments. (G) At the ERGIC and trans-Golgi network, WHAMM and JMY activate the Arp2/3 complex to promote membrane tubulation or transport. The small G-protein Rab1 recruits WHAMM to membranes but limits its nucleation-promoting activity. Note that other G-proteins and kinases (not shown) can activate the WASP-family members by multiple mechanisms, and organelles/proteins are not drawn to scale.
Two major types of plasma membrane protrusions are filopodia and lamellipodia [Fig. 1A–C]. Filopodia are narrow structures utilized for environmental exploration, sensing, and adhesion (Fischer et al., 2019; Gallop, 2020). A network of branched actin filaments at the cell periphery often underlies the filopodial base, whereas linear bundled filaments comprise the filopodial shaft [Fig. 1B]. Because WASP expression is restricted to hematopoietic cells (Massaad et al., 2013), several filopodia models have utilized its closest relative, the ubiquitously-expressed N-WASP. In fact, the initial characterizations of N-WASP described how its overexpression can induce filopodia biogenesis (Miki et al., 1998a, 1996).
Studies on both WASP and N-WASP, in addition to providing insights into filopodia, have revealed key mechanisms governing the inhibition and activation of actin nucleation factors. WASP and N-WASP can adopt inactive auto-inhibited conformations, where intramolecular interactions physically sequester the C-terminal WCA domain, preventing it from activating the Arp2/3 complex (Padrick and Rosen, 2010). The N-termini of WASP and N-WASP also form stable complexes with actin-binding proteins from the WIP family (Sokolik et al., 2020). Conformational changes stemming from interactions between several internal domains of WASP/N-WASP with signaling molecules such as small GTPases, phosphoinositides, SH3 domain-containing adaptor proteins, and tyrosine kinases can relieve the auto-inhibited arrangements of these proteins and enable potent Arp2/3 activation (Alekhina et al., 2017). Moreover, oligomerization of WASP and N-WASP stimulates their nucleation-promoting activities (Campellone et al., 2008a; Cheng et al., 2008; Padrick et al., 2008; Sallee et al., 2008). Multivalent complexes formed with receptors and adaptors also control their phase separation into biomolecular condensates, which regulates actin polymerization kinetics (Banani et al., 2017). Among these signaling mechanisms, the G-protein-based regulation of N-WASP may be indirectly relevant to filopodial biogenesis.
In one model for filopodia assembly, termed convergent elongation, small Rho-family GTPases such as Cdc42 bind and activate N-WASP to stimulate the Arp2/3 complex to generate a dendritic actin network, from which some filaments can be bundled and elongated by other actin-binding proteins (Fischer et al., 2019). While overexpression and reconstituted in vitro systems are compatible with a foundational Cdc42-N-WASP-Arp2/3-based model for actin assembly (Dobramysl et al., 2021; Lee et al., 2010; Lim et al., 2008; Snapper et al., 2001), this pathway may not be the quintessential driver of filopodial biogenesis because filopodia can form in some cells lacking Cdc42, N-WASP, or Arp2/3 complex functions (Czuchra et al., 2005; Lommel et al., 2001; Steffen et al., 2006). In the distinct tip nucleation model for filopodia assembly, Cdc42 and other G-proteins relieve the auto-inhibition of actin nucleators belonging to the Formin family, such as mDia1/2/3 and FMNL2/3, which directly polymerize and elongate filaments at the growing tip (Dimchev et al., 2021; Young et al., 2015). Actin polymerases from the Ena/VASP family also function in filament elongation (Faix and Rottner, 2022). Heterogeneity in the factors capable of promoting filopodia formation is further exhibited in reconstitutions of filopodia-like structures using a variety of combinations of actin nucleation factors and regulators (Dobramysl et al., 2021; Jarsch et al., 2020). Given the existence of multiple mechanisms for actin polymerization within filopodia, these models may reflect the fact that different populations of filopodia are capable of forming under distinct conditions or in diverse cell types.
In contrast to filopodia, lamellipodia are broad, flat cellular protrusions driven forward by the polymerization of branched and short, rather than linear and long, actin filaments (Alexandrova et al., 2020; Innocenti, 2018). The Arp2/3 complex is crucial for generating lamellipodia, and within these structures it is activated by members of the SCAR/WAVE sub-group (WAVE1, WAVE2, or WAVE3) of nucleation factors (Fig. 1C). Each WAVE protein acts within a heteropentameric complex consisting of WAVE1, 2, or 3 with the canonical subunits Sra1 (Cyfip1), Nap1 (Hem2), Abi1, and Brk1 (Hspc300) (Eden et al., 2002; Gautreau et al., 2004). Instead of an auto-inhibitory regulatory mechanism, the basal inactivity of the WAVEs is maintained in trans, as structural studies show that the WH2 and C regions of the WCA domain are bound to Sra1, thus blocking Arp2/3 interaction (Chen et al., 2010; Ismail et al., 2009). Several of the subunits (WAVE1/2/3 themselves, Cyfip1/2, Hem1/2, Abi1/2/3) also exist in multiple isoforms, giving rise to a variety of WAVE regulatory complexes with potentially different activities and functional properties (Dubielecka et al., 2011; Pathania et al., 2014; Polesskaya et al., 2022; Suetsugu et al., 2003; Tang et al., 2020).
Like WASP and N-WASP, the localization and activation of the WAVE complexes are regulated by numerous signaling molecules like G-proteins, phospholipids, and protein kinases. This extensive repertoire of WAVE regulatory complex binding partners is detailed elsewhere (Kramer et al., 2022). In the simplest model for WAVE complex activation during lamellipodial protrusion, the G-protein Rac1 binds to two sites on Sra1, promoting reorganization of the complex, exposing the WCA domain, and thus allowing it to stimulate the Arp2/3 complex (Chen et al., 2017, 2014, 2010). The G-protein Arf1 can cooperate with Rac1 in activating the WAVE complex (Koronakis et al., 2011; Singh et al., 2019), and was recently shown to interact with Sra1 at a site distinct from where Rac1 binds (Yang et al., 2022).
Lamellipodia-like membrane ruffles also form following activation of other WASP-family members including N-WASP (Legg et al., 2007), JMY (Coutts et al., 2009; Zuchero et al., 2009), and WHIMP (Kabrawala et al., 2020). These factors may work in experimental context-, stimulus-, cell type-, and/or organism-specific manners. Among such WASP-family members, WHIMP is the most recently discovered and is related to the WAVEs, but is only present in a subset of animals and appears to be the least potent Arp2/3 activator (Kabrawala et al., 2020). In mouse cells, WHIMP can promote motility and hyper-ruffling phenotypes through an additional ability to stimulate Src-family kinase signaling, which may amplify Arp2/3 activation via the other WAVEs (Fig. 1C). Although WHIMP appears to be absent from the most recent assembly of the human genome (Nurk et al., 2022), its existence in other animals reinvigorates questions regarding how organisms can evolutionarily lose members of the actin regulatory machinery yet maintain finely-tuned processes like membrane protrusion and movement (Kollmar et al., 2012; Veltman and Insall, 2010).
While protrusion is one necessary step in cell motility, adhesion using substrate contacts embedded within lamellipodia is often crucial as well. The ultrastructure and function of focal adhesions are reviewed in greater depth elsewhere (Burridge and Guilluy, 2016; Case and Waterman, 2015). Briefly, nascent adhesions form near the leading edge in many cell types, where the force of actin polymerization and retrograde actin flow activates integrins that simultaneously associate with intracellular F-actin-binding proteins like talin and α-actinin and the extracellular matrix (ECM) (Gardel et al., 2010; Romero et al., 2020). As adhesions mature, so do the pools of factors that are organized in layers to coordinate signaling and force transduction (Kanchanawong et al., 2010). Force can be further modulated by actin arcs connected to focal adhesions by dorsal stress fibers (Burnette et al., 2014). Phosphotyrosine signal transduction during adhesion is carried out by kinases like FAK and Src-family members, the same molecules that phosphorylate and interact with N-WASP and the WAVEs during Arp2/3 complex-mediated protrusion (Ardern et al., 2006; Cory et al., 2002; LeClaire et al., 2008; Padrick and Rosen, 2010; Suetsugu et al., 2002; Torres and Rosen, 2006; Wu et al., 2004). Furthermore, depletion of WASP, the WAVE complex, or the Arp2/3 complex in phagocytes or N-WASP in fibroblasts impairs cell adherence (Kumar et al., 2012; Misra et al., 2007; Rotty et al., 2017; Stahnke et al., 2021).
Actin assembly in adhesions can also be modulated by variations of the core nucleation machinery, as non-canonical Arp2/3 complexes consisting of Arp2, Arp3, ArpC2/3, and vinculin can be formed (Chorev et al., 2014; Pizarro-Cerdá et al., 2017). These observations not only bring to light the potential relevance of hybrid Arp2/3 complexes containing specialized factors but serve as a reminder that the canonical Arp2/3 complex subunits themselves exhibit diversity that may affect activity. In recent years, the functional consequences of isoform diversity within the Arp2/3 complex have begun to emerge.
Using Vaccinia virus-infected cells as a model system, different isoforms of ArpC1 and ArpC5 were analyzed. Strikingly, Vaccinia actin tails are longer when cells are depleted of ArpC5 or ArpC1A, and are shorter after depletion of the ArpC5L or ArpC1B isoforms (Abella et al., 2016). Complexes containing distinct ArpC1 and ArpC5 isoforms also show differences in actin nucleation activity in vitro (Abella et al., 2016). These results suggest that Vaccinia, which utilizes an N-WASP-dependent mechanism of Arp2/3 activation, may have a molecular preference for a particular composition of Arp2/3 complex during actin-based motility. Subsequent studies have examined how the structure of Arp2/3 complex isoforms can influence nucleation activity, filament elongation, filament disassembly, branch junction stability, and cell cycle progression (Fäßler et al., 2023; Galloni et al., 2021; Molinie et al., 2019; von Loeffelholz et al., 2020). Future avenues of exploration may reveal how the many possible Arp2/3 complex isoforms behave in cell- or tissue-specific manners, respond to distinct WASP-family members, or act in different pathological situations.
Given that the Arp2/3 complex and WASP-family members are so important for protrusion, adhesion, and motility, it is not surprising that genetic mutations or epigenetic alterations that change the molecular activities or expression levels of such nucleation factors are associated with diseases (Kramer et al., 2022). For the Arp2/3 complex, frameshift mutations in ARPC1B result in immunodeficiency phenotypes such as vasculitis and poor wound healing (Kahr et al., 2017; Kuijpers et al., 2017; Volpi et al., 2019). Lack of a functional ArpC1B subunit leads to deficiencies in T-cell spreading, synapse formation, and proliferation (Brigida et al., 2018; Randzavola et al., 2019; Somech et al., 2017). These studies describe a role for the Arp2/3 complex in immune disorders and emphasize the importance of the ArpC1B isoform in particular. Changes in expression and mutations in Arp2/3 complex subunits, as well as WASP-family members, have additionally been observed in a variety of cancers, as reviewed elsewhere (Biber et al., 2020; Molinie and Gautreau, 2018) and discussed further below.
Disruptions of plasma membrane remodeling, signaling, and motility arising from mutations in WASP-family genes have also been associated with immunodeficiencies. Loss-of-function point mutations and microdeletions in the X-linked gene encoding WASP were the first to be connected to disease and found to cause Wiskott-Aldrich Syndrome, related immune system disorders, and bleeding conditions (Bouma et al., 2009; Thrasher, 2009). Such mutations may result in the absence of functional WASP, as is historically the case with the most severe cases of Wiskott-Aldrich Syndrome (Thrasher and Burns, 2010). In contrast, pseudo-gain-of-function mutations that disrupt WASP auto-inhibition lead to neutropenia. Since WASP is important for so many hematopoietic cell processes, including T-cell maturation and signaling, phagocytosis, and the formation of invadosomal protrusions that allow for invasive migration and degradation of the ECM (Linder, 2009; Murphy and Courtneidge, 2011), WASP mutants have pleiotropic effects in patients. Given that pathogenic WAS mutations are so well catalogued and are X-linked, it is unsurprising that they are prime candidates for gene therapy (Aiuti et al., 2013; Rai et al., 2020), leading to the possibility that patient prognoses may eventually be greatly improved.
More recent work indicates that mutations in the autosomal WASP-family genes may give rise to illnesses beyond immune disorders. For example, neurodevelopmental diseases have been attributed to autosomal dominant de novo mutations in the gene encoding WAVE1 (Ito et al., 2018; Srivastava et al., 2021). Taken together with the observations that numerous CYFIP1/2 variants are associated with other neurological conditions (Kramer et al., 2022), these new results suggest that additional alterations in the WAVE complexes which adversely affect brain function will likely be revealed by more genomic testing.
As ongoing research clarifies human disease phenotypes and clinical outcomes concerning the functions of different WASP-family members, it seems likely that the parallel use of animal models deficient in one or more of these proteins, both globally and in a tissue-specific manner (Aloisio and Barber, 2022; Gligorijevic et al., 2012; Juin et al., 2019; Kim et al., 2020; Liu et al., 2021; Morris et al., 2018; Papalazarou et al., 2020; Gruenbaum-Cohen et al., 2012; Zhou et al., 2015), would provide a great benefit. Such models could allow for the visualization of pathogenesis and the effects of potential therapeutic interventions at the organismal level. Furthermore, the study of cell morphogenesis and migration in animal models continues to expand beyond immune responses and cancers, as deletion of either Arp3 or N-WASP in conditional knockout mice negatively influences kidney function through disrupted development, adhesion, and protrusion of podocytes, cells which are crucial for forming the filtration barrier in the glomerulus (Schell et al., 2018, 2013). With the body of work on nucleation factor-deficient mouse models becoming more prevalent, so too will the connections between the actin assembly machinery, tissue-specific processes, development, and disease.
3. Macropinocytosis, endocytosis, and endocytic trafficking
Plasma membrane protrusions, in addition to their key roles in cell movement and exploration of the environment, also allow cells to internalize extracellular materials (Fig. 1D). Protrusions in the form of membrane ruffles can sample the surroundings and engulf microbes, nutrients, or other molecules. In one type of ingestion, termed macropinocytosis, actin-driven ruffles project out into the milieu, enclose, and internalize, creating a fluid-filled vacuole known as a macropinosome (Kay, 2021; Swanson and King, 2019). N-WASP, WAVE1, WAVE2, and WHIMP have all been localized to peripheral, dorsal, or pathogen-induced membrane ruffles or analogous sites of macropinocytosis (Fig. 1D). N-WASP localizes to dorsal ruffles, and some N-WASP-deficient mouse embryonic fibroblasts exhibit aberrant ruffle formation (Legg et al., 2007). Distinct WAVE subfamily members are also enriched at ruffle structures in fibroblasts, with WAVE1 important dorsally, and WAVE2 more prominent in peripheral ruffling (Suetsugu et al., 2003), although dorsal ruffles can form in some cells lacking WAVE1 (Legg et al., 2007). In any case, internalization of fluid-phase molecules appears to be more dependent on components of the WAVE complex than N-WASP (Innocenti et al., 2005). As with WAVE complex-mediated lamellipodial protrusion, macropinocytic protrusion is positively regulated by G-proteins from the Rac and Arf sub-families. Finally, murine WHIMP localizes to dorsal and peripheral ruffles, and its expression increases macropinocytosis (Kabrawala et al., 2020). Collectively, these observations underscore the idea that cells from different tissues or organisms are versatile in their capacity to form ruffles and undergo macropinocytosis.
While macropinocytic protrusions are one way in which the actin assembly machinery can contribute to the internalization of extracellular material, the Arp2/3 complex and WASP-family members are also key players in membrane invagination processes including receptor- and clathrin-mediated endocytosis (Fig. 1E). During these processes, actin is assembled in an Arp2/3- and N-WASP-dependent manner as the membrane deformation turns inward to the cytoplasm to form the endocytic pit (Chakrabarti et al., 2021; Kaksonen and Roux, 2018). Multiple loss-of-function approaches including dominant negative proteins, gene knockouts, and RNA interference indicate that N-WASP is important for the internalization of several receptors and cargo (Benesch et al., 2005; Bu et al., 2010; Innocenti et al., 2005; Merrifield et al., 2004). Branched filaments appear at the neck of the pit and can also surround the pit itself. More recent high-resolution imaging and modeling reveals that N-WASP- and Arp2/3-mediated actin networks can create asymmetric forces for internalization of some endocytic vesicles (Akamatsu et al., 2020; Jin et al., 2022; Serwas et al., 2022).
Actin dynamics are not only important for the internalization step of endocytosis, but also for the subsequent movement and trafficking of endocytic vesicles and their cargo (Fig. 1F). Internalized vesicles associated with N-WASP and the Arp2/3 complex move via actin tail rocketing motility (Benesch et al., 2002; Southwick et al., 2003; Taunton et al., 2000). Once endosomes are within the cell, encapsulated cargo must be properly sorted and directed through the endomembrane system. The destination of endocytic components is initially determined at an early or sorting endosome, from which tubules emerge to form different endosomal compartments, where some receptors can be directed back to the plasma membrane through recycling endosomes. Other receptors and ligands may proceed to the Golgi or distinct organelles for further modification and sorting, or develop into late endosomes that mature and acidify into lysosomes (MacDonald et al., 2020; Wang et al., 2018). Within the WASP-family, WASH is the central figure in endocytic trafficking.
Originally discovered to be encoded by numerous human subtelomeric genes (Linardopoulou et al., 2007), WASH is evolutionarily conserved in many animals and protists, but not fungi (Kollmar et al., 2012; Velle and Fritz-Laylin, 2019; Veltman and Insall, 2010). The relevance of the human WASH paralogs is unclear, as most mammalian studies have focused on mouse WASH or human WASH1 (both referred to simply as WASH here) and have been reviewed elsewhere (Alekhina et al., 2017; Fokin and Gautreau, 2021). WASH is part of a heteropentameric protein complex that is structurally and functionally similar to that of the WAVE regulatory complex (Derivery et al., 2009; Gomez and Billadeau, 2009; Jia et al., 2010). The core subunits are Fam21 (WashC2), Ccdc53 (WashC3), Swip (WashC4), and Strumpellin (WashC5). Several early papers demonstrated that WASH localizes to multiple endosomal populations (Derivery et al., 2009; Duleh and Welch, 2010; Gomez and Billadeau, 2009; Park et al., 2013; Zech et al., 2011). WASH is essential in mice (Gomez et al., 2012; Xia et al., 2013), and at the cellular level, WASH inactivation or depletion disrupts numerous endosomal cargo sorting events. Furthermore, WASH maintains the integrity, distribution, and size of the lysosome population (Gomez et al., 2012; Park et al., 2013), extending WASH’s influence over the entire endolysosomal system.
The importance of WASH appears to lie in its ability to promote Arp2/3-mediated actin assembly, whereby the actin filaments enable endosomal tubule scission (Derivery et al., 2009; Gomez and Billadeau, 2009). The recruitment of WASH to endosomal membranes is primarily achieved through interactions of Fam21 with the retromer complex, a key constituent of the endosomal sorting apparatus (Seaman, 2021; Simonetti and Cullen, 2019). Newer studies have further revealed a connection between WASH and the retromer-related retriever complex, which is recruited to endosomal membranes through interactions with subunits of yet another multi-protein complex, the CCC complex (McNally et al., 2017; Phillips-Krawczak et al., 2015; Singla et al., 2019). The WASH complex also coordinates with the F-actin capping machinery and microtubule-associated factors, as Fam21 interacts with CapZ to perform an anti-capping function and create seed filaments based on remodeling of the dynactin complex (Fokin et al., 2021; Jia et al., 2010). In contrast to the many studies on how WASH complex binding partners regulate its recruitment to endosomal membranes, the activation of the complex by signaling molecules is less understood. Mammalian WASH does not appear to be activated by Cdc42 or Rac1, but it is ubiquitinated, which appears to be important for Arp2/3-mediated actin assembly on endosomes (Hao et al., 2013). More work is needed to elucidate the other molecular mechanisms underlying WASH complex activation.
It is also notable that endocytic trafficking pathways can intersect with biosynthetic pathways, and several WASP-family members influence ER-to-Golgi transport, membrane tubulation, trans-Golgi dynamics, and other secretory remodeling events (Fig. 1G). These nucleation factors and topics have been discussed elsewhere (Blom et al., 2015; Campellone et al., 2008b; Chakrabarti et al., 2021; Luna et al., 2002; Matas et al., 2004; Russo et al., 2016; Schlüter et al., 2014).
Disruption of the endolysosomal network has long been connected to disease, but our understanding of the contribution of WASP-family members is now beginning to come into play (Kramer et al., 2022). In pancreatic cancer, N-WASP may enable ductal adenocarcinoma cell migration via its role in endocytic recycling of a chemotaxis-related receptor (Juin et al., 2019). Mutant forms of WASH complex subunits can disrupt endolysosomal integrity and cargo sorting in the context of neurological disorders (Courtland et al., 2021; Ropers et al., 2011; Valdmanis et al., 2007). The importance of WASH in membrane trafficking processes that affect protein homeostasis or turnover is also particularly relevant to neurodegenerative diseases, as explained below. While characterizing phenotypic alterations in cellular processes like endosomal trafficking are crucial to understanding many aspects of disease, in cases involving nucleation factors like N-WASP and WASH that have numerous functions, it remains challenging to identify which defective processes are the primary drivers of pathogenesis. As our ability to more precisely sort out the molecular basis of cellular dysfunction improves, the paths for designing therapeutic interventions will likely become clearer.
4. Proteostasis and autophagy
Following the discovery of WASH and two other WASP-family members, WHAMM and JMY, cellular studies on the functions of the Arp2/3 complex and its nucleation-promoting factors began to branch out in new directions. In particular, investigations into how the actin assembly machinery contributes to maintaining several aspects of cellular homeostasis have emerged [Table 1]. Protein homeostasis, termed proteostasis, is the maintenance of appropriate protein concentrations within the cell amidst changing conditions (Hipp et al., 2019; Yerbury et al., 2016). Proteostasis is sustained through several coordinated processes [Fig. 2A], including protein synthesis, folding, trafficking, and degradation [Fig. 2B–E]. Disruption of one or more of these activities can impair multiple intracellular behaviors and reduce cell viability. Among the different arms of the proteostasis network, those pertaining to protein trafficking and degradation currently have the strongest connections to the actin cytoskeleton.
Table 1.
Functions for the ubiquitous mammalian WASP-family proteins in cellular homeostasis and stress responses.
Fig. 2.
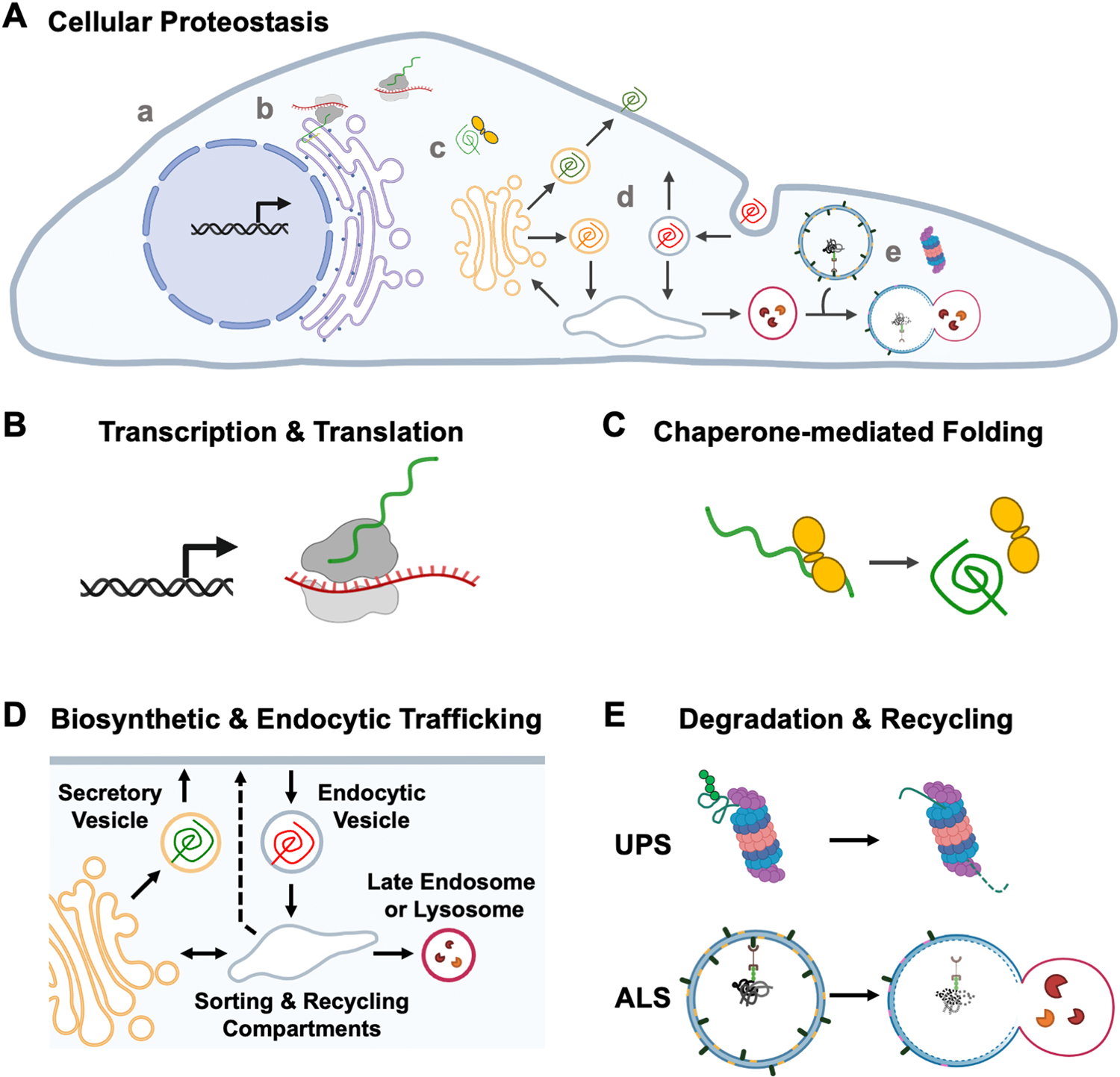
Cellular processes that impact proteostasis. (A) Cellular proteostasis is maintained through the coordinated synthesis and turnover of proteins. In (a) a generic mammalian cell, the processes that influence proteostasis include (b) transcription and translation, (c) protein folding, (d) membrane protein trafficking, and (e) protein degradation. (B) Protein production is regulated at the gene expression level in the nucleus, where transcription is modulated by internal and external stimuli. It is also controlled at the polypeptide synthesis level in the cytoplasm, where translation of mRNA by ribosomes in the cytosol or at the ER creates new proteins. Note that co/post-transcriptional regulation (e.g., splicing, nucleo-cytoplasmic transport) and post-translational modifications (e.g., phosphorylation, ubiquitination) are not depicted. (C) During and after synthesis, polypeptide folding may be mediated by chaperone proteins that help the protein achieve its final functional structure. Molecular chaperones are also vital in quality control and preventing the accumulation of toxic protein species, as the chaperones can attempt to refold a misfolded protein, modulate its aggregation, or direct it towards a degradation pathway if it cannot adopt a satisfactory conformation. (D) Proteins are sorted and trafficked to their appropriate destinations through the biosynthetic (e.g., conventional secretory/anterograde) and endolysosome (e.g., endocytic/retrograde) systems. They may be directed to different organelles or structures within the cytoplasm, and to or from the plasma membrane. (E) Proper protein levels are maintained by degradation pathways that remove proteins which are misfolded, no longer needed, nonfunctional, or targeted for elimination. Degradation occurs through two primary mechanisms: the ubiquitin-proteasome system (UPS) or the autophagosome-lysosome system (ALS). Both systems can be used to degrade ubiquitinated proteins. Proteasome-mediated proteolysis requires protein unfolding, whereas autophagosomes participating in selective autophagy can accommodate larger protein aggregates, defective organelles, or microbial pathogens prior to digestion by lysosomes.
Targeted protein degradation in cells takes place via two primary mechanisms: the ubiquitin-proteasome system (UPS) and the autophagosome-lysosome system (ALS) (Fig. 2E). Both pathways operate constitutively, but can be upregulated in response to several stimuli (Korolchuk et al., 2010; Yin et al., 2020). Though these pathways differ in their targets and steps leading to degradation, both involve the process of ubiquitination. Ubiquitin is a 76 amino acid polypeptide containing several lysine (K) residues that can be conjugated to other protein substrates at their lysines, either as a single moiety rendering the substrate monoubiquitinated or as a polyubiquitin chain. These modifications designate targets for degradation or promote other non-proteolytic processes (Ji and Kwon, 2017). The fates of the target proteins are influenced by the types of ubiquitin-lysine linkages. For example, K48 linkages commonly lead to proteasomal degradation, while K63 linkages can be detected by autophagy receptors (Yau and Rape, 2016). The K48 polyubiquitin chains are recognized by the 26S proteasome, composed of a 19S regulatory particle acting as a lid to the proteolytic 20S core, where degradation of the unfolded proteins takes place (Livneh et al., 2016). The relationship between the UPS and the actin cytoskeleton is not well understood, although actin and actin-binding proteins can be degraded by the proteasome, and actin filaments have been shown to interact with proteasomes in yeast (Haarer et al., 2011).
In contrast to UPS-mediated cytosolic digestion of proteins, the ALS degrades and recycles cellular components through delivery of protein and other macromolecular cargo to lysosomes (Ballabio and Bonifacino, 2020). Autophagy is formally categorized into 3 distinct degradation processes: microautophagy, chaperone-mediated autophagy, and macroautophagy. Microautophagy involves the direct uptake of cytosolic cargo into lysosomes via membrane invagination (Park and Cuervo, 2013). Chaperone-mediated autophagy targets proteins possessing KFERQ-like motifs for transport by the chaperone HSC70 to lysosomes, where they dock at the membrane receptor LAMP2A and are translocated into the lysosomal lumen (Kaushik and Cuervo, 2018). Macroautophagy (often and hereafter referred to simply as autophagy) is defined by the sequestration of cytosolic components within double-membrane-bound vesicles known as autophagosomes followed by autophagosomal fusion with and digestion by lysosomes (Morishita and Mizushima, 2019; Zhao and Zhang, 2019). It is also the degradation pathway in which the actin cytoskeleton has been most studied (Fig. 3A–D).
Fig. 3.
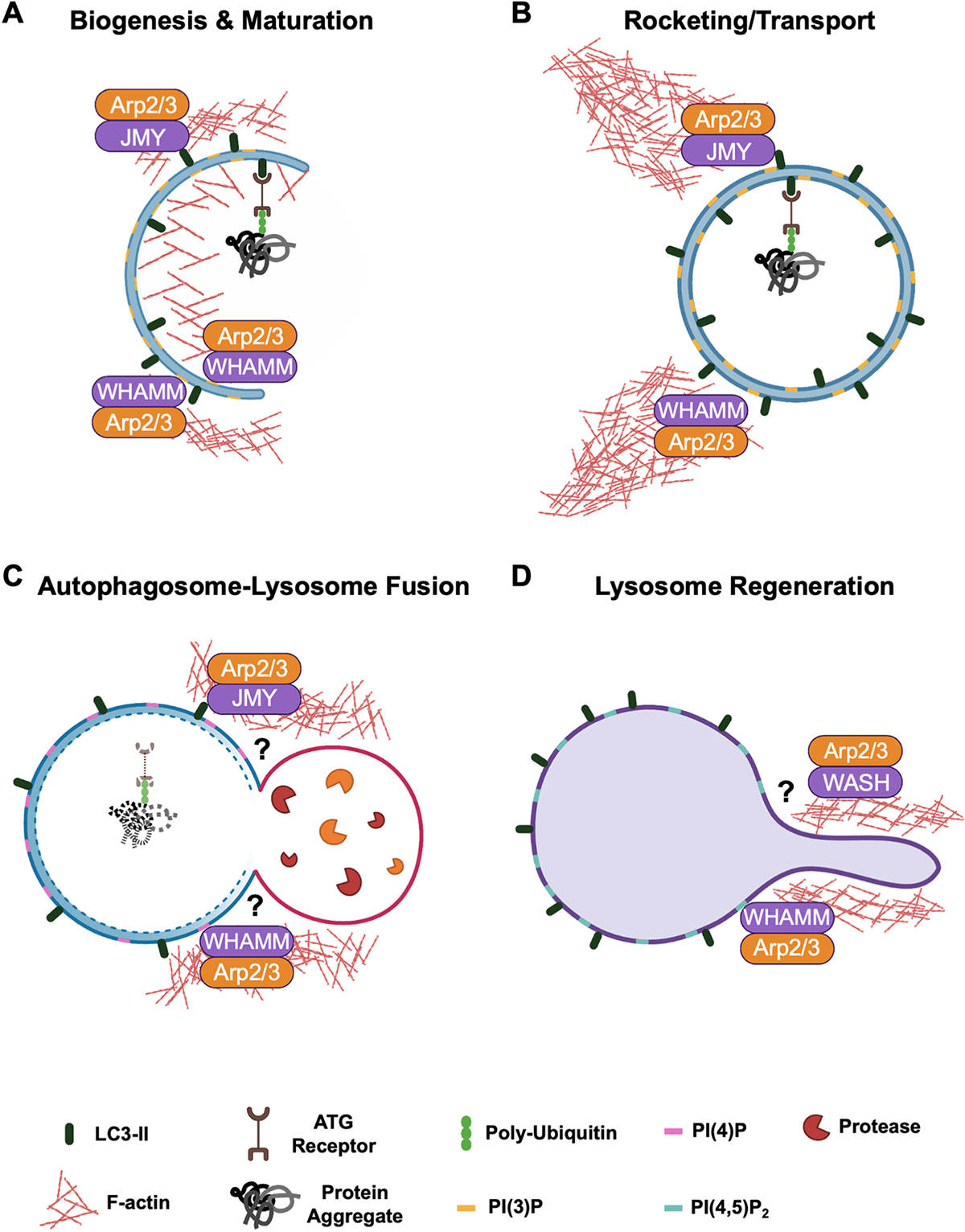
Multiple steps of autophagy rely on actin nucleation factors. (A) During autophagosome biogenesis, actin is polymerized inside and outside of the PI(3)P-rich phagophore (nascent autophagosome) to shape the membrane and allow incorporation of lipidated LC3. WHAMM is recruited to forming autophagosomes through its interaction with the phospholipid PI(3)P, while JMY is localized by binding LC3. Both WHAMM and JMY activate the Arp2/3 complex. (B) Autophagosome movement is driven by the force generated by F-actin comet tails derived from JMY- and WHAMM-mediated Arp2/3 complex activation. (C) Autolysosomes contain WHAMM and JMY (and Cortactin; not shown) on their surface, although the extent to which the nucleation factors affect the autophagosome-lysosome membrane fusion step (?) are not well understood. (D) During lysosome regeneration, WHAMM is recruited by PI(4,5)P2 on the autolysosome, where it activates Arp2/3-mediated actin assembly to promote membrane tubulation. WASH is also localized to tubules, although its mechanisms of recruitment to specific autolysosomal membranes (?) is unclear. Collaboration with microtubules, motor proteins, and scission factors (not depicted) enables the reformation of lysosomes.
Autophagy is a fundamental mechanism for catabolizing and recycling intracellular components in response to multiple stressors, including nutrient starvation (Ohsumi, 2014). Autophagosomes can envelop bulk cytoplasm in a nonselective process, or they can isolate specific, often ubiquitinated, cargo in processes collectively called selective autophagy (Gatica et al., 2018; Hansen et al., 2018). Autophagosome biogenesis is frequently initiated by the ULK1 (ATG1) protein kinase complex that phosphorylates key factors which participate in the autophagy pathway, such as components of the Beclin- and Vps34-based PI3 kinase complex that produce the phospholipid PI(3)P at sites of nascent autophagosome, or phagophore, formation (Palamiuc et al., 2020; Sridharan et al., 2011). A significant number of these isolation membranes are formed in association with endoplasmic reticulum (ER) structures known as omegasomes (Roberts and Ktistakis, 2013). The formation of PI(3)P-binding complexes allows for the assembly of the core ATG machinery, a ubiquitin-like conjugation system that incorporates autophagosomal membrane proteins from the ATG8 family into the developing phagophore, which elongates, matures, and eventually closes to form an autophagosome surrounding the cargo (Carlsson and Simonsen, 2015; Nakatogawa, 2020).
In mammals, the ATG8 family consists of the LC3 (LC3A/B/C) and GABARAP (GABARAP, GABARAP-L1/L2) subfamilies, which exist in cytosolic forms (e.g., LC3-I or GABARAP-I) that can be lipidated with phosphatidylethanolamine to create their autophagosomal membrane-associated forms (e.g., LC3-II or GABARAP-II) (Nguyen and Lazarou, 2022). For completion of autophagy, autophagosomes fuse with lysosomes to create autolysosomes, where the contents are degraded and/or recycled to meet the metabolic and biosynthetic needs of the cell (Nakamura and Yoshimori, 2017). Autolysosomes may then undergo tubulation and scission to form protolysosomes in the process of lysosome regeneration. The protolysosomes mature into lysosomes, serving to maintain the intracellular lysosomal population (Yang and Wang, 2021).
Selective autophagy, as the name implies, targets specific cargo for degradation. Autophagy receptors act as the link between the target and the autophagosome, utilizing a ubiquitin-binding domain to recognize the ubiquitinated cargo, and an LC3 Interacting Region (LIR) to bind LC3 embedded in the autophagosomal membrane (Morishita and Mizushima, 2019). Forms of selective autophagy are typically named for their intended targets: aggrephagy for protein aggregates, mitophagy for mitochondria, xenophagy for microbes, and several others (Lamark and Johansen, 2021). Some autophagy receptors, including SQSTM1/p62, can participate broadly in several types of selective autophagy, while others such as OPTN, NDP52, NBR1, and TAX1BP1 may be used preferentially with different cargoes (Kirkin et al., 2009; Lazarou et al., 2015; Pankiv et al., 2007; Sarraf et al., 2020; Thurston et al., 2009; Wild et al., 2011; Wong and Holzbaur, 2014).
Selective autophagy is especially relevant to neurodegenerative disorders like Huntington’s, Alzheimer’s, and Parkinson’s Disease, in which proteins aggregate and proteostasis is disrupted. More specifically, changes in mitophagy significantly impact neurodegeneration, as the hallmark players in neuronal mitophagy – the protein kinase PINK1, phosphorylated ubiquitin, and the ubiquitin ligase Parkin – are often dysregulated in Parkinson’s Disease patients (Killackey et al., 2020; Kitada et al., 1998; Pickrell and Youle, 2015; Valente et al., 2004). PINK/Parkin-mediated mitophagy is coordinated with mitochondrial remodeling, as mitochondria cluster into mitoaggregates until they can be disassembled in an actin-associated fashion to allow for encapsulation by an autophagosome (Hsieh and Yang, 2019). Irrespective of the particular molecular cargo, the actin cytoskeleton then appears to play multiple roles in the pathway of autophagosome biogenesis, transport, and turnover (Fig. 3A–D).
For a long time, little was known about how the actin cytoskeleton might impact autophagic processes. Many years ago, actin filaments were found to be necessary for autophagosome formation in mammalian cells during nutrient starvation, as blocking polymerization with cytochalasin D reduced the number of autophagic vacuoles (Aplin et al., 1992). It was later shown that the branching factor Cortactin is recruited to ubiquitinated protein aggregates destined for autophagic degradation, and that Cortactin depletion results in less actin at these aggregates, a decrease in autophagosome-lysosome fusion, and an increased accumulation of autophagosomes (Lee et al., 2010). The connections between various forms of autophagy and the actin nucleation machinery then began to come into focus.
While the Arp2/3 complex had previously been localized to autophagosomal structures in yeast (Monastyrska et al., 2008), it was several years until the mammalian complex, namely the ArpC1B subunit, was observed to localize to LC3-positive autophagosomes in rat kidney epithelial cells (Mi et al., 2015). In the latter system, an autophagosomal membrane remodeling pathway is dependent on the phospholipid PI(3) P and the F-actin capping protein CapZ to stimulate actin assembly within developing phagophores and positively control their size and shape (Mi et al., 2015). Several subsequent studies revealed that multiple nucleation-promoting factors can also affect the autophagosome biogenesis process.
Among WASP-family members, WASP itself is important for multiple aspects of non-selective autophagy in some types of myeloid cells (Lee et al., 2017; Rivers et al., 2020), but much of what we know about the cytoskeletal mechanisms that govern several forms of autophagy in diverse cell types come from studies on WHAMM, JMY, and WASH (Fig. 3A–D). Early in the constitutive and starvation-induced autophagic pathways, WHAMM localizes to omegasomes and sites of autophagosome biogenesis, in part through its ability to bind PI(3)P (Kast et al., 2015; Mathiowetz et al., 2017). Deletion of WHAMM or removal of its WCA domain results in decreases in LC3-I to LC3-II conversion and autophagosome abundance, and mutation of a PI(3)P-binding motif within its PX-like domain abrogates WHAMM localization to autophagosomes (Mathiowetz et al., 2017). The expression of WHAMM positively correlates with autophagosome size and number, and this relationship requires its capacity to activate the Arp2/3 complex (Kast et al., 2015). WHAMM can also be recruited to endomembranes by the small G-protein Rab1 (Russo et al., 2016), a known regulator of autophagy (Huang et al., 2011; Kakuta et al., 2017; Thomas et al., 2014; Tremel et al., 2021; Zoppino et al., 2010), but it is not known if Rab1 controls WHAMM localization or activity during autophagosome formation.
JMY additionally promotes autophagosome formation (Fig. 3A), as JMY physically interacts with LC3 via a LIR motif, and depletion of JMY results in decreases in LC3-I and LC3-II levels (Coutts and La Thangue, 2015). The physical motility of mature autophagosomes is also influenced by both WHAMM and JMY [Fig. 3B], as these two WASP-family members can promote Arp2/3-mediated actin-based rocketing of autophagosomes (Hu and Mullins, 2019; Kast et al., 2015). LC3 binding activates JMY-driven actin assembly during this process (Hu and Mullins, 2019). While WHAMM possesses an N-terminal LIR homologous to the one found in JMY, the mechanisms of WHAMM activation and the biological purposes of autophagosomal rocketing are unclear.
Actin assembly is also important later in the canonical autophagy pathway (Fig. 3C–D), as WHAMM possesses two PI(4,5)P2-binding motifs that mediate localization to autolysosomes, where WHAMM promotes Arp2/3-dependent actin assembly during membrane tubulation for lysosomal regeneration (Dai et al., 2019). WHAMM is further capable of interacting with BLOC1S1, a component of the BLOC-1 lysosome biogenesis complex, which is thought to participate in recruiting WHAMM during lysosomal tubule formation (Wu et al., 2021). This membrane remodeling activity of WHAMM is analogous to that of WHAMM in driving actin assembly to tubulate membranes that transport cargo through the ER-Golgi secretory pathway (Campellone et al., 2008b; Russo et al., 2016; Shen et al., 2012). The molecular similarities and differences underlying WHAMM-mediated tubulation of secretory versus autophagic organelles remain to be determined.
Apart from WHAMM and JMY, WASH is another WASP-family member that participates in autophagy. Given its role in the sorting of cargo through the multiple endocytic trafficking pathways mentioned earlier (Figs. 1F, 2 D), it is not surprising that depletion or disruption of the WASH regulatory complex impacts the lysosomal functions that enable autophagic flux and turnover of autolysosomal cargo [Fig. 3D]. Like WHAMM, WASH appears to interact with the BLOC-1 lysosome biogenesis complex (Monfregola et al., 2010; Ryder et al., 2013). However, conflicting findings have led to debate on the net positive versus negative effects that WASH has on autophagy. Mouse fibroblasts lacking WASH show an increased conversion of LC3-I to LC3-II and degradation of p62, while cells overexpressing WASH show decreased Beclin-1 ubiquitination, which diminishes its association with Vps34, suggesting that WASH may normally suppress autophagy (Xia et al., 2013). On the other hand, depletion of WASH in HeLa or neuronal cells decreases LC3-II levels and alters the localization of ATG9 to LC3-positive membranes, suggesting that WASH positively influences autophagosome formation (Zavodszky et al., 2014). WASH-associated autophagy is also impaired in Parkinson’s Disease patients with a D620N mutation in Vps35, a component of the retromer complex. This mutation weakens the association between retromer and WASH, and also results in a decrease of LC3-II (Zavodszky et al., 2014). It seems likely that different populations of WASH act in different steps of autophagy, or that its separate functions may be cell or tissue specific. More work is needed to bring the role of WASH in autophagy into focus.
On the topic of proteostasis and disease, especially related to neurodegenerative disorders, the accumulation of protein aggregates and intracellular defects in their removal via the UPS or ALS is undeniable (Mizushima and Levine, 2020). From a cytoskeletal standpoint, disruptions in endolysosomal trafficking and alterations in WASH complex subunits have been implicated in disease. Several mutations in the gene encoding Strumpellin/WashC5 are found in patients with severe hereditary spastic paraplegia (Valdmanis et al., 2007). Moreover, in a form of autosomal recessive intellectual disability, a single amino acid substitution in SWIP/WashC4 diminishes the stability of the WASH complex, leading to impaired lysosome formation and accumulation of lipid-like inclusions in motor cortex neurons (Courtland et al., 2021; Ropers et al., 2011). It is clear that investigating the presence of mutations within WASP-family genes, especially those with functions tied to maintaining proteostasis, is an avenue worth exploring to further understand the effects of the actin assembly machinery on disease.
One such mutation occurs in WHAMM in a subset of patients with Galloway-Mowat Syndrome (GMS), an autosomal recessive disorder characterized by neurodevelopmental delay and progressive renal failure. The genetic basis of GMS has been attributed to mutations in many different genes, including WDR73, NUP107, PRDM15, WDR4, and subunits comprising the KEOPS complex such as OSGEP, LAGE3, TP53RK, and TPRKB (Ben-Omran et al., 2015; Braun et al., 2018, 2017; Colin et al., 2014; Domingo-Gallego et al., 2019; Jinks et al., 2015; Mann et al., 2021; Rosti et al., 2017; Vodopiutz et al., 2015). The underlying relationship between GMS pathogenesis and the functions of the proteins encoded by these genes is unclear, as the proteins participate in numerous cellular processes, including cell proliferation, nucleo-cytoplasmic transport, tRNA modification, and transcriptional activation. Interestingly, a cohort of patients within the Amish community afflicted with this syndrome was found to possess a homozygous loss-of-function mutation in WHAMM in addition to a homozygous mutation in WDR73 (Jinks et al., 2015; Mathiowetz et al., 2017). The GMS-associated WHAMM mutation results in altered WHAMM mRNA splicing and the production of frameshifted WHAMM protein variants that lack a WCA domain and are unable to activate the Arp2/3 complex (Mathiowetz et al., 2017). GMS patient fibroblasts expressing these truncated versions of WHAMM show profound defects in autophagosome biogenesis that can be rescued by re-introduction of full-length WHAMM, but not WDR73, highlighting the importance of WHAMM in mammalian autophagy (Mathiowetz et al., 2017). Nevertheless, the specific contributions of each genetic mutation to neuronal or kidney phenotypes remain unresolved.
As we come to understand the roles of WASP-family members in proteostasis and autophagy, there is still much to be learned about their molecular modes of action and how their activities are coordinated. For example, while WHAMM binds phospholipids and Rab1, and JMY binds LC3 and has genetic interactions with the G-protein RhoD, the spatiotemporal relationships between WHAMM and JMY functions are unknown. Moreover, whether these two factors work in multi-subunit complexes or are activated by signaling molecules in ways resembling how the WASP, WAVE, and WASH subfamilies are regulated has yet to be elucidated. More broadly, the ways in which the UPS and ALS degradation pathways themselves are coordinated, and how they may compensate for one another in cells with defective WASP-family members remains an open area for investigation. Finally, while work in cultured cell lines has been critical for deciphering the basic functions of WASP-family members and linking them to human diseases, future studies utilizing animal models will truly begin to uncover the impact of specific nucleation factors on different physiological systems, tissues, and relevant cell types. Expanding our characterizations of these factors in vivo will be necessary for understanding their influence on development and disease.
5. Apoptosis
In addition to autophagy, another fundamental cellular response to multiple kinds of stress – but in this case lethal stimuli – is apoptosis (Kerr et al., 1972). In contrast to the homeostatic and pro-survival functions of autophagy, apoptosis is a programmed form of cell death regulated by signaling cascades that culminate in organized proteolysis, nuclear and DNA fragmentation, and cytoskeleton-mediated morphogenic changes (Fig. 4A–C). Apoptotic cell suicide can be driven by intrinsic mitochondria-mediated or extrinsic receptor-mediated pathways that involve distinct initial signaling events but converge on a shared terminal execution pathway to cause an irreversible deterioration of intracellular functions and loss of cellular integrity (Tait and Green, 2013; von Karstedt et al., 2017).
Fig. 4.
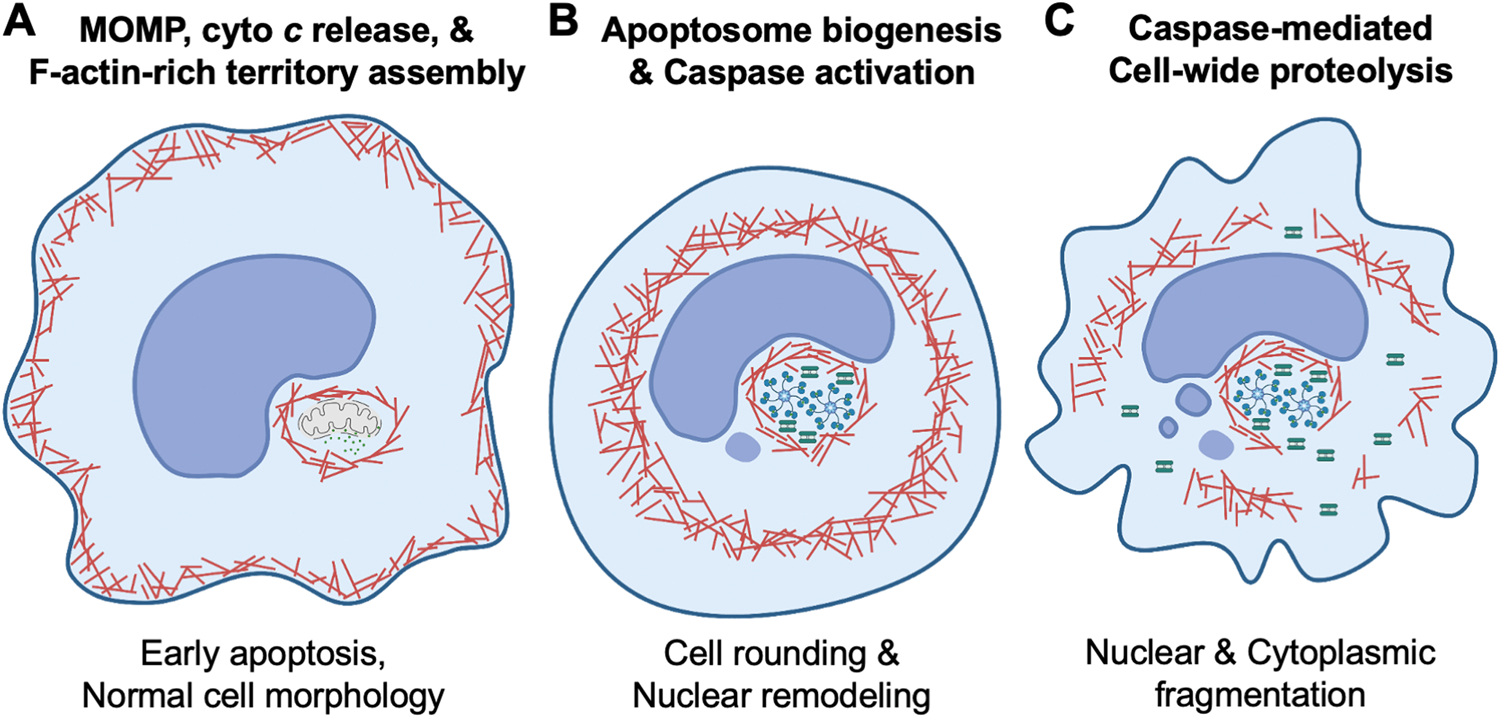
F-actin rearrangements and cellular morphogenesis during apoptosis. (A) Early in intrinsic apoptosis, mitochondrial outer membrane permeabilization (MOMP) enables cytochrome c (cyto c) release into the cytosol, and F-actin is polymerized and reorganized at a juxtanuclear site containing the permeabilized mitochondria. These F-actin-rich territories promote the sequestration of cytosolic cyto c and facilitate interactions between apoptosome components. (B) As apoptosis progresses, cell rounding and chromatin condensation occur. The juxtanuclear F-actin-rich territory functions as a subcellular compartment that clusters multiple pro-apoptotic constituents and serves to couple the apoptosome biogenesis process to the activation of the caspase cascade. (C) Executioner caspases clustered within the territories are physically protected from caspase inhibitors and become enzymatically active. They eventually reach quantities that can overcome the inhibitors, escape the territory, and execute a sudden breakdown of the cell leading to the characteristic morphological features of apoptosis such as nuclear fragmentation and cortical membrane collapse. Note that the stoichiometry of organelles and macromolecules shown in the figure (e.g., mitochondria, cyto c, apoptosomes, caspases) are not drawn to scale.
Intrinsic apoptosis pathways are triggered in response to developmental programs or intracellular damage and are characterized by mitochondrial outer membrane permeabilization (MOMP), the export of apoptogenic proteins from mitochondria to the cytosol, and the subsequent formation of macromolecular signaling platforms called apoptosomes (Bock and Tait, 2020). In contrast, extrinsic apoptosis pathways are initiated in response to external stimuli when extracellular ligands bind to transmembrane death receptors that promote the intracellular formation of multi-protein complexes called death-inducing signaling complexes (DISCs) (Dickens et al., 2012; Yang, 2015). The key enzymes that orchestrate the apoptotic death signaling cascades are cysteine-dependent aspartate proteases known as caspases (Julien and Wells, 2017). Caspase zymogens, or procaspases, are inactive precursors with limited enzymatic activity that must undergo proteolytic processing to form active caspase enzymes. The 9 mammalian caspases involved in apoptosis are divided into two groups, initiators and executioners. The initiator caspases differ between the intrinsic versus extrinsic pathways, and the executioner caspases are common between both mechanisms. From an actin cytoskeletal perspective, more is understood about intrinsic mitochondrial pathways (Fig. 5).
Fig. 5.
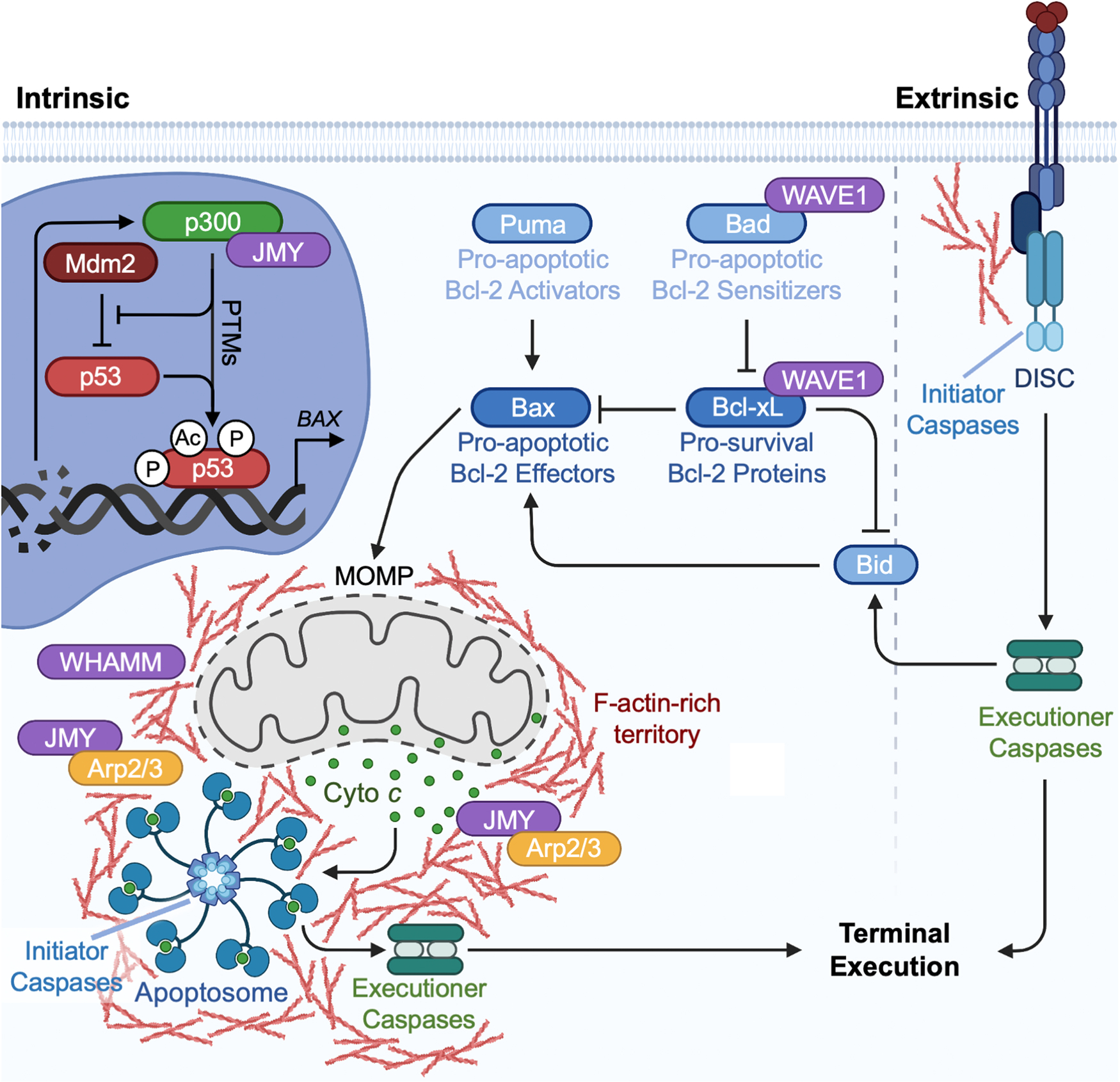
Intrinsic and extrinsic apoptosis pathways. Apoptosis is initiated by either intrinsic (left) or extrinsic (right) pathways that converge on a terminal execution program. Under normal conditions, p53 is negatively regulated by Mdm2. Intrinsic stressors, such as DNA damage, are detected by cofactors that promote p53 stabilization and activation through post-translational modifications (PTMs) including phosphorylation (P) and acetylation (Ac). p53 then binds response elements of specific target genes to promote their transcription. JMY can promote cell death by enhancing the p53-mediated transcription of BAX, which encodes a pro-apoptotic Bcl-2-family effector protein. The intrinsic pathway is driven by Bcl-2 effector oligomerization, resulting in MOMP and the release of the apoptogenic factor cyto c from the mitochondrial intermembrane space. WAVE1 functions in intrinsic apoptosis by influencing the localization or modification of multiple Bcl-2-family proteins. JMY and WHAMM activate the Arp2/3 complex to polymerize actin into a dense F-actin-rich territory surrounding the juxtanuclear site of mitochondrial permeabilization. This territory creates a microenvironment where cytosolic cyto c is sequestered and can efficiently trigger the assembly of Apaf-1 into mature apoptosomes. The apoptosomes bind and activate initiator procaspases, and the resulting active initiator caspases (e.g., caspase-9) process executioner procaspases. The resulting active executioner caspases (e.g., caspase-3) accumulate within the territory, which provides temporary physical protection against caspase inhibitors. The F-actin-rich territory thus coordinates the spatiotemporal progression of intrinsic apoptotic signaling from the step of apoptosome biogenesis to the activation of the caspase cascade. Active executioner caspases eventually escape the territory using an unknown mechanism and go on to cleave many proteins to drive terminal execution. The extrinsic pathway is initiated by death receptor ligation at the plasma membrane, causing the recruitment of adaptor proteins and initiator procaspases to form the death-inducing signaling complex (DISC). Actin influences the aggregation of death receptors and is important for DISC formation. Upon recruitment to the DISC, initiator caspases are activated and can cleave executioner caspases. The intrinsic and extrinsic pathways are interconnected, as active caspases cleave the Bcl-2 effector Bid to an active form that can stimulate MOMP. Note that the stoichiometry of organelles and macromolecules shown in the figure (e.g., mitochondria, apoptosomes, proteins) are not drawn to scale.
Mitochondria are dynamic organelles that synthesize ATP but also produce reactive oxygen species and, upon dysfunction or damage, must be isolated from the healthy population to prevent cellular injury and maintain homeostasis. They exist as tubular chains or individual organelles, and their fusion and fission are controlled by several components of the actin assembly machinery (Ji et al., 2015; Korobova et al., 2014, 2013; Li et al., 2015; Liu et al., 2021; Manor et al., 2015). The actin cytoskeleton additionally appears to monitor the health of the mitochondrial network, as actin filaments cyclically associate with subpopulations of mitochondria (Moore et al., 2016). The Arp2/3 complex and Formin nucleators cause the rapid formation of cytoskeletal cages that surround and segregate damaged mitochondria during interphase and also promote actin tail assembly and mitochondrial mixing in mitotic cells (Kruppa et al., 2018; Moore et al., 2021). Multiple actin nucleation pathways are involved in these processes. After calcium induction, the Formin INF2 polymerizes actin, whereas following acute mitochondrial depolarization, Cdc42-activated FMNL-family Formins trigger linear actin polymerization, and Rac1-activated WAVE complexes stimulate Arp2/3-mediated branched actin assembly (Fung et al., 2022, 2019). A subset of these pathways may segue into mitophagy, as described above.
Mitochondrial-mediated intrinsic apoptosis occurs in response to a variety of internal stressors, including DNA damage, mitotic defects, and mitochondrial injury. The main arbitrator of the cellular fates that follow stress is the transcription factor and tumor suppressor protein p53 (Hafner et al., 2019; Kastenhuber and Lowe, 2017). In unstressed cells, p53 is negatively regulated through interactions with the ubiquitin ligase Mdm2, which binds p53 to block its transcriptional activity and ubiquitinates p53 to promote its degradation via the proteasome (Haupt et al., 1997; Kubbutat et al., 1997). In response to stressors, p53 is stabilized and activated by stimulus-dependent cofactor-mediated post-translational modifications including serine/threonine phosphorylation and acetylation (Fig. 5). Once stimulated, p53 then behaves as a decision-maker by integrating these signals and orchestrating transcriptional programs that govern a variety of cellular responses like cell cycle arrest, DNA repair, senescence, or apoptosis (Andrysik et al., 2017; Sullivan et al., 2018). Genes that are upregulated by p53 include cell cycle inhibitors and mitochondrial permeabilization factors.
Upon initiation of intrinsic apoptotic signaling, MOMP is driven by pro-apoptotic Bcl-2 family proteins such as Bax and Bak (Fig. 5). In a process coordinated by other Bcl-2 family members, these factors oligomerize into pores in the mitochondrial outer membrane to enable the export of pro-apoptotic proteins from the intermembrane space into the cytosol (Kale et al., 2018; Kalkavan and Green, 2018). Some evidence indicates that pores initially form in only a few mitochondria, and that permeabilization propagates in a wave-like fashion across the mitochondrial network (Bhola et al., 2009; Lartigue et al., 2008; Rehm et al., 2009). However, it is unclear what signals determine the initial area of pore formation.
Cytochrome c (cyto c) is a key mitochondrial protein that is released into the cytosol through the apoptotic pores (Fig. 5). Once cytosolic, cyto c instigates the assembly of apoptosomes, wheel-like macromolecular structures that contain the scaffolding protein Apaf-1 and serve as platforms to mediate the multimerization and activation of initiator procaspase-9 (Dorstyn et al., 2018; Kim et al., 2005; Yuan et al., 2010; Zhou et al., 2015; Zou et al., 1999). Active caspase-9 can interact with apoptosomes, forming holo-apoptosomes that process executioner procaspase-3 and -7 and release them as cleaved caspases (Hu et al., 2014; Li et al., 2017; Rodriguez and Lazebnik, 1999; Yin et al., 2006; Yuan et al., 2011). Those enzymatically active executioner caspases then drive the terminal death pathway by targeting numerous cellular substrates for proteolytic cleavage (Fuentes-Prior and Salvesen, 2004).
Early cytoskeletal studies of apoptosis focused primarily on the actin filament rearrangements and disassembly that take place during the late execution phase and contribute to changes in cell adherence and morphology (Desouza et al., 2012; Gourlay and Ayscough, 2005). Unsurprisingly, manipulating actin dynamics by treating cells with drugs that stabilize (Cioca and Kitano, 2002; Odaka et al., 2000; Posey and Bierer, 1999; Rao et al., 1999; White et al., 2001) or depolymerize (Genescà et al., 2006; Martin and Leder, 2001; Morley et al., 2003; Paul et al., 2002; Suria et al., 1999) actin filaments cause the appearance of many morphological and some biochemical hallmarks of apoptosis. While these studies suggest that alterations in actin dynamics can influence apoptotic signaling cascades, the broadly disruptive nature of such drugs makes it difficult to interpret specific mechanisms of action for the actin cytoskeleton in cell death pathways. Some of the first investigations on actin dynamics in intrinsic apoptosis therefore focused on the roles of the actin turnover machinery, eventually including several proteins with specific activities at mitochondria.
Dying cells undergo dramatic morphological alterations during the execution phase of the apoptotic process (Fig. 4B–C). Actin-binding proteins have been identified as targets for caspase processing, and caspase-mediated cleavage of such structural proteins instigates a loss of focal adhesions and the formation of contractile Myosin-F-actin rings, causing cells to adopt a rounded and shrunken morphology (Charras et al., 2006; Levkau et al., 1998; Rosenblatt et al., 2001). The actin-severing protein Gelsolin is also a caspase target, and full-length Gelsolin may play a pro-survival role under normal circumstances (Ahn et al., 2003; Kusano et al., 2000; Ohtsu et al., 1997). However, Gelsolin cleavage by caspases yields an N-terminal fragment with unregulated F-actin severing activity that reduces the overall stability of the actin cytoskeleton and contributes to the morphological features of apoptosis (Chhabra et al., 2005; Kothakota et al., 1997). Earlier in the intrinsic apoptosis pathway, at around the time of mitochondrial permeabilization, actin and the actin depolymerizing protein Cofilin are recruited to mitochondria (Rehklau et al., 2012; Tang et al., 2006; Wang et al., 2008). Cofilin interacts with the GTPase Drp1 to promote both mitochondrial fission and cyto c release (Chua et al., 2003; Hoffmann et al., 2021; Hu et al., 2020; Klamt et al., 2009; Rehklau et al., 2012; Zhu et al., 2006). Taken together, such studies on Gelsolin and Cofilin indicate that the actin disassembly machinery is important for multiple apoptotic events which take place between the time of mitochondrial permeabilization and the overall collapse of the cell.
While actin turnover proteins partake in apoptosis, it eventually became apparent that some actin assembly proteins also function at mitochondria during apoptosis (Fig. 5). WAVE1 has effects on apoptosis and was the first WASP-family protein shown to influence the localization or modification of Bcl-2-family proteins. In hepatocytes, WAVE1 associates with the pro-apoptotic sensitizer protein Bad to promote mitochondrial pore formation (Danial et al., 2003), and in neuronal cells WAVE1 engages the anti-apoptotic protein Bcl-xL to allow mitochondrial recruitment of the pro-apoptotic family member Bax (Cheng et al., 2007). In contrast, in leukemia cells exposed to chemotherapeutic stimuli, WAVE1 interacts with pro-survival Bcl-2 members to reduce apoptotic signaling (Kang et al., 2010). In yet another cell type, fibroblasts, a permanent knockout of WAVE1 does not significantly affect the number of cells that undergo apoptosis following acute DNA damage (King et al., 2021). Why WAVE1, but not other members of the WAVE subfamily, modulates apoptotic signaling in certain contexts is not understood. Thus, defining the mechanisms by which WAVE1 imparts its cell type-specific effects on Bcl-2 family protein homeostasis requires further investigation.
Another WASP-family protein that functions in apoptosis is JMY. In fact, JMY was discovered to participate in apoptosis a full decade before it was realized to be an actin nucleation factor (Shikama et al., 1999; Zuchero et al., 2009). Under normal cellular growth conditions, JMY can be found in both the cytoplasm and nucleus (Fig. 5). It is regulated in the cytoplasm through interactions with Mdm2, the same ubiquitin ligase that controls p53 (Coutts et al., 2007). Upon genotoxic stress, JMY binds importins and accumulates in the nucleus (Coutts et al., 2009; Zuchero et al., 2012), where it associates with p53, the acetyltransferase p300, and stress response protein STRAP (Demonacos et al., 2001; Shikama et al., 1999). There, JMY can promote pro-apoptotic signaling by enhancing the transcription of BAX (Coutts et al., 2009; Shikama et al., 1999).
To systematically evaluate the impact of actin nucleation factors on apoptosis, a panel of human fibroblasts genetically lacking each Arp2/3 complex activator was created and exposed to etoposide, an acute genotoxic agent (King et al., 2021). All of the cell lines underwent rapid p53-dependent apoptosis with two exceptions – JMY knockouts and WHAMM knockouts (King et al., 2021). Given the findings on p53-mediated transcription a decade earlier, the identification of JMY as an important player in apoptosis was not surprising. The recognition of WHAMM also makes sense, since it is the WASP-family member with the greatest homology to JMY. Unexpectedly, however, the effect of JMY as a transcriptional modulator in the nucleus was minimal in these studies. By comparison, the capacity of JMY to act as an Arp2/3-activating factor in the cytoplasm was crucial for its pro-apoptotic function (King et al., 2021). Both JMY and WHAMM likely play multiple roles in intrinsic apoptotic signaling within the cytoplasm, because the propagation of mitochondrial cyto c release, processing of initiator caspase-9, and cleavage/activation of executioner caspase-3 are all delayed or blocked in cells lacking JMY, WHAMM, or both family members (King et al., 2021).
The specific functions of JMY and WHAMM became clearer when their localizations in relation to actin filaments, cyto c, and the caspases were examined (King and Campellone, 2023). At a single juxtanuclear cluster of permeabilized mitochondria, actin is polymerized and reorganized into a structure called an F-actin-rich territory (Figs. 4 and 5). JMY, WHAMM, and the Arp2/3 complex localize throughout the territory, while the branchpoint stabilizer Cortactin is found at the territory perimeter (King and Campellone, 2023). In contrast, STRAP, a known inhibitor of JMY-mediated actin assembly (Hu and Mullins, 2019), and microtubules, which can repress WHAMM-mediated Arp2/3 activation (Shen et al., 2012), are excluded from the apoptotic F-actin-rich regions. Mechanistically, JMY and WHAMM polymerize actin into dense filamentous territories to mediate the early sequestration of permeabilized mitochondria and cyto c, and to create microenvironments that are conducive to apoptosome assembly (King and Campellone, 2023).
The F-actin-rich territories also provide a temporary physical barrier against caspase inhibitors to allow for an efficient activation of the caspase cascade, thus coordinating the spatiotemporal progression of intrinsic apoptotic signaling (King and Campellone, 2023). By compartmentalizing the transitions from cyto c release to apoptosome assembly to caspase activation, the actin cytoskeleton provides a cytoprotective function until active executioner caspases accumulate to levels sufficient to escape the territory and cause a sudden demolition of the rest of the cell (Figs. 4 and 5). The caspases transmit the cell death signal by cleaving a multitude of cytosolic and nuclear proteins including other caspases, kinases, endonucleases, nuclear lamins, and structural proteins. These cleavage events create proteins with gain- or loss-of-function activities that cause the characteristic morphological and molecular features of apoptosis such as cell shrinkage, membrane blebbing, and nuclear fragmentation.
While the roles of actin nucleation factors in intrinsic apoptosis are beginning to emerge, the influence of actin rearrangements on extrinsic pathways is less well understood. Actin filaments affect the aggregation of death receptors and DISC formation (Algeciras-Schimnich et al., 2002; Fais et al., 2005; Luciani et al., 2004), but it is unclear specifically how the actin cytoskeleton regulates signaling progression. Interestingly, the extrinsic and intrinsic pathways appear to be interconnected, as canonical extrinsic caspases can translocate to mitochondria where they promote permeabilization events, release of apoptogenic proteins, and apoptosome-mediated activation of intrinsic caspases (Jost et al., 2009; Li et al., 1998; Luo et al., 1998). This apoptotic amplification loop results in more caspase activation and rapid progression to cell death. It has not been determined how the actin cytoskeleton may modulate this process either.
It is also important to note that the cytoskeleton itself is a target of caspases, as actin has been identified as a substrate for caspase processing. Caspases cleave actin monomers, resulting in two fragments that are incapable of polymerizing into filaments (Mashima et al., 1997, 1995). Transient transfection of the smaller actin fragment can promote the appearance of apoptotic morphologies (Kayalar et al., 1996; Mashima et al., 1999; Utsumi et al., 2003). In the future, it would be interesting to define the relationship between caspase-mediated actin cleavage and F-actin-rich territory breakdown.
Since apoptosis plays pivotal roles in tissue organization, turnover, and homeostasis, further studies on how WAVE1, JMY, WHAMM, and the Arp2/3 complex impact programmed cell death will deepen our understanding of actin nucleation factors in organismal development. Furthermore, the importance of apoptosis in human health and disease is highlighted by the fact that the improper regulation of cell suicide pathways is an underlying contributor to, or even cause of, many pathological conditions, such as tumorigenesis, autoimmunity, and neurodegeneration. Many anti-cancer therapeutics are designed to target various aspects of apoptosis pathways, including transcriptional regulation, Bcl-2-family homeostasis, and caspase activation (Boice and Bouchier-Hayes, 2020; Carneiro and El-Deiry, 2020; Diepstraten et al., 2022). As mentioned above, genes encoding regulators of actin dynamics also frequently exhibit expression changes in cancerous cells (Molinie and Gautreau, 2018). Thus, whereas the functions of WASP-family proteins in cell motility fit with proto-oncogenic or pro-metastasis roles, the findings that some components of the actin nucleation machinery can promote cell death hold promise for their potential as mediators of tumor suppression.
6. The nuclear options
All of the previously discussed functions for the Arp2/3 complex and WASP-family members take place in the cytoplasm, either beneath the inner leaflet of the plasma membrane, at the surface of an organelle, or elsewhere in the cytosol to create a novel cytoskeletal compartment. However, other work in recent years has revealed that the most understudied location of actin – the nucleus – is ripe for new discoveries of cytoskeletal functions. For a long time, the mere presence of actin, as well as its polymerization and organization in the nucleus, were debated (Pederson, 2008). Now, functions for actin in the nucleus are not only accepted, but studies on nucleation factors around and inside the nucleus have become more common.
The nucleus is a dynamic organelle, and the actin cytoskeleton influences multiple nuclear processes (Fig. 6). On the cytoplasmic side, the actin cytoskeleton is mechanically connected to the nucleus through transmembrane complexes that allow it to transfer forces across the nuclear envelope, resulting in changes in morphology and positioning, as well as alterations in chromatin organization and gene expression (Davidson and Cadot, 2021; Lusk and King, 2017; Makhija et al., 2016; Mishra and Levy, 2022; Seirin-Lee et al., 2019; Ulferts et al., 2021). Actin and actin-binding proteins also shuttle between the cytosol and nucleus, where they participate in several functions such as transcription, repair of damaged DNA, and chromatin dynamics, the latter of which takes place during both interphase and mitosis. While Arp2/3 complex subunits and WASP-family members can be found in the nucleus and contain nuclear localization and export signals (Suetsugu and Takenawa, 2003; Taylor et al., 2010; Verboon et al., 2015; Zuchero et al., 2012), their specific roles in these diverse nuclear processes are just beginning to be uncovered.
Fig. 6.
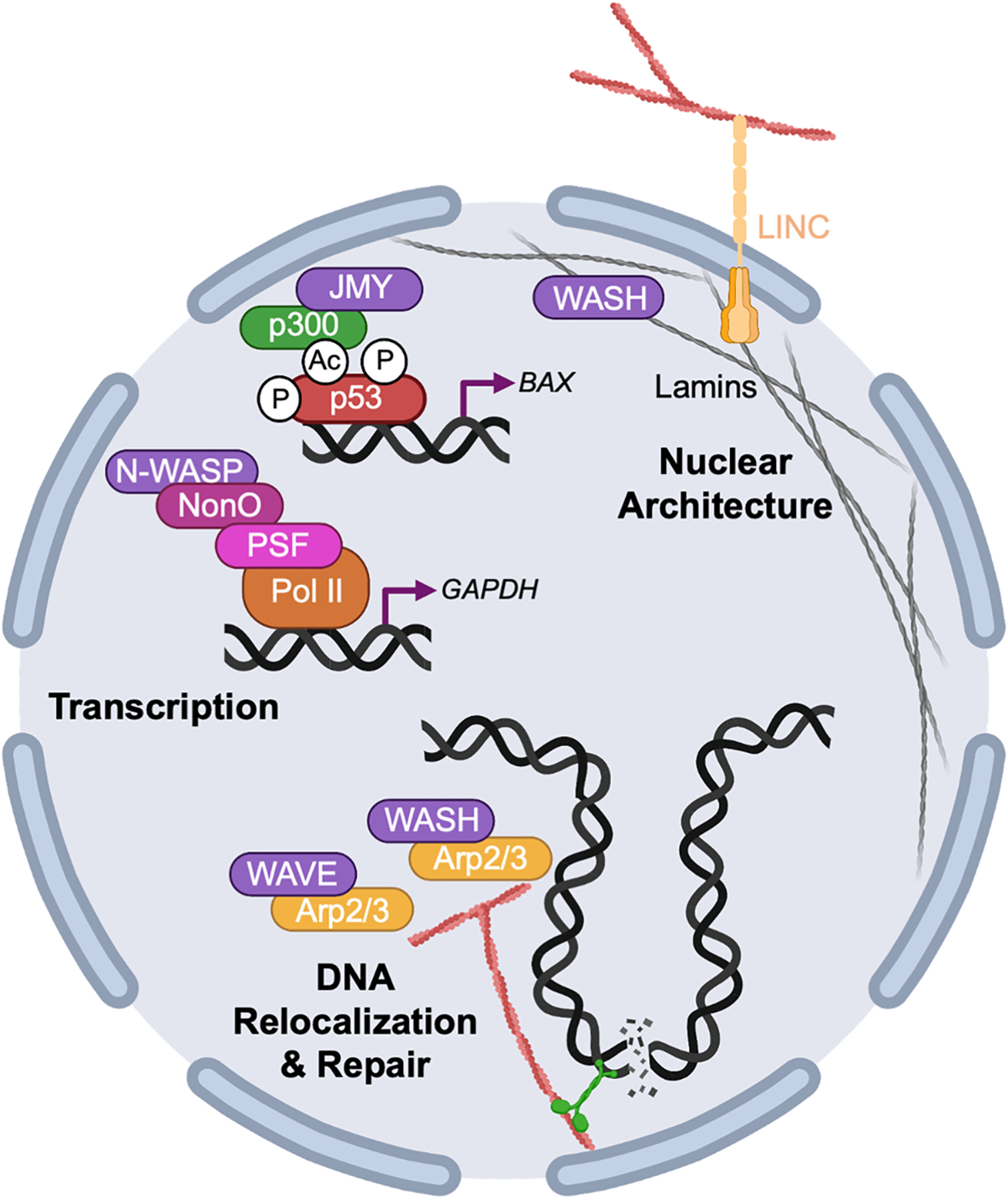
Nuclear architecture, transcription, and DNA repair are controlled by actin nucleation factors. The actin cytoskeleton and actin-binding proteins influence nuclear architecture, organization, and function. The cytoplasmic actin cytoskeleton is connected to the nucleus through transmembrane complexes that allow it to transfer mechanical forces to the nuclear envelope, resulting in changes in nuclear morphology and positioning. WASH can interact with nuclear lamins to change nuclear architecture and thereby alter chromatin organization and gene expression. N-WASP and JMY more directly contribute to transcriptional regulation by interacting with chromatin remodeling complexes, RNA polymerases, and transcription factors. WASH and WAVE collaborate with the Arp2/3 complex and myosin motor proteins to drive the relocalization of damaged DNA to sites of repair in the nuclear periphery.
Nuclear actin contributes to transcriptional regulation by interacting with chromatin remodeling complexes, RNA polymerases, and transcription factors (Hofmann et al., 2004; Hu et al., 2004; Kapoor and Shen, 2014; Miralles and Visa, 2006; Percipalle, 2013; Philimonenko et al., 2004; Vartiainen et al., 2007). WASP, the titular member of the WASP-family, accumulates in the nuclei of differentiating T-helper cells, where it associates with chromatin remodeling complexes to influence histone modifications and transcription through apparently Arp2/3-independent mechanisms (Sarkar et al., 2014; Taylor et al., 2010). Alterations in chromatin organization and gene expression are also found in cells from patients with Wiskott-Aldrich Syndrome (Sadhukhan et al., 2014; Sarkar et al., 2014).
The more ubiquitously-expressed family members N-WASP, WAVE1, WASH, and JMY also participate in transcriptional regulation (Fig. 6). N-WASP was the first to be directly associated with transcription, as it interacts with RNA polymerase II to affect gene expression, and the Arp2/3-activating capacity of N-WASP appears to be important for its function in transcriptional regulation (Suetsugu and Takenawa, 2003; Wu et al., 2006). More recently, N-WASP was found to be required for RNA polymerase II and nuclear actin clustering, which drives serum-induced transcriptional programs (Wei et al., 2020). In Xenopus, WAVE1 localizes to nuclei and impacts the expression of developmental genes during nuclear reprogramming and embryogenesis (Miyamoto et al., 2013, 2011). Whether WAVE1 or the other WAVEs play similar roles in mammals is unclear. In mouse hematopoietic stem cells, WASH interacts with nucleosome remodeling complexes to affect chromatin accessibility and gene expression (Xia et al., 2014). JMY, as stated above, was discovered as a p53 and p300 cofactor that increases transcription of the pro-apoptotic gene BAX (Shikama et al., 1999). To further define the connection between the actin nucleation machinery and transcriptional regulation, it would be interesting to decipher which WASP-family members associate with different RNA polymerase complexes during promoter-specific gene expression under distinct experimental conditions.
The actin cytoskeleton may also alter gene expression indirectly by controlling intranuclear organization (Fig. 6). Evidence from Drosophila indicates that WASH interacts with nuclear lamins to govern the global architecture of the nucleus, with the depletion or expression of mutant forms of WASH leading to a multitude of nuclear deformation phenotypes (Verboon et al., 2015). The observation that WASH depletion also changes histone modification patterns suggests that chromatin accessibility may be altered, opening new avenues of investigation into how actin assembly factors control gene expression via several distinct mechanisms. It also remains to be seen if the actions of WASH in nuclear organization involve the actin cytoskeleton and whether the findings in flies are echoed in mammalian systems.
One of the most exciting new nuclear functions for actin nucleators, nucleation-promoting factors, and even myosin motor proteins has come from studies of DNA damage and repair (Caridi et al., 2019). Technical advances using fluorescent fusion proteins to visualize nuclear G- and F-actin spurred the discovery of actin filament formation inside the nucleus in response to several types of stress (Belin et al., 2014; Melak et al., 2017). Following DNA damage, the nucleators Formin-2, Spire1, and Spire2 assemble nuclear actin filaments (Belin et al., 2015), suggesting an important role for nucleoplasmic actin polymerization in recognizing, reorganizing, and/or repairing damaged DNA. Studies in Drosophila and mammalian cells next showed that nuclear DNA damage responses also rely on the Arp2/3 complex (Fig. 6).
Chemical inhibition, transient depletion, or permanent deletion of the Arp2/3 complex leads to significant defects in the cellular reactions to DNA damage, such as the apoptotic responses described above and DNA repair pathways (Caridi et al., 2018; Schrank et al., 2018). To enable DNA repair in both insect and mammalian cells following ionizing radiation, endonuclease activation, or exposure to other genotoxic agents, the Arp2/3 complex polymerizes actin in the nucleus (Caridi et al., 2018; Schrank et al., 2018). In Drosophila cells, the Arp2/3 complex assembles actin in the vicinity of heterochromatic double-stranded DNA (dsDNA) break sites to provide tracks for their myosin-dependent relocalization to the nuclear periphery where repair takes place (Caridi et al., 2018). This movement appears to involve the Drosophila orthologs of WAVE and WASH, but not WASP, and depletion of the former factors or the Arp2/3 complex in larvae causes genomic instability and aneuploidy (Caridi et al., 2018).
In contrast to the clear findings in the fly model, the identities of the WASP-family proteins that enable Arp2/3-dependent DNA repair in mammalian cells are debated. In U2OS osteosarcoma cells, WASP was reported to cooperate with the Arp2/3 complex for clustering DNA containing double-stranded breaks (Schrank et al., 2018). However, the expression of WASP in non-hematopoietic cells is controversial, and in other human cell lines WASH was found to promote repair of etoposide-induced DNA breaks by interacting with components of the non-homologous end joining machinery (Wang et al., 2022).
Despite this uncertainty upstream of Arp2/3, it is noteworthy that the complex is not only important for repair of the severe damage that is created by acute experimental manipulations like radiation exposures or chemical treatments. The Arp2/3 complex is also crucial for repair of endogenously-generated DNA breaks in mouse fibroblasts (Haarer et al., 2023). This function of the Arp2/3 complex in preventing the accumulation of damaged DNA in mammalian cells is emphasized by the findings that deletion of the complex from mouse fibroblasts results in dsDNA breaks, chromosome segregation abnormalities, chromatin fragments that incorporate into micronuclei, and ultimately cellular senescence (Haarer et al., 2023).
Some of the latter observations are reminders that the Arp2/3 complex has key functions not just during interphase, but throughout the cell cycle. For example, complexes containing the ArpC1B isoform are important for the progression of G1 to S phase (Molinie et al., 2019). This transition appears to be dependent on the WAVE complex, although other WASP-family members may also contribute (Molinie et al., 2019). Actin and Arp2/3 influence chromosome dynamics during meiosis and mitosis as well. Actin filaments are found at meiotic spindles in mouse oocytes where they control spindle migration and affect chromosome alignment and segregation (Azoury et al., 2008; Mogessie and Schuh, 2017). The Arp2/3 complex directs microtubule spindle positioning in mouse oocytes via actin filament-dependent cytoplasmic streaming (Sun et al., 2011b; Yi et al., 2011). WAVE2, JMY, and WHAMM seem to be important in this system, as depletion of each factor leads to changes in Arp2/3 localization as well as alterations in spindle positioning, stability, and migration (Huang et al., 2013; Jo et al., 2021; Liu et al., 2012; Sun et al., 2011a, 2011c). The Arp2/3 complex also localizes to F-actin structures at centrosomes in multiple mammalian cell types, and inhibition of the complex reduces centrosomal actin levels and perturbs microtubule spindle assembly (Farina et al., 2019, 2016; Inoue et al., 2019; Plessner et al., 2019). Further, deletion of the complex causes chromosome segregation defects and increased micronucleus formation (Caridi et al., 2018; Haarer et al., 2023). While it appears that the Arp2/3 complex is a major player in multiple aspects of meiosis and mitosis, the mechanisms underlying how the specific WASP-family members and their upstream regulators coordinate these processes remain less understood. The pathways of nucleation factor recruitment and activation that drive proper chromatin dynamics and nuclear division will hopefully be uncovered in the future.
When considering the functions of actin dynamics in health and disease, it goes without saying that the inability to repair damaged DNA or to properly segregate chromosomes would expectedly have major deleterious effects on cells and, consequently, organisms. The relationships between aneuploidy and cancer are well discussed (Gordon et al., 2012; Li and Zhu, 2022), as are the connections between senescence and the pathologies of organismal aging (Di Micco et al., 2021; Gasek et al., 2021; Gorgoulis et al., 2019; McHugh and Gil, 2018). Furthermore, alterations in the concentration and organization of nuclear actin have been linked to cancers, neurodegenerative diseases, and myopathies. Shifts in the balance between cytosolic and nuclear actin pools can affect tumorigenesis, where increased levels of nuclear actin contribute to the ability to escape quiescence and acquire cancerous characteristics (Fiore et al., 2017; Spencer et al., 2011). In addition, the accumulation of F-actin rods in the nuclei of neurons or muscle cells is a common cell stress response and has been observed in samples from patients with neurodegenerative and muscle disorders (Bamburg et al., 2010; Bathe et al., 2007; Clarkson et al., 2004; Costa et al., 2004; Domazetovska et al., 2007; Minamide et al., 2000; Munsie et al., 2011).
The discoveries of actin cytoskeletal functions in the nucleus provide important contributions to discussions about the potentially opposing roles of actin nucleation factors in pro-metastasis (motility) and pro-survival (autophagy) mechanisms versus anti-proliferative (apoptosis, senescence) processes. The observations that DNA repair pathways and proper transcriptional responses to environmental insults can safeguard cells against the mutations and genomic instability that lead to cancer further support the tumor suppressive perspective. Therefore, continuing to define the activities of the actin assembly machinery in aspects of nuclear biology like chromatin dynamics, gene expression, and DNA repair could offer new insights into tumorigenesis and provide innovative means for understanding and modulating cellular stress response mechanisms.
7. Conclusion
The Arp2/3 complex and WASP-family proteins are well-established players in driving cell motility and shaping the endolysosome system. The more recent emergence of their functions in autophagy, apoptosis, and chromatin dynamics has created new branching-off points for experimental exploration. Undoubtedly, there is still much to be learned about the molecular, cellular, and organismal functions of the nucleation factors. By characterizing the intersections between the actin assembly machinery and the diverse macromolecular complexes that govern proteostasis, apoptotic pathways, and nuclear organization, we will gain a more complete picture of the mechanisms underlying such fundamental cellular activities. Finally, the study of natural variants, pathogenic mutations, alternatively-spliced isoforms, combinatorial complexity, and post-translational modifications also holds great promise for the future. Deciphering how such structural alterations in the Arp2/3 complex and WASP-family members precisely affect their functions in specific cellular processes will provide great clarity in our understanding of human health and disease.
Acknowledgments
We thank Alyssa Coulter and Corey Theodore for their comments on this manuscript. KGC was supported by National Institutes of Health grants R01-GM107441 and K02-AG050774. Portions of the figures were created using BioRender.
Abbreviations:
- Arp2/3
Actin-related protein 2/3
- ALS
Autophagosome-Lysosome System
- Cyto c
Cytochrome c
- DISC
Death-Inducing Signaling Complex
- JMY
Junction Mediating and regulatorY protein
- MOMP
Mitochondrial Outer Membrane Permeabilization
- N-WASP
Neuronal Wiskott-Aldrich Syndrome Protein
- UPS
Ubiquitin-Proteasome System
- WASP
Wiskott-Aldrich Syndrome Protein
- WASH
WASP and SCAR Homolog
- WAVE
WASP family VErprolin homolog
- WHAMM
WASP Homolog associated with Actin, Membranes, and Microtubules
- WHIMP
WAVE Homology In Membrane Protrusions
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
CRediT authorship contribution statement
Kenneth G. Campellone, Nadine M. Lebek, Virginia L. King: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. Kenneth G. Campellone: Funding acquisition, Project administration, Supervision.
References
- Abella JVG, Galloni C, Pernier J, Barry DJ, Kjaer S, Carlier M-F, Way M, 2016. Isoform diversity in the Arp2/3 complex determines actin filament dynamics. Nat. Cell Biol. 18, 76–88. 10.1038/ncb3286. [DOI] [PubMed] [Google Scholar]
- Ahn JS, Jang I-S, Kim D-I, Cho KA, Park YH, Kim K, Kwak CS, Chul Park S, 2003. Aging-associated increase of gelsolin for apoptosis resistance. Biochem. Biophys. Res. Commun. 312, 1335–1341. 10.1016/j.bbrc.2003.11.061. [DOI] [PubMed] [Google Scholar]
- Aiuti A, Biasco L, Scaramuzza S, Ferrua F, Cicalese MP, Baricordi C, Dionisio F, Calabria A, Giannelli S, Castiello MC, Bosticardo M, Evangelio C, Assanelli A, Casiraghi M, Di Nunzio S, Callegaro L, Benati C, Rizzardi P, Pellin D, Di Serio C, Schmidt M, Von Kalle C, Gardner J, Mehta N, Neduva V, Dow DJ, Galy A, Miniero R, Finocchi A, Metin A, Banerjee PP, Orange JS, Galimberti S, Valsecchi MG, Biffi A, Montini E, Villa A, Ciceri F, Roncarolo MG, Naldini L, 2013. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science 341, 1233151. 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu M, Vasan R, Serwas D, Ferrin MA, Rangamani P, Drubin DG, 2020. Principles of self-organization and load adaptation by the actin cytoskeleton during clathrin-mediated endocytosis. eLife 9, e49840. 10.7554/eLife.49840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekhina O, Burstein E, Billadeau DD, 2017. Cellular functions of WASP family proteins at a glance. J. Cell Sci. 130, 2235–2241. 10.1242/jcs.199570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrova AY, Chikina AS, Svitkina TM, 2020. Actin cytoskeleton in mesenchymal-to-amoeboid transition of cancer cells. Int. Rev. Cell Mol. Biol. 356, 197–256. 10.1016/bs.ircmb.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algeciras-Schimnich A, Shen L, Barnhart BC, Murmann AE, Burkhardt JK, Peter ME, 2002. Molecular ordering of the initial signaling events of CD95. Mol. Cell. Biol. 22, 207–220. 10.1128/MCB.22.1.207-220.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloisio FM, Barber DL, 2022. Arp2/3 complex activity is necessary for mouse ESC differentiation, times formative pluripotency, and enables lineage specification. Stem Cell Rep. 17, 1318–1333. 10.1016/j.stemcr.2022.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrysik Z, Galbraith MD, Guarnieri AL, Zaccara S, Sullivan KD, Pandey A, MacBeth M, Inga A, Espinosa JM, 2017. Identification of a core TP53 transcriptional program with highly distributed tumor suppressive activity. Genome Res 27, 1645–1657. 10.1101/gr.220533.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aplin A, Jasionowski T, Tuttle DL, Lenk SE, Dunn WA, 1992. Cytoskeletal elements are required for the formation and maturation of autophagic vacuoles. J. Cell. Physiol. 152, 458–466. 10.1002/jcp.1041520304. [DOI] [PubMed] [Google Scholar]
- Ardern H, Sandilands E, Machesky LM, Timpson P, Frame MC, Brunton VG, 2006. Src-dependent phosphorylation of Scar1 promotes its association with the Arp2/3 complex. Cell Motil. Cytoskelet. 63, 6–13. 10.1002/cm.20101. [DOI] [PubMed] [Google Scholar]
- Azoury J, Lee KW, Georget V, Rassinier P, Leader B, Verlhac M-H, 2008. Spindle positioning in mouse oocytes relies on a dynamic meshwork of actin filaments. Curr. Biol. 18, 1514–1519. 10.1016/j.cub.2008.08.044. [DOI] [PubMed] [Google Scholar]
- Ballabio A, Bonifacino JS, 2020. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 21, 101–118. 10.1038/s41580-019-0185-4. [DOI] [PubMed] [Google Scholar]
- Bamburg JR, Bernstein BW, Davis RC, Flynn KC, Goldsbury C, Jensen JR, Maloney MT, Marsden IT, Minamide LS, Pak CW, Shaw AE, Whiteman I, Wiggan O, 2010. ADF/Cofilin-actin rods in neurodegenerative diseases. Curr. Alzheimer Res. 7, 241–250. 10.2174/156720510791050902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK, 2017. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298. 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bathe FS, Rommelaere H, Machesky LM, 2007. Phenotypes of myopathy-related actin mutants in differentiated C2C12 myotubes. BMC Cell Biol. 8, 2. 10.1186/1471-2121-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Rawls JF, Saxe CL, 1998. SCAR, a WASP-related Protein, Isolated as a Suppressor of Receptor Defects in Late Dictyostelium Development. J. Cell Biol. 142, 1325–1335. 10.1083/jcb.142.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin BJ, Goins LM, Mullins RD, 2014. Comparative analysis of tools for live cell imaging of actin network architecture. Bioarchitecture 4, 189–202. 10.1080/19490992.2014.1047714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin BJ, Lee T, Mullins RD, 2015. DNA damage induces nuclear actin filament assembly by Formin −2 and Spire-½ that promotes efficient DNA repair. eLife 4, e07735. 10.7554/eLife.07735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benesch S, Lommel S, Steffen A, Stradal TEB, Scaplehorn N, Way M, Wehland J, Rottner K, 2002. Phosphatidylinositol 4,5-biphosphate (PIP2)-induced vesicle movement depends on N-WASP and involves Nck, WIP, and Grb2. J. Biol. Chem. 277, 37771–37776. 10.1074/jbc.M204145200. [DOI] [PubMed] [Google Scholar]
- Benesch S, Polo S, Lai FPL, Anderson KI, Stradal TEB, Wehland J, Rottner K, 2005. N-WASP deficiency impairs EGF internalization and actin assembly at clathrin-coated pits. J. Cell Sci. 118, 3103–3115. 10.1242/JCS.02444. [DOI] [PubMed] [Google Scholar]
- Ben-Omran T, Fahiminiya S, Sorfazlian N, Almuriekhi M, Nawaz Z, Nadaf J, Khadija KA, Zaineddin S, Kamel H, Majewski J, Tropepe V, 2015. Nonsense mutation in the WDR73 gene is associated with Galloway-Mowat syndrome. J. Med. Genet 52, 381–390. 10.1136/jmedgenet-2014-102707. [DOI] [PubMed] [Google Scholar]
- Bhola PD, Mattheyses AL, Simon SM, 2009. Spatial and temporal dynamics of mitochondrial membrane permeability waves during apoptosis. Biophys. J. 97, 2222–2231. 10.1016/j.bpj.2009.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biber G, Ben-Shmuel A, Sabag B, Barda-Saad M, 2020. Actin regulators in cancer progression and metastases: From structure and function to cytoskeletal dynamics. Int. Rev. Cell Mol. Biol. 356, 131–196. 10.1016/bs.ircmb.2020.05.006. [DOI] [PubMed] [Google Scholar]
- Blom M, Reis K, Nehru V, Blom H, Gad AKB, Aspensträm P, 2015. RhoD is a Golgi component with a role in anterograde protein transport from the ER to the plasma membrane. Exp. Cell Res. 333, 208–219. 10.1016/j.yexcr.2015.02.023. [DOI] [PubMed] [Google Scholar]
- Bock FJ, Tait SWG, 2020. Mitochondria as multifaceted regulators of cell death. Nat. Rev. Mol. Cell Biol. 21, 85–100. 10.1038/s41580-019-0173-8. [DOI] [PubMed] [Google Scholar]
- Boice A, Bouchier-Hayes L, 2020. Targeting apoptotic caspases in cancer. Biochim. Biophys. Acta Mol. Cell Res. 1867, 118688 10.1016/j.bbamcr.2020.118688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouma G, Burns SO, Thrasher AJ, 2009. Wiskott–Aldrich Syndrome: Immunodeficiency resulting from defective cell migration and impaired immunostimulatory activation. Immunobiology 214, 778–790. 10.1016/j.imbio.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DA, Rao J, Mollet G, Schapiro D, Daugeron M-C, Tan W, Gribouval O, Boyer O, Revy P, Jobst-Schwan T, Schmidt JM, Lawson JA, Schanze D, Ashraf S, Boddaert N, Collinet B, Martin G, Liger D, Lovric S, Furlano M, Guerrera IC, Sanchez-Ferras O, Menten B, Vergult S, De Rocker N, Airik M, Hermle T, Shril S, Widmeier E, Gee HY, Choi W-I, Sadowski CE, Pabst WL, Warejko J, Daga A, LeBerre TB, Matejas V, Behnam B, Beeson B, Begtrup A, Bruce M, Ch’ng G-S, Lin S-P, Chang J-H, Chen C-H, Cho MT, Gipson PE, Hsu C-H, Kari JA, Ke Y-Y, Kiraly-Borri C, Lai W, Lemyre E, Littlejohn RO, Masri A, Moghtaderi M, Nakamura K, Praet M, Prasad C, Prytula A, Roeder E, Rump P, Schnur RE, Shiihara T, Sinha M, Soliman NA, Soulami K, Sweetser DA, Tsai W-H, Tsai J-D, Vester U, Viskochil DH, Vatanavicharn N, Waxler JL, Wolf MTF, Wong S-N, Poduri A, Truglio G, Mane S, Lifton RP, Bouchard M, Kannu P, Chitayat D, Magen D, Calleweart B, van Tilbeurgh H, Zenker M, Antignac C, Hildebrandt F, 2017. Mutations in the evolutionarily highly conserved KEOPS complex genes cause nephrotic syndrome with microcephaly. Nat. Genet 49, 1529–1538. 10.1038/ng.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DA, Shril S, Sinha A, Schneider R, Tan W, Ashraf S, Hermle T, Jobst-Schwan T, Widmeier E, Majmundar AJ, Daga A, Warejko JK, Nakayama M, Schapiro D, Chen J, Airik M, Rao J, Schmidt JM, Hoogstraten CA, Hugo H, Meena J, Lek M, Laricchia KM, Bagga A, Hildebrandt F, 2018. Mutations in WDR4 as a new cause of Galloway-Mowat syndrome. Am. J. Med. Genet. A 176, 2460–2465. 10.1002/ajmg.a.40489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Goode BL, 2013. Formins at a glance. J. Cell Sci. 126, 1–7. 10.1242/jcs.107250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigida I, Zoccolillo M, Cicalese MP, Pfajfer L, Barzaghi F, Scala S, Oleaga-Quintas C, Álvarez-Álvarez JA, Sereni L, Giannelli S, Sartirana C, Dionisio F, Pavesi L, Benavides-Nieto M, Basso-Ricci L, Capasso P, Mazzi B, Rosain J, Marcus N, Lee YN, Somech R, Degano M, Raiola G, Caorsi R, Picco P, Moncada Velez M, Khourieh J, Arias AA, Bousfiha A, Issekutz T, Issekutz A, Boisson B, Dobbs K, Villa A, Lombardo A, Neven B, Moshous D, Casanova J-L, Franco JL, Notarangelo LD, Scielzo C, Volpi S, Dupré L, Bustamante J, Gattorno M, Aiuti A, 2018. T-cell defects in patients with ARPC1B germline mutations account for combined immunodeficiency. Blood 132, 2362–2374. 10.1182/blood-2018-07-863431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu W, Lim KB, Yu YH, Chou AM, Sudhaharan T, Ahmed S, 2010. Cdc42 interaction with N-WASP and Toca-1 regulates membrane tubulation, vesicle formation and vesicle motility: implications for endocytosis. PloS One 5, e12153. 10.1371/journal.pone.0012153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette DT, Shao L, Ott C, Pasapera AM, Fischer RS, Baird MA, Der Loughian C, Delanoe-Ayari H, Paszek MJ, Davidson MW, Betzig E, Lippincott-Schwartz J, 2014. A contractile and counterbalancing adhesion system controls the 3D shape of crawling cells. J. Cell Biol. 205, 83–96. 10.1083/jcb.201311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K, Guilluy C, 2016. Focal adhesions, stress fibers and mechanical tension. Exp. Cell Res. 343, 14–20. 10.1016/j.yexcr.2015.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD, 2010. A nucleator arms race: Cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 11, 237–251. 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Cheng H-C, Robbins D, Siripala AD, McGhie EJ, Hayward RD, Welch MD, Rosen MK, Koronakis V, Leong JM, 2008a. Repetitive N-WASP-binding elements of the enterohemorrhagic Escherichia coli effector EspF(U) synergistically activate actin assembly. PLoS Pathog. 4, e1000191 10.1371/journal.ppat.1000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Webb NJ, Znameroski EA, Welch MD, 2008b. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell 134, 148–161. 10.1016/j.cell.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Yonis A, Vaghela M, Barriga EH, Chugh P, Smith MB, Maufront J, Lavoie G, Méant A, Ferber E, Bovellan M, Alberts A, Bertin A, Mayor R, Paluch EK, Roux PP, Jégou A, Romet-Lemonne G, Charras G, 2020. SPIN90 associates with mDia1 and the Arp2/3 complex to regulate cortical actin organization. Nat. Cell Biol. 22, 803–814. 10.1038/s41556-020-0531-y. [DOI] [PubMed] [Google Scholar]
- Caridi CP, D’Agostino C, Ryu T, Zapotoczny G, Delabaere L, Li X, Khodaverdian VY, Amaral N, Lin E, Rau AR, Chiolo I, 2018. Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature 559, 54–60. 10.1038/s41586-018-0242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caridi CP, Plessner M, Grosse R, Chiolo I, 2019. Nuclear actin filaments in DNA repair dynamics. Nat. Cell Biol. 21, 1068–1077. 10.1038/s41556-019-0379-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson SR, Simonsen A, 2015. Membrane dynamics in autophagosome biogenesis. J. Cell Sci. 128, 198–205. 10.1242/jcs.141036. [DOI] [PubMed] [Google Scholar]
- Carneiro BA, El-Deiry WS, 2020. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 17, 395–417. 10.1038/s41571-020-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, Waterman CM, 2015. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat. Cell Biol. 17, 955–963. 10.1038/ncb3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R, Lee M, Higgs HN, 2021. Multiple roles for actin in secretory and endocytic pathways. Curr. Biol. 31, R603–R618. 10.1016/J.CUB.2021.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charras GT, Hu C-K, Coughlin M, Mitchison TJ, 2006. Reassembly of contractile actin cortex in cell blebs. J. Cell Biol. 175, 477–490. 10.1083/jcb.200602085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Brinkmann K, Chen Z, Pak CW, Liao Y, Shi S, Henry L, Grishin NV, Bogdan S, Rosen MK, 2014. The WAVE Regulatory Complex Links Diverse Receptors to the Actin Cytoskeleton. Cell 156, 195. 10.1016/J.CELL.2013.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Chou H-T, Brautigam CA, Xing W, Yang S, Henry L, Doolittle LK, Walz T, Rosen MK, 2017. Rac1 GTPase activates the WAVE regulatory complex through two distinct binding sites. eLife 6, e29795. 10.7554/eLife.29795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Borek D, Padrick SB, Gomez TS, Metlagel Z, Ismail AM, Umetani J, Billadeau DD, Otwinowski Z, Rosen MK, 2010. Structure and control of the actin regulatory WAVE complex, 4687323 468 Nature 2010, 533–538. 10.1038/nature09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Arumugam TV, Liu D, Khatri RG, Mustafa K, Kwak S, Ling H-P, Gonzales C, Xin O, Jo D-G, Guo Z, Mark RJ, Mattson MP, 2007. Pancortin-2 interacts with WAVE1 and Bcl-xL in a mitochondria-associated protein complex that mediates ischemic neuronal death. J. Neurosci. 27, 1519–1528. 10.1523/JNEUROSCI.5154-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H-C, Skehan BM, Campellone KG, Leong JM, Rosen MK, 2008. Structural Mechanism of WASP Activation by the Enterohaemorrhagic E. coli Effector EspFU. Nature 454, 1009–1013. 10.1038/nature07160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra D, Nosworthy NJ, dos Remedios CG, 2005. The N-terminal fragment of gelsolin inhibits the interaction of DNase I with isolated actin, but not with the cofilin-actin complex. Proteomics 5, 3131–3136. 10.1002/pmic.200401127. [DOI] [PubMed] [Google Scholar]
- Chorev DS, Moscovitz O, Geiger B, Sharon M, 2014. Regulation of focal adhesion formation by a vinculin-Arp2/3 hybrid complex. Nat. Commun. 5, 3758. 10.1038/ncomms4758. [DOI] [PubMed] [Google Scholar]
- Chua BT, Volbracht C, Tan KO, Li R, Yu VC, Li P, 2003. Mitochondrial translocation of cofilin is an early step in apoptosis induction. Nat. Cell Biol. 5, 1083–1089. 10.1038/ncb1070. [DOI] [PubMed] [Google Scholar]
- Cioca DP, Kitano K, 2002. Induction of apoptosis and CD10/neutral endopeptidase expression by jaspamide in HL-60 line cells. Cell. Mol. Life Sci. 59, 1377–1387. 10.1007/s00018-002-8515-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson E, Costa CF, Machesky LM, 2004. Congenital myopathies: diseases of the actin cytoskeleton. J. Pathol. 204, 407–417. 10.1002/path.1648. [DOI] [PubMed] [Google Scholar]
- Colin E, Huynh Cong E, Mollet G, Guichet A, Gribouval O, Arrondel C, Boyer O, Daniel L, Gubler M-C, Ekinci Z, Tsimaratos M, Chabrol B, Boddaert N, Verloes A, Chevrollier A, Gueguen N, Desquiret-Dumas V, Ferré M, Procaccio V, Richard L, Funalot B, Moncla A, Bonneau D, Antignac C, 2014. Loss-of-Function Mutations in WDR73 Are Responsible for Microcephaly and Steroid-Resistant Nephrotic Syndrome: Galloway-Mowat Syndrome. Am. J. Hum. Genet 95, 637–648. 10.1016/j.ajhg.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory GOC, Garg R, Cramer R, Ridley AJ, 2002. Phosphorylation of tyrosine 291 enhances the ability of WASp to stimulate actin polymerization and filopodium formation. Wiskott-Aldrich Syndrome protein. J. Biol. Chem. 277, 45115–45121. 10.1074/jbc.M203346200. [DOI] [PubMed] [Google Scholar]
- Costa CF, Rommelaere H, Waterschoot D, Sethi KK, Nowak KJ, Laing NG, Ampe C, Machesky LM, 2004. Myopathy mutations in alpha-skeletal-muscle actin cause a range of molecular defects. J. Cell Sci. 117, 3367–3377. 10.1242/jcs.01172. [DOI] [PubMed] [Google Scholar]
- Courtemanche N, 2018. Mechanisms of formin-mediated actin assembly and dynamics. Biophys. Rev. 10, 1553–1569. 10.1007/s12551-018-0468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtland JL, Bradshaw TW, Waitt G, Soderblom EJ, Ho T, Rajab A, Vancini R, Kim IH, Soderling SH, 2021. Genetic disruption of WASHC4 drives endo-lysosomal dysfunction and cognitive-movement impairments in mice and humans. eLife 10. 10.7554/ELIFE.61590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutts AS, La Thangue NB, 2015. Actin nucleation by WH2 domains at the autophagosome. Nat. Commun. 6. 10.1038/ncomms8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutts AS, Boulahbel H, Graham A, La Thangue NB, 2007. Mdm2 targets the p53 transcription cofactor JMY for degradation. EMBO Rep. 8, 84–90. 10.1038/sj.embor.7400855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou SZ, Chatterjee M, Pollard TD, 2022, Mechanism of actin filament branch formation by Arp2/3 complex revealed by a high-resolution cryo-EM structure of the branch junction. Proc. Natl. Acad. Sci. U. S. A. 119, e2206722119. 10.1073/pnas.2206722119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutts AS, Weston L, La Thangue NB, 2009, A transcription co-factor integrates cell adhesion and motility with the p53 response. Proc. Natl. Acad. Sci. U. S. A. 106, 19872–19877. 10.1073/pnas.0906785106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czuchra A, Wu X, Meyer H, van Hengel J, Schroeder T, Geffers R, Rottner K, Brakebusch C, 2005. Cdc42 is not essential for filopodium formation, directed migration, cell polarization, and mitosis in fibroblastoid cells. Mol. Biol. Cell 16, 4473–4484. 10.1091/mbc.E05-01-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai A, Yu L, Wang HW, 2019. WHAMM initiates autolysosome tubulation by promoting actin polymerization on autolysosomes. Nat. Commun. 10. 10.1038/S41467-019-11694-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danial NN, Gramm CF, Scorrano L, Zhang C-Y, Krauss S, Ranger AM, Datta SR, Greenberg ME, Licklider LJ, Lowell BB, Gygi SP, Korsmeyer SJ, 2003. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature 424, 952–956. 10.1038/nature01825. [DOI] [PubMed] [Google Scholar]
- Davidson PM, Cadot B, 2021. Actin on and around the Nucleus. Trends Cell Biol. 31, 211–223. 10.1016/j.tcb.2020.11.009. [DOI] [PubMed] [Google Scholar]
- Demonacos C, Krstic-Demonacos M, La Thangue NB, 2001. A TPR motif cofactor contributes to p300 activity in the p53 response. Mol. Cell 8, 71–84. 10.1016/s1097-2765(01)00277-5. [DOI] [PubMed] [Google Scholar]
- Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A, 2009. The Arp2/3 Activator WASH Controls the Fission of Endosomes through a Large Multiprotein Complex. Dev. Cell 17, 712–723. 10.1016/J.DEVCEL.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Derry JM, Ochs HD, Francke U, 1994. Isolation of a Novel Gene Mutated in Wiskott-Aldrich Syndrome. Cell 76, 635–644. [DOI] [PubMed] [Google Scholar]
- Desouza M, Gunning PW, Stehn JR, 2012. The actin cytoskeleton as a sensor and mediator of apoptosis. Bioarchitecture 2, 75–87. 10.4161/bioa.20975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Krizhanovsky V, Baker D, d’Adda di Fagagna F, 2021. Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 22, 75–95. 10.1038/s41580-020-00314-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickens LS, Powley IR, Hughes MA, MacFarlane M, 2012. The “complexities” of life and death: death receptor signalling platforms. Exp. Cell Res. 318, 1269–1277. 10.1016/j.yexcr.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Diepstraten ST, Anderson MA, Czabotar PE, Lessene G, Strasser A, Kelly GL, 2022. The manipulation of apoptosis for cancer therapy using BH3-mimetic drugs. Nat. Rev. Cancer 22, 45–64. 10.1038/s41568-021-00407-4. [DOI] [PubMed] [Google Scholar]
- Dimchev V, Lahmann I, Koestler SA, Kage F, Dimchev G, Steffen A, Stradal TEB, Vauti F, Arnold H-H, Rottner K, 2021. Induced Arp2/3 Complex Depletion Increases FMNL2/3 Formin Expression and Filopodia Formation. Front. Cell Dev. Biol. 9, 634708 10.3389/fcell.2021.634708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B, Narvaez-Ortiz HY, Singh Y, Hocky GM, Chowdhury S, Nolen BJ, 2022, Structure of Arp2/3 complex at a branched actin filament junction resolved by single-particle cryo-electron microscopy. Proc. Natl. Acad. Sci. U. S. A. 119, e2202723119. 10.1073/pnas.2202723119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobramysl U, Jarsch IK, Inoue Y, Shimo H, Richier B, Gadsby JR, Mason J, Szałapak A, Ioannou PS, Correia GP, Walrant A, Butler R, Hannezo E, Simons BD, Gallop JL, 2021. Stochastic combinations of actin regulatory proteins are sufficient to drive filopodia formation. J. Cell Biol. 220, e202003052 10.1083/jcb.202003052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazetovska A, Ilkovski B, Kumar V, Valova VA, Vandebrouck A, Hutchinson DO, Robinson PJ, Cooper ST, Sparrow JC, Peckham M, North KN, 2007. Intranuclear rod myopathy: molecular pathogenesis and mechanisms of weakness. Ann. Neurol. 62, 597–608. 10.1002/ana.21200. [DOI] [PubMed] [Google Scholar]
- Domingo-Gallego A, Furlano M, Pybus M, Barraca D, Martínez AB, Mora Muñoz E, Torra R, Ars E, 2019. Novel homozygous OSGEP gene pathogenic variants in two unrelated patients with Galloway-Mowat syndrome: case report and review of the literature. BMC Nephrol. 20, 126. 10.1186/s12882-019-1317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R, 2016. The WH2 domain and actin nucleation: necessary but insufficient. Trends Biochem. Sci. 41, 478–490. 10.1016/j.tibs.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorstyn L, Akey CW, Kumar S, 2018. New insights into apoptosome structure and function. Cell Death Differ. 25, 1194–1208. 10.1038/s41418-017-0025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubielecka PM, Ladwein KI, Xiong X, Migeotte I, Chorzalska A, Anderson KV, Sawicki JA, Rottner K, Stradal TE, Kotula L, 2011. Essential role for Abi1 in embryonic survival and WAVE2 complex integrity. Proc. Natl. Acad. Sci. U. S. A. 108, 7022–7027. 10.1073/pnas.1016811108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duleh SN, Welch MD, 2010. WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskelet 67, 193. 10.1002/CM.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW, 2002. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418, 790–793. 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- Fais S, De Milito A, Lozupone F, 2005. The role of FAS to ezrin association in FAS-mediated apoptosis. Apoptosis 10, 941–947. 10.1007/s10495-005-0478-2. [DOI] [PubMed] [Google Scholar]
- Faix J, Rottner K, 2022. Ena/VASP proteins in cell edge protrusion, migration and adhesion. J. Cell Sci. 135, jcs259226 10.1242/jcs.259226. [DOI] [PubMed] [Google Scholar]
- Farina F, Gaillard J, Guérin C, Couté Y, Sillibourne J, Blanchoin L, Théry M, 2016. The centrosome is an actin-organizing centre. Nat. Cell Biol. 18, 65–75. 10.1038/ncb3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina F, Ramkumar N, Brown L, Samandar Eweis D, Anstatt J, Waring T, Bithell J, Scita G, Thery M, Blanchoin L, Zech T, Baum B, 2019. Local actin nucleation tunes centrosomal microtubule nucleation during passage through mitosis. EMBO J. 38, e99843 10.15252/embj.201899843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fäßler F, Javoor MG, Datler J, Döring H, Hofer FW, Dimchev G, Hodirnau V-V, Faix J, Rottner K, Schur FKM, 2023. ArpC5 isoforms regulate Arp2/3 complex-dependent protrusion through differential Ena/VASP positioning. Sci. Adv. 9, eadd6495. 10.1126/sciadv.add6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore APZP, Spencer VA, Mori H, Carvalho HF, Bissell MJ, Bruni-Cardoso A, 2017. Laminin-111 and the Level of Nuclear Actin Regulate Epithelial Quiescence via Exportin-6. Cell Rep. 19, 2102–2115. 10.1016/j.celrep.2017.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer RS, Lam PY, Huttenlocher A, Waterman CM, 2019. Filopodia and focal adhesions: An integrated system driving branching morphogenesis in neuronal pathfinding and angiogenesis. Dev. Biol. 451, 86. 10.1016/J.YDBIO.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokin AI, Gautreau AM, 2021. Assembly and Activity of the WASH Molecular Machine: Distinctive Features at the Crossroads of the Actin and Microtubule Cytoskeletons. Front. Cell Dev. Biol. 9, 658865 10.3389/fcell.2021.658865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokin AI, David V, Oguievetskaia K, Derivery E, Stone CE, Cao L, Rocques N, Molinie N, Henriot V, Aumont-Nicaise M, Hinckelmann M-V, Saudou F, Le Clainche C, Carter AP, Romet-Lemonne G, Gautreau AM, 2021. The Arp1/11 minifilament of dynactin primes the endosomal Arp2/3 complex. Sci. Adv. 7, eabd5956 10.1126/sciadv.abd5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Prior P, Salvesen GS, 2004. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem. J. 384, 201–232. 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung TS, Ji W-K, Higgs HN, Chakrabarti R, 2019. Two distinct actin filament populations have effects on mitochondria, with differences in stimuli and assembly factors. J. Cell Sci. 132, jcs234435. 10.1242/jcs.234435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung TS, Chakrabarti R, Kollasser J, Rottner K, Stradal TEB, Kage F, Higgs HN, 2022. Parallel kinase pathways stimulate actin polymerization at depolarized mitochondria. e8 Curr. Biol. CB 32, 1577–1592. 10.1016/j.cub.2022.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloni C, Carra D, Abella JVG, Kjær S, Singaravelu P, Barry DJ, Kogata N, Guérin C, Blanchoin L, Way M, 2021. MICAL2 enhances branched actin network disassembly by oxidizing Arp3B-containing Arp2/3 complexes. J. Cell Biol. 220, e202102043 10.1083/jcb.202102043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallop JL, 2020. Filopodia and their links with membrane traffic and cell adhesion. Semin. Cell Dev. Biol. 102, 81–89. 10.1016/J.SEMCDB.2019.11.017. [DOI] [PubMed] [Google Scholar]
- Gardel ML, Schneider IC, Aratyn-Schaus Y, Waterman CM, 2010. Mechanical Integration of Actin and Adhesion Dynamics in Cell Migration. Annu. Rev. Cell Dev. Biol. 26, 315–333. 10.1146/annurev.cellbio.011209.122036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasek NS, Kuchel GA, Kirkland JL, Xu M, 2021. Strategies for Targeting Senescent Cells in Human Disease. Nat. Aging 1, 870–879. 10.1038/s43587-021-00121-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatica D, Lahiri V, Klionsky DJ, 2018. Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 20, 233–242. 10.1038/s41556-018-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautreau A, Ho HH, Li J, Steen H, Gygi SP, Kirschner MW, 2004, Purification and architecture of the ubiquitous Wave complex. Proc. Natl. Acad. Sci. U. S. A. 101, 4379–4383. 10.1073/pnas.0400628101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautreau AM, Fregoso FE, Simanov G, Dominguez R, 2022. Nucleation, stabilization, and disassembly of branched actin networks. Trends Cell Biol. 32, 421–432. 10.1016/j.tcb.2021.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genescá M, Sola A, Hotter G, 2006. Actin cytoskeleton derangement induces apoptosis in renal ischemia/reperfusion. Apoptosis 11, 563–571. 10.1007/s10495-006-4937-1. [DOI] [PubMed] [Google Scholar]
- Gligorijevic B, Wyckoff J, Yamaguchi H, Wang Y, Roussos ET, Condeelis J, 2012. N-WASP-mediated invadopodium formation is involved in intravasation and lung metastasis of mammary tumors. J. Cell Sci. 125, 724–734. 10.1242/jcs.092726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TS, Billadeau DD, 2009. A FAM21-Containing WASH Complex Regulates Retromer-Dependent Sorting. Dev. Cell 17, 699. 10.1016/J.DEVCEL.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TS, Gorman JA, de Narvajas AAM, Koenig AO, Billadeau DD, 2012. Trafficking defects in WASH-knockout fibroblasts originate from collapsed endosomal and lysosomal networks. Mol. Biol. Cell 23, 3215. 10.1091/MBC.E12-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DJ, Resio B, Pellman D, 2012. Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 13, 189–203. 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- Gorgoulis V, Adams PD, Alimonti A, Bennett DC, Bischof O, Bishop C, Campisi J, Collado M, Evangelou K, Ferbeyre G, Gil J, Hara E, Krizhanovsky V, Jurk D, Maier AB, Narita M, Niedernhofer L, Passos JF, Robbins PD, Schmitt CA, Sedivy J, Vougas K, von Zglinicki T, Zhou D, Serrano M, Demaria M, 2019. Cellular Senescence: Defining a Path Forward. Cell 179, 813–827. 10.1016/j.cell.2019.10.005. [DOI] [PubMed] [Google Scholar]
- Gourlay CW, Ayscough KR, 2005. The actin cytoskeleton: a key regulator of apoptosis and ageing. Nat. Rev. Mol. Cell Biol. 6, 583–589. 10.1038/nrm1682. [DOI] [PubMed] [Google Scholar]
- Gruenbaum-Cohen Y, Harel I, Umansky K-B, Tzahor E, Snapper SB, Shilo B-Z, Schejter ED, 2012, The actin regulator N-WASp is required for muscle-cell fusion in mice. Proc. Natl. Acad. Sci. U. S. A. 109, 11211–11216. 10.1073/pnas.1116065109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer B, Aggeli D, Viggiano S, Burke DJ, Amberg DC, 2011. Novel Interactions between Actin and the Proteasome Revealed by Complex Haploinsufficiency. PLoS Genet 7, e1002288. 10.1371/JOURNAL.PGEN.1002288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer EL, Theodore CJ, Guo S, Frier RB, Campellone KG, 2023. Genomic instability caused by Arp2/3 complex inactivation results in micronucleus biogenesis and cellular senescence. PLOS Genet 19, e1010045. 10.1371/journal.pgen.1010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner A, Bulyk ML, Jambhekar A, Lahav G, 2019. The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 20, 199–210. 10.1038/s41580-019-0110-x. [DOI] [PubMed] [Google Scholar]
- Hansen M, Rubinsztein DC, Walker DW, 2018. Autophagy as a promoter of longevity: insights from model organisms. Nat. Rev. Mol. Cell Biol. 19, 579–593. 10.1038/s41580-018-0033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y-H, Doyle JM, Ramanathan S, Gomez TS, Jia D, Xu M, Chen ZJ, Billadeau DD, Rosen MK, Potts PR, 2013. Regulation of WASH-Dependent Actin Polymerization and Protein Trafficking by Ubiquitination. Cell 152, 1051–1064. 10.1016/j.cell.2013.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M, 1997. Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299. 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Helgeson LA, Nolen BJ, 2013. Mechanism of synergistic activation of Arp2/3 complex by cortactin and N-WASP. eLife 2, e00884. 10.7554/eLife.00884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp MS, Kasturi P, Hartl FU, 2019. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 20, 421–435. 10.1038/s41580-019-0101-y. [DOI] [PubMed] [Google Scholar]
- Hoffmann L, Waclawczyk MS, Tang S, Hanschmann E-M, Gellert M, Rust MB, Culmsee C, 2021. Cofilin1 oxidation links oxidative distress to mitochondrial demise and neuronal cell death. Cell Death Dis. 12, 953. 10.1038/s41419-021-04242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, Kysela K, Goodrich JA, Lessard JL, Hope TJ, Hozak P, de Lanerolle P, 2004. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat. Cell Biol. 6, 1094–1101. 10.1038/ncb1182. [DOI] [PubMed] [Google Scholar]
- Hsieh CW, Yang WY, 2019. Omegasome-proximal PtdIns(4,5)P2 couples F-actin mediated mitoaggregate disassembly with autophagosome formation during mitophagy. Nat. Commun. 10. 10.1038/S41467-019-08924-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Zhang H, Li J, Jiang X, Zhang Y, Wu Q, Shen L, Shi J, Gao N, 2020. ROCK1 activation-mediated mitochondrial translocation of Drp1 and cofilin are required for arnidiol-induced mitochondrial fission and apoptosis. J. Exp. Clin. Cancer Res. CR 39, 37. 10.1186/s13046-020-01545-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Wu S, Hernandez N, 2004. A role for beta-actin in RNA polymerase III transcription. Genes Dev. 18, 3010–3015. 10.1101/gad.1250804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Wu D, Chen W, Yan Z, Yan C, He T, Liang Q, Shi Y, 2014, Molecular determinants of caspase-9 activation by the Apaf-1 apoptosome. Proc. Natl. Acad. Sci. U. S. A. 111, 16254–16261. 10.1073/pnas.1418000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Mullins RD, 2019. LC3 and STRAP regulate actin filament assembly by JMY during autophagosome formation. J. Cell Biol. 218, 251–266. 10.1083/jcb.201802157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Birmingham CL, Shahnazari S, Shiu J, Zheng YT, Smith AC, Campellone KG, Heo WD, Gruenheid S, Meyer T, Welch MD, Ktistakis NT, Kim PK, Klionsky DJ, Brumell JH, 2011. Antibacterial autophagy occurs at PI (3)P-enriched domains of the endoplasmic reticulum and requires Rab1 GTPase. Autophagy 7, 17–26. 10.4161/auto.7.1.13840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Ding L, Pan R, Ma P-F, Cheng P-P, Zhang C-H, Shen Y-T, Xu L, Liu Y, He X-Q, Qi Z-Q, Wang H-L, 2013. WHAMM is required for meiotic spindle migration and asymmetric cytokinesis in mouse oocytes. Histochem. Cell Biol. 139, 525–534. 10.1007/s00418-012-1051-z. [DOI] [PubMed] [Google Scholar]
- Innocenti M, 2018. New insights into the formation and the function of lamellipodia and ruffles in mesenchymal cell migration. Cell Adhes. Migr. 12, 401–416. 10.1080/19336918.2018.1448352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti M, Gerboth S, Rottner K, Lai FPL, Hertzog M, Stradal TEB, Frittoli E, Didry D, Polo S, Disanza A, Benesch S, Di Fiore PP, Carlier M-F, Scita G, 2005. Abi1 regulates the activity of N-WASP and WAVE in distinct actin-based processes. Nat. Cell Biol. 7, 969–976. 10.1038/ncb1304. [DOI] [PubMed] [Google Scholar]
- Inoue D, Obino D, Pineau J, Farina F, Gaillard J, Guerin C, Blanchoin L, Lennon-Duménil A-M, Théry M, 2019. Actin filaments regulate microtubule growth at the centrosome. EMBO J. 38, e99630 10.15252/embj.201899630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail AM, Padrick SB, Chen B, Umetani J, Rosen MK, 2009. The WAVE Regulatory Complex is Inhibited. Nat. Struct. Mol. Biol. 16, 561. 10.1038/NSMB.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Carss KJ, Duarte ST, Hartley T, Keren B, Kurian MA, Marey I, Charles P, Mendonça C, Nava C, Pfundt R, Sanchis-Juan A, van Bokhoven H, van Essen A, van Ravenswaaij-Arts C, Kernohan KM, Dyack S, K.D., Raymond FL, NIHR BioResourceCare4Rare Canada Consortium, Boycott, 2018. De Novo Truncating Mutations in WASF1 Cause Intellectual Disability with Seizures. Am. J. Hum. Genet 103, 144–153 10.1016/j.ajhg.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarsch IK, Gadsby JR, Nuccitelli A, Mason J, Shimo H, Pilloux L, Marzook B, Mulvey CM, Dobramysl U, Bradshaw CR, Lilley KS, Hayward RD, Vaughan TJ, Dobson CL, Gallop JL, 2020. A direct role for SNX9 in the biogenesis of filopodia. J. Cell Biol. 219, e201909178 10.1083/jcb.201909178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji CH, Kwon YT, 2017. Crosstalk and interplay between the ubiquitin-proteasome system and autophagy. Mol. Cells 40, 441–449. 10.14348/molcells.2017.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W, Hatch AL, Merrill RA, Strack S, Higgs HN, 2015. Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. eLife 4, e11553. 10.7554/eLife.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Gomez TS, Metlagel Z, Umetani J, Otwinowski Z, Rosen MK, Billadeau DD, 2010, WASH and WAVE actin regulators of the Wiskott–Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proc. Natl. Acad. Sci. 107, 10442–10447. 10.1073/pnas.0913293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Shirazinejad C, Wang B, Yan A, Schöneberg J, Upadhyayula S, Xu K, Drubin DG, 2022. Branched actin networks are organized for asymmetric force production during clathrin-mediated endocytosis in mammalian cells. Nat. Commun. 13, 3578. 10.1038/s41467-022-31207-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks RN, Puffenberger EG, Baple E, Harding B, Crino P, Fogo AB, Wenger O, Xin B, Koehler AE, McGlincy MH, Provencher MM, Smith JD, Tran L, Al Turki S, Chioza BA, Cross H, Harlalka GV, Hurles ME, Maroofian R, Heaps AD, Morton MC, Stempak L, Hildebrandt F, Sadowski CE, Zaritsky J, Campellone K, Morton DH, Wang H, Crosby A, Strauss KA, 2015. Recessive nephrocerebellar syndrome on the Galloway-Mowat syndrome spectrum is caused by homozygous protein-truncating mutations of WDR73. Brain 138, 2173–2190. 10.1093/brain/awv153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo Y-J, Kwon J, Jin Z-L, Namgoong S, Kwon T, Yoon S-B, Lee D-H, Kim J-S, Kim N-H, 2021. WHAMM is essential for spindle formation and spindle actin polymerization in maturing mouse oocytes. Cell Cycle 20, 225–235. 10.1080/15384101.2020.1867791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost PJ, Grabow S, Gray D, McKenzie MD, Nachbur U, Huang DCS, Bouillet P, Thomas HE, Borner C, Silke J, Strasser A, Kaufmann T, 2009. XIAP discriminates between type I and type II FAS-induced apoptosis. Nature 460, 1035–1039. 10.1038/nature08229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juin A, Spence HJ, Martin KJ, McGhee E, Neilson M, Cutiongco MFA, Gadegaard N, Mackay G, Fort L, Lilla S, Kalna G, Thomason P, Koh YWH, Norman JC, Insall RH, Machesky LM, 2019. N-WASP Control of LPAR1 Trafficking Establishes Response to Self-Generated LPA Gradients to Promote Pancreatic Cancer Cell Metastasis. Dev. Cell 51, 431. 10.1016/J.DEVCEL.2019.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien O, Wells JA, 2017. Caspases and their substrates. Cell Death Differ. 24, 1380–1389. 10.1038/cdd.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabrawala S, Zimmer MD, Campellone KG, 2020. WHIMP links the actin nucleation machinery to Src-family kinase signaling during protrusion and motility. PLOS Genet 16, e1008694. 10.1371/JOURNAL.PGEN.1008694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadzik RS, Homa KE, Kovar DR, 2020. F-Actin Cytoskeleton Network Self-Organization Through Competition and Cooperation. Annu. Rev. Cell Dev. Biol. 36, 35–60. 10.1146/annurev-cellbio-032320-094706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahr WHA, Pluthero FG, Elkadri A, Warner N, Drobac M, Chen CH, Lo RW, Li L, Li R, Li Q, Thoeni C, Pan J, Leung G, Lara-Corrales I, Murchie R, Cutz E, Laxer RM, Upton J, Roifman CM, Yeung RSM, Brumell JH, Muise AM, 2017. Loss of the Arp2/3 complex component ARPC1B causes platelet abnormalities and predisposes to inflammatory disease. Nat. Commun. 8 10.1038/NCOMMS14816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M, Roux A, 2018. Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 19, 313–326. 10.1038/nrm.2017.132. [DOI] [PubMed] [Google Scholar]
- Kakuta S, Yamaguchi J, Suzuki C, Sasaki M, Kazuno S, Uchiyama Y, 2017. Small GTPase Rab1B is associated with ATG9A vesicles and regulates autophagosome formation. FASEB J. 31, 3757–3773. 10.1096/fj.201601052R. [DOI] [PubMed] [Google Scholar]
- Kale J, Osterlund EJ, Andrews DW, 2018. BCL-2 family proteins: changing partners in the dance towards death. Cell Death Differ. 25, 65–80. 10.1038/cdd.2017.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkavan H, Green DR, 2018. MOMP, cell suicide as a BCL-2 family business. Cell Death Differ. 25, 46–55. 10.1038/cdd.2017.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM, 2010. Nanoscale architecture of integrin-based cell adhesions. Nature 468, 580–584. 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Tang D, Yu Y, Wang Z, Hu T, Wang H, Cao L, 2010. WAVE1 regulates Bcl-2 localization and phosphorylation in leukemia cells. Leukemia 24, 177–186. 10.1038/leu.2009.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor P, Shen X, 2014. Mechanisms of nuclear actin in chromatin-remodeling complexes. Trends Cell Biol. 24, 238–246. 10.1016/j.tcb.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast DJ, Zajac AL, Holzbaur ELF, Ostap EM, Dominguez R, 2015. WHAMM Directs the Arp2/3 Complex to the ER for Autophagosome Biogenesis through an Actin Comet Tail Mechanism. Curr. Biol. 25, 1791–1797. 10.1016/j.cub.2015.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenhuber ER, Lowe SW, 2017. Putting p53 in Context. Cell 170, 1062–1078. 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM, 2018. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 19, 365–381. 10.1038/s41580-018-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay RR, 2021. Macropinocytosis: Biology and mechanisms. Cells Dev. 168, 203713 10.1016/J.CDEV.2021.203713. [DOI] [PubMed] [Google Scholar]
- Kayalar C, Ord T, Testa MP, Zhong LT, Bredesen DE, 1996, Cleavage of actin by interleukin 1 beta-converting enzyme to reverse DNase I inhibition. Proc. Natl. Acad. Sci. U. S. A. 93, 2234–2238. 10.1073/pnas.93.5.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JF, Wyllie AH, Currie AR, 1972. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 26, 239–257. 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killackey SA, Philpott DJ, Girardin SE, 2020. Mitophagy pathways in health and disease. J. Cell Biol. 219, e202004029 10.1083/JCB.202004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H-E, Du F, Fang M, Wang X, 2005. Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc. Natl. Acad. Sci. U. S. A. 102, 17545–17550. 10.1073/pnas.0507900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IH, Kim N, Kim S, Toda K, Catavero CM, Courtland JL, Yin HH, Soderling SH, 2020. Dysregulation of the Synaptic Cytoskeleton in the PFC Drives Neural Circuit Pathology, Leading to Social Dysfunction. Cell Rep. 32, 107965 10.1016/j.celrep.2020.107965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King VL, Leclair NK, Coulter AM, Campellone KG, 2021. The actin nucleation factors JMY and WHAMM enable a rapid Arp2/3 complex-mediated intrinsic pathway of apoptosis. PLoS Genet 17, e1009512. 10.1371/journal.pgen.1009512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou Y-S, Bjørkøy G, Nunn JL, Bruun J-A, Shvets E, Mcewan DG, Clausen TH, Wild P, Bilusic I, Theurillat J-P, Øvervatn A, Ishii T, Elazar Z, Komatsu M, Dikic I, Johansen T, 2009. A Role for NBR1 in Autophagosomal Degradation of Ubiquitinated Substrates. Mol. Cell 33, 505–516. 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N, 1998. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608. 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Klamt F, Zdanov S, Levine RL, Pariser A, Zhang Y, Zhang B, Yu L-R, Veenstra TD, Shacter E, 2009. Oxidant-induced apoptosis is mediated by oxidation of the actin-regulatory protein cofilin. Nat. Cell Biol. 11, 1241–1246. 10.1038/ncb1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P, 1992. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell 68, 521–531. 10.1016/0092-8674(92)90188-I. [DOI] [PubMed] [Google Scholar]
- Kollmar M, Lbik D, Enge S, 2012. Evolution of the eukaryotic ARP2/3 activators of the WASP family: WASP, WAVE, WASH, and WHAMM, and the proposed new family members WAWH and WAML. BMC Res. Notes 5, 88. 10.1186/1756-0500-5-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F, Ramabhadran V, Higgs HN, 2013. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339, 464–467. 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F, Gauvin TJ, Higgs HN, 2014. A role for myosin II in mammalian mitochondrial fission. Curr. Biol. CB 24, 409–414. 10.1016/j.cub.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk VI, Menzies FM, Rubinsztein DC, 2010. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 584, 1393–1398. 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- King VL, Campellone KG, 2023, F-actin-rich territories coordinate apoptosome assembly and caspase activation during DNA damage-induced intrinsic apoptosis. Mol Biol Cell in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronakis V, Hume PJ, Humphreys D, Liu T, Hørning O, Jensen ON, McGhie EJ, 2011, WAVE regulatory complex activation by cooperating GTPases Arf and Rac1. Proc. Natl. Acad. Sci. U. S. A. 108, 14449–14454. 10.1073/PNAS.1107666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothakota S, Azuma T, Reinhard C, Klippel A, Tang J, Chu K, McGarry TJ, Kirschner MW, Koths K, Kwiatkowski DJ, Williams LT, 1997. Caspase-3-generated fragment of gelsolin: effector of morphological change in apoptosis. Science 278, 294–298. 10.1126/science.278.5336.294. [DOI] [PubMed] [Google Scholar]
- Kramer DA, Piper HK, Chen B, 2022. WASP family proteins: Molecular mechanisms and implications in human disease. Eur. J. Cell Biol. 101, 151244 10.1016/J.EJCB.2022.151244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruppa AJ, Kishi-Itakura C, Masters TA, Rorbach JE, Grice GL, Kendrick-Jones J, Nathan JA, Minczuk M, Buss F, 2018. Myosin VI-Dependent Actin Cages Encapsulate Parkin-Positive Damaged Mitochondria. Dev. Cell 44 (484–499), e6. 10.1016/j.devcel.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH, 1997. Regulation of p53 stability by Mdm2. Nature 387, 299–303. 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Kuijpers TW, Tool ATJ, van der Bijl I, de Boer M, van Houdt M, de Cuyper IM, Roos D, van Alphen F, van Leeuwen K, Cambridge EL, Arends MJ, Dougan G, Clare S, Ramirez-Solis R, Pals ST, Adams DJ, Meijer AB, van den Berg TK, 2017. Combined immunodeficiency with severe inflammation and allergy caused by ARPC1B deficiency. e10 J. Allergy Clin. Immunol. 140, 273–277. 10.1016/J.JACI.2016.09.061. [DOI] [PubMed] [Google Scholar]
- Kumar S, Xu J, Perkins C, Guo F, Snapper S, Finkelman FD, Zheng Y, Filippi M-D, 2012. Cdc42 regulates neutrophil migration via crosstalk between WASp, CD11b, and microtubules. Blood 120, 3563–3574. 10.1182/blood-2012-04-426981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano H, Shimizu S, Koya RC, Fujita H, Kamada S, Kuzumaki N, Tsujimoto Y, 2000. Human gelsolin prevents apoptosis by inhibiting apoptotic mitochondrial changes via closing VDAC. Oncogene 19, 4807–4814. 10.1038/sj.onc.1203868. [DOI] [PubMed] [Google Scholar]
- Lamark T, Johansen T, 2021. Mechanisms of Selective Autophagy. Annu. Rev. Cell Dev. Biol. 37, 143–169. 10.1146/ANNUREV-CELLBIO-120219-035530. [DOI] [PubMed] [Google Scholar]
- Lappalainen P, Kotila T, Jégou A, Romet-Lemonne G, 2022. Biochemical and mechanical regulation of actin dynamics. Nat. Rev. Mol. Cell Biol. 23, 836–852. 10.1038/s41580-022-00508-4. [DOI] [PubMed] [Google Scholar]
- Lartigue L, Medina C, Schembri L, Chabert P, Zanese M, Tomasello F, Dalibart R, Thoraval D, Crouzet M, Ichas F, De Giorgi F, 2008. An intracellular wave of cytochrome c propagates and precedes Bax redistribution during apoptosis. J. Cell Sci. 121, 3515–3523. 10.1242/jcs.029587. [DOI] [PubMed] [Google Scholar]
- Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ, 2015. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309–314. 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClaire LL, Baumgartner M, Iwasa JH, Mullins RD, Barber DL, 2008. Phosphorylation of the Arp2/3 complex is necessary to nucleate actin filaments. J. Cell Biol. 182, 647–654. 10.1083/jcb.200802145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, Pandey UB, Kaushik S, Tresse E, Lu J, Taylor JP, Cuervo AM, Yao TP, 2010. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 29, 969. 10.1038/EMBOJ.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Gallop JL, Rambani K, Kirschner MW, 2010. Self-Assembly of Filopodia-Like Structures on Supported Lipid Bilayers. Science 329, 1341–1345. 10.1126/science.1191710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PP, Lobato-Márquez D, Pramanik N, Sirianni A, Daza-Cajigal V, Rivers E, Cavazza A, Bouma G, Moulding D, Hultenby K, Westerberg LS, Hollinshead M, Lau YL, Burns SO, Mostowy S, Bajaj-Elliott M, Thrasher AJ, 2017. Wiskott-Aldrich syndrome protein regulates autophagy and inflammasome activity in innate immune cells. Nat. Commun. 8. 10.1038/S41467-017-01676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legg JA, Bompard G, Dawson J, Morris HL, Andrew N, Cooper L, Johnston SA, Tramountanis G, Machesky LM, 2007. N-WASP Involvement in Dorsal Ruffle Formation in Mouse Embryonic Fibroblasts. Mol. Biol. Cell 18, 678. 10.1091/MBC.E06-06-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkau B, Herren B, Koyama H, Ross R, Raines EW, 1998. Caspase-mediated cleavage of focal adhesion kinase pp125FAK and disassembly of focal adhesions in human endothelial cell apoptosis. J. Exp. Med 187, 579–586. 10.1084/jem.187.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhu H, Xu CJ, Yuan J, 1998. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94, 491–501. 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- Li R, Zhu J, 2022. Effects of aneuploidy on cell behaviour and function. Nat. Rev. Mol. Cell Biol. 23, 250–265. 10.1038/s41580-021-00436-9. [DOI] [PubMed] [Google Scholar]
- Li S, Xu S, Roelofs BA, Boyman L, Lederer WJ, Sesaki H, Karbowski M, 2015. Transient assembly of F-actin on the outer mitochondrial membrane contributes to mitochondrial fission. J. Cell Biol. 208, 109–123. 10.1083/jcb.201404050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhou M, Hu Q, Bai X-C, Huang W, Scheres SHW, Shi Y, 2017, Mechanistic insights into caspase-9 activation by the structure of the apoptosome holoenzyme. Proc. Natl. Acad. Sci. U. S. A. 114, 1542–1547. 10.1073/pnas.1620626114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KB, Bu W, Goh WI, Koh E, Ong SH, Pawson T, Sudhaharan T, Ahmed S, 2008. The Cdc42 effector IRSp53 generates filopodia by coupling membrane protrusion with actin dynamics. J. Biol. Chem. 283, 20454–20472. 10.1074/jbc.M710185200. [DOI] [PubMed] [Google Scholar]
- Linardopoulou EV, Parghi SS, Friedman C, Osborn GE, Parkhurst SM, Trask BJ, 2007. Human Subtelomeric WASH Genes Encode a New Subclass of the WASP Family. PLoS Genet 3, 2477–2485. 10.1371/JOURNAL.PGEN.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder S, 2009. Invadosomes at a glance. J. Cell Sci. 122, 3009–3013. 10.1242/jcs.032631. [DOI] [PubMed] [Google Scholar]
- Liu A, Kage F, Higgs HN, 2021. Mff oligomerization is required for Drp1 activation and synergy with actin filaments during mitochondrial division. ar5 Mol. Biol. Cell 32. 10.1091/mbc.E21-04-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang Q-C, Wang F, Duan X, Dai X-X, Wang T, Liu H-L, Cui X-S, Kim N-H, Sun S-C, 2012. Nucleation promoting factors regulate the expression and localization of Arp2/3 complex during meiosis of mouse oocytes. PloS One 7, e52277. 10.1371/journal.pone.0052277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang J, Pinder BD, Liu Q, Wang D, Yao H, Gao Y, Toker A, Gao J, Peterson A, Qu J, Siminovitch KA, 2021. WAVE2 suppresses mTOR activation to maintain T cell homeostasis and prevent autoimmunity. Science 371, eaaz4544. 10.1126/science.aaz4544. [DOI] [PubMed] [Google Scholar]
- Livneh I, Cohen-Kaplan V, Cohen-Rosenzweig C, Avni N, Ciechanover A, 2016. The life cycle of the 26S proteasome: From birth, through regulation and function, and onto its death. Cell Res 26, 869–885. 10.1038/cr.2016.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommel S, Benesch S, Rottner K, Franz T, Wehland J, Kühn R, 2001. Actin pedestal formation by enteropathogenic Escherichia coli and intracellular motility of Shigella flexneri are abolished in N-WASP-defective cells. EMBO Rep. 2, 850–857. 10.1093/embo-reports/kve197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciani F, Matarrese P, Giammarioli AM, Lugini L, Lozupone F, Federici C, Iessi E, Malorni W, Fais S, 2004. CD95/phosphorylated ezrin association underlies HIV-1 GP120/IL-2-induced susceptibility to CD95(APO-1/Fas)-mediated apoptosis of human resting CD4(+)T lymphocytes. Cell Death Differ. 11, 574–582. 10.1038/sj.cdd.4401374. [DOI] [PubMed] [Google Scholar]
- Luna A, Matas OB, Martínez-Menárguez JA, Mato E, Durán JM, Ballesta J, Way M, Egea G, 2002. Regulation of Protein Transport from the Golgi Complex to the Endoplasmic Reticulum by CDC42 and N-WASP. Mol. Biol. Cell 13, 866–879. 10.1091/mbc.01-12-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Budihardjo I, Zou H, Slaughter C, Wang X, 1998. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 94, 481–490. 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- Lusk CP, King MC, 2017. The nucleus: keeping it together by keeping it apart. Curr. Opin. Cell Biol. 44, 44–50. 10.1016/j.ceb.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald E, Savage B, Zech T, 2020. Connecting the dots: combined control of endocytic recycling and degradation. Biochem. Soc. Trans. 48, 2377. 10.1042/BST20180255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Insall RH, 1998. Scar1 and the related Wiskott–Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 8, 1347–1356. 10.1016/S0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- Machesky LM, Atkinson SJ, Ampe C, Vandekercldaove J, Pollard TD, 1994. Purification of a Cortical Complex Containing Two Unconventional Actins from Acanthamoeba by Affinity Chromatography on Profilin-Agarose. J. Cell Biol. 127, 107–115. 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Reeves E, Wientjes F, Mattheyse FJ, Grogan A, Totty NF, Burlingame AL, Justin J, ‡r H, Segal AW, 1997. Mammalian actin-related protein 2/3 complex localizes to regions of lamellipodial protrusion and is composed of evolutionarily conserved proteins. Biochem J. 328, 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhija E, Jokhun DS, Shivashankar GV, 2016, Nuclear deformability and telomere dynamics are regulated by cell geometric constraints. Proc. Natl. Acad. Sci. U. S. A. 113, E32–40. 10.1073/pnas.1513189113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann N, Mzoughi S, Schneider R, Kühl SJ, Schanze D, Klämbt V, Lovric S, Mao Y, Shi S, Tan W, Kühl M, Onuchic-Whitford AC, Treimer E, Kitzler TM, Kause F, Schumann S, Nakayama M, Buerger F, Shril S, van der Ven AT, Majmundar AJ, Holton KM, Kolb A, Braun DA, Rao J, Jobst-Schwan T, Mildenberger E, Lennert T, Kuechler A, Wieczorek D, Gross O, Ermisch-Omran B, Werberger A, Skalej M, Janecke AR, Soliman NA, Mane SM, Lifton RP, Kadlec J, Guccione E, Schmeisser MJ, Zenker M, Hildebrandt F, 2021. Mutations in PRDM15 Are a Novel Cause of Galloway-Mowat Syndrome. J. Am. Soc. Nephrol. 32, 580–596. 10.1681/ASN.2020040490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor U, Bartholomew S, Golani G, Christenson E, Kozlov M, Higgs H, Spudich J, Lippincott-Schwartz J, 2015. A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. eLife 4, e08828. 10.7554/eLife.08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SS, Leder P, 2001. Human MCF10A mammary epithelial cells undergo apoptosis following actin depolymerization that is independent of attachment and rescued by Bcl-2. Mol. Cell. Biol. 21, 6529–6536. 10.1128/MCB.21.19.6529-6536.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashima T, Naito M, Fujita N, Noguchi K, Tsuruo T, 1995. Identification of actin as a substrate of ICE and an ICE-like protease and involvement of an ICE-like protease but not ICE in VP-16-induced U937 apoptosis. Biochem. Biophys. Res. Commun. 217, 1185–1192. 10.1006/bbrc.1995.2894. [DOI] [PubMed] [Google Scholar]
- Mashima T, Naito M, Noguchi K, Miller DK, Nicholson DW, Tsuruo T, 1997. Actin cleavage by CPP-32/apopain during the development of apoptosis. Oncogene 14, 1007–1012. 10.1038/sj.onc.1200919. [DOI] [PubMed] [Google Scholar]
- Mashima T, Naito M, Tsuruo T, 1999. Caspase-mediated cleavage of cytoskeletal actin plays a positive role in the process of morphological apoptosis. Oncogene 18, 2423–2430. 10.1038/sj.onc.1202558. [DOI] [PubMed] [Google Scholar]
- Massaad MJ, Ramesh N, Geha RS, 2013. Wiskott-Aldrich syndrome: A comprehensive review. Ann. N. Y. Acad. Sci. 1285, 26–43. 10.1111/NYAS.12049. [DOI] [PubMed] [Google Scholar]
- Matas OB, Martínez-Menárguez J, Egea G, 2004. Association of Cdc42/N-WASP/Arp2/3 Signaling Pathway with Golgi Membranes. Traffic 5, 838–846. 10.1111/j.1600-0854.2004.00225.x. [DOI] [PubMed] [Google Scholar]
- Mathiowetz AJ, Baple E, Russo AJ, Coulter AM, Carrano E, Brown JD, Jinks RN, Crosby AH, Campellone KG, 2017. An Amish founder mutation disrupts a PI (3)P-WHAMM-Arp2/3 complex-driven autophagosomal remodeling pathway. Mol. Biol. Cell 28, 2492–2507. 10.1091/mbc.E17-01-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh D, Gil J, 2018. Senescence and aging: Causes, consequences, and therapeutic avenues. J. Cell Biol. 217, 65–77. 10.1083/jcb.201708092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally KE, Faulkner R, Steinberg F, Gallon M, Ghai R, Pim D, Langton P, Pearson N, Danson CM, Nägele H, Morris LM, Singla A, Overlee BL, Heesom KJ, Sessions R, Banks L, Collins BM, Berger I, Billadeau DD, Burstein E, Cullen PJ, 2017. Retriever, a multiprotein complex for retromer-independent endosomal cargo recycling. Nat. Cell Biol. 19, 1214–1225. 10.1038/ncb3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melak M, Plessner M, Grosse R, 2017. Actin visualization at a glance. J. Cell Sci. 130, 525–530. 10.1242/jcs.189068. [DOI] [PubMed] [Google Scholar]
- Merrifield CJ, Qualmann B, Kessels MM, Almers W, 2004. Neural Wiskott Aldrich Syndrome Protein (N-WASP) and the Arp2/3 complex are recruited to sites of clathrin-mediated endocytosis in cultured fibroblasts. Eur. J. Cell Biol. 83, 13–18. 10.1078/0171-9335-00356. [DOI] [PubMed] [Google Scholar]
- Mi N, Chen Y, Wang S, Chen M, Zhao M, Yang G, Ma M, Su Q, Luo S, Shi J, Xu J, Guo Q, Gao N, Sun Y, Chen Z, Yu L, 2015. CapZ regulates autophagosomal membrane shaping by promoting actin assembly inside the isolation membrane. Nat. Cell Biol. 17, 1112–1125. 10.1038/ncb3215. [DOI] [PubMed] [Google Scholar]
- Miki H, Miura K, Takenawa T, 1996. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO J. 15, 5326–5335. 10.1002/j.1460-2075.1996.tb00917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Sasaki T, Takai Y, Takenawa T, 1998a. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature 391, 93–96. 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- Miki H, Suetsugu S, Takenawa T, 1998b. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 17, 6932–6941. 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamide LS, Striegl AM, Boyle JA, Meberg PJ, Bamburg JR, 2000. Neurodegenerative stimuli induce persistent ADF/cofilin-actin rods that disrupt distal neurite function. Nat. Cell Biol. 2, 628–636. 10.1038/35023579. [DOI] [PubMed] [Google Scholar]
- Miralles F, Visa N, 2006. Actin in transcription and transcription regulation. Curr. Opin. Cell Biol. 18, 261–266. 10.1016/j.ceb.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Mishra S, Levy DL, 2022. Nuclear F-actin and Lamin A antagonistically modulate nuclear shape. J. Cell Sci. 135, jcs259692 10.1242/jcs.259692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra A, Lim RPZ, Wu Z, Thanabalu T, 2007. N-WASP plays a critical role in fibroblast adhesion and spreading. Biochem. Biophys. Res. Commun. 364, 908–912. 10.1016/j.bbrc.2007.10.086. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Pasque V, Jullien J, Gurdon JB, 2011. Nuclear actin polymerization is required for transcriptional reprogramming of Oct4 by oocytes. Genes Dev. 25, 946–958. 10.1101/gad.615211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Teperek M, Yusa K, Allen GE, Bradshaw CR, Gurdon JB, 2013. Nuclear Wave1 is required for reprogramming transcription in oocytes and for normal development. Science 341, 1002–1005. 10.1126/science.1240376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, 2020. Autophagy in Human Diseases. N. Engl. J. Med 383, 1564–1576. 10.1056/NEJMra2022774. [DOI] [PubMed] [Google Scholar]
- Mogessie B, Schuh M, 2017. Actin protects mammalian eggs against chromosome segregation errors. Sci. 357, eaal1647. 10.1126/science.aal1647. [DOI] [PubMed] [Google Scholar]
- Molinie N, Gautreau A, 2018. The Arp2/3 regulatory system and its deregulation in cancer. Physiol. Rev. 98, 215–238. 10.1152/PHYSREV.00006.2017. [DOI] [PubMed] [Google Scholar]
- Molinie N, Rubtsova SN, Fokin A, Visweshwaran SP, Rocques N, Polesskaya A, Schnitzler A, Vacher S, Denisov EV, Tashireva LA, Perelmuter VM, Cherdyntseva NV, Biéche I, Gautreau AM, 2019. Cortical branched actin determines cell cycle progression. Cell Res 29, 432–445. 10.1038/s41422-019-0160-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monastyrska I, He C, Geng J, Hoppe AD, Li Z, Klionsky DJ, 2008. Arp2 Links Autophagic Machinery with the Actin Cytoskeleton. Mol. Biol. Cell 19, 1962. 10.1091/MBC.E07-09-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monfregola J, Napolitano G, D’Urso M, Lappalainen P, Ursini MV, 2010. Functional characterization of Wiskott-Aldrich syndrome protein and scar homolog (WASH), a bi-modular nucleation-promoting factor able to interact with biogenesis of lysosome-related organelle subunit 2 (BLOS2) and gamma-tubulin. J. Biol. Chem. 285, 16951–16957. 10.1074/jbc.M109.078501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AS, Wong YC, Simpson CL, Holzbaur ELF, 2016. Dynamic actin cycling through mitochondrial subpopulations locally regulates the fission-fusion balance within mitochondrial networks. Nat. Commun. 7, 12886. 10.1038/ncomms12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AS, Coscia SM, Simpson CL, Ortega FE, Wait EC, Heddleston JM, Nirschl JJ, Obara CJ, Guedes-Dias P, Boecker CA, Chew T-L, Theriot JA, Lippincott-Schwartz J, Holzbaur ELF, 2021. Actin cables and comet tails organize mitochondrial networks in mitosis. Nature 591, 659–664. 10.1038/s41586-021-03309-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita H, Mizushima N, 2019. Diverse Cellular Roles of Autophagy. Annu. Rev. Cell Dev. Biol. 35, 453–475. 10.1146/annurev-cellbio-100818-125300. [DOI] [PubMed] [Google Scholar]
- Morley SC, Sun G-P, Bierer BE, 2003. Inhibition of actin polymerization enhances commitment to and execution of apoptosis induced by withdrawal of trophic support. J. Cell. Biochem 88, 1066–1076. 10.1002/jcb.10449. [DOI] [PubMed] [Google Scholar]
- Morris HT, Fort L, Spence HJ, Patel R, Vincent DF, Park JH, Snapper SB, Carey FA, Sansom OJ, Machesky LM, 2018. Loss of N-WASP drives early progression in an Apc model of intestinal tumourigenesis. J. Pathol. 245, 337. 10.1002/PATH.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD, 1998, The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. U. S. A. 95, 6181–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munsie L, Caron N, Atwal RS, Marsden I, Wild EJ, Bamburg JR, Tabrizi SJ, Truant R, 2011. Mutant huntingtin causes defective actin remodeling during stress: defining a new role for transglutaminase 2 in neurodegenerative disease. Hum. Mol. Genet 20, 1937–1951. 10.1093/hmg/ddr075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DA, Courtneidge SA, 2011. The “ins” and “outs” of podosomes and invadopodia: characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12, 413–426. 10.1038/nrm3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Yoshimori T, 2017. New insights into autophagosome–lysosome fusion. J. Cell Sci. 130, 1209–1216. 10.1242/jcs.196352. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, 2020. Mechanisms governing autophagosome biogenesis. Nat. Rev. Mol. Cell Biol. 21, 439–458. 10.1038/s41580-020-0241-0. [DOI] [PubMed] [Google Scholar]
- Nguyen TN, Lazarou M, 2022. A unifying model for the role of the ATG8 system in autophagy. J. Cell Sci. 135, jcs258997. 10.1242/jcs.258997. [DOI] [PubMed] [Google Scholar]
- Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, Vollger MR, Altemose N, Uralsky L, Gershman A, Aganezov S, Hoyt SJ, Diekhans M, Logsdon GA, Alonge M, Antonarakis SE, Borchers M, Bouffard GG, Brooks SY, Caldas GV, Chen N-C, Cheng H, Chin C-S, Chow W, de Lima LG, Dishuck PC, Durbin R, Dvorkina T, Fiddes IT, Formenti G, Fulton RS, Fungtammasan A, Garrison E, Grady PGS, Graves-Lindsay TA, Hall IM, Hansen NF, Hartley GA, Haukness M, Howe K, Hunkapiller MW, Jain C, Jain M, Jarvis ED, Kerpedjiev P, Kirsche M, Kolmogorov M, Korlach J, Kremitzki M, Li H, Maduro VV, Marschall T, McCartney AM, McDaniel J, Miller DE, Mullikin JC, Myers EW, Olson ND, Paten B, Peluso P, Pevzner PA, Porubsky D, Potapova T, Rogaev EI, Rosenfeld JA, Salzberg SL, Schneider VA, Sedlazeck FJ, Shafin K, Shew CJ, Shumate A, Sims Y, Smit AFA, Soto DC, Sovíc I, Storer JM, Streets A, Sullivan BA, Thibaud-Nissen F, Torrance J, Wagner J, Walenz BP, Wenger A, Wood JMD, Xiao C, Yan SM, Young AC, Zarate S, Surti U, McCoy RC, Dennis MY, Alexandrov IA, Gerton JL, O’Neill RJ, Timp W, Zook JM, Schatz MC, Eichler EE, Miga KH, Phillippy AM, 2022. The complete sequence of a human genome. Science 376, 44–53. 10.1126/science.abj6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odaka C, Sanders ML, Crews P, 2000. Jasplakinolide induces apoptosis in various transformed cell lines by a caspase-3-like protease-dependent pathway. Clin. Diagn. Lab. Immunol. 7, 947–952. 10.1128/CDLI.7.6.947-952.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y, 2014. Historical landmarks of autophagy research. Cell Res 24, 9–23. 10.1038/cr.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsu M, Sakai N, Fujita H, Kashiwagi M, Gasa S, Shimizu S, Eguchi Y, Tsujimoto Y, Sakiyama Y, Kobayashi K, Kuzumaki N, 1997. Inhibition of apoptosis by the actin-regulatory protein gelsolin. EMBO J. 16, 4650–4656. 10.1093/emboj/16.15.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Rosen MK, 2010. Physical Mechanisms of Signal Integration by WASP Family Proteins. Annu. Rev. Biochem. 79, 707–735. 10.1146/annurev.biochem.77.060407.135452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Cheng H-C, Ismail AM, Panchal SC, Doolittle LK, Kim S, Skehan BM, Umetani J, Brautigam CA, Leong JM, Rosen MK, 2008. Hierarchical Regulation of WASP/WAVE Proteins. Mol. Cell 32, 426–438. 10.1016/j.molcel.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palamiuc L, Ravi A, Emerling BM, 2020. Phosphoinositides in autophagy: current roles and future insights. FEBS J. 287, 222–238. 10.1111/febs.15127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun J-A, Outzen H, Øvervatn A, Bjørkøy G, Johansen T, 2007. p62/SQSTM1 Binds Directly to Atg8/LC3 to Facilitate Degradation of Ubiquitinated Protein Aggregates by Autophagy. J. Biol. Chem. 282, 24131–24145. 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Papalazarou V, Swaminathan K, Jaber-Hijazi F, Spence H, Lahmann I, Nixon C, Salmeron-Sanchez M, Arnold HH, Rottner K, Machesky LM, 2020. The Arp2/3 complex is crucial for colonisation of the mouse skin by melanoblasts. Development 147. 10.1242/DEV.194555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C, Cuervo AM, 2013. Selective Autophagy: Talking with the UPS. Cell Biochem. Biophys. 67, 3–13. 10.1007/s12013-013-9623-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park L, Thomason PA, Zech T, King JS, Veltman DM, Carnell M, Ura S, Machesky LM, Insall RH, 2013. Cyclical Action of the WASH Complex: FAM21 and Capping Protein Drive WASH Recycling, Not Initial Recruitment. Dev. Cell 24, 169–181. 10.1016/j.devcel.2012.12.014. [DOI] [PubMed] [Google Scholar]
- Pathania M, Davenport EC, Muir J, Sheehan DF, López-Doménech G, Kittler JT, 2014. The autism and schizophrenia associated gene CYFIP1 is critical for the maintenance of dendritic complexity and the stabilization of mature spines. Transl. Psychiatry 4, e374. 10.1038/tp.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul C, Manero F, Gonin S, Kretz-Remy C, Virot S, Arrigo A-P, 2002. Hsp27 as a negative regulator of cytochrome C release. Mol. Cell. Biol. 22, 816–834. 10.1128/MCB.22.3.816-834.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T, 2008. As functional nuclear actin comes into view, is it globular, filamentous, or both? J. Cell Biol. 180, 1061–1064. 10.1083/jcb.200709082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P, 2013. Co-transcriptional nuclear actin dynamics. Nucl 4, 43–52. 10.4161/nucl.22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philimonenko VV, Zhao J, Iben S, Dingová H, Kyselá K, Kahle M, Zentgraf H, Hofmann WA, de Lanerolle P, Hozák P, Grummt I, 2004. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat. Cell Biol. 6, 1165–1172. 10.1038/ncb1190. [DOI] [PubMed] [Google Scholar]
- Phillips-Krawczak CA, Singla A, Starokadomskyy P, Deng Z, Osborne DG, Li H, Dick CJ, Gomez TS, Koenecke M, Zhang J-S, Dai H, Sifuentes-Dominguez LF, Geng LN, Kaufmann SH, Hein MY, Wallis M, McGaughran J, Gecz J, van de Sluis B, Billadeau DD, Burstein E, 2015. COMMD1 is linked to the WASH complex and regulates endosomal trafficking of the copper transporter ATP7A. Mol. Biol. Cell 26, 91–103. 10.1091/mbc.E14-06-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell AM, Youle RJ, 2015. The Roles of PINK1, Parkin and Mitochondrial Fidelity in Parkinson’s Disease. Neuron 85, 257. 10.1016/J.NEURON.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro-Cerdá J, Chorev DS, Geiger B, Cossart P, 2017. The Diverse Family of Arp2/3 Complexes. Trends Cell Biol. 27, 93–100. 10.1016/j.tcb.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessner M, Knerr J, Grosse R, 2019. Centrosomal Actin Assembly Is Required for Proper Mitotic Spindle Formation and Chromosome Congression. iScience 15, 274–281. 10.1016/j.isci.2019.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya A, Boutillon A, Yang S, Wang Y, Romero S, Liu Y, Lavielle M, Molinie N, Rocques N, Fokin A, Guérois R, Chen B, David NB, Gautreau AM, 2022. Restrained activation of CYFIP2-containing WAVE complexes controls membrane protrusions and cell migration. 10.1101/2020.07.02.184655. [DOI] [Google Scholar]
- Pollard TD, 2016. Actin and Actin-Binding Proteins. Cold Spring Harb. Perspect. Biol. 8, a018226 10.1101/cshperspect.a018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, 2018. Evolution of research on cellular motility over five decades. Biophys. Rev. 10, 1503–1508. 10.1007/s12551-018-0473-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posey SC, Bierer BE, 1999. Actin stabilization by jasplakinolide enhances apoptosis induced by cytokine deprivation. J. Biol. Chem. 274, 4259–4265. 10.1074/jbc.274.7.4259. [DOI] [PubMed] [Google Scholar]
- Rai R, Romito M, Rivers E, Turchiano G, Blattner G, Vetharoy W, Ladon D, Andrieux G, Zhang F, Zinicola M, Leon-Rico D, Santilli G, Thrasher AJ, Cavazza A, 2020. Targeted gene correction of human hematopoietic stem cells for the treatment of Wiskott - Aldrich Syndrome. Nat. Commun. 11, 4034. 10.1038/s41467-020-17626-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randzavola LO, Strege K, Juzans M, Asano Y, Stinchcombe JC, Gawden-Bone CM, Seaman MNJ, Kuijpers TW, Griffiths GM, 2019. Loss of ARPC1B impairs cytotoxic T lymphocyte maintenance and cytolytic activity. J. Clin. Invest 129, 5600. 10.1172/JCI129388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao JY, Jin YS, Zheng Q, Cheng J, Tai J, Hemstreet GP, 1999. Alterations of the actin polymerization status as an apoptotic morphological effector in HL-60 cells. J. Cell. Biochem 75, 686–697. [PubMed] [Google Scholar]
- Rehklau K, Gurniak CB, Conrad M, Friauf E, Ott M, Rust MB, 2012. ADF/cofilin proteins translocate to mitochondria during apoptosis but are not generally required for cell death signaling. Cell Death Differ. 19, 958–967. 10.1038/cdd.2011.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm M, Huber HJ, Hellwig CT, Anguissola S, Dussmann H, Prehn JHM, 2009. Dynamics of outer mitochondrial membrane permeabilization during apoptosis. Cell Death Differ. 16, 613–623. 10.1038/cdd.2008.187. [DOI] [PubMed] [Google Scholar]
- Rivers E, Rai R, Lötscher J, Hollinshead M, Markelj G, Thaventhiran J, Worth A, Cavazza A, Hess C, Bajaj-Elliott M, Thrasher AJ, 2020. Wiskott Aldrich syndrome protein regulates non-selective autophagy and mitochondrial homeostasis in human myeloid cells. eLife 9, 1–23. 10.7554/ELIFE.55547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R, Ktistakis NT, 2013. Omegasomes: PI3P platforms that manufacture autophagosomes. Essays Biochem 55, 17–27. 10.1042/bse0550017. [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Lazebnik Y, 1999. Caspase-9 and APAF-1 form an active holoenzyme. Genes Dev. 13, 3179–3184. 10.1101/gad.13.24.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero S, Clainche CL, Gautreau A, 2020. Actin polymerization downstream of integrins: signaling pathways and mechanotransduction. Biochem. J. 477, 1. 10.1042/BCJ20170719. [DOI] [PubMed] [Google Scholar]
- Ropers F, Derivery E, Hu H, Garshasbi M, Karbasiyan M, Herold M, Nürnberg G, Ullmann R, Gautreau A, Sperling K, Varon R, Rajab A, 2011. Identification of a novel candidate gene for non-syndromic autosomal recessive intellectual disability: the WASH complex member SWIP. Hum. Mol. Genet. 20, 2585–2590. 10.1093/HMG/DDR158. [DOI] [PubMed] [Google Scholar]
- Rosenblatt J, Raff MC, Cramer LP, 2001. An epithelial cell destined for apoptosis signals its neighbors to extrude it by an actin- and myosin-dependent mechanism. Curr. Biol. 11, 1847–1857. 10.1016/s0960-9822(01)00587-5. [DOI] [PubMed] [Google Scholar]
- Rosti RO, Sotak BN, Bielas SL, Bhat G, Silhavy JL, Aslanger AD, Altunoglu U, Bilge I, Tasdemir M, Yzaguirrem AD, Musaev D, Infante S, Thuong W, Marin-Valencia I, Nelson SF, Kayserili H, Gleeson JG, 2017. Homozygous mutation in NUP107 leads to microcephaly with steroid-resistant nephrotic condition similar to Galloway-Mowat. Syndr. J. Med. Genet. 54, 399–403. 10.1136/jmedgenet-2016-104237. [DOI] [PubMed] [Google Scholar]
- Rotty JD, Wu C, Bear JE, 2013. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat. Rev. Mol. Cell Biol. 14, 7–12. 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- Rotty JD, Brighton HE, Craig SL, Asokan SB, Cheng N, Ting JP, Bear JE, 2017. Arp2/3 complex is required for macrophage integrin functions but is dispensable for FcR phagocytosis and in vivo motility. Dev. Cell 42 (498–513), e6. 10.1016/j.devcel.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo AJ, Mathiowetz AJ, Hong S, Welch MD, Campellone KG, 2016. Rab1 recruits WHAMM during membrane remodeling but limits actin nucleation. Mol. Biol. Cell 27, 967–978. 10.1091/mbc.E15-07-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder PV, Vistein R, Gokhale A, Seaman MN, Puthenveedu MA, Faundez V, 2013. The WASH complex, an endosomal Arp2/3 activator, interacts with the Hermansky-Pudlak syndrome complex BLOC-1 and its cargo phosphatidylinositol-4-kinase type IIα. Mol. Biol. Cell 24, 2269–2284. 10.1091/mbc.E13-02-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadhukhan S, Sarkar K, Taylor M, Candotti F, Vyas YM, 2014. Nuclear role of WASp in gene transcription is uncoupled from its ARP2/3-dependent cytoplasmic role in actin polymerization. J. Immunol. 1950 (193), 150–160. 10.4049/jimmunol.1302923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallee NA, Rivera GM, Dueber JE, Vasilescu D, Mullins RD, Mayer BJ, Lim WA, 2008. The Pathogen Protein EspFU Hijacks Actin Polymerization Using Mimicry and Multivalency. Nature 454, 1005–1008. 10.1038/nature07170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar K, Sadhukhan S, Han S-S, Vyas YM, 2014. Disruption of hSWI/SNF complexes in T cells by WAS mutations distinguishes X-linked thrombocytopenia from Wiskott-Aldrich syndrome. Blood 124, 3409–3419. 10.1182/blood-2014-07-587642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf SA, Shah HV, Kanfer G, Pickrell AM, Holtzclaw LA, Ward ME, Youle RJ, 2020. Loss of TAX1BP1-Directed Autophagy Results in Protein Aggregate Accumulation in the Brain. e10 Mol. Cell 80, 779–795. 10.1016/j.molcel.2020.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaks M, Giannone G, Rottner K, 2019. Actin dynamics in cell migration. Essays Biochem 63, 483. 10.1042/EBC20190015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell C, Baumhakl L, Salou S, Conzelmann AC, Meyer C, Helmstädter M, Wrede C, Grahammer F, Eimer S, Kerjaschki D, Walz G, Snapper S, Huber TB, 2013. N-WASP is required for stabilization of podocyte foot processes. J. Am. Soc. Nephrol. 24, 713–721. 10.1681/ASN.2012080844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell C, Sabass B, Helmstaedter M, Geist F, Abed A, Yasuda-Yamahara M, Sigle A, Maier JI, Grahammer F, Siegerist F, Artelt N, Endlich N, Kerjaschki D, Arnold HH, Dengjel J, Rogg M, Huber TB, 2018. ARP3 Controls the Podocyte Architecture at the Kidney Filtration Barrier. Dev. Cell 47, 741. 10.1016/J.DEVCEL.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter K, Waschbüsch D, Anft M, Hügging D, Kind S, Hänisch J, Lakisic G, Gautreau A, Barnekow A, Stradal TEB, 2014. JMY is involved in anterograde vesicle trafficking from the trans-Golgi network. Eur. J. Cell Biol. 93, 194–204. 10.1016/j.ejcb.2014.06.001. [DOI] [PubMed] [Google Scholar]
- Schnoor M, Stradal TE, Rottner K, 2018. Cortactin: cell functions of a multifaceted actin-binding protein. Trends Cell Biol. 28, 79–98. 10.1016/j.tcb.2017.10.009. [DOI] [PubMed] [Google Scholar]
- Schrank BR, Aparicio T, Li Y, Chang W, Chait BT, Gundersen GG, Gottesman ME, Gautier J, 2018. Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature 559, 61–66. 10.1038/s41586-018-0237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MNJ, 2021. The retromer complex: from genesis to revelations. Trends Biochem. Sci. 46, 608–620. 10.1016/j.tibs.2020.12.009. [DOI] [PubMed] [Google Scholar]
- Seirin-Lee S, Osakada F, Takeda J, Tashiro S, Kobayashi R, Yamamoto T, Ochiai H, 2019. Role of dynamic nuclear deformation on genomic architecture reorganization. PLoS Comput. Biol. 15, e1007289 10.1371/journal.pcbi.1007289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SenGupta S, Parent CA, Bear JE, 2021. The principles of directed cell migration. Nat. Rev. Mol. Cell Biol. 22, 529. 10.1038/S41580-021-00366-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwas D, Akamatsu M, Moayed A, Vegesna K, Vasan R, Hill JM, Schoneberg J, ¨ Davies KM, Rangamani P, Drubin DG, 2022. Mechanistic insights into actin force generation during vesicle formation from cryo-electron tomography. e5 Dev. Cell 57, 1132–1145. 10.1016/j.devcel.2022.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaaban M, Chowdhury S, Nolen BJ, 2020. Cryo-EM reveals the transition of Arp2/3 complex from inactive to nucleation-competent state. Nat. Struct. Mol. Biol. 27, 1009–1016. 10.1038/s41594-020-0481-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar S, Pernier J, Carlier M-F, 2016. Regulators of actin filament barbed ends at a glance. J. Cell Sci. 129, 1085–1091. 10.1242/jcs.179994. [DOI] [PubMed] [Google Scholar]
- Shen QT, Hsiue PP, Sindelar CV, Welch MD, Campellone KG, Wang HW, 2012. Structural insights into WHAMM-mediated cytoskeletal coordination during membrane remodeling. J. Cell Biol. 199, 111–124. 10.1083/jcb.201204010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikama N, Lee CW, France S, Delavaine L, Lyon J, Krstic-Demonacos M, La Thangue NB, 1999. A novel cofactor for p300 that regulates the p53 response. Mol. Cell 4, 365–376. 10.1016/s1097-2765(00)80338-x. [DOI] [PubMed] [Google Scholar]
- Simonetti B, Cullen PJ, 2019. Actin-dependent endosomal receptor recycling. Curr. Opin. Cell Biol. 56, 22–33. 10.1016/j.ceb.2018.08.006. [DOI] [PubMed] [Google Scholar]
- Singh V, Davidson AC, Hume PJ, Humphreys D, Koronakis V, 2019. Arf GTPase interplay with Rho GTPases in regulation of the actin cytoskeleton. Small GTPases 10, 411–418. 10.1080/21541248.2017.1329691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla A, Fedoseienko A, Giridharan SSP, Overlee BL, Lopez A, Jia D, Song J, Huff-Hardy K, Weisman L, Burstein E, Billadeau DD, 2019. Endosomal PI(3)P regulation by the COMMD/CCDC22/CCDC93 (CCC) complex controls membrane protein recycling. Nat. Commun. 10, 4271. 10.1038/s41467-019-12221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapper SB, Takeshima F, Antón I, Liu C-H, Thomas SM, Nguyen D, Dudley D, Fraser H, Purich D, Lopez-Ilasaca M, Klein C, Davidson L, Bronson R, Mulligan RC, Southwick F, Geha R, Goldberg MB, Rosen FS, Hartwig JH, Alt FW, 2001. N-WASP deficiency reveals distinct pathways for cell surface projections and microbial actin-based motility. Nat. Cell Biol. 3, 897–904. 10.1038/ncb1001-897. [DOI] [PubMed] [Google Scholar]
- Sokolik CG, Qassem N, Chill JH, 2020. The Disordered Cellular Multi-Tasker WIP and Its Protein-Protein Interactions: A Structural View. Biomolecules 10, 1084. 10.3390/biom10071084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somech R, Lev A, Lee YN, Simon AJ, Barel O, Schiby G, Avivi C, Barshack I, Rhodes M, Yin J, Wang M, Yang Y, Rhodes J, Marcus N, Garty B-Z, Stein J, Amariglio N, Rechavi G, Wiest DL, Zhang Y, 2017. Disruption of thrombocyte and T lymphocyte development by a mutation in ARPC1B. J. Immunol. 1950 (199), 4036–4045. 10.4049/jimmunol.1700460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwick FS, Li W, Zhang F, Zeile WL, Purich DL, 2003. Actin-based endosome and phagosome rocketing in macrophages: activation by the secretagogue antagonists lanthanum and zinc. Cell Motil. Cytoskelet. 54, 41–55. 10.1002/cm.10083. [DOI] [PubMed] [Google Scholar]
- Spencer VA, Costes S, Inman JL, Xu R, Chen J, Hendzel MJ, Bissell MJ, 2011. Depletion of nuclear actin is a key mediator of quiescence in epithelial cells. J. Cell Sci. 124, 123–132. 10.1242/jcs.073197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan S, Jain K, Basu A, 2011. Regulation of Autophagy by Kinases. Cancers 3, 2630–2654. 10.3390/cancers3022630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Macke EL, Swanson LC, Coulter D, Klee EW, Mullegama SV, Xie Y, Lanpher BC, Bedoukian EC, Skraban CM, Villard L, Milh M, Leppert MLO, Cohen JS, 2021. Expansion of the Genotypic and Phenotypic Spectrum of WASF1-Related Neurodevelopmental Disorder. Brain Sci. 11, 931. 10.3390/brainsci11070931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahnke S, Döring H, Kusch C, de Gorter DJJ, Dütting S, Guledani A, Pleines I, Schnoor M, Sixt M, Geffers R, Rohde M, Müsken M, Kage F, Steffen A, Faix J, Nieswandt B, Rottner K, Stradal TEB, 2021. Loss of Hem1 disrupts macrophage function and impacts migration, phagocytosis, and integrin-mediated adhesion. e8 Curr. Biol. 31, 2051–2064. 10.1016/J.CUB.2021.02.043. [DOI] [PubMed] [Google Scholar]
- Steffen A, Faix J, Resch GP, Linkner J, Wehland J, Small JV, Rottner K, Stradal TEB, 2006. Filopodia Formation in the Absence of Functional WAVE- and Arp2/3-Complexes. Mol. Biol. Cell 17, 2581–2591. 10.1091/MBC.E05-11-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S, Takenawa T, 2003. Translocation of N-WASP by nuclear localization and export signals into the nucleus modulates expression of HSP90. J. Biol. Chem. 278, 42515–42523. 10.1074/jbc.M302177200. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Miki H, Takenawa T, 1999. Identification of two human WAVE/SCAR homologues as general actin regulatory molecules which associate with the Arp2/3 complex. Biochem. Biophys. Res. Commun. 260, 296–302. 10.1006/bbrc.1999.0894. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Hattori M, Miki H, Tezuka T, Yamamoto T, Mikoshiba K, Takenawa T, 2002. Sustained activation of N-WASP through phosphorylation is essential for neurite extension. Dev. Cell 3, 645–658. 10.1016/s1534-5807(02)00324-6. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Yamazaki D, Kurisu S, Takenawa T, 2003. Differential Roles of WAVE1 and WAVE2 in Dorsal and Peripheral Ruffle Formation for Fibroblast Cell Migration. Dev. Cell 5, 595–609. 10.1016/S1534-5807(03)00297-1. [DOI] [PubMed] [Google Scholar]
- Sullivan KD, Galbraith MD, Andrysik Z, Espinosa JM, 2018. Mechanisms of transcriptional regulation by p53. Cell Death Differ. 25, 133–143. 10.1038/cdd.2017.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S-C, Sun Q-Y, Kim N-H, 2011a. JMY is required for asymmetric division and cytokinesis in mouse oocytes. Mol. Hum. Reprod. 17, 296–304. 10.1093/molehr/gar006. [DOI] [PubMed] [Google Scholar]
- Sun S-C, Wang Z-B, Xu Y-N, Lee S-E, Cui X-S, Kim N-H, 2011b. Arp2/3 complex regulates asymmetric division and cytokinesis in mouse oocytes. PloS One 6, e18392. 10.1371/journal.pone.0018392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S-C, Xu Y-N, Li Y-H, Lee S-E, Jin Y-X, Cui X-S, Kim N-H, 2011c. WAVE2 regulates meiotic spindle stability, peripheral positioning and polar body emission in mouse oocytes. Cell Cycle 10, 1853–1860. 10.4161/cc.10.11.15796. [DOI] [PubMed] [Google Scholar]
- Suria H, Chau LA, Negrou E, Kelvin DJ, Madrenas J, 1999. Cytoskeletal disruption induces T cell apoptosis by a caspase-3 mediated mechanism. Life Sci. 65, 2697–2707. 10.1016/s0024-3205(99)00538-x. [DOI] [PubMed] [Google Scholar]
- Svitkina T, 2018. The Actin Cytoskeleton and Actin-Based Motility. Cold Spring Harb. Perspect. Biol. 10, a018267. 10.1101/cshperspect.a018267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney KF, Li R, 2016. Function and regulation of the Arp2/3 complex during cell migration in diverse environments. Curr. Opin. Cell Biol. 42, 63–72. 10.1016/j.ceb.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson JA, King JS, 2019. The breadth of macropinocytosis research. Philos. Trans. R. Soc. B Biol. Sci. 374, 20180146. 10.1098/rstb.2018.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons M, Derry JMJ, Karlak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A, 1996. Wiskott–Aldrich Syndrome Protein, a Novel Effector for the GTPase CDC42Hs, Is Implicated in Actin Polymerization. Cell 84, 723–734. 10.1016/S0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- Tait SWG, Green DR, 2013. Mitochondrial regulation of cell death. Cold Spring Harb. Perspect. Biol. 5, a008706. 10.1101/cshperspect.a008706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang HL, Le A-HP, Lung HL, 2006. The increase in mitochondrial association with actin precedes Bax translocation in apoptosis. Biochem. J. 396, 1–5. 10.1042/BJ20060241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Schaks M, Koundinya N, Yang C, Pollard LW, Svitkina TM, Rottner K, Goode BL, 2020. WAVE1 and WAVE2 have distinct and overlapping roles in controlling actin assembly at the leading edge. Mol. Biol. Cell 31, 2168–2178. 10.1091/mbc.E19-12-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taunton J, Rowning BA, Coughlin ML, Wu M, Moon RT, Mitchison TJ, Larabell CA, 2000. Actin-dependent propulsion of endosomes and lysosomes by recruitment of N-WASP. J. Cell Biol. 148, 519–530. 10.1083/jcb.148.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MD, Sadhukhan S, Kottangada P, Ramgopal A, Sarkar K, D’Silva S, Selvakumar A, Candotti F, Vyas YM, 2010. Nuclear role of WASp in the pathogenesis of dysregulated TH1 immunity in human Wiskott-Aldrich syndrome. Sci. Transl. Med. 2, 37ra44. 10.1126/scitranslmed.3000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JD, Zhang Y-J, Wei Y-H, Cho J-H, Morris LE, Wang H-Y, Zheng XFS, 2014. Rab1A is an mTORC1 activator and a colorectal oncogene. Cancer Cell 26, 754–769. 10.1016/j.ccell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrasher AJ, 2009. New insights into the biology of Wiskott-Aldrich syndrome (WAS. Hematology 2009, 132–138. 10.1182/ASHEDUCATION-2009.1.132. [DOI] [PubMed] [Google Scholar]
- Thrasher AJ, Burns SO, 2010. WASP: a key immunological multitasker. Nat. Rev. Immunol. 103 (10), 182–192. 10.1038/nri2724. [DOI] [PubMed] [Google Scholar]
- Thurston TLM, Ryzhakov G, Bloor S, von Muhlinen N, Randow F, 2009. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 10, 1215–1221. 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- Torres E, Rosen MK, 2006. Protein-tyrosine kinase and GTPase signals cooperate to phosphorylate and activate Wiskott-Aldrich syndrome protein (WASP)/neuronal WASP. J. Biol. Chem. 281, 3513–3520. 10.1074/jbc.M509416200. [DOI] [PubMed] [Google Scholar]
- Tremel S, Ohashi Y, Morado DR, Bertram J, Perisic O, Brandt LTL, von Wrisberg M-K, Chen ZA, Maslen SL, Kovtun O, Skehel M, Rappsilber J, Lang K, Munro S, Briggs JAG, Williams RL, 2021. Structural basis for VPS34 kinase activation by Rab1 and Rab5 on membranes. Nat. Commun. 12, 1564. 10.1038/s41467-021-21695-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulferts S, Prajapati B, Grosse R, Vartiainen MK, 2021. Emerging Properties and Functions of Actin and Actin Filaments Inside the Nucleus. Cold Spring Harb. Perspect. Biol. 13, a040121. 10.1101/cshperspect.a040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsumi T, Sakurai N, Nakano K, Ishisaka R, 2003. C-terminal 15 kDa fragment of cytoskeletal actin is posttranslationally N-myristoylated upon caspase-mediated cleavage and targeted to mitochondria. FEBS Lett. 539, 37–44. 10.1016/s0014-5793(03)00180-7. [DOI] [PubMed] [Google Scholar]
- Valdmanis PN, Meijer IA, Reynolds A, Lei A, MacLeod P, Schlesinger D, Zatz M, Reid E, Dion PA, Drapeau P, Rouleau GA, 2007. Mutations in the KIAA0196 Gene at the SPG8 Locus Cause Hereditary Spastic Paraplegia. Am. J. Hum. Genet 80, 152. 10.1086/510782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MMK, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, González-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B, Auburger G, Wood NW, 2004. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 304, 1158–1160. 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- Vartiainen MK, Guettler S, Larijani B, Treisman R, 2007. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 316, 1749–1752. 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- Velle KB, Fritz-Laylin LK, 2019. Diversity and evolution of actin-dependent phenotypes. Curr. Opin. Genet. Dev. 58–59, 40–48. 10.1016/j.gde.2019.07.016. [DOI] [PubMed] [Google Scholar]
- Veltman DM, Insall RH, 2010. WASP family proteins: their evolution and its physiological implications. Mol. Biol. Cell 21, 2880–2893. 10.1091/mbc.E10-04-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboon JM, Rincon-Arano H, Werwie TR, Delrow JJ, Scalzo D, Nandakumar V, Groudine M, Parkhurst SM, 2015. Wash interacts with lamin and affects global nuclear organization. Curr. Biol. CB 25, 804–810. 10.1016/j.cub.2015.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodopiutz J, Seidl R, Prayer D, Khan MI, Mayr JA, Streubel B, Steiß JO, Hahn A, Csaicsich D, Castro C, Assoum M, Müller T, Wieczorek D, Mancini GMS, Sadowski CE, Lévy N, Mégarbané A, Godbole K, Schanze D, Hildebrandt F, Delague V, Janecke AR, Zenker M, 2015. WDR73 Mutations Cause Infantile Neurodegeneration and Variable Glomerular Kidney Disease. Hum. Mutat. 36, 1021–1028. 10.1002/HUMU.22828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpi S, Cicalese MP, Tuijnenburg P, Tool ATJ, Cuadrado E, Abu-Halaweh M, Ahanchian H, Alzyoud R, Akdemir ZC, Barzaghi F, Blank A, Boisson B, Bottino C, Brigida I, Caorsi R, Casanova JL, Chiesa S, Chinn IK, Dückers G, Enders A, Erichsen HC, Forbes LR, Gambin T, Gattorno M, Karimiani EG, Giliani S, Gold MS, Jacobsen EM, Jansen MH, King JR, Laxer RM, Lupski JR, Mace E, Marcenaro S, Maroofian R, Meijer AB, Niehues T, Notarangelo LD, Orange J, Pannicke U, Pearson C, Picco P, Quinn PJ, Schulz A, Seeborg F, Stray-Pedersen A, Tawamie H, van Leeuwen EMM, Aiuti A, Yeung R, Schwarz K, Kuijpers TW, 2019. A combined immunodeficiency with severe infections, inflammation, and allergy caused by ARPC1B deficiency. J. Allergy Clin. Immunol. 143, 2296. 10.1016/J.JACI.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Karstedt S, Montinaro A, Walczak H, 2017. Exploring the TRAILs less travelled: TRAIL in cancer biology and therapy. Nat. Rev. Cancer 17, 352–366. 10.1038/nrc.2017.28. [DOI] [PubMed] [Google Scholar]
- von Loeffelholz O, Purkiss A, Cao L, Kjaer S, Kogata N, Romet-Lemonne G, Way M, Moores CA, 2020. Cryo-EM of human Arp2/3 complexes provides structural insights into actin nucleation modulation by ARPC5 isoforms. Biol. Open 9, bio054304. 10.1242/bio.054304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AR, Luan Q, Liu S-L, Nolen BJ, 2013. Dip1 defines a class of Arp2/3 complex activators that function without preformed actin filaments. Curr. Biol. CB 23, 1990–1998. 10.1016/j.cub.2013.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhou G-L, Vedantam S, Li P, Field J, 2008. Mitochondrial shuttling of CAP1 promotes actin- and cofilin-dependent apoptosis. J. Cell Sci. 121, 2913–2920. 10.1242/jcs.023911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Fedoseienko A, Chen B, Burstein E, Jia D, Billadeau DD, 2018. Endosomal Receptor Trafficking: Retromer and Beyond. Traffic 19, 578. 10.1111/TRA.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Du X-H, Hong Y, Hong X, Fan L, Zhou J-W, Sun H, Ge J, Billadeau DD, Deng Z-H, 2022. WASH interacts with Ku to regulate DNA double-stranded break repair. iScience 25, 103676. 10.1016/j.isci.2021.103676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M, Fan X, Ding M, Li R, Shao S, Hou Y, Meng S, Tang F, Li C, Sun Y, 2020. Nuclear actin regulates inducible transcription by enhancing RNA polymerase II clustering. Sci. Adv. 6, eaay6515. 10.1126/sciadv.aay6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, DePace AH, Verma S, Iwamatsu A, Mitchison TJ, 1997. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J. Cell Biol. 138, 375–384. 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, Rosenblatt J, Skoble J, Portnoy DA, Mitchison TJ, 1998. Interaction of Human Arp2/3 Complex and the Listeria monocytogenes ActA Protein in Actin Filament Nucleation. Science 281, 105. [DOI] [PubMed] [Google Scholar]
- White SR, Williams P, Wojcik KR, Sun S, Hiemstra PS, Rabe KF, Dorscheid DR, 2001. Initiation of apoptosis by actin cytoskeletal derangement in human airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 24, 282–294. 10.1165/ajrcmb.24.3.3995. [DOI] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dötsch V, Bumann D, Dikic I, 2011. Phosphorylation of the Autophagy Receptor Optineurin Restricts Salmonella Growth. Science 333, 228–233. 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YC, Holzbaur ELF, 2014, Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc. Natl. Acad. Sci. U. S. A. 111, E4439–E4448. 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Seylani A, Wu J, Wu X, Bleck CKE, Sack MN, 2021. BLOC1S1/GCN5L1/BORCS1 is a critical mediator for the initiation of autolysosomal tubulation. Autophagy 17, 3707–3724. 10.1080/15548627.2021.1894759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Suetsugu S, Cooper LA, Takenawa T, Guan J-L, 2004. Focal adhesion kinase regulation of N-WASP subcellular localization and function. J. Biol. Chem. 279, 9565–9576. 10.1074/jbc.M310739200. [DOI] [PubMed] [Google Scholar]
- Wu X, Yoo Y, Okuhama NN, Tucker PW, Liu G, Guan J-L, 2006. Regulation of RNA-polymerase-II-dependent transcription by N-WASP and its nuclear-binding partners. Nat. Cell Biol. 8, 756–763. 10.1038/ncb1433. [DOI] [PubMed] [Google Scholar]
- Xia P, Wang S, Du Y, Zhao Z, Shi L, Sun L, Huang G, Ye B, Li C, Dai Z, Hou N, Cheng X, Sun Q, Li L, Yang X, Fan Z, 2013. WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J. 32, 2685–2696. 10.1038/emboj.2013.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P, Wang S, Huang G, Zhu P, Li M, Ye B, Du Y, Fan Z, 2014. WASH is required for the differentiation commitment of hematopoietic stem cells in a c-Myc-dependent manner. J. Exp. Med 211, 2119–2134. 10.1084/jem.20140169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Wang X, 2021. Lysosome biogenesis: Regulation and functions. J. Cell Biol. 220 10.1083/JCB.202102001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JK, 2015. Death effecter domain for the assembly of death-inducing signaling complex. Apoptosis Int. J. Program. Cell Death 20, 235–239. 10.1007/s10495-014-1060-6. [DOI] [PubMed] [Google Scholar]
- Yang S, Tang Y, Liu Y, Brown AJ, Schaks M, Ding B, Kramer DA, Mietkowska M, Ding L, Alekhina O, Billadeau DD, Chowdhury S, Wang J, Rottner K, Chen B, 2022. Arf GTPase activates the WAVE regulatory complex through a distinct binding site. Sci. Adv. 8, eadd1412. 10.1126/sciadv.add1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau R, Rape M, 2016. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 186 (18), 579–586. 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- Yerbury JJ, Ooi L, Dillin A, Saunders DN, Hatters DM, Beart PM, Cashman NR, Wilson MR, Ecroyd H, 2016. Walking the tightrope: Proteostasis and neurodegenerative disease. J. Neurochem 137, 489–505. 10.1111/jnc.13575. [DOI] [PubMed] [Google Scholar]
- Yi K, Unruh JR, Deng M, Slaughter BD, Rubinstein B, Li R, 2011. Dynamic maintenance of asymmetric meiotic spindle position through Arp2/3-complex-driven cytoplasmic streaming in mouse oocytes. Nat. Cell Biol. 13, 1252–1258. 10.1038/ncb2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Park HH, Chung JY, Lin S-C, Lo Y-C, da Graca LS, Jiang X, Wu H, 2006. Caspase-9 holoenzyme is a specific and optimal procaspase-3 processing machine. Mol. Cell 22, 259–268. 10.1016/j.molcel.2006.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Popelka H, Lei Y, Yang Y, Klionsky DJ, 2020. The Roles of Ubiquitin in Mediating Autophagy. Cells 9, 2025. 10.3390/cells9092025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LE, Heimsath EG, Higgs HN, 2015. Cell type-dependent mechanisms for formin-mediated assembly of filopodia. Mol. Biol. Cell 26, 4646–4659. 10.1091/mbc.E15-09-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Yu X, Topf M, Ludtke SJ, Wang X, Akey CW, 2010. Structure of an apoptosome-procaspase-9 CARD complex. Structure 1993 (18), 571–583. 10.1016/j.str.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Yu X, Asara JM, Heuser JE, Ludtke SJ, Akey CW, 2011. The holo-apoptosome: activation of procaspase-9 and interactions with caspase-3. Structure 1993 (19), 1084–1096. 10.1016/j.str.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavodszky E, Seaman MNJ, Moreau K, Jimenez-Sanchez M, Breusegem SY, Harbour ME, Rubinsztein DC, 2014. Mutation in VPS35 associated with Parkinson’s disease impairs WASH complex association and inhibits autophagy. Nat. Commun. 5 10.1038/ncomms4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zech T, Calaminus SDJ, Caswell P, Spence HJ, Carnell M, Insall RH, Norman J, Machesky LM, 2011. The Arp2/3 activator WASH regulates α5β1-integrin-mediated invasive migration. J. Cell Sci. 124, 3753–3759. 10.1242/jcs.080986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YG, Zhang H, 2019. Autophagosome maturation: An epic journey from the ER to lysosomes. J. Cell Biol. 218, 757–770. 10.1083/jcb.201810099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Sumigray KD, Lechler T, 2015. The Arp2/3 complex has essential roles in vesicle trafficking and transcytosis in the mammalian small intestine. Mol. Biol. Cell 26, 1995–2004. 10.1091/mbc.E14-10-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Li Y, Hu Q, Bai X-C, Huang W, Yan C, Scheres SHW, Shi Y, 2015. Atomic structure of the apoptosome: mechanism of cytochrome c- and dATP-mediated activation of Apaf-1. Genes Dev. 29, 2349–2361. 10.1101/gad.272278.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Fukada K, Zhu H, Kyprianou N, 2006. Prohibitin and cofilin are intracellular effectors of transforming growth factor beta signaling in human prostate cancer cells. Cancer Res 66, 8640–8647. 10.1158/0008-5472.CAN-06-1443. [DOI] [PubMed] [Google Scholar]
- Zimmet A, Van Eeuwen T, Boczkowska M, Rebowski G, Murakami K, Dominguez R, 2020. Cryo-EM structure of NPF-bound human Arp2/3 complex and activation mechanism. Sci. Adv. 6, eaaz7651 10.1126/sciadv.aaz7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoppino FCM, Militello RD, Slavin I, Álvarez C, Colombo MI, 2010. Autophagosome formation depends on the small GTPase rab1 and functional ER exit sites. Traffic 11, 1246–1261. 10.1111/j.1600-0854.2010.01086.x. [DOI] [PubMed] [Google Scholar]
- Zou H, Li Y, Liu X, Wang X, 1999. An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J. Biol. Chem. 274, 11549–11556. 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
- Zuchero JB, Coutts AS, Quinlan ME, Thangue NBL, Mullins RD, 2009. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat. Cell Biol. 11, 451–459. 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchero JB, Belin B, Mullins RD, 2012. Actin binding to WH2 domains regulates nuclear import of the multifunctional actin regulator JMY. Mol. Biol. Cell 23, 853–863. 10.1091/mbc.E11-12-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]


