Abstract
Potassium can stabilize the formation of chair- or edge-type quadruplex DNA structures and appears to be the only naturally occurring cation that can do so. As quadruplex DNAs may be important in the structure of telomere, centromere, triplet repeat and other DNAs, information about the details of the potassium–quadruplex DNA interactions are of interest. The structures of the 1:1 and the fully saturated, 2:1, potassium–DNA complexes of d(GGTTGGTGTGGTTGG) have been determined using the combination of experimental NMR results and restrained molecular dynamics simulations. The refined structures have been used to model the interactions at the potassium binding sites. Comparison of the 1:1 and 2:1 potassium:DNA structures indicates how potassium binding can determine the folding pattern of the DNA. In each binding site potassium interacts with the carbonyl oxygens of both the loop thymine residues and the guanine residues of the adjacent quartet.
INTRODUCTION
Quadruplex DNA may play a role in telomere structure and function (1,2), and in the activities of fragile X (3–6), immunoglobin switch regions and centromere DNAs as well as in other biological systems (7–13). Quadruplex DNAs are also being investigated as lead molecules in drug design (14–16). The selective binding of porphyrins and other molecules to quadruplex DNAs has become, in rather a short time, an area of considerable interest with a number of intriguing results and hypotheses being presented (17–21). This interest is partially based on the biological importance of telomerase and telomere DNA and their potential as therapeutic targets.
The structure and stability of quadruplex DNAs are known to be dependent on the concentration and type of monovalent cation present in the solution (1,22–27). These and other studies (28–32) have shown that the effects of monovalent cations on quadruplex DNA are distinct from those observed for duplex DNA, tRNA or pseudoknot RNA (26,33–35). There have also been reports that certain divalent and trivalent ions interact with quadruplex DNAs (27,36,37). We have shown that manganese preferentially binds to the narrow grooves of two distinct types of quadruplex DNA (38,39), as does europium (40).
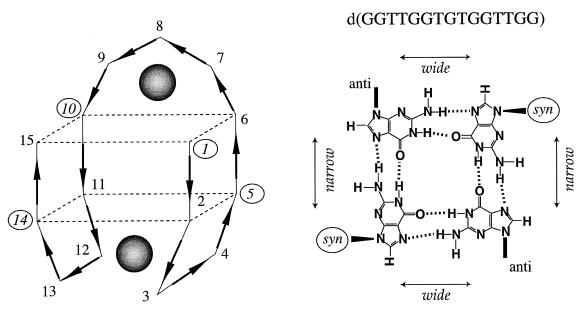
Three general types of quadruplex DNA structure based on dG quartets are known. In the chair- or edge-type structure the residues of each quartet alternate syn-anti-syn-anti as depicted in Figure 1. In the basket, or crossover-type structure (41,42) the residues alternate syn-syn-anti-anti within each quartet. The quadruplex structures formed by four parallel strands (43–47) have four medium width grooves with all the residues in the quartets anti. A classification scheme has been proposed for the potassium binding of these three types of DNA quadruplex structure (48). The proposal is that only the chair-type quadruplex structure requires potassium and that the potassium binds between the loops and the adjacent quartets. All of the quadruplex DNAs with quartets of dG residues examined to date follow this pattern (48).
Figure 1.
The structure of the chair-type quadruplex structure of d(GGTTGGTGTGGTTGG) is shown along with the positions of the two strong potassium binding sites. The positions of the potassium binding sites are indicated by the shaded spheres. On the right is shown the syn-anti-syn-anti arrangement of the dG residues in each of the two quartets of the chair-type structure.
The potassium binding that is essential for a specific type of quadruplex structure to be formed is the main interest here. Another class of weaker binding is evidenced by the modest structural changes that are induced by the presence of potassium relative to the sodium form of a DNA quadruplex (22). In this latter case, the addition of potassium to a basket-type quadruplex DNA in the presence of sodium induces modest structural changes but does not change the type of quadruplex structure. Ammonia can also bind to basket-type quadruplex structures with modest effects on the structure of the DNA (49,50). In the crystal structure of a parallel strand quadruplex structure, sodium was found between adjacent quartets (45).
The DNA d(GGTTGGTGTGGTTGG) has been shown to adopt a chair-type quadruplex structure in the presence of potassium (51–53). Potassium is required for this DNA to adopt the structure needed for the inhibition of thrombin (15,52,53). NMR has been used to monitor the titration of this DNA with potassium and the results showed that the structural changes are complete upon the addition of two potassium ions per DNA. The complex is formed in a sequential, stepwise fashion with the binding of a single potassium per DNA allowing a chair-type structure to form. The binding of the second potassium induces additional structural changes with the DNA remaining in a chair-type structure. In this study, NMR and restrained molecular dynamics have been used to obtain refined structures of both the 1:1 and 2:1 potassium:DNA complexes. The structures of the DNA in these complexes have been used to determine the locations of and interactions at the potassium binding sites.
MATERIALS AND METHODS
DNA purification and NMR sample preparation
The 15mer d(GGTTGGTGTGGTTGG) was obtained from Integrated DNA Technologies, Inc. (Coralville, IA), purified by HPLC, lyophilized and ethanol precipitated three times. The sample was examined by NMR and HPLC methods, and neither method detected any impurity in the sample.
The NMR samples each consisted of 500 µl 1.4 mM DNA, 100 A260, in 140 mM NaCl and 20 mM perdeuteriated Tris buffer at pH 7.0 in grade-6 NMR tubes (Scientific Glassware, Vineland, NJ). Each sample was dried in the NMR tube using N2 gas and then dissolved with 500 µl 2H2O. The pH was checked and adjusted, if necessary, to 7.0. The extinction coefficient of the DNA is 143 303 M–1cm–1 (53).
KCl was added to one of the DNA NMR samples to produce a 1:1 potassium:DNA ratio using 9.979 ± 0.0004 mM KCl whose concentration was determined by atomic absorption (Galbraith Laboratories, Inc., Knoxville, TN). The DNA was annealed after KCl titration. The other sample contained 5 mM KCl.
NMR procedures
The one-dimensional 1H and 31P-NMR experiments on the samples in 2H2O were carried out on a Varian 400 MHz UnityPlus spectrometer with the samples at 15°C. The one-dimensional proton spectra were obtained with 512 transients, a spectral width of 6000 Hz and a delay time of 1 s. The one-dimensional 31P experiments were obtained with 5000 transients, a spectral width of 9000 and a delay time of 1 s.
BASHD-TOCSY experiments (54) were also conducted on the Varian 400 MHz Unity Plus spectrometer. BASHD-TOCSY spectra were acquired at 15°C using a delay time of 1 s, a spectral width of 4000 Hz in the proton dimension and 1500 Hz in the phosphorus dimension. 120 increments of t1 were used with 196 transients for each increment, and with a mixing time of 70 ms as previously described. The data were transformed into a 2048 × 2048 real point matrix.
One-dimensional NMR experiments with watergate suppression were carried out on a Varian Inova 500 MHz spectrometer. Sixteen thousand complex points were collected with 2048 transients with the samples at 5°C. A spectral width of 20 000 Hz and a delay time of 1 s were used.
ROESY, NOESY, NOESY with watergate suppression and PE-COSY experiments were conducted using the Varian Inova 500 MHz spectrometer. ROESY spectra were carried out with the samples at 15°C using a delay time of 1 s, a mixing time of 55 ms and a spectral width of 5000 Hz in both dimensions; 128 transients were used for each of the 250 increments of t1. The data were zero filled to 2048 × 2048 real points.
NOESY experiments were carried out with the samples at 15°C using a mixing time of 250 or 100 ms, a delay time of 1 s and a spectral width of 5000 Hz in both dimensions; 4096 complex points in t2 were collected along with 128 transients for each of 300 increments of t1. The data were zero filled to 4096 × 4096 real points.
Watergate NOESY spectra were carried out with the samples at 5°C, a delay time of 1 s, a mixing time of 250 ms, a spectral width of 12 000 Hz in both dimensions and 128 transients for each of 400 increments of t1. The data were zero filled to 4096 × 4096 real points.
PE-COSY experiments were carried out with the samples at 15°C, 31P decoupling during t1, a delay time of 1 s and a spectral width of 7500 Hz in both dimensions; 2048 complex points in t2 were used and 128 transients were averaged for each of 300 increments of t1. The data was zero filled to 2048 × 2048 real points.
All the NMR data were processed with Gaussian weighting functions in both dimensions using Varian VNMR 6.1 software.
Assignments and quantification of NOE cross-peaks
The spectra of both samples were assigned based on methods previously described (38,39,52,53,55). The NOE cross-peak volumes of both structures obtained at 500 MHz with mixing times of 100 and 250 ms were quantified using both FELIX 97.0 and VNMR 6.1 software
Structure determination and refinement
Structure refinement of the DNA saturated with potassium was performed using complete relaxation matrix, restrained molecular dynamics, essentially as previously described (39,48,56). There were 303 NOE volume constraints from the 100 ms and 498 from the 250 ms NOESY data. In addition 10 NOE constraints involving the imino protons of residues G1, G5, G6, G8, G10, G14 and G15 were used as distance constraints. Four hydrogen bonds were used for each guanine involved in a quartet. Experimental coupling constants were used to generate 45 dihedral constraints, 15 from BASHD-TOCSY experiments (H3′-31P), and 30 from PE-COSY data (H1′-C1′-C2′-H2′ and H1′-C1′-C2′-H2″). Additional constraints were used to keep the purine rings planar. The starting structure used was the structure obtained in a previous refinement (39). Additional detail is included in the Supplementary Material.
PDB and BMRB file information
The coordinates of the structures have been deposited. The file for the structure of the 2:1 complex coordinates is 1C32, and the structure with the single potassium is 1C34. The file for the structure of the 1:1 complex coordinates is 1C35, and the structure with the potassium is 1C38. The 10 structures were extracted from the trajectory at 2 ps intervals from 80 to 100 ps in each file. All of the NMR data relevant to the structure determinations have been deposited at the BioMagRes Bank as file BMRB-4372.
RESULTS AND DISCUSSION
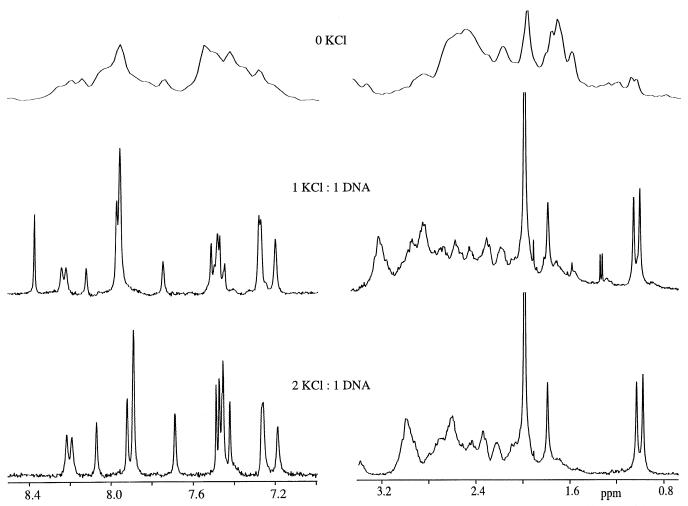
Potassium induces striking changes in the chemical shifts of the resonances of the non-exchangeable protons of the 15mer, as demonstrated by the results in Figure 2. The proton NMR spectrum of the sample in the presence of 140 mM sodium does not exhibit sharp, resolved resonances in any spectral region. At the level of one potassium ion per DNA, a spectrum with one sharp resonance from each non-exchangeable proton in all spectral regions is obtained, as shown in Figure 2. This is indicative of a single, stable structure. This spectrum contains resonances that are not present in the spectrum of either the potassium-free or the potassium-saturated samples. The clearest example is the most downfield resonance, from the H8 of dG11, at 8.35 p.p.m., that is observed only for the intermediate (1:1) case. The presence of unique resonances indicates that the intermediate state has structural features distinct from those of both the potassium-free and the potassium-saturated states.
Figure 2.
The one-dimensional proton spectra of the 15mer sample in the presence of zero, one or two potassiums are shown. The spectra were obtained with the sample at 15°C.
When there are two potassium ions per DNA present, the proton spectrum is well resolved and contains one sharp resonance from each non-exchangeable proton in all spectral regions, as the results in Figure 2 show. Essentially, all of the proton resonances and all of the 31P resonances have been assigned (38,39,51–53,57). The addition of the second potassium also changes the chemical shifts and linewidths of the imino protons as shown in the Supplementary Material. The addition of more than two potassiums per DNA does not induce appreciable additional change in either the chemical shifts or linewidths of the proton resonances (48).
The titration of the DNA with potassium was also monitored by 31P-NMR. The 31P-NMR spectrum of the sample with one potassium per DNA contains resonances that are not observed from either the potassium-free or the potassium-saturated samples, as shown in the Supplementary Material. The largest differences in 31P chemical shifts between the 1:1 and saturated complexes are those of the phosphate between residues 7 and 8 and the one between residues 8 and 9. The addition of more than two potassium ions per DNA does not induce appreciable additional change in the chemical shifts or linewidths of the 31P resonances. The 31P results on the samples were found to be entirely complementary to the proton NMR data.
Qualitative analysis of the titration data
These titration results suggest that the presence of potassium induces a significant change in the structure of the DNA and that most of the changes occur upon the addition of the first potassium. Two potassium ions per DNA are needed to induce the entire structural change and there is sequential binding of the two potassium ions, as previously reported (48). Prior results obtained with 1.5 potassium ions per DNA showed that there is slow exchange between the 1:1 and saturated forms on the NMR time scale (48). In the presence of 0.5 potassiums per DNA the spectrum was found to be a mixture of sharp and broad resonances indicating the presence of slow exchange under these conditions (48).
While the changes in the chemical shifts and linewidths that are observed are consistent with there being structural differences between the 1:1 and 2:1 forms of the DNA, they do not provide sufficient information to determine the nature of the structural changes. To examine the details of the structural differences between the 1:1 and 2:1 complexes, their structures were determined. The structures were then used to model the energy of interaction with potassium to determine the locations of the potassium binding sites.
Structure determination of 1:1 and 2:1 complexes
The structure determinations generally followed the procedures we have used previously for quadruplex and duplex DNAs (39,56,58,59). NOE cross-peak volumes were determined from data obtained at multiple mixing times, both for non-exchangeable and exchangeable protons, and the volumes were used as constraints. Homonuclear and heteronuclear scalar couplings were also determined and these were used as additional constraints via the use of the appropriate Karplus equations. Distance constraints were used for those heavy atoms involved in hydrogen bonds and for the NOEs involving exchangeable protons. The data set used here for the 2:1 sample contains about twice as many NOE constraints as were available for our prior structure determinations (39,53). More NOE volumes were determined due to access to higher quality data. The current data sets of the NOEs of the exchangeable protons were also much better defined due to the availability of superior water suppression technology using pulsed field gradients. The presence of structural disorder in one loop of the 1:1 structure led to fewer constraints being available than for the 2:1 case. The assignments of the resonances are given in the Supplementary Material as a list of the largest NOE differences between the two forms. Lists of the NOE cross-peak volumes, the scalar couplings and the coordinates of the structures have been deposited as described above.
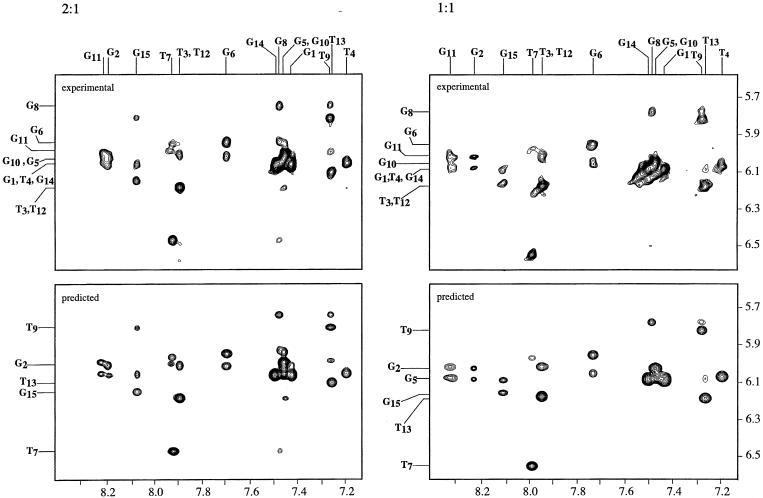
Regions of the NOESY data for the DNA under both sets of conditions are shown in Figures 3 and 4 along with the assignments of many of the crosspeaks. The overall features of the NOE data in the two cases are generally similar. In both cases there are four syn dG residues and the NOE connectivities are those of a chair-type quadruplex structure. Two of the larger differences obtained under the two sets of conditions are highlighted in Figure 3. Many of the largest differences in the NOE volumes involve dT7 or dG8.
Figure 3.
A region of the 250 ms mixing time, 500 MHz NOESY spectra of the 15mer is shown. The region shown contains the connectivities between the aromatic protons and those of the H1′. The top left contains the experimental data for the 2:1 complex and the bottom left the same spectral region predicted from the 10 structures obtained in the restrained molecular dynamics trajectory. The spectra were backcalculated from each of the ten structures and averaged. The top right contains the experimental data for the 1:1 complex and the bottom left the same spectral region predicted from the 10 structures obtained in the restrained molecular dynamics trajectory.
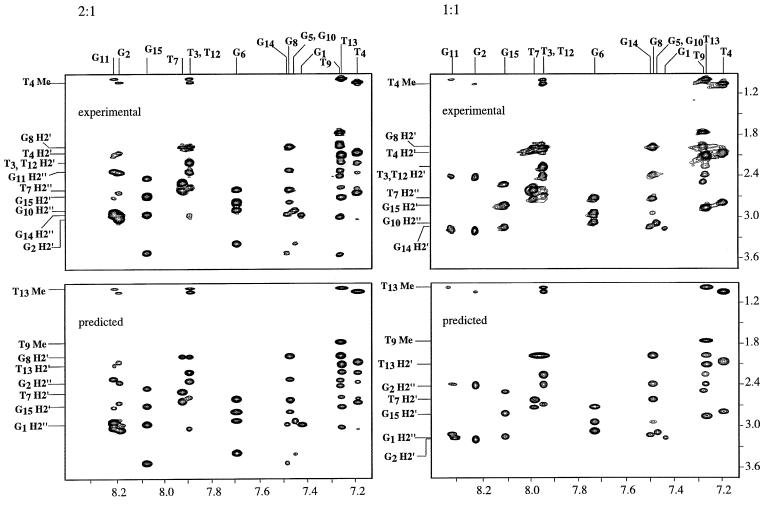
Figure 4.
A region of the 250 ms mixing time, 500 MHz NOESY spectra of the 15mer is shown. The region shown contains the aromatic-H2′, H2″, methyl connectivities. The top left contains the experimental data for the 2:1 complex and the bottom left the same spectral region predicted from the 10 structures obtained in the restrained molecular dynamics trajectory. The spectra were backcalculated from each of the 10 structures and averaged. The top right contains the experimental data for the 1:1 complex and the bottom left the same spectral region predicted from the 10 structures obtained in the restrained molecular dynamics trajectory.
The predicted spectra, those backcalculated from the refined structures, are also shown in Figures 3 and 4. The predicted and experimental spectra are in excellent agreement. The predicted spectra contain all of the crosspeaks of the experimental spectra with similar intensities. The predicted spectra do not contain any crosspeaks that are not observed in the experimental data.
The regions of the NOE maps obtained with the samples in H2O containing the crosspeaks of the methyl of dT9 to imino protons are shown in Figure 5. NOEs from the methyl group of dT9 to the imino protons of residues 1, 6, 10 and 15 are observed for the 2:1 sample but only to those of 1 and 10 for the 1:1 sample. Similarly, the NOEs of the H1′ of dT9 and dT8 to the imino protons of the dG residues of the top quartet change substantially upon addition of the second potassium, as the NOESY spectra in the Supplementary Material show.
Figure 5.
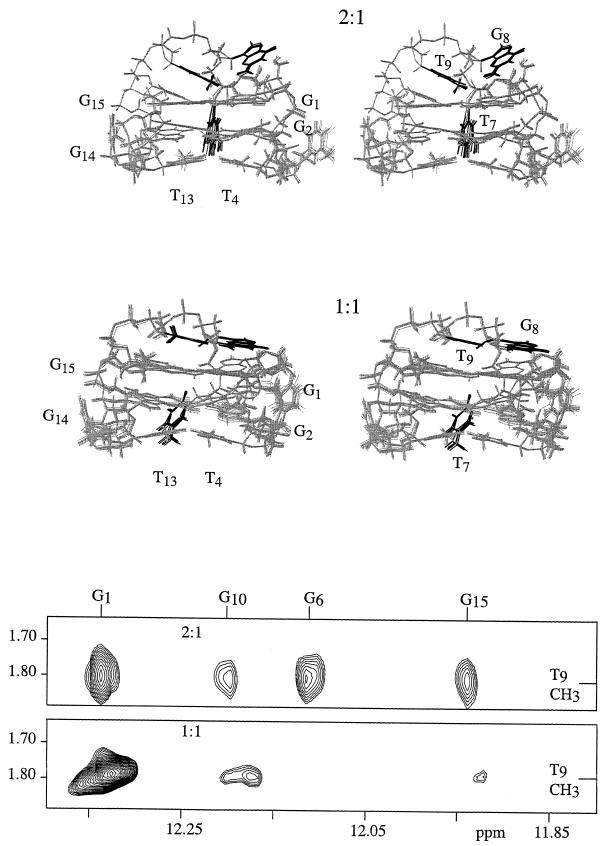
The 10 structures extracted from the trajectory of the 2:1 complex at 2 ps intervals, from 80 to 100 ps, are shown at the top of the figure. The 10 structures extracted from the trajectory of the 1:1 complex at 2 ps intervals, from 80 to 100 ps, are shown in the middle of the figure. The structures are shown in stereo mode. The aromatic bases of residues 7, 8 and 9 are depicted in black and the rest of the molecule in gray. Selected residues are labeled. A portion of the 100 ms mixing time, 500 MHz NOESY spectra of the 15mer in H2O is shown at the bottom of the figure. The region contains the NOE crosspeaks between the methyl protons of dT9 and the imino protons of residues 1, 6, 10 and 15. The spectra indicate that the position of the methyl of dT9 is closer to the imino protons of residues 6, 10 and 15 when the DNA is in the presence of two potassium than in the presence of one potassium.
A qualitative interpretation of the one-dimensional proton and phosphorus results and of the NOESY data indicates that most of the structural differences between the 1:1 and potassium-saturated forms of the DNA are in residues 7, 8 and 9 which make up the top loop. To allow examination of the details of the differences between the 1:1 and 2:1 forms, both structures have been determined.
Analysis of the structures of the 2:1 and 1:1 complexes
The overlay of 10 structures from the trajectory, at 2 ps intervals from 80 to 100 ps for both the 1:1 and 2:1 forms of the DNA, are shown in Figure 5. The overlay mode of representation indicates the range of structures that are consistent with the experimental data and shows that there is a relatively narrow range of structures consistent with the experimental results. This is a narrower range than we have found for prior structural determinations of quadruplex DNAs and reflects the relatively large number of experimental constraints that are now available.
The comparison of the structures of the 1:1 and 2:1 forms, shown in Figure 5, indicates that the quartets are nearly the same in the 1:1 and 2:1 cases. The structures of the two ‘bottom’ loops, containing dT3, dT4 and dT12, dT13, are also very similar in the 1:1 and 2:1 cases. The structure of the top loop, containing dT7, dG8 and dT9, is quite different in the two cases, the most striking difference between the two structures is the position of the base of dT7.
To determine the locations of the potassium binding sites the DNA structures were held fixed, potassium ions were added to the system and the energy of the DNA–potassium system was minimized. The minimum energy positions have been found to be independent of the starting positions of the potassium ions in the simulation as discussed in the Materials and Methods. The same basic approach was also used to predict the binding sites of manganese and europium that were found to be in excellent agreement with the experimentally determined binding sites (39,40). Unlike potassium, manganese and europium bind most strongly to the narrow grooves of the quadruplex DNAs (39,40).
Locations of the potassium binding sites
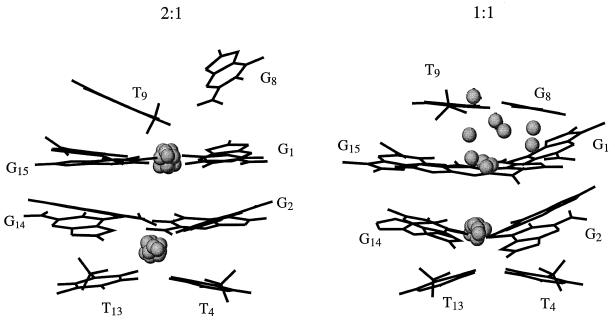
The positions of the potassium binding sites were determined for each of the 10 structures of the 2:1 and 1:1 complexes. Each of these structures, extracted from the trajectory at 2 ps intervals, was individually used to predict the positions of the potassium binding sites. The two lowest energy positions for potassium for each of the 10 structures are shown in Figure 6. The Supplementary Material contains figures that contain overlays of all of the structures and all of the potassium binding sites. It is seen that the predicted positions are all very close to one another. These positions are nearly identical to those determined using the previously refined structure (48). The modeling has the potassium ions binding to the residues of a quartet and to the dT residues in the adjacent loops. In each binding site the potassium ion can interact with the carbonyl atoms of both dT and dG residues as discussed previously (48). This model predicts that the substitution of a purine for a pyrimidine at any of the dT positions involved in the binding of the potassium will disrupt the binding, and this prediction has been experimentally confirmed (48).
Figure 6.
The structure of the 2:1 complex is shown on the left. The structure of the DNA is that obtained at 100 ps in the trajectory. Only some of the bases of the DNA are shown for clarity. The spheres indicate the positions of the potassiums that were found for each of the 10 structures extracted from the trajectory from 80 to 100 ps. The spheres have one-third, 0.65 Å, the van der Waals radius of potassium, 1.96 Å. The analogous structures are shown on the right for the 1:1 case.
The same protocol has been applied to the 1:1 structure with the predicted binding sites shown in Figure 6. The positions of the potassium binding sites were determined for each of the 10 structures of the 1:1 complex that are shown in overlay mode in Figure 5. The minimization places the strong binding site for potassium between the bottom loops and the adjacent quartet. This binding site is as well defined as was found for the 2:1 case, being nearly the same for all of the 10 structures extracted from the trajectory. The situation for the weaker site is quite different. Each of the 10 structures predicts a distinct binding site for the potassium near the top loop. All of these predicted sites are of higher energy (average energy of 1135 with the highest energy 1348, lowest 1087 and a standard deviation of 186 kcal/mol; RMSD of potassium positions 0.230 Å) than that of the bottom loop (average of 1067 with the highest energy 1073, lowest 1061 and a standard deviation of 9 kcal/mol; RMSD of potassium positions 0.010 Å). These energy calculations indicate that there is not a single, minimum energy site for potassium binding to the top loop and top quartet.
The predicted energy of the bottom loop for the 2:1 case is lower (average of 880 with the highest energy 885, lowest 876 and a standard deviation of 5 kcal/mol; RMSD of potassium positions 0.037 Å), within the confidence level of these calculations, than that of the analogous case in the 1:1 case. This indicates that the binding of the second potassium increases the stability of the first site. The energy of the top loop-quartet binding site in the saturated case (average of 918 with the highest energy 927, lowest 915 and a standard deviation of 7 kcal/mol; RMSD of potassium positions 0.031 Å) is not as strong as the bottom-loop quartet binding site.
Nature of special interaction between chair-type quadruplex structures and potassium
The simulations predict that there is only one well-defined potassium binding site for the structure determined using data for the DNA in the presence of one potassium ion per DNA. The simulations on the structure obtained for the DNA in the presence of a saturating amount of potassium has two binding sites, in agreement with our prior results, those shown in Figures 2–5 and those in the Supplementary Material. Thus, the simulations based on the structures agree with the experimental results on the number of strong binding sites. The structure of the DNA in the presence of one potassium is, however, very similar in most respects to that in the presence of two potassiums per DNA. The formation of the 1:1 complex is sufficient to obtain the overall global fold of the DNA. The binding of the second potassium primarily alters the structure of the top loop and perhaps its flexibility as the range of structures consistent with the experimental results is a bit narrower for the saturated case.
These results are also consistent with our prior examination of the interactions of potassium with a series of DNA of varying loop size and sequence. The results showed that for some sequences saturation occurs with one potassium per DNA and the chair-type global fold is formed (48). The relative strengths of the two binding sites are dependent on the loop sequences (48).
DNA quadruplexes with crossover loops, also known as basket structures, do not have a potassium requirement. Basket structures have more residues in their loops than do chair-type structures which have loops on the edge of the quartets. The short length and orientation of the bottom loops of chair structures appear to be the key elements in determining the potassium requirement. The bottom loops of chair-type structures can consist of two or three residues. At this length the dT residues in the loops can orient towards the carbonyl oxygens of the dG residues in the bottom quartet to form a high affinity binding site for potassium. The results presented here show that the structure of this binding site is not significantly altered by the binding of the additional potassium to the residues of the top loop and top quartet.
Summary and biological implications
There are two classes of potassium interactions with quadruplex DNAs that are based on quartets of dG residues (48). In one class of interaction the potassium binds to the quadruplex DNA but does not alter its structural type, and the same structural type can be formed in the presence of sodium alone as in the presence of potassium and sodium. This is the case for all known basket and parallel-stranded quadruplex DNAs based on dG quartets. A potassium concentration of ∼50 mM is needed to fully induce the modest structural changes of a crossover-type structure (22). The second class of effect is the requirement of chair-type structures for the presence of potassium. While the binding constant of chair-type structures has not been determined, the potassium affinity is >1 mM (48).
The potassium binding sites of crossover-type quadruplex structures have been proposed to be between adjacent quartets with the potassium interacting with the carbonyls of dG residues of both quartets (22,60–62). It has also been proposed that the locations of the binding sites of ammonia and of potassium to crossover-type quadruplex DNAs are the same (49,60). This may well be the case for the relatively weak binding of potassium to crossover-type quadruplex structures but is not the case for basket-type structures. We have found that even in the presence of 50 mM ammonia the 15mer does not adopt the structure induced by potassium.
Chair-type structures based on dG quartets have only been observed in the presence of potassium. Changes in the potassium concentration have pronounced effects on DNAs that can form chair-type structures. As the chair-type structures will only form in the presence of potassium, changes in the effective potassium concentration could switch such processes as replication, transcription and telomerase replication on and off. Regulation of the structural change could be obtained by competition between protein and potassium binding to the DNA with high potassium levels driving the formation of chair-type quadruplex structures by telomere, centromere, triplet repeat, integration site or other DNA. The total potassium concentration in the nucleus is >50 mM but the concentration of potassium that is not bound to anions or otherwise sequestered is not known (63,64). This suggests that the effects that require high levels, in the order of 50 mM, of potassium may not be biologically relevant.
The quadruplex structure of a head-to-tail dimer that contains both G-C-G-C and G-G-G-G quartets has been obtained for the sample in the presence of 100 mM potassium (65). This DNA is similar to the 15mer in that the dG residues alternate syn-anti-syn-anti in the G-G-G-G quartets, and that the loops are of the chair variety. This structure is consistent with a total of five binding sites for potassium. One of the proposed binding sites is made up entirely of four loop residues. The other potassium binding sites are primarily in the grooves. One of the potential binding sites between two G-G-G-G quartets may exist. The relative energies of the proposed sites were not presented (65). The potassium-free and potassium-bound forms of this DNA are of the same quadruplex type. The titration of the DNA with potassium indicates that fast averaging, on the NMR time scale, between the two forms can occur. This is in distinct contrast to what is observed for the 15mer which exhibits slow exchange and only adopts the chair quadruplex form in the presence of potassium.
The presence of potassium is essential for the formation of the chair-type quadruplex structure by d(GGTTGGTGTGGT-TGG) that is needed to inhibit thrombin. The structure of the top loop, which is stabilized by the binding of the second potassium, has been shown to be important in thrombin inhibition (52,53,66).
SUPPLEMENTARY MATERIAL
Five figures and one table containing the phosphorus NMR spectra, additional structures, selected NOSEY data obtained on the samples in H2O buffer and a description of the details of the structure determination methods are available as Supplementary Material at NAR Online. The table contains a listing of the largest percentage differences in the intraresidue and interresidue NOEs between the saturated 2:1 and intermediate 1:1 forms of the 15mer.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Dr Richard Beger for his assistance with the structure determination, and Haribabu Arthanari and Kevin McConnell for use of their structure and NOE averaging programs. This research was supported, in part, by grant GM 51298 from the National Institutes of Health. The 400 MHz NMR spectrometer was purchased with support from the National Science Foundation BIR 93-03077. The 500 MHz spectrometer was purchased with support from the National Science Foundation BIR-95-12478, and from the Camille and Henry Dreyfus Foundation.
NDB/PDB accession nos+ To whom correspondence should be addressed. Tel: +1 860 685 2668; Fax: +1 860 685 2211; Email: pbolton@wesleyan.edu 1C32, 1C34, 1C35, 1C38
REFERENCES
- 1.Feigon J., Dieckmann,T. and Smith,F.W. (1996) Chem. Biol., 3, 611–617. [DOI] [PubMed] [Google Scholar]
- 2.Williamson J.R. (1994) Annu. Rev. Biophys. Biomol. Struct., 23, 703–730. [DOI] [PubMed] [Google Scholar]
- 3.Kettani A., Kumar,R.A. and Patel,D.J. (1995) J. Mol. Biol., 254, 638–656. [DOI] [PubMed] [Google Scholar]
- 4.Fry M. and Loeb,L.A. (1994) Proc. Natl Acad. Sci. USA, 91, 4950–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mariappan S.V., Catasti,P., Chen,X., Ratliff,R., Moyzis,R.K., Bradbury,E.M. and Gupta,G. (1996) Nucleic Acids Res., 24, 784–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith S.S., Laayoun,A., Lingeman,R.G., Baker,D.J. and Riley,J. (1994) J. Mol. Biol., 243, 143–151. [DOI] [PubMed] [Google Scholar]
- 7.deLange T. (1994) Proc. Natl Acad. Sci. USA, 91, 2882–2885.8159672 [Google Scholar]
- 8.Blackburn E.H. (1994) Cell, 77, 621–623. [DOI] [PubMed] [Google Scholar]
- 9.Greider C.W. and Blackburn,E.H. (1996) Sci. Am., 274, 92–97. [DOI] [PubMed] [Google Scholar]
- 10.Marx J. (1994) Science, 265, 1656–1658. [DOI] [PubMed] [Google Scholar]
- 11.Schippen D.E. (1993) Curr. Opin. Genet. Dev., 5, 759–763. [DOI] [PubMed] [Google Scholar]
- 12.Sen D. and Gilbert,W. (1992) Methods Enzymol., 211, 191–199. [DOI] [PubMed] [Google Scholar]
- 13.Williamson J.R. (1993) Proc. Natl Acad. Sci. USA, 90, 3124.7682693 [Google Scholar]
- 14.Bock L.C., Griffin,L.C., Latham,J.A., Vermaas,E.H. and Toole,J.J. (1992) Nature, 355, 564–566. [DOI] [PubMed] [Google Scholar]
- 15.Paborsky L.R., McCurdy,S.N., Griffin,L.C., Toole,J.J. and Leung,L.L. (1993) J. Biol. Chem., 268, 20808–20811. [PubMed] [Google Scholar]
- 16.Tasset D.M., Kubik,M.F. and Steiner,W. (1997) J. Mol. Biol., 272, 688–698. [DOI] [PubMed] [Google Scholar]
- 17.Shafer R.H. (1998) Prog. Nucleic Acid Res. Mol. Biol., 59, 55–94. [DOI] [PubMed] [Google Scholar]
- 18.Wheelhouse R.T., Sun,D., Han,H., Han,F.X. and Hurley,L.H. (1998) J. Am. Chem. Soc., 120, 3261–3262. [Google Scholar]
- 19.Arthanari H., Basu,S., Kawano,T.L. and Bolton,P.H. (1998) Nucleic Acids Res., 26, 3724–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anantha N.V., Azam,M. and Sheardy,R.D. (1998) Biochemistry, 37, 2709–2714. [DOI] [PubMed] [Google Scholar]
- 21.Fedoroff O.Y., Salazar,M., Han,H., Chemeris,V.V., Kerwin,S.M. and Hurley,L.H. (1998) Biochemistry, 37, 12367–12374. [DOI] [PubMed] [Google Scholar]
- 22.Hud N.V., Smith,F.W., Anet,F.A. and Feigon,J. (1996) Biochemistry, 35, 15383–15390. [DOI] [PubMed] [Google Scholar]
- 23.Hardin C.C., Corregan,M.J., Lieberman,D.V. and Brown,B.A.,II (1997) Biochemistry, 36, 15428–15450. [DOI] [PubMed] [Google Scholar]
- 24.Ross W.S. and Hardin,C.C. (1994) J. Am. Chem. Soc., 116, 6070–6080. [Google Scholar]
- 25.Lee J.S. (1990) Nucleic Acids Res., 20, 6057–6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardin C.C., Henderson,E., Watson,T. and Prosser,J.K. (1991) Biochemistry, 30, 4460–4472. [DOI] [PubMed] [Google Scholar]
- 27.Hardin C.C., Watson,T., Corregan,M. and Bailey,C. (1992) Biochemistry, 31, 833–841. [DOI] [PubMed] [Google Scholar]
- 28.Nagesh N. and Chatterji,D. (1995) J. Biochem. Biophys. Methods, 30, 1–8. [DOI] [PubMed] [Google Scholar]
- 29.Raghuraman M.K. and Cech,T.R. (1990) Nucleic Acids Res., 18, 4543–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braunlin W.H. and Nordenskiöld,L. (1984) Eur. J. Biochem., 142, 133–137. [DOI] [PubMed] [Google Scholar]
- 31.Anwander E.H.S., Probst,M.M. and Rode,B.M. (1990) Biopolymers, 29, 757–769. [DOI] [PubMed] [Google Scholar]
- 32.Record M.T., Anderson,C.F. and Lohman,T.M. (1978) Q. Rev. Biophys., 11, 102–178. [Google Scholar]
- 33.Sen D. and Gilbert,W. (1990) Nature, 344, 410–414. [DOI] [PubMed] [Google Scholar]
- 34.Williamson J.R., Raghuraman,M.K. and Cech,T.R. (1989) Cell, 59, 871–880. [DOI] [PubMed] [Google Scholar]
- 35.Guschlbauer W., Chantot,J.F. and Thiele,D. (1990) J. Biomol. Struc. Dyn., 3, 491–511. [DOI] [PubMed] [Google Scholar]
- 36.Nagesh N., Bhargava,P. and Chatterji,D. (1992) Biopolymers, 32, 1421–1424. [DOI] [PubMed] [Google Scholar]
- 37.Blume S.W., Guarcello,V., Zacharias,W. and Miller,D.M. (1997) Nucleic Acids Res., 25, 617–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang K.Y., Gerena,L., Swaminathan,S. and Bolton,P.H. (1995) Nucleic Acids Res., 23, 844–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marathias V.M., Wang,K.Y., Kumar,S., Pham,T.Q., Swaminathan,S. and Bolton,P.H. (1996) J. Mol. Biol., 260, 378–394. [DOI] [PubMed] [Google Scholar]
- 40.Beger R.D., Marathias,V.M., Volkman,B.F. and Bolton,P.H. (1998) J. Magn. Reson., 135, 256–259. [DOI] [PubMed] [Google Scholar]
- 41.Smith F.W. and Feigon,J. (1992) Nature, 356, 164–168. [DOI] [PubMed] [Google Scholar]
- 42.Kang C., Zhang,X., Ratliff,R., Moyzis,R. and Rich,A. (1992) Nature, 356, 126–131. [DOI] [PubMed] [Google Scholar]
- 43.Sen D. and Gilbert,W. (1988) Nature, 334, 364–366. [DOI] [PubMed] [Google Scholar]
- 44.Aboul-ela F., Murchie,A.L.H. and Lilley,D.M.J. (1992) Nature, 360, 280–282. [DOI] [PubMed] [Google Scholar]
- 45.Phillips K., Dauter,Z., Murchie,A.I., Lilley,D.M. and Luisi,B. (1997) J. Mol. Biol., 273, 171–182. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y. and Patel,D.J. (1993) J. Mol. Biol., 234, 1171–1183. [DOI] [PubMed] [Google Scholar]
- 47.Laughlan G., Murchie,A.I.H., Norman,D.G., Moore,M.H., Moody,C.E., Lilley,D.M.J. and Luisi,B. (1994) Science, 265, 520–524. [DOI] [PubMed] [Google Scholar]
- 48.Marathias V.M. and Bolton,P.H. (1999) Biochemistry, 38, 4355–4364. [DOI] [PubMed] [Google Scholar]
- 49.Hud N.V., Schultze,P., Sklenar,V. and Feigon,J. (1999) J. Mol. Biol., 285, 233–243. [DOI] [PubMed] [Google Scholar]
- 50.Schultze P., Hud,N.V., Smith,F.W. and Feigon,J. (1999) Nucleic Acids Res., 27, 3018–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Macaya R.F., Schultze,P., Smith,F.W., Roe,J.A. and Feigon,J. (1993) Proc. Natl Acad. Sci. USA, 90, 3745–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang K.Y., Krawczyk,S.H., Bischofberger,N., Swaminathan,S. and Bolton,P.H. (1993) Biochemistry, 32, 11285–11295. [DOI] [PubMed] [Google Scholar]
- 53.Wang K.Y., McCurdy,S., Shea,R.G., Swaminathan,S. and Bolton,P.H. (1993) Biochemistry, 32, 1899–1904. [DOI] [PubMed] [Google Scholar]
- 54.Krishnamurthy V.V. (1996) J. Magn. Reson. B., 113, 46–52. [DOI] [PubMed] [Google Scholar]
- 55.Wang K.Y., Goljer,I. and Bolton,P.H. (1994) J. Magn. Reson. B., 103, 192–196. [DOI] [PubMed] [Google Scholar]
- 56.Beger R.D. and Bolton,P.H. (1998) J. Biol. Chem., 273, 15565–15573. [DOI] [PubMed] [Google Scholar]
- 57.Macaya R.F., Waldron,J.A., Beutel,B.A., Gao,H., Joesten,M.E., Yang,M., Patel,R., Bertelsen,A.H. and Cook,A.F. (1995) Biochemistry, 34, 4478–4492. [DOI] [PubMed] [Google Scholar]
- 58.Kung H.C. and Bolton,P.H. (1997) J. Biol. Chem., 272, 9227–9236. [DOI] [PubMed] [Google Scholar]
- 59.Wang K.Y., Parker,S.A., Goljer,I. and Bolton,P.H. (1997) Biochemistry, 36, 11629–11639. [DOI] [PubMed] [Google Scholar]
- 60.Hud N.V., Sklenar,V. and Feigon,J. (1999) J. Mol. Biol., 286, 651–660. [DOI] [PubMed] [Google Scholar]
- 61.Jing N., Gao,X., Rando,R.F. and Hogan,M.E. (1997) J. Biomol. Struct. Dyn., 15, 573–585. [DOI] [PubMed] [Google Scholar]
- 62.Jing N., Rando,R.F., Pommier,Y. and Hogan,M.E. (1997) Biochemistry, 36, 12498–12505. [DOI] [PubMed] [Google Scholar]
- 63.Century T.J., Fenichel,I.R. and Horowitz,S.B. (1970) J. Cell Sci., 7, 5–13. [DOI] [PubMed] [Google Scholar]
- 64.Cameron I.L., Hardman,W.E., Hunter,K.E., Haskin,C., Smith,N.K. and Fullerton,G.D. (1990) Scanning Microsc., 4, 89–100. [PubMed] [Google Scholar]
- 65.Bouaziz S., Kettani,A. and Patel,D.J. (1998) J. Mol. Biol., 282, 637–652. [DOI] [PubMed] [Google Scholar]
- 66.Latham J.A., Johnson,R. and Toole,J.J. (1994) Nucleic Acids Res., 22, 2817–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.