SUMMARY
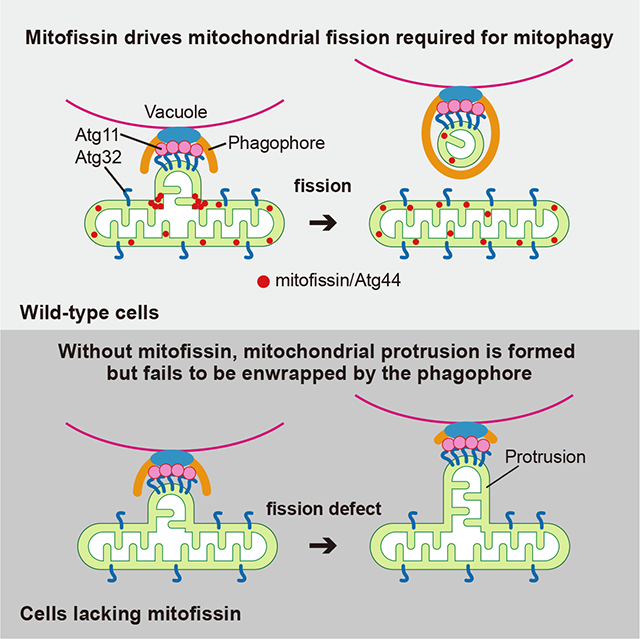
Mitophagy plays an important role in mitochondrial homeostasis by selective degradation of mitochondria. During mitophagy, mitochondria should be fragmented to allow engulfment within autophagosomes, whose capacity is exceeded by the typical mitochondria mass. However, the known mitochondrial fission factors, dynamin-related proteins Dnm1 in yeasts and DNM1L/Drp1 in mammals, are dispensable for mitophagy. Here, we identify Atg44 as a mitochondrial fission factor that is essential for mitophagy in yeasts, and therefore term Atg44 and its orthologous proteins as mitofissin. In mitofissin-deficient cells, a part of the mitochondria is recognized by the mitophagy machinery as cargo but cannot be enwrapped by the autophagosome precursor, the phagophore, due to a lack of mitochondrial fission. Furthermore, we show that mitofissin directly binds to lipid membranes and brings about lipid membrane fragility to facilitate membrane fission. Taken together, we propose that mitofissin acts directly on lipid membranes to drive mitochondrial fission required for mitophagy.
eTOC blurb
Fukuda et al. identify yeast Atg44 as a membrane fission protein that resides in the mitochondrial intermembrane space, and named it mitofissin. Mitofissin drives Dnm1/DNM1L-independent mitochondrial fission that is essential for mitopahgy. Mitofissin has the ability to bind directly to lipid membranes and bring about membrane fragility, promoting membrane fission.
Graphical Abstract:
INTRODUCTION
Autophagy is a catabolic process that is induced by various cellular stresses and recycles cytoplasmic components through vacuolar/lysosomal degradation. Upon the induction of autophagy, cup-shaped membranous structures, called phagophores, emerge in the cytosol, expand, and engulf cytoplasmic proteins and organelles to form autophagosomes. The autophagosome fuses with the vacuole in yeast and plants or the lysosome in metazoans, leading to the degradation of its sequestered materials by resident hydrolases.1 Mitophagy is the selective autophagic degradation of mitochondria, which plays an important role in maintaining mitochondrial quality and quantity.2 Mitophagy shares most of the molecular processes with non-selective autophagy. However, mitophagy requires at least two additional steps: 1. The selection of particular mitochondrial regions as cargo; and 2. The adjustment of the cargo morphology to fit the shape and size of the phagophore and autophagosome.
The cargo selection process of mitophagy relies on receptor proteins localized on the outer mitochondrial membrane (OMM). In Saccharomyces cerevisiae (Sc), Atg32 is the sole mitophagy receptor,3,4 which is phosphorylated upon mitophagy induction enabling it to interact with the cytosolic scaffold/adaptor protein Atg115,6 and focally accumulates on the mitochondrion that is targeted for degradation.7 Subsequently, Atg11 interacts with the Atg1 complex consisting of core autophagy proteins that assemble at the phagophore assembly site (PAS) to initiate phagophore formation.3,8 In mammals, several mitophagy receptors, such as BNIP3L/NIX, BNIP3, FUNDC1, BCL2L13, and FKBP8 have been identified. These receptors interact with mammalian Atg8-family proteins located on the phagophore membrane to ensure selective entrapment of mitochondria as autophagy cargo.2 Besides receptor-mediated mitophagy, ubiquitin-mediated mitophagy, in which mitochondrial cargo tagged by PINK1- and PRKN/parkin-mediated ubiquitination is recognized by the autophagy machinery, serves as another subset of mitophagy pathways in mammals.2
Because mitochondria are typically larger than the autophagosome, it was suggested that mitochondrial fission generates mitochondrial fragments of appropriate size in advance of their engulfment by a phagophore.9–12 However, well-studied mitochondrial fission factors, such as dynamin-related proteins Dnm1 in yeasts and DNM1L/Drp1 in mammals,13 are dispensable for receptor-mediated mitophagy,14–16 raising the possibility that an unidentified machinery/mechanism also acts to promote mitochondrial fission. In this study, we identified Atg44 in fission yeast and budding yeast as a mitochondrial intermembrane space (IMS) protein that drives Dnm1/DNM1L-independent mitochondrial fission essential for receptor-mediated mitophagy. As Atg44-like proteins can be found in fungi, algae, and some plants, we termed this family of proteins as mitofissin. In vitro reconstitution and structural analysis suggested that the yeast mitofissin Atg44 has the ability to directly bind to lipid membranes and brings about lipid membrane fragility to facilitate membrane fission. Our study sheds light on the process in which mitochondrial fission contributes to efficient engulfment of mitochondrial cargo by a phagophore and provides insights into the molecular basis for the function of mitofissin in mitochondrial membrane fission.
RESULTS
Atg44 is essential for mitophagy
To identify genes required for mitophagy, we screened the fission yeast Schizosaccharomyces pombe (Sp) genome-wide deletion library for mitophagy-deficient mutants and found SPAC26A3.14C, an uncharacterized gene, to be essential for mitophagy. According to the unified nomenclature for autophagy-related genes in yeast,17 we named this gene atg44+. This gene encodes a protein (Atg44) that is composed of 73 amino acids without any known functional domains (Figure S1A). In S. pombe, nitrogen starvation induces mitophagy, which can be monitored by the vacuolar processing of a chimeric mitochondrial protein Tuf1-RFP or Sdh2-GFP to produce free RFP or GFP18 (Figures 1A and S1B). In atg44Δ cells, mitophagy was completely blocked similar to cells lacking Atg1, a core autophagy protein (Figures 1A and S1B). Atg44 is conserved among fungi, algae, and some plants (Figure S1A), and Sc-Atg44 has recently been reported as a mitochondrial protein of unknown function (designated as Mco8).19 Exogenous expression of Sc-ATG44 can rescue mitophagy in Sp-atg44Δ cells, suggesting functional conservation (Figure 1B). Consistently, in S. cerevisiae, deletion of Sc-ATG44 resulted in loss of mitophagy, which was rescued by exogenous expression of Sp-Atg44 or Atg44 from Komagataella phaffii/Pichia pastoris (Kp-ATG44) (Figures 1C and 1D). The deletion of ATG44 in either S. pombe or S. cerevisiae did not or only marginally affected non-selective macroautophagy as measured by GFP/RFP processing (Figures S1C, S1D, and S1F) or the Pho8Δ60 assay (Figure 1E), or other types of selective autophagy including the Cvt pathway which delivers the precursor form of the hydrolase aminopeptidase I to the vacuole (Figure S1G), endoplasmic reticulum-phagy/reticulophagy (Figures S1E and S1H), and pexophagy (Figure S1I), suggesting that Atg44 is specifically required for mitophagy.
Figure 1. The mitochondrial protein Atg44 is essential for mitophagy.
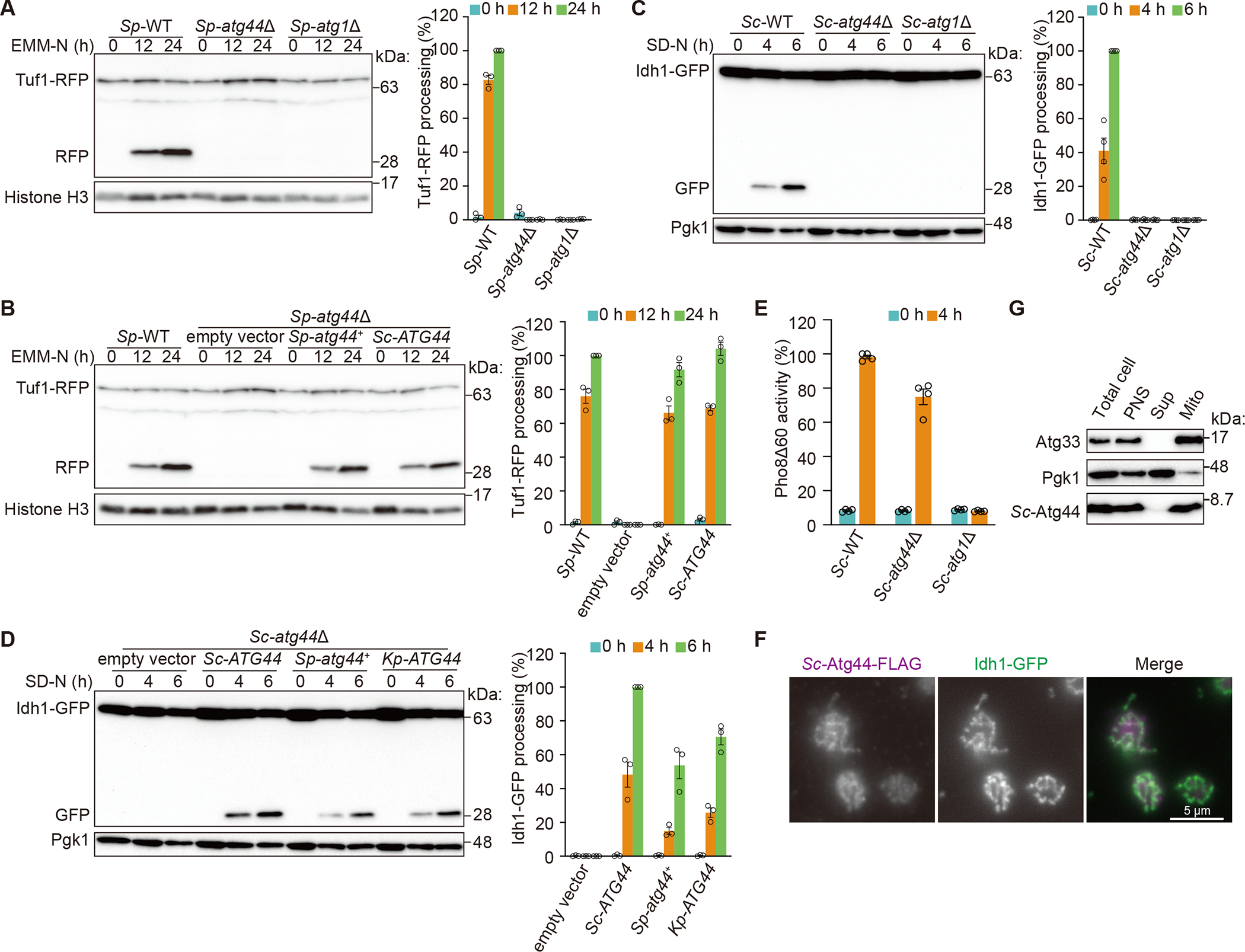
(A) The indicated S. pombe strains expressing Tuf1-RFP were grown in EMM and shifted to nitrogen-starvation medium (EMM-N). Cells were collected at the indicated time points and Tuf1-RFP processing was monitored by immunoblotting.
(B) Mitophagy was examined for the wild-type strain and the atg44Δ mutant exogenously expressing Sp-atg44+ or Sc-ATG44 by the Tuf1-RFP processing assay as in (A).
(C) The indicated S. cerevisiae strains expressing Idh1-GFP were grown in YPL and shifted to nitrogen-starvation medium (SD-N). Cells were collected at the indicated time points and Idh1-GFP processing was monitored by immunoblotting.
(D) Mitophagy in S. cerevisiae atg44Δ cells exogenously expressing Sc-ATG44, Sp-atg44+, or Kp-ATG44 was monitored using the Idh1-GFP processing assay as shown in (C).
(E) The indicated S. cerevisiae strains expressing Pho8Δ60 were grown in YPD and shifted to SD-N for 4 h. Samples were collected, and the protein extracts were assayed for Pho8Δ60 activity. The results represent the mean and standard deviation (SD) of four experiments.
(F) S. cerevisiae cells expressing Sc-Atg44-FLAG and Idh1-GFP were cultured in SMD and analyzed by immunofluorescence microscopy using an anti-FLAG antibody. Scale bar, 5 μm.
(G) Subcellular fractionation was conducted using S. cerevisiae. The total cell homogenate (Total cell) was fractionated by centrifugation to obtain a post-nuclear supernatant (PNS), a mitochondria-enriched pellet (Mito) and a supernatant (Sup). Atg33 and Pgk1 were detected as mitochondrial and cytosolic markers, respectively.
Histone H3 (A and B) and Pgk1 (C and D) were used as loading controls. The ratio of RFP to total RFP (Tuf1-RFP and RFP) (A and B) or GFP to total GFP (Idh1-GFP and GFP) (C and D) were quantified. The value of the wild-type strain at the 24-h time point (A and B), wild-type strain at the 6-h time point (C), or Sc-ATG44-expressing strain at the 6-h time point (D) was set to 100%. The results represent the mean and SD of three (A, B, and D) or four (C) experiments.
See also Figures S1 and S2.
Atg44 resides in the mitochondrial intermembrane space (IMS)
To investigate the localization of Atg44, we expressed Sc-Atg44-FLAG, whose functionality was verified (Figure S1J), in S. cerevisiae, and found that Sc-Atg44-FLAG colocalized with the mitochondrial protein Idh1-GFP (Figure 1F). By cell fractionation, Sc-Atg44 was collected in the 6500 × g pellet fraction where mitochondrial Atg33 was enriched (Figure 1G). These findings suggest that Atg44 is a mitochondrial protein. We then treated isolated mitochondria with 0.1 M sodium carbonate (pH 11) and separated the membrane and supernatant fractions, and found that Atg44 was released into the supernatant along with Atp2, the beta subunit of the F1Fo ATPase (Figure S1K). Finally, we isolated mitochondria from S. cerevisiae or S. pombe and studied the intramitochondrial localization of Atg44 using a proteinase K (ProK) protection assay. Treatment with ProK alone caused degradation of the OMM protein Atg33 or Tom70-GFP but not of Sc-Atg44-FLAG or Sp-Atg44. However, ProK treatment under the hypotonic condition that disrupts the OMM but maintains the inner mitochondrial membrane (IMM) caused the degradation of Sc-Atg44-FLAG or Sp-Atg44 as well as the IMS protein Mia40-GFP or Mic60-FLAG, but not the matrix protein Atp2 or Tuf1-RFP (Figures S1L and S1M); the latter were degraded when all mitochondrial membranes were disrupted with Triton X-100. Based on these results, we conclude that Atg44 localizes in the IMS and is not a transmembrane protein.
Atg44 is required for engulfment of mitochondria by the phagophore
Next, we examined a role for Atg44 in mitophagy using S. cerevisiae. We initially tested whether Sc-Atg44 is required for the cargo selection process that involves the phosphorylation, Atg11 interaction, and focal accumulation of Atg32.5–7 The phosphorylation status of Atg32 in Sc-atg44Δ cells upon nitrogen starvation was the same as that of wild-type cells (Figure S2A). The Atg32-Atg11 interaction was also unaffected in the absence of Sc-Atg44 (Figure S2B). After interacting with Atg11, Atg32 accumulates at one or multiple sites on mitochondria where mitophagy is expected to proceed.7 This accumulation of Atg32 was comparable between Sc-atg44Δ and wild-type cells (Figure S2C). These results suggest that the absence of Atg44 does not affect the cargo selection processes.
We then tested whether Atg44 is required for formation of autophagosomes that contain mitochondria as cargo (hereafter referred to as mitophagosomes). Autophagosomes/mitophagosomes are accumulated in cells lacking Ypt7, a Rab GTPase-family protein required for fusion of autophagosomes/mitophagosomes with vacuoles.16,20 Upon mitophagy induction, Sc-ypt7Δ cells accumulated mitophagosomes, which were observed as punctate structures labeled with Idh1-GFP that overlapped with RFP-Atg8, a marker for the phagophore and autophagosome membranes (Figure S2D). By contrast, mitophagosomes were completely absent in Sc-atg44Δ Sc-ypt7Δ cells (Figure S2D), suggesting that Atg44 is required for the process between cargo selection and mitophagosome formation.
Notably, the Sc-atg44Δ Sc-ypt7Δ mutant exhibited mitochondrial protrusions that extended towards the center of the cell (Figure 2A). Such protrusions were also observed in the Sc-atg44Δ single mutant labeled with Idh1-GFP (matrix protein), Om14-RFP (OMM protein), or MitoTracker (membrane potential-dependent dye) (Figures 2B, S2E, and S2F). Formation of the protrusions required the core autophagy proteins Atg1 and Atg8 as well as mitophagy factors such as Atg32 and Atg11 (Figure 2B), indicating that the mitochondrial protrusion formation depends on the mitophagy process. We found that GFP-Atg32 accumulated around the protrusions and that GFP-Atg8 and GFP-Atg11 puncta colocalized with the protrusions with high frequency (Figure 2C), suggesting that the protrusions correspond to the mitochondrial regions that are destined to be delivered by mitophagy. These results also suggested the possible interaction between Atg44 and Atg32 at the sites of Atg32 accumulation, but such interaction was not detected under our experimental conditions (Figure S2G).
Figure 2. Mitochondrial protrusions were formed in atg44Δ cells upon mitophagy induction dependent on mitophagy factors.
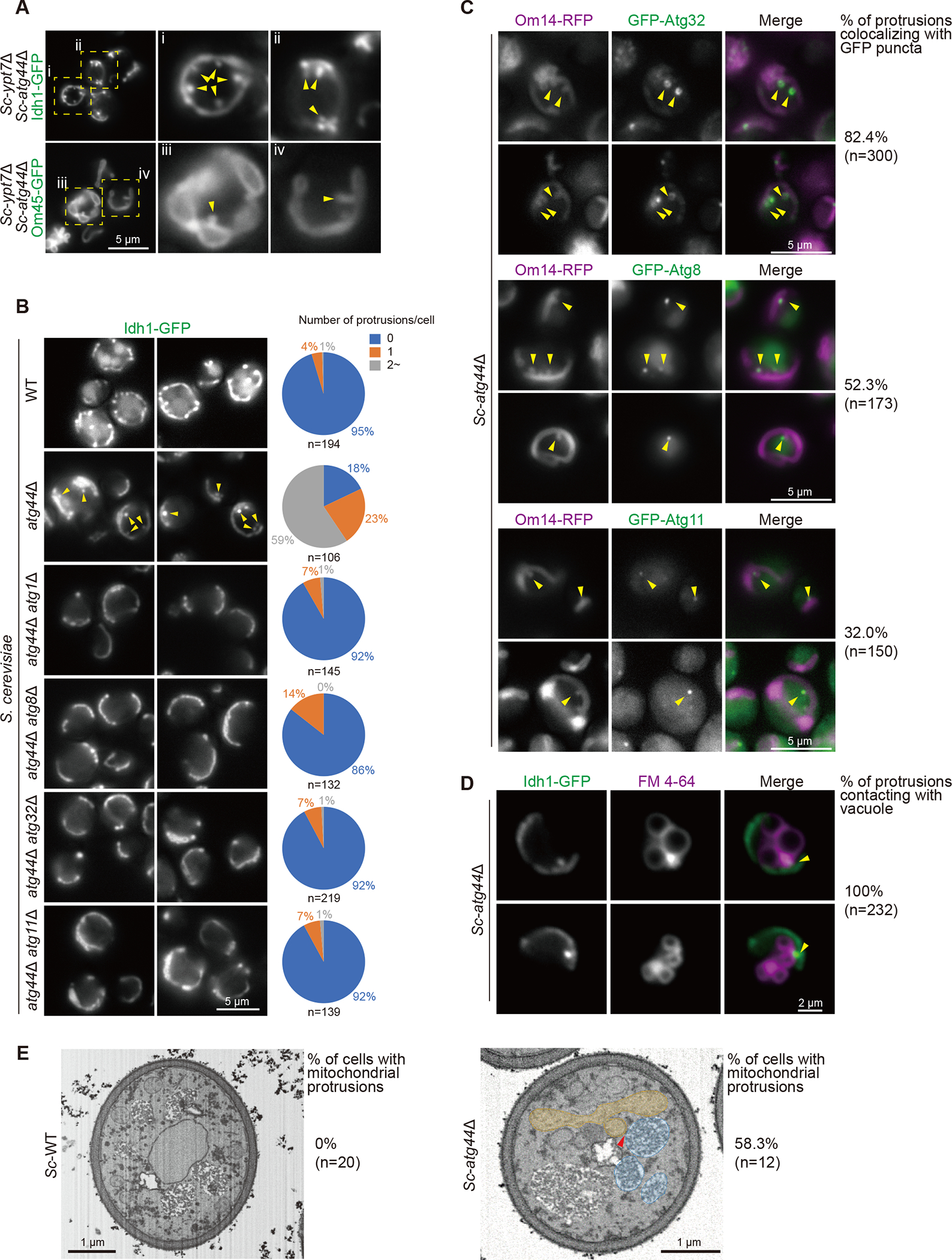
(A and B) The indicated S. cerevisiae cells expressing a mitochondrial protein Idh1-GFP (A and B) or Om45-GFP (A) were grown in YPL and shifted to SD-N medium for 6 h and then analyzed by fluorescence microscopy. Arrowheads indicate mitochondrial protrusions. The number of mitochondrial protrusions per cell was counted (B).
(C) The indicated S. cerevisiae cells expressing a mitochondrial protein Om14-RFP and GFP-Atg32, GFP-Atg8, or GFP-Atg11 were grown in YPL and shifted to SD-N medium for 6 h and then analyzed by fluorescence microscopy. Arrowheads indicate accumulation of GFP-Atg32, GFP-Atg8, or GFP-Atg11 on the mitochondrial protrusions, and the percentage of their colocalization was quantified.
(D) S. cerevisiae atg44Δ cells expressing Idh1-GFP were grown in YPL and shifted to SD-N for 6 h in the presence of FM 4–64 and then analyzed by fluorescence microscopy. Arrowheads indicate the contact between a mitochondrial protrusion and the vacuole.
(E) S. cerevisiae WT and atg44Δ cells were grown in YPL and shifted to SD-N for 6 h. Cells were collected and processed for FIB-SEM. The arrowhead in atg44Δ cell indicates the tip of mitochondrial protrusion close to the vacuole. Yellow, mitochondria; blue, vacuole. See also Videos S1 and S2. The percentage of cells with mitochondrial protrusions is indicated.
The value of n represents the number of observed cells (B and E) and protrusions (C and D). Scale bars and their lengths are provided in each panel.
See also Figure S2.
We also found that the protrusions made contacts with the vacuolar surface presumably at the PAS, the site of phagophore membrane formation (Figure 2D). Detailed analysis with focused ion beam scanning electron microscopy (FIB-SEM) revealed that the mitochondrial protrusion, which was detectable specifically in atg44Δ cells, was formed from a part of the mitochondria, and its tip was in close proximity to the vacuole (Figure 2E; Videos S1 and S2). The cristae structure appeared to be substantially maintained inside the mitochondrial protrusion (Figure 2E; Video S2).
Taken together, these findings confirm that the autophagy machinery assembles at particular sites on mitochondria and these portions are targeted to the vacuolar surface as mitophagy proceeds. In the absence of Atg44, however, these parts remain connected to the main body of the mitochondrion, resulting in formation of the mitochondrial protrusions that are not enwrapped by the phagophore.
Atg44 acts in mitochondrial fission
During our analysis of Sc-atg44Δ cells, we realized that their mitochondria were spherically enlarged and similar to those in dnm1Δ cells, which are defective in mitochondrial fission (Figures 3A, 3B, and S3A). As atg44Δ cells show a defect in the engulfment of mitochondria by the phagophore, we hypothesized that Atg44 is involved in mitochondrial fission that produces fragments suitable for phagophore engulfment.
Figure 3. Atg44 has a role in mitochondrial fission.
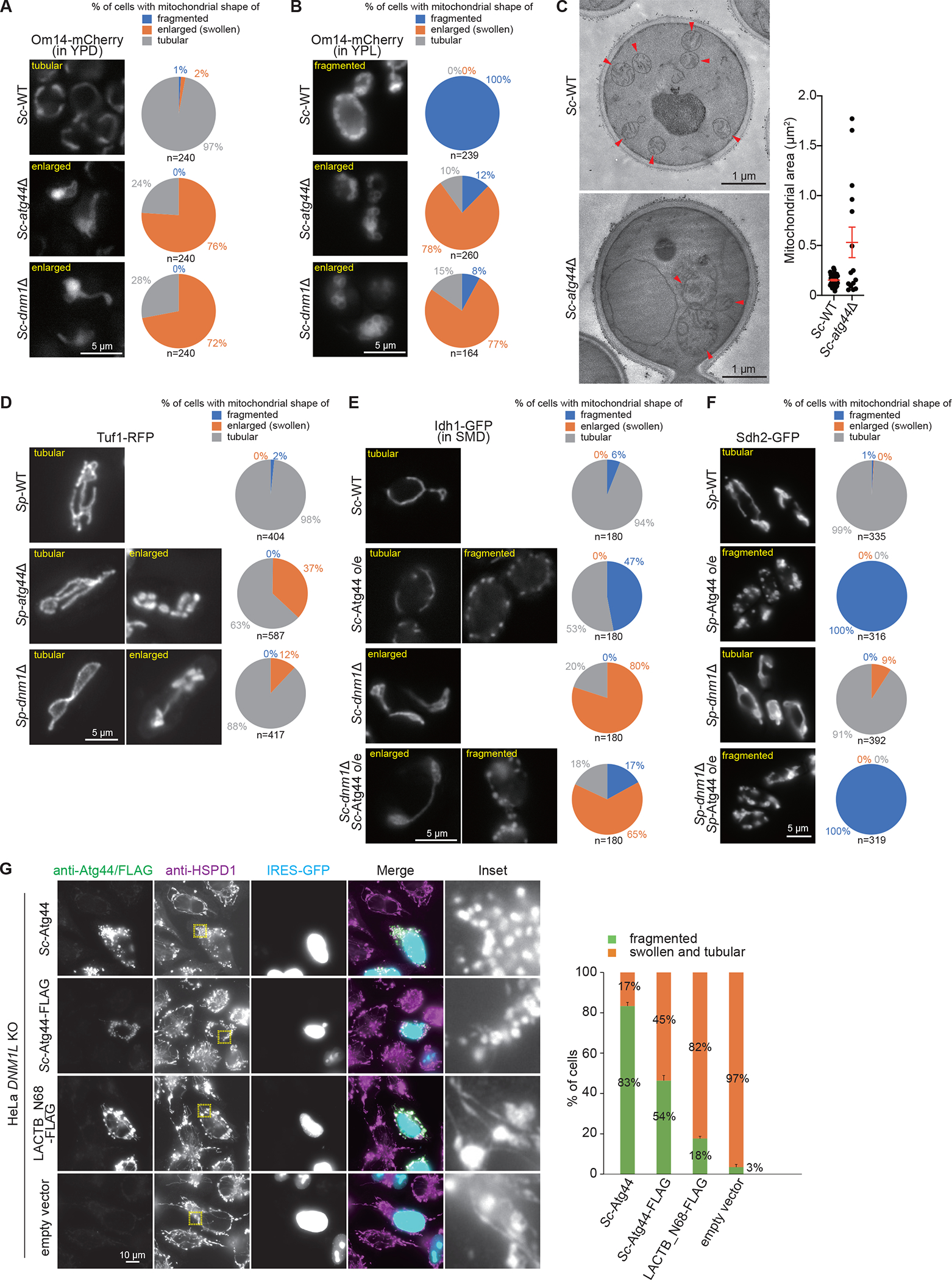
(A, B, and D–F) The indicated S. cerevisiae cells expressing Om14-mCherry (A and B) or Idh1-GFP (E) were grown in YPD (A), YPL (B), or SMD-Ura (E) and S. pombe cells expressing Tuf1-RFP (D) or Sdh2-GFP (F) were grown in EMM, and analyzed by fluorescence microscopy. Cells were classified according to their mitochondrial morphology (fragmented, enlarged, tubular in shape), and their ratio was quantified. o/e, overexpression.
(C) The indicated S. cerevisiae cells were grown in YPL. Cells were collected and processed for transmission electron microscopy. Mitochondrial area was quantified and plotted. The error bars represent the mean ± SEM. Arrowheads indicate mitochondria. See also Videos S3 and S4.
(G) DNM1L/Drp1-knockout HeLa cells were transfected with the IRES-GFP-NLS vector to express Sc-Atg44, Sc-Atg44-FLAG, or LACTB_N68-FLAG. The expressed proteins and the mitochondrial morphology were analyzed by immunofluorescence microscopy using the anti-Sc-Atg44 (Sc-Atg44), anti-FLAG (Sc-Atg44-FLAG, LACTB_N68-FLAG, and empty vector), and anti-HSPD1/HSP60 antibodies. Fluorescence images were shown with a same image scaling. Nuclear GFP-expressing cells were classified according to whether their mitochondria were fragmented or swollen and tubular in shape, and their ratio was quantified. The results represent the mean and SD of three experiments. More than 65 cells were analyzed for each experiment.
The value of n represents the number of observed cells. Scale bars and their lengths are provided in each panel.
See also Figure S3.
To further examine the involvement of Atg44 in mitochondrial morphology, we observed mitochondria in Sc-atg44Δ cells by electron microscopy. Consistent with the results of fluorescence microscopy, mitochondria in Sc-atg44Δ cells were spherically enlarged, although the cristae structure was unaffected (Figure 3C; Videos S3 and S4). Similarly, loss of Atg44 in S. pombe affected mitochondrial morphology; some of the Sp-atg44Δ cells showed spherically enlarged mitochondria like Sp-dnm1Δ cells (Figure 3D). Because enlarged mitochondria are a critical feature of a deficiency in mitochondrial fission, these findings support the hypothesis that Atg44 has a positive role in mitochondrial fission.
To further confirm that Atg44 acts in mitochondrial fission, Atg44 was overexpressed in S. cerevisiae and S. pombe cells. Overexpression of Atg44 in both species caused mitochondrial fragmentation not only in wild-type cells but also in Dnm1-deficient cells (Figures 3E and 3F). We next expressed Sc-Atg44 in HeLa cells to test whether Atg44 causes mitochondrial fission in mammalian cells. Another mitochondrial protein with a similar size as Atg44, LACTB_N68-FLAG (N-terminal 68 amino acids of LACTB with a FLAG tag),21 was used as a control. We confirmed that LACTB_N68 localizes in the IMS (Figure S3B). Unexpectedly, HeLa cells expressing any of these proteins exhibited fragmented mitochondria (Figure S3C). We assumed that the observed fragmentation was attributed to the mitochondrial division that occurred in response to mitochondrial stress caused by overloading exogenous proteins in the IMS. To prevent such a type of mitochondrial division, we performed the same analysis using mitochondrial fission-deficient DNM1L/Drp1 knockout (DNM1L KO) HeLa cells. DNM1L KO cells exhibited tubular or spherically enlarged mitochondria due to the loss of mitochondrial fission (Figure 3G).22 Expression of Sc-Atg44 and Sc-Atg44-FLAG, but not LACTB_N68-FLAG, brought about fragmentation of mitochondria (Figure 3G). These findings suggest that Atg44 promotes mitochondrial fission without a need for DNM1L/Dnm1. Involvement of Atg44 in mitochondrial fission supports our hypothesis that Atg44 produces fragments that are dimensionally suitable for efficient engulfment during mitophagy. As Dnm1 is not essential for mitophagy (Figures S3D and S3E),15,16 we infer that Atg44 and Dnm1 mediate mitochondrial fission events with distinct physiological roles.
To test whether mitophagy could take place without Atg44 if mitochondria were artificially divided into fragments small enough for phagophore engulfment, we utilized cells lacking the mitochondrial fusion factor Mgm1 or Fzo1. In the absence of Mgm1-mediated mitochondrial fusion, mitochondrial fission predominates over fusion, leading to increased fragmentation (Figures S3F and S3G).23 As expected, in S. pombe atg44Δ cells lacking Mgm1, some of the mitochondria became fragmented and mitophagy was partially rescued (Figures 4A and S3F). Similarly, in S. cerevisiae, the absence of Mgm1 or Fzo1 in atg44Δ cells rescued mitophagy to levels comparable to cells deficient for only Mgm1 or Fzo1 (Figures 4B and S3G). These findings further support the concept that Atg44 is a mitochondrial fission factor that generates mitochondrial fragments suitable for mitophagy.
Figure 4. Atg44 and Dnm1 mediate mitochondrial fission with different properties.
(A and B) Mitophagy in the indicated S. pombe (A) or S. cerevisiae (B) strains was monitored using the Tuf1-RFP or Idh1-GFP processing assay as shown in Figure 1A or 1C, respectively. The results represent the mean and SD of three experiments.
(C and D) The indicated S. cerevisiae (C) or S. pombe (D) cells were grown in YPD or EMM, respectively, until the early log growth phase, and serial dilutions of each culture were spotted on the indicated agar plates and grown at 30°C.
See also Figure S3.
Atg44 and Dnm1 mediate mitochondrial fission with different properties
The maintenance of mitochondrial morphology by balancing mitochondrial fusion and fission is important for proper mitochondrial function.13 Fusion-deficient S. cerevisiae cells, such as mgm1Δ or fzo1Δ, frequently lose mitochondrial DNA, resulting in a respiratory growth defect. This phenotype can be rescued by the deletion of the DNM1 gene, suggesting that Dnm1-mediated mitochondrial fission counterbalances Mgm1- and Fzo1-mediated mitochondrial fusion.23 Accordingly, we tested whether Atg44-mediated fission can counterbalance Mgm1-mediated fusion. As reported previously, the lack of Dnm1 suppressed mitochondrial fragmentation as well as the respiratory growth defect of mgm1Δ in S. cerevisiae (Figures 4C and S3G).23 However, the absence of Atg44 did not substantially rescue the respiratory growth of S. cerevisiae mgm1Δ cells (Figure 4C). A similar lack of rescue was observed in S. pombe with a loss of Atg44 (Figures 4D and S3F). Collectively, the two fission factors play specialized roles; Atg44 is required for the mitochondrial fission that facilitates mitophagy, whereas Dnm1 predominantly promotes the fission that balances the fusion activity of Mgm1, maintaining mitochondrial function under homeostatic conditions.
Atg44 accumulates on lipid nanotubes and mediates their fission in vitro
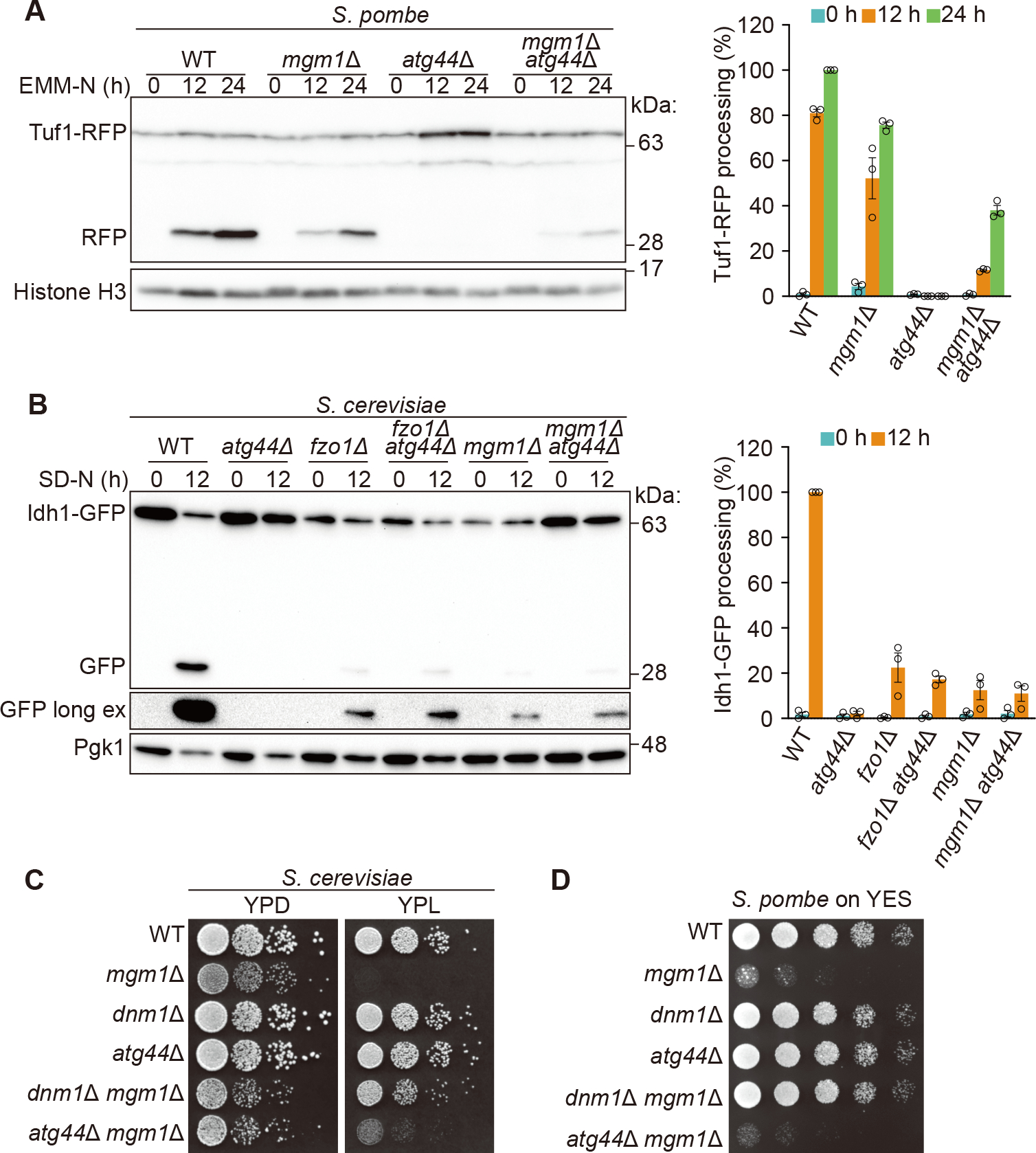
To address whether Atg44 itself acts on lipid membranes to promote fission, we prepared recombinant Sp-Atg44 and MBP-Sc-Atg44 for in vitro studies (Figures S4A–S4D). Size-exclusion chromatography (SEC) coupled with multi-angle light scattering analysis estimated the molar mass of soluble Sp-Atg44 to be 71±4 kDa (Figure S4B) despite the calculated molecular mass of 9 kDa, indicating that recombinant Sp-Atg44 forms an octamer in solution. Using the recombinant Sp-Atg44, we first studied whether Sp-Atg44 binds lipid membranes. Alexa Fluor 647 (AF647)-labeled Sp-Atg44 accumulated on a giant unilamellar vesicle (GUV) derived from a lipid film with a phospholipid composition similar to that of the IMM (Figure 5A), indicating that Sp-Atg44 has an ability to bind the lipid membrane. We then prepared lipid nanotubes fluorescently labeled with liss Rhod PE and used them to analyze the membrane fission activity of Sp-Atg44 (Figures 5B and 5C). Upon application of Sp-Atg44, most nanotubes underwent fission at multiple sites (Figure 5B; Video S5). The diameter of the tubes, estimated from the fluorescence intensity (Figure S4E),24 indicates that the efficiency of the Sp-Atg44-induced fission was higher for thinner nanotubes (Figures 5B, 5D, and S4F). We confirmed that negative controls, GFP and AF647 alone, were not able to fission the nanotubes (Figures 5C and 5D). Moreover, accumulation of Sp-Atg44 on the lipid nanotubes was also observed (Figures 5E and S4G). These data demonstrate that Sp-Atg44 can bind with lipid membranes and mediate membrane fission without requiring any other factors.
Figure 5. Membrane fission by Atg44 in vitro.
(A) Membrane binding of Sp-Atg44. Sp-Atg44 was applied to GUVs produced from a lipid film containing phospholipids of the IMM-like composition. AF647-Sp-Atg44 and liss Rhod PE in the membrane were observed by confocal laser scanning microscopy. DIC, differential interference contrast.
(B) Membrane fission by Sp-Atg44. Sp-Atg44 was applied to lipid nanotubes. AF647-Sp-Atg44 and liss Rhod PE in the membrane were observed. Positions of fission are marked with blue arrowheads. See also Video S5.
(C) Control experiments of (B). GFP and AF647 were applied to lipid nanotubes.
(D) Tube radius dependency of fission by Sp-Atg44. Fission events were counted for 10 min after sample application to lipid nanotubes. A total of 37, 22 and 7 lipid nanotubes were examined for Sp-Atg44, GFP and AF647, respectively.
(E) Fluorescence profile of Sp-Atg44 on lipid nanotubes. Each micrograph shows average intensity for 1 min.
(F) Cryo-EM observation of MBP-Sc-Atg44 on the membrane. Particles corresponding to MBP-Sc-Atg44 are marked with arrowheads. Black dots are a fiducial marker of 10 nm.
(G–I) HS-AFM images of lipid nanorod only (G), with Sp-Atg44 (H) or MBP-Sc-Atg44
(I) are shown. Height profiles at red lines were analyzed.
Scale bars and their lengths are provided in each panel.
Atg44 tends to bind to membranes with high positive curvature
We next performed a cryogenic electron microscopy (cryo-EM) to examine the binding of Atg44 to lipid membranes in more detail. MBP-Sc-Atg44 formed clusters irrespective of the presence of lipid membranes, and bound to the surface of liposomes and nanotubes in the form of clusters (Figure 5F). In the case of the lipid nanotubes, MBP-Sc-Atg44 tended to bind to the edges with high positive curvature (Figure 5F). We also performed high-speed atomic force microscopy (HS-AFM) and found that Sp-Atg44 and MBP-Sc-Atg44 again bound to the lipid nanotubes as a cluster, often at their edges (Figures 5G–5I). Thus, we hypothesized that Atg44 preferentially binds membranes with high positive curvature. To further test this possibility, we performed a flotation assay to evaluate the membrane binding of Atg44 and found that Sp-Atg44 binds more efficiently to small liposomes having an IMM-like lipid composition than to large ones (Figure S5A and S5B). We also found that this binding was attenuated under high salt concentration or by removing cardiolipin from the liposomes (Figures S5B and S5C). From these findings, we conclude that Atg44 preferentially binds to lipid membranes with high positive curvature in an electrostatic interaction-dependent manner.
Atg44 binds to lipid membranes and causes membrane fragility
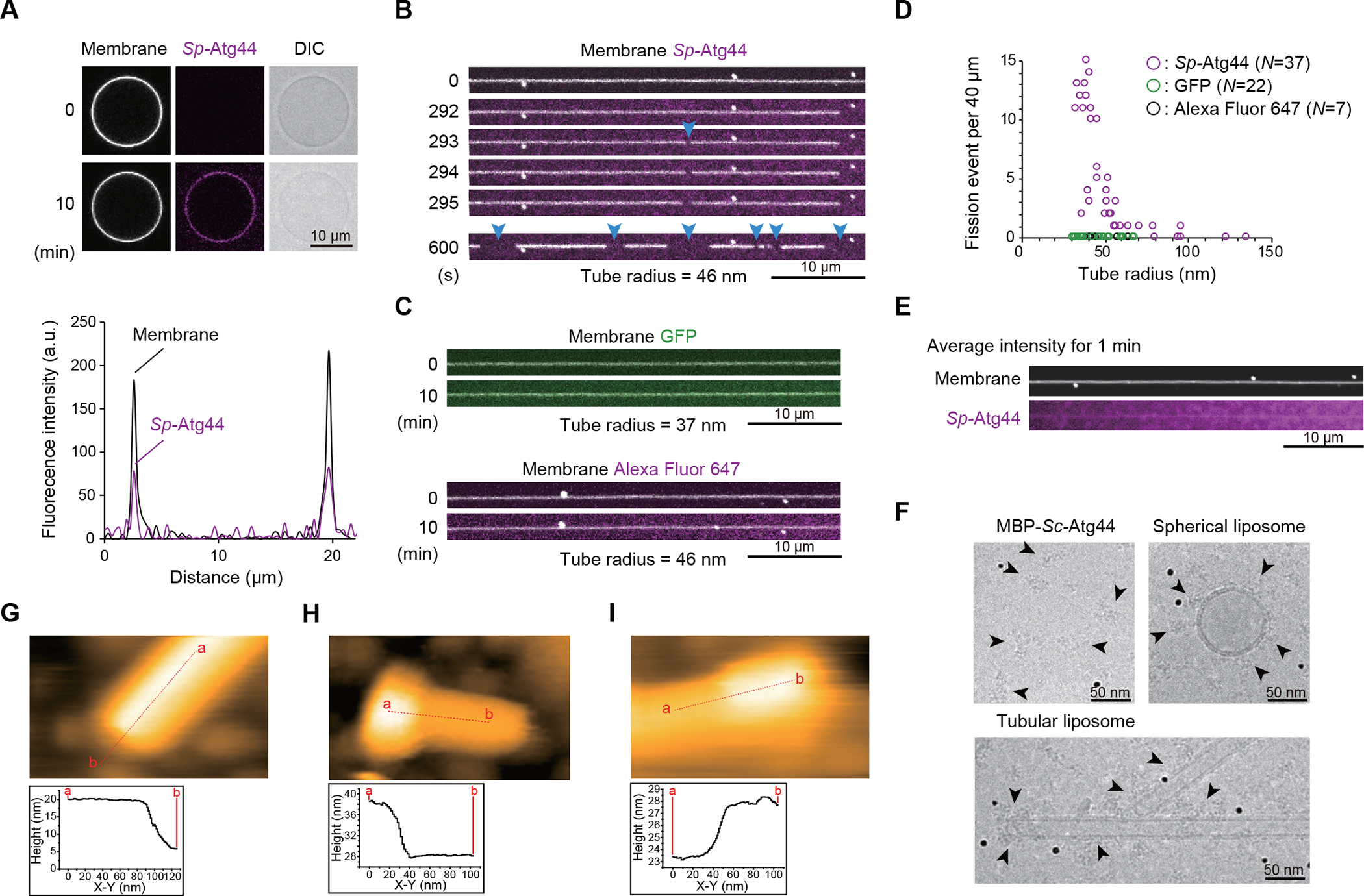
To elucidate the function of Atg44 at the molecular level, we determined the crystal structure of Sp-Atg44 at a resolution of 1.58 Å (Figures 6A and S5D–S5G; Table 1). The Atg44 protomer consists of three α-helices that interact with each other to form a globular fold. One side of the fold is hydrophilic whereas the other side is hydrophobic, implying that the protomer would be unstable in solution. Consistently, Sp-Atg44 behaved as an octamer in solution (Figure S4B) and crystallized as an octamer (Figure 6B), where the hydrophobic surfaces were hidden inside the octamer and constituted four hydrophobic cavities, each accommodating two phospholipids (Figures 6B and S5D–S5G).
Figure 6. Crystal structure of Atg44 and its membrane-bound models.
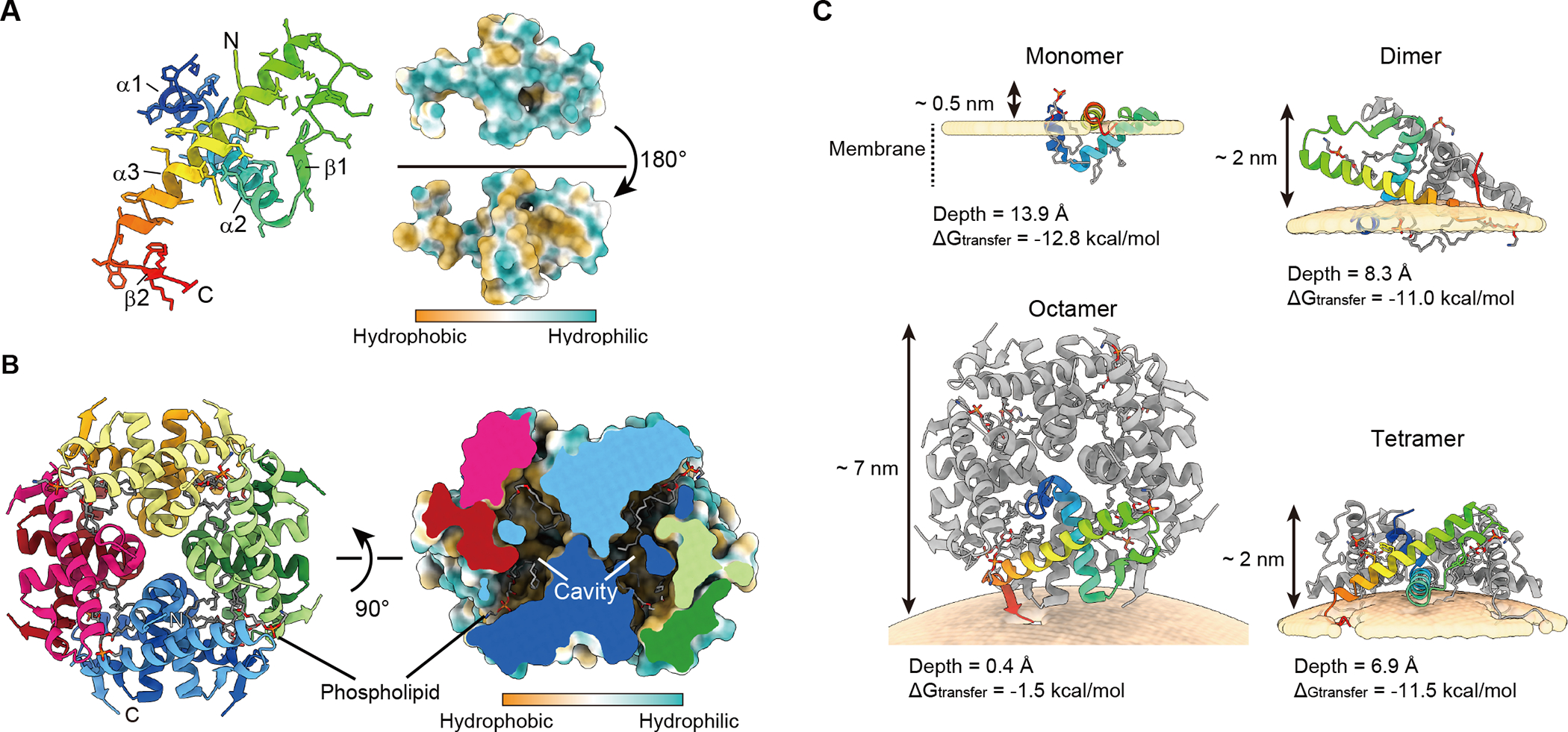
(A) Ribbon model and hydrophobicity surface model of the Sp-Atg44 protomer. Side chains are shown in a stick model. Hydrophobic and hydrophilic properties are colored in brown and sea green, respectively.
(B) Ribbon model and hydrophobicity surface model of the octameric architecture of Sp-Atg44. Phospholipids modeled as dioleoylphosphatidylcholine (DOPE) are shown as a stick model. The surface in the cross-section panel is colored according to hydrophilicity.
(C) Computational model of Sp-Atg44 on the membrane. The model structures of the Sp-Atg44 monomer, dimer, tetramer, and octamer are obtained from the PPM server. ΔG indicates transfer free energies of protein structures from water to the membrane-like environment and the depth indicates the distance from the deepest membrane-embedded protein portion to the membrane surface.
See also Figures S5 and S6.
Table 1.
Data collection, phasing and refinement statistics, related to Figure 6
| Native SpAtg44 | Selenomethionine-labeled SpAtg44V42M | |
|---|---|---|
|
| ||
| Data collection statistics | ||
| Beamline | SPring-8 BL32XU | SPring-8 BL32XU |
| Wavelength (Å) | 1.0000 | 0.9790 |
| Space group | P4122 | F432 |
| Cell dimensions | ||
| a, b, c (Å) | 82.364, 82.364, 113.427 | 166.322, 166.322, 166.322 |
| a, b, c (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Resolution range (Å) | 46.71–1.58(1.64–1.58) | 50.16–2.18(2.25–2.18) |
| R merge (I) | 0.260(1.124) | 0.266(1.749) |
| I / σ(I) | 5.2(0.5) | 8.7(0.7) |
| Completeness (%) | 97.3 (78.8) | 100.0(100.0) |
| Phasing statistics | ||
| Resolution range (Å) | 41.58–2.18 | |
| No. of sites | 3 | |
| Mean figure of merit | 0.27 | |
| Refinement statistics | ||
| Resolution range (Å) | 46.71–1.58 | |
| No. of reflections | 52606 | |
| Rwork / Rfree | 0.196/0.226 | |
| r.m.s.d.a from ideality | ||
| Bond lengths (Å) | 0.006 | |
| Bond angles (°) | 0.823 | |
root-mean-square deviation
Atg44 binds to lipid membranes in vitro (Figures 5A and 5E) and the cryo-EM and HS-AFM analyses (Figures 5F–5I) suggest that Atg44 tends to bind to lipid membranes with high curvature. We thus positioned Sp-Atg44 structures on the membrane with curvature and calculated binding energy using the PPM server.25 While the Sp-Atg44 octamer structure did not show a stable binding with the lipid membrane, the monomer, dimer, and tetramer structures showed tight interaction with the lipid membrane; i.e., the monomer binds to the planar membrane, whereas the others bind to positively-curved membranes (Figures 6C and S6). To further investigate how Atg44 interacts with the lipid membrane, Sp-Atg44 was observed using HS-AFM. In the absence of lipid membranes, Sp-Atg44 was observed at a size corresponding to an octamer in solution (Figure 7A). Next, a lipid bilayer was prepared on mica, and Sp-Atg44 was applied to the lipid bilayer and observed by HS-AFM (Figures 7B and 7C). Most of the Sp-Atg44 proteins were bound to the edge of the lipid bilayer as a dot that was much smaller than the octamer, and the average thickness of the bound Sp-Atg44 above the membrane surface was approximately 1.4 nm, suggesting that Sp-Atg44 was bound to membranes not as an octamer (Figure 7C). Although it is difficult to determine the precise oligomerization state of Sp-Atg44 on the membrane, these data, together with the membrane binding model in Figure 6C, suggest that Sp-Atg44 binds to membranes either as a monomer, dimer, or tetramer, and not as an octamer. Unexpectedly, we noticed that addition of Sp-Atg44 caused fragmentation of lipid bilayers on the mica (compare Figures 7B and 7C) and the fragmented lipid bilayers abundant with Sp-Atg44 underwent division and fusion (Figure 7D; Video S6). These observations suggest that Sp-Atg44 has the ability to cause membrane fragility through physical interaction.
Figure 7. Atg44 binds to lipid-bilayer membranes and causes membrane fragility.
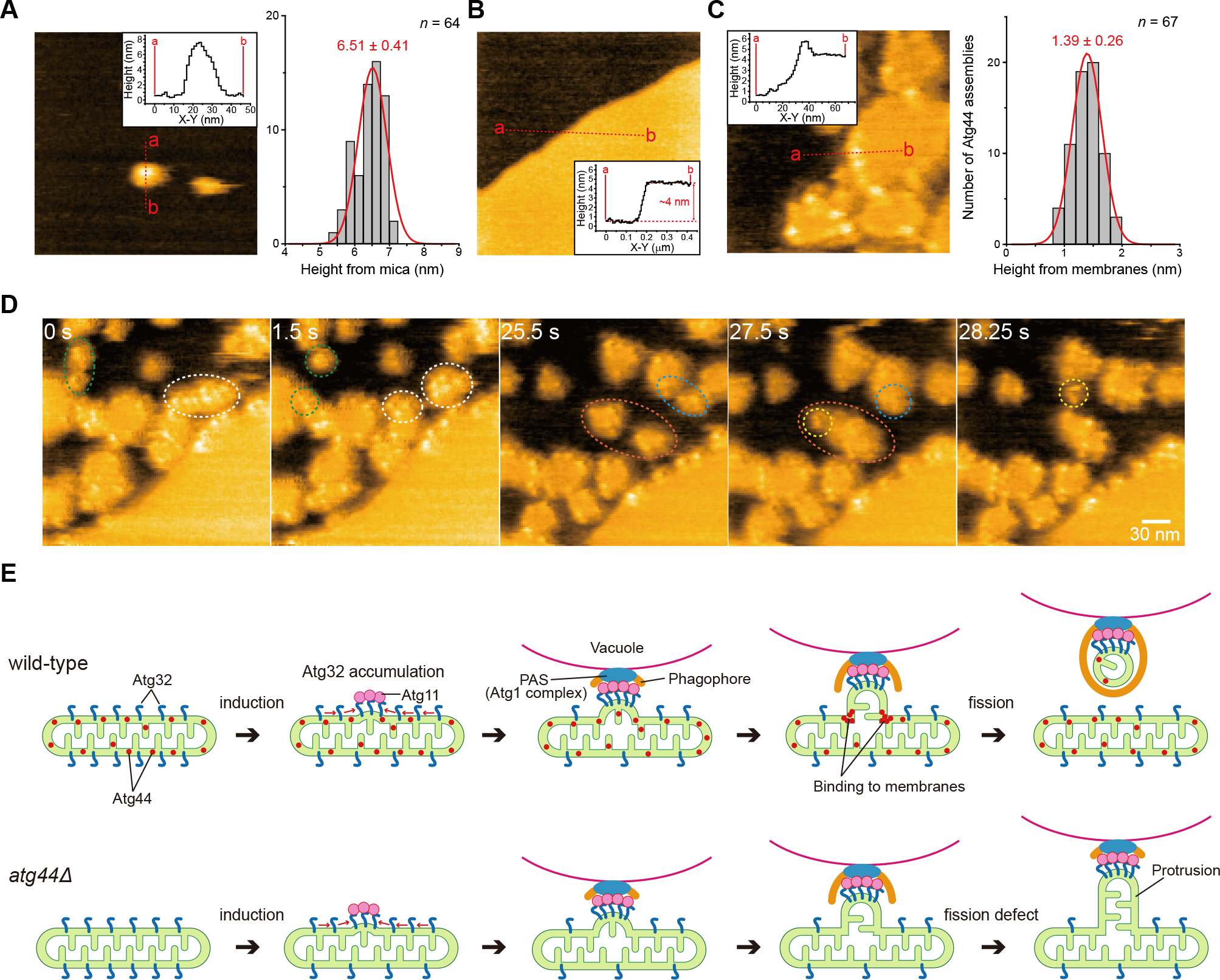
(A–C) HS-AFM images of Sp-Atg44 on the mica surface (A), the planar lipid bilayers covering mica (B), or Sp-Atg44 bound to the planar lipid bilayers (C). Inset shows height profile of the red line in each image. Histograms in (A) and (C) show the height distribution of Sp-Atg44 assemblies on the mica surface (n = 64) and membrane-bound Sp-Atg44 assemblies (n = 67), respectively.
(D) Successive HS-AFM images of division and fusion of the fragmented lipid bilayers by Sp-Atg44. Scale bar, 30 nm. See also Video S6.
(E) Schematic model of Atg44-dependent mitochondrial fission during mitophagy.
See also Figure S7.
Finally, to elucidate how Atg44 acts on the mitochondrial membrane, we analyzed DNM1L KO HeLa cells expressing Atg44 by immuno-electron microscopy against this protein. As shown in Figure S7, Atg44 was detected in mitochondria as clusters mostly localized to the IMM and the IMS. In the areas where the Atg44 clusters resided, the IMM showed abnormal morphology, possibly due to the membrane fragility caused by Atg44 (Figure S7, arrowheads). These findings suggest that Atg44 accumulates and exerts its function mainly on the IMM.
Taken together, we conclude that Atg44 binds to mitochondrial membranes, especially those with positive curvature, resulting in membrane fragility that facilitates membrane fission.
DISCUSSION
Atg44 possesses unique characteristics that differentiate it from dynamin-like DNM1L/Dnm1; Atg44 is a small protein of only 73 amino acids with neither a GTPase nor ATPase domain. Indeed, our HS-AFM and in vitro analyses have shown that recombinant Atg44 alone is sufficient to cause membrane fragility and the fragmentation of lipid-nanotubes, respectively, without GTP or ATP supplementation (Figures 5B and 7D). These findings suggest that Atg44 contains a membrane fission activity that requires neither nucleotides nor any other protein cofactors, in contrast to DNM1L/Dnm1, which requires GTP as well as its specific receptors on the mitochondrial surface.26,27 Furthermore, unlike Dnm1, which acts on the OMM from outside the mitochondria, Atg44 acts in the IMS. Atg44 thus exhibits unique properties and Atg44-like proteins are conserved in algae and bryophytes, as well as among a wide range of fungi. Therefore, we term this family of mitochondrial fission proteins as mitofissin. In addition to mitophagy, mitofissin may be involved in divergent cellular processes that require membrane remodeling of mitochondria in eukaryotes, because yeast cells lacking mitofissin exhibit abnormal mitochondrial morphology under vegetative growth conditions (Figures 3A–3D and S3A).
Based on our results, we propose a model of the mitophagy process as shown in Figure 7E. Upon mitophagy induction, Atg32 interacts with Atg11 and focally accumulates on the mitochondrial surface.7 Subsequently, Atg11 recruits and tethers the mitochondrial region to the PAS from which the phagophore extends. Almost simultaneously, the yeast mitofissin Atg44 binds to mitochondrial membranes and drives their fission, although the precise timing of the fission remains undetermined. Eventually, the mitochondrial fragment generated by the fission is engulfed by the phagophore. In the absence of mitofissin, however, the Atg32-accumulated region of the mitochondria is connected to the PAS without a separation from the main body of the mitochondria, resulting in the formation of the mitochondrial protrusion that fails to be enwrapped by the phagophore.
The mechanism by which mitofissin cleaves lipid membranes could be inferred from the structural feature of mitofissin. The crystal structure revealed that the protomer structure of the yeast mitofissin Atg44 is amphiphilic and that it assembles into a stable octamer by burying the hydrophobic portions inside the octamer (Figures 6A and 6B). On the other hand, HS-AFM indicates that Atg44 on the membrane behaves either as a monomer, dimer, or tetramer with the hydrophobic portions exposed to the outside (Figures 7A–7C). Based on these observations, we speculate that Atg44 interacts with the lipid bilayer of the membrane in a less-assembled state by inserting the exposed hydrophobic portions as predicted in Figures 6C and S6. Insertion of proteins into the outer layer creates the membrane-area difference between the two leaflets, which is known to induce spontaneous membrane curvature and, in some cases, drive fission of bilayer membranes.28 Therefore, mitofissin may promote membrane fission by creating the membrane-area difference. Another possible mechanism for the mitofissin-mediated membrane fission is the imposition of steric pressure on the membrane by protein crowding.29 Because mitofissin preferentially binds to membranes with high positive curvature, local accumulation of mitofissin on the membrane area with positive curvature may generate protein crowding that drives mitochondrial fission. The two possible mechanisms are not exclusive but rather might cooperate to induce efficient mitochondrial fission during mitophagy.
Mitofissin theoretically has access to both the IMM and the OMM because it resides in the IMS. The majority of yeast mitofissin is localized to the IMM, and the IMM exhibits abnormal morphology in the areas where mitofissin clusters (Figure S7). Furthermore, mitofissin binds to liposomes with IMM-like lipid composition in a manner dependent on cardiolipin, an IMM-specific phospholipid (Figure S5C). We also found that the membrane binding of mitofissin is attenuated under high salt conditions (Figure S5B). Based on these results, we propose the following model: mitofissin first accesses the IMM surface using electrostatic interactions that act over long distances, and then partially buries itself in the IMM using hydrophobic interactions. Further analysis is needed to support this model.
In summary, our work has identified mitofissin as a mitochondrial fission factor, discovered an unprecedented type of mitochondrial fission in which mitofissin directly binds to lipid membranes and causes membrane fragility, and revealed a role of mitofissin-mediated mitochondrial fission in adjusting the size of mitochondria to fit the capacity of the phagophore and autophagosome during mitophagy. Currently, mitofissin is not found in metazoans and higher plants, but its orthologous proteins or a mitochondrial process equivalent to the mitofissin-mediated mitochondrial fission may exist in those species, because mammalian mitophagy is clearly associated with DNM1L-independent mitochondrial fission.
Limitations of the study
We leave multiple open questions that should be addressed in the future studies. First, we have not found the amino acid residues that are crucial for the in vivo and in vitro function of Atg44. Their identification, together with their position within the structure and evolutionary conservation, is important for understanding the nature of mitofissin. Second, the role of Atg44 in mitochondrial fission that takes place independently of mitophagy is still elusive. As the morphological abnormality of mitochondria in cells lacking mitofissin is somewhat similar to that in dnm1Δ cells, it is possible that mitofissin cooperates with Dnm1 in coordinating the mitochondrial fission process under homeostatic conditions. Third, we have not determined the structure of membrane-bound Atg44, including its oligomeric state, and the relationship between the oligomeric state of Atg44 and its activity, which will provide further insights into the mechanism of mitofissin-mediated membrane fission. Finally, analysis of the functional conservation of mitofissin beyond fungi will be challenging but very important.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further inquiries and requests for materials should be directed to the lead contact, Tomotake Kanki (kanki@med.niigata-u.ac.jp)
Materials Availability
All requests for resources and reagents including plasmids and cell lines are available from the Lead Contact subject to a Materials Transfer Agreement.
Data and code availability
Coordinates and diffraction data of Sp-Atg44 as well as unprocessed images (microscopy, gels, and blots) were deposited in the Protein Data Bank (PDB) and Mendeley data respectively and are publicly available as of the date of publication. Accession numbers and DOI are listed in the key resources table.
This paper does not report any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Sc-Atg44 | This study | N/A |
| Guinea pig polyclonal anti-Sp-Atg44 | This study | N/A |
| Rabbit polyclonal anti-Atg33 | This study | N/A |
| Rabbit polyclonal anti-RFP | MBL | Cat# PM005; RRID: AB_591279 |
| Mouse monoclonal anti-GFP (JL-8) | Takara Bio | Cat# 632380; RRID: AB_10013427 |
| Mouse monoclonal anti-FLAG | Sigma-Aldrich | Cat# F1804; RRID: AB_262044 |
| Rabbit polyclonal anti-DDDDK | MBL | Cat# PM020; RRID: AB_591224 |
| Rabbit monoclonal anti-DYKDDDDK tag | Cell Signaling Technology | Cat# 14793; RRID: AB_2572291 |
| Rabbit polyclonal anti-ATPB | Abcam | Cat# ab128743; RRID: AB_2810299 |
| Rabbit polyclonal anti-Histone H3 | Abcam | Cat# ab1791; RRID: AB_302613 |
| Mouse monoclonal anti-HSPD1/HSP60 | Proteintech | Cat# 66041-1-Ig; RRID: AB_11041709 |
| Mouse monoclonal anti-Tim23 | BD Biosciences | Cat# 611222; RRID: AB_398754 |
| Rabbit polyclonal anti-Tom20 | Proteintech | Cat# 21090-1-AP; RRID: AB_10734583 |
| Rabbit polyclonal anti-CHCHD4 | Santa Cruz Biotechnology | Cat# sc-11415; RRID: AB_2207533 |
| AF594 F(ab’)2-goat anti-rabbit IgG (H+L) | Thermo Fisher Scientific | Cat# A-11072; RRID: AB_2534116 |
| AF594 F(ab’)2-goat anti-mouse IgG (H+L) | Thermo Fisher Scientific | Cat# A-11020; RRID: AB_141974 |
| AF647 goat anti-rabbit IgG (H+L) | Thermo Fisher Scientific | Cat# A-21244; RRID: AB_2535812 |
| Mouse monoclonal anti-HA | Sigma-Aldrich | Cat# H9658; RRID: AB_260092 |
| Mouse monoclonal anti-Pgk1 | Thermo Fisher Scientific | Cat# 459250; RRID: AB_2532235 |
| Chicken polyclonal anti-protein A | GeneTex | Cat# GTX77595; RRID: AB_379859 |
| Mouse monoclonal anti-Cox2 | MitoScience | Cat# MS419; RRID: AB_1618187 |
| Rabbit polyclonal anti-Ape1 | Kim et al. (1997)34 | N/A |
| Rabbit polyclonal anti-Atg32 | Aoki et al. (2011)6 | N/A |
| AF488 FluoroNanogold conjugated goat anti-rabbit IgG | Nanoprobes | Cat# 7204; RRID: AB_2868458 |
| Goat anti-mouse IgG-HRP | Merck Millipore | Cat# AP124P; RRID: AB_90456 |
| Goat anti-rabbit IgG-HRP | Jackson ImmunoResearch | Cat# 111-035-003; RRID: AB_2313567 |
| Bacterial and virus strains | ||
| Escherichia coli C41 (DE3) | Sigma-Aldrich | Cat# CMC0017 |
| Escherichia coli BL21(DE3) | Sigma-Aldrich | Cat# CMC0014 |
| Chemicals, peptides, and recombinant proteins | ||
| Zymolyase 100T | Nacalai tesque | Cat# 07665-55 |
| Lysing Enzymes from Trichoderma harzianum | Sigma-Aldrich | Cat# L1412 |
| Proteinase K, recombinant, PCR Grade | Roche | Cat# 03115836001 |
| MitoTracker Red CMXRos | Thermo Fisher Scientific | Cat# M7512 |
| FM 4-64 | Molecular Probes | Cat# T3166 |
| Block Ace | DS Pharma Biomedical | Cat# UKB80 |
| Amylose resin | New England Biolabs | Cat# E8021 |
| Ni-NTA Superflow | QIAGEN | Cat# 30410 |
| COSMOGEL GST-Accept | Nacalai tesque | Cat# 09277-56 |
| Alexa Fluor 647 C2 Maleimide | Invitrogen | Cat# A20347 |
| Alexa Fluor 647 hydrazide | Invitrogen | Cat# A20502 |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) | Avanti Polar Lipids | Cat# 850457 |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) | Avanti Polar Lipids | Cat# 850757 |
| L-α-phosphatidylinositol (PI) | Avanti Polar Lipids | Cat# 840042 |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS) | Avanti Polar Lipids | Cat# 840034 |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate (POPA) | Avanti Polar Lipids | Cat# 840857 |
| 1’,3’-bis[1,2-dioleoyl-sn-glycero-3-phospho]-glycerol (CL) | Avanti Polar Lipids | Cat# 710335 |
| 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (liss Rhod PE) | Avanti Polar Lipids | Cat# 810150 |
| D-galactosyl-β-1,1’ N-nervonoyl-D-erythro-sphingosine (GC) | Avanti Polar Lipids | Cat# 860546 |
| Silica beads (50 μm) | Nanocs | Cat# Si01-50u-1 |
| BSA tracer | Aurion | Cat# 210.033 |
| Tamavidin 2 solution | FUJIFILM Wako | Cat# 201-21271 |
| Critical commercial assays | ||
| NEBuilder HiFi DNA Assembly Master Mix | New England Biolabs | Cat# E2621 |
| PrimeSTAR Mutagenesis Basal Kit | Takara Bio | Cat# R046A |
| IgG Sepharose 6 Fast Flow | GE Healthcare | Cat# 17096901 |
| EzWestLumi One | ATTO | Cat# WSE-7110 |
| EzWestLumi plus | ATTO | Cat# WSE-7120 |
| Clarity Max Western ECL Substrate | Bio-Rad | Cat# 1705062 |
| FuGENE HD Transfection Reagent | Promega | Cat# E2311 |
| Oriole Fluorescent Gel Stain | Bio-Rad | Cat# 1610496 |
| Deposited data | ||
| Crystal structure of Sp-Atg44 | This study | PDB: 7YDO |
| Unprocessed images (microscopy, gels, and blots) | This study | Mendeley data: http://dx.doi.org/10.17632/92p3bx6nnz.1 |
| Experimental models: Cell lines | ||
| Human: HeLa (WT) | Yamashita et al. (2016)16 | N/A |
| Human: HeLa (DNM1L KO) | Yamashita et al. (2016)16 | N/A |
| Experimental models: Organisms/strains | ||
| S. cerevisiae strains, see Table S1 | This study | N/A |
| S. pombe strains, see Table S2 | This study | N/A |
| Software and algorithms | ||
| Image Lab | Bio-Rad | https://www.bio-rad.com/en-jp/product/image-lab-software |
| MetaMorph 7 | Molecular Devices | https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy |
| Auto Slice & View (version 5.6.0) | Thermo Fisher Scientific | https://www.thermofisher.com/jp/ja/home/electron-microscopy/products/software-em-3d-vis/auto-slice-view-4-software.html |
| Fiji | Schindelin et al. (2012)42 | http://fiji.sc/Fiji |
| Image J | National Institutes of Health | https://imagej.nih.gov/ij |
| Amira (version 5.4.5) | Thermo Fisher Scientific | https://thermofisher.com/amira-avizo |
| Zen 2.1 SP1 | Carl Zeiss | https://www.zeiss.com/microscopy/en/products/software/zeiss-zen.html |
| Huygens Professional | Scientific Volume Imaging | https://svi.nl/Huygens-Professional |
| Astra | Wyatt Technology | https://www.wyatt.com/products/software/astra.html |
| DigitalMicrograph | Gatan | https://www.gatan.com/resources/digitalmicrograph-scripts |
| XDS Program Package | Kabsch (2010)48 | https://xds.mr.mpg.de/ |
| PHENIX | Adams et al. (2010)49 | https://phenix-online.org/ |
| COOT | Emsley et al. (2010)50 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| UCSF ChimeraX (version 1.3) | Pettersen et al. (2021)51 | https://www.rbvi.ucsf.edu/chimerax |
| PPM 3.0 | Lomize et al. (2022)25 | https://opm.phar.umich.edu/ppm_server3_cgopm |
| Kodec (version 4.4.7.39) | Ngo et al. (2015)53 | https://doi.org/10.7554/eLife.04806.031 |
| Prism 9 (version 9.5.1) | GraphPad Software | https://www.graphpad.com/ |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Yeast strains
S. cerevisiae and S. pombe strains used in this study are shown in Tables S1 and S2, respectively. Cells were cultured on solid agar plates or in liquid culture as described in the method details.
HeLa cells
WT and DNM1L KO HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS and antibiotics. The cells were maintained at 37°C in a humidified atmosphere of 95% air and 5% CO2.
METHOD DETAILS
Yeasts culture conditions
S. cerevisiae cells were cultured at 30°C in rich medium (YPD: 1% yeast extract, 2% peptone, and 2% glucose), lactate medium (YPL: 1% yeast extract, 2% peptone, and 2% lactate), oleic acid medium (YTO: 0.67% yeast nitrogen base without amino acids, 0.1% Tween-40, and 0.1% oleic acid), or synthetic minimal medium with glucose (SMD: 0.67% yeast nitrogen base, 2% glucose, and amino acids) or lactate (SML: 0.67% yeast nitrogen base, 2% lactate, and amino acids). Nitrogen starvation experiments were performed in synthetic minimal medium lacking nitrogen (SD-N: 0.17% yeast nitrogen base without amino acids and ammonium sulfate, and 2% glucose). S. pombe cells were cultured at 30°C in complete medium (YES: 0.5% yeast extract and 3% glucose) or synthetic minimal EMM medium (Formedium). Nitrogen starvation experiments were performed in EMM lacking nitrogen (EMM-N) (Formedium).
For growth assays, S. cerevisiae or S. pombe cells grown to early-log phase in YPD or EMM were spotted onto YPD, YPL, or YES agar plates and incubated at 30°C.
Plasmids
To construct Sc-Atg44, Sc-Atg44-FLAG, Sp-Atg44, and Kp-Atg44 expression plasmids under the control of the CUP1 promoter to transform in S. cerevisiae, their coding regions were amplified by PCR and cloned into the EcoRI-XhoI, BamHI-XhoI, EcoRI-XhoI, and BamHI-SalI sites of pCu416,30 respectively. To construct an Sc-Atg44 overexpression plasmid, the TDH3 promoter region (SacI-BamHI) and Sc-ATG44 region (BamHI-XhoI) were ligated and cloned into the SacI-XhoI sites of pRS416.31
The plasmids to express PA-Atg32, GFP-Atg32, HA-Atg11, and RFP-Atg8 (pCu416-PA-ATG32, pCu416-GFP-ATG32, pCu414-HA-ATG11, and pRS416-RFP-ATG8) were previously reported.16,32 To construct Sp-Atg44 and Sc-Atg44 expression plasmids to transform in S. pombe, their coding regions were amplified by PCR and cloned into the NdeI-BamHI sites of pNATZA11 (NBRP ID: FYP2875). To construct Sc-Atg44 expression plasmids in HeLa cells, coding regions of Sc-Atg44 were amplified by PCR and cloned into the BamHI-XhoI sites of the IRES-GFP-NLS vector.33 To construct Sc-Atg44-FLAG and LACTB_N68-FLAG expression plasmids in HeLa cells, coding region of Sc-Atg44 and the N-terminal mitochondrial IMS targeting sequence of LACTB were amplified by PCR with the reverse primer containing the FLAG sequence and cloned into BamHI-XhoI sites of the IRES-GFP-NLS vector. To construct recombinant Sc-Atg44 and Sp-Atg44 protein expression vectors, the genes encoding Sc-Atg44 and Sp-Atg44 were amplified by PCR and assembled into an in-house modified pET15b vector (Novagen) encoding maltose-binding protein (MBP) and PreScission protease recognition sequences by NEbuilder HiFi DNA Assembly (New England Biolabs). A linker sequence, GyGlyGlySerGlyGlyGlySer was inserted between PreScission protease recognition and Atg44 sequences using a PrimeSTAR Mutagenesis Basal Kit (Takara Bio). Mutations were introduced into each vector with the PrimeSTAR Mutagenesis Basal Kit using wild-type plasmid as a template. To construct a recombinant GFP expression vector, the gene encoding GFP was amplified by PCR and assembled into the pGEX6P vector (GE Healthcare) using the NEbuilder HiFi DNA Assembly. The A207K mutation was introduced into GFP using the PrimeSTAR Mutagenesis Basal Kit to produce monomeric GFP.
Antibodies
Anti-Sc-Atg44 and anti-Sp-Atg44 antibodies were produced by immunizing rabbits and guinea pigs with recombinant His-tagged Sc-Atg44-Sc-Atg44 (two full-length Sc-Atg44 in a tandem repeat) and recombinant His-tagged Sp-Atg44-Sp-Atg44, respectively, followed by affinity purification with recombinant His-tagged Sc-Atg44 conjugated to AminoLink Plus Coupling Resin (Thermo Fisher Scientific, 20501) and recombinant His-tagged Sp-Atg44-Sp-Atg44 transferred on a polyvinylidene difluoride membrane. The anti-Sc-Atg33 serum was produced by immunizing rabbits with recombinant His-tagged full-length Atg33. Anti-RFP (MBL, PM005, RRID:AB_591279), anti-GFP (Takara Bio, 632380, RRID:AB_10013427), anti-FLAG (Sigma, F1804, RRID:AB_262044), anti-DDDDK (MBL, PM020, RRID:AB_591224), anti-DYKDDDDK tag (Cell Signaling Technology, 14793, RRID:AB_2572291), anti-Atp2 (Abcam, ab128743, RRID:AB_2810299), anti-histone H3 (Abcam, ab1791, RRID:AB_302613), anti-HSPD1/HSP60 (Proteintech, 66041–1-Ig, RRID:AB_11041709), anti-Tim23 (BD Biosciences, 611222, RRID:AB_398754), anti-CHCHD4 (Protentech, 21090–1-AP, RRID:AB_10734583), anti-Tom20 (Santa Cruz Biotechnology, sc-11415, RRID:AB_2207533), Alexa Fluor 594 (AF594) F(ab’)2-goat anti-rabbit IgG (H+L) (Thermo Fisher Scientific, A11072, RRID:AB_2534116), AF594 F(ab’)2-goat anti-mouse IgG (H+L) (Thermo Fisher Scientific, A11020, RRID:AB_141974), AF647 goat anti-rabbit IgG (H+L) (Thermo Fisher Scientific, A21244, RRID:AB_2535812), anti-HA (Sigma-Aldrich, H9658, RRID:AB_260092), anti-Pgk1 (Thermo Fisher Scientific, 459250, RRID:AB_2532235), anti-protein A (PA; GeneTex, GTX77595, RRID:AB_379859), anti-Cox2 (MitoScience, MS419, RRID:AB_1618187), anti-Ape134, and anti-Atg326 were used for immunoblotting and immunofluorescence microscopy. AF488 FluoroNanogold conjugated goat anti-rabbit IgG (Nanoprobes, 7204, RRID: AB_2868458) was used for immuno-electron microscopy.
Autophagy assays in S. cerevisiae
To monitor mitophagy, the Idh1-GFP processing assay was performed as previously described.35 In brief, cells were cultured in YPL or SML medium until mid-log phase, and then shifted to SD-N for nitrogen starvation. The cells were collected at multiple time points, and the cell lysates were subjected to immunoblotting analysis. To monitor macroautophagy in S. cerevisiae, the phosphatase activity of Pho8Δ60 was measured or the GFP-Atg8 processing assay was performed as previously described.36,37 Pex14-GFP and Sec63-GFP processing assays were performed to monitor pexophagy and reticulophagy, respectively, as previously described.38,39
Autophagy assays in S. pombe
To monitor mitophagy, the Tuf1-RFP and Sdh2-GFP processing assays were performed.18 Logarithmically growing cells in EMM medium were transferred to EMM-N for nitrogen starvation. The cells were collected at multiple time points and the cell lysates were subjected to immunoblotting analysis. To monitor bulk autophagy and reticulophagy, strains expressing Pgk1-GFP or Tdh1-RFP and Yop1-GFP, respectively, were processed in the same way as for mitophagy.18
Screen for mitophagy-defective mutants in S. pombe
The haploid S. pombe deletion mutant library (Bioneer) was used for the genetic screen.18 Mutant strains were transformed with a DNA fragment encoding Tuf1-RFP, making a Tuf1-RFP-expressing mutant library. Mutant strains were grown in YES medium, and shifted to EMM-N. Following overnight culturing, cells were subjected to immunoblotting for the mitophagy assay.
Immunoblotting analysis
Protein samples from S. cerevisiae cells were resuspended in sodium dodecyl sulfate (SDS) sample buffer (50 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 5% 2-mercaptoethanol, and 0.1% bromophenol blue), incubated at 42°C for 60 min or at 100°C for 5 min, and subjected to tris-glycine or tris-tricine SDS-polyacrylamide gel electrophoresis (PAGE). For S. pombe cells, C buffer (8 M urea, 5% SDS, 40 mM tris-HCl, pH 6.8, 0.1 mM EDTA, and 10% 2-mercaptoethanol) was used instead of SDS sample buffer. Proteins were transferred from polyacrylamide gels to polyvinylidene difluoride (PVDF) membranes (Merck Millipore) using transfer buffer (25 mM tris, 192 mM glycine, 20% methanol). The membranes were blocked with phosphate-buffered saline (PBS: 10 mM PO43-, pH 7.4, 140 mM NaCl, and 2.7 mM KCl) containing 0.05% Tween-20 (PBS-T) and 5% skim milk for 1 h. The membranes were incubated with primary antibodies in PBS-T containing 2% skim milk overnight at 4°C and washed 3 times with PBS-T. The membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies in PBS-T containing 2% skim milk for 1 h at room temperature, and washed 3 times with PBS-T. Chemiluminescence signals were detected using ChemiDoc™ XRS+ (Bio-Rad) and analyzed using Image Lab software (Bio-Rad).
Protein A affinity-isolation assay
S. cerevisiae cells expressing PA-tagged Atg32 and HA-tagged Atg11 or Atg44 under the control of the CUP1 promoter were cultured in SMD medium until the early-log growth phase, and then shifted to SD-N for 1 h. Cells were collected and lysed with glass beads in IP buffer (PBS, 0.2% Triton X-100, 1 mM PMSF, and proteinase inhibitors), and the lysates were centrifuged at 10,000 × g for 10 min at 4°C. The supernatant was mixed with IgG-Sepharose at 4°C for 4 h. The Sepharose was washed with ice cold wash buffer (PBS containing 0.2% Triton X-100) five times, and the sample was eluted with SDS sample buffer. The elution samples were observed by immunoblotting with anti-protein A, anti-HA, and anti-Atg44 antibodies.
Isolation of mitochondria, sodium carbonate treatment and ProK protection assay
S. cerevisiae cells expressing Sc-Atg44-FLAG and Mia40-GFP or S. pombe cells expressing Tom20-GFP, Mic60-FLAG, and Tuf1-RFP were cultured in YPD or YES, respectively, until the mid-log growth phase and resuspended in DTT buffer (10 mM dithiothreitol, 0.1 M tris-HCl, pH 9.3) for 30 min at 30°C. Cells were collected and converted to spheroplasts in sorbitol buffer (1.2 M sorbitol, 20 mM KH2PO4, pH 7.4) with Zymolyase 100T (Nacalai tesque). Lysing enzymes from Trichoderma harzianum (Sigma) were also added for S. pombe cells. Spheroplasts were collected by centrifugation (1000 × g for 10 min at 4°C) and resuspended in ice-cold homogenization buffer (0.6 M sorbitol, 20 mM HEPES, pH 7.4) and homogenized in a Potter-Elvehjem homogenizer. The cell homogenate was centrifuged at 1000 × g for 10 min at 4°C. The supernatant was centrifuged at 6500 × g for 10 min at 4°C. The pellet was collected as the mitochondrial fraction. Isolated mitochondria were separately suspended in ice-cold 0.1 M sodium carbonate, pH 11.0, incubated for 30 min on ice and then centrifuged at 100,000 × g for 30 min at 4°C. Proteins in the pellet and supernatant fractions were precipitated by adding 10% TCA. Isolated mitochondria were separately suspended in ice-cold homogenization buffer, hypotonic buffer (20 mM HEPES, pH 7.4), or hypotonic buffer with 0.5% Triton X-100 and treated with ProK (200 μg/ml) for 30 min on ice. The ProK reaction was stopped by adding 10% trichloroacetic acid (TCA). TCA-precipitated proteins were washed with acetone and subjected to immunoblotting.
HeLa cells expressing Sc-Atg44-FLAG or LACTB_N68-FLAG were suspended in homogenization buffer (250 mM sucrose, 20 mM HEPES, pH 7.4) and homogenized by passing through a 26G needle 10 times. The cell homogenate was centrifuged at 800 × g for 5 min at 4°C. The supernatant was centrifuged at 10,000 × g for 10 min at 4°C. The pellet was collected as the mitochondrial fraction. A protease protection assay of the isolated mitochondria was performed by the same method as used with yeast.
Fluorescence microscopy of yeasts and HeLa cells
S. cerevisiae or S. pombe cells expressing the indicated fluorescent proteins were cultured in the indicated media. To stain the mitochondria, cells were incubated with 50 nM MitoTracker Red CMXRos (Thermo Fisher Scientific) for 30 or 60 min. To stain the vacuole, cells were incubated with 1.6 μg/ml N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenylhexatrienyl) pyridinium dibromide (FM 4–64; Molecular Probes) for 30 min. For immunofluorescence of S. cerevisiae, cells expressing Idh1-GFP and Sc-Atg44-FLAG were cultured in YPD to early log phase and were fixed with 4% paraformaldehyde for 30 min at room temperature. Cells were collected and converted to spheroplasts in spheroplast buffer (1.2 M sorbitol, 100 mM KH2PO4, pH 7.4, 1% 2-mercaptoethanol, 100 μg/ml Zymolyase 100T) and incubated for 15 min at room temperature. Cells were permeabilized with 0.5% Triton X-100 for 10 min, then blocked with 1% bovine serum albumin (BSA) for 30 min. Immunofluorescence staining was performed with primary antibodies against GFP or FLAG, and secondary antibodies conjugated with Alexa Fluor 488 (AF488) or AF594.
HeLa cells were cultured on coverslips and transfected with IRES-GFP-NLS-empty, - Sc-ATG44, -Sc-ATG44-FLAG, or -LACTB_N68-FLAG vectors using FuGENE HD (Promega) according to the manufacturer’s instructions. After 24 h, cells were fixed with 4% paraformaldehyde for 15 min at room temperature. Fixed cells were permeabilized with 0.2% Triton X-100 for 5 min, then blocked with 1% BSA for 30 min. Immunofluorescence staining was performed with primary antibody against Sc-Atg44, FLAG, or HSPD1/HSP60 and secondary antibody conjugated with AF594 or AF647.
Fluorescence images were taken with an Olympus IX73 microscope with a UPlanSApo 100× oil objective lens and a cooled charge-coupled device (CCD) camera (Exi Blue, Qimaging) or Nikon Ti2 eclipse with a Plan Apo Lambda 100× oil objective lens and a CCD camera (MD-695, Molecular Devices). Fluorescence images were analyzed using MetaMorph 7 software (Molecular Devices). Super resolution images were taken with a ZEISS LSM880 Airyscan microscope system (super resolution mode) with a Plan-APOCHROMAT 63×/1.4 Oil DIC and analyzed using Zen 2.1 SP1 (Carl Zeiss) and Huygens Professional (Scientific Volume Imaging). Yeast and HeLa cells were classified according to whether their mitochondria were fragmented, enlarged, tubular, or a mixture in shape, and their ratio was quantified.
Electron microscopy
S. cerevisiae cells grown in the indicated conditions were fixed with a 2% solution of glutaraldehyde in 50 mM phosphate buffer (pH 7.2) for 2 h at room temperature. The cells were then washed with 50 mM phosphate buffer (pH 7.2) and further fixed with a 1.2% solution of potassium permanganate (KMnO4) in distilled water at 4°C overnight.40,41 The cells were then washed with distilled water and collected by centrifugation at 2,000 × g at room temperature for 3 min. The pellets were embedded in 2% low-melting agar (Sigma Aldrich), and the agar containing the cells was cut into pieces of approximately 1–2 mm, and dehydrated through a graded ethanol series (50, 60, 70, 80, 90, 95, and 100%). The samples were infiltrated with QY-1 (Nissin EM Co. Ltd.), embedded in epoxy resin mixture (Quetol 812: Nissin EM Co. Ltd.), and polymerized at 60°C for 2 days. These resin blocks containing the samples were used for transmission electron microscopy (TEM), serial block-face scanning electron microscopy (SBF-SEM), and focused ion beam scanning electron microscopy (FIB-SEM). For TEM, ultrathin sections (approximately 70-nm thick) were prepared with a diamond knife and mounted on formvar-coated Cu single-slot grids. The sections were contrasted by staining with 2% uranyl acetate and lead citrate, and examined with a TEM (JEM1010, JEOL). For SBF-SEM, the resin blocks were trimmed and glued onto an aluminum rivet with a conductive epoxy resin (SPI Conductive Silver Epoxy: SPI Supplies and Structure Prove, Inc.), and then coated with gold using an ion coater (IB-3: Eiko Engineering). SBF-SEM (MERLIN: Carl Zeiss Microscopy; 3View: Gatan Inc., Pleasanton) equipped with an OnPoint BSE detector (Gatan Inc., Pleasanton) was used to slice and image the specimen. The scanning electron microscope was operated at a low accelerating voltage of 1.3 kV. The serial images were automatically acquired by using Gatan Digital Micrograph software. All images were taken at an image size of 8192 × 8192 pixels (pixel size = 2.3 nm) with a thickness of 70 nm. For FIB-SEM, the resin blocks were trimmed and glued on to an aluminum 45° inclined specimen stage with a conductive paste (Silver paste: EM Japan Co., LTD.), and then coated with gold using an ion coater (MSP-1S Magnetron Sputter: Vacuum Device Inc., Japan). FIB-SEM (Aquilos 2: Thermo Fisher Scientific) equipped with a focused ion beam for slicing and a segmented lower (T1) detector for imaging. The focused ion beam was operated at a high voltage 30 kV with beam current 300 pA, and the scanning electron microscope was operated at a low accelerating voltage of 1.5 kV with beam current 50 pA. The serial images were automatically acquired by Auto Slice & View software version 5.6.0. All images were taken at an image size of 2203 × 1951 pixels (pixel size = 10 nm) with a thickness of 20 nm. The image series was automatically aligned using Fiji42 and cropped around each cell. Segmentation of the volume data was performed with Amira version 5.4.5 (Thermo Fisher Scientific).
Immuno-electron microscopy
DNM1L/Drp1 KO HeLa cells were cultured in 4-well chamber slides (ThermoFisher Scientific, 177437) and transfected with the Sc-ATG44 vector. The detailed procedure of pre-embedding immuno-electron microscopy analysis was described previously.43 Briefly, the HeLa cells in the chamber-slides were fixed with 4% PFA, followed by the incubation with the blocking solution containing 5.0% BlockAce (DS Pharma Biomedical) with 0.05% saponin in 0.1 M phosphate buffer (pH 7.4) for 30 min at 25°C. Fixed cells were exposed to a rabbit anti-Sc-Atg44 antibody (1:500) in blocking solution for 96 h at 4°C, followed by the incubation with an AF488 FluoroNanogold conjugated goat anti-rabbit secondary antibody (1:100, Nanoprobes, 7204) for 24 h at 4°C. The target cells were confirmed by fluorescence microscopy (LSM880, CarlZeiss) in which the AF488 signals were identified. After 2.5% glutaraldehyde fixation in 0.1 M phosphate buffer (pH 7.4), washed 0.1 M phosphate buffer (pH 7.4), 50 mM HEPES (pH 5.8) 3 times for 10 min, washed with diluted water for 1 min, and nanogold signals were enhanced with silver enhancement solution for 2 min at 25°C. The gold-labeled sections were post-fixed with 1.0% osmium tetroxide (TAAB Laboratories) for 90 min at 4°C, en bloc staining with uranyl acetate for 20 min at 25°C, and dehydrated through diluted ethanol (2 times of 50, 70, 80, 90, 100% EtOH for 5 min each), with a graded epoxy resin series (epoxy resin:ethanol = 1:3, 1:1, 3:1 for 30 min each), and 100% epoxy resin for 48 h at 4°C for enhance infiltration, and finally embedded into 100% polymerizing epoxy resin. After completing the polymerization for 72 h at 65°C, resin blocks were trimmed and were ultrathin-sectioned at 70-nm thickness with an ultramicrotome (UC7, Leica) using a diamond knife (Ultra, DiATOME). The ultrathin-sections were collected on the copper grids, stained with uranyl acetate and lead citrate. The ultrathin-sections were imaged with a TEM (H-7650, Hitachi) at 80 keV.
Preparation of recombinant Sc-Atg44 and Sp-Atg44
Sc-Atg44 and Sp-Atg44 were overexpressed as MBP fusion proteins in Escherichia coli C41(DE3) cells. Cells were grown in Luria broth (LB) medium at 37°C and protein expression was induced with 0.5 mM isopropyl-β-D-glucoside (IPTG). After overnight culture at 20°C, cells were harvested by centrifugation, suspended in PBSN (PBS supplemented with 1.0 M NaCl) and frozen until purification. For purification, the cell suspension was supplemented with 1.0 mM PMSF. After sonication and centrifugation, the supernatant was loaded on amylose resin (New England Biolabs) pre-equilibrated with PBSN. The resin was extensively washed, and then bound proteins were eluted with PBSN supplemented with 10 mM maltose. The eluate was concentrated by using a centrifugal filter device, 10-kDa cut-off (Millipore), and then subjected to SEC using a Superose 6 10/300 column (GE Healthcare) in HEPES buffer (20 mM HEPES-NaOH, pH 7.0 and 150 mM NaCl). For Sc-Atg44, the eluates containing MBP-Sc-Atg44 were merged, concentrated and frozen until used in an experiment. For Sp-Atg44, the eluates containing MBP-Sp-Atg44 were merged and treated with house-made PreScission protease overnight. The resultant solution was purified with Ni-NTA resin (QIAGEN) pre-equilibrated with HEPES buffer supplemented with 10 mM imidazole. The resin was washed, and then bound protein was eluted with HEPES buffer supplemented with 200 mM imidazole. The eluate was further purified by SEC using a Superdex 75 Increase 10/300 GL column (GE Healthcare) in HEPES buffer. The eluates containing Sp-Atg44 were merged, concentrated and frozen until used in an experiment. For the preparation of selenomethionine (SeMet)-labeled Sp-Atg44V42M, transformed E. coli C41(DE3) cells were cultured in minimal medium supplemented with SeMet. SeMet-labeled Sp-Atg44V42M and mutant forms of Sp-Atg44 were purified in a similar manner to the wild-type protein.
GFP was overexpressed as a GST fusion protein in E. coli BL21(DE3) cells. Cells were grown in LB medium and protein expression was induced by the addition of 0.1 mM IPTG. After overnight culture at 16°C, cells were pelleted, suspended in PBS buffer and frozen until purification. For purification, the cell suspension was supplemented with 1.0 mM PMSF. After sonication and centrifugation, the supernatant was loaded on GST-Accept resin (Nacalai Tesque) pre-equilibrated with PBS buffer. The resin was extensively washed with PBS buffer and the bound protein was eluted with PBS buffer supplemented with 10 mM reduced glutathione. The eluate was desalted and treated with PreScission protease overnight. The solution was passed through GST-Accept resin pre-equilibrated with PBS buffer, concentrated and then subjected to SEC using a Superdex 75 Increase 10/300 GL column in HEPES buffer.
Size-exclusion chromatography coupled to multi-angle light scattering (SEC-MALS)
SEC-MALS was performed with a Shimadzu chromatography system which was connected in-line to a DAWN HELEOS II (Wyatt) for light scattering and RI-501 (Shodex) for differential refractive index. Purified MBP-Sc-Atg44 was subjected to SEC using a Superose 6 10/300 column. Sp-Atg44 was subjected to SEC using a Superdex 75 Increase 10/300 GL column. The system was pre-equilibrated with HEPES buffer at room temperature. Obtained data were analyzed using Astra (Wyatt).
Fluorescent labeling
For site-selective fluorescent labeling, a Cys residue was introduced between PreScission protease recognition and linker sequences in the Sp-Atg44 construct. Maleimide-modified AF647 (Invitrogen) dissolved in dimethyl sulfoxide (DMSO) was mixed with Cys-Sp-Atg44 at an equivalent molar ratio for 30 min at room temperature. The mixture was then dialyzed against HEPES buffer using a Mini Dialysis Kit, 8-kDa cut-off (GE Healthcare) to remove the remaining fluorescent dye.
GUV experiment
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), L-α-phosphatidylinositol (PI), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate (POPA), 1’,3’-bis[1,2-dioleoyl-sn-glycero-3-phospho]-glycerol (CL) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (liss Rhod PE) were purchased from Avanti Polar Lipids. GUVs were prepared by the natural swelling method as described previously.44 In detail, 100 nmol of phospholipids containing POPC:POPE:PI:POPS:POPA:CL:liss Rhod PE=25:24:10:10:10:20:1 (mol%) in chloroform was gently dried under nitrogen gas and put in a vacuum overnight. The dried lipid film was pre-hydrated with water at ~55°C for 7 min, soaked in HEPES buffer supplemented with 100 mM sucrose and then incubated at 60°C for 2 h. GUVs were spontaneously produced in this condition. The GUV solution was cooled at room temperature and freshly used for experiments.
GUV solution was deposited in HEPES buffer supplemented with 100 mM glucose in a BSA-precoated handmade chamber. The mixture of 90 μM Sp-Atg44 and 10 μM AF647-Sp-Atg44 was continuously applied to GUVs via a glass micropipette installed on a three-dimensional micromanipulator (Narishige) on the stage of an FV3000RS (Olympus) at room temperature. The sample application was hydraulically controlled, and the pressure was monitored by digital meter. The confocal images were acquired using FV31S-SW (Olympus) and analyzed with Fiji.
Membrane fission experiment
Excess membrane reservoir was prepared as described previously.45 Briefly, 100 nmol lipid film containing POPC:POPE:PI:POPS:POPA:CL:liss Rhod PE=25:24:10:10:10:20:1 (mol%) was hydrated in water, vortexed and sonicated with a tip sonicator until transparent to create liposomes. Silica beads with ~50-μm diameter in water (Nanocs) were added to 200 μl of 200 μM lipid solution in 1.0 M NaCl and incubated at room temperature with intermittent vertexing for 30 min to produce silica beads covered with membrane; this was called the excess membrane reservoir. The excess membrane reservoir was washed by repeats of the addition of water and centrifugation, and freshly used for experiments.
To produce lipid nanotubes, excess membrane reservoir was deposited in HEPES buffer in the BSA-coated handmade chamber. The chamber was then tilted to allow the excess membrane reservoir to roll slowly, which produced lipid nanotubes with various radii. The mixture of 90 μM Sp-Atg44 and 10 μM AF647-Sp-Atg44 in HEPES buffer was continuously applied to lipid nanotubes via a glass micropipette on the stage of an FV3000RS. Twenty μM GFP or 10 μM AF647 hydrazide in HEPES buffer were used for the control experiments. Confocal images were acquired using FV31S-SW and analyzed with Fiji. The radius of lipid nanotubes, r was estimated by the equations, IROI=αA, r=IROI/2πLα, where IROI is the total fluorescence intensity in the ROI, α is the correlation factor, A is the ROI area and L is the tube length.24 To estimate the α value, excess membrane reservoir was deposited in HEPES buffer in the chamber without BSA coating. The excess membrane was then spilled out on the glass surface to produce the supported lipid bilayer. The total fluorescence intensity of the supported lipid bilayer in the ROI was measured after subtracting background and plotted against the ROI area to calculate the α value. Fluorescence intensity and tube length were measured with Fiji.
Flotation assay
Dried lipid film was hydrated in a flotation buffer (20 mM HEPES-NaOH, pH 7.0, 150 mM or 500 mM NaCl and 100 mM glucose) and vigorously agitated by vortex mixer. The suspension was subjected to 5 freeze-thaw cycles using liquid N2 and a block incubator at 42°C. For the preparation of small liposomes, the suspension was sonicated by using a hand-type tip sonicator until transparent. For the preparation of large liposomes, the suspension was passed through a membrane filter (Whatman) with the pore size of 200 nm more than 21 times using a mini-extruder (Avanti Polar Lipids). The prepared solution was centrifuged at 18,000 × g for 15 min to remove debris and the supernatant containing liposomes was used for the flotation assay as follows: 100 μl of liposome solution consisting of 1.0 mM phospholipids whose compositions were indicated in the figures and legends was incubated with 40 μM Sp-Atg44 for more than 1 h at room temperature. For the control sample, a flotation buffer was instead incubated with Sp-Atg44. The mixture was gently merged with 100 μl of a flotation buffer supplemented with 60% (w:v) sucrose and loaded in an open-top thin wall ultra-clear tube of 0.8 ml (Beckman Coulter). The solution was then overlaid with 350 μl of a flotation buffer supplemented with 25% (w:v) sucrose and finally mounted with ~150 μl of a flotation buffer. The sample was centrifuged at 200,000 × g for 1 h at 20°C on SW55Ti swinging-bucket rotor (Beckman Coulter), and the top 150 μl fraction and the bottom 200 μl fraction were analyzed by SDS-PAGE. The gel was stained with oriole (Bio-Rad) and visualized using a Gel Doc EZ Imager (Bio-Rad). The densitometry was performed after subtracting background by Fiji.
Cryo-EM observation
Lipid nanorods were prepared by using D-galactosyl-β-1,1’ N-nervonoyl-D-erythro-sphingosine (GC, Avanti Polar Lipids).46 Briefly, dried lipid film containing POPC:POPE:PI:POPS:POPA:CL:GC=15:15:6:6:6:12:40 (mol%) was hydrated in HEPES buffer, vortexed and sonicated in a bath sonicator for 30 min. MBP-Sc-Atg44 (20 μM) was mixed with 2.0 mM lipid solution and incubated at room temperature overnight. An aliquot (3 μl) of the mixture supplemented with a fiducial marker (BSA tracer 10 nm, Aurion) was loaded on a glow-discharged Quantifoil (R1.2/1.3 Cu 200 mesh) grid, left for 2 min and then blotted and plunge-frozen in liquid ethane held on liquid N2 using the FEI Vitrobot Mark IV system (ThermoFisher Scientific) set to 20°C and 100% humidity. The grid was loaded on the Elsa cryo-transfer holder (Gatan) and transferred to a JEM-1400Flash electron microscope (JEOL) running at 120 kV equipped with a Gatan Rio 16 camera. Images were acquired using Serial EM47 and DigitalMicrograph (Gatan).
X-ray data collection
Purified Sp-Atg44 was concentrated above 2.0 mg/ml and subjected to the initial screening for crystallization. Optimized crystallization was performed by the sitting drop vapor diffusion method at 293 K. Crystals of native Sp-Atg44 and SeMet-labeled Sp-Atg44V42M were obtained in a reservoir solution containing 0.10 M citrate, pH 5.5, 20% PEG3350 and 0.10% n-octyl-β-D-glucoside. The crystals were soaked in the reservoir solution supplemented with 15% glycerol, flash-cooled and then kept under a stream of nitrogen gas at 90 K during data collection. All diffraction data were collected at the SPring-8 beamline BL32XU. Multiwavelength anomalous diffraction (MAD) data were collected from a single crystal of SeMet-labeled Sp-Atg44V42M. Each data set was independently indexed and integrated using the XDS program package.48
Structure determination and refinement
The structure of SpAtg44V42M was determined by the MAD phasing method using PHENIX.49 Three out of 4 selenium sites were found and approximately 90% of the residues were automatically modeled. Further modeling was manually performed with COOT50 and then refined using PHENIX. The structure of Sp-Atg44 wild-type was determined by the molecular replacement method in PHENIX using the structure of Sp-Atg44V42M as a search model. Further modeling was manually performed by COOT and refined by PHENIX. Coordinates and diffraction data were deposited in the Protein Data Bank (PDB) with accession code 7YDO. 3D structures were visualized using UCSF ChimeraX.51
Liposomes and lipid-nanorods preparation
Lipid nanorods were prepared as described previously46. Briefly, 100 nmol lipid film (galactosyl ceramide:POPC:POPE:PI:POPS:POPA:CL:biotinyl cap PE = 40:15:5:6:6:6:12:10) was hydrated in 100 μL of imaging buffer A (150 mM NaCl, 20 mM HEPES-NaOH, pH 7.0) and vortexed. The solution (1 mM lipid mixture) was sonicated in a bath sonicator for 10 min at 23°C and further sonicated with a tip sonicator for 5 s. For preparation of liposomes, 100 nmol lipid film (POPC:POPE:PI:POPS:POPA:CL = 5:5:2:2:2:4) was hydrated in 100 μL water and vortexed. The solution (1 mM lipid mixture) was stored at −20°C until use. After addition of 9 μL of imaging buffer B (100 mM NaCl, 50 mM HEPES-NaOH, pH 8.0, 20 mM MgCl2) to 1 μL of lipid mixture in a 0.5-mL tube, the solution was subjected to water bath sonication for 10 min at 23°C.
HS-AFM
The procedure for HS-AFM observation has already been described.52 HS-AFM images were acquired in tapping mode using a sample scanning HS-AFM instrument (MS-NEX, Research Institute of Biomolecule Metrology Co., Ltd.). We used cantilevers measuring ~7-μm long, ~2-μm wide and ~0.08-μm thick with electron beam deposited/EBD tips (tip radius < 10 nm) (USC-F1.2-k0.15, NanoWorld). Their resonant frequency and spring constant were 1.2 MHz in air and 0.15 N/m, respectively. Imaging conditions were as follows: scan size, 200 × 120 nm2 (Figures 5G and 5H), 240 × 144 nm2 (Figure 5I), 150 × 150 nm2 (Figures 7A and 7C), 800 × 800 nm2 (Figure 7B), or 240 × 240 nm2 (Figure 7D); pixel size, 200 × 120 pixels (Figures 5G and 5H), 150 × 90 pixels (Figure 5I), 120 × 120 pixels (Figures 7A, 7C and 7D), or 150 × 150 pixels (Figure 7B); imaging rate, 2 frames/s (Figures 5G and 5H), 3.13 frames/s (Figure 5I), 4 frames/s (Figures 7A, 7C and 7D), or 1 frame/s (Figure 7B). Imaging was performed at 23°C. HS-AFM images were viewed and analyzed using the softwares Kodec4.4.7.3953 and ImageJ.
Sample preparation for HS-AFM imaging
For imaging of lipid nanorods (Figure 5G), tamavidin 2 2D crystals covering the mica surface54,55 were used as a support for immobilization of the biotin-containing lipid nanorods. Tamavidin 2 solution (Wako) was diluted to 0.14 mg/ml in buffer C (400 mM NaCl, 20 mM HEPES-NaOH, pH 8.5, 200 mM MgCl2, 20% [w:v] PEG6000). After incubation for 20 min for 23°C, a drop (2 μl) of the diluted tamavidin 2 solution was deposited onto freshly cleaved mica attached to the top of a glass stage (diameter, 1.5 mm; height, 2 mm) and incubated for 5 min. After rinsing the mica surface with 0.01% glutaraldehyde diluted with buffer D (20 mM HEPES-NaOH, pH 8.5, 100 mM MgCl2), the last drop was kept on mica for 5 min and rinsed with imaging buffer A. After confirmation of tamavidin 2 2D crystals covering the mica surface with HS-AFM observation, a drop of 0.1 mM nanorods was deposited onto the tamavidin 2 2D crystals. After 5-min incubation, the mica was rinsed with imaging buffer A. For imaging of lipid nanorods with Sp-Atg44 (Figure 5H) or MBP-Sc-Atg44 (Figure 5I), a drop of 40 μM Sp-Atg44 or 30 μM MBP-Sc-Atg44 diluted with imaging buffer A was deposited onto lipid nanorods immobilized on mica-supported tamavidin 2 2D crystals. After 5-min incubation, the mica was immersed in the liquid cell containing ~90 μl of imaging buffer A without rinse.
For imaging of Sp-Atg44 on the mica surface (Figure 7A), a drop of Sp-Atg44 (0.5 μM) diluted with imaging buffer E (100 mM NaCl, 50 mM HEPES-NaOH, pH 8, 1 mM MgCl2) was deposited onto bare mica. After a 3-min incubation, the mica was rinsed with imaging buffer E. For imaging of lipid bilayers covering the mica surface (Figure 7B), a drop of 0.1 mM liposomes were deposited onto bare mica. After 2-min incubation, the mica surface was rinsed with imaging buffer B. For imaging of Sp-Atg44 bound to lipid bilayers (Figures 7C and 7D; Video S6), a drop of 0.5 μM Sp-Atg44 was deposited onto mica covered by the lipid bilayers. After 3-min incubation, the mica was immersed in the liquid cell containing imaging buffer B without rinse.
QUANTIFICATION AND STATISTICAL ANALYSIS
Autophagy assay
Quantification of the immunoblotting was performed with Image Lab software and GraphPad Prism 9. Protein band intensities were quantified by drawing a rectangle along the gel lane and obtaining the lane profile. The area of the peak in the profile was taken as a value of the band intensity. The intensity of free GFP or RFP was divided by the total GFP or RFP intensity, and the resulting values are presented relative to the control. The assay was independently performed at least three times. Representative blotting images are also shown.
Membrane fission assay
Quantification of the membrane fission assay was performed with Fiji software. The parts of fission were counted based on fluorescence. Tube radius was estimated from tube length, fluorescence intensity, and the correlation factor, which was calculated by plotting fluorescence intensities of the ROI against the ROI area. The assay was independently performed at least three times and representative results are shown.
Flotation assay
Quantification of the flotation assay was performed with Fiji software. Protein band intensities were quantified by drawing a rectangle along the gel lane and obtaining the lane profile. The area of the peak in the profile was taken as a value of the band intensity. The amount of floated Sp-Atg44 was estimated by calculating the ratio of the band intensity in the top fraction to that in the sum of the top and bottom fractions. The average value is shown with plots of three independent experiments. Representative gel images are also shown.
Supplementary Material
Video S1. Three-dimensional representation of S. cerevisiae WT cells upon mitophagy induction, related to Figure 2E
Three-dimensional representation of S. cerevisiae WT cells upon mitophagy induction, reconstructed from serial-sectioned images obtained using FIB-SEM.
Video S2. Three-dimensional representation of S. cerevisiae atg44Δ cells upon mitophagy induction, related to Figure 2E
Three-dimensional representation of S. cerevisiae atg44Δ cells upon mitophagy induction, reconstructed from serial-sectioned images obtained using FIB-SEM. Yellow: mitochondria; blue: vacuoles.
Video S3. Three-dimensional representation of whole S. cerevisiae WT cells grown in YPL, related to Figure 3C
Three-dimensional representation of whole S. cerevisiae WT cells grown in YPL, reconstructed from serial-sectioned images obtained using SBF-SEM. Brown: mitochondria; red: nucleus; blue: vacuoles.
Video S4. Three-dimensional representation of whole S. cerevisiae atg44Δ cells grown in YPL, related to Figure 3C
Three-dimensional representation of whole S. cerevisiae atg44Δ cells grown in YPL reconstructed from serial-sectioned images obtained using SBF-SEM. Brown: mitochondria; red: nucleus; blue: vacuoles.
Video S5. Time-lapse imaging of Sp-Atg44-mediated fission of lipid nanotubes, related to Figure 5B
Video S6. HS-AFM observation of lipid bilayer membrane containing Sp-Atg44, related to Figure 7D
Highlights.
Mitofissin is a mitochondrial fission factor that resides in the IMS
The yeast mitofissin Atg44 promotes mitochondrial fission essential for mitophagy
Atg44 directly introduces membrane fragility to facilitate membrane fission
Mechanisms and roles of membrane fission by Atg44 are different from that by Dnm1
ACKNOWLEDGMENTS
We thank Dr. Eri Hirata and Dr. Kuninori Suzuki (University of Tokyo) and the National Bio-Resource Project (NBRP, Japan) for providing plasmids. This work was supported in part by the Japan Society for the Promotion of Science KAKENHI grants 22H02615, 19H05712, 18H04858, 18H04691, 17H03671, and 15KK0253 (to T.K.), 21K05500 and 23H04255 (to K.F.), 20K06552 (to T.F.), 19H05707 (to N.N.N.), 16H06280 (to K.M.), 19H05794, 22H04926 (to Y. Okada); AMED grants JP21gm6110013 (to T.K.), JP22bm0704046 (to S.S.); JST grants JPMJCR20E3 (to N.N.N.), JPMJMS2025–14, JPMJCR20E2 (to Y. Okada), JPMJCR20E5 (to K. Ikeda); the Takeda Science Foundation (to T.F., K.F., and N.N.N.); the Noda Institute of Scientific Research (to K.F.); NIH grant GM131919 (to D.J.K.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Nakatogawa H (2020). Mechanisms governing autophagosome biogenesis. Nat. Rev. Mol. Cell Biol. 21, 439–458. 10.1038/s41580-020-0241-0. [DOI] [PubMed] [Google Scholar]
- 2.Pickles S, Vigie P, and Youle RJ (2018). Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance. Curr. Biol. 28, R170–R185. 10.1016/j.cub.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanki T, Wang K, Cao Y, Baba M, and Klionsky DJ (2009). Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell 17, 98–109. 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okamoto K, Kondo-Okamoto N, and Ohsumi Y (2009). Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 17, 87–97. 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 5.Kanki T, Kurihara Y, Jin X, Goda T, Ono Y, Aihara M, Hirota Y, Saigusa T, Aoki Y, Uchiumi T, and Kang D (2013). Casein kinase 2 is essential for mitophagy. EMBO Rep. 14, 788–794. 10.1038/embor.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aoki Y, Kanki T, Hirota Y, Kurihara Y, Saigusa T, Uchiumi T, and Kang D (2011). Phosphorylation of Serine 114 on Atg32 mediates mitophagy. Mol. Biol. Cell 22, 3206–3217. 10.1091/mbc.E11-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furukawa K, Fukuda T, Yamashita SI, Saigusa T, Kurihara Y, Yoshida Y, Kirisako H, Nakatogawa H, and Kanki T (2018). The PP2A-like Protein Phosphatase Ppg1 and the Far Complex Cooperatively Counteract CK2-Mediated Phosphorylation of Atg32 to Inhibit Mitophagy. Cell Rep. 23, 3579–3590. 10.1016/j.celrep.2018.05.064. [DOI] [PubMed] [Google Scholar]
- 8.Yorimitsu T, and Klionsky DJ (2005). Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol. Biol. Cell 16, 1593–1605. 10.1091/mbc.e04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashrafi G, and Schwarz TL (2013). The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 20, 31–42. 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liesa M, and Shirihai OS (2013). Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 17, 491–506. 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sebastian D, Palacin M, and Zorzano A (2017). Mitochondrial Dynamics: Coupling Mitochondrial Fitness with Healthy Aging. Trends Mol. Med. 23, 201–215. 10.1016/j.molmed.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Shirihai OS, Song M, and Dorn GW 2nd (2015). How mitochondrial dynamism orchestrates mitophagy. Circ. Res. 116, 1835–1849. 10.1161/CIRCRESAHA.116.306374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wai T, and Langer T (2016). Mitochondrial Dynamics and Metabolic Regulation. Trends Endocrinol. Metab. 27, 105–117. 10.1016/j.tem.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Burman JL, Pickles S, Wang C, Sekine S, Vargas JNS, Zhang Z, Youle AM, Nezich CL, Wu X, Hammer JA, and Youle RJ (2017). Mitochondrial fission facilitates the selective mitophagy of protein aggregates. J. Cell Biol. 216, 3231–3247. 10.1083/jcb.201612106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendl N, Occhipinti A, Muller M, Wild P, Dikic I, and Reichert AS (2011). Mitophagy in yeast is independent of mitochondrial fission and requires the stress response gene WHI2. J. Cell Sci. 124, 1339–1350. 10.1242/jcs.076406. [DOI] [PubMed] [Google Scholar]
- 16.Yamashita SI, Jin X, Furukawa K, Hamasaki M, Nezu A, Otera H, Saigusa T, Yoshimori T, Sakai Y, Mihara K, and Kanki T (2016). Mitochondrial division occurs concurrently with autophagosome formation but independently of Drp1 during mitophagy. J. Cell Biol. 215, 649–665. 10.1083/jcb.201605093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klionsky DJ, Cregg JM, Dunn WA Jr., Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, and Ohsumi Y (2003). A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5, 539–545. 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 18.Fukuda T, Ebi Y, Saigusa T, Furukawa K, Yamashita SI, Inoue K, Kobayashi D, Yoshida Y, and Kanki T (2020). Atg43 tethers isolation membranes to mitochondria to promote starvation-induced mitophagy in fission yeast. eLife 9. 10.7554/eLife.61245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgenstern M, Stiller SB, Lubbert P, Peikert CD, Dannenmaier S, Drepper F, Weill U, Hoss P, Feuerstein R, Gebert M, et al. (2017). Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale. Cell Rep. 19, 2836–2852. 10.1016/j.celrep.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, and Ohsumi Y (1999). Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147, 435–446. 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polianskyte Z, Peitsaro N, Dapkunas A, Liobikas J, Soliymani R, Lalowski M, Speer O, Seitsonen J, Butcher S, Cereghetti GM, et al. (2009). LACTB is a filament-forming protein localized in mitochondria. Proc. Natl. Acad. Sci. U S A 106, 18960–18965. 10.1073/pnas.0906734106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otera H, Miyata N, Kuge O, and Mihara K (2016). Drp1-dependent mitochondrial fission via MiD49/51 is essential for apoptotic cristae remodeling. J. Cell Biol. 212, 531–544. 10.1083/jcb.201508099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sesaki H, Southard SM, Yaffe MP, and Jensen RE (2003). Mgm1p, a dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane. Mol. Biol. Cell 14, 2342–2356. 10.1091/mbc.e02-12-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Espadas J, Pendin D, Bocanegra R, Escalada A, Misticoni G, Trevisan T, Velasco Del Olmo A, Montagna A, Bova S, Ibarra B, et al. (2019). Dynamic constriction and fission of endoplasmic reticulum membranes by reticulon. Nat. Commun 10, 5327. 10.1038/s41467-019-13327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molecular AL, Todd SC, and Pogozheva ID (2022). Spatial arrangement of proteins in planar and curved membranes by PPM 3.0. Protein Sci. 31, 209–220. 10.1002/pro.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalia R, Wang RY, Yusuf A, Thomas PV, Agard DA, Shaw JM, and Frost A (2018). Structural basis of mitochondrial receptor binding and constriction by DRP1. Nature 558, 401–405. 10.1038/s41586-018-0211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mears JA, Lackner LL, Fang S, Ingerman E, Nunnari J, and Hinshaw JE (2011). Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat. Struct. Mol. Biol. 18, 20–26. 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McMahon HT, and Gallop JL (2005). Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438, 590–596. 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 29.Snead WT, Hayden CC, Gadok AK, Zhao C, Lafer EM, Rangamani P, and Stachowiak JC (2017). Membrane fission by protein crowding. Proc. Natl. Acad. Sci. U S A 114, E3258–E3267. 10.1073/pnas.1616199114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labbe S, and Thiele DJ (1999). Copper ion inducible and repressible promoter systems in yeast. Methods Enzymol. 306, 145–153. 10.1016/s0076-6879(99)06010-3. [DOI] [PubMed] [Google Scholar]
- 31.Sikorski RS, and Hieter P (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanki T, Wang K, Baba M, Bartholomew CR, Lynch-Day MA, Du Z, Geng J, Mao K, Yang Z, Yen WL, and Klionsky DJ (2009). A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol. Biol. Cell 20, 4730–4738. 10.1091/mbc.e09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, and Mihara K (2010). Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 191, 1141–1158. 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J, Scott SV, Oda MN, and Klionsky DJ (1997). Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 137, 609–618. 10.1083/jcb.137.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanki T, and Klionsky DJ (2008). Mitophagy in yeast occurs through a selective mechanism. J. Biol. Chem. 283, 32386–32393. 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noda T, Matsuura A, Wada Y, and Ohsumi Y (1995). Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 210, 126–132. 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- 37.Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA Jr., and Klionsky DJ (2001). Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 153, 381–396. 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reggiori F, Monastyrska I, Shintani T, and Klionsky DJ (2005). The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 16, 5843–5856. 10.1091/mbc.e05-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mochida K, Oikawa Y, Kimura Y, Kirisako H, Hirano H, Ohsumi Y, and Nakatogawa H (2015). Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 522, 359–362. 10.1038/nature14506. [DOI] [PubMed] [Google Scholar]
- 40.Osumi M, Shimoda C, and Yanagishima N (1974). Mating reaction in Saccharomyces cerevisiae. V. Changes in the fine structure during the mating reaction. Arch. Mikrobiol. 97, 27–38. [PubMed] [Google Scholar]
- 41.Miyazaki N, Esaki M, Ogura T, and Murata K (2014). Serial block-face scanning electron microscopy for three-dimensional analysis of morphological changes in mitochondria regulated by Cdc48p/p97 ATPase. J. Struct. Biol. 187, 187–193. 10.1016/j.jsb.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 42.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shibata S, Iseda T, Mitsuhashi T, Oka A, Shindo T, Moritoki N, Nagai T, Otsubo S, Inoue T, Sasaki E, et al. (2019). Large-Area Fluorescence and Electron Microscopic Correlative Imaging With Multibeam Scanning Electron Microscopy. Front. Neural. Circuits. 13, 29. 10.3389/fncir.2019.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamazaki M (2008). Chapter 5 The Single Guv Method to Reveal Elementary Processes of Leakage of Internal Contents from Liposomes Induced by Antimicrobial Substances. Adv. Planar Lipid Bilayers Liposomes 7, 121–142. 10.1016/s1554-4516(08)00005-7. [DOI] [Google Scholar]
- 45.Pucadyil TJ, and Schmid SL (2010). Supported bilayers with excess membrane reservoir: a template for reconstituting membrane budding and fission. Biophys. J. 99, 517–525. 10.1016/j.bpj.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colom A, Redondo-Morata L, Chiaruttini N, Roux A, and Scheuring S (2017). Dynamic remodeling of the dynamin helix during membrane constriction. Proc. Natl. Acad. Sci. U S A 114, 5449–5454. 10.1073/pnas.1619578114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51. 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 48.Kabsch W (2010). Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, and Ferrin TE (2021). UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82. 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uchihashi T, Kodera N, and Ando T (2012). Guide to video recording of structure dynamics and dynamic processes of proteins by high-speed atomic force microscopy. Nat. Protoc. 7, 1193–1206. 10.1038/nprot.2012.047. [DOI] [PubMed] [Google Scholar]
- 53.Ngo KX, Kodera N, Katayama E, Ando T, and Uyeda TQ (2015). Cofilin-induced unidirectional cooperative conformational changes in actin filaments revealed by high-speed atomic force microscopy. eLife 4. 10.7554/eLife.04806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noshiro D, and Ando T (2018). Substrate protein dependence of GroEL-GroES interaction cycle revealed by high-speed atomic force microscopy imaging. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373. 10.1098/rstb.2017.0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kawasaki Y, Ariyama H, Motomura H, Fujinami D, Noshiro D, Ando T, and Kohda D (2020). Two-State Exchange Dynamics in Membrane-Embedded Oligosaccharyltransferase Observed in Real-Time by High-Speed AFM. J. Mol. Biol. 432, 5951–5965. 10.1016/j.jmb.2020.09.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Three-dimensional representation of S. cerevisiae WT cells upon mitophagy induction, related to Figure 2E
Three-dimensional representation of S. cerevisiae WT cells upon mitophagy induction, reconstructed from serial-sectioned images obtained using FIB-SEM.
Video S2. Three-dimensional representation of S. cerevisiae atg44Δ cells upon mitophagy induction, related to Figure 2E
Three-dimensional representation of S. cerevisiae atg44Δ cells upon mitophagy induction, reconstructed from serial-sectioned images obtained using FIB-SEM. Yellow: mitochondria; blue: vacuoles.
Video S3. Three-dimensional representation of whole S. cerevisiae WT cells grown in YPL, related to Figure 3C
Three-dimensional representation of whole S. cerevisiae WT cells grown in YPL, reconstructed from serial-sectioned images obtained using SBF-SEM. Brown: mitochondria; red: nucleus; blue: vacuoles.
Video S4. Three-dimensional representation of whole S. cerevisiae atg44Δ cells grown in YPL, related to Figure 3C
Three-dimensional representation of whole S. cerevisiae atg44Δ cells grown in YPL reconstructed from serial-sectioned images obtained using SBF-SEM. Brown: mitochondria; red: nucleus; blue: vacuoles.
Video S5. Time-lapse imaging of Sp-Atg44-mediated fission of lipid nanotubes, related to Figure 5B
Video S6. HS-AFM observation of lipid bilayer membrane containing Sp-Atg44, related to Figure 7D
Data Availability Statement
Coordinates and diffraction data of Sp-Atg44 as well as unprocessed images (microscopy, gels, and blots) were deposited in the Protein Data Bank (PDB) and Mendeley data respectively and are publicly available as of the date of publication. Accession numbers and DOI are listed in the key resources table.
This paper does not report any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Sc-Atg44 | This study | N/A |
| Guinea pig polyclonal anti-Sp-Atg44 | This study | N/A |
| Rabbit polyclonal anti-Atg33 | This study | N/A |
| Rabbit polyclonal anti-RFP | MBL | Cat# PM005; RRID: AB_591279 |
| Mouse monoclonal anti-GFP (JL-8) | Takara Bio | Cat# 632380; RRID: AB_10013427 |
| Mouse monoclonal anti-FLAG | Sigma-Aldrich | Cat# F1804; RRID: AB_262044 |
| Rabbit polyclonal anti-DDDDK | MBL | Cat# PM020; RRID: AB_591224 |
| Rabbit monoclonal anti-DYKDDDDK tag | Cell Signaling Technology | Cat# 14793; RRID: AB_2572291 |
| Rabbit polyclonal anti-ATPB | Abcam | Cat# ab128743; RRID: AB_2810299 |
| Rabbit polyclonal anti-Histone H3 | Abcam | Cat# ab1791; RRID: AB_302613 |
| Mouse monoclonal anti-HSPD1/HSP60 | Proteintech | Cat# 66041-1-Ig; RRID: AB_11041709 |
| Mouse monoclonal anti-Tim23 | BD Biosciences | Cat# 611222; RRID: AB_398754 |
| Rabbit polyclonal anti-Tom20 | Proteintech | Cat# 21090-1-AP; RRID: AB_10734583 |
| Rabbit polyclonal anti-CHCHD4 | Santa Cruz Biotechnology | Cat# sc-11415; RRID: AB_2207533 |
| AF594 F(ab’)2-goat anti-rabbit IgG (H+L) | Thermo Fisher Scientific | Cat# A-11072; RRID: AB_2534116 |
| AF594 F(ab’)2-goat anti-mouse IgG (H+L) | Thermo Fisher Scientific | Cat# A-11020; RRID: AB_141974 |
| AF647 goat anti-rabbit IgG (H+L) | Thermo Fisher Scientific | Cat# A-21244; RRID: AB_2535812 |
| Mouse monoclonal anti-HA | Sigma-Aldrich | Cat# H9658; RRID: AB_260092 |
| Mouse monoclonal anti-Pgk1 | Thermo Fisher Scientific | Cat# 459250; RRID: AB_2532235 |
| Chicken polyclonal anti-protein A | GeneTex | Cat# GTX77595; RRID: AB_379859 |
| Mouse monoclonal anti-Cox2 | MitoScience | Cat# MS419; RRID: AB_1618187 |
| Rabbit polyclonal anti-Ape1 | Kim et al. (1997)34 | N/A |
| Rabbit polyclonal anti-Atg32 | Aoki et al. (2011)6 | N/A |
| AF488 FluoroNanogold conjugated goat anti-rabbit IgG | Nanoprobes | Cat# 7204; RRID: AB_2868458 |
| Goat anti-mouse IgG-HRP | Merck Millipore | Cat# AP124P; RRID: AB_90456 |
| Goat anti-rabbit IgG-HRP | Jackson ImmunoResearch | Cat# 111-035-003; RRID: AB_2313567 |
| Bacterial and virus strains | ||
| Escherichia coli C41 (DE3) | Sigma-Aldrich | Cat# CMC0017 |
| Escherichia coli BL21(DE3) | Sigma-Aldrich | Cat# CMC0014 |
| Chemicals, peptides, and recombinant proteins | ||
| Zymolyase 100T | Nacalai tesque | Cat# 07665-55 |
| Lysing Enzymes from Trichoderma harzianum | Sigma-Aldrich | Cat# L1412 |
| Proteinase K, recombinant, PCR Grade | Roche | Cat# 03115836001 |
| MitoTracker Red CMXRos | Thermo Fisher Scientific | Cat# M7512 |
| FM 4-64 | Molecular Probes | Cat# T3166 |
| Block Ace | DS Pharma Biomedical | Cat# UKB80 |
| Amylose resin | New England Biolabs | Cat# E8021 |
| Ni-NTA Superflow | QIAGEN | Cat# 30410 |
| COSMOGEL GST-Accept | Nacalai tesque | Cat# 09277-56 |
| Alexa Fluor 647 C2 Maleimide | Invitrogen | Cat# A20347 |
| Alexa Fluor 647 hydrazide | Invitrogen | Cat# A20502 |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) | Avanti Polar Lipids | Cat# 850457 |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) | Avanti Polar Lipids | Cat# 850757 |
| L-α-phosphatidylinositol (PI) | Avanti Polar Lipids | Cat# 840042 |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS) | Avanti Polar Lipids | Cat# 840034 |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate (POPA) | Avanti Polar Lipids | Cat# 840857 |
| 1’,3’-bis[1,2-dioleoyl-sn-glycero-3-phospho]-glycerol (CL) | Avanti Polar Lipids | Cat# 710335 |
| 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (liss Rhod PE) | Avanti Polar Lipids | Cat# 810150 |
| D-galactosyl-β-1,1’ N-nervonoyl-D-erythro-sphingosine (GC) | Avanti Polar Lipids | Cat# 860546 |
| Silica beads (50 μm) | Nanocs | Cat# Si01-50u-1 |
| BSA tracer | Aurion | Cat# 210.033 |
| Tamavidin 2 solution | FUJIFILM Wako | Cat# 201-21271 |
| Critical commercial assays | ||
| NEBuilder HiFi DNA Assembly Master Mix | New England Biolabs | Cat# E2621 |
| PrimeSTAR Mutagenesis Basal Kit | Takara Bio | Cat# R046A |
| IgG Sepharose 6 Fast Flow | GE Healthcare | Cat# 17096901 |
| EzWestLumi One | ATTO | Cat# WSE-7110 |
| EzWestLumi plus | ATTO | Cat# WSE-7120 |
| Clarity Max Western ECL Substrate | Bio-Rad | Cat# 1705062 |
| FuGENE HD Transfection Reagent | Promega | Cat# E2311 |
| Oriole Fluorescent Gel Stain | Bio-Rad | Cat# 1610496 |
| Deposited data | ||
| Crystal structure of Sp-Atg44 | This study | PDB: 7YDO |
| Unprocessed images (microscopy, gels, and blots) | This study | Mendeley data: http://dx.doi.org/10.17632/92p3bx6nnz.1 |
| Experimental models: Cell lines | ||
| Human: HeLa (WT) | Yamashita et al. (2016)16 | N/A |
| Human: HeLa (DNM1L KO) | Yamashita et al. (2016)16 | N/A |
| Experimental models: Organisms/strains | ||
| S. cerevisiae strains, see Table S1 | This study | N/A |
| S. pombe strains, see Table S2 | This study | N/A |
| Software and algorithms | ||
| Image Lab | Bio-Rad | https://www.bio-rad.com/en-jp/product/image-lab-software |
| MetaMorph 7 | Molecular Devices | https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy |
| Auto Slice & View (version 5.6.0) | Thermo Fisher Scientific | https://www.thermofisher.com/jp/ja/home/electron-microscopy/products/software-em-3d-vis/auto-slice-view-4-software.html |
| Fiji | Schindelin et al. (2012)42 | http://fiji.sc/Fiji |
| Image J | National Institutes of Health | https://imagej.nih.gov/ij |
| Amira (version 5.4.5) | Thermo Fisher Scientific | https://thermofisher.com/amira-avizo |
| Zen 2.1 SP1 | Carl Zeiss | https://www.zeiss.com/microscopy/en/products/software/zeiss-zen.html |
| Huygens Professional | Scientific Volume Imaging | https://svi.nl/Huygens-Professional |
| Astra | Wyatt Technology | https://www.wyatt.com/products/software/astra.html |
| DigitalMicrograph | Gatan | https://www.gatan.com/resources/digitalmicrograph-scripts |
| XDS Program Package | Kabsch (2010)48 | https://xds.mr.mpg.de/ |
| PHENIX | Adams et al. (2010)49 | https://phenix-online.org/ |
| COOT | Emsley et al. (2010)50 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| UCSF ChimeraX (version 1.3) | Pettersen et al. (2021)51 | https://www.rbvi.ucsf.edu/chimerax |
| PPM 3.0 | Lomize et al. (2022)25 | https://opm.phar.umich.edu/ppm_server3_cgopm |
| Kodec (version 4.4.7.39) | Ngo et al. (2015)53 | https://doi.org/10.7554/eLife.04806.031 |
| Prism 9 (version 9.5.1) | GraphPad Software | https://www.graphpad.com/ |


