Abstract
Knowledge of the kinds and numbers of nuclear point mutations in human tissues is essential to the understanding of the mutation mechanisms underlying genetic diseases. However, nuclear point mutant fractions in normal humans are so low that few methods exist to measure them. We have now developed a means to scan for point mutations in 100 bp nuclear single copy sequences at mutant fractions as low as 10–6. Beginning with about 108 human cells we first enrich for the desired nuclear sequence 10 000-fold from the genomic DNA by sequence-specific hybridization coupled with a biotin–streptavidin capture system. We next enrich for rare mutant sequences 100-fold against the wild-type sequence by wide bore constant denaturant capillary electrophoresis (CDCE). The mutant-enriched sample is subsequently amplified by high fidelity PCR using fluorescein-labeled primers. Amplified mutant sequences are further enriched via two rounds of CDCE coupled with high fidelity PCR. Individual mutants, seen as distinct peaks on CDCE, are then isolated and sequenced. We have tested this approach by measuring N-methyl-N ′-nitro-N-nitrosoguanidine (MNNG)-induced point mutations in a 121 bp sequence of the adenomatous polyposis coli gene (APC) in human lymphoblastoid MT1 cells. Twelve different MNNG-induced GC→AT transitions were reproducibly observed in MNNG-treated cells at mutant fractions between 2 × 10–6 and 9 × 10–6. The sensitivity of this approach was limited by the fidelity of Pfu DNA polymerase, which created 14 different GC→TA transversions at a mutant fraction equivalent to ~10–6 in the original samples. The approach described herein should be general for all DNA sequences suitable for CDCE analysis. Its sensitivity and capacity would permit detection of stem cell mutations in tissue sectors consisting of ~108 cells.
INTRODUCTION
Most human inherited diseases and cancers are known to be caused by mutations in nuclear genes. But the primary causes of these mutations are still unknown. Determination of mutational spectra in disease-related genes in non-tumorous tissues may provide direct evidence as to whether a specific mutagenic agent or pathway is involved in a particular human cancer. However, the methodologies used in all previous studies of mutational spectra were based on phenotypic selection, in which rare mutant cells were recognized and isolated on the basis of an altered phenotype conferring the ability to grow under selective conditions (1–4). Such methods preclude analysis of genes for which selective conditions cannot be devised in single cell systems. They also preclude analysis of any gene in human tissues the cells of which cannot yet be grown in vitro.
To overcome these limitations to mutational spectrometry, we have been developing means to ‘select’ rare mutants based on differences in the cooperative melting behavior between wild-type and mutant DNA sequences (5,6). These efforts are built upon the original demonstration of Fischer and Lerman that small differences in DNA melting temperatures created clear and discernible differences in the electrophoretic mobility of DNA upon moving through a gel matrix under partially denaturing conditions (7). We have extended their approach to constant denaturant capillary electrophoresis (CDCE) which, in combination with high fidelity PCR, has enabled us to scan for point mutations in a 100 bp mitochondrial DNA sequence in human cells and tissues (8,9). Reproducible mitochondrial point mutational hot-spots were discovered at frequencies of from 10–5 to greater than 10–4 (9–11). Remarkably, the same set of mitochondrial hot-spots was found in all of the samples analyzed, indicating that human mitochondrial point mutations are spontaneous in origin (9,10). Our next goal is to extend our study to the nuclear genome, where most of the disease-related genes reside.
The criterion for an assay of sufficient sensitivity to detect nuclear mutations differs among tissues. Studies in peripheral T lymphocytes have shown that the average mutant fraction in the nuclear HPRT and HLA loci is ~10–8 per bp in middle-aged humans (12,13). However, mutations are not randomly distributed over DNA sequences but occur as ‘hot-spots’ which have mutant fractions 10 or 100 times higher than the average for all base pairs (1). One would thus expect these nuclear hot-spots to occur at mutant fractions of 10–7 to somewhat greater than 10–6. Tissue organization must also be considered in developing our approach to mutation measurements in human tissues. Cell replacement in adult mammalian tissues is believed to take place via discrete units of proliferation termed ‘turnover units’ (14). A turnover unit is defined by one stem cell and its descendant transition and terminal cells. Mutations that occur in a stem cell would be both maintained in the stem cell and subsequently transmitted to the descendant cells, thus giving rise to a mutant colony with the size of a turnover unit. We have estimated the turnover unit size to be about 128 cells in rat mammary glands and about 256 cells in human lung epithelium (unpublished data). We expect that different tissues will be found to have turnover units of different sizes. For mutational analysis anatomically distinct sectors can be excised from most tissues and analyzed in series (10). If the turnover unit consists of 100 or more cells and each tissue sector is dissected to contain about 0.5 × 108 cells (or 108 gene copies), the expected mutant fraction would be 10–6. Thus a mutation assay with a sensitivity of 10–6 would permit the detection of mutations that have occurred in stem cells.
However, the application of our previous CDCE/high fidelity PCR approach to nuclear genes was not straightforward. Both the cellular copy numbers and the point mutant fractions of nuclear genes are several hundred-fold lower than those of the mitochondrial DNA (9,12,13,15). In practical terms, this means working with a few micrograms of genomic DNA to reproducibly detect mitochondrial mutational hot-spots at fractions of 10–5–10–4 (8–11), but several milligrams of genomic DNA to reproducibly detect nuclear hot-spots at fractions of 10–6. It was clear that the mutational analysis of nuclear genes required significant technical improvements in both the sample handling capacity and the detection sensitivity of our previous approach.
We have now made these improvements by employing new strategies to first isolate the desired nuclear sequence from genomic DNA and then to achieve a higher enrichment of mutants against the wild-type sequences prior to PCR. As an initial sequence for the developmental work, we chose the cDNA bp 8434–8704 region of the human adenomatous polyposis coli gene (APC) (16). The chosen 271 bp fragment had the necessary contiguous high and low isomelting domains for separation of mutants from wild type (7) as well as conveniently located restriction sites. The target sequence in which mutations can be detected comprises 121 bp (APC cDNA bp 8543–8663) within the low melting domain. We demonstrate herein the sensitivity and reproducibility of our improved approach in measuring N-methyl-N′-nitro-N-nitrosoguanidine (MNNG)-induced point mutations in the near diploid human MT1 cells.
MATERIALS AND METHODS
Human cell line and MNNG treatment
Human lymphoblastoid MT1 cells are resistant to the toxicity but not the mutagenicity of MNNG, putatively because they are knockout mutants for the hMSH6 gene for mismatch repair (17–19). An exponentially growing culture consisting of 8 × 107 MT1 cells was treated with 4 µM MNNG (Sigma Chemicals, St Louis, MO) for 45 min. The procedures for cell culturing and treatment were as previously described (17,20). After treatment the cells were grown for 30 generations with daily dilution to dilute any unrepaired premutagenic lesions which could be mistaken for mutations by our procedures. Untreated cells grown in parallel cultures served as controls. Using a microtiter plate mutation assay and selecting for 6-thioguanine-resistant (6TGR) colonies (21), the MNNG-induced mutant fraction in the HPRT gene was measured to be 8 × 10–3 in this study.
Preparation of cellular DNA enriched for the desired APC gene fragment
Genomic DNA was isolated from over 3 × 108 MNNG-treated and untreated MT1 cells without exposing the DNA to either phenol or anion exchange resins (8). The genomic DNA was digested with two ‘inexpensive’ endonucleases, HaeIII and XbaI (New England Biolabs, Beverly, MA), at 1 U enzyme/µg DNA and 2–3 mg DNA/ml overnight to liberate the target sequence embedded in a 482 bp fragment representing the APC cDNA bp 8422–8903.
The initial copy numbers of the APC target sequence and a 205 bp human mitochondrial DNA sequence (mitochondrial bp 10011–10215) (used as a reference sequence) in the restriction digestion were measured based on competitive PCR followed by CDCE separation (8). In brief, a small aliquot of the sample was mixed with known copies of an artificial mutant and subjected to PCR amplification (see below). The PCR products were separated by CDCE (see below). The areas under the wild-type, mutant and heteroduplex peaks were measured. The ratio of the amount of the wild-type sequence to the artificial mutant in the PCR products was used to calculate the initial copy number of the target sequence. The APC artificial mutant used represented the APC cDNA bp 8422–8913 containing an AT→GC transition at bp 8652. The mitochondrial artificial mutant represented the mitochondrial bp 10011–10215 with a TA→CG at bp 10072. Based on the target copy number measured in the digestion sample, the APC artificial mutant was added to the sample at a precise mutant fraction to serve as an internal standard.
To enrich for the desired APC gene fragment, two 5′-biotinylated 30mer probes, BP1 (5′-CAA AAC TGA CAG CAC AGA ATC CAG TGG AAC-3′; APC cDNA bp 8472–8501) and BP2 (5′-AAG ACC CAG AAT GGC GCT TAG GAC TTT GGG-3′; complementary to APC cDNA bp 8501–8530) (Synthetic Genetics, San Diego, CA), were added to the genomic DNA digestion at a probe/target molar ratio of 5 × 104 each. The positions of the two probes in the APC restriction fragment are shown in Figure 1A. The probes were PAGE purified and the biotin moiety was linked to each probe by an 18 carbon spacer arm. The sample was boiled for 2 min and immediately chilled in an ice bath for 10 min. The hybridization was then performed at 58°C for 2 h in 6× SSPE (1.08 M NaCl, 60 mM sodium phosphate, pH 7.4, 6 mM EDTA). The probe–target hybrids were captured by mixing with streptavidin-coated controlled pore glass paramagnetic beads (CPG, Lincoln Park, NJ) at 0.4 mg beads/108 target copies at 50°C for 1 h. The beads were washed four times with washing buffer (1 M NaCl, 10 mM Tris–HCl pH 7.6, 2 mM EDTA) at 10 mg beads/ml at 50°C for 5 min. The desired probe-bound DNA was recovered by twice washing the beads with deionized H2O at 20 mg beads/ml at 70°C for 2 min. The copy numbers of the APC target sequence and the mitochondrial DNA sequence in the elution were measured to estimate the yield and enrichment of the target sequence.
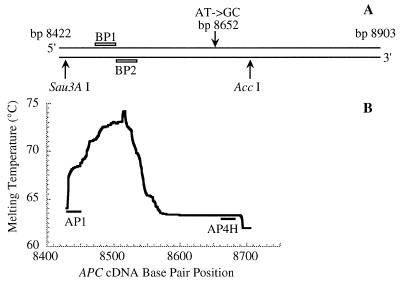
Figure 1.
(A) The 482 bp APC gene fragment (APC cDNA bp 8422–8903) released by digestion of human genomic DNA with endonucleases HaeIII and XbaI. The positions of the two biotinylated 30mer probes (BP1 and BP2) for enriching this fragment from genomic DNA are indicated by open bars. The positions of the AT→GC transition at APC cDNA bp 8652 carried by the internal standard and the recognition sites of endonucleases Sau3AI and AccI used to liberate the 271 bp APC gene fragment (APC cDNA bp 8434–8704) are indicated by arrows. (B) The melting profile of the 271 bp APC gene fragment was calculated according to the melting algorithm of Lerman and Silverstein (23). The positions of the primers (AP1 and AP4H) used in high fidelity PCR are indicated by filled bars.
The elution was concentrated to 10 µl by Speed-Vac centrifugation and re-annealed at 55°C for 16 h in 0.2 M NaCl, 10 mM Tris–HCl, pH 7.6, 2 mM EDTA buffer. This re-annealing converted all rare mutant sequences into mutant/wild-type heteroduplexes in the presence of an excess of wild-type sequences, which was necessary for efficient separation of mutants from wild-type DNA based on differences in the melting temperature (22). The re-annealed DNA was digested with two ‘expensive’ endonucleases, Sau3AI and AccI (New England Biolabs), to excise the desired 271 bp APC fragment (APC cDNA bp 8434–8704) suitable for CDCE analysis (Fig. 1B). The sample was then desalted and concentrated through ultrafiltration using a Microcon-50 device (Amicon, Beverley, MA).
Constant denaturant capillary electrophoresis
The CDCE instrument using a laser-induced fluorescence detection system was applied as previously described (6,8,24). The inner surfaces of fused silica capillaries (Polymicro Technologies, Phoenix, AZ) were coated with 6% linear polyacrylamide. Electrophoresis was performed in a piece of coated capillary (21–33 cm) filled with a 5% linear polyacrylamide matrix in 1× TBE (89 mM Tris–borate, 1 mM EDTA, pH 8.3). The matrix was replaced before each run. A portion of the capillary near its inlet was heated with a temperature-controlled water jacket (6–10 cm) where DNA sequences with even a single base pair substitution could be separated based on differences in the melting temperature. The detection window on the capillary was positioned 7 cm from its outlet (the anode end).
The target-enriched cellular DNA sample was separated by CDCE using a 19.5 cm long, 540 µm i.d. capillary and a 6 cm water jacket at a temperature of 66.0°C. CDCE-purified wild-type APC fragments were also run on CDCE to serve as a negative control. The DNA was electrokinetically loaded onto the column through tightly mounted Teflon tubing (24). Electrophoresis was performed at a constant current of 80 µA. The desired mutant/wild-type heteroduplex fraction was electroeluted from the capillary outlet into 10 µl collecting buffer consisting of 0.8× TBEB (71 mM Tris–borate, pH 8.3, 0.8 mM EDTA, 0.2 mg/ml BSA). The collected heteroduplex fraction from the cellular DNA sample was desalted by drop dialysis against 0.1× TBE (25) and subjected to CE separation at room temperature. The fraction containing the 271 bp APC restriction fragment was electroeluted. The efficiency of mutant enrichment was determined by measuring the copy number of the target sequence loaded onto the capillary and that eluted in the mutant-enriched fraction.
PCR products were separated by CDCE using a 75 µm i.d. capillary. About 108 copies of the target sequence were electroinjected. To separate the heteroduplexes from wild-type homoduplex, CDCE was performed in a 21 cm capillary at a constant current of 9 µA. A 6 cm water jacket at a temperature of 64.6°C was used. The heteroduplex fraction was electroeluted into 10 µl of 0.1× TBEB buffer. To separate mutant homoduplexes, CDCE was run in a 33 cm capillary using a 19 cm water jacket. The initial mutant fraction of each mutant was determined by comparison of the area under the mutant peak with that of the internal standard. Mutant peaks were individually purified (through CDCE collection followed by PCR) and sequenced.
High fidelity PCR
High fidelity PCR was performed in 10–50 µl capillary tubes using native Pyrococcus furiosus (Pfu) DNA polymerase with an associated 3′→5′ exonuclease activity (26) (Stratagene, La Jolla, CA) and an Air Thermo-Cycler™ (Idaho Technology, Idaho Falls, ID). The PCR mixture contained 20 mM Tris–HCl, pH 8.0, 2 mM MgCl2, 10 mM KCl, 6 mM (NH4)2SO4, 0.1% Triton X-100, 0.1 mg/ml BSA, 0.1 mM dNTPs, 0.2 µM each primer and 0.1 U/µl Pfu.
The primer sequences to amplify the APC gene target sequence were 5′-fluorescein-labeled AP1 (5′-GAA TAA CAA CAC AAA GAA GC-3′; APC cDNA bp 8441–8460) and AP4H (5′-AAC AAA AAC CCT CTA ACA AG-3′; complementary to APC cDNA bp 8664–8683) (Synthetic Genetics). The reaction conditions were as follows: 94°C for 2 min; an appropriate number of cycles of 10 s at 94°C, 20 s at 50°C and 20 s at 72°C; 72°C for 2 min. The PCR products were then incubated with additional Pfu (0.05 U/µl) at 72°C for 5 min (to remove interfering by-products). To create mutant/wild-type heteroduplexes, a sufficient number of PCR cycles (based on a PCR efficiency of 0.5–0.6) was used to deplete the primers such that the mutants would re-anneal with excess wild-type sequence during the final cycles. To create mutant homoduplexes for the final CDCE display, aliquots of the PCR products were subjected to 3–6 cycles of PCR such that an excess of primers was still present in the final reaction.
The PCR conditions for amplification of the 205 bp human mitochondrial DNA sequence (mitochondrial bp 10011–10215) were as previously described (8). The primer sequences were CW7 (5′-ACC GTT AAC TTC CAA TTA AC-3′; mitochondrial bp 10011–10031) and 5′-fluorescein-labeled J3 (5′-GCG GGC GCA GGG AAA GAG GT-3′; complementary to mitochondrial bp 10196–10215).
RESULTS
Enrichment of the desired APC gene fragment from human genomic DNA
A major technical challenge for detecting low frequency point mutations in single copy nuclear genes has been the large quantity of genomic DNA required. This impedes the performance of most mutation detection methods (27). In this study, we significantly reduced the amount of DNA by employing a sequence-specific enrichment strategy to highly enrich the desired nuclear gene fragment, a 482 bp APC gene sequence, from digested human genomic DNA. In this strategy, the genomic DNA is heat denatured and hybridized simultaneously with excess biotin-labeled oligonucleotide probes targeted to the Watson–Crick strands of the desired sequence. The hybrids are captured by streptavidin-coated paramagnetic beads and magnetically separated from the bulk DNA solution. The target fragments are then eluted from the beads by heating and subsequently re-annealed to form double strands.
By monitoring the copy numbers of the APC target fragment and a 205 bp mitochondrial DNA sequence used as a reference, we adjusted the experimental conditions (including the hybridization temperature, the molar ratio of probe/target, the quantities of paramagnetic beads and the hybrid–bead binding conditions) to obtain both a high level of target sequence yield and enrichment. The copy numbers of the two sequences were measured in the initial genomic DNA sample and the final target-enriched sample based on competitive PCR followed by CDCE separation. After the enrichment procedure under the optimal conditions (see Materials and Methods), the molar ratio of the APC target sequence to the mitochondrial DNA sequence was found to increase from 1/(321 ± 55) (95% confidence interval) in the initial genomic DNA samples to 32 ± 12 in the final elutions, resulting in ~104-fold enrichment of the target relative to the mitochondrial DNA sequence. The yield of the target sequence was measured and found to be 74 ± 7%.
The target-enriched sample was restriction digested to release the 271 bp APC fragment suitable for CDCE separation (Fig. 1B). The sample was then desalted and concentrated by ultrafiltration in order to be completely electroinjected onto the capillary column. This procedure produced a yield of 50–60%.
Thus, an initial sample of 3 mg genomic DNA from 3 × 108 MT1 cells was reduced to <1 µg containing about 2 × 108 copies of the target sequence (6 × 108 × 74% × 50%). Based on the yield of the subsequent CE separations of almost 100% and the estimated efficiency of the first cycle PCR of 0.5, any mutant at an initial mutant fraction of 10–6 in the sample would have been represented by a minimum of 100 mutant copies in our procedures. A sample of this size should permit measurement of the cellular mutants at or above 10–6 with an expected numerical variation of about ±20% (Poisson distribution, 95% confidence limits).
Enrichment and amplification of rare mutants through capillary electrophoresis coupled with high fidelity PCR
Approximately 2 × 108 total target copies enriched from MT1 genomic DNA were subjected to CDCE using a 540 µm i.d. capillary. Such a wide bore capillary has sufficient loading capacity (up to 5 µg DNA) to permit the separation of heteroduplexes from the wild-type homoduplex in the presence of residual cellular DNA and other impurities in the sample. We have previously demonstrated that excellent CDCE resolution can be achieved in wide bore capillaries at low electric field strengths to avoid the effect of Joule heating (24). The CDCE was performed under conditions for which all heretoduplexes co-eluted within a single fraction well separated from the wild-type homoduplex fraction. The heteroduplex fraction was electroeluted as a fraction enriched in mutant sequences. The mutant enrichment efficiency was estimated based on the ratio of the total target copies loaded on the column to that eluted in the heteroduplex fraction. The enrichment was found to be 20-fold for the target-enriched cellular DNA samples and over 200-fold for the CDCE-purified wild-type DNA sample.
As a means to obtain superior enrichment of cellular DNA samples, the heteroduplex collection of the cellular DNA sample was then run on CE at room temperature, which separated true heteroduplexes from the residual wild-type sequences that migrated slower than the 271 bp wild-type homoduplex fragment. These uncharacterized wild-type sequences may have been generated in the earlier procedures due to incomplete restriction digestion and/or inappropriate re-annealing. An additional 5- to 10-fold mutant enrichment was obtained by collecting the fraction containing the desired 271 bp APC fragment. Taken together, through the combination of CDCE and CE at room temperature, the mutants in the target-enriched cellular DNA were enriched 100- to 200-fold in total.
The mutant-enriched samples were next amplified by high fidelity PCR using native Pfu DNA polymerase and fluorescein-labeled primers to yield 2.5 × 1012 copies of the target sequence. Owing to the pre-PCR mutant enrichment, the true cellular mutants at an initial fraction of 10–6 would be present at 1–2 × 10–4 in the PCR products. This can be compared with the expected fractions of PCR-introduced mutations. The Pfu error rate has been reported to be ~10–6 errors/bp/doubling (26,28). The PCR-induced mutations after 21 doublings (from 106 to 2.5 × 1012 copies) would occur at a mutant fraction of 2 × 10–5 per bp on average, or 2 × 10–4 for hot-spots with a 10-fold increase in mutant fraction.
The amplified PCR products were subjected to post-PCR mutant enrichment via CDCE in a regular 75 µm i.d. capillary (Fig. 2). The heteroduplex fraction was collected separately from the wild-type homoduplex (Fig. 2A), allowing an additional 20-fold enrichment of mutants. The collected heteroduplexes were PCR amplified and run on a second CDCE. As shown in Figure 2B, the two heteroduplex peaks of the internal standard at an initial mutant fraction of 10–5 were clearly identifiable at this stage, each constituting 2% of the wild-type peak. To observe mutants at lower mutant fractions, the heteroduplex collection was repeated to further enrich the mutants an additional ~5-fold.
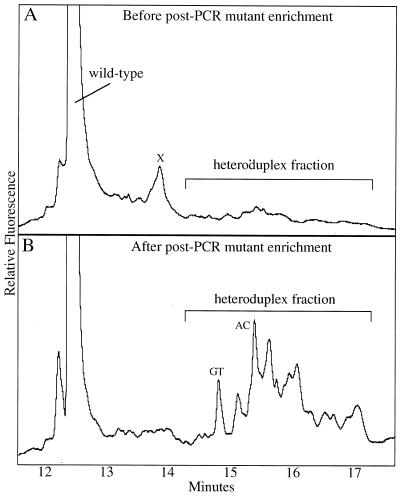
Figure 2.
Post-PCR enrichment of mutant sequences by CDCE coupled with PCR. Depicted are CDCE separations of PCR products derived from the MNNG-treated MT1 cells. The horizontal axis represents the time when the peak reaches the detector after the beginning of the run. The vertical axis represents the relative intensity of fluorescence, which is proportional to the molecular number. The wild-type peak in both electropherograms is shown at 1/10 of its full height. (A) The mutant-enriched fraction from wide bore CE separations was subjected to 40 cycles of high fidelity PCR. The PCR products were separated by CDCE and the heteroduplex fraction was collected. X marks a PCR by-product. (B) The collected heteroduplex fraction was subjected to 38 cycles of PCR and separated by CDCE. GT and AC mark the two heteroduplexes derived from the internal standard introduced at an initial mutant fraction of 10–5. The heteroduplex fraction was collected and amplified to further enrich the mutants.
Mutational spectra in MNNG-treated and untreated MT1 cells
The final CDCE separations of mutant homoduplexes derived from two MNNG-treated and two untreated MT1 cultures, as well as the CDCE-purified wild-type DNA serving as a PCR noise control, are shown in Figure 3. As compared to the purified wild-type DNA and untreated samples, both MNNG-treated samples contained a distinct set of peaks nearly all of which migrated after the wild-type peak (Fig. 3A), indicating that they were mutant homoduplex sequences with melting temperatures (Tm) lower than the wild-type homoduplex. As no strong mutant peaks appeared with melting temperatures higher than the wild-type, our analysis focused on the low Tm mutants.
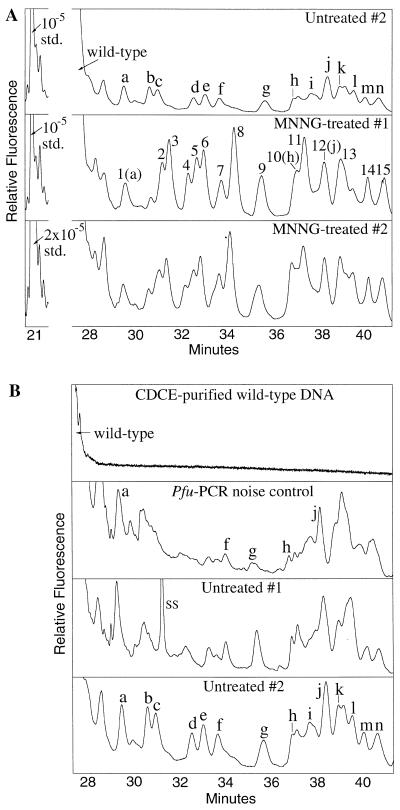
Figure 3.
CDCE separations of mutant homoduplexes derived from two MNNG-treated and two untreated MT1 cultures and the CDCE-purified wild-type DNA. The axes are as in Figure 2. (A) Comparison of mutants derived from two MNNG-treated and one untreated samples. The wild-type peak and the internal standard (std.) at its initial mutant fraction are indicated. The std. peak is shown at half its height in the Untreated #2 and MNNG-treated #1 samples and at a quarter of its height in MNNG-treated #2. (B) Comparison of mutants derived from the CDCE-purified wild-type DNA (Pfu PCR noise control) and two untreated samples. Each mutant peak that was isolated and sequenced is designated by a letter or number. SS, single-stranded DNA.
Each mutant peak that has been identified is designated by a letter in the untreated sample or a number in the MNNG-treated sample (Fig. 3A). The migration times and mutant fractions of each set of peaks in the replicate experiments exhibited excellent reproducibility when CDCE was run at several different water jacket temperatures (data not shown). Although co-migration on CDCE is not sufficient proof of identity, in our previous studies over 90% of the peaks identified by co-migration with mutant standards were indeed verified by either direct sequencing or on-column hybridization (8,10). In this study, several mutant peaks derived from the CDCE-purified wild-type DNA sample (Fig. 3B) were confirmed to be identical to their co-migrating counterparts in the untreated MT1 samples by direct sequencing.
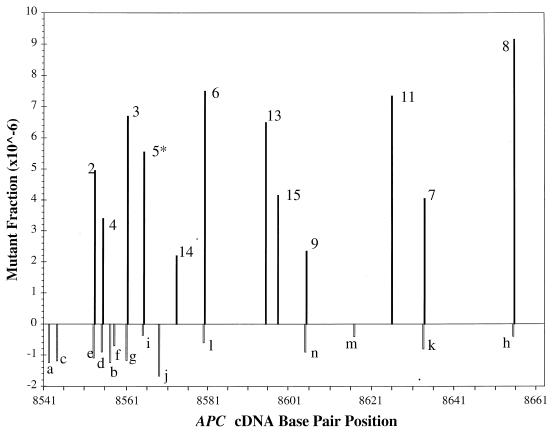
Figure 4 shows the position, type of mutation and sequence context for all of the mutants identified in MNNG-treated and untreated samples. All of the 14 mutants found in the untreated samples contained a GC→TA transversion. Their mutant fractions ranged from 4 × 10–7 to 1.8 × 10–6 as determined by comparison with the internal standard (Fig. 5). In contrast, 12 of the 15 mutants in the MNNG-treated samples contained a GC→AT transition at a mutant fraction of between 2.2 × 10–6 and 9.2 × 10–6 (Fig. 5). The three exceptions (peaks 1, 10 and 12) were found to be identical to three mutants in the untreated sample (peaks a, h and j, respectively), suggesting that they belong to the set of background mutations.
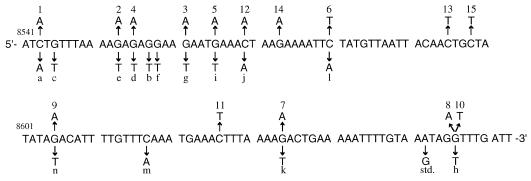
Figure 4.
Distribution of base substitutions in the APC gene target sequence (APC cDNA bp 8543–8663) in MNNG-treated and untreated MT1 cells. Mutants from the treated cells are shown above the wild-type sequence, while those from the untreated are below the sequence. Mutant labels correspond to those in Figure 3. The base substitution of the internal standard (std.) is also indicated.
Figure 5.
MNNG-induced and background point mutational spectra in the APC gene target sequence in MT1 cells. The horizontal axis represents the APC cDNA base pair position of the target sequence. The height of each vertical bar represents the average fraction of each mutant in the replicate samples. Each mutant is labeled with a number or letter corresponding to those in Figures 3 and 4. Open bars represent background GC→TA transversions; filled bars represent MNNG-induced GC→AT transitions. Except for peak 5 (indicated by *), all transitions occurred at guanine residues preceded (5′) by a purine. The mutant fraction was measured by comparing the area under each mutant peak with that under the internal standard in a CDCE electropherogram. To obtain an accurate measurement, CDCE was performed at different water bath temperatures (between 64.8 and 65.5°C). For each peak to be measured, the appropriate electropherogram showing the best separation of this peak from its adjacent ones was chosen.
The observation of the 12 GC→AT transitions in the MNNG-treated MT1 cultures is consistent with the predicted miscoding potential of O6-methylguanine, which is proposed to be the major premutagenic lesion produced by MNNG (29,30). An analysis of the influence of the neighboring base sequences on MNNG-induced mutation is shown in Table 1, which includes all of the 29 guanine sites in the APC target sequence. On average, guanine residues preceded 5′ by a purine are seven times more likely to be mutated than those preceded 5′ by a pyrimidine. In addition, the 3′ flanking base also seems to exert a slight influence on mutation in favor of GPu 3′ sites. As shown in Table 1, the average frequency of MNNG-induced GC→AT transitions appears to decrease in the order 5′-PuGPu-3′ > 5′-PuGPy-3′ > 5′-PyGPu-3′ > 5′-PyGPy-3′.
Table 1. Influence of the 5′ and 3′ flanking bases on MNNG-induced GC→AT transitions.
| Sequence (5′→3′) | No. of guanine sites | No. of mutated sites | Total mutant fraction (×106) | Average mutant fraction per site (×106) |
|---|---|---|---|---|
| PuG | 17 | 11 | 59 | 3.5 |
| PyG | 12 | 1 | 6 | 0.5 |
| GPu | 15 | 8 | 37 | 2.5 |
| GPy | 14 | 4 | 27 | 1.9 |
| PuGPu | 10 | 7 | 47 | 4.7 |
| PuGPy | 7 | 4 | 18 | 2.6 |
| PyGPu | 5 | 1 | 6 | 1.2 |
| PyGPu | 7 | 0 | 0 | 0 |
DISCUSSION
The means to measure point mutational spectra of nuclear single copy genes without phenotypic selection
Based on two crucial technical improvements, we have extended our previously developed CDCE/high fidelity PCR approach to mutational spectrometry of nuclear genes, exemplified here by a 121 bp APC gene sequence. First, sequence-specific hybridization coupled with a biotin–streptavidin capture system was employed to enrich the target sequence from genomic DNA by ~104-fold. Using this strategy, several milligrams of genomic DNA, which are required for reproducible observations of nuclear gene mutations at mutant fractions as low as 10–6, were reduced to <1 µg containing over 70% of the desired target copies. This large reduction in sample size permitted the subsequent mutational analysis. Furthermore, we have successfully applied this method to enrich four other nuclear sequences located in the human p53, Ki-ras and HPRT genes from the same genomic DNA samples for mutational analysis (X.-C.Li-Sucholeiki et al., unpublished results).
Our second technical improvement was wide bore capillary electrophoresis to enrich for mutant sequences in the presence of the residual 1 µg of cellular DNA. Compared to the CDGE technique used in our earlier studies (8–11), CDCE using a 540 µm i.d. column provided a large sample loading capacity without compromising resolution or target sequence yield (almost 100%). A greater than 100-fold mutant enrichment was achieved through two consecutive separations of CDCE and CE at room temperature. The high efficiency of mutant enrichment in combination with subsequent high fidelity PCR using Pfu DNA polymerase, which exhibits the lowest error rate among all commercially available thermostable DNA polymerases (28,31,32), created a means to measure nuclear point mutations at frequencies at or above 10–6.
Our approach should be general for any 100 bp isomelting DNA sequences juxtaposed to a naturally occurring high Tm domain, which renders them suitable for CDCE analysis. While nearly all genes contain such sequences, they represent only ~9% of the human genome (A.Kim, personal communication). Nevertheless, this general approach can be extended to 98% of the remaining gene sequences by attachment of an artificial GC clamp (33,34; A.Kim, personal communication). Such attachment could be carried out through strategies such as hybridization to clamp-containing probes or high efficiency clamp ligation after the target fragment is enriched from the genomic DNA (33,35).
Limit to sensitivity: PCR-induced noise
The background mutant peaks observed in the untreated MT1 samples were also found in the Pfu PCR noise control using CDCE-purified wild-type DNA. Independent PCR experiments with CDCE-purified wild-type DNA and Pfu DNA polymerase further demonstrated that the mutant fraction of the set of background mutations increased linearly with the number of PCR doublings, indicating that these mutations arose predominantly from the PCR process itself, as opposed to DNA lesions generated prior to PCR (data not shown). The total background mutant fraction in the final PCR products of the untreated MT1 samples was measured to be 4.6 × 10–3, given that the internal standard was present at 10–3 after pre-PCR mutant enrichment. The Pfu error rate in the 121 bp APC target sequence was thus estimated to be 1.8 × 10–6 mutations/bp/doubling [= 4.6 × 10–3 mutations/(121 bp × 21 doublings)]. A further increase in sensitivity of our approach will require an improved pre-PCR mutant enrichment and/or fidelity of PCR.
Interestingly, the 14 identified low Tm background mutations, which represented ~50% of the total PCR-induced mutations in the APC target sequence, were exclusively GC→TA transversions (Fig. 4). Transversions have been previously identified as the predominant Pfu-induced mutational hot-spots in a 100 bp human mitochondrial DNA sequence (28). The Pfu mutational spectra observed in these two templates are markedly different from the spectra reported for other DNA polymerases (including Taq, Klenow fragment, Vent, T4 and modified T7) which are dominated by transition mutations (31,36). This difference may reflect the properties of Pfu with respect to misinsertion, proofreading and/or mispair extension. Future studies on the mechanistic aspects of Pfu-induced mutations may provide insight to improve PCR fidelity.
MNNG-induced point mutational spectrum in the APC gene target sequence in MT1 cells
We tested the validity of our approach in a study of MNNG-induced point mutations in the 121 bp APC target sequence in MT1 cells. Distinct from the background mutations in the untreated controls, 12 GC→AT transitions were reproducibly observed in the MNNG-treated cells at mutant fractions ranging from 2.2 × 10–6 to 9.2 × 10–6 (Figs 3–5). The transitions were predominantly found at guanine residues flanked by a purine, particularly at their 5′ position (Table 1). It is unlikely that our CDCE-based method could preferentially enrich and/or separate transitions at these sites, because no bias in favor of 5′-PuG sites is seen for the spontaneous GC→AT transitions detected in a 100 bp human mitochondrial DNA sequence using a similar approach (9,10).
Our results with regard to the kind of mutation induced by MNNG and its site specificity agree with previous observations in Escherichia coli, yeast and other human systems (20,37–39). It has been suggested that the neighboring nucleotides may influence the distribution of O6-methylguanine lesions by modifying the reactivity of the O6 position of guanine and/or replication and repair process (37). However, because all previous studies were based on phenotypic selection, it was also argued that the non-random distribution of MNNG-induced mutations in some targets could be a reflection of the influence of the mutation on protein structure and function (40,41). For example, a GC→AT transition in the Gly (5′-GGN-3′) and Trp (5′-UGG-3′) codon sequences is more likely to be selected as a hot-spot because Gly residues are often strategic to protein structure and a transition in the Trp coding sequence results in a translation termination signal. Our results clearly show that the observed site specificity of MNNG-induced mutations is independent of phenotypic selection.
We note that transitions were not detected at six of the 17 5′-PuG sites (including three of the 10 5′-PuGPu sites) in the APC target sequence. This suggests that in addition to the influence of the neighboring sequence context, other factors (such as the structural and physical properties of the chromatin) might also modulate the accessibility of the DNA sites to reactive chemical species and/or replication and repair proteins.
In summary, our demonstration that human genomic point mutations can be detected in both mitochondrial and nuclear DNA at a level or above 10–6 seems complete. The application of this technology to the discovery and enumeration of rare point mutations in human tissues or large pooled samples of human populations seems not only possible but also a realistic reachable goal.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Ms Jacklene Goodluck-Griffith for assistance in cell culture. We acknowledge our collaborators Dr B. Karger (Northeastern University) and Dr K. Khrapko (Harvard University) whose contributions to the development of CE and CDCE and collegial discussions have made this work possible. This work was supported by grants from the National Institute for Environmental Health Sciences: NIEHS Center (P30-ESO2109), Genetics and Toxicology (P01-ESO3926), Mutagenic Effects of Air-Borne Toxicants (P01-ESO7168) and Superfund Basic Research (P42-ESO4675).
REFERENCES
- 1.Benzer S. and Freese,E. (1958) Proc. Natl Acad. Sci. USA, 44, 112–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller J.H. (1983) Annu. Rev. Genet., 17, 215–238. [DOI] [PubMed] [Google Scholar]
- 3.Fuchs R.P.P., Schwartz,N. and Daune,M.D. (1981) Nature, 294, 657–659. [DOI] [PubMed] [Google Scholar]
- 4.Thilly W.G. (1990) Annu. Rev. Pharmacol. Toxicol., 30, 369–385. [DOI] [PubMed] [Google Scholar]
- 5.Khrapko K., André,P., Cha,R., Hu,G. and Thilly,W.G. (1994) Prog. Nucleic Acid Res. Mol. Biol., 49, 285–311. [DOI] [PubMed] [Google Scholar]
- 6.Khrapko K., Hanekamp,J.S., Thilly,W.G., Belenkii,A., Foret,F. and Karger,B.L. (1994) Nucleic Acids Res., 22, 364–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer S.G. and Lerman,L.S. (1983) Proc. Natl Acad. Sci. USA, 80, 1579–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khrapko K., Coller,H., André,P., Li,X.-C., Foret,F., Belenky,A., Karger,B.L. and Thilly,W.G. (1997) Nucleic Acids Res., 25, 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khrapko K., Coller,H.A., André,P.C., Li,X.-C., Hanekamp,J.S. and Thilly,W.G. (1997) Proc. Natl Acad. Sci. USA, 94, 13798–13803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coller H.A., Khrapko,K., Torres,A., Frampton,M.W., Utell,M.J. and Thilly,W.G. (1998) Cancer Res., 58, 1268–1277. [PubMed] [Google Scholar]
- 11.Marcelino L.A., André,P.C., Khrapko,K., Coller,H.A., Griffith,J. and Thilly,W.G. (1998) Cancer Res., 58, 2857–2862. [PubMed] [Google Scholar]
- 12.Grist S.A., McCarron,M., Kutlaca,A., Turner,D.R. and Morley,A.A. (1992) Mutat. Res., 266, 189–196. [DOI] [PubMed] [Google Scholar]
- 13.Robinson D.R., Goodall,K., Albertini,R.J., O’Neill,J.P., Finette,B., Sala-Trepat,M., Moustacchi,E., Tates,A.D., Beare,D.M., Green,M.H.L. and Cole,J. (1994) Mutat. Res., 313, 227–247. [DOI] [PubMed] [Google Scholar]
- 14.Potten C.S. and Loeffler,M. (1990) Development, 110, 1001–1020. [DOI] [PubMed] [Google Scholar]
- 15.Robin E.D. and Wong,R. (1988) J. Cell Physiol., 136, 507–513. [DOI] [PubMed] [Google Scholar]
- 16.Groden J., Thliveris,J., Samowitz,W., Carlson,M., Gelbert,L., Albertsen,H., Joslyn,G., Stevens,J., Spirio,L., Robertson,M. et al. (1991) Cell, 66, 589–600. [DOI] [PubMed] [Google Scholar]
- 17.Goldmacher V.S., Cuzick,R.A. and Thilly,W.G. (1986) J. Biol. Chem., 261, 12462–12471. [PubMed] [Google Scholar]
- 18.Kat A., Thilly,W.G., Fang,W., Longley,M.J., Li,G.-M. and Modrich,P. (1993) Proc. Natl Acad. Sci. USA, 90, 6424–6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papadopoulos N., Nicolaides,N.C., Liu,B., Parsons,R., Lengauer,C., Palombo,F., D’Arrigo,A., Markowitz,S., Willson,J.K.V., Kinzler,K.W., Jiricny,J. and Vogelstein,B. (1995) Science, 268, 1915–1917. [DOI] [PubMed] [Google Scholar]
- 20.Kat A.G (1992) PhD thesis, Massachusetts Institute of Technology, Cambridge, MA.
- 21.Furth E.E., Thilly,W.G., Penman,B.W., Liber,H.L. and Rand,W.M. (1981) Anal. Biochem., 110, 1–8. [DOI] [PubMed] [Google Scholar]
- 22.Thilly W.G. (1985) In Huberman,E. and Barr,S.H. (eds), Carcinogenesis. Raven Press, New York, NY, Vol. 10, pp. 511–528.
- 23.Lerman L.S. and Silverstein,K. (1987) Methods Enzymol., 155, 482–501. [DOI] [PubMed] [Google Scholar]
- 24.Li X.-C. and Thilly,W.G. (1996) Electrophoresis, 17, 1884–1889. [DOI] [PubMed] [Google Scholar]
- 25.Marusyk R. and Sergeant,A. (1980) Anal. Biochem., 105, 403–404. [DOI] [PubMed] [Google Scholar]
- 26.Lundberg K.S., Shoemaker,D.D., Adams,M.W., Short,J.M., Sorge,J.A. and Mathur,E.J. (1991) Gene, 108, 1–6. [DOI] [PubMed] [Google Scholar]
- 27.Parsons B.L. and Heflich,R.H. (1997) Mutat. Res., 387, 97–121. [DOI] [PubMed] [Google Scholar]
- 28.André P., Kim,A., Khrapko,K. and Thilly,W.G. (1997) Genome Res., 7, 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loveless A. (1969) Nature, 223, 206–207. [DOI] [PubMed] [Google Scholar]
- 30.Loechler E.L., Green,C.L. and Essigmann,J.M. (1984) Proc. Natl Acad. Sci. USA, 81, 6271–6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keohavong,P. and Thilly W.G. (1989) Proc. Natl Acad. Sci. USA, 86, 9253–9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eckert K.A. and Kunkel,T.A. (1991) PCR Methods Appl., 1, 17–24. [DOI] [PubMed] [Google Scholar]
- 33.Myers R.M., Fischer,S.G., Maniatis,T. and Lerman,L.S. (1985) Nucleic Acids Res., 13, 3111–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cariello N.F., Keohavong,P., Kat,A.G. and Thilly,W.G. (1990) Mutat. Res., 231, 165–176. [DOI] [PubMed] [Google Scholar]
- 35.Abrams E.S., Murdaugh,S.E. and Lerman,L.S. (1990) Genomics, 7, 463–475. [DOI] [PubMed] [Google Scholar]
- 36.Keohavong P., Ling,L., Dias,C. and Thilly,W.G. (1993) PCR Methods Appl., 2, 288–292. [DOI] [PubMed] [Google Scholar]
- 37.Burns P.A., Gordon,A.J.E. and Glickman,B.W. (1987) J. Mol. Biol., 194, 385–390. [DOI] [PubMed] [Google Scholar]
- 38.Kohalmi S.E. and Kunz,B.A. (1988) J. Mol. Biol., 204, 561–568. [DOI] [PubMed] [Google Scholar]
- 39.Yang J.-L., Hu,M.-C. and Wu,C.-W. (1991) J. Mol. Biol., 221, 421–430. [DOI] [PubMed] [Google Scholar]
- 40.Gordon A.J.E. and Glickman,B.W. (1988) Mutat. Res., 208, 105–108. [DOI] [PubMed] [Google Scholar]
- 41.Horsfall M.J., Gordon,A.J.E., Burns,P.A., Zielenska,M., van der Vliet,G.M.E. and Glickman,B.W. (1990) Environ. Mol. Mutagen., 15, 107–122. [DOI] [PubMed] [Google Scholar]