Abstract
Background
Insulin, secreted from pancreatic islets of Langerhans, is of critical importance in regulating glucose homeostasis. Defective insulin secretion and/or the inability of tissues to respond to insulin results in insulin resistance and to several metabolic and organ alterations. We have previously demonstrated that BAG3 regulates insulin secretion. Herein we explored the consequences of beta-cells specific BAG3 deficiency in an animal model.
Methods
We generated a beta-cells specific BAG3 knockout mouse model. Glucose and insulin tolerance tests, proteomics, metabolomics, and immunohistochemical analysis were used to investigate the role of BAG3 in regulating insulin secretion and the effects of chronic exposure to excessive insulin release in vivo.
Results
Beta-cells specific BAG3 knockout results in primary hyperinsulinism due to excessive insulin exocytosis finally leading to insulin resistance. We demonstrate that resistance is mainly muscle-dependent while the liver remains insulin sensitive. The chronically altered metabolic condition leads in time to histopathological alterations in different organs. We observe elevated glycogen and lipid accumulation in the liver reminiscent of non-alcoholic fatty liver disease as well as mesangial matrix expansion and thickening of the glomerular basement membrane, resembling the histology of chronic kidney disease.
Conclusion
Altogether, this study shows that BAG3 plays a role in insulin secretion and provides a model for the study of hyperinsulinemia and insulin resistance.
Keywords: Glucose homeostasis, BAG3, Primary hyperinsulinism, Insulin resistance, Chronic kidney disease
Highlights
-
•
BAG3 is involved in insulin release.
-
•
Beta-Cells specific BAG3 knockout results in primary hyperinsulinism.
-
•
Chronic hyperinsulinemia leads to insulin resistance in this mouse model.
-
•
Beta-Cells loss of BAG3 deficient mice developed multiple organ alterations.
1. Introduction
Insulin, secreted from pancreatic beta cells, is a crucial hormone in regulating glucose metabolism. Insulin release and action must precisely meet the metabolic demand, defective insulin secretion by pancreatic β-cells and/or the inability of tissues to respond to insulin results in insulin resistance and eventually in the onset of type 2 diabetes (T2D) [[1], [2], [3], [4]]. Identification of proteins involved in insulin release, as well as understanding the underlying mechanisms, is therefore of uttermost importance and can shed light on the pathogenesis of several metabolic disorders such as congenital hyperinsulinism, metabolic syndrome, T2D, nonalcoholic fatty liver disease, cardiovascular disease, and kidney disease [[5], [6], [7]].
BCl2-associated athanogene 3 (BAG3) is a member of a family of co-chaperones that interacts with heat shock protein 70. It is involved in several cellular functions including development, cytoskeleton organization, cell death, and autophagy. BAG3 is expressed constitutively only in few tissues, while it is induced in many cell types in response to various stimuli [[8], [9], [10], [11]]. We have previously shown that BAG3 is constitutively expressed in pancreatic beta cells and regulates insulin secretion, being part of the t-SNARE complex that regulates the trafficking of insulin-containing granules [12]. Indeed, silencing BAG3 in β-TC-6 mouse insulinoma cells results in decreased intracellular insulin content, suggesting a defect in retaining insulin-containing granules and increasing insulin secretion in response to glucose stimulation [12]. Moreover, Youn et al. have shown that BAG3KO mice, which die 4 weeks after birth because of marked degeneration of the diaphragm, intercostal muscles, heart, and other skeletal muscles, also show hypoglycemia and fatty livers. Moreover, in these mice, quantitative analysis of the hepatic mRNA encoding for enzymes involved in glucose and lipid metabolism suggests an increase in gluconeogenesis and β-oxidation as well as a decrease in lipogenesis, indicating a typical adaptive response to malnutrition, which was supported by serum profiles of metabolites as well as the scarcity of peripheral fat [13].
Here we selectively knocked out BAG3 in pancreatic beta cells (BAG3βKO) by Cre-mediated recombination expressing the Cre recombinase under the control of the insulin 1 promoter. BAG3βKO mice develop normally, show a normal life span, and display a good glucose tolerance but show impairment in insulin sensitivity, secrete more insulin after glucose stimulation and become overtly insulin resistant at 64 weeks of age. In addition, these mice show histopathological features of chronic kidney (CKD) disease and glycogen and lipids accumulation in the liver. Overall, our data confirm that BAG3 plays a crucial role in modulating insulin secretion; its knockout in pancreatic islets of Langerhans results in increased insulin release that over time determines insulin resistance and a complex metabolic disorder finally leading to multiorgan disfunctions.
2. Materials and methods
2.1. Animals and maintenance
All animal experimentation procedures were conducted according to protocols approved by the Institutional Animal Healthcare of the University of Chieti and in accordance with the institutional guidelines of the Italian Ministry of Health (Protocol No. 204/2022-PR). All mice were housed in a pathogen-free environment (22 °C, 12-h/12-h light/dark cycle), where they accessed freely to food and water. To generate beta cell-specific BAG3 knockout mice the Cre-loxP mediated recombination technology was used. Briefly, BAG3 homozygous floxed mice (BAG3tm1c(EUCOMM)Hmgu), BAG3flox/flox, which were a gift from the laboratory of Dr. Michael Hanhe, were crossed with mice expressing Cre recombinase under the mouse insulin 1 promoter B6(Cg)-Ins1tm1.1(cre)Thor/J, INS1cre+/+, (stock no. 026801; The Jackson Laboratory). For experiments, control mice had two floxed BAG3 alleles and no INS1cre alleles (BAG3fl/fl), or only INS1cre alleles (INS1cre+/+) whereas beta-cells specific BAG3 knockout mice harbored two floxed alleles and INS1cre+/+ at the same time (BAG3βKO). The mice were divided into different cages according to their genotypes at 4 weeks of age; the body weight was monitored weekly. Aged-matched control and BAG3βKO male mice were used for the experiments, being known that female mice are protected from metabolic decline [14].
2.2. DNA extraction and genotyping
Genomic DNA was extracted from a small ear punch and amplified by polymerase chain reaction (PCR) using the Xpert directXtract PCR Kit (GRiSP, Porto, Portugal). Genotyping for INS1cre is detailed on The Jackson Laboratory's Web site (http://jaxmice.jax.org/). Genotyping for BAG3 was performed using the following primers: BAG3-471 forward – ACTATCTGCTGTCTACACTGC; flanked LoxP BAG3-459 reverse – TGAGCGGCACTTTAAGGTTT. PCR products were resolved in a 2% agarose gel and were either only wild-type DNA (product ≈300 bp) or wild-type DNA with the upstream LoxP site included (mutant product ≈450 bp). The genotyping was performed on a MJ Mini 48-Well Personal Thermal Cycler (Bio-Rad, Hercules, CA, USA) and the following amplification scheme was used: 95 °C for 3 min for 1 cycle; 95 °C for 15 s, 55 °C for 15 s, 72 °C for 2 s for 40 cycles; 72 °C for 3 min for 1 cycle and then 4 °C to soak.
2.3. Islets isolation
Mouse pancreatic islets were isolated as previously described with a few adaptations [15]. Briefly, 1.2 mg/mL collagenase solution (#C5138, Sigma–Aldrich, Steinheim, Germany) was injected into the common bile duct of a euthanized animal. The pancreas was then excised and digested at 37 °C for 15 min. Afterward, islets were recovered and purified using a Histopaque gradient (Histopaque®-1077 and Histopaque®-1119, Sigma–Aldrich) handpicked and employed for further experiments.
2.4. Immunofluorescence analysis
Freshly isolated islets were subjected to cytocentrifugation on glass slides for 3 min at 500 rpm. Next, the cytospin preparations were fixed with 4% paraformaldehyde (Santa Cruz Biotechnology) and incubated with rabbit polyclonal anti-BAG3 (1/200 dilution; NBP2-27398, Novus Biologicals, Milan, Italy) and mouse monoclonal anti-Insulin B (C-12) (1/1000 dilution; sc-377,071, Santa Cruz Biotechnology) antibodies overnight, followed by anti-rabbit Alexa fluor 488 (A-11008, Thermo Fisher Scientific, Waltham, MA, USA) and anti-mouse Alexa fluor 546 (A-11030, Thermo Fisher Scientific, Waltham, MA, USA) secondary antibodies for 1 h at RT. For immunofluorescence staining of pancreatic tissues, slides were deparaffinized, serially rehydrated, and after antigen retrieval procedure for 10 min on the microwave with EnVision FLEX Target Retrieval Solution Low pH buffer (K8005, Dako), incubated overnight with mouse monoclonal anti-Insulin B (C-12) antibody (1/1000 dilution; sc-377,071, Santa Cruz Biotechnology), rabbit polyclonal anti-Glucagon antibody (1/200 dilution; PA5-88091, Thermo Fisher Scientific), and rat monoclonal anti-somatostatin antibody (1/300 dilution; MA5-16987, Thermo Fisher Scientific). After incubation, slides were stained with anti-mouse Alexa fluor-plus 647 (A-32728, Thermo Fisher Scientific), anti-rabbit Alexa fluor 546 (A-11010, Thermo Fisher Scientific) and anti-rat Alexa fluor 488 (A-11006, Thermo Fisher Scientific) secondary antibodies for 1 h at RT. Nuclei were counterstained with Dapi (D9542, Sigma) for 15 min, and images were acquired by Zeiss LSM800 confocal microscope (ZEISS, Oberkochen, Germany).
2.5. RNA extraction and qPCR
Total RNA was extracted from islets or livers by using the RNeasy kit (Qiagen, Valencia, CA). RNA was then quantified by measuring absorbance with a NanoDrop (Thermo Fisher Scientific) and was transcribed to cDNA using TaqMan™ Reverse Transcription Reagents (Applied Biosystems, Thermo Fisher Scientific). Quantitative PCR was performed using the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) using a Bio-Rad CFX96 Touch System. Transcript levels were normalized to actin and expressed as ratios of control levels. Primer sequences used are listed in Table S2.
2.6. Western blotting
Tissues were homogenized using ice-cold lysis buffer (TRIS–HCl 50 mM pH 8, NaCl 150 mM, Triton X-100 1%, NaF 100 mM, EDTA 1 mM, MgCl2 1 mM, Glycerol 10%) with a protease inhibitor cocktail (Sigma–Aldrich, St. Louis, MO, USA) and a phosphatase inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). Bradford method was used to quantify proteins (Bio-Rad). Sample aliquots containing 20–30 μg protein were loaded on SDS-polyacrylamide gel electrophoresis gels (10%) and subsequently transferred to a nitrocellulose membrane (GE Healthcare Life Science, Piscataway, NJ, USA). Equal amounts of proteins were separated on an SDS-PAGE and detected after being transferred to a nitrocellulose membrane (GE Healthcare Life Science). The membranes were incubated overnight at 4 °C with primary antibodies: anti-BAG3 used 1:2000 (Novus Biologicals, Milan, Italy), anti-INSR used 1:1000, anti-AKT used 1:1000, anti-phospho-AKT used 1:1000, anti-ERK1/2 used 1:1000, anti-phospho-ERK1/2 and anti-GAPDH used 1:2000, all supplied by Cell Signaling Technology (Danvers, MA, USA), anti-GLUT4 Rabbit polyclonal Ab used 1:1000 (Antibodies.com, Cat. #95872), anti p62/SQSTM1 used 1:1000 (BD Bioscience, San Jose, CA, USA), anti-Insulin B (C-12) used 1:500 (Santa Cruz Biotechnology) and anti-β-actin used 1:40,000 (Sigma–Aldrich, Milan, Italy). Following that, the membranes were washed and incubated for 1 h at RT with the secondary antibody, diluted 1:20,000 (Bio-Rad, Milan, Italy). Bound antibodies were detected using the enhanced chemiluminescent (ECL) method (PerkinElmer Italia, Milan, Italy).
2.7. Blood glucose measurements
Blood glucose levels were measured under fed and fasting conditions using a Contour Next glucometer (Ascensia Diabetes Care, Milan, Italy) from tail vein bleeds.
2.8. Metabolic studies
An intraperitoneal glucose tolerance test (IPGTT) was performed after an overnight fast. Blood glucose levels, obtained from the tail vein, were measured using a glucometer (Contour Next, Ascensia Diabetes Care) at time 0 and at 15, 30, 60, 90, and, 120 min after injection with glucose (2 g/kg of body weight). For the Intraperitoneal insulin tolerance test (IPITT), mice were deprived of food for 4 h before insulin (0.75 U/kg of body weight) was injected intraperitoneally; blood glucose levels were measured at time 0 and 15, 30, 60, 90 and 120 min after insulin injection, using a glucometer. IPGTTs and IPITTs were performed on 12-, 24- and 64-weeks old mice.
2.9. Plasma insulin and glucagon measurements
Plasma insulin and glucagon concentrations were quantified using enzyme-linked immunosorbent (ELISA) assays following the manufacturer's instructions. Ultra-Sensitive Mouse Insulin ELISA and Mouse Glucagon ELISA Kit were purchased from Crystal Chem (Crystal Chem Inc., Downers Grove, IL, USA).
2.10. Homeostasis model assessment of insulin resistance
To assess the onset of insulin resistance, the Homeostasis Model Assessment of Insulin Resistance (HOMA-IR) was used. HOMA-IR was calculated by the formula: fasting insulin (μU/dL) × fasting blood glucose (mmol/L)/22.5 [16].
2.11. Biochemical analysis
Mice at 12, 24, and 64 weeks of age were fasted overnight. Blood samples were collected in Microvette® 200 Lithium heparin (Sarstedt, Nümbrecht, Germany), by cardiac puncture from euthanized mice. 100 ul of lithium-heparin collected blood samples obtained from cardiac puncture were centrifuged for 10 min at 4 °C for plasma separation. Triglycerides, Total Cholesterol, HDL, and LDL were measured on a VChemyS Automatic Biochemical Analyzer (VODEN medical instruments Spa, Monza-Brianza, Italy) according to the manufacturer's instructions.
2.12. Metabolomics
Mice at 12, 24, and 64 weeks of age were fasted overnight. A drop of blood, obtained from the tail vein, was spotted onto an absorbent filter paper and air dried at room temperature for a minimum of 2 h. Dried blood spot (DBS) disks (of approximately 3.2 μL whole blood) were punched out into 3.2 mm-disks and extracted for the determination of 13 Ammino acids (AAs), 34 acylcarnitines (ACs), free carnitine, Adenosine (ADO) and Deoxyadenosine (D-ADO) and 4 Lyso Phosphatidylcholines (LPCs), by using the NeoBase 2 Non-Derivatized MSMS Kit (Perkin Elmer Life and Analytical Sciences, Turku, Finland), as already described [17,18]. In particular, 125 μL of extraction solution containing isotopically labeled internal standard, were added into DBS disks and extracted at 45 °C for 30 min, mixed at 700 rpm. 100 μL of extracted solution were transferred into a 96-well plate and were subjected to targeted metabolomics approach by Flow-Injection Analysis Tandem Mass Spectrometry (FIA-MS/MS). The FIA-MS/MS system consists of an Acquity UPLC I-Class coupled to a Xevo TQD tandem quadrupole mass spectrometer (Waters Corporation, Milford, MA, USA). The system operates in positive electrospray ionization mode by multiple reaction monitoring (MRM) acquisition. The capillary voltage was set to 3.50 kV, the source temperature was 120 °C, the desolvation temperature was 350 °C, and the collision cell gas pressure was 3.0 × 10−3 mbar Argon. 10 μL are injected into the ion source and the run time is 1.1 min, injection-to-injection. Data are finally processed by MassLynx V4.2 and NeoLynx Software (Waters Corporation, Milford, MA, USA).
2.13. NAD+/NADH ratio measurement
Total NAD+ and NADH levels were measured according to the manufacturer's instructions (Abcam, Cambridge, UK). Briefly, 25 μl of NAD/NADH control buffer or NADH extraction buffer was added to the whole liver lysate sample and then incubated at 37 °C for 15 min. 75 μl of NAD/NADH reaction mixture was added into each sample and incubated at RT for 1 h. Fluorescence signals were measured on a Synergy H1 microplate reader (BioTek, Winooski, VT, USA) at Excitation/Emission 540/590.
2.14. Histological and immunohistochemical analysis
Mice at 12, 24, and 64 weeks of age were fasted overnight and sacrificed. Necroscopy was performed and all organs were harvested and fixed in 10% neutral buffered formalin (05-01005Q, Bio-Optica), embedded in paraffin and then sectioned at 5-μm thickness. Sections were stained with Hematoxylin (05–06002/L, Bio-Optica) and Eosin (05-M11007, Bio-Optica) for histological analysis. Slides were independently evaluated by two pathologists to evaluate the presence of histological changes in the tissues harvested. Immunohistochemical staining on formalin-fixed paraffin-embedded (FFPE) tissues was performed using the following primary antibodies: mouse monoclonal anti-Insulin B (C-12) antibody (1/1000 dilution; sc-377,071, Santa Cruz Biotechnology) and rabbit polyclonal anti-Glucagon antibody (1/200 dilution; PA5-88091, Thermo Fisher Scientific). Antigen retrievals were performed by microwaving slides for 10 min in EnVision FLEX Target Retrieval Solution Low pH buffer (K8005, Dako). Sections were then incubated with the primary antibodies mentioned above, overnight. Envision anti-rabbit (K4003, Dako) or anti-mouse (K4001, Dako) IgG were used as secondary antibodies for 30 min, followed by 3-3′ tetrahydrochloride diaminobenzidine DAB (K3468, Dako) as the chromogenic agent. After chromogen incubation, slides were counterstained in Hematoxylin (Cat Hematoxylin, CATHE-MM, Biocare Medical) and images were acquired by Leica DMRD optical microscope (Leica).
For histological assessment of glycogen deposition and lipids accumulation, PAS staining was performed on liver sections using the PAS Periodic Acid Schiff Kit (04–130,802, Bio-Optica), according to the manufacturer's instructions. For histological and immunohistochemical evaluations, whole slide sections were scanned with a Nanozoomer scanner from Hamamatsu and analyzed with Qu-Path v0.3.2 software using the following tools: area annotation tool for islet area, positive cell detection tool for insulin and glucagon quantifications (% of positive cells) and pixel classifier tool for PAS evaluation. Microvesicular steatosis was assessed and scored from 0 to 3 based on the percentage of the total area affected, into the following categories: 0 (<5%), 1 (5–33%), 2 (34–66%) and 3 (>66%) according to the steatosis grading score used by the NASH Clinical Research Network Scoring System [19]. Insulin and Glucagon secretion index was calculated using the following formula: % of positive cells X area of islets X number of islets.
2.15. Label-free proteomics analysis and bioinformatics
Liver and muscle tissues from BAG3insKO and WT mice were treated according to the FASP (Filter Aided Sample Preparation) method after mechanical homogenization through a turrax mixer (1-minute, highest speed) and ultrasonication on ice (Sonicator U200S control, IKA Labortechnik, Staufen, Germany) at 80% amplitude in a lysis buffer (urea 6 M in 100 mM Tris/HCl, pH = 7.5) and centrifuged to remove tissues debris. A total of 50 μg of proteins for each sample, quantified by Bradford Protein Assay (Bio-Rad, Hercules, CA, USA) using Bovine Serum Albumin (BSA, Sigma–Aldrich, St. Louis, MI, USA) as the standard calibration curve, were digested by trypsin (Promega, Madison, WI, USA). To perform the protein label identification and quantification, tryptic digest from each liver and muscle sample was analyzed in triplicate with LC-MS/MS through the UltiMate™ 3000 UPLC (Thermo Fischer Scientific, Waltham, MA, USA) chromatographic system coupled to the Orbitrap Fusion™ Tribrid™ (Thermo Fischer Scientific, Waltham, MA, USA) mass spectrometer. Peptides were loaded on the Trap Cartridge C18 (0.3 mm ID, 5 mm L, 5 μm PS, Thermo Fisher Scientific, Waltham, MA, USA) and then separated on an EASY-spray AcclaimTM PepMapTM C18 (75 μm ID, 25 cm L, 2 μm PS, Thermo Fisher Scientific Waltham, MA, USA) nanoscale chromatographic column according to the method previously described for liver samples [20]. Proteomics raw data on liver and muscle were processed by MaxQuant version 1.6.10.50 (Max-Planck Institute for Biochemistry, Martinsried, Germany). The UniProt database “released 2019_11, taxonomy Mus Musculus, 21,990 entries)” was used in Andromeda for picking and peptide search. Processing parameters were set as already described in our previous work [20]. In particular, a univariate statistical analysis, visualizing results as a Volcano Plot (FDR = 0.003 and S0 = 0.1), was performed to individuate differential proteins significantly expressed between two experimental conditions (p-value threshold = 0.05) and subsequently the protein ratios (BAG3insKO livers/Control) were uploaded to Ingenuity Pathway Analysis tool (IPA, Qiagen, Hilden, Germany) in order to perform a Gene Ontology and functional enrichment analysis (z-scores ≥2.0 and z-scores ≤ −2.0 for activation and inhibition of transcriptional regulators or downstream effects for the loaded dataset based on published literature, respectively). Finally, to perform a robust protein quantification, we evaluated as quantified proteins those with an abundance ratio variability [%] less than 50.00 and at least two peptides identified. The STRING database (http://String-db.org/, accessed on 27 July 2022) was used to analyze PPI (protein–protein interaction) networks. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [21] partner repository with the dataset identifier PXD038843.
2.16. Statistical analysis
Experimental results were expressed as means ± standard error (SEM). GraphPad Prism v.9 (GraphPad Software, Inc.) was used to perform statistical analyses. Student's t-test or Mann–Whitney U test were used after the determination of distribution by the Shapiro–Wilk normality test. When more than two groups were compared analysis of variance (ANOVA) or Kruskal–Wallis test were used, followed by post-hoc Tukey or Dunn's, respectively. Unless specified otherwise, asterisks in the figures indicate statistical significance ∗: p < 0.05, ∗∗: p < 0.01, ∗∗∗: p < 0.001, ∗∗∗∗: p < 0.0001.
3. Results
3.1. Generation of beta-cell specific BAG3 knockout mice
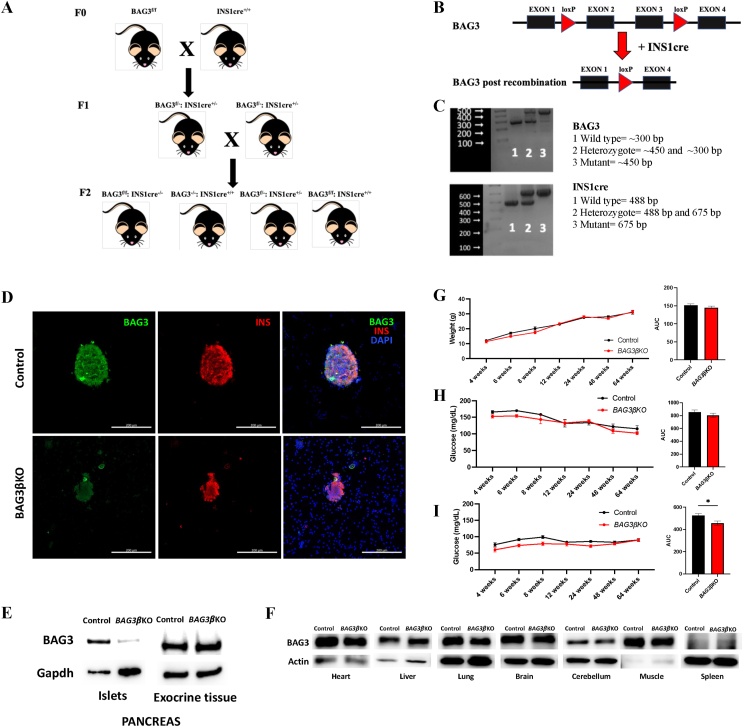
We have previously shown that silencing BAG3 in insulinoma cells leads to altered insulin secretion. We, therefore, generated pancreatic beta-cell specific BAG3 knockout (BAG3βKO) mice to evaluate the effect of BAG3 deletion on glucose homeostasis in vivo. We used INS1cre mice, harboring a Cre allele under the control of the endogenous Insulin 1 promoter, which, unlike Insulin 2 promoters, drives specific expression in beta-cells [22]. INS1cre homozygous mice were crossed with BAG3fl/fl mice (F1 heterozygous generation); BAG3fl/-:INS1cre+/− were inbred to obtain the F2 generation with different genotypes, such as BAG3f/f: INS1cre−/− and BAG3−/−: INS1cre+/+ (Control); BAG3f/-: Ins1cre+/− and BAG3f/f: INS1cre+/+ (BAG3βKO) as represented in Figure 1A. Cre recombinase expression resulted in the deletion of BAG3 floxed exon 2 and exon 3 (Figure 1B). Genotyping of F1 and F2 generations are shown in Figure 1C and was performed on genomic DNA isolated from ear punch. The genotype of BAG3βKO mice displayed together floxed and cre alleles. Immunofluorescence staining for BAG3 (Figure 1D) and Western blot analysis of isolated islets of Langerhans (Figure 1E) confirm the lack of expression in BAG3βKO mice islets, while its expression is comparable to control mice in all other tissues tested by Western blot (Figure 1F). While mice with a complete deficiency of BAG3 died 4 weeks after birth [13], BAG3βKO mice showed normal fecundity and litter size, normal longevity, and exhibited no macroscopic phenotypic abnormalities (data not shown). No differences in body weight were seen over time (Figure 1G). We then evaluated blood glucose levels under fed and fasting conditions and found no differences between fed BAG3βKO and control mice (Figure 1H). In contrast, the glycemia of BAG3βKO mice was significantly lower than controls when blood glucose levels were measured after 16 h of fasting, as displayed by the AUC in Figure 1I, particularly within the first 12 weeks after birth.
Figure 1.
Generation and validation of BAG3βKO mouse model. (A) Breeding scheme for generating beta-cell specific BAG3 knockout mice. Only the 4 genotypes of interest are shown in the scheme i) BAG3f/f: INS1cre−/− and BAG3−/−: INS1cre+/+ (Control), ii) BAG3f/-: Ins1cre+/− and iii) BAG3f/f: INS1cre+/+ (BAG3βKO). (B) Schematic image of floxed BAG3 allele and the deletion product after INS1cre recombination; for simplicity, only one allele has been shown. (C) PCR genotyping results of BAG3 floxed mice and INS1cre recombinase mice from DNA obtained from ear punch samples: 1-Wild type (BAG3−/−and INS1cre−/−); 2-heterozygote (BAG3f/- and Ins1cre+/−) and 3-Mutant (BAG3f/fand INS1cre+/+). (D) Representative immunofluorescence images of BAG3 and insulin staining in control and BAG3βKO mice isolated islets. Scale bar: 200 μm. (E) Western blot analysis of pancreatic islets and exocrine tissue to assess BAG3 specific knockout, while (F) no Cre recombination was found in the heart, liver, lung, brain, cerebellum, muscle, and spleen of BAG3βKO mice. (G) The growth curve of the body weight and area under the curve (AUC) monitored over time. (H) Fed and (I) fasting blood glucose levels monitored over time and their AUC. Data are presented as mean ± SEM (n = 6–7 for each group), ∗p < 0.05.
3.2. BAG3βKO mice show impaired insulin sensitivity and develop insulin resistance
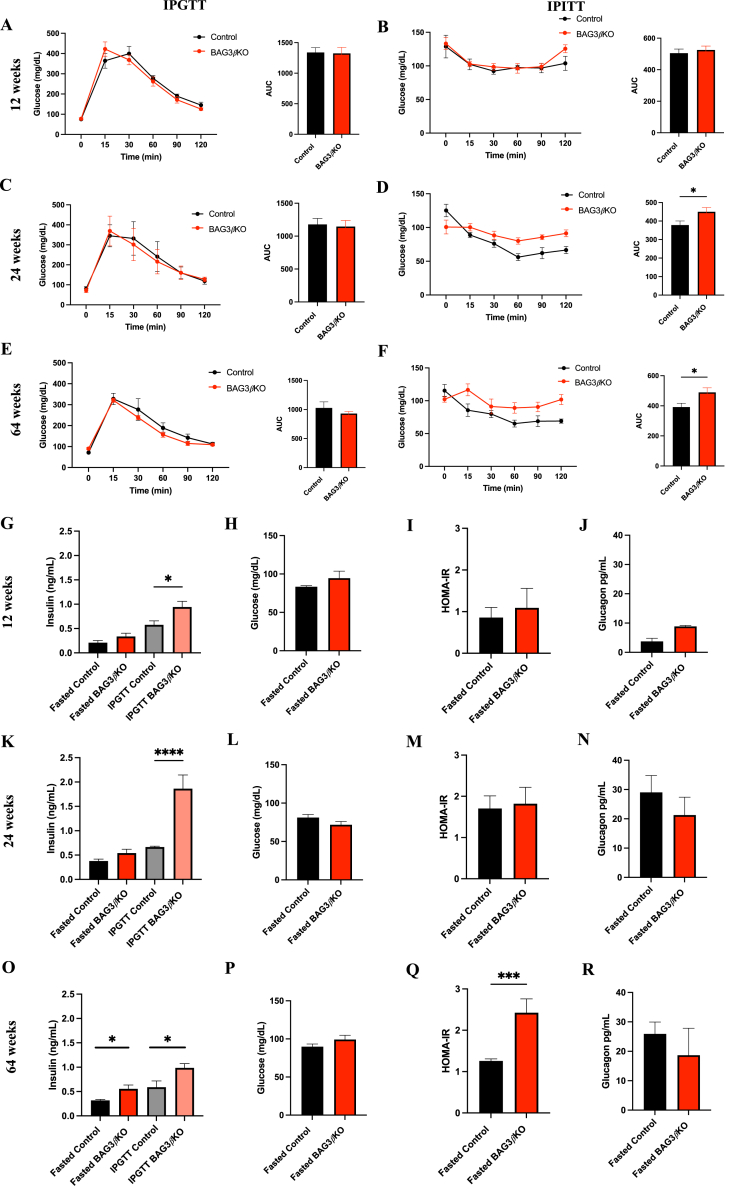
We performed an intraperitoneal glucose tolerance test (IPGTT) and an intraperitoneal insulin tolerance test (IPITT) in BAG3βKO mice and controls at 12, 24, and 64 weeks of age. No difference between the two groups in IPGTT was observed at any time points examined (Figure 2A,C and 2E). On the contrary, significant differences were observed in response to insulin injection (0.75 U/Kg) administered intraperitoneally in mice starved for 4 h (IPITT). At 12 weeks of age the BAG3βKO mice response was identical to that of control animals (Figure 2B), while at 24 and 64 weeks of age they showed a significantly lower decrease in glucose levels indicating impaired response to insulin (Figure 2D,F). We then evaluated basal insulin level under fasting conditions in BAG3βKO mice at 12 weeks of age (when the IPITT was normal) and at 24 and 64 weeks of age (when the IPITT was altered) (Figure 2G,K, and 2O). At 12 and at 24 weeks basal insulin concentration show a modest and not significant increase in fasted BAG3βKO mice compared to the control group. No difference in fasting glucose levels at these two time points was observed in the two groups (Figure 2H,L); consistently, the homeostasis model assessment for insulin resistance (HOMA-IR) index showed no differences (Figure 2I,M). However, at 64 weeks of age, fasted BAG3βKO mice showed significantly higher insulin level (Figures 2O), while blood glucose concentrations were still similar between the two groups (Figure 2P). Consistently, BAG3βKO mice showed a significant increased HOMA-IR (Figure 2Q). No differences in plasma glucagon concentrations were observed in fasted BAG3βKO mice (Figures 2J, 2 N, and 2 R). Importantly, upon glucose stimulation, BAG3βKO mice secreted significantly more insulin (Figure 2G, K and 2O). Altogether these data suggest that mice are initially, at least up to 12 weeks of age, insulin sensitive and become progressively resistant, being overtly resistant at 64 weeks of age, as proven by the HOMA-IR.
Figure 2.
Insulin sensitivity impairment in BAG3βKO mice leads to insulin resistance. IPGTT (A, C, E) and IPITT (B, D, F) in control vs BAG3βKO mice on 12 (A, B), 24 (C, D), 64 (E, F) weeks of age, and the respective AUCs. IPGTTs were performed after 16 h of fasting (overnight) and IPITTs after 4 h of fasting. Glucose (2 mg/kg) and insulin (0.75 units/kg) were injected intraperitoneally, and glycemia was measured at different times after injection (n = 4–10 for each group, data are presented as mean values ± SEM; ∗p < 0.05.). Circulating insulin (G, K and O) was measured after 16 h of fasting and after a 15-minute glucose challenge (2 g/kg) in control vs BAG3βKO mice at 12 (G) 24 (K) and 64 (O) weeks of age. Fasting blood glucose levels at 12 (H), 24 (L) and 64 weeks (P). HOMA-IR after 12 (I), 24 (M) and 64 weeks (Q). Fasting glucagon levels at 12 (J), 24 (N), and 64 weeks (R). Data are presented as mean ± SEM (n = 7–9 for each group); ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001.
3.3. Increased insulin secretion is associated with changes in pancreatic islets morphology
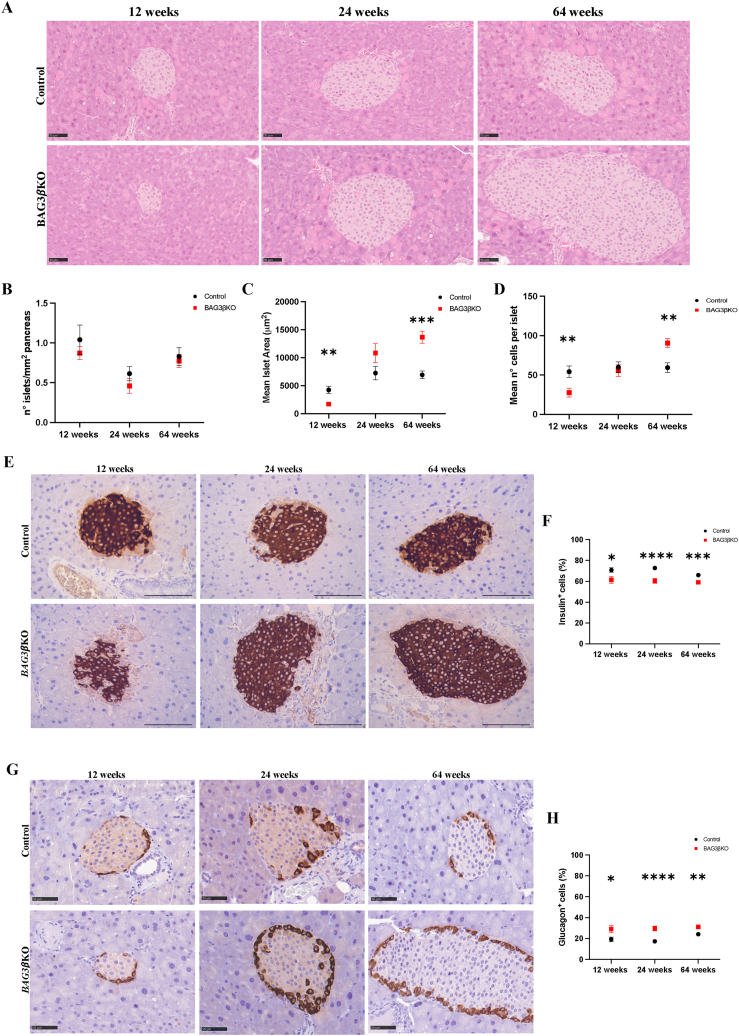
Pancreatic islets are known to adapt to insulin resistance mainly through β-cell hyperplasia and hypertrophy [23]. Histological analysis of pancreatic tissue in BAG3βKO and control mice (Figure 3A,F and 3I) shows that the number of endocrine islets remained almost constant at any age in both groups (Figure 3B); on the contrary, islets' size varied significantly, increasing with age in BAG3βKO mice. In 12-weeks old BAG3βKO mice the mean area of islets was smaller, possibly reflecting a developmental deficit resulting from constitutive deletion of BAG3 in β-cells. However, at later time points islets appeared to recover a normal size and morphology and at 64-weeks of age the size was more than doubled most likely adapting to insulin resistance (Figure 3C). The mean of the total cell number per islet reflected the same trend (Figure 3D). Over time, a more detailed analysis of islets' dimensions showed an increase in the proportion of large islets and a corresponding decrease in the number of smaller islets in BAG3βKO mice (Figure. S2A). Then, we performed multiple immunofluorescence staining to detect insulin-secreting β-cells, glucagon-secreting α-cells, and somatostatin-secreting δ-cells (Figure. S2B). The islets’ architecture was similar in the two groups, with the core formed by β-cells and the periphery populated by α- and δ-cells. BAG3βKO displayed overall conservation of normal cell distribution but in some BAG3βKO mice rare α- and δ-cells were intermingled with β-cells resembling the histological features found in pancreatic islets of obese and diabetic mice. Conversely, the fraction of β- and α-cells within the islets varied between BAG3βKO and control mice (Figure 3E,G). To better quantify these differences, we performed image analysis on pancreatic tissues stained with antibodies for insulin and glucagon, and we plotted the number of positive cells against the total number of islet cells. The percentage of insulin and glucagon-producing cells remains constant over time, with a significantly higher percentage of insulin-producing cells (Figure 3F) and a lower percentage of glucagon-producing cells (Figure 3H) in control mice compared to BAG3βKO mice. Finally, insulin content of isolated islets appears to be reduced in BAG3βKO (Figure. S3). Although not quantitative, this result is in line whit what observed in the insulinoma model [12].
Figure 3.
Pancreatic islets morphology evaluation at 12, 24, and 64 weeks of age in BAG3βKO mice compared to control mice. (A) Representative images of hematoxylin-eosin-stained (H&E) pancreatic sections. Scale bar: 50 μm. (B) The number of Langerhans islets per section of pancreas. (C) Mean area of islets. (D) Mean number of cells per islet. (E) Representative images of insulin-stained pancreatic sections. Scale bar: 100 μm (F) Percentage of insulin-positive cells. (G) Representative images of glucagon-stained pancreatic sections. Scale bar: 100 μm. (H) Percentage of glucagon-positive cells. Data are presented as mean values ± SEM; n = 3 for 12 weeks group, n = 8–7 for 24 weeks group, n = 16–14 for 64 weeks group ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
3.4. Functional proteomics of the liver and muscle reveals peripheral insulin resistance
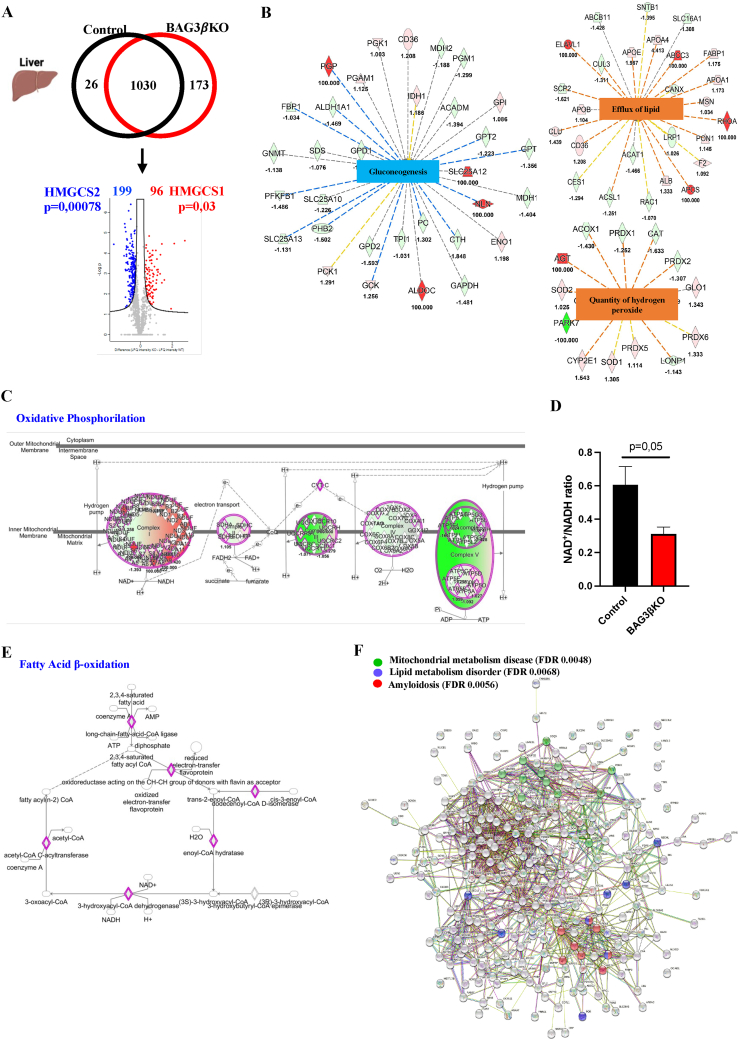
We used a proteomic approach to identify altered metabolic pathways in the muscle and liver of 64-weeks-old BAG3βKO mice when BAG3βKO mice showed a significantly increased HOMA-IR. Pooled homogenates muscle and liver taken from BAG3βKO and control mice muscle and liver were analyzed with nanoLC-Orbitrap-MS/MS. A total of 1229 proteins were identified in livers (Table S1). Among the 1030 common proteins, statistical analysis revealed 295 differentially expressed proteins (DEPs), 96 were up-regulated and 199 were down-regulated in BAG3βKO mice (Figure 4A). Among DEPs we observed decreased levels of hydroxymethylglutaryl CoA synthase 2, mitochondrial (HMGCS2) (p = 0.00078), involved in ketone body production. In contrast, the isoenzymes hydroxymethylglutaryl-CoA synthase 1 (HMGCS1), located in the cytoplasm and involved in sterol biosynthesis, was significantly increased (p = 0.03) (Figure 4A). Ingenuity Pathway Analysis (IPA) was used to identify predicted altered pathways (Table S2) by using all the quantified proteins. The downstream analysis revealed a potent inhibition of gluconeogenesis (z score = −2.745). At the same time, the efflux of lipids (z score = 2) and the quantity of hydrogen peroxide (z score = 2.097) were significantly increased (Figure 4B). Moreover, among the predicted canonical pathways, oxidative phosphorylation (z score = −2,47) was significantly inhibited in BAG3βKO mice (Figure 4C). It is known that this inhibition increases NADH concentration. To validate the predicted oxidative phosphorylation decrease, we quantified the NAD+/NADH ratio in hepatic tissue extracts of BAG3βKO and control mice. As expected, the livers of BAG3βKO mice presented more NADH than control mice (Figure 4D). The altered NAD+/NADH ratio is known to inhibit fatty acid oxidation [24], a canonical pathway also predicted to be significantly repressed (z score = −2.55), promoting fat accumulation in the liver (Figure 4E). Then, we used the STRING tool to rebuild interactions that occurred among the 173 proteins found only in BAG3βKO livers showing, oxidation-reduction process (FDR = 0.00089), intracellular transport (FDR = 0.0012) and regulation of cellular response to insulin stimulus (FDR = 0.022) interaction clusters (PPI p = 2.53 × 10−09) (Figure. S3A). We then integrated the 173 unique proteins with the up-regulated DEPs and found amyloidosis (FDR = 0.0056), lipid metabolism disorders (FDR = 0.0068) and mitochondrial metabolism disease (FDR = 0.0048) as disease–gene associations clusters (Figure 4F). When we performed the same analysis on the 26 unique proteins of the control group, no significant clusters of interaction were found (Figure. S3B). Taken together the findings in BAG3βKO are in line with liver properly responding to insulin in a situation (starvation) where insulin should not be present confirming a situation of hyperinsulinemia.
Figure 4.
Liver proteomics of BAG3βKO and Control mice. (A) Venn diagram and volcano plot showing the number of proteins and DEPs detected in livers of 64-weeks old control and BAG3βKO mice. Down-regulated and up-regulated DEPs identified by MS/MS analysis are highlighted in blue and red, respectively. HMGCS2 and HMGCS1 are highlighted. (B) Significantly inhibited and increased downstream functions predicted by IPA analysis of DEPs. Orange nodes and edges indicate activation, while blue nodes and edges indicate inhibition. Yellow edges reveal an inconsistent relationship between the findings and the state of the downstream node; instead, the gray ones indicate no predicted effects (C) Oxidative phosphorylation as decreased canonical pathway predicted by IPA. (D) NAD+/NADH ratio was calculated from liver extracts of control and BAG3βKO mice; data are presented as mean values ± SEM; n = 5 for control and n = 6 for BAG3βKO mice. (E) Fatty acid oxidation as decreased canonical pathway predicted by IPA. (F) Disease-gene association identified by STRING among the unique proteins found in BAG3βKO in addition to the significant upregulated DEPs.
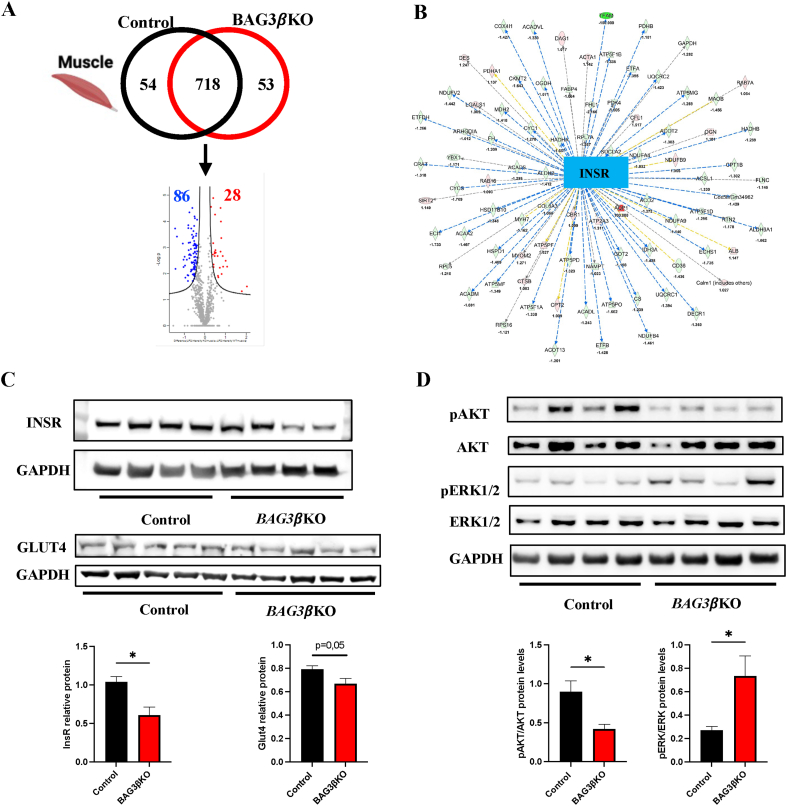
Muscle proteomic profiling identified a total of 825 proteins (Table S3). Among the 718 common proteins, 114 were differentially expressed, in particular, 86 were up-regulated and 28 down-regulated (Figure 5A). Interestingly, IPA predicted the inhibition of the insulin receptor (INSR) (z score = −5.404) as shown in Figure 5B (Table S4). Indeed, Western blot analysis showed reduced expression of INSR and of one of its direct targets, the glucose transporter type 4 (GLUT4), in the muscles of BAG3βKO mice compared to control mice (Figure 5C). Moreover, we examined the changes in the levels of (Protein kinase B) AKT phosphorylation. AKT is known to play a central role in the in the transduction pathway finally leading to GLUT4 translocation [25]. As shown by Western blot analysis, BAG3βKO mice showed a significant reduction in AKT phosphorylation than control mice (Figure 5D). Furthermore, we analyzed the phosphorylation state of extracellular-signal-regulated kinase (ERK)1/2 protein. Interestingly we found a significant increase of ERK1/2 phosphorylation in BAG3βKO mice muscle (Figure 5D). These results suggest a condition of muscle insulin resistance [26]. Notably, proteomic profiling of the liver did not show inhibition of the insulin receptor pathway, and no significant reduction of INSR expression is observed in the livers of BAG3βKO mice (Figure. S4), suggesting that the liver is still insulin responsive at this stage.
Figure 5.
BAG3βKO mice displayed peripheral but not hepatic insulin resistance. (A) Venn diagram and volcano plot showing the number of proteins and DEPs detected in muscles of 64-weeks old control and BAG3βKO mice. Down-regulated and up-regulated DEPs identified by MS/MS analysis are highlighted in blue and red, respectively. (B) Significantly inhibited upstream predicted by IPA analysis. (C, D) Representative Western blot of muscle INSR, GLUT4 phospho-AKT/AKT, and phospho-ERK1/2/ERK1/2 expression in control and BAG3βKO mice. Relative protein expression data are presented as mean values ± SEM; n = 7–8 for each group; ∗p < 0.05.
3.5. BAG3βKO mice show altered circulating branched-chain amino acids (BCAAs) profile
Using a mass spectrometry-based metabolite profiling platform, we evaluated the changes in the profile of blood acyl-carnitines, free carnitines and amino acids of fasted control and BAG3βKO mice at 12, 24, and 64 weeks of age as reported in Table 1. Data were analyzed using two-way ANOVA and Bonferroni multiple comparisons test. Changes in acyl-carnitines and free carnitines were not significant. Conversely, blood leucine (LEU), isoleucine (ILE), and valine (VAL) concentrations, which belong to the BCAAs group [27], were significantly lower in BAG3βKO mice when compared to control mice at 12 weeks of age (LEU\ILE\PRO-OH p < 0.0001 and VAL p = 0.0166) and at 24 weeks of age (LEU\ILE\PRO-OH p = 0.0344) in line with a primary hyperinsulinism [28]. Circulating BCAAs concentrations were significantly increased in BAG3βKO mice at 64 weeks of age (LEU\ILE\PRO-OH p < 0.0001; VAL p < 0.0001) in accordance with the knowledge that elevated circulating BCAAs are associated with insulin resistance [29,30]. Normalization of the results by calculating the ratio between the concentrations of BCAAs and alanine and the ratio between the BCAAs and phenylalanine, an aromatic amino acid, confirms that the changes in BCAAs levels were significant. Moreover, at 24 weeks of age, BAG3βKO mice also presented lower levels of glycine (p = 0.0008). However, at 64 weeks, also glycine resulted significantly increased in BAG3βKO mice (GLY p = 0.0003).
Table 1.
Metabolomic signature of BAG3βKO mice.
| 12 weeks |
24 weeks |
64 weeks |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Control | BAG3βKO | p | Control | BAG3βKO | p | Control | BAG3βKO | p | |
| ALA | 122 ± 4.5 | 128.8 ± 7.2 | ns | 132.7 ± 5.92 | 124.2 ± 9.7 | ns | 141.9 ± 13.3 | 173.5 ± 11.6 | ns |
| ARG | 84.2 ± 1.93 | 80.4 ± 3.05 | ns | 80.07 ± 3.24 | 72.7 ± 3.3 | ns | 85.0 ± 6.8 | 94.5 ± 6.8 | ns |
| CIT | 31.8 ± 1.88 | 35.3 ± 2.65 | ns | 33.7 ± 1.67 | 36.02 ± 3.9 | ns | 33.5 ± 3.0 | 37.72 ± 2.6 | ns |
| GLN\LYS | 360.3 ± 18.65 | 352.06 ± 20.02 | ns | 455.5 ± 27.4 | 470.4 ± 23.3 | ns | 503.77 ± 47.33 | 504.65 ± 38.77 | ns |
| GLU | 64.1 ± 4.32 | 62.6 ± 4.04 | ns | 82.1 ± 4.1 | 83.4 ± 3.5 | ns | 98.8 ± 5.8 | 107.51 ± 9,0 | ns |
| GLY | 196.7 ± 9.43 | 198.8 ± 8.62 | ns | 243.5 ± 19.3 | 212.64 ± 18.12 | 0.0008∗∗∗ | 219.62 ± 7.49 | 265.18 ± 19.6 | 0.0003∗∗∗ |
| LEU\ILE\PRO-OH | 220.2 ± 8.59 | 180.2 ± 9.54 | < 0.0001∗∗∗∗ | 210.96 ± 10.29 | 186.72 ± 15.72 | 0.0344∗ | 167.6 ± 14.7 | 235.6 ± 8.3 | < 0.0001∗∗∗∗ |
| MET | 20.7 ± 1.17 | 21.7 ± 1.24 | ns | 27.22 ± 2.8 | 24.8 ± 1.80 | ns | 21.4 ± 0.83 | 21.3 ± 1.6 | ns |
| ORN | 23.5 ± 1.15 | 23.01 ± 0.88 | ns | 24.6 ± 2.3 | 26.7 ± 3.5 | ns | 31.0 ± 1.9 | 43.8 ± 1.32 | ns |
| PHE | 52.80 ± 1.51 | 54.2 ± 1.99 | ns | 56.2 ± 2.8 | 53.42 ± 2.4 | ns | 56.9 ± 3.9 | 68.99 ± 2.2 | ns |
| PRO | 30.64 ± 1.84 | 33.90 ± 2.79 | ns | 35.2 ± 2.3 | 36.67 ± 2.87 | ns | 41.66 ± 2.6 | 49.06 ± 2.63 | ns |
| SA | 0.50 ± 0.04 | 0.61 ± 0.05 | ns | 0.53 ± 0.05 | 0.46 ± 0.05 | ns | 0.45 ± 0.03 | 0.51 ± 0.04 | ns |
| TYR | 38.7 ± 1.71 | 40.5 ± 5.22 | ns | 41.9 ± 3.8 | 44.0 ± 7.3 | ns | 56.2 ± 3.7 | 71.32 ± 4.22 | ns |
| VAL | 161.9 ± 5.014 | 144.3 ± 6.57 | 0.0166∗ | 157.1 ± 6.08 | 136.8 ± 10.2 | ns | 125.85 ± 10.60 | 200.9 ± 10.6 | < 0.0001∗∗∗∗ |
| ASA-Total | 0.24 ± 0.03 | 0.31 ± 0.04 | ns | 0.13 ± 0.04 | 0.16 ± 0.03 | ns | 0.318 ± 0.05 | 0.27 ± 0.03 | ns |
| ADO | 0.16 ± 0.012 | 0.15 ± 0.02 | ns | 0.201 ± 0.01 | 0.23 ± 0.02 | ns | 0.18 ± 0.02 | 0.25 ± 0.05 | ns |
| C0 | 10.7 ± 0.88 | 10.2 ± 0.65 | ns | 9.83 ± 0.38 | 9.71 ± 0.68 | ns | 14.52 ± 1.3 | 15.90 ± 0.65 | ns |
| C10 | 0.04 ± 0.002 | 0.042 ± 0.003 | ns | 0.045 ± 0.002 | 0.039 ± 0.002 | ns | 0.039 ± 0.002 | 0.038 ± 0.002 | ns |
| C10:1 | 0.05 ± 0.001 | 0.05 ± 0.004 | ns | 0.06 ± 0.001 | 0.061 ± 0.005 | ns | 0.049 ± 0.002 | 0.046 ± 0.001 | ns |
| C10:2 | 0.0042 ± 0.0002 | 0.0052 ± 0.0005 | ns | 0.005 ± 0.001 | 0.005 ± 0.0007 | ns | 0.0037 ± 0.0004 | 0.003 ± 0.0004 | ns |
| C2 | 26.08 ± 1.8 | 24.49 ± 1.03 | ns | 22.9 ± 0.63 | 25.5 ± 1.9 | ns | 29.68 ± 0.89 | 33.60 ± 0.88 | ns |
| C3 | 1.21 ± 0.13 | 1.12 ± 0.14 | ns | 1.33 ± 0.04 | 0.92 ± 0.12 | ns | 1.46 ± 0.21 | 1.52 ± 0.09 | ns |
| C3DC\C4OH | 0.32 ± 0.018 | 0.32 ± 0.03 | ns | 0.30 ± 0.02 | 0.30 ± 0.01 | ns | 0.43 ± 0.04 | 0.51 ± 0.04 | ns |
| C4 | 0.29 ± 0.04 | 0.33 ± 0.06 | ns | 0.13 ± 0.004 | 0.14 ± 0.013 | ns | 0.53 ± 0.03 | 0.83 ± 0.07 | ns |
| C4DC\C5OH | 0.14 ± 0.004 | 0.14 ± 0.005 | ns | 0.14 ± 0.003 | 0.14 ± 0.007 | ns | 0.17 ± 0.01 | 0.17 ± 0.003 | ns |
| C5 | 0.07 ± 0.004 | 0.068 ± 0.006 | ns | 0.09 ± 0.007 | 0.08 ± 0.005 | ns | 0.11 ± 0.01 | 0.131 ± 0.008 | ns |
| C5:1 | 0.001 ± 0.0002 | 0.002 ± 0.0004 | ns | 0.0032 ± 0.001 | 0.0028 ± 0.0005 | ns | 0.003 ± 0.0004 | 0.003 ± 0.0005 | ns |
| C5DC\C6OH | 0.073 ± 0.005 | 0.069 ± 0.007 | ns | 0.073 ± 0.004 | 0.065 ± 0.003 | ns | 0.08 ± 0.008 | 0.106 ± 0.006 | ns |
| C6 | 0.077 ± 0.007 | 0.079 ± 0.006 | ns | 0.101 ± 0.006 | 0.081 ± 0.004 | ns | 0.11 ± 0.008 | 0.129 ± 0.007 | ns |
| C6DC | 0.044 ± 0.004 | 0.06 ± 0.006 | ns | 0.044 ± 0.003 | 0.039 ± 0.003 | ns | 0.067 ± 0.007 | 0.043 ± 0.003 | ns |
| C8 | 0.046 ± 0.004 | 0.045 ± 0.004 | ns | 0.055 ± 0.003 | 0.049 ± 0.005 | ns | 0.054 ± 0.002 | 0.053 ± 0.003 | ns |
| C8:1 | 0.012 ± 0.001 | 0.012 ± 0.0005 | ns | 0.011 ± 0.0002 | 0.011 ± 0.001 | ns | 0.010 ± 0.001 | 0.011 ± 0.0007 | ns |
| D-ADO | 0.012 ± 0.005 | 0.004 ± 0.002 | ns | 0.01 ± 0.004 | 0.011 ± 0.005 | ns | 0.012 ± 0.002 | 0.012 ± 0.002 | ns |
| C12 | 0.06 ± 0.004 | 0.065 ± 0.004 | ns | 0.071 ± 0.003 | 0.053 ± 0.002 | ns | 0.060 ± 0.002 | 0.061 ± 0.006 | ns |
| C12:1 | 0.036 ± 0.002 | 0.039 ± 0.002 | ns | 0.042 ± 0.003 | 0.035 ± 0.002 | ns | 0.037 ± 0.001 | 0.034 ± 0.0006 | ns |
| C14 | 0.22 ± 0.011 | 0.26 ± 0.02 | ns | 0.26 ± 0.01 | 0.21 ± 0.01 | ns | 0.22 ± 0.01 | 0.24 ± 0.028 | ns |
| C14:1 | 0.11 ± 0.007 | 0.13 ± 0.006 | ns | 0.15 ± 0.008 | 0.12 ± 0.003 | ns | 0.125 ± 0.003 | 0.12 ± 0.01 | ns |
| C14:2 | 0.033 ± 0.003 | 0.035 ± 0.002 | ns | 0.05 ± 0.003 | 0.039 ± 0.001 | ns | 0.031 ± 0.001 | 0.033 ± 0.003 | ns |
| C14OH | 0.018 ± 0.001 | 0.021 ± 0.002 | ns | 0.02 ± 0.001 | 0.019 ± 0.002 | ns | 0.0227 ± 0.003 | 0.026 ± 0.003 | ns |
| C16 | 1.53 ± 0.11 | 1.6 ± 0.08 | ns | 1.59 ± 0.08 | 1.54 ± 0.13 | ns | 1.31 ± 0.06 | 1.48 ± 0.11 | ns |
| C16:1 | 0.17 ± 0.008 | 0.21 ± 0.009 | ns | 0.24 ± 0.02 | 0.201 ± 0.007 | ns | 0.22 ± 0.02 | 0.228 ± 0.02 | ns |
| C16:1OH\C17 | 0.05 ± 0.002 | 0.057 ± 0.003 | ns | 0.076 ± 0.006 | 0.0604 ± 0.003 | ns | 0.056 ± 0.004 | 0.053 ± 0.006 | ns |
| C16OH | 0.078 ± 0.002 | 0.076 ± 0.007 | ns | 0.061 ± 0.005 | 0.054 ± 0.002 | ns | 0.072 ± 0.01 | 0.066 ± 0.010 | ns |
| C18 | 0.27 ± 0.02 | 0.29 ± 0.015 | ns | 0.32 ± 0.018 | 0.292 ± 0.03 | ns | 0.24 ± 0.01 | 0.260 ± 0.02 | ns |
| C18:1 | 0.56 ± 0.032 | 0.62 ± 0.03 | ns | 0.77 ± 0.04 | 0.69 ± 0.03 | ns | 0.70 ± 0.04 | 0.801 ± 0.08 | ns |
| C18:1OH | 0.06 ± 0.002 | 0.059 ± 0.006 | ns | 0.06 ± 0.006 | 0.055 ± 0.005 | ns | 0.06 ± 0.01 | 0.06 ± 0.01 | ns |
| C18:2 | 0.27 ± 0.01 | 0.29 ± 0.01 | ns | 0.43 ± 0.02 | 0.38 ± 0.01 | ns | 0.24 ± 0.02 | 0.30 ± 0.029 | ns |
| C18:2OH | 0.023 ± 0.001 | 0.027 ± 0.003 | ns | 0.03 ± 0.004 | 0.029 ± 0.002 | ns | 0.023 ± 0.003 | 0.024 ± 0.005 | ns |
| C18OH | 0.017 ± 0.001 | 0.019 ± 0.002 | ns | 0.019 ± 0.002 | 0.015 ± 0.001 | ns | 0.018 ± 0.002 | 0.020 ± 0.003 | ns |
| C20 | 0.19 ± 0.013 | 0.32 ± 0.03 | ns | 0.026 ± 0.002 | 0.027 ± 0.002 | ns | 0.012 ± 0.001 | 0.018 ± 0.003 | ns |
| C20:0-LPC | 1.25 ± 0.05 | 0.88 ± 0.11 | ns | 2.38 ± 0.25 | 1.82 ± 0.27 | ns | 0.93 ± 0.08 | 0.998 ± 0.07 | ns |
| C22 | 0.007 ± 0.001 | 0.007 ± 0.0005 | ns | 0.012 ± 0.002 | 0.013 ± 0.001 | ns | 0.008 ± 0.0004 | 0.009 ± 0.001 | ns |
| C22:0-LPC | 0.91 ± 0.04 | 0.71 ± 0.05 | ns | 1.42 ± 0.09 | 1.53 ± 0.04 | ns | 0.66 ± 0.04 | 0.65 ± 0.06 | ns |
| C24 | 0.01 ± 0.001 | 0.010 ± 0.0007 | ns | 0.017 ± 0.003 | 0.018 ± 0.0013 | ns | 0.011 ± 0.001 | 0.014 ± 0.001 | ns |
| C24:0-LPC | 1.60 ± 0.05 | 1.29 ± 0.09 | ns | 2.46 ± 0.2 | 2.41 ± 0.21 | ns | 1.16 ± 0.11 | 1.31 ± 0.08 | ns |
| C26 | 0.003 ± 0.0008 | 0.005 ± 0.001 | ns | 0.0032 ± 0.0008 | 0.0042 ± 0.001 | ns | 0.007 ± 0.001 | 0.008 ± 0.0005 | ns |
| C26:0-LPC | 0.37 ± 0.05 | 0.38 ± 0.019 | ns | 0.503 ± 0.04 | 0.37 ± 0.04 | ns | 0.26 ± 0.04 | 0.30 ± 0.03 | ns |
| LEU-ILE-PRO-OH/ALA | 1.82 ± 0.11 | 1.42 ± 0.11 | 0.037∗ | 1.60 ± 0.09 | 1.54 ± 0.17 | ns | 1.19 ± 0.05 | 1.38 ± 0.09 | ns |
| VAL/ALA | 1.34 ± 0.07 | 1.14 ± 0.092 | ns | 1.19 ± 0.05 | 1.13 ± 0.11 | ns | 0.89 ± 0.06 | 1.18 ± 0.08 | 0.038∗ |
| LEU-ILE-PRO-OH/PHE | 4.17 ± 0.14 | 3.35 ± 0.24 | 0.018∗ | 3.76 ± 0.11 | 3.49 ± 0.2 | ns | 2.95 ± 0.16 | 3.42 ± 0.08 | 0.022∗ |
| VAL/PHE | 3.07 ± 0.101 | 2.68 ± 0.19 | ns | 2.80 ± 0.05 | 2.56 ± 0.14 | ns | 2.22 ± 0.15 | 2.91 ± 0.13 | 0.0093∗∗ |
Values are mean ± SEM; n = 5 for 12 weeks group, n = 5 for 24 weeks group, n = 4–6 for 64 weeks group.
3.6. BAG3βKO mice show histopathological in liver and kidney
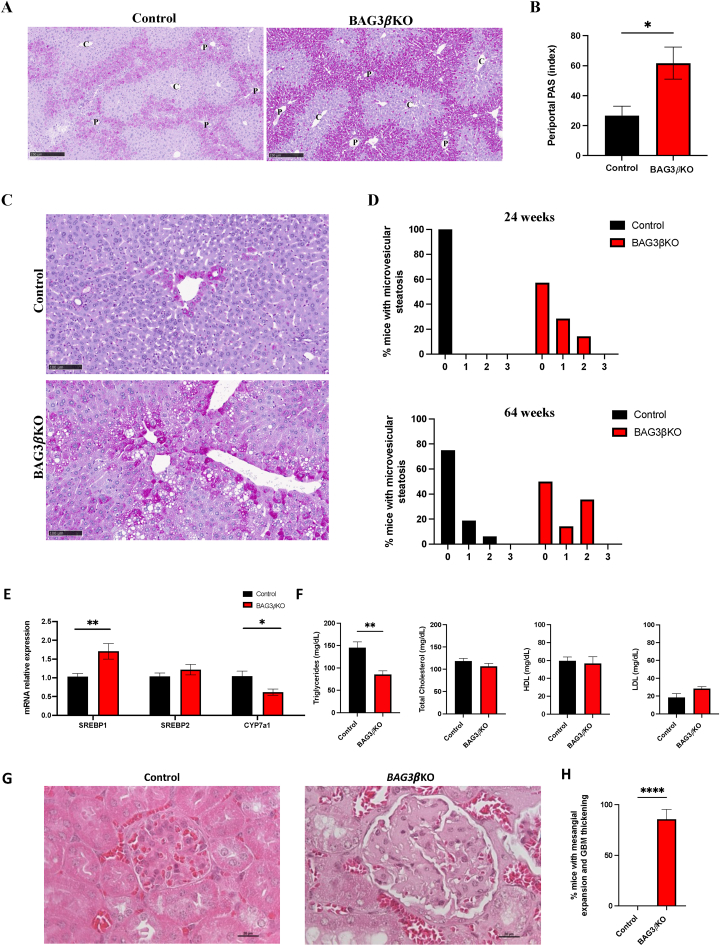
It is known that tight regulation of glycogen deposition and degradation plays an important role in blood glucose control [31]. We, therefore, investigated glycogen storage by Periodic Acid Schiff (PAS) staining of liver sections. The analysis of glycogen distribution in mouse liver after 16 h of fasting showed, at 12 weeks of age, the almost complete disappearance of glycogen from the liver of both control and BAG3βKO mice, with only scattered, sporadic PAS positive hepatocytes starting to re-accumulate glycogen around the central vein (Figures. S5A, S5B, S5C and S5D). At 24 weeks of age, the recovery of glycogen stores in pericentral hepatocytes appears to be more relevant in both groups with a slight, non-statistically significant increase in BAG3βKO mice (Figure. S5C). At 64 weeks of age, the glycogen content of the liver was further increased both in control and KO mice but appeared higher in BAG3βKO mice (Figure. S5D). Ten out of 16 (62.5%) control mice and 6 out of 14 (43%) BAG3βKO mice showed a similar amount of glycogen accumulated in pericentral hepatocytes. Notably, six out of 16 (37.5%) mice in the control group and 8 out of 14 (57%) mice in the BAG3βKO group showed a periportal glycogen accumulation (Figure 6A), significantly more pronounced in BAG3βKO mice (Figure 6B), underlining a major difference in glycogen metabolism, in line with the observed insulin sensitivity in liver. Next, we analyzed the liver proteomic data focusing on the enzymes involved in glycogen metabolism and whether the different protein expression could explain the increase in periportal glycogen accumulation. No significant alterations were found in glycogen synthase, glycogen phosphorylase and 1,4-alpha-glucan-brancing enzyme protein expression (Figure. S5E) suggesting that the difference in periportal glycogen accumulation is probably due to the different activity of these enzymes. PAS staining also highlighted the presence of a more prominent microvesicular steatosis in BAG3βKO livers compared to controls (Figure 6C). Indeed, at 24 weeks of age no steatosis is observed in control mice, while 43% of BAG3βKO mice showed steatosis of levels 1 and 2 of severity evaluated as described in material and methods (Figure 6D, upper histogram). At 64 weeks of age 25% of control mice presented steatosis of levels 1 and 2, while this percentage reached 50% in BAG3βKO mice (Figure 6D, lower histogram). Evaluation of expression of genes encoding for key regulators of lipid metabolism, as well as evaluation of circulating lipid levels in 64-weeks-old BAG3βKO mice compared to controls, support the biochemical and morphological alterations described above. In particular, we observed: overexpression of SREBP1 but not of SREBP2 and downregulation of CYP7a1 (Figure 6E); significantly lower plasma triglycerides and no differences in concentrations of total cholesterol, high-density lipoproteins (HDL) and low-density lipoproteins (LDL) (Figure 6F). These results are again in line with insulin responsiveness.
Figure 6.
Histopathological evaluation of liver and kidney in the new BAG3βKO mouse model. (A) Representative images of PAS-stained livers of 64 weeks old BAG3βKO and control mice: portal triad (P), central vein (C). Scale bar: 250 μm. (B) Quantification of liver glycogen accumulation around the portal triads in BAG3βKO mice compared to control mice. Data are presented as mean values ± SEM; n = 16–14 for 64 weeks group; ∗p < 0.05. (C) PAS-stained liver tissue sections of control and BAG3βKO mice at 64 weeks of age showing microvesicular liver steatosis in a BAG3βKO mouse. Scale bar: 100 μm. (D) Histograms showing the percentage of BAG3βKO and control mice presenting microvesicular steatosis at 24 and 64 weeks of age. Microvesicular steatosis was scored from 0 to 3 based on the percentage of the total area affected: 0 (<5%), 1 (5–33%), 2 (34–66%) and 3 (>66%). € mRNA levels evaluated by real-time PCR for SREBP1, SREBP2, and CYP7a. Data are presented as mean values ± SEM; n = 4 for each group; ∗p < 0.05; ∗∗p < 0.01. (F) Levels of plasma triglycerides, total cholesterol, HDL, and LDL in 64 weeks old BAG3βKO and control mice. Data are presented as mean values ± SEM; n = 7 for each group; ∗∗p < 0.01. (G) Representative H&E-stained kidney sections from control and BAG3βKO mice showing mesangial matrix expansion and thickening of the glomerular basement membrane in 64 weeks old BAG3βKO mouse. Scale bar: 20 mm. (H) Percentage of mice, showing mesangial matrix expansion and/or thickening of the glomerular basement membrane. Data are presented as mean values ± SEM; n = 14–16 for each group; ∗∗∗∗p ≤ 0.0001.
We also detected morphological alterations in kidneys from BAG3βKO mice. While the histological tissue features of renal glomeruli and tubules of BAG3βKO mice were not distinguishable from their control littermates at 12 and 24 weeks of age (data not shown), at 64 weeks of age, 12 out of 15 (85.7%) BAG3βKO mice showed mesangial matrix expansion and/or thickening of the glomerular basement membrane (Figure 6G,H). Mesangial matrix expansion is a well-known pathological hallmark of CKD, a common complication in insulin resistance [32,33]. Moreover, though only in one BAG3βKO mouse, we observed tubular atrophy and tubulointerstitial fibrosis (Figure. S6).
4. Discussion
Following our previous work showing the involvement of BAG3, as part of the SNARE complex, in insulin secretion we knocked out BAG3 selectively in pancreatic beta cells of mice. BAG3βKO mice breed normally and appear to have normal development and a regular growth rate compared to their control counterpart, however, show increased circulating insulin levels. Under fasting conditions circulating insulin levels progressively increased in BAG3βKO compared to control mice during aging. Glucose levels of BAG3-deficient mice appear normal under fed conditions while appear to be lower than controls during the first 12 weeks of age, suggesting an exposure to higher levels of insulin in line with what we previously showed in an insulinoma model [12]. Later, compensation mechanisms occur, and at 24 weeks glucose levels in starved KO mice are comparable to those of controls. A IPGTT test confirms the ability of BAG3βKO to maintain a normal glucose homeostasis. However, the IPITT test showed impaired response to insulin starting from 24 weeks of age, suggesting that mice are becoming insulin resistant, possibly to compensate for increased insulin secretion. Indeed, BAG3βKO mice at 64 weeks of age show an increased HOMA-IR index, strongly suggestive of insulin resistance.
In addition to altering the SNARE complex, loss of BAG3 may result in impaired autophagy and this can contribute to the observed phenotype [34]. Indeed, we observed reduced levels of p62/SQSTM1 a key player in autophagy (Figure. S2) [35,36]. Autophagy is necessary to maintain structure, mass, and function of pancreatic beta cells [37,38] and is known to be regulated by BAG3 in concert with the ubiquitin-binding protein p62/SQSTM1 [39]. The decrease of p62/SQSTM1 strongly suggests a role for altered autophagy in this model, though further work is required to fully elucidate this potential mechanism.
Analysis of muscle and liver, show important differences suggesting that at least up to 64 weeks of age, the liver is still appropriately responding to insulin while muscle has become resistant. Insulin acts on liver metabolism mainly by activating glycogen synthesis, increasing lipogenic gene expression, and decreasing gluconeogenic gene expression [[40], [41], [42]]. Proteomics analysis of BAG3βKO mice livers revealed an inhibition of gluconeogenesis, beta-oxidation, oxidative phosphorylation, and ketone body production, whereas the lipid efflux and sterol biosynthesis were predicted to be activated. These changes found in BAG3βKO are associated with liver properly responding to insulin stimulus, despite the analysis was performed in mice under starving conditions, in line with the finding of higher basal insulin levels. On the contrary proteomic profiling of muscle cells indicates a reduced activity of the insulin receptor pathway, indeed INSR and GLUT4 expression, as well as AKT phosphorylation, are decreased in muscle. Moreover, we found higher ERK1/2 phosphorylation in BAG3βKO muscles. The activation of ERK1/2 pathway has been seen to dramatically suppress INSR expression [43]. Exposure to higher levels of insulin both in basal conditions and in response to glucose intake, however, results in abnormal accumulation of glycogen and lipids in the liver and reduced levels of circulating triglycerides. It is well known that diabetic liver accumulates more glycogen than healthy liver under normal and fasting states [44]. Accordingly, we found elevated periportal glycogen accumulation in BAG3βKO mice, probably due to structural variations under fasting conditions that affect glycogen storage [45]. Lipid metabolism and homeostasis in the liver are mainly regulated by sterol regulatory-element binding proteins (SREBPs), a family of transcription factors that control the expression of enzymes required for endogenous cholesterol, fatty acid, triglycerides, and phospholipid synthesis [46,47]. The SREBP family comprises two SREBP1 splice variants, SREBP1a and SREBP1c, and SREBP2. The livers of BAG3βKO mice exhibit increased mRNA levels of SREBP1, which mainly increases transcription of genes involved in fatty acid and triglycerides synthesis, but not in SREBP2 mRNA levels, which primarily targets genes involved in the cholesterol biosynthetic pathway. Furthermore, Cyp7a1 gene expression, the rate-limiting enzyme in the process of hepatic bile acids synthesis, was repressed. At the same time, it has been seen that CYP7a1 overexpression in the liver prevents high-fat diet-induced obesity, fatty liver, and insulin resistance in mice [48]. Collectively our data on BAG3βKO mice liver metabolism indicate that these could be signs of an early stage of non-alcoholic fatty liver disease (NAFLD). Our proteomic data also suggest activation of complex I and a block of the other oxidative phosphorylation complexes leading to a decreased NAD+/NADH ratio that we confirm in the livers of BAG3βKO mice. The altered NAD+/NADH ratio inhibits fatty acid oxidation, promoting fat accumulation in the liver. Accumulation of fat in the liver can predispose to subsequent oxidative damage already present due to the NAD+/NADH redox imbalance in mitochondria, thus creating a vicious circle. This supports studies suggesting that increased NAD+ levels may exhibit ameliorative effects on metabolic disorders, such as T2D, metabolic syndrome, and NAFLD [49].
The unregulated insulin secretion from pancreatic β-cells leading to hyperinsulinemic hypoglycemia is the basis of congenital hyperinsulinism (CHI), a heterogenous and complex disorder [50,51]. To date mutations in at least 11 different key genes that are involved in regulating insulin secretion have been found responsible for CHI [52]. CHI occurs also in some congenital disorders of glycosylation and syndromic conditions, however in many cases the underlying molecular defects remains unclear. Therefore, defects of BAG3 might be involved in the pathogenesis of CHI, since the phenotype of KO mice show primary hyperinsulinism at an early age. Presumably the need for additional insulin production when mice become insulin resistant, results in an increase in the size of pancreatic islets in BAG3βKO mice, in line with other studies showing that pancreatic islets undergo morphological and metabolic changes to maintain normoglycemia [23,53,54]. Reduced levels of circulating BCAAs at 12 weeks of age are also in line with primary hyperinsulinism, as a result of insulin-inhibited gluconeogenesis with consequent amino acid release by skeletal muscle 39 [55]. Conversely, as mice become insulin resistant, they display increased circulating levels of BCAAs, accordingly to this condition 18 [27]. BAG3βKO mice also displayed high levels of circulating glycine, in contrast to what has been reported in association with obesity, T2D, and NAFLDs 40 [56]. However, our mice show an insulin resistant state, but they do not present obesity, T2D or a clear NAFLDs.
The altered metabolic situation described above also results in a renal glomerulopathy, characterized by mesangial matrix expansion and thickening of the glomerular basement membrane, which are the most important renal structural changes in type 1 diabetic patients with diabetic nephropathy and hallmarks of a Diabetic Kidney Disease (DKD) 41 [57]. Intriguingly, the association of severe insulin resistance (SIR) with features of DKD has been observed in T2D patients 42 [58]. Thickening of the glomerular basement membrane and mesangial expansion are also pathological features of CKD. The typical histopathological changes associated with CKD include glomerulosclerosis and tubulointerstitial fibrosis, which result in the loss of normal renal architecture, microvascular capillary rarefaction, hypoxia, and tubular atrophy. It is indeed known that IR can trigger CKD even in the absence of diabetes [[59], [60], [61]].
5. Conclusions
In conclusion, we showed that selective knockout of BAG3 in pancreatic β-cells leads to impaired insulin secretion both in basal conditions and in response to glucose administration. At an early stage, primary hyperinsulinism is manifest with lower plasma glucose levels, but peripheral insulin-resistance develops overtime. Under conditions of insulin resistance, high insulin levels are required to enhance cellular glucose uptake and inhibit hepatic glucose production to avoid hyperglycemia, thus leading to a further increase of insulin levels in a vicious circle. This chronically impaired metabolic condition leads to organ damage. Remarkably, we observed impaired glycogen and lipids accumulation in the liver and renal alterations leading to renal glomerulopathy. The present work highlights the role of BAG3 in insulin secretion and shows the genesis of insulin resistance in BAG3βKO mice. Silencing of BAG3 expressed in beta cells causes early primary hyperinsulinism followed by insulin resistance over time. Therefore, BAG3 could represent a new gene involved in the pathogenesis of congenital hyperinsulinism. Furthermore, the BAG3βKO mouse represents a novel model for the study of pathogenetic mechanisms leading to multiple organ damage linked to exposure to increased insulin levels and to insulin resistance.
CRediT authorship contribution statement
V.D. and V.D.L. conceived and planned the experiment. V.D., A.L., I·C., M.C·C., F.D.L., carried out the experiments and F.D.M. was responsible for the bioinformatic analyses and B.D. contributed to samples preparation. R.L, M.I. and A.L. performed histological and immunohistochemical analysis. M.H. and M.C.T. provided reagents. P.D.B., D.M., and M.I. contributed to the interpretation of the results. M.F., A.M., C.D.-V. provided expertise and feedback and critically reviewed the manuscript. V.D. and V.D.L wrote the paper with input from all authors. V.D.L. secured funding.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2023.101752.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
I have shared the link to the data
References
- 1.Fujimoto W.Y. The importance of insulin resistance in the pathogenesis of type 2 diabetes mellitus. Am J Med. 2000;108(Suppl 6a):9–14. doi: 10.1016/S0002-9343(00)00337-5. [DOI] [PubMed] [Google Scholar]
- 2.Galicia-Garcia U., Benito-Vicente A., Jebari S., Larrea-Sebal A., Siddiqi H., Uribe K.B., et al. Pathophysiology of type 2 diabetes mellitus. Int J Mol Sci. 2020;21:1–34. doi: 10.3390/IJMS21176275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groop L. Pathogenesis of type 2 diabetes: the relative contribution of insulin resistance and impaired insulin secretion. Int J Clin Pract Suppl. 2000:3–13. [PubMed] [Google Scholar]
- 4.Roden M., Shulman G.I. The integrative biology of type 2 diabetes. Nature. 2019;576:51–60. doi: 10.1038/S41586-019-1797-8. [DOI] [PubMed] [Google Scholar]
- 5.Yan T., Huang Y., Wu J.H.Y., Zhuang X.-D., Pan X.-F. Editorial: insulin resistance, metabolic syndrome, and cardiovascular disease. Front Cardiovasc Med. 2022;9 doi: 10.3389/FCVM.2022.959680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perdomo C.M., Garcia-Fernandez N., Escalada J. Diabetic kidney disease, cardiovascular disease and non-alcoholic fatty liver disease: a new triumvirate? J Clin Med. 2021;10 doi: 10.3390/JCM10092040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seo D.H., Suh Y.J., Cho Y., Ahn S.H., Seo S., Hong S., et al. Advanced liver fibrosis is associated with chronic kidney disease in patients with type 2 diabetes mellitus and nonalcoholic fatty liver disease. Diabetes Metab J. 2022;46:630–639. doi: 10.4093/dmj.2021.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marzullo L., Turco M.C., de Marco M. The multiple activities of BAG3 protein: mechanisms. Biochim Biophys Acta Gen Subj. 2020:1864. doi: 10.1016/J.BBAGEN.2020.129628. [DOI] [PubMed] [Google Scholar]
- 9.Rosati A., Graziano V., de Laurenzi V., Pascale M., Turco M.C. BAG3: a multifaceted protein that regulates major cell pathways. Cell Death Dis. 2011;2 doi: 10.1038/cddis.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sherman M.Y., Gabai V. The role of Bag3 in cell signaling. J Cell Biochem. 2022;123 doi: 10.1002/jcb.30111. [DOI] [PubMed] [Google Scholar]
- 11.Behl C. Breaking BAG: the Co-chaperone BAG3 in Health and disease. Trends Pharmacol Sci. 2016;37 doi: 10.1016/j.tips.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Iorio V., Festa M., Rosati A., Hahne M., Tiberti C., Capunzo M., et al. BAG3 regulates formation of the SNARE complex and insulin secretion. Cell Death Dis. 2015;6 doi: 10.1038/cddis.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Youn D.Y., Lee D.H., Lim M.H., Yoon J.S., Ji H.L., Seung E.J., et al. Bis deficiency results in early lethality with metabolic deterioration and involution of spleen and thymus. Am J Physiol Endocrinol Metab. 2008;295 doi: 10.1152/ajpendo.90704.2008. [DOI] [PubMed] [Google Scholar]
- 14.Stone A.C., Noland R.C., Mynatt R.L., Velasquez S.E., Bayless D.S., Ravussin E., et al. Female mice are protected from metabolic decline associated with lack of skeletal muscle hur. Biology. 2021;10 doi: 10.3390/BIOLOGY10060543/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carter J.D., Dula S.B., Corbin K.L., Wu R., Nunemaker C.S. A practical guide to rodent islet isolation and assessment. Biol Proced Online. 2009;11:3–31. doi: 10.1007/S12575-009-9021-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tahapary D.L., Pratisthita L.B., Fitri N.A., Marcella C., Wafa S., Kurniawan F., et al. Challenges in the diagnosis of insulin resistance: focusing on the role of HOMA-IR and Tryglyceride/glucose index. Diabetes Metabol Syndr: Clin Res Rev. 2022;16 doi: 10.1016/J.DSX.2022.102581. [DOI] [PubMed] [Google Scholar]
- 17.Rossi C., Cicalini I., Zucchelli M., di Ioia M., Onofrj M., Federici L., et al. Metabolomic signature in sera of multiple sclerosis patients during pregnancy. Int J Mol Sci. 2018;19 doi: 10.3390/IJMS19113589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rossi C., Cicalini I., Rizzo C., Zucchelli M., Consalvo A., Valentinuzzi S., et al. A false-positive case of methylmalonic aciduria by tandem mass spectrometry newborn screening dependent on maternal malnutrition in pregnancy. Int J Environ Res Publ Health. 2020;17 doi: 10.3390/ijerph17103601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleiner D.E., Brunt E.M., van Natta M., Behling C., Contos M.J., Cummings O.W., et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/HEP.20701. [DOI] [PubMed] [Google Scholar]
- 20.Madonna R., Moscato S., Polizzi E., Pieragostino D., Cufaro M.C., del Boccio P., et al. 2021. (Connexin 43 and connexin 26 involvement in the ponatinib-induced cardiomyopathy: sex-related differences in a murine model). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Riverol Y., Bai J., Bandla C., García-Seisdedos D., Hewapathirana S., Kamatchinathan S., et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50 doi: 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thorens B., Tarussio D., Maestro M.A., Rovira M., Heikkilä E., Ferrer J. Ins1 Cre knock-in mice for beta cell-specific gene recombination. Diabetologia. 2015;58 doi: 10.1007/s00125-014-3468-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mezza T., Muscogiuri G., Sorice G.P., Clemente G., Hu J., Pontecorvi A., et al. Insulin resistance alters islet morphology in nondiabetic humans. Diabetes. 2014;63:994–1007. doi: 10.2337/DB13-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan L., Cacicedo J.M., Ido Y. Impaired nicotinamide adenine dinucleotide (NAD+) metabolism in diabetes and diabetic tissues: implications for nicotinamide-related compound treatment. J Diabetes Investig. 2020;11:1403–1419. doi: 10.1111/JDI.13303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernandez R., Teruel T., Lorenzo M. Akt mediates insulin induction of glucose uptake and up-regulation of GLUT4 gene expression in brown adipocytes. FEBS Lett. 2001;494:225–231. doi: 10.1016/S0014-5793(01)02353-5. [DOI] [PubMed] [Google Scholar]
- 26.Leto D., Saltiel A.R. Regulation of glucose transport by insulin: traffic control of GLUT4. Nat Rev Mol Cell Biol 2012 13:6. 2012;13:383–396. doi: 10.1038/nrm3351. [DOI] [PubMed] [Google Scholar]
- 27.Ramzan I., Ardavani A., Vanweert F., Mellett A., Atherton P.J., Idris I. The association between circulating branched chain amino acids and the temporal risk of developing type 2 diabetes mellitus: a systematic review & meta-analysis. Nutrients. 2022;14 doi: 10.3390/nu14204411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaussain J.L., Georges P., Gendrel D., Donnadieu M., Job J.C. Serum branched-chain amino acids in the diagnosis of hyperinsulinism in infancy. J Pediatr. 1980;97:923–926. doi: 10.1016/S0022-3476(80)80421-5. [DOI] [PubMed] [Google Scholar]
- 29.Lynch C.J., Adams S.H. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol 2014 10:12. 2014;10:723–736. doi: 10.1038/nrendo.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon M.S. The emerging role of branched-chain amino acids in insulin resistance and metabolism. Nutrients. 2016;8 doi: 10.3390/nu8070405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nawaz A., Zhang P., Li E., Gilbert R.G., Sullivan M.A. The importance of glycogen molecular structure for blood glucose control. iScience. 2020;24 doi: 10.1016/J.ISCI.2020.101953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spoto B., Pisano A., Zoccali C. Insulin resistance in chronic kidney disease: a systematic review. Am J Physiol Ren Physiol. 2016;311:F1087–F1108. doi: 10.1152/AJPRENAL.00340.2016/ASSET/IMAGES/LARGE/ZH20161680750005. [JPEG] [DOI] [PubMed] [Google Scholar]
- 33.Steffes M.W., Chavers B., Mauer S.M., Osterby R. Mesangial expansion as a central mechanism for loss of kidney function in diabetic patients. Diabetes. 1989;38 doi: 10.2337/diab.38.9.1077. [DOI] [PubMed] [Google Scholar]
- 34.Stürner E., Behl C. The role of the multifunctional bag3 protein in cellular protein quality control and in disease. Front Mol Neurosci. 2017;10 doi: 10.3389/FNMOL.2017.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamark T., Svenning S., Johansen T. Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem. 2017;61:609–624. doi: 10.1042/EBC20170035. [DOI] [PubMed] [Google Scholar]
- 36.Kumar A.V., Mills J., Lapierre L.R. Selective autophagy receptor p62/SQSTM1, a pivotal player in stress and aging. Front Cell Dev Biol. 2022;10 doi: 10.3389/FCELL.2022.793328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riahi Y., Wikstrom J.D., Bachar-Wikstrom E., Polin N., Zucker H., Lee M.S., et al. Autophagy is a major regulator of β-cell insulin homeostasis. Diabetologia. 2016;59:1480. doi: 10.1007/S00125-016-3868-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ebato C., Uchida T., Arakawa M., Komatsu M., Ueno T., Komiya K., et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metabol. 2008;8:325–332. doi: 10.1016/J.CMET.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 39.Gamerdinger M., Hajieva P., Kaya A.M., Wolfrum U., Hartl F.U., Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28:889–901. doi: 10.1038/EMBOJ.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel B.M., Goyal R.K. Liver and insulin resistance: new wine in old bottle. Eur J Pharmacol. 2019:862. doi: 10.1016/J.EJPHAR.2019.172657. [DOI] [PubMed] [Google Scholar]
- 41.Cherrington A.D., Edgerton D., Sindelar D.K. The direct and indirect effects of insulin on hepatic glucose production in vivo. Diabetologia. 1998;41:987–996. doi: 10.1007/s001250051021. [DOI] [PubMed] [Google Scholar]
- 42.Petersen M.C., Shulman G.I. Mechanisms of insulin action and insulin resistance. Physiol Rev. 2018;98:2133–2223. doi: 10.1152/PHYSREV.00063.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujishiro M., Gotoh Y., Katagiri H., Sakoda H., Ogihara T., Anai M., et al. Three mitogen-activated protein kinases inhibit insulin signaling by different mechanisms in 3T3-L1 adipocytes. Mol Endocrinol. 2003;17 doi: 10.1210/me.2002-0131. [DOI] [PubMed] [Google Scholar]
- 44.De Oliveira D.S., Amado C.A.B., Martini M.C., Suzuki-Kemmelmeier F., Bracht A. Glycogen levels and energy status of the liver of fasting rats with diabetes types 1 and 2. Braz Arch Biol Technol. 2007;50:785–791. doi: 10.1590/S1516-89132007000500006. [DOI] [Google Scholar]
- 45.Wang L., Liu Q., Wang M., Du Y., Tan X., Xu B., et al. Effects of fasting on liver glycogen structure in rats with type 2 diabetes. Carbohydr Polym. 2020;237 doi: 10.1016/J.CARBPOL.2020.116144. [DOI] [PubMed] [Google Scholar]
- 46.Shimano H., Sato R. SREBP-regulated lipid metabolism: convergent physiology — divergent pathophysiology. Nat Rev Endocrinol 2017 13:12. 2017;13:710–730. doi: 10.1038/nrendo.2017.91. [DOI] [PubMed] [Google Scholar]
- 47.DeBose-Boyd R.A., Ye J. SREBPs in lipid metabolism, insulin signaling, and beyond. Trends Biochem Sci. 2018;43:358–368. doi: 10.1016/J.TIBS.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li T., Owsley E., Matozel M., Hsu P., Novak C.M., Chiang J.Y.L. Transgenic expression of cholesterol 7α-hydroxylase in the liver prevents high-fat diet–induced obesity and insulin resistance in mice. Hepatology. 2010;52:678–690. doi: 10.1002/HEP.23721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie N., Zhang L., Gao W., Huang C., Huber P.E., Zhou X., et al. NAD+ metabolism: pathophysiologic mechanisms and therapeutic potential. Signal Transduct Targeted Ther 2020 5:1. 2020;5:1–37. doi: 10.1038/s41392-020-00311-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maiorana A., Dionisi-Vici C. Hyperinsulinemic hypoglycemia: clinical, molecular and therapeutical novelties. J Inherit Metab Dis. 2017;40:531–542. doi: 10.1007/S10545-017-0059-X. [DOI] [PubMed] [Google Scholar]
- 51.Galcheva S., Demirbilek H., Al-Khawaga S., Hussain K. The genetic and molecular mechanisms of congenital hyperinsulinism. Front Endocrinol. 2019;10 doi: 10.3389/fendo.2019.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maiorana A., Lepri F.R., Novelli A., Dionisi-Vici C. Hypoglycaemia metabolic gene panel testing. Front Endocrinol. 2022;13 doi: 10.3389/fendo.2022.826167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mezza T., Cinti F., Cefalo C.M.A., Pontecorvi A., Kulkarni R.N., Giaccari A. B-cell fate in human insulin resistance and type 2 diabetes: a perspective on islet plasticity. Diabetes. 2019;68:1121–1129. doi: 10.2337/db18-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kehm R., König J., Nowotny K., Jung T., Deubel S., Gohlke S., et al. Age-related oxidative changes in pancreatic islets are predominantly located in the vascular system. Redox Biol. 2018;15:387–393. doi: 10.1016/J.REDOX.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu J., Xie G., Jia W., Jia W. Insulin resistance and the metabolism of branched-chain amino acids. Front Med. 2013;7:53–59. doi: 10.1007/S11684-013-0255-5. [DOI] [PubMed] [Google Scholar]
- 56.Alves A., Bassot A., Bulteau A.L., Pirola L., Morio B. Glycine metabolism and its alterations in obesity and metabolic diseases. Nutrients. 2019;11 doi: 10.3390/NU11061356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tervaert T.W.C., Mooyaart A.L., Amann K., Cohen A.H., TerenceCook H., Drachenberg C.B., et al. Pathologic classification of diabetic nephropathy. J Am Soc Nephrol. 2010;21:556–563. doi: 10.1681/ASN.2010010010. [DOI] [PubMed] [Google Scholar]
- 58.Ahlqvist E., Storm P., Käräjämäki A., Martinell M., Dorkhan M., Carlsson A., et al. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol. 2018;6:361–369. doi: 10.1016/S2213-8587(18)30051-2. [DOI] [PubMed] [Google Scholar]
- 59.Artunc F., Schleicher E., Weigert C., Fritsche A., Stefan N., Häring H.U. The impact of insulin resistance on the kidney and vasculature. Nat Rev Nephrol 2016 12:12. 2016;12:721–737. doi: 10.1038/nrneph.2016.145. [DOI] [PubMed] [Google Scholar]
- 60.Dave N., Wu J., Thomas S. Chronic kidney disease-induced insulin resistance: current state of the field. Curr Diabetes Rep. 2018;18 doi: 10.1007/S11892-018-1010-8. [DOI] [PubMed] [Google Scholar]
- 61.Cosmo S.D., Menzaghi C., Prudente S., Trischitta V. Role of insulin resistance in kidney dysfunction: insights into the mechanism and epidemiological evidence. Nephrol Dial Transplant. 2013;28:29–36. doi: 10.1093/NDT/GFS290. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
I have shared the link to the data