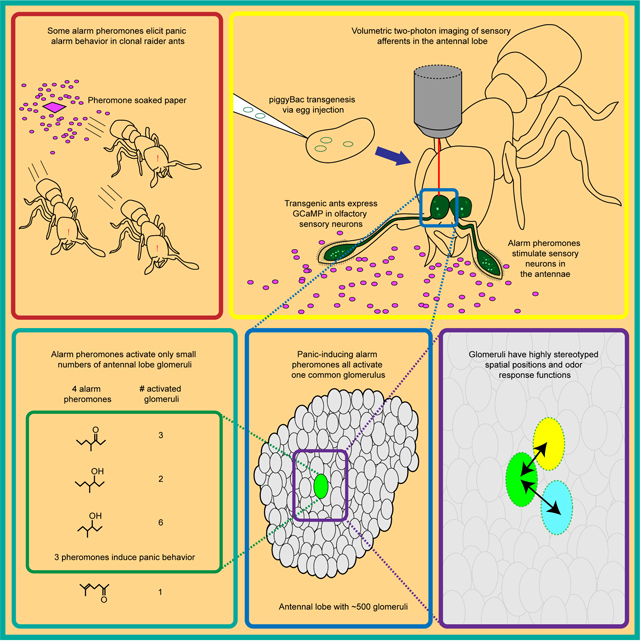
Summary
Ants communicate via large arrays of pheromones and possess expanded, highly complex olfactory systems, with antennal lobes in the brain comprising up to ~500 glomeruli. This expansion implies that odors could activate hundreds of glomeruli, which would pose challenges for higher order processing. To study this problem, we generated transgenic ants, expressing the genetically encoded calcium indicator GCaMP in olfactory sensory neurons. Using two-photon imaging, we mapped complete glomerular responses to four ant alarm pheromones. Alarm pheromones robustly activated ≤6 glomeruli, and activity maps for the three pheromones inducing panic-alarm in our study species converged on a single glomerulus. These results demonstrate that, rather than using broadly tuned combinatorial encoding, ants employ precise, narrowly tuned, and stereotyped representation of alarm pheromones. The identification of a central sensory hub glomerulus for alarm behavior suggests that a simple neural architecture is sufficient to translate pheromone perception into behavioral outputs.
Keywords: antennal lobe, calcium imaging, chemosensation, clonal raider ant, communication, GCaMP, odor coding, olfaction, Ooceraea biroi, pheromone
Graphical Abstract
In Brief:
A transgenesis method for the clonal raider ant, yielding expression of the genetically encoded calcium indicator GCaMP6s in all olfactory sensory neurons, has shown how ants use spatially precise encodings for alarm pheromones.
Introduction
Eusocial insects, like ants and honeybees, use vast arrays of pheromones to communicate information with conspecifics and to regulate colony life. These adaptations correspond to elaborations of the chemosensory system, which are particularly striking in ants. Insect olfactory systems have a conserved organization, with olfactory sensory neurons (OSNs) in peripheral sensory organs innervating glomeruli in the antennal lobes (ALs) in the brain.1–3 Much of the detailed knowledge of insect olfactory system development, anatomy, and neural function comes from studies of the vinegar fly Drosophila melanogaster. However, ants express an order of magnitude more odorant receptor genes (ORs) in their antennae, and possess an order of magnitude more AL glomeruli, than Drosophila.4–14 In Drosophila, the ~50 AL glomeruli each receive input from a functional class of OSNs and have stereotyped positions across individuals, which allowed the creation of atlases mapping odor-evoked response functions for each glomerulus.2,15–18 By contrast, little is known about the wiring of OSN subpopulations, OR expression patterns at the level of individual OSNs, or how odors are represented in the more complex olfactory system of ants, which contains up to ~500 AL glomeruli.
Here we focus on the neural representation of alarm pheromones, “danger” signals that are chemically well characterized across several ant species. Ant alarm pheromones are typically produced in the mandibular-, poison-, or Dufour’s gland, and stored in a glandular reservoir. The pheromone is then released into the surrounding air in response to danger.19 Stimulating individuals with volatile alarm pheromones is experimentally simple and quickly elicits behavioral responses, which makes these pheromones attractive models for studying the neurobiological basis of chemical communication. Upon perception of the pheromone, locomotion usually increases, and the subsequent behavioral responses are often grouped into two major categories: “aggression” and “panic”.20 Panic alarm responses involve fast movements either away from the alarm source or without a clear direction and can culminate in nest evacuation, where ants leave the nest carrying brood.20–22 Specific features of alarm behavior vary with context, species, and specific mixtures and concentrations of chemicals, and in addition to increased locomotor speed and alertness, can include changes in the posture of antennae, mandibles, and the sting.20,23–24
Alarm pheromone representation has been investigated using calcium dyes to record activity from subsections of the AL in several carpenter ant species25–27 and honeybees.28–34 These studies found broad, multi-glomerular activation patterns without evidence for specialized glomerulus clusters, similar to the combinatorial representation of general odorants in Drosophila.18,28,32,35–37 Such a combinatorial model in which many glomeruli respond to a given odorant implies that odor mixtures could potentially activate combinations of hundreds of glomeruli in the expanded ant AL. Because the number of potential combinations of glomeruli grows super-linearly with each additional glomerulus in the AL, this scenario poses much bigger challenges for higher order neurons in ants vs. Drosophila with respect to decoding multicomponent olfactory signals, detecting and identifying pheromones, and activating appropriate behavioral responses. In contrast, if most odorants only activate a small number of glomeruli, this could simplify the neural architecture necessary for processing odor information in the complex olfactory environment of an ant colony and ensure that chemical signals can be rapidly and accurately perceived.
The ant olfactory system also differs from that of Drosophila in several developmental properties that might be linked to its increased complexity.11,12,38–39 Based on these differences, it has been suggested that ants, similar to mice but unlike flies, might rely on intrinsic features of ORs for OSN axon guidance and AL patterning.11,39 This in turn could translate to increased developmental plasticity in the olfactory system. In both mice and Drosophila, olfactory glomeruli receiving input from a defined class of OSNs are consistently located in the same anatomical region, but at the local scale, homologous mouse glomeruli vary substantially in their spatial position across individuals, and even across the left/right axis within a single individual.40–43 Whether the level of anatomical-functional stereotypy of the ant olfactory glomeruli more closely resembles Drosophila or mice has not been assessed. However, the number of glomeruli in ants varies with sex, caste, and worker body size,4–5,9,44 suggesting that stereotypy may be low.
We studied the representation of alarm pheromones in the clonal raider ant Ooceraea biroi, an experimentally tractable species that lives in small colonies, reproduces asexually, and preys on other ants.12,45–46 We implemented neurogenetic tools in ants by developing a piggyBac transgenesis protocol to generate a line that expresses the genetically encoded calcium indicator GCaMP6s in OSNs. We then examined the relationships between behavioral outputs of alarm pheromone stimuli and single glomerulus-resolution, whole-AL calcium responses for four ant alarm pheromones.
Results
Alarm pheromones elicit a range of behavioral responses
The alarm pheromones 4-methyl-3-heptanone and 4-methyl-3-heptanol have previously been extracted from clonal raider ants and verified to elicit panic alarm responses, with ants rapidly leaving the nest pile and evacuating the nest chamber.22 These compounds induce panic both alone and as a 9:1 blend that mimics their relative abundance in ant head extracts (Fig. 1A–B, Table S1).22 These pheromones are exclusively found in the head and likely derive from the mandibular gland.22 We decided to also study the effects of two alarm pheromones of other ant species, 4-methyl-3-hexanol and 6-methyl-5-hepten-2-one. These compounds were not found in clonal raider ant chemical extracts but share chemical similarity to the clonal raider ant alarm pheromones (Fig. 1A, Table S1).22,47–55 Because clonal raider ants are specialized predators of a variety of other ants,46 they are likely exposed to the alarm pheromones of their prey species during raids, potentially including 4-methyl-3-hexanol and 6-methyl-5-hepten-2-one.
Figure 1. Behavioral responses to four ant alarm pheromones.
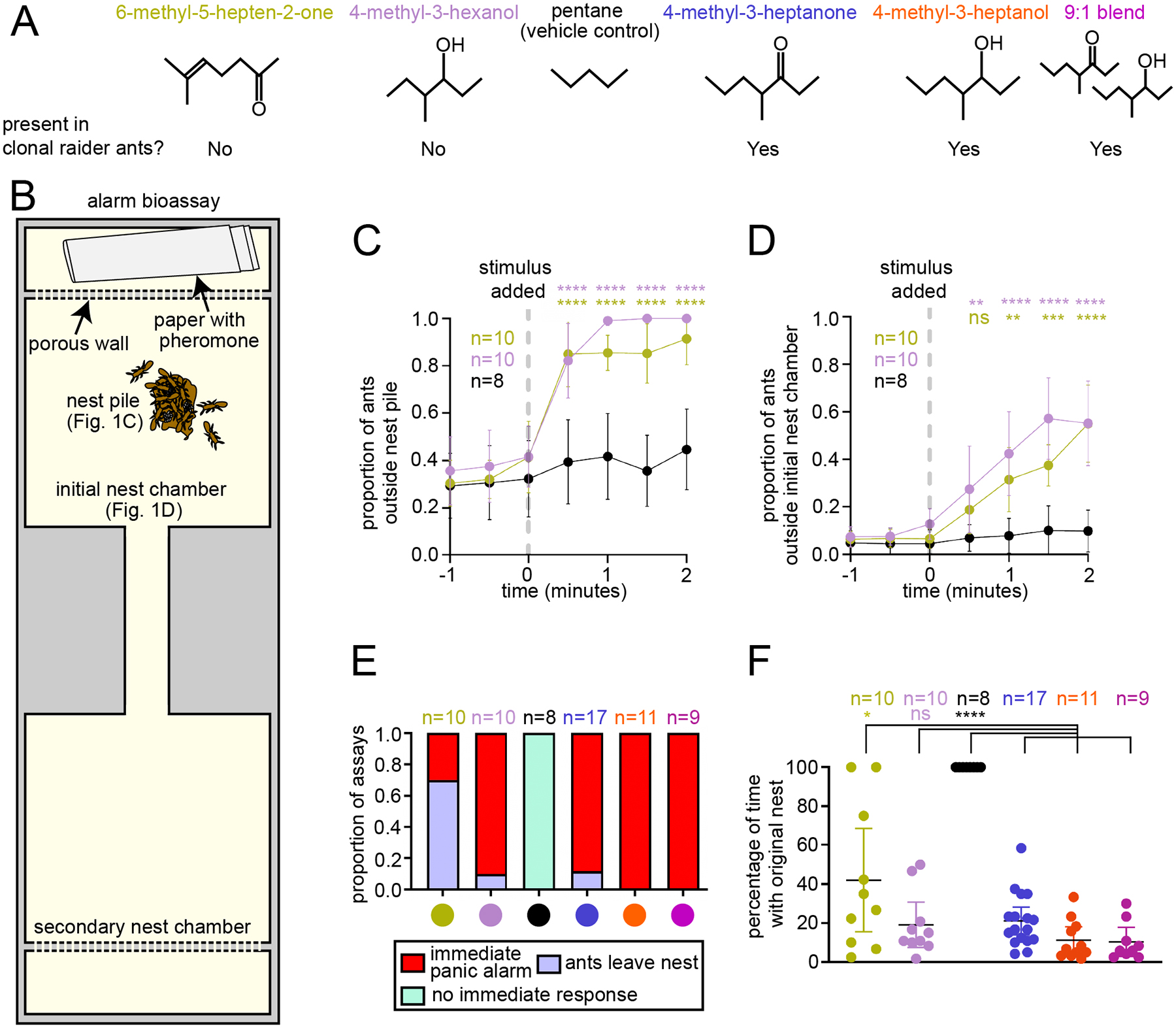
(A) Chemical structures of four ant alarm pheromones and the vehicle control used in this study, obtained from the PubChem database (National Institute for Biotechnology Information: https://pubchem.ncbi.nlm.nih.gov). (B) Experimental design for the colony alarm bioassay.22 The features used for analyses in (C-D) are indicated. (C-D) Time series of colony responses to the alarm pheromones 6-methyl-5-hepten-2-one and 4-methyl-3-hexanol vs. control (mean±SEM). See Fig. S2 for time series plots for 4-methyl-3-heptanone and 4-methyl-3-heptanol in wild type and GCaMP6s ants. (E) Categorical analysis of major behavioral responses to alarm pheromone stimuli. (F) Quantification of the length of time that the original nest pile remained intact for two minutes post stimulus in the bioassays from (C-D); mean±95% CI; see Table S2 for details. * = p<0.05; ** = p<0.01; *** = p<0.001; **** = p<0.0001, compared to vehicle control for (C-D); non-O. biroi alarm pheromones and the vehicle control were compared to known O. biroi alarm pheromones for (F); see Table S2 for details. The color code for chemical compounds in (A) applies to all figure panels.
Using the same bioassay and analyses that we previously used to study 4-methyl-3-heptanone and 4-methyl-3-heptanol (Fig. 1B),22 we characterized the behavioral response to 4-methyl-3-hexanol and 6-methyl-5-hepten-2-one. Both compounds caused ants to leave the initial nest pile (consisting of at least 2 adult workers and at least 1 egg) and the initial nest chamber (Fig. 1C–D). However, the behavioral responses were qualitatively distinct from one another, prompting additional analyses. Blinded categorization of the major behavioral response to each pheromone (see STAR Methods), including re-analysis of videos from our previous study,22 showed that 4-methyl-3-heptanone, 4-methyl-3-heptanol, the 4-methyl-3-heptanone/4-methyl-3-heptanol blend, and 4-methyl-3-hexanol all caused the ants to rapidly leave and disassemble the nest pile (which was defined as persisting as long as it contained at least one egg and two workers) within one minute after exposure in at least 80% of trials. We call this response “immediate panic alarm”. The most common response to 6-methyl-5-hepten-2-one was for the majority of ants to slowly walk away from the nest, while the nest pile persisted for more than one minute. We call this response “ants leave nest” (Fig. 1E, Supplemental Video 1).
In many of our behavioral trials, the original nest pile was disassembled, which is consistent with nest evacuation as part of a panic alarm response. In other cases, the ants moved away from the nest pile while leaving it at least partially intact, which reflects a disturbance among the ants but not a clear evacuation or panic response. We analyzed the length of time that the original nest remained intact for each odorant and found that treatment with 4-methyl-3-hexanol led to similarly rapid disassembly of the nest as 4-methyl-3-heptanone, 4-methyl-3-heptanol, and the blend (Fig. 1F, Table S2). In contrast, treatment with 6-methyl-5-hepten-2-one produced a wide range of outcomes, and the average response was significantly different from responses to clonal raider ant alarm pheromones (Fig. 1F, Table S2).22 In summary, 4-methyl-3-hexanol elicits panic alarm behavior similar to the native clonal raider ant alarm pheromones 4-methyl-3-heptanone and 4-methyl-3-heptanol. 6-methyl-5-hepten-2-one, on the other hand, lacks panic alarm activity and does not normally cause nest evacuation. The occasional alarm responses to 6-methyl-5-hepten-2-one could represent secondary responses, in which an ant emits actual alarm pheromone in response to the stimulus compound.
Creation of transgenic ants
GCaMP has been used to study olfaction in several insect species, including honeybees.18,56–57 We reasoned that targeting GCaMP to ant OSNs using a promoter from the odorant receptor co-receptor Orco could allow optical recording of neural activity in sensory afferents in the ALs, similar to other insects.18,56,58–59 We therefore cloned a 2.4 kb genomic fragment upstream of the O. biroi Orco gene which presumably contained promoter and enhancer elements sufficient to drive specific expression in all clonal raider ant OSNs (fragment ObirOrco), following Orco’s expression pattern.11,12 We then constructed a piggyBac vector plasmid where ObirOrco drives expression of GCaMP6s60 using the QF2 and 15xQUAS binary expression driver and effector elements in tandem to amplify transgene expression (Fig. 2A).61 Because we did not know if GCaMP6s would be detectable in live animals, we included an expression construct with the baculovirus-derived ie1 enhancer/promoter element to drive expression of the red fluorescent protein DsRed, based on similar designs used in other insects (Fig. 2A).62–64 We injected ant eggs with a mix of plasmid DNA and transposase mRNA65 and reared the resulting G0 individuals using protocols modified from Trible et al.12 (see STAR Methods for details; Table 1). Although we generated several separate transgenic lines, we recovered a large and stable population only from one of them, which we used for all later experiments (first four rows, Table 1). Henceforth, we refer to these ants as “GCaMP6s ants”.
Figure 2. Transgene construct and GCaMP6s expression.
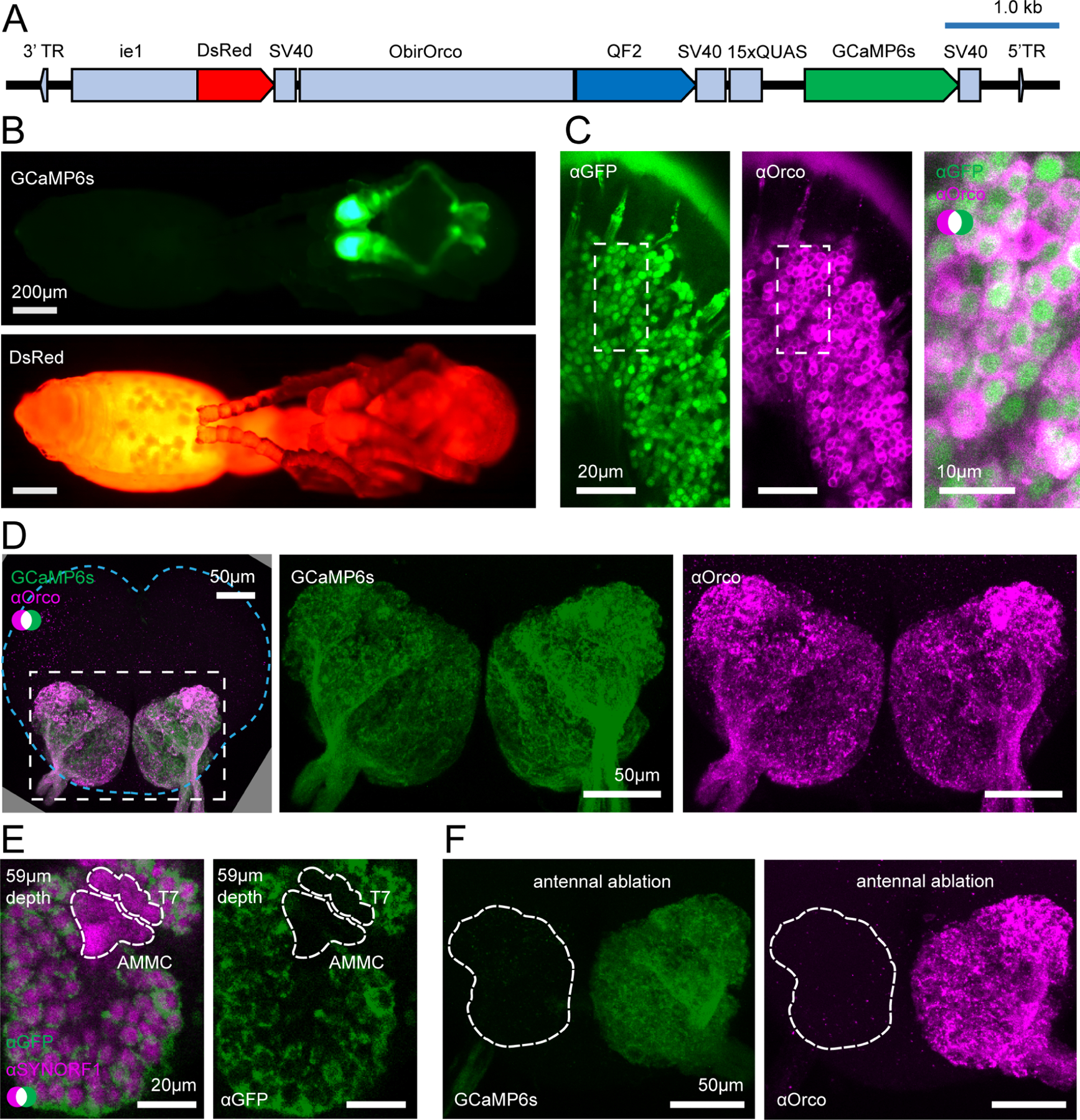
(A) Construct design. (B) Transgenic pupa under epifluorescence: GCaMP6s (top); DsRed (bottom); see Fig. S1 for comparisons with wild types. (C) Anti-GFP (green, cytoplasmic) and anti-Orco (magenta, membrane bound) densely label OSNs in the antennal club (max z-projection through 3 1μm slices). (D) GCaMP6s and anti-Orco signal co-localize in the ALs (max z-projection through the AL); brain contour is shown with cyan line. (E) Anti-SYNORF1 (magenta; neuropil) and anti-GFP (green) staining from a single optical slice in the AL. T7: T7 cluster of glomeruli; AMMC: antennal mechanosensory and motor center. (F) Unilateral ablation of the antenna eliminates GCaMP6s (green, left) and anti-Orco signal (magenta, right) from the AL (max z-projections; white outline indicates the AL boundary as determined from phalloidin stain). See Figs. S1–S2 for additional characterization of GCaMP6s ant brains. See Fig. S3 for genomic analyses of GCaMP6s ants.
Table 1.
Generation of transgenic clonal raider ants expressing GCaMP6s.
| Treatment |
Egg age at injection (hours) | # eggs injected | # G0 eggs hatched | # G0 adults eclosed | # G0 adults with fluorescence | Minimum # of lines generated | Overall efficiency | Transformation efficiency |
|---|---|---|---|---|---|---|---|---|
| 27.8/110 | <5 | 1945 | 155 (8.0%) | 14 | 3 | 1 | 0.00021 | 0.026 |
| 27.8/220 | <5 | 1367 | 72 (5.3%) | 16 | 0 | |||
| 27.8/440 | <5 | 739 | 6 (0.8%) | 0 | 0 | |||
| 27.8/110 | <3 | 637 | 15 (2.4%) | 8 | 5 | |||
| 27.8/110 | <3 | 353 | 44 (12.5%) | 17 | 2 | 1 | 0.0028 | 0.059 |
The “Treatment” column indicates injection mix concentrations of plasmid DNA and transposase mRNA. G0 adults from the first four treatments were reared as a group, and we therefore cannot determine which treatment generated the line that was propagated from that group. Overall efficiency was calculated by dividing the minimum number of lines generated by the number of eggs injected; transformation efficiency was calculated by dividing the minimum number of lines generated by the number of G0 adults eclosed.
Characterization of transgenic ants
We characterized transgene expression in our transgenic line to determine if it would be useful for imaging odor-evoked calcium responses. Transgenic pupae had detectable GCaMP6s fluorescence in the antennae, consistent with expression in OSNs, and DsRed was broadly visible under epifluorescence (Fig. 2B, Fig. S1A). DsRed is expressed at a low level in the ALs, possibly due to leaky expression from ObirOrco (Fig. S1B). We assessed GCaMP6s expression in OSNs in the antennal club using immunohistochemistry and found that GCaMP6s labels the great majority of Orco-positive cells (Fig. 2C). In the ALs, high levels of GCaMP6s co-localized with Orco, which labels OSN afferents (Fig. 2D).11 GCaMP6s is also expressed in parts of the sub-esophageal zone and central complex (Fig. S1C–D). These fluorescence patterns were all consistent across individuals.
To validate that our transgenic ants were good candidates for the study of AL function, we performed anatomical reconstructions of the ALs of two GCaMP6s ants using two different staining methods that label all brain neuropil. From the first AL, we reconstructed 505 glomeruli using anti-SYNORF1 signal (Fig. S2A). From the second AL, using phalloidin, we reconstructed 508 glomeruli (Fig. S2B). The total numbers of glomeruli are within the published range of wild type ants (493–509 glomeruli),9,11,12 showing that the gross AL anatomy of transgenic ants is normal. Next, we looked at the expression of GCaMP and Orco within the AL. Using our second reconstruction, which was co-stained with anti-Orco, we counted 502 Orco-positive glomeruli, all of which were also GCaMP6s-positive (Fig. S2B). The 6 glomeruli of the T7 cluster were the only Orco-negative glomeruli, consistent with previous studies.9,11 Weak GCaMP6s signal was detected in 4 of the 6 T7 glomeruli (Fig. S2B).
We then further investigated GCaMP expression using confocal images of brains stained with anti-GFP and anti-SYNORF1. While glomeruli in T1-T6 were always robustly labeled by anti-GFP, signal in the T7 glomeruli was consistently weaker than in other glomeruli and was often undetectable (Fig. 2E). The antennal mechanosensory and motor center, another adjacent Orco-negative sensory structure,11,66 was consistently unlabeled by GCaMP6s or anti-GFP in our confocal stacks (Fig. 2E). Together, this indicated that our transgenic line is a good candidate for detecting calcium responses from all olfactory glomeruli of the AL (about 99% of total glomeruli).
To see whether GCaMP6s is expressed by cells other than OSNs in the ALs, we performed unilateral antennal ablations on transgenic animals to sever the antennal nerve and examined their brains after allowing GCaMP6s and Orco to be cleared. While GCaMP6s signal in the sub-esophageal zone and central complex was unaffected (Fig. S1E), GCaMP6s and anti-Orco signals were greatly reduced across the entire AL connected to the ablated antenna, and no clear glomerular labeling remained (Fig. 2F). This indicates that GCaMP6s signal in the AL derives from the antennae and is likely to be exclusive to sensory neuron axons.
Expression of genetically encoded calcium indicators can alter cellular calcium buffering and affect behavior.67–68 We therefore examined whether the GCaMP6s ants had defects in alarm behavior by subjecting them to our alarm behavior bioassay. The ants left the nest cluster in response to 4-methyl-3-heptanone, 4-methyl-3-heptanol and the blend, similar to wild types (Fig. S2C–D). The effect on leaving the nest chamber was only significantly different from control for 4-methyl-3-heptanone and the blend (Fig. S2E–F). Crucially, GCaMP6s ants perceive both alarm pheromones, and their behavioral response is qualitatively similar to wild types.
Finally, non-targeted transgene insertions can disrupt endogenous sequences,69 and we therefore sequenced the genome of a GCaMP6s ant. The line contains a single, haploid transgene insertion on the 2nd chromosomal scaffold (Fig. S3A–B). The insertion occurred at location Chr2:3,870,844–3,870,847, within an intron of the gene trace amine-associated receptor 9 (Fig. S3C). Since the insertion is haploid and not within a coding region, and because GCaMP6s animals have normal AL anatomy and robust behavioral responses, these animals are well-suited for functional studies of the clonal raider ant olfactory system.
Recording calcium responses to general odorants
We developed an in vivo two-photon imaging preparation for clonal raider ants, where animals are head-fixed, and a small imaging window is excised from the cuticle covering the ALs (Fig. 3A–B). Ants are then exposed to reproducible odor stimuli via a computer-controlled olfactometer18,25,70 and the resulting changes in GCaMP6s fluorescence are captured at 27.5fps, imaging the volume containing the entire AL every 1.2s (33 z-planes at 5μm increments; Fig. 3C–E, Supplemental Video 2). Because most clonal raider ant glomeruli are 10–20μm in diameter, they are all sampled in multiple imaging planes. Individual glomeruli were often discernible from baseline GCaMP6s fluorescence and always from calcium responses due to spatially clustered pixels with time-correlated responses (Fig. 3C). While previous studies of olfactory function in eusocial insects were limited to small subsets of ORs,71–72 a few olfactory sensilla73–74 or neurons,75 or restricted subsections of the AL,25–34 our volumetric GCaMP imaging approach allowed us to record from all olfactory glomeruli throughout the entire AL during single odor stimulus trials, without possible confounding signals from projection neurons, lateral interneurons, or glia, and without bias toward particular AL regions (Fig. 3E).
Figure 3. Imaging odor-evoked calcium responses in the antennal lobe.
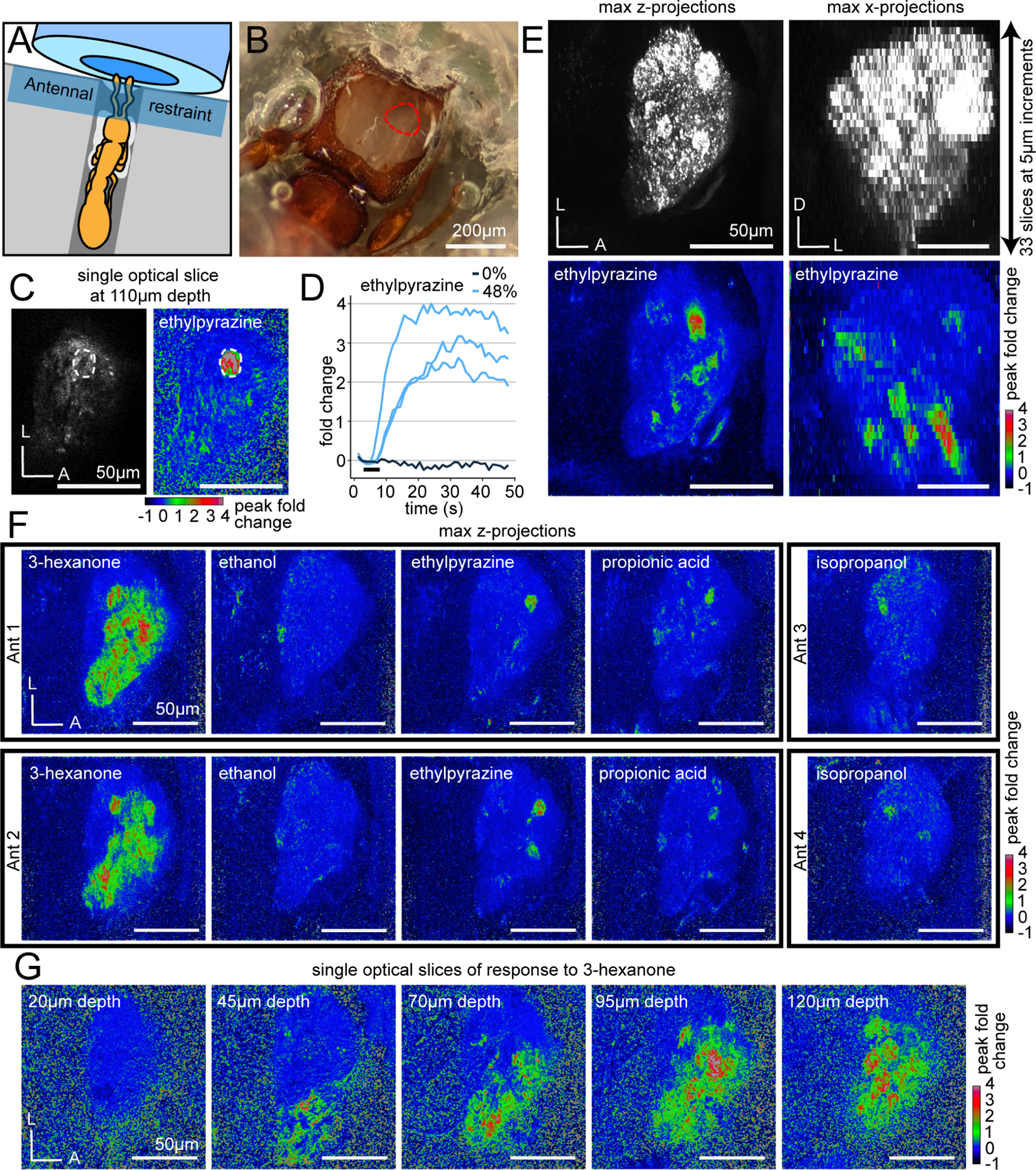
(A) Ant adhered to a plastic base with glue (white). A Parafilm strip restrains the antennae in front of the air tube. (B) Cuticle and glandular tissue are removed to expose the AL (red outline). (C) Single optical slice through the AL using two-photon microscopy showing raw fluorescence (left, brightness and contrast enhanced) and the peak fold change of fluorescence after a 5s odor presentation at 48% concentration (right). A single glomerulus of interest is circled. (D) Time series of calcium responses in the glomerulus from (C) from trials with ethylpyrazine or paraffin oil vehicle (0%); black bar indicates the 5s odor presentation. (E) Volumetric imaging of clonal raider ant ALs. Raw GCaMP6s fluorescence (top) in max z-projection (left) and max x-projection (right); responding glomeruli are visible throughout the volume (bottom) after presentation with ethylpyrazine (48%). (F) Responses to general odorants in different individuals. (G) Responses to 3-hexanone in five different optical slices. See Table S3 for vapor pressures of general odorants. D: dorsal; L: lateral; A: anterior.
To obtain a basic overview of odor representation, we presented ants (n=6) with a panel of five general (non-pheromone) volatile odorants selected from the DoOR database of olfactory studies in Drosophila,37 studies of OR function in other ants,71 and soil volatiles76 (Table S3). To simplify the display of calcium responses while considering the entire AL, we calculated the peak fold change of fluorescence in each slice of the volumetric videos and then flattened them using max z-projection. Viewed this way, it was apparent that the ant AL exhibits properties of odor encoding that have been shown in other insects:18,30 each odorant activated a unique combination of glomeruli, and responses to the same odorant occurred in similar regions of the AL in different individuals, indicating that odor representation is qualitatively similar across individuals (Fig. 3F). We also found that the breadth of glomerular responses varied dramatically across odorants, with most odorants activating a few glomeruli, while 3-hexanone activated large regions of the ventral/medial AL (Fig. 3F–G). This demonstrates that our imaging approach can detect both sparse and broad calcium responses, if they occur.
Pheromone representation is sparse, and alarm-inducing compounds activate a single shared glomerulus
To study the encoding of alarm pheromones, we first imaged responses to 4-methyl-3-hexanol in both ALs simultaneously (n=3 ants) and found that response patterns were bilaterally symmetrical (Fig. S4A). Given the equivalence between the two ALs, we then performed additional experiments imaging only the right AL. We presented each ant (n=13 ants) with the four alarm pheromones at a range of concentrations (Fig. 4A). Sparse, unique subsets of AL glomeruli responded to all pheromones, while the paraffin oil vehicle did not generate responses (Fig. 4B–C). Fluorescence increases were frequently large (1–2-fold change) and lasted longer than the 5s odor presentation. We did not observe any fluorescence decreases in response to odor, although we did detect small, non-specific decreases in fluorescence due to minor shifts in AL position and photobleaching. This artifact did not affect our ability to detect calcium responses, which remained robust after normalization for the duration of the experiment (Fig. S4B–C). Comparison of calcium traces from two adjacent glomeruli showed high specificity of the response functions, without evidence for weak or transient calcium responses that might not be visible from analysis of peak fold change (Fig. S4B–C). The response patterns to the same alarm pheromone in different individuals were qualitatively similar, in accordance with what we observed for general odorants (Fig. 3F, Fig. 4D).
Figure 4. The representation of alarm pheromones in the antennal lobe.
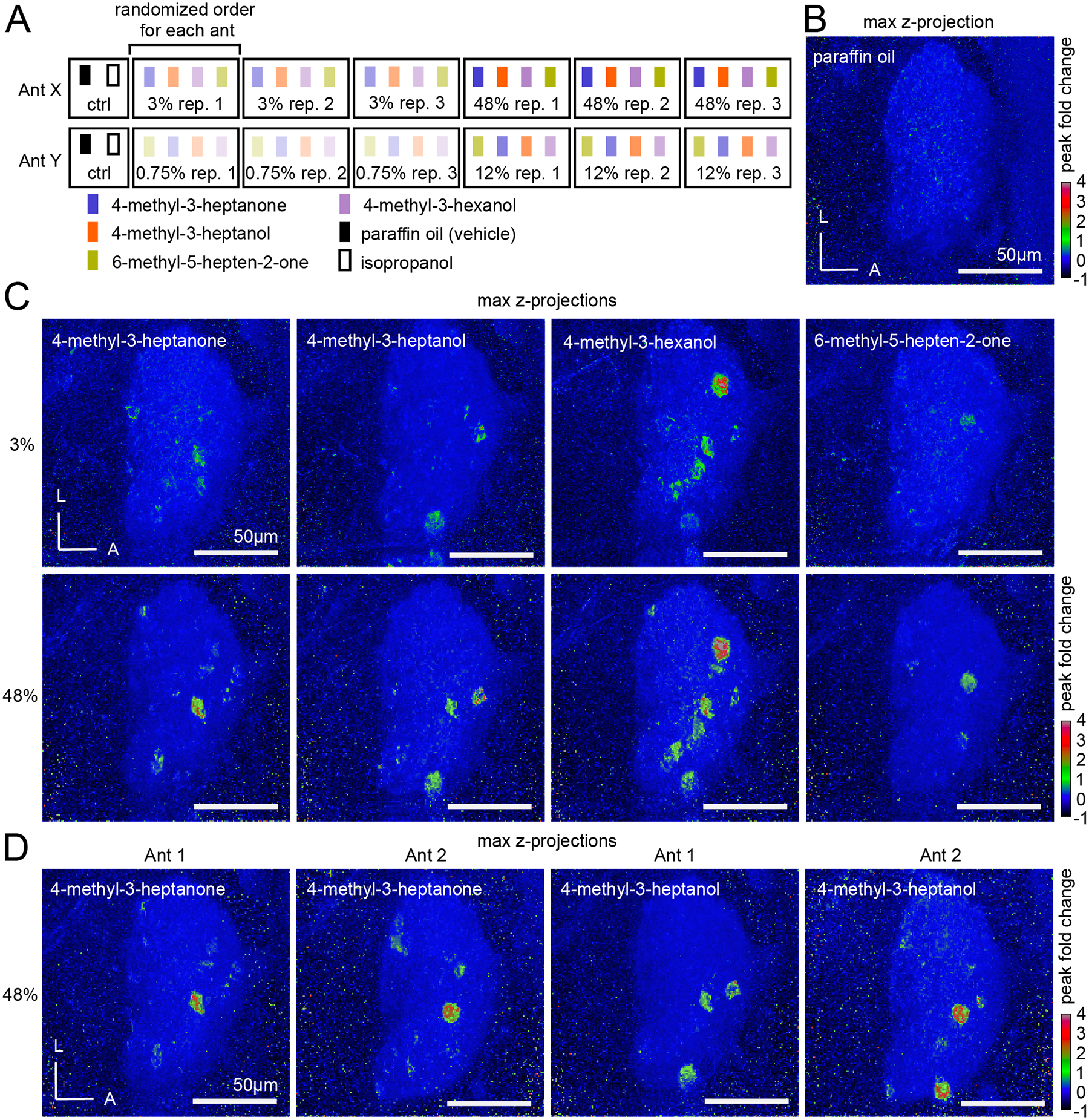
(A) Cartoon of the odor stimulus regime, with two ants shown as examples. Four alarm pheromone concentrations were tested in total (0.75%, 3.0%, 12.0%, and 48.0% v/v), but each individual ant was exposed to only two out of the four possible concentrations. (B) The paraffin oil vehicle does not generate calcium responses. (C) Representative max z-projections of peak fold changes from a single ant in response to four alarm pheromones at 3% and 48% concentrations. (D) Two different individuals stimulated with 4-methyl-3-heptanone (left) and 4-methyl-3-heptanol (right) at high (48%) concentration. See Fig. S4 for additional characterization of calcium responses. See Fig. S5 for quantification of numbers of responding glomeruli. See Table S3 for vapor pressures of alarm pheromones. L: lateral; A: anterior.
We sought to determine how many of the ~500 glomeruli responded to each alarm pheromone by examining the max z-projections of the calcium responses. We identified all regions of interest corresponding to glomeruli activated in response to any of the four analyzed pheromones, quantified the mean peak fold change in response to each pheromone/concentration, and used a threshold of ≥0.2 mean peak fold change to find robust odor-evoked responses (Fig. S5A). Higher concentrations produced more robust responses, with a few more glomeruli passing the threshold, but overall spatial response patterns were similar across concentrations (Figs. 4C, S4B–C, S5A). Even at the highest concentration tested, the four pheromones activated a median of at most 6 glomeruli (Fig. S5A). Despite the small number of responding glomeruli, we observed consistent partial overlap in the response patterns activated by the three compounds eliciting panic alarm responses, 4-methyl-3-heptanone, 4-methyl-3-heptanol, and 4-methyl-3-hexanol, with a single glomerulus activated by all three (Fig. S5B). We refer to this glomerulus as the “panic glomerulus, broad” (PGb). This finding is consistent with the expectation that these pheromones, which can elicit slightly different forms of alarm behavior,22 might share sensory pathways while also activating distinct sets of glomeruli. In contrast, while we sometimes observed responses to 6-methyl-5-hepten-2-one and either 4-methyl-3-heptanone or 4-methyl-3-hexanol in an overlapping region, those occurrences were rare and inconsistent (Fig. S5C).
Alarm pheromone-responsive glomeruli are spatially stereotyped
To better understand the level of stereotypy in the ant AL, we decided to localize PGb and characterize its local environment. The raw recordings revealed that PGb is consistently located in the anterior part of the ventral AL hemi-lobe, adjacent to a gap containing no glomeruli (Fig. 5A–B). This gap is distinct from the T7 glomerulus cluster, which is not reliably labeled by GCaMP6s. PGb is located approximately halfway between the dorsal and ventral AL surfaces, and is neighbored by two additional glomeruli that respond to alarm pheromones, with all three visible in the same optical plane (Fig. 5A–B). While PGb responds to 4-methyl-3-heptanone, 4-methyl-3-heptanol, and 4-methyl-3-hexanol, a nearby glomerulus responds to 6-methyl-5-hepten-2-one, which we refer to as the “6-methyl-5-hepten-2-one glomerulus” (6G). Both glomeruli were identified in 13/13 individuals. In 11/13 individuals, we identified a third neighboring glomerulus that responds to 4-methyl-3-heptanol and 4-methyl-3-hexanol, which we termed the “panic glomerulus, alcohol” (PGa). Examination of the position of the three glomeruli in the z-stacks and comparison with a previous segmentation of the AL9 showed that they are part of the T6 glomerulus cluster, which is innervated by OSNs from basiconic sensilla on the ventral surface of the ant antennal club that typically express members of the 9-exon OR subfamily (Fig. 5B).9 In gross anatomy, PGb, PGa, and 6G resemble typical O. biroi AL glomeruli and do not show obvious differences in shape or size. To validate our initial finding that these three glomeruli are functionally distinct from one another, we aligned them across individuals and quantified glomerulus-specific odor responses. This demonstrated that, while PGb, PGa, and 6G are spatially adjacent, they each reliably respond to unique combinations of odorants, with several pheromone/glomerulus combinations producing no detectable responses (Figs. 5C, S6). Importantly for its potential role in mediating alarm behavior, PGb did not respond to 6-methyl-5-hepten-2-one, showing selectivity in its receptive tuning (Figs. 5B–C, S6).
Figure 5. A glomerular cluster with stereotyped spatial organization and robust responses to alarm pheromones.
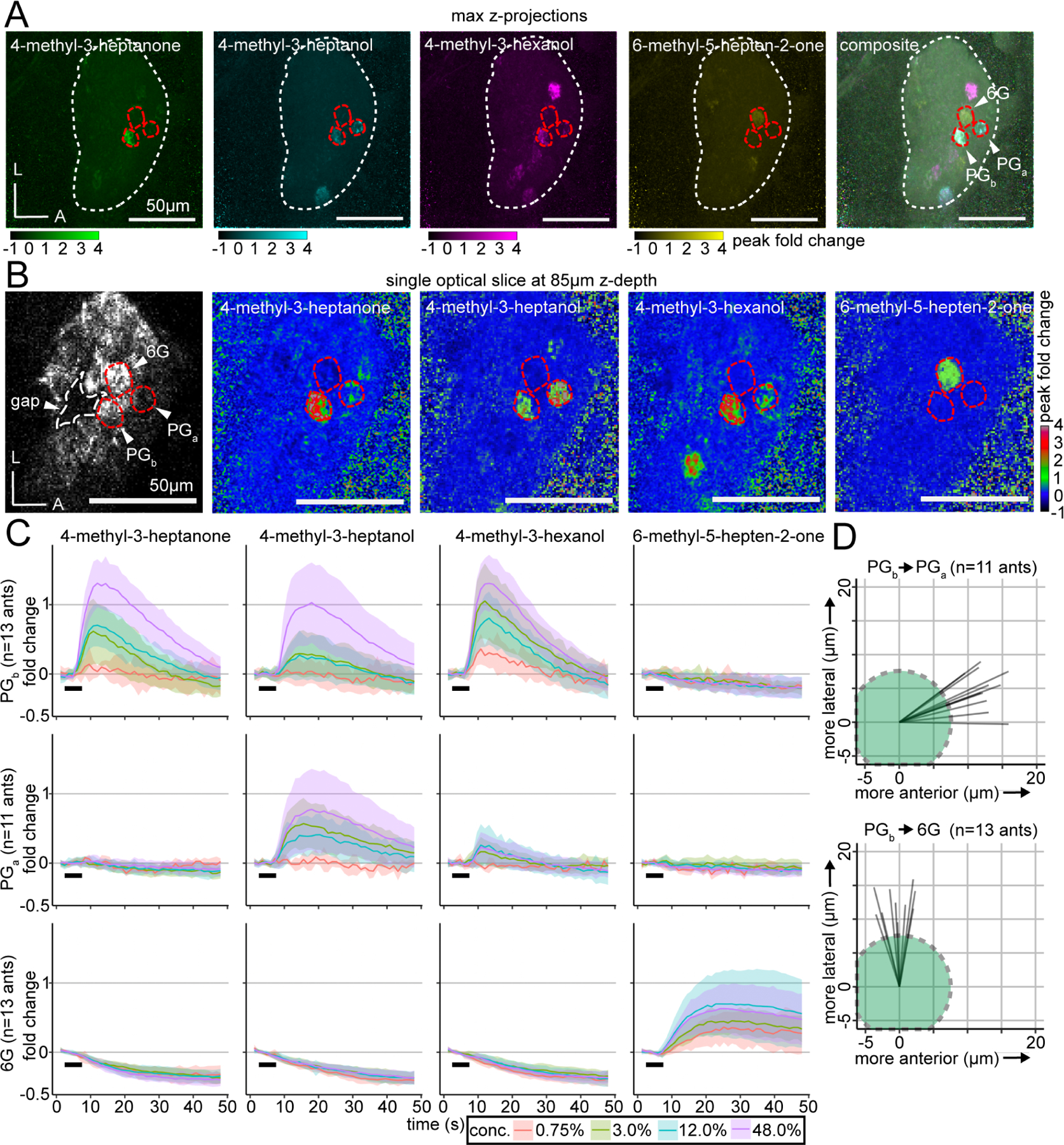
(A) Whole-AL activation patterns for alarm pheromones overlap in several glomeruli. Three focal glomeruli are outlined in red. (B) Single optical slice through the AL with the three focal glomeruli, which are adjacent to an AL region lacking glomeruli (“gap”; outlined in white). Fluorescence with enhanced brightness/contrast (left). Peak fold change in response to odors (right four panels). See Fig. S5 for quantifications of responding glomerulus numbers at different concentrations, and Fig. S6 for peak calcium response quantifications. (C) Time series of calcium responses in PGb (top), PGa (middle), and 6G (bottom); mean±SD. Black bars indicate the 5s odor presentation. See Fig. S7 for extended time series of responses to 6-methyl-5-hepten-2-one in 6G. (D) Vectors of the spatial displacement between the centers of the PGb and PGa (top), and between the PGb and 6G (bottom) glomeruli. Green circles represent a typical 15μm-diameter glomerulus, for scale. L: lateral; A: anterior.
Calcium responses had slow temporal dynamics, and in some cases calcium signals remained elevated above baseline for the duration of a single 48s recording trial. Examination of the temporal dynamics showed that while responses in PGb and PGa peaked and then declined close to baseline by the end of the recording, responses in 6G were extremely slow, with a fluorescence plateau of tens of seconds (Fig. 5C). We therefore performed additional odor presentations with 6-methyl-5-hepten-2-one with an extended recording period (144s) and found that calcium responses did eventually return to baseline, although this sometimes took >100s (Fig. S7A). Quantifying time to response onset and time to response maximum for the different pheromones in the three focal glomeruli showed that different combinations had distinct temporal dynamics, as has been shown in other species (Fig. S7B–C).35–36,77–78
Our analyses thus far show that alarm pheromones evoke bilaterally symmetrical, qualitatively similar calcium responses across individuals, and that the number of activated glomeruli is consistent for a given odor. However, they do not answer the question of whether the activated glomeruli are located in fixed positions within the AL as in Drosophila, or whether there is significant local variation as in mice. To quantify the level of stereotypy, we examined the relative spatial positioning between PGb, PGa, and 6G along the medial-lateral and anterior-posterior axes (spatial resolution along the dorsal-ventral axis was insufficient for this analysis, especially given that these glomeruli are located at similar z-depths). We found that PGa was always located anterior (mean distance between centers: 12.9±1.9SD μm), and slightly lateral (mean distance: 5.1±2.9SD μm) to PGb (Fig. 5D). In comparison, 6G was always lateral to PGb (mean distance: 13.1±2.6SD μm), and in a similar position along the anterior-posterior axis (mean distance: 0.6±2.2SD μm) (Fig. 5D). The standard deviation values are much smaller than the typical diameter of a glomerulus (10–20μm). We therefore conclude that these three glomeruli occupy stereotyped positions even within their local glomerular cluster and show stereotyped odor response functions across individuals.
The median number and positions of responding glomeruli for each pheromone, in combination with the pheromones’ behavioral outputs, allowed us to outline a conceptual schematic of alarm pheromone representation in the ant AL (Fig. 6). The three pheromones with overlapping calcium response patterns all robustly elicited panic alarm behavior, while 6-methyl-5-hepten-2-one did not elicit panic alarm behavior and generated a non-overlapping response (Fig. 6). These findings point to a shared pathway for eliciting panic alarm behavior, centered on PGb.
Figure 6. Conceptual schematic for the representation of alarm pheromones in the clonal raider ant AL.
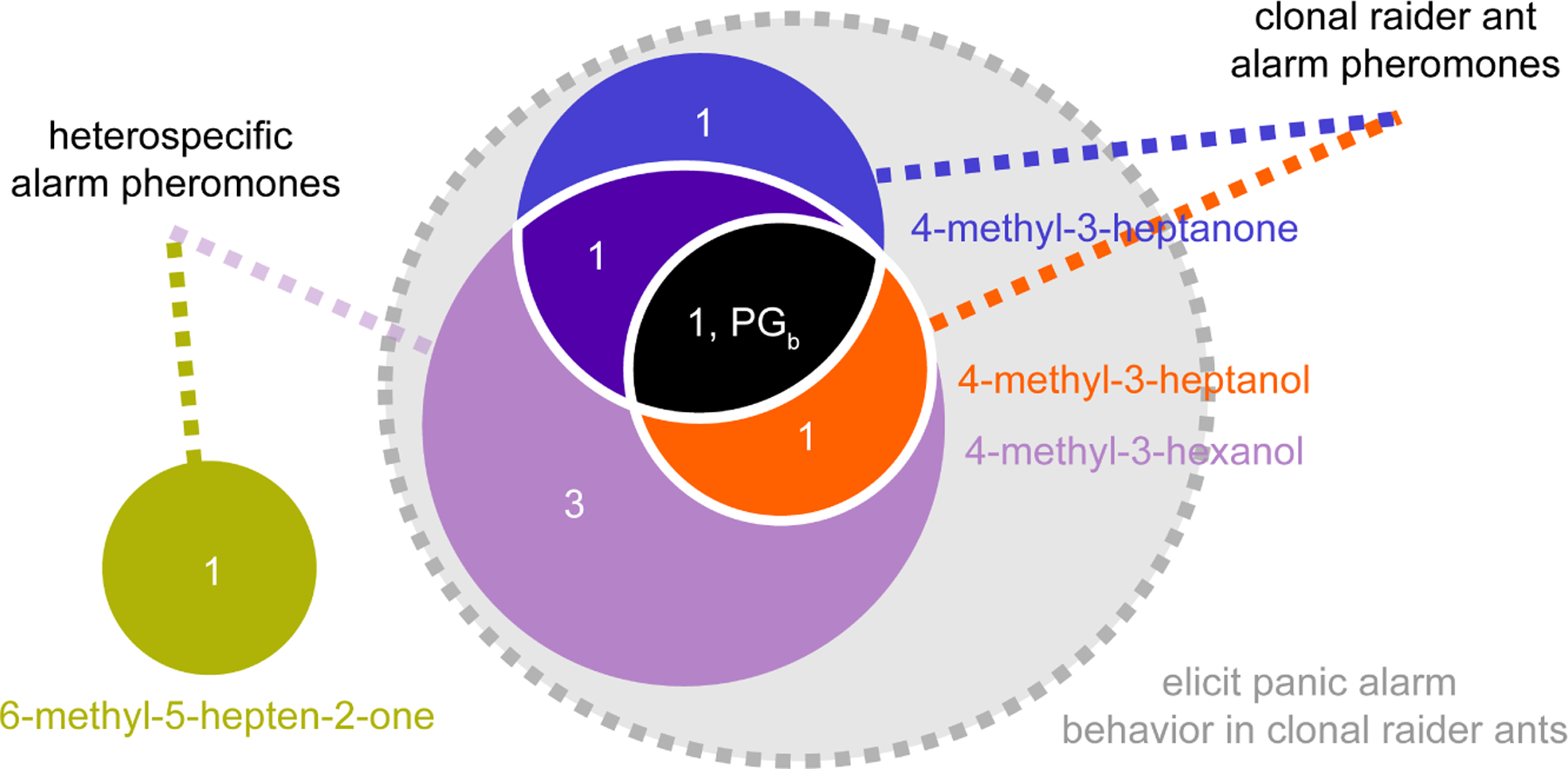
Numbers show the median number of responding glomeruli for each pheromone combination at the highest concentration tested (48%; n=8 ants). PGb is indicated on the diagram according to its response function.
Discussion
In this study, we pioneered the combination of GCaMP with volumetric two-photon imaging to study social insect neurobiology. Because our stable transgenic line propagates clonally in the lab, these resources can be maintained indefinitely, adapted, and expanded to study many topics related to ant olfaction. To our knowledge, this has so far not been feasible in other eusocial insects due to challenges associated with transgenesis, performing crosses, and maintaining genetically modified strains. We employed our transgenic line to address long-standing questions about pheromone representation in the ant AL. To study stereotypy in the ant olfactory system, we mapped a cluster of three AL glomeruli across individual clonal raider ants and found that they have consistent positions, spatial organization, and odor-evoked response functions. Ant ALs thus possess a high degree of spatial conservation at the scale of individual glomeruli, suggesting that, similar to Drosophila, axon targeting by OSNs can be stereotyped, despite the vastly increased complexity of the olfactory system. However, additional work is required to determine whether this level of stereotypy is conserved across other parts of the AL.
The proportion of glomeruli that robustly responded to any alarm pheromone was very small, with a maximum of only 6 glomeruli displaying robust activation out of ~500 total. Contrary to previous studies on social insects,25–34 this sparse activation shows that alarm pheromones are in fact encoded by small numbers of glomeruli, similar to ecologically relevant chemicals in Drosophila and moths, such as sex pheromones79–83 and aversive compounds including CO284 and the microbial odorant geosmin.56 This sparse encoding logic could simplify the neuronal computation required to respond to molecules indicative of danger, despite the complex olfactory environment of an ant colony.
With the exception of 3-hexanone, the general odorants tested here also only activated small numbers of glomeruli. Unlike in Drosophila,35–37 narrow glomerular tuning might thus be a more general property of the ant AL, and could help compensate for the potentially much greater complexity of odor encoding implied by an expanded olfactory system. Using sparse encoding for sensory signals could decrease the probability of odor mixtures activating hundreds of glomeruli simultaneously, reducing the need for vast numbers of neural connections for decoding highly combinatorial signals. Sparse glomerular encoding could emerge from a simple organizational model where each glomerulus is innervated by a single OSN class that expresses a single narrowly tuned OR, and at least some ant ORs are indeed narrowly tuned.71–72
Alternatively, ant glomerular tuning properties could emerge from more complex patterns of OR expression or OSN connectivity, potentially via lateral inhibition85–87 or chemoreceptor co-expression.88–89 Collecting single-cell resolution data on OR expression and OSN connectivity in ants will be key for determining how ant olfactory coding properties arise. We also found that the temporal dynamics of calcium responses differed by odor and glomerulus. These features provide additional information that olfactory systems can use to interpret sensory inputs, including mixtures of odors.35–36,77–78
Two of the alarm pheromones we studied are produced by the clonal raider ant, but we also investigated two additional alarm pheromones from other ant species. All four compounds share some structural features, including a methylated main carbon chain of six or seven carbons and alcohol or ketone functional groups. Of the two non-native compounds, 4-methyl-3-hexanol elicits panic alarm behavior and activates glomeruli that overlap with the two native alarm pheromones, 4-methyl-3-heptanone and 4-methyl-3-heptanol. This overlap can potentially be explained by the substantial structural similarity of these three compounds, including the similar arrangement of functional groups. It also suggests that the precise chemical structure of alarm pheromones could evolve rapidly across species, while the corresponding neural architecture underlying chemosensation and behavior is conserved.
A main difference between sex- and alarm pheromone detection systems is their level of specificity. Behavioral and neural responses to sex pheromones are highly specific, and close structural analogs elicit greatly reduced activity or even act as antagonists, a feature of the circuit logic that has been exploited in pest control.90–91 In contrast, compounds structurally similar to ant alarm pheromones usually elicit strong alarm responses, similar to the native pheromones.92 Our finding that some alarm pheromone-sensitive glomeruli respond to multiple panic alarm-inducing compounds provides a neural mechanism for these behavioral observations, and shows that sex- and alarm pheromone detection systems can differ substantially in the chemical specificity of the pheromone-sensitive glomeruli. This difference could reflect the different selective pressures acting on these two systems. The high specificity of sex pheromones is a key contributor to prezygotic isolation in sympatric species.93–94 In the case of alarm pheromones, on the other hand, a circuit logic like that of sex pheromone perception would open the door for predators and parasites to block alarm signaling within an ant colony using chemical antagonists or inhibitors. This could be catastrophic, leaving colonies defenseless to exploitation. The relatively broad excitability of the alarm pheromone detection system could thus confer protection against inhibitors. While “appeasement allomones” have been described in ants, these chemicals are structurally unrelated and likely function through distinct sensory mechanisms from alarm pheromones.95–97 More often, rather than suppressing alarm signaling, ant predators and parasites manipulate their target species by inducing alarm responses,96,98–100 in some cases via pheromone mimicry.101–102
In contrast to the other alarm pheromones, 6-methyl-5-hepten-2-one does not robustly cause panic alarm behavior in clonal raider ants. The glomerular response pattern is distinct from those of the panic inducing alarm pheromones, which aligns with previous work showing that compounds with different behavioral activity are usually detected through distinct olfactory channels.103–104 Interestingly, an ant-hunting spider uses 6-methyl-5-hepten-2-one to locate its prey, the meat ant Iridomyrmex purpureus.105 Given that O. biroi is a specialized predator of other ants, our results raise the possibility that O. biroi may also employ alarm pheromones as cues to detect prey.
In both mice and Drosophila, olfactory glomeruli with similar chemical receptive ranges are clustered into functional subdomains, a pattern that can result from the duplication and gradual divergence of ancestral chemosensory receptors and their associated glomeruli.106–108 In our experiments, all four pheromones, which share structural similarities, activated combinations of spatially adjacent glomeruli. This suggests that the ant olfactory system also tends to map proximity in chemical space to actual spatial proximity in the AL. Here we focused on glomeruli in the T6 cluster, which are mostly innervated by OSNs expressing ORs in the 9-exon subfamily.9 This subfamily is particularly highly expanded via gene duplications and undergoes rapid evolution in ants.9–10 Our results are thus consistent with a model in which recently duplicated ORs are not only activated by chemically related compounds but are expressed in OSNs innervating adjacent AL glomeruli. An electrophysiological study of subsets of randomly selected olfactory projection neurons in carpenter ants also found spatially clustered responses. However, these responses came from two chemically distinct alarm pheromone components, suggesting that spatial patterning in the ant AL may also reflect pheromone social functions in addition to chemical similarity.75
Ant pheromone communication employs diverse chemical substrates, including compound mixtures.109–110 These mixtures can be complex, as is the case for the cuticular hydrocarbon blends that serve as nest membership gestalt odors.111 While ant ALs could in principle use broad encoding to represent such complex blends, insect olfactory systems can have an impressive capacity to reduce the complexity of ecologically relevant signal inputs. Mosquito ALs, for example, encode critical features of complex host odor mixtures using only a few glomeruli.59 Future work should investigate whether the sparse encoding we report here for alarm pheromones holds true for nestmate recognition cues and other types of pheromones used by ants. Indeed, previous studies in leaf-cutting ants suggested that a trail pheromone component may be detected primarily via a specialized macroglomerulus found only in large workers.44,112 This will help develop a general understanding of how glomerular tuning evolves in the context of chemical cues with high ecological relevance, complex chemical communication, and expanded olfactory systems.
Limitations of the Study
We tested a relatively small panel of general odorants and alarm pheromones, and these compounds did not systematically span chemical space, limiting inferences about general tuning properties of the ant olfactory system. General odorants with low vapor pressures generated no calcium responses at the tested concentrations, implying that different delivery methods will be required for these odorants.72 Furthermore, because it is generally challenging to determine the amount of odorant that an animal is exposed to, and because our behavioral- and GCaMP-imaging experiments differed in many aspects of odorant delivery, we do not know whether the experienced amounts were comparable across the two assays. Similarly, we do not know what pheromone concentrations the animals encounter under naturalistic conditions, a common limitation that applies to the vast majority of pheromones. The general odorant and alarm pheromone imaging experiments were performed separately and in different individuals, preventing us from matching glomeruli between individuals across the two experiments, or determining if the alarm pheromone-sensitive glomeruli also responded to general odorants. So far, we lack methods to functionally manipulate individual AL glomeruli in behaving ants and therefore cannot formally test whether specific glomeruli generate alarm behavior. These are all promising avenues for future studies of the ant olfactory system.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Daniel J. C. Kronauer (dkronauer@rockefeller.edu).
Materials availability
The plasmid used for generating the GCaMP6s ants has been deposited to Addgene (accession # 200400).
Data and code availability
DNA sequence data have been deposited to NCBI BioProject. Confocal microscopy data have been deposited to the Brain Image Library. Calcium imaging data have been deposited to the DANDI Archive. Accession numbers are listed in the key resources table.
All original code has been deposited to GitHub and the repository is listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken polyclonal anti-GFP | Abcam | Cat#ab13970; RRID: AB_300798 |
| Rabbit polyclonal anti-RFP | Rockland | Cat#600-401-379; RRID: AB_2209751 |
| Mouse monoclonal anti-SYNORF1 | Developmental Systems Hybridoma Bank | DSHB: 3C11; RRID: AB_528479 |
| Mouse monoclonal anti-Orco | Gift from Vanessa Ruta; Butterwick et al.136 | clone 20F7 |
| Goat anti-Chicken Alexa 488 | Invitrogen | Cat#A-11039; RRID: AB_2534096 |
| Donkey anti-Mouse Alexa 647 | Invitrogen | Cat#A32787; RRID: AB_2762830 |
| Donkey anti-Rabbit Alexa 594 | Invitrogen | Cat# A-21207; RRID: AB_141637 |
| Chemicals, peptides, and recombinant proteins | ||
| AlexaFluor 555 phalloidin | Invitrogen | Cat#A34055 |
| DAPI | Invitrogen | Cat#D1306 |
| Paraffin oil | Hampton Research | Cat#HR3-421 |
| 100% pentane | Sigma Aldrich | Cat#236705 |
| 96% 4-methyl-3-heptanone | Pfaltz and Bauer | Cat#M19160 |
| ≥99% 4-methyl-3-heptanol | Sigma-Aldrich | Cat#M48309 |
| 99% 6-methyl-5-hepten-2-one | Sigma-Aldrich | Cat#M48805-100ML |
| 95% 4-methyl-3-hexanol | Enamine | Cat# 615-29-2 |
| 98% 3-hexanone | Aldrich Chemistry | Cat#103020-10G |
| 98% ethylpyrazine | Sigma-Aldrich | Cat#250384-5G |
| 99% propionic acid | Sigma-Aldrich | Cat#W292419-SAMPLE-K |
| 100% ethanol | Decon Laboratories | Cat#2716 |
| ≥99.5% isopropanol | Fisher Chemical | Cat#A416SK-4 |
| RNAClean SPRI XPBeads | Beckman Coulter | Cat#A63987 |
| Critical commercial assays | ||
| Endotoxin-free midiprep kit | Machery-Nagel | Cat#740420.10 |
| HiScribe T7 Arca mRNA kit (with tailing) | New England Biolabs | Cat#E2060S |
| Experimental models: Organisms/strains | ||
| O. biroi clonal line A wild type | Kronauer Lab | N/A |
| O. biroi clonal line B wild type | Kronauer Lab | N/A |
| O. biroi clonal line B [ie1-DsRed-ObirOrco-QF2-15xQUAS-GCaMP6s]; “GCaMP6s ant” | This paper | N/A |
| Deposited data | ||
| O. biroi reference genome v5.4 | McKenzie and Kronauer10 | GCA: 003672135.1 |
| Whole-genome sequence of GCaMP6s ant | This paper | BioProject ID: PRJNA947257 |
| Confocal microscopy data | This paper | Brain Image Library IDs: 7b20a5b168a92088; 626fd578e97289bc; c2031d8218de058b |
| Calcium imaging data | This paper | DANDI Archive ID: 000467 |
| Oligonucleotides | ||
| Primers for plasmid construction; see Table S4 | This paper | N/A |
| Recombinant DNA | ||
| plasmid pGL3-IE1 | Gift from Zach Adelman; Anderson et al.62 | Addgene, ID 52894 |
| plasmid pBAC-ECFP-15xQUAS_TATA-SV40 | Gift from Christopher Potter; Riabinina et al.122 | Addgene, ID 104875 |
| plasmid pBac-DsRed-ORCO_9kbProm-QF2 | Gift from Christopher Potter; Riabinina et al.122 | Addgene, ID 104877 |
| plasmid pGP-CMV-GCaMP6s | Gift from Douglas Kim & GENIE Project; Chen et al.60 | Addgene 40753 |
| plasmid hyPBapis | Gift from Martin Beye; Otte et al.65 | N/A |
| plasmid pBAC-ie1-DsRed-ObirOrco-QF2-15xQUAS-GCaMP6s | This paper | Addgene, ID 200400 |
| Software and algorithms | ||
| ImageJ (Fiji) version 2.0.0 | Schindelin et al.123 | https://fiji.sc/ |
| LABKIT plugin for ImageJ | Arzt et al.124 | https://github.com/juglab/labkit-ui |
| Trimmomatic 0.36 | Bolger et al.125 | http://www.usadellab.org/cms/?page=trimmomatic |
| bwa mem | Li126 | https://github.com/lh3/bwa |
| Picard | Broad Institute | http://broadinstitute.github.io/picard/ |
| samtools | Li et al.127 | https://github.com/samtools/samtools |
| R | R Core Team128 | www.R-project.org. |
| ggplot2 | Wickam129 | https://ggplot2.tidyverse.org |
| Integrative Genomics Viewer | Robinson et al.130 | https://software.broadinstitute.org/software/igv/ |
| msa | Bodenhofer et al.131; Larkin et al.132 | https://bioconductor.org/packages/release/bioc/html/msa.html |
| BLAST | Morgulis et al.133 | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| karyoploteR | Gel and Serra134 | http://bioconductor.org/packages/release/bioc/html/karyoploteR.html |
| Prism | GraphPad Prism | https://www.graphpad.com |
| Image Stabilizer plugin for ImageJ | Li135 | http://www.cs.cmu.edu/~kangli/code/Image_Stabilizer.html |
| napari | napari contributors | Zenodo DOI: 10.5281/zenodo.3555620 |
| Other | ||
| Additional scripts | This study | https://github.com/Social-Evolution-and-Behavior/HartKronauer2023 |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Ant husbandry and maintenance.
Ants were kept at 25°C in nests constructed by lining 5cm diameter Petri dishes with plaster of Paris. Nests were kept humidified and supplied with frozen fire ant pupae as food ~3 times per week during the brood care phase. Petri dishes held 20–80 workers each. GCaMP6s ants were propagated by cross-fostering GCaMP6s eggs into colonies with clonal line A adults,12 which were then separated into isogenic GCaMP6s colonies after eclosion. Isogenic colonies can easily be assembled in this species because O. biroi reproduces clonally.45,113 We separated transgenic animals at the G1 stage and returned all offspring of a particular G1 individual to the same nest as their parent. For live imaging experiments, stock colonies for experiments were assembled by moving cohorts of cross-fostered GCaMP6s ants that eclosed within 2 weeks of one another into fresh Petri dish nests. Adult female ants were selected from stock colonies for GCaMP imaging experiments. The age of experimental ants was 55–60- and 90–104 days post eclosion for the general odorant and alarm pheromone imaging experiments, respectively. Individuals with eyespots (indicative of intercastes)114–115 were excluded from our imaging study.
METHOD DETAILS
Behavior
Alarm pheromones.
We purchased 96% 4-methyl-3-heptanone from Pfaltz and Bauer (Item #: M19160), and ≥99% 4-methyl-3-heptanol and 99% 6-methyl-5-hepten-2-one from SigmaAldrich (Item numbers M48309 and M48805–100ML, respectively). 95% 4-methyl-3-hexanol was purchased from Enamine (CAS# 615–29-2), and paraffin oil from Hampton Research (cat. #HR3–421). We also initially tested the compound undecane, which functions as an alarm pheromone in several other ant species and is found in clonal raider ant extracts.22,116–119 However, undecane has a lower volatility / vapor pressure than the other alarm pheromones (Table S3), and only elicited non-specific walking behavior and no robust calcium responses in our experimental paradigms. We therefore did not investigate undecane further.
General odorants.
98% 3-hexanone was purchased from Aldrich Chemistry (Item number 103020–10G). 98% ethylpyrazine and 99% propionic acid were purchased from Sigma-Aldrich (Item numbers 250384–5G and W292419-SAMPLE-K, respectively). 100% ethanol was purchased from Decon Laboratories (Item #: 2716), and ≥99.5% isopropanol from Fisher Chemical (Item #: A416SK-4). We initially also tested six additional general odorants with lower volatility / vapor pressure (Table S3). However, these odorants did not elicit robust calcium responses in our experimental paradigm and were therefore not studied further.
Colony alarm bioassay.
Alarm behavior assays were performed as described previously.22 For experiments with 4-methyl-3-hexanol and 6-methyl-5-hepten-2-one, 30 mixed-age ants from clonal line B were introduced without brood into each arena. Trials were also performed with undecane, which only induced non-specific walking behavior. For behavioral experiments with GCaMP6s ants, due to limited numbers, 15–20 ants were introduced into each arena. Prior to behavioral experiments, ants were allowed to settle for at least 5 days, until they had laid eggs and spent most of their time within a tightly packed nest pile.
Each compound (pure compounds for 4-methyl-3-heptanone, 4-methyl-3-heptanol, 4-methyl-3-hexanol, 6-methyl-5-hepten-2-one, or a 9:1 4-methyl-3-heptanone:4-methyl-3-heptanol blend) was freshly diluted 1:20 with 100% pentane each day of experiments. After recording baseline activity for 4 minutes and 30 seconds, 50 μL of each compound was added to a ~1 cm2 piece of filter paper and allowed to evaporate for 30 seconds before folding and placing into the stimulus chamber. Behavioral responses were recorded for another 5 minutes.
Data were analyzed as described previously, scoring three metrics of interest by hand: (1) the number of ants outside the nest pile, (2) the number of ants outside the nest chamber, and (3) the number of ants touching the mesh wall. We limited statistical analyses to the time window starting 1 minute prior to adding the stimulus and 2 minutes after. To evaluate the effect of the stimulus over time, we performed a two-way repeated measures ANOVA, and to determine the effect of the stimulus at each timepoint we used Dunnett’s multiple comparisons test.
Categorical analysis of the major behavioral response to each odorant (4-methyl-3hexanol, 6-methyl-5-hepten-2-one, and the vehicle control, plus reanalysis of responses to 4methyl-3-heptanone, 4-methyl-3-heptanol, and the blend from experiments in a previous study22) was performed by visually classifying each video as one of the following in a blinded manner: “Immediate panic alarm”: The nest pile was disassembled within one minute following stimulus exposure. “Ants leave nest”: The nest pile persisted for at least one minute following stimulus exposure, but over half the ants left the nest pile within the first minute. “No immediate response”: The nest pile persisted for at least one minute following stimulus exposure, and fewer than half the ants left the nest pile within the first minute. We also identified the time when the initial nest pile disappeared after addition of the stimulus. Because we could not discern the removal of single eggs from the nest pile from the video, for the purposes of this analysis the nest pile was considered to persist as long as the same area continued to contain at least one egg and two adult ants. We calculated the percentage of time during which the initial nest pile persisted for the first two minutes after addition of the stimulus. We evaluated the effect of the compounds on the nest pile dissipating using a one-way ANOVA and Šidák’s multiple comparisons test to compare each additional alarm pheromone to each of the two known O. biroi alarm pheromones (4-methyl-3-heptanone, 4-methyl-3-heptanol, and a 9:1 blend of the two compounds). Statistical analyses on behavioral data were performed using GraphPad Prism Version 9.4.0 for Windows, GraphPad Software, San Diego, California USA (https://www.graphpad.com).
Generation of transgenic ants
Cloning and plasmid assembly.
We assembled plasmid pBAC-ie1-DsRed-ObirOrco-QF2–15xQUAS-GCaMP6s using multiple rounds of PCR for generating fragments, restriction digestion with gel purification for backbones, and Gibson assembly cloning.120–121 Following each Gibson assembly step, correct assembly was verified using restriction digests and by sequencing PCR amplicons spanning across each of the fragment boundaries. Primer sequences for plasmid construction are listed together in Table S4. A plasmid schematic was made using MacVector software, MacVector Inc, Apex, North Carolina, USA (https://macvector.com).
ObirOrco: A 2.4kb promoter/enhancer fragment, including intergenic sequence and the entire 5’ UTR, amplified from clonal raider ant genomic DNA, clonal line B (NCBI LOC105284785) (primers: forward, 5’- tagttgtggtttgttgttcgcacaTATGTCACGTAATCAGCTTTTGACG −3’, lowercase shows Gibson homology region; reverse 5’- gcgcttgggtggcatgttgcaTCATATGTCTGCGAGCAAATGGAACG −3’).
piggyBac backbone from pBAC-ECFP-15xQUAS_TATA-SV40 (Addgene, ID #104875),122 from double restriction digest with SpeI (New England Biolabs [NEB] #R3133S) and EcoRV (NEB #R0195S).
ie1-A: An enhancer/promoter from pGL3-IE1 (Addgene ID #52894)62 (primers: forward 5’- ttatcgaattcctgcagcccgggggatccaACTAGTTGTTCGCCGAGCTCTTACGCGC −3’, reverse 5’- ctcggaggaggccatCCGCGGCGAACAGGTCACTTGGTTGTTCACGATCTTG −3’).
DsRed from pBac-DsRed-ORCO_9kbProm-QF2 (Addgene ID #104877)122 (primers: forward 5’- acctgttcgccgcggATGGCCTCCTCCGAGAA −3’, reverse 5’- ttattatatatatattttcttgttatagatGGCGCGCCCGAACACATATGCGAACAACAAACCACAACTAGAATGCAGTG −3’).
QF2 from pBac-DsRed-ORCO_9kbProm-QF2 (primers: forward 5’- aaccaagtgacctgttcgggccggACATATGCAACATGCCACCCAA −3’, reverse 5’- acccagtgacacgtgaccgCGAGCGCTGGATCTAAACGAGTTTTTAAGC −3’).
15xQUAS from pBAC-ECFP-15xQUAS_TATA-SV40 (primers: forward 5’- cggtcacgtgtcact −3’, reverse 5’- tgagaacccatcgaacaagcGTTTAAACAGATCTGTTAACGAATTGATC −3’).
GCaMP6s from pGP-CMV-GCaMP6s (Addgene ID # 40753)60 (primers: forward 5- gggccggcctgttcgAGCGCTTGTTCGATGGGTTCTCATCATCATC −3’, reverse 5’- atatattttcttgttatagatggCGCGCCGTAGCCCTAAGATACATTGATGAGTTTG −3’)
pBAC-ie1-DsRed from Gibson assembly of piggyBac backbone, ie1-A, and DsRed fragments, transformed into NEB 10-beta competent cells (item # C3019H).
ie1-B from pBAC-ie1-DsRed, (primers: forward 5’- ctgcattctagttgtggtttgttgttcgcaCATATGTGTTCGCCGAGCTCTTACGCG −3’, reverse 5’- catcgaacaagcgctcgaacaggccggcccGAACAGGTCACTTGGTTGTTCAC −3’)
pBAC-ie1-DsRed-ie1-GCaMP6s from Gibson assembly of pBAC-ie1-DsRed (linearized using double restriction digest with NdeI [NEB #R0111S] and AscI [NEB #R0558S]), ie1-B, and GCaMP6s.
pBAC-ie1-DsRed-ie1-QF2–15xQUAS-GCaMP6s from Gibson assembly of pBAC-ie1-DsRed-ie1-GCaMP6s (linearized using double restriction digest with FseI [NEB #R0588S] and AfeI [NEB # R0652S]), QF2, and 15xQUAS.
pBAC-ie1-DsRed-ObirOrco-QF2–15xQUAS-GCaMP6s from Gibson assembly of pBAC-ie1-DsRed-ie1-GCaMP6s (linearized and second ie1 copy removed using restriction digest with NdeI) and ObirOrco.
Preparation of injection mixes.
Plasmid DNA for injection was purified using a Machery-Nagel endotoxin-free midiprep kit (item #740420.10). The final pellet was washed under RNAse-free conditions and dissolved in nuclease-free water. To remove precipitated DNA from injection mixes, the dissolved plasmid mix was spun in a microcentrifuge at top speed for 5 minutes, and the top 90% of the supernatant was recovered. This step was repeated at least 5 times to produce injectable mix with negligible precipitate, which was stored at −20°C until injection.
We generated mRNA from the hyperactive piggyBac variant hyPBaseapis.65 A DNA template was generated by PCR amplification of the transposase coding sequence, with addition of a T7 promoter on the forward PCR primer, then purified using Beckman Coulter RNAClean SPRI XPBeads (item #A63987). In vitro transcription was performed with the NEB HiScribe T7 Arca mRNA kit (with tailing) (item #E2060S) to produce poly(A) tailed mRNA encoding hyPBaseapis. The mRNA was purified using RNAClean beads (using 1.5x volume of beads compared to the reaction mix) and stored in nuclease-free water at −80°C. Template and RNA were handled under RNAse-free conditions, and a sample of mRNA was examined on an Agilent Bioanalyzer to verify RNA length and confirm absence of degradation. All DNA and RNA concentrations were measured using a Thermofisher Nanodrop.
Egg collection, microinjection, and larval rearing.
Eggs were collected as described previously,12 with a modified schedule for treatments with eggs <3 hours old. We tested the effect of injecting even younger eggs than our previous protocol which used eggs <5 hours old12 so that hyPBaseapis mRNA could be translated into active transposase while embryos still had very few nuclei, potentially reducing mosaicism. For these treatments, old eggs were removed from nests from 9am-10am, and eggs for injection were collected from 11am-11:30am, 1pm1:30pm, 3pm-3:30pm, and 5pm-5:30pm. Injections were performed from 11:30am-12:30pm, 1:30pm-2:30pm, 3:30pm-4:30pm, and 5:30pm-6:30pm. This schedule meant that the vast majority of eggs were less than 3 hours old when injected.
Microinjections were performed as described previously,12 with the following changes: On each injection day, final injection mixes were produced by thawing and combining stored aliquots of plasmid DNA and hyPBaseapis mRNA under RNAse-free conditions in nuclease-free water, into a final concentration of 27.8fmol/μL plasmid and the desired concentration of hyPBaseapis. The injected plasmid had a length of 12,025bp. The final mix was spun at top speed in a microcentrifuge for 5 minutes, and the top 90% of supernatant was used for injection. The initial mix was split into 4 aliquots and kept on ice for the day. A different aliquot was used for each round of injections. On occasions where the needle clogged, the mix was spun at top speed in a microcentrifuge before loading a new needle. The injection pressure was initially set to 3600kpa but was adjusted throughout the course of injections to maintain a consistent flow of liquid into the embryos. We varied the age of eggs and the concentration of transposase mRNA in the injection mix. Higher rates of fluorescent G0s were obtained when eggs were <3 hours old rather than <5 hours old at the time of injection. Mixes with >110ng/μL mRNA concentrations produced low hatch rates and no fluorescent G0s (Table 1).
Larvae were reared as described previously.12 Briefly, G0 larvae were hatched and placed in small colonies housed in 5cm diameter Petri dishes with a moist plaster of Paris floor to be reared by adult ants from clonal line A, which we refer to as “chaperones” when we use them to rear offspring transferred from other colonies.12 Colonies were examined under an epifluorescence microscope to confirm that some larvae expressed DsRed, indicating uptake of the plasmid.
Rearing initial transgenic populations.
G0 individuals were reared to adulthood. For cohorts of sufficient size (~20 individuals), chaperones were removed. When the number of G0s was too small to form a robust colony, they were supplemented with wild type clonal line A ants to obtain a population of ~20 individuals. One hind leg was removed from each wild type ant to reduce their egg-laying rate compared to the G0 ants in the nest. Then, the colonies were allowed to produce G1 eggs, which were usually collected twice a week. Collected eggs were transferred to a small colony of ~20 chaperones. G1 individuals were reared to adulthood in these nests and were examined for fluorescence. Different G1 individuals potentially resulted from independent transgene insertion events. To ensure that future transgenic populations were genetically homogeneous, each fluorescent G1 adult was separated soon after eclosion, and transferred to a new transgenic line-founding colony with ~19 clonal line A ants. Eggs were collected about twice a week from these nests and given to chaperones. Fluorescent adults produced from these colonies were then returned to the transgenic line-founding colony of origin. Through several cycles of this process, genetically homogenous transgenic populations were raised and non-fluorescent individuals were removed, yielding pure colonies.
Phenotyping transgenic ants
Fluorescence microscopy.
Confocal microscopy of antibody-stained tissue was conducted using Zen image acquisition software on a Zeiss LSM 880 and a Zeiss LSM 900 equipped with 405nm, 488nm, 561nm and 633nm laser lines. Images were obtained using either a Zeiss LD LCI Plan-Apochromat 40X / 1.2NA or a Zeiss LD LCI Plan-Apochromat 25X / 0.8NA multi-immersion objective lens depending on the tissue sample and Zeiss Immersol G immersion medium (Zeiss # 462959–9901-000). Z-projection images were produced from stacks taken at 1μm steps using ImageJ/FIJI.123 Two-photon fluorescence microscopy was performed using a Bruker Investigator with a Coherent Axon laser tuned to 920nm, equipped with dual GaAsP detectors, resonant scanning galvanometer, Z-piezo module for high-speed Z-positioning, PrairieView software, and an Olympus 40X 0.9NA water-immersion objective. Images of transgenic pupae (Fig. 1B) were produced on an Olympus SZX16 epifluorescent microscope equipped with an X-Cite XYLIS light source, Olympus EP50 camera, and the appropriate filter cubes.
Immunohistochemistry.
Antibody staining of ant brains was performed as reported previously.9 Briefly, the brains of female ants of a single-age cohort were dissected in cold phosphatebuffered saline (PBS) and fixed in 4% paraformaldehyde for 2 hours at room temperature. For antenna staining, a small section of cuticle was mechanically separated prior to fixation to enhance access. Blocking was performed for at least 2 hours using fresh PBS containing 0.1% or 0.5% Triton X-100 and 5% donkey serum albumin. Samples were incubated with the appropriate dilution of primary antibody in fresh blocking solution on an orbital shaker table at room temperature. Following primary incubation, samples were washed and incubated with fluorescently tagged secondary antibody diluted in fresh blocking solution. The following antibodies were used: chicken anti-GFP (Abcam #ab13970), rabbit anti-RFP (Rockland #600–401-379), mouse anti-SYNORF1 (DSHB #3C11), mouse anti-Orco (gift from V. Ruta), goat anti-chicken Alexa 488 (Invitrogen #A-11039), donkey anti-mouse Alexa 647 (Invitrogen #A32787), and donkey anti-rabbit Alexa 594 (Invitrogen # A-21207). For some experiments, DAPI (Invitrogen #D1306) and fluorescently tagged phalloidin (Invitrogen #A34055) were included during the secondary antibody incubation step. Stained tissue was mounted in SlowFade mounting medium on silane-coated microscopy slides (VWR #63411–01) and stored at 4°C. For the high quality anatomical AL reconstruction, a confocal stack of the right AL from a GCaMP6s-positive brain stained with anti-SYNORF1 was manually segmented using the LABKIT plugin for ImageJ, at 1μm z-axis resolution.123–124 To quantify overlap between GCaMP and Orco expression, a second AL reconstruction was performed using the left AL of a GCaMP6s-positive brain stained with anti-Orco and phalloidin. All glomeruli were reconstructed and then checked for GCaMP6s/anti-Orco/phalloidin signal. The T7 glomerulus cluster was identified by its anatomical position and lack of Orco antibody staining, as described previously.11 Renders of the reconstructed ALs (Fig. S2A–B) were generated using napari (Zenodo DOI: 10.5281/zenodo.3555620). While examining brains one month after unilateral antennal ablation, we noticed that the AL associated with the ablated antenna appeared smaller than the AL associated with the intact antenna. We therefore reconstructed AL volumes from several brains (n=3) and found a non-significant trend toward smaller volumes on the ablated side (means of 165,360μm3 and 209,412μm3 for ablated vs. non-ablated ALs, respectively; p=0.073, paired values T-test). We did not investigate this phenomenon further.
Genome sequencing and genomic analyses.
A single GCaMP6s ant was disrupted with a Qiagen TissueLyser II, and genomic DNA was extracted using a Qiagen QIAmp DNA Micro Kit. Libraries were prepared using Nextera Flex, and paired end, 150 base pair reads were sequenced on an Illumina NovaSeq S1 Flow Cell. Raw reads were trimmed using Trimmomatic 0.36125 and aligned using bwa mem126 to both the O. biroi reference genome (Obir_v5.4, GenBank assembly accession: GCA_003672135.1)10 and a linearized plasmid reference genome created by “cutting open” the plasmid sequence at an arbitrary location on the backbone, and pasting 150 bp from the end at the front of the sequence and 150 bp from the front at the end of the sequence to accommodate any reads that might align to the vicinity of the “cut”. Reads were sorted and deduplicated using Picard (http://broadinstitute.github.io/picard/), and read depth was recorded at all sites using “samtools depth -aa”127 (obtaining approximately 44x coverage). To infer the read depth of well-assembled genomic regions, we obtained all heterozygous SNPs with read depth less than 2x the genome-wide median, which excluded the fewer than 0.5% of such SNPs which likely resulted from errors in genome assembly. We then randomly selected an equal number of heterozygous SNPs as the number of base pairs in the transgene insert, and calculated read depth at those sites, and separately along both the portion of the transgene insert sequence that aligned to ObirOrco and the rest of the transgene insert. Data in Fig. S3A were plotted using the R package ggplot2.128–129
Junction reads that aligned to both the transgene insert and the O. biroi reference genome were identified using the Integrative Genomics Viewer,130 and alignments were queried by each junction read name using “samtools view”.127 We performed multiple sequence alignment on these junction reads from each end of the insert using CLUSTAL 2.1 in the R package msa128,131,132 and generated consensus sequences. To obtain the sequence of the insertion site in the reference genome, the portion of the sequence that was identical to the end of the transgene insert sequence was removed from the junction read consensus sequences. BLAST133 searches of the partial consensus sequence identified a position consistent with the position these junction reads had aligned to in the O. biroi reference genome. Fig. S3B was generated using the R package karyoploteR.134 The insertion locus was examined in the NCBI genome data viewer (Obir_v5.4, GenBank assembly accession: GCA_003672135.1)10 to check for the presence of predicted gene models.
In vivo calcium imaging
Specimen preparation.
Ants for live imaging were anesthetized on ice for ~3 minutes and then fastened to a custom two-photon imaging mount using blue-light curable glue. The antennae were restrained with a thin strip of Parafilm to decrease motion artifacts. A sheet of Parafilm with a hole for the ant’s head was applied on top of the preparation, and a watertight seal was created around the border of the head using additional glue. The preparation was then bathed with fresh ant saline (127 mM NaCl, 7 mM KCl, 1.5 mM CaCl2, 0.8 mM Na2HPO4, 0.4 mM KH2PO4, 4.8 mM TES, 3.2 mM Trehalose, pH 7.0)26 and suffused for the duration of the imaging session with additional ant saline to prevent desiccation, before excising a small imaging window in the cuticle using a sterile hypodermic needle and sharp forceps. The window was positioned above the brain, and connective and glandular tissue were removed to reveal the antennal lobes. We always imaged the right antennal lobe. Care was taken to keep the antennae and antennal nerves intact. In some cases, a muscle between the ALs and near the esophagus was severed, which reduced the amount of brain motion. This was advantageous for imaging, but not always feasible due to the small distance between the ALs and slight differences in the accessibility of the muscle from ant to ant.
Two-photon recording.
Antennal lobe volumes were recorded at 2X optical zoom and a resolution of 512×512×33 voxels (XYZ) with 5μm Z steps, resulting in a volume with dimensions of 148μm × 148μm × 165μm, large enough to capture calcium transients from the entire AL which has approximate dimensions of 65μm x125μm x150μm. As glomeruli are typically spheroid with a diameter of 10–20μm, each glomerulus was captured in many voxels in all three dimensions. Recordings were obtained at 27.5 frames per second, resulting in 0.83 volumes per second. At the beginning of each imaging experiment, we located the dorsal surface of the AL and set that as the top of the imaging volume. We could clearly detect the boundary at the ventral surface of the AL where GCaMP6s signal disappeared, indicating that we imaged all GCaMP6s-positive glomeruli. Laser power and gain were adjusted for each ant so that all glomeruli were visible, but signal was unsaturated. Because we imaged at different depths, we compensated for loss of signal through tissue by increasing the laser power at greater depth using an exponential function. We regularly re-calibrated the position of the imaging volume, laser power, and gain in case there were any changes in baseline fluorescence or brain position during the experiment. The bilateral calcium imaging experiment was conducted at 1X optical zoom, resulting in an imaging volume with dimensions of 296μm × 296μm × 165μm.
Stimulus presentation.
Odors were presented using a custom-built olfactometer on 600mL/min of filtered, medical-grade air regulated with a pair of digital mass flow controllers (AliCat# MC-1SLPM-D-IPC/5M). A constant ‘carrier’ air stream (200mL/min) was presented to the ant for the duration of the imaging session to reduce mechanical stimulation of the antennae resulting from air turbulence, while a ‘stimulus’ portion of the air stream (400mL/min) was diverted and perfumed before rejoining the carrier stream at a manifold immediately upstream of the imaging preparation. By default, stimulus air bypassed control and odor vials and entered the manifold directly. During stimulus presentation, the air was perfumed by triggering high-speed three-way valves (Grainger# 6JJ52) controlled by an Arduino Uno and custom MatLab scripts, which directed the air to control or odor vials. Imaging and stimulus trials were synchronized in time using Bruker PrairieView software (i.e., the same TTL signal initiated both imaging and odor stimulation). Odors were dissolved in paraffin oil vehicle to a total volume of 300μL (concentrations represent v/v in the vial), were stored in 4mL amber glass vials with PTFE/silicone septa and connected to valves and the odor manifold via sterile hypodermic needles and nylon Luer tapers. Odor vials were prepared at the beginning of each day of imaging experiments. The air stream was directed onto the ant’s antennae using flexible PVC/vinyl tubing with an internal diameter of 1.588mm (United States Plastic Corp. Item #: 54411) from a distance of approximately 1mm.
All odor presentations had a 3s lead time and lasted for 5s. Before odor presentation, we presented the ant with the paraffin oil vehicle as a negative control and confirmed the absence of fluorescence changes before continuing the experiment. For the general odorant imaging experiment, each ant was then presented with a randomized sequence of 7–9 general odorants (48.0% concentration) which was repeated for three trials. Each of the odorants in the panel was tested in 2–6 ants. Odorants: 3-hexanone, butyric acid, dodecyl acetate, ethanol, ethylpyrazine, geranyl acetate, isopropanol, linalool, propionic acid, terpineol, and (+)-valencene. Only responses to the 5 odorants that generated robust calcium responses that were consistent across ants are shown in Fig. 3. We sometimes observed calcium activity from the other odorants, but responses were weak and not reproducible across trials in different ants. For the alarm pheromone imaging experiment, we first presented each ant with the paraffin oil vehicle and then with a positive control isopropanol stimulus. We only continued experiments with animals that showed calcium responses to the positive control but not the negative control. Each ant was presented with the four alarm pheromones in a random sequence which was first repeated for three trials at the lower concentration, followed by three additional trials at the higher concentration (for a total of 24 pheromone presentations per animal). Additional trials were performed with undecane, but these trials were not analyzed further due to absence of robust calcium responses. To reduce the impact of habituation to stimulus, each ant was presented with odors at two concentrations out of four concentrations tested (n=13 ants total, 3 ants presented with 0.75% and 12.0% odor concentrations; 2 ants with 3.0% and 12.0%; 3 ants with 12.0% and 48.0%; and 5 ants with 3.0% and 48.0%). In rare cases, we observed large motion artifacts during a recording, in which case the trial was repeated. For the bilateral calcium imaging experiment, ants were presented only with the paraffin oil vehicle and 4-methyl-3-hexanol at 48.0%. Vials and caps were reused after cleaning as follows: removal of remaining liquid, 2x wash with 100% ethanol alternating with 2x rinse in distilled water, 2x wash with 3% Alconox alternating with 2x rinse in distilled water, 2x rinse in distilled water, air dry.
Image processing and analysis.
Image processing was done in Fiji/ImageJ.123 To initially characterize response to odorants, we loaded recordings, used the “Deinterleave” function to separate them into 33 slices corresponding to videos of each recording depth, ran the Image Stabilizer plugin,135 applied the “Gaussian Blur” filter with 1-sigma, calculated F0 from the mean of frames 1–5 (before any calcium changes were detected) and calculated ΔF/F0 by subtracting and then dividing the image stack from F0. The peak fold change was calculated using the “Z Project” function set to average the ΔF/F0 from frames 9–14, when the calcium responses typically peaked. After applying a pseudocolor LUT, we examined the peak fold change at all 33 depth positions to get a sense of the organization of glomerular responses across the ALs. We determined that all responses were positive and responding glomeruli were generally well-separated in the x/y axes. We performed additional analyses using max z-projections. Z-projections were generated by running image stabilization on each imaging plane,135 computing ΔF/F0, running the “Minimum” filter with 2-pixel radius to reduce noise, applying the “Z Project” function through all slices with maximum setting, and changing all values >4 to 4 or <−1 to −1 using the “changeValues” function, to equalize the LUT range. To analyze glomerular response patterns across the whole AL, we examined all max z-projection images at the highest odor concentration for each ant and drew regions of interest (ROIs) around every glomerular region that responded to any odor in at least two trials (a small number of trials were excluded due to large motion artifacts that were only apparent after generating max projections). We then quantified the peak fold change across all trials for a particular odor and concentration and designated an ROI as responding if the value was ≥0.2. In cases where two odors activated ROIs that overlapped in the max z-projection, we examined the z-stacks to determine if the responses occurred at the same z-depth and excluded overlaps if the responses occurred at different depths. For visualizing the imaging volume (Fig. 3E, top), we used the first frame of a recording, and generated max z-projections using the z-project function. For the x-projection, we used the “Re-slice” function starting from the left to re-order the pixels, and then used the max z-project function. To visualize calcium responses throughout the imaging volume (Fig. 3E, bottom), the max z-projections of calcium responses were generated as before, but because imaging noise was more apparent in the x-projections due to higher resolution in that axis compared to the z-axis, the minimum filter was set to a 3-pixel radius.
For analyses of single glomeruli, we visually identified the z-plane containing the center of the glomerulus of interest for each trial, generated a max z-projection across 3 adjacent imaging planes (to reduce the impact of brain motion in the z-axis), and then calculated ΔF/F0. Peak fold change was quantified by averaging the ΔF/F0 over frames 9–14, the time range during which most odor-evoked calcium responses peaked.
Spatial relationships between PGb, PGa, and 6G were quantified by examining a video z-plane in which all three glomeruli were visible, placing a marker at the center of each glomerulus, and calculating the vector connecting the centers, with PGb at (0,0). In two individuals, the spatial relationship between PGb and PGa was not quantified because PGa could not be identified.
Statistical analyses of odor responses.
We analyzed the responses of the three glomeruli PGb, PGa, and 6G to different odors and concentrations. For every glomerulus/odor combination, peak fold change values from all trials were loaded into R, and a linear regression model was fit for the peak calcium response as a function of odor concentration, with a random effect for individual, using the glm function.128 Model predictions were generated and plotted with ggplot2129 with 95% confidence intervals.
To examine temporal dynamics in the three focal glomeruli, normalized calcium response traces from each glomerulus were loaded into R.128 The first five recorded frames were used as the baseline, and calcium response onset was defined as the latency between the start of the stimulus presentation and the time point where ΔF/F0 exceeded the mean of the baseline + 3SD of the baseline. The time to response maximum was defined as the latency between the start of the stimulus presentation and the timepoint with the maximum value of ΔF/F0. Traces where ΔF/F0 never exceeded the mean of the baseline + 3SD of the baseline were excluded. Only glomerulus/pheromone combinations with typically robust responses were included (4-methyl-3-heptanone in PGb; 4-methyl-3-heptanol and 4-methyl-3-hexanol in PGb and PGa; 6-methyl-5-hepten-2-one in 6G). Data were plotted with ggplot2.129 To test for effects of pheromone and glomerulus identity on calcium response temporal dynamics, we performed statistical analyses on the subset of data for which the same pheromone caused responses in more than one focal glomerulus, i.e., responses in PGb and PGa from trials with 4-methyl-3-heptanol and 4-methyl-3-hexanol. For each temporal parameter, we built a linear mixed effects model using the lme function in R,128 modeling the effects of pheromone, glomerulus, and an interaction of pheromone/glomerulus, with a random effect for trial ID nested within ant ID.
QUANTIFICATION AND STATISTICAL ANALYSIS
Analyses of behavioral data were performed using PRISM (GraphPad) (Figs. 1C–F, S2C–F; Table S2). Analyses of genomic data were performed using software as described and cited above (Fig. S3). Analyses of calcium imaging data were performed using R and the packages as described and cited above (Figs. 3D, 5C-D, S4C, S5, S6, S7). Data are presented as mean±SEM, mean±SD, or in another format as indicated in the relevant figure legends.
Supplementary Material
Supplemental Video 1. Colony alarm bioassay, related to Fig. 1. Representative videos of each of the three categories of major behavioral responses. Videos show 1 minute prior to stimulus, time skips while stimuli are put in place, then 2 minutes after stimulus (filter paper + compound). The presence of the red circle on the upper right indicates that stimulus is in place. Sped up 8x.
Supplemental Video 2. Odor-evoked calcium response in the ant antennal lobe, related to Fig. 3. Representative two-photon recording of the calcium response to 5s presentation with ethylpyrazine (48%). A single slice corresponding to 110μm z-depth was extracted from the volumetric recording, and image stabilization was applied [S21]. The “ON” label indicates the 5s odor presentation period. Sped up 3x.
Figure S1. Additional characterization of GCaMP6s ants, related to Fig. 2. (A) The same two clonal line B pupae, one wild type and one transgenic, imaged under bright field (left) and epifluorescence, with filters set to detect GCaMP6s (middle) and DsRed (right). Pupae were imaged 10 days after pupation. (B) Anti-DsRed (magenta) labels the ALs, indicating co-expression of DsRed with GCaMP6s (green; endogenous fluorescence) in ants carrying [ie1-DsRed, ObirOrco-QF2, 15xQUAS-GCaMP6s]. Phalloidin stains actin (cyan). (C) GCaMP6s fluorescence (green) is detectable in the subesophageal zone (SEZ). (D) GCaMP6s fluorescence (green) is also visible in processes innervating part of the central complex (CC) as well as in a nearby cluster of somas. Images show max z-projections through the imaged brain regions. (E) After unilateral ablation of the antenna (from the scape), bilaterally symmetrical GCaMP6s signal is still detectable in the central complex (CC), as well as the subesophageal zone (SEZ). No anti-Orco signal was detected in these brain regions. Images show max z-projections through the imaged brain regions.
Figure S2. GCaMP6s ants have normal antennal lobes and respond to alarm pheromones, related to Figs. 1–2. (A) 505 glomeruli reconstructed from the right AL of a GCaMP6s ant using anti-SYNORF1 staining. (B) 508 glomeruli reconstructed from the left AL of a GCaMP6s ant using GCaMP6s signal, anti-Orco staining, and phalloidin staining. 502 glomeruli stained positive for phalloidin, GCaMP6s, and Orco; 4 glomeruli stained positive for phalloidin and GCaMP6s, but not Orco; and 2 glomeruli stained positive for phalloidin, but not GCaMP6s and Orco. The AL in (B) comes from a different ant than that in (A) and is shown in mirror image to align more closely with the image in (A). (C-F) Colony alarm bioassay, showing mean±SEM. (C-D) GCaMP6s ants and wild type ants leave the nest in response to 4-methyl-3-heptanone, 4-methyl-3-heptanol, and a 9:1 blend of the two compounds. (E-F) GCaMP6s ants and wild type ants leave the nest chamber in response to 4-methyl-3-heptanone and the blend; wild type ants also leave the nest chamber in response to 4-methyl-3-heptanol, while this response was not statistically significant in the GCaMP6s ants. Analyses for wild type ants (D, F) are based on previously published data (Fig. 3, 2600μg and vehicle control treatments in Lopes et al.22). For the experiment in GCaMP6s ants (C, E), all pheromone treatments were compared to the same set of vehicle control trials. For the experiments in wild type ants (D, F), each pheromone treatment was conducted alongside its own set of vehicle control trials. Treatments and controls from the same experiment were statistically compared to each other and are plotted using identical symbol shapes. *: p<0.05; **: p<0.01; ***: p<0.001; ****: p<0.0001. L: lateral; A: anterior.
Figure S3. Genomic analyses of the transgenic line used for imaging, related to Fig. 2. (A) Normalized read depth for reads aligning to a panel of heterozygous SNPs, the ObirOrco promoter, and the non-ObirOrco portion of the transgene. Normalized read depth for ObirOrco is ~1.5, corresponding to a single additional copy of ObirOrco inserted into the genome (added to the two endogenous copies). Normalized read depth of ~0.5 at the rest of the insert is also consistent with a single copy (haploid) insertion. (B) The transgene insert was localized to a site on the 2nd chromosomal scaffold. Black bars indicate breaks between contigs. (C) Close-up of the transgene insertion locus within an intron of the gene trace amine-associated receptor 9.
Figure S4. Calcium responses are bilaterally symmetrical and remain robust across trials, related to Fig. 4. (A) Max z-projections of both antennal lobes, imaged bilaterally. Both in raw fluorescence (left) and peak fold change of calcium response to presentation with 4-methyl-3-hexanol (right), the antennal lobes appear bilaterally symmetrical. (B) Max z-projections of peak fold change from a single ant after presentation with 4-methyl-3-hexanol. Three trials were performed at 3.0% concentration (top), and three additional trials were performed at 48.0% concentration (bottom). Timestamps for each trial demonstrate that responses are robust over the duration of a full experiment. Two adjacent focal glomeruli are circled. (C) Time series of calcium responses from each trial in (B) for the two adjacent glomeruli; responses in the left glomerulus are shown as alternating short and long dashes and responses in the right glomerulus are shown as solid lines; black bars indicate the 5s odor presentations. Responses are quantified from max z-projections of three slices centered on 105μm z-depth. L: lateral; A: anterior.
Figure S5. Increased odor concentration results in more responding glomeruli, related to Figs. 4–5. Counts of the number of responding glomeruli from max z-projections; boxes enclose the first to third quartile range, with bold line showing the median and whiskers enclosing the min and max values that fall within 1.5x the interquartile range. Data points show the mean number of responding glomeruli for a given ant across all trials for a particular odorant/concentration. n=13 ants total, 3 ants presented with 0.75% and 12.0% odor concentrations; 2 ants with 3.0% and 12.0%; 3 ants with 12.0% and 48.0%; and 5 ants with 3.0% and 48.0%. Single pheromones each activated a small number of glomeruli (A), and 4-methyl-3-heptanone, 4-methyl-3-heptanol, and 4-methyl-3-hexanol activated overlapping sets of glomeruli (B). 6-methyl-5-hepten-2-one only rarely activated glomeruli shared with the other pheromones (C). All pheromones were presented separately, rather than as blends. Only pheromones and pheromone combinations that activated at least one glomerulus are shown.
Figure S6. Quantification of peak fold change in PGb (top), PGa (middle), and 6G (bottom), related to Fig. 5. Three trials per ant for each odorant/concentration; n=13 ants total, 3 ants presented with 0.75% and 12.0% odor concentrations; 2 ants with 3.0% and 12.0%; 3 ants with 12.0% and 48.0%; and 5 ants with 3.0% and 48.0%. PGa could not be identified in two ants. Graphs show outputs of linear models (with 95% confidence intervals) for dose/response to each odorant in each glomerulus, with a random effect for individual. Concentrations were log transformed to show the linear relationship.
Figure S7. Temporal dynamics in three focal glomeruli, related to Fig. 5. (A) Extended time series for the response to 6-methyl-5-hepten-2-one in 6G. Data were collected separately from Fig. 5C. Shown are single trials each from five different individuals at 48% concentration (colored traces); mean±SD (black line and gray ribbon). Fluorescence plateaued for ~30s before declining and returning to baseline ~80s after odor presentation. (B) Two parameters of temporal dynamics extracted from glomerulus-specific calcium response traces. (C) Quantification of time to response onset (top) and time to response maximum (bottom) in the three focal glomeruli PGb, PGa, and 6G in response to stimuli at 48% concentration (n=8 ants, three trials per condition per ant). Only glomerulus/pheromone combinations with typically robust responses are shown. Boxes enclose the first to third quartile range, with bold lines showing the median and whiskers enclosing the min and max values that fall within 1.5x the interquartile range. For PGb and PGa responses to 4-methyl-3-heptanol and 4-methyl-3-hexanol, we used linear mixed effects models to test for effects of pheromone, glomerulus, and a pheromone/glomerulus interaction on the time parameters. Time to response onset: significant effects of pheromone (p=0.0034), glomerulus (p<0.0001), and the interaction (p=0.0030). Time to response maximum: significant effects of pheromone (p<0.0001) and glomerulus (p=0.047), but not the interaction (p=0.88).
Highlights.
An efficient transgenesis method for the clonal raider ant
Generation of a transgenic line expressing GCaMP6s in olfactory sensory neurons
Calcium imaging reveals highly stereotyped odor encoding in the ant antennal lobe
Alarm pheromones are sparsely encoded and activate a core glomerulus
Acknowledgments
We thank members of the Kronauer, Ruta, and Vosshall labs at Rockefeller University for helpful advice and discussions. Ben Matthews provided samples of the hyPBaseapis, Pbac-ECFP-15xQUAS_TATA-SV40, and pBac-DsRed-ORCO_9kbProm-QF2 plasmids. We thank Martin Beye for permission to use the hyPBaseapis plasmid, and Chris Potter for permission to use the pBAC-ECFP-15xQUAS_TATA-SV40 and pBac-DsRed-ORCO_9kbProm-QF2 plasmids prior to their appearance in publications. We thank Rob Harrell for sharing an alternate [ie1-DsRed] plasmid that was used in test injections. The anti-SYNORF1 antibody developed by E. Buchner was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology. We thank Meg Younger for initial training and access to a two-photon microscope. We thank Daniel Pastor for sharing his protocol for immunohistochemistry in the whole-mounted antennal club. DNA sequencing was performed at the Rockefeller University Genomics Resource Center. Some confocal microscopy was performed at the Rockefeller University Bio-Imaging Resource Center. This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R35GM127007, as well as the National Institute of Neurological Disorders and Stroke under Award Number R01NS123899 to D.J.C.K. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Additional support was provided by a Faculty Scholars Award from the Howard Hughes Medical Institute to D.J.C.K. T.H. was supported by an NSF Graduate Research Fellowship under award number 1946429 and a project grant from the Kavli Neural Systems Institute at The Rockefeller University. D.D.F. is an Open Philanthropy Fellow of the Life Sciences Research Foundation. L.E.L. was supported by an NSF Graduate Research Fellowship under award number DGE 194642. This work was supported in part by a grant to The Rockefeller University from the Howard Hughes Medical Institute through the James H. Gilliam Fellowships for Advanced Study program (to L.E.L. and D.J.C.K.). D.J.C.K. is an investigator of the Howard Hughes Medical Institute. This is Clonal Raider Ant Project paper number 28.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interests
The authors declare no competing interests.
References
- 1.Strausfeld NJ and Hildebrand JG (1999). Olfactory systems: common design, uncommon origins? Curr. Opin. Neurobiol 9, 634–639. 10.1016/S0959-4388(99)00019-7. [DOI] [PubMed] [Google Scholar]
- 2.Vosshall LB, Wong AM, and Axel R (2000). An olfactory sensory map in the fly brain. Cell 102, 147–159. 10.1016/S0092-8674(00)00021-0. [DOI] [PubMed] [Google Scholar]
- 3.Zhao Z and McBride CS (2020). Evolution of olfactory circuits in insects. J. Comp. Physiol. A 206, 353–367. 10.1007/s00359-020-01399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mysore K, Subramanian KA, Sarasij RC, Suresh A, Shyamala BV, VijayRaghavan K, and Rodrigues V (2009). Caste and sex specific olfactory glomerular organization and brain architecture in two sympatric ant species Camponotus sericeus and Camponotus compressus (Fabricius, 1798). Arthropod Struct. Dev 38, 458–497. 10.1016/j.asd.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Kelber C, Rössler W, and Kleineidam CJ (2010). Phenotypic plasticity in number of glomeruli and sensory innervation of the antennal lobe in leaf-cutting ant workers (A. vollenweideri). Dev. Neurobiol 70, 222–234. 10.1002/dneu.20782. [DOI] [PubMed] [Google Scholar]
- 6.Smith CD, Zimin A, Hold C, Abouheif E, Benton R, Cash E, Croset V, Currie CR, Elhaik E, Elsik E et al. (2011). Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile). Proc. Natl. Acad. Sci. U. S. A 108, 5673–5678. 10.1073/pnas.1008617108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou X, Slone JD, Rokas A, Berger SL, Liebig J, Ray A, Reinberg D, and Zwiebel LJ (2012). Phylogenetic and transcriptomic analyses of chemosensory receptors in a pair of divergent ant species reveals sex-specific signatures of odor coding. PLoS Genet 8, e1002930. 10.1371/journal.pgen.1002930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou X, Rokas A, Berger SL, Liebig J, Ray A, and Zwiebel LJ (2015). Chemoreceptor evolution in Hymenoptera and its implications for the evolution of eusociality. Genome Biol. Evol 10, 2490–2500. 10.1093/gbe/evy131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKenzie SK, Fetter-Pruneda I, Ruta V, and Kronauer DJC (2016). Transcriptomics and neuroanatomy of the clonal raider ant implicate an expanded clade of odorant receptors in chemical communication. Proc. Natl. Acad. Sci. U. S. A 113, 14091–14096. 10.1073/pnas.1610800113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKenzie SK and Kronauer DJC (2018). The genomic architecture and molecular evolution of ant odorant receptors. Genome Res. 28, 1757–1765. 10.1101/gr.237123.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryba AR, McKenzie SK, Olivos-Cisneros L, Clowney EJ, Pires PM, and Kronauer DJC (2020). Comparative development of the ant chemosensory system. Curr. Biol 30, 3223–3230.e4. 10.1016/j.cub.2020.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trible W, Olivos-Cisneros L, McKenzie, Saragosti J, Chang N-CSK, Matthews BJ, Oxley PR, and Kronauer DJC (2017). orco mutagenesis causes loss of antennal lobe glomeruli and impaired social behavior in ants. Cell 170, 727–735. 10.1016/j.cell.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferguson ST, Bakis I, and Zwiebel LJ (2021). Advances in the study of olfaction in eusocial ants. Insects 12, 252. 10.3390/insects12030252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benton R (2022). Drosophila olfaction: past, present, and future. Proc. R. Soc. B 289, 20222054. 10.1098/rspb.2022.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao Q, Yuan B, and Chess A (2000). Convergent projections of Drosophila olfactory neurons to specific glomeruli in the antennal lobe. Nat. Neurosci 3, 780–785. 10.1038/77680. [DOI] [PubMed] [Google Scholar]
- 16.Stocker RF (1994). The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res. 275, 3–26. 10.1007/BF00305372. [DOI] [PubMed] [Google Scholar]
- 17.Stocker RF, Lienhard MC, Borst A, and Fischbach K-F (1990). Neuronal architecture of the antennal lobe in Drosophila melanogaster. Cell Tissue Res. 262, 9–34. 10.1007/BF00327741. [DOI] [PubMed] [Google Scholar]
- 18.Wang JW, Wong AM, Flores J, Vosshall LB, and Axel R (2003). Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell 112, 271–282. 10.1016/S0092-8674(03)00004-7. [DOI] [PubMed] [Google Scholar]
- 19.Duffield RM, Blum MS, and Wheeler JM (1976). Alkylpyrazine alarm pheromones in primitive ants with small colonial units. Comp. Biochem. Physiol 54, 439–440. 10.1016/0305-0491(76)90116-4. [DOI] [PubMed] [Google Scholar]
- 20.Wilson EO and Regnier FEJ (1971). The evolution of the alarm-defense system in the formicine ants. Am. Nat 105, 279–289. 10.1086/282724. [DOI] [Google Scholar]
- 21.Smith AA and Haight KL (2008). Army ants as research and collection tools. J. Insect. Sci 8, 71. 10.1673/031.008.7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopes LE, Frank ET, Kárpáti Z, Schmitt T, and Kronauer DJC (2023). The alarm pheromone and alarm response of the clonal raider ant. J. Chem. Ecol 49, 1–10. 10.1007/s10886-023-01407-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blum MS (1969). Alarm pheromones. Annu. Rev. Entomol 14, 57–81. 10.1146/annurev.en.14.010169.000421. [DOI] [Google Scholar]
- 24.Vander Meer RK, and Alonso LE (1998). Pheromone directed behavior in ants. In Pheromone Communication in Social Insects, Vander Meer RK, Breed MD, Espelie KE, and Winston M, ed. (Boulder, CO, USA: Westview Press; ), pp. 159–192. [Google Scholar]
- 25.Galizia CG, Menzel R, and Hölldobler B (1999). Optical imaging of odor-evoked glomerular activity patterns in the antennal lobes of the ant Camponotus rufipes. Naturwissenschaften 86, 533–537. 10.1007/s001140050669. [DOI] [PubMed] [Google Scholar]
- 26.Zube C, Kleineidam CJ, Kirschner S, Neef J, and Rössler W (2008). Organization of the olfactory pathway and odor processing in the antennal lobe of the ant Camponotus floridanus. J. Comp. Neurol 506, 425–441. 10.1002/cne.21548. [DOI] [PubMed] [Google Scholar]
- 27.Brandstaetter AS, Rössler W, and Kleineidam CJ (2011). Friends and foes from an ant brain’s point of view -- neuronal correlates of colony odors in a social insect. PLoS One 6, e21383. 10.1371/journal.pone.0021383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joerges J, Küttner A, Galizia CG, and Menzel R (1997). Representations of odours and odour mixtures visualized in the honeybee brain. Nature 387, 285–288. 10.1038/387285a0. [DOI] [Google Scholar]
- 29.Galizia CG, Nägler K, Hölldobler B, and Menzel R (1998). Odour coding is bilaterally symmetrical in the antennal lobes of honeybees (Apis mellifera). Eur. J. Neurosci 10, 2964–2974. 10.1111/j.1460-9568.1998.00303.x. [DOI] [PubMed] [Google Scholar]
- 30.Sachse S, Rappert A, and Galizia CG (1999). The spatial representation of chemical structures in the antennal lobe of honeybees: steps towards the olfactory code. Eur. J. Neurosci 11, 3970–3982. 10.1046/j.1460-9568.1999.00826.x. [DOI] [PubMed] [Google Scholar]
- 31.Guerrieri F, Schubert M, Sandoz JC, and Giurfa M (2005). Perceptual and neural olfactory similarity in honeybees. PLoS Biol. 3, 0718–0732. 10.1371/journal.pbio.0030060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carcaud J, Giurfa M, and Sandoz J-C (2015). Differential combinatorial coding of pheromones in two olfactory subsystems of the honey bee brain. J. Neurosci 35, 4157–4167. 10.1523/JNEUROSCI.0734-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paoli M, and Galizia GC (2021). Olfactory coding in honeybees. Cell Tissue Res. 383, 35–58. 10.1007/s00441-020-03385-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haase A, Rigosi E, Trona F, Anfora G, Vallortigara G, Antolini R, and Vinegoni C (2011). In-vivo two-photon imaging of the honey bee antennal lobe. Biomed. Opt. Express 2, 131–138. 10.1364/boe.2.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laurent G (1999). A systems perspective on early olfactory coding. Science 286, 723–728. 10.1126/science.286.5440.723. [DOI] [PubMed] [Google Scholar]
- 36.Hallem EA and Carlson JR (2006). Coding of odors by a receptor repertoire. Cell 125, 143–160. 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 37.Münch D and Galizia CG (2016). DoOR 2.0 - Comprehensive mapping of Drosophila melanogaster odorant responses. Sci. Rep 6, 21841. 10.1038/srep21841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan H, Opachaloemphan C, Mancini G, Yang H, Gallitto M, Mlejnek J, Leibholz A, Haight K, Ghaninia M, Huo L, et al. (2017). An engineered orco mutation in ants produces aberrant social behavior and defective neural development. Cell 170, 736–747. 10.1016/j.cell.2017.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duan Q, and Volkan PC (2020). Ant olfaction: smells like an insect, develops like a mammal. Curr. Biol 30, R950–R952. 10.1016/j.cub.2020.06.074. [DOI] [PubMed] [Google Scholar]
- 40.Lodovichi C, and Belluscio L (2012). Odorant receptors in the formation of the olfactory bulb circuitry. Physiology 27, 200–212. 10.1152/physiol.00015.2012. [DOI] [PubMed] [Google Scholar]
- 41.Schaefer ML, Finger TE, and Restrepo D (2001). Variability of position of the P2 glomerulus within a map of the mouse olfactory bulb. J. Comp. Neurol 436, 351–362. 10.1002/cne.1072. [DOI] [PubMed] [Google Scholar]
- 42.Strotmann J, Conzelmann S, Beck A, Feinstein P, Breer H, and Mombaerts P (2000). Local permutations in the glomerular array of the mouse olfactory bulb. J. Neurosci 20, 6927–6938. 10.1523/jneurosci.20-18-06927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zapiec B, and Mombaerts P (2015). Multiplex assessment of the positions of odorant receptor-specific glomeruli in the mouse olfactory bulb by serial two-photon tomography. Proc. Natl. Acad. Sci. U. S. A 112, E5873–E5882. 10.1073/pnas.1512135112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuebler LS, Kelber C, and Kleineidam CJ (2010). Distinct antennal lobe phenotypes in the leaf-cutting ant (Atta vollenweideri). J. Comp. Neurol 518, 352–365. 10.1002/cne.22217. [DOI] [PubMed] [Google Scholar]
- 45.Oxley PR, Ji L, Fetter-Pruneda I, McKenzie SK, Li C, Hu H, Zhang G, and Kronauer DJC (2014). The genome of the clonal raider ant Cerapachys biroi. Curr. Biol 24, 451–458. 10.1016/j.cub.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chandra V, Gal A, and Kronauer DJC (2021). Colony expansions underlie the evolution of army ant mass raiding. Proc. Natl. Acad. Sci. U. S. A 118, e2026534118. 10.1073/pnas.2026534118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bernardi R, Cardani C, Ghiringelli D, Selva A, Baggini A, and Pavan M (1967). On the components of secretion of mandibular glands of the ant Lasius (Dendrolasius) fuliginosus. Tetrahedron Lett. 40, 3893–3896. 10.1016/S0040-4039(01)89747-1. [DOI] [Google Scholar]
- 48.Duffield RM, Brand JM, and Blum MS (1977). 6-methyl-5-hepten-2-one in Formica species: identification and function as an alarm pheromone (Hymenoptera: Formicidae). Ann. Entomol. Soc. Am 70, 309–310. 10.1093/aesa/70.3.309. [DOI] [Google Scholar]
- 49.Han S, Chen W, and Elgar MA (2022). An ambiguous function of an alarm pheromone in the collective displays of the Australian meat ant, Iridomyrmex purpureus. Ethology 128, 70–76. 10.1111/eth.13241. [DOI] [Google Scholar]
- 50.Keegans SJ, Billen J, Morgan DE, and Gökcen OA (1993). Volatile glandular secretions of three species of new world army ants, Eciton burchelli, Labidus coecus, and Labidus praedator. J. Chem. Ecol 19, 2705–2719. 10.1007/BF00980702. [DOI] [PubMed] [Google Scholar]
- 51.McGurk DJ (1968). I. Studies of volatile compounds from ants II. Degradation studies and structure proof of cis, cis-nepetalactone. Oklahoma State University. [Google Scholar]
- 52.Morgan ED, Jackson BD, Keegans SJ, Nicholls DJ, Ali MF, and Cammaerts R (1992). Alkanols in the mandibular gland secretion of the ant Tetramorium caespitum. Belg. J. Zool 122, 69–74. [Google Scholar]
- 53.Oldham NJ, Morgan ED, Gobin B, Schoeters E, and Billen J (1994). Volatile secretions of old world army ant Aenictus rotundatus and chemotaxonomic implications of army ant dufour gland chemistry. J. Chem. Ecol 20, 3297–3305. 10.1007/BF02033727. [DOI] [PubMed] [Google Scholar]
- 54.Pasteels JM, Verhaeghe JC, Braekman JC, Daloze D, and Tursch B (1980). Caste-dependent pheromones in the head of the ant Tetramorium caespitum. J. Chem. Ecol 6, 467–472. 10.1007/BF01402923. [DOI] [Google Scholar]
- 55.Pasteels JM, Verhaeghe JC, Ottinger R, Braekman JC, and Daloze D (1981). Absolute configuration of (3R,4S)-4-methyl-3-hexanol-A pheromone from the head of the ant Tetramorium impurum foerster. Insect Biochem. 11, 675–678. 10.1016/0020-1790(81)90057-3. [DOI] [Google Scholar]
- 56.Stensmyr MC, Dweck HKM, Farhan A, Ibba I, Strutz A, Mukunda L, Linz J, Grabe V, Steck K, Lavista-Llanos S, et al. (2012). A conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila. Cell 151, 1345–1357. 10.1016/j.cell.2012.09.046. [DOI] [PubMed] [Google Scholar]
- 57.Carcaud J, Otte M, Grünewald B, Haase A, Sandoz J-C, and Beye M (2023). Multisite imaging of neural activity using a genetically encoded calcium indicator in the honey bee. PLoS Biol. 21, e3001984. 10.1371/journal.pbio.3001984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stökl J, Strutz A, Dafni A, Svatos A, Doubsky J, Knaden M, Sachse S, Hansson BS, and Stensmyr MC (2010). A deceptive pollination system targeting drosophilids through olfactory mimicry of yeast. Curr. Biol 20, 1846–1852. 10.1016/j.cub.2010.09.033. [DOI] [PubMed] [Google Scholar]
- 59.Zhao Z, Zung JL, Hinze A, Kriete AL, Iqbal A, Younger MA, Matthews BJ, Merhof D, Thiberge S, Ignell R et al. (2022). Mosquito brains encode unique features of human odour to drive host seeking. Nature 605, 706–712. 10.1038/s41586-022-04675-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Riabinina O, Luginbuhl D, Marr E, Liu S, Wu MN, Luo L, and Potter CJ (2015). Improved and expanded Q-system reagents for genetic manipulations. Nat. Methods 12, 37–54. 10.1016/bs.mcb.2015.01.016.Observing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anderson MAE, Gross TL, Myles KM, and Adelman ZN (2010). Validation of novel promoter sequences derived from two endogenous ubiquitin genes in transgenic Aedes aegypti. Insect Mol. Biol 19, 441–449. 10.1111/j.1365-2583.2010.01005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suzuki MG, Funaguma S, Kanda T, Tamura T, and Shimada T (2003). Analysis of the biological functions of a doublesex homologue in Bombyx mori. Dev. Genes Evol 213, 345–354. 10.1007/s00427-003-0334-8. [DOI] [PubMed] [Google Scholar]
- 64.Masumoto M, Ohde T, Shiomi K, Yaginuma T, and Niimi T (2012). A baculovirus immediate-early gene, ie1, promoter drives efficient expression of a transgene in both Drosophila melanogaster and Bombyx mori. PLoS One 7, e49323. 10.1371/journal.pone.0049323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Otte M, Netschitailo O, Kaftanoglu O, Wang Y, Page RE Jr., Beye M (2018). Improving genetic transformation rates in honeybees. Sci. Rep 8, 16534. 10.1038/s41598-018-34724-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Habenstein J, Amini E, Grübel K, el Jundi B, and Rössler W (2020). The brain of Cataglyphis ants: neuronal organization and visual projections. J. Comp. Neurol 528, 3479–3506. 10.1002/cne.24934. [DOI] [PubMed] [Google Scholar]
- 67.Ferkey DM, Hyde R, Haspel G, Dionne HM, Hess HA, Suzuki H, Schafer WR, Koelle MR, and Hart AC (2007). C. elegans G protein regulator RGS-3 controls sensitivity to sensory stimuli. Neuron 53, 39–52. 10.1016/j.neuron.2006.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tian L, Hires SA, and Looger LL (2012). Imaging neuronal activity with genetically encoded calcium indicators. Cold Spring Harb. Protoc 6, 647–656. 10.1101/pdb.top069609. [DOI] [PubMed] [Google Scholar]
- 69.Bellen HJ, Levis RW, He Y, Carlson JW, Evans-Holm M, Bae E, Kim J, Metaxakis A, Savakis C, Schulze KL et al. (2011). The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics 188, 731–743. 10.1534/genetics.111.126995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Galizia CG, Joerges J, Küttner A, Faber T, and Menzel R (1997). A semi-in-vivo preparation for optical recording of the insect brain. J. Neurosci. Methods 76, 61–69. 10.1016/S0165-0270(97)00080-0. [DOI] [PubMed] [Google Scholar]
- 71.Slone JD, Pask GM, Ferguson ST, Millar JG, Berger SL, Reinberg D, Liebig J, Ray A, and Zwiebel JL (2017). Functional characterization of odorant receptors in the ponerine ant, Harpegnathos saltator. Proc. Natl. Acad. Sci. U. S. A 114, 8586–8591. 10.1073/pnas.1704647114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pask GM, Slone JD, Millar JG, Das P, Moreira JA, Zhou X, Bello J, Berger SL, Bonasio R, Desplan C, et al. (2017). Specialized odorant receptors in social insects that detect cuticular hydrocarbon cues and candidate pheromones. Nat. Commun 8, 297. 10.1038/s41467-017-00099-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ozaki M, Wada-Katsumata A, Fujikawa K, Iwasaki M, Yokohari F, Satoji Y, Nisimura T, and Yamaoka R (2005). Ant nestmate and non-nestmate discrimination by a chemosensory sensillum. Science 309, 311–314. 10.1126/science.1105244. [DOI] [PubMed] [Google Scholar]
- 74.Sharma KR, Enzmann BL, Schmidt Y, Moore D, Jones GR, Parker J, Berger SL, Reinberg D, Zweibel LJ, Breit B, et al. (2015). Cuticular hydrocarbon pheromones for social behavior and their coding in the ant antenna. Cell Rep. 12, 1261–1271. 10.1016/j.celrep.2015.07.031. [DOI] [PubMed] [Google Scholar]
- 75.Yamagata N, Nishino H, and Mizunami M (2006). Pheromone-sensitive glomeruli in the primary olfactory centre of ants. Proc. R. Soc. B Biol. Sci 273, 2219–2225. 10.1098/rspb.2006.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Insam H and Seewald MSA (2010). Volatile organic compounds (VOCs) in soils. Biol. Fertil. Soils 46, 199–213. 10.1007/s00374-010-0442-3. [DOI] [Google Scholar]
- 77.Hallem EA, Ho MG, and Carlson JR (2004). The molecular basis of odor coding in the Drosophila antenna. Cell 117, 965–979. 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 78.Su C-Y, Martelli C, Emonet T, and Carlson JR (2011). Temporal coding of odor mixtures in an olfactory receptor neuron. Proc. Natl. Acad. Sci. U. S. A 108, 5075–5080. 10.1073/pnas.1100369108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kurtovic A, Widmer A, and Dickson BJ (2007). A single class of olfactory neurons mediates behavioral responses to a Drosophila sex pheromone. Nature 446, 542–546. 10.1038/nature05672. [DOI] [PubMed] [Google Scholar]
- 80.Christensen TA and Hildebrand JG (1987). Male-specific, sex pheromone-selective projection neurons in the antennal lobes of the moth Manduca sexta. J. Comp. Physiol. A 160, 553–569. 10.1007/BF00611929. [DOI] [PubMed] [Google Scholar]
- 81.Sakurai T, Nakagawa T, Mitsuno H, Mori H, Endo Y, Tanoue S, Yasukochi Y, Touhara K, and Nishioka T (2004). Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc. Natl. Acad. Sci. U. S. A 101, 16653–16658. 10.1073/pnas.040759610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hildebrand JG and Shepherd GM (1997). Mechanisms of olfactory discrimination: converging evidence for common principles across phyla. Annu. Rev. Neurosci 20, 595–631. 10.1146/annurev.neuro.20.1.595. [DOI] [PubMed] [Google Scholar]
- 83.Dweck HKM, Ebrahim SAM, Thoma M, Mohamed AAM, Keesey IW, Trona F, Lavista-Llanos S, Svatoš A, Sachse S, Knaden M et al. (2007). Pheromones mediating copulation and attraction in Drosophila. Proc. Natl. Acad. Sci. U. S. A 112, E2829–E2835. 10.1073/pnas.1504527112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jones WD, Cayirlioglu P, Kadow IG, and Vosshall LB (2007). Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 445, 86–90. 10.1038/nature05466. [DOI] [PubMed] [Google Scholar]
- 85.Su C-Y, Menuz K, Reisert J, and Carlson JR (2012). Non-synaptic inhibition between grouped neurons in an olfactory circuit. Nature 492, 66–71. 10.1038/nature11712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Root CM, Masuyama K, Green DS, Enell LE, Nässel DR, Lee C-H, and Wang JW (2008). A presynaptic gain control mechanism fine-tunes olfactory behavior. Neuron 59, 311–321. 10.1016/j.neuron.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Olsen SR and Wilson RI (2008). Lateral presynaptic inhibition mediates gain control in an olfactory circuit. Nature 452, 956–960. 10.1038/nature06864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Task D, Lin C-C, Vulpe A, Afify A, Ballou S, Brbic M, Schlegel P, Raji J, Jefferis G, et al. (2022). Chemoreceptor co-expression in Drosophila melanogaster olfactory neurons. Elife 11, e72599. 10.7554/eLife.72599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Herre M, Goldman OV, Lu T-C, Caballero-Vidal G, Qi Y, Gilbert ZN, Gong Z, Morita T, Rahiel S, Ghaninia M, et al. (2022). Non-canonical odor coding in the mosquito. Cell 185, 3104–3123.e28. 10.1016/j.cell.2022.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tumlinson JH, Klein MG, Doolittle RE, Ladd TL, and Proveaux AT (1977). Identification of the female Japanese beetle sex pheromone: inhibition of male response by an enantiomer. Science 197, 789–792. 10.1126/science.197.4305.789. [DOI] [PubMed] [Google Scholar]
- 91.Renou M and Guerrero A (2000). Insect parapheromones in olfaction research and semiochemical-based pest control strategies. Annu. Rev. Entomol 48, 605–630. 10.1146/annurev.ento.45.1.605. [DOI] [PubMed] [Google Scholar]
- 92.Amoore JE, Palmieri G, Wanke E, and Blum MS (1969). Ant alarm pheromone activity: correlation with molecular shape by scanning computer. Science 165, 1266–1269. 10.1126/science.165.3899.1266. [DOI] [PubMed] [Google Scholar]
- 93.Roelofs WL (1982). Pheromones and evolutionary relationships of Tortricidae. Annu. Rev. Ecol. Syst 13, 395–422. 10.1146/annurev.es.13.110182.002143. [DOI] [Google Scholar]
- 94.Adams SA and Tsutsui ND (2020). The evolution of species recognition labels in insects. Philos. Trans. R. Soc. B 375, 20190476. 10.1098/rstb.2019.0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.D’Ettorre P, Errard C, Ibarra F, Francke W, and Hefetz A (2000). Sneak in or repel your enemy: Dufour’s gland repellent as a strategy for successful usurpation in the slave-maker Polyergus rufescens. Chemoecol. 10, 135–142. 10.1007/PL00001815. [DOI] [Google Scholar]
- 96.Lenoir A, D’Ettorre P, and Hefetz A (2001). Chemical ecology and social parasitism in ants. Annu. Rev. Entomol 46, 573–599. 10.1146/annurev.ento.46.1.573. [DOI] [PubMed] [Google Scholar]
- 97.Menzel F, Blüthgen N, Tolasch T, Conrad J, Beifuß, Beuerle T, and Schmitt T (2013). Crematoenes - a novel substance class exhibited by ants functions as appeasement signal. Front. Zool 10, 32. 10.1186/1742-9994-10-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Regnier FE and Wilson EO (1971). Chemical communication and “propaganda” in slave-making ants. Science 172, 267–269. 10.1126/science.172.3980.267. [DOI] [PubMed] [Google Scholar]
- 99.Brandt M, Heinze J, Schmitt T, and Foitzik S (2006). Convergent evolution of the Dufour’s gland secretion as a propaganda substance in the slave-making ant genera Protomognathus and Harpagoxenus. Insect. Soc 53, 291–299. 10.1007/s00040-006-0871-z. [DOI] [Google Scholar]
- 100.Dejean A, Corbara B, Roux O, and Orivel J (2014). The antipredatory behaviours of Neotropical ants towards army ant raids (Hymenoptera: Formicidae). Myrmecol. News 19, 17–24. [Google Scholar]
- 101.Lloyd HA, Schmuff NR, and Hefetz A (1986). Chemistry of the anal glands of Bothriomyrmex syrius Forel. Olfactory mimetism and temporary social parasitism. Comp. Biochem. Physiol. B, Biochem 83B, 71–73. 10.1016/0305-0491(86)90333-0. [DOI] [PubMed] [Google Scholar]
- 102.Stoeffler M, Maier TS, Tolasch T, and Steidle JL (2007). Foreign-language skills in rove-beetles? Evidence for chemical mimicry of ant alarm pheromones in myrmecophilous Pella beetles (Coleoptera: Staphylinidae). J. Chem. Ecol 33, 1382–1392. 10.1007/s10886-007-9315-0. [DOI] [PubMed] [Google Scholar]
- 103.Cattaneo AM, Gonzalez F, Bengtsson JM, Corey EA, Jacquin-Joly E, Montagné N, Salvagnin U, Walker WB, Witzgall P, Anfora G, et al. (2017). Candidate pheromone receptors of codling moth Cydia pomonella respond to pheromones and kairomones. Sci. Rep 7, 41105. 10.1038/srep41105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Isogai Y, Si S, Pont-Lezica L, Tan T, Kapoor V, Murthy VN, and Dulac C (2011). Molecular organization of vomeronasal chemoreception. Nature 478, 241–245. 10.1038/nature10437.Molecular. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Allan RA, Elgar MA, and Capon RJ (1996). Exploitation of an ant chemical alarm signal by the zodariid spider Habronestes bradleyi Walckenaer. Proc. R. Soc. B Biol. Sci 263, 69–73. 10.1098/rspb.1996.0012. [DOI] [Google Scholar]
- 106.Uchida N, Takahashi YK, Tanifuji M, and Mori K (2000). Odor maps in the mammalian olfactory bulb: domain organization and odorant structural features. Nat. Neurosci 3, 1035–1043. 10.1038/79857. [DOI] [PubMed] [Google Scholar]
- 107.Fishilevich E and Vosshall LB (2005). Genetic and functional subdivision of the Drosophila antennal lobe. Curr. Biol 15, 1548–1553. 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- 108.Prieto-Godino LL, Rytz R, Cruchet S, Bargeton B, Abuin L, Silbering AF, Ruta V, Dal Peraro M, and Benton R (2017). Evolution of acid-sensing olfactory circuits in drosophilids. Neuron 93, 661–676.e6. 10.1016/j.neuron.2016.12.024. [DOI] [PubMed] [Google Scholar]
- 109.Hölldobler B (1995). The chemistry of social regulation: multicomponent signals in ant societies. Proc. Natl. Acad. Sci. U. S. A 92, 19–22. 10.1073/pnas.92.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Morgan ED (2009). Trail pheromones of ants. Physiol. Entomol 34, 1–17. 10.1111/j.1365-3032.2008.00658.x. [DOI] [Google Scholar]
- 111.Bonavita-Cougourdan A, Clément JL, and Lange C (1987). Nestmate recognition: the role of cuticular hydrocarbons in the ant Camponotus vagus Scop. J. Entomol. Sci 22, 1–10. 10.18474/0749-8004-22.1.1. [DOI] [Google Scholar]
- 112.Kleineidam CJ, Obermayer M, Halbich W, and Rössler W (2005). A macroglomerulus in the antennal lobe of leaf-cutting ant workers and its possible functional significance. Chem. Senses 30, 383–392. 10.1093/chemse/bji033. [DOI] [PubMed] [Google Scholar]
- 113.Ravary F and Jaisson P (2004). Absence of individual sterility in thelytokous colonies of the ant Cerapachys biroi Forel (Formicidae, Cerapachyinae). Insectes Soc. 51, 67–73. 10.1007/s00040-003-0724-y. [DOI] [Google Scholar]
- 114.Teseo S, Châline N, Jaisson P, and Kronauer DJC (2014). Epistasis between adults and larvae underlies caste fate and fitness in a clonal ant. Nat. Commun 5, 3363. 10.1038/ncomms4363(2014). [DOI] [PubMed] [Google Scholar]
- 115.Kronauer DJC, Tsuji K, Pierce NE, and Keller L (2013). Non-nest mate discrimination and clonal colony structure in the parthenogenetic ant Cerapachys biroi. Behav. Ecol 24, 617–622. 10.1093/beheco/ars227. [DOI] [Google Scholar]
- 116.Regnier FE and Wilson EO (1968). The alarm-defence system of the ant Acanthomyops claviger. J. Insect Physiol 14, 955–970. 10.1016/0022-1910(68)90006-1. [DOI] [Google Scholar]
- 117.Regnier FE and Wilson EO (1969). The alarm-defence system of the ant Lasius alienus. J. Insect Physiol 15, 893–898. 10.1016/0022-1910(69)90129-2. [DOI] [Google Scholar]
- 118.Ayre GL, and Blum MS (1971). Attraction and alarm of ants (Camponotus spp.: Hymenoptera: Formicidae) by pheromones. Physiol. Zool 44, 77–83. 10.1086/physzool.44.2.30155558. [DOI] [Google Scholar]
- 119.Lenz EL, Krasnec MO, and Breed MD (2013). Identification of undecane as an alarm pheromone of the ant Formica argentea. J. Insect Behav 26, 101–108. 10.1007/s10905-012-9337-5. [DOI] [Google Scholar]
- 120.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, and Smith HO (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 121.Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, et al. (2010). Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329, 52–56. 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 122.Riabinina O, Task D, Marr E, Lin C-C, Alford R, O’Brochta DA, and Potter CJ (2016). Organization of olfactory centres in the malaria mosquito Anopheles gambiae. Nat. Commun 7, 13010. 10.1038/ncomms13010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Arzt M, Deschamps J, Schmied C, Pietzsch T, Schmidt D, Tomancak P, Haase R, and Jug F (2022). LABKIT: labeling and segmentation toolkit for big image data. Front. Comput. Sci 4, 777728. 10.3389/fcomp.2022.777728. [DOI] [Google Scholar]
- 125.Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li H (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at arXiv, 1303.3997 [q-bio.GN]. 10.48550/arXiv.1303.3997. [DOI] [Google Scholar]
- 127.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.R Core Team (2021). R: A language and environment for statistical computing. http://www.R-project.org.
- 129.Wickam H (2016). ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag. [Google Scholar]
- 130.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol 29, 24–26. 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bodenhofer U, Bonatesta E, Horejš-Kainrath C, and Hochreiter S (2015). msa: an R package for multiple sequence alignment. Bioinformatics 31, 3997–3999. 10.1093/bioinformatics/btv494. [DOI] [PubMed] [Google Scholar]
- 132.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 133.Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, and Schäffer AA (2008). Database indexing for production MegaBLAST searches. Bioinformatics 24, 1757–1764. 10.1093/bioinformatics/btn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gel B and Serra E (2017). karyoploteR: an R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics 33, 31–33. 10.1093/bioinformatics/btx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li K (2008). The image stabilizer plugin for ImageJ. http://www.cs.cmu.edu/~kangli/code/Image_Stabilizer.html.
- 136.Butterwick JA, del Mármol J, Kim KH, Kahlson MA, Rogow JA, Walz T, and Ruta V (2018). Cryo-EM structure of the insect olfactory receptor Orco. Nature 560, 447–452. 10.1038/s41586-018-0420-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Video 1. Colony alarm bioassay, related to Fig. 1. Representative videos of each of the three categories of major behavioral responses. Videos show 1 minute prior to stimulus, time skips while stimuli are put in place, then 2 minutes after stimulus (filter paper + compound). The presence of the red circle on the upper right indicates that stimulus is in place. Sped up 8x.
Supplemental Video 2. Odor-evoked calcium response in the ant antennal lobe, related to Fig. 3. Representative two-photon recording of the calcium response to 5s presentation with ethylpyrazine (48%). A single slice corresponding to 110μm z-depth was extracted from the volumetric recording, and image stabilization was applied [S21]. The “ON” label indicates the 5s odor presentation period. Sped up 3x.
Figure S1. Additional characterization of GCaMP6s ants, related to Fig. 2. (A) The same two clonal line B pupae, one wild type and one transgenic, imaged under bright field (left) and epifluorescence, with filters set to detect GCaMP6s (middle) and DsRed (right). Pupae were imaged 10 days after pupation. (B) Anti-DsRed (magenta) labels the ALs, indicating co-expression of DsRed with GCaMP6s (green; endogenous fluorescence) in ants carrying [ie1-DsRed, ObirOrco-QF2, 15xQUAS-GCaMP6s]. Phalloidin stains actin (cyan). (C) GCaMP6s fluorescence (green) is detectable in the subesophageal zone (SEZ). (D) GCaMP6s fluorescence (green) is also visible in processes innervating part of the central complex (CC) as well as in a nearby cluster of somas. Images show max z-projections through the imaged brain regions. (E) After unilateral ablation of the antenna (from the scape), bilaterally symmetrical GCaMP6s signal is still detectable in the central complex (CC), as well as the subesophageal zone (SEZ). No anti-Orco signal was detected in these brain regions. Images show max z-projections through the imaged brain regions.
Figure S2. GCaMP6s ants have normal antennal lobes and respond to alarm pheromones, related to Figs. 1–2. (A) 505 glomeruli reconstructed from the right AL of a GCaMP6s ant using anti-SYNORF1 staining. (B) 508 glomeruli reconstructed from the left AL of a GCaMP6s ant using GCaMP6s signal, anti-Orco staining, and phalloidin staining. 502 glomeruli stained positive for phalloidin, GCaMP6s, and Orco; 4 glomeruli stained positive for phalloidin and GCaMP6s, but not Orco; and 2 glomeruli stained positive for phalloidin, but not GCaMP6s and Orco. The AL in (B) comes from a different ant than that in (A) and is shown in mirror image to align more closely with the image in (A). (C-F) Colony alarm bioassay, showing mean±SEM. (C-D) GCaMP6s ants and wild type ants leave the nest in response to 4-methyl-3-heptanone, 4-methyl-3-heptanol, and a 9:1 blend of the two compounds. (E-F) GCaMP6s ants and wild type ants leave the nest chamber in response to 4-methyl-3-heptanone and the blend; wild type ants also leave the nest chamber in response to 4-methyl-3-heptanol, while this response was not statistically significant in the GCaMP6s ants. Analyses for wild type ants (D, F) are based on previously published data (Fig. 3, 2600μg and vehicle control treatments in Lopes et al.22). For the experiment in GCaMP6s ants (C, E), all pheromone treatments were compared to the same set of vehicle control trials. For the experiments in wild type ants (D, F), each pheromone treatment was conducted alongside its own set of vehicle control trials. Treatments and controls from the same experiment were statistically compared to each other and are plotted using identical symbol shapes. *: p<0.05; **: p<0.01; ***: p<0.001; ****: p<0.0001. L: lateral; A: anterior.
Figure S3. Genomic analyses of the transgenic line used for imaging, related to Fig. 2. (A) Normalized read depth for reads aligning to a panel of heterozygous SNPs, the ObirOrco promoter, and the non-ObirOrco portion of the transgene. Normalized read depth for ObirOrco is ~1.5, corresponding to a single additional copy of ObirOrco inserted into the genome (added to the two endogenous copies). Normalized read depth of ~0.5 at the rest of the insert is also consistent with a single copy (haploid) insertion. (B) The transgene insert was localized to a site on the 2nd chromosomal scaffold. Black bars indicate breaks between contigs. (C) Close-up of the transgene insertion locus within an intron of the gene trace amine-associated receptor 9.
Figure S4. Calcium responses are bilaterally symmetrical and remain robust across trials, related to Fig. 4. (A) Max z-projections of both antennal lobes, imaged bilaterally. Both in raw fluorescence (left) and peak fold change of calcium response to presentation with 4-methyl-3-hexanol (right), the antennal lobes appear bilaterally symmetrical. (B) Max z-projections of peak fold change from a single ant after presentation with 4-methyl-3-hexanol. Three trials were performed at 3.0% concentration (top), and three additional trials were performed at 48.0% concentration (bottom). Timestamps for each trial demonstrate that responses are robust over the duration of a full experiment. Two adjacent focal glomeruli are circled. (C) Time series of calcium responses from each trial in (B) for the two adjacent glomeruli; responses in the left glomerulus are shown as alternating short and long dashes and responses in the right glomerulus are shown as solid lines; black bars indicate the 5s odor presentations. Responses are quantified from max z-projections of three slices centered on 105μm z-depth. L: lateral; A: anterior.
Figure S5. Increased odor concentration results in more responding glomeruli, related to Figs. 4–5. Counts of the number of responding glomeruli from max z-projections; boxes enclose the first to third quartile range, with bold line showing the median and whiskers enclosing the min and max values that fall within 1.5x the interquartile range. Data points show the mean number of responding glomeruli for a given ant across all trials for a particular odorant/concentration. n=13 ants total, 3 ants presented with 0.75% and 12.0% odor concentrations; 2 ants with 3.0% and 12.0%; 3 ants with 12.0% and 48.0%; and 5 ants with 3.0% and 48.0%. Single pheromones each activated a small number of glomeruli (A), and 4-methyl-3-heptanone, 4-methyl-3-heptanol, and 4-methyl-3-hexanol activated overlapping sets of glomeruli (B). 6-methyl-5-hepten-2-one only rarely activated glomeruli shared with the other pheromones (C). All pheromones were presented separately, rather than as blends. Only pheromones and pheromone combinations that activated at least one glomerulus are shown.
Figure S6. Quantification of peak fold change in PGb (top), PGa (middle), and 6G (bottom), related to Fig. 5. Three trials per ant for each odorant/concentration; n=13 ants total, 3 ants presented with 0.75% and 12.0% odor concentrations; 2 ants with 3.0% and 12.0%; 3 ants with 12.0% and 48.0%; and 5 ants with 3.0% and 48.0%. PGa could not be identified in two ants. Graphs show outputs of linear models (with 95% confidence intervals) for dose/response to each odorant in each glomerulus, with a random effect for individual. Concentrations were log transformed to show the linear relationship.
Figure S7. Temporal dynamics in three focal glomeruli, related to Fig. 5. (A) Extended time series for the response to 6-methyl-5-hepten-2-one in 6G. Data were collected separately from Fig. 5C. Shown are single trials each from five different individuals at 48% concentration (colored traces); mean±SD (black line and gray ribbon). Fluorescence plateaued for ~30s before declining and returning to baseline ~80s after odor presentation. (B) Two parameters of temporal dynamics extracted from glomerulus-specific calcium response traces. (C) Quantification of time to response onset (top) and time to response maximum (bottom) in the three focal glomeruli PGb, PGa, and 6G in response to stimuli at 48% concentration (n=8 ants, three trials per condition per ant). Only glomerulus/pheromone combinations with typically robust responses are shown. Boxes enclose the first to third quartile range, with bold lines showing the median and whiskers enclosing the min and max values that fall within 1.5x the interquartile range. For PGb and PGa responses to 4-methyl-3-heptanol and 4-methyl-3-hexanol, we used linear mixed effects models to test for effects of pheromone, glomerulus, and a pheromone/glomerulus interaction on the time parameters. Time to response onset: significant effects of pheromone (p=0.0034), glomerulus (p<0.0001), and the interaction (p=0.0030). Time to response maximum: significant effects of pheromone (p<0.0001) and glomerulus (p=0.047), but not the interaction (p=0.88).
Data Availability Statement
DNA sequence data have been deposited to NCBI BioProject. Confocal microscopy data have been deposited to the Brain Image Library. Calcium imaging data have been deposited to the DANDI Archive. Accession numbers are listed in the key resources table.
All original code has been deposited to GitHub and the repository is listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken polyclonal anti-GFP | Abcam | Cat#ab13970; RRID: AB_300798 |
| Rabbit polyclonal anti-RFP | Rockland | Cat#600-401-379; RRID: AB_2209751 |
| Mouse monoclonal anti-SYNORF1 | Developmental Systems Hybridoma Bank | DSHB: 3C11; RRID: AB_528479 |
| Mouse monoclonal anti-Orco | Gift from Vanessa Ruta; Butterwick et al.136 | clone 20F7 |
| Goat anti-Chicken Alexa 488 | Invitrogen | Cat#A-11039; RRID: AB_2534096 |
| Donkey anti-Mouse Alexa 647 | Invitrogen | Cat#A32787; RRID: AB_2762830 |
| Donkey anti-Rabbit Alexa 594 | Invitrogen | Cat# A-21207; RRID: AB_141637 |
| Chemicals, peptides, and recombinant proteins | ||
| AlexaFluor 555 phalloidin | Invitrogen | Cat#A34055 |
| DAPI | Invitrogen | Cat#D1306 |
| Paraffin oil | Hampton Research | Cat#HR3-421 |
| 100% pentane | Sigma Aldrich | Cat#236705 |
| 96% 4-methyl-3-heptanone | Pfaltz and Bauer | Cat#M19160 |
| ≥99% 4-methyl-3-heptanol | Sigma-Aldrich | Cat#M48309 |
| 99% 6-methyl-5-hepten-2-one | Sigma-Aldrich | Cat#M48805-100ML |
| 95% 4-methyl-3-hexanol | Enamine | Cat# 615-29-2 |
| 98% 3-hexanone | Aldrich Chemistry | Cat#103020-10G |
| 98% ethylpyrazine | Sigma-Aldrich | Cat#250384-5G |
| 99% propionic acid | Sigma-Aldrich | Cat#W292419-SAMPLE-K |
| 100% ethanol | Decon Laboratories | Cat#2716 |
| ≥99.5% isopropanol | Fisher Chemical | Cat#A416SK-4 |
| RNAClean SPRI XPBeads | Beckman Coulter | Cat#A63987 |
| Critical commercial assays | ||
| Endotoxin-free midiprep kit | Machery-Nagel | Cat#740420.10 |
| HiScribe T7 Arca mRNA kit (with tailing) | New England Biolabs | Cat#E2060S |
| Experimental models: Organisms/strains | ||
| O. biroi clonal line A wild type | Kronauer Lab | N/A |
| O. biroi clonal line B wild type | Kronauer Lab | N/A |
| O. biroi clonal line B [ie1-DsRed-ObirOrco-QF2-15xQUAS-GCaMP6s]; “GCaMP6s ant” | This paper | N/A |
| Deposited data | ||
| O. biroi reference genome v5.4 | McKenzie and Kronauer10 | GCA: 003672135.1 |
| Whole-genome sequence of GCaMP6s ant | This paper | BioProject ID: PRJNA947257 |
| Confocal microscopy data | This paper | Brain Image Library IDs: 7b20a5b168a92088; 626fd578e97289bc; c2031d8218de058b |
| Calcium imaging data | This paper | DANDI Archive ID: 000467 |
| Oligonucleotides | ||
| Primers for plasmid construction; see Table S4 | This paper | N/A |
| Recombinant DNA | ||
| plasmid pGL3-IE1 | Gift from Zach Adelman; Anderson et al.62 | Addgene, ID 52894 |
| plasmid pBAC-ECFP-15xQUAS_TATA-SV40 | Gift from Christopher Potter; Riabinina et al.122 | Addgene, ID 104875 |
| plasmid pBac-DsRed-ORCO_9kbProm-QF2 | Gift from Christopher Potter; Riabinina et al.122 | Addgene, ID 104877 |
| plasmid pGP-CMV-GCaMP6s | Gift from Douglas Kim & GENIE Project; Chen et al.60 | Addgene 40753 |
| plasmid hyPBapis | Gift from Martin Beye; Otte et al.65 | N/A |
| plasmid pBAC-ie1-DsRed-ObirOrco-QF2-15xQUAS-GCaMP6s | This paper | Addgene, ID 200400 |
| Software and algorithms | ||
| ImageJ (Fiji) version 2.0.0 | Schindelin et al.123 | https://fiji.sc/ |
| LABKIT plugin for ImageJ | Arzt et al.124 | https://github.com/juglab/labkit-ui |
| Trimmomatic 0.36 | Bolger et al.125 | http://www.usadellab.org/cms/?page=trimmomatic |
| bwa mem | Li126 | https://github.com/lh3/bwa |
| Picard | Broad Institute | http://broadinstitute.github.io/picard/ |
| samtools | Li et al.127 | https://github.com/samtools/samtools |
| R | R Core Team128 | www.R-project.org. |
| ggplot2 | Wickam129 | https://ggplot2.tidyverse.org |
| Integrative Genomics Viewer | Robinson et al.130 | https://software.broadinstitute.org/software/igv/ |
| msa | Bodenhofer et al.131; Larkin et al.132 | https://bioconductor.org/packages/release/bioc/html/msa.html |
| BLAST | Morgulis et al.133 | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| karyoploteR | Gel and Serra134 | http://bioconductor.org/packages/release/bioc/html/karyoploteR.html |
| Prism | GraphPad Prism | https://www.graphpad.com |
| Image Stabilizer plugin for ImageJ | Li135 | http://www.cs.cmu.edu/~kangli/code/Image_Stabilizer.html |
| napari | napari contributors | Zenodo DOI: 10.5281/zenodo.3555620 |
| Other | ||
| Additional scripts | This study | https://github.com/Social-Evolution-and-Behavior/HartKronauer2023 |


