Significance
Plasma cell-free DNA (cfDNA) is a noninvasive biomarker for cell death of all organs. Deciphering the tissue origin of cfDNA can reveal abnormal cell death because of diseases, which has great clinical potential in disease detection and monitoring. To fully exploit this potential, we present one of the largest comprehensive and high-quality tissue methylation atlases, constructed from 521 noncancer tissue samples spanning 29 major human tissues. Based on this rich data, we develop the first deep-learning-powered model, cfSort, for tissue deconvolution. We demonstrated that cfSort has superior sensitivity and accuracy compared to existing methods. We validated cfSort in patients with cirrhosis and cancer. Our atlas and cfSort shall have broad research and clinical applications in disease detection and monitoring.
Keywords: cell-free DNA, DNA methylation, tissue deconvolution, disease diagnosis, disease monitoring
Abstract
Plasma cell-free DNA (cfDNA) is a noninvasive biomarker for cell death of all organs. Deciphering the tissue origin of cfDNA can reveal abnormal cell death because of diseases, which has great clinical potential in disease detection and monitoring. Despite the great promise, the sensitive and accurate quantification of tissue-derived cfDNA remains challenging to existing methods due to the limited characterization of tissue methylation and the reliance on unsupervised methods. To fully exploit the clinical potential of tissue-derived cfDNA, here we present one of the largest comprehensive and high-resolution methylation atlas based on 521 noncancer tissue samples spanning 29 major types of human tissues. We systematically identified fragment-level tissue-specific methylation patterns and extensively validated them in orthogonal datasets. Based on the rich tissue methylation atlas, we develop the first supervised tissue deconvolution approach, a deep-learning-powered model, cfSort, for sensitive and accurate tissue deconvolution in cfDNA. On the benchmarking data, cfSort showed superior sensitivity and accuracy compared to the existing methods. We further demonstrated the clinical utilities of cfSort with two potential applications: aiding disease diagnosis and monitoring treatment side effects. The tissue-derived cfDNA fraction estimated from cfSort reflected the clinical outcomes of the patients. In summary, the tissue methylation atlas and cfSort enhanced the performance of tissue deconvolution in cfDNA, thus facilitating cfDNA-based disease detection and longitudinal treatment monitoring.
Dying cells from all tissues release their DNA into the bloodstream as cell-free DNA (cfDNA) (1–3). The development and treatment of many diseases, such as cancer (4–9), autoimmune diseases (10), and sepsis (11), can influence cell death rates, thus impacting the fractions of cfDNA from respective tissues in blood (11, 12). Therefore, the abnormal tissue-derived cfDNA fractions can reveal altered tissue homeostasis due to diseases and collateral tissue damage due to treatments (13). As a result, cfDNA provides a noninvasive and comprehensive profile of wellness across all tissues in the body. Deciphering the tissue origin of cfDNA, i.e., tissue deconvolution of cfDNA, holds great clinical potential in aiding disease diagnosis, prognosis, and treatment monitoring. Despite the great promise, tissue deconvolution of cfDNA faces unique challenges: 1) cfDNA from solid organs comprises only a minor fraction of cfDNA with an overwhelming background cfDNA (~85%) from blood cells (14–16). The signal from the pathologic organs is usually weak in cfDNA. 2) All tissues in the body can release cfDNA (3, 15), requiring the joint deconvolution of as many tissue types as possible, not just a few tissue types, for an accurate tissue deconvolution.
Thanks to the tissue specificity of DNA methylation, cfDNA can be traced back to the tissues they originated from based on its methylation pattern (14). Several studies proposed methylation-based tissue deconvolution methods to estimate the proportions of tissue-specific cfDNA, including nonnegative least square methods (14, 15) and likelihood-based methods (17, 18). Although these methods have demonstrated the viability of methylation-based tissue deconvolution, their sensitivity and accuracy are still inadequate to detect a minor fraction of tissue-derived cfDNA, due to three major limitations: 1) They used unsupervised deconvolution approaches, which are known to be inferior to supervised approaches in terms of power, generalizability, and robustness to noises. 2) Only one or a few methylation profiles were available for a single tissue type, which cannot sufficiently represent inter-individual variances. 3) High-resolution methylation profiles were only available for limited tissue types, which cannot permit comprehensive characterization of tissue-specific methylation and joint deconvolution of all tissue types.
Here, we present a comprehensive and high-resolution tissue methylation atlas and a deep neural network (DNN)-based model named cfSort for quantifying tissue composition sensitively and robustly in cfDNA in a supervised manner. The methylation atlas resolves the knowledge gap in tissue-specific methylation, and the cfSort addresses the technical limitations of the existing deconvolution methods. Thus, they can improve the sensitivity and accuracy of tissue deconvolution in cfDNA, enhancing the clinical utility of the tissue-derived cfDNA.
To construct the tissue methylation atlas, we systematically identified tissue-specific methylation signatures from high-resolution Reduced Representative Bisulfite Sequencing (RRBS) data of 521 samples covering 29 major types of noncancerous human tissue, including 8 tissue types that are not covered in ref. 16. These signatures and data constitute one of the largest base-resolution tissue methylation atlases. The traditional tissue-specific methylation signatures were discovered at the population level using the average methylation level of all DNA fragments in genomic bins (14, 15, 17). However, because tissue samples usually comprise DNA from heterogeneous cell types, the average methylation across all DNA can blur the tissue-specific signals that appear in a minor cell proportion (15, 19). To address the tissue heterogeneity issue, here we analyze methylation signals at the individual DNA fragment level, in order to sensitively pick up signatures present even in minor cell populations (19). In addition, we carefully validated the methylation signature atlas in independent methylation datasets, orthogonal epigenomic markers, and transcription regulatory elements.
Here we develop the first supervised method, cfSort, to sensitively and accurately quantify the tissue composition in cfDNA. Taking advantage of the rich tissue methylation data, we generated large-scale diverse training samples of in silico tissue mixtures, which fully exploit the experimental and interindividual variance and ensure the robustness of cfSort. Therefore, compared to the existing unsupervised methods, cfSort intrinsically has advantages in accuracy (20). Combining the comprehensive methylation atlas and cfSort, we demonstrated a more sensitive and accurate estimation of tissue composition compared to the existing methods. In addition, we showed that cfSort was robust against the tissue epigenetic variability, interindividual difference, and experimental noise. We further demonstrated the clinical utilities of cfSort with two potential applications: 1) aiding disease diagnosis and 2) monitoring treatment side effects. For disease diagnosis, we applied the cfSort to plasma cfDNA from healthy individuals and diseased patients, including cancer patients and cirrhosis patients, where cfSort effectively identified a significantly elevated proportion of cfDNA from the affected tissue in those patients, even with methylation data generated by different platforms. For treatment monitoring, we applied cfSort to serial plasma samples from non-small cell lung cancer (NSCLC) patients who received anti-PD-1 immunotherapy. The tissue fractions estimated by the cfSort consistently reflected the organ damages in agreement with biochemical test results. The results of these two clinical scenarios demonstrated the applicability of cfSort in noninvasively disease diagnosis and monitoring.
Results
Building a Comprehensive Tissue Methylation Atlas.
We generated base-resolution methylation data (RRBS) for 521 tissue samples of noncancer participants from the Genotype-Tissue Expression (GTEx) project (21). These tissue samples covered 29 major types of human tissues (SI Appendix, Table S1). From these RRBS data, we performed the analysis at the DNA fragment level and systematically discovered tissue-specific methylation markers. Briefly, we quantified methylation levels in individual DNA fragments, in contrast to the conventional marker discovery using average methylation levels of all DNA fragments within large genomic bins (14, 15). Using the fragment-level methylation, we then identified genomic regions as tissue-specific methylation markers if DNA fragments with tissue-specific methylation patterns (namely tissue-specific DNA fragments) nearly exclusively exist in one group of tissue types, regardless of the fraction, but not in another group of tissue types (Materials and Methods). Therefore, the fragment-level marker discovery is robust to the heterogeneity in the tissue samples (17).
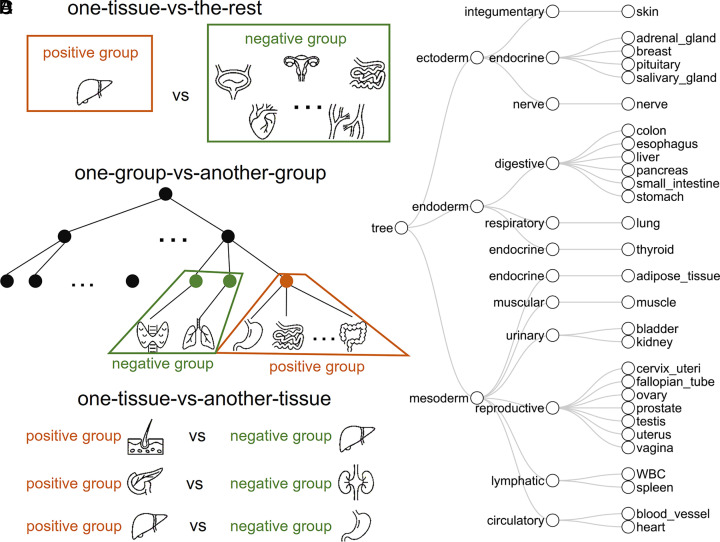
As shown in previous studies (14, 15, 22), different marker discovery strategies focus on different differential methylation patterns between tissues (e.g., one tissue type vs. other tissue types), which can lead to different types of tissue markers. To build a comprehensive tissue methylation atlas, we employed three marker discovery strategies, resulting in three marker types that can cover nearly all differential tissue patterns. Specifically, they include: 1) The one-tissue-vs.-the-rest strategy identifies the Type I markers, with differential methylation signatures between one tissue type and all the other tissue types (Fig. 1A). 2) The one-group-vs.-the-another-group strategy identifies the Type II markers (Fig. 1B), with differential methylation between two tissue groups (tissue group defined by the tissue phylogeny in early development (23), e.g., between the digestive system and lymphatic system, Fig. 1C). 3) The one-tissue-vs.-another-tissue strategy identifies Type III markers, with differential methylation between two tissue types (Fig. 1D), which can help distinguish similar tissue types from adjacent organs, such as the esophagus and stomach. For each strategy, the markers were ranked by their consistency across tissue samples, i.e., the number of samples showing the tissue-specific methylation pattern (Materials and Methods). The top-ranked 100 Type I markers, the top-ranked 200 Type II markers, and the top-ranked 50 Type III markers from each comparison were utilized in the tissue deconvolution (Fig. 2A), in total 51,035 markers (3,775 Type I markers, 6,660 Type II markers, and 40,600 Type III markers). The three types of tissue-specific methylation markers were complementary to each other. Over 70% of markers were unique for each marker type (90.4% for Type I, 73.5% for Type II, and 81.4% for Type III). Therefore, we combined these markers to construct the tissue marker atlas.
Fig. 1.
Three strategies to select the tissue-specific methylation signatures. Illustration of the tissue comparisons in the one-tissue-vs.-the-rest strategy (A), the one-group-vs.-another-group strategy (B) following the tissue development phylogeny (C), and the one-tissue-vs.-another-tissue strategy (D). The fragment-level methylation in a genomic region was compared between the negative group and the positive group. The phylogenetic tree (C) constructed was based on early tissue development (23). The first layer corresponded to the three germ layers in early embryo development. The second layer corresponded to the function systems. The third layer contained the 29 tissue types in our deconvolution model.
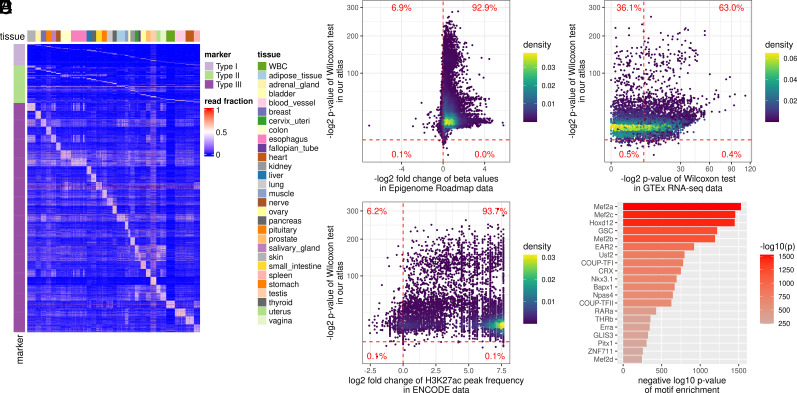
Fig. 2.
Construction and validation of the tissue-specific methylation atlas. (A) Heatmap of three types of tissue-specific markers used in the tissue deconvolution (i.e., the top-ranked tissue markers in the methylation atlas). The methylation atlas consists of the tissue markers that distinguish 29 human tissues. The tissue markers were identified by three strategies (Materials and Methods): Type I markers from the one-tissue-vs.-the-rest strategy (Top); Type II markers from the one-group-vs.-the-another-group strategy using the tissue phylogeny (Middle), and Type III markers from the one-tissue-vs.-another-tissue strategy (Bottom). The color in the heatmap showed the fraction of the tissue-specific fragments out of all fragments at a marker (referred to as the read fraction). (B) Validation of the reproducibility of the identified tissue markers in Epigenome Roadmap data. For each marker, from the RRBS data of our tissue samples, we performed the one-sided Wilcoxon rank-sum test between the corresponding tissues (comparing lowly with highly methylated tissues); on the WGBS data from the Epigenome Roadmap project, we calculated the fold change of the beta values between the corresponding tissues. Each point in the figure corresponds to a marker. The vertical dashed line showed a fold change of 1. The points on the right side of the vertical dashed line represented the markers with fold change <1, indicating a consistent methylation pattern with our RRBS data. The horizontal dashed line indicated a significant P value (<0.01). (C) Marker association with tissue-specific H3K27ac modification. For each marker, on the H3K27ac ChIP-seq data from the ENCODE project, we calculated the fold change of the H3K27ac peak frequency between the corresponding tissues. Each point in the figure corresponds to a marker. The vertical dashed line indicated that the fold change was 1. The horizontal dashed line indicated a significant P value (<0.01). (D) Marker association with tissue-specific transcription. For each marker, on the RNA-seq data from the GTEx project, we performed the Wilcoxon rank-sum test between the corresponding tissues. Each point in the figure corresponds to a marker. The vertical and horizontal dashed lines indicated a significant P value (<0.01). (E) Marker association with tissue-specific transcription regulation. We analyzed the enrichment of transcription factor binding motifs at the marker regions using HOMER. The top 20 enriched motifs and their P values were shown in the figure.
Validation of the Tissue Specificity of the Tissue Methylation Atlas.
We validated the tissue marker atlas with independent data sources from four aspects (SI Appendix): 1) The reproducibility of the tissue-specific methylation. 92.9% of our tissue markers showed consistent tissue-specific methylation in the whole-genome bisulfite sequencing (WGBS) data of the Epigenome Roadmap projects (24) (Fig. 2B and SI Appendix, Fig. S1A). This indicated that the tissue-marker atlas captured real tissue-specific methylation patterns that were reproducible in the independent data with a different cfDNA methylation assay (i.e., WGBS). 2) The association with the tissue-specific histone modification. We focused on the H3K27ac modification, which has the most abundant data in the ENCODE project (25). We observed consistent tissue-specific H3K27ac modification at 93.7% of the tissue markers (Fig. 2C and SI Appendix, Fig. S1B). Hypomethylated regions for a tissue type usually correspond to a tissue-specific elevation of H3K27ac modification, consistent with previous studies (24, 26). 3) The association with the tissue-specific gene expression. In the RNA-seq data in the GTEx project (21), 63.0% of tissue markers had increased gene transcription levels when the corresponding promoter regions were hypomethylated in the tissue types (Fig. 2D), implying that tissue-specific methylation may impact the tissue-specific gene expression. 4) The association with tissue-specific transcription regulation. We performed the enrichment analysis of transcription factor binding motifs at the tissue markers using HOMER (27). The enriched motifs mostly relate to development, differentiation, and tissue-specific expression (Fig. 2E and SI Appendix, Table S2). For example, Homeobox protein Hox-D12 plays an important role in morphogenesis and myocyte enhancer factor-2 also contributes to the development of many tissues (28, 29). These results indicated that the tissue markers of our atlas are indeed involved in tissue-specific biological processes. Through these four validations using independent datasets, we showed that the tissue markers in our atlas were tissue-specific and biologically meaningful.
Overview of cfSort.
Due to the limited quantity and quality of available tissue methylation data, existing cfDNA deconvolution methods relied on unsupervised models, such as non-negative least squares (14, 15) and likelihood-based models (17, 18). Without learning from the ground truth tissue composition, these unsupervised models were intrinsically less powerful compared to the supervised models (20). The major challenge of applying supervised models is the lack of cfDNA data with ground-truth tissue compositions for training and evaluation because it is impossible to know the actual tissue compositions for real cfDNA samples. To address this challenge, we generated a large number of in silico cfDNA data to comprehensively cover the landscape of cfDNA tissue compositions. A similar framework has been proven successful to predict cell composition from tissue expression profiles (30). Using the large in silico cohort, we developed a DNN-based model, cfSort, for cfDNA tissue deconvolution, by considering the cfDNA properties in key components of the DNN constructions, including 1) data generation, 2) feature construction, 3) network architecture, and 4) model training.
Data generation.
We used the RRBS data of the 521 tissue samples to generate in silico cfDNA methylation data with predefined tissue compositions and at different depths of coverage as the training, validation, and testing data (Fig. 3A). As the majority of cfDNA comes from white blood cells (WBC) (3, 14, 15), we required the WBC always to be the major contributor in an in silico cfDNA sample. Specifically, we split the original tissue samples into three groups (SI Appendix, Fig. S2): training (75%), validation (10%), and testing (15%). The in silico cfDNA data for the model training, validation, and testing were generated from the tissue samples in the corresponding group respectively. The data of an in silico cfDNA sample was generated in four steps (SI Appendix, Fig. S3 and details see Materials and Methods): 1) randomly select contributing tissue types; 2) randomly choose one original tissue sample from each tissue type selected in step 1; 3) generate random tissue composition for the in silico cfDNA sample following symmetric Dirichlet distribution under the cfDNA-specific constraint (i.e., WBC as the major contributor); 4) subsample DNA fragments from each tissue sample selected in step 2, following uniform distribution based on the tissue composition (generated in step 3) and mix them to generate the in silico cfDNA sample. A large number of our tissue samples allowed us to generate 295,484 diverse training samples and thousands of validation and testing samples, which fulfilled the requirement of the DNN training and evaluation (31). In addition, in the data generation, we fully exploited the combination of different tissue types, different samples, and different tissue compositions, exploring the possible noise and bias in the data. As a result, cfSort would learn robust tissue-specific features from these data.
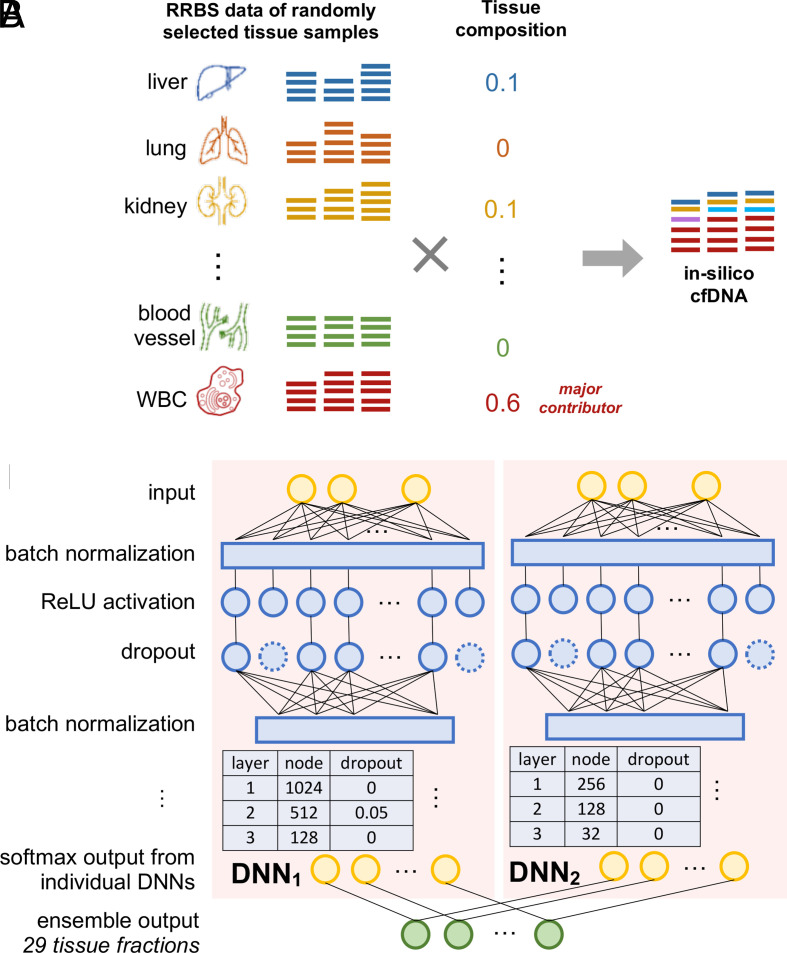
Fig. 3.
Overview of in silico cfDNA data generation and the DNN of the cfSort. (A) Illustration of in silico cfDNA data generation. The data were generated by in silico mixing of the data of tissue samples (Materials and Methods). For a sample, we randomly selected the original tissue samples and generated a tissue composition where the WBC was always the major contributor. Then we uniformly and randomly sampled DNA fragments from the RRBS data of the selected tissue samples based on the corresponding tissue fraction in the tissue composition. The sampled DNA fragments from every tissue sample were pooled together as the simulated sample. The tissue composition was regarded as the ground truth. (B) Illustration of cfSort. cfSort is an ensemble of two component DNNs, which have three dense hidden layers with the ReLU activation. We applied a batch normalization layer before each dense hidden layer and a dropout layer after each hidden layer. The output layer of each DNN contained 29 nodes corresponding to the 29 tissue types in the deconvolution. We utilized the softmax activation function in the output layer. The final output of cfSort is the average of the output from the two component DNNs.
Feature construction and profile generation.
We constructed the input features of cfSort by leveraging the tissue markers in our atlas. Note that cfDNAs protected by nucleosomes generally have higher abundances than those not protected (32). Because different tissue types have different nucleosome positioning (33), the tissue composition in individual genomic regions can deviate from the overall tissue composition, thus resulting in locally unstable cfDNA methylation levels. However, such effects cannot be reflected by our tissue-derived in silico cfDNA data. To address this problem, we designed a marker clustering strategy to merge individual tissue markers into a marker cluster that is robust against the impact of nucleosome positioning. We performed the constrained K-means clustering (34) on the individual markers based on their methylation profiles across training samples, allowing four to seven individual markers in a cluster. This resulted in 10,183 marker clusters. A marker cluster (covering approximately 400 bp to 1,000 bp) has a much larger size compared to the DNA wrapped around a nucleosome (~160 bp). For every marker cluster, we derived a numerical feature as the fraction of tissue-specific DNA fragments at all markers within the cluster. Therefore, for each sample, we derived 10,183 numerical features to form an input feature profile for the DNN. For details see SI Appendix, section S5.1.
DNN architecture.
The basic structure of cfSort is an ensemble of two DNNs. The ensemble helps to reduce the prediction variance (31). Each DNN takes the feature profiles as input and outputs the predicted tissue compositions of the 29 tissue types (Fig. 3B). For each DNN, we constructed three dense hidden layers with a decreased number of nodes (1,024, 512, 128, and 256, 128, 32 respectively) and used the rectified linear unit (ReLU) (35) as the activation function. The hidden nodes can automatically learn weights that prioritize the input features and make the DNN resistant to noises in the data. Considering the size and diversity of our training data, we added a batch normalization layer before each dense layer to stabilize and accelerate the training process (36). In addition, we apply a dropout layer after each dense layer (with dropout rate 0, 0.05, 0, and 0, 0, 0, respectively) to regularize the DNN to increase model robustness and avoid overfitting (37). We calculated the final predicted tissue composition as the averaged predictions from the two DNNs.
Model training.
We applied the state-of-the-art optimizer Adam (38) with a learning rate of 0.001 and a batch size of 32. We used the mean absolute error between the estimated tissue composition and the ground truth as the loss function. To avoid overfitting, we applied two strategies in the training process: 1) Early stopping, i.e., to stop training the DNN when the performance drops on the validation data, i.e., when the model starts to overfit the training data. This strategy has proven effective in cell-type proportion estimation with gene expression data (30). 2) Independent validation data from the training to fairly evaluate the validation loss during the training process. Through these two strategies, overfitting can be effectively avoided.
Analytical Performance of cfSort.
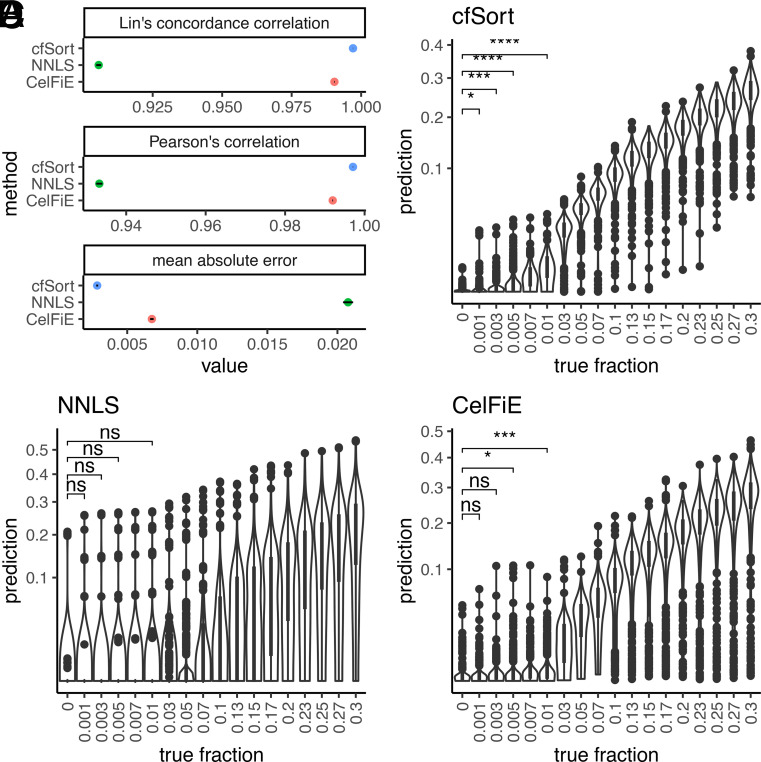
We tested cfSort on an independent testing set of the in silico cfDNA samples (n = 3,660, see Materials and Methods). We compared its performance to two existing tissue deconvolution methods (SI Appendix): the non-negative least square method (NNLS) (14–16) and the CelFiE (17). We evaluated the accuracy of the methods using mean absolute error, Lin’s concordance correlation coefficient, and Pearson’s correlation between the estimated tissue fraction and ground truth. cfSort outperformed NNLS and CelFiE on all three metrics (Fig. 4A). cfSort achieved a lower mean absolute error [0.00286, 95% CI = (90.00279, 0.00293)] than NNLS [0.02076, 95% CI = (0.02040, 0.02112)] and CelFiE [0.00676, 95% CI = (0.00664, 0.00688)], and higher Pearson’s correlation and Lin’s concordance correlation [0.99707, 95% CI = (0.99704, 0.99711) and 0.99707, 95% CI = (0.99704, 0.99711), respectively] than NNLS [0.93323, 95% CI = (0.93245, 0.93400) and 0.90557, 95% CI = (0.90467, 0.90645)] and CelFiE [0.99197, 95% CI = (0.99188, 0.99207) and 0.99038, 95% CI = (0.99028, 0.99049)]. These results indicated that cfSort achieved higher accuracy in estimating tissue compositions than the two competing methods.
Fig. 4.
Analytical performance of the cfSort and comparisons with the existing methods. (A) The accuracy of the estimated tissue composition from the cfSort, NNLS, and CelFiE on the independent testing set. The accuracy was measured by Lin’s concordance correlation, Pearson’s correlation, and mean absolute error between the estimated tissue composition and the ground truth. The dots indicated the metric values, and the line segments indicated the 95% CI. (B–D) The detection limit of the cfSort (B), NNLS (C), and CelFiE (D) were evaluated on the testing dilution series. The detection limit was measured by the statistical significance of a one-sided Student’s t test between the estimated tissue fractions of the samples at every dilution level and the control samples (i.e., 0% tissue fraction). The statistical significance in the figures indicated the P values of the one-sided Student’s t tests at 0.1%, 0.3%, 0.5%, and 1%: “ns” means not statistically significant (P value > 0.05); “*” means P value < 0.05; “**” means P value < 0.01; “***” means P value < 0.001; “****” means P value < 0.0001.
Deconvolution methods need to have a high detection limit to detect tissue-derived cfDNA at low proportions. To assess cfSort’s detection limit, we utilized a widely used approach based on in silico dilution series (n = 20,960). For each sample in the dilution series, we mixed a single tissue sample with a WBC sample in the testing set with known tissue fractions (0%, 0.1%, 0.3%, 0.5%, 0.7%, 1%, 3%, 5%, 7%, 10%, 13%, 15%, 17%, 20%, 23%, 25%, 27%, and 30%) and at different depths of coverage (ranging from 20× to 120×, see Materials and Methods). All tissue samples from all tissue types in the testing group were used to generate the dilution series. Therefore, the evaluation of the detection limit shall reflect the overall performance of cfSort across all tissue types. We determined the detection limit for tissue-derived cfDNA at a specific tissue fraction using one-sided Student t tests against the control samples with 0% tissue fraction (Materials and Methods). At 20×, cfSort detected tissue-derived cfDNA at 0.1% tissue fraction (P value = 0.028, Fig. 4B), while NNLS detected it at 5% (P value = 0.009, Fig. 4C) and CelFiE at 0.5% (P value = 0.010, Fig. 4D and SI Appendix, Table S3). As the depth of coverage increased, cfSort showed improved detection of tissue-derived cfDNA (SI Appendix, Fig. S4). These results demonstrate that cfSort has a better detection limit than the two competing methods.
Robustness of cfSort.
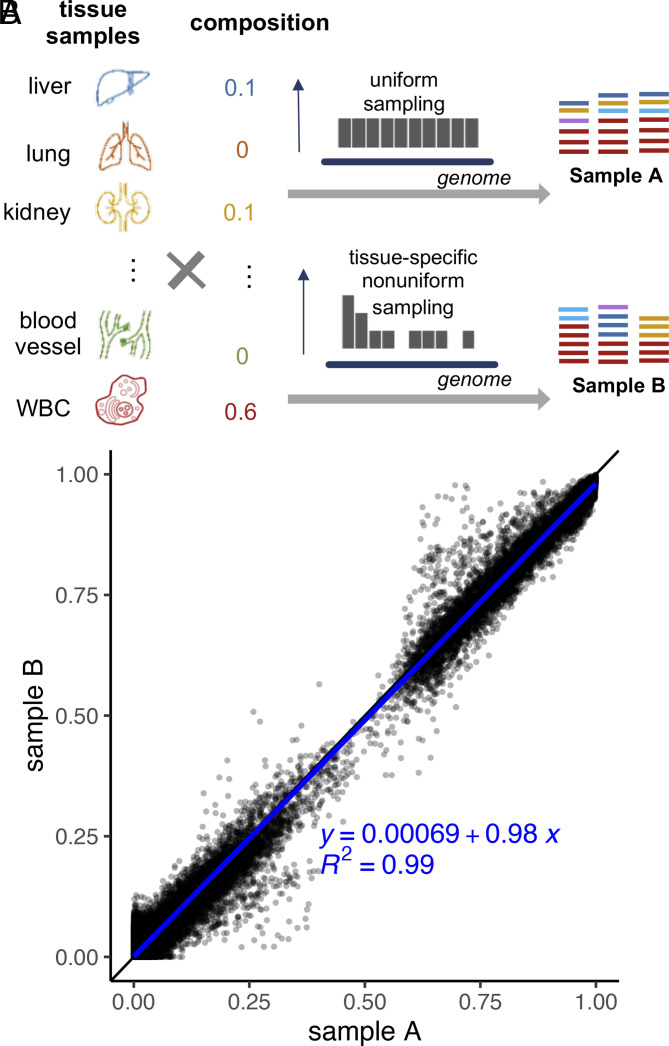
As aforementioned, the nucleosome positioning and other factors (e.g., experimental noise) can cause local fluctuation of the tissue-derived cfDNA fraction. To assess cfSort’s robustness against this local fluctuation, we compared the consistency of estimated tissue composition from in silico cfDNA samples with the same overall tissue composition but different local compositions. We generated in silico cfDNA testing sample pairs (n = 9,023) from the same test tissue samples with different sampling distributions of sequencing reads (Fig. 5A). For sample A, tissue DNA fragments were randomly selected with a uniform distribution, while for sample B, fragments were sampled following a non-uniform tissue-specific distribution generated through random permutation of the average read distribution in cfDNA from 167 healthy individuals (Materials and Methods). Different sampling distributions led to different probabilities of sampling a tissue DNA fragment in a local genomic region, mimicking the epigenetic impact in different tissues. Thus, samples A and B had different local tissue compositions despite having the same overall tissue composition.
Fig. 5.
Evaluation of robustness of the cfSort. (A) Generation of the simulated testing sample pairs for the evaluation of robustness. We generated a testing sample pair (A and B) using the same tissue composition and the same original tissue samples but with different sequencing read sampling distributions. For sample A, we randomly sampled DNA fragments from the original tissue samples following a uniform distribution. For sample B, we used a nonuniform distribution to sample DNA fragments from the original tissue samples. The non-uniform distribution was randomly generated for each tissue type, and the distribution was different for different tissue types. (B) Robustness of the cfSort. The robustness was evaluated by the intercept, slope, and of the fitted linear regression model between the tissue fractions estimated from the testing sample pairs.
The cfSort model was trained on data generated in the same process as sample A and then applied to the testing sample pairs. Therefore, the consistency of the estimated tissue fraction from the testing sample pairs can illustrate the robustness of cfSort under the fluctuation of the local tissue compositions. We measured the consistency using the intercept, slope, and of the fitted linear regression model between the estimated tissue fractions of samples A and B. cfSort demonstrated remarkable robustness, as evidenced by a regression equation close to a perfect diagonal line (intercept = 0.00069 and slope = 0.98) with on the simulated testing sample pairs (Fig. 5B). This result demonstrated that cfSort can estimate tissue fractions with high accuracy even when faced with unexpected randomness not present in the training process. It showed the general applicability of cfSort to the cfDNA samples from diverse individuals.
Clinical Application: Elevated Tissue Fraction in Diseased Patients.
Diseases, including cancers, can impact cell death in affected tissues (3). Decomposing the tissue composition in cfDNA can reveal altered homeostasis in affected tissues (13). In this study, we applied cfSort to cfDNA methylome data [i.e., cfMethyl-Seq data (19) and WGBS data (39)] of healthy and diseased individuals to investigate if the tissue fractions in cfDNA can indicate the incidence of diseases (SI Appendix, Fig. S5). We collected the cfMethyl-Seq data of the plasma cfDNA samples from 100 healthy individuals, 21 cirrhosis patients, and 201 cancer patients (98 lung, 27 liver, and 47 colorectal, 29 stomach cancer patients) (19). The cfMethyl-Seq technology is a revised RRBS technology, specifically adapted to cost-effectively profiling cfDNA methylome (19). Additionally, we curated the WGBS data of the plasma cfDNA samples from 32 healthy individuals and 24 liver cancer patients (39) for cross-platform validation.
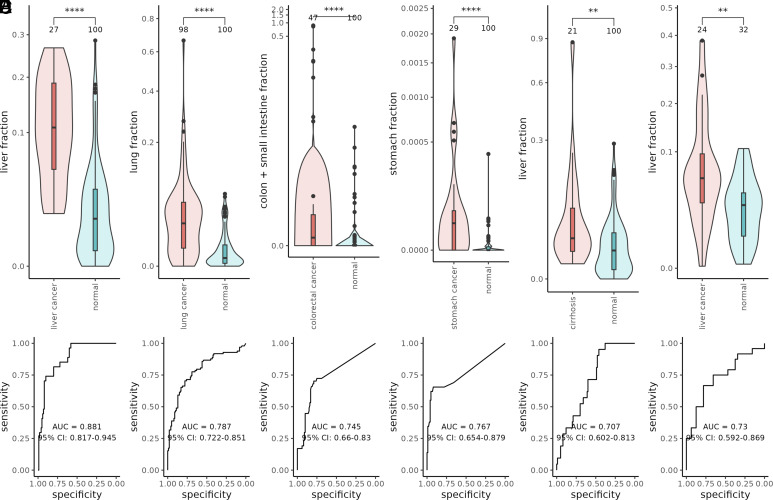
We compared the affected tissue fraction in diseased and healthy cohorts using Student’s t tests. In all comparisons, we observed a significantly higher affected tissue fraction in the diseased patients than in the healthy individuals (Fig. 6 A–E, Wilcoxon rank sum test P value = 1.408e-09, 3.132e-12, 1.468e-07, 1.141e-06, and 0.0029, for liver cancer, lung cancer, colorectal cancer, stomach cancer, and cirrhosis respectively). In addition, the affected tissue fraction increased with cancer stages (SI Appendix, Fig. S6). For colon cancer patients, we included both the small intestine and the colon as the affected tissues. The colon tissues in the GTEx project were collected only from the middle and end parts of the colon, which cannot represent a full picture of the colon (21). Therefore, we utilized the small intestine tissues collected near the start of the colon to complement the incomplete profile of the colon (21). We also evaluated the performance of disease detection using the affected tissue fraction as the sole predictor (Fig. 6 A–E). Our results demonstrated that cfSort can detect elevated cfDNA originating from diseased tissues, indicating the broad clinical utility of cfSort in disease detection and monitoring. However, for a specific disease (e.g., cancer), the integration of tissue-specific and disease-specific (e.g., cancer-specific) methylation patterns, if known, shall lead to the best detection results (19). Our results on the cfDNA WGBS data of liver cancer and healthy individuals (Fig. 6F, Wilcoxon rank sum test P value = 0.0030) further validated the applicability of cfSort on methylation data from different platforms to reveal disease-caused tissue composition changes.
Fig. 6.
The tissue-derived cfDNA fractions of the affected tissue in the diseased and normal individuals. (A) The liver-derived cfDNA fractions from the cfMethyl-Seq data of liver cancer patients and normal individuals. (B) The lung-derived cfDNA fractions from the cfMethyl-Seq data of the lung cancer patients and normal individuals. (C) The intestine-derived cfDNA fractions (including colon and small intestine) from the cfMethyl-Seq data of the colon cancer patients and normal individuals. (D) The stomach-derived cfDNA fractions from the cfMethylSeq data of the stomach cancer patients and normal individuals. (E) The liver-derived cfDNA fractions from the cfMethyl-Seq data of the cirrhosis patients and normal individuals. (F) The liver-derived cfDNA fractions from the WGBS data of the liver cancer patients and normal individuals. The difference between the diseased and normal individuals was evaluated by the Wilcoxon rank sum tests between the estimated fractions of affected-tissue-derived cfDNA. The statistical significance of the tests was indicated by the asterisks: “**” means P value < 0.01; “***” means P value < 0.001; “****” means P value < 0.0001. The receiver operating characteristic (ROC) curve and the area under ROC curve (AUC) showed the performance of disease detection using the tissue-derived cfDNA fractions of the affected tissue as a sole predictor. The number at the top of each violin showed the number of samples.
Clinical Application: Tissue Fraction Changes Reflecting Tissue Damage during Anti-PD-1 Immunotherapy.
The rapidly developing cancer treatments have been improving the survival of cancer patients (40). However, side effects commonly arise from these cancer treatments, often causing tissue damage (41). Considering the large patient population with a history of cancer, the management of side effects is an essential part of their care, affecting the completion of treatment and quality of life (41). The standard evaluation of side effects is based on biochemical tests on tissue-specific markers (42), e.g., alanine transaminase for the liver. Since tissue damage can lead to abnormal cfDNA levels of the affected tissue, tissue-derived cfDNA levels theoretically can provide a novel path to monitor side effects to all tissues in the body.
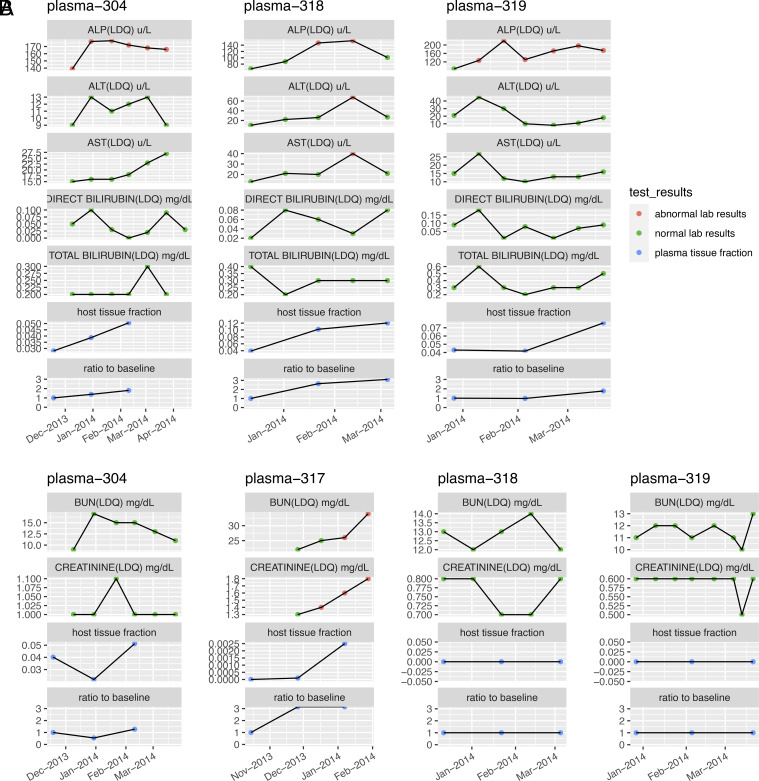
To investigate the clinical potential of tissue-derived cfDNA potential in detecting side effects, we applied cfSort to cfDNA series from 4 NSCLC patients receiving anti-PD-1 immunotherapy (SI Appendix, Fig. S5). Their plasma cfDNA was collected at 0, 6, and 12 wk from the start of treatment. The side effects of anti-PD-1 immunotherapy often impact liver and kidney functions for lung cancer patients, so we focused on the liver and kidney cfDNA fractions. Standard biochemical markers for the liver (including alkaline phosphatase, alanine transaminase, aspartate aminotransferase, direct bilirubin, and total bilirubin) and kidney (blood urea nitrogen and creatinine) were measured alongside the treatment to compare tissue fraction changes. We regarded a patient to have tissue damage if a biochemical marker had abnormal values for at least two consecutive time points, or multiple biochemical markers had abnormal values at the same time; we regarded a patient to have no tissue damage if no biochemical markers had abnormal values at any time, and we regarded a patient as unanalyzable if the patient had only transient abnormal test results at one time point.
For liver damage, three of the four patients were analyzable, and their biochemical tests indicated the presence of liver damage. Consistent with their biochemical test results, all three patients showed an increased level of liver-derived cfDNA (Fig. 7A). For kidney damage (Fig. 7B), one of the four patients (plasma-317) had kidney damage and showed an increased level of kidney-derived cfDNA. In contrast, the other three patients (plasma-304, plasma-318, and plasma-319) did not have kidney damage. Two of them (plasma-318 and plasma-319) have undetectable kidney-derived cfDNA. The other one of the three patients (plasma-304) showed an unstable kidney-derived cfDNA level. In addition, we observed a consistently strong correlation (average Pearson’s correlation = 0.899) between the abnormal biochemical test results and the affected tissue fractions for the side effects on both liver and kidney (SI Appendix, Table S4). In general, the tissue-derived cfDNA levels changed consistently with the biochemical test results, indicating potential tissue damage from cancer treatments. Although further validation in large patient cohorts is needed, our case study showed the first indication of detecting side effects on noncancer tissues of cancer patients using cfDNA. The results implied the potential of tissue-derived cfDNA in comprehensive side effect monitoring during cancer treatments, which is especially meaningful for those organs without standard biochemical markers.
Fig. 7.
The tissue-derived cfDNA fractions and the biochemical marker levels of four NSCLC cancer patients who received anti-PD-1 immunotherapy. (A) The liver-derived cfDNA fractions and the levels of biochemical markers indicating liver functions. (B) The kidney-derived cfDNA fractions and the levels of biochemical markers indicating kidney functions. The plasma cfDNA samples were collected at the 0 wk, 6 wk, and 12 wk, measured starting from the beginning of the treatment. The biochemical markers were tested during the treatment. The affected tissue fraction was estimated by cfSort; the ratio to baseline was the ratio between the affected tissue fraction at a certain time point and the fraction at the 0 wk.
Discussion
We presented a comprehensive high-resolution tissue methylation atlas and the first supervised tissue deconvolution method for cfDNA, namely cfSort. They enabled sensitive and robust quantification of tissue fractions in cfDNA. We validated the atlas of tissue markers by multiple independent datasets, and we showed that these markers were associated with tissue development, tissue differentiation, and tissue-specific transcription.
We developed the supervised tissue deconvolution method for cfDNA, cfSort, by using the tissue methylation atlas to simulate large-scale training data with ground truth, facilitating supervised learning. To ensure the robustness of tissue deconvolution, we generated diverse training samples to exploit the potential experimental noise and individual variance in the population. To model the complicated relationship of tissue methylation and prioritize the tissue-specific markers, we applied the nonlinear hidden layers and dropouts in the DNN. We have shown that the cfSort outperformed the existing methods in terms of accuracy and detection limit: making more accurate tissue fraction estimation and distinguishing a lower level of tissue-derived cfDNA. In addition, the cfSort demonstrated nearly perfect robustness toward the unseen local fluctuations of tissue compositions, indicating its wide applicability to diverse individuals.
While preparing this manuscript, a large WGBS dataset of human cell types has been published (16). While our dataset is on tissue samples, this other dataset is on cell types. Note that a tissue may contain many cell types. Thus, our dataset provides more comprehensive coverage of tissue characteristics, while the other dataset provides more homogenous profiles of specific cell types. In addition, our atlas contains more samples for each tissue (median 15 in our dataset vs. 3 in the other dataset) and covers eight more tissue types. Although our dataset uses RRBS profiling, we showed that the RRBS data covered the majority of the cell-type–specific markers identified by WGBS in this study (SI Appendix, Table S5). Therefore, both datasets are complementary in covering the human tissue atlas. Note that this other study utilized the unsupervised NNLS for deconvolution, while we proposed the supervised DNN deconvolution.
The cfSort is a general tool for quantifying tissue composition in cfDNA, which could be widely used in all tissue-related applications. In this study, we presented two clinical applications that 1) identified elevated tissue fractions in cancer and cirrhotic patients compared to controls and 2) detected tissue fraction changes consistent with liver and kidney damages in NSCLC patients treated with anti-PD-1 immunotherapy. In addition, we showed that the cfSort was directly applicable to the cross-platform data (i.e., WGBS), although it was trained on the data of different platforms (e.g., enriched methylome sequencing data, such as RRBS and cfMethyl-Seq). With these results, we demonstrated the potential clinical utilities of the cfSort. We believe that the cfSort will facilitate and advance cfDNA-based disease detection, therapy prognosis, and longitudinal treatment monitoring.
Materials and Methods
Data Collection.
We collected the cfMethyl-Seq data of the plasma samples from 100 healthy individuals, 21 cirrhosis patients, and 201 cancer patients (98 lung, 27 liver, and 47 colon, 29 stomach cancer patients) under the accession code EGAS00001006020 in the European Genome-Phenome Archive (19). We also collected the WGBS data of the plasma cfDNA samples from 32 healthy individuals and 24 liver cancer patients under the accession code EGAS00001000566 in the European Genome-Phenome Archive (39). These data were used as an example of applications for cfSort. We curated orthogonal validation data for the tissue methylation atlas from public databases (SI Appendix, Table S6), including the WGBS data from the Epigenome Roadmap projects (24), the RNA-seq data from the GTEx project (21), and the chromatin immunoprecipitation sequencing (ChIP-seq) data from the ENCODE project (25).
Human Subjects.
We collected 12 plasma cfDNA samples from 4 NSCLC patients treated with anti-PD-1 immunotherapy at the University of California, Los Angeles for KEYNOTE-001 under clinical trial registration ClinicalTrials.gov number NCT01295827. All patients provided written consent before any study-related procedures were performed. The plasma samples were collected from each patient at 0 wk, 6 wk, and 12 wk, measured at the start of the treatment. We collected 521 genomic DNA samples, including 464 non-WBC tissue samples from the GTEx project (21) and 57 WBC samples from UCLA hospitals. This project was approved by the Institutional Review Board of the University of California, Los Angeles (IRB# 12-001891, IRB# 11-003066, IRB#19-000618, IRB#19-000230, IRB#19-001488, IRB#16-000659, IRB#17-000985, and IRB# 13-00394). Our research complies with all relevant ethical regulations. All participants gave their written informed consent. The RRBS libraries of the genomic DNA were constructed following the standard RRBS protocol (43); the cfMethyl-Seq libraries of the serial plasma cfDNA samples from the four NSCLC patients were constructed following the standard protocol (19) (SI Appendix). The preprocessing of the RRBS and cfMethyl-Seq data followed a standard procedure described in ref. 19 (SI Appendix).
DNA Fragment Level Discovery of Methylation Markers.
To conquer the cell-type heterogeneity in the tissue samples (15), we employed a DNA-fragment-level marker discovery framework to stratify tissue-specific DNA fragments from background DNA fragments (tissue-invariant DNA fragments) for capturing tissue signals sensitively and specifically (19). In this method, we utilized our previously proposed fragment-level methylation measurement (so-called α-value), defined as the fraction of methylated CpGs out of all CpGs on a DNA fragment (19). This fragment-level measurement has been utilized in several studies to identify cancer-specific methylation markers (19, 44). In brief, we identified the tissue markers between two groups (namely positive and negative groups) of tissue samples at the fragment level. We first generated the α -value distribution of the DNA fragments in a genomic region for the tissue samples in the positive and negative groups respectively. The α value distribution was defined as a function of α-value ( ). For a given α, was calculated as the number of tissue samples containing fragments whose α-value was less than α. Then we identified markers as the genomic regions where the α-value distribution in the samples of the positive group (namely positive samples) has a well-separated component from those of the negative group (namely negative samples). Specifically, for a genomic region, we looked for a threshold such that a number of positive samples (denoted as ) contained DNA fragments with α-values but nearly no negative samples (denoted as ) contained such DNA fragments. If an can be found for a genomic region, we treated that genomic region as a tissue marker for the positive group with a tissue-specific α-value-threshold , i.e., all DNA fragments with α-value were treated as tissue-specific DNA fragments from the positive group. The more tissue samples with tissue-specific DNA fragments in the positive group (i.e., the larger ), the higher quality and more stable the markers are. Therefore, the identified markers were ranked by . The DNA-fragment-level marker discovery was applied to all three strategies of identifying tissue-specific methylation markers.
Construction of the Tissue Marker Atlas.
We constructed the tissue methylation atlas by using three strategies for identifying tissue-specific methylation markers: one-tissue-vs.-the-rest comparisons (Type I markers), one-group-vs.-another-group comparisons (Type II markers), and one-tissue-vs.-another-tissue comparisons (Type III markers). The three strategies complementarily identified different types of tissue-specific methylation markers. In the one-tissue-vs.-the-rest comparisons, we used the samples from one tissue type as the positive group and all samples from other tissue types as the negative group (Fig. 1A) and then applied the DNA-fragment-level marker discovery framework to identify markers. Therefore, we discovered the unique methylation patterns for each tissue type by this strategy. Because the negative group contained hundreds of samples, we required of the selected markers to be less than 20. In the one-group-vs.-another-group comparisons, we discovered markers following the tissue phylogeny (Fig. 1B) in early human development. We constructed this phylogeny based on the literature (23). The comparison was conducted at every tree level, and every node was compared against its siblings (Fig. 1C). In other words, when all samples under a node were regarded as the positive group, all samples under its sibling nodes were regarded as the negative group in the comparison. In this way, we discovered markers following the tissue differentiation trajectory by employing the DNA-fragment-level marker discovery framework on the defined negative and positive groups. We required of the selected markers to be less than 10. In the one-tissue-vs.-another-tissue comparisons, we only considered two tissue types at a time. We used the samples from one tissue type as the positive group and the samples from the other tissue type as the negative group (Fig. 1D). Using this strategy together with the DNA-fragment-level marker discovery framework, we can identify differential methylation patterns that distinguish two similar tissue types, e.g., vagina and uterus. Considering the size of the negative group, we required of the selected markers to be less than 2. In the tissue deconvolution, we used the top 100 Type I markers (29 comparisons), the top 200 Type II markers (38 comparisons), and the top 50 Type III markers per comparison (812 comparisons). Ties were included. In total, 51,035 individual tissue markers were used in the tissue deconvolution.
Simulation of Tissue Mixtures from the RRBS Data of Tissue Samples.
Because cfMethyl-Seq and RRBS profile the exact same genomic regions (19), we can directly utilize the RRBS data of tissues to generate the simulated cfMethyl-Seq data of plasma cfDNA with known tissue compositions. Specifically, the simulated cfMethyl-Seq data of an in silico cfDNA sample was generated in four steps (SI Appendix, Fig. S3). In step 1, we chose a number of tissue types that contributed to the simulated sample with positive fractions. In addition to WBC, we randomly selected a certain number (ranging from 1 to 9) of non-WBC tissue types. Note that we did not use 29 tissue types altogether, because the number of combinations went up exponentially as the number of tissue types increased. In addition, the tissue fraction of most tissue types can be quite low if we mix 29 tissue types altogether. Therefore, it will be computationally impractical to obtain enough samples with tissue fractions spanning a desirable range. In step 2, we randomly chose a real tissue sample for each selected tissue type and WBC. Note that we pooled multiple real WBC samples together, if a single real WBC sample contained inadequate reads to generate the simulated data. The corresponding RRBS datasets of these selected tissue samples were to be used as the data sampling source for the simulated tissue mixture. In step 3, we created random tissue composition for the simulated sample. Briefly, a random positive integer was chosen for each selected tissue type and WBC, and then this number was divided by the sum of all random numbers to ensure the tissue fractions added up to 1. The tissue fraction of a tissue type was calculated as , where was the random number generated for the tissue . We specifically required the WBC to always have a large fraction (on average 75%) in the tissue composition. This ensured that the WBC was the main contributor, consistent with the characteristics of real cfDNA samples. We assigned zero as the tissue fraction if a tissue type was not selected in step 1. Therefore, we generated the ground truth composition for the simulated sample. In step 4, we randomly sampled sequencing reads from the selected tissue samples (from Step 2) based on the tissue composition (generated in Step 3). We generated the simulated cfMethyl-Seq data at 20x, 40x, 60x, 90x, and 120x coverage, equivalent to approximately 20, 40, 60, 90, and 120 million paired-end sequencing reads. Therefore, for a tissue type , we randomly sampled read pairs from the RRBS dataset of the selected tissue sample, where is the total number of reads to be sampled. Finally, we mixed the sampled read pairs from all tissue types as the in silico cfMethyl-Seq data to represent a simulated cfDNA sample. Due to the limited depth of the original WBC data, we pooled multiple WBC samples together to generate in silico cfDNA samples at high depths (>20x).
We split the original tissue samples into three groups (SI Appendix, Fig. S2): training (75%), validation (10%), and testing (15%). Then we applied the above simulation procedure to each of the three groups and generated the corresponding simulated training, validation, and testing cfDNA data with known tissue compositions.
Evaluation of Robustness Toward Nonrandom Fragmentation.
To evaluate the robustness of cfSort, we created the simulated testing sample pairs (n = 9,023) with different tissue-specific read distributions, but with the same tissue composition and the same source of tissue fragments (Fig. 5A). We generated a pair of testing samples (samples A and B) following Steps 1 to 3 described above. For sample A, we followed Step 4 described above. For sample B, we used the same selected tissue samples and the same tissue composition. Instead of sampling reads using a uniform distribution, we enforced the sampled tissue-specific reads to follow a tissue-specific read distribution. To generate the tissue-specific read distribution, we calculated the average read count per million (RCPM) at each region in the cfMethylSeq data of 167 healthy plasma samples. We directly used the average RCPM as the read distribution for WBC; while for each non-WBC tissue type, we permutated the average RCPM in the regions as the read distribution. Then for a region, the number of reads sampled from the original tissue sample was proportional to the respective tissue-specific read distribution in the region. The robustness of cfSort was evaluated as the consistency of the estimated tissue compositions between the testing sample pairs, i.e., the intercept, slope, and of the fitted linear regression model.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by the National Cancer Institute (grant no. U01CA230705 to S.W.F., S.-H.B.H., and X.J.Z. grant no. R01CA246329 to W.L., S.M.D, and X.J.Z. grant no. U01CA237711 to W.L., grant no. R01CA264864 to X.J.Z., grant no. R01CA246304 to V.G.A., grant no. R01CA253651 to V.G.A.), the NSF (grant no. DMS1952386 for Q.L. and W.H.W.), and the NIH (grant no. R01HG010359 for Q.L. and W.H.W., and P50HG007735 for Q.L. and W.H.W.). This work was supported by the V Foundation Translational Award to X.J.Z. and a gift from the Connie Frank Foundation to X.J.Z. This work was kindly supported by funds from the Integrated Diagnostics Program, Department of Radiological Sciences & Department of Pathology and Laboratory Medicine, David Geffen School of Medicine at University of California at Los Angeles. The Genotype-Tissue Expression (GTEx) project kindly supported our study by providing tissue DNA samples. The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 08/26/2019 and/or dbGaP accession number phs000424.v8.p2. The GTEx Project was supported by the Common Fund of the Office of the Director of the NIH, and by National Cancer Institute, National Human Genome Research Institute, National Heart, Lung, and Blood Institute, National Institute on Drug Abuse, National Institute of Mental Health, and National Institute of Neurological Disorders and Stroke.
Author contributions
S.L., W.H.W., and X.J.Z. designed research; S.L., W.Z., X.N., Q.L., W.L., M.L.S., Y.Z., A.G., K.K., P.A., D.S.L., S.S.R., W.H., D.R.A., C.E.M., S.W.F., S.-H.B.H., E.B.G., V.G.A., W.H.W., S.M.D., and X.J.Z. performed research; S.L. and M.L.S. analyzed data; W.Z. and Y.Z. performed experiments and generated data; X.N., Q.L., W.L., E.B.G., W.H.W., and S.M.D. provided scientific advice; A.G., K.K., P.A., W.H., D.R.A., C.E.M., S.W.F., S.-H.B.H., E.B.G., and V.G.A. provided patient samples and clinical data; D.S.L., S.S.R., E.B.G, and S.M.D. provided clinical guidance; X.J.Z. supervised the overall study; and S.L., W.Z., W.L., and X.J.Z. wrote the paper.
Competing interests
W.L., W.H.W., and X.J.Z. are co-founders of EarlyDiagnostics Inc. X.N. and M.L.S. are employees at EarlyDiagnostics Inc. S.L. is a former employee at EarlyDiagnostics Inc. S.L., W.Z., X.N., W.L., M.L.S., Y.Z., W.H.W., S.M.D., and X.J.Z. own stocks of EarlyDiagnostics Inc. The other authors declare no competing interests.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Wing Hung Wong, Email: whwong@stanford.edu.
Steven M. Dubinett, Email: SDubinett@mednet.ucla.edu.
Xianghong Jasmine Zhou, Email: XJZhou@mednet.ucla.edu.
Data, Materials, and Software Availability
cfSort is implemented in Python and is freely available for academic and research usage through the GitHub repository, https://github.com/jasminezhoulab/cfSort (45). The raw sequencing data of the tissue samples and the plasma samples is deposited at the European Genome-phenome Archive (EGA) under the accession number EGAS00001007213 (46). DNA methylation data of 521 tissue samples are available in bed format (methylation level at individual CpG sites) and in pat format (fragment-level information, including CpG starting index, methylation pattern of all covered CpGs and number of fragments with exact multiCpG pattern) at GEO under the accession number GSE233417 (47).
Supporting Information
References
- 1.Wan J., et al. , Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Stroun M., et al. , The origin and mechanism of circulating DNA. Ann. N. Y. Acad. Sci. 906, 161–168 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Kustanovich A., et al. , Life and death of circulating cell-free DNA. Cancer Biol. Therapy 20, 1057–1067 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li S., et al. , cfTrack: A method of exome-wide mutation analysis of cell-free DNA to simultaneously monitor the full spectrum of cancer treatment outcomes including MRD, recurrence, and evolutioncfTrack: Comprehensive cancer monitoring using cfDNA. Clin. Cancer Res. 28, 1841–1853 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li S., et al. , Sensitive detection of tumor mutations from blood and its application to immunotherapy prognosis. Nat. Commun. 12, 1–14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li S., et al. , cfSNV: a software tool for the sensitive detection of somatic mutations from cell-free DNA. Nat. Protoc. 18, 1563–1583 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W., et al. , CancerDetector: ultrasensitive and non-invasive cancer detection at the resolution of individual reads using cell-free DNA methylation sequencing data. Nucleic Acids Res. 46, e89–e89 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen S. Y., et al. , Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 563, 579–583 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Liu M. C., et al. , Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 31, 745–759 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tug S., et al. , Correlation between cell free DNA levels and medical evaluation of disease progression in systemic lupus erythematosus patients. Cell Immunol. 292, 32–39 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Lehmann-Werman R., et al. , Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl. Acad. Sci. U.S.A. 113, E1826–E1834 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jahr S., et al. , DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 61, 1659–1665 (2001). [PubMed] [Google Scholar]
- 13.Barefoot M. E., et al. , Detection of cell types contributing to cancer from circulating, cell-free methylated DNA. Front. Genet. 12, 671057 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun K., et al. , Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl. Acad. Sci. U.S.A. 112, E5503–E5512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moss J., et al. , Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 9, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loyfer N., et al. , A DNA methylation atlas of normal human cell types. Nature 613, 1–10 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caggiano C., et al. , Comprehensive cell type decomposition of circulating cell-free DNA with CelFiE. Nat. Commun. 12, 1–13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vecchio D., et al. , Cell-free DNA methylation and transcriptomic signature prediction of pregnancies with adverse outcomes. Epigenetics 16, 642–661 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stackpole M. L., et al. , Cost-effective methylome sequencing of cell-free DNA for accurately detecting and locating cancer. Nat. Commun. 13, 1–12 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Talaei Khoei T., Kaabouch N., A comparative analysis of supervised and unsupervised models for detecting attacks on the intrusion detection systems. Information 14, 103 (2023). [Google Scholar]
- 21.Lonsdale J., et al. , The genotype-tissue expression (GTEx) project. Nat. Genet. 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lokk K., et al. , DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome Biol. 15, 1–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pansky B., Review of Medical Embryology (Macmillan, 1982). [Google Scholar]
- 24.Kundaje A., et al. , Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sloan C. A., et al. , ENCODE data at the ENCODE portal. Nucleic Acids Res. 44, D726–D732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsankov A. M., et al. , Transcription factor binding dynamics during human ES cell differentiation. Nature 518, 344–349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heinz S., et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stelzer G., et al. , The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr. Protocols Bioinf. 54, 1–30 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Safran M., et al. , “The genecards suite” in Practical Guide Life Science Databases (Springer, Singapore, 2021), vol. 6, pp. 27–56. [Google Scholar]
- 30.Menden K., et al. , Deep learning–based cell composition analysis from tissue expression profiles. Sci. Adv. 6, eaba2619 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodfellow I., Bengio Y., Courville A., Deep Learning (MIT press, 2016). [Google Scholar]
- 32.Snyder M. W., et al. , Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 164, 57–68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oruba A., Saccani S., van Essen D., Role of cell-type specific nucleosome positioning in inducible activation of mammalian promoters. Nat. Commun. 11, 1075 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bradley P. S., Bennett K. P., Demiriz A., “Constrained k-means clustering” (Microsoft Research Technical Report 2000.65; ), vol. 20, pp. 0–8. [Google Scholar]
- 35.Hahnloser R. H. R., et al. , Digital selection and analogue amplification coexist in a cortex-inspired silicon circuit. Nature 405, 947–951 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Ioffe S., Szegedy C., “Batch normalization: Accelerating deep network training by reducing internal covariate shift” in Proceedings of the 32nd International Conference on Machine Learning (PMLR) (Lille, France, 2015). [Google Scholar]
- 37.Srivastava N., et al. , Dropout: A simple way to prevent neural networks from overfitting. J. Machine Learning Res. 15, 1929–1958 (2014). [Google Scholar]
- 38.Kingma D. P., Ba J., “Adam: A method for stochastic optimization” in 3rd International Conference for Learning Representations, Y. Bengio and Y. LeCun, Eds. (arXiv.org, Ithaca, NY, 2014). [Google Scholar]
- 39.Allen Chan K. C., et al. , Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc. Natl. Acad. Sci. U.S.A. 110, 18761–18768 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pizzoli S. F. M., et al. , From life-threatening to chronic disease: Is this the case of cancers? A systematic review. Cogent. Psychol. 6, 1577593 (2019). [Google Scholar]
- 41.Miller K. D., et al. , Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 69, 363–385 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Garon E. B., et al. , Five-year overall survival for patients with advanced non-small-cell lung cancer treated with pembrolizumab: Results from the phase I KEYNOTE-001 study. J. Clin. Oncol. 37, 2518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meissner A., et al. , Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 33, 5868–5877 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J., et al. , DISMIR: Deep learning-based noninvasive cancer detection by integrating DNA sequence and methylation information of individual cell-free DNA reads. Briefings Bioinf. 22, bbab250 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li S., Li W., Stackpole M.L., Zhou X.J., cfSort. GitHub. https://github.com/jasminezhoulab/cfSort. Deposited 1 May 2023.
- 46.Li S., et al. , Deep-learning-powered tissue deconvolution for cfDNA. EGA. https://ega-archive.org/studies/EGAS00001007213. Deposited 19 May 2023. [Google Scholar]
- 47.Li S., et al. , A comprehensive DNA methylation atlas for noncancer human tissue types. GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE233417. Deposited 25 May 2023. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
cfSort is implemented in Python and is freely available for academic and research usage through the GitHub repository, https://github.com/jasminezhoulab/cfSort (45). The raw sequencing data of the tissue samples and the plasma samples is deposited at the European Genome-phenome Archive (EGA) under the accession number EGAS00001007213 (46). DNA methylation data of 521 tissue samples are available in bed format (methylation level at individual CpG sites) and in pat format (fragment-level information, including CpG starting index, methylation pattern of all covered CpGs and number of fragments with exact multiCpG pattern) at GEO under the accession number GSE233417 (47).