Significance
EMT is generally considered to be a cell-intrinsic process that augments the metastatic capacity of cancer cells by activating a polarity-axis switch that enhances cell motility and invasiveness. Here, we showed that lung adenocarcinoma (LUAD) cells gain metastatic capacity through an EMT-activated secretory program that coordinates cancer cell invasion and immunosuppression in the tumor microenvironment. Importantly, we demonstrated that secretory blockade reverses acquired resistance to anti-PD-L1 checkpoint blockade, an emerging clinical problem. These findings represent a conceptual leap that EMT drives LUAD metastasis through a cell-extrinsic mechanism, and they provide a foundation for clinical strategies designed to overcome immunotherapy resistance.
Keywords: epithelial-mesenchymal transition (EMT), membrane trafficking, lung cancer
Abstract
Epithelial-to-mesenchymal transition (EMT) underlies immunosuppression, drug resistance, and metastasis in epithelial malignancies. However, the way in which EMT orchestrates disparate biological processes remains unclear. Here, we identify an EMT-activated vesicular trafficking network that coordinates promigratory focal adhesion dynamics with an immunosuppressive secretory program in lung adenocarcinoma (LUAD). The EMT-activating transcription factor ZEB1 drives exocytotic vesicular trafficking by relieving Rab6A, Rab8A, and guanine nucleotide exchange factors from miR-148a-dependent silencing, thereby facilitating MMP14-dependent focal adhesion turnover in LUAD cells and autotaxin-mediated CD8+ T cell exhaustion, indicating that cell-intrinsic and extrinsic processes are linked through a microRNA that coordinates vesicular trafficking networks. Blockade of ZEB1-dependent secretion reactivates antitumor immunity and negates resistance to PD-L1 immune checkpoint blockade, an important clinical problem in LUAD. Thus, EMT activates exocytotic Rabs to drive a secretory program that promotes invasion and immunosuppression in LUAD.
Metastasis is a complex process by which the tumor microenvironment (TME) is skewed toward an immunosuppressive state, thereby permitting cancer cells to invade and disseminate to distant sites (1). Cancer cells inactivate tumor-infiltrating CD8+ T cells through immune checkpoints mediated by, among other ligands, PD-L1 (2). Clinical strategies designed to target immune checkpoints have improved survival in patients bearing a variety of different tumor types, including lung adenocarcinoma (LUAD) (3). However, only a minority of cancer patients benefit from checkpoint inhibitor therapies, and most responding patients eventually acquire resistance to this treatment modality (4, 5). Thus, strategies to overcome resistance to checkpoint inhibitor therapies are urgently needed.
Paracrine signals emanating from cancer cells establish an immunosuppressive TME that prompts a decrease in tumor-infiltrating CD8+ T cells and an increase in the infiltration of myeloid-derived suppressor cells (6, 7), which is in line with a proposed tumor-as-organizer model in which the primary source of TME-molding signals is hypersecretory cancer cells (8). Proteins destined for secretion are synthesized in the endoplasmic reticulum and transferred to the Golgi apparatus, where cargos are packaged into secretory vesicles that are transported to the plasma membrane through post-Golgi traffic (9). Secretory vesicle biogenesis and trafficking are coordinated by exocytotic Rabs, including Rab6A, which promotes fission of exocytotic vesicles from Golgi membranes and activates vesicle trafficking toward the cell surface, and Rab8A, which promotes the docking and fusion of exocytotic vesicles at the plasma membrane (10). These Rabs cycle through active (GTP-bound) and inactive (GDP-bound) states through the coordinated actions of guanine nucleotide exchange factors (GEFs) and GTP-activating proteins (11–13). For example, Rgp1 and Rabin8 are GEFs that recruit Rab6A to the Golgi and polarized Rab8A+ vesicular transport to cell protrusions, respectively (14), and have been implicated in cancer progression (15–17). However, the way in which cancer cells activate exocytotic Rabs to acquire hypersecretory states remains unclear.
Epithelial-to-mesenchymal transition (EMT) causes cancer cells to switch their axis of polarity from apical–basal to front-rear, which orients organelles and actin-based cytoskeletal structures toward the leading edge of the cell to facilitate directional motility and metastasis (18). EMT is driven by transcription factor families (e.g., ZEB, Twist, Snail) that control large transcriptomes, in part owing to their capacities to silence a wide range of microRNAs (19). In metastatic cancer cells, high ZEB1 levels generate a compact, polarized Golgi organelle that establishes a front-rear polarity axis with focal adhesions (FAs) that anchor the leading edge to the substratum and are disassembled, endocytosed, and transported back to the leading edge as cancer cells move forward (20, 21), a process that is facilitated by Rab8A (22).
Here, we hypothesized that EMT activates exocytotic vesicular trafficking networks to coordinate multiple prometastatic functions, including immunosuppression and tumor cell invasion. To test this hypothesis, we utilized LUAD cell lines derived from humans and genetically engineered mice that develop metastatic LUAD from the expression of mutant K-ras and p53 (20, 23, 24) (SI Appendix, Table S1). The murine LUAD cell lines (hereafter termed KP cells) display variable metastatic activity; highly metastatic KP cells undergo a partial EMT or the reverse process in response to extracellular cues, a plasticity governed by mutual antagonism between miR-200 family members and the EMT-activating transcription factor ZEB1, whereas poorly metastatic KP cells exhibit epithelial properties that do not change in response to extracellular cues (24). The findings presented herein support a model in which ZEB1 accelerates post-Golgi exocytotic trafficking to activate a secretory program that coordinates multiple prometastatic functions and might be targeted to overcome immune checkpoint inhibitor resistance.
Results
ZEB1 Activates Exocytotic Vesicular Transport.
To test our hypothesis, we initially surveyed the expression levels of Rab family members in an epithelial KP cell line (393P) subjected to ectopic ZEB1 expression (393P_ZEB1) and found that ZEB1 up-regulated the mRNA levels of Rabs involved in exocytosis (Rab6A and Rab8A), lysosomal trafficking (Rab7), and Golgi-to-endosome trafficking (Rab14 and Rab40B) (25, 26) (SI Appendix, Fig. S1A). Given that exocytotic Rabs are key secretory drivers, we selected Rab6A and Rab8A for further analysis.
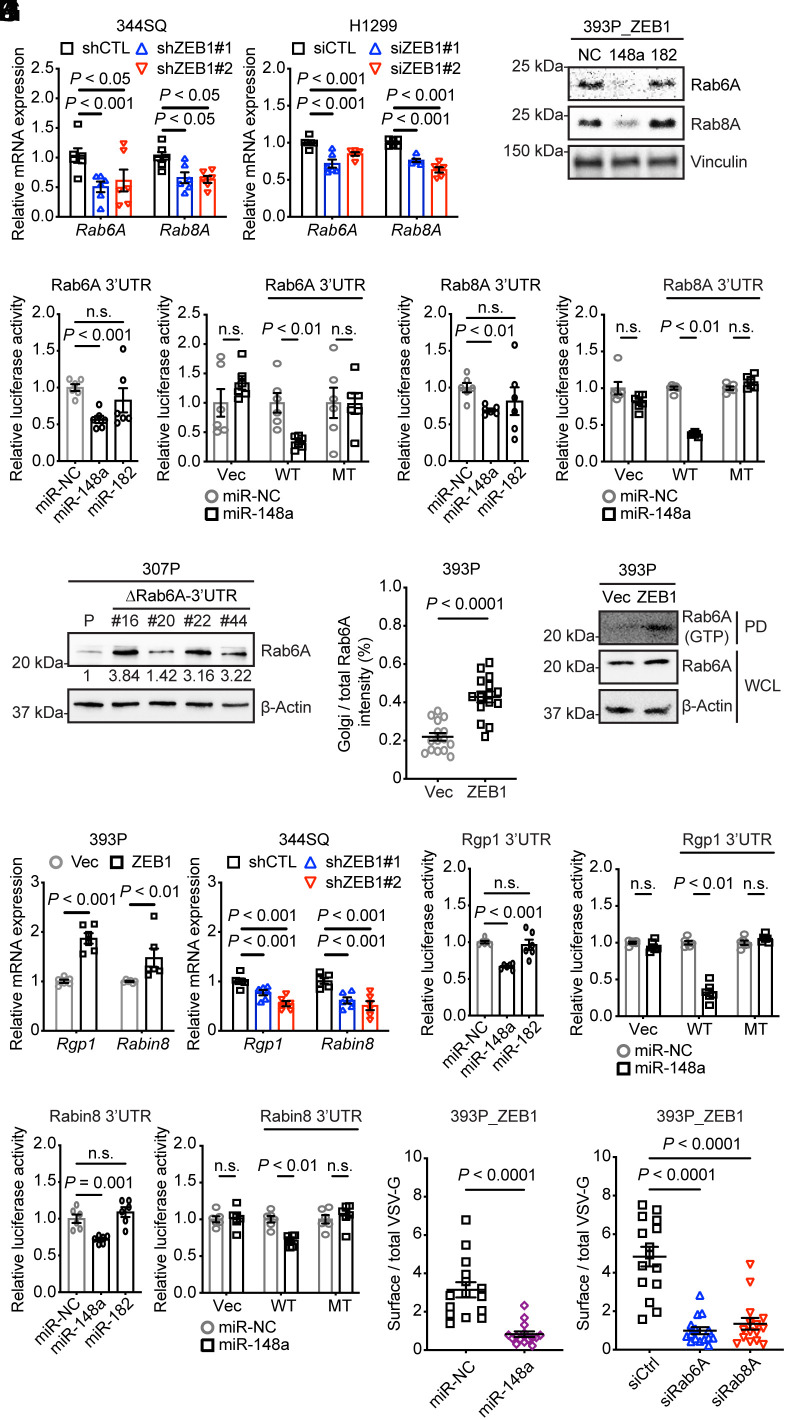
After confirming that their mRNA and protein levels changed in a corresponding fashion following ZEB1 gain- or loss-of-function (Fig. 1A and SI Appendix, Fig. S1B), we asked whether ZEB1 up-regulates Rab6A and Rab8A through a common intermediate. Analysis of their 3′-untranslated regions (3′-UTRs) (www.targetscan.org) identified predicted binding sites for miR-148a and miR-182 (SI Appendix, Table S2), which are ZEB1-silenced miRNAs in KP cells (20, 23). Treatment with miR-148a mimics decreased Rab6A and Rab8A levels in human and murine lung cancer cells, whereas miR-182 mimics did not have this effect (Fig. 1B and SI Appendix, Fig. S1C). Treatment with miR-148a mimics suppressed the activity of human and murine Rab6A and Rab8A reporters containing wild-type but not mutant 3′-UTRs lacking predicted miR-148a binding sites (Fig. 1 C and D and SI Appendix, Fig. S1 D and E), and Rab6A levels increased following CRISPR/Cas9-mediated mutation of the miR-148a-binding site in an epithelial KP cell line (307P) (Fig. 1E and SI Appendix, Fig. S1F). Ectopic ZEB1 expression increased Rab6A recruitment to the Golgi and enhanced Rab6A binding to the Rab6A effector bicaudalD2 (Fig. 1 F and G and SI Appendix, Fig. S2 A and B), which are indications of increased Rab6A activity (13). The levels of Rgp1 and Rabin8, which are GEFs for Rab6A and Rab8A, respectively (12, 14), were increased by ectopic ZEB1 expression in an epithelial KP cell (393P_ZEB1) and reduced by short hairpin RNA (shRNA)- or small interfering RNA (siRNA)-mediated ZEB1 depletion in mesenchymal murine and human LUAD cells (344SQ_shZEB1, H1299_siZEB1) (Fig. 1H and SI Appendix, Fig. S2 C–F). In 3′-UTR reporter assays, Rgp1 and Rabin8 were found to be functional miR-148 targets (Fig. 1 I and J and SI Appendix, Fig. S2G and Table S2). In temperature-sensitive mutant vesicular stomatitis virus (VSV-G) assays, ZEB1-dependent VSV-G transport from Golgi to plasma membrane was attenuated by miR-148a mimics or siRNA-mediated depletion of Rab6A or Rab8A (Fig. 1 K and L and SI Appendix, Fig. S3 A and B). Thus, ZEB1 activated exocytotic vesicular transport by relieving Rab6A, Rab8A, and their associated GEFs from miR-148a-mediated silencing.
Fig. 1.
ZEB1 activates exocytotic Rabs by silencing miR-148a. (A) Q-PCR analysis of Rab6A and Rab8A mRNA expression levels in ZEB1 shRNA (shZEB1)- or siRNA (siZEB1)-transfected cells (n = 6 replicates per condition). Control shRNA or siRNA sequences (CTL). (B) WB analysis of Rab6A and Rab8A expressions in cells transfected with miR-148a mimics, miR-182 mimics, or noncoding control (NC). (C and D) Mouse Rab6A (C) and mouse Rab8A (D) 3′-UTR reporter assays. Luciferase activities in 344SQ cells transiently cotransfected with pre-miRs and mouse Rab6A 3′UTR (C) or mouse Rab8A 3′UTR (D) reporters containing wild-type (WT) or mutant 3′-UTRs lacking predicted miR-binding sites (MT). Empty reporters (Vec). Results represent means ± SEM (n = 6 replicates per condition). (E) WB analysis of Rab6A levels in parental 307P cells (P) and mutant 307P cells bearing a deletion in the miR-148a-binding site in the Rab6A 3′-UTR (ΔRab6A-3′-UTR). Mutant clones (#16, #20, #22, #44). P values were determined using two-tailed Student’s t test. (F) The dot plot quantifies Golgi-localized Rab6A per cell (dot) based on % of total Rab6A that colocalizes with Golgi (GM130 channel) (n = 15 cells per group). (G) GTP-bound Rab6A assay. WB analysis of Rab6A associated with BICD2, a Rab6A effector, in BICD2 pull-down samples (PD) or in whole-cell lysates (WCL). (H) Q-PCR analysis of Rgp1 and Rabin8 mRNA levels (n = 6 replicates per condition). (I and J) Mouse Rgp1 (I) and mouse Rabin8 (J) 3′-UTR reporter assays. Luciferase activities in 344SQ cells transiently cotransfected with pre-miRs and mouse Rgp1 3′UTR (I) or mouse Rabin8 3′UTR (J) reporters containing wild-type (WT) or mutant 3′-UTRs lacking predicted miR-binding sites. Empty reporter (Vec). (n = 6 replicates per condition). (K and L) The scatter plots represent the ratio of surface VSV-G to total VSV-G in each cell (dots, n = 15 cells per group) for each condition. P values were determined using two-tailed Student’s t-test.
Exocytotic Rabs Drive LUAD Progression.
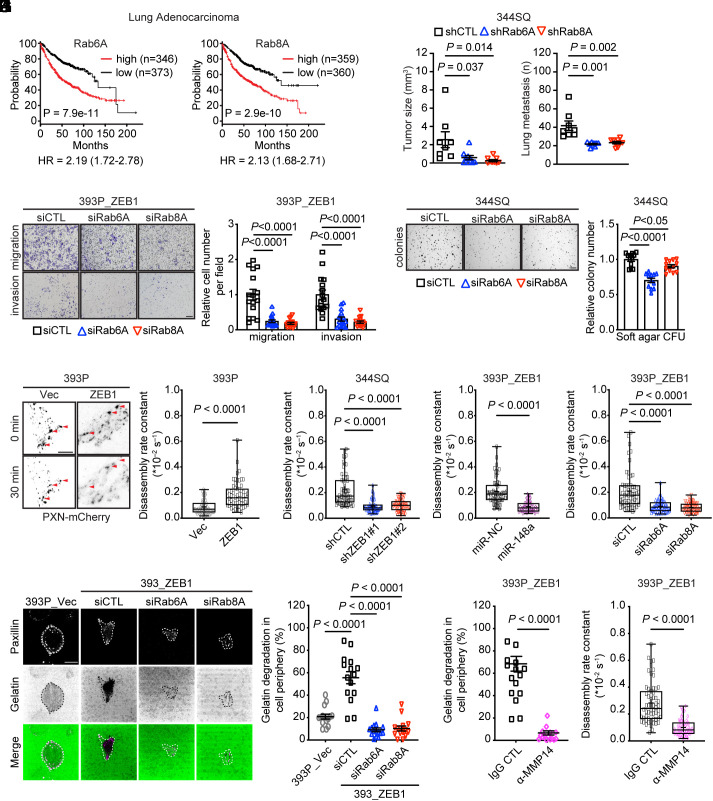
EMT-related gene expression signatures are associated with advanced disease and a worse clinical outcome in LUAD (27). In human LUAD cohorts, high Rab6A and Rab8A mRNA levels are correlated with shorter survival durations (Fig. 2A) and with a z-normalized, 16-gene EMT score (P = 1.05e−12) that we reported (28, 29). In an orthotopic LUAD model generated in immunocompetent mice, shRNA-mediated depletion of Rab6A or Rab8A (SI Appendix, Fig. S4A) reduced primary tumor size and metastases to mediastinal lymph nodes and contralateral lung (Fig. 2B). Because metastasis suppression in the Rab-deficient tumor cohorts may have resulted, in part, from primary tumor growth retardation, we assessed metastatic properties in vitro and found that KP cells deficient in Rab6A or Rab8A had sharply reduced migratory and invasive activities in Boyden chambers (Fig. 2C) and decreased colony formation in soft agar (Fig. 2D), whereas proliferation in monolayer culture was unaffected (SI Appendix, Fig. S4B). FA disassembly rates, which are accelerated in migratory cells (30), were hastened by ectopic ZEB1 expression and retarded by ZEB1 depletion or treatment with miR-148a mimics or siRNAs against Rab6A or Rab8A (Fig. 2 E–H and SI Appendix, Fig. S4 C–F). In line with evidence that matrix metalloproteinase-14 (MMP14) initiates extracellular matrix degradation and FA disassembly (22), we found that surface-exposed MMP14 levels (SI Appendix, Fig. S4G) and gelatin degradation (Fig. 2I) were increased by ectopic ZEB1 expression and reduced by Rab6A or Rab8A depletion, but MMP14 expression levels did not change (SI Appendix, Fig. S4H), suggesting that ZEB1 initiated exocytotic MMP14 transport through a Rab6A/8A-dependent mechanism. Furthermore, MMP14 neutralization retarded ZEB1-dependent gelatin degradation and FA disassembly (Fig. 2 J and K and SI Appendix, Fig. S4 I and J). Thus, Rab6A and Rab8A regulate promigratory FA dynamics to promote LUAD metastasis.
Fig. 2.
Rab6A and Rab8A drive LUAD progression. (A) Kaplan–Meier survival analysis of LUAD patients on the basis of Rab6A and Rab8A mRNA levels above (high) or below (low) the median value. (B) Orthotopic lung tumor size (left dot plot) and numbers of metastases to contralateral lung and mediastinal lymph nodes (right dot plot). Syngeneic, immunocompetent mice were injected with shRNA-transfected 344SQ cells. Control shRNA (shCTL). (C) Bright-field micrographs of crystal violet-stained, siRNA-transfected cells that migrated through Matrigel-coated (invasion) or noncoated (migration) filters in Boyden chambers. (Scale bar, 200 μm.) Replicates (16 fields from four replicates) quantified 16 h after seeding and normalized to siCTL. (D) Colonies formed in soft agarose by siRNA-transfected 344SQ cells were imaged and quantified. (Scale bar, 2 mm.) Replicates (12 fields from four replicates) quantified and normalized to siCTL. (E–H) Representative TIRF micrographs of KP cells subjected to ectopic ZEB1 expression (E, Left). Disassembling FAs in the indicated cells detected by paxillin–mCherry (arrowheads). (Scale bars, 10 μm.) FA disassembly rate constants (dot plots) quantified over the entire 60 min time-lapse sequences in the indicated transfectants (E–H). Each dot represents a single FA (n = 60 FAs per group). P values were determined using two-tailed Student’s t-test. (I) Confocal micrographs of paxillin-CFP-transfected cells seeded on Oregon Green-488–gelatin and imaged 90 min later. Cells outlined (dotted lines, paxillin channel). The absence of green in the merged channel indicates degraded matrix. (Scale bar, 10 μm.) Gelatin degradation per cell (dot) quantified based on the percentage of cell periphery lacking gelatin (dot plot). (J and K) Gelatin degradation (J) and disassembling FAs (K) in the KP cells treated for 16 h with IgG or anti-MMP14 neutralizing antibody.
ZEB1 Activates a Proinvasive Autocrine Loop.
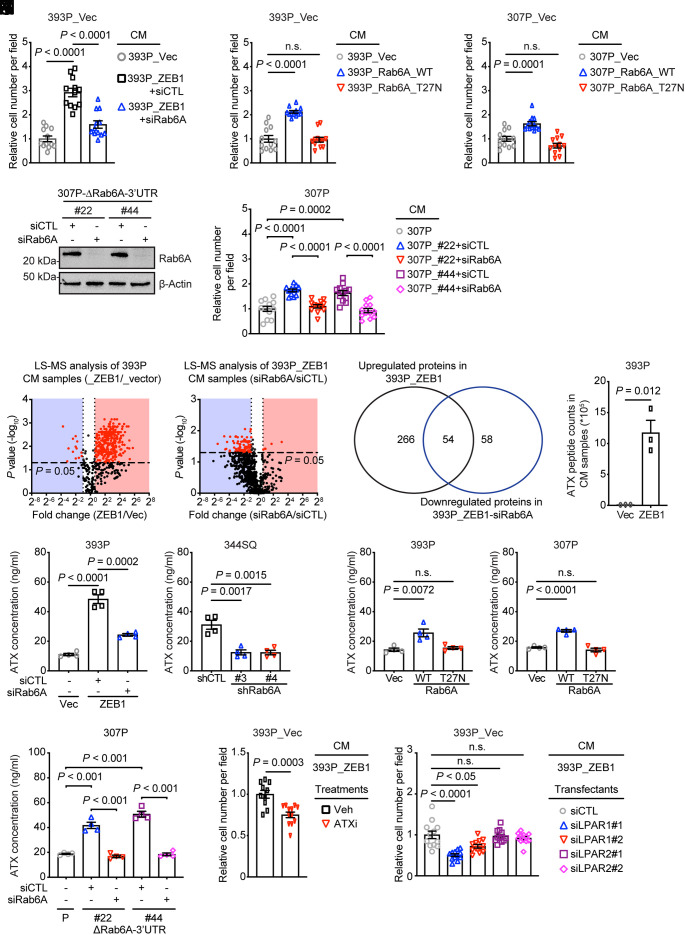
In Boyden chambers, the invasive activity of 393P_vector cells was enhanced by conditioned medium (CM) samples from Rab6A-replete but not deficient 393P_ZEB1 cells (Fig. 3A and SI Appendix, Fig. S5A), epithelial KP cells that ectopically express wild-type but not enzymatically dead Rab6A (Fig. 3 B and C and SI Appendix, Fig. S5 B and C), and Rab6A 3′_UTR-mutant but not wild-type 307P cells (Fig. 3 D and E and SI Appendix, Fig. S5D), suggesting that ZEB1 coordinates Rab6A to activate a proinvasive secretory process. To identify candidate secretory mediators, we carried out liquid chromatography/mass spectrometry analysis on CM samples from 393P_vector cells and Rab6A-deficient or replete 393P_ZEB1 cells and identified 54 proteins that were up-regulated by ectopic ZEB1 expression and down-regulated by Rab6A depletion (Fig. 3 F and G and Datasets S1 and S2), whereas Rab8A depletion decreased the secretion of only 10 proteins (SI Appendix, Fig. S5E and Dataset S3). These findings are in line with evidence that Rab6A serves as a master regulator of post-Golgi secretory vesicle trafficking (10). Rab6A-dependent secreted proteins included multiple ligands, extracellular matrix molecules, and peptidases (Dataset S2). Of particular interest was autotaxin (ATX), a Rab6A-regulated protein that was present at 54-fold higher concentrations in 393P_ZEB1 cells than 393P_vector cells (Fig. 3H). Enzyme-linked immunosorbent assays on CM samples confirmed that ATX levels in CM samples were reduced by Rab6A depletion and enhanced by mutagenesis of the Rab6A 3′-UTR or ectopic expression of ZEB1 or Rab6A (Fig. 3 I–L), whereas ATX expression was not affected (SI Appendix, Fig. S5 F–H), suggesting that ZEB1 activates Rab6A-dependent ATX secretion.
Fig. 3.
ZEB1 activates a proinvasive autocrine loop. (A–C) Quantification of cells that invaded through Matrigel-coated filters in Boyden chambers. Upper chambers loaded with indicated CM samples. Wild-type (WT) or enzymatically dead mutant (T27N) Rab6A. Replicates (12 fields from four replicates) were averaged (bar graph). (D) WB analysis of Rab6A levels in siRNA-transfected 307P cells. CRISPR/Cas-9 mutagenesis carried out to generate a deletion in the miR-148a-binding site in the Rab6A 3′-UTR (ΔRab6A-3′-UTR). (E) Quantification of invasive cells in Matrigel-coated Boyden chambers. Upper chambers contained CM from the indicated cells. Replicates (12 fields from four replicates) were averaged (bar graph). P values were determined using two-tailed Student’s t-test. (F) Volcano plot of proteins identified by LC–MS analysis of CM samples. P values (y axis) and fold-change (x axis). Proteins at significantly different concentrations (red dots, P < 0.05) in 393P_ZEB1 cells and 393P_vector cells (left plot) and in Rab6A-deficient and replete 393P_ZEB1 cells (right plot). (G) Venn diagram illustration of differentially expressed proteins identified in F. (H) ATX peptide counts by LC–MS analysis. (I–L) ELISA of ATX concentrations in CM samples (n = 4 replicates per sample) from KP cells subjected to ectopic ZEB1 expression (I), Rab6A depletion (J), ectopic expression of wild-type (WT) or enzyme-dead mutant (T27N) Rab6A (K), or deletion of the miR-148a binding site in the Rab6A 3′-UTR (ΔRab6A-3′UTR) (L). Parental (P) and mutant clones (#22, #44) (L). (M) Quantification of invasive cells in Matrigel-coated Boyden chambers. Upper chambers contained 393P_ZEB1 cell-derived CM and vehicle or ATX inhibitor (ATXi). (N) Quantification of invasive cells in Matrigel-coated Boyden chambers. Cells were transfected with distinct siRNAs (#1 or #2) against LPAR1 or LPAR2. Upper chambers loaded with CM from 393P_ZEB1 cells. P values were determined using two-tailed Student’s t-test.
ATX converts extracellular lysophospholipids into lysophosphatidic acid (LPA) and stimulates cancer cell invasion through LPA receptors (LPARs) (31–34). To assess ATX enzymatic activity, we quantified LPA in CM samples and found that LPA concentrations were increased by ectopic ZEB1 expression or mutagenesis of the Rab6A-3′-UTR and decreased by siRNA-mediated Rab6A depletion or treatment with an ATX-selective small molecule antagonist (PF-8380) (SI Appendix, Fig. S5 I–K). In Boyden chambers, 393P_vector cell invasive activity was reduced by ATX inhibition in the presence of CM from 393P_ZEB1 cells (Fig. 3M and SI Appendix, Fig. S5L). To determine whether ATX promotes invasion via an autocrine mechanism, we carried out invasion assays on 393P_vector cells that are deficient or replete in the LPA receptor (LPAR)-1 or -2 (SI Appendix, Fig. S5M), which are the most commonly expressed LPAR family members in epithelial cancer cells (33, 34), and found that invasion was inhibited by depletion of LPAR1 (Fig. 3N and SI Appendix, Fig. S5N). Thus, ZEB1 activates an ATX-dependent autocrine loop that promotes LUAD cell invasion.
Secretion Drives Immunosuppression in the TME.
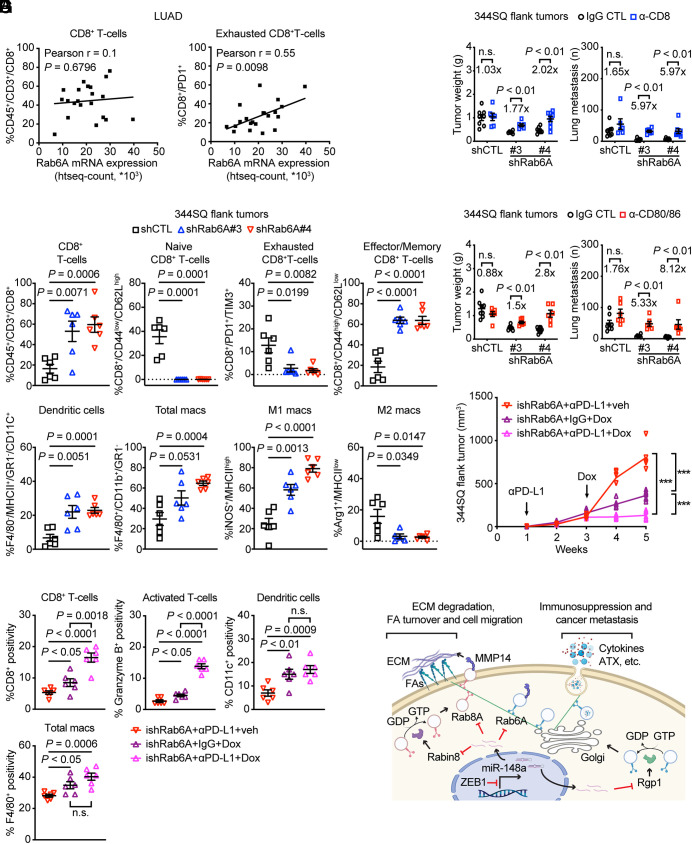
Rab6A mRNA levels were correlated with increased percentages of exhausted, but not total, CD8+ T cells in a human LUAD cohort (Fig. 4A), and multiplexed antibody-based cytokine analysis identified numerous immunomodulatory proteins that were present at higher levels in CM samples from 393P_ZEB1 cells than 393P_vector cells and were diminished by Rab6A depletion (SI Appendix, Fig. S6 A–C), including immunosuppressive cytokines (e.g., G-CSF, GM-CSF, IL-6, IL-10, IP-10, KC, MCP-1, RANTES). Therefore, we reasoned that ZEB1-dependent secretion may create an immunosuppressive TME and addressed this possibility by carrying out flow cytometry on flank tumors generated by Rab6A-deficient or replete KP cells in syngeneic, immunocompetent mice, which were killed 3 wk after injection when tumor sizes were similar in the two cohorts (SI Appendix, Fig. S6D). We found that Rab6A deficiency sharply increased total and effector/memory CD8+ T cells, extinguished naive and exhausted CD8+ T cells, and skewed intratumoral macrophages toward an M1 polarized state (Fig. 4B), suggesting a profound reactivation of antitumor immunity. Relative to the effects of IgG control, an anti-CD8 neutralizing antibody to deplete CD8+ T cells caused a twofold increase in primary tumor size and a fivefold increase in metastasis numbers in mice bearing Rab6A-deficient tumors but had negligible effects on Rab6A-replete tumors (Fig. 4C), suggesting that CD8+ T cell activation contributed to the antitumor effect of Rab6A depletion.
Fig. 4.
A Rab6A-dependent immunosuppressive secretory process. (A) Correlation of Rab6A mRNA levels with percentages of total CD8+ T cells (A, Left) and exhausted CD8+ T cells (A, Right) quantified by flow cytometric analysis in a human LUAD cohort. (B) Flow cytometric analysis of Rab6A-deficient (shRab6A#3 and #4) or replete (shCTL) 344SQ flank tumors isolated from syngeneic, immunocompetent mice (n = 6 tumors per cohort). Percentages of T cells (CD8+ T cells, CD44low/CD62Lhigh naive CD8+ T cells, PD1+/TIM3+ exhausted CD8+ T cells and CD44high/CD62Llow effector/memory CD8+ T cells) and antigen-presenting cells (dendritic cells, total macrophages, M1 macrophages, and M2 macrophages) were quantified. (C and D) Flank tumor weight and lung metastasis numbers in syngeneic, immunocompetent mice (dots) injected with Rab6A-deficient (shRab6A#3 and #4) or replete (shCTL) 344SQ cells and treated with IgG CTL or anti-CD8 (C) or anti-CD80/CD86 antibodies (D). Fold-changes in tumor size and lung metastasis numbers were calculated for each cohort based on the mean values in anti-CD8-, anti-CD80/86, and IgG-treated groups. (E) Subcutaneous tumor growth curves in syngeneic, immunocompetent mice (dots). Cohorts are color-coded. Mean ± SEM. ***P < 0.0005. (F) CD8-, granzyme B-, CD11c-, and F4/80-positive cells in flank tumors generated by injection of 344SQ cells into syngeneic, immunocompetent mice. The percentage of positive cells in each cohort (plot). (G) Schematic illustration of a working model. ZEB1 relieves Rab6A, Rab8A, and their associated GEFs from miR-148a-dependent silencing, thereby accelerating Rab6A- and Rab8A-dependent exocytotic trafficking of vesicles carrying cargos that enhance FA turnover (e.g., MMP14) and establish an immunosuppressive TME (e.g., cytokines and ATX) to promote cancer metastasis.
Given that dendritic cells and macrophages were more numerous in Rab6A-deficient than replete tumors, we reasoned that antigen-presenting cells could activate antitumor immunity through CD80/CD86-mediated engagement of the costimulatory T cell receptor (35). In support of this possibility, treatment of tumor-bearing mice with anti-CD80 and anti-CD86 neutralizing antibodies increased primary tumor size and metastasis numbers to a greater extent in Rab6A-deficient than replete tumors (Fig. 4D).
Because ATX has immunomodulatory functions (36), we carried out studies on murine splenocytes cocultured with syngeneic KP cells in the presence or absence of the ATX inhibitor (PF-8380) and found that ATX antagonism recapitulated the effect of Rab6A-deficient KP cells on naive and exhausted CD8+ T cells, dendritic cells, and M1 and M2 macrophages (SI Appendix, Fig. S7), suggesting that ATX is an immunosuppressive mediator of ZEB1.
Secretory Blockade Abrogates Resistance to Immune Checkpoint Inhibition. Given that EMT underlies acquired resistance to multiple types of targeted therapies, including immune checkpoint inhibition (37, 38), our model predicts that secretory blockade can reverse checkpoint inhibitor resistance. To address this possibility, we utilized mice bearing subcutaneous KP tumors that acquire drug resistance following prolonged exposure to anti-PD-L1 antibody treatment (7) (SI Appendix, Fig. S8A). To initiate secretory blockade at a timepoint that corresponds to the onset of anti-PD-L1 resistance, we injected mice subcutaneously with KP cells that express doxycycline (dox)-inducible shRNAs (ishCTL or ishRab6A) and initiated dox (or vehicle) treatment 2 wk after the first dose of anti-PD-L1 (or IgG) antibody. Tumors were measured weekly and removed at the time of treatment completion to confirm Rab6A depletion in dox-treated mice (SI Appendix, Fig. S8B) and to quantify intratumoral immune cell populations (total CD8+ T cells, granzyme B+ T cells, CD11c+ dendritic cells, and F4/80+ macrophages) by immunohistochemical analysis (SI Appendix, Fig. S8C). Rab6A depletion sharply decreased tumor size (Fig. 4E) and increased intratumoral immune cell infiltration (Fig. 4F). While PD-L1 inhibition alone had no detectable effect, PD-L1 inhibition cooperated with Rab6A depletion to reduce tumor size and increase intratumoral CD8+ T cells and granzyme B+ T cells (Fig. 4 E and F and SI Appendix, Fig. S8D). These findings suggest that secretory blockade mediated by Rab6A depletion reactivates antitumor immunity to abrogate resistance to anti-PD-L1 treatment in this model.
Discussion
To initiate metastasis, cancer cells coordinate biologically disparate cellular functions, including detachment from the substratum and immunosurveillance escape (39). How these disparate biologic activities are coordinated in space and time is a matter of ongoing debate. Based on their circuitry and dynamics, transcription factor regulatory networks are ideally suited to coordinate disparate biologic activities (38). The transcriptomes of EMT-activating transcription factors are particularly broad owing to their unique enrichment in miRs that have functionally diverse target genes, including EMT-activating transcription factors themselves, creating a feedforward loop that drives EMT (24, 40). Here, we show that ZEB1 activates promigratory FA dynamics and a secretory process that drives immunosuppression in the TME by silencing a single miR that targets two post-Golgi exocytotic Rab family members and their associated GEFs (Fig. 4G). Thus, miR-148a is a central node within an EMT-activated transcriptional network that coordinates disparate biologic activities.
Several shortcomings of our study warrant discussion. While our findings are relevant to acquired resistance to immune checkpoint blockade in Kras/p53-mutant LUAD (7, 41, 42), our study did not address other genetic contexts, such as KRAS/LKB1-mutant LUAD, which demonstrates immunologically cold phenotypes (43). Furthermore, we did not address how hypersecretion relates to acquired resistance to immune checkpoint blockade in other tumor types or to EMT-independent mechanisms of acquired resistance (44). Several issues limit the translational relevance of our findings. We did not determine the extent to which ATX mediates resistance to immune checkpoint blockade, and there are currently no clinically available strategies to target Rab6A or ATX. However, building on recent successes in targeting mutant KRAS, strategies to selectively target Rab family members are under consideration (45–47), and selective ATX and LPAR antagonists are at early stages of clinical development (48, 49).
Materials and Methods
Animal Husbandry.
All mouse studies were approved by the Institutional Animal Care and Use Committee at The University of Texas MD Anderson Cancer Center (Houston, Texas). Mice underwent standard care and were killed at predetermined time points or at first signs of morbidity according to the standards set forth by the Institutional Animal Care and Use Committee. To generate orthotopic lung tumors, we intrathoracically injected 344SQ cells (0.5 × 106 cells in 50 µL PBS) phosphate-buffered saline into the left lungs of syngeneic, immunocompetent mice (n = 8 to 10 per cohort) and carried out necropsies 7 to 10 d later to quantify primary tumor size and metastases to mediastinal lymph nodes and contralateral lung as we previously described (50). To generate subcutaneous tumors, we injected 344SQ cells (0.1 × 106 cells in 100 µL PBS for PD-L1 blockade or 0.5 × 106 cells in 100 µL PBS for others) into the right flanks of syngeneic, immunocompetent mice (n = 8 to 10 per cohort) and carried out necropsies 5 to 8 wk later to quantify primary tumor weights and lung metastases on the pleural surfaces as we described (20). For T cell depletion, mice were intraperitoneally pretreated with anti-CD8 antibody (#BE0061, BioXCell; 400 μg per mouse) 1 wk before tumor cell injection. Anti-CD8 antibody (200 μg per mouse) was injected intraperitoneally once weekly beginning 1 d after subcutaneous cancer cell injection as previously described (42). For CD80/86 blockade, 300 μg of anti-CD80 (#BE0024, BioXCell) and 300 μg of anti-CD86 (#BE0025, BioXCell) per mouse were intraperitoneally administered 1 wk before tumor cell injection and then once a week to maintain the blockade as previously described (51). The IgG controls (IgG2a, #BE0089 and IgG2b, #BE0090, BioXCell) were intraperitoneally administered in parallel. For PD-L1 blockade, mice were treated with anti-PD-L1 antibody (#BE0101, BioXCell) or IgG control (IgG2b, #BE0090, BioXCell) (200 μg; intraperitoneally) once a week for 4 wk beginning 7 d after tumor cell injection. Doxycycline in drinking water (2 mg/mL was begun 2 wk after initiating anti-PD-L1 treatment as previously described (7). Tumors were measured weekly.
Reagents.
Murine LUAD cell lines were derived previously (24). We purchased human H1299 cell lines from ATCC; fetal bovine serum (FBS), live-cell imaging solution, PBS, RPMI-1640, Alexa Fluor-tagged secondary antibodies, Oregon Green 488-gelatin, Pierce™ glutathione agarose, paraformaldehyde, bovine serum albumin (BSA), DAPI, and Triton X-100 from Thermo Fisher Scientific; jetPRIME transfection reagent from Polyplus Transfection; puromycin from InvivoGene; transwell and Matrigel-coated Boyden chambers from BD Biosciences; glass-bottom dishes and multiwell plates from MatTek; Trypsin from Corning; qScript cDNA superMix and SYBR Green FastMix from Quanta Biosciences; 10× cell lysis buffer and protease/phosphatase inhibitor cocktail from Cell Signaling Technologies; CCK-8 (#K1018) from APExBIO; ATX inhibitor (PF-8380) (#S8218) from Selleck Chemicals; RNeasy Plus Mini Kit (#74136) and AllStars Negative Control siRNA (#1027281) from QIAGEN; Murine miR-148a mimics (#PM10263) and Murine miR-182 mimics (#PM13088) from Thermo Fisher Scientific; human ZEB1 siRNA #1 (#SASI_Hs01_0011_1599) and #2 (#SASI_ Hs01_0011_1598), murine Rab6A siRNA pool (#EMU084591), murine Rab8A siRNA pool (#EMU038621), murine Rab6A shRNA #3 (#TRCN0000295355) and #4 (#TRCN0000295357), murine Rgp1 siRNA #1 (#SASI_Mm02_00344827) and #2 (#SASI_Mm01_00095501), murine LPAR1 siRNA #1 (#SASI_Mm02_00318340) and #2 (#SASI_Mm01_00021095), LPAR2 siRNA #1 (#SASI_Mm01_00165280) and #2 (#SASI_Mm01_00165282) from Millipore-Sigma; SMARTvector inducible lentiviral control (#VSC11651) and murine shRab6A (#V3SM11253-233022802) from Horizon; primary antibodies against ATX (#ab77104), MMP14 (#ab51074) from Abcam; Rab6A (#9625), Rab8A (#6975), β-actin (#4967), ZEB1 (#3396) from Cell Signaling Technologies; LPAR1 (#NBP1-03363) from Novus Biologicals; LPAR2 (#PA5-102074), Rgp1 (#PA5-20519), Rabin8 (#12321-1-AP) from Thermo Fisher Scientific; Alexa Fluor-488 Mouse anti-GM130 (#560257) from BD Biosciences; VSV-G (#EB0012) from Kerafast; MMP14 neutralizing antibody (#MAB3328) from Millipore-Sigma; purified anti-mouse CD3ε antibody (#100302) and purified anti-mouse CD28 Antibody (#102102) from BioLegend; pmCherry-Paxillin (#50526), CFP-Paxillin (#50510), wild-type Rab6A-EGFP (#49469) and T27N mutant Rab6A-EGFP (#49484) from Addgene; 3′-UTR luciferase reporters of Rab6A (#3835708), Rab8A (#3836408), Rgp1 (#3945708), and Rabin8 (#3834008) from Applied Biological Materials; pRL Renilla-CMV Luciferase Control Reporter (#E2261) and pGL3 Firefly Luciferase Reporter (Basic) (#E1751) from Promega.
Cell Culture.
Murine and human LUAD cell lines were cultured in RPMI-1640 with 10% FBS. Cells were maintained at 37 °C in an incubator with a humidified atmosphere containing 5% CO2. Cells were transfected with vectors using jetPRIME transfection reagent, and stable transfectants were selected for 2 wk using puromycin.
CRISPR/Cas9.
CRISPR-Cas9–mediated Rab6A-3′UTR KO 307P cells were generated in the Cell-Based Assay Screening Service Core Facility (Baylor College of Medicine) using the following guide RNA sequences: 5′- caaagggctgggttagactg and 5′- attacttagaccttctttga.
Live and Fixed Cell Imaging.
Cells were seeded on type I collagen-coated cover glass (#1.5). Live cells were imaged in live-cell imaging media within incubation chambers. For fixed-cell imaging, cells were fixed using 4% paraformaldehyde for 10 min, permeabilized using 0.1% Triton X-100 for 5 min, and blocked with 5% BSA for 30 min. Primary antibody incubation was performed in blocking buffer for 1 h at room temperature or overnight at 4 °C, followed by Alexa Fluor–conjugated secondary antibodies (1:500) in blocking buffer for 1 h at room temperature. Nuclei were counterstained with DAPI, and a cover glass was mounted using Vectashield (#H-1000). Each step was followed by washing 3 times with PBS, which was also used as the solvent in all steps. For surface exposure of MMP14 and VSV-G assay, the cells were not permeabilized.
Microscopy.
Total Internal Reflection Fluorescence (TIRF) imaging was performed on the OMX Blaze V4 SIM Super-Resolution microscope equipped with a TIRF objective. Confocal imaging was performed on an A1+ platform (Nikon Instruments) equipped with 63×/1.4 NA Oil, 100×/1.45 NA Oil, and 20×/0.75 NA Air objectives; 405/488/561 nm laser lines; GaAsP detectors, and Okolab stage top incubator. Images were acquired using NIS-Elements software (Nikon instruments). Routine wide-field fluorescence, bright field, and phase-contrast imaging were performed on an IX71 microscope (Olympus).
Image Processing and Quantitative Analysis.
For fixed cell imaging, the raw images were processed and fluorescent intensity was analyzed in Fiji/ImageJ (https://imagej.nih.gov/ij/download.html). For live cell imaging, the raw images were processed/analyzed in Imaris 9.6 (Bitplane software, Oxford instruments) with MATLAB XTensions and Fiji/ImageJ. Time-lapse videos were analyzed for FA disassembly in Imaris spots tracking module in Surpass mode, and the disassembly rate constant was calculated according to a previous study (30).
qRT-PCR.
Total RNA was isolated from cells using the RNeasy Plus Mini Kit and subjected to reverse transcription and qPCR analysis as described (20, 52). mRNA levels were normalized based on ribosomal protein L32 (Rpl32) mRNA. MicroRNA levels were quantified using stem-loop RT-PCR assays as described (20, 52). Primer sequences are listed in SI Appendix, Table S3.
Luciferase Reporter Assays.
For 3′-UTR activity assays, 344SQ cells were cotransfected with luciferase reporters driven by 3′-UTR sequences (100 ng) or pGL3 Firefly Luciferase Reporter (Basic) (100 ng), pRL Renilla-CMV Luciferase Control Reporter (2 ng) and microRNA mimics or control mimics (50 nM). After 48 h, firefly and Renilla luciferase activities were measured with the Dual-Luciferase Reporter Assay System (Promega).
Mutagenesis.
The Q5 Site-Directed Mutagenesis Kit (#E0554S, NEB) was used to generate mutants according to the manufacturer’s instructions. The primer sequences utilized for mutagenesis are 5′- TTTTATAAATTACTTAGACCTTCT-3′ and 5′- AAAATCAAGAATAATTTTGGACATT-3′ for mouse Rab6A 3′UTR; 5′-TTTCAGAAAATTAGATTAAAGTGC-3′ and 5′- AAACTAGTCAGTGTAAACATGGTA-3′ for mouse Rab8A 3′UTR; 5′-TTTTATAAATTACGGTCTTCTTTG-3′ and 5′-AAAATCAAGAATAATTTTGGACATC-3′ for human Rab6A 3′UTR; 5′-ACATTTATCGATGTAAACAGAGCA-3′ and 5′-TTTTAAGGAAAACAAACAAATTAA-3′ for human Rab8A 3′UTR; 5′-ACTCCTTTGCTTTTATCTAGATATC-3′ and 5′-TTTTAGCAGGGGCCCCCAGGTA-3′ for mouse Rgp1 3′UTR; 5′-AAATAACATGAAGCCAGGATGCTCA-3′ and 5′-TTTAGCATCATCATGGAATAGAA-3′ for mouse Rabin 3′UTR. All primers were purchased from Millipore-Sigma.
Western blotting.
Cells cultured in each well of a 6-well plate at 80% confluency were washed three times with PBS and harvested/resuspended in 150 to 200 μL of 2× Laemmli buffer (Bio-Rad). The cell lysate was boiled for 10 min and loaded onto an SDS gel. After transferring to a nitrocellulose membrane (Bio-Rad), membranes were blocked with 5% milk in TBST buffer and were probed with primary antibodies diluted in 5% BSA in TBST buffer. Horseradish peroxidase (HRP)-conjugated secondary antibodies were used according to the manufacturer’s instructions. Quantitative analysis was performed using Fiji/ImageJ.
Rab6A Activity Assay.
GST-tagged recombinant bicaudal D homolog 2(Bicd2) (#CSB-EP835631MO, Cusabio), a Rab6A effector was incubated with Pierce™ Glutathione Agarose (#16100, Thermo Fisher Scientific) for 1 h at 4 °C with shaking. The slurry was further incubated with total protein extracts from KP cells for 1 h with 10 μM GTP. The active effector complexes were harvested/resuspended in 2× Laemmli buffer and subjected to western blotting analysis.
Gelatin Degradation Assay.
CFP-Paxillin-transfected KP cells were seeded on Oregon Green 488-gelatin-coated glass-bottom dishes for 90 min and then imaged in live-cell imaging media within incubation chambers. Gelatin degradation was determined by a loss of fluorescence intensity relatively to nondegraded matrix, using Fiji/ImageJ (53).
VSV-G Assay.
The VSV-G transport assay was performed as described previously (20, 50). In brief, cells were transiently transfected with EGFP-VSV-G (ts045), transferred to the restrictive temperature of 40 °C for 20 h, and then transferred to the permissive temperature of 32 °C for 1 h in the presence of 100 mg/mL cycloheximide, at which point the cells were fixed. In nonpermeabilized cells, exofacial and total VSV-G were detected by staining with an anti-VSV-G antibody and by measurement of EGFP signal intensity, respectively. VSV-G trafficking to the plasma membrane was measured based on the ratio of exofacial (surface) VSV-G fluorescence signal to the EGFP (total) signal intensity.
Cell Proliferation Assay.
Cells were seeded on 96-well plates at a density of 2 × 103 cells per well on day 0. Relative cell density was measured by using the CCK-8 Counting Kit according to the manufacturer’s instructions. Briefly, cells were incubated with culture medium containing the CCK-8 solution for 2 h at 37 °C, and the absorbance was read at 450 nm.
Cell Migration and Invasion Assays in Boyden Chambers.
As described previously (20), 1 × 105 cells were seeded in the upper wells of Transwell (BD Biosciences) for migration assays or Matrigel-coated Boyden chambers (BD Biosciences) for invasion assays. Cells were allowed to migrate toward 10% FBS in the bottom wells. After 16 h of incubation, migrating or invading cells were stained with 0.1% crystal violet, photographed, and counted. For conditioned medium (CM) transfer experiments, CM samples were isolated, filtered through a 0.45-μm filter, and applied to cells that had been seeded in the upper wells.
Liquid Chromatography/Mass Spectrometry.
The samples were prepared similar to as described (54). Briefly, 25 μg of conditioned media was solubilized with 40 μL 5% SDS, 50 mM TEAB, pH 7.55, and incubated at RT for 30 min. The supernatant containing proteins of interest was then transferred to a new tube and reduced by making the solution 10 mM TCEP (Thermo, #77720) and incubated at 65 °C for 10 min. The sample is then cooled to RT and 3.75 μL 1M iodoacetamide acid added and allowed to react for 20 min in the dark after which 0.5 μL 2M DTT is added to quench the reaction. To the 50 μL protein solution, 5 μL 12% phosphoric acid is added. Then, 350 μL of binding buffer (90% methanol, 100 mM TEAB final; pH 7.1) is added to the solution. The resulting solution is added to the S-Trap spin column (protifi.com) and passed through the column using a bench-top centrifuge (30 s spin at 4,000 g). The spin column is washed with 400 μL binding buffer and centrifuged. This is repeated three times. Trypsin is added to the protein mixture in a ratio of 1:25 in 50 mM TEAB, pH = 8, and incubated at 37 °C for 4 h. Peptides were eluted with 80 μL of 50 mM TEAB, followed by 80 μL 0.2% formic acid, and finally 80 μL 50% acetonitrile and 0.2% formic acid. The combined peptide solution is then dried in a speed vac and resuspended in 2% acetonitrile, 0.1% formic acid, and 97.9% water and placed in an autosampler vial.
Peptide mixtures were analyzed by nanoflow liquid chromatography–tandem mass spectrometry (nanoLC–MS/MS) using a nano-LC chromatography system (UltiMate 3,000 RSLCnano, Dionex), coupled online to a Thermo Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific, San Jose, CA) through a nanospray ion source (Thermo Scientific). A trap-and-elute method was used. All LC–MS/MS data were acquired using XCalibur, version 2.5 (Thermo Fisher Scientific) in positive ion mode using a top-speed data-dependent acquisition method with a 3-s cycle time. Tandem mass spectra were extracted and charge state deconvoluted by Proteome Discoverer (Thermo Fisher, version 2.2.0388). Deisotoping is not performed. All MS/MS spectra were searched against a UniProt mouse database (version 04-04-2018) using Sequest. Searches were performed with a parent ion tolerance of 5 ppm and a fragment ion tolerance of 0.60 Da. Trypsin is specified as the enzyme, allowing for two missed cleavages. Fixed modification of carbamidomethyl (C) and variable modifications of oxidation (M) and deamidation were specified in Sequest.
Scaffold (version Scaffold_4.11.1, Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (55). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters.
ATX and LPA Measurement.
The Autotaxin Sandwich ELISA Kit (#K-5600, Echelon Biosciences) and General Lysophosphatidic Acid (LPA) ELISA Kit (#MBS2700658, MyBioSource) were used to measure the ATX and LPA concentrations in the conditioned medium samples, respectively, according to the manufacturer’s instructions.
Immunology Multiplex Assay.
As described previously (50), CM samples were collected and frozen until analysis. Each sample was thawed, prepared, and analyzed using a multiplex magnetic bead-based assay (Luminex 200 System, Luminex, and Multiplex Analysis 5.1 software, Millipore-Sigma) to quantify cytokine concentrations in CM samples.
Immune Cell Profiling in Tumor Tissues.
As described previously (50), 3.5 wk after subcutaneous injection of 1 × 106 344SQ_shCTL or 344SQ_shRab6A into the flank of wild-type mice, subcutaneous tumors were processed using the MACS Miltenyl Biotec tumor dissociation kit. Digestion was performed using collagenase I (3 mg/mL) and dispase II (4 mg/mL). Spleens were processed by grinding tissues using a 40-micron nylon filter. After single-cell suspensions were obtained, RBCs were lysed using 1 × RBC lysis buffer (BioLegend) following manufacturer instructions. Single-cell suspension was stained according to standard protocols with the following antibodies: CD3-PE-594 (#100246, BioLegend), CD45-Pacific Blue (#103126, BioLegend), CD4-APCCy7 (#100526, BioLegend), CD8-PE-Cy7 (#100721, BioLegend), CD278-PE (#117406, BioLegend), CD25-BUV395 (#564022, BD Biosciences), PD1-BV-505 (#135220, BioLegend), CD62L-FITC (#35-0621-U500, Thermo Fisher Scientific), CD44-BV-711 (#103057, BioLegend), TIM3-APC (#134007, BioLegend), FoxP3-PerCP-Cy5.5 (#45-5773-82, Invitrogen), GR1-BV-711 (#106443, BioLegend), CD11b-BV-650 (#101239, BioLegend), CD11c-BV-785 (#117335, BioLegend), F4/80-APC (#204801-U100, Tonbo), PDL1-PE-Dazzle-594 (#124323, BioLegend), CD86-APC-Cy7 (#105030, BioLegend), MCHII-PE-Cy7 (#107629, BioLegend), CD80-BV-605 (#104729, BioLegend), CD68-PerCP-Cy5.5 (#137009, BioLegend), iNOS-PE (#125920-80, Invitrogen), Arg1-FITC (IC5868F, R&D Systems), CD31-BV-786 (#740870, BD Biosciences), and the live/dead cell marker Ghost Violet-BV-510 (#10-0870-T100, VWR). For intracellular staining, cells were fixed and permeabilized using the intracellular staining perm wash buffer (BioLegend) according to manufacturer instructions. Data were acquired on a Fortessa X20 analyzer (BD Biosciences) and analyzed using FlowJo software (version 7.6; Tree Star).
Analysis of Kaplan–Meier Survival Data.
NSCLC patient survival data were downloaded from the Kaplan–Meier plotter database (56). Analysis of NSCLC patients was performed in Rab6A or Rab8A high- and low-expression cohorts. The P value was calculated using the log-rank test (56).
Clinical Datasets.
All study protocols were approved by the Institutional Review Board at The University of Texas MD Anderson Cancer Center (Houston, Texas). All subjects provided informed consent according to the standards set forth by the Institutional Review Board. Available data from the Immune Genomic Profiling of NSCLC (ICON) cohort (MD Anderson Cancer Center [MDACC] PA15-1112) were evaluated. The correlation of Rab6A mRNA expression and intratumoral immune cells was analyzed using the following platforms: 1) differential gene expression using RNA microarray and 2) tumor-infiltrating lymphocyte (TIL) immune profiling using flow cytometry (57, 58).
Immunohistochemistry.
All mouse tumors were fixed in 10% formalin and subjected to immunohistochemistry by MDACC DVMS Veterinary Pathology Services using the Leica Bond three automatic immuno stainer (Leica Biosystem). Primary antibodies for CD8 (1:400, #98941), CD11c (1:150, #97585), F4/80 (1:600, #70076), and granzyme B (1:100, #44153) from Cell Signaling Technologies were validated in the mouse lymph nodes as positive controls, and omission of the primary antibody was used as a negative control. All slides were scanned using an Aperio Digital Pathology Slide Scanner (Aperio AT2 DX System, Leica Biosystem). Immunohistochemistry images were analyzed by Aperio ImageScope (v12.4.6, Leica Biosystem) using the Positive Pixel Count v9 algorithm for the quantification of positivity.
Statistics.
Unless mentioned otherwise, the results shown are representative of replicated experiments and are the means ± SEM from triplicate samples or randomly chosen cells within a microscopic field. Statistical evaluations were carried out with Prism 9 (GraphPad Software, Inc.). Unpaired two-tailed Student t-tests or ANOVA was used to compare means for two or more groups, respectively, and P values < 0.05 were considered statistically significant.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Acknowledgments
This work was supported by the (NIH) through R01 CA181184 (to J.M.K.), R01 CA2111125 (to J.M.K.), K99 CA249048 (to G.-Y.X), NIH Lung Cancer SPORE grant P50 CA70907 (to J.M.K.) and by CPRIT-MIRA RP160652. J.M.K. holds the Gloria Lupton Tennison Distinguished Professorship in Lung Cancer. The work was also supported by the generous philanthropic contributions to The University of Texas MD Anderson Lung Cancer Moon Shots Program, the Gil and Dody Weaver Foundation and Bill and Katie Weaver Charitable Trust, and the MD Anderson Cancer Center Support Grant P30 CA016672. This study was supported by the Translational Molecular Pathology-Immunoprofiling Moonshots Platform (TMP-IL) at the Department of Translational Molecular Pathology, the University of Texas MD Anderson Cancer Center.
Author contributions
G.-Y.X. designed research; G.-Y.X., X.T., B.L.R., X.L., J.Y., M.E.V., J.X., W.K.R., C.H., Y.L., J.Z., L.S., and I.I.W. performed research; G.-Y.X. and J.X. contributed new reagents/analytic tools; G.-Y.X., X.T., B.L.R., D.L.G., S.W., C.W., M.E.V., H.T.T., and W.K.R. analyzed data; J.M.K. conceived and supervised the project and contributed to the design and interpretation of all experiments; and G.-Y.X. and J.M.K. wrote the paper.
Competing interests
J.M.K. has received consulting fees from Halozyme.
Footnotes
This article is a PNAS Direct Submission. D.A.T. is a guest editor invited by the Editorial Board.
Contributor Information
Guan-Yu Xiao, Email: Guan-Yu.Xiao@uky.edu.
Jonathan M. Kurie, Email: jkurie@mdanderson.org.
Data, Materials, and Software Availability
All study data are included in the article and/or supporting information.
Supporting Information
References
- 1.Valkenburg K. C., de Groot A. E., Pienta K. J., Targeting the tumour stroma to improve cancer therapy. Nat. Rev. Clin. Oncol. 15, 366–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pardoll D. M., The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tumeh P. C., et al. , PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koyama S., et al. , Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat. Commun. 7, 10501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma P., Hu-Lieskovan S., Wargo J. A., Ribas A., Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wellenstein M. D., de Visser K. E., Cancer-cell-intrinsic mechanisms shaping the tumor immune landscape. Immunity 48, 399–416 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Chen L., et al. , CD38-mediated immunosuppression as a mechanism of tumor cell escape from PD-1/PD-L1 blockade. Cancer Discov. 8, 1156–1175 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J., Stanger B. Z., The tumor as organizer model. Science 363, 1038–1039 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Stalder D., Gershlick D. C., Direct trafficking pathways from the Golgi apparatus to the plasma membrane. Semin. Cell Dev. Biol. 107, 112–125 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grigoriev I., et al. , Rab6, Rab8, and MICAL3 cooperate in controlling docking and fusion of exocytotic carriers. Curr. Biol. 21, 967–974 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Grigoriev I., et al. , Rab6 regulates transport and targeting of exocytotic carriers. Dev. Cell 13, 305–314 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Pusapati G. V., Luchetti G., Pfeffer S. R., Ric1-Rgp1 complex is a guanine nucleotide exchange factor for the late Golgi Rab6A GTPase and an effector of the medial Golgi Rab33B GTPase. J. Biol. Chem. 287, 42129–42137 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergbrede T., et al. , Biophysical analysis of the interaction of Rab6a GTPase with its effector domains. J. Biol. Chem. 284, 2628–2635 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Hattula K., Furuhjelm J., Arffman A., Peranen J., A Rab8-specific GDP/GTP exchange factor is involved in actin remodeling and polarized membrane transport. Mol. Biol. Cell 13, 3268–3280 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makowski M., Archer K. J., Generalized monotone incremental forward stagewise method for modeling count data: Application predicting micronuclei frequency. Cancer Inform. 14, 97–105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pang H., Ebisu K., Watanabe E., Sue L. Y., Tong T., Analysing breast cancer microarrays from african americans using shrinkage-based discriminant analysis. Hum. Genomics 5, 5–16 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kakiuchi S., et al. , Prediction of sensitivity of advanced non-small cell lung cancers to gefitinib (Iressa, ZD1839). Hum. Mol. Genet. 13, 3029–3043 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-Boulan E., Macara I. G., Organization and execution of the epithelial polarity programme. Nat. Rev. Mol. Cell Biol. 15, 225–242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brabletz T., EMT and MET in metastasis: Where are the cancer stem cells? Cancer Cell 22, 699–701 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Tan X., et al. , Epithelial-to-mesenchymal transition drives a pro-metastatic Golgi compaction process through scaffolding protein PAQR11. J. Clin. Invest. 127, 117–131 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banerjee P., et al. , The EMT activator ZEB1 accelerates endosomal trafficking to establish a polarity axis in lung adenocarcinoma cells. Nat. Commun. 12, 6354 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bravo-Cordero J. J., et al. , A novel high-content analysis tool reveals Rab8-driven cytoskeletal reorganization through Rho GTPases, calpain and MT1-MMP. J. Cell Sci. 129, 1734–1749 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Ahn Y. H., et al. , ZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression. J Clin. Invest. 122, 3170–3183 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibbons D. L., et al. , Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes. Dev. 23, 2140–2151 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stenmark H., Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Jin H., et al. , Rab GTPases: Central coordinators of membrane trafficking in cancer. Front. Cell Dev. Biol. 9, 648384 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larsen J. E., et al. , ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J. Clin. Invest. 126, 3219–3235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibbons D. L., Creighton C. J., Pan-cancer survey of epithelial-mesenchymal transition markers across the cancer genome atlas. Dev. Dyn. 247, 555–564 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee J. M., Dedhar S., Kalluri R., Thompson E. W., The epithelial-mesenchymal transition: New insights in signaling, development, and disease. J. Cell Biol. 172, 973–981 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stehbens S. J., et al. , CLASPs link focal-adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nat. Cell Biol. 16, 561–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu S., Murph M., Panupinthu N., Mills G. B., ATX-LPA receptor axis in inflammation and cancer. Cell Cycle 8, 3695–3701 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aiello S., Casiraghi F., Lysophosphatidic acid: Promoter of cancer progression and of tumor microenvironment development. A promising target for anticancer therapies? Cells 10, 1390 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato K., et al. , Opposite roles of LPA1 and LPA3 on cell motile and invasive activities of pancreatic cancer cells. Tumour. Biol. 33, 1739–1744 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Yu S., et al. , Lysophosphatidic acid receptors determine tumorigenicity and aggressiveness of ovarian cancer cells. J. Natl. Cancer Inst. 100, 1630–1642 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greenwald R. J., Freeman G. J., Sharpe A. H., The B7 family revisited. Annu. Rev. Immunol. 23, 515–548 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Knowlden S., Georas S. N., The autotaxin-LPA axis emerges as a novel regulator of lymphocyte homing and inflammation. J. Immunol. 192, 851–857 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chong C. R., Janne P. A., The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat. Med. 19, 1389–1400 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu W., Kang Y., Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev. Cell 49, 361–374 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiss F., Lauffenburger D., Friedl P., Towards targeting of shared mechanisms of cancer metastasis and therapy resistance. Nat. Rev. Cancer 22, 157–173 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y., Yang P., Wang X. F., Microenvironmental regulation of cancer metastasis by miRNAs. Trends Cell Biol. 24, 153–160 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L., et al. , Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 5, 5241 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen L., et al. , Growth and metastasis of lung adenocarcinoma is potentiated by BMP4-mediated immunosuppression. Oncoimmunology 5, e1234570 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skoulidis F., et al. , STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 8, 822–835 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoenfeld A. J., Hellmann M. D., Acquired resistance to immune checkpoint inhibitors. Cancer Cell 37, 443–455 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marchwicka A., Kaminska D., Monirialamdari M., Blazewska K. M., Gendaszewska-Darmach E., Protein prenyltransferases and their inhibitors: Structural and functional characterization. Int. J. Mol. Sci. 23, 5424 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malolepsza J., et al. , Rational design, optimization, and biological evaluation of novel alpha-Phosphonopropionic acids as covalent inhibitors of Rab geranylgeranyl transferase. J. Enzyme. Inhib. Med. Chem. 37, 940–951 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gendaszewska-Darmach E., Garstka M. A., Blazewska K. M., Targeting small GTPases and their prenylation in diabetes mellitus. J. Med. Chem. 64, 9677–9710 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zulfikar S., Mulholland S., Adamali H., Barratt S. L., Inhibitors of the autotaxin-lysophosphatidic acid axis and their potential in the treatment of interstitial lung disease: Current perspectives. Clin. Pharmacol. 12, 97–108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin Y. H., Lin Y. C., Chen C. C., Lysophosphatidic acid receptor antagonists and cancer: The current trends, clinical implications, and trials. Cells 10, 1629 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan X., et al. , PI4KIIIbeta is a therapeutic target in chromosome 1q-amplified lung adenocarcinoma. Sci. Transl. Med. 12, eaax3772 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kamphorst A. O., et al. , Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science 355, 1423–1427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan X., et al. , The epithelial-to-mesenchymal transition activator ZEB1 initiates a prometastatic competing endogenous RNA network. J. Clin. Invest. 128, 1267–1282 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wiesner C., El Azzouzi K., Linder S., A specific subset of RabGTPases controls cell surface exposure of MT1-MMP, extracellular matrix degradation and three-dimensional invasion of macrophages. J. Cell Sci. 126, 2820–2833 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Baillat D., Russell W. K., Wagner E. J., CRISPR-Cas9 mediated genetic engineering for the purification of the endogenous integrator complex from mammalian cells. Protein Expr. Purif. 128, 101–108 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Nesvizhskii A. I., Keller A., Kolker E., Aebersold R., A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 (2003). [DOI] [PubMed] [Google Scholar]
- 56.Gyorffy B., Surowiak P., Budczies J., Lanczky A., Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 8, e82241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Federico L., et al. , Distinct tumor-infiltrating lymphocyte landscapes are associated with clinical outcomes in localized non-small-cell lung cancer. Ann. Oncol. 33, 42–56 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmidt S. T., et al. , Shared nearest neighbors approach and interactive browser for network analysis of a comprehensive non-small-cell lung cancer data set. JCO Clin. Cancer Inform. 6, e2200040 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Data Availability Statement
All study data are included in the article and/or supporting information.