Significance
In plants, MP (MONOPTEROS)-mediated auxin signaling is essential for organ initiation. We suggest a molecular framework for auxin in the robustness control of organ initiation in the meristem. We demonstrate that MP interacts with DRNL (DORNROSCHEN-LIKE) to trigger organ initiation by limiting cytokinin accumulation and activating AINTEGUMENTA, AINTEGUMENTA-LIKE6, TARGET OF MONOPTEROS 3, and FILAMENTOUS FLOWERS expression. Although DRNL and its paralog DRN are not coexpressed, they act redundantly during organ initiation. We show that in drnl mutants, DRN transcripts are ectopically activated in organ initiation sites to compensate for the functional deficiency of drnl in organ initiation. Our work suggests that a spatial gene compensation–based safety strategy in auxin signaling participates to the genetic robustness control of organ initiation.
Keywords: auxin, organ initiation, DORNROSCHEN-LIKE, spatial gene compensation, robustness control
Abstract
Auxin signaling is essential for organ initiation in plants. How genetic robustness controls auxin output during organ initiation is largely unknown. Here, we identified DORNROSCHEN-LIKE (DRNL) as a target of MONOPTEROS (MP) that plays essential roles in organ initiation. We demonstrate that MP physically interacts with DRNL to inhibit cytokinin accumulation by directly activating ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN 6 and CYTOKININ OXIDASE 6. DRN, the paralogous gene of DRNL, acts redundantly with DRNL but is not coexpressed with DRNL in the organ founder cells in which DRNL is expressed. We demonstrate that DRNL directly inhibits DRN expression in the peripheral zone, whereas DRN transcripts are ectopically activated in drnl mutants and fully restore the functional deficiency of drnl in organ initiation. Our results provide a mechanistic framework for the robust control of auxin signaling in organ initiation through paralogous gene-triggered spatial gene compensation effects.
Unlike animals, plants have the ability to develop new organs nearly throughout their entire lifespan. This ensures that sessile plants can survive various external pressures, such as environmental stresses, diseases, and predators. This vital ability depends on the activity of populations of stem cells in meristems, which have the capacity to self-renew as well as to give rise to daughter cells for lateral organ formation in both the shoot and root (1–4). The generation of the aboveground organs of higher plants depends on the maintenance and continuous differentiation of the shoot apical meristem (SAM), which is controlled by a complex regulatory network of signaling molecules (1, 3, 5, 6). Among these, phytohormones account for a large proportion, including auxin and cytokinin, which have been shown to be essential for SAM regulation (7–13). Cytokinin is enriched in the central zone (CZ), which harbors stem cells, and acts synergistically with auxin to maintain stem cell fate by activating WUSCHEL expression (8, 14, 15).
Auxin mainly accumulates in the differentiated peripheral zone (PZ), functioning in lateral organ initiation (7, 9, 16). The position and timing of organ initiation depends on the local accumulation of auxin maximum in the PZ by PINFORMED1 (PIN1)-mediated polar auxin transport (7, 16–19). Consistently, the loss-of-function mutants of pin1 or pinoid (pid) fail to initiate organs during the reproductive stage (20, 21). In auxin signaling, AUXIN RESPONSE FACTOR5 (ARF5)/MONOPTEROS (MP) has been shown to be a key transcription factor that relays auxin signals during organ initiation, whose mutation also shows “pin-like” inflorescence as pin1 and pid (8, 22). Given this critical role, multiple targets have been shown to be under direct control by MP during organ initiation, including LEAFY (LFY), AINTEGUMENTA (ANT), ANT-LIKE6 (AIL6), TARGET OF MONOPTEROS 3 (TMO3), and FILAMENTOUS FLOWERS (FIL) (10, 23). However, how MP-mediated auxin signaling robustly controls organ initiation remains poorly understood. Here, we identified DORNROSCHEN-LIKE (DRNL) as a direct target of MP in the PZ. DRNL interacts with MP and forms a complex mediating cytokinin–auxin cross talk during lateral organ initiation. We demonstrate that most known MP targets during organ initiation are also under direct control by DRNL. Although we observed functional redundancy between DRNL and its paralog DORNROSCHEN (DRN), DRN is not coexpressed with DRNL in organ founder cells. Surprisingly, we observed that DRN transcripts that were originally located in the CZ were ectopically activated in organ founder cells in the drnl mutant and fully restored the functional deficiency of drnl during organ initiation. We further demonstrate that DRN expression in the PZ is under direct negative control by DRNL and that DRNL-triggered spatial paralogous gene compensation mediates the robust control of auxin signaling during organ initiation.
Results
DRNL Acts Downstream of Auxin in Lateral Organ Initiation.
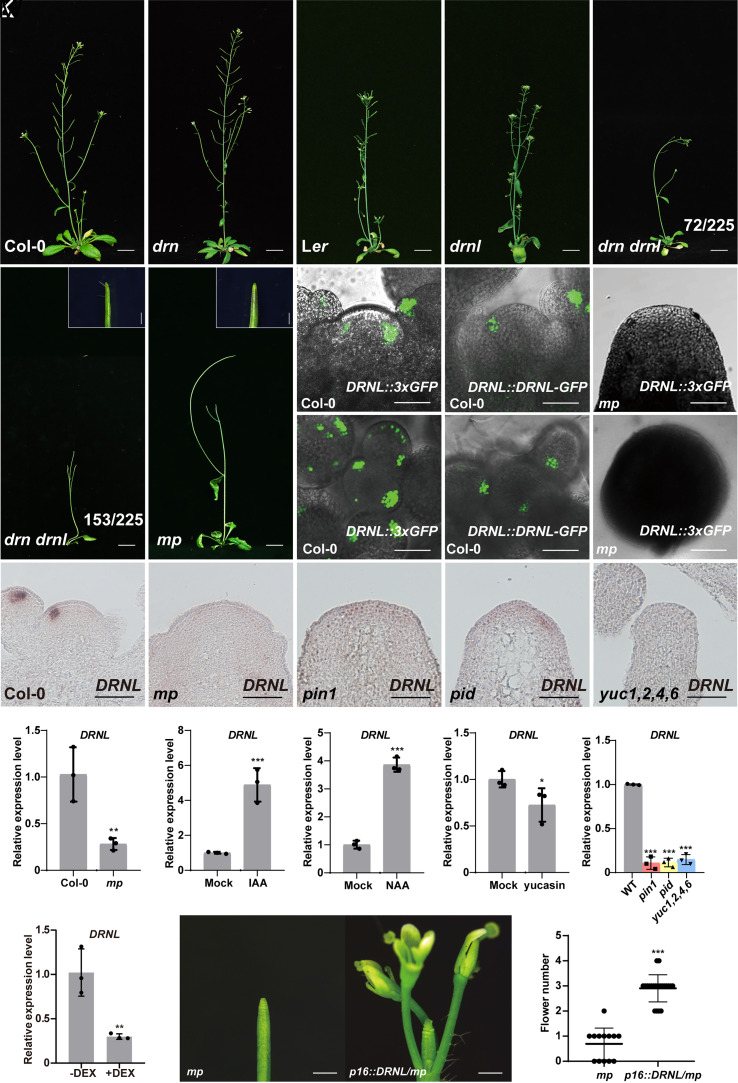
DRN and its paralogous gene DRNL are previously shown to be involved in the regulation of the plant stem-cell pool under the direct control of auxin signaling (24). Interestingly, the mutation in LEAFLESS (LFS), the single ortholog of DRN and DRNL in tomato that can be induced by auxin, shows “pin-like” shoots (25). Consistently, in Arabidopsis, we observed that the drn drnl double mutant also showed severe defects in lateral organ initiation with 68% (153 out of 225) of the double mutant eventually forming a “pin-like” inflorescence (Fig. 1 A–F and SI Appendix, Fig. S1 A–F). We further quantified the silique numbers in the main inflorescence of double mutants and observed a significantly reduced number in the double mutant compared to either single mutants or wild-type plants (SI Appendix, Fig. S2), suggesting that DRN and DRNL act redundantly in controlling lateral organ initiation in the SAM. Conversely, overexpression of DRNL caused a large increase in the number of siliques (SI Appendix, Fig. S3). Although DRN is not expressed in the PZ where lateral organs are initiated in wild-type plants (24, 26, 27), we did find that DRNL was expressed in the outer PZ where organs were initiated (Fig. 1N and SI Appendix, Fig. S4), as previously shown (24, 26–29), which was further confirmed by DRNL::3×GFP transgenic plants (Fig. 1 H and K). Furthermore, we generated DRNL::DRNL-GFP transgenic plants with the upstream sequences of DRNL that were previously reported (30) and observed that DRNL proteins were specifically expressed during organ initiation (Fig. 1 I and L).
Fig. 1.
DRNL acts downstream of auxin signaling in lateral organ initiation. (A–G) Six-week-old Col-0 (A), drn (B), Ler (C), drnl (D), mp (G), and drn drnl (E and F) plants. The number of drn drnl mutants with different phenotypes is indicated (E and F). (Scale bars, 2 cm.) Magnification of the “pin-like” inflorescence of drn drnl (F) and mp (G) is shown in the upper right corner with scale bars 1 mm. (H–M) DRNL expression patterns in the SAM were detected by DRNL::3×GFP in Col-0 (H and K) and mp (J and M) plants. DRNL protein distribution patterns in DRNL::DRNL-GFP transgenic plants (I and L). (K–M) show the Top view of (H–J); n ≥ 10 shoot apexes per genotype were observed with similar results. (Scale bars, 50 μm.) (N–R) DRNL expression patterns in the SAM were detected by RNA in situ hybridization in Col-0 (N), mp (O), pin1 (P), pid (Q) and yuc1,2,4,6 (R) plants. n ≥ 12 shoot apexes per genotype were observed with similar results. (Scale bars, 50 μm.) (S–W) DRNL expression levels in the SAM of mp (S), pin1pid and yuc1,2,4,6 (W) mutants, and IAA (T), NAA (U), yucasin (V) treatments measured by qRT–PCR. The data are shown as mean ± SD; n = 3 biological replicates, two-tailed Student’s t tests, *P < 0.05, **P < 0.01, ***P < 0.001. (X) The expression levels of DRNL in the SAM of RPS5A::GR-bdl transgenic plants with or without DEX induction in the presence of cycloheximide. The data are shown as mean ± SD; n = 3 biological replicates, two-tailed Student’s t tests, **P < 0.01. (Y) The “pin-like” inflorescence of mp is partially rescued by overexpressing DRNL under the p16 promoter. (Scale bars, 1 mm.) (Z) Quantification of flower numbers of (Y). mp plants (n = 13); p16::DRNL/mp plants (n = 21), two-tailed Student’s t tests, ***P < 0.001.
DRN, a paralogous gene of DRNL, has been shown to be under direct control of the key transcription factor ARF5/MP in auxin signaling (24, 31), and the drn drnl double mutant showed “pin-like” inflorescence phenocopying the weak allele of mp-S319 (8) (Fig. 1G and SI Appendix, Fig. S1G). We therefore tested whether DRNL expression was regulated by MP-mediated auxin signaling. We crossed the DRNL::3×GFP transgenic plants with the mp-S319 mutant and observed a dramatic decrease in DRNL expression in the mp mutant (Fig. 1 H, K, J, and M), which was further confirmed using in situ hybridization (Fig. 1 N and O) and quantitative reverse transcription PCR (qRT–PCR) on wild-type and mp mutant plants (Fig. 1S). This suggested that DRNL is under positive control by MP. Support for this idea came from the observation that the transcript and protein accumulation domains of DRNL overlapped with MP in the PZ during organ initiation (SI Appendix, Fig. S5), and two auxin response elements (AuxREs) have been shown to be necessary for DRNL expression in the SAM (28). Consistent with the early observation that DRNL is regulated by auxin (32), we observed that DRNL transcripts were significantly increased after treatment with indole-3-acetic acid (IAA) and naphthalene acetic acid (NAA) for 2 h (Fig. 1 T and U).
To determine whether endogenous auxin contributes to the regulation of DRNL expression, we analyzed the yuc1 yuc2 yuc4 yuc6 quadruple mutant lacking essential auxin biosynthesis genes (33) and observed a dramatic decrease in DRNL transcripts in the quadruple mutant (Fig. 1 N, R, and W). Consistently, using the chemical treatment of yucasin to reduce the endogenous auxin levels by inhibiting the expression of YUCCA genes (34), we observed that the expression of DRNL decreased significantly (Fig. 1V). The local accumulation of auxin in the PZ is essential for new organ initiation, which is achieved by auxin polar transport. In pinformed1 (pin1) and pinoid (pid) mutants with compromised auxin polar transport, we observed a dramatic decrease in DRNL expression (Fig. 1 N, P, Q, and W). To avoid effects on expression from morphological defects in pin1 and pid that fail to initiate organs during the reproductive stage, we treated plants with the auxin transport inhibitor N-1-naphthylphthalamic acid (NPA). DRNL expression was decreased just 1 d after treatment and showed continued declines with increasing treatment time (SI Appendix, Fig. S6). Because the first visible phenotypes occurred only 5 d after treatment with NPA (8), we concluded that the reduction in DRNL transcripts was directly caused by the loss of local auxin accumulation in the SAM.
Given that DRNL expression was significantly elevated upon auxin treatment, we then tested whether DRNL expression in the SAM was under direct control by auxin response factors. To this end, we performed dexamethasone (DEX) induction on RPS5A::GR-bdl transgenic plants, in which mutated bdl (BODENLOS) proteins inactivated several ARFs, including MP, by direct binding, which cannot be degraded by intracellular auxin (35). We observed that DRNL expression drastically decreased upon DEX induction accompanied by treatment with the protein biosynthesis inhibitor cycloheximide (Fig. 1X). As DRNL expression was decreased in the mp mutant, DRNL is likely under direct positive control by MP. To test the interaction between MP and DRNL genetically, we expressed DRNL from the p16 promoter (36) in mp mutants, whose promoter has been shown to be highly active in the SAM. Concomitant with a repression of DRNL in the mp mutant, we observed that overexpressed DRNL partially rescued the primordium initiation defects in mp mutants (Fig. 1 Y and Z), demonstrating that activation of DRNL transcription is a relevant aspect of MP functions in priming lateral organ initiation.
Cytokinin Signaling Was Disturbed in the drn drnl Mutant.
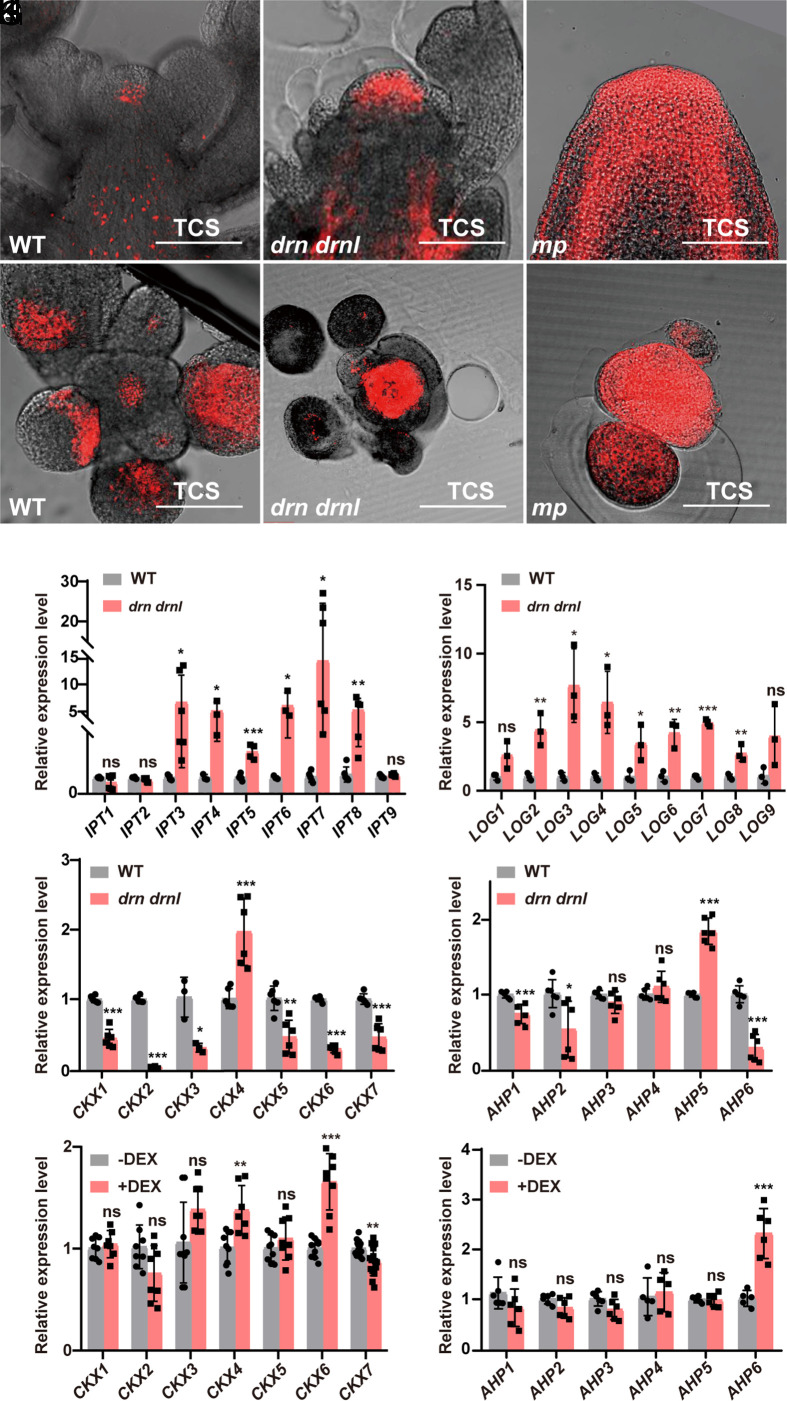
A-type ARABIDOPSIS RESPONSE REGULATORs (ARRs) are primary response genes that can be rapidly induced by cytokinin (37). Previously, the expression of three ARRs, including ARR4, ARR5, and ARR6, was shown to be reduced in 35S::DRNL transgenic plants (38). As drn drnl double mutants showed severe defects in organ initiation, it raises the possibility that DRNL was involved in the regulation of cytokinin signaling in the PZ that contributed to lateral organ formation. To test this hypothesis, we examined the activity of cytokinin signaling in the SAM by introducing the two-component-output sensor (TCS)::dTomato into drn drnl mutant plants. We observed extremely enlarged fluorescence signals of TCS::dTomato in almost the entire meristem of drn drnl plants (Fig. 2 A, B, D, and E), which was consistent with the early observation in tomato lfs mutants (25). This demonstrated that DRNL/DRN negatively regulates cytokinin signaling in the SAM. Given that DRNL was under direct positive control by MP-mediated auxin signaling (Fig. 1), we observed an even stronger accumulation of cytokinin in the SAM of mp mutants (Fig. 2 A, C, D, and F), suggesting that the MP-DRNL/DRN module mediates cytokinin–auxin cross talk during lateral organ initiation.
Fig. 2.
Cytokinin signaling is disrupted in drn drnl and mp mutants. (A–F) Cytokinin signaling detected by TCS::dTomato in the inflorescence apexes of Col-0 (A and D), drn drnl (B and E), and mp (C and F) plants with longitudinal (A–C) and transverse sections (D–F). n ≥ 20 shoot apexes per genotype were observed with similar results. (Scale bars, 100 μm.) (G–J) Expression levels of cytokinin-related genes in biosynthesis, degradation, and signal transduction pathways in the SAM of the drn drnl mutant, including IPTs (G), LOGs (H), CKXs (I) and AHPs (J). The data are shown as mean ± SD; n ≥ 3 biological replicates, two-tailed Student’s t tests, *P < 0.05, **P < 0.01, ***P < 0.001; ns, no significant difference. (K and L) Expression levels of CKXs (K) and AHPs (L) in the inflorescence apexes of UBQ10::DRNL-GR plants with or without DEX induction in the presence of cycloheximide using qRT–PCR. The data are shown as mean ± SD; n ≥ 5 biological replicates, two-tailed Student’s t tests, **P < 0.01, ***P < 0.001; ns, no significant difference.
To shed light on the mechanism by which DRNL negatively regulates cytokinin, we examined the expression of cytokinin-related genes in biosynthesis, degradation and signal transduction in the SAM of the drn drnl mutant. Consistent with the increased cytokinin signaling, we observed that expression of the most cytokinin biosynthesis genes, including ISOPENTENYL TRANSFERASEs (IPTs) and LONELY GUYs (LOGs), was largely activated in drn drnl mutants (Fig. 2 G and H). Among the genes involved in cytokinin degradation, we observed that six of seven CYTOKININ OXIDASE (CKX) genes, except CKX4, were drastically reduced in the double mutant (Fig. 2I). Moreover, the inhibitor of cytokinin signaling ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN 6 (AHP6) was also observed to be repressed in drn drnl mutants (Fig. 2J). Similarly, the transcripts of most A-type ARRs that respond to cytokinin were significantly induced (SI Appendix, Fig. S7A).
DRNL Represses Cytokinin Signaling by Activating AHP6 and CKX6.
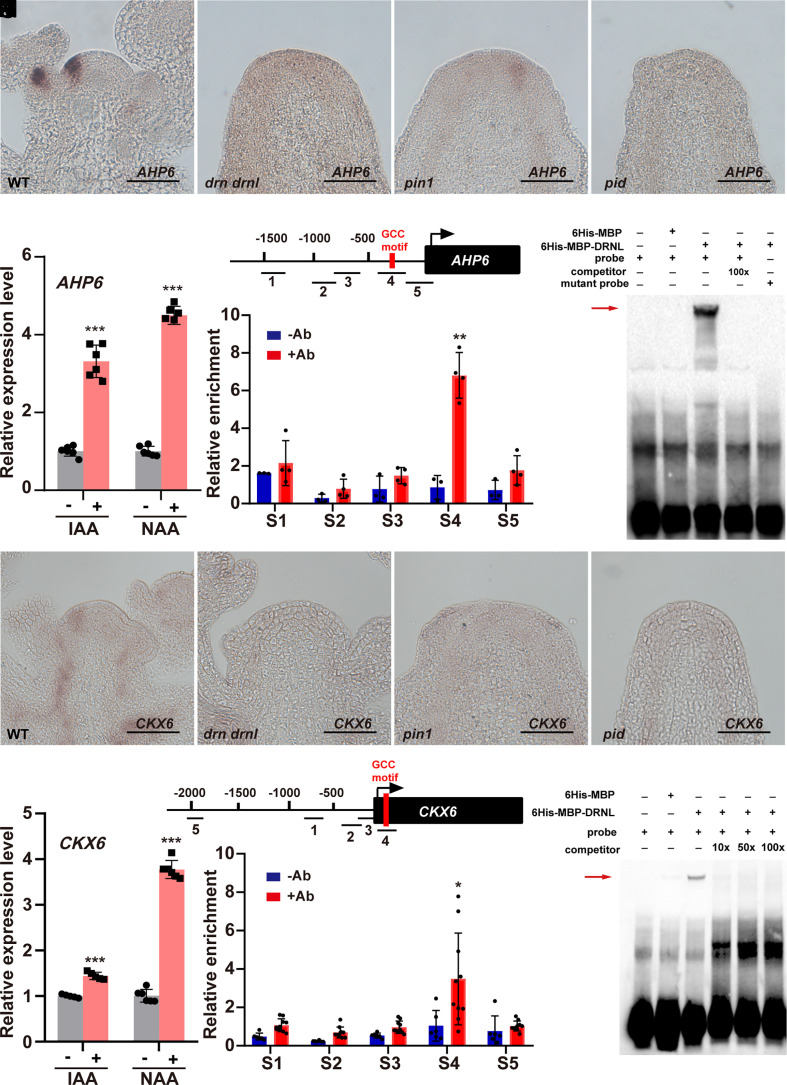
To identify the direct targets of DRNL in cytokinin signaling, we performed DEX induction on UBQ10::DRNL-GR transgenic plants combined with cycloheximide treatment. Among the genes that negatively regulate cytokinin accumulation and whose expression was reduced in the drn drnl mutant, we observed that the expression of CKX6 and AHP6 was significantly elevated upon DEX induction (Fig. 2 K and L). A-type ARRs (SI Appendix, Fig. S7B) and LOGs (SI Appendix, Fig. S8B) did not respond to induction. Although the expression levels of IPT4, IPT6, and IPT8 were also reduced by DEX induction (SI Appendix, Fig. S8A), we failed to find any putative DRNL-binding site in their promoters. We therefore focused on AHP6 and CKX6, two negative regulators of cytokinin. As shown in a previous study (39), we observed that AHP6 was specifically expressed during organ initiation (Fig. 3A), and its expression pattern was similar to that of DRNL (Fig. 1N). Consistent with the notion that AHP6 was activated by MP (39) and DRNL (Fig. 2L), AHP6 transcripts were observed to be decreased in the mp (39), drn drnl, pin1 and pid mutants (Fig. 3 A–D) but activated by IAA and NAA treatments (Fig. 3E). We further observed that the expression patterns of CKX6 were similar to those of DRNL and AHP6 (Fig. 3H), whose transcripts were also largely reduced in the mp (SI Appendix, Fig. S9), drn drnl, pin1 and pid mutants (Fig. 3 H–K) but increased by auxin treatment (Fig. 3L). To further investigate whether DRNL could directly associate with the AHP6 and CKX6 promoters, we performed ChIP assays with inflorescence apexes of UBQ10::DRNL-GR transgenic plants and observed the highest enrichment of DRNL in both the AHP6 and CKX6 promoters with fragments containing putative DRNL-binding sites of the GCC box (40) (Fig. 3 F and M). Moreover, using EMSAs, we demonstrated the direct binding of DRNL to exactly the ChIP-positive GCC box-containing fragments in the AHP6 and CKX6 promoters (Fig. 3 G and N). Thus, we concluded that DRNL directly activates AHP6 and CKX6 expression and mediates cytokinin–auxin cross talk during organ initiation.
Fig. 3.
DRNL directly activates AHP6 and CKX6 transcription in the shoot apical meristem. (A–D) AHP6 expression patterns in WT (A), drn drnl (B), pin1 (C), and pid (D) plants using RNA in situ hybridization; n ≥ 11 shoot apexes per genotype were observed with similar results. (Scale bars, 50 μm.) (E) Detection of AHP6 expression levels in the SAM under IAA and NAA treatment using qRT–PCR. The data are shown as mean ± SD; n = 6 biological replicates, two-tailed Student’s t tests, ***P < 0.001. (F) Enrichment of AHP6 promoter fragments after ChIP using the inflorescence apexes of UBQ10::DRNL-GR plants; −Ab, no antibody control; +Ab, with GR antibody; red box, GCC motif; n = 4 biological replicates, two-tailed Student’s t tests, **P < 0.01. (G) EMSA shows that DRNL specifically binds to the GCC motif of the AHP6 promoter in vitro. The red arrow indicates the specific interactions. Two independent experiments were performed with similar results. (H–K) CKX6 expression patterns in WT (H), drn drnl (I), pin1 (J) and pid (K) plants using RNA in situ hybridization. n ≥ 8 shoot apexes per genotype were observed with similar results. (Scale bars, 50 μm.) (L) Detection of CKX6 expression levels in the SAM under IAA and NAA treatment using qRT–PCR. The data are shown as mean ± SD; n = 5 biological replicates, two-tailed Student’s t tests, ***P < 0.001. (M) Enrichment of CKX6 promoter fragments after ChIP using the inflorescence apexes of UBQ10::DRNL-GR plants; −Ab, no antibody control; +Ab, with GR antibody; red box, GCC motif; n = 6 biological replicates, two-tailed Student’s t tests, *P < 0.05. (N) EMSA shows that DRNL specifically binds to the GCC motif of the CKX6 promoter in vitro. The red arrow indicates the specific interactions. Two independent experiments were performed with similar results.
To test this interaction genetically, we expressed CKX6 and AHP6 from the MP promoter in drn drnl mutants, which drives expression in the PZ. Concomitant with the decreases in CKX6 and AHP6 in the drn drnl mutant during organ initiation, we observed that the proportion of “pin-like” plants significantly decreased in both MP::AHP6/drn drnl and MP::CKX6/drn drnl transgenic plants compared with that of drn drnl mutants (SI Appendix, Fig. S10), suggesting that activation of AHP6 or CKX6 in the PZ partially rescued the primordia initiation defects in drn drnl.
MP and DRNL Form a Complex That Mediates Cytokinin–Auxin Cross Talk during Organ Initiation.
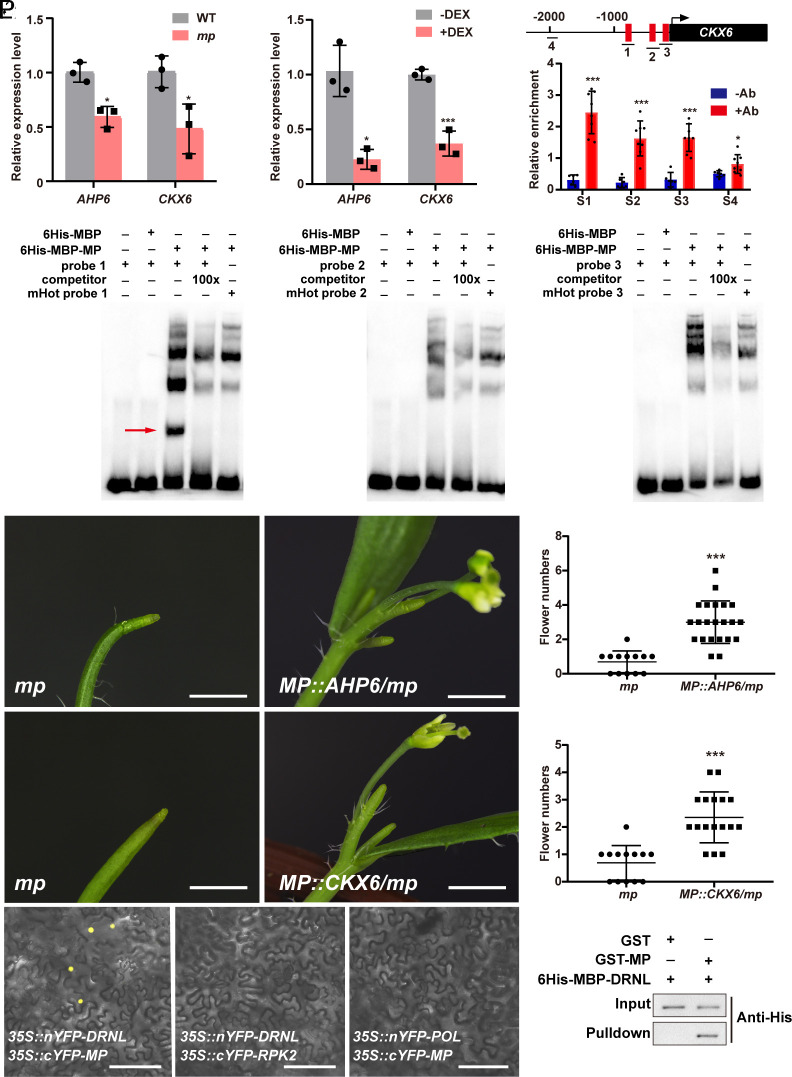
Given that AHP6 is under the direct control of both MP (39) and DRNL (Fig. 3 F and G), we then tested whether CKX6 was also directly controlled by MP. To this end, we first examined CKX6 expression in the mp mutant and observed a significant reduction similar to AHP6 (39) (Fig. 4A). Using DEX induction on RPS5A::GR-bdl transgenic plants with cycloheximide treatment, we observed that both CKX6 and AHP6 were repressed by the induction (Fig. 4B). We then tested whether MP could also associate with the CKX6 promoter by performing ChIP assays with inflorescence apexes of MP::MP-GFP/mp-rescued plants. We observed significant enrichment of MP in three putative AuxREs in the CKX6 promoter (Fig. 4C). By EMSAs, we mapped the binding of MP to the highest ChIP-positive AuxRE in the CKX6 promoter (Fig. 4 D–F), suggesting that MP directly activates the expression of both AHP6 and CKX6. Genetically, we expressed CKX6 and AHP6 in the mp mutant from the MP promoter and observed that the elevation of AHP6 or CKX6 partially rescued the organ initiation defects in mp mutants (Fig. 4 G–L). Our data demonstrate that AHP6 and CKX6, at least in part, mediate auxin signaling in the PZ in priming organ initiation.
Fig. 4.
The genetic interaction of the MP-DRNL module and AHP6 and CKX6 during organ initiation. (A and B) The expression levels of AHP6 and CKX6 in the SAM are decreased in mp (A) and RPS5A::GR-bdl plants with DEX and cycloheximide induction (B). The data are shown as mean ± SD; n = 3 biological replicates, two-tailed Student’s t tests, *P < 0.05, ***P < 0.001. (C) Enrichment of CKX6 promoter fragments after ChIP using the inflorescence apexes of MP::MP-GFP/mp plants; −Ab, no antibody control; +Ab, with GFP antibody, red box, AuxRE motif; n = 6 biological replicates, two-tailed Student’s t tests, *P < 0.05, ***P < 0.001. (D−F) EMSA showing that MP binds to the first AuxRE motif of the CKX6 promoter (D) in vitro. The red arrow indicates the specific interactions. Two independent experiments were performed with similar results. (G−L) The organ initiation defects in mp (G and J) were partially rescued in MP::AHP6/mp (H and I) and MP::CKX6/mp (K and L) plants, whose phenotypes were quantified by the numbers of flowers at 7 days after bolting (I and L). (Scale bars, 1 mm.) mp (I), n = 13; MP::AHP6/mp, n = 22; mp (L), n = 13; MP::CKX6/mp, n = 17; two-tailed Student’s t tests, ***P < 0.001. (M−P) MP interacts with DRNL in vivo in tobacco leaves by BiFC (M) and in vitro by pull-down assays (P). 35S::cYFP-RPK2 (N) and 35S::nYFP-POL (O) were used as negative controls for BiFC, n ≥ 6 for each of three independent experiments. Two independent experiments were performed for pull-down assays (P) with similar results. (Scale bars, 50 μm.)
Because AHP6 and CKX6 were under direct positive control by both MP and DRNL, we hypothesized that these two transcription factors might form a complex to regulate the expression of AHP6 and CKX6. To explore this possibility, we performed bimolecular fluorescence complementation (BiFC) experiments in tobacco leaves and observed an interaction between MP and DRNL in vivo (Fig. 4 M–O). To test their physical interaction in vitro, we conducted pull-down assays and observed that 6xHis-MBP-DRNL bound to GST-MP beads but not GST beads (Fig. 4P). Previously, AIL6, ANT, FIL, LFY, and TMO3 were shown to be directly activated by MP during organ initiation (10, 23). We therefore tested whether these genes were also under direct control by DRNL. By performing DEX induction on UBQ10::DRNL-GR transgenic plants combined with cycloheximide treatment, we observed that four of five of these genes except LFY were significantly induced by DEX induction (SI Appendix, Fig. S11A). Our data demonstrate that MP activates the transcription of DRNL in the PZ and forms a complex to direct organ initiation by activating AIL6, ANT, FIL, and TMO3 and repressing cytokinin accumulation in the PZ.
DRNL-Triggered Spatial Gene Compensation Mediates Auxin Signaling Robustness during Organ Initiation.
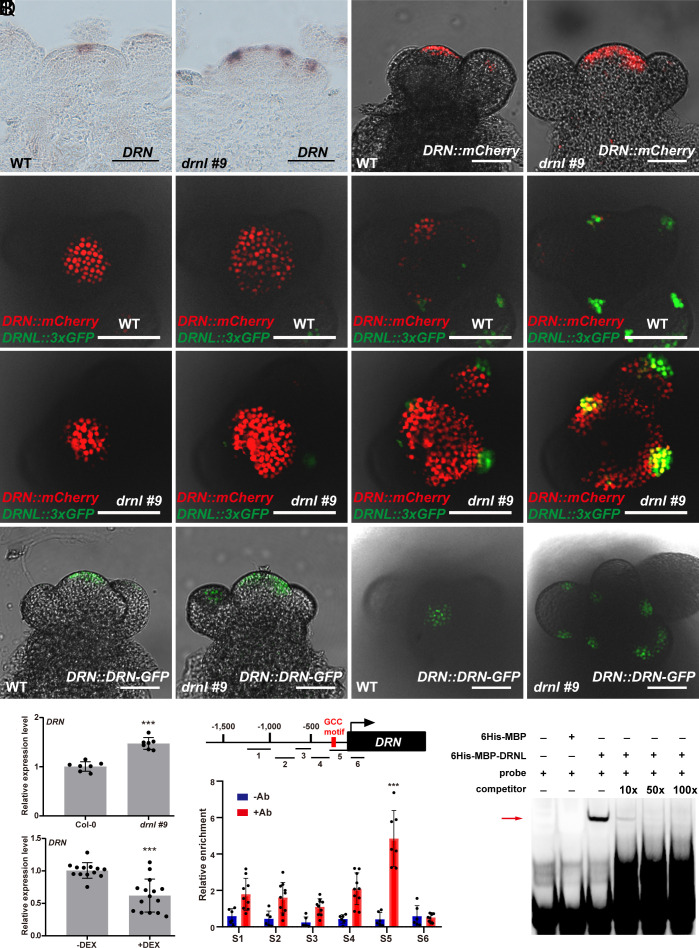
We have shown above that DRN and DRNL are functionally redundant in regulating organ initiation. However, the expression domains of DRN (Fig. 5A) and DRNL (Fig. 1N) in the SAM did not overlap in the wild-type plant (24, 26–28, 41). This was further confirmed using transgenic plants with both DRN::mCherry and DRNL::3×GFP reporters (Fig. 5 E–H). This raises a critical issue regarding how these two paralogs fulfill their redundancy in organ initiation. As DRN was not expressed in the organ founder cells in the wild type, we therefore examined DRN expression in the drnl mutant. To separate effects on expression from different genetic backgrounds, we generated a drnl #9 mutant using the CRISPR/Cas9 system (42) in the Col-0 background with only the first eight amino acids remaining correct (SI Appendix, Fig. S12). In contrast to the wild type, in which DRN is mainly expressed in the CZ (Fig. 5A), we observed an ectopic activation of DRN in the PZ where the lateral organ initiated in drnl #9 mutant plants (Fig. 5B), which was further confirmed by the distribution of the DRN promoter reporter (Fig. 5 C and D) and DRN proteins (Fig. 5 M–P and SI Appendix, Fig. S13). To carry out a direct comparison, we crossed both DRN::mCherry and DRNL::3×GFP in the drnl #9 mutant and observed colocalization of both genes in the primordia (Fig. 5 I–L and SI Appendix, Figs. S15 and S16), and the patterns were distinct from those in the wild-type plants (Fig. 5 E–H and SI Appendix, Figs. S14 and S16).
Fig. 5.
DRNL-triggered spatial gene compensation mediated auxin signaling robustness during organ initiation. (A–D) DRN expression patterns in the SAM of WT Col-0 (A and C) and the drnl #9 mutant (B and D) detected by RNA in situ hybridization (A and B) and DRN::mCherry transgenic plants (C and D). n = 15 shoot apexes per genotype were observed with similar results (A and B); n ≥ 6 shoot apexes per genotype were observed with similar results (C and D). (Scale bars, 50 μm.) (E–L) Top view of DRNL::3×GFP and DRN::mCherry in WT Col-0 (E–H) and drnl #9 mutant (I–L) inflorescences in serial transverse sections imaged using an Olympus FV3000 confocal microscope. n ≥ 20 shoot apexes per genotype were observed with similar results (Scale bars, 50 μm.) (M–P) DRN protein distribution patterns in the inflorescences of DRN::DRN-GFP plants in the WT Col-0 (M and O) and drnl #9 (N and P) backgrounds; n ≥ 16 shoot apexes per genotype were observed with similar results. (Scale bars, 50 μm.) (Q and R) Detection of DRN expression levels in the SAM using the drnl #9 mutant (Q) and UBQ10::DRNL-GR plants with DEX and cycloheximide induction (R). The data are shown as mean ± SD; (Q), n = 7 biological replicates; (R), n ≥ 13 biological replicates; two-tailed Student’s t tests, ***P < 0.001. (S) Enrichment of DRN promoter fragments after ChIP using the inflorescence apexes of UBQ10::DRNL-GR plants; −Ab, no antibody control; +Ab, with GR antibody; red box, GCC motif; n ≥ 6 biological replicates, two-tailed Student’s t tests, ***P < 0.001. (T) EMSA shows that DRNL specifically binds to the DRN promoter in vitro. The red arrow indicates the specific interactions. Two independent experiments were performed with similar results.
Consistent with the observation that DRN was activated ectopically in the organ founder cells in drnl mutants, we observed a significant increase in DRN transcripts in drnl mutant plants (Fig. 5Q). To examine whether DRNL directly repressed DRN at the transcriptional level, we performed DEX induction with cycloheximide on UBQ10::DRNL-GR plants and observed a significant reduction in DRN transcripts (Fig. 5R), suggesting that DRN is under direct negative control by DRNL. Thus, we tested whether DRNL associates with the DRN promoter by ChIP and found an interaction with fragments that contain the GCC element in the DRN promoter (Fig. 5S). Using EMSAs, we observed that DRNL bound to the ChIP-positive fragment specifically in vitro (Fig. 5T), indicating a direct role of DRNL in the negative regulation of DRN transcription.
If DRN was ectopically expressed in the drnl mutant and compensated for the functional deficiency of DRNL during organ initiation, we would expect that DRN should also directly activate AHP6 and CKX6 expression. Indeed, we observed that AHP6 and CKX6 expression levels were significantly increased upon DEX induction in 35S::DRN-GR transgenic plants (SI Appendix, Fig. S17). Likely, the expression levels of AIL6, ANT, FIL, and TMO3 were also increased in 35S::DRN-GR transgenic plants upon induction (SI Appendix, Fig. S11B). Moreover, BiFC experiments in tobacco leaves (SI Appendix, Fig. S18A) and pull-down assays (SI Appendix, Fig. S18B) demonstrated that DRN can also directly interact with MP in plants. Our data demonstrate that DRNL-triggered spatial gene compensation is the molecular basis of the functional redundancy of DRNL and DRN in the PZ. This gene compensation–based safety strategy of DRNL participates in the genetic robustness of auxin signaling during organ initiation.
Our previous study showed that DRN expression was repressed by MP-mediated auxin signaling in the CZ (24). Given that MP proteins were highly accumulated in the PZ (SI Appendix, Fig. S5B), we then examined, in the drnl mutant background, whether ectopically expressed DRN in the PZ was activated or repressed by auxin signaling. Given that DRN did not affect its own expression (SI Appendix, Fig. S19A), we then examined DRN expression in RPS5A::GR-bdl/drn drnl plants with or without DEX induction. As the drn-1 mutant contains a dSpm element insertion (41), we therefore designed primers upstream of the insertion site for qRT–PCR and subsequent in situ hybridization experiments. After DEX induction, we observed that DRN expression was significantly reduced using qRT–PCR and in situ hybridization (SI Appendix, Fig. S19 B–D), suggesting that ectopically expressed DRN in the drnl mutant was still activated by auxin signaling. MP and DRNL seem to be versatile transcriptional regulators that either activate or repress downstream genes in a tissue-specific manner or even in the same tissues. One possible mechanism underlying these effects could be that these versatile transcriptional regulators recruit different cofactors tissue specifically or target them specifically to direct the expression of downstream genes in opposite directions.
As DRN and DRNL also act redundantly in stem cells where DRN is expressed (24), we further tested whether DRNL also showed gene compensation effects in stem cells with DRN. Although DRNL expression levels were significantly increased in drn mutants (SI Appendix, Fig. S20A), we did not observe any direct effects of DRN in repressing DRNL expression in 35S::DRN-GR plants (SI Appendix, Fig. S20B). Using in situ hybridization or reporter lines, we failed to detect any DRNL transcript, protein, or promoter activity in the CZ (SI Appendix, Fig. S20 C–H). A possible mechanism underlying these effects could be that the redundancy of DRN and DRNL in stem cells was indirectly mediated by an unknown mobile factor.
Discussion
Auxin and cytokinin play essential roles in the regulation of the SAM. In the CZ, which harbors undifferentiated stem cells, the functions of auxin and cytokinin are synergistic to maintain stem cell fate by activating WUS (8). Here, we showed that in the differentiated PZ, these two phytohormones are antagonistic in promoting lateral organ initiation. We demonstrated that DRNL is under positive control by MP-mediated auxin signaling. MP physically interacts with DRNL to inhibit cytokinin accumulation during organ initiation by directly activating AHP6 and CKX6, whose genes are involved in both cytokinin signaling and degradation pathways to synergistically limit cytokinin levels in organ founder cells (Fig. 6). Consistently, AHP6 (39) and CKX6 (43) have also been shown to be induced by auxin in the regulation of phyllotaxis or developing leaf primordia under low red/far-red conditions. Because activation of AHP6 or CKX6 in the PZ could partially rescue the organ initiation defects in both mp and drn drnl mutants (Fig. 4 G–L and SI Appendix, Fig. S10), repression of cytokinin accumulation is a relevant aspect of MP function in the PZ. Support for this idea came from the observation that cytokinin is also overaccumulated in the “pin-like” shoot of lfs mutants in tomato (25). This suggests that DRNL/DRN-mediated cross talk between cytokinin and auxin is crucial for primordium initiation in the SAM. In addition to cytokinin, DRNL also directly activates the expression of AIL6, ANT, FIL, and TMO3, which are the known targets of MP in organ initiation, suggesting that MP-DRNL is a key module in auxin-mediated organ initiation.
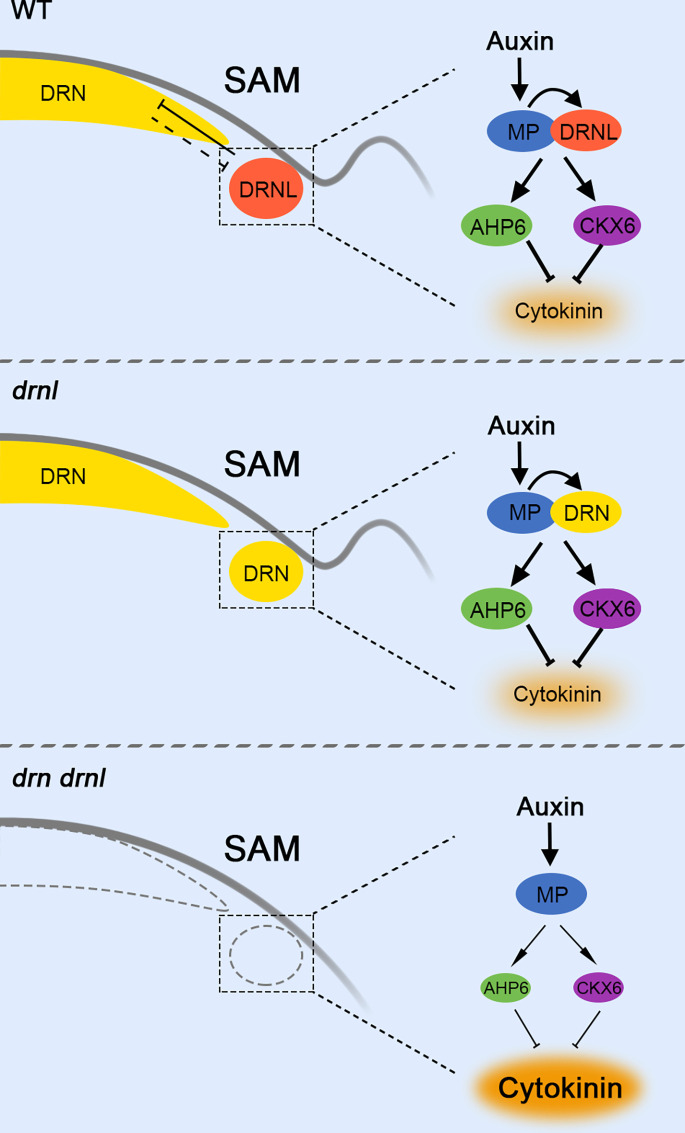
Fig. 6.
Auxin-cytokinin cross talk during organ initiation mediated by DRNL-triggered spatial gene compensation. In the wild type, DRNL directly interacts with MP in the organ initiation cells in the PZ to inhibit cytokinin accumulation by activating AHP6 and CKX6, which is essential for organ initiation. Although the expression and function of DRN, a paralog of DRNL, is mainly in the CZ, DRN transcripts are ectopically activated in the PZ in drnl mutants and fully restore the functional deficiency of drnl during organ initiation. In the drn drnl double mutant, cytokinin levels are highly accumulated resulting in severe defects in organ initiation. This spatial gene compensation triggered by DRNL provides a robust basis for auxin to promote organ initiation.
DRN and DRNL both belong to the largest subclass of the AP2/ERF gene family in Arabidopsis with only a single AP2 domain (44). These two paralogs are closely related with 91% similarity in the AP2 domain (SI Appendix, Fig. S21A) and act redundantly in embryonic development (45), shoot regeneration (30), floral development (27), axillary meristem formation (46), and stem cell maintenance (24). However, in the SAM, they show distinct spatial expression patterns, with DRN mainly in the CZ (Fig. 5A) and DRNL in the PZ (Fig. 1N). In this study, our data support a hypothesis of functional redundancy of the DRN and DRNL in regulating organ initiation. Although DRN was not expressed in the organ initiation site in the wild type, we demonstrate that DRN transcripts are ectopically activated in the drnl mutant to compensate for the loss of DRNL and restore the functional deficiency of drnl in organ initiation (Fig. 6). This gene compensation effect through spatial activation of the paralogous gene provides a molecular basis for auxin in the robustness control of organ initiation. Likely, in the rib zone of the SAM, the redundancy between CLAVATA1 (CLV1) and BARELY ANY MERISTEM (BAM) also relies on the ectopic expression of BAM genes to compensate for the loss of CLV1 (47).
In the progress of evolution, how homologous genes generate and functionally diversify is a key question in understanding the origination of new genes and functions. During embryonic development, the expression of DRN is first observed in the two- to four-cell stage, while DRNL is expressed much later in the early globular embryo (41, 45, 48). Interestingly, from the globular stage to the heart stage, DRN and DRNL share similar expression patterns but diverge afterward (45, 48). We wondered whether this sequential expression difference of two paralogs in ontogeny might also reflect functional divergence during evolution. Support for this idea came from the observation that the origin of DRN was predicted to be 306 My while DRNL was 113 My by GenOrigin (http://genorigin.chenzxlab.cn/) (49) (SI Appendix, Fig. S21 B and C). A plausible scenario would then be that DRNL originated from a gene duplication event from the DRN and showed redundant functions in both stem cells and differentiated cells immediately following the duplication (SI Appendix, Fig. S22). During evolution, the functions of these two paralogous genes began to diverge. DRN expression is restricted in the CZ for stem cell maintenance by the direct repression of DRNL, whereas the function of DRNL is limited to differentiating cells indirectly by DRN (SI Appendix, Fig. S22). Despite their distinct expression patterns and biological functions, these two paralogs still show redundancy in both auxin-mediated stem cell maintenance and differentiation. The finding that DRN is directly repressed by DRNL in the PZ but ectopically reactivated in the drnl mutant fits well with the well-established “active compensation” model (50), which allows robust control of auxin during organ initiation.
Materials and Methods
Plant Materials and Growth Conditions.
The Arabidopsis thaliana Columbia-0 (Col-0) ecotype was used except for drnl (drnl-2, Ler background). The seeds of drn (drn-1), drnl (drnl-2), drn drnl (drn-1 drnl-2), mp-S319, RPS5A::GR-bdl, DRN::mCherry-N7, DRN::DRN-GFP, DRNL::3×GFP-N7 and MP::MP-GFP/mp-rescued plants have been described previously (24). The TCS::dTomato (51) transgenic plant was kindly provided by Yulin Jiao (Peking University). The drnl #9 mutant in the Col-0 background was generated by CRISPR/Cas9 (42). All transgenic plants were generated in the Col-0 ecotype. All seeds were sterilized by 70% ethanol and 0.5% Tween 20 for 10 min, followed by washing two times with 96% ethanol and air drying. Plants were grown on 1/2 Murashige and Skoog (MS) medium plates with 1% sucrose or on soil at 22 °C under long-day conditions (16 h light/8 h dark).
Chemical and Hormone Treatments.
For DEX induction, inflorescence apexes were treated with 15 μM DEX and 50 μM cycloheximide in 1/2 MS liquid medium for 2 h. For hormone treatments, 1/2 MS liquid medium containing 50 μM IAA, 50 μM NAA, 50 μM NPA, or 100 μM yucasin was used for the treatment for 2 h supplied with 0.01% Silwet L-77, except where noted. For controls, 0.1% ethanol (mock) and 0.01% Silwet L-77 were used.
Plasmid Construction.
For DRNL::DRNL-GFP, a 4.3-kb upstream sequence before the ATG of DRNL was used as a promoter according to a previous study (30). To generate DEX-inducible constructs, DRNL and DRN coding sequences (CDS) were subcloned downstream of the UBQ10 and 35S promoters to obtain UBQ10::DRNL-GR and 35S::DRN-GR. To generate p16::DRNL, the DRNL coding sequence was subcloned downstream of a p16 promoter, which is highly active in proliferating cells (36). For MP::AHP6 and MP::CKX6, the full-length genomic sequences of AHP6 and CKX6 were cloned under the 4.1-kb promoter of MP. The primer sequences used in the plasmid construction are listed in SI Appendix, Table S1.
Total RNA Isolation and Quantitative RT–PCR.
The inflorescence apexes of plants at 7 d after bolting were dissected as previously described and were immediately transferred to liquid nitrogen (52). Total RNA was isolated using TRI reagent (Sigma, T9424). The PrimeScript™ RT Reagent Kit (TaKaRa, RR047A) was used for cDNA synthesis. Quantitative PCR was performed using the ChamQ Universal SYBR qPCR Master Mix (Vazyme, Q711) in a Roche LightCycle 96 real-time PCR system with the following conditions: Step 1 to 95 °C for 5 min; Step 2 to 40 cycles of 95 °C for 10 s, followed by 57 °C for 30 s and 72 °C for 30 s; Step 3 to 72 °C for 10 min. The relative expression level of each gene was normalized to the housekeeping gene TUBULIN. The primer sequences used in qRT–PCR are listed in SI Appendix, Table S1.
Chromatin Immunoprecipitation (ChIP).
ChIP was performed on the inflorescence apexes of UBQ10::DRNL-GR and MP::MP-GFP/mp plants, and 500-mg apexes were used for each independent experiment. ChIP was performed as previously described (8, 24) with minor modifications. A Diagenode Bioruptor UCD-200 was used for sonication (30 s on, 30 s off, medium, 15-min duration; sonication buffer: 10 mM Na3PO4, 100 mM NaCl, 10 mM EDTA, 0.5% sarkosyl, 1 mM PMSF, 1 tablet per 10 mL, pH 7). Anti-GR antibodies (Santa-sc-393232X) and anti-GFP antibodies (Abcam, ab290) were used to precipitate chromatin, and no antibody was used as a negative control. The bound DNA fragments were then analyzed using quantitative PCR. The primers used in the ChIP assays are listed in SI Appendix, Table S1.
Electrophoretic Mobility Shift Assay.
The electrophoretic mobility shift assays (EMSAs) were performed as previously described (24, 53, 54). The CDS of DRNL and MP were cloned following a 6xHis-MBP tag to produce the recombinant proteins that were expressed in Escherichia coli strain Rosetta and purified with Nickel Sepharose™ 6 Fast Flow (GE Healthcare, 17-5318-01). The DNA probes were labeled with 5′-biotin, and unlabeled (cold) probes were used as specific competitors. A Light Shift Chemiluminescent EMSA kit (Thermo Scientific 20148) was used for the binding reactions. The primer sequences used in EMSAs are listed in SI Appendix, Table S1.
RNA In Situ Hybridization.
RNA in situ hybridization was performed according to standard protocols as previously described (1, 55). The inflorescence apexes were harvested and fixed with FAA (50% ethanol, 5% acetic acid, 3.7% formaldehyde). After embedding in wax, sectioning was performed using Leica RM2235. Templates of RNA probes were amplified from cDNAs with gene-specific primers containing the T7 or T3 promoter sequence at the 5′ end. RNA probes were synthesized using T7/T3 polymerase and labeled with digoxin-UTP (Roche, 11277073910). The primer sequences are listed in SI Appendix, Table S1.
Confocal Microscopy.
For the detection of fluorescence signals in the SAM, the inflorescence apexes were fixed and sectioned as previously described (24, 54). An Olympus FV3000 confocal microscope was used to obtain images in Fig. 5 and SI Appendix, Figs. S13–S16. The remaining confocal images were obtained using a Zeiss LSM710, except for TCS::dTomato signals, for which images were obtained using Olympus FV1200. To detect the GFP signals, a 488-nm laser was used for excitation, and a 500 to 550-nm emission spectrum was used for detection. mCherry was excited at 594 nm and detected at wavelengths between 590 and 632 nm. dTomato was excited at 554 nm and detected at wavelengths between 550 and 590 nm.
Nicotiana Benthamiana Infiltration.
Agrobacterium tumefaciens harboring relevant constructs was cultured at 28 °C for 2 d. The bacteria were then harvested by centrifugation at 3,500 rpm for 10 min and resuspended in infiltration buffer (10 mM MES, 10 mM MgCl2, 150 μM acetosyringone, pH 5.8). The cells with different constructs were incubated for 2 h at room temperature and mixed with different combinations to infiltrate the abaxial surface of leaves in 3-wk-old N. benthamiana using an injector. Approximately 48 to 72 h after infiltration, the fluorescence signals were imaged with a Zeiss LSM710.
BiFC.
For BiFC, A. tumefaciens containing plasmids of interest were transiently transformed into leaves of N. benthamiana and then detected using a Zeiss LSM710. The binary vectors 35S::nYFP-DRNL, 35S::cYFP-MP, and 35S::nYFP-DRN were used to examine the protein–protein interaction, and 35S::nYFP-POL and 35S::cYFP-RPK2 were used as negative controls.
Pull-Down Assays.
Full-length DRNL CDS and the protein–protein interaction domain of DRN (residues 1 to 200) were cloned behind a 6xHis-MBP tag. The full-length MP CDS were cloned into the pGEX-6P-1 vector to generate the GST-MP construct. The 6xHis-MBP-fusion proteins were purified with Nickel Sepharose™ 6 Fast Flow (GE Healthcare, 17-5318-01). The GST and GST-MP proteins were purified with Glutathione Sepharose™ 4B (GE Healthcare, 17-0756-01), and beads were incubated with soluble 6xHis-MBP-fusion proteins at 4 °C overnight. The beads were washed six to eight times with a solution containing 20 mM Tris-HCl, pH 8.0; 200 mM NaCl; 1 mM EDTA, pH 8.0; 0.25% NP-40, and 25 ng/μL PMSF and then separated on an sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS–PAGE gel) and immunoblotted using an anti-His antibody (Proteintech, 66005-1-Ig) at a 1:1,000 dilution.
Statistical Analysis.
Differences between groups were identified using Student’s t test, and the P value level was set at 5%.
Graph Drawing.
Graphs with dot plots (individual data points) were drawn using GraphPad Prism 8.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Prof. Yulin Jiao (Peking University) for sharing the TCS::dTomato transgenic plant. This work was supported by grants to Z.Z. from the National Natural Science Foundation of China (32130009, 31870264); University of Science and Technology of China Research Funds of the Double First-Class Initiative (YD9100002025); and the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB27030105).
Author contributions
Y.D., L.L., and Z.Z. designed research; Y.D. and L.L. performed research; Y.D., L.L., and Z.Z. analyzed data; and Y.D., L.L., and Z.Z. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Weigel D., Jurgens G., Stem cells that make stems. Nature 415, 751–754 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Barton M. K., Twenty years on: The inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 341, 95–113 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Aichinger E., Kornet N., Friedrich T., Laux T., Plant stem cell niches. Annu. Rev. Plant Biol. 63, 615–636 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Heidstra R., Sabatini S., Plant and animal stem cells: Similar yet different. Nat. Rev. Mol. Cell Biol. 15, 301–312 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Scheres B., Stem cells: A plant biology perspective. Cell 122, 499–504 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Williams L., Fletcher J. C., Stem cell regulation in the Arabidopsis shoot apical meristem. Curr. Opin. Plant Biol. 8, 582–586 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Reinhardt D., et al. , Regulation of phyllotaxis by polar auxin transport. Nature 426, 255–260 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Zhao Z., et al. , Hormonal control of the shoot stem-cell niche. Nature 465, 1089–1092 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Vernoux T., et al. , The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 7, 508 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaguchi N., et al. , A molecular framework for auxin-mediated initiation of flower primordia. Dev. Cell 24, 271–282 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Cheng Z. J., et al. , Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3. Plant Physiol. 161, 240–251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaller G. E., Bishopp A., Kieber J. J., The yin-yang of hormones: Cytokinin and auxin interactions in plant development. Plant Cell 27, 44–63 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee Z. H., Hirakawa T., Yamaguchi N., Ito T., The roles of plant hormones and their interactions with regulatory genes in determining meristem activity. Int. J. Mol. Sci. 20, 4065 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayer K. F. X., et al. , Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Gordon S. P., Chickarmane V. S., Ohno C., Meyerowitz E. M., Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. U.S.A. 106, 16529–16534 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reinhardt D., Mandel T., Kuhlemeier C., Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12, 507–518 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benkova E., et al. , Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Heisler M. G., et al. , Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15, 1899–1911 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Vernoux T., Besnard F., Traas J., Auxin at the shoot apical meristem. Cold Spring Harb. Perspect. Biol. 2, a001487 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okada K., Ueda J., Komaki M. K., Bell C. J., Shimura Y., Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3, 677–684 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennett S. R. M., Alvarez J., Bossinger G., Smyth D. R., Morphogenesis in pinoid mutants of Arabidopsis-thaliana. Plant J. 8, 505–520 (1995). [Google Scholar]
- 22.Przemeck G. K. H., Mattsson J., Hardtke C. S., Sung Z. R., Berleth T., Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 200, 229–237 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Wu M. F., et al. , Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. Elife 4, e09269 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo L. J., Zeng J., Wu H. J., Tian Z. X., Zhao Z., A molecular framework for auxin-controlled homeostasis of shoot stem cells in Arabidopsis. Mol. Plant 11, 899–913 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Capua Y., Eshed Y., Coordination of auxin-triggered leaf initiation by tomato LEAFLESS. Proc. Natl. Acad. Sci. U.S.A. 114, 3246–3251 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandler J. W., Jacobs B., Cole M., Comelli P., Werr W., DORNRA-SCHEN-LIKE expression marks Arabidopsis floral organ founder cells and precedes auxin response maxima. Plant Mol. Biol. 76, 171–185 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Chandler J. W., Werr W., DORNROSCHEN, DORNROSCHEN-LIKE, and PUCHI redundantly control floral meristem identity and organ initiation in Arabidopsis. J. Exp. Bot. 68, 3457–3472 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Comelli P., Glowa D., Chandler J. W., Werr W., Founder-cell-specific transcription of the DORNROSCHEN-LIKE promoter and integration of the auxin response. J. Exp. Bot. 67, 143–155 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Prunet N., Yang W. B., Das P., Meyerowitz E. M., Jack T. P., SUPERMAN prevents class B gene expression and promotes stem cell termination in the fourth whorl of Arabidopsis thaliana flowers. Proc. Natl. Acad. Sci. U.S.A. 114, 7166–7171 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuo N., Makino M., Banno H., Arabidopsis ENHANCER OF SHOOT REGENERATION (ESR)1 and ESR2 regulate in vitro shoot regeneration and their expressions are differentially regulated. Plant Sci. 181, 39–46 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Cole M., et al. , DORNROSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development 136, 1643–1651 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Ram H., et al. , An integrated analysis of cell-type specific gene expression reveals genes regulated by REVOLUTA and KANADI1 in the Arabidopsis shoot apical meristem. PLoS Genet. 16, e1008661 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng Y. F., Dai X. H., Zhao Y. D., Auxin synthesized by the YUCCA flavin Monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19, 2430–2439 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishimura T., et al. , Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis (vol 77, pg 352, 2014). Plant J. 81, 649–649 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Weijers D., et al. , Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev. Cell 10, 265–270 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Schuster C., et al. , A regulatory framework for shoot stem cell control integrating metabolic, transcriptional, and phytohormone signals. Dev. Cell 28, 438–449 (2014). [DOI] [PubMed] [Google Scholar]
- 37.To J. P. C., et al. , Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16, 658–671 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikeda Y., Banno H., Niu Q. W., Howell S. H., Chua N. H., The ENHANCER OFSHOOT REGENERATION 2 gene in Arabidopsis regulates CUP-SHAPED COTYLEDON 1 at the transcriptional level and controls cotyledon development. Plant Cell Physiol. 47, 1443–1456 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Besnard F., et al. , Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature 505, 417–421 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Banno H., Mase H., Maekawa K., Analysis of functional domains and binding sequences of Arabidopsis transcription factor ESR1. Plant Biotechnol. 23, 303–308 (2006). [Google Scholar]
- 41.Kirch T., Simon R., Grunewald M., Werr W., The DORNROSCHEN/ENHANCER OF SHOOT REGENERATION1 gene of Arabidopsis acts in the control of meristem cell fate and lateral organ development. Plant Cell 15, 694–705 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z. P., et al. , Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16, 144 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carabelli M., et al. , Canopy shade causes a rapid and transient arrest in leaf development through auxin-induced cytokinin oxidase activity. Genes Dev. 21, 1863–1868 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alonso J. M., et al. , Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Chandler J. W., Cole M., Flier A., Grewe B., Werr W., The AP2 transcription factors DORNROSCHEN and DORNROSCHEN-LIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development 134, 1653–1662 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Zhang C., et al. , Spatiotemporal control of axillary meristem formation by interacting transcriptional regulators. Development 145, dev158352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nimchuk Z. L., Zhou Y., Tarr P. T., Peterson B. A., Meyerowitz E. M., Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development 142, 1043–1049 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chandler J. W., Cole M., Jacobs B., Comelli P., Werr W., Genetic integration of DORNROSCHEN and DORNROSCHEN-LIKE reveals hierarchical interactions in auxin signalling and patterning of the Arabidopsis apical embryo. Plant Mol. Biol. 75, 223–236 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Tong Y. B., et al. , GenOrigin: A comprehensive protein-coding gene origination database on the evolutionary timescale of life. J. Genet. Genomics 48, 1122–1129 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Diss G., Ascencio D., DeLuna A., Landry C. R., Molecular mechanisms of paralogous compensation and the robustness of cellular networks. J. Exp. Zool. B Mol. Dev. Evol. 322, 488–499 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Zurcher E., et al. , A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol. 161, 1066–1075 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmid M., et al. , Dissection of floral induction pathways using global expression analysis. Development 130, 6001–6012 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Wu H., et al. , WUSCHEL triggers innate antiviral immunity in plant stem cells. Science 370, 227–231 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Zeng J., et al. , Endogenous stress-related signal directs shoot stem cell fate in Arabidopsis thaliana. Nat. Plants 7, 1276–1287 (2021), 10.1038/s41477-021-00985-z. [DOI] [PubMed] [Google Scholar]
- 55.Andersen S. U., et al. , Requirement of B2-type cyclin-dependent kinases for meristem integrity in Arabidopsis thaliana. Plant Cell 20, 88–100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.