Abstract
Studies of protein–protein interactions in membranes are very important to fully understand the biological function of a cell. The extraction of proteins from the native membrane environment is a critical step in the preparation of membrane proteins that might affect the stability of protein complexes. In this work, we used the amphiphilic diisobutylene/maleic acid copolymer to extract the membrane proteome of the opportunistic pathogen Pseudomonas aeruginosa, thereby creating a soluble membrane-protein library within a native-like lipid-bilayer environment. Size fractionation of nanodisc-embedded proteins and subsequent mass spectrometry enabled the identification of 3358 proteins. The native membrane-protein library showed a very good overall coverage compared to previous proteome data. The pattern of size fractionation indicated that protein complexes were preserved in the library. More than 20 previously described complexes, e.g. the SecYEG and Pili complexes, were identified and analyzed for coelution. Although the mass-spectrometric dataset alone did not reveal new protein complexes, combining pulldown assays with mass spectrometry was successful in identifying new protein interactions in the native membrane-protein library. Thus, we identified several candidate proteins for interactions with the membrane phosphodiesterase NbdA, a member of the c-di-GMP network. We confirmed the candidate proteins CzcR, PA4200, SadC, and PilB as novel interaction partners of NbdA using the bacterial adenylate cyclase two-hybrid assay. Taken together, this work demonstrates the usefulness of the native membrane-protein library of P. aeruginosa for the investigation of protein interactions and membrane-protein complexes. Data are available via ProteomeXchange with identifiers PXD039702 and PXD039700.
Keywords: Pseudomonas aeruginosa, membrane proteome, c-di-GMP, native nanodiscs, complexes, soluble library
A soluble membrane-protein library of Pseudomonas aeruginosa was generated and applied to investigate protein–protein interactions and membrane-protein complexes.
Introduction
Bacterial cell membranes are dynamic, fluid lipid bilayers that consist of a heterogeneous matrix of lipids and lipid-soluble proteins. These membrane proteins integrated into the two-dimensional fluid are the bridge between the aqueous compartments that are separated by the membrane. They adapt their native conformation in the bipolar environment of the membrane as they are in contact with both the non-polar and the polar phases in an intact cell. Thereby, the lipid environment not only stabilizes the proteins but also affects their folding, structure, and function (Thoma and Burmann 2020). Membrane proteins serve many crucial functions for the cell such as transport of molecules, energy storage, sensing of environmental signals, motility, or cellular defense. Many of them interact with other proteins to fulfil their biological function (Carlson et al. 2019). The membrane integrity is therefore crucial for these complexes (Sachs and Engelman 2006, Guérin et al. 2020).
The membrane proteome of bacteria has been the focus of research for decades (Filip et al. 1973, Carlone et al. 1986, Blonder et al. 2004, Papanastasiou et al. 2013). With the elucidation of large membrane-protein complexes, the understanding of cellular processes has significantly advanced (Komar et al. 2016). However, the dynamics and interaction of membrane proteins is still intriguing, since not all protein interactions are stable for in vitro elucidation. For instance, lipid molecules were shown to modulate the oligomerization of membrane-protein complexes (Sun et al. 2018, Sobti et al. 2020). Thus, the extraction of membrane proteins from the lipid bilayer might affect the stability and native conformation (Jodaitis et al. 2021).
Frequently, membrane proteins are extracted from the membrane with the aid of detergents and protein complexes have been investigated successfully (Heide et al. 2012, Walian et al. 2012, Van Strien et al. 2019). However, in the process of protein solubilization, the lipid environment of the membrane protein is replaced by the detergent. This treatment often comes along with a significant loss of stability (Yang et al. 2014). Reconstitution of the membrane proteins into liposomes, peptidiscs, or membrane scaffold protein (MSP) nanodiscs can improve stability and pave the way for more detailed studies (Carlson et al. 2019, Thoma and Burmann 2020). By reconstituting the membrane proteins into detergent-free scaffolds, the exposure of the extracted proteins to detergent is strongly reduced. The result is improved stability of membrane proteins and protein complexes, which can also positively affect activity (Bayburt et al. 2006, Alami et al. 2007). However, an initial extraction of the membrane proteins by detergents is still required. By contrast, amphiphilic copolymers can directly extract proteins from membranes together with their lipid environment by forming ‘native nanodiscs’ (Knowles et al. 2009, Dörr et al. 2014, Oluwole et al. 2017). Several membrane proteins, e.g. rhomboid proteases (Barniol-Xicota and Verhelst 2018) and bacteriorhodopsin (Ueta et al. 2020) as well as protein complexes, e.g. the SecYEG/SecDF/YajC/YidC complex (Komar et al. 2016), have been successfully extracted from native membranes and showed enhanced stability (Swainsbury et al. 2014).
We therefore asked whether polymer-encapsulated nanodiscs can be used to detect interactions of membrane proteins within a lipid-bilayer environment. Since polymer nanodiscs extract membrane proteins along with their natural lipid-bilayer environment, we expected that native protein–protein interactions within a nanodisc would be preserved as well. To test this hypothesis, we used the amphiphilic diisobutylene/maleic acid copolymer (DIBMA) (Oluwole et al. 2017) to generate a soluble library containing the membrane proteins of the biofilm model organism Pseudomonas aeruginosa PAO1 within a native-like lipid-bilayer environment.
Our model organism P. aeruginosa strives in many environments, but a major research interest focuses on its ability to form biofilms. The formation of biofilms goes along with an increased resistance against antibiotics (Rasamiravaka et al. 2015). Especially immunocompromised patients suffer from chronic infections by P. aeruginosa that lead to an increased morbidity and mortality (Stover et al. 2000, Donlan and Costerton 2002, Rasamiravaka et al. 2015). Bacterial biofilms represent surface-attached communities that are enclosed in a matrix of self-produced extracellular polymeric substances (Flemming et al. 2016). The formation of biofilms is controlled by the intracellular concentration of the nucleotide second messenger 3′,5′-cyclic diguanylic acid, or c-di-GMP (Ha and O'Toole 2015). Synthesis and degradation of c-di-GMP is mediated by two types of enzymes. The diguanylate cyclases (DGC) containing a GGDEF motif synthesize the cyclic dinucleotide from two molecules of GTP. For the degradation of c-di-GMP, two types of phosphodiesterases (PDE), containing either an EAL- or a HD-GYP motif, are described (Ha and O'Toole 2015).
The genome of P. aeruginosa PAO1 codes for more than 40 putative c-di-GMP modulating proteins, containing either a single DGC or PDE domain or tandem proteins with both domains (Kulasakara et al. 2006, Römling et al. 2013, Ha and O'Toole 2015). Furthermore, c-di-GMP receptor proteins, so-called effectors, that can sense and transmit the c-di-GMP concentration into a cellular response were identified (Düvel et al. 2012, Nesper et al. 2012, Banerjee et al. 2021). The redundant enzyme activities are tightly regulated on transcriptional and posttranslational levels. The majority of the c-di-GMP modulating proteins possess specialized sensory or regulatory domains that regulate enzymatic activity depending on environmental cues (Ha and O'Toole 2015, Schirmer 2016). Diverse mechanisms have been reported for the regulation of DGC activity, e.g. phosphorylation of WspR or binding of interaction partners such as HsbD/HsbA, and SiaD/SiaC in P. aeruginosa (Hickman et al. 2005, Valentini et al. 2016, Chen and Liang 2020, Chen et al. 2021). The study of interactions between soluble proteins has been successfully demonstrated many times. By contrast, the interactions of membrane proteins with regulatory function have remained elusive. In P. aeruginosa, about 33 out of 43 deduced c-di-GMP-modulating proteins are predicted to reside in the membrane (www.pseudomonas.com, (Winsor et al. 2016)).
To investigate membrane-associated proteins involved in c-di-GMP signaling and their interaction partners, we created a native membrane-protein library representing the membrane proteome of P. aeruginosa PAO1. The library was characterized by testing for reproducible and stable extraction of membrane proteins, coverage of the membrane proteome, and the capability to detect protein–protein interactions within a lipid-bilayer environment as well as membrane-bound protein complexes.
Material and methods
Growth of P. aeruginosa cells
Pseudomonas aeruginosa PAO1 was cultured in a baffled Erlenmeyer flask. OD600 was initially set to 0.01 from an overnight culture in Lurina-Bertani (LB) medium and grown at 37°C and 100 rpm for 7 h (to stationary phase). Cells were harvested by centrifugation at 17 500 × g for 15 min. Pellets were stored at −20°C. For nbdA expression, P. aeruginosa PAO1 carrying a plasmid for production of NbdA-Strep or untagged NbdA as a control were grown in LB media supplemented with 150 µg/ml tetracycline (Tab. S1). OD600 was initially set to 0.05 from an overnight culture in LB medium and grown at 37°C and 100 rpm. When cells reached OD600 of 0.5, gene expression was induced with 0.1% l-Arabinose and cells were further grown for 5 h. Cells were harvested at 17 500 × g for 15 min, and pellets were stored at −20°C.
Membrane preparation
Cell pellets were thawed at 4°C and lysis buffer (50 mM Tris HCl, 300 mM NaCl, pH 8, with cOmplete Protease inhibitor mix (Roche, Grenzach-Wyhlen, Germany) was added in a ratio 1:2 gram per cell wet weight. Subsequently, cells were treated with lysozyme and DNAse I for 20 min on ice. Cell disruption was performed by sonication (Bandelin, Berlin, Germany) for 3 min total, with 20 s pulse intervals followed by 30 s cooling. The crude extract was centrifuged for 15 min at 7000 × g at 4°C. Membranes were isolated by ultracentrifugation at 100 000 × g for 1 h at 4°C. Membrane pellets were resuspended in solubilization buffer [50 mM Tris HCl, 300 mM NaCl, 10 mM MgCl2, pH 6.8, with cOmplete® Protease inhibitor mix (Roche, Grenzach-Wyhlen, Germany)] to 200 mg/ml membrane wet weight.
Solubilization
Polymer stocks were prepared by dialyzing 10% (w/v) DIBMA (Glycon Biochemicals, Luckenwalde, Germany) in solubilization buffer in 300-fold volume twice for 36 h (membrane: MWCO 3.5 kDa). Polymer solutions were filtered (0.45 µm pore size), and concentration was measured using an Abbemat 500 refractometer (Anton Paar, Graz, Austria) as previously described (Grethen et al. 2017). Solubilization for complexome analysis was carried out using 40 mg/ml membrane wet weight and a DIBMA concentration at 3.5% (w/v). Samples were incubated at room temperature overnight with gentle shaking. Non-solubilized material was removed by ultracentrifugation (200 000 × g for 10 min at 4°C), and the supernatant was filtered (0.45 µm PVDF).
Determination of protein content
Protein content was measured using a Pierce BCA protein assay kit (Thermo Fisher Scientific, Waltham, USA) according to the manufacturer's instructions.
Size exclusion chromatography
Polymer nanodiscs were separated by size exclusion chromatography on an ÄKTA Purifier 10 system with a Superose 6 Increase GL 10/300 column (GE Healthcare, Chicago, USA) in solubilization buffer (50 mM Tris HCl, 300 mM NaCl, 10 mM MgCl2, pH 6.8). A total of 200 µl sample (300–400 mg protein) was run with a flow rate of 0.2 ml/min at 4°C and 250 µl fractions were collected. Fractions from 8.22 ml to 10.22 ml were pooled to a 1 ml fraction size, and between 17.22 ml and 23.22 ml fractions were pooled to a fraction size of 500 µl. Fractions between 8.22 ml and 23.22 ml elution volume were subjected to mass spectrometry analysis. The apparent molecular weight (app. MW) was calculated based on a calibration curve with soluble marker proteins: Apoferritin (443 kDa), alcohol dehydrogenase (150 kDa), albumin (66 kDa), and carbonic anhydrase (29 kDa). The molecular weight of the free membrane proteins was derived from the Pseudomonas Genome database (DB) (Winsor et al. 2016). The app. MW in the size exclusion chromatography (SEC) was divided by the calculated molecular weight for each protein to estimate the x-fold increase of size by nanodisc formation (MWapp/MWcalc).
Protein precipitation
Protein fractions were precipitated with six volumes of acetone at −80°C overnight. Samples were centrifuged at 20 000 × g for 20 min at 4°C and protein pellets were washed in five volumes of 80% acetone and centrifuged again for 20 000 × g for 25 min at 4°C. Pellets were dried and resuspended in 25 µl urea buffer (8 M urea, 25 mM ammonium bicarbonate, MS grade).
Sample preparation and mass spectrometry
In solution, digest of samples was carried out as previously described (Glueck et al. 2022). Tryptic peptides were desalted on C18-StageTips according to a published protocol (Rappsilber et al. 2007) and analyzed on a Q Exactive HF™ Mass Spectrometer coupled in-line to EASY-nLC 1200 ultra-high pressure chromatography system (both Thermo Fisher Scientific, Waltham, USA). For mass spectrometry (MS) analysis, desalted peptides were separated on a 50 cm reverse phase column with an inner diameter of 75 μm (New Objective, Woburn, USA) packed in-house with 1.8 μm ReproSil-Pur 120 C18-AQ particles (Dr. Maisch GmbH, Ammerbruch-Entrigen, Germany) using a 90 min non-linear gradient of 2%–95% buffer B (0.1% (v/v) formic acid, 80% (v/v) acetonitrile) at a flow rate of 250 nl/min. All MS data were recorded with a data-dependent acquisition strategy. Survey scans were acquired with a resolution of 60′000 at m/z = 200. The top 15 most abundant precursors with charge >2 were selected for fragmentation. MS/MS scans were acquired with a resolution of 15′000 at m/z = 200. All other parameters can be obtained from raw files available at the ProteomeXchange repository (PXD039702 and PXD039700). MS data were processed with the MaxQuant software (version 2.0.1.0). Peak lists were searched against protein sequences derived from the Pseudomonas Genome DB [version 20.2, (Winsor et al. 2016)] applying a false-discovery rate (FDR) of 0.01 for peptides and proteins, a minimal peptide length of seven amino acids and at least two peptides for quantification. ‘Match between run’ was disabled.
MS data analysis for nanodisc library
Raw data are available via ProteomeXchange with the identifier PXD039702. Mass spectrometry data were analyzed with Perseus (version 1.6.15.0) (Tyanova et al. 2016). The intensity-based absolute quantification (IBAQ) values were calculated by dividing the total peptide intensities by the number of theoretically observable peptides (Schwanhäusser et al. 2011). It is highly correlated to the abundance of a protein (Krey et al. 2014). Relative IBAQ values, scaled from 0 to 1, were used to compare individual elution profiles of proteins in the dataset. For clustering, Pearson correlation was used as a distance measure. Clustering was performed for each replicate individually. The remaining data analysis was carried out in the Excel® (version 2112, Microsoft, Redmond, USA). Correlation coefficients were calculated with the Excel® data analysis plugin. Plots were made with OriginLab (version 2022 SR, OriginLab, Northampton, USA).
Pulldown of tagged NbdA
For pulldown analysis, P. aeruginosa PAO1 carrying a plasmid for the expression of NbdA-Strep or untagged NbdA as a control (Tab. S1) were grown, membranes were isolated and solubilized into a nanodisc library as described above. The nanodisc library was incubated for 4 h under gentle shaking at 4°C with Strep-Tactin®XT 4Flow® beads (IBA Lifesciences GmbH, Göttingen, Germany). Unbound proteins were removed by gravity flow, and beads were washed twice with 20 column volumes of wash buffer (100 mM Tris, 150 mM NaCl, 1 mM EDTA, pH 8). Proteins bound to the beads were eluted by a wash buffer containing 50 mM biotin. Samples were digested and analyzed by MS as described above.
MS data analysis for pulldown
Raw data are available via ProteomeXchange with the identifier PXD039700. Mass spectrometry data were analyzed with Perseus (version 1.6.15.0) (Tyanova et al. 2016). For pulldown data analysis, label-free quantitation (LFQ) intensities were log2 transformed and filtered to have four valid values in all four replicates of the NbdA pull-down samples. Missing values in the control pull-downs were replaced with values drawn from a normal distribution centered around the detection limit of the MS instrument with a width of 0.3 and a downshift of 1.8 with respect to the standard deviation and mean of all protein intensities of each sample (Tyanova et al. 2016). Two sample t-tests with permutation-based FDR control were carried out in Perseus. Two significance cut-offs were applied to call Class A (S0 = 4, FDR <0.01) and Class B (S0 = 4, FDR between 0.05 and 0.01) interactors.
Bacterial adenylate cyclase two-hybrid assay
Bacterial adenylate cyclase two-hybrid (BACTH) protein-protein interaction assays were carried out as previously described (Karimova et al. 1998, Claessen et al. 2008, Ouellette et al. 2017). Escherichia coli BTH101 was co-transformed with plasmid p25N carrying nbdA and plasmid pUT18 carrying the target genes (Tab. S1). Cells were resuspended in LB medium and dropped on MacConkey agar plates supplemented with 1% maltose, 0.5mM IPTG, 50 µg/ml kanamycin, and 100 µg/ml ampicillin and incubated for 24 h at 37°C followed by 24–48 h at room temperature.
Results
Creating a soluble library reflecting the P. aeruginosa membrane proteome
For the detergent-free extraction of the membrane proteome of P. aeruginosa PAO1, we employed the amphiphilic copolymer DIBMA. DIBMA extracts membrane proteins directly from the membrane, forming soluble nanodiscs that preserve a native-like lipid-bilayer environment, thereby generating a soluble membrane-protein library. To obtain detailed insight into the efficiency and coverage, we first fractionated the library by size exclusion chromatography and subsequently analyzed it by mass spectrometry (LC-MS/MS) (Fig. 1). The native nanodisc library showed a broad elution profile from the SEC column (100–3000 kDa) (Fig. S1). Proteins were detected over the entire elution range, with a uniform distribution indicating a stable solubilization. Due to the formation of nanodisc particles, the app. MW in size exclusion chromatography increases around 11 times compared to the calculated molecular weight of the free membrane protein. In the chromatograms of the library, one elution peak was observed in the void volume of the SEC column, indicating to large particles derived from the original membrane material (Fig. S1). Three independent biological replicates were analyzed by mass spectrometry (LC-MS/MS). In total, 3358 proteins were identified in the native nanodisc library, corresponding to 60% of the annotated proteins in P. aeruginosa PAO1 (a total of 5570 proteins, Fig. 2A, Tab. S2).
Figure 1.
Workflow for preparing native membrane-protein libraries reflecting the P. aeruginosa membrane proteome. Pseudomonas aeruginosa membrane fractions were harvested from cell lysates by ultracentrifugation. Membrane proteins were solubilized from the membranes with the polymer DIBMA, spontaneously forming nanodiscs containing membrane proteins embedded in a lipid bilayer. The nanodiscs were separated and fractionated by size exclusion chromatography. Proteins from the collected fractions were precipitated and subsequently analyzed by mass spectrometry. The relative abundance of the identified proteins was plotted against the elution volume in chromatographic separation.
Figure 2.
Analysis of the composition of the proteome library from P. aeruginosa. (A) Venn diagram of the three library replicates depicting numbers of all identified proteins. (B) Pie chart showing the distribution of the identified proteins among the different subcellular compartments as predicted for each protein by the Pseudomonas Genome DB (Winsor et al. 2016). Only the highest confidence for the localization was considered. (C) Violin plot showing the distribution of peak elution fractions (a fraction with the highest measured intensity) for each protein grouped by their predicted subcellular localization. Averages from three replicates were used. Box plots indicate a range of 25%–75% of the data, whiskers the range of 10%–90%. Median (square) and average (circle) of the maximal peak elution fraction for the different subcellular localized proteins are also shown. Violines show the smoothing via the Kernel equation with equal areas.
To determine the composition of the library, each identified protein was assigned the annotated subcellular localization from the Pseudomonas Genome DB (Winsor et al. 2016). Only the highest confidence class for the annotated localization was considered. Approximately half of the identified proteins were classified as cytosolic proteins (Fig. 2B). Thus, the cytosolic proteins are clearly overrepresented in the dataset. This is due to the preparation of the membrane fractions without stringent washing steps. The second largest group of the library consisted of 580 predicted cytoplasmic membrane proteins, corresponding to 48% of the annotated membrane proteins (a total of 1194). About 575 proteins were detected whose subcellular localization is unknown. Furthermore, 50 proteins from the outer membrane and 307 proteins predicted to reside in outer membrane vesicles were identified. We suspect that the outer membrane vesicle proteins were detected before vesicles are formed, as they are constantly produced and released from P. aeruginosa (Lee et al. 2008, Choi et al. 2011).
Interestingly, in size exclusion chromatography, proteins of different subcellular localizations show distinct elution patterns (Fig. 2C). Cytosolic proteins, periplasmic proteins, and extracellular proteins were identified mostly in later fractions (>18), whereas proteins from the cytoplasmic membrane, the outer membrane, and outer membrane vesicles were identified in early fractions (<18). Elution in the early fractions corresponds to a larger hydrodynamic radius, ergo particle size. As the membrane proteins are embedded in a shell of lipids into the nanodisc particles they show a larger apparent weight. Soluble proteins eluting in this volume range are likely to be present in large complexes or interact with nanodiscs. The elution profiles of all 3358 identified proteins were plotted in a heatmap (Fig. 3). Therefore, IBAQ values were calculated and used as a measure for protein abundance (Schwanhäusser et al. 2011, Krey et al. 2014). Each column of the plot shows the elution profile of an individual protein from size exclusion chromatography. The relative abundance of the protein is color coded. The elution profiles for almost all proteins are distinct but also very broad. On average, a protein was detected in 17 out of 40 fractions with high abundance (upper 90% of its IBAQ value). The figure shows further the elution of proteins along the molecular separation range of the column (5 000 000–1000 Da) lacking a clear size separation of individual complexes. This is due to the size heterogeneity inherent to flexible polymer-encapsulated lipid-bilayer nanodiscs (Oluwole et al. 2017, Glueck et al. 2022).
Figure 3.
Heatmap of all identified proteins from one library replicate (rep 3). For all identified proteins (3358 proteins, x-axis) the individual elution profile from size exclusion chromatography (y-axis) is shown. The relative abundance of the proteins is color coded, whereby the fraction with the highest amount of protein (elution maximum) is dark red/brown in the color gradient. Identified proteins were sorted by the fraction with the maximal relative abundance. Apparent molecular weight (app. MW) was calculated based on a calibration curve with soluble marker proteins.
Membrane-protein complexes are preserved in the native nanodisc library
To test whether native membrane-protein complexes are preserved in the nanodisc library, we compared the elution profiles of protein components of several well-described protein complexes. To this end, more than 20 described membrane-protein complexes (Tab. S3) were selected, and the elution profiles of the individual components were analyzed. We found that the protein components from each tested complex co-occurred in several fractions for all the tested complexes. One example of a bona fide membrane complex is the cytochrome oxidase cbb3-1 (Comolli and Donohue 2004, Kawakami et al. 2010). This complex is part of the respiratory chain and consists of four membrane-integrated protein subunits, CcoNOPQ (Comolli and Donohue 2004, Jo et al. 2017). In our dataset, all proteins from this complex were identified, and the proteins showed largely overlapping elution profiles (Fig. 4A). All proteins eluted with the highest abundance at an elution volume of 13–16 ml.
Figure 4.
Elution profiles of membrane protein complexes. Well-described membrane protein complexes were analyzed for coelution. The relative abundance of each protein based on the IBAQ value was plotted against the elution volume from size exclusion chromatography. Graphs show data from a single replicate. (A) Cytochrome oxidase cbb3-1 of P. aeruginosa (CcoOPQN1). (B) Secretion machinery (SEC-SRP). (C) Pilus assembly subcomplex PilMNOP with major pilin PilA. (D) NADH dehydrogenase complex (NuoABDEFGHILMN).
We also tested complexes that consist of a membrane part as well as cytosolic or periplasmic-associated proteins. One example is the SEC-SRP complex, consisting of a membrane channel SecYEG and membrane-associated helper proteins: SecB is the chaperone that binds the unfolded protein and SecA is an ATPase that drives the export through the SecYEG channel. YidC, SecDF, and YajC facilitate protein export. The proteins FtsY and Ffh help with the insertion of proteins into the membrane (Müller et al. 2001, Ma et al. 2003). Again, most proteins showed a largely overlapping elution profiles, with maximum abundances at elution volumes of 13.5–16 ml (Fig. 4B). The chaperone SecB eluted in later fractions, independently of the SecYEG complex, corresponding to a smaller hydrodynamic size.
Another protein complex tested for coelution is the type IV pili membrane alignment subcomplex PilMNOP (Ayers et al. 2009). This complex connects the membrane motor in the cytoplasmic membrane to the outer membrane subcomplexes (e.g. secretin pore subcomplex PilQ and PilF) and also interacts with the major pilin subunit PilA (Burrows 2012, Tammam et al. 2013). All proteins of the complex were detected in the library and showed a similar elution profile, with high relative abundances at elution volumes of 12–18 ml (Fig. 4C). We also analyzed large complexes, such as the NADH-dehydrogenase (NuoABDEFGHIJKLMN) involved in the respiratory chain: The NADH-dehydrogenase complex consists of three subunits, namely, the dehydrogenase domain, the connecting domain, and the membrane domain (Berrisford et al. 2016). All identified proteins of the complex showed high relative abundances in 12–14 ml elution volume, indicating the existence of a large protein complex (Fig. 4D). However, with increasing elution volume, the relative abundance of the membrane part (blue colors) decreased, whereas the hydrophilic dehydrogenase subunit and the connecting subunit remained at high relative abundances until 16.5 ml elution volume (red colors). This indicates that the library contains both a large NADH dehydrogenase complex as well as several subcomplexes.
When proteins from a complex were identified, they always co-occurred in the same fractions as their interacting partners. This holds true for both pure membrane-protein complexes and mixed complexes that contain both integral membrane proteins as well as associated proteins. These results indicate that non-covalent protein–protein interactions are preserved in the native nanodisc library.
Analysis of complexes in the c-di-GMP signaling network of P. aeruginosa
Next, we focused on the complex network of c-di-GMP-regulating proteins and their interaction partners. In total, 29 of the 43 predicted GGDEF/EAL/HD-GYP domain-containing proteins were identified, of which 24 proteins are predicted to be membrane proteins (Table 1). Additionally, several c-di-GMP effector proteins were detected in this study, such as FleQ, the master regulator of motility and biofilm formation (Table 1) (Hueso-Gil et al. 2020). To our knowledge, this is the first report where 73% of c-di-GMP modulating membrane proteins were identified in a proteome study.
Table 1.
Proteins involved in c-di-GMP signaling.
| Locus tag | Name | Localization | Identified in replicate | |
|---|---|---|---|---|
| GGDEF domain containing | ||||
| PA0290 | – | cm | 2, 3 | |
| PA0847 | – | cm | 2, 3 | |
| PA1107 | RoeA | cm | 3 | |
| PA1120 | YfiN,TtpbB | cm | 1, 2, 3 | |
| PA1851 | – | cm | 2 | |
| PA2771 | Dcsbis | cm | 2, 3 | |
| PA2870 | – | cm | 2, 3 | |
| PA3343 | HsbD | cm | 1, 2, 3 | |
| PA3702 | WspR | c | 1, 2, 3 | |
| PA4332 | SadC | cm | 3 | |
| PA4843 | AdcA, GcbA | c | 1, 3 | |
| PA4929 | NicD | cm | 1, 3 | |
| PA5487 | DgcH | c | 1, 2, 3 | |
| GGDEF-EAL domain containing | ||||
| PA0285 | PipA | cm | 1, 2, 3 | |
| PA0575 | RmcA | cm | 1, 3 | |
| PA0861 | RbdA | cm | 1, 2, 3 | |
| PA1181 | – | cm | 1, 2, 3 | |
| PA1433 | LapD | cm | 1, 2, 3 | |
| PA2072 | – | cm | 1, 2, 3 | |
| PA2567 | – | cm | 1, 2, 3 | |
| PA3311 | NbdA | cm | 1, 2, 3 | |
| PA4367 | BifA | cm | 1, 2, 3 | |
| PA4601 | MorA | cm | 3 | |
| PA4959 | FimX | cm | 1, 2, 3 | |
| PA5017 | DipA | cm | 2 | |
| EAL domain containing | ||||
| PA2818 | Arr | cm | 1, 2, 3 | |
| PA3825 | – | cm | 1, 2, 3 | |
| HD-GYP domain | ||||
| PA2572 | – | c | 1, 2, 3 | |
| PA4781 | – | c | 1, 2, 3 | |
| Effector proteins | ||||
| PA1097 | FleQ | c | 1, 2, 3 | |
| PA2799 | – | c | 2 | |
| PA2960 | PilZ | u | 1, 2, 3 | |
| PA2989 | – | c | 1, 2, 3 | |
| PA3353 | FlgZ | c | 1, 2, 3 | |
| PA4608 | MapZ | c | 1, 2, 3 | |
| PA4878 | BrlR | c | 2, 3 | |
| PA4958 | FimW | c | 1, 2, 3 | |
| PA5346 | SadB | c | 1, 2, 3 | |
Identified proteins with GGDEF/EAL/HD-GYP domains and c-di-GMP effectors. Predicted localization of the proteins taken from Pseudomonas Genome DB [www.pseudomonas.com (Winsor et al. 2016)], where only the highest confidence class was considered. CM = cytoplasmic membrane, C = cytoplasmic. List of described c-di-GMP effectors from Valentini and Filloux (2016) and Banerjee et al. (2021) (Valentini and Filloux 2016, Banerjee et al. 2021).
It has been suspected and, in some cases, already demonstrated that c-di-GMP-producing and -degrading proteins are localized near their cellular targets for a specific local regulation (Sarenko et al. 2017). We, therefore, tested the library for co-occurrence and co-elution of described protein-target groups. As a first example, we tested the Wsp chemosensory-like signal transduction complex (Güvener and Harwood 2007). The activation of this system eventually leads to an increase of c-di-GMP levels in the cell. This system consists of a core complex (WspABDE), the methyltransferase WspC, the methyl esterase WspF, and the diguanylate cyclase WspR, which produces c-di-GMP in response to surface contact (Hickman et al. 2005, Güvener and Harwood 2007, O'Neal et al. 2022). In our nanodisc library, the proteins of the core complex WspABDE and, furthermore, WspF and WspR were reproducibly identified. The methyltransferase WspC was not detected in our study. The elution profiles of the individual proteins were overall similar but revealed increasing differences in later fractions, again indicating the presence of subcomplexes lacking one or several protein components (Fig. 5A). The methyl esterase WspF eluted in a clearly different profile from the rest of the complex. This protein is required to turn off the sensory system by demethylation of WspA (Hickman et al. 2005). Therefore, WspF might not be recruited to the complex in our experimental conditions.
Figure 5.
Elution profiles for proteins and complexes known to be involved in c-di-GMP signaling are shown. Data from one replicate are depicted. (A) Elution profiles are shown for the surface sensing Wsp system with the associated diguanylate cyclase WspR. (B) The Redox/membrane perturbation recognizing system YfiNBR including the diguanylate cyclase YfiN. YfiR does not show coelution with YfiN or YfiB. (C) The interaction partners DgcH (DgcP in PA14) and the landmark protein FimV show sparsely overlapping profiles. (D) The elution profile of the c-di-GMP specific effector protein FimX and the ATPase PilB involved in type IV pilus extension partially overlap.
Another membrane-associated complex, including a c-di-GMP modulating protein is the tripartite periplasmic signaling system YfiNBR. In this dynamic system, the periplasmic protein YfiR can bind and thereby inactivate the cytoplasmic membrane diguanylate cyclase YfiN. In the presence of an unknown signal, the outer membrane component YfiB competes for binding of the periplasmic protein YfiR and, thereby, releases and activates the diguanylate cyclase YfiN (Malone et al. 2012, Giardina et al. 2013). In our library, all components of the system were detected (in two out of three replicates), but individual proteins showed very different elution profiles (Fig. 5B). The diguanylate cyclase YfiN was most abundant in fraction 11 (13.2 ml elution volume), where most proteins from the cytoplasmic membrane eluted (Figs 2 and 5B). YfiB was detected in the first fraction, and at elution volumes of 13–17 ml. The early elution of the outer membrane protein YfiB in fraction 1 might indicate aggregate formation, while the later elution fractions likely contain the soluble form of YfiB. The periplasmic protein YfiR eluted in much later fractions with a very irregular profile. All three components of the YfiNBR system were detected independently, indicating that this system experiences only weak interactions or that the nanodiscs did not preserve the complex in the library.
A stable interaction was reported for the diguanylate cyclase DgcP and the landmark protein FimV in P. aeruginosa PA14. The correct localization for DgcP to the cell pole depends on FimV (Nicastro et al. 2020). Both proteins (alias DgcH and FimV in PAO1) showed an overlapping elution peak in fractions with 11.7–13.5 ml elution volume, but more pronounced deviations in the later fractions (Fig. 5C).
An example of described protein interactions of a c-di-GMP effector with its cellular target is the protein FimX. This protein was reported to interact with the type IV pilus ATPase PilB that fuels pilus function (Jain et al. 2017). In our library, both proteins FimX and PilB were detected and showed overlapping elution in the first 15 fractions (elution volume of 10–14 ml), while PilB showed a broader elution profile, probably due to other interactions with other components of the pilus machinery. In conclusion, GGDEF/EAL proteins and their described interaction partners were detected in the nanodisc library. They show similar or overlapping elution profiles, suggesting the preservation of these complexes. Known interactions can be tracked, but the co-elution is not sufficient to predict novel interaction partners in the c-di-GMP modulating network.
Global correlation analysis for identified protein complexes
In the above analysis, we observed that proteins in complexes showed similar elution profiles in our library. In the next step, we tested if unknown interacting proteins could be identified from this dataset. To this end, we analyzed individual elution profiles for similarities, as this might indicate an interaction. We focused on co-occurrence in early fractions, corresponding to a large app. MW (3000–50 kDa), expected for protein complexes. The elution fractions of 19–24 ml correspond to an apparent relative size of 30–5 kDa. These fractions more likely contain subcomplexes, small complexes, or individual proteins.
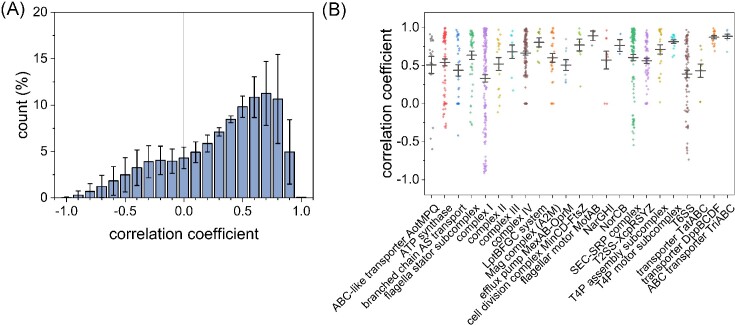
In the nanodisc library, most proteins showed broad elution profiles extending over multiple fractions. The overall similarity of the profiles limits the possibility of detecting coeluting proteins. Therefore, the correlation of the profiles across the entire dataset was tested by global correlation analysis (Fig. S2). Hereby, each protein was compared pairwise to all other proteins by linear regression. Subsequently, a histogram of all correlation coefficients was created to reveal the distribution of the correlation coefficient (Fig. 6A). The correlation coefficients are not normally distributed but shifted toward high correlation. This confirms that a high proportion of proteins show a similar elution profile. Therefore, even high correlation coefficients must be treated with caution and need to be interpreted in the context of the whole dataset.
Figure 6.
Global correlation analysis. (A) All identified proteins were assayed for linear regression toward all other proteins. Correlation was calculated by the Excel data analysis plugin. Correlation coefficients ranging from −1 for negative correlation to 1 for correlation. Histogram depicts the distribution of all calculated correlation coefficients for the three replicates. Error bars indicate the standard deviation. (B) More than 20 described protein complexes were analyzed for co-elution and correlation. Correlation coefficients of all protein pairs for each complex in the three replicates are depicted as dots. Median (black line) and standard deviation.
When analyzing the correlation of known protein complexes, we found that the distribution mirrors the total distribution of the entire dataset (Fig. 6B). The known complexes show no significantly higher correlation toward each other than to random proteins, as the correlation among the whole dataset is shifted to higher similarity. Therefore, prediction of protein–protein interactions from our dataset was not possible with the method used. In addition, a global cluster analysis with Pearson distance measurement was performed, and clusters were analyzed for the described complexes (data not shown). However, the complexes were not sorted more often in the same clusters than random proteins.
Pulldown assay of the nanodisc embedded membrane protein NbdA
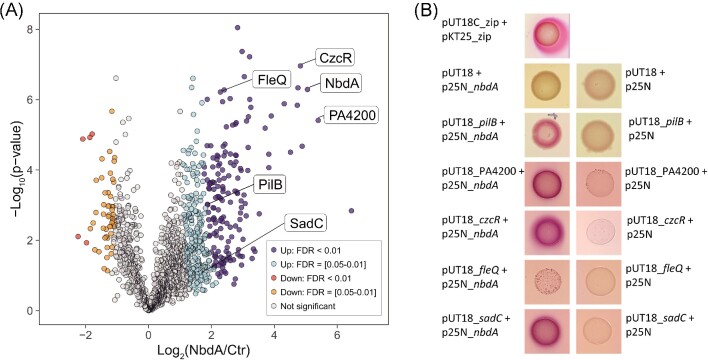
Although global correlation analysis was unable to identify new interaction partners, we could use the soluble library of membrane proteins to identify direct protein–protein interactions using a pulldown assay. As first target protein, we chose the membrane-integrated phosphodiesterase NbdA. The protein consists of an N-terminal MHYT domain consisting of seven transmembrane helices followed by a degenerated GGDEF domain and an active EAL domain in the cytosol. The MHYT domain was postulated to bind diatomic gases such as NO, CO, or O2 (Galperin et al. 2001, Li et al. 2013). The protein was expressed ectopically with a C-terminal strep-tag in P. aeruginosa and used as a bait. Proteins that exhibited a strong enrichment versus the untagged control were designated as candidate interactors (Fig. 7A). In the pulldown sample, we found many proteins specifically enriched (Tab. S4). From 163 significantly enriched proteins, 45% are predicted cytoplasmic proteins, 31% are predicted to reside in the inner membrane, and 10% have unknown localization. Proteins from different COG categories (clusters of orthologous groups) were identified, for instance, energy production and conversion, signal transduction, and transport. Several transcriptional regulators (e.g. CzcR, LasR, and RhlR) involved in regulation, antibiotic resistance and quorum sensing were detected (Gambello et al. 1993, Gilbert et al. 2009, Dieppois et al. 2012). Furthermore, we detected in total 15 proteins involved in the c-di-GMP signaling network (Tab. S4). Among them, the effector FleQ and the GGDEF/EAL protein RcmA, are highly enriched (P > .01). Two membrane proteins, PA4332 (SadC) and PA0861 (RbdA), were found significantly enriched (P > .05). To confirm the protein interaction, BACTH assays were performed. Therefore, NbdA and the candidate protein were genetically fused to two independent subunits of the adenylate cyclase of Bordetella pertussis and co-transformed into the E. coli test strain (Karimova et al. 1998). Robust interaction of two fusion proteins was indicated by an elevated cAMP content in E. coli, resulting in purple colonies on McConkey agar. From the list of highly enriched proteins CzcR, a transcription factor involved in swarming motility, and the hypothetical protein PA4200 were selected for BACTH analyses. A strong positive interaction with NbdA was observed for both proteins (Fig. 7).
Figure 7.
Pulldown approach for protein interactions. (A): MS analysis of NbdA-enriched libraries reveals potential interactors. A Strep-tagged version of NbdA was produced in P. aeruginosa, and a proteome library was prepared from the membrane fraction. NbdA-Strep and interacting proteins were enriched from the library on Strep-Tactin XT resin and compared to controls from strains expressing untagged NbdA. The volcano plot shows the results of a modified Student's t-test with permutation-based FDR control calculated from four replicates in each group (Tusher et al. 2001). Two significance thresholds were applied to call significantly changed proteins (Class A: S0 = 4, FDR <0.01, dark blue and red; Class B: S0 = 4, FDR = [0.05–0.01], light blue and orange). (B): Bacterial adenylate cyclase two-hybrid analysis for the interaction of NbdA and target proteins PilB, PA4200, CzcR, FleQ, and SadC. The target genes of P. aeruginosa were cloned in reporter plasmids generating a gene fusion to the T18 or T25 fragment of the bacterial adenylate cyclase. Escherichia coli BTH101 was cotransformed with these plasmids and monitored for interaction. Purple colonies indicate positive interaction. Plasmids expressing T18 and T25 fragments alone were used as negative controls, pUT18C_zip and pKT25_zip were used as positive controls.
Significantly enriched in the pulldown assay although not among the top hits was the c-di-GMP-responsive transcription factor FleQ (Hickman and Harwood 2008). For this candidate the interaction with NbdA could not be confirmed in the BACTH system (Fig. 7). Among the moderately enriched proteins in the pulldown assay, we also detected the diguanylate cyclase SadC. The BACTH analysis validated the positive interaction of SadC with NbdA (Fig. 7).
Notably, among the potential NbdA interactors we also found several type IV pili components (Tab. S5). One of them was PilB, the type IV pilus ATPase known to promote polymerization of the pilus subunits during attachment and twitching motility of P. aeruginosa (Whitchurch et al. 1991, Turner et al. 1993, Mattick 2002). Although not being one of the top enriched proteins, PilB was robustly detected with 20 peptides and its levels were more than 3.5-fold enriched over those in the untagged control strain. The BACTH test could clearly confirm PilB as an interactor of NbdA (Fig. 7). To demonstrate the reliability of this method, we also tested for interaction with non-enriched proteins. In the same library, the effector protein FimV was detected but was not specifically enriched in the pulldown sample. In the bacterial two-hybrid assay, no interaction was detected between FimV and NbdA (Fig. S3). In summary, these results show that the soluble nanodisc library of the P. aeruginosa membrane proteome can be used to co-purify interaction partners of membrane proteins and, thereby, reveal novel protein-interaction networks.
Discussion
Creating a native membrane-protein library of P. aeruginosa
Membrane proteins and their complexes have been investigated successfully with the use of detergents and liposomes, e. g. the bacterial β-barrel assembly machinery (BAM) complex, and ATP transporters (Hollenstein et al. 2007, Xu et al. 2013, Iadanza et al. 2016, Hu 2021). In the past two decades, nanodiscs have evolved as powerful tools to stabilize membrane proteins in solution, preserving their lipid-bilayer environment. In contrast with nanodiscs formed by membrane scaffolding proteins, amphiphilic polymers can extract membrane proteins directly from cellular membranes without the need of detergents (Knowles et al. 2009, Oluwole et al. 2017). Previous work has shown the increased stability of nanodisc-embedded membrane proteins (Swainsbury et al. 2014, Oluwole et al. 2017, Barniol-Xicota and Verhelst 2018, Pollock et al. 2018, Dilworth et al. 2021). This allowed the elucidation of functions, structures, and interactions of a fast-growing number of membrane proteins and complexes (Dörr et al. 2014, Thoma and Burmann 2020, Dilworth et al. 2021, Janson et al. 2022). Recently, stable proteome libraries from prokaryotic and eukaryotic cell membranes have been generated in polymer nanodiscs (Carlson et al. 2019, Glueck et al. 2022).
The proteome of the cell envelope of P. aeruginosa PAO1 has been thoroughly studied by conventional methods (Nouwens et al. 2000, Blonder et al. 2004, Peng et al. 2005, Imperi et al. 2009, Choi et al. 2011, Dé et al. 2011, Düvel et al. 2012, Casabona et al. 2013, Kumari et al. 2014, Magnowska et al. 2014, Motta et al. 2020). In this work, we used the polymer DIBMA to extract membrane proteins directly from the membrane fraction of P. aeruginosa to generate a stable and soluble nanodisc library. The aim of this approach was to extract membrane proteins along with all peripheral and associated proteins within a lipid-bilayer environment as a basis for the study of complexes and protein–protein interactions under native-like conditions. Therefore, rigorous washing and separation of membrane fractions were omitted. This approach enriches membrane proteins but also results in a high abundance of soluble proteins. The library was then subjected to size exclusion chromatography with subsequent characterization by mass spectrometry. The protein content of the library was highly reproducible, as we found a very good match of data from three biological replicates regarding the identified proteins and co-fractionation pattern.
Comparing the native nanodisc library with results from previous works, the proteome coverage is very similar to data obtained by conventional methods. Of note, not all available proteome data represent the same growth conditions of the cells, though some variation is expected when compared against the previous data. Casabona et al. (2013), identified 991 proteins from inner membrane fractions prepared by sucrose centrifugation and subsequent mass spectrometry, of which 964 proteins were also found in our library. In very similar growth conditions, Kumari et al. identified 2965 proteins from membrane fractions after extraction with harsh detergents and organic solvents (Kumari et al. 2014). In our native nanodisc library, about 86% of these proteins (2556 proteins) were detected and stably extracted. In an in-depth proteomic analysis of P. aeruginosa PAO1, about 2539 membrane-associated proteins were detected from cells in the exponential growth phase (Motta et al. 2020). We were able to identify about 69% of those proteins (1769 out of 2539) in our library and, additionally, 1568 proteins that had not been detected in the previous work. These results show the good extraction capacity of DIBMA for membrane proteins of P. aeruginosa PAO1, enabling a high coverage of the entire membrane proteome within native nanodiscs. The major advantage of the nanodisc library over previous proteome work is that the soluble library preserves a nanoscale lipid-bilayer environment around the extracted proteins but nevertheless is suitable for downstream experimental analyses.
The subcellular localization of the identified proteins was partly reflected in the elution profile from the size exclusion chromatography. In general, it became apparent that all proteins in the library eluted with rather broad size distributions. Furthermore, proteins from the outer membrane eluted very early in SEC, indicating incomplete extraction. The broad size distributions and incomplete extraction into DIBMA nanodiscs have previously been observed and are considered the major bottleneck for the use of this polymer in SEC studies (Glueck et al. 2022). More recent, partially glycosylated polymers such as Glyco-DIBMA (Danielczak et al. 2022), electroneutral polymers such as Sulfo-DIBMA (Glueck et al. 2022, Janson et al. 2022), and certain small-molecule amphiphiles (Mahler et al. 2021) form nanodiscs having much narrower size distributions and, thus, might overcome this limitation in future studies. Nevertheless, the elution profiles of individual components from several known membrane-protein complexes, e.g. the secretion apparatus (SEC-SRP), and the type IV pilus assembly subcomplex (Fig. 4 and 6) were strongly correlated in the present study. Still, global correlation analysis revealed an overall shift of the correlation coefficients to high values (Fig. 6). Thus, the elution profiles from this library could not be used for complexome profiling or de novo interactome prediction since well-correlated protein pairs cannot be distinguished from non-interacting proteins in the dataset. However, the app. MW of nanodiscs in size exclusion chromatography indicates that the proteome library contains very large nanodisc particles likely to contain membrane-protein complexes. Thus, the proteome library enabled the targeted search for membrane-protein interactions by affinity purification combined with mass spectrometry. Previously, a soluble peptidisc library of the E. coli membrane proteome has successfully been used to profile binary interactions in a targeted approach (Carlson et al. 2019).
Identifying interacting proteins in the c-di-GMP signaling network of P. aeruginosa.
It has previously been observed in E. coli that many c-di-GMP modulating proteins are controlled by the formation of regulatory supermodules (Sarenko et al. 2017, Hengge 2021). These supermodules are mainly formed by the c-di-GMP-producing or degrading enzymes, specific effectors, and cellular target structures, e.g. flagellar system (Sarenko et al. 2017, Dahlstrom et al. 2018, Richter et al. 2020). We identified about 29 proteins potentially involved in c-di-GMP synthesis or degradation in the nanodisc library of P. aeruginosa and a further seven c-di-GMP binding effector proteins. Most of the identified proteins are membrane-integrated or likely to be membrane-associated (Table 1). To our knowledge, the dataset presented in this work is thus far the largest set of proteins detected from the c-di-GMP network of P. aeruginosa (Blonder et al. 2004, Düvel et al. 2012, Casabona et al. 2013, Kumari et al. 2014, Bense et al. 2022). In our nanodiscs library, GGDEF/EAL proteins were detected together with their described interaction partner(s) in the overlapping fractions, indicating preservation of these interactions. However, elution peaks were broad and correlation coefficients were upshifted for the whole dataset, which limits the feasibility for de novo predictions.
In the analysis of protein–protein interactions, the stability of the complex is the most critical factor. When dealing with membrane proteins, the extraction process proper and the lipid environment of the membrane proteins after extraction affect the stability of membrane-protein complexes the most (Overduin and Esmaili 2019). To provide experimental evidence for the preservation of protein complexes in the native nanodisc library, we used a pulldown approach coupled with mass spectrometry. Novel candidates for interaction with the phosphodiesterase NbdA could thus be enriched. It should be noted, that with this strategy some of the hits are false positives, e.g. proteins that interact with the membrane material or the polymer itself. The interaction of NbdA with the enriched proteins CzcR, PA4200, SadC, and PilB could be further verified by the BACTH assay. The interaction partner PA4200 is a homologue of the YtnP lipase from B. subtilis. This protein was shown to catalyze the formation and cleavage of long-chain acylglycerols, important for pathogenicity and biofilm formation (Jaeger et al. 1999, Stehr et al. 2003). The novel interaction partner CzcR is part of a two-component system regulating heavy metal efflux and virulence (Dieppois et al. 2012). The physiological role of the interaction of these proteins with NbdA has to be further studied.
NbdA has been reported to be involved in biofilm dispersal, swimming motility, and rhamnolipid production of P. aeruginosa (Li et al. 2013, Xin et al. 2019, Liu et al. 2022). These functions are directly related to the nucleotide second messenger c-di-GMP and the enzymatic activity of NbdA. The diguanylate cyclase SadC is also involved in the complex regulatory network of c-di-GMP. Previously, the interaction of the homologues of SadC and NbdA in P. fluorescens Pf01 was shown by BACTH analyses (Dahlstrom et al. 2018). Furthermore, SadC interacts directly with MotC, the flagellar motor protein and PilT, and the motor for pili retraction. Interestingly, we identified as NbdA interactors SadC as well as the pili assembly motor PilB.
However, whether the interaction of NbdA with these proteins is relevant for the biofilm formation, swimming, pili function, or a different output, will be the subject of further investigation.
Overall, the results in this work demonstrate the usefulness of native nanodisc libraries combining DIBMA extraction and affinity-based pulldown coupled to mass spectrometry for identifying novel protein–protein interactions. With this work, we detected a large number of candidate interactors for the membrane phosphodiesterase NbdA, especially in the c-di-GMP network and verified four candidates by BACTH analyses.
Supplementary Material
Acknowledgements
Miriam Haak, Isabel Schultheiss, Martina Rüger, and Selma Beganović are acknowledged for the help with cloning experiments.
Contributor Information
Anna Scherhag, Department of Microbiology, RPTU Kaiserslautern-Landau, Kaiserslautern 67655, Germany.
Markus Räschle, Department of Molecular Genetics, RPTU Kaiserslautern-Landau, Kaiserslautern 67655, Germany.
Niklas Unbehend, Department of Microbiology, RPTU Kaiserslautern-Landau, Kaiserslautern 67655, Germany.
Benedikt Venn, Department of Computational Systems Biology, RPTU Kaiserslautern-Landau, Kaiserslautern 67655, Germany.
David Glueck, Department of Biophysics, Institute of Molecular Biosciences (IMB), NAWI Graz, University of Graz, Graz 8010, Austria; Department of Field of Excellence BioHealth, University of Graz, Graz 8010, Austria; BioTechMed-Graz, Graz 8010, Austria.
Timo Mühlhaus, Department of Computational Systems Biology, RPTU Kaiserslautern-Landau, Kaiserslautern 67655, Germany.
Sandro Keller, Department of Biophysics, Institute of Molecular Biosciences (IMB), NAWI Graz, University of Graz, Graz 8010, Austria; Department of Field of Excellence BioHealth, University of Graz, Graz 8010, Austria; BioTechMed-Graz, Graz 8010, Austria.
Eugenio Pérez Patallo, Department of Microbiology, RPTU Kaiserslautern-Landau, Kaiserslautern 67655, Germany.
Susanne Zehner, Department of Microbiology, RPTU Kaiserslautern-Landau, Kaiserslautern 67655, Germany.
Nicole Frankenberg-Dinkel, Department of Microbiology, RPTU Kaiserslautern-Landau, Kaiserslautern 67655, Germany.
Funding
This work has been funded by the priority program SPP1879 of the German research foundation to NFD (FR1478/12–2) and by the Landesforschungsinitiative BioComp by the state of Rhineland-Palatine, Germany.
Conflict of interest statement
The authors declare no competing interests.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al. 2022) partner repository with the dataset identifier PXD039702 (nanodisc library) and PXD039700 (pulldown).
References
- Alami M, Dalal K, Lelj-Garolla Bet al. Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. Embo J. 2007;26:1995–2004. 10.1038/sj.emboj.7601661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers M, Sampaleanu LM, Tammam Set al. PilM/N/O/P proteins form an inner membrane complex that affects the stability of the Pseudomonas aeruginosa type IV pilus secretin. J Mol Biol. 2009;394:128–42. 10.1016/j.jmb.2009.09.034. [DOI] [PubMed] [Google Scholar]
- Banerjee P, Sahoo PK, Sheenu AAet al. Molecular and structural facets of c-di-GMP signalling associated with biofilm formation in Pseudomonas aeruginosa. Mol Aspects Med. 2021;81:101001. 10.1016/j.mam.2021.101001. [DOI] [PubMed] [Google Scholar]
- Barniol-Xicota M, Verhelst SHL. Stable and functional rhomboid proteases in lipid nanodiscs by using diisobutylene/maleic acid copolymers. J Am Chem Soc. 2018;140:14557–61. 10.1021/jacs.8b08441. [DOI] [PubMed] [Google Scholar]
- Bayburt TH, Grinkova YV, Sligar SG. Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch Biochem Biophys. 2006;450:215–22. 10.1016/j.abb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Bense S, Witte J, Preuße Met al. Pseudomonas aeruginosa post-translational responses to elevated c-di-GMP levels. Mol Microbiol. 2022;117:1213–26. 10.1111/mmi.14902. [DOI] [PubMed] [Google Scholar]
- Berrisford JM, Baradaran R, Sazanov LA. Structure of bacterial respiratory complex I. Biochim Biophys Acta (BBA)—Bioenergetics. 2016;1857:892–901. 10.1016/j.bbabio.2016.01.012. [DOI] [PubMed] [Google Scholar]
- Blonder J, Goshe MB, Xiao Wet al. Global analysis of the membrane subproteome of Pseudomonas aeruginosa using liquid chromatography-tandem mass spectrometry. J Proteome Res. 2004;3:434–44. 10.1021/pr034074w. [DOI] [PubMed] [Google Scholar]
- Burrows LL. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu Rev Microbiol. 2012;66:493–520. 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- Carlone GM, Thomas ML, Rumschlag HSet al. Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J Clin Microbiol. 1986;24:330–2. 10.1128/jcm.24.3.330-332.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson ML, Stacey RG, Young JWet al. Profiling the Escherichia coli membrane protein interactome captured in peptidisc libraries. Elife. 2019;8. 10.7554/eLife.46615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casabona MG, Vandenbrouck Y, Attree Iet al. Proteomic characterization of Pseudomonas aeruginosa PAO1 inner membrane. Proteomics. 2013;13:2419–23. 10.1002/pmic.201200565. [DOI] [PubMed] [Google Scholar]
- Chen G, Liang H. A novel c-di-GMP signal system regulates biofilm formation in Pseudomonas aeruginosa. MIC. 2020;7:160–1. 10.15698/mic2020.06.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Zhou J, Zuo Yet al. Structural basis for diguanylate cyclase activation by its binding partner in Pseudomonas aeruginosa. Elife. 2021;10:10. 10.7554/eLife.67289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DS, Kim DK, Choi SJet al. Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa. Proteomics. 2011;11:3424–9. 10.1002/pmic.201000212. [DOI] [PubMed] [Google Scholar]
- Claessen D, Emmins R, Hamoen LWet al. Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol Microbiol. 2008;68:1029–46. 10.1111/j.1365-2958.2008.06210.x. [DOI] [PubMed] [Google Scholar]
- Comolli JC, Donohue TJ. Differences in two Pseudomonas aeruginosa cbb3 cytochrome oxidases. Mol Microbiol. 2004;51:1193–203. 10.1046/j.1365-2958.2003.03904.x. [DOI] [PubMed] [Google Scholar]
- Dahlstrom KM, Collins AJ, Doing Get al. A multimodal strategy used by a large c-di-GMP network. J Bacteriol. 2018;200:e00703–17. 10.1128/jb.00703-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielczak B, Rasche M, Lenz J. et al. “A bioinspired glycopolymer for capturing membrane proteins in native-like lipid-bilayer nanodiscs. Nanoscale. 2022;14:1855–67. 10.1039/d1nr03811g. [DOI] [PubMed] [Google Scholar]
- Dé E, Cosette P, Coquet Let al. Membrane proteomes of Pseudomonas aeruginosa and Acinetobacter baumannii. Pathol Biol (Paris). 2011;59:e136–9. 10.1016/j.patbio.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Dieppois G, Ducret V, Caille Oet al. The transcriptional regulator CzcR modulates antibiotic resistance and quorum sensing in Pseudomonas aeruginosa. PLoS One. 2012;7:e38148. 10.1371/journal.pone.0038148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth MV, Findlay HE, Booth PJ. Detergent-free purification and reconstitution of functional human serotonin transporter (SERT) using diisobutylene maleic acid (DIBMA) copolymer. Biochim Biophys Acta (BBA)—Biomembranes. 2021;1863:183602. 10.1016/j.bbamem.2021.183602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–93. 10.1128/cmr.15.2.167-- 193. 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr JM, Koorengevel MC, Schäfer Met al. Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: the power of native nanodiscs. Proc Natl Acad Sci USA. 2014;111:18607–12. 10.1073/pnas.1416205112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düvel J, Bertinetti D, Möller Set al. A chemical proteomics approach to identify c-di-GMP binding proteins in Pseudomonas aeruginosa. J Microbiol Methods. 2012;88:229–36. 10.1016/j.mimet.2011.11.015. [DOI] [PubMed] [Google Scholar]
- Filip C, Fletcher G, Wulff JLet al. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J Bacteriol. 1973;115:717–22. 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming HC, Wingender J, Szewzyk Uet al. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14:563–75. 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- Galperin MY, Gaidenko TA, Mulkidjanian AYet al. MHYT, a new integral membrane sensor domain. FEMS Microbiol Lett. 2001;205:17–23. 10.1111/j.1574-6968.2001.tb10919.x. [DOI] [PubMed] [Google Scholar]
- Gambello MJ, Kaye S, Iglewski BH. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect Immun. 1993;61:1180–4. 10.1128/iai.61.4.1180-1184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina G, Paiardini A, Fernicola Set al. Investigating the allosteric regulation of YfiN from Pseudomonas aeruginosa: clues from the structure of the catalytic domain. PLoS One. 2013;8:e81324. 10.1371/journal.pone.0081324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert KB, Kim TH, Gupta Ret al. Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol Microbiol. 2009;73:1072–85. 10.1111/j.1365-2958.2009.06832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glueck D, Grethen A, Das Met al. Electroneutral polymer nanodiscs enable interference-free probing of membrane proteins in a lipid-bilayer environment. Small. 2022;18:2202492. 10.1002/smll.202202492. [DOI] [PubMed] [Google Scholar]
- Grethen A, Oluwole AO, Danielczak Bet al. Thermodynamics of nanodisc formation mediated by styrene/maleic acid (2:1) copolymer. Sci Rep. 2017;7:11517. 10.1038/s41598-017-11616-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérin A, Sulaeman S, Coquet Let al. Membrane proteocomplexome of Campylobacter jejuni using 2-D blue native/SDS-PAGE combined to bioinformatics analysis. Front Microbiol. 2020;11:530906. 10.3389/fmicb.2020.530906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güvener ZT, Harwood CS. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol. 2007;66:1459–73. 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha DG, O'Toole GA. c-di-GMP and its effects on biofilm formation and dispersion: a Pseudomonas aeruginosa review. Microbiol Spectr. 2015;3:Mb–0003-2014. 10.1128/microbiolspec.MB-0003-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heide H, Bleier L, Steger Met al. Complexome profiling identifies TMEM126B as a component of the mitochondrial complex I assembly complex. Cell Metab. 2012;16:538–49. 10.1016/j.cmet.2012.08.009. [DOI] [PubMed] [Google Scholar]
- Hengge R. High-specificity local and global c-di-GMP signaling. Trends Microbiol. 2021;29:993–1003. 10.1016/j.tim.2021.02.003. [DOI] [PubMed] [Google Scholar]
- Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008;69:376–89. 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci USA. 2005;102:14422–7. 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenstein K, Frei DC, Locher KP. Structure of an ABC transporter in complex with its binding protein. Nature. 2007;446:213–6. 10.1038/nature05626. [DOI] [PubMed] [Google Scholar]
- Hu J. Toward unzipping the ZIP metal transporters: structure, evolution, and implications on drug discovery against cancer. Febs J. 2021;288:5805–25. 10.1111/febs.15658. [DOI] [PubMed] [Google Scholar]
- Hueso-Gil Á, Calles B, de Lorenzo V. The Wsp intermembrane complex mediates metabolic control of the swim-attach decision of Pseudomonas putida. Environ Microbiol. 2020;22:3535–47. 10.1111/1462-2920.15126. [DOI] [PubMed] [Google Scholar]
- Iadanza MG, Higgins AJ, Schiffrin Bet al. Lateral opening in the intact β-barrel assembly machinery captured by cryo-EM. Nat Commun. 2016;7:12865. 10.1038/ncomms12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperi F, Ciccosanti F, Perdomo ABet al. Analysis of the periplasmic proteome of Pseudomonas aeruginosa, a metabolically versatile opportunistic pathogen. Proteomics. 2009;9:1901–15. 10.1002/pmic.200800618. [DOI] [PubMed] [Google Scholar]
- Jaeger KE, Dijkstra BW, Reetz MT. Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu Rev Microbiol. 1999;53:315–51. 10.1146/annurev.micro.53.1.315. [DOI] [PubMed] [Google Scholar]
- Jain R, Sliusarenko O, Kazmierczak BI. Interaction of the cyclic-di-GMP binding protein FimX and the Type 4 pilus assembly ATPase promotes pilus assembly. PLoS Pathog. 2017;13:e1006594. 10.1371/journal.ppat.1006594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson K, Kyrilis FL, Tüting Cet al. Cryo-electron microscopy snapshots of eukaryotic membrane proteins in native lipid-bilayer nanodiscs. Biomacromolecules. 2022;23:5084–94. 10.1021/acs.biomac.2c00935. [DOI] [PubMed] [Google Scholar]
- Jo J, Cortez KL, Cornell WCet al. Elife. 2017;6:e30205. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodaitis L, van Oene T, Martens C. Assessing the role of lipids in the molecular mechanism of membrane proteins. IJMS. 2021,7267, 22. 10.3390/ijms22147267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann Aet al. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A. 1998;95:5752–6. 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Kuroki M, Ishii Met al. Differential expression of multiple terminal oxidases for aerobic respiration in Pseudomonas aeruginosa. Environ Microbiol. 2010;12:1399–412. 10.1111/j.1462-2920.2009.02109.x. [DOI] [PubMed] [Google Scholar]
- Knowles TJ, Finka R, Smith Cet al. Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J Am Chem Soc. 2009;131:7484–5. 10.1021/ja810046q. [DOI] [PubMed] [Google Scholar]
- Komar J, Alvira S, Schulze RJet al. Membrane protein insertion and assembly by the bacterial holo-translocon SecYEG-SecDF-YajC-YidC. Biochem J. 2016;473:3341–54. 10.1042/bcj20160545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krey JF, Wilmarth PA, Shin JBet al. Accurate label-free protein quantitation with high- and low-resolution mass spectrometers. J Proteome Res. 2014;13:1034–44. 10.1021/pr401017h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulasakara H, Lee V, Brencic Aet al. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci USA. 2006;103:2839–44. 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari H, Murugapiran SK, Balasubramanian Det al. LTQ-XL mass spectrometry proteome analysis expands the Pseudomonas aeruginosa AmpR regulon to include cyclic di-GMP phosphodiesterases and phosphoproteins, and identifies novel open reading frames. J Proteomics. 2014;96:328–42. 10.1016/j.jprot.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EY, Choi DS, Kim KPet al. Proteomics in gram-negative bacterial outer membrane vesicles. Mass Spectrom Rev. 2008;27:535–55. 10.1002/mas.20175. [DOI] [PubMed] [Google Scholar]
- Li Y, Heine S, Entian Met al. NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by an MHYT domain-coupled phosphodiesterase. J Bacteriol. 2013;195:3531–42. 10.1128/jb.01156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Xu A, Xie Bet al. Priority changes between biofilm exopolysaccharides synthesis and rhamnolipids production are mediated by a c-di-GMP-specific phosphodiesterase NbdA in Pseudomonas aeruginosa. Iscience. 2022;25:105531. 10.1016/j.isci.2022.105531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Zhai Y, Schneider JCet al. Protein secretion systems of Pseudomonas aeruginosa and P. fluorescens. Biochim Biophys Acta. 2003;1611:223–33. 10.1016/s0005-2736(03)00059-2. [DOI] [PubMed] [Google Scholar]
- Magnowska Z, Hartmann I, Jänsch Let al. Membrane proteomics of Pseudomonas aeruginosa. Methods Mol Biol. 2014;1149:213–24. 10.1007/978-1-4939-0473-0_18. [DOI] [PubMed] [Google Scholar]
- Mahler F, Meister A, Vargas Cet al. Self-assembly of protein-containing lipid-bilayer nanodiscs from small-molecule amphiphiles. Small. 2021;17:2103603. 10.1002/smll.202103603. [DOI] [PubMed] [Google Scholar]
- Malone JG, Jaeger T, Manfredi Pet al. The YfiBNR signal transduction mechanism reveals novel targets for the evolution of persistent Pseudomonas aeruginosa in cystic fibrosis airways. PLoS Pathog. 2012;8:e1002760. 10.1371/journal.ppat.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- Motta S, Vecchietti D, Martorana AMet al. The landscape of Pseudomonas aeruginosa membrane-associated proteins. Cells. 2020,2421, 9. 10.3390/cells9112421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Koch HG, Beck Ket al. Protein traffic in bacteria: multiple routes from the ribosome to and across the membrane. Prog Nucleic Acid Res Mol Biol. 2001;66:107–57. 10.1016/s0079-6603(00)66028-2. [DOI] [PubMed] [Google Scholar]
- Nesper J, Reinders A, Glatter Tet al. A novel capture compound for the identification and analysis of cyclic di-GMP binding proteins. J Proteomics. 2012;75:4874–8. 10.1016/j.jprot.2012.05.033. [DOI] [PubMed] [Google Scholar]
- Nicastro GG, Kaihami GH, Pulschen AAet al. c-di-GMP-related phenotypes are modulated by the interaction between a diguanylate cyclase and a polar hub protein. Sci Rep. 2020;10:3077. 10.1038/s41598-020-59536-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouwens AS, Cordwell SJ, Larsen MRet al. Complementing genomics with proteomics: the membrane subproteome of Pseudomonas aeruginosa PAO1. Electrophoresis. 2000;21:3797–809. . [DOI] [PubMed] [Google Scholar]
- Oluwole AO, Klingler J, Danielczak Bet al. Formation of lipid-bilayer nanodiscs by diisobutylene/maleic acid (DIBMA) copolymer. Langmuir. 2017;33:14378–88. 10.1021/acs.langmuir.7b03742. [DOI] [PubMed] [Google Scholar]
- O'Neal L, Baraquet C, Suo Zet al. The Wsp system of Pseudomonas aeruginosa links surface sensing and cell envelope stress. Proc Natl Acad Sci USA. 2022;119:e2117633119. 10.1073/pnas.2117633119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette SP, Karimova G, Davi Met al. Analysis of membrane protein interactions with a bacterial adenylate cyclase-based two-hybrid (BACTH) technique. Curr Protoc Mol Biol. 2017;118:20.12.1–20.12.24. 10.1002/cpmb.36. [DOI] [PubMed] [Google Scholar]
- Overduin M, Esmaili M. Structures and interactions of transmembrane targets in native nanodiscs. SLAS Discovery. 2019;24:943–52. 10.1177/2472555219857691. [DOI] [PubMed] [Google Scholar]
- Papanastasiou M, Orfanoudaki G, Koukaki Met al. The Escherichia coli peripheral inner membrane proteome. Mol Cell Proteomics. 2013;12:599–610. 10.1074/mcp.M112.024711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Xu C, Ren Het al. Proteomic analysis of the sarcosine-insoluble outer membrane fraction of Pseudomonas aeruginosa responding to ampicilin, kanamycin, and tetracycline resistance. J Proteome Res. 2005;4:2257–65. 10.1021/pr050159g. [DOI] [PubMed] [Google Scholar]
- Perez-Riverol Y, Bai J, Bandla Cet al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50:D543–52. 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock NL, Lee SC, Patel JHet al. Structure and function of membrane proteins encapsulated in a polymer-bound lipid bilayer. Biochim Biophys Acta (BBA)—Biomembranes. 2018;1860:809–17. 10.1016/j.bbamem.2017.08.012. [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–906. 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- Rasamiravaka T, Labtani Q, Duez Pet al. The formation of biofilms by Pseudomonas aeruginosa: a review of the natural and synthetic compounds interfering with control mechanisms. Biomed Res Int. 2015;2015:759348. 10.1155/2015/759348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter AM, Possling A, Malysheva Net al. Local c-di-GMP signaling in the control of synthesis of the E. coli biofilm exopolysaccharide pEtN-cellulose. J Mol Biol. 2020;432:4576–95. 10.1016/j.jmb.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. 10.1128/mmbr.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs JN, Engelman DM. Introduction to the membrane protein reviews: the interplay of structure, dynamics, and environment in membrane protein function. Annu Rev Biochem. 2006;75:707–12. 10.1146/annurev.biochem.75.110105.142336. [DOI] [PubMed] [Google Scholar]
- Sarenko O, Klauck G, Wilke FMet al. More than enzymes that make or break cyclic di-GMP-local signaling in the interactome of GGDEF/EAL domain proteins of Escherichia coli. mBio. 2017;8. 10.1128/mBio.01639-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer T. C-di-GMP synthesis: structural aspects of evolution, catalysis and regulation. J Mol Biol. 2016;428:3683–701. 10.1016/j.jmb.2016.07.023. [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li Net al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42. 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Sobti M, Walshe JL, Wu Det al. Cryo-EM structures provide insight into how E. coli F(1)F(o) ATP synthase accommodates symmetry mismatch. Nat Commun. 2020;11:2615. 10.1038/s41467-020-16387-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehr F, Kretschmar M, Kröger Cet al. Microbial lipases as virulence factors. J Mol Catal B: Enzym. 2003;22:347–55. https://doi.org/. [Google Scholar]
- Stover CK, Pham XQ, Erwin ALet al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–64. 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Sun C, Benlekbir S, Venkatakrishnan Pet al. Structure of the alternative complex III in a supercomplex with cytochrome oxidase. Nature. 2018;557:123–6. 10.1038/s41586-018-0061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swainsbury DJ, Scheidelaar S, van Grondelle Ret al. Bacterial reaction centers purified with styrene maleic acid copolymer retain native membrane functional properties and display enhanced stability. Angew Chem Int Ed. 2014;53:11803–7. 10.1002/anie.201406412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammam S, Sampaleanu LM, Koo Jet al. PilMNOPQ from the Pseudomonas aeruginosa type IV pilus system form a transenvelope protein interaction network that interacts with PilA. J Bacteriol. 2013;195:2126–35. 10.1128/jb.00032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma J, Burmann BM. Fake it 'till you make it—The pursuit of suitable membrane mimetics for membrane protein biophysics. Int J Mol Sci. 2020;22. 10.3390/ijms22010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner LR, Lara JC, Nunn DNet al. Mutations in the consensus ATP-binding sites of XcpR and PilB eliminate extracellular protein secretion and pilus biogenesis in Pseudomonas aeruginosa. J Bacteriol. 1993;175:4962–9. 10.1128/jb.175.16.4962-4969.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–21. 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn Pet al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13:731–40. 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- Ueta T, Kojima K, Hino Tet al. Applicability of styrene-maleic acid copolymer for two microbial rhodopsins, RxR and HsSRI. Biophys J. 2020;119:1760–70. 10.1016/j.bpj.2020.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini M, Filloux A. Biofilms and cyclic di-GMP (c-di-GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria. J Biol Chem. 2016;291:12547–55. 10.1074/jbc.R115.711507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini M, Laventie BJ, Moscoso Jet al. The diguanylate cyclase HsbD intersects with the HptB regulatory cascade to control Pseudomonas aeruginosa biofilm and motility. PLoS Genet. 2016;12:e1006354. 10.1371/journal.pgen.1006354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Strien J, Guerrero-Castillo S, Chatzispyrou IAet al. COmplexome Profiling ALignment (COPAL) reveals remodeling of mitochondrial protein complexes in Barth syndrome. Bioinformatics. 2019;35:3083–91. 10.1093/bioinformatics/btz025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walian PJ, Allen S, Shatsky Met al. High-throughput isolation and characterization of untagged membrane protein complexes: outer membrane complexes of Desulfovibrio vulgaris. J Proteome Res. 2012;11:5720–35. 10.1021/pr300548d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch CB, Hobbs M, Livingston SPet al. Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene. 1991;101:33–44. 10.1016/0378-1119(91)90221-v. [DOI] [PubMed] [Google Scholar]
- Winsor GL, Griffiths EJ, Lo Ret al. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44:D646–53. 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin L, Zeng Y, Sheng Set al. Regulation of flagellar motor switching by c-di-GMP phosphodiesterases in Pseudomonas aeruginosa. J Biol Chem. 2019;294:13789–99. 10.1074/jbc.RA119.009009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Zhang M, Zhao Qet al. Crystal structure of a folate energy-coupling factor transporter from Lactobacillus brevis. Nature. 2013;497:268–71. 10.1038/nature12046. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wang C, Zhou Qet al. Membrane protein stability can be compromised by detergent interactions with the extramembranous soluble domains. Protein Sci. 2014;23:769–89. 10.1002/pro.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al. 2022) partner repository with the dataset identifier PXD039702 (nanodisc library) and PXD039700 (pulldown).