Abstract
We convened an ad hoc International Working Group for Antibody Validation in order to formulate the best approaches for validating antibodies used in common research applications and to provide guidelines that ensure antibody reproducibility. We recommend five conceptual ‘pillars’ for antibody validation to be used in an application-specific manner.
Antibodies are among the most frequently used tools in basic science research and in clinical assays. Despite their widespread use, as well as extensive and valuable discourse in the literature1–6, a comprehensive scientific framework for antibody validation across research applications is lacking. As a result, the quality and consistency of data generated through the use of antibodies vary greatly. This poses an impediment to the rigor and reproducibility that are the cornerstones of the advancement of science.
The extensive discussion of antibody validation in the literature indicates a collective need for standards to validate antibody specificity and reproducibility, as well as a need for adequate reporting practices. For example, in 2010, Bourbeillon and colleagues4 introduced the minimum information about a protein affinity reagent (MIAPAR) proposal. This proposal was meant to formalize a standard for how to report information about affinity binder reagents so that the correct reagent for a particular target could be selected for a specific application. The MIAPAR proposal is a useful guide for this purpose; however, it does not include explicit recommendations for the experimental approaches best suited to support validation of antibody specificity in particular applications.
Immunoreagents are used in a range of applications. According to the antibody reagent portal Antibodypedia (http://www.antibodypedia.com; Supplementary Fig. 1), their most common application is in western blot assays (immunoblotting), followed by immunohistochemistry and immunocytochemistry. In addition, the sandwich assay (e.g., ELISA), although it encompasses only a low percentage of overall antibody use, is an important application from a clinical perspective.
It is essential to note that samples are treated substantially differently in preparation for different antibody-based assays (Supplementary Table 1). Proteins are typically in near-native form for flow cytometry and sandwich assays, but they are wholly or partly denatured for western blot assays, immunohistochemistry, and immunocytochemistry. Because of differences in protein conformation and target accessibility, antibodies that perform well in one context may perform inadequately in others. In addition, the ratio of the target protein to other proteins in a sample may lead to significantly different levels of off-target binding. This is true even if the antibody’s affinity for such proteins is much lower than its affinity for the target protein. Given this complexity it is challenging, if not impossible, to identify a simple and single benchmark for characterizing antibody performance for the full range of possible applications. Indeed, extensive characterization of antibody performance in western blotting may indicate nothing about the performance of the same antibody in an ELISA assay, where the antibody must recognize the epitope within the protein’s native conformation. Likewise, an antibody may specifically recognize a cell surface protein in unfixed hematopoietic cells in flow cytometry but fail to bind the same protein in fixed liver tissue processed for immunohistochemistry. Therefore, approaches for antibody validation must be carried out in an application- and context-specific manner.
The International Working Group for Antibody Validation (IWGAV) was convened as an ad hoc committee of international scientists with diverse research interests but the shared goal of improving standards for antibody use and validation. Here, we propose a set of standard guidelines for validating antibodies, guidelines that may be used in an application-specific manner and that in part take advantage of technologies recently introduced by the genomics and proteomics communities. We suggest five conceptual pillars for validation of antibodies: (i) genetic strategies, (ii) orthogonal strategies, (iii) independent antibody strategies, (iv) expression of tagged proteins, and (v) immunocapture followed by mass spectrometry (MS). We suggest that at least one of these pillars should be used as a minimum criterion for claiming that a particular antibody has been adequately validated for a specific application. The use of multiple strategies would further strengthen this conclusion. We discuss the potential for implementation of our proposal in the concluding remarks.
While we have developed our proposal for conventional immunoreagents, such as polyclonal and monoclonal antibodies, it is also applicable to other recombinant or synthetic binders. We have focused on antibodies toward protein targets, but antibodies directed against post-translationally modified peptides or nonpeptide antigens also constitute an important class of immunoreagents. In some cases, the pillars described here may also be applicable for such reagents. However, we caution that they may require a unique set of strategies for confirming antibody specificity.
Determining antibody specificity
A highly specific antibody recognizes its target with minimal crossreactivity (off-target binding) within a given application and experimental context. The proposed conceptual pillars should provide evidence that an antibody binds its target, and in most cases they should also allow evaluation of potential crossreactivity under the conditions tested. Each strategy will be best suited to a particular series of applications (Table 1). We illustrate how these validation pillars may apply to antibodies used in western blot (Fig. 1). The five proposed conceptual pillars, their characteristics, and their suggested use are described in more detail below.
Table 1 |.
Proposed conceptual pillars for validation of antibodies
| Validation strategy | Genetic | Orthogonal | Independent antibody | Tagged protein expression | IMS |
|---|---|---|---|---|---|
| Validation principle | The expression of the target protein is eliminated or significantly reduced by genome editing or RNA interference | Expression of the target protein is compared with an antibody-independent method | Expression of the target protein is compared using two antibodies with nonoverlapping epitopes | The target protein is expressed using a tag, preferably expressed at endogenous levels | The target protein is captured using an antibody and analyzed using MS |
| Validation criteria | Elimination or significant reduction in antibody labeling after gene disruption or mRNA knockdown | Significant correlation of protein levels detected by an antibody and an orthogonal method (e.g., MS) | Significant correlation of protein levels detected by two different antibodies recognizing independent regions of the same target protein | Significant correlation between antibody labeling and detection of the epitope tag | Target protein peptides among the most abundant detected by MS following immunocapture |
| Suitable for these applications | WB, IHC, ICC, FS, SA, IP/ChIP, RP | WB, IHC, ICC, FS, SA, RP | WB, IHC, ICC, FS, SA, IP/ChIP, RP | WB, IHC, ICC, FS | IP/ChIP |
WB, western blot; IHC, immunohistochemistry; ICC, immunocytochemistry, including immunofluorescence microscopy; FS, flow sorting and analysis of cells; SA, sandwich assays, including ELISA; IP, immunoprecipitation; ChIP, chromatin immunoprecipitation; and RP, reverse-phase protein arrays.
Figure 1 |.
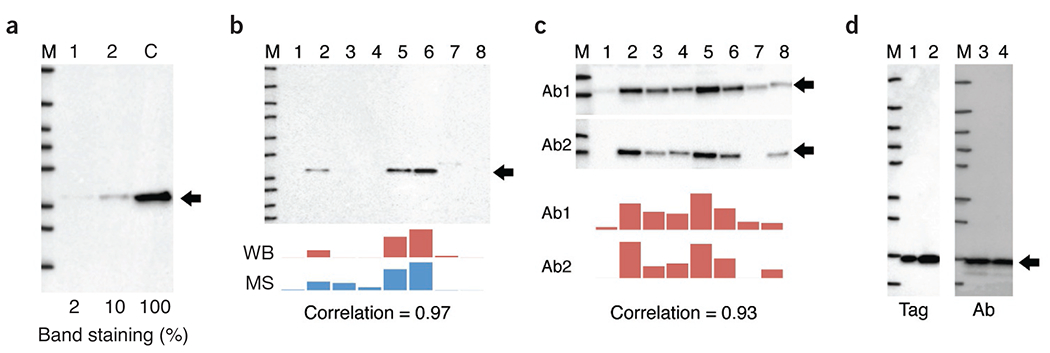
Validation of antibodies in western blot applications using genetic strategies (a), orthogonal strategies (b), independent antibody strategies (c) and tagged protein expression (d). (a) The western blot against human C9orf78 compares siRNA knockdown of protein in cell line (labeled 1 and 2) with control (without siRNA, labeled C). Percent staining of the band is indicated. M, size marker, (b) Simultaneous analysis of human STOM abundance in eight cell lines (labeled 1–8) using western blot (WB) and targeted proteomics (MS, mass spectrometry) resulting in a Pearson correlation of 0.97 (P = 0.00006). (c) Simultaneous western blot analysis of human PRKCA in eight cell lines using two independent antibodies (Ab1 and Ab2, 1 and 2) resulting in a Pearson correlation of 0.93 (P = 0.0009). (d) A fusion of human IL-8 with a peptide tag was stained with either the antibody to be validated (Ab) or a tag-specific antibody (Tag) in cell line samples (labeled 1–4). The black arrows indicate the theoretical sizes of the target protein.
Genetic strategies.
Antibody specificity can be assessed by measuring the relevant signal in control cells or tissues in which the target gene or epitope has been knocked out or knocked down using techniques such as CRISPR–Cas9 or RNA interference (RNAi)1,7–9. In this way, the expression of the target protein is either eliminated or reduced; any signal observed after substantial reduction of protein levels indicates crossreactivity. An example of this strategy is shown in Figure 1a. The protein levels of human C9orf78 are reduced by two independent siRNA molecules; this protein level reduction correlates with substantial reduction in antibody staining (90% and 98% reduction, respectively) in the western blot assay, thus providing evidence for specificity of the antibody. In this approach, the use of genome editing techniques is preferred since they may result in complete loss of protein expression. But gene silencing is also useful, particularly for essential genes.
Genetic approaches are powerful because they provide a direct link between the gene, the target protein, and its detection by the antibody. They are particularly useful for examining antibody specificity for proteins that come from related genes (i.e., members of multigene families). However, genetic strategies cannot be used for some applications and types of samples; in particular, they cannot be used for human tissue samples and body fluids, such as plasma and serum.
Orthogonal strategies.
A general approach to validating antibody specificity is to use an antibody-independent method for target quantification across many samples and then to examine the correlation of this approach with antibody-based target quantification. For example, targeted proteomics approaches using labeled internal standards may be used to quantify relative target protein expression across samples10,11. Antibody labeling should then correlate strongly with protein abundance measured in this way. For this approach, it is important to examine a set of samples with variable expression of the target protein to be confident of specificity and to allow for reliable statistical analysis. For example, the staining by an antibody putatively recognizing the protein STOM in western blot can be compared with measurement of STOM protein abundance from an MS-based targeted proteomics approach (Fig. 1b). The results are highly correlated across eight cell lines, thus validating the antibody for western blot analysis.
Independent antibody strategies.
Two (or more) independent antibodies that recognize the same target maybe used to assess antibody specificity in a range of assays. This approach requires that the expression pattern generated by the two antibodies correlate within a given application environment6. ‘Independent’ means that the two antibodies are able to bind to different regions of the protein; they thus have different epitopes, minimizing the likelihood of off-target binding to the same unrelated protein. Again, it is important to examine a set of samples with variable expression of the target protein, possibly also including cells subjected to knockout or knockdown procedures, to confirm specificity and allow for reliable statistical analysis. Alternatively, samples with a quantifiable spatial expression pattern can be used for imaging-based applications.
One specific example of independent antibody strategies is the proximity ligation assay (PLA), which uses two (or more) antibodies (or other binding reagents) conjugated to complementary DNA probes. If the antibodies bind to the same target within the sample, their proximity allows for ligation, amplification, and quantification via realtime polymerase chain reaction12. Proximity extension assays are based on a similar concept to that underlying PLAs13. Since proximity assays are sensitive and minimize background effects resulting from antibody crossreactivity, they are an attractive option for antibody-based sandwich assays14.
In an example of independent antibody strategies applied to an antibody used for western blotting (Fig. 1c), two antibodies toward the protein PRKCA yield correlated signals across a panel of eight cell lines, suggesting that both antibodies recognize the intended target. It is important to note that for the western blot validation assay, the whole gel should be shown, not only the area around the size of the target proteins. In this way, possible crossreactive bands of sizes other than that of the intended target are also displayed.
Expression of tagged proteins.
Antibodies may be validated by expressing a protein containing an affinity tag (such as FLAG, V5, etc.) or a fluorescent protein (such as green fluorescent protein). This will allow for parallel detection with additional, well-validated immunoreagents or direct observation, respectively. The detection pattern of the antibody being validated must then match the pattern demonstrated by the anti-affinity tag antibody or the fluorescent signal. Substantial discord between these patterns would suggest crossreactivity. For example, the result of probing a tagged fusion protein with an antibody toward IL-8 produces a detection pattern that is similar to that of the tag-specific antibody, thus demonstrating the specificity of the antibody under these conditions (Fig. 1d).
Because antibody performance is highly dependent on target protein concentration, tagged proteins should be expressed at endogenous levels; overexpression of the target protein might mask the detection of off-target binding events. It is therefore preferable to tag the endogenous gene using genome editing tools9,15,16. The key limitations of this method are similar to those of the genetic approaches described above. One must also avoid potential artifacts introduced by the tag itself, such as altered subcellular localization or affected protein activity1,17.
Immunocapture followed by mass spectrometry.
Immunocapture isolates a protein from a solution through binding with a target-specific antibody. This technique may be coupled with MS analysis (IMS) to identify proteins that interact directly with the purified antibody, as well as additional proteins that interact indirectly with the target protein18. Following immunocapture, proteins bound to the purified antibody may be directly digested off the bead, followed by peptide analysis by MS to identify target-specific peptides. When using this approach, we recommend that an antibody be considered specific if the top three peptides are derived from the expected target protein, in accordance with the threshold defined by the Structural Genomics Consortium18.
While IMS is one of the best methods for identifying off-target protein binding (assuming the experiments are carried out quantitatively with appropriate controls), the main limitation is the difficulty in distinguishing direct interactors with the antibody versus proteins that form relevant complexes with the target protein. We recommend IMS as an appropriate strategy to validate antibodies used in applications involving immunocapture (Table 1). Marcon and colleagues18 have reported that IMS may also identify antibodies that can be used in immunofluorescence applications, but it is important to note that some of the antibodies validated to the highest standard with IMS still do not perform adequately in immunofluorescence assays.
Additional methods for antibody characterization.
There are additional methods of antibody characterization that provide biochemical and immunological information about the antibody, including affinity measurements, determination of the DNA sequence of the gene encoding the antibody, isotype determination, epitope mapping, and the use of protein microarrays to confirm target binding (Supplementary Table 2)19. These analyses yield useful information and serve as good starting points for estimating antibody performance. However, they do not directly gauge antibody specificity in an application- or context-specific manner and, as a result, we do not recommend them as primary characterization methods. Another validation method commonly used in immunohisto chemistry applications is adsorption20, in which the antigen is preincubated with the antibody before the assay. Although useful to show target binding, this method will not rule out crossreactivity with proteins containing similar epitopes as the intended target.
We recognize that other experimental approaches may also provide evidence that an antibody binds the intended target. For example, functional assays may provide strong evidence that an antibody interacts with a given target. However, functional analysis does not provide insight into the etiology of off-target effects. While many assays provide information that characterizes antibodies used in research, we recommend using the pillars described above to validate antibodies for use in various applications. A list of references relevant for validation of antibodies can be found in Supplementary Table 3.
Antibody reproducibility
Many factors contribute to the reproducibility of antibody function in research and industrial applications. Ideally, researchers at the bench should be able to identify whether a failure to reproduce published data in their laboratories is based on a valid difference in experimental findings or on the result of changes in production or distribution of research tools. To ensure reproducibility, the reporting of research reagents must be complete and unambiguous.
The Resource Identification Initiative has been created to standardize the reporting of the key resources used in work published in biomedical journals3. The initiative assigns each antibody (including the lot number) with a unique identifier that provides sufficient information for precise identification (https://scicrunch.org/resources/). We recommend the use of Research Resource Identifiers (RRIDs), since they are free to generate, are machine readable, and may be used consistently across publishers and journals. We hope that a wider adoption of this approach may lead to significant improvements in experimental reproducibility.
Another useful identifier for monoclonal antibodies and similar affinity reagents is the sequence of the antibody itself. This would be analogous to known gene or mRNA sequences used in nucleic acid research5. Reporting of sequences would facilitate the unambiguous identification of antibodies and consequently the reproducibility of experimental results. In addition, any reagents with published sequences could be recreated and used in perpetuity. However, the use of fully sequenced reagents will not circumvent the need for diligent characterization and validation, since knowledge of sequence is clearly independent of antibody specificity.
Suggestions for providers and users
The functionality of an antibody is dependent on both application and context. Thus, validation data using a particular cell or tissue extract cannot necessarily be used to prove that the antibody performs equivalently in another cellular context. We therefore recommend that users of antibodies carry out at least one of the validation strategies described here in their own particular application or sample context. We also recommend that users adhere to appropriate reporting guidelines and, at a minimum, include catalog number, lot number, and perhaps RRID3 to ensure that any antibodies used in their research can be unambiguously identified. The current author guidelines for the Journal of Comparative Neurology provide an example of enforcing standards for antibody user reporting21. This journal requires a subsection within the Methods section named “Antibody Characterization,” in which there is a brief description for each antibody used in the reported study that explains how the antibody was characterized (http://www.nature.com/authors/policies/availability.html).
For antibody providers it is preferable to validate each antibody with at least one of the conceptual pillars described here in each application of interest and to cover as many cellular contexts as possible. Providers should also include as much additional information regarding the antibody as possible, such as antibody concentration in characterization assays, concentration of cellular extract or of the immunogen, details of the immunogen used for antibody production, antibody–epitope affinity, antibody isotype, buffer formulation, and the material data sheet. When possible, the standard operating procedures used for validation should be provided, since changing a parameter such as a blocking buffer or detergent may alter reagent performance. Producers should repeat validation experiments for each new lot to ensure that data remain relevant. Antibody identity (such as RRIDs) should supplement the catalog number and lot number3.
Concluding remarks
In this report, we have described a series of scientifically sound approaches for antibody validation. While each of these approaches provides sufficient evidence for validation in a particular application, combinations of multiple validation strategies may increase the confidence in antibody specificity. Our proposal is intended to enable the development of comprehensive guidelines for antibody use. Such guidelines could extend our recommendations by defining experimental best practices for antibody use in specific applications, establishing criteria for the interpretation of data generated using antibodies and setting standards for training students on how to perform antibody-based applications. Although establishing comprehensive guidelines for antibody use may be a longterm goal, adoption and enforcement of our proposal by scientific publishers and funding agencies can, in the near future, help improve research reproducibility and increase the use of well-validated antibodies that specifically recognize their intended target.
We recognize that input from all stakeholders, including funders, publishers, antibody providers, and the research community, will facilitate widespread adoption of our recommendations. Specifically, this wider community can provide critical insight into the timing for adoption of new proposals, the mechanisms used to enforce recommendations, and the specific responsibilities of each stakeholder as recommendations are implemented. Ultimately, we believe that through continued engagement of all stakeholders, comprehensive guidelines that improve the reproducibility of biomedical studies and reduce the amount of time and resources spent on inappropriate immunoreagents are on the horizon.
Supplementary Material
ACKNOWLEDGMENTS
The authors are responsible for all content and editorial derisions and received no honoraria related to the development of this publication. Editorial assistance in the preparation of this publication was provided by Phase Five Communications, supported by Thermo Fisher Scientific, which had no other involvement in the development of this publication.
COMPETING FINANCIAL INTERESTS
M.U. is cofounder of Atlas Antibodies, Affibody Medical, and Antibodypedia. S.C. is a consultant to Biogen and PTM BioLabs Inc. A.B. is the founder of SciCrunch Inc., the technology backing for the Resource Identification Initiative. E.L. acknowledges formal links to Atlas Antibodies. D.L.R. is a consultant to Amgen, Applied Cellular Diagnostics, AstraZeneca, Agendia, Bethyl Labs, Biocept, BMS, Cernostics, FivePrime, Genoptix/Novartis, Metamark Genetics, MDAgree, OptraScan, and Perkin Elmer; he has received honoraria from Genentech/Roche and Ventana; and he acknowledges research support from Cepheid, Genoptix, Gilead Sciences, Kolltan, Perkin Elmer, and Nantomics. D.L.R. holds equity in MD Agree.
Footnotes
Any Supplementary Information and Source Data files are available in the online version of the paper.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Bordeaux J et al. Biotechniques 48, 197–209 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker M Nature 527, 545–551 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Bandrowski A et al. Neuroinformatics 14, 169–182 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourbeillon J et al. Nat. Biotechnol. 28, 650–653 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Bradbury A & Plückthun A Nature 518, 27–29 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Saper CB J. Comp. Neurol 493, 477–478 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Stadler C et al. J. Proteomics 75, 2236–2251 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Olds W & Li J F1000 Res. 5, 308 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanchiswamy CN, Maffei M, Malnoy M, Velasco R & Kim JS Trends Biotechnol. 34, 562–574 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Carr SA et al. Mol. Cell. Proteomics 13, 907–917 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abbatiello SE et al. Mol. Cell. Proteomics 14, 2357–2374 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebai T, Kamali-Moghaddam M & Landegren U Curr. Protoc. Mol. Biol 109, 20.10.1–20.10.25 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Lundberg M, Eriksson A, Tran B, Assarsson E & Fredriksson S Nucleic Acids Res. 39, e102 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juncker D, Bergeron S, Laforte V & Li H Curr. Opin. Chem. Biol 18, 29–37 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Mahen R et al. Mol. Biol. Cell 25, 3610–3618 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sander JD & Joung JK Nat. Biotechnol 32, 347–355 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stadler C et al. Nat. Methods 10, 315–323 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Marcon E et al. Nat. Methods 12, 725–731 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Sjöberg R et al. N. Biotechnol 33, 582–592 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Jositsch G et al. Naunyn Schmiedebergs Arch. Pharmacol 379, 389–395 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Author guidelines for J. Comp. Neurol 10.1002/(ISSN)1096-9861/homepage/ForAuthors.html (accessed 11 April 2016). [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


