Abstract
Lymphoma risk is elevated for relatives with common non-Hodgkin lymphoma (NHL) subtypes, suggesting shared genetic susceptibility across subtypes. To evaluate the extent of mutual heritability among NHL subtypes and discover novel loci shared among subtypes, we analyzed data from eight genome-wide association studies within the InterLymph Consortium, including 10,629 cases and 9,505 controls. We utilized Association analysis based on SubSETs (ASSET) to discover loci for subsets of NHL subtypes and evaluated shared heritability across the genome using Genome-wide Complex Trait Analysis (GCTA) and polygenic risk scores. We discovered 17 genome-wide significant loci (P<5 × 10−8) for subsets of NHL subtypes, including a novel locus at 10q23.33 (HHEX) (P = 3.27 × 10−9). Most subset associations were driven primarily by only one subtype. Genome-wide genetic correlations between pairs of subtypes varied broadly from 0.20 to 0.86, suggesting substantial heterogeneity in the extent of shared heritability among subtypes. Polygenic risk score analyses of established loci for different lymphoid malignancies identified strong associations with some NHL subtypes (P<5 × 10−8), but weak or null associations with others. Although our analyses suggest partially shared heritability and biological pathways, they reveal substantial heterogeneity among NHL subtypes with each having its own distinct germline genetic architecture.
Introduction
Non-Hodgkin lymphoma (NHL) is the most common hematological malignancy worldwide, representing 2.8% of all cancers diagnosed.1 It is comprised of over fifty subtypes with distinct morphologic, genetic, and clinical features.2 Although all lymphomas arise from lymphocytic clones, they have arrested at different stages of development, and the etiology of different subtypes may be similar in some aspects and quite unique in others. Epidemiologic studies show that some environmental, medical, and lifestyle factors are shared across subtypes, but there is also significant heterogeneity in etiology.3 For example, human immunodeficiency virus (HIV) infection is strongly associated with an elevated risk of NHL, especially AIDS-defining NHL subtypes, such as diffuse large B-cell lymphoma (DLBCL), whereas it is not associated with risk of other subtypes, such as mantle cell lymphoma and chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL).4 Family history of lymphoid malignancy is a consistent risk factor for all common NHL subtypes, suggesting a shared genetic component.5 Stronger associations have been observed for first degree relatives with the same NHL subtype, which could reflect some subtype specificity in risk.6
To date, genome-wide association studies (GWAS) have identified over 60 susceptibility loci for specific NHL subtypes, including CLL, DLBCL, follicular lymphoma (FL), and marginal zone lymphoma (MZL).7–18 These studies suggest some common genetic susceptibility regions among subtypes. For example, genetic variants within the human leukocyte antigen (HLA) region are associated with multiple lymphoma subtypes.7, 9, 13, 18–22 The HLA-B*08:01 allele, which is associated with other immune-related diseases,23, 24 is associated with an increased risk of both DLBCL and MZL.7, 9 Outside the HLA region, some genetic loci appear to be shared by NHL subtypes (e.g., chromosome 18q21.33 (BCL2) for CLL and FL),8, 10 but the extent of pleiotropy and shared heritability is unclear. Some NHL loci are also located in regions where variants have been reported for other lymphoid malignancies, such as Hodgkin lymphoma (HL), acute lymphoblastic leukemia (ALL), and multiple myeloma (MM). For example, germline variants at chromosome 9p21.3 (CDKN2A) have been linked to CLL, MM, and ALL,10, 25–27 SNPs within the same linkage disequilibrium block at chromosome 8q24 have been identified for HL, FL, and DLBCL,7, 8, 21 , and a SNP at 6p25.3 (EXOC2) was discovered to be associated with both DLBCL and Waldenström’s macroglobulinemia.7, 28 These observations suggest that there may be shared genetic factors across lymphoid malignancies.
We sought to explore pleiotropy and shared heritability among four common NHL subtypes (CLL, DLBCL, FL, and MZL) and to discover new loci that may be associated with subgroups of NHL or lymphoid malignancies more generally using data from GWAS.7–10 Specifically, we sought to identify new loci that previously had not been identified for any lymphoma subtype, perhaps because they failed to reach genome-wide significance for any one subtype. We also sought to determine the extent to which different NHL subtypes share the same underlying genetic susceptibility. Understanding the genetic architecture of NHL subtypes can provide insight into common biological mechanisms as well as pathways specific to individual subtypes.
Methods
Study Population
To explore pleiotropy across NHL subtypes and discover new loci for NHL susceptibility, we utilized data from eight previous GWAS of NHL within the InterLymph Consortium (Supplementary Table 1).7–10, 13, 18, 29–32 NHL subtype was harmonized centrally at the InterLymph Data Coordinating Center according to the hierarchical classification proposed by the InterLymph Pathology Working Group based on the World Health Organization (WHO) classification (2008).33 Across the eight GWAS, there were 3,100 CLL cases, 3,857 DLBCL cases, 2,847 FL cases, and 825 MZL cases, and 9,505 controls of European ancestry (Supplementary Table 2), providing adequate power to detect moderate effects. All studies obtained informed consent from participants, and the study was approved by the appropriate Institutional Review Boards at each institution. 7–10, 13, 18, 29–32
Genotyping
Genotyping for the eight GWAS was done using Illumina and Affymetrix arrays, and standard quality control metrics were applied to each GWAS (Supplementary Table 3).7–9, 11 Samples with poor call rates, gender discordance, abnormal heterozygosity, or of non-European ancestry were excluded, and SNPs with low call rates or Hardy-Weinberg equilibrium p-value < 1 × 10−6 were removed. Principal components analysis was used to evaluate population stratification for each GWAS (Supplementary Figure 1), and outliers were removed. Imputation was conducted separately for each GWAS using the 1000 Genomes Project version 3 (March 2012 release) as the reference panel. Poorly imputed SNPs (INFO score <0.3) and SNPs with minor allele frequency <1% were excluded from each study, leaving roughly ~8.5 million SNPs for analysis. The genotype data for the NCI NHL GWAS is available at dbGap (phs000802.v2.p1).
Association Testing
Association testing was conducted for each NHL subtype and each GWAS separately using SNPTEST version 2, adjusting for age, sex (except for UCSF1/NHS), and significant principal components (P<0.05 in null model with age and sex). Lambdas for each study are provided in Supplementary Table 3. For NHL subtype with more than one available GWAS, meta-analyses were performed using the fixed effects inverse variance method based on the beta estimates and standard errors from each GWAS. For each previously published susceptibility SNP, we evaluated the risk across the four NHL subtypes.
ASSET: Discovery and Replication of New NHL Loci
To explore pleiotropy among four common NHL subtypes and discover novel loci for unique subsets of NHL subtypes, we utilized Association analysis based on SubSETs (ASSET) analysis, which explores all possible combinations of subsets and chooses the subset with the maximum test statistic (e.g., most significant p-value).34 The statistical significance of the best subset is then adjusted for the optimization (e.g., multiple testing). For the ASSET analysis, we used the summary statistics from the subtype-specific analysis or meta-analysis. We limited the analysis to SNPs with info score (>0.6) and minor allele frequency ≥ 1% and adjusted for the use of shared controls within the analysis. For the discovery, SNPs that were at least 500kb from the index SNP of an established locus for any NHL subtype with a P<1×10−6 were considered potentially novel loci.
Four potential novel loci from the ASSET analysis with P<1×10−6 (rs11187157, rs12127426, rs34517439, rs9421684) were taken forward for replication using TaqMan custom genotyping assays (Applied Biosystems). All four SNPs were well imputed in the discovery with average info scores of 0.78–0.99 across the different SNP arrays. Independent replication of the SNPs was undertaken in 4,468 additional cases, including 1,404 CLL, 1,259 DLBCL, 1,351 FL and 454 MZL cases, and 2,185 controls of European ancestry from four different studies (Supplementary Tables 4 and 5). Genotyping was conducted separately at each study center with appropriate quality control metrics. For each study, association testing was conducted for each subtype and for each subset identified from ASSET, adjusting for age and sex (and Ashkenazi ancestry for MSKCC). The subtype- and subset-specific results from the replication studies were meta-analyzed together and with the discovery results using an inverse variance fixed effects model.
Meta-Analyses: NHL and other lymphoid malignancies
To discover additional loci for NHL and lymphoid malignancies, we conducted a meta-analysis of available GWAS, including the eight NHL GWAS. For GWAS with multiple NHL subtypes using the same set of controls (e.g., NCI NHL, UCSF2), association testing was conducted for all NHL subtypes combined, adjusting for age, sex, and significant principal components, in a single analysis and then meta-analyzed with the other GWAS. In addition, we obtained association results from previous GWAS meta-analyses of MM and HL.35, 36 The MM results included 1,318 cases and 1,480 controls of European ancestry, imputed using the 1000 Genomes Project reference panel. The HL results included 1,816 cases and 7,877 controls of European ancestry, imputed using the HapMap Phase III and 1000 Genomes Project reference panels. The NHL, MM, and HL GWAS were then meta-analyzed using a fixed effects meta-analysis.
Heritability Analyses
To estimate the heritability based on common SNPs (both known and unknown) for individual NHL subtypes and the shared heritability between NHL subtypes, we utilized Genome-wide Complex Trait Analysis (GCTA),37, 38 which quantifies the contribution of a set of SNPs to the heritability of a trait on the liability threshold scale. For this analysis, we used all genotyped SNPs in the NCI NHL GWAS. Additional quality control metrics were implemented to limit cryptic relatedness, and the analysis was adjusted for age, sex, and principal components. For interpretability, we transformed our estimates of heritability on the liability threshold scale to familial relative risks.39
Biological Pathways
To explore potential underlying biological pathways, we used Data-driven Expression Prioritized Integration for Complex Traits (DEPICT),40 which is a method that systematically prioritizes genes, tissues/cell types, and pathways for associated genetic loci based on co-regulation and gene expression data from a large compilation of microarrays. For each of the four NHL subtypes, we used the most significant independent loci with P < 1 × 10−5 from the genome-wide summary statistics (e.g., meta-analysis) and tested for gene, tissue/cell type, and pathway enrichment. We used Functional element Overlap analysis of the Results of Genome Wide Association Study (GWAS) Experiments 2 (FORGE2)41 to evaluate cell type-specific enrichment for regulatory elements for NHL subtype. FORGE2 utilizes epigenetic data from ENCODE, BLUEPRINT and Roadmap and tests for enrichment for overlap with putative functional elements among GWAS SNPs compared to a matched set of background SNPs.
Polygenetic risk score analysis
To further explore pleiotropy across NHL subtypes and other lymphoid malignancies, we generated polygenic risk scores using the established loci for each lymphoid malignancy and tested for association within the eight NHL GWAS. A list of the 119 established loci used for generating the polygenic risk scores can be found in Supplementary Table 6. The polygenic risk scores were calculated by multiplying the reported beta coefficient for each known SNP by the allelic dosage for the SNP and then summing these products across all established SNPs for each subtype or lymphoid malignancy. Logistic regression was used to test the association between each polygenic risk score and each of the four NHL subtypes, adjusting for age, sex, and principal components. Analyses were done separately by subtype and GWAS and then meta-analyzed using a fixed effects model.
Results
ASSET: Discovery and Replication
To evaluate pleiotropy across the four NHL subtypes (CLL, DLBCL, FL, and MZL) and discover new loci, we conducted an analysis using ASSET34 and data from eight genome-wide association studies, including 10,629 cases, and 9,505 controls of European ancestry. We discovered enrichment for small p-values for the best subsets at each SNP (Supplementary Figure 2). This enrichment was driven largely by the established loci for specific subtypes, and removal of SNPs within +/− 500kb of the established loci of the four subtypes resulted in substantial attenuation. A total of 17 loci reached genome-wide significance (P<5 × 10−8) in the ASSET analysis, many of which were driven primarily by one subtype and had been previously reported for that subtype (Supplementary Table 7). The 10q23.1 locus, which was identified for the subset of DLBCL, FL, and MZL, was novel (P=2.40×10−8). Three other promising novel loci with lower significance (P<1×10−6) were also noted at 1p31.1, 1q44 and 10q23.33.
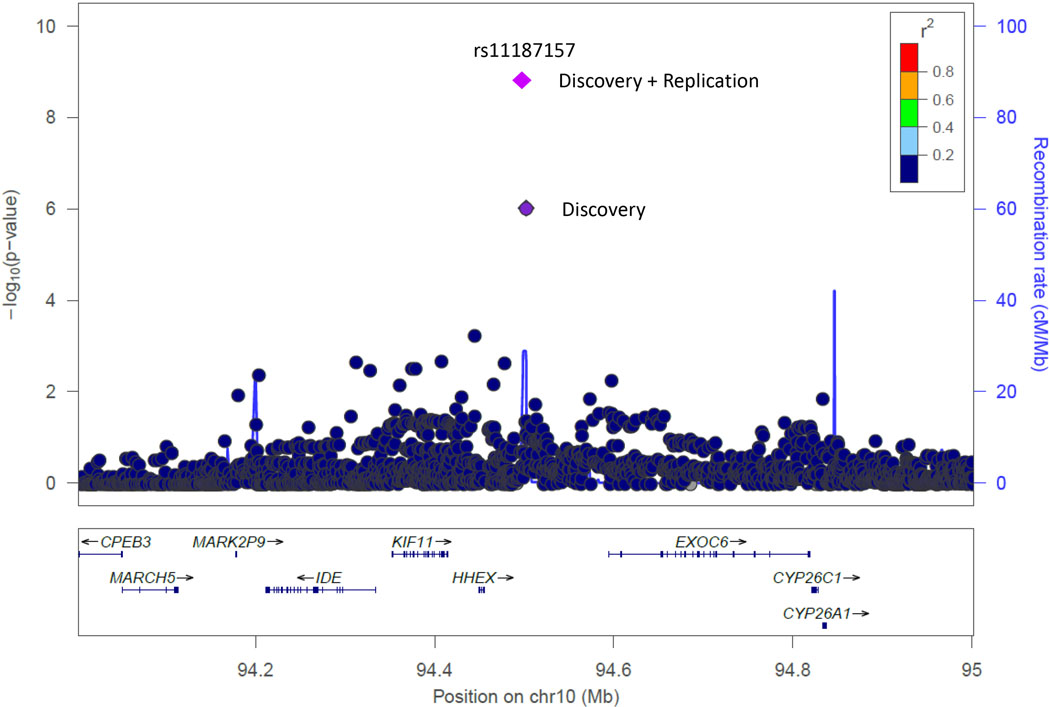
The 10q23.1 locus and the three other promising novel loci (P<1×10−6) were taken forward for replication in an independent set of 4,468 additional cases and 2,185 controls of European ancestry (Supplementary Table 8). Of the four loci, only the 10q23.33 locus (rs11187157) replicated and achieved genome-wide significance in the combined discovery and replication analysis for the identified subset (OR=1.15, 95%CI: 1.10–1.21, P=3.27×10−9) (Table 1, Figure 1). Although ASSET identified the subset containing the three subtypes, CLL, FL, and MZL, as the most significant subset, the replication results suggested that the association was largely driven by CLL (OR=1.27, 95%CI: 1.14–1.40, P=4.36×10−6). Associations for FL and MZL were weaker (OR=1.06, 95% CI: 0.95–3.19, P=0.30, and OR=1.06, 95%C: 0.90–3.41, P=0.47, respectively). The 10q23.33 locus reached genome-wide significance for CLL, independently of the other subtypes, in the combined discovery and replication analysis (OR=1.19, 95% CI: 1.13–1.26, P=2.05×10−10), making it a newly discovered locus for CLL.
Table 1.
Novel pleiotropic locus for a subset of three NHL subtypes (CLL, FL, and MZL)
| Discovery | Replication | Discovery + Replication | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||||
| Cytoband | SNP | Position (GRCh37/hg19) | Risk/Other allele | Subtype | No. Cases/ No. controls | OR | P | No. Cases/ No. controls | OR | P | No. Cases/ No. controls | OR | P |
|
| |||||||||||||
| 10q23.33 | rs11187157 | 94502244 | C/T | CLL | 3,097/7,664 | 1.16 | 4.00E-06 | 1,376/2,142 | 1.27 | 4.36E-06 | 4,473/9,806 | 1.19 | 2.05E-10 |
| FL | 2,845/8,105 | 1.15 | 5.36E-05 | 1,336/2,002 | 1.06 | 0.30 | 4,181/10,107 | 1.12 | 7.84E-05 | ||||
| MZL | 825/6,221 | 1.15 | 0.01 | 431/2,002 | 1.06 | 0.47 | 1,256/8,223 | 1.12 | 0.01 | ||||
| Combined | 6,767/8,800 | 1.16 | 9.70E-07 | 3,143/2,142 | 1.15 | 0.0009 | 9,910/10,942 | 1.15 | 3.27E-09 | ||||
Abbreviations: non-Hodgkin lymphoma (NHL), chronic lymphocytic leukemia (CLL), follicular lymphoma (FL), marginal zone lymphoma (MZL), odds ratio (OR)
Figure 1. Regional association plot of novel locus at chromosome 10q23.33 (rs11187157) for the NHL subset of CLL, FL, and MZL.
Shown are the -log10 association P values from the discovery log-additive genetic model for all SNPs in the region (dots) and rs11187157 (diamond). The lead SNP is shown with results from both the discovery (dark purple diamond) and combined discovery and replication (light purple diamond) analyses. Estimated recombination rates from the 1000 Genome Project are plotted in blue. Locations of recombination hotspots are depicted by peaks corresponding to the rate of recombination. The SNPs surrounding the most significant SNP are color-coded to reflect their r2 correlation with the lead SNP. Pairwise r2 values are from European ancestry subjects in the 1000 Genomes Project. Genes, position of exons and direction of exons and direction of transcription from UCSC genome browser are denoted. Plot was generated using LocusZoom.
Meta-analyses
Although ASSET has greater statistical power if there is heterogeneity among the subtypes, it can have less power than a standard meta-analysis if the associations across subtypes are homogeneous.34 To identify additional new loci for NHL that may have been missed in the ASSET analysis, we conducted a standard meta-analysis of the four NHL subtypes. We identified 15 loci that reached genome-wide significance using this approach (Supplementary Table 9), which is slightly less than what we discovered using ASSET. Thirteen of these loci had been previously reported for at least one NHL subtype, and two had been identified earlier through the ASSET analysis but failed to replicate.
To discover loci for lymphoid malignancies more generally, we further meta-analyzed our NHL results with summary results for MM and HL. We discovered 15 genome-wide significant loci in this larger meta-analysis (Supplementary Table 10). Twelve of these loci had reached genome-wide significance in our NHL meta-analysis, and 13 had been previously reported for at least one lymphoid malignancy. The remaining two loci had been discovered previously through the ASSET analysis but failed to replicate (Supplementary Table 8).
Shared heritability
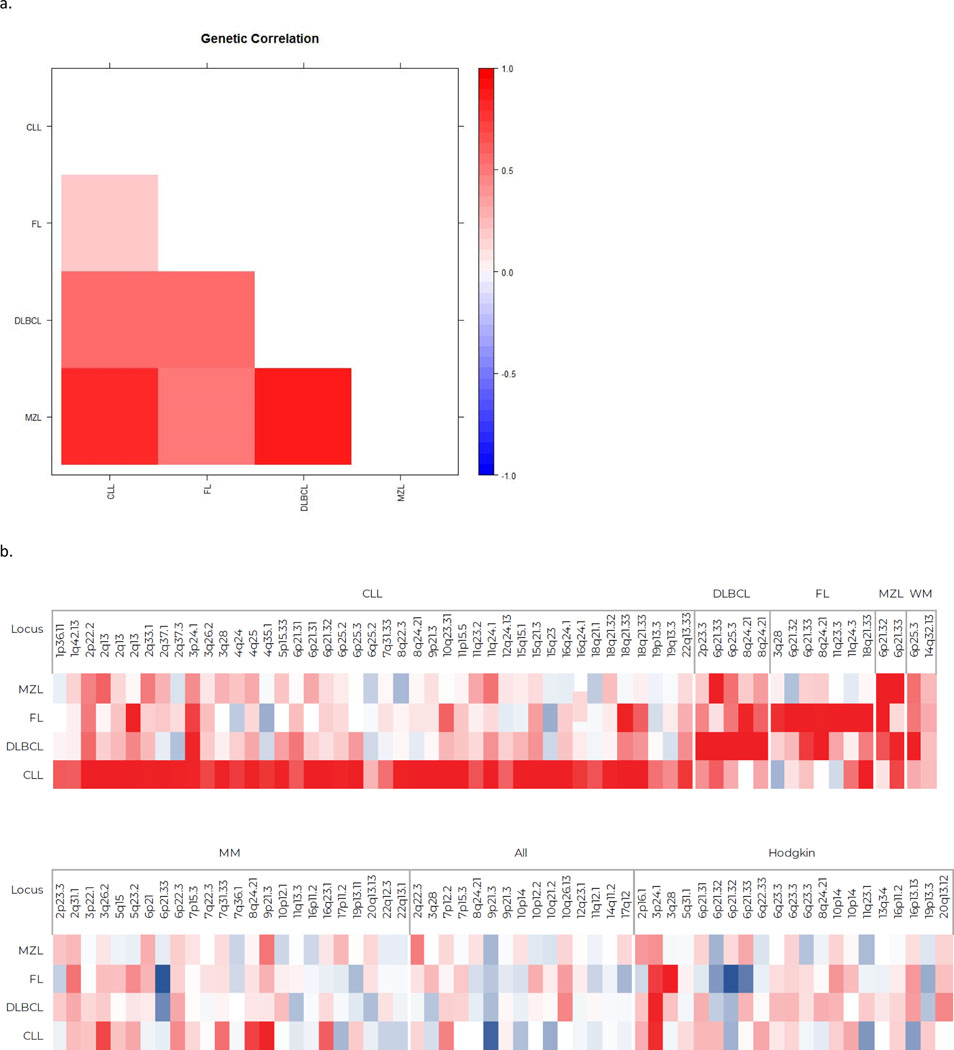
Using GCTA,37, 38 we estimated the heritability based on common SNPs of each of the four NHL subtypes and NHL overall. The estimated heritability ranged from 0.24 (95%CI: 0.18–0.30) for CLL to 0.08 (95% CI: 0–0.19) for MZL with an estimate of 0.10 (95% CI: 0.07–0.14) for the four NHL subtypes combined (Table 2). We transformed our heritability estimates to familial relative risks (FRR) and observed FRRs from 2.47 (95% CI: 2.01–3.00) for CLL to 1.40 (95% CI: 1.15–1.69) for DLBCL. No significant differences in heritability were observed by sex. Common variants (MAF > 20%) contributed more to the heritability for CLL than for MZL (Supplementary Figure 3). Examination of the genetic correlations among the four NHL subtypes revealed a broad range of correlations from 0.20 to 0.86 (Figure 2a, Supplementary Table 11). Significant positive correlations were observed between CLL and MZL (rG=0.70; SE=0.33) and between CLL and DLBCL (rG=0.54; SE=0.26).
Table 2.
Heritability and familial relative risk estimates* for four NHL subtypes, individually and combined
| hL2 (95% CI) | FRR (95% CI) | |
|---|---|---|
|
| ||
| NHL Subtype | ||
|
| ||
| CLL | 0.24 (0.18–0.30) | 2.47 (2.01–3.00) |
| FL | 0.16 (0.10–0.22) | 1.92 (1.54–2.38) |
| DLBCL | 0.09 (0.04–0.15) | 1.40 (1.15–1.69) |
| MZL | 0.08 (0–0.19) | 1.46 (0.84–2.43) |
|
| ||
| NHL Overall | 0.10 (0.07–0.14) | 1.35 (1.21–1.49) |
hL2 is the estimated heritability based on the liability scale. FFR is the estimated familial relative risk.
Abbreviations: non-Hodgkin lymphoma (NHL), chronic lymphocytic leukemia (CLL), follicular lymphoma (FL), diffuse large B-cell lymphoma (DLBCL), marginal zone lymphoma (MZL)
Figure 2. Shared genetic correlations and pleiotropy among four NHL subtypes (CLL, DLBCL, FL, and MZL).
a) Shared genetic correlations based on GCTA analysis. b) Heat plot of directional Z-scores of associations with sentinel SNPs at established genetic loci for individual lymphoid malignancies [e.g., chronic lymphocytic leukemia (CLL), diffuse large B-cell lymphoma (DLBCL), follicular lymphoma (FL), marginal zone lymphoma (MZL), multiple myeloma (MM), acute lymphocytic leukemia (ALL), Hodgkin lymphoma (HL)]. Red color indicates positive association/correlation, and blue indicates inverse or negative association/correlation.
Biological pathways
To explore common biological pathways across the four NHL subtypes, we used DEPICT40 and FORGE2.41 Using FORGE2, we discovered CD20+ cells were enriched for DNase I hotspots for FL and CLL, but a different subgroup of B-cells displayed enrichment for DLBCL and MZL (Supplementary Figure 4), suggesting that distinct subgroups of regulatory elements from different cell types inform NHL etiology. Patterns of gene expression by cell/tissue type also varied across NHL subtypes (Supplementary Figure 5). We observed enrichment for gene expression in multiple cell types and tissues for CLL, including cells in blood and immune system (FDR<0.01). Although we did not find significant cell/tissue enrichment for the other NHL subtypes, nominal associations were observed for T lymphocytes for DLBCL and spleen tissue for MZL among others. When we tested for gene sets using DEPICT, we discovered enrichment for gene sets related to negative T-cell and thymic selection for DLBCL (FDR<0.05) (Supplementary Table 12). No significant gene set enrichment was seen with CLL, FL, or MZL, but nominal associations were observed for antigen processing and presentation, MHC class I receptor activity and apoptosis for CLL.
Pleiotropy Across Lymphoid Malignancies
To explore pleiotropy among NHL subtypes, we examined the associations between the established loci for individual lymphoid malignancies and risk of the four NHL subtypes (e.g. CLL, FL, DLBCL, and MZL). Figure 2b shows a heat plot of the associations with the sentinel SNP at each established locus based on directional z-scores. Apart from CLL (P=0.30), the results for DLBCL, FL and MZL showed more loci with the same direction of effect as previously reported for a different lymphoid malignancy than would be expected by chance (P=1.47×10−7, P=8.98×10−5, and P=0.0002, respectively). All four subtypes displayed more SNPs with the same direction of effect and P<0.05 than expected by chance (P=0.0002, P=4.46×10−7, P=3.26×10−7, and P=0.01 for CLL, DLBCL, FL, and MZL, respectively). After adjustment for multiple testing, 21 SNPs were found to be significantly associated with at least one other NHL subtype in addition to the lymphoid malignancy originally reported (Supplementary Table 6); however, in most cases, this was because the SNP was located near an established locus for that subtype and in linkage disequilibrium. Some potentially novel associations for future follow-up include chromosome 3p24.1 (rs3806624, EOMES) and FL (OR=1.15, 95%CI: 1.08–1.22, P=2.46×10−5) and chromosome 16q23.1 (rs7193541, RFWD3) and CLL (OR=1.11, 95%CI: 1.05–1.19, P=0.0006).
To further explore across lymphoid malignancies, we generated polygenic risk scores comprised of the established loci for each lymphoid malignancy (Supplementary Table 6) and tested for association with risk for the four NHL subtypes (Table 3). Association testing revealed significant shared genetic risk among the DLBCL, FL, and MZL subtypes in particular, but no associations with MM or acute lymphoblastic leukemia. Genome-wide significant positive associations (P<5×10−8) were observed for polygenic risk scores based on the known loci for DLBCL and the risk of FL and MZL and for polygenic risk scores of CLL, FL, MZL, and Waldenström Macroglobulinemia (WM) and the risk of DLBCL. The polygenic risk score comprised of HL loci was inversely associated with FL risk (P=8.20×10−6), largely due to the strong negative association between the HLA risk alleles and FL risk.
Table 3.
Risk of four NHL subtypes associated with polygenic risk scores* for eight lymphoid malignancies
| NHL Subtype | ||||
|---|---|---|---|---|
|
|
||||
| CLL | DLBCL | FL | MZL | |
| OR (95%CI) | OR (95%CI) | OR (95%CI) | OR (95%CI) | |
|
| ||||
| PRS based on 43 CLL SNPs | 2.17 (2.07–2.28) | 1.17 (1.12–1.22) | 1.12 (1.07–1.17) | 1.15 (1.07–1.24) |
| 3.42E-222 | 1.26E-14 | 1.01E-06 | 0.0002 | |
| PRS based on 5 DLBCL SNPs | 1.33 (1.14–1.54) | 2.69 (2.35–3.08) | 1.66 (1.42–1.94) | 2.10 (1.63–2.72) |
| 0.0002 | 2.53E-46 | 2.02E-10 | 1.25E-08 | |
| PRS based on 7 FL SNPs | 1.07 (0.98–1.17) | 1.28 (1.19–1.39) | 2.77 (2.52–3.04) | 0.94 (0.81–1.09) |
| 0.12 | 4.97E-10 | 2.15E-100 | 0.41 | |
| PRS based on 2 MZL SNPs | 1.26 (1.09–1.46) | 1.53 (1.34–1.75) | 1.39 (1.21–1.61) | 2.43 (1.93–3.06) |
| 0.002 | 7.27E-10 | 6.48E-06 | 4.00E-14 | |
| PRS based on 2 WM SNPs | 1.07 (1.01–1.14) | 1.24 (1.18–1.31) | 1.12 (1.05–1.19) | 1.18 (1.07–1.30) |
| 0.02 | 3.35E-16 | 0.0007 | 0.0009 | |
| PRS based on 24 MM SNPs | 1.09 (1.02–1.16) | 0.98 (0.93–1.04) | 1.01 (0.95–1.09) | 1.05 (0.94–1.18) |
| 0.01 | 0.56 | 0.70 | 0.38 | |
| PRS based on 15 ALL SNPs | 0.95 (0.90–1.00) | 0.99 (0.94–1.04) | 1.01 (0.95–1.06) | 1.02 (0.93–1.12) |
| 0.06 | 0.62 | 0.87 | 0.62 | |
| PRS based on 21 HL SNPs | 1.02 (0.97–1.08) | 1.07 (1.02–1.13) | 0.88 (0.83–0.93) | 1.05 (0.96–1.16) |
| 0.43 | 0.006 | 8.20E-06 | 0.28 | |
Polygenic risk scores (PRS) based on previously reported loci for eight lymphoid malignancies [chronic lymphocytic leukemia (CLL), diffuse large B-cell lymphoma (DLBCL), follicular lymphoma (FL), marginal zone lymphoma (MZL), Waldenström’s macroglobulinemia (WM), multiple myeloma (MM), pediatric acute lymphoblastic leukemia (ALL), and Hodgkin lymphoma (HL)]. Odds ratios, 95% confidence intervals and p-values are provided. Bold indicates significance after adjustment for multiple testing.
Discussion
In this large international collaborative effort within the InterLymph Consortium, we provide the first comprehensive evaluation of pleiotropy among four common NHL subtypes. We demonstrate that there is some pleiotropy and shared heritability among NHL subtypes; however, each subtype appears to have its own distinct genetic architecture. None of the genetic loci identified to date appear to be associated with all four NHL subtypes. Analyses including other common lymphoid malignancies, MM and HL, further support the hypothesis that genetic susceptibility varies by subtype.
We identified one novel locus at chromosome 10q23.33 (rs11187157) for a subset of NHL subtypes; however, the association was strongest and genome-wide significant for CLL risk. rs11187157 is located approximately 42kb downstream of the hematopoietically expressed homeobox (HHEX) gene and 88kb upstream of the exocyst complex component 6 (EXOC6) gene. In animal models, HHEX is an important regulator of hematopoietic development and is necessary for the maturation and proliferation of the earliest definitive hematopoietic progenitors.42 HHEX has been shown to be critical in lymphopoiesis43 and differentially active in naïve B-cells, germinal center B-cells, and memory B-cells.44 HHEX is overexpressed in leukemia45 and lymphoma44 cell lines. Studies in acute myeloid leukemia suggest its aberrant expression may contribute to disease pathogenesis through multiple mechanisms including differentiation blockade and by fostering epigenetic repression of the CDKN2A tumor suppressor locus.46 Although rs11187157 may not be the functional genetic variant responsible for the association, it lies in a DNase 1 hypersensitivity site for multiple cell lines, including CD20+ (normal B cell), CD14+ (monocytes), mobilized CD34+ hematopoietic progenitor cells, many HapMap B-cell lymphoblastoid lines, and 3 leukemia cells lines (CLL, HL-60 and NB4 promyelocytic leukemia). Rs11187157 is significantly associated with HHEX gene expression in lymphoblastoid cell lines47 and blood.48 In addition, it resides in a transcription binding site for many transcription factors, including IRF4 in B-lymphocyte lymphoblastoid lines, and SNPs in IRF4 have previously been identified as associated with CLL and HL.15, 49 Moderate signals for histones H3K4Me1 and H3K27ac in the general region indicate the possibility for an enhancer role.
Our findings of distinctly different patterns of association with some shared heritability are consistent with observational studies of environmental and lifestyle risk factors, which suggest some common risk factors but substantial heterogeneity among NHL subtypes with some risk factors being subtype-specific.3 Although 17 loci reached genome-wide significance in our ASSET analysis and 15 loci were genome-wide significant in our meta-analysis, most of these were driven primarily by one subtype. Those with nominally significant contributions by more than one subtype included 2q13 (ACOXL/BCL2L11), 3p24.1 (EOMES), 3q13.33 (CD86), 6p21.32 (HLA-DQA1), 8q24.21 (PVT1), and 18q21.33 (BCL2). Although the 2q13 locus had been previously identified for CLL, the ASSET analysis revealed that the subset including MZL was significant. The association with MZL may be spurious; however, BCL2L11, which encodes the pro-apoptotic protein Bim, has been shown to be deregulated in CLL and MZL.50 The most significant SNP at 3q13.33 in our ASSET analysis was rs2681416, which failed to replicate for both DLBCL and FL in our previous GWAS.7, 8 Another SNP at 3q13.33, rs9831894, which is only modestly correlated with rs2681416 (r2=0.23), also reached genome-wide significance (P=1.93×10−9) in our ASSET analysis with both DLBCL and FL contributing to risk. We recently replicated the observed association between rs9831894 at 3q13.33 and DLBCL risk in an independent set of cases and controls.51 rs9831894 is located near CD86, which encodes a member of the immunoglobulin superfamily that negatively regulates T-cell activation by binding to cytotoxic T-lymphocyte-associated protein 4 and augments B-cell activity.52
Similar to our study, Law et al. used ASSET to examine pleiotropy between CLL, multiple myeloma, and Hodgkin lymphoma and reported one novel locus associated with opposing risk associations for CLL and Hodgkin lymphoma.53 We did not observe evidence for this locus for CLL (rs11715604, P=0.69) or Hodgkin lymphoma (rs13075615, r2=0.81, P=0.92) in our study; no association was observed for the other NHL subtypes (Supplementary Table 6). We were unable to include MM and HL in our ASSET analysis; however, we were able to conduct a meta-analysis of MM, HL, and four common NHL subtypes. Our meta-analysis yielded 15 genome-wide significant loci for lymphoid malignancies. Most loci had previously been identified for at least one subtype, suggesting little discovery gain by combining subtypes.
Examination of individual associations with published loci for lymphoid malignancies showed more SNPs with the same direction of effect and P<0.05 than would be expected by chance for the four NHL subtypes. These findings are consistent with the study by Went et al. that suggested shared risk loci between CLL and MM may be enriched for B-cell regulatory elements.54 Polygenic risk score analyses with established NHL loci showed genome-wide significant associations for multiple NHL subtypes, suggesting significant pleiotropy; however, the magnitude of the risks varied among subtypes. We observed very little or no association with risk scores based on the established loci for ALL, HL, and MM, suggesting more limited pleiotropy with other lymphoid malignancies.
Heritability analyses revealed a broad range of genetic correlations between NHL subtype pairs ranging from 0.20 to 0.86, suggesting some shared heritability among subtypes, but substantial etiologic differences as well. If the genetic etiology of all four NHL subtypes was highly shared, one might expect all genetic correlations to be >0.7 or 0.8, but we did not find this to be true. The positive genetic correlations were statistically significant between CLL and MZL and between CLL and DLBCL, the latter of which was previously reported.39 These findings suggest that there may be some shared biological pathways for these subtypes. We were unable to estimate the shared genetic correlation with other lymphoid malignancies using LD score regression due to relatively small sample sizes (N<10,000 cases), but partitioning heritability by regulatory markers might yield additional insight.
Our analysis was limited to participants of European ancestry, so the results may not be generalizable to other populations. A previous GWAS reported an association at chromosome 3q27 and risk of B-cell lymphoma in Chinese.55 We observed a nominal association between rs6773854 and B-cell lymphoma risk in our meta-analysis of four NHL subtypes (P=0.01). Our analysis of NHL was limited to four common B-cell subtypes and may not be reflective of B-cell lymphoma risk in the general population. However, these four subtypes comprise the vast majority of NHL cases, so the bias is likely small. Our study also suggests that many loci are subtype-specific and so unbiased estimates of associations for B-cell lymphoma may be of less importance. Finally, our results assume that these four subtypes are homogeneous; however, there may etiologically distinct molecular or biologic subtypes in these groups, such as by cell of origin or MYC status for DLBCL. Further subtyping may reveal additional heterogeneity in etiology.
In conclusion, our evaluation of the genetic etiology of NHL demonstrated that there is shared heritability and pleiotropy among common NHL subtypes (i.e., CLL, FL, DLBCL, MZL); however, many of the loci identified in our ASSET analysis and B-cell meta-analyses appeared to be driven primarily by one specific subtype. Indeed, the novel locus we discovered for a subset of NHL subtypes at chromosome 10q23.33 was strongly associated with CLL, in particular. Although additional studies are needed to fully elucidate the genetic architecture of NHL, our study suggests that genetic susceptibility to NHL is complex with some overlapping loci but with substantial heterogeneity among subtypes for common variants. This is consistent with studies of environment and lifestyle risk factors and specific NHL subtypes. Future studies are needed to further clarify which exogenous and genetic risk factors contribute to the etiology of multiple NHL subtypes, which are subtype-specific, and what are the underlying biological mechanisms of each pattern. Further, larger studies will be able to investigate pleiotropy with rarer variants and rarer NHL subtypes.
Supplementary Material
Acknowledgements
This study was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH. The funders had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication. The authors thank Mr. William Wheeler (Information Management Services, Inc.) for his analytic support. A complete list of funding sources and acknowledgements for individual studies is listed in the Supplementary Material.
Footnotes
Disclaimers
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. Where authors are identified as personnel of the International Agency for Research on Cancer / World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer / World Health Organization.
Competing Interests:
Tait Shanafelt received research support to his institution from Genetech, Pharacyclics, AbbVie, Cephalon, Hospira, GlaxoSmithKline, Polyphenon E International, Merck, and Celgene and holds a patent (US14/292,075) on green tea extract epigallocatechin gallate in combination with chemotherapy for chronic lymphocytic leukemia. Karin Smedby received research funding from Janssen Pharmaceuticals AB for research unrelated to this project. Corinne Haioun received honoraria from Novartis, Amgen, Servier/Pfizer, and Gilead Sciences, acted as a consultant or advisor to Roche, Celgene, Janssen-Cilag, Gilead Sciences, Takeda, Miltenyi Biotec, Abbvie, and ADC Therapeutics, and received travel, accommodations and/or expenses from Roche, Celgene, and Amgen. Kenan Onel is currently a full-time employee at Sema4. The other authors declare no competing interests.
Data Availability
Genotype data from the NCI NHL GWAS is available on dbGaP (phs000801.v2.p1) for research purposes in accordance with dbGaP data access policies. Other data in this manuscript is available for shared research purposes through the InterLymph Consortium upon approval in accordance with institutional review boards and general data protection regulations.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2.Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morton LM, Slager SL, Cerhan JR, Wang SS, Vajdic CM, Skibola CF, et al. Etiologic heterogeneity among non-Hodgkin lymphoma subtypes: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monogr. 2014;2014(48):130–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibson TM, Morton LM, Shiels MS, Clarke CA, Engels EA. Risk of non-Hodgkin lymphoma subtypes in HIV-infected people during the HAART era: a population-based study. AIDS. 2014;28(15):2313–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang SS, Slager SL, Brennan P, Holly EA, De Sanjose S, Bernstein L, et al. Family history of hematopoietic malignancies and risk of non-Hodgkin lymphoma (NHL): a pooled analysis of 10 211 cases and 11 905 controls from the International Lymphoma Epidemiology Consortium (InterLymph). Blood. 2007;109(8):3479–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sud A, Chattopadhyay S, Thomsen H, Sundquist K, Sundquist J, Houlston RS, et al. Analysis of 153 115 patients with hematological malignancies refines the spectrum of familial risk. Blood. 2019;134(12):960–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerhan JR, Berndt SI, Vijai J, Ghesquieres H, McKay J, Wang SS, et al. Genome-wide association study identifies multiple susceptibility loci for diffuse large B cell lymphoma. Nat Genet. 2014;46(11):1233–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skibola CF, Berndt SI, Vijai J, Conde L, Wang Z, Yeager M, et al. Genome-wide association study identifies five susceptibility loci for follicular lymphoma outside the HLA region. Am J Hum Genet. 2014;95(4):462–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vijai J, Wang Z, Berndt SI, Skibola CF, Slager SL, de SS, et al. A genome-wide association study of marginal zone lymphoma shows association to the HLA region. Nat Commun. 2015;6:5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berndt SI, Skibola CF, Joseph V, Camp NJ, Nieters A, Wang Z, et al. Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat Genet. 2013;45(8):868–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berndt SI, Camp NJ, Skibola CF, Vijai J, Wang Z, Gu J, et al. Meta-analysis of genome-wide association studies discovers multiple loci for chronic lymphocytic leukemia. Nat Commun. 2016;7:10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Law PJ, Berndt SI, Speedy HE, Camp NJ, Sava GP, Skibola CF, et al. Genome-wide association analysis implicates dysregulation of immunity genes in chronic lymphocytic leukaemia. Nat Commun. 2017;8:14175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conde L, Halperin E, Akers NK, Brown KM, Smedby KE, Rothman N, et al. Genome-wide association study of follicular lymphoma identifies a risk locus at 6p21.32. Nat Genet. 2010;42(8):661–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crowther-Swanepoel D, Broderick P, Di Bernardo MC, Dobbins SE, Torres M, Mansouri M, et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat Genet. 2010;42(2):132–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Bernardo MC, Crowther-Swanepoel D, Broderick P, Webb E, Sellick G, Wild R, et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2008;40(10):1204–10. [DOI] [PubMed] [Google Scholar]
- 16.Slager SL, Skibola CF, Di Bernardo MC, Conde L, Broderick P, McDonnell SK, et al. Common variation at 6p21.31 (BAK1) influences the risk of chronic lymphocytic leukemia. Blood. 2012;120(4):843–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Speedy HE, Di Bernardo MC, Sava GP, Dyer MJ, Holroyd A, Wang Y, et al. A genome-wide association study identifies multiple susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2014;46(1):56–60. [DOI] [PubMed] [Google Scholar]
- 18.Slager SL, Rabe KG, Achenbach SJ, Vachon CM, Goldin LR, Strom SS, et al. Genome-wide association study identifies a novel susceptibility locus at 6p21.3 among familial CLL. Blood. 2011;117(6):1911–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skibola CF, Bracci PM, Halperin E, Conde L, Craig DW, Agana L, et al. Genetic variants at 6p21.33 are associated with susceptibility to follicular lymphoma. Nat Genet. 2009;41(8):873–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chubb D, Weinhold N, Broderick P, Chen B, Johnson DC, Forsti A, et al. Common variation at 3q26.2, 6p21.33, 17p11.2 and 22q13.1 influences multiple myeloma risk. Nat Genet. 2013;45(10):1221–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enciso-Mora V, Broderick P, Ma Y, Jarrett RF, Hjalgrim H, Hemminki K, et al. A genome-wide association study of Hodgkin’s lymphoma identifies new susceptibility loci at 2p16.1 (REL), 8q24.21 and 10p14 (GATA3). Nat Genet. 2010;42(12):1126–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urayama KY, Jarrett RF, Hjalgrim H, Diepstra A, Kamatani Y, Chabrier A, et al. Genome-wide association study of classical Hodgkin lymphoma and Epstein-Barr virus status-defined subgroups. J Natl Cancer Inst. 2012;104(3):240–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanscombe KB, Morris DL, Noble JA, Dilthey AT, Tombleson P, Kaufman KM, et al. Genetic fine mapping of systemic lupus erythematosus MHC associations in Europeans and African Americans. Hum Mol Genet. 2018;27(21):3813–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller FW, Chen W, O’Hanlon TP, Cooper RG, Vencovsky J, Rider LG, et al. Genome-wide association study identifies HLA 8.1 ancestral haplotype alleles as major genetic risk factors for myositis phenotypes. Genes Immun. 2015;16(7):470–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell JS, Li N, Weinhold N, Forsti A, Ali M, van Duin M, et al. Genome-wide association study identifies multiple susceptibility loci for multiple myeloma. Nat Commun. 2016;7:12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherborne AL, Hosking FJ, Prasad RB, Kumar R, Koehler R, Vijayakrishnan J, et al. Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nat Genet. 2010;42(6):492–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu H, Zhang H, Yang W, Yadav R, Morrison AC, Qian M, et al. Inherited coding variants at the CDKN2A locus influence susceptibility to acute lymphoblastic leukaemia in children. Nat Commun. 2015;6:7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McMaster ML, Berndt SI, Zhang J, Slager SL, Li SA, Vajdic CM, et al. Two high-risk susceptibility loci at 6p25.3 and 14q32.13 for Waldenstrom macroglobulinemia. Nat Commun. 2018;9(1):4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smedby KE, Foo JN, Skibola CF, Darabi H, Conde L, Hjalgrim H, et al. GWAS of follicular lymphoma reveals allelic heterogeneity at 6p21.32 and suggests shared genetic susceptibility with diffuse large B-cell lymphoma. PLoS Genet. 2011;7(4):e1001378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schumacher FR, Berndt SI, Siddiq A, Jacobs KB, Wang Z, Lindstrom S, et al. Genome-wide association study identifies new prostate cancer susceptibility loci. Hum Mol Genet. 2011;20(19):3867–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siddiq A, Couch FJ, Chen GK, Lindstrom S, Eccles D, Millikan RC, et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum Mol Genet. 2012;21(24):5373–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Vivo I, Prescott J, Setiawan VW, Olson SH, Wentzensen N, Australian National Endometrial Cancer Study G, et al. Genome-wide association study of endometrial cancer in E2C2. Hum Genet. 2014;133(2):211–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner JJ, Morton LM, Linet MS, Clarke CA, Kadin ME, Vajdic CM, et al. InterLymph hierarchical classification of lymphoid neoplasms for epidemiologic research based on the WHO classification (2008): update and future directions. Blood. 2010;116(20):e90–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhattacharjee S, Rajaraman P, Jacobs KB, Wheeler WA, Melin BS, Hartge P, et al. A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. Am J Hum Genet. 2012;90(5):821–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rand KA, Song C, Dean E, Serie DJ, Curtin K, Sheng X, et al. A Meta-analysis of Multiple Myeloma Risk Regions in African and European Ancestry Populations Identifies Putatively Functional Loci. Cancer Epidemiol Biomarkers Prev. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cozen W, Timofeeva MN, Li D, Diepstra A, Hazelett D, Delahaye-Sourdeix M, et al. A meta-analysis of Hodgkin lymphoma reveals 19p13.3 TCF3 as a novel susceptibility locus. Nat Commun. 2014;5:3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28(19):2540–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sampson JN, Wheeler WA, Yeager M, Panagiotou O, Wang Z, Berndt SI, et al. Analysis of Heritability and Shared Heritability Based on Genome-Wide Association Studies for Thirteen Cancer Types. J Natl Cancer Inst. 2015;107(12):djv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pers TH, Karjalainen JM, Chan Y, Westra HJ, Wood AR, Yang J, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. 2015;6:5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Breeze CE, Haugen E, Reynolds A, Teschendorff A, van Dongen J, Lan Q, et al. Integrative analysis of 3604 GWAS reveals multiple novel cell type-specific regulatory associations. Genome Biol. 2022;23(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paz H, Lynch MR, Bogue CW, Gasson JC. The homeobox gene Hhex regulates the earliest stages of definitive hematopoiesis. Blood. 2010;116(8):1254–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jackson JT, Nasa C, Shi W, Huntington ND, Bogue CW, Alexander WS, et al. A crucial role for the homeodomain transcription factor Hhex in lymphopoiesis. Blood. 2015;125(5):803–14. [DOI] [PubMed] [Google Scholar]
- 44.Nagel S, MacLeod RAF, Meyer C, Kaufmann M, Drexler HG. NKL homeobox gene activities in B-cell development and lymphomas. PLoS One. 2018;13(10):e0205537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song JH, Kim HJ, Lee CH, Kim SJ, Hwang SY, Kim TS. Identification of gene expression signatures for molecular classification in human leukemia cells. Int J Oncol. 2006;29(1):57–64. [PubMed] [Google Scholar]
- 46.Jackson JT, Ng AP, Shields BJ, Haupt S, Haupt Y, McCormack MP. Hhex induces promyelocyte self-renewal and cooperates with growth factor independence to cause myeloid leukemia in mice. Blood Adv. 2018;2(4):347–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501(7468):506–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vosa U, Claringbould A, Westra HJ, Bonder MJ, Deelen P, Zeng B, et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet. 2021;53(9):1300–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Broderick P, Cunningham D, Vijayakrishnan J, Cooke R, Ashworth A, Swerdlow A, et al. IRF4 polymorphism rs872071 and risk of Hodgkin lymphoma. Br J Haematol. 2010;148(3):413–5. [DOI] [PubMed] [Google Scholar]
- 50.Tessoulin B, Papin A, Gomez-Bougie P, Bellanger C, Amiot M, Pellat-Deceunynck C, et al. BCL2-Family Dysregulation in B-Cell Malignancies: From Gene Expression Regulation to a Targeted Therapy Biomarker. Front Oncol. 2018;8:645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kleinstern G, Yan H, Hildebrandt MAT, Vijai J, Berndt SI, Ghesquieres H, et al. Inherited variants at 3q13.33 and 3p24.1 are associated with risk of diffuse large B-cell lymphoma and implicate immune pathways. Hum Mol Genet. 2020;29(1):70–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suvas S, Singh V, Sahdev S, Vohra H, Agrewala JN. Distinct role of CD80 and CD86 in the regulation of the activation of B cell and B cell lymphoma. J Biol Chem. 2002;277(10):7766–75. [DOI] [PubMed] [Google Scholar]
- 53.Law PJ, Sud A, Mitchell JS, Henrion M, Orlando G, Lenive O, et al. Genome-wide association analysis of chronic lymphocytic leukaemia, Hodgkin lymphoma and multiple myeloma identifies pleiotropic risk loci. Sci Rep. 2017;7:41071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Went M, Sud A, Speedy H, Sunter NJ, Forsti A, Law PJ, et al. Genetic correlation between multiple myeloma and chronic lymphocytic leukaemia provides evidence for shared aetiology. Blood Cancer J. 2018;9(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan DE, Foo JN, Bei JX, Chang J, Peng R, Zheng X, et al. Genome-wide association study of B cell non-Hodgkin lymphoma identifies 3q27 as a susceptibility locus in the Chinese population. Nat Genet. 2013;45(7):804–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genotype data from the NCI NHL GWAS is available on dbGaP (phs000801.v2.p1) for research purposes in accordance with dbGaP data access policies. Other data in this manuscript is available for shared research purposes through the InterLymph Consortium upon approval in accordance with institutional review boards and general data protection regulations.