Summary
Tet2 is a member of the Ten-eleven translocation (Tet1/2/3) family of enzymes and is expressed in embryonic stem cells (ESCs). It demethylates DNA (catalytic functions) and partners with chromatin modifiers (noncatalytic functions) to regulate genes. However, the significance of these functions in ESCs is less defined. Using Tet2 catalytic mutant (Tet2m/m) and knockout (Tet2−/−) ESCs, we identified Tet2 target genes regulated by its catalytic dependent versus independent roles. Tet2 was enriched at their active enhancers and promoters to demethylate them. We also identified the histone deacetylase component Sin3a as a Tet2 partner, co-localizing at promoters and active enhancers. Tet2 deficiency diminished Sin3a at these regions. Tet2 and Sin3a co-occupancy overlapped with Tet1. Combined loss of Tet1/2, but not of their catalytic activities, reduced Sin3a at active enhancers. These findings establish Tet2 catalytic and noncatalytic functions as regulators of DNA demethylation and Sin3a recruitment at active enhancers in ESCs.
Subject areas: Biological sciences, Genetics, Molecular biology
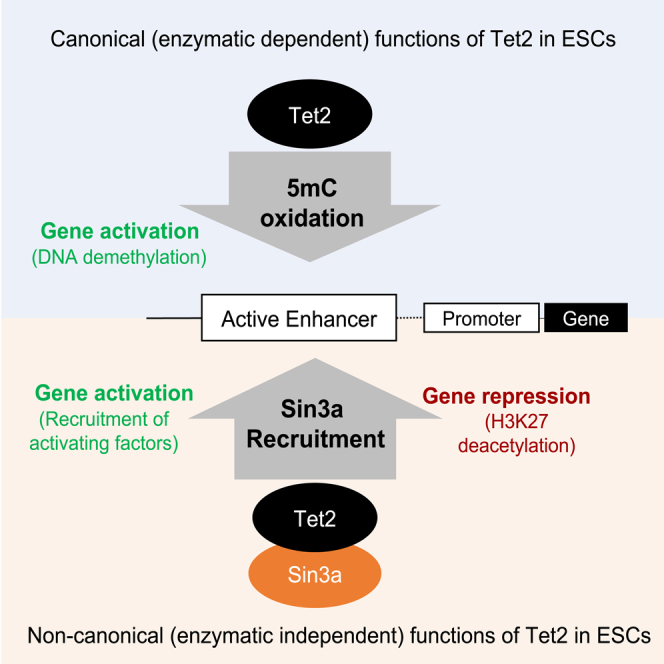
Graphical abstract
Highlights
-
•
Tet2 has distinct catalytic and noncatalytic target genes in ESCs
-
•
Tet2 demethylates enhancers and promoters of its target genes
-
•
Tet2 and Sin3a interact in ESCs and co-occupy active enhancers
-
•
Tet2 noncatalytically facilitates Sin3a enrichment at active enhancers
Biological sciences; Genetics; Molecular biology
Introduction
The Ten-eleven translocation (Tet) family of enzymes (Tet1, Tet2, and Tet3) is an important epigenetic regulator of gene expression during mammalian development. These enzymes promote DNA demethylation by catalyzing the iterative oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC).1,2,3,4 These oxidized bases can be recognized and removed by thymine DNA glycosylase (TDG) and the DNA repair machinery to promote active DNA demethylation.5 5hmC is a stable epigenetic mark that is recognized by chromatin-modifying proteins and it can promote passive demethylation by interfering with the recruitment of the maintenance methyltransferase Dnmt1 during replication.6,7 Tet enzymes are dynamically expressed in embryonic stem cells (ESCs) and during development where they play essential roles in regulating ESC biology and lineage specification.2 Tet1 and Tet2 are highly expressed in mouse ESCs.8,9 Tet3 is not expressed in ESCs but is induced on differentiation as Tet1 and Tet2 levels decline.10 Individual deletion of Tet enzymes in ESCs does not impair self-renewal or pluripotency but compromises gene expression programs leading to aberrant differentiation in vitro.8,9,11,12,13 Combined deficiency of all three Tets has more robust effects on the developmental potential of ESCs, suggesting some redundancy amongst Tet enzymes.10 Consistently, mouse embryos lacking all three Tet enzymes cannot complete gastrulation.14
Tet enzymes are large proteins with a C-terminal catalytic domain that is conserved among them. This domain alone is sufficient to catalyze 5mC oxidation both in vitro and in vivo.1,2,6 The catalytic functions of Tet enzymes have been well-studied in various contexts, including in ESCs. Tet1 contributes to ∼30% and Tet2 to ∼70% of 5hmC in ESCs.8,10,13 Tet1 deficient ESCs have lower 5hmC levels particularly at gene promoters, whereas Tet2 knockout ESCs have lower 5hmC levels at gene bodies and exon-intron junctions of highly expressed genes.15 Loss of all three Tet genes in ESCs results in global hypermethylation, including at gene regulatory regions such as promoters and enhancers.16 In addition to their canonical enzymatic roles in promoting DNA demethylation, Tet enzymes also possess noncanonical enzymatic-independent functions by interacting with chromatin-modifying proteins to influence chromatin dynamics in ESCs and other cell types.6 We and others have shown that Tet1, independent of its catalytic activity, forms complexes with Sin3a and PRC2 to facilitate their recruitment to promoters of bivalent genes for H3K27 deacetylation and trimethylation.11,12,17,18 Tet1-mediated recruitment of Sin3a has also been implicated in gene activation.19 Tet2 interacts with several transcription factors and chromatin-modifying enzymes including O-liked N-acetylglucosamine transferase (Ogt), Klf4, Parp1, Hdac1/2, Nono, and Pspc1 in ESCs and other cell types.20,21,22,23,24 In hematopoietic stem cells, Tet2 catalytic functions are important for the regulation of the myeloid lineage, and the noncatalytic functions are critical for the lymphoid lineage.25 Tet2 also interacts with Hdac2 in immune cells and facilitates the deacetylation of histone H3 to silence pro-inflammatory genes.21
Given the dual catalytic and noncatalytic roles of Tet enzymes in gene regulation, an active area of research has been defining their respective contributions to ESC biology. Both Tet1 and Tet2 are highly expressed in ESCs.8,9 However, the functions of Tet2, which contributes to 70% of 5hmC in ESCs8,13 and partners with many proteins, are less studied in ESCs. Here, using Tet2 catalytic mutant (Tet2m/m) and knockout (Tet2−/−) ESCs we have identified the catalytic and noncatalytic target genes of Tet2. The majority of Tet2 target genes are directly bound by Tet2 at promoters and active enhancers, which are robustly hypermethylated in Tet2m/m and Tet2−/− ESCs. We also find that Tet2, like Tet1, interacts with Sin3a, and co-occupy a large number of promoters and active enhancers. Loss of Tet2 diminished Sin3a enrichment at regulatory regions of downregulated genes. Our findings show that Tet2 not only regulates DNA demethylation but also Sin3a targeting at promoters and active enhancers, thus contributing in both catalytic dependent and independent manners to ESC gene regulation.
Results
Generation and characterization of Tet2 catalytic-mutant (Tet2m/m) and knockout (Tet2−/−) ESCs
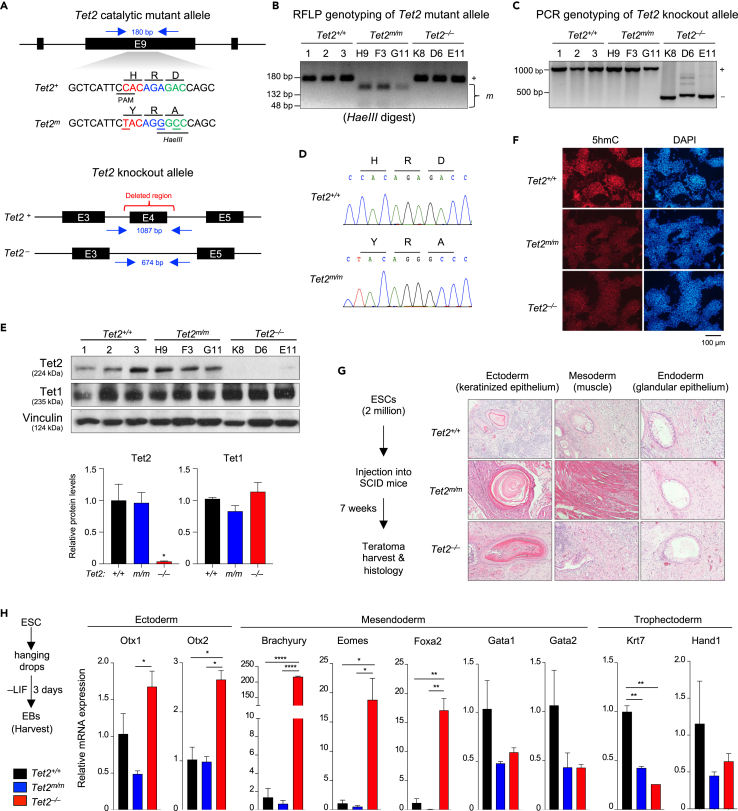
To study the catalytic-dependent and independent roles of Tet2 in ESCs, we generated ESC lines that lacked only the Tet2 catalytic activity (Tet2m/m) or the entire Tet2 protein (Tet2−/−) (n = 3 of each) (Figure 1A). For generating Tet2m/m ESCs, we introduced point mutations in exon 9 of Tet2 for amino acid substitutions H1367Y and D1369A in the iron-binding pocket of Tet2 using CRISPR/Cas9-based gene editing in V6.5 mouse ESCs (Figure 1A). These mutations are previously shown to completely abrogate the catalytic activity of Tet2 without any dominant-negative effects.2,25 For generating Tet2−/− ESCs, we deleted exon 4 of Tet2 using a pair of flanking gRNAs (Figure 1A). Genotypes of properly targeted ESC lines were confirmed by RFLP and PCR (Figures 1B and 1C) and verified by Sanger sequencing (Figure 1D). Complete loss of Tet2 protein in Tet2−/− ESCs, as well as normal expression of Tet2 catalytic mutant protein in Tet2m/m ESCs, were confirmed by western blot (Figure 1E). Consistent with loss of Tet2 catalytic activity, Tet2m/m ESCs had substantially reduced levels of 5hmC, similar to the 5hmC levels in Tet2−/− ESCs (Figure 1F). In agreement with previous reports showing that loss of Tet2 does not affect ESC pluripotency, both the Tet2m/m and Tet2−/− ESC lines maintained normal pluripotency, forming teratomas containing tissue types of three embryonic germ layers (Figure 1G). However, when Tet2m/m and Tet2−/− ESC were differentiated to embryoid bodies (EBs), we found increased expression of some ectodermal and mesendodermal markers in Tet2−/−, but not in Tet2m/m, EBs suggesting that Tet2 noncatalytic activities may play subtle roles in committing to cell types of these germ layers (Figure 1H). Given that Tet2−/− ESCs lack all functions of the protein and the Tet2m/m ESCs only lack the enzymatic activity of the protein, these lines serve as a valuable tool for distinguishing the catalytic-dependent and independent requirements of Tet2 in ESC gene regulation and biology.
Figure 1.
Generation and validation of Tet2 catalytic mutant (Tet2m/m) and knockout (Tet2−/−) ESCs
(A) Schematic of gene targeting strategy for generating Tet2m/m ESCs (top) and Tet2−/− ESCs (bottom).
(B) Validating genotypes of properly targeted Tet2m/m ESCs by RFLP (restriction fragment-length polymorphism) using HaeIII enzyme. Correctly targeted (mutated) allele bands after digestion are 132 bp + 48 bp. Allele not carrying the mutation is 180 bp. (3 independent clones were generated H9, F3, and G11).
(C) Genotyping of properly targeted Tet2−/− ESCs by PCR. Amplification of a shorter fragment (∼674 bp) confirms the correct deletion of exon 4. (3 independent clones were generated K8, D6, and E11).
(D) Sanger sequencing confirms the correct introduction of H1367Y and D1369A mutations in the Tet2 catalytic mutant allele.
(E) Quantification of Tet1 and Tet2 protein levels in ESCs of indicated genotypes by Western blot (top). Signal intensity normalized to the loading control Vinculin (average of three lines) is plotted (bottom). Note the complete loss of Tet2 protein in Tet2−/− ESCs and normal expression of catalytic mutant Tet2 in Tet2m/m ESCs. Tet1 protein levels are not changed in either genotype compared to wildtype (∗p < 0.05 versus wildtype, one-way ANOVA).
(F) Immunostaining for 5hmC in ESC of indicated genotypes using an anti-5hmC antibody. Nuclei are stained with DAPI.
(G) Hematoxylin and Eosin (H&E) staining of sections of teratomas derived from ESCs of indicated genotypes.
(H) Quantification of mRNA levels of germ layer markers in day 3 embryoid bodies (EBs) derived from wildtype, Tet2m/m and Tet2−/− ESCs by RT-qPCR. Data normalized to Gapdh. Error bars = SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Loss of Tet2 versus loss of its catalytic activity alone leads to distinct gene expression changes in ESCs
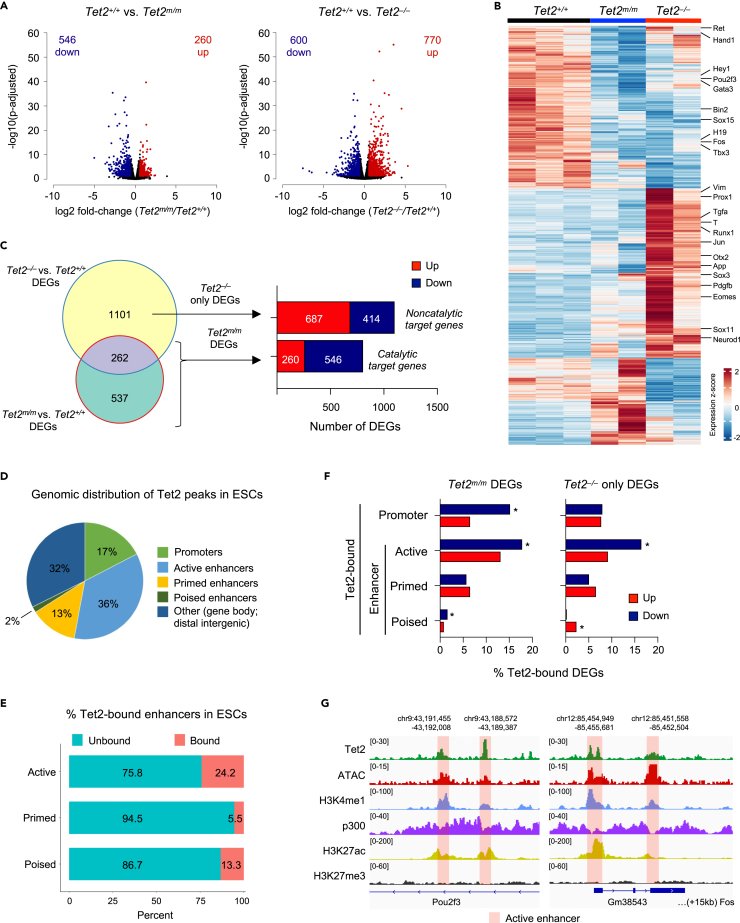
To identify genes regulated by the catalytic versus noncatalytic functions of Tet2 in ESCs we compared the transcriptome of Tet2m/m, Tet2−/−, and wildtype ESCs by RNA-seq (Figures 2 and S1A). Principal component analysis (PCA) and clustering by Euclidean distance revealed that the biological replicates clustered together, and the three genotypes were well separated, with Tet2m/m ESCs clustering closer to wildtype ESCs than Tet2−/− ESCs (Figures S1B and S1C). We found 806 (546 down and 260 upregulated) differentially expressed genes (DEGs) in Tet2m/m ESCs (versus wildtype), and 1,370 (600 down and 770 upregulated) DEGs in Tet2−/− ESCs (versus wildtype) (Figures 2A and 2B). To distinguish genes regulated by the catalytic versus noncatalytic functions of Tet2, we overlapped DEGs affected in Tet2m/m and Tet2−/− ESCs (Figure 2C). In contrast to the 806 DEGs (546 down, 260 up) affected in Tet2m/m ESCs (i.e., catalytic genes) there were 1101 DEGs (414 down, 687 up) affected only in Tet2−/− ESCs (noncatalytic genes) (Figure 2C). Gene ontology (GO) analysis revealed significant enrichment for important developmental processes, such as nervous system, cardiovascular and embryonic development, in both up and downregulated DEGs (Figure S1D). These genes included important lineage specifiers such as Hand1, Gata3, Sox17 and Bin2 that were downregulated in both Tet2m/m and Tet2−/− ESCs, and Brachyury (T), Otx2, Sox11 and Eomes that were upregulated in Tet2−/− ESCs only (Figure 2B). Together, these data suggest that Tet2 influences gene expression of ESCs via both its catalytic and noncatalytic functions.
Figure 2.
Identification of Tet2 catalytic and noncatalytic target genes in mouse ESCs
(A) Volcano plots showing the number of differentially expressed genes (DEGs, fold-change >1.5, FDR <0.05) between Tet2+/+ versus Tet2m/m ESCs (left) and Tet2+/+ versus Tet2−/−ESCs (right). (Two independent Tet2m/m and Tet2−/− ESCs and three independent Tet2+/+ ESCs used in this analysis).
(B) Heatmap of all DEGs (n = 1900) in Tet2m/m and Tet2−/− ESCs. Selected lineage genes are shown to the right. The color key represents the relative expression extracted from normalized counts.
(C) Venn diagram showing the overlap of DEGs in Tet2m/m and Tet2−/− ESCs and revealing genes deregulated in Tet2m/m, and in Tet2−/− (left). The number of up and downregulated DEGs in each category is plotted (right).
(D) Genomic distribution of Tet2 ChIP-seq peaks in wildtype mouse ESCs. Note that Tet2 is highly enriched at active enhancers. Tet2 ChIP-seq data used from Rasmussen et al., 2019. Enhancer subtypes were annotated using Cruz-Molina et al., 2017. Promoters were defined as +/−2kb of TSS.
(E) Percent of Tet2-bound and unbound enhancer subtypes in wildtype mouse ESCs is plotted.
(F) Percent of DEGs in Tet2m/m and Tet2−/− ESCs that are bound by Tet2 at their promoters or enhancers is plotted. Genes that are downregulated in Tet2m/m ESCs are significantly enriched for genes bound by Tet2 at promoters and active enhancers (∗p < 0.05 by hypergeometric test).
(G) Genome browser tracks showing enrichment of Tet2 at active enhancers of selected genes that are down-regulated in Tet2m/m ESCs. Tet2 ChIP-seq tracks are from Rasmussen et al., 2019. ATAC-seq data from Chronis et al., 2017. p300, H3K4me1, H3K27ac, and H2K27me3 ChIP-seq tracks are from Cruz-Molina et al., 2017.
Tet2 is enriched at promoters and active enhancers of downregulated differentially expressed genes
To identify the direct target genes of Tet2 in ESCs (i.e. genes bound by Tet2), we re-analyzed a previously published Tet2 ChIP-seq dataset in wildtype mouse ESCs26 to map the genomic distribution of Tet2 across genes and gene regulatory regions. We found 8,261 Tet2 peaks genome-wide, of which 1,438 (17%) mapped to promoters (+/− 2kb of TSS) and the remaining vast majority mapped to gene bodies and distal intergenic regions (Figure S2A). Because these regions harbor enhancer elements and Tet2 has been previously associated with enhancers,26 we assessed Tet2 occupancy at different enhancer subtypes in more detail. We overlapped Tet2 non-promoter peaks to previously published maps of active (H3K4me1+, H3K27ac+, H3K27me3–), primed (H3K4me1+, H3K27ac–, H3K27me3–) and poised (H3K4me1+, H3K27ac–, H3K27me3+) enhancers27 and accessibility data28 in ESCs. We found that 36% (2,936/8,261) of Tet2 peaks mapped to active enhancers, 13% (1,079/8,261) to primed enhancers, and only 2% (135/8,261) to poised enhancers (Figures 2D, S2B, and S2C). Importantly, 24% of all active enhancers found in wildtype ESCs (2,936/12,142) were bound by Tet2 (Figure 2E) suggesting that Tet2 is associated with a large fraction of active enhancers and may play a role in their regulation. We confirmed enrichment of Tet2 at selected gene regulatory regions (Nanog enhancer 2 and Oct4 promoter 1) in endogenously flag-tagged Tet2m/m;flag/flag and Tet2+/+;flag/flag ESCs by ChIP-qPCR, which also showed that catalytic mutant and wildtype Tet2 were comparably enriched at these regions (Figure S2D). Finally, to identify genes directly regulated by Tet2 in ESCs, we overlapped genes bound by Tet2 at promoters and enhancers with DEGs in Tet2m/m and Tet2−/− ESCs. We found that a significant fraction of genes bound by Tet2 at promoters or active enhancers were downregulated in Tet2m/m ESCs (Figures 2F and 2G). Together, these data suggest that Tet2 is preferentially associated with active enhancers and to a lesser extent with promoters in ESCs and facilitates activation of its catalytic target genes.
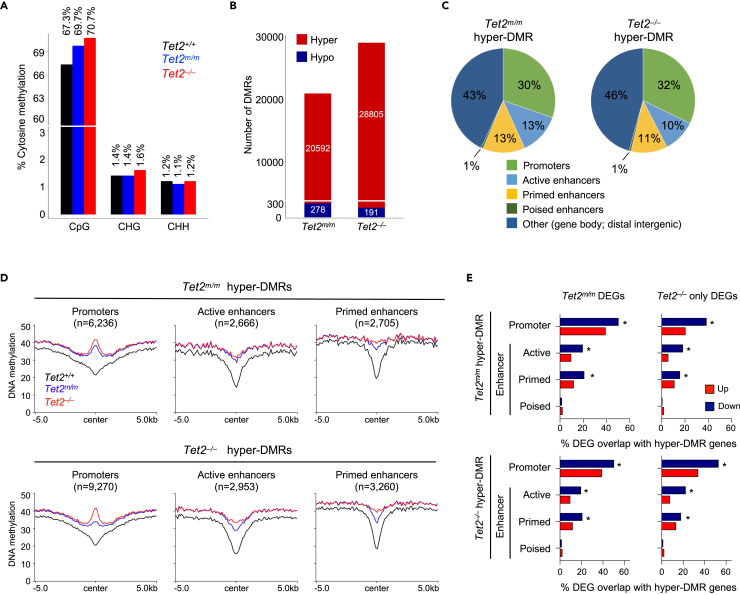
Loss of Tet2 or loss of its catalytic activity alone leads to promoter and enhancer hypermethylation which correlates with gene downregulation
Because Tet2 promotes DNA demethylation, we examined the global levels of DNA methylation (5mC) in wildtype, Tet2m/m and Tet2−/− ESCs by whole-genome bisulfite sequencing (WGBS) (Figure S3A). We found that global 5mC levels were subtly but comparably increased in both Tet2m/m (69.7%) and Tet2−/− (70.7%) ESCs versus wildtype (67.3%) ESCs, mainly in the context of CpG dinucleotides (Figure 3A). We next performed differential methylation analysis and found 20,870 and 28,996 differentially methylated regions (DMRs) in Tet2m/m and Tet2−/− ESCs when compared to wildtype ESCs, respectively (DMRs defined as regions >5 CpG, DNA methylation difference >20% versus wildtype, FDR <0.05) (Figure 3B). Consistent with the catalytic role of Tet2 in DNA demethylation, the majority of the DMRs were hypermethylated (hyper-DMRs) (Tet2m/m = 20,592/20,870 (98.7%), Tet2−/− = 28,805/28,996 (99.3%)) (Figure 3B). There was a strong overlap between hyper-DMRs in Tet2m/m and Tet2−/− ESCs (∼90%) as well as between genes annotated to hyper-DMRs in Tet2m/m and Tet2−/− ESCs (∼95%) (Figure S3B). We also found that promoters, active and primed enhancers were highly enriched in hyper-DMRs of Tet2m/m and Tet2−/− ESCs (Figure 3C). The levels of DNA methylation at hyper-DMRs in gene regulatory regions were robustly high in both Tet2m/m and Tet2−/− ESCs, with the largest difference at the center of the DMR (Figure 3D). Of interest, Tet2−/− ESCs had slightly more DNA methylation at the center of the DMR than Tet2m/m ESCs. Consistent with a role for Tet2 catalytic activity in DNA hydroxylation, the levels of 5hmC at selected CpG-rich genomic regions containing hyper-DMRs were equally and robustly reduced in both Tet2m/m and Tet2−/− ESCs (Figure S3C). Hyper-DMRs associated with promoters were enriched for motifs of pluripotency factors Klf5 and Esrrb whereas hyper-DMRs associated with active enhancers were enriched for Klf5, Esrrb, Nr5a2 and Sox2 (Figure S3D). Overlapping hyper-DMR-associated genes with DEGs revealed a significant enrichment for downregulated genes in Tet2m/m and Tet2−/− ESCs that contained either promoter or active/primed enhancer hyper-DMRs (Figure 3E). Integrating hyper-DMRs with Tet2 occupancy data revealed that active enhancers selectively had a very high number of Tet2-bound hyper-DMRs (∼24%) in both Tet2m/m and Tet2−/− ESCs (Figure S3E), which were associated with downregulated DEGs in Tet2m/m and Tet2−/− ESCs (Figure S3F). Collectively, these data suggest that Tet2 maintains the expression of its target genes by demethylating their promoters and active enhancers.
Figure 3.
Genome-wide analysis of DNA methylation in Tet2m/m and Tet2−/− ESCs reveals hypermethylation of gene regulatory elements
(A) Genome-wide percent CpG, CHG and CHH methylation levels in Tet2+/+, Tet2m/m, and Tet2−/− ESCs measured by whole-genome bisulfite sequencing (WGBS).
(B) Quantification of differentially methylated regions (DMRs = >5 CpGs, methylation difference >20%, and FDR <0.05) in Tet2m/m and Tet2−/− ESCs compared to wildtype ESCs. Note that the majority of DMRs are hypermethylated in both cell types.
(C) Assignment of hypermethylated (hyper-) DMRs in Tet2m/m and Tet2−/− ESCs to genomic regions. Note that the majority of DMRs are at promoters and active or primed enhancers.
(D) Profile plots of DNA methylation levels at hyper-DMRs located at promoters and enhancers in Tet2m/m and Tet2−/− ESCs.
(E) Percent of DEGs in Tet2m/m and Tet2−/− ESCs that overlap with hyper-DMR-associated genes. Note the significant association between downregulated genes and hypermethylated promoters, active and primed enhancers (∗p < 0.05 by hypergeometric test).
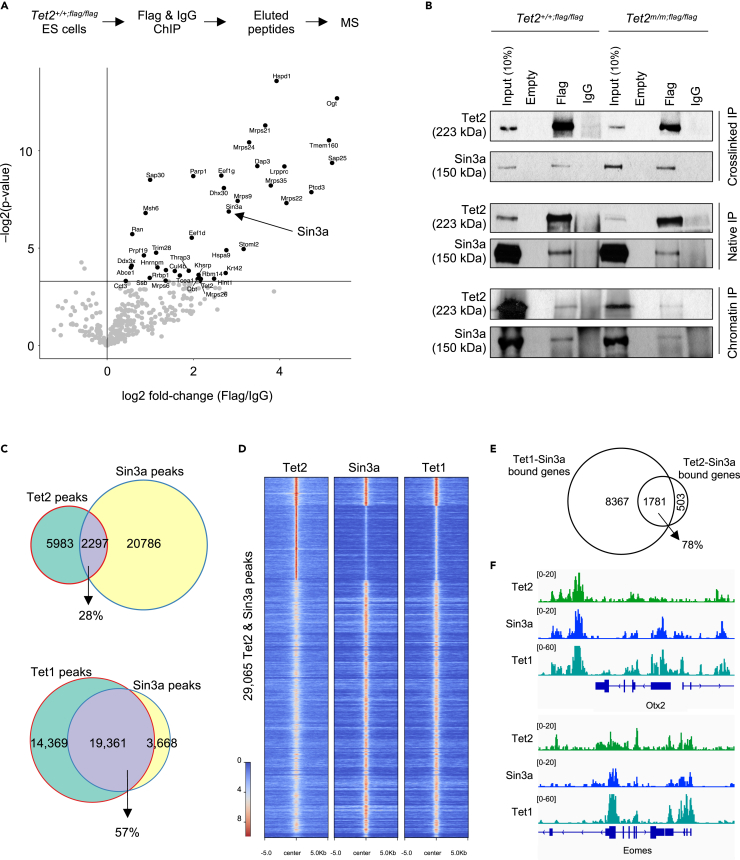
Tet2 and Sin3a are in a complex and co-occupy promoters and enhancers in ESCs
To gain insights into how Tet2 regulates genes independent of its catalytic activity, we sought to identify its chromatin binding partners using Chromatin immunoprecipitation (ChIP) followed by mass spectrometry (MS). Owing to a lack of commercially available Tet2 ChIP-grade antibodies, we generated endogenously Flag-tagged Tet2+/+;flag/flag and Tet2m/m;flag/flag ESC lines by knocking in a 3x-Flag tag before the stop codon of Tet2 (Figures S4A–S4C) to facilitate ChIP using an anti-Flag antibody. We subjected the Tet2+/+;flag/flag ESCs to an anti-Flag ChIP-MS. Among the top hits identified in this analysis were some of the previously known Tet2 binding partners such as Ogt and Parp1,20,22 as well as three novel binding partners, including the histone deacetylase component Sin3a and the Sin3a-associated proteins Sap25 and Sap30 (Figure 4A and Table S1). Sin3a has been shown to interact with Tet1 in ESCs to promote both gene activation and repression,18,19 but its partnership with Tet2 and potential relevance in gene regulation in ESCs have not been previously reported. We validated the Tet2-Sin3a complex formation in ESCs by subjecting Tet2+/+;flag/flag and Tet2m/m;flag/flag ESC lysates to co-immunoprecipitation (co-IP) using anti-Flag antibody followed by western blot for Sin3a. Sin3a co-immunoprecipitated with wildtype and catalytic mutant Tet2 in both native and crosslinked IP lysates as well as in chromatin IP (Figure 4B). This suggests that Tet2 is stably in a complex with Sin3a in ESCs and that this interaction is independent of Tet2 catalytic activity. Of note Sin3a protein levels were unaffected in Tet2m/m and Tet2−/− ESCs (Figure S4D) and its interactions with wildtype or catalytic mutant Tet2 were comparable (Figure 4B).
Figure 4.
Tet2 is in a complex with the histone deacetylase Sin3a in ESCs
(A) Schematic of ChIP-MS strategy for identification of endogenous binding partners of Tet2 using a Tet2flag/flag ESC line and anti-Flag antibody (top). Enrichment scores (log2 fold-change x -log2(p value)) of proteins that immunoprecipitated with Tet2 as quantified by MS is plotted (bottom). Note that several known partners of Tet2 (i.e. Ogt and Parp1) as well as new partners (i.e. Sin3a, Sap25 and Sap30) were identified.
(B) Validation of Tet2 interaction with Sin3a by immunoprecipitation (IP) of native and crosslinked nuclear lysates, and ChIP of Tet2+/+;flag/flag and Tet2m/m; flag/flag ESCs using anti-Flag antibodies and Western blot of Sin3a. Anti-IgG was used as negative control.
(C) Venn diagram showing overlap of Tet2 ChIP-seq peaks with Sin3a peaks in ESCs (top), and venn diagram showing the overlap of Tet1 and Sin3a peaks from published datasets (Chrysanthou et al., 2022) (bottom).
(D) Heatmap showing enrichment of Tet2, Sin3a, and Tet1 signal at Tet2 and Sin3a peaks (29,065 peaks total).
(E) Venn diagram showing overlap of Tet2-Sin3a co-bound genes with Tet1-Sin3a co-bound genes from our previously published datasets (Chrysanthou et al., 2022).
(F) Genome browser tracks showing the overlap of Tet2, Sin3a and Tet1 peaks at selected developmental genes.
Next, we examined if Tet2 and Sin3a co-occupy similar genomic loci in ESCs and how that compares to Tet1 occupancy. We used published Tet2 ChIP-seq datasets26 as well as Sin3a and Tet1 CUT&Tag datasets12 in wildtype ESCs to compare their global genomic distributions. We found that 28% of all Tet2 peaks overlapped with Sin3a peaks, in contrast to 57% of Tet1 peaks that overlapped with Sin3a peaks (Figure 4C). The majority of Tet2 peaks that overlapped with Sin3a also overlapped with Tet1 occupancy (Figure 4D) which suggests that Tet1 and Tet2 may cooperate with Sin3a to regulate a subset of their target genes. Consistently, a vast majority (78%) of Tet2-Sin3a co-bound genes overlapped with Tet1-Sin3a co-bound genes (Figure 4E) and included several developmental DEGs such as Otx2 and Eomes (Figure 4F). Together, these data show that Tet2, independent of its catalytic activity, can bind to Sin3a and that their genomic distributions overlap in a large number of loci.
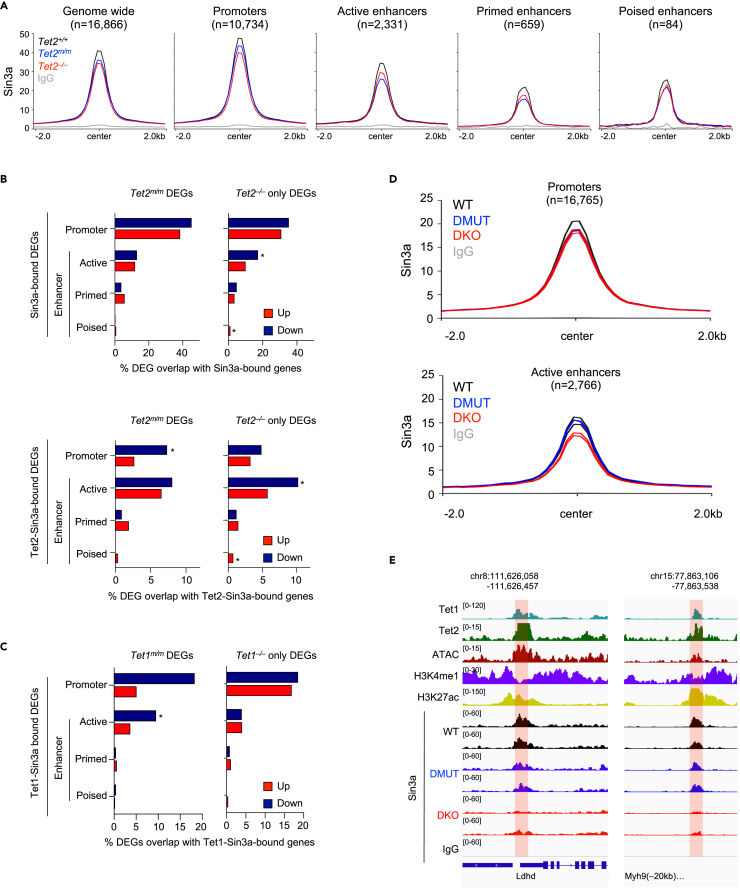
Loss of Tet2 reduces Sin3a levels at promoters and enhancers
The complex formation and genomic co-occupancy of Tet2 with Sin3a prompted us to test whether Tet2 facilitates enrichment of Sin3a to the chromatin. To this end, we mapped the genome-wide distribution of Sin3a by Cleavage Under Targets and Tagmentation (CUT&Tag) in Tet2m/m, Tet2−/−, and wildtype ESCs (Figures 5 and S5). This analysis identified ∼16K peaks for Sin3a (Figure S5A). We found that Sin3a levels were decreased genome-wide and across promoters and enhancers in both Tet2m/m and Tet2−/− ESCs compared to wildtype ESCs (Figure 5A). To examine how Sin3a occupancy influenced gene expression, we overlapped Sin3a-bound genes and Sin3a-Tet2 bound genes with DEGs in Tet2m/m and Tet2−/− ESCs. We found that many DEGs, in particular downregulated genes bound by Tet2 and Sin3a at promoters and active enhancers, were significantly enriched (Figure 5B). This suggests that Sin3a may have an activating role in the regulation of these genes, rather than its well-known repressive role in H3K27 deacetylation. To gain further insight into this, we mapped H3K27ac levels in Tet2m/m and Tet2−/− ESCs by CUT&Tag (Figure S5B). We found that the distribution of H3K27ac peaks across genomic regions was largely comparable in Tet2m/m and Tet2−/− ESCs (Figure S5C). We also assessed H3K27ac levels at all Sin3a-Tet2-bound regions in Tet2m/m, Tet2−/−, and wildtype ESCs. H3K27ac levels were not increased but rather mildly decreased at promoters in both Tet2m/m and Tet2−/− ESCs, whereas at active enhancers H3K27ac levels were decreased mainly in Tet2−/− ESCs (Figure S5D). This decrease is consistent with the increased DNA hypermethylation at these regions (Figure 3D) compromising proper H3K27 deacetylation. Thus, the reduced levels of Sin3a at Tet2-bound regions do not lead to increased H3K27ac levels.
Figure 5.
Tet2 facilitates Sin3a chromatin enrichment at gene regulatory regions
(A) Enrichment of Sin3a CUT&Tag signal in wildtype, Tet2m/m, and Tet2−/− ESCs genome-wide and at gene regulatory regions. Note the mild reduction in Sin3a signal in Tet2m/m and Tet2−/− ESCs.
(B) Percent of Sin3a-bound DEGs (top) and Tet2-Sin3 co-bound DEGs (bottom). Note that genes that are downregulated are significantly enriched for genes bound by Sin3a and/or Tet2 at promoters and active enhancers (∗p < 0.05 by hypergeometric test).
(C) Percent of Tet1-Sin3 co-bound DEGs in Tet1m/m, and Tet1−/− ESCs calculated using Tet1 and Sin3a occupancy and DEGs from our previously published datasets (Chrysanthou et al. 2022). Note that genes that are downregulated are significantly enriched for genes bound by Sin3a and Tet1 at promoters (∗p < 0.05 by hypergeometric test).
(D) Sin3a levels at promoters and active enhancers in Tet1m/m;Tet2m/m double mutant (DMUT), Tet1−/−;Tet2−/− double knockout (DKO) and wildtype ESCs. Note that at active enhancers Sin3a is reduced specifically in DKO ESCs.
(E) Genome browser tracks showing enrichment of Sin3a levels at active enhancers (shaded area) of selected genes in wildtype, DMUT, and DKO ESCs. Tet1 tracks are from Chrysanthou et al., 2022. Tet2 tracks are from Rasmussen et al., 2019. ATAC-seq data from Chronis et al., 2017. H3K4me1 and H3K27ac tracks from Cruz-Molina et al., 2017.
Finally, given that both Tet1 and Tet2 are expressed in ESCs and contribute to the proper enrichment of Sin3a to regulatory regions, we examined how Tet1-Sin3a versus Tet2-Sin3a complexes regulate genes. To this end, using our previously published datasets we identified Tet1-Sin3a bound DEGs in Tet1m/m and Tet1−/− ESCs (Figure 5C) and compared them to the Tet2-Sin3a bound DEGs in Tet2m/m and Tet2−/− ESCs (Figure 5B lower panel). We found that there are more Tet1-Sin3a bound DEGs in Tet1m/m and Tet1−/− ESCs compared to Tet2-Sin3a bound DEGs in Tet2m/m and Tet2−/− ESCs. This suggests that Tet1-Sin3a regulates more genes than Tet2-Sin3a. We also found that DEGs in Tet1m/m and Tet1−/− ESCs were co-bound by Tet1 and Sin3a mostly at promoters whereas DEGs in Tet2m/m and Tet2−/− ESCs were co-bound by Tet2 and Sin3a mostly at enhancers and at promoters. This suggests that Tet1-Sina3 influence gene regulation largely via promoters, whereas Tet2-Sin3a largely via enhancers and promoters. This is consistent with the literature showing Tet1 mostly enriched at promoters and Tet2 at enhancers.18,26 Lastly, both the Tet1-Sin3a bound DEGs in Tet1m/m and Tet1−/− ESCs and the Tet2-Sin3a bound DEGs in Tet2m/m and Tet2−/− ESCs were predominantly downregulated. This suggests that Tet1-Sin3a and Tet2-Sin3a complexes facilitate gene expression, which is consistent with a gene activating role for Sin3a.19,29 To better understand the dynamics of Tet1/2 and Sin3a in gene regulation, we examined how combined loss of Tet1 and Tet2 affects Sin3a targeting to the chromatin. To this end, we generated Tet1 and Tet2 double catalytic mutant (DMUT), and knockout (DKO) ESCs by re-targeting our previously published Tet1m/m and Tet1−/− ESCs12 to mutate or delete Tet2 (Figures S6A and S6B). Mapping the genomic distribution of Sin3a in wildtype, DMUT, and DKO ESCs by CUT&Tag (Figures S6C and S6D) revealed that while at promoters Sin3a levels were decreased in both DMUT and DKO ESCs compared to wildtype ESCs, at active enhancers Sin3a levels were only decreased in DKO ESCs (Figures 5D and 5E), suggesting that Tet1 and Tet2 noncatalytic functions are important for proper Sin3a enrichment at active enhancers.
Discussion
Tet2 is highly expressed in ESCs where it regulates gene expression by DNA demethylation and partnering with several transcription factors and chromatin modifiers. However, these functions of Tet2 in ESC gene expression programs are not well studied and only a handful of Tet2 binding partners in ESCs have been identified. Here, using Tet2 catalytic deficient and knockout ESCs, we have: (1) distinguished Tet2 target genes that are regulated by its catalytic versus noncatalytic functions, (2) established Tet2 enrichment at promoters and active enhancers of its target genes where it is critical for their demethylation, (3) identified a novel complex formation between Tet2 and Sin3a and their co-occupancy at active enhancers where deficiency of Tet2 diminishes Sin3a enrichment. These findings suggest that the catalytic functions of Tet2 in DNA demethylation as well as its noncatalytic roles in partnering with Sin3a contribute to regulating gene promoters and enhancers in ESCs.
We show that of the ∼1900 genes that are influenced by Tet2 in ESCs, ∼800 are regulated by the catalytic (i.e. affected in Tet2m/m ESCs), and ∼1100 are regulated by the noncatalytic functions of Tet2 (i.e. affected only in Tet2−/− ESCs). Tet2 is enriched at the promoters and active enhancers of a large subset of DEGs. Our work strongly ties promoter and enhancer hypermethylation in Tet2m/m and Tet2−/− ESCs to a large fraction of downregulated genes. The deregulation of noncatalytic target genes is likely a combination of demethylation-independent functions of Tet2 entailing the many partnerships of Tet2 with chromatin repressive complexes such as Hdac1/2 as well as Sin3a which we identify as a novel binding partner of Tet2 in ESCs.
Consistent with a role for Tet2 in DNA demethylation, the vast majority of DMRs found in both Tet2m/m and Tet2−/− ESCs were hypermethylated. The slight increase in the number of DMRs in Tet2−/− ESCs could be an indirect consequence of the noncatalytic functions of Tet2. It is also possible that Tet2, like Tet1,30 can protect unmethylated CpGs from aberrant methylation at certain regions. Regions bound by Tet2 as well as regions hypermethylated in Tet2m/m and Tet2−/− ESCs were strongly enriched for motifs of several pluripotency transcription factors (TFs) and contained promoters and enhancers. Because DNA methylation can influence TF binding,31 a catalytic role for Tet2 is the demethylation of promoters and enhancers for regulating TF recruitment as is shown for some TFs.32 Future studies are warranted to investigate the effects of hypermethylation of promoters and enhancers in Tet2m/m and Tet2−/− ESCs on the enrichment of pluripotency TFs and its impact on gene dysregulation. Our findings that 36% of Tet2 peaks are at active enhancers (versus 17% at promoters) and that a quarter of all active enhancers in ESCs are bound by Tet2 strongly suggest that Tet2 has regulatory roles at active enhancers. Consistently, Tet2-occupied active enhancers were more robustly hypermethylated in Tet2m/m and Tet2−/− ESCs and were associated with downregulated genes. These findings are in agreement with other work implicating Tet2 in the regulation of enhancers in embryonic and hematopoietic stem cells26 and Tet enzymes regulating developmental enhancers during post-gastrulation development.33
Tet2 is known to interact with several transcription factors and epigenetic modifiers in ESCs and in other cell types. Our ChIP followed by MS analysis identified several of these known partners of Tet2 including Ogt and Parp1.20,22 However, of interest was the identification of Sin3a and the Sin3a-associated proteins Sap25 and Sap30 as new partners of Tet2 in ESCs. Sin3a has both gene activating and repressive functions. In a complex with Hdacs, Sin3a promotes H3K27 deacetylation and gene repression. Sin3a also forms gene activating complexes, such as with Fam60a or Nanog, to promote gene expression.19,34,35 Tet1 is a well-established binding partner of Sin3a in ESCs. It facilitates Sin3a chromatin enrichment for H3K27 deacetylation and silencing of developmental genes.12,18 It also interacts with Sin3a to facilitate expression of self-renewal and pluripotency genes.19 However, Tet2 has not been associated with Sin3a. Our findings strongly establish that wildtype or catalytic mutant Tet2 is in a complex with Sin3a in ESCs. Deficiency of Tet2 diminished Sin3a levels at promoter and enhancers. Of interest, H3K27ac levels were not increased in Tet2m/m and Tet2−/− ESCs, but rather marginally decreased which could be because of increased DNA methylation at enhancers. Our findings that Sin3a-bound genes and Sin3a-Tet2 bound genes overlapped particularly with downregulated DEGs in Tet2m/m and Tet2−/− ESCs suggests that Sin3a has an activating role in the regulation of these genes, rather than a repressive role in H3K27 deacetylation. This is also supported by the fact that the reduced Sin3a levels at promoters and active enhancers in Tet2m/m and Tet2−/− ESCs did not lead to increased H3K27ac levels as noted earlier. This raises the question how the Tet2-Sin3a partnership promotes gene activation. One possibility can be that Tet2 recruits Sin3a to its target and that allows for formation of the typical Sin3a activating complexes with other transcription factors like Nanog or Fam60a.34,35 In addition to this, Tet2 could also promote DNA demethylation which would further promote transcription at the Tet2-Sin3a co-bound genes. This would entail both the catalytic dependent and independent roles of Tet2 working together to activate genes. A similar model has been proposed for Tet1-Sin3a mediated gene activation.12,19 Identifying additional components of the Tet2-Sin3a complex in ESCs could further elaborate on which transcription factors and mechanisms precisely mediate gene activation by Tet2-Sin3a. Our findings that 57% of Tet1 peaks and 28% of Tet2 peaks overlap with Sin3a peaks suggest that Tet1 is likely a more prominent partner of Sin3a in ESCs than Tet2. Consistently, all Tet2 and Sin3a co-bound regions contained Tet1. This suggests that Tet1 and Tet2 cooperate at a subset of Sin3a targets. Of interest, at active enhancers Sin3a levels were specifically more reduced in double knockout ESCs than in double catalytic mutant ESCs, indicating that the noncatalytic roles of Tet1/2 influence Sin3a levels at active enhancers.
We conclude that the catalytic functions of Tet2 regulate DNA demethylation at promoters and active enhancers whereas its noncatalytic roles facilitate Sin3a recruitment at active enhancers. This work distinguishes the enzymatic and nonenzymatic target genes of Tet2 in ESCs. It associates Tet2-mediated DNA demethylation and Sin3a enrichment at active enhancers to regulation of a subset of these genes. However, how all Tet2 noncatalytic targets are regulated requires more investigation. Tet2 has many binding partners including TFs as well as activating and repressive proteins. Thus, future studies using our platform of Tet2m/m and Tet2−/− ESCs will establish how each of these partners of Tet2 is affected in the absence of Tet2 or its enzymatic activity alone. Although Sin3a enrichment is only mildly affected because of Tet2 loss, it is possible that Ogt, Parp1, Sall4, and other binding partners of Tet220,21,22,23,24,36,37 are influenced more profoundly and distinctly by the dual functions of Tet2. This study builds the foundation for further dissecting Tet2 functions beyond ESCs with implications in development and human diseases.
Complete loss of Tet2 in ESCs does not affect ESC pluripotency,8,9 so it is expected that loss of its catalytic activity alone also does not affect pluripotency as we have shown here. However, Tet2 may have distinct catalytic dependent and independent roles during ESC differentiation to specific lineages. Differentiation of Tet2m/m and Tet2−/− ESCs to EBs alludes to potential noncatalytic functions of Tet2 in preventing the aberrant upregulation of ectoderm and mesendoderm programs and hence proper lineage specification. Similar noncatalytic roles have been attributed to Tet1 in preventing untimely expression of mesendoderm and trophectoderm genes in ESCs.12 Expression of some mesoderm and trophectoderm markers were equally dysregulated Tet2m/m and Tet2−/− EBs which suggests that the catalytic functions of Tet2 may be more important in regulation of these lineages. Of note, in teratoma assay (which is merely a qualitative assay and not quantitative) both Tet2m/m and Tet2−/− ESCs form cell types of the three germ layers (Figure 1G). Together, these findings suggest that although loss of Tet2 or loss of its catalytic activity does not block differentiation to any particular germ layer, Tet2 catalytic dependent and independent functions can have subtle unique effects in fine-tuning commitment to each germ layer as found in our EB formation assay. Overall, the lack of self-renewal or pluripotency defects in Tet2m/m and Tet2−/− ESCs is in agreement with the overtly normal development of Tet2m/m and Tet2−/− mice25 and may underlie potential compensation by Tet1 which is highly expressed in ESCs and Tet3 which is induced on differentiation and is dynamically expressed during development.8,10 Of interest, loss of Tet2 blocks reprogramming of fibroblasts to iPSCs, a defect rescued only by re-expression of catalytically active Tet2 which demethylates pluripotency genes in fibroblasts.22 Previously, we have shown that Tet2 catalytic and noncatalytic functions regulate myeloid and lymphoid lineages respectively in the bone marrow.25 Together, our findings in the context of these studies highlight that the catalytic and noncatalytic functions of Tet2 impact various lineages and biological processes differently and it will be of interest to study these functions using lineage-specific differentiation approaches.
Although Tet2 is a key partner of Sin3a in ESC, Tet2m/m and Tet2−/− ESCs do not phenocopy Sin3a knockout ESCs where both self-renewal and pluripotency are severely compromised.19,29,38 This is not surprising as Sin3a has broader roles in gene regulation29 beyond partnering with Tet2. This is supported by our findings that only a subset of Sin3a peaks (∼28%) overlap with Tet2 peaks in ESCs. Likewise, Tet1 or Tet1/2 DKO ESCs, which have normal self-renewal but skewed differentiation toward trophectoderm lineages,8,10,12 are not phenocopying Sin3a knockout ESCs either. This reiterates a more global and critical involvement of Sin3a in regulating ESC self-renewal and pluripotency programs beyond its interaction with Tet1 or Tet2. It highlights that its partnership with Tet1 or Tet2 entails a subset of its activities, which if compromised do not impact ESC biology as severely as complete loss of Tets. Tet1, but not Tet2, has a Sin3a-interatcting-domain (SID) which if mutated prevents interaction with Sin3a and silencing of mesoderm genes.39 It would be interesting to map Tet2 domains essential for complex formation with Sin3a and specifically abrogate the Tet2-Sin3a complex formation. This will allow for defining the precise significance of this interaction in ESC gene regulation and biology, and uncouple it from the broader biological functions of Tet2 and Sin3a independent of this interaction.
Limitations of the study
This study does not map and compare the genome-wide distribution of catalytic mutant Tet2 in Tet2m/m ESCs to that of wildtype Tet2 in Tet2+/+ ESCs. Our limited analysis of two regulatory regions of pluripotency genes found a comparable enrichment of catalytic mutant and wildtype Tet2 in Tet2m/m and Tet2+/+ ESCs, respectively (Figure S2D). Although we anticipate the same at the global level, a comparative genome-wide mapping of wildtype and catalytic mutant Tet2 in ESCs will be needed to better assess similarities and differences in their global distribution. Based on our recent studies catalytic mutant and wildtype Tet1 have similar genomic distribution and enrichment levels in Tet1m/m and Tet1+/+ ESCs.12 We predict the same for catalytic mutant and wildtype Tet2. Any deviations may suggest that catalytic mutant Tet2 has altered binding affinity to DNA. Likewise, although our locus-specific analyses of 5hmC levels in Tet2m/m and Tet2−/− ESCs found comparable robust reductions in both lines, future comparisons of genome-wide mapping of 5hmC in Tet2m/m and Tet2−/− ESCs can better assess the presence of any differentially hydroxy-methylated sites between these two genotypes. That may also require taking into consideration whether Tet1 compensates for loss of Tet2 during 5hmC deposition in ESCs warranting further 5hmC mapping in Tet1 and Tet2 single and double catalytic deficient and knockout ESCs.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Tet2 | Abcam | ab124297 |
| Anti-Tet1 | Genetex | GTX125888 |
| Anti-Flag (for Western Blot) | Sigma | F1804 |
| Anti-Flag (for ChIP) | CST | 2368 |
| Anti-Sin3a | Abcam | ab3479 |
| Anti-H3K27ac | Abcam | ab4729 |
| Rabbit IgG isotype control DA1E | CST | 3900 |
| Guinea pig anti-rabbit | Antibodies Online | ABIN101961 |
| Anti-beta-actin | Abcam | AC-15 |
| Anti-Vinculin | Proteintech | 66305 |
| Anti-5hmC | Active Motif | 39069 |
| Goat Anti-Rabbit IgG-HRP | Millipore | 401393 |
| Goat Anti-Mouse IgG-HRP | Millipore | 401253 |
| Alexa Flour 594 anti-rabbit | Life Technologies | A21207 |
| Critical commercial assays | ||
| Omega E.Z.N.A. Total RNA kit | Omega | R6834-02 |
| Quick-DNA miniprep kit | Zymo | D3024 |
| Superscript III first strand | Invitrogen | 18080-400 |
| XtremeGene 9 DNA transfection reagent | Roche | 06365787001 |
| EpiQuik hMeDIP kit | Epigentek | P-1038-24 |
| Deposited data | ||
| RNA-seq | This paper | GEO:GSE213398 |
| CUT&Tag | This paper | GEO:GSE213398 |
| WGBS | This paper | GEO:GSE213398 |
| Experimental models: Cell lines | ||
| Tet2–/– mouse ESC | This paper | N.A. |
| Tet2m/m mouse ESC | This paper | N.A. |
| Tet2+/+;flag/flag mouse ESC | This paper | N.A. |
| Tet2m/m;flag/flag mouse ESC | This paper | N.A. |
| Oligonucleotides | ||
| RT-qPCR primers, see Table S2 | This paper | N.A. |
| Genotyping primers, see Table S2 | This paper | N.A. |
| gRNA oligos for gene targeting, see Table S2 | This paper | N.A. |
| Recombinant DNA | ||
| Topo pcr2.1 Tet2 mutant donor vector | This paper | N.A. |
| pX330-Tet2-gRNA | This paper | N.A. |
| Software and algorithms | ||
| Trim galore v0.6.5 | Babraham Institute | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| STAR v2.7.3a | N.A. | https://github.com/alexdobin/STAR |
| DESeq2 v1.20.0 | N.A. | http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html |
| DAVID 6.8 | N.A. | https://david.ncifcrf.gov |
| Bismark v0.22.3 | N.A. | https://github.com/FelixKrueger/Bismark |
| Methpipe v3.4.3 | N.A. | http://smithlabresearch.org/software/methpipe/ |
| HOMER v4.7 | UCSD | http://homer.ucsd.edu/homer/motif/ |
| ChIPseeker v1.30.3 | Github | https://guangchuangyu.github.io/software/ChIPseeker/ |
| Bedtools v2.30.0 | N.A. | https://bedtools.readthedocs.io/en/latest/ |
| deepTools v3.5.1 | N.A. | https://deeptools.readthedocs.io/en/develop/index.html |
| Bowtie2 v2.4.5 | John Hopkins University | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Integrative Genomics Viewer v2.12.3 | Broad Institute | http://software.broadinstitute.org/software/igv/ |
| MACS v2.2.7.1 | Github | https://github.com/macs3-project/MACS |
| GraphPad Prism 9 v9.3.1 | GraphPad | https://www.graphpad.com/ |
| ImageJ v1.53 | N.A. | https://imagej.nih.gov/ij/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author, Meelad M. Dawlaty (meelad.dawlaty@einsteinmed.edu).
Materials availability
This study generates Tet2 knockout and catalytic deficient mouse ESCs as well as endogenously Tet2-flag tagged mouse ESCs which are available upon request from the corresponding author.
Experimental model and study participant details
Cell lines
Generation of Tet2m/m, Tet2−/− and Tet2flag/flag ESCs
Tet2 catalytic-mutant (Tet2m/m), knockout (Tet2–/–), and Flag-tagged (Tet2flag/flag) ESCs (V6.5, mixed 129/B6, male) were generated following our previously published CRISPR/Cas9 gene editing protocols.40 Sequences of gRNAs, oligonucleotides and donor vectors used are shown in Table S1. To generate Tet2m/m ESCs, a gRNA targeting exon 9 was cloned into a px330-GFP vector. A gene block containing the amino acid substitutions H1367Y and D1369A in the catalytic domain of Tet2, silent mutations to introduce a unique HaeIII site, and flanking homology arms was synthesized and cloned into a Topo PCR2.1 vector. 1.5 μg of gRNA vector and 3.5 μg of donor vector were transfected into wildtype V6.5 mouse ESCs. Correctly targeted clones were confirmed by Sanger sequencing. For generating Tet2–/– ESCs, two gRNAs flanking exon 4 (Table S2) were designed and cloned into px330-Cherry vectors. 2.5 μg of each gRNA was transfected into wildtype ESCs and processed as just described. Correctly targeted clones were screened by PCR and loss of Tet2 mRNAs and protein were confirmed by qPCR (primers in exon 3 and 4), and Western blot (Tet2 Abcam 124297), respectively. To generate Tet2flag/flag ESCs, 3.5 μg of a ssDNA oligo containing a GGSG linker and a 3x-Flag sequence immediately before the stop codon of Tet2, and 1.5 μg of px330-GFP vector containing gRNA targeting the stop codon in exon 11 were transfected into wildtype and Tet2m/m ESCs. Correctly targeted clones were confirmed by PCR, Sanger sequencing and Western blot using Flag (Sigma M2 F1804) and Tet2 (Abcam 124297) antibodies. To generate Tet1, Tet2 double catalytic-mutant (DMUT), and double knockout (DKO) ESCs, we targeted the Tet2 locus in our previously published Tet1m/m and Tet1–/– ESCs12 using the same strategies described above and validated properly targeted clones by PCR, RFLP and Western blot.
Method details
Mouse embryonic stem cell (ESC) culture
ESCs (V6.5 mixed 129/B6 background, male) were cultured in 6-well plates on irradiated feeders in media containing serum and LIF (DMEM supplemented with 10% FBS, 2 mM glutamine, 1x non-essential amino acids, 100 U/mL penicillin, 100 g/mL streptomycin, 0.02 μg/mL LIF, 50 mM β-mercaptoethanol). For all experiments, ESCs were trypsinized, pre-plated for 45 mins to remove feeders, seeded onto gelatin-coated plates for 24 hrs, and then harvested for transcriptomic and epigenomic analyses. For differentiation to EBs, ESCs were pre-plated for 45 mins on gelatin to remove feeders and then suspended in hanging drops in the absence of LIF and cultured for 3 days as described before.12
Teratoma assay
ESCs were plated overnight on gelatin-coated 6-well plates and 2 million cells were harvested and injected subcutaneously on the flanks of SCID mice following our published protocols.12 Mice were euthanized and tumors were harvested 7 weeks later, fixed in formalin for 2 days, paraffin-embedded, sectioned, and stained with Hematoxylin and Eosin following standard procedures at Einstein Histopathology Core. Slides were imaged under light microscopy.
RT-qPCR, ChIP-qPCR, hMeDIP-qPCR
Real Time quantitative PCR (RT-qPCR) was performed as described before.12 Briefly, ESCs were cultured on feeder-free gelatin-coated plates for 1 day and RNA was extracted using Omega E.Z.N.A. Total RNA kit (R6834-02). RNA was subjected to cDNA synthesis using SuperScript III kit (Invitrogen, 18080-400), according to manufacturer’s instructions. cDNA was subjected to Real Time quantitative PCR (RT-qPCR) using SYBR green and primers (Table S2) in a BD Applied Biosystems StepOneTM Real-Time PCR System. For quantification of germ layer markers in EBs, RNA was extracted from day 3 EBs and RT-qPCR was performed as described above using primers from our previous studies.12 Data were normalized to Gapdh and wildtype controls. A one-way ANOVA test was used for statistical analysis. ChIP-qPCR for Tet2 in ESCs was performed using anti-Flag antibody (Sigma M2 F1804) and primers targeting Nanog enhancer 2, Oct4 promoter 1 and a negative region as described before.12,41 Data was normalized to IgG and the negative control region, and plotted as fold change. For hMeDIP-qPCR, ESCs were grown for 1 day in feeder-free gelatin-coated plates and pelleted. DNA was extracted by Quick-DNA miniprep kit (Zymo, D3024) following the manufacturer’s instructions. DNA was subjected to Hydroxymethylated DNA immunoprecipitation (hMeDIP) using EpiQuik hMeDIP kit (P-1038-24) following manufacturer’s instructions. RT-qPCR was performed as described above using primers targeting two loci (Bend3 and Ecat1) from our previous studies.13 Data was normalized to IgG and the negative control region, and plotted as fold change.
Western blot
Western blots were performed as described before12 using antibodies listed in Table S2. Briefly, ESCs were cultured on feeder-free gelatin-coated plates for 1 day and protein was extracted using RIPA buffer (50 mM Tris-HCl, pH 7.4, 250 mM NaCl, 2% Nonidet-P40, 2.5 mM EDTA, 0.1% SDS, 0.5% DOC) supplemented with Halt PIC (Thermo, 78430) and quantified with BSA assay (Thermo Scientific 23227). 20 μg of protein lysate was mixed with 2x Laemmli buffer and resolved on 7-9% SDS-PAGE gel (Mini-PROTEAN electrophoresis chamber, Bio-Rad), and transferred to PVDF membranes (Mini Trans-Blot apparatus, Bio-Rad) in 10% methanol transfer buffer following the manufacturer’s instructions. Membranes were blocked in 5% milk in PBS with 0.1% Tween-20 (PBS-T) and incubated overnight at 4°C with primary antibodies (anti-Tet2 1:1000 Abcam ab124297; anti-Tet1 1:3000 GeneTex, GTX125888; anti-Vinculin 1:1000 Proteintech 66305; anti-Flag 1:1000 Sigma M2 F1804; anti-Sin3a 1:1000 Abcam ab3479; anti-beta actin 1:30000 Abcam ab6276). Next day, membranes were washed twice with PBS-T (10min each) and incubated with secondary antibody (goat anti-mouse HRP 401253, or goat anti-rabbit HRP, 401393, Millipore 1:2500) for 1 hr at room temperature. Protein bands were detected using ECL chemiluminescence reagent (Amersham RPN2106) and standard radiography (Konica SRX-101A).
Immunostaining
Immunostainings were performed as described before.10 Briefly, ESCs were cultured on feeder-free gelatin-coated plates for 1 day, washed with PBS, fixed with 4% formaldehyde for 15 mins at room temperature, and washed with PBS. For detection of 5hmC, cells were first permeabilized (0.1% Triton X-100 in PBS) for 15 mins at room temperature, DNA was denatured by 2N HCl treatment for 30 mins, washed with 100 mM Tric-HCl for 5 mins, and blocked with 0.2% Triton X-100, 5% donkey serum for 30 mins. Cells were incubated with primary antibody (5hmC 1:100 Active Motif 39769) at 4°C overnight. The next day cells were washed three times PBS-T and then incubated with secondary antibody (Alexa Flour 594 anti-rabbit 1:500 Life tech A21207) for 1 hr at room temperature. Nuclei were counter-stained with DAPI (1:1000, 5 μg/ml stock). Cells were imaged using an inverted fluorescence microscope.
Immunoprecipitation (IP)
All IPs were performed as described before.12,41 Briefly, ESCs were expanded and cultured in 15-cm feeder-free gelatin-coated dishes for 1 day. Cells were washed and scraped in ice-cold PBS supplemented with Halt PIC (Thermo 78430). For crosslinked IPs, cells were incubated with 1% formaldehyde at room temperature for 8 mins, quenched with glycine (final concentration of 0.1 M), and washed with PBS before scrapping. Nuclear extracts were prepared and quantified with BSA assay (Thermo Scientific 23227). 1 mg of protein was incubated with 2 μg of antibody (anti-Flag CST, 2368; Rabbit-IgG CST, 3900) crosslinked to Protein G-conjugated magnetic beads (Dynabeads protein G, Invitrogen) overnight at 4°C. IPs were washed with buffer containing 20 mM HEPES, pH 7.6, 10% glycerol, 100 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA. Proteins were eluted by resuspending beads in Laemmli buffer and incubating them at 95°C for 5 mins and detected by Western blot as described above.
Chromatin immunoprecipitation (ChIP) for mass spectrometry (MS)
ChIP followed by MS was performed as described before.42,43 ESCs were grown in 15-cm feeder-free gelatin-coated dishes for 1 day and then crosslinked with 1% formaldehyde in serum-free media for 8 mins and quenched with glycine (to a final concentration of 0.1 M). Cells were washed with PBS, scraped with ice-cold PBS containing protease inhibitors (Halt PIC Thermo 78430), and pelleted. Pellets were resuspended in lysis buffer 1 (50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100) and incubated for 10 min at 4°C on a rotator. Lysate was centrifuged and the pellet was resuspended in lysis buffer 2 (10 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA) and incubated for 5 min at 4°C on a rotator. Lysate was cleared by centrifugation and pellet was resuspended in lysis buffer 3 (10 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% sodium deoxycholate, 0.5% N-lauroylsarcosine) and sonicated at 4C to breakdown DNA to a size range of 200-600 bp (Diagenode, Bioruptor UCD-200TM-EX; conditions: high setting, 30 sec ON/OFF cycles for 15 mins). Chromatin was quantified by BCA assay and 1 mg of chromatin was incubated with antibody-bound beads overnight (anti-Flag CST 2368; Rabbit-IgG CST, 3900; Protein G Dynabeads Invitrogen). The next day beads were washed, and proteins were eluted with 5% SDS in 50 mM ammonium bicarbonate (pH = 8) by incubating them at 95°C for 10 mins. Eluted proteins were incubated with 5 mM DTT for 30 mins at 54°C, followed by incubation with 20 mM iodoacetamide for 30 mins at room temperature in the dark. Afterward, phosphoric acid was added at a final concentration of 1.2%. Samples were diluted in six volumes of binding buffer (90% methanol and 10 mM ammonium bicarbonate, pH 8.0). After mixing, the protein solution was loaded to an S-trap filter (Protifi C02-micro) and spun at 500 g for 30 sec. The sample was washed twice with binding buffer. Finally, 1 μg of sequencing grade trypsin (Promega), diluted in 50 mM ammonium bicarbonate, was added into the S-trap filter and samples were digested at 37°C for 18 h. Peptides were eluted in three steps: (i) 40 μl of 50 mM ammonium bicarbonate, (ii) 40 μl of 0.1% TFA and (iii) 40 μl of 60% acetonitrile and 0.1% TFA. The peptide solution was pooled, spun at 1,000 g for 30 sec, and dried in a vacuum centrifuge. Prior to mass spectrometry analysis, samples were desalted using a 96-well plate filter (Orochem) packed with 1 mg of Oasis HLB C-18 resin (Waters). Briefly, the samples were resuspended in 100 μl of 0.1% TFA and loaded onto the HLB resin, which was previously equilibrated using 100 μl of the same buffer. After washing with 100 μl of 0.1% TFA, the samples were eluted with a buffer containing 70 μl of 60% acetonitrile and 0.1% TFA and then dried in a vacuum centrifuge.
Liquid chromatography followed by tandem mass spectrometry (LC-MS/MS)
Samples were resuspended in 10 μl of 0.1% TFA and loaded onto a Dionex RSLC Ultimate 300 (Thermo Scientific), coupled online with an Orbitrap Fusion Lumos (Thermo Scientific). Chromatographic separation was performed with a two-column system, consisting of a C-18 trap cartridge (300 μm ID, 5 mm length) and a picofrit analytical column (75 μm ID, 25 cm length) packed in-house with reversed-phase Repro-Sil Pur C18-AQ 3 μm resin. To analyze the proteome, peptides were separated using a 60 min gradient from 4-30% buffer B (buffer A: 0.1% formic acid, buffer B: 80% acetonitrile + 0.1% formic acid) at a flow rate of 300 nl/min. The mass spectrometer was set to acquire spectra in a data-dependent acquisition (DDA) mode. Briefly, the full MS scan was set to 300-1200 m/z in the orbitrap with a resolution of 120,000 (at 200 m/z) and an AGC target of 5x10e5. MS/MS was performed in the ion trap using the top speed mode (2 secs), an AGC target of 1x10e4 and an HCD collision energy of 35. Proteome raw files were searched using Proteome Discoverer software (v2.4, Thermo Scientific) using SEQUEST search engine and the SwissProt mouse database. The search for total proteome included variable modification of N-terminal acetylation, and fixed modification of carbamidomethyl cysteine. Trypsin was specified as the digestive enzyme with up to 2 missed cleavages allowed. Mass tolerance was set to 10 pm for precursor ions and 0.2 Da for product ions. Peptide and protein false discovery rate was set to 1%. Following the search, data was processed as described before.42 Briefly, proteins were log2 transformed, normalized by the average value of each sample and missing values were imputed using a normal distribution 2 standard deviations lower than the mean. Statistical regulation was assessed using heteroscedastic t-test (if p-value <0.05). Complete list of proteins identified is provided in Table S1.
RNA-seq and data analysis
RNA-seq was performed as described before.12 Briefly, ESCs (2 clones per genotype) were cultured on feeder-free, gelatin-coated plates overnight and total RNA was extracted using Omega E.Z.N.A. Total RNA kit I (R6834). Library preparation and mRNA sequencing were performed at Novogene using their Illumina Novoseq 6000 platform. Adaptors were trimmed using trim galore (v0.6.5) and clean reads were mapped to the mouse genome (mm10) using STAR (v2.7.3a) with default parameters. Gene counts were extracted from mapped reads using featureCounts with --largestOverlap parameter. Raw counts were used to identify differentially expressed genes with DESeq2 (FDR <0.05 and fold-change >1.5), following the standard package documentation. Gene ontology terms and KEGG pathways were identified on selected DEGs using DAVID (https://david.ncifcrf.gov/). All plots were made in R using custom scripts. A hypergeometric test was used to identify statistically significant (p <0.05) enrichment between different gene sets (16,756 genes with >10 counts in wildtype ESCs were assumed to be expressed and used as background).
WGBS and data analysis
Whole-genome bisulfite sequencing (WGBS) was performed as described previously.12 ESCs were grown for 1 day in feeder-free gelatin-coated plates and pelleted. DNA was extracted by Quick-DNA miniprep kit (Zymo, D3024) following the manufacturer’s instructions. Bisulfite conversion and sequencing were performed at BGI Genomics (https://en.genomics.cn/). Lamda DNA spike-in confirmed a >99.4% bisulfite conversion efficiency. The libraries were subjected to 100 bp pair-end sequencing on a HiSeq 4000 Illumina platform. The raw reads were filtered by SOAPnuke (v1.5.5) (https://github.com/BGI-flexlab/SOAPnuke) with the parameters -n 0.001 -l 20 -q 0.4 -A 0.25 -Q 2 -G to remove adaptors and filter out low-quality reads. Clean reads were mapped to mouse genome mm10 using Bismark (v0.22.3) with default parameters. Duplicated reads were removed using deduplicate_bismark and methylation status of each cytosine extracted with bismark_methylation_extractor. Differentially methylated regions (DMRs) were identified between Tet2m/m vs. Tet2+/+, and Tet2–/– vs. Tet2+/+ using MethPipe (v3.4.3) with standard parameters (regions >5 CpGs, methylation difference >20%, and FDR <0.05). DMR were annotated to genomic features with R package ChIPseeker (v1.30.3). Motif analysis of DMRs was performed by HOMER (v4.7). Methylation line plots at DMRs were generated using plotProfile function of deepTools (v3.5.1). For visualization on Integrative Genome Browser (IGV), bedGraph files were converted to bigwig using bedGraphToBigWig from bedtools (v2.30.0).
Cleavage under target & tagmentation (CUT&Tag) and data analysis
To map the genomic distribution of Sin3a, and H3K27ac, CUT&Tag was performed as described previously.12 Briefly, ESCs were cultured on feeder-free, gelatin-coated plates overnight, harvested, and counted. 500,000 cells (per genotype, per condition) were washed in PBS, crosslinked with 0.5% formaldehyde for 5 mins, quenched with glycine to final concentration of 375 mM and washed. Cells were bound to Concavalin A-coated beads, permeabilized, and incubated with primary antibodies (anti-Sin3a Abcam ab3479, anti-H3K27ac Abcam ab4729, and rabbit IgG isotype control CST 3900) overnight at 4°C. The next day, samples were incubated with secondary antibody (guinea pig anti-rabbit Antibodies Online ABIN101961) at room temperature followed by incubation with pre-loaded pA-Tn5. Transposase was activated by incubation with tagmentation buffer containing magnesium and incubated at 37°C. DNA was isolated by phenol-chloroform isoamyl alcohol extraction. DNA was amplified and sequencing libraries were generated using NEBNext HiFi 2x PCR Master mix and cleaned up with AMPure XP beads (#A63880). Libraries were subjected to 75bp paired-end sequencing using Illumina NextSeq 500 platform at the Einstein Epigenomics Core. Sequencing reads were mapped to the mouse genome (mm10) using bowtie2 (v2.4.5) with the following parameters: -local -very-sensitive-local -no-unal -no-mixed -no-discordant -I 10 -X 700, and removed of duplicates using Picard tools MarkDuplicates function. The resulting BAM files were balanced to sample with the lowest reads using samtools. Balanced BAM files were used to call peaks using MACS2 (v2.2.7.1) with the following parameters: -p 0.00001 -f BAMPE --keep-dup all. BED files were then intersected using bedtools with default parameters. Resulting BED files with genomic coordinates were annotated using ChIPseeker (v1.30.3). Line plots and heatmaps were generated using deepTools. Barplots were generated with custom scripts in R.
Identification of Tet2-bound enhancers in ESCs
To identify enhancers bound by Tet2, we annotated previously published coordinates of high-confidence Tet2 ChIP-seq peaks in wildtype mouse ESCs26 to genomic regions using ChIPseeker (v1.30.3), and excluded peaks at promoters (+/–2kb TSS). Next, we overlapped Tet2 non-promoter peaks to previously published coordinates of active, primed, and poised enhancers27 using beedtools (v2.30.0) intersect function with default parameters. We confirmed the enrichment of appropriate chromatin marks at Tet2-bound enhancers using published ATAC-seq,28 p300 and histone ChIP-seq tracks.27 The resulting coordinates and annotated genes were integrated with RNA-seq, WGBS, and CUT&Tag data described above.
Quantification and statistical analysis
We used One way ANOVA test or unpaired t-test in GraphPad Prism 7 to calculate statistical significance. Statistical methods used for genomewide studies and mass spectrometry experiments are explained in detail under the respective methods sub-sections.
Acknowledgments
We thank the Einstein Epigenomics Cores for help with next-generation sequencing. We are grateful to the Skoultchi lab at Einstein for providing reagents for CUT&Tag. We thank Masako Suzuki and Deyou Zheng for their advice on bioinformatics analyses. We are grateful to Ian MacArthur and other members of the Dawlaty laboratory for helpful suggestions and discussions. This work was supported by funds to M.M.D from NIH R01GM122839 (ESC work and gene regulation mechanisms) and NYSDOH/NYSTEM Contract C34877GG (DNA methylation analyses). M.M.D is also supported by NIH R01HL148852, Hirschle Trust Funds and funds from Albert Einstein College of Medicine Stem Cell Institute and Genetics Department. J.C.F. supported by NIH F31 predoctoral fellowship award F31GM140554. S.S. is supported by AFAR (Sagol Network GerOmics award), Deerfield (Xseed award), Relay Therapeutics, Merck, the NIH Office of the Director (1-S10-OD030286-01), the Einstein-Mount Sinai Diabetes Research Center, and the Einstein Cancer Center (P30-CA013330-47).
Author contributions
J.C.F. generated the cell lines, performed the experiments, analyzed data, and prepared the figures. S.S. performed mass spectrometry. M.M.D. and J.C.F. designed the study and wrote the manuscript with input from S.S. M.M.D. supervised the study and secured funding.
Declaration of interests
The authors declare no competing financial or other conflicts of interest.
Published: June 17, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107170.
Supplemental information
Data and code availability
-
•
The RNA-seq, CUT&Tag and WGBS data have been deposited in the Gene Expression Omnibus (GEO) database and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ito S., D'Alessio A.C., Taranova O.V., Hong K., Sowers L.C., Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y. Tet Proteins Can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu S.C., Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pastor W.A., Aravind L., Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bachman M., Uribe-Lewis S., Yang X., Williams M., Murrell A., Balasubramanian S. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem. 2014;6:1049–1055. doi: 10.1038/nchem.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawlaty M.M., Breiling A., Le T., Raddatz G., Barrasa M.I., Cheng A.W., Gao Q., Powell B.E., Li Z., Xu M., et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev. Cell. 2013;24:310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koh K.P., Yabuuchi A., Rao S., Huang Y., Cunniff K., Nardone J., Laiho A., Tahiliani M., Sommer C.A., Mostoslavsky G., et al. Tet1 and Tet2 Regulate 5-Hydroxymethylcytosine Production and Cell Lineage Specification in Mouse Embryonic Stem Cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawlaty M.M., Breiling A., Le T., Barrasa M.I., Raddatz G., Gao Q., Powell B.E., Cheng A.W., Faull K.F., Lyko F., Jaenisch R. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev. Cell. 2014;29:102–111. doi: 10.1016/j.devcel.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chrysanthou S., Flores J.C., Dawlaty M.M. Tet1 Suppresses p21 to Ensure Proper Cell Cycle Progression in Embryonic Stem Cells. Cells. 2022;11:1366. doi: 10.3390/cells11081366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chrysanthou S., Tang Q., Lee J., Taylor S.J., Zhao Y., Steidl U., Zheng D., Dawlaty M.M. The DNA dioxygenase Tet1 regulates H3K27 modification and embryonic stem cell biology independent of its catalytic activity. Nucleic Acids Res. 2022;50:3169–3189. doi: 10.1093/nar/gkac089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dawlaty M.M., Ganz K., Powell B.E., Hu Y.C., Markoulaki S., Cheng A.W., Gao Q., Kim J., Choi S.W., Page D.C., Jaenisch R. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai H.Q., Wang B.A., Yang L., Chen J.J., Zhu G.C., Sun M.L., Ge H., Wang R., Chapman D.L., Tang F., et al. TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling. Nature. 2016;538:528–532. doi: 10.1038/nature20095. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y., Chavez L., Chang X., Wang X., Pastor W.A., Kang J., Zepeda-Martínez J.A., Pape U.J., Jacobsen S.E., Peters B., Rao A. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2014;111:1361–1366. doi: 10.1073/pnas.1322921111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu F., Liu Y., Jiang L., Yamaguchi S., Zhang Y. Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev. 2014;28:2103–2119. doi: 10.1101/gad.248005.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu H., D'Alessio A.C., Ito S., Xia K., Wang Z., Cui K., Zhao K., Sun Y.E., Zhang Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams K., Christensen J., Pedersen M.T., Johansen J.V., Cloos P.A.C., Rappsilber J., Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu F., Zhu Q., Ye D., Zhang Q., Yang Y., Guo X., Liu Z., Jiapaer Z., Wan X., Wang G., et al. Sin3a-Tet1 interaction activates gene transcription and is required for embryonic stem cell pluripotency. Nucleic Acids Res. 2018;46:6026–6040. doi: 10.1093/nar/gky347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Q., Chen Y., Bian C., Fujiki R., Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Q., Zhao K., Shen Q., Han Y., Gu Y., Li X., Zhao D., Liu Y., Wang C., Zhang X., et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature. 2015;525:389–393. doi: 10.1038/nature15252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doege C.A., Inoue K., Yamashita T., Rhee D.B., Travis S., Fujita R., Guarnieri P., Bhagat G., Vanti W.B., Shih A., et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature. 2012;488:652–655. doi: 10.1038/nature11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sardina J.L., Collombet S., Tian T.V., Gómez A., Di Stefano B., Berenguer C., Brumbaugh J., Stadhouders R., Segura-Morales C., Gut M., et al. Transcription Factors Drive Tet2-Mediated Enhancer Demethylation to Reprogram Cell Fate. Cell Stem Cell. 2018;23:905–906. doi: 10.1016/j.stem.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guallar D., Bi X., Pardavila J.A., Huang X., Saenz C., Shi X., Zhou H., Faiola F., Ding J., Haruehanroengra P., et al. RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nat. Genet. 2018;50:443–451. doi: 10.1038/s41588-018-0060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito K., Lee J., Chrysanthou S., Zhao Y., Josephs K., Sato H., Teruya-Feldstein J., Zheng D., Dawlaty M.M., Ito K. Non-catalytic Roles of Tet2 Are Essential to Regulate Hematopoietic Stem and Progenitor Cell Homeostasis. Cell Rep. 2019;28:2480–2490.e4. doi: 10.1016/j.celrep.2019.07.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasmussen K.D., Berest I., Keβler S., Nishimura K., Simón-Carrasco L., Vassiliou G.S., Pedersen M.T., Christensen J., Zaugg J.B., Helin K. TET2 binding to enhancers facilitates transcription factor recruitment in hematopoietic cells. Genome Res. 2019;29:564–575. doi: 10.1101/gr.239277.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cruz-Molina S., Respuela P., Tebartz C., Kolovos P., Nikolic M., Fueyo R., van Ijcken W.F.J., Grosveld F., Frommolt P., Bazzi H., Rada-Iglesias A. PRC2 Facilitates the Regulatory Topology Required for Poised Enhancer Function during Pluripotent Stem Cell Differentiation. Cell Stem Cell. 2017;20:689–705.e9. doi: 10.1016/j.stem.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Chronis C., Fiziev P., Papp B., Butz S., Bonora G., Sabri S., Ernst J., Plath K. Cooperative Binding of Transcription Factors Orchestrates Reprogramming. Cell. 2017;168:442–459.e20. doi: 10.1016/j.cell.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams G.E., Chandru A., Cowley S.M. Co-repressor, co-activator and general transcription factor: the many faces of the Sin3 histone deacetylase (HDAC) complex. Biochem. J. 2018;475:3921–3932. doi: 10.1042/BCJ20170314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams K., Christensen J., Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2011;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin Y., Morgunova E., Jolma A., Kaasinen E., Sahu B., Khund-Sayeed S., Das P.K., Kivioja T., Dave K., Zhong F., et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017;356:eaaj2239. doi: 10.1126/science.aaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Domcke S., Bardet A.F., Adrian Ginno P., Hartl D., Burger L., Schübeler D. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature. 2015;528:575–579. doi: 10.1038/nature16462. [DOI] [PubMed] [Google Scholar]
- 33.Cheng S., Mittnenzweig M., Mayshar Y., Lifshitz A., Dunjić M., Rais Y., Ben-Yair R., Gehrs S., Chomsky E., Mukamel Z., et al. The intrinsic and extrinsic effects of TET proteins during gastrulation. Cell. 2022;185:3169–3185.e20. doi: 10.1016/j.cell.2022.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Streubel G., Fitzpatrick D.J., Oliviero G., Scelfo A., Moran B., Das S., Munawar N., Watson A., Wynne K., Negri G.L., et al. Fam60a defines a variant Sin3a-Hdac complex in embryonic stem cells required for self-renewal. EMBO J. 2017;36:2216–2232. doi: 10.15252/embj.201696307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saunders A., Huang X., Fidalgo M., Reimer M.H., Jr., Faiola F., Ding J., Sánchez-Priego C., Guallar D., Sáenz C., Li D., Wang J. The SIN3A/HDAC Corepressor Complex Functionally Cooperates with NANOG to Promote Pluripotency. Cell Rep. 2017;18:1713–1726. doi: 10.1016/j.celrep.2017.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiong J., Zhang Z., Chen J., Huang H., Xu Y., Ding X., Zheng Y., Nishinakamura R., Xu G.L., Wang H., et al. Cooperative Action between SALL4A and TET Proteins in Stepwise Oxidation of 5-Methylcytosine. Mol. Cell. 2016;64:913–925. doi: 10.1016/j.molcel.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 37.Vella P., Scelfo A., Jammula S., Chiacchiera F., Williams K., Cuomo A., Roberto A., Christensen J., Bonaldi T., Helin K., Pasini D. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol. Cell. 2013;49:645–656. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 38.McDonel P., Demmers J., Tan D.W.M., Watt F., Hendrich B.D. Sin3a is essential for the genome integrity and viability of pluripotent cells. Dev. Biol. 2012;363:62–73. doi: 10.1016/j.ydbio.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chandru A., Bate N., Vuister G.W., Cowley S.M. Sin3A recruits Tet1 to the PAH1 domain via a highly conserved Sin3-Interaction Domain. Sci. Rep. 2018;8:14689. doi: 10.1038/s41598-018-32942-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravichandran M., Lei R., Tang Q., Zhao Y., Lee J., Ma L., Chrysanthou S., Lorton B.M., Cvekl A., Shechter D., et al. Rinf Regulates Pluripotency Network Genes and Tet Enzymes in Embryonic Stem Cells. Cell Rep. 2019;28:1993–2003.e5. doi: 10.1016/j.celrep.2019.07.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aguilan J.T., Kulej K., Sidoli S. Guide for protein fold change and p-value calculation for non-experts in proteomics. Mol. Omics. 2020;16:573–582. doi: 10.1039/d0mo00087f. [DOI] [PubMed] [Google Scholar]
- 43.Mohammed H., Taylor C., Brown G.D., Papachristou E.K., Carroll J.S., D'Santos C.S. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat. Protoc. 2016;11:316–326. doi: 10.1038/nprot.2016.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The RNA-seq, CUT&Tag and WGBS data have been deposited in the Gene Expression Omnibus (GEO) database and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.