Abstract
Although aptamers have shown excellent target specificity in preclinical and clinical studies either by themselves or as aptamer‐drug conjugates, their in vivo tissue pharmacokinetic (PK) analysis is still problematic. We aimed to examine the utility of image‐based positron emission tomography (PET) to evaluate in vivo tissue PK, target specificity, and applicability of oligonucleotides. For this, fluorine‐18‐labeled aptamers with erb‐b2 receptor tyrosine kinase 2 (ERBB2)‐specific binding were synthesized by base‐pair hybridization using a complementary oligonucleotide platform. To investigate the PKs and properties of in vivo tissue, usefulness of in vivo PET imaging in the development of an oligonucleotide‐based drug as an assessment tool was evaluated in normal and tumor xenografted mice. ERBB2‐cODN‐idT‐APs‐[18F]F ([18F]1), injected intravenously showed significant and rapid uptake in most tissues except for the initial brain and muscle; the uptake was highest in the heart, followed by kidneys, liver, lungs, gall bladder, spleen, and stomach. The main route of excretion was through the renal tract ~77.8%, whereas about 8.3% was through the biliary tract of the total dose. The estimated effective dose for an adult woman was 0.00189 mGy/MBq, which might be safe. ERBB2‐positive tumor could be well visualized in the KPL4 xenograft animal model by in vivo PET imaging. Consequently, the distribution in each organ including ERBB2 expression could be well determined and quantified by PET with fluorine‐18‐labeled aptamers. In vivo PK parameters such as terminal half‐life, time to maximum concentration, area under the curve, and maximum concentration, were also successfully estimated. These results suggest that image‐based PET with radioisotope‐labeled aptamers could be provide valuable information on properties of oligonucleotide‐based drugs in drug discovery of targeted therapeutics against various diseases.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Image‐based serial positron emission tomography (PET) in living subjects with radioisotope‐labeled aptamers may potentially be useful during the development process and could provide valuable information to visualize and quantify the tissue pharmacokinetic (PK) properties for the characterization of oligonucleotide‐based drug candidates.
WHAT QUESTION DID THIS STUDY ADDRESS?
What is the tissue PK profile of oligonucleotide‐based drug candidates when administered with ERBB2‐cODN‐idT‐APs‐[18F]F ([18F]1) to normal and ERBB2‐positive tumor xenografted mice?
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
Quantitative real‐time polymerase chain reaction currently used to evaluate the PKs of aptamers is controversial because the concentration of the drug in the blood may not reflect the concentration in the tissue. Tissue‐blood PK modeling by image‐based PET can provide valuable information on properties of oligonucleotide‐based drug candidates in the context of various diseases.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
PET/computed tomography data using radiolabeled aptamers can accelerate translational studies by providing information on in vivo properties by exploring tissue PKs in early drug development.
INTRODUCTION
Aptamers are artificial nucleic acid ligands derived from single‐stranded DNA/RNA with tertiary structures ranging from 20 to 100 base pairs in length. 1 Aptamers include short nucleic acid sequences, which exhibit specific high‐affinity molecule binding. This has enabled the systematic development of ligands by exponential enrichment (SELEX) to achieve high‐affinity target binding with binding affinities in the picomolar range comparable to antibodies or small molecules. 2 , 3 Therefore, aptamers have been explored as therapeutic drug candidates for the treatment of various diseases. 3 , 4 Simultaneously, there is a growing demand for viable methods for evaluating and validating aptamers that have already been or are under development. 3 The advantage of aptamers is that they are chemically synthesized polymer materials that can be produced without animal testing and without involving potentially unsafe moieties, such as antigenic proteins or viruses in the development stage. 5 , 6 Large‐scale production of aptamers is also much easier and cheaper than antibody production, despite the nucleotide analogs of antibodies. 7 , 8 Thus, aptamers can provide a new paradigm for developing novel therapeutic as well as diagnostic modalities. 9
The pharmacokinetic (PK) parameters of a drug, such as the duration of action, are very important in preclinical and clinical efficacy studies. However, the methodology for evaluating the PK/PD of aptamers in preclinical and clinical studies remains controversial. 10 , 11 Although small molecule‐based drug candidates can be analyzed using methods such as autoradiography or mass balance, it is necessary to establish a research method for macromolecular biopharmaceuticals such as high‐molecular‐weight antibodies. If aptamers are to be approved as new drugs, it is necessary to establish PK and pharmacodynamic (PD) evaluation methods for these molecules. PK characteristics describe the absorption, distribution, metabolism, and excretion (ADME) of drug candidates in living subjects. Real‐time quantitative polymerase chain reaction (qPCR) is still used as an established method for analyzing aptamers. 12 , 13 However, evaluating the PK and PD parameters of aptamers by qPCR is controversial because the drug concentrations in the blood may not reflect those in tissues. If blood and tissue concentrations are available, tissue–blood PK modeling can help further optimize drug dosing regimens.
Positron emission tomography (PET) is a molecular imaging modality that enables the noninvasive, real‐time visualization of biochemical events at the cellular and molecular level in living subjects. 14 PET can be used to study the specific and selective binding of radioisotope‐labeled drug candidates to a target, and has advantages over conventional PK studies. 15 , 16 , 17 , 18 The superior sensitivity of PET and the exceptional temporal and spatial resolution of PET images allows for the visualization and quantification of tissue PK properties of drug candidates in organs and lesions of interest. 19 , 20 , 21 , 22 , 23 Thus, image‐based PET can be used for the identification of biochemical or pathological phenomena, diagnosis of diseases, treatment prognosis, and treatment planning. 24 , 25
Herein, we aimed to evaluate the in vivo tissue PKs and target specificity of an oligonucleotide as a new drug candidate (K d = 3.1 nM; INTEROligo Corporation), which is developing for the treatment of erb‐b2 receptor tyrosine kinase 2 (ERBB2, also known as HER2)‐positive breast cancer by PET. We present data on preclinical PK parameters, such as terminal half‐life (t 1/2), time to maximum concentration (T max), area under the curve (AUC), and maximum concentration (C max) for each organ in normal and ERBB2‐positive tumor‐xenografted mice; we also determined the excretion route. Several direct and indirect labeling methods on oligonucleotides with PET or single photon emission computed tomography (SPECT) isotopes, such as C‐11, F‐18, Cu‐64, Tc‐99m, or Br‐76, have been reported and molecular imaging trials in experimental animals have been investigated. 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 However, a labeling method that can be commonly applied to various aptamers still is a challenging process because reaction conditions can vary depending on the heterogeneity of the aptamers. We, in present study, selected radioisotope fluorine‐18, which is the most frequently used radioisotope in PET radiopharmaceuticals. Of the nuclides mentioned above, fluorine‐18 has an ideal half‐life, well‐established and versatile chemistry along with relatively high molar activity, and thermally stable and oxidation resistant of carbon‐fluorine bond. 37 In addition, ERBB2‐specific and selective binding aptamers labeled with fluorine‐18 (ERBB2‐cODN‐idT‐APs‐[18F]F, [18F]1) were synthesized by base‐pair hybridization using a complementary oligonucleotide platform to label water‐friendly aptamers. We thus show that the organ‐specific detection and quantification of fluorine‐18‐labeled target‐specific aptamers is feasible, most likely safe, and can be used for the determination of PK parameters of similar potential drug candidates.
MATERIALS AND METHODS
Materials
ERBB2‐specific binding aptamers, intermediates, and standard aptamers for radiosynthesis were provided by the INTEROligo Corporation. ERBB2‐APs originally developed are 40‐mer DNA aptamers (molecular weight: 14,067.80, K d = 3.1 nM) and its 3′ acid group has been modified with complementary oligonucleotides (cODN, CAG CCA CAC CAC CAG) and inverted‐deoxythymidine (idT) to conjugate fluorine‐18 and to expect the enhancement of blood circulation, respectively, gave ERBB2‐cODN‐idT‐APs (6, molecular weight: 18,909.91, K d = 8.2 nM). The 11‐azido‐3,6,9‐trioxa‐1‐undecanol mesylate (N3‐PEG3‐OMs, 2) was purchased from FutureChem Co. Sep‐Pak Accell Plus QMA Carbonate Plus Light Cartridges and Sep‐Pak C18 Plus Short Cartridge were purchased from Waters. All other chemicals and solvents were commercially obtained from Sigma‐Aldrich and were used without further modification or purification.
Radiosynthesis of ERBB2‐cODN‐idT‐APs‐[ 18F]F ([ 18F]1)
The ERBB2‐specific and selective binding aptamer labeled with the radioisotope, fluorine‐18, (ERBB2‐cODN‐idT‐APs‐[18F]F, [18F]1) was prepared by base‐pair hybridization using a complementary oligonucleotide platform, according to the literature with minor modifications. 38 A schematic illustration of ERBB2‐cODN‐idT‐APs‐[18F]F, ([18F]1) is shown in Figure 1. Details of the reaction conditions and chromatogram of each step (Figures S1–S11) are described in Appendix S1.
FIGURE 1.
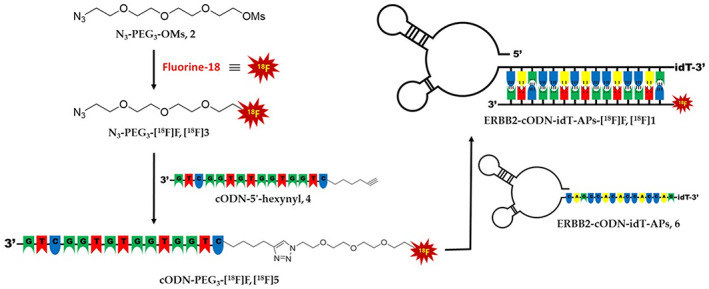
Schematic illustration of erb‐b2 receptor tyrosine kinase 2 (ERBB2)‐targeting aptamers labeled with fluorine‐18.
Animals
ICR mice (female, 8 weeks old, 35.9 ± 0.5 g, n = 4) and BALB/c nude mice (female, 9 weeks, 26.6 ± 1.6 g, n = 4) were purchased from Orient Bio Inc. All animals were maintained at a controlled temperature (24 ± 2°C), humidity (40 ± 2%), and 12 h light–dark cycle. The animals were acclimated for 1 week with the provision of sufficient water and food. The ICR mice were used as normal subjects to evaluate the in vivo tissue PK characteristics of ERBB2‐cODN‐idT‐APs‐[18F]F ([18F]1) and the internal absorbed dose, whereas the BALB/c nude mice were used to construct a breast cancer xenograft model by implanting KPL4 cells (2.0 × 107 cells) on the sides of the mice. The animals were proceeded for PET/computed tomography (CT) imaging once the breast tumor xenografts reached ~281.2 ± 97 mm3 (~14–18 days postinjection). The study was conducted in accordance with institutional guidelines, and the protocol was approved by the Ewha Womans University College of Medicine Institutional Animal Care and Use Committee (EWHA MEDIACUC past‐021, January 25, 2021).
PET/CT imaging
All mice were fasted for 6 h before imaging with PET/CT. Mice were anesthetized with a mixture of 2.5% isoflurane and 7:3 N2/O2 during imaging with a small‐animal‐dedicated PET/CT scanner (nanoScan PET/CT; Mediso Medical Imaging Systems, Budapest, Hungary). To investigate the in vivo tissue PKs of ERBB2‐cODN‐idT‐APs‐[18F]F ([18F]1), [18F]1 was intravenously injected into ICR mice (female, 8 weeks old, 35.9 ± 0.5 g, n = 4) at a single dose of 6.9 ± 0.2 MBq in 200 μL of saline. Whole‐body PET/CT images in the list mode were obtained concomitantly with the intravenous injection of [18F]1 for 90 min. CT scans were used for attenuation correction and anatomic localization of the PET signals. The acquired PET images were reconstructed using the three‐dimensional Adjoint Monte Carlo method with scatter and random correction. Whole‐body PET images were used to determine the biodistribution of [18F]1 and time‐radioactivity profiles were generated for each organ of interest. Canvas images of volume‐of‐interest (VOI) drawings were chosen from the CT and PET images according to organ visibility and mobility. VOIs for the brain, heart, kidneys, liver, lungs, spleen, stomach, and muscles were drawn in a slice‐by‐slice manner on CT images of individual subjects to analyze the uptake over time, whereas those for the gallbladder, intestines, and urinary bladder were drawn on averaged PET images by adjusting the isocontour VOIs. In addition, using dynamic whole‐body PET scans during 90 min, the blood PK was applied a factor analysis approach to extract the temporal changes in blood distribution. 39 The uptake in each organ was expressed as the percentage of injected dose (ID)/g using the following formula: % ID/g = ([total amount of radioactivity in organ]/[amount of radioactivity intravenously administered]) × volume of organ × 100. The PET images were quantitatively analyzed using the InterView Fusion software (version 3.03.089.0000; Mediso Medical Imaging Systems). The representative PK parameters of [18F]1 in each organ were quantitatively assessed based on the time–concentration profiles of the organs of interest and calculated using GraphPad Prism version 5.0 (GraphPad Software). The following PK parameters were obtained: C max, T max, t 1/2, and AUC from time 0 to 5400 s (AUC0–5400 s). For evaluating target tissue PK, BALB/c nude mice (female, 9 weeks, 26.6 ± 1.6 g, n = 4) bearing ERBB2‐amplified KPL4 breast cancer cells were fasted for 6 h before PET image acquisition. The [18F]1 was intravenously injected to the disease model animals at a single dose of 9.4 ± 0.2 MBq in 200 μL of saline while maintaining respiratory anesthesia using isoflurane; subsequently, whole‐body PET/CT images in the list mode were acquired using nanoScan PET/CT for 90 min. PET/CT data were reconstructed into 3D images and analyzed using the InterView Fusion software (Mediso Medical Imaging Systems). The radioactivity uptake of [18F]1 in the tumor tissue was calculated over time, and PK parameters, such as AUC, t 1/2, C max, and T max, were analyzed using a time‐concentration profile. The data for C max, t 1/2, and AUC are presented in geometrical mean ± geometrical standard deviation and T max as median (minimum–maximum).
Internal dosimetry of experimental animals
Internal dosimetry of the experimental animals was performed using an image‐based approach with Geant4 Monte Carlo N‐particle codes according to previously described methods. 22 , 40 PET/CT images were acquired for 90 min in the list mode after intravenous injection of [18F]1 and additionally acquired at 150, 240, and 360 min postinjection (Figure S12). The residence times were calculated based on the time‐activity curves of the acquired VOI PET data. The estimated human absorbed dose of [18F]1 was acquired from the calculated small‐animal data; the S value for adult female mice and the corresponding residence times were used for this purpose.
Immunocytochemistry
KPL4 (a human breast cancer cell line) cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum (v/v, Hyclone; Thermo Fisher Scientific) and 1% penicillin–streptomycin (v/v, Gibco; Thermo Fisher Scientific). Culture flasks were maintained in a humidified atmosphere of 95% air and 5% carbon dioxide environment at 37°C. KPL4 cells were seeded at 1 × 104 cells/well in a confocal dish. The cells were incubated with 5% bovine serum albumin and anti‐ERBB2 antibody (Abcam) for 1 h at 4°C. The cells were then incubated with Alexa Fluor 594‐conjugated secondary antibody (excitation: 561 nm, emission: 594 nm) for 1 h at room temperature. The nuclei were stained with 4′,6‐diamidino‐2‐phenylindole. The cells were visualized using a fluorescence microscope (Laser scanning microscope 780; Zeiss).
RESULTS
Radiosynthesis of ERBB2‐cODN‐idT‐APs‐[ 18F]F, [ 18F]1
To conjugate fluorine‐18 to the aptamers, ERBB2‐cODN‐idT‐APs‐[18F]F ([18F]1) was prepared via three steps, as shown in Figure 1: (1) radiofluorination, (2) Cu‐mediated click reaction, and (3) hybridization of aptamers using cODN. First, the precursor, N3‐PEG3‐OMs (2) and fluorine‐18 were subjected to conventional nucleophilic radiofluorination conditions with tetra‐n‐butylammonium bicarbonate as a phase‐transfer catalyst, to obtain N3‐PEG3‐[18F]F ([18F]3). The radioactivity yield of N3‐PEG3‐[18F]F ([18F]3) was 32.5 ± 2.8% (n = 44, non‐decay corrected [n.d.c.]) with over 99% of the radiochemical purity. Next, the 5′‐hexynyl‐functionalized complementary oligonucleotide (4, cODN‐5′‐hexynyl) was conjugated with N3‐PEG3‐[18F]F ([18F]3) via a Cu‐mediated click reaction with a conversion yield of ~80%; cODN‐PEG3‐[18F]F ([18F]5) was obtained with a radioactivity yield of 29.2 ± 4.3% (n = 29, n.d.c.); this was calculated from the initial radioactivity of N3‐PEG3‐[18F]F ([18F]3). The radiochemical purity of [18F]5 was found to be greater than 99%, and each fluorine‐18 labeled compound was confirmed by co‐injection high‐performance liquid chromatography (HPLC) with the corresponding standard compound. Finally, a high hybridization efficiency of over 95% was achieved at 100 nmol concentration of ERBB2‐cODN‐idT‐APs (6). The total elapsed time for the synthesis of ERBB2‐cODN‐idT‐APs‐[18F]F, [18F]1, was ~4.5 h, including the two HPLC separation processes. The final radioactivity yield was ~4.7 ± 1.2% (n = 16, n.d.c.), and the radiochemical purity as determined by HPLC was found to be greater than 95%.
Preclinical in vivo tissue PKs of ERBB2‐cODN‐idT‐APs‐[ 18F]F ([ 18F]1) by PET/CT
To evaluate the in vivo tissue PKs of ERBB2‐cODN‐idT‐APs‐[18F]F in normal mice, hybridized aptamers, [18F]1, was intravenously administered to ICR mice. Whole‐body PET/CT images in the list mode were obtained concomitantly with injection. The acquired PET images were reconstructed into three‐dimensional images by combining with CT images. Representative PET/CT images as time and time–radioactivity profiles generated for each organ of interest determined from whole‐body PET images are shown in Figures 2 and 3, respectively. In addition, PK parameters, such as t 1/2, T max, AUC, and C max, were analyzed based on the quantified biodistribution data of [18F]1 on each organ over time (Table S1); and the excretion routes were also estimated. The ratio of AUCs for various organs to the blood were also plotted to estimate the distribution of aptamers in tissues relative to the plasma over a late time‐interval (70–90 min; Figure S13).
FIGURE 2.
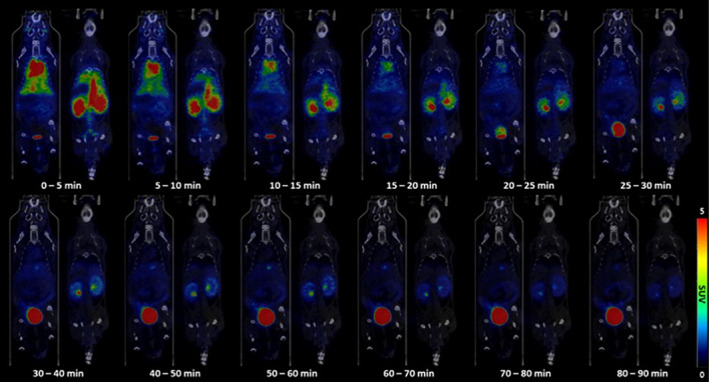
Representative positron emission tomography/computed tomography images of ERBB2‐cODN‐idT‐APs‐[18F]F ([18F]1) in various organs of the body over time in healthy mice.
FIGURE 3.
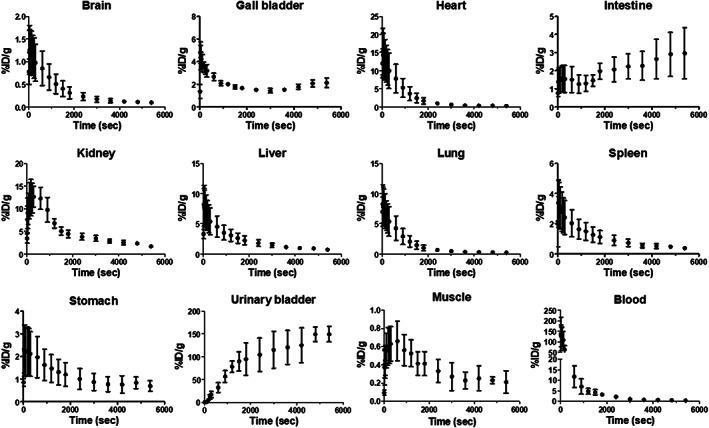
Time‐activity curves of ERBB2‐cODN‐idT‐APs‐[18F]F ([18F]1) calculated from whole‐body positron emission tomography images.
Initially, intravenously administered [18F]1 showed significant and rapid uptake in most tissues except for the initial brain and muscle; the uptake was highest in the heart, followed by kidneys, liver, lungs, gall bladder, spleen, and stomach. The [18F]1 was found in the initial heart (C max = 13.9 ± 1.7% ID/g and T max = 30.0 [30.0–30.0] s), liver (C max = 7.6 ± 1.7% ID/g and T max = 30.0 [30.0–30.0] s), and lungs (C max = 7.7 ± 1.7% ID/g and T max = 30.0 [30.0–45.0] s); organs with high uptake over time showed rapidly elimination of the aptamer (t 1/2; heart = 505.8 ± 1.3 s, liver = 628.1 ± 1.1 s, and lungs = 497.1 ± 1.3 s). In the brain (C max = 1.2 ± 1.7% ID/g) and muscles (C max = 0.6 ± 1.4% ID/g), there was negligible uptake of [18F]1; the stomach (C max = 2.1 ± 1.7% ID/g) and spleen (C max = 3.1 ± 1.9% ID/g) showed an initial moderate uptake but subsequently exhibited slow excretion of the aptamer over time (t 1/2; stomach = 1025.0 ± 1.7 s and spleen = 697.2 ± 1.2 s).
A significant amount of radioactivity was distributed to the kidneys and urinary bladder, and moderate radioactivity was observed in the gallbladder, liver, and intestines. Most of the intravenously administered [18F]1 was probably shown to be excreted via the renal route (AUC0‐5400 s; kidneys = 26,519.2 ± 1.2% ID/g s and urinary bladder = 524,726.6 ± 1.3% ID/g s), whereas moderate hepatic metabolism was observed via the biliary route (AUC0‐5400 s; intestines = 11,090.9 ± 1.3% ID/g s and gall bladder = 10,420.8 ± 1.1 %ID/g s). Of the total amount of intravenously administered [18F]1, 77.8% (=AUC0–5400 s, urinary bladder + kidneys/AUC0–5400 s, total organs × 100%) was excreted via the renal excretion route, whereas ~8.3% (=AUC0–5400 s, intestine + gall bladder/AUC0–5400 s total organs × 100%) was excreted via the biliary route.
Immunocytochemistry
KPL4 tumor cells are characterized by the overexpression of the ERBB2 protein. Immunocytochemistry was performed to determine whether ERBB2 was overexpressed in KPL4 cells used in this study with specific antibody. As shown in Figure 4, immunocytochemistry images confirmed the high expression of ERBB2 protein in the cells and a xenograft model was constructed by implanting KPL4 cells for in vivo target tissue PKs by PET/CT.
FIGURE 4.
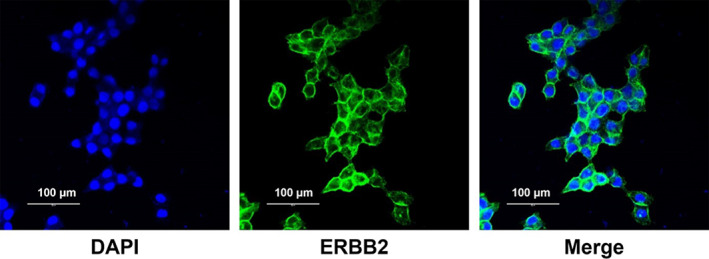
Expression analysis of ERBB2 in KPL4 cells.
In vivo target tissue PKs of ERBB2‐cODN‐idT‐APs‐[ 18F]F ([ 18F]1) by PET/CT in ERBB2 overexpressed KPL4 transplanted mice
To investigate target‐specific accumulation, [18F]1 was evaluated in ERBB2‐positive tumor (KPL4)‐bearing mice by PET/CT. Figure 5 shows representative PET/CT images of [18F]1 in the disease model. The distribution in the major organs of the intravenously administered ERBB2‐cODN‐idT‐APs‐[18F]F ([18F]1) throughout the body over time in the disease model appeared to be similar to that in the control group (data not shown). This is likely due to relatively lower tumor uptake. High uptake was observed in the kidneys, lungs, intestines, heart, and liver, and this caused poor tumor‐to‐background contrast in the maximum‐intensity projection. The maximum uptake value in the tumor tissue was observed immediately after administration (T max = 210.0 [120.0–300.0] s and C max = 0.86 ± 1.06% ID/g); excretion of the aptamer was observed subsequently over time (t 1/2 = 3666.3 ± 1.8 s; Table S2).
FIGURE 5.
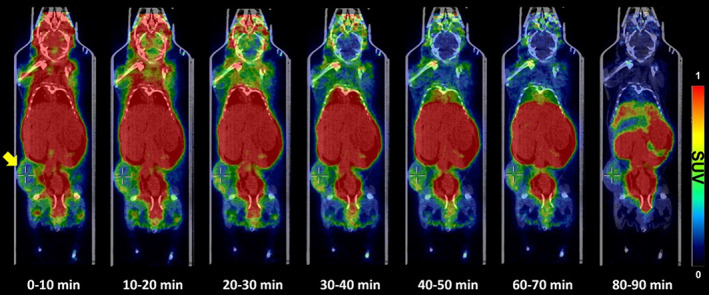
Representative positron emission tomography/computed tomography images of ERBB2‐cODN‐idT‐APs‐[18F]F ([18F]1) distributed throughout the body over time in ERBB2‐positive tumor (KPL4, yellow arrow)‐bearing mice. SUV, standard uptake value.
Internal absorbed dose calculation
The internal absorbed dose in humans was estimated using the values obtained in mice. Using the calculated S‐value and residence time for the internal absorbed dose in mice, the actual dose received by the mouse was estimated to be 1.83 mSv/MBq throughout the body, as summarized in Table S3. The organ‐level absorbed dose in adult women was evaluated using the residence time of [18F]1 in animals and the S‐value in adult women; the effective dose for adult women was calculated to be 0.00189 mGy/MBq as summarized in Table 1. A high absorbed dose was detected in the lower large intestine wall, stomach wall, liver, lungs, ovaries, and the urinary bladder wall.
TABLE 1.
Internal absorbed dose in adult women (mGy/MBq) estimated using small‐animal biodistribution measurements.
| Organs | Absorbed dose (mGy/MBq) |
|---|---|
| Adrenal glands | 3.94E‐06 |
| Brain | 2.80E‐06 |
| Breasts | 1.14E‐05 |
| Gallbladder wall | 0.00E+00 |
| LLI wall | 1.03E‐04 |
| Small intestine | 1.45E‐05 |
| Stomach wall | 1.10E‐04 |
| ULI wall | 4.53E‐06 |
| Heart wall | 0.00E+00 |
| Kidneys | 2.92E‐05 |
| Liver | 1.79E‐04 |
| Lungs | 2.30E‐04 |
| Muscle | 3.73E‐06 |
| Ovaries | 1.95E‐04 |
| Pancreas | 3.73E‐06 |
| Red marrow | 5.08E‐05 |
| Osteogenic cells | 3.23E‐06 |
| Skin | 2.04E‐06 |
| Spleen | 9.66E‐06 |
| Urinary bladder wall | 9.14E‐04 |
| Uterus | 7.10E‐06 |
| Effective dose | 1.89E‐03 |
Abbreviations: LLI, lower large intestine; ULI, upper large intestine.
DISCUSSION
One of the most important properties of aptamers is that they are chemically synthesized single‐stranded RNA or DNA oligonucleotides and have the properties of high specificity and affinity for their targets; the targets range from small ions, peptides, and proteins, to tissues. Aptamers are also termed as chemical antibodies because they bind to their targets through specific three‐dimensional structures and usually exhibit pico‐ to nano‐molar binding affinity. 41 More importantly, due to their high thermal stability, non‐immunogenic properties, and well‐established synthesis protocols and chemical modification techniques, aptamers have become promising tools for targeted therapy in a variety of diseases. One such drug, Macugen (Pegaptanib), has been approved by the US Food and Drug Administration in 2004 for the treatment of wet age‐related macular degeneration, and another aptamer drug, AS1411, has shown potential as an anticancer agent in a wide range of cancers. 42 , 43 , 44 In addition, various studies on aptamers have highlighted their potential applicability in a variety of biomedical fields including therapeutics. Aptamers were first introduced 2 decades ago and due to their unique chemical properties and biological functions, they have been used in diverse areas, such as targeted therapeutics (as aptamers alone or as aptamer‐drug conjugates), in vitro and in vivo imaging, and nanotechnology. However, the methods to study the PK and PD properties of aptamers in living subjects remains a controversial issue. Information on the ADME of aptamers is essential for understanding their PK properties after administration, and thus is crucial for their success as new drug candidates.
The main objective of the present study was to evaluate the preclinical PK characteristics, target efficacy, and absorbed radiation dose of an oligonucleotide‐based ERBB2‐specific binding aptamer drug candidate using PET. To accomplish this goal, the oligonucleotide‐based drug candidate was labeled with the radioisotope fluorine‐18 in three steps: radiofluorination, Cu‐mediated click reaction, and hybridization. Radiofluorination and copper‐mediated click reaction steps provided a relatively high radioactivity yield, at ~30% (n.d.c.). The radiochemical purity of the desired intermediates at each step was found to be greater than 99%. Through this, the optimized reaction conditions and purification method, including column, solvent, and pretreatment process before HPLC purification in the present study are expected to be applicable to various oligonucleotide reactions. Although Cu‐free click reactions can be used, they show efficient results when relatively large amounts of counter compound should be required and reaction volume is also small during click reaction.
To investigate the optimal hybridization efficiency at minimal concentrations of ERBB2‐cODN‐idT‐APs (6) with cODN‐PEG3‐[18F]F ([18F]5) isolated from HPLC, the hybridization of aptamers was tested at various concentrations (1, 2, 3, 5, 10, 50, 100, and 200 nmol) of 6. The hybridization yield increased in proportion to the increase in the amounts of ERBB2‐cODN‐idT‐APs (6; 24% at 1 nmol, 45% at 2 nmol, 54% at 3 nmol, 71% at 5 nmol, 78% at 10 nmol, 89% at 50 nmol, >95% at 100 nmol, and >95% at 200 nmol of 6), as shown in Figure S14. Optimal hybridization efficiency of greater than 95% was achieved with 100 nmol of 6. The final solution was diluted in saline and used for PET imaging studies without further purification. This optimization process showing the hybridization efficiency of 95% or more is an essential and important because it is very difficult to separate between the oligonucleotides used for labeling and the desire product, not only by the base‐pair hybridization labeling method using a complementary oligonucleotide platform, but also by other labeling methods, in particular, if high molar activity is required.
The excretion route of intravenously administered ERBB2‐cODN‐idT‐APs‐[18F]F ([18F]1) mainly excreted through the renal route (77.8%) of the total dose, as most aptamers have molecular weights between 30 and 50 kDa. ERBB2‐cODN‐idT‐APs in this study included idT to enhance blood circulation; they showed rapid distribution through the bloodstream and rapid renal excretion. Contrary to expectations following the introduction of idT, there was no significant difference. These features are advantageous for molecules that are applied as diagnostic radioligands. However, it is believed that these features could be major pitfalls for molecules that are applied as therapeutic drugs. Nevertheless, the specific organ (and target organ/tumor lesion) distribution of the 18F‐labeled‐aptamers could be easily monitored by PET‐based visualization and quantification of the change in radioactivity in each organ in the disease animal model. The excretion routes and PK parameters in vivo were also successfully determined, and its PK profile similar to those of commonly known aptamers were found. In addition, the tumor targeted aptamers reported to date still showed a relatively lower uptake, so the estimated internal absorbed dose values have not yet been known. To predict whether aptamers with a typical PK profile are problematic for the internal absorbed dose in human subject, this parameter was estimated for an adult woman using an image‐based approach based on the residence time in animals and the S‐value in humans. The effective dose in adults of [18F]1 estimated herein and that of 2‐[18]fluoro‐2‐deoxy‐D‐glucose ([18F]FDG), the most widely used molecule in nuclear medicine, are 1.89E‐03 mSv/MBq and 4.64E‐02 mSv/MBq, respectively. 45 The effective radiation dose of [18F]1 was relatively lower than that of [18F]FDG; thus, [18F]1 may be safe at a dose similar to that used for [18F]FDG. Based on our results, we conclude that PET/CT, a key three‐dimensional medical imaging technique, together with radioisotope‐labeled aptamers, can successfully provide in vivo biodistribution data for each organ and the target organ with quantitative information that cannot be obtained from conventional blood PK studies.
Nevertheless, our study had several limitations. First, our study did not include metabolite analysis to confirm the accumulation of the aptamers in the target tissue. However, even if the tumor was isolated and analyzed, the identity of the 55‐mer aptamer ([18F]1) would have been difficult to confirm by HPLC. Additional methods that can accurately distinguish molecular identities should be implemented in future studies. Second, there is no data for the blood (% plasma/total blood, metabolites). The data may be essential to consider because of the property of oligonucleotides that stick to plasma proteins. Finally, the [18F]1 used in our study was not the desired aptamer, but a compound containing a triazole moiety and poly(ethylene glycol) (PEG). Even a very small modification relative to the desired 55‐mer aptamers has the potential to affect the binding affinity. In future studies, if a drug candidate is designed such that the radioisotope can be introduced without modifying the original aptamer structure at the early stage of drug development, it is expected that the in vivo PET PKs of the original drug candidate can be evaluated successfully.
CONCLUSIONS
Our methodology provides an advanced approach to study the biodistribution and PK properties, including the excretion pathways and internal absorbed doses, of radiolabeled oligonucleotides applied as therapeutic drug candidates by serial PET/CT imaging in living subjects. The present results suggest that image‐based PET with radioisotope‐labeled aptamers may potentially be useful for the development process and could provide valuable information on properties of oligonucleotide‐based drug candidates in the context of various diseases. Nevertheless, more accurate and useful data could be provided if the blood data through blood sampling were complemented.
AUTHOR CONTRIBUTIONS
S.M.P., S.B., K.P.L., and B.S.M. wrote the manuscript. K.P.L. and B.S.M. designed the research. S.M.P., S.B., S.K.W., T.S.L., Y.‐K.K., S.Y.K., M.Y.Y., H.‐J.Y., K.P.L., and B.S.M. performed the research. S.M.P., S.B., S.K.W., T.S.L., H.S.P., K.P.L., and B.S.M. analyzed the data. S.M.P., S.B., J.H.L., S.K.W., T.S.L., J.L., B.S.K., K.P.L., and B.S.M. contributed new reagents/analytical tools.
FUNDING INFORMATION
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea, funded by the Ministry of Science and ICT (2021R1I1A1A01049147) and the R&D grant from Union Korea Pharm.
CONFLICT OF INTEREST STATEMENT
J.H.L. and J.L. are employed by INTEROligo Corporation and K.P.L. and S.B. are employed by UMUST R&D Corporation. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The funders had no role in the design of the study; the collection, analyses, or interpretation of data; the writing of the manuscript; or in the decision to publish the results.
Supporting information
Appendix S1
Park SM, Baek S, Lee JH, et al. In vivo tissue pharmacokinetics of ERBB2‐specific binding oligonucleotide‐based drugs by PET imaging. Clin Transl Sci. 2023;16:1186‐1196. doi: 10.1111/cts.13522
Sun Mi Park and Suji Baek contributed equally to this study.
Contributor Information
Bom Sahn Kim, Email: kbomsahn@ewha.ac.kr.
Kang Pa Lee, Email: umustrnd@naver.com.
Byung Seok Moon, Email: bsmoon@ewha.ac.kr.
REFERENCES
- 1. Zhu G, Chen X. Aptamer‐based targeted therapy. Adv Drug Deliv Rev. 2018;134:65‐78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kinghorn AB, Fraser LA, Lang S, et al. Aptamer bioinformatics. Int J Mol Sci. 2017;18:2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Park KS. Nucleic acid aptamer‐based methods for diagnosis of infections. Biosens Bioelectron. 2018;102:179‐188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marrazza G. Aptamer sensors. Biosensors. 2017;7:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhou J, Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discov. 2017;16:181‐202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bouchard PR, Hutabarat RM, Thompson KM. Discovery and development of therapeutic aptamers. Annu Rev Pharmacol Toxicol. 2010;50:237‐257. [DOI] [PubMed] [Google Scholar]
- 7. Conrad RC, Giver L, Tian Y, et al. In vitro selection of nucleic acid aptamers that bind proteins. Methods Enzymol. 1996;267:336‐367. [DOI] [PubMed] [Google Scholar]
- 8. Kulbachinskiy AV. Methods for selection of aptamers to protein targets. Biochemistry (Mosc). 2007;72:1505‐1518. [DOI] [PubMed] [Google Scholar]
- 9. Kaur H, Bruno JG, Kumar A, Sharma TK. Aptamers in the therapeutics and diagnostics pipelines. Theranostics. 2018;8:4016‐4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yin W, Rogge M. Targeting RNA: a transformative therapeutic strategy. Clin Transl Sci. 2019;12:98‐112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kovacevic KD, Gilbert JC, Jilma B. Pharmacokinetics, pharmacodynamics and safety of aptamers. Adv Drug Deliv Rev. 2018;134:36‐50. [DOI] [PubMed] [Google Scholar]
- 12. Mackay IM, Arden KE, Nitsche A. Real‐time PCR in virology. Nucleic Acids Res. 2002;30:1292‐1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kubista M, Andrade JM, Bengtsson M, et al. The real‐time polymerase chain reaction. Mol Aspects Med. 2006;27:95‐125. [DOI] [PubMed] [Google Scholar]
- 14. Saha GB, MacIntyre WJ, Go RT. Cyclotrons and positron emission tomography radiopharmaceuticals for clinical imaging. Semin Nucl Med. 1992;22:150‐161. [DOI] [PubMed] [Google Scholar]
- 15. Lau J, Rousseau E, Kwon D, Lin KS, Bénard F, Chen X. Insight into the development of PET radiopharmaceuticals for oncology. Cancers. 2020;12:1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hong YS, Kim KP, Lim HS, et al. A phase I study of DHP107, a mucoadhesive lipid form of oral paclitaxel, in patients with advanced solid tumors: crossover comparisons with intravenous paclitaxel. Invest New Drugs. 2013;31:616‐622. [DOI] [PubMed] [Google Scholar]
- 17. Bauer M, Wagner CC, Langer O. Microdosing studies in humans: the role of positron emission tomography. Drugs R D. 2008;9:73‐81. [DOI] [PubMed] [Google Scholar]
- 18. Bergström M, Grahnén A, Långström B. Positron emission tomography microdosing: a new concept with application in tracer and early clinical drug development. Eur J Clin Pharmacol. 2003;59:357‐366. [DOI] [PubMed] [Google Scholar]
- 19. Baek S, Kim J, Moon BS, et al. Camphene attenuates skeletal muscle atrophy by regulating oxidative stress and lipid metabolism in rats. Nutrients. 2020;12:3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim D, Moon BS, Park SM, et al. Feasibility of TSPO‐specific positron emission tomography radiotracer for evaluating paracetamol‐induced liver injury. Diagnostics. 2021;11:1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. An HH, Moon BS, Park HS, et al. Comparative study in different filters for efficient sterile filtration. B Kor Chem Soc. 2020;41:824‐828. [Google Scholar]
- 22. Moon BS, Park HS, Sunwoo J, et al. Tissue pharmacokinetics of DHP107, a novel lipid‐based oral formulation of paclitaxel, in mice and patients by positron emission tomography. Clin Transl Sci. 2021;14:1747‐1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park SM, Kim J, Baek S, et al. Feasibility of 18F‐fluorocholine PET for evaluating skeletal muscle atrophy in a starved rat model. Diagnostics. 2022;12:1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kitajima K, Murphy RC, Nathan MA, Sugimura K. Update on positron emission tomography for imaging of prostate cancer. Int J Urol. 2014;21:12‐23. [DOI] [PubMed] [Google Scholar]
- 25. Kazakauskaitė E, Žaliaduonytė‐Pekšienė D, Rumbinaitė E, Keršulis J, Kulakienė I, Jurkevičius R. Positron emission tomography in the diagnosis and management of coronary artery disease. Medicina (Kaunas). 2018;54:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rockey WM, Huang L, Kloepping KC, Baumhover NJ, Giangrande PH, Schultz MK. Synthesis and radiolabeling of chelator‐RNA aptamer bioconjugates with copper‐64 for targeted molecular imaging. Bioorg Med Chem. 2011;19:4080‐4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kobori N, Imahori Y, Mineura K, Ueda S, Fujii R. Visualization of mRNA expression in CNS using 11C‐labeled phosphorothioate oligodeoxynucleotide. Neuroreport. 1999;10:2971‐2974. [DOI] [PubMed] [Google Scholar]
- 28. Liu ZF, Ye QN, Yang J, Yang M, Pan DH, Dong MJ. Preclinical evaluation of [68Ga]Ga‐MALAT‐1‐antisense oligonu‐cleotides for specific PET imaging of MALAT‐1 expressing tumours. Nucl Med. Communi. 2021;42:782‐791. [DOI] [PubMed] [Google Scholar]
- 29. Hedberg E, Långström B. Synthesis of 4‐([18F]fluoromethyl)phenyl isothiocyanate and its use in labelling oligonucleotides. Acta Chemical Scandinavica. 1997;51:1236‐1240. [Google Scholar]
- 30. Roivainen A, Tolvanen T, Salomäki S, et al. 68Ga‐labeled oligonucleotides for in vivo imaging with PET. J Nucl Med. 2004;45:347‐355. [PubMed] [Google Scholar]
- 31. Correa CR, de Barros AL, Ferreira Cde A, et al. Aptamers directly radiolabeled with technetium‐99 m as a potential agent capable of identifying carcinoembryonic antigen (CEA) in tumor cells T84. Bioorg Med Chem Lett. 2014;24:1998‐2001. [DOI] [PubMed] [Google Scholar]
- 32. Yngve U, Hedberg E, Lövqvist A, et al. Synthesis of N‐succinimidyl 4‐[76Br]bromobenzoate and its use in conjugation labeling of macromolecules. Acta Chemica Scandinavica. 1999;53:508‐512. [Google Scholar]
- 33. Wu F, Yngve U, Hedberg E, et al. Distribution of 76Br‐labeled antisense oligonucleotides of different length determined ex vivo in rats. Eur J Pharm Sci. 2000;10:179‐186. [DOI] [PubMed] [Google Scholar]
- 34. de Vries EF, Vroegh J, Dijkstra G, et al. Synthesis and evaluation of a fluorine‐18 labeled antisense oligonucleotide as a potential PET tracer for iNOS mRNA expression. Nucl Med Biol. 2004;31:605‐612. [DOI] [PubMed] [Google Scholar]
- 35. Kühnast B, Dollé F, Terrazzino S, et al. General method to label antisense oligonucleotides with radioactive halogens for pharmacological and im‐aging studies. Bioconjug Chem. 2000;11:627‐636. [DOI] [PubMed] [Google Scholar]
- 36. Tanaka K, Fukase K. PET (positron emission tomography) imaging of biomolecules using metal‐DOTA complexes: a new collaborative challenge by chemists, biologists, and physicians for future diagnostics and exploration of in vivo dynamics. Org Biomol Chem. 2008;6:815‐828. [DOI] [PubMed] [Google Scholar]
- 37. Jacobson O, Kiesewetter DO, Chen X. Fluorine‐18 radiochemistry, labeling strategies and synthetic routes. Bioconjug Chem. 2015;26:1‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Park JY, Lee TS, Song IH, et al. Hybridization‐based aptamer labeling using complementary oligonucleotide platform for PET and optical imaging. Biomaterials. 2016;100:143‐151. [DOI] [PubMed] [Google Scholar]
- 39. Wimberley C, Nguyen DL, Truillet C, et al. Longitudinal mouse‐PET imaging: a reliable method for estimating binding parameters without a reference region or blood sampling. Eur J Nucl Med Mol Imaging. 2020;47:2589‐2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Woo SK, Jang SJ, Seo MJ, et al. Development of 64Cu‐NOTA‐trastuzumab for HER2 targeting: a radiopharmaceutical with improved pharmacokinetics for human studies. J Nucl Med. 2019;60:26‐33. [DOI] [PubMed] [Google Scholar]
- 41. Sun H, Zhu X, Lu PY, Rosato RR, Tan W, Zu Y. Oligonucleotide aptamers: new tools for targeted cancer therapy. Mol Ther Nucleic Acids. 2014;3:e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Que‐Gewirth NS, Sullenger BA. Gene therapy progress and prospects: RNA aptamers. Gene Ther. 2007;14:283‐291. [DOI] [PubMed] [Google Scholar]
- 43. Sundaram P, Kurniawan H, Byrne ME, Wower J. Therapeutic RNA aptamers in clinical trials. Eur J Pharm Sci. 2013;48:259‐271. [DOI] [PubMed] [Google Scholar]
- 44. Ireson CR, Kelland LR. Discovery and development of anticancer aptamers. Mol Cancer Ther. 2006;5:2957‐2962. [DOI] [PubMed] [Google Scholar]
- 45. Kaushik A, Jaimini A, Tripathi M, et al. Estimation of radiation dose to patients from 18FDG whole body PET/CT investigations using dynamic PET scan protocol. Indian J Med Res. 2015;142:721‐731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1


