Abstract
Fasting triggers diverse physiological adaptations including increases in circulating fatty acids and mitochondrial respiration to facilitate organismal survival. The mechanisms driving mitochondrial adaptations and respiratory sufficiency during fasting remain incompletely understood. Here we show that fasting or lipid availability stimulates mTORC2 activity. Activation of mTORC2 and phosphorylation of its downstream target NDRG1 at serine 336 sustains mitochondrial fission and respiratory sufficiency. Time-lapse imaging shows that NDRG1, but not the phosphorylation-deficient NDRG1Ser336Ala mutant, engages with mitochondria to facilitate fission in control cells, as well as in those lacking DRP1. Using proteomics, a small interfering RNA screen, and epistasis experiments, we show that mTORC2-phosphorylated NDRG1 cooperates with small GTPase CDC42 and effectors and regulators of CDC42 to orchestrate fission. Accordingly, RictorKO, NDRG1Ser336Ala mutants and Cdc42-deficient cells each display mitochondrial phenotypes reminiscent of fission failure. During nutrient surplus, mTOR complexes perform anabolic functions; however, paradoxical reactivation of mTORC2 during fasting unexpectedly drives mitochondrial fission and respiration.
Subject terms: Energy metabolism, Nutrient signalling
Martinez-Lopez et al. show that fasting or lipid availability stimulates mTORC2 activity in the liver, leading to phosphorylation of NDRG1 and NDRG1–CDC42-mediated mitochondrial fission.
Main
Dynamic mitochondrial networks are essential for mitochondrial function and organismal wellbeing1. Mitochondrial networking is in turn controlled by coordinated fission and fusion events regulated by proteins localized at endoplasmic reticulum (ER)–mitochondria contacts (mitochondria-associated membranes, MAMs)2. Indeed, blocking fission by deleting dynamin-related protein 1 (DRP1) (refs. 3,4) or mitochondrial fission factor (MFF)4 or inhibiting fusion by silencing optic atrophy 1 (OPA1) (ref. 5) and mitofusin (MFN) proteins6 alters mitochondrial morphology and function. Although, recent work has identified a role of AMPK in mitochondrial fission7, we do not completely understand how dietary stressors such as fasting influence mitochondrial dynamics in intact organisms. Since nutrient signalling is coupled to healthspan, it remains critical to understand how impairment in these processes lead to age-related diseases.
In this Article, we show that nutrient-responsive mTORC2 is paradoxically reactivated by fasting to stimulate mitochondrial fission. We show that the mTORC2–SGK1 cascade phosphorylates a known target NDRG1 (ref. 8) at Ser336, which then engages with mitochondria to drive fission. NDRG1, but not the phosphorylation-deficient NDRG1Ser336Ala mutant, interacts with CDC42 (ref. 9), a cytokinetic protein with intrinsic GTP hydrolysis activity10, to drive fission. mTORC2, NDRG1 and CDC42 each localize to MAMs, and silencing Rictor, Ndrg1 or Cdc42 or identified CDC42 effectors blocks fission. Thus, paradoxical reactivation of an mTORC2–NDRG1Ser336–CDC42 axis drives mitochondrial fission during fasting.
Results
Fasting or lipid availability activates mTORC1/2 signalling
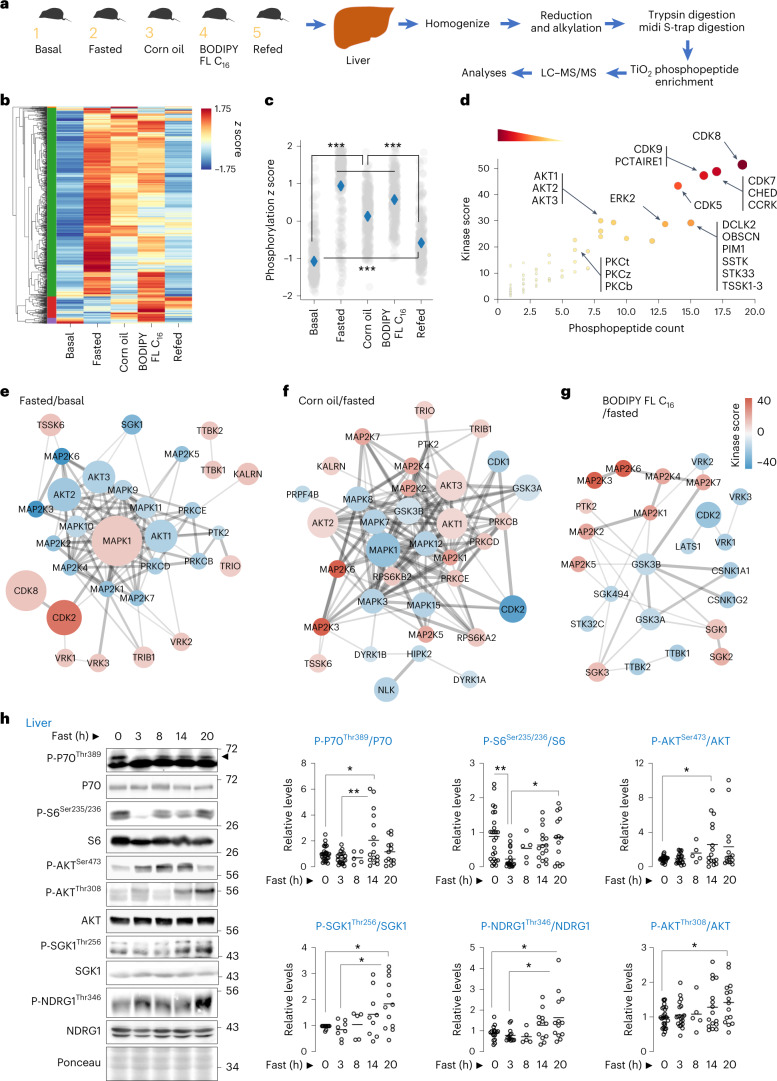
Fasting increases circulating free fatty acids (FFAs), which undergo mitochondrial oxidation to support organismal sustenance11. To understand the mechanisms driving metabolic adaptations during fasting or fatty acid availability, we sought to identify the signalling cascades that are activated under these conditions. To this purpose, we performed unbiased quantitative phosphoproteomics in livers of mice that were (1) basal fed; (2) overnight (14–16 h) fasted; or fasted overnight and then gavaged with (3) dietary triglycerides as corn oil; or (4) BODIPY FL C16/palmitic acid; or (5) refed a high-fat diet (Fig. 1a). Corn oil or BODIPY FL C16 groups served as models for exogenous lipid availability, while refeeding served as a control to simulate physiological feeding. Corn oil is absorbed as FFA and repackaged and secreted by enterocytes as lipoproteins and subsequently delivered to liver as FFA. Delivery of BODIPY FL C16 to livers was confirmed by direct fluorescence of liver slices (Extended Data Fig. 1a). Phosphoproteomics in the five groups identified 2,160 phosphosites across 942 phosphoproteins, of which 863 phosphosites (39.95%) were significantly modulated. Unsupervised hierarchical clustering analyses grouped basal and refed cohorts into one cluster, while lipid-exposed groups (that is, fasted, corn oil and BODIPY FL C16) clustered into the second group (Extended Data Fig. 1b and Supplementary Table 1). A second clustering analysis to determine phosphoproteins that are coordinately modulated revealed a major ‘green cluster’ encompassing 86.9% of significantly modulated phosphosites (Fig. 1b and Supplementary Table 2). The average normalized abundance of phosphosites belonging to the green cluster (to better appreciate group-to-group modulation rather than phosphoprotein expression differences) was significantly higher in lipid-exposed groups, when compared with basal and refed groups (Fig. 1c). Interestingly, despite strong reduction in phosphorylations in the refed group, the green cluster-normalized abundance was relatively higher in the refed cohort compared with the basal fed cohort, indicating qualitative differences in phosphopeptides between basal fed and refed groups (Fig. 1c).
Fig. 1. Phosphoproteomics reveal kinases that respond to fasting or lipids.
a, Phosphoproteomics in livers as per plan in cartoon. b, Heat map and hierarchical clustering of phosphosites across groups indicated in a. c, Phosphoproteome-wide comparisons via z score normalization of phosphosites in green cluster. Grey dots represent individual phosphosites. Blue diamonds represent group means. ***P < 0.001, non-parametric ANOVA (Kruskal–Wallis statistic 797.3, P < 0.0001) followed by Dunn’s multiple comparisons test. d, iGPS prediction of upstream individual kinases that respond to lipids across groups indicated and identified in the green cluster in b. e–g, Pairwise comparisons between indicated groups (fasted versus basal (e), corn oil versus fasted (f), and BODIPY FL C16 versus fasted (g)), showing upregulated or downregulated kinase networks. For a–g, n = 4 mice. For d, darker colour intensity reflects higher kinase score. h, Immunoblots (IB) and quantification for indicated proteins in livers of 2–10-month-old male and female mice that were fed or fasted for indicated durations. N values for number of mice analysed at each timepoint for individual phosphoproteins are indicated in parentheses. P-P70Thr389/P70: 0 h (n = 27), 3 h (n = 21), 8 h (n = 5), 14 h (n = 16) and 20 h (n = 14); P-S6Ser235/236/S6: 0 h (n = 26), 3 h (n = 20), 8 h (n = 5), 14 h (n = 16) and 20 h (n = 14); P-AKTSer473/AKT: 0 h (n = 26), 3 h (n = 21), 8 h (n = 5), 14 h (n = 16) and 20 h (n = 15); P-SGK1Thr256/SGK1: 0 h (n = 11), 3 h (n = 9), 8 h (n = 4), 14 h (n = 9) and 20 h (n = 12); P-NDRG1Thr346/NDRG1: 0 h (n = 17), 3 h (n = 14), 8 h (n = 5), 14 h (n = 12) and 20 h (n = 13); and P-AKTThr308/AKT: 0 h (n = 27), 3 h (n = 20), 8 h (n = 5), 14 h (n = 16) and 20 h (n = 15). Ponceau is loading control. Individual replicates and means are shown. *P < 0.05 and **P < 0.01, one-way ANOVA followed by Tukey’s multiple comparisons test (h). Please refer to Supplementary Table 10 statistical summary, and Supplementary Tables 1 and 2. Source numerical data are available in Source Data Extended Data Table 1, and unprocessed blots are available in the Source Data for this figure.
Extended Data Fig. 1. Kinases responsive to fasting or fatty acids.
(a) Direct BODIPY fluorescence in liver slices of 8 mo-old C57BL/6 male mice subjected to 30 min gavage with vehicle or 10 mg/kg of BODIPY FL C16 after 14–16 h fasting (n = 5 mice). (b) Dendrogram of hierarchical clustering analysis (Euclidean distance) between experimental groups based on phosphorylation status of 863 phosphosites. (c) Prediction for kinases that putatively target the phosphosites identified within the green cluster in main Fig. 1b. Darker color intensity reflects higher kinase score. (d) Pairwise comparisons showing upregulated (red) or downregulated (blue) kinase networks in mice refed high fat diet (HFD) for 30 min after 14–16 h fasting. For b-d (n = 4 mice). (e) Representative IB, and (f) quantification for the indicated proteins in livers of 2–8 mo-old C57BL/6 male or female mice kept fasted for 14–16 h or treated with corn oil or BODIPY FL C16 or refed a HFD for 30 min after fasting. N values for number of mice analyzed for individual proteins and conditions are indicated in parentheses. RAPTOR and P-AMPKThr172/AMPK: n = 4 per group; RICTOR: fasted (n = 8), corn oil (n = 8), BODIPY FL C16 (n = 4), refed (n = 4); P-P70Thr389/P70: fasted (n = 23), corn oil (n = 22), BODIPY FL C16 (n = 8), refed (n = 4); P-S6Ser235/236/S6 fasted (n = 23), corn oil (n = 24), BODIPY FL C16 (n = 9), refed (n = 4); P-AKTSer473/AKT: fasted (n = 17), corn oil (n = 13), BODIPY FL C16 (n = 9), refed (n = 4); P-AKTThr308/AKT: fasted (n = 4), corn oil (n = 3), BODIPY FL C16 (n = 4), refed (n = 4). Ponceau is the loading control. (g) Circulating free fatty acids (FFA) in 2–7 mo-old male mice fasted for: 0 h (n = 20 mice), 3 h (n = 9 mice), 14 h (n = 3 mice) or 20 h (n = 15 mice). Individual replicates and mean values are shown. *P < 0.05, ***P < 0.001 and ****P < 0.0001, One-way ANOVA followed by Šídák’s multiple comparisons test (f); One-way ANOVA followed by Tukey’s multiple comparisons test (g). Please refer to Supplementary Table 10_statistical summary, and Supplementary Tables 1, 2. Source numerical data are available in SourceData_Table 1, and unprocessed blots are available in Source Data Extended Data Fig. 1.
To predict the kinases putatively modulating the phosphosites in the green cluster, we used GPS algorithm with the interaction filter, or in vivo GPS (iGPS)12, which revealed that these phosphosites are targets of cyclin-dependent kinases (CDKs), Ca2+/calmodulin-dependent kinases, mitogen-activated protein kinases (MAPKs) and Ser/threonine (Thr) cAMP-dependent, cGMP-dependent and protein kinase C (AGC) kinases (Fig. 1d, Extended Data Fig. 1c and Supplementary Table 2). Pairwise comparisons showed upregulation of CDK2/CDK8 and MAPK1 during fasting when compared with basal group (Fig. 1e). Corn oil, which is devoid of proteins, and BODIPY FL C16, each perturbed a number of kinase groups albeit to a lesser extent when compared with fasting. Interestingly, iGPS revealed enrichment of downstream substrates of mTORC1/2 signalling, that is, RPS6KA2/B2 (ref. 13), AKT1-3 (ref. 14), PRKCB/D (refs. 15,16) and SGK1-3 (ref. 17) in livers of corn oil-gavaged mice, and SGK1-3 (ref. 17) in livers of BODIPY FL C16-treated mice, indicating lipid-driven mTORC1/2 activation (Fig. 1f,g). Refeeding reduced the overall kinase network, but expectedly activated nutrient-sensitive kinases, for example, AKT1-3 and SGK1-3, and suppressed those induced by fasting, for example, CDK2 (Extended Data Fig. 1d).
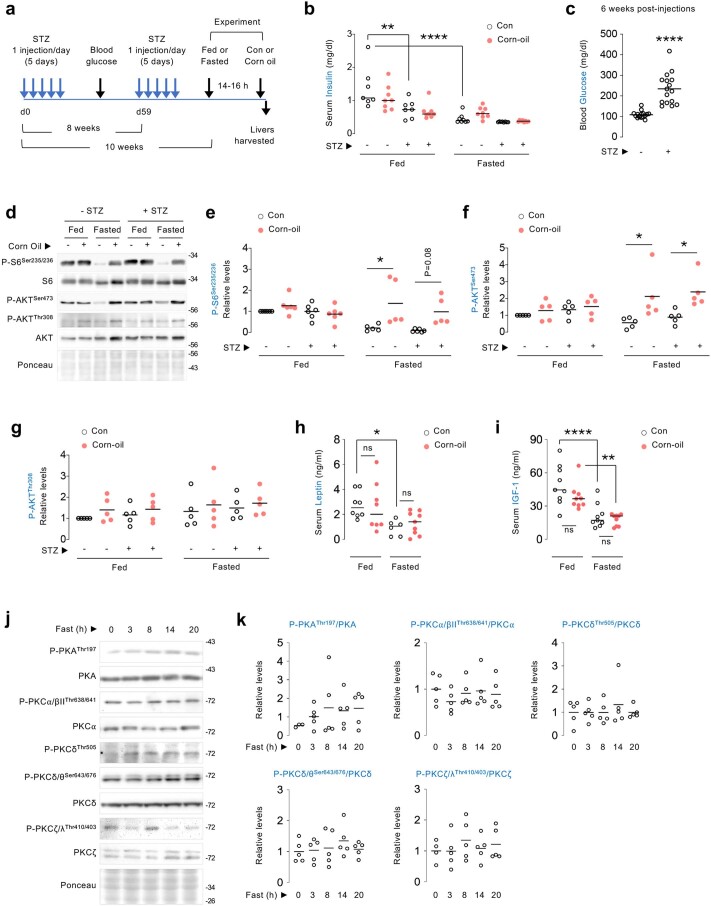
Confirming that dietary triglycerides stimulate mTORC1/2 signalling, immunoblotting showed higher levels of P-P70Thr389 and P-S6Ser235/236 (mTORC1 markers) and P-AKTSer473 (mTORC2 marker) in livers in response to corn oil without perturbing other nutrient-sensitive kinases, for example, AMPK18 (Extended Data Fig. 1e,f). Since insulin activates mTOR, we asked if insulin drives mTOR activation in response to corn oil. Interestingly, low-dose streptozotocin (STZ), which depletes insulin, causing hyperglycaemia due to β-cell destruction (Extended Data Fig. 2a–c), failed to block lipid-driven mTORC1 (P-S6Ser235/236), mTORC2 (P-AKTSer473) and AKT (P-AKTThr308) activation (Extended Data Fig. 2d–g). Furthermore, corn oil per se did not elicit insulin secretion (Extended Data Fig. 2b) excluding a role for insulin in lipid-driven mTOR activation. Similarly, equivalent serum levels of adipokine leptin or IGF-1 in corn-oil-treated and untreated mice excluded their role in lipid-driven mTOR activation (Extended Data Fig. 2h,i).
Extended Data Fig. 2. Protein kinases A and Cs are not activated by fasting in liver, and lipid-driven mTORC1/C2 activation is leptin, IGF-1 or insulin-independent.
(a) Experimental plan for generation of insulin-deficient diabetic mice using streptozotocin (STZ). (b) Serum insulin levels in 3 mo-old STZ-treated C57BL/6 male mice that were fed or fasted for 14–16 h and then gavaged with corn oil for 30 min. N values for number of mice in each group are in parenthesis. Fed (n = 7); fed + STZ (n = 7); fed + corn oil (n = 8); fed + STZ + corn oil (n = 8); fasted (n = 8); fasted + STZ (n = 8); fasted + corn oil (n = 8); fasted + STZ + corn oil (n = 8). (c) Blood glucose levels 6 weeks after the first injection with STZ (n = 16 mice). (d) Representative IB, and (e-g) quantification for indicated proteins normalized to corresponding total protein in livers of 3 mo-old STZ-treated C57BL/6 male mice that were fed or fasted for 14–16 h and then gavaged with corn oil for 30 min. For e, fed (n = 6); fed + STZ (n = 6); fed + corn oil (n = 6); fed + STZ + corn oil (n = 6); fasted (n = 5); fasted + STZ (n = 6); fasted + corn oil (n = 5); fasted + STZ + corn oil (n = 5). For f and g, all indicated groups consist of n = 5 mice each. (h, i) Serum (h) leptin and (i) IGF-1 levels in 3 mo-old C57BL/6 male mice that were fed or fasted for 14–16 h and then gavaged with corn oil for 30 min. For h, fed (n = 8); fed + corn oil (n = 8); fasted (n = 6); fasted + corn oil (n = 9). For i, all indicated groups consist of n = 9 mice each. (j) IB and (k) quantification for the indicated proteins in livers of 2–10 mo-old mice fed or fasted for the indicated durations. N values for number of mice analyzed at each time-point for individual phosphoproteins are indicated in parentheses. P-PKAThr197/PKA: 0 h (n = 3), 3 h-20 h (n = 5); P-PKCα/βIIThr638/641/PKCα (all time-points are n = 5); P-PKCδThr505/PKCδ (all time-points are n = 5); P-PKCδ/θSer643/676/PKCδ (all time-points are n = 5); and P-PKCζ/λThr410/403/PKCζ: 0 h (n = 4), 3 h-20 h (n = 5). Ponceau is the loading control. Individual replicates and mean values are shown. *P < 0.05, **P < 0.01 and ****P < 0.0001, 2-way ANOVA followed by Tukey’s multiple comparisons test (b, e, f, h and i); two-tailed unpaired Student’s t-test (c). ns=not significant. Please refer to Supplementary Table 10_statistical summary. Source numerical data are available in SourceData_Table 1, and unprocessed blots are available in Source Data Extended Data Fig. 2.
Since fasting redistributes FFA from adipose tissue to liver11,19, we asked whether fasting-induced increases in FFA (Extended Data Fig. 1g) reactivate mTORC1/2 signalling in liver, as noted with corn oil exposure. Interestingly, fasting for 14 h reactivates mTORC1/2 signalling in liver, indicated by phosphorylation of their respective targets20, P70Thr389 (mTORC1) and AKTSer473, SGK1Thr256 and NDRG1Thr346 (mTORC2) (Fig. 1h). By contrast, fasting did not affect phosphorylated levels of PKAThr197, PKCα/βIIThr638/641, PKCδThr505, PKCδ/θSer643/676 and PKCζ/λThr410/403 in liver (Extended Data Fig. 2j,k), suggesting that fasting specifically reactivates the AKT and SGK1, but not PKA/PKC, arms of mTORC2 signalling. Hence, exogenous lipids or endogenous FFA availability during fasting activates mTORC1/2 signalling.
mTORC2 supports mitochondrial respiration during fasting
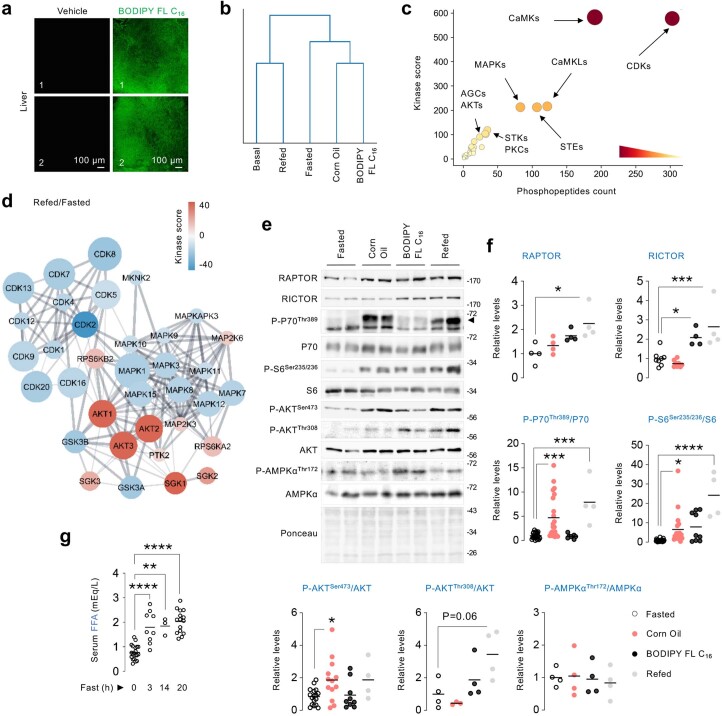
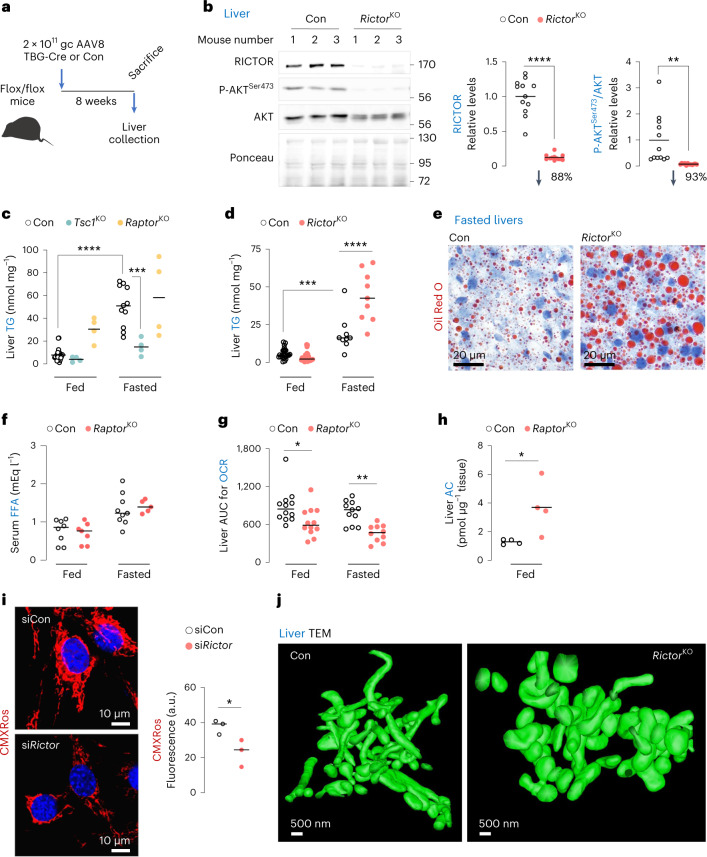
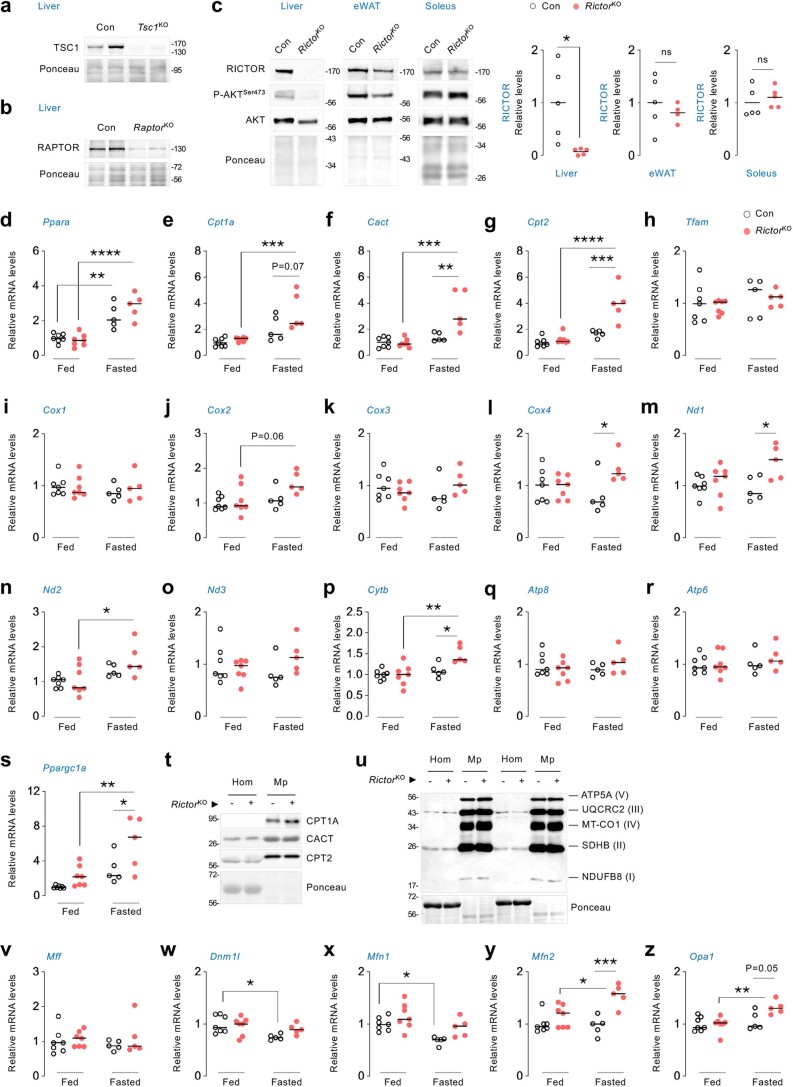
To determine the physiological roles of mTOR reactivation during fasting, we inactivated mTORC1 or mTORC2 or hyperactivated mTORC1 by knocking out Raptor21, Rictor22 or Tsc1 (ref. 23), respectively, using liver-restricted AAV8-TBG-iCre (Fig. 2a,b and Extended Data Fig. 3a,b). Loss of Rictor/mTORC2 activity in liver, and not fat or muscle, was confirmed by reduced AKTSer473 phosphorylation (Extended Data Fig. 3c). Since fasted livers accumulate triglyceride, we examined the effect of loss of each gene on fasting-induced increases in liver triglycerides. While control and mTORC1 inactivated (RaptorKO) livers showed equivalent liver triglycerides during fasting, hyperactivation of mTORC1 (Tsc1KO) lowered liver triglycerides (Fig. 2c) consistent with the role of mTORC1 in VLDL secretion24. Surprisingly, in contrast to the established triglyceride lowering effect of Rictor loss in fed/obesogenic states25, inactivating mTORC2 (RictorKO) markedly increased liver triglycerides and lipid droplet content during fasting (Fig. 2d,e) without affecting circulating FFA (Fig. 2f). Interestingly, RictorKO livers displayed lower oxygen consumption rates (OCRs) (Fig. 2g) and accumulation of substrates for mitochondrial respiration, acyl carnitines (Fig. 2h), which in conjunction with reduced mitochondrial membrane potential in siRictor cells (Fig. 2i) indicate mitochondrial insufficiency. Decreased OCR was not due to impaired expression of FFA oxidation or electron transport genes. In fact, fasted RictorKO livers displayed increased expression of genes involved in fat oxidation (Ppara, Cpt1a, Cpt1b, Cact and Cpt2), electron transport (Cox4, Nd1 and Cytb) and mitochondrial biogenesis (Ppargc1a) (Extended Data Fig. 3d–s). Levels of mitochondrial fatty acid uptake proteins (CPT1A/CPT2/CACT) and electron transport chain components, NDUFB8 (complex I), SDHB (II), UQCRC2 (III), MT-CO1 (IV) and ATP5A (V) were also comparable in control and RictorKO livers (Extended Data Fig. 3t,u). Interestingly, loss of Rictor led to increased expression of mitochondrial fusion genes Mfn2 and Opa1 during fasting without affecting fission genes Mff and Dnm1l (DRP1) (Extended Data Fig. 3v–z). Consistent with mitochondrial insufficiency, 3D transmission electron microscopy (TEM) revealed mitochondrial distention and blunted networking (Fig. 2j and Supplementary Videos 1 and 2) suggesting that reactivation of mTORC2 during fasting supports mitochondrial dynamics.
Fig. 2. mTORC2 supports mitochondrial respiration and triglyceride disposal during fasting.
a, Generation of liver-specific knockout (KO) of Rictor, Tsc1 or Raptor. b, Representative immunoblots to validate deletion of Rictor gene in livers of 4–6-month-old RictorKO male and female mice. Quantifications and percentage reduction of protein levels for RICTOR and P-AKTSer473/AKT in RictorKO livers are shown (n = 12 mice). c,d, Liver triglyceride (TGs) in 3–7-month-old Con, Tsc1KO and RaptorKO (c) or 4–5-month-old Con and RictorKO (d) male and female mice that were fed or fasted for 14–16 h. N values for number of mice per group are indicated in parentheses. For c, fed Con (n = 27), fasted Con (n = 11) and fed or fasted Tsc1KO or RaptorKO mice (n = 4). For d, fed Con (n = 24), fasted Con (n = 13), fed RictorKO (n = 23) and fasted RictorKO (n = 12). e, Representative Oil Red O stains in livers of 6-month-old Con or RictorKO male mice fasted for 14–16 h (n = 5 mice). f, Serum FFA levels in fed or 14–16 h fasted 3–6-month-old Con or RictorKO male mice. Fed Con (n = 8), fasted Con (n = 9), fed RictorKO (n = 7) and fasted RictorKO (n = 5) mice. g, Area under curve (AUC) for OCRs in Con and RictorKO livers from fed or 14–16 h fasted mice. Fed Con (n = 12), fasted Con (n = 11), fed RictorKO (n = 12) and fasted RictorKO (n = 10) mice. h, Acylcarnitine (AC) content in livers of 4–5-month-old Con or RictorKO male mice (n = 4 mice). i, MitoTracker CMXRos fluorescence in serum-deprived and OA-treated siControl (siCon) or siRictor NIH3T3 cells (siCon 59 cells and siRictor 59 cells from n = 3 independent experiments). j, Representative 3D-TEM images of mitochondria in livers of 4–5-month-old Con or RictorKO male mice (n = 3 mice). Please refer to Supplementary Video 1 (Con) and Supplementary Video 2 (RictorKO). Ponceau is loading control. Individual replicates and means are shown. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001, two-tailed unpaired Student’s t-test (b, h and i); two-way ANOVA followed by Tukey’s multiple comparisons test (c, d and g). Please refer to Supplementary Table 10 statistical summary. Source numerical data are available in Source Data Extended Data Table 1, and unprocessed blots are available in the Source Data for this figure.
Extended Data Fig. 3. Effect of loss of mTORC2 on expression of genes and proteins related to mitochondrial oxidative metabolism and dynamics.
(a, b) Representative IB to validate deletion of the indicated genes in livers of 4–6 mo-old Con, Tsc1KO or RaptorKO male and female mice. Blots for TSC1 and RAPTOR are representative of n = 7 Con or corresponding KO mice obtaining similar results. (c) IB for mTORC2 signaling and quantification of RICTOR protein levels in liver (n = 5 mice), epididymal white adipose tissue (eWAT) (n = 5 Con and n = 4 RictorKO mice) and soleus (n = 5 mice) of 4–6 mo-old Con and RictorKO male mice fasted for 14–16 h. (d-s and v-z) Relative mRNA expression of indicated genes in livers of 4–6 mo-old Con and RictorKO male mice that were fed or fasted for 14–16 h (n = 7 fed Con or RictorKO mice, and n = 5 fasted Con or RictorKO mice). (t) Representative IB for proteins involved in mitochondrial fatty acid uptake, and (u) OXPHOS in whole homogenates (Hom) and pure mitochondrial (Mp) fractions from livers of 5–6 mo-old Con and RictorKO male mice after 14–16 h fasting (n = 3 Con and n = 5 RictorKO mice). Ponceau is the loading control. Individual replicates and mean values are shown. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001, two-tailed unpaired Student’s t-test (c); 2-way ANOVA followed by Tukey’s multiple comparisons (d-s and v-z). ns=not significant. Please refer to Supplementary Table 10_statistical summary. Source numerical data are available in SourceData_Table 1, and unprocessed blots are available in Source Data Extended Data Fig. 3.
Loss of mTORC2 blocks fasting-induced mitochondrial fission
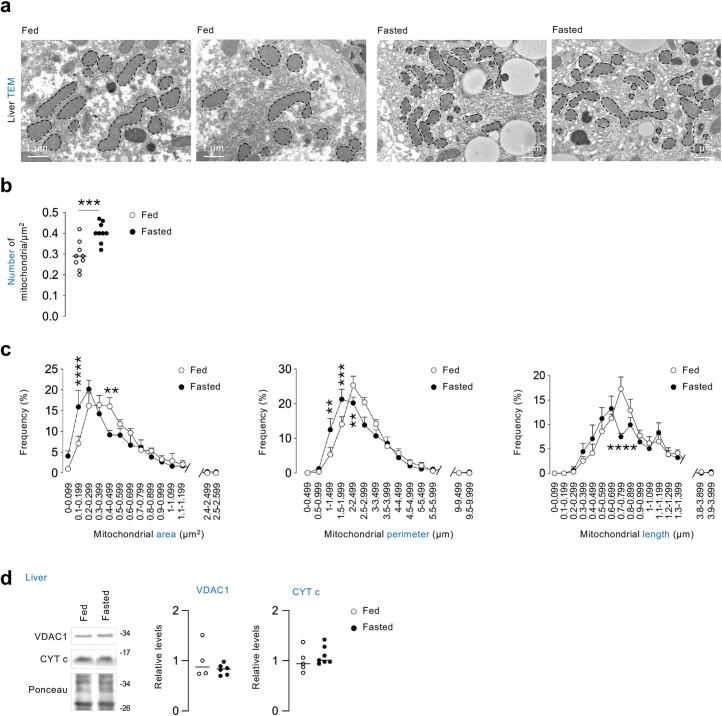
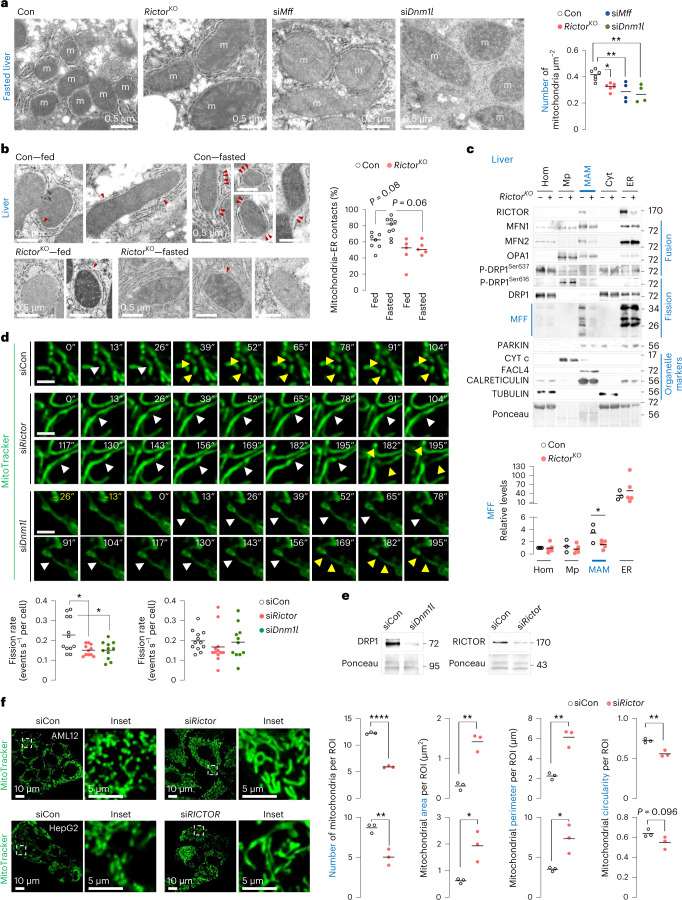
To determine whether mTORC2 regulates mitochondrial dynamics during fasting, we first determined how fasting impacts mitochondrial dynamics. TEM of fasted livers revealed increased mitochondrial number and higher frequency of mitochondria with reduced area, perimeter and length (Extended Data Fig. 4a–c), reflecting increased fission. Increased mitochondrial number during fasting occurred independently of changes in mitochondrial mass indicated by similar VDAC1 and CYT c levels in fed or fasted livers (Extended Data Fig. 4d). By contrast, RictorKO livers and livers silenced for Mff26 or Dnm1l27, each failed to increase their mitochondrial number during fasting (Fig. 3a) and displayed increased mitochondrial area and perimeter (Extended Data Fig. 5a–d) indicating fission failure. Consistently, TEM of fasted RictorKO livers showed reduced mitochondrial–ER contacts (MAMs) (Fig. 3b), which are contact sites regulating mitochondrial fission4.
Extended Data Fig. 4. Fasting stimulates mitochondrial fission in livers.
(a-c) (a) Liver TEM of fed or 14–16 h fasted 4–7-mo-old male mice. Quantification for mitochondrial number is shown in b. Frequency histograms depicting distribution of mitochondrial area, perimeter and length are shown in c. Mean ± SEM is shown. For a-c (n = 9 mice). (d) Representative IB and quantification for indicated mitochondrial markers in livers of fed or 14–16 h fasted mice. N values for number of mice analyzed for individual proteins and conditions are indicated in parentheses. VDAC1: fed (n = 4) and fasted (n = 6); CYT c: fed (n = 5) and fasted (n = 7). Ponceau is the loading control. Individual replicates and mean values are shown in b and d. **P < 0.01, ***P < 0.001 and ****P < 0.0001, two-tailed unpaired Student’s t-test (b); 2-way ANOVA followed by Tukey’s multiple comparisons test (c). Please refer to Supplementary Table 10_statistical summary. Source numerical data are available in SourceData_Table 1, and unprocessed blots are available in Source Data Extended Data Fig. 4.
Fig. 3. Loss of mTORC2 blocks fasting-induced mitochondrial fission.
a, Conventional TEM in 14–16 h fasted Con, RictorKO, siMff or siDnm1l livers of 4–9-month-old male mice. N values for number of mice per group are indicated in parentheses. Con or RictorKO mice (n = 6), and siMff or siDnm1l mice (n = 4). Quantification for mitochondrial number is shown. b, TEM in fed or 14–16 h fasted Con and RictorKO livers of 4–9-month-old male mice. Fed Con (n = 7), fasted Con (n = 9) and fed or fasted RictorKO mice (n = 5). Quantification for percentage of mitochondria–ER contacts is shown. Red arrowheads depict contact sites. c, Immunoblots and quantification of indicated proteins in homogenates (Hom), pure mitochondria (Mp), MAMs, cytosol (Cyt) and ER fractions from 14–16 h fasted Con (n = 3) and RictorKO livers (n = 5). Two livers were pooled to generate one sample. d, Live cell imaging and quantification for fission and fusion rates in siCon, siRictor or siDnm1l NIH3T3 cells cultured in serum-free medium for 30 min in presence of MitoTracker green to stain for mitochondria (siCon 12 cells, siRictor 11 cells (fission rate) and n = 12 cells (fusion rate), and siDnm1l 11 cells from n = 8 independent experiments; each cell was tracked on an independent plate). White arrowheads depict mitochondrial constriction sites. Yellow arrowheads depict daughter mitochondria arising from fission at a mitochondrial constriction. Scale bar, 2 µm. Please refer to Supplementary Videos 3 (siCon cells), 4 (siRictor cells) and 5 (siDnm1l cells). e, Representative IB for indicated proteins in siCon, siDnm1l and siRictor NIH3T3 cells. Blots are representative of n = 8 (DRP1) and n = 4 (RICTOR) independent experiments obtaining similar results. f, Representative confocal images of (top) AML12 and (bottom) HepG2 cells knocked down for Rictor, and corresponding controls in serum-free medium for 30 min in presence of MitoTracker green to stain for mitochondria. Magnified insets are shown. Quantifications for mitochondrial number and mitochondrial size/shape descriptors (area, perimeter and circularity) are shown (AML12 45 siCon or siRictor cells; HepG2 35 siCon and 38 siRICTOR cells from n = 3 independent experiments each). Ponceau is loading control. Individual replicates and means are shown. *P < 0.05, **P < 0.01 and ****P < 0.0001, one-way ANOVA followed by Tukey’s multiple comparisons test (a and d); two-way ANOVA followed by Tukey’s multiple comparisons test (b); two-tailed unpaired Student’s t-test (c and f). Please refer to Supplementary Table 10 statistical summary. Source numerical data are available in Source Data Extended Data Table 1, and unprocessed blots are available in the Source Data for this figure.
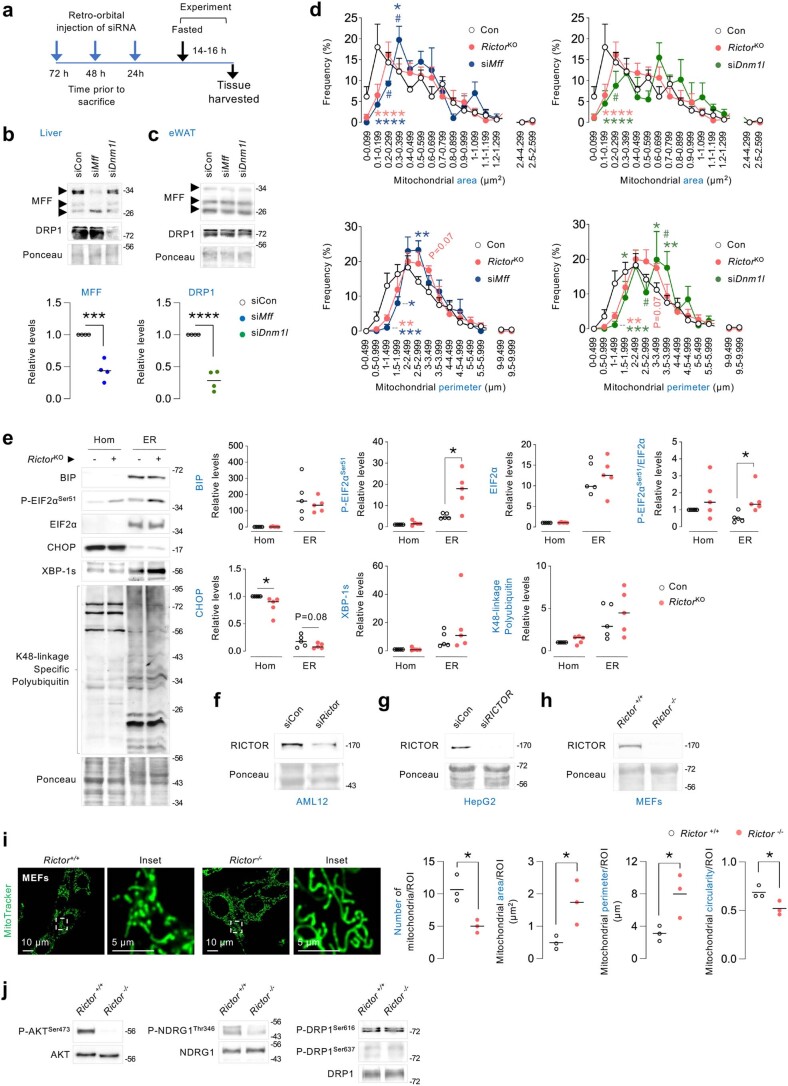
Extended Data Fig. 5. Fasting-induced mitochondrial fission is mTORC2-dependent.
(a) Experimental plan and (b-c) representative IB for indicated proteins in (b) livers and (c) eWAT of 4–9 mo-old male mice injected with siCon, siMff or siDnm1l and fasted for 14–16 h (n = 4 mice). Quantifications for loss of MFF and DRP1 protein levels in liver are shown. (d) Frequency histograms depicting distribution of mitochondrial area and perimeter in Con, RictorKO and siMff or siDnm1l livers. N values for number of mice are in parenthesis: Con (n = 6), RictorKO (n = 6), siMff (n = 4), and siDnm1l (n = 4). Mean ± SEM is shown. (e) IB and quantification for indicated ER-stress and proteostasis markers in Hom and ER fractions from livers of 6 mo-old Con and RictorKO mice fasted for 14–16 h (n = 5 mice). (f-h) Representative IB for RICTOR in siCon or siRictor (f) AML12 or (g) HepG2 cells, or in (h) Rictor +/+ and Rictor -/- MEFs. RICTOR blots are representative of independent experiments obtaining similar results in AML12 (n = 4), HepG2 (n = 3), and MEF (n = 7) cells. (i) Representative confocal images of Rictor +/+ and Rictor -/- MEFs cultured in serum-free medium for 30 min in presence of MitoTracker green. Magnified insets are shown. Quantification for mitochondrial number and mitochondria size/shape descriptors is shown (Rictor+/+ 36 cells and Rictor-/- 43 cells from n = 3 independent experiments). (j) Representative IB for indicated proteins in Rictor+/+ and Rictor-/- MEFs (n = 3 independent experiments). Ponceau is loading control. Individual replicates and mean values are shown. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 are versus Con; #P < 0.05 versus RictorKO, two-tailed unpaired student’s t-test (b, e and i); One-way ANOVA followed by Tukey’s multiple comparisons test (d). Please refer to Supplementary Table 10_statistical summary. Source numerical data are available in SourceData_Table 1, and unprocessed blots are available in Source Data Extended data Fig. 5.
Since perturbations in membrane lipids or ER stress could alter mitochondrial dynamics, we tested if these changes associate with impaired fission in our model. Loss of Rictor mildly affected lipid composition of MAMs (Supplementary Table 3) without inducing ER stress or proteostasis failure (Extended Data Fig. 5e), excluding their contribution to impaired fission. Because mTORC2 localizes to MAMs to regulate Ca2+ homeostasis and apoptosis28, we envisioned that mTORC2 at MAMs also regulates fission. Indeed, MAMs from 14–16 h fasted control livers revealed the presence of RICTOR, fission proteins4 MFF and DRP1, and fusion proteins OPA1 (ref. 29), MFN1 and MFN2 (Fig. 3c). By contrast, MAMs from fasted RictorKO livers showed markedly reduced MFF levels without affecting P-DRP1Ser616, P-DRP1Ser637, DRP1, MFN1, MFN2 or OPA1 levels (Fig. 3c) supporting that loss of mTORC2 impairs mitochondrial fission.
Confirming the role of mTORC2 in fission, time-lapse microscopy revealed markedly reduced fission rates in siRictor cells that were comparable to fission failure in siDnm1l cells (Fig. 3d,e and Supplementary Videos 3–5). Fusion rates were identical in siCon and siRictor cells, excluding the contribution of excessive fusion to the mitochondrial phenotype in siRictor cells. Furthermore, AML12 and HepG2 hepatocytes silenced for Rictor/RICTOR, and Rictor−/− mouse fibroblasts (Fig. 3f and Extended Data Fig. 5f–i), each showed decreased mitochondrial number and circularity, and markedly increased mitochondrial area and perimeter, demonstrating that mTORC2 drives fission in diverse cell types. Since mTORC1 stimulates mitochondrial fission via the fission protein MTFP1 (ref. 30), we examined whether impaired fission in RictorKO livers is due to altered mTORC1 signalling. Loss of Rictor did not affect levels of P-P70Thr389 (mean ± s.e.m.: Con (n = 6 mice) versus RictorKO (n = 10 mice): 0.77 ± 0.17 versus 0.81 ± 0.13; P = 0.85) or MTFP1 (Con (n = 7 mice) versus RictorKO (n = 10 mice): 1.00 ± 0.20 versus 1.06 ± 0.12; P = 0.80) or affect levels of DRP1 or DRP1Ser616 and DRP1Ser637 phosphorylation (Extended Data Fig. 5j), which regulate mitochondrial dynamics31. Hence, reactivation of mTORC2 during fasting drives fission, independent of mTORC1 signalling.
mTORC2–SGK1 phosphorylates NDRG1 at Ser336
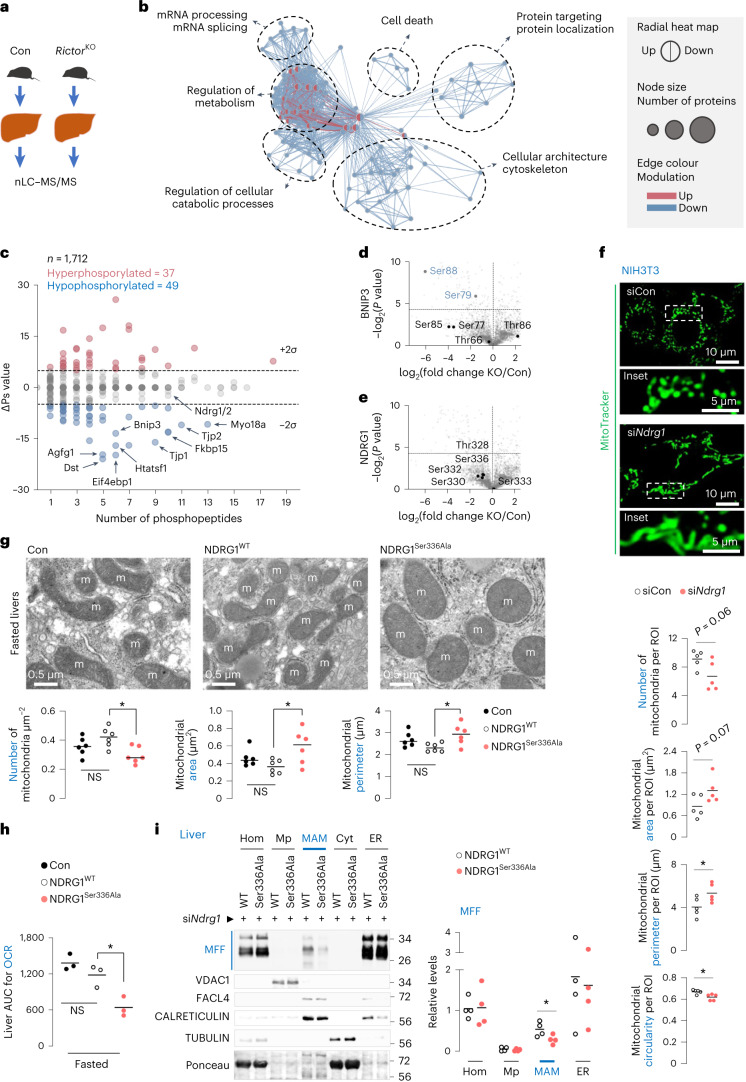
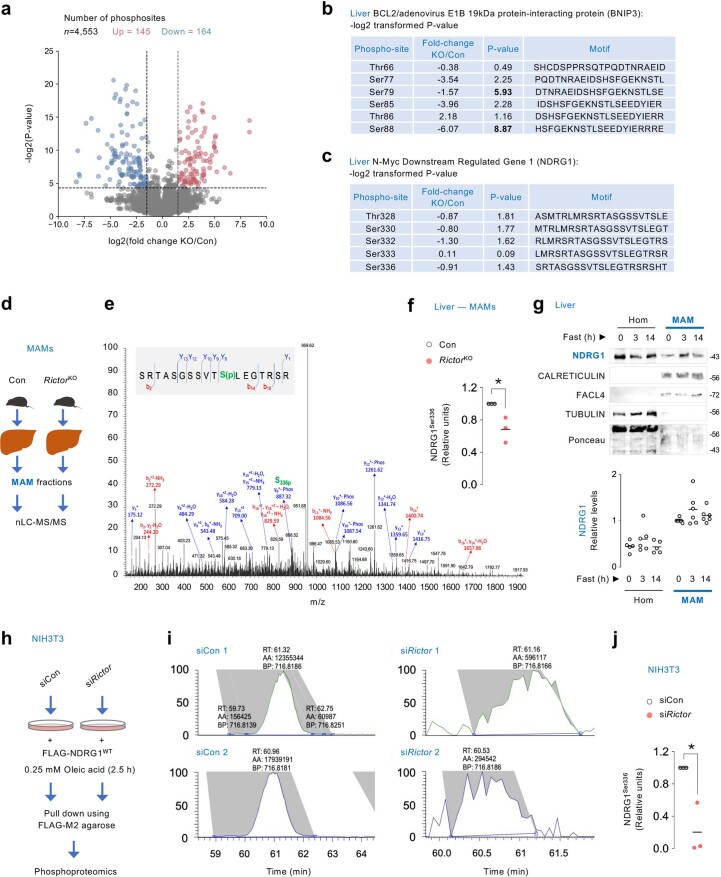
To determine whether phosphorylated targets of mTORC2 (ref. 32) support fission, we used quantitative nano liquid chromatography coupled online with tandem mass spectrometry (nLC–MS/MS) in control and RictorKO livers (Fig. 4a), which identified 4,553 phosphosites from 1,712 phosphoproteins. Of these, 309 phosphosites (145 upregulated and 164 downregulated) (6.79%) on 212 phosphoproteins (12.38%) were significantly modulated in RictorKO livers (Extended Data Fig. 6a and Supplementary Table 4). Gene Ontology and enrichment map network analysis33 revealed that the hypophosphorylated clusters in RictorKO livers were related to cytoskeleton and cellular architecture, mRNA processing and splicing, protein targeting and regulation of cellular catabolic processes (Fig. 4b and Supplementary Tables 2 and 5). The denser cluster populated by both upregulated and downregulated phosphoproteins contained the term ‘regulation of metabolism’. Since protein function is modulated by site-specific phosphorylation or cumulative phosphorylation of multiple phosphosites34, we measured the overall phosphorylation status (∆Ps) of phosphoproteins in our dataset, which revealed hyperphosphorylation (∆Ps > 2σ) in 37 phosphoproteins and hypophosphorylation (∆Ps < −2σ) in 49 phosphoproteins (Fig. 4c). Interestingly, phosphorylation of mitophagy receptor BNIP3 at Ser79 and Ser88 was significantly reduced in RictorKO livers (Fig. 4d and Extended Data Fig. 6b) with no known roles assigned to BNIP3Ser79/Ser88. We also focused on NDRG1 (Fig. 4e), which is phosphorylated on its C-terminus by the mTORC2 target SGK1 (refs. 8,35), and regulates lipid droplet content36. Although NDRG1 showed trends towards hypophosphorylation in RictorKO total homogenates (Fig. 4e and Extended Data Fig. 6c), phosphoproteomics from fasted livers showed marked NDRG1Ser336 hypophosphorylation in RictorKO MAMs when compared with controls (Extended Data Fig. 6d–f and Supplementary Table 6). Since NDRG1 is present in MAMs (Extended Data Fig. 6g), we sought to confirm that mTORC2–SGK1 indeed phosphorylates NDRG1 at Ser336. Accordingly, phosphoproteomics and relative quantification of extracted ion chromatogram of peptide SRTASGSSVTS(p)LEGTRSR, corresponding to Flag–NDRG1, from siCon or siRictor cells (Extended Data Fig. 6h–j and Supplementary Table 7) revealed reduced enrichment in siRictor cells compared with siCon cells, confirming that mTORC2 phosphorylates NDRG1 at Ser336.
Fig. 4. mTORC2 signalling drives mitochondrial fission via NDRG1Ser336 phosphorylation.
a, Experimental plan for b–e. b, Enrichment map-based network visualization of Gene Ontology enrichment for differentially modulated phosphosites. Blue edges show similarity between decreased phosphosites, and red nodes show similarity between increased phosphosites. Node size indicates the number of proteins per node; major clusters are circled. Associated name represents the major functional association. c, Global ∆Ps analyses of phosphoproteins. Hyperphosphorylated and hypophosphorylated peptides in each comparison are shown. Labels indicate the genes encoding the proteins. Dotted lines: ∆Ps = ±2σ. d,e, Volcano plot for BNIP3 (d) or NDRG1 (e) phosphorylation in RictorKO versus Con livers fasted for 14–16 h. For a–e, n = 3 mice. f, Representative MitoTracker green fluorescence in serum-deprived siCon and siNdrg1 NIH3T3 cells (siCon 84 cells and siNdrg1 91 cells from n = 5 independent experiments). Quantifications for mitochondrial number and mitochondrial size/shape descriptors are shown. g–i, TEM with mitochondrial quantifications (n = 6 mice) (g), AUC for liver OCR (n = 3 mice) (h) and immunoblots and quantification (i) for indicated proteins in indicated fractions from livers of 3–4-month-old fasted (14–16 h) male mice expressing NDRG1WT or NDRG1Ser336Ala after silencing endogenous Ndrg1 by siRNAs (n = 4 mice). Ponceau is loading control. Individual replicates and means are shown. *P < 0.05, two-tailed unpaired Student’s t-test (c–f and i); one-way ANOVA followed by Tukey’s comparisons test (g and h). NS, not significant. Please refer to Supplementary Table 10 statistical summary, and Supplementary Tables 4 and 5. Source numerical data are available in Source Data for Extended Data Table 1, and unprocessed blots are available in the Source Data for this figure.
Extended Data Fig. 6. Phosphoproteomics reveal mTORC2 phosphorylation of NDRG1 at Ser336 in MAMs.
(a) Volcano plot for phosphoproteomics in 14–16 h fasted Con and RictorKO livers. The numbers indicate total phosphosites. Red and blue dots represent significantly increased (P < 0.05 and log2(fold change)>1.5) and decreased (P < 0.05 and log2(fold change)<1.5) phosphosites, respectively. (b, c) Tables showing fold-change and log2 transformed P-values for indicated phosphorylations on liver (b) BNIP3, and (c) NDRG1 from 3–4-mo-old liver-specific RictorKO mice fasted for 14–16 h. For a-c (n = 3 mice). (d) MAMs of 4–6 mo-old Con and RictorKO livers from 14–16 h fasted mice were subjected to phosphoproteomics. (e) Annotated MS/MS spectrum of phosphopeptide SRTASGSSVTS(p)LEGTRSR in NDRG1, wherein S(p) represents phosphorylated Ser336. (f) Quantification for SRTASGSSVTS(p)LEGTRSR peptide in NDRG1 in MAMs is shown, with relative abundance of SRTASGSSVTS(p)LEGTRSR in Con or RictorKO MAMs. For d-f (n = 3 samples wherein 2 livers were pooled to generate 1 sample). (g) Representative IB and quantifications for indicated proteins in Hom and MAM fractions from livers of 2–10 mo-old male mice fed or fasted for indicated time points. N values for number of mice at each time point are in parenthesis: Hom 0 h and 14 h (n = 5); Hom 3 h (n = 6); and all time points for MAMs (n = 6). Ponceau is the loading control. (h) Experimental plan to pulldown FLAG-NDRG1WT in siCon or siRictor NIH3T3 cells co-transfected with FLAG-NDRG1WT plasmid for assessment of phosphorylation of FLAG-NDRG1WT via phosphoproteomics. FLAG-NDRG1WT pulled-down from total lysates of serum-deprived siCon or siRictor NIH3T3 cells in presence of OA for 2.5 h. (i) Representative extracted ion chromatograms of SRTASGSSVTS(p)LEGTRSR in FLAG-NDRG1 in siCon and siRictor NIH3T3 cells, and (j) quantification for relative abundance of SRTASGSSVTS(p)LEGTRSR peptide from pulled-down FLAG-NDRG1 from siCon and siRictor cells expressing FLAG-NDRG1WT. For h-j (n = 3 independent experiments). Individual replicates and means are shown. *P < 0.05, two-tailed unpaired Student’s t-test (a-c, f and j). p=phosphorylation. Please refer to Supplementary Table 10_statistical summary. Please refer to Supplementary Table 4(a-c), Supplementary Table 6(e and f), and Supplementary Table 7(i and j). Source numerical data are available in SourceData_Table 1, and unprocessed blots are available in Source Data Extended Data Fig. 6.
Phosphorylated NDRG1Ser336 drives mitochondrial fission
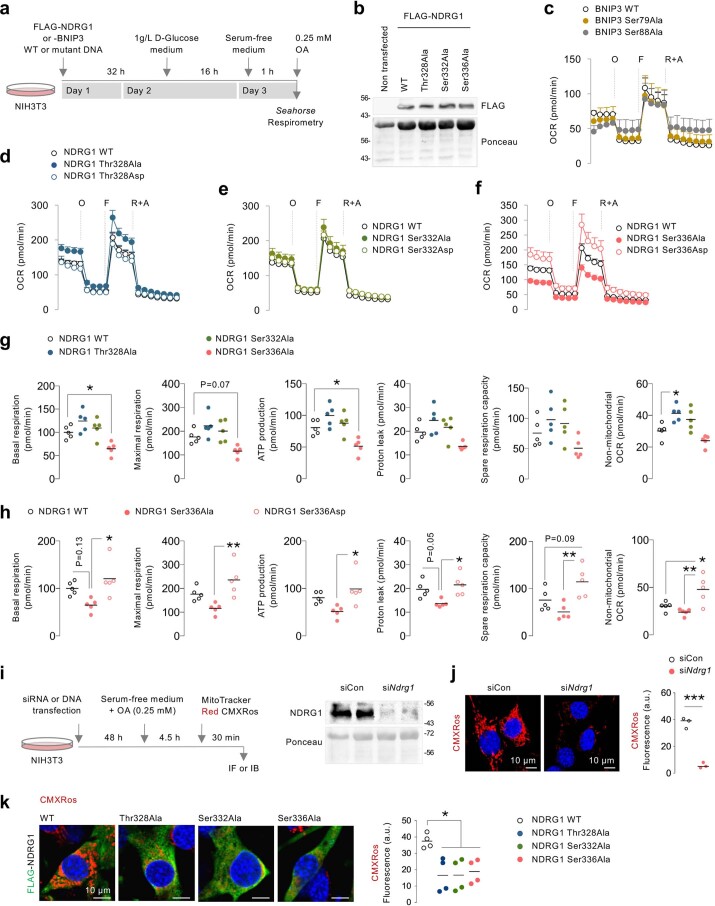
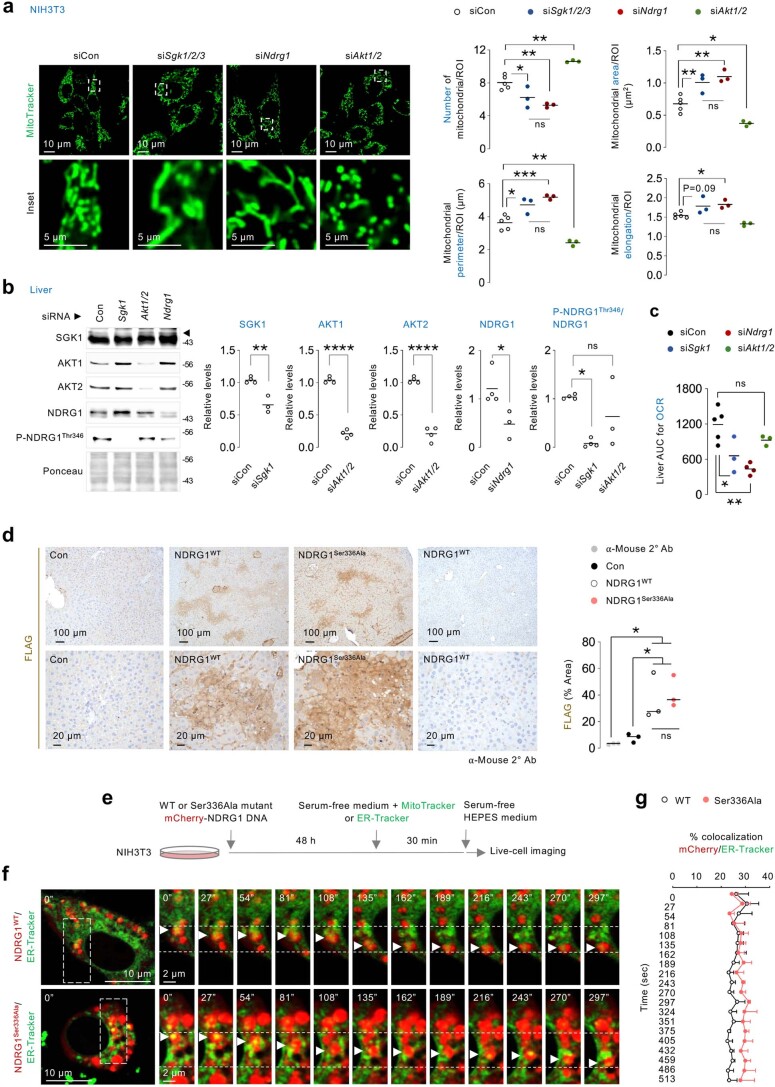
To determine whether mTORC2 drives fission by phosphorylating BNIP3 at Ser79 or Ser88 or NDRG1 at Thr328, Ser332 or Ser336, we used in vitro Seahorse-based mito-stress screens. We expressed Flag-tagged phosphorylation-deficient or phosphomimetic BNIP3 or NDRG1 mutants by switching these Ser or Thr residues to Ala or Asp, respectively (Extended Data Fig. 7a,b). Serum-deprived and oleic acid (OA)-treated cells (to emulate fasting) expressing BNIP3WT (wild type, WT) or BNIP3Ser79Ala or BNIP3Ser88Ala showed equivalent mitochondrial respiration (Extended Data Fig. 7c), eliminating that P-BNIP3Ser79/Ser88 regulates mitochondrial function. Expressing NDRG1Thr328Ala, NDRG1Ser332Ala, NDRG1Thr328Asp or NDRG1Ser332Asp mutants also failed to impact respiration; however, blocking NDRG1Ser336 phosphorylation reduced basal and maximal respiration and ATP production (Extended Data Fig. 7d–g), while phosphomimetic NDRG1Ser336Asp stimulated respiration compared with NDRG1WT (Extended Data Fig. 7f,h). Furthermore, silencing Ndrg1 (Extended Data Fig. 7i,j) or expressing each phosphorylation-deficient NDRG1 mutant (Extended Data Fig. 7k) substantially lowered mitochondrial membrane potential, suggesting that mTORC2 supports mitochondrial function via NDRG1Ser336 phosphorylation. Indeed, the mTORC2–SGK1 axis mediates fission via NDRG1Ser336 phosphorylation, since silencing Sgk1-3 or Ndrg1, but not Akt1/2, significantly reduced mitochondrial number and increased mitochondrial area, perimeter and elongation (Fig. 4f and Extended Data Fig. 8a) as observed in siDnm1l37 or siMff3 cells. Consistently, silencing Sgk1 and Ndrg1, but not Akt1/2, reduced cellular respiration in vivo (Extended Data Fig. 8b,c).
Extended Data Fig. 7. Phosphorylation of NDRG1 at Ser336 supports mitochondrial respiration and membrane potential.
(a) Experimental plan for Seahorse mitochondrial stress tests shown through c-h. (b) Representative IB for FLAG in NIH3T3 cells expressing the indicated FLAG-tagged NDRG1 wild type (WT) or FLAG-tagged Ser/Thr>Ala mutant of NDRG1 (n = 3 independent experiments). (c) Seahorse mitochondrial stress test in serum-deprived NIH3T3 cells expressing FLAG-tagged BNIP3 WT or FLAG-tagged Ser79Ala or Ser88Ala mutant BNIP3 in presence of 0.25 mM OA, followed by sequential addition of oligomycin (O), FCCP (F), and rotenone + antimycin (R + A) to assess mitochondrial respiratory function (n = 5 independent experiments). (d-f) Seahorse mitochondrial stress tests in NIH3T3 cells expressing the indicated FLAG-tagged NDRG1 WT or Ser/Thr>Ala or Ser/Thr>Asp mutant of NDRG1. Quantifications for mitochondrial respiratory function in cells expressing (g) FLAG-tagged NDRG1 WT or Ser/Thr>Ala mutants of NDRG1, or (h) FLAG-tagged NDRG1 WT or Ser336Ala and Ser336Asp mutants of NDRG1 are shown. For d-h (n = 5 independent experiments). (i) Cartoon depicting experimental plan for confocal microscopy performed in j and k. Representative blots for NDRG1 in siCon and siNdrg1 NIH3T3 cells are shown in i, right (n = 5 independent experiments). (j) MitoTracker CMXRos red fluorescence in siCon or siNdrg1 cells (siCon 58 cells and siNdrg1 62 cells from n = 3 independent experiments). (k) MitoTracker CMXRos red fluorescence in FLAG (green fluorescence)-tagged WT or phosphorylation-deficient mutants of NDRG1 (NDRG1WT 37 cells, NDRG1Thr328Ala 34 cells, NDRG1Ser332Ala 32 cells, and NDRG1Ser336Ala 33 cells from n = 4 independent experiments). Ponceau is loading control. For c-f, values are mean ± SEM. For g, h, j and k, individual replicates and means are shown. *P < 0.05, **P < 0.01 and ***P < 0.001, One-way ANOVA followed by Tukey’s multiple comparisons test (g, h and k); two-tailed unpaired Student’s t-test (j). Please refer to Supplementary Table 10_statistical summary. Source numerical data are available in SourceData_Table 1, and unprocessed blots are available in Source Data Extended Data Fig. 7.
Extended Data Fig. 8. Silencing NDRG1 and SGK1, but not AKT1/2, recapitulates mitochondrial fission failure observed in Rictor silenced cells.
(a) Representative confocal images of NIH3T3 cells silenced for Sgk1/2/3, Ndrg1 or Akt1/2 and cultured in serum-free medium for 30 min in presence of MitoTracker green. Magnified insets are shown. Quantifications for mitochondrial number and mitochondrial size/shape descriptors are shown (siCon 71 cells, siSgk1/2/3 36 cells, siNdrg1 62 cells, and siAkt1/2 36 cells from n = 5 (siCon) and n = 3 (siSgk1/2/3, siNdrg1, and siAkt1/2) independent experiments). (b) IB and quantifications for indicated proteins in livers of 3–4 mo-old male mice injected with siRNAs against Sgk1, Akt1/2 or Ndrg1 and subjected to 14–16 h fasting. N values for number of mice analyzed for indicated proteins are in parentheses. SGK1: siCon (n = 4) and siSgk1 (n = 3); AKT1 and 2: siCon (n = 4) and siAkt1/2 (n = 4); NDRG1: siCon (n = 4) and siNdrg1 (n = 3); P-NDRG1Thr346/NDRG1: siCon (n = 4), siSgk1 (n = 4) and siAkt1/2 (n = 3). (c) AUC for OCR in livers of 14–16 h fasted Con (n = 5), siSgk1 (n = 3), siNdrg1 (n = 4), and siAkt1/2 (n = 3) mice. (d) Immunohistochemistry and quantification for equivalent FLAG expression in livers silenced for endogenous Ndrg1 and then injected with FLAG-tagged WT or Ser336Ala NDRG1 plasmid (n = 3 mice). 2° Ab-only control is shown. (e) Cartoon depicting experimental plan performed in Fig. 5a and Extended Data Fig. 8f. (f) Representative live-cell imaging of mCherry-NDRG1WT or mCherry-NDRG1Ser336Ala and ER-Tracker green in NIH3T3 cells cultured in serum-free medium for 30 min. Magnified insets are shown. White arrowheads: NDRG1 (mCherry)/ER (ER-Tracker) contacts. (h) Quantification for % colocalization of mCherry with ER-tracker in g. Values are mean ± SEM (n = 3 independent experiments). Please refer to Supplementary Video 7 (mCherry-NDRG1WT/ER-Tracker), and Supplementary Video 8 (mCherry-NDRG1Ser336Ala/ER-Tracker). Ponceau is loading control. Individual replicates and means are shown. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001, One-way ANOVA followed by Tukey’s multiple comparisons test (a, c and d); two-tailed unpaired Student’s t-test (b); 2-way ANOVA followed by Tukey’s multiple comparisons test (g). ns=not significant. Please refer to Supplementary Table 10_statistical summary. Source numerical data are available in SourceData_Table 1, and unprocessed blots are available in Source Data Extended Data Fig. 8.
NDRG1Ser336Ala mutant livers exhibit fission failure
To determine whether NDRG1Ser336Ala mutant livers recapitulate the mitochondrial phenotype of RictorKO livers, we expressed Flag–NDRG1WT or Flag–NDRG1Ser336Ala in livers silenced for endogenous Ndrg1 and confirmed equivalent Flag expression by immunohistochemistry (Extended Data Fig. 8d). Consistent with our observations in RictorKO livers, fasted NDRG1Ser336Ala livers showed enlarged mitochondria with reduced mitochondrial number, and increased area and perimeter when compared with NDRG1WT and untransfected livers (Con) (Fig. 4g), reflecting impaired fission. As observed in RictorKO livers, fasted NDRG1Ser336Ala livers showed reduced cellular respiration (Fig. 4h). Furthermore, when compared with corresponding controls, MAMs from RictorKO (Fig. 3c) and NDRG1Ser336Ala livers (Fig. 4i), each showed lower levels of MFF without affecting levels of total and phosphorylated DRP1Ser616 and DRP1Ser637, which modulate dynamics31 (Fig. 3c). Hence, our data support a role for the mTORC2–NDRG1Ser336 axis in driving mitochondrial fission.
NDRG1 requires MFF, but not DRP1, for mitochondrial fission
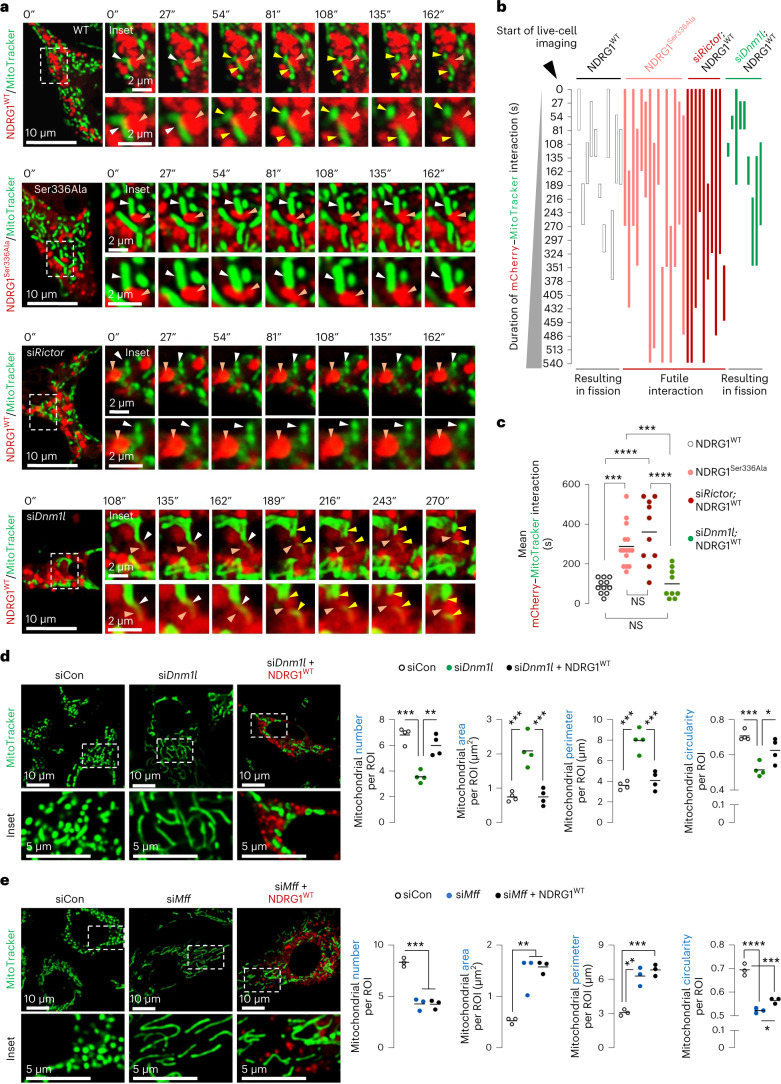
To determine how NDRG1 facilitates fission, we used time-lapse microscopy to test if NDRG1WT interacts with mitochondria. Interestingly, NDRG1WT frequently co-localized with a constricted region of mitochondria, culminating in fission (Fig. 5a–c and Supplementary Video 6). Quantifications revealed that, while NDRG1WT–mitochondrial interactions caused fission within ~90.8 ± 11.1 s of contact (Fig. 5a–c and Supplementary Video 6), NDRG1Ser336Ala maintained its co-localization with ER (Extended Data Fig. 8e–g and Supplementary Videos 7 and 8) but exhibited extended futile interactions (~289.8 ± 26.7 s) with mitochondria that did not lead to fission (Fig. 5a–c and Supplementary Video 9). Consistently, silencing Rictor led to extended futile interactions of NDRG1WT with mitochondria (409.5 ± 63.7 s) and blocked the ability of NDRG1WT to divide mitochondria (Fig. 5a–c and Supplementary Video 10), linking mTORC2-driven NDRG1Ser336 phosphorylation to mitochondrial fission. To determine whether NDRG1-mediated fission requires DRP1, we attempted to KO Dnm1l using CRISPR, but failed to generate viable healthy cells, and therefore this limit in interpretation remains. However, upon using small interfering RNAs (siRNAs) to deplete Dnm1l, mitochondrial division via NDRG1WT remained intact in siDnm1l cells (Fig. 5a–c and Supplementary Video 11). Indeed, despite >90% loss of Dnm1l, NDRG1WT continued to engage with mitochondria resulting in fission in ~102 ± 24.6 s. In fact, expressing NDRG1WT completely restored the altered mitochondrial number, area, perimeter and circularity in siDnm1l cells (Fig. 5d and Extended Data Fig. 9a). By contrast, NDRG1WT failed to restore the alterations in mitochondrial number, area, perimeter and circularity in siMff cells (Fig. 5e and Extended Data Fig. 9b) suggesting that, although DRP1 is a key regulator of fission, it appears to not influence mitochondrial fission via the mTORC2–NDRG1 axis. By contrast, the mTORC2–NDRG1 axis requires MFF for fission, supported by data showing reduced MFF enrichment in MAMs from RictorKO (Fig. 3c) and NDRG1Ser336Ala livers (Fig. 4i) and that siMff cells resist NDRG1WT-mediated fission (Fig. 5e).
Fig. 5. Phosphorylated NDRG1Ser336 requires MFF, but not DRP1, for mitochondrial fission.
a, Live cell imaging of mCherry–NDRG1WT or mCherry–NDRG1Ser336Ala and MitoTracker green in siCon NIH3T3 cells or live cell imaging of mCherry–NDRG1WT and MitoTracker in siRictor or siDnm1l NIH3T3 cells cultured in serum-free medium for 30 min. Orange arrowheads: NDRG1 (mCherry). White arrowheads: mCherry/MitoTracker contact reflecting NDRG1/mitochondria contact before fission. Yellow arrowheads: divided mitochondria after scission by NDRG1 (mCherry). Magnified insets are shown. Please refer to Supplementary Video 6 (mCherry–NDRG1WT/MitoTracker), Supplementary Video 9 (mCherry–NDRG1Ser336Ala/MitoTracker), Supplementary Video 10 (siRictor; mCherry–NDRG1WT/MitoTracker) and Supplementary Video 11 (siDnm1l; mCherry–NDRG1WT/MitoTracker). b, Quantification for duration/fate (fission versus no fission) of interaction between NDRG1 (mCherry) and mitochondria (MitoTracker). Quantifications are also shown for duration of NDRG1 (mCherry)-mitochondrial (MitoTracker) interaction events and whether each interaction led to fission (useful) or not (futile) as recorded via live cell imaging. The X axis represents time in seconds—reflecting duration of contact of NDRG1 (mCherry) with mitochondria (MitoTracker). Each individual-coloured bar on the Y axis represents one interaction per individual cell. The length of each coloured bar represents the time from the initiation of interaction of NDRG1 (mCherry) with mitochondria (MitoTracker) till end of interaction. c, Quantification for mean duration of mCherry–NDRG1/mitochondria (MitoTracker) interaction is shown. For b and c (NDRG1WT 11 cells, NDRG1Ser336Ala 15 cells, siRictor/NDRG1WT 10 cells and siDnm1l/NDRG1WT 9 cells from n = 4 independent experiments; each tracked cell was monitored on an independent plate). d,e, Representative images of siDnm1l (n = 4 independent experiments) (d) or siMff (n = 3 independent experiments) (e) NIH3T3 cells expressing mCherry–NDRG1WT or not and cultured in serum-free medium for 30 min in presence of MitoTracker green. Magnified insets are shown. Quantifications for mitochondrial number and mitochondrial size/shape descriptors are shown. Individual replicates and means are shown. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001, one-way ANOVA followed by Tukey’s multiple comparisons test. Please refer to Supplementary Table 10 statistical summary. Source numerical data are available in the Source Data to Extended Data Table 1.
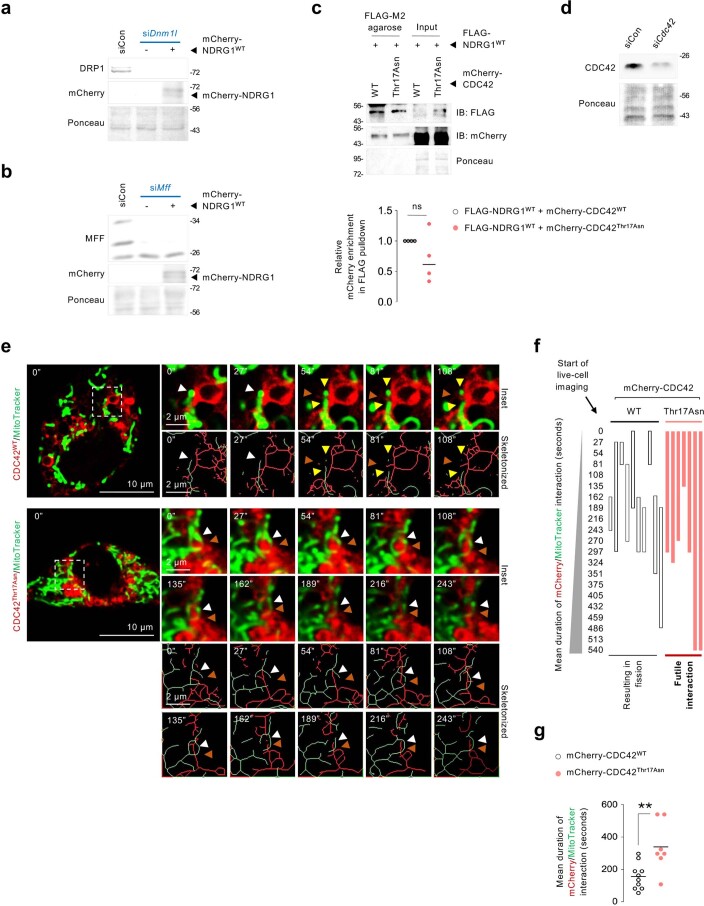
Extended Data Fig. 9. Validating silencing of Dnml1, Mff and Cdc42 in NIH3T3 cells, and time-lapse imaging showing loss of fission in cells expressing the mcherry-CDC42Thr17Asn GTP-binding mutant.
(a, b) Representative IB for indicated proteins in NIH3T3 cells silenced for (a) Dnml1 or (b) Mff and expressing mCherry-tagged NDRG1WT. Blots are representative of n = 3 independent experiments obtaining similar results. (c) IB for FLAG and mCherry in OA-treated (2.5 h) NIH3T3 cells co-expressing FLAG-NDRG1WT and mCherry-CDC42WT or mCherry-CDC42Thr17Asn mutant and subjected to pulldown of FLAG using FLAG M2 agarose (n = 4 independent experiments). Quantification for relative enrichment of mCherry in FLAG pulldowns was calculated by normalizing the densitometry value of mCherry to the densitometry value of FLAG. (d) Representative IB for indicated proteins in NIH3T3 cells silenced for Cdc42 and expressing mCherry-tagged NDRG1WT. CDC42 blot is representative of n = 3 independent experiments obtaining similar results. (e) Representative live-cell imaging in cells expressing mcherry-CDC42WT or dominant negative mcherry-CDC42Thr17Asn mutant in presence of MitoTracker green to label mitochondria. Red arrowheads: CDC42 (mCherry). White arrowheads: CDC42 (mCherry)/mitochondria (MitoTracker) contacts prior to fission. Yellow arrowheads: divided mitochondria after contact with CDC42 (mCherry). Magnified insets are shown. Please refer to Supplementary Video 14 (mCherry-CDC42WT/MitoTracker), and Supplementary Video 15 (mCherry-CDC42Thr17Asn/MitoTracker). (f) Graphical representation for duration of interaction between mCherry-CDC42WT or mCherry-CDC42Thr17Asn and mitochondria (MitoTracker), and whether interactions lead to division or are futile. (g) Quantification for mean duration of interaction (mCherry-CDC42WT 10 cells, and mCherry-CDC42Thr17Asn 7 cells from n = 3 independent experiments. Each tracked cell was monitored on an independent plate). Ponceau is loading control. Individual replicates and means are shown. **P < 0.01, two-tailed unpaired Student’s t-test (c and g). ns=not significant. Please refer to Supplementary Table 10_statistical summary. Source numerical data are available in SourceData_Table 1, and unprocessed blots are available in Source Data Extended Data Fig. 9.
Phosphorylated NDRG1Ser336 binds CDC42 to drive fission
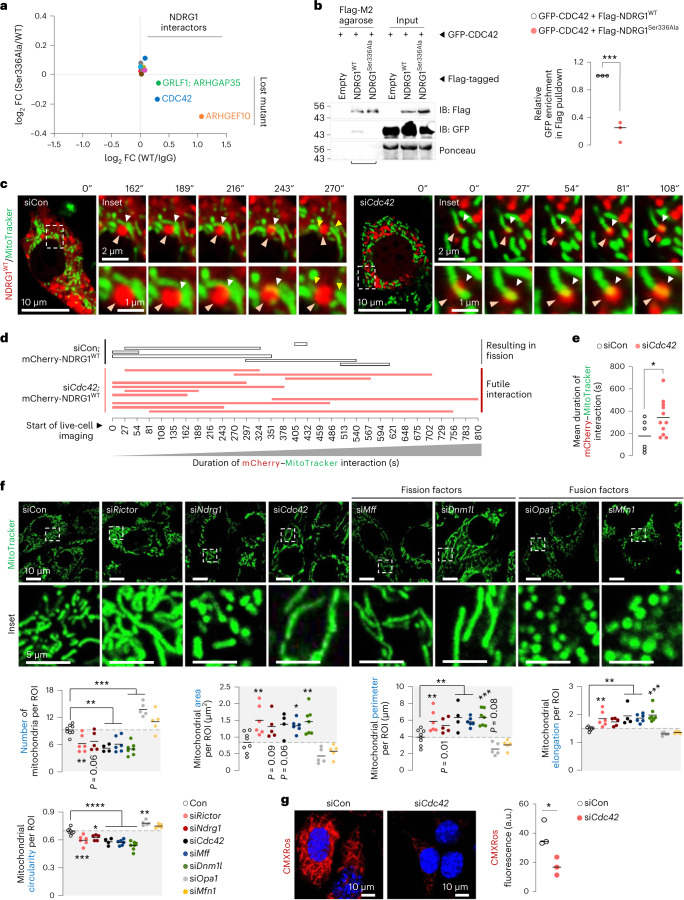
Since NDRG1 does not exhibit the intrinsic GTPase activity required for membrane scission38,39, we asked whether P-NDRG1Ser336 engages with proteins with intrinsic GTPase activity to facilitate fission. Accordingly, proteomics to identify proteins bound to Flag-tagged NDRG1WT, but not NDRG1Ser336Ala, revealed interaction with CDC42, a RHO GTPase that regulates actin cytoskeleton40 and cytokinesis9 (Fig. 6a and Supplementary Table 8). Indeed, when compared with NDRG1WT, NDRG1Ser336Ala displayed reduced, albeit modest, binding to CDC42, ARHGEF10 (RHO GEF that activates RHO GTPases by stimulating GDP/GTP exchange)41, and ARHGAP35 (RHO GAP that facilitates GTP hydrolysis to inactivate RHO GTPases)41. Consequently, we hypothesized that GTPase CDC42 mediates the effects of mTORC2–NDRG1Ser336 on mitochondrial fission. Supporting this hypothesis, co-immunoprecipitation (co-IP) confirmed that exogenously expressed GFP–CDC42 interacts with Flag–NDRG1WT but fails to interact with mutant NDRG1Ser336Ala (Fig. 6b). Furthermore, Flag–NDRG1WT interacts with both mCherry–CDC42WT and mutant CDC42Thr17Asn, which is expected to be predominantly GDP-bound (Extended Data Fig. 9c), indicating that CDC42–GTP loading is not required for CDC42–NDRG1 interaction. However, NDRG1Ser336 phosphorylation and CDC42–GTP loading are each critical for mitochondrial fission. Indeed, silencing Cdc42 (Extended Data Fig. 9d) led to prolonged and futile engagement of NDRG1WT with mitochondria and blocked fission (Fig. 6c–e and Supplementary Videos 12 and 13). Furthermore, while CDC42WT engaged with, and divided, mitochondria in ~157 ± 26 s, mutant CDC42Thr17Asn exhibited prolonged (~339 ± 58 s) and futile interactions with mitochondria (Extended Data Fig. 9e–g and Supplementary Videos 14 and 15). Supporting that the mTORC2–NDRG1–CDC42 axis drives fission, knocking down Rictor or Ndrg1 or Cdc42 each reduced mitochondrial number and circularity, and increased mitochondrial area, perimeter and elongation (Fig. 6f), recapitulating the fission failure phenotype of siMff or siDnm1l cells (Fig. 6f). Consistently, silencing Cdc42 reduced mitochondrial membrane potential (Fig. 6g), suggesting that CDC42 cooperates with P-NDRG1Ser336 to support mitochondrial division.
Fig. 6. Phosphorylated NDRG1Ser336 interacts with CDC42 to drive mitochondrial fission.
a, The log2-transformed fold change (FC) of interaction of Flag–NDRG1WT or Flag–NDRG1Ser336Ala with CDC42, ARHGAP35 and ARHGEF10 (n = 3 independent experiments). b, Pulldowns of Flag (using Flag M2 agarose) and immunoblots and quantification for GFP and Flag levels in OA-treated (2.5 h) NIH3T3 cells co-expressing Flag–NDRG1WT or Flag–NDRG1Ser336Ala and GFP–CDC42WT (n = 3 independent experiments). Flag-tagged empty vector is the negative control. Ponceau is loading control. Quantification for relative enrichment of GFP in Flag pulldowns was calculated by normalizing the densitometric value of GFP to the densitometric value of Flag. c, Representative live-cell imaging of mCherry–NDRG1WT and MitoTracker green in siCon or siCdc42 NIH3T3 cells. Magnified insets are shown. Orange arrowhead: NDRG1 (mCherry) mediating fission. White arrowhead: mCherry/MitoTracker reflecting NDRG1/mitochondrial co-localization before fission. Yellow arrowheads: divided mitochondria after fission. Please refer to Supplementary Video 12 (siCon cells; mCherry–NDRG1WT) and Supplementary Video 13 (siCdc42 cells; mCherry–NDRG1WT). d, Graphical representation for duration of interaction between mCherry–NDRG1 and mitochondria (MitoTracker) in siCon or siCdc42 cells, and whether interactions lead to division or are futile. e, Quantification for mean duration of mCherry–NDRG1WT/mitochondria (MitoTracker) interaction is shown (siCon 6 cells and siCdc42 11 cells from n = 3 independent experiments; each tracked cell was monitored on an independent plate). f, Representative confocal images of NIH3T3 cells transfected with indicated siRNAs and cultured in serum-free medium with MitoTracker green for 30 min. Magnified insets are shown. Quantifications for mitochondria number and mitochondrial size/shape descriptors are shown (siCon 142 cells, siRictor 105 cells, siNdrg1 91 cells, siCdc42 64 cells, siMff 106 cells, siDnm1l 128 cells, siOpa1 86 cells and siMfn1 83 cells from n = 8 (siCon), n = 6 (siRictor and siMff), n = 5 (siNdrg1, siOpa1 and siMfn1), n = 4 (siCdc42) and n = 7 (siDnm1l) independent experiments). Grey areas indicate mitochondria fission-deficient models. g, MitoTracker CMXRos fluorescence in siCon and siCdc42 cells cultured in serum-free medium in presence of OA for 5 h (siCon 102 cells and siCdc42 116 cells from n = 3 independent experiments). Individual replicates and means are shown. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001, two-tailed unpaired Student’s t-test (a, b, e and g); one-way ANOVA and Dunnett’s multiple comparisons test (f). Please refer to Supplementary Table 10 statistical summary, and Supplementary Table 8. Source numerical data are available in Source Data Extended Data Table 1, and unprocessed blots are available in the Source Data for this figure.
CDC42 regulators and effectors modulate fission
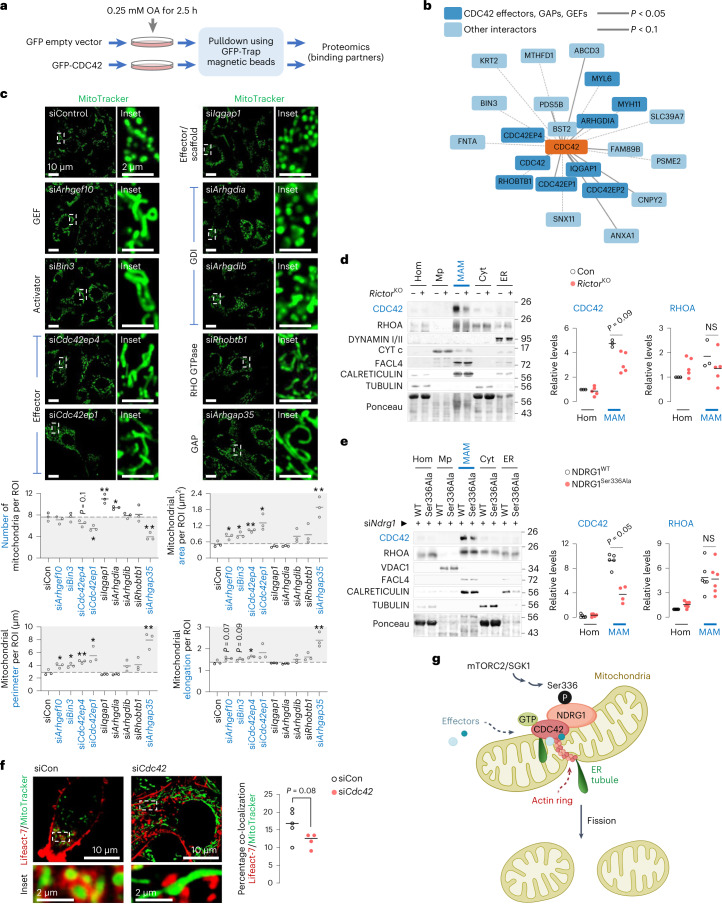
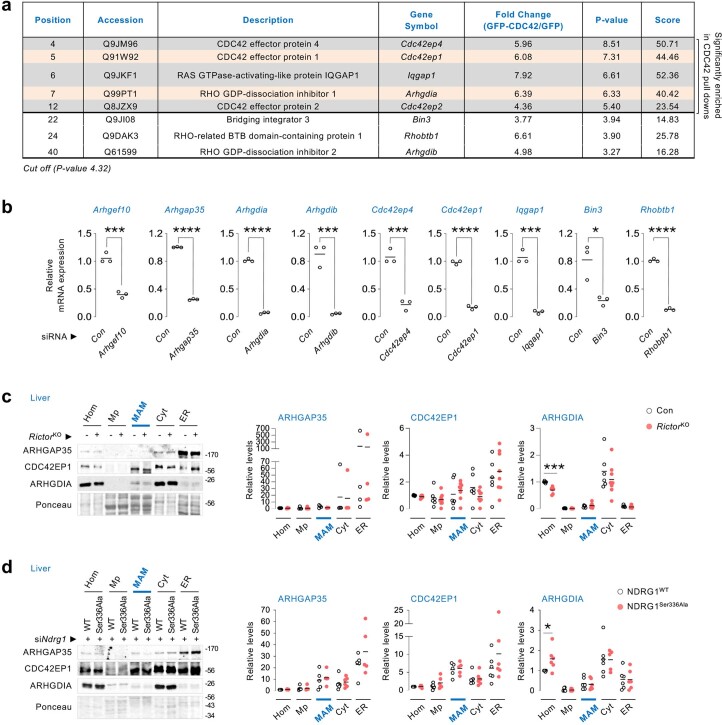
Since CDC42 activity is tightly orchestrated by regulators or effectors, we used proteomics to identify CDC42 interactors that may potentially regulate fission (Fig. 7a and Supplementary Table 9). Using NIH3T3 cells expressing GFP–CDC42 or GFP–empty vector as negative control and analysing fold-change interaction (using cut-off P value of 4.32), we short-listed 11 proteins that were significantly enriched in GFP–CDC42 pulldowns when compared with empty vector (Fig. 7b and Extended Data Fig. 10a). Of note, the five enriched targets were CDC42 effectors (CDC42EP4/BORG4, CDC42EP1/BORG5 and CDC42EP2/BORG21), RHO GTPase inhibitor RHO GDP Dissociation Inhibitor (GDI) alpha (ARHGDIA/RHOGDI1) and IQ motif-containing GTPase-activating protein 1 (IQGAP1), which is a downstream effector and upstream scaffold protein for CDC42 (ref. 42) (Fig. 7b and Extended Data Fig. 10a). We also observed enrichment (albeit insignificant) in GFP–CDC42 pulldowns of a known driver of CDC42 in muscle cells, bridging integrator 3 (BIN3), the atypical RHO GTPase/RHO-Related BTB Domain Containing 1 (RHOBTB1) and RHO GTPase inhibitor beta (ARHGDIB/RHOGDI2) (Fig. 7b and Extended Data Fig. 10a). In addition, we included to screen for two identified NDRG1 binding partners, ARHGEF10 and ARHGAP35, which serve as GEF and GAP41, respectively (Fig. 6a).
Fig. 7. CDC42 regulators and effectors modulate fission.
a, Experimental plan to pull down GFP-tagged CDC42 to identify interactors that regulate mitochondrial fission. b, Cartoon representing significantly (P < 0.05) enriched interacting partners of CDC42 (in bold) identified via proteomics, some of which belong to the RHO family of GTPases (n = 4 independent experiments). c, Representative images of NIH3T3 cells knocked-down for indicated CDC42-binding partners and cultured in serum-free medium for 30 min in presence of MitoTracker green. Magnified insets are shown. Quantifications for mitochondrial number and mitochondrial size/shape descriptors are shown (siCon, siArhgef10, siIqgap, siArhgdib, siRhobtb1 and siArhgap35 39 cells, siBin3 and siCdc42ep4 40 cells, siCdc42ep1 43 cells and siArhgdia 37 cells from n = 3 independent experiments). Grey areas indicate mitochondria fission-deficient models. d,e, Immunoblots and quantification for indicated proteins in indicated fractions from livers of 14–16 h-fasted 5–6-month-old Con (n = 3) and RictorKO (n = 5) mice (d), and 3–4-month-old mice expressing NDRG1WT or NDRG1Ser336Ala plasmids after silencing endogenous Ndrg1 with siRNAs (e). N values for number of mice per fraction are indicated in parentheses. CDC42: all fractions from NDRG1WT and NDRG1Ser336Ala mice are n = 5 except for NDRG1Ser336Ala MAMs where n = 4; RHOA: n = 6 mice for all groups. Ponceau is loading control. f, Representative confocal images of siCon or siCdc42 NIH3T3 cells expressing mCherry–Lifeact-7 and cultured in serum-free medium for 30 min in presence of MitoTracker green. Magnified insets are shown. Quantification for percentage co-localization of mCherry–Lifeact-7 with mitochondria is shown (siCon 33 cells and siCdc42 30 cells from n = 5 (siCon) or n = 4 (siCdc42) independent experiments). g, Reactivation of mTORC2 during fasting phosphorylates NDRG1 at Ser336, which engages with mitochondria and recruits CDC42 to mitochondria–ER contact sites wherein CDC42 and its effector proteins orchestrate fission. Individual replicates and means are shown. *P < 0.05 and **P < 0.01, two-tailed unpaired Student’s t-test. NS, not significant. Please refer to Supplementary Table 10 statistical summary, and Supplementary Table 9. Source numerical data are available in Source Data Extended Data Table 1, and unprocessed blots are available in the Source Data for this figure.
Extended Data Fig. 10. An siRNA screen to target interacting partners of CDC42 reveals effectors and regulators of CDC42 controlling mitochondrial dynamics.
(a) Significantly enriched interacting partners of CDC42 that belong to the RHO family of GTPases. A log2 transformed P-value cut-off of 4.32 was used as threshold (n = 4 independent experiments). (b) qPCR in NIH3T3 cells to validate the silencing of selected CDC42-binding partners identified by proteomics (n = 3 independent experiments). (c, d) IB and quantification for indicated proteins in Hom, Mp, MAMs, Cyt, and ER fractions from livers of (c) 5–6 mo-old Con or RictorKO male mice, and (d) 3–4 mo-old NDRG1WT or NDRG1Ser336Ala male mice co-injected with siRNA against endogenous Ndrg1 and fasted for 14–16 h. N values for number of mice analyzed for individual proteins in indicated fractions are in parentheses. For c, ARHGAP35: all fractions from Con (n = 4) and RictorKO (n = 4) mice; CDC42EP1: all fractions, Con (n = 6) and RictorKO (n = 8) mice except RictorKO Hom where n = 7 mice; ARHGDIA: all fractions from Con (n = 6) mice except Mp, MAMs and ER where n = 5 mice, and all fraction from RictorKO (n = 8) mice except Hom and ER where n = 7 mice. For d, ARHGAP35: all fractions from NDRG1WT and NDRG1Ser336Ala mice are n = 6 except Mp, MAMs and ER from NDRG1WT mice where n = 5; CDC42EP1: all fractions from NDRG1WT and NDRG1Ser336Ala mice are n = 6 except MAMs in both groups where n = 5; ARHGDIA: all fractions from NDRG1WT and NDRG1Ser336Ala mice are n = 6 except Cyt in NDRG1Ser336Ala mice where n = 5. Ponceau is loading control. Individual replicates and means are shown. *P < 0.05, ***P < 0.001 and ****P < 0.0001, two-tailed unpaired Student’s t-test (a-d). GAP: GTPase activating protein; GDI: GDP dissociation inhibitors. Please refer to Supplementary Table 10_statistical summary, and Supplementary Table 9. Source numerical data are available in SourceData_Table 1, and unprocessed blots are available in Source Data Extended Data Fig. 10.
To examine if the identified candidates regulate mitochondrial fission, we transfected NIH3T3 cells with siRNAs against each target (Extended Data Fig. 10b), except Cdc42ep2 since silencing it severely reduced viability. Our results indicate that CDC42 and its family of effectors/regulators control mitochondrial fission, since deleting Cdc42 or CDC42 activators, Arhgef10 and Bin3, or CDC42 downstream effectors, Cdc42ep4/BORG4 or Cdc42ep1/BORG5, each resulted in increased mitochondrial area, perimeter and elongation, reflecting impaired fission (Fig. 7c). RHOGDI proteins can act as negative regulators of RHO GTPases by retaining RHO GTPases in the cytosol, inhibiting their GTPase activity, and preventing their interaction with GEFs, GAPs and effectors43,44. Accordingly, we suspect that silencing Arhgdia/RHOGDI1 releases CDC42 from the inhibitory effect of ARHGDIA/RHOGDI1, leading to fission. Consistently, silencing Arhgdia/RHOGDI1 (but not Arhgdib/RHOGDI2) increased mitochondrial number (Fig. 7c), reflecting increased fission. Not all RHO GTPases impact mitochondrial dynamics, since depleting the RHO GTPase Rhobtb1 gene had no effect on mitochondrial morphology, suggesting specificity of RHO GTPase CDC42 towards fission. We also found that silencing IQGAP1, which regulates CDC42 as an upstream scaffold and as a downstream effector of CDC42 (ref. 42), increased mitochondrial number (Fig. 7c). Indeed, as a scaffold protein, IQGAP1 provides a molecular link between Ca2+/calmodulin and CDC42-mediated processes45, while as a downstream effector, CDC42 enhances the F-actin-cross-linking activity of IQGAP1 during actin reorganization46. Since ARHGAP35 inactivates GTPases, we anticipated that depleting Arhgap35 would stimulate CDC42, leading to fission; however, knocking down Arhgap35 decreased mitochondrial number, reflecting fission failure (Fig. 7c). This probably reflects the complex regulation of CDC42 requiring subsequent inactivation to complete its function47,48, as well as specificity among the different effectors and regulators in stimulating fission. Alternatively, ARHGAP35 is perhaps not active towards CDC42 and affects, instead, an antagonistic GTPase. Consistent with these findings, in addition to CDC42, we detected the presence in MAMs of CDC42 effector, CDC42EP1/BORG5, and ARHGAP35 and ARHGDIA/RHOGDI1 (Extended Data Fig. 10c,d). Interestingly, levels of ARHGAP35, CDC42EP1/BORG5 and ARHGDIA/RHOGDI1 in MAMs from RictorKO (Extended Data Fig. 10c) and NDRG1Ser336Ala expressing livers (Extended Data Fig. 10d) were comparable to those in corresponding controls, indicating that fission is probably regulated at the level of recruitment of CDC42 to MAMs.
Since CDC42 action is regulated by membrane binding49 and GTP loading50, we suspect that mTORC2-driven NDRG1Ser336 phosphorylation is a signal to recruit CDC42 to MAMs for its activation to drive fission. Indeed, CDC42 and RHOA51 (Fig. 7d), but not dynamins, were abundantly present in MAMs from fasted livers. By contrast, MAMs from fasted RictorKO (Fig. 7d) and NDRG1Ser336Ala livers (Fig. 7e) each showed markedly reduced CDC42 levels without affecting RHOA levels, suggesting that the mTORC2–NDRG1 axis recruits CDC42 to MAMs. Given the enrichment of CDC42 in MAMs in an mTORC2- and NDRG1-sensitive manner, it is likely that CDC42 governs local downstream mechanisms that control fission. Since dynamic cycling of actin through populations of mitochondria controls fission52,53, we asked whether CDC42 mediates the effect of mTORC2–NDRG1 on mitochondrial fission by remodelling actin cytoskeleton. Indeed, in control cells, actin assembled around mitochondria to generate ring-like structures consistent with maintained fission53 (Fig. 7f). By contrast, silencing Cdc42 decreased co-localization of actin with mitochondria, which correlated with elongated mitochondria (Fig. 7f), suggesting that CDC42 facilitates the organization of actin around mitochondria to enable fission.
Discussion
In sum, we show that the typically nutrient-responsive mTORC2 is paradoxically reactivated by fasting to regulate NDRG1Ser336 phosphorylation, which serves to recruit CDC42 to MAMs to drive mitochondrial fission. In support of this model (Fig. 7g), NDRG1 engages with mitochondrial constrictions to facilitate fission, and that fission events are blocked in cells expressing phosphorylation-deficient NDRG1Ser336Ala or in cells lacking Rictor or Cdc42 or the identified CDC42 effector/regulators (Fig. 7g), thus revealing an mTORC2–NDRG1–CDC42 axis facilitating mitochondrial fission during fasting.
Fasting and feeding are hormonally distinct physiological states19. While nutrient deprivation in cultured cells blocks mitochondrial fission to preserve ATP synthesis and cell viability54,55, cultured cells do not completely recapitulate the complex physiology of intact organisms. In fact, we show that the highly integrated liver exhibits marked increases in fission during an acute fast. Indeed, switching between nutrient availability and deprivation modulates mitochondrial cristae and ER contacts, which per se impact mitochondrial dynamics56,57. In keeping with this, we suspect that fasting-induced increases in adipose lipolysis and increased availability of lipids reactivate mTOR during fasting. Indeed, cholesterol58 and phosphatidic acid59 activate mTORC1 in vitro, and we show here that exposure to dietary corn oil or fasting each activates mTORC2 in liver, as has been shown for mTORC1 in starved cultured cells60. Although no function has been assigned to fasting-induced reactivation of mTOR, we demonstrate that paradoxical reactivation of mTORC2 during fasting is required for mitochondrial remodelling to possibly support the increased energetic demands of fasting. In fact, enzymatic reactions, for example, those part of the Krebs cycle, appear to be sensitive to changes in mitochondrial shape, volume and connectivity61. Consistently, not only does loss of Rictor impact mitochondrial fission, we also noted marked accumulation of acylcarnitines, a metabolic signature consisted with dampened mitochondrial respiration.
Mitochondrial division is tightly orchestrated by recruitment of dynamin-related GTPase DRP1 from the cytosol to MAMs by the mitochondrial outer membrane receptor MFF4. DRP1 oligomerization and interaction with actin filaments promote scission in a GTP hydrolysis-dependent manner. Indeed, overexpression of MFF fails to restore fission in cells co-expressing the assembly-defective DRP1 mutant, indicating that MFF acts via DRP1 (ref. 4). Yet, our data show that DRP1 is dispensable for mTORC2–NDRG1-mediated mitochondrial fission, since overexpressing NDRG1WT restores mitochondrial fission in DRP1-deficient cells, and perhaps most surprisingly, NDRG1WT fails to restore fission when MFF is depleted. These data suggest that mTORC2–NDRG1-mediated fission is dependent on MFF but appears to not require DRP1. Supporting this possibility, loss of Rictor or expressing the NDRG1Ser336Ala mutant each markedly reduced MFF levels in MAMs without affecting DRP1 levels. These findings suggest that roles for MFF in fission are complex and not restricted to merely serving as a receptor for DRP1 recruitment4.
How then does NDRG1 drive mitochondrial fission? Since NDRG1 lacks GTP hydrolysis activity, a requirement for fission38,39, we examined whether NDRG1 engages with additional GTPases to facilitate fission. Here we identify the small GTPase CDC42 as a binding partner of NDRG1 that fails to interact with the phosphorylation-deficient NDRG1Ser336Ala mutant. Indeed, time-lapse imaging revealed that NDRG1 and CDC42 both engage at mitochondrial constrictions to facilitate fission, and that in absence of Cdc42 or presence of inactive GDP-bound CDC42Thr17Asn mutant, NDRG1 fails to cut mitochondria. Furthermore, we detected the presence of CDC42 in MAMs, the enrichment of which appears to depend on an intact functional mTORC2–NDRG1 axis since loss of Rictor or expressing the NDRG1Ser336Ala mutant each markedly reduced CDC42 levels in MAMs. Given that CDC42 modulates the actin cytoskeleton40, it is tempting to speculate that recruitment of CDC42 to MAMs by the mTORC2–NDRG1 axis orchestrates a local interplay between actin tubules and ER in driving scission, although careful future assessments are needed to conclusively demonstrate the same. Since Rictor insufficiency shortens lifespan62,63, and given the age-related impairment in mitochondrial function, it is also tempting to speculate that stimulation of mTORC2 to sustain mitochondrial fission could potentially delay age-related diseases in which defective mitochondrial dynamics play a part.
Methods
This research complies with all relevant ethical regulations including animal protocol approval from the IACUC of Albert Einstein College of Medicine (protocol number 00001051).
Animal models
C57BL/6 (000664), Rictorflox/flox (020649), Raptorflox/flox (013188) and Tsc1flox/flox (005680) mice were from Jackson Laboratory. Studies were performed in 2–10-month-old male and female mice fed regular chow (5058; Lab Diet) and maintained in barrier facility at 22–23 °C under 40–60% humidity and a 12 h:12 h light/dark cycle. Liver-specific RictorKO, RaptorKO or Tsc1KO mice were generated via retro-orbital injections of 2 × 1011 genome copies per mouse of AAV8-TBG-iCre adenovirus (Vector Biolabs, VB1724) and mice were humanely killed after 8 weeks64. AAV8-TBG-eGFP (Vector Biolabs, VB1743)-injected mice were controls. Mice were fasted with free access to water and were compared with ad libitum mice. Fasted mice were treated with: oral gavage of (1) corn oil (400 μl; Sigma-Aldrich, 8267), (2) BODIPY FL C16 (10 mg kg−1; Invitrogen, D3821) or (3) refed high-fat diet (60% kcal in fat; Research Diets, D12492) for 30 min. Mice were mock-gavaged for 5 days before experiment, and control animals were gavaged with vehicle (10% dimethyl sulfoxide–saline solution) to match for volume and distension. STZ (40 mg kg−1; Sigma-Aldrich, S0130) was injected intraperitoneally once a day for 5 consecutive days. The protocol was repeated after 8 weeks, and tissues were collected 2 weeks after the last injection.
Corn oil
Corn oil (Sigma-Aldrich, 8267) is protein free, and provides fatty acids and monoacylglycerols, which are absorbed by the gut and delivered systemically. The composition of corn oil is detailed in Supplementary Table 11.
Cell culture
NIH3T3 (ATCC, CRL-1658) and HepG2 (ATCC, HB-8065) cells were cultured in high-glucose Dulbecco’s modified Eagle medium (DMEM) (Gibco, 11965118) supplemented with 10% (v/v) foetal bovine serum (FBS) (Sigma-Aldrich, 12106C) and 1% (v/v) penicillin–streptomycin (Gibco, 15140). AML12 cells (ATCC, CRL-2254) were cultured in DMEM/F-12 medium (Gibco, 11320033) supplemented with 10% FBS, 1% insulin–transferrin–selenium (Gibco, 41400-045), 40 ng ml−1 dexamethasone (Sigma-Aldrich, D4902) and 1% penicillin–streptomycin. Cells were maintained at 37 °C in 5% CO2. Wherever indicated, NIH3T3 cells were washed once with PBS and incubated in serum-free DMEM/P/S in presence of 0.25 mM OA (Sigma-Aldrich, O3008) for indicated durations.
Primary mouse embryonic fibroblasts were isolated from Rictorflox/flox mice as described65. Rictorflox/flox cells were plated at ~80% confluency and infected with 50 multiplicity of infection of adenoviral-null Ad(RGD)-fLuc (Vector Biolabs, 9999) or Ad(RGD)-CMV-iCre (Vector Biolabs, 1769) in serum-free medium for 24 h. The virus-containing medium was replaced by 10% FBS medium, and 72 h post-infection cells were used for experiments.
Plasmid DNAs
Mouse NDRG1_OMu19504D, BNIP3_OMu13517D and CDC42_OMu16203C_cDNA expression plasmids were synthesized by GenScript USA. NDRG1_OMu19504D and BNIP3_OMu13517D were each cloned into pcDNA3.1+/C-(K)-DYK vector. CDC42_OMu16203C was cloned into pcDNA3.1(+)-N-eGFP vector. cDNA encoding NDRG1Thr328Ala, NDRG1Thr328Asp, NDRG1Ser332Ala, NDRG1Ser332Asp, NDRG1Ser336Ala, NDRG1Ser336Asp, BNIP3Ser79Ala or BNIP3Ser88Ala_pcDNA3.1+/C-(K)-DYK mutants was generated by site-directed mutagenesis. pcDNA3.1+/C-(K)-DYK or pcDNA3.1(+)-N-eGFP vectors were negative controls. For live-cell imaging, mouse NDRG1_OMu19504D WT, mutant NDRG1Ser336Ala and CDC42_OMu16203C WT plasmids were each cloned into a pcDNA3.1(+)-mCherry vector. cDNA encoding CDC42Thr17Asn_pcDNA3.1(+)-mCherry mutant was generated by site-directed mutagenesis. mCherry–Lifeact-7 was a gift from M. Davidson (Addgene, 54491).
In vitro transfections of nucleic acids
In vitro transfections were performed using Lipofectamine 3000 (Invitrogen, L3000). For expression of DNA plasmids, 120,000 NIH3T3 cells ml−1 of growth medium were transfected with 1 μg of DNA and plated in 12-well plate dishes for 48 h. For gene silencing, 120,000 cells ml−1 of growth medium were transfected with siRNA for 48 h (Supplementary Table 12). Scrambled RNA was used as negative control (siCon). Silencing efficiency was confirmed by western blotting or qPCR.
In vivo delivery of nucleic acids
In vivo delivery of plasmid DNAs was performed via in vivo-jetPEI (Polyplus-transfection SA, 201-50G) as per the manufacturer’s instructions. Briefly, 100 μg of NDRG1WT or NDRG1Ser336Ala_pcDNA3.1+/C-(K)-DYK was diluted in glucose solution and combined with 7 μl of in vivo-jetPEI for 15 min at room temperature. Then 200 μl of transfection mix was administered retro-orbitally to C57BL/6 mice in a single injection 24 h before tissue collection. Transfection efficiency was determined by immunohistochemistry. Livers from non-transfected mice were used as negative controls. In vivo siRNA delivery was performed using Invivofectamine 3.0 Reagent (Invitrogen, IVF3005) as per the manufacturer’s instructions. Briefly, 50 μg of siRNAs was mixed with complexation buffer, added to Invivofectamine 3.0 Reagent (1:1 ratio) and incubated for 30 min at 50 °C. The mix was diluted in PBS (pH 7.4), and 200 μl of siRNA mix was administered retro-orbitally to C57BL/6 mice every 24 h for 3 consecutive days before tissue collection.
RNA isolation and real-time PCR
mRNA expression was performed as described66 using M-MLV Reverse Transcriptase (Invitrogen, 28025). The primers are detailed in Supplementary Table 13.
Western blotting
Total cell lysates from cells in culture were prepared using lysis buffer (20 mM Tris pH 7.5, 50 mM NaCl, 0.5%, 1 mM EDTA, 1 mM EGTA and 1% Triton X-100) supplemented with complete EDTA-free protease inhibitor (Roche, 11873580001) and phosphatase inhibitor cocktails 2 and 3 (Sigma-Aldrich, P5726 and P0044). Total protein from liver or epididymal adipose tissue was isolated in RIPA buffer (50 mM Tris pH 8.0, 150 mM NaCl, 0.5% sodium deoxycholate, 1% SDS and 1% NP-40) supplemented with protease/phosphatase inhibitors. Total protein from soleus muscle was isolated as described67. Lysates were centrifuged at 17,000g for 30 min at 4 °C, and supernatants were immunoblotted by denaturing 20–30 μg of protein at 95 °C for 5 min in 3× Laemmli buffer. For analysis of OXPHOS, samples were boiled at 50 °C for 5 min and resolved by SDS–PAGE as described68. Protein bands were normalized to Ponceau S and quantified by ImageJ (National Institutes of Health, NIH). Antibodies are detailed in Supplementary Table 14.
Subcellular fractionation
Fresh livers were fractionated for isolation of MAMs, pure mitochondria, cytosol and ER fractions as described69. Cytochrome c (CYT c) and Voltage Dependent Anion Channel 1 (VDAC1) were used as enrichment/purity markers for mitochondria, long-chain fatty acid coenzyme A ligase 4 (FACL4) as marker for MAM, calreticulin as marker for MAMs and ER, and tubulin as marker for cytoplasm.
Co-IP
For Fig. 6b, lysates (1,000 μg) from NIH3T3 cells co-expressing Flag–NDRG1WT or Flag–NDRG1Ser336Ala with GFP–CDC42 were incubated with 30 μl of Anti-Flag M2 Affinity Gel (Sigma-Aldrich, A2220) and eluted with 3× Flag peptide (Sigma-Aldrich, F4799) by incubating for 2 h at 4 °C in rotation. Co-IP eluents were immunoblotted. Cells expressing Flag empty vector were negative controls. For Extended Data Fig. 9c, lysates (1,000 μg) from NIH3T3 cells co-expressing mCherry–CDC42WT or mCherry–CDC42Thr17Asn with Flag–NDRG1WT were subjected to Flag pulldowns as above. For identification of Flag–NDRG1WT or Ser336Ala-interacting partners in Fig. 6a, lysates (700 μg) from NIH3T3 cells expressing Flag–NDRG1WT or Flag–NDRG1Ser336Ala were subjected to Flag pulldowns, and eluents were subjected to mass spectrometry using S-trap columns and nLC–MS/MS as below. Non-transfected cells were negative controls. For identification of CDC42-interacting partners in Fig. 7b, lysates (700 μg) from NIH3T3 cells expressing GFP–CDC42 or GFP–empty vector were incubated with 25 μl of GFP-Trap Magnetic Agarose (Chromotek, gtma) for 2 h at 4 °C in rotation after which beads were washed and subjected to on-beads digestion for mass spectrometry using S-trap columns and nLC–MS/MS as below.
Sample preparation for phosphoproteomics
Liver (500 μg), liver MAM fractions (700 μg) or co-IP eluents from Flag pulldowns performed in total cell lysates (700 μg) of siCon or siRictor NIH3T3 cells co-expressing Flag–NDRG1WT were homogenized in 2% SDS/5 mM dithiothreitol (with protease/phosphatase inhibitors) to retrieve proteins in solution and incubated for 1 h at room temperature for disulfide bond reduction. Proteins were alkylated using 20 mM iodoacetamide for 30 min in the dark. Protein digestion was performed utilizing S-trap mini cartridges (ProtiFi) as per the manufacturer’s instructions. Phosphorylated peptides were enriched from the S-trap eluate using titanium dioxide beads (TiO2, GL Sciences) as described70. Following TiO2 enrichment, peptides were concentrated with a speed vac, desalted in HLB resin (Waters) and concentrated in a speed vac once more before analysing peptides by nLC–MS/MS.
nLC–MS/MS acquisition
Samples were resuspended in 10 μl of water/0.1% trifluoroacetic acid and loaded onto a Dionex RSLC Ultimate 300 (Thermo Scientific) coupled online with an Orbitrap Fusion Lumos (Thermo Scientific). The two-column chromatographic separation system consisted of a C18 trap cartridge (300 μm internal diameter (ID), 5 mm length) and a picofrit analytical column (75 μm ID, 30 cm length) packed in-house with reversed-phase Repro-Sil Pur C18-AQ 3 μm resin. Peptides were separated using a 180 min gradient from 2% to 28% buffer B (buffer A: 0.1% formic acid; buffer B: 80% acetonitrile/0.1% formic acid) at a flow rate of 300 nl min−1. The mass spectrometer acquired spectra in a data-dependent acquisition mode. Briefly, the full MS scan was set to 300–1,200 m/z in the Orbitrap with a resolution of 120,000 (at 200 m/z) and an AGC target of 5 × 105. MS/MS was performed in the ion trap using top speed mode (2 s), an AGC target of 10 × 104 and higher collisional dissociation (HCD) collision energy of 30. Two additional targeted scans were added in each instrument duty cycle to detect the low-abundance NDRG1Ser336 peptide: a selected ion monitoring scan for the intact mass quantification and a targeted MS/MS scan for identification of the peptide.
Phosphoproteomics data analysis was conducted using Proteome Discoverer v2.4 (Thermo Scientific) at standard settings for tolerances, modifications and filters, and phosphorylation on Ser/Thr/tyrosine as dynamic modifications. SwissProt mouse proteome database was used (downloaded August 2019). Peptide abundance was obtained using the intensity of the extracted ion chromatogram; values were log2 transformed and normalized, and missing values were imputed as described71. Comparisons between groups were performed in a binary manner; each sample type was compared with the fasted condition utilizing a two-tails heteroscedastic t-test (significant, if P value < 0.05). The data distribution was assumed to be normal.
Significantly modified proteins were selected by Benjamini–Hochberg correction (P < 0.05). When false discovery rate correction led to no hit, inspection of uncorrected P value distribution was performed: if an anti-conservative distribution was observed, we applied an alternative method of false discovery rate control by combining threshold for significance (P < 0.05) with fold-change cut-off (fold-change >1.5) as suggested72. Phosphorylation state change (∆Ps) for individual proteins was calculated as described34, as the sum of log2(fold change) value of all phosphopeptides with statistically significant changes (P < 0.05) compared to control. If none of the phosphopeptide P values is below 0.05, the ∆Ps value will be zero. We applied a stringent cut-off for ∆Ps value at two standard deviations (2σ) to represent the concept of cumulative phosphorylation. Gene Ontology was performed using BINGO or Enrichr73. In the enrichment map-based network visualization of Gene Ontology enrichment of differentially modulated phosphosites, blue edges show similarity between decreased phosphosites while red nodes show similarity between increased phosphosites; node size indicates the number of proteins per node; major clusters are circled, and the associated names represent major functional associations. The enrichment map was generated in Cytoscape (3.8.1) using Enrichment map plugin (3.3.0) (ref. 33) using P value <0.05, false discovery rate <0.001. Data handling and statistical analyses were performed using Python (Python software foundation; v.3.7.4 available at https://www.python.org/) and Scientific Python Stack: SciPy (v.1.3.1) (ref. 74), NumPy (v.1.17.2) (ref. 75) and Matplotlib (v.3.1.1). Phosphosites showing significant regulation between groups were used to predict the kinase responsible for their catalysis using the iGPS software12. Significantly regulated phosphorylation events were used to predict the kinases responsible for their catalysis using iGPS12. Positive kinase scores represent most confident and frequent predictions for upregulated phosphosites, with blue representing downregulated phosphosites. The higher the cumulative score retrieved from iGPS, the more intense the colour coding of the bubbles in the network. Upregulated and downregulated refers to numerator and denominator as defined in the header of each panel. The bubble size is scaled on the basis of the number of phosphorylation events predicted to be catalysed by the given kinase. The connector lines represent previously associated genetic interactions between listed proteins retrieved from the database STRING v.11 (ref. 76). The network was displayed using Cytoscape77.
Biochemical analyses
Blood glucose was measured using Ascensia Contour glucometer (Bayer). Serum insulin (ALPCO, 80-INSNS-E01), leptin (R&D Systems, DY49805), IGF-1 (R&D Systems, DY791), FFAs (FUJIFILM, NEFA-HR (2)), serum triglyceride (Sigma-Aldrich, T2449, F6428) and liver triglycerides (BioVision, K622) were evaluated as per the manufacturer’s instructions.
Seahorse XF Cell Mito Stress Test was performed as per the manufacturer’s instructions (Agilent Technologies). Briefly, 12,000 NIH3T3 cells per 200 μl of medium were transfected with DNAs and/or siRNAs and seeded onto a Seahorse XF96 Cell Culture Microplate (Agilent Technologies, 101085-004) for 32 h. After 16 h of stress in low-glucose medium (1 g l−1; Agilent Technologies, 103577), cells were washed with PBS, cultured in 165 μl of XF Base Medium (Agilent Technologies, 103335) supplemented with 1 g l−1 d-glucose, 2 mM sodium pyruvate (Gibco, 11360) and 4 mM l-glutamine (Gibco, 25030) and incubated at 37 °C without CO2 for 1 h. OA (0.25 mM) was added, and the microplate was loaded into XF Analyzer. Basal OCR measurements were recorded four times (mix 3 min, wait 2 min, measure 3 min) after sequential injections of oligomycin (1 μM), carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP; 20 μM) and rotenone/antimycin (1 μM) with four readings (mix 3 min, wait 2 min, measure 3 min) after each injection. OCR was normalized to cell number estimated with CyQUANT Cell Proliferation Assay (Invitrogen, C7026) as per the manufacturer’s instructions. Mitochondrial parameters were calculated as per the manufacturer’s instructions. OCRs of liver explants were performed as described66.
Histological analyses
Flag was detected using a Mouse on Mouse (MOM) ImmPRESS HRP (Peroxidase) Polymer Kit (Vector Laboratories, MP-2400). Paraffin-embedded livers were cut into 5 μm sections, deparaffinated in xylene and rehydrated in a series of graded alcohols and water. For antigen unmasking, sections were incubated in citrate-based antigen unmasking solution (pH 6.0; Vector Laboratories, H-3300) at high temperature for 20 min. After blocking in BLOXALL Endogenous Blocking Solution (Vector Laboratories, SP-6000) for 10 min and a 1 h incubation in MOM Mouse IgG Blocking Reagent, sections were stained with mouse monoclonal anti-DYKDDDDK Tag antibody (1:100; Cell Signaling Technology, 8146) in 2.5% normal horse serum MOM solution overnight at 4 °C. DYKDDDDK signal was revealed by incubation with MOM ImmPRESS Reagent for 10 min and enhanced with ImmPACT DAB EqV Peroxidase (HRP) Substrate (Vector laboratories, SK-4103) for 1 min. Sections were counterstained with haematoxylin, dehydrated, mounted with Permount mounting medium (Fisher, SP15) and imaged in a Zeiss Axiolab 5 microscope/Axiocam 305 colour camera (Carl Zeiss Microscopy). Quantification of Flag percentage area was performed as described78.
Oil Red O staining
Oil Red O staining was performed as described79.
Confocal microscopy
Was performed as described68. For Flag detection, DYKDDDDK Tag Rabbit antibody was used at 1:100 dilution (Cell Signaling Technology, 14793). Where indicated, 30 min before fixation with 4% paraformaldehyde, cells were incubated with 100 nM MitoTracker Red CMXRos (Invitrogen, M7512) to assess mitochondrial membrane potential. Mounted coverslips were imaged on a Leica TCS SP8 Confocal Laser Scanning Microscope (Leica Microsystems) with ×63 objective and 1.4 numerical aperture. Quantification of MitoTracker Red CMXRos fluorescence intensity per cell was performed using ImageJ (NIH) and expressed as mean integrated density. For detection of BODIPY FL C16 in vivo, sections from freshly isolated livers were mounted with Fluoromount-G medium (SouthernBiotech, 0100) and imaged on Leica TCS SP8 Confocal Laser Scanning Microscope with ×10 objective and 1.4 numerical aperture.
Live cell imaging
Cells were transfected with siRNA and/or DNA as above and seeded onto a glass-bottom 35 mm culture dish (MatTek Corporation, P35G-1.5-14-C) for 48 h. After PBS washing, cells were incubated in serum-free DMEM in presence of MitoTracker Green FM (500 nM; Invitrogen, M7514) or ER-Tracker Green (500 nM; Invitrogen, E34251) for 30 min to stain mitochondria and ER, respectively. Cells were washed once with PBS and incubated in red phenol-free DMEM (Gibco, 31053) with 4 mM l-glutamine and 12 mM HEPES (pH 7.4), and imaged using a Leica TCS SP8 confocal laser scanning microscope (Leica Microsystems) and single planes were acquired with ×63 objective and 1.4 numerical aperture. For time-lapse imaging, cells were tracked at a rate of one frame per 13 or 27 s (for single or simultaneous dual-channel acquisition, respectively) over 10 min.
Image analysis was done with ImageJ (NIH). Individual frames were denoised by applying Gaussian filter and a region of interest (ROI) of 35 μm2 was selected across the different experimental conditions. After image auto-thresholding, quantification of mitochondrial number and morphology parameters was performed using the ‘analyze particles’ macro as described80. Mitochondrial elongation was calculated as the inverse of circularity81. Mitochondrial fission and fusion frequency were calculated as described82 and expressed as number of events per cell per second. Percentage co-localization was calculated using the JACoP plugin as described68.
TEM
Freshly isolated livers were fixed with 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M sodium cacodylate, post-fixed with 2% osmium tetroxide, 1.5% potassium ferrocyanide, 0.15 M sodium cacodylate, 2 mM CaCl2, followed by 1% thiocarbohydrazide, and then 2% osmium tetroxide, en bloc stained with 1% uranyl acetate and further stained with lead aspartate. Samples were dehydrated through graded series of ethanol and embedded in LX112 resin (LADD Research Industries). Ultrathin (55 nm) sections were cut on a Leica ARTOS 3D ultramicrotome and collected onto silicon wafers. Sections were examined on Zeiss Supra 40 Field Emission Scanning Electron Microscope (Carl Zeiss Microscopy, LLC North America) in backscatter mode using an accelerating voltage of 8.0 kV. The number of mitochondria was counted manually in an ROI of 71.2 μm2. Quantification of mitochondrial shape descriptors was performed by manual tracing of individual mitochondria using freehand tool. Contact sites between mitochondria and ER (defined to be at 10–30 nm distance83) were quantified, normalized to total number of mitochondria and expressed as percentage. For 3D reconstruction, regions of interest were collected using ATLAS 5.0, with a pixel size of 6.0 and dwell time of 6 µs. Stacks were aligned, and segmentation was done using IMOD84. Tomographic reconstruction was performed as described85.
Lipidomic analyses
Lipid extracts from liver homogenates, MAM, pure mitochondria and ER fractions were prepared using modified Bligh and Dyer method, spiked with appropriate internal standards, and analysed on an Agilent 1260 Infinity HPLC integrated to Agilent 6490 A QQQ mass spectrometer controlled by Masshunter v 7.0 (Agilent Technologies). Glycerophospholipids and sphingolipids were separated with normal-phase HPLC as described86, with a few modifications. An Agilent Zorbax Rx-Sil column (2.1 × 100 mm, 1.8 µm) at 25 °C was used under the following conditions: mobile phase A (chloroform:methanol:ammonium hydroxide, 89.9:10:0.1, v/v) and mobile phase B (chloroform:methanol:water:ammonium hydroxide, 55:39:5.9:0.1, v/v); 95% A for 2 min, decreased linearly to 30% A over 18 min and further decreased to 25% A over 3 min, before returning to 95% over 2 min and held for 6 min. Separation of sterols and glycerolipids was carried out on a reverse phase Agilent Zorbax Eclipse XDB-C18 column (4.6 × 100 mm, 3.5 µm) using an isocratic mobile phase, chloroform:methanol:0.1 M ammonium acetate (25:25:1) at a flow rate of 300 μl min−1.
Quantification of lipid species was accomplished using multiple reaction monitoring transitions86,87 under positive and negative ionization modes and using internal standards: phosphatidic acid (PA) 14:0/14:0, phosphatidylcholine (PC) 14:0/14:0, phosphatidylethanolamine (PE) 14:0/14:0, phosphatidylgylcerol (PG) 15:0/15:0, phosphatidylinositol (PI) 17:0/20:4, phosphatidylserine (PS) 14:0/14:0, bis[monoacylglycero]phosphate (BMP) 14:0/14:0, acylphosphatidyl glycerol (APG) 14:0/14:0, lysophosphatidylcholine (LPC) 17:0, lysophosphatidylethanolamine (LPE) 14:0, lysophosphatidylinositol (LPI) 13:0, ceramide (Cer) d18:1/17:0, sphingomyelin (SM) d18:1/12:0, dihydrosphingomyelin (dhSM) d18:0/12:0, galactosylceramide (GalCer) d18:1/12:0, glucosylceramide (GluCer) d18:1/12:0, lactosylceramide (LacCer) d18:1/12:0, D7-cholesterol, cholesterol ester (CE) 17:0, monoglyceride (MG) 17:0, 4ME 16:0 diether DG, D5-TG 16:0/18:0/16:0 (Avanti Polar Lipids). Lipids per sample were calculated by summing total moles of all lipid species measured by all three LC–MS methodologies, and normalizing to mol % (Supplementary Table 3).
Data collection and analysis softwares
The following devices and softwares were used: (1) Zeiss Axiolab 5 microscope with Axiocam 305 colour camera for immunohistochemistry (Zeiss ZEN v3.7), (2) Leica TCS SP8 confocal laser scanning microscope (LAS X v.5.7.23225), (3) Zeiss Supra 40 Field Emission Scanning Electron Microscope to acquire transmission electron microscope (Zeiss SmartSEM v6.0) and 3D Modeling (3DMOD v4.9.10), (4) XF96 and X24 Seahorse analyzers (Agilent Technologies) to collect OCRs (WAVE Pro v10.0.1.84; v2.6.1.56, respectively), (5) Synergy HTX (BioTek) multi-mode plate reader (Gen5 v3.12), (6) StepOnePlus Real-Time PCR System (Applied Biosystems) for mRNA expression (StepOne v2.3), (7) Microsoft Excel v16.48, Microsoft Word v16.48, Microsoft PowerPoint v16.47, (8) Prism v8.4.3, (9) Endnote X9.3.3 and (10) ImageJ v2.0.0-rc-69/1.52p. Enrichment map was generated in Cytoscape v3.8.1, Enrichment map plugin v3.3.0. Handling and analyses of proteomics data were performed via Python v.3.7.4 and Scientific Python Stack: SciPy v.1.3.1, NumPy v.1.17.2 and Matplotlib v.3.1.1. Phosphosites showing significant regulation between groups were used to predict the kinase responsible for their catalysis using the iGPS software.
Illustration
The proposed model in Fig. 7g was created with BioRender (BioRender.com).
Statistics
All data are mean of a minimum of three independent experiments unless otherwise stated. Statistical significance was assessed by two-tailed unpaired Student’s t-test, one-way or two-way analyses of variance (ANOVAs) followed by Tukey’s, Šídák’s or Dunnett’s multiple-comparisons test. n numbers indicate biological replicates. Statistical summary is presented in Supplementary Table 10. Raw source data are presented in Source Data Extended Data Table 1.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41556-023-01163-3.
Supplementary information
Supplementary Table 1. Phosphoproteomics in the five groups, that is, livers of basal fed; overnight fasted; or fasted overnight and gavaged with corn oil or BODIPY FL C16 or refed a high-fat diet. Supplementary Table 2. Phosphoproteomics identifying phosphoproteins that are coordinately modulated in the five groups, that is, livers of basal fed; overnight fasted; or fasted overnight and gavaged with corn oil or BODIPY FL C16 or refed a high-fat diet. Supplementary Table 3. Lipidomics from fractions in Con or RictorKO livers (mol %). Supplementary Table 4. Phosphoproteomics showing changes in protein phosphorylation in RictorKO livers compared with control livers. Supplementary Table 5. Enrichment map analyses for phosphoproteomics in RictorKO livers compared with control livers. Supplementary Table 6. Phosphoproteomics identifying reduced NDRG1Ser336 phosphorylation in RictorKO MAMs. Supplementary Table 7. Phosphoproteomics identifying reduced NDRG1Ser336 phosphorylation in siRictor cells. Supplementary Table 8. Proteomics identifying binding partners of NDRG1WT that show altered binding to NDRG1Ser336Ala. Supplementary Table 9. Proteomics identifying CDC42 binding partners. Supplementary Table 10. Statistical summary. Supplementary Table 11. Corn oil composition. Supplementary Table 12. siRNAs. Supplementary Table 13. qPCR primers. Supplementary Table 14. Antibodies.
Movie of Con Liver 3D (refers to Fig. 2j).
Movie of RictorKO Liver 3D (refers to Fig. 2j).
Video of siControl + MitoTracker (refers to Fig. 3d).
Video of siRictor + MitoTracker (refers to Fig. 3d).
Video of siDnm1l + MitoTracker (refers to Fig. 3d).
Video of NDRG1WT + MitoTracker (refers to Fig. 5a).
Video of NDRG1WT + ER-Tracker (refers to Extended Data Fig. 8f).
Video of NDRG1Ser336Ala + ER-Tracker (refers to Extended Data Fig. 8f).
Video of NDRG1Ser336Ala + MitoTracker (refers to Fig. 5a).
Video of siRictor - NDRG1WT + MitoTracker (refers to Fig. 5a).
Video of siDnm1l - NDRG1WT + MitoTracker (refers to Fig. 5a).
Video of siCon - NDRG1WT + MitoTracker (refers to Fig. 6c).
Video of siCdc42 - NDRG1WT + MitoTracker (refers to Fig. 6c).
Video of Cdc42WT + MitoTracker (refers to Extended Data Fig. 9e).
Video of Cdc42Thr17Asn + MitoTracker (refers to Extended Data Fig. 9e).
Acknowledgements
This work was supported by RF1AG043517, R01DK123327, R01AG065985 and P01AG031782 (R.S.); NIH P30 CA013330 47 (S.S. and J.T.A.); R56AG072794 and R56AG062271 (L.B.J.M); P30CA013330 and SIG #1S10OD016214-01A1 (Einstein Analytical Imaging Facility). This study was supported by the Biomarkers Core Laboratory at the Irving Institute for Clinical and Translational Research, home to Columbia University’s Clinical and Translational Science Award. We thank K. Kulej for assistance with phosphoproteomic analyses. We thank S. Kaushik for suggestions for time-lapse imaging.
Extended data
Source data
Unprocessed western blots for Fig. 1.
Unprocessed western blots for Fig. 2.
Unprocessed western blots for Fig. 3.
Unprocessed western blots for Fig. 4.
Unprocessed western blots for Fig. 6.
Unprocessed western blots for Fig. 7.
Unprocessed western blots for Extended Data Fig. 1.
Unprocessed western blots for Extended Data Fig. 2.
Unprocessed western blots for Extended Data Fig. 3.
Unprocessed western blots for Extended Data Fig. 4.
Unprocessed western blots for Extended Data Fig. 5.
Unprocessed western blots for Extended Data Fig. 6.
Unprocessed western blots for Extended Data Fig. 7.
Unprocessed western blots for Extended Data Fig. 8.
Unprocessed western blots for Extended Data Fig. 9.
Unprocessed western blots for Extended Data Fig. 10.
Numerical source data.
Author contributions
Conceptualization, R.S. and N.M.-L.; methodology, R.S. and N.M.-L.; investigation, N.M.-L., P.M., M.T., H.B., M.K., M.L.A. and M.S.; writing—original draft, R.S.; revised draft, R.S. and N.M.-L.; data analyses, R.S., N.M.-L., S.S. and M.B.; funding acquisition, R.S.; resources, L.G.-C., F.P.M., L.B.J.M, J.T.A. and S.S.; supervision, R.S.
Peer review
Peer review information
Nature Cell Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE88 partner repository, and data are available via ProteomeXchange with identifier PXD041696. Data from this study are available at 10.6084/m9.figshare.22670575. Source data have been provided with this paper. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code availability
All the Python codes used in proteomic analyses are fully available on GitHub (https://github.com/MathieuBo/mTorc2_mito_fission).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
is available for this paper at 10.1038/s41556-023-01163-3.
Supplementary information
The online version contains supplementary material available at 10.1038/s41556-023-01163-3.
References
- 1.Sharma A, Smith HJ, Yao P, Mair WB. Causal roles of mitochondrial dynamics in longevity and healthy aging. EMBO Rep. 2019;20:e48395. doi: 10.15252/embr.201948395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barazzuol L, Giamogante F, Cali T. Mitochondria associated membranes (MAMs): architecture and physiopathological role. Cell Calcium. 2021;94:102343. doi: 10.1016/j.ceca.2020.102343. [DOI] [PubMed] [Google Scholar]
- 3.Loson OC, Song Z, Chen H, Chan DC. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell. 2013;24:659–667. doi: 10.1091/mbc.e12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otera H, et al. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 2010;191:1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ge, Y. et al. Two forms of Opa1 cooperate to complete fusion of the mitochondrial inner-membrane. eLife10.7554/eLife.50973 (2020). [DOI] [PMC free article] [PubMed]
- 6.Chen H, et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toyama EQ, et al. Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science. 2016;351:275–281. doi: 10.1126/science.aab4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mason JA, et al. SGK1 signaling promotes glucose metabolism and survival in extracellular matrix detached cells. Cell Rep. 2021;34:108821. doi: 10.1016/j.celrep.2021.108821. [DOI] [PubMed] [Google Scholar]
- 9.Drechsel DN, Hyman AA, Hall A, Glotzer M. A requirement for Rho and Cdc42 during cytokinesis in Xenopus embryos. Curr. Biol. 1997;7:12–23. doi: 10.1016/S0960-9822(06)00023-6. [DOI] [PubMed] [Google Scholar]
- 10.Johnson DI. Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol Mol. Biol. Rev. 1999;63:54–105. doi: 10.1128/MMBR.63.1.54-105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cahill GF., Jr. Fuel metabolism in starvation. Annu. Rev. Nutr. 2006;26:1–22. doi: 10.1146/annurev.nutr.26.061505.111258. [DOI] [PubMed] [Google Scholar]
- 12.Song C, et al. Systematic analysis of protein phosphorylation networks from phosphoproteomic data. Mol. Cell Proteom. 2012;11:1070–1083. doi: 10.1074/mcp.M111.012625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sridharan, S. & Basu, A. Distinct roles of mTOR targets S6K1 and S6K2 in breast cancer. Int. J. Mol. Sci.10.3390/ijms21041199 (2020). [DOI] [PMC free article] [PubMed]
- 14.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 15.Basu A, Sridharan S, Persaud S. Regulation of protein kinase C delta downregulation by protein kinase C epsilon and mammalian target of rapamycin complex 2. Cell Signal. 2009;21:1680–1685. doi: 10.1016/j.cellsig.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baffi, T. R. et al. mTORC2 controls the activity of PKC and Akt by phosphorylating a conserved TOR interaction motif. Sci. Signal10.1126/scisignal.abe4509 (2021). [DOI] [PMC free article] [PubMed]
- 17.Garcia-Martinez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) Biochem. J. 2008;416:375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- 18.Egan DF, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geisler CE, Hepler C, Higgins MR, Renquist BJ. Hepatic adaptations to maintain metabolic homeostasis in response to fasting and refeeding in mice. Nutr. Metab. 2016;13:62. doi: 10.1186/s12986-016-0122-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 21.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarbassov DD, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 23.Kwiatkowski DJ, et al. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum. Mol. Genet. 2002;11:525–534. doi: 10.1093/hmg/11.5.525. [DOI] [PubMed] [Google Scholar]
- 24.Quinn WJ, 3rd, et al. mTORC1 stimulates phosphatidylcholine synthesis to promote triglyceride secretion. J. Clin. Invest. 2017;127:4207–4215. doi: 10.1172/JCI96036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagiwara A, et al. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab. 2012;15:725–738. doi: 10.1016/j.cmet.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 26.Takeichi Y, et al. Non-alcoholic fatty liver disease in mice with hepatocyte-specific deletion of mitochondrial fission factor. Diabetologia. 2021;64:2092–2107. doi: 10.1007/s00125-021-05488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, et al. Disruption of mitochondrial fission in the liver protects mice from diet-induced obesity and metabolic deterioration. Diabetologia. 2015;58:2371–2380. doi: 10.1007/s00125-015-3704-7. [DOI] [PubMed] [Google Scholar]
- 28.Betz C, et al. Feature Article: mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc. Natl Acad. Sci. USA. 2013;110:12526–12534. doi: 10.1073/pnas.1302455110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frezza C, et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 30.Morita M, et al. mTOR controls mitochondrial dynamics and cell survival via MTFP1. Mol. Cell. 2017;67:922–935 e925. doi: 10.1016/j.molcel.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 31.Chang CR, Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J. Biol. Chem. 2007;282:21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- 32.Cameron AJ, Linch MD, Saurin AT, Escribano C, Parker PJ. mTORC2 targets AGC kinases through Sin1-dependent recruitment. Biochem. J. 2011;439:287–297. doi: 10.1042/BJ20110678. [DOI] [PubMed] [Google Scholar]
- 33.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS ONE. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, et al. Quantitative phosphoproteomic analysis of the molecular substrates of sleep need. Nature. 2018;558:435–439. doi: 10.1038/s41586-018-0218-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park KC, et al. Identification of differential phosphorylation and sub-cellular localization of the metastasis suppressor, NDRG1. Biochim. Biophys. Acta, Mol. Basis Dis. 2018;1864:2644–2663. doi: 10.1016/j.bbadis.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 36.Sevinsky CJ, et al. NDRG1 regulates neutral lipid metabolism in breast cancer cells. Breast Cancer Res. 2018;20:55. doi: 10.1186/s13058-018-0980-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fonseca TB, Sanchez-Guerrero A, Milosevic I, Raimundo N. Mitochondrial fission requires DRP1 but not dynamins. Nature. 2019;570:E34–E42. doi: 10.1038/s41586-019-1296-y. [DOI] [PubMed] [Google Scholar]
- 38.Kamerkar SC, Kraus F, Sharpe AJ, Pucadyil TJ, Ryan MT. Dynamin-related protein 1 has membrane constricting and severing abilities sufficient for mitochondrial and peroxisomal fission. Nat. Commun. 2018;9:5239. doi: 10.1038/s41467-018-07543-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mears JA, et al. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat. Struct. Mol. Biol. 2011;18:20–26. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erickson JW, Cerione RA, Hart MJ. Identification of an actin cytoskeletal complex that includes IQGAP and the Cdc42 GTPase. J. Biol. Chem. 1997;272:24443–24447. doi: 10.1074/jbc.272.39.24443. [DOI] [PubMed] [Google Scholar]
- 41.Muller PM, et al. Systems analysis of RhoGEF and RhoGAP regulatory proteins reveals spatially organized RAC1 signalling from integrin adhesions. Nat. Cell Biol. 2020;22:498–511. doi: 10.1038/s41556-020-0488-x. [DOI] [PubMed] [Google Scholar]
- 42.Choi S, Anderson RA. IQGAP1 is a phosphoinositide effector and kinase scaffold. Adv. Biol. Regul. 2016;60:29–35. doi: 10.1016/j.jbior.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dovas A, Couchman JR. RhoGDI: multiple functions in the regulation of Rho family GTPase activities. Biochem. J. 2005;390:1–9. doi: 10.1042/BJ20050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho, H. J., Kim, J. T., Baek, K. E., Kim, B. Y. & Lee, H. G. Regulation of Rho GTPases by RhoGDIs in human cancers. Cells10.3390/cells8091037 (2019). [DOI] [PMC free article] [PubMed]
- 45.Ho YD, Joyal JL, Li Z, Sacks DB. IQGAP1 integrates Ca2+/calmodulin and Cdc42 signaling. J. Biol. Chem. 1999;274:464–470. doi: 10.1074/jbc.274.1.464. [DOI] [PubMed] [Google Scholar]
- 46.Fukata M, et al. Regulation of cross-linking of actin filament by IQGAP1, a target for Cdc42. J. Biol. Chem. 1997;272:29579–29583. doi: 10.1074/jbc.272.47.29579. [DOI] [PubMed] [Google Scholar]
- 47.Atkins BD, et al. Inhibition of Cdc42 during mitotic exit is required for cytokinesis. J. Cell Biol. 2013;202:231–240. doi: 10.1083/jcb.201301090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Onishi M, Ko N, Nishihama R, Pringle JR. Distinct roles of Rho1, Cdc42, and Cyk3 in septum formation and abscission during yeast cytokinesis. J. Cell Biol. 2013;202:311–329. doi: 10.1083/jcb.201302001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson JL, Erickson JW, Cerione RA. C-terminal di-arginine motif of Cdc42 protein is essential for binding to phosphatidylinositol 4,5-bisphosphate-containing membranes and inducing cellular transformation. J. Biol. Chem. 2012;287:5764–5774. doi: 10.1074/jbc.M111.336487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hong L, et al. Characterization of a Cdc42 protein inhibitor and its use as a molecular probe. J. Biol. Chem. 2013;288:8531–8543. doi: 10.1074/jbc.M112.435941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee JE, Westrate LM, Wu H, Page C, Voeltz GK. Multiple dynamin family members collaborate to drive mitochondrial division. Nature. 2016;540:139–143. doi: 10.1038/nature20555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hatch AL, Gurel PS, Higgs HN. Novel roles for actin in mitochondrial fission. J. Cell Sci. 2014;127:4549–4560. doi: 10.1242/jcs.153791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moore AS, Wong YC, Simpson CL, Holzbaur EL. Dynamic actin cycling through mitochondrial subpopulations locally regulates the fission-fusion balance within mitochondrial networks. Nat. Commun. 2016;7:12886. doi: 10.1038/ncomms12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl Acad. Sci. USA. 2011;108:10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Castro-Sepulveda M, et al. The fasting-feeding metabolic transition regulates mitochondrial dynamics. FASEB J. 2021;35:e21891. doi: 10.1096/fj.202100929R. [DOI] [PubMed] [Google Scholar]
- 57.Sood A, et al. A Mitofusin-2-dependent inactivating cleavage of Opa1 links changes in mitochondria cristae and ER contacts in the postprandial liver. Proc. Natl Acad. Sci. USA. 2014;111:16017–16022. doi: 10.1073/pnas.1408061111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Castellano BM, et al. Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science. 2017;355:1306–1311. doi: 10.1126/science.aag1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Menon D, et al. Lipid sensing by mTOR complexes via de novo synthesis of phosphatidic acid. J. Biol. Chem. 2017;292:6303–6311. doi: 10.1074/jbc.M116.772988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu L, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lizana L, Bauer B, Orwar O. Controlling the rates of biochemical reactions and signaling networks by shape and volume changes. Proc. Natl Acad. Sci. USA. 2008;105:4099–4104. doi: 10.1073/pnas.0709932105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 2009;23:496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lamming DW, et al. Depletion of Rictor, an essential protein component of mTORC2, decreases male lifespan. Aging Cell. 2014;13:911–917. doi: 10.1111/acel.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hammerschmidt P, et al. CerS6-derived sphingolipids interact with Mff and promote mitochondrial fragmentation in obesity. Cell. 2019;177:1536–1552 e1523. doi: 10.1016/j.cell.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 65.Durkin, M. E., Qian, X., Popescu, N. C. & Lowy, D. R. Isolation of mouse embryo fibroblasts. Bio Protoc.10.21769/bioprotoc.908 (2013). [DOI] [PMC free article] [PubMed]
- 66.Martinez-Lopez N, et al. Autophagy in the CNS and periphery coordinate lipophagy and lipolysis in the brown adipose tissue and liver. Cell Metab. 2016;23:113–127. doi: 10.1016/j.cmet.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Milan G, et al. Regulation of autophagy and the ubiquitin-proteasome system by the FoxO transcriptional network during muscle atrophy. Nat. Commun. 2015;6:6670. doi: 10.1038/ncomms7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martinez-Lopez N, Athonvarangkul D, Mishall P, Sahu S, Singh R. Autophagy proteins regulate ERK phosphorylation. Nat. Commun. 2013;4:2799. doi: 10.1038/ncomms3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wieckowski MR, Giorgi C, Lebiedzinska M, Duszynski J, Pinton P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat. Protoc. 2009;4:1582–1590. doi: 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]
- 70.Kulej K, et al. Time-resolved global and chromatin proteomics during herpes simplex virus type 1 (HSV-1) infection. Mol. Cell Proteom. 2017;16:S92–S107. doi: 10.1074/mcp.M116.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aguilan JT, Kulej K, Sidoli S. Guide for protein fold change and p-value calculation for non-experts in proteomics. Mol. Omics. 2020;16:573–582. doi: 10.1039/D0MO00087F. [DOI] [PubMed] [Google Scholar]
- 72.Pascovici D, Handler DC, Wu JX, Haynes PA. Multiple testing corrections in quantitative proteomics: a useful but blunt tool. Proteomics. 2016;16:2448–2453. doi: 10.1002/pmic.201600044. [DOI] [PubMed] [Google Scholar]
- 73.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Virtanen P, et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harris CR, et al. Array programming with NumPy. Nature. 2020;585:357–362. doi: 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Szklarczyk D, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Crowe, A. R. & Yue, W. Semi-quantitative determination of protein expression using immunohistochemistry staining and analysis: an integrated protocol. Bio Protoc.10.21769/BioProtoc.3465 (2019). [DOI] [PMC free article] [PubMed]
- 79.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marchi S, Bonora M, Patergnani S, Giorgi C, Pinton P. Methods to assess mitochondrial morphology in mammalian cells mounting autophagic or mitophagic responses. Methods Enzymol. 2017;588:171–186. doi: 10.1016/bs.mie.2016.09.080. [DOI] [PubMed] [Google Scholar]
- 81.Wiemerslage L, Lee D. Quantification of mitochondrial morphology in neurites of dopaminergic neurons using multiple parameters. J. Neurosci. Methods. 2016;262:56–65. doi: 10.1016/j.jneumeth.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chacko LA, Ananthanarayanan V. Quantification of mitochondrial dynamics in fission yeast. Bio Protoc. 2019;9:e3450. doi: 10.21769/BioProtoc.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Friedman JR, et al. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 85.Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- 86.Chan RB, et al. Comparative lipidomic analysis of mouse and human brain with Alzheimer disease. J. Biol. Chem. 2012;287:2678–2688. doi: 10.1074/jbc.M111.274142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gulati S, et al. Profiling the essential nature of lipid metabolism in asexual blood and gametocyte stages of Plasmodium falciparum. Cell Host Microbe. 2015;18:371–381. doi: 10.1016/j.chom.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Perez-Riverol Y, et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50:D543–D552. doi: 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Phosphoproteomics in the five groups, that is, livers of basal fed; overnight fasted; or fasted overnight and gavaged with corn oil or BODIPY FL C16 or refed a high-fat diet. Supplementary Table 2. Phosphoproteomics identifying phosphoproteins that are coordinately modulated in the five groups, that is, livers of basal fed; overnight fasted; or fasted overnight and gavaged with corn oil or BODIPY FL C16 or refed a high-fat diet. Supplementary Table 3. Lipidomics from fractions in Con or RictorKO livers (mol %). Supplementary Table 4. Phosphoproteomics showing changes in protein phosphorylation in RictorKO livers compared with control livers. Supplementary Table 5. Enrichment map analyses for phosphoproteomics in RictorKO livers compared with control livers. Supplementary Table 6. Phosphoproteomics identifying reduced NDRG1Ser336 phosphorylation in RictorKO MAMs. Supplementary Table 7. Phosphoproteomics identifying reduced NDRG1Ser336 phosphorylation in siRictor cells. Supplementary Table 8. Proteomics identifying binding partners of NDRG1WT that show altered binding to NDRG1Ser336Ala. Supplementary Table 9. Proteomics identifying CDC42 binding partners. Supplementary Table 10. Statistical summary. Supplementary Table 11. Corn oil composition. Supplementary Table 12. siRNAs. Supplementary Table 13. qPCR primers. Supplementary Table 14. Antibodies.
Movie of Con Liver 3D (refers to Fig. 2j).
Movie of RictorKO Liver 3D (refers to Fig. 2j).
Video of siControl + MitoTracker (refers to Fig. 3d).
Video of siRictor + MitoTracker (refers to Fig. 3d).
Video of siDnm1l + MitoTracker (refers to Fig. 3d).
Video of NDRG1WT + MitoTracker (refers to Fig. 5a).
Video of NDRG1WT + ER-Tracker (refers to Extended Data Fig. 8f).
Video of NDRG1Ser336Ala + ER-Tracker (refers to Extended Data Fig. 8f).
Video of NDRG1Ser336Ala + MitoTracker (refers to Fig. 5a).
Video of siRictor - NDRG1WT + MitoTracker (refers to Fig. 5a).
Video of siDnm1l - NDRG1WT + MitoTracker (refers to Fig. 5a).
Video of siCon - NDRG1WT + MitoTracker (refers to Fig. 6c).
Video of siCdc42 - NDRG1WT + MitoTracker (refers to Fig. 6c).
Video of Cdc42WT + MitoTracker (refers to Extended Data Fig. 9e).
Video of Cdc42Thr17Asn + MitoTracker (refers to Extended Data Fig. 9e).
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE88 partner repository, and data are available via ProteomeXchange with identifier PXD041696. Data from this study are available at 10.6084/m9.figshare.22670575. Source data have been provided with this paper. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
All the Python codes used in proteomic analyses are fully available on GitHub (https://github.com/MathieuBo/mTorc2_mito_fission).