Summary
Alveolar macrophages (AMs) are resident innate immune cells that play vital roles in maintaining lung physiological functions. However, the effects of aging on their dynamics, heterogeneity, and transcriptional profiles remain to be fully elucidated. Through single cell RNA sequencing (scRNA-seq), we identified CBFβ as an indispensable transcription factor that ensures AM self-renewal. Intriguingly, despite transcriptome similarities of proliferating cells, AMs from aged mice exhibited reduced embryonic stem cell–like features. Aged AMs also displayed compromised DNA repair abilities, potentially leading to obstructed cell cycle progression and an elevation of senescence markers. Consistently, AMs from aged mice exhibited impaired self-renewal ability and reduced sensitivity to GM-CSF. Decreased CBFβ was observed in the cytosol of AMs from aged mice. Similar senescence-like phenotypes were also found in human AMs. Taken together, these findings suggest that AMs in aged hosts demonstrate senescence-like phenotypes, potentially facilitated by the abrogated CBF activity.
Subject areas: Bioinformatics, Omics, Transcriptomics
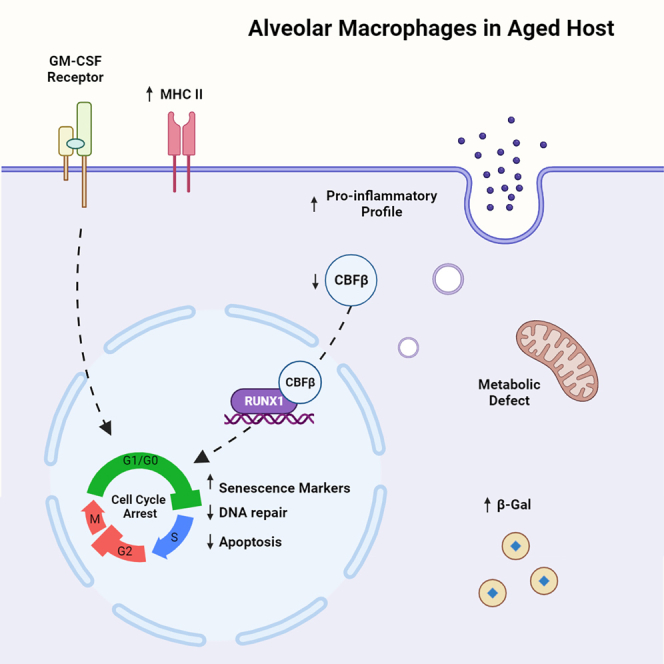
Graphical abstract
Highlights
-
•
scRNA-seq revealed alveolar macrophage (AM) heterogeneity and self-renewal
-
•
CBFβ is associated with AM cell cycle and facilitates AM self-renewal
-
•
AMs displayed senescence-like phenotypes during physiological aging
-
•
Disrupted CBFβ expression may affect senescence-like phenotypes in aged AMs
Bioinformatics; Omics; Transcriptomics
Introduction
Alveolar macrophages (AMs) are the major immune cells in the alveoli during steady state. AMs function as the first line of defense against microbial infections.1,2 Meanwhile, they play important roles in maintaining tissue homeostasis by regulating other immune cells, participating in surfactant turnover as well as phagocytosis of apoptotic cells and debris.1,2 Thus, it is important to understand the mechanism underlying the maintenance of the AM compartment and the regulation of its function. Both human and murine AMs are derived from fetal precursors.3,4 On maturation in a steady state, they remain embryonic stem cell (ESC)–like features,5 which enable the self-maintenance of an AM pool without the need for input from adult hematopoietic stem cells.6 This notion is supported by the persistence of donor AMs in lung transplant recipients 3.5 years after surgery.7 To this end, AMs depend on GM-CSF and autocrine TGF-β signaling for their maturation, proliferation, and maintenance.3,8,9 Transcription factors including PPAR-γ,10 BHLHE40 and BHLHE41,11 Bach2,12 Hem-113 as well as EGR214 have been reported to be indispensable for the generation and/or maintenance of the AM compartment. Furthermore, mitochondrial-dependent oxidative metabolism is also vital for AM self-renewal and maintenance.15,16 However, the exact molecule program regulating AM cell-cycle progression, proliferation, and self-renewal remains largely undefined.
Cellular senescence describes a cellular state marked by irreversible cell-cycle arrest in response to internal and external stress.17 It is characterized by unresolved DNA damage, the inability of cell-cycle progression and, meanwhile, the resistance against apoptosis.18 Cells that undergo senescence express an increased level of factors involved in cell-cycle arrest, most notably p16 (encoded by Cdkn2a), and a secretome termed senescence-associated secretory phenotype (SASP), resembled by the expression of innate effector molecules at baseline.18 Alterations in cellular metabolism and the epigenetic landscape are also important components in cellular senescence.17,18 Physiological aging leads to increased features of senescence in the lung,19 accompanied by accumulation of pro-inflammatory cytokines, complement components, and molecules indicative of oxidative stress in the alveolar lining fluid.20 It has been shown that the senescent immune cells, particularly T cells, can independently drive tissue damage and organ aging in non-lymphoid tissues including lung tissue.19 However, it is not known whether tissue-resident macrophages exhibit senescent-like phenotypes during aging. To this end, previous research has described a dysfunctional phenotype in AMs derived from aged hosts.21,22 Nevertheless, whether AMs undergo cellular senescence during aging remains to be explored and, if so, what the exact cellular and molecular mechanisms underlying such phenomenon would be.
The core binding factor β subunit (CBFβ) is a non-DNA-binding transcription factor that requires its DNA-binding partner, RUNX1, RUNX2, or RUNX3, to facilitate its function.23 Core binding factors play an essential role in hematopoiesis during an embryonic stage where their deficiency can lead to defects in the generation of hematopoietic stem cells in fetal liver.24,25 Besides the establishment of definitive hematopoiesis, CBFβ is also important for the maturation of cells in the myeloid lineage from adult hematopoietic stem cells.26 For instance, deficiency of a CBFβ-RUNX1-binding enhancer of IRF8 skewed the fate of myeloid progenitor cells toward differentiation into monocytes.27 In the respiratory tract, myeloid deficiency of CBFβ caused defects in the AM compartment despite normal development of monocytes, neutrophils, DCs, and interstitial macrophages (IMs).28 However, the underlying mechanisms by which CBFβ regulates AM homeostasis remains to be explored.
Here, we generated a single cell RNA sequencing (scRNA-seq) dataset that contains a large number of AMs from young (12,358 cells) or aged mice (5,269 cells). The abundant cell number facilitated the study of AM heterogeneity with detailed resolution. Identification of actively proliferating AM populations enabled the insight into mechanisms governing the process of AM self-renewal. We observed that AMs from the aged mice exhibited reduced proliferation and self-renewal capacity. Moreover, they displayed features of cellular senescence including cell-cycle arrest, Cdkn2a (p16) upregulation, insensitivity to growth factor stimulation, and SASP. With the premise of scRNA-seq, a transcription factor, Cbfb, was found to be preferentially associated with proliferating AMs. Moreover, it was validated in vitro and in vivo that CBFβ was indispensable in AM proliferation. Importantly, protein level of CBFβ decreased in AMs from the aged hosts when compared to that from young hosts, indicating its potential role in the senescence-like phenotypes. Such hypothesis was supported by enhanced Cdkn2a (p16) expression in CBFβ-deficient AMs. Of interest, similar senescence-like phenotypes were also found in AMs from humans. Taken together, our data suggests that AMs in aged hosts possess senescence-like phenotypes, which could be partially attributed to the reduced expression of CBFβ in AMs during aging.
Results
scRNA-seq reveals the functional heterogeneity of AMs in young hosts
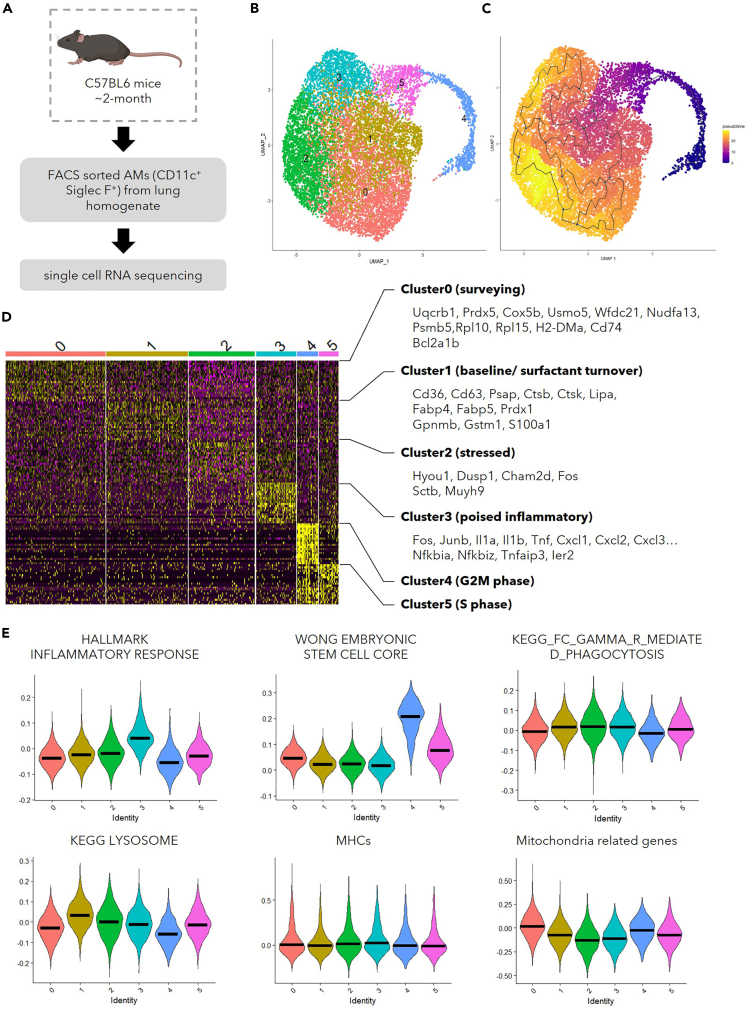
AMs have been previously characterized with technologies that possess single cell resolution.22,29 However, the granularity of AMs in these data was restrained by the limited number of cells presented. To gain further insights into the functional heterogeneity within the AM population, a large number of sorted CD11c+Siglec-F+ cells from the lung homogenate of C57BL/6 mice at the age of 2 months were put through scRNA-seq (Figure 1A). Compared to other myeloid cells in lung, AMs possess unique molecular features.30 Such knowledge was utilized to exclude the contaminating myeloid cells in our library. Cells in cluster 8 were in lack of AM characteristics, and they were enriched in molecule profile of other mononuclear phagocytes (Figure S1A). We thus included only the cells that were likely to be AMs (cluster 1–7) for further analysis. With K-nearest neighbor (KNN) graph, 6 different clusters were discovered with Louvain algorithm, a modularity optimization technique, in Seurat31 from the 12,358 AMs that passed quality control (Figure 1B). As a proof of concept, we performed trajectory analysis using Monocle332 (Figure 1C). Consistent with previous reports that AMs could maintain themselves through self-renewal during homeostasis,6 AMs from the young mice have been inferred in the same branch (Figure 1C). In addition, the top 50 featured genes in each cluster (Figure 1D, Table S1) and the enrichment scores of relevant pathways (Figures 1E and S1B) were utilized to obtain knowledge of functional heterogeneity within these AMs. AMs in cluster 0 were characterized by the expression of genes involved in antigen presentation and oxidative phosphorylation (OXPHOS) (Figures 1D and S1B). Elevated expression of scavenger receptor genes like Cd36 was found in cluster 1, which had the highest score in the lysosome-related pathway (Figures 1D and 1E). Such transcriptome suggested that these AMs may be preferentially responsible for surfactant turn-over, which is further supported by the enhanced expression of genes related to lipid metabolism and antioxidants (Figure 1D). Cells in cluster 2 possessed heightened expression of genes involved in intracellular or environmental stress responses (Figure 1D). Cluster 3 appeared to consist of cells poised for inflammation, for they were enriched with expression of genes encoding pro-inflammatory signaling pathways, transcription factors, cytokines, and chemokines (Figures 1D and S1B). AMs have been reported to possess embryonic stem cell (ESC) like features.5 Such a profile33 was enriched in cluster 4 and cluster 5, the proliferating AM clusters. Together, these data demonstrate that the AM compartment is functionally heterogeneous during homeostasis.
Figure 1.
scRNA-seq reveals functional heterogeneity in AMs
(A) Schematic display the library preparation for scRNA-seq. AMs (CD11c+ Siglec-F+) were FACS-sorted from lung homogenate combined from young female mice (∼2-month-old, n = 3).
(B) UMAP displaying the clustering of 12,358 AMs that passed quality control.
(C) Visualization of the trajectory analysis with UMAP.
(D) Heatmap demonstrating top 50 genes featured in each cluster with selected genes shown on the right.
(E) Violin plots showing the module scores evaluating gene lists of inflammatory response (MSigDB M5932), ESC-like features (MSigDB M7079), phagocytosis (MSigDB M16121), lysosome related genes (MSigDB M11266), MHC molecules, and mitochondria-related genes.36
The bar in (E) represents the median of module scores generated with gene lists indicated among the cells in each cluster as shown in (B). See also Figures S1A and S1B.
scRNA-seq identifies CBFβ as a critical regulator of AM proliferation
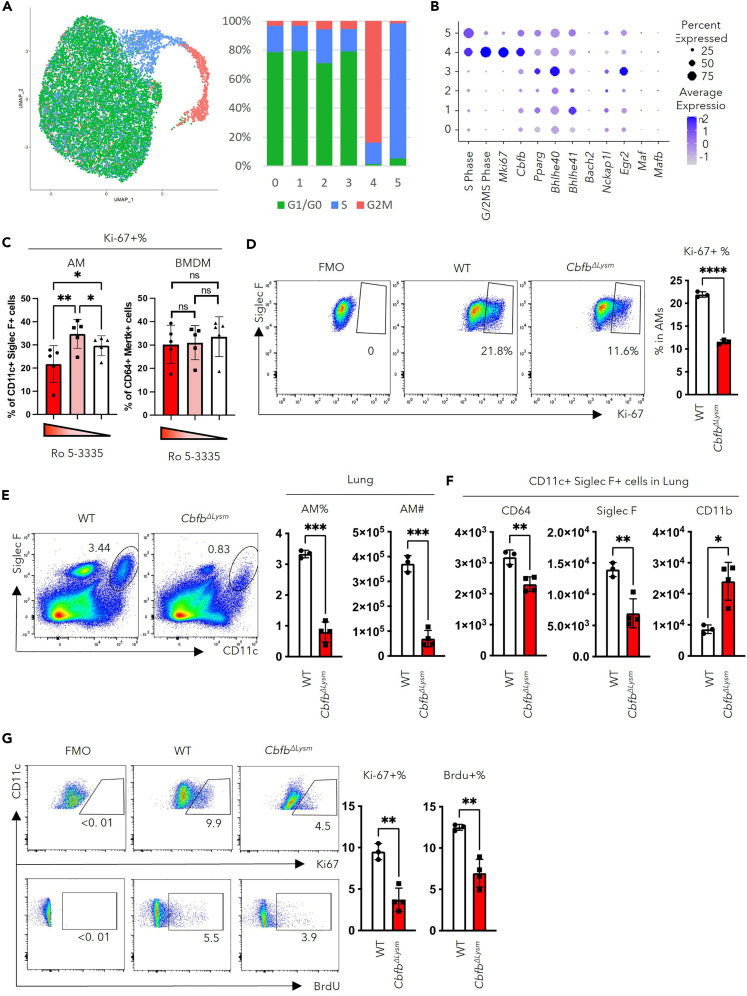
During homeostasis, the AM compartment is maintained by self-renewal and proliferation. However, the transcriptional regulation of AM cell cycle progression remains poorly understood. To investigate the mechanisms underlying the self-renewal ability of AMs, we first identified actively proliferating cells using previously reported gene lists for G2/M or S phase markers.34 We found that cells in cluster 4 were mainly in G2/M phase whereas cells in cluster 5 were mainly in S phase (Figure 2A). The transcription factors enriched in cluster 4 and cluster 5 were examined to elucidate the transcriptional control of AM proliferation. Notably, a transcription factor named Cbfb, was found to be highly enriched in clusters 4 and 5 (Figure 2B). Furthermore, Cbfb, but not any of the other transcription factors known to be important for AM development and/or proliferation,10,11,12,13,14 correlated with the pattern of G2/M phase score and the expression of proliferation marker, Mki67 (Figure 2B).
Figure 2.
CBFβ regulates AM proliferation and self-renewal
(A) Cell cycle analysis of AMs from the young hosts using list of genes associated with S or G2/M phases of cell cycle. UMAP (left) and quantification (right) of the cells predicted to be in each cell-cycle phase with respect clustering shown in Figure 1B.
(B) Dot plot revealing the score for S-Phase or G/2M-Phase related genes, transcription factors related to proliferation and identity of AMs, and transcription factors associated with the suppression of the ESC-like feature of AMs.
(C) Percentage of Ki-67 expressing macrophages in response to its major growth factor culture on treatment with different concentrations of Ro 5–3335 (100μM, 50μM, and 0 μM respectively, each dot represents one biological replicate).
(D) Adherent cells in BAL from CbfbΔLysm (n = 3) or wild-type (n = 3) mice were stimulated with 10 ng/ml GM-CSF in vitro. The representative FACS plot (left) and quantification (right) of Ki-67 expressing AMs following 24 h in culture. (E to G) Bone-marrow-chimeric mice were constructed by transferring bone marrow from CbfbΔLysm or wild-type (WT) mice to age and gender matched WT mice (each dot represents one mouse).
(E) Representative FACS plot (left) and quantification (right) of AMs in the lungs of CbfbΔLysm or wild-type BM chimeric mice.
(F) Geometry mean fluorescence intensity (gMFI) of markers for AM identity, CD64, Siglec-F, and CD11b, in AMs (defined as CD11c+Siglec-F+ cells).
(G) BrdU was injected 24h before euthanization of the mice. Representative FACS plot (left) and the quantification (right) of the percentage of AMs expressing Ki-67 or having BrdU incorporated in lungs from CbfbΔLysm or wild-type BM chimeric mice.
RM one-way ANOVA with the Geisser-Greenhouse correction and multiple comparisons (C) and unpaired Student’s t test with Welch’s correction (D-G) were utilized for statistical evaluation. Data were shown as mean ± SD ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001. Data in (B) were displayed in dots with diameter representing the percent of cells in the cluster (as shown in Figure 1B) that expressed the gene (or list of genes) indicated, with the depth of color indicating the average expression level. See also Figures S1C–S1E.
It has been reported that CBFβ deletion led to defects in the AM compartment,28 but the underlying mechanisms remain unexplored. To this end, we examined the role of CBFβ in the proliferation of AMs using in vitro and in vivo models. CBFβ requires RUNX family members which possess DNA-binding domains23 to confer its functions. With data from ImmGen,35 AMs were found to express mainly Runx1 (Figure S1C). A small molecule drug, Ro 5–3335, was thus utilized to inhibit the interaction between CBFβ and RUNX1. AMs were isolated from BAL with minimum contamination from blood (Figures S1D and S1E). Using bone marrow–derived macrophages (BMDMs) as control, the isolated AMs were treated in vitro with different concentrations of Ro 5–3335.16,36 Despite similar viability of cells (Figure S1F), treatment with 100μM Ro 5–3335 inhibited the proliferation of AMs but not BMDMs (Figure 2C). To exclude the possibility for off-target effects from Ro 5–3335, mice with myeloid-specific deletion of CBFβ (CbfbΔLysm) were utilized to evaluate whether loss of CBFβ would result in diminished AM proliferation. Upon GM-CSF stimulation in vitro, CBFβ-deficient AMs displayed a significantly decreased proportion of Ki-67 expressing cells compared to that of WT AMs, suggesting that CBFβ is critical for AM proliferation in response to growth factors (Figure 2D). We next examined whether CBFβ is important for AM proliferation in vivo. As Lyz2 may also be expressed in alveolar type II (AT II) cells,37 bone-marrow chimeric mice were generated to avoid potential contribution from the Cbfb-expressing stromal cells.37 Bone marrow from CbfbΔLysm mice or control mice was transferred into lethally irradiated WT mice. Examination of the AMs’ number and proliferation was performed 8 weeks post-irradiation. Deficiency in CBFβ led to decreased number of AMs in both lungs (Figure 2E) and BAL (Figure S1G). These AMs also displayed immature AM phenotype, evidenced by decreased expression of Siglec-F and increased expression of CD11b (Figure 2F). Consistent with our in vitro findings, the proliferation, as shown by Ki-67 expression or BrdU incorporation, decreased in AMs lacking CBFβ (Figure 2G). Taken together, we identify CBFβ as a potential important transcription factor for AMs’ identity and self-renewal ability.
scRNA-seq identified distinct phenotypes of AMs from young and aged mice
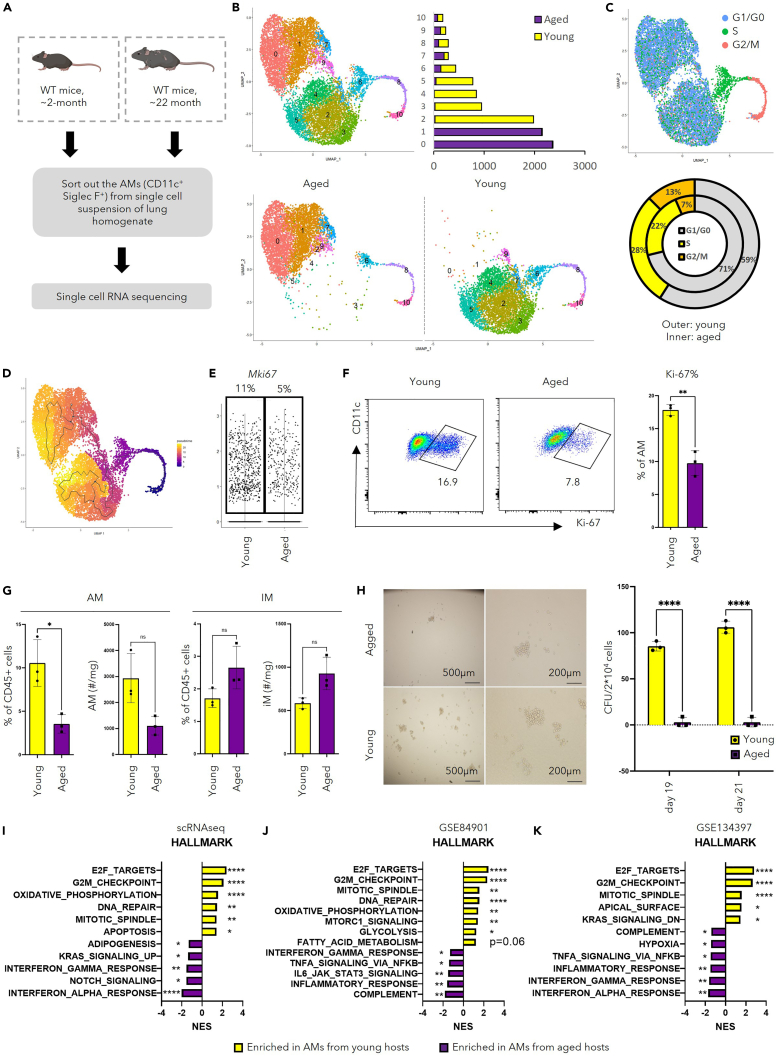
To investigate how aging influences cellular and molecular features of AMs, scRNA-seq library prepared from AMs in young (∼2-month-old) and aged (∼22-month-old) mice were combined for analysis (Figure 3A). The same number of cells from each group were put into the detection of differentially expressed genes, ensuring equal input from both groups. Moreover, other myeloid cells (cluster 10) were removed based on their low expression of AM markers and their heightened expression of other myeloid features (Figure S2A). Using the same method described in the previous session, re-clustering of the remaining cells, as well as further cell cycle and trajectory analysis, were then performed (Figures 3C and 3D, Table S2). Cluster 6 was enriched in a gene profile of cells in the S phase, whereas clusters 8 and 10 were enriched in a gene profile of cells in G2/M phase (Figures 3B and 3C). Because of the self-renewing nature of AMs, the proliferating cells were chosen as “stem cells” for trajectory analysis. Although “upstream” proliferating cells (clusters 6, 8, and 10) had similar transcriptional profiles, AMs from young and aged mice gradually branched into 2 different streams with cluster 0 or cluster 4 at the end of the AMs from aged mice or young mice respectively (Figures 3B and 3D). Both cluster 0 and cluster 4 can be defined as poised inflammatory cells because both pro-inflammatory and anti-inflammatory genes were enriched (Figures S2D and S2F). During scRNA-seq library construction (Figure 3A), each step was performed with young and aged mice side by side to ensure minimum batch effect, and we combined 3 mice in each group to homogenize individual differences within the group. Despite these, to minimize the possibility that the phenotypic differences in non-proliferating AMs observed between young and aged hosts were artifacts brought by library preparation steps or individual differences, publicly available database GSE12487237 was utilized for further analysis and comparison. Identification of AMs was performed based on featured genes for AMs in scRNA-seq30 (Figure S2C), followed by re-clustering of “isolated” AMs. Consistent with the observation in our scRNA-seq dataset (Figures 2B and 2C), in GSE124872,37 aside from the proliferating clusters identified by cell-cycle analysis (Figure S2B, upper right), spatial distribution of AMs from young and aged hosts remained distinct (Figure S2B). These data collectively indicate distinct transcriptomes of non-proliferating AMs in young and aged hosts.
Figure 3.
AMs from the aged mice display reduced self-renewal ability
(A) Schematic display of library preparation for scRNA-seq. AMs (CD11c+ Siglec-F+) were FACS-sorted from lung homogenate combined from young (∼2-month-old, n = 3) or aged (∼24-month-old, n = 3) female mice.
(B) UMAP displaying overall clustering of 10,531 AMs that passed quality control (upper left), split view with regard to their hosts (lower), and bar graph demonstrating quantification of cell number in each cluster (upper right).
(C) Cells were denoted to indicated phase in the cell cycle based on PCA analysis using genes sets of S phase and G2M phase, as displayed in UMAP (top) and quantification of cells in each phase (bottom).
(D) UMAP showing the trajectory analysis using Monocle3 with the proliferating cells as “stem cells”.
(E) Violin plot displaying Mki67 expression in AMs in young or aged mice from scRNA-seq. Quantification of percentage of cells expressing Mki67 was shown on the top.
(F and G) Flow cytometry of macrophages in the lung of young (n = 3) and aged mice (n = 3). F. Percentage of AMs expressing Ki-67 from the lung of young and aged mice shown by representative FACS plot (left) and quantification (right). G. Quantification of percentage and absolute number of AMs (left) and IMs (right).
(H) Colony formation assay using AMs from young (n = 3) and aged mice (n = 3). Demonstrated by representative pictures of the colony (left) and quantification of colony unit number (right).
(I) Bar graph showing selected results of pathway analysis comparing AMs from young and aged mice using GSEA.
(J) Bar graph showing selected results of pathway analysis comparing AMs from young and aged mice in published microarray data (GSE84901 using GSEA).
(K) Bar graph showing selected results of pathway analysis comparing AMs from young and aged mice in published bulk RNA sequencing data (GSE134397 using GSEA).
Unpaired Student’s t test with Welch’s correction (F, G and H) or GSEA (I, J and K) were utilized for statistical evaluation. Data were shown as mean ± SD ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001. See also Figure S2.
Clusters 2, 3, and 4 were mainly derived from young mice and followed an order of 3 => 2 => 4 in pseudotime (Figure 3D). Cluster 3 had a relatively higher score in ESC-like features (Figure S2F) and expressed transcription factors known to facilitate anti-apoptotic functions, i.e., Bcl2a1b and Bcl2a1d (Figure S2D). Moreover, cluster 3 was enriched in mitochondria related profile and OXPHOS related genes (Figure S2E), which is consistent with previous reports that OXPHOS is required for the proliferative function of AMs.16,36 Cluster 2 appeared to be an intermediate stage between cluster 3 and cluster 4. Meanwhile, clusters representing AMs from the aged mice, cluster 0 and cluster 1, were enriched in genes for MHC II molecules (Figure S2F). Such result was validated by flow cytometry (Figure S2G). Together with the trajectory analysis (Figure 3D), cluster 1 is likely to represent AMs differentiating from cluster 0. In relatively upstream of the pseudotime (Figure 3D), cluster 7 and cluster 9 were enriched in genes associated with lysosome activity and lipid metabolism (Figures S2E and S2F), suggesting their preferential role in surfactant turnover. Lastly, AMs in cluster 5 may undergo apoptosis as they were enriched solely in genes for cytoskeleton compartments and genes indicative of mitochondria leakage like mt-Atp8 (Figure S2D). Taken together, the combined dataset of AMs from young and aged mice revealed functional heterogeneity in AMs as well as marked distinction between the two transcriptomes.
Consistent with previous reports that AMs in the aged hosts exhibited decreased ability for self-renewal,21,22 the proportion of cells denoted to the S phase or G2/M phase decreased in the aged hosts (Figure 3C). This is confirmed by the decreased percentage of Mki67-expressing cells in the aged mice revealed by scRNA-seq analysis (Figure 3E). Such a notion was further validated by flow cytometry (Figure 3F). The reduced proliferation of AMs led to a significant reduction in number of AMs but not IMs in the lungs of aged mice (Figure 3G). To directly evaluate the self-renewal ability of AMs in addition to proliferation, we performed the colony-formation assay, where AMs from young and aged mice were sparsely seeded on matrix gels and the colonies derived from these cells were examined after in vitro culture. AMs from aged hosts displayed decreased number and size of colonies (Figure 3H), suggesting diminished potential for self-renewal. Pathway analysis was performed with gene set enrichment analysis (GSEA)38,39 comparing AMs from young and aged mice with datasets including 1000 cells randomized out from each group in our scRNA-seq dataset, a published microarray dataset (GSE84901),21 and a published bulk RNA-seq dataset (GSE134397).22 Consistently, AMs from young hosts were enriched in the proliferation-related transcriptome including pathways related to cell cycle checkpoints, cell replication machinery, DNA repair, and telomere capping (Figures 3I–3K and S3B). Furthermore, we found that AMs from aged mice exhibited diminished expression of genes involved in ESC-like features (Figure S3A).33 Taken together, these data demonstrate that aging leads to a reduction of ESC-like features and self-renewal ability of AMs.
AMs in the aged mice exhibit senescence-like phenotypes
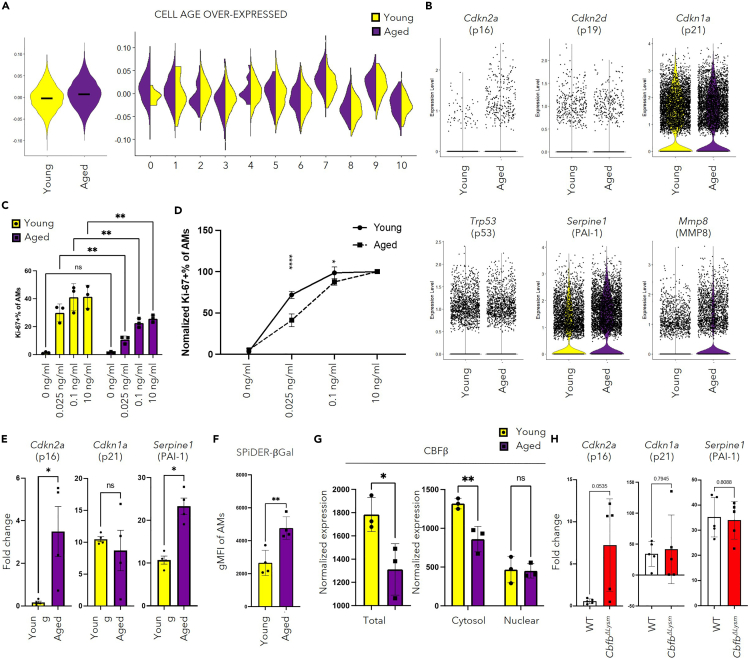
Cellular senescence describes a state of permanent cell-cycle arrest together with increased senescent markers and senescence-associated secretory phenotype (SASP).17,18 Aside from the decreased self-renewal ability, AMs in aged hosts have decreased apoptosis-related profiles and enhanced inflammation-related profiles (Figures 3I–3K and S3B). We thus hypothesize that AMs from aged hosts possess senescence-like phenotypes. Indeed, although we were not able to observe a specific subset of AMs from the aged mice that were enriched in senescence-like phenotypes, AMs from aged hosts showed elevated score of the reported list of genes associated with senescence40 when compared to that of the young (Figure 4A). To further confirm such phenotype, markers associated with cellular senescence, which included genes that emphasize cell-cycle regulation and inflammation, were analyzed in our scRNA-seq data. With regards to cell-cycle regulation related markers, aging led to increased Cdkn2a (p16) but not Cdkn2d (p19), Cdkn1a (p21), or Trp53 (p53) expression in AMs (Figure 4B), which was validated by qPCR in young and aged mice from the same source (JAX) (Figure 4E). Moreover, the elevation of Cdkn2a (p16) in the AMs from the aged mice was not only observed in the cells denoted to S phase or G2/M phase, but also in cells attributed to G1/G0 phase (Figure S3C). Meanwhile, the proportion of Cdkn1a (p21) positive cells was similar between young and aged mice in each cell-cycle phase (Figure S3D). Upon in vitro stimulation with GM-CSF, AMs from aged mice displayed decreased proliferation capacity, evaluated by the proportion of cells expressing Ki-67 (Figures 4C and 4D). Even on saturated concentration of GM-CSF (10nullng/mL), AMs from aged mice had reduced percentage of Ki-67-expressing cells, indicating that a significant proportion of them experienced irreversible cell-cycle arrest (Figure 4C). When normalized to the 10nullng/mL group in AMs from young and aged mice, AMs from aged mice were more sensitive to the drop in GM-CSF concentration (Figure 4D). Increased expression of other senescence-related markers including Serpine1 (PAI1) (Figures 4B and 4E) and Mmp8 (MMP8) were also observed (Figure 4B). Both PAI-1 and MMP8 have been implicated in the pathogenesis of lung fibrosis.41,42 In addition, TGF-β1/p53/PAI-1 signaling cascade has been reported to be involved in cellular senescence.43,44,45 Enhanced lysosomal activity, indicated by elevated senescence-associated β-galactosidase (SA-β-gal) activity, has been considered to be a hallmark for cellular senescence.46,47 Evaluation of SPiDER-βGal staining evaluated by flow cytometry confirmed that AMs in the aged lung could be undergoing age-associated processes like cellular senescence (Figures 4F and S3F). It has been reported that the senescent cells could promote the development of lung fibrosis,48 and the accelerated immunosenescence mouse model has suggested a potential role of immunosenescence in solid organ aging.19 Previous studies have shown collagen accumulation in lungs of both aged mice49 and humans.50 As the major immune cell population in the lung during homeostasis, AMs from aged hosts have increased expression of mRNA for Collagen IV and Collagen XIV (Figure S3E), raising the possibility of direct collagen deposition in the aged lung.50
Figure 4.
AMs from the aged mice possess senescence-like phenotypes
(A) Module scores of cellular-senescence-related genes were calculated for the AMs.40 Violin Plot comparing the scores between AMs from young and aged mice (left), and the scores among the clusters as shown in Figure 3B (right).
(B) Violin plots displaying selected senescence markers with gene names and protein names shown on the top respectively. (C and D) AMs were isolated from young and aged mice, followed by in vitro culture with different concentrations of GM-CSF (each dot represents one biological replicate).
(C) Bar graph displaying the percentage of Ki-67 expressing AMs on in vitro stimulation with indicated concentration of GM-CSF.
(D) Normalized percentage of Ki-67 expressing cells with that on saturated concentration of GM-CSF (10ng/mL) of AMs from young or aged mice.
(E) Quantification of relative expression of selected markers for cellular senescence evaluated by qPCR. Each dot represents one biological individual.
(F) Geometric mean of Fluorescence Intensity (gMFI) of SPiDER-βGal staining in AMs from lung homogenate of young (n = 4) or aged mice (n = 4).
(G) Quantification of CBFβ in BAL cells from young (n = 3) and aged (n = 3) mice with western blot. The protein level was normalized to the total protein stain. Each dot represents one biological individual.
(H) qPCR was performed with the cells in BAL from CbfbΔLysm or control mice. Data combined from 2 independent experiments. Each dot represents one biological replicate. Relative expression of selected markers for cellular senescence was displayed.
two-Way ANOVA with multiple comparisons (C, D, and G, right) and unpaired Student’s t test with Welch’s correction (E, F, H and G, left) were utilized for statistical evaluation. Data were shown as mean ± SD ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗∗, p < 0.0001. The bar in (A) represents the median of module scores calculated with cellular-senescence-associated genes.40 See also Figures S3 and S4.
To further investigate the potential molecular mechanisms underlying this phenotype, the scRNA-seq analysis of AMs from young and aged mice was explored. The expression of growth factor receptors, autocrine Tgfb1,9 and transcription factors known to be important for the proliferation of AMs was similar in AMs from young and aged hosts (Figures S4A and S4B). Moreover, the expression levels of Maf (cMAF) and Mafb (MAFB), two transcription factors considered to be inhibitory to the ESC-like features of AMs, were found to be similar between the groups5 (Figure S4C). Given the potential role of CBFβ in promoting the proliferation of AMs from young mice, the role of CBFβ in the senescence-like phenotypes observed in AMs from the aged mice was evaluated. There were no significant differences in the expression of Cbfb and its binding partners between AMs from young and aged mice (Figures S4E and S4F). However, the protein level of CBFβ in AMs from aged mice was significantly lower than that of the young mice (Figures 4G and S4G), suggesting that reduced CBFβ protein expression may contribute to the diminished proliferation and increased senescence-related features observed in AMs from aged mice. In support of this idea, deficiency of CBFβ in AMs led to an increase in Cdkn2a (p16), but not Cdkn1a (p21) or Serpine1 (PAI1) (Figure 4H). Taken together, these data suggest that impaired CBFβ expression may at least partially contribute to diminished proliferation and increased senescence in AMs during aging.
scRNA-seq revealed conserved phenotypes in AMs from human
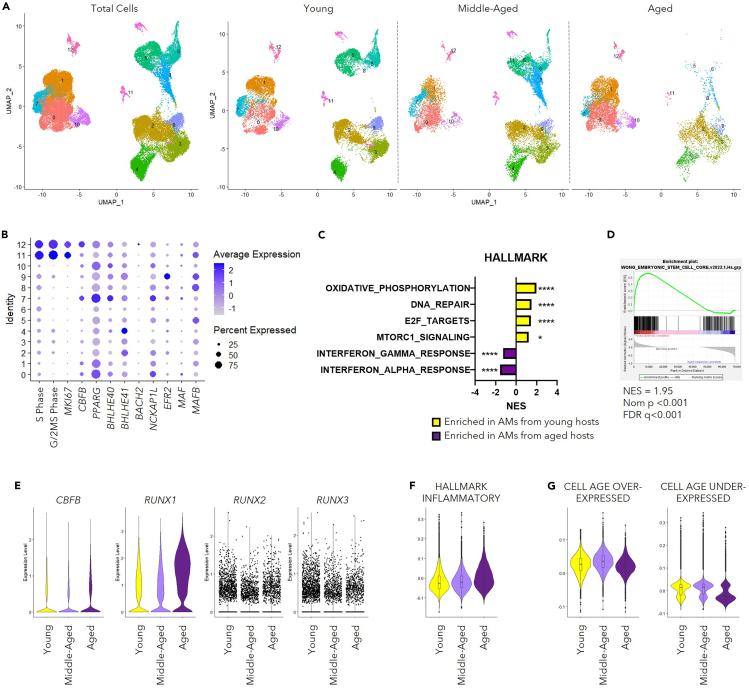
To investigate whether similar phenotypes could be observed in AMs from humans as those of mouse AMs, we utilized published scRNA-seq datasets in GEO database on lung homogenate from “Healthy” Donors of GSE135893,51 GSE122960,52 GSE128033,53 GSE136831,54 and one unidentified dataset stored as “GSE135893_ILD_annotated_fullsize.rds.gz” in GEO database (Table S3). We defined individuals aged <40 years old as “Young” (35 samples), individuals aged between 40 and 60 years old as “Middle-Aged” (27 samples), and individuals aged ≥60 years old as “Aged” (17 samples). AMs were identified with the expression of their featured markers30 and the lack of the expression of markers indicating other lineages (Figure S5A). A graph-based integration algorism75 was performed for clustering to overcome the batch effect between datasets (Figure 5A). Consistent with AMs from mice, the expression of CBFB was enriched in proliferating human AM populations (Figure 5B). In addition, pathway analysis enabled by GSEA38,39 revealed enhanced pro-inflammatory pathways, altered metabolism-related features, as well as abrogated capacity for proliferation and DNA repair in AMs from “Aged” individuals (Figure 5C). Importantly, consistent with our findings in mice, AMs in “Aged” individuals also showed decreased ESC-like characteristics (Figure 5D). To note, the mRNA level of the growth factor receptors and proliferation-related transcription factors including CBFB was similar between the “Young” and “Aged” groups (Figures 5E, S5B and S5D). Such observation stayed true for inhibitory transcription factors (MAF and MAFB) for ESC-like phenotype and autocrine TGFB1 (Figures S5C and S5E). Module score revealed enhanced pro-inflammatory phenotype of AMs in the “Aged” individuals during homeostasis (Figure 5F). Furthermore, among the senescence-associated genes that had been identified from various human cell types (∼40% fibroblasts),40 there was a moderate decline of genes that were downregulated (under-expressed) in senescent cells in AMs from aged individuals compared to those from young individuals (Figure 5G). Taken together, these data suggest that the senescence-like phenotypes were conserved between human and mice during aging.
Figure 5.
AMs from the elderly possess senescence-like phenotypes
Data of human AMs were extracted from combined scRNA-seq profiling pneumocytes of “Healthy” donors (GSE135893,51GSE122960,52GSE128033,53GSE13683154). Samples were divided into “Young” (<40-years old), “Middle-Aged” (40-60- years old) and “Aged” (≥60-years old) groups. In total, 79 samples with 35 “Young”, 27 “Middle-Aged”, and 17 ″Aged” were included.
(A) UMAP displaying the clustering of AMs as total cells (left) and in separate groups (right).
(B) Dot plot revealing the score for S-Phase or G/2M-Phase related genes, transcription factors related to proliferation and identity of AMs, and transcription factors associated with the suppression of the ESC-like feature of AMs.
(C) Bar graph displaying selected pathways identified with GSEA using gene sets from HALLMARK database comparing individuals from “Young” and “Aged” groups.
(D) GSEA result analysis with a list of genes associated with ESC-like features (MSigDB M7079).
(E) Violin Plots displaying mRNA level of CBFB and RUNX family members (RUNX1, RUNX2, and RUNX3).
(F) Violin Plot displaying module scores calculated with a gene list featuring inflammatory response (MSigDB M5932).
(G) Violin plots revealing module scores calculated with gene lists associated with cellular senescence.40
Data in (B) were displayed in dots with diameter representing the percent of cells in the cluster (as shown in A) that expressed the gene (or list of genes) indicated, with the depth of color indicating the average expression level. Data in (C and D) were analyzed with GSEA, shown as ∗, p < 0.05; ∗∗∗∗, p < 0.0001. The boxplots within the Violin Plots in (F) and (G) displayed the 25, 50 and 75 percentiles of the module scores calculated with genes lists indicated. See also Figure S5.
Discussion
In this study, the scRNA-seq library with abundant cell counts ensures the statistical strength needed to reveal functional heterogeneity within AMs. The transcription factor, CBFβ, was identified to be associated with AMs undergoing active cell cycle progression and was essential for AM proliferation and self-renewal. We then described the senescence-like phenotypes in AMs from aged mice and humans. Moreover, our data revealed that diminished CBFβ expression during aging at least partially contributes to the observed phenotype.
Consistent with precious reports,21,22 abrogated proliferation and self-renewal of AMs in the aged hosts was observed. Pathway analysis revealed their decreased ESC-like features as well. However, the mechanisms underlying the acquisition of this phenotype during aging remain unclear. McQuattie-Pimentel et al. have attributed this to the aging lung environment.22 Indeed, it has been shown that Alveolar type II (AT II) cells could promote the proliferation of AM by providing GM-CSF55 and through synchronization of Ca2+ flow.56 The lung environment could also regulate AM function, namely the cell cycle regulatory genes like Ccnd1 and Mdm2, through epigenetic alterations.57,58 Moreover, the pro-inflammatory molecules in the alveolar lining fluid20,59 may promote the pro-inflammatory phenotype of AM even during homeostasis. However, cell intrinsic mechanisms could account for the age-associated phenotypes found in AMs as well. In the study from McQuattie-Pimentel et al., AMs from aged mice were transferred into young mice or vice versa,22 among the differential expressed genes analyzed with the bulk-RNA-sequencing data of, a cluster of genes remained unchanged regardless of the alteration of lung environment. In addition, adoptive transfer of AMs isolated from aged mice to Csf2ra−/− neonatal mice displayed abrogated protection against influenza challenge in adulthood when compared to AMs from the young mice.60 Such phenotype suggested that there may be a cell autonomous mechanism involved in the altered AM phenotypes from the aged hosts. It has been suggested that AMs from aged mice had an increased proportion of cells derived from monocyte origin and exhibited heightened pro-inflammatory characteristics specifically in those monocyte-derived AMs.59 Such a notion is also supported by evidence from a Ms4a3-Cre-driven lineage tracing model.61 In the same report, it was shown that monocyte-derived AMs could compose up to ∼60% of total AMs at the age of 36-week-old. However, such data were not supported by long-term parabiosis (5 months of pairing)6 and bone-marrow-chimera with thoracic shield experiments (∼20 months post reconstitution).22 Constant challenges, i.e. long-term exposure to PM 2.5, experienced by the human lung throughout life could “open” the “niche” for AMs. Although there is a lack of studies on the “dirty” mouse models during aging for mimicry, it would be conceivable to consider the differences potentially brought by ontology in such a setting. Taken together, it is possible that both cell autonomous and non-autonomous factors could contribute to the declined self-renewal ability of AMs during aging, but the exact mechanism remains to be explored.
Enhanced cellular senescence has been described in the lungs of both aged mice62 and human.50 Previous reports attributed the accumulation of senescent cells in the lung predominately to the stromal cell compartment.49 Our data suggest that AMs, the self-renewing resident macrophage, potentially also undergo cellular senescence during aging. Specifically, we have shown that AMs from aged mice displayed cell-cycle arrest, decreased profile for apoptosis, enhanced pro-inflammatory profile at baseline, and elevated expression of senescence markers, i.e., SA-β-gal staining and Cdkn2a (p16) expression. During scRNA-seq library construction, mice from both C57BL/6J (young) and C57BL/6N (aged) backgrounds were utilized. Such strategy may introduce differences brought by the default mutation in nicotinamide nucleotide transhydrogenase (Nnt) gene in C57BL/6J but not C57BL/6N mice. This hypothesis could be plausible given the importance of Nnt in redox metabolism and mitochondrial function.63 Defects in mitochondrial function could serve as a hallmark for cellular senescence.64,65 However, even hypothetically the mutation had functioned, the differences in senescence-like phenotypes would be blurred due to the lack of this gene in young C57BL/6 J mice, instead of further causing the distinction between AMs from young and aged mice. In addition, we have validated our findings in both C57BL/6N young vs C57BL/6N aged mice (Figures 3I and 3J), as well as C57BL/6J young vs C57BL/6J aged mice (Figure 4E). Taken together, these data suggest that the senescence-like phenotypes we observed in in aged mice are not likely to stem from differences brought by genetic backgrounds.
The concept of macrophage senescence is still controversial. It was suggested by Ogrodnik et al.66 that p16 and SA-β-gal upregulation is a normal feature of macrophage activation rather than “true” cellular senescence. In both in vitro and in vivo experiments, the upregulation of p16 was proposed to be positively associated with the M−2 like phenotype in peritoneal macrophages. Such phenotype was shown to be reversible on stimuli promoting “M−1 Macrophage” polarization such as Poly I:C.68 However, pathway analysis with datasets generated by various groups revealed that AMs from the aged hosts would fit into the pro-inflammatory or “M-1” phenotype of macrophages. Furthermore, it is reasonable to discuss cellular senescence in AMs, which possess ESC-like features and exhibit self-renewal capacity in vitro and in vivo.5 To this end, we found that AMs from aged mice had decreased proliferation in vivo and were less responsive to proliferation following growth factor treatment in vitro. They also displayed decreased self-renew ability in the colony formation assay, confirming potential senescence-like phenotypes. These phenotypes in accompany by the increased p16 expression and β-gal upregulation strongly suggest their “senescence-like” phenotypes, although additional experiments are needed for further confirmation.
Notably, previous studies have suggested that senescent fibroblasts48,49 and ATII cells43 may contribute to increased collagen deposition in aged lungs. However, senescence in immune cells may also be important for the aging of lung.19 For instance, adoptive transfer of the splenocytes from aged, but not young mice could induce enhanced senescence in the lung of recipient mice,19 suggesting a prominent role for aged immune cells in shaping the aged lung environment. However, the presence of senescent immune cells and their physiological role in the lung remains to be explored. Because AMs represent the primary immune cells present in the alveoli during homeostasis, it is plausible to hypothesize that the senescence of AMs may contribute to increased lung tissue fibrosis during aging. Such a possibility warrants further studies. Furthermore, we found that AMs from aged mice exhibited increased expression of Col4a1, Col4a2, and Col14a1 when compared to young mice. Thus, aged AMs may also directly contribute to lung fibrosis through the deposition of collagen.
Understanding the mechanisms underlying the dysfunctional phenotype of AMs during aging may help to identify factors and therapeutic targets to promote healthy aging of lung. Our data suggest that impaired CBFβ expression in AMs may at least in part contribute to the diminished proliferation and enhanced cellular senescence in AMs during aging. Thus, augmenting CBFβ expression and/or its transcriptional activities may facilitate AM self-renewal and mitigate the development of senescence during aging. Future studies focusing on refined characterization of the role of CBFβ in the development, maintenance, and senescence of AMs are needed.
Limitations of the study
Although we described the senescence-like phenotypes of AMs during aging, we were not able to address the acquisition of such phenotypes. Furthermore, our studies did not address the physiological function of AM senescence in lungs during aging. In addition, the description of senescence-associated profiles remains to be refined with consideration of different cell types. The datasets (“Cell Age Overexpressed” and “Cell Age Under-expressed”)40 we used to evaluate the senescence-like phenotypes in AMs were identified from manipulative experiments with different human cell types (∼40% of these experiments were performed with human fibroblasts). In both mouse and human data, we have observed only marginal differences between AMs from young and aged hosts using gene sets identified in this way. The technical limitations from the “drop-out” issue during mapping of scRNA-seq library could also contribute to such results. For human datasets specifically, there is so far no efficient way to correct the batch effect brought by individual differences within donors, different digestion protocols, various library preparation procedures, and different sequencing methods etc. Another caviar of this study lies in limited functional validation of cellular senescence. Furthermore, despite that most of our findings derived from in vivo experiments, we could not eliminate the phenotypic alterations brought by in vitro culture of AMs when describing differences of sensitivity toward GM-CSF stimulation comparing young and aged AMs. Meanwhile, the senescence-related features in AMs could be unique, especially considering their functions as innate cells. A refined portfolio and functional description of the cellular senescence with respect to specific cell types during homeostasis and disease state is in urgent need for identification of promising intervention strategies for cellular aging.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti SiglecF-BV421 (clone E50-2440) | BD Biosciences | Cat# 562681; RRID: AB_2722581 |
| Anti SiglecF-PE (clone E50-2440) | BD Biosciences | Cat# 552126; RRID: AB_394341 |
| Anti CD11b-PerCP-Cy5.5 (clone M1/70 | Biolegend | Cat# 101228; RRID: AB_893232 |
| Anti CD11c-BV510 (clone N418) | Biolegend | Cat# 117338; RRID: AB_2562016 |
| Anti MHCII-FITC (clone M5/114.15.2) | Biolegend | Cat# 107606; RRID: AB_313321 |
| Anti Ly6C-BV711 (clone HK1.4) | Biolegend | Cat# 128037; RRID: AB_2562630 |
| Anti CD64-BV711 (clone X54-5/7.1) | Biolegend | Cat# 139311; RRID: AB_2563846 |
| Anti CD64-PE (clone X54-5/7.1) | Biolegend | Cat# 139304; RRID: AB_10612740 |
| Anti MerTK- APC (clone 2B10C42) | Biolegend | Cat# 151508; RRID: AB_2650739 |
| Anti MerTK- FITC (clone 2B10C42) | Biolegend | Cat# 151504; RRID: AB_2617035 |
| Anti Ki67-FITC (clone SolA15) | eBioscience | Cat# 11-5698-82; RRID: AB_11151330 |
| Anti Ki67-APC (clone SolA15) | eBioscience | Cat# 17-5698-82; RRID: AB_2688057 |
| Anti CD45-APC-Cy7 (clone 30-F11) | Biolegend | Cat# 103116; RRID: AB_312981 |
| Anti CBFb (clone EPR6322) | Abcam | Cat# ab133600; RRID: |
| IRDye® 800CW Goat anti-Rabbit IgG Secondary Antibod | LI-COR | Cat# 926-32211 |
| IRDye® 680RD Donkey anti-Mouse IgG Secondary Antibody | LI-COR | Cat# 926-68072 |
| Chemicals, peptides, and recombinant proteins | ||
| Zombie NIR™ Fixable Viability Kit | Biolegend | Cat# 423106 |
| Zombie Aqua™ Fixable Viability Kit | Biolegend | Cat# 423102 |
| Recombinant mouse GM-CSF | Biolegend | Cat# 576308 |
| Recombinant mouse M-CSF | Biolegend | Cat# 574804 |
| Ro 5-3335 | TargetMol | Cat# T4687 |
| Protease Inhibitor Cocktail | Sigma | Cat# P8340-5ML |
| Critical commercial assays | ||
| Cellular Senescence Detection Kit - SPiDER-βGal | Donjindo | SG04-01 |
| GenElute Mammalian Total RNAMiniprep Kit | Sigma-Aldrich | Cat# RTN350 |
| BD PharMingen BrdU Flow Kits | BD Biosciences | Cat# 552598 |
| eBioscience Foxp3 / TranscriptionFactor Staining Buffer Set | Thermo Fisher Scientific | Cat# 00-5523-00 |
| Pierce BCA Protein Assay Kit | Thermo Scientific | Cat# 23225 |
| NE-PER™ Nuclear and Cytoplasmic Extraction Reagents | Thermofisher | Cat# 78833 |
| Revert™ 700 Total Protein Stain Kits for Western Blot Normalization | LI-COR | Cat# 926-11010 |
| Deposited data | ||
| Raw and analyzed scRNAseq of AMs from young and aged mice | This paper | GEO: GSE205982 |
| Microarray of AMs from young and aged mice (Figure 3J) | (Wong et al., 2017)21 | GEO: GSE84901 |
| Bulk-RNA-seq of AMs from young and aged mice (Figure 3K) | (McQuattie-Pimentel et al., 2021)22 | GEO: GSE134397 |
| sc-RNA-seq of pneumoncytes of "Healthy" individuals (Figures 5 and S5) | (Habermann et al., 2020)51 | GEO: GSE135893 |
| sc-RNA-seq of pneumoncytes of "Healthy" individuals (Figures 5 and S5) | (Reyfman et al., 2019)52 | GEO: GSE122960 |
| sc-RNA-seq of pneumoncytes of rejected organ donors (Figures 5 and S5) | (Morse et al., 2019)53 | GEO: GSE128033 |
| sc-RNA-seq of pneumoncytes of "Healthy" individuals (Figures 5 and S5) | (Adams et al., 2020)54 | GEO: GSE136831 |
| Bulk-RNA-seq of different macrophages (Figure S1C) | (Heng et al., 2008)35 | ImmGen |
| sc-RNA-seq of pneumoncytes of young (3-month) and aged (24-month) mice (Figure S2B and S2C) | (Simon LM et al., 2019)37 | GEO: GSE124872 |
| Experimental models: organisms/strains | ||
| C57BL/6J | The Jackson Laboratory | Cat# 000664 |
| C57BL/6N | National Institutes of Aging | N/A |
| Lyz2-cre | The Jackson Laboratory | Cat# 004781 |
| Cbfbfl/fl | Laboratory of Dr. Thomas J. Braciale | (Cardani et al., 2017) |
| Oligonucleotides | ||
| Cdkn1a-F qPCR | GTCAGGCTGGTCTGCCTCCG | N/A |
| Cdkn1a-R qPCR | CGGTCCCGTGGACAGTGAGCAG | N/A |
| Cdkn2a-F qPCR | CCCAACGCCCCGAACT | N/A |
| Cdkn2a-R qPCR | GCAGAAGAGCTGCTACGTGAA | N/A |
| Serpine1-F qPCR | GACACCCTCAGCATGTTCATC | N/A |
| Serpine1-R qPCR | AGGGTTGCACTAAACATGTCAG | N/A |
| mHprt qPCR-F | CTCCGCCGGCTTCCTCCTCA | N/A |
| mHprt qPCR-R | ACCTGGTTCATCATCGCTAATC | N/A |
| Software and algorithms | ||
| GraphPad Prism 9 | GraphPad Software | http://www.graphpad.com |
| FlowJo (version 10.3) | LLC | https://www.flowjo.com/ |
| Image Studio™ Lite Quantification Software | LI-COR | N/A |
| Cell Ranger v2.0.0 | 10X Genomics | |
| R project | https://www.r-project.org/ | |
| R studio | Posit | https://posit.co/ |
| Seurat | Satija lab | https://satijalab.org/seurat/ |
| Monocle3 | Trapnell lab | https://cole-trapnell-lab.github.io/monocle3/ |
| Gene Set Enrichment Anlaysis (GSEA) | UC San Diego and Broad Institute | http://www.gsea-msigdb.org/gsea/index.jsp |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jie Sun (js6re@virginia.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
Mice and bone marrow chimera
Female aged C57BL/6 mice were received from National Institutes of Aging or the Jackson lab (Figure 3A) at the age around 20-21 month. Their young control, around 2-month-old female mice, were bred in house from breeders set up with C57BL/6 mice purchased from Jackson Laboratory (Harbor, ME). Beddings of the cages harboring young and aged mice were exchanged once per week for at least 4 consecutive weeks to ensure the homogenization of microbiota. For experiments comparing young and aged mice, female mice were utilized. Both male and female CbfbΔLysm mice were utilized in relevant experiments. All animal experiments were performed in animal housing facilities at the Mayo Clinic (Rochester, MN) or the University of Virginia (UVA, Charlottesville, VA). The animal experiments were approved by the the Mayo Clinic or UVA Institutional Animal Care and Use Committees (IACUC).
To generate bone marrow chimera, bone marrow cells were isolated from CbfbΔLysm or Control mice as with procedures modified from a published protocol.67 In brief, after long bones were isolated and cleaned, one of the ends of each bone was cut open. The bones, which had their cut-open end facing down, were then put into 0.5ml Eppendorf tubes with holes created utilizing 18G needle. The 0.5ml Eppendorf Tubes harboring the bones were then put into a 1.5ml Eppendorf tube with 100ml culture media. The bone marrow was isolated by centrifuge at 10,000g for 15 seconds at 4°C, followed by filtering the cells through 70μm mesh (Falcon) and lysis of red blood cells using ammonium-chloride-potassium (ACK) buffer (deionized water with 0.15M NH4Cl, 1mM KHCO3, and 0.1mM Na2EDTA). Subsequently, the prepared cells were i.v. injected into the irradiated (1100Rads) WT mice at the quantity of ∼4 million cells/mice. The chimera mice were sacrificed 10 weeks post reconstitution.
Method details
Broncho-alveolar lavage (BAL) fluid
After euthanization of the mouse with an overdose of Ketamine/xylazine, the trachea was exposed with incision on the skin and blunt dissection of the thyroid as well as connective tissue. A small incision was made between thyroid cartilage and Cricoid cartilage, followed by insertion of pipette tip (20ml tip topping on 1000ml tip). Using p1000 pipette, BAL fluid was obtained by flushing the airway five times with a single inoculum of 1000μl sterile PBS (with 2% FBS and 1X Pen/Strep/Glutamate) each time. Cells in BAL fluid were spun down at 1600rpm for 5 minutes followed by lysis of red blood cells using ammonium-chloride-potassium (ACK) buffer. The cells were then palleted (1600rpm for 5 minutes) and resuspended for flow cytometry or cell culture.
Single cell suspension from Lung homogenate
Animals were euthanized with an overdose of Ketamine/xylazine. Lungs were perfused with 10ml PBS from the right ventricle of heart through pulmonary circulation. Following the harvest, lung tissue was minced and put in digestion buffer (IMDM with 183 U/ml type 2 collagenase (Worthington)). After 30 minutes of incubation at 37 C°, the homogenate was put through the “m_lung_02” program on a gentleMACS tissue disrupter (Miltenyi). Specifically, for Spider-βGal staining, to avoid stress from the process, manual homogenization with metal mesh was used in this step. A 70μm mesh (Falcon) was used to filter the suspension. The flow through will be washed using MACS buffer (PBS with 2% FBS and 2 mM/L Na2EDTA). The cells were then pelleted at 1600rpm for 5 minutes, followed by lysis of red blood cells using ammonium-chloride-potassium (ACK) buffer (deionized water with 0.15M NH4Cl, 1mM KHCO3, and 0.1mM Na2EDTA). The single cell suspension would be utilized to perform flow cytometry or FACS sorting.
In vitro GM-CSF stimulation experiment
Cells from BAL were processed as described in relevant session. They were then put into a 12-well plate in complete medium (RPMI 1640 with 10% FBS and 1X Pen/Strep/Glutamate) at 37 C° in 5% CO2. The non-adherent cells were washed off with warm PBS after 2 hours, and AMs would be purified from these cells by its adherent nature. The remaining adherent cells were cultured overnight in complete medium without GM-CSF, followed by culture in complete media supplemented with 10ng/ml (unless specified) GM-CSF for 24 hours. AMs were then detached with 0.25% Tyrosine-EDTA solution (Gibco) and put through flow cytometry to evaluate its proliferation.
In vitro inhibition of CBFβ-RUNX1 interaction
Cells of BAL fluid and bone marrow were harvested from the same mice. AMs were purified in 12-well plates as described in the last section. Followed overnight culture in complete medium without GM-CSF, AMs were treated with 100μM, 50μM or 0μM Ro 5-3335 (TargetMol) in complete medium supplemented with 10 ng/ml GM-CSF for 24 hours.
Bone Marrow was isolated as previously described.67 After filtering the cells through 70μm mesh (Falcon), lysis of red blood cells was performed with ACK buffer. The cells were then resuspended in culture media (DMEM with 50 ng/ml M-CSF, and 1X Pen/Strep/Glutamate) and seeded in the 10cm Petri dish (Falcon). The day of seeding was set as day 0, cells were then cultured at 37 C° with 5% CO2 for 7 days with change for fresh media on day 4. The BMDMs were detached and re-seed in 12-well plates on day7. After resting the cells in culture media overnight, BMDMs were treated with 100 μM, 50 μM or 0 μM Ro 5-3335 in culture media for 24 hours.
0.25% Trypsin-EDTA solution (Gibco) was then used to detach AMs/BMDMs from the plate, followed by evaluation of proliferation using flow cytometry.
Colony formation assay
Colony formation assays were performed with cells in BAL from indicated groups with protocol modified from the procedures published by Soucie, E. L. et al.5 Briefly, freshly isolated cells from BAL were seeded at 20,000 cells in 35 mm culture dishes. These cells were then cultured in MethoCult medium (M3231, Stem Cell Technologies) supplemented with 10 ng/ml GM-CSF, 50 mg/ml penicillin/streptomycin, and 2 mM glutamine. The number of colonies were counted on day 19 and day 21 after plating.
Flow cytometry
Detached AMs or cells from lung homogenate were prepared for flow cytometry. Cells were then incubated in PBS with viability dye (Zombie NIR™ Fixable Viability Kit, Cat# 423106; Zombie Aqua™ Fixable Viability Kit, Cat# 423102) at 1:1000 dilution. The Fc receptor was blocked with α-mouse CD16/CD32 (BioXcell). Cells were then incubated in solutions with antibodies targeting surface makers on ice for 30min. When intracellular proteins of interest were not involved, the samples would be ready after washing off the antibodies.
When intracellular staining was needed, cells were washed with MACS buffer twice after staining for surface markers. The cells were fixed with fixation butter (Invitrogen) at room temperature for 30 minutes, followed by 1 hour incubation in permeabilization buffer (Invitrogen) on ice after washing off the fixation buffer. And staining of intracellular markers was then performed on ice for 1 hour. The samples would be ready after washing off the antibodies.
Samples were put in MACS buffer and put through FACS Attune. Analysis of the data was enabled with FlowJo.
BrdU incorporation
BrdU (Sigma, 1 mg/mouse in sterile PBS) was i.p. injected 1 day before harvest. And flow cytometry was used to evaluate the Ki-67 and BrdU incorporation of AMs.
SPiDER-bGal staining
The cells in the single cell suspension from lung homogenate were stained with extracellular markers for flow before putting through SPiDER-βGal (Dojindo) staining. Cells were palleted and resuspended in 1ml warn RPMI-1640 with Bafilomycin A1 (1:1000), which was then incubated at 37 C° in 5% CO2 for 1 hour. Another 1ml warm RPMI-1640 with SPiDER-bGal (1:1000, the final concentration in the working solution is 1:2000) and Bafilomycin A1 (1:1000) was added in. The dye was washed off after another 30min incubation 37 C° in 5% CO2. These cells were stained with viability dye (Biolegend, Zombie Aqua™ Fixable Viability Kit, Cat# 423102) to evaluate the viability of the cell. For quantification of SPiDER-βGal staining, geometry mean was taken in the live AMs (Zombie-CD64+Mertk+CD11c+Siglec-F+).
Nuclear extraction and Western Blot
NE-PER™ Nuclear and Cytoplasmic Extraction Reagents (ThermoFisher) were used for nuclear extraction of AMs. Following the protocol, around 0.5∗106 Cells from BAL were spin down and resuspend in 50ml pre-cooled CRE I solution by vertexing for 15 seconds. After 10-minute incubation on ice, 2.75ml pre-cooled CRE II solution was added in. Start and end with 5-second vertexing, 1-minute incubation was done on ice, followed by 5-minute centrifugation at maximum speed in a microcentrifuge at 4 C°. The supernatant (cytoplasmic extract) was transferred to pre-chilled Eppendorf Tube on ice. 25ml pre-cooled NER solution was used to resuspend the pallet by repeated vertexing for 15 seconds every 10 minutes for a total of 40 minutes. Centrifugation was performed at the highest speed (4 C°) for 10 minutes, and the supernatant (nuclear extract) was collected in pre-chilled Eppendorf Tube.
Protein denaturationof both the cytoplasmic extract and nuclear extract was performed utilizing 4X Protein Sample Loading Buffer (LI-COR) with β-mercaptoethanol. Samples were boiled at 100 C° for 5 minutes. Electrophoresis was performed with pre-casted NuPAGE® Novex 4-12% Bis-Tris gel (Invitrogen) in MES buffer. Transfer was done overnight in transfer buffer (1XTris/Glycine buffer with 10% methanol) at 30V, 4 C°. Total protein was then stained using REVERT™ Total Protein Stain (LI-COR). After imaging the membrane in the 700nm channel in Odyssey® imaging system, protein stain was washed off using destaining solution in the kit. The membrane was put in Intercept® (TBS) Blocking Buffer (LI-COR) for 24 hours to ensure the removal of destaining solution. Blocking was done using the same buffer, and the membrane was incubated in primary antibody of CBFβ (Abcam, 1:1000) overnight. Secondary antibody incubation was performed in room temperature in Blocking buffer with anti-Rabbit antibody (LI-COR, 1:15000) 0.1% Tween 20 and 0.01%SDS. After washing the membrane in PBST (PBS +0.1% Tween20), the membrane was dried in dark at room temperature overnight. And the membrane was then visualized in 800nm channel in Odyssey® imaging system. ImageStudio was utilized for quantification. Expression of CBFβ was calculated as:
Quantitative RT-PCR
RNA extraction of freshly isolated cells in BAL or detached AMs was performed using total RNA purification kit (Sigma) together with DNase I (invitrogen) following the protocol in the kit. Reverse transcription for cDNA was then executed with Random primers (Invitrogen) and Moloney murine leukemia virus (MMLV) reverse transcriptase (Invitrogen), followed by RT-PCR with Fast SYBR Green PCR Master Mix (Applied Biosystems) in QuantStudio3 (Applied Bioscience). Relative expression of the target gene was calculated with respect to house-keeping gene, hypoxanthine phosphoribosyl transferase (Hprt).
scRNAseq analysis
The primary data was annotated and aligned using CellRanger 2.0. Using R project and the package of Seurat, we were able to further process the data. Genes that have been detected in at least 3 cells were included. And cells that has 200-4000 total genes detected, with <5% mitochondrial genes were considered to pass quality control. To better display the functional heterogeneity of AMs, For data shown in both Figures 1 and 3, clustering was modified from published workflow31 and the original vignette can be found in https://satijalab.org/seurat/articles/pbmc3k_tutorial.html. In brief, Principal Component Analysis was performed using differential expressed genes found across the dataset. Clustering was then conducted based on K-nearest neighbor (KNN) graph constructed. Of note, since the number AMs from the young mice were almost twice as many as that of AMs from the aged mice, as shown in Figure 3, the same number of AMs from the young mice was randomized out for the combined analysis to ensure the equal input in finding the differential expressed genes. Contaminating myeloid cells were excluded as described in the manuscript. And clustering was performed again using the AM population.
Cell-cycle analysis was performed based on published gene sets featuring S phase and G2/M phase.34 Principle-component- analysis (PCA)-based scoring was then used to assign each cell as “S phae”, “G2/M phase” or “G1/G0 phase”. Module score was calculated using the AddModuleScore() function of Seurat with gene lists as indicated. Trajectory analysis was performed using Monocle3 in conjunction with Signac, an extension of Seurat, as described in the paper,32,68 and the vignette can be found in https://satijalab.org/signac/articles/monocle.html. The proliferating cells were chosen as the “stem cells”. For pathway analysis, 1000 cells from each group were randomized out and the scaled data were put through Gene Set Enrichment Analysis as described.38,39
For scRNAseq data in Figures S2B and S2C, original data was accessed from GEO database with identification number of GSE124872.37 The age of mice was revealed in the “orig.ident” in the meta.data stored in the original data downloaded. Identification of individual mice was enabled by extraction of mouse number in the string of cell barcoding. Inclusion criteria of AMs includes expressed of feature genes for AMs,30 whereas exclusion criteria are expression of markers of other lineages or other myeloid cells.30 Cells with 200-1500 genes and <5% mitochondria-associated genes were considered to pass quality control. Further clustering and cell cycle analysis were performed as described in previous paragraphs.
For human data in Figures 5 and S5, the annotated and aligned data, “GSE135893_ILD_annotated_fullsize.rds.gz”, from GSE135893 in GEO database with scRNAseq from GSE135893,51 GSE122960,52 GSE128033,53 GSE136831,54 and one unidentified dataset was utilized. 79 libraries with “Healthy” in “Status” and information of “Age” were involved. The samples were then stratified with “Age”: individuals aged <40-years old were grouped as “Young”, individuals aged 40-60 years old were grouped as “Middle-Aged”, and individuals aged ≥ 60-years old were grouped as “Aged”. Cells with 200-8000 genes and <10% mitochondria-associated genes were considered to pass quality control. Workflow for integration of scRNAseq profile from the 3 groups was then adapted from published graph-based method69 to minimize the batch effect. The vignette can be found in https://satijalab.org/seurat/articles/integration_introduction.html. Briefly, differentially expressed genes were identified independently within each group. The shared genes were then used as “anchors” to enable graph-based integration of 3 datasets. AM population was defined as described in the manuscript. For the input data for further pathway analysis, samples with less than 20 cells were excluded. The average expression of genes within each sample was then calculated and used as input data for GSEA.
Pathway analysis
Gene Set Enrichment Analysis (GSEA), a software developed by UCSD and Broad Institute38,39 (http://www.gsea-msigdb.org/gsea/index.jsp), was utilized to conduct pathway analysis of data from both our scRNAseq dataset and publicly available datasets. In brief, genes of input were evaluated by a rank-based scoring system. Gene sets in HALLMARK database and Gene Ontology database were then used for comparison between AMs from young and aged hosts.
Acknowledgments
This work is supported by the US National Institutes of Health grants AI147394, AG069264, AI112844, AI154598 and AI176171 to J.S. The scholarship for Graduate School was awarded by Mayo Clinic Foundation to Y.W. We appreciate intellectual input from Drs. Daniel D. Billadeau, Hirohito Kita, Joao Passos, Wai Kee Eddie Ip, and Hu Li. We thank Drs. Jinyi Tang and Arka Sen Chaudhuri for proofreading. Graphical Abstract was created with BioRender.com.
Author contributions
Conception, Y.W., B.Z., and J.S.; data acquisition (wet lab), Y.W., R.Z., N.G., X.G., H.N., and A.S.; data acquisition (dry lab), Y.W., S.H., Y.C., and Y.L.; manuscript wiring, Y.W. and J.S.; reagents, mice, and manuscript editing, A.S., C.Z., H.D., and T.B.; funding acquisition, J.S.
Declaration of interests
J.S. is a consultant for the Teneofour company and receives research funding from Icosavax, which do not directly involve with this project.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: June 23, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107197.
Contributor Information
Bibo Zhu, Email: zhubibohzau@gmail.com.
Jie Sun, Email: js6re@virginia.edu.
Supplemental information
Data and code availability
The scRNAseq was deposited in GEO (https://www.ncbi.nlm.nih.gov/geo/) with accession number GSE205982.
This paper did not generate any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Mowat A.M., Scott C.L., Bain C.C. Barrier-tissue macrophages: functional adaptation to environmental challenges. Nat. Med. 2017;23:1258–1270. doi: 10.1038/nm.4430. [DOI] [PubMed] [Google Scholar]
- 2.Hussell T., Bell T.J. Alveolar macrophages: plasticity in a tissue-specific context. Nat. Rev. Immunol. 2014;14:81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- 3.Guilliams M., De Kleer I., Henri S., Post S., Vanhoutte L., De Prijck S., Deswarte K., Malissen B., Hammad H., Lambrecht B.N. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J. Exp. Med. 2013;210:1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evren E., Ringqvist E., Doisne J.M., Thaller A., Sleiers N., Flavell R.A., Di Santo J.P., Willinger T. CD116+ fetal precursors migrate to the perinatal lung and give rise to human alveolar macrophages. J. Exp. Med. 2022;219 doi: 10.1084/jem.20210987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soucie E.L., Weng Z., Geirsdóttir L., Molawi K., Maurizio J., Fenouil R., Mossadegh-Keller N., Gimenez G., Vanhille L., Beniazza M., et al. Lineage-specific enhancers activate self-renewal genes in macrophages and embryonic stem cells. Science. 2016;351:aad5510. doi: 10.1126/science.aad5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hashimoto D., Chow A., Noizat C., Teo P., Beasley M.B., Leboeuf M., Becker C.D., See P., Price J., Lucas D., et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nayak D.K., Zhou F., Xu M., Huang J., Tsuji M., Hachem R., Mohanakumar T. Long-term persistence of donor alveolar macrophages in human lung transplant recipients that influences donor-specific immune responses. Am. J. Transplant. 2016;16:2300–2311. doi: 10.1111/ajt.13819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Draijer C., Penke L.R.K., Peters-Golden M. Distinctive effects of GM-CSF and M-CSF on proliferation and polarization of two major pulmonary macrophage populations. J. Immunol. 2019;202:2700–2709. doi: 10.4049/jimmunol.1801387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu X., Buttgereit A., Lelios I., Utz S.G., Cansever D., Becher B., Greter M. The cytokine TGF-β promotes the development and homeostasis of alveolar macrophages. Immunity. 2017;47:903–912.e4. doi: 10.1016/j.immuni.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Schneider C., Nobs S.P., Kurrer M., Rehrauer H., Thiele C., Kopf M. Induction of the nuclear receptor PPAR-γ by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat. Immunol. 2014;15:1026–1037. doi: 10.1038/ni.3005. [DOI] [PubMed] [Google Scholar]
- 11.Rauschmeier R., Gustafsson C., Reinhardt A., A-Gonzalez N., Tortola L., Cansever D., Subramanian S., Taneja R., Rossner M.J., Sieweke M.H., et al. Bhlhe40 and Bhlhe41 transcription factors regulate alveolar macrophage self-renewal and identity. EMBO J. 2019;38:e101233. doi: 10.15252/embj.2018101233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamura A., Ebina-shibuya R., Itoh-nakadai A., Muto A., Shima H., Saigusa D., Aoki J., Ebina M., Nukiwa T., Igarashi K. Transcription repressor Bach2 is required for pulmonary surfactant homeostasis and alveolar macrophage function. J. Exp. Med. 2013;210:2191–2204. doi: 10.1084/jem.20130028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suwankitwat N., Libby S., Liggitt H.D., Avalos A., Ruddell A., Rosch J.W., Park H., Iritani B.M. The actin-regulatory protein Hem-1 is essential for alveolar macrophage development. J. Exp. Med. 2021;218 doi: 10.1084/JEM.20200472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mccowan J., Fercoq F., Kirkwood P.M., T'Jonck W., Hegarty L.M., Mawer C.M., Cunningham R., Mirchandani A.S., Hoy A., Humphries D.C., et al. The transcription factor EGR2 is indispensable for tissue-specific imprinting of alveolar macrophages in health and tissue repair. Sci. Immunol. 2021;6:eabj2132. doi: 10.1126/sciimmunol.abj2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu B., Wu Y., Huang S., Zhang R., Son Y.M., Li C., Cheon I.S., Gao X., Wang M., Chen Y., et al. Uncoupling of macrophage inflammation from self-renewal modulates host recovery from respiratory viral infection. Immunity. 2021;54:1200–1218.e9. doi: 10.1016/j.immuni.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao X., Zhu B., Wu Y., Li C., Zhou X., Tang J., Sun J. TFAM-dependent mitochondrial metabolism is required for alveolar macrophage maintenance and homeostasis. J. Immunol. 2022;208:1456–1466. doi: 10.4049/jimmunol.2100741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herranz N., Gil J., Herranz N., Gil J. Mechanisms and functions of cellular senescence. J. Clin. Invest. 2018;128:1238–1246. doi: 10.1172/JCI95148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernandez-Segura A., Nehme J., Demaria M. Hallmarks of cellular senescence. Trends Cell Biol. 2018;28:436–453. doi: 10.1016/j.tcb.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Yousefzadeh M.J., Flores R.R., Zhu Y., Schmiechen Z.C., Brooks R.W., Trussoni C.E., Cui Y., Angelini L., Lee K.-A., McGowan S.J., et al. An aged immune system drives senescence and ageing of solid organs. Nature. 2021;594:100–105. doi: 10.1038/s41586-021-03547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moliva J.I., Rajaram M.V.S., Sidiki S., Sasindran S.J., Guirado E., Pan X.J., Wang S.H., Ross P., Lafuse W.P., Schlesinger L.S., et al. Molecular composition of the alveolar lining fluid in the aging lung. Age. 2014;36:9633. doi: 10.1007/s11357-014-9633-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong C.K., Smith C.A., Sakamoto K., Kaminski N., Koff J.L., Goldstein D.R., Weiss R. Aging impairs alveolar macrophage phagocytosis and increases influenza-induced mortality in mice. J. Immunol. 2017;199:1060–1068. doi: 10.4049/jimmunol.1700397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McQuattie-Pimentel A.C., Ren Z., Joshi N., Watanabe S., Stoeger T., Chi M., Lu Z., Sichizya L., Aillon R.P., Chen C.-I., et al. The lung microenvironment shapes a dysfunctional response of alveolar macrophages in aging. J. Clin. Invest. 2021;131 doi: 10.1172/JCI140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Speck N.A., Gilliland D.G. Core-binding factors in haematopoiesis and leukaemia. Nat. Rev. Cancer. 2002;2:502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 24.Wang Q., Stacy T., Miller J.D., Lewis A.F., Gu T.L., Huang X., Bushweller J.H., Bories J.C., Alt F.W., Ryan G., et al. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 25.Kundu M., Chen A., Anderson S., Kirby M., Xu L., Castilla L.H., Bodine D., Liu P.P. Role of Cbfb in hematopoiesis and perturbations resulting from expression of the leukemogenic fusion gene Cbfb-MYH11. Blood. 2002;100:2449–2456. doi: 10.1182/blood-2002-04-1064. [DOI] [PubMed] [Google Scholar]
- 26.Kundu M., Liu P.P. CBFβ is involved in maturation of all lineages of hematopoietic cells during embryogenesis except erythroid. Blood Cells Mol. Dis. 2003;30:164–169. doi: 10.1016/S1079-9796(03)00030-5. [DOI] [PubMed] [Google Scholar]
- 27.Murakami K., Sasaki H., Nishiyama A., Kurotaki D., Kawase W., Ban T., Nakabayashi J., Kanzaki S., Sekita Y., Nakajima H., et al. A RUNX–CBFβ-driven enhancer directs the Irf8 dose-dependent lineage choice between DCs and monocytes. Nat. Immunol. 2021;22:301–311. doi: 10.1038/s41590-021-00871-y. [DOI] [PubMed] [Google Scholar]
- 28.Cardani A., Boulton A., Kim T.S., Braciale T.J. Alveolar macrophages prevent lethal influenza pneumonia by inhibiting infection of type-1 alveolar epithelial cells. PLoS Pathog. 2017;13:10061400–e1006225. doi: 10.1371/journal.ppat.1006140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mould K.J., Moore C.M., McManus S.A., McCubbrey A.L., McClendon J.D., Griesmer C.L., Henson P.M., Janssen W.J. Airspace macrophages and monocytes exist in transcriptionally distinct subsets in healthy adults. Am. J. Respir. Crit. Care Med. 2021;203:946–956. doi: 10.1164/RCCM.202005-1989OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aegerter H., Lambrecht B.N., Jakubzick C.V. Biology of lung macrophages in health and disease. Immunity. 2022;55:1564–1580. doi: 10.1016/j.immuni.2022.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong D.J., Liu H., Ridky T.W., Cassarino D., Segal E., Chang H.Y. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kowalczyk M.S., Tirosh I., Heckl D., Rao T.N., Dixit A., Haas B.J., Schneider R.K., Wagers A.J., Ebert B.L., Regev A. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015;25:1860–1872. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heng T.S.P., Painter M.W., Immunological Genome Project Consortium. Project G. The Immunological Genome Project : networks of gene expression in immune cells. Nat. Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 36.Zhu B., Wu Y., Huang S., Zhang R., Son Y.M., Li C., Cheon I.S., Gao X., Wang M., Chen Y., et al. Uncoupling of macrophage inflammation from self- renewal modulates host recovery from respiratory viral infection Uncoupling of macrophage inflammation from self-renewal modulates host recovery from respiratory viral infection. Immunity. 2021;54:1200–1218.e9. doi: 10.1016/j.immuni.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Angelidis I., Simon L.M., Fernandez I.E., Strunz M., Mayr C.H., Greiffo F.R., Tsitsiridis G., Ansari M., Graf E., Strom T.M., et al. An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat. Commun. 2019;10:963. doi: 10.1038/s41467-019-08831-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mootha V.K., Lindgren C.M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 39.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chatsirisupachai K., Palmer D., Ferreira S., de Magalhães J.P. A human tissue-specific transcriptomic analysis reveals a complex relationship between aging, cancer, and cellular senescence. Aging Cell. 2019;18:e13041. doi: 10.1111/acel.13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu H., Degen J.L., Sisson T.H., Hattori N., Moore B.B., Pandrangi R.G., Simon R.H., Drew A.F., Pandrangi R.G., Simon R.H., et al. Bleomycin-induced pulmonary fibrosis in fibrinogen-null mice Find the latest version : in fibrinogen-null mice. J. Clin. Invest. 2000;106:1341–1350. doi: 10.1172/JCI10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.García-Prieto E., González-López A., Cabrera S., Astudillo A., Gutiérrez-Fernández A., Fanjul-Fernandez M., Batalla-Solís E., Puente X.S., Fueyo A., López-Otín C., Albaiceta G.M. Resistance to bleomycin-induced lung fibrosis in MMP-8 deficient mice is mediated by interleukin-10. PLoS One. 2010;5:e13242. doi: 10.1371/journal.pone.0013242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang C., Liu G., Luckhardt T., Antony V., Zhou Y., Carter A.B., Thannickal V.J., Liu R.M. Serpine 1 induces alveolar type II cell senescence through activating p53-p21-Rb pathway in fibrotic lung disease. Aging Cell. 2017;16:1114–1124. doi: 10.1111/acel.12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khan S.S., Shah S.J., Klyachko E., Baldridge A.S., Eren M., Place A.T., Aviv A., Puterman E., Lloyd-Jones D.M., Heiman M., et al. A null mutation in SERPINE1 protects against biological aging in humans. Sci. Adv. 2017;3:eaao1617. doi: 10.1126/sciadv.aao1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samarakoon R., Higgins S.P., Higgins C.E., Higgins P.J. The TGF-β1/p53/PAI-1 signaling axis in vascular senescence: role of Caveolin-1. Biomolecules. 2019;9:341–412. doi: 10.3390/biom9080341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee B.Y., Han J.A., Im J.S., Morrone A., Johung K., Goodwin E.C., Kleijer W.J., DiMaio D., Hwang E.S. Senescence-associated β-galactosidase is lysosomal β-galactosidase. Aging Cell. 2006;5:187–195. doi: 10.1111/j.1474-9726.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- 47.Ivanov A., Pawlikowski J., Manoharan I., van Tuyn J., Nelson D.M., Rai T.S., Shah P.P., Hewitt G., Korolchuk V.I., Passos J.F., et al. Lysosome-mediated processing of chromatin in senescence. J. Cell Biol. 2013;202:129–143. doi: 10.1083/jcb.201212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schafer M.J., White T.A., Iijima K., Haak A.J., Ligresti G., Atkinson E.J., Oberg A.L., Birch J., Salmonowicz H., Zhu Y., et al. Cellular senescence mediates fibrotic pulmonary disease. Nat. Commun. 2017;8:14532. doi: 10.1038/ncomms14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Calhoun C., Shivshankar P., Saker M., Sloane L.B., Livi C.B., Sharp Z.D., Orihuela C.J., Adnot S., White E.S., Richardson A., Le Saux C.J. Senescent cells contribute to the physiological remodeling of aged lungs. J. Gerontol. A Biol. Sci. Med. Sci. 2016;71:153–160. doi: 10.1093/gerona/glu241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee S., Islam M.N., Boostanpour K., Aran D., Jin G., Christenson S., Matthay M.A., Eckalbar W.L., DePianto D.J., Arron J.R., et al. Molecular programs of fibrotic change in aging human lung. Nat. Commun. 2021;12:6309. doi: 10.1038/s41467-021-26603-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Habermann A.C., Gutierrez A.J., Bui L.T., Yahn S.L., Winters N.I., Calvi C.L., Peter L., Chung M.-I., Taylor C.J., Jetter C., et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci. Adv. 2020;6:eaba1972. doi: 10.1126/sciadv.aba1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reyfman P.A., Walter J.M., Joshi N., Anekalla K.R., McQuattie-Pimentel A.C., Chiu S., Fernandez R., Akbarpour M., Chen C.I., Ren Z., et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2019;199:1517–1536. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morse C., Tabib T., Sembrat J., Buschur K.L., Bittar H.T., Valenzi E., Jiang Y., Kass D.J., Gibson K., Chen W., et al. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur. Respir. J. 2019;54 doi: 10.1183/13993003.02441-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams T.S., Schupp J.C., Poli S., Ayaub E.A., Neumark N., Ahangari F., Chu S.G., Raby B.A., Deiuliis G., Januszyk M., et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci. Adv. 2020;6:eaba1983. doi: 10.1126/sciadv.aba1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gschwend J., Sherman S.P.M., Ridder F., Feng X., Liang H.E., Locksley R.M., Becher B., Schneider C. Alveolar macrophages rely on GM-CSF from alveolar epithelial type 2 cells before and after birth. J. Exp. Med. 2021;218 doi: 10.1084/jem.20210745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Westphalen K., Gusarova G.A., Islam M.N., Subramanian M., Cohen T.S., Prince A.S., Bhattacharya J. Sessile alveolar macrophages communicate with alveolar epithelium to modulate immunity. Nature. 2014;506:503–506. doi: 10.1038/nature12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Svedberg F.R., Brown S.L., Krauss M.Z., Campbell L., Sharpe C., Clausen M., Howell G.J., Clark H., Madsen J., Evans C.M., et al. The lung environment controls alveolar macrophage metabolism and responsiveness in type 2 inflammation. Nat. Immunol. 2019;20:571–580. doi: 10.1038/s41590-019-0352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Subramanian S., Busch C.J.L., Molawi K., Geirsdottir L., Maurizio J., Vargas Aguilar S., Belahbib H., Gimenez G., Yuda R.A.A., Burkon M., et al. Long-term culture-expanded alveolar macrophages restore their full epigenetic identity after transfer in vivo. Nat. Immunol. 2022;23:458–468. doi: 10.1038/s41590-022-01146-w. [DOI] [PubMed] [Google Scholar]
- 59.Lafuse W.P., Rajaram M.V.S., Wu Q., Moliva J.I., Torrelles J.B., Turner J., Schlesinger L.S. Identification of an increased alveolar macrophage subpopulation in old mice that displays unique inflammatory characteristics and is permissive to Mycobacterium tuberculosis infection. J. Immunol. 2019;203:2252–2264. doi: 10.4049/jimmunol.1900495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li F., Piattini F., Pohlmeier L., Feng Q., Rehrauer H., Kopf M. Monocyte-derived alveolar macrophages autonomously determine severe outcome of respiratory viral infection. Sci. Immunol. 2022;7:eabj5761. doi: 10.1126/sciimmunol.abj5761. [DOI] [PubMed] [Google Scholar]
- 61.Liu Z., Gu Y., Chakarov S., Bleriot C., Kwok I., Chen X., Shin A., Huang W., Dress R.J., Dutertre C.A., et al. Fate mapping via ms4a3-expression history traces monocyte-derived cells. Cell. 2019;178:1509–1525.e19. doi: 10.1016/j.cell.2019.08.009. [DOI] [PubMed] [Google Scholar]
- 62.Omori S., Wang T.-W., Johmura Y., Kanai T., Nakano Y., Kido T., Susaki E.A., Nakajima T., Shichino S., Ueha S., et al. Generation of a p16 reporter mouse and its use to characterize and target p16high cells in vivo. Cell Metabol. 2020;32:814–828.e6. doi: 10.1016/j.cmet.2020.09.006. [DOI] [PubMed] [Google Scholar]
- 63.Ronchi J.A., Figueira T.R., Ravagnani F.G., Oliveira H.C.F., Vercesi A.E., Castilho R.F. A spontaneous mutation in the nicotinamide nucleotide transhydrogenase gene of C57BL/6J mice results in mitochondrial redox abnormalities. Free Radic. Biol. Med. 2013;63:446–456. doi: 10.1016/j.freeradbiomed.2013.05.049. [DOI] [PubMed] [Google Scholar]
- 64.Martini H., Passos J.F. Cellular senescence: all roads lead to mitochondria. FEBS J. 2023;290:1186–1202. doi: 10.1111/febs.16361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.López-Otín C., Blasco M.A., Partridge L., Serrano M., Kroemer G. Hallmarks of aging: an expanding universe. Cell. 2023;186:243–278. doi: 10.1016/j.cell.2022.11.001. [DOI] [PubMed] [Google Scholar]
- 66.Ogrodnik M., Salmonowicz H., Jurk D., Passos J.F. Expansion and cell-cycle arrest: common denominators of cellular senescence. Trends Biochem. Sci. 2019;44:996–1008. doi: 10.1016/j.tibs.2019.06.011. [DOI] [PubMed] [Google Scholar]
- 67.Amend S.R., Valkenburg K.C., Pienta K.J. Murine hind limb long bone dissection and bone marrow isolation. J. Vis. Exp. 2016;110 doi: 10.3791/53936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNAseq was deposited in GEO (https://www.ncbi.nlm.nih.gov/geo/) with accession number GSE205982.
This paper did not generate any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.