Summary
Background
COVID-19 vaccines have been critical for protection against severe disease following infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) but gaps remain in our understanding of the immune responses that contribute to controlling subclinical and mild infections.
Methods
Vaccinated, active-duty US military service members were enrolled in a non-interventional, minimal-risk, observational study starting in May, 2021. Clinical data, serum, and saliva samples were collected from study participants and were used to characterise the humoral immune responses to vaccination and to assess its impact on clinical and subclinical infections, as well as virologic outcomes of breakthrough infections (BTI) including viral load and infection duration.
Findings
The majority of VIRAMP participants had received the Pfizer COVID-19 vaccine and by January, 2022, N = 149 had a BTI. The median BTI duration (PCR+ days) was 4 days and the interquartile range was 1–8 days. Participants that were nucleocapsid seropositive prior to their BTI had significantly higher levels of binding and functional antibodies to the spike protein, shorter median duration of infections, and lower median peak viral loads compared to seronegative participants. Furthermore, levels of neutralising antibody, ACE2 blocking activity, and spike-specific IgA measured prior to BTI also correlated with the duration of infection.
Interpretation
We extended previous findings and demonstrate that a subset of vaccine-induced humoral immune responses, along with nucleocapsid serostatus are associated with control of SARS-CoV-2 breakthrough infections in the upper airways.
Funding
This work was funded by the DoD Joint Program Executive Office for Chemical, Biological, Radiological and Nuclear Defense (JPEO-CBRND) in collaboration with the Defense Health Agency (DHA) COVID-19 funding initiative for the VIRAMP study.
Keywords: SARS-CoV-2, COVID-19 vaccine, Antibody responses, Breakthrough infection duration
Research in context.
Evidence before this study
There are currently four COVID-19 vaccines authorized for emergency use or approved by the United States Food and Drug Administration. The principle immunogen delivered by all four vaccines is the spike protein. Global transmission and rapid virus evolution have resulted in the emergence of SARS-CoV-2 variants of concern with spike protein mutations that improve virus fitness and can escape vaccine immunity. Neutralising antibodies are the primary immune correlate of protection against SARS-CoV-2 but studies have also highlighted the importance of other antibody mediated effector functions and cellular immune responses for protection. Several studies have indicated that COVID-19 vaccines provide durable protection against severe disease but are less effective at preventing infection. However, multiple studies have reported that vaccinated individuals have reduced viral loads and increased rates of virus clearance compared to unvaccinated individuals. The nucleocapsid protein is not a component of authorized vaccines but prior studies have reported a lower incidence of disease in nucleocapsid antibody seropositive individuals. Despite a growing body of literature covering SARS-CoV-2 breakthrough infections and correlates of protection, gaps still remain in our understanding of the immune responses that are critical for controlling and eliminating subclinical and mild infections.
Added value of this study
The Vaccine Effectiveness and Immune Response of SARS-CoV-2 Vaccines in Active Military Personnel (VIRAMP) study is an observational study that started enrolling active-duty military service members in May, 2021. Clinical data and prospective longitudinal sampling of serum and saliva were used to characterise the humoral immune responses to vaccination and to assess protection from confirmed clinical and subclinical infections. Additionally, virologic outcomes of breakthrough infections including viral load and infection duration were captured. This study design allowed for comprehensive characterisation of humoral immune responses prior to breakthrough infections. Furthermore, frequent prospective sampling of saliva and PCR testing allowed for early detection of infections and good resolution of infection duration and peak viral load. These virologic outcomes were compared and correlated with a large panel of binding, neutralising, and other functional antibody responses to better delineate the humoral immune responses that are important for controlling SARS-CoV-2 infections.
Implications of all the available evidence
Our results from VIRAMP are in agreement with results from other clinical studies that have characterised breakthrough infections in vaccinated and unvaccinated cohorts. We extended previous findings that COVID-19 vaccination increases the rate of virus clearance during breakthrough infections by defining specific humoral immune responses that correlate with this function. We also expand on previous observations that nucleocapsid antibody serostatus prior to breakthrough infection correlates with disease protection by showing that seropositive individuals have a significantly increased rate of virus clearance and lower viral loads compared to seronegatives. Additional research will be required to determine if immune responses to the nucleocapsid protein itself are important for protection, or whether these responses are a sentinel for other protective cellular and humoral responses elicited by natural infection. Inclusion of non-spike viral proteins is likely to be important for next generation COVID-19 vaccines.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19) and was declared a global pandemic in 2020. There are currently four COVID-19 vaccines authorized for emergency use or approved by the United States Food and Drug Administration (FDA) (https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/covid-19-vaccines, accessed in January, 2023). The vaccines include Pfizer-BioNTech COVID-19 vaccine (BNT162b2, now marketed as Comirnaty), Moderna COVID-19 vaccine (mRNA-1273, now marketed as Spikevax), Janssen COVID-19 vaccine (Ad26.COV2.S), and Novavax COVID-19 vaccine (NVX-CoV2373). The immunogen delivered by all four vaccines is the spike (S) protein of SARS-CoV-2.
Neutralising antibodies are the primary immune correlate of protection against SARS-CoV-2 and both authentic virus and pseudovirus neutralisation assays have been widely used to assess COVID-19 vaccines.1,2 Animal models have highlighted a role for other antibody mediated effector functions in control of viral replication mediated by vaccines and neutralising monoclonal antibodies.3, 4, 5, 6, 7, 8 Cellular immunity, including CD4+ T cells that augment antibody responses and CD8+ T cells that help control virus replication by killing infected cells also contribute to protection from SARS-CoV-2 infections.9
Continuous global transmission of SARS-CoV-2 has resulted in a large number of mutations that have accumulated throughout the virus genome, including many in the S protein gene that can improve transmission and mediate immune-escape. This process has given rise to variants of concern including Alpha, Beta, Delta, and most recently a constellation of Omicron lineages.10, 11, 12, 13 Authorized COVID-19 vaccines have provided better and more durable effectiveness against severe disease than against infection and mild symptoms, particularly with the more genetically and antigenically divergent Omicron variants.14, 15, 16 Vaccine efficacy against infection and symptomatic disease decreases substantially by 6-months post full vaccination.16 COVID-19 vaccines have been shown to be effective at reducing viral load of breakthrough infections (BTI) in a relatively short time-period of up to 3 months post vaccination compared to unvaccinated individuals.17, 18, 19 Furthermore, multiple studies have examined the duration of BTI in vaccinated and unvaccinated individuals and have demonstrated that vaccination increases the rate of virus clearance.20, 21, 22, 23, 24 However, these studies did not characterise the pre-BTI immune responses that contributed to clearing the virus more rapidly.
VIRAMP (Vaccine Effectiveness and Immune Response of SARS-CoV-2 Vaccines in Active Military Personnel) is an observational study that started enrolling active-duty service members in May of 2021. Clinical data, serum, and saliva samples were collected from study participants in order to characterise the humoral immune responses to vaccination and assess protection against confirmed clinical and subclinical infections, as well as virologic outcomes of BTI including viral load and infection duration. In this report, we focused on characterizing the humoral immune responses of VIRAMP participants that had a BTI. Serum samples collected pre-BTI at enrollment and 6-months post COVID-19 vaccination were interrogated by a panel of binding and functional antibody assays and comparisons were made with virologic outcome. The results of this study contribute to our understanding of the types of humoral immunity that modulate the course of SARS-CoV-2 breakthrough infections.
Methods
Study design
This was a non-interventional, minimal-risk, observational study to document clinical, virologic, and immunologic experience over a 12–13 month interval in approximately 1000 healthy adults who received at least one dose of a COVID-19 vaccine under emergency use authorization (EUA) or Biologic License Application (BLA) authorization. Volunteers were recruited starting in May of 2021 from the military population at US Joint Base San Antonio (JBSA) and Fort Hood, Texas and study activities were conducted at Brooke Army Medical Center (BAMC), Wilford Hall Ambulatory Surgical Center (WHASC), and Fort Hood. Enrolled subjects were required to self-report vaccination history, and were asked to provide a weekly report (via brief email questionnaire) of health status, with focus on febrile and/or respiratory signs and symptoms. The Enrolled Population included subjects who met all the inclusion and exclusion criteria. Inclusion criteria were active-duty service members that were 18–65 years of age, assigned at JBSA, and had received a COVID-19 vaccination. Exclusion criteria were evidence of infection at time of enrollment, known allergy/adverse reaction to study materials/procedures for blood and saliva collection, current participation in any study involving treatment or prophylaxis for COVID-19, SARS, MERS, and anticipated military separation or deployment within 6 months of enrollment. The Per-Protocol Population consisted of the enrolled population excluding subjects that experienced at least one major protocol deviation.
Sample size
Enrollment of 1000 participants was estimated to result in the identification of approximately 70 SARS-CoV-2 breakthrough infections over a 1 year time-period. This was estimated using 80% COVID-19 vaccine efficacy and a community point prevalence of 1%.
Ethics
We adhered to the policies for protection of human subjects, as prescribed in Army Regulation (AR) 70-25. This study was approved by the US Joint Base San Antonio (JBSA), Brooke Army Medical Center (BAMC) and Wilford Hall Ambulatory Surgical Center (WHASC) IRB (reference no. C.2021.067/eIRB no. 947879). The Walter Reed Army Institute of Research (WRAIR) IRB deferred to the forementioned sites and approved a non-human subjects determination. Informed consent was obtained from participants (or his/her legally authorized representative).
Saliva collection and testing
Saliva was self-collected according to the COVID-19 at-home saliva testing instructions (vaulthealth.com/covid) by study subjects twice weekly to monitor for SARS-CoV-2 infection. Saliva samples were shipped to Vault Health per kit instructions and were divided into 3 aliquots that were frozen at −80 °C. SARS-CoV-2 RNA was extracted and purified from saliva using Chemagen kits and a Chemagic 360 instrument (PerkinElmer). One aliquot was pooled with aliquots collected from other study subjects for batch testing using the Infinity BiologiX TaqPath SARS-CoV-2 Assay (https://www.fda.gov/media/137776/download). The remaining saliva aliquots were used for testing of individual samples in positive saliva pools using a quantitative Infinity BiologiX TaqPath SARS-CoV-2 Assay and by Illumina next-generation sequencing (NGS) at Vault Health. For saliva sampling triggered by illness or COVID-19 exposure, samples were shipped to Vault Health per kit instructions and were tested individually by RT-qPCR and NGS. The SARS-CoV-2 variant and lineage determinations were conducted by Vault Health.
Venous blood collection and testing
20 mL of blood in serum separator tubes (SST) were obtained at enrollment, and at 6 and 12 months following the last COVID-19 vaccination dose received. The blood was processed for serum separation, aliquots made, and stored at −20 °C until shipment to WRAIR or the Tasso Care laboratory for testing using the Elecsys Anti-SARS-CoV-2 immunoassay for the qualitative detection of antibodies against SARS-CoV-2 (Roche, cat. no. 09203095190) and was done according to the manufacturer’s instructions.
Tasso capillary blood collection and testing
Two Tasso SSTs (2 × 300 μl) per time-point and total volume of 600 μl of blood samples were self-collected by study subjects every month using a Tasso self-collection system (Tasso SST and/or Tasso+ device) according to the manufacturer’s instructions (https://www.tassoinc.com/). The samples were shipped according to kit instructions to the Tasso Care laboratory and were tested using the Elecsys Anti-SARS-CoV-2 immunoassay (Roche, cat. no. 09203095190) according to the manufacturer’s instructions.
Questionnaires
Were administered monthly to assess social and community exposure risk and weekly to assess for symptoms and exposure. Individuals with exposure to persons with documented SARS-CoV-2 infection, or who became ill and/or tested positive for SARS-CoV-2 locally at the medical treatment facility, were asked to complete a more comprehensive questionnaire detailing clinical symptoms and exposure history, and to collect additional saliva specimens every other day for 10 days after exposure/illness onset to assess SARS-CoV-2 viral load.
SARS-CoV-2 pseudovirus neutralisation
This assay has been described previously.25 The S expression plasmid sequences for SARS-CoV-2 were codon optimized and modified to remove the last 18 amino acids of the cytoplasmic tail to improve S incorporation into pseudovirions (PSV). PSV were produced by cotransfection of HEK293T/17 cells (RRID:CVCL_1926; ATCC cat. no. CRL-11268) with either a SARS-CoV-2 S plasmid, derived from the Wuhan-Hu-1 genome sequence (GenBank accession number MN908947.3) and an HIV-1 pNL4-3 luciferase reporter plasmid (pNL4-3.Luc.R-E-, NIH AIDS Reagent Program, cat. no. ARP-3418). S expression plasmids for SARS-CoV-2 variants (GenScript) were similarly codon optimized and modified, and included the following mutations: B.1.617.2 or Delta (E156G, D614G, P681R, T19R, T478K, L452R, D950N, 157–158 del), B.1.1.529 or Omicron (A67V, 69–70 del, T95I, G142D, 143–145 del, N211I, 212 del, G339D, S371L, S373P, S375F, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, N969K, Q954H, L981F). Infectivity and neutralisation titers were determined using ACE2-expressing HEK293 target cells (Integral Molecular, cat. no. C-HA102). Virions pseudotyped with the vesicular stomatitis virus G protein were used as a nonspecific control. Test sera were diluted 1:40 in growth medium and serially diluted; then 25 μl per well was added to a white 96-well plate. An equal volume of diluted PSV was added to each well, and plates were incubated for 1 h at 37 °C. Target cells were added to each well (40,000 cells per well), and plates were incubated for an additional 48 h. Relative light units (RLU) were measured with the Synergy Neo2 Hybrid Multi-Mode Microplate Reader (Agilent BioTek) using the Bright-Glo Luciferase Assay System (Promega, cat. no. E2650). Neutralisation dose–response curves were fitted by nonlinear regression using GraphPad Prism. Final titers are reported as the reciprocal of the serum dilution necessary to achieve 50%, 80%, or 90% inhibition of SARS-CoV-2 infectivity (ID50, ID80, and ID90, respectively). Assay equivalency was established by participation in the SARS-CoV-2 Neutralising Assay Concordance Survey run by the Virology Quality Assurance Program and External Quality Assurance Program Oversight Laboratory at the Duke Human Vaccine Institute.
Opsonization assay
SARS-CoV-2 spike-expressing FreeStyle™ 293-F (RRID:CVCL_D603; Invitrogen cat. no. R79007) cells were generated by transfection with linearized plasmid (pcDNA3.1) encoding codon-optimized full-length SARS-CoV-2 spike protein (GenScript) matching the amino acid sequence having mutations E156G, D614G, P681R, T19R, T478K, L452R, D950N, and 157–158 del for Delta. Stable transfectants were single-cell sorted and selected to obtain a high-level Spike surface expressing clone (293F-Spike-Delta). 293F-Spike-Delta cells were incubated with 100 μl of 4-fold serial dilutions of plasma starting at 100-fold for 30 min at 4 °C. Cells were washed twice and stained with anti-human IgG PE, anti-human IgM Alexa Fluor 647, and anti-human IgA FITC (Southern Biotech, Birmingham, AL, USA, cat. nos. 2040-09, 2020-31, and 2050-02). Cells were then fixed with 4% formaldehyde solution (Tousimis, Rockville, MD, USA, cat. no. 1008B) and fluorescence was evaluated on a LSRII (BD Bioscience).
Antibody-dependent cellular phagocytosis (ADCP) assay
This assay was described previously.26 Briefly, biotinylated SARS-CoV-2 Delta spike trimer (ECD; SinoBiological, Beijing, China, cat. no. 40589-V08H10) was incubated with yellow-green neutravidin-fluorescent beads (Molecular Probes, Eugene, OR, USA, cat. no. F8776) for 2 h at 37 °C. 10 μl of a 100-fold dilution of beads–protein was incubated 2 h at 37 °C with 100 μl of diluted plasma (900-fold) before addition of 25,000 cells per well THP-1 cells (RRID:CVCL_0006; Millipore Sigma, Burlington, MA, USA, cat. no. 88081201). After 19 h incubation at 37 °C, the cells were fixed with 2% formaldehyde solution (Tousimis, Rockville MD USA, cat. no. 1008B) and fluorescence was evaluated on a LSRII (BD Bioscience). The phagocytic score was calculated by multiplying the percentage of bead-positive cells by the geometric mean fluorescence intensity (MFI) of the bead-positive cells and dividing by 104.
Antibody-dependent complement deposition (ADCD) assay
This assay was described previously.27 Briefly, 293F-Spike-Delta cells, described above, were incubated with 100 μl of diluted plasma (10-fold) for 30 min at 4 °C. Cells were washed twice and resuspended in R10 media. Lyophilized guinea pig complement (CL4051, Cedarlane, Burlington, Canada, cat. no. CL4051) was reconstituted per the manufacturer’s instructions in 1 mL cold water and centrifuged to remove aggregates for 5 min at 4 °C. Cells were washed with PBS and resuspended in 200 μl of guinea pig complement, which was prepared at a 1:50 dilution in Gelatin Veronal Buffer with Ca2+ and Mg2+ (Sigma-Aldrich, St. Louis, MO, USA, cat. no. G6514). After incubation at 37 °C for 20 min, cells were washed in PBS 15 mM EDTA (ThermoFisher Scientific Baltics UAB, Vilnius, Lithuania, cat. no. AM9260G) and stained with an anti-guinea pig complement C3 FITC (polyclonal, MPBiomedicals, Solon, OH, USA, cat. no. 0855385). Cells were fixed with 4% formaldehyde solution (Tousimis, Rockville, MD, USA, cat. no. 1008B) and fluorescence was evaluated on a LSRII (BD Bioscience).
Antibody-dependent cellular cytotoxicity (ADCC) CD16 reporter assay
SARS-CoV-2 spike-expressing CEM-NKR cells (RRID:CVCL_X622; NIH AIDS Reagent Program cat. no. ARP-458) were generated by transfection with linearized plasmid (pcDNA3.1) encoding codon-optimized full-length SARS-CoV-2 spike protein (GenScript) matching the amino acid sequence having mutations E156G, D614G, P681R, T19R, T478K, L452R, D950N, and 157–158 del for Delta. Stable transfectants were single-cell sorted and selected to obtain a high-level spike surface expressing clone (CEM-Spike-Delta). CEM-Spike-Delta cells were plated at 100,000 per well in round bottom 96-well plates and incubated with 100 μl of diluted plasma (100-fold) for 30 min at 4 °C. Cells were washed and 200,000 Jurkat-Lucia NFAT-CD16 cells (RRID:CVCL_A7ZT; Invivogen, San Diego, CA, USA, cat. no. jktl-nfat-cd16) were added to each well in 100 μl of IMDM (Gibco, Burlington, ON, Canada, cat. no. 12440-053) 10% FBS. The cells were then centrifuge for 1 min at low speed and co-cultured for 24 h at 37 °C. 50 μl of Quanti-Luc (Invivogen, cat. no. rep-qlc2) was added to 20 μl of co-culture and luminescence was measured immediately on a luminometer (2104 Multilabel reader, PerkinElmer).
Binding and inhibitory antibodies
SARS-CoV-2–specific binding IgG and IgA antibodies as well as ACE2-inhibitory antibodies were measured using MULTI-SPOT 96-well V-PLEX SARS-Cov-2 Plate 13 kit from MSD (MSD, IgG, cat. no. K15463U; IgA, cat. no. K15465U; ACE2, cat. no. K15466U).25 The three respective kits contained all of the required reagents. Multiplex wells were coated with SARS-CoV-2 spike antigens from different variants (WA; P2; B1.617.1; B1.617.2; B1.617.3; B.1.617; P1; B.1.1.7; B.1.351 and B.526.1); at a concentration of 200–400 ng/mL. Plates were blocked with MSD Blocker A buffer for 1 h at RT while shaking at 700 rpm. Plates were washed with wash buffer before the addition of samples, reference standard and controls. Serum samples were diluted 1:1000 to 1:100,000 for IgG detection and 1:100–1:10,000 for IgA detection. Plates were incubated for 2 h at RT while shaking at 700 rpm, and then washed. MSD SULFO-TAG (anti-IgG or anti-IgA) antibody was added to each well. Plates were incubated for 1 h at RT with shaking at 700 rpm and washed, and then MSD GOLD Read buffer B was added to each well. Plates were read by the MESO SECTOR S600 Reader. IgG or IgA concentration was calculated using DISCOVERY WORKBENCH MSD Software and reported as binding antibody units per milliliter (BAU/mL). For antibodies that block S binding to ACE2, antigen-coated plates were blocked and washed as described above. Assay calibrator and samples were diluted at 1:25 to 1:1000 in MSD Diluent buffer, then added to the wells. Plates were incubated for 1 h at RT while shaking at 700 rpm. ACE2 protein conjugated with MSD SULFO-TAG was added, and plates were incubated for 1 h at RT while shaking at 700 rpm and washed and read as described above. ACE2 inhibitory antibody was reported as arbitrary units per milliliter (AU/mL).
Statistical analyses
All statistical analyses were performed using GraphPad Prism 8.1.0. Comparisons among groups were done using a Mann–Whitney test for unpaired groups, a Wilcoxon test for paired groups, and for three or more groups a Kruskal–Wallis test with Dunn’s correction for multiple comparisons was done. Spearman correlations were performed. Age and body-mass index were not associated with virologic outcomes by Spearman correlation and stratification of virologic outcome by sex yielded similar medians and interquartile ranges (IQR), therefore, the analyses did not adjust for these factors. The type of statistical test used and the number of participants is indicated in all Figure legends. Participants with data values missing (e.g., a sample was not available for testing) for specific assays were excluded from the corresponding analyses. Medians and IQR (Q1–Q3) are reported where applicable.
Role of funders
The DoD Joint Program Executive Office for Chemical, Biological, Radiological and Nuclear Defense (JPEO-CBRND) had a role in the study design; collection, analysis, and interpretation of data; writing of the manuscript; and in the decision to publish.
Results
Study population
VIRAMP is an observational study being conducted at JBSA that started enrolling participants in May of 2021 and by the end of January of 2022 there were a total of 889 active duty service members that had been enrolled in the study for at least two weeks. Of these 889 participants, N = 746 (83.9%) received Pfizer vaccine, N = 32 (3.6%) received Moderna vaccine, N = 12 (1.3%) received AstraZeneca vaccine, N = 10 (1.1%) received Janssen vaccine, and N = 89 (10.0%) were vaccinated but did not report a manufacturer. Clinical data, serum and saliva samples were collected from study participants in order to characterise the humoral immune responses elicited by vaccination and to assess protection against confirmed clinical and subclinical infections, as well as virologic outcomes of infection. There were N = 149 participants (16.8% of the study population) that experienced a BTI after enrollment, in a 6-month period between July 2021 and January 2022. The characteristics of these participants are listed in Table 1. The median age of participants with a BTI was 36 years (IQR: 29–41 years) and this was moderately higher than the median age of the overall cohort (median = 32 years, IQR: 27–38 years). The majority of participants with a BTI were male (N = 97, 65.1%), indicated their race to be white (N = 105, 70.5%), and received the Pfizer COVID-19 vaccine (N = 130, 87.2%). This matched the overall cohort that was majority male (N = 557, 62.7%) and indicated their race to be white (N = 622, 70.0%). The median time between first and second dose of the primary vaccination series, if applicable, for participants with a BTI was 21 days (IQR: 21–24 days) and this was similar to the overall cohort (median = 21 days, IQR: 21–25 days). Only 27/149 (18.1%) of participants with a BTI had received a booster vaccination by January 2022, and the majority (26/27, 96.3%) received a Pfizer COVID-19 vaccine booster. This was similar to the overall cohort with 178/889 (20.0%) participants that received a booster vaccination and 173/178 (97.2%) received a Pfizer COVID-19 vaccine booster by the end of January 2022. Roughly half of participants with a BTI (N = 71, 47.6%) were healthcare workers and the remainder (N = 78, 52.3%) had other occupations including administrative support, education, training, pilot/aircrew, public health, veterinary medicine, first responder, security forces, communications, and intelligence. The overall cohort was 53.0% healthcare workers (N = 471) and 47% (N = 418) had other occupations.
Table 1.
Characteristics of study participants that had a breakthrough infection (N = 149).
| Sex | N | % |
|---|---|---|
| Male | 97 | 65.1 |
| Median | IQR | |
|---|---|---|
| Age (years) | 36 | 29–41 |
| N | % | |
|---|---|---|
| Race | ||
| American Indian or Alaska Native | 1 | 0.7 |
| Asian | 11 | 7.4 |
| Black or African American | 22 | 14.8 |
| Native Hawaiian or other Pacific Islander | 2 | 1.3 |
| White | 105 | 70.5 |
| Other | 8 | 5.4 |
| Primary vaccination series | ||
| Pfizer vaccine | 130 | 87.2 |
| Moderna vaccine | 4 | 2.7 |
| AstraZeneca vaccine | 3 | 2.0 |
| Johnson and Johnson/Janssen | 3 | 2.0 |
| Othera | 9 | 6.0 |
| Total | 149 | 100.0 |
| Booster vaccination | ||
| Pfizer vaccine | 26 | 17.4 |
| Moderna vaccine | 1 | 0.7 |
| Total | 27 | 18.1 |
Eight volunteers did not provide the manufacturer of the vaccine that they received.
One volunteer received a 1st dose of Pfizer vaccine and a 2nd dose of Moderna vaccine.
Breakthrough infection characteristics
None of the N = 149 participants with a BTI were hospitalized but clinical symptoms were captured in questionnaires. The most common symptoms, in descending order of frequency, were dry cough (N = 48, 32.2%), fatigue (N = 34, 22.8%), sore throat (N = 32, 21.5%), malaise (N = 30, 20.1%), headache (N = 22, 14.8%), muscle aches (N = 22, 14.8%), shortness of breath or difficulty breathing (N = 11, 7.4%), loss of taste or smell (N = 9, 6.0%), diarrhea (N = 5, 3.4%), and nausea or vomiting (N = 5, 3.4%). A total of N = 97 (65.1%) reported COVID-19 symptoms, while N = 48 (32.2%) reported no COVID-19 symptoms, and N = 4 (2.7%) did not report this information.
There were N = 34 (22.8%) of BTI that had sufficient genome sequence coverage to identify the SARS-CoV-2 variant. There were N = 13 identified as Delta (lineages AY.103 [N = 1], AY.12 [N = 4], AY.3 [N = 2], AY.39 [N = 1], AY.4 [N = 1], AY.44 [N = 1], and B.1.617.2 [N = 3]) and N = 21 identified as Omicron (lineages BA.1 [N = 10] and BA.1.1 [N = 11]). The Delta BTIs occurred from July to November, 2021 and the Omicron BTIs occurred in December, 2021 and January, 2022.
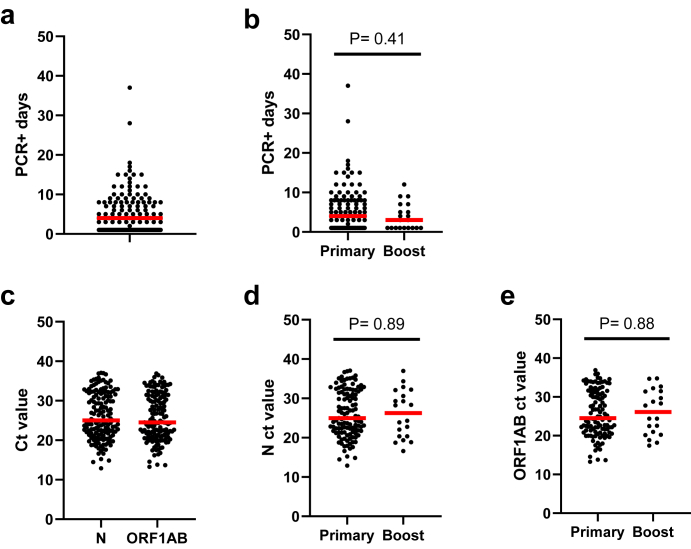
The median duration (PCR+ days) of BTIs in this population (N = 149) was 4 days and the IQR was 1–8 days (Fig. 1a). The duration of infection among participants that had completed a primary vaccination series and received two vaccine doses (Pfizer or Moderna) or one Janssen dose (N = 117) was a median of 4 days and those that received a booster third vaccine dose of Pfizer or Moderna (N = 20) was similar with a median of 3 days (Fig. 1b). There were N = 12 participants excluded from these analyses because they had a BTI before their second dose of vaccine. The peak viral load was also examined using the lowest measured N and ORF1AB gene ct values (Fig. 1c). The N gene median ct value was 25.0 and the ORF1AB gene median ct value was 24.5. There was a very strong correlation between N and ORF1AB ct values (Supplemental Figure S1a). Peak BTI viral load among participants that had completed a primary vaccination series (N ct median = 25.0, ORF1AB ct median = 24.5) was similar to those that received a booster dose (median N ct = 26.3, median ORF1AB ct = 26.1) (Fig. 1d and e). We also examined the relationship between the duration of infection and peak viral load and there were weak to moderate, negative correlations between N or ORF1AB ct values and the duration of infection, with longer duration infections also having higher peak viral loads (Supplemental Figure S1b and S1c). However, we did not observe correlations when comparing the duration of infection or peak viral load (N or ORF1AB ct values) with time (number of days) post-vaccination with our dataset (Supplemental Figure S1d–S1f).
Fig. 1.
Viral load and duration of breakthrough infections. Quantitative PCR for N and ORF1AB genes was done on saliva samples collected during breakthrough infections. Panel a) shows all participants (N = 149) duration of breakthrough infection calculated by the number of contiguous days they were qPCR positive (+). Panel b) shows a comparison of the duration of breakthrough infection in participants post primary vaccination series (N = 117) and post a booster vaccination (N = 20). Panel c) shows the viral load (Ct value) for N and ORF1AB gene targets for all participants (N = 149). Panels d and e) show a viral load comparison among participants post primary vaccination series (N = 117) and post a booster vaccination (N = 20) for N and ORF1AB gene targets, respectively. The red bar in all graphs is the median and statistical comparisons between groups were done using a Mann–Whitney test. P values are indicated for each comparison.
Enrollment sample antibody responses
A range of neutralising, binding, and other functional antibody responses against SARS-CoV-2 wild-type, Delta, and Omicron were detected in participants at enrollment, prior to their BTI (Supplemental Figure S2). Neutralising antibody was measured for N = 141 participants using a pseudovirus neutralisation assay. There were 82.3%, 70.9% and 58.2% of participants that had a detectable ID50, ID80, and ID90 values, respectively, to wild-type. Similarly, 86.5%, 72.3%, and 63.1% of participants had detectable ID50, ID80, and ID90 values, respectively, to Delta. Whereas, only 13.5%, 7.8%, and 5.7% of participants had detectable ID50, ID80, and ID90 values, respectively, to Omicron. The median neutralising antibody ID50, ID80, and ID90 titers for wild-type were 653, 160, and 74, respectively, and were higher than those for the Delta variant, which were 175 (3.7-fold lower), <40 (>32-fold lower), and <40 (>14.8-fold lower), respectively (Supplemental Figure S2a–S2c). The median neutralising antibody ID50, ID80, and ID90 for the Omicron variant were <40 and were substantially lower than for wild-type. ACE2 blocking (N = 125) and spike reactive IgG (N = 126) and IgA (N = 126) responses were measured by MSD. The median ACE2 blocking was similar for wild-type (11.8 AU/mL) and Delta (12.2 AU/mL) (Supplemental Figure S2d). The median spike reactive IgG BAU/mL for wild-type (556 BAU/mL) was moderately higher than Delta (282 BAU/mL) (Supplemental Figure S2e). The median spike reactive IgA BAU/mL for wild-type (169 BAU/mL) was similar to Delta (128 BAU/mL) (Supplemental Figure S2f). Several different binding and functional antibody responses were also measured against the Delta variant spike-expressing HEK293F cells, including ADCP (N = 135), ADCD (N = 134), ADCC (N = 119), IgG MFI (1:100 dilution) (N = 119), IgA MFI (1:100 dilution) (N = 119), as well as IgG and IgA 50% opsonization (OP50) titers (N = 119) (Supplemental Figure S2g–k). Notably, the median IgG OP50 titer was 904-fold higher than the IgA OP50 titer. By comparison, the MSD assay binding of wild-type and Delta variant spike-reactive IgG was only a modest 3.3-fold and 2.2-fold higher than IgA, respectively.
We also examined correlations among the different enrollment antibody responses. Neutralising antibody is an accepted correlate of protection for SARS-CoV-2 therefore we looked at correlations between other binding and functional antibody response to wild-type and/or Delta and neutralising antibody ID50 values. All of the anti-spike IgG and IgA binding data and the opsonizing IgG and IgA, ACE2 blocking, ADCP, ADCD, and ADCC data had moderate to strong correlations with neutralising antibody ID50 values (Supplemental Figure S3). We also looked at the relationship between enrollment antibody responses and time post-vaccination. There were weak to moderate negative correlations, consistent with waning antibody levels to wild-type and/or Delta for neutralisation, ACE2 blocking, IgG, IgA, ADCD, and opsonizing IgG and IgA (Supplemental Figure S4).
Impact of nucleocapsid serostatus at enrollment
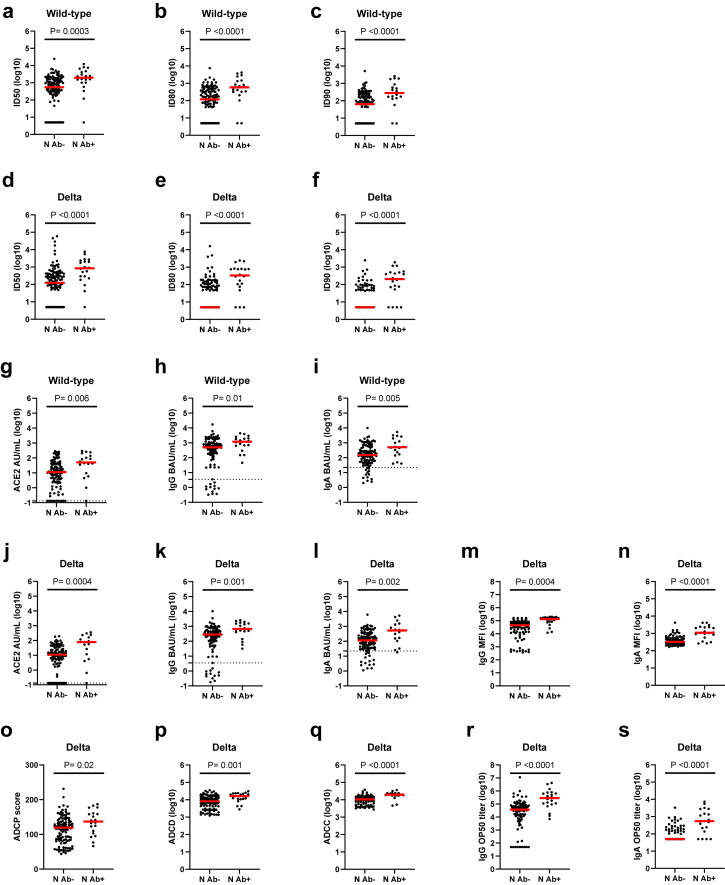
Of participants with a BTI, there were N = 23 that were nucleocapsid (N) antibody positive (Ab+) at enrollment, N = 121 that were N antibody negative (Ab−), and N = 5 that did not have a result for this test. We examined if there were differences in the levels of humoral immunity between these participants based on their N Ab serostatus at enrollment (Fig. 2). The ID50, ID80, and ID90 neutralising antibody levels to wild-type and Delta were significantly higher for participants that were N Ab+ at enrollment (Fig. 2a–f). The N Ab+ participants had median neutralising antibody levels that were 4–5 fold higher for wild-type and 7-fold to >67-fold higher for Delta compared to N Ab− participants. Differences between N Ab+ compared to N Ab− participants were also seen for the median ACE2 blocking (2–5 fold higher), anti-spike binding and opsonizing IgG and IgA (2–7 fold higher), ADCP (1.1-fold higher), ADCD (2-fold higher), ADCC (2-fold higher), and IgG and IgA OP50 titers (8–11 fold higher) (Fig. 2g–s). Thus, N Ab serostatus at enrollment had a significant impact on the levels of binding, neutralising, and other functional antibody responses.
Fig. 2.
Enrollment nucleocapsid serostatus and antibody levels. Comparisons between nucleocapsid (N) seronegative (Ab−) and seropositive (Ab+) participants are shown for a) ID50 wild-type (Ab− N = 117, Ab+ N = 20), b) ID80 wild-type (Ab− N = 117, Ab+ N = 20), c) ID90 wild-type (Ab− N = 117, Ab+ N = 20), d) ID50 Delta (Ab− N = 117, Ab+ N = 20), e) ID80 Delta (Ab− N = 117, Ab+ N = 20), f) ID90 Delta (Ab− N = 117, Ab+ N = 20), g) wild-type ACE2 blocking (Ab− N = 103, Ab+ N = 18), h) wild-type spike-reactive IgG (Ab− N = 104, Ab+ N = 18), i) wild-type spike-reactive IgA (Ab− N = 104, Ab+ N = 18), j) Delta ACE2 blocking (Ab− N = 103, Ab+ N = 18), k) Delta spike-reactive IgG (Ab− N = 104, Ab+ N = 18), l) Delta spike-reactive IgA (Ab− N = 104, Ab+ N = 18), m) Delta spike-expressing cell opsonizing IgG MFI (Ab− N = 95, Ab+ N = 20), n) Delta spike-expressing cell opsonizing IgA MFI (Ab− N = 95, Ab+ N = 20), o) Delta ADCP (Ab− N = 111, Ab+ N = 20), p) Delta ADCD (Ab− N = 110, Ab+ N = 20), q) Delta ADCC (Ab− N = 95, Ab+ N = 20), r) Delta spike-expressing cell 50% IgG opsonization titer (Ab− N = 95, Ab+ N = 20), and s) Delta spike-expressing cell 50% IgA opsonization titer (Ab− N = 95, Ab+ N = 20). The red bar in all graphs is the median and statistical comparisons between N Ab− and N Ab+ participants were done using a Mann–Whitney test and the P values are indicated for each comparison.
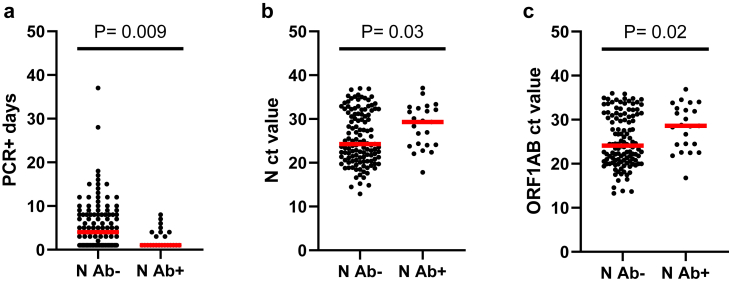
Next we examined the impact of enrollment N Ab serostatus on BTI virologic outcomes. The N Ab+ participants had a shorter median duration of infection of 1 day compared to N Ab− participants who had a median duration of infection of 4 days (Fig. 3a). Similarly, N Ab+ participants had significantly lower viral loads (i.e., higher ct values) with median N and ORF1AB ct values of 29.3 and 28.6, respectively, compared to median ct values of 24.3 and 24.1, respectively, for N Ab− participants (Fig. 3b and c). These observations suggest that enrollment N Ab+ participants were better able to control their BTI. Interestingly, of the N = 121 participants that tested N Ab− at enrollment, only N = 25 (20.7%) seroconverted after their BTI, which indicates that natural SARS-CoV-2 infection is not guaranteed to result in N seroconversion. The median duration of infection for participants that seroconverted was 7 days and was longer than the median of 3.5 days for participants that did not seroconvert (Supplemental Figure S5a). Thus, longer duration infections may increase the chance of N seroconversion but additional data would be required to confirm this observation. The median N and ORF1AB ct values for participants that seroconverted was 24.0 and 23.8, respectively, and was only slightly lower than the median ct values for participants that did not seroconvert (25.0 and 24.7, respectively) (Supplemental Figure S5b and S5c). The proportion of participants that reported symptoms was similar between those that seroconverted to N (73.9% symptomatic) and those that did not seroconvert to N (70.5% symptomatic), suggesting that this was not a factor.
Fig. 3.
Enrollment nucleocapsid serostatus, viral load, infection duration. Comparisons between nucleocapsid (N) seronegative (Ab−) (N = 121) and seropositive (Ab+) (N = 23) participants are shown for a) breakthrough infection duration (PCR+ days), b) N ct value, and c) ORF1AB ct value. The red bar in all graphs is the median and statistical comparisons between N Ab− and N Ab+ participants were done using a Mann–Whitney test and the P values are indicated for each comparison.
Correlations between humoral immune responses and virologic outcomes of infection
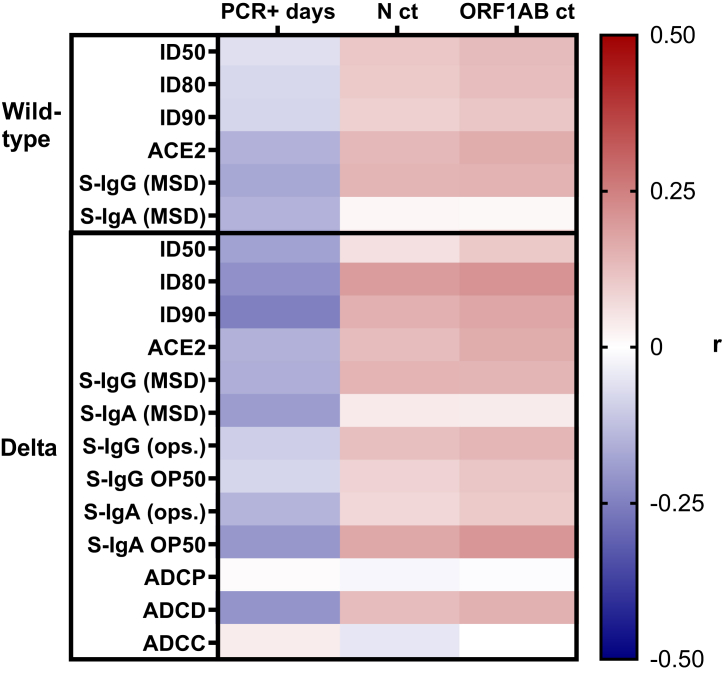
We compared the relationships between the pre-BTI levels of humoral immunity and the virologic outcomes including infection duration (PCR+ days) and peak viral load (lowest N and ORF1AB ct values). Enrollment sample antibody correlations were evaluated for N = 88 participants that had a sample taken at least 14 days post primary vaccination series and before the date of their BTI. There were N = 61 participants that were excluded from enrollment sample antibody correlations because either they completed their primary vaccination series <14 days before the enrollment sample was taken, or after the enrollment sample was taken, and/or a sample collection/vaccination date was missing. Correlations were not done with Omicron ID50, ID80, or ID90 because most participants had responses below the limit of detection of the assay. Fig. 4 and Supplemental Figure S6 show heat maps of Spearman r values for different enrollment sample antibody responses and BTI virologic outcomes. There were weak, negative correlations between enrollment sample antibody responses and the duration of infection (PCR+ days) with notable examples being: Delta neutralising antibody ID80 (r = −0.22, P = 0.04) and ID90 (r = −0.25, P = 0.02); ACE2 blocking with B.1.351 (r = −0.23, P = 0.04); and spike-reactive IgA for B.1.351 (r = −0.23, P = 0.04), B.1.617.3 (r = −0.25, P = 0.03), B.1.617.1 (r = −0.26, P = 0.02), and P1 (r = −0.26, P = 0.02). There were also weak, positive correlations between many enrollment sample antibody responses and peak viral load (N and ORF1AB ct values). The Spearman r values, 95% CI, and P values for all enrollment sample correlations are in Supplemental Table S1.
Fig. 4.
Correlations between enrollment antibody responses, viral load, and infection duration. The heat map (N = 88 participants) shows the Spearman r values for correlations between infection duration (PCR+ days) or viral load (N or ORF1AB ct values) and wild-type or Delta ID50, ID80, ID90, ACE2 blocking, spike-reactive IgG and IgA, as well as Delta spike-expressing cell opsonizing IgG and IgA MFI, ADCP, ADCD, ADCC, and spike-expressing cell 50% IgG or IgA opsonization titer. The Spearman r values, 95% CI, and P values are in Supplemental Table S1.
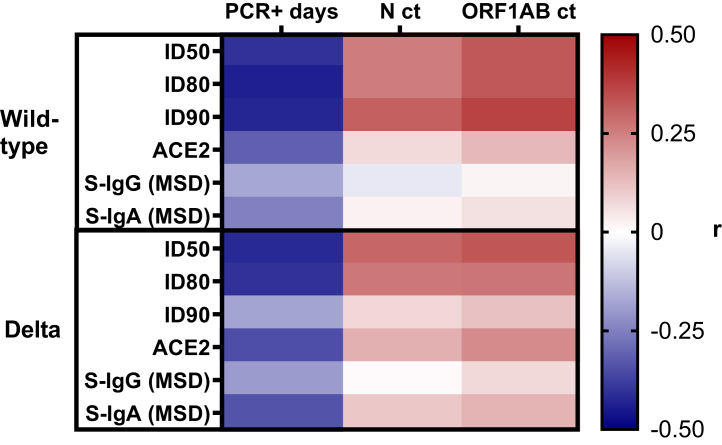
The 6-month sample antibody correlations were evaluated for N = 48 participants that had a sample taken 6-months post primary vaccination series and before the date of their BTI. Fig. 5 and Supplemental Figure S7 show heat maps of Spearman r values for different 6-month sample antibody responses and BTI virologic outcomes. The 6-month sample correlations were stronger than the enrollment sample correlations with BTI outcomes. There were moderate, negative correlations between 6-month sample antibody responses and the duration of infection with notable examples being: wild-type ID50 (r = −0.40, P = 0.004), ID80 (r = −0.44, P = 0.002), ID90 (r = −0.43, P = 0.002), and ACE2 blocking (r = −0.31, P = 0.03); and Delta ID50 (r = −0.42, P = 0.003), ID80 (r = −0.41, P = 0.004), ACE2 blocking (r = −0.35, P = 0.01), and spike-reactive IgA (r = −0.34, P = 0.02). All 10 variant spike proteins used for measuring ACE2 blocking and spike-reactive IgA by MSD had notable, moderate negative correlations with the duration of infection. There were also moderate, positive correlations between 6-month sample antibody responses and peak viral load with notable examples being: wild-type ID90 (r = 0.31, P = 0.03) and Delta ID50 (r = 0.30, P = 0.04) with N ct values; wild-type ID50 (r = 0.33, P = 0.02), ID80 (r = 0.33, P = 0.02), ID90 (r = 0.37, P = 0.01) with ORF1AB ct values; and Delta ID50 (r = 0.33, P = 0.02) with ORF1AB ct values. The Spearman r values, 95% CI, and P values for all 6-month sample correlations are in Supplemental Table S2. Overall, higher levels of functional antibody responses including neutralisation and ACE2 blocking as well as higher levels of spike-specific IgA were associated with a shorter duration of infection. Higher levels of neutralising antibody were also associated with lower peak viral load.
Fig. 5.
Correlations between 6-month post vaccination antibody responses, viral load, and infection duration. The heat map (N = 48 participants) shows the Spearman r values for correlations between infection duration (PCR+ days) or viral load (N or ORF1AB ct values) and wild-type or Delta ID50, ID80, ID90, ACE2 blocking, and spike-reactive IgG and IgA. The Spearman r values, 95% CI, and P values are in Supplemental Table S2.
Discussion
In this preliminary report from the VIRAMP clinical study, we focused on N = 149 COVID-19 vaccinated military service members that had a SARS-CoV-2 BTI between July 2021 and January 2022. The VIRAMP study design allowed us to investigate the humoral immune responses elicited by COVID-19 vaccination and to assess their impact on subsequent breakthrough infection dynamics. Sustained global transmission of SARS-CoV-2 has resulted in the emergence of variants of concern including Alpha, Delta, and Omicron.10, 11, 12, 13 Relative to wild-type SARS-CoV-2, the Delta variant had 9 spike protein mutations, the initial Omicron variant had more than 30 spike protein mutations, and emerging Omicron sub-lineages have accumulated additional spike protein mutations that have substantially reduced effectiveness of current vaccines.28,29 Previous reports indicated that neutralisation titers against Delta were a modest 2–4 fold lower than wild-type, while neutralisation titers for Omicron variants can be greater than 15-fold lower than wild-type and booster immunizations can significantly improve neutralisation potency against both variants.28, 29, 30, 31, 32, 33, 34, 35 In our study, the reductions in neutralising antibody ID50s for Delta and Omicron are in agreement with previous reports.
Several studies have indicated that COVID-19 vaccines provide more effective and durable protection against severe disease than against mildly symptomatic or subclinical infections and that vaccine efficacy decreases substantially by 6-months post immunization due to waning immunity.14, 15, 16 We observed similar outcomes in the VIRAMP study. A wide range of binding and functional antibody responses were measured in participants that went on to have a BTI and these responses waned significantly with time post-vaccination. In our study, more than half of the participants had their BTI >6-months post full vaccination and more than half of all participants with a BTI reported COVID-19 symptoms.
Previous studies have shown that COVID-19 vaccination can modify SARS-CoV-2 infection dynamics. Multiple studies have demonstrated that vaccination reduces viral load in a 3 month time-period post full vaccination, compared to unvaccinated individuals.17, 18, 19 The majority of participants in our study had a BTI infection well beyond this time period, yet we still observed some significant correlations between pre-BTI neutralising antibody responses and peak viral load. COVID-19 vaccination has also been shown to increase the rate of virus clearance (decrease infection duration) relative to unvaccinated individuals.20, 21, 22, 23, 24 An important finding from our study is that multiple different humoral immune readouts including neutralising antibody levels, MSD ACE2 blocking activity, and MSD spike-specific IgA levels inversely correlated with the duration of infection (rate of virus clearance), particularly in samples collected closer to the time of the BTI. Notably, these immune readouts relied on serum from circulation and not on samples taken from the site of infection in the upper airways. Other groups have measured SARS-CoV-2 specific IgG, IgA, and neutralising antibodies from various respiratory samples and saliva and these antibody responses correlated with antibody levels in circulation, but they tended to be less abundant and were therefore more difficult to detect than antibodies found in serum.36, 37, 38, 39, 40, 41 Additionally, SARS-CoV-2 mucosal antibody levels were higher in individuals that were naturally exposed prior to vaccination compared to vaccination alone.36,37,41,42 In our study, MSD spike-specific IgA levels, but not spike-specific IgG levels, correlated with the duration of infection, suggesting that spike-specific IgA in circulation is a useful correlate of protection from infection in the upper airways.
Natural SARS-CoV-2 infection can elicit anti-nucleocapsid antibodies that have been used as a serological marker of natural exposure, but these responses have been observed to wane substantially over time.43, 44, 45, 46, 47, 48 The N protein is not a component of vaccines currently used in the US and therefore detection of N antibody has been used to discriminate between natural exposures and vaccine immunity. Nucleocapsid antibody serostatus has also been observed to impact subsequent SARS-CoV-2 infections, with a lower incidence of disease observed in those that were N antibody seropositive.49 In our study, enrollment N antibody seropositive participants had significantly higher binding and functional antibody responses to SARS-CoV-2 and they also had significantly shorter BTI durations and lower peak viral loads in accordance with disease protection observed by other groups. A recent report indicated that N antibody seroconversion rates are significantly reduced in Moderna (mRNA-1273) vaccinated (40% N seropositive) compared to unvaccinated (93% N seropositive) individuals.50 This is in agreement with our results that showed 20.7% N antibody seroconversion following a BTI in baseline seronegatives that predominately received the Pfizer (BNT162b2) vaccine. Importantly, this indicates that vaccination modifies SARS-CoV-2 infections in a way that reduces de novo immune responses to the N protein and possibly other viral antigens that could contribute to more comprehensive and protective immunity.
The current study has several limitations. The study is based on a US military population that only reflects a segment of the general population that may be younger, healthier, and have less comorbidities. We did not measure T cell responses or mucosal immune responses because those samples were not collected or not available but may be important for protection. We also did not measure levels of infectious virus in respiratory samples, which could have been useful for assessing the impact of humoral immunity on virus shedding and the potential for transmission. Also saliva was tested by RT-qPCR instead of nasopharyngeal swab specimens that have been shown to be more sensitive for SARS-CoV-2 detection in some studies. Furthermore, the Omicron reagents for the MSD assays were not widely available when the samples were tested, but future testing will include this variant, which will likely improve correlations with the outcome of Omicron BTIs. Also, the results described here were derived from samples collected before COVID-19 booster immunizations were widely accessible and work is currently on-going to evaluate the impact of humoral immune responses in VIRAMP study participants on more contemporary waves of Omicron breakthrough infections.
Overall, this preliminary report from the VIRAMP study corroborates results from other clinical studies that have characterised breakthrough infections in vaccinated and unvaccinated cohorts. Antibody reactivity and function was significantly lower with SARS-CoV-2 Delta and Omicron compared to wild-type and this was exacerbated by waning humoral immunity that correlated with time post-vaccination. We expanded on previous findings that COVID-19 vaccination increases the rate of virus clearance during breakthrough infections by delineating some of the humoral immune responses that are involved. Furthermore, we extended previous observations that nucleocapsid serostatus prior to breakthrough infection correlates with disease protection by showing that seropositive individuals have an increased rate of virus clearance and lower peak viral loads compared to seronegatives. More work is needed to determine if immune responses to the nucleocapsid protein, which are elicited by natural infection and not vaccination, are important for protection or if cellular and humoral immune responses to other viral proteins are involved.
Contributors
Conceptualization: GDG, JRC, JO, KM, DPP, JC, and KP; Data Curation: GDG, CMC, SM, JK, IS, MKM, JRC, DPP, JC, and KP; Writing – Original Draft: GDG, CMC, DPP, and KP; Writing – Review & Editing: GDG, CMC, SM, JK, IS, MKM, DC, JE, CS, JD, SP, DE, KM, JRC, JO, DPP, JC, and KP. GDG, CMC, MKM, JRC, and DPP verified the underlying data. All authors read and approved the final version of the manuscript.
Data sharing statement
Data will be available upon reasonable request.
Declaration of interests
KM (Allucent) was contracted by the US Department of Defense to conduct the VIRAMP clinical study, to perform testing on clinical study samples, and to collect epidemiologic data. JC served as the Acting Chief Medical Officer for the Vaccine Acceleration Program at the Joint Program Executive Office for Chemical, Biological, Radiological and Nuclear Defense that funded the VIRAMP clinical study, and was involved with clinical study design and establishing collaborations for analysis of samples. The other authors declare no conflicts of interest.
Acknowledgements
This work was funded by the US Department of Defense (DoD) Joint Program Executive Office for Chemical, Biological, Radiological and Nuclear Defense (JPEO-CBRND) in collaboration with the Defense Health Agency (DHA) COVID-19 funding initiative for the VIRAMP study. The authors would like to thank April Griggs and Hanna Kim for critical research support. This work was supported by a cooperative agreement (W81XWH-18-2-0040) between the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., and the U.S. DoD as well as supported in part by the US Army Medical Research and Development Command under Contract No. W81-XWH-18-C-0337.
Disclaimer: Material has been reviewed by the Walter Reed Army Institute of Research and the US Department of Defense Joint Program Executive Office. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the author, and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense or the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104683.
Contributor Information
Gregory D. Gromowski, Email: gregory.d.gromowski.civ@health.mil.
Jessica Cowden, Email: jessica.cowden.mil@afrims.org.
Appendix A. Supplementary data
Supplemental Figure 1. Correlations between viral load, infection duration, and time post vaccination. Panel a) shows the correlation between ORF1AB and N gene Ct values (N = 149). Panels b and c) show the correlation between N or ORF1AB ct values, respectively, and the duration of infection (N = 149). Panels d–f) show correlations between breakthrough infection duration (PCR+ days), N ct value, or ORF1AB ct value, respectively, and the number of days post vaccination (N = 117). Spearman correlations were done and the r and P values are indicated.
Supplemental Figure 2. Enrollment sample antibody responses. Neutralising antibody ID50, ID80, and ID90 are shown in panels a–c), respectively, for wild-type, Delta, and Omicron (N = 141). A Kruskal–Wallis test with Dunn’s correction for multiple comparisons was done. MSD assay ACE2 blocking (N = 125), spike-reactive IgG (N = 126), and spike-reactive IgA (N = 126) are shown in panels d–f), respectively, for wild-type and Delta. A Wilcoxon test was done. Panels g) ADCP score (N = 135), h) ADCD (N = 134), i) ADCC (N = 119), j) spike-expressing, cell opsonizing IgG and IgA MFI (N = 119), and k) 50% cell opsonizing IgG and IgA titers (N = 119), for Delta. The red bar in all graphs is the median.
Supplemental Figure 3. Correlations between neutralising antibody titers and other functional or binding antibody responses. Correlations between neutralising antibody ID50 and a) wild-type ACE2 blocking (N = 125), b) wild-type spike-reactive IgG (N = 126), c) wild-type spike-reactive IgA (N = 126), d) Delta ACE2 blocking (N = 125), e) Delta spike-reactive IgG (N = 126), f) Delta spike-reactive IgA (N = 126), g) Delta ADCP (N = 135), h) Delta ADCD (N = 134), i) Delta ADCC (N = 119), j) Delta spike-expressing cell opsonizing IgG MFI (N = 119), k) Delta spike-expressing cell opsonizing IgA MFI (N = 119), l) Delta 50% cell opsonizing IgG titers (N = 119), and m) Delta 50% cell opsonizing IgA titers (N = 119). Spearman correlations were done and the r and P values are indicated.
Supplemental Figure 4. Correlations between antibody responses and time post vaccination. Correlations between time post vaccination and a) wild-type ID50 (N = 141), b) wild-type ID80 (N = 141), c) wild-type ID90 (N = 141), d) Delta ID50 (N = 141), e) Delta ID80 (N = 141), f) Delta ID90 (N = 141), g) wild-type ACE2 blocking (N = 125), h) wild-type spike-reactive IgG (N = 126), i) wild-type spike reactive IgA (N = 126), j) Delta ACE blocking (N = 125), k) Delta spike-reactive IgG (N = 126), l) Delta spike-reactive IgA (N = 126), m) Delta spike-expressing cell opsonizing IgG MFI (N = 119), n) Delta spike-expressing cell opsonizing IgA MFI (N = 119), o) Delta ADCP (N = 135), p) Delta ADCD (N = 134), q) Delta ADCC (N = 119), r) Delta spike-expressing cell 50% IgG opsonization titer (N = 119), and s) Delta spike-expressing cell 50% IgA opsonization titer (N = 119). Spearman correlations were done and the r and P values are indicated.
Supplemental Figure 5. Post breakthrough infection nucleocapsid serostatus, viral load, and infection duration. Comparisons between nucleocapsid (N) seronegative (Ab−) (N = 96) and seropositive (Ab+) (N = 25) participants following the breakthrough infection are shown for a) infection duration (PCR+ days), b) N ct value, and c) ORF1AB ct value. The red bar in all graphs is the median and statistical comparisons between N Ab− and N Ab+ participants were done using a Mann–Whitney test and the P values are indicated for each comparison.
Supplemental Figure 6. Correlations between enrollment antibody responses to additional SARS-CoV-2 variants, viral load, and infection duration. The heat map (N = 88 participants) shows the Spearman r values for correlations between infection duration (PCR+ days) or viral load (N or ORF1AB ct values) and ACE2 blocking and spike-reactive IgG and IgA. The Spearman r values, 95% CI, and P values are in Supplemental Table S1.
Supplemental Figure 7. Correlations between 6-month post vaccination antibody responses to additional SARS-CoV-2 variants, viral load, and infection duration. The heat map (N = 48 participants) shows the Spearman r values for correlations between infection duration (PCR+ days) or viral load (N or ORF1AB ct values) and ACE2 blocking and spike-reactive IgG and IgA. The Spearman r values, 95% CI, and P values are in Supplemental Table S2.
References
- 1.Khoury D.S., Cromer D., Reynaldi A., et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. 2021;27(7):1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 2.DeGrace M.M., Ghedin E., Frieman M.B., et al. Defining the risk of SARS-CoV-2 variants on immune protection. Nature. 2022;605(7911):640–652. doi: 10.1038/s41586-022-04690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mercado N.B., Zahn R., Wegmann F., et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586(7830):583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu J., Tostanoski L.H., Peter L., et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369(6505):806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamin R., Jones A.T., Hoffmann H.H., et al. Fc-engineered antibody therapeutics with improved anti-SARS-CoV-2 efficacy. Nature. 2021;599(7885):465–470. doi: 10.1038/s41586-021-04017-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dussupt V., Sankhala R.S., Mendez-Rivera L., et al. Low-dose in vivo protection and neutralization across SARS-CoV-2 variants by monoclonal antibody combinations. Nat Immunol. 2021;22(12):1503–1514. doi: 10.1038/s41590-021-01068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winkler E.S., Gilchuk P., Yu J., et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021;184(7):1804–1820.e16. doi: 10.1016/j.cell.2021.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ullah I., Prevost J., Ladinsky M.S., et al. Live imaging of SARS-CoV-2 infection in mice reveals that neutralizing antibodies require Fc function for optimal efficacy. Immunity. 2021;54(9):2143–2158.e15. doi: 10.1016/j.immuni.2021.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldblatt D., Alter G., Crotty S., Plotkin S.A. Correlates of protection against SARS-CoV-2 infection and COVID-19 disease. Immunol Rev. 2022;310(1):6–26. doi: 10.1111/imr.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plante J.A., Mitchell B.M., Plante K.S., Debbink K., Weaver S.C., Menachery V.D. The variant gambit: COVID-19's next move. Cell Host Microbe. 2021;29(4):508–515. doi: 10.1016/j.chom.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiegand T., Nemudryi A., Nemudraia A., et al. The rise and fall of SARS-CoV-2 variants and ongoing diversification of Omicron. Viruses. 2022;14(9):2009. doi: 10.3390/v14092009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLean G., Kamil J., Lee B., et al. The impact of evolving SARS-CoV-2 mutations and variants on COVID-19 vaccines. mBio. 2022;13(2) doi: 10.1128/mbio.02979-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Q., Zhang J., Wang P., Zhang Z. The mechanisms of immune response and evasion by the main SARS-CoV-2 variants. iScience. 2022;25(10) doi: 10.1016/j.isci.2022.105044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feikin D.R., Abu-Raddad L.J., Andrews N., et al. Assessing vaccine effectiveness against severe COVID-19 disease caused by Omicron variant. Report from a meeting of the World Health Organization. Vaccine. 2022;40(26):3516–3527. doi: 10.1016/j.vaccine.2022.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andrews N., Tessier E., Stowe J., et al. Duration of protection against mild and severe disease by covid-19 vaccines. N Engl J Med. 2022;386(4):340–350. doi: 10.1056/NEJMoa2115481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feikin D.R., Higdon M.M., Abu-Raddad L.J., et al. Duration of effectiveness of vaccines against SARS-CoV-2 infection and COVID-19 disease: results of a systematic review and meta-regression. Lancet. 2022;399(10328):924–944. doi: 10.1016/S0140-6736(22)00152-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levine-Tiefenbrun M., Yelin I., Alapi H., et al. Viral loads of delta-variant SARS-CoV-2 breakthrough infections after vaccination and booster with BNT162b2. Nat Med. 2021;27(12):2108–2110. doi: 10.1038/s41591-021-01575-4. [DOI] [PubMed] [Google Scholar]
- 18.Levine-Tiefenbrun M., Yelin I., Alapi H., et al. Waning of SARS-CoV-2 booster viral-load reduction effectiveness. Nat Commun. 2022;13(1):1237. doi: 10.1038/s41467-022-28936-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puhach O., Adea K., Hulo N., et al. Infectious viral load in unvaccinated and vaccinated individuals infected with ancestral, delta or Omicron SARS-CoV-2. Nat Med. 2022;28(7):1491–1500. doi: 10.1038/s41591-022-01816-0. [DOI] [PubMed] [Google Scholar]
- 20.Kissler S.M., Fauver J.R., Mack C., et al. Viral dynamics of SARS-CoV-2 variants in vaccinated and unvaccinated persons. N Engl J Med. 2021;385(26):2489–2491. doi: 10.1056/NEJMc2102507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Kampen J.J.A., van de Vijver D., Fraaij P.L.A., et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19) Nat Commun. 2021;12(1):267. doi: 10.1038/s41467-020-20568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolfel R., Corman V.M., Guggemos W., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 23.Singanayagam A., Hakki S., Dunning J., et al. Community transmission and viral load kinetics of the SARS-CoV-2 delta (B.1.617.2) variant in vaccinated and unvaccinated individuals in the UK: a prospective, longitudinal, cohort study. Lancet Infect Dis. 2022;22(2):183–195. doi: 10.1016/S1473-3099(21)00648-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson M.G., Burgess J.L., Naleway A.L., et al. Prevention and attenuation of covid-19 with the BNT162b2 and mRNA-1273 vaccines. N Engl J Med. 2021;385(4):320–329. doi: 10.1056/NEJMoa2107058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King H.A.D., Joyce M.G., Lakhal-Naouar I., et al. Efficacy and breadth of adjuvanted SARS-CoV-2 receptor-binding domain nanoparticle vaccine in macaques. Proc Natl Acad Sci U S A. 2021;118(38) doi: 10.1073/pnas.2106433118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ackerman M.E., Moldt B., Wyatt R.T., et al. A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J Immunol Methods. 2011;366(1–2):8–19. doi: 10.1016/j.jim.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischinger S., Fallon J.K., Michell A.R., et al. A high-throughput, bead-based, antigen-specific assay to assess the ability of antibodies to induce complement activation. J Immunol Methods. 2019;473 doi: 10.1016/j.jim.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurhade C., Zou J., Xia H., et al. Low neutralization of SARS-CoV-2 Omicron BA.2.75.2, BQ.1.1 and XBB.1 by parental mRNA vaccine or a BA.5 bivalent booster. Nat Med. 2022;29(2):344–347. doi: 10.1038/s41591-022-02162-x. [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann M., Behrens G.M.N., Arora P., et al. Effect of hybrid immunity and bivalent booster vaccination on Omicron sublineage neutralisation. Lancet Infect Dis. 2023;23(1):25–28. doi: 10.1016/S1473-3099(22)00792-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Servellita V., Syed A.M., Morris M.K., et al. Neutralizing immunity in vaccine breakthrough infections from the SARS-CoV-2 Omicron and delta variants. Cell. 2022;185(9):1539–1548.e5. doi: 10.1016/j.cell.2022.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185(3):457–466.e4. doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Planas D., Saunders N., Maes P., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602(7898):671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 33.Lu L., Mok B.W.Y., Chen L.L., et al. Neutralization of severe acute respiratory syndrome coronavirus 2 Omicron variant by sera from BNT162b2 or CoronaVac vaccine recipients. Clin Infect Dis. 2022;75(1):e822–e826. doi: 10.1093/cid/ciab1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carreno J.M., Alshammary H., Tcheou J., et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602(7898):682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- 35.Muik A., Lui B.G., Wallisch A.K., et al. Neutralization of SARS-CoV-2 Omicron by BNT162b2 mRNA vaccine-elicited human sera. Science. 2022;375(6581):678–680. doi: 10.1126/science.abn7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russell M.W., Mestecky J. Mucosal immunity: the missing link in comprehending SARS-CoV-2 infection and transmission. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.957107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Longet S., Hargreaves A., Healy S., et al. mRNA vaccination drives differential mucosal neutralizing antibody profiles in naive and SARS-CoV-2 previously-infected individuals. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.953949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li D., Calderone R., Nsouli T.M., Reznikov E., Bellanti J.A. Salivary and serum IgA and IgG responses to SARS-CoV-2-spike protein following SARS-CoV-2 infection and after immunization with COVID-19 vaccines. Allergy Asthma Proc. 2022;43(5):419–430. doi: 10.2500/aap.2022.43.220045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang J., Zeng C., Cox T.M., et al. Respiratory mucosal immunity against SARS-CoV-2 after mRNA vaccination. Sci Immunol. 2022;7 doi: 10.1126/sciimmunol.add4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan R.W.Y., Chan K.C.C., Lui G.C.Y., et al. Mucosal antibody response to SARS-CoV-2 in paediatric and adult patients: a longitudinal study. Pathogens. 2022;11(4):397. doi: 10.3390/pathogens11040397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darwich A., Pozzi C., Fornasa G., et al. BNT162b2 vaccine induces antibody release in saliva: a possible role for mucosal viral protection? EMBO Mol Med. 2022;14(5) doi: 10.15252/emmm.202115326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sano K., Bhavsar D., Singh G., et al. SARS-CoV-2 vaccination induces mucosal antibody responses in previously infected individuals. Nat Commun. 2022;13(1):5135. doi: 10.1038/s41467-022-32389-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Elslande J., Oyaert M., Ailliet S., et al. Longitudinal follow-up of IgG anti-nucleocapsid antibodies in SARS-CoV-2 infected patients up to eight months after infection. J Clin Virol. 2021;136 doi: 10.1016/j.jcv.2021.104765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shrotri M., Harris R.J., Rodger A., et al. Persistence of SARS-CoV-2 N-antibody response in healthcare workers, London, UK. Emerg Infect Dis. 2021;27(4):1155–1158. doi: 10.3201/eid2704.204554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garritsen A., Scholzen A., van den Nieuwenhof D.W.A., et al. Two-tiered SARS-CoV-2 seroconversion screening in the Netherlands and stability of nucleocapsid, spike protein domain 1 and neutralizing antibodies. Infect Dis. 2021;53(7):498–512. doi: 10.1080/23744235.2021.1893378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paul G., Strnad P., Wienand O., et al. The humoral immune response more than one year after SARS-CoV-2 infection: low detection rate of anti-nucleocapsid antibodies via Euroimmun ELISA. Infection. 2022;11:397. doi: 10.1007/s15010-022-01830-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gallais F., Gantner P., Bruel T., et al. Evolution of antibody responses up to 13 months after SARS-CoV-2 infection and risk of reinfection. EBioMedicine. 2021;71 doi: 10.1016/j.ebiom.2021.103561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dan J.M., Mateus J., Kato Y., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371(6529) doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kohler P., Gusewell S., Seneghini M., et al. Impact of baseline SARS-CoV-2 antibody status on syndromic surveillance and the risk of subsequent COVID-19-a prospective multicenter cohort study. BMC Med. 2021;19(1):270. doi: 10.1186/s12916-021-02144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Follmann D., Janes H.E., Buhule O.D., et al. Antinucleocapsid antibodies after SARS-CoV-2 infection in the blinded phase of the randomized, placebo-controlled mRNA-1273 COVID-19 vaccine efficacy clinical trial. Ann Intern Med. 2022;175(9):1258–1265. doi: 10.7326/M22-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Correlations between viral load, infection duration, and time post vaccination. Panel a) shows the correlation between ORF1AB and N gene Ct values (N = 149). Panels b and c) show the correlation between N or ORF1AB ct values, respectively, and the duration of infection (N = 149). Panels d–f) show correlations between breakthrough infection duration (PCR+ days), N ct value, or ORF1AB ct value, respectively, and the number of days post vaccination (N = 117). Spearman correlations were done and the r and P values are indicated.
Supplemental Figure 2. Enrollment sample antibody responses. Neutralising antibody ID50, ID80, and ID90 are shown in panels a–c), respectively, for wild-type, Delta, and Omicron (N = 141). A Kruskal–Wallis test with Dunn’s correction for multiple comparisons was done. MSD assay ACE2 blocking (N = 125), spike-reactive IgG (N = 126), and spike-reactive IgA (N = 126) are shown in panels d–f), respectively, for wild-type and Delta. A Wilcoxon test was done. Panels g) ADCP score (N = 135), h) ADCD (N = 134), i) ADCC (N = 119), j) spike-expressing, cell opsonizing IgG and IgA MFI (N = 119), and k) 50% cell opsonizing IgG and IgA titers (N = 119), for Delta. The red bar in all graphs is the median.
Supplemental Figure 3. Correlations between neutralising antibody titers and other functional or binding antibody responses. Correlations between neutralising antibody ID50 and a) wild-type ACE2 blocking (N = 125), b) wild-type spike-reactive IgG (N = 126), c) wild-type spike-reactive IgA (N = 126), d) Delta ACE2 blocking (N = 125), e) Delta spike-reactive IgG (N = 126), f) Delta spike-reactive IgA (N = 126), g) Delta ADCP (N = 135), h) Delta ADCD (N = 134), i) Delta ADCC (N = 119), j) Delta spike-expressing cell opsonizing IgG MFI (N = 119), k) Delta spike-expressing cell opsonizing IgA MFI (N = 119), l) Delta 50% cell opsonizing IgG titers (N = 119), and m) Delta 50% cell opsonizing IgA titers (N = 119). Spearman correlations were done and the r and P values are indicated.
Supplemental Figure 4. Correlations between antibody responses and time post vaccination. Correlations between time post vaccination and a) wild-type ID50 (N = 141), b) wild-type ID80 (N = 141), c) wild-type ID90 (N = 141), d) Delta ID50 (N = 141), e) Delta ID80 (N = 141), f) Delta ID90 (N = 141), g) wild-type ACE2 blocking (N = 125), h) wild-type spike-reactive IgG (N = 126), i) wild-type spike reactive IgA (N = 126), j) Delta ACE blocking (N = 125), k) Delta spike-reactive IgG (N = 126), l) Delta spike-reactive IgA (N = 126), m) Delta spike-expressing cell opsonizing IgG MFI (N = 119), n) Delta spike-expressing cell opsonizing IgA MFI (N = 119), o) Delta ADCP (N = 135), p) Delta ADCD (N = 134), q) Delta ADCC (N = 119), r) Delta spike-expressing cell 50% IgG opsonization titer (N = 119), and s) Delta spike-expressing cell 50% IgA opsonization titer (N = 119). Spearman correlations were done and the r and P values are indicated.
Supplemental Figure 5. Post breakthrough infection nucleocapsid serostatus, viral load, and infection duration. Comparisons between nucleocapsid (N) seronegative (Ab−) (N = 96) and seropositive (Ab+) (N = 25) participants following the breakthrough infection are shown for a) infection duration (PCR+ days), b) N ct value, and c) ORF1AB ct value. The red bar in all graphs is the median and statistical comparisons between N Ab− and N Ab+ participants were done using a Mann–Whitney test and the P values are indicated for each comparison.
Supplemental Figure 6. Correlations between enrollment antibody responses to additional SARS-CoV-2 variants, viral load, and infection duration. The heat map (N = 88 participants) shows the Spearman r values for correlations between infection duration (PCR+ days) or viral load (N or ORF1AB ct values) and ACE2 blocking and spike-reactive IgG and IgA. The Spearman r values, 95% CI, and P values are in Supplemental Table S1.
Supplemental Figure 7. Correlations between 6-month post vaccination antibody responses to additional SARS-CoV-2 variants, viral load, and infection duration. The heat map (N = 48 participants) shows the Spearman r values for correlations between infection duration (PCR+ days) or viral load (N or ORF1AB ct values) and ACE2 blocking and spike-reactive IgG and IgA. The Spearman r values, 95% CI, and P values are in Supplemental Table S2.