Abstract
Mice with deletion of Cyp2c70 have a human-like bile acid composition, display age- and sex-dependent signs of hepatobiliary disease and can be used as a model to study interactions between bile acids and the gut microbiota in cholestatic liver disease. In the present study, we rederived Cyp2c70−/− mice as germ-free (GF) and colonized them with a human or a mouse microbiota to investigate whether the presence of a microbiota can be protective in cholangiopathic liver disease associated with Cyp2c70-deficiency. GF Cyp2c70−/− mice showed reduced neonatal survival, liver fibrosis, and distinct cholangiocyte proliferation. Colonization of germ-free breeding pairs with a human or a mouse microbiota normalized neonatal survival of the offspring, and particularly colonization with mouse microbiota from a conventionally raised mouse improved the liver phenotype at 6–10 weeks of age. The improved liver phenotype in conventionalized (CD) Cyp2c70−/− mice was associated with increased levels of tauro-ursodeoxycholic acid (TUDCA) and UDCA, resulting in a more hydrophilic bile acid profile compared with GF and humanized Cyp2c70−/− mice. The hydrophobicity index of biliary bile acids of CD Cyp2c70−/− mice was associated with changes in gut microbiota, liver weight, liver transaminases, and liver fibrosis. Hence, our results indicate that neonatal survival of Cyp2c70−/− mice seems to depend on the establishment of a gut microbiota at birth, and the improved liver phenotype in CD Cyp2c70−/− mice may be mediated by a larger proportion of TUDCA/UDCA in the circulating bile acid pool and/or by the presence of specific bacteria.
Keywords: bile acids, cyp2c70, faecal microbiota transplantation, gut microbiota, hepatobiliary disease, ursodeoxycholic acid
Introduction
Interactions between bile acids and the gut microbiota affect host physiology and metabolism and may influence the development and progression of liver disease, but underlying mechanisms are still unknown. The use of mouse models to study the cross-talk between bile acids and the gut microbiota is limited by differences in bile acid profile between mice and humans. In addition to the human primary bile acids cholic acid (CA) and chenodeoxycholic (CDCA), mice also synthesize very hydrophilic 6β-hydroxylated primary bile acids such as α-muricholic acid (αMCA) and β-muricholic acid (βMCA) [1,2]. CYP2C70 is the enzyme responsible for the formation of 6β-hydroxylated murine bile acids [3] and mice with deletion of the Cyp2c70 gene have a human-like bile acid pool and may be a more relevant model to study bile acid metabolism [4,5]. Cyp2c70 knockout mice display increased hepatic inflammation, fibrosis and transient neonatal cholestasis [6] and could therefore constitute a model for cholestatic liver disease such as primary sclerosing cholangitis (PSC), in which interactions between gut microbiota and bile acids have been implicated [7,8]. Similarly, studies with multidrug resistance gene 2 (Mdr2 or Abcb4)-deficient mice, another mouse model of genetically induced cholestatic liver disease, have shown that the absence of a gut microbiota worsens cholestasis and liver phenotype [9] and leads to increased lethality [10]. Thus, we have previously suggested that the gut microbiota might have a protective role in cholestatic liver diseases [11]. Therefore, rederiving Cyp2c70-deficient mice as germ-free (GF) and colonizing them with a human microbiota may constitute a more physiological model to investigate how the gut microbiota interacts with bile acids to modulate liver disease.
Methods
Animal experiments
Generation of GF Cyp2c70−/− mice and colonization experiment
Conventionally raised wild-type C57/Bl6 females were crossed with conventionally raised Cyp2c70−/− (C57Bl/6J-Cyp2c70em3Umcg) males provided by Folkert Kuipers’ lab in Groningen, The Netherlands (www.labpediatricsrug.nl). Cyp2c70+/− mice were rederived as GF by caesarian-section. The uterine packages from pregnant donor mice were dissected and transferred into a sterile isolator via a transfer chamber filled with chlorine bleach. The pups were removed from the uterine package and placed in a cage with a GF Swiss Webster dam. The GF Cyp2c70+/− mice were then housed in sterile isolators and crossed to generate GF Cyp2c70−/−, Cyp2c70+/−, and Cyp2c70+/+ mice. For humanization, GF Cyp2c70+/− breeding pairs were transferred to a new isolator and colonized with fresh human faeces from a 47-year-old healthy (no medications or nutritional supplements) male donor. For conventionalization, GF Cyp2c70+/− breeding pairs were transferred to our open facility and colonized with caecal content from a conventionally raised wild-type C57/Bl6 mouse. Colonization was performed by diluting the human faecal or mouse caecal samples (approximately 0.5 g) in 5 ml reduced PBS containing 0.2 g/l Na2S and 0.5 g/l cysteine as reducing agents in Hungate tubes. About 0.2 ml of this suspension was introduced by gavage into each GF mouse. The samples were obtained shortly before colonization and immediately (within 5 min) diluted into the PBS and introduced into the GF mice within 2 h after dilution. The humanized (HUM) mice were gavaged a second time after two days with inoculum from a new fresh faecal sample from the same human donor.
The GF and the HUM mice were maintained in flexible plastic gnotobiotic isolators under a strict 12 h light cycle and fed an autoclaved chow diet (LabDiet 5021, St Louis, MO) ad libitum. GF isolators were routinely tested for sterility by culturing and PCR analysis of faeces amplifying the 16S rRNA gene. The conventionalized (CD) mice were maintained in conventional cages under a strict 12 h light cycle and fed the same autoclaved chow diet as the GF and HUM mice ad libitum.
All animal experiments were performed at Experimental Biomedicine, University of Gothenburg, using protocols approved by the Gothenburg Animal Research Ethics Committee (Ethical number 4805/23).
Tissue collection
Blood was collected from the inferior vena cava under deep isoflurane-induced anesthesia following 4 h fasting. The mice were euthanized by cervical dislocation and tissues (liver, gallbladder, and caecum) were collected and immediately frozen in liquid nitrogen and stored at −80°C until further processing. In addition, liver biopsies were fixed in 4% paraformaldehyde at 4°C for at least 48 h and then washed with PBS and stored in 70% ethanol at 4°C until paraffine-embedding.
Histology
Liver sections (5 µm) were processed as previously described [12]. Sections were stained for collagen using Sirius Red (Sigma-Aldrich, 366548) and analyzed with Python (3.9.15) using Napari (0.4.17), pyclesperanto (0.22.0), SimpleITK (2.2.1) [13], and APOC (0.12.0) using the devbio-napari (0.8.1) package. Images were acquired with a 10× objective on a Zeiss Apotome Axioplan 2. For Sirius Red quantification, random forest decision trees were trained on an image and applied to all images. Labeled pixels were quantified and the total pixel count was normalized to the image resolution. Labeled pixels were mapped to an image for manual control of segmentation.
Immunohistochemistry
Liver sections were stained for cytokeratin 19 (CK19) using a rabbit antibody (Abcam, ab-52625, 1:300). The sections were incubated overnight at 4°C followed by staining using a secondary Goat Anti-Rabbit Alexa 594 antibody (Thermo Fischer Scientific, A-11072, 1:300). The sections were stained for nuclei using Hoechst solution (1:5000). Images were acquired with a 10× objective using a Nikon Eclipse Ni-E.
Images of sections stained with CK19 antibody were analyzed with Python (3.9.15) using Napari (0.4.17), pyclesperanto (0.22.0) and SimpleITK (2.2.1) using the devbio-napari (0.8.1) package. More specifically, a Top-hat filter was applied to remove background using default settings, and a median filter was applied for noise reduction using default settings. Stained areas were labeled using the Renyi entropy threshold. The labeled pixels were quantified, and the total pixel count was normalized to the image resolution. Labeled pixels were mapped to an image for manual control of labeling.
Liver function tests
Liver transaminases, aspartate transaminase (AST), and alanine transaminase (ALT) were measured in 100 µl of serum from the inferior caval vein. Analyses were performed at Clinical Chemistry Department, Sahlgrenska University Hospital, using the system Alinity ci 1303, software version 3.4.0 (Abbott laboratories).
Quantitative real-time PCR
Approximately 30 mg of liver was homogenized using Tissuelyser II (Qiagen) and total RNA was isolated using RNeasy mini kit (Qiagen, 74106). High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, 4368813) was used to synthesize 20 µl cDNA templates from 500 ng purified RNA using random hexamer primers, and the products were diluted 7x before use in subsequent reactions. 1× iQ™ SYBR® Green Supermix (Bio-Rad, 1708886) was used for qRT-PCR at final reaction volumes of 10 µl. 900 nM gene-specific primers were used in each reaction, and all gene expression data were normalized to TATA-binding protein (Tbp) gene expression (forward: 5′-CGA ACA GAG TTT GAC AGA GA-3′; reverse: 5′-CAT CTT CAC CAG GAT CAG C-3′). Analyzed genes and primers were chemokine ligand 2 (Ccl2) (forward: 5′-CCC CAA GAA GGA ATG GGT CC-3′; reverse: 5′-GGT TGT GGA AAA GGT AGT GG-3′), neutrophil membrane glycoprotein NB1 (Cd177) (forward: 5′-CCG GAC AGG GAC TTC TGT AA-3′; reverse: 5′-ACA CAG CTG CCG GTT GAA-3′), adhesion G protein-coupled receptor E1 (Adgre1 or F4/80) (forward: 5′-CTT TGG CTA TGG GCT TCC AGT C-3′; reverse: 5′-GCA AGG AGG ACA GAG TTT ATC GTG-3′), and tumor necrosis factor α (Tnfα) (forward: 5′-CCC CAA AGG GAT GAG AAG TT-3′; reverse: 5′-CTC CTC CAC TTG GTG GTT TG-3′). Assays were performed in a CFX-Connect Real-Time System (Bio-Rad), and data were analyzed using the ΔΔCT analysis method.
Bile acid analysis
Gallbladder, liver, caecum, and serum from the inferior caval vein were collected for bile acid analysis. Tissue samples (∼50–100 mg) and the whole gallbladder were extracted with methanol, containing d4-TCA, d4-CA, d4-CDCA, and d4-LCA (2.5 µM of each) as internal standards. The samples were homogenized with ceramic beads for 10 min using the Tissuelyser II (Qiagen). After 10 min of centrifugation at 20,000 g, 20 μl of supernatant was diluted with 980 μl of methanol:water 1:1. Serum samples (25 µl) were extracted with 10 volumes of methanol containing 50 nM of respective deuterated internal standards. After 5 min of vortex and 10 min of centrifugation at 20 000 g, supernatants were evaporated using a stream of nitrogen and reconstituted in 100 µl of methanol: water [1:1]. Approximately 5 µl of the samples were used for the bile acid analysis.
Bile acids were analyzed using ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS). After injection the bile acids were separated on a C18 column (1.7 µm, 2.1 × 100 mm; Kinetex, Phenomenex, U.S.A.) using water with 7.5 mM ammonium acetate and 0.019% formic acid (mobile phase A) and acetonitrile with 0.1% formic acid (mobile phase B). The chromatographic separation started with 1-min isocratic separation at 20% B. The B-phase was then increased to 35% for 4 min. During the next 10 min, the B-phase was increased to 100%, and it was held at 100% for 3.5 min before returning to 20%. The total runtime was 20 min. Bile acids were detected using multiple reaction monitoring (MRM) in negative mode on a QTRAP 5500 mass spectrometer (Sciex, Concord, Canada), and quantification was made against appropriate deuterated internal standards with adjustments by using external individual standard curves. Heuman’s method was used to determine hydrophobicity indices of biliary bile acids [14].
16S rRNA microbiota analysis
Genomic DNA was extracted using the Nucleospin Soil Kit (Macherey-Nagel, 740780.50). Mouse caecum (∼50–100 mg) was aliquoted and extracted using two rounds of chemical lysis and bead-beating. For each round, samples were put into SL2 buffer with SX enhancer and subsequently incubated at 90°C for 10 min. The samples were then sheared for three runs of bead beating at 5.5 m/s for 60 s in a FastPrep-24 Instrument (MP Biomedicals) with 5 min on ice in between. The V4 region of the 16S rRNA gene was amplified with a PCR program of denaturation for 3 min at 94°C, with 25 cycles of denaturation for 45 s at 94°C, annealing for 60 s at 52°C, and elongation for 90 s at 72°C, concluding with elongation for 10 min at 72°C. Reaction volumes were 25 μl and contained 200 nM of 515F and 806R primers [15] with added 1× 5PRIME HotMasterMix (5PRIME), 0.4 mg/ml BSA, and 5% dimethylsulfoxide, and duplicate reactions were run. Duplicate reactions were combined and Quant-iT PicoGreen dsDNA kit (Invitrogen, P11496) was used to determine concentration of DNA. The NucleoSpin Gel and PCR Clean-Up kit (Macherey-Nagel, 740609.250) was used for purification of the combined reactions and samples were subsequently diluted to 10 ng/µl and pooled in equal amounts. The pools were repurified with AMPure XP Kit (Agencourt, A63880) for removal of primer-dimers and short products. The repurified pools were run on a 2100 Bioanalyzer (Agilent) to certify that short amplification product was not present. The pooled amplicons were sequenced on an in-house MiSeq instrument (RTA (v.1.18.54.0) with MCS (v.2.6.2.1; Illumina) with the V2 kit (2 × 250 bp paired-ended reads; Illumina).
Reads were merged, filtered, de-duplicated, de-noised and aligned to Zero-radius operational taxonomic units (zOTUs) using Usearch 11.0.067 [16]. During merging a maximum of five mismatches were allowed in the alignment with a merge length between 251 and 257. Merged reads were filtered and reads with > 1 expected error were excluded, and remaining reads were subsequently de-duplicated. The filtered reads were de-noised using the UNOISE3 algorithm [17], and reads were aligned to each zOTUs’ sequence concluding pre-processing with Usearch. Each zOTUs’ sequence was then taxonomically assigned using DADA2's assignTaxonomy(minBoot = 50) [18,19] with a trained SILVA database v. 138 [20,21]. Prior to analysis, zOTUs that contributed less than 0.002% of the total amount of reads were filtered out and remaining zOTUs were normalized by resampling with replacement so that all samples had the same library size.
Microbiota analysis was performed using the software R (4.2.2). The phyloseq package (1.42.0) was used for handling zOTU data [22]. Correlation analysis was performed on zOTUs aggregated at the lowest taxonomic rank they were annotated at using Spearman’s correlation and visualized with either ggplot (3.4.0) or the ComplexHeatmap package (2.14.0). Prior to correlation analysis the zOTU counts were first centered log-ratio transformed filling zeroes with half of the minimum value. Principal Coordinate Analysis (PCoA) was performed using weighted UniFrac distance matrix and was also used for all other distance-based methods. Microbiota composition dissimilarity was tested with 9999 permutations with the adonis2 function from the vegan package (2.6.4) [23].
Statistical analyses
Mouse physiology data is presented with bar plots ± SEM and tested with unpaired two-tailed Student’s t-test. Genotype ratios were tested against a Mendelian ratio (1:2:1) with a Chi-square goodness of fit test. Comparisons of survival curves were made using Log-rank (Mantel-Cox) tests. Comparisons of the degree of Sirius Red and CK19 staining were made between Cyp2c70+/+ and Cyp2c70−/− females and males separately and only within the same group (GF, HUM and CD) and tested with Wilcoxon rank sum tests. Bile acid data are presented with pie-charts with bile acids contributing to ≥3% of the mean of the sum of bile acids. Comparisons of bile acid concentrations were made between Cyp2c70+/+ and Cyp2c70−/− females and males separately and only within the same group (GF, HUM and CD) and tested with Wilcoxon rank sum tests using Benjamini and Hochberg correction for multiple testing adjustment of individual bile acids. Differences in biliary hydrophobicity indices were tested using Kruskal–Wallis H tests. Following rejection of the null hypothesis, the Conover–Iman test was used for multiple comparisons using Benjamini and Hochberg correction for multiple testing adjustment. Differences in gene expression and liver transaminases were tested with Wilcoxon rank sum tests, and comparisons were made between Cyp2c70+/+ and Cyp2c70−/− females and males separately and only within the same group (GF, HUM and CD). Correlations between Cd177, F4/80, Tnfα, Ccl2, AST, or ALT and biliary hydrophobicity indices of bile acids were performed using Spearman’s correlation. Differences in α-diversity were tested with Wilcoxon rank sum tests between Cyp2c70+/+ and Cyp2c70−/− females and males separately and only within the same group (HUM and CD). Differences in abundance of Desulfovibrio and P. excrementihominis were tested with Wilcoxon rank sum tests and comparisons were made between early and late timepoints in CD Cyp2c70−/− mice. Correlations between bacterial taxa and biliary hydrophobicity indices of bile acids were tested using Spearman’s correlation and adjusted for multiple testing using Benjamini and Hochberg correction.
Results
Deletion of Cyp2c70 is partially lethal in the absence of a gut microbiota
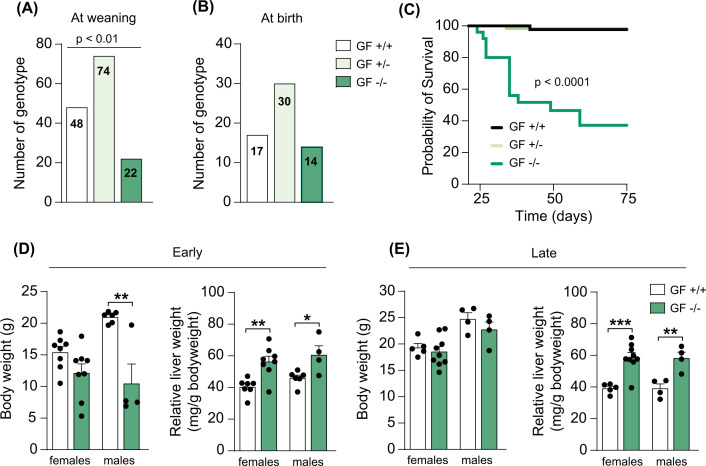
We rederived Cyp2c70+/− mice as GF by caesarian-section and bred GF Cyp2c70+/− females with Cyp2c70+/− males to obtain GF Cyp2c70−/− mice and observed that the offspring did not display the expected Mendelian ratio (1:2:1) at three weeks of age. From 144 pups, we obtained 48 Cyp2c70+/+ mice, 74 Cyp2c70+/− mice and 22 Cyp2c70−/− mice, indicating that complete deletion of the Cyp2c70 gene affects survival in a GF setting (Figure 1A). No differences were found between females and males. To investigate if the detrimental effects of Cyp2c70 deletion occur in utero or if the mice are actually born at the expected Mendelian ratio, we set up new breeding pairs and genotyped 9 litters at birth. From 61 pups, we obtained 17 Cyp2c70+/+ mice, 30 Cyp2c70+/− mice, and 14 Cyp2c70−/− mice, which corresponded to a Mendelian ratio (Figure 1B). This indicated that the reduced number of Cyp2c70−/− mice at 3 weeks was due to mortality during the neonatal period and not in utero. We then assessed overall survival in a subset of mice, from day 21 until day 75, and found that the median survival of the GF Cyp2c70−/− mice was 49 days (Figure 1C). Importantly, mice displaying severe worsening of the general condition such as weakness, rugged fur, and passivity were euthanized and considered not viable. Of note, the median survival for GF Cyp2c70+/+ or Cyp2c70+/− littermate controls could not be determined since only one mouse in each group had died during this period.
Figure 1. Reduced survival and increased relative liver weight in GF Cyp2c70−/− mice.
(A,B) Number of GF mice genotyped at weaning (three weeks of age) (A) and immediately after birth (B). (C) Kaplan–Meier estimator of survival over time in GF Cyp2c70 mice (Cyp2c70+/+, n=46, Cyp2c70+/−, n=61, Cyp2c70−/−, n=25) analyzed with Log-rank (Mantel-Cox) test. (D,E) Body weight and relative liver weight in GF Cyp2c70+/+ and Cyp2c70−/− mice at the early timepoint (D) (n=4–8 mice per group) and the late timepoint (E) (n=4–9 mice per group). Data are presented as mean ± SEM, *P<0.05, **P<0.01, ***P<0.001 indicate differences between female or male GF Cyp2c70+/+ and GF Cyp2c70−/− mice analyzed with two-tailed unpaired t-tests.
Next, to further investigate the impact of Cyp2c70 deletion under GF conditions we euthanized Cyp2c70−/− mice and their Cyp2c70+/+ littermate controls to evaluate phenotypical parameters. We euthanized mice at an early timepoint, i.e., 4–5 weeks of age, and at a later timepoint, i.e., at 6–10 weeks of age. At the early timepoint, most of the Cyp2c70−/− mice were small and hunchbacked. Furthermore, male Cyp2c70−/− mice had a lower body weight and both female and male Cyp2c70−/− mice showed an increased relative liver weight compared with their Cyp2c70+/+ littermate controls (Figure 1D). At the later timepoint, there were no differences in body weight anymore, but the relative liver weights were still significantly higher in both female and male Cyp2c70−/− mice compared with their Cyp2c70+/+ littermate controls (Figure 1E).
Colonization with a human or a mouse microbiota improves neonatal survival
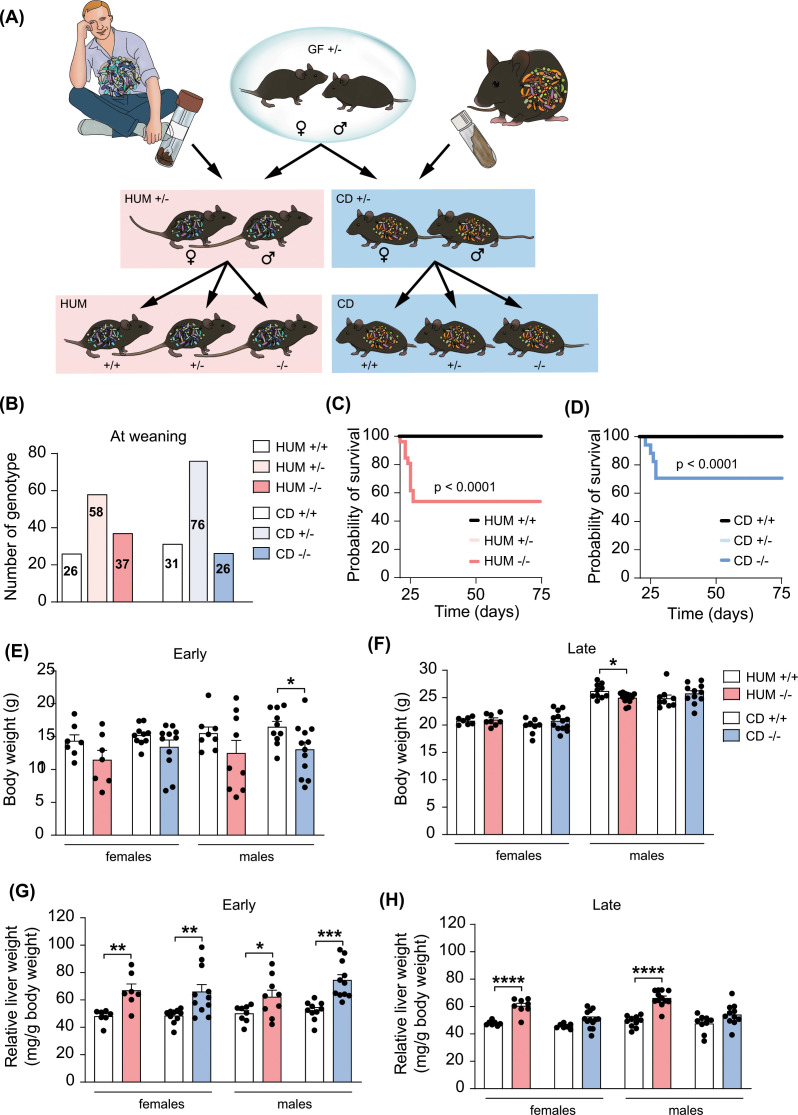
To test our hypothesis that a gut microbiota can improve survival and the hepatobiliary phenotype in Cyp2c70−/− mice, we colonized mice with human or mouse microbiota. First, Cyp2c70+/− mice were transferred into a new isolator and colonized with human microbiota from a healthy donor and 5 breeding pairs were set up to generate humanized (HUM) Cyp2c70−/− mice and HUM Cyp2c70+/+ littermate controls (Figure 2A). We obtained four to five litters from each breeding pair, generating 121 pups in total. Genotyping at three weeks showed the following distribution: 26 HUM Cyp2c70+/+, 58 HUM Cyp2c70+/− and 37 HUM Cyp2c70−/− mice (Figure 2B), which corresponds to a Mendelian ratio, indicating that there was no increased mortality during the first three neonatal weeks. However, a subset of the Cyp2c70−/− mice died or needed to be euthanized between day 21 and 26; thus, the mortality was not completely eliminated (Figure 2C).
Figure 2. Colonization of Cyp2c70+/− mice with human or mouse microbiota.
(A) Graphical illustration of the experimental design for the colonization experiments. (B) Number of HUM and CD mice genotyped at weaning (3 weeks of age). (C,D) Kaplan–Meier estimator of survival over time in HUM mice (C) (Cyp2c70+/+, n=21; Cyp2c70+/−, n=45; Cyp2c70−/−, n= 26) and CD Cyp2c70 mice (D) (Cyp2c70+/+, n=25, Cyp2c70+/−, n=37, Cyp2c70−/−, n=17) analyzed with Log-rank (Mantel-Cox) tests. (E,F) Body weight in HUM and CD Cyp2c70+/+ and Cyp2c70−/− mice at the early timepoint (E) (HUM n=7–9 mice per group, CD n=8–11 mice per group) and the late timepoint (F) (HUM n=7–9 mice per group, CD n=8–13 mice per group). (G,H) Relative liver weight of HUM and CD Cyp2c70+/+ and Cyp2c70−/− mice at the early timepoint (G) (same mice as in panel E) and the late timepoint (H) (same mice as in panel F). Data are presented as mean ± SEM; *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 indicate differences between female or male Cyp2c70+/+ and Cyp2c70−/− mice within each group (HUM or CD) analyzed with two-tailed unpaired t-tests.
Next, to generate conventionalized (CD) mice, we transferred GF Cyp2c70+/− mice into our barrier facility and colonized them with caecal content from a conventionally raised mouse and set up three breeding pairs to obtain CD Cyp2c70−/− mice (Figure 2A). The genotype distribution at 3 weeks was: 31 CD Cyp2c70+/+, 76 CD Cyp2c70+/−, and 26 CD Cyp2c70−/− mice, which is consistent with the expected Mendelian ratio (Figure 2B). Similar to HUM mice, the overall survival of CD Cyp2c70−/− mice was improved (Figure 2D).
Colonization with a mouse microbiota ameliorates the liver phenotype of Cyp2c70−/− mice
HUM and CD mice were euthanized at similar timepoints as the GF mice. At the early timepoint, male CD Cyp2c70−/− mice had a decreased body weight compared with their CD Cyp2c70+/+ littermate controls (Figure 2E) while genotype did not affect body weight in CD females or HUM mice. At the later timepoint, body weight was slightly lower in male HUM Cyp2c70−/− mice compared to their HUM Cyp2c70+/+ littermate controls (Figure 2F) whereas there were no differences in HUM females or CD mice. Relative liver weight was increased in both HUM and CD Cyp2c70−/− mice compared with their Cyp2c70+/+ counterparts at the early timepoint (Figure 2G), while at the later timepoint, relative liver weight was still higher in HUM Cyp2c70−/− mice, but not in the CD Cyp2c70−/− mice (Figure 2H).
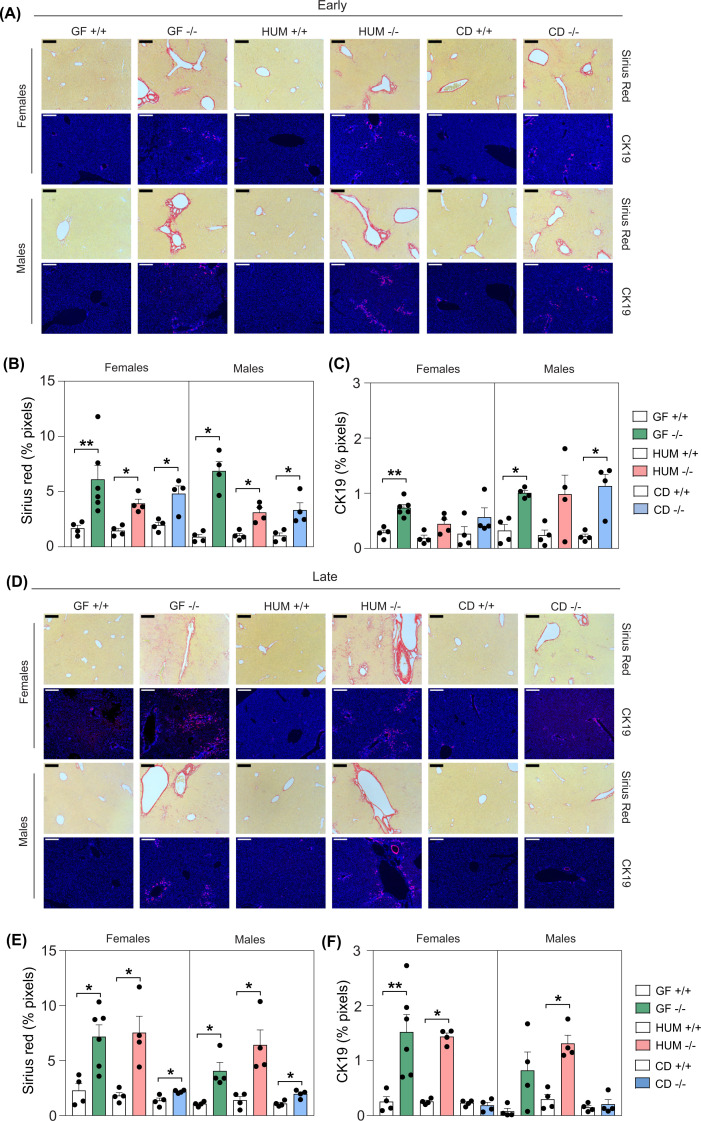
Sirius Red and CK19 staining showed that GF, HUM and CD Cyp2c70−/− mice had increased liver fibrosis and cholangiocyte proliferation at the early timepoint (Figure 3A–C). At the later timepoint, there were still clear features of liver fibrosis and cholangiocyte proliferation in GF and HUM Cyp2c70−/− mice, but not in CD Cyp2c70−/− mice (Figure 3D–F). In addition, qPCR analyses of liver tissue showed increased expression of immune cell (Cd177 and F4/80) and inflammation (Tnfα and Ccl2) markers in GF, HUM and CD Cyp2c70−/− mice at the early time point (Supplementary Figure S1A). At the late timepoint, there were still increased levels of infiltration and inflammation in the GF and HUM Cyp2c70−/− mice, while the levels in the CD Cyp2c70−/− mice were similar to CD Cyp2c70+/+ mice, which is consistent with an improved liver phenotype. Liver function tests in serum showed elevated liver transaminases (AST and ALT) in GF and HUM Cyp2c70−/− mice at both timepoints and the CD Cyp2c70−/− mice showed increased levels at the early timepoint, but normalized levels at the late timepoint mainly in the male mice (Supplementary Figure S1B).
Figure 3. Ameliorated liver phenotype in CD Cyp2c70−/− mice.
(A–F) Histological and immunological staining and quantification showing Sirius Red and CK19 in livers from female and male GF, HUM and CD Cyp2c70+/+ and Cyp2c70−/− mice at the early timepoint (A–C) and at the late timepoint (D–F). 200 µm indicators are shown. To increase visibility and brightness for the visual presentation of CK19-stained area the γ was set to 0.8 and gain to 1.7 (Blue: Hoechst, Red: CK19). Data are presented as mean ± SEM; *P<0.05 and **P<0.01 indicate differences between female or male Cyp2c70+/+ and Cyp2c70−/− mice within each group (GF, HUM or CD) analyzed with Wilcoxon rank sum tests.
In summary, colonization with a mouse or human microbiota improved neonatal survival of Cyp2c70−/− mice, and the presence of a mouse microbiota also ameliorated the liver phenotype at 6–10 weeks of age.
Colonization with a mouse microbiota increases production of TUDCA/UDCA
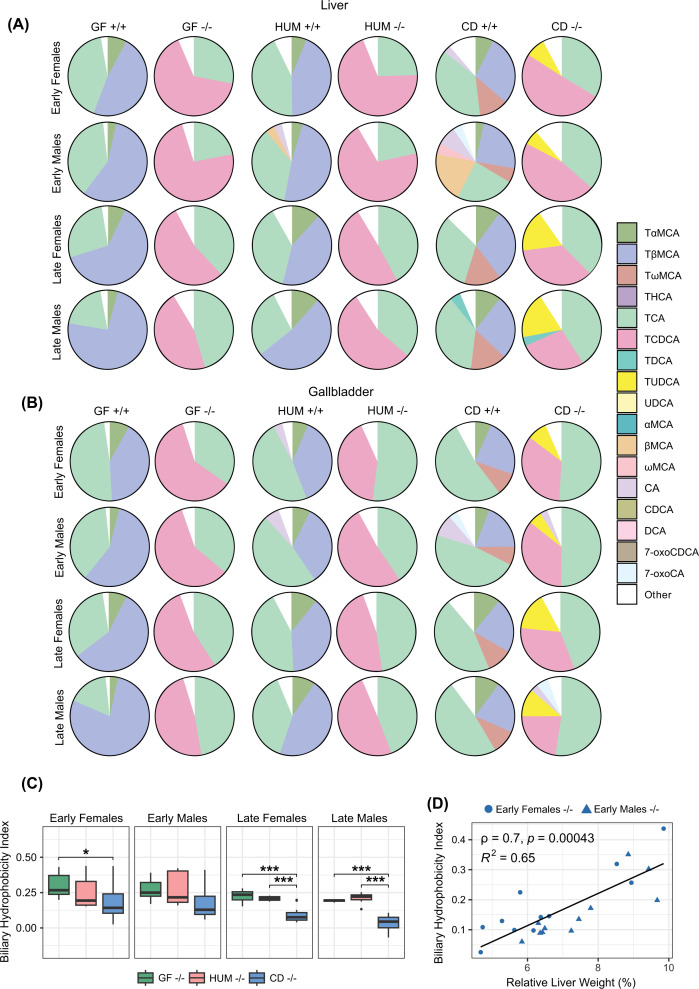
Bile acid analyses showed that deletion of Cyp2c70 decreased the levels of primary murine bile acids, TαMCA/αMCA and TβMCA/βMCA, and increased the proportion of TCDCA/CDCA in liver and gallbladder, resulting in a human-like primary bile acid profile (Figure 4A,B and Supplementary Table S1A,B). These observations are in line with previous findings [4–6,24]. We found traces of murine bile acids in some of the Cyp2c70−/− mice. This has previously been observed and may be explained by the fact that mice are coprophagous and Cyp2c70−/− mice were co-housed together with their Cyp2c70+/+ littermates [6].
Figure 4. Bile acid profiles in liver and gallbladder from GF, HUM, and CD mice.
(A,B) Bile acid profiles in liver (A) and gallbladder (B) from female and male GF, HUM, and CD Cyp2c70+/+ and Cyp2c70−/− mice at early and late timepoints. (C) Biliary hydrophobicity index of female and male GF, HUM, and CD Cyp2c70−/− mice at the early and late timepoints presented with a box plot. The lower (quartile 1) and upper (quartile 3) quartiles form the box with median represented by the central line. Whiskers reach to 1.5× the inner quartile range. Outliers are presented with visible points beyond the whiskers. (D) Scatterplot with a linear model fit and Spearman’s correlation analysis of biliary hydrophobicity index, and relative liver weight in CD Cyp2c70−/− mice at the early timepoint. *P<0.05, **P<0.01, ***P<0.001 indicate differences between female or male GF, HUM and CD Cyp2c70−/− mice analyzed with Kruskal–Wallis H tests with the Conover-Iman test as post hoc using Benjamini and Hochberg correction for multiple testing adjustment. Females, n = 5–12 mice per group; males, n = 4–12 mice per group. αMCA, α-muricholic acid; βMCA, β-muricholic acid; ωMCA, ω-muricholic acid; 7-oxoCDCA, 7-oxochenodeoxycholic acid; 7-oxoCA, 7-oxocholic acid; CA, cholic acid; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; TαMCA, tauro-α-muricholic acid; TβMCA, tauro-β-muricholic acid; TωMCA, tauro-ω-muricholic acid; TCA, tauro-cholic acid; TCDCA, tauro-chenodeoxycholic acid; TDCA, tauro-deoxycholic acid; THCA, tauro-hyocholic acid; TUDCA, tauro-ursodeoxycholic acid; UDCA, ursodeoxycholic acid.
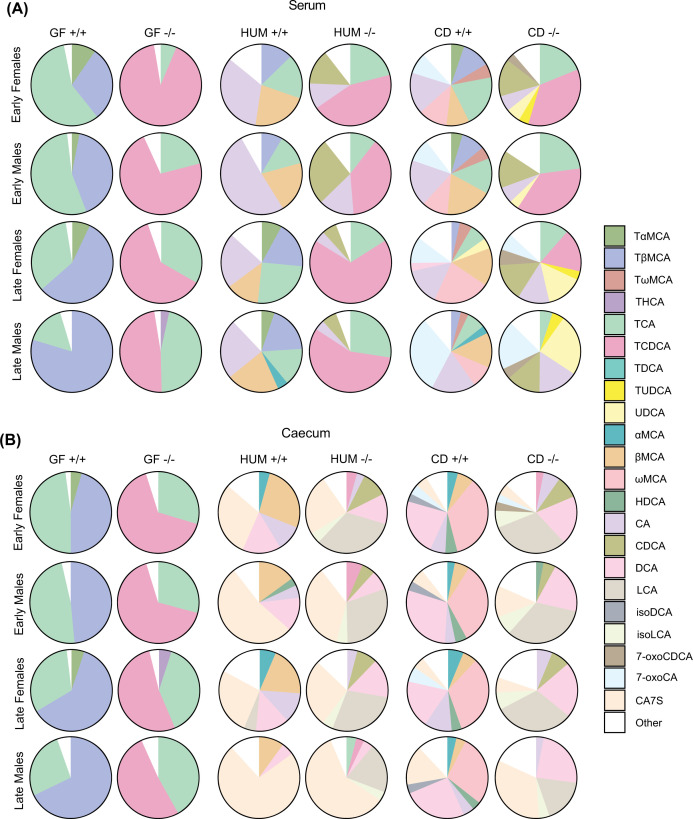
In GF Cyp2c70−/− mice, bile acid profiles in liver, gallbladder, serum and caecum contained almost exclusively TCA and TCDCA, while the profile in GF Cyp2c70+/+ mice was dominated by TCA and TβMCA (Figures 4A,B and 5A,B and Supplementary Table S1A–D). Hence, GF mice did not produce secondary bile acids, which is in line with our previous findings [1,25].
Figure 5. Bile acid profiles in serum and caecum from GF, HUM, and CD mice.
(A,B) Bile acid profiles in serum (A) and caecum (B) from female and male GF, HUM, and CD Cyp2c70+/+ and Cyp2c70−/− mice at the early and late timepoint. Females, n=4–13 mice per group; males, n=4–13 mice per group. αMCA, α-muricholic acid; βMCA, β-muricholic acid; ωMCA, ω-muricholic acid; 7-oxoCDCA, 7-oxochenodeoxycholic acid; 7-oxoCA, 7-oxocholic acid; CA7S, cholic acid-7-sulfate; CA, cholic acid; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; HDCA, hyodeoxycholic acid; isoDCA, iso-deoxycholic acid (3β-deoxycholic acid); isoLCA, iso-lithocholic acid (3β-lithocholic acid); LCA, lithocholic acid; TαMCA, tauro-α-muricholic acid; TβMCA, tauro-β-muricholic acid; TωMCA, tauro-ω-muricholic acid; TCA, tauro-cholic acid; TCDCA, tauro-chenodeoxycholic acid; TDCA, tauro-deoxycholic acid; THCA, tauro-hyocholic acid; TUDCA, tauro-ursodeoxycholic acid; UDCA, ursodeoxycholic acid.
HUM Cyp2c70−/− mice had a high proportion of TCA and TCDCA in liver and gallbladder, while HUM Cyp2c70+/+ mice had a high proportion of TCA and TβMCA. Thus, the bile acid profiles in liver and gallbladder of HUM mice were similar to those of GF mice (Figure 4A,B and Supplementary Table S1A,B). In addition to the bile acids found in HUM and GF mice, we found a large proportion of TUDCA in liver and gallbladder of CD Cyp2c70−/− mice and of tauro-ω-muricholic acid (TωMCA) in the CD Cyp2c70+/+ mice (Figure 4A,B and Supplementary Table S1A,B). UDCA and ωMCA can be produced by microbial transformation of CDCA and βMCA respectively [2]. These unconjugated bile acids are then transported to the liver where they can be conjugated to taurine, which is in line with the enrichment of TUDCA and TωMCA in liver and gallbladder of CD mice.
To compare the biochemical properties of the bile acid pools in Cyp2c70−/− mice, we calculated the hydrophobicity indices of biliary bile acids in GF, HUM and CD Cyp2c70−/− mice. At the early timepoint, there was a slight decrease in the biliary hydrophobicity index in CD Cyp2c70−/− females compared to GF Cyp2c70−/− females but no difference between the males. However, at the later timepoint the biliary hydrophobicity index was significantly lower in both female and male CD Cyp2c70−/− mice compared with their GF and HUM Cyp2c70−/− counterparts, largely due to higher amounts of hydrophilic TUDCA, and lower amounts of hydrophobic TCDCA (Figure 4C). The biliary hydrophobicity index of CD Cyp2c70−/− mice correlated with relative liver weight (Figure 4D), hepatic gene expression of Cd177, F4/80, Tnfα, and Ccl2, and serum levels of AST, and ALT (Supplementary Figure S2), indicating that a more hydrophilic bile acid profile is associated with an improved liver phenotype.
Serum bile acid profiles of HUM mice were dominated by primary bile acids, while CD mice had a more diverse profile including TωMCA/ωMCA in CD Cyp2c70+/+ mice and TUDCA/UDCA in CD Cyp2c70−/− mice (Figure 5A and Supplementary Table S1C). Bile acid profiles in caecum showed that all groups of colonized mice produced the secondary bile acid deoxycholic acid (DCA), which is generated from CA, and both HUM and CD Cyp2c70−/− mice, but not Cyp2c70+/+ mice, produced the secondary bile acid lithocholic acid (LCA), which is generated from CDCA (Figure 5B and Supplementary Table S1D). Additionally, we found the sulphated bile acid CA7S in all groups of colonized mice with particularly high levels in HUM mice (Figure 5B and Supplementary Table S1D), which is in line with our previous observations [25].
Cyp2c70−/− mice showed a tendency toward increased total bile acid levels in serum and decreased levels in caecum and gallbladder compared with Cyp2c70+/+ littermate controls (Supplementary Figure S3 and Supplementary Table S1B–D). This may be interpreted as an indication of decreased biliary bile acid secretion and thus cholestasis, yet, reduced biliary bile acid secretion was not observed in earlier studies [6]. Notably, apart from a small increase in total hepatic bile acid levels in male HUM Cyp2c70−/− mice at the early timepoint, deletion of Cyp2c70 did not affect bile acid levels in the liver, indicating that the liver phenotype of Cyp2c70−/− mice was not driven by elevated hepatic bile acid levels (Supplementary Table S1A and Supplementary Figure S3). Noteworthy, the male CD mice had low total bile acid levels in both liver and serum at the later timepoint, without differences between the genotypes (Supplementary Figure S3) indicating normalization of liver function upon conventionalization.
Taken together, we show that deletion of Cyp2c70 in a GF setting diminished the production of TαMCA and TβMCA and increased the contribution of TCDCA to the bile acid pool. Additionally, Cyp2c70−/− mice colonized with human or mouse microbiota generated unconjugated primary bile acids and the secondary bile acids DCA and LCA. Interestingly, colonization with a mouse but not with a human microbiota resulted in increased proportions of TUDCA/UDCA, translating into a more hydrophilic bile acid profile.
Alterations in the gut microbiota are linked to a more hydrophilic bile acid profile and improved liver phenotype
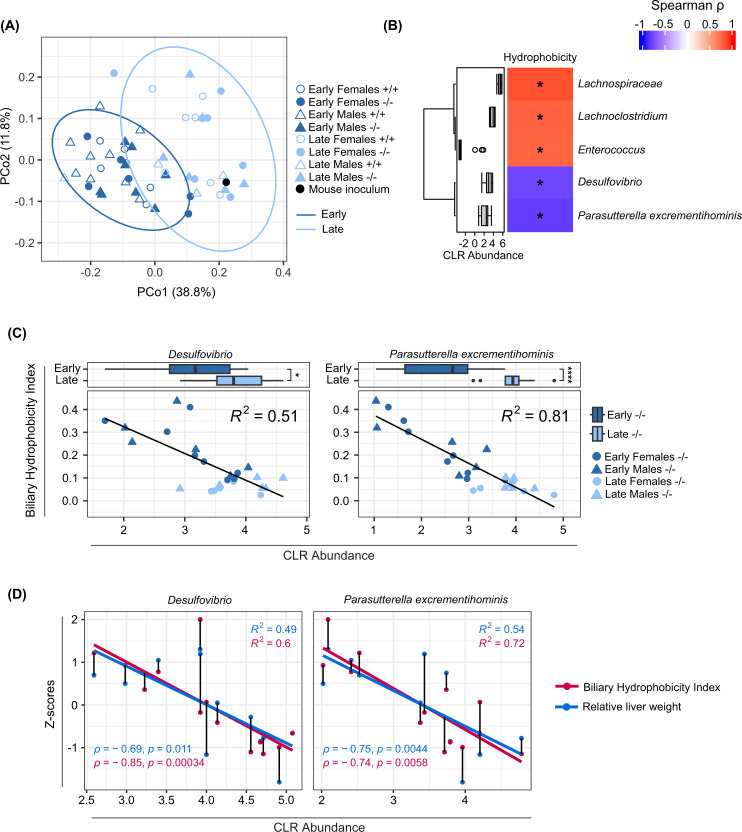
In line with previous findings that transfer of human microbiota to mice is generally less efficient than transfer of mouse microbiota [25–28], we found that the α-diversity was lower in HUM mice compared with CD mice (Supplementary Figure S4A). The difference in α-diversity was also larger between the human inoculum and HUM recipient mice than between the mouse inoculum and CD recipient mice. Deletion of Cyp2c70 did not affect α-diversity in neither HUM nor CD mice. Principle coordinate analysis (PCoA) showed that the mouse donor sample clustered together with CD recipient mice while human donor samples and HUM recipient mice were separated, indicating reduced colonization of human microbiota compared with mouse microbiota (Supplementary Figure S4B).
We next performed PCoA of the microbiota from CD mice to evaluate differences in composition between genotypes and timepoints and found that the microbiota composition was mainly influenced by timepoint (Figure 6A). However, permutational ANOVA showed that the microbiota composition differed between CD Cyp2c70+/+ and Cyp2c70−/− mice at the early timepoint, and between early and late timepoints in both Cyp2c70+/+ and Cyp2c70−/− mice (Supplementary Table S2A). Hence, there was a genotype-induced difference at the early timepoint. To assess whether there was a link between bile acid profile composition and the gut microbiota, we performed a permutational ANOVA between the biliary hydrophobicity index and the gut microbiota composition in the CD Cyp2c70−/− mice at the early timepoint and found that the biliary hydrophobicity index could explain 19.9% of the variation in the gut microbiota composition (Supplementary Table S2B). Next, we performed correlation analyses between the biliary hydrophobicity index and bacterial taxa in CD Cyp2c70−/− mice at the early timepoint and found that Desulfovibrio and Parasutterella excrementihominis correlated negatively with the biliary hydrophobicity index (Figure 6B). The abundance of these bacterial taxa was higher in the CD Cyp2c70−/− mice at the late timepoint, and was associated with a lower hydrophobicity index (Figure 6C). Finally, a more hydrophilic bile acid profile, as well as a lower relative liver weight was associated with increased abundance of both Desulfovibrio and P. excrementihominis (Figure 6D).
Figure 6. Caecal microbiota analyses in CD mice.
(A) Microbiota analysis by 16S rRNA sequencing of caecum from CD mice showing first and second Principal Coordinates of the weighted UniFrac distance matrix of CD Cyp2c70+/+ and Cyp2c70−/− mice at early and late timepoints and the mouse inoculum. Ellipses indicate 95% confidence assuming a multivariate t-distribution. (B) Spearman's correlation between biliary hydrophobicity index and caecal bacterial taxa of CD Cyp2c70−/− mice at the early timepoint. Distribution of bacterial abundance is presented as centered log-ratio (CLR) abundance with a box plot. The lower (quartile 1) and upper (quartile 3) quartiles form the box with the median represented by the central line. Whiskers reach to 1.5× the inner quartile range. Outliers are presented with visible points beyond the whiskers. (C) Scatterplots with linear models fitted of biliary hydrophobicity index and bacterial taxa presented as CLR abundance for CD Cyp2c70−/− mice at early and late timepoints. Distribution of bacterial abundance is presented with a box plot. The lower (quartile 1) and upper (quartile 3) quartiles form the box with the median represented by the central line. Whiskers reach to 1.5× the inner quartile range. Outliers are presented with visible points beyond the whiskers. (D) Scatterplots with linear models fitted of bacterial taxa presented as CLR abundance that correlate with biliary hydrophobicity index and relative liver weight in CD Cyp2c70−/− mice at the early timepoint. Lines between points signify that the observations are from the same mouse; *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 indicate differences between Cyp2c70−/− mice at early and late timepoints analyzed with Wilcoxon rank sum tests.
Discussion
In this study we used the novel Cyp2c70−/− mouse model to investigate whether a gut microbiota can be protective in the development of hepatobiliary disease. We rederived Cyp2c70−/− mice as GF and demonstrated that deletion of Cyp2c70 in the absence of bacteria led to a markedly reduced neonatal survival. To examine the protective role of the gut microbiota, we colonized GF Cyp2c70+/− mice with either a human or a mouse microbiota and showed that the presence of a microbiota improved neonatal survival of Cyp2c70−/− mice, independent of donor origin, while signs of liver fibrosis, immune cell infiltration and expression of inflammatory markers as well as ductular reactions only improved in Cyp2c70−/− mice colonized with a mouse microbiota.
The increased mortality of GF Cyp2c70−/− mice may be caused by the substantially elevated levels of the hydrophobic, and thus cytotoxic, bile acid TCDCA [14]. TCDCA cannot be metabolized in a GF setting and this could lead to toxic effects during the neonatal state. However, our bile acid analyses showed that bile acid profiles in liver and gallbladder were very similar in HUM and GF Cyp2c70−/− mice, yet neonatal survival was improved in the HUM Cyp2c70−/− mice. This suggests that there are other developmental changes occurring after birth, which are microbiota-dependent and could be crucial for coping with toxic bile acids, such as formation of bile ducts or structures in the gastrointestinal tract [29–31].
HUM Cyp2c70−/− mice had decreased microbial diversity compared with CD Cyp2c70−/− mice. Still the bile acid profile in caecum was rather similar in HUM and CD Cyp2c70−/− mice, indicating that the decreased diversity was not associated with a decreased capacity to produce secondary bile acids in general. On the contrary, bile acid profiles in liver and gallbladder differed substantially between HUM and CD Cyp2c70−/− mice. CD Cyp2c70−/− mice had a large proportion of TUDCA, while HUM and GF Cyp2c70−/− mice had similar profiles dominated by TCDCA and TCA. A small proportion of TUDCA was detected in the GF mice, which confirms our previous finding that UDCA is a primary bile acid in mice [1]. However, there was a robust increase in TUDCA and UDCA production in the CD mice suggesting that UDCA primarily is a secondary bile acid produced by specific bacteria. The most likely route for UDCA production is 7β-epimerization from CDCA via the intermediate 7-oxoCDCA [32,33]. However, we cannot determine whether all steps in the transformation were carried out by bacteria in the gut or if some modifications occurred in the liver. It has previously been shown in an in vitro study using mouse liver microsomes that 7-oxoCDCA can be converted to CDCA and, to a lesser extent, to UDCA [34]. Interestingly, the levels of UDCA were low in caecum suggesting that UDCA was produced in the upper part of the gut and efficiently absorbed into the enterohepatic circulation, or alternatively, that it was produced in the liver. The HUM Cyp2c70−/− mice had high levels of the precursor CDCA yet very low UDCA levels, indicating that some microbial enzymes essential for 7β-epimerization of CDCA were missing in the HUM Cyp2c70−/− mice. This could be due to lack of specific bacteria in the human donor sample or due to limited transfer of human bacteria into the recipient mice, which has been reported in previous studies with GF or antibiotic-treated wild-type mice [25–28]. We hypothesized that colonization of Cyp2c70−/− mice with a human microbiota, which have a human-like bile acid profile, would be a more physiological model to study bile acid–microbiota interactions in liver disease. However, the reduced neonatal survival of GF Cyp2c70−/− mice prohibited us from colonizing adult or adolescent GF Cyp2c70−/− mice and instead we colonized heterozygous breeding pairs, which is a limitation of the model. If Cyp2c70−/− mice could be colonized directly, the transfer of human bacteria might be improved, and it would also facilitate colonization and comparison between different human donors.
The proportion of TUDCA/UDCA was increased at the later timepoint, which was reflected by a decreased biliary hydrophobicity index, and we believe that the improved hepatobiliary phenotypes in CD Cyp2c70−/− mice at this timepoint could be, at least in part, attributed to microbial production of hydrophilic, and thus hepatoprotective, UDCA. Indeed, feeding of female Cyp2c70−/− mice with a UDCA-enriched diet (0.1% wt/wt) fully restored cholangiopathy within a 6-week time frame [6], while the hydrophobic FXR agonist obeticholic acid (OCA) was not effective [35]. We found that a lower hydrophobicity index was associated with lower relative liver weight, and that this was associated with increased abundance of Desulfovibrio and P. excrementihominis. However, there is no evidence that these are UDCA-producing bacteria.
In accordance with our results, previous studies on the cholestatic mouse model induced by deletion of Mdr2/Abcb4, have also demonstrated a worsened hepatobiliary phenotype in GF conditions, suggesting that a commensal microbiota may be protective against bile duct injury [9,36]. One important difference between the Mdr2−/− mouse model and our model, is that Mdr2−/− mice have a murine primary bile acid profile but lack phospholipids in bile [37], while Cyp2c70−/− mice have a human-like primary bile acid profile, which is more hydrophobic, and hence toxic, and a higher biliary phospholipid content [6]. Interestingly, one study found no increased mortality in GF Mdr2−/− mice during the first 60 days [9], while another study found no neonatal mortality, but 100% mortality at 8 weeks. Furthermore, treatment with broad spectrum antibiotics in adult Mdr2−/− mice, which result in a GF-like condition, increased the lethality [10].
One common feature of the Mdr2−/− and Cyp2c70−/− models is that UDCA seems to be protective against the somewhat similar hepatobiliary phenotypes. Previous studies have shown that UDCA feeding reverses the adult-onset of a cholestatic liver phenotype in conventionally raised Cyp2c70−/− mice at 27 weeks of age [6] and improves liver pathology in Mdr2−/− mice [38,39]. Another study showed that perinatal exposure to UDCA prevented neonatal cholestasis of conventionally raised Cyp2c70−/− mice, but there were no long-lasting effects on liver pathophysiology after discontinuation of UDCA treatment [40].
In line with our findings that a gut microbiota has a protective role in hepatobiliary disease, colonization experiments with GF Mdr2−/− mice showed that liver-related mortality was reduced when mice were colonized at 3–4 weeks of age, but not when colonization was performed at 5–7 weeks [36]. However, additional experiments identified some bacteria, such as Enterococcus faecalis and Escherichia coli, with detrimental effects on disease development, which underscores that the role of the microbiota in hepatobiliary disease is complex and needs further investigation.
In conclusion, we show that the presence of a gut microbiota is important for neonatal survival of Cyp2c70−/− mice. We also show that microbially induced production of TUDCA/UDCA results in a hydrophilic bile acid profile, which correlates with improved liver phenotype in Cyp2c70−/− mice. Our model emphasizes that there is an important link between the gut microbiota and hepatobiliary disease and a future strategy could be to identify UDCA-producing bacteria and test their hepatoprotective ability.
Clinical perspectives
Conventionally raised Cyp2c70−/− mice with a human-like bile acid composition have shown promise as a model for hepatobiliary disease, however, the impact of the gut microbiota in disease development and progression is unknown.
We rederived Cyp2c70−/− mice as GF and showed that the absence of a gut microbiota reduced neonatal survival, and that colonization with a human or mouse microbiota improved neonatal survival while only mouse microbiota induced TUDCA/UDCA production, which was reflected by a lower hydrophobicity index of biliary bile acids and improved liver phenotype.
Our findings indicate that microbially induced production of TUDCA/UDCA may protect against hepatobiliary disease in Cyp2c70−/− mice.
Supplementary Material
Acknowledgments
The authors thank Anna Hallén, Carina Arvidsson, Louise Helldén and Gabriéla Rydén for technical assistance and Anna Hallén for assistance with figures and artwork and the authors thank Barbara Becattini for providing primers for Ccl2, F4/80, and Tnfα.
Abbreviations
- αMCA
α-muricholic acid
- βMCA
β-muricholic acid
- ωMCA
ω-muricholic acid
- 7-oxoCA
7-oxocholic acid
- 7-oxoCDCA
7-oxochenodeoxycholic acid
- CA7S
cholic acid-7-sulfate
- CA
cholic acid
- CDCA
chenodeoxycholic acid
- DCA
deoxycholic acid
- HDCA
hyodeoxycholic acid
- isoDCA
iso-deoxycholic acid (3β-deoxycholic acid)
- isoLCA
iso-lithocholic acid (3β-lithocholic acid)
- LCA
lithocholic acid
- TαMCA
tauro-α-muricholic acid
- TβMCA
tauro-β-muricholic acid
- TωMCA
tauro-ω-muricholic acid
- TCA
tauro-cholic acid
- TCDCA
tauro-chenodeoxycholic acid
- TDCA
tauro-deoxycholic acid
- THCA
tauro-hyocholic acid
- TUDCA
tauro-ursodeoxycholic acid
- UDCA
ursodeoxycholic acid
Contributor Information
Fredrik Bäckhed, Email: Fredrik.Backhed@wlab.gu.se.
Hanns-Ulrich Marschall, Email: hanns-ulrich.marschall@gu.se.
Data Availability
16S rRNA sequencing data have been deposited to the European Nucleotide Archive under accession number PRJEB61888 (https://www.ebi.ac.uk/ena/browser/view/PRJEB61888). The random forest model for Sirius Red histological analysis is available at Github (https://github.com/wilhelmsjoland/Cyp2c70).
Competing Interests
F.B. receives research funding from Biogaia AB, is co-founder and shareholder of Roxbiosens Inc and Implexion AB, and is on the scientific advisory board of Bactolife A/S. The other authors declare no competing financial interests of relevance for this manuscript.
Funding
This study was supported by the Swedish Research Council [grant number 2019-01599 (to F.B.) and 2019-01069 (to H.-U.M.)]; Transatlantic Networks of Excellence Award from the Leducq Foundation [grant number 17CVD01]; Novo Nordisk Fonden [grant number NNF21OC0070298]; Swedish Heart Lung Foundation [grant number 20210366]; Knut and Alice Wallenberg Foundation [grant number 2017.0026]; grants from the Swedish state under the agreement between the Swedish government and the county councils; the ALF-agreement [grant numbers ALFGBG-718101 (to F.B.) and ALFGBG-964970 (to H.-U.M.)]; and by the Sahlgrenska Academy, University of Gothenburg, project-linked PhD-project (to H.-U.M.and W.S.]. F.B. is Wallenberg Scholar and Torsten Söderberg Professor in Medicine. F.K. is supported by the Netherlands Heart Foundation (IN CONTROL II, CVON 2018-27) and the Noaber Foundation, Lunteren, The Netherlands. The computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under project SNIC 2019-3-384.
Open Access
Open access for this article was enabled by the participation of University Of Gothenburg in an all-inclusive Read & Publish agreement with Portland Press and the Biochemical Society under a transformative agreement with Individual.
CRediT Author Contribution
Wilhelm Sjöland: Conceptualization, Software, Formal analysis, Investigation, Visualization, Methodology, Writing—original draft, Writing—review & editing. Annika Wahlström: Conceptualization, Formal analysis, Supervision, Investigation, Visualization, Writing—original draft, Writing—review & editing. Kassem Makki: Investigation, Methodology, Writing—review & editing. Marc Schöler: Methodology, Writing—review & editing. Antonio Molinaro: Methodology. Lisa Olsson: Software, Formal analysis, Methodology, Writing—review & editing. Thomas Uwe Greiner: Methodology, Writing—review & editing. Robert Caesar: Writing—review & editing. Jan Freark de Boer: Conceptualization, Writing—review & editing. Folkert Kuipers: Conceptualization, Writing—review & editing. Fredrik Bäckhed: Conceptualization, Resources, Supervision, Funding acquisition, Writing—review & editing. Hanns-Ulrich Marschall: Conceptualization, Resources, Supervision, Funding acquisition, Writing—review & editing.
References
- 1.Sayin S.I., Wahlstrom A., Felin J., Jantti S., Marschall H.U., Bamberg K.et al. (2013) Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17, 225–235 10.1016/j.cmet.2013.01.003 [DOI] [PubMed] [Google Scholar]
- 2.Wahlstrom A., Sayin S.I., Marschall H.U. and Backhed F. (2016) Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 24, 41–50 10.1016/j.cmet.2016.05.005 [DOI] [PubMed] [Google Scholar]
- 3.Takahashi S., Fukami T., Masuo Y., Brocker C.N., Xie C., Krausz K.W.et al. (2016) Cyp2c70 is responsible for the species difference in bile acid metabolism between mice and humans. J. Lipid Res. 57, 2130–2137 10.1194/jlr.M071183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honda A., Miyazaki T., Iwamoto J., Hirayama T., Morishita Y., Monma T.et al. (2020) Regulation of bile acid metabolism in mouse models with hydrophobic bile acid composition. J. Lipid Res. 61, 54–69 10.1194/jlr.RA119000395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Boer J.F., Verkade E., Mulder N.L., de Vries H.D., Huijkman N., Koehorst M.et al. (2020) A human-like bile acid pool induced by deletion of hepatic Cyp2c70 modulates effects of FXR activation in mice. J. Lipid Res. 61, 291–305 10.1194/jlr.RA119000243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Boer J.F., de Vries H.D., Palmiotti A., Li R., Doestzada M., Hoogerland J.A.et al. (2021) Cholangiopathy and biliary fibrosis in Cyp2c70-deficient mice are fully reversed by ursodeoxycholic acid. Cell Mol. Gastroenterol. Hepatol. 11, 1045–1069 10.1016/j.jcmgh.2020.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tabibian J.H., Talwalkar J.A. and Lindor K.D. (2013) Role of the microbiota and antibiotics in primary sclerosing cholangitis. Biomed. Res. Int. 2013, 389537 10.1155/2013/389537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hov J.R. and Karlsen T.H. (2023) The microbiota and the gut-liver axis in primary sclerosing cholangitis. Nat. Rev. Gastroenterol. Hepatol. 20, 135–154 10.1038/s41575-022-00690-y [DOI] [PubMed] [Google Scholar]
- 9.Tabibian J.H., O'Hara S.P., Trussoni C.E., Tietz P.S., Splinter P.L., Mounajjed T.et al. (2016) Absence of the intestinal microbiota exacerbates hepatobiliary disease in a murine model of primary sclerosing cholangitis. Hepatology 63, 185–196 10.1002/hep.27927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider K.M., Candels L.S., Hov J.R., Myllys M., Hassan R., Schneider C.V.et al. (2021) Gut microbiota depletion exacerbates cholestatic liver injury via loss of FXR signalling. Nat. Metab. 3, 1228–1241 10.1038/s42255-021-00452-1 [DOI] [PubMed] [Google Scholar]
- 11.Marschall H.U. and Backhed F. (2016) Could gut microbiota protect against sclerosing cholangitis? Hepatology 63, 26–27 10.1002/hep.28135 [DOI] [PubMed] [Google Scholar]
- 12.Liang W., Menke A.L., Driessen A., Koek G.H., Lindeman J.H., Stoop R.et al. (2014) Establishment of a general NAFLD scoring system for rodent models and comparison to human liver pathology. PLoS ONE 9, ARTN e115922 10.1371/journal.pone.0115922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yaniv Z., Lowekamp B.C., Johnson H.J. and Beare R. (2019) Correction to: SimpleITK image-analysis notebooks: a collaborative environment for education and reproducible research. J. Digit. Imaging 32, 1118 10.1007/s10278-018-0165-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heuman D.M. (1989) Quantitative estimation of the hydrophilic-hydrophobic balance of mixed bile salt solutions. J. Lipid Res. 30, 719–730 10.1016/S0022-2275(20)38331-0 [DOI] [PubMed] [Google Scholar]
- 15.Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K. and Schloss P.D. (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar R.C. (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 17.Edgar R.C. (2016) UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. bioRxiv 081257 10.1101/081257 [DOI] [Google Scholar]
- 18.Wang Q., Garrity G.M., Tiedje J.M. and Cole J.R. (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J. and Holmes S.P. (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yilmaz P., Parfrey L.W., Yarza P., Gerken J., Pruesse E., Quast C.et al. (2014) The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–D648 10.1093/nar/gkt1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P.et al. (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMurdie P.J. and Holmes S. (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oksanen J., Blanchet F.G., Kindt R., Legendre P., Minchin P.R., O'hara R.B.et al. (2013) Community ecology package. R Package Version 2, 321–326 [Google Scholar]
- 24.Truong J.K., Bennett A.L., Klindt C., Donepudi A.C., Malla S.R., Pachura K.J.et al. (2022) Ileal bile acid transporter inhibition in Cyp2c70 KO mice ameliorates cholestatic liver injury. J. Lipid Res. 63, 100261 10.1016/j.jlr.2022.100261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wahlstrom A., Kovatcheva-Datchary P., Stahlman M., Khan M.T., Backhed F. and Marschall H.U. (2017) Induction of farnesoid X receptor signaling in germ-free mice colonized with a human microbiota. J. Lipid Res. 58, 412–419 10.1194/jlr.M072819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turnbaugh P.J., Ridaura V.K., Faith J.J., Rey F.E., Knight R. and Gordon J.I. (2009) The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1, 6ra14 10.1126/scitranslmed.3000322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staley C., Kaiser T., Beura L.K., Hamilton M.J., Weingarden A.R., Bobr A.et al. (2017) Stable engraftment of human microbiota into mice with a single oral gavage following antibiotic conditioning. Microbiome 5, 87 10.1186/s40168-017-0306-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L., Bahl M.I., Roager H.M., Fonvig C.E., Hellgren L.I., Frandsen H.L.et al. (2017) Environmental spread of microbes impacts the development of metabolic phenotypes in mice transplanted with microbial communities from humans. ISME J. 11, 676–690 10.1038/ismej.2016.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanimizu N., Kaneko K., Itoh T., Ichinohe N., Ishii M., Mizuguchi T.et al. (2016) Intrahepatic bile ducts are developed through formation of homogeneous continuous luminal network and its dynamic rearrangement in mice. Hepatology 64, 175–188 10.1002/hep.28521 [DOI] [PubMed] [Google Scholar]
- 30.Lemaigre F.P. (2003) Development of the biliary tract. Mech. Dev. 120, 81–87 10.1016/S0925-4773(02)00334-9 [DOI] [PubMed] [Google Scholar]
- 31.Frazer L.C. and Good M. (2022) Intestinal epithelium in early life. Mucosal. Immunol. 15, 1181–1187 10.1038/s41385-022-00579-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salen G., Verga D., Batta A.K., Tint G.S. and Shefer S. (1982) Effect of 7-ketolithocholic acid on bile acid metabolism in humans. Gastroenterology 83, 341–347 10.1016/S0016-5085(82)80326-0 [DOI] [PubMed] [Google Scholar]
- 33.Lee J.Y., Arai H., Nakamura Y., Fukiya S., Wada M. and Yokota A. (2013) Contribution of the 7beta-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon. J. Lipid Res. 54, 3062–3069 10.1194/jlr.M039834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Penno C.A., Morgan S.A., Vuorinen A., Schuster D., Lavery G.G. and Odermatt A. (2013) Impaired oxidoreduction by 11beta-hydroxysteroid dehydrogenase 1 results in the accumulation of 7-oxolithocholic acid. J. Lipid Res. 54, 2874–2883 10.1194/jlr.M042499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li R., Hovingh M.V., Koehorst M., de Blaauw P., Verkade H.J., de Boer J.F.et al. (2022) Short-term obeticholic acid treatment does not impact cholangiopathy in Cyp2c70-deficient mice with a human-like bile acid composition. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 1867, 159163 10.1016/j.bbalip.2022.159163 [DOI] [PubMed] [Google Scholar]
- 36.Awoniyi M., Wang J., Ngo B., Meadows V., Tam J., Viswanathan A.et al. (2023) Protective and aggressive bacterial subsets and metabolites modify hepatobiliary inflammation and fibrosis in a murine model of PSC. Gut 72, 671–685 10.1136/gutjnl-2021-326500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smit J.J., Schinkel A.H., Oude Elferink R.P., Groen A.K., Wagenaar E., van Deemter L.et al. (1993) Homozygous disruption of the murine mdr2 P-glycoprotein gene leads to a complete absence of phospholipid from bile and to liver disease. Cell 75, 451–462 10.1016/0092-8674(93)90380-9 [DOI] [PubMed] [Google Scholar]
- 38.Van Nieuwkerk C.M., Elferink R.P., Groen A.K., Ottenhoff R., Tytgat G.N., Dingemans K.P.et al. (1996) Effects of Ursodeoxycholate and cholate feeding on liver disease in FVB mice with a disrupted mdr2 P-glycoprotein gene. Gastroenterology 111, 165–171 10.1053/gast.1996.v111.pm8698195 [DOI] [PubMed] [Google Scholar]
- 39.Meng F., Kennedy L., Hargrove L., Demieville J., Jones H., Madeka T.et al. (2018) Ursodeoxycholate inhibits mast cell activation and reverses biliary injury and fibrosis in Mdr2(-/-) mice and human primary sclerosing cholangitis. Lab. Invest. 98, 1465–1477 10.1038/s41374-018-0101-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Vries H.D., Palmiotti A., Li R., Hovingh M.V., Mulder N.L., Koehorst M.et al. (2023) Perinatal exposure to UDCA prevents neonatal cholestasis in Cyp2c70(-/-) mice with human-like bile acids. Pediatr. Res. 93, 1582–1590 10.1038/s41390-022-02303-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S rRNA sequencing data have been deposited to the European Nucleotide Archive under accession number PRJEB61888 (https://www.ebi.ac.uk/ena/browser/view/PRJEB61888). The random forest model for Sirius Red histological analysis is available at Github (https://github.com/wilhelmsjoland/Cyp2c70).