Abstract
Oxidative stress has been identified as a major cause of cellular injury in a variety of neurodegenerative disorders. This study aimed to investigate the cytoprotective effects of piceatannol on hydrogen peroxide (H2O2)-induced pheochromocytoma-12 (PC-12) cell damage and explore the underlying mechanisms. Our findings indicated that piceatannol pre-treatment significantly attenuated H2O2-induced PC-12 cell death. Furthermore, piceatannol effectively improved mitochondrial content and mitochondrial function, including enhancing mitochondrial reactive oxygen species (ROS) elimination capacity and increasing mitochondrial transcription factor (TFAM), peroxisome-proliferator-activated receptor-γ coactivator-1α (PGC-1α) and mitochondria Complex IV expression. Meanwhile, piceatannol treatment inhibited mitochondria-mediated autophagy as demonstrated by restoring mitochondrial membrane potential, reducing autophagosome formation and light chain 3B II/I (LC3B II/I) and autophagy-related protein 5 (ATG5) expression level. The protein expression level of SIRT3 was significantly increased by piceatannol in a concentration-dependent manner. However, the cytoprotective effect of piceatannol was dramatically abolished by sirtuin 3 (SIRT3) inhibitor, 3-(1H-1,2,3-Triazol-4-yl) pyridine (3-TYP), which led to an exacerbated mitochondrial dysfunction and autophagy in PC-12 cells under oxidative stress. In addition, the autophagy activator (rapamycin) abrogated the protective effects of piceatannol on PC-12 cell death. These findings demonstrated that piceatannol could alleviate PC-12 cell oxidative damage and mitochondrial dysfunction by inhibiting autophagy via the SIRT3 pathway.
Keywords: piceatannol, oxidative damage, mitochondrial function, autophagy, SIRT3
1. Introduction
Oxidative stress, a hallmark of aging, acts as the main risk factor for most neurodegenerative diseases including Alzheimer’s disease (AD) and Parkinson’s disease (PD) [1,2]. Reactive oxygen species (ROS) can be generated from the impairment of metabolic function under oxidative stress [3]. Excessive ROS from oxidative stress led to mitochondrial dysfunction, causing neuron injury [4]. The mitochondrion is essential for the maintenance of cellular homeostasis involved in cell processes relevant to neuronal function, including autophagy [5]. Cai et al. found that mitochondrial ROS generation could decrease mitochondrial membrane potential (MMP), activating cell autophagy [6]. Moreover, ROS participates as a signal molecule associated with autophagy, considered as the main mechanism of oxidative stress-caused neurodegenerative disorders [7]. Autophagy is a highly conserved cellular process of the degradation of intracellular organelles and components by lysosomes [8]. The autophagic pathway plays an important role in the differentiation, growth, maintenance and homeostasis of neuronal cells. Glycine was demonstrated to attenuate oxidative injury in neurons via reducing autophagy both in vitro and in vivo [6]. Meanwhile, the dysregulation of autophagy has been shown to contribute to the pathogenesis of neurological diseases [9]. Thus, the regulation of autophagy could be a potential strategy to prevent neurological disease processes.
Sirtuin 3 (SIRT3), a nicotinamide adenine dinucleotide (NAD+)-dependent enzyme, is highly expressed in the nervous system and vital to physiological neuronal processes and brain functions, which modulates mitochondrial function, metabolism and oxidation responses [10,11]. Shulyakova et al. showed that SIRT3 overexpression protected differentiated pheochromocytoma-12 (PC-12) cells against H2O2-induced cell death [12]. Moreover, SIRT3 plays an important role in autophagy [13]. SIRT3 exhibits inhibitory or promotive effects on hepatic cell autophagy through different mechanisms in distinct physiological contexts [14]. Similarly, in neuronal cells, the regulatory role of SIRT3 on the autophagy process is different in different cell models [15,16].
Several clinical studies have implicated neuroprotective diets enriched with plant polyphenols, which promoted memory, cognition, and other brain functions via suppressing oxidative stress and neuroinflammation from neurotoxin damage [17]. Piceatannol (3,3’,4’,5-tetrahydroxy-trans-stilbene), a naturally occurring stilbene, has been evidenced with many bioactivities, such as antioxidant, anti-inflammation, anti-diabetes, anti-cancer, cardio-protection and neuroprotection [18,19,20,21,22,23,24,25]. Previous studies showed that piceatannol displayed effective cytoprotection of nerve cells against H2O2-induced oxidative stress [22]. However, the underlying neuroprotection mechanism for the therapeutic role of piceatannol needs to be fully elucidated. We previously found that piceatannol attenuated oxidative damage and mitochondrial-mediated apoptosis, exerting a cytoprotective effect in PC-12 cells through SIRT3/Forkhead box O3 (FOXO3a) signaling pathway [23]. Meanwhile, piceatannol improved the cognitive function of AD mice by reducing oxidative stress, neuroinflammation and beta-amyloid (Aβ) production in the hippocampus [24]. However, whether piceatannol plays the cytoprotective effects through SIRT3-mediated autophagy in PC-12 cells needs to be further clarified.
In the present study, the neuroprotective effects of piceatannol against H2O2-induced mitochondrial dysfunction and autophagy overactivation in PC-12 cells were investigated. 3-(1H-1,2,3-Triazol-4-yl) pyridine (3-TYP), SIRT3 inhibitor, and rapamycin (autophagy activator) were used to explore the underlying mechanism of piceatannol-treated PC-12 cells viability under oxidative stress. The current research could provide new insights into the prevention and management of neurodegenerative diseases by dietary supplementation of piceatannol.
2. Materials and Methods
2.1. Materials and Reagents
Piceatannol, dimethyl sulfoxide (DMSO), and bovine serum albumin (BSA) were obtained from Sigma-Aldrich (St. Louis, MO, USA). RPMI 1640 medium, horse serum (HS), fetal bovine serum (FBS), Hank’s balanced salt solution (HBSS) and penicillin/streptomycin solution were purchased from Gibco (Grand Island, NY, USA). 3-TYP and Cell-Counting-Kit-8 (CCK8) were purchased from Med Chem Express (Monmouth Junction, NJ, USA). MitoSOX™ Red, SYTO™ 9 and MitoTracker™ Red CMXRos were obtained from Invitrogen (Grand Island, NY, USA). Bicinchoninic acid (BCA) protein assay kits were purchased from Thermo Fisher Scientific (Waltham, MA, USA). JC-1 Mitochondrial Membrane Potential Assay Kit was purchased from Abcam (Cambridge, UK). Autophagy Staining Assay Kits with monodansylcadavrine (MDC) and propidium iodide (PI) were obtained from Beyotime Biotechnology (Shanghai, China). Antibodies against mitochondrial transcription factor (TFAM) (A3173), Complex IV (A17889), peroxisome-proliferator-activated receptor-γ coactivator-1α (PGC-1α) (A20995), light chain 3B (LC3B) (A19665), autophagy-related protein 5 (ATG5) (A19677) and goat anti-rabbit IgG H&L (HRP) (AS014) were obtained from ABclonal Technology Co., Ltd. (Wuhan, China).
2.2. Cell Culture
Rat pheochromocytoma cells (PC-12 cells) were purchased from American Type Culture Collection (ATCC, Manassas, MD, USA). PC-12 cells were cultured in RPMI 1640 medium supplemented with 10% HS, 5% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin and incubated in a humidified atmosphere at 37 °C containing 5% CO2.
2.3. Cell Viability
PC-12 cells were seeded in a flat-bottom 96-well plate at a density of 2 × 104 cells/well for 24 h. PC-12 cells were incubated with different concentrations of piceatannol for 24 h and induced by H2O2 for another 24 h. Then, cell viability was investigated using CCK8 assay which was a sensitive colorimetric assay for the determination of cell viability. CCK8 solution (10 μL) was added to each well and incubated with PC-12 cells at 37 °C for 1 h according to the manufacturer’s protocol. Subsequently, the cell viability was detected by a microplate reader (Spark, Tecan, Switzerland) at 450 nm wavelength.
2.4. Measurement of Mitochondrial Superoxide Level
The accumulation of mitochondrial superoxide was detected using MitoTracker™ Red CMXRos according to the manufacturer’s protocol. PC-12 cells were seeded in 12-well plates at a density of 2 × 105 cells/well for 24 h. After treatments, the cells were washed by PBS and then MitoSOX™ reagent was used to stain the mitochondrial ROS for 20 min in the dark. The nucleic acids were stained with SYTO™ 9 reagent for another 2 min at 37 °C. After incubation, the fluorescence intensities were measured using a microplate reader (Spark, Tecan, Switzerland) with Excitation/Emission wavelength at 581/644 nm for MitoSOX™ and 485/498 nm for SYTO™ 9. The fluorescence image was observed under a fluorescence microscope (IX71, Olympus, Japan).
2.5. Measurements of Mitochondrial Membrane Potential (MMP)
The MMP of PC-12 cells was detected using JC-1 Mitochondrial Membrane Potential Assay Kit. PC-12 cells were placed in 12-well plates for 24 h and treated with piceatannol for 24 h, followed by incubating with 250 μM H2O2 for another 24 h. Then, 1000 μL JC-1 dye loading solution was pipetted to each well and incubated with PC-12 cells for 20 min at 37 °C. Subsequently, the MMP was determined at a concentration of 2 × 105 cells/mL using a flow cytometry (CytoFLEX S, Beckman, CA, USA) and analyzed by CytExpert software 2.4.0.28 (Beckman, CA, USA).
2.6. Detection of Autophagy
The autophagy of PC-12 cells was detected using Autophagy Staining Assay Kit (Beyotime, Shanghai, China) according to the manufacturer’s protocol. Briefly, PC-12 cells were pretreated with piceatannol for 24 h followed by incubating with 250 μM H2O2 for 24 h. Then, PC-12 cells were stained with the MDC and PI dye-loading solution for another 1 h. After staining, the fluorescence of PC-12 cells at a concentration of 2 × 105 cells/mL was detected using flow cytometry (CytoFLEX S, Beckman, CA, USA) and analyzed by CytExpert software (Beckman, CA, USA).
2.7. Western Blotting
PC-12 cells (2 × 105 cells/mL) were seeded in 12-well plates for 24 h. After treatment with piceatannol for 24 h, cells were treated with or without 3-TYP or rapamycin for 4 h, followed by H2O2 treatment for another 24 h, PC-12 cells were lysed and collected with radio-immunoprecipitation assay buffer (RIPA) mixed with phosphatase inhibitor and protease inhibitor cocktail (Sigma Aldrich, St. Louis, MO, USA). Whole-cell protein extracts were measured by BCA protein assay kits (Thermo Fisher Scientific, Waltham, MA, USA) and denatured at 95 °C for 5 min. Subsequently, 40 μg protein of each sample was uploaded and fractionated by a 12% SDS-PAGE gel and then transferred to an NC membrane (Pall Life Sciences, Port Washington, NY, USA). After washing with Tris-buffered saline containing 0.1% Tween-20 (0.1% TBST), the NC membrane was blocked with 5% (w/w) skim milk in TBST for 1 h. Then the membrane was incubated with primary antibodies (1:1000) at 4 °C overnight and secondary antibodies at 37 °C for 2 h. Protein bands were visualized and analyzed using a Bio-Rad imaging system (Bio-Rad Laboratories, Hercules, CA, USA).
2.8. Statistical Analysis
The data in this study were expressed as the mean ± standard deviation (SD). Statistical analyses were carried out by one-way ANOVA using SPSS V. 26 (SPSS, Chicago, IL, USA). p < 0.05 was considered to demonstrate statistically significant differences.
3. Results
3.1. Piceatannol Attenuated PC-12 Cells Mitochondrial Dysfunction Induced by H2O2
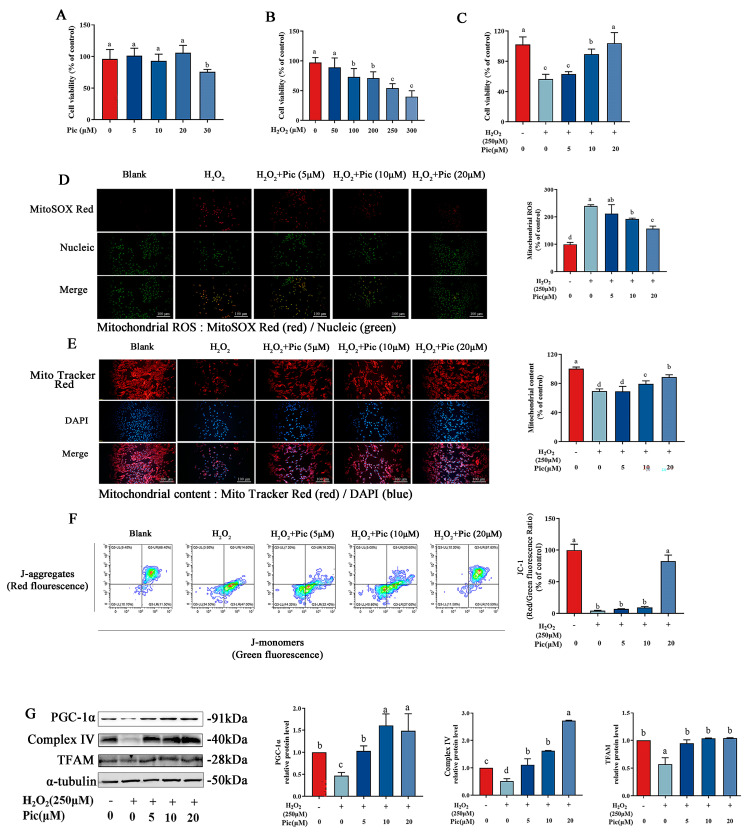
To determine the toxicity effect of H2O2 and piceatannol on PC-12 cells, cell viability was evaluated after 24 h exposure to different concentrations of H2O2 or piceatannol by using CCK8 assay. As shown in Figure 1A, piceatannol showed no cytotoxic effect on PC-12 cells at concentrations up to 20 μM (p > 0.05). However, the cell viability significantly decreased following incubation with piceatannol at a concentration over 30 μM. The cell viability of PC-12 dramatically decreased in a concentration-dependent manner after H2O2 treatment (50–300 μM) (Figure 1B). In addition, H2O2 treatment (250 μM) almost led to 50% cell death, which was used for the subsequent experiments to evaluate the protective effect of piceatannol. To further explore the protective effect of piceatannol on H2O2-induced PC-12 cell oxidative damage, PC-12 cells were incubated with piceatannol (5–20 μM), followed by exposure with 250 μM H2O2 for another 24 h. The results revealed that piceatannol pretreatment significantly increased cell viability in a concentration-dependent manner, compared to cells induced by H2O2 alone. Pre-incubation with 5, 10 and 20 μM of piceatannol prevented cell death under oxidative stress to 63%, 90% and 104%, respectively (Figure 1C). Moreover, H2O2 treatment significantly upregulated mitochondrial ROS fluorescent signal in PC-12 cells compared to the control group, while pretreatment with piceatannol significantly lowered mitochondrial ROS production (Figure 1D). According to MitoTracker Red CMXRos staining result, the piceatannol supplement group gained a higher level of red fluorescence than cells treated with H2O2 alone, indicating that piceatannol attenuated mitochondrial content loss (Figure 1E). Furthermore, the flow cytometry results showed that H2O2 notably decreased the red/green fluorescence ratio in PC-12 cells, indicating impaired mitochondrial integrity. Whereas, pretreatment with piceatannol dramatically promoted the red/green fluorescence ratio in PC-12 cells in a concentration-dependent manner (Figure 1F). Results of Western blot revealed that H2O2 dramatically decreased the protein expression of PGC-1α, Complex IV and TFAM in PC-12 cells, which was upregulated by piceatannol pretreatment (Figure 1G). These results illustrated that piceatannol could ameliorate the mitochondrial dysfunction of PC-12 cells under oxidative stress.
Figure 1.
Piceatannol attenuated mitochondrial dysfunction of PC-12 cells under oxidative stress. (A) Cell viability of PC-12 cells treated with different concentrations of piceatannol (Pic) (0–30 μM). (B) Cell viability of PC-12 cells treated with different concentrations of H2O2 (0–300 μM). (C) Cell viability of PC-12 cells preincubated with piceatannol (0, 5, 10 and 20 μM) for 24 h, followed by incubation with or without 250 μM H2O2 for another 24 h. Mitochondrial ROS (D) and mitochondrial content (E) observed by fluorescence microscope (Magnification, ×100; Scale bar = 100 μm). (F) Mitochondrial membrane potential measured by flow cytometer. (G) The protein expression level of PGC-1α, Complex IV and TFAM measured by Western blotting assay. The data for the experiments were represented as the means ± standard deviation (SD). Results marked with the same letters were not significantly different (p < 0.05).
3.2. Piceatannol Decreased H2O2-Induced Autophagy in PC-12 Cells
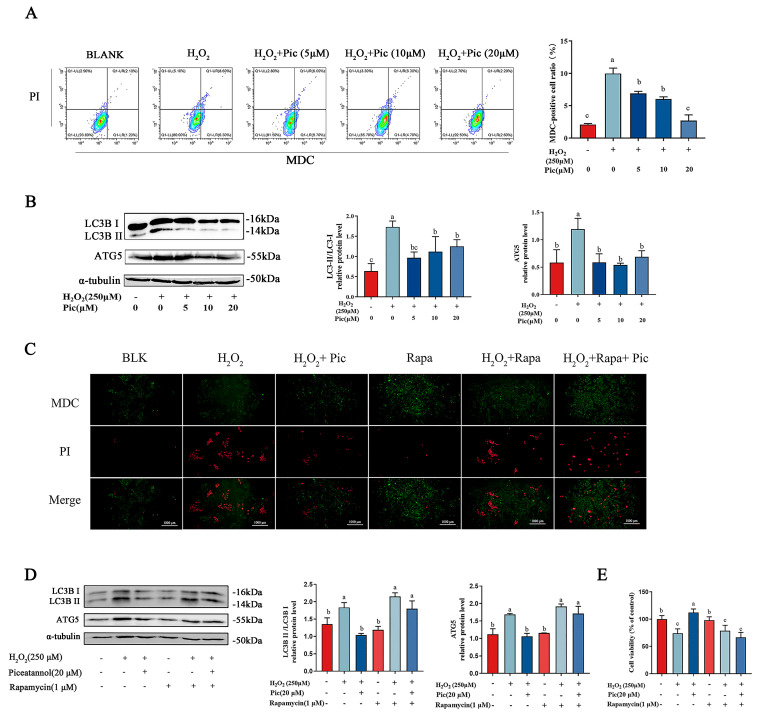
Autophagy plays a prominent role in cellular damage and cell death [26,27]. The autophagy of PC-12 cells was determined by using MDC, a specific acidic vesicular organelles (AVOs) fluorescence staining pigment, which was often used to observe intracellular autophagosomes. Moreover, PI staining was used to determine the cellular death of PC-12. As shown in Figure 2A, the flow cytometry result indicated an elevated percentage of PC-12 cells with MDC and PI signaling after exposure to H2O2, while pre-incubation with piceatannol significantly reduced the ratio of positive MDC and PI signaling. These results suggested that piceatannol markedly inhibited the overactive autophagy of PC-12 cells. Furthermore, compared to the control group, the expression level of LC3B II/I and ATG5 were notably upregulated in PC-12 cells induced by H2O2. While the expression level of these proteins was markedly downregulated in piceatannol-pretreated groups, compared to H2O2 treatment alone (Figure 2B).
Figure 2.
Piceatannol attenuated PC-12 cell death and autophagy induced by H2O2. (A) PC-12 cells were pretreated with piceatannol for 24 h, exposed to 250 μM H2O2 for another 24 h. Then cells were stained with MDC and PI fluorescence probe and detected by flow cytometer. (B) The protein expression level of LC3B II/I and ATG5 measured by Western blotting. PC-12 cells were incubated with piceatannol for 24 h, then treated with rapamycin (Rapa) for 4 h and H2O2 for another 24 h. (C) The MDC/PI probe staining observed by fluorescence microscope (Magnification, ×100; Scale bar = 100 μm). (D) The protein expression of LC3B II/I and ATG5 measured by Western blotting assay. (E) The cell viability detected by CCK8 assay. The data for the experiments were represented as the means ± standard deviation (SD). Results marked with the same letters were not significantly different (p < 0.05).
To investigate the mechanism of the inhibitory effect of piceatannol, an autophagy activator rapamycin was applied in PC-12 cells cultured with H2O2. According to MDC/PI fluorescent intensity, rapamycin significantly enhanced the autophagosome formation which was reduced by piceatannol (Figure 2C). Western blotting results revealed that piceatannol pre-incubation significantly down-regulated the ratios of LC3B-II/I and reduced the expression of ATG5 in H2O2-induced PC-12 cells, while rapamycin significantly promoted the expression of LC3B-II/I and ATG5 even after piceatannol pretreatment (Figure 2D). These findings suggested that rapamycin alleviated autophagic inhibition by piceatannol in PC-12 cells under oxidative stress. Furthermore, the cell viability results indicated that piceatannol could distinctly prevent the cell death of PC-12 cells induced by H2O2. However, rapamycin eliminated the protective effect of piceatannol on H2O2-induced PC-12 cells (Figure 2E). Consequently, these results illustrated that piceatannol reverted the impairment of PC-12 cell viability induced by H2O2 through downregulating autophagy.
3.3. Piceatannol Improved Mitochondrial Function of PC-12 Cells by Inhibiting Autophagy
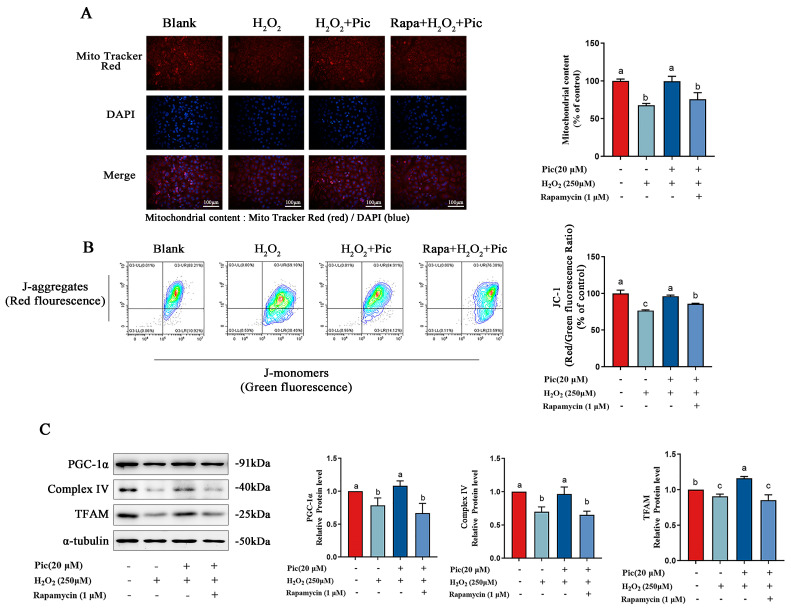
To investigate whether autophagy was involved in the regulation of mitochondrial function by piceatannol, autophagy activator rapamycin was applied in H2O2-induced PC-12 cells. The result of the fluorescence staining showed that activation of autophagy abolished the promotion of mitochondrial content by piceatannol (Figure 3A). Meanwhile, the protective effect of piceatannol on mitochondrial membrane potential was also counteracted in the presence of rapamycin (Figure 3B). In addition, mitochondrial function-related protein expression (PGC-1α, Complex IV, and TFAM) enhanced by piceatannol were significantly reduced after co-treatment with rapamycin (Figure 3C). These results demonstrated that the activator of autophagy could reverse the protection of piceatannol on mitochondrial function in PC-12 cells.
Figure 3.
Piceatannol improved mitochondrial function of PC-12 cells by inhibiting autophagy. PC-12 cells were pretreated with piceatannol for 24 h, then treated with rapamycin for 4 h and H2O2 for another 24 h. (A) Mitochondrial content observed by fluorescence microscope (Magnification, ×100; Scale bar = 100 μm). (B) Mitochondrial membrane potential observed by flow cytometer. (C) The protein expression level of PGC-1α, Complex IV and TFAM measured by Western blotting assay. The data for the experiments were represented as the means ± standard deviation (SD). Results marked with the same letters were not significantly different (p < 0.05).
3.4. Piceatannol Protected PC-12 Cells against Mitochondrial Dysfunction by Inhibiting Autophagy via SIRT3 Pathway
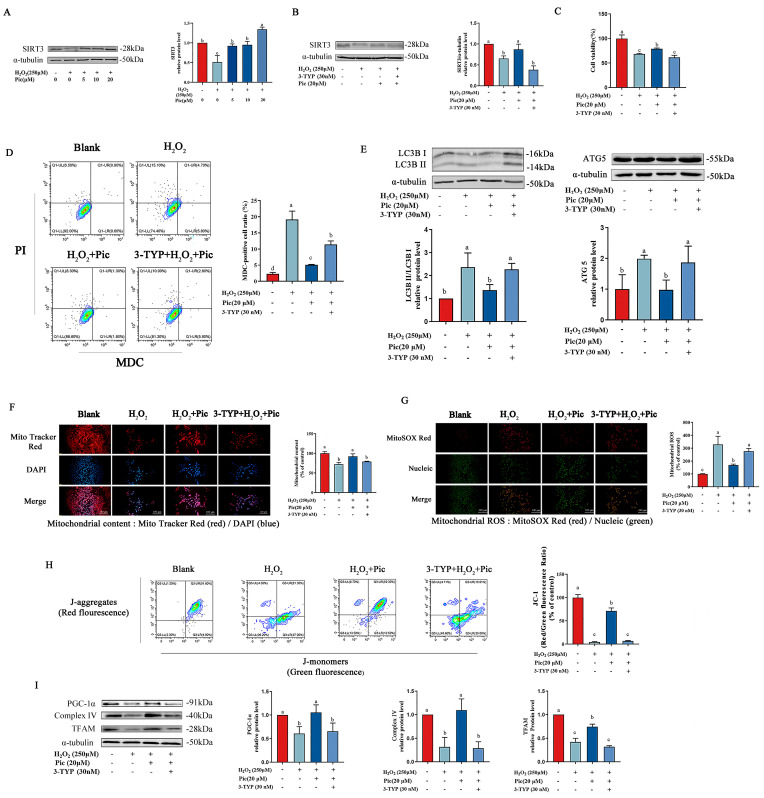
As shown in Figure 4A, H2O2 exposure significantly downregulated the SIRT3 protein level compared with the control group, while pretreatment with piceatannol successfully restored the SIRT3 expression level. To investigate whether SIRT3 was involved in the protective effect of piceatannol on mitochondrial function, 3-TYP (inhibitor of SIRT3) was used to disturb SIRT3 signaling pathway in PC-12 cells. Compared with the piceatannol-pretreated group, the protein expression level of SIRT3 was significantly downregulated in PC-12 cells after co-treatment with piceatannol and 3-TYP (Figure 4B). Furthermore, inhibition of SIRT3 by 3-TYP dramatically diminished the protective effect of piceatannol (Figure 4C). Meanwhile, PC-12 cells co-treated with 3-TYP and piceatannol exhibited increased autophagosomes under oxidative stress, compared with the piceatannol treatment group (Figure 4D). 3-TYP significantly increased the protein expression level of LC3B-II/I and ATG5 which were downregulated by piceatannol (Figure 4E).
Figure 4.
Piceatannol improved mitochondrial function of PC-12 cells by inhibiting autophagy via SIRT3 signaling pathway. (A) PC-12 cells were preincubated with 0, 5, 10 and 20 μM piceatannol for 24 h, followed by incubation with or without 250 μM H2O2 for another 24 h. The protein expression level of SIRT3. (B) PC-12 cells were pretreated with piceatannol for 24 h, then treated with 3-TYP for 4 h and H2O2 for another 24 h. The protein expression level of SIRT3 measured by Western blotting assay. (C) The cell viability detected by CCK8 assay. (D) The MDC/PI probe staining observed by flow cytometer. (E) The protein expression of LC3B II/I and ATG5 measured by Western blotting assay. The mitochondrial content (F) and mitochondrial ROS (G) observed by fluorescence microscope (Magnification, ×100; Scale bar = 100 μm). (H) Mitochondrial membrane potential measured by flow cytometer. (I) The protein expression level of PGC-1α, Complex IV and TFAM measured by Western blotting assay. The data for the experiments were represented as the means ± standard deviation (SD). Results marked with the same letters were not significantly different (p < 0.05).
Additionally, the protective effect of piceatannol on mitochondrial function in PC-12 cells was measured after SIRT3 inhibitor treatment. The results revealed that 3-TYP dramatically reduced mitochondrial content while elevating mitochondrial ROS levels in PC-12 cells compared to the piceatannol treatment group (Figure 4F,G). Similarly, 3-TYP markedly decreased MMP protected by piceatannol in PC-12 cells under oxidative stress (Figure 4H). In addition, piceatannol-upregulated protein (PGC-1α, Complex IV and TFAM) expression levels were also significantly reduced by 3-TYP (Figure 4I). These results illustrated that the SIRT3 inhibitor significantly inhibited the protective effect of piceatannol, suggesting the mitochondrial protective role of piceatannol in autophagy inhibition was via the SIRT3 signaling pathway.
4. Discussion
Mitochondria plays a crucial role in neuronal cell function which acts as a regulator of energy metabolism, while mitochondrial dysfunction leads to abnormal energy metabolism which is associated with neuronal cell death [21,28]. Moreover, mitochondrial dysfunction caused by oxidative stress is one of the factors contributing to many neurodegenerative diseases [29]. Therefore, a potential therapy for neural diseases is to maintain and reinforce mitochondrial function. Mitochondrial quality is characterized by mitochondrial biogenesis [30] and mitochondrial oxidative phosphorylation (OXPHOS) system [31]. PGC-1α regulates the production of various ROS-detoxifying enzymes and plays a key role in mitochondrial biogenesis and oxidative phosphorylation [32]. Previous studies implied that PGC-1α overexpression protected neural cells from oxidative stress-induced death, while PGC-1α deficiency affected mitochondrial structure, leading to elevated mitochondrial ROS levels and promoting cellular senescence [33,34]. Established studies showed that piceatannol promoted the transcription and activation of PGC-1α in hepatic cells and renal cells [35], and also possessed a cytoprotective effect on improving ROS-induced mitochondrial dysfunction in PC-12 cells [23]. Our findings showed that piceatannol eliminated the content of mitochondrial ROS, associated with upregulating PGC-1α expression level. In addition, mitochondrial biogenesis was regulated by TFAM, while TFAM downregulation causes mitochondrial OXPHOS deficiency, resulting in increased ROS production [36]. Additionally, TFAM overexpression significantly alleviated mtDNA oxidative damage [37]. Increasing evidence has proved that the upregulation of TFAM is a feasible intervention to alleviate neurodegenerative disease [38]. In this study, piceatannol was found to inhibit mitochondria content loss through the upregulation of TFAM. Moreover, mitochondrial respiratory chain complexes, including Complex I-V are essential for maintaining mitochondrial function in highly metabolic tissues. For instance, piceatannol was reported to reduce hepatocyte apoptosis by enhancing the activities of mitochondria Complex I, II, III, and V [21]. Additionally, piceatannol exerted a protective effect against hypoxia-induced toxicity in H9c2 cardiomyocytes by increasing the expression of the mitochondrial antioxidant enzyme [39]. Our study also showed that piceatannol enhanced the expression of mitochondria Complex IV, indicating the possible protective role of piceatannol for mitochondrial function.
Autophagy is a process of the degradation of intracellular organelle and proteins in lysosomes that is regulated by a variety of signal molecules and pathways. Many studies have reported that autophagy disorder was involved in the development of neurodegenerative diseases [40]. In neuronal cells, the overproduction of reactive oxygen species promoted autophagy flux and resulted in cell death, while the inhibition of autophagy protected the neurons from cell death [41]. It has been reported that H2O2-induced PC-12 cell autophagy was via increasing LC3B II/LC3B I ratio and decreasing P62 expression [42,43]. LC3B is located on the membrane surfaces of pre-autophagic and autophagic vacuoles, reflecting autophagy activity to some extent [44]. ATG5 is a key molecule in the elongation phase of autophagy and knockout of ATG5 blocks autophagy [45]. In this study, H2O2 significantly promoted the autophagic protein (LC3B and ATG5) expression level in PC-12 cells, which was consistent with those reported studies. Meanwhile, piceatannol pretreatment markedly reversed the upregulation of LC3B and ATG5 and significantly reduced the intracellular autophagosome number in PC-12 cells under oxidative stress. Moreover, rapamycin markedly inhibited the protective effect of piceatannol on autophagy and cell death, suggesting that piceatannol promoted cell viability by alleviating exaggerated autophagy. Other phytochemicals were found to display similar functions. For example, berberine protected PC-12 cells from oxidative damage by inhibiting hydrogen peroxide-induced elevation of autophagy [46]. It was verified that the modulation of mitochondrial dysfunction and autophagy in PC-12 cells exhibited a protective effect on cell survival and improved redox-mediated neurological disorders [27].
SIRT3, a nicotinamide adenine dinucleotide (NAD+)-dependent enzyme, was reported to play a key role in the modulation of mitochondrial function and is considered a therapeutic target of neurodegeneration diseases [47]. It has been illustrated that SIRT3 activation presented the neuroprotective effect, reducing the beta-amyloid (Aβ) and Tau accumulation which contributed to oxidative neurotoxicity in the brain tissues of AD animals [48]. Meanwhile, Ying et al. found that SIRT3 showed neuroprotective effects by eliminating mitochondrial ROS [49]. However, SIRT3 deficiency exacerbated neuronal degeneration through the progressive accumulation of ROS caused by abnormal autophagy, cell apoptosis and mitochondrial dysfunction [50,51]. Piceatannol was proven to enhance the expression level of sirtuins (SIRT) including SIRT1, SIRT3, SIRT6 and the downstream target of SIRT3, PGC-1α, which improved inflammatory effect and insulin resistance in high-fat-diet fed rats [52]. In a previous study, piceatannol protected neurons against oxidative damage via upregulating anti-oxidative enzyme expression and modulating the protein expression of SIRT3 as well as its downstream FOXO3a signaling in PC-12 cells [23]. Here, we found that 3-TYP dramatically reversed the protective effect of piceatannol on mitochondrial function via downregulating SIRT3 expression. Furthermore, 3-TYP dramatically enhanced mitochondrial ROS production and aggravated mitochondrial content loss in PC-12 cells with piceatannol treatment. Meanwhile, our findings showed that 3-TYP downregulated the expression of mitochondria-related proteins (TFAM, PGC-1α and Complex IV) in PC-12 cells treated with piceatannol, revealing that the protection by piceatannol against oxidative stress was via activating SIRT3 signaling pathway.
Furthermore, the results of this study indicated that SIRT3 was involved in the neuroprotective effects of piceatannol on mitochondria-mediated autophagy. In line with this, Rodriguez-Enriquez et al. found that mitochondrial ROS were primarily generated by damaged mitochondria, typically the ones with abnormal mitochondrial membrane potential which acted as an indicator of mitochondria-mediated autophagy [53]. For neuronal cells, SIRT3 exhibited different regulatory effects on autophagy depending on the inducing conditions. For instance, the plant-derived bioactive component, scopoletin, was reported to exert neuroprotective effects by inhibiting the autophagy of SH-SY5Y cells via SIRT3 signaling [15]. Resveratrol increased autophagy by promoting SIRT3 expression to protect neuronal HT22 cells under tunicamycin-induced endoplasmic reticulum stress [16]. In our study, the inhibitory effect of piceatannol on the autophagy of PC-12 cells was alleviated when the SIRT3 signaling was inhibited. These findings indicated that piceatannol could mitigate autophagy by improving mitochondrial function mediated by SIRT3 signaling.
5. Conclusions
In conclusion, the cytoprotection of piceatannol against oxidative damage was verified in PC-12 cells. The findings in this study indicated that piceatannol could significantly inhibit the cell death induced by H2O2 by ameliorating the overactivated autophagy. Moreover, the activation of the SIRT3 signaling pathway by piceatannol significantly reduced H2O2-induced mitochondrial dysfunction and autophagy. These findings suggest that piceatannol holds the promise of a potential bioactive component for neuron protection against oxidative stress, contributing to the development of piceatannol-based products to prevent neurodegenerative disorders.
Abbreviations
Aβ, beta amyloid; AD, Alzheimer’s disease; ATG5, autophagy-related protein 5; ATP, adenosine triphosphate; AVOs, acidic vesicular organelles; FOXO3a, Forkhead box O3; LC3B, light chain 3B; MDC, monodansylcadavrine; MMP, mitochondrial membrane potential; NAD, nicotinamide adenine dinucleotide; OXPHOS, oxidative phosphorylation; PC-12, Pheochromocytoma-12; PD, Parkinson’s disease; PGC-1α, peroxisome-proliferator-activated receptor-γ coactivator-1α; PI, propidium iodide; Pic, piceatannol; Rapa, rapamycin; ROS, reactive oxygen species; SIRT3, sirtuin 3; TFAM, mitochondrial transcription factor A; 3-TYP, 3-(1H-1,2,3-Triazol-4-yl) pyridine.
Author Contributions
J.L.: Conceptualization; Writing—original draft. P.M.: Data curation; Software. Z.Y.: Data curation; Software. Z.W. (Zongwei Wang): Methodology; Validation. W.Y.: Project administration. Z.W. (Ziyuan Wang): Writing review and editing; Project administration. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare that there are no conflict of interest.
Funding Statement
This research was financially supported by the Project of Cultivation for young top-notch Talents of Beijing Municipal Institutions (BPHR202203044), Key R&D Program of Shandong Province (2021CXGC010807-03), and National Key Research and Development Program of China (2022YFF1100605).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Giasson B.I., Duda J.E., Murray I.V., Chen Q., Souza J.M., Hurtig H.I., Ischiropoulos H., Trojanowski J.Q., Lee V.M. Oxidative damage linked to neurodegeneration by selective alpha-synuclein nitration in synucleinopathy lesions. Science. 2000;290:985–989. doi: 10.1126/science.290.5493.985. [DOI] [PubMed] [Google Scholar]
- 2.Sbodio J.I., Snyder S.H., Paul B.D. Redox Mechanisms in Neurodegeneration: From Disease Outcomes to Therapeutic Opportunities. Antioxid. Redox Signal. 2019;30:1450–1499. doi: 10.1089/ars.2017.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hidalgo C., Donoso P. Crosstalk between calcium and redox signaling: From molecular mechanisms to health implications. Antioxid. Redox Signal. 2008;10:1275–1312. doi: 10.1089/ars.2007.1886. [DOI] [PubMed] [Google Scholar]
- 4.Imbriani P., Martella G., Bonsi P., Pisani A. Oxidative stress and synaptic dysfunction in rodent models of Parkinson’s disease. Neurobiol. Dis. 2022;173:105851. doi: 10.1016/j.nbd.2022.105851. [DOI] [PubMed] [Google Scholar]
- 5.Mahadevan H.M., Hashemiaghdam A., Ashrafi G., Harbauer A.B. Mitochondria in Neuronal Health: From Energy Metabolism to Parkinson’s Disease. Adv. Biol. 2021;5:2100663. doi: 10.1002/adbi.202100663. [DOI] [PubMed] [Google Scholar]
- 6.Cai C.C., Zhu J.H., Ye L.X., Dai Y.Y., Fang M.C., Hu Y.Y., Pan S.L., Chen S., Li P.J., Fu X.Q., et al. Glycine Protects against Hypoxic-Ischemic Brain Injury by Regulating Mitochondria-Mediated Autophagy via the AMPK Pathway. Oxidative Med. Cell. Longev. 2019;2019:4248529. doi: 10.1155/2019/4248529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Redza-Dutordoir M., Averill-Bates D.A. Interactions between reactive oxygen species and autophagy Special issue: Death mechanisms in cellular homeostasis. BBA-Mol. Cell Res. 2021;1868:119041. doi: 10.1016/j.bbamcr.2021.119041. [DOI] [PubMed] [Google Scholar]
- 8.Gomes L.R., Menck C.F.M., Cuervo A.M. Chaperone-mediated autophagy prevents cellular transformation by regulating MYC proteasomal degradation. Autophagy. 2017;13:928–940. doi: 10.1080/15548627.2017.1293767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Talebi M., Mohammadi Vadoud S.A., Haratian A., Talebi M., Farkhondeh T., Pourbagher-Shahri A.M., Samarghandian S. The interplay between oxidative stress and autophagy: Focus on the development of neurological diseases. Behav. Brain Funct. 2022;18:3. doi: 10.1186/s12993-022-00187-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J., Xiang H.G., Liu J., Chen Y., He R.R., Liu B. Mitochondrial Sirtuin 3: New emerging biological function and therapeutic target. Theranostics. 2020;10:8315–8342. doi: 10.7150/thno.45922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Almalki W.H., Alzahrani A., El-Daly M., Ahmed A. The emerging potential of SIRT-3 in oxidative stress-inflammatory axis associated increased neuroinflammatory component for metabolically impaired neural cell. Chem. Biol. Interact. 2021;333:109328. doi: 10.1016/j.cbi.2020.109328. [DOI] [PubMed] [Google Scholar]
- 12.Shulyakova N., Sidorova-Darmos E., Fong J., Zhang G., Mills L.R., Eubanks J.H. Over-expression of the Sirt3 sirtuin Protects neuronally differentiated PC12 Cells from degeneration induced by oxidative stress and trophic withdrawal. Brain Res. 2014;1587:40–53. doi: 10.1016/j.brainres.2014.08.066. [DOI] [PubMed] [Google Scholar]
- 13.Shumin C., Wei X., Yunfeng L., Jiangshui L., Youguang G., Zhongqing C., Tao L. Genipin alleviates vascular hyperpermeability following hemorrhagic shock by up-regulation of SIRT3/autophagy. Cell Death Discov. 2018;4:52. doi: 10.1038/s41420-018-0057-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho C.S., Lombard D.B., Lee J.H. SIRT3 as a regulator of hepatic autophagy. Hepatology. 2017;66:700–702. doi: 10.1002/hep.29271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suwanjang W., Sirisuwat C., Srisung S., Isarankura-Na-Ayudhya C., Pannengpetch S., Prachayasittikul S. Protective Efficacy of Spilanthes acmella Murr. Extracts and Bioactive Constituents in Neuronal Cell Death. Rejuv. Res. 2022;25:2–15. doi: 10.1089/rej.2021.0002. [DOI] [PubMed] [Google Scholar]
- 16.Yan W.J., Liu R.B., Wang L.K., Ma Y.B., Ding S.L., Deng F., Hu Z.Y., Wang D.B. Sirt3-Mediated Autophagy Contributes to Resveratrol-Induced Protection against ER Stress in HT22 Cells. Front. Neurosci. 2018;12:116. doi: 10.3389/fnins.2018.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan L., Guo M.S., Zhang Y., Yu L., Wu J.M., Tang Y., Ai W., Zhu F.D., Law B.Y.K., Chen Q., et al. Dietary Plant Polyphenols as the Potential Drugs in Neurodegenerative Diseases: Current Evidence, Advances, and Opportunities. Oxidative Med. Cell. Longev. 2022;2022:5288698. doi: 10.1155/2022/5288698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao Y., Smith W., Yan L., Kong L.B. Overview of Cellular Mechanisms and Signaling Pathways of Piceatannol. Curr. Stem. Cell Res. Ther. 2020;15:4–10. doi: 10.2174/1574888x14666190402100054. [DOI] [PubMed] [Google Scholar]
- 19.Hao Y., Liu J., Wang Z., Yu L., Wang J. Piceatannol Protects Human Retinal Pigment Epithelial Cells against Hydrogen Peroxide Induced Oxidative Stress and Apoptosis through Modulating PI3K/Akt Signaling Pathway. Nutrients. 2019;11:1515. doi: 10.3390/nu11071515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Storniolo C.E., Moreno J.J. Resveratrol Analogs with Antioxidant Activity Inhibit Intestinal Epithelial Cancer Caco-2 Cell Growth by Modulating Arachidonic Acid Cascade. J. Agr. Food Chem. 2019;67:819–828. doi: 10.1021/acs.jafc.8b05982. [DOI] [PubMed] [Google Scholar]
- 21.Jia P., Ji S., Zhang H., Chen Y., Wang T. Piceatannol Ameliorates Hepatic Oxidative Damage and Mitochondrial Dysfunction of Weaned Piglets Challenged with Diquat. Animals. 2020;10:1239. doi: 10.3390/ani10071239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Son Y., Byun S.J., Pae H.O. Involvement of heme oxygenase-1 expression in neuroprotection by piceatannol, a natural analog and a metabolite of resveratrol, against glutamate-mediated oxidative injury in HT22 neuronal cells. Amino Acids. 2013;45:393–401. doi: 10.1007/s00726-013-1518-9. [DOI] [PubMed] [Google Scholar]
- 23.Yang W., Wang Y., Hao Y.M., Wang Z.Y., Liu J., Wang J. Piceatannol alleviate ROS-mediated PC-12 cells damage and mitochondrial dysfunction through SIRT3/FOXO3a signaling pathway. J. Food Biochem. 2022;46:e13820. doi: 10.1111/jfbc.13820. [DOI] [PubMed] [Google Scholar]
- 24.Wei Y., Zongwei W., Ziyuan W., Jie L., Min Z., Jing W. Improvement effect of piceatannol on cognitive function of mice with Alzheimer’s disease. J. Food Sci. Technol. 2023;41:126–134. doi: 10.12301/spxb202200175. [DOI] [Google Scholar]
- 25.Kwon J.Y., Kershaw J., Chen C.Y., Komanetsky S.M., Zhu Y., Guo X., Myer P.R., Applegate B., Kim K.H. Piceatannol antagonizes lipolysis by promoting autophagy-lysosome-dependent degradation of lipolytic protein clusters in adipocytes. J. Nutr. Biochem. 2022;105:108998. doi: 10.1016/j.jnutbio.2022.108998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song K.S., Kim J.S., Yun E.J., Kim Y.R., Seo K.S., Park J.H., Jung Y.J., Park J.I., Kweon G.R., Yoon W.H., et al. Rottlerin induces autophagy and apoptotic cell death through a PKC-delta-independent pathway in HT1080 human fibrosarcoma cells: The protective role of autophagy in apoptosis. Autophagy. 2008;4:650–658. doi: 10.4161/auto.6057. [DOI] [PubMed] [Google Scholar]
- 27.Gao J., Deng Y., Yin C., Liu Y., Zhang W., Shi J., Gong Q. Icariside II, a novel phosphodiesterase 5 inhibitor, protects against H2O2-induced PC12 cells death by inhibiting mitochondria-mediated autophagy. J. Cell. Mol. Med. 2017;21:375–386. doi: 10.1111/jcmm.12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bento A.C., Bippes C.C., Kohler C., Hemion C., Frank S., Neutzner A. UBXD1 is a mitochondrial recruitment factor for p97/VCP and promotes mitophagy. Sci. Rep. 2018;8:12415. doi: 10.1038/s41598-018-30963-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cardoso S.M., Pereira C.F., Moreira P.I., Arduino D.M., Esteves A.R., Oliveira C.R. Mitochondrial control of autophagic lysosomal pathway in Alzheimer’s disease. Exp. Neurol. 2010;223:294–298. doi: 10.1016/j.expneurol.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Z., Li H., Chen W., Cui Y., Huang A., Qi X. Perindopril Improves Cardiac Function by Enhancing the Expression of SIRT3 and PGC-1 alpha in a Rat Model of Isoproterenol-Induced Cardiomyopathy. Front. Pharmacol. 2020;11:94. doi: 10.3389/fphar.2020.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gurunathan S., Jeyaraj M., Kang M.H., Kim J.H. Anticancer Properties of Platinum Nanoparticles and Retinoic Acid: Combination Therapy for the Treatment of Human Neuroblastoma Cancer. Int. J. Mol. Sci. 2020;21:6792. doi: 10.3390/ijms21186792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spiegelman B.M. Mitochondrial Biology: New Perspectives: Novartis Foundation Symposium 287. Volume 287. John Wiley & Sons, Ltd.; Chichester, UK: 2007. Transcriptional control of mitochondrial energy metabolism through the PGC1 coactivators; pp. 60–63; discussion 63–69. [PubMed] [Google Scholar]
- 33.Kim S.B., Heo J.I., Kim H., Kim K.S. Acetylation of PGC1 alpha by Histone Deacetylase 1 Downregulation Is Implicated in Radiation-Induced Senescence of Brain Endothelial Cells. J. Gerontol. Ser. A-Biol. Sci. Med. Sci. 2019;74:787–793. doi: 10.1093/gerona/gly167. [DOI] [PubMed] [Google Scholar]
- 34.St-Pierre J., Drori S., Uldry M., Silvaggi J.M., Rhee J., Jager S., Handschin C., Zheng K., Lin J., Yang W., et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 35.Lee H.J., Kang M.G., Cha H.Y., Kim Y.M., Lim Y., Yang S.J. Effects of Piceatannol and Resveratrol on Sirtuins and Hepatic Inflammation in High-Fat Diet-Fed Mice. J. Med. Food. 2019;22:833–840. doi: 10.1089/jmf.2018.4261. [DOI] [PubMed] [Google Scholar]
- 36.Zhao M., Wang Y.Z., Li L., Liu S.Y., Wang C.S., Yuan Y.J., Yang G., Chen Y.N., Cheng J.Q., Lu Y.R., et al. Mitochondrial ROS promote mitochondrial dysfunction and inflammation in ischemic acute kidney injury by disrupting TFAM-mediated mtDNA maintenance. Theranostics. 2021;11:1845–1863. doi: 10.7150/thno.50905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oka S., Leon J., Sakumi K., Ide T., Kang D., LaFerla F.M., Nakabeppu Y. Human mitochondrial transcriptional factor A breaks the mitochondria-mediated vicious cycle in Alzheimer’s disease. Sci. Rep. 2016;6:37889. doi: 10.1038/srep37889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang I., Chu C.T., Kaufman B.A. The mitochondrial transcription factor TFAM in neurodegeneration: Emerging evidence and mechanisms. FEBS Lett. 2018;592:793–811. doi: 10.1002/1873-3468.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boccellino M., Donniacuo M., Bruno F., Rinaldi B., Quagliuolo L., Ambruosi M., Pace S., De Rosa M., Olgac A., Banoglu E., et al. Protective effect of piceatannol and bioactive stilbene derivatives against hypoxia-induced toxicity in H9c2 cardiomyocytes and structural elucidation as 5-LOX inhibitors. Eur. J. Med. Chem. 2019;180:637–647. doi: 10.1016/j.ejmech.2019.07.033. [DOI] [PubMed] [Google Scholar]
- 40.Chen W., Zhang L., Zhang K., Zhou B., Kuo M.L., Hu S., Chen L., Tang M., Chen Y.-R., Yang L., et al. Reciprocal regulation of autophagy and dNTP pools in human cancer cells. Autophagy. 2014;10:1272–1284. doi: 10.4161/auto.28954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yue J., Wang X.S., Feng B., Hu L.N., Yang L.K., Lu L., Zhang K., Wang Y.T., Liu S.B. Activation of G-Protein-Coupled Receptor 30 Protects Neurons against Excitotoxicity through Inhibiting Excessive Autophagy Induced by Glutamate. ACS Chem. Neurosci. 2019;10:4227–4236. doi: 10.1021/acschemneuro.9b00287. [DOI] [PubMed] [Google Scholar]
- 42.Zhong L., Fang S., Wang A.Q., Zhang Z.H., Wang T., Huang W., Zhou H.X., Zhang H., Yin Z.S. Identification of the Fosl1/AMPK/autophagy axis involved in apoptotic and inflammatory effects following spinal cord injury. Int. Immunopharmacol. 2022;103:108492. doi: 10.1016/j.intimp.2021.108492. [DOI] [PubMed] [Google Scholar]
- 43.He X., Zhang J.N., Guo Y.S., Yang X.W., Huang Y.F., Hao D.J. LncRNA MIAT Promotes Spinal Cord Injury Recovery in Rats by Regulating RBFOX2-Mediated Alternative Splicing of MCL-1. Mol. Neurobiol. 2022;59:4854–4868. doi: 10.1007/s12035-022-02896-2. [DOI] [PubMed] [Google Scholar]
- 44.Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin T.C., Chen Y.R., Kensicki E., Li A.Y.J., Kong M., Li Y., Mohney R.P., Shen H.M., Stiles B., Mizushima N., et al. Resetting glutamine-dependent metabolism and oxygen consumption. Autophagy. 2012;8:1477–1493. doi: 10.4161/auto.21228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z.M., Jiang T., Lu Q., Xu K., He J.P., Xie L., Chen Z.F., Zheng Z.L., Ye L.X., Xu K.B., et al. Berberine attenuated the cytotoxicity induced by t-BHP via inhibiting oxidative stress and mitochondria dysfunction in PC-12 cells. Cell. Mol. Neurobiol. 2020;40:587–602. doi: 10.1007/s10571-019-00756-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anamika, Khanna A., Acharjee P., Acharjee A., Trigun S.K. Mitochondrial SIRT3 and neurodegenerative brain disorders. J. Chem. Neuroanat. 2019;95:43–53. doi: 10.1016/j.jchemneu.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 48.Liu Y., Cheng A.W., Li Y.J., Yang Y., Kishimoto Y., Zhang S., Wang Y., Wan R.D., Raefsky S.M., Lu D.Y., et al. SIRT3 mediates hippocampal synaptic adaptations to intermittent fasting and ameliorates deficits in APP mutant mice. Nat. Commun. 2019;10:1886. doi: 10.1038/s41467-019-09897-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ying Y., Lu C., Chen C., Liu Y., Liu Y.U., Ruan X., Yang Y. SIRT3 Regulates Neuronal Excitability of Alzheimer’s Disease Models in an Oxidative Stress-Dependent Manner. Neuromol. Med. 2022;24:261–267. doi: 10.1007/s12017-021-08693-9. [DOI] [PubMed] [Google Scholar]
- 50.Ha D.T., Chen Q.C., Hung T.M., Youn U.J., Ngoc T.M., Thuong P.T., Kim H.J., Seong Y.H., Min B.S., Bae K. Stilbenes and oligostilbenes from leaf and stem of Vitis amurensis and their cytotoxic activity. Arch. Pharm. Res. 2009;32:177–183. doi: 10.1007/s12272-009-1132-2. [DOI] [PubMed] [Google Scholar]
- 51.Pi H., Xu S., Reiter R.J., Guo P., Zhang L., Li Y., Li M., Cao Z., Tian L., Xie J., et al. SIRT3-SOD2-mROS-dependent autophagy in cadmium-induced hepatotoxicity and salvage by melatonin. Autophagy. 2015;11:1037–1051. doi: 10.1080/15548627.2015.1052208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen M.C., Ryoo S. Intravenous administration of piceatannol, an arginase inhibitor, improves endothelial dysfunction in aged mice. Korean J. Physiol. Pharmacol. 2017;21:83–90. doi: 10.4196/kjpp.2017.21.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodriguez-Enriquez S., He L.H., Lemasters J.J. Role of mitochondrial permeability transition pores in mitochondrial autophagy. Int. J. Biochem. Cell Biol. 2004;36:2463–2472. doi: 10.1016/j.biocel.2004.04.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.