Abstract
Background
Immune checkpoint inhibitor (ICI) therapy has substantially improved the overall survival (OS) in patients with non-small-cell lung cancer (NSCLC); however, its response rate is still modest. In this study, we developed a machine learning-based platform, namely the Cytokine-based ICI Response Index (CIRI), to predict the ICI response of patients with NSCLC based on the peripheral blood cytokine profiles.
Methods
We enrolled 123 and 99 patients with NSCLC who received anti-PD-1/PD-L1 monotherapy or combined chemotherapy in the training and validation cohorts, respectively. The plasma concentrations of 93 cytokines were examined in the peripheral blood obtained from patients at baseline (pre) and 6 weeks after treatment (early during treatment: edt). Ensemble learning random survival forest classifiers were developed to select feature cytokines and predict the OS of patients undergoing ICI therapy.
Results
Fourteen and 19 cytokines at baseline and on treatment, respectively, were selected to generate CIRI models (namely preCIRI14 and edtCIRI19), both of which successfully identified patients with worse OS in two completely independent cohorts. At the population level, the prediction accuracies of preCIRI14 and edtCIRI19, as indicated by the concordance indices (C-indices), were 0.700 and 0.751 in the validation cohort, respectively. At the individual level, patients with higher CIRI scores demonstrated worse OS [hazard ratio (HR): 0.274 and 0.163, and p<0.0001 and p=0.0044 in preCIRI14 and edtCIRI19, respectively]. By including other circulating and clinical features, improved prediction efficacy was observed in advanced models (preCIRI21 and edtCIRI27). The C-indices in the validation cohort were 0.764 and 0.757, respectively, whereas the HRs of preCIRI21 and edtCIRI27 were 0.141 (p<0.0001) and 0.158 (p=0.038), respectively.
Conclusions
The CIRI model is highly accurate and reproducible in determining the patients with NSCLC who would benefit from anti-PD-1/PD-L1 therapy with prolonged OS and may aid in clinical decision-making before and/or at the early stage of treatment.
Keywords: Immune Checkpoint Inhibitors; Non-Small Cell Lung Cancer; Cytokines; Biomarkers, Tumor; Biostatistics
WHAT IS ALREADY KNOWN ON THIS TOPIC.
Current biomarkers are insufficient to accurately predict therapeutic outcomes and patient overall survival (OS) in response to immune checkpoint inhibitor (ICI) therapy. Therefore, there is an urgent need to identify robust, accurate, and routine clinical biomarkers for predicting the responsiveness to ICI therapy, particularly with respect to the OS, in a minimally invasive manner.
WHAT THIS STUDY ADDS
Our study developed a machine learning-based platform, namely the Cytokine-based ICI Response Index (CIRI), to predict the ICI response of patients with non-small-cell lung cancer (NSCLC) with respect to OS.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
CIRI is highly accurate and reproducible in identifying and predicting patients with NSCLC who would benefit from anti-PD-1/PD-L1 therapy with prolonged OS, and may aid in clinical decisions made before and/or at the early stage of treatment.
Introduction
Immune checkpoint inhibitor (ICI) therapy targeting programmed death-1/ligand-1 (PD-1/PD-L1) and/or cytotoxic T-lymphocyte antigen-4 (CTLA-4) has substantially improved the survival in patients with non-small-cell lung cancer (NSCLC), particularly in those without targetable oncogenic driver mutations.1 2 The bottlenecks of ICI therapy in clinical practice, such as low response rate, immune-related adverse events, primary and acquired resistance, and economic burden, underscore the importance of stratifying patients responsive to ICI therapy.3 Currently, PD-L1 expression is a US Food and Drug Administration (FDA)-approved biomarker for predicting the responsiveness to ICI therapy, and four PD-L1 immunohistochemistry assays have been approved for clinical application in lung cancer.4 However, accumulating evidence has shown that the PD-L1 expression levels are insufficient to accurately predict therapeutic outcomes and patient survival in response to ICI therapy in NSCLC, especially in combination with chemotherapy.5–9 More recently, tumor mutational burden (TMB) has been proven to predict the objective response rate and progression-free survival (PFS) in response to ICI therapy in NSCLC, irrespective of the PD-L1 expression level.10–13 However, the clinical value of TMB as a potential biomarker for evaluating the response to ICI therapy in NSCLC is challenging owing to limited evidence concerning the association between TMB and overall survival (OS) and the lack of standardization and need for more tissue samples and incurred cost in TMB determination.1 Therefore, there is an urgent need to identify robust, accurate, and routine clinical biomarkers for predicting the responsiveness to ICI therapy in NSCLC, particularly with respect to the OS, in a minimally invasive manner.
Cytokines are soluble mediators of host immune activity and play critical roles in cancer immunotherapy by promoting tumor infiltration and reactivation of the effector lymphocytes in the tumor microenvironment.14 Several studies have revealed that circulating cytokines are promising predictive biomarkers that can be employed for evaluating the prognosis of ICI therapy in various cancers, such as renal cell carcinoma15 and melanoma.16 17 Until recently, only a few selected cytokines, such as interleukin (IL)-6,18 19 IL-8,20–22 IL-10,22 IL-11,21 interferon (IFN)-γ,22 23 transforming growth factor (TGF)-β1,24 and tumor necrosis factor-alpha (TNFα),18 22 have been individually evaluated for the prediction of responsiveness to ICI therapy in small samples of patients with NSCLC. In contrast, few studies have explored the predictive potential of the overall signature of circulating cytokine profiles in peripheral blood for the efficiency of ICI therapy in NSCLC. Machine learning-based feature extraction with a combination of non-linear high-dimensional data has been proven to be a flexible and powerful approach in patient stratification and treatment selection for personalized cancer immunotherapy.25–27 However, the performance of machine learning in predicting the response to ICI therapy in NSCLC based on circulating cytokine levels remains unexplored.
Therefore, this study aimed to develop machine learning models to extract and combine the optimal features of circulating cytokine profiles, and identify predictive biomarkers associated with the OS in patients with NSCLC in response to anti-PD-1/PD-L1 therapy.
Materials and methods
Study design
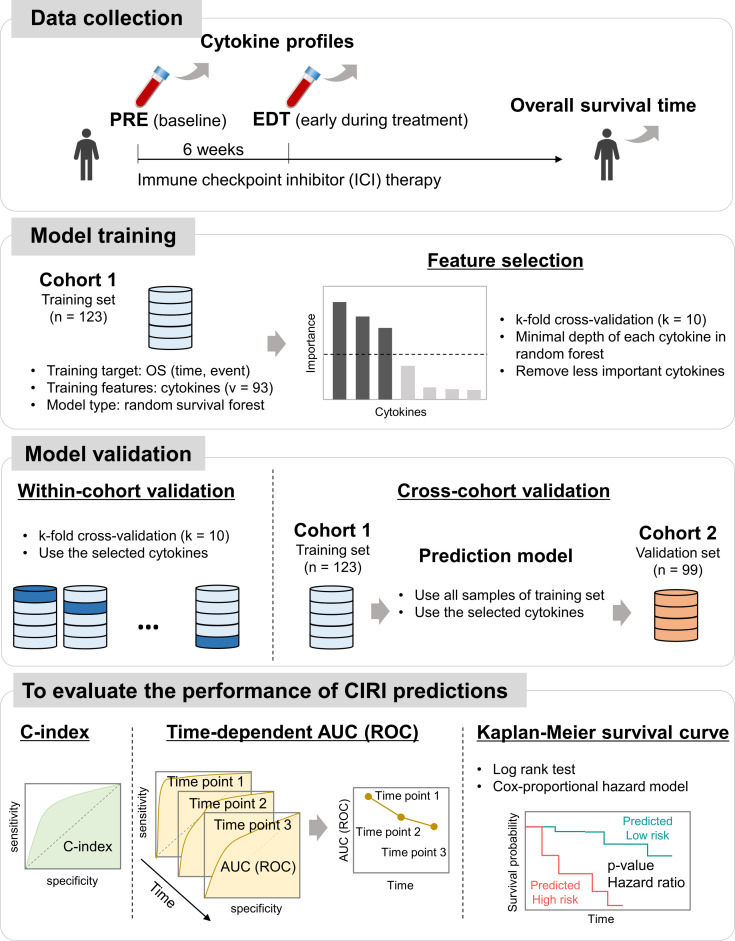
An overview of the study design is shown in figure 1. Therapeutic outcomes in response to anti-PD-1/PD-L1 immunotherapy were determined as the true endpoint OS of patients with NSCLC, instead of surrogate endpoints, such as PFS or change in tumor size. The concentrations of 93 circulating cytokines were examined in the plasma obtained from patients at baseline (named as ‘pre’) and 6 weeks after anti- PD-1/PD-L1 therapy (early during treatment, named as ‘edt’). Ensemble learning random survival forest (RSF) classifiers were developed to select feature cytokines related to the OS and integrate these selected cytokines into a single score, the Cytokine-based ICI Response Index (CIRI), which reflects the immunotherapy outcome. To overcome overfitting of machine learning, within-cohort and cross-cohort validations were performed in training (cohort 1; n=123) and completely independent validation (cohort 2; n=99) sets, respectively. The predictive utility of the CIRI was confirmed using the concordance index (C-index), the time-dependent area under the curve (AUC) of the receiver operating characteristic (ROC) at the population level, and the Kaplan-Meier method with both the log rank test and the Cox-proportional hazard regression at the individual level.
Figure 1.
Overview of a circulating cytokine-based machine learning approach for prediction of immunotherapeutic outcome in patients with non-small-cell lung cancer (NSCLC). AUC, area under the curve; CIRI, Cytokine-based ICI Response Index; OS, overall survival; ROC, receiver operating characteristic.
Patients and data collection
Patients with advanced/recurrent/metastatic NSCLC treated with anti-PD-1 antibody (nivolumab or pembrolizumab) or anti-PD-L1 (atezolizumab) alone or in combination with chemotherapy at Kurume University Hospital (Kurume, Japan) or Kanagawa Cancer Center (Yokohama, Japan) were included in this study. Cohort 1, which included 123 patients enrolled between September 2016 and February 2020, was used as the training set for discovery; cohort 2, which included 99 patients enrolled between February 2020 and February 2021 and was completely independent from cohort 1, was used as the validation set. Patient characteristics are summarized in table 1. Tumor PD-L1 expression was determined by immunohistochemistry of paraffin-embedded tissue sections with an anti-PD-L1 monoclonal antibody (clone E1L3N; Cell Signaling Technology, Danvers, Massachusetts, USA; clone 22C3; Agilent Technologies/Dako, Carpinteria, California, USA). In most of the patients, PD-L1 expression was measured with tumor tissues obtained before the first-line treatment, even in those treated with ICI therapy as a second-line or later-line of treatment. The best clinical response was determined according to the Response Evaluation Criteria in Solid Tumors (RECIST) v.1.1.
Table 1.
Summary of patient characteristics
| Patient characteristics | Training set: cohort 1 | Validation set: cohort 2 | P value* |
| Total (n=123) | Total (n=99) | ||
| Age (years), mean (SD) | 68.8 (8.6) | 68.2 (10.0) | 0.62 |
| Sex, n (%) | 0.75 | ||
| Female | 30 (24.4) | 26 (26.3) | |
| Male | 93 (75.6) | 73 (73.7) | |
| BMI, mean (SD) | 21.9 (3.9) | 22.3 (3.9) | 0.36 |
| Smoking, n (%) | 0.44 | ||
| Former | 95 (77.2) | 81 (81.8) | |
| Never | 27 (22.0) | 18 (18.2) | |
| Unknown | 1 (0.8) | – | |
| Stages, n (%) | 0.04 | ||
| Stage III | 26 (21.1) | 11 (11.1) | |
| Stage IV | 73 (59.3) | 74 (74.7) | |
| Recurrence/metastasis | 24 (19.5) | 14 (14.1) | |
| Histology, n (%) | 0.03 | ||
| Non-squamous | 89 (72.4) | 64 (64.6) | |
| Squamous | 34 (27.6) | 30 (30.3) | |
| Others | – | 5 (5.1) | |
| Driver mutation, n (%) | 0.39 | ||
| Wild type | 104 (84.6) | 83 (83.8) | |
| EGFR | 18 (14.6) | 13 (13.1) | |
| ALK | 1 (0.8) | 3 (3.0) | |
| Tumor PD-L1 expression, n (%) | <0.01 | ||
| <1% | 44 (35.8) | 18 (18.2) | |
| 1%–49% | 32 (26.0) | 41 (41.4) | |
| >50% | 47 (38.2) | 30 (30.3) | |
| Unknown | – | 10 (10.1) | |
| Treatment line, n (%) | <0.01 | ||
| First line | 51 (41.5) | 65 (65.7) | |
| Second line | 49 (39.8) | 17 (17.2) | |
| Third line | 13 (10.6) | 6 (6.1) | |
| Further lines | 10 (8.1) | 11 (11.1) | |
| ECOG PS, n (%) | 0.03 | ||
| 0–1 | 99 (80.5) | 90 (90.9) | |
| 2–3 | 24 (19.5) | 9 (9.1) | |
| Metastasis, n (%) | |||
| Brain | 16 (13.0) | 16 (16.2) | 0.34 |
| Liver | 19 (15.5) | 12 (12.1) | 0.33 |
| Treatment option, n (%) | <0.01 | ||
| Monotherapy | 96 (78.1) | 34 (34.3) | |
| Combination therapy | 27 (22.0) | 65 (65.7) | |
| Blood test, mean (SD) | |||
| baseline | |||
| Albumin (g/dL) | 3.44 (0.60) | 3.54 (0.60) | 0.22 |
| NLR | 5.07 (3.75) | 6.41 (6.43) | 0.07 |
| 6 weeks after treatment | |||
| Albumin (g/dL) | 3.42 (0.68) | 3.64 (0.59) | 0.02 |
| NLR | 4.48 (4.74) | 4.85 (8.89) | 0.73 |
| Best clinical response (RECIST), n (%) | 0.15 | ||
| Partial response | 43 (35.0) | 34 (34.3) | |
| Stable disease | 30 (24.4) | 34 (34.3) | |
| Progressive disease | 47 (38.2) | 26 (26.3) | |
| Unknown | 3 (2.4) | 5 (5.1) | |
| Progression-free survival (days), median (95% Cl)† | 113 (84 to 196) | 169 (140 to 233) | 0.55 |
| Overall survival (days), median (95% CI)† | 404 (347 to 552) | 682 (359 to undefined) | 0.22 |
*Continuous variables: t-test; categorical variables: χ2; survival time: log rank test.
†Kaplan-Meier method.
ALK, anaplastic lymphoma kinase; BMI, body mass index; ECOG PS, Eastern Cooperative Oncology Group Performance Status; EGFR, epidermal growth factor receptor; NLR, neutrophil to lymphocyte ratio; RECIST, Response Evaluation Criteria in Solid Tumors.
Analysis of circulating cytokines
Peripheral blood samples were obtained before (pre) and at 6 weeks after (edt) the initiation of therapy. Plasma was separated from the whole blood samples collected in tubes containing heparin as an anticoagulant by centrifugation. A profile of 93 cytokines was quantified in 50 µL aliquots of 4-fold diluted plasma with the exception of TGF-β1, which used 16-fold-diluted plasma after oxidation, using a bead-based multiplex assay (Bio-Plex 200 system; Bio-Rad Laboratories, Hercules, California, USA) in accordance with the manufacturer’s instructions. For this assay, analyte kits from Bio-Rad Laboratories were used to measure 93 cytokines (online supplemental table S1). Cytokine concentrations were normalized to avoid potential batch effects.
jitc-2023-006788supp001.pdf (947.5KB, pdf)
Machine learning
We implemented an RSF for survival analysis using randomForestSRC package in R (v.4.1.3; https://www.r-project.org/) software. The cytokine profiles were used in their measured concentrations. For feature selection, datasets including all the 93 cytokines were considered, and an RSF minimal depth filter was used to score the feature importance of each cytokine: the random forest minimal depth evaluates the level at which a feature (in this case, cytokine) is split in the decision tree, which indicates how many branches a decision tree has to go through before reaching a particular feature.28 A smaller minimal depth indicates closer to the root node of the decision tree, and a more direct influence on the final prediction. Therefore, the smaller the minimal depth, the more important the cytokine is in determining the outcome of the model. We selected cytokines with a minimal depth less than the overall mean value as important feature cytokines, and used the reciprocal of the minimal depth (defined as the ‘importance score’) to visualize the importance of the variables. Fourteen and 19 cytokines were selected as the feature cytokines of the pre and edt cytokine profiles, respectively. The hyperparameters of feature selection were as follows: number of splits of k-fold cross-validation, 10; number of trees, 1000; size of node, 3; and rule of split, ‘logrank’. Selected cytokine profiles were used to build the prediction models. For the within-cohort validation, k-fold cross-validation was applied for the training set. For the cross-cohort validation, all the samples in the training set were used to train a model, and the completely independent validation set was used to test the predictive performances.
Statistical analyses
C-indices and time-dependent AUC of the ROC were calculated using randomForestSRC and survivalROC packages. The cut-off value of the CIRI for risk-predictive classification was optimized using the survival status at 1 year time point using Youden’s method using the survival package; a predicted CIRI score higher than the cut-off was defined as the high-risk group with poor treatment response and poor prognosis; otherwise, it was considered as the low-risk group with good treatment response and good prognosis. Kaplan-Meier plots, log rank p values, and Cox-proportional hazard ratios (HRs) were generated using the survival and survminer packages. The Wilcoxon signed-rank test, Student’s t-test, and χ2 test were performed using the stats package. Spearman’s rank correlation coefficient was calculated using cor and cor.test R-functions. All the aforementioned packages and functions are included in the R programming language.
Results
Patient characteristics
As shown in table 1, no difference was observed in the age, sex, body mass index (BMI), smoking, driver mutation (epidermal growth factor receptor and anaplastic lymphoma kinase), brain and liver metastasis, albumin level (pre), and neutrophil to lymphocyte ratio (NLR; pre and edt) between cohorts 1 and 2. There were significant differences in the stage, histology, tumor PD-L1 level, treatment line, Eastern Cooperative Oncology Group performance status, and albumin level (edt). There was a greater number of patients with monotherapy (78.1%) and combination therapy (65.7%) in cohorts 1 and 2, respectively; however, there was no significant difference in the patient response determined by RECIST, PFS, and OS following immunotherapy between these two independent cohorts.
Prediction of therapeutic outcomes by baseline cytokine profiles
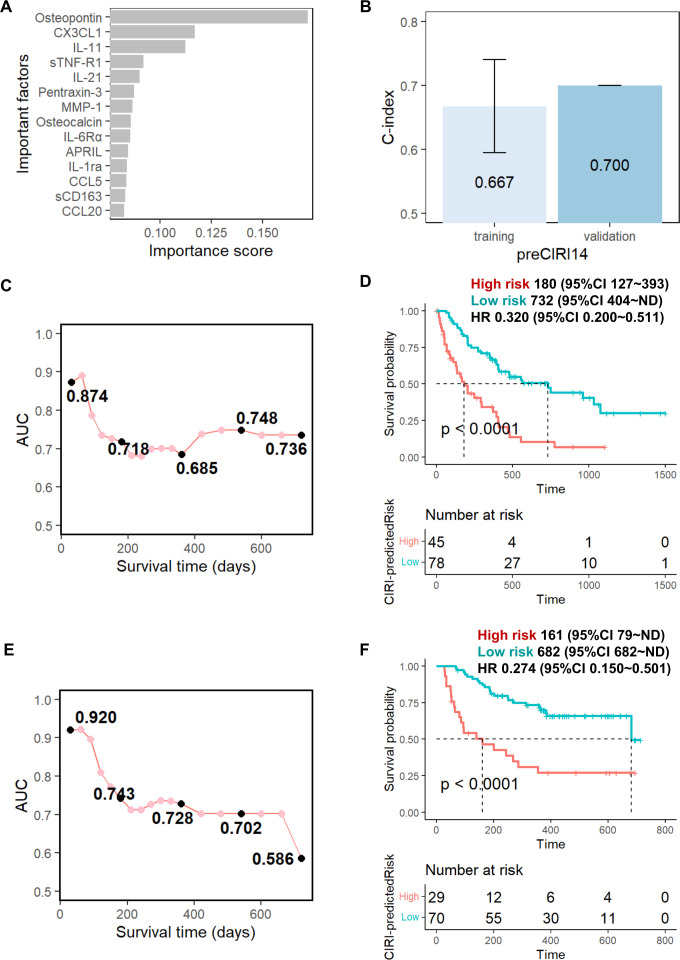
We initially developed an ensemble learning random forest classifier for the OS prediction of anti-PD-1/PD-L1 immunotherapy with baseline cytokine profiles. First, the 93 baseline cytokines of the training set were scored and arranged by their importance in the OS prediction models according to their minimal depth in the random forest algorithm.28 As shown in figure 2A, 14 cytokines (osteopontin, CX3CL1, IL-11, sTNF-R1, IL-21, pentraxin-3, MMP-1, osteocalcin, IL-6Rα, APRIL, IL-1ra, CCL5, sCD163, and CCL20) were identified as the feature cytokines and selected to build the OS prediction model (hereafter called ‘preCIRI14’). The preCIRI14 model achieved superior performance, as indicated by a C-index of 0.667 in predicting the OS of patients following treatment in the training set (figure 2B). Next, the preCIRI14 was applied to predict the occurrence of events at each time point. Time-dependent AUC of the ROC curve estimation demonstrated that the AUC remained above 0.8 during 1–2 months after treatment, and then decreased to approximately 0.7 over time (figure 2C). To examine the prediction accuracy at the individual level, patients were scored based on the predicted risk of prognosis using preCIRI14. Then, the optimized cut-off value of the predicted preCIRI14 score was determined according to the ROC curve at the time point of 1 year after treatment using Youden’s method (online supplemental figure S1), which was used to divide the patients into high-risk and low-risk groups. As shown in figure 2D, 45 and 78 patients in the training set were classified into the high-risk and low-risk groups, respectively. The OS rates estimated using the Kaplan-Meier method and the log rank test showed that patients in the high-risk group had a significantly worse prognosis than those in the low-risk group (HR 0.320; 95% CI 0.200 to 0.511; p<0.0001). Notably, the median survival of the patients in the high-risk group was 180 days, whereas that of the low-risk group was 732 days. Finally, the performance of the preCIRI14 score in risk stratification of immunotherapy in terms of OS was evaluated in 99 patients with NSCLC in the validation set (cohort 2). At the population level, the prediction accuracy of preCIRI14, as indicated by the C-index, was 0.700 in the validation set, which was slightly higher than that of the training set (figure 2B). The time-dependent ROC curve estimation of the predicted probability also confirmed that the AUC remained above 0.7 within 1.5 years following treatment, and subsequently decreased to 0.586 at time point of 2 years in the validation set (figure 2E). At the individual level, 29 and 70 patients in the validation set were classified into the high-risk and low-risk groups, respectively. The Kaplan-Meier survival analysis confirmed that patients in the high-risk group had a significantly worse prognosis than those in the low-risk group (HR 0.274; 95% CI 0.150 to 0.501; p<0.0001). The median survival of the patients in the high-risk group was 161 days, while that of the low-risk group was 682 days (figure 2F).
Figure 2.
Prediction performance of preCIRI14 model using baseline circulating cytokine signature. (A) Feature cytokines selected using random survival forest (RSF) minimal depth filter: the importance score was defined as 1/(minimal depth); (B) Concordance (C)-indices of preCIRI14 model for the training and validation sets; time-dependent area under the curve AUC of receiver operating characteristic curves (ROCs) at each time point for the (C) training and (E) validation sets; overall survival time of ‘high risk’ and ‘low risk’ groups predicted by preCIRI14 for (D) training and (F) validation sets using cut-off value optimized by training set (see online supplemental figure S1). The CIRI scores indicate risk of event occurrence, whereas ‘high risk’ indicates patients with high CIRI scores and ‘low risk’ indicates patients with low CIRI scores.
Since tumor PD-L1 gene expression is an FDA-approved biomarker for clinical use to predict the immunotherapy efficacy of NSCLC before treatment, we examined the utility of baseline protein expression of tumor PD-L1 in risk stratification of ICI therapy. As shown in online supplemental figure S2, no significant association was observed between tumor PD-L1 protein expression and patient survival in response to anti-PD-1/PD-L1 immunotherapy in either the training or validation sets.
Prediction of the therapeutic outcomes based on the early during treatment cytokine profiles
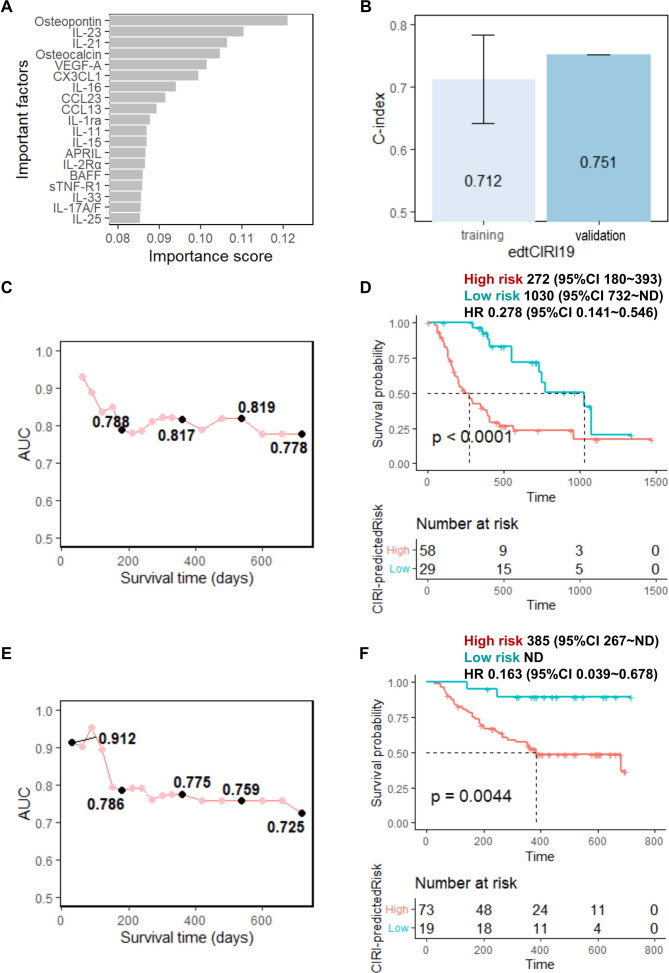
Thereafter, we examined the utility of the cytokine profiles at 6 weeks after treatment in the stratification of patients in response to anti-PD-1/PD-L1 immunotherapy. Similar to the analysis of the baseline cytokine profiles, 19 feature cytokines (osteopontin, IL-23, IL-21, osteocalcin, VEGF-A, CX3CL1, IL-16, CCL23, CCL13, IL-1ra, IL-11, IL-15, APRIL, IL-2Rα, BAFF, sTNF-R1, IL-33, IL-17A/F, and IL-25) (figure 3A) were identified and selected to build the edt cytokine profile-based OS prediction model (hereafter called ‘edtCIRI19’). Compared with the predictive performance of preCIRI14, the cytokine response in the early phase after treatment initiation showed a stronger correlation with patient survival in response to immunotherapy. The C-indices of the edtCIRI19 model were 0.712 and 0.751 for predicting the OS of patients following treatment in the training and validation sets, respectively (figure 3B). The ROC curves at each time point also showed a similar predictive performance of edtCIRI19 in the training and validation sets, where the AUC remained above 0.778 and 0.725 for the training and validation sets, respectively, until 24 months following treatment (figure 3C,E). At the individual level, the patients were classified into poor-prognosis and good-prognosis groups based on the predicted scores of the edtCIRI19 model using the optimized cut-off value according to the ROC curve at the time point of 1 year after treatment in the training set (online supplemental figure S3). The Kaplan-Meier analysis showed that patients in the high-risk group had a significantly worse prognosis than those in the low-risk group in both the training and validation sets (HR 0.278; 95% CI 0.141 to 0.546; p<0.0001 in the training set; HR 0.163; 95% CI 0.039 to 0.678; p=0.0044 in the validation set; figure 3D,F).
Figure 3.
Prediction performance of edtCIRI19 model using circulating cytokine signature early during treatment. (A) Feature cytokines selected using RSF minimal depth filter: the importance score was defined as 1/(minimal depth); (B) C-indices of edtCIRI19 model for training and validation sets; time-dependent AUC of ROC at each time points for the (C) training and (E) validation sets; overall survival time of ‘high risk’ (high CIRI) and ‘low risk’ (low CIRI) groups predicted by edtCIRI19 for the (D) training and (F) validation sets using cut-off values optimized by the training set (see online supplemental figure S3). AUC, area under the curve; CIRI, Cytokine-based ICI Response Index; ROC, receiver operating characteristic; RSF, random survival forest.
We also examined whether the dynamic changes in the circulating cytokine profiles before and 6 weeks after treatment were associated with patient survival in response to anti-PD-1/PD-L1 immunotherapy. Based on the difference (subtraction) and fold change in cytokine concentrations, 17 features cytokines (osteopontin, VEGF-A, IL-33, IL-21, IL-15, IL-31, IL-2Ra, CCL11, IL-17A/F, CCL22, IL-16, CCL7, CX3CL1, IL-11, CXCL11, CXCL5, and CCL24; online supplemental figure S4) and 20 cytokines (osteopontin, VEGF-A, CXCL5, TGF-β1, Osteocalcin, CCL22, IL-2Rα, IL-15, IL-31, CCL8, FGF-basic, CCL23, IL-17A, IL-16, IL-33, IL-21, IL-9, IL-27, IL-22, and CCL11; online supplemental figure S5) were selected to build the OS prediction model (hereafter called ‘diffCIRI17’ and ‘foldCIRI20’, respectively). As shown in online supplemental figures S4 and S5, reduced performance in terms of the prediction accuracy and reproducibility in the validation cohort of diffCIRI17 and foldCIRI20 in OS prediction following anti-PD-1/PD-L1 immunotherapy was observed in comparison with that of edtCIRI19 (figure 3).
Advanced prediction models
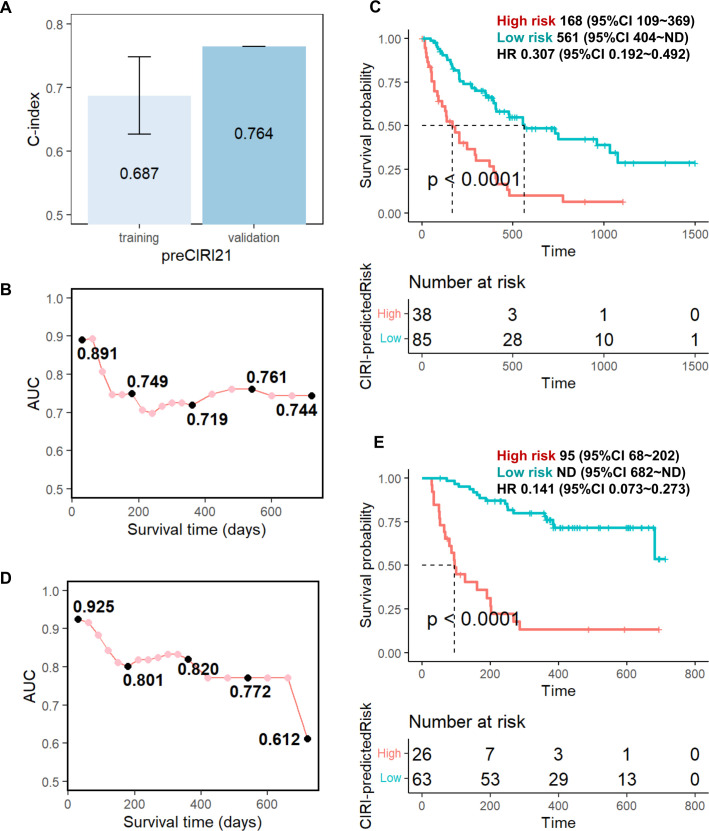
Finally, we developed advanced prediction models for patient survival in response to anti-PD-1/PD-L1 immunotherapy by combining cytokines with other circulating factors and clinical features that have been proven to be related to immunotherapy response in clinical studies; we included factors, such as age,29 sex, stage,30 BMI,31 32 blood albumin,25 NLR,25 33–36 tumor PD-L1 expression, and treatment option (only for the edtCIRI model) in the advanced prediction models (hereafter called ‘preCIRI21’ and ‘edtCIRI27’, respectively). As shown in figures 4 and 5, as well as online supplemental figures S6 and S7, improved prediction efficacy was observed with preCIRI21 and edtCIRI27 compared with circulating cytokines alone in both the training and validation sets. In the validation set, the C-indices of the preCIRI21 and edtCIRI27 models were 0.764 and 0.757, respectively, in predicting the OS of patients after treatment. At the individual level, the patients classified as high-risk groups based on the predicted scores of the preCIRI21 and edtCIRI27 models had a significantly worse prognosis than those in the low-risk group (HR 0.141; 95% CI 0.073 to 0.273; p<0.0001 in the preCIRI21 model; HR 0.158; 95% CI 0.021 to 1.170; p=0.038 in the edtCIRI27 model).
Figure 4.
Prediction performance of the preCIRI21 model. Fourteen feature cytokines of preCIRI14 model and 7 clinical and circulating factors (age, sex, stage, BMI, blood albumin of pre, NLR of pre, and tumor PD-L1 expression) were used in preCIRI21. (A) C-indices of preCIRI21 model for the training and validation sets; time-dependent AUC of ROC at each time points for the (B) training and (D) validation sets; overall survival time of ‘high risk’ (high CIRI) and ‘low risk’ (low CIRI) groups predicted by preCIRI21 for the (C) training and (E) validation sets using optimized cut-off values (see online supplemental figure S6). AUC, area under the curve; BMI, body mass index; CIRI, Cytokine-based ICI Response Index; NLR, neutrophil-to-lymphocyte ratio; ROC, receiver operating characteristic.
Figure 5.
Prediction performance of the edtCIRI27 model. Nineteen feature cytokines of edtCIRI21 model and 8 clinical and circulating factors (age, sex, stage, BMI, blood albumin of edt, NLR of edt, tumor PD-L1 expression, and treatment option) were used in edtCIRI27. (A) C-indices of edtCIRI27 model for the training and validation sets; time-dependent AUC of ROC at each time points for the (B) training and (D) validation sets; overall survival time of ‘high risk’ (high CIRI) and ‘low risk’ (low CIRI) groups predicted by preCIRI21 for the (C) training and (E) validation sets using optimized cut-off values (see online supplemental figure S7). AUC, area under the curve; BMI, body mass index; CIRI, Cytokine-based ICI Response Index; edt, early during treatment; NLR, neutrophil-to-lymphocyte ratio; ROC, receiver operating characteristic.
Association between feature cytokine and predicted prognosis
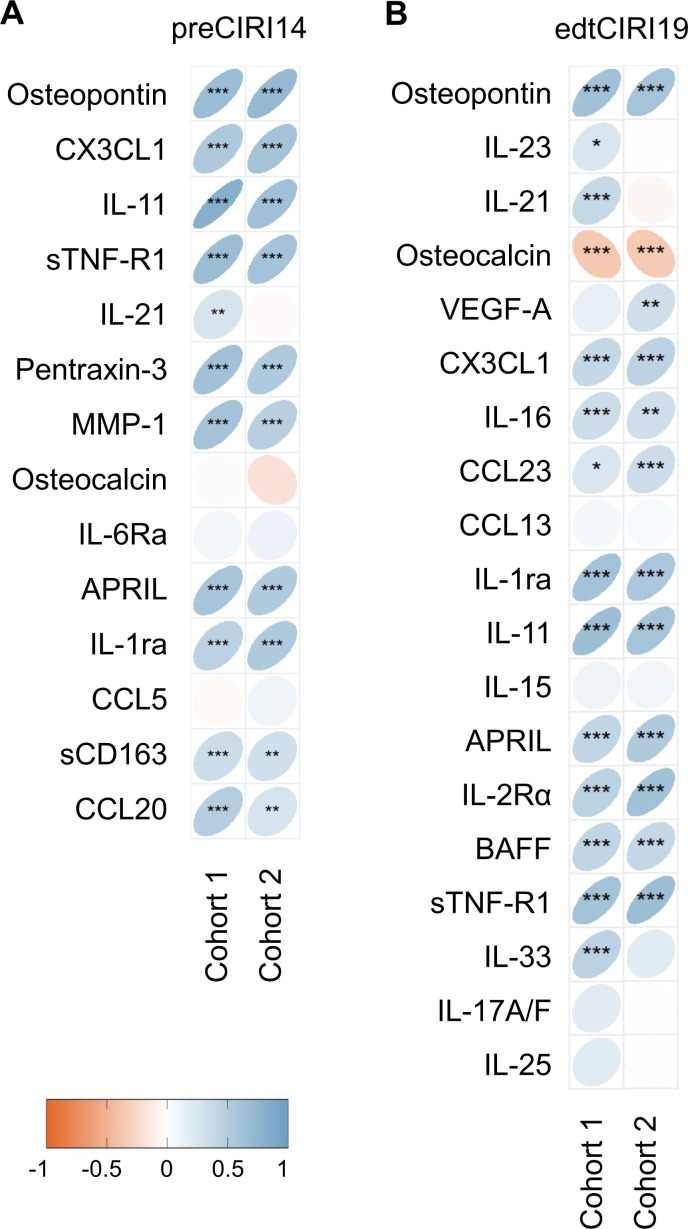
Next, Spearman’s rank correlation coefficient was calculated between the feature cytokines levels and CIRI scores at the individual level. Significant correlations were observed between the levels of most feature cytokines and the CIRI scores in the two independent cohorts (figure 6). In the preCIRI14 model, a significant positive association was observed between the baseline osteopontin, CX3CL1, IL-11, sTNF-R1, pentraxin-3, MMP-1, APRIL, IL-1ra, sCD163, and CCL20 levels and the CIRI score of patients in both the training and validation sets (figure 6A). In the edtCIRI19 model, a significant positive association was observed between the osteopontin, CX3CL1, IL-16, CCL23, IL-1ra, IL-11, APRIL, IL-2Rα, BAFF, and sTNF-R1 levels 6 weeks after treatment and the CIRI score of patients in both the training and validation sets (figure 6B). In contrast, the osteocalcin 6 weeks after treatment negatively correlated with the CIRI score (figure 6B). In addition, the distribution of features in the preCIRI21 and edtCIRI27 models among low-risk and high-risk groups of the combined cohorts 1 and 2 is shown in online supplemental figure S8. In the high-risk group of the preCIRI21, lower levels of Alb and BMI, higher levels of NLR, and a higher proportion of female patients were observed. On the other hand, in the high-risk group of the edtCIRI27, lower levels of Alb and higher levels of NLR and PD-L1 were observed.
Figure 6.
Association between feature cytokines and predicted Cytokine-based ICI Response Index (CIRI) scores. Spearman’s rank correlation coefficient between the feature cytokine levels and CIRI scores predicted by the (A) preCIRI14 and (B) edtCIRI19 models of cohort 1 and cohort 2. Significance levels: *p≤0.05, **p≤0.01, ***p≤0.001. ICI, immune checkpoint inhibitor.
Discussion
In this study, a machine learning approach was applied to circulating cytokine signatures to extract OS-associated feature cytokines and establish OS predictive models for patients with NSCLC in response to anti-PD-1/PD-L1 immunotherapy. Fourteen and 19 cytokines at baseline and on treatment, respectively, were selected to generate CIRI models (namely, preCIRI14 and edtCIRI19, respectively), both of which successfully identified patients with poor long-term OS in two completely independent cohorts. Importantly, the baseline expression of tumor PD-L1 was not related to the OS of patients, highlighting the potential of the preCIRI14 score in identifying and predicting patients with NSCLC who would benefit from anti-PD-1/PD-L1 immunotherapy with prolonged survival and may aid in taking clinical decisions at an early stage before treatment. In addition, the efficacy of OS prediction was improved by combining circulating cytokine signatures with other circulating factors and clinical features, suggesting that circulating cytokine-based OS prediction holds promise for clinical practice as a ‘plug-in’ approach to identify a subset of patients with an elevated likelihood of poor therapeutic outcomes in response to immunotherapy.
Compared with cohort 2, a relatively greater number of patients received ICI therapy as second-line and third-line treatment in cohort 1. Previous treatment, such as cytotoxic chemotherapy, could have affected the expression level of PD-L1 before ICI therapy37; therefore, the stratification performance of PD-L1 expression level in response to ICI therapy could be more disturbed in cohort 1. In addition, a relatively greater number of patients received combination therapy with ICI and chemotherapy in cohort 2. The tumor PD-L1 expression and survival probability in cohort 2 was inconsistent; patients with tumor PD-L1 expression of <1% showed a higher survival probability at the later stage compared with that of patients with tumor PD-L1 expression of 1%–49%, which is probably attributed to the effect of chemotherapy. Besides, it is known that the heterogeneity in PD-L1 expression due to the difference in the disease site and time point of biopsy may affect its performance in predicting immunotherapy efficacy of NSCLC patients.38 Collectively, these data indicate the limitation of tumor PD-L1 expression in the stratification of patients in response to ICI therapy combined with chemotherapy.
Our data showed reduced cross-cohort performance in the prediction of patient survival in response to anti-PD-1/PD-L1 immunotherapy, with dynamic changes in the circulating cytokine profiles compared with those before and 6 weeks after treatment. We presume that this is owing to the difference in the dynamic changes of the circulating cytokines in response to ICI monotherapy and combination therapy with ICI and chemotherapy. Chemotherapy reportedly might also induce a change in the circulating cytokine profiles.39 40 We acknowledge that the effect of chemotherapy on circulating cytokine profiles is one of the reasons why the treatment-induced changes, as indicated in our diffCIRI and foldCIRI models, can only demonstrate good predictivity in the training cohort with monotherapy but cannot be reproduced in the validation cohort with combined ICI treatment and chemotherapy. Notably, the preCIRI and edtCIRI models, based on levels but not changes of circulating cytokines, can overcome this difference in treatment option and show consistent performance in both the training and validation cohorts. The function of cytokines is dynamic and context-dependent, with a cytokine having opposite target responses under different conditions. Different cytokines may act coordinately or counteractively, and the collective outcome represents the strength of immune responses.41 Therefore, the circulating cytokine signature as the mediator and indicator of host immune activity demonstrated improved predictive performance of patient survival in response to ICI therapy irrespective of the differences in patient characteristics, such as treatment line and treatment option. Our study shows that the machine learning-generated CIRI score, based on the profile of a set of circulating cytokines, could represent the achieved status of the immune system, which showed advanced performance in the OS prediction of patients following ICI treatment, regardless of different backgrounds and treatment options. We hypothesized that the status attained, rather than the dynamic changes in the circulating cytokines during treatment, was associated with the therapeutic outcomes of anti-PD-1/PD-L1 immunotherapy in patients with NSCLC.
Feature importance ranking in ensemble RSF classifiers identified osteopontin as the most important feature cytokine for the performance of all the CIRI prediction models in predicting the OS of patients with NSCLC in response to anti-PD-1/PD-L1 immunotherapy. The osteopontin levels at baseline and 6 weeks after treatment were significantly positively associated with the CIRI scores in both the training and validation cohorts, suggesting an unfavorable role of osteopontin in anti-PD-1/PD-L1 immunotherapy in NSCLC. Osteopontin is a multifunctional phosphoglycoprotein encoded by the secreted phosphoprotein 1 (SPP1) gene and plays a critical role in tumor immune evasion by regulating macrophage polarization, recruitment, and inhibition of T cell activation in the tumor microenvironment.42 43 SPP1 knockout in colon tumor cells increased tumor-specific cytotoxic T cell-mediated cytotoxicity in vitro and resulted in decreased tumor growth in vivo, indicating the potential of osteopontin blockade in immunotherapy.44 It was reported that overexpressed osteopontin is often detected in the tumor microenvironment and elevated serum osteopontin levels are correlated with poor-prognosis of patients with NSCLC.45–47 However, insufficient performance in the stratification of patients with NSCLC was reported in response to ICI therapy with circulating osteopontin alone.48 49 Our data demonstrated an improvement in the prediction performance with the circulating cytokine signatures than that with the level of a single cytokine.
Beyond osteopontin, significant positive associations were also observed between the levels of other feature cytokines, such as CX3CL1 and IL-11, at baseline and 6 weeks after treatment and the CIRI scores of patients in both the training and validation sets in the preCIRI14 and edtCIRI19 models. A previous study showed that inhibition of CX3CL1 in human NSCLC tissues and cell lines led to decreased tumor growth and cell invasion through the inactivation of the ERK/AKT pathway.50 In a small cohort study of lung adenocarcinoma with 42 patients, the CX3CL1 level was considered an independent risk factor for the OS, and patients with poor-prognosis showed increased CX3CL1 expression.51 IL-11 is a stromal-cell-derived pleiotropic cytokine with tumor-promoting properties by activating STAT3 and promoting BCL2 and survivin-mediated anti-apoptotic pathways in NSCLC,52 and the increased level of circulating IL-11 was related with worse prognosis of patients with NSCLC in response to immunotherapy.21 Notably, the osteocalcin levels 6 weeks after treatment but not at baseline negatively correlated with the CIRI score, suggesting a better prognosis of patients with increased levels of osteocalcin at the early stage of treatment. Osteocalcin-expressing osteoblastic cells in bones reportedly promote lung tumors by producing SiglecFhigh neutrophils,53 and increased circulating osteocalcin levels were associated with longer OS in head and neck squamous cell carcinoma in response to anti-PD-L1 immunotherapy.54 Collectively, these data suggest that the CIRI models in our study can extract feature cytokines that play critical roles in antitumor immunity.
Several studies have shown that TGF-β expression in the tumor microenvironment is associated with immune escape and poor response to ICI treatment.55–58 A study of 33 patients with NSCLC treated with ICI monotherapy also observed a significant association between baseline circulating TGF-β1 levels and OS.24 However, these findings were not confirmed in our study cohorts. Possible explanations for this discrepancy include the heterogeneous clinical characteristics of patients in our cohorts, which included both monotherapy and a combination of ICI and chemotherapy, as well as the difference in ethnic background of patients. It is also important to note that machine learning methods are data driven and evaluate variables based on their distribution characteristics rather than biological significance, which indicates that the selected cytokine signatures have a greater impact in predictive performance when evaluated as a whole, and that the association between baseline TGF-β1 levels and OS may not be as strong as other important cytokines.
Although our study provides important insights into the prognostic value of cytokine signatures in ICI-treated patients with NSCLC, there are several limitations that need to be considered. First, the sample size needs to be increased to further improve the overall predictive accuracy and robustness of the model. A larger sample size would also enable more detailed subgroup analysis to evaluate the impact of patient heterogeneity (such as treatment lines, treatment options, oncogene-addicted and non-oncogene-addicted tumors, different levels of PD-L1 expression). For example, by separating the ICI monotherapy group from the combination chemotherapy group and modeling the changes (difference and fold change) before and after treatment, we can better understand the changes in cytokine signatures under different treatment methods and their association with prognosis, thus aiding treatment decision-making. Second, cytokine levels can fluctuate over time, and thus multiple time point measurements may be necessary to better capture the dynamic changes in cytokine signatures. Additionally, our study was retrospective, and therefore, the data were subject to potential bias, missing data, and confounding factors that may affect the results. Future studies could also aim to validate the CIRI models using independent prospective cohorts to address concerns of model overfitting and ensure generalizability. Third, while the machine learning models used in this study are effective at identifying prognostic cytokine signatures unnoticed so far, they do not provide mechanistic insights into the underlying biology. Future studies could consider using complementary experimental approaches to explore the biological mechanisms behind the identified cytokine signatures. We believe the CIRI score-based patient stratification strategy may provide a powerful way to deepen the understanding of the differences between the high- and low-risk groups. In conclusion, our data suggest that CIRI is highly accurate and reproducible in identifying and predicting patients with NSCLC who would benefit from anti-PD-1/PD-L1 therapy with prolonged OS and may aid in clinical decisions made before and/or at the early stage of treatment. Our study also suggests further endeavors to explore the potential of cytokine-based therapy, such as inhibition of osteopontin, to improve the host immune response status and thereby, the long-term efficacy of anti-PD-1/PD-L1 therapy in NSCLC.
Acknowledgments
The authors would like to acknowledge and thank Akiko Orikasa and Makoto Wakatsuki (Kanagawa Cancer Center Research Institute) and Kimiko Inoue (Kurume University) for sample handling.
Footnotes
Contributors: Conception and design: FW and TS. Provision of study materials or patients: KA, YN, HS, NM, TTagami, TKouro, YI, TKato, SM, RU, TTokito, HH, SH, KT, TB, TTamura, YM and TS. Collection and assembly of data: KA, YN, NM, TTagami, KM and TS. Data analysis and interpretation: FW and TS. Manuscript writing: all authors. Final approval of manuscript: all authors. Responsible for the overall content as guarantors: FW and TS.
Funding: This work was supported by Japan Society for the Promotion of Science Grant to FW: Grant-in-Aid for Scientific Research (C): 22K08724; AMED Grant to TS: JP19ae0101076; and The Cell Science Research Foundation Fellowship to FW.
Competing interests: KA received honoraria from AstraZeneka, MSD, Bristol Myers Squibb, Ono and Chugai. YN received honoraria from Ono, Takeda, Eli Lilly, Kyowa Kirin, Boehringer Ingelheim, and AstraZeneca, Bristol Myers Squibb, and research funds from Bristol Myers Squibb. HS received honoraria from Boehringer Ingelheim, Eli Lilly, Pfizer, AstraZeneca, Bristol Myers Squibb, Chugai, and Ono, and research funds from AstraZeneca, Bristol Myers Squibb, Chugai, and Ono. TTagami is an employee of Ajinomoto Co. TKato’s spouse is an employee of Eli Lilly, and he received honoraria from AstraZeneca, Boehringer Ingelheim, Bristol Myers Squibb, Chugai, Daiichi-Sankyo, Eli Lilly, Merck Biopharma, MSD, Novartis, Ono, Pfizer, and Roche, consulting fees from Abbvie, Amgen, AstraZeneca, Beigene, Chugai, Daiichi-Sankyo, Eli Lilly, Glaxo, Merck Biopharma, MSD, Nippon Kayaku, Novartis, Ono, Pfizer, Roche, Taiho, and Takeda, and research funds from Abbvie, Amgen, AstraZeneca, Blueprint, Chugai, Eli Lilly, Haihe, Merck Biopharma, MSD, Novartis, Pfizer, Regeneron, and Takeda. TKondo received honoraria from AstraZeneca, Daiichi-Sankyo, and Otsuka, and research funds from AstraZeneca, Chugai, and Taiho. SM received honoraria from AstraZeneca, Chugai, Boehringer Ingelheim, Taiho, Pfizer, MSD, and Ono. KM received honoraria from AstraZeneka and Chugai. TS received honoraria from Chugai and Bristol Myers Squibb, and research funds from Taiho and BrightPath Biotherapeutics.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by Institutional Review Boards of Kurume University Hospital (approval numbers: 15210 and 19240) and Kanagawa Cancer Center (approval number: 2019-131). Participants gave informed consent to participate in the study before taking part.
References
- 1. Doroshow DB, Sanmamed MF, Hastings K, et al. Immunotherapy in non-small cell lung cancer: facts and hopes. Clin Cancer Res 2019;25:4592–602. 10.1158/1078-0432.CCR-18-1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Reck M, Remon J, Hellmann MD. First-line Immunotherapy for non-small-cell lung cancer. J Clin Oncol 2022;40:586–97. 10.1200/JCO.21.01497 [DOI] [PubMed] [Google Scholar]
- 3. Xia L, Liu Y, Wang Y. PD-1/PD-L1 blockade therapy in advanced non-small-cell lung cancer: Current status and future directions. Oncologist 2019;24(Suppl 1):S31–41. 10.1634/theoncologist.2019-IO-S1-s05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Büttner R, Gosney JR, Skov BG, et al. Programmed death-ligand 1 immunohistochemistry testing: A review of Analytical assays and clinical implementation in non-small-cell lung cancer. JCO 2017;35:3867–76. 10.1200/JCO.2017.74.7642 [DOI] [PubMed] [Google Scholar]
- 5. Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus Docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med 2015;373:123–35. 10.1056/NEJMoa1504627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in advanced Nonsquamous non-small-cell lung cancer. N Engl J Med 2015;373:1627–39. 10.1056/NEJMoa1507643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med 2016;375:1823–33. 10.1056/NEJMoa1606774 [DOI] [PubMed] [Google Scholar]
- 8. Carbone DP, Reck M, Paz-Ares L, et al. First-line Nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med 2017;376:2415–26. 10.1056/NEJMoa1613493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garassino MC, Cho B-C, Kim J-H, et al. Durvalumab as third-line or later treatment for advanced non-small-cell lung cancer (ATLANTIC): an open-label, single-arm, phase 2 study. Lancet Oncol 2018;19:521–36. 10.1016/S1470-2045(18)30144-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ready N, Hellmann MD, Awad MM, et al. First-line Nivolumab plus Ipilimumab in advanced non-small-cell lung cancer (Checkmate 568): outcomes by programmed death ligand 1 and tumor mutational burden as biomarkers. J Clin Oncol 2019;37:992–1000. 10.1200/JCO.18.01042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hellmann MD, Ciuleanu T-E, Pluzanski A, et al. Nivolumab plus Ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med 2018;378:2093–104. 10.1056/NEJMoa1801946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gandara DR, Paul SM, Kowanetz M, et al. Blood-based tumor mutational burden as a Predictor of clinical benefit in non-small-cell lung cancer patients treated with Atezolizumab. Nat Med 2018;24:1441–8. 10.1038/s41591-018-0134-3 [DOI] [PubMed] [Google Scholar]
- 13. Rizvi NA, Hellmann MD, Snyder A, et al. Cancer Immunology. mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124–8. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Berraondo P, Sanmamed MF, Ochoa MC, et al. Cytokines in clinical cancer Immunotherapy. Br J Cancer 2019;120:6–15. 10.1038/s41416-018-0328-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang R, Zheng J, Shao X, et al. Development of a Prognostic composite cytokine signature based on the correlation with Nivolumab clearance: Translational PK/PD analysis in patients with renal cell carcinoma. J Immunother Cancer 2019;7:348. 10.1186/s40425-019-0819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tarhini AA, Zahoor H, Lin Y, et al. Baseline circulating IL-17 predicts toxicity while TGF-Beta1 and IL-10 are Prognostic of relapse in Ipilimumab Neoadjuvant therapy of Melanoma. J Immunother Cancer 2015;3:39. 10.1186/s40425-015-0081-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koguchi Y, Hoen HM, Bambina SA, et al. Serum Immunoregulatory proteins as predictors of overall survival of metastatic Melanoma patients treated with Ipilimumab. Cancer Res 2015;75:5084–92. 10.1158/0008-5472.CAN-15-2303 [DOI] [PubMed] [Google Scholar]
- 18. Ozawa Y, Amano Y, Kanata K, et al. Impact of early inflammatory cytokine elevation after commencement of PD-1 inhibitors to predict efficacy in patients with non-small cell lung cancer. Med Oncol 2019;36:33. 10.1007/s12032-019-1255-3 [DOI] [PubMed] [Google Scholar]
- 19. Keegan A, Ricciuti B, Garden P, et al. Plasma IL-6 changes correlate to PD-1 inhibitor responses in NSCLC. J Immunother Cancer 2020;8:e000678. 10.1136/jitc-2020-000678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sanmamed MF, Perez-Gracia JL, Schalper KA, et al. Changes in serum Interleukin-8 (IL-8) levels reflect and predict response to anti-PD-1 treatment in Melanoma and non-small-cell lung cancer patients. Ann Oncol 2017;28:1988–95. 10.1093/annonc/mdx190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Agulló-Ortuño MT, Gómez-Martín Ó, Ponce S, et al. Blood predictive biomarkers for patients with non-small-cell lung cancer associated with clinical response to Nivolumab. Clin Lung Cancer 2020;21:75–85. 10.1016/j.cllc.2019.08.006 [DOI] [PubMed] [Google Scholar]
- 22. Kauffmann-Guerrero D, Kahnert K, Kiefl R, et al. Systemic inflammation and pro-inflammatory cytokine profile predict response to Checkpoint inhibitor treatment in NSCLC: a prospective study. Sci Rep 2021;11:10919. 10.1038/s41598-021-90397-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hirashima T, Kanai T, Suzuki H, et al. The levels of interferon-gamma release as a biomarker for non-small-cell lung cancer patients receiving immune Checkpoint inhibitors. Anticancer Res 2019;39:6231–40. 10.21873/anticanres.13832 [DOI] [PubMed] [Google Scholar]
- 24. de Miguel-Perez D, Russo A, Gunasekaran M, et al. Baseline extracellular Vesicle TGF-beta is a predictive biomarker for response to immune Checkpoint inhibitors and survival in non-small cell lung cancer. Cancer 2023;129:521–30. 10.1002/cncr.34576 [DOI] [PubMed] [Google Scholar]
- 25. Chowell D, Yoo S-K, Valero C, et al. Improved prediction of immune Checkpoint blockade efficacy across multiple cancer types. Nat Biotechnol 2022;40:499–506. 10.1038/s41587-021-01070-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jiang P, Zhang Y, Ru B, et al. Systematic investigation of cytokine signaling activity at the tissue and single-cell levels. Nat Methods 2021;18:1181–91. 10.1038/s41592-021-01274-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kong J, Ha D, Lee J, et al. Network-based machine learning approach to predict Immunotherapy response in cancer patients. Nat Commun 2022;13:3703. 10.1038/s41467-022-31535-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ishwaran H, Kogalur UB, Gorodeski EZ, et al. High-dimensional variable selection for survival data. Journal of the American Statistical Association 2010;105:205–17. 10.1198/jasa.2009.tm08622 [DOI] [Google Scholar]
- 29. Ikeguchi A, Machiorlatti M, Vesely SK. Disparity in outcomes of Melanoma adjuvant Immunotherapy by demographic profile. Melanoma Manag 2020;7:MMT43. 10.2217/mmt-2020-0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuai J, Yang F, Li G-J, et al. In vitro-activated tumor-specific T lymphocytes prolong the survival of patients with advanced gastric cancer: a retrospective cohort study. Onco Targets Ther 2016;9:3763–70. 10.2147/OTT.S102909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Z, Aguilar EG, Luna JI, et al. Paradoxical effects of obesity on T cell function during tumor progression and PD-1 Checkpoint blockade. Nat Med 2019;25:141–51. 10.1038/s41591-018-0221-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sanchez A, Furberg H, Kuo F, et al. Transcriptomic signatures related to the obesity paradox in patients with clear cell renal cell carcinoma: a cohort study. Lancet Oncol 2020;21:283–93. 10.1016/S1470-2045(19)30797-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Valero C, Lee M, Hoen D, et al. Pretreatment neutrophil-to-lymphocyte ratio and mutational burden as biomarkers of tumor response to immune Checkpoint inhibitors. Nat Commun 2021;12:729. 10.1038/s41467-021-20935-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jaillon S, Ponzetta A, Di Mitri D, et al. Neutrophil diversity and plasticity in tumour progression and therapy. Nat Rev Cancer 2020;20:485–503. 10.1038/s41568-020-0281-y [DOI] [PubMed] [Google Scholar]
- 35. Park W, Lopes G. Perspectives: neutrophil-to-lymphocyte ratio as a potential biomarker in immune Checkpoint inhibitor for non-small-cell lung cancer. Clin Lung Cancer 2019;20:143–7. 10.1016/j.cllc.2018.12.003 [DOI] [PubMed] [Google Scholar]
- 36. Maymani H, Hess K, Groisberg R, et al. Predicting outcomes in patients with advanced non-small cell lung cancer enrolled in early phase Immunotherapy trials. Lung Cancer 2018;120:137–41. 10.1016/j.lungcan.2018.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sakai H, Takeda M, Sakai K, et al. Impact of cytotoxic chemotherapy on PD-L1 expression in patients with non-small cell lung cancer negative for EGFR Mutation and ALK fusion. Lung Cancer 2019;127:59–65. 10.1016/j.lungcan.2018.11.025 [DOI] [PubMed] [Google Scholar]
- 38. Hong L, Negrao MV, Dibaj SS, et al. Programmed death-ligand 1 heterogeneity and its impact on benefit from immune Checkpoint inhibitors in NSCLC. J Thorac Oncol 2020;15:1449–59. 10.1016/j.jtho.2020.04.026 [DOI] [PubMed] [Google Scholar]
- 39. Pusztai L, Mendoza TR, Reuben JM, et al. Changes in plasma levels of inflammatory Cytokines in response to paclitaxel chemotherapy. Cytokine 2004;25:94–102. 10.1016/j.cyto.2003.10.004 [DOI] [PubMed] [Google Scholar]
- 40. Galluzzi L, Humeau J, Buqué A, et al. Immunostimulation with chemotherapy in the era of immune Checkpoint inhibitors. Nat Rev Clin Oncol 2020;17:725–41. 10.1038/s41571-020-0413-z [DOI] [PubMed] [Google Scholar]
- 41. Altan-Bonnet G, Mukherjee R. Cytokine-mediated communication: a quantitative appraisal of immune complexity. Nat Rev Immunol 2019;19:205–17. 10.1038/s41577-019-0131-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Klement JD, Paschall AV, Redd PS, et al. An Osteopontin/Cd44 immune Checkpoint controls Cd8+ T cell activation and tumor immune evasion. J Clin Invest 2018;128:5549–60. 10.1172/JCI123360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wei J, Marisetty A, Schrand B, et al. Osteopontin mediates glioblastoma-associated macrophage infiltration and is a potential therapeutic target. J Clin Invest 2019;129:137–49. 10.1172/JCI121266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Klement JD, Poschel DB, Lu C, et al. Osteopontin blockade Immunotherapy increases cytotoxic T lymphocyte Lytic activity and suppresses colon tumor progression. Cancers (Basel) 2021;13:1006. 10.3390/cancers13051006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang J, Takahashi K, Takahashi F, et al. Differential Osteopontin expression in lung cancer. Cancer Lett 2001;171:215–22. 10.1016/s0304-3835(01)00607-3 [DOI] [PubMed] [Google Scholar]
- 46. Moorman HR, Poschel D, Klement JD, et al. Osteopontin: A key regulator of tumor progression and Immunomodulation. Cancers (Basel) 2020;12:3379. 10.3390/cancers12113379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Donati V, Boldrini L, Dell’Omodarme M, et al. Osteopontin expression and Prognostic significance in non-small cell lung cancer. Clin Cancer Res 2005;11:6459–65. 10.1158/1078-0432.CCR-05-0541 [DOI] [PubMed] [Google Scholar]
- 48. Carbone F, Grossi F, Bonaventura A, et al. Baseline serum levels of Osteopontin predict clinical response to treatment with Nivolumab in patients with non-small cell lung cancer. Clin Exp Metastasis 2019;36:449–56. 10.1007/s10585-019-09984-z [DOI] [PubMed] [Google Scholar]
- 49. Rud AK, Boye K, Oijordsbakken M, et al. Osteopontin is a Prognostic biomarker in non-small cell lung cancer. BMC Cancer 2013;13:540. 10.1186/1471-2407-13-540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tang W, Jia P, Zuo L, et al. Suppression of Cx3Cl1 by miR-497-5p inhibits cell growth and invasion through Inactivating the ERK/AKT pathway in NSCLC cells. Cell Cycle 2022;21:1697–709. 10.1080/15384101.2022.2067438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li S, Liu S, Xun J, et al. Expression of Cx3Cl1 and Ccl28 in spinal metastases of lung adenocarcinoma and their correlation with clinical features and prognosis. J Healthc Eng 2022;2022:2580419. 10.1155/2022/2580419 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52. Leung JH, Ng B, Lim W-W. Interleukin-11: A potential biomarker and molecular therapeutic target in non-small cell lung cancer. Cells 2022;11:2257. 10.3390/cells11142257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Engblom C, Pfirschke C, Zilionis R, et al. Osteoblasts remotely supply lung tumors with cancer-promoting Siglecf(High) neutrophils. Science 2017;358:eaal5081. 10.1126/science.aal5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Arends R, Guo X, Baverel PG, et al. Association of circulating protein biomarkers with clinical outcomes of Durvalumab in head and neck squamous cell carcinoma. Oncoimmunology 2021;10:1898104. 10.1080/2162402X.2021.1898104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tauriello DVF, Palomo-Ponce S, Stork D, et al. Tgfbeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018;554:538–43. 10.1038/nature25492 [DOI] [PubMed] [Google Scholar]
- 56. Chakravarthy A, Khan L, Bensler NP, et al. TGF-beta-associated extracellular matrix genes link cancer-associated fibroblasts to immune evasion and Immunotherapy failure. Nat Commun 2018;9:4692. 10.1038/s41467-018-06654-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mariathasan S, Turley SJ, Nickles D, et al. Tgfbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544–8. 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bialkowski L, Van der Jeught K, Bevers S, et al. Immune Checkpoint blockade combined with IL-6 and TGF-beta inhibition improves the therapeutic outcome of mRNA-based Immunotherapy. Int J Cancer 2018;143:686–98. 10.1002/ijc.31331 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2023-006788supp001.pdf (947.5KB, pdf)
Data Availability Statement
Data are available on reasonable request.