Abstract
The genetic circuits that allow cancer cells to evade immune killing via epithelial mesenchymal plasticity remain poorly understood. Here, we showed that mesenchymal-like (Mes) KPC3 pancreatic cancer cells were more resistant to cytotoxic T lymphocyte (CTL)–mediated killing than the parental epithelial–like (Epi) cells and used parallel genome-wide CRISPR screens to assess the molecular underpinnings of this difference. Core CTL-evasion genes (such as IFN-γ pathway components) were clearly evident in both types. Moreover, we identified and validated multiple Mes-specific regulators of cytotoxicity, such as Egfr and Mfge8. Both genes were significantly higher expressed in Mes cancer cells, and their depletion sensitized Mes cancer cells to CTL-mediated killing. Notably, Mes cancer cells secreted more Mfge8 to inhibit proliferation of CD8+ T cells and production of IFN-γ and TNFα. Clinically, increased Egfr and Mfge8 expression was correlated with a worse prognosis. Thus, Mes cancer cells use Egfr-mediated intrinsic and Mfge8-mediated extrinsic mechanisms to facilitate immune escape from CD8+ T cells.
Genetic screens elucidate distinct properties of epithelial and mesenchymal pancreatic cancer cells to evade immune killing.
INTRODUCTION
The emergence of immune checkpoint blockade (ICB) therapies [including anti–cytotoxic T lymphocyte antigen 4 (CTLA-4) and anti–programmed death protein 1 or ligand 1 (PD-1/PD-L1)] has led anticancer treatment into a new era, but achieving effective and durable responses is still challenging in most patients with cancer. To harness the vulnerability of tumors capable of evading immune-mediated killing more effectively, additional immune resistance mechanisms are being characterized and exploited (1), and tumor epithelial-mesenchymal transition or plasticity (EMT or EMP) is an emerging mechanism (2–4). During this process, epithelial (Epi) cancer cells lose cell-cell adhesion properties and undergo a morphological transition into mesenchymal-like (Mes) cancer cells. EMT coincides with decreased expression of epithelial markers (such as E-cadherin), gain of mesenchymal marker expression (such as N-cadherin and vimentin), loss of apical-basal polarity, and acquisition of migratory and invasive properties (5). Compared to epithelial cancer cells, mesenchymal cancer cells are more prone to metastasize (6) and are more resistant to chemotherapy and immune-targeted therapy (7–9). EMT is a reversible and highly dynamic multistep process, and the term EMP was recently coined to describe the dynamic and reversible spectrum of phenotypic states between the fully or partially epithelial and fully or partially mesenchymal states occurring during EMT or mesenchymal-epithelial transition (5, 10). EMP has been identified as a critical determinant for cancer cell stemness, metastasis, and chemo- and radiotherapy resistance (11–13) and is emerging as a clinically relevant mechanism of immune evasion (2–4, 9, 14, 15). Clinically, mesenchymal features in patients with cancer show a negative correlation with the response to ICB therapy (9). The mechanisms by which mesenchymal versus epithelial tumors promote an immunosuppressive microenvironment and the identities of executors are not clear.
An elegant study by Dongre et al. (8, 16) demonstrated that EMT contributed to immunosuppression in breast carcinomas. Tumors formed from mesenchymal cell lines were found to be more resistant to immune attack than those formed from epithelial cell lines after the cells were implanted orthotopically into immunocompetent syngeneic mice. One of the underlying mechanisms was that the mesenchymal tumors suppressed infiltration of CD8+ T cells and increased the numbers of CD25+, transcription factor forkhead box protein 3–positive regulatory T cells (Tregs), and M2 macrophages, providing cross-protection to the nearby epithelial cancer cells to prevent immune-mediated elimination. This mechanism was also observed in prostate cancer, lung cancer, and melanoma (17–20). Moreover, in vivo data showed that mesenchymal breast tumors did not respond to anti–CTLA-4 immunotherapy, whereas epithelial tumors responded well (8, 16). Direct ectopic expression of the EMT-inducing transcription factor zinc finger E-box binding homeobox 1 (ZEB1) in melanoma cells triggered resistance, while depletion of ZEB1 improved the response to anti–PD-1 immunotherapy (20). In addition, a number of studies have demonstrated that EMT is associated with inflammatory signals and elevated expression of multiple immune checkpoint molecules (15, 21–24). A recent study revealed that the secretion or expression of several factors [ecto-5′-nucleotidase (CD73) and colony stimulating factor 1] or secreted phosphoprotein 1 by mesenchymal cancer cells regulated macrophages to directly or indirectly suppress immune attack (8). Together, these studies indicate that cancer cells can use EMT to evade immune surveillance.
While the differences in the tumor microenvironment are well studied between epithelial and mesenchymal cells, which genetic factors expressed or suppressed by mesenchymal cells govern these alterations are mostly unclear. We developed an inducible EMT system in pancreatic cancer KPC3 cells that we used to uncover the genetic basis for the decreased sensitivity of mesenchymal cells to CTL-mediated killing. Using parallel genome-wide CRISPR-Cas9 knockout screens in these epithelial and mesenchymal KPC3 cells, our findings revealed a systematic profile of enriched or depleted single-guide RNAs (sgRNAs) that are involved in different signaling pathways. Although core CTL evasion genes [such as interferon gamma receptor 1/2 (Ifngr1/2), Janus kinase 1/2 (Jak1/2), and protein tyrosine phosphatase nonreceptor type 11 (Ptpn11)] were clearly evident in both types, Mes cancer cells showed less sensitivity to interferon-γ (IFN-γ) signaling. Furthermore, upon validating several candidates, we identified two genes, epidermal growth factor receptor (Egfr) and milk fat globule-EGF factor 8 protein (Mfge8, also known as lactadherin), that particularly stood out in the CRISPR screens combined with RNA transcriptional profiling as critical determinants for CD8+ T cell–mediated Mes cancer cell killings. Functional follow-up studies combined with bioinformatic analysis of clinical cancer datasets confirmed the unique and distinct roles of these genes in EMT-mediated resistance to anticancer immunity.
RESULTS
Mes pancreatic cancer cells are more resistant to CD8+ T cell–mediated killing
To investigate the role of tumor EMP in CD8+ T cell cytotoxicity, we generated two types of tumor cells with Epi or Mes features through an inducible EMT system (Fig. 1A). The parental KPC3 pancreatic cancer cell line, derived from a female KrasG12D/+; Trp53R172H/+; Pdx-1-Cre mouse used as a pancreatic ductal adenocarcinoma model (25), was stably transduced with green fluorescent protein (GFP) and ovalbumin (OVA); this cell line can be specifically killed by matched OT-I CD8+ T cells, which are isolated form OT-I T cell receptor transgenic mice that contain transgenic inserts for mouse Tcra-V2 and Tcrb-V5 genes and recognize major histocompatibility complex (MHC) class I presented OVA antigen on cancer cells. KPC3 cells underwent EMT when exposed to transforming growth factor–β (TGF-β), a major driver of EMT, and in order to avoid the influence of TGF-β on T cells while maintaining stable Mes characteristics (26, 27), we transduced a doxycycline (Dox)–induced constitutively kinase-active TGF-β type I receptor, also termed ALK5 CA (threonine-204 mutated to aspartic acid), into KPC3 cells (28, 29), which were then named KPC3-OVA :: Tet-On ALK5 CA cells. As expected from (30), after treatment with Dox, only the kinase-active variant ALK5 CA, but not the kinase-dead mutant ALK5 DN (lysine-232 mutated to arginine), activated phosphorylated Smad2 (p-Smad2), and this activation was blocked when treated with the selective ALK5 kinase inhibitor SB431542 (fig. S1A). Ectopic expression of ALK5 CA was found insufficient to stimulate prominent EMT in a short period of time (fig. S1, B and C), which is why we first treated cells with TGF-β for 2 days (to boost a prominent EMT), then removed TGF-β, and thereafter added Dox to activate ALK5 CA to maintain the Mes state in the absence of exogenous TGF-β (Fig. 1A).
Fig. 1. Mes pancreatic cancer cells are more resistant to CD8+ T cell–mediated killing.
(A) Schematic representation of inducible constitutively active ALK5 (ALK5 CA) using the Tet-On system. On the basis of it, KPC3-OVA cancer cells with an Epi and Mes status were established. (B) Western blot analysis of the protein expression of EMT markers and p-Smad2 in the epithelial or mesenchymal state (Epi or Mes). (C) Reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis of the mRNA expression of the TGF-β target gene Pai-1 and EMT-associated marker genes. The results are quantified as mean ± SD by an unpaired two-sided Student’s t test (**P < 0.01 and ***P < 0.001). (D) Heatmap of the normalized mRNA expression of EMT marker genes obtained from RNA-seq analysis of Mes and Epi cancer cells. The significantly DEGs are listed; the cutoff was |fold change| > 1.5, P adjusted < 0.001. (E) Gene set enrichment analysis (GSEA) plot of EMT signature. The P values were calculated on the basis of a hypergeometric distribution. The normalized enrichment scores (NES) indicate enrichment in the corresponding function. (F to H) Epi and Mes KPC-OVA cells were challenged with increasing ratios of OT-I CD8+ T cells. E:T, effector T cell:target cancer cell (F) Representative live images of green fluorescence at the 48-hour time point (n = 3). (G) Quantification of the intensity images as shown in (F) (n = 3). Statistical significance was determined using two-way analysis of variance (ANOVA) with Sidak’s multiple comparisons test (***P < 0.001). (H) Cancer cells were fixed and stained with crystal violet after 3 days of coculture with OT-I CD8+ T cells.
The occurrence of EMT and maintenance of the mesenchymal state upon TGF-β challenge and Dox-mediated ALK5 expression were validated by measuring the protein and RNA expression of epithelial and mesenchymal markers. Immunofluorescence staining showed decreased E-cadherin expression and increased stress filamentous (F)–actin fibers in Mes cancer cells (fig. S1D), which were accompanied by protein and mRNA expression changes in EMT markers [including E-cadherin, Snail family transcriptional repressor 1 (Snail) and Zeb1/2] (Fig. 1, B and C). Furthermore, the mesenchymal phenotype could be maintained for more than 16 days; this was more than sufficient for our following experiments conducted on a time scale of 12 days (fig. S1, B and C). To further investigate the transcriptional differences between Epi and Mes cancer cells, we performed mRNA sequencing. Principal components analysis (PCA) displayed clear separation between the two types of cancer cells (fig. S1E); the levels of epithelial marker genes were significantly decreased, while those of mesenchymal marker genes were increased in Mes compared to Epi cancer cells (Fig. 1D). Gene set enrichment analysis (GSEA) demonstrated that Mes cancer cell–enriched genes positively correlated with up-regulated EMT gene sets (Fig. 1E and fig. S1F) and negatively correlated with down-regulated EMT gene sets (fig. S1G). The proliferation of these two types of cells, as measured by either MTS cell viability assay or confluency measured with IncuCyte live cell imager, was not significantly different (fig. S1, H and I). Since this approach to convert Epi state into Mes state was driven by TGF-β signaling, we detected the concentration of TGF-β secreted by Epi or Mes cancer cells using a HepG2-Smad3–dependent transcriptional CAGA-GFP reporter (30); analysis of conditioned medium from both cell types revealed no differences in either the latent or active form of TGF-β (fig. S1, J and K). This ruled out the influence of differential TGF-β expression on CD8+ T cell–mediated cancer cell killing (31). Thus, we successfully established an in vitro pancreatic cancer model, which could be used to explore what differences in the EMP state affect CTL killing activity.
To test differential sensitivity, we cocultured Epi or Mes KPC3-OVA : GFP cancer cells with increasing numbers of OT-I CD8+ T cells and monitored the percentage of surviving KPC3 cells. Through live imaging of GFP (Fig. 1, F and G) or crystal violet staining (Fig. 1H), we found that Mes cancer cells were more resistant to CD8+ T cell–mediated killing than Epi cancer cells in cytotoxicity assays. Those results are in line with, and extend previous results that the acquisition of Mes features confer cancer cells more resistant to CD8+ T cell cytotoxicity (4).
Parallel genome-wide screens identify regulators of EMT-mediated resistance to CD8+ T cell cytotoxicity in KPC3 cancer cells
To identify genes modulating EMT-mediated immune evasion in KPC3 cancer cells in an unbiased manner, we performed parallel genome-scale screens (Fig. 2A). Epi and Mes cancer cells were individually infected with the Brie CRISPR Knockout Library (32) and challenged with two rounds of OT-I CD8+ T cells, each resulting in a 50% survival rate, allowing for the identification of both immune resistance– and sensitization-related genes in those two types of cancer cells. sgRNAs were then recovered from the surviving cells and analyzed by deep sequencing (table S1). The results showed sufficient library representation, and strong correlations between replicates were observed (fig. S2A). Moreover, enriched sgRNAs targeting the IFN-γ signaling pathway [such as Ifngr1/2, Jak1/2, and signal transducer and activator of transcription 1 (Stat1)] and depleted sgRNAs involved in the tumor necrosis factor (TNF) pathway were significantly identified in both the Epi and Mes screen (fig. S2, B and D); these pathways are well-characterized and important to T cell killing activity (33–40). Together, our screening results exhibited robustness and effectiveness, convincing us that the screens were performed in such a way that the results could be compared between these two cell types.
Fig. 2. Parallel genome-wide screens identify regulators of EMT-mediated resistance to CD8+ T cell cytotoxicity in KPC3 cancer cells.
(A) Schematic overview of the genome-wide parallel CRISPR-Cas9 knockout screens of Epi and Mes KPC3-OVA cancer cells challenged with OT-I CD8+ T cells. (B and C) Robust ranking aggregation (RRA) score and delta log fold change (Delta LFC) plot showing specifically enriched genes (red) or depleted genes (green) in Mes (B) and Epi cells (C). RRA scores were −log10-transformed from MAGeCK RRA scores. Delta LFCs were log2-transformed from the difference of the fold change between the Mes and Epi screens. The dashed line indicates the cutoff of RRA score at 3 or −3 (shown as |RRA| < 3) and the cutoff of Delta LFC at log2(1.5) or −log2(1.5) [shown as diff (fold change) > 1.5]. The P value of sgRNA from the MAGeck algorithm < 0.001 was used as the cutoff. (D and E) GO term GSEA (D) and STRING network analysis (E) showing biological processes or gene interactions of the specifically enriched and depleted genes in either the Epi or Mes cancer cells from (B) and (C). GO:BP, Biological Processes of Gene Ontology term; STRING, Search Tool for the Retrieval of Interacting Genes/Proteins.
We next investigated the differences of screening results in Epi and Mes types, respectively. By plotting robust ranking aggregation (RRA) scores in the Mes screen, the fold enrichment in Mes versus Epi (already normalized to the respective untreated control group), we uncovered numerous sgRNAs that were specifically gained or lost in Mes screens (Fig. 2B). Similar analysis methods were also applied in Epi screens (Fig. 2C). To further explore the gene networks involved in regulating cancer-intrinsic immune evasion in these two different states, we converged all specific sgRNAs of either Epi or Mes status and performed GSEA on Gene Ontology (GO) (Fig. 2D) or WikiPathways sets (fig. S2C). These analyses demonstrated the diversity of signaling pathways involved in evading CTL-mediated killing by these two types of cancer cells; IFN-γ signaling, mitochondrial electron transport, and adenosine triphosphate or oxidation metabolic process were highly enriched in Epi cells, whereas the Egfr signaling pathway and interleukin-5 (IL-5) and IL-2 signaling pathways were highly depleted in Mes cells (Fig. 2D and fig. S2C). Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) network analysis further confirmed that the more depleted sgRNAs in Mes cells were significantly connected with the Egfr signaling pathway [false discovery rate (FDR) = 0.00571], and the more enriched sgRNAs in Epi cells were significantly related to the IFN-γ signaling pathway (FDR = 8.8 × 10−05) (Fig. 2E). In addition, some particularly depleted sgRNAs, such as sgRNAs targeting Egfr, phosphatidylinositol-4-phosphate 5-kinase type 1 alpha (Pip5k1a), Mfge8, calcineurin binding protein 1 (Cabin1), PPFIA binding protein 2 (Ppfibp2), aldehyde dehydrogenase 18 family member a1(Aldh18a1), and GATA zinc finger domain containing 2a (Gatad2a), represented significant changes in Mes cancer cells, while Epi cancer cells were not significantly affected by their loss (fig. S2, D and E). These results suggested that Mes cancer cells gain resistance by using multiple pathways, such as increasing the activity of the Egfr signaling pathway and lowering the activity of the IFN-γ signaling pathway.
To validate the screening results, we focused on sgRNAs that were specifically depleted in Mes cancer cells, as we hypothesized that if these genes were genetically ablated, Mes cancer cells would become more sensitized to CTL-mediated killing, similar to Epi cancer cells. Epi and Mes cancer cells were transduced with a control sgRNA (sgCtrl) and individual candidate sgRNAs, and the effects of specific gene depletion on the responses in a CD8+ T cell cytotoxicity assay and competition assay were determined. Pip5k1a, Aldh18a1, Ppfibp2, Egfr, Gata2a, or Mfge8 ablation all sensitized Mes cancer cells to killing by CTLs (fig. S3A). Furthermore, these genes were much more essential for Mes cancer cells to evade CTL killing than for Epi cancer cells, as the Mes cells exhibited significantly higher fold changes of cell death after gene ablation (fig. S3B). To further validate our results, we performed competition assays in which two groups of cancer cells were labeled with different fluorophores and then mixed at 1:1 and subsequently challenged with CD8+ T cells (fig. S3C). We first confirmed the competition difference between Epi and Mes control cells (fig. S3, D and E); as expected, flow cytometry analysis showed that more Mes control cells than Epi control cells survived. Next, we compared CellTrace Violet (CTV)–labeled Mes control cancer cells (sgCtrl) with cancer cells transduced with individual sgRNA targeting indicated genes. Flow cytometry analysis validated the candidates that we tested (fig. S3F). Together, our approach identified genes specifically required for CTL-mediated killing of mesenchymal KPC3 cancer cells.
Down-regulation of Ifngr2 inhibits Mes cancer cells from sensing IFN-γ signaling
Given that our Epi and Mes cancer cells are of the same origin, the differences in susceptibility to immune attack should be a consequence of altered gene expression. Therefore, we integrated the results of our genome-wide CRISPR-Cas9 screens with those of the transcriptional profiles of the Epi and Mes states.
Upon analysis of RNA sequencing (RNA-seq) data, we found large transcriptional differences between Epi and Mes cancer cells; 1670 genes were significantly up-regulated and 1204 genes were down-regulated in Mes cancer cells compared to Epi cancer cells (fig. S4A). In addition to the activated EMT signature, the interferon signatures were suppressed in the Mes state (fig. S4B). Although sgRNA-targeting IFN-γ–related genes were enriched in either Epi or Mes state (fig. S2B), these genes were more depleted in the Epi CRISPR screens (Fig. 2, D and E, and fig. S2C), arguing that Epi and Mes KPC3 cells respond differently to IFN-γ signaling.
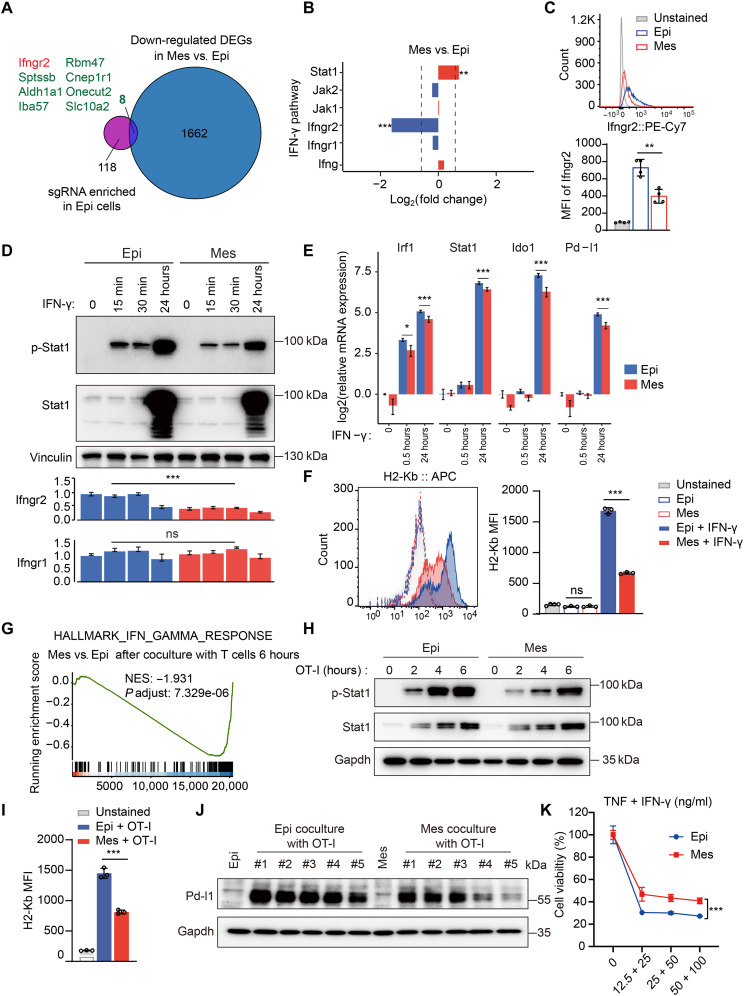
When we overlapped the down-regulated differentially expressed genes (DEGs; the significantly DEGs were cut off at |fold change| >1.5, P adjusted < 0.001) (1670) in Mes versus Epi cancer cells and the sgRNAs specifically enriched in Epi cells (126), many interesting candidates were identified (Fig. 3A). We focused on Ifngr2 because it associates with Ifngr1 to form the heterodimeric IFN-γ receptor, and it was the only down-regulated gene in the IFN-γ pathway (Fig. 3B; the |fold change| >1.5, P adjusted < 0.001 was used as the cutoff). This finding prompted us to suspect that the loss of Ifngr2 in Mes cancer cells disrupted the IFN-γ response. We first confirmed that the membrane protein level of Ifngr2 was robustly decreased in Mes cancer cells (Fig. 3C). Challenging Epi and Mes cancer cells with IFN-γ revealed that the level of Stat1 phosphorylation was lower in Mes cells than in Epi cells (Fig. 3D). This was further confirmed by examining the expression levels of IFN-γ signaling target genes [such as interferon regulatory factor 1 (Irf1), Stat1, indoleamine 2,3-dioxygenase 1 (Ido1), and Pd-l1] in response to IFN-γ, as the levels were lower in Mes cells than Epi cells (Fig. 3E). Because of the loss of IFN-γ sensing, the protein levels of MHC class I (H2Kb) and Pd-l1 were also decreased in Mes cells (Fig. 3F and fig. S4C). Together, these results showed an attenuated IFN-γ response was present in Mes cancer cells compared to Epi cells, probably mediated by down-regulation of Ifngr2.
Fig. 3. Down-regulation of Ifngr2 inhibits Mes cancer cells from sensing IFN-γ signaling.
(A) RNA-seq data analysis using Venn diagrams revealed the overlap of down-regulated DEGs (1670) in Mes versus Epi cancer cells and specifically enriched sgRNAs (126) in Epi cancer cells. (B) RNA-seq analysis of IFN-γ signaling components in Mes versus Epi cancer cells. (C) Flow cytometry analysis of Ifngr2 expression in Epi or Mes cancer cells (n = 4). (D) Western blot analysis of p-Stat1 and Stat1 (top part) and RT-qPCR analysis of Ifngr2 and Ifngr1 mRNA expression (bottom part) in Epi or Mes cancer cells after IFN-γ treatment (n = 3). (E) RT-qPCR analysis of Irf1, stat1, Ido1, and Pd-l1 mRNA expression in Epi or Mes cancer cells after IFN-γ treatment. (F) Flow cytometry analysis of H2-Kb in Epi or Mes cancer cells after 24-hour treatment with IFN-γ (n = 3). (G) GSEA showing a decreased IFN-γ response in Mes versus Epi cancer cells after OT-I T cell coculture. (H) Western blot analysis of p-Stat1 and Stat1 expression in Epi or Mes cancer cells after OT-I CD8+ T cell treatment. (I) Flow cytometry analysis of H2-Kb in Epi or Mes cancer cells co-cultured with OT-I CD8+ T cells. (J) Western blot analysis of Pd-l1 in Epi or Mes cancer cells (un)treated with OT-I CD8+ T cells (n = 5). (K) Cell viability analysis of Epi or Mes cancer cells treated with (different doses) TNF and IFN-γ (n = 3). Two-sided Student’s t test in (C), (D), (E), (F), and (I) and two-way ANOVA with Sidak’s multiple comparisons test in (K) (*P < 0.05, **P < 0.01, and ***P < 0.001). For all experiments, bars represent mean ± SD. ns, not significant. MFI, mean fluorescence intensity.
Next, we examined whether this differential sensitivity to IFN-γ also occurred in Mes versus Epi cancer cells after coculture with CD8+ T cells. We cocultured Epi and Mes cancer cells with OT-I CD8+ T cells for 0, 6, or 24 hours and performed RNA-seq analysis. PCA showed clear separation of the Mes and Epi cancer cells after OT-I T cell treatment (fig. S4D). The IFN-γ signaling responses were significantly attenuated in Mes cancer cells compared to Epi cancer cells after T cell treatment (Fig. 3G and fig. S4E). Consistent with the IFN-γ stimulation results, Mes cancer cells had reduced Stat1 activation (Fig. 3H) and lower levels of H2kb and Pd-l1 responsiveness after T cell treatment compared to Epi cancer cells (Fig. 3, I and J). Furthermore, we used the coculture system to detect the production of cytokines, mainly including IFN-γ and TNFα, which are established effectors of CD8+ T cell anticancer function (36). We found that OT-I T cells cocultured with Mes cancer cells secreted less IFN-γ after 24 hours and less TNFα during 2 to 16 hours (fig. S4F), which indicated that compared to Epi cancer cells, Mes cancer cells partially blocked the activity of T cells. Given the direct antitumor functions of IFN-γ and TNFα, we used them alone or in combination to treat Epi or Mes cancer cells. Unexpectedly, both Mes and Epi cancer cells were extremely resistant to single cytokine IFN-γ or TNFα treatment, as well as treatment with TNF Superfamily Member 10 (TRAIL), another ligand that induces apoptosis (fig. S4G). However, the combination of IFN-γ and TNF killed both Epi and Mes cancer cells, and Mes cancer cells were significantly more resistant than Epi cancer cells (Fig. 3K). This result was also confirmed in a competitive T cell killing assay using CTV staining (fig. S4H). These results are consistent with the previously proposed synergistic effect of TNFα and IFN-γ in triggering cancer cell death (26–28). Together, these results demonstrated that Mes cancer cells escaped T cell cytotoxicity in part by losing sensitivity to IFN-γ signaling via down-regulation of IFNGR2.
Mes cancer cells up-regulate Egfr to evade CD8+ T cell–mediated killing
Further combined analysis of RNA-seq data with our screen results revealed three significantly up-regulated genes, Egfr, Mfge8, and Cabin1, by overlapping the up-regulated DEGs (1204) in Mes versus Epi cancer cells and sgRNAs specifically depleted in Mes cancer cells (Fig. 4A; the significantly DEGs were cut off at |fold change| > 1.5, P adjusted < 0.001). This made us speculate that these three genes are the executors of the acquisition of resistance specifically in Mes cancer cells.
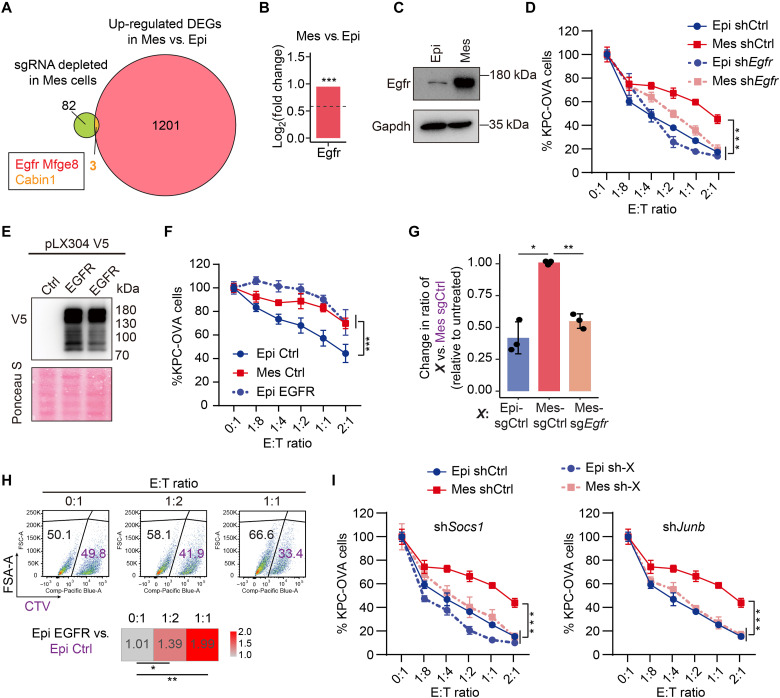
Fig. 4. Mes cancer cells up-regulate Egfr to evade CD8+ T cell–mediated killing.
(A) RNA-seq data analysis using Venn diagram plots revealed the overlap of up-regulated DEGs (1204) in Mes versus Epi cells and specifically depleted sgRNAs (85) in Mes cells. (B and C) RNA-seq analysis (B) and Western blot analysis (C) of Egfr expression in Epi and Mes cells. (D) Epi or Mes cells expressing shCtrl) or shEgfr were challenged with OT-I CD8+ T cells (two biological replicates, each with two technical replicates). (E) Western blot analysis of EGFR-V5 expression in Epi cells. (F) Epi cells expressing EGFR (Epi EGFR) or control (Epi Ctrl) and control Mes cells (Mes Ctrl) were challenged with OT-I CD8+ T cells (three biological replicates, each with two technical replicates). (G) Competition assays performed with Mes cancer cells expressing sgCtrl or sgEgfr or Epi sgCtrl cells treated with OT-I CD8+ T cells, analyzed by flow cytometry (see fig. S5C). The change in the ratio between cancer cells expressing the indicated sgCtrl or sgEgfr (black) versus Mes sgCtrl cancer cells (purple) is presented relative to cancer cells without T cell challenge (n = 3). (H) Competition assays with Epi cells expressing Egfr (Epi Egfr) or a control vector (Epi Ctrl), treated with OT-I CD8+ T cells and analyzed by flow cytometry. The ratio change of Epi Egfr versus Epi sgCtrl was calculated relative to cancer cells without T cells challenge. (I) Epi or Mes cells expressing shCtrl or indicated genes (sh-X) were challenged with OT-I CD8+ T cells (two biological replicates, each with two technical replicates). Two-way ANOVA with Sidak’s multiple comparisons test in (D), (F), and (I) and a two-sided Student’s t test in (G) and (H) (*P < 0.05, **P < 0.01, ***P < 0.001).
Egfr was indeed highly expressed at the mRNA and protein levels in Mes cancer cells compared with Epi cancer cells (Fig. 4, B and C), which was also observed in the coculture system containing cancer cells and OT-I T cells (fig. S5A). Consistent with the knockout results (fig. S3, A and B), Egfr knockdown via short hairpin RNA (fig. S5B) greatly augmented the sensitivity of Mes cancer cells to CD8+ T cell–mediated killing but not that of Epi cancer cells (Fig. 4D). This suggests a potential role for Egfr in the ability of Mes cancer cells to evade CTL-mediated killing.
Next, we explored whether overexpressing EGFR alone in Epi cancer cells could make these cells more resistant to CTL-mediated killing. Overexpression of EGFR (Fig. 4E) conferred a more resistant phenotype to Epi cancer cells in the context of CD8+ T cell–mediated killing (Fig. 4F). We used competition assays to evaluate whether up-regulation of EGFR is a cancer-intrinsic mechanism for evading killing by T cells. Flow cytometry analysis showed that both genetic ablation and mRNA knockdown of Egfr sensitized Mes cancer cells to CD8+ T cell–mediated killing (Fig. 4G and figs. S3F and S5C). In contrast, also in this assay, ectopic expression of EGFR enhanced the immune resistance of Epi cancer cells (Fig. 4H).
On the basis of STRING network analysis of depleted sgRNAs in Mes cancer cells (Fig. 2E), we suspected that other related genes in the Egfr signaling pathway may also participate in the evasion of CTL killing activity. We selected two genes. One was suppressor of cytokine signaling 1 (Socs1), which is recruited by the Egfr C-tail to block Stat activation (41), and the other was Junb, a downstream adapter in Egfr signaling (42). Similar to the results for Egfr, knockdown of Socs1 or JunB proto-oncogene, AP-1 transcription factor subunit (Junb) sensitized Mes cancer cells to CD8+ T cells (Fig. 4I). These results indicate that engagement of the Egfr signaling pathway is a cancer-intrinsic immune evasion mechanism that is used by Mes cancer cells.
Cancer cell–secreted Mfge8 directly inhibits the anticancer activity of CD8+ T cells
Another candidate that we explored functionally was Mfge8. It has been reported that Mfge8 is a secreted glycoprotein derived from human or bovine milk (43) that promotes mesenchymal features in melanoma cells by inducing twist family bHLH transcription factor 1 (Twist) and Snail (44). In addition, it plays a critical role in ameliorating inflammation, such as promoting the engulfment of apoptotic cells by immune cells (45, 46) and inhibiting Foxp3+ Tregs (47, 48). These characteristics of Mfge8 prompted us to hypothesize that it can be secreted by Mes cancer cells and then directly act on immune cells to suppress their killing activity against cancer cells.
We first validated both at the mRNA and protein level that Mes cancer cells expressed higher levels of Mfge8 than Epi cancer cells (Fig. 5, A and B). Mfge8 expression was increased in both cell types after coculture with T cells but maintained higher in Mes cancer cells than in Epi cancer cells (Fig. 5B). Furthermore, through harvesting conditioned medium from Epi or Mes tumor cells, we found that the KPC3-OVA cancer cells secreted Mfge8, which could be suppressed by knocking out Mfge8 (Fig. 5C). Consistently, Mes cancer cells secreted much more Mfge8 than Epi cancer cells (Fig. 5C). We also checked the level of Mfge8 in the coculture system and found that the secreted Mfge8 level was increased after T cell treatment and persistently higher in Mes cancer cells than in Epi cancer cells (fig. S6A). These findings and previous data demonstrated that genetic ablation of Mfge8 sensitized Mes cancer cells to T cell–mediated killing (fig. S3, A and B), which raised the interesting possibility that Mes cancer cells might secrete Mfge8 to impair T cell function. To test this hypothesis, we collected conditioned medium from different states of cancer cells and added those medium into the coculture system (Fig. 5D). When the conditioned medium from Mes cancer cells (Mes-conditioned medium) was added to the Epi cancer cell and T cell coculture system, the Epi cancer cells exhibited better survival than those in the normal culture medium treatment group (Fig. 5E).
Fig. 5. Cancer cell–secreted Mfge8 directly inhibits the anticancer activity of CD8+ T cells.
(A) RNA-seq analysis of mfge8 mRNA expression in Epi and Mes KPC3-OVA cells. ***P adjusted < 0.001, fold change > 1.5. (B) Western blot analysis of Mfge8 in Epi and Mes cells cultured alone or with OT-I cells. (C) Enzyme-linked immunosorbent assay (ELISA) of Mfge8 expression in the conditioned medium (CM) of Epi or Mes cells expressing sgCtrl or sgMfge8 (n = 3). (D) Schematic of CM exchange assay. (E) CM derived from Epi or Mes cancer cells was added to the Epi or Mes cancer cells and OT-I killing system. (F) CM from the Mes cancer cells expressing sgRNAs targeting control (sgCtrl) or Mfge8 (sgMfge8) was added into the Epi cancer cells and OT-I killing system. (G) CM from cancer cells ectopically expressing control vector (Ctrl) or Mfge8 (Mfge8 OE) was added into the Epi cancer cells and OT-I killing system. N = 2 independent donors, each with two replicates. (H) ELISA detection of IFN-γ and TNF secreted by OT-I cells cultured with CM from Epi or Mes cancer cells expressing sgCtrl or sgMfge8. N = 3 independent donors, each with two replicates. (I) Proliferation quantification of OT-I cells using CTV prestaining and anti-CD3 activation after treatment with different CM sources. N = 3 independent donors, each with two replicates. (J) ELISA for IFN-γ and TNF secretion by OT-I cells cultured with CM from epi Ctrl or Mfge8 OE cells. N = 3 independent donors. (K) OT-I CD8+ T cell proliferation quantification (CTV prestained, anti-CD3 activated) and treated with CM from cells expressing a control vector (Ctrl) or Egfr (Egfr OE). N = 2 independent donors, each with two replicates. Two-sided Student’s t test in (C), (H), (I), and (K) or two-way ANOVA with Sidak’s multiple comparisons test in (E) to (G) and (J). (*P < 0.05, **P < 0.01, and ***P < 0.001).
Using two complementary approaches, a conditioned medium exchange approach was then used to test the effect of secreted Mfge8. We adjusted the concentration of Mfge8 in the supernatant by removing Mfge8 through genetic depletion of Mfge8 (sgMfge8) or by increasing the Mfge8 level through ectopic expression of Mfge8 (Mfge8 OE). After removal of Mfge8 from the Mes cancer cell–derived supernatant (Fig. 5C), T cells were more efficient in killing Epi cancer cells as compared to the intact supernatant (Fig. 5F). On the other hand, after increasing the Mfege8 level in the supernatant (fig. S6B), T cells were less efficient in killing Epi cancer cells (Fig. 5G). To further dissect whether cancer cell–secreted Mfge8 directly affected the function of CD8+ T cells, we examined its effect on cytokine (IFN-γ and TNFα) secretion and proliferation of CD8+ T cells. Compared with conditioned medium from Epi control cells, conditioned medium from Mes control cells inhibited the secretion of both TNFα and IFN-γ by CD8+ T cells, which were found to be rescued after removing Mfge8 especially in Mes cancer cells (Fig. 5H). Consistently, the downstream activation of p-Stat1 in cancer cells was increased after genetic ablation of Mfge8, suggesting that the level of IFN-γ secretion by CD8+ T cells was indeed affected by Mfge8 and altered their killing efficiency (fig. S6C). In addition, we used CTV staining to monitor the division of T cells by assessing dye dilution and found that the proliferation of T cells was suppressed by conditioned medium from cancer cells; conditioned medium from Mes cancer cells showed the strongest inhibitory effect compared to either normal medium or conditioned medium from Epi cancer cells, and this difference was partially eliminated by removing Mfge8 (Fig. 5I). In contrast, after treatment with Mfge8-supplemented conditioned medium (Mfge8 OE supernatant), the secretion of TNFα and IFN-γ by CD8+ T cells and the proliferation of T cells were further inhibited (Fig. 5, J and K). These results are in line with previous reports showing that Mfge8 promoted cancer growth by suppressing anticancer immune responses in a mouse model (47) and extend that observation by indicating that cancer cell–secreted Mfge8 directly inhibits CD8+ T cells by limiting their cytokine secretion and proliferation.
High EGFR and MFGE8 expression is associated with a poor prognosis
The above mechanistic data demonstrated that the up-regulation of Egfr and Mfge8 in Mes KPC3 pancreatic cancer cells rendered them more resistant to CD8+ T cell–mediated killing. As the fourth leading cause of cancer-related death, pancreatic adenocarcinoma (PAAD) is quite refractory to immunotherapy (49, 50). We therefore investigated whether pancreatic cancer patients with high EGFR and MFGE8 expression would have a relatively poor prognosis.
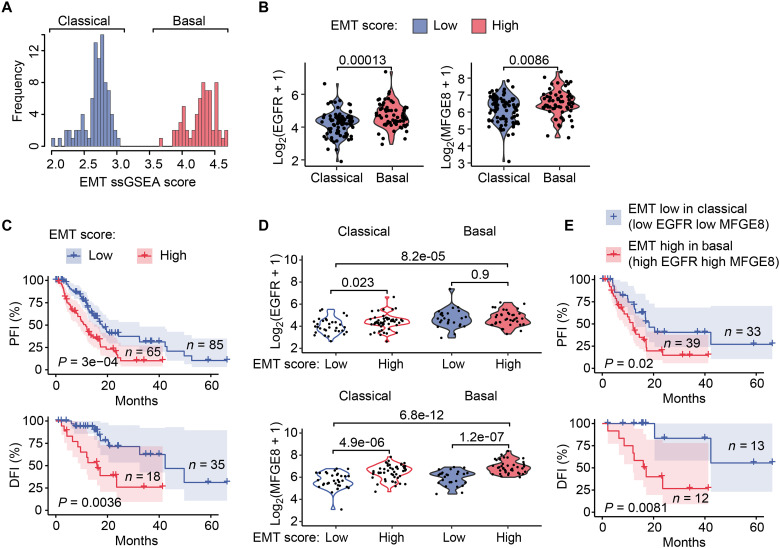
Given that bulk RNA-seq data contain other noncancer cells, especially stromal cells, this inclusion increases the mesenchymal score calculation, which may affect the EMT score (51, 52). We therefore used a deconvolution method developed by Tyler and Tirosh (52), which yielded a series of EMT signatures for different cancer types/subtypes that robustly decoupled EMT from stromal profiles to calculate the “real” EMT score of cancer samples in The Cancer Genome Atlas (TCGA)–PAAD cohort. Using this method, we successfully separated PAAD samples into two subtypes based on the EMT score (Fig. 6A), the “classical” and “basal” subtypes, which were consistent with previously identified molecular characterizations (53–55). The classical subtype exhibits an epithelial state and is more frequently resectable with a higher level of differentiation, and the basal subtype exhibits a quasi-mesenchymal or squamous state with worse clinical outcome and loss of differentiation (55, 56). Within each subtype, EMT scores were also well correlated with the expression of EMT signature genes (fig. S7A). We found that EGFR, as well as MFGE8, had higher expression levels in EMT-high cancers (with a high EMT score) than in EMT-low cancers (with a low EMT score) (Fig. 6B). Notably, we did not observe a difference in IFNGR1 or IFNGR2 (fig. S7B). Therefore, we focused on EGFR and MFGE8 for the following analysis, as they were significantly positively associated with EMT scores in the TCGA-PAAD cohort (fig. S7D). Consistent with previous reports (9, 52), we observed that pancreatic cancer patients with a high EMT score had a significantly worse progression-free interval (PFI) or disease-free interval (DFI) than patients with a low EMT score (Fig. 6C). A similar trend was also seen in an overall survival analysis (fig. S7C).
Fig. 6. High EGFR and MFGE8 expression is associated with a poor prognosis.
(A) The distribution of EMT scores for patients with pancreatic cancer from the TCGA (TCGA-PAAD) calculated by the single-sample GSEA (ssGSEA) method using gene set variation analysis (GVSA). Samples are annotated with subtype information (“classic” and “basic” subtypes). (B) The mRNA expression [log2(TPM +1)] of EGFR or MFGE8 in TCGA-PAAD cancer patients grouped by EMT score and annotated by subtype. Statistical significance was computed by the Wilcoxon test, P values were labeled. (C) Kaplan-Meier PFI or DFI analysis of TCGA-PAAD patients divided into “EMT-low” and “EMT-high” groups based on the EMT score in (A). The P value was computed by the log-rank test. (D) The mRNA expression [log2(TPM +1)] of EGFR or MFGE8 in the two TCGA-PAAD subtypes (classical and basal), which were each further subdivided into EMT-low and EMT-high groups respectively. EMT-low was defined as a z-score < 0 for the EMT score, while EMT-high was defined as a z-score > 0 for the EMT score. Statistical significance was computed by the Wilcoxon test, P values were labeled. (E) Kaplan-Meier PFI or DFI analysis of TCGA-PAAD patients in the EMT-low group for the classical subtype versus those in the EMT-high group for the basal subtype. The P value was computed by the log-rank test.
To further investigate the roles of EGFR and MFGE8, we subdivided the patients into EMT-high and EMT-low groups for each subtype based on the EMT score (fig. S7A). In either the classical or basal subtype, MFGE8 was expressed at higher levels in the EMT-high group than EMT-low group (Fig. 6D). Similarly, EGFR was already highly expressed in patients with the classical subtype with high EMT score (Fig. 6D). The expression of EFGR and MFEGE8 showed the greatest differences between the classical subtype with a low EMT score and the basal subtype with a high EMT score (Fig. 6D). We further used these two groups to perform survival analysis and found that patients with pancreatic cancer expressing low levels of EGFR and MFGE8 had a significantly improved PFI and DFI compared with cancer patients with high levels of EGFR and MFGE8 (Fig. 6E). Overall, the clinical analysis demonstrated that the expression of EGFR and MFGE8 was up-regulated in pancreatic cancer patients with Mes features and was associated with a poor clinical prognosis.
DISCUSSION
Over the past few years, many efforts have focused on identifying the mechanisms that render cancers unresponsive to ICB therapy or unable to achieve durable responses (57–62). These mechanisms are generally divided into two categories: (i) cancer cell–intrinsic mechanisms, such as loss of tumor antigens or antigen presentation and IFN-γ sensing deficiency, and (ii) cancer cell–extrinsic mechanisms, which promote an immunosuppressive microenvironment, including reductions in the function or number of CD8+ T cells, an increase in the number of Tregs or M2-like tumor–associated macrophages, and the production of immunosuppressive cytokines. In addition to these elucidated mechanisms, other mechanisms must exist to explain many cases for which the cause of insensitivity remains unknown (59, 63–71). Our results are not only consistent with previous reports but also expand on the proposed roles for EMP as an important mechanism of immune evasion (4). We demonstrated that EMT in KPC3 pancreatic cancer cells mediated direct resistance to CD8+ T cell cytotoxicity in both cytotoxicity or competition assays and conditioned medium exchange assays, indicating that cancer cell–intrinsic factors contribute to the increased resistance. To understand precisely how Mes cancers exert immunosuppressive effects, we performed parallel genome-wide CRISPR-Cas9 screens using an induced EMT system to identify genes whose inactivation or activation specifically sensitized Mes cancer cells to CD8+ T cells but not Epi cancer cells.
Although multiple genome-wide CRISPR-Cas9 screens were performed with a cancer cell–immune cell coculture system to identify sensitizers or resistors to immune attack (35, 36, 72–74), those screens did not account for the intrinsic resistance caused by cellular plasticity. Many reports, as well as our data, have demonstrated an important role for EMP and EMT in sensitivity to CTLs, and our work defines several pathways by which mesenchymal cells downplay their response. Identifying the critical determinants of EMT-mediated immune evasion is particularly important, as most human primary tumors are heterogeneous with diverse EMP states, and the underlying mechanisms would also apply to other cancer types with active EMT, highlighting potential therapeutic targets to enhance ICB therapy.
To overcome the genetic differences brought by using the respective epithelial and mesenchymal cell line, we made use of the ability of a Dox-inducible ALK5 CA system to induce a robust EMT response, allowing us to screen in the same genetic background. Using this approach, we identified a core conserved set of genes and pathways for each cancer status, with down-regulation of IFN-γ pathway, EGFR signaling, and secretion of MFGE8 as key drivers of immune resistance in mesenchymal cancer cells.
Previous reports found that loss of the IFN-γ pathway is a common immunotherapy resistance mechanism and that JAK1/JAK2 mutation usually occurs in ICB nonresponders (72, 75–77). Extending these notions, we demonstrated that Ifngr2 was down-regulated in Mes cancer cells and that decreased Ifngr2 expression conferred Mes cancer cells less sensitive to IFN-γ, thereby evading IFN-γ- and TNFα-mediated killing and T cell cytotoxicity. Recent findings show that the IFN-γ pathway may have a suppressive effect in vivo because IFN-γ also increases the expression of several immune checkpoint molecules, inhibiting enzymes and MHC components in the late phase, which may limit the anticancer activity of T cells or natural killer cells (33, 73, 74). We observed less induced expression of Pd-l1 and H2-Kb, showing a defective transduction of IFN-γ in Mes cancer cells. In vivo data reported by Gao et al. (76) revealed that knockdown of Ifngr1 promoted tumor growth and reduced survival in response to anti–CTLA-4 therapy, suggesting that down-regulation of Ifngr2 is also one way in which the EMT program contributes to immunosuppression in vivo. Notably, a recent study found that mesenchymal-transitioned human small airway epithelial cells could inhibit interferon regulatory factor 1 (IRF1) expression, thereby suppressing the interferon pathway during airway fibrosis (78). This could be an additional mechanism or a consequence of reduced STAT1 activation and hereby reduced IRF1 expression.
Combined with the differential gene expression analysis, we identified two additional genes that were differentially expressed in cancer cells with the Epi or Mes status, and their ablation also exhibited differential susceptibility to killing by CD8+ T cells. Egfr and Mfge8 were highly expressed in EMT-induced cancer cells, and their sgRNAs were significantly depleted after CD8+ T cell challenge. Knockout or knockdown of these genes resensitized Mes cancer cells to T cell–mediated killing. In contrast, ectopic expression of EGFR strongly protected Epi cancer cells from T cell–mediated killing. Although Egfr activation also supports the EMT process (79, 80), we did not observe EMT in KPC3 cancer cells when only EGFR was ectopically expressed. These results suggest that EGFR expression is a downstream consequence of EMT and acts directly on immune resistance. Recently, a high-throughput screen was performed to identify kinase inhibitors that inhibited or enhanced T cell–mediated killing of cancer cells and found that erlotinib, an EGFR inhibitor, was effective in enhancing T cell–mediated killing of ID8 murine serous ovarian cancer cells (81). We also tested this inhibitor in our setting and found that treatment with a low dose (10 or 50 nM) of erlotinib enhanced the T cell–mediated killing of Mes KPC3 cancer cells but had no effect on Epi cancer cells. Thus, combining this information with our findings, it provides promising possibilities to target EGFR to enhance immunotherapy, as many non–small cell lung cancer patients with EGFR alterations fail to benefit from ICB treatment (82).
MFGE8 regulated immune resistance in a different way. We demonstrated that this secreted protein is highly expressed in Mes cancer cells compared to Epi cancer cells, and its levels further increased after cocultured with CD8+ T cells. Using a conditioned medium transfer approach, we found that removing Mfge8 from conditioned medium (by depleting it from Mfge8-producing Mes cells) augmented the killing efficiency of T cells, whereas increasing the Mfge8 level in conditioned medium reduced cancer cell killing by CD8+ T cells. Furthermore, when CD8+ T cells were directly treated with conditioned medium, their proliferation and cytokine secretion (IFN-γ and TNFα) were inhibited. Thus, our findings reveal a notable mechanism whereby mesenchymal-transformed cancer cells secrete Mfge8 to suppress the anticancer activity of CD8+ T cells.
To extend our findings to the clinic, TCGA-PAAD data were analyzed and revealed that the expression of both EGFR and MFGE8 was up-regulated in pancreatic cancer patients with Mes features and was also associated with a poor clinical prognosis. As the available clinical data of pancreatic cancer patients treated with ICB therapies is limited, the role of EGFR or MFGE8 in ICB treatment still needs to be further investigated. However, our data about the immunosuppressive function for MFGE8 are in line with others, where MFGE8 expression in esophageal cancer was found to correlate with decreased CD8+ T cell infiltration, and MFGE8 impaired T cell activation in the context of transplantation (83). Furthermore, a recent study demonstrated a critical role for EGFR in creating an immunosuppressive environment in breast cancer (84).
Above all, our work systematically delineates carcinoma cell–intrinsic factors that render cancers with an active EMT program more resistant to CD8+ T cell cytotoxicity, partially explaining how EMT contributes to immunosuppression and immunotherapy resistance, at least in relation to CD8+ T cells. We identified and highlighted three key factors, Ifngr2, Egfr, and Mfge8, and the underlying mechanisms that Mes cancer cells use to escape T cell–mediated killing (Fig. 7). Our results provide notable and attractive pharmacological targets that may be used, especially in cancers with an active EMT program, to empower immunotherapy.
Fig. 7. Model of the tumor-intrinsic factors used by EMP to evade CD8+ T cell–mediated killing.
Mes mouse KPC3 pancreatic cancer cells are more resistant to CD8+ T cell cytotoxicity than Epi parental cancer cells. EMT was induced and maintained by exposing the KPC3 epithelial cells to 2 days of TGF-β followed by a Dox treatment to induce expression of a constitutively active TGF-β type I receptor (ALK5 CA) (using a rTA-Tet-on–inducible ALK5 CA expression construct). The increased resistance of Mes versus Epi cells was attributed to several underlying mechanisms, including the down-regulation of Ifngr2 to inhibit susceptibility to IFN-γ signaling, the up-regulation of Egfr to evade T cell–mediated killing, and the increased secretion of Mfge8 to dampen the anticancer activity of CD8+ T cells.
MATERIALS AND METHODS
Cell lines
Human embryonic kidney (HEK) 293T cells [CRL3216, American Type Culture Collection (ATCC), Manassas, VA, USA] and human liver cancer cells (HepG2) (HB-8065, ATCC) were obtained from ATCC. The mouse pancreatic cancer cell line KPC3-OVA was provided by F. Ossendorp’s laboratory; this line was derived from the primary tumor of a female KrasG12D/+;Trp53R172H/+;Pdx-1-Cre mouse (25) and then transduced with an OVA-GFP expression construct. All cells were cultured in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific; catalog no. 41965062) containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific; catalog no. 16000044) and penicillin-streptomycin (100 U/ml; Thermo Fisher Scientific; catalog no. 15140163) and grown at 37°C in a humidified atmosphere containing 5% CO2. All cells were free of rodent viruses and mycoplasma, as determined by routine polymerase chain reaction (PCR) tests. Low passage number cultures from stock vials were used for all experiments.
Isolation and in vitro activation of OT-I CD8+ T cells
OT-I CD8+ T cells were isolated from the spleen of C57BL/6-Tg (TcraTcrb)1100Mjb mice (40), also referred to as OT-I mice (both female amd male mice were used, age 8 to 14 weeks), which were bred and housed at the Leiden University Medical Centre (LUMC) animal facility under specific pathogen–free conditions; these cells recognize OVA257–264 in the context of H2Kb. The animal experiments were designed according to the Code of Practice of the Dutch Animal Ethical Commission and subjected to the regulations as stated in project AVD11600202013796. The title of the project is Immunotherapy and vaccination against cancer.
Spleens from OT-I mice were mechanically dissociated, filtered through a 70-μm filter, and incubated in 2 ml of red blood cell lysis buffer per spleen for 5 min. The lysis reaction was quenched with 10 × the volume of phosphate-buffered saline (PBS) + 2% FBS + 5 mM EDTA. CD8+ T cells were enriched using the Mouse CD8+ T Lymphocyte Enrichment Set-DM (BD IMagTM #558471) according to the manufacturer’s protocol. Freshly isolated CD8+ T cells were stimulated with anti-CD3e/CD28 (BD Biosciences; #553057 and #553294) beads at concentrations of 1 and 5 μg/ml for 48 hours.
Pre-activated OT-I CD8+ T cells were cultured in RPMI 1640 medium (Gibco) supplemented with 10% FBS (Thermo Fisher Scientific, catalog no. 16000044), 1% penicillin/streptomycin (Thermo Fisher Scientific, catalog no. 15140163), 50 μM β-mercaptoethanol (Gibco, catalog no. 21985), IL-2 (100 ng/ml; PeproTech, #200-02), IL-7 (10 ng/ml; ImmunoTools, #12340073), and IL-15 (10 ng/ml; ImmunoTools, #12340153).
Generation of Epi or Mes KPC3-OVA cells
Parental KPC3-OVA cells were transduced with a constitutively kinase-active TGF-β type I receptor (ALK5 CA), which was constructed by Nakao et al. and Itoh et al. (28, 29). We further reengineered it into a Dox-induced system in our laboratory. In this system, we first treated cells with TGF-β (5 ng/ml) for 2 days to induce EMT and thereafter removed the TGF-β (as it may have other effects on the immune cells with which the cancer cells were cocultured) before adding Dox to sustain activated ALK5 signaling in the cancer cells in the absence of the exogenous ligand, thereby maintaining the mesenchymal features. Analysis of EMT marker expression revealed that mesenchymal phenotype was maintained for more than 16 days, which was sufficient time to perform all our experiments.
Lentiviral transduction and generation of stable cell lines
Lentiviruses were generated by HEK293T cell transfection with lentiCRISPR-v2 sgRNA plasmids (Addgene, #52961) or the indicated overexpressed cDNAs and the helper plasmids pCMV-VSVG, pMDLg-RRE (gag-pol), and pRSV-REV using polycation polyethylenimine transfection reagent. The cell supernatants were harvested 48 hours posttransfection, filtered through a 0.45-μm filter, and either used to infect cells or stored at −80°C. To obtain stable cell lines, cells were infected at low cell culture confluence (20%) with a lentivirus for 24 hours using normal culture medium supplemented with polybrene (8 μg/ml; Sigma-Aldrich). After 48 hours of infection, the cells were selected with antibiotics for at least 1 week and then passaged before use.
Knockout cells were generated using a puromycin-selectable variant of lentiCRISPR-v2 for each sgRNA. sgRNAs were cloned into the lentiCRISPR-v2 plasmid by the SAM target sgRNA cloning protocol (85). The sgRNA sequences used were as follows: sgCtrl: 5′-GCGAGGTATTCGGCTCCGCG-3′, sgMfge8 #1: 5′ACTCAAACTTGCGTCCGTCG-3′, sgMfge8 #2: 5′-AGTGCTTACCGGTTTCACAG-3′, sgEgfr #1: 5′ ACCAGACAGTCACTCTCTCG-3′, sgEgfr #2: 5′-CATGAATAGGCCAATCCCAA-3′, sgPip5k1a #1: 5′-GTGAGTGATGCCTAACTGGA-3′, sgPip5k1a #2: 5′-ACAGCTATGGAATCCATCCA-3′, sgAldh18a1 #1: 5′-CGATGGGGACGATGTTCATG-3′, sgAldh18a1 #2: 5′-CGTCCGAGAGGACAATCAAG-3′, sgPpfibp2 #1: 5′-ACAGTCACAGTACCTAATAC-3′, sg Ppfibp2 #2: 5′-CAGTCCCAGGTAAATCACCA-3′, sgGatad2a#1: 5′-TCAGACAGCATTCCTCTACG-3′, and sgGatad2a#2: 5′-GGGCGCACGAACCTGAAGTG-3′. For stable overexpression cells, the following expression constructs were used: Egfr cDNA (NM_005228) was inserted into the pLX304-V5 blast vector (Addgene, #25890), and the Mfge8 plasmid was purchased from Addgene (#46847).
Compound and recombinant proteins
TGF-β3 was a gift from A. Hinck, University of Pittsburgh, and was generally used at a concentration of 1 ng/ml (dissolved in 4 mM HCl/0.1% recombinant bovine serum albumin) when added to cells. The selective small molecule TβRI/ALK5 kinase inhibitor SB505124 (#3263) was used at 1 mM (#3263, Tocris, Bristol, UK).
Genome-wide CRISPR-Cas9 knockout screen and analysis
The genome-wide Brie CRISPR-knockout (KO) library (four sgRNAs per gene; ~78,637 sgRNAs) was purchased from Addgene (#73632) (32) and amplified according to the supplier’s protocol. The Epi and Mes KPC3-OVA cells were individually infected with the Brie CRISPR-KO library viruses at a multiplicity of infection of 0.3 while maintaining 500× coverage of the library. After 24 hours of transduction, the cells were selected with puromycin (1.5 μg/ml) for 5 days to achieve a stable phenotype after gene knockout. The CRISPR-Cas9 library–transduced KPC3-OVA cells were challenged with activated OT-I CD8+ T cells at an effector:target (E:T) ratio of 1:1 until reaching approximately 50% killing (determined by microscopic evaluation), and selection was conducted twice. The OT-I CD8+ T cells were derived from two different mice, and the treatment was performed independently on three samples. Untreated cells were cultured alongside as control groups. The surviving cells were harvested, and genomic DNA was isolated using the QIAamp DNA Blood Maxi Kit (51194, Qiagen).
sgRNA sequences were recovered by PCR using NEBNext High-Fidelity 2x PCR Master Mix (M0541L, New England BioLabs) with primers as described in https://addgene.org/browse/article/16517/. Sequencing was performed using the Illumina NovaSeq6000.
The obtained sequence reads were aligned to the Brie CRISPR-KO library, and the counts per sgRNA were generated, where reads containing mismatches in common and sgRNA sequences were excluded from the analysis. Enrichment and depletion of sgRNAs and genes in the epithelial and mesenchymal groups were determined relative to the corresponding untreated groups and analyzed using the MAGeCK algorithm (v0.5.9.4) (86). The MAGeCK results of genome-wide screen in KPC3-OVA pancreatic cancer cells (epithelial or mesenchymal types) challenged with or without OT-1 CD8+ T are shown in table S1. The −log10 transformed MAGeCK RRA scores, P values, FDR, and log2 fold change were used to assess which genes were specifically depleted or enriched in mesenchymal-type cells or epithelial-type cells. We selected specific screen sgRNAs in either mesenchymal-type or epithelial-type cancer cells by selecting depleted or enriched genes with a fold change difference between the mesenchymal-type screen and epithelial-type screen > 1.5 [labeled as diff (fold change) > 1.5] and |RRA score| > 3 with P value <0.01. The derived specifically depleted or enriched genes in mesenchymal-type or epithelial-type cells were further analyzed by GSEA using g:Profiler (https://biit.cs.ut.ee/gprofiler/gost) on GO and WikiPathways pathways and STRING network analysis (https://apps.cytoscape.org/apps/stringapp).
In vitro CD8+ T cell cytotoxicity assay
KPC3-OVA GFP cells were seeded in 12-well (1 × 105 cells per well) or 96-well (4 × 103 cells per well) plates. After overnight attachment, the cells were treated with activated OT-I CD8+ T cells at different effector:target (E:T) ratios. The GFP intensity of live cancer cells was recorded via the IncuCyte Life Cell Imaging system. After 48 hours of coculture, the CD8+ T cells and dead cancer cells were gently washed off using PBS, and then the remaining cells were fixed and stained using a solution containing 0.1% crystal violet (Sigma-Aldrich) and 50% methanol (Honeywell). After removing culture medium, the cell viability was measured by the CellTiter-Blue Viability Assay (G8081, Promega). In all cases, cell viability was calculated relative to the untreated group, and each group contained at least three replicates.
In vitro competition assay
Cells with the indicated gene modification were stained with the CTV Cell Proliferation Kit (C34571, Thermo Fisher Scientific) following the manufacturer’s instructions. Labeled cells were mixed at a 1:1 ratio and then challenged with activated CD8+ T cells or left untreated for 48 hours.
For cytokine treatment, IFN-γ, TNFα, or TRAIL rather than T cells was added to the culture medium. The cells were then washed with PBS twice and cultured for another day before analysis by fluorescence-activated cell sorting (FACS). The sensitivity was calculated by comparison of the proportions of CTV-negative versus CTV-positive cells in each group and then normalized to the data for the control group.
In vitro cytokine cytotoxicity assay
A total of 4 × 103 cancer cells were seeded per well in 96-well plates and then treated with different doses of recombinant murine IFN-γ (PeproTech, #315-05), murine TNFα (ImmunoTools, #12343014) or murine TRAIL (PeproTech, #315-19) at the indicated concentrations for 3 days. The cell viability was read using the Cell Titer Blue Viability Assay (G8081, Promega) according to the manufacturer’s instructions.
Western blot analysis
Cells were washed with PBS, and whole cell lysates were prepared in radioimmunoprecipitation assay buffer [50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, and 0.1% SDS] containing a freshly added complete protease inhibitor cocktail (Roche, catalog no. 11836153001). The BCA protein assay (Bio-Rad, catalog no. 5000111) was used for protein quantification. Samples were loaded on 8 to 15% SDS–polyacrylamide gel electrophoresis gels and transferred to 0.45-μm polyvinylidene difluoride membranes (Merck Millipore, catalog no. IPVH00010). The membranes were blocked in 5% nonfat dry milk dissolved in tris-buffered saline with 0.1% Tween 20 (TBST) for 1 hour at room temperature and then incubated at 4°C overnight with primary antibodies. After washing with TBST, secondary antibodies were applied for 1 hour at room temperature. The Clarity Western ECL Substrate (Bio-Rad, catalog no. 1705060) and the ChemiDoc Imaging System (Bio-Rad, catalog no. 17001402) were used to detect signals.
The primary antibodies used in this study are listed in table S2. The secondary antibodies used in this study were anti–immunoglobulin G (Sigma-Aldrich, catalog no. NA931V) and anti-rabbit (Cell Signaling, catalog no. 7074 S). All results were derived from at least three independent biological replicates, and representative results are shown. Levels of tubulin, Gapdh, or vinculin as determined by Western blotting or Ponceau S staining of the membrane were used for loading controls. Protein levels were quantified by densitometric analysis using ImageJ (National Institutes of Health, USA).
Flow cytometry
Cells were harvested and stained with fluorescent-conjugated antibodies targeting surface molecules of interest according to the manufacturer’s protocol and directly measured on a BD LSRII (BD Biosciences, Franklin Lakes, NJ, USA). Flow cytometry data were analyzed using FlowJoTM (v10, BD Biosciences, Franklin Lakes, NJ, USA). The FACS antibodies used in this study are listed in table S2.
RT-qPCR
Total RNA was extracted using the NucleoSpin RNA II Kit (Macherey-Nagel, Duren, Germany). A total of 1 μg of RNA was reverse-transcribed into cDNA using the Revert Aid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Quantitative PCR was performed on a CFX Connect Real-Time PCR detection system (Bio-Rad, Hercules, CA, USA) using SYBR GoTaq qPCR master mix (Promega, Madison, WI, USA) and 0.5 mM primers. The experiment was conducted three independent biological times with three technical repeats. All target gene expression levels were normalized to the Gapdh levels. The reverse transcription quantitative polymerase chain reaction (RT-qPCR) primer sequences used to detect target gene expression are listed in table S3.
RNA isolation and RNA-seq
A total of 2 × 106 KPC3-OVA cells (epithelial or mesenchymal type) were cocultured with 1 × 106 OT-I T cells for 0, 6, or 24 hours. Each group contained three independent biological replicates (OT-I T cells derived from three different donors). The OT-I T cells and culture medium were removed, and the remaining cells were washed three times with ice-cold PBS. The cancer cells were collected for RNA sample preparation with a NucleoSpin RNA kit. After mRNA enrichment by Oligo dT selection and subsequent library preparation, RNA-seq was performed on the DNBseq platform (Beijing Genomics Institute, BGI, Hong Kong).
RNA-seq data analysis
The RNA-seq raw dataset generated here is available in the Sequence Read Archive (SRA) repository and can be accessed using GEO accession numbers GSE215803 (https://ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE215803). RNA-seq files were afterward processed using the open source BIOWDL RNAseq pipeline v5.0.0 (https://zenodo.org/record/5109461#.Ya2yLFPMJhE) developed at the LUMC. This pipeline includes FASTQ preprocessing (including quality control, quality trimming, and adapter clipping), RNA-seq alignment, read quantification, and optional transcript assembly.
FASTQ files were quantified by FastQC (v0.11.9). The low-quality sequences and adapters were removed, and then the sequences were mapped to the mouse reference genome GRCm39 (mm39) using STAR (v2.7) (87) with default settings. Gene reads were counted using HTSeq2 (v0.11.1) (88). Count data were analyzed in R 4.2 and RStudio (v2022.07.1) with DESeq2 (v1.36.0) (89).
RNA-seq count data were normalized using rlog transformation as implemented in DESeq2. DEGs were calculated using DESeq2, and P adjusted <0.001 and fold change > 1.5 were used as the cutoffs. GSEA was performed in R using the clusterProfiler package (v4.1.4) (90) with the preranked log2 fold change in gene expression as a metric and using the C2-CP subcollection from MSigDB (91). GSEA plots were redrawn using the gseaplot2 function of the enrichplot package (https://github.com/YuLab-SMU/enrichplot).
Clinical data preparation and analysis
The TCGA bulk RNA-seq data (https://gdc.cancer.gov/about-data) for the cancer types considered in this study were downloaded using the R package TCGAbiolinks (v2.24.3), in which STAR count–derived RNA-seq count and transcripts per million (TPM) data were used. Nontumor samples (derived from normal tissue) were excluded. The TCGA-PAAD tumor subtype information (basal and classical types) and clinical data were downloaded alongside as rda format data and then retrieved by column name “paper_mRNA.Moffitt.clusters..All.150.Samples..1basal..2classical” (53–55).
To calculate the EMT signature score, the TPM data were passed on as an input for single-sample GSEA (92) in the gene set variation analysis RNA-seq normalized mode (93) using previously experimentally validated pancreatic cancer gene signatures: (i) the PAAD basal EMT signature and (ii) PAAD classical EMT signature, which was created and optimized to decouple EMT-specific genes from stromal cells by integrative expression analysis (52). The derived EMT scores were normalized. The derived EMT scores successfully distinguished PAAD patients between the basal and classical subtypes for identification of patients with high and low EMT scores. Next, we analyzed the two subtypes separately. Within each subtype, we normalized EMT scores using the Z-scale. On the basis of Z score normalized EMT scores, we further subdivided the classical and basal subtypes into “EMT-high or EMT-low in the classical subtype” and “EMT-high or EMT-low in the basal subtype.” The log2 (TMP + 1) expression of genes of interest was plotted and analyzed between different groups. Statistical significance computed by the Wilcoxon test is annotated by the number of stars (*P < 0.05; **P < 0.01; ***P < 0.001). Kaplan-Meier analysis was conducted using the R package survival (v3.4.0) and plotted using survminer (v0.4.9). The P value was computed by the log-rank test.
Quantification and statistical analysis
Either a two-tailed Student’s t test or one-way analysis of variance (ANOVA) followed by Dunnett’s correction test for multiple comparisons was used to compare the means of two groups. For two factors with multiple groups, selected groups were compared with two-way ANOVA followed by Sidak’s multiple comparisons test. Each experiment was repeated with at least two independent biological repeats including two to three technical replicates. For OT-I CD8+ T cell coculture assays, three biologically independent donors were used, and each donor has two replicates (error bars in the figures indicate the SD). To compare gene expression in the clinical dataset, the Wilcoxon rank-sum test was performed using the R function wilcox.test. GSEA was performed using the R package clusterProfiler (v4.1.4). Kaplan-Meier analysis was performed using the R packages survival (v3.4.0) and survminer (v0.4.9), and the P value was quantified by the log-rank Mantel-Cox test. For exceptions to these statistical methods, more details for each figure can be found in the corresponding figure legends. Analyses were performed using Prism 8 (GraphPad Software Inc., v8.0.2) or R (v4.2) and RStudio (2022.07.1). Unless otherwise indicated, P values less than 0.05 were considered statistically significant, and significance levels were set as follows: *P < 0.05, **P < 0.01, and ***P < 0.001.
Acknowledgments
We thank all members of our laboratories for helpful discussions and M. van Dinther for technical assistance. We thank the research facilities of LUMC for support. We thank N. P. van Montfoort (Department of Gastroenterology–Hepatology at Leiden University Medical Center) for the gift of KPC3-OVA cell line. We thank D. L. Marvin for providing the vector with Dox inducible of constitutively active ALK5 expression plasmid.
Funding: This work was supported by ZonMW grant (09120012010061) and Cancer Genomics Center Netherlands (CGC.NL.).
Author contributions: Conceptualization: Y.G., Z.Z., and P.t.D. Methodology: Y.G., M.G.M.C., F.O., and Z.Z. Investigation: Y.G., Z.Z., and R.H.W. Visualization: Y.G. and Z.Z. Supervision: P.t.D. Writing—original draft: Y.G. and Z.Z. Writing—review and editing: Y.G., Z.Z., R.H.W., and P.t.D.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary materials. The materials generated in this study did not generate substantial unique reagents. The RNA-seq raw dataset can be accessed using GEO accession numbers GSE215803 (https://ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE215803).
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S3
REFERENCES AND NOTES
- 1.C. M. Fares, E. M. Van Allen, C. G. Drake, J. P. Allison, S. Hu-Lieskovan, Mechanisms of resistance to immune checkpoint blockade: Why does checkpoint inhibitor immunotherapy not work for all patients? Am. Soc. Clin. Oncol. Educ. Book 39, 147–164 (2019). [DOI] [PubMed] [Google Scholar]
- 2.A. Dongre, R. A. Weinberg, New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 20, 69–84 (2019). [DOI] [PubMed] [Google Scholar]
- 3.L. A. Horn, K. Fousek, C. Palena, Tumor plasticity and resistance to immunotherapy. Trends Cancer 6, 432–441 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.R. D. Z. Mullins, A. Pal, T. F. Barrett, M. E. Heft Neal, S. V. Puram, Epithelial-mesenchymal plasticity in tumor immune evasion. Cancer Res. 82, 2329–2343 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.J. Yang, P. Antin, G. Berx, C. Blanpain, T. Brabletz, M. Bronner, K. Campbell, A. Cano, J. Casanova, G. Christofori, S. Dedhar, R. Derynck, H. L. Ford, J. Fuxe, A. García de Herreros, G. J. Goodall, A.-K. Hadjantonakis, R. J. Y. Huang, C. Kalcheim, R. Kalluri, Y. Kang, Y. Khew-Goodall, H. Levine, J. Liu, G. D. Longmore, S. A. Mani, J. Massagué, R. Mayor, D. McClay, K. E. Mostov, D. F. Newgreen, M. A. Nieto, A. Puisieux, R. Runyan, P. Savagner, B. Stanger, M. P. Stemmler, Y. Takahashi, M. Takeichi, E. Theveneau, J. P. Thiery, E. W. Thompson, R. A. Weinberg, E. D. Williams, J. Xing, B. P. Zhou, G. Sheng; EMT International Association (TEMTIA) , Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 21, 341–352 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.N. M. Aiello, Y. Kang, Context-dependent EMT programs in cancer metastasis. J. Exp. Med. 216, 1016–1026 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.F. Lüönd, N. Sugiyama, R. Bill, L. Bornes, C. Hager, F. Tang, N. Santacroce, C. Beisel, R. Ivanek, T. Bürglin, S. Tiede, J. van Rheenen, G. Christofori, Distinct contributions of partial and full EMT to breast cancer malignancy. Dev. Cell 56, 3203–3221.e11 (2021). [DOI] [PubMed] [Google Scholar]
- 8.A. Dongre, M. Rashidian, E. N. Eaton, F. Reinhardt, P. Thiru, M. Zagorulya, S. Nepal, T. Banaz, A. Martner, S. Spranger, R. A. Weinberg, Direct and indirect regulators of epithelial-mesenchymal transition-mediated immunosuppression in breast carcinomas. Cancer Discov. 11, 1286–1305 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.G. Wang, D. Xu, Z. Zhang, X. Li, J. Shi, J. Sun, H.-Z. Liu, X. Li, M. Zhou, T. Zheng, The pan-cancer landscape of crosstalk between epithelial-mesenchymal transition and immune evasion relevant to prognosis and immunotherapy response. NPJ Precis. Oncol. 5, 56 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.I. Pastushenko, C. Blanpain, EMT transition states during tumor progression and metastasis. Trends Cell Biol. 29, 212–226 (2019). [DOI] [PubMed] [Google Scholar]
- 11.W. Lu, Y. Kang, Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev. Cell 49, 361–374 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.V. da Silva-Diz, L. Lorenzo-Sanz, A. Bernat-Peguera, M. Lopez-Cerda, P. Muñoz, Cancer cell plasticity: Impact on tumor progression and therapy response. Semin. Cancer Biol. 53, 48–58 (2018). [DOI] [PubMed] [Google Scholar]
- 13.X. Zhuang, H. Zhang, G. Hu, Cancer and microenvironment plasticity: Double-edged swords in metastasis. Trends Pharmacol. Sci. 40, 419–429 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.J. K. Tiwari, S. Negi, M. Kashyap, S. Nizamuddin, A. Singh, A. Khattri, Pan-cancer analysis shows enrichment of macrophages, overexpression of checkpoint molecules, inhibitory cytokines, and immune exhaustion signatures in EMT-high tumors. Front. Oncol. 11, 793881 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Y. Lou, L. Diao, E. R. P. Cuentas, W. L. Denning, L. Chen, Y. H. Fan, L. A. Byers, J. Wang, V. A. Papadimitrakopoulou, C. Behrens, J. C. Rodriguez, P. Hwu, I. I. Wistuba, J. V. Heymach, D. L. Gibbons, Epithelial-mesenchymal transition is associated with a distinct tumor microenvironment including elevation of inflammatory signals and multiple immune checkpoints in lung adenocarcinoma. Clin. Cancer Res. 22, 3630–3642 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A. Dongre, M. Rashidian, F. Reinhardt, A. Bagnato, Z. Keckesova, H. L. Ploegh, R. A. Weinberg, Epithelial-to-mesenchymal transition contributes to immunosuppression in breast carcinomas. Cancer Res. 77, 3982–3989 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.K. Kolijn, E. I. Verhoef, M. Smid, R. Böttcher, G. W. Jenster, R. Debets, G. J. L. H. van Leenders, Epithelial-mesenchymal transition in human prostate cancer demonstrates enhanced immune evasion marked by IDO1 expression. Cancer Res. 78, 4671–4679 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Y. Guo, X. Lu, Y. Chen, B. Rendon, R. A. Mitchell, M. Cuatrecasas, M. Cortés, A. Postigo, Y. Liu, D. C. Dean, Zeb1 induces immune checkpoints to form an immunosuppressive envelope around invading cancer cells. Sci. Adv. 7, eabd7455 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.C. Kudo-Saito, H. Shirako, T. Takeuchi, Y. Kawakami, Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell 15, 195–206 (2009). [DOI] [PubMed] [Google Scholar]
- 20.M. Plaschka, V. Benboubker, M. Grimont, J. Berthet, L. Tonon, J. Lopez, M. Le-Bouar, B. Balme, G. Tondeur, A. de la Fouchardière, L. Larue, A. Puisieux, Y. Grinberg-Bleyer, N. Bendriss-Vermare, B. Dubois, C. Caux, S. Dalle, J. Caramel, ZEB1 transcription factor promotes immune escape in melanoma. J. Immunother. Cancer 10, e003484 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.V. Aggarwal, C. A. Montoya, V. S. Donnenberg, S. Sant, Interplay between tumor microenvironment and partial EMT as the driver of tumor progression. iScience 24, 102113 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.I. Akalay, B. Janji, M. Hasmim, M. Z. Noman, J. P. Thiery, F. Mami-Chouaib, S. Chouaib, EMT impairs breast carcinoma cell susceptibility to CTL-mediated lysis through autophagy induction. Autophagy 9, 1104–1106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Y. K. Chae, S. Chang, T. Ko, J. Anker, S. Agte, W. Iams, W. M. Choi, K. Lee, M. Cruz, Epithelial-mesenchymal transition (EMT) signature is inversely associated with T-cell infiltration in non-small cell lung cancer (NSCLC). Sci. Rep. 8, 2918 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.S. Funaki, Y. Shintani, E. Fukui, Y. Yamamoto, R. Kanzaki, N. Ose, T. Kanou, M. Minami, E. Mori, M. Okumura, The prognostic impact of programmed cell death 1 and its ligand and the correlation with epithelial-mesenchymal transition in thymic carcinoma. Cancer Med. 8, 216–226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.J. W. Lee, C. A. Komar, F. Bengsch, K. Graham, G. L. Beatty, Genetically engineered mouse models of pancreatic cancer: The KPC model (LSL-KrasG12D/+;LSL-Trp53R172H/+;Pdx-1-Cre), its variants, and their application in immuno-oncology drug discovery. Curr. Protoc. Pharmacol. 73, 14.39.1–14.39.20 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.L. Yang, Y. Pang, H. L. Moses, TGF-β and immune cells: An important regulatory axis in the tumor microenvironment and progression. Trends Immunol. 31, 220–227 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.E. Batlle, J. Massagué, Transforming growth factor-β signaling in immunity and cancer. Immunity 50, 924–940 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.S. Itoh, M. Thorikay, M. Kowanetz, A. Moustakas, F. Itoh, C.-H. Heldin, P. ten Dijke, Elucidation of Smad requirement in transforming growth factor-β type I receptor-induced responses. J. Biol. Chem. 278, 3751–3761 (2003). [DOI] [PubMed] [Google Scholar]
- 29.A. Nakao, T. Imamura, S. Souchelnytskyi, M. Kawabata, A. Ishisaki, E. Oeda, K. Tamaki, J. Hanai, C. H. Heldin, K. Miyazono, P. ten Dijke, TGF-β receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 16, 5353–5362 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D. L. Marvin, L. You, L. Bornes, M. van Dinther, N. Peters, H. Dang, S. K. Hakuno, M. Hornsveld, O. Kranenburg, J. van Rheenen, J. H. T. Rohling, M.-P. Chien, P. ten Dijke, L. Ritsma, Dynamic visualization of TGF-β/SMAD3 transcriptional responses in single living cells. Cancer 14, 2508 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D. A. Thomas, J. Massagué, TGF-β directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell 8, 369–380 (2005). [DOI] [PubMed] [Google Scholar]
- 32.J. G. Doench, N. Fusi, M. Sullender, M. Hegde, E. W. Vaimberg, K. F. Donovan, I. Smith, Z. Tothova, C. Wilen, R. Orchard, H. W. Virgin, J. Listgarten, D. E. Root, Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.K. A. Lawson, C. M. Sousa, X. Zhang, E. Kim, R. Akthar, J. J. Caumanns, Y. Yao, N. Mikolajewicz, C. Ross, K. R. Brown, A. A. Zid, Z. P. Fan, S. Hui, J. A. Krall, D. M. Simons, C. J. Slater, V. De Jesus, L. Tang, R. Singh, J. E. Goldford, S. Martin, Q. Huang, E. A. Francis, A. Habsid, R. Climie, D. Tieu, J. Wei, R. Li, A. H. Y. Tong, M. Aregger, K. S. Chan, H. Han, X. Wang, P. Mero, J. H. Brumell, A. Finelli, L. Ailles, G. Bader, G. A. Smolen, G. A. Kingsbury, T. Hart, C. Kung, J. Moffat, Functional genomic landscape of cancer-intrinsic evasion of killing by T cells. Nature 586, 120–126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.S. J. Patel, N. E. Sanjana, R. J. Kishton, A. Eidizadeh, S. K. Vodnala, M. Cam, J. J. Gartner, L. Jia, S. M. Steinberg, T. N. Yamamoto, A. S. Merchant, G. U. Mehta, A. Chichura, O. Shalem, E. Tran, R. Eil, M. Sukumar, E. P. Guijarro, C.-P. Day, P. Robbins, S. Feldman, G. Merlino, F. Zhang, N. P. Restifo, Identification of essential genes for cancer immunotherapy. Nature 548, 537–542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.C. J. Kearney, S. J. Vervoort, S. J. Hogg, K. M. Ramsbottom, A. J. Freeman, N. Lalaoui, L. Pijpers, J. Michie, K. K. Brown, D. A. Knight, V. Sutton, P. A. Beavis, I. Voskoboinik, P. K. Darcy, J. Silke, J. A. Trapani, R. W. Johnstone, J. Oliaro, Tumor immune evasion arises through loss of TNF sensitivity. Sci. Immunol. 3, eaar3451 (2018). [DOI] [PubMed] [Google Scholar]
- 36.D. W. Vredevoogd, T. Kuilman, M. A. Ligtenberg, J. Boshuizen, K. E. Stecker, B. de Bruijn, O. Krijgsman, X. Huang, J. C. N. Kenski, R. Lacroix, R. Mezzadra, R. Gomez-Eerland, M. Yildiz, I. Dagidir, G. Apriamashvili, N. Zandhuis, V. van der Noort, N. L. Visser, C. U. Blank, M. Altelaar, T. N. Schumacher, D. S. Peeper, Augmenting immunotherapy impact by lowering tumor TNF cytotoxicity threshold. Cell 178, 585–599.e15 (2019). [DOI] [PubMed] [Google Scholar]
- 37.X. G. Zhu, A. Chudnovskiy, L. Baudrier, B. Prizer, Y. Liu, B. N. Ostendorf, N. Yamaguchi, A. Arab, B. Tavora, R. Timson, S. Heissel, E. de Stanchina, H. Molina, G. D. Victora, H. Goodarzi, K. Birsoy, Functional genomics in vivo reveal metabolic dependencies of pancreatic cancer cells. Cell Metab. 33, 211–221.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Z. Zhang, X. Kong, M. A. Ligtenberg, S. E. van Hal-van Veen, N. L. Visser, B. de Bruijn, K. Stecker, P. W. van der Helm, T. Kuilman, E. P. Hoefsmit, D. W. Vredevoogd, G. Apriamashvili, B. Baars, E. E. Voest, S. Klarenbeek, M. Altelaar, D. S. Peeper, RNF31 inhibition sensitizes tumors to bystander killing by innate and adaptive immune cells. Cell Rep. Med. 3, 100655 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.N. Frey, L. Tortola, D. Egli, S. Janjuha, T. Rothgangl, K. F. Marquart, F. Ampenberger, M. Kopf, G. Schwank, Loss of Rnf31 and Vps4b sensitizes pancreatic cancer to T cell-mediated killing. Nat. Commun. 13, 1804 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.K. A. Hogquist, S. C. Jameson, W. R. Heath, J. L. Howard, M. J. Bevan, F. R. Carbone, T cell receptor antagonist peptides induce positive selection. Cell 76, 17–27 (1994). [DOI] [PubMed] [Google Scholar]
- 41.L. Xia, L. Wang, A. S. Chung, S. S. Ivanov, M. Y. Ling, A. M. Dragoi, A. Platt, T. M. Gilmer, X.-Y. Fu, Y. E. Chin, Identification of both positive and negative domains within the epidermal growth factor receptor COOH-terminal region for signal transducer and activator of transcription (STAT) activation. J. Biol. Chem. 277, 30716–30723 (2002). [DOI] [PubMed] [Google Scholar]
- 42.K. Kani, C. Garri, K. Tiemann, P. D. Malihi, V. Punj, A. L. Nguyen, J. Lee, L. D. Hughes, R. M. Alvarez, D. M. Wood, A. Y. Joo, J. E. Katz, D. B. Agus, P. Mallick, JUN-mediated downregulation of EGFR signaling is associated with resistance to gefitinib in EGFR-mutant NSCLC cell lines. Mol. Cancer Ther. 16, 1645–1657 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R. L. Ceriani, K. Thompson, J. A. Peterson, S. Abraham, Surface differentiation antigens of human mammary epithelial cells carried on the human milk fat globule. Proc. Natl. Acad. Sci. U.S.A. 74, 582–586 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.M. Jinushi, Y. Nakazaki, D. R. Carrasco, D. Draganov, N. Souders, M. Johnson, M. C. Mihm, G. Dranoff, Milk fat globule EGF-8 promotes melanoma progression through coordinated Akt and twist signaling in the tumor microenvironment. Cancer Res. 68, 8889–8898 (2008). [DOI] [PubMed] [Google Scholar]
- 45.R. Hanayama, M. Tanaka, K. Miyasaka, K. Aozasa, M. Koike, Y. Uchiyama, S. Nagata, Autoimmune disease and impaired uptake of apoptotic cells in MFG-E8-deficient mice. Science 304, 1147–1150 (2004). [DOI] [PubMed] [Google Scholar]
- 46.R. Hanayama, M. Tanaka, K. Miwa, A. Shinohara, A. Iwamatsu, S. Nagata, Identification of a factor that links apoptotic cells to phagocytes. Nature 417, 182–187 (2002). [DOI] [PubMed] [Google Scholar]
- 47.M. Jinushi, M. Sato, A. Kanamoto, A. Itoh, S. Nagai, S. Koyasu, G. Dranoff, H. Tahara, Milk fat globule epidermal growth factor–8 blockade triggers tumor destruction through coordinated cell-autonomous and immune-mediated mechanisms. J. Exp. Med. 206, 1317–1326 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.J.-Y. Zhao, X.-L. Ma, Z.-M. Li, R. Deng, S.-M. Wang, G.-B. Shen, J. Zhang, F.-T. Wang, B.-L. Zhang, Y.-Q. Wei, Down-regulation of MFG-E8 by RNA interference combined with doxorubicin triggers melanoma destruction. Clin. Exp. Med. 15, 127–135 (2015). [DOI] [PubMed] [Google Scholar]
- 49.M. Yarchoan, A. Hopkins, E. M. Jaffee, Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 377, 2500–2501 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.S. Wang, Z. He, X. Wang, H. Li, X.-S. Liu, Antigen presentation and tumor immunogenicity in cancer immunotherapy response prediction. eLife 8, e49020 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.K. Yoshihara, M. Shahmoradgoli, E. Martínez, R. Vegesna, H. Kim, W. Torres-Garcia, V. Treviño, H. Shen, P. W. Laird, D. A. Levine, S. L. Carter, G. Getz, K. Stemke-Hale, G. B. Mills, R. G. W. Verhaak, Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 4, 2612 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.M. Tyler, I. Tirosh, Decoupling epithelial-mesenchymal transitions from stromal profiles by integrative expression analysis. Nat. Commun. 12, 2592 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.E. A. Collisson, P. Bailey, D. K. Chang, A. V. Biankin, Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 16, 207–220 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Cancer Genome Atlas Research Network , Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 32, 185–203.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.R. A. Moffitt, R. Marayati, E. L. Flate, K. E. Volmar, S. G. H. Loeza, K. A. Hoadley, N. U. Rashid, L. A. Williams, S. C. Eaton, A. H. Chung, J. K. Smyla, J. M. Anderson, H. J. Kim, D. J. Bentrem, M. S. Talamonti, C. A. Iacobuzio-Donahue, M. A. Hollingsworth, J. J. Yeh, Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 47, 1168–1178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.R. Nicolle, Y. Blum, L. Marisa, C. Loncle, O. Gayet, V. Moutardier, O. Turrini, M. Giovannini, B. Bian, M. Bigonnet, M. Rubis, N. Elarouci, L. Armenoult, M. Ayadi, P. Duconseil, M. Gasmi, M. Ouaissi, A. Maignan, G. Lomberk, J.-M. Boher, J. Ewald, E. Bories, J. Garnier, A. Goncalves, F. Poizat, J.-L. Raoul, V. Secq, S. Garcia, P. Grandval, M. Barraud-Blanc, E. Norguet, M. Gilabert, J.-R. Delpero, J. Roques, E. Calvo, F. Guillaumond, S. Vasseur, R. Urrutia, A. de Reyniès, N. Dusetti, J. Iovanna, Pancreatic adenocarcinoma therapeutic targets revealed by tumor-stroma cross-talk analyses in patient-derived xenografts. Cell Rep. 21, 2458–2470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.J. M. Pitt, M. Vétizou, R. Daillère, M. P. Roberti, T. Yamazaki, B. Routy, P. Lepage, I. G. Boneca, M. Chamaillard, G. Kroemer, L. Zitvogel, Resistance mechanisms to immune-checkpoint blockade in cancer: Tumor-intrinsic and -extrinsic factors. Immunity 44, 1255–1269 (2016). [DOI] [PubMed] [Google Scholar]
- 58.P. Sharma, S. Hu-Lieskovan, J. A. Wargo, A. Ribas, Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.A. J. Schoenfeld, M. D. Hellmann, Acquired resistance to immune checkpoint inhibitors. Cancer Cell 37, 443–455 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.S. Bagchi, R. Yuan, E. G. Engleman, Immune checkpoint Inhibitors for the treatment of cancer: Clinical impact and mechanisms of response and resistance. Annu. Rev. Pathol. 16, 223–249 (2021). [DOI] [PubMed] [Google Scholar]
- 61.S. Kalaora, A. Nagler, J. A. Wargo, Y. Samuels, Mechanisms of immune activation and regulation: Lessons from melanoma. Nat. Rev. Cancer 22, 195–207 (2022). [DOI] [PubMed] [Google Scholar]
- 62.M. D. Vesely, T. Zhang, L. Chen, Resistance mechanisms to anti-PD cancer immunotherapy. Annu. Rev. Immunol. 40, 45–74 (2022). [DOI] [PubMed] [Google Scholar]
- 63.H. Rizvi, F. Sanchez-Vega, K. La, W. Chatila, P. Jonsson, D. Halpenny, A. Plodkowski, N. Long, J. L. Sauter, N. Rekhtman, T. Hollmann, K. A. Schalper, J. F. Gainor, R. Shen, A. Ni, K. C. Arbour, T. Merghoub, J. Wolchok, A. Snyder, J. E. Chaft, M. G. Kris, C. M. Rudin, N. D. Socci, M. F. Berger, B. S. Taylor, A. Zehir, D. B. Solit, M. E. Arcila, M. Ladanyi, G. J. Riely, N. Schultz, M. D. Hellmann, Molecular determinants of response to anti–programmed cell death (PD)-1 and anti–programmed death-ligand 1 (PD-L1) blockade in patients with non–small-cell lung cancer profiled with targeted next-generation sequencing. JCO 36, 633–641 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.V. Gopalakrishnan, B. A. Helmink, C. N. Spencer, A. Reuben, J. A. Wargo, The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell 33, 570–580 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.S. Middha, R. Yaeger, J. Shia, Z. K. Stadler, S. King, S. Guercio, V. Paroder, D. D. B. Bates, S. Rana, L. A. Diaz, L. Saltz, N. Segal, M. Ladanyi, A. Zehir, J. F. Hechtman, Majority of B2M-mutant and -deficient colorectal carcinomas achieve clinical benefit from immune checkpoint inhibitor therapy and are microsatellite instability-high. JCO Precis. Oncol. 3, PO.18.00321 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.M. Gromeier, M. C. Brown, G. Zhang, X. Lin, Y. Chen, Z. Wei, N. Beaubier, H. Yan, Y. He, A. Desjardins, J. E. Herndon II, F. S. Varn, R. G. Verhaak, J. Zhao, D. P. Bolognesi, A. H. Friedman, H. S. Friedman, F. McSherry, A. M. Muscat, E. S. Lipp, S. K. Nair, M. Khasraw, K. B. Peters, D. Randazzo, J. H. Sampson, R. E. McLendon, D. D. Bigner, D. M. Ashley, Very low mutation burden is a feature of inflamed recurrent glioblastomas responsive to cancer immunotherapy. Nat. Commun. 12, 352 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.D. J. McGrail, P. G. Pilié, N. U. Rashid, L. Voorwerk, M. Slagter, M. Kok, E. Jonasch, M. Khasraw, A. B. Heimberger, B. Lim, N. T. Ueno, J. K. Litton, R. Ferrarotto, J. T. Chang, S. L. Moulder, S.-Y. Lin, High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann. Oncol. 32, 661–672 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.M. Platten, W. Wick, B. J. Van den Eynde, Tryptophan catabolism in cancer: Beyond IDO and tryptophan depletion. Cancer Res. 72, 5435–5440 (2012). [DOI] [PubMed] [Google Scholar]
- 69.G. Parajuli, M. Tekguc, J. B. Wing, A. Hashimoto, D. Okuzaki, T. Hirata, A. Sasaki, T. Itokazu, H. Handa, H. Sugino, Y. Nishikawa, H. Metwally, Y. Kodama, S. Tanaka, H. Sabe, T. Yamashita, S. Sakaguchi, T. Kishimoto, S. Hashimoto, Arid5a promotes immune evasion by augmenting tryptophan metabolism and chemokine expression. Cancer Immunol. Res. 9, 862–876 (2021). [DOI] [PubMed] [Google Scholar]
- 70.S. M. Lightman, J. L. Peresie, L. M. Carlson, G. A. Holling, M. M. Honikel, C. A. Chavel, M. J. Nemeth, S. H. Olejniczak, K. P. Lee, Indoleamine 2,3-dioxygenase 1 is essential for sustaining durable antibody responses. Immunity 54, 2772–2783.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.I. Pérez-Núñez, C. Rozalén, J. Á. Palomeque, I. Sangrador, M. Dalmau, L. Comerma, A. Hernández-Prat, D. Casadevall, S. Menendez, D. D. Liu, M. Shen, J. Berenguer, I. R. Ruiz, R. Peña, J. C. Montañés, M. M. Albà, S. Bonnin, J. Ponomarenko, R. R. Gomis, J. M. Cejalvo, S. Servitja, D. M. Marzese, L. Morey, L. Voorwerk, J. Arribas, B. Bermejo, M. Kok, L. Pusztai, Y. Kang, J. Albanell, T. Celià-Terrassa, LCOR mediates interferon-independent tumor immunogenicity and responsiveness to immune-checkpoint blockade in triple-negative breast cancer. Nat. Cancer 3, 355–370 (2022). [DOI] [PubMed] [Google Scholar]
- 72.A. S. Dighe, E. Richards, L. J. Old, R. D. Schreiber, Enhanced in vivo growth and resistance to rejection of tumor cells expressing dominant negative IFN gamma receptors. Immunity 1, 447–456 (1994). [DOI] [PubMed] [Google Scholar]
- 73.J. Dubrot, P. P. Du, S. K. Lane-Reticker, E. A. Kessler, A. J. Muscato, A. Mehta, S. S. Freeman, P. M. Allen, K. E. Olander, K. M. Ockerman, C. H. Wolfe, F. Wiesmann, N. H. Knudsen, H.-W. Tsao, A. Iracheta-Vellve, E. M. Schneider, A. N. Rivera-Rosario, I. C. Kohnle, H. W. Pope, A. Ayer, G. Mishra, M. D. Zimmer, S. Y. Kim, A. Mahapatra, H. Ebrahimi-Nik, D. T. Frederick, G. M. Boland, W. N. Haining, D. E. Root, J. G. Doench, N. Hacohen, K. B. Yates, R. T. Manguso, In vivo CRISPR screens reveal the landscape of immune evasion pathways across cancer. Nat. Immunol. 23, 1495–1506 (2022). [DOI] [PubMed] [Google Scholar]
- 74.J. B. Williams, S. Li, E. F. Higgs, A. Cabanov, X. Wang, H. Huang, T. F. Gajewski, Tumor heterogeneity and clonal cooperation influence the immune selection of IFN-γ-signaling mutant cancer cells. Nat. Commun. 11, 602 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.J. M. Zaretsky, A. Garcia-Diaz, D. S. Shin, H. Escuin-Ordinas, W. Hugo, S. Hu-Lieskovan, D. Y. Torrejon, G. Abril-Rodriguez, S. Sandoval, L. Barthly, J. Saco, B. Homet Moreno, R. Mezzadra, B. Chmielowski, K. Ruchalski, I. P. Shintaku, P. J. Sanchez, C. Puig-Saus, G. Cherry, E. Seja, X. Kong, J. Pang, B. Berent-Maoz, B. Comin-Anduix, T. G. Graeber, P. C. Tumeh, T. N. M. Schumacher, R. S. Lo, A. Ribas, Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med. 375, 819–829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.J. Gao, L. Z. Shi, H. Zhao, J. Chen, L. Xiong, Q. He, T. Chen, J. Roszik, C. Bernatchez, S. E. Woodman, P.-L. Chen, P. Hwu, J. P. Allison, A. Futreal, J. A. Wargo, P. Sharma, Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 167, 397–404.e9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.D. H. Kaplan, V. Shankaran, A. S. Dighe, E. Stockert, M. Aguet, L. J. Old, R. D. Schreiber, Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc. Natl. Acad. Sci. U.S.A. 95, 7556–7561 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.J. Yang, B. Tian, H. Sun, R. P. Garofalo, A. R. Brasier, Epigenetic silencing of IRF1 dysregulates type III interferon responses to respiratory virus infection in epithelial to mesenchymal transition. Nat. Microbiol. 2, 17086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.H.-W. Lo, S.-C. Hsu, W. Xia, X. Cao, J.-Y. Shih, Y. Wei, J. L. Abbruzzese, G. N. Hortobagyi, M.-C. Hung, Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 67, 9066–9076 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.A. Clapéron, M. Mergey, T. H. Nguyen Ho-Bouldoires, D. Vignjevic, D. Wendum, Y. Chrétien, F. Merabtene, A. Frazao, V. Paradis, C. Housset, N. Guedj, L. Fouassier, EGF/EGFR axis contributes to the progression of cholangiocarcinoma through the induction of an epithelial-mesenchymal transition. J. Hepatol. 61, 325–332 (2014). [DOI] [PubMed] [Google Scholar]
- 81.P. H. Lizotte, R.-L. Hong, T. A. Luster, M. E. Cavanaugh, L. J. Taus, S. Wang, A. Dhaneshwar, N. Mayman, A. Yang, M. Kulkarni, L. Badalucco, E. Fitzpatrick, H.-F. Kao, M. Kuraguchi, M. Bittinger, P. T. Kirschmeier, N. S. Gray, D. A. Barbie, P. A. Jänne, A high-throughput immune-oncology screen identifies EGFR inhibitors as potent enhancers of antigen-specific cytotoxic T-lymphocyte tumor cell killing. Cancer Immunol. Res. 6, 1511–1523 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.S. Kumagai, S. Koyama, H. Nishikawa, Antitumour immunity regulated by aberrant ERBB family signalling. Nat. Rev. Cancer 21, 181–197 (2021). [DOI] [PubMed] [Google Scholar]
- 83.T. Kanemura, H. Miyata, T. Makino, K. Tanaka, K. Sugimura, M. Hamada-Uematsu, Y. Mizote, H. Uchida, Y. Miyazaki, T. Takahashi, Y. Kurokawa, M. Yamasaki, H. Wada, K. Nakajima, S. Takiguchi, M. Mori, Y. Doki, H. Tahara, Immunoregulatory influence of abundant MFG-E8 expression by esophageal cancer treated with chemotherapy. Cancer Sci. 109, 3393–3402 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.X. Wang, T. Semba, G. C. Manyam, J. Wang, S. Shao, F. Bertucci, P. Finetti, S. Krishnamurthy, L. T. H. Phi, T. Pearson, S. J. Van Laere, J. K. Burks, E. N. Cohen, J. M. Reuben, F. Yang, H. Min, N. Navin, V. N. Trinh, T. Iwase, H. Batra, Y. Shen, X. Zhang, D. Tripathy, N. T. Ueno, EGFR is a master switch between immunosuppressive and immunoactive tumor microenvironment in inflammatory breast cancer. Sci. Adv. 8, eabn7983 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.H. Nishimasu, F. A. Ran, P. D. Hsu, S. Konermann, S. I. Shehata, N. Dohmae, R. Ishitani, F. Zhang, O. Nureki, Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.W. Li, H. Xu, T. Xiao, L. Cong, M. I. Love, F. Zhang, R. A. Irizarry, J. S. Liu, M. Brown, X. S. Liu, MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.A. Dobin, C. A. Davis, F. Schlesinger, J. Drenkow, C. Zaleski, S. Jha, P. Batut, M. Chaisson, T. R. Gingeras, STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.S. Anders, P. T. Pyl, W. Huber, HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.M. I. Love, W. Huber, S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.G. Yu, L.-G. Wang, Y. Han, Q.-Y. He, clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.A. Subramanian, P. Tamayo, V. K. Mootha, S. Mukherjee, B. L. Ebert, M. A. Gillette, A. Paulovich, S. L. Pomeroy, T. R. Golub, E. S. Lander, J. P. Mesirov, Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.D. A. Barbie, P. Tamayo, J. S. Boehm, S. Y. Kim, S. E. Moody, I. F. Dunn, A. C. Schinzel, P. Sandy, E. Meylan, C. Scholl, S. Fröhling, E. M. Chan, M. L. Sos, K. Michel, C. Mermel, S. J. Silver, B. A. Weir, J. H. Reiling, Q. Sheng, P. B. Gupta, R. C. Wadlow, H. Le, S. Hoersch, B. S. Wittner, S. Ramaswamy, D. M. Livingston, D. M. Sabatini, M. Meyerson, R. K. Thomas, E. S. Lander, J. P. Mesirov, D. E. Root, D. G. Gilliland, T. Jacks, W. C. Hahn, Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.S. Hänzelmann, R. Castelo, J. Guinney, GSVA: Gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics 14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 to S3