SUMMARY
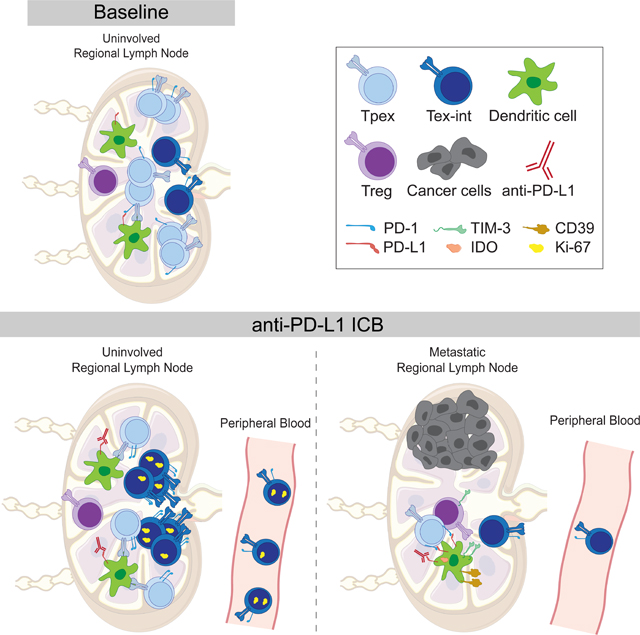
CD8+ T cell responses are critical for anti-tumor immunity. While extensively profiled in the tumor microenvironment, recent studies in mice identified responses in lymph nodes (LN) as essential; however, the role of LN in human cancer patients remains unknown. We examined CD8+ T cells in human head and neck squamous cell carcinomas, regional LN, and blood using mass cytometry, single-cell genomics, and multiplexed ion beam imaging. We identified progenitor exhausted CD8+ T cells (Tpex) that were abundant in uninvolved LN and clonally related to terminally exhausted cells in the tumor. After anti-PD-L1 immunotherapy, Tpex in uninvolved LN reduced in frequency but localized near dendritic cells and proliferating intermediate-exhausted CD8+ T cells (Tex-int), consistent with activation and differentiation. LN responses coincided with increased circulating Tex-int. In metastatic LN, these response hallmarks were impaired, with immunosuppressive cellular niches. Our results identify important roles for LN in anti-tumor immune responses in humans.
Graphical Abstract
INTRODUCTION
Immune checkpoint blockade (ICB) immunotherapy targeting the PD-1/PD-L1 axis has revolutionized oncology, but the underlying mechanisms remain incompletely understood, especially in humans. CD8+ T cells are central effector cells that mediate the efficacy of ICB and have been extensively studied in the tumor microenvironment (TME). However, recent studies found that CD8+ T cells in the periphery, such as in secondary lymphoid organs (SLOs) including tumor-draining (td) lymph nodes (LN), are integral for ICB response in mouse models. During the initiation of CD8+ T cell responses, naïve CD8+ T cells are typically primed by dendritic cells (DCs) in the LN before trafficking through the blood to the tumor.1 Blocking egress of lymphocytes from SLOs using the S1PR-blocking agent FTY720 abrogates ICB efficacy,2,3 and targeted administration of anti-PD-L1 to the tdLN is sufficient to control tumor growth in mice.4 In humans, analysis of CD8+ T cell receptor clonotypes in patient blood and tumors revealed the expansion of novel clones in the tumor after ICB that did not exist pretreatment, supporting a model where new clones are recruited from the periphery.5–8 Despite the potential importance of the tdLN in ICB-driven CD8+ T cell responses, a lack of LN sampling in clinical datasets leaves many unanswered questions about the relationship between immune responses in the LN and tumor in human cancer patients.
In settings of chronic antigen exposure, such as cancer and chronic infection, antigen-specific CD8+ T cells can become exhausted or dysfunctional, exhibiting elevated expression of inhibitory receptors such as PD-1.9 In mice, progenitor or precursor exhausted cells (Tpex) expressing the transcription factor TCF-1 undergo self-renewal and have high proliferative capacity.10,11 Tpex can differentiate into transitional intermediate exhausted cells (Tex-int) and subsequently into terminally exhausted cells (Tex-term), losing proliferative capacity and effector functions as they differentiate.12–17 In mice, Tpex are necessary for the maintenance of antigen-specific CD8+ T cell responses in settings of chronic antigen stimulation.10,18 In response to ICB in mice, Tpex are the primary exhausted subset that expands and differentiates into Tex-term, driving an increase of Tex-term in the tumor.12,14 Tex-term themselves do not substantially proliferate in mice with ICB treatment12,14,19, though they may become more activated.20 Of note, the tdLN is the primary site where Tpex are maintained and stimulated in mouse models of cancer,21,22 suggesting that ICB-responsive CD8+ T cells may reside in the tdLN.
It remains unclear to what extent these findings translate to humans. Tpex and Tex-term have been identified within tumor infiltrating lymphocytes (TILs) in multiple cancer types.23–28 A higher ratio of TCF-1+ CD8+ TILs to TCF-1- CD8+ TILs in melanoma was associated with longer survival after anti-PD-1 treatment.24 However, the abundance of tissue-resident memory T cells (Trm) in human tumors has also been associated with improved overall survival.29–31 Furthermore, in addition to exhausted cells, potential tumor-reactive CD8+ T cells in human tumors and peripheral blood have been identified as Trm and Temra cells, respectively.28 Given that most patients do not currently exhibit durable responses to ICB,32 identifying which CD8+ T cell subsets are targeted by ICB in patients, and where activation takes place anatomically, will be necessary to improve therapeutic efficacy.
In this study, we examined the relationship between CD8+ T cell responses across key anatomic sites, including TME, tdLN, and blood in human cancer patients. Patients with advanced head and neck squamous cell carcinoma (HNSCC) are routinely treated with surgery, including resection of regional LN, providing a unique opportunity to address these open questions. Through a combination of single-cell analyses by mass cytometry (CyTOF), single-cell RNA-sequencing and TCR-sequencing (sc-RNA+TCR-seq), sc-RNA- and protein-sequencing (CITE-Seq), and multiplexed ion beam imaging (MIBI), we examined CD8+ T cells across tissues from patients treated with surgery as standard of care (SOC) and patients treated with perioperative anti-PD-L1 ICB (atezolizumab) enrolled in a clinical trial at UCSF Medical Center (NCT03708224). We identified a subset of Tpex in uninvolved, regional lymph nodes (uiLN) that were clonally related to Tex-term in the TME. Patients treated with ICB shortly before surgery exhibited a decrease in Tpex frequency in uiLN with a concomitant increase in more differentiated Tex-int, which were localized in proximity to DCs. Changes in uiLN correlated with an increase in proliferating Tex-int in peripheral blood. Moreover, regional lymph nodes with tumor metastasis (metLN) exhibited an impairment in these responses to ICB associated with immunosuppressive cellular niches around Tpex and a reduction in the circulating CD8+ T cell response. These results highlight a central role for uiLN in mediating responses to ICB, which may create new opportunities for next-generation immunotherapies focused on optimally harnessing these responses.
RESULTS
Tpex are increased in uninvolved lymph nodes of HNSCC patients
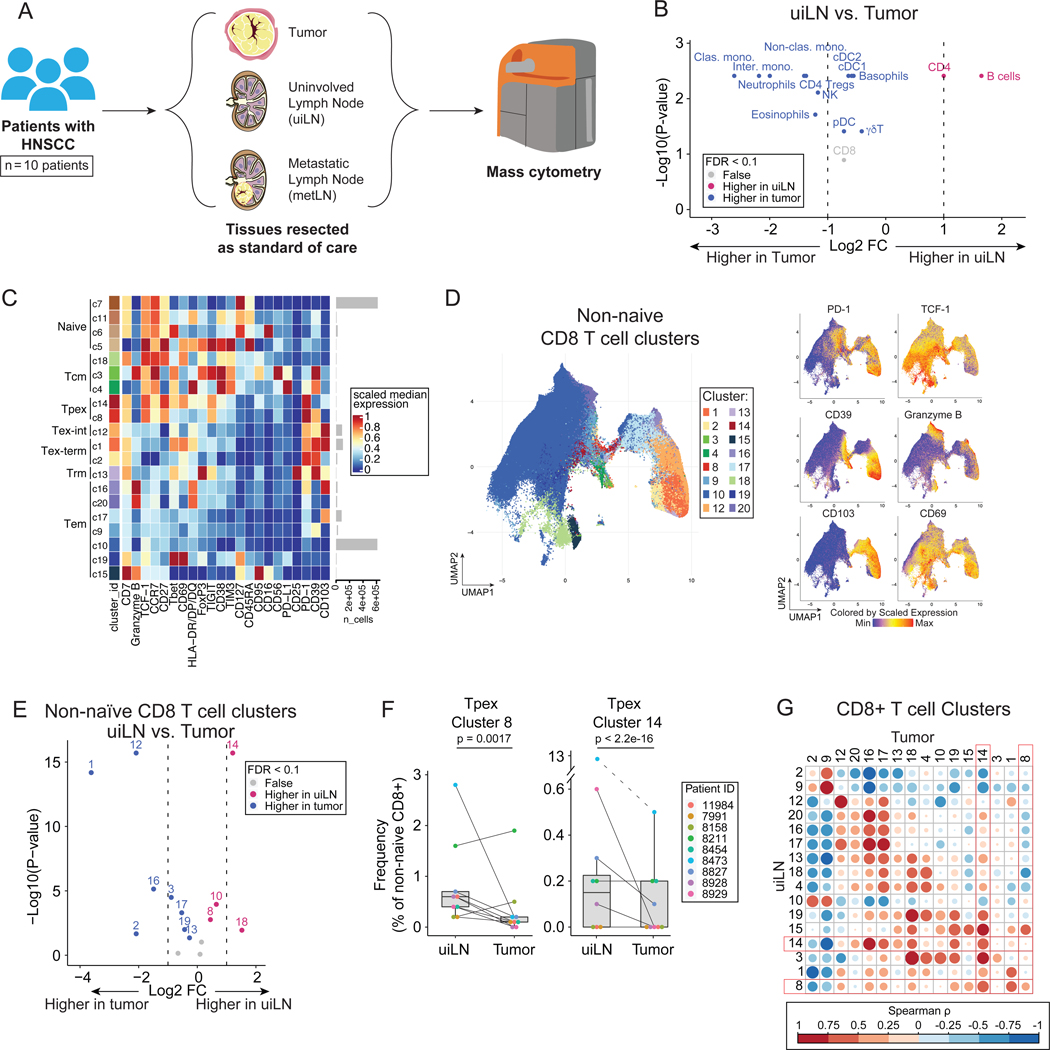
Using mass cytometry, we profiled immune cells in the TME and LN of 10 patients with locally metastatic HNSCC who had both tumor and matched uiLN and/or metLN available for analysis (Fig. 1A; Table S1). All tissue specimens were collected immediately following tumor resection and neck dissection for each patient. To understand the immune composition present within each tissue, we manually gated major immune cell subsets (Fig. S1A–B). As expected, CD4+ T cells and B cells were more frequent within uiLN, while other immune cell subsets were more frequent in the tumor as a percentage of total immune cells (Fig. 1B). CD8+ T cells were present at similar frequencies in both tissues (Fig. 1B).
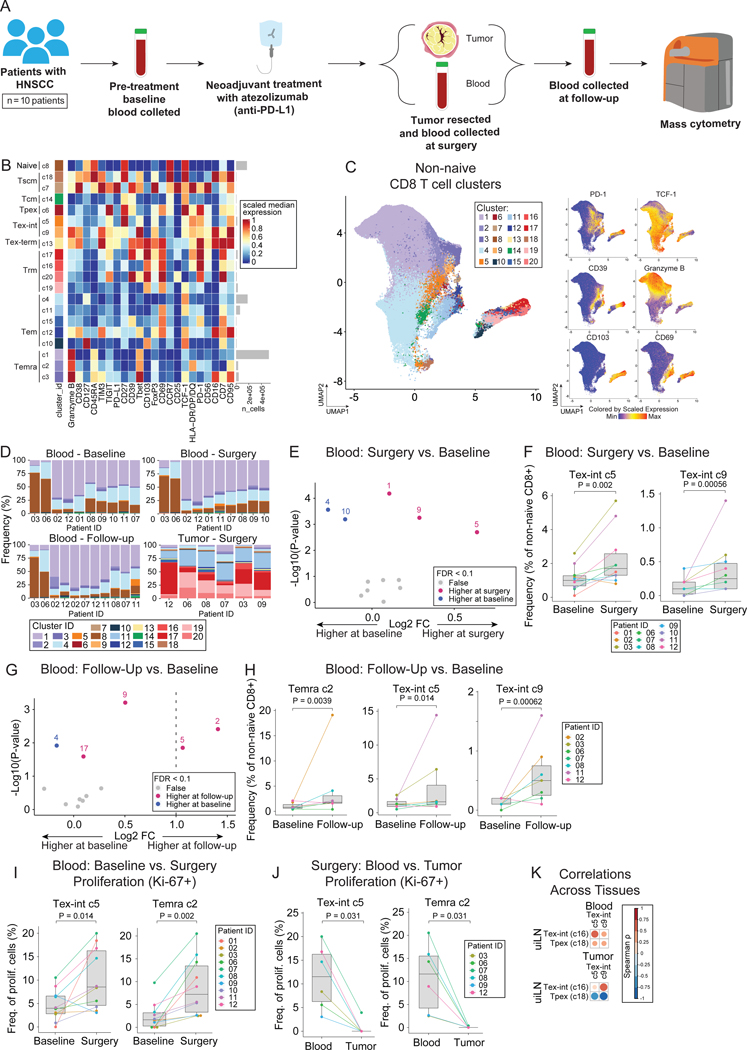
Figure 1: Tpex cells are increased in uninvolved lymph nodes of HNSCC patients.
A) Overview of cohort. Paired tumor and lymph node samples (uninvolved; uiLN and/or metastatic; metLN) were obtained from patients with HNSCC (n = 10). Samples were analyzed by mass cytometry. B) Paired differential abundance (DA) of main immune cell populations between uiLN and tumor (n = 9; paired Wilcoxon Rank Sum Test). The log2 fold changes are plotted against the negative log10 (nominal p-values). Colors indicate if cell populations are significantly more abundant in uiLN (purple), tumor (blue), or not differentially abundant (False, grey) after Benjamini-Hochberg correction, FDR < 0.1. C) Heatmap of markers used for CD8+ T cell clustering. Scaled median expression per marker is shown for cluster annotation. D) UMAP of non-naïve CD8+ T cell clusters and expression for a subset of markers. E) Paired differential abundance analysis of non-naïve CD8+ T cell subsets between uiLN and tumor (n = 9; generalized linear mixed models). See color scheme for Figure 1B. F) Cluster 8 and 14 abundances (as percentage of non-naïve CD8+ T cells) in paired samples from uiLN and tumor. P-values obtained by generalized linear mixed models. G) Spearman correlation between clusters in uiLN and clusters in tumors. Tpex clusters are highlighted in red boxes. See also Figure S1.
We hypothesized that the functional states of CD8+ T cells might differ between the tumor and uiLN. We performed unsupervised clustering of CD8+ T cells by FlowSOM33, which partitioned cells into 20 clusters based on protein expression (Fig. 1C–D, S1C–D). In addition to naïve (CD45RA+ CCR7+), central memory (Tcm: CD45RA- CCR7+), and effector memory (Tem: CD45RA- CCR7-) cells, we also identified Tpex (PD-1+ TCF-1+), Tex-int (PD-1+ TCF-1- CD69-), and Tex-term (PD-1+ TCF-1- CD69+) (Fig. 1C).16 Tpex cells expressed CD27 and TIGIT as well as variable expression of CD39 and CD69 (Fig. 1C–D). After dimensionality reduction by UMAP, the Tpex clusters were positioned between abundant Tem cells, including a cluster expressing the protein CD39 associated with tumor-reactivity,34,35 and cells expressing the integrin CD103 associated with Trm (Fig 1D). Amongst non-naïve CD8+ T cells, differential abundance analysis revealed that both Tpex clusters (c8, c14) were significantly more frequent in uiLN compared with matched tumors (Fig. 1E–F). In contrast, clusters representing Tex (c1, c12) and Tem (c16, c19) were enriched within the tumor (Fig. 1E). Tpex clusters in uiLN were positively correlated with Tpex clusters in matched tumors as well as a Tex cluster (c1), various Tem clusters (c16, c17, c19) and a Tcm cluster (c18) (Fig. 1G). These results show that uiLN in HNSCC patients harbor increased frequencies of CD8+ Tpex cells, while CD8+ Tex cells are present at greater frequencies within primary tumors. Together with the literature, these results suggest that this reservoir of Tpex cells in the LN may play an important role in human therapeutic responses.
Tpex in LN are clonally related to Tex within the TME
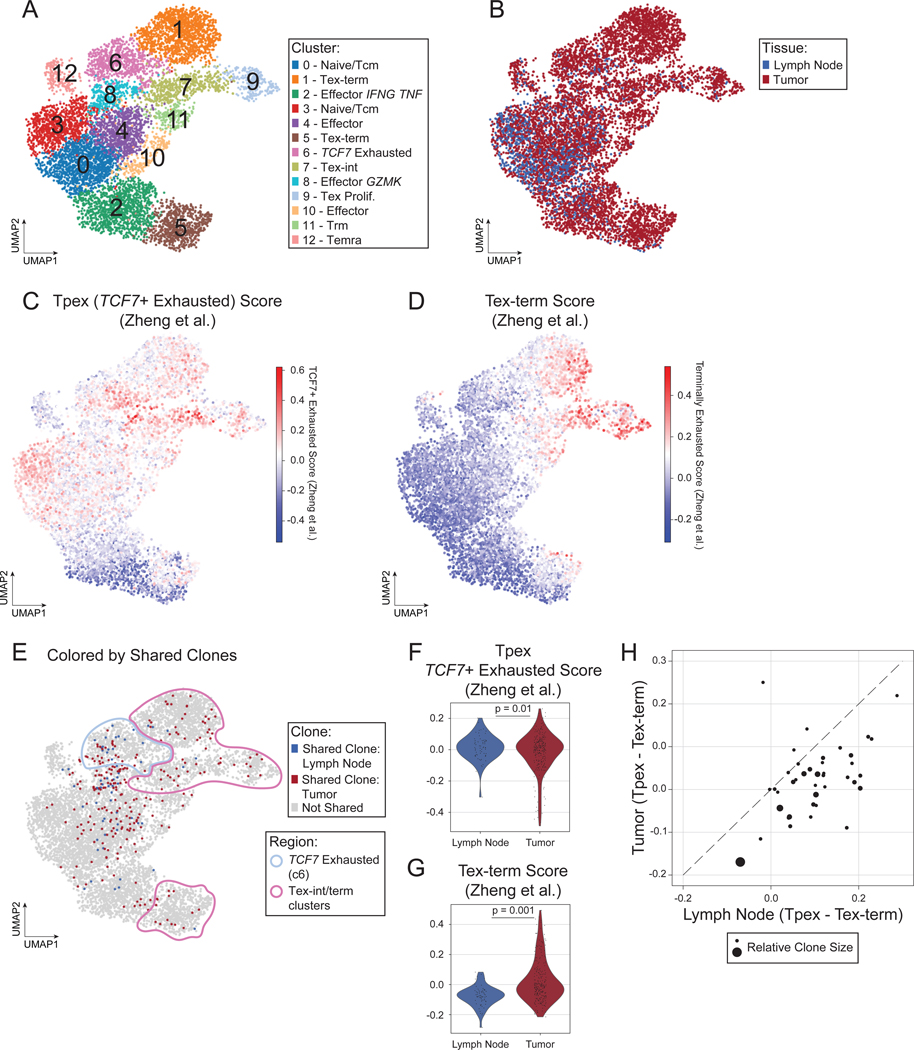
If anti-tumor CD8+ T cell responses are coordinated across tissues, we would expect clonally related CD8+ T cells to be present in both LN and tumor. Therefore, we hypothesized that CD8+ Tpex cells in the LN are clonally related (i.e., share the same T cell receptor (TCR) sequence) to more exhausted CD8+ T cells within the TME. To assess CD8+ T cell clonality, paired tumor and LN tissues surgically resected as SOC treatment from 5 patients with HNSCC (Table S2) were subjected to sc-RNA-seq, of which paired TCR-seq data were generated from 4 patients and paired single-cell protein expression data (CITE-Seq) were generated from 3 patients. Unsupervised clustering identified major cell populations in the TME and LN (Fig. 2SA), annotated based on the expression of canonical marker genes (Fig. S2B). We identified CD8+ T cells based on a combination of RNA expression, protein expression (when available), and cluster annotation and subclustered these cells (Fig. 2A, S2C–F). Clustering and visualization of the CD8+ T cells revealed phenotypes that were shared between the TME and LN and phenotypes specifically enriched in the TME (Fig. 2B, S2G). To confirm the identity of exhausted cell subsets, we computed gene expression scores derived in a recent meta-analysis of sc-RNA-seq data from human CD8+ TILs, which identified TCF7+ exhausted cells (resembling Tpex; the TCF7 gene encodes the transcription factor TCF-1) and Tex-term (Fig. 2C–D, S2H).28 Indeed, our cluster annotations for Tpex and Tex mirror the clusters with the highest expression of these scores, respectively. In addition, cluster 7 exhibited higher expression of both scores, reminiscent of Tex-int, which was confirmed by analysis of differentially expressed genes (Table S3).
Figure 2: CD8+ T cells in the tumor and LN are clonally related.
A-E) UMAP of 8,245 CD8+ T cells from 5 paired tumor and LN samples (10 samples total) colored by (A) Leiden cluster, (B) tissue of origin, (C) TCF7 exhaustion score, (D) Tex-term score, or (E) highlighting shared clones between the tumor (red) and lymph node (blue). F-G) Violin plot of expression of (F) TCF7 exhaustion score and (G) Tex-term score in CD8+ T cells with shared clones in the LN versus tumor. P-values obtained by generalized linear mixed models. H) Scatter plot of average TCF7 exhaustion score – average terminal exhaustion score for each shared clone in the LN (x-axis) versus tumor (y-axis). Dashed line is the identity line. Dots are sized according to the number of cells in LN and tumor for the clone. See also Figure S2.
Based on TCR CDR3 sequences, we identified clonally-related CD8+ T cells (termed “shared clones”) in the LN and TME of each participant with available TCR-seq data (3–29 clones per participant; Fig. 2E, S2I). Shared clones from cells localized in the LN were more likely to belong to the TCF7+ exhausted cluster (cluster 6), while their counterparts in the TME were more likely to belong to the Tex-int/Tex-term clusters (P = 0.0005, odds ratio = 5.71 by Fisher’s exact test) (Fig. 2E). Moreover, cells belonging to shared clones in the LN exhibited significantly higher TCF7+ exhausted gene set scores (Fig. 2F), while their counterparts in the TME exhibited significantly higher Tex-term gene set scores (Fig. 2G). At the individual clone level, the difference between the average TCF7+ exhausted (Tpex) and average Tex-term scores was higher for cells located in the LN compared to those in the TME for the majority of clones (Fig. 2H). Collectively, these results indicate that shared CD8+ T cell clones exist in the LN and TME across patients, albeit in different functional states. CD8+ T cells in the LN were more likely to be in a state resembling Tpex, while their counterparts in the TME were more likely to be in a Tex-term state. Because CD8+ T cells lose functional capacity as they differentiate from Tpex to Tex-term,12–17 these results underscore the importance of the LN in maintaining a reservoir of functional anti-tumor CD8+ T cell clones in humans, which could represent an important target for immunotherapies.
Tpex are primarily localized in the T cell zone in human uiLN
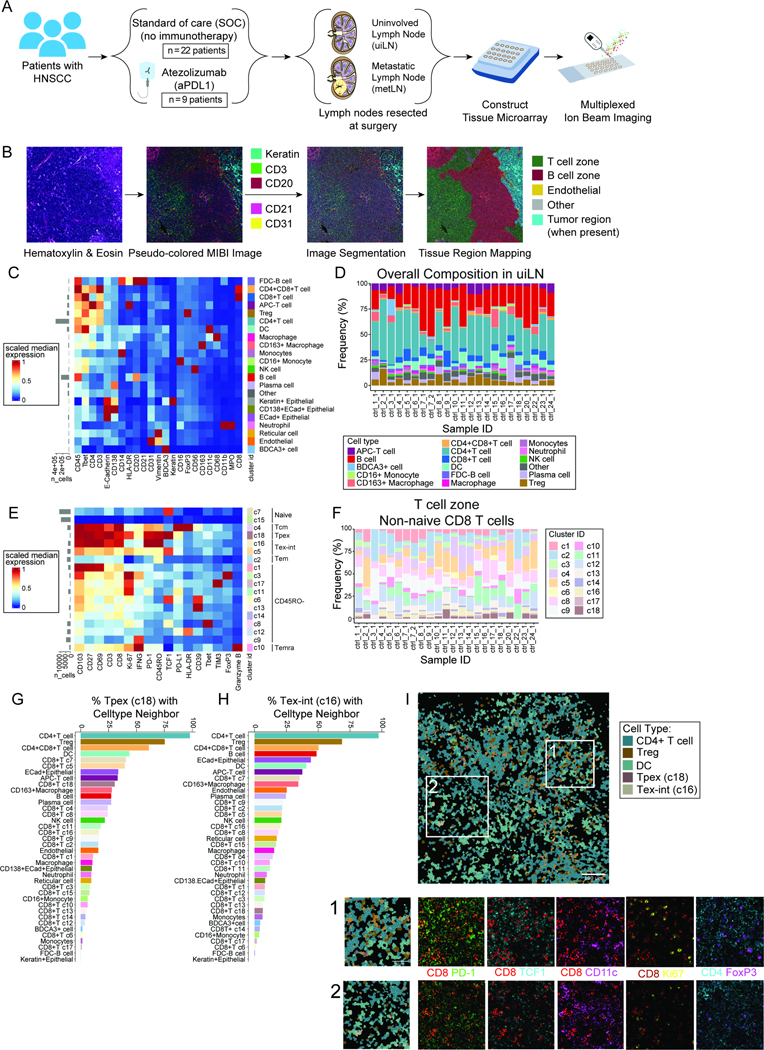
We next sought to understand the Tpex architectural niche in the uiLN as well as what effect ICB might have on their organization. We recently treated 10 patients with human papilloma virus (HPV)-negative HNSCC with 1 to 2 cycles of the PD-L1 inhibitor atezolizumab prior to surgery through clinical trial NCT03708224 (Table S4). We obtained uiLN specimens from 9 of these patients and LN with tumor metastasis (metLN) from 4 patients. Additionally, we obtained uiLN resected as SOC treatment from HPV-negative HNSCC patients who were not treated with immunotherapy (n = 22). LN tissue cores were randomized across two tissue microarrays, of which sections were stained and analyzed by multiplexed ion beam imaging (MIBI) (Fig. 3A, Table S5). Following image acquisition, pixels were clustered according to their intensity values for each measured parameter (Fig. S3A), individual cells were segmented using the Mesmer algorithm,36 and LN tissue regions were identified, including B cell follicles/B cell rich zones, T cell rich zones, and metastatic tumor regions, when present (Fig. 3B). Single cells were then clustered by FlowSOM based on their overall clustered pixel composition (Fig. S3B), identifying major cell populations (Fig. 3C–D, S3C–D).
Figure 3: Localization of Tpex in human uiLN.
A) Overview of cohort. uiLN samples were obtained from patients with HNSCC that received standard of care (SOC; n = 23) or 1 to 2 cycles of anti-PD-L1 treatment prior to surgery (n = 9). For anti-PD-L1 treated patients that had metastatic disease (n = 4), metLN samples were also obtained. Tissue cores were distributed across two tissue microarrays for analysis by multiplexed ion beam imaging (MIBI). B) Overview of the image analysis pipeline. C) Heatmap of scaled median marker expression for cell lineage assignments. D) Relative abundance of main immune cell types in uiLN (global, n = 23) from SOC treated patients (n = 22). Sample ID represents patient ID followed by LN number. E) Heatmap of markers used for CD8+ T cell clustering. Scaled median expression per marker is shown for cluster annotation. F) Relative abundance of non-naïve CD8+ T cell clusters in uiLN (T cell zone, n = 23) from SOC treated patients (n = 22). G-H) Percentage of (G) cluster 18 and (H) cluster 16 cells with a specific cell type as its neighbor in uiLN (T cell zone) from SOC treated patients. I) Representative image of an uiLN from a SOC treated patient (patient 23) showing the spatial localization of CD4 T cells, Tregs, DCs, Tpex (cluster 18), and Tex-int (cluster 16). Cell identity overlaid onto the segmentation mask. Highlighted regions 1 and 2 are colored by the expression of CD8+ (red), PD-1 (green), TCF-1 (cyan), CD11c (purple), Ki-67 (yellow), CD4 (cyan), and FoxP3 (purple). See also Figure S3.
We first focused our analysis on CD8+ T cells within untreated uiLN. CD8+ T cells were subclustered by FlowSOM and partitioned into 18 clusters based on their protein expression (Fig. 3E–F, S3E). Consistent with our prior results, a population of PD-1+ TCF-1+ Tpex (c18) was identified in the uiLN in addition to two clusters of Tex-int (c16, c5), which were distinguished by their differential expression levels of PD-1 and TCF-1. Leveraging this high-dimensional spatial information, we assessed where these cells were localized in uiLN and with which other cell types they were adjacent. Tpex cells can express CCR7,14 which promotes migration to T cell zones of LN,37 as well as CXCR5,12 which in contrast drives migration toward B cell zones.38 Tpex were more likely to be localized in the T cell zones, though a fraction were also localized in B cell zones (Fig. S3F). Within the T cell zone, Tpex (c18) and Tex-int (c16) were often found neighboring CD4+ T cells, Tregs, CD4+ CD8+ (double-positive) T cells, and DCs, which was readily observable in the MIBI images (Fig. 3G–I).
anti-PD-L1 ICB impacts the frequency of Tpex and Tex-int cells and their cellular neighborhoods in uiLN
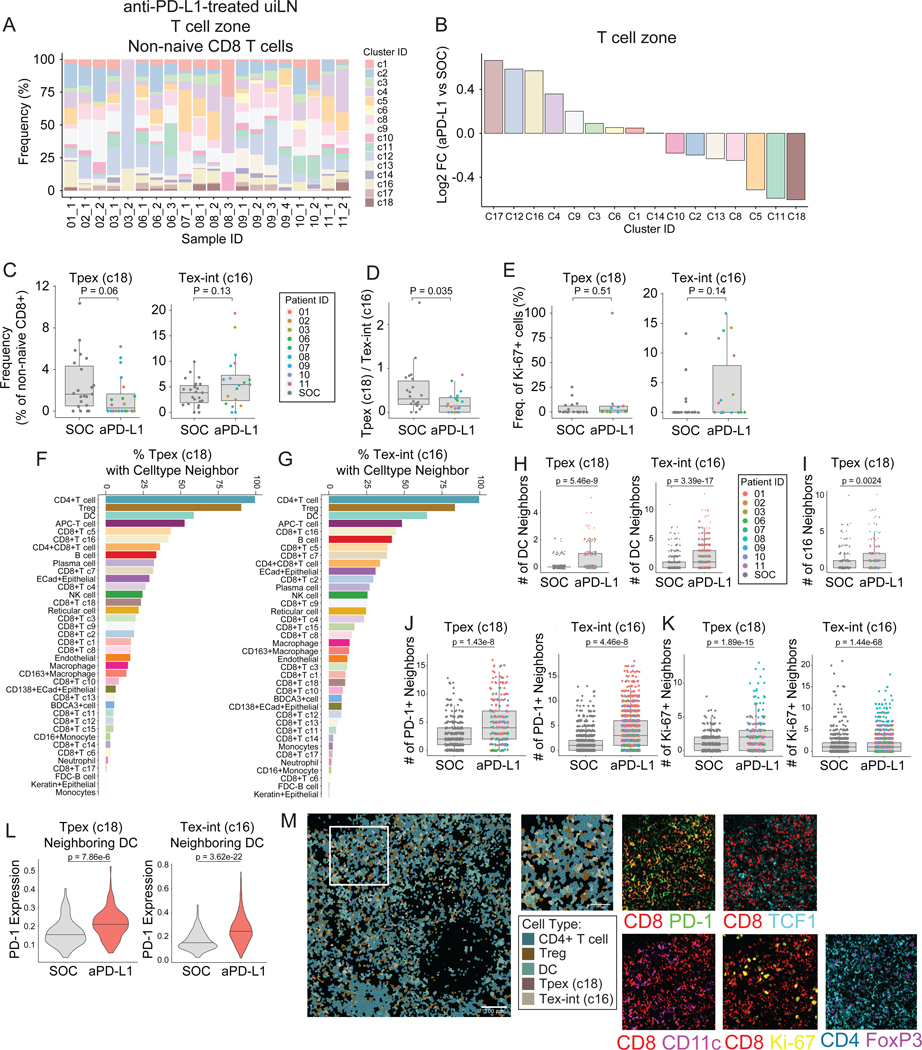
The identification of Tpex in uiLN prompted the question of whether and how Tpex and their surrounding cell neighbors respond to ICB in humans. Therefore, we also evaluated uiLN tissues from patients who received anti-PD-L1 ICB prior to surgery. Compared with the SOC uiLN (Fig. 3D, S3D), the frequency of major cell populations was not dramatically altered by therapy (Fig. S4A). However, changes in the composition of CD8+ T cell subsets were clearly evident (Fig. 4A–B, S4B). We therefore hypothesized that anti-PD-L1 ICB may change the frequency and activation state of Tpex and Tex-int in uiLN. Indeed, in the T cell zone, Tpex (c18) had the largest decrease in frequency among CD8+ T cell clusters in anti-PD-L1 treated patients compared to SOC treated patients (Fig. 4B–C). This change was specific to the T cell zone, as Tpex frequencies were not meaningfully altered in the B cell zone (Fig. S4C). In contrast to Tpex, Tex-int (c16) had one of the largest increases amongst CD8+ T cell clusters in the T cell zone following treatment, though frequencies were somewhat variable across patients and LNs (Fig. 4B–C). As a result of these effects, the ratio of Tpex to Tex-int significantly decreased in uiLN from ICB-treated patients (Fig. 4D), consistent with a shift toward more differentiated phenotypes (e.g., Tex-int) within the exhausted CD8+ T cell compartment after ICB. Other clusters with similar increases included a CD45RO- TIM3+ PD-1- cluster (c17) and a CD45RO- HLA-DR+ cluster (c12), the latter of which may represent recently-activated CD8+ T cells (Fig. 4B). While the proportion of Tpex that were proliferating (Ki-67+) did not change for most patients post-treatment, strikingly, 100% of Tpex were Ki-67+ in an uiLN resected from the only patient who experienced a major pathological response to therapy (patient 11) (Fig. 4E). In contrast, Tex-int exhibited a trend toward increased proliferation in patients treated with anti-PD-L1 (Fig. 4E).
Figure 4: anti-PD-L1 ICB impacts Tpex and Tex-int in uiLN.
A) Relative abundance of non-naïve CD8+ T cell clusters in uiLN (T cell zone, n = 20) from anti-PD-L1 treated patients (n = 9). Sample ID represents patient ID followed by LN number. B) Non-naïve CD8+ T cell cluster ratios represented as log2 fold changes between uiLN (T cell zone, n = 20) from anti-PD-L1 treated patients and uiLN (T cell zone, n = 22) from SOC treated patients. C) Cluster 18 and 16 abundances (as percentage of non-naïve CD8+ T cells) in the T cell zone of uiLN from SOC and anti-PD-L1 treated patients. D) Cluster 18/cluster 16 ratio in uiLN (T cell zone) from SOC and anti-PD-L1 treated patients. E) Percentage of cluster 18 and 16 proliferating cells in uiLN (T cell zone) from SOC and anti-PD-L1 treated patients. F-G) Percentage of (F) cluster 18 and (G) cluster 16 cells with a specific cell type as its neighbor in uiLN (T cell zone) from anti-PD-L1 treated patients. H) Number of DC neighbors for cluster 18 and cluster 16 cells in uiLN (T cell zone) from SOC and anti-PD-L1 treated patients. I) Number of cluster 16 neighbors for cluster 18 cells in uiLN (T cell zone) from SOC and anti-PD-L1 treated patients. J-K) Number of (J) PD-1 positive and (K) Ki-67+ neighbors for cluster 18 and cluster 16 cells in uiLN (T cell zone) from SOC and anti-PD-L1 treated patients. L) Expression of PD-1 on cluster 18 and cluster 16 cells with a DC neighbor in uiLN (T cell zone) from SOC and anti-PD-L1 treated patients. M) Representative image of an uiLN from an anti-PD-L1 treated patient (patient 01) showing the spatial localization of CD4 T cells, Tregs, DCs, Tpex (cluster 18), and Tex-int (cluster 16). Cell identity overlaid onto the segmentation mask. Highlighted region is colored by the expression of CD8+ (red), PD-1 (green), TCF-1 (cyan), CD11c (purple), Ki-67 (yellow), CD4 (cyan), and FoxP3 (purple). C-E and H-L) P-values obtained by Wilcoxon Rank Sum Test. See also Figure S4.
Next, we evaluated if the spatial localization of Tpex and Tex-int cells was altered within uiLN following ICB by assessing each cell’s local neighborhood (STAR methods).39,40 Notably, both Tpex (c18) and Tex-int (c16) were more frequently localized in proximity to DCs (Fig. 4F–G). Several clusters of CD8+ T cells had significantly higher numbers of DC neighbors after anti-PD-L1 treatment (Fig. S4D), including Tpex and Tex-int, which exhibited the largest fold-change and the most significant p-value, respectively (Fig. 4H, S4D). These data are consistent with mouse cancer models in which DCs in the LN mediate responses to ICB.41 However, the frequency of DCs was not different after anti-PD-L1 treatment (Fig. S4E), nor did DC frequencies significantly correlate with Tpex or Tex-int frequencies (Fig. S4F–G). Additionally, Tpex were more likely to be near Tex-int after treatment, consistent with ongoing activation and differentiation into a Tex-int functional state (Fig. 4I).
Interestingly, both Tpex and Tex-int had more neighboring PD-1+ and Ki-67+ cells around them after treatment (Fig. 4J–K), suggestive of cellular niches containing activated and proliferating cells. Indeed, Tpex and Tex-int in proximity to DCs exhibited higher expression of PD-1 after treatment, suggesting they were activated and/or differentiating, which was evident when visualizing MIBI images (Fig. 4L–M). In summary, uiLN resected after anti-PD-L1 ICB treatment exhibited a decrease in Tpex frequency with a concomitant increase in Tex-int that were localized close to Tpex, raising the possibility that treatment caused Tpex to transition to Tex-int. Furthermore, both Tpex and Tex-int were more likely to be located near DCs and exhibited elevated PD-1 expression compared to patients who had not received anti-PD-L1 immunotherapy, supporting a role for DCs in uiLN in mediating anti-tumor CD8+ T cell responses.
Tex-int increase in the blood following ICB
Given the significant CD8+ T cell responses within uiLN of treated patients, we wanted to determine if similar changes could be detected within the blood and tumor, as an expansion of activated CD8+ T cells in the blood has been previously associated with clinical response to ICB across several types of solid tumors,6,7,42–45 though the source of these cells has remained speculative. For all 10 patients treated with neoadjuvant anti-PD-L1 ICB, blood was collected at baseline just prior to their first infusion as well as on the day of surgery, and resected tumor tissue was available for analysis from 6 patients (Fig. 5A, S5A). For 6 patients, blood was also collected at a follow-up visit approximately one month following their surgery. Fresh uiLN tissue was also available from one patient (11), collected during surgery (Fig. S5A). As expected, mass cytometry analysis revealed clear differences in the frequency of major immune cell populations between matched tumor and blood from the same patients, regardless of when blood was collected, as well as in the uiLN sample (Fig. S5B–C).
Figure 5: Tex-int are present at higher levels in the blood following ICB.
A) Overview of cohort. Blood samples were collected pre-treatment (baseline, 2–29 days before surgery), at time of surgery, and at follow-up (27–38 days post-surgery) from patients with HNSCC that received anti-PD-L1 treatment prior to surgery (n = 10). Additionally, tumor samples were collected at time of surgery. For a few patients, additional samples were collected (see complete overview in Figure S6A). Samples were analyzed by mass cytometry. B) Heatmap of markers used for CD8+ T cell clustering. Scaled median expression per marker is shown for cluster annotation. C) UMAP of non-naïve CD8+ T cell clusters and expression for a subset of markers. D) Frequencies of non-naïve CD8+ T cell clusters. E) Paired differential abundance analysis of non-naïve CD8+ T cell subsets between blood at time of surgery and baseline (n = 10) (generalized linear mixed models). The log2 fold changes are plotted against the negative log10(nominal p-values). Colors indicate if cell populations are significantly more abundant in blood at time of surgery (purple) or baseline (blue) or not differentially abundant (False, grey) after Benjamini-Hochberg correction, FDR < 0.1. F) Cluster 5 and 9 abundances (as percentage of non-naïve CD8+ T cells) in paired blood samples at time of surgery and baseline. P-values obtained by generalized linear mixed models. G) Paired differential abundance analysis of non-naïve CD8+ T cell subsets between blood at time of follow-up and at baseline (n = 7) (generalized linear mixed models). See color scheme for Figure 6C. H) Cluster 2, 5, and 9 abundances (as percentage of non-naïve CD8+ T cells) in paired blood samples at follow-up and baseline. P-values obtained by generalized linear mixed models. I-J) Percentage of cluster 2 and 5 proliferating cells in (I) paired blood samples at time of surgery and baseline and (J) in blood and tumor at time of surgery. P-values obtained by paired Wilcoxon Rank Sum Test. K) Spearman correlation between cluster 16 and 18 in LN (MIBI data) and cluster 5 and 9 in blood (CyTOF data) at time of surgery. See also Figure S5.
Unsupervised clustering of CD8+ T cells from all samples partitioned cells into 20 clusters, which reflected distinct differentiation states (Fig. 5B–C). Cluster 6 expressed the highest levels of TCF-1 among the PD-1+ clusters, corresponding to Tpex, and was present in the uiLN sample; however, it was only detected at low levels in blood and tumor samples (Fig. 5B–C, Fig. S5E), implying that the decreased frequency of Tpex observed in treated uiLN was not due to Tpex egress from LN. In contrast, Tex-int clusters (c5, c9) were detectable in the blood (Fig 5D) and significantly increased after treatment (at the time of surgery) compared to baseline, along with a Temra cluster (c1) (Fig. 5E–F). At the 1 month follow-up, similar changes were observed (Fig. 5G–H). While most patients had some increase in Tex-int frequency at follow-up compared with baseline, two patients (3 and 11) had substantially larger increases (Fig. 5H). Although the neoadjuvant clinical trial design precluded assessment of clinical response by radiology (RECIST criteria), patient 11 was the only patient with significant tumor treatment response microscopically (i.e., most of the tumor was necrotic) and is still alive nearly a year following surgery. Additionally, patient 3 has not had signs of tumor recurrence and is still alive nearly three years following surgery.
Tex-int cells express Ki-67 in the blood following anti-PD-L1 ICB
To further evaluate the activation state of Tex-int (c5, c9) and Temra (c2) in the blood, we assessed the proportion of cells expressing the proliferation marker Ki-67. Consistent with their increased frequencies within blood after treatment, Tex-int (c5) and Temra (c2) exhibited significant increases in their proliferating fraction after ICB (Fig. 5I), while the other Tex-int cluster (c9) did not (Fig. S5F). Comparing the phenotypes of the Tex-int clusters, c5 expressed higher levels of the memory-associated co-stimulatory receptor CD27, while c9 expressed higher levels of the effector-associated transcription factor Tbet and TIM3, suggesting that c9 may be at a later differentiation state compared to c5 (Fig. 5B).
It is plausible that Ki-67+ Tex-int in circulation could originate in uiLN or the tumor. Because we previously found increased proliferation in the uiLN, we also investigated whether there was evidence of proliferating Tex-int in the TME. Tex-int (c5) were significantly more proliferative in the blood than in the tumor of the same patients, where little proliferation was evident (Fig. 5J). This was also true for the proliferative Temra population (c2) observed in the blood (Fig. 5J). Consistent with a peripheral origin of Tex-int, these cells were also more frequent in blood compared to matched tumors after treatment (Fig. S5G–H). Finally, we assessed whether these populations were correlated across uiLN, blood, and tumor from the same patients (Fig. S5I–K). Indeed, both Tpex (c18) and Tex-int (c16) in the uiLN were positively correlated with both Tex-int clusters (c5, c9) in the blood (Fig. 5K, S5J). While Tex-int (c16) in the uiLN were also positively correlated with Tex-int clusters in the tumor (c5, c9), Tpex (c18) in the uiLN exhibited negative correlations with these clusters in the tumor (Fig. 5K, S5K). Therefore, increased frequencies of Tex-int in the tumor after treatment were associated with fewer Tpex within the treated uiLN and an increase in the frequency of Tex-int in the uiLN. These data are consistent with a model in which Tpex differentiate to Tex-int within the uiLN following treatment, which then transit through the blood to the TME.
Metastases within LNs alter immune composition
As with many solid tumors, HNSCC frequently metastasize to regional lymph nodes. Having established an important role for uiLN in anti-tumor CD8+ T cell responses, we asked whether metastases in LN (metLN) would impact this response. Using mass cytometry, we profiled the immune cells present in paired metLN and uiLN from the same SOC patients with locally advanced disease, overlapping with the patients we had previously analyzed (n = 7). Paired metLN and uiLN were regional and ipsilateral to the same primary tumor; distant metastases were not identified at the time of surgery. Immune composition within metLN was very similar to the tumor and distinct from paired uiLN from the same individuals (Fig. S6A–C). This pattern extended to CD8+ T cells, in which Tpex were almost absent in metLN, while Tex-term were more abundant (Fig. 6A–B). In contrast, no significant differences were observed between CD8+ T cell composition in metLN and paired tumors (Fig. S6D). Thus, metLN exhibit a distinct immune environment as compared to paired uiLN but recapitulate a similar immune microenvironment to the primary tumor from which they originated.
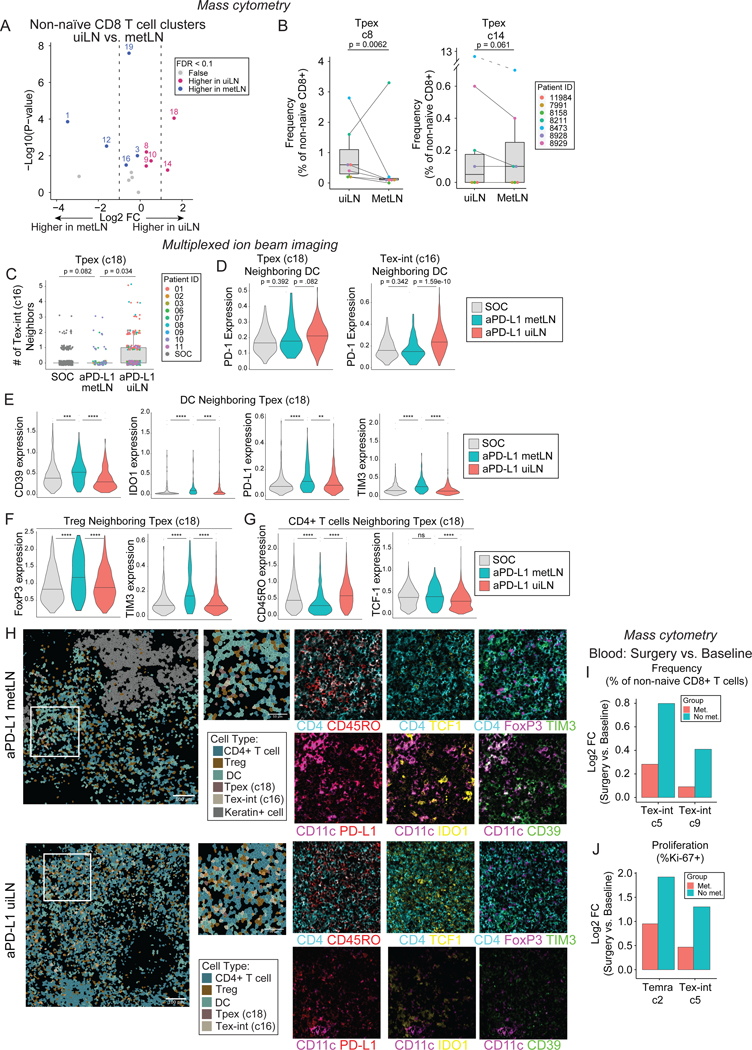
Figure 6: Immunosuppressive niches surround Tpex and Tex-int in metLN after anti-PD-L1 ICB.
A) Paired differential abundance (DA) analysis of main immune cell populations between uiLN and metLN (n = 9; paired Wilcoxon Rank Sum Test). The log2 fold changes are plotted against the negative log10 (nominal p-values). Colors indicate if cell populations are significantly more abundant in uiLN (purple), tumor (blue), or not differentially abundant (False, grey) after Benjamini-Hochberg correction, FDR < 0.1. B) Cluster 8 and 14 abundances (as percentage of non-naïve CD8+ T cells) in paired samples from uiLN and metLN. P-values obtained by generalized linear mixed models. C) Number of cluster 16 neighbors for cluster 18 cells in ui-cores (global) and met-cores (global) from SOC and anti-PD-L1 treated patients. D) Expression of PD-1 on cluster 18 and cluster 16 cells with a DC neighbor in ui-cores (global) and met-cores (global) from SOC and anti-PD-L1 treated patients. E) Expression of CD39, IDO1, PD-L1, and TIM3 on DCs neighboring Tpex (cluster 18 cells) in ui-cores (global) and met-cores (global) from SOC and anti-PD-L1 treated patients. F) Expression of FOXP3 and TIM3 on Tregs neighboring Tpex (cluster 18 cells) in ui-cores (global) and met-cores (global) from SOC and anti-PD-L1 treated patients. G) Expression of CD45RO and TCF-1 on CD4+ T cells neighboring Tpex (cluster 18 cells) in ui-cores (global) and met-cores (global) from SOC and anti-PD-L1 treated patients. H) Representative images of a metLN (patient 10) and uiLN (patient 01) from anti-PD-L1 treated patients showing the spatial localization of CD4 T cells, Tregs, DCs, Tpex (cluster 18), Tex-int (cluster 16), and keratin+ cells. Cell identity overlaid onto the segmentation mask. Highlighted region is colored by the expression of CD4+ (cyan), CD45RO (red), CD11c (purple), PD-L1 (red), TCF-1 (yellow), IDO1 (yellow), FoxP3 (purple), TIM-3 (green), and CD39 (green). I) Log2 fold changes of cluster 5 and 9 abundances at time of surgery vs baseline stratified into patients with metastatic LN (Met) and patients without metastatic LN (No met). J) Log2 fold changes of cluster 2 and 5 frequencies of proliferating cells at time of surgery vs baseline stratified into patients with metastatic LN (Met) and patients without metastatic LN (No met). C-G) P-values obtained by Wilcoxon Rank Sum Test. See also Figure S6.
Having established that uiLN are an important site for CD8+ T cell responses to ICB, we evaluated whether these responses remained intact or differed in metLN. Of the 9 patients treated with anti-PD-L1 from whom uiLN were collected, 4 patients also had at least one LN with evidence of metastasis, with metastatic cores (met-cores) from 3 patients available for MIBI analysis. Differences in major cell population frequencies mirrored the results from mass cytometry (Fig. S6E–F). Within the non-naïve CD8+ T cell compartment, Tpex (c18) and Tex-int (c16) were also amongst the clusters with the largest relative reductions in frequency within met-cores as compared to ui-cores after treatment (Fig. S6G–H). Though these differences were not statistically significant, perhaps due to the smaller sample size, median Tpex (c18) abundance was reduced in met-cores compared to ui-cores from either immunotherapy-treated or -naive patients, and the median frequency of Tex-int (c16) was even lower in met-cores from treated patients compared to ui-cores of patients who had not received immunotherapy (Fig. S6I).
metLN exhibit immunosuppressive niches surrounding Tpex and Tex-int following ICB
We next evaluated the cellular neighbors of these CD8+ T cell clusters of interest in met-cores after treatment. Similar to uiLN, Tpex and Tex-int were most likely to be localized next to CD4+ T cells, Tregs, and DCs (Fig. S6J–K). However, in contrast to uiLN, Tpex were less likely to be near Tex-int in met-cores after treatment (Fig. 6C), and neither of these populations upregulated PD-1 when in proximity to DCs (Fig. 6D). These results are consistent with a failure of Tpex to become activated and differentiate into Tex-int in the metLN. Therefore, we further evaluated whether their neighboring cells may exhibit more immunosuppressive properties. Indeed, DCs neighboring both Tpex and Tex-int in met-cores after treatment expressed significantly higher levels of the regulatory molecules CD39, IDO, TIM3, and PD-L1 (Fig. 6E, Fig. S6L), all associated with a tolerogenic DC state.46–49 Moreover, Tregs neighboring both Tpex and Tex-int exhibited higher expression of the master regulator transcription factor, FoxP3, elevated levels of CD39 and TIM3 (Fig. 6F, Fig. S6M) associated with enhanced suppressive function,50–54, and increased Ki-67. In addition, neighboring CD4+ T cells expressed significantly lower levels of CD45RO and PD-1 along with higher levels of TCF-1, indicative of a more naïve or quiescent phenotype (Fig. 6G, Fig. S6N). These results collectively indicate that Tpex and Tex-int are localized in more immunosuppressive niches in metLN compared to uiLN after ICB (Fig. 6H).
Given these differences, we hypothesized that perhaps patients with metLN would exhibit muted CD8+ T cell responses in the blood after ICB. Indeed, patients with metLN exhibited more modest increases in the frequencies of circulating Tex-int (c5, c9) as compared to patients with no evidence of LN metastases (Fig. 6I). Moreover, patients with LN metastases also exhibited smaller changes in Ki-67+ Tex-int (c5, c9) and Temra (c2) cells in the blood after treatment (Fig. 6J, S6O). Collectively, these data indicate that the changes in Tpex frequencies and cellular neighborhoods in metLN are associated with blunted responses to ICB in the blood.
DISCUSSION
It has remained unclear whether regional LN are important hubs of anti-tumor immunity and ICB response in patients with cancer. This information is critical to understanding the mechanistic basis of current immunotherapies in humans and identifying new opportunities to improve responses by optimally harnessing the right cell subsets.
Given prior work showing the proliferative burst of Tpex observed in the tdLN and tumor of mice after ICB treatment,12,14,18 we originally hypothesized that the frequency of Tpex in the uiLN of human patients might increase after ICB treatment. Surprisingly, the frequency of Tpex in the uiLN decreased after immunotherapy in patients. Our data support a model in which ICB causes Tpex in human uiLN to differentiate further into Tex-int, ultimately resulting in a decrease in the frequency of Tpex in the uiLN at the post-treatment time points analyzed in this study. ICB has been shown to drive the differentiation of Tpex into Tex-int and/or Tex-term cells in mice,12–17 and in humans, CD8+ T cells in the TME have been observed to adopt Tpex, Tex-int, or Tex-term fates.25,27 Indeed, we observed an increase in the frequency of PD-1+ TCF-1- CD8+ T cells with low expression of inhibitory receptors, resembling Tex-int, with increased Ki-67 expression in the uiLN after ICB. Based on the kinetics of Ki-67 expression as a function of the cell cycle,55 these cells may be actively proliferating or have recently differentiated from dividing cells, such as Tpex. Consistent with this notion, Tpex and Tex-int were in closer proximity to one another in uiLN after ICB. In blood samples, we also observed a corresponding increase in the frequency of Tex-int, but not Tpex, between baseline and post-treatment surgery timepoints. Tpex themselves were rare in blood, while Tex-int were not proliferative in paired tumors. Thus, our data suggest a model in human cancer patients in which ICB drives Tpex in the uiLN to differentiate into Tex-int, which then transit through the blood and ultimately arrive at the tumor, where they become more terminally exhausted.
Because Tpex are critical to sustain endogenous and ICB-mediated CD8+ T cell responses, determining the cellular niche that regulates Tpex in human patients will be critical for harnessing them more reproducibly across cancer patients. Our data suggest that Tpex and Tex-int are localized closely with DCs in the uiLN, and this interaction increases with ICB treatment. These observations parallel previous studies demonstrating that Tpex and Tex-int in the TME reside near antigen-presenting cells in human and mouse tumors.25,56,57 In mouse studies of chronic infection, DCs promote Tpex maintenance in the spleen58. CD28 expression, the ligands of which are typically expressed on DCs, is necessary for the proliferative burst of CD8+ T cells after ICB.59 DC expression of PD-L1 also restricts CD8+ T cell responses in mouse tumor models.41 Thus, in human cancer patients, our data support the notion that ICB results in greater interactions between Tpex and DCs, promoting proliferation and differentiation of Tex-int cells.
Our data also add to a developing understanding of the impact of metastases in LN on the anti-tumor immune response. In a mouse model of melanoma, LN metastasis drove immunosuppression, including induction of Tregs and alterations in DC phenotypes, resulting in impaired CD8+ T cell responses.60 Moreover, in human melanoma patients, metLN exhibited higher expression of immunosuppressive genes and reduced lymphocyte activation associated with distant recurrence.60,61 Our data indicate that cellular niches surrounding Tpex and Tex-int become more immunosuppressive in metLN from recently treated patients, including elevated expression of inhibitory proteins on DCs and Tregs as well as more naïve and quiescent CD4+ T cells. Patients with metLN also experienced weaker CD8+ T cell responses in the blood following treatment, supporting an important role for LN in generating circulating CD8+ T cell responses that associate with clinical response.
In summary, our data highlight the important role of CD8+ T cell responses in human LN at steady-state and after ICB immunotherapy while also revealing the disruption of these key processes by LN metastasis. These results lay a foundation for the future development of immunotherapies that optimally harness anti-tumor immunity in human LN and inform immune monitoring strategies for cancer patients treated with ICB.
Limitations of the Study:
While our results indicate that key CD8+ T cell subsets in human LN have more DC neighbors after PD-L1 blockade, we have not yet determined the mechanisms responsible. It is possible that DC migration from the tumor into the LN is enhanced or that these cells change their relative localization with one another after treatment. Because we did not observe an increase in the total frequency of DCs after anti-PD-L1 treatment (Fig. S4E), we favor the latter possibility. Additionally, the specific chemokines or adhesion molecules that regulate this remain to be determined. Several studies of Tpex in mouse models of cancer and chronic infection identified their expression of the gene Xcl1,62–64 which encodes a DC chemoattractant that could play a role. The clusters annotated as Tex-int in the mass cytometry analysis and MIBI analysis had minor differences in their expression of some proteins, though the key proteins used to annotate these cell subsets were consistent. These differences may be attributable to distinct antibody clones and sample preparation methods between the assays; for instance, intracellular protein stores are accessible for staining by MIBI, while cell-surface antigens are stained for mass cytometry before cell permeabilization. Moreover, the molecular mechanisms that result in the observed reduction of Tpex and suppression of Tpex and Tex-int responses after ICB in metLN remain to be determined.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Matthew Spitzer (matthew.spitzer@ucsf.edu).
Materials availability
No new materials were generated during the course of this study.
Data and code availability
All mass cytometry and multiplexed ion beam imaging data are available on Mendeley Data under doi:10.17632/2zgppyr2rr.1. Single-cell genomics data have been deposited into GEO under accession number GSE212797.
This paper does not report original code, however available software packages used for analysis are described in the STAR methods section and Key Resource Table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| For a list of antibodies used for analysis by mass cytometry (CyTOF), multiplexed ion beam imaging (MIBI), and CITE-seq, please see Supplemental Table S5. | Various | Various |
| Human TruStain FcX (Fc Receptor Blocking Solution) |
Biolegend | Cat#422301 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Collagenase IV | Worthington | Cat#LS004188 |
| Deoxyribonuclease I | Sigma-Aldrich | Cat#DN25 |
| cis- Diammineplatinum(II) dichloride (Cisplatin) |
Sigma-Aldrich | Cat#P4394 |
| Paraformaldehyde aqueous solution | Electron Microscopy Sciences |
Cat#15710 |
| Zombie Aqua Fixable Viability Dye |
Thermo Fisher | Cat#L34966 |
| Candor PBS Antibody Stabilization solution |
Candor Bioscience |
Cat#13150 |
| Maxpar 10X Barcode Perm Buffer | Fluidigm | Cat#201057 |
| Cell-ID Intercalator-IR | Fluidigm | Cat#201192A |
| Maxpar Cell Acquisition Solution |
Fluidigm | Cat#201240 |
| Target Retrieval Solution | Dako Agilent | Cat#S1699 |
| PBS IHC Wash Buffer + Tween 20 |
Cell Marque | Cat#934B-06 |
| EQ Four Element Calibration Beads | Fluidigm | Cat#201078 |
| ACK Lysing Buffer | Thermo Fisher | Cat#A1049201 |
| Cell Staining Buffer | Biolegend | Cat#420201 |
| Normal Donkey Serum | Sigma-Aldrich | Cat#566460 |
| Critical Commercial Assays | ||
| Chromium Next GEM Single Cell 5’ Library |
10X Genomics | Cat#PN-1000265 |
| Kit | ||
| Chromium Single Cell V(D)J Enrichment kit |
10X Genomics | Cat#PN-1000005 |
| 5’ Feature Barcode Kit |
10X Genomics | Cat#PN-1000256 |
| MaxPar X8 Antibody Labeling Kits | Fluidigm | Various |
| Metal-Loaded MIBItags | IonPath | Various |
| Avidin/Biotin Blocking System |
Biolegend | Cat#927301 |
| Deposited Data | ||
| Single-cell genomics data | Generated by current investigators | Deposited into GEO under accession number GSE212797 |
| Mass cytometry and multiplexed ion beam imaging data | Generated by current investigators | Deposited into Mendeley Data under doi:10.17632/2zgppyr2rr.1 |
| Software and Algorithms | ||
| R Core Team (v.4.0.5) |
The R Project for Statistical
Computing |
https://www.r-project.org/ |
| RStudio | Posit | https://posit.co/ |
| Python (v.3.88) | Python Software Foundation. Python Language Reference, version 3.8. |
http://www.python.org |
| ParkerICI/Premessa | Parker Institute for Cancer
Immunotherapy |
https://github.com/ParkerICI/premessa |
| CellEngine | CellCarta | https://cellcarta.com/cellenginesoftware/ |
| CytoNorm | Van Gassen et al., 2020 | https://github.com/saeyslab/CytoNorm |
| Flowcore | Ellis et al., 2022 | https://bioconductor.org/packages/release/bioc/html/flowCore.html |
| CATALYST | Nowicka et al., 2017; Chevrier et al., 2018 | https://github.com/HelenaLC/CATALYST |
| FlowSOM | Van Gassen et al, 2015 | https://github.com/SofieVG/FlowSOM |
| diffcyt | Weber et al., 2019 | https://github.com/lmweber/diffcyt |
| Mesmer | Greenwald et al., 2021 | https://github.com/vanvalenlab/kiosk-console |
| scanpy (v.1.7.1) | Wolf et al., 2018 | https://github.com/scverse/scanpy |
| Harmony (v.0.05) | Korsunsky et al., 2019 | https://github.com/immunogenomics/harmony |
| sklearn (v.0.24.1) | Pedregosa et al., 2011 | https://github.com/scikit-learn/scikit-learn |
| corrplot | Simko, 2021 | https://github.com/taiyun/corrplot |
| scipy (v.1.6.1) | Virtanen et al., 2020 | https://github.com/scipy/scipy |
| pandas (v.1.2.3) |
McKinney,
2010; Pandas Development Team |
https://github.com/pandas-dev/pandas |
| numpy (v.3.3.4) | Harris et al., 2020 | https://github.com/numpy/numpy |
| matplotlib (v.3.3.4) | Hunter, 2007 | https://github.com/matplotlib/matplotlib |
| statsmodels (v.0.12.2) | Seabold and Perktold, 2010 | https://github.com/statsmodels/statsmodels |
| dplyr | Wickham et al., 2021 | https://github.com/tidyverse/dplyr |
| reshape2 | Wickham, 2007 | https://github.com/cran/reshape2 |
| ggplot2 | Wickham, 2009 | https://github.com/tidyverse/ggplot2 |
| RColorBrewer | Neuwirth, 2014 | https://cran.r-hub.io/web/packages/RColorBrewer/index.html |
| Other | ||
| gentleMacs C Tubes | Miltenyi Biotec | Cat#130–093-237 |
| Gold-Coated Slides | IonPath | 567001 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Subject consent and biospecimen collection
Subjects were enrolled using a consecutive sampling approach and provided informed consent under UCSF IRB approved protocols (UCSF IRB# 14–15342 and IRB# 18–25114) for collection of blood before and after surgery as well as collection of tumor, lymph node, and blood on the day of their surgery. Tissue samples were obtained from tumor resection specimens on the day of surgery by UCSF Pathology Assistants; blood was collected in EDTA coated vacutainer tubes in the operating room prior to surgery. Tissue specimens were placed in ice cold Leibovitz’s L-15 medium in a 50 mL conical tube and along with blood samples immediately transported on ice to the laboratory for preparation for either mass cytometry or single-cell RNA sequencing and TCR sequencing (see additional tissue and blood processing details below). Demographic information, including age and sex, and all clinical information for each subject cohort can be found in Table S1, S2, and S4.
Pathologic review of formalin fixed paraffin embedded tissues
All formalin fixed paraffin embedded tissues used in this study were reviewed by a board certified oral and maxillofacial pathologist (K.B.J.).
METHOD DETAILS
Tumor, lymph node, and blood processing for mass cytometry
Tumor and lymph node samples were finely minced and digested in Leibovitz’s L-15 medium with 800 U/ml collagenase IV and 0.1 mg/ml DNase I with gentle agitation for 45 minutes at 37°C. After digestion, cells were filtered through a 70μm filter into PBS/5mM EDTA solution, spun down at 500g for 5 minutes at 4˚C, the supernatant aspirated, and resuspended in fresh PBS/EDTA solution and kept on ice.
Blood samples were mixed with Ammonium-Chloride-Potassium Lysis Buffer at room temperature for 3 to 5 minutes to lyse red blood cells, centrifuged at 300g for 5 minutes at 4°C, the supernatant aspirated, and resuspended in fresh PBS/EDTA solution and kept on ice.
Cells from both tissue and blood were then washed with PBS/EDTA and re-suspended 1:1 with PBS/EDTA and 50 μM cisplatin for 60 seconds at room temperature before quenching 1:1 with PBS/5mM EDTA/0.5% BSA to determine viability as previously described65. Cells were centrifuged at 500g for 5 min at 4 °C and re-suspended in PBS/EDTA/BSA at a density between 1 × 10^6 and 10 × 10^6 cells per ml. Suspensions were fixed for 10 min at room temperature using 1.6% paraformaldehyde (PFA) and frozen at −80 °C until ready to be run for CyTOF.
Tumor and lymph node processing for single-cell sequencing
Tumor and lymph node samples were thoroughly minced with surgical scissors and transferred to GentleMACs C Tubes containing 800 U/ml Collagenase IV and 0.1 mg/ml DNase I in L-15/2% FCS per 0.3 g tissue. GentleMACs C Tubes were then installed onto the GentleMACs Octo Dissociator (Miltenyi Biotec) and incubated for 20 min (lymph node) or 35 min (tumor) according to the manufacturer’s instructions. Samples were then quenched with 15 mL of sort buffer (PBS/2% FCS/2mM EDTA), filtered through 100μm filters and spun down. Red blood cell lysis was performed with 175 mM ammonium chloride, if needed.
Cells were then incubated with Human TruStain FcX Receptor Blocking Solution to prevent non-specific antibody binding before staining with Zombie Aqua Fixable Viability Dye and anti-human CD45 antibody in PBS/2%FCS/2mM EDTA/0.01% sodium azide and incubated for 25 minutes on ice in the dark. Live CD45+ and CD45- cells were sorted on a BD FACSAria Fusion. CD45+ and CD45-cells were pelleted and resuspended at 1×10^3 cells/ml in 0.04%BSA/PBS buffer before mixing in an 8:2 CD45+:CD45- ratio and loaded onto the Chromium Controller (10X Genomics) to generate 5’ v1.1 gel beads-in-emulsions (GEM). For CITE-Seq samples, pooled 8:2 CD45+:CD45- cells were resuspended in Cell Staining Buffer (BioLegend) and stained with a pool of 137 TotalSeq-C antibodies (Table S5) according to the manufacturer’s protocol before loading onto the Chromium Controller (10X Genomics) for GEM generation. The cDNA libraries were generated using all or a subset of Chromium Next GEM Single Cell 5’ Library Kit for gene expression (GEX), Chromium Single Cell V(D)J Enrichment kit (10X Genomics) for T cell receptor (TCR), and Chromium Single Cell 5’ Feature Barcode Library kit for antibody derived tag (ADT) according to the manufacturer’s instructions. The libraries were subsequently sequenced on a Novaseq S4 sequencer (Illumina) to generate fastqs with the following mean reads per cell: 42,000 (GEX), 34,000 (TCR), and 5,700 (ADT).
Tissue microarray fabrication for multiplexed ion beam imaging
For all patient lymph node samples used for MIBI analysis, hematoxylin and eosin (H&E) stained tissue sections prepared by the UCSF Department of Pathology as part of normal patient care were retrieved and reviewed by K.B.J. In general, lymph nodes used in the study were selected from neck levels most likely to represent areas of lymphatic drainage from patient tumors. 2mm diameter cores in both the cortex and paracortex regions of each lymph node were manually annotated on the H&E slides. In lymph nodes with metastatic disease, 2mm cores were also obtained from the tumor-lymph node interface. The slides were then used to select 2mm cores from their corresponding paraffin blocks using a 3D-Histech Tissue Microarray Master II robot computer assisted/image-guided production system. We created two, 5 × 12 tissue microarrays following the manufacturer’s instructions. Cores of a single sample of normal human tonsil were also included in duplicate as controls on each microarray.
Antibody heavy metal conjugation for mass cytometry
The sources for all mass cytometry antibodies can be found in Table S5. Antibodies were conjugated to their associated metals with MaxPar X8 labeling reagent kits according to manufacturer instructions, diluted with Candor PBS Antibody Stabilization solution supplemented with 0.02% sodium azide, and filtered through an UltrafreeMC 0.1-mm centrifugation filter (Millipore) before storage at 4°C. Surface and intracellular master antibody cocktails were made and kept at −80°C.
Antibody heavy metal conjugation for multiplexed ion beam imaging
Antibodies were conjugated to metal-loaded MIBItags (Ionpath) according to manufacturer instructions, and stored at 4°C prior to use.
Mass-tag cellular barcoding for mass cytometry
Prior to antibody staining, mass tag cellular barcoding of prepared samples was performed by incubating cells with distinct combinations of isotopically-purified palladium ions chelated by isothiocyanobenzyl-EDTA as previously described.66 After counting, 1 × 10^6 cells were barcoded with distinct combinations of stable Pd isotopes for 15 min at room temperature on a shaker in Maxpar Barcode Perm Buffer. Cells were washed twice with cell staining media (PBS with 0.5% BSA and 0.02% NaN3), and pooled into a single 15 mL tube for subsequent staining and washing steps.
Mass cytometry staining
Barcoded cells were stained with Human TruStain FcX Receptor Blocking Solution at 20 mg/mL for 5 min at RT on a shaker. Surface antibody cocktail was then added with a 500ul final reaction volume for 30 min at RT on a shaker. Following staining, cells were washed twice with cell staining media. Before intracellular staining, cells were permeabilized for 10 min with methanol at 4°C. Methanol was then removed by washing the cells 2 times with cell staining media. The intracellular cocktail was then added to the cells for a 500uL final reaction volume for 1 h at RT on a shaker. Cells were washed twice in cell staining media to remove unbound antibodies and then stained with 1mL of 1:4000 191/193Ir Cell-ID Intercalator Solution diluted in PBS with 4% PFA overnight. Before mass cytometry was run, cells were washed once with cell staining media, and twice with Cell Acquisition Solution.
Multiplexed ion beam imaging tissue section staining
Tissue sections (5 μm thick) were cut from FFPE blocks of the tissue microarrays and mounted on gold- and tantalum-sputtered microscope slides (IonPath). Slides were baked at 70°C for 1 hr followed by deparaffinization and rehydration with sequential washes in xylene (2x), 100% ethanol (2x), 95% ethanol (2x), 80% ethanol (1x), 70% ethanol (1x), and ddH2O (2x) with a Leica ST4020 Linear Stainer (Leica Biosystems) programmed for 30 s each. Tissues next underwent antigen retrieval by submerging slides in Target Retrieval Solution (pH 9) and incubating them at 97°C for 40 min and cooled down to 65ºC in a Lab Vision PT Module (Thermo Fisher Scientific). Slides were further cooled to room temperature and washed in 1x phosphate-buffered saline (PBS) IHC Washer Buffer with Tween 20. Next, endogenous biotin and avidin proteins were successively blocked using an Avidin/Biotin Blocking System, for 10 min each. Tissues were then washed with wash buffer and blocked for 1 h at room temperature with 1x PBS IHC Wash Buffer with Tween 20 and 5% (v/v) normal donkey serum (Sigma-Aldrich). Two antibody panels were prepared. The first antibody cocktail was prepared in 1x PBS IHC Wash Buffer with Tween 20 with 5% (v/v) normal donkey serum and 0.5 mM EDTA and filtered through a 0.1-mm centrifugal filter (Millipore) prior to incubation with tissue overnight at 4° C in a humidity chamber. Following the overnight incubation, slides were washed twice for 5 min in wash buffer. The secondary antibody cocktail was prepared as described above and incubated with the tissues for 1 h at room temperature in a humidity chamber. Slides were dried under vacuum prior to imaging.
Mass cytometry data acquisition
Mass cytometry samples were diluted in Cell Acquisition Solution containing bead standards to approximately 1 × 10^6 cells/mL and then analyzed on a Helios mass cytometer (Fluidigm) equilibrated with Cell Acquisition Solution. A minimum of 10 × 10^6 cell events were collected for each barcoded set of samples at an event rate of 400–500 events/second.
Multiplexed ion beam imaging data acquisition
Imaging was performed using a MIBI-TOF instrument (IonPath) with a Hyperion ion source. Xe+ primary ions were used to sequentially sputter pixels for a given field of view. The following imaging parameters were used: acquisition setting: 80 kHz; field size: 800×800 μm, 2048 × 2048 pixels; dwell time: 1 ms; median gun current on tissue: 5.5 nA Xe+.
Data normalization and de-barcoding for mass cytometry
Bead standard data normalization and de-barcoding of the pooled samples into their respective conditions was performed using the R package from the PICI institute available at https://github.com/ParkerICI/premessa.
QUANTIFICATION AND STATISTICAL ANALYSIS
Mass cytometry batch normalization
Each group of barcoded samples was run with a control sample (replicates of a single normal human tonsil) to validate staining and for normalization between groups of barcoded samples. Bead standard data normalization and de-barcoding of the pooled samples into their respective conditions was performed using the R package Premessa from the PICI institute available at https://github.com/ParkerICI/premessa. All manually gated live, intact, single cells were downloaded as FCS files from CellEngine (CellCarta, Montreal, Canada). CytoNorm67 (https://github.com/saeyslab/CytoNorm) was utilized to correct for batch effects. All markers were used for batch effect normalization.
Mass cytometry manual gating
Batch effect normalized FCS files were uploaded to CellEngine for manual gating (Figure S1A).
Mass cytometry clustering
Manually gated CD8+ T cells were downloaded as FCS files from CellEngine, and flowCore68 was used to import FCS files into R. The FlowSOM clustering algorithm33, available through the CATALYST R/Bioconductor package69,70, was used to generate clusters based on CD8+ T cell specific markers (Granzyme B, CD38, CD127, CD45RA, TIM3, TIGIT, PD-L1, CD27, CD39, Tbet, CD103, FoxP3, CD69, CCR7, CD25, TCF-1, Pan-HLA-DR, PD-1, CD56, CD16, CD7, CD95). We ran FlowSOM with a 10 × 10 grid and utilized the built-in ConsensusClusterPlus71 metaclustering step to obtain 20 CD8+ T cell clusters. Clusters were visualized using UMAPs via the umap package72 and heatmaps via the ComplexHeatmap package73, both available through CATALYST. For paired differential abundance analyses, we used generalized linear mixed models implemented in the diffcyt R package74.
Low level image processing for multiplexed ion beam imaging
To remove background generated from gold, oxides and adducts, as well as compensate for channel cross talk. We used the Rosetta algorithm, which uses a flow-cytometry style compensation approach to remove spurious signals. All compensation parameters were first evaluated in a subset of 10 images to ensure contaminant signal was removed while preserving target signal. Next, finalized parameters were applied to the full image set.
Region Masks for multiplexed ion beam imaging
Region masks were generated to define histologic regions of each FOV including the T-cell-zone, B-cell-zone, tumor, endothelium, or other. The tumor mask was first generated by applying smoothing (Gaussian blur, radius 2 px) and a pixel thresholder to Keratin signal. To generate the B-cell-zone mask, tumor mask was subtracted from the FOV. In the remaining area, smoothing and pixel thresholder were applied to CD20 signal. T-cell zone and Endothelium masks were similarly generated using CD3 and CD31 signal, respectively and any remaining area of the FOV was classified as “Other”. For each region, all holes were filled.
Single-cell segmentation for multiplexed ion beam imaging
To delineate the location of single cells in MIBI-TOF images, we performed cell segmentation using the pre-trained Mesmer convolutional neural network architecture36. We used dsDNA as the nuclear marker and HLA Class 1, ABC as the membrane marker as input to the network. The output of Mesmer is the location of each cell in the image. Cells with area larger than 200 μm2 were frequently due to out-of-focus regions of an image and therefore excluded from analysis. Additionally, FOVs with fewer than 3500 total cells were excluded from analysis.
Single-cell phenotyping for multiplexed ion beam imaging
Single cell phenotyping was accomplished using a previously described method75. Pre-processed MIBI-TOF images were first Gaussian blurred using a standard deviation of 2 for the Gaussian kernel. To account for both technical and biological confounders, pixels were normalized by their total expression, such that the total expression of each pixel was equal to 1. A 99.9% normalization was applied for each marker. Pixels were clustered into 100 clusters using FlowSOM33 based on the expression of 24 phenotypic markers: Foxp3, CD11b, CD11c, CD138, CD14, CD16, CD163, CD20, CD21, CD3, CD31, CD4, CD45, CD56, CD68, CD8, E-Cadherin, HLADR, Keratin, MPO, PD-1, T-bet, Vimentin, BDCA3. The average expression of each of the 100 pixel clusters was found and the z-score for each marker across the 100 pixel clusters was computed. Using these z-scored expression values, the 100 pixel clusters were meta-clustered using consensus hierarchical clustering. These meta-clusters were manually inspected and adjusted by comparing with the MIBI-TOF images. Next, by applying the segmentation masks that delineate the boundaries of all cells in the images, we counted the number of each of the pixel clusters in each cell. This resulted in a pixel cluster by cell count table. These counts were then normalized by cell size. Using these frequency measurements as the feature vector, the cells were clustered using FlowSOM into 100 cell clusters. Similarly to the pixel clusters, the average expression of each of the 100 cell clusters was found and the z-score was computed. Using these z-scored values, the 100 cell clusters were meta-clustered using consensus hierarchical clustering. Each of the cell meta-clusters was then manually inspected and adjusted by comparison with the images, then annotated with its cell phenotype. Clustering in this manner resulted in better cluster definition than clustering using integrated expression.75 Cell populations were refined by inspecting biaxial plots of integrated marker expression for each single cell.
CD8+ T cells were further subclustered using integrated marker expression for each single cell normalized by cell size. The following markers were used: CD103, CD16, CD27, CD39, CD45RO, CD56, CD69, Foxp3, Granzyme B, HLADR, IFNG, PD-1, PD-L1, T-bet, TCF-1, TIM3. Cells were clustered using FlowSOM into 100 clusters and meta-clustered using consensus hierarchical clustering. The final meta-clusters were manually inspected and adjusted by comparing with MIBI-TOF images.
Neighborhood analysis for multiplexed ion beam imaging
Cell neighborhoods were produced as previously described.40 A cell neighbor matrix was generated, in which each row represents an index cell and columns indicate the number of each cell phenotype within a 50-pixel radius of the index cell. A similar approach was used to generate a cell neighbor matrix of cells positive for functional markers. Functional marker thresholds were determined by manually visualizing images. Each cell in the dataset was determined to be positive or negative using these manual thresholds, then the number of cells positive for each functional marker within a 50-pixel radius of each cell was determined.
Alignment of single-cell sequencing libraries
The raw fastq files were aligned using Cell Ranger v.3.0.2 and v.6.0.2 software with the default settings to the hg38 transcriptome and the TotalSeq-C Feature Reference provided by BioLegend for the GEX and ADT fastqs, respectively. The raw TCR fastq files for each participant from the lymph node and tumor were aligned jointly in order to call shared clonotypes across the tissues using Cell Ranger v.7.0.0 to the vdj GRCh38 v.7.0.0 reference.
Demultiplexing pooled single-cell samples
The GEX BAM alignment files from each of the pools were used as the input for dsc-pileup v.0.1beta to generate pileup files which were then used as input for freemuxlet v.0.1beta for doublet identification, inferred genotypes for individuals in each pool, and singlet assignment to those inferred genotypes. The percentage of shared single nucleotide polymorphism (SNP) assignments from the freemuxlet genotypes was used to match samples from the two individuals in the lymph node pool and the tumor pool to each other. The two samples in each pool were from a male and a female participant so the sex assignment (based on XIST expression and the percentage of total counts based on unique molecular identifiers (UMIs) mapping to Y chromosome genes; Fig. S2J) was used to match the freemuxlet IDs to the clinical trial participant IDs.
Single-cell sequencing full dataset clustering
We filtered out 747 cells with less than 100 or more than 3,000 genes detected and filtered out 4,883 genes detected in less than 3 cells. We also filtered out 1,943 cells with more than 10% of total counts (UMIs) mapping to mitochondrial genes and 70 cells identified as red blood cells (based on HBB expression) or platelets (based on PF4 expression). For the multiplexed samples, we also filtered out genetic doublets (629 cells) identified by freemuxlet. The raw counts were normalized to 10,000 counts per cell and log(count + 1) transformed. We identified 1,098 highly variable genes which were scaled and used with the default settings in scanpy v.1.7.176 for PCA analysis. We used Harmony v.0.0.577 for batch correction in the PC space with each sample as a batch. The top 20 corrected PCs from Harmony were used for nearest neighbor detection followed by leiden78 clustering and UMAP79 projection. This analysis identified 22 clusters which we collapsed into 13 cell types based on marker gene expression.
Single-cell sequencing CD8 T cell clustering
For the ADT data, we filtered out isotype control antibodies (7 total in the 137 panel) and normalized the raw counts with CLR (count + 1) by cell. To identify CD8 T cells by protein, we used a Gaussian mixture model tool from sklearn v.0.24.1 to create positive and negative gates for CD3 protein, CD8 protein, and CD4 protein on the normalized expression for each marker. We assigned CD8 T cells by including 1) cells that expressed CD3 (CD3E or CD3D) and CD8 (CD8A or CD8B) by RNA and did not express CD4 by RNA or protein, 2) cells gated as CD8 T cells by protein [CD3+CD8+CD4-], and 3) cells in the clusters annotated “CD8 T cell” that did not express CD4 by RNA or protein. After removing 11 tumor cells (based on KRT14 expression) from the resulting cells, we subsetted the raw count matrix from the CD8 T cells (8,245 cells) and removed 9,251 genes that were not expressed in at least three cells. The counts from the remaining 13,305 genes were normalized to 10,000 counts per cell, log (count + 1) transformed, and scaled. We used the default settings in scanpy v.1.7.1 for PCA dimensionality reduction. We iteratively used Harmony v.0.0.5 for batch correction in the PC space with sex, cell cycle phase, and sample as the batch IDs. The top 20 corrected PCs were used for nearest neighbor detection followed by leiden clustering and UMAP projection. This identified 13 clusters which we collapsed into 9 CD8 cell sub-types based on marker gene expression.
CD8 T cell gene set scoring
We used the 43 gene S phase gene set excluding 1 gene (MLF1IP) and the 54 gene G2M phase gene set excluding 2 genes (FAM64A, HN1) to assign cells to S, G1, and G2M cell cycle phase with the score_genes_cell_cycle function from scanpy v.1.7.1. We used the 399 gene TCF7+ Tex gene set excluding 15 genes (LHFP, IGFL2, C1orf228, UNQ6494, SEPT6, C16orf45, ATP5G2, GPX1, NTRK2, ATP5A1, TMEM2, C6orf48, C20orf196, TMEM256-PLSCR3, ATP5D) to assign single cell Tpex scores and the 1,261 gene terminal Tex gene set excluding 24 genes (UNQ6494, C16orf45, LHFP, ATP5G3, ATP5J2, ATP5C1, SEPT2, ASNA1, ATP5G1, ATP5J, ATP5A1, ATP5G2, NTRK2, ATP5D, PLA2G16, RTFDC1, C19orf24, ATP5B, SEPT7, FAM96A, ATPIF1, FAM96B, C20orf24, C17orf62) to assign single cell Tex-term scores with the score_genes function from scanpy v.1.7.1. All excluded genes were not detected in our CD8 T cells. S phase gene set and G2M phase gene set were from 80 TCF7+ Tex gene set and terminal Tex gene set were from 28.
Shared clone assignment
Cells with <=2 TRA chains and <=1 TRB chains were used in the TCR clonotype analyses.
Clonotypes were assigned to CD8 T cells for the four paired tumor and lymph node samples with a TCR library. Cells were marked as having a “shared clone” status if the cells from a paired lymph node and tumor were assigned the same clonotype by Cell Ranger.
Statistical analysis
For CyTOF and MIBI data, all statistical tests were performed in R.81,82 The non-parametric Wilcoxon rank sum test was utilized to compare samples from anti-PD-L1 treated patients to standard of care treated patients and metLN to uiLN for MIBI data. The paired Wilcoxon rank sum test and generalized linear mixed models were used for paired CyTOF data. We used Spearman’s correlation to measure the correlation between cluster abundances in paired data, which were plotted using the corrplot package.83 For multiple testing corrections, we applied Benjamini-Hochberg correction and statistical differences were declared significant at FDR < 0.1. For hypothesis testing, statistical differences were declared significant at P < 0.05.
The R packages dplyr84 and reshape285 were used for data manipulation. Most plots were produced with ggplot286 and RColorBrewer.87
For single cell sequencing data, all statistical tests were performed in python v.3.8.8.88 We compared Tpex scores and Tex-term scores in shared clones in the tumor versus lymph node using a linear mixed effect model from statsmodels v.0.12.289 with tissue origin as a fixed effect and clonotype ID as a random effect. We used a Fisher’s exact test from scipy v.1.6.190 to test for an association between tissue origin (lymph node or tumor) and cell sub-type assignment (cluster or Tex-term) for the cells in those cell sub-types that were shared clones. We used pandas v.1.2.391,92 and numpy v.3.3.493 were used for data manipulation in python. We used scanpy v.1.7.1 and matplotlib v.3.3.4 (Matplotlib: A 2D Graphics Environment) for creating plots in python.
Statistical parameters (e.g., sample size [n], etc.) are detailed in the main text and figure legends for each dataset.
ADDITIONAL RESOURCES
All samples used from subjects treated with atezolizumab were from clinical trial NCT03708224. Additional information about this clinical trial can be viewed at https://clinicaltrials.gov/ct2/show/NCT03708224.
Supplementary Material
Related to Figure 1 and STAR methods.
Related to Figure 2 and STAR methods.
Related to Figure 2 and STAR methods.
Related to Figures 3, 4 and 6 and STAR methods.
Related to STAR Methods.
Related to Figure 1. A) Gating strategy. B) Relative abundance of main immune cell types. C) UMAP of all CD8 T cell clusters. D) Relative abundance of non-naive CD8 T cell clusters. Numbers below indicate the number of Tpex and non-naive CD8 T cells analyzed from each sample.
Related to Figure 2. A) UMAP of 35,551 cells from 5 paired tumor and draining lymph node samples (10 samples total). Cells are colored according to annotated cell type. B) Dot plot of standard scale (subtract minimum and divide by maximum) column normalized expression for the indicated genes for each annotated cell type from all cells indicating percentage of cells with expression greater than zero (dot size) and mean expression for cells with nonzero expression (color). C) UMAP of 8,245 CD8 T cells from 5 paired tumor and draining lymph node samples (10 samples total) colored by annotated cell subset. D) Heatmap of gene expression by cluster. E) UMAP of all cells with CITE-seq data colored by normalized expression of CD3 protein, CD8 protein, and CD4 protein. F) Heatmap of CITE-seq protein expression by cluster. G) Frequencies of non-naive CD8 T cell clusters by tissue. H) Gene set scores from Zheng et al. (2021) by cluster. I) Bar plots of percentage of total CD8 T cells with TCR clonotype assigned for each sample. J) Violin plots of XIST expression and percentage of counts from Y chromosome genes for sample demultiplexing.
Related to Figure 3. A) Average marker intensity for each pixel cluster. B) Average pixel assigmnent for each cell cluster. C) Representative single-parameter images for each antibody used for MIBI analysis. D) Relative abundance of main immune cell types per LN in the T cell zone or B cell zone. Sample ID represents patient ID followed by LN number. E) Relative abundance of CD8 T cell clusters per LN in the B cell zone and global. F) Relative frequency of Tpex (c18) in the B cell zone regions and T cell zone regions of the uiLN. P-value obtained by two-tailed Wilcoxon Rank Sum Test.
Related to Figure 4. A) Relative abundance of main immune cell types in uiLN (global, T cell zone, and B cell zone) from anti-PD-L1 treated patients. Sample ID represents patient ID followed by LN number. B) Relative abundance of non-naïve CD8 T cell clusters in uiLN (global and B cell zone) from anti-PD-L1 treated patients. Sample ID represents patient ID followed by LN number. C) Percent Tpex (c18) of non-naive CD8 T cells in the B cell zone from untreated versus anti-PD-L1 treated patients. P-values obtained by Wilcoxon Rank Sum Test. D) Difference in the number of DC neighbors between untreated and anti-PD-L1 treated patients for each cluster of CD8 T cells (Wilcoxon Rank Sum Test). Colors indicate if DC neighbors are significantly more abundant in anti-PD-L1 (purple) or SOC (blue) or not differentially abundant (False, grey) after Benjamini-Hochberg correction, FDR < 0.05. E) Percent DC of total immune cells from untreated versus anti-PD-L1 treated patients. F-G) Spearman correlation between % DC of total immune cells and F) % Tpex (c18) or G) % Tex-int (c16) of total non-naive CD8 T cells in samples from patients receiving SOC or PD-L1 blockade.
Related to Figure 5. A) Samples collected from each patient. Time (days) are relative to the first sample collected. B) Relative abundance of main immune cell types. C) Principal component analysis of gated main immune cell populations. D) UMAP of all CD8 T cell clusters. E) Frequencies of non-naïve CD8 T cell clusters. F) Percentage of cluster 9 proliferating cells in paired samples from blood at time of surgery and baseline. P-value obtained by paired Wilcoxon Rank Sum Test. G) Paired differential abundance analysis of non-naïve CD8 T cell subsets between tumor and blood at time of surgery (n = 6) (generalized linear mixed models). See color scheme for Figure 6C. H) Cluster 5 abundance (as percentage of non-naïve CD8 T cells) in paired samples from blood and tumor at time of surgery. P-values obtained by generalized linear mixed models. I) Correlation between clusters in tumor and blood at time of surgery (Spearman correlation). J-K) Correlation between clusters in LN (MIBI data) and clusters in blood (CyTOF data) (J) and clusters in LN (MIBI data) and clusters in tumor (CyTOF data) (K) at time of surgery (Spearman correlation).
Related to Figure 6. A) Principal component analysis of gated main immune cell populations in uninvolved lymph nodes (uiLN), metastatic lymph nodes (metLN), and tumors. B) Relative abundance of main immune cell types for patients 11084 and 8158. C) Paired differential abundance (DA) analysis of main immune cell populations between uiLN and metLN (n = 7; paired Wilcoxon Rank Sum Test). The log2 fold changes are plotted against the negative log10(nominal p-values). Colors indicate if cell populations are significantly more abundant in uiLN (purple) or tumor/metLN (blue) or not differentially abundant (False, grey) after Benjamini-Hochberg correction, FDR < 0.1. D) Paired differential abundance analysis of non-naïve CD8 T cell subsets between metLN and tumor (n = 8) (generalized linear mixed models). Grey: not differentially abundant after Benjamini-Hochberg correction, FDR < 0.1. E) Relative abundance of main immune cell types in met-cores (global) from anti-PD-L1 treated patients. Sample ID represents patient ID followed by LN number and core number. F) Main immune cell type ratios represented as log2 fold changes between met-cores (global) and ui-cores (global) from antiPD-L1 treated patients. G) Relative abundance of non-naïve CD8+ T cell clusters in metastatic cores (global, n = 5) from anti-PD-L1 treated patients (n = 3). Sample ID indicates patient ID followed by LN number and core number. H) Non-naïve CD8+ T cell cluster ratios represented as log2 fold changes between met-cores (global, n = 5) and ui-cores (global, n = 39) from anti-PD-L1 treated patients. I) Cluster 18 and 16 abundances (as percentage of non-naïve CD8 T cells) in met-cores (global) and ui-cores (global) from anti-PD-L1 treated patients, and ui-cores (global) from SOC treated patients. P-values obtained by Wilcoxon Rank Sum Test. J-K) Percentage of cluster 18 (J) and cluster 16 (K) cells with a specific cell type as its neighbor in met-cores (global) from anti-PD-L1 treated patients. L) Expression of CD39, IDO1, PD-L1, and TIM3 on DCs neighboring cluster 16 cells in ui-cores (global) and met-cores (global) from SOC and anti-PD-L1 treated patients. P-values obtained by Wilcoxon Rank Sum Test. M) Expression of CD39 and Ki-67 on Tregs neighboring cluster 18 cells (left) and expression of CD39, FoxP3, Ki-67, and TIM-3 on Tregs neighboring cluster 16 cells (right) in ui-cores (global) and met-cores (global) from SOC and anti-PD-L1 treated patients. P-values obtained by Wilcoxon Rank Sum Test. N) Expression of PD1 on CD4+ T cells neighboring cluster 18 cells (left) and expression of CD45RO, PD-1, and TCF-1 on CD4+ T cells neighboring cluster 16 cells (right) in ui-cores (global) and met-cores (global) from SOC and anti-PD-L1 treated patients. P-values obtained by Wilcoxon Rank Sum Test. O) Log2 fold changes of cluster 9 frequencies of proliferating cells at time of surgery vs baseline stratified into patients with metastatic disease (Met) and patients without metastatic disease (No met).
Highlights.
Tpex are abundant in uiLN and clonally related to Tex-term in the TME
After anti-PD-L1 treatment, activated Tpex and Tex-int localize near DCs in uiLN
uiLN anti-PD-L1 responses coincide with increased circulating Tex-int
Tpex and Tex-int responses to anti-PD-L1 therapy are disrupted in metLN
A study of CD8+ T cell populations in human head and neck squamous cell carcinomas identifies a role for lymph nodes in an modulating antitumor responses in response to immunotherapy.
ACKNOWLEDGMENTS
This study was supported by funding from NIH grants DP5 OD023056 and R01 DE032033 to M.H.S., a Cancer Research Institute Lloyd J. Old STAR Award to M.H.S., a Junior Cancer Research Award from UCSF Helen Diller Family Comprehensive Cancer Center to M.H.S., NIH grants S10 OD025187 and S10 OD018040 to procure the MIBIscope and CyTOF, NIH/NIDCR grant K23DE029239 to K.B.J., a Carlsberg Foundation Internationalization Fellowship (CF19-0493) to T.L.H.O, an NSF GRFP fellowship to J.L.Y., Roche/Genentech through the imCORE consortium to M.H.S., A.P.A., P.H., and L.F., and the UCSF Immunoprofiler Consortium to M.F.K. and M.H.S.
We thank D. Sandel, E.F. McCaffrey, N.F. Greenwald, S. Dodgson, and members of the Spitzer lab for helpful discussions. We thank all patients who participated in this study.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
DECLARATION OF INTERESTS
W.E.G. and K.B.J. are employees of Genentech/Roche. M.F.K. is senior advisor and founder of Foundery Immune Studios. M.A. is an inventor on patents related to MIBI technology. M.A. is a consultant, board member, and shareholder in Ionpath Inc. M.H.S. is founder and a board member of Teiko.bio and has received a speaking honorarium from Fluidigm Inc. M.F.K., L.F., A.P.A., P.H., and M.H.S. received research funding from Roche/Genentech.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hiam-Galvez KJ, Allen BM, and Spitzer MH (2021). Systemic immunity in cancer. Nat. Rev. Cancer 21, 345–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spitzer MH, Carmi Y, Reticker-Flynn NE, Kwek SS, Madhireddy D, Martins MM, Gherardini PF, Prestwood TR, Chabon J, Bendall SC, et al. (2017). Systemic Immunity Is Required for Effective Cancer Immunotherapy. Cell 168, 487–502.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fransen MF, Schoonderwoerd M, Knopf P, Camps MG, Hawinkels LJ, Kneilling M, van Hall T, and Ossendorp F. (2018). Tumor-draining lymph nodes are pivotal in PD-1/PD-L1 checkpoint therapy. JCI Insight 3. 10.1172/jci.insight.124507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dammeijer F, van Gulijk M, Mulder EE, Lukkes M, Klaase L, van den Bosch T, van Nimwegen M, Lau SP, Latupeirissa K, Schetters S, et al. (2020). The PD-1/PD-L1-Checkpoint Restrains T cell Immunity in Tumor-Draining Lymph Nodes. Cancer Cell 38, 685–700.e8. [DOI] [PubMed] [Google Scholar]
- 5.Yost KE., Satpathy AT., Wells DK., Qi Y., Wang C., Kageyama R., McNamara KL., Granja JM., Sarin KY., Brown RA., et al. (2019). Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med. 25, 1251–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu TD, Madireddi S, de Almeida PE, Banchereau R, Chen Y-JJ, Chitre AS, Chiang EY, Iftikhar H, O’Gorman WE, Au-Yeung A, et al. (2020). Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature 579, 274–278. [DOI] [PubMed] [Google Scholar]
- 7.Luoma AM, Suo S, Wang Y, Gunasti L, Porter CBM, Nabilsi N, Tadros J, Ferretti AP, Liao S, Gurer C, et al. (2022). Tissue-resident memory and circulating T cells are early responders to pre-surgical cancer immunotherapy. Cell 185, 2918–2935.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagasaki J, Inozume T, Sax N, Ariyasu R, Ishikawa M, Yamashita K, Kawazu M, Ueno T, Irie T, Tanji E, et al. (2022). PD-1 blockade therapy promotes infiltration of tumor-attacking exhausted T cell clonotypes. Cell Rep. 38, 110331. [DOI] [PubMed] [Google Scholar]
- 9.McLane LM, Abdel-Hakeem MS, and Wherry EJ (2019). CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer. Annu. Rev. Immunol. 37, 457–495. [DOI] [PubMed] [Google Scholar]
- 10.Utzschneider DT, Charmoy M, Chennupati V, Pousse L, Ferreira DP, Calderon-Copete S, Danilo M, Alfei F, Hofmann M, Wieland D, et al. (2016). T Cell Factor 1-Expressing Memory-like CD8(+) T Cells Sustain the Immune Response to Chronic Viral Infections. Immunity 45, 415–427. [DOI] [PubMed] [Google Scholar]
- 11.Wu T, Ji Y, Moseman EA, Xu HC, Manglani M, Kirby M, Anderson SM, Handon R, Kenyon E, Elkahloun A, et al. (2016). The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci Immunol 1. 10.1126/sciimmunol.aai8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH, et al. (2016). Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hudson WH, Gensheimer J, Hashimoto M, Wieland A, Valanparambil RM, Li P, Lin J-X, Konieczny BT, Im SJ, Freeman GJ, et al. (2019). Proliferating Transitory T Cells with an Effector-like Transcriptional Signature Emerge from PD-1 Stem-like CD8 T Cells during Chronic Infection. Immunity 51, 1043–1058.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, Yates KB, Lako A, Felt K, Naik GS, et al. (2019). Subsets of exhausted CD8 T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20, 326–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zander R, Schauder D, Xin G, Nguyen C, Wu X, Zajac A, and Cui W. (2019). CD4 T Cell Help Is Required for the Formation of a Cytolytic CD8 T Cell Subset that Protects against Chronic Infection and Cancer. Immunity 51, 1028–1042.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beltra J-C, Manne S, Abdel-Hakeem MS, Kurachi M, Giles JR, Chen Z, Casella V, Ngiow SF, Khan O, Huang YJ, et al. (2020). Developmental Relationships of Four Exhausted CD8 T Cell Subsets Reveals Underlying Transcriptional and Epigenetic Landscape Control Mechanisms. Immunity 52, 825–841.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zehn D, Thimme R, Lugli E, de Almeida GP, and Oxenius A. (2022). “Stem-like” precursors are the fount to sustain persistent CD8 T cell responses. Nat. Immunol. 23, 836–847. [DOI] [PubMed] [Google Scholar]
- 18.Siddiqui I, Schaeuble K, Chennupati V, Fuertes Marraco SA, Calderon-Copete S, Pais Ferreira D, Carmona SJ, Scarpellino L, Gfeller D, Pradervand S, et al. (2019). Intratumoral Tcf1PD-1CD8 T Cells with Stem-like Properties Promote Tumor Control in Response to Vaccination and Checkpoint Blockade Immunotherapy. Immunity 50, 195–211.e10. [DOI] [PubMed] [Google Scholar]
- 19.Schietinger A, Philip M, Krisnawan VE, Chiu EY, Delrow JJ, Basom RS, Lauer P, Brockstedt DG, Knoblaugh SE, Hämmerling GJ, et al. (2016). Tumor-Specific T Cell Dysfunction Is a Dynamic Antigen-Driven Differentiation Program Initiated Early during Tumorigenesis. Immunity 45, 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Tuong ZK, Dean I, Willis C, Gaspal F, Fiancette R, Idris S, Kennedy B, Ferdinand JR, Peñalver A, et al. (2022). In vivo labeling reveals continuous trafficking of TCF-1+ T cells between tumor and lymphoid tissue. J. Exp. Med. 219. 10.1084/jem.20210749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Connolly KA, Kuchroo M, Venkat A, Khatun A, Wang J, William I, Hornick NI, Fitzgerald BL, Damo M, Kasmani MY, et al. (2021). A reservoir of stem-like CD8 T cells in the tumor-draining lymph node preserves the ongoing antitumor immune response. Sci Immunol 6, eabg7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schenkel JM., Herbst RH., Canner D., Li A., Hillman M., Shanahan S-L., Gibbons G., Smith OC., Kim JY., Westcott P., et al. (2021). Conventional type I dendritic cells maintain a reservoir of proliferative tumor-antigen specific TCF-1 CD8 T cells in tumor-draining lymph nodes. Immunity 54, 2338–2353.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brummelman J, Mazza EMC, Alvisi G, Colombo FS, Grilli A, Mikulak J, Mavilio D, Alloisio M, Ferrari F, Lopci E, et al. (2018). High-dimensional single cell analysis identifies stem-like cytotoxic CD8 T cells infiltrating human tumors. J. Exp. Med. 215, 2520–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, Lieb DJ, Chen JH, Frederick DT, Barzily-Rokni M, et al. (2018). Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 175, 998–1013.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jansen CS, Prokhnevska N, Master VA, Sanda MG, Carlisle JW, Bilen MA, Cardenas M, Wilkinson S, Lake R, Sowalsky AG, et al. (2019). An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature 576, 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galletti G, De Simone G, Mazza EMC, Puccio S, Mezzanotte C, Bi TM, Davydov AN, Metsger M, Scamardella E, Alvisi G, et al. (2020). Two subsets of stem-like CD8 memory T cell progenitors with distinct fate commitments in humans. Nat. Immunol. 21, 1552–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eberhardt CS, Kissick HT, Patel MR, Cardenas MA, Prokhnevska N, Obeng RC, Nasti TH, Griffith CC, Im SJ, Wang X, et al. (2021). Functional HPV-specific PD-1 stem-like CD8 T cells in head and neck cancer. Nature 597, 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng L, Qin S, Si W, Wang A, Xing B, Gao R, Ren X, Wang L, Wu X, Zhang J, et al. (2021). Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science 374, abe6474. [DOI] [PubMed] [Google Scholar]
- 29.Ganesan A-P, Clarke J, Wood O, Garrido-Martin EM, Chee SJ, Mellows T, Samaniego-Castruita D, Singh D, Seumois G, Alzetani A, et al. (2017). Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat. Immunol. 18, 940–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duhen T, Duhen R, Montler R, Moses J, Moudgil T, de Miranda NF, Goodall CP, Blair TC, Fox BA, McDermott JE, et al. (2018). Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. Nat. Commun. 9, 2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anadon CM, Yu X, Hänggi K, Biswas S, Chaurio RA, Martin A, Payne KK, Mandal G, Innamarato P, Harro CM, et al. (2022). Ovarian cancer immunogenicity is governed by a narrow subset of progenitor tissue-resident memory T cells. Cancer Cell 40, 545–557.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ribas A, and Wolchok JD (2018). Cancer immunotherapy using checkpoint blockade. Science 359, 1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Gassen S, Callebaut B, Van Helden MJ, Lambrecht BN, Demeester P, Dhaene T, and Saeys Y. (2015). FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645. [DOI] [PubMed] [Google Scholar]
- 34.Simoni Y, Becht E, Fehlings M, Loh CY, Koo S-L, Teng KWW, Yeong JPS, Nahar R, Zhang T, Kared H, et al. (2018). Bystander CD8 T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 557, 575–579. [DOI] [PubMed] [Google Scholar]
- 35.Hanada K-I, Zhao C, Gil-Hoyos R, Gartner JJ, Chow-Parmer C, Lowery FJ, Krishna S, Prickett TD, Kivitz S, Parkhurst MR, et al. (2022). A phenotypic signature that identifies neoantigen-reactive T cells in fresh human lung cancers. Cancer Cell 40, 479–493.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greenwald NF., Miller G., Moen E., Kong A., Kagel A., Dougherty T., Fullaway CC., McIntosh BJ., Leow KX., Schwartz MS., et al. (2022). Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. Nat. Biotechnol. 40, 555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reif K, Ekland EH, Ohl L, Nakano H, Lipp M, Förster R, and Cyster JG (2002). Balanced responsiveness to chemoattractants from adjacent zones determines B-cell position. Nature 416, 94–99. [DOI] [PubMed] [Google Scholar]
- 38.Ansel KM, Ngo VN, Hyman PL, Luther SA, Förster R, Sedgwick JD, Browning JL, Lipp M, and Cyster JG (2000). A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature 406, 309–314. [DOI] [PubMed] [Google Scholar]
- 39.McCaffrey EF, Donato M, Keren L, Chen Z, Delmastro A, Fitzpatrick MB, Gupta S, Greenwald NF, Baranski A, Graf W, et al. (2022). Author Correction: The immunoregulatory landscape of human tuberculosis granulomas. Nat. Immunol. 23, 814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Risom T, Glass DR, Averbukh I, Liu CC, Baranski A, Kagel A, McCaffrey EF, Greenwald NF, Rivero-Gutiérrez B, Strand SH, et al. (2022). Transition to invasive breast cancer is associated with progressive changes in the structure and composition of tumor stroma. Cell 185, 299–310.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oh SA, Wu D-C, Cheung J, Navarro A, Xiong H, Cubas R, Totpal K, Chiu H, Wu Y, Comps-Agrar L, et al. (2020). PD-L1 expression by dendritic cells is a key regulator of T-cell immunity in cancer. Nat Cancer 1, 681–691. [DOI] [PubMed] [Google Scholar]
- 42.Huang AC, Postow MA, Orlowski RJ, Mick R, Bengsch B, Manne S, Xu W, Harmon S, Giles JR, Wenz B, et al. (2017). T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 545, 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fairfax BP, Taylor CA, Watson RA, Nassiri I, Danielli S, Fang H, Mahé EA, Cooper R, Woodcock V, Traill Z, et al. (2020). Peripheral CD8 T cell characteristics associated with durable responses to immune checkpoint blockade in patients with metastatic melanoma. Nat. Med. 26, 193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valpione S, Galvani E, Tweedy J, Mundra PA, Banyard A, Middlehurst P, Barry J, Mills S, Salih Z, Weightman J, et al. (2020). Immune-awakening revealed by peripheral T cell dynamics after one cycle of immunotherapy. Nat Cancer 1, 210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carlisle JW, Jansen CS, Cardenas MA, Sobierajska E, Reyes AM, Greenwald R, Del Balzo L, Prokhnevska N, Kucuk O, Carthon BC, et al. (2022). Clinical outcome following checkpoint therapy in renal cell carcinoma is associated with a burst of activated CD8 T cells in blood. J Immunother Cancer 10. 10.1136/jitc-2022-004803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshida O, Kimura S, Jackson EK, Robson SC, Geller DA, Murase N, and Thomson AW (2013). CD39 expression by hepatic myeloid dendritic cells attenuates inflammation in liver transplant ischemia-reperfusion injury in mice. Hepatology 58, 2163–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mellor AL, and Munn DH (2004). IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat. Rev. Immunol. 4, 762–774. [DOI] [PubMed] [Google Scholar]
- 48.de Mingo Pulido Á, Gardner A, Hiebler S, Soliman H, Rugo HS, Krummel MF, Coussens LM, and Ruffell B. (2018). TIM-3 Regulates CD103 Dendritic Cell Function and Response to Chemotherapy in Breast Cancer. Cancer Cell 33, 60–74.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dixon KO, Tabaka M, Schramm MA, Xiao S, Tang R, Dionne D, Anderson AC, Rozenblatt-Rosen O, Regev A, and Kuchroo VK (2021). TIM-3 restrains anti-tumour immunity by regulating inflammasome activation. Nature 595, 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng Y, Arvey A, Chinen T, van der Veeken J, Gasteiger G, and Rudensky AY (2014). Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell 158, 749–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borsellino G, Kleinewietfeld M, Di Mitri D, Sternjak A, Diamantini A, Giometto R, Höpner S, Centonze D, Bernardi G, Dell’Acqua ML, et al. (2007). Expression of ectonucleotidase CD39 by Foxp3+ Treg cells: hydrolysis of extracellular ATP and immune suppression. Blood 110, 1225–1232. [DOI] [PubMed] [Google Scholar]
- 52.Gu J, Ni X, Pan X, Lu H, Lu Y, Zhao J, Guo Zheng S, Hippen KL, Wang X, and Lu L. (2017). Human CD39 regulatory T cells present stronger stability and function under inflammatory conditions. Cell. Mol. Immunol. 14, 521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gautron A-S, Dominguez-Villar M, de Marcken M, and Hafler DA (2014). Enhanced suppressor function of TIM-3+ FoxP3+ regulatory T cells. Eur. J. Immunol. 44, 2703–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Banerjee H., Nieves-Rosado H., Kulkarni A., Murter B., McGrath KV., Chandran UR., Chang A., Szymczak-Workman AL., Vujanovic L., Delgoff GM., et al. (2021). Expression of Tim-3 drives phenotypic and functional changes in Treg cells in secondary lymphoid organs and the tumor microenvironment. Cell Rep. 36, 109699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller I, Min M, Yang C, Tian C, Gookin S, Carter D, and Spencer SL (2018). Ki67 is a Graded Rather than a Binary Marker of Proliferation versus Quiescence. Cell Rep. 24, 1105–1112.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stoltzfus CR, Sivakumar R, Kunz L, Olin Pope BE, Menietti E, Speziale D, Adelfio R, Bacac M, Colombetti S, Perro M, et al. (2021). Multi-Parameter Quantitative Imaging of Tumor Microenvironments Reveals Perivascular Immune Niches Associated With Anti-Tumor Immunity. Front. Immunol. 12, 726492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Di Pilato M, Kfuri-Rubens R, Pruessmann JN, Ozga AJ, Messemaker M, Cadilha BL, Sivakumar R, Cianciaruso C, Warner RD, Marangoni F, et al. (2021). CXCR6 positions cytotoxic T cells to receive critical survival signals in the tumor microenvironment. Cell 184, 4512–4530.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dähling S, Mansilla AM, Knöpper K, Grafen A, Utzschneider DT, Ugur M, Whitney PG, Bachem A, Arampatzi P, Imdahl F, et al. (2022). Type 1 conventional dendritic cells maintain and guide the differentiation of precursors of exhausted T cells in distinct cellular niches. Immunity 55, 656–670.e8. [DOI] [PubMed] [Google Scholar]
- 59.Kamphorst AO, Wieland A, Nasti T, Yang S, Zhang R, Barber DL, Konieczny BT, Daugherty CZ, Koenig L, Yu K, et al. (2017). Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science 355, 1423–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reticker-Flynn NE, Zhang W, Belk JA, Basto PA, Escalante NK, Pilarowski GOW, Bejnood A, Martins MM, Kenkel JA, Linde IL, et al. (2022). Lymph node colonization induces tumor-immune tolerance to promote distant metastasis. Cell 185, 1924–1942.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Krimpen A, Gerretsen VIV, Mulder EEAP, van Gulijk M, van den Bosch TPP, von der Thüsen J, Grünhagen DJ, Verhoef C, Mustafa D, Aerts JG, et al. (2022). Immune suppression in the tumor-draining lymph node corresponds with distant disease recurrence in patients with melanoma. Cancer Cell 40, 798–799. [DOI] [PubMed] [Google Scholar]
- 62.Andreatta M, Corria-Osorio J, Müller S, Cubas R, Coukos G, and Carmona SJ (2021). Interpretation of T cell states from single-cell transcriptomics data using reference atlases. Nat. Commun. 12, 2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Herrera FG, Ronet C, Ochoa de Olza M, Barras D, Crespo I, Andreatta M, Corria-Osorio J, Spill A, Benedetti F, Genolet R, et al. (2022). Low-Dose Radiotherapy Reverses Tumor Immune Desertification and Resistance to Immunotherapy. Cancer Discov. 12, 108–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsui C, Kretschmer L, Rapelius S, Gabriel SS, Chisanga D, Knöpper K, Utzschneider DT, Nüssing S, Liao Y, Mason T, et al. (2022). MYB orchestrates T cell exhaustion and response to checkpoint inhibition. Nature 609, 354–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spitzer MH, Gherardini PF, Fragiadakis GK, Bhattacharya N, Yuan RT, Hotson AN, Finck R, Carmi Y, Zunder ER, Fantl WJ, et al. (2015). IMMUNOLOGY. An interactive reference framework for modeling a dynamic immune system. Science 349, 1259425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zunder ER, Finck R, Behbehani GK, Amir E-AD, Krishnaswamy S, Gonzalez VD, Lorang CG, Bjornson Z, Spitzer MH, Bodenmiller B, et al. (2015). Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat. Protoc. 10, 316–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Gassen S, Gaudilliere B, Angst MS, Saeys Y, and Aghaeepour N. (2020). CytoNorm: A Normalization Algorithm for Cytometry Data. Cytometry A 97, 268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meur Le (2020). flowCore: flowCore: Basic structures for flow cytometry data. [Google Scholar]
- 69.Chevrier S, Crowell HL, Zanotelli VRT, Engler S, Robinson MD, and Bodenmiller B. (2018). Compensation of Signal Spillover in Suspension and Imaging Mass Cytometry. Cell Syst 6, 612–620.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nowicka M., Krieg C., Crowell HL., Weber LM., Hartmann FJ., Guglietta S., Becher B., Levesque MP., and Robinson MD. (2017). CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000Res. 6, 748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilkerson MD, and Hayes DN (2010). ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26, 1572–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Healy MLA UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. [Google Scholar]
- 73.Gu Z, Eils R, and Schlesner M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. [DOI] [PubMed] [Google Scholar]
- 74.Weber LM, Nowicka M, Soneson C, and Robinson MD (2019). diffcyt: Differential discovery in high-dimensional cytometry via high-resolution clustering. Commun Biol 2, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu CC, Greenwald NF, Kong A, McCaffrey EF, Leow KX, Mrdjen D, and Angelo M. (2022). Robust phenotyping of highly multiplexed tissue imaging data using pixel-level clustering. bioRxiv, 2022.08.16.504171. 10.1101/2022.08.16.504171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh P-R, and Raychaudhuri S. (2019). Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Traag V, Waltman L, and van Eck NJ (2018). From Louvain to Leiden: guaranteeing well-connected communities. 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McInnes L, Healy J, and Melville J. (2018). UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. 10.48550/arXiv.1802.03426. [DOI] [Google Scholar]
- 80.Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. (2016). Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.R Core Team (2022). R: A language and environment for statistical. [Google Scholar]
- 82.Team RStudio (2016). RStudio: Integrated Development Environment for R. [Google Scholar]
- 83.Simko TWA (2021). R package “corrplot”: Visualization of a Correlation Matrix. [Google Scholar]
- 84.Hadley Wickham and Romain François and Lionel Henry and Kirill Müller (2021). dplyr: A Grammar of Data Manipulation. [Google Scholar]
- 85.Wickham H. (2007). Reshaping data with the reshape package. J. Stat. Softw. 21. [Google Scholar]
- 86.Wickham H. (2009). ggplot2: Elegant Graphics for Data Analysis (Springer Science & Business Media; ). [Google Scholar]
- 87.Neuwirth E. (2014). RColorBrewer: ColorBrewer Palettes. [Google Scholar]
- 88.Kluyver T., Ragan-Kelley B., Pérez F., Granger B., Bussonnier M., Frederic J., Kelley K., Hamrick J., Grout J., Corlay S., et al. (2016). Jupyter Notebooks – a publishing format for reproducible computational workflows. In Positioning and Power in Academic Publishing: Players, Agents and Agendas, Loizides F. and Scmidt B, eds. (IOS Press; ), pp. 87–90. [Google Scholar]
- 89.Seabold S, and Perktold J. (2010). Statsmodels: Econometric and Statistical Modeling with Python. Proceedings of the Python in Science Conference. 10.25080/majora-92bf1922-011. [DOI] [Google Scholar]
- 90.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, et al. (2020). SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.The pandas development team (2020). pandas-dev/pandas: Pandas (Zenodo) 10.5281/zenodo.3509134. [DOI] [Google Scholar]
- 92.McKinney W. (2010). Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference (SciPy). 10.25080/majora-92bf1922-00a. [DOI] [Google Scholar]
- 93.Harris CR, Millman KJ, van der Walt SJ, Gommers R, Virtanen P, Cournapeau D, Wieser E, Taylor J, Berg S, Smith NJ, et al. (2020). Array programming with NumPy. Nature 585, 357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related to Figure 1 and STAR methods.
Related to Figure 2 and STAR methods.
Related to Figure 2 and STAR methods.
Related to Figures 3, 4 and 6 and STAR methods.
Related to STAR Methods.
Related to Figure 1. A) Gating strategy. B) Relative abundance of main immune cell types. C) UMAP of all CD8 T cell clusters. D) Relative abundance of non-naive CD8 T cell clusters. Numbers below indicate the number of Tpex and non-naive CD8 T cells analyzed from each sample.
Related to Figure 2. A) UMAP of 35,551 cells from 5 paired tumor and draining lymph node samples (10 samples total). Cells are colored according to annotated cell type. B) Dot plot of standard scale (subtract minimum and divide by maximum) column normalized expression for the indicated genes for each annotated cell type from all cells indicating percentage of cells with expression greater than zero (dot size) and mean expression for cells with nonzero expression (color). C) UMAP of 8,245 CD8 T cells from 5 paired tumor and draining lymph node samples (10 samples total) colored by annotated cell subset. D) Heatmap of gene expression by cluster. E) UMAP of all cells with CITE-seq data colored by normalized expression of CD3 protein, CD8 protein, and CD4 protein. F) Heatmap of CITE-seq protein expression by cluster. G) Frequencies of non-naive CD8 T cell clusters by tissue. H) Gene set scores from Zheng et al. (2021) by cluster. I) Bar plots of percentage of total CD8 T cells with TCR clonotype assigned for each sample. J) Violin plots of XIST expression and percentage of counts from Y chromosome genes for sample demultiplexing.
Related to Figure 3. A) Average marker intensity for each pixel cluster. B) Average pixel assigmnent for each cell cluster. C) Representative single-parameter images for each antibody used for MIBI analysis. D) Relative abundance of main immune cell types per LN in the T cell zone or B cell zone. Sample ID represents patient ID followed by LN number. E) Relative abundance of CD8 T cell clusters per LN in the B cell zone and global. F) Relative frequency of Tpex (c18) in the B cell zone regions and T cell zone regions of the uiLN. P-value obtained by two-tailed Wilcoxon Rank Sum Test.
Related to Figure 4. A) Relative abundance of main immune cell types in uiLN (global, T cell zone, and B cell zone) from anti-PD-L1 treated patients. Sample ID represents patient ID followed by LN number. B) Relative abundance of non-naïve CD8 T cell clusters in uiLN (global and B cell zone) from anti-PD-L1 treated patients. Sample ID represents patient ID followed by LN number. C) Percent Tpex (c18) of non-naive CD8 T cells in the B cell zone from untreated versus anti-PD-L1 treated patients. P-values obtained by Wilcoxon Rank Sum Test. D) Difference in the number of DC neighbors between untreated and anti-PD-L1 treated patients for each cluster of CD8 T cells (Wilcoxon Rank Sum Test). Colors indicate if DC neighbors are significantly more abundant in anti-PD-L1 (purple) or SOC (blue) or not differentially abundant (False, grey) after Benjamini-Hochberg correction, FDR < 0.05. E) Percent DC of total immune cells from untreated versus anti-PD-L1 treated patients. F-G) Spearman correlation between % DC of total immune cells and F) % Tpex (c18) or G) % Tex-int (c16) of total non-naive CD8 T cells in samples from patients receiving SOC or PD-L1 blockade.
Related to Figure 5. A) Samples collected from each patient. Time (days) are relative to the first sample collected. B) Relative abundance of main immune cell types. C) Principal component analysis of gated main immune cell populations. D) UMAP of all CD8 T cell clusters. E) Frequencies of non-naïve CD8 T cell clusters. F) Percentage of cluster 9 proliferating cells in paired samples from blood at time of surgery and baseline. P-value obtained by paired Wilcoxon Rank Sum Test. G) Paired differential abundance analysis of non-naïve CD8 T cell subsets between tumor and blood at time of surgery (n = 6) (generalized linear mixed models). See color scheme for Figure 6C. H) Cluster 5 abundance (as percentage of non-naïve CD8 T cells) in paired samples from blood and tumor at time of surgery. P-values obtained by generalized linear mixed models. I) Correlation between clusters in tumor and blood at time of surgery (Spearman correlation). J-K) Correlation between clusters in LN (MIBI data) and clusters in blood (CyTOF data) (J) and clusters in LN (MIBI data) and clusters in tumor (CyTOF data) (K) at time of surgery (Spearman correlation).
Related to Figure 6. A) Principal component analysis of gated main immune cell populations in uninvolved lymph nodes (uiLN), metastatic lymph nodes (metLN), and tumors. B) Relative abundance of main immune cell types for patients 11084 and 8158. C) Paired differential abundance (DA) analysis of main immune cell populations between uiLN and metLN (n = 7; paired Wilcoxon Rank Sum Test). The log2 fold changes are plotted against the negative log10(nominal p-values). Colors indicate if cell populations are significantly more abundant in uiLN (purple) or tumor/metLN (blue) or not differentially abundant (False, grey) after Benjamini-Hochberg correction, FDR < 0.1. D) Paired differential abundance analysis of non-naïve CD8 T cell subsets between metLN and tumor (n = 8) (generalized linear mixed models). Grey: not differentially abundant after Benjamini-Hochberg correction, FDR < 0.1. E) Relative abundance of main immune cell types in met-cores (global) from anti-PD-L1 treated patients. Sample ID represents patient ID followed by LN number and core number. F) Main immune cell type ratios represented as log2 fold changes between met-cores (global) and ui-cores (global) from antiPD-L1 treated patients. G) Relative abundance of non-naïve CD8+ T cell clusters in metastatic cores (global, n = 5) from anti-PD-L1 treated patients (n = 3). Sample ID indicates patient ID followed by LN number and core number. H) Non-naïve CD8+ T cell cluster ratios represented as log2 fold changes between met-cores (global, n = 5) and ui-cores (global, n = 39) from anti-PD-L1 treated patients. I) Cluster 18 and 16 abundances (as percentage of non-naïve CD8 T cells) in met-cores (global) and ui-cores (global) from anti-PD-L1 treated patients, and ui-cores (global) from SOC treated patients. P-values obtained by Wilcoxon Rank Sum Test. J-K) Percentage of cluster 18 (J) and cluster 16 (K) cells with a specific cell type as its neighbor in met-cores (global) from anti-PD-L1 treated patients. L) Expression of CD39, IDO1, PD-L1, and TIM3 on DCs neighboring cluster 16 cells in ui-cores (global) and met-cores (global) from SOC and anti-PD-L1 treated patients. P-values obtained by Wilcoxon Rank Sum Test. M) Expression of CD39 and Ki-67 on Tregs neighboring cluster 18 cells (left) and expression of CD39, FoxP3, Ki-67, and TIM-3 on Tregs neighboring cluster 16 cells (right) in ui-cores (global) and met-cores (global) from SOC and anti-PD-L1 treated patients. P-values obtained by Wilcoxon Rank Sum Test. N) Expression of PD1 on CD4+ T cells neighboring cluster 18 cells (left) and expression of CD45RO, PD-1, and TCF-1 on CD4+ T cells neighboring cluster 16 cells (right) in ui-cores (global) and met-cores (global) from SOC and anti-PD-L1 treated patients. P-values obtained by Wilcoxon Rank Sum Test. O) Log2 fold changes of cluster 9 frequencies of proliferating cells at time of surgery vs baseline stratified into patients with metastatic disease (Met) and patients without metastatic disease (No met).
Data Availability Statement
All mass cytometry and multiplexed ion beam imaging data are available on Mendeley Data under doi:10.17632/2zgppyr2rr.1. Single-cell genomics data have been deposited into GEO under accession number GSE212797.
This paper does not report original code, however available software packages used for analysis are described in the STAR methods section and Key Resource Table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| For a list of antibodies used for analysis by mass cytometry (CyTOF), multiplexed ion beam imaging (MIBI), and CITE-seq, please see Supplemental Table S5. | Various | Various |
| Human TruStain FcX (Fc Receptor Blocking Solution) |
Biolegend | Cat#422301 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Collagenase IV | Worthington | Cat#LS004188 |
| Deoxyribonuclease I | Sigma-Aldrich | Cat#DN25 |
| cis- Diammineplatinum(II) dichloride (Cisplatin) |
Sigma-Aldrich | Cat#P4394 |
| Paraformaldehyde aqueous solution | Electron Microscopy Sciences |
Cat#15710 |
| Zombie Aqua Fixable Viability Dye |
Thermo Fisher | Cat#L34966 |
| Candor PBS Antibody Stabilization solution |
Candor Bioscience |
Cat#13150 |
| Maxpar 10X Barcode Perm Buffer | Fluidigm | Cat#201057 |
| Cell-ID Intercalator-IR | Fluidigm | Cat#201192A |
| Maxpar Cell Acquisition Solution |
Fluidigm | Cat#201240 |
| Target Retrieval Solution | Dako Agilent | Cat#S1699 |
| PBS IHC Wash Buffer + Tween 20 |
Cell Marque | Cat#934B-06 |
| EQ Four Element Calibration Beads | Fluidigm | Cat#201078 |
| ACK Lysing Buffer | Thermo Fisher | Cat#A1049201 |
| Cell Staining Buffer | Biolegend | Cat#420201 |
| Normal Donkey Serum | Sigma-Aldrich | Cat#566460 |
| Critical Commercial Assays | ||
| Chromium Next GEM Single Cell 5’ Library |
10X Genomics | Cat#PN-1000265 |
| Kit | ||
| Chromium Single Cell V(D)J Enrichment kit |
10X Genomics | Cat#PN-1000005 |
| 5’ Feature Barcode Kit |
10X Genomics | Cat#PN-1000256 |
| MaxPar X8 Antibody Labeling Kits | Fluidigm | Various |
| Metal-Loaded MIBItags | IonPath | Various |
| Avidin/Biotin Blocking System |
Biolegend | Cat#927301 |
| Deposited Data | ||
| Single-cell genomics data | Generated by current investigators | Deposited into GEO under accession number GSE212797 |
| Mass cytometry and multiplexed ion beam imaging data | Generated by current investigators | Deposited into Mendeley Data under doi:10.17632/2zgppyr2rr.1 |
| Software and Algorithms | ||
| R Core Team (v.4.0.5) |
The R Project for Statistical
Computing |
https://www.r-project.org/ |
| RStudio | Posit | https://posit.co/ |
| Python (v.3.88) | Python Software Foundation. Python Language Reference, version 3.8. |
http://www.python.org |
| ParkerICI/Premessa | Parker Institute for Cancer
Immunotherapy |
https://github.com/ParkerICI/premessa |
| CellEngine | CellCarta | https://cellcarta.com/cellenginesoftware/ |
| CytoNorm | Van Gassen et al., 2020 | https://github.com/saeyslab/CytoNorm |
| Flowcore | Ellis et al., 2022 | https://bioconductor.org/packages/release/bioc/html/flowCore.html |
| CATALYST | Nowicka et al., 2017; Chevrier et al., 2018 | https://github.com/HelenaLC/CATALYST |
| FlowSOM | Van Gassen et al, 2015 | https://github.com/SofieVG/FlowSOM |
| diffcyt | Weber et al., 2019 | https://github.com/lmweber/diffcyt |
| Mesmer | Greenwald et al., 2021 | https://github.com/vanvalenlab/kiosk-console |
| scanpy (v.1.7.1) | Wolf et al., 2018 | https://github.com/scverse/scanpy |
| Harmony (v.0.05) | Korsunsky et al., 2019 | https://github.com/immunogenomics/harmony |
| sklearn (v.0.24.1) | Pedregosa et al., 2011 | https://github.com/scikit-learn/scikit-learn |
| corrplot | Simko, 2021 | https://github.com/taiyun/corrplot |
| scipy (v.1.6.1) | Virtanen et al., 2020 | https://github.com/scipy/scipy |
| pandas (v.1.2.3) |
McKinney,
2010; Pandas Development Team |
https://github.com/pandas-dev/pandas |
| numpy (v.3.3.4) | Harris et al., 2020 | https://github.com/numpy/numpy |
| matplotlib (v.3.3.4) | Hunter, 2007 | https://github.com/matplotlib/matplotlib |
| statsmodels (v.0.12.2) | Seabold and Perktold, 2010 | https://github.com/statsmodels/statsmodels |
| dplyr | Wickham et al., 2021 | https://github.com/tidyverse/dplyr |
| reshape2 | Wickham, 2007 | https://github.com/cran/reshape2 |
| ggplot2 | Wickham, 2009 | https://github.com/tidyverse/ggplot2 |
| RColorBrewer | Neuwirth, 2014 | https://cran.r-hub.io/web/packages/RColorBrewer/index.html |
| Other | ||
| gentleMacs C Tubes | Miltenyi Biotec | Cat#130–093-237 |
| Gold-Coated Slides | IonPath | 567001 |