Abstract
Blood vessels in different vascular beds vary in lumen diameter, which is essential for their function and fluid flow along the vascular network. Molecular mechanisms involved in the formation of a vascular lumen of appropriate size, or tubulogenesis, are still only partially understood. Src homology 2 domain containing E (She) protein was previously identified in a screen for proteins that interact with Abelson (Abl)-kinase. However, its biological role has remained unknown. Here we demonstrate that She and Abl signaling regulate vascular lumen size in zebrafish embryos and human endothelial cell culture. Zebrafish she mutants displayed increased endothelial cell number and enlarged lumen size of the dorsal aorta (DA) and defects in blood flow. Vascular endothelial specific overexpression of she resulted in a reduced diameter of the DA lumen, which correlated with the reduced arterial cell number and lower endothelial cell proliferation. Chemical inhibition of Abl signaling in zebrafish embryos caused a similar reduction in the DA diameter and alleviated the she mutant phenotype, suggesting that She acts as a negative regulator of Abl signaling. Enlargement of the DA lumen in she mutants correlated with an increased endothelial expression of claudin 5a and 5b (cldn5a / cldn5b), which encode proteins enriched in tight junctions. Inhibition of cldn5a expression partially rescued the enlarged DA in she mutants, suggesting that She regulates DA lumen size, in part, by promoting cldn5a expression. SHE knockdown in human endothelial umbilical vein cells resulted in a similar increase in the diameter of vascular tubes, and also increased phosphorylation of a known ABL downstream effector CRKL. These results argue that SHE functions as an evolutionarily conserved inhibitor of ABL signaling and regulates lumen size during vascular tubulogenesis.
Keywords: blood vessel, vascular, lumen, zebrafish, tubulogenesis
Introduction
Formation of a properly sized vascular lumen, or tubulogenesis, is one of the critical steps during vascular development. Blood vessels vary in size in different vascular beds, which is essential to allow the flow of fluids and blood cells along the vascular network. Various genetic mechanisms have been implicated in establishing and maintaining vascular lumen of a proper size. A family of Rho GTPases which include Rac1, Cdc42 and RhoA have well established roles in regulating cell cytoskeleton rearrangements that take place during vascular tubulogenesis [1–8]. Activity of these Rho GTPases is known to be regulated by integrin-fibronectin signaling [9]. Several other pathways, including VEGF and Notch signaling are also thought to contribute to regulation of vascular lumen size directly [10]. However, our understanding of molecular mechanisms that regulate vascular tubulogenesis is still incomplete.
Abelson (Abl) kinase signaling has diverse roles during morphogenesis such as regulating cytoskeletal organization that is important for cellular protrusions, cell migration, morphogenesis, adhesion, endocytosis and phagocytosis [11]. Chromosomal translocation of human ABL1 to the breakpoint cluster region (BCR) gene results in production of the BCR-ABL1 fusion protein in patients with different types of leukemias [12]. While numerous studies have focused on the role of BCR-ABL1 in leukemias, the role of ABL signaling during developmental morphogenesis is less understood. Intriguingly, Abl signaling is known to regulate cell-cell adhesion through Rho and Rac GTPases. Chemical inhibition of Abl signaling, or mutation of the two ABL homologs Abl1 and Arg in epithelial cell culture, results in activation of RhoA and increased actomyosin contractility, disrupting adherens junctions [13–16] In addition, Abl signaling is known to activate Rac1 in cultured human embryonic kidney cells [17]. Abl signaling is also known to promote cell proliferation in diverse cell types, such as smooth muscle cells and fibroblasts [18, 19]. Previously established roles of Abl signaling in vascular endothelium include regulating angiogenesis, endothelial cell (EC) survival and vascular permeability [20–22]. Recent data also indicate that ABL signaling is increased in a cell culture model of venous malformations (VMs) and that inhibition of ABL signaling reduces lumen diameter in a VM model in vivo [23]. However, the role of ABL signaling in the formation of the vascular lumen during embryogenesis has not been previously investigated.
ABL signaling is known to be regulated by various effector proteins. In a yeast two hybrid screen, two novel Abl-interacting proteins Shd and She were identified [24]. Together with a related Shb protein they comprise a separate family of SH2 proteins. Shd is known to be phosphorylated by Abl kinase in vitro, while She phosphorylation has not been previously investigated [24]. However, biological roles of these proteins remain largely unknown.
We have previously identified she as a novel vascular endothelial specific gene, which functions downstream of the ETS transcription factor Etv2 / Etsrp in zebrafish embryos [25]. She expression is enriched in arterial vessels, such as the dorsal aorta. Its protein sequence is highly conserved between different vertebrates. However, its biological function has not been previously investigated in any model system.
Here we used genome editing approaches to generate zebrafish she genetic mutants. We show that she mutants display enlarged dorsal aorta (DA), followed by a failure of circulation and embryonic lethality. In contrast, She overexpression caused a reduction in DA lumen size. Chemical inhibition of Abl signaling caused a similar reduction of the DA size and reversed enlarged aorta observed in she mutants. Our data further suggest that She functions in part by regulating expression of claudin 5a in the DA. Knockdown of She in human umbilical vein endothelial cells (HUVECs) resulted in a similar dilation of vascular tubes arguing that She function is evolutionarily conserved. In summary, our results identify novel roles for Abl signaling and She in regulating vascular tubulogenesis during embryonic development.
Results
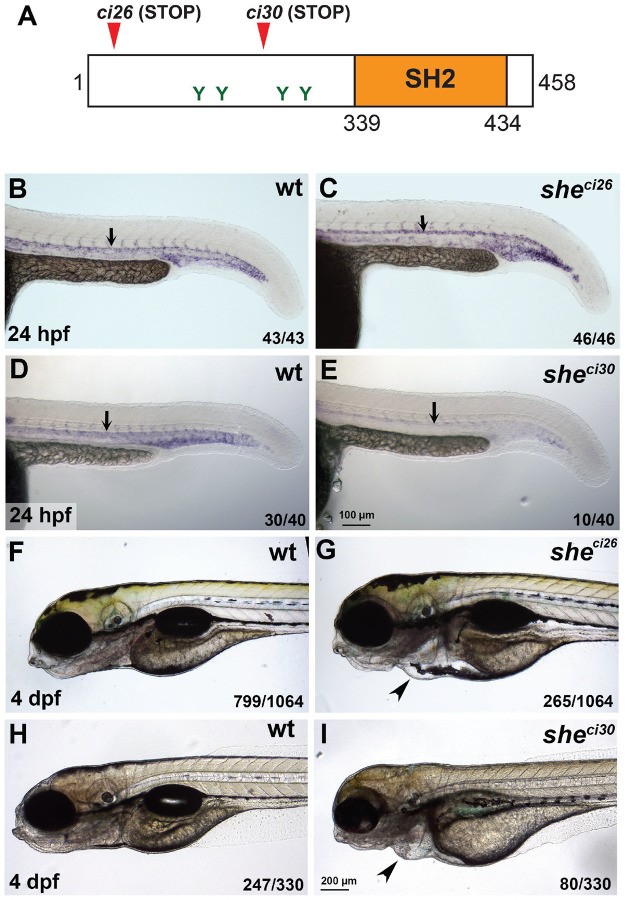
To determine the function of zebrafish She in cardiovascular development, we used TALEN and CRISPR/Cas9 genome editing to generate two loss-of-function she mutant alleles (Suppl. Fig. S1). The sheci26 allele has a 7 bp deletion, which is predicted to result in a frameshift after amino acid 4 and is expected to lead to premature translation termination. The sheci30 allele has a deletion of 575 bp and an insertion of 24 bp which is predicted to lead to a frameshift and a premature stop codon after amino acid 243 (Fig. 1A, Suppl. Fig. S1). she mRNA expression in sheci26 mutants was not significantly affected, while she mRNA expression in sheci30 mutant embryos was significantly decreased (Fig. 1B–E), indicating a possibility of nonsense-mediated RNA decay.
Figure 1. she mutants display pericardial edema and loss of blood circulation.
(A) Zebrafish SHE protein diagram. she ci26 and ci30 mutant alleles are predicted to result in a frameshift and premature stop codons. SH2 domain and consensus tyrosine ABL phosphorylation sites are shown.
(B-E) In situ hybridization analysis of she expression in she mutants at 24 hpf. Note that she expression is unaffected in sheci26 embryos while it is strongly reduced in sheci30 embryos. Homozygous sheci26 mutant embryos were obtained by an incross of sheci26−/−; fli1a:she-2A-mCherry; kdrl:GFP parents (which are viable due to fli1a:she-2A-mCherry rescue) and selected for mCherry negative embryos. wt control embryos were obtained by incross of sibling wt; fli1a:she-2A-mCherry parents and selected for mCherry- embryos. sheci30 embryos were obtained by incross of sheci30+/− parents and genotyped after in situ hybridization. 25% (10 out of 40) embryos showed strong reduction in she expression which correlated with the mutant phenotype.
(F-I) Brightfield images of sheci26 and sheci30 mutant embryos and their siblings (wild-type and or heterozygous) at 4 dpf. Note the pericardial edema (arrowheads) in the mutant embryos. Embryos were obtained by the incross of heterozygous parents in kdrl:GFP background. 24.9% (265 out of 1064) and 24.2% (80 out of 330) embryos obtained in sheci26 or sheci30 incross, respectively, showed this phenotype. A subset of embryos was genotyped to confirm the correlation between the phenotype and genotype.
Homozygous she mutant embryos were then obtained by an incross of respective heterozygous adult carriers and analyzed for morphological defects. The embryos appeared morphologically normal until 4 days-post-fertilization (dpf). At 4–6 dpf both sheci26 and sheci30 mutants developed pericardial edema, and their blood circulation slowed down or completely stopped (Fig. 1F–I, Movies 1,2). As a result, mutant embryos for both alleles were lethal by approximately 7 dpf. sheci26 mutants showed edema and blood circulation failure at a slightly earlier stage (4 dpf) compared to the sheci30 allele (5–6 dpf). sheci26 and sheci30 mutants failed to complement each other, and in the complementation cross 58 out of 262 embryos (22.1%) developed pericardial edema and greatly reduced or absent blood flow by 6 dpf. Because sheci26 allele showed a slightly more severe phenotype, it was used for all further studies.
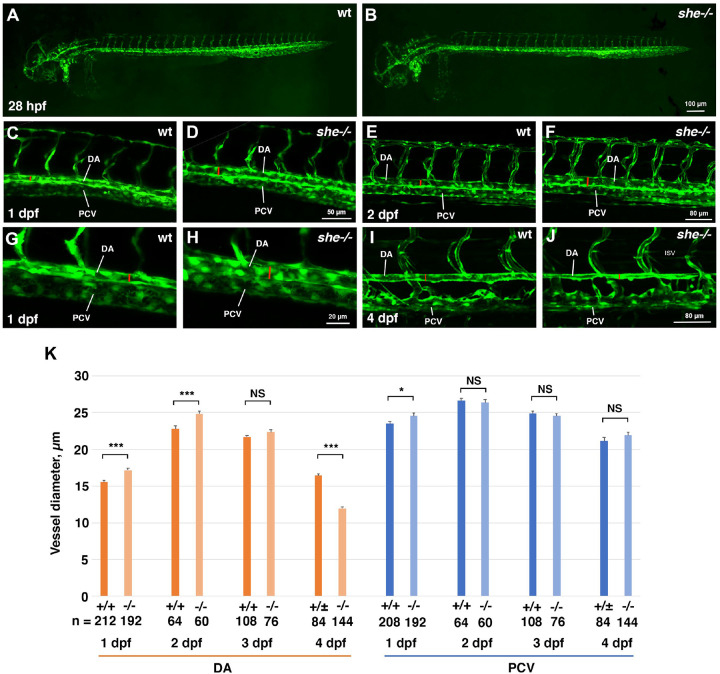
To analyze potential vascular defects, she mutants were crossed into vascular endothelial reporter kdrl:GFP background. Overall vascular patterning was normal and unaffected at 1–4 dpf, when analyzed by confocal imaging for GFP expression (Fig. 2A,B). In addition, arterial and venous trunk intersegmental vessel sprouting appeared normal in mutant and wild-type sibling embryos (Suppl. Fig. S2). There was also no effect on endothelial marker kdrl, arterial marker flt1 or venous marker dab2 expression when analyzed by in situ hybridization at 24 hpf (Suppl. Fig. S3). However, the dorsal aorta (DA) was wider in she mutants compared with their sibling wild-type (wt) embryos at both 1 and 2 dpf (Fig. 2C–H,K). Notably, no circulation defects were apparent in she mutants at these stages. In contrast, the DA was narrower in she mutants at 4 dpf when compared to their siblings (Fig. 2I–K). It is likely that the DA collapse apparent at this stage is a secondary consequence of failing blood circulation. The diameter of the posterior cardinal vein (PCV) was not significantly different between she mutant and wild-type embryos at 2–4 dpf, however at 1 dpf, a slight enlargement of the PCV was observed in mutant embryos (Fig. 2K).
Figure 2. she mutants show enlarged diameter of the dorsal aorta.
(A,B) Overall vascular patterning of she−/− mutants is normal when compared to their sibling wild-type embryos at 28 hpf. Embryos are in kdrl:GFP background.
(C-H) A wider DA is observed in she mutant embryos compared to their wild-type (she+/+) siblings at 1 and 2 dpf (28 and 48 hpf respectively). Red line indicates DA diameter. (G,H) shows higher magnification image of trunk vasculature at 1 dpf.
(I,J) DA is narrower in she mutants at 4 dpf compared to their siblings.
(K) Diameter of the DA and PCV at 1–4 dpf in she mutants and their wild-type siblings. * p<0.05; ***p<0.001, NS – not significant, Student t-test. Error bars show SEM.
At all stages, she mutant and sibling embryos were obtained by in-crossing sheci26+/−; kdrl:GFP carriers. Embryos at 1 and 2 dpf were genotyped after imaging. Embryos at 4 dpf were separated based on the phenotype, and wild-type siblings include she+/+ and she+/− embryos at this stage.
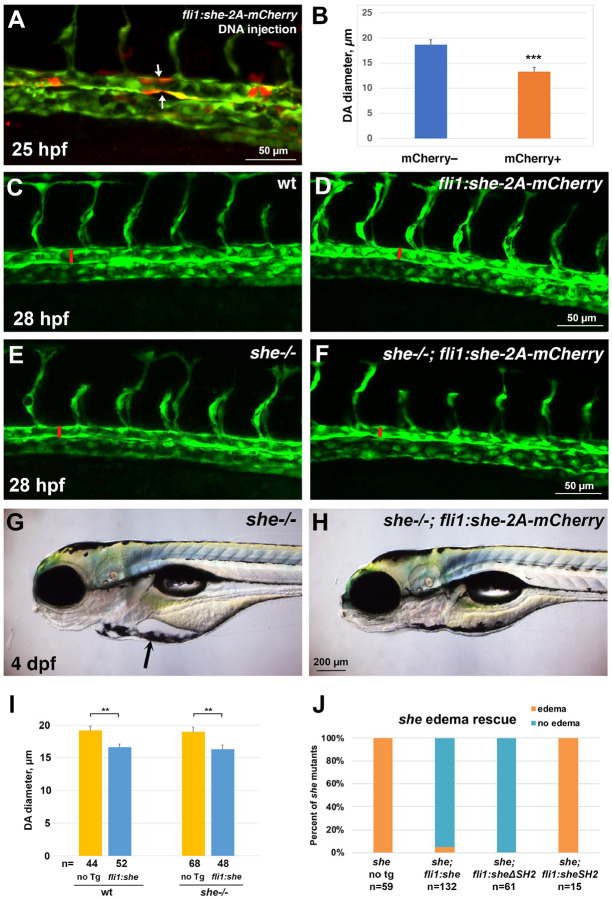
This suggests that She may function to restrict the DA size during vascular development. To test if She function is sufficient to limit the size of the DA, we made a construct which contains the coding sequence for the zebrafish She driven by the vascular endothelial fli1a promoter, followed by the self-cleaving peptide 2A and mCherry reporter. Injection of this DNA construct in combination with Tol2 mRNA into zebrafish embryos resulted in mosaic overexpression of she-2A-mCherry in vascular endothelial cells. Interestingly, segments of the DA with multiple mCherry-positive cells displayed narrower lumen compared with adjacent mCherry-negative regions of the DA (Fig. 3A,B). A stable zebrafish transgenic reporter line was then established using Tol2-mediated transgenesis and used for further studies (Suppl. Fig. S4A–C). fli1:she-2A-mCherry embryos were viable, yet they exhibited a reduction in the DA diameter at 28 hpf, when compared to sibling embryos without the transgene (Fig. 3C,D,I). To test if vascular endothelial she expression could rescue the she mutant phenotype, the fli1:she-2A-mCherry line was crossed into the sheci26 mutant background. Remarkably, she−/−; fli1:she-2A-mCherry embryos were viable and had smaller DA diameter when compared with she−/− sibling embryos without the transgene (Fig. 3E,F,I). They did not exhibit pericardial edema and showed normal blood circulation at 4 dpf (Fig. 3G,H,J). Altogether, these results show that She functions cell-autonomously in the vascular endothelium to regulate the DA size.
Figure 3. she overexpression in vasculature reduces the diameter of the dorsal aorta and rescues the she mutant phenotype.
(A) Injection of fli1:she-2A-mCherry DNA plasmid together with Tol2 mRNA results in mosaic expression of she-2A-mCherry in endothelial cells. Note that mCherry positive segments of the DA (arrows, A) show reduced diameter compared to adjacent mCherry-negative segments. (B) Quantification of the DA diameter in mCherry+ and mCherry– DA segments in embryos from two independent experiments. n denotes the number of embryos analyzed. *** p<0.001 (Student’s t-test).
(D-F) Trunk vasculature of wild-type, fli1:she-2A-mCherry, she−/− and she−/−; fli1:she-2A-mCherry embryos in kdrl:GFP background at 28 hpf. Confocal imaging of GFP expression in live embryos obtained from stable transgenic lines; the diameter of the DA is marked with red line.
(G,H) Pericardial edema in she−/− mutants (E) is rescued in she−/−; fli1:she-2A-mCherry embryos at 4 dpf.
(I) DA diameter is reduced in fli1:she-2A-mCherry embryos compared to wild-type non-transgenic (no Tg) siblings. Similarly, DA diameter is reduced in she−/−; fli1:she-2A-mCherry embryos compared to she siblings without the transgene. Embryos were analyzed at 28 hpf in kdrl:GFP background. Embryos were obtained by incross of either she+/+ (referred to as wild-type); fli1:she-2A-mCherry+/− or she−/−; fli1:she-2A-mCherry+/− parents and separated based on mCherry expression. Note that wild-type and she−/− embryos cannot be directly compared in this experiment because they came from different parents and are not siblings; embryos from different pairs can show significant variability in the DA diameter. Error bars show SEM. **p<0.01, Student’s t-test.
(J) Percentage of she mutant embryos showing pericardial edema at 4 dpf. Note that vascular endothelial expression of full length She (fli1:she-2A-mCherry) and the construct carrying a deletion of the SH2 domain in She protein (fli1:sheDSH2–2A-mCherry) rescues the mutant phenotype, while overexpression of She construct with SH2 domain alone (fli1:sheSH2–2A-mCherry) fails to rescue the phenotype.
SH2 domains are known to bind to phosphorylated tyrosines [26]. To test the functional requirement for the SH2 domain within the zebrafish She protein, we created She deletion constructs (Suppl. Fig. S4D) and generated stable zebrafish transgenic lines using vascular endothelial fli1 promoter. Intriguingly, the construct that lacked the SH2 domain was sufficient to rescue the she mutant phenotype, and no pericardial edema or circulation defects were observed in fli1:sheΔSH2–2A-mCherry; she−/− embryos, which were viable through adulthood (Fig. 3H). In contrast, the construct containing only the SH2 domain was insufficient to rescue the she mutant phenotype (Fig. 3H).
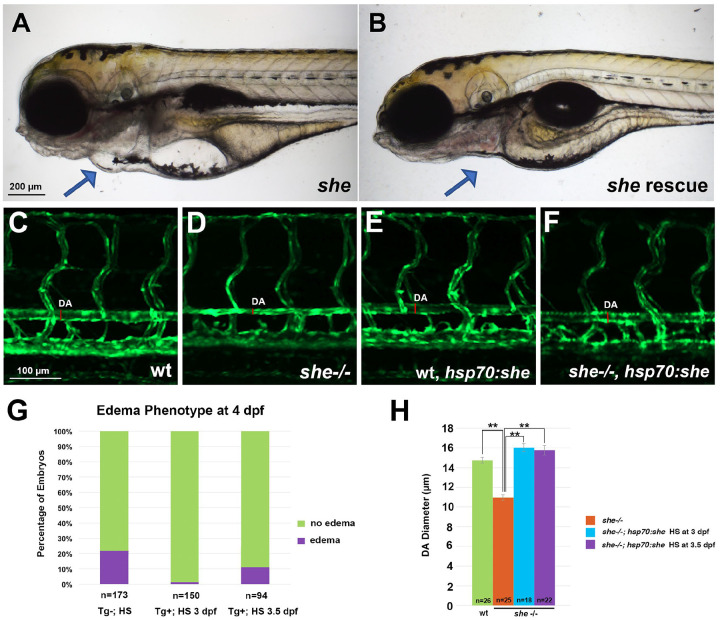
To determine when She function is required, we made a construct which encodes a full-length zebrafish She under a heat-shock inducible hsp70 promoter followed by 2A-mCherry and established stable zebrafish lines using Tol2 transgenesis. Heat-shock at 3 or 3.5 dpf was sufficient to rescue the pericardial edema phenotype fully or partially in she mutant embryos, positive for the hsp70:she-2A-mCherry transgene (Fig. 4A,B,G). This also prevented the collapse of DA at 4 dpf (Fig. 4C–G). This suggests that She has a critical functional requirement between 3 and 4 dpf.
Figure 4. Inducible She expression under a heat-shock promoter hsp70 rescues the she mutant phenotype.
(A,B) Pericardial edema is observed in she mutant embryos at 4 dpf (A), while embryos positive for hsp70:she-2A-mCherry do not show pericardial edema and have normal blood circulation after heat-shock was performed at 3 dpf (B). Embryos were obtained from the cross of hsp70:she-2A-mCherry; cryaa:dsRed +/−; she+/− X she+/− parents in kdrl:GFP background. Embryos were sorted for dsRed in the lens (driven by the lens specific cryaa promoter which correlates with the presence of hsp70:she-2A-mCherry transgene) and mCherry in the body. Representative embryos with and without edema are shown for each group.
(C-F) Dorsal aorta in she mutants is narrower or collapsed at 4 dpf, while heat-shock at 3 dpf restores normal DA size in she−/−; hsp70:she-2A-mCherry embryos.
(G) Percentage of total embryos showing the pericardial edema phenotype at 4 dpf. Embryos were obtained by crossing hsp70:she-2A-mCherry; cryaa:dsRed +/−; she+/− X she+/− adults in kdrl:GFP background and subjected to heat-shock (HS) at 3 or 3.5 dpf. Embryos with dsRed in the lens (Tg+) or transgene negative controls (Tg-) were selected for the analysis. Tg- embryos were also subjected to heat shock at 3 or 3.5 dpf (both groups were combined).
(H) The diameter of the dorsal aorta at 4 dpf in wild-type siblings (includes heterozygous and wild-type embryos), she mutant embryos without the transgene, and she−/−; hsp70:she-2A-mCherry embryos heat-shocked at 3 or 3.5 dpf. ** p<0.01, Student’s t-test.
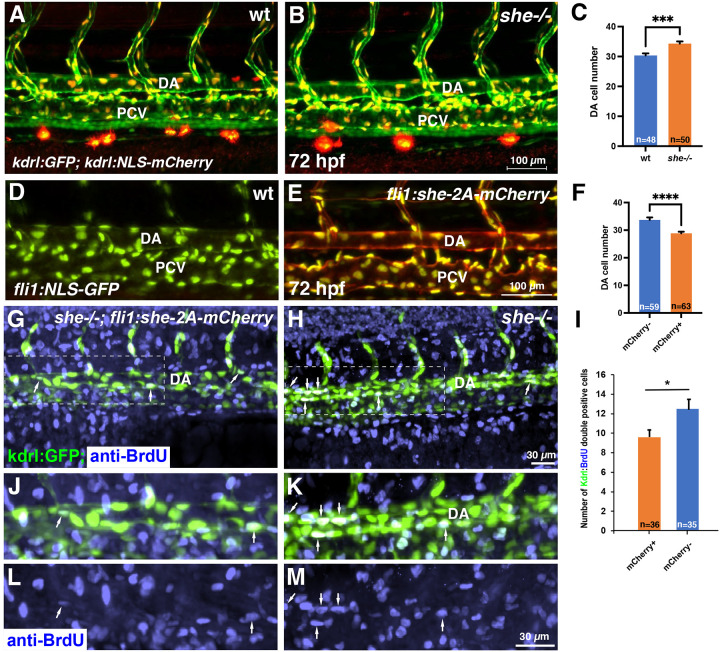
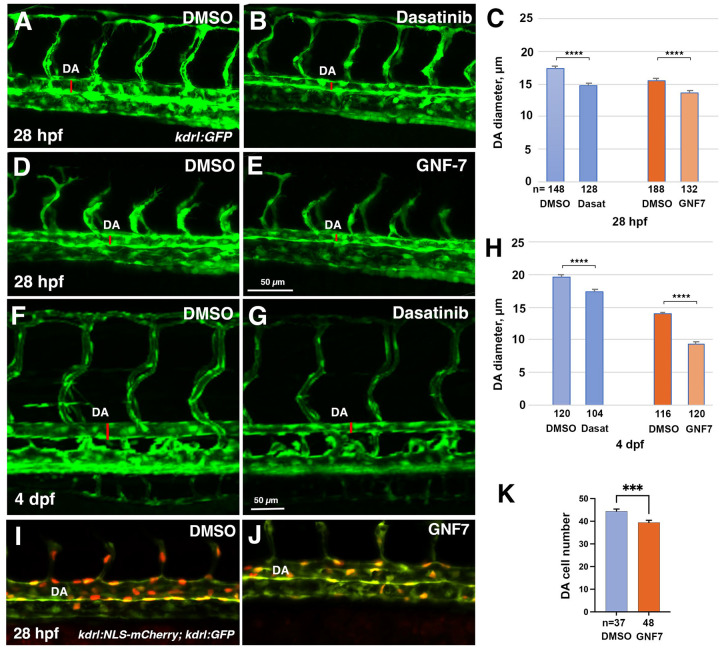
To determine the cellular cause of the enlarged aorta observed in she mutants, we analyzed the endothelial cell number. The number of cells in the DA was significantly increased in she mutants crossed to the kdrl:NLS-mCherry endothelial nuclear reporter line when compared with their wild-type siblings at 72 hpf (Fig. 5A–C). In contrast, fli1:she-2A-mCherry embryos, which overexpress she in endothelial cells, had significantly fewer cells in the DA compared to their sibling mCherry negative embryos in the nuclear fli1:NLS-GFP reporter background (Fig. 5D–F). As analyzed by BrdU incorporation assay at 30 hpf, fli1:she-2A-mCherry embryos in she mutant background (which are phenotypically normal) showed reduced cell proliferation within the DA compared with mCherry-negative she−/− sibling embryos (Fig. 5G–M). These results argue that she restricts the DA size by inhibiting endothelial cell proliferation. We also analyzed mural cell formation by performing fluorescent ISH analysis for pdgfrb expression. However, no significant change pdgfrb expression was observed, suggesting that the changes in the DA size are not caused by defects in mural cell formation (Suppl. Fig. S5). It has been previously demonstrated that SHE interacts with ABL kinase in vitro [24]. To test a potential role for ABL signaling in regulating vascular lumen size, we treated zebrafish embryos with chemical inhibitors of ABL signaling, dasatinib and GNF-7 [27, 28], starting at 6 hpf. Embryos treated with either ABL inhibitor displayed reduction in the DA size at 28 hpf (Fig. 6A–E). No other morphological defects were observed at this stage. By 4 dpf dasatinib and GNF-7-treated embryos developed pericardial edema and their blood circulation slowed down or stopped, while the DA diameter remained reduced at this stage (Fig. 6F–H). Importantly, the number of endothelial cells in the DA was reduced in GNF7 treated embryos at 28 hpf, compared to DMSO-treated controls (Fig. 6I–K).
Figure 5. she affects cell number and inhibits endothelial cell proliferation.
(A-C) Analysis of cell number in the DA of she mutant and wild-type (she+/+) sibling embryos at 72 hpf. Embryos were obtained from an incross of she+/−; fli1:GFP; kdrl:NLS-mCherry parents, imaged by confocal microscopy and subsequently genotyped. Note increased cell number in she mutant embryos. Data combined from 2 independent experiments. ***p<0.001, Student’s t-test. (D-F) Analysis of cell number in the DA of fli1:she-2A-mCherry; fli1:NLS-GFP embryos and their mCherry negative (wt) siblings at 72 hpf. Note the reduced cell number in mCherry+ embryos. Data combined from 2 independent experiments. ****p<0.0001, Student’s t-test. (G-M) Cell proliferation analysis using BrdU incorporation assay in she−/−; fli1:she-2A-mCherry embryos (phenotypically normal) and their sibling mCherry negative she−/− embryos in kdrl:GFP background. Note the increased number of BrdU and kdrl:GFP double positive cells within the DA in mCherry-negative she mutant embryos. Data combined from 2 independent experiments. *p<0.05, Student’s t-test.
Figure 6. Inhibitors of Abl signaling reduce DA diameter and cell number in zebrafish embryos.
(A-E) Dorsal aorta diameter at 28 hpf is reduced in kdrl:GFP embryos treated with 5 μM Dasatinib or 0.5 μM GNF-7 compared to controls treated with 0.1% DMSO.
(F-H) Embryos treated with 5 μM Dasatinib exhibit pericardial edema (arrow) and narrower DA at 4 dpf compared to controls treated with 0.1% DMSO.
(I-K) The number of cells in the DA is reduced in embryos treated with 1 μM GNF-7 compared to control embryos treated with 0.1% DMSO. Error bars show SEM in all graphs. ***p<0.001, ****p<0.0001, Student’s t-test. Data are combined from at least two independent experiments.
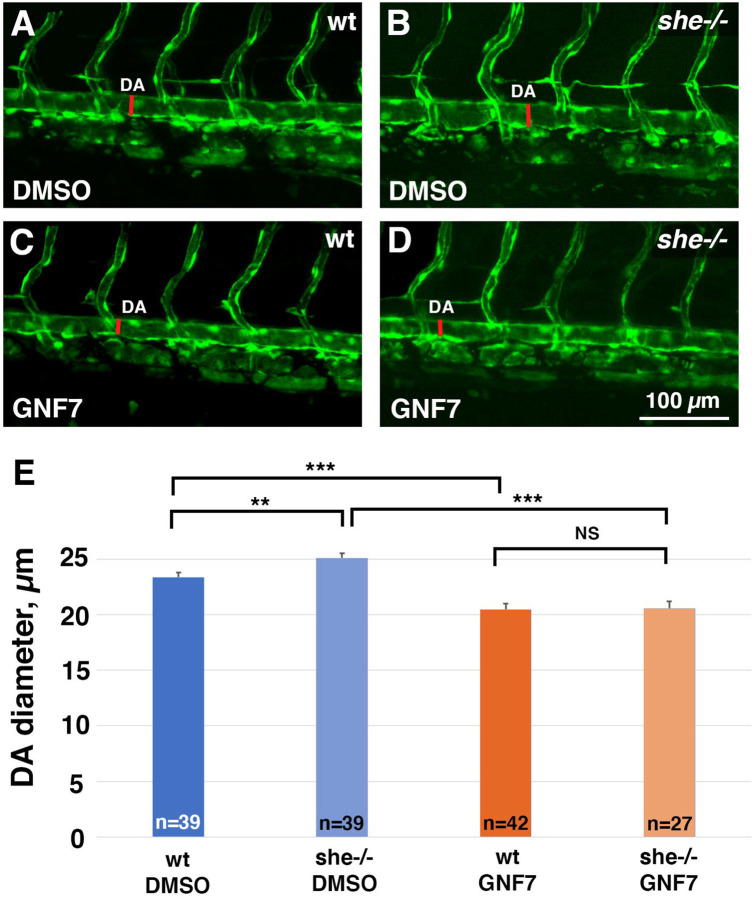
These results show that both She overexpression and inhibition of Abl signaling resulted in similar phenotypes of reduced DA diameter and lowered arterial cell number. We tested if inhibition of Abl signaling may reverse the enlarged DA observed in she mutants. she mutants and wild-type sibling embryos were treated with GNF-7 and analyzed for the DA diameter at 2 dpf. Indeed, while she mutants showed enlarged DA, GNF-7 treatment reversed this phenotype and resulted in a reduced width DA (Fig. 7).
Figure 7. Inhibition of Abl signaling reverses enlarged DA in she mutant embryos.
(A-D) she+/−; kdrl:GFP adults were crossed to obtain she mutant embryos. Embryos were treated starting at 6 hpf with either 0.5 μM GNF-7 or 0.1% DMSO. Embryos were imaged at approximately 55 hpf and subsequently genotyped. DA measurements were performed blinded. Mid-trunk region is shown, anterior is to the left. Note the slightly wider DA (red line) in she−/− mutant embryos compared to wild-type (she+/+) siblings. DA is reduced in both wild-type and she mutant embryos treated with GNF-7.
(E) Quantification of DA diameter in wild-type or she mutant embryos treated with GNF-7 or DMSO. Error bars show SEM. **p<0.01, ***p<0.001, NS – not significant, Student’s t-test.
Summary of two independent experiments.
It has been previously shown that ABL kinase can phosphorylate a related protein SHD in vitro [24]. The SHE sequence has four consensus tyrosine kinase phosphorylation sites with the YXXP motif (Suppl. Fig. S6A), which are conserved between different vertebrates and between SHE and SHD. Therefore, we hypothesized that ABL kinase may similarly target SHE protein for phosphorylation. To test the in vivo requirement for the consensus phosphorylation sites, we performed site directed mutagenesis on the zebrafish She to substitute all four consensus tyrosines into phenylalanine (YXXP –> FXXP). A zebrafish line was established that expressed the mutated She FXXP sequence under fli1 promoter. Expression of the mutant construct failed to rescue pericardial edema and blood circulation defects in she mutant embryos (Suppl. Fig. S6B). In contrast to overexpression of wild-type fli1:she-2A-mCherry construct which results in narrower DA, fli1:sheFXXP-2A-mCherry embryos showed DA enlargement at 28 hpf stage (Suppl. Fig. S6C). Overexpression of the mutant SheFXXP may interfere with the wild-type She protein, resulting in a dominant negative effect, which can explain the DA enlargement observed in fli1:sheFXXP embryos. Altogether, these results argue that the consensus YXXP phosphorylation sites are required for SHE function.
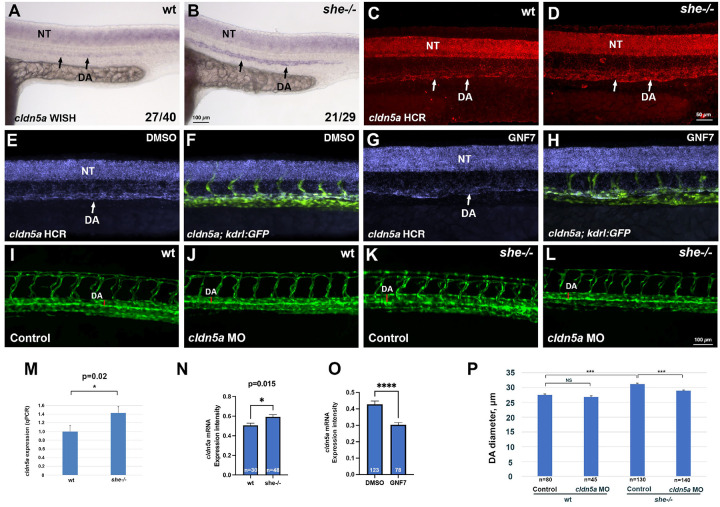
Recently it has been reported that zebrafish mutants for claudin cldn5a, the homolog of the mammalian Cldn5, exhibit reduced DA [29]. Therefore, we tested if cldn5a expression was altered in she mutants. Indeed, she mutants exhibited increased cldn5a expression within the DA at 24 hpf when analyzed by standard chromogenic and fluorescent hybridization chain reaction (HCR) in situ hybridization approaches (Fig. 8A–D). Average cldn5a fluorescence intensity was increased by 17% in she mutants compared to wild-type embryos (Fig. 8N). Expression of cldn5a in the neural tube was unaffected in she mutants. To verify this result, we purified all vascular endothelial cells using FACS sorting from kdrl:GFP line in she mutants and wild-type embryos at 24 hpf and performed qPCR. 1.4-fold increase in cldn5a expression was observed using this approach (Fig. 8M). Arterial expression of a related homolog cldn5b was also increased in she mutants (Suppl. Fig. S7), and so was Cldn5 protein expression (both Cldn5a and Cldn5b are recognized by the same Cldn5 antibody) (Suppl. Fig. S8). In contrast, inhibition of Abl signaling by treating wild-type kdrl:GFP embryos with GNF7 resulted in a reduced cldn5a expression in the DA (Fig. 8E–H, O). To test if reduction in cldn5a level could restore the DA size, we knocked down cldn5a function in she mutants and sibling embryos using a previously validated Cldn5a morpholino (MO) [29]. A subphenotypic dose of MO was used which did not result in a significant reduction of DA width in wild-type embryos. Intriguingly, the same dose of cldn5a MO resulted in a reduction of DA diameter in she mutants leading to a partial rescue of the she phenotype (Fig. 8I–L, P).
Figure 8. She regulates vascular lumen size by inhibiting cldn5a expression.
(A-D) Chromogenic whole mount in situ hybridization (WISH) (A,B) and fluorescent in situ hybridization analysis using hybridization chain reaction (HCR) (C,D) analysis for cldn5a expression in she mutants and sibling wild-type (she+/+) embryos at 24 hpf. Note increased cldn5a expression in the dorsal aorta (DA) in she mutants while neural tube (NT) expression is unaffected. Quantification of cldn5a expression in the DA is shown in (N). Embryos were obtained by incross of heterozygous she parents and genotyped after WISH or HCR. (E-H) HCR analysis for cldn5a expression in control 0.01% DMSO (E,F) or 1 μM GNF7 treated kdrl:GFP embryos (G,H). cldn5a (purple) and merged cldn5a; kdrl:GFP channels are shown in the trunk region, anterior is to the left. Note the reduction in cldn5a expression in the DA while neural tube expression is unaffected. Quantification is shown in (O).
(I-L) Inhibition of Cldn5a function using cldn5a MO injection reduces enlarged DA in she mutants. Embryos were obtained by an incross of she+/−; kdrl:GFP parents and imaged for GFP expression at 2 dpf. Mid-trunk region is shown, anterior is to the left. Quantification is shown in
(P). Note the increase in DA width (red line) in she mutants (K), which is reduced upon cldn5a MO injection (L).
(M) qPCR analysis of cldn5a expression in vascular endothelial cells obtained from FACS sorted she mutant and wild-type embryos at 24 hpf. Embryos were obtained by incross of she−/−; fli1:she-2A-mCherry+/− or she+/+ (wild-type); fli1:she-2A-mCherry+/− parents in kdrl:GFP background and sorted for the absence of mCherry. Error bars show SEM.
(N) Quantification of cldn5a expression in the DA of she mutants and wild-type siblings after HCR fluorescent in situ hybridization at 24 hpf. To reduce the staining variability between different embryos, expression values in the DA were normalized to the neural tube expression in each embryo. Expression analysis was performed blindly without knowledge of embryo genotypes.
(O) Quantification of cldn5a expression in embryos treated with 0.01% DMSO or 1 μM GNF7 at 24 hpf. Relative expression in the DA was calculated by dividing the intensity in the DA over the expression intensity in the neural tube. Student’s t-test, error bars show SEM in (N,O).
(P) Quantification of DA diameter at 2 dpf in wild-type and she mutant embryos injected with cldn5a MO. Error bars show SEM. *** p<0.001, NS – not significant.
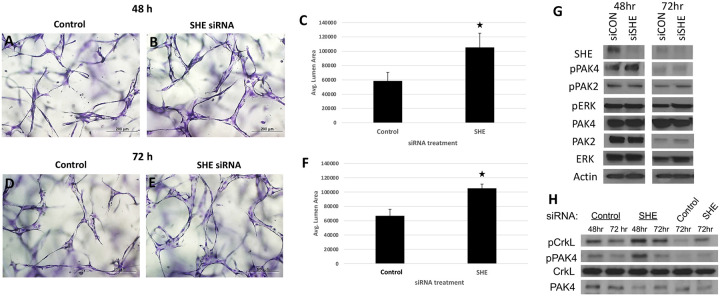
To test the conservancy of She function in mammalian cells, we inhibited She function in human umbilical vein endothelial cells (HUVECs) using siRNA. Cells were transfected with either She or control siRNA, and tubulogenesis was assayed using 3D collagen matrix assay under serum-free defined conditions. siRNA transfected cells showed enlarged vascular lumens compared to control cells, reproducibly observed both at 48 and 72 hours after transfection (Fig. 9A–F). Assay of intracellular signaling kinases by Western blot showed increased phosphorylation of pPAK4 (Fig. 9G,H), which is a known signaling effector acting downstream of Cdc42 during vascular tubulogenesis [7, 8, 30]. Importantly, phosphorylation of a known ABL signaling effector CRKL was also increased in SHE inhibited cells, while pERK phosphorylation was not affected (Fig. 9G,H). These results argue that She function is conserved between vertebrates and suggest that She restricts lumen size by inhibiting Abl and Cdc42 activities.
Figure 9. Inhibition of SHE in HUVECs results in enlarged tubulogenesis.
(A-F) HUVEC cells were transfected with either control or SHE siRNA and analyzed in 3D collagen matrix assay at 48 (A-C) or 72 h (D-F) after transfection. The values (± standard deviation) are derived from analyzed fields (n=15) where total EC tube area was measured. Asterisks indicate significance at p<0.05 compared to control conditions.
(G,H) Western blots for expression of SHE and phosphorylation of PAK4, PAK2, ERK and CRKL in HUVEC cells transfected with a control or SHE siRNA. Note greatly reduced SHE expression and increased pPAK4 and pCRKL in cells transfected with siSHE RNA. (H) shows replicate experiments for pPAK4 and pCRKL.
Discussion
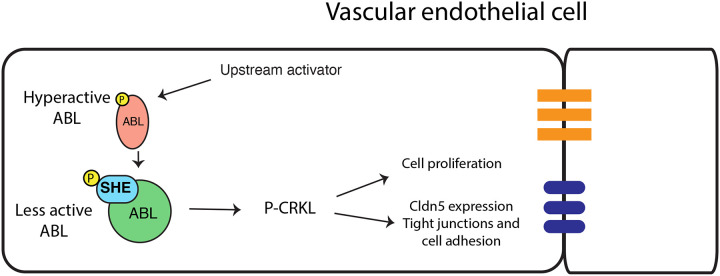
Our results demonstrate that She functions as a negative regulator of vascular lumen size. Loss of function she zebrafish mutants exhibit enlarged DA and have more arterial cells, while vascular endothelial she overexpression resulted in a reduced size DA with fewer cells. Inhibition of Abl signaling resulted in a similar reduction in DA size and cell number in wild-type embryos and reversed DA enlargement observed in she mutants, suggesting that She functions as a negative regulator of Abl signaling. A previous study demonstrated a direct interaction between She and Abl kinase in a yeast two hybrid assay [24]. Abl binding partners are often also substrates and/or activators of Abl kinase. Based on our data, we hypothesize that she functions as an adaptor protein to inhibit Abl activity. According to this model, Abl phosphorylates She, which then acts as a negative regulator to reduce Abl activity (Fig. 10). This type of feedback inhibition would allow for more accurate and sensitive regulation of vascular lumen size and could be modulated by She expression or its phosphorylation level. In support of this model, zebrafish She has four YXXP motifs that correspond to predicted Abl tyrosine phosphorylation sites [31], all of which are conserved in human and mouse. She mutant variant YXXP -> FXXP with substitutions of tyrosines to phenylalanines failed to rescue the she mutant phenotype, demonstrating the importance of all four consensus phosphorylation sites for She function. Notably, these ABL phosphorylation sites are located outside of the SH2 domain, which is consistent with our results showing that the She protein lacking the SH2 domain is sufficient to rescue she mutant embryos. These data strongly suggest that Abl kinase phosphorylates She on these consensus sites, which is then critical for She activity.
Figure 10. A proposed model for SHE and ABL signaling during vascular tubulogenesis.
Activated ABL promotes enlarged vascular lumen through a downstream effector P-CRKL which increases endothelial cell proliferation and increases Cldn5 expression, thus affecting tight junctions and cell adhesion. Activated ABL phosphorylates SHE, which then interacts with ABL to dampen its activity resulting in the lumen of appropriate size.
Although the role of Abl signaling in normal vascular tubulogenesis has not been previously known, recent work has established the role of ABL signaling during pathogenesis of venous malformations (VM). In a VM cell culture model which overexpresses activating Tie2 receptor with Tie2-L914F mutation, enlarged vascular tubes and increased phosphorylation of c-ABL was observed [23]. Inhibition of ABL signaling reduced vascular lumen size and caused VM regression. Our data suggest that ABL signaling may play a similar role during embryonic tubulogenesis by promoting enlargement of vascular lumen, while the upstream initiating signal remains unclear.
Abl kinase is known to signal through multiple cytoplasmic effector proteins in different cellular contexts. In endothelial cells, Abl can enhance Angiopoietin-mediated activation of the AKT and ERK signaling pathways [20]. It is also known to promote Rac1 activation and cell-cell adhesion by phosphorylating Crk and Crkl proteins, which are considered specific substrates of Abl kinase [15, 32, 33]. Our results show that Crkl phosphorylation is increased in She knockdown HUVEC cells, which suggests that Crkl functions as a downstream effector of She-Abl signaling in vascular tubulogenesis.
While our results strongly suggest that She functions in the Abl signaling pathway, it is important to note that the chemical inhibitors may also inhibit additional signaling pathways, and their specificity in vivo has not been fully determined. Therefore, we cannot exclude the possibility that the DA decrease in GNF7 and dasatinib treated embryos may be caused by inhibition of other signaling pathways. Obtaining additional genetic evidence will be important to fully validate the role of Abl signaling in vascular tubulogenesis.
Rho GTPases (Rac1, Cdc42 and RhoA) have been previously implicated as key regulators of actomyosin network during vascular lumen formation [1–5]. Rac1 and Cdc42 stimulate cytoskeletal remodeling to establish cell polarity including localization and stabilization of cell junctions to regulate endothelial tubulogenesis, while RhoA has an opposing activity [3, 8, 34]. Abl signaling has been implicated in regulating Rac1 activity in different non-endothelial cell types [13–16], therefore it is likely that She and Abl signaling regulate vascular lumen size by modulating the activity of Cdc42 and Rac1 or other GTPases. In support of this model, our data demonstrate that phosphorylation of PAK4, a downstream effector of Cdc42 [7, 30], is increased in SHE knockdown endothelial cells.
In addition to regulating cytoskeletal remodeling, Abl signaling is known to promote cell proliferation in different cell types, including smooth muscle cells and fibroblasts [18, 19]. Our data argue that Abl signaling also promotes vascular endothelial cell proliferation in the DA, while She acts as an inhibitor of this process. It is quite likely that downstream effectors affecting cell proliferation and cytoskeletal rearrangements are different, which needs to be investigated further in the context of vascular tubulogenesis.
Intriguingly, she expression is primarily enriched in the arterial vasculature, including the DA [25]. she mutants had little if any change in the PCV diameter, while DA diameter was enlarged as early as 28 hpf. Abl inhibition did not cause a significant change in the PCV diameter, while the DA was reduced. This suggests that Abl activity primarily regulates size of the arterial vasculature. Although vascular diameter in older embryos and adults is regulated by smooth muscle contraction, there is no smooth muscle cell coverage yet at 28 hpf. Therefore, DA enlargement is unlikely to be caused by defects in smooth muscle cells. Our results show that cldn5a and cldn5b expression, which encode zebrafish homologs of Claudin 5, a major component of the tight junctions, is upregulated in she mutants. It is possible that She-Abl signaling directly represses cldn5a/b expression to ensure an optimal amount of Cldn5a/b. Alternatively, cldn5a/b expression may be upregulated as a secondary consequence of other changes in cell shape, junctions, or polarity. cldn5a mutants display reduction in the DA lumen size [29]. Our results show that downregulation of cldn5a expression reverses the phenotype of enlarged DA observed in she mutant embryos, suggesting the DA enlargement is mediated at least partially by an increase in cldn5a expression. In addition to mediating vascular permeability, Cldn5 has been implicated in cell motility and proliferation of endothelial and tumor cells [29, 35, 36]. It is possible that increased cell proliferation in the DA may be a direct or indirect consequence of upregulated Cldn5 expression. Alternatively, increase in Cldn5 expression may be independent from She effect on cell proliferation.
The SHE protein sequence is highly conserved between zebrafish and other vertebrates. Intriguingly, siRNA knockdown of human SHE in HUVECs resulted in a similar increase in vascular tube diameter. There is no blood flow or smooth muscle / pericyte cells in this 3D collagen assay, therefore the observed tube enlargement must be cell-autonomous to vascular endothelial cells. Endothelial-specific expression of She rescued the she loss-of-function phenotype in zebrafish embryos, further supporting cell autonomous role for SHE.
In summary, our results demonstrate novel roles for SHE and ABL signaling in regulating vascular lumen size. These findings will contribute to our understanding of molecular mechanisms that regulate vascular tubulogenesis and eventually may facilitate development of novel therapeutic approaches for pathological disorders, such as venous malformations which display ectatic vessels with enlarged lumens.
Materials and Methods
Zebrafish lines. Generation of sheci26 line.
All zebrafish work was performed after approval of USF and CCHMC IACUC committees. TALEN genome editing was used to create the sheci26 allele. TALENs were designed using the TAL Effector Nucleotide Targeter software (https://tale-nt.cac.cornell.edu/) [37]. The TALEN target sequence TGTGCGTATTCGTCATGGCgaagtggtttaaagaTTTTCCAACTAGTTTGAAGA is located in exon 1 of the she gene. The lowercase letters represent the spacer sequence, which contains MseI restriction site TTAA. TALENs were assembled using Golden Gate assembly [38], and mRNA for the left and right arms was synthesized using the T3 mMessage mMachine kit (ThermoFisher). 100 pg of each mRNA was injected into one-cell stage wild type embryos. she_F1 and she_R1 primers were used to test TALEN efficacy and identify sheci26 adult carriers (primer sequences listed in Suppl. Table S1). These primers amplify a 333 bp product, which can then be digested with MseI.
Generation of sheci30 line.
CRISPR/Cas9 editing was used to create the sheci30 allele. Guide RNAs were designed using ChopChop (http://chopchop.cbu.uib.no). Two separate guide RNA templates were generated by PCR as described [39], using she_gRNA1, she_gRNA2 and the common she_gRNA_R1 primer (Suppl. Table S1). gRNAs were synthesized using the MEGAshortscript T7 kit (Life Technologies). The two gRNAs were injected together with Cas9-NLS protein (New England Biolabs) into one-cell stage embryos. The injected animals were outcrossed and PCR was performed on pooled embryos to identify carriers of she mutations, using She_gRNA left and She_gRNA right primers (Suppl. Table S1). These primers span the large deletion in sheci30, and thus generate an approximately 200 bp PCR product only from the mutant allele. To identify the wild type allele, a separate PCR reaction was performed using She_gRNA midR reverse primer (Suppl. Table S1). This primer is located within the deleted region, thus will amplify a 234 bp product only from the wild type allele.
Generation of fli1:she-2A-mCherry and hsp70:she-2A-mCherry lines and DNA overexpression analysis.
To create full length over-expression constructs, we amplified the she full length coding sequence from wild type cDNA using she_attB_F1 and she_attB_R3 primers (Suppl. Table S1) for Gateway cloning (Invitrogen). The PCR product was recombined via BP reaction into pDONR221. To make the fli1:she-2A-mCherry construct, the resulting middle entry vector (pDONR221-she) was then recombined via LR reaction with p5E-fli1ep and p3E-2AmCherry into the destination vector pDestTol2pA2 [40]. To make the hsp70-she-2A-mCherry construct, pDONR221-she, p5E-hsp70, and p3E-2A-mCherry were recombined via LR reaction into the destination vector pDestTol2pA; cryaa:dsRed. Induction of the hsp70:she-2A-mCherry transgene was confirmed by ubiquitous mCherry fluorescence, while expression of fli1:she-2A-mCherry was observed in vascular endothelial cells (Suppl. Fig. S4).
To analyze mosaic overexpression, embryos were injected with 30 ng of fli1:she-2A-mCherry DNA construct and 225 ng of Tol2 mRNA mixture. Embryos with endothelial mCherry expression were chosen for confocal imaging and further analysis. mCherry positive and negative segments of the DA (1 segment per embryo) were chosen for measurements in Fiji software. 3 measurements were performed for each segment analyzed, and measurements were then averaged to obtain a single data point used for further calculations.
To create She deletion constructs, we amplified portions of she from wild type cDNA. To generate sheDSH2 which contains SH2 domain deletion, she_attB_F1 and she_deltaSH2_attB_R primers were used. To generate sheSH2 construct which contains SH2 domain only, sheSH2_attB_F and she_attB_R3 primers were used (Suppl. Table S1). Each product was recombined via BP reaction into pDONR221. The resulting middle entry vectors were each then recombined via LR reaction with p5E-fli1ep and p3E-2AmCherry into the destination vector pDestTol2pA2. All constructs were confirmed by sequencing. All final constructs were injected with Tol2 mRNA into one-cell stage embryos derived from a cross of sheci26 heterozygous animals. Injected embryos were raised to adulthood and outcrossed to produce embryos to screen for germline integration. Transgenic lines were identified based on vascular endothelial mCherry expression.
To create the she-FXXP construct, we performed sequential site-directed mutagenesis on each YXXP site starting with the pDONR221-she construct, using the QuikChange II Site-directed Mutagenesis Kit (Agilent). Each construct was fully sequenced before proceeding with the next round of mutagenesis. The primer pairs sheYXXP1-P4 used for mutagenesis of each site are listed in Suppl. Table S1. This construct was then combined via LR reaction with the p5E-fli1ep and p3E-2A-mCherry into the pDestTol2pA2 destination vector, then injected as described above. All she lines were maintained in kdrl:GFPs843 background [41].
Other lines.
The following lines were used in the study: kdrl:GFPs843 [41], Tg(kdrl:NLS-mCherry)is4Tg [42], Tg(fli1:nGFP)y7Tg [43].
Heat-shock induction.
Embryos at the indicated stages were placed in 96 well PCR plates in embryo media with 5–10 embryos per well. To induce transgene expression, embryos were incubated at 37°C for one hour in a thermocycler. Following heat shock, embryos were transferred from PCR tubes to fresh media in petri dishes and sorted for dsRed expression in the lens or mCherry fluorescence in the body to indicate presence of the transgene. For DA diameter analysis, embryos were genotyped following confocal imaging and analysis.
Abl inhibitor treatments.
Dasatinib (Selleckchem) and GNF-7 (Selleckchem) were resuspended in DMSO to a concentration of 10 mM. Stocks were further diluted into embryo water to the appropriate working concentrations as noted. DMSO was diluted 1:1,000 or 1:10,000 in embryo water as a control. Pools of 20 dechorionated kdrl:GFP or she−/−; kdrl:GFPs843 embryos were placed in 1 mL diluted inhibitor or diluted DMSO in glass vials starting at 6 hpf. Embryos were placed on a nutator in the dark until 24–28 hpf, when they were imaged for analysis.
In situ hybridization and hybridization chain reaction analysis.
Chromogenic whole mount in situ hybridization was performed as described [44]. she DIG-labeled antisense mRNA probe was synthesized as previously described [45]. To make ISH probe for cldn5a, 897 bp fragment corresponding to the coding sequence of cldn5a was amplified by PCR from zebrafish 24 hpf embryonic cDNA. The PCR product was subcloned into pCRII-TOPO using TOPO TA cloning kit (ThermoFisher). The construct was linearized using XhoI restriction endonuclease (ThermoFisher), and antisense DIG-labeled RNA was synthesized using SP6 RNA polymerase (Promega).
Fluorescent in situ hybridization using hybridization chain reaction (HCR) for cldn5a was performed as previously described [46, 47]. Embryos were obtained from an incross of she+/−; kdrl:GFP parents, imaged using 10x or 20x objective on a Nikon A1R or Nikon Eclipse confocal microscope and subsequently genotyped. To measure fluorescence intensity of cldn5a staining, a small rectangular area which included a portion of the DA was selected using Fiji (Image J). Three measurements at different locations of the DA (using the same size rectangular) in the trunk region were performed for each embryo. In addition, a similar size area was selected in the cldn5a expression domain within the neural tube (for normalization) and in the embryo region which did not show any specific cldn5a expression (for background subtraction). Integrated density was calculated for each area. Subsequently, background subtraction was performed by subtracting integrated density values obtained for the area which did not show specific cldn5a expression. All DA expression values were then normalized by dividing them over the neural tube expression of the same embryo. This procedure reduced variation due to staining intensity variability between different embryos. cldn5a expression in the neural tube was not expected to be affected, therefore it served as a reference point. Student’s t-test was used to calculate statistical significance between normalized integrated density values of cldn5a expression in the DA of wild-type and she mutant sibling embryos. All measurements were performed blindly without a prior knowledge of embryo genotypes.
A similar approach was used to measure cldn5b intensity. HCR probes and fluorescent 647nm hairpins were obtained from Molecular Instruments Inc (Los Angeles, CA). Because cldn5b only stains DA but not the neural tube, normalization was not performed in this case. All measurement values (3 per embryo at different locations of the DA) were used for statistical analysis of cldn5a and cldn5b expression.
HCR for pdgfrb expression in she; kdrl:GFP embryos was performed under a similar protocol. pdgfrb probe was obtained from Molecular Instruments and used with 594nm hairpins. Fiji / Image J was used to measure the mean intensity of pdgfrb expression within the DA region that spans approximately 4 ISVs in each embryo. Data were combined from 3 separate replicate experiments. Because the overall staining intensity varied between separate experiments, relative fluorescence intensity was normalized within each experiment. For this purpose, mean pdgfrb fluorescence intensity values were calculated for wild-type embryos within each experiment. Then the measured intensity values for each wild-type or mutant embryo were divided over the same mean wt intensity value calculated for each experiment. The resulting relative expression values were used for the graph and statistical calculations.
Cldn5 immunostaining.
Cldn5 immunostaining was performed using a previously established protocol [48]. Embryos were obtained from a cross of she+/−; kdrl:GFP parents, imaged using confocal microscopy and subsequently genotyped. To measure fluorescence intensity of Cldn5 immunostaining, a small rectangular area which included a portion of the DA was selected using Fiji (Image J). 10 measurements at different locations of the DA (using the same size rectangular) in the trunk region were performed for each embryo. In addition, a similar size area was selected in the Cldn5 expression domain within the neural tube (for normalization) and in the embryo region which did not show any specific Cldn5 expression (for background subtraction). Mean fluorescence intensity was measured for each area. Subsequently, background subtraction was performed by subtracting mean fluorescence values obtained for the area which did not show specific Cldn5 expression. All DA expression values were then normalized by dividing them over the neural tube expression of the same embryo. This procedure reduced variation due to staining intensity variability between different embryos. Student’s t-test was used to calculate statistical significance between all normalized values of Cldn5 expression in the DA of wild-type / heterozygous (she+/+ and she+/− embryos were combined into a single group) and she−/− sibling embryos. All measurements were performed blindly without prior knowledge of embryo genotypes.
Morpholino injection.
4 ng of a previously validated cldn5a MO [29](GeneTools Inc, sequence AGGCCATCGCTTTCTTTTCCCACTC) was injected into embryos at 1-cell stage obtained from a cross of she+/−; kdrl:GFP parents. Embryos from injected and control uninjected groups were imaged at 2 dpf using confocal microscopy (Nikon Eclipse) and genotyped. DA lumen diameter was analyzed as described above. All analysis was performed blindly without knowing embryo genotypes.
qPCR analysis using FACS-sorted endothelial cells.
Wild-type and she mutant embryos were obtained from respective incrosses of she+/+; fli1:she-2A-mCherry; kdrl:GFP and she−/−; fli1:she-2A-mCherry; kdrl:GFP parent fish (which were siblings). 15–20 mCherry-negative embryos were obtained for each group at 24 hpf stage. Single-cell suspension was obtained as previously described [49]. FACS sorting for GFP-positive cells was performed at the CCHMC FACS Core facility. Total RNA was purified using RNA purification kit (Norgen Biotek). cDNA was synthesized at CCHMC Gene Expression Core facility. Quantitative real-time PCR (qRT-PCR) was carried out using PowerUp SYBR Green Master Mix (Applied Biosystems) in a StepOnePlus Real-Time PCR System. Each reaction was repeated twice, and two biological replicates were used. qPCR amplification for cldn5a and EF1α was performed using the following primers: cldn5a-F (GACAACGTGAAAGCGCGGG), cldn5a-R (AGGAGCAGCAGAGTATGCTTCCC), EF1a-F (TCACCCTGGGAGTGAAACAGC) and EF1a-R (ACTTGCAGGCGATGTGAGCAG). Fold changes were calculated using a relative standard curve methods and normalized to EF1a expression.
Microscopy imaging.
For brightfield imaging, sheci30 and sheci26 embryos were obtained by respective crosses of heterozygous parents. Live embryos were anesthetized in 0.016% Tricaine solution, mounted in 2% methylcellulose and imaged using Zeiss Axioimager (10x objective). Embryos were subsequently genotyped. For brightfield imaging of embryos after in situ hybridization, they were mounted in 0.6% low-melting-point agarose and imaged using 10x objective on a Nikon Eclipse compound microscope.
Fluorescent imaging of live embryos and fixed embryos after HCR was performed using Nikon confocal microscope (A1R or Eclipse). Embryos were mounted in 0.6% low melting-point-agarose. Nikon Elements AR Denoise.ai algorithm was used to remove noise from images with high background signal. Maximum intensity projections were obtained using Nikon Elements or Fiji (Image J) software. Image levels were adjusted using Adobe Photoshop 2021 to maximize brightness and contrast; in all cases similar adjustments in control and experimental embryos were performed.
Measurements of DA and PCV diameter.
To analyze the DA and PCV diameter of sheci26 mutants, embryos were obtained by a cross of heterozygous parents. Live embryos were anesthetized in Tricaine, mounted in 0.6% LMP agarose and imaged using Nikon A1 confocal microscope. Subsequently embryos were retrieved and genotyped. Measurements were performed in Fiji (Image J). Four measurements across the DA or PCV were taken in different areas of the mid-trunk for each embryo. Measurements were performed at the midpoint between two ISVs, every other ISV. Because the diameter of the DA (or PCV) can vary within each embryo, each measurement was treated as an independent datapoint for statistical calculations. Measurements were performed blindly without knowledge of embryo genotypes. DA diameter was measured similarly in embryos treated with Abl inhibitors. In the experiment where she mutant and control embryos were treated with GNF7 (Fig. 6), 3 measurements within the DA in each embryo were performed (embryos were imaged at higher magnification, and thus a smaller region of DA in the trunk was available for analysis). In the experiment where she and mutant embryos were injected with cldn5a MO (Fig. 7E–H), 5 measurements within the DA in each embryo were performed (lower magnification imaging was used for this analysis covering a larger region of an embryo).
Cell number analysis.
To analyze arterial cell number in she mutants, embryos were obtained from an incross of she+/−; kdrl:GFP; kdrl:NLS-mCherry adults and imaged using confocal microscopy under a 20x objective at 72 hpf. Embryos were then extracted from the LMP agarose and genotyped. Nuclei cell counts analysis of the DA region was performed within a segment between 5 ISVs using Imaris 7.2 software. First, the region of interest was selected to limit the counts within the DA. An estimated diameter was calculated by measuring the size of selected cell nuclei within the Imaris. Then, the preliminary results were manually corrected to remove or add nuclei based on the confocal 3D image. Similar approach was used for cell counts in GNF7 treated embryos and DMSO controls using kdrl:GFP; kdrl:NLS-mCherry line.
To count arterial cells in She overexpressing embryos, double transgenic fli1:she-2A-mCherry; fli1:NLS-GFP embryos were obtained from a cross of separate transgenic parents. Confocal imaging at 72 hpf and cell counts were performed in GFP channel as described above.
Cell proliferation analysis.
Embryos were obtained through an incross of she−/−; fli1:she-2A-mCherry; kdrl:GFP adult zebrafish. mCherry positive and negative embryos were segregated by screening under a fluorescent microscope. The embryos were treated with BrdU (Millipore-Sigma, Cat No. B5002) in a 24-well plate (20 embryos per well in a 200μl of 15 mM BrdU solution in 15% DMSO) from 29 to 29.5 hpf. At 29.5 hpf, the embryos were allowed to recover in embryo water for 30 minutes and fixed afterwards at 30hpf. The embryos were fixed in BT-fix for 2 hours at room temperature (RT). Fixed embryos were walked with gradual changes into 100% methanol and kept overnight at −20°C. The next day, embryos were walked into PBT (PBS + 0.1% Tween 20). The embryos were washed with PBT for 3X15 mins. This was followed by incubation in 2N HCL for 1 hour at RT. The embryos were washed with PBT for 3X5 mins. The embryos were then placed in blocking buffer for 2.5 hours. This was followed by overnight incubation in primary anti-BrdU antibody (ab6326, abcam, 1:250 dilution). The embryos were washed the next day with PBT for 6X15 mins. This was followed by overnight incubation in the goat anti-rat Alexa Fluor 647 secondary antibody (A21247, ThermoFisher, 1:500). The embryos were washed with PBT 4X15 min and kept at 4°C overnight. The embryos were imaged the next day on a Nikon Eclipse confocal microscope with 20X water immersion objective. The entire trunk region was imaged in separate segments (3 images / embryo). The double labelled cells were counted using the Imaris software 9.1 (Bitplane). A region of interest was drawn to encompass only the DA. Separate spots were created for kdrl:GFP positive cells and anti-BrdU labeled cells, respectively. Co-localization was defined as spots by no more than 4μm. Colocalization and cell identity was confirmed by manual analysis of the image. The data were obtained from two independent experiments.
Intersegmental vessel analysis.
To count arterial and venous connections, sheci26+/−; kdrl:GFP adults were crossed to obtain a mixture of she homozygous, heterozygous and wild-type embryos, which were then mounted in 0.6% LMP agarose and imaged by confocal microscopy using a 10x objective at 72 hpf. Embryos were then extracted from the agarose and genotyped. Obtained images were analyzed blindly without knowing embryo genotype to determine arterial or venous connections of 10 consecutive pairs (20 ISVs per embryo.)
EC tube formation assays in 3D collagen matrices.
Human umbilical vein endothelial cells (ECs) were grown and used from passages 3–6 as previously described [50]. Human ECs were seeded in 3D collagen matrices in the presence of a combination of recombinant growth factors, stem cell factor (SCF), interleukin-3 (IL-3), stromal-derived factor-1 alpha (SDF-1α), and fibroblast growth factor-2 (FGF-2), which were added to Medium 199 containing a 1:250 dilution of reduced serum supplement-2 [50], which has insulin as a component. The recombinant growth factors were obtained from R&D Systems and were added at the following concentrations: SCF, IL-3, and SDF-1α at 40 ng/ml and FGF-2 at 50 ng/ml. The details of the serum-free defined assay system set-up have been described previously [51, 52]. siRNA suppression of ECs was performed as described [50], using two rounds of transfection using either a control siRNA or an siRNA directed to She. Silencer select siRNAs (Invitrogen) were utilized for these experiments with a control siRNA (Negative control #2- catalog number 4390896) and a She siRNA (catalog number s43069). After siRNA treatment, ECs were seeded in 3D collagen matrices in the presence of the recombinant growth factors, and after 48 or 72 hr, cultures were either fixed with 3% glutaraldehyde in phosphate-buffered saline or collagen gels were plucked and lysates were prepared using SDS-PAGE Laemmli 1.5X sample buffer. Western blots were run and probed with antibodies directed to phospho-Pak2, phospho-Pak4, phospho-Erk, phospho-Crkl (Tyr207, Cat No. 3181S), total Pak2, total Pak4, total Erk and total Crkl (Cat No. 38710S) (Cell Signaling Technology). The anti-She antibody was from Novus and the anti-actin antibody was from Calbiochem. Fixed cultures were stained with 0.1% toluidine blue, and cultures were photographed using an Inverted Olympus microscope, and tube area was traced and quantitated using Metamorph software. Representative experiments were analyzed, and the data was obtained from triplicate wells while lumen area was measured from n=15 fields. Statistical analysis utilized a Student’s t-test and statistical significance was established with a p<0.05.
Reagent availability.
All DNA plasmids and zebrafish lines are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
We thank Matt Kofron and CCHMC Confocal Core for assistance with imaging and CCHMC FACS core for assistance with cell sorting.
Funding
This work was supported by NIH R01 HL163161 award to S.S. and the Trustee award to J.A.S. from the Cincinnati Children’s Research Foundation.
Funding Statement
This work was supported by NIH R01 HL163161 award to S.S. and the Trustee award to J.A.S. from the Cincinnati Children’s Research Foundation.
Footnotes
Competing interests
No competing interests declared.
References
- 1.Sit ST, Manser E. Rho GTPases and their role in organizing the actin cytoskeleton. J Cell Sci. 2011;124(Pt 5):679–83. Epub 2011/02/16. doi: 10.1242/jcs.064964. [DOI] [PubMed] [Google Scholar]
- 2.Rogers KK, Jou TS, Guo W, Lipschutz JH. The Rho family of small GTPases is involved in epithelial cystogenesis and tubulogenesis. Kidney Int. 2003;63(5):1632–44. Epub 2003/04/05. doi: 10.1046/j.1523-1755.2003.00902.x. [DOI] [PubMed] [Google Scholar]
- 3.Bayless KJ, Davis GE. The Cdc42 and Rac1 GTPases are required for capillary lumen formation in three-dimensional extracellular matrices. J Cell Sci. 2002;115(Pt 6):1123–36. Epub 2002/03/09. [DOI] [PubMed] [Google Scholar]
- 4.Barry DM, Koo Y, Norden PR, Wylie LA, Xu K, Wichaidit C, et al. Rasip1-Mediated Rho GTPase Signaling Regulates Blood Vessel Tubulogenesis via Nonmuscle Myosin II. Circ Res. 2016;119(7):810–26. Epub 20160802. doi: 10.1161/CIRCRESAHA.116.309094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayless KJ, Davis GE. Microtubule depolymerization rapidly collapses capillary tube networks in vitro and angiogenic vessels in vivo through the small GTPase Rho. J Biol Chem. 2004;279(12):11686–95. Epub 2003/12/31. doi: 10.1074/jbc.M308373200. [DOI] [PubMed] [Google Scholar]
- 6.Davis GE, Stratman AN, Sacharidou A, Koh W. Molecular basis for endothelial lumen formation and tubulogenesis during vasculogenesis and angiogenic sprouting. Int Rev Cell Mol Biol. 2011;288:101–65. doi: 10.1016/B978-0-12-386041-5.00003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Norden PR, Kim DJ, Barry DM, Cleaver OB, Davis GE. Cdc42 and k-Ras Control Endothelial Tubulogenesis through Apical Membrane and Cytoskeletal Polarization: Novel Stimulatory Roles for GTPase Effectors, the Small GTPases, Rac2 and Rap1b, and Inhibitory Influence of Arhgap31 and Rasa1. PLoS One. 2016;11(1):e0147758. Epub 20160126. doi: 10.1371/journal.pone.0147758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barry DM, Xu K, Meadows SM, Zheng Y, Norden PR, Davis GE, et al. Cdc42 is required for cytoskeletal support of endothelial cell adhesion during blood vessel formation in mice. Development. 2015;142(17):3058–70. Epub 20150807. doi: 10.1242/dev.125260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bayless KJ, Salazar R, Davis GE. RGD-dependent vacuolation and lumen formation observed during endothelial cell morphogenesis in three-dimensional fibrin matrices involves the alpha(v)beta(3) and alpha(5)beta(1) integrins. Am J Pathol. 2000;156(5):1673–83. Epub 2000/05/04. doi: 10.1016/s0002-9440(10)65038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iruela-Arispe ML, Davis GE. Cellular and molecular mechanisms of vascular lumen formation. Dev Cell. 2009;16(2):222–31. Epub 2009/02/17. doi: 10.1016/j.devcel.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khatri A, Wang J, Pendergast AM. Multifunctional Abl kinases in health and disease. J Cell Sci. 2016;129(1):9–16. Epub 2016/01/06. doi: 10.1242/jcs.175521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong S, Witte ON. The BCR-ABL story: bench to bedside and back. Annu Rev Immunol. 2004;22:247–306. Epub 2004/03/23. doi: 10.1146/annurev.immunol.22.012703.104753. [DOI] [PubMed] [Google Scholar]
- 13.Adams DM, Trenor CC 3rd, Hammill AM, Vinks AA, Patel MN, Chaudry G, et al. Efficacy and Safety of Sirolimus in the Treatment of Complicated Vascular Anomalies. Pediatrics. 2016;137(2):e20153257. Epub 2016/01/20. doi: 10.1542/peds.2015-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li R, Pendergast AM. Arg kinase regulates epithelial cell polarity by targeting beta1-integrin and small GTPase pathways. Curr Biol. 2011;21(18):1534–42. Epub 2011/09/13. doi: 10.1016/j.cub.2011.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zandy NL, Playford M, Pendergast AM. Abl tyrosine kinases regulate cell-cell adhesion through Rho GTPases. Proc Natl Acad Sci U S A. 2007;104(45):17686–91. Epub 2007/10/30. doi: 10.1073/pnas.0703077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li R, Knight JF, Park M, Pendergast AM. Abl Kinases Regulate HGF/Met Signaling Required for Epithelial Cell Scattering, Tubulogenesis and Motility. PLoS One. 2015;10(5):e0124960. Epub 2015/05/07. doi: 10.1371/journal.pone.0124960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sini P, Cannas A, Koleske AJ, Di Fiore PP, Scita G. Abl-dependent tyrosine phosphorylation of Sos-1 mediates growth-factor-induced Rac activation. Nat Cell Biol. 2004;6(3):268–74. Epub 2004/03/25. doi: 10.1038/ncb1096. [DOI] [PubMed] [Google Scholar]
- 18.Mitra S, Beach C, Feng GS, Plattner R. SHP-2 is a novel target of Abl kinases during cell proliferation. J Cell Sci. 2008;121(Pt 20):3335–46. Epub 20080930. doi: 10.1242/jcs.035691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia L, Wang R, Tang DD. Abl regulates smooth muscle cell proliferation by modulating actin dynamics and ERK1/2 activation. Am J Physiol Cell Physiol. 2012;302(7):C1026–34. Epub 20120201. doi: 10.1152/ajpcell.00373.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chislock EM, Ring C, Pendergast AM. Abl kinases are required for vascular function, Tie2 expression, and angiopoietin-1-mediated survival. Proc Natl Acad Sci U S A. 2013;110(30):12432–7. Epub 2013/07/11. doi: 10.1073/pnas.1304188110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raimondi C, Fantin A, Lampropoulou A, Denti L, Chikh A, Ruhrberg C. Imatinib inhibits VEGF-independent angiogenesis by targeting neuropilin 1-dependent ABL1 activation in endothelial cells. J Exp Med. 2014;211(6):1167–83. Epub 2014/05/28. doi: 10.1084/jem.20132330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chislock EM, Pendergast AM. Abl family kinases regulate endothelial barrier function in vitro and in mice. PLoS One. 2013;8(12):e85231. Epub 2013/12/25. doi: 10.1371/journal.pone.0085231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Cai Y, Goines J, Pastura P, Brichta L, Lane A, et al. Ponatinib Combined With Rapamycin Causes Regression of Murine Venous Malformation. Arterioscler Thromb Vasc Biol. 2019;39(3):496–512. Epub 2019/01/11. doi: 10.1161/ATVBAHA.118.312315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oda T, Kujovich J, Reis M, Newman B, Druker BJ. Identification and characterization of two novel SH2 domain-containing proteins from a yeast two hybrid screen with the ABL tyrosine kinase. Oncogene. 1997;15(11):1255–62. Epub 1997/10/07. doi: 10.1038/sj.onc.1201299. [DOI] [PubMed] [Google Scholar]
- 25.Wong KS, Proulx K, Rost MS, Sumanas S. Identification of vasculature-specific genes by microarray analysis of etsrp/etv2 overexpressing zebrafish embryos. Dev Dyn. 2009;238(7):1836–50. Epub 2009/06/09. doi: 10.1002/dvdy.21990. [DOI] [PubMed] [Google Scholar]
- 26.Liu BA, Engelmann BW, Nash PD. The language of SH2 domain interactions defines phosphotyrosine-mediated signal transduction. FEBS Lett. 2012;586(17):2597–605. Epub 2012/05/10. doi: 10.1016/j.febslet.2012.04.054. [DOI] [PubMed] [Google Scholar]
- 27.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305(5682):399–401. Epub 2004/07/17. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 28.Choi HG, Ren P, Adrian F, Sun F, Lee HS, Wang X, et al. A type-II kinase inhibitor capable of inhibiting the T315I “gatekeeper” mutant of Bcr-Abl. J Med Chem. 2010;53(15):5439–48. Epub 2010/07/08. doi: 10.1021/jm901808w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z, Wu S, Fontana F, Li Y, Xiao W, Gao Z, et al. The tight junction protein Claudin-5 limits endothelial cell motility. J Cell Sci. 2021;134(1). Epub 2020/12/17. doi: 10.1242/jcs.248237. [DOI] [PubMed] [Google Scholar]
- 30.Koh W, Mahan RD, Davis GE. Cdc42- and Rac1-mediated endothelial lumen formation requires Pak2, Pak4 and Par3, and PKC-dependent signaling. J Cell Sci. 2008;121(Pt 7):989–1001. Epub 20080304. doi: 10.1242/jcs.020693. [DOI] [PubMed] [Google Scholar]
- 31.Cujec TP, Medeiros PF, Hammond P, Rise C, Kreider BL. Selection of v-abl tyrosine kinase substrate sequences from randomized peptide and cellular proteomic libraries using mRNA display. Chem Biol. 2002;9(2):253–64. Epub 2002/03/07. [DOI] [PubMed] [Google Scholar]
- 32.Feller SM, Knudsen B, Hanafusa H. c-Abl kinase regulates the protein binding activity of c-Crk. EMBO J. 1994;13(10):2341–51. Epub 1994/05/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greulich H, Reichman C, Hanafusa H. Delay in serum stimulation of Erk activity caused by oncogenic transformation. Oncogene. 1996;12(8):1689–95. Epub 1996/04/18. [PubMed] [Google Scholar]
- 34.Duquette PM, Lamarche-Vane N. Rho GTPases in embryonic development. Small GTPases. 2014;5(2):8. Epub 2014/12/09. doi: 10.4161/sgtp.29716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akizuki R, Shimobaba S, Matsunaga T, Endo S, Ikari A. Claudin-5, −7, and −18 suppress proliferation mediated by inhibition of phosphorylation of Akt in human lung squamous cell carcinoma. Biochim Biophys Acta Mol Cell Res. 2017;1864(2):293–302. Epub 20161121. doi: 10.1016/j.bbamcr.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 36.Ma SC, Li Q, Peng JY, Zhouwen JL, Diao JF, Niu JX, et al. Claudin-5 regulates blood-brain barrier permeability by modifying brain microvascular endothelial cell proliferation, migration, and adhesion to prevent lung cancer metastasis. CNS Neurosci Ther. 2017;23(12):947–60. Epub 20170929. doi: 10.1111/cns.12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, Vandyk JK, et al. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40(Web Server issue):W117–22. Epub 2012/06/14. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cermak T, Starker CG, Voytas DF. Efficient design and assembly of custom TALENs using the Golden Gate platform. Methods Mol Biol. 2015;1239:133–59. Epub 2014/11/20. doi: 10.1007/978-1-4939-1862-1_7. [DOI] [PubMed] [Google Scholar]
- 39.Talbot JC, Amacher SL. A streamlined CRISPR pipeline to reliably generate zebrafish frameshifting alleles. Zebrafish. 2014;11(6):583–5. Epub 2014/12/04. doi: 10.1089/zeb.2014.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villefranc JA, Amigo J, Lawson ND. Gateway compatible vectors for analysis of gene function in the zebrafish. Dev Dyn. 2007;236(11):3077–87. Epub 2007/10/20. doi: 10.1002/dvdy.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin SW, Beis D, Mitchell T, Chen JN, Stainier DY. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development. 2005;132(23):5199–209. Epub 2005/10/28. doi: 10.1242/dev.02087. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Kaiser MS, Larson JD, Nasevicius A, Clark KJ, Wadman SA, et al. Moesin1 and Ve-cadherin are required in endothelial cells during in vivo tubulogenesis. Development. 2010;137(18):3119–28. doi: 10.1242/dev.048785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roman BL, Pham VN, Lawson ND, Kulik M, Childs S, Lekven AC, et al. Disruption of acvrl1 increases endothelial cell number in zebrafish cranial vessels. Development. 2002;129(12):3009–19. doi: 10.1242/dev.129.12.3009. [DOI] [PubMed] [Google Scholar]
- 44.Jowett T. Analysis of protein and gene expression. Methods Cell Biol. 1999;59:63–85. Epub 1999/01/19. doi: 10.1016/s0091-679x(08)61821-x. [DOI] [PubMed] [Google Scholar]
- 45.Sumanas S, Jorniak T, Lin S. Identification of novel vascular endothelial-specific genes by the microarray analysis of the zebrafish cloche mutants. Blood. 2005;106(2):534–41. Epub 2005/04/02. doi: 10.1182/blood-2004-12-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chestnut B, Casie Chetty S, Koenig AL, Sumanas S. Single-cell transcriptomic analysis identifies the conversion of zebrafish Etv2-deficient vascular progenitors into skeletal muscle. Nat Commun. 2020;11(1):2796. Epub 2020/06/05. doi: 10.1038/s41467-020-16515-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, et al. Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development. 2018;145(12). Epub 20180626. doi: 10.1242/dev.165753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Leeuwen LM, Evans RJ, Jim KK, Verboom T, Fang X, Bojarczuk A, et al. A transgenic zebrafish model for the in vivo study of the blood and choroid plexus brain barriers using claudin 5. Biol Open. 2018;7(2). Epub 20180202. doi: 10.1242/bio.030494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manoli M, Driever W. Fluorescence-activated cell sorting (FACS) of fluorescently tagged cells from zebrafish larvae for RNA isolation. Cold Spring Harb Protoc. 2012;2012(8). Epub 2012/08/03. doi: 10.1101/pdb.prot069633. [DOI] [PubMed] [Google Scholar]
- 50.Koh W, Stratman AN, Sacharidou A, Davis GE. In vitro three dimensional collagen matrix models of endothelial lumen formation during vasculogenesis and angiogenesis. Methods Enzymol. 2008;443:83–101. doi: 10.1016/S0076-6879(08)02005-3. [DOI] [PubMed] [Google Scholar]
- 51.Stratman AN, Davis MJ, Davis GE. VEGF and FGF prime vascular tube morphogenesis and sprouting directed by hematopoietic stem cell cytokines. Blood. 2011;117(14):3709–19. Epub 20110114. doi: 10.1182/blood-2010-11-316752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davis GE, Kim DJ, Meng CX, Norden PR, Speichinger KR, Davis MT, et al. Control of vascular tube morphogenesis and maturation in 3D extracellular matrices by endothelial cells and pericytes. Methods Mol Biol. 2013;1066:17–28. doi: 10.1007/978-1-62703-604-7_2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.