Abstract
Background and Objectives:
Single nucleotide variants near TMEM106B associate with risk of frontotemporal lobar dementia with TDP-43 inclusions (FTLD-TDP) and Alzheimer’s disease (AD) in genome-wide association studies (GWAS), but the causal variant at this locus remains unclear. Here we asked whether a novel structural variant on TMEM106B is the causal variant.
Methods:
An exploratory analysis identified structural variants on neurodegeneration-related genes. Subsequent analyses focused on an Alu element insertion on the 3’UTR of TMEM106B. This study included data from longitudinal aging and neurogenerative disease cohorts at Stanford University, case-control cohorts in the Alzheimer’s Disease Sequencing Project (ADSP), and expression and proteomics data from Washington University in St. Louis (WUSTL). 432 individuals from two Stanford aging cohorts were whole-genome long-read and short-read sequenced. 16,906 samples from ADSP were short-read sequenced. Genotypes, transcriptomics, and proteomics data were available in 1,979 participants from an aging and dementia cohort at WUSTL. Selection criteria were specific to each cohort. In primary analyses, the linkage disequilibrium between the TMEM106B locus variants in the FTLD-TDP GWAS and the 3’UTR insertion was estimated. We then estimated linkage by ancestry in the ADSP and evaluated the effect of the TMEM106B lead variant on mRNA and protein levels.
Results:
The primary analysis included 432 participants (52.5% females, age range 45–92 years old). We identified a 316 bp Alu insertion overlapping the TMEM106B 3’UTR tightly linked with top GWAS variants rs3173615(C) and rs1990622(A). In ADSP European-ancestry participants, this insertion is in equivalent linkage with rs1990622(A) (R2=0.962, D’=0.998) and rs3173615(C) (R2=0.960, D’=0.996). In African-ancestry participants, the insertion is in stronger linkage with rs1990622(A) (R2=0.992, D’=0.998) than with rs3173615(C) (R2=0.811, D’=0.994). In public datasets, rs1990622 was consistently associated with TMEM106B protein levels but not with mRNA expression. In the WUSTL dataset, rs1990622 is associated with TMEM106B protein levels in plasma and cerebrospinal fluid, but not with TMEM106B mRNA expression.
Discussion:
We identified a novel Alu element insertion in the 3’UTR of TMEM106B in tight linkage with the lead FTLD-TDP risk variant. The lead variant is associated with TMEM106B protein levels, but not expression. The 3’UTR insertion is a lead candidate for the causal variant at this complex locus, pending confirmation with functional studies.
Introduction
Genome-wide association studies (GWAS) using imputed micro-array data or short-read next-generation sequencing (NGS) have identified single nucleotide variants (SNVs) associated with neurodegenerative diseases. Variants associated with disease risk are frequently intergenic or intronic and rarely exonic. Among significant variants at a given locus, one may be causal or in linkage with a nearby genetic feature that is the true disease risk-modifying variant. Identifying the true causal variant at a GWAS locus is critical for elucidating disease pathogenesis and for developing candidate therapeutics. Structural variants (SVs) – which include large insertions, deletions, duplications, and other genomic features greater than 50 base pairs (bp) in length – are a source of genetic diversity whose impact on protein function is often readily interpretable due to their large size. SVs are challenging to identify with short-read NGS due to the typical 150 bp read length. SVs exceeding this length are detected in NGS data by analyzing paired and split-read evidence as well as changes in sequencing depth1–3. Emerging long-read sequencing (LRS) technology utilizes reads typically averaging 10 to 20 kilobases, enabling large SVs to be directly sequenced and correctly aligned to the genome4. LRS greatly enhances SV discovery over short-read NGS, identifying more than twice as many SVs as ensemble methods operating on short-read NGS data. Up to 83% of insertions identified by LRS are not detected by NGS algorithms5.
We performed LRS and SV calling for participants enrolled in Stanford’s Iqbal Farrukh and Asad Jamal Alzheimer’s Disease Research Center (ADRC) and the Stanford Aging and Memory Study (SAMS)6. We undertook an initial exploratory analysis looking for SVs overlapping exons, the 5’UTR or the 3’UTR of 4579 genes linked in Gene Ontology to neurodegenerative disorders. This analysis revealed a 316 base-pair insertion on the 3’UTR of TMEM106B which—given the strong risk-modifying effects of a TMEM106B locus against frontotemporal lobar dementia with TAR DNA-binding protein pathology (FTLD-TDP)—we explored further7,8.
The initial FTLD-TDP GWAS7 identified three significant SNVs (rs1990622, rs6966915, rs1020004) associated with reduced risk and in high linkage disequilibrium (LD) with one another, all on or near TMEM106B7. Subsequent work showed a pronounced effect on age-at-onset in GRN mutation carriers with autosomal dominant FTLD-TDP9. The mechanism by which variants at the TMEM106B locus affect FTLD-TDP risk remains unclear because the significant variants are intronic or intergenic7. The only coding SNV in LD with rs1990622 is the missense variant rs3173615, which results in a p.T185S amino acid change in exon 6 of TMEM106B. A cell-based assay suggested that rs3173615 may hasten protein degradation10. However, an in vivo study using a GRN−/− mouse model homozygous for TMEM106B*S186 (the conserved residue in mice) revealed no change in TMEM106B protein levels relative to wild-type, nor amelioration of the pathological lysosomal phenotype11. Furthermore, a meta-analysis of FTLD-TDP GWAS evaluating both rs1990622 and rs3173615 found that rs1990622 was the most significant SNV at the locus12. As such, the evidence for rs3173615 as the causative variant on TMEM106B remains mixed. Other studies have nominated an intergenic variant near rs1990622 that alters chromatin architecture and modulates TMEM106B expression13 or 3’UTR variants that affect microRNA binding sites14. Recently, a large Alzheimer’s disease (AD) GWAS identified what appears to be the same TMEM106B locus. As in FTLD-TDP, the minor allele in Europeans was associated with reduced risk for AD, though the effect size was much smaller. Given the uncertainty regarding the causal variant at this locus, we examined whether this newly identified TMEM106B 3’UTR insertion may mediate the observed effect on FTLD-TDP and AD risk.
Methods
PARTICIPANTS AND SOURCES OF DATA
The Stanford ADRC is a cohort of healthy older controls and patients with AD and related neurological disorders (n=323 with LRS and short-read NGS, age range 45–92 years old, 169 females and 154 males, healthy controls = 150, mild cognitive impairment individuals = 60, AD cases = 30, other diagnoses = 83). The Stanford Aging and Memory Study (SAMS) is a cohort of cognitively unimpaired older individuals (n=109 with LRS and short-read NGS, age range 60–88 years old, 58 females and 51 males). The WUSTL participants were recruited as part of the Knight-ADRC and included longitudinally assessed community-dwelling adults enrolled via prospective studies of memory and aging (n=1979 with short-read NGS, transcriptomics and/or CSF and/or plasma proteomics, age range 18–103 years old, 1034 females and 945 males, healthy controls = 1005, AD cases = 858, other diagnoses = 116). Details on eligibility and assessments of participants in all datasets are provided in eMethods.
LONG-READ SEQUENCING, ALIGNMENT, AND SV CALLING
High molecular weight DNA was extracted from primary blood mononuclear cells stored at −80C using a Puregene kit (Qiagen, Germany). DNA was sheared using a G-tube (Covaris LLC, Massachusetts). Sequencing libraries were prepared using Nanopore LSK-110 and sequenced on the PromethION48 (Oxford Nanopore Technologies, United Kingdom). An average of 50.4 gigabases were sequenced per sample, with a read length N50 of 18 kb. Sequencing data were base called using Guppy (High Accuracy, version 6.3), and aligned to hg38 using Minimap215. Structural variants were called using Sniffles216 in population mode. SVs overlapping exons, the 5’UTR or the 3’UTR of 4579 genes linked in Gene Ontology to neurodegenerative disorders and extracted using the following keywords: [“neuro”, “alzheimer”, “apolipo”, “amyloid”].
SHORT-READ NEXT GENERATION SEQUENCING
TMEM106B SNV genotypes were determined from short-read NGS performed at either the Beijing Genomics Institute (BGI) in Shenzhen, China on DNBseq platform (T10 and T7), or as part of the Stanford Extreme Phenotypes in Alzheimer’s Disease project with sequencing performed at the Uniformed Services University of the Health Sciences (USUHS) on an Illumina HiSeq platform. Among the 432 participants, 29 participants were sequenced via USUHS and 403 via BGI. The whole-genome targeted coverage for both platforms was 30x and the read length was 150 bp. The Genome Analysis Toolkit (GATK) workflow germline short variant discovery was used to map genome sequencing data to the reference genome (hg38) and to produce high-confidence variant calls using joint-calling17.
3’UTR INSERTION AND SNV CALLS VALIDATION WITH IGV
The genotypes of rs1990622, rs3173615, and the 3’UTR insertion were extracted for participants with both LRS and short-read NGS available. 18 individuals with discordant doses of the three variants in LRS – where the dose of any of the three variants differed from any other – were identified for validation with IGV. The manual validation protocol is described in eMethods.
COLOCALIZATION ANALYSIS
Colocalization was performed using the R package coloc18. We report the posterior probability of colocalization (PP4) between AD19 and FTLD-TDP7. Colocalization between the two GWAS results was visualized with locuscompareR20. Similarly, coloc analysis was performed between the plasma pQTL GWAS 21 22 and the two neurodegenerative diseases19,7.
ADSP LINKAGE DISEQUILIBRIUM ANALYSIS
Alzheimer’s disease sequencing project (ADSP) R.3 SNVs, Manta1, and Biograph23 SV calls were downloaded from NIAGADS (https://dss.niagads.org/datasets/ng00067/#data-releases). SNVs were subset to rs3173615 (7:12229791:C:G) and rs1990622 (7:12244161:A:G) using Plink 1.924. SNV genotyping and SV genotypes were available in 16,906 samples. After detailed curation of the SV genotypes (eMethods), 16,582 unique participants in ADSP had a robust calling of the 3’UTR insertion.
To identify European and African ancestry individuals in the ADSP, the ancestries of all ADSP individuals were determined using SNPWeights v225 with reference populations from the 1000 Genomes Consortium26. Individuals with greater than 75% African global ancestry were classified as African ancestry27, and similarly for European ancestry28.
eQTL AND pQTL ASSOCIATIONS WITH THE TMEM106B Locus
Expression quantitative trait locus (eQTL) and protein quantitative trait locus (pQTL) effect sizes and p-values were queried for rs1990622 from summary statistics (GTEx29, MetaBrain30, eQTLGen31 for eQTLs and ARIC22, DECODE32, Wingo33 for pQTLs).
Additionally, we analyzed a novel dataset collected at WUSTL, which included 1,979 participants with NGS (1,979 participants), blood bulk RNASeq (428 European participants, all healthy controls), plasma (1,150 European and 200 African participants, with 711 European and 120 African healthy controls), and CSF (1,210 European participants, with 588 healthy controls) proteomics obtained using SomaScan aptamers. Global ancestry was estimated using principal component analysis using the 1000 Genomes ancestry as anchors, and individuals were separated into European and African ancestry groups (eTable 1 provides the demographics by analyses). Details on the ‘omics analysis methods are provided in eMethods. These were adjusted for age and sex, due to the association of TMEM106B and GRN protein levels with these two demographic factors (eFigures 1–3).
Standard Protocol Approvals, Registrations, and Patient Consents
Participants or their caregivers provided written informed consent in the original studies. Study protocols were approved by the Institutional Review Boards at Stanford University and Washington University in St. Louis (WUSTL).
Data Availability
Data included in this paper will be provided in anonymized form upon reasonable request made via email to the corresponding author and completion of a Material Transfer Agreement.
RESULTS
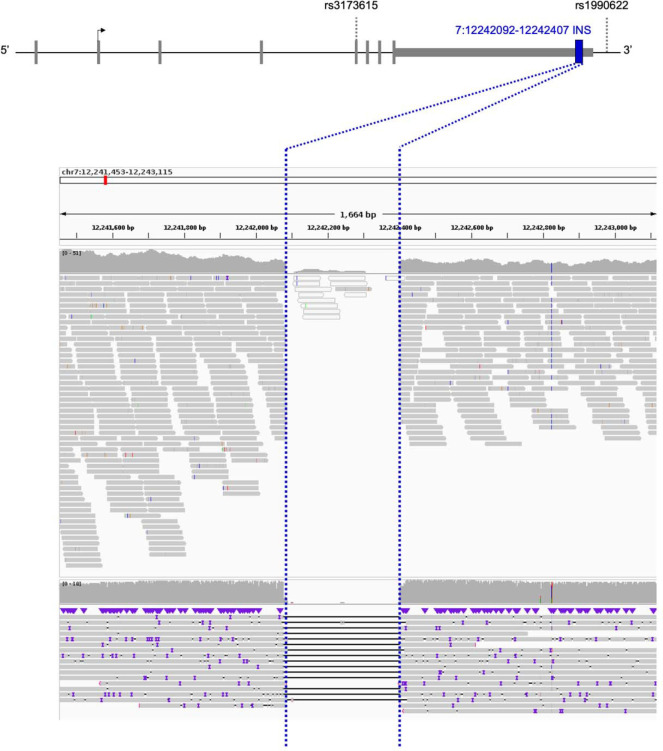
We carried out whole-genome LRS and SV calling for 432 participants enrolled in the Stanford ADRC or SAMS. In total, we identified 208 SVs overlapping an exon, 3’UTR, or 5’UTR site of one of the 4579 genes considered. Among these, we identified a common 322 bp deletion on the 3’UTR of TMEM106B for further analysis given the interest in this locus (Figure 1a)34. This SV overlaps a 316 bp AluYb8 mobile element that is prevalent in European-ancestry individuals and is included in the hg38 reference genome35. Our analysis pipeline therefore detected this SV as a 322 bp deletion when comparing individual genomes to the hg38 reference. We will refer to the SV, hereafter, as an insertion.
Figure 1. TMEM106B locus landscape and 3’ UTR insertion location.
(Top) TMEM106B 3’ UTR insertion position relative to SNVs associated with FTLD-TDP. (Bottom) The TMEM106B 3’ UTR insertion, detected as a deletion compared to the reference genome, was seen in both next-generation sequencing (upper panel) and long-read sequencing (lower panel). The region corresponding to the structural variant is delineated with blue dotted lines. Both sequencing modalities are displayed here for a representative cohort participant without the TMEM106B 3’ UTR insertion.
The insertion is highly prevalent with allele frequency (AF)=0.535 in our LRS dataset, comparable to the major allele frequency, in European non-Finnish ancestry, in gnomAD (v3.1.2) of rs1990622(A) (AF=0.586) and rs3173615(C) (AF=0.587). The insertion was detectable in both LRS and short-read NGS (Figure 1b). We linked LRS data to high coverage short-read NGS data to evaluate the LD between the 3’UTR insertion, rs1990622(A) and rs3173615(C). The TMEM106B 3’UTR insertion was in perfect LD with rs1990622(A) and rs3173615(C) in all but two Stanford individuals. For the sake of consistency, we will display all SNV data with the risk allele that is most often co-inherited with the insertion. This is the A allele for rs1990622 and the C allele for rs3173615. Note that the insertion, rs1990722(A) and rs3173615(C) are the major alleles in European-ancestry individuals (AF ~0.59) but the minor alleles in African-ancestry individuals (AF ~0.27).
The linkage between the TMEM106B 3’UTR insertion, rs1990622(A), and rs3173615(C) was then established in a large cohort by querying the ADSP database. 10,265 European ancestry and 2,212 African ancestry participants’ samples were genotyped at both SNVs and the SV, with respective AFs for the Alu insertion of AFEUR=0.591 and AFAFR=0.272. In European ancestry individuals, The Alu insertion is in comparable LD with rs1990622 (R2=0.962, D’=0.998) and rs3173615 (R2=0.960, D’=0.996). In African ancestry individuals in ADSP, the Alu insertion is in stronger LD with rs1990622 (R2=0.992, D’=0.998) than with rs3173615 (R2=0.811, D’=0.994).
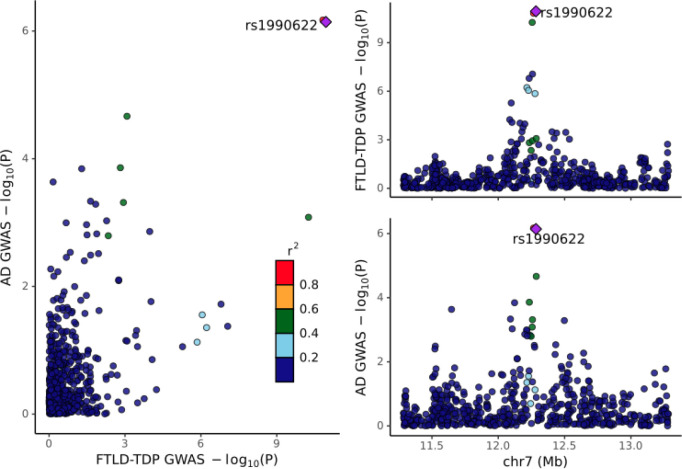
A colocalization analysis (Figure 2) demonstrated that the signals identified in the FTLD-TDP GWAS7and AD GWAS 19 at the TMEM106B locus have the same linkage structure (PP4 = 99.5%), suggesting that the same genetic signal is driving effects across both disorders.
Figure 2. Colocalization between the AD19 and FTLD-TDP7 genome-wide association studies.
The posterior probability of colocalization (PP4) = 99.5%. Note that the FTLD-TDP GWAS only included directly genotyped variants, i.e., not imputed, and thus the intersection of the two GWAS on this 2MB window is somewhat sparse, including only 607 variants.
We next evaluated the effect of rs1990622 on TMEM106B expression and protein levels in eQTL and pQTL datasets (Table 1). In a large brain eQTL dataset (European ancestry - Metabrain; n=6,601), rs1990622(A) was not associated with TMEM106B expression. In a smaller brain eQTL dataset (African ancestry - Metabrain; n=1,016), rs1990622(A) was significantly associated with increased TMEM106B expression. In two plasma eQTL datasets (GTEx, n=670; and eQTLGen, n=31,247), rs1990622(A) was significantly associated with decreased TMEM106B expression. In three large plasma pQTL datasets (deCODE; n=35,371; ARIC European; n=7,213; ARIC African; n=1,871), rs1990622(A) was significantly associated with increased TMEM106B protein levels. In a smaller brain pQTL dataset (Wingo; n=722), rs1990622(A) was not significantly associated with TMEM106B protein levels (p=0.1) but the direction of the effect was consistent with the plasma pQTL results. In addition, colocalization analyses showed that the plasma-based pQTL association with TMEM106B level in deCODE and ARIC colocalizes with the signal in AD and FTLD-TDP at the TMEM106B locus. Notably, all posterior probabilities of colocalization between the densely imputed AD GWAS and TMEM106B protein level associations in plasma were above 80% (PP4 with deCODE = 81.1%; PP4 with ARIC European = 86.3%; PP4 with ARIC African = 91.6%) (eFigure 4).
Table 1. Effect of rs1990622 as a TMEM106B expression quantitative trait locus (eQTL) or protein quantitative trait locus (pQTL).
All effect sizes are reported for the tested A allele with the G allele set as reference at rs1990622. Parameter estimates are those reported by respective studies and may be on different scales given different normalization procedures for transcriptomic and proteomic data.
| Dataset | Tissue | Sample size | Effect size | p-value |
|---|---|---|---|---|
| Expression (e)QTL | ||||
| eQTLGen | Blood | 31,427 | - * | 0.0174 |
| GTEx | Blood | 670 | −0.25 | 4.00E-30 |
| Metabrain EA | Brain (cortex) | 6,601 | +0.0268 | 0.3115 |
| Metabrain AA | Brain (cortex) | 1,016 | +0.2053 | 0.0031 |
| Protein (p)QTL | ||||
| deCODE | Plasma | 35,371 | +0.1106 | 3.69E-43 |
| ARIC EA | Plasma | 7,213 | +0.2547 | 7.33E-53 |
| ARIC AA | Plasma | 1,871 | +0.3147 | 1.50E-19 |
| Wingo | Brain (cortex) | 722 | +0.0773 | 0.1005 |
Negative Z-value indicating that A allele is associated with decreased TMEM106B expression. Abbreviations: EA: European Ancestry, AA: African Ancestry.
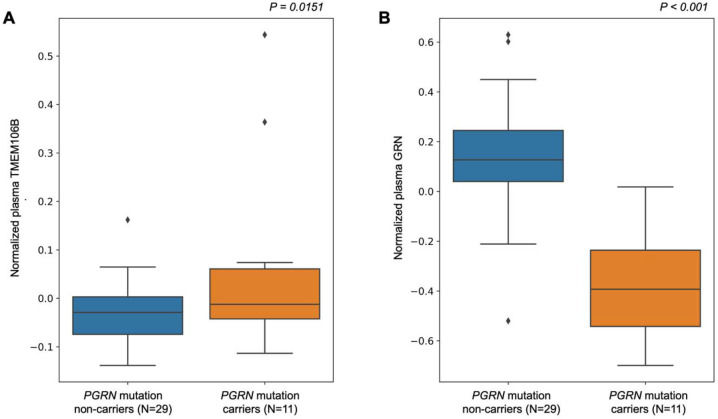
To expand beyond these eQTL and pQTL results from publicly available databases, we tested the association between rs1990622 and TMEM106B expression, TMEM106B protein levels, and GRN protein levels in the WUSTL Neurogenomics dataset36,37,38. First, we examined GRN and TMEM106B protein levels in a group of 11 carriers of FTLD-TDP-causing progranulin (PGRN) mutations. The PGRN mutation carriers have significantly higher plasma levels of TMEM106B compared to non-carriers from the same families (p < 0.05, Figure 3a). GRN levels were significantly lower in PRGN mutation carriers compared to non-carriers from the same families (p < 0.001, Figure 3b). With only 11 PGRN mutation carriers we could not determine if the TMEM106B locus variants impacted TMEM106B or GRN levels. When comparing AD cases versus older controls we found that plasma TMEM106B levels were significantly lower in AD (β = −0.011; 95% CI [−0.019; −0.004]; p = 4.25×10−3) as were CSF TMEM106B levels (β = −0.186; 95% CI [−0.277; −0.095]; p = 6.35×10−5). We also found a significant increase in plasma (β = 0.016; 95% CI [0.002; 0.031]; p =0.021) and CSF GRN levels (β = 0.0969; 95% CI [0.004; 0.19]; p = 0.04) when comparing AD cases with controls.
Figure 3. Differential abundance analysis of PGRN mutation carriers versus non-carriers on plasma TMEM106B and GRN protein levels.
A) Boxplot of the normalized plasma protein TMEM106B levels between PGRN mutation carriers and non-carriers. Mutation carriers have significantly higher TMEM106B levels (p = 0.0151).
B) Boxplot of normalized plasma protein GRN levels between PGRN mutation carriers and non-carriers. Mutation carriers have significantly lower GRN levels (p = 2.89×10−8), Analyses were adjusted for age, sex, and proteomic principal components 1 and 2.
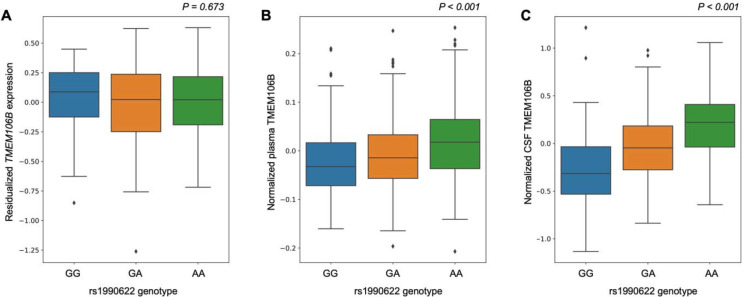
In the large WUSTL cohort of healthy older European-ancestry controls, rs1990622 was not associated with TMEM106B expression levels (p = 0.87) in blood but was associated with TMEM106B protein levels in both plasma and CSF (Figure 4). rs1990622(A) was significantly associated with increased levels of TMEM106B in plasma (β = 0.0227; 95% CI [0.015;0.031]; p = 5.6×10−8) and in CSF (β = 0.280; 95% CI [0.232;0.327]; p = 3.9×10−27), with a roughly 10-fold larger effect size in CSF. rs1990622(A) was also associated with TMEM106B protein levels in plasma of African-ancestry controls (β = 0.0290; 95% CI [0.002;0.057]; p = 0.0387 and eTable 2). There were not enough African-ancestry CSF samples to perform similar analyses. In examining GRN levels, we did not find a significant association of rs1990622(A) with GRN levels in plasma or CSF.
Figure 4. Effects of rs1990622 on TMEM106B gene expression in blood and TMEM106B protein levels in plasma and cerebrospinal fluid of European-ancestry controls.
A) Boxplot of the residualized TMEM106B gene expression in healthy older control blood samples across the rs1990622 genotypes (beta = 0.00481, p = 0.673).
B) Boxplot of normalized TMEM106B protein levels from plasma in healthy older controls across the rs1990622 genotypes (beta = 0.0215, p = 6.3×10−11).
C) Boxplot of normalized TMEM106B protein levels from cerebrospinal fluid in healthy older controls across the rs1990622 genotypes (beta = 0.283, p = 2.5×10−32). All analyses were adjusted for age and sex.
Since rs1990622 and rs3173615 were called directly from NGS in the WUSTL data we could compare the effect sizes of the two SNVs directly. Results were nearly identical with a numerically larger effect of rs1990622 in plasma and CSF of European-ancestry subjects and a numerically larger effect of rs3173615 in plasma of African-ancestry subjects (eTable 2, eFigure 5).
DISCUSSION
The TMEM106B 3’UTR AluYb8 insertion is a previously unreported variant that may mediate the effect of the TMEM106B locus on FTLD-TDP risk and AD risk. At present, the candidates for the causal variant at this locus are (1) the TMEM106B 3’UTR insertion described here; (2) rs3173615, the only coding SNV in this linkage block; (3) rs1990622, the leading GWAS hit; or (4) another variant in this linkage block.
Here we used publicly available expression and protein datasets to establish that rs1990622 is a consistent pQTL but an inconsistent eQTL and then confirmed this in a large, novel transcriptomics and proteomics dataset from WUSTL. This suggests that the effect at the TMEM106B locus is likely mediated by a genetic variant that acts after transcription to impact TMEM106B protein levels. Because intronic and intergenic variants are not incorporated into the processed mRNA molecule it is less likely that such variants, including rs1990622, are directly mediating the effect of this locus. Furthermore, the pQTL findings, coupled with the finding of increased TMEM106B protein levels in PGRN mutation carriers, suggest that the TMEM106B locus exerts its effect by altering protein availability rather than by altering protein function due to an amino acid change. This reduces the likelihood that rs3173615 is the causal variant. A prior in vitro study suggested that the TMEM106B p.S185 protein is less stable than wild-type and may therefore result in reduced protein levels10, but recent in vivo mouse model work suggests that the rs3173615 missense variant is not causal11. Moreover, rs1990622 was found to be more significant than rs3173615 in an FTLD-TDP GWAS meta-analysis12, which would be unexpected if the protective effect of the rs1990622 minor allele were due to its linkage with rs3173615.
We found in ADSP that the TMEM106B 3’UTR insertion is in higher LD with rs1990622(A) than rs3173615(C), which is consistent with a model in which the respective significance of these two SNVs is related to their linkage with the TMEM106B 3’UTR insertion. The proteomics data from WUSTL showed that the effect sizes of these variants are almost identical for CSF and plasma TMEM106B levels with a minimally larger effect for rs1990622 in European-ancestry subjects and a minimally larger effect for rs3173615 in African-ancestry individuals. We emphasize here that the differences between the two variants on protein levels are exceedingly small and should not be considered significant. Directly comparing the effect size of the two SNVs in the same set of subjects, would be more powerful in a large sample of African-ancestry AD cases and controls or a larger sample of African-ancestry proteomics dataset given that roughly 6% of African-ancestry haplotypes are discordant across these two SNVs compared to only ~0.5% of European-ancestry haplotypes. With the growing focus on enrolling more underrepresented minority participants in genetics studies, such an analysis should be feasible in the near future. Given the high LD across the locus, such analyses will be more reliable and definitive when carried out on short-read NGS or LRS datasets rather than on imputed SNV array datasets.
There are several possibilities as to how the TMEM106B 3’UTR Alu insertion could increase risk in FTLD-TDP and AD. The pQTL evidence indicates that the major risk allele at the TMEM106B locus increases TMEM106B protein levels both in plasma and in CSF. In cortex, the trend is in the same direction but did not reach significance (p = 0.10) with this relatively smaller sample size (Table 1). In a small group of PGRN mutation carriers, we replicate prior work 9,39,40 showing reduced plasma levels of GRN and demonstrate, for the first time, that TMEM106B levels are increased. As FTLD-TDP due to PGRN mutations is thought to result from haploinsufficiency of GRN34,41,42 a compelling hypothesis is that the Alu insertion on the 3’UTR results in an increase of TMEM106B levels in the brain which further depletes GRN levels. These two proteins are both major components of the endolysosomal pathway and so are well-positioned to interact37,43 Further evidence that the two proteins interact comes from recent work demonstrating that TMEM106B fibrils form aggregates in a variety of neurodegenerative disorders and as a function of age, but that these TMEM106B fibrils are far more prominent in PGRN mutation carriers with FTLD-TDP44–47. In AD we found the opposite pattern (reduced TMEM106B levels and increased GRN levels in plasma and in CSF). This is less readily interpretable given that we do not understand GRN’s role in AD. One possibility is that the findings in AD reflect compensatory changes in the setting of pathogenesis rather than causal effects driving pathogenesis.
The mechanism by which the insertion may impact TMEM106B levels remains uncertain. The insertion may result in selective enrichment of an alternate transcript polyadenylation site, changing the 3’UTR. Such a change in the 3’UTR could affect protein binding, which may in turn impact translational efficiency, alter the subcellular localization of the RNA, or impair protein routing to the endoplasmic reticulum48. Identifying a clear-cut mechanism linking the insertion to increased TMEM106B protein levels is still required to confirm that this is the causal variant at the locus.
In summary, we report a 316 bp Alu insertion on TMEM106B that is in tight LD with rs1990622(A) and rs3173615(C) in a large LRS dataset. LRS provides a valuable tool for the detection of large genomic variants that can aid in the interpretation of GWAS results and elucidate genetic drivers of disease. Ongoing functional work by our group and others will elaborate on the mechanisms connecting the 3’UTR AluYb8 insertion to pathogenesis in FTLD-TDP and AD.
Supplementary Material
FUNDING AND ACKNOWLEDGMENTS
This work was supported by the following NIH grants to Stanford investigators: RO1AG060747; R35AG072290; R01AG048076; R01AG074339; P30AG066515; and K99AG075238.This work was also supported by the Alzheimer’s Association (AARF-20-683984).
Multiomics data generated by the NeuroGenomics and Informatics Funding was supported by grants from the NIH:R01AG044546; P01AG003991; RF1AG053303: RF1AG058501; U01AG058922;, RF1AG074007; the Chan Zuckerberg Initiative (CZI); the Michael J. Fox Foundation; the Department of Defense (W81XWH2010849); the Alzheimer’s Association Zenith Fellows Award (ZEN-22-848604); and an Anonymous foundation. The recruitment and clinical characterization of research participants at Washington University were supported by NIH P30AG066444; P01AG03991; and P01AG026276. This work was supported by access to equipment made possible by the Hope Center for Neurological Disorders, the Neurogenomics and Informatics Center (NGI: https://neurogenomics.wustl.edu/), and the Departments of Neurology and Psychiatry at Washington University School of Medicine. The following investigators assisted in the preparation of the WUSTL ‘omics dataset Matt Johnson, Maulikkumar Patel, Maria Victoria Fernandez, Jessie Sanford, Devin Dikec, Ellen Liu, Dan Western, Thomas Marsh, Priyanka Gorijala, Jigyasha Timsina, and Judy Lihua Wang.
References
- 1.Chen X, Schulz-Trieglaff O, Shaw R, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32(8):1220–1222. [DOI] [PubMed] [Google Scholar]
- 2.Pedersen BS, Quinlan AR. Duphold: scalable, depth-based annotation and curation of high-confidence structural variant calls. Gigascience. 2019;8(4). doi: 10.1093/gigascience/giz040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahmoud M, Gobet N, Cruz-Dávalos DI, Mounier N, Dessimoz C, Sedlazeck FJ. Structural variant calling: the long and the short of it. Genome Biol. 2019;20(1):246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Logsdon GA, Vollger MR, Eichler EE. Long-read human genome sequencing and its applications. Nat Rev Genet. 2020;21(10):597–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaisson MJP, Sanders AD, Zhao X, et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat Commun. 2019;10(1):1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trelle AN, Carr VA, Guerin SA, et al. Hippocampal and cortical mechanisms at retrieval explain variability in episodic remembering in older adults. Elife. 2020;9. doi: 10.7554/eLife.55335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Deerlin VM, Sleiman PMA, Martinez-Lage M, et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet. 2010;42(3):234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z, Farias FHG, Dube U, et al. The TMEM106B FTLD-protective variant, rs1990621, is also associated with increased neuronal proportion. Acta Neuropathol. 2020;139(1):45–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruchaga C, Graff C, Chiang HH, et al. Association of TMEM106B gene polymorphism with age at onset in granulin mutation carriers and plasma granulin protein levels. Arch Neurol. 2011;68(5):581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicholson AM, Finch NA, Wojtas A, et al. TMEM106B p.T185S regulates TMEM106B protein levels: implications for frontotemporal dementia. J Neurochem. 2013;126(6):781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cabron AS, Borgmeyer U, Richter J, et al. Lack of a protective effect of the Tmem106b “protective SNP” in the Grn knockout mouse model for frontotemporal lobar degeneration. Acta Neuropathol Commun. 2023;11(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pottier C, Zhou X, Perkerson RB 3rd, et al. Potential genetic modifiers of disease risk and age at onset in patients with frontotemporal lobar degeneration and GRN mutations: a genome-wide association study. Lancet Neurol. 2018;17(6):548–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallagher MD, Posavi M, Huang P, et al. A Dementia-Associated Risk Variant near TMEM106B Alters Chromatin Architecture and Gene Expression. Am J Hum Genet. 2017;101(5):643–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen-Plotkin AS, Unger TL, Gallagher MD, et al. TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J Neurosci. 2012;32(33):11213–11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34(18):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smolka M, Paulin LF, Grochowski CM, et al. Comprehensive Structural Variant Detection: From Mosaic to Population-Level. bioRxiv. Published online April 5, 2022:2022.04.04.487055. doi: 10.1101/2022.04.04.487055 [DOI] [Google Scholar]
- 17.Poplin R, Ruano-Rubio V, DePristo MA, et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv. Published online July 24, 2018:201178. doi: 10.1101/201178 [DOI] [Google Scholar]
- 18.Giambartolomei C, Vukcevic D, Schadt EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10(5):e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bellenguez C, Küçükali F, Jansen IE, et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat Genet. 2022;54(4):412–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu B, Gloudemans MJ, Rao AS, Ingelsson E, Montgomery SB. Abundant associations with gene expression complicate GWAS follow-up. Nat Genet. 2019;51(5):768–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beyter D, Ingimundardottir H, Oddsson A, et al. Long-read sequencing of 3,622 Icelanders provides insight into the role of structural variants in human diseases and other traits. Nat Genet. 2021;53(6):779–786. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Dutta D, Köttgen A, et al. Plasma proteome analyses in individuals of European and African ancestry identify cis-pQTLs and models for proteome-wide association studies. Nat Genet. 2022;54(5):593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.English AC, McCarthy N, Flickenger R, et al. Leveraging a WGS compression and indexing format with dynamic graph references to call structural variants. bioRxiv. Published online April 25, 2020:2020.04.24.060202. doi: 10.1101/2020.04.24.060202 [DOI] [Google Scholar]
- 24.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen CY, Pollack S, Hunter DJ, Hirschhorn JN, Kraft P, Price AL. Improved ancestry inference using weights from external reference panels. Bioinformatics. 2013;29(11):1399–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.1000 Genomes Project Consortium, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Guen Y, Raulin AC, Logue MW, et al. Association of African Ancestry-Specific APOE Missense Variant R145C With Risk of Alzheimer Disease. JAMA. 2023;329(7):551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Guen Y, Belloy ME, Grenier-Boley B, et al. Association of Rare APOE Missense Variants V236E and R251G With Risk of Alzheimer Disease. JAMA Neurol. 2022;79(7):652–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Consortium GTEx. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369(6509):1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Klein N, Tsai EA, Vochteloo M, et al. Brain expression quantitative trait locus and network analyses reveal downstream effects and putative drivers for brain-related diseases. Nat Genet. 2023;55(3):377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Võsa U, Claringbould A, Westra HJ, et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet. 2021;53(9):1300–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beyter D, Ingimundardottir H, Oddsson A, et al. Long read sequencing of 3,622 Icelanders provides insight into the role of structural variants in human diseases and other traits. bioRxiv. Published online December 14, 2020:848366. doi: 10.1101/848366 [DOI] [PubMed] [Google Scholar]
- 33.Wingo T, Liu Y, Gerasimov ES, et al. Integrating Human Brain Proteomes With GWAS Results to Identify Causal Brain Proteins for the Major Psychiatric Disorders. Biol Psychiatry. 2023;93(9, Supplement):S55–S56. [Google Scholar]
- 34.Chemparathy A, Guen YL, Zeng Y, et al. A 3’ UTR Deletion Is a Leading Candidate Causal Variant at the TMEM106B Locus Reducing Risk for FTLD-TDP. medRxiv. Published online July 8, 2023. doi: 10.1101/2023.07.06.23292312 [DOI] [Google Scholar]
- 35.Salazar A, Tesi N, Hulsman M, et al. An AluYb8 mobile element further characterises a risk haplotype of TMEM106B associated in neurodegeneration. bioRxiv. Published online July 16, 2023. doi: 10.1101/2023.07.16.23292721 [DOI] [Google Scholar]
- 36.Wang C, Western D, Yang C, et al. Unique genetic architecture of CSF and brain metabolites pinpoints the novel targets for the traits of human wellness. Res Sq. Published online June 9, 2023. doi: 10.21203/rs.3.rs-2923409/v1 [DOI] [Google Scholar]
- 37.Cruchaga C, Western D, Timsina J, et al. Proteogenomic analysis of human cerebrospinal fluid identifies neurologically relevant regulation and informs causal proteins for Alzheimer’s disease. Res Sq. Published online June 9, 2023. doi: 10.21203/rs.3.rs-2814616/v1 [DOI] [Google Scholar]
- 38.Phillips B, Western D, Wang L, et al. Proteome wide association studies of LRRK2 variants identify novel causal and druggable proteins for Parkinson’s disease. NPJ Parkinsons Dis. 2023;9(1):107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finch N, Baker M, Crook R, et al. Plasma progranulin levels predict progranulin mutation status in frontotemporal dementia patients and asymptomatic family members. Brain. 2009;132(Pt 3):583–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meeter LHH, Patzke H, Loewen G, et al. Progranulin Levels in Plasma and Cerebrospinal Fluid in Granulin Mutation Carriers. Dement Geriatr Cogn Dis Extra. 2016;6(2):330–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baker M, Mackenzie IR, Pickering-Brown SM, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442(7105):916–919. [DOI] [PubMed] [Google Scholar]
- 42.Gass J, Cannon A, Mackenzie IR, et al. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum Mol Genet. 2006;15(20):2988–3001. [DOI] [PubMed] [Google Scholar]
- 43.Brady OA, Zheng Y, Murphy K, Huang M, Hu F. The frontotemporal lobar degeneration risk factor, TMEM106B, regulates lysosomal morphology and function. Hum Mol Genet. 2013;22(4):685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang YX, Cao Q, Sawaya MR, et al. Amyloid fibrils in FTLD-TDP are composed of TMEM106B and not TDP-43. Nature. 2022;605(7909):304–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang A, Xiang X, Wang J, et al. Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases. Cell. 2022;185(8):1346–1355.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schweighauser M, Arseni D, Bacioglu M, et al. Age-dependent formation of TMEM106B amyloid filaments in human brains. Nature. 2022;605(7909):310–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perneel J, Neumann M, Heeman B, et al. Accumulation of TMEM106B C-terminal fragments in neurodegenerative disease and aging. Acta Neuropathol. 2023;145(3):285–302. [DOI] [PubMed] [Google Scholar]
- 48.Mitschka S, Mayr C. Context-specific regulation and function of mRNA alternative polyadenylation. Nat Rev Mol Cell Biol. 2022;23(12):779–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in this paper will be provided in anonymized form upon reasonable request made via email to the corresponding author and completion of a Material Transfer Agreement.