Abstract
DNA polymerase θ (Pol θ) is a DNA repair enzyme widely conserved in animals and plants. Pol θ uses short DNA sequence homologies to initiate repair of double-strand breaks by theta-mediated end joining. The DNA polymerase domain of Pol θ is at the C terminus and is connected to an N-terminal DNA helicase–like domain by a central linker. Pol θ is crucial for maintenance of damaged genomes during development, protects DNA against extensive deletions, and limits loss of heterozygosity. The cost of using Pol θ for genome protection is that a few nucleotides are usually deleted or added at the repair site. Inactivation of Pol θ often enhances the sensitivity of cells to DNA strand–breaking chemicals and radiation. Since some homologous recombination–defective cancers depend on Pol θ for growth, inhibitors of Pol θ may be useful in treating such tumors.
Keywords: DNA polymerases, mutations, DNA double-strand breaks, DNA helicase, translesion DNA synthesis, DNA repair
1. INTRODUCTION
DNA polymerases exist in every organism, not only to accomplish replication of genomic, mitochondrial, and plastid genomes but also to preserve the integrity of these genomes by operating during DNA repair and recombination (14a, 30, 46, 51, 86). This review focuses on the biological functions of DNA polymerase theta (Pol θ). Distinctive properties of this multi-activity enzyme allow it to protect genomes by initiating a specific mechanism for end-joining repair of double-strand breaks (DSBs) in DNA. In some situations, Pol θ may participate in additional reactions of DNA repair and damage tolerance.
Double-strand break (DSB):
DNA damage arising when the phosphodiester backbone is broken in close proximity on both strands of the helix by reactive oxygen species, other chemical reactions, or enzymes
2. POL θ IS WIDELY CONSERVED IN EUKARYOTES
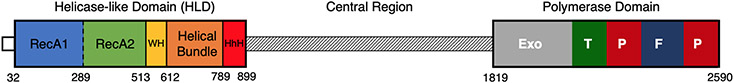
The first discovery of a gene for Pol θ emanated from studies of the Drosophila melanogaster mutant mus308, and investigations in that organism continue to make leading contributions in the field (10, 122). Based on hypersensitivity of the mutant to DNA-damaging agents, mus308 was suggested to play a role in DNA repair (2, 14). Sequencing of the mus308 gene revealed a multidomain configuration, with the C-terminal part of the protein encoding a DNA polymerase, and the N-terminal part of the protein encoding a helicase-like domain (HLD) (39). In human cells, Pol θ is a 290-kDa protein, which includes an extensive central connecting region (Figure 1). Pol θ orthologs are, consequently, relatively large enzymes (95, 96, 122). The gene encoding Pol θ is generally named POLQ.
Figure 1.
DNA polymerase θ three-domain architecture and depiction of the human protein, encoding a 2,590–amino acid polypeptide. The helicase-like domain includes five main structural elements (77): two RecA-like subdomains (RecA1 and RecA2), a winged helix subdomain (WH), a helical bundle, and a helix-hairpin-helix subdomain (HhH). The central region is predicted to be mostly disordered (12, 95) and has a variable length between organisms. The polymerase domain consists of an A-family DNA polymerase with a nonfunctional editing exonuclease domain (Exo) and the fingers (F), palm (P), and thumb (T) regions characteristic of DNA polymerases (124).
The HLD–polymerase domain arrangement of Pol θ is preserved from a common eukaryotic ancestor 1.5 billion years ago (104), strongly indicating that this domain connection has a conserved function. Such genes are found throughout most of the eukaryotic lineage in both animals and plants (104). Even the smallest free-living eukaryote, the unicellular green alga Ostreococcus tauri, contains POLQ. The gene appears to have been lost in the evolution of the fungal kingdom (10, 122). In the excavate protists and some other organisms, there are separate gene products homologous to the two enzymatic domains (HLD and polymerase) of POLQ (104). For example, there are genes in trypanosomes that encode proteins similar to the polymerase domain of POLQ, and some of these genes affect sensitivity to DNA damage (15, 60).
3. POL θ SUPPORTS GENOME INTEGRITY
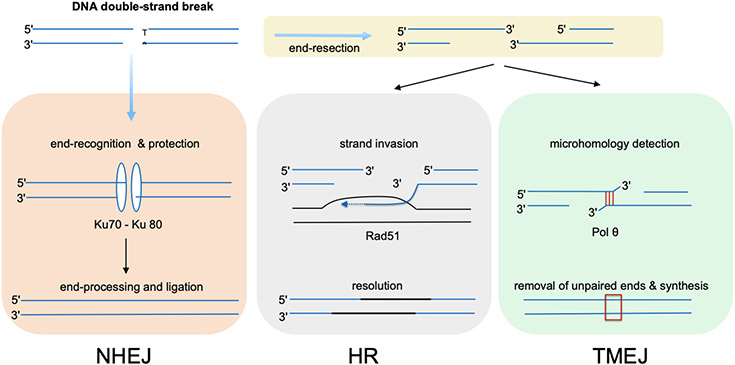
The primary function of Pol θ is the repair of DNA DSBs, as first shown in Drosophila (20). The process is referred to as theta-mediated end joining (TMEJ) (90). Among the strategies that cells use to repair DSBs (Figure 2), the central feature of TMEJ is the use of short sequence homologies (see the sidebar titled Mechanisms of Double-Strand Break Repair). These short homologies are used by Pol θ to join two 3′ single-stranded ends exposed by enzymatic resection DNA near a break. TMEJ does not require an undamaged copy of the sequence for repair. Consequently, TMEJ is distinct from the microhomology-mediated break-induced replication process analyzed in several organisms, which involves the polymerase-mediated restart of stalled replication forks (40, 92). Pol θ helps maintain the integrity of genomes in diverse organisms throughout nature (Table 1).
Figure 2.
Three major strategies for repair of a DNA double-strand break. In nonhomologous end joining (NHEJ, left), the ends of the break are bound and protected by Ku protein homologs, and then reactions proceed to join the ends, often involving limited nuclease trimming and DNA synthesis to repair broken DNA before ligation. Alternatively, 5′ ends at the break may be processed by nuclease-mediated resection. Resection generates 3′ single-stranded DNA tails. In the process of homologous recombination repair (HR, middle), a 3′ tail pairs with an intact sister strand in a recombination process dependent on Rad51. The primary HR repair pathway is synthesis-dependent strand annealing in which one strand is extended for some distance by DNA synthesis so that it can be paired with its original partner and repair completed. In some cases, 3′ ends are joined by theta-mediated end joining (TMEJ, right). Pol θ locates a microhomology, typically of 2–6 base pairs in the 3′ single strands. Processing of the 3′ ends of the DNA will usually have to occur to remove unpaired regions and allow completion of synthesis (see Figure 4). In general, TMEJ creates short deletions or templated insertions in the DNA (red box).
Table 1.
Examples of genome-protective functions of DNA polymerase θ
| Function | Examples of organisms studied |
|---|---|
| Reduces the frequency of unrepaired chromosome breaks, both spontaneous and radiation-induceda | Human, mouse |
| Resistance to agents causing double-strand breaksb | Animals, plants |
| Protection against large deletionsc | Nematode, Drosophila |
| Rescuing lethality of double-strand breaks during developmentd | Zebrafish, Drosophila, plants |
| Suppression of mutagenic interhomolog recombination and mitotic crossovere | Mammalian cells, Drosophila |
Theta-mediated end joining (TMEJ):
DSB repair process mediated by Pol θ that accounts for much of the repair that is also known as microhomology-mediated or alternative end joining in cells
MECHANISMS OF DOUBLE-STRAND BREAK REPAIR
There are multiple mechanisms of double-strand break (DSB) repair. Three of the major ones are depicted in Figure 2: nonhomologous end joining (NHEJ), homologous recombination (HR), and theta-mediated end joining (TMEJ). The latter two processes occur if 3′-ended tails at the break are generated by nuclease-mediated end resection, a process that is finely regulated by many protein factors, including chromatin components (19a, 113). HR repair is the process by which two homologous segments of DNA can interact with one another to provide missing information by copying a damaged strand (72a). The main recombinase in eukaryotes is called RAD51 (13). In mammalian cells, HR involves many other factors, including the breast cancer-associated proteins BRCA1 and BRCA2 (83a). A few other strategies for DSB repair are not shown in the figure. If the break occurs near sequences containing a longer homology, ends can be joined by RAD52- and HELQ-dependent single-strand annealing (SSA) with deletion of the intervening DNA between the repeats (5, 54). An additional process of Pol θ–independent but HELQ-dependent microhomology-mediated end-joining can also join some breaks (54). These processes take place only in animals, as HELQ is absent in plants (54, 104). The break-induced replication (BIR) mechanism is another way to reactivate broken DNA replication forks by copying from undamaged DNA (40, 92).
3.1. Pol θ in Animals
Pol θ–dependent repair is necessary to mend a significant fraction of DSBs arising during normal DNA replication, as well as breaks caused by exposure to DNA-damaging agents. A genetic screen of mutagenized mice revealed that disruption of mouse Polq elevates the frequency of micronuclei in circulating reticulocytes (100). An increase in frequency of micronuclei also occurs in Polq-deficient cultures of mouse bone marrow stromal cells (36) and human cells (85). Exposure of cells or animals to a sublethal dose of ionizing radiation increases the frequency of micronuclei by about eight- to tenfold at the next mitosis (36, 100). Mutational disruption of Pol θ in mouse and human cells also increases the lethal toxicity of some chemical compounds that induce DSBs in DNA, such as topoisomerase II inhibitors (21, 110, 123).
In Drosophila, a defect in Pol θ (Mus308) causes extreme hypersensitivity to ionizing radiation when homologous recombination (HR) repair is impaired by mutation of Rad51/spn-A (20) (see the sidebar titled Mechanisms of Double-Strand Break Repair). This was the initial demonstration that Pol θ is extraordinarily important in the absence of HR. Cell survival measurements show moderately enhanced sensitivity of Polq mutant mouse bone marrow stromal cells to X-rays, with about a twofold difference in dose yielding 10% survival (123). In cultured human cells, Pol θ disruption leads to unrepaired breaks and enhanced sensitivity to ionizing radiation but to a lesser degree (43).
A major function of Pol θ is in the protection of cells against potentially catastrophic large deletions, compellingly illustrated in studies of how Caenorhabditis elegans deals with DNA sequences that can form G-quadruplex (G4) structures in the genome. G4 structures cause stalling of DNA replication and frequently result in DSBs (37). In C. elegans cells where the FancJ-related gene dog-1 is inactive, disruption of Pol θ has a profound effect. In this case, large deletions (5 to greater than 30 kb) are formed in the absence of Pol θ (54, 90). An interpretation is that TMEJ repairs a fraction of breaks that form during stalled replication of G4 structures, and this repair avoids extensive loss of genomic DNA. As with Drosophila, mutations in the polq-1 gene increase the sensitivity of C. elegans to interstrand crosslink-inducing agents (74).
G-quadruplex (G4):
a secondary structure formed by guanine-rich sequences of nucleic acids, in which base-paired guanine tetrads are stacked into helical structures formed from one, two, or four strands
TMEJ is important for the repair of DSBs occurring during early development. A striking example of this was found by introducing a targeted DSB into one-cell-stage zebrafish embryos (105). Wild-type embryos repaired the break and developed, but polq mutant embryos did not survive. Pol θ was also required for the survival of 128-cell embryos exposed to ionizing radiation (105). These experiments show that TMEJ is an essential and dominant DSB repair pathway during early fish development.
In Drosophila, a screen was done to discover genes needed for hindgut papillar organogenesis after cells were subjected to DSBs from I-CreI treatment or ionizing radiation (25). Pol θ was a strong hit in this screen, but components of nonhomologous end joining (NHEJ) were not. The results demonstrate that Pol θ is also crucial for repairing DSBs that occur during fly development. TMEJ may be better suited than other repair pathways to join breaks in the rapidly proliferating cells characteristic of embryonic development. Extension of these investigations to other organisms is warranted.
Pol θ is also fundamentally important during gene amplification in Drosophila. This became apparent after the discovery that fertilized eggs laid by mus308/Polq mutant females have only ~5% hatching frequency, compared to 85% for eggs from wild-type mothers (3). The surviving eggs frequently have patchy, thin eggshells. The reason for these defects is that follicle cells use rereplication to amplify genomic regions that include genes encoding abundant eggshell proteins. If this process is impaired, insufficient proteins are available to form a normal eggshell. However, the complex structures generated during rereplication are vulnerable to occasional DSBs. TMEJ is needed during rereplication in follicle cells, and other DNA repair pathways do not compensate for its absence. At two regions of rereplication, Polq mutants have normal origin firing but reduced rates of DNA replication fork progression (3).
3.2. Pol θ in Plants
Pol θ defends plant DNA against chemical agents that lead to DSBs. For instance, an insertional mutagenesis screen for sensitivity to DNA damage was done in the unicellular green alga Chlamydomonas reinhardtii (82). A mutant strain was isolated that was highly sensitive to Zeocin (phleomycin D1, a DSB-inducing antibiotic), with little enhanced sensitivity to ultraviolet (UV) radiation, hydroxyurea, methyl methanesulfonate, or mitomycin C. This strain was identified as having a deletion in Polq, rendering the alga unable to grow in 50 μg/L of Zeocin, a condition that allowed proliferation of normal strains. In the bryophyte moss Physcomitrella patens, a knockout of the Polq gene exhibited hypersensitivity to bleomycin, a natural compound in the same family as Zeocin (53).
In flowering plants, Pol θ is also important for genome repair—particularly during developmental and regenerative processes. Rice carrying a disruption of Polq had severely impaired regeneration of plants from root segments (79). In Arabidopsis thaliana, the POLQ gene was dubbed TEBICHI (TEB). The first reported teb mutants showed morphological defects, including shorter roots, serrated leaves, and defective patterns of cell division in the meristem (49). One allele, teb-1, conferred enhanced sensitivity to mitomycin C and methyl methanesulfonate as measured by the relative elongation of roots and leafing of aerial shoots. Double mutants teb-1 rad51d and teb-2 xrcc2 showed more pronounced developmental impairment (48).
Arabidopsis plants with mutations in POLQ have visible phenotypes of variable severity (48, 49, 78, 79, 107). In one study, mutant plants showed growth and developmental defects in tissue culture and were intolerant to growth in bleomycin (79). Another investigation revealed a variable or partially penetrant phenotype for teb mutants (78). Here, the aerial parts of 10–15% of teb2 or teb5 plants had severe developmental defects. Evidence was provided that DNA damage encountered during DNA replication can exacerbate developmental defects in rapidly dividing tissues. It would be valuable to understand the sources of environmentally associated DNA damage that could account for the variable phenotypes of Pol θ–defective teb mutant plants. Bleomycin and other bioactive molecules are natural products of soil bacteria (85a). Local soil microbiomes producing distinct mutagenic compounds might help account for some of the differences between plants and laboratory locations.
3.3. Theta-Mediated End Joining and Chromosome Translocations
Translocations arise when DSBs in two different chromosomes are misjoined by DNA repair. These are infrequent events, but they can activate oncogenes in mammals. Both NHEJ and alternative end-joining mechanisms can mediate translocations (35, 112). CRISPR-Cas9 breakage of two chromosomes in mouse cells can also induce a low frequency of translocations, which in one study was decreased in the absence of Pol θ (68) and in another case increased (117). Pol θ protects against a type of translocation that occurs during immunoglobulin class switching in the mouse. Class switching occurs by intrachromosomal joining of DSBs, and TMEJ is one of the pathways employed (123). The action of TMEJ in class switching is evident from the Pol θ–dependent sequence insertions found in approximately 10% of joins (123). Rarely, a gene in the heavy chain locus on chromosome 12 is translocated adjacent to the myc gene on mouse chromosome 15, comparable to the human Burkitt lymphoma translocation. This translocation frequency was increased by fourfold in Polq knockout mice compared to wild-type mice (123). During class switching, the immunoglobulin heavy chain region is peppered with DSBs initiated by the AIDCA deaminase. Multiple local DSBs might be a particularly dangerous situation in which TMEJ acts to join correct chromosome fragments and limits misjoining elsewhere.
3.4. Pol θ Suppresses Mutagenic Interhomolog Recombination and Crossing Over
Mitotic recombination between chromosome homologs can be a mutagenic form of DNA repair when it leads to loss of heterozygosity (LOH) because this can leave cells with only a deleterious disease-contributing allele. Pol θ helps protect cells against at least two routes that lead to LOH.
When a DSB, or a nick, is placed on each chromosome homolog by CRISPR-Cas9, a process of interhomolog recombination can occur (29). This process of recombination is proposed to proceed by reciprocal end joining rather than Holliday junction–mediated HR. If Pol θ is present, DSBs are joined in cis without LOH. Normally, interhomolog recombination is rare, but its frequency is elevated when POLQ is disrupted (29).
Pol θ also suppresses mitotic crossing over that is mediated by HR. Such crossing over can potentially generate LOH that can affect whole chromosome arms. When a DSB was introduced into the Drosophila ry gene, repair of this break was equally common by either end joining or HR (17). In wild-type flies, mitotic crossovers occurred at a frequency of only 0.2%. In Pol θ–deficient flies, mitotic crossovers were elevated 18-fold (17).
In Drosophila, the preferred HR pathway is synthesis-dependent strand annealing (SDSA). When this process fails, repair then proceeds to either a pathway involving double Holliday junction resolution or TMEJ using Pol θ (Figure 2). If both pathways are inactivated by disabling Pol θ as well as Holliday junction resolvases, cells cannot survive (17). This appears to explain the inviability of a polq slx4 gen1 mutant in Drosophila. Similarly, POLQ shows synthetic lethality with SLX4 in mammalian cells in culture (17).
In summary, TMEJ has manifold protective functions for genomes. It maintains integrity by precluding more detrimental forms of repair, such as those resulting in larger deletions (16), interhomolog recombination after a break is made in both homologs (29), chromosome translocations in a region containing many DSBs (123), and LOH during Holliday junction resolution (17). The damaging potential of such events is evidently more harmful to cells than the short indels that are a frequent consequence of TMEJ, as discussed in Section 6.
4. MECHANISM OF THETA-MEDIATED END JOINING
As described below, major insights have emerged from the crystal structures obtained for the polymerase domain (124) and HLD of human Pol θ (77). A further productive advance for the study of TMEJ in mammalian cells has been the development of assays for joining synthesized DNA substrates that model DNA break ends (87). Following transfection into cells with different genetic backgrounds, the yield, sequence, and genetic dependence of break repair can be studied (16, 117, 123).
4.1. DNA Polymerase Domain
The function of Pol θ in TMEJ is to begin the extension of a primer from microhomologies of only 2–6 bp. Enzymatic resection at a DSB produces two single-stranded tails. From this perspective, a sequence in one single strand functions as a primer, and the template is contributed by the other single strand captured by Pol θ in the opposite polarity. Some structural characteristics of the polymerase domain (116, 124) and its biochemical properties help explain how Pol θ can carry out this acrobatic reaction.
4.1.1. A tight grasp on the primer.
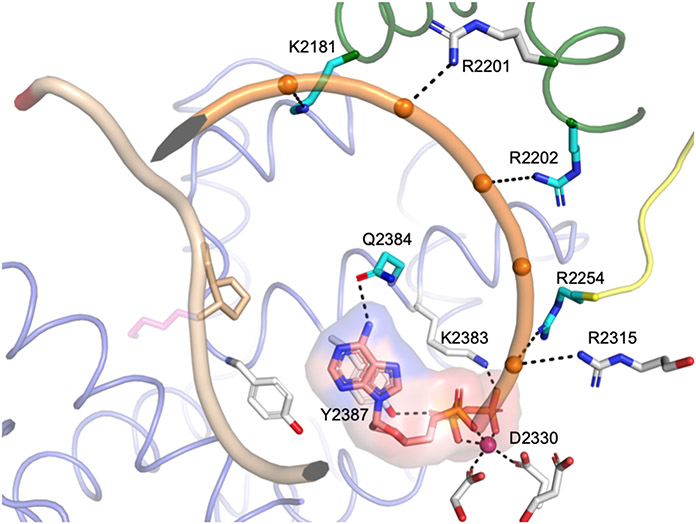
The polymerase domain of Pol θ belongs to DNA polymerase family A, showing sequence and structural similarity to Escherichia coli DNA pol I (39, 67), with the characteristic polymerase fold including the subdomains designated as thumb, palm, and fingers. Distinctive and unique features of Pol θ were unveiled, however, by a crystal structure of the Pol θ DNA polymerase domain in a ternary complex with DNA primer/template and incoming nucleotide. The structure revealed the locations of five insert regions, relative to bacterial family A polymerases, in the polymerase domain of human Pol θ (45, 96, 124). These inserts are present in vertebrate Pol θ but are much shorter or absent in the Pol θ genes of invertebrates. The structure further showed that five positively charged arginine (Arg) and lysine (Lys) residues make specific contacts with phosphates in the primer DNA backbone (Figure 3). Additional unique contacts are made to the incoming nucleotide. Gln2384 contacts the major groove side of the base, and Tyr2387 stacks onto the incoming nucleotide and contacts its triphosphate tail.
Figure 3.
Pol θ makes unique contacts that grasp the primer strand and the incoming nucleotide. A view of the ternary complex of the polymerase domain bound to furan-containing DNA and ddATP (PDB ID 4X0P) (126). The finger, palm, thumb, and insert 2 regions are depicted in blue, red, green, and yellow, respectively. The DNA backbones of the template and primer strands are shown in pale yellow and orange, respectively, with the 3′ terminus becoming red. The divalent cation (metal A) coordinated in the polymerase active site is depicted as a magenta sphere. Of the five positively charged residues contacting the primer backbone, two contacts are conserved in all A-family polymerases (shown with white carbons), and three of them are unique residues in Pol θ (cyan carbons), with Arg2254 emerging from the distinctive insert 2 (yellow). Abbreviations: ddATP, 2′,3′-dideoxyadenosine-5′-triphosphate; PDB ID, Protein Data Bank identification; Pol θ, DNA polymerase θ.
This unusually tight grasp on the primer and coordination with the incoming base are likely to account for some biochemical behavior of Pol θ. For example, when Pol θ copies a template, a common error is a frameshift mutation in a run of identical template bases (6). This sort of error occurs when the primer realigns or slips relative to the template. Pol θ extends a 3′ terminal mismatch with an efficiency approaching that of a normal base pair (41, 97). In addition, Pol θ can incorporate a nucleotide opposite a noncoding template base, such as an abasic (AP) site, thymine glycol, or some UV radiation–induced photoproducts (45, 57, 96, 97). By realignment of the template, deletion of one or two bases can occur during bypass (57).
4.1.2. Templated extension of single-stranded DNA.
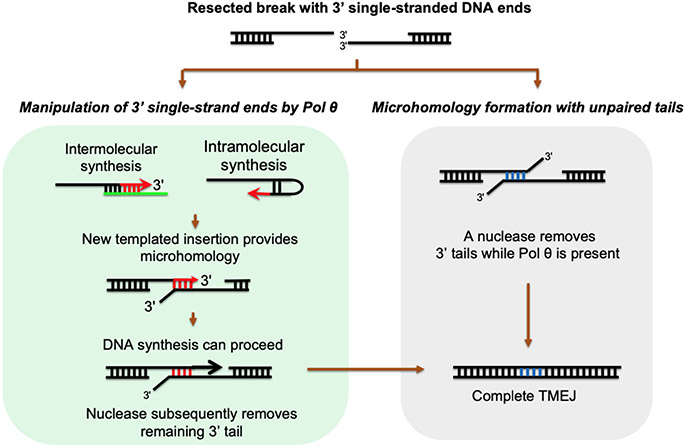
Pol θ possesses an activity on single-stranded DNA oligonucleotides that is not shared by other known A-family DNA polymerases: templated extension of single-stranded DNA, which arises from the ability of Pol θ to reconfigure single-stranded DNA within the pol domain (Figure 4).
Figure 4.
The task of TMEJ is to join a break by making use of microhomologies within single-stranded, 3′-ended DNA tails. In the general case, preexisting microhomologies are internal. If one or both tails can be removed by a nuclease so that Pol θ can extend from a microhomology, as shown in the box at the right, then repair could be completed. Another route is shown in the box at the left. Pol θ can manipulate 3′ single-stranded DNA by engaging in one or more short tracts of inter- or intramolecular synthesis, usually using nearby DNA as a template. This produces the templated insertions that are characteristic of TMEJ. A terminus is generated with a sequence different from the original, allowing iterative microhomology searches. In principle, the immediate extension of only one 3′ end is sufficient to generate stable joining of the break. A single remaining 3′ tail could then be removed by a nuclease after the DNA polymerase has dissociated, at which point further extension and sealing of the break by a DNA ligase can occur. Abbreviation: TMEJ, theta-mediated end joining.
Pol θ extends single-stranded DNA oligonucleotides by incorporating bases that are usually complementary to the template (12, 44, 123). For example, extension of homopolymeric oligonucleotides in the presence of Mg2+ occurs most efficiently by providing the complementary base (44). Like other A-family polymerases, Pol θ can use deoxyadenosine triphosphate (dATP) to add an additional A residue a blunt DNA end, but otherwise shows predictable template requirements in a first round of DNA synthesis (41). In the presence of Mn2+, Pol θ can carry out promiscuous synthesis arising from a high frequency of mismatch incorporation (88), which should not be mistaken for a bona fide terminal transferase activity.
4.2. Helicase-Like Domain
The HLD of Pol θ is a member of the Ski2-like subfamily in helicase superfamily 2 (108). The HLD is composed of five domains, two of which are RecA-like catalytic cores (Figure 1). Related archaeal helicases can unwind duplex DNA by using a ratchet mechanism of translocation (33, 38, 80). While the Pol θ HLD can hydrolyze ATP, it does not efficiently unwind double-stranded DNA or RNA under standard conditions (77, 93, 95). In the presence of an oligonucleotide trap, displacement of short oligonucleotides from a complementary strand was observed (81, 93).
Both the polymerase domain and the HLD are necessary to fully execute TMEJ in human cells, as measured by nonhomologous integration of plasmid DNA or by TMEJ-mediated chromosome translocations (69, 128). Both domains are also needed for Pol θ function in Drosophila (9). One ATP-driven function of the HLD may be to prepare ends for joining by displacing proteins bound to single- or double-stranded DNA. Another function may be to aid the search for a site of microhomology in the two 3′ tails (77).
4.2.1. Protein displacement.
Single-stranded DNA is bound tightly in the cell by Replication protein A (RPA). The HLD can remove RPA from single-stranded DNA, in an ATP-dependent manner (69, 93). The related human HELQ protein also has this property (5). The homologous Helq-1 protein in C. elegans can displace Rad51 from double-stranded DNA but not single-stranded DNA (111). The purified HLD of Pol θ can displace RAD51 from single-stranded DNA, but less efficiently than it can displace RPA (93).
4.2.2. Annealing and search for microhomology.
In human cells, helicases containing RecA catalytic cores similar to the Pol θ HLD include the DNA repair–related enzymes RECQ1, RECQ4, RECQ5, BLM, WRN, and HELQ (11, 109). In addition to their abilities to unwind paired DNA structures (5, 52, 72, 83, 103), BLM and WRN can promote branch migration of Holliday junctions, which involves unwinding and reannealing reactions (27, 55). Perhaps relevant to the activities of pol θ HLD, RECQ subfamily members and HELQ can catalyze the annealing of complementary single-stranded DNA (23, 34, 65, 66, 98). This ability to perform the annealing reaction appears to be related to oligomerization. RECQ1 and BLM both have strand-annealing activities that are associated with the assembly of higher-order complexes, including tetramers (23, 56, 75). Pol θ HLD can form a tetramer, both when unliganded and when bound to ADP or an ATP analog (77).
The ability of the HLD to promote annealing of complementary single-stranded DNA is beginning to be investigated. In a process depending on ATP hydrolysis, the HLD can anneal DNA substrates that are prebound to RPA (69). The HLD is also able to stably bridge noncomplementary single DNA strands (93). One hypothesis is that the HLD acts in TMEJ to both physically displace RPA from single-stranded DNA and facilitate a search for microhomologies (69, 93).
4.2.3. ATPase-dependent and ATPase-independent functions.
The ATPase function of DNA helicases can be inactivated by mutation of the Lys in the highly conserved GKT sequence in motif A. In POLQ, a K121M mutation inactivates DNA-dependent ATPase activity (77, 81, 95). This affects some, but not all, biological and biochemical endpoints. Pol θ–dependent random integration of plasmid DNA into mouse embryonic stem cells is abolished by mutation of this Lys (128). Another assay examined ionizing radiation–induced nuclear foci of RAD51 in human U2OS cells. These RAD51 foci may mark aborted recombination intermediates that are channeled into repair by TMEJ (19). Expression of wild-type POLQ complementary DNA in the U2OS cells reduced the frequency of RAD51 foci, while only partial rescue was conferred by a mutant POLQ with inactivated ATPase function (19).
A mutation inactivating the Polq ATPase increased the sensitivity of Drosophila larvae to nitrogen mustard, but not to ionizing radiation, in a spn-A (Rad51) background (9). This implies that the HLD ATPase is needed in some DNA repair settings but not others (9). In this same genetic background, repair was examined for a transposase-induced break with 17-nucleotide (nt) single-stranded ends. An ATPase defect did not affect the frequency of end joining but changed the distribution of the types of end-joining events, with a higher proportion having short microhomologies. A 17-nt single-stranded end may not require an ATPase-driven RPA removal function of Pol θ because RPA requires about 30 nt for stable binding to single-stranded DNA. At this locus, Pol θ protects against deletions larger than 1.5 kb. The ATPase-inactivated mutant and wild-type Pol θ were equally effective at this protection.
Both domains of Pol θ have some role in controlling rare translocations in human cells (69). In a biochemical study, the joining of two single-stranded DNA oligonucleotides with 6-nt terminal homologies was not impaired by using the purified human Pol θ K121M variant (12).
4.2.4. Central domain.
The central domain of Pol θ is predicted to be largely devoid of secondary structure. It is about 900 residues long in vertebrate cells but only about 200 amino acids long in C. elegans (122). In human and other vertebrate genomes, most of the central domain is encoded by one exon, which does not have homology to the shorter central domains in invertebrates.
Based on peptide array binding experiments, the central domain of human Pol θ was suggested to contain Rad51-binding sites with biological relevance (19). For two of the proposed binding sites, there is little evidence for functionality and conservation across organisms. The most conserved proposed binding site (residues 847–894) is actually located in subdomain 5 of the HLD. RAD51 was proposed to interact with full-length pol θ to suppress HR (19). Subsequent studies found, however, that a Pol θ defect does not significantly change HR frequency in human U2OS cells (47) and Polq-deficient murine cells have normal levels of HR (69, 128). TMEJ is sometimes engaged when other major DSB repair pathways are impaired but may not directly inhibit these other pathways (117).
4.2.5. Coordination between domains.
The two ends of a DSB in cells will rarely provide a terminal microhomology by chance. Instead, Pol θ must identify embedded complementary sequences through a scanning step (16). It seems probable that the Pol θ polymerase domain and HLD cooperate to locate suitable microhomologies. A 2–6-bp patch of microhomology is likely to be found in a region within 15 bp of each 3′ terminus at a resected DSB (16). The Pol θ HLD may make initial contact with 3′ single-stranded DNA to capture the two tails. It could then displace bound RPA in an ATP-dependent reaction. Concomitant movement of DNA with respect to the HLD may assist the polymerase domain in locating a suitable microhomology.
Terminal unpaired bases outside the microhomology would be expected to impair extension from a 3′ end. If a nuclease can gain access to DNA before or after Pol θ has engaged with a microhomology, then removal of unpaired bases would be possible (Figure 4). Although Pol θ contains structural vestiges of a proofreading domain, it lacks the conserved catalytic amino acids and other elements necessary for 3′→5′ exonuclease activity (18, 124). Pol θ does not remove terminal nucleotides by pyrophosphorolysis either (126).
Pol θ manipulates the 3′ end of the single-stranded DNA in the active site, forming a transitory structure that serves as the intermediate for extension of the 3′ end (125, 126). A new 3′ end could lead to successful extension and would influence the selection of microhomologies. Each 3′ end has the potential to be processed independently through multiple cycles.
One study proposed that catalysis of both extension and nuclease activity could take place from the same active site of Pol θ (126). However, subsequent research from the authors’ laboratories led to a new interpretation of the experiments. DNA synthesis by Pol θ on short GC-rich oligonucleotides can yield products which migrate faster on denaturing polyacrylamide gels. These arise by production of stable stem-loop structures rather than from nuclease activity (D. Carvajal-Maldonado, S. Doublié, RD Wood, unpublished observations).
If an extendable 3′ end is not found, cycles of exonuclease action and/or self-templated synthesis may iteratively change the 3′ primer end until an extendable end is produced (Figure 4) (18, 126). After one of the 3′ ends serves as a primer, Pol θ could extend far enough to establish a region of stable base pairing. DNA synthesis by Pol θ has moderate processivity (~ 100 nt), in line with other A-family DNA polymerases (6). Another DNA polymerase may then take over. The other end may still have unpaired bases at the 3′ end, and these could be trimmed by one of the 3′ nucleases or editing activities in cells (Figure 4).
5. REGULATION AND USE OF THETA-MEDIATED END JOINING IN MAMMALIAN CELLS
5.1. Expression of Pol θ
Although TMEJ is clearly beneficial in many biological settings, it might be excessively mutagenic if used as a default repair pathway. In fact, it appears that Pol θ amounts are under tight control. POLQ messenger RNA (mRNA) is expressed at some level in most human and mouse tissues. Bone marrow and lymphoid tissues are among the highest expressing cell types (https://www.proteinatlas.org/ENSG00000051341-POLQ/tissue). However, Pol θ protein is not relatively abundant. In many human or mouse cells, Pol θ is one or two orders of magnitude less abundant than the catalytic subunits of pol α, pol δ, or pol β (https://pax-db.org).
Determining the expression of Pol θ protein in mammalian cells has been challenging because commercial antibodies, while apt at identifying overexpressed Pol θ, do not authentically detect Pol θ in cell extracts. In fact, many published immunoblots should be taken cum grano salis regarding the identification of Pol θ because the bands have the wrong molecular weight and do not disappear in genetic knockout controls.
5.2. Pol θ in Cancer Cells
When the level of expression of POLQ mRNA is used to divide breast cancer patients into two groups, the cohort with higher POLQ expression has a lower chance of long-term survival (42, 61). An explanation for this finding is now emerging: POLQ is more highly expressed in a substantial subset of triple-negative breast cancers (TNBCs), and individuals with TNBC have a lower chance of long-term survival.
Triple-negative breast cancer (TNBC):
a type of breast cancer in which there is no expression of estrogen or progesterone receptors and where amplification of the HER2 gene is absent, making the cancer more challenging to treat
One characteristic of TNBC is a higher expression, at the mRNA level, of a group of genes including POLQ (116). Intriguingly, POLQ is the only DNA polymerase gene in the most highly correlated group (116). The likely explanation for this is that most TNBCs have HR defects (24, 102) and consequently depend on Pol θ for survival, as discussed below.
TNBCs can generally be divided into basal-like and claudin-low subtypes. An antibody that can authentically detect higher amounts of Pol θ was used to show that levels of Pol θ were generally higher in cell lines and tumors of the basal-like category and lower in the claudin-low category (85). Claudin-low tumors express the transcription factor ZEB1. Consequently, there is a strong negative correlation between ZEB1 and POLQ expression in breast cancers. ZEB1 appears to be a direct negative regulator of POLQ expression. There are several ZEB1 consensus binding sites in the human POLQ gene promoter, and the activity of the promoter in a reporter system can be suppressed by ZEB1 (85).
The transforming growth factor β (TGF-β) pathway is also involved in the regulation of TMEJ. Head and neck cancer cell lines with higher TGF-β expression show lower mRNA expression of POLQ, PARP1, and the ligase LIG1. Experiments using a chromosomal reporter show that Ku-independent alternative end joining is lower in TGF-β-expressing cancer cell lines (62).
5.3. Origins of Damage Repaired by Theta-Mediated End Joining
Normal DNA replication of a genome includes events that give rise to DNA DSBs. Replication forks can stall at difficult-to-replicate sequences, leading to breaks (37). RNA transcription complexes can result in fragile structures, easily broken after collision with DNA replication forks or proteins trapped on DNA (28).
There is good evidence that some of the DNA DSBs that originate during DNA replication are eventually repaired by TMEJ. As mentioned earlier, G4-forming DNA sequences can cause DNA replication to stall, and proper repair of the resulting breaks requires TMEJ (90). Gene amplification by rereplication gives rise to breaks that require TMEJ for repair (3). Some Pol θ is targeted to mitochondria in human cells, where it helps sustain replication in damaged mitochondrial genomes (115).
Single-stranded nicks that occur near DNA replication forks can also be converted to DNA DSBs. One route is through an active process of fork collapse, with some of these events requiring Pol θ for repair. For example, inhibition of the ATR protein kinase causes replication fork collapse and ensuing DSBs (99). Pol θ–defective cells have elevated sensitivity to ATR inhibition (110).
Topoisomerases are constantly active during DNA replication. A Pol θ defect causes enhanced sensitivity of mouse (123) and human (110) cells to topoisomerase I inhibitors. Inhibition of topoisomerase I results in single-stranded nicks, some of which are converted to DSBs if replication of the nicked region occurs before repair. Pol θ–defective cells have some sensitivity to Cas9 single-strand nickase (110) but not to single-strand breaks created by the clinically active deoxycytidine analog CNDAC (63).
Pol θ is also important in plants subjected to increased replication stress. A Pol2a mutant in Arabidopsis, conferring impaired replication, had highly compromised root development and increased unrepaired DSBs when combined with the teb mutation (78). This indicates that TMEJ can repair DSBs that occur when DNA replication is disturbed in developing tissues. Mutation of atr5/6 in Arabidopsis causes defects in a specific histone H3 methylation. This gives rise to Pol θ–dependent amplification of heterochromatin regions in leaf nuclei (28a). The amplification arises because the normal pathway for resolving broken DNA replication forks is defective in atr5/6 mutants, so that TMEJ is mobilized to repair breaks (28a).
Some repair by Pol θ can occur during mitosis. DSBs arising at a replication fork are one-ended and repaired primarily by HR (1, 7). End-joining activity can be toxic for one-ended DSBs (1, 76). One study examined the action of TMEJ in HR-defective BRCA2 mutant cells (64). Under these conditions, TMEJ acted after the initiation of the mitosis following DNA replication. This was found for endogenous DSBs arising in unperturbed cells and for those induced by ionizing radiation and camptothecin. A second break end is generated from an approaching replication fork, creating the possibility for TMEJ to join ends correctly. Delaying TMEJ until the onset of mitosis avoids toxic end joining of one-ended DSBs. Chromatin condensation may also help juxtapose the correct ends (64). Following DNA replication in Xenopus egg extracts, stalled or incomplete replication forks are broken when extracts are driven into a mitotic state (29a). Pol θ participates in rejoining some of these broken forks in mitotic extracts, sometimes yielding aberrant products (29a).
5.4. Theta-Mediated End Joining and Homologous Recombination
Pol θ–mediated TMEJ is especially important in situations where other DSB repair pathways are compromised by mutation. Inactivation of Pol θ in HR-defective cells greatly enhances sensitivity to DSBs (20). Inactivation of HR, such as with inactivation of mammalian BRCA1 or BRCA2, can make some cells essentially reliant on Pol θ (19, 68). A Pol θ defect can also impair cellular fitness when combined with disruption of other genes, including those encoding 53BP1 and some NHEJ components (31, 117). These observations have encouraged a search for small molecule inhibitors of Pol θ that may be used clinically to help kill BRCA-mutated or other HR-defective tumors (32, 127, 130). Such inhibition works by a different mechanism than the inhibition of poly (ADP-ribose) polymerase (PARP) enzymes, so Pol θ inhibition may be useful in combination with PARP inhibitors or in cases of PARP inhibitor resistance (26, 127, 130). PARP is highly abundant in mammalian cellular nuclei, at a concentration of approximately 1 per 20 nucleosomes (73). It may not be specifically necessary as a functional component of any DNA repair pathway, but PARP modifies the activity of multiple DNA repair enzymes and helps to target repair factors to damaged sites. Inhibition of PARP activity can interfere with DNA repair.
Despite the many genome-protective functions of Pol θ, unchallenged adult cells and organisms survive without POLQ, at least for a few generations. Pol θ appears to be most important in protecting against environmental DNA damage during development and in replicating cells. Pol θ inhibitors used in a synthetic lethality approach would thus not be expected to have major toxic effects on normal cells. Caution might be warranted, however, to guard against developmental damage during pregnancy.
6. GENOMIC CHANGES INDUCED BY THETA-MEDIATED END JOINING IN NORMAL AND CANCER CELLS
The role of Pol θ in TMEJ in preserving genome integrity in many biological situations has ensured its widespread conservation through 1.5 billion years of eukaryotic evolution. However, the use of TMEJ to join breaks that are otherwise unrepairable has a price. A few nucleotides are often lost or added at the site of the DNA break. Much of the DNA in multicellular eukaryotes is noncoding, so many of these indel mutations may have no immediate functional consequence. Some of the mutations, however, will alter gene function.
Short deletions at the break repair site will arise because TMEJ utilizes microhomologies to initiate repair, and most microhomologies will not be located at the terminus of a DNA break (Figure 4). Short deletions are one expense of using TMEJ, and their extent will vary depending on the particular DNA break (90, 117). Insertions are generally templated, as Pol θ copies a short region of local template to create a new terminus before the repair is completed. DNA sequencing analysis of repaired breaks shows that several rounds of copying from different templates may occur. These templated insertions have been dubbed TINs, and their relevance to genome changes in cancers is covered in depth elsewhere (16, 47, 94). Pol θ–mediated deletions and templated insertions are an important source of global genetic diversification in C. elegans (90).
The processing of DNA ends largely explains the mutational signature of TMEJ in cancer genomes (16, 47, 94). For example, in a genome-wide analysis of mutations in BRCA-defective cancers expressing higher levels of POLQ, the most enriched mutation signature was the Catalogue of Somatic Mutations in Cancer (COSMIC) small insertion and deletion signature 6 (ID6) (47). This ID6 signature is characterized as including 1 to ≥5-bp deletions, commonly overlapping with ≥2-bp microhomology at breakpoint junctions (4). This corresponds well with the outcomes of TMEJ.
Another way in which TMEJ influences genome evolution is by repairing breaks during integration or excision of foreign DNA. In Drosophila, P element transposase creates a DSB. When HR is not active, these breaks are repaired by an end-joining process (71). This joining depends on Pol θ (70), and the sequences of repair junctions show the TMEJ signature of templated insertions and deletions. A separate model study found that integration of group II intron RNA sequences into the Drosophila genome depended on Pol θ and left a characteristic TMEJ signature (114). Such integration requires reverse transcription. Similar to some other DNA polymerases (18), Pol θ can copy from an RNA template as well as from DNA (22).
When exogenous plasmid DNA is introduced into cells by transfection, the majority of it is integrated randomly in the genome. This integration occurs by NHEJ or TMEJ. If both processes are eliminated by mutation (for example, in a lig4 polq double mutant), then the frequency of random integration is reduced to near zero in both murine (128) and human cells (91). The temporary inhibition of both NHEJ and TMEJ may be practically useful in suppressing nonhomologous integration. In the moss P. patens and the algae C. reinhardtii, TMEJ is important for mediating CRISPR-Cas9-mediated mutagenesis and for random DNA integration (66a, 101). The TMEJ pathway is also important in some instances of CRISPR-Cas9-mediated gene targeting by synthesis-directed oligonucleotide repair, as noted in experiments with C. reinhardtii (101) and human cells (56a).
In the cases described here, integration of foreign DNA is facilitated by end-joining pathways. Introduction of large genome segments yields raw material for some evolution, such as the introduction of new introns providing mutable linkers between protein domains.
No DSB repair pathway is wholly accurate. NHEJ and TMEJ are together responsible for most mutagenesis caused by CRISPR-Cas9 (66a, 91, 129). DNA breaks caused by ionizing radiation rarely have ligatable 3′-OH and 5′-phosphate ends, so removal of a few bases is often necessary, leading to deletions. Short deletions or insertions may be the price to pay for the execution of these repair pathways to allow rejoining of the proper regions of the chromosome and cellular survival. Mammalian genomes mostly comprise DNA that is noncoding, and there is likely little phenotypic consequence for the majority of indels. HR is also not necessarily error free; it can be initiated between sequences with incomplete homologies. Deletions can result, and LOH can occur after crossing over, as discussed above.
7. CAN ALL PHENOTYPES OF POLQ DISRUPTION BE EXPLAINED BY A THETA-MEDIATED END JOINING DEFECT?
In addition to the TMEJ activities described in this review, Pol θ can mediate additional reactions that are relevant to other modes of DNA damage tolerance.
5′-Deoxyribose phosphate (dRP) lyase activity is present in Pol θ (84). Removal of a 5′ sugar-phosphate by such activity is a step in base excision repair following cleavage of a DNA chain at an abasic site. Lys residue 2383 is critical for abasic site lyase activity in human Pol θ, and this residue is also critical for DNA polymerase activity (59). In many organisms, the major DNA polymerase utilized during base excision repair is Pol β. In vitro, Pol θ can substitute in BER reactions (58, 84). Some organisms, including C. elegans and plants, do not contain Pol β. There is evidence that Pol θ can participate in BER in C. elegans (8).
One of the first biochemical activities recognized for human Pol θ was its ability to efficiently bypass some forms of template DNA damage in vitro by translesion DNA synthesis (TLS). Full-length Pol θ, as well as the isolated polymerase domain, can efficiently insert an A to bypass an abasic site in a template (57, 96). Thymine glycol (96, 121), UV radiation–induced photoproducts (97, 120), and 1,N6-ethenodeoxyadenosine (119) can also be bypassed. Whether such TLS activities of Pol θ are regularly used in vivo is not certain. Based on reporter gene experiments, one study proposed that Pol θ bypasses UV radiation–induced photoproducts in human cells (120). An unusual aspect of these observations is that only the Pol domain of Pol θ protein was needed for the TLS activity, in contrast to other activities where Pol θ coordinates its helicase-like and polymerase functions. The same study suggested that Pol θ–defective mouse cells are more sensitive than normal cells to UV radiation. In zebrafish, Pol θ–defective embryos are not hypersensitive to UV radiation (105). A Pol θ defect in C. elegans only confers increased sensitivity to UV radiation if cells are additionally defective in nucleotide excision repair and Pol η, the major TLS enzyme for UV radiation–induced photoproducts. In this situation, the UV radiation sensitivity of the triple mutant can arise from a defect in TMEJ rather than a bypass of DNA damage (106).
An interesting possibility, yet to be explored in depth, is that Pol θ might use its TLS activity as an auxiliary action while engaged in TMEJ. Ionizing radiation produces clusters of DNA-damaging hydroxyl radicals, so some of the bases surrounding a DSB will be damaged by oxidation. The TLS activity of Pol θ could be beneficial because lesions such as thymine glycol are likely to be found within the resected 3′ single-stranded tails that Pol θ uses as templates.
Some of the activities of Pol θ are unique, and more must be learned about their biological functions. These activities might serve as alternative targets for small molecule inhibitors for use in the therapy of HR-defective cancers. In any case, it is increasingly clear that TMEJ is the primary DSB pathway for some situations. In many organisms, Pol θ is essential when strand breaks arise during development and in cases where other repair pathways are compromised.
SUMMARY POINTS.
DNA polymerase theta (Pol θ) is the defining enzyme for a pathway of DNA double-strand break repair that uses short microhomologies to join ends.
Even though Pol θ often creates indels, its repair activity prevents catastrophic events and thus enhances genome stability.
Pol θ is crucial for the maintenance of damaged genomes during development and limits loss of heterozygosity.
Pol θ has an unusual three-domain architecture, in which the C-terminal polymerase domain is linked to an N-terminal helicase-like domain via a central linker.
Pol θ harbors multiple activities including DNA polymerase, DNA-dependent ATPase, templated single-strand extension, and 5′-deoxyribose phosphate (dRP) lyase.
Some cancer cells depend on theta-mediated end joining (TMEJ) to repair double-strand breaks, making Pol θ a compelling drug target.
FUTURE ISSUES.
How can researchers obtain a fuller understanding of the regulation of Pol θ in normal and cancer cells?
Are there protective roles for Pol θ during the development of mammals?
What are the mechanistic steps of the self-templated synthesis that gives rise to templated insertions?
How are the polymerase and helicase-like domains coordinated during TMEJ?
In what situations does the translesion DNA synthesis activity of Pol θ have biological relevance?
What factors influence the operation of TMEJ in different stages of the cell cycle?
How is TMEJ/Pol θ involved in DNA interstrand crosslink repair?
What more can we learn from the phylogenetic distribution and sequence features of Pol θ in eukaryotes?
ACKNOWLEDGMENTS
For comments on the manuscript and help with figures, we thank Kei-ichi Takata, Mélanie Prodhomme, Denisse Carvajal, Yuzhen Li, Scott Vanson, Adele Guerin, Martha Scannell, and Karl Zahn. We appreciate ongoing discussions with our colleagues Dale Ramsden, Gaorav Gupta, Eli Rothenberg, and their teams. Funding for research on Pol θ in our laboratories was provided by National Institutes of Health grants R01 CA052040 (to S.D.); P01 CA193124 (to R.D.W.); and P01 CA247773 (to S.D. and R.D.W.) and by the J. Ralph Meadows Chair in Carcinogenesis Research (to R.D.W.).
Footnotes
DISCLOSURE STATEMENT
R.D.W. is a member of the Environmental Mutagenesis and Genomics Society and owns shares in Repare Therapeutics. S.D. was a recent member of the Board of Scientific Counselors of the National Institute of Environmental Health Sciences. R.D.W. and S.D. are paid consultants for Schrödinger, Inc.
LITERATURE CITED
- 1.Adachi N, So S, Koyama H. 2004. Loss of nonhomologous end joining confers camptothecin resistance in DT40 cells. Implications for the repair of topoisomerase I-mediated DNA damage. J. Biol. Chem 279:37343–48 [DOI] [PubMed] [Google Scholar]
- 2.Aguirrezabalaga I, Sierra LM, Comendador MA. 1995. The hypermutability conferred by the mus308 mutation of Drosophila is not specific for cross-linking agents. Mutat. Res 336:243–50 [DOI] [PubMed] [Google Scholar]
- 3.Alexander JL, Beagan K, Orr-Weaver TL, McVey M. 2016. Multiple mechanisms contribute to double-strand break repair at rereplication forks in Drosophila follicle cells. PNAS 113:13809–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, et al. 2020. The repertoire of mutational signatures in human cancer. Nature 578:94–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anand R, Buechelmaier E, Belan O, Newton M, Vancevska A, et al. 2022. HELQ is a dual-function DSB repair enzyme modulated by RPA and RAD51. Nature 601:268–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. 2008. Low-fidelity DNA synthesis by human DNA polymerase θ. Nucleic Acids Res. 36:3847–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnaudeau C, Lundin C, Helleday T. 2001. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol 307:1235–45 [DOI] [PubMed] [Google Scholar]
- 8.Asagoshi K, Lehmann W, Braithwaite EK, Santana-Santos L, Prasad R, et al. 2012. Single-nucleotide base excision repair DNA polymerase activity in C. elegans in the absence of DNA polymerase β. Nucleic Acids Res. 40:670–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beagan K, Armstrong RL, Witsell A, Roy U, Renedo N, et al. 2017. Drosophila DNA polymerase theta utilizes both helicase-like and polymerase domains during microhomology-mediated end joining and interstrand crosslink repair. PLOS Genet. 13:e1006813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beagan K, McVey M. 2016. Linking DNA polymerase theta structure and function in health and disease. Cell. Mol. Life Sci 73:603–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernstein KA, Gangloff S, Rothstein R. 2010. The RecQ DNA helicases in DNA repair. Annu. Rev. Genet 44:393–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Black SJ, Ozdemir AY, Kashkina E, Kent T, Rusanov T, et al. 2019. Molecular basis of microhomology-mediated end-joining by purified full-length Polθ. Nat. Commun 10:4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonilla B, Hengel SR, Grundy MK, Bernstein KA. 2020. RAD51 Gene Family Structure and Function. Annu. Rev. Genet 54:25–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyd JB, Sakaguchi K, Harris PV. 1990. mus308 mutants of Drosophila exhibit hypersensitivity to DNA cross-linking agents and are defective in a deoxyribonuclease. Genetics 125:813–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Burgers PMJ, Kunkel TA. 2017. Eukaryotic DNA replication fork. Annu. Rev. Biochem 86:417–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burton P, McBride DJ, Wilkes JM, Barry JD, McCulloch R. 2007. Ku heterodimer-independent end joining in Trypanosoma brucei cell extracts relies upon sequence microhomology. Eukaryot. Cell 6:1773–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvajal-Garcia J, Cho J-E, Carvajal-Garcia P, Feng W, Wood RD, et al. 2020. Mechanistic basis for microhomology identification and genome scarring by polymerase theta. PNAS 117:8476–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carvajal-Garcia J, Crown KN, Ramsden DA, Sekelsky J. 2021. DNA polymerase theta suppresses mitotic crossing over. PLOS Genet. 17:e1009267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carvajal-Maldonado D, Drogalis Beckham L, Wood RD, Doublié S. 2022. When DNA polymerases multitask: functions beyond nucleotidyl transfer. Front. Mol. Biosci 8:815845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, et al. 2015. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 518:258–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Cejka P, Symington LS. 2021. DNA End Resection: Mechanism and Control. Annu. Rev. Genet 55: 285–307 [DOI] [PubMed] [Google Scholar]
- 20.Chan SH, Yu AM, McVey M. 2010. Dual roles for DNA polymerase θ in alternative end-joining repair of double-strand breaks in Drosophila. PLOS Genet. 6:e1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandramouly G, Liao SR, Rusanov T, Borisonnik N, Calbert ML, et al. 2021. Polθ promotes the repair of 5′-DNA-protein crosslinks by microhomology-mediated end-joining. Cell Rep. 34:108820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandramouly G, Zhao J, McDevitt S, Rusanov T, Hoang T, et al. 2021. Pol θ reverse transcribes RNA and promotes RNA-templated DNA repair. Sci. Adv 7:eabf1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheok CF, Wu L, Garcia PL, Janscak P, Hickson ID. 2005. The Bloom’s syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res. 33:3932–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chopra N, Tovey H, Pearson A, Cutts R, Toms C, et al. 2020. Homologous recombination DNA repair deficiency and PARP inhibition activity in primary triple negative breast cancer. Nat. Commun 11:2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clay DE, Bretscher HS, Jezuit EA, Bush KB, Fox DT. 2021. Persistent DNA damage signaling and DNA polymerase theta promote broken chromosome segregation. J. Cell Biol 220:e202106116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cong K, Peng M, Kousholt AN, Lee WTC, Lee S, et al. 2021. Replication gaps are a key determinant of PARP inhibitor synthetic lethality with BRCA deficiency. Mol. Cell 81:3128–44.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, et al. 2000. Werner’s syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 1:80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cristini A, Gromak N, Sordet O. 2020. Transcription-dependent DNA double-strand breaks and human disease. Mol. Cell. Oncol 7:1691905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Davarinejad H, Huang Y-C, Mermaz B, LeBlanc C, Poulet A, et al. 2022. The histone H3.1 variant regulates TONSOKU-mediated DNA repair during replication. Science 375:1281–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis L, Khoo KJ, Zhang Y, Maizels N. 2020. POLQ suppresses interhomolog recombination and loss of heterozygosity at targeted DNA breaks. PNAS 117:22900–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Deng L, Wu RA, Sonneville R, Kochenova OV, Labib K, et al. 2019. Mitotic CDK promotes replisome disassembly, fork breakage, and complex DNA rearrangements. Mol. Cell 73:915–29.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doublié S, Zahn KE. 2014. Structural insights into eukaryotic DNA replication. Front. Microbiol 5:444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng W, Simpson DA, Carvajal-Garcia J, Price BA, Kumar RJ, et al. 2019. Genetic determinants of cellular addiction to DNA polymerase theta. Nat. Commun 10:4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng W, Smith CM, Simpson DA, Gupta GP. 2022. Targeting non-homologous and alternative end joining repair to enhance cancer radiosensitivity. Semin. Radiat. Oncol 32:29–41 [DOI] [PubMed] [Google Scholar]
- 33.Fujikane R, Shinagawa H, Ishino Y. 2006. The archaeal Hjm helicase has recQ-like functions, and may be involved in repair of stalled replication fork. Genes Cells 11:99–110 [DOI] [PubMed] [Google Scholar]
- 34.Garcia PL, Liu Y, Jiricny J, West SC, Janscak P. 2004. Human RECQ5β, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 23:2882–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghezraoui H, Piganeau M, Renouf B, Renaud J-B, Sallmyr A, et al. 2014. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol. Cell 55:829–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goff JP, Shields DS, Seki M, Choi S, Epperly MW, et al. 2009. Lack of DNA polymerase θ (POLQ) radiosensitizes bone marrow stromal cells in vitro and increases reticulocyte micronuclei after total-body irradiation. Radiat. Res 172:165–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guiblet WM, DeGiorgio M, Cheng X, Chiaromonte F, Eckert KA, et al. 2021. Selection and thermostability suggest G-quadruplexes are novel functional elements of the human genome. Genome Res. 31:1136–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guy CP, Bolt EL. 2005. Archaeal Hel308 helicase targets replication forks in vivo and in vitro and unwinds lagging strands. Nucleic Acids Res. 33:3678–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris PV, Mazina OM, Leonhardt EA, Case RB, Boyd JB, Burtis KC. 1996. Molecular cloning of Drosophila mus308, a gene involved in DNA cross-link repair with homology to prokaryotic DNA polymerase I genes. Mol. Cell. Biol 16:5764–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hastings PJ, Ira G, Lupski JR. 2009. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLOS Genet. 5:e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He P, Yang W. 2018. Template and primer requirements for DNA Pol θ-mediated end joining. PNAS 115:7747–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Higgins GS, Harris AL, Prevo R, Helleday T, McKenna WG, Buffa FM. 2010. Overexpression of POLQ confers a poor prognosis in early breast cancer patients. Oncotarget 1:175–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Higgins GS, Prevo R, Lee YF, Helleday T, Muschel RJ, et al. 2010. A small interfering RNA screen of genes involved in DNA repair identifies tumor-specific radiosensitization by POLQ knockdown. Cancer Res. 70:2984–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hogg M, Sauer-Eriksson AE, Johansson E. 2012. Promiscuous DNA synthesis by human DNA polymerase θ. Nucleic Acids Res. 40:2611–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hogg M, Seki M, Wood RD, Doublié S, Wallace SS. 2011. Lesion bypass activity of DNA polymerase θ (POLQ) is an intrinsic property of the pol domain and depends on unique sequence inserts. J. Mol. Biol 405:642–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoitsma NM, Whitaker AM, Schaich MA, Smith MR, Fairlamb MS, Freudenthal BD. 2020. Structure and function relationships in mammalian DNA polymerases. Cell. Mol. Life Sci 77:35–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hwang T, Reh S, Dunbayev Y, Zhong Y, Takata Y, et al. 2020. Defining the mutation signatures of DNA polymerase θ in cancer genomes. NAR Cancer 2:zcaa017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inagaki S, Nakamura K, Morikami A. 2009. A link among DNA replication, recombination, and gene expression revealed by genetic and genomic analysis of TEBICHI gene of Arabidopsis thaliana. PLOS Genet. 5:e1000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inagaki S, Suzuki T, Ohto M-A, Urawa H, Horiuchi T, et al. 2006. Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell 18:879–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jain R, Aggarwal AK, Rechkoblit O. 2018. Eukaryotic DNA polymerases. Curr. Opin. Struct. Biol 53:77–87 [DOI] [PubMed] [Google Scholar]
- 52.Kamath-Loeb A, Loeb LA, Fry M. 2012. The Werner syndrome protein is distinguished from the Bloom syndrome protein by its capacity to tightly bind diverse DNA structures. PLOS ONE 7:e30189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kamisugi Y, Whitaker JW, Cuming AC. 2016. The transcriptional response to DNA-double-strand breaks in Physcomitrella patens. PLOS ONE 11:e0161204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kamp JA, Lemmens BBLG, Romeijn RJ, Changoer SC, van Schendel R, Tijsterman M. 2021. Helicase Q promotes homology-driven DNA double-strand break repair and prevents tandem duplications. Nat. Commun 12:7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karow JK, Constantinou A, Li J-L, West SC, Hickson ID. 2000. The Bloom’s syndrome gene product promotes branch migration of Holliday junctions. PNAS 97:6504–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karow JK, Newman RH, Freemont PS, Hickson ID. 1999. Oligomeric ring structure of the Bloom’s syndrome helicase. Curr. Biol 9:597–600 [DOI] [PubMed] [Google Scholar]
- 56a.Kelso AA, Lopezcolorado FW, Bhargava R, Stark JM. 2019. Distinct roles of RAD52 and POLQ in chromosomal break repair and replication stress response. PLOS Genet 15:e1008319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laverty DJ, Averill AM, Doublié S, Greenberg MM. 2017. The A-rule and deletion formation during abasic and oxidized abasic site bypass by DNA polymerase θ. ACS Chem. Biol 12:1584–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laverty DJ, Greenberg MM. 2018. Expanded substrate scope of DNA polymerase θ and DNA polymerase β: lyase activity on 5′-overhangs and clustered lesions. Biochemistry 57:6119–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laverty DJ, Mortimer IP, Greenberg MM. 2018. Mechanistic insight through irreversible inhibition: DNA polymerase θ uses a common active site for polymerase and lyase activities. J. Am. Chem. Soc 140:9034–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leal AZ, Schwebs M, Briggs E, Weisert N, Reis H, et al. 2020. Genome maintenance functions of a putative Trypanosoma brucei translesion DNA polymerase include telomere association and a role in antigenic variation. Nucleic Acids Res. 48:9660–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lemée F, Bergoglio V, Fernandez-Vidal A, Machado-Silva A, Pillaire M-J, et al. 2010. DNA polymerase θ up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. PNAS 107:13390–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Q, Palomero L, Moore J, Guix I, Espín R, et al. 2021. Loss of TGFβ signaling increases alternative end-joining DNA repair that sensitizes to genotoxic therapies across cancer types. Sci. Transl. Med 13:eabc4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu X, Jiang Y, Takata K-i, Nowak B, Liu C, et al. 2019. CNDAC-induced DNA double-strand breaks cause aberrant mitosis prior to cell death. Mol. Cancer Ther 18:2283–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Llorens-Agost M, Ensminger M, Le HP, Gawai A, Liu J, et al. 2021. POLθ-mediated end joining is restricted by RAD52 and BRCA2 until the onset of mitosis. Nat. Cell Biol 23:1095–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Machwe A, Lozada EM, Xiao L, Orren DK. 2006. Competition between the DNA unwinding and strand pairing activities of the Werner and Bloom syndrome proteins. BMC Mol. Biol 7:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Macris MA, Krejci L, Bussen W, Shimamoto A, Sung P. 2006. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair 5:172–80 [DOI] [PubMed] [Google Scholar]
- 66a.Mara K, Charlot F, Guyon-Debast A, Schaefer DG, Collonnier C, et al. 2019. POLQ plays a key role in the repair of CRISPR/Cas9-induced double-stranded breaks in the moss Physcomitrella patens. New Phytol. 222:1380–91 [DOI] [PubMed] [Google Scholar]
- 67.Marini F, Kim N, Schuffert A, Wood RD. 2003. POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J. Biol. Chem 278:32014–19 [DOI] [PubMed] [Google Scholar]
- 68.Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A. 2015. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 518:254–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mateos-Gomez PA, Kent T, Deng SK, McDevitt S, Kashkina E, et al. 2017. The helicase domain of Polθ counteracts RPA to promote alt-NHEJ. Nat. Struct. Mol. Biol 24:1116–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McVey M. 2010. Strategies for DNA interstrand crosslink repair: Insights from worms, flies, frogs, and slime molds. Environ. Mol. Mutagen 51:646–58 [DOI] [PubMed] [Google Scholar]
- 71.McVey M, Radut D, Sekelsky JJ. 2004. End-joining repair of double-strand breaks in Drosophila melanogaster is largely DNA ligase IV independent. Genetics 168:2067–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mohaghegh P, Karow JK, Brosh RM Jr., Bohr VA, Hickson ID. 2001. The Bloom’s and Werner’s syndrome proteins are DNA structure–specific helicases. Nucleic Acids Res. 29:2843–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72a.Morrical SW. 2015. DNA-pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harb. Perspect. Biol 7: a016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Muthurajan UM, Hepler MRD, Hieb AR, Clark NJ, Kramer M, et al. 2014. Automodification switches PARP-1 function from chromatin architectural protein to histone chaperone. PNAS 111:12752–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muzzini DM, Plevani P, Boulton SJ, Cassata G, Marini F. 2008. Caenorhabditis elegans POLQ-1 and HEL-308 function in two distinct DNA interstrand cross-link repair pathways. DNA Repair 7:941–50 [DOI] [PubMed] [Google Scholar]
- 75.Muzzolini L, Beuron F, Patwardhan A, Popuri V, Cui S, et al. 2007. Different quaternary structures of human RECQ1 are associated with its dual enzymatic activity. PLOS Biol. 5:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nakamura K, Kustatscher G, Alabert C, Hodl M, Forne I, et al. 2021. Proteome dynamics at broken replication forks reveal a distinct ATM-directed repair response suppressing DNA double-strand break ubiquitination. Mol. Cell 81:1084–99.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Newman JA, Cooper CDO, Aitkenhead H, Gileadi O. 2015. Structure of the helicase domain of DNA polymerase theta reveals a possible role in the microhomology-mediated end-joining pathway. Structure 23:2319–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nisa M, Bergis C, Pedroza-Garcia J-A, Drouin-Wahbi J, Mazubert C, et al. 2021. The plant DNA polymerase theta is essential for the repair of replication-associated DNA damage. Plant J. 106:1197–207 [DOI] [PubMed] [Google Scholar]
- 79.Nishizawa-Yokoi A, Saika H, Hara N, Lee L-Y, Toki S, Gelvin SB. 2021. Agrobacterium T-DNA integration in somatic cells does not require the activity of DNA polymerase θ. New Phytol. 229:2859–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oyama T, Oka H, Mayanagi K, Shirai T, Matoba K, et al. 2009. Atomic structures and functional implications of the archaeal RecQ-like helicase Hjm. BMC Struct. Biol 9:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ozdemir AY, Rusanov T, Kent T, Siddique LA, Pomerantz RT. 2018. Polymerase θ-helicase efficiently unwinds DNA and RNA-DNA hybrids. J. Biol. Chem 293:5259–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Plecenikova A, Slaninova M, Riha K. 2014. Characterization of DNA repair deficient strains of Chlamydomonas reinhardtii generated by insertional mutagenesis. PLOS ONE 9:e105482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Popuri V, Bachrati CZ, Muzzolini L, Mosedale G, Costantini S, et al. 2008. The human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J. Biol. Chem. 283:17766–76 [DOI] [PubMed] [Google Scholar]
- 83a.Prakash R, Zhang Y, Feng W, Jasin M. 2015. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol 7: a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Prasad R, Longley MJ, Sharief FS, Hou EW, Copeland WC, Wilson SH. 2009. Human DNA polymerase θ possesses 5′-dRP lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic Acids Res. 37:1868–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Prodhomme MK, Pommier RM, Franchet C, Fauvet F, Bergoglio V, et al. 2021. EMT transcription factor ZEB1 represses the mutagenic POLθ-mediated end-joining pathway in breast cancers. Cancer Res. 81:1595–606 [DOI] [PubMed] [Google Scholar]
- 85a.Quinn GA, Banat AM, Abdelhameed AM, Banat IM. 2020. Streptomyces from traditional medicine: sources of new innovations in antibiotic discovery. J Med. Microbiol 69:1040–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Raia P, Delarue M, Sauguet L. 2019. An updated structural classification of replicative DNA polymerases. Biochem. Soc. Trans 47:239–49 [DOI] [PubMed] [Google Scholar]
- 87.Ramsden DA, Carvajal-Garcia J, Gupta GP. 2022. Mechanism, cellular functions and cancer roles of polymerase-theta-mediated DNA end joining. Nat. Rev. Mol. Cell Biol 23:125–40 [DOI] [PubMed] [Google Scholar]
- 88.Randrianjatovo-Gbalou I, Rosario S, Sismeiro O, Varet H, Legendre R, et al. 2018. Enzymatic synthesis of random sequences of RNA and RNA analogues by DNA polymerase theta mutants for the generation of aptamer libraries. Nucleic Acids Res. 46:6271–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Roerink SF, van Schendel R, Tijsterman M. 2014. Polymerase θ-mediated end joining of replication-associated DNA breaks in C. elegans. Genome Res. 24:954–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Saito S, Maeda R, Adachi N. 2017. Dual loss of human POLQ and LIG4 abolishes random integration. Nat. Commun 8:16112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sakofsky CJ, Malkova A. 2017. Break induced replication in eukaryotes: mechanisms, functions, and consequences. Crit. Rev. Biochem. Mol. Biol 52:395–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schaub JM, Soniat MM, Finkelstein IJ. 2022. Polymerase theta-helicase promotes end joining by stripping single-stranded DNA-binding proteins and bridging DNA ends. Nucleic Acids Res. 50:3911–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schimmel J, van Schendel R, den Dunnen JT, Tijsterman M. 2019. Templated insertions: a smoking gun for polymerase theta-mediated end joining. Trends Genet. 35:632–44 [DOI] [PubMed] [Google Scholar]
- 95.Seki M, Marini F, Wood RD. 2003. POLQ (Pol θ), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 31:6117–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, et al. 2004. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 23:4484–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Seki M, Wood RD. 2008. DNA polymerase θ (POLQ) can extend from mismatches and from bases opposite a (6-4) photoproduct. DNA Repair 7:119–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sharma S, Sommers JA, Choudhary S, Faulkner JK, Cui S, et al. 2005. Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J. Biol. Chem 280:28072–84 [DOI] [PubMed] [Google Scholar]
- 99.Shastri N, Tsai YC, Hile S, Jordan D, Powell B, et al. 2018. Genome-wide identification of structure-forming repeats as principal sites of fork collapse upon ATR inhibition. Mol. Cell 72:222–38.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shima N, Hartford SA, Duffy T, Wilson LA, Schimenti KJ, Schimenti JC. 2003. Phenotype-based identification of mouse chromosome instability mutants. Genetics 163:1031–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sizova I, Kelterborn S, Verbenko V, Kateriya S, Hegemann P. 2021. Chlamydomonas POLQ is necessary for CRISPR/Cas9-mediated gene targeting. G3 11:jkab114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Staaf J, Glodzik D, Bosch A, Vallon-Christersson J, Reutersward C, et al. 2019. Whole-genome sequencing of triple-negative breast cancers in a population-based clinical study. Nat. Med 25:1526–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sun H, Karow JK, Hickson ID, Maizels N. 1998. The Bloom’s syndrome helicase unwinds G4 DNA. J. Biol. Chem 273:27587–92 [DOI] [PubMed] [Google Scholar]
- 104.Takata K-I, Reh S, Yousefzadeh MJ, Zelazowski MJ, Bhetawal S, et al. 2017. Analysis of DNA polymerase ν function in meiotic recombination, immunoglobulin class-switching, and DNA damage tolerance. PLOS Genet. 13:e1006818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thyme SB, Schier AF. 2016. Polq-mediated end joining is essential for surviving DNA double-strand breaks during early zebrafish development. Cell Rep. 15:707–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.van Bostelen I, Tijsterman M. 2017. Combined loss of three DNA damage response pathways renders C. elegans intolerant to light. DNA Repair 54:55–62 [DOI] [PubMed] [Google Scholar]
- 107.van Kregten M, de Pater S, Romeijn R, van Schendel R, Hooykaas PJ, Tijsterman M. 2016. T-DNA integration in plants results from polymerase-θ-mediated DNA repair. Nat. Plants 2:16164. [DOI] [PubMed] [Google Scholar]
- 108.Vanson S, Li Y, Wood RD, Doublié S. 2022. Probing the structure and function of polymerase θ helicase-like domain. DNA Repair 116:103358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vindigni A, Marino F, Gileadi O. 2010. Probing the structural basis of RecQ helicase function. Biophys. Chem 149:67–77 [DOI] [PubMed] [Google Scholar]
- 110.Wang Z, Song Y, Li S, Kurian S, Xiang R, et al. 2019. DNA polymerase θ (POLQ) is important for repair of DNA double-strand breaks caused by fork collapse. J. Biol. Chem 294:3909–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ward JD, Muzzini DM, Petalcorin MI, Martinez-Perez E, Martin JS, et al. 2010. Overlapping mechanisms promote postsynaptic RAD-51 filament disassembly during meiotic double-strand break repair. Mol. Cell 37:259–72 [DOI] [PubMed] [Google Scholar]
- 112.Wei P-C, Chang AN, Kao J, Du Z, Meyers RM, et al. 2016. Long neural genes harbor recurrent DNA break clusters in neural stem/progenitor cells. Cell 164:644–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Whelan DR, Rothenberg E. 2021. Super-resolution mapping of cellular double-strand break resection complexes during homologous recombination. PNAS 118:e2021963118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.White TB, Lambowitz AM. 2012. The retrohoming of linear group II intron RNAs in Drosophila melanogaster occurs by both DNA ligase 4-dependent and -independent mechanisms. PLOS Genet. 8:e1002534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wisnovsky S, Sack T, Pagliarini DJ, Laposa RR, Kelley SO. 2018. DNA polymerase θ increases mutational rates in mitochondrial DNA. ACS Chem. Biol 13:900–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wood R, Doublié S. 2016. DNA polymerase θ (POLQ), double-strand break repair, and cancer. DNA Repair 44:22–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wyatt DW, Feng W, Conlin MP, Yousefzadeh MJ, Roberts SA, et al. 2016. Essential roles for polymerase θ-mediated end joining in the repair of chromosome breaks. Mol. Cell 63:662–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yoon J-H, Johnson RE, Prakash L, Prakash S. 2019. DNA polymerase θ accomplishes translesion synthesis opposite 1,N6-ethenodeoxyadenosine with a remarkably high fidelity in human cells. Genes Dev. 33:282–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yoon J-H, McArthur MJ, Park J, Basu D, Wakamiya M, et al. 2019. Error-prone replication through UV lesions by DNA polymerase θ protects against skin cancers. Cell 176:1295–309.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yoon J-H, Roy Choudhury J, Park J, Prakash S, Prakash L. 2014. A role for DNA polymerase θ in promoting replication through oxidative DNA lesion, thymine glycol, in human cells. J. Biol. Chem 289:13177–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yousefzadeh MJ, Wood R. 2013. DNA polymerase POLQ and cellular defense against DNA damage. DNA Repair 12:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yousefzadeh MJ, Wyatt DW, Takata K, Mu Y, Hensley SC, et al. 2014. Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLOS Genetics 10:e1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zahn KE, Averill AM, Aller P, Wood RD, Doublié S. 2015. Human DNA polymerase θ grasps the primer terminus to mediate DNA repair. Nat. Struct. Mol. Biol 22:304–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zahn KE, Jensen RB. 2021. Polymerase θ coordinates multiple intrinsic enzymatic activities during DNA repair. Genes 12:1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zahn KE, Jensen RB, Wood RD, Doublié S. 2021. Human DNA polymerase θ harbors DNA end-trimming activity critical for DNA repair. Mol. Cell 81:1534–47.e4 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 127.Zatreanu D, Robinson HMR, Alkhatib O, Boursier M, Finch H, et al. 2021. Polθ inhibitors elicit BRCA-gene synthetic lethality and target PARP inhibitor resistance. Nat. Commun 12:3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zelensky AN, Schimmel J, Kool H, Kanaar R, Tijsterman M. 2017. Inactivation of Pol θ and C-NHEJ eliminates off-target integration of exogenous DNA. Nat. Commun 8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhao B, Rothenberg E, Ramsden DA, Lieber MR. 2020. The molecular basis and disease relevance of non-homologous DNA end joining. Nat. Rev. Mol. Cell Biol 21:765–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhou J, Gelot C, Pantelidou C, Li A, Yücel H, et al. 2021. A first-in-class polymerase theta inhibitor selectively targets homologous-recombination-deficient tumors. Nat. Cancer 2:598–610 [DOI] [PMC free article] [PubMed] [Google Scholar]